Significance
RNase E performs an indispensable function in gram-negative bacteria by initiating breakdown of cellular RNAs and providing the scaffold for the primary RNA decay machine, the RNA degradosome. Additionally, RNase E executes gene regulation by cleaving mRNAs that are flagged for decay upon binding to small regulatory RNAs (sRNAs) often associated with the RNA chaperone Hfq. However, a detailed mechanistic understanding of the interactions between these four components remains elusive. Although sRNA binding is known to promote mRNA decay, our work presented here indicates that sRNAs and Hfq are recruited indirectly through bound target mRNAs to the second arginine-rich RNA-binding region (AR2) within the C-terminal domain of RNase E. Consequently, sRNAs are degraded, which broadly impacts bacterial physiology and survival.
Keywords: RNase E, sRNA, small RNA, polynucleotide phosphorylase, Hfq
Abstract
The C-terminal domain (CTD) of the major endoribonuclease RNase E not only serves as a scaffold for the central RNA decay machinery in gram-negative bacteria but also mediates coupled degradation of small regulatory RNAs (sRNAs) and their cognate target transcripts following RNA chaperone Hfq–facilitated sRNA–mRNA base pairing. Despite the crucial role of RNase E CTD in sRNA-dependent gene regulation, the contribution of particular residues within this domain in recruiting sRNAs and mRNAs upon base pairing remains unknown. We have previously shown that in Escherichia coli, the highly conserved 3′-5′-exoribonuclease polynucleotide phosphorylase (PNPase) paradoxically stabilizes sRNAs by limiting access of RNase E to Hfq-bound sRNAs and by degrading target mRNA fragments that would otherwise promote sRNA decay. Here, we report that in the absence of PNPase, the RNA-binding region AR2 in the CTD is required for RNase E to initiate degradation of the Hfq-dependent sRNAs CyaR and RyhB. Additionally, we show that introducing mutations in either hfq that disrupts target mRNA binding to Hfq or the AR2 coding region of rne impairs RNase E binding to sRNAs. Altogether, our data support a model where sRNAs are recruited via bound mRNA targets to RNase E by its AR2 domain after Hfq catalyzes sRNA–mRNA pairing. These results also support our conclusion that in a PNPase-deficient strain, more rapid decay of sRNAs occurs due to accelerated pairing with mRNA targets as a consequence of their accumulation. Our findings provide insights into the mechanisms by which sRNAs and mRNAs are regulated by RNase E.
Small regulatory RNAs (sRNAs) are ubiquitous posttranscriptional regulators that broadly impact gene expression across a wide range of bacteria. The sRNAs play crucial roles in ensuring increased survival of bacterial species including pathogens under a myriad of stress conditions by rapidly reprogramming gene expression (1). In Escherichia coli and other gram-negative bacteria, the best-characterized sRNAs are encoded in trans. These riboregulators function in conjunction with Hfq RNA chaperone to recognize target transcripts through base pairing interactions leading to alterations in mRNA transcription, translation, or stability (2). In addition to Hfq, recent discoveries have identified other RNA-binding proteins including the FinO family protein ProQ (3) and the well-conserved 3′-5′-exoribonuclease polynucleotide phosphorylase (PNPase) (4, 5) as global regulators of sRNA stability and function.
Apart from its established role in RNA degradation, PNPase stabilizes Hfq-binding sRNAs and promotes their function in E. coli (4, 6–8). Recent cryo-EM studies on the Hfq–sRNA–PNPase ternary complex have yielded insights into one mechanism by which PNPase protects sRNAs from cleavage by RNase E. Collectively, these resolved cryo-EM structures illustrate how E. coli PNPase interacts with Hfq and client substrate sRNAs (9). Within these RNA carrier complex structures, Hfq appears to cooperate with the KH and S1 RNA-binding domains of PNPase to capture the sRNA. Interactions of the 3′-end of the sRNA with Hfq preclude it from entering the central catalytic channel of PNPase, where 3′-to-5′-exonucleolytic degradation occurs (6, 9). Additionally, these proteins occlude accessible regions of the sRNA that would otherwise be susceptible to RNase E cleavage. Very recently, we have also demonstrated that the enzymatic activity of PNPase is required for the degradation of short mRNA-derived fragments that originate either from general RNA decay pathways or sRNA-dependent regulation (10). Interestingly, we have discovered that most of these fragments can interact with Hfq and/or possess sRNA-pairing sites. Therefore, our recent findings indicate that PNPase can also promote sRNA stability by degrading short mRNA-derived fragments, which can otherwise base pair with sRNAs in an Hfq-dependent manner to initiate sRNA decay by recruiting RNase E (10).
The major endoribonuclease RNase E initiates degradation of most mRNAs and noncoding RNAs including sRNAs and organizes the RNA degradosome, a large, multienzyme, RNA decay machine (11). The highly conserved N-terminal domain (NTD) of RNase E contains the catalytic core of the degradosome and recognizes and cleaves single-stranded RNA transcripts. Moreover, the enzymatic activity of RNase E can be activated by the 5′-monophosphate group of substrate RNAs, which is detected by the 5′-sensor pocket in the NTD (12). The natively disordered and relatively variable C-terminal domain (CTD) of this enzyme supplies a scaffold for the degradosome assembly through its five small, distinct, linear recognition motifs (microdomains). Three of these microdomains provide binding sites for three separate proteins, the DEAD-box helicase RhlB, glycolytic enzyme enolase, and PNPase. The remaining two microdomains, ARRBD and AR2, harbor arginine-rich RNA-binding sites that specifically recognize RNA substrates (13, 14). Prior work has established that Hfq can also associate with the CTD (15, 16), and interactions between Hfq and the CTD of RNase E directly impact sRNA function by facilitating transcript cleavage as a consequence of sRNA-mediated negative regulation (16–19). Ablation of the CTD leads to defects in sRNA regulation by either reducing the target cleavage rates as previously shown for the SgrS–ptsG sRNA–mRNA pair (20) or diminishing the efficiency of the coupled degradation rates for the RyhB–sodB RNA pair (21). Importantly, these findings and recent structural studies examining association of Hfq with RNase E highlight a role of the CTD in presenting RNA duplexes to the catalytic core of RNase E to initiate RNA decay (22). Despite the crucial role of the CTD in modulating Hfq-dependent sRNA regulation, the molecular mechanisms underlying the Hfq–sRNA–mRNA complex (Hfq–RNA complex) recognition by RNase E CTD in vivo remain unknown. Indeed, there has been an increasing body of evidence suggesting that association of Hfq with the CTD of RNase E likely occurs in the presence of RNA (15, 22–24).
Here, we investigated the mechanism by which RNase E CTD recognizes the Hfq–RNA complex in an effort to further understand how PNPase stabilizes E. coli sRNAs from RNase E–mediated decay following target pairing. To address this, we explored the contributions of distinct microdomains in the C terminus of RNase E toward recruitment of the Hfq–RNA complex, negative regulation of target mRNAs by sRNAs, and sRNA decay in the presence or absence of PNPase. We found evidence supporting a model in which the microdomain AR2 RNA-binding region mediates interactions between Hfq and RNase E by recognizing the bound target RNA, which leads to rapid decay of cognate sRNAs when PNPase is absent. Not only have we defined a specific region within the CTD of RNase E that regulates sRNA turnover in vivo but also uncovered the mechanism by which the exoribonuclease PNPase protects Hfq-binding sRNAs from RNase E–mediated decay.
Results
CTD of RNase E Is Required for sRNA Decay in the Absence of PNPase but Not Hfq.
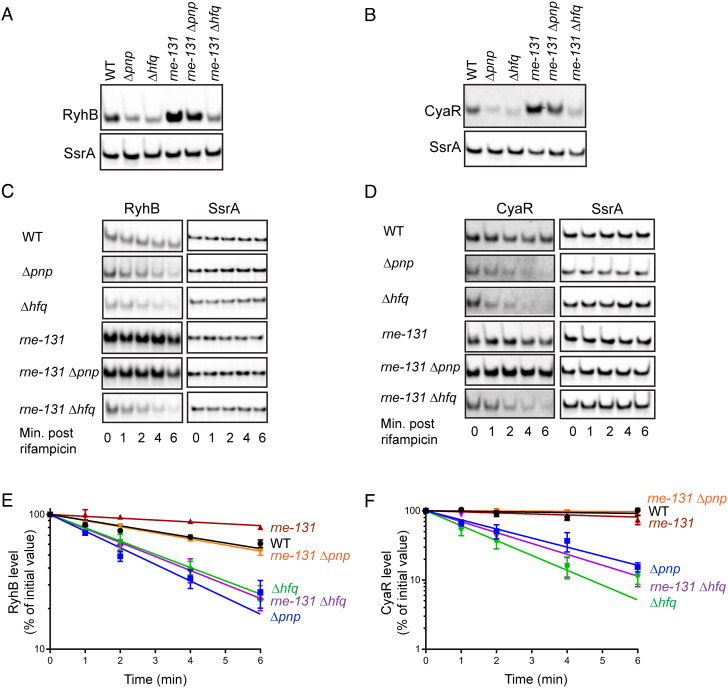
We have recently shown that in the absence of PNPase, sRNA–mRNA base pairing interactions drive degradation of Hfq-dependent sRNAs in E. coli (10). Based on several prior studies, we also know that target pairing–mediated decay of sRNAs can be suppressed by introducing an internal deletion in rne (rne-131), which generates a truncated form of RNase E lacking the CTD (4, 21, 25, 26). Therefore, we first assessed the ability of an rne-131 mutation to suppress the instability of Hfq-binding sRNAs RyhB and CyaR that was previously observed in a PNPase-deficient strain (4, 10). Introduction of rne-131 mutation in a Δpnp strain did suppress defects in both steady-state levels and stabilities of RyhB (Fig. 1 A, C, and E) and CyaR (Fig. 1 B, D, and F) sRNAs, restoring their half-life values to those observed in a corresponding pnp+ wild-type (WT) parent strain (Table 1). However, the rne-131 mutation failed to restore the steady-state levels and stability of both RyhB and CyaR sRNAs in a Δhfq strain (Fig. 1 and Table 1). These results indicate that the CTD of RNase E does not play a critical role in initiating degradation of sRNAs not associated with Hfq. Taken together, our data confirm that in the absence of PNPase, accelerated decay of Hfq-dependent sRNAs following target pairing is mediated by the CTD of RNase E.
Fig. 1.
The CTD of RNase E facilitates decay of Hfq-dependent sRNAs in the absence of PNPase. (A and B) Representative northern blots corresponding to RyhB and CyaR steady-state levels in WT E. coli parent (WT: NRD1138) and derived isogenic mutants (Δpnp: NRD1139, Δhfq: DS021, rne-131: DS102, rne-131 Δpnp: NRD1143, and rne-131 Δhfq: DS130). RyhB and CyaR expression was induced for 15 min by the addition of 2, 2′-dipyridyl or cAMP, respectively, and total RNA was extracted from early-exponential phase cultures for northern blot analysis to determine corresponding levels of sRNAs and SsrA (loading control). (C and D) Representative northern blots corresponding to RNA stability time course experiment to determine half-lives of RyhB and CyaR sRNAs in the abovementioned set of strains. Total RNA was extracted from early-exponential phase cultures 15 min after RyhB (C) or CyaR (D) induction at indicated time points after rifampicin addition. Blots were intensity adjusted to improve visibility of faint bands. (E and F) RyhB and CyaR signal intensities were quantified and normalized to their corresponding loading controls (SsrA). The sRNA decay curves were generated by fitting the normalized signal intensities for each time point. Points and error bars in the curves represent the means and the SEMs of at least three independent experiments. RyhB and CyaR half-life measurements corresponding to RNA stability curves are shown in Table 1.
Table 1.
Half-lifea measurements
| sRNA | Strain | Average half-life(min) ± SE |
|---|---|---|
| RyhB | WT | 6.6 ± 1.0 |
| Δpnp | 2.5 ± 0.2 | |
| Δhfq | 3.2 ± 0.3 | |
| rne-131 | 22 ± 3.3 | |
| Δpnp rne-131 | 6.7 ± 0.4 | |
| Δhfq rne-131 | 2.9 ± 0.3 | |
| rneΔARRBD | 16 ± 3.1† | |
| rneΔAR2 | 4.9 ± 0.2 | |
| Δpnp rneΔARRBD | 2.7 ± 0.4 | |
| Δpnp rneΔAR2 | 4.0 ± 0.4 | |
| Δhfq rneΔARRBD | 3.2 ± 0.3 | |
| Δhfq rneΔAR2 | 3.3 ± 0.2 | |
| CyaR | WT | 8.3 ± 1.3† |
| Δpnp | 1.7 ± 0.1† | |
| Δhfq | 1.4 ± 0.3† | |
| rne-131 | 20 ± 4.8 | |
| Δpnp rne-131 | > 26 | |
| Δhfq rne-131 | 1.9 ± 0.2 | |
| rneΔARRBD | 10 ± 2.8† | |
| rneΔAR2 | 3.8 ± 0.4† | |
| Δpnp rneΔARRBD | 2.9 ± 0.6† | |
| Δpnp rneΔAR2 | 11 ± 3.2† | |
| Δhfq rneΔARRBD | 2.9 ± 0.6† | |
| Δhfq rneΔAR2 | 1.8 ± 0.4† |
Each half-life measurement represents the mean and SEM of at least three independent experiments.
aHalf-lives were determined as described in SI Appendix.
†Exponential decay curves best-fitted RNA stability data corresponding to the first four time points (SI Appendix). Half-lives and standard error were derived from the best-fit curves of the combined replicate data.
AR2 Region within the CTD Facilitates RNase E–Mediated Decay of sRNAs.
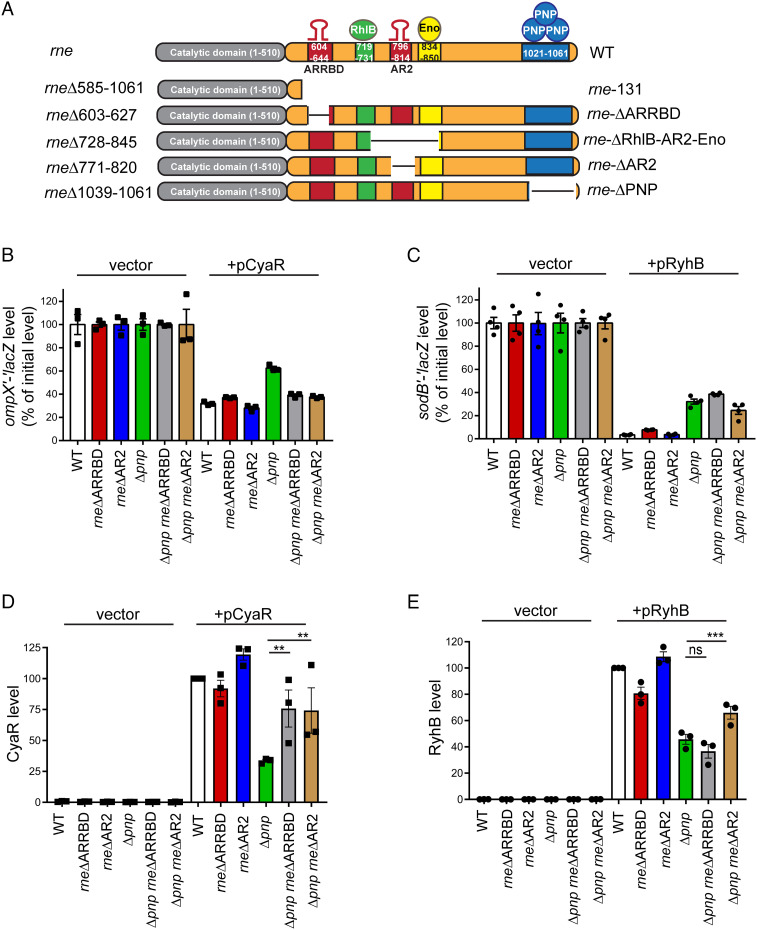
The CTD (amino acids 511–1,061) of RNase E is characterized by the presence of two RNA-binding microdomains, ARRBD (CTD: amino acids 604–644) and AR2 (CTD: amino acids 796–814), and three protein-binding microdomains, which are known to recruit the RNA helicase RhlB (CTD: amino acids 719–731), the metabolic enzyme enolase (CTD: amino acids 834–850), and PNPase (CTD: amino acids 1,021–1,061) (11, 13, 17, 27). We next sought to identify the particular microdomains within the RNase E CTD that are responsible for facilitating decay of RyhB and CyaR sRNAs in a Δpnp mutant. To accomplish this, we created a set of CTD truncation mutants deleted for either one or more microdomains including ARRBD (rne ΔARRBD: rneΔ603–627), AR2 (rne ΔAR2: rneΔ771–820), PNPase (rne ΔPNP: rneΔ1039–1061), and RhlB- and enolase-binding regions (rne ΔRhlB-AR2-Eno: rneΔ728–845) (Fig. 2A). We subsequently used these mutants in a β-galactosidase (β-gal)–based assay to screen for the ability of these truncations to suppress the previously observed defect in target gene regulation by sRNAs RyhB and CyaR in a Δpnp mutant, which is due to their fast turnover (4, 28). In these experiments, we measured the impact of expression of RyhB or CyaR from a plasmid on production of β-gal from lacZ translational reporter fusions to their respective target mRNAs, i.e., sodB′–′lacZ or ompX′–′lacZ (Fig. 2 B and C), in a Δpnp or a WT (pnp+; WT) strain that also contained one of the abovementioned CTD truncations. Consistent with previous studies, we observed a greater than 3-fold repression of ompX′–′lacZ reporter by CyaR in the WT strain compared to 1.6-fold repression of the same translational fusion in an isogenic Δpnp strain (Fig. 2B). Expectedly, introduction of rne-131 mutation in a Δpnp strain completely suppressed the observed defect in ompX′–′lacZ regulation by CyaR (SI Appendix, Fig. S1 A and B). Interestingly, we noticed that either the ARRBD or AR2 deletion when introduced into a Δpnp strain increased CyaR-dependent ompX′–′lacZ repression from 1.6- to 2.6-fold indicating that these RNA-binding regions within the RNase E CTD may be involved in recruitment of CyaR to RNase E for degradation (Fig. 2B and SI Appendix, Fig. S1 A and B). In contrast, removal of the RhlB and enolase (rne ΔRhlB-AR2-Eno) or PNPase-binding sites (rne ΔPNP) from RNase E had no significant impact on ompX′–′lacZ fusion expression in either a pnp+ or Δpnp strain (SI Appendix, Fig. S1 A and B). Next, we evaluated the repercussions of these mutations on CyaR expression in these strains. Consistently, we observed decreases in CyaR steady-state levels in the Δpnp mutant that were almost completely reversed by introduction of ARRBD or AR2 deletions (Fig. 2D and SI Appendix, Fig. S2A).
Fig. 2.
Impact of RNase E CTD deletions on sRNA-mediated target regulation. (A) Schematic representation of RNase E comprising the N-terminal catalytic domain and the C-terminal degradosome scaffold domain (top diagram) followed by illustrations depicting specific deletions within the CTD that were further analyzed in this study. ARRBD and AR2 refer to two distinct arginine-rich RNA-binding regions, while Eno and PNP are abbreviations for glycolytic enzyme enolase and exoribonuclease PNPase, respectively. (B and C) β-gal assay to determine the impact of removal of the ARRBD or AR2 RNA-binding regions from RNase E on sRNA-mediated regulation of target mRNA translation. (B) To assess CyaR-mediated regulation of ompX, β-gal assays were performed on early-exponential phase cultures of a WT strain (WT: NRD377) or derived isogenic mutants (Δpnp: NRD677, rneΔ603-627: NRD1015, rneΔ771-820: NRD1035, Δpnp rneΔ603-627: NRD1025, and Δpnp rneΔ771-820: NRD1037) containing a PBAD::ompX′–′lacZ fusion. Strains either harbored an empty vector (vector) or expressed CyaR from a plasmid (pCyaR). (C) To assess RyhB-mediated regulation of sodB, β-gal assays were performed on early-exponential phase cultures of a WT strain (WT: NRD1041) or derived isogenic mutants (Δpnp: NRD1064, rneΔ603-627: NRD1053, rneΔ771-820: NRD1055, Δpnp rneΔ603-627: NRD1061, and Δpnp rneΔ771-820: NRD1063) containing a PBAD::sodB′–′lacZ fusion. Strains either harbored an empty vector (vector) or expressed RyhB from a plasmid (pRyhB). The amount of β-gal activity produced was normalized to the empty plasmid vector in each background. Points, bars, and error bars represent the value for each replicate, mean, and SEM of at least three independent experiments. (D and E) Northern blots were used to determine CyaR and RyhB steady-state levels in abovementioned sets of strains at the same time points samples were taken for assaying β-gal activity in panels B and C, respectively. The sRNA levels were first normalized to those of SsrA loading controls. CyaR and RyhB levels in WT strains expressing either CyaR or RyhB from a plasmid (WT + pCyaR or WT + pRyhB) were set to 100%, and the sRNA amount for the rest of the samples was scaled to that level. Results represent the mean of at least three independent experiments, and error bars indicate SEM. **P < 0.005 and ***P < 0.0005; ns, not significant.
Based on these findings, we subsequently tested the impact of rne ΔARRBD and rne ΔAR2 mutations on RyhB-dependent regulation of a sodB′–′lacZ translational fusion in the presence (pnp+ strain; WT) or absence of PNPase (Δpnp). We observed an approximately 29-fold repression of sodB′–′lacZ reporter by RyhB in a pnp+ strain, while it was only 3-fold in the Δpnp mutant, again consistent with previous findings (4, 28) (Fig. 2C). However, introduction of rne ΔAR2 mutation in a Δpnp strain resulted in a modest improvement in RyhB-dependent sodB′–′lacZ repression from 3-fold (Δpnp strain) to 4-fold (Δpnp rne ΔAR2), whereas rne ΔARRBD had no impact on sodB′–′lacZ regulation by RyhB in the absence of PNPase (Fig. 2C). When we assayed for RyhB expression in these strains, we discovered reduced amounts in the Δpnp strain. Introduction of rne ΔAR2 into this mutant strain led to a rise in RyhB levels, whereas deletion of ARRBD had no significant effect (Fig. 2E and SI Appendix, Fig. S2B).
These results prompted us to further examine the ability of rne ΔARRBD and rne ΔAR2 mutations to suppress the previously observed defect in RyhB-mediated negative regulation of a different target transcript, sdhCDAB, in a Δpnp strain. We took advantage of a Δfur strain background, where RyhB is constitutively expressed and accumulates under protection of Hfq. In turn, Hfq promotes base pairing with the sdhCDAB mRNA, blocking expression of succinate dehydrogenase complex, which results in an inability of a fur mutant to grow on succinate as the sole carbon source (29). Inactivation of pnp or hfq can cause rapid turnover of RyhB leading to upregulation of the sdhCDAB transcript, which can then allow a Δfur strain to grow on succinate as the sole carbon source (4). Consistent with prior studies, we found that deleting pnp in a Δfur background allowed growth on succinate minimal medium with a yield similar to that observed for a WT (fur+) strain at the end of 24 h (SI Appendix, Fig. S1C). Remarkably, the rne ΔAR2 but not rne ΔARRBD mutation caused significant suppression of the succinate growth phenotype observed for a Δfur Δpnp strain as indicated by a reduced growth yield of the Δfur Δpnp rneΔAR2 triple mutant in succinate minimal medium (SI Appendix, Fig. S1C).
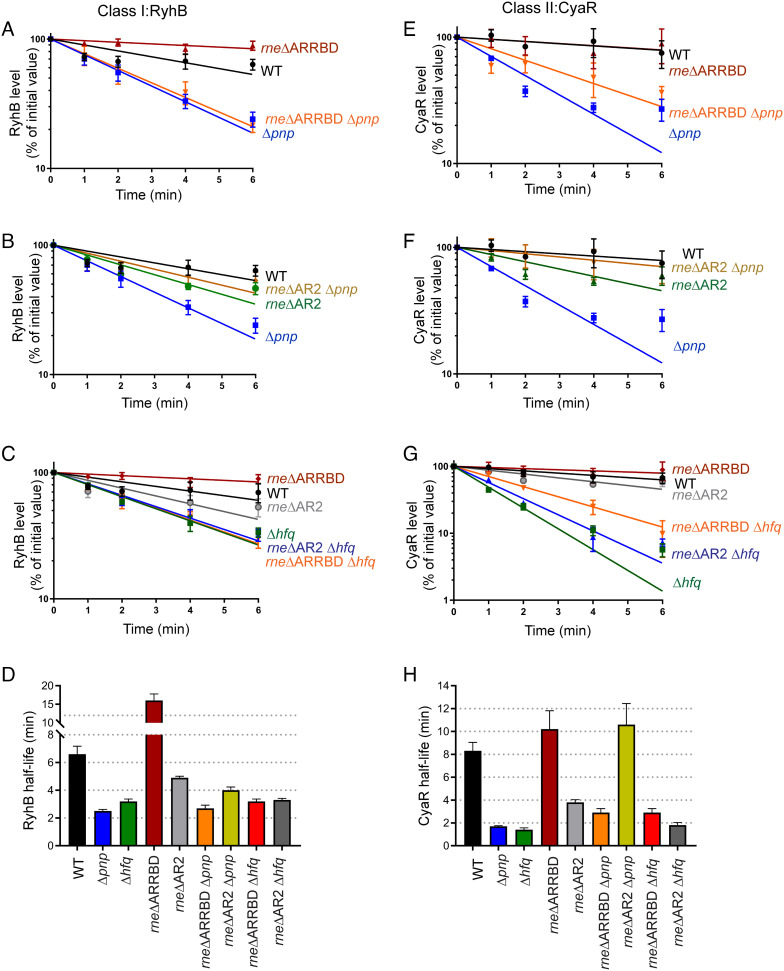
Based on these observations, we predicted that the AR2 domain of RNase E mediates degradation of Hfq-binding sRNAs. However, it was possible that the ARRBD domain may also contribute to this function. To directly test whether removal of the AR2 or ARRBD RNA-binding regions from RNase E reduced or eliminated the rapid turnover of RyhB and CyaR sRNAs that occurs in a Δpnp mutant (Fig. 1), we measured the turnover (half-lives) of these sRNAs in a pnp+, Δpnp strain and derived mutants expressing versions of RNase E lacking the ARRBD or AR2 domain. As anticipated, the half-lives of RyhB and CyaR increased, respectively, from 2.5 min and 2.0 min in a Δpnp strain to 4.0 min and 11 min in an rneΔAR2 Δpnp double mutant, which was comparable to their respective half-lives in the WT strain (Fig. 3 B, D, F, and H, SI Appendix, Fig. S3, and Table 1). These results suggest that the AR2 RNA-binding region in the RNase E CTD has a function in recruiting Hfq-dependent sRNAs for decay in the absence of PNPase, which is evaluated in more detail below. In contrast, the half-lives of both RyhB and CyaR sRNAs were comparable between Δpnp and Δpnp rne ΔARRBD mutants (Fig. 3 A, D, E, and H, SI Appendix, Fig. S3, and Table 1) demonstrating that the ARRBD region likely does not play a role in sRNA degradation by RNase E in a PNPase-deficient strain. Consistent with our observations for an rne131 Δhfq double mutant, both rne ΔARRBD and rne ΔAR2 CTD truncation mutations failed to suppress the stability defects of RyhB or CyaR sRNA in the absence of Hfq (Fig. 3 C, D, G, and H, SI Appendix, Fig. S3, and Table 1). Together, these results indicate that the AR2 RNA-binding region plays an important role in facilitating the rapid decay of Hfq-bound sRNAs in the absence of PNPase leading to a consequential defect in sRNA-mediated target regulation.
Fig. 3.
AR2 RNA-binding region within the CTD of RNase E drives sRNA decay in the absence of PNPase. Stability curves of RyhB (A–C) and CyaR (E–G) in a WT strain (WT: NRD1138) or derived isogenic mutants (Δpnp: NRD1139, rneΔ603-627: NRD1591, rneΔ771-820: NRD1592, Δpnp rneΔ603-627: NRD1593, Δpnp rneΔ771-820: NRD1594, Δhfq: DS021, Δhfq rneΔ603-627: DS199, and Δhfq rneΔ771-820: NRD195). Expression of RyhB and CyaR was induced by the addition of 2, 2′-dipyridyl or cAMP, respectively, and total RNA was extracted from early-exponential phase cultures 15 min after induction at indicated time points after rifampicin addition. The sRNA signals were assessed by northern blot and normalized to that of SsrA loading control. Lines indicate best-fit exponential decay curves of three replicates, and error bars represent SEM of each time point. (D and H) Graphs showing mean sRNA half-life values corresponding to RyhB and CyaR stability curves. The sRNA half-lives are tabulated in Table 1.
AR2 Region Recruits Hfq-Dependent sRNAs to RNase E for Decay.
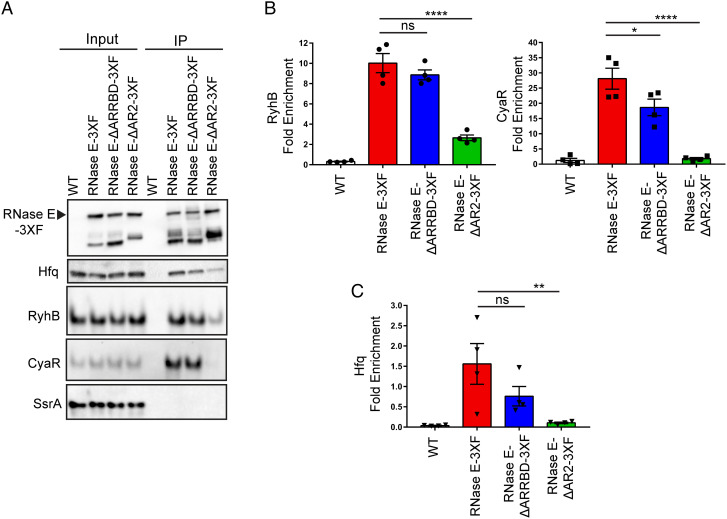
From our results described above, we hypothesized that the RNA-binding regions within the CTD are responsible for facilitating interactions between Hfq-dependent sRNAs and RNase E following target pairing leading to sRNA decay, which occurs to a greater extent in the absence of PNPase. To address this, we introduced a C-terminal 3X-FLAG (3XF) epitope tag at the native chromosomal locus of rne both in WT (RNase E-3XF) and mutant strains that lacked either AR2 (RNase E ΔAR2-3XF) or ARRBD (RNase E ΔARRBD-3XF) RNA-binding regions. RNase E expression was comparable between strains expressing WT (RNase E-3XF) or mutant forms of RNase E (i.e., RNase E ΔAR2-3XF and RNase E ΔARRBD-3XF), which indicated that the RNA-binding regions within the CTD do not regulate RNase E protein levels (SI Appendix, Fig. S4 A and B). Next, we evaluated the contribution of the AR2 and ARRBD regions to sRNA binding by RNase E in vivo in a coimmunoprecipitation assay using these strains that express RNase E, Hfq, and sRNAs from their cognate promoters at their native chromosomal loci. Interestingly, deletion of the AR2 region significantly decreased the ability of RNase E to bind RyhB and CyaR sRNAs (Fig. 4 A and B). Contrasted with our observations for an RNase E-3XF strain, RyhB and CyaR sRNA enrichment with RNase E decreased by 3.8- and 15.8-fold, respectively, in the absence of the AR2 RNA-binding region (Fig. 4B and SI Appendix, Fig. S5). But no significant difference was observed between the amount of RyhB coimmunoprecipitated with RNase E ΔARRBD-3XF and RNase E-3XF (Fig. 4B and SI Appendix, Fig. S5A). However, we did notice a slight but significant 1.5-fold reduction in CyaR enrichment with RNase E lacking the ARRBD region (Fig. 4 A and B and SI Appendix, Fig. S5B). Next, we assayed for Hfq coimmunoprecipitation with RNase E from lysates of the aforementioned set of strains. Notably, we observed Hfq enrichment with RNase E to be significantly reduced by 14.8-fold in the absence of AR2, contrasted to a modest 2.1-fold reduction when the ARRBD RNA-binding region was absent (Fig. 4 A and C). These findings indicate that the AR2 region within the CTD is critical for recruiting Hfq-bound sRNAs to RNase E for degradation.
Fig. 4.
AR2 region within the CTD facilitates interactions between Hfq-dependent sRNAs and RNase E. (A) Cell extracts prepared from late-exponential phase cultures of E. coli strains expressing untagged RNase E (WT: NRD1138) or FLAG-tagged constructs of full-length or truncated forms of RNase E (RNase E-3XF: DS196, RNase E-ΔARRBD-3XF: DS228, and RNase E-ΔAR2-3XF: DS229) were used to assess coprecipitation of sRNAs and Hfq with RNase E by northern blot and immunoblot, respectively. (B) Fold enrichment of a given RNA upon immunoprecipitation was determined by first calculating the signal intensity per microgram of RNA for the input and the elution from northern blots in (A). (C) Fold enrichment of Hfq upon immunoprecipitation was determined by first normalizing Hfq band intensities to corresponding RNase E band signals in the input and elution fractions from western blots in (A). sRNA and Hfq fold enrichments were then calculated by dividing normalized elution signal by the input signal. Points, bars, and error bars represent the value of each replicate, means, and SEM of at least four independent experiments. *P < 0.05, **P < 0.01, and ****P < 0.0001; ns, not significant. An untagged WT strain (WT) was used as a control.
AR2 Region Recognizes Cognate Target RNAs to Initiate RNase E–Mediated Degradation of Hfq-Dependent sRNAs.
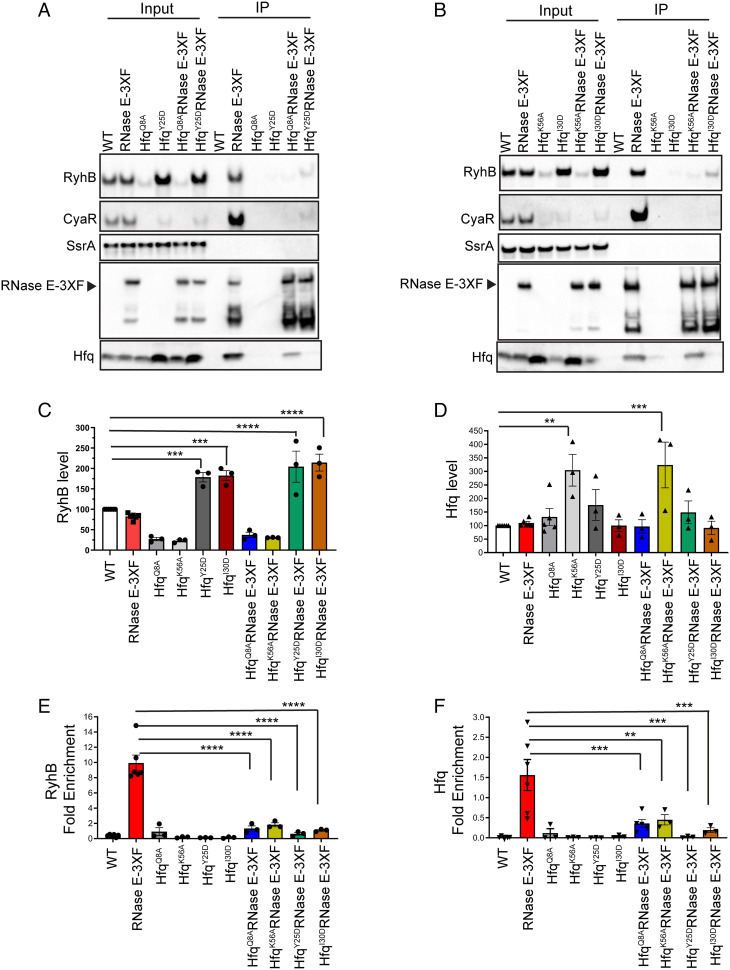
Next, we investigated the mechanism by which the AR2 RNA-binding region recognizes Hfq-bound sRNAs to initiate sRNA decay. Recent studies directed toward understanding Hfq–RNA interactions have identified four distinct surfaces on Hfq: the proximal face, distal face, rim, and C-terminal tail, each possessing unique structural characteristics, which promote binding of different RNAs in particular configurations (30–33). Hfq-dependent sRNAs are further categorized according to which faces these riboregulators interact with. Class I sRNAs (e.g., RyhB) bind the rim and the proximal face of Hfq, and their mRNA targets contact the distal face; in contrast, class II sRNAs (e.g., CyaR) associate with the distal and proximal faces, whereas their mRNA targets bind the rim (31, 32). We hypothesized that the AR2 region recruits Hfq to RNase E either by interacting with sRNAs bound to Hfq or by recognizing cognate target transcripts of Hfq-dependent sRNAs. To distinguish between these possibilities, we assessed the impact of substituting key residues of Hfq on the proximal (HfqQ8A and HfqK56A) or distal (HfqY25D and HfqI30D) face on RNase E binding to Hfq and the sRNAs RyhB and CyaR. Specifically, we introduced HfqQ8A, HfqK56A, HfqY25D, or HfqI30D mutations into a strain expressing 3XF–tagged form of WT RNase E (RNase E-3XF) from rne chromosomal locus. Next, we tested the abilities of these Hfq variants and RyhB and CyaR sRNAs to coimmunoprecipitate with RNase E. As expected, RyhB levels were significantly reduced in both the proximal face mutants HfqQ8A and HfqK56A, respectively, by 2.3- and 2.7-fold (Fig. 5 A–C) due to impaired Hfq binding. Whereas RyhB levels were significantly increased by 2.4- and 2.6-fold, respectively, in the distal face mutants, HfqY25D and HfqI30D, due to protection from mRNA target pairing–mediated decay (Fig. 5 A–C). Interestingly, RyhB enrichment with immunoprecipitated RNase E decreased by 16- and 8-fold in HfqY25D and HfqI30D mutants, respectively, despite significantly higher levels of the sRNA in corresponding input fractions (Fig. 5 A–C and E). Furthermore, these distal face substitutions that have been shown to interfere with the binding of cognate targets of RyhB (SI Appendix, Fig. S6) and other class I sRNAs to Hfq (31) also resulted in drastic defects (68- and 8.2-fold reductions for HfqY25D and HfqI30D, respectively) in the ability of Hfq to coprecipitate with RNase E (Fig. 5 A, B, and F). In addition, both the proximal face mutants (HfqQ8A and HfqK56A) exhibited approximately 4-fold decreased coprecipitation with RNase E (Fig. 5 A, B, and F). Hfq expression was comparable between input fractions of strains expressing WT and mutant forms of Hfq, except for the HfqK56A variant where Hfq levels were significantly increased by 3-fold (Fig. 5D). These results indicate that the decreased ability of Hfq mutants to interact with RNase E is not due to reduced expression. Finally, as expected, CyaR steady-state levels were significantly reduced in the input fractions of all four Hfq variants (Fig. 5 A and B and SI Appendix, Fig. S7A) that resulted in concomitant decreases in the amount of CyaR that pulled down with RNase E relative to WT Hfq (Fig. 5 A and B and SI Appendix, Fig. S7B). Altogether, our results are consistent with a model where the AR2 RNA-binding region plays an important role in recruiting Hfq and Hfq-dependent sRNAs to RNase E through its interactions with the chaperone-bound mRNA targets (Fig. 6 and discussed further below).
Fig. 5.
Class I mRNA binding distal face residue of Hfq is critical for its association with RNase E. (A and B) RNase E-3XF pulldowns were performed using cell extracts prepared from late-exponential phase cultures of E. coli strains containing a FLAG-tagged construct of RNase E and expressing Hfq (RNase E-3XF: DS196) or Hfq mutants with substitutions in its proximal (Q8A and K56A) or distal (Y25D and I30D) faces (HfqQ8A RNase E-3XF: DS233, HfqK56A RNase E-3XF: DS240, HfqY25D RNase E-3XF: DS234, and HfqI30D RNase E-3XF: DS241). Control pulldowns were carried out with cell extracts from strains expressing an untagged RNase E and WT Hfq (WT, NRD1138) or mutant Hfq proteins (HfqQ8A: DS058, HfqK56A: DS239, HfqY25D: NRD1410, and HfqI30D: NRD1411). Coprecipitation of sRNAs and protein was analyzed by northern blot and immunoblot, respectively. Representative images of blots from a total of three independent RNase E coimmunoprecipitation assays are shown. (C and D) Determination of steady-state levels of RyhB and Hfq in the input fractions corresponding to Fig. 5 A and B. Northern and western blots were used to determine RyhB and Hfq amounts. RyhB and Hfq levels were normalized to those of SsrA and DnaK loading controls, respectively. Results represent the mean of at least three independent experiments, and error bars indicate SEM. **P < 0.002, ***P < 0.001, and ****P < 0.0001. RyhB levels calculated in (C) for HfqQ8A, HfqK56A, HfqQ8A RNase E-3XF, and HfqK56A RNase E-3XF were significantly different from that in the WT strain (**P < 0.002). (E and F) RyhB and Hfq fold enrichments were calculated as described in the legend of Fig. 4. Points, bars, and error bars represent the value of each replicate, means, and SEM of at least three independent experiments. **P < 0.01, ***P < 0.001, and ****P < 0.0001. RyhB fold enrichment calculated for HfqK56A RNase E-3XF was significantly different from that for HfqY25D RNase E-3XF (*P < 0.05).
Fig. 6.
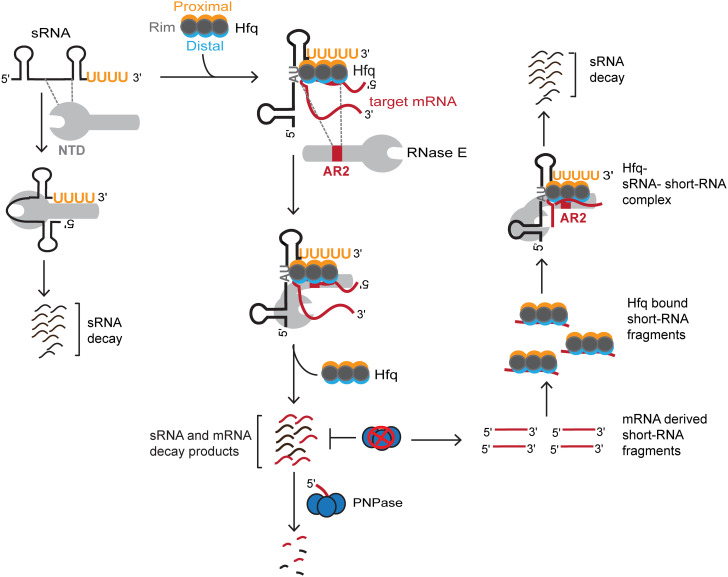
A model depicting the mechanism by which RNase E regulates sRNAs and target mRNAs following Hfq-mediated base pairing. (Left) In the absence of Hfq, the NTD of RNase E recognizes the naked sRNA to activate its decay independent of the CTD. (Middle) AR2 RNA-binding region within the CTD of RNase E recognizes the Hfq-bound target mRNAs to subsequently recruit Hfq-dependent sRNAs to RNase E. In case of sRNA-mediated negative regulation, RNase E activates sequential degradation of target mRNA and its cognate sRNA. Exoribonuclease activity of PNPase is required for clearing of the decay products generated by RNase E. (Right) In the absence of PNPase, mRNA-derived short RNA fragments accumulate, which can base pair with sRNAs via Hfq. AR2 RNA-binding region within the CTD of RNase E subsequently recognizes the bound mRNA-derived short RNA fragments in the Hfq–sRNA–short RNA ternary complex to recruit Hfq along with the bound cognate sRNAs. These interactions ultimately activate RNase E catalytic function to deplete Hfq-dependent sRNAs through accelerated decay.
Discussion
The CTD of RNase E plays an important role in sRNA function through its interactions with Hfq RNA chaperone. However, our knowledge of the specific contributions of distinct protein- and RNA-binding microdomains within the CTD of RNase E toward recognizing Hfq and its bound RNAs to impact sRNA-dependent target regulation in vivo has remained limited. Here, we investigated the mechanism by which the CTD mediates interactions between Hfq and RNase E in the gram-negative model bacterium E. coli to promote degradation of Hfq-dependent sRNAs, which is increased in the absence of PNPase. We discovered that in a Δpnp mutant, the second arginine-rich RNA-binding region AR2 within the RNase E CTD is necessary to drive degradation of both class I (RyhB) and class II (CyaR) Hfq-binding sRNAs following chaperone-assisted base pairing with cognate target transcripts (Figs. 1 and 3 B and F). The AR2 region carries out this function through interactions with Hfq-bound target transcripts complementary to sRNAs to recruit them to RNase E for degradation (Figs. 4 and 5; for discussion, see below). The quick decay of several Hfq-dependent sRNAs including RyhB and CyaR in a PNPase-deficient strain results as a consequence of increased accumulation of mRNA-derived fragments that possess sRNA-pairing sites and/or Hfq-binding ability (10). Taken together, these results elucidate a mechanism by which PNPase, a well-known conserved exoribonuclease, paradoxically protects Hfq-binding sRNAs from RNase E–mediated decay. Specifically, we show that in the absence of PNPase, interactions between the Hfq–target mRNA complex and the AR2 region within the CTD of RNase E trigger degradation of sRNAs following target pairing (Fig. 6).
Hfq Binding to RNase E Is Indirect through RNAs.
A long-standing debate has been whether the interaction between Hfq and the CTD of RNase E is direct or mediated by RNAs (15, 17, 22, 23). It was originally articulated that RNase E directly associates with Hfq based on results showing that Hfq immunoprecipitated with RNase E in cell extracts treated with micrococcal nuclease (17). While the authors clearly demonstrated that micrococcal nuclease treatment eliminated SgrS sRNA in these samples, it is possible that other bridging RNAs that were better protected within the RNase E–Hfq complex remained (17). Contrasted to those observations, recent in vitro studies using purified, RNA-free Hfq and RNase E have found that association of these proteins requires the addition of RNA (15, 22). These results clearly indicate that in the absence of RNA interaction, Hfq–RNase E association is unlikely to occur in vivo. Interestingly, one of the in vitro studies used biophysical experiments to analyze interactions between the Hfq–sRNA complex and a 248-residue segment of the CTD of RNase E (amino acids 603-850) referred to as the recognition core that included the two RNA-binding regions (ARRBD and AR2) and the binding sites for RhlB and enolase (22). Those experiments provided further evidence in support of a model where highly flexible and dynamic RNA-binding regions, ARRBD and AR2, adopt more rigid structures upon binding of RhlB and enolase to the recognition core. As a consequence, ARRBD and AR2 regions acquire a smaller set of conformations that are predicted to interact with Hfq via the bound sRNA (22). Following sRNA binding, the catalytic function of RNase E is activated either through 5′-monophosphate sensing by its NTD or through recognition of RNA structural elements presented in trans by the Hfq-bound sRNA to initiate cleavage of cognate target transcripts (19). These data are consistent with earlier studies, which demonstrated that sRNA-induced efficient cleavage of target mRNAs by RNase E requires both Hfq and a functional RNA degradosome (21, 25). Additionally, it was previously shown that target mRNA decay subsequently drives the sequential degradation of the paired sRNAs in certain cases (20, 21, 24). However, the mechanism by which the CTD recognizes Hfq in vivo to initiate RNase E–dependent degradation of the mRNA–sRNA hybrid remained elusive.
Our data presented here support a model in which RNAs act as a bridge between Hfq and RNase E rather than a model in which these proteins directly interact. First, we discovered by coimmunoprecipitation experiments using cell lysates prepared from strains expressing a 3XF–tagged RNase E that removal of a small, 50–amino acid segment comprising the arginine-rich AR2, an established RNA-binding region (22), significantly reduced binding of Hfq and RyhB and CyaR sRNAs to RNase E (Fig. 4). Second, we found that substitution of key RNA-binding residues of Hfq disrupted its association with RNase E (Fig. 5). In particular, substitution of Y25 and I30, residues involved in binding the targets of class I sRNAs, caused the greatest disruption of RNase E–Hfq interactions. Altogether, these results highlight possible interactions between the AR2 domain and Hfq-associated RNAs in facilitating Hfq binding to RNase E in vivo and further points to the important function of this microdomain in Hfq recruitment.
The AR2 Domain of RNase E Recognizes mRNAs, Not sRNAs Bound to Hfq.
Here, we have used several in vivo techniques including coimmunoprecipitation experiments to unravel the mechanism by which Hfq-binding sRNAs RyhB and CyaR are recruited to RNase E. In our experiments, deleting the AR2 region within the CTD was sufficient to suppress defects in RyhB and CyaR sRNA functions and stabilities observed in a Δpnp mutant (Figs. 2 B and C and 3 B and F) suggesting that the AR2 region is involved in facilitating cleavage of Hfq-dependent sRNAs by RNase E. Importantly, loss of the AR2 region significantly reduced binding of Hfq and RyhB and CyaR sRNAs to RNase E (Fig. 4). When either a Q8A or K56A substitution was introduced into Hfq, which abrogates its ability to bind most if not all sRNAs including RyhB and CyaR (31), but not target mRNAs (Fig. 5 A–C and SI Appendix, Fig. S6), we observed a 4-fold decrease in the ability of Hfq to interact with RNase E (Fig. 5F). This observed difference in Hfq fold enrichments (4-fold) in HfqQ8A and HfqK56A mutants correlated with corresponding decreases in CyaR and RyhB levels and their abilities to interact with RNase E (Fig. 5 C and E and SI Appendix, Fig. S7). We observed even stronger defects (8- to 16-fold) in Hfq binding to RNase E in both distal face mutants, HfqI30D and HfqY25D (Fig. 5 A, B, and F and SI Appendix, Fig. S6) where RyhB steady-state levels significantly increased in abundance, but the chaperone’s ability to interact with cognate target mRNAs corresponding to class I Hfq-binding sRNAs (31) has been disrupted. These results further indicate that the AR2 region plays an important role in recognizing Hfq primarily by interacting with the bound target transcript instead of its cognate sRNA. Finally, the residual binding of CyaR to RNase E in an HfqY25D variant (Fig. 5A) may be through class II mRNA targets that associate with the rim. Thus, we predict that recruitment of both CyaR and Hfq to RNase E may also be reduced by introducing substitutions in key residues along the rim of Hfq.
Contrary to the classical model where sRNAs are proposed to primarily facilitate interactions between Hfq and the C-terminal scaffolding domain of the RNA degradosome to activate target mRNA decay (16, 17, 19, 22), we provide in vivo evidence supporting an alternative model where Hfq interacts with the AR2 RNA-binding region in the CTD through interactions with the bound target mRNA that in turn recruit Hfq-dependent sRNAs to induce sRNA decay by RNase E (Fig. 6). Indeed, there have been past studies suggesting that interactions of RNA substrates with the arginine-rich regions in the CTD can favor their recognition by the NTD (13, 34). Moreover, the interplay between the NTD and CTD is indicated by the synthetic lethal phenotype that results from combining a CTD truncation with mutations in the NTD that impede the ability of RNase E to sense 5′-ends of RNA substrates (35). However, we did observe noticeable interactions between RyhB and RNase E (Fig. 4 A and B) that occurred in the absence of the AR2 region, which may be due to the recruitment of sRNAs not associated with Hfq via the NTD. This idea is consistent with our observation of rapid decay of Hfq-dependent sRNAs in Δhfq mutants that could not be suppressed by deleting the CTD of RNase E (Fig. 1). This is also in accordance with results from a previous study that clearly demonstrated that the RNase E NTD (amino acids 1–529) rapidly cleaves Hfq-dependent sRNAs in the absence of Hfq in vitro, whereas this degradation activity was impaired by its presence (36).
A Mechanism of PNPase-Mediated sRNA Protection.
We have identified a mechanism where the enzymatic function of PNPase is crucial for protecting Hfq-dependent sRNAs from RNase E–mediated degradation by diminishing illegitimate Hfq-facilitated target pairing (Fig. 6), and this mechanism is distinct from the previously proposed protective role of PNPase that results as a consequence of RNA carrier complex formation (6, 9). In addition to being supported by the data presented here, this model (Fig. 6) is also consistent with our previous discovery that a Y25D substitution in Hfq, that blocks class I target mRNAs from binding, impedes the decay of RyhB in cells lacking PNPase (10). However, there remains a possibility that the two distinct mechanisms by which PNPase stabilizes sRNAs may work in concert for some sRNAs, whereas only one pathway might be sufficient for stabilizing other sRNAs. But what particular mRNA-derived short fragments could be improperly driving RyhB and CyaR decay via the AR2 region in the absence of PNPase still remains unknown. Comparing our earlier observations with the previously published RIL-seq analysis (37), we do know that among the 106 short mRNA-derived fragments that increased the abundance in the absence of PNPase, 12 and 17 fragments can base pair with RyhB and CyaR, respectively (10). Future studies will resolve which transcript fragments up-regulated in the absence of PNPase can interact with the AR2 region upon binding of Hfq to initiate degradation of these sRNAs. We also do not know whether these mRNA-derived fragments that possess pairing sites for Hfq-dependent sRNAs remain stuck to Hfq in the absence of PNPase or can repeatedly form transient RNA complexes with Hfq. Prior kinetic studies have revealed that Hfq-facilitated sRNA–mRNA complex formations are highly dynamic in nature, and the binding affinities of different target transcripts to Hfq vary considerably, which subsequently dictate the regulatory outcome of a cognate sRNA (20). Furthermore, it has been proposed that upon sRNA–mRNA annealing, the CTD of Hfq plays an important role in clearing the sRNA–mRNA pairs from this chaperone (38). However, other work has recently shown that removal of the CTD of Hfq does not lead to dramatic impacts on sRNA-based regulation in E. coli (39). Perhaps, this is due to the presence of PNPase in vivo, whereas the studies previously performed with purified components were done in its absence.
Notably, we have previously reported that the deletion of Hfq can eliminate, decrease, or alter the sizes of bands observed for certain mRNA-derived fragments that accumulated in a PNPase-deficient strain and coprecipitated with Hfq, further suggesting a role of Hfq in either generation or stabilization of these specific RNA fragments (10). Recent reports also demonstrate that the Hfq distal face facilitates sRNA-independent interactions between the chaperone and cellular mRNAs in E. coli during exponential growth (23). A functional RNase E–based degradosome is required to initiate mRNA degradation to subsequently release Hfq from the bound cellular mRNAs for facilitating sRNA regulation under specific stress conditions (23). Therefore, we speculate that PNPase as a component of the RNA degradosome performs an essential function in removing via degradation the bound mRNA fragments from Hfq following the initial endonucleolytic cleavage by RNase E.
Materials and Methods
Bacterial Strains and Plasmids.
Bacterial strains, plasmids, and oligonucleotides used in this study are listed in SI Appendix, Tables S1 and S2. Details about strain and plasmid construction are provided in SI Appendix, Supplementary Materials and Methods.
Bacterial Growth.
Bacterial growth was monitored in LB-rich or M9 minimal medium supplemented with 0.001% vitamin B1 and 0.2% glucose or succinate and appropriate antibiotics as described in SI Appendix.
RNA Stability Experiments, Northern Analysis, and Immunoblot.
Following RNA stability assays, specific RNAs were detected by northern analysis. Proteins were detected by immunoblot analysis as described in detail in SI Appendix.
Coimmunoprecipitation Assays.
Protein and RNA interactions with RNase E were tested via a coimmunoprecipitation assay using a 3XF epitope–tagged construct of RNase E, as described in detail in SI Appendix.
β-Gal Assays.
To determine β-gal activity, assays were carried out as described in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by the McGovern Medical School Startup funds and NIGMS grant R01GM121368 to N.R.D.
Author contributions
D.S. and N.R.D. designed research; performed research; analyzed data; and wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Wagner E. G. H., Romby P., Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 90, 133–208 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Hör J., Matera G., Vogel J., Gottesman S., Storz G., Trans-acting small rnas and their effects on gene expression in Escherichia coli and Salmonella enterica. EcoSal Plus 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmqvist E., Berggren S., Rizvanovic A., RNA-binding activity and regulatory functions of the emerging sRNA-binding protein ProQ. Biochim. Biophys. acta. Gene Regul. Mech. 1863, 194596 (2020). [DOI] [PubMed] [Google Scholar]
- 4.De Lay N., Gottesman S., Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA 17, 1172–1189 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron T. A., Matz L. M., De Lay N. R., Polynucleotide phosphorylase: Not merely an RNase but a pivotal post-transcriptional regulator. PLoS Genet. 14, e1007654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bandyra K. J., Sinha D., Syrjanen J., Luisi B. F., De Lay N. R., The ribonuclease polynucleotide phosphorylase can interact with small regulatory RNAs in both protective and degradative modes. RNA 22, 360–372 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viegas S. C., et al. , Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 35, 7651–7664 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrade J. M., Pobre V., Matos A. M., Arraiano C. M., The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq. RNA 18, 844–855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dendooven T., et al. , A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. Mol. Cell 81, 2901–2913.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cameron T. A., Matz L. M., Sinha D., De Lay N. R., Polynucleotide phosphorylase promotes the stability and function of Hfq-binding sRNAs by degrading target mRNA-derived fragments. Nucleic Acids Res. 47, 8821–8837 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bandyra K. J., Luisi B. F., RNase E and the high-fidelity orchestration of RNA metabolism. Microbiol. Spectr. 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callaghan A. J., et al. , Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437, 1187–1191 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Callaghan A. J., et al. , Studies of the RNA degradosome-organizing domain of the Escherichia coli ribonuclease RNase E. J. Mol. Biol. 340, 965–979 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Dendooven T., et al. , Multi-scale ensemble properties of the Escherichia coli RNA degradosome. Mol. Microbiol. 117, 102–120 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Worrall J. A. R., et al. , Reconstitution and analysis of the multienzyme Escherichia coli RNA degradosome. J. Mol. Biol. 382, 870–883 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda Y., Yagi M., Morita T., Aiba H., Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol. Microbiol. 79, 419–432 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Morita T., Maki K., Aiba H., RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 19, 2176–2186 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prévost K., Desnoyers G., Jacques J.-F., Lavoie F., Massé E., Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 25, 385–396 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bandyra K. J., et al. , The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol. Cell 47, 943–953 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fei J., et al. , RNA biochemistry. Determination of in vivo target search kinetics of regulatory noncoding RNA. Science 347, 1371–1374 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massé E., Escorcia F. E., Gottesman S., Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17, 2374–2383 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruce H. A., et al. , Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes. Nucleic Acids Res. 46, 387–402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park S., et al. , Dynamic interactions between the RNA chaperone Hfq, small regulatory RNAs, and mRNAs in live bacterial cells. Elife 10, e64207 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalaouna D., et al. , Binding of the RNA chaperone Hfq on target mRNAs promotes the small RNA RyhB-induced degradation in Escherichia coli. Non-coding RNA 7, 64 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desnoyers G., Massé E., Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev. 26, 726–739 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinha D., Matz L., Cameron T., De Lay N. R., Poly(A) polymerase is required for RyhB sRNA stability and function in Escherichia coli. RNA 24, 1496–1511 (2018), 10.1261/rna.067181.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanzo N. F., et al. , Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 12, 2770–2781 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cameron T. A., De Lay N. R., The phosphorolytic exoribonucleases polynucleotide phosphorylase and RNase PH stabilize sRNAs and facilitate regulation of their mRNA targets. J. Bacteriol. 198, 3309–3317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massé E., Gottesman S., A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 4620–4625 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel J., Luisi B. F., Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang A., Schu D. J., Tjaden B. C., Storz G., Gottesman S., Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J. Mol. Biol. 425, 3678–3697 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schu D. J., Zhang A., Gottesman S., Storz G., Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 34, 2557–2573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Updegrove T. B., Zhang A., Storz G., Hfq: The flexible RNA matchmaker. Curr. Opin. Microbiol. 30, 133–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaberdin V. R., Walsh A. P., Jakobsen T., McDowall K. J., von Gabain A., Enhanced cleavage of RNA mediated by an interaction between substrates and the arginine-rich domain of E. coli ribonuclease E. J. Mol. Biol. 301, 257–264 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Garrey S. M., Mackie G. A., Roles of the 5’-phosphate sensor domain in RNase E. Mol. Microbiol. 80, 1613–1624 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Chao Y., et al. , In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways. Mol. Cell 65, 39–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melamed S., et al. , Global mapping of small RNA-target interactions in bacteria. Mol. Cell 63, 884–897 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santiago-Frangos A., Kavita K., Schu D. J., Gottesman S., Woodson S. A., C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc. Natl. Acad. Sci. U.S.A. 113, E6089–E6096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kavita K., et al. , Multiple in vivo roles for the C-terminal domain of the RNA chaperone Hfq. Nucleic Acids Res. 50, 1718–1733 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.