Summary
Genomics-driven drug discovery is indispensable for accelerating the development of novel therapeutic targets. However, the drug discovery framework based on evidence from genome-wide association studies (GWASs) has not been established, especially for cross-population GWAS meta-analysis. Here, we introduce a practical guideline for genomics-driven drug discovery for cross-population meta-analysis, as lessons from the Global Biobank Meta-analysis Initiative (GBMI). Our drug discovery framework encompassed three methodologies and was applied to the 13 common diseases targeted by GBMI (Nmean = 1,329,242). Individual methodologies complementarily prioritized drugs and drug targets, which were systematically validated by referring previously known drug-disease relationships. Integration of the three methodologies provided a comprehensive catalog of candidate drugs for repositioning, nominating promising drug candidates targeting the genes involved in the coagulation process for venous thromboembolism and the interleukin-4 and interleukin-13 signaling pathway for gout. Our study highlighted key factors for successful genomics-driven drug discovery using cross-population meta-analyses.
Keywords: genomics-driven drug discovery, genome-wide association study, cross-population meta-analysis, gene prioritization, Mendelian randomization, genetically regulated gene expression
Graphical abstract

Highlights
-
•
Applying genomics-driven drug discovery to cross-population GWAS meta-analyses
-
•
A drug discovery framework consisting of three complementary methodologies
-
•
Integration of the three methodologies synergistically prioritizes drug targets
-
•
Cross-population GWAS meta-analyses expand the catalog of drug candidates
Namba et al. present a drug discovery framework that enables comprehensive in silico screening of drugs and drug targets with supporting evidence to facilitate the translation of GWAS findings to therapeutic targets. They demonstrate that enhanced sample sizes of cross-population GWAS meta-analyses contribute to the acceleration of novel drug discovery.
Introduction
Efficient screening of novel therapeutic targets is an essential process to accelerate drug discovery. Despite the enormous effort to develop novel drugs, the overall success rate of clinical application has been decreasing because of the considerable increase in both the cost and the duration.1 Genomics-driven drug discovery is one of the promising solutions, as drug targets with human genetic support are more likely to be successful in clinical development.2,3 In particular, rare-variant studies for Mendelian diseases have led to drug development, such as PSCK9 inhibitors for low-density lipoprotein cholesterol.4 For common diseases, genome-wide association studies (GWAS) have provided valuable opportunities for drug discovery; nevertheless, drug discovery based on GWAS remains challenging.5 Few bioinformatics tools directly prioritize candidate drugs,6 and there exist no practical guidelines regarding how to conduct genomics-driven drug discovery.
Recently, an increasing number of large-scale GWAS meta-analyses of multiple populations have been carried out. These have revealed key insights into the biological processes underlying complex diseases,7,8 thus affording the possibility of in-depth application of genomics-driven drug discovery. However, the majority of previous genomics-driven drug discovery projects were carried out for GWAS of a single ancestry of Europeans,6 and there are few successful applications to cross-population GWAS meta-analyses. The global heterogeneity in genetic background (e.g., different allele frequencies and linkage disequilibrium [LD]) among populations makes it difficult to perform downstream analyses such as gene expression prediction9 and colocalization analysis.10 In addition, causal effect sizes are population specific especially in functionally important regions.11 Therefore, a specialized drug discovery framework is required for cross-population GWAS meta-analyses.
In this study, we introduce a practical guideline as lessons from the Global Biobank Meta-analysis Initiative (GBMI).7 GBMI meta-analyzed GWASs of global biobanks from diverse ancestries incorporating several recruitment strategies (e.g., population-based or hospital-based biobanks), including up to 1.8 million participants from the 18 biobanks in four continents (341,000 East Asians [EAS]; 31,000 Central and South Asians; 33,000 Africans; 18,000 admixed Americans; 1,600 Middle Easterners; 156,000 Finns; and 1,220,000 non-Finnish Europeans [NFE]), serving as a gold standard cross-population GWAS meta-analysis.
We propose a cross-population drug discovery framework comprising three major methodologies. First, overlap enrichment of disease risk genes with targets of existing drugs12, 13, 14 identifies drug repurposing opportunities. Second, endophenotype Mendelian randomization (MR), and subsequent quality controls, including colocalization analyses,15 establishes causal links between proteins and disease processes. Finally, screening of negative correlations between genetically regulated disease case-control gene expression (GReX) and compound-regulated gene expression profiles16 can be used to identify compounds that might correct disease-related alterations in gene expression.
We applied our framework to the 13 common and relatively rare disease GWAS included in GBMI: asthma, primary open-angle glaucoma (POAG), gout, chronic obstructive pulmonary disease (COPD), venous thromboembolism (VTE), thyroid cancer (ThC), abdominal aortic aneurysm (AAA), heart failure (HF), idiopathic pulmonary fibrosis (IPF), stroke, uterine cancer (UtC), acute appendicitis (AcApp), and hypertrophic cardiomyopathy (HCM). We validated individual methodologies by examining the overlap enrichment of the prioritized results in the disease-relevant medication categories. The integration of the candidate drugs and compounds across methodologies nominated 266 drug/compound-disease pairs as a comprehensive catalog for repositioning. Our results demonstrated the utility of genomics-driven drug discovery using a cross-population GWAS meta-analysis and suggest key factors for successful drug discovery.
Results
Overview of the genomics-driven drug discovery framework
Various types of omics-based approaches have been proposed for novel target identification and drug repositioning from GWAS summary statistics.6 Each type of omics data necessitates specialized methodologies. Furthermore, no simple method has emerged that enables the interpretation of genomics discoveries for drug discovery. Therefore, our framework was composed of three parts, in which each component utilized different external omics data and knowledge bases to obtain biological insights from GWAS summary statistics (Figure 1). First, we performed an overlap enrichment analysis of the disease risk genes with the target genes of existing drugs.12, 13, 14 Gene prioritization tools summarized variant-level p values at the gene level, and the pharmacological agents targeting the prioritized genes served as drug candidates. Genes that are modulated by approved drugs are known to be enriched in disease-relevant clinical classifications, such as drug medication categories (i.e., Anatomical Therapeutic Chemical Classification System [ATC]) and disease categories (i.e., International Statistical Classification of Diseases and Related Health Problems [ICD-10]),13,14 which were utilized to assess the validity of the drug candidates. Second, we performed endophenotype MR and subsequent quality controls, including colocalization analyses.15 MR is a method that is used for estimating the causal effect of one trait (exposure) on another trait (outcome) using genetic variants as instrument variables (IVs).17 We used the lead variants of the protein quantitative trait loci (pQTL) as IVs to examine the disease-causing effects of the proteins. Subsequent quality controls were effective in avoiding false-positive causalities.18 Particularly, colocalization analysis is an important step for the exclusion of confounding by LD. Finally, we performed a screening of negative correlations between case-control GReX and compound-regulated gene expression profiles.16 Transcriptome-wide association studies (TWAS) use expression QTL (eQTL) acting in cis to impute disease-specific GReX from the GWAS summary statistics.9 The compounds that have inverse effects on gene expression against case-control GReX serve as candidates for the disease of interest.16,19 We imputed GReX for the tissues included in the Genotype-Tissue Expression project (GTEx) v.720 and used compound-induced gene expression profiles for thousands of compounds in various conditions and cell lines collected in one of the largest public databases available, the Library of Integrated Network-based Cellular Signatures project (LINCS) L1000 connectivity map.21
Figure 1.
Overview of the genomics-driven drug discovery framework
The framework consisted of three components. Each component utilizes the summary statistics of genome-wide association analyses and external resources to prioritize candidate drugs. GBMI, Global Biobank Meta-analysis Initiative; ATC, Anatomical Therapeutic Chemical Classification System; ICD-10, International Statistical Classification of Diseases and Related Health Problems; pQTL, protein quantitative trait loci; LD, linkage disequilibrium; MR, Mendelian randomization; IV, instrumental variable; GTEx, the Genotype-Tissue Expression project; TWAS, transcriptome-wide association study; LINCS, the Library of Integrated Network-based Cellular Signatures project.
Overlap enrichment of disease risk genes in medication categories
The first component prioritized disease risk genes by calculating gene scores or p values, and examined whether the prioritized genes are enriched in drug-target genes of specific medication categories. There exist several gene prioritization tools, although it remains unclear which tool is best optimized for drug discovery. Therefore, we evaluated four tools in parallel, i.e., MAGMA,22 DEPICT,23 Priority index (Pi),5 and Polygenic Priority Score (PoPS),24 by applying to the summary statistics of GBMI cross-population GWAS meta-analyses.
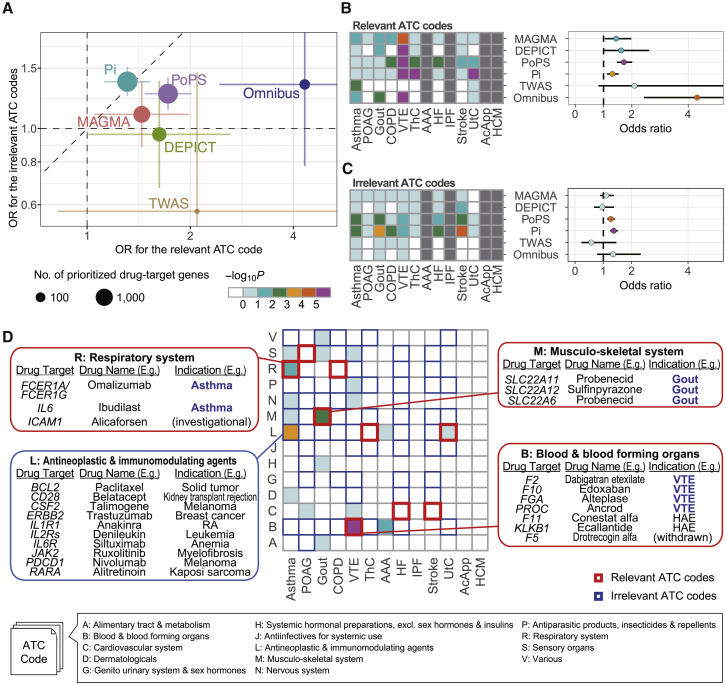
MAGMA is a simple method that is used to summarize variant-level p values according to gene positions and LD structure. The prioritized genes by MAGMA were nominally overlapped with drug-target genes in the disease-relevant ATC codes for Gout, COPD, and VTE (p < 0.05; Figure S1A). DEPICT, which uses co-regulated gene expression for gene prioritization,23 showed clearer enrichment than did MAGMA for gout and VTE; however, no genes were prioritized for diseases with a relatively small number of genome-wide significant loci, such as IPF and UtC (Figure S1B). Pi is a scoring system that was designed for drug development of immune-related diseases, and integrates multiple annotations, including eQTL, chromatin interaction, and genes implicated in immune functions.5 The genes targeted by antineoplastic and immunomodulating agents were enriched for all diseases using Pi (Figure S1C), suggesting that Pi could specifically provide enrichment of the immune genes regardless of the disease categories, even including non-immune diseases. PoPS estimates responsible genes using various gene features, such as cell-type-specific gene expression and biological pathways.24 We observed enrichment of drug-target genes for broad ATC codes, suggesting that PoPS provided relatively high scores for the entire drug-target gene collection, regardless of their medication categories (Figure S1D). We replicated the analyses using ICD-10 and observed a similar pattern of enrichment as ATC codes (Figure S2).
In addition to the four gene prioritization tools, TWAS also prioritizes disease-risk genes by incorporating cis-eQTL information. Therefore, we also evaluated the enrichment of the genes prioritized by TWAS in the medication categories. Because a large sample size and population-specific LD structure are required for robust in silico estimation of GReX by TWAS,9 we applied TWAS to the summary statistics of GBMI NFE-specific and EAS-specific GWAS meta-analyses and subsequently combined the results (STAR Methods). TWAS successfully captured the disease-relevant drug targets for asthma (p = 3.0 × 10−3 for the “respiratory system” category) (Figure S1E). For other diseases, the prioritized genes were concentrated on the diseases with a relatively large number of genome-wide significant loci (asthma, POAG, gout, COPD, and VTE).
Next, we summarized the overlap enrichment into disease-relevant and disease-irrelevant medication categories across the diseases. For both ATC and ICD-10 codes, all tools confirmed enrichment in the relevant codes, although a relatively high enrichment in the irrelevant codes was also observed for Pi and PoPS (Figures 2A–2C and S3), reflecting biased enrichment in immune genes and non-specific enrichment in drug-target genes, respectively. DEPICT yielded a lower enrichment in irrelevant codes than did MAGMA. As a sensitivity analysis, we sequentially changed the thresholds of the gene scores and p values. Among the five tools, TWAS showed the largest enrichment for relevant codes and the smallest enrichment for irrelevant codes. However, TWAS also showed the largest 95% confidence intervals, reflecting the small number of prioritized genes. The pattern of the overall enrichment was robust for a wide range of thresholds (Figure S4). The enrichment of disease genes prioritized by DEPICT in the relevant ATC codes became smaller with the stringent thresholds. DEPICT calculates p values for the genes in the genome-wide significant loci exclusively by default; therefore, liberal thresholds such as a false discovery rate (FDR) of 0.2 might be suitable for DEPICT.
Figure 2.
Enrichment of prioritized drug-target genes in the disease-relevant medication categories
(A) Overall enrichment of drug-target genes nominated by five gene prioritization tools and their omnibus results. The error bars represent 95% confidence intervals.
(B and C) Enrichment of the prioritized drug-target genes in the disease-relevant ATC codes (B) and the disease-irrelevant ATC codes (C). The diseases are sorted in the descending order of the number of genome-wide significant loci determined in GBMI GWAS.
(D) Enrichments for the omnibus results per disease and ATC code. OR, odds ratio; POAG, primary open-angle glaucoma; COPD, chronic obstructive pulmonary disease; VTE, venous thromboembolism; ThC, thyroid cancer; AAA, abdominal aortic aneurysm; HF, heart failure; IPF, idiopathic pulmonary fibrosis; UtC, uterine cancer; AcApp, acute appendicitis; HCM, hypertrophic cardiomyopathy; RA, rheumatoid arthritis; HAE, acute attacks of hereditary angioedema.
Given that the five tools separately prioritized genes according to the different methodologies, we hypothesized that omnibus integration of the five methods could efficiently improve the enrichment of disease-relevant drug-target genes. While each tool was previously constructed for gene prioritization, the effectiveness of combining these methods has not been explored in the context of drug discovery. As an omnibus approach, we selected 177 gene-disease pairs that were prioritized by at least 4 of the 5 tools (Table S1), and showed a twice larger odds ratio (OR) than those of any single tool for the relevant ATC codes (OR = 4.14, p = 2.2 × 10−5) (Figures 2A–2C). Conversely, the enrichment in the irrelevant ATC codes (OR = 1.33, p = 0.19) was close to that of the single tools, indicating an advantage of combining multiple gene prioritization methods to confine the prioritized genes to the highly disease-relevant genes while retaining controlled type 1 errors. We also confirmed the efficacy of the omnibus strategy using ICD-10 (Figure S3). In addition, we found that the omnibus approach successfully incorporated multiple gene prioritization features, including proximity to the lead variants, tagging of the functional variants, protein-protein interaction with the nearest genes, gene functions, and tissue-specific expression status (Data S1; Figure S5).
The examination of the omnibus results for each disease revealed an enrichment in the relevant ATC codes for asthma, gout, and VTE (Figure 2D), which corresponded to 154 drugs in total (Table S1). For asthma and gout, all prioritized genes in the disease-relevant ATC codes were targeted by the drugs with approved indication, except for ICAM1 for asthma, which was targeted by investigational drugs. The genes prioritized for VTE were involved in the coagulation cascade, and four of them (F2, F10, FGA, and PROC) were the approved drug targets for VTE. The prioritized genes for asthma were also enriched in antineoplastic and immunomodulating agents. Although the drugs in this category have not been indicated for asthma, their target genes involved immune genes, such as IL1R1, suggesting that the omnibus approach correctly prioritized genes related to asthma.
We conducted two sensitivity analyses. First, instead of using the summary statistics of cross-population GWAS meta-analyses, we applied individual gene prioritization tools to the summary statistics of population-specific GWAS meta-analyses and subsequently meta-analyzed the results. The gene scores of individual tools were more similar to those calculated from NFE-specific GWAS summary statistics than those from cross-population GWAS summary statistics (Figure S6). This was possibly because NFE were the majority even in GBMI, and the population-specific GBMI GWAS summary statistics were currently not available for some non-NFE populations (Table S2). In GBMI, the population-specific summary statistics were currently not available to avoid distributing biobank-specific summary statistics if only one biobank corresponds to a particular single population (e.g., Finnish GWAS results from FinnGen). The omnibus gene prioritization nominated 90 gene-disease pairs in the approach using the population-specific dataset, much fewer than the approach using cross-population summary statistics (Figure S7A). The smaller number of participants included in the population-specific dataset might explain the difference in the number of prioritized genes. The enrichment of the prioritized genes in the medication categories was not apparently different between the two approaches (Figures S7B–S7D). Next, we examined whether the overlap enrichment analysis was affected by potential confounders, such as gene-gene correlations and gene length. The enrichment in the disease-relevant ATC codes was consistent after accounting for confounders (STAR Methods; Figure S8).
Endophenotype MR
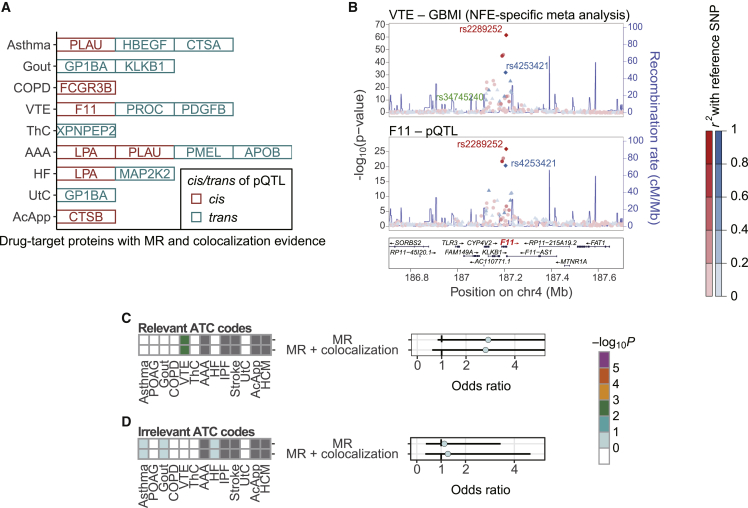
Understanding protein regulation is essential for drug discovery. Therefore, we conducted MR analyses to infer disease-causing proteins.15,25 Although the current eQTL studies yield larger sample sizes than pQTL,26 there exists low correlation between transcript expression and protein abundance.27 Here, we utilized the lead variants summarized from five European pQTL studies27, 28, 29, 30, 31 as IVs for MR. To avoid confounding factors, such as horizontal pleiotropy, we restricted IVs to those with low heterogeneity between studies and low pleiotropy between proteins, whereas we used both cis- and trans-pQTL to target a wide range of proteins, as described previously18 (STAR Methods). We conducted an MR analysis using the 894 IVs associated with the 818 proteins, to test the causal effects on the 13 diseases. After applying multiple test corrections, 64 protein-disease pairs, including 24 pairs with drug-target proteins, showed significant causality (FDR < 0.05; Table S3). Twenty-six pairs were derived from cis-pQTL, whereas the remaining ones were derived from trans-pQTL.
To protect against false positives from the results of the MR analysis, we applied two quality-control metrics, i.e., colocalization analysis and concordance of directional effects. The colocalization analysis was used to check whether two signals were equally distributed on the local LD structure. Therefore, we used the GBMI GWAS of the NFE-specific meta-analysis for MR and colocalization analysis, rather than the all-ancestry meta-analysis, to match the population background to the pQTL studies. All pairs passed the directionality check, while colocalization was confirmed for 34 pairs, including 18 pairs with drug-target proteins (Figure 3A). The colocalization of F11-VTE is shown in Figure 3B as an illustrative example. Most of the colocalization methods assume only a single causal variant per locus; however, recent methodological advances enabled us to address the possibility of multiple causal variants in one locus.32 We applied coloc33 to conditionally independent signals decomposed by SuSiE.34 SuSiE detected three and two signals for the GWAS of VTE and pQTL of F11, respectively, of which two signals were inferred to be colocalized.
Figure 3.
Endophenotype Mendelian randomization
(A) Drug-target proteins with significant causal effects inferred by Mendelian randomization and with colocalization between GBMI GWAS and protein quantitative trait loci (pQTL).
(B) LocusZoom35 plots showing colocalization between GWAS for VTE and pQTL for F11. The fine-mapped variants are shown with their rsID. Only the variants shared between GBMI GWAS and pQTL summary statistics are shown for visualization purposes.
(C and D) Enrichment of the prioritized drug-target proteins in the disease-relevant ATC codes (B) and the disease-irrelevant ATC codes (C). The error bars represent 95% confidence intervals.
To validate the prioritized proteins in the context of drug discovery, we examined the overlap enrichment analysis of the prioritized proteins in the medication categories. We observed relative enrichment in the disease-relevant codes (OR = 2.91, p = 0.11; Figure 3C) and not in the disease-irrelevant codes (OR = 1.13, p = 0.51; Figure 3D), supporting the validity of the MR-prioritized proteins. Restricting the analysis to the proteins supported by the colocalization had little effect on the enrichment.
Next, we assessed whether the inferred causal relationships were consistent with clinical implications and experimental evidence. We found literature-based support of the causal signs of the MR effect sizes for six drug-target protein-disease pairs (Table S4). For example, lipoprotein(a) (LPA) for AAA and PDGFB for VTE have been reported to be disease biomarkers.36,37 Similarly, ApoB-containing lipoproteins were associated with angiotensin II-induced AAA in a mouse model.38 In contrast, we found that the negative sign of the MR effect size for PROC-VTE was not consistent with the knowledge that protein C, which is encoded by PROC, itself, is used for the treatment of VTE. The pQTL of PROC was identified as a trans-pQTL, which might confound the sign because PROC may not be the direct target of the pQTL effects.
For the prioritized proteins with colocalization evidence, we curated drugs from the four major drug databases: DrugBank,39 Therapeutic Target Database,40 PharmGKB,41 and the Open Targets Platform,42 resulting in 83 drugs for 14 protein-disease pairs (Table S4). These drugs included MAP2K inhibitors for HF, which experimentally ameliorate cardiac hypertrophy and cardiomyopathy.43,44 Regarding the F11-VTE pair, an F11 inhibitor, abelacimab, showed efficacy for the prevention of VTE in a phase II trial.45 In addition, as an agonist of PLAU for asthma, the urokinase-type plasminogen activator was reported to reduce airway remodeling in a mouse model.46
Negative correlation tests between genetically determined and compound-regulated gene expression
Finally, we performed an in silico screening of negative correlations between TWAS-based disease case-control GReX and compound-regulated gene expression profiles16 to identify compounds with a potentially beneficial effect for treatment of each disease. By matching the cell and tissue specificity between TWAS (based on the GTEx tissues) and compound-regulated gene expression profiles (cell lines collected in the LINCS L1000 library), we tested the negative correlation for 308,872 compound-tissue-condition pairs per disease (STAR Methods). As mentioned earlier, a large sample size and population-specific LD structure are required for robust in silico estimation of GReX by TWAS.9 Therefore, we restricted our analysis to the results of the population-specific meta-analyses of EAS and NFE. An EAS-specific meta-analysis was not performed in GBMI for three diseases, i.e., ThC, AAA, and AcApp, which were excluded. We calculated correlation coefficients in the two populations separately and subsequently combined them in a random-effect meta-analysis framework. We obtained 31 compound-disease pairs with an FDR < 0.05 with no apparent inflation (inflation factors [lambda] ranged from 0.86 [POAG] to 1.18 [COPD]) (Figure 4A). In particular, while most of the compounds with FDR < 0.05 were prioritized for gout, inflation was not observed for gout (lambda = 0.97). Despite our thorough investigation, we observed no features significantly correlated with the inflation factors (Data S2; Figures S9 and S10).
Figure 4.
Negative correlation tests between genetically determined and compound-regulated gene expression profiles
(A) Quantile-quantile plots of the negative correlation tests between genetically determined and compound-regulated gene expression profiles. Compounds with false discovery rates (FDR) < 0.05 are indicated by larger dots. The compound names are shown for at most three significant compounds, for visualization purposes.
(B and C) Enrichment of the prioritized compounds in the disease-relevant ATC codes (B) and the disease-irrelevant ATC codes (C). No compound was prioritized for HF, IPF, stroke, UtC, and HCM (colored in gray). The error bars represent 95% confidence intervals, and the confidence interval was infinite for the compounds with FDR < 0.05 in (B).
The negative correlation tests can be applied even to compounds without known targets when the compound-induced gene expression changes are assayed. In fact, most of the prioritized compounds (14 out of 31) were understudied or had no known targets. These compounds were valuable, because they could be therapeutic drugs with different modes of action from existing drugs. Nevertheless, several prioritized compounds were well studied and had supporting evidence. A histone deacetylase (HDAC) inhibitor, vorinostat, was prioritized for asthma; concordantly, HDAC inhibition was an effective treatment in an animal model of asthma.47
To validate the concept of the negative correlation tests, we compared the negative correlations of the approved compound-indication pairs to those of non-approved pairs. The approved pairs had larger negative correlations consistently for all diseases (Table S5), supporting the concept of this approach. We further examined whether the prioritized compounds were enriched in the disease-relevant medication categories. However, ATC codes were assigned only for 6 of the 31 compounds because most of the compounds were understudied. We then used 123 unique compound-disease pairs with marginal significance (FDR < 0.1). There were 72 compounds with known mechanistic actions, and ATC codes were assigned for 35 compounds. We observed enrichment in the disease-relevant codes (OR = 3.40, p = 9.0 × 10−3; Figure 4B) and not in the disease-irrelevant codes (OR = 1.38, p = 0.12; Figure 4C), again validating the compounds prioritized by the negative correlation tests.
When we expanded our literature search also for the compound-disease pairs with FDR < 0.1, the supporting literature was identified for 26 pairs. Indacaterol (a beta-2 adrenergic receptor [ADRB2] agonist) and masatinib (a proto-oncogene c-Src [SRC] inhibitor) have undergone phase III clinical trials for asthma.48,49 There are approved drugs (acetazolamide, fluorouracil, and naproxen and indomethacin, respectively) with the same mechanistic action as cianidanol (a carbonic anhydrase inhibitor) and raltitrexed (a thymidylate synthase inhibitor) for POAG,50 and piketoprofen and diclofenac (cyclooxygenase 1/2 inhibitors) for gout, respectively. We also prioritized several compounds currently under investigation, including phosphatidylinositol 3-kinase inhibitors for asthma,51 a sodium channel inhibitor for POAG,52 and a cyclooxygenase-2 inhibitor for VTE.53 We comprehensively summarized the screened drug list and their relevant evidence in Table S6, which should provide genetic support to the compounds under investigation and the understudied compounds. In addition, we searched for structurally similar compounds (similarity > 0.85) for the compounds without known targets by using BindingDB.54 We identified potential targets for seven compounds (Table S7), which would help investigate those compounds as therapeutic candidates.
Combining the three approaches for in-depth drug discovery
We summarize the representative drugs and their targets, which were prioritized through the three drug discovery approaches, in Table 1. The overlap enrichment analysis of the omnibus gene prioritization and the endophenotype MR analysis identified 154 and 83 drugs for 14 and 14 drug targets, respectively, and 31 compounds were nominated by the negative correlation tests. Eight drug-target genes were prioritized by both the omnibus gene prioritization and the endophenotype MR (Table S8), providing further support for these genes. In addition, one gene prioritized by the omnibus approach (HSP90AA1 for gout) was also targeted by a compound marginally prioritized by the negative correlation tests (ganetespib, FDR < 0.1). Multiple components nominated drug candidates for asthma, gout, COPD, and VTE, indicating that the three components were complementarily for these diseases. These nominated diseases had a relatively large number of case sample sizes and genome-wide significant loci. Conversely, no drugs were nominated for diseases with a relatively small number of loci by either the overlap enrichment analysis or negative correlation tests (e.g., IPF, UtC, and HCM). As these two components require relatively large numbers of GWAS signals in their schemes, further accumulation of samples is warranted.
Table 1.
Drug targets and representative drugs prioritized in this study
| Disease | Overlap enrichment in disease-relevant ATC codes | Endophenotype MR | Negative correlation tests |
|---|---|---|---|
| Asthma | ibudilast (IL6); omalizumab (FCER1A/FCER1G); alicaforsen (ICAM1) | SAR164653 (CTSA); KHK-2866 (HBEGF); amediplase (PLAU) | donepezil (ACHE); vorinostat (HDAC family); BRD-K50748030 (unknown) |
| POAG | – | – | LY-288513 (unknown) |
| Gout | probenecid (SLC22A6/SLC22A11); lesinurad (SLC22A11/SLC22A12) | anfibatide (GP1BA); berotralstat (KLKB1) | mesoridazine (DRD2/HTR2A); CP-724714 (unknown)a |
| COPD | – | GMA-161 (FCGR3B) | BRD-K48950795 (unknown); RAN-29 (unknown) |
| VTE | alteplase (FGA); edoxaban (F10); ancrod (PROC); dabigatran etexilate (F2); abelacimab (F11); ecallantide (KLKB1); drotrecogin alfa (F5) | abelacimab (F11); CR-002 (PDGFB) | WH-4025 (unknown) |
| ThC | – | tosedostat (XPNPEP2) | (excluded from the analysis) |
| AAA | – | MG-1102 (LPA); amediplase (PLAU) | (excluded from the analysis) |
| HF | – | MG-1102 (LPA) | – |
| IPF | – | – | – |
| Stroke | – | – | – |
| UtC | – | Anfibatide (GP1BA) | – |
| AcApp | – | GNF-PF-5434 (CTSB) | (excluded from the analysis) |
| HCM | – | – | – |
The diseases are sorted in the decreasing order of the number of the genome-wide significant loci in GBMI GWAS.
Drugs without known target genes were not shown except for the drug with the lowest p value, CP-724714. Full results can be found in Table S2.
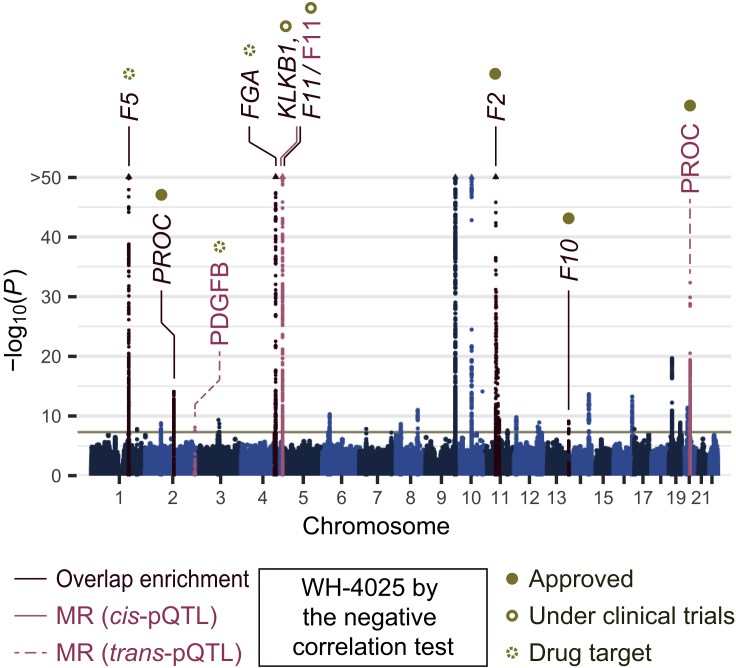
We noted that the VTE GWAS was most successful in the screening of the drug targets: it prioritized drugs corresponding to the eight drug-target genes and one compound in total (Figure 5). All drug targets but PDGFB were involved in the coagulation cascade. PDGFB has been reported to induce the expression of a tissue factor that triggers the coagulation cascade,55 which underscores the strong enrichment of candidate drug targets in coagulation-related genes. Of the eight drug targets, three (PROC, F2, and F10) were targeted by the approved drugs, and two (KLKB1 and F11) were targeted by drugs under clinical trials for VTE, thus supporting the validity of genomics-driven drug discovery and repositioning for VTE.
Figure 5.
Drug discovery nominated plausible candidate drugs and target genes for VTE
Drug-target genes nominated by omnibus gene prioritization and proteins nominated by Mendelian randomization (MR) are highlighted in a Manhattan plot of GBMI GWAS for VTE. The compound with a significant negative correlation between genetically regulated and compound-regulated gene expression profiles is also shown.
We further evaluated the overlap of the prioritized gene-disease pairs at the pathway level. We observed significant enrichment for 163 pathways (Table S9), and the enrichment for 20 pathways became significant when combining three approaches. One such pathway was the interleukin-4 (IL-4) and IL-13 signaling pathway for gout (FDR = 5.4 × 10−3) (Figure 6). This pathway included five genes separately prioritized by the three approaches (HSP90AA1 by the omnibus approach, SAA1 and CCL12 by endophenotype MR, and MAOA and STAT3 by the negative correlation tests of gene expression profiles), illustrating that the three approaches prioritized drug targets synergistically.
Figure 6.
The three drug discovery approaches complementarily prioritized drug targets for gout
The genes prioritized for gout by the three drug discovery approaches were connected to each other if their protein-protein interaction scores56 were larger than 0.3. Out of the 50 prioritized genes, 43 genes formed a large cluster and shown in the figure. Line thickness represents the strength of protein-protein interaction. The prioritized genes were enriched in six pathways, and these pathways are overlayed on the gene network.
The enhanced sample sizes and power of cross-population GWAS meta-analyses contributed to genomics-driven drug discovery
The enhanced sample sizes and power of the GWAS for a wide range of phenotypes in the framework of global biobank collaboration should contribute to the acceleration of novel drug discovery. The novel loci in GBMI7 indeed led to the prioritization of PROC for VTE (via the overlap enrichment analysis) and PLAU for AAA (via the endophenotype MR). To further evaluate the effect of the enhanced GWAS sample sizes and power on genomics-driven drug discovery, we compared the prioritized results with the identical analyses conducted with European-only GWAS summary statistics. We note that we here focused on the European population because Europeans are generally the primary population in GWAS. When using GBMI NFE-specific GWAS summary statistics (Nmean = 859,137) and the GBMI summary statistics of the NFE participants in the UK Biobank (Nmean = 396,227), smaller numbers of genes, proteins, and compounds were prioritized for all three approaches (Figure S11). We also applied our framework to seven publicly available GWAS summary statistics (primarily consisting of Europeans, Nmean = 435,450) (Table S2). The number of prioritizations using the public dataset was generally smaller than those using the GBMI dataset except for some diseases for which the public dataset had larger GWAS power than the GBMI dataset. For such diseases, we anticipate that cross-biobank meta-analysis approaches will surpass most of the disease-oriented studies in sample sizes in the near future. Together, these results clearly show that the enhanced GWAS sample sizes and power of global biobank collaboration also increased the statistical power of genomics-driven drug discovery as well.
Discussion
In this study, we present a practical framework that combines three approaches for in-depth genomics-driven drug discovery, and demonstrate its utility through application to the GBMI GWAS meta-analysis. Each approach has specific advantages. By focusing on the genes prioritized in the disease-relevant medication codes, we obtained a list of candidate drugs, most of which were indicated for the diseases. Endophenotype MR and subsequent quality controls estimated the causality of proteins regarding diseases, which could not be inferred by gene prioritization. Finally, the negative correlation tests of gene expression profiles were able to nominate compounds with or without known target genes. Our study demonstrated the importance of combining the three components for the thorough evaluation of candidate drugs.
Multi-ethnic GWAS meta-analyses incorporated populations with diverse genetic backgrounds and architectures. Matching of genetic ancestry is important to follow-up functional interpretation of the GWAS results with omics information, including drug discovery. To address the difference in the LD structure between populations, we used the GWAS summary statistics from the population-specific meta-analysis for endophenotype MR and the negative correlation tests, whereas we used those from the all-population meta-analyses for gene prioritization. The differences in ancestry-matching strategies among components depended on the current availability of the corresponding omics resource requested for each analysis. Further accumulation of public resources with diverse ancestry should be expected.25,57
A large number of GWAS loci were required for genomics-driven drug discovery, especially for the overlap enrichment analysis and the negative correlation tests. Global biobank collaborations, such as GBMI, have the potential to improve the power of genetic association studies to detect novel GWAS signals by incorporating diverse populations with large sample sizes, and will facilitate genomics-driven drug discovery. Of note, our framework successfully nominated drug candidates and their target genes, particularly for VTE, which had the fifth-largest number of GWAS loci among the 13 diseases. There may be additional factors other than the number of GWAS loci that are important for the success of drug discovery in VTE. Among the traits with a large number of loci, VTE showed a relatively modest polygenicity,7 which might be beneficial for pinpointing the disease-relevant genes. The genes implicated in the VTE GWAS were mainly centered at the coagulation cascade.58 The drugs targeting coagulation factors have been under active development,59 and these drugs can be promising candidates immediately repositioned to coagulation disorders other than the disease for which the drugs were originally developed. The further evaluation of the suitability of drug discovery for a broader range of phenotypes, including cancer, autoimmune diseases, and coagulation disorders, would be an interesting direction for future research.
In conclusion, our drug discovery framework practically afforded the in silico screening of abundant drugs and targets with supporting evidence. It enables the routine to conduct the post-GWAS genomics-driven drug discovery in the era of cross-population GWAS meta-analysis, which would further facilitate the translation of GWAS findings to therapeutic targets.
Limitations of the study
There are several potential limitations for each drug discovery component. Drug-target and drug classification information need to be annotated to conduct the overlapping enrichment analysis in medication categories. Endophenotype MR could be applied to proteins targeted by pQTL studies; however, the number of proteins in the pQTL studies is currently limited because of the technological difficulty of proteomics.60 Regarding the negative correlation tests, the LINCS L1000 compound library does not contain gene expression profiles for all pairs of compounds and cell lines.21 Therefore, we used all tissue types in GTEx, regardless of disease relevance. In addition, the methodological limitations of TWAS9,61 might affect drug discovery using GReX. Finally, although we validated individual approaches by referring known drug-disease relationships, direct in vivo or in vitro experimentation should be warranted as a next step for the prioritized genes and compounds in this study.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources should be directed to the lead contact, Yukinori Okada (yokada@sg.med.osaka-u.ac.jp).
Materials availability
This study did not generate new unique reagents.
Method details
GBMI GWAS meta-analysis
GBMI GWAS is a meta-analysis of 18 biobanks incorporating up to 1.8 million participants with diverse ancestries (341,000 EAS; 31,000 Central and South Asians; 33,000 Africans; 18,000 Admixed Americans; 1,600 Middle Easterners; 156,000 Finns; and 1,220,000 NFE).7 We used the GBMI GWAS of 13 common diseases: asthma, POAG, gout, COPD, VTE, ThC, AAA, HF, IPF, stroke, UtC, AcApp, and HCM. Although a GBMI GWAS was also conducted for appendectomy, we excluded this trait because it was a procedure endpoint rather than a disease. We defined genome-wide significant loci in the same way as the GBMI flagship paper.7 Specifically, we iteratively spanned the ±500 kb region around the most significant variant and merging overlapping regions until no genome-wide significant variants were detected within ±500 kb. We estimated the polygenicity and the SNP heritability by applying SBayesS64 with default parameters to the summary statistics of the variants with minor allele frequency >0.01.
Gene prioritization
We used four tools for gene prioritization from GWAS summary statistics, i.e., MAGMA,22 DEPICT,23 Pi,5 and PoPS.24 We also used TWAS for gene prioritization, and the methods for TWAS were described later. We used the default settings, unless otherwise stated.
MAGMA is a simple method that summarizes variant-level p-values into gene-level p-values according to gene positions and LD structure. For MAGMA, we used the “—gene-model snp-wise = mean” option. DEPICT aims to capture a set of genes within associated loci so that the genes share functional annotations. Briefly, DEPICT maps lead variants to proximal genes and then prioritize the genes with gene set memberships similar to genes from other associated loci. Variants with p-values < 1.0 × 10−5 were used as input for DEPICT. Pi is a scoring system designed for drug development of immune-related diseases. Pi first maps lead variants to genes using proximity, chromatin interaction, and eQTL. These “seed” genes were annotated for immune-related functions, phenotypes, and diseases. Then, higher p-value-like scores are assigned for genes with higher network connectivity to the seed genes. The scores are combined using Fisher’s method and rescaled to 0–5. We followed the original paper for the setup of Pi.5 Specifically, we used the lead variants with p < 5.0 × 10−8 as input, eQTL in the peripheral blood and immune cells, chromatin interaction in immune cells, topologically associating domain boundary in the GM12878 cell line, and the STRING56 protein–protein interaction network, with a high confidence score. PoPS is a tool that assigns high scores to genes with features characteristic of the genes with strong gene-level associations. PoPS models MAGMA gene-level association scores by gene features derived from cell-type specific gene expression, biological pathways, and protein–protein interactions. Then, association scores predicted by the model, not the original ones, are reported. PoPS was originally developed to pinpoint one responsible gene per locus, and chooses the gene with the highest score at the locus of interest.24 Here, we used top-ranked genes, rather than pinpointed genes, to incorporate multiple genes per locus for drug discovery.
We used the summary statistics of the all-population GWAS meta-analyses as input and the European subset of the 1000 Genomes Project (Phase 3) as a reference, given that more than half of the GBMI samples were NFE.7 We note that the annotations internally used in the individual tools were mainly derived from European samples. We prioritized genes with conventional thresholds, i.e., FDR <0.05 for MAGMA, and top 5% of the genes in the descending order of gene scores for Pi and PoPS. We used an FDR <0.2 for DEPICT, as DEPICT calculates p-values only for the genes in the pre-featured target loci, by default. When we examined sequentially changed thresholds as a sensitivity analysis, we used FDR thresholds of 0.001, 0.005, 0.01, 0.05, 0.1, 0.2, and 0.5 for MAGMA and DEPICT. Pi and PoPS calculate the gene score instead of FDR. We used the top 1%, 5%, and 10% of the genes with the highest gene scores. In addition, a Pi score >1.5, 2.0, 2.5, and 3.0 for Pi, and a PoPS score >0, 0.5, 1.0, 1.5, and 2 for PoPS were also examined.
Meta-analysis of gene prioritization results
As a sensitivity analysis, we applied gene prioritization tools to population-specific GWAS summary statistics and subsequently meta-analyzed the results. The four gene prioritization tools calculate different metrics from each other. MAGMA calculates Z-scores and p-values, and we meta-analyzed the Z-scores by Stouffer’s method using the function implemented in MAGMA. DEPICT calculates p-values only for the genes in the genome-wide significant loci. Therefore, we imputed the missing p-values with 0.5 and converted the p-values into Z-scores using the inverse normal distribution. Subsequently, we meta-analyzed the Z-scores by Stouffer’s method. For Pi and PoPS, we took the median scores weighted by the squared root of the GWAS effective sample sizes.
Transcriptome-wide association study
We conducted probabilistic TWAS fine-mapping using FOCUS.62 FOCUS is a Bayesian approach that takes GWAS summary statistics and eQTL weights as inputs and estimates causal effects of genes mediated by gene expression changes. We used the same parameters used internally in Trans-Phar.16 Specifically, we used default parameters and priors, except that we set the p-value threshold to 1 to target genome-wide regions. We ran FOCUS repeatedly for 44 GTEx v7 tissues and took the median of their posterior inclusion probabilities (PIP). We further combined the PIP calculated from population-specific GWAS summary statistics by taking the median weighted by the squared root of the GWAS effective sample sizes. We prioritized genes with PIP >0.1, and used PIP thresholds of 0.01, 0.05, 0.1, and 0.2 for the sensitivity analysis. We note that the GTEx project largely contains European participants, which might lead to potential LD mismatch for EAS-specific GWAS meta-analyses. We used European and East Asian participants in the 1000 genomes project as the LD reference for NFE and EAS, respectively.
Gene prioritization features
We compiled 50 features for protein-coding genes and performed one-sided Fisher’s exact test to evaluate their enrichment in the genes prioritized by the omnibus approach. Out of the 50 features, we calculated seven locus-to-gene features for each disease (the genes nearest to the lead variants, the genes with protein–protein interaction [PPI] to the nearest genes, and the genes for which the GWAS loci tagged functional variants with p < 1.0 × 10−4 in five categories: putative loss-of-function [pLoF] variants, non-synonymous variants, exonic variants, promoter-region variants, and enhancer-region variants). We also curated five gene features (gene length, amino-acid [AA] length, transcription factors, immune-related genes, cancer-related genes) and the genes expressed specifically to 38 tissues from the GTEx project.65 Two numeric gene features (gene length and AA length) were converted to binary features by dividing the genes into the top-5% longest genes and the other. To examine whether the features were associated with the prioritized genes conditioned on the nearest genes, we conducted logistic regression by jointly modelling the nearest genes and other features. We calculated p-values by the one-sided Wald test.
Mendelian randomization of the pQTL signals
We used two-sided Wald ratio tests for MR analyses. The lead variants of the five European protein QTL studies27, 28, 29, 30, 31 were evaluated as described previously.18 For all evaluated proteins, including those currently not targeted by drugs, we used the lead variants classified as tier 1 as instrumental variables. The tier 1 variants were defined as the variants that were associated with no more than five proteins and did not show heterogeneity in the five studies. When the lead variant of the pQTL was missing in the GBMI GWAS summary statistics, we used a proxy variant with the largest R2, if the R2 was larger than 0.8. We checked the directionality of causal relationships using Steiger filtering.66 All MR analyses were performed using the TwoSampleMR R package.63
Colocalization analysis
To test colocalization in the presence of multiple causal variants, we applied coloc to the signals decomposed by SuSiE32 for each locus, including the variants located within ±500 kb of the lead variant (coloc + SuSiE). If SuSiE34 did not converge in 100 iterations for either pQTL or GBMI GWAS, we instead used coloc.33 Coloc + SuSiE and coloc were performed using their default parameters. The summary statistics were not publicly available for two pQTL studies30,31; therefore, we compared R2 between the lead variants of the pQTL and the GBMI GWAS for those studies. We considered that the signals of pQTL and GBMI GWAS were colocalized if the maximum posterior probability of colocalization (i.e., PP.H4 for coloc and coloc + SuSiE) was larger than 0.8, or the R2 between the lead variants was larger than 0.8.
Negative correlation tests of gene expression
We utilized Trans-Phar16 for the negative correlation tests. Specifically, we performed a series of negative correlation tests between disease case–control GReX and compound-regulated gene expression profiles for 308,872 compound–tissue–condition pairs per disease. Trans-Phar internally used FOCUS62 to infer disease case–control GReX for 44 GTEx v7 tissues based on the GWAS summary statistics. Compound-regulated gene expression profiles were obtained from the LINCS L1000 library.21 We calculated Spearman’s rho between GReX and compound-regulated gene expression profiles using the GReX inferred from the NFE-specific and EAS-specific GWAS meta-analyses, separately. The correlation coefficients were combined by a random-effect model using the metacor R package and p-values were calculated by the one-sided tests. In the LINCS L1000 library, some compounds had multiple compound-regulated gene expression profiles evaluated with different conditions. When we compared the negative correlations between approved and non-approved compounds, we kept one condition per compound by selecting the compound-regulated gene expression profiles that showed the largest negative correlation with genetically determined gene expression profiles. The target-gene information of the LINCS L1000 library was curated from its official database via Google BigQuery (https://github.com/cmap/cmapBQ) and the drug repurposing hub (https://clue.io/repurposing),67 in addition to the four major drug databases (DrugBank,39 TTD,40 PharmGKB,41 and the Open Targets Platform42).
Filtering of GWAS summary statistics
Because of the diverse ancestries in the GWASs for meta-analysis, there was remarkable heterogeneity in the effective sample sizes across the genome-wide variants of the GBMI results of each phenotype. This heterogeneity affected the performance of downstream analyses, including those of polygenic risk score.68 Therefore, we excluded variants with effective sample sizes <50% of the maximum effective sample size from the GWAS summary statistics of each phenotype.
Quantification and statistical analysis
Enrichment analysis in medication categories
We used the list of prioritized genes as input to perform a series of Fisher’s exact tests for ATC or ICD-10 codes, to test the enrichment of drug-target genes in particular codes. We used the drug-target database provided by GREP,14 which was constructed by curating two major drug databases, Drug Bank and TTD. The ICD-10 codes were summarized into the 21 large categories, as shown in Figure S2. Because ICD-10 is a disease-classification system, we simply defined the relevant ICD-10 category as the category that contained the disease. Conversely, ATC is a drug-classification system, and the approved drugs can belong to multiple ATC codes. Therefore, we defined the disease-relevant ATC code as the ATC code with the largest number of approved drugs for the disease-relevant ICD-10 category. We defined the disease-irrelevant ATC codes as the ATC codes without any approved drugs for the disease-relevant ICD-10 category. Regarding ATC, we limited the enrichment analyses to the diseases for which there were more than four approved drugs in the disease-relevant ATC codes. As a result, four diseases (i.e., AAA, IPF, AcApp, and HCM) were excluded. We also utilized MAGMA gene set enrichment analysis to evaluate the enrichment accounting for potential confounders. MAGMA gene set enrichment analysis is a linear regression model of gene-level Z-scores. We used the default covariates: gene length, gene density, and inverse minor allele counts, and their logarithmic values. Technically, MAGMA gene set enrichment analysis uses all analyzed genes as the background genes, but the background genes of the overlap enrichment analysis in medication categories should be restricted to the drug-target genes. To address that, we created the subset of the output file MAGMA gene analysis containing only drug-target genes and used it for MAGMA gene set enrichment analysis. p-values were calculated by one-sided tests for both Fisher’s exact tests and MAGMA gene set enrichment analysis.
Pathway enrichment analysis
We conducted one-sided hypergeometric tests implemented in the ReactomePA R package69 for the enrichment analysis of the prioritized genes in the Reactome pathways. Following the default settings, we considered the pathways as significant if p-values < 0.05 after being adjusted by the Benjamini-Hochberg method and Q-values < 0.2. We subsequently filtered out the pathways with less than three prioritized genes to ensure that multiple genes in the pathways were prioritized.
Acknowledgments
The authors thank Drs. Saori Sakaue and Masahiro Kanai for thoughtful discussions on genomics-driven drug discovery. The authors also thank Drs. Benjamin M. Neale, Sarah Graham, Jie Zheng, and Jibril Hirbo for the review within GBMI. S.N. was supported by Takeda Science Foundation. T.K. is an employee of Japan Tobacco Inc. W.Z. was supported by the National Human Genome Research Institute of the National Institutes of Health under award no. T32HG010464. Y.O. was supported by JSPS KAKENHI (22H00476), AMED (JP21gm4010006, JP22km0405211, JP22ek0410075, JP22km0405217, and JP22ek0109594), JST Moonshot R&D (JPMJMS2021 and JPMJMS2024), Takeda Science Foundation, and Bioinformatics Initiative of Osaka University Graduate School of Medicine, Osaka University.
Author contributions
S.N., T.K., and Y.O. designed the study and wrote the manuscript. S.N., T.K., K.-H.W., and W.Z. conducted data analysis. Y.O. supervised the study.
Declaration of interests
The authors declare no competing interests.
Published: October 12, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2022.100190.
Supplemental information
Data and code availability
The GBMI GWAS summary statistics are publicly available at https://www.globalbiobankmeta.org/resources. The genomics-driven drug discovery analysis was conducted using the following publicly available tools: MAGMA (https://ctg.cncr.nl/software/magma), DEPICT (https://data.broadinstitute.org/mpg/depict/), PoPS (https://github.com/FinucaneLab/pops), FOCUS (https://github.com/bogdanlab/focus), GREP (https://github.com/saorisakaue/GREP), Trans-Phar (https://github.com/konumat/Trans-Phar), LocusZoom (http://locuszoom.org/), and the Pi, TwoSampleMR, coloc, susieR, and metacor R packages.
References
- 1.Hay M., Thomas D.W., Craighead J.L., Economides C., Rosenthal J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014;32:40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 2.Nelson M.R., Tipney H., Painter J.L., Shen J., Nicoletti P., Shen Y., Floratos A., Sham P.C., Li M.J., Wang J., et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 3.King E.A., Davis J.W., Degner J.F. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 2019;15:e1008489. doi: 10.1371/JOURNAL.PGEN.1008489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabatine M.S., Giugliano R.P., Keech A.C., Honarpour N., Wiviott S.D., Murphy S.A., Kuder J.F., Wang H., Liu T., Wasserman S.M., et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med. 2017;376:1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 5.Chen L., ULTRA-DD Consortium. Knezevic B., Burnham K.L., Sanniti A., Lledó Lara A., De Cesco S., Wegner J.K., McCann F.E., Fang H., Handunnetthi L., et al. A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat. Genet. 2019;51:1082–1091. doi: 10.1038/s41588-019-0456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reay W.R., Cairns M.J. Advancing the use of genome-wide association studies for drug repurposing. Nat. Rev. Genet. 2021;22:658–671. doi: 10.1038/s41576-021-00387-z. [DOI] [PubMed] [Google Scholar]
- 7.Zhou W., Kanai M., Wu K.-H.H., Humaira R., Tsuo K., Hirbo J.B., Wang Y., Bhattacharya A., Zhao H., Namba S., et al. Global Biobank Meta-analysis Initiative: power genetic discovery for human diseases with > 2.6 million samples across diverse ancestries. medRxiv. 2021 doi: 10.1101/2021.11.19.21266436. Preprint at. [DOI] [Google Scholar]
- 8.Sakaue S., Kanai M., Tanigawa Y., Karjalainen J., Kurki M., Koshiba S., Narita A., Konuma T., Yamamoto K., Akiyama M., et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 2021;53:1415–1424. doi: 10.1038/s41588-021-00931-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wainberg M., Sinnott-Armstrong N., Mancuso N., Barbeira A.N., Knowles D.A., Golan D., Ermel R., Ruusalepp A., Quertermous T., Hao K., et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 2019;51:592–599. doi: 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuchenbaecker K., Telkar N., Reiker T., Walters R.G., Lin K., Eriksson A., Gurdasani D., Gilly A., Southam L., Tsafantakis E., et al. The transferability of lipid loci across African, Asian and European cohorts. Nat. Commun. 2019;10:4330. doi: 10.1038/s41467-019-12026-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi H., Gazal S., Kanai M., Koch E.M., Schoech A.P., Siewert K.M., Kim S.S., Luo Y., Amariuta T., Huang H., et al. Population-specific causal disease effect sizes in functionally important regions impacted by selection. Nat. Commun. 2021;12:1098. doi: 10.1038/s41467-021-21286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2013;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik R., Chauhan G., Traylor M., Sargurupremraj M., Okada Y., Mishra A., Rutten-Jacobs L., Giese A.K., Van Der Laan S.W., Gretarsdottir S., et al. Multiancestry genome-wide association study of 520, 000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakaue S., Okada Y. GREP: genome for REPositioning drugs. Bioinformatics. 2019;35:3821–3823. doi: 10.1093/bioinformatics/btz166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou S., Butler-Laporte G., Nakanishi T., Morrison D.R., Afilalo J., Afilalo M., Laurent L., Pietzner M., Kerrison N., Zhao K., et al. A Neanderthal OAS1 isoform protects individuals of European ancestry against COVID-19 susceptibility and severity. Nat. Med. 2021;27:659–667. doi: 10.1038/s41591-021-01281-1. [DOI] [PubMed] [Google Scholar]
- 16.Konuma T., Ogawa K., Okada Y. Integration of genetically regulated gene expression and pharmacological library provides therapeutic drug candidates. Hum. Mol. Genet. 2021;30:294–304. doi: 10.1093/HMG/DDAB049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonehara K., Okada Y. Genomics-driven drug discovery based on disease-susceptibility genes. Inflamm. Regen. 2021;41:8. doi: 10.1186/s41232-021-00158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng J., Haberland V., Baird D., Walker V., Haycock P.C., Hurle M.R., Gutteridge A., Erola P., Liu Y., Luo S., et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat. Genet. 2020;52:1122–1131. doi: 10.1038/s41588-020-0682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.So H.-C., Chau C.K.-L., Chiu W.-T., Ho K.-S., Lo C.-P., Yim S.H.-Y., Sham P.-C. Analysis of genome-wide association data highlights candidates for drug repositioning in psychiatry. Nat. Neurosci. 2017;20:1342–1349. doi: 10.1038/nn.4618. [DOI] [PubMed] [Google Scholar]
- 20.GTEx Consortium. Statistical Methods groups—Analysis Working Group. Enhancing GTEx eGTEx groups. NIH Common Fund. Biospecimen Collection Source Site—NDRI Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., Gould J., Davis J.F., Tubelli A.A., Asiedu J.K., et al. A next generation connectivity map: L1000 platform and the first 1, 000, 000 profiles. Cell. 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/JOURNAL.PCBI.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pers T.H., Karjalainen J.M., Chan Y., Westra H.-J., Wood A.R., Yang J., Lui J.C., Vedantam S., Gustafsson S., Esko T., et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weeks E.M., Ulirsch J.C., Cheng N.Y., Trippe B.L., Fine R.S., Miao J., Patwardhan T.A., Kanai M., Nasser J., Fulco C.P., et al. Leveraging polygenic enrichments of gene features to predict genes underlying complex traits and diseases. medRxiv. 2020 doi: 10.1101/2020.09.08.20190561. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H., Rasheed H., Nøst T.H., Cho Y., Liu Y., Bhatta L., Bhattacharya A., Initiative G.B.M., Hemani G., Davey Smith G., et al. Proteome-wide Mendelian randomization in global biobank meta-analysis reveals trans-ancestry drug targets for common diseases. medRxiv. 2022 doi: 10.1101/2022.01.09.21268473. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Võsa U., Claringbould A., Westra H.-J., Bonder M.J., Deelen P., Zeng B., Kirsten H., Saha A., Kreuzhuber R., Yazar S., et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 2021;53:1300–1310. doi: 10.1038/s41588-021-00913-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun B.B., Maranville J.C., Peters J.E., Stacey D., Staley J.R., Blackshaw J., Burgess S., Jiang T., Paige E., Surendran P., et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suhre K., Arnold M., Bhagwat A.M., Cotton R.J., Engelke R., Raffler J., Sarwath H., Thareja G., Wahl A., DeLisle R.K., et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 2017;8:14357. doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Folkersen L., Fauman E., Sabater-Lleal M., Strawbridge R.J., Frånberg M., Sennblad B., Baldassarre D., Veglia F., Humphries S.E., Rauramaa R., et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet. 2017;13:e1006706. doi: 10.1371/JOURNAL.PGEN.1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emilsson V., Ilkov M., Lamb J.R., Finkel N., Gudmundsson E.F., Pitts R., Hoover H., Gudmundsdottir V., Horman S.R., Aspelund T., et al. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361:769–773. doi: 10.1126/science.aaq1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao C., Chen G., Song C., Keefe J., Mendelson M., Huan T., Sun B.B., Laser A., Maranville J.C., Wu H., et al. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat. Commun. 2018;9:3268. doi: 10.1038/s41467-018-05512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallace C. A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet. 2021;17:e1009440. doi: 10.1371/journal.pgen.1009440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/JOURNAL.PGEN.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G., Sarkar A., Carbonetto P., Stephens M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J. R. Stat. Soc. B. 2020;82:1273–1300. doi: 10.1111/rssb.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/BIOINFORMATICS/BTQ419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotani K., Sahebkar A., Serban M.-C., Ursoniu S., Mikhailidis D.P., Mariscalco G., Jones S.R., Martin S., Blaha M.J., Toth P.P., et al. Lipoprotein(a) levels in patients with abdominal aortic aneurysm. Angiology. 2017;68:99–108. doi: 10.1177/0003319716637792. [DOI] [PubMed] [Google Scholar]
- 37.Bruzelius M., Iglesias M.J., Hong M.-G., Sanchez-Rivera L., Gyorgy B., Souto J.C., Frånberg M., Fredolini C., Strawbridge R.J., Holmström M., et al. PDGFB, a new candidate plasma biomarker for venous thromboembolism: results from the VEREMA affinity proteomics study. Blood. 2016;128:e59–e66. doi: 10.1182/blood-2016-05-711846. [DOI] [PubMed] [Google Scholar]
- 38.Liu J., Lu H., Howatt D.A., Balakrishnan A., Moorleghen J.J., Sorci-Thomas M., Cassis L.A., Daugherty A. Associations of ApoAI and ApoB–containing lipoproteins with AngII–induced abdominal aortic aneurysms in mice. Arterioscler. Thromb. Vasc. Biol. 2015;35:1826–1834. doi: 10.1161/ATVBAHA.115.305482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X., Ji Z.L., Chen Y.Z. TTD: therapeutic target database. Nucleic Acids Res. 2002;30:412–415. doi: 10.1093/nar/30.1.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whirl-Carrillo M., Huddart R., Gong L., Sangkuhl K., Thorn C.F., Whaley R., Klein T.E. An evidence-based framework for evaluating pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2021;110:563–572. doi: 10.1002/cpt.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochoa D., Hercules A., Carmona M., Suveges D., Gonzalez-Uriarte A., Malangone C., Miranda A., Fumis L., Carvalho-Silva D., Spitzer M., et al. Open Targets Platform: supporting systematic drug–target identification and prioritisation. Nucleic Acids Res. 2021;49:D1302–D1310. doi: 10.1093/NAR/GKAA1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sala V., Gallo S., Gatti S., Medico E., Vigna E., Cantarella D., Fontani L., Natale M., Cimino J., Morello M., et al. Cardiac concentric hypertrophy promoted by activated Met receptor is mitigated in vivo by inhibition of Erk1, 2 signalling with Pimasertib. J. Mol. Cell. Cardiol. 2016;93:84–97. doi: 10.1016/j.yjmcc.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 44.Muchir A., Wu W., Sera F., Homma S., Worman H.J. Mitogen-activated protein kinase kinase 1/2 inhibition and angiotensin II converting inhibition in mice with cardiomyopathy caused by lamin A/C gene mutation. Biochem. Biophys. Res. Commun. 2014;452:958–961. doi: 10.1016/j.bbrc.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verhamme P., Yi B.A., Segers A., Salter J., Bloomfield D., Büller H.R., Raskob G.E., Weitz J.I., ANT-005 TKA Investigators Abelacimab for prevention of venous thromboembolism. N. Engl. J. Med. 2021;385:609–617. doi: 10.1056/NEJMoa2105872. [DOI] [PubMed] [Google Scholar]
- 46.Kuramoto E., Nishiuma T., Kobayashi K., Yamamoto M., Kono Y., Funada Y., Kotani Y., Sisson T.H., Simon R.H., Nishimura Y. Inhalation of urokinase-type plasminogen activator reduces airway remodeling in a murine asthma model. Am. J. Physiol. Lung Cell Mol. Physiol. 2009;296:L337–L346. doi: 10.1152/ajplung.90434.2008. [DOI] [PubMed] [Google Scholar]
- 47.Ren Y., Su X., Kong L., Li M., Zhao X., Yu N., Kang J. Therapeutic effects of histone deacetylase inhibitors in a murine asthma model. Inflamm. Res. 2016;65:995–1008. doi: 10.1007/s00011-016-0984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Zyl-Smit R.N., Krüll M., Gessner C., Gon Y., Noga O., Richard A., de los Reyes A., Shu X., Pethe A., Tanase A.-M., et al. Once-daily mometasone plus indacaterol versus mometasone or twice-daily fluticasone plus salmeterol in patients with inadequately controlled asthma (PALLADIUM): a randomised, double-blind, triple-dummy, controlled phase 3 study. Lancet Respir. Med. 2020;8:987–999. doi: 10.1016/S2213-2600(20)30178-8. [DOI] [PubMed] [Google Scholar]
- 49.Chanez P., Israel E., Davidescu L., Ursol G., Korzh O., Deshmukh V., Kuryk L., Nortje M.-M., Godlevska O., Devouassoux G., et al. B93. LATE BREAKING CLINICAL TRIALS IN AIRWAY DISEASES. 2020. Masitinib significantly decreases the rate of asthma exacerbations in patients with severe asthma uncontrolled by oral corticosteroids: a phase 3 multicenter study; p. A4210. [DOI] [Google Scholar]
- 50.Faro V.L., Bhattacharya A., Zhou W., Zhou D., Wang Y., Läll K., Kanai M., Lopera-Maya E., Straub P., Pawar P., et al. Genome-wide association meta-analysis identifies novel ancestry-specific primary open-angle glaucoma loci and shared biology with vascular mechanisms and cell proliferation. medRxiv. 2021 doi: 10.1101/2021.12.16.21267891. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoo E.J., Ojiaku C.A., Sunder K., Panettieri R.A. Phosphoinositide 3-kinase in asthma: novel roles and therapeutic approaches. Am. J. Respir. Cell Mol. Biol. 2017;56:700–707. doi: 10.1165/rcmb.2016-0308TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hains B.C., Waxman S.G. Neuroprotection by sodium channel blockade with phenytoin in an experimental model of glaucoma. Invest. Ophthalmol. Vis. Sci. 2005;46:4164–4169. doi: 10.1167/iovs.05-0618. [DOI] [PubMed] [Google Scholar]
- 53.Anderson D.R., Dunbar M., Murnaghan J., Kahn S.R., Gross P., Forsythe M., Pelet S., Fisher W., Belzile E., Dolan S., et al. Aspirin or rivaroxaban for VTE prophylaxis after hip or knee arthroplasty. N. Engl. J. Med. 2018;378:699–707. doi: 10.1056/NEJMoa1712746. [DOI] [PubMed] [Google Scholar]
- 54.Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44:D1045–D1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gebhard C., Akhmedov A., Mocharla P., Angstenberger J., Sahbai S., Camici G.G., Lüscher T.F., Tanner F.C. PDGF-CC induces tissue factor expression: role of PDGF receptor α/β. Basic Res. Cardiol. 2010;105:349–356. doi: 10.1007/s00395-009-0060-0. [DOI] [PubMed] [Google Scholar]
- 56.Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/NAR/GKQ973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolford B.N., Zhao Y., Surakka I., Wu K.-H.H., Yu X., Richter C.E., Bhatta L., Brumpton B., Desch K., Thibord F., et al. Multi-ancestry GWAS for venous thromboembolism identifies novel loci followed by experimental validation in zebrafish. medRxiv. 2021 doi: 10.1101/2022.06.21.22276721. Preprint at. [DOI] [Google Scholar]
- 59.Grover S.P., Mackman N. Intrinsic pathway of coagulation and thrombosis. Arterioscler. Thromb. Vasc. Biol. 2019;39:331–338. doi: 10.1161/ATVBAHA.118.312130. [DOI] [PubMed] [Google Scholar]
- 60.Pietzner M., Wheeler E., Carrasco-Zanini J., Cortes A., Koprulu M., Wörheide M.A., Oerton E., Cook J., Stewart I.D., Kerrison N.D., et al. Mapping the proteo-genomic convergence of human diseases. Science. 2021;374:eabj1541. doi: 10.1126/science.abj1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Leeuw C., Werme J., Savage J.E., Peyrot W.J., Posthuma D. Reconsidering the validity of transcriptome-wide association studies. bioRxiv. 2021 doi: 10.1101/2021.08.15.456414. [DOI] [Google Scholar]
- 62.Mancuso N., Freund M.K., Johnson R., Shi H., Kichaev G., Gusev A., Pasaniuc B. Probabilistic fine-mapping of transcriptome-wide association studies. Nat. Genet. 2019;51:675–682. doi: 10.1038/s41588-019-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hemani G., Zheng J., Elsworth B., Wade K.H., Haberland V., Baird D., Laurin C., Burgess S., Bowden J., Langdon R., et al. The MR-base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408–e34429. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeng J., Xue A., Jiang L., Lloyd-Jones L.R., Wu Y., Wang H., Zheng Z., Yengo L., Kemper K.E., Goddard M.E., et al. Widespread signatures of natural selection across human complex traits and functional genomic categories. Nat. Commun. 2021;12:1164. doi: 10.1038/s41467-021-21446-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sonawane A.R., Platig J., Fagny M., Chen C.-Y., Paulson J.N., Lopes-Ramos C.M., DeMeo D.L., Quackenbush J., Glass K., Kuijjer M.L. Understanding tissue-specific gene regulation. Cell Rep. 2017;21:1077–1088. doi: 10.1016/j.celrep.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hemani G., Tilling K., Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. doi: 10.1371/JOURNAL.PGEN.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corsello S.M., Bittker J.A., Liu Z., Gould J., McCarren P., Hirschman J.E., Johnston S.E., Vrcic A., Wong B., Khan M., et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat. Med. 2017;23:405–408. doi: 10.1038/nm.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y., Namba S., Lopera-Maya E.A., Kerminen S., Tsuo K., Lall K., Kanai M., Zhou W., Wu K.-H.H., Fave M.-J., et al. Global biobank analyses provide lessons for computing polygenic risk scores across diverse cohorts. medRxiv. 2021 doi: 10.1101/2021.11.18.21266545. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu G., He Q.-Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016;12:477–479. doi: 10.1039/C5MB00663E. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GBMI GWAS summary statistics are publicly available at https://www.globalbiobankmeta.org/resources. The genomics-driven drug discovery analysis was conducted using the following publicly available tools: MAGMA (https://ctg.cncr.nl/software/magma), DEPICT (https://data.broadinstitute.org/mpg/depict/), PoPS (https://github.com/FinucaneLab/pops), FOCUS (https://github.com/bogdanlab/focus), GREP (https://github.com/saorisakaue/GREP), Trans-Phar (https://github.com/konumat/Trans-Phar), LocusZoom (http://locuszoom.org/), and the Pi, TwoSampleMR, coloc, susieR, and metacor R packages.