Abstract
There is still much to uncover regarding the molecular details of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. As the most abundant protein, coronavirus nucleocapsid (N) protein encapsidates viral RNAs, serving as the structural component of ribonucleoprotein and virion, and participates in transcription, replication, and host regulations. Virus–host interaction might give clues to better understand how the virus affects or is affected by its host during infection and identify promising therapeutic candidates. Considering the critical roles of N, we here established a new cellular interactome of SARS-CoV-2 N by using a high-specific affinity purification (S-pulldown) assay coupled with quantitative mass spectrometry and immunoblotting validations, uncovering many N-interacting host proteins unreported previously. Bioinformatics analysis revealed that these host factors are mainly involved in translation regulations, viral transcription, RNA processes, stress responses, protein folding and modification, and inflammatory/immune signaling pathways, in line with the supposed actions of N in viral infection. Existing pharmacological cellular targets and the directing drugs were then mined, generating a drug–host protein network. Accordingly, we experimentally identified several small-molecule compounds as novel inhibitors against SARS-CoV-2 replication. Furthermore, a newly identified host factor, DDX1, was verified to interact and colocalize with N mainly by binding to the N-terminal domain of the viral protein. Importantly, loss/gain/reconstitution-of-function experiments showed that DDX1 acts as a potent anti-SARS-CoV-2 host factor, inhibiting the viral replication and protein expression. The N-targeting and anti-SARS-CoV-2 abilities of DDX1 are consistently independent of its ATPase/helicase activity. Further mechanism studies revealed that DDX1 impedes multiple activities of N, including the N–N interaction, N oligomerization, and N–viral RNA binding, thus likely inhibiting viral propagation. These data provide new clues to better depiction of the N–cell interactions and SARS-CoV-2 infection and may help inform the development of new therapeutic candidates.
Keywords: SARS-COV-2, nucleocapsid (N) protein, virus–host interaction, mass spectrometry, interactome, antiviral drug, DDX1, host restriction factor
Graphical Abstract
Highlights
-
•
S-pulldown and MS establishes a new cellular interactome of SARS-CoV-2 N.
-
•
The interactome is well linked to N actions and shows new clues to the virus biology.
-
•
New anti-SARS-CoV-2 drugs are identified based on the interactome.
-
•
The NTD domain of N is the major target of DDX1, a representative N interactor.
-
•
DDX1 blocks multiple critical N activities, thus inhibiting SARS-CoV-2 infection.
In Brief
By S-pulldown and quantitative proteomics, we established a new cellular interactome of SARS-CoV-2 nucleocapsid (N) protein, which is well linked to the supposed N actions and shows many valuable clues to SARS-CoV-2 biology. Accordingly, we identified new anti-SARS-CoV-2 drugs. An interaction of N with a representative host factor, DDX1, was detailedly characterized. In-depth function/mechanism studies uncovered that the N N-terminal domain is a major target of DDX1, and consistently, DDX1 blocks multiple critical N activities, thus inhibiting SARS-CoV-2 infection.
The ongoing coronavirus disease-2019 pandemic has caused unprecedented impacts on the social and economic development and public health around the world. The causative pathogen severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the β-coronavirus genus and shares high homology to the SARS-CoV that emerged in 2003 (1). The viral genome is encapsulated by the nucleocapsid (N) protein, a major structural protein, to form the ribonucleoprotein (RNP) complex that is the virion structural core (2, 3, 4). SARS-CoV-2 N that is produced at high levels in infected cells is essential for viral replication and also can enhance viral transcription efficiency (5). In addition, it has been reported that N proteins of coronaviruses including SARS-CoV-2 likely regulate or are regulated by host proteins and the corresponding biological processes (BPs), leading to wide effects on viral infection and pathogenesis (6, 7, 8). Accordingly, the N protein has been also exploited as one of the therapeutic targets for antiviral research (9). Thus, deciphering the interactions of SARS-CoV-2 N with host cells is important for not only the understanding of viral infection but also development of antiviral strategies.
Previously, by using affinity purification mass spectrometry (AP-MS), Gordan et al. (10) have provided potential interaction networks of SARS-CoV-2 viral proteins with host proteins and identified 15 host proteins as possible N-interacting factors; subsequently, several studies also reported potential cellular interactions of N by AP-MS (11, 12, 13, 14) or by complementary proximity-based labeling MS method (BioID-MS) (15, 16, 17, 18, 19). However, experimental validations of the proposed interactions were often lacking in these studies. Moreover, the results are largely divergent, particularly with very few host proteins reported from these studies are consistent (10, 12, 14). Given the multiple and critical roles of N in viral infection, further determination of its cellular interactions is obviously warranted. Therefore, we here analyzed the SARS-CoV-2 N˗cell interaction proteome by a distinct AP strategy, that is, S-pulldown assay of high affinity and specificity, combined with quantitative mass spectrometry (MS). Consequently, we established a new cellular interactome of SARS-CoV-2 N, in which most proteins are for the first time identified as N-interacting factors. Series of bioinformatics analyses and experimental validations showed that the N interactome established here not only well supports the supposed actions of N but also provides many new implications regarding the virus–host interactions and potential antiviral intervention. Accordingly, we indeed identified several new small molecules (especially anisomycin and dasatinib) with antiviral activities against SARS-CoV-2 by in vitro antiviral experiments. Moreover, as a proof of principle, we detailedly validated and characterized the interaction of N with a newly identified cellular protein by the interactome analysis, DDX1, and thus functionally and mechanistically uncovered the role of DDX1 as a notable host restriction factor against SARS-CoV-2.
Experimental Procedures
Cell and Virus
HEK293T and Huh7 cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (Gibco). HEK293, Vero E6, and Caco-2 cells were grown in Eagle's minimum essential medium supplemented with 10% fetal bovine serum. All the cell lines were purchased from American Type Culture Collection. To generate Huh7-ACE2 cells, Huh7 cells were transduced with pLVX-hACE2-IRES-Puro lentivirus and selected using 2.5 μg/ml puromycin, followed by monoclonal screening as previously described (20). SARS-CoV-2 (WIV04, GenBank: MN996528.1, CSTR: 16533.06. IVCAS 6.7512) was expanded in Vero E6 cells in a biosafety level 3 laboratory. Virus titers were determined by 50% tissue culture infectious dose method. The DDX1-KO HEK293 cell lines were generated by using CRISPR–Cas9 system as previously described (21), and the gene-specific guide sequence was GCTGAACTGAAATTTAACTT. Cell viability was detected by using Cell Counting Kit-8 (MCE).
Antibody and Reagent
Primary antibodies were purchased from the indicated manufacturers as follows: mouse monoclonal antibody against SARS-CoV-2 N (ABclonal; catalog no.: A20142), β-actin (Beyotime; catalog no.: AA128), and FLAG (Sigma–Aldrich; catalog no.: F1804); rabbit monoclonal antibodies against SARS-COV-2 N (Sino Biological, Inc; catalog nos.: 40143-R019 and 40143-R001), HSPH1 (ABclonal; catalog no.: A4687), and RPS6 (Cell Signaling Technology; catalog no.: 2217); polyclonal antibodies to S tag (Abcam; catalog no.: ab183674), HNRNPU (ABclonal; catalog no.: A3917), DDX1 (Biorbyt; catalog no.: orb341115), DHX9 (Proteintech; catalog no.: 17721-1-AP), LSG1 (catalog no.: DF15179; Affinity Biosciences), PABPC1 (ABclonal; catalog no.: A14872), IGF2BP1 (ABclonal; catalog no.: A1517), LARP1 (Proteintech; catalog no.: 13708-1-AP), RPS8 (ABclonal; catalog no.: A18377), RPS12 (ABclonal; catalog no.: A5890), RPL7 (ABclonal; catalog no.: A5932), RPLP1 (ABclonal; catalog no.: A6725), HSPA5 (ABclonal; catalog no.: A11366), and HSPA4 (Proteintech; catalog no.: 21206-1-AP). Fluorescence-labeled secondary antibodies used were Alexa Fluor-488 goat antimouse immunoglobulin G (IgG) (Abcam; catalog no.: ab150113), and Alexa Fluor-647 goat anti-rabbit IgG (Abcam; catalog no.: ab150079). Horseradish peroxidase–labeled goat anti-rabbit (catalog no.: A6154) or anti-mouse IgG antibodies (catalog no.: A4416) were purchased from Sigma–Aldrich. MiniBEST Viral RNA Extraction Kit and PrimeScript RT reagent Kit with genomic DNA Eraser and SYBR Green Realtime PCR Master Mix were purchased from TAKARA. Disuccinimidyl suberate, RiboLock RNase inhibitor, Lipofectamine RNAiMAX, and Lipofectamine 3000 were purchased from Thermo Fisher. JetPRIME (Polyplus), Hoechst 33258 (Beyotime), Trizol (Invitrogen), and S-protein agarose (MBL) were purchased from the indicated manufacturers. Dasatinib, remdesivir, and phenyllactic acid were purchased from TargetMol. Anisomycin, 4-hydroxyphenylpyruvic acid, and etidronic acid were purchased from MedChemExpress.
Plasmid Construct and siRNA
Expression plasmids for human DDX1, DDX1 mutants (K52A, S295E, and R296E), SARS-CoV-2 N, and ACE2 without or with an indicated tag were constructed by standard molecular biology techniques. The siRNAs targeting human DDX1 were designed and synthesized by GenePharma. The target sequences of siRNAs are listed in Table 1.
Table 1.
List of siRNA target sequences for knockdown
| Target | RNAi plasmid | Target sequence (sense) |
|---|---|---|
| Human DDX1 | siDDX1-1# | GCUGAACUGAAAUUUAACU(TT) |
| siDDX1-2# | GAGCCACAUUAGAACUGAU(TT) | |
| siDDX1-3# | GGAGUUAGCUGAACAAACU(TT) | |
| Negative control | UUCUCCGAACGUGUCACGU(TT) |
Real-Time qRT–PCR
To analyze the viral or cellular RNA levels, real-time qRT–PCR was performed as previously described (21, 22). The primers used are listed in Table 2.
Table 2.
List of primers for real-time qPCR
| Primer namea | Primer sequence (5′ to 3′) |
|---|---|
| N-F | CGAATTCGTGGTGGTGACGG |
| N-R | TGCGGGTGCCAATGTGATCT |
| DDX1-F | TTTCCAAGGCACCGGATGGT |
| DDX1-R | ACTCCCGGGAAGGTTCAACA |
| GAPDH-F | ACCACAGTCCATGCCATCAC |
| GAPDH-R | TCCACCACCCTGTTGCTGTA |
F, the forward primer; R, the reverse primer.
Immunofluorescence and Confocal Microscopy
Briefly, cells were fixed with 4% paraformaldehyde and then permeabilized with 0.5% Triton X-100 in PBS. After blocking with 5% bovine serum albumin (Biosharp), cells were successively incubated with indicated primary antibodies and fluorescence-labeled secondary antibodies, followed by nucleus staining with Hoechst 33258. Images were taken and analyzed on an Andor Dragonfly 202 microscope (Oxford Instruments).
S-Pulldown and Silver Staining
S-tag pulldown assay (S-pulldown) was used to analyze the interactions of S-tagged proteins with other proteins as described previously (20, 21, 22, 23, 24). Briefly, at 48 h post-transfection, cells were lysed with a lysis buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) containing protease inhibitor cocktail (Roche Applied Science). After centrifugation, the cell lysate supernatants were incubated with S-protein agarose slurry (Merck Novagen) at 4 °C for 6 h. After extensive washing, part of the beads (1/20) were boiled in 1× SDS loading buffer to elute precipitated proteins, which were subjected to SDS-PAGE and then silver staining (Thermo Fisher) or Western blotting. The other beads are delivered to MS analysis.
AP-MS
HEK293T cells (1.5 × 107 cells) were transfected with pCAGGSP7-N.STAG or the control empty vector pCAGGSP7 by Lipofectamine 3000 (in total of six samples including three biological replicates for both the N and control groups). AP was performed to enrich N and interacted proteins by using S-pulldown assay as described previously. Bead-bound proteins were denatured, reduced, alkylated, and eluted by one-step treatment in the reaction buffer (1% sodium deoxycholate/100 mM Tris–HCl, pH 8.5/10 mM Tris(2-carboxyethyl)phosphine/40 mM chloroacetamide) at 95 °C for 10 min. The supernatants were then diluted with equal volume of H2O and then subjected to trypsin digestion at 37 °C overnight, followed by adding TFA to stop the digestion. After centrifugation, obtained supernatant peptides were desalted and lyophilized for LC–MS/MS analysis.
LC–MS/MS data acquisition was carried out on a hybrid quadrupole-TOF LC–MS/MS mass spectrometer (TripleTOF 5600; SCIEX) coupled with an Eksigent microLC 415 system (SCIEX) via a microspray source. In detail, peptides were dissolved in MS loading buffer (0.1% formic acid), loaded onto a C18 trap column (5 μm, 5 × 0.3 mm; Agilent Technologies) through autosampler, and then eluted into a C18 analytical column (300 μm × 150 mm, 3 μm particle size, 120 Å pore size; Eksigent). Mobile phase A (3% dimethyl sulfoxide, 97% H2O, and 0.1% formic acid) and mobile phase B (3% dimethyl sulfoxide, 97% acetonitrile, and 0.1% formic acid) were mixed to establish a 30 min gradient at a constant flow of 5 μl/min for peptide separation, which was started from 6% to 25% mobile phase B over 19 min, followed by an increase to 45% B over 4 min and an increase to 80% B over 0.5 min, and then held at 80% B for 6.5 min. Separated peptides were then directly ionized by electrospray ionization and transferred into the mass spectrometer (spray voltage, 2.3 kV; curtain gas, 20 PSI; interface heater temperature, 150 °C). MS runs were performed in a data-dependent acquisition mode including repeating cycles of one full-scan mass spectrum (with m/z ranging from 350 to 1500, ion accumulation time 250 ms), followed by 40 MS/MS events (m/z ranging from 100 to 1500, ion accumulation time 50 ms). The threshold for MS/MS acquisition activation was set to 120 cps for +2∼+5 precursors. Former target ion exclusion was set for 18 s. Before the next run, a blank run was carried to wash and check the equipment condition of remained sample. The raw data have been deposited to the Science Data Bank.
Raw Data Processing, Protein Identification, and Quantification
Raw data from TripleTOF 5600 were analyzed with MaxQuant software (version 1.6.2.10, Max Planck Institute of Biochemistry) using the Andromeda database search algorithm. Spectra files were searched against the UniProt reference human proteome database (containing 74,489 entries, downloaded on May 16, 2019) supplemented with SARS-CoV-2 protein sequences using the following parameters: type, standard; mode, label-free quantification (LFQ); digestion, trypsin specificity required with cleavage at C-terminal region after K or R and up to two missed cleavages per peptide were allowed; fixed modifications, carbamidomethylation (Cys); variable modifications, oxidation (Met), acetyl (protein N-term), and deamidation (Asn or Gln). As no labeling was performed, multiplicity was set to 1. The MS1 match tolerance (mass tolerances for precursor ions) was set as 20 ppm for the first search and 4.5 ppm for the main search; the MS2 tolerance (mass tolerances for fragment ions) was set as 20 ppm. The search results were filtered with 1% false positive rate at both protein and peptide levels. Normalized LFQ values were obtained by applying the in-built MaxLFQ algorithm. The relevant quantification data are available in supplemental Table S4, and spectra for single unique peptide-based protein identifications were uploaded as supplemental spectra (supplemental Fig. S5).
Experimental Design and Statistical Rationale
S-pulldown AP samples from three biological replicates for each group were trypsin digested and analyzed using label-free data-dependent acquisition nano-LC-MS/MS (LFQ nLC-MS/MS), followed by LFQ proteomics analysis using MaxQuant software. The “proteingroups.txt” file produced by MaxQuant was used for further analysis. First, hits to the reverse database, contaminants and proteins only identified with modified peptides were eliminated. Then the LFQ intensities were logarithmized, and identifications were filtered for proteins having at least two valid values in at least one replicate group. Missing values were imputed with values representing a normal distribution around the detection limit of the mass spectrometer. To that end, mean and standard deviation of the distribution of the real intensities were determined, and then a new distribution with a downshift of 1.8 standard deviations and a width of 0.3 standard deviations was created. The total matrix was imputed using these values, enabling statistical analysis. A Student’s t test was performed comparing the bait pull-downs to the control pull-downs. Significant changed proteins were screened out according to the fold changes and p values of the proteins. In this study, criteria with a p value <0.05 and fold change >2 were taken for selection. Additional details were provided as described previously.
Bioinformatic Analysis
Bioinformatic analyses were conducted as described previously (20). In Brief, a Venn analysis was performed through an online website tool (http://jvenn.toulouse.inra.fr/app/example.html, https://bioinfogp.cnb.csic.es/tools/venny/) (25) to overlap the coronavirus N-related targets. The STRING database (version 11.0, https://string-db.org/) coupled with Cytoscape software (version 3.8) (26, 27) was used to retrieve molecular networks of the identified cellular proteins, to group proteins into similar functional classes, and to construct a PPI network. GO analysis was used to determine molecular function and BP attributes of the PPI network by using the enricher function of clusterProfiler package in R with default parameters (28). Significant GO terms (adjusted p < 0.05) were identified and further refined by the simplify method (cutoff = 0.5, by = “p.adjust”) to cut redundant gene sets. To identify potential druggable targets, N-interacting host factors were searched against the DrugBank databases (29). Cytoscape software was then utilized to visualize the drug–host target network (26).
Statistical Analysis
Statistical analyses were performed by GraphPad Prism 8 (GraphPad Software) using the ANOVA method followed by Dunnett's multiple comparisons test or unpaired two-tailed Student’s t test. All results were expressed as mean ± SD. p < 0.05 was considered statistically significant.
Results and Discussion
Establishment of a New Cellular Interactome of SARS-CoV-2 N in Human Embryonic Kidney 293T Cells
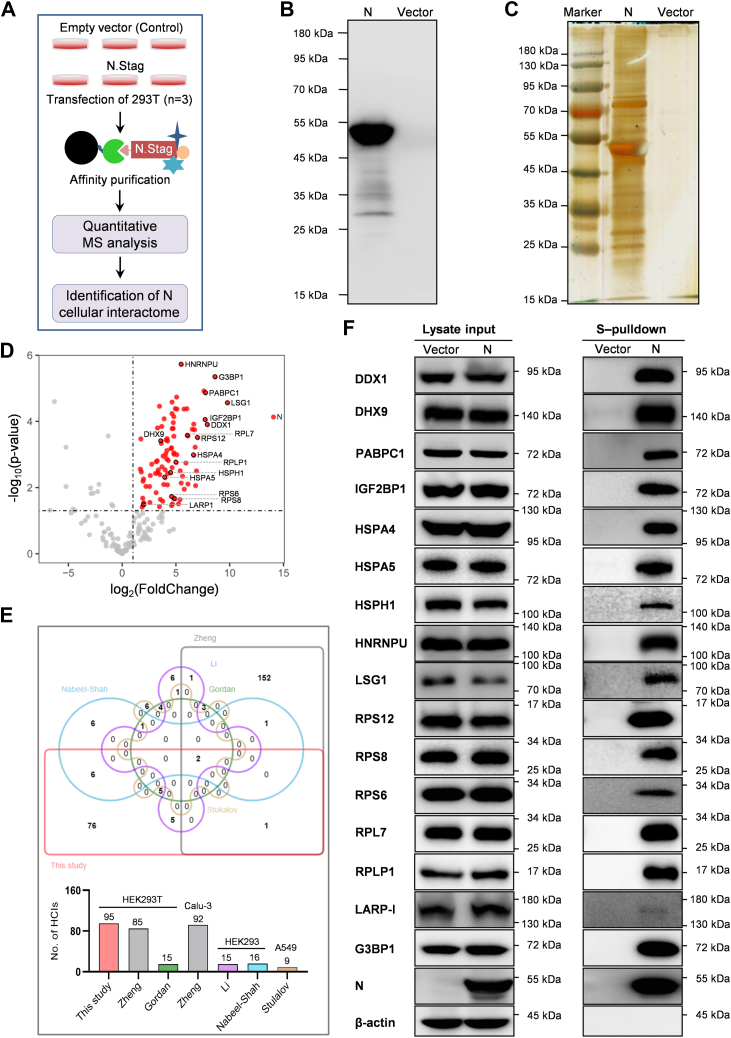
To further comprehensively analyze the cellular interactions of SARS-CoV-2 N, we determined the cellular interactome of N by utilizing S-pulldown AP strategy coupled with label-free MS quantitation (Fig. 1A) (20, 23). Immunoblot and silver staining analyses showed that in addition to the efficient precipitation of the bait N protein, many specific bands of cellular proteins coprecipitated with N were observed, validating the efficient and specific S-pulldown strategy (Fig. 1, B and C). Based on the strategy, pulldown samples were next delivered to LC–MS/MS to identify proteins, followed by quantitative proteomics analysis using the MaxQuant software (Max Planck Institute of Biochemistry). Of the 298 cellular proteins identified, 199 were quantified. Then, 95 cellular proteins with fold change >2 and p value <0.05 were taken as high-confidence N interactors (HCIs) (Fig. 1D and supplemental Table S1). As shown in the Venn diagrams (Fig. 1E and supplemental Fig. S1), previously reported host factors potentially interacting with N are greatly discrepant among different studies (few common elements repeatably detected between any two studies), whereas 7 of the 15 host proteins identified respectively by Gordan et al. or by Li et al. (i.e., 46.7% from each report) and 8 of the 16 host proteins identified by Nabeel-Shah et al. (i.e., 50%) were also found in the present study, despite of different experimental strategies. However, furthermore, most of the other host proteins in our interactome (76/95) are newly discovered here (Fig. 1E and supplemental Table S2), manifesting a new cellular interactome of SARS-CoV-2 N established by AP-MS.
Fig. 1.
Establishment of a new cellular interactome of SARS-CoV-2 N.A, schematic representation of the strategy for analyzing cellular interacting proteins of SARS-COV-2 N via S-pulldown. HEK293T cells were transfected with plasmid DNA encoding N.STAG or the empty vector (control). Thirty-six hours post-transfection, supernatants of the cell lysates were harvested and subjected to S-pulldown affinity purification assays, followed by quantitative mass spectrometry (MS) analysis and protein identification. B and C, immunoblot and silver staining analyses of N and the coprecipitated host proteins. S-pulldown products from (A) were also delivered to Western blotting with anti-N monoclonal antibody (mAb) (B) and silver staining of SDS-polyacrylamide gel (C). D, Volcano diagram of the identified protein targets. Points with fold change (FC) >2 and p value <0.05 are highlighted in red as N-interacting targets of high confidence. Targets marked with a black circle represent those validated by immunoblotting in the following. E, Venn diagram analysis of potential N interactors identified from different studies. Size of each list together with the cell types used in the studies was indicated in the histogram below. See also supplemental Fig. S1. F, validation of the SARS-CoV-2 N interactome data obtained in this study with immunoblotting. The interactions of N with representative host proteins identified here were confirmed by independent pulldown assays and Western blotting analyses with specific antibodies against the indicated proteins. HEK293T, human embryonic kidney 293 cell line; N protein, nucleocapsid protein; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
In addition to AP-MS, BioID-MS that has the potential to capture insoluble, weak, or transient protein–protein interactions (PPIs) is also often used for interactome analysis (30, 31). However, interaction candidates identified by BioID-MS could include spatially adjacent but not interacting proteins without biological significance, decreasing the specificity and instructive value for the following research. In addition, the BirA (R118G)-derived BioID/BioID2/TurboID/mini Turbo tags (28–35 kDa) fused with the baits in BioID are larger, compared with the tags used in AP (e.g., the S tag of 1.75 kDa), which may lead to adverse effects on the bait conformation and function. Thus, both AP-MS and BioID-MS have their advantages and disadvantages. We also compared our interactome with the data based on BioID-MS or AP-MS combined with BioID-MS (supplemental Table S2), which produces 9 and 29 common elements with our 95 HCIs, respectively. It suggests that even in comparison with the BioID-MS reports, most of the HCIs in the present study is newly identified, significantly enriching the knowledge of the SARS-CoV-2 N–cell interactions.
The low overlap between different studies as seen in Figure 1E was probably because of the differences of experimental materials and methods including the MS quantification methods used. In order to validate our proteomic data, the interactions of SARS-CoV-2 N with a series of representative host proteins chosen from different clusters in the following STRING analysis were analyzed by independent pulldown and immunoblot analysis. In total, 16 of the 95 HCIs (i.e., 16.8%), including those exclusively discovered in our AP-MS interactome, such as DDX1, DHX9, IGF2BP1, LSG1, HNRNPU, RPS6, RPS8, RPS12, RPL7, and RPLP1, were subjected to the validation by immunoblotting. As shown in Figure 1F, well consistent with the MS results, all the host proteins were coprecipitated with N, further supporting the new SARS-CoV-2 N interactome.
Biological Implications of the Interactome for N–Cell Interactions, Viral Biology, and Antiviral Intervention
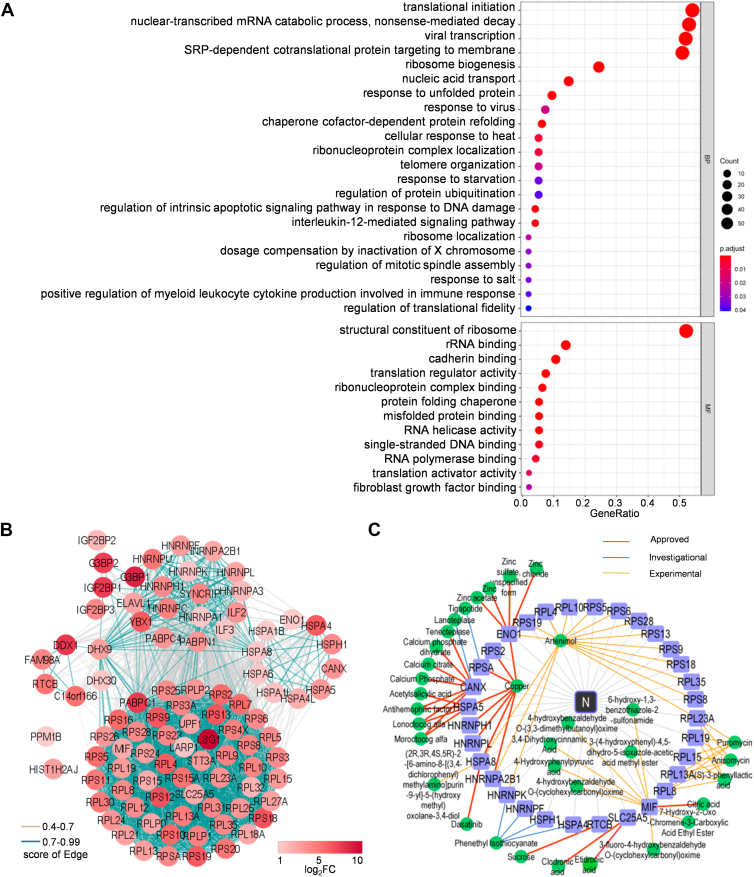
Next, Gene Ontology (GO) analyses were performed to determine the basic molecular function and BP attributes of the identified N-interacting cellular proteins. As shown in Figure 2A, these host factors are mainly enriched in translation regulations, viral transcription, RNA processes, stress responses, protein folding and modification, response to virus, and inflammatory and immune signaling pathways. N and its assembled viral RNPs are essential machineries participating in and regulating viral transcription, replication, and assembly. Thus, the enriched physiological processes are well linked to the supposed activities of N in viral infection. STRING analysis of PPI network then identified 94 nodes and 2004 interactions (edges), with several main clusters or subnetworks including translation ribosomal proteins, stress body/P-body proteins, HSPs, HNRNPs, and RNA helicases (Fig. 2B).
Fig. 2.
Gene Ontology (GO), interaction network, and druggable target analyses of the N interactome.A, GO analysis was used to determine the biological process (BP) and molecular function (MF) attributes of the identified N-interacting cellular proteins by clusterProfiler and significant GO terms (adjusted p < 0.05). B, interaction network analysis of the identified cellular proteins interacting with N. Interaction networks of the cellular proteins (fold change [FC] >2, p < 0.05) were analyzed by STRING and visualized with Cytoscape. Proteins were clustered into five units through Markov clustering based on their molecular action. Disconnected nodes (HST1H2AJ and PPM1B) are also displayed in the network. The edge color is based on confidence (score) cutoff values (light gray, 0.4–0.7; blue, >0.7). The node color proportional to the rank of log2(FC) values. C, target–drug network. Pharmacological targets of the N-interacting host factors were identified by data mining. The druggable proteins along with the targeting drugs were then visualized in the network by Cytoscape. See also Figure 3 for the data-guided drug testing. N protein, nucleocapsid protein.
Interactions of viral proteins with ribosomal proteins have been proposed to have significant biological effects on some coronavirus infections. Previous studies have shown that coronavirus nsp1 proteins including the nsp1 of SARS-CoV-2 can bind with ribosomal proteins to shut off global host cell translation but spare the translation of the viruses themselves. In addition, the N of mouse hepatitis virus (MHV, another β-CoV belonging to the Embevovirus subgenus) was also shown to favor the translation of viral over cellular mRNA (32, 33). Here, we found enriched ribosome proteins as HCIs of SARS-CoV-2 N, of which multiple were validated by the independent immunoblot analyses (Fig. 1F). However, few ribosomal proteins were identified in the interactomes of SARS-CoV-2 nsp13 in our previous studies using the same S-pulldown and MS methods (20), indicating the specificity of the SARS-CoV-2 N-ribosomal protein interactions. Interestingly, the N of infectious bronchitis virus (a γ-CoV) was also shown to cosedimentate a large amount of ribosomal proteins (49 in 143 HCIs) by using stable isotope labeling with amino acid–based MS and GFP-Nanotrap pulldown methodologies (34). Given the interactions of N with ribosomal proteins and the BP enrichment of translation regulations, it will be interesting to test whether SARS-CoV-2 N may thus also modulate cellular or viral translation, apart from its critical activities in viral replication and transcription. Also notably, PABPC1, identified here in the ribosomal protein cluster, is a multifaceted factor promoting efficient translation by facilitating multiple steps in the process and has been reported to be targeted by SARS-CoV-2 RNA to control messenger RNP activity (35, 36, 37); IGF2BP1, an m(6)A reader, has been verified to stabilize viral RNA (vRNA) and augment SARS-CoV-2 translation (38). These previously reported molecular phenomena are in line with the present findings as well. In view of these, more investigations are needed to better unravel the possible regulations of viral or cellular translation by SARS-CoV-2 and its N protein by following the clues revealed here.
Furthermore, HSPs, HNRNPs, and stress body proteins, mainly related to the GO terms of responses to various stresses including viral infection, are three other clusters found in the present N-interaction map (Fig. 2, A and B). Here, HNRNPs and stress granule markers G3BP1 and G3BP2, together with several HSPs and the 40S ribosome subunit, all could be conditionally localized in cytoplasmic P bodies and stress granules (39, 40, 41). Very interestingly, SARS-CoV-2 N has been reported by several groups to disrupt the formation of stress granules and phase separate with stress granule markers G3BP1/G3BP2 and HNRNPA2 (5, 6, 7, 42). The phase separation of N has been proposed for critical functions of N in facilitating viral replication, viral genomic RNA packaging, and modulation of host–cell responses to infection (6, 7, 42). It will be merited to explore whether and how other N-interacting cellular factors identified here, especially the HSPs and other HNRNPs, also contribute to N phase separation and hence viral replication.
Several RNA helicases enriched by N, such as DDX1, DHX9, and DHX30 identified in this study (Fig. 2, A and B) and previously reported DDX21 (10), attracted attention as well. Many RNA helicases have been demonstrated to positively or negatively regulate virus infections. On one hand, viruses have an intimate need for RNA helicases encoded by themselves and/or by host cells in their reproduction. For SARS-COV-2, in addition to its own helicase nsp13, some host RNA helicases may be needed to regulate and maintain the large viral genome and related RNA processes. On the other hand, some cellular RNA helicases have been reported to act as host restriction factors inhibiting viral infection by directly targeting viral components or by promoting host antiviral immune signaling pathways (43, 44, 45, 46). The roles of the currently identified RNA helicases in N functions and SARS-CoV-2 infection are also worthwhile to be analyzed. In the present study, we validated the interaction of DDX1 with N in details and showed its negative impact on SARS-CoV-2 infection (see later).
In addition, cytokine immune/inflammatory regulations (including interleukin-12-mediated signaling pathway and positive regulation of myeloid leukocyte cytokine production involved in immune response) were highlighted as top N-interactor-involved BPs (Fig. 2A). Inflammatory responses are pivotal aspects of SARS-CoV-2 pathogenesis. The results may suggest potential roles of N in viral interactions with cytokine inflammatory pathways, such as interleukin-12-mediated signaling. Taken together, these findings are not only well consistent with the previously uncovered or proposed molecular events in coronavirus infection but also reveal many new clues for further functional exploration and mechanism explanation of SARS-CoV-2 N interplays with host processes upon the viral infection and pathogenesis.
Furthermore, potential druggable targets in the N-interacting host proteins together with their targeting drugs were identified by database mining with the new interactome. Consequently, 32 pharmacological cellular targets and 34 corresponding drugs with 18 approved in at least one jurisdiction anywhere at some point in time, 13 in experimental phase, and 3 in investigational phase were uncovered (Fig. 2C and supplemental Table S3), yielding a host target–drug network. Interestingly, multiple drugs suggested in this network, including acetylsalicylic acid, copper, and artenimol, indeed have been shown to inhibit SARS-CoV-2 replications (47, 48, 49), supporting the instructive value of the present findings. Moreover, our findings suggest that the N–host factor interaction could be a target of these anti-SARS-CoV-2 agents.
Experimental Screening Based on the Interactome Identifies Anisomycin and Dasatinib as Anti-SARS-CoV-2 Agents
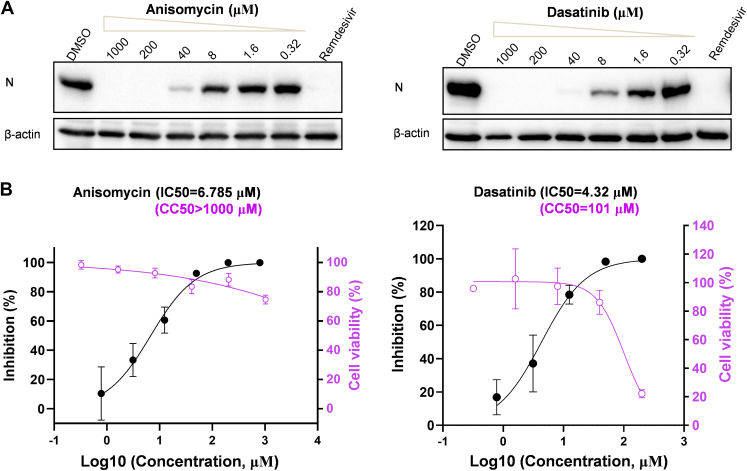
Importantly, as a proof of principle, we here tested the anti-SARS-CoV-2 activities of several new agents, including anisomycin, dasatinib, 4-hydroxyphenylpyruvic acid, etidronic acid, and (S)-3-phenyllactic acid, which have not been experimentally examined previously. As shown in Figure 3A and supplemental Fig. S2, two of these tested small molecules, anisomycin and dasatinib, indeed exhibited obvious anti-SARS-CoV-2 effects, whereas the others seemed to have no or only marginal activities, based on the inhibition of the viral protein expression. Thus, further quantitative analyses were performed for anisomycin and dasatinib. As shown in the following quantitative PCR (qPCR) analyses of the viral replication, anisomycin ([IC50] = 6.785 μM, half-cytotoxic concentration [CC50] >1000 μM, and selectivity index >147.3) and dasatinib (IC50 = 4.32 μM, CC50 = 101 μM, and selectivity index = 23.3) both showed notable anti-SARS-CoV-2 activities (Fig. 3B). Together, based on the interactome data, we identified new anti-SARS-CoV-2 agents, and these findings further support the reliability and instructive value of the interactome for development of specific antiviral intervention strategy targeting the N–cell interaction interface in the future.
Fig. 3.
Anisomycin and dasatinib, identified with the N interactome, exhibit notable anti-SARS-CoV-2 activities.A and B, Huh7-ACE2 cells were pretreated with different doses of the indicated drugs for 1 h, followed by SARS-CoV-2 infection (0.05 moi) in the presence of the drugs. At 24 h postinfection, cells were collected for Western blot (WB) analyses of the expression levels of SARS-CoV-2 N proteins by using antibodies against the indicated proteins (A) or for the following quantitative PCR (qPCR) analyses of viral RNA (B, black curves), respectively. Remdesivir (10 μM) was used as a control. The left Y-axes in (B) represent mean percent inhibition of virus yield determined by qPCR (black curves). The right Y-axes represent mean percent cell viability of Huh7-ACE2 cells treated with different doses of the indicated drugs for 24 h, which was determined by CCK8 kit (purple curves). Graphs show mean ± SD, n ≥ 3 biological replicates. The IC50 and half-cytotoxic concentration (CC50) were calculated by GraphPad Prism 8.3. ACE2, angiotensin-converting enzyme 2; CCK8, Cell Counting Kit-8; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
The Colocalization and Interaction of DDX1 With SARS-CoV-2 N
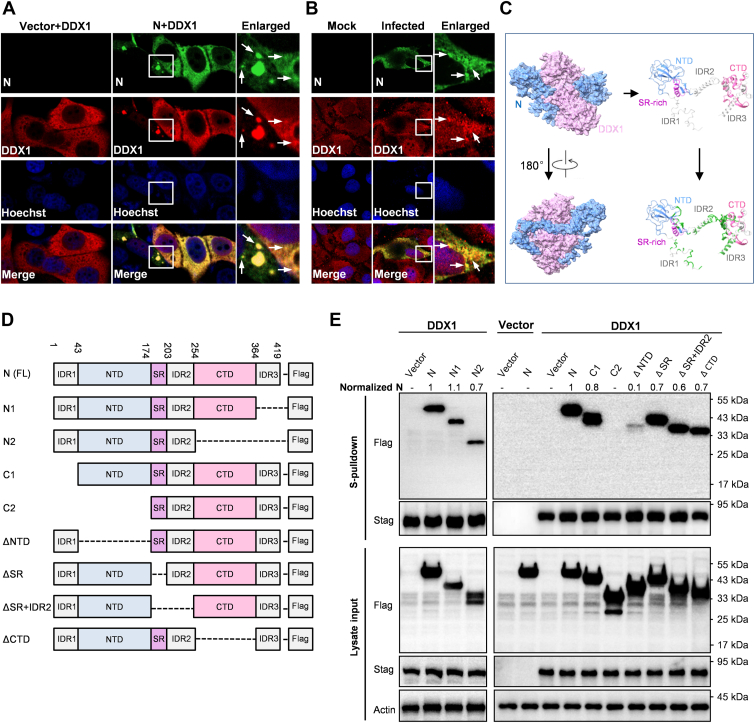
DDX1 has been reported to play negative or positive regulatory roles in some virus infections. Given its strong interaction with SARS-CoV-2 N (the third most enriched protein in N coprecipitates, fold change = 244), we hypothesized that DDX1 might also have a significant role in SARS-CoV-2 infection. First, immunofluorescence assay and confocal microscopy analyses showed an interesting colocalization of DDX1 with N in cytosol, and N condensates in the contexts of ectopic expression and SARS-CoV-2 infection, consistent with the protein interaction (Fig. 4, A and B and supplemental Fig. S3).
Fig. 4.
The colocalization and interaction of DDX1 with SARS-CoV-2 N.A, colocalization of ectopically expressed DDX1 with N. Huh7 cells were transfected with the DDX1 and N expression plasmids for 24 h, followed by fixation and immunofluorescence assay (IFA) analysis. B, colocalization of endogenous DDX1 with N upon SARS-CoV-2 infection. CaCo-2 cells were infected with SARS-COV-2 for 24 h, followed by fixation and IFA analysis. Nuclei were stained with Hoechst. Arrows indicate some typical N-condensates. Consistent with the previous studies (6), N-condensates could be more notable in some cells. Colocalization of DDX1 with N could be also observed in these condensate puncta. See also supplemental Fig. S3. C, predication of the DDX1–N interaction by AlphaFold2. AlphaFold2 predication reveals that DDX1 (purple) and N (blue) may form an intertwined dimer (left panels). The potential interaction interfaces on N were marked in green (right panels). D, linear representation of the domain organization for FLAG-tagged full-length, N-terminal- or C-terminal-truncated, or specific domain-deleted N, named as indicated. E, mapping of N domains targeted by DDX1. Huh7-ACE2 were cotransfected with the DDX1.STAG expression plasmid and the plasmids encoding full-length or truncated N proteins fused with FLAG or the corresponding control vectors as indicated. At 36 h post-transfection, protein interactions were analyzed by S-pulldown assays. Cell lysates (input) and pulldown products (S-pulldown) were subjected to Western blot (WB) analyses with specific antibodies against the indicated proteins. Relative coprecipitation ratios of N were calculated by normalization of the band intensities to those of the inputs and precipitated DDX1. The coprecipitation ratio of the full-length N in the same membrane was set as 1. Band intensities were measured using ImageJ. CTD, C-terminal oligomerization domain; IDR, putatively intrinsically disordered region; N, nucleocapsid protein; NTD, N-terminal domain; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SR, serine/arginine-rich region.
Furthermore, we predicted the possible complex structure formed by DDX1 and N by using deep learning–based AlphaFold2 program (DeepMind). Interestingly, as indicated in Figure 4C, DDX1 and N seemed to be intertwined together in the predicated complex structure, exhibiting a DDX1-N heterodimer of high predication confidence. Accordingly, there are several potential interaction interfaces that locate in multiple domains of N and may be involved in the binding with DDX1. In order to experimentally map the critical N domain(s) targeted by DDX1, we constructed a series of N mutants (Fig. 4D). Because some mutations led to extreme reduction of the protein expression or stability, affecting the judgment of the protein interaction, we chose the mutants that could be readily detected and adopted a relative quantification method (50) of N coprecipitation for the following analyses. As demonstrated in Figure 4E, the deletion of the C-terminal IDR3 of N (N1) did not impair the interaction with DDX1, but further deletion of the C-terminal domain (CTD) (N2) seemed to result in a weak reduction, suggesting that the CTD may have a small contribution to the DDX1–N interaction. Consistently, the interaction was largely retained after the deletion of the CTD alone (ΔCTD) (Fig. 4E). Then, the N-terminal truncation assays showed that a mutant with IDR1 deleted (C1) still kept a strong coprecipitation with DDX1, whereas further removal of NTD (C2) completely deprived the protein of the ability to interact with DDX1 (Fig. 4E), indicating that NTD plays a major role in the N–DDX1 interaction, although IDR1 may also have a weak effect. In agreement, even deleting the NTD alone (ΔNTD) mostly disrupted the targeting by DDX1. Finally, mutants with deletion of SR or both SR and IDR2 still had substantial interactions with DDX1, despite possible partial decrease based on the relative coprecipitation ratio of N. Taken together, these results suggest that although multiple domains of N may contribute to the interaction between N and DDX1, the NTD likely plays a dominant role in DDX1 targeting of N. Currently, the structures of full-length N and DDX1 have not been solved. Further structural resolution by biochemical and physical approaches are merited to validate the predicted model of the DDX1–N complex and understand the more detailed molecular fundamental of the DDX1–N interaction. The understanding will be valuable for the design of intervention molecules, for example, those mimicking DDX1 action, in the future.
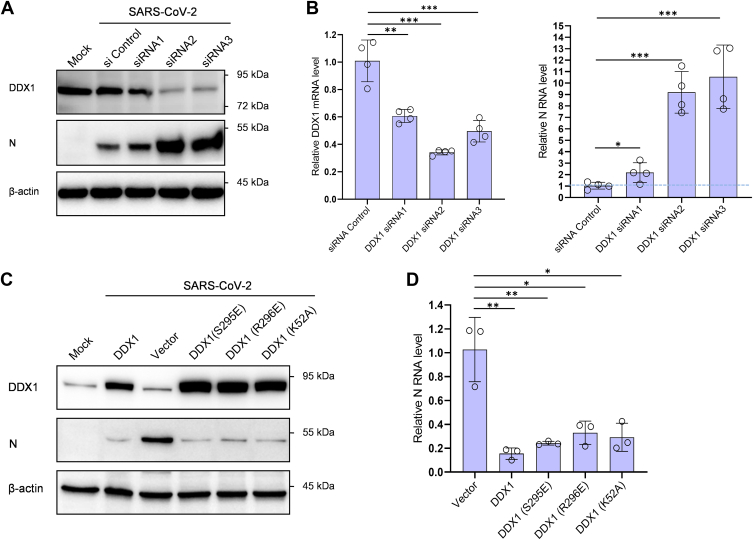
DDX1 Is a Host Cell Restriction Factor Against SARS-CoV-2 Infection and Acts Independently Its ATPase/Helicase Activities
Given the essential roles of N in virus life cycle, we considered that the targeting of SARS-CoV-2 N by DDX1 may affect the viral infection. To test the activity of DDX1 in SARS-CoV-2 infection, we first investigated the effect of DDX1 knockdown (KD) on the levels of vRNA and protein expression. Three DDX1-targeting siRNA constructs and a control were generated and used to be transfected into Huh7 cells stably expressing human angiotensin-converting enzyme 2 (Huh7-ACE2 cells), followed by SARS-CoV-2 infection. At 24 h postinfection, vRNA and protein levels were analyzed by qPCR and Western blotting. As shown in Figure 5, A and B, DDX1 KD significantly increased vRNA accumulation and viral protein expression, manifesting a strong restriction role of DDX1 against SARS-CoV-2 infection. Moreover, the siRNA with relatively lower silence efficiency (siRNA1), consistently, exhibited weaker activity to boost the viral infection in comparison with the other two RNAi constructs, showing dose dependence. Similarly, SARS-CoV-2 infection was boosted by DDX1 KD in human embryonic kidney 293 (HEK293) cells expressing ACE2 (supplemental Fig. S4A). Then, DDX1 KO HEK293 cells were generated by CRISPR–Cas9 editing. Consistently, enhancement of SARS-CoV-2 infection was also observed in the DDX1-KO cells expressing ACE2 (supplemental Fig. S4, B and C). Furthermore, overexpression of DDX1 was shown to inhibit viral protein and RNA production, which is consistent with the results of loss-of-function assays and corroborates the inhibitory role of DDX1 in SARS-CoV-2 infection (Fig. 5, C and D). Moreover, reconstitution of DDX1 expression in the DDX1-KO cells conferred dramatic resistance to SARS-CoV-2 (supplemental Fig. S4D).
Fig. 5.
DDX1 acts as a cellular restriction factor against SARS-CoV-2 infection, independently of its ATPase/helicase activities.A and B, DDX1 knockdown promotes SARS-CoV-2 infection. Huh7-ACE2 cells were transfected with specific DDX1 siRNAs or siRNA control. At 48 h post-transfection, cells were infected with SARS-CoV-2 (0.05 moi) for 24 h, followed by Western blot (WB) or quantitative PCR (qPCR) analyses of the expression levels of DDX1 or SARS-CoV-2 N proteins (A) or RNAs (B), respectively. Cells that were not transfected or infected (mock) were also analyzed by WB as additional controls. See also supplemental Fig. S4 for the similar results obtained in HEK293 cells with DDX1 knockdown or KO using CRISPR–Cas9 editing. C and D, DDX1 or mutant overexpression inhibits SARS-CoV-2 infection. Huh7-ACE2 cells were transfected with DDX1 or DDX1 mutants or the control vector. At 24 h post-transfection, cells were infected with SARS-CoV-2 (0.05 moi) for 24 h, followed by WB or qPCR analyses of the expression levels of the cellular or viral proteins or RNAs, respectively. WB analyses were conducted by using antibodies against the indicated proteins. Graphs show mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. See also supplemental Fig. S4 for the similar results obtained with reconstitution of DDX1 and its mutants in DDX1-KO HEK293 cells and supplemental Fig. S4E showing that the targeting of N by DDX1 is consistently independent of the ATPase/helicase activities of DDX1. HEK293, human embryonic kidney 293 cell line; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
DDX1 is well known for its ATP-dependent RNA helicase activity. In order to evaluate the role of the RNA helicase activity in its inhibition to SARS-CoV-2 infection, we constructed three ATPase/helicase-deficient mutants, K52A (the ATPase-deficient Walker A-motif mutant), S295E, and R296E (losing the helicase activity) (51, 52). Interestingly, all the mutants showed similar anti-SARS-CoV-2 effect compared with the wildtype DDX1 in Huh7-ACE2 cells (Fig. 5, C and D). Then, reconstitution of the mutants in the KO HEK293 cells provided the resistance against SARS-CoV-2 infection, similar to that of the wildtype protein (supplemental Fig. S4D). Consistently, by additional pulldown experiments, we further found that the mutants retain the capability to interact with N (supplemental Fig. S4E). These results suggest that the N-targeting ability and the resulting anti-SARS-CoV-2 activity of DDX1 are independent of its ATPase and helicase activities.
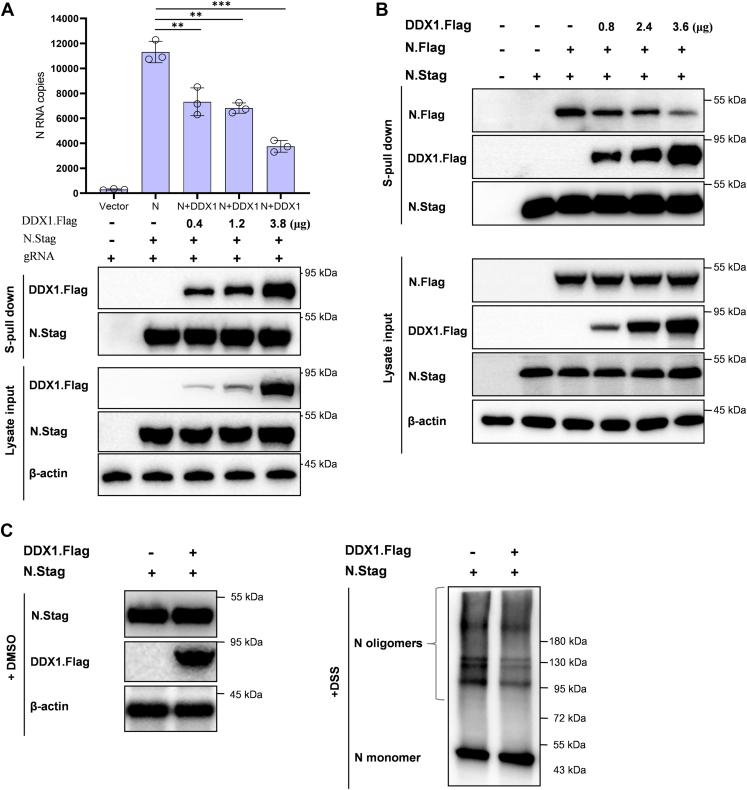
DDX1 Impedes the N–Genome RNA Binding, N–N Interaction, and N Oligomerization
By interactions with RNA, N encapsidates the viral genome RNA (gRNA) into the RNP complex and participates in the viral transcription and replication. Moreover, the NTD is speculated to contribute to the RNA-binding activity of N (53, 54, 55). In order to gain further mechanistical insights into DDX1 targeting of N and inhibition of SARS-CoV-2, we first tested the effect of DDX1 on N–gRNA interaction by N pulldown assays of gRNA. As shown in Figure 6A, viral gRNA was efficiently coprecipitated by N, which however, was significantly inhibited by DDX1 expression. The result indicates that the N–vRNA interaction is hampered by DDX1, which is also consistent with DDX1 targeting of the N NTD.
Fig. 6.
DDX1 can inhibit the N–genome RNA (gRNA) binding, N–N interaction, and N oligomerization, the critical activities of N in SARS-CoV-2 propagation.A, the effect of DDX1 on the N–gRNA interaction. Huh7-ACE2 cells were transfected with the N.Stag-expressing plasmid, along with different amounts of the DDX1.Flag expression plasmid or control vector. At 48 h post-transfection, the supernatants of the cell lysates were incubated with RNase inhibitor (1 U/μl) and viral gRNA (∼2 × 105 copies) extracted from virus stock, followed by S-pulldown assays. The N coprecipitates of RNAs and proteins were respectively analyzed by quantitative PCR (qPCR) or Western blot (WB) using antibodies against the indicated tags (or β-actin protein). Graphs show mean ± SD. ∗∗p < 0.01; ∗∗∗p < 0.001. B, DDX1 impedes the N–N interaction. Huh7-ACE2 cells were cotransfected with the plasmids expressing N proteins fused with Stag or Flag (N.Stag or N.Flag) together with different amounts of the plasmid encoding Flag-tagged DDX1 (DDX1.Flag) or the corresponding control vectors. At 48 h post-transfection, cells were lysed for S-pulldown assays. The cell lysates and N.Stag coprecipitates were respectively analyzed by Western blot (WB) using antibodies against the indicated tags (or β-actin protein). C, DDX1 weakens the N oligomerization. Huh7-ACE2 cells were transfected with the DDX.Flag-expressing plasmid or control vector, together with the plasmid encoding N. At 48 h post-transfection, cells were harvested and washed by PBS, followed by crosslinking with disuccinimidyl suberate (DSS, 0.5 mM) that enables the fixation of oligomerized N in situ. After crosslinking at 37 °C for 30 min, the reaction was quenched by 1 M Tris buffer (pH 7.5), and samples were delivered to SDS-PAGE and WB analyses of the N oligomerization and protein expression. ACE2, angiotensin-converting enzyme 2; N, nucleocapsid protein; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
In addition to the N–vRNA interaction, it is considered that N–N interaction and the further oliomerization of CoV N proteins also play an essential role to drive the virus assembly and propagation (56). Thus, we next investigated whether the N–N interaction and N oligomerization can be affected by DDX1. Interestingly, by an additional pulldown assay, we observed that the N–N interaction was blocked by DDX1 as shown in Figure 6B. Furthermore, we evaluated the oligomerization of SARS-CoV-2 N proteins by using chemical crosslinking with disuccinimidyl suberate that enables the fixation of oligomerized N in situ (57). Consistently, the N oligomerization was likely attenuated by DDX1 (Fig. 6C), substantiating the hinderance of the critical activities of N by DDX1. Taken together, these data suggest that by the strong targeting of N and particularly the NTD, DDX1 obstructs the N–gRNA binding, N–N interaction, and N oligomerization, interfering with SARS-CoV-2 infection.
Previous studies have shown that DDX1 can regulate transition of MHV from discontinuous to continuous transcription, promoting longer subgenomic mRNA synthesis but having no effect on N expression (43). In addition, DDX1 was reported to interact with infectious bronchitis virus nsp14 and hence likely positively regulate viral replication (45); however, in a study by Zhou et al. (58), it was demonstrated that DDX1 interacts with the nsp14 of another coronavirus, transmissible gastroenteritis virus, and is required for transmissible gastroenteritis virus nsp14-induced antiviral interferon response, thus conferring antiviral activity on host cells. These studies suggest that DDX1 seems to play differential (positive or negative) roles in infections of different coronaviruses. We noted that a study by Ariumi (8) suggested a possible proviral activity of DDX1 in SARS-CoV-2 infection of HEK293T cells by an RNAi KD screening assay using a single shRNA-encoding lentiviral vector and puromycin-resistant pooled cells after the lentiviral transduction. However, in our study, RNAi assays with transfection of different siRNA constructs consistently demonstrated the restrictive activity of DDX1 that moreover exhibited a dose-dependent manner. Similar results were observed in different cells and also validated in the context of DDX1 KO by CRISPR–Cas9 gene editing. The reason for the discrepancy remains unknown. However, it is possible that the infection of the lentiviral vector and the following superinfection of SARS-CoV-2 may lead to some unpredictable effects. Moreover, DDX1 also play multiple roles in lentivirus infection itself (59), which may further increase the complexity of the results. In addition, regarding the role of DDX1, it remains to be explored whether DDX1 can target other components of SARS-CoV-2. Likewise, it remains unknown whether DDX1 and its interaction with N have roles in modulation of SARS-CoV-2 genomic RNA or subgenomic mRNAs, as seen upon MHV infection. Furthermore, the currently established interactome also identifies several other helicases and their interaction partners, many of which (e.g., DHX30 and DHX9) have been documented as regulatory factors in virus infections. Their roles in SARS-CoV2 infection are worth further studies as well.
In summary, we here established a new cellular interactome of SARS-CoV-2 N, a core structural protein of the virus, by a high-specific AP combined with quantitative LC–MS/MS and immunoblotting validations, uncovering many N-interacting host factors previously unrecognized and multiple host processes likely regulating or being regulated by N and hence the virus infection. Accordingly, we also identified series of potential druggable targets and corresponding drugs and presented a drug–host target network, meriting further testing and screening for antiviral research. Based on the network, we indeed identified several small molecules as new antiviral drugs against SARS-CoV-2 infection in vitro. Furthermore, the interaction of DDX1 with N was corroborated and characterized in details. Although multiple domains of N may be involved in the binding by DDX1, the N NTD likely plays a major role in the DDX1 targeting of N. Moreover, in line with its targeting of N (especially the NTD), DDX1 was then demonstrated to act as a restriction factor against SARS-CoV-2 infection. The inhibition of SARS-CoV-2 by DDX1 is independent of the ATPase/helicase activities of DDX1, whereas multiple critical actions of N, including the N–gRNA binding, N–N interaction, and N polymerization, are likely hindered by the DDX1 interaction with N, showing further mechanism details. These findings provide new insights into SARS-CoV-2 N interaction with host cells and may help guide further studies on the viral infection mechanisms and design of antiviral intervention strategies.
Data Availability
The raw MS proteomics data have been deposited to the Science Data Bank, and the private access link for review is https://www.scidb.cn/s/E3qAfe. Other data are contained within the article.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant number: 32170171), the National Key Research and Development Program of China (grant nos.: 2022YFC2303300 and 2018YFA0507202), the Youth Innovation Promotion Association of Chinese Academy of Sciences, and Guangzhou Laboratory (grant no.: EKPG21-30-2). We thank the Core Facility and Technical Support of Wuhan Institute of Virology, National Virus Resource Center, Wuhan Institute of Virology, and the running team of National Biosafety Laboratory, Wuhan, Chinese Academy of Sciences for technical assistants. We thank Mr Tong Liu, Dr Yizi Liu, Dr Qiangqiang Han, Dr Jun Shang, and Miss Xin Zhou from SpecAlly for technical support in MS raw data preparation.
Author contributions
Y.-J. N. conceptualization; Y.-Q. M. methodology; Y. J. software; Y.-Q. M. and M. H. validation; Y.-Q. M. formal analysis; Y.-Q. M., M. H., and K. F. investigation; X. S. and Y.-J. N. resources; Y.-Q. M. data curation; Y.-Q. M. writing–original draft; X. S. and Y.-J. N. writing–review & editing; Y.-Q. M. visualization; X. S. supervision; X. S. and Y.-J. N. funding acquisition.
Contributor Information
Xiulian Sun, Email: sunxl@wh.iov.cn.
Yun-Jia Ning, Email: nyj@wh.iov.cn.
Supplemental Data
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao C., Cai Z., Xiao X., Rao J., Chen J., Hu N., et al. The architecture of the SARS-CoV-2 RNA genome inside virion. Nat. Commun. 2021;12:3917. doi: 10.1038/s41467-021-22785-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021;12:1936. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cascarina S.M., Ross E.D. Phase separation by the SARS-CoV-2 nucleocapsid protein: consensus and open questions. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.101677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perdikari T.M., Murthy A.C., Ryan V.H., Watters S., Naik M.T., Fawzi N.L. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 2020;39 doi: 10.15252/embj.2020106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ariumi Y. Host cellular RNA helicases regulate SARS-CoV-2 infection. J. Virol. 2022;96 doi: 10.1128/jvi.00002-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng Y., Du N., Lei Y.Q., Dorje S., Qi J.X., Luo T.R., et al. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020;39 doi: 10.15252/embj.2020105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon D.E., Jang G.M., Bouhaddou M., Xu J.W., Obernier K., White K.M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nabeel-Shah S., Lee H., Ahmed N., Burke G.L., Farhangmehr S., Ashraf K., et al. SARS-CoV-2 nucleocapsid protein binds host mRNAs and attenuates stress granules to impair host stress response. iScience. 2002;25:103562. doi: 10.1016/j.isci.2021.103562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng X., Sun Z., Yu L., Shi D., Zhu M., Yao H., et al. Interactome analysis of the nucleocapsid protein of SARS-CoV-2 virus. Pathogens. 2021;10:1155. doi: 10.3390/pathogens10091155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stukalov A., Girault V., Grass V., Karayel O., Bergant V., Urban C., et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594:246–252. doi: 10.1038/s41586-021-03493-4. [DOI] [PubMed] [Google Scholar]
- 14.Li J., Guo M., Tian X., Wang X., Yang X., Wu P., et al. Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med. 2021;2:99–112.e7. doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.May D.G., Martin-Sancho L., Anschau V., Liu S., Chrisopulos R.J., Scott K.L., et al. A BioID-derived proximity interactome for SARS-CoV-2 proteins. Viruses. 2022;14:611. doi: 10.3390/v14030611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X., Huuskonen S., Laitinen T., Redchuk T., Bogacheva M., Salokas K., et al. SARS-CoV-2-host proteome interactions for antiviral drug discovery. Mol. Syst. Biol. 2021;17 doi: 10.15252/msb.202110396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z., Wang C., Feng X., Nie L., Tang M., Zhang H., et al. Interactomes of SARS-CoV-2 and human coronaviruses reveal host factors potentially affecting pathogenesis. EMBO J. 2021;40 doi: 10.15252/embj.2021107776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samavarchi-Tehrani P., Abdouni H., Knight J.D.R., Astori A., Samson R., Lin Z.-Y., et al. A SARS-CoV-2–host proximity interactome. bioRxiv. 2020 doi: 10.1101/2020.09.03.282103. [preprint] [DOI] [Google Scholar]
- 19.Laurent E.M.N. Global BioID-based SARS-CoV-2 proteins proximal interactome unveils novel ties between viral polypeptides and host factors involved in multiple COVID19-associated mechanisms. bioRxiv. 2020 doi: 10.1101/2020.08.28.272955. [preprint] [DOI] [Google Scholar]
- 20.Feng K., Min Y.Q., Sun X., Deng F., Li P., Wang H., et al. Interactome profiling reveals interaction of SARS-CoV-2 NSP13 with host factor STAT1 to suppress interferon signaling. J. Mol. Cell Biol. 2021;13:760–762. doi: 10.1093/jmcb/mjab068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min Y.Q., Ning Y.J., Wang H., Deng F. A RIG-I-like receptor directs antiviral responses to a bunyavirus and is antagonized by virus-induced blockade of TRIM25-mediated ubiquitination. J. Biol. Chem. 2020;295:9691–9711. doi: 10.1074/jbc.RA120.013973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ning Y.J., Feng K., Min Y.Q., Cao W.C., Wang M.L., Deng F., et al. Disruption of type I interferon signaling by the nonstructural protein of severe fever with thrombocytopenia syndrome virus via the hijacking of STAT2 and STAT1 into inclusion bodies. J. Virol. 2015;89:4227–4236. doi: 10.1128/JVI.00154-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ning Y.J., Wang M., Deng M., Shen S., Liu W., Cao W.C., et al. Viral suppression of innate immunity via spatial isolation of TBK1/IKKepsilon from mitochondrial antiviral platform. J. Mol. Cell Biol. 2014;6:324–337. doi: 10.1093/jmcb/mju015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Min Y.Q., Shi C., Yao T., Mo Q., Deng F., Wang H., et al. The nonstructural protein of guertu virus disrupts host defenses by blocking antiviral interferon induction and action. ACS Infect. Dis. 2020;6:857–870. doi: 10.1021/acsinfecdis.9b00492. [DOI] [PubMed] [Google Scholar]
- 25.Bardou P., Mariette J., Escudie F., Djemiel C., Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics. 2014;15:293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu G.C., Wang L.G., Han Y.Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunham W.H., Mullin M., Gingras A.C. Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics. 2012;12:1576–1590. doi: 10.1002/pmic.201100523. [DOI] [PubMed] [Google Scholar]
- 31.Lambert J.P., Tucholska M., Go C., Knight J.D.R., Gingras A.C. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. J. Proteomics. 2015;118:81–94. doi: 10.1016/j.jprot.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tahara S.M., Dietlin T.A., Nelson G.W., Stohlman S.A., Manno D.J. Mouse hepatitis virus nucleocapsid protein as a translational effector of viral mRNAs. Adv. Exp. Med. Biol. 1998;440:313–318. doi: 10.1007/978-1-4615-5331-1_41. [DOI] [PubMed] [Google Scholar]
- 33.Tahara S.M., Dietlin T.A., Bergmann C.C., Nelson G.W., Kyuwa S., Anthony R.P., et al. Coronavirus translational regulation: leader affects mRNA efficiency. Virology. 1994;202:621–630. doi: 10.1006/viro.1994.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emmott E., Munday D., Bickerton E., Britton P., Rodgers M.A., Whitehouse A., et al. The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. J. Virol. 2013;87:9486–9500. doi: 10.1128/JVI.00321-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt N., Lareau C.A., Keshishian H., Ganskih S., Schneider C., Hennig T., et al. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat. Microbiol. 2021;6:339–353. doi: 10.1038/s41564-020-00846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spagnolo J.F., Hogue B.G. Host protein interactions with the 3' end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 2000;74:5053–5065. doi: 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattijssen S., Kozlov G., Fonseca B.D., Gehring K., Maraia R.J. LARP1 and LARP4: up close with PABP for mRNA 3' poly(A) protection and stabilization. RNA Biol. 2021;18:259–274. doi: 10.1080/15476286.2020.1868753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S.J., Huang W.Z., Ren L.L., Ju X.H., Gong M.L., Rao J., et al. Comparison of viral RNA-host protein interactomes across pathogenic RNA viruses informs rapid antiviral drug discovery for SARS-CoV-2. Cell Res. 2022;32:9–23. doi: 10.1038/s41422-021-00581-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lloyd R.E. Regulation of stress granules and P-bodies during RNA virus infection. Wires RNA. 2013;4:317–331. doi: 10.1002/wrna.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kedersha N., Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 41.Protter D.S.W., Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savastano A., Ibanez de Opakua A., Rankovic M., Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020;11:6041. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C.H., Chen P.J., Yeh S.H. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe. 2014;16:462–472. doi: 10.1016/j.chom.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z.Q., Kim T., Bao M.S., Facchinetti V., Jung S.Y., Ghaffari A.A., et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu L.H., Khadijah S., Fang S.G., Wang L., Tay F.P.L., Liu D.X. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J. Virol. 2010;84:8571–8583. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye P.Y., Liu S.F., Zhu Y.P., Chen G.F., Gao G.X. DEXH-Box protein DHX30 is required for optimal function of the zinc-finger antiviral protein. Protein Cell. 2010;1:956–964. doi: 10.1007/s13238-010-0117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geiger N., Konig E.M., Oberwinkler H., Roll V., Diesendorf V., Fahr S., et al. Acetylsalicylic acid and salicylic acid inhibit SARS-CoV-2 replication in precision-cut lung slices. Vaccines (Basel) 2022;10:1619. doi: 10.3390/vaccines10101619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nair M.S., Huang Y., Fidock D.A., Polyak S.J., Wagoner J., Towler M.J., et al. Artemisia annua L. extracts inhibit the in vitro replication of SARS-CoV-2 and two of its variants. J. Ethnopharmacol. 2021;274:114016. doi: 10.1016/j.jep.2021.114016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andreou A., Trantza S., Filippou D., Sipsas N., Tsiodras S. COVID-19: the potential role of copper and N-acetylcysteine (NAC) in a combination of candidate antiviral treatments against SARS-CoV-2. In Vivo. 2020;34:1567–1588. doi: 10.21873/invivo.11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng K., Deng F., Hu Z.H., Wang H.L., Ning Y.J. Heartland virus antagonizes type I and III interferon antiviral signaling by inhibiting phosphorylation and nuclear translocation of STAT2 and STAT1. J. Biol. Chem. 2019;294:9503–9517. doi: 10.1074/jbc.RA118.006563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lamichhane R., Hammond J.A., Pauszek R.F., Anderson R.M., Pedron I., van der Schans E., et al. A DEAD-box protein acts through RNA to promote HIV-1 Rev-RRE assembly. Nucleic Acids Res. 2017;45:4632–4641. doi: 10.1093/nar/gkx206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kellner J.N., Reinstein J., Meinhart A. Synergistic effects of ATP and RNA binding to human DEAD-box protein DDX1. Nucleic Acids Res. 2015;43:2813–2828. doi: 10.1093/nar/gkv106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein--forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bai Z., Cao Y., Liu W., Li J. The SARS-CoV-2 nucleocapsid protein and its role in viral structure, biological functions, and a potential target for drug or vaccine mitigation. Viruses. 2021;13:1115. doi: 10.3390/v13061115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu W., Cheng Y., Zhou H., Sun C., Zhang S. The SARS-CoV-2 nucleocapsid protein: its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostics. Virol. J. 2023;20:6. doi: 10.1186/s12985-023-01968-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cong Y.Y., Kriegenburg F., de Haan C.A.M., Reggiori F. Coronavirus nucleocapsid proteins assemble constitutively in high molecular oligomers. Sci. Rep. 2017;7:5740. doi: 10.1038/s41598-017-06062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mo Q., Xu Z., Deng F., Wang H.L., Ning Y.J. Host restriction of emerging high-pathogenic bunyaviruses via MOV10 by targeting viral nucleoprotein and blocking ribonucleoprotein assembly. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Y.R., Wu W., Xie L.L., Wang D., Ke Q.Y., Hou Z.Z., et al. Cellular RNA helicase DDX1 is involved in trasmissible gastroenteritis virus nsp14-induced interferon-beta production. Front. Immunol. 2017;8:940. doi: 10.3389/fimmu.2017.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgcomb S.P., Carmel A.B., Naji S., Ambrus-Aikelin G., Reyes J.R., Saphire A.C.S., et al. DDX1 is an RNA-dependent ATPase involved in HIV-1 rev function and virus replication. J. Mol. Biol. 2012;415:61–74. doi: 10.1016/j.jmb.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw MS proteomics data have been deposited to the Science Data Bank, and the private access link for review is https://www.scidb.cn/s/E3qAfe. Other data are contained within the article.