Abstract
Aging is the greatest risk factor for a multitude of age-related diseases including sarcopenia—the loss of skeletal muscle mass and strength—which occurs at remarkable rates each year. There is an unmet need not only to understand the mechanisms that drive sarcopenia but also to identify novel therapeutic strategies. Given the ease and affordability of husbandry, along with advances in genomics, genome editing technologies, and imaging capabilities, teleost models are increasingly used for aging and sarcopenia research. Here, we explain how teleost species such as zebrafish, African turquoise killifish, and medaka recapitulate many of the classical hallmarks of sarcopenia, and discuss the various dietary, pharmacological, and genetic approaches that have been used in teleosts to understand the mechanistic basis of sarcopenia.
Keywords: Age-related pathology, Fish models, Skeletal muscle, Sarcopenia
Aging is a naturally occurring and universal phenomenon resulting in a progressive decline in the ability of organs to perform their physiological functions. One such organ is skeletal muscle, which, despite being indispensable for movement and a critical metabolic and storage organ (1), undergoes striking atrophy and a reduction in strength—known as sarcopenia—at 1% and 3% in humans each year, respectively (2,3). Although there is a lack of consensus on the clinical definition of sarcopenia, with no clear established diagnostic criteria for it, what is well appreciated is that sarcopenia is a complex and multifaceted neuromuscular condition. It is characterized by numerous cellular and molecular changes including muscle fiber atrophy, stem cell dysfunction, telomere attrition, epigenetic alterations, mitochondrial dysfunction, and metabolic alterations among others (reviewed in (4)). Despite being extensively characterized at a cellular level, pharmacotherapies to treat sarcopenia have thus far not been identified, with the most promising management strategies limited to exercise and healthy nutrition, which, although beneficial, have their own limitations—primarily in a form of limited compliance and tolerance (reviewed in (5)). One of the major challenges that has hindered the identification of suitable pharmacotherapies is the limited understanding of the molecular, physiological, and metabolic mechanisms underlying muscle wasting. Although a multitude of hallmarks of sarcopenia have been identified, we currently do not understand which of these are pathogenic and directly contribute to muscle wasting, and those that are simply a consequence of the aging process having no impact on muscle function. Further to that, it is currently unknown if these pathogenic hallmarks can be pharmacologically attenuated or reversed to ameliorate sarcopenia. Research addressing the mechanistic basis of sarcopenia is therefore needed to inform more targeted development of therapies. This is particularly important given the ever-growing aged population worldwide, which has resulted in an increased prevalence of sarcopenia, subsequently placing a considerable social and economic burden on the healthcare system.
To study the mechanistic basis of sarcopenia, several animal models have been routinely used including nematodes, flies, and rodents such as rats, mice, and guinea pigs. Although the invertebrate models have short lifespans and are remarkably easy and cheap to maintain, they do not fully recapitulate the human aging and sarcopenic pathologies (6). In contrast, rodent models are more commonly used given that they share several physiological characteristics with humans and therefore recapitulate not only sarcopenic pathology but also other diseases. However, due to their longer lifespan, differences in metabolic stability, cost, and labor-intensive husbandry requirements, alternative models that can address these experimental challenges are required (7,8). Although teleosts do not address all these limitations, they have relatively short lifespans and are easier and more affordable to maintain and use for research. This, coupled with advances in genomics, genome editing technologies, and improved imaging capabilities, has made teleosts a popular model for sarcopenia and aging research. This review will highlight the sarcopenic pathologies observed in teleost species, with a primary focus on zebrafish, African turquoise killifish and medaka; discuss how they have contributed to our understanding of the fundamental biology of sarcopenia; and highlight the implications of these findings for the identification of suitable pharmacotherapies.
Fish Models Most Commonly Used for Sarcopenia Research
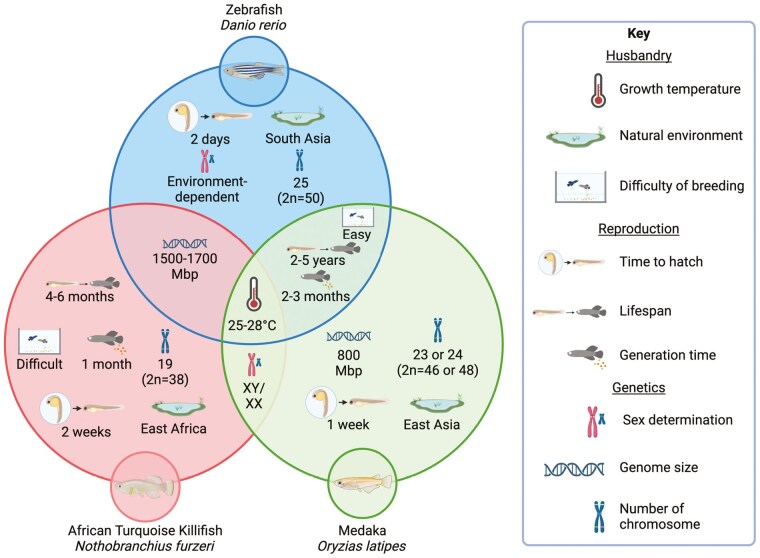
Zebrafish (Danio rerio) are a tropical freshwater fish native to South Asia and a prominent model for understanding vertebrate biology and disease (Figure 1). Although they have successfully been used to model myopathies and muscular dystrophies, they have recently attracted attention as a model for sarcopenia research. In line with the alterations observed in aged mammals, aged zebrafish display skeletal muscle degeneration and reduced muscle mass, accompanied with increased senescence-associated β-galactosidase activity (SA-β-Gal), mitochondrial dysfunction, oxidized protein accumulation, spinal curvature, and display reduced swimming performance with increasing age, indicative of a decline in muscle function (9–12). However, given that the average lifespan of zebrafish is 2 to 5 years (10,13), the use of wild-type strains is not highly feasible for aging studies, and instead mutant models that display aging phenotypes are commonly used to study the biology of sarcopenia.
Figure 1.
Characteristics of the zebrafish, African turquoise killifish, and medaka.
The African turquoise killifish (Nothobranchius furzeri) are the shortest-lived vertebrates that can be bred in captivity and are an emerging model for the investigation of aging (14). The African turquoise killifish inhabit ephemeral ponds in Zimbabwe and Mozambique. During the dry season, African turquoise killifish eggs deposited in the muddy substrate are arrested in a dormant state, and at the start of the rainy season, they exit this arrested state and undergo rapid maturation reaching sexual maturity within 4 to 5 weeks (15). The median lifespan of these fish is 4 months to 1 year depending on the strain (Figure 1). During their lifespan, African turquoise killifish not only undergo remarkable levels of growth but they also exhibit hallmarks of aging such as appearance of neoplastic lesions in the liver, kidney, heart, and gonads (16), reduced regenerative capacity of the fin (17), decreased mitochondrial DNA copy number and function (18), and shortening of telomeres (19). In the context of muscle, aged African turquoise killifish display moderate reduction in fiber cross-sectional area, a reduced number of muscle stem cells, muscle denervation associated with neuromuscular junction degeneration, and striking alterations in lipid metabolism (20). Further to this, aged African turquoise killifish show reduced locomotion highlighting a reduction in muscle function (21). Given that African turquoise killifish display many of the typical signs of muscle aging and have a lifespan that is comparable to many invertebrate models, they provide a unique system to study the biology of sarcopenia and muscle aging. Notably, while N furzeri is the most commonly used killifish species for research, several additional species including Nothobranchius kadleci, Nothobranchius rachovii, and Nothobranchius guentheri have also been studied and are discussed in this review.
Medaka (Oryzias latipes), commonly known as Japanese rice fish, has been used as a model species for research since the early 1900s (22). Medaka inhabits both brackish and freshwater environments in areas of East Asia (Figure 1) (23). Although they are similar in size, and have similar anatomy and physiology to zebrafish, they are highly tolerant to inbreeding making them an optimal laboratory model. Aged medaka have been shown to display age-associated degeneration of multiple tissues including skin, liver, and heart but no age-associated skeletal muscle alterations were observed, although this could be due to the limited assays (lipofuscin and BrDU) performed (22). The sarcopenic pathologies of aged medaka therefore remain to be characterized.
Multiple other fish species have been embraced for biomedical research and while we do not discuss them in any detail, they have been studied in the context of telomere biology and, as such, have been briefly introduced in Table 1.
Table 1.
Telomere Length in Human, Mouse and Teleosts
| Species | Common Name | Average Lifespan | Telomere Length (kb) | Tissue(s) Examined | References |
|---|---|---|---|---|---|
| Homo sapiens | Humans | 70–80 years | 5–15 | Blood, sperm, kidney, and placental DNA | (24,25) |
| Mus musculus | Mouse | 2–3 years | 20–150 | Liver, bone marrow cells, skin fibroblasts, spleen, and kidney | (26–28) |
| Danio rerio | Zebrafish | 2–5 years | 15–20 | Whole animal cell suspension | (29,30) |
| Nothobranchius furzeri, Nothobranchius kadleci, Nothobranchius rachovii | Annual killifish | 4–12 months | 5–10, 6–8 (GRZ strain) | Muscle, skin, caudal fin, whole animal cell suspension | (19,31,32) |
| Oryzias melastigma | Marine medaka | 2–3 months | 0.5–12 | Whole fry cell suspension, liver | (33,34) |
| Oryzias latipes | Japanese rice fish (medaka) | 2–5 years | 7–15 kb | Whole embryo cell suspension, whole fry cell suspension, liver, kidney, intestine, muscle, gonad, heart, brain, spleen, and gill | (33,35,36) |
| Oncorhynchus mykiss | Rainbow Trout | 11 years | 17–20 | Head kidney | (37,38) |
| Xiphophorus maculatus, X helleri and X couchianus | Platyfish | 1–4 years | 2–6 | Liver, gill, brain, eyes, testis, and ovaries | (39) |
| Menidia menidia | Atlantic Silverside | 2 years | Relative telomere length reported | Larvae, muscle, and brain | (40) |
| Gadus morhua | Atlantic Cod | 25 years | Relative telomere length reported | Liver | (41) |
| Gambusia holbrooki | Eastern Mosquito fish | 1–2 years | Relative telomere length reported | Tail muscle | (42) |
| Merluccius merluccius | European Hake | 15–30 years | Relative telomere length reported | Brain, kidney, liver, and muscle | (43) |
| Cyprinus carpio | Common Carp | 47 years | 0.6–0.8 | Muscle tissue and fin clips | (44) |
Molecular Hallmarks of Sarcopenia and Insights From Teleosts
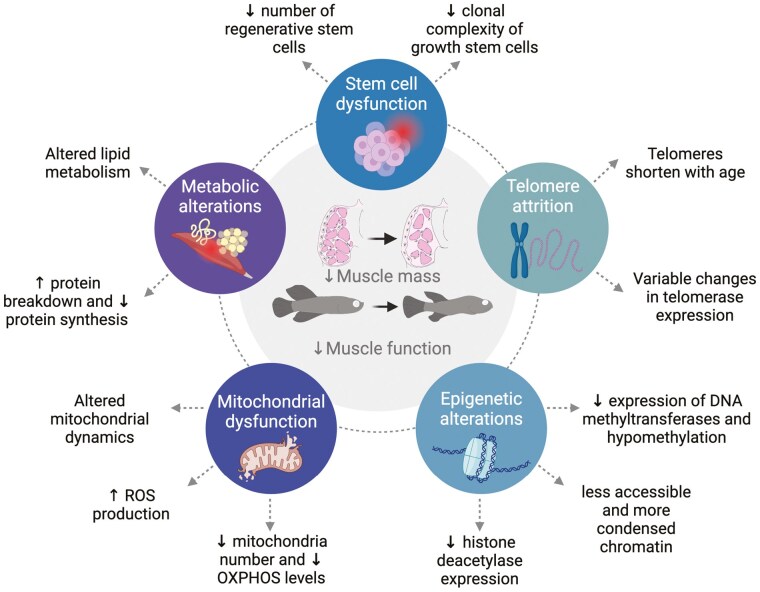
In this section, we will discuss the significant contributions made using teleost models to understand 6 of the main hallmarks of sarcopenia—muscle fiber atrophy and fiber composition, muscle stem cell function in growth and regeneration, telomere attrition, epigenetic alterations, mitochondrial dysfunction, and metabolic changes (Figure 2). Notably, several genetic mutants have been created as models of premature aging (45,46), particularly in zebrafish, and while they have contributed to our understanding of sarcopenia, they are not the focus of this review. Instead, we have focused on insights gained from the natural aging process in teleosts.
Figure 2.
Summary of the hallmarks of sarcopenia pathologies in teleosts.
Muscle Fiber Atrophy and Fiber Composition
The formation of skeletal muscle in fish is a highly conserved and well-characterized process of myogenesis, which has been reviewed several times (47,48). Briefly, during segmentation paraxial mesodermal cells form a monolayer against the notochord, termed adaxial cells. Upon activation of Hedgehog signaling these cells migrate to the lateral end of the myotome to form the superficial slow muscle layer, with the remaining nonadaxial cells of the posterior somite differentiating to form the fast muscle. Notably, a small number of adaxial cells remain at the notochord to form a teleost-specific slow muscle cell type known as the muscle pioneer cell (49). This process of muscle development results in 2 distinct compartments of muscle fibers, with the oxidative, type I slow fibers located superficially, and the rest of the myotome composed of glycolytic, type II fast muscle fibers. Although the compartmentalization of slow and fast muscle cells is unique to, and typical of teleosts, not all of them follow the same profile. For example, in tuna, as an adaptation to their high performance and pelagic lifestyle, slow muscle is medially located adjacent to the spinal cord (50).
In humans, sarcopenia is characterized by the reduction in size (atrophy), primarily of the fast, type 2, muscle fibers and to a lesser extent, a reduction in number (hypoplasia) of both slow and fast muscle cells (reviewed in (51)). As in humans, aged zebrafish show reduced fiber cross-sectional area, although it is unknown if this is fiber type-specific (11,12). Aged African turquoise killifish also show muscle fiber atrophy, specifically of the medium-sized fast muscle fibers localized deeper within the myotome, with no evidence of hypoplasia (20). Importantly, in humans, sarcopenia is often associated with a shift from type II, fast muscle fibers to a type I, slow phenotype (52,53). Although this has not been thoroughly investigated in fish, this phenomenon of fiber type switching does not appear to occur in the African turquoise killifish (20), suggesting that fish muscle may display distinct changes in metabolic profiles during aging.
Muscle Stem Cell Function in Growth and Regeneration
A unique feature of fish muscle is its remarkable ability to expand and grow severalfold as the animal approaches its adult size. Unlike in mammals, whereby postnatal muscle growth occurs only via hypertrophy, defined as the increase in size of existing cells, muscle growth in fish also occurs via hyperplasia—the addition of new fibers, which is particularly prevalent in the embryonic and larval stages. In adults however, there is a plateau in hyperplastic growth and hypertrophy becomes the primary muscle growth strategy (54).
The ability to undergo hyperplasia is attributed to the presence of a unique muscle stem cell population, marked by the transcription factors pax3 and meox1, found within a superficially located, extracellular matrix rich region termed the external cell layer (ECL) (54–56). Here, the stem cells are arrested in the G2 phase of the cell cycle, which has been hypothesized to prime them for growth stimuli (54). Lineage tracing experiments have been used to examine how these stem cells are deployed for hyperplastic muscle growth. These experiments have revealed that while multiple independent stem cells randomly contribute to myofiber generation during the early growth phase, a single stem cell clone comes to dominate growth in adult muscle over successive self-renewal events (54). This phenomenon, termed clonal drift, is characterized by the stochastic loss and reciprocal expansion of the progeny of individual stem cells. This ultimately leads to a pool of clonally related stem cells responsible for replenishing the entire stem cell population, which in turn generates clonally related differentiated tissues. Importantly, while this loss in muscle stem cell clonality has been observed in adult zebrafish muscle, it is uncertain if the same occurs in aged skeletal muscle. In mice, muscle stem cells have been shown to maintain clonal complexity with homeostatic aging, although reduced functional heterogeneity is observed (57). The differences in clonal complexity in zebrafish and mice may be explained by the difference in stem cell types, with the zebrafish experiments focusing on heterogeneity of ECL localized stem cell, and the mouse experiments focused on the classical satellite cell population (discussed below); and the differences in growth strategies used—with zebrafish utilizing hyperplastic and hypertrophic strategies and mice displaying only hypertrophic growth. In any case, although the mechanisms regulating clonal drift and the implications of it remain unknown, it can be postulated that drift may be a consequence of the selection of the most optimal stem cell clone required for growth. If this is the case, an understanding of what makes this stem cell clone the most optimal, and the processes that mediate its selection will be highly invaluable for managing sarcopenia, whereby inducing the formation of new muscle cells is desirable.
In addition to the ECL-localized stem cells, teleosts also have the classical muscle stem cell population termed the satellite cells, which are dispersed in the myotome. As in amniotes, satellite cells in teleosts are marked by pax3, pax7, and met and are required for muscle regeneration. Although the role of satellite cells in muscle regeneration in the larval setting is well-characterized (reviewed in (58)), their role in adult muscle is not well-studied. It is known that in adult zebrafish, satellite cells proliferate and form new muscle following injury, with pax3/pax7 double mutants displaying an impairment in this process (59). However, how this process is altered during aging, and the role of muscle stem cells in regenerating aged muscle tissue remain to be investigated. A recent study in African turquoise killifish has revealed a significant decline in muscle stem cell number in extremely old African turquoise killifish with normal stem cell number in the aged cohort (20). Interestingly, this decrease in stem cell number was associated with increased expression of myogenic genes. It is therefore possible that the reduced stem cell number could be due to increased fusion to muscle cells to enable the maintenance of muscle size, suggesting that alterations in cell number may be a consequence of the aging process, rather than a cause. The role of stem cells in the pathogenesis of sarcopenia has also been questioned in mammalian models, and further research in this area is warranted.
Telomere Attrition
Telomeres are repetitive DNA sequences (TTAGGGn) that protect the ends of chromosomes from fusion events (60). As linear chromosomes cannot be fully replicated by replicative DNA polymerases, the presence of telomerase, a specialized reverse transcriptase enzyme that is able to elongate chromosomes is necessary (61). However, telomerase has limited expression in most human somatic cells, including muscle cells, such that telomere attrition occurs with increasing age (62,63). When telomeres are shortened to a critical length, DNA damage response pathways are activated, leading to cellular senescence (reviewed in (64)).
Several studies have investigated the length of telomeres in teleosts and shown similarities to those seen in humans (Table 1). In muscle, telomere length varies between teleosts with zebrafish muscle telomeres being the largest at around 17 kb (29,30), and medaka and African turquoise killifish muscle telomeres ranging from 8.7 to 13.9 kb (33,35,36) and 6.2 to 6.6 kb (19), respectively. Despite the differences in telomere length, telomeres have been shown to shorten with age in each of the 3 species (19,30,35). Notably, unlike in humans, whereby telomere shortening has been shown to occur in multiple tissues regardless of proliferative capacity (65), in zebrafish, telomeres shorten only in the gut and muscle, and this has been shown to systemically drive the aging process (66). Consistent with this, muscle cells also undergo telomere shortening in African turquoise killifish, although this is strain-specific. That is, while the longer-lived African turquoise killifish strain (MZM-0403) displays telomere shortening in muscle with increasing age, the shorter-lived strain (GRZ) shows no significant telomere attrition, despite the high conservation in telomerase gene expression between the 2 strains (19). This suggests that, at least in the short-lived strain, telomere attrition does not contribute to the pathogenesis of sarcopenia. Notably, while the sarcopenic pathologies in the long-lived strain have been characterized (20), the same has not been done for the short-lived strain, and therefore it is not possible to draw conclusions on the role of telomere attrition in sarcopenia pathogenesis in teleosts. In any case, the differences in telomere biology may provide a valuable tool to tease out the role of telomere attrition in sarcopenia and aging.
Although the functional domains of the enzyme telomerase are highly conserved between humans and teleosts (67–69), the activity profile is significantly different. That is, in humans, telomerase is active in germline cells but not in somatic cells (70), whereas in teleosts, telomerase activity is detected in most tissues, with moderate levels in muscle (19,68,69,71). The relationship between telomerase expression in muscle cells and age has been investigated in both zebrafish and African turquoise killifish. In zebrafish (30) and the short-lived African turquoise killifish strain (19), telomerase expression and activity were not altered with age, whereas in the long-lived African turquoise killifish strain telomerase expression and activity increased with age (19). Although the functional consequences of this variation in telomerase expression remain to be determined, the impact of telomerase deficiency has been examined. Telomerase-deficient zebrafish (tert-/-) have shorter telomeres than their wild-type counterparts and display p53-dependent age-associated decay including a reduction in body mass, cachexia, and sarcopenia, which resembles the sarcopenic pathology seen in humans (29,72). Interestingly, rescue of telomerase expression in the gut of telomerase-deficient zebrafish was shown to not only rescue gut senescence and restore gut integrity but also to reduce the degeneration of various tissues including muscle, and increase the lifespan of the fish (71). This highlights the systemic nature of aging and sarcopenia, and the importance of studying tissues as a collection rather than as individual modalities.
Epigenetic Alterations
Epigenetics is the study of heritable and reversible alterations to DNA, without altering its sequence, with the 2 most common forms being DNA methylation and histone modification. Within the human genome, approximately 70%–80% of methylation occurs at the C5 position of cytosine (5mC) in CpG dinucleotides, catalyzed by DNA methyltransferases (DNMTs), whereas demethylation is mediated by the ten-eleven translocation methylcytosine dioxygenase (TET) family of enzymes. Phylogenetic analyses have revealed a high degree of conservation of both DNMT and TET genes in teleosts, although teleosts have multiple paralogues that emerged from genome duplication events. Importantly, studies have shown clear tissue- and age-specific expression and function of DNMT and TET genes (73,74). In the context of sarcopenia, muscle from aged African turquoise killifish displayed age-dependent decrease in DNMT1 and DNMT3, with no significant alterations in expression of the TET family of genes (74). Consistent with this reduction in methylation enzymes, analysis of global DNA methylation in skeletal muscle from African turquoise killifish showed a significant negative correlation of 5mC with age (74). Although the impact of this hypomethylation on gene expression and sarcopenia remains to be examined, studies in other fish models have identified differentially methylated positions associated with muscle atrophy and quality (75) and with muscle fiber type (76). It would therefore be interesting to determine if any of these known markers are hypomethylated in aged muscle as this may reveal novel mechanisms driving atrophy in sarcopenia.
The development of human epigenetic clocks and their strong association with chronological age in muscle, and aging-related phenotypes is exciting and holds great potential for identifying novel mechanisms regulating sarcopenia, aging, and longevity. Epigenetic clocks utilize machine learning algorithms to determine DNA methylation sites that correlate with age (77). Epigenetic clocks have been developed for 2 fish species—European seabass and zebrafish—with their main purpose being age determination without the requirement for lethal sampling (78,79). Notably, both clocks were developed using a small number of CpG sites—with the European seabass clock based upon 48 CpG sites from 4 genes in muscle samples (78), and the zebrafish clock utilizing 26 sites in caudal fin tissue (79). Although both epigenetic clocks are associated with chronological age, the functional impact of the DNA methylation changes identified and their potential role in sarcopenia have thus far not been examined. That is, are the observed DNA methylation changes a consequence of the aging process, or do they directly cause sarcopenia? Further research in this field is required.
In addition to DNA methylation, changes in histone modification (methylation and acetylation) also contribute to sarcopenia. Histone acetylation is regulated by the enzymes histone acetyltransferases and histone deacetylases (HDACs). Consistent with the broad expression patterns of HDAC genes in mammals, African turquoise killifish also show ubiquitous expression in embryonic stages (80). However, during aging, there is a significant downregulation of HDAC1 expression and an inverse upregulation of HDAC3 in aged skeletal muscle (80). Although the implications of this change on histone acetylation were not determined, Cencioni et al. (2019) have since characterized the epigenetic profile of skeletal muscle from aged African turquoise killifish tissue (81). Aged African turquoise killifish muscles displayed an increase in histone marks associated with heterochromatin (H3K27me3, H3K9me3, and H4K20me3), and a reduction in euchromatin histone marks (H3K9ac and H4K16ac), which is consistent with alterations in expression of the histone acetyltransferase enzymes. ChIP-seq analysis for H3K27me3 and H3K9ac in combination with RNA-seq was subsequently used to identify promoters that may be epigenetically regulated during aging. These studies revealed a downregulation of genes involved in cell cycle, differentiation, and DNA repair and an upregulation of inflammation and senescence genes (81). Although functional studies have not been performed to validate the involvement of these pathways in sarcopenia, the use of such epigenetic profiles can lead to the identification of novel mechanisms driving sarcopenia.
Mitochondrial Dysfunction
Mitochondria are the powerhouse of the cell and although they are primarily involved in synthesizing energy in the form of ATP through oxidative phosphorylation, they also have important roles in ion homeostasis, cellular metabolism, programmed cell death, and the production and consumption of reactive oxygen species (ROS). Aging and sarcopenia are characterized by an impairment in mitochondrial integrity, reduced mitochondrial biogenesis, and an overall reduction in mitochondrial function subsequently resulting in excessive ROS production, which induces further mitochondrial and cellular deterioration (82). Consistent with these hallmarks of sarcopenia, aged zebrafish display a reduction in mitochondrial number, an increase in mitochondrial rupture and lysis of the matrix, increased vacuolization, and a loss of cristae (12). Additionally, aged zebrafish displayed increased ROS production and striking alterations in mitochondrial dynamics, as evidenced by the reduction in mitochondrial biogenesis and fusion-related genes and upregulation in mitochondrial fission-related factors, which may collectively result in impaired mitochondrial function, although this has not been tested (12). Remarkably, exercise is sufficient to activate the AMPK/SIRT1/PGC-1α pathway, subsequently reversing many of the defects in mitochondrial dynamics, and also ameliorating age-related muscle atrophy (12). These results not only highlight the beneficial effects of exercise in sarcopenia but also demonstrate that the biological processes regulating mitochondrial dysfunction in aged zebrafish are highly conserved with those identified in humans.
In agreement with the zebrafish findings, aged African turquoise killifish also display a reduction in mitochondrial number and function, including reduced abundance of OXPHOS complexes and an accumulation of mitochondrial DNA damage in skeletal muscle (18,20). Notably, while reduced OXPHOS complexes are generally thought to be detrimental, in several organisms including African turquoise killifish, it has been shown to be beneficial by triggering ROS-mediated mitohormesis and extending lifespan (83–86). Consistent with this idea, the reduction in OXPHOS complexes in skeletal muscle of aged African turquoise killifish has been proposed to trigger ROS-mediated mitohormesis and induce the expression of SIRT1 and PGC-1α in extremely old animals culminating in their increased lifespan and overall reduction in mortality rates (20). The regulation of mitochondrial biology during aging is complex, and there may be a fine balance between mitochondrial dysfunction contributing to muscle wasting, and triggering stress-responsive pathways subsequently improving muscle health and lifespan. Given that teleosts accurately model many of the mitochondrial changes seen in aging humans, they may provide a useful system to investigate these processes to better understand the role of mitochondria in sarcopenia pathogenesis.
Metabolic Alterations
In addition to being indispensable for locomotion, skeletal muscle is an essential metabolic organ mediating systemic metabolism and influencing energy homeostasis (87). Recent studies have utilized teleost models to identify metabolic pathways associated with sarcopenia. Using a comparative approach, Maslov et al. (2019) (88) examined skeletal muscle metabolites in fish with differing rates of aging, which included muscle samples from the negligible senescent species pike and sterlet, biosamples of gradually senescent species (zander and perch), and muscle samples from rapidly senescent species (chum salmon and pink salmon). Untargeted metabolomics, which enables the unbiased identification of metabolites present in a sample, was performed on muscle from each of these 3 groups, following which the most overrepresented metabolites were characterized in each group (88). Using this strategy, it was reported that muscle from negligible senescent species is associated with high levels of amino acids and biogenic amines (88). Although the functional implications of these increased levels were not examined, elevated amounts of amino acids are hypothesized to improve muscle growth and energy metabolism, provide antioxidant protection, enhance protein synthesis, and inhibit proteolysis, which collectively prevent the presentation of sarcopenia. The rapidly aging salmonoid species on the other hand, were associated with an accumulation of lipid metabolites including medium- and long-chain acylcarnitines, intermediates of the citric acid cycle and intermediates of sugar metabolism (88). Given the lack of available literature on the role of lipids in regulating sarcopenia, the enrichment of lipid metabolites in the rapidly aging salmonoid species was thought to be a result of external factors independent of the aging process. Collectively, although this work reveals a potential role of protein and lipid metabolism in the regulation of sarcopenia, the differences in the biology of each of the species including factors such as age, environment, diet, and feeding regime present as significant confounding factors. Functional evidence is therefore required to determine if the metabolic pathways identified are an inconsequential read-out of the biology of the animal or are true regulators of sarcopenia.
More recently, untargeted metabolomics experiments were performed in the African turquoise killifish to characterize the metabolome in skeletal muscle from young, aged, and extremely old animals. Using a systems biology approach, Ruparelia et al. (2023) revealed that although most metabolites are unaffected during the aging process, of those that are significantly altered, there is an enrichment of protein and lipid metabolites (20). With regards to the alterations in protein metabolism, Ruparelia et al. (2023) report an increase in peptide species in the aged cohort, which are downregulated in the extremely old animals (20). Peptide species are associated with proteolysis, and their increased abundance in the aged cohort suggests increased protein breakdown. Consistent with this argument, aged muscle was found to express high levels of atrogin-1, an E3 ubiquitin ligase known to be upregulated in numerous muscle wasting conditions including sarcopenia, where it ubiquitinates substrate proteins and targets them for degradation, subsequently resulting in atrophy (20). In extremely old animals, on the other hand, reduced abundance of peptide species was associated with reduced expression of atrogin-1 and stabilization of muscle fiber size, supporting the hypothesis that protein metabolic changes identified are most likely a reflection of changes in proteolysis (20). Studies in zebrafish have also reported similar changes in protein metabolism, whereby aged zebrafish display increased expression of the atrogenes atrogin-1 and murf1, and a downregulation of genes of the IGF-1/PI3K/Akt/mTOR signaling pathway known to regulate protein synthesis, subsequently leading to muscle atrophy (12). Remarkably, an 8-week swimming exercise regime in zebrafish was sufficient to not only restore the expression of both protein synthesis and degradation genes back to levels seen in younger animals but also restore muscle mass (12), highlighting the importance of protein metabolism in sarcopenia.
In addition to protein metabolism, lipid metabolism has also been implicated in regulating muscle aging and longevity in African turquoise killifish. Ruparelia et al. (2023) revealed that during aging, increased lipolysis results in the depletion of triglycerides, an important energy reserve in the cell (20). Although the mechanisms for this reduction and the consequence of muscle wasting were not investigated, the authors suggest that the reduction in triglycerides during aging mimics a state of calorie restriction. This triggers increased ROS production, subsequently activating the stress-responsive pathway mitohormesis in extremely old animals, as evident by the increased expression of sirt1 and PGC1α (20). One proposed beneficial effect of this is the restoration of triglyceride levels to those seen in young animals, and alteration in lipid profile dynamics to enable more efficient utilization of energy (20). Consistent with this hypothesis, fat maintenance has been shown to predict longevity in wild-type mice strains (89), and increased consumption of specific types of dietary fats extends lifespan and improves mitochondrial dynamics in aged murine skeletal muscle (90), collectively highlighting the role of adiposity in sarcopenia and longevity. Although there is correlative evidence that the same occurs in teleosts, functional studies are needed to better understand this process and leverage this knowledge for developing improved nutritional and pharmacological strategies to manage sarcopenia.
Lifespan and Healthspan Extension Strategies, and Its Impact on Muscle Health
In recent years, several dietary, pharmacological, and genetic approaches have been used in teleosts to identify novel mechanisms regulating muscle health and lifespan, and strategies to improve and extend them respectively. These have been summarized in Tables 2 and 3 and are discussed below.
Table 2.
Outcomes From Dietary Interventions in Teleosts
| Intervention | Species | Concentration | Period of Treatment | Observed Alterations | Number of Fish | References |
|---|---|---|---|---|---|---|
| Dietary restriction | N. furzeri | DR fish were fed twice a day, every alternate day so that no uneaten food was left after 30 minutes | From 4 weeks | Increased maximum lifespan in long-lived and short-lived strains; however, increased baseline mortality in long-lived strain. DR ameliorated age-related impairment in memory and learning. DR reduced expression of neurodegeneration markers (brain) and lipofuscin (liver). | n = 10 for assays | (91) |
| Dietary restriction | N furzeri | 15 mg per day, in 3 feedings of 5 mg over 2 h | From 4 weeks | Increase in median lifespan for males only. | n = 21–25 per group | (92) |
| Dietary restriction | D. rerio | 90 mg of dry food every alternate day and artemia once per week. | 12-week period of DR in 9-month- and 20-month-old fish | Reduction in BMI. Old DR animals had increased protein expression of PSD95 (brain). Young DR animals had increased protein expression of GluR2/3 (brain). Overall these results show DR increases elements of synaptic integrity and excitatory neurotransmission. |
n = 12–13 per group | (93) |
| Dietary restriction | D rerio | 90 mg of dry food every alternate day and artemia once a week | 10-week period of DR in 8–8.5-month- and 26–32.5-month-old fish | Reduction in weight. Shortening of telomeres (brain). No change in telomere length (skeletal muscle, spleen, and tail). |
n = 27–34 per group | (94) |
| Dietary restriction | G aculeatus | 2% bodyweight of bloodworms daily | 300 days old until death | No change in median lifespan. Produced significantly fewer clutches. |
n = 32–44 | (95) |
| Calorie restriction | D rerio | HF-LD (overfed): 3 × 100 mL paramecia from 5–13 days post fertilization (dpf), and 3 × 10 drops of artemia from 14 dpf onwards LF-HD (CR): daily feeding of 3 × 100 mL paramecia from 5–13 dpf, and 3 × 10 drops of artemia from 14 dpf onwards |
2 weeks to 18 months | When compared to overfed fish: Reduced body length. Slower mineralization of scales. Reduced number and size of adipocytes. Reduced oocyte growth rate. When compared to control feeding: No change in BMI. Elevated cholesterol. No change in triglycerides. No change in ovarian weight. |
n = 2–3 per experiment | (96) |
| Calorie restriction | D rerio | 100 mg of dry food every alternate day and artemia once a week | 12-week period of CR in 5-month- and 25-month-old fish | Reduced BMI. Reduction in RNA expression of stem cell marker SOX2 in calorie restricted fish when compared to overfed fish (brain). | n = 6 for each age group | (97) |
| Intermittent food restriction | N guentheri | 2 consecutive days per week fed 10 mg dried worms/g fish weight/day while the other 5 days there was no restriction (40 mg dried worms/g fish weight/day | 9-month-old male fish | Significant reduction in body weight in 38–50-week-old fish. Increase in mean and maximum lifespan. Reduced lipofuscin staining (gills). Reduced protein oxidation and lipid peroxidation (muscle). Increased antioxidant enzyme activity (muscle). |
n = 50 for lifespan observations, n = 3 for assays | (98) |
| Intermittent fasting | G aculeatus | Starvation for 5 consecutive days then 15% bodyweight of bloodworms for 9 days | 300 days old until death | Reduction in median and maximum lifespan. Same investment in reproduction as control fish. |
n = 32–44 | (95) |
| Intermittent fasting | D rerio | 90 mg of dry food every alternate day and artemia once a week | 8-week period of IF in 6–10-month- and 26–31-month-old fish | Reduction in body weight, BMI and glucose levels (whole body). Suppressed mTOR activity (brain). Increase in neurogenic marker DCAMKL1 (brain). |
n = 4–16 | (99) |
Table 3.
Outcomes From Pharmacological Interventions in Teleosts
| Intervention | Species | Concentration | Period of Treatment | Observed Alterations | Number of Fish | References |
|---|---|---|---|---|---|---|
| Resveratrol | N furzeri | 600 µg/g/food 120 µg/g/food 24 µg/g/food |
From 4 weeks old (sexual maturity) until death | Dose-dependent lifespan extension. Retarded onset of age-dependent cognitive and locomotive deficit. Prevented neurofibrillary degeneration (brain). | n = 20, n = 60 and n = 30, respectively | (100) |
| Resveratrol | N furzeri | 96 µg/g/food | 4-week treatment from 33 to 37 weeks of age | A shift in metabolic profile with resveratrol-treated males displaying significant increase in triglyceride levels and changes in lipid localization (muscle). |
n = 6 | (20) |
| Resveratrol | N guentheri | 12 µg resveratrol/fish/day | From 12 weeks old (sexual maturity) until death | Extension of median and maximum lifespan. Prevention of loss of radial glia (brain). |
n = 72 | (101) |
| Resveratrol | N guentheri | 200 µg/g/food | From 16 weeks old until death | Extension of mean and maximum lifespan. Reduction in ROS (skin, bone, and muscle). Increase in antioxidant enzyme activity (liver) and reduced protein oxidation (skin, bone, and muscle). Reduction in lipofuscin accumulation in gill epithelia. | n = 76 | (102) |
| Resveratrol | N guentheri | 200 µg/g/food | From 16 weeks old until death | Extension of mean and maximum lifespan. No alteration in body size Enhanced cognitive performance and locomotor activity. Reduction in neurodegeneration (brain), lipofuscin staining (liver) and SA-β-Gal staining (skin). | n = 38 | (103) |
| Resveratrol | N guentheri | 200 µg/g/food | From 16 weeks old until sacrifice at either 6, 9, or 12 months of age | Reduction in the number of SA-β-gal positive cells (thymus and kidney). Reduced protein expression of SIRT1 and SIRT3, and increased protein expression of NF-κB (thymus and kidney). | n = 5 | (104) |
| Resveratrol | N guentheri | 200 µg/g/food | From 16 weeks old until sacrifice at either 6, 9, or 12 months of age | Reduced SA-β-Gal staining (gut). Amelioration of the SASP phenotype (gut). Resveratrol treatment prevented as much reduction in intestinal stem cells and epithelial cells over lifespan. |
n = 3 for staining, ELISA, but n = 20–30 for qPCR | (105) |
| Resveratrol | N guentheri | 600 µg/g/food | From 10–15 weeks old for 20 weeks | Increased embryo production and increased NAMPT protein expression suggesting a positive effect on female fertility. | n = 8 per group | (106) |
| Resveratrol | N guentheri | 200 µg/g/food | From 16 weeks old until sacrifice at either 6, 9, or 12 months of age | Reduced SA-β-Gal staining (ovary). Reduced lipofuscin staining (ovary). Improved cell proliferation and ovarian reserve (ovary). Reduced inflammation and ER stress (ovary). |
n = 3 | (107) |
| Metformin | N guentheri | 2 mg/g/food | From 16 weeks old (sexual maturity) | Increase in the mean survival in male fish. Significant reduction in body weight and body length. Significant reduction in cholesterol and triglyceride (serum). Reduction in SA-β-gal and lipofuscin staining (skin). Enhanced locomotor and cognitive ability. Reduced neurodegeneration and inflammation (brain). |
n = 48 | (108) |
| Metformin | N guentheri | 2 mg/g/food | From 16 weeks | Extension in mean and maximum lifespan. Reduction in SA-β-gal staining (gut). Reduced inflammation (gut). | n = 6 | (109) |
| Rapamycin | D rerio | 0.1 nM final concentration in the tank | 8-week period of treatment in 6–10-month- and 26–31-month-old fish | No difference in BMI, body weight or glucose levels (whole body). Inhibitory effect on mTOR pathway in young animals (brain). Decreased proliferation (PCNA), astrocyte activation (GFAP), postsynaptic transmission (GEP) and autophagy (LC3-II/LC3-I ratio) seen in young animals only (brain). |
n = 4–16 | (99) |
| Melatonin | D. rerio | 100 nM final concentration in the tank | 30-minute treatment in 1- and 4-year-old fish | Counteracted the sleep alterations, reduced intrinsic circadian rhythm of activity, and reduction in cognitive performance in aged zebrafish. | n = 8–10 | (110) |
| Melatonin | N. korthausae | 5mg melatonin/kg body weight/day | 72-week-old male fish | Improved the regularity, fragmentation and amplitude of the rest-activity rhythm, and sleep efficiency. | n = 10 | (111) |
| Diosgenin | N. guentheri | 13.8 μg/g body weight/day | 9-month-old male fish | Extension of mean and maximum lifespan in male fish. Reduced accumulation of lipofuscin (gills). Reduced accumulation of SA-β-gal (caudal fin). Lowered protein oxidation, lipid peroxidation and ROS accumulation (muscle). |
n = 50 for lifespan observations, n = 3 for assays | (112) |
Dietary Interventions
The most well-studied dietary intervention is dietary/calorie restriction (DR), defined as an overall reduction in total caloric intake without targeting a specific dietary component, which has been shown to increase maximum lifespan in multiple teleost models (Table 2). In the African turquoise killifish, however, strain differences have been reported. Although DR increases the maximum lifespan in both the shorter-lived, inbred strain and the longer-lived, outbred strain, in the latter, increased baseline mortality was also observed (91). These strain-specific responses to DR may be attributed to genetic variation, in that the DR regime is detrimental to some individuals and beneficial to others (91). Although such variants have thus far not been identified, recent work has implicated differences in gene function between individuals in explaining these distinct responses to DR. African turquoise killifish males have previously been shown to display increased median lifespan following DR, with the lifespan of females being unchanged (92), suggesting that DR acts in a sexually dimorphic manner. In line with this, a reduction in the AMP biosynthesis gene APRT has been shown to increase nutrient sensitivity, specifically in males (113). These results implicate AMP biosynthesis in regulating longevity in males and highlight the potential use of teleosts to uncover mechanisms by which genes or genetic variants regulate interspecies, intraspecies, and/or sex differences in longevity.
In addition to genetic variation, differences in responses to DR may also be explained by the idea that DR may act as a stressor, and while most individuals die as a result of stress, some trigger stress-defence mechanisms, which increases resistance to age-induced damage subsequently selecting for their survival. Indeed, a similar mechanism has recently been proposed in fish that naturally live longer. Although based on descriptive data, it has recently been suggested that the depletion of triglycerides, the main energy reserves in the cell mimics a state of calorie restriction resulting in increased production of ROS. In some animals, this triggers mitohormesis, which results in the maintenance of nutrient homeostasis, subsequently leading to the deceleration of mortality rates and the manifestation of the “extremely old, late-life” phase. Further studies that functionally validate this mechanism and determine why certain animals selectively respond to stress, and subsequently live longer are needed.
Notably, the timepoint at which dietary interventions are initiated is particularly important in determining its impact on health and longevity. Using the short-lived African turquoise killifish, it was recently shown that fasting results in reduced expression of the AMPK γ-subunit Prkag1 and a suppression of energy metabolism, protein synthesis, and cellular proliferation in several tissues including muscle, which is reversed following refeeding (114). In young animals, the fasting-refeeding cycles result in oscillatory expression of Prkag1, and potentially other targets, leading to improved energy metabolism, metabolic health, and a subsequent extension of lifespan (114). Aged animals, however, despite being fed normal diets, show a transcriptional profile that is similar to fasted animals, including a reduction in Prkag1 expression (114). This constitutive state of fasting in aged animals prevents the restoration of Prkag1 levels, and this has been suggested to contribute to sarcopenic pathologies and aging. The introduction of fasting and other dietary interventions in aged animals may therefore have no impact on improving muscle health or longevity and may in fact have deleterious effects. Teleosts provide a model to not only explore the mechanisms by which dietary interventions function but also to identify the optimal timepoints in which maximum therapeutic benefit can be achieved.
Pharmacological Interventions
The 2 most well-studied lifespan/healthspan modulating pharmacotherapies that have been examined in the context of muscle in teleosts are resveratrol and metformin. Additional compounds such as rapamycin, melatonin, and diosgenin have also been shown to modify lifespan in teleosts but given their impact on muscle has not been explicitly explored, we have limited their discussion to Table 3. Metformin treatment of sexually mature killifish N guentheri has been shown to not only prolong lifespan but also reduce the presentation of typical aging pathologies including reduced lipofuscin in the liver, lower SA-β-gal activity in the skin, inhibition of inflammatory response, and improved motor, learning, and memory skills—highlighting a conserved mechanism of action of metformin in teleosts lifespan (108). In line with this, resveratrol treatment of multiple additional killifish (N furzeri (100) and N guentheri (101–103)) has also been shown to extend both mean and maximum lifespan and improve locomotor capacity, evident by an increase in average time active and average velocity of swimming (100,101). Although this could be attributed to the reduced neurodegeneration following resveratrol treatment, it could also be due to improved muscle innervation and pathology, although this was not examined. More recently, a 4-week resveratrol treatment regime on aged African turquoise killifish was shown to not only increase sirt1 levels but also increase skeletal muscle triglyceride levels and improve lipid droplet dynamics in muscle fibers, which is hypothesized to contribute to the extension of lifespan (20). Overall, these results demonstrate that therapeutic interventions can extend lifespan in teleosts, although further studies that examine the impact on muscle health are required.
Genetic Approaches to Study Sarcopenia
The availability of annotated reference genomes for zebrafish (115,116), medaka (117), and African turquoise killifish (118,119), along with development of genome editing tools, has enabled the generation of genetic models to study sarcopenia. Many of these have been reviewed elsewhere (45,46) and as such, here we limit our discussion to mutants that displayed muscle phenotypes. As part of a forward genetic screen in zebrafish, which used SA-β-Gal accumulation in early life stage as an outcome measure for premature senescence, several key regulators of aging sarcopenia were identified (120). This includes identification and characterization of the terf1a mutant, which showed enlarged telomere speckles and abnormal nuclear shapes, along with a reduction in mean lifespan as expected due to the role of this gene in telomere protection (120), and the nrs mutant line had increased SA-β-Gal staining in the skin and trunk and increased lipofuscin in the skeletal muscle along with a reduction in lifespan. Additional mutants, which showed reduced SA-β-Gal staining and increased lifespan, were subsequently described using similar genetic screens (121). Multiple additional screens have been performed (122) and they collectively demonstrate the capacity of forward genetic screens in fish models to identify novel genes and mechanisms regulating longevity, aging, and sarcopenia.
Although many mutations in fish have been characterized, there are relatively few that show premature signs of aging outside the context of known diseases. One mutant that has attracted attention in the field of sarcopenia and aging is the αklotho mutant. αKlotho plays an essential role in discharging excess phosphate from blood to prevent unwanted ectopic calcification and mice deficient in it show signs of aging at about 4 weeks of age and die prematurely at 8–9 weeks of age (123). In line with the mouse mutant phenotypes, zebrafish deficient in αklotho display premature aging from about 5 months of age including emaciation, protruding eyes, a reduction in locomotor activity, and premature death (124,125). At a cellular level, widespread calcification of blood vessels in skeletal muscle, as well as degeneration and fibrosis of skeletal muscle, is evident in the αklotho zebrafish mutant (125). Interestingly, in zebrafish (126) and the African turquoise killifish (127), αklotho has been shown to be primarily expressed in the liver and kidney, with low/no expression evident in other tissues such as skeletal muscle. Despite this, the loss of αklotho results in striking skeletal muscle phenotypes. These results highlight a role of nonautonomous mechanisms in driving sarcopenia and reiterate the need to consider these in the quest for therapeutics.
Transgenesis techniques have also been employed in teleosts and have been especially invaluable in the development of tools to enable in vivo imaging and lineage tracing. Examples that are particularly relevant for sarcopenia research in teleosts include: Musclebow and Killibow systems for lineage tracing (128,129), a transparent African turquoise killifish line klara, which enables observation of processes such as regeneration (130), and FUCCI lines for investigating cell cycle dynamics (131). The application of these tools to sarcopenia and aging studies will provide further understanding of the molecular mechanisms underlying these processes.
Finally, it would be remiss not to mention the advances in multiomic technologies including single-cell omics and spatial transcriptomics. The recent release of a comprehensive RNA-sequencing data set for muscle (among other tissues) of the shorter-lived strain of the African turquoise killifish at varying life stages (132) can be examined to understand tissue-specific alterations linked to the aging process. Single-cell resolution transcriptomics atlases in zebrafish are also available, although they currently do not include aged cohorts (133,134). Finally, while spatial transcriptomic approaches offer great promise to uncover cell types and pathways regulating sarcopenia and aging and have been used to examine muscle biology in teleosts, the resolution is limited (135). Therefore, improved spatial transcriptomics resources along with availability of more muscle-targeted scRNA-seq (and other omics-based) data sets in aged teleosts will allow a more comprehensive understanding of alterations in muscle tissue and provide insight into the molecular mechanisms of sarcopenia.
Concluding Remarks
In this review, we have highlighted the sarcopenic pathologies observed in the commonly used teleost species and discussed how they contribute to our understanding of the mechanistic basis of sarcopenia. Although teleosts recapitulate many of the hallmarks of sarcopenia, making them invaluable for sarcopenia research, it is important to acknowledge some of their limitations. There are obvious biological and physiological differences between fish and mammalian models that need to be considered. For example, teleosts are ectothermic, which means that their body temperature and therefore biological processes are regulated by external stimuli. Alterations in husbandry protocols including diet (136), temperature (21,137), reproductivity (138), and exposure to stressors (139) can significantly affect their development, growth, and aging trajectories, and as such it is imperative to record and report husbandry protocols to promote reproducibility and standardization efforts of the field. Toward this end, several husbandry protocols have recently been published, at least for the African turquoise killifish, including use of an automated feeding system (92,140). Further to this, there is substantial evidence suggesting that there are differences in the biology of captive and wild strains. For example, while male and female African turquoise killifish bred in captivity have comparable lifespans, male counterparts in the wild have been shown to die sooner (141). Consistent with this idea, at the molecular level, wild and captive Nile tilapia strains have been shown to display significant differences in global DNA methylation levels and expression of immune, metabolic, and muscle development genes (142). Given these differences, cross-species comparative approaches will be required to confirm the translatability of findings to human aging. Despite these limitations, teleosts are increasingly used for sarcopenia and aging research, and given their unique advantages, they have the potential to drive remarkable discoveries in the sarcopenia and aging fields.
Acknowledgments
All figures were created using biorender.com.
Contributor Information
Tuyen K Quach, Centre for Muscle Research, Department of Anatomy and Physiology, School of Biomedical Sciences, Faculty of Medicine, Dentistry, and Health Sciences, University of Melbourne, Melbourne, Victoria, Australia.
Megan F Taylor, Australian Regenerative Medicine Institute, Monash University, Clayton, Victoria, Australia.
Peter D Currie, Australian Regenerative Medicine Institute, Monash University, Clayton, Victoria, Australia; EMBL Australia, Victorian Node, Monash University, Clayton, Victoria, Australia.
Nir Eynon, Australian Regenerative Medicine Institute, Monash University, Clayton, Victoria, Australia.
Avnika A Ruparelia, Centre for Muscle Research, Department of Anatomy and Physiology, School of Biomedical Sciences, Faculty of Medicine, Dentistry, and Health Sciences, University of Melbourne, Melbourne, Victoria, Australia; Australian Regenerative Medicine Institute, Monash University, Clayton, Victoria, Australia (Biological Sciences Section).
Funding
This work was supported by a Medical Research Future Fund (MRFF) grant to P.D.C and A.A.R.
Conflict of Interest
None.
References
- 1. Frontera WR, Ochala J. Skeletal muscle: a brief review of structure and function. Calcif Tissue Int. 2015;96(3):183–195. https://doi.org/ 10.1007/s00223-014-9915-y [DOI] [PubMed] [Google Scholar]
- 2. Yuan S, Larsson SC. Epidemiology of sarcopenia: prevalence, risk factors, and consequences. Metabolism. 2023;144(155533):155560. https://doi.org/ 10.1016/j.metabol.2023.155560 [DOI] [PubMed] [Google Scholar]
- 3. Goodpaster BH, Park SW, Harris TB, et al. The loss of skeletal muscle strength, mass, and quality in older adults: the Health, Aging and Body Composition Study. J Gerontol A Biol Sci Med Sci. 2006;61(10):1059–1064. https://doi.org/ 10.1093/gerona/61.10.1059 [DOI] [PubMed] [Google Scholar]
- 4. Hardee JP, Lynch GS. Current pharmacotherapies for sarcopenia. Expert Opin Pharmacother. 2019;20(13):1645–1657. https://doi.org/ 10.1080/14656566.2019.1622093 [DOI] [PubMed] [Google Scholar]
- 5. Lynch GS. Identifying the challenges for successful pharmacotherapeutic management of sarcopenia. Expert Opin Pharmacother. 2022;23(11):1233–1237. https://doi.org/ 10.1080/14656566.2022.2076593 [DOI] [PubMed] [Google Scholar]
- 6. Murthy M, Ram JL. Invertebrates as model organisms for research on aging biology. Invertebr Reprod Dev. 2015;59(suppl 1):1–4. https://doi.org/ 10.1080/07924259.2014.970002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Christian CJ, Benian GM. Animal models of sarcopenia. Aging Cell. 2020;19(10):e13223. https://doi.org/ 10.1111/acel.13223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Demetrius L. Of mice and men. EMBO Rep. 2005;6 Spec No(S1):S39–S44. https://doi.org/ 10.1038/sj.embor.7400422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gilbert MJH, Zerulla TC, Tierney KB. Zebrafish (Danio rerio) as a model for the study of aging and exercise: physical ability and trainability decrease with age. Exp Gerontol. 2014;50:106–113. https://doi.org/ 10.1016/j.exger.2013.11.013 [DOI] [PubMed] [Google Scholar]
- 10. Kishi S, Uchiyama J, Baughman AM, Goto T, Lin MC, Tsai SB. The zebrafish as a vertebrate model of functional aging and very gradual senescence. Exp Gerontol. 2003;38(7):777–786. https://doi.org/ 10.1016/s0531-5565(03)00108-6 [DOI] [PubMed] [Google Scholar]
- 11. Rutkove SB, Callegari S, Concepcion H, et al. Electrical impedance myography detects age-related skeletal muscle atrophy in adult zebrafish. Sci Rep. 2023;13:7191. https://doi.org/ 10.1038/s41598-023-34119-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun C-C, Yang D, Chen Z-L, et al. Exercise intervention mitigates zebrafish age-related sarcopenia via alleviating mitochondrial dysfunction. FEBS J. 2023;290(6):1519–1530. https://doi.org/ 10.1111/febs.16637 [DOI] [PubMed] [Google Scholar]
- 13. Gerhard GS, Kauffman EJ, Wang X, et al. Life spans and senescent phenotypes in two strains of Zebrafish (Danio rerio). Exp Gerontol. 2002;37(8-9):1055–1068. https://doi.org/ 10.1016/s0531-5565(02)00088-8 [DOI] [PubMed] [Google Scholar]
- 14. Valdesalici S, Cellerino A. Extremely short lifespan in the annual fish Nothobranchius furzeri. Proc Biol Sci. 2003;270(Suppl 2):S189–S191. https://doi.org/ 10.1098/rsbl.2003.0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu C-K, Brunet A. The African turquoise killifish: a research organism to study vertebrate aging and diapause. Aging Cell. 2018;17(3):e12757. https://doi.org/ 10.1111/acel.12757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Di Cicco E, Tozzini ET, Rossi G, Cellerino A. The short-lived annual fish Nothobranchius furzeri shows a typical teleost aging process reinforced by high incidence of age-dependent neoplasias. Exp Gerontol. 2011;46(4):249–256. https://doi.org/ 10.1016/j.exger.2010.10.011 [DOI] [PubMed] [Google Scholar]
- 17. Wendler S, Hartmann N, Hoppe B, Englert C. Age-dependent decline in fin regenerative capacity in the short-lived fish Nothobranchius furzeri. Aging Cell. 2015;14(5):857–866. https://doi.org/ 10.1111/acel.12367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hartmann N, Reichwald K, Wittig I, et al. Mitochondrial DNA copy number and function decrease with age in the short-lived fish Nothobranchius furzeri. Aging Cell. 2011;10(5):824–831. https://doi.org/ 10.1111/j.1474-9726.2011.00723.x [DOI] [PubMed] [Google Scholar]
- 19. Hartmann N, Reichwald K, Lechel A, et al. Telomeres shorten while Tert expression increases during ageing of the short-lived fish Nothobranchius furzeri. Mech Ageing Dev. 2009;130(5):290–296. https://doi.org/ 10.1016/j.mad.2009.01.003 [DOI] [PubMed] [Google Scholar]
- 20. Ruparelia AA, Salavaty A, Barlow CK, et al. The African killifish: a short-lived vertebrate model to study the biology of sarcopenia and longevity. Aging Cell. 2023;23:e13862. https://doi.org/ 10.1111/acel.13862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Valenzano DR, Terzibasi E, Cattaneo A, Domenici L, Cellerino A. Temperature affects longevity and age-related locomotor and cognitive decay in the short-lived fish Nothobranchius furzeri. Aging Cell. 2006;5(3):275–278. https://doi.org/ 10.1111/j.1474-9726.2006.00212.x [DOI] [PubMed] [Google Scholar]
- 22. Ding L, Kuhne WW, Hinton DE, Song J, Dynan WS. Quantifiable biomarkers of normal aging in the Japanese medaka fish (Oryzias latipes). PLoS One. 2010;5(10):e13287. https://doi.org/ 10.1371/journal.pone.0013287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hori H. A glance at the past of medaka fish biology. In: Naruse K, Tanaka M, Takeda H, eds. Medaka. Springer; 2011:1–16. https://doi.org/ 10.1007/978-4-431-92691-7_1 [DOI] [Google Scholar]
- 24. de Lange T, Shiue L, Myers RM, et al. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10(2):518–527. https://doi.org/ 10.1128/mcb.10.2.518-527.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moyzis RK, Buckingham JM, Cram LS, et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988;85(18):6622–6626. https://doi.org/ 10.1073/pnas.85.18.6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kipling D, Cooke HJ. Hypervariable ultra-long telomeres in mice. Nature. 1990;347(6921):400–402. https://doi.org/ 10.1038/347400a0 [DOI] [PubMed] [Google Scholar]
- 27. Starling JA, Maule J, Hastie ND, Allshire RC. Extensive telomere repeat arrays in mouse are hypervariable. Nucleic Acids Res. 1990;18(23):6881–6888. https://doi.org/ 10.1093/nar/18.23.6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mark J, Zijlmans JM, Martens UM, et al. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc Natl Acad Sci U S A. 1997;94(14):7423–7428. https://doi.org/ 10.1073/pnas.94.14.7423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henriques CM, Carneiro MC, Tenente IM, Jacinto A, Ferreira MG. Telomerase is required for zebrafish lifespan. PLoS Genet. 2013;9(1):e1003214. https://doi.org/ 10.1371/journal.pgen.1003214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anchelin M, Murcia L, Alcaraz-Pérez F, García-Navarro EM, Cayuela ML. Behaviour of telomere and telomerase during aging and regeneration in zebrafish. PLoS One. 2011;6(2):e16955. https://doi.org/ 10.1371/journal.pone.0016955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hsu C-Y, Chiu Y-C, Hsu W-L, Chan Y-P. Age-related markers assayed at different developmental stages of the annual fish Nothobranchius rachovii. J Gerontol A Biol Sci Med Sci. 2008;63(12):1267–1276. https://doi.org/ 10.1093/gerona/63.12.1267 [DOI] [PubMed] [Google Scholar]
- 32. Reichard M, Giannetti K, Ferreira T, et al. Lifespan and telomere length variation across populations of wild-derived African killifish. Mol Ecol. 2022;31(23):5979–5992. https://doi.org/ 10.1111/mec.16287 [DOI] [PubMed] [Google Scholar]
- 33. Au DWT, Mok HOL, Elmore LW, Holt SE. Japanese medaka: a new vertebrate model for studying telomere and telomerase biology. Comp Biochem Physiol C Toxicol Pharmacol. 2009;149(2):161–167. https://doi.org/ 10.1016/j.cbpc.2008.08.005 [DOI] [PubMed] [Google Scholar]
- 34. Yip BW, Mok HO, Peterson DR, et al. Sex-dependent telomere shortening, telomerase activity and oxidative damage in marine medaka Oryzias melastigma during aging. Mar Pollut Bull. 2017;124(2):701–709. https://doi.org/ 10.1016/j.marpolbul.2017.01.021 [DOI] [PubMed] [Google Scholar]
- 35. Hatakeyama H, Nakamura K-I, Izumiyama-Shimomura N, et al. The teleost Oryzias latipes shows telomere shortening with age despite considerable telomerase activity throughout life. Mech Ageing Dev. 2008;129(9):550–557. https://doi.org/ 10.1016/j.mad.2008.05.006 [DOI] [PubMed] [Google Scholar]
- 36. Hatakeyama H, Yamazaki H, Nakamura K-I, et al. Telomere attrition and restoration in the normal teleost Oryzias latipes are linked to growth rate and telomerase activity at each life stage. Aging (Milano). 2016;8(1):62–76. https://doi.org/ 10.18632/aging.100873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Panasiak L, Szubert K, Polonis M, Ocalewicz K. Telomere length variation does not correspond with the growth disturbances in the rainbow trout (Oncorhynchus mykiss). J Appl Genet. 2022;63(1):133–139. https://doi.org/ 10.1007/s13353-021-00669-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Panasiak L, Dobosz S, Ocalewicz K. Telomere dynamics in the diploid and triploid rainbow trout (Oncorhynchus mykiss) assessed by Q-FISH analysis. Genes. 2020;11(7):786. https://doi.org/ 10.3390/genes11070786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Downs KP, Shen Y, Pasquali A, et al. Characterization of telomeres and telomerase expression in Xiphophorus. Comp Biochem Physiol C Toxicol Pharmacol. 2012;155(1):89–94. https://doi.org/ 10.1016/j.cbpc.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gao J, Munch SB. Does reproductive investment decrease telomere length in Menidia menidia? PLoS One. 2015;10(5):e0125674. https://doi.org/ 10.1371/journal.pone.0125674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carney Almroth B, Sköld M, Sköld HN. Gender differences in health and aging of Atlantic cod subject to size selective fishery. Biol Open. 2012;1(9):922–928. https://doi.org/ 10.1242/bio.20121446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rollings N, Miller E, Olsson M. Telomeric attrition with age and temperature in Eastern mosquitofish (Gambusia holbrooki). Naturwissenschaften. 2014;101(3):241–244. https://doi.org/ 10.1007/s00114-014-1142-x [DOI] [PubMed] [Google Scholar]
- 43. López de Abechuco E, Soto M, Pardo MA, Haussmann M, Diez G. Suitability of real-time quantitative PCR to estimate the relative telomere length in European Hake (Merluccius merluccius Linnaeus, 1758). Rev Investig Mar AZTI-Tecnalia. 2013;20:29–36. [Google Scholar]
- 44. Izzo C, Bertozzi T, Gillanders BM, Donnellan SC. Variation in telomere length of the common carp, Cyprinus carpio (Cyprinidae), in relation to body length. Copeia. 2014;2014(1):87–94. https://doi.org/ 10.1643/ci-11-162 [DOI] [Google Scholar]
- 45. Daya A, Donaka R, Karasik D. Zebrafish models of sarcopenia. Dis Model Mech. 2020;13(3):dmm042689. https://doi.org/ 10.1242/dmm.042689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ichii S, Matsuoka I, Okazaki F, Shimada Y. Zebrafish models for skeletal muscle senescence: lessons from cell cultures and rodent models. Molecules. 2022;27(23):8625. https://doi.org/ 10.3390/molecules27238625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keenan SR, Currie PD. The developmental phases of zebrafish myogenesis. J Dev Biol. 2019;7(2):12. https://doi.org/ 10.3390/jdb7020012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rossi G, Messina G. Comparative myogenesis in teleosts and mammals. Cell Mol Life Sci. 2014;71(16):3081–3099. https://doi.org/ 10.1007/s00018-014-1604-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Felsenfeld AL, Curry M, Kimmel CB. The fub-1 mutation blocks initial myofibril formation in zebrafish muscle pioneer cells. Dev Biol. 1991;148(1):23–30. https://doi.org/ 10.1016/0012-1606(91)90314-s [DOI] [PubMed] [Google Scholar]
- 50. Cromie Lear MJ, Millard M, Gleiss AC, et al. Biomechanical analysis of the slow-twitch (Red) muscle force transmission pathways in tunas. Physiol Biochem Zool. 2020;93(3):185–198. https://doi.org/ 10.1086/708247 [DOI] [PubMed] [Google Scholar]
- 51. Deschenes MR. Effects of aging on muscle fibre type and size. Sports Med. 2004;34(12):809–824. https://doi.org/ 10.2165/00007256-200434120-00002 [DOI] [PubMed] [Google Scholar]
- 52. Larsson L, Sjödin B, Karlsson J. Histochemical and biochemical changes in human skeletal muscle with age in sedentary males, age 22–65 years. Acta Physiol Scand. 1978;103(1):31–39. https://doi.org/ 10.1111/j.1748-1716.1978.tb06187.x [DOI] [PubMed] [Google Scholar]
- 53. Lee W-S, Cheung W-H, Qin L, Tang N, Leung K-S. Age-associated decrease of type IIA/B human skeletal muscle fibers. Clin Orthop Relat Res. 2006;450:231–237. https://doi.org/ 10.1097/01.blo.0000218757.97063.21 [DOI] [PubMed] [Google Scholar]
- 54. Nguyen PD, Gurevich DB, Sonntag C, et al. Muscle stem cells undergo extensive clonal drift during tissue growth via Meox1-mediated induction of G2 cell-cycle arrest. Cell Stem Cell. 2017;21(1):107–119.e6. https://doi.org/ 10.1016/j.stem.2017.06.003 [DOI] [PubMed] [Google Scholar]
- 55. Devoto SH, Stoiber W, Hammond CL, et al. Generality of vertebrate developmental patterns: evidence for a dermomyotome in fish. Evol Dev. 2006;8(1):101–110. https://doi.org/ 10.1111/j.1525-142X.2006.05079.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hollway GE, Bryson-Richardson RJ, Berger S, Cole NJ, Hall TE, Currie PD. Whole-somite rotation generates muscle progenitor cell compartments in the developing zebrafish embryo. Dev Cell. 2007;12(2):207–219. https://doi.org/ 10.1016/j.devcel.2007.01.001 [DOI] [PubMed] [Google Scholar]
- 57. Tierney MT, Stec MJ, Rulands S, Simons BD, Sacco A. Muscle stem cells exhibit distinct clonal dynamics in response to tissue repair and homeostatic aging. Cell Stem Cell. 2018;22(1):119–127.e3. https://doi.org/ 10.1016/j.stem.2017.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Manneken JD, Dauer MVP, Currie PD. Dynamics of muscle growth and regeneration: lessons from the teleost. Exp Cell Res. 2022;411(2):112991. https://doi.org/ 10.1016/j.yexcr.2021.112991 [DOI] [PubMed] [Google Scholar]
- 59. Berberoglu MA, Gallagher TL, Morrow ZT, et al. Satellite-like cells contribute to pax7-dependent skeletal muscle repair in adult zebrafish. Dev Biol. 2017;424(2):162–180. https://doi.org/ 10.1016/j.ydbio.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shay JW, Wright WE. Telomeres and telomerase: three decades of progress. Nat Rev Genet. 2019;20(5):299–309. https://doi.org/ 10.1038/s41576-019-0099-1 [DOI] [PubMed] [Google Scholar]
- 61. Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59(3):521–529. https://doi.org/ 10.1016/0092-8674(89)90035-4 [DOI] [PubMed] [Google Scholar]
- 62. Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345(6274):458–460. https://doi.org/ 10.1038/345458a0 [DOI] [PubMed] [Google Scholar]
- 63. Huffman KE, Levene SD, Tesmer VM, Shay JW, Wright WE. Telomere shortening is proportional to the size of the G-rich telomeric 3ʹ-overhang. J Biol Chem. 2000;275(26):19719–19722. https://doi.org/ 10.1074/jbc.M002843200 [DOI] [PubMed] [Google Scholar]
- 64. Rossiello F, Jurk D, Passos JF, d’Adda di Fagagna F. Telomere dysfunction in ageing and age-related diseases. Nat Cell Biol. 2022;24(2):135–147. https://doi.org/ 10.1038/s41556-022-00842-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Daniali L, Benetos A, Susser E, et al. Telomeres shorten at equivalent rates in somatic tissues of adults. Nat Commun. 2013;4:1597. https://doi.org/ 10.1038/ncomms2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Carneiro MC, Henriques CM, Nabais J, Ferreira T, Carvalho T, Ferreira MG. Short telomeres in key tissues initiate local and systemic aging in zebrafish. PLoS Genet. 2016;12(1):e1005798. https://doi.org/ 10.1371/journal.pgen.1005798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Harel I, Benayoun BA, Machado B, et al. A platform for rapid exploration of aging and diseases in a naturally short-lived vertebrate. Cell. 2015;160(5):1013–1026. https://doi.org/ 10.1016/j.cell.2015.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lau BW-M, Wong AO-L, Tsao GS-W, So K-F, Yip HK-F. Molecular cloning and characterization of the zebrafish (Danio rerio) telomerase catalytic subunit (telomerase reverse transcriptase, TERT). J Mol Neurosci. 2008;34(1):63–75. https://doi.org/ 10.1007/s12031-007-0072-x [DOI] [PubMed] [Google Scholar]
- 69. Pfennig F, Kind B, Zieschang F, Busch M, Gutzeit HO. Tert expression and telomerase activity in gonads and somatic cells of the Japanese medaka (Oryzias latipes). Dev Growth Differ. 2008;50(3):131–141. https://doi.org/ 10.1111/j.1440-169X.2008.00986.x [DOI] [PubMed] [Google Scholar]
- 70. Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet. 1996;18(2):173–179. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 71. El Maï M, Bird M, Allouche A, et al. Gut-specific telomerase expression counteracts systemic aging in telomerase-deficient zebrafish. Nat Aging. 2023;3(5):567–584. https://doi.org/ 10.1038/s43587-023-00401-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Anchelin M, Alcaraz-Pérez F, Martínez CM, Bernabé-García M, Mulero V, Cayuela ML. Premature aging in telomerase-deficient zebrafish. Dis Model Mech. 2013;6(5):1101–1112. https://doi.org/ 10.1242/dmm.011635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rai K, Jafri IF, Chidester S, et al. Dnmt3 and G9a cooperate for tissue-specific development in zebrafish. J Biol Chem. 2010;285(6):4110–4121. https://doi.org/ 10.1074/jbc.M109.073676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zupkovitz G, Kabiljo J, Kothmayer M, et al. Analysis of methylation dynamics reveals a tissue-specific, age-dependent decline in 5-methylcytosine within the genome of the vertebrate aging model Nothobranchius furzeri. Front Mol Biosci. 2021;8(627143):627143. https://doi.org/ 10.3389/fmolb.2021.627143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Salem M, Al-Tobasei R, Ali A, Kenney B. Integrated analyses of DNA methylation and gene expression of rainbow trout muscle under variable ploidy and muscle atrophy conditions. Genes. 2022;13(7):1151. https://doi.org/ 10.3390/genes13071151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pan Y, Chen L, Cheng J, et al. Genome-wide DNA methylation profiles provide insight into epigenetic regulation of red and white muscle development in Chinese perch Siniperca chuatsi. Comp Biochem Physiol B Biochem Mol Biol. 2021;256:110647. https://doi.org/ 10.1016/j.cbpb.2021.110647 [DOI] [PubMed] [Google Scholar]
- 77. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):3156. https://doi.org/ 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Anastasiadi D, Piferrer F. A clockwork fish: age prediction using DNA methylation-based biomarkers in the European seabass. Mol Ecol Resour. 2020;20:387–397. https://doi.org/ 10.1111/1755-0998.13111 [DOI] [PubMed] [Google Scholar]
- 79. Mayne B, Korbie D, Kenchington L, Ezzy B, Berry O, Jarman S. A DNA methylation age predictor for zebrafish. Aging (Milano). 2020;12(24):24817–24835. https://doi.org/ 10.18632/aging.202400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zupkovitz G, Lagger S, Martin D, et al. Histone deacetylase 1 expression is inversely correlated with age in the short-lived fish Nothobranchius furzeri. Histochem Cell Biol. 2018;150(3):255–269. https://doi.org/ 10.1007/s00418-018-1687-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cencioni C, Heid J, Krepelova A, et al. Aging triggers H3K27 trimethylation hoarding in the chromatin of Nothobranchius furzeri skeletal muscle. Cells. 2019;8(10):1169. https://doi.org/ 10.3390/cells8101169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bellanti F, Buglio AL, Vendemiale G. Mitochondrial impairment in sarcopenia. Biology. 2021;10(1):31. https://doi.org/ 10.3390/biology10010031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Baumgart M, Priebe S, Groth M, et al. Longitudinal RNA-Seq analysis of vertebrate aging identifies mitochondrial complex I as a small-molecule-sensitive modifier of lifespan. Cell Syst. 2016;2(2):122–132. https://doi.org/ 10.1016/j.cels.2016.01.014 [DOI] [PubMed] [Google Scholar]
- 84. Copeland JM, Cho J, Lo T, et al. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19(19):1591–1598. https://doi.org/ 10.1016/j.cub.2009.08.016 [DOI] [PubMed] [Google Scholar]
- 85. Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144(1):79–91. https://doi.org/ 10.1016/j.cell.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lee SS, Lee RYN, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33(1):40–48. https://doi.org/ 10.1038/ng1056 [DOI] [PubMed] [Google Scholar]
- 87. Baskin KK, Winders BR, Olson EN. Muscle as a “Mediator” of systemic metabolism. Cell Metab. 2015;21(2):237–248. https://doi.org/ 10.1016/j.cmet.2014.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Maslov DL, Trifonova OP, Mikhailov AN, et al. Comparative analysis of skeletal muscle metabolites of fish with various rates of aging. Fishes. 2019;4(2):25. https://doi.org/ 10.3390/fishes4020025 [DOI] [Google Scholar]
- 89. Liao C-Y, Rikke BA, Johnson TE, Gelfond JAL, Diaz V, Nelson JF. Fat maintenance is a predictor of the murine lifespan response to dietary restriction. Aging Cell. 2011;10(4):629–639. https://doi.org/ 10.1111/j.1474-9726.2011.00702.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gutiérrez-Casado E, Khraiwesh H, López-Domínguez JA, et al. The impact of aging, calorie restriction and dietary fat on autophagy markers and mitochondrial ultrastructure and dynamics in mouse skeletal muscle. J Gerontol A Biol Sci Med Sci. 2019;74(6):760–769. https://doi.org/ 10.1093/gerona/gly161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Terzibasi E, Lefrançois C, Domenici P, Hartmann N, Graf M, Cellerino A. Effects of dietary restriction on mortality and age-related phenotypes in the short-lived fish Nothobranchius furzeri. Aging Cell. 2009;8(2):88–99. https://doi.org/ 10.1111/j.1474-9726.2009.00455.x [DOI] [PubMed] [Google Scholar]
- 92. McKay A, Costa EK, Chen J, et al. An automated feeding system for the African killifish reveals the impact of diet on lifespan and allows scalable assessment of associative learning. eLife. 2022;11:e69008. https://doi.org/ 10.7554/eLife.69008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Karoglu-Eravsar ET, Tuz-Sasik MU, Adams MM. Short-term dietary restriction maintains synaptic plasticity whereas short-term overfeeding alters cellular dynamics in the aged brain: evidence from the zebrafish model organism. Neurobiol Aging. 2021;106:169–182. https://doi.org/ 10.1016/j.neurobiolaging.2021.06.010 [DOI] [PubMed] [Google Scholar]
- 94. Arslan-Ergul A, Erbaba B, Karoglu ET, Halim DO, Adams MM. Short-term dietary restriction in old zebrafish changes cell senescence mechanisms. Neuroscience. 2016;334:64–75. https://doi.org/ 10.1016/j.neuroscience.2016.07.033 [DOI] [PubMed] [Google Scholar]
- 95. Inness CLW, Metcalfe NB. The impact of dietary restriction, intermittent feeding and compensatory growth on reproductive investment and lifespan in a short-lived fish. Proc Biol Sci. 2008;275(1644):1703–1708. https://doi.org/ 10.1098/rspb.2008.0357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Leibold S, Hammerschmidt M. Long-term Hyperphagia and caloric restriction caused by low- or high-density husbandry have differential effects on zebrafish postembryonic development, somatic growth, fat accumulation and reproduction. PLoS One. 2015;10(3):e0120776. https://doi.org/ 10.1371/journal.pone.0120776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Erbaba B, Macaroglu D, Ardic-Avci NI, Arslan-Ergul A, Adams MM. Caloric restriction reinforces the stem cell pool in the aged brain without affecting overall proliferation status. Gene. 2023;851:147026. https://doi.org/ 10.1016/j.gene.2022.147026 [DOI] [PubMed] [Google Scholar]
- 98. Wang X, Du X, Zhou Y, Wang S, Su F, Zhang S. Intermittent food restriction initiated late in life prolongs lifespan and retards the onset of age-related markers in the annual fish Nothobranchius guentheri. Biogerontology. 2017;18(3):383–396. https://doi.org/ 10.1007/s10522-017-9699-3 [DOI] [PubMed] [Google Scholar]
- 99. Celebi-Birand D, Ardic NI, Karoglu-Eravsar ET, Sengul GF, Kafaligonul H, Adams MM. Dietary and pharmacological interventions that inhibit mammalian target of rapamycin activity alter the brain expression levels of neurogenic and glial markers in an age-and treatment-dependent manner. Rejuvenation Res. 2020;23(6):485–497. https://doi.org/ 10.1089/rej.2019.2297 [DOI] [PubMed] [Google Scholar]
- 100. Valenzano DR, Terzibasi E, Genade T, Cattaneo A, Domenici L, Cellerino A. Resveratrol prolongs lifespan and retards the onset of age-related markers in a short-lived vertebrate. Curr Biol. 2006;16(3):296–300. https://doi.org/ 10.1016/j.cub.2005.12.038 [DOI] [PubMed] [Google Scholar]
- 101. Genade T, Lang DM. Resveratrol extends lifespan and preserves glia but not neurons of the Nothobranchius guentheri optic tectum. Exp Gerontol. 2013;48(2):202–212. https://doi.org/ 10.1016/j.exger.2012.11.013 [DOI] [PubMed] [Google Scholar]
- 102. Liu T, Qi H, Ma L, et al. Resveratrol attenuates oxidative stress and extends life span in the annual fish Nothobranchius guentheri. Rejuvenation Res. 2015;18(3):225–233. https://doi.org/ 10.1089/rej.2014.1618 [DOI] [PubMed] [Google Scholar]
- 103. Yu X, Li G. Effects of resveratrol on longevity, cognitive ability and aging-related histological markers in the annual fish Nothobranchius guentheri. Exp Gerontol. 2012;47(12):940–949. https://doi.org/ 10.1016/j.exger.2012.08.009 [DOI] [PubMed] [Google Scholar]
- 104. Hou Y, Li S, Zhu H, Qiao M, Sun X, Li G. Development of the thymus and kidney and effects of resveratrol on their aging in a short-lived fish. J Gerontol A Biol Sci Med Sci. 2023;78(9):1550–1557. https://doi.org/ 10.1093/gerona/glad089 [DOI] [PubMed] [Google Scholar]
- 105. Liu S, Zheng Z, Ji S, et al. Resveratrol reduces senescence-associated secretory phenotype by SIRT1/NF-κB pathway in gut of the annual fish Nothobranchius guentheri. Fish Shellfish Immunol. 2018;80:473–479. https://doi.org/ 10.1016/j.fsi.2018.06.027 [DOI] [PubMed] [Google Scholar]
- 106. Lee Y, Drake AC, Thomas NO, Ferguson LG, Chappell PE, Shay KP. Dietary resveratrol increases mid-life fecundity of female Nothobranchius guentheri. Comp Biochem Physiol C Toxicol Pharmacol. 2018;208:71–76. https://doi.org/ 10.1016/j.cbpc.2017.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhu H, Li X, Qiao M, Sun X, Li G. Resveratrol alleviates inflammation and ER stress through SIRT1/NRF2 to delay ovarian aging in a short-lived fish. J Gerontol A Biol Sci Med Sci. 2023;78(4):596–602. https://doi.org/ 10.1093/gerona/glad009 [DOI] [PubMed] [Google Scholar]
- 108. Wei J, Qi H, Liu K, Zhao C, Bian Y, Li G. Effects of metformin on life span, cognitive ability, and inflammatory response in a short-lived fish. J Gerontol A Biol Sci Med Sci. 2020;75(11):2042–2050. https://doi.org/ 10.1093/gerona/glaa109 [DOI] [PubMed] [Google Scholar]
- 109. Li S, Hou Y, Liu K, et al. Metformin Protects against inflammation, oxidative stress to delay poly I:C-induced aging-like phenomena in the gut of an annual fish. J Gerontol A Biol Sci Med Sci. 2022;77(2):276–282. https://doi.org/ 10.1093/gerona/glab298 [DOI] [PubMed] [Google Scholar]
- 110. Zhdanova IV, Yu L, Lopez-Patino M, Shang E, Kishi S, Guelin E. Aging of the circadian system in zebrafish and the effects of melatonin on sleep and cognitive performance. Brain Res Bull. 2008;75(2-4):433–441. https://doi.org/ 10.1016/j.brainresbull.2007.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lucas-Sánchez A, Almaida-Pagán PF, Martinez-Nicolas A, Madrid JA, Mendiola P, de Costa J. Rest-activity circadian rhythms in aged Nothobranchius korthausae. The effects of melatonin. Exp Gerontol. 2013;48(5):507–516. https://doi.org/ 10.1016/j.exger.2013.02.026 [DOI] [PubMed] [Google Scholar]
- 112. Song L, Li C, Wu F, Zhang S. Dietary intake of diosgenin delays aging of male fish Nothobranchius guentheri through modulation of multiple pathways that play prominent roles in ROS production. Biogerontology. 2022;23(2):201–213. https://doi.org/ 10.1007/s10522-022-09955-0 [DOI] [PubMed] [Google Scholar]
- 113. Astre G, Atlan T, Goshtchevsky U, et al. Genetic perturbation of AMP biosynthesis extends lifespan and restores metabolic health in a naturally short-lived vertebrate. Dev Cell. 2023;58(15):1350–1364.e10. https://doi.org/ 10.1016/j.devcel.2023.05.015 [DOI] [PubMed] [Google Scholar]
- 114. Ripa R, Ballhysa E, Steiner JD, et al. Refeeding-associated AMPKγ1 complex activity is a hallmark of health and longevity. Nat Aging. 2023;3(12):1544–1560. https://doi.org/ 10.1038/s43587-023-00521-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Howe K, Clark MD, Torroja CF, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 7446;496:498–503. https://doi.org/ 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Chernyavskaya Y, Zhang X, Liu J, Blackburn J. Long-read sequencing of the zebrafish genome reorganizes genomic architecture. BMC Genomics. 2022;23(1):116. https://doi.org/ 10.1186/s12864-022-08349-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kasahara M, Naruse K, Sasaki S, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447(7145):714–719. https://doi.org/ 10.1038/nature05846 [DOI] [PubMed] [Google Scholar]
- 118. Valenzano DR, Benayoun BA, Singh PP, et al. The African turquoise killifish genome provides insights into evolution and genetic architecture of lifespan. Cell. 1539;163(6):1539–1554. https://doi.org/ 10.1016/j.cell.2015.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Reichwald K, Petzold A, Koch P, et al. Insights into sex chromosome evolution and aging from the genome of a short-lived fish. Cell. 2015;163(6):1527–1538. https://doi.org/ 10.1016/j.cell.2015.10.071 [DOI] [PubMed] [Google Scholar]
- 120. Kishi S, Bayliss PE, Uchiyama J, et al. The identification of zebrafish mutants showing alterations in senescence-associated biomarkers. PLoS Genet. 2008;4(8):e1000152. https://doi.org/ 10.1371/journal.pgen.1000152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Sasaki T, Kishi S. Molecular and chemical genetic approaches to developmental origins of aging and disease in zebrafish. Biochim Biophys Acta. 2013;1832(9):1362–1370. https://doi.org/ 10.1016/j.bbadis.2013.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Li C, Barton C, Henke K, et al. celsr1a is essential for tissue homeostasis and onset of aging phenotypes in the zebrafish. eLife. 2020;9:e50523. https://doi.org/ 10.7554/eLife.50523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kuro-o M, Matsumura Y, Aizawa H, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390(6655):45–51. https://doi.org/ 10.1038/36285 [DOI] [PubMed] [Google Scholar]
- 124. Ogura Y, Kaneko R, Ujibe K, Wakamatsu Y, Hirata H. Loss of αklotho causes reduced motor ability and short lifespan in zebrafish. Sci Rep. 2021;11(15090). https://doi.org/ 10.1038/s41598-021-93909-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Singh AP, Sosa MX, Fang J, et al. αKlotho regulates age-associated vascular calcification and lifespan in zebrafish. Cell Rep. 2019;28(11):2767–2776.e5. https://doi.org/ 10.1016/j.celrep.2019.08.013 [DOI] [PubMed] [Google Scholar]
- 126. Mangos S, Amaral AP, Faul C, Jüppner H, Reiser J, Wolf M. Expression of fgf23 and αklotho in developing embryonic tissues and adult kidney of the zebrafish, Danio rerio. Nephrol Dial Transplant. 2012;27(12):4314–4322. https://doi.org/ 10.1093/ndt/gfs335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zupkovitz G, Kabiljo J, Martin D, Laffer S, Schöfer C, Pusch O. Phylogenetic analysis and expression profiling of the Klotho gene family in the short-lived African killifish Nothobranchius furzeri. Dev Genes Evol. 2018;228(6):255–265. https://doi.org/ 10.1007/s00427-018-0619-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Rozenberg I, Atlan T, Franek R, et al. Exploring life-long tissue homeostasis through lineage tracing and cell transplantation. BioRxiv. 2023:2023.05.01.538839. https://www.biorxiv.org/content/10.1101/2023.05.01.538839v1
- 129. Gurevich DB, Nguyen PD, Siegel AL, et al. Asymmetric division of clonal muscle stem cells coordinates muscle regeneration in vivo. Science. 2016;353(6295):aad9969. https://doi.org/ 10.1126/science.aad9969 [DOI] [PubMed] [Google Scholar]
- 130. Krug J, Perner B, Albertz C, Mörl H, Hopfenmüller VL, Englert C. Generation of a transparent killifish line through multiplex CRISPR/Cas9mediated gene inactivation. eLife. 2023;12:e81549. https://doi.org/ 10.7554/eLife.81549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Dolfi L, Ripa R, Antebi A, Valenzano DR, Cellerino A. Cell cycle dynamics during diapause entry and exit in an annual killifish revealed by FUCCI technology. EvoDevo. 2019;10:29. https://doi.org/ 10.1186/s13227-019-0142-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Xu A, Teefy BB, Lu RJ, et al. Transcriptomes of aging brain, heart, muscle, and spleen from female and male African turquoise killifish. Sci Data. 2023;10:695. https://doi.org/ 10.1038/s41597-023-02609-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Jiang M, Xiao Y, Weigao E, et al. Characterization of the zebrafish cell landscape at single-cell resolution. Front Cell Dev Biol. 2021;9:743421. https://doi.org/ 10.3389/fcell.2021.743421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lange M, Granados A, VijayKumar S, et al. Zebrahub—multimodal zebrafish developmental atlas reveals the state-transition dynamics of late-vertebrate pluripotent axial progenitors. BioRxiv. 2023. https://www.biorxiv.org/content/10.1101/2023.03.06.531398v2 [DOI] [PubMed]
- 135. Liu G, Ito T, Kijima Y, et al. Zebrafish Danio rerio myotomal muscle structure and growth from a spatial transcriptomics perspective. Genomics. 2022;114(5):110477. https://doi.org/ 10.1016/j.ygeno.2022.110477 [DOI] [PubMed] [Google Scholar]
- 136. Beardall CH, Johnston IA. Muscle atrophy during starvation in a marine teleost. Eur J Cell Biol. 1983;29(2):209–217. [PubMed] [Google Scholar]
- 137. Lu C-Y, Hsu C-Y. Ambient temperature reduction extends lifespan via activating cellular degradation activity in an annual fish (Nothobranchius rachovii). Age. 2015;37(2):33. https://doi.org/ 10.1007/s11357-015-9775-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Graf M, Cellerino A, Englert C. Gender separation increases somatic growth in females but does not affect lifespan in Nothobranchius furzeri. PLoS One. 2010;5(8):e11958. https://doi.org/ 10.1371/journal.pone.0011958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Tsai SB, Tucci V, Uchiyama J, et al. Differential effects of genotoxic stress on both concurrent body growth and gradual senescence in the adult zebrafish. Aging Cell. 2007;6(2):209–224. https://doi.org/ 10.1111/j.1474-9726.2007.00278.x [DOI] [PubMed] [Google Scholar]
- 140. Chen J, Khondker RC, Brunet A. Breeding and reproduction of the African turquoise killifish Nothobranchius furzeri. Cold Spring Harb Protoc. 2023;2023(6):pdb.prot107816. https://doi.org/ 10.1101/pdb.prot107816 [DOI] [PubMed] [Google Scholar]
- 141. Cui R, Willemsen D, Valenzano DR. Nothobranchius furzeri (African turquoise killifish). Trends Genet. 2020;36(7):540–541. https://doi.org/ 10.1016/j.tig.2020.01.012 [DOI] [PubMed] [Google Scholar]
- 142. Konstantinidis I, Sætrom P, Mjelle R, Nedoluzhko AV, Robledo D, Fernandes JMO. Major gene expression changes and epigenetic remodelling in Nile tilapia muscle after just one generation of domestication. Epigenetics. 2020;15(10):1052–1067. https://doi.org/ 10.1080/15592294.2020.1748914 [DOI] [PMC free article] [PubMed] [Google Scholar]