ABSTRACT
tRNA modifications play important roles in maintaining translation accuracy in all domains of life. Disruptions in the tRNA modification machinery, especially of the anticodon stem loop, can be lethal for many bacteria and lead to a broad range of phenotypes in baker’s yeast. Very little is known about the function of tRNA modifications in host-pathogen interactions, where rapidly changing environments and stresses require fast adaptations. We found that two closely related fungal pathogens of humans, the highly pathogenic Candida albicans and its much less pathogenic sister species, Candida dubliniensis, differ in the function of a tRNA-modifying enzyme. This enzyme, Hma1, exhibits species-specific effects on the ability of the two fungi to grow in the hypha morphology, which is central to their virulence potential. We show that Hma1 has tRNA-threonylcarbamoyladenosine dehydratase activity, and its deletion alters ribosome occupancy, especially at 37°C—the body temperature of the human host. A C. albicans HMA1 deletion mutant also shows defects in adhesion to and invasion into human epithelial cells and shows reduced virulence in a fungal infection model. This links tRNA modifications to host-induced filamentation and virulence of one of the most important fungal pathogens of humans.
IMPORTANCE
Fungal infections are on the rise worldwide, and their global burden on human life and health is frequently underestimated. Among them, the human commensal and opportunistic pathogen, Candida albicans, is one of the major causative agents of severe infections. Its virulence is closely linked to its ability to change morphologies from yeasts to hyphae. Here, this ability is linked—to our knowledge for the first time—to modifications of tRNA and translational efficiency. One tRNA-modifying enzyme, Hma1, plays a specific role in C. albicans and its ability to invade the host. This adds a so-far unknown layer of regulation to the fungal virulence program and offers new potential therapeutic targets to fight fungal infections.
KEYWORDS: tRNA modification, Candida albicans, host-pathogen interactions, virulence regulation
INTRODUCTION
tRNA molecules present the structural interface between the RNA and protein worlds. In addition to their decoding function, tRNAs play important roles in translation accuracy and dynamics, and in primary metabolism (1) as well as stress responses (2). These functions depend on a plethora of post-transcriptional chemical modifications of the tRNA molecule itself. For example, a typical baker’s yeast exhibits roughly 25 different types of modifications on 36 different locations in its cytoplasmatic tRNA molecules (3), thus making tRNAs the most heavily modified of all RNA species. The anticodon loop is a hot spot of modifications that are critical for translational efficiency and fidelity (4). The purine base 37 is frequently modified by a variety of chemical groups (5). tRNA modifications are introduced by a diverse group of enzymes. Prominent examples are the elongator complex (6) and the URM1 pathway (7), which catalyze wobble uridine modifications (mcm5s2U34) and the threonyl-carbamoyl transferase complex, comprising Tcs1-Tcs7 in Saccharomyces cerevisiae (8, 9), for adenosine N6-threonylcarbamoyladenosine (t6A37) modifications at the anticodon stem loop (ASL). Most common and well conserved are t6A, N6-isopentenyladenosine (i6A), and 1-methylguanosine (m1G), which facilitate mRNA-to-tRNA binding and prevent misreading. Furthermore, translation-frame selection is affected by t6A during decoding of the start codon (1, 10, 11). t6A, first described in 1974 (12), was later shown to be a hydrolyzation product of cyclic threonylcarbamoyladenosine (ct6A), which is commonly found in bacteria, fungi, and plants (13). Defects in modifications of the ASL are lethal for most bacteria (14) and result in a broad range of phenotypes in yeast, including increased susceptibility to diverse stressors (15), defects in translation, and aggregation of misfolded proteins (9, 16).
Finally, tRNAs act as signaling molecules for the nutritional state of cells. Uncharged tRNAs accumulate under amino acid depletion and are recognized by the Gcn2 kinase (17). Gcn2 subsequently activates transcription factors for the expression of amino acid biosynthesis genes. This Tor1 kinase-dependent pathway is known as the general amino acid control and has been associated with tRNA modifications of the ASL: A S. cerevisiae tcs2Δ mutant, defective in t6A modification of ANN codon-tRNAs, lacks Gcn2-dependent Gcn4 activation (18), and cells that miss members of the URM1 pathway are sensitive to rapamycin (19). As yeast requires thiolated tRNAs to balance metabolic carbon and amino acid fluxes (20), its nitrogen starvation-induced pseudohyphal growth is blocked by a lack of tRNA thiolation at the ASL (21).
Candida species are fungal pathogens which frequently cause bloodstream infections with high mortality rates. For these (often) nosocomial infections, Candida albicans is by far the leading cause (22, 23). In contrast, its closest relative, Candida dubliniensis, rarely causes life-threatening infections (24), even though it is detectable in up to one-third of oral candidosis in HIV-infected patients (25). Although the vast majority of C. dubliniensis genes share sequence identities of over 80% with their C. albicans orthologs, certain pathogenicity-related genes differ markedly in sequence or are even absent in C. dubliniensis (26). These genomic differences are reflected in in vitro conditions, where C. dubliniensis is less robust against oxidative, osmotic, and temperature stresses (27).
Both C. albicans and C. dubliniensis are polymorphic fungi that rely on yeast-to-hyphae transitions for tissue invasion and dissemination within their host (25, 28). However, the induction of hyphae by nutrient availability differs significantly between the two species, implying differences in Tor1-dependent regulation (29). Furthermore, on reconstituted human epithelium, C. dubliniensis fails to produce germ tubes and cannot invade the tissue—in stark contrast to the highly invasive C. albicans. The overall lower virulence of C. dubliniensis also manifests in animal models like mice (30) or embryonated chicken eggs (31). Therefore, investigating the genetic differences between C. albicans and C. dubliniensis is an excellent model to gain key insights into species-specific stress responses, regulators of morphogenesis, and pathogenicity-related pathways in human infections.
In this study, a cross-species approach reveals a novel role for the C. albicans tRNA-modifying enzyme, Hma1, during starvation-induced morphogenesis and links it to the TOR pathway. We confirm the molecular function of Hma1 by quantifying t6A levels as a proxy for Hma1 activity and determine its role in translation by codon-specific ribosome-profiling analyses. Finally, we show the strong impact of HMA1 on C. albicans pathogenesis in vitro and in vivo.
MATERIALS AND METHODS
Reagents
Antibodies were acquired from Acris (rabbit α-Candida, #BP1006) and Abcam (goat α-rabbit, Alexa Fluor 488-coupled, #ab150077). Kits were bought from GE Healthcare (ECL Direct Nucleic Acid Labeling and Detection Kit, #RPN3001) and Takara Bio Europe (InFusion Cloning Kit, #638916). Specific chemicals were obtained from Sigma-Aldrich, Germany: antimycin A (#A8674), caffeine (#C0750), calcofluor white (#910090), cycloheximide (#01810), L-homoserine (#8.14359), menadione (#M5625), oligomycin (#O4876), and rapamycin (#R8781). Nourseothricin was bought from Werner Bioagents, Germany (#5.010.000). The AXIO Observer.Z1 was bought from Carl Zeiss, GermanyI.
Biological resources
All Candida albicans and Candida dubliniensis strains in this work are listed in Table 1. The TR146 cell line was originally obtained from the European Collection of Cell Cultures as lot number 13D016. Identity of the cell line and absence of mycoplasm contamination were tested regularly before, during, and after the experiments described here. Fertilized chicken eggs (breed “weiße Leghorn”) were obtained from local producers (32).
TABLE 1.
Candida strains used in this study
| Strain | Parent | Genotype | Reference |
|---|---|---|---|
| SC5314 | C. albicans wild-type strain | (33) | |
| CaCSP1-GFP | SC5314 | orf19.3512/orf19.3512-GFP-TACT1 | (34) |
| HMA1/SAT1 A/B | SC5314 | orf19.2115Δ::SAT1-FLIP/orf19.2115 | This study |
| HMA1/hma1Δ A/B | HMA1/SAT1 A/B | orf19.2115Δ::FRT/orf19.2115 | This study |
| hma1Δ/SAT1 A/B | HMA1/hma1Δ A/B | orf19.2115Δ::FRT/orf19.2115Δ::SAT1-FLIP | This study |
| hma1Δ/Δ A/B | hma1Δ/SAT1 A/B | orf19.2115Δ::FRT/orf19.2115Δ::FRT | This study |
| HMA1KS1 | hma1Δ/ΔB | orf19.2115Δ::FRT/orf19.2115Δ::orf19.2115-TACT1-SAT1-FLIP | This study |
| hma1Δ/HMA1 | HMA1KS1 | orf19.2115Δ::FRT/orf19.2115Δ::orf19.2115-TACT1-FRT | This study |
| Wü284 | C. dubliniensis wild-type strain | (35) | |
| CdCSP1-GFP | Wü284 | CD36_30750/CD36_30750-GFP-TACT1 | (34) |
| CdUM4A | Wü284 | Cdura3Δ::FRT/Cdura3Δ::FRT | (36) |
| CdINT1 | CdUM4A | Cdura3Δ::FRT/Cdura3::pCdINT1 | (37) |
| CdINT4 | CdUM4A | Cdura3Δ::FRT/Cdura3::pCdINT1-Ca21chr2:22964-27892W | This study |
| Cd2113 | CdUM4A | Cdura3Δ::FRT/Cdura3Δ::pCdINT1-orf19.2113 | This study |
| Cd2114 | CdUM4A | Cdura3Δ::FRT/Cdura3Δ::pCdINT1-orf19.2114 | This study |
| Cd2115 | CdUM4A | Cdura3Δ::FRT/Cdura3Δ::pCdINT1-orf19.2115 | This study |
| CdHMA1/SAT1 A/B | Wü284 | CD36_15150Δ::SAT1-FLIP/CD36_15150 | This study |
| CdHMA1/Cdhma1Δ A/B | CdHMA1/SAT1 A/B /B | CD36_15150Δ::FRT/CD36_15150 | This study |
| Cdhma1Δ/SAT1 A/B | CdHMA1/Cdhma1Δ A/B | CD36_15150Δ::FRT/CD36_15150Δ::SAT1-FLIP | This study |
| Cdhma1Δ/Δ A/B | Cdhma1Δ/SAT1 A/B | CD36_15150Δ::FRT/CD36_15150Δ::FRT | This study |
| CdHMA1KS1 | Cdhma1Δ/Δ | CD36_15150Δ::FRT/CD36_15150Δ::CD36_15150-TACT1-SAT1-FLIP | This study |
| Cdhma1Δ/HMA1 | CdHMA1KS1 | CD36_15150Δ::FRT/CD36_15150Δ::CD36_15150-TACT1-FRT | This study |
| Cd2115_CdHMA1/SAT1 A/B | Cd2115 | Cdura3Δ::FRT/Cdura3Δ:: pCdINT1-orf19.2115 CD36_15150Δ::SAT1-FLIP/CD36_15150 | This study |
| Cd2115_CdHMA1/Cdhma1Δ A/B | CdHMA1M1A/B | Cdura3Δ::FRT/Cdura3Δ:: pCdINT1-orf19.2115 CD36_15150Δ::FRT/CD36_15150 | This study |
| Cd2115_Cd A/B | CdHMA1M2A/B | Cdura3Δ::FRT/Cdura3Δ::pCdINT1-orf19.2115 CD36_15150Δ::FRT/CD36_15150Δ::SAT1-FLIP | This study |
| Cd2115_Cdhma1Δ/Δ A/B | CdHMA1M3A/B | Cdura3Δ::FRT/Cdura3Δ:: pCdINT1-orf19.2115 CD36_15150Δ::FRT/CD36_15150Δ::FRT | This study |
Statistical analyses
For all experiments, P < 0.05 was considered statistically significant. Lower P-values are indicated by asterisks, with *, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. For data with expected normal distribution, two-sided t-tests were used with a biological replicate count n ≥ 3 (indicated in the figure legends). All data are shown as mean with standard deviation of biological (independent) replicates. In case of multiple testing, an appropriate correction was used (Benjamini-Hochberg or Šidák as indicated). For the infection experiments, the Mantel-Cox test was used to detect statistical significance of the difference between survival curves, with n = 20 for each group.
Culture conditions
Candida strains were routinely propagated on YPD agar (20 g peptone, 10 g yeast extract, 20 g glucose, 15 g agar per liter) at 30°C and stored as frozen stocks in YPD medium with 15% (vol/vol) glycerol at –80°C.
Morphological tests
Chlamydospore formation
Chlamydospore production by C. dubliniensis was induced on solid synthetic low-ammonium dextrose (SLAD) agar (0.17% Yeast Nitrogen Base without Amino Acids and Ammonium Sulfate; BD, Heidelberg, Germany) (38) without any additional nitrogen source but with 2% glucose and 2% agar (Kobe I, Carl Roth GmbH + Co. KG, Germany). Formation of chlamydospores by C. albicans was induced on corn meal-Tween 80 agar (BD, Heidelberg, Germany). The plates were incubated at 28°C for 2–7 days in darkness, and chlamydospore formation was monitored microscopically (Axiovert, Zeiss, Germany).
Germ tube assays
For hyphae induction in liquid media, Candida strains were pre-grown in liquid YPD overnight (30°C, 180 rpm), washed with phosphate-buffered saline (PBS), and 105 cells were transferred into 500 µL + H2O plus 10% (vol/vol) fetal calf serum (FCS) and the morphology was analyzed microscopically (Axiovert, Zeiss, Germany). ECE1 transcript levels were determined using the same medium. RNA was isolated from yeasts (0 h) and hyphae (6 h) using a glass bead method with the Qiagen RNeasy Kit. RNA quality and quantity were measured using an Agilent BioAnalyzer and a Nanodrop instrument, respectively. cDNA was synthesized using Invitrogen Superscript III following the protocol from reference (39).
Filamentation assay
Candida cells from YPD overnight cultures were washed, and 5 µL of 2 × 107 cells/mL was spotted on the following agar plates: YPD, Spider, water agar, SLAD, or boiled blood (chocolate) agar (40). Colony filamentation was investigated after incubation at 37°C in either atmospheric or 5% CO2-enriched air after 5 days incubation, using a binocular (Stemi 2000-C, Zeiss, Germany).
Stress assays
Resistance of Candida strains to stressors was tested by growth curve assays or by serial drop dilution tests on solid agar plates. In both cases, Candida cells from YPD overnight cultures (30°C, 180 rpm) were initially washed twice with water. For growth in liquid medium, cells were diluted to OD600 = 0.01 in YPD or in synthetic minimal media: SD (Yeast Nitrogen Base with Ammonium Sulfate, MP, Santa Ana, CA, USA) or YCB medium (Yeast Carbon Base, BD, Heidelberg, Germany).
Stress tests were conducted with indicated concentrations of supplements. YPD medium contained 5 or 7.5 nM rapamycin, 6 or 8 mM caffeine, 2.5 or 5 mg/mL cycloheximide, 100 mM menadione, 2 µM antimycin A, 2 µM oligomycin, 1 µg/mL calcofluor white, and 1M NaCl. YCB medium contained 1 mg/mL L-homoserine and 5 mg/mL ammonium sulfate; SD medium was used as growth control (All stressors Sigma-Aldrich GmbH, Germany). Growth cultures were incubated at 30°C was determined after 30-s shaking every 20 min over 65 h.
For drop dilution tests on solid agar plate, Candida cultures were adjusted to 2 × 108 cells/mL and successively diluted by factor 10. A volume of 5 µL of each concentration was spotted onto solid agar plates, and colony growth was followed for 2 or 3 days under the indicated conditions. Furthermore, 2% glucose was used as standard carbon source of YP (20 g peptone, 10 g yeast extract, and 15 g agar per liter) plates and was replaced with 2% glycerol or ethanol (Sigma-Aldrich GmbH, Germany) to induce respiratory growth. Images were taken with an imaging system (Vilber-Lourmat GmbH, Germany).
Adhesion/invasion assay
The capability of C. albicans cells to adhere and invade host cell layers was assayed by infecting monolayers of human oral epithelial TR146. Cell cultivation, infection, and fixation procedures were performed as previously described (41) using an MOI of 0.4. Three hours post infection, non-adherent cells were rinsed with PBS, and samples were fixed with Histofix (Carl Roth GmbH + Co. KG, Germany). Adherent Candida cells on the epithelium were marked with primary anti-Candida antibody (Acris Anti Candida, rabbit, Herford, Germany) and detected with an Alexa Fluor 488-coupled secondary anti-rabbit antibody. Subsequent permeabilization of the TR146 cells with 0.5% Triton X-100 allowed staining of invasive hyphae parts with calcofluor white (Sigma-Aldrich GmbH, Germany). Hyphal length and invasiveness were determined for 100 hyphae of each strain in triplicates using fluorescence microscopy (Zen2 pro, Zeiss, Germany).
Screening of a C. albicans genomic library and identification of the C. albicans genomic fragments inserted in C. dubliniensis
The construction of a genomic C. albicans library in C. dubliniensis was described before by Staib and Morschhäuser (37). Approximately 20,000 library clones were screened on SLAD agar under chlamydospore-inducing conditions, and colonies with altered colony morphology were confirmed on SLAD agar. The C. albicans-derived DNA fragment from the transformant CdINT4 was amplified and sequenced using the primers CdUra14 and M13rev. Sequence analysis demonstrated that CdINT4 contained C. albicans sequences corresponding to chromosomal coordinates 22964–27892 of chromosome 2. Three complete C. albicans ORFs, orf19.2113, orf19.2114, and orf19.2115, are located within this region. Each ORF, including native promotor and terminator regions, was amplified via PCR using the corresponding primer pairs (IF2113_1/2, IF2114_1/2, IF2115_1/2) and cloned into ApaI/XhoI-cut plasmid pcdINT1 using the InFusion Cloning Kit (Takara Bio Company). The resulting plasmids were transformed (see Candida transformant construction) into C. dubliniensis CdUM4A (36), and their morphology was investigated.
Plasmid construction
The deletion cassette for orf19.2115 was constructed as follows: An ApaI-XhoI fragment with orf19.2115 upstream sequences was cloned after amplification by PCR with the primers 2115_3 and 2115_4 (Table S1) using genomic DNA from C. albicans SC5314 as template. A SacII-SacI fragment containing orf19.2115 downstream sequences was obtained with the primers 2115_5 and 2115_6. These orf19.2115 upstream and downstream fragments were used to replace SSU2 upstream and downstream fragments in plasmid pSSU2M2 (42) via the introduced restriction sites, to result in p2115M2, in which the SAT1 flipper cassette is flanked by orf19.2115 sequences. The gene deletion cassette pcd15150M2 for CD36_15150 in C. dubliniensis was constructed analogously using the primer pairs 15150_3 and 15150_4 or 15150_5 and 15150_6, respectively, with gDNA from C. dubliniensis Wü284 as template.
The orf19.2115 gene was amplified for genetic reconstitution in C. albicans using the primers 2115_3 and 2115_7. The ApaI/BamHI-cut DNA fragment was integrated into pSAP2KS1 (43) resulting in p2115KS1. The orf19.2115 downstream DNA fragment was amplified using the primers 2115_8 and 2115_9, and the NotI/SacII-cut DNA fragment was ligated into p2115KS1. For the CD36_15150 revertant in C. dubliniensis, the whole CD36_15150 gene was amplified using the primer pair 15150_3 and 15150_7, and the downstream DNA fragment using the primers 15150_5 and 15150_6 c. The ApaI/SmaI- or SacII/SacI-cut fragments were integrated into pSAP2KS1.
Candida transformant construction
Chemically competent C. albicans or C. dubliniensis cells were transformed with linear DNA fragments by heat shock at 44°C (44, 45), and resulting clones were selected on YPD plates containing 50–200 µg/mL nourseothricin (Werner Bioagents, Jena, Germany). The SAT1 flipper strategy allows the recycling of the selection marker (46). The insertion locus of the DNA fragment was confirmed by Southern Blot analyses.
Southern blot
Ten micrograms of isolated genomic DNA were digested with an appropriate restriction enzyme. After DNA separation on an agarose gel (1%), DNA was transferred onto a nylon membrane using a vacuum blot system. UV-linked DNA was hybridized with chemiluminescence-enabled probes and detected via the Amersham ECL Direct Nucleic Acid Labeling and Detection (GE Healthcare, Braunschweig, Germany) according to the manufacturer’s instructions.
Reversed phase nano liquid chromatography mass spectrometry of ribonucleosides
Candida strains were grown overnight in liquid YPD cultures (30°C, 180 rpm), washed, and diluted to an OD600 of 0.2 in 50 mL YPD medium. Cultures were grown for additional 4 h at 30°C or 37°C. Cells were harvested by centrifugation, frozen in liquid nitrogen, and stored at −80°C until lysis. Total RNA was isolated according to the protocol by Miyauchi et al. (13) using acidic phenol-TRIzol (Life Technologies GmbH, Darmstadt, Germany) extraction. The samples were kept on ice whenever possible. The tRNA was subsequently isolated by gel extraction from denaturing 8 M urea 8% polyacrylamide gels (47). For each biological replicate, we prepared two to three technical replicates depending on the amount of tRNA that we extracted from the gel (50 samples in total for C. albicans wt and revertant, 50 for C. albicans deletion mutants, and 27 for C. dubliniensis wt; Table S2). The gel-extracted tRNAs were enzymatically digested under acidic conditions and dephosphorylation into single ribonucleosides as described (48). The digested ribonucleosides were dissolved in 5 mM NH4HCO2, pH 4.8, spiked with 15N-labeled ribonucleosides from Chlamydomonas reinhardtii, and subsequently subjected to reversed phase chromatography using a self-packed C18 (Synergi 4 µm Hydro-RP 80 Å; Phenomenex Ltd., Aschaffenburg, Germany) capillary column (75 µm ID × 430 mm) coupled to a Proxeon EASY nLC (Thermo Fisher Scientific GmbH, Dreieich, Germany). A multi-step gradient (0%–38% B in 31 min, 38%–50% B in 6 min, 50%–100% B in 6 min, hold at 100% B for 16 min) with a solvent system consisting of NH4HCO2, pH 4.8 (solvent A) and 40% acetonitrile (solvent B), and a flow rate of 250 nL/min was applied. On-column temperature was set to 20°C. Electrospray ionization mass spectrometry analysis was performed using a Q Exactive mass spectrometer (Thermo Finnigan LLC, San Jose, CA) operated in the positive mode at a resolution of 70,000, the AGC (automated gain control) target value set to 3 × 106, and the fill time to 50 ms. Full MS spectra (m/z 100–700) and Top5 ddMS2 spectra were recorded. Immediately before or after the measurements of the biological samples, we measured chemical standards of t6A and ct6A to determine their elution time from the gradient. The identity of the chemical standards was verified by MS2 fragmentation.
Quantitative analysis of LC-MS/MS data
Acquired Thermo RAW files were converted to mzML format (49) using msconvert as part of Proteowizard (version 3.0.11799) (50). Data analysis was carried out using MORAST (Leufken et al., unpublished data) using default parameters, and the fragmentation method set to “stepped_low_hcd.” Within MORAST, spectra were accessed and centroided, if necessary, by pymzML (version 2.0.2) (51). Furthermore, pyQms (version 0.5.0) was used for quantification of all known ribonucleosides (52) [data obtained from MODOMICS; (53)], in MS1 (5 ppm accuracy) and MS2 (20 ppm accuracy) spectra. Metabolic labels were defined for nitrogen using either an enrichment of 0% (unlabeled) or 99% (fully labeled) of 15N. LC-MS/MS runs were corrected for systematic m/z errors, if necessary. Spiked-in 15N-labeled nucleosides were used for internal quality control and normalization. The identities of quantified ribonucleosides were verified by their specific fragmentation patterns in MS2 and by predetermined chromatographic elution orders of structural isomers (48). Abundances for all ribonucleosides were determined using the area under curve for each chromatographic peak. Furthermore, MORAST uses the Python packages: SciPy (54), Scikit-learn (55), and statsmodels (56). pyQms (57) calculates an accurate isotope pattern for each molecule and generates a quality score for each MS1 measurement. Therefore, it does not only use the m/z value of a given molecule but also extracts the chemical sum formula of each ion. The ct6A signal in the biological measurements was so low that the mass spectrometer did not select the ion for MS2 fragmentation. Therefore, we report the detection of a weak ct6A signal in cases where pyQms detected an ion with the sum formula of ct6A (C15H18N6O7) and a high pyQms quality score at the same retention time as the ct6A chemical standard.
Ribosome profiling
Ribosome profiling was conducted essentially as described (47, 58, 59). C. albicans and C. dubliniensis cells were grown to OD600≈ 0.4, harvested by vacuum filtration using a 0.45-µm cellulose nitrate filter (GE Healthcare), and flash frozen in liquid nitrogen. The cells were lyzed in a Freezer-Mill (SPEX SamplePrep) using two cycles at 5 CPS and an intermittent cooling step of 2 min. The lysates were thawed in lysis buffer (20 mM Tris-HCl pH 7.4, 5 mM MgCl2, 100 mM NaCl, 1% Triton, 2 mM DTT, 100 µg/mL cycloheximide) and clarified by two rounds of centrifugation (5 min; 4°C; 10,000 g). And, 10 A260 units of the lysates were digested for 1 h at 22°C using 600 U RNase I (Ambion, Thermo Fisher). Subsequently, the reaction was stopped by 15 µL SuperaseIn (Thermo Fisher), monosomes were isolated, and 27–29-nt-long footprints were excised from an acrylamide gel. During library generation, 3′-adapters introduced four randomized positions to reduce ligation biases (47). Following sequencing, the adapter sequences were clipped, and the four randomized nucleotides removed using the FASTX-Toolkit. The processed reads were mapped to ORFs (cgdGene) using bowtie (60). Reference ORFs (C_albicans_SC5314_version_A22, allele A) were extended by 18 nt into the UTRs. A-site codons were mapped according to the frame of the 5′ end of footprints—excluding reads that mapped to the first or last 15 codons of each transcript—and an appropriate offset was defined (16). Differential gene expression analysis was performed using DESeq2 (61). For altered transcripts, the P values were adjusted with Benjamini-Hochberg correction, and a-threshold was set to 0.05. To identify unaltered transcripts, the althypothesis function was set to “lessAbs.”
Gene ontology analysis
Significant differences in gene expression of the C. albicans hma1Δ/Δ mutant (cutoff Padj <0.05 and log2FC ± 1) were analyzed for enriched biological process using the CGD gene ontology (GO) term finder online tool with default settings [Candida Genome Database http://www.candidagenome.org/ (62) access in April 2021] and summarized by removing redundant terms using REVIGO (63). Enrichment of regulated GO terms was calculated as the ratio of cluster frequency to background frequency.
Chicken embryo infection model
The embryonated chicken infection model was used to study virulence as described previously (32). Briefly, overnight cultures of yeasts were washed with PBS and adjusted to 108 or 106 cells/mL. A high (107 yeasts/egg) or low inoculum (105 yeasts/egg) was applied onto the chorio-allantoic membrane at developmental day 10 via an artificial air chamber. In each experiment, the viability of 20 eggs per group (Candida or PBS control) was evaluated for 7 days by daily candling. Experiments were performed in duplicate. Surviving embryos were humanely terminated by chilling on ice at the end of the experiment. All experiments were performed in compliance with the German animal protection law. According to this, no specific approval was needed for work performed in avian embryos. All experiments were terminated on developmental day 18 at the latest.
RESULTS
Identification of a novel factor promoting species-specific chlamydospore formation
To uncover genes that contribute to the obvious differences in the virulence of Candida albicans and Candida dubliniensis, we screened a library of C. albicans genomic fragments integrated into a C. dubliniensis recipient strain. We especially screened for novel morphology-associated C. albicans genes, based on a finding that SLAD agar induces extensive chlamydospore production only in C. dubliniensis after prolonged incubation at 27–30°C [Fig. 1A and B, and (64)]. This was confirmed with chlamydospore-specific GFP reporter strains (34) (Fig. S1).
Fig 1.
Screening of a C. albicans genomic library on SLAD agar. Cells were cultivated in liquid SD medium overnight (30°C, 180 rpm) and spread onto SLAD medium. Agar plates were incubated for 2 days at 30°C in the dark. (A) The C. albicans wild-type strain SC5314 grew in smooth colonies comprised yeasts, whereas (B) C. dubliniensis Wü284 formed pseudohyphae with end-terminal chlamydospores, identifiable by the rough colony morphology. (C) By screening the integrative genomic C. albicans library in C. dubliniensis, CdINT4 was isolated as a transformant, which resembled the smooth colony morphology of C. albicans. (D) Integration of C. albicans orf19.2115 (HMA1) into C. dubliniensis resulted in the strain Cd2115 that phenocopied the C. albicans-specific colony morphology. The scale bar represents 100 µm.
In total, about 20,000 transformants were tested under these conditions. We found smooth colonies of C. dubliniensis transformants that phenocopied the C. albicans colony appearance. One clone, CdINT4, showed a strong and reproducible reduction in sporulation and filamentation (Fig. 1C). Sequence analysis of the integrated C. albicans DNA fragment revealed an assembly of three adjacent open reading frames (orf19.2113, orf19.2114, and orf19.2115). To determine which orf was responsible for the phenotype, each (including up- and downstream regions) was separately integrated into C. dubliniensis. Only orf19.2115 (strain Cd2115) changed the colony morphology to mimic the original integrant strain on SLAD agar (Fig. 1D). The S. cerevisiae ortholog of orf19.2115, TCD2, encodes a tRNA-threonylcarbamoyladenosine dehydratase (13), which like many genes in S. cerevisiae has a paralog, TCD1. No paralogs of orf19.2115 seem to exist in C. albicans or C. dubliniensis. As the gene name TCD2 is already in use in C. albicans, and considering the results described in the next sections, we named this gene HMA1 (for Hypermodification of Adenosine) and its C. dubliniensis ortholog (CD36_15150) CdHMA1 in this text. These two genes are syntenic, and their proteins are nearly identical, with 95.6% identities and 97.7% similarities in a BLASTP search (alignment in Fig. S2). We then created the deletion mutants hma1Δ/Δ and Cdhma1Δ/Δ and the revertants hma1Δ/HMA1 (C. albicans) and Cdhma1Δ/CdHMA1 (C. dubliniensis) by integration of one allele into the native locus. Finally, CdHMA1 was deleted in Cd2115 to obtain a C. dubliniensis CdHMA1-deficient strain which carries only the C. albicans HMA1 in one copy (Cd2115_Cdhma1Δ/Δ). All strains were verified by Southern blotting (Fig. S3).
Hma1 is a regulator of morphogenesis in C. albicans and interacts with the TOR pathway
We had identified HMA1 on a nitrogen-poor medium. The TOR pathway plays a central role in nutritional stress in eukaryotes (65). Hence, we tested the growth of the C. albicans wild-type SC5314, hma1Δ/Δ, and hma1Δ/HMA1 in the presence of sublethal concentrations of the TOR antagonists rapamycin and caffeine. In YPD without additives, all strains grew fast and reached a plateau within 16 h (Fig. 2A). Addition of 7.5 nM rapamycin or 8 mM caffeine (Fig. 2B) increased the generation times in all strains, but the C. albicans hma1Δ/Δ strains were significantly less affected than the wild type. The complemented strain bearing one HMA1 allele showed an intermediate growth. All tested C. dubliniensis strains grew equally well in YPD media (Fig. 2A), but in direct contrast to C. albicans hma1Δ/Δ, C. dubliniensis Cdhma1Δ/Δ strains exhibited an increased sensitivity to rapamycin and caffeine (Fig. 2B). Together, these findings indicate that Hma1 function interacts with Tor1-mediated signaling.
Fig 2.
Stress resistances test of C. albicans hma1Δ/Δ and C. dubliniensis Cdhma1Δ/Δ strains. Candida strains (C. albicans: SC5314, hma1Δ/Δ, hma1Δ/HMA1 and C. dubliniensis: Wü284, Cdhma1Δ/Δ, Cdhma1Δ/HMA1) were grown overnight in YPD medium at 30°C. Growth of C. albicans strains was monitored over 65 h in (A) YPD and YCB plus 5 mg/mL ammonium sulfate, (B) YPD plus 7.5 nM rapamycin, YPD plus 8 mM caffeine, (C) YPD plus 5 mg/mL cycloheximide, YCB plus 4 mg/mL L-homoserine. Stressor concentrations were adapted for growth of C. dubliniensis strains to YPD plus 5 nM rapamycin, 6 mM caffeine, or 2.5 mg/mL cycloheximide. The temperature was set at 30°C for all conditions. Graphs show the mean ± SD of three independent biological replicates and two independently created hma1Δ/Δ strains for both species.
The TOR pathway is crucial for responding to the cellular nutritional status. Hence, we tested growth on the non-fermentable carbon sources glycerol and ethanol but found no effect of the HMA1 deletion in either Candida species (Fig. S4A). Similarly, C. albicans hma1Δ/Δ showed no phenotypic effect when tested under oxidative (menadione), antifungal (oligomycin, antimycin A), cell wall (calcofluor white), and osmotic (NaCl) stress (Fig. S4B). This hints toward a specific role for HMA1 in TOR-related morphology and nutrient signaling, rather than a global effect, similar to the phenotypes of t6A-deficient S. cerevisiae (9).
Correct tRNA modifications optimize translational accuracy and consequently growth. Addition of the translation inhibitor cycloheximide in sublethal levels resulted in a strong growth reduction of hma1Δ/Δ (Fig. 2C), suggesting an important role for Hma1 in translation. C. dubliniensis was generally more susceptible to cycloheximide, and no differences were evident between the effect on the wild type and the Cdhma1Δ/Δ mutants (Fig. 2C).
The threonine analog L-homoserine suppresses the growth defect of t6A tRNA modification-defective S. cerevisiae mutants (9). Similarly, hma1Δ/Δ grew better than the wild type and the complemented strain when 4 mg/mL L-homoserine was the sole nitrogen source, while the deletion of CdHMA1 in C. dubliniensis wild type had no effect (Fig. 2C). The control condition with ammonium sulfate supplementation revealed similar growth of mutants and parental strains (Fig. 2A). Metabolism of homoserine hyper-activates the protein degradation response (66), and our data therefore hint toward a possible link of C. albicans HMA1 to this pathway.
Many tRNA modifications are mediators of heat stress responses (67, 68). In-line with these findings, a slight decrease in growth was observed for the mutants in both species at elevated temperatures (Fig. S5A and B). Therefore, we assume a species-independent role for HMA1 during heat stress.
C. albicans HMA1 encodes a tRNA-modifying enzyme
Tcd2, a tRNA threonylcarbamoyladenosine dehydratase in S. cerevisiae, shows high similarity with C. albicans Hma1 (identities 60.2%, similarities 77.0%) and C. dubliniensis CdHma1 (61.4%, 78.4%). The overall structure of Tcd2 and Hma1 is also similar (Fig. 3A), suggesting similar functions. This enzyme targets t6A residues at position 37 (t6A37) specifically in tRNAs bearing an NNU anticodon, where it catalyzes the formation of an oxazolone ring (ct6A37) using ATP (Fig. 3B) (13).
Fig 3.
Quantification of t6A37 modification in Candida species. (A) Structure of the proteins Tcd2 and Hma in S. cerevisiae and C. albicans, respectively. Important regions are highlighted to illustrate the overall structure. (B) Proposed model of cyclic N6-threonylcarbamoyladenosine formation by Hma1 reaction in C. albicans (designed with ISIS/Draw2.1.4). (C) Overnight cultures of Candida strains (C. albicans: SC5314, hma1Δ/Δ A/B, hma1Δ/HMA1, and C. dubliniensis Wü284) were inoculated in fresh YPD medium (OD600 of 0.2) and incubated for 4 h at 30°C or 37°C, cells were harvested, and RNA was isolated. Quantitative liquid chromatography mass spectrometry analysis of tRNA isolated from Candida strains was performed, and results of biological triplicates with their means were plotted relative to the spike in. The levels of t6A revealed a significant increase in the hma1Δ/Δ A and B mutants compared to the C. albicans wild-type SC5314 in both test conditions. t6A levels of C. dubliniensis Wü284 and C. albicans hma1Δ/HMA1 revertant remained at C. albicans wild-type level. Experiments were performed in biological triplicates, and statistical analyses used t-test with Šidák corrections for multiple comparisons (**P ≤ 0.01 and ***P ≤ 0.001).
To uncover the molecular function of Hma1, we cultivated C. albicans wild-type, hma1Δ/Δ, and revertant strains, as well as C. dubliniensis wild type in YPD at 30°C and 37°C and isolated tRNA from these strains. Subsequently, we analyzed the presence of t6A derivates by RNA mass spectrometry (48). Since ct6A is very labile (13), we determined the levels of its educt, t6A relative to a 15N-labeled spike-in (Fig. 3C). We found a striking increase (>50%) of t6A in both independent hma1Δ/Δ strains. However, we did not observe a difference among the growth conditions or between the wild types of the two species. Importantly, t6A levels of the revertants were similar to the wild-type strains under both conditions. The total amount of ct6A was insufficient to obtain quantitative results due to its limited stability. However, we detected a weak MS1 signal indicating the presence of ct6A in 75 of 77 samples [49/50 (98%) in C. albicans and 27/27 (100%) in C. dubliniensis, over all biological and technical replicates and conditions; Table S2]) from the wild types and the revertant but only in 2 of 50 (4%) samples taken from the deletion mutants. This signal was observed at the same retention time as a chemically synthesized nucleoside standard for ct6A. Finally, the total levels of adenosine and other nucleosides remained essentially unchanged in all tRNA samples (Fig. S6). Altogether, these findings provide evidence for a lack of conversion of t6A37 to ct6A37 upon deletion of HMA1 in C. albicans. This is in-line with findings in S. cerevisiae and suggests that Hma1 enables the conversion of t6A to ct6A (Fig. 3B).
Hma1 plays a species-specific role during hyphae formation
Given the role in morphology we have established for HMA1, we next assayed the filamentation abilities of all strains with minimal or nutrient-deprived (water and SLAD) agar and complex media (YPD, Spider, and chocolate agar, and water + 10% fetal calf serum liquid medium).
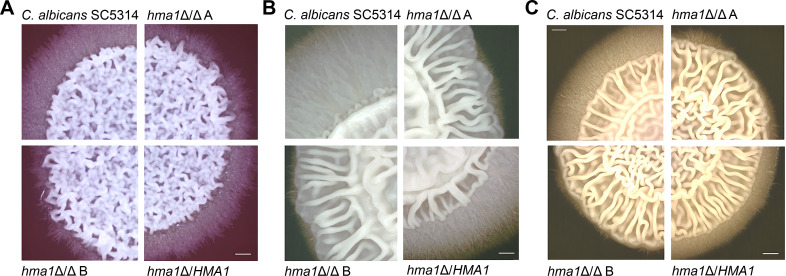
The C. albicans hma1Δ/Δ deletion mutants showed clearly impaired hyphae fringes at the colony borders on chocolate, Spider, and YPD agar (Fig. 4A through C). The phenotype was nearly fully restored in the hma1Δ/HMA1 revertant. In contrast, in all strains, the central wrinkling of the colonies was comparable to the wild type (Fig. 4A through C). This indicates a specific reduction in longer hyphal elements, which invade the agar, rather than a general loss of filamentation. Similarly, using an XTT assay, we found no difference in the biofilm formation by the C. albicans hma1Δ/Δ mutants (data not shown). During formation of biofilms, hyphae growth is important, but no invasion takes place.
Fig 4.
Filamentation of C. albicans strains on solid agar. C. albicans cells from a YPD overnight culture were washed, and 105 cells of each strain were spotted on (A) chocolate agar, (B) Spider agar, or (C) YPD agar. Plates were incubated at 37°C for 5 days. Each medium induced wrinkling in the center of the colonies and extensive hyphae formation by the C. albicans wild-type SC5314. The hma1Δ/Δ mutant strains were severely impaired in formation of the hyphal fringe, but central wrinkles remained. The scale bar represents 0.5 cm.
In general, C. dubliniensis shows little to no hyphae formation on standard agar plates. Only on minimal medium agar at 37°C, a late robust hyphae formation without central wrinkling was observed (Fig. S7A through C). However, the deletion, revertant, and transformant mutants exhibited no difference to the C. dubliniensis wild type, indicating that CdHma1 is dispensable in C. dubliniensis filamentation on agar plates.
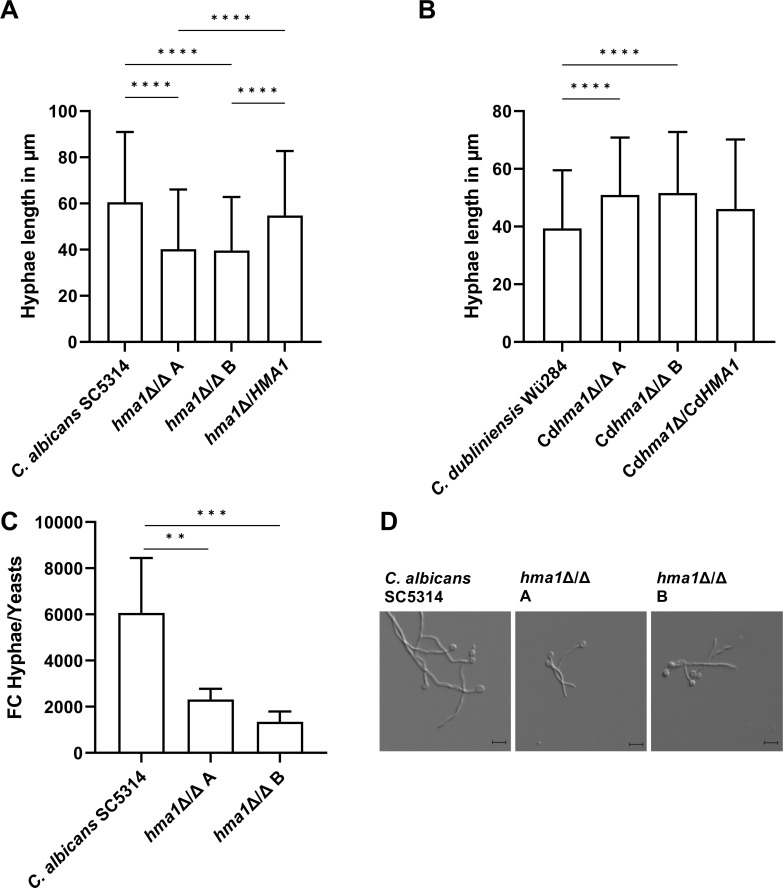
We also tested both species for filamentation in liquid medium using water with 10% FCS. Hyphae length of C. albicans wild type reached on average 60.5 µM after 4 h, whereas hma1Δ/Δ hyphae were significantly shorter (40.2 and 39.6 µm for the two independent mutants; Fig. 5A). The complemented strain reached almost wild-type levels (54.8 µm). In stark contrast, the hyphae length of Cdhma1Δ/Δ in water-serum medium increased significantly (129% and 131%), which was largely reversed by re-introducing CdHMA1 (Fig. 5B). This shows that the effect of Hma1 differs strikingly between the two species and that C. albicans Hma1 promotes growth of long hyphae, whereas C. dubliniensis Hma1 suppresses it. When we investigated the transcription of ECE1, the gene coding for the C. albicans toxin candidalysin (69), we found a strong reduction in its upregulation upon hyphae formation (Fig. 5C). As ECE1 transcript levels are strongly correlated with hyphal length (70, 71), these data support our morphological findings (Fig. 5D). Since hyphae formation and the secretion of ECE1-encoded candidalysin are central to C. albicans pathogenesis, we continued to investigate Hma1’s role in virulence.
Fig 5.
Length of hyphae of C. albicans and C. dubliniensis strains incubated in water plus 10% serum. In total, 104 Candida cells were incubated in water plus 10% FCS for 4 h at 37°C, and atmospheric CO2 content and hyphal length were measured microscopically. (A) C. albicans hma1Δ/Δ mutants had significantly shorter hyphae than the wild-type strain. (B) C. dubliniensis Cdhma1Δ/Δ mutants had longer filaments compared to the wild type. The column bars show mean values with standard deviation. (C) Transcriptional upregulation of the toxin-coding ECE1 gene upon hyphae formation. The transcript levels are significantly lower in the mutant, in-line with (D) the reduced hyphae length of the mutants as seen in micrographs. Statistical analysis used one-way ANOVA test with Tukey’s multiple comparisons test (****P ≤ 0.0001). Sample size (N) is 3 biological replicates (A–C) with at least 50 individual measurements (A, B).
HMA1 deletion reduces adherence and invasiveness into oral epithelial cells
To investigate adhesion and invasion as the two primary steps in Candida infections, we infected the human oral epithelial TR146 cell line with C. albicans wild type, hma1Δ/Δ, and revertant strains. After 3 h, 25.7% of the wild-type cells, but only 13.0% and 15.4% of the two independent hma1Δ/Δ strains were found adhered to host cells, a reduction by roughly half. This phenotype was mostly reversed in hma1Δ/HMA1, at 21.2% adherence (Fig. 6A).
Fig 6.
C. albicans hma1Δ/Δ mutants are defective in adhesion and invasion of oral epithelial cells. C. albicans cells were washed, and an MOI of 0.4 was used to infect 2-day-old TR146 cells. After co-incubation of fungal and human cells for 3 h, non-adherent cells were rinsed off, and samples were fixed for staining and microscopic analysis. C. albicans hma1Δ/Δ mutants were (A) less adherent and (B) less invasive. (C) The total hyphae length was reduced for hma1Δ/Δ strains but (D) unaltered for the invasive parts of hyphae. The column bars show mean values with standard deviation. Statistical analysis used one-way ANOVA test with Tukey’s multiple comparisons test (**P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001). Sample size (N) corresponds to 3 biological replicates with 36 individual measurements of image sections for A and 24 for B and 50 individual measurements of hyphae lengths for C and D. (For D, hyphae without an invasive part were not included.)
Furthermore, we determined invasiveness, defined here as the proportion of hyphae with an invasive part relative to all adherent fungal cells. , 36.0% of the C. albicans wild-type cells were invasive, while the hma1Δ/Δ mutants were significantly reduced (24.0% and 26.5%; Fig. 6B). The complemented strain regained wild-type levels of invasiveness at 41.3%. Similarly, the total hyphal length in the hma1Δ/Δ strains was reduced to 76.2% and 80.6% of wild type (Fig. 6C), in agreement with our in vitro water-serum medium data. Interestingly, if a hypha had successfully invaded epithelial cells, the length of the invasive part did not differ significantly (Fig. 6D). This indicates that adhered growth and initial invasion, but not subsequent elongation, inside the host cell were inhibited.
Lack of Hma1 attenuates virulence of C. albicans in vivo
Given the in vitro effects of the HMA1 deletion on hyphae, we predicted an impact of the loss of Hma1 (and hence tRNA modifications) on C. albicans virulence. To test this hypothesis, we employed an embryonated chicken egg infection model (32), where we infected eggs with high or low doses of C. albicans wild type, hma1Δ/Δ, or hma1Δ/HMA1. Vitality of the avian embryos acted as the read-out for fungal virulence over 7 days.
As expected, infection with high doses of C. albicans resulted in high lethality within 3 days with all strains (Fig. S8A). However, at a lower infection dose where fungal growth became more relevant, significant differences were observed. Infection resulted in a mortality of 92.5% after day 7 (Fig. 7) for the wild type, but only 27.5% for one hma1Δ/Δ strain, whose isogenic hma1Δ/HMA1 heterozygous strain reverted to an intermediate 52.5%. A second, independent hma1Δ/Δ mutant (strain A) was similarly severely attenuated in virulence, albeit at a slightly lower overall level (Fig. 7). Together, these data show that the Hma1 is required for full virulence of C. albicans.
Fig 7.
Virulence of the C. albicans wild-type strain SC5314, hma1Δ/Δ mutants, and the hma1Δ/HMA1 revertant in infected chicken embryos. Survival after infection is depicted as Kaplan-Meyer plots. There were 20 chicken embryos per group per experiment, and the combined results of two independent experiments are shown. The hma1Δ/Δ mutants exhibited significantly attenuated virulence (***P < 0.001) compared with the wild type. The reconstituted mutant hma1Δ/HMA1 did not fully revert the virulent phenotype but led to a significantly (***P < 0.001) reduced survival compared to its parent, hma1Δ/Δ (B). All significances were calculated by log-rank (Mantel-Cox) test.
Ribosome profiling reveals translational stalling on ANN and non-ANN codons in a hma1∆/∆ mutant
We found that the loss of Hma1 affects ct6A37 tRNA modification formation in C. albicans. Therefore, we performed ribosome profiling to assess mRNA translation in wild type and hma1∆/∆. When cultivated at 30°C—a temperature non-conducive to hyphae formation—we did not observe any difference in codon occupancy patterns for both strains (Fig. 8A). However, incubation at 37°C led to an increase of ribosome occupancy of several codons for the hma1∆/∆ mutant. Half of the 16 codons with predicted t6A modifications differed significantly between wild-type and hma1∆/∆ mutant, which supports the idea that Hma1 acts in the formation of ct6A37 (Fig. 8B). Surprisingly, a large subset of non-ANN codons was also affected. In particular codons which code for arginine or histidine exhibited increased ribosomes occupancy in the hma1∆/∆ strain, as did non-canonical start codons (CTG, GTG, TTG). This may indicate an increased number of non-AUG translation initiation events.
Fig 8.
Ribosome profiling of C. albicans hma1∆/∆ mutant vs wild-type SC5314. C. albicans strains were incubated in YPD medium at 30°C and 37°C, and samples were processed for ribosomal profiling to analyze the codon-specific ribosome stalling and for ribosome sequencing. (A, B) Codon-specific changes in ribosome occupancy in C. albicans hma1∆/∆ in comparison to wild-type SC5314 (geometric mean ± GSD; n = 3). Symbol color reflects statistically significant changes in codon occupancy (two-tailed student’s t-test, Padj <0.05). Selected tRNA isoacceptors were labeled for arginine charged in violet, histidine charged in turquoise, and gray highlights non-canonical start codons. (C, D) Ribosome-protected fragments of the C. albicans hma1∆/∆ mutant and the wild-type SC5314 (both cultivated at 37°C) were sequenced to determine differentially abundant mRNA transcripts. For these, enrichment of gene ontology terms was determined (log2FC ± 1, Padj <0.05) and summarized using the REVIGO tool (63). Log10 P-values, enrichment scores, and gene list sizes for enriched GO terms with less (C) or more (D) abundant transcripts in the hma1∆/∆ vs the wild-type SC5314 are shown.
Finally, we analyzed differences in the overall translatome between hma1Δ/Δ and wild type. At 37°C, we found 731 transcripts to be differentially translated between wild type and hma1∆/∆. Of these, 182 transcripts showed more ribosome reads in the wild type, with an enrichment of the GO processes “filamentation,” “aggregation,” and “biofilm formation” (Fig. 8C). These processes are therefore likely facilitated by Hma1 function. Among the more abundantly found transcripts are those of the adhesin gene ALS1 and the G1 cyclin-related gene HGC1, which is involved in hyphal morphogenesis. Transcripts that were less abundant in the wild-type translatome were enriched for metabolic categories, especially lipid oxidation, glyoxylate cycle, and respiration (Fig. 8D).
DISCUSSION
Cross-species comparison is a powerful tool to elucidate virulence factors
We started our investigation by exploring the genetic differences between C. albicans and its closely related sister species, C. dubliniensis. During evolution, (sub)populations of microorganisms adapt to novel niches, and new, beneficial genetic attributes can get fixed in the population (72), such as the acquisition of specific pathogenic features in response to an adaptation to a new host. Cross-species comparisons therefore can be a powerful tool to obtain novel insights into the genetic basis of species-specific lifestyles of related microorganisms.
While C. albicans appears to have acquired a number of specific virulence factors like hydrolytic enzymes or hyphae-associated genes (27), the less pathogenic C. dubliniensis underwent reductive evolution (26). Similarly, a cross-species transcriptome comparison of these Candida species has revealed differences in expression of orthologous, virulence-associated genes like ECE1 (69, 73). In our cross-species morphological screen (37), we verified SLAD agar as an improved alternative to the often ill-defined classical chlamydospore induction media (28, 74). Its consistent and reproducible composition makes this synthetic medium an excellent candidate for discriminative Candida diagnostics.
The approach to integrate a genomic C. albicans library into C. dubliniensis was first used by Staib and Morschhäuser to discover the role of Nrg1 as a major C. albicans chlamydospore repressor (37). In fact, we found a similar phenotype by integration of the previously uncharacterized C. albicans gene HMA1. The resulting strain was impaired in chlamydospore and pseudohyphae formation. However, Hma1 is not a transcription factor but instead showed high similarities with the S. cerevisiae tRNA threonylcarbamoyladenosine dehydratase Tcd2. Interestingly, a previous large-scale screen in S. cerevisiae revealed that overexpression of TCD2 increased invasive and pseudohyphal growth on nitrogen-sufficient medium (75), a phenotype that is to that of C. albicans hma1Δ/Δ mutants.
Wide-ranging consequences of t6A modifications
The importance of tRNA modifications for cellular processes has been extensively studied in baker’s yeast and humans, as many genetic diseases, including cancer, diabetes, and several neuropathies, are caused by defective tRNA modifications (76, 77), yet the effector mechanisms are poorly understood. Miyauchi et al. discovered the cyclic t6A37 and showed its unexpected wide prevalence in bacteria, fungi, and plants (13). Further investigations into this condensed compound are lacking, but the structural and steric similarities to the t6A precursor molecule allow for functional assumptions. The t6A modification is one of the few conserved tRNA modifications which occurs in all domains of life and is found in all eukaryotic initiator tRNAs (78). Defects in the t6A synthesis pathway are lethal in many prokaryotes (79), including the pathogens Mycoplasma pulmonis (80), Salmonella Typhi (81), and Staphylococcus aureus (82). In fact, strategies to target t6A biosynthesis for antibiotic therapy have been considered (10). Similarly, t6A-defective mutants in S. cerevisiae are retarded in growth (9), while ct6A37-defective strains grow normally (13, 83). This was also reflected by the robust growth that we observed in hma1Δ/Δ mutants in C. albicans and C. dubliniensis. However, treatment with the translation inhibitor cycloheximide reduced the growth of C. albicans hma1Δ/Δ mutants, indicating its function in correct translation, which was not observed in C. dubliniensis. Species-specific responses to translational perturbations have been described previously in yeasts and may contribute to this observation (59).
The molecular function of Hma1 as a tRNA threonylcarbamoyladenosine dehydratase of C. albicans was demonstrated by the accumulation of the ct6A precursor, t6A in the deletion mutants. Direct detection of ct6A, the final product of the proposed Hma1 reaction, could not be achieved quantitatively due to its low stability and its inefficient ionization during mass spectrometric measurements (13). However, we were able to detect trace amounts in the wild type and the revertant but not in the deletion mutant. Based on these findings, we conclude that Hma1 catalyzes the formation of ct6A from t6A in C. albicans—similar to Tcd2, its S. cerevisiae homolog—and is thus critical for correct tRNA modifications in this fungus. The ct6A modification is thought to facilitate proper translation of ANN codon stretches in S. cerevisiae. Surprisingly, numerous codons were slowed down in translation including ANN and non-ANN codons. The highly affected tRNA isoacceptors were those charged with arginine and histidine, which could be a secondary effect due to a general diminished translational activity. The enrichment of non-canonical start codons like CTG could reflect an alternative translational scanning mechanism in response to cellular stress (84). Alternatively, unmodified initiator tRNA may interfere with the decoding of near-cognate start codons.
In S. cerevisiae, Tcd2 is associated with the KEOPS complex, and the knockout of members of this complex blocks or diminishes t6A formation (83). Threonine, in addition to carbonate and ATP, is a substrate in the initial steps of t6A formation, and availability of threonine is therefore a determinant of t6A modification levels (10). Supplementation with L-homoserine, an isomer of threonine, consequently increased the growth rate of S. cerevisiae tcd2Δ (9). In C. albicans, hma1Δ/Δ mutants even outperformed the parental strain when exposed to L-homoserine. Key phenotypes of S. cerevisiae tcd2Δ mutants include elevated sensitivity to heat stress (37°C) (85) and the inability to use non-fermentable carbon sources (13, 86). As shown before, the connection between heat resistance and tRNA modification differs among yeast species depending on their evolutionary history and ecological niche (68). However, the effect was observable in the Candida hma1Δ/Δ deletion mutants as a growth defect at elevated, fever-like temperatures. Moreover C. albicans codon occupancy was remarkably different in the hma1Δ/Δ mutant at 37°C, indicating an important role in normal protein biosynthesis of C. albicans at this temperature, with evident consequences for colonization and infections of endothermic hosts like humans.
Baker’s yeast and Crabtree-negative Candida species differ in their primary carbon catabolism (87), and we consequently observed no correlation between ct6A formation and respiratory growth for the latter. Nevertheless, ribosome profiling showed that genes implicated in lipid and purine metabolism were differentially translated in the hma1Δ/Δ mutant. Finally, levels of many tRNA modifications have been proposed as possible internal markers for exposure to cellular stressors (15, 88). However, we did not observe growth defects for C. albicans hma1Δ/Δ mutants in the presence of osmotic, oxidative, or cell wall stressors. This suggests that Hma1 is not a mediator of the global stress response.
tRNA modification levels affect the TOR pathway and influence morphology
TOR signaling directs complex cellular networks in response to nutrient availability (89). Generally, C. dubliniensis was more sensitive to rapamycin in our growth assays. The reason is not clear, but we note that differences in TOR activation have been linked to the lower rate of filamentation in C. dubliniensis compared to C. albicans (27). Potentially, the basal TOR activity level or the downstream signaling—including modulation by Hma1—may differ between the two fungi (27), making C. dubliniensis more sensitive to perturbations.
The link between the TOR pathway and tRNA modifications like t6A is conserved among the eukaryotes: A loss of modifications at the ASL decreases rapamycin resistance in S. cerevisiae (11, 19), KEOPS complex mutants in Drosophila resemble TOR pathway-deficient mutants (90), disturbance of tRNA-modifying enzymes reduces the TOR-driven cell growth in Arabidopsis thaliana (91), and a hypomodified adenosine residue at position 37 of tRNAs is considered a risk factor for type 2 diabetes in humans as a consequence of mTOR overactivation (92, 93). Interestingly, loss of CdHMA1 in C. dubliniensis elicited rapamycin sensitivity similar to S. cerevisiae t6A-deficient strains (9). The opposite was found for the hma1Δ/Δ gene deletion in C. albicans, which even enhanced resistance to TOR-antagonistic substances. Hence, Hma1 acts—in interplay with the TOR-pathway—in a species-specific manner in these sister species. Whether this is due to intrinsic differences in the enzymatic activity between the two orthologs, different expression pattern or the underlying biological differences between the two species, is an interesting open question for future research.
In addition to the generally conserved TOR functions in nutrient signaling, reduced Tor1 activity contributes to C. albicans yeast-to-hyphae transition via nucleosome repositioning that allows expression of hyphae-associated genes (94). This was reflected by the role of Hma1 in polymorphism: A function in morphological transitions was initially indicated by the integration of C. albicans HMA1 into C. dubliniensis, which changed its morphology under nitrogen and amino acid deficiency. Deletion of HMA1 in C. albicans drastically decreased the conversion of t6A into ct6A, concomitant with a severe decrease in hyphae length and invasiveness into oral epithelial cells. Interestingly, this effect was speciesspecific like the rapamycin response, as C. dubliniensis Cdhma1Δ/Δ mutants formed unaltered hyphae on solid media and even longer filaments in the presence of serum. It cannot be excluded that in addition to the TOR pathway, other regulators of hyphae formation are affected by Hma1 directly or indirectly. Especially the TOR pathway can affect negatively and positively acting transcription factors of hyphae-associated genes, like Nrg1 or Efg1 (64). Future studies may help to disentangle the relative contributions of these regulators to the tRNA modification-driven phenotype.
Filamentation responses of C. albicans are known to depend on the medium used and often differ significantly between liquid and solid media depending on their composition (95). On (semi-) solid media, TOR pathway activity can increase the hyphae formation of C. albicans (64). In contrast, in liquid medium, Tor1 activity favors growth in yeast form of both C. albicans and C. dubliniensis, but hyphae elongation can be enhanced by supplementing rapamycin (29, 94, 96). How does Hma1 compare to these Tor1 effects? In our C. albicans experiments, Hma1 was required for the formation of a hyphal colony fringe specifically on nutrient-rich agar, where rapamycin is known to have no effect (64). Therefore, C. albicans Hma1 appears to be required under conditions where Tor1 activity is dispensable for hyphal growth. Based on these data, we suggest opposing roles for Tor1 and Hma1-mediated ct6A modification in C. albicans. Interestingly, in C. dubliniensis Cdhma1Δ/Δ mutants, both the effect on rapamycin resistance and on hyphae formation was reversed in comparison to C. albicans HMA1 deletion. It further points toward an evolutionary difference in the mutual relationship and the relative contributions of the HMA1 and TOR pathway activity in the two species. The generally higher propensity of C. albicans to form hyphae, compared to C. dubliniensis, can thus be attributed in part to the differences in Hma1 activity, as demonstrated by our gene deletion and transfer experiments. Previously published data show that there is no difference in transcript levels of the HMA1 orthologs between C. albicans and C. dubliniensis, even under conditions where only C. albicans filaments (73). Together with our gene transfer experiments, this indicates that Hma1 function, rather than expression level, may be at the core of the species specificity. It cannot be excluded, of course, that expression levels vary under specific conditions like nutrient availability, with consequences for tRNA modification levels and hyphae formation.
The influence of translation efficiency on C. albicans filamentation is barely understood. Isolated data point to a possible link between tRNA modification and morphology in fungi: low modification levels at the tRNA wobble anticodon in S. cerevisiae elicit defects in starvation-induced agar invasion (97), and genes coding for tRNA-modifying enzymes (UBA4, URM1, NCS2, NCS6) were enriched in a systematic screening of S. cerevisiae deletion mutants with altered colony morphology on solid agar (98). The integrity of tRNA modifications can affect the development of morphological structures in other organisms, from filamentous morphologies in Salmonella (99) to alterations in the auxin-regulated morphogenesis in Arabidopsis (100). Our ribosome profiling in C. albicans identified numerous morphology-associated transcripts enriched in wild-type translation, including key regulators for filamentation and biofilm formation: Hgc1, Brg1, Ras1, and Tec1 (94, 101–103). Recently, a reduced translational efficiency of highly expressed hyphae-associated genes of C. albicans has been reported, indicating a fine-tuning mechanism for translation of these virulence-related genes (104). This process could be regulated in part by tRNA modification patterns, such as ct6A.
In fact, overexpression of the HMA1 ortholog TCD2 in S. cerevisiae is known to result in enhanced pseudohyphal growth as well as increased invasion into nitrogen-sufficient agar (75). Similarly, the reduced hyphae length of C. albicans hma1Δ/Δ was accompanied by impaired invasiveness on agar plates and, importantly, on human epithelium, a phenotype that is linked to general virulence potential (41).
tRNA modifications are required for full virulence
In most bacteria, a defect in t6A modification is lethal (14), and altered levels of prenylation of i6A37 were found it to be crucial to the fitness and virulence of extraintestinal pathogenic Escherichia coli (105). Modifications that target the tRNA wobble bases are known to be essential for the expression of virulence factors of gram-negative bacteria like Pseudomonas aeruginosa (106). In Samonella Typhimurium, disrupted tRNA modification reduces expression of flagellar and secretion type III genes (99). Little is known on the role of tRNA modifications for fungal pathogenesis, but the plant pathogen Colletotrichum lagenarium failed to penetrate host tissue if 7-methylguanosine formation was defective (107)—a phenotype analogous to our C. albicans hma1Δ/Δ mutants. Furthermore, the levels of wobble uridine tRNA modification were recently shown to modulate the virulence of infectious strains of S. cerevisiae and C. albicans (108). Recently, C. albicans Hma1 appeared as a candidate in a large phenotypic screening for damage factors of intestinal epithelia cells (109). Our finding that ECE1 transcript levels are reduced in the mutant, in-line with the shorter hyphae, offers an explanation for this effect. Lower ECE1 transcription likely results in less secretion of candidalysin, the C. albicans toxin that is mainly responsible for epithelial cell damage (39, 69). Together with the reduced adhesion and invasion properties during infection of human oral epithelia in our experiments, this suggests a general virulence defect of these mutants. As such, this would add another layer to the intricate regulation of hyphae formation, which is considered to be a central virulence mechanism of C. albicans. In addition to the established transcriptional and post-translational regulation, we here saw a co-translational regulation—a layer that differentiates the less pathogenic C. dubliniensis from C. albicans.
We hence used an alternative infection model to test the virulence potential of our tRNA modification-defective mutants: embryonated chicken eggs, a model which is known to correlate well with mice infections and which is suitable to predict progress of infection in mammals (110). In this avian model infection with ct6A-defective strains significantly reduced mortality rates, supporting our concept of a role for tRNA modifications for fully expressing Candida virulence factors. Importantly, our in silico analyses showed Hma1 to be fungal specific without a human ortholog. Therefore, tRNA-modifying enzymes could potentially serve as future antifungal targets.
Conclusion
tRNAs, with their modifications, link the transcriptome and the proteome of the cell. So far, tRNA modifications have not been implicated in the virulence and morphogenesis of human fungal pathogens. Based on our data, we propose a model (Fig. 9) where the interplay of ct6A tRNA modification levels and TOR pathway activity influence the filamentation of C. albicans and C. dubliniensis via a common route but with species-specific outcomes. Central in this model is the tRNA-modifying enzyme Hma1, which influences the translation efficiency of many hyphae- and pathogenicity-associated genes. Together with the output of the Tor1-dependent signaling, the combined effect has a strong impact on morphogenesis, invasion, and in consequence virulence of C. albicans. To the best of our knowledge, this is the first study to directly link tRNA modifications to fungal pathogenicity as a co-translational factor in morphogenesis.
Fig 9.
Model presenting the impact of ct6A37 tRNA modification levels on pathogenicity-related features in C. albicans and C. dubliniensis. Environmental signals like high abundance of nutrient sources or a temperature of 37°C increase ct6A tRNA modification levels and Tor1 activity. The latter affects filamentation intensity of C. albicans and C. dubliniensis via transcriptional and post-translational regulation (64), but addition of rapamycin or caffeine inhibits this pathway. Hma1 interacts with the TOR pathway in a species-specific manner and impacts the translation of morphology-, metabolism-, and virulence-associated factors. The interplay of all pathways positively controls filamentation of C. albicans in ct6A-replete cells and is necessary for proper adhesion to and invasion into surfaces like epithelia. Via these pathways, tRNA modifications strongly influence the biologically relevant outcome, virulence—as seen by the correlation between ct6A levels in C. albicans and chicken embryo mortality.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joachim Morschhäuser for providing the genetic library, Rita Müller for help with the ECE1 transcript analyses, our students and technical assistants for their supporting work, and Bernhard Hube for helpful discussions.
This work was supported in part by the Deutsche Forschungsgemeinschaft [SPP2225 to S.B., STA1147/1-1 to B.B., TRR124 to S.V. and S.A.]; the Bundesminsterium für Bildung und Forschung [03Z22JN11 to S.V.]; and the European Research Council [ERC-2012-StG 310489-tRNAmodi to S.A.L.].
Contributor Information
Sascha Brunke, Email: sascha.brunke@leibniz.hki.de.
Gustavo H. Goldman, Universidade de Sao Paulo, Sao Paulo, Brazil
Attila Gacser, Department of Microbiology, University of Szeged, Szeged, Hungry.
Gary Moran, The University of Dublin Trinity College, Dublin, Ireland.
DATA AVAILABILITY
The data sets are available in the following databases: Sequencing data from Candida albicans ribosome profiling experiments – Gene Expression Omnibus GSE199421; Data from RNA mass spectrometry – MetaboLights database MTBLS4439.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.04255-22.
Supplementary Figures S1-S8 and supplementary Tables S1 and S2.
An accounting of the reviewer comments and feedback.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Lamichhane TN, Blewett NH, Crawford AK, Cherkasova VA, Iben JR, Begley TJ, Farabaugh PJ, Maraia RJ. 2013. Lack of tRNA modification isopentenyl-A37 alters mRNA decoding and causes metabolic deficiencies in fission yeast. Mol Cell Biol 33:2918–2929. doi: 10.1128/MCB.00278-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gu C, Begley TJ, Dedon PC. 2014. tRNA modifications regulate translation during cellular stress. FEBS Lett 588:4287–4296. doi: 10.1016/j.febslet.2014.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Phizicky EM, Hopper AK. 2015. tRNA processing, modification, and subcellular dynamics: past, present, and future. RNA 21:483–485. doi: 10.1261/rna.049932.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. El Yacoubi B, Bailly M, de Crécy-Lagard V. 2012. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet 46:69–95. doi: 10.1146/annurev-genet-110711-155641 [DOI] [PubMed] [Google Scholar]
- 5. Konevega AL, Soboleva NG, Makhno VI, Semenkov YP, Wintermeyer W, Rodnina MV, Katunin VI. 2004. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA 10:90–101. doi: 10.1261/rna.5142404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shigi N. 2014. Biosynthesis and functions of sulfur modifications in tRNA. Front Genet 5:67. doi: 10.3389/fgene.2014.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schaffrath R, Leidel SA. 2017. Wobble uridine modifications-a reason to live, a reason to die?! RNA Biol 14:1209–1222. doi: 10.1080/15476286.2017.1295204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, Karzai AW, Sternglanz R. 2011. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J 30:873–881. doi: 10.1038/emboj.2010.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thiaville PC, Legendre R, Rojas-Benítez D, Baudin-Baillieu A, Hatin I, Chalancon G, Glavic A, Namy O, de Crécy-Lagard V. 2016. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb Cell 3:29–45. doi: 10.15698/mic2016.01.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deutsch C, El Yacoubi B, de Crécy-Lagard V, Iwata-Reuyl D. 2012. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J Biol Chem 287:13666–13673. doi: 10.1074/jbc.M112.344028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rojas-Benitez D, Thiaville PC, de Crécy-Lagard V, Glavic A. 2015. The levels of a universally conserved tRNA modification regulate cell growth. J Biol Chem 290:18699–18707. doi: 10.1074/jbc.M115.665406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elkins BN, Keller EB. 1974. The enzymatic synthesis of N-(purin-6-ylcarbamoyl)threonine, an anticodon-adjacent base in transfer ribonucleic acid. Biochemistry 13:4622–4628. doi: 10.1021/bi00719a024 [DOI] [PubMed] [Google Scholar]
- 13. Miyauchi K, Kimura S, Suzuki T. 2013. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem Biol 9:105–111. doi: 10.1038/nchembio.1137 [DOI] [PubMed] [Google Scholar]
- 14. Thiaville PC, El Yacoubi B, Köhrer C, Thiaville JJ, Deutsch C, Iwata-Reuyl D, Bacusmo JM, Armengaud J, Bessho Y, Wetzel C, Cao X, Limbach PA, RajBhandary UL, de Crécy-Lagard V. 2015. Essentiality of threonylcarbamoyladenosine (t6A), a universal tRNA modification, in bacteria. Mol Microbiol 98:1199–1221. doi: 10.1111/mmi.13209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan CTY, Deng W, Li F, DeMott MS, Babu IR, Begley TJ, Dedon PC. 2015. Highly predictive reprogramming of tRNA modifications is linked to selective expression of codon-biased genes. Chem Res Toxicol 28:978–988. doi: 10.1021/acs.chemrestox.5b00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nedialkova DD, Leidel SA. 2015. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161:1606–1618. doi: 10.1016/j.cell.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zaborske JM, Narasimhan J, Jiang L, Wek SA, Dittmar KA, Freimoser F, Pan T, Wek RC. 2009. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J Biol Chem 284:25254–25267. doi: 10.1074/jbc.M109.000877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin CA, Ellis SR, True HL. 2010. The Sua5 protein is essential for normal translational regulation in yeast. Mol Cell Biol 30:354–363. doi: 10.1128/MCB.00754-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leidel S, Pedrioli PGA, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. 2009. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458:228–232. doi: 10.1038/nature07643 [DOI] [PubMed] [Google Scholar]
- 20. Gupta R, Walvekar AS, Liang S, Rashida Z, Shah P, Laxman S. 2019. A tRNA modification balances carbon and nitrogen metabolism by regulating phosphate homeostasis. Elife 8:e44795. doi: 10.7554/eLife.44795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laxman S, Tu BP. 2011. Multiple TORC1-associated proteins regulate nitrogen starvation-dependent cellular differentiation in Saccharomyces cerevisiae. PLoS One 6:e26081. doi: 10.1371/journal.pone.0026081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946 [DOI] [PubMed] [Google Scholar]
- 23. Almirante B, Rodríguez D, Park BJ, Cuenca-Estrella M, Planes AM, Almela M, Mensa J, Sanchez F, Ayats J, Gimenez M, Saballs P, Fridkin SK, Morgan J, Rodriguez-Tudela JL, Warnock DW, Pahissa A, Barcelona Candidemia Project Study Group . 2005. Epidemiology and predictors of mortality in cases of Candida bloodstream infection: results from population-based surveillance, Barcelona, Spain, from 2002 to 2003. J Clin Microbiol 43:1829–1835. doi: 10.1128/JCM.43.4.1829-1835.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khan Z, Ahmad S, Joseph L, Chandy R. 2012. Candida dubliniensis: an appraisal of its clinical significance as a bloodstream pathogen. PLoS One 7:e32952. doi: 10.1371/journal.pone.0032952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sullivan D, Coleman D. 1998. Candida dubliniensis: characteristics and identification. J Clin Microbiol 36:329–334. doi: 10.1128/JCM.36.2.329-334.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jackson AP, Gamble JA, Yeomans T, Moran GP, Saunders D, Harris D, Aslett M, Barrell JF, Butler G, Citiulo F, et al. 2009. Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res 19:2231–2244. doi: 10.1101/gr.097501.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moran GP, Coleman DC, Sullivan DJ. 2012. Candida albicans versus Candida dubliniensis: why is C. albicans more pathogenic? Int J Microbiol 2012:205921. doi: 10.1155/2012/205921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Staib P, Morschhäuser J. 1999. Chlamydospore formation on Staib agar as a species-specific characteristic of Candida dubliniensis. Mycoses 42:521–524. doi: 10.1046/j.1439-0507.1999.00516.x [DOI] [PubMed] [Google Scholar]
- 29. Sullivan DJ, Moran GP. 2011. Differential virulence of Candida albicans and C. dubliniensis: a role for Tor1 kinase? Virulence 2:77–81. doi: 10.4161/viru.2.1.15002 [DOI] [PubMed] [Google Scholar]
- 30. Stokes C, Moran GP, Spiering MJ, Cole GT, Coleman DC, Sullivan DJ. 2007. Lower filamentation rates of Candida dubliniensis contribute to its lower virulence in comparison with Candida albicans. Fungal Genet Biol 44:920–931. doi: 10.1016/j.fgb.2006.11.014 [DOI] [PubMed] [Google Scholar]
- 31. Böttcher B, Palige K, Jacobsen ID, Hube B, Brunke S. 2015. Csr1/Zap1 maintains zinc homeostasis and influences virulence in Candida dubliniensis but Is not coupled to morphogenesis. Eukaryot Cell 14:661–670. doi: 10.1128/EC.00078-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jacobsen ID, Grosse K, Berndt A, Hube B. 2011. Pathogenesis of Candida albicans infections in the alternative chorio-allantoic membrane chicken embryo model resembles systemic murine infections. PLoS One 6:e19741. doi: 10.1371/journal.pone.0019741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5'-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198:179–182. doi: 10.1007/BF00328721 [DOI] [PubMed] [Google Scholar]
- 34. Palige K, Linde J, Martin R, Böttcher B, Citiulo F, Sullivan DJ, Weber J, Staib C, Rupp S, Hube B, Morschhäuser J, Staib P. 2013. Global transcriptome sequencing identifies chlamydospore specific markers in Candida albicans and Candida dubliniensis. PLoS One 8:e61940. doi: 10.1371/journal.pone.0061940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morschhäuser J, Ruhnke M, Michel S, Hacker J. 1999. Identification of CARE-2-negative Candida albicans isolates as Candida dubliniensis. Mycoses 42:29–32. doi: 10.1046/j.1439-0507.1999.00259.x [DOI] [PubMed] [Google Scholar]
- 36. Staib P, Moran GP, Sullivan DJ, Coleman DC, Morschhäuser J. 2001. Isogenic strain construction and gene targeting in Candida dubliniensis. J Bacteriol 183:2859–2865. doi: 10.1128/JB.183.9.2859-2865.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Staib P, Morschhäuser J. 2005. Differential expression of the NRG1 repressor controls species-specific regulation of chlamydospore development in Candida albicans and Candida dubliniensis. Mol Microbiol 55:637–652. doi: 10.1111/j.1365-2958.2004.04414.x [DOI] [PubMed] [Google Scholar]
- 38. Biswas K, Morschhäuser J. 2005. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol Microbiol 56:649–669. doi: 10.1111/j.1365-2958.2005.04576.x [DOI] [PubMed] [Google Scholar]
- 39. Mogavero S, Sauer FM, Brunke S, Allert S, Schulz D, Wisgott S, Jablonowski N, Elshafee O, Krüger T, Kniemeyer O, Brakhage AA, Naglik JR, Dolk E, Hube B. 2021. Candidalysin delivery to the invasion pocket is critical for host epithelial damage induced by Candida albicans. Cell Microbiol 23:e13378. doi: 10.1111/cmi.13378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sheth CC, Johnson E, Baker ME, Haynes K, Mühlschlegel FA. 2005. Phenotypic identification of Candida albicans by growth on chocolate agar. Med Mycol 43:735–738. doi: 10.1080/13693780500265998 [DOI] [PubMed] [Google Scholar]
- 41. Wächtler B, Citiulo F, Jablonowski N, Förster S, Dalle F, Schaller M, Wilson D, Hube B. 2012. Candida albicans-epithelial interactions: dissecting the roles of active penetration, induced endocytosis and host factors on the infection process. PLoS One 7:e36952. doi: 10.1371/journal.pone.0036952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hennicke F, Grumbt M, Lermann U, Ueberschaar N, Palige K, Böttcher B, Jacobsen ID, Staib C, Morschhäuser J, Monod M, Hube B, Hertweck C, Staib P. 2013. Factors supporting cysteine tolerance and sulfite production in Candida albicans. Eukaryot Cell 12:604–613. doi: 10.1128/EC.00336-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Staib P, Lermann U, Blass-Warmuth J, Degel B, Würzner R, Monod M, Schirmeister T, Morschhäuser J. 2008. Tetracycline-inducible expression of individual secreted aspartic proteases in Candida albicans allows isoenzyme-specific inhibitor screening. Antimicrob Agents Chemother 52:146–156. doi: 10.1128/AAC.01072-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Köhler GA, White TC, Agabian N. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J Bacteriol 179:2331–2338. doi: 10.1128/jb.179.7.2331-2338.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hoffman CS, Winston F. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267–272. doi: 10.1016/0378-1119(87)90131-4 [DOI] [PubMed] [Google Scholar]
- 46. Reuss O, Vik A, Kolter R, Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. doi: 10.1016/j.gene.2004.06.021 [DOI] [PubMed] [Google Scholar]
- 47. Lecanda A, Nilges BS, Sharma P, Nedialkova DD, Schwarz J, Vaquerizas JM, Leidel SA. 2016. Dual randomization of oligonucleotides to reduce the bias in ribosome-profiling libraries. Methods 107:89–97. doi: 10.1016/j.ymeth.2016.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sarin LP, Kienast SD, Leufken J, Ross RL, Dziergowska A, Debiec K, Sochacka E, Limbach PA, Fufezan C, Drexler HCA, Leidel SA. 2018. Nano LC-MS using capillary columns enables accurate quantification of modified ribonucleosides at low femtomol levels. RNA 24:1403–1417. doi: 10.1261/rna.065482.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martens L, Chambers M, Sturm M, Kessner D, Levander F, Shofstahl J, Tang WH, Römpp A, Neumann S, Pizarro AD, Montecchi-Palazzi L, Tasman N, Coleman M, Reisinger F, Souda P, Hermjakob H, Binz P-A, Deutsch EW. 2011. mzML—a community standard for mass spectrometry data. Mol Cell Proteomics 10:R110.000133. doi: 10.1074/mcp.R110.000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, et al. 2012. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30:918–920. doi: 10.1038/nbt.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kösters M, Leufken J, Schulze S, Sugimoto K, Klein J, Zahedi RP, Hippler M, Leidel SA, Fufezan C. 2018. pymzML v2. 0: introducing a highly compressed and seekable gzip format. Bioinformatics 34:2513–2514. doi: 10.1093/bioinformatics/bty046 [DOI] [PubMed] [Google Scholar]
- 52. Leufken J, Niehues A, Sarin LP, Wessel F, Hippler M, Leidel SA, Fufezan C. 2017. pyQms enables universal and accurate quantification of mass spectrometry data. Mol Cell Proteomics 16:1736–1745. doi: 10.1074/mcp.M117.068007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46:D303–D307. doi: 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, et al. 2020. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17:261–272. doi: 10.1038/s41592-019-0686-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Varoquaux G, Buitinck L, Louppe G, Grisel O, Pedregosa F, Mueller A. 2015. Scikit-learn: machine learning without learning the machinery. GetMobile Mob Comput Commun 19:29–33. doi: 10.1145/2786984.2786995 [DOI] [Google Scholar]
- 56. Seabold S, Perktold J. Proceedings of the 9th Python in science conference, Vol. 57, p 61, Austin, TX [Google Scholar]
- 57. Leufken J, Niehues A, Sarin LP, Wessel F, Hippler M, Leidel SA, Fufezan C. 2017. pyQms enables universal and accurate quantification of mass spectrometry data. Mol Cell Proteomics 16:1736–1745. doi: 10.1074/mcp.M117.068007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. 2012. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7:1534–1550. doi: 10.1038/nprot.2012.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sharma P, Nilges BS, Wu J, Leidel SA. 2019. The translation inhibitor cycloheximide affects ribosome profiling data in a species-specific manner. bioRxiv. doi: 10.1101/746255 [DOI]
- 60. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Inglis DO, Skrzypek MS, Arnaud MB, Binkley J, Shah P, Wymore F, Sherlock G. 2013. Improved gene ontology annotation for biofilm formation, filamentous growth, and phenotypic switching in Candida albicans. Eukaryot Cell 12:101–108. doi: 10.1128/EC.00238-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLOS ONE 6:e21800. doi: 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bastidas RJ, Heitman J, Cardenas ME. 2009. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog 5:e1000294. doi: 10.1371/journal.ppat.1000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Takahara T, Maeda T. 2013. Evolutionarily conserved regulation of TOR signalling. J Biochem 154:1–10. doi: 10.1093/jb/mvt047 [DOI] [PubMed] [Google Scholar]
- 66. Kingsbury JM, McCusker JH. 2010. Homoserine toxicity in Saccharomyces cerevisiae and Candida albicans homoserine kinase (thr1Δ) mutants. Eukaryot Cell 9:717–728. doi: 10.1128/EC.00044-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Damon JR, Pincus D, Ploegh HL. 2015. tRNA thiolation links translation to stress responses in Saccharomyces cerevisiae. Mol Biol Cell 26:270–282. doi: 10.1091/mbc.E14-06-1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Alings F, Sarin LP, Fufezan C, Drexler HCA, Leidel SA. 2015. An evolutionary approach uncovers a diverse response of tRNA 2-thiolation to elevated temperatures in yeast. RNA 21:202–212. doi: 10.1261/rna.048199.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Höfs S, Gratacap RL, Robbins J, Runglall M, et al. 2016. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532:64–68. doi: 10.1038/nature17625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Birse CE, Irwin MY, Fonzi WA, Sypherd PS. 1993. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect Immun 61:3648–3655. doi: 10.1128/iai.61.9.3648-3655.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Garbe E, Thielemann N, Hohner S, Kumar A, Vylkova S, Kurzai O, Martin R. 2023. Functional analysis of the Candida albicans ECE1 Promoter. Microbiol Spectr 11:e0025323. doi: 10.1128/spectrum.00253-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Boero F. 2015. From Darwin’s origin of species toward a theory of natural history. F1000Prime Rep 7:49. doi: 10.12703/P7-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Grumaz C, Lorenz S, Stevens P, Lindemann E, Schöck U, Retey J, Rupp S, Sohn K. 2013. Species and condition specific adaptation of the transcriptional landscapes in Candida albicans and Candida dubliniensis. BMC Genomics 14:212. doi: 10.1186/1471-2164-14-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Al Mosaid A, Sullivan DJ, Coleman DC. 2003. Differentiation of Candida dubliniensis from Candida albicans on Pal's agar. J Clin Microbiol 41:4787–4789. doi: 10.1128/JCM.41.10.4787-4789.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shively CA, Eckwahl MJ, Dobry CJ, Mellacheruvu D, Nesvizhskii A, Kumar A. 2013. Genetic networks inducing invasive growth in Saccharomyces cerevisiae identified through systematic genome-wide overexpression. Genetics 193:1297–1310. doi: 10.1534/genetics.112.147876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Torres AG, Batlle E, Ribas de Pouplana L. 2014. Role of tRNA modifications in human diseases. Trends Mol Med 20:306–314. doi: 10.1016/j.molmed.2014.01.008 [DOI] [PubMed] [Google Scholar]
- 77. Sarin LP, Leidel SA. 2014. Modify or die?-RNA modification defects in metazoans. RNA Biol 11:1555–1567. doi: 10.4161/15476286.2014.992279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Asano K. 2014. Why is start codon selection so precise in eukaryotes? Translation (Austin) 2:e28387. doi: 10.4161/trla.28387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset J-P, Iwata-Reuyl D, Murzin AG, de Crécy-Lagard V. 2011. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J 30:882–893. doi: 10.1038/emboj.2010.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. French CT, Lao P, Loraine AE, Matthews BT, Yu H, Dybvig K. 2008. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol Microbiol 69:67–76. doi: 10.1111/j.1365-2958.2008.06262.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Langridge GC, Phan M-D, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, Pleasance SJ, Peters SE, Maskell DJ, Charles IG. 2009. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH). BMC Genomics 10:291. doi: 10.1186/1471-2164-10-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Thiaville PC, Iwata-Reuyl D, de Crécy-Lagard V. 2014. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t6A), a universal modification of tRNA. RNA Biol 11:1529–1539. doi: 10.4161/15476286.2014.992277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kearse MG, Wilusz JE. 2017. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev 31:1717–1731. doi: 10.1101/gad.305250.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sinha H, David L, Pascon RC, Clauder-Münster S, Krishnakumar S, Nguyen M, Shi G, Dean J, Davis RW, Oefner PJ, McCusker JH, Steinmetz LM. 2008. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics 180:1661–1670. doi: 10.1534/genetics.108.092932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Qian W, Ma D, Xiao C, Wang Z, Zhang J. 2012. The genomic landscape and evolutionary resolution of antagonistic pleiotropy in yeast. Cell Rep 2:1399–1410. doi: 10.1016/j.celrep.2012.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Askew C, Sellam A, Epp E, Hogues H, Mullick A, Nantel A, Whiteway M. 2009. Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog 5:e1000612. doi: 10.1371/journal.ppat.1000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dedon PC, Begley TJ. 2014. A system of RNA modifications and biased codon use controls cellular stress response at the level of translation. Chem Res Toxicol 27:330–337. doi: 10.1021/tx400438d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Loewith R, Hall MN. 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189:1177–1201. doi: 10.1534/genetics.111.133363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rojas-Benítez D, Ibar C, Glavic Á. 2013. The Drosophila EKC/KEOPS complex: roles in protein synthesis homeostasis and animal growth. Fly (Austin) 7:168–172. doi: 10.4161/fly.25227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Leiber R-M, John F, Verhertbruggen Y, Diet A, Knox JP, Ringli C. 2010. The TOR pathway modulates the structure of cell walls in Arabidopsis. Plant Cell 22:1898–1908. doi: 10.1105/tpc.109.073007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Blagosklonny MV. 2013. TOR-centric view on insulin resistance and diabetic complications: perspective for endocrinologists and gerontologists. Cell Death Dis 4:e964. doi: 10.1038/cddis.2013.506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wei F-Y, Tomizawa K. 2011. Functional loss of Cdkal1, a novel tRNA modification enzyme, causes the development of type 2 diabetes. Endocr J 58:819–825. doi: 10.1507/endocrj.ej11-0099 [DOI] [PubMed] [Google Scholar]
- 94. Lu Y, Su C, Liu H. 2012. A GATA transcription factor recruits Hda1 in response to reduced Tor1 signaling to establish a hyphal chromatin state in Candida albicans. PLoS Pathog 8:e1002663. doi: 10.1371/journal.ppat.1002663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Azadmanesh J, Gowen AM, Creger PE, Schafer ND, Blankenship JR. 2017. Filamentation involves two overlapping, but distinct, programs of filamentation in the pathogenic fungus Candida albicans. G3 (Bethesda) 7:3797–3808. doi: 10.1534/g3.117.300224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Su C, Lu Y, Liu H. 2013. Reduced TOR signaling sustains hyphal development in Candida albicans by lowering Hog1 basal activity. Mol Biol Cell 24:385–397. doi: 10.1091/mbc.E12-06-0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Goehring AS, Rivers DM, Sprague Jr GF. 2003. Urmylation: a ubiquitin-like pathway that functions during invasive growth and budding in yeast. Mol Biol Cell 14:4329–4341. doi: 10.1091/mbc.e03-02-0079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Voordeckers K, De Maeyer D, van der Zande E, Vinces MD, Meert W, Cloots L, Ryan O, Marchal K, Verstrepen KJ. 2012. Identification of a complex genetic network underlying Saccharomyces cerevisiae colony morphology. Mol Microbiol 86:225–239. doi: 10.1111/j.1365-2958.2012.08192.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Shippy DC, Heintz JA, Albrecht RM, Eakley NM, Chopra AK, Fadl AA. 2012. Deletion of glucose-inhibited division (gidA) gene alters the morphological and replication characteristics of Salmonella enterica Serovar typhimurium. Arch Microbiol 194:405–412. doi: 10.1007/s00203-011-0769-7 [DOI] [PubMed] [Google Scholar]
- 100. Leitner J, Retzer K, Malenica N, Bartkeviciute R, Lucyshyn D, Jäger G, Korbei B, Byström A, Luschnig C. 2015. Meta-regulation of Arabidopsis auxin responses depends on tRNA maturation. Cell Rep 11:516–526. doi: 10.1016/j.celrep.2015.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138. doi: 10.1016/j.cell.2011.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Feng Q, Summers E, Guo B, Fink G. 1999. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J Bacteriol 181:6339–6346. doi: 10.1128/JB.181.20.6339-6346.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zheng X, Wang Y, Wang Y. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J 23:1845–1856. doi: 10.1038/sj.emboj.7600195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Mundodi V, Choudhary S, Smith AD, Kadosh D, Dudley A. 2021. Global translational landscape of the Candida albicans morphological transition. G3 (Bethesda) 11:jkaa043. doi: 10.1093/g3journal/jkaa043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Fleming BA, Blango MG, Rousek AA, Kincannon WM, Tran A, Lewis AJ, Russell CW, Zhou Q, Baird LM, Barber AE, Brannon JR, Beebout CJ, Bandarian V, Hadjifrangiskou M, Howard MT, Mulvey MA. 2022. A tRNA modifying enzyme as a tunable regulatory nexus for bacterial stress responses and virulence. Nucleic Acids Res 50:7570–7590. doi: 10.1093/nar/gkac116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Shippy DC, Fadl AA. 2014. tRNA modification enzymes GidA and MnmE: potential role in virulence of bacterial pathogens. Int J Mol Sci 15:18267–18280. doi: 10.3390/ijms151018267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Takano Y, Takayanagi N, Hori H, Ikeuchi Y, Suzuki T, Kimura A, Okuno T. 2006. A gene involved in modifying transfer RNA is required for fungal pathogenicity and stress tolerance of Colletotrichum lagenarium. Mol Microbiol 60:81–92. doi: 10.1111/j.1365-2958.2006.05080.x [DOI] [PubMed] [Google Scholar]
- 108. Alings F, Scharmann K, Eggers C, Böttcher B, Sokołowski M, Shvetsova E, Sharma P, Roth J, Rashiti L, Glatt S, Brunke S, Leidel SA. 2023. Ncs2* mediates in vivo virulence of pathogenic yeast through sulphur modification of cytoplasmic transfer RNA. Nucleic Acids Res 51:8133–8149. doi: 10.1093/nar/gkad564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Allert S, Förster TM, Svensson C-M, Richardson JP, Pawlik T, Hebecker B, Rudolphi S, Juraschitz M, Schaller M, Blagojevic M, Morschhäuser J, Figge MT, Jacobsen ID, Naglik JR, Kasper L, Mogavero S, Hube B. 2018. Candida albicans-induced epithelial damage mediates translocation through intestinal barriers. mBio 9:e00915-00918. doi: 10.1128/mBio.00915-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jacobsen ID, Grosse K, Hube B. 2012. Embryonated chicken eggs as alternative infection model for pathogenic fungi. Methods Mol Biol 845:487–496. doi: 10.1007/978-1-61779-539-8_34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-S8 and supplementary Tables S1 and S2.
An accounting of the reviewer comments and feedback.
Data Availability Statement
The data sets are available in the following databases: Sequencing data from Candida albicans ribosome profiling experiments – Gene Expression Omnibus GSE199421; Data from RNA mass spectrometry – MetaboLights database MTBLS4439.