Inhibiting autophagy in pancreatic cancer cells enhances intracellular accumulation of tumor antigens to induce dendritic cell activation and synergizes with immunotherapy to markedly inhibit the growth of pancreatic ductal adenocarcinoma.
Abstract
The effect of immune checkpoint inhibitors is extremely limited in patients with pancreatic ductal adenocarcinoma (PDAC) due to the suppressive tumor immune microenvironment. Autophagy, which has been shown to play a role in antitumor immunity, has been proposed as a therapeutic target for PDAC. In this study, single-cell RNA sequencing of autophagy-deficient murine PDAC tumors revealed that autophagy inhibition in cancer cells induced dendritic cell (DC) activation. Analysis of human PDAC tumors substantiated a negative correlation between autophagy and DC activation signatures. Mechanistically, autophagy inhibition increased the intracellular accumulation of tumor antigens, which could activate DCs. Administration of chloroquine, an autophagy inhibitor, in combination with Flt3 ligand–induced DC infiltration inhibited tumor growth and increased tumor-infiltrating T lymphocytes. However, autophagy inhibition in cancer cells also induced CD8+ T-cell exhaustion with high expression of immune checkpoint LAG3. A triple-therapy comprising chloroquine, Flt3 ligand, and an anti-LAG3 antibody markedly reduced tumor growth in orthotopic syngeneic PDAC mouse models. Thus, targeting autophagy in cancer cells and activating DCs sensitize PDAC tumors to immune checkpoint inhibitor therapy, warranting further development of this treatment approach to overcome immunosuppression in pancreatic cancer.
Significance: Inhibiting autophagy in pancreatic cancer cells enhances intracellular accumulation of tumor antigens to induce dendritic cell activation and synergizes with immunotherapy to markedly inhibit the growth of pancreatic ductal adenocarcinoma.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a lethal cancer with a much lower 5-year survival rate (13%) than that of other solid cancers (1). Although the recent development of immune checkpoint inhibitors (ICI) has markedly advanced the treatment of cancer (2, 3), the therapeutic effects of ICIs are extremely limited in patients with PDAC due to the suppressive tumor immune microenvironment (TIME), lack of target expression (e.g., PD-L1), and lack of antigens for recognition (4, 5). However, it was reported that patients with microsatellite instability–high pancreatic cancer responded exceptionally well to ICIs, and patients with PDAC with higher levels of neoantigens and tumor-infiltrating CD8+ T cells exhibited longer survival rates (6, 7). In addition, several studies have reported that the association between infiltration of T cells and patient outcomes was dependent on the TIME, particularly immunosuppressive granulocytes and macrophages (8, 9). These reports suggest that ICIs could effectively treat patients with PDAC under an antitumorigenic TIME.
Dendritic cells (DC) are the most important antigen-presenting cells (APC) for T-cell priming, which is essential in the initiation phase of antitumor adaptive immunity (10, 11). Recent studies have revealed that DCs are often dysfunctional in the PDAC TIME, meaning that they are unable to effectively infiltrate the tumor or present tumor antigens to T cells (12, 13), which contributes to the formation of the suppressive TIME observed in PDAC. These studies provide strong evidence that improving DC infiltration into the tumor and reversing DC dysfunction could be promising approaches in the development of effective PDAC immunotherapies.
Autophagy, a highly conserved self-degradation process, which is critical for maintaining cellular homeostasis under conditions of stress, protects cancer cells from various stressors and promotes their survival (14, 15). In preclinical models, inhibiting cancer cell autophagy has been reported as a promising therapeutic target (16–18). Indeed, several clinical trials have shown the efficacy of hydroxychloroquine in combination with chemotherapy as a preoperative treatment for patients with PDAC (19–21); however, an autophagy inhibitor has not yet been approved for clinical use, with a lack of clear evidence of survival improvements. Recently, autophagy has also been reported to be associated with tumor immunity. For instance, inhibiting autophagy in cancer cells enhances antitumor immunity by increasing the expression of MHC class I (MHC I) on the tumor surface (22), sensitizing MHC I–deficient cancer cells to apoptosis by T-cell–derived cytokines (23), or causing tumors to secrete NK cell–activating chemokines (24). These reports have led to increased interest in autophagy inhibitors as candidates for use as part of antitumor combination immunotherapies, and an ongoing study is attempting multi-agent combinations with ICIs (NCT04214418). However, the effects of autophagy inhibition on the TIME have not been yet fully revealed, and the best way to combine autophagy inhibition with immunotherapy has not yet been established.
In this study, we performed single-cell RNA sequencing (scRNA-seq) of autophagy-deficient murine syngeneic PDAC tumors to further evaluate the impact of autophagy inhibition on the TIME and discovered that the expression of genes associated with DC function was increased in autophagy-deficient tumors. We hypothesized that inhibiting autophagy in cancer cells may lead to the intracellular accumulation of proteins, which may then serve as cancer antigens, thereby increasing the antigenicity of cancer cells. Thus, we sought to determine the correlation between autophagy in cancer cells and the cross-presentation capacity of DCs. Additionally, we aimed to identify a novel immune activation mechanism, which could be exploited to develop new autophagy inhibitor–compatible combination immunotherapies for patients with PDAC.
Materials and Methods
Cells
Primary human PDAC cells were collected from a patient with PDAC with ascites during the treatment process at the Department of Cancer Immunotherapy, Fukuoka General Cancer Clinic. Written informed consent from the patient was obtained. Murine PDAC cells and cancer-associated fibroblasts (CAF) were established from primary pancreatic tumors of LSL-KrasG12D/+, LSL-Trp53R172H/+, Pdx-1-Cre (KPC) mice using the outgrowth method described previously (25). The human and mouse cell lines were cryopreserved after being established in our laboratories. All cell lines were used within eight passages after revival from frozen stocks and were certified Mycoplasma-free using PCR performed by International Council for Laboratory Animal Science (ICLAS) Monitoring Center, Central Institution for Experimental Medicine and Life Science. These cell lines did not undergo authentication.
Human PDAC cells, murine PDAC cells, and murine CAFs were maintained in DMEM (Sigma-Aldrich) supplemented with 10% FBS, streptomycin (100 mg/mL), and penicillin (100 mg/mL) at 37°C in a humidified atmosphere containing 10% CO2. DCs and T cells were maintained in RPMI 1640 medium supplemented with 10% FBS, streptomycin (100 mg/mL), and penicillin (100 mg/mL) at 37°C in a humidified atmosphere containing 5% CO2.
Mice
Five to eight weeks old female and male C57BL/6 mice and BALB/c AJcl nu/nu mice were purchased from CLEA Inc. The generations of KPC mice and OT-1 transgenic mice have been described previously (26, 27). All animal experiment protocols were approved by the Ethics Committee of Kyushu University and the guidelines of the Institutional Animal Committee of Kyushu University (approval numbers: A22-128, A22-160, A22-161, and A22-339).
For the s.c. syngeneic models, KPC cells were suspended in 50 µL 50% Matrigel (in a 1:1 Matrigel to PBS ratio) per mouse and injected subcutaneously into the flank of 6-week-old female syngeneic C57BL/6 mice. For the immunodeficient models, KPC cells were injected into the flank of BALB/c-nu mice.
For the orthotopic syngeneic models, KPC cells were suspended in 50 µL 50% Matrigel per mouse and injected into the pancreas of 6-week-old female and male C57BL/6 mice.
In vivo tumor experiments
For the s.c. syngeneic PDAC models, 2.5 × 105 KPC1-shNC/shATG7 cells were suspended in 50 µL 50% Matrigel (in a 1:1 Matrigel to PBS ratio) per mouse and injected subcutaneously into the flank of 6-week-old female syngeneic C57BL/6 mice.
For the s.c. immunodeficient PDAC models, 1.0 × 105 KPC1-shNC/shATG7 cells were suspended in 50 µL 50% Matrigel and injected into the flank of BALB/c-nu mice.
For the orthotopic syngeneic PDAC models, 1.0 × 105 KPC1-shNC/shATG7, 5.0 × 105 KPC2-shNC/shATG7 cells, 1.0 × 105 KPC1-shNC/shATG5 cells, 5.0 × 105 KPC2-shNC/shATG5 cells, or 5.0 × 105 KPC2-shNC/shATG4B cells were suspended in 50 µL 50% Matrigel per mouse and injected into the pancreas of 6-week-old female C57BL/6 mice.
The orthotopic syngeneic PDAC models were used for the treatment experiments. For the chloroquine (CQ) and Flt3 ligand (Flt3L) combination therapy experiments, mice were treated with CQ (60 mg/kg dissolved in 50 µL PBS, i.p., daily, #C6628, Sigma-Aldrich) or vehicle (50 µL PBS) and/or Flt3L (10 µg, i.p., daily for 9 days, #BE0098, Bio X Cell) or vehicle (IgG isotype control, BE0096, #Bio X Cell). For the CQ, Flt3L, and anti-LAG3 (aLAG3) triple-therapy experiments, female and male mice were treated with CQ or vehicle, and/or Flt3L or vehicle, and/or aLAG3 (50 µg, i.p., every 3 days, #BE0174, Bio X Cell) or vehicle (IgG isotype control, #BE0088, Bio X Cell).
The s.c. syngeneic PDAC models were used for the experiment of CD8+ T-cell depletion. Mice received anti-CD8a (100 µg, i.p. on days −2, 0, 7, and 14, #BE0061, Bio X Cell) or vehicle (IgG2b isotype control, #BE0090, Bio X Cell).
Tumor volume was calculated using the following formula: π/6 × L × W × W, in which L is the largest tumor diameter and W is the smallest.
In all mouse experiments, the animals were euthanized if they lost more than 20% of their body weight in 7 days, showed evidence of abdominal distention due to tumor growth or ascites, had rapid breathing, had difficulty feeding or ingesting water, or presented with external findings with no sign of recovery. In the s.c. syngeneic PDAC models, mice were promptly euthanized when the tumor reached a maximum diameter of 20 mm. All euthanasia and surgical procedures were performed under general anesthesia with medetomidine 0.3 mg/kg + midazolam 4.0 mg/kg + butorphanol 5.0 mg/kg. In the treatment experiments, the mice were randomized into the treatment groups. In a preliminary experiment using three mice, the mean and SD of tumor sizes were estimated to decide the correct sample size. All mouse experiments, except for the preliminary ones using subcutaneously implanted models, were initiated with six mice per condition; mice that met the above conditions during observation but died unexpectedly the day after transplantation or had numerous peritoneal nodules due to leakage during transplantation were excluded. Outcome measures, tumor volume, and tumor weight were confirmed by multiple experimenters.
Analysis of immunofluorescent formalin-fixed, paraffin-embedded sections from human PDAC samples
Human PDAC tissue samples used in this study were obtained from 39 patients who underwent pancreatectomy for pancreatic cancer at the Department of Surgery and Oncology, Kyushu University Hospital, between 2012 and 2020. Only patients who had never received any preoperative therapy were selected, to exclude the effect of chemotherapy or radiation on the tumor microenvironment. Written informed consent was acquired from all patients before surgery. The study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of Kyushu University (approval numbers: 2022-100 and 2023-3).
Establishment of autophagy-deficient cell lines
Five Atg7 small-hairpin RNA (shRNA) lentiviral vectors (#TRCN92166, #TRCN305928, #TRCN375444, #TRCN375421, and #TRCN305991; Sigma-Aldrich), three Atg5 shRNA lentiviral vectors (#TRCN99431, #TRCN99432, and #TRCN375819; Sigma-Aldrich), and three Atg4B shRNA lentiviral vectors (#TRCN30940, #TRCN30941, and #TRCN30942) were transduced into the KPC1 and KPC2 cell lines according to the manufacturer’s instructions. A non-targeting shRNA (#SHC016V, Sigma-Aldrich) was transduced as a control (shNC). Puromycin (#631305; Takara) was used to select shRNA-transduced clones. Knockdown efficiency was confirmed by Western blotting, as described in the relevant section. According to their knockdown efficiency, two types of cell lines were selected for each target gene: shATG7#1, #2; shATG5#1, #3; and shATG4B#1, #2.
Establishment of KPC cells expressing ovalbumin
KPC1 cells were transduced with pLV[Exp]-CMV>mOVA-EF1A>EGFP/Neo lentiviral vector and were sorted based on their enhanced GFP (EGFP) expression by flow cytometry. KPC cells expressing stable EGFP were also established by transducing KPC1 cells with a pLV[Exp]-EGFP:T2A:Puro-EF1A>mCherry lentiviral vector. The lentiviral vectors used to express ovalbumin (OVA) or EGFP in our study were constructed and packaged by VectorBuilder Inc. The vectorbuilder.com vector IDs were VB221104-1064htn (OVA) and VB010000-9298rtf (EGFP), which can be used to retrieve detailed information about these vectors. OVA expression was confirmed by qRT-PCR, and EGFP expression was confirmed by examination under a BZ-X800 fluorescence microscope (KEYENCE).
RNA extraction and qRT-PCR
Total RNA was extracted from KPC cells using High Pure RNA Isolation Kit (#11828665001, Roche) according to the manufacturer’s instructions. qRT-PCR was performed on triplicate samples using iTaq Universal SYBR Green One-Step Kit (#170-8892; Bio-Rad Laboratories) and the CFX96 Real-Time PCR System (Bio-Rad Laboratories). The expression value of a target gene in a given sample was normalized to Gapdh expression. Primers for OVA and Gapdh were purchased from Takara Bio Inc.
Western blotting
KPC cells were washed twice with cold PBS, and then total protein was extracted using PRO-PREP Protein Extraction Solution (#17081, iNtRON Biotechnology) according to the manufacturer’s instructions. The protein concentration was normalized to 20 µg per 30-µL sample using a protein assay dye reagent.
For SDS-PAGE, samples were boiled at 95°C for 5 minutes, loaded onto and separated on 4%–15% Mini-PROTEAN TGX Precast Protein Gels (#456-1084, Bio-Rad Laboratories), and transferred to Trans-Blot Turbo Mini PVDF Transfer Packs (#170-4156, Bio-Rad Laboratories) using the Trans-Blot Turbo Transfer Starter System (Bio-Rad Laboratories). After transfer, membranes were blocked with 5% skim milk in TBST buffer (0.1% Tween 20) for 1 hour. The membranes were then incubated with prediluted primary antibodies at 4°C overnight. After washing three times with TBST buffer, the membranes were probed with horseradish peroxidase–conjugated secondary antibodies and then developed using Amersham Enhanced Chemiluminescence Select reagent. The membranes were imaged using the ChemiDoc XRS System (Bio-Rad Laboratories) and analyzed using Quantity One software (Bio-Rad Laboratories).
Cell proliferation assay
KPC cells were seeded in 96-well polystyrene cell culture microplates (#655083, Greiner Bio-One International) at 1 × 103 cells per well. After 24 hours, the medium was aspirated and replaced with fresh complete DMEM with or without 20 µmol/L CQ. Cell proliferation was evaluated using the Cell Titer-Glo luminescent cell viability assay (#G7570, Promega) according to the manufacturer’s instructions. Fluorescence was measured after 0, 12, 36, and 48 hours. The emission value was measured on a microplate reader (Infinite F200; Tecan) following the manufacturer’s instructions. Each experiment was performed at least in triplicate and repeated twice.
Flow cytometry
Fresh murine PDAC tumors were finely sliced into 0.5- to 1.0-mm fragments and dissolved using reagents from Tumor Dissociation Kit (#130-960-730, Miltenyi Biotec) according to the manufacturer’s instructions. After filtering through a 70-μm cell strainer (#431751, Corning) to remove cell aggregates, single cells were pelleted by centrifugation and resuspended in PBS. To analyze the immune cells cultured in vitro, cells were collected from the U-bottomed 96-well culture plates by pipetting. Harvested cells were incubated with anti-mouse CD16/CD32 antibody (#101319, BioLegend) for 10 minutes at 4°C. The cells were then stained with the indicated antibodies for 30 minutes at 4°C. After washing off unbound antibodies with PBS, the cells were acquired on a FACSAria Fusion flow cytometer (BD Biosciences). For each experiment, single-stained controls were prepared to select the appropriate voltage and compensation settings. For the analysis of DC activation, a fluorescence minus one sample was used as a negative control to differentiate between positive and negative signals. However, for MHC II staining, cells expressing both CD11c and MHC II were identified as DCs, and the population that expressed MHC II more strongly was labeled as MHC II+ DCs. All flow cytometry data were analyzed using FlowJo software (ver.10.5.3, BD).
IHC
Paraffin-embedded murine pancreatic tissues were sliced into 4-µm thick sections. The sections were deparaffinized in xylene and dehydrated using an ethanol gradient. Antigen retrieval was achieved in sodium citrate buffer (pH 6.0) or Tris-EDTA buffer (pH 9.0) using a microwave or a pressure cooker. After blocking with 3% BSA in PBS, the sections were incubated with the primary antibodies anti-CD8a (1:400, #98941, Cell Signaling Technology), anti-CD11c (1:400, #97585, Cell Signaling Technology), anti-LC3AB (1:200, #12741, Cell Signaling Technology), anti-p62 (1/1,000, #PM045, MBL), and anti–pan cytokeratin (pCK; 1/1,000, #ab308262, Abcam) diluted in 1% BSA overnight at 4°C. Next, they were labeled with EnVision+ System horseradish peroxidase–labeled polyclonal anti-rabbit (#K4003, Dako) or anti-mouse (#K4001, Dako) antibodies for 1 hour at room temperature and visualized with a 3,3′-diaminobenzidine kit (Sigma–Aldrich, #D5537-5G). Counterstaining was performed with hematoxylin.
The sequential immunoperoxidase labeling and erasing method was performed to evaluate the colocalizations of LC3AB/pCK or p62/pCK, as previously reported (28). Briefly, the detection of the antibodies was performed with 3-amino-9-ethylcarbazole (AEC) Substrate Kit (#ab64252, Abcam), and slides were cover-slipped in an aqueous mounting medium. After scanning, slides were decoverslipped in water and dehydrated in 95% ethanol until no visible AEC reaction product remained. The tissue was then restained for another antibody, beginning with the blocking step. To make multicolor composite images, digital snapshots of tissue sections, which were applied pseudo-colors to replace the AEC precipitate color, were overlaid using the BZ-X800 analyzer (ver.1.1.2.4, KEYENCE).
Immunofluorescence
For the immunofluorescence (IF) assay of tumor tissues, the IHC method was used up to the 3% BSA blocking procedure. After blocking, the sections of murine PDAC tissues were stained with anti-CD11c (1:500) and anti-MHC II (I-A/I-E; 1:500, #14-5321-85, Invitrogen), or anti-CD8a (1:400) and anti-granzyme B (1:500, #14-8822-82, Invitrogen), or anti-CD8a and anti-perforin (1:500, ab16074, Abcam), or anti-CD8a and anti–LAG-3 (1:500, AF3328, Novus Biologicals) antibodies, diluted in 1% BSA, overnight at 4°C.
For the staining of human PDAC samples, the following primary antibodies were used to evaluate autophagy levels in cancer cells and the tumor infiltration of activated DCs: anti-CD11c (1:500, #ab52632, Abcam) and anti-MHC II (1:500, #MA1-25914, Invitrogen), or anti-CD11c (1:500) and anti-MHC I (1:500, #ab70328, Abcam), or anti-LC3AB (1:500) and anti-CK19 (1:500, #M0888, Agilent). All antibodies were diluted in 1% BSA and incubated with the tissue samples overnight at 4°C.
The tissues were then stained with 4',6-diamidino-2-phenylindole (DAPI; 1:1,000) and the following secondary antibodies: Alexa Fluor 546–conjugated anti-rabbit IgG (1:400, #A-10040, Invitrogen), Alexa Fluor 488–conjugated anti-rat IgG (1:400, #A-21208, Invitrogen), Alexa Fluor 488–conjugated anti-mouse IgG (1:400, #A-21202, Invitrogen), and Alexa Fluor 488–conjugated anti-goat IgG (1:400, #A-11055, Invitrogen) in 0.1% BSA for 1 hour at room temperature. Autofluorescence was blocked using Vector TrueVIEW Autofluorescence Quenching Kit (#SP-8400, Vector Laboratories) according to the manufacturer’s instructions. Images were acquired on a fluorescence microscope, BZ-X800 (KEYENCE). Whole-tissue slide scans were performed at ×4 magnification; fine scanning of at least three different representative areas was then performed at ×20 magnification. Image analysis involved counting the positively stained cells and quantifying the extent of fluorochrome colocalization using the BZ-X800 analyzer (ver.1.1.2.4, KEYENCE).
For the assessment of autophagy levels in cancer cells, the patients with PDAC were ranked according to their ratios of LC3+CK19+ cells to CK19+ cells; patients with ratios above the median were defined as the “autophagy-high” group, whereas those with ratios below the median were defined as the “autophagy-low” group. For the analysis of activated DCs, the patients with PDAC were ranked according to their ratios of MHC I+CD11c+ cells to DAPI+ cells or MHC II+CD11c+ cells to DAPI+ cells; patients with ratios above the mean were defined as the “high” group, whereas those with ratios below the mean were defined as the “low” group. All survival analyses were conducted using the Kaplan–Meier method, and the survival curves were compared using the log-rank test. Correlations between cancer cell LC3 expression and the extent of activated DC infiltration were conducted using the Spearman correlation coefficient.
For the intracellular staining of OVA in autophagy-deficient KPC cells, KPC cells treated with CQ (20 µmol/L) and 3-methyladenine (3-MA; 2.0 mmol/L, #M9281, Sigma-Aldrich) were stained with anti-OVA (1/500, #ab306591, Abcam), followed by Alexa Fluor 546–conjugated anti-rabbit IgG (1/200). Fixation with 4% paraformaldehyde, permeabilization with 0.1% Triton X-100, and blocking with 3% BSA were performed before the staining.
siRNA-mediated gene silencing
KPC cells were transfected with siAtg7 (#SI00900515, #SI00900522; Qiagen), and human PDAC cells were transfected with siATG5 (#SI02633946, #SI02655310; Qiagen) and siATG7 (#SI04952339, #SI02655373; Qiagen). Each cell type was also separately transfected with a negative control siRNA (siNC; #1027310, Qiagen), which was included as a control in every assay. The cells were transfected with siRNAs by electroporation using the Nucleofector System (Lonza) according to the manufacturer’s instructions. Knockdown efficiency was confirmed by Western blotting 48 hours after transfection. Transfected cells were used in subsequent experiments 48 to 72 hours after transfection.
DC generation
Mouse DC generation was performed as described previously (29, 30). Briefly, bone marrow (BM) cells were harvested from the femurs and tibias of 8-week-old female C57BL/6 mice. The BM was flushed to remove cells, which were then centrifuged at 350 × g for 5 minutes. Blood cells were lysed with Lysing Buffer (#555899, BD Biosciences) for 8 minutes at room temperature. The cells were filtered sequentially through 70- and 40-µm cell strainers (#431751 and #431750, Corning) to remove cell aggregates. After determination of the cell numbers, 1.0 × 107 cells were cultured in a T75 flask (#156499, Thermo Fisher Scientific) in complete RPMI 1640 medium containing 5.0 ng/mL recombinant murine GM-CSF (#077-04674, Wako) and 200 ng/mL recombinant murine Flt3L (#250-31L, PeproTech). On day 3, fresh medium with GM-CSF and Flt3L was added to the cultured BM-derived DCs. On day 6, the medium was replaced with new medium containing fresh cytokines. DCs were harvested on day 9 or 10 and used in the assays.
Human DC generation was performed as described previously (31). Peripheral blood mononuclear cells (PBMC) were obtained using the leukapheresis procedure and isolated on a Ficoll-Hypaque gradient. After isolation, the PBMCs were frozen and stored at −80°C. To generate human DCs from PBMCs, the PBMCs were thawed and then cultured in 6-well plates (BD Falcon) in complete medium containing 1% autologous serum for 30 minutes. After removing nonadherent cells and washing with complete RPMI medium, adherent cells were cultured in complete DC medium containing recombinant human GM-CSF (PMC-GM-300, Primmune Inc.) and iIL-4 (PMC-IL4-100, Primmune Inc.). On day 6, the immature DCs were harvested and used in assays.
Cancer cell and DC coculture assay
For the coculture of KPC cells and murine DCs, 1.0 × 105 KPC cells (KPC1 shNC/shATG5#1/shATG5#3, KPC2 shNC/shATG5#1/shATG5#3, KPC1 shNC/shATG7#1/shATG7#2, KPC1 shNC/shATG4B#1/shATG4B#2, or KPC2 shNC/shATG4B#1/shATG4B#2) were cocultured with 1.0 × 105 murine BM-derived DCs in U-bottomed 96-well plates. The cells were maintained in complete RPMI medium with or without 20 µmol/L CQ at 37°C with 5% CO2. Twenty four hours later, DCs were analyzed by flow cytometry for the expression of DC activation and maturation markers CD80, CD86, MHC I, and MHC II. For the coculture of apoptotic cancer cells and DCs, KPC cells were irradiated with germicidal UV light, at room temperature and in the absence of growth media, to induce apoptosis. The LUNA-II automated cell counter (#L40001, Logos Biosystems) was used to confirm of apoptosis induction and count the apoptotic cells following trypan blue staining. For the coculture of necrotic cancer cells and DCs, necrosis of KPC cells was induced by three cycles of freezing (at −80°C) and thawing (at 37°C). After induction of necrosis, the KPC cells were centrifuged at 500 × g for 5 minutes and harvested to use in assays. For 3D coculture, 1.0 × 105 KPC cells and 1.0 × 105 DCs were diluted in 10% Matrigel (at a Matrigel to RPMI ratio of 1:9, #3445, Bio-Techne) and then centrifuged at 1,000 × g for 1 minute in U-bottomed 96-well plates. For the coculture experiments to verify the impact of type 1 IFN, anti-IFNα/β receptor 1–blocking antibody (#BP0241, Bio X Cell) or an IgG1 isotype control (BE0088, Bio X Cell) were added at 50 μg/mL to the culture medium at the start of the coculture experiment.
For the coculture of human PDAC cells and human DCs, 1.0 × 105 PDAC cells (PDAC siNC/siATG5#1/siATG5#2/siATG7#1/siATG7#2) were cocultured with 1.0 × 105 human PBMC-derived DCs. The culture conditions were the same as those in the murine coculture assays. DCs were analyzed by flow cytometry for the expression of the DC activation and maturation markers CD40, CD80, CD86, MHC I, and MHC II.
T-cell and DC cross-priming assay
Coculture assays of OT-1 CD8+ T cells and DCs were performed as described previously (29, 30), with some modifications. Briefly, BM-derived DCs were cocultured with KPC cells for 24 hours and then positively isolated using a PE-conjugated anti-mouse CD11c antibody (#553802, BD Biosciences) and anti-PE MicroBeads (#130-048-801, Miltenyi Biotec), according to the manufacturer’s instructions. Naïve CD8+ T cells were isolated from the spleens of OT-1 mice using Naïve CD8+ T cell Isolation Kit (#130-096-543, Miltenyi Biotec). Purified CD8+ T cells were labeled with 2.0 mmol/L carboxyfluorescein diacetate succinimidyl ester (CFSE; #4238011, BioLegend) by incubating at room temperature for 10 minutes and washing three times. A measure of 1.0 × 105 DCs were cocultured with 2.5 × 105 carboxyfluorescein diacetate succinimidyl ester–labeled naïve CD8+ T cells in RPMI 1640 medium (10% FBS) supplemented with rIL2 (30 U/mL, #212-12, PeproTech) in U-bottomed 96-well plates at 37°C and 5% CO2. Forty eight hours later, CD8+ T-cell proliferation was analyzed by flow cytometry.
DC antigen capture assay
For the assessment of antigen capture by DCs, murine DCs were cocultured with dye-labeled KPC cells (Cell Tracking Dye Kit–Green-Cytopainter, ab138891, Abcam) or KPC cells expressing EGFP, as described above. DCs were analyzed by flow cytometry for the acquisition of fluorescence. DCs cocultured with nonlabeled KPC cells were used as a negative control to demarcate between the positive and negative fluorescent signals.
In vivo vaccination
To assess the immunogenic effects of autophagy inhibition, a vaccination experiment was performed as described previously (29, 32), with some modifications. On day 7, 1.0 × 105 KPC1-shNC/shATG7/shATG5 cells were suspended in 50 µL DMEM (per mouse) and injected subcutaneously into the left flank of 6-week-old female C57BL/6 mice. Control mice received a s.c. injection of 50 µL DMEM (vehicle control). Tumors were resected 7 days later (day 0). On the same day (day 0), 5.0 × 106 KPC1 cells + 5.0 × 106 murine CAFs were injected subcutaneously into the right flanks of mice as a second tumor challenge. The tumor-bearing mice were sacrificed 21 days later (on day 20), and the secondary challenge tumors were resected to measure their volumes and weights. Mice with inadequate resection of the primary tumor in the left flank were excluded. Resected secondary challenge tumors were also evaluated by flow cytometry to analyze the tumor-infiltrating T cells.
Assessment of antigen-specific CD8+ T cells
The induction of antigen-specific CD8+ T cells by autophagy-deficient cancer cells was determined as described previously (33). At first, 1.0 × 105 KPC1-OVA shNC/shATG7 cells suspended in 50 µL 50% Matrigel (per mouse) were injected orthotopically into the pancreas of 6-week-old female C57BL/6 mice. Two weeks later, the spleens from the tumor-bearing mice were collected. Following blood cell removal by incubation with Lysing Buffer (#555899, BD Biosciences) for 8 minutes at room temperature, the splenocytes were analyzed by flow cytometry. H-2Kb OVA Tetramer-SIINFEKL-PE (#TS5001, MBL Life Science) was used to detect antigen-specific CD8+ T cells. DCs presenting OVA-derived peptides were detected using APC/Fire 750–conjugated anti-mouse antibody recognizing the H-2Kb–bound SIINFEKL epitope of OVA (#141614, BioLegend).
scRNA-seq
Sample preparation
Fresh murine PDAC tumors (KPC1 shNC/shATG7, n = 3/3) were finely sliced into 0.5- to 1.0-mm fragments and dissolved using reagents from Tumor Dissociation Kit (#130-960-730, Miltenyi Biotec) according to the manufacturer’s instructions. After filtering through a 70-μm cell strainer to remove cell aggregates, the cells were collected by centrifugation and resuspended in cold PBS containing 0.1% BSA. Following dead cell removal using Dead Cell Removal Kit (#130-090-101, Miltenyi Biotec) according to the manufacturer’s instructions, the cells were filtered through a 40-µm cell strainer. Next, the cells were stained with BV421-conjugated anti-CD45 (#563890, BD Biosciences) and propidium iodide, and then live immune (propidium iodide− CD45+) cells were sorted using a BD FACSAria Fusion flow cytometer, as described in the section “Flow cytometry.” All experimental manipulations were performed on ice to preserve cell quality. Finally, 2,141 cells from KPC1 shNC tumors and 2,316 cells from KPC1 shATG7 tumors were subjected to the analysis.
scRNA-seq library preparation and sequencing
Single cells were encapsulated in droplets using the 10X Genomics Chromium Controller, and the cDNA library was prepared using Chromium Next GEM Single Cell 5′ Reagent Kits v2 (10X Genomics) by following the manufacturer’s instructions. A target of profiling 10,000 cells per library was set. The resultant libraries were sequenced on the DNBSEQ-G400 sequencing instrument (MGI Tech), and the sequence read number was set at 40,000 reads per cell.
scRNA-seq data analysis: filtering, normalization, clustering, and Uniform Manifold Approximation and Projection
Raw sequencing reads were aligned to the mouse reference genome, mm10, and processed into a matrix, which quantifies the number of unique molecular identifiers (UMI) per gene per cell for all valid 10X barcodes, using Cell Ranger v5.0.0 (10x Genomics). The output files were analyzed with the R v3.6.3 package “Seurat v3.1.5” (34). Low-quality cells (<100 UMI/cell, >6,000 UMI/cell, and >50% mitochondrial genes) were excluded; doublets were filtered out using the DoubletFinder tool (35). The “CellCycleScoring” function was used to mitigate the effects of cell-cycle heterogeneity by calculating cell-cycle phase scores based on canonical markers and regressing them out of the data. The “SCTransform” package, which is designed for the normalization and variance stabilization of molecular count data, was used to merge data from all samples. Next, a linear dimensional reduction by principal component analysis of the top 2,000 most variable genes was performed. Thirty principal components (PC) were then selected to generate the shared nearest neighbor graph using the “FindNeighbors” function. To visualize the data according to the Uniform Manifold Approximation and Projection (UMAP) dimensional reduction method, the cells were clustered using the “FindClusters” function, with a resolution of 0.4. Subsequently, 11 clusters were identified using well-known markers of immune cell populations.
scRNA-seq analysis of DCs
C5, C9, and C10 clusters, which expressed a canonical DC signature (H2-Ab1, Itgax, and Flt3), were identified as DCs; they were then reclustered (as described above but with a resolution of 0.3) into four clusters. The “FindAllMarkers” function was used to identify the differentially expressed genes (DEG) for each cluster compared with all other clusters, provided that the genes were expressed in more than 25% of cells and underwent a ≥0.25-fold difference in expression. To identify the clusters, the DC cluster DEGs were manually compared with the DC signatures previously reported in the literature (36–38). “Antigen-presenting score,” “IFN-induced score,” “maturation score,” “migration score,” and “regulatory score” were calculated to compare the transcriptional differences between tumors originating from KPC1 shNC cells or KPC1 shATG7 cells. The gene sets for each score were identified by referencing the study in which they had been previously reported (38–40).
scRNA-seq analysis of CD8+ T cells
C0, C2, C4, and C6 clusters, which expressed CD3e, were identified as T cells; T cells expressing CD8a were further labeled as CD8+ T cells. CD8+ T cells were reclustered (as described above but with a resolution of 0.2) into three clusters. DEG analysis was performed as described above. To identify the clusters, the CD8+ T-cell cluster DEGs were manually compared with the T-cell signatures previously reported in the literature (41, 42). “Effector signature score,” “proliferation score,” and “exhaustion score” were calculated using the gene sets referenced in previously published studies (43, 44).
Secondary analysis of a scRNA-seq dataset
scRNA-seq data from patients with PDAC (24 primary PDAC tumors and 10 control pancreases) were obtained from a public dataset (45), which is available at the Genome Sequence Archive under the accession number CRA001160. The downloaded data were normalized using the “SCTransform” package. Clustering and UMAP generation were performed as described above. Among the 12 clusters identified, C0 and C1 were isolated as “ductal cells,” and each sample was assigned “autophagy signature scores,” which were calculated from the expression of the autophagy-related genes derived from the WP4923 “Autophagy (Homo sapiens)” dataset. The 24 PDAC samples were then divided into the “autophagy-high” and “autophagy-low” groups by the autophagy signature scores of ductal cells. In the DC analysis, clusters “0,” “5,” and “8” from the reclustered myeloid cell cluster (C4) were then identified as DC populations. In the T-cell analysis, we identified C3 as T cells and then further classified them into seven clusters. Among them, clusters “0,” “1,” “5,” and “6” were identified as CD8+ T cells, whereas “4” was identified as regulatory T cells (Tregs), based on the expression of CD4, FOXP3, CTLA4, and IL2RB. The transcriptional differences in DCs and CD8+ T cells between the two groups were compared, as described above, using the gene sets referenced in published literature (43, 46).
Data quantification and statistical analysis
All statistical analyses and graph generation (except for scRNA-seq) were performed using GraphPad Prism (ver.9.3.1, GraphPad). The unpaired Student t test or Mann–Whitney test were used to compare two independent groups. One-way ANOVA was used for comparisons of multiple (>2) groups. Survival analyses were conducted using the Kaplan–Meier method, and the curves were compared using the log-rank test. Correlations between the autophagy level of cancer cells and the extent of DC activation were evaluated using the Spearman correlation coefficient. For scRNA-seq, statistical analyses and graph generation were performed using the Seurat package of R. The “stat_compare_means” function was used to compare two groups via the Wilcoxon rank-sum test. Differences with the following P values were considered statistically significant: ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; and ∗∗∗∗, P < 0.0001.
Key resources
Key resources (antibodies, chemicals, oligonucleotides, and software and algorithms) are listed in Supplementary Table S1.
Data availability
The data generated in this study are available upon request from the corresponding author. The scRNA-seq data have been deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). The accession number is GSE233663. Previously published scRNA-seq data from patients with PDAC analyzed in this study were obtained from the Genome Sequence Archive under the accession number CRA001160.
Results
Negative correlation between cancer cell autophagy and DC activation signatures in human PDAC tumors
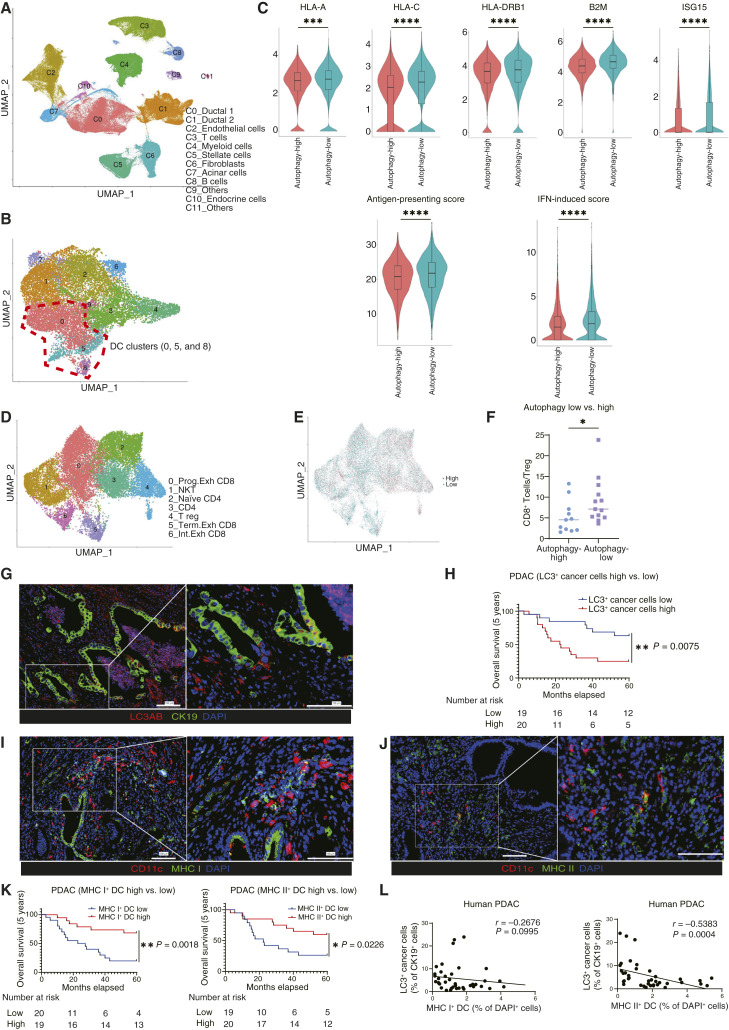
To investigate the relationship between autophagy in cancer cells and the TIME in PDAC tumors, we analyzed a publicly available scRNA-seq dataset of patients with PDAC generated by Peng and colleagues (45). The cells were classified into 12 clusters (Fig. 1A). At first, we assessed the expression of autophagy-related genes in ductal cells (Supplementary Tables S2 and S3) and found that the autophagy signature scores were significantly higher in the PDAC samples than in the normal pancreas samples (Supplementary Fig. S1A). Next, we divided the 24 PDAC samples into the “autophagy-high” and “autophagy-low” groups based on the autophagy signature scores of ductal cells and assessed the effect of autophagy on the activation of tumor-infiltrated DCs. Myeloid cell clusters (C4) were reclustered, and clusters “0,” “5,” and “8” were identified as DC populations (Fig. 1B; Supplementary Fig. S1B). The expression of antigen-presenting related genes and IFN-induced genes was significantly higher in the autophagy-low group than in the autophagy-high group (Fig. 1C; Supplementary Fig. S1C; Supplementary Table S4). Furthermore, the analysis of T-cell population composition revealed that the “autophagy-low” group had a significantly higher CD8+ T-cell to Treg ratio than the “autophagy-high” group (Fig. 1D–F; Supplementary Fig. S1D).
Figure 1.
The autophagy level of cancer cells is negatively correlated with the extent of DC activation in patients with PDAC. A, Analysis of a publicly available scRNA-seq dataset generated by Peng and colleagues (CRA001160; ref. 45), which contains data from 24 primary human PDAC tumors and 10 control pancreases. B, The myeloid cluster (C4) was reclustered into nine clusters. Clusters of “0,” “5,” and “8” were identified as DCs. C, The expression levels of gene sets related to DC functions were analyzed in the overall DC cluster. The expression levels of representative genes are shown as violin plots. The “antigen-presenting score” and “IFN-induced score” were calculated for the comparison of the “autophagy-high” and “autophagy-low” groups. D, The T-cell cluster (C3) was reclustered into seven clusters. E, T cells were divided into cells derived from the autophagy-high group (red) and those derived from the autophagy-low group (green). F, The ratio of CD8+ T cells to Tregs (%) in the autophagy-high and autophagy-low groups. G, IF analysis of 39 PDAC patient samples. Autophagy levels in cancer cells were evaluated by staining for LC3AB (red) and CK19 (green). Representative images are shown. H, Kaplan–Meier overall survival analysis of patients with PDAC according to their cancer cell autophagy levels, defined by the ratio of LC3AB+ CK19+ cells to CK19+ cells. I and J, Representative IF images of activated DCs, expressing CD11c (red) and MHC I/II (green). K, Kaplan–Meier overall survival analysis of activated DCs, defined by the ratio of MHC I+ CD11c+ cells to DAPI+ cells from I and MHC II+CD11c+ cells to DAPI+ cells from J. L, Spearman correlation between the autophagy levels of cancer cells (% LC3AB+ CK19+ cells/CK19+ cells) and activated DCs (% MHC I+CD11c+ cells/DAPI+ cells or % MHC II+ CD11c+ cells/DAPI+ cells). Scale bars, 100 μm (G, I, and J). Bars, median; boxplots show a centerline, median; box limits, upper and lower quartiles; whiskers that extend up to 1.5× the IQR beyond the upper and lower quartiles (C). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; analyzed using the Wilcoxon rank-sum test (C), Student t test (F), and log-rank test (H and K).
Next, to examine the clinical significance of autophagy levels in cancer cells and DCs infiltrated in PDAC tumors, we analyzed human PDAC tissues resected at the Department of Surgery and Oncology, Kyushu University, Japan. Immunofluorescent staining for the evaluation of autophagy in cancer cells revealed that autophagy-low patients with PDAC, with a lower LC3+CK19+ cell to CK19+ cell ratio, had a significantly better prognosis than patients in the autophagy-high group (Fig. 1G and H). The relationship between cancer cell LC3 expression and clinicopathologic factors is shown in Supplementary Table. S5. The evaluation of DC activation showed that patients with PDAC with higher numbers of activated tumor-infiltrating DCs with the MHC I+CD11c+ or MHC II+CD11c+ signatures had a significantly better prognosis than those who had lower numbers of these cells (Fig. 1I–K; Supplementary Fig. S1E and S1F). Moreover, the number of activated DCs was higher in the autophagy-low group than in the autophagy-high group of patients with PDAC (Fig. 1L). Thus, autophagy signatures in cancer cells are negatively correlated with DC activation signatures in human PDAC.
scRNA-seq of murine PDAC tumors revealed that the expression of genes associated with DC function is increased in autophagy-deficient tumors
To investigate the effect of autophagy ablation in cancer cells on tumor immunity, we used murine PDAC models. We established mouse PDAC cell lines (KPC1 and KPC2) from the primary PDAC tumors of KPC mice (26), and autophagy-deficient KPC cells were generated by lentiviral transduction with shRNAs targeting the autophagy-related genes Atg7, Atg5, and Atg4b (Supplementary Fig. S2A–S2C). Autophagy inhibition by shRNA did not affect the proliferation of KPC cells in vitro (Supplementary Fig. S2D). To verify whether autophagy regulates antitumor adaptive immunity, we subcutaneously implanted ATG7-depleted KPC cells (KPC1 shATG7) into syngeneic immunocompetent C57BL/6 mice or T-cell–depleted immunodeficient BALB/c-nu mice. Although ATG7 depletion slightly suppressed PDAC tumor growth in BALB/c-nu mice, as previously reported (18, 47), this inhibitory effect was more prominent in C57BL/6 mice, which was associated with the increased number of tumor-infiltrating CD8a+ cells and CD11c+ cells (Supplementary Fig. S2E–S2G). In addition, CD8+ T-cell depletion was performed in immunocompetent models (Supplementary Fig. S2H), and we discovered that the tumor suppression by autophagy inhibition was reduced in mice treated with anti-CD8a antibodies, which revealed the dependency of CD8+ T cells (Supplementary Fig. S2I). Next, when autophagy-defective KPC cells (KPC1 shATG7 or KPC1 shATG5) were orthotopically implanted in the pancreas of C57BL/6 mice, tumor growth was significantly suppressed compared with control cells (KPC1 shNC; Fig. 2A and B). These results are consistent with the previous report that autophagy inhibition enhances antitumor immune responses with the expansion of T cells (22).
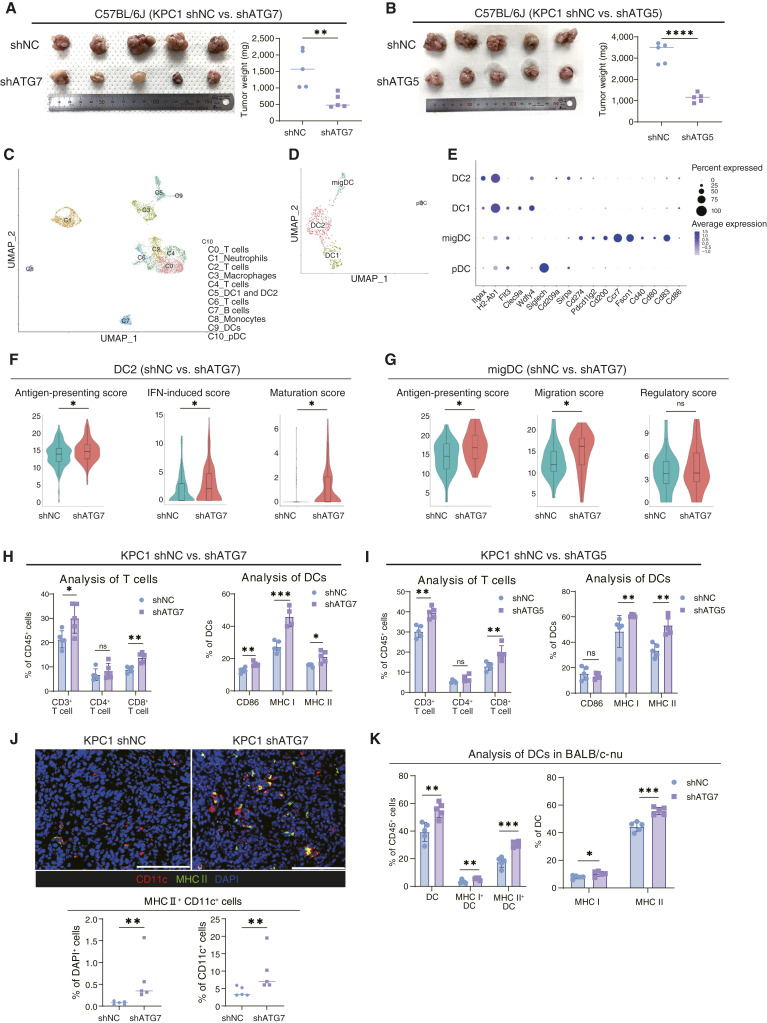
Figure 2.
Orthotopic syngeneic PDAC tumors revealed that the expression of genes associated with DC functions was increased in autophagy-deficient tumors. A and B, KPC1 shNC/shATG7 (A) and KPC1 shNC/shATG5 (B) cells were orthotopically transplanted into C57BL/6 to compare tumor growth. C, scRNA-seq was used to assess the transcriptional dynamics induced by autophagy inhibition in cancer cells. The UMAP technique was used to visualize cell clusters identified within the population of PI− CD45+ cells sorted from orthotopic KPC1 shNC/shATG7 syngeneic tumors. D, The DC clusters (C5, C9, and C10) were reclustered into four clusters; cell count: 480 cells. E, Expression of key signature genes used to identify specific DC clusters. F and G, The expression levels of gene sets related to DC function were analyzed in the DC2 (F) and migDC (G) subsets. The following gene sets were selected for the comparison of the two groups of tumors: “antigen-presenting genes,” “IFN-induced genes,” “maturation genes,” “migration genes,” and “regulatory genes”; the gene sets corresponding to each function are listed in dot plots (Supplementary Fig. S3C and S3D). H and I, Tumor-infiltrating T cells and DCs were analyzed by flow cytometry in KPC1 shNC/shATG7 tumors (H) and KPC1 shNC/shATG5 tumors (I). J, IF was used to assess orthotopic syngeneic KPC1 shNC/shATG7 tumors for the presence of activated DCs (red, expressing CD11c; green, MHC II). Representative images are shown. The graph shows the proportions of MHC II+ CD11c+ cells/DAPI+ cells (%) and MHC II+ CD11c+ cells/CD11c+ cells (%). K, KPC1 shNC/shATG7 cells were subcutaneously implanted into BALB/c-nu. DC activation was evaluated by flow cytometry. Scale bars, 100 μm (J). Error bars, mean ± SD; bars, median. Boxplots show a centerline, median; box limits, upper and lower quartiles; whiskers that extend up to 1.5× the IQR beyond the upper and lower quartiles (F and G). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant; analyzed using the Wilcoxon rank-sum test (F and G) and Student t test or Mann–Whitney test (A, B, and H–K). pDC, plasmacytoid DC.
To validate the negative correlation between autophagy and DC activation observed in human PDAC tumors, we next examined the effects of autophagy depletion on the transcriptional landscape of CD45+ immune cells in murine orthotopic PDAC tumors by scRNA-seq. We sorted live CD45+ cells from the tumors of KPC1 shNC and KPC1 shATG7 cells (n = 3 per condition). Clustered scRNA-seq data were displayed using the UMAP dimensional reduction technique, and 11 clusters were identified (Fig. 2C; Supplementary Fig. S3A). We identified clusters C5, C9, and C10 as DCs, and the DC clusters were then reclustered into four clusters, namely, type1 classical DC (DC1), type2 classical DC (DC2), migratory DC (migDC), and plasmacytoid DC subsets, referring to the literature (Fig. 2D and E; Supplementary Fig. S3B; Supplementary Table S6; refs. 38, 40, 46). We compared the functional signatures of DCs between the KPC1 shATG7 and KPC1 shNC tumor groups (Supplementary Table S7; refs. 38, 40) and found that the DC2 and migDC clusters in KPC1 shATG7 tumors expressed significantly higher levels of genes involved in antigen presentation, IFN response, maturation, and migration than the same DC cluster in KPC1 shNC tumors (Fig. 2F and G; Supplementary Fig. S3C and S3D). Moreover, although migDCs are considered to contain immunosuppressive DC subtypes characterized by expression of the genes involved in immunoregulation (38), there was no significant difference in the “regulatory score” between the two groups (Fig. 2G), suggesting that inhibiting autophagy in cancer cells enhanced only DC activation without inducing immunosuppressive DCs. In the DC1 cluster, there was no significant difference in the functional signatures between the two tumor groups (Supplementary Fig. S3E).
We validated DC activation by flow cytometry in several orthotopic syngeneic PDAC models (Supplementary Fig. S4A). In all autophagy-deficient tumor models (KPC2 shATG7, KPC2 shATG5, and KPC2 shATG4B), tumor growth was significantly suppressed compared with autophagy-competent tumors, and flow cytometry analysis revealed that the expression of DC activation and maturation markers, such as CD86, MHC I, and MHC II, was significantly higher in autophagy-deficient tumors than in the controls, suggesting that DCs were activated (Fig. 2H and I; Supplementary Fig. S4B–S4D). IF analysis also confirmed that the numbers and ratios of activated tumor-infiltrated DCs increased in the autophagy-deficient PDAC tumors (Fig. 2J; Supplementary Fig. S4E). Autophagy inhibition in cancer cells was confirmed by sequential IHC revealing both the decrease of LC3AB and the accumulation of p62 in pCK-expressing cells in autophagy-deficient PDAC tumors (Supplementary Fig. S4F). On the other hand, activation of DCs may be considered to be due to increased cytotoxicity of T cells elicited by elevated expression of MHC I in cancer cells (22). Also, in our study, the expression of MHC I of CD45− increased in autophagy-deficient tumors, as previously reported (Supplementary Fig. S4G; ref. 22). However, our flow cytometry analysis of the tumors in BALB/c-nu mice showed that autophagy-defective cancer cells induce DC activation in the absence of T cells (Fig. 2K). Thus, the primary cause of DC activation in our murine KPC tumor models is not likely MHC I induction in cancer cells.
Autophagy ablation increases antigen accumulation in cancer cells, thereby activating DCs through enhanced antigen presentation capacity
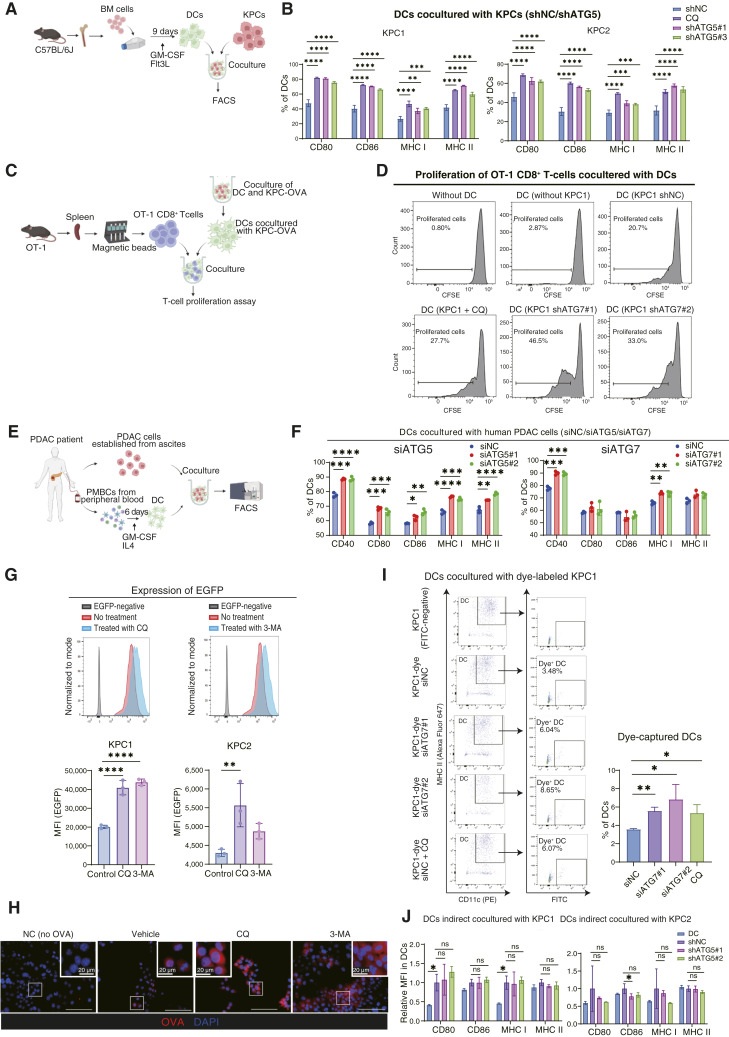
We next investigated whether pharmacologic or genetic inhibition of autophagy in cancer cells directly activates DCs in coculture assay in vitro (Fig. 3A; Supplementary Fig. S5A). DCs were cocultured with autophagy-deficient KPC cells or with KPC cells in the presence of CQ, an autophagy inhibitor. In coculture with autophagy-ablated KPC cells, DCs expressed higher levels of the activation and maturation markers than DCs cocultured with control KPC cells, indicating that inhibition of autophagy in cancer cells directly promotes DC activation (Fig. 3B; Supplementary Fig. S5B–S5F). Importantly, there is no effect of CQ on the induction of CD80, CD86, and MHC genes in DCs without coculture with KPC cells (Supplementary Fig. S5G). In addition, neither CQ treatment nor autophagy inhibition by shRNA altered the proliferation of KPC cells (Supplementary Figs. S2D and S6A), and the activation of DCs was not induced by coculture with UV-irradiated apoptotic cancer cells (Supplementary Fig. S6B), excluding the possibility that DCs were simply activated by dead cancer cells. Next, we evaluated the effect of type I IFN secreted from KPC cells because autophagy inhibition was previously reported to induce the secretion of type 1 IFN involved in DC activation (48, 49). However, coculture experiments with a blocking antibody against the IFN α/β receptor did not affect DC activation, suggesting the activation of DC was not due to increased secretion of type1 IFN (Supplementary Fig. S6C). In addition, in the 3D culture system mimicking the TIME, autophagy ablation in cancer cells also induced DC activation (Supplementary Fig. S6D). We next investigated whether activated DCs present antigens to CD8+ T cells (Fig. 3C). To this end, we used OVA-expressing KPC cells (KPC-OVA cells) and OT-1 CD8+ T cells, which specifically recognize the OVA-derived peptide SIINFEKL loaded on the MHC I on the cell surface (Supplementary Fig. S6E). When DCs were initially cocultured with autophagy-defective KPC-OVA shATG7 cells and then mixed with OT-1 CD8+ T cells, KPC-OVA shATG7 cell–cocultured DCs exhibited more potent effects on T-cell proliferation than DCs cocultured with autophagy-competent KPC-OVA shNC cells (Fig. 3D), indicating that autophagy-deficient cancer cells promote antigen presentation in DCs. Moreover, when human DCs isolated from PDAC PBMCs were cocultured with autologous PDAC cells (Fig. 3E), the expression of activation and maturation markers was also increased in human DCs cocultured with autophagy-deficient human PDAC cells (Fig. 3F).
Figure 3.
Autophagy-deficient cancer cells induce DC activation and T-cell priming via increased accumulation of intracellular antigens. A, Schematic outline of the coculture assay involving BM-derived DCs and KPC cells. B, Flow cytometry analysis of the expression of the activation and maturation markers, CD80, CD86, MHC I, MHC II on DCs cocultured with KPC1/2 shNC, KPC1/2 + 20 µmol/L CQ, or KPC1/2 shATG5#1, #2 cells. C, Schematic outline of the coculture assay involving DCs and OT-1 cells. D, DCs were pre-cocultured with KPC1 shNC, KPC1 + 20 µmol/L CQ, or KPC1 shATG7#1, #2 cells and then cocultured with OT-1 cells (extracted from the spleen of the OT-1 mouse). Flow cytometry was used to evaluate the proliferation of CFSE-labeled OT-1 cells. E, Schematic outline of the coculture assay involving autologous human PBMC–derived DCs and PDAC cells. F, Flow cytometry analysis of the activation and maturation markers on human DCs cocultured with autologous PDAC cells transfected with siNC; siATG5#1, #2; or siATG7#1, #2. G, KPC1-EGFP and KPC2-EGFP were treated with the autophagy inhibitors CQ (20 µmol/L) or 3-MA (2.0 mmol/L). Flow cytometry was used to assess the changes in EGFP expression. Representative histograms (top) and the corresponding quantification of EGFP expression in KPC1-EGFP and KPC2-EGFP (bottom) are shown. H, KPC1-OVA cells were treated with CQ (20 µmol/L) or 3-MA (2.0 mmol/L). IF analysis was performed to assess the accumulation of intracellular OVA. I, DCs were cocultured with KPC1 cells (no dye), KPC1-dye siNC, KPC1-dye, KPC1-dye + 20 µmol/L CQ, or KPC1-dye siATG7#1, #2. Flow cytometry was used to detect the signal of green dye in DCs to compare the amount of antigen captured by DCs. Representative dot plots are shown. J, Flow cytometry analysis of DCs indirect cocultured with KPC1/2 shNC or KPC1/2 shATG5#1, #2 cells. Error bars, mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001; analyzed using one-way ANOVA. FACS, flow cytometry analysis. (A,C, and E, Created with BioRender.com.)
We reasoned that autophagy ablation increased the antigenicity of cancer cells potentially through impaired degradation of antigenic proteins. To assess the impacts of tumor antigenic proteins on DC activation, we conducted a coculture assay using freeze–thawed cancer cells, which are cell membrane permeabilized and thus release intracellular antigens extracellularly (50). As a result, freeze–thawed KPC1 shATG7 cells activated cocultured DCs more potently than freeze–thawed KPC1 shNC cells (Supplementary Fig. S6F). To examine the effects of autophagy inhibition on the degradation of antigenic proteins, we established transgenic KPC cells expressing EGFP as a reporter of intracellular antigen (KPC1-EGFP and KPC2-EGFP). When the KPC1/2-EGFP cells were treated with autophagy inhibitors CQ and 3-MA, EGFP expression was markedly upregulated (Fig. 3G). IF analysis of KPC-OVA cells revealed the intracellular accumulation of OVA in KPC-OVA cells treated with CQ and 3-MA, suggesting the increased accumulation of tumor antigens (Fig. 3H). Next, to assess the amount of antigen captured by DCs, we cocultured DCs with dye-labeled KPC1 cells and measured fluorescence-positive, i.e., antigen-phagocytosed DCs. DCs cocultured with autophagy-deficient cancer cells captured more antigens than those cocultured with autophagy-competent cells (Fig. 3I). The increase of antigen-captured DCs by autophagy-deficient cancer cells was also revealed in KPC1-EGFP cells (Supplementary Fig. S6G). In addition, DCs indirectly cocultured with autophagy-deficient KPC cells in transwells did not change the expression of DC activation markers, suggesting that immunostimulating cytokines and chemokines other than type 1 IFN and immunosuppressive secreted factors represented by IL10, which may be impacted by autophagy inhibition, do not affect DC activation induced by autophagy-deficient cancer cells (Fig. 3J). Thus, DCs are activated through direct intercellular communication with autophagy-defective cancer cells.
Therefore, these results indicate that autophagy is involved in the degradation of intracellular proteins, which seem to serve as antigens in cancer cells; consequently, autophagy inhibition enhances cancer cell antigenicity.
Autophagy-defective cancer cell–primed DCs efficiently expand antigen-specific CD8+ T cells in vivo
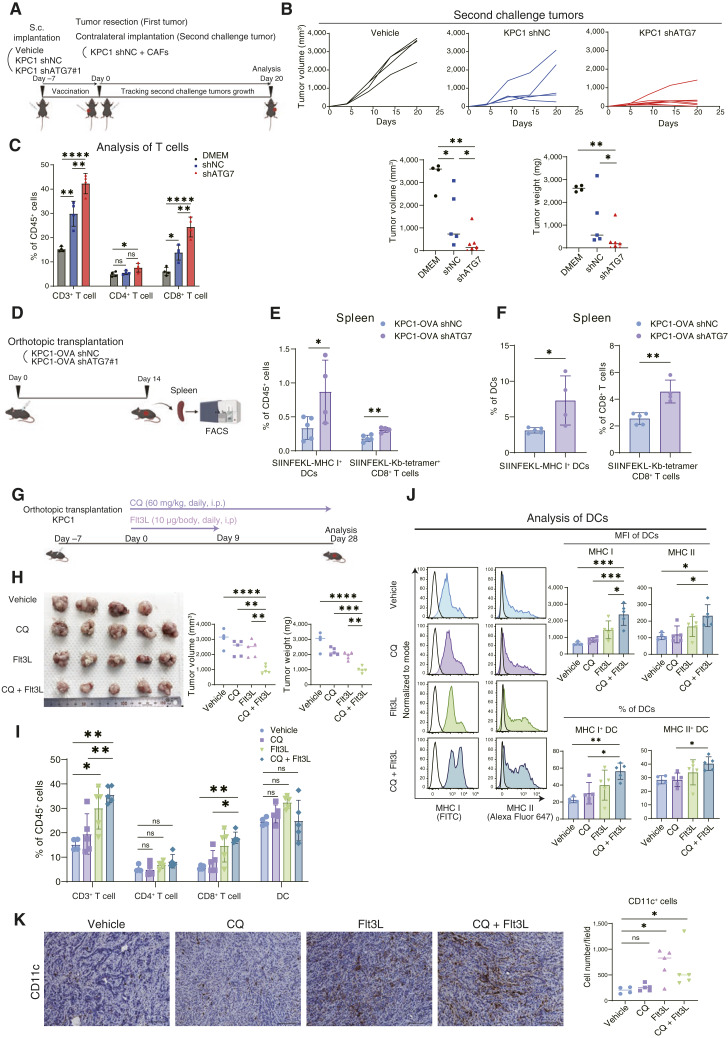
To address whether autophagy inhibition affects DC-mediated activation of T cells in vivo, we conducted vaccination experiments with tumor transplantation, removal, and subsequent second tumor challenge. First, KPC1 shNC or KPC1 shATG7 cells were subcutaneously implanted into the left flanks of C57BL/6 mice. One week after implantation, the first tumors were resected, and all the mice were rechallenged with second untreated KPC1 cells together with CAFs in the contralateral flanks (Fig. 4A). When the mice were vaccinated with autophagy-deficient KPC1 shATG7 cells, tumor growth of the second tumors of untreated KPC1 cells were prohibited more potently than that of the tumors in the mice vaccinated with KPC1 shNC cells (Fig. 4B). This result was replicated in the mice vaccinated with KPC1 shATG5 cells (Supplementary Fig. S7A). Flow cytometry analysis revealed that the number of tumor-infiltrating CD8+ T cells in the second-challenged tumors was significantly higher in the mice vaccinated with autophagy-deficient cancer cells than in those vaccinated with control cells (Fig. 4C), indicating that autophagy inhibition in cancer cells increased the ability of DCs to effectively prime CD8+ T cells.
Figure 4.
Combination of autophagy inhibition and DC induction synergistically suppresses tumor growth. A, Schematic outline of the in vivo vaccination experiment. B, Individual tumor growth curves and the corresponding quantification (on day 20) of the secondary challenge tumors from mice vaccinated with vehicle, KPC1 shNC, or KPC1 shATG7. C, Flow cytometry analysis of tumor-infiltrating lymphocytes in the secondary challenge tumors. D, Schematic outline of the experimental procedure used to evaluate antigen-specific CD8+ T cells in vivo. E and F, KPC1-OVA shNC or shATG7 tumors were orthotopically transplanted into C57BL/6 mice. Fourteen days later, the spleens were resected and analyzed by flow cytometry; DCs presenting the OVA-derived peptide SIINFEKL and antigen-specific CD8+ T cells bound to the H-2Kb OVA tetramer-SIINFEKL were quantified. The percentages of SIINFEKL-MHC I+ DCs and antigen-specific CD8+ T cells within the total CD45+ cell population were evaluated (E). The percentages of SIINFEKL-MHC I+ DCs/total DCs and antigen-specific CD8+ T cells/total CD8+ T cells were also assessed (F). G, Schematic outline of the treatment experiment used to assess the synergy between CQ and Flt3L in vivo. H–K, KPC1 tumors were orthotopically transplanted into C57BL/6 mice. Tumor-bearing mice were treated with vehicle, CQ (60 mg/kg), Flt3L (10 µg), or CQ + Flt3L. After 28 days of treatment, the tumor volumes and weights were measured (H). Flow cytometry was then used to determine the extent of tumor-infiltrating T-cell (I) and DC activation (J). The mean fluorescence intensities (MFI) of MHC I/II in DCs and the percentage of MHC I/II+ DCs (% of DCs) are shown (J). IHC analysis for CD11c was performed to assess the absolute number of DCs infiltrating in tumors (K). Scale bars, 100 μm (K). Bars, median; Error bars, mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001; analyzed using the one-way ANOVA (C and H–J), Student t test or Mann–Whitney test (B, E, F, and K). (A,D, and G, Created with BioRender.com.)
We next examined whether antigen-specific T cells are induced in the spleens of C57BL/6 mice implanted with KPC1-OVA cells (Fig. 4D). The number of DCs presenting the SIINFEKL on the MHC I was significantly increased in the mice transplanted with KPC1-OVA shATG7 cells compared with those in the mice transplanted with KPC1-OVA shNC cells. The number of SIINFEKL-H-Kb tetramer+ CD8+ T cells was significantly increased in the spleens of KPC1-OVA shATG7 cell–transplanted mice compared with that of control cell–transplanted mice (Fig. 4E and F). Thus, autophagy inhibition in cancer cells expands antigen-specific CD8+ T cells through enhancing antigen presentation in DCs.
CQ and Flt3L combination therapy suppresses tumor growth
We examined whether the pharmacologic blockade of autophagy with CQ suppresses tumor growth in the orthotopic tumor model. Autophagy was successfully blocked by CQ with the findings of the accumulation of LC3AB and p62 in the IHC analysis; however, monotherapy of CQ did not affect tumor growth, as previously reported (Fig. 4G; Supplementary Fig. S7B; refs. 12, 13, 22). We reasoned that recruitment of DCs with CQ only is not sufficient to induce tumor immunity sufficient to exhibit antitumor effects in the orthotopic model, and thus we investigated whether accelerated recruitment of DCs with the feline McDonough sarcoma-related receptor tyrosine kinase 3 ligand (Flt3L) may induce antitumor effects in orthotopic tumors (Fig. 4G). Flt3L is crucial for DC development and has been reported to promote DC infiltration into tumors (12, 13). Although a single treatment with Flt3L did not affect tumor growth, as previously reported (12, 13, 22), a combination of CQ and Flt3L significantly inhibited tumor growth in the pancreas (Fig. 4H). Flow cytometry analysis showed that CQ plus Flt3L significantly increased the number of tumor-infiltrating CD8+ T cells in the tumors, as well as MHC I and MHC II expressions on the surface of DCs, compared with single treatment with each agent alone (Fig. 4I and J). We assessed the absolute number of tumor-infiltrating DCs by IHC analysis and confirmed that the administration of Flt3L promotes DC infiltration into the tumors (Fig. 4K). Thus, the combination of pharmacologic autophagy blockade and accelerated DC infiltration with CQ and Flt3L exhibits tumor-suppressive effects in orthotopic tumors, whereas its potency is not sufficient to eliminate tumors completely.
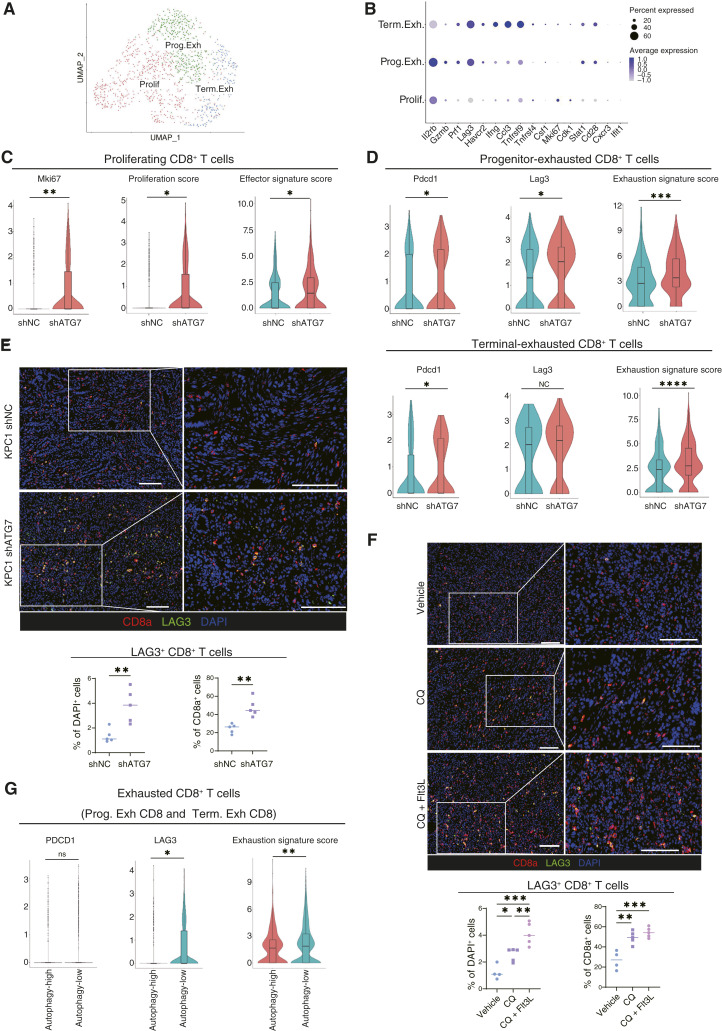
Autophagy inhibition in cancer cells induces CD8+ T-cell exhaustion, characterized by increased expression of Lag3
To search for any other factors that hamper autophagy inhibition to exert tumor immunity sufficient to shrink mouse PDAC tumors in the orthotopic model, we analyzed the T-cell population in our scRNA-seq data of autophagy-defective tumors. We extracted C0, C2, C4, and C6 as T-cell clusters from the cluster of CD45+ immune cells classified by UMAP dimensional reduction (Fig. 2C). T cells expressing Cd8a were further identified within these clusters, and these CD8+ T-cell clusters were reclustered into three clusters of terminal exhausted (Term. Exh), progenitor exhausted (Prog. Exh), and proliferating (Prolif) CD8+ T cells according to the expression of genes previously used in the classification of these CD8+ T-cell subsets (Fig. 5A and B; Supplementary Fig. S8A; Supplementary Table S8). Although we found no significant differences in the proportions of those three CD8+ T-cell clusters between autophagy-deficient tumors (shAtg7) and control tumors (shNC; Supplementary Fig. S8B and S8C), the effector and proliferation signature scores of Prolif CD8+ T cells were significantly higher in the autophagy-deficient tumors than in the control tumors (Fig. 5C; Supplementary Fig. S8D; Supplementary Table S9). On the other hand, the exhaustion scores of the Prog. Exh and Term. Exh CD8+ T cells were significantly higher in the autophagy-deficient tumors than in the control tumors (Fig. 5D; Supplementary Table S9). Interestingly, lymphocyte-activation gene 3 (Lag3) was identified as the most highly upregulated gene in the autophagy-defective tumors among the immunosuppressive molecules, including Pdcd1 (Supplementary Fig. S8E). When we performed If analysis of the tumor sections, the number of cytotoxic CD8+ T cells (i.e., granzyme B+ and perforin+) increased in the autophagy-deficient tumors as expected (Supplementary Fig. S8F and S8G). Consistent with scRNA-seq, both the number and proportion of Lag3+CD8+ T cells were significantly higher in the shATG7 tumors than in the shNC controls (Fig. 5E; Supplementary Fig. S8H). Immunofluorescent staining of the tumors treated with CQ and Flt3L also showed that the combination therapy significantly increased the number of tumor-infiltrating Lag3+CD8+ T cells compared with that in the control tumors (Fig. 5F).
Figure 5.
Autophagy inhibition in cancer cells induces exhaustion of tumor-infiltrating CD8+ T cells, characterized by high LAG3 expression. A, scRNA-seq analysis of CD8+ T cells from the immune cell populations shown in Fig. 1A. CD8+ T cells were reclustered into three clusters. B, Expression of key signature genes used to identify the specific CD8+ T-cell clusters. C and D, The expression levels of gene sets related to CD8+ T-cell function was analyzed in the Prolif (C), Prog. Exh, and Term. Exh (D) CD8+ T cells. “Proliferation score,” “effector signature score” (C), and “exhaustion signature score” (D) were calculated using the gene sets listed in Supplementary Table S9. E and F, Representative IF images of orthotopic syngeneic KPC1 shNC/shATG7 tumors (E) and orthotopic syngeneic KPC1 tumors treated with the combination therapy (CQ ± Flt3L; F). Red, CD8a; green, LAG3. Quantification of LAG3+ CD8a+ cells/DAPI+ cells (%) and LAG3+ CD8a+ cells/CD8a+ cells (%) is shown. G, The expression levels of featured genes related to T-cell exhaustion were analyzed in exhausted CD8+ T-cell clusters (“5” and “6” in Fig. 1D) in human PDAC scRNA-seq data. Scale bars, 100 µm (E and F). Bars, median; boxplots show a centerline, median; box limits, upper and lower quartiles; whiskers that extend up to 1.5× the IQR beyond the upper and lower quartiles (C, D, and G). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; analyzed using the Wilcoxon rank-sum test (C, D, and G), Student t test (E), and one-way ANOVA (F).
Furthermore, we analyzed the scRNA-seq data of patients with PDAC to examine if any correlation between autophagy in cancer cells and T-cell exhaustion in human PDAC tumors. The clusters of Prog. Exh and Term. Exh CD8+ T cells were identified within the CD8+ T-cell population (Fig. 1E); the expression of exhaustion-related genes in these two clusters was significantly higher in the autophagy-low group than in the autophagy-high group (Fig. 5G). Consistent with murine PDAC tumors, LAG3 expression was highly upregulated in the autophagy-low group more significantly than PDCD1 expression.
Therefore, although autophagy-targeted therapy enhances antitumor adaptive immune response by promoting DC activation, it also leads to CD8+ T-cell exhaustion associated with increased expression of LAG3, implicating that Lag3 inhibition may be useful to boost the antitumor effects of autophagy inhibitor plus DC maturation agents.
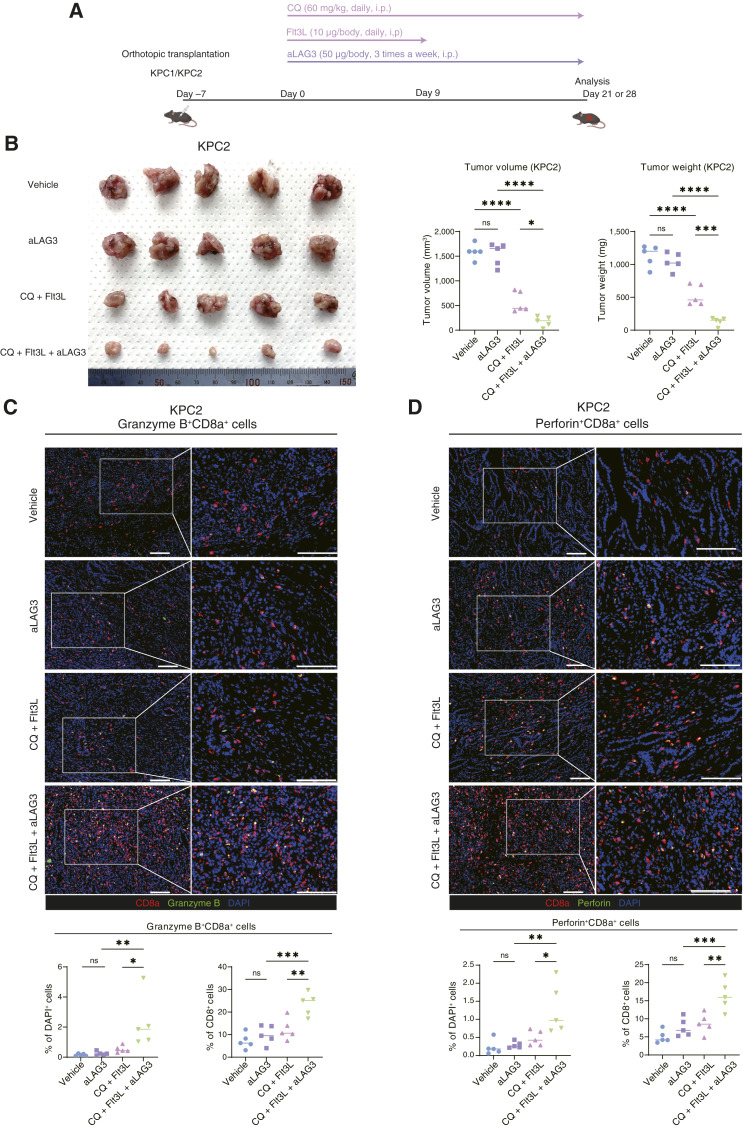
Triple-therapy with CQ, Flt3L, and an anti-LAG3 antibody markedly decreases tumor growth in murine syngeneic PDAC models
We next evaluated the therapeutic efficacy of a triple-therapy comprising CQ, Flt3L, and an anti-LAG3 antibody (aLAG3; Fig. 6A). Although aLAG3 monotherapy did not have any effects on tumor size, the triple-combination therapy with CQ, Flt3L, and aLAG3 markedly reduced the tumor growth of KPC2 cells, indicating the existence of an obvious synergistic relationship between CQ, Flt3L, and aLAG3 (Fig. 6B). These synergistic effects were reproducible when using the other cell line KPC1 (Supplementary Fig. S9A and S9B). IF staining showed that the number and proportion of activated cytotoxic CD8+ T cells in the tumors were significantly higher in the triple-treatment group than in the control group (Fig. 6C and D; Supplementary Fig. S9C and S9D). In addition, we confirmed that Flt3L is essential to fully exert the tumor-suppressive effect of the triple-combination therapy, demonstrating the need for the induction of DC infiltration in this new combination therapy (Supplementary Fig. S9E). These results demonstrate that triple-therapy with CQ, Flt3L, and aLAG3 is a potent inducer of antitumor immunity to break through the suppressive TIME of PDAC.
Figure 6.
Triple-therapy consisting of CQ, Flt3L, and aLAG3 markedly reduces the growth of orthotopic syngeneic PDAC tumors. A, Schematic outline of the triple treatment protocol involving CQ, Flt3L, and aLAG3. B–D, KPC2 cells were orthotopically transplanted into C57BL/6 mice. Tumor-bearing mice were treated with vehicle, aLAG3 (50 µg), CQ (60 mg/kg) + Flt3L (10 µg), or CQ + Flt3L + aLAG3. After 28 days of treatment, the tumor volumes and weights were measured (B). IF was used to characterize cytotoxic CD8+ T cells as granzyme B+ CD8a+ T cells (red, CD8a; green, granzyme B; C) and perforin+ CD8a+ T cells (red, CD8a; green, perforin; D). Representative images are shown. Quantification of granzyme B+ CD8a+ cells/DAPI+ cells (%) and granzyme B+ CD8a+ cells/CD8a+ cells (%; C); and perforin+ CD8a+ cells/DAPI+ cells (%) and perforin+ CD8a+ cells/CD8a+ cells (%; D) is shown in the bar graphs under the corresponding IF images. Scale bars, 100 µm (C and D). Bars, median; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant; analyzed using one-way ANOVA (B–D). (A, Created with BioRender.com.)
Discussion
In this study, we elucidated the novel immunostimulatory mechanism by autophagy inhibition, showing that inhibition of autophagy in cancer cells induces DC activation via increased accumulation of intracellular tumor antigen. On the other hand, the scRNA-seq analysis revealed that autophagy inhibition induced significant exhaustion of CD8+ T cells, characterized by the increased expression of LAG3, and the triple-therapy with CQ, Flt3L, and aLAG3 markedly reduced the tumors in orthotopic syngeneic PDAC models.
To the best of our knowledge, this is the first study to demonstrate how the inhibition of autophagy in cancer cells altered the transcriptional dynamics of the TIME using scRNA-seq. Our scRNA-seq analysis of DCs revealed that the expression of genes related to DC functions was significantly increased following autophagy inhibition in cancer cells, especially in the DC2 and migDC subsets. It was previously thought that cross-presentation to CD8+ T cells was predominantly a functional characteristic of the DC1 subset; however, it was recently discovered that the DC2 subset plays an important role in cross-presentation (40, 51). Duong and colleagues (40) recently reported that type 1 IFN, secreted by cancer cells, induced the activation of CD11b+ conventional DCs (i.e., the DC2 subset), which were characterized by the expression of interferon-stimulated genes and the capacity to present intact tumor-derived peptide–MHC I complexes. These findings led us to initially hypothesize that the activation of the DC2 subset by autophagy-deficient cancer cells may have been due to the increased type 1 IFN secretion by cancer cells. However, the present study revealed that autophagy-deficient cancer cells induced DC activation independently of cancer cell–derived type 1 IFN. We subsequently identified the accumulation of tumor antigens (antigenicity) as a mechanism of DC activation by autophagy-deficient cancer cells. The result that indirect coculture did not induce DC activation also supports the mechanism that autophagy-deficient cancer cells induce DC activation not by secreted factors, such as cytokines and chemokines, but by taking up more antigens in contact with cancer cells.
Following the result that autophagy-deficient cancer cells induced DC activation, we investigated the antitumor efficacy of the combination of CQ and Flt3L. Although there was a clear synergy between these agents, they were unable to achieve tumor clearance. The scRNA-seq analysis of CD8+ T cells revealed that autophagy inhibition in cancer cells not only promoted CD8+ T-cell activation but also markedly contributed to their exhaustion. Interestingly, the dominant CD8+ T-cell exhaustion phenotype was high LAG3 expression. Given that PDAC is largely resistant to the effects of anti-PD-1 (aPD-1) or anti-CTLA4 (aCTLA4) antibodies (52, 53), we wondered whether a combination immunotherapy comprising aLAG3 could modify the suppressive TIME in PDAC. Indeed, the CQ, Flt3L, and aLAG3 triple-therapy markedly reduced the PDAC tumor volume. LAG3, an immune checkpoint receptor expressed on the plasma membrane of both activated cytotoxic T cells and Tregs, has received much attention as the third most promising targeted immune checkpoint molecule (54). LAG3 induces T-cell dysfunction by strongly binding to stable peptide-loaded MHC II molecules on the APCs, thereby preventing the binding of the T-cell receptor to the same peptide–MHC II site (55). Thus, blocking LAG3 in our experiments may have enabled T cells to productively engage with DCs by binding to free peptide–MHC II molecules. Recently, Gulhati and colleagues (56) reported that 93% of patients with PDAC have LAG3-expressing T cells, explaining why aLAG3 exerted a markedly better tumor suppressive effect in orthotopic syngeneic PDAC models than aPD-1 or aCTLA4 and advocating LAG3 as a promising therapeutic target in PDAC. Although a previous study reported the combined effect of CQ and aPD-1/aCTLA4 (22), our triple-therapy focusing on increasing DC activation and maturation (by inhibiting autophagy in cancer cells and administering Flt3L, respectively) and alleviation of T-cell exhaustion (by blocking LAG3) could potentially elicit superior outcomes in PDAC. Although further validation is needed, the triple therapy of CQ, Flt3L, and aLAG3 is certainly promising therapeutic candidate, which could potentially overcome the immunosuppressive TME of PDAC.
Although several studies have already shown that autophagy in cancer cells can be therapeutically targeted to enhance antitumor immunity (22, 24, 48), we are the first to demonstrate that the inhibition of cancer cell autophagy triggered DC activation by increasing the accumulation of antigens. A study by Yamamoto and colleagues (22) showed that the inhibition of autophagy increased MHC I expression on cancer cells, which promoted their recognition and attack by cytotoxic T cells. Thus, DC activation may be considered as a secondary consequence of enhanced antitumor immunity. Meanwhile, the results of our in vitro coculture and in vivo experiments in T-cell–deficient BALB/c-nu mice showed that autophagy-deficient cancer cells directly induced DC activation. Specifically, we demonstrated that the increase in DC activation was due to the accumulation, and autophagy-deficient cancer cells elicited better vaccination outcomes and were more efficient at priming CD8+ T cells against the experimental OVA antigen than the same cancer cells with normal levels of autophagy. These results revealed a novel immunostimulatory mechanism of autophagy inhibition in cancer cells in a T-cell–independent manner, which suggests that the increase in immunogenicity of cancer cells by autophagy inhibition may occur in a prior step to the blocking MHC I degradation previously identified.
In this study, we focused on autophagy in cancer cells; however, the systemic effects of autophagy inhibition on other cells have not been fully evaluated. Although some studies have indicated that autophagy in DCs may influence the cross-presentation capacity of DCs, this relationship remains controversial (57–59). Our results showed that CQ administration in cancer cell and DC coculture assays, which inhibited autophagy in both of these cell types, increased the expression of DC activation and maturation genes, suggesting that systemic autophagy inhibition by CQ did not attenuate DC function. Other reports have indicated that autophagy in other cell populations, such as macrophages and CAFs, can also affect tumor progression (18, 60). Therefore, determining how inhibiting autophagy in other cell populations affects the TME remains a challenge for the future. Furthermore, establishing the predictive markers that might be associated with a better response to autophagy inhibition is also one of the important issues. We expect that the expressions of LC3 and p62 in cancer cells might be one of the predictive markers based on the hypothesis that autophagy inhibition has a potent effect in patients with high autophagy flux. Further investigation would be needed to identify patients highly responsive to this therapy.
The present study has several limitations. First, the increase of DC function in DC1 cluster was not found in our scRNA-seq analysis. The evaluation of transcriptional level by scRNA-seq alone might not be definitive to show the activation or dysfunction of DC. We consider that functional evaluations by coculture assay or cross-presentation assay using each separated cluster, such as DC1, DC2, and migDC, are necessary to confirm the difference in their activation in the future. Second, only investigation with artificial antigens such as OVA and EGFP is not sufficient. Investigating the recognition of endogenous antigens in cancer cells after autophagy inhibition will be important subjects of future work.
In summary, the present study elucidated the mechanisms underlying how autophagy inhibition in cancer cells synergizes with immunotherapy to significantly reduce tumor growth. The mechanistic insights described here provide a strong rationale for progressing toward clinical trials of the triple-therapy comprising an autophagy inhibitor to evaluate its clinical impact on patients with PDAC.
Supplementary Material
Analysis of human PDAC tumors reveals a negative correlation between autophagy in cancer cells and DC activation
Autophagy inhibition in cancer cells triggers T-cell-mediated immune response
scRNA-seq analysis of orthotopic syngeneic KPC1 shNC and shATG7 tumors
Autophagy inhibition in cancer cells induces the activation of DCs in several murine autophagy-deficient PDAC models
In vitro coculture assays reveal that autophagy-deficient cancer cells directly induce DC activation
Inhibiting autophagy in cancer cells increases the accumulation of intracellular antigens
Vaccination experiments with KPC1 shNC/shATG5 and confirmation of the effect of CQ treatment
scRNA-seq analysis and immunofluorescence images revealed that CD8+ T cells in KPC-shATG7 tumors express high LAG3 levels
Triplet therapy comprising CQ, Flt3L, and aLAG3 markedly suppresses tumor growth
Lists of autophagy-related genes used to calculate the autophagy signature scores of ductal cells
Autophagy signature scores for each sample from the public PDAC patient scRNA-seq dataset
Gene lists and sources of each gene sets used in the analysis of public PDAC patient scRNA-seq data
Relationship Between Cancer Cell-LC3 Expression and Clinicopathologic Factors
Top 30 DEGs for four DC clusters identified from scRNA-seq of CD45+ immune cells in KPC1 shNC / shATG7 syngeneic PDAC tumors
Gene lists and sources for each DC-associated gene set used in the scRNA-seq analysis of CD45+ immune cells from the KPC1 shNC/shATG7 syngeneic PDAC tumors
Top 30 DEGs for three CD8+ T cell clusters identified from scRNA-seq of CD45+ immune cells in KPC1 shNC / shATG7 transplanted syngeneic PDAC tumors
Gene lists and sources for each CD8+ T-cell-associated gene set used in the scRNA-seq analysis of CD45+ immune cells in the KPC1 shNC/shATG7 syngeneic PDAC tumors
Key Resources
Acknowledgments
We would like to thank S. Sadatomi, E. Manabe, and N. Torata (Department of Surgery and Oncology, Kyushu University) and members of The Research Support Center, Research Center for Human Disease Modeling, Kyushu University Graduate School of Medical Sciences, for their technical assistance. We are grateful to N. Koya and M. Umebayashi (Department of Cancer Immunotherapy, Fukuoka General Cancer Clinic) for their help with human PDAC cell and DC coculture assays. We would like to thank Anya Lissina, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript. This work was supported by JSPS KAKENHI grant numbers JP22H00480 (M. Nakamura), JP22H02922 (N. Ikenaga), JP22K15560 (T. Abe), JP21K19530 (K. Ohuchida), JP23H02982 (K. Nakata), and JP23KJ1748 (K. Oyama).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors’ Disclosures
No disclosures were reported.
Authors’ Contributions
K. Oyama: Conceptualization, data curation, formal analysis, validation, investigation, methodology, writing–original draft. K. Nakata: Conceptualization, supervision, funding acquisition, methodology, writing–original draft, project administration, writing–review and editing. C. Tsutsumi: Formal analysis, investigation. M. Hayashi: Investigation. B. Zhang: Investigation. Y. Mochida: Investigation. T. Shinkawa: Investigation. K. Hirotaka: Validation, investigation. P. Zhong: Investigation. S. Date: Investigation. H. Luo: Investigation. A. Kubo: Investigation. N. Higashijima: Investigation. Y. Yamada: Investigation. T. Abe: Funding acquisition, writing–review and editing. N. Ideno: Writing–review and editing. K. Koikawa: Writing–review and editing. C. Iwamoto: Writing–review and editing. N. Ikenaga: Funding acquisition, writing–review and editing. K. Ohuchida: Funding acquisition, project administration, writing–review and editing. H. Onishi: Writing–review and editing. T. Morisaki: Resources, writing–review and editing. K. Kuba: Writing–review and editing. Y. Oda: Resources. M. Nakamura: Supervision, funding acquisition, project administration, writing–review and editing.
References
- 1. Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin 2024;74:12–49. [DOI] [PubMed] [Google Scholar]
- 2. Larkin J, Chiarion-Sileni V, Gonzalez R, Grob J-J, Rutkowski P, Lao CD, et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 2019;381:1535–46. [DOI] [PubMed] [Google Scholar]
- 3. Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med 2015;372:2521–32. [DOI] [PubMed] [Google Scholar]
- 4. Karamitopoulou E. Tumour microenvironment of pancreatic cancer: immune landscape is dictated by molecular and histopathological features. Br J Cancer 2019;121:5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017;377:2500–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 2017;551:512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liudahl SM, Betts CB, Sivagnanam S, Morales-Oyarvide V, da Silva A, Yuan C, et al. Leukocyte heterogeneity in pancreatic ductal adenocarcinoma: phenotypic and spatial features associated with clinical outcome. Cancer Discov 2021;11:2014–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dias Costa A, Väyrynen SA, Chawla A, Zhang J, Väyrynen JP, Lau MC, et al. Neoadjuvant chemotherapy is associated with altered immune cell infiltration and an anti-tumorigenic microenvironment in resected pancreatic cancer. Clin Cancer Res 2022;28:5167–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Böttcher JP, Reis e Sousa C. The role of type 1 conventional dendritic cells in cancer immunity. Trends Cancer 2018;4:784–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wculek SK, Cueto FJ, Mujal AM, Melero I, Krummel MF, Sancho D. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol 2020;20:7–24. [DOI] [PubMed] [Google Scholar]
- 12. Hegde S, Krisnawan VE, Herzog BH, Zuo C, Breden MA, Knolhoff BL, et al. Dendritic cell paucity leads to dysfunctional immune surveillance in pancreatic cancer. Cancer Cell 2020;37:289–307.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin JH, Huffman AP, Wattenberg MM, Walter DM, Carpenter EL, Feldser DM, et al. Type 1 conventional dendritic cells are systemically dysregulated early in pancreatic carcinogenesis. J Exp Med 2020;217:e20190673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000;290:1717–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell 2019;176:11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med 2019;25:2521–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med 2019;25:628–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Endo S, Nakata K, Ohuchida K, Takesue S, Nakayama H, Abe T, et al. Autophagy is required for activation of pancreatic stellate cells, associated with pancreatic cancer progression and promotes growth of pancreatic tumors in mice. Gastroenterology 2017;152:1492–506.e24. [DOI] [PubMed] [Google Scholar]
- 19. Zeh HJ, Bahary N, Boone BA, Singhi AD, Miller-Ocuin JL, Normolle DP, et al. A randomized phase II preoperative study of autophagy inhibition with high-dose hydroxychloroquine and gemcitabine/nab-paclitaxel in pancreatic cancer patients. Clin Cancer Res 2020;26:3126–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boone BA, Bahary N, Zureikat AH, Moser AJ, Normolle DP, Wu WC, et al. Safety and biologic response of pre-operative autophagy inhibition in combination with gemcitabine in patients with pancreatic adenocarcinoma. Ann Surg Oncol 2015;22:4402–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karasic TB, O’Hara MH, Loaiza-Bonilla A, Reiss KA, Teitelbaum UR, Borazanci E, et al. Effect of gemcitabine and nab-paclitaxel with or without hydroxychloroquine on patients with advanced pancreatic cancer: a phase 2 randomized clinical trial. JAMA Oncol 2019;5:993–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yamamoto K, Venida A, Yano J, Biancur DE, Kakiuchi M, Gupta S, et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nat Res 2020;581:100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ito Y, Pan D, Zhang W, Zhang X, Juan TY, Pyrdol JW, et al. Addressing tumor heterogeneity by sensitizing resistant cancer cells to T cell-secreted cytokines. Cancer Discov 2023;13:1186–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Noman MZ, Parpal S, van Moer K, Xiao M, Yu Y, Viklund J, et al. Inhibition of Vps34 reprograms cold into hot inflamed tumors and improves anti-PD-1/PD-L1 immunotherapy. Sci Adv 2020;6:eaax7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bachem MG, Schünemann M, Ramadani M, Siech M, Beger H, Buck A, et al. Pancreatic carcinoma cells induce fibrosis by stimulating proliferation and matrix synthesis of stellate cells. Gastroenterology 2005;128:907–21. [DOI] [PubMed] [Google Scholar]
- 26. Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005;7:469–83. [DOI] [PubMed] [Google Scholar]
- 27. Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell 1994;76:17–27. [DOI] [PubMed] [Google Scholar]
- 28. Glass G, Papin JA, Mandell JW. SIMPLE: a sequential immunoperoxidase labeling and erasing method. J Histochem Cytochem 2009;57:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marin I, Boix O, Garcia-Garijo A, Sirois I, Caballe A, Zarzuela E, et al. Cellular senescence is immunogenic and promotes antitumor immunity. Cancer Discov 2023;13:410–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu C, Guan J, Lu S, Jin Q, Rousseau B, Lu T, et al. DNA sensing in mismatch repair-deficient tumor cells is essential for anti-tumor immunity. Cancer Cell 2021;39:96–108.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morisaki T, Hikichi T, Onishi H, Morisaki T, Kubo M, Hirano T, et al. Intranodal administration of neoantigen peptide-loaded dendritic cell vaccine elicits epitope-specific T cell responses and clinical effects in a patient with chemorefractory ovarian cancer with malignant ascites. Immunol Invest 2020;50:562–79. [DOI] [PubMed] [Google Scholar]
- 32. Liu X, Jiang J, Liao Y-P, Tang I, Zheng E, Qiu W, et al. Combination chemo-immunotherapy for pancreatic cancer using the immunogenic effects of an irinotecan silicasome nanocarrier plus anti-PD-1. Adv Sci (Weinh) 2021;8:2002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ferris ST, Durai V, Wu R, Theisen DJ, Ward JP, Bern MD, et al. cDC1 prime and are licensed by CD4+ T cells to induce anti-tumour immunity. Nature 2020;584:624–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst 2019;8:329–37.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 2019;50:1317–34.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bosteels C, Neyt K, Vanheerswynghels M, van Helden MJ, Sichien D, Debeuf N, et al. Inflammatory type 2 cDCs acquire features of cDC1s and macrophages to orchestrate immunity to respiratory virus infection. Immunity 2020;52:1039–56.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maier B, Leader AM, Chen ST, Tung N, Chang C, LeBerichel J, et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 2020;580:257–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baldominos P, Barbera-Mourelle A, Barreiro O, Huang Y, Wight A, Cho JW, et al. Quiescent cancer cells resist T cell attack by forming an immunosuppressive niche. Cell 2022;185:1694–708.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duong E, Fessenden TB, Lutz E, Dinter T, Yim L, Blatt S, et al. Type I interferon activates MHC class I-dressed CD11b+ conventional dendritic cells to promote protective anti-tumor CD8+ T cell immunity. Immunity 2022;55:308–23.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 2019;20:326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Beltra J-C, Manne S, Abdel-Hakeem MS, Kurachi M, Giles JR, Chen Z, et al. Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 2020;52:825–41.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bhatt D, Kang B, Sawant D, Zheng L, Perez K, Huang Z, et al. STARTRAC analyses of scRNAseq data from tumor models reveal T cell dynamics and therapeutic targets. J Exp Med 2021;218:e20201329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oliveira G, Stromhaug K, Klaeger S, Kula T, Frederick DT, Le PM, et al. Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature 2021;596:119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peng J, Sun B-F, Chen C-Y, Zhou J-Y, Chen Y-S, Chen H, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res 2019;29:725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dixon KO, Tabaka M, Schramm MA, Xiao S, Tang R, Dionne D, et al. TIM-3 restrains anti-tumour immunity by regulating inflammasome activation. Nature 2021;595:101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev 2011;25:717–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jiang H, Courau T, Borison J, Ritchie AJ, Mayer AT, Krummel MF, et al. Activating immune recognition in pancreatic ductal adenocarcinoma via autophagy inhibition, MEK blockade and CD40 agonism. Gastroenterology 2022;162:590–603.e14. [DOI] [PubMed] [Google Scholar]
- 49. Gui X, Yang H, Li T, Tan X, Shi P, Li M, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 2019;596:262–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Herr W, Ranieri E, Olson W, Zarour H, Gesualdo L, Storkus WJ. Mature dendritic cells pulsed with freeze-thaw cell lysates define an effective in vitro vaccine designed to elicit EBV-specific CD4+ and CD8+ T lymphocyte responses. Am Soc Hematol 2000;96:1857–64. [PubMed] [Google Scholar]
- 51. He M, Roussak K, Ma F, Borcherding N, Garin V, White M, et al. CD5 expression by dendritic cells directs T cell immunity and sustains immunotherapy responses. Science 1979;2023:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O’Reilly EM, Oh DY, Dhani N, Renouf DJ, Lee MA, Sun W, et al. Durvalumab with or without tremelimumab for patients with metastatic pancreatic ductal adenocarcinoma: a phase 2 randomized clinical trial. JAMA Oncol 2019;5:1431–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wainberg ZA, Hochster HS, Edward J-HK, George B, Kalyan A, Chiorean EG, et al. Phase I study of nivolumab (Nivo) + nab-paclitaxel (nab-P) + gemcitabine (Gem) in advanced pancreatic cancer (APC). J Clin Oncol 2019;37:298. [Google Scholar]
- 54. Lui Y, Davis SJ. LAG-3: a very singular immune checkpoint. Nat Immunol 2018;19:1278–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maruhashi T, Okazaki I-M, Sugiura D, Takahashi S, Maeda TK, Shimizu K, et al. LAG-3 inhibits the activation of CD4+ T cells that recognize stable pMHCII through its conformation-dependent recognition of pMHCII. Nat Immunol 2018;19:1415–26. [DOI] [PubMed] [Google Scholar]
- 56. Gulhati P, Schalck A, Jiang S, Shang X, Wu C-J, Hou P, et al. Targeting T cell checkpoints 41BB and LAG3 and myeloid cell CXCR1/CXCR2 results in antitumor immunity and durable response in pancreatic cancer. Nat Cancer 2023;4:62–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ravindran R, Khan N, Nakaya HI, Li S, Loebbermann J, Maddur MS, et al. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science 2014;343:313–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tey SK, Khanna R. Autophagy mediates transporter associated with antigen processing- independent presentation of viral epitopes through MHC class I pathway. Blood 2012;120:994–1004. [DOI] [PubMed] [Google Scholar]
- 59. Ho NI, Camps MGM, Verdoes M, Münz C, Ossendorp F. Autophagy regulates long-term cross-presentation by murine dendritic cells. Eur J Immunol 2021;51:835–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cunha LD, Yang M, Carter R, Guy C, Harris L, Crawford JC, et al. LC3-Associated phagocytosis in myeloid cells promotes tumor immune tolerance. Cell 2018;596:429–41.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of human PDAC tumors reveals a negative correlation between autophagy in cancer cells and DC activation
Autophagy inhibition in cancer cells triggers T-cell-mediated immune response
scRNA-seq analysis of orthotopic syngeneic KPC1 shNC and shATG7 tumors
Autophagy inhibition in cancer cells induces the activation of DCs in several murine autophagy-deficient PDAC models
In vitro coculture assays reveal that autophagy-deficient cancer cells directly induce DC activation
Inhibiting autophagy in cancer cells increases the accumulation of intracellular antigens
Vaccination experiments with KPC1 shNC/shATG5 and confirmation of the effect of CQ treatment
scRNA-seq analysis and immunofluorescence images revealed that CD8+ T cells in KPC-shATG7 tumors express high LAG3 levels
Triplet therapy comprising CQ, Flt3L, and aLAG3 markedly suppresses tumor growth
Lists of autophagy-related genes used to calculate the autophagy signature scores of ductal cells
Autophagy signature scores for each sample from the public PDAC patient scRNA-seq dataset
Gene lists and sources of each gene sets used in the analysis of public PDAC patient scRNA-seq data
Relationship Between Cancer Cell-LC3 Expression and Clinicopathologic Factors
Top 30 DEGs for four DC clusters identified from scRNA-seq of CD45+ immune cells in KPC1 shNC / shATG7 syngeneic PDAC tumors
Gene lists and sources for each DC-associated gene set used in the scRNA-seq analysis of CD45+ immune cells from the KPC1 shNC/shATG7 syngeneic PDAC tumors
Top 30 DEGs for three CD8+ T cell clusters identified from scRNA-seq of CD45+ immune cells in KPC1 shNC / shATG7 transplanted syngeneic PDAC tumors
Gene lists and sources for each CD8+ T-cell-associated gene set used in the scRNA-seq analysis of CD45+ immune cells in the KPC1 shNC/shATG7 syngeneic PDAC tumors
Key Resources
Data Availability Statement
The data generated in this study are available upon request from the corresponding author. The scRNA-seq data have been deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). The accession number is GSE233663. Previously published scRNA-seq data from patients with PDAC analyzed in this study were obtained from the Genome Sequence Archive under the accession number CRA001160.