Abstract
Hydrogels are useful drug release systems and tissue engineering scaffolds. However, synthetic hydrogels often require harsh gelation conditions and can contain toxic by‐products while naturally derived hydrogels can transmit pathogens and in general have poor mechanical properties. Thus, there is a need for a hydrogel that forms under ambient conditions, is non‐toxic, xeno‐free, and has good mechanical properties. A recombinant spider silk protein‐derived hydrogel that rapidly forms at 37 °C is recently developed. The temperature and gelation times are well‐suited for an injectable in situ polymerising hydrogel, as well as a 3D cell culture scaffold. Here, it is shown that the diffusion rate and the mechanical properties can be tuned by changing the protein concentration and that human fetal mesenchymal stem cells encapsulated in the hydrogels show high survival and viability. Furthermore, mixtures of recombinant spider silk proteins and green fluorescent protein (GFP) form gels from which functional GFP is gradually released, indicating that bioactive molecules are easily included in the gels, maintain activity and can diffuse through the gel. Interestingly, encapsulated ARPE‐19 cells are viable and continuously produce the growth factor progranulin, which is detected in the cell culture medium over the study period of 31 days.
Keywords: arpe19, biomaterials, encapsulation, mechanical properties, mesenchymal stem cells, polymerization, progranulin, tissue engineering
Recombinant spider silk based hydrogels can encapsulate and release bioactive molecules and works as a cell culture scaffold. Polymerization of the gel is simply triggered by incubation at 37 °C, and the mechanical properties and diffusion rate are easily tuned to fit those of different tissues.

1. Introduction
Biomaterials are crucial for tissue engineering as they provide structural support for cells to attach, grow, proliferate, and differentiate.[ 1 , 2 , 3 , 4 , 5 ] In general, hydrogels are known to: i) mimic the fibrillar network of the extracellular matrix,[ 6 , 7 ] ii) allow diffusion of nutrients and waste products,[ 8 ] and iii) be versatile as they can be modified to contain cell adhesion sites, or biopharmaceutics such as growth factors, hormones, enzymes or cytokines.[ 9 , 10 , 11 , 12 , 13 ] In some applications, hydrogel encapsulation of bioactive molecules is essential to avoid their degradation or to provide cues to surrounding cells. In addition, hydrogels can often be used for 3D bioprinting which enables the manufacture of specific and complex structures, in some cases even containing live cells.[ 14 , 15 , 16 , 17 , 18 ]
Hydrogel formation is generally induced by either chemical crosslinking that covalently link polymers,[ 19 ] or by physical crosslinking whereby temperature, vortexing, or sonication induce chain entanglements, hydrophobic interactions, and/or hydrogen bonding.[ 20 , 21 , 22 ] A major drawback of both chemical and physical crosslinking is the risk of damaging incorporated cells and/or bioactive substances, as well as surrounding tissue if performed in vivo.
Hydrogels can be made from synthetic polymers, which in general are well‐defined and economically affordable. However, their synthesis may include the use of organic solvents and if the polymerization is incomplete, tedious and expensive purification protocols are needed to remove toxic components and crosslinkers.[ 23 , 24 , 25 ] In addition, toxic degradation products and the lack of cell‐attachment sites may cause problems with cell viability. Naturally derived polymers like collagen, gelatin, and chitosan are generally biodegradable and provide good support for cells.[ 26 ] However, also naturally derived materials are associated with drawbacks, e.g., product impurities, batch‐to‐batch variations, and inferior mechanical properties. Recombinant spider silk protein hydrogels are emerging as a new promising biomaterial that can address these problems.[ 22 , 27 , 28 , 29 , 30 ] However, poor mechanical properties and long gelation times remain major challenges.
Artificial spider silk is a promising biomaterial, provided that it can be manufactured from proteins produced in heterologous hosts. Previously, a biomimetic spider silk production method was developed, which enables the production of artificial spider silk proteins and fibers without involving harsh organic solvents.[ 31 ] This was made possible by the design of a small spider silk protein (spidroin) called NT2RepCT that is as soluble as native spider silk proteins (30–50% w/v) in aqueous buffers and recapitulates important features of native spider silk dope.[ 31 , 32 , 33 , 34 ] A big advantage of NT2RepCT is that it can be produced in large quantities (14.5 g purified protein per liter culture in bioreactors) and is possible to purify in a semi‐automated process.[ 35 ] Recently, we discovered that NT2RepCT and a wide range of additional recombinant spidroins form hydrogels within minutes if incubated at 37 °C, i.e., without the use of crosslinkers.[ 27 ] The temperature and time range for NT2RepCT gel formation are compatible with most bioactive agents and living cells, which make the gels interesting for the development of injectables intended to polymerize in situ and/or for 3D bioprinting under physiological conditions.
Herein, we characterize the NT2RepCT hydrogels and explore their use as cell culture scaffolds and drug release systems (Figure 1 ). We find that diffusion rates and mechanical properties can be tuned by changing the NT2RepCT concentration. Human fetal mesenchymal stem cells (hfMSC) showed high survival and viability when encapsulated for 21 days and 7 days, respectively. Green fluorescent protein (GFP) can be mixed with the NT2RepCT solution and remain active after gelation, and upon incubation in buffer, GFP diffuses out of the gel. Furthermore, we show that progranulin overexpressing ARPE‐19 cells, a spontaneously immortalized retinal pigment epithelia (RPE) cell line,[ 36 ] could be encapsulated in the hydrogels and continued to produce progranulin which was released from the gels for at least 31 days. These data, together with the gels’ mechanical properties indicate that they are good candidates for the development of drug release systems and 3D cell culture scaffolds.
Figure 1.

Production of bioactive hydrogels. NT2RepCT is produced at high yields in a bioreactor and purified with a semi‐automated FPLC system. To form bioactive gels, the NT2RepCT proteins are mixed with cells or GFP and incubated at 37 °C for 120 min to encapsulate the cells or GFP, respectively, within the hydrogel. Created with BioRender.com.
2. Results and Discussion
2.1. Mechanical Properties of NT2RepCT Hydrogels can be Tuned by Varying the Protein Concentration
A key strategy in tissue engineering is to design biomaterials with mechanical properties that can mimic different extracellular environments, as tissue stiffness has various effects on cells including morphology and differentiation fate.[ 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 ] To characterize the mechanical properties of the hydrogels, we performed unconfined compression tests on NT2RepCT hydrogel disks (1 cm height x 1 cm diameter), (Figure 2 ; Figure S1, Supporting Information). The stress at fracture ranged from 4 kPa (50 mg mL−1 hydrogel) to 123 kPa (200 mg mL−1 hydrogel) positively correlating with the protein concentration (Figure 2a; Table S1, Supporting Information). Notably, the strength of the 200 mg mL−1 hydrogels is at least 2 times higher than that of collagen hydrogels.[ 48 , 49 ] The strain at break decreased with increased protein concentration (Figure 2b; Table S1, Supporting Information), but was always higher than mammalian skeletal muscle that typically shows strain at break values of 20%.[ 50 ] The compression modulus, which is indicative of the stiffness of the gel, increased from 14 kPa (50 mg mL−1 hydrogel) to 100 kPa for the 100 mg mL−1 gel, to 317 kPa for the 150 mg mL−1 gel, and to 792 kPa for the 200 mg mL−1 gel (Figure 2c; Movie S1–S4, Supporting Information). The compression modulus of the 50 mg mL−1 hydrogel is in the range of an elastic hydrogel (1–20 kPa) and in line with muscle tissue (25 kPa).[ 51 ]The more concentrated gel samples had a stiffness that was more similar to cardiac muscle (100 kPa).[ 51 , 52 ] and cartilage (240–850 kPa), (Figure 2d).[ 53 ] Thus, the mechanical properties of NT2RepCT hydrogels are tunable and in general superior to other protein‐based hydrogels.[ 48 , 49 , 54 , 55 ]
Figure 2.

Mechanical properties of NT2RepCT hydrogels at 50, 100, 150, and 200 mg mL−1 determined by unconfined compression tests. a) Strength, b) Strain at break, c) Compressive modulus and d) Comparison of the compression modulus of hydrogels characterized in this study and the compression modulus of different tissues.[ 51 , 52 ] The error bars represent the standard error of mean. *p<0.05; **p < 0.01; no asterisks are shown for p > 0.05.
2.2. Recombinant Spider Silk Hydrogels have Tunable Diffusion Rates and form A Dense Fibrillar Network
Efficient diffusion through hydrogels is important as it ensures the exchange of nutrients, oxygen, growth factors, and waste products. To determine the diffusion coefficients, we used fluorescence recovery after photobleaching (FRAP), which is based on photobleaching of a fluorescent probe with a laser, after which neighboring fluorescent molecules diffuse into the bleached area at a rate that depends on the diffusion coefficient.[ 56 ] We applied FRAP on gels formed from NT2RepCT solutions with concentrations of 50, 100, 200, and 300 mg mL−1 in the presence of fluorescein isothiocyanate (FITC)‐labeled dextran of different molecular weights (40, 70, and 250 kDa). The results show that the size of FITC‐dextrans and NT2RepCT protein concentration negatively correlated with the diffusion coefficient, i.e., higher molecular weight FITC‐dextrans and higher protein concentrations result in lower diffusion coefficients (Figure 3a; Table S2, Supporting Information). Hence, diffusion can be tuned by varying the NT2RepCT concentration to fit the intended application. The diffusion coefficients in NT2RepCT hydrogels are lower than in many other hydrogels[ 57 , 58 , 59 , 60 , 61 , 62 , 63 ] which would be ideal for the development of a sustained drug release system or for encapsulating cells.[ 64 ] The decreased macromolecular mobility in high concentration hydrogels and the relatively slow diffusion are likely a consequence of the dense NT2RepCT nano‐fibrillar network in the gels (Figure 3b).[ 27 ]
Figure 3.

Diffusion and morphology of NT2RepCT hydrogels. a) Diffusion coefficients determined with FRAP of 50, 100, 200, and 300 mg mL−1 gels with FITC‐dextrans of different molecular weights (40, 70, and 250 kDa). Three separate samples for each combination of protein concentration and FITC‐dextran size were each measured three times. b) SEM images of NT2RepCT hydrogels at 50, 100, 200, and 300 mg mL−1 (scale bars are 1 µm). c) Release profiles from GFP (1, 30, and 50 mg mL−1 final protein concentration) encapsulated in NT2RepCT hydrogels (50 mg mL−1). Gels were incubated in 20 mM Tris‐HCl buffer (pH 8) and samples were taken after 1, 5, 24, and 48 h. Y‐axis shows % equilibrium where 100% is full equilibrium with the surrounding buffer. Whiskers show standard deviation. d) SDS‐PAGEs of GFP release from NT2RepCT (50 mg mL−1) hydrogels after 1, 5, 24, and 48 h at 37 °C. GFP was encapsulated in NT2RepCT hydrogels at final concentration of 1, 30, and 50 mg mL−1.
Since NT2RepCT gels form under mild conditions we speculated that other proteins may be mixed with the NT2RepCT solution and maintain their activity after gelation. To test this, we mixed GFP at different concentrations with NT2RepCT at 50 mg mL−1 final concentration, formed gels, added 20 mM Tris‐HCl, pH 8, and monitored the release of GFP over time. By measuring the fluorescence intensity of released GFP, we found that at all concentrations tested, GFP encapsulated in NT2RepCT hydrogels reached equilibrium with the medium after 24 h (Figure 3c), indicating that GFP maintains its fold and function after gelation and that GFP does not interact with the gel, i.e., is not retained. Furthermore, no unpolymerized NT2RepCT (33 kDa) was released from the hydrogels as seen by SDS‐PAGE (Figure 3d; Figure S2, Supporting Information) suggesting stability and complete polymerization of the gels.
2.3. Human Mesenchymal Stem Cells Survive and are Viable in NT2RepCT Hydrogels
To test the hydrogels’ suitability as cell culture scaffolds, we tested if mesenchymal stem cells (MSC) survive in the hydrogels. Treatments with MSC hold therapeutic relevance in regenerative medicine as they can differentiate into bone, cartilage, muscle, and connective tissue.[ 65 ] Therefore, human fetal MSC (hfMSC)[ 66 ] were encapsulated in NT2RepCT hydrogels formed at 50 mg mL−1. To optimize culture conditions, cell seeding densities of 0.1, 1.0, 2.5, and 5.0 × 104 cells mL−1 were mixed with the protein solutions. The cell‐protein mix was plated in 48‐well plates and either the bottom of the well was covered (100 µL) or the mixture was deposited as a drop in the center of the well (10 µL). After gelation, the cell culture medium was added to the wells, and an AlamarBlue assay was performed on the following day (Figure 4a). The fluorescence intensity measured in cell culture medium from wells with cells encapsulated in 10 µL gels positively correlated with the cell seeding density. Cells encapsulated in 100 µL gels had lower fluorescence intensity than cells encapsulated in 10 µL gels which might be due to shorter diffusion distance and proportionally larger contact surface to the surrounding cell culture medium in 10 µL gels compared to the 100 µL gels.
Figure 4.

hfMSC proliferation in 50 mg/mL NT2RepCT hydrogels. a) Cell seeding density versus fluorescence intensity of AlamarBlue in 10 and 100 µL hydrogels. b) hfMSC proliferation over 21 days of two different cell seeding densities (2.5 and 5.0 × 104 cells µL−1) in 10 µL hydrogels. Whiskers show standard deviation.
Cell proliferation was therefore monitored for 21 days using 10 µL gels (50 mg mL−1) at 2.5 and 5.0 × 104 cells mL−1 cell seeding densities (Figure 4b). Fluorescence intensity remained mostly at the same level for the 21‐day culture period, which means that hfMSC survive in the gels, but that proliferation likely is limited. To investigate if the protein concentration and thereby the stiffness of the gels affects cell proliferation, we followed hfMSC grown in 50, 100, and 150 mg mL−1 gels for up to 7 days, but could detect no difference in proliferation rate (Figure S3, Supporting Information). A possible explanation for this is that the cells are physically entrapped by the dense fibrillar network of the scaffolds (Figure 3b),[ 27 ] which could restrict the cells and prevent them from proliferating and migrating through the gels. Notably, NT2RepCT hydrogels maintained their structural integrity during the entire culture period and remained adherent to the cell culture well (Figure S4, Supporting Information). A negligible number of cells grew outside the gels (Figure S5, Supporting Information).
We next visualized hfMSC in the hydrogels after 7 days of culture using live/dead staining (Calcein AM in green for live cells and DRAQ7 in magenta for dead cells) by laser scanning confocal microscopy (Figure 5 ). The hfMSC encapsulated in 50 mg mL−1 NT2RepCT hydrogels showed a spherical morphology (Figure 5a), again likely due to the restricted space in the fibrillar network and possibly also due to the lack of cell adhesion motifs in the gels[ 67 , 68 ] as has been observed before for several other hydrogels and cell types.[ 29 , 68 , 69 , 70 , 71 , 72 , 73 ] The number of live and dead cells was quantified and plotted as the cell viability on day 7 (Figure 5b). The mean viability was 53% and 67% for cell seeding densities 2.5 and 5.0 × 104 cells mL−1, respectively which is lower compared to other established hydrogels like collagen, gelatin, or alginate (98%, 97%, and 80% respectively),[ 68 , 74 , 75 ] but in line with previously described silk hydrogels (70% viability of fibroblasts after 48 h).[ 29 ] Future studies addressing the suitability of the gels for applications in regenerative medicine should explore the differentiation capacity of the encapsulated hfMSC.
Figure 5.
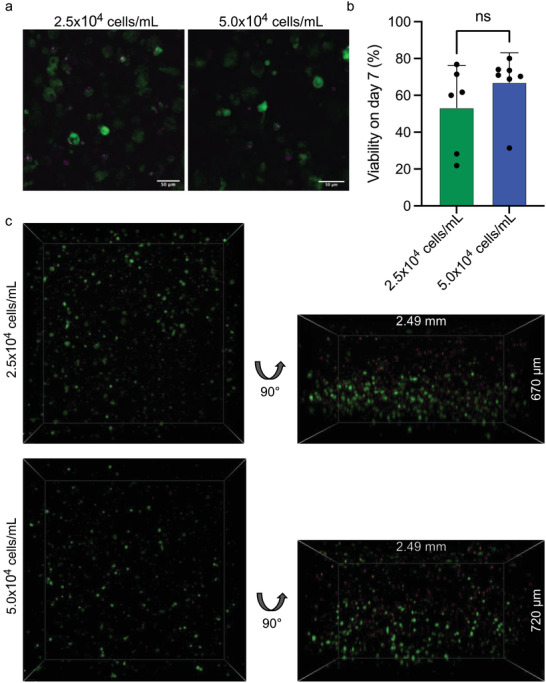
hfMSC show good viability when encapsulated in NT2RepCT hydrogels. a) Representative image of hfMSC stained with Calcein AM (green, live cells) and DRAQ7 (magenta, dead cells) for cell seeding densities 2.5 and 5.0 × 104 cells mL−1. Scale bars are 50 µm. b) Quantification of cell viability for the two different cell seeding densities. Whiskers show standard deviation. c) 3D representations of hfMSC in hydrogels.
3D visualization of the cells in the hydrogels showed that cells were well dispersed in the gels without cell clustering or aggregation (Movie S5, Supporting Information). However, there was a tendency for higher cell densities in the lower parts of some gels, which means that the cells likely sink toward the bottom prior to gelation (Figure 5c). A majority of the cells were alive, in line with the gentle encapsulation process. Some dead cells were located mainly in the upper part of the gel (Movie S6, Supporting Information). Since the gel‐formation and encapsulation procedure demands that the gels are incubated in air for 2 h, we speculate that the cells closest to the gel surface could have dried out. To decrease the time required for polymerization, recombinant silk proteins that are designed to be more prone to form amyloid‐like fibrils should be tested for their ability to form hydrogels faster.[ 76 ]
2.4. ARPE‐19 Cells Encapsulated in NT2RepCT Gels Secrete Human Progranulin
Encouraged by the findings that macromolecules are intact after gelation and can diffuse out of the gel and that hfMSC survive in the hydrogels, we investigated if cells that secrete bioactive molecules can be encapsulated in the hydrogels. ARPE‐19 cells secrete human progranulin (PGRN) which is an ≈80 kDa glycosylated growth factor associated with embryonic development, tissue repair, and inflammation.[ 36 , 77 , 78 ] Due to its additional roles that involve the regulation of neuronal survival and lysosomal function, it is being investigated as a therapeutic target for neurodegenerative diseases.[ 79 ] One therapeutic approach could be to encapsulate PGRN producing cells in implantable medical devices, as has been used for the delivery of nerve growth factors to the brain.[ 80 ] Here, the ARPE‐19 cells were encapsulated in the NT2RepCT hydrogels and cultured for 31 days during which the levels of PGRN released during a 4 h period were measured on days 3, 7, 14, and 31 by sandwich ELISA (Figure 6 ). The cells continued to secrete PGRN that, despite its relatively large size, diffused through the gels and was released into the medium where it could be detected. PGRN levels increased over time and at the same cell seeding density, the levels of PGRN released from encapsulated cells and control cells that were cultured under standard conditions (on cell culture plastic surfaces), were slightly lower on day 7 and 14 but on the same level on day 3 and day 31, indicating that encapsulation has little negative effect on the cells. Possibly, this approach could be useful in applications where cells should be encapsulated and protected in the gels over long periods to continuously release proteins. In line with this idea, the use of silk gels for drug release was recently studied by Trossmann et al who used human embryonic kidney 293 cells that secreted a fusion protein composed of tumor necrosis factor receptor 2 (TNFR2) and the Fc domain of IgG1.[ 73 ]
Figure 6.

PGRN release from ARPE‐19 cells alone or encapsulated in 50 mg mL−1 NT2RepCT hydrogels are similar. The release of progranulin from ARPE‐19 cells at the same seeding density during a 4 h pulse experiment was quantified with ELISA on day 3, 7, 14, and 31. Whiskers show standard deviation. *p<0.05; **p < 0.01; ***p<0.001; ****p < 0.0001; ns–not significant.
3. Conclusions
Herein, we report the development of a hydrogel that works as a macromolecule release system as well as a cell culture scaffold. The optical transparency and absence of autofluorescence facilitates the evaluation of cells by microscopy. Primary human cells show high survival after being mixed with the protein solution and encapsulated in the gels by incubation at 37 °C. This indicates that the procedure does not harm the cells, and that the gel is non‐toxic and allows diffusion of nutrients and waste products. Moreover, cells can be entrapped in the gels forming a living material that produces biologically active molecules that are continuously released to the surrounding. Finally, by varying the protein concentration, the mechanical properties of the gels can be tailored to fit different tissues, and can even match those of, e.g., cartilage.
4. Experimental Section
Protein Expression and Purification
The expression of NT2RepCT was carried out using a fed‐batch culture, exactly as described previously.[ 35 ] Briefly, a day culture of E. coli BL21 (DE3) transformed with pT7‐ NT2RepCT was grown in LB‐medium (Kanamycin 50 µg mL−1) using a baffled shake flask at 37 °C to OD600 5–10. The pre‐culture was used to inoculate 1 L of sterile semi‐complex medium originally described by da Silva and coworkers (experiment #4),[ 81 ] in a 3 L single‐walled glass vessel, which was connected to a BioBench modular controller unit (Solida Biotech, Munich, Germany). After inoculation, the culture was grown at 29 °C until the OD600 approached 60. Then, the culture was cooled to 20 °C, before the expression of NT2RepCT was induced by the addition of IPTG to a final concentration of 150 µM. Feeding with a semi‐complex medium containing 40% glycerol according to da Silva et al.[ 81 ] was initiated once a spike in the pO2 profile was seen, which indicated that the initial carbon source was depleted. Up to 500 mL of feed was consumed using a feeding profile, which consisted of a linear increase from 11 mL to 30 mL h−1 for 10 h, followed by constant feeding. The pH level was adjusted continuously to pH 7 either by the addition of 25% NH3 or 3 M H3PO4. The pO2 level was set to 30%, by adjusting the stirrer speed between 200–1200 rpm. 22 h post‐induction, the culture was harvested by centrifugation at 4000 xg. Finally, the supernatant was discarded, and the wet cell pellet was diluted to a ratio of 1:2 in 20 mM Tris‐HCl, pH 8, by gentle agitation, and stored at −20 °C.
The thawed cell pellet resuspension was diluted with 20 mM Tris‐HCl, pH 8, to obtain a dilution ratio wet cell pellet to buffer of 1:10, before adding DNAse I to a final concentration of 10 µg mL−1. Cell lysis was executed with a Cell disruptor (Constant Systems, Daventry, United Kingdom). The lysed cell suspension was centrifugated at 25,0000 xg, before the supernatant was filtered through a 0.45 µm Filtropur S (Sarstedt, Nümbrecht, Germany). Then, NT2RepCT was purified from the lysate with an Äkta Explorer liquid chromatographic system at 6 °C (GE Healthcare, Uppsala, Sweden) and 2 × 20 mL HisPrep FF 16/10 columns (Cytiva, Uppsala). To maximize the productivity and process up to 2.5 L of lysate, the instrument was programmed with Unicorn 5.1 to automate repeated loading and elution, using the loop function. During each cycle, 250 mL lysate was first passed over the column at 5 mL min−1 (loading), followed by washing with 3.5 column volumes (CV) 20 mM Tris‐HCl buffer, pH 8, and washing with 4 CV with Tris‐buffer containing 2 mM imidazole. NT2RepCT was eluted with 2 CV 200 mM imidazole. The combined eluate was dialyzed against 20 mM Tris‐HCl pH 8, at 4 °C overnight, using a Spectra/Por dialysis membrane with a 6–8 kDa molecular weight cutoff.
His‐NT‐QG‐GFP was supplemented with 1 mM dithiothreitol and cleaved using 3C protease in weight‐to‐weight ratio 50:1 overnight with rotation at 4 °C. Cleaved protein was dialyzed against 20 mM Tris‐HCl pH 8, at 4 °C overnight, using a Spectra/Por dialysis membrane with a 6–8 kDa molecular weight cutoff. Reverse immobilized metal affinity chromatography (RIMAC) was performed using Hisprep™ Fast Flow 16/10 Cytiva column. Cleaved GFP was collected in the flowthrough fraction.
Protein concentrations were determined with a spectrophotometer and the purity was determined by SDS‐polyacrylamide (4–20%) gel electrophoresis and Coomassie Brilliant Blue staining. Proteins were concentrated to the indicated concentrations using centrifugal filter units (VivaSpin 20, GE healthcare) with a 10 kDa molecular weight cutoff at 4000 xg in rounds of 20 min.
The purified protein obtained by this method was used in all further experiments related to the NT2RepCT hydrogels. The details regarding the mechanisms and procedure of gel formation have been described previously by Arndt T, Jaudzems K et al.[ 27 ]
Uniaxial Unconfined Compression Tests
Gels were prepared at 37 °C in steel cylinders with an inner diameter of 1 cm and a height of 5 cm. Gels were removed with a pistil and cut with a scalpel into 1 × 1 cm cylinders. The 50 mg mL−1 gels shrank to 8 mm in diameter. A texture Analyzer HDi (Stable Micro Systems, UK) with a 500 N load cell and a 50 mm cylindrical aluminum probe and a single column Instron‐5940 with a load cell of 100 N were used for compression tests. Gels were compressed at a rate of 1 mm s−1 to 80% of their original size. Engineering stress and strain were calculated considering the initial gauge length and the nominal area. The compressive modulus was calculated by the slope of the linear part in the stress strain curve. For each sample at least seven compression tests were performed. Average and standard error of mean values were calculated.
Fluorescence Recovery after Photo Bleaching (FRAP)
Ten microliters of the NT2RepCT solutions at varying concentrations (50, 100, 200, and 300 mg mL−1) were mixed with 150 ppm FITC‐dextrans and pipetted onto imaging dishes (Zell‐Kontakt, Germany), covered with cover slips, sealed with mineral oil and incubated at 37 °C for gelation. An inverted Nikon Eclipse Ti confocal microscope equipped with a 10x objective was used to image the gels. At least three samples of every gel concentration and FITC‐dextran size were prepared and each sample was imaged in triplicates with sufficient distance between the bleaching regions. Measurements were performed in the center of the gel (y‐direction) and the intensity of three regions of interests (ROI's) were measured; the actual bleaching region (ROI 1), a reference region within the gel (ROI 2) to account for bleaching due to imaging and a reference region outside the gel (ROI 3) for noise and background subtraction. Images were collected with 512 × 512 pixels at zoom factor 5 yielding a pixel size of 0.49 mm. Each measurement consisted of, 20 prebleach images, 2 bleach frames (100% laser power, 150 msec), and a recovery time of 2 to 10 min depending on the gel concentration and FITC‐dextran size. Analysis was performed with EasyFRAP‐web in which the curves were averaged, double normalized and fitted with the double term exponential equation.[ 82 ]
Scanning Electron Microscopy (SEM)
Gels were cut into cubes of ≈2 × 2 × 2 mm and incubated in 2 mL 2.5% glutaraldehyde and 0.1% ruthenium red over night at 4 °C. Gel cubes were washed 3 times for 10 min in 20 mM Tris‐HCl pH 8, fixed in 1% Osmium for 2 h and washed 3 times for 10 min in dH2O. The water present in the gels were exchanged to increasing concentration of ethanol which was then removed in a critical point dryer to avoid shrinking of the gels. The fixated gels were fractured, mounted on specimen stub using carbon tape and sputter coated with gold for 5 × 30 s. The gels were then imaged with a FlexSEM 1000 II (Hitachi, Tokyo, Japan) at 50 k X magnification under high vacuum, with an acceleration voltage of 7 kV and a working distance of 6 mm.
GFP Release Assay
NT2RepCT and GFP were mixed such that the final concentration of NT2RepCT was 50 mg mL−1 and that of GFP was 1, 30 and 50 mg mL−1 in the respective samples. One hundred microliters of each sample were added per well of a 96‐well plate (Nunc, standard black opaque microwell plates). In addition, 100 µL of NT2RepCT (50 mg mL−1) and 100 µL of GFP (1, 30, and 50 mg mL−1) were plated individually as controls. All dilutions were done in 20 mM Tris‐HCl pH 8. The plates were then incubated at 37 °C, overnight. The incubator was kept humidified by placing a tray of water inside it to prevent the gels from drying. The next day, 100 µL of 20 mM Tris‐HCl pH 8 was added to each well and the plate was kept at 37 °C. After 1, 5, 24, and 48 h, 70 µL of the buffer/supernatant was removed and transferred to another 96 well plate (Nunc, standard black opaque microwell plates). Individual sets of samples were prepared for each time point. The fluorescence intensity of the supernatant was measured using a microplate reader (SpectraMax) at excitation 470 nm and emission 510 nm. To calculate when the equilibrium between gel and media was reached (100% equilibrium) the following formula was used:
| (1) |
Each sample was done in triplicates and the experiment was repeated thrice.
Cell Culture
Cell Culture—ARPE‐19 cell culture and ELISA of PGRN
ARPE‐19 cells, a spontaneously immortalized retinal pigment epithelia (RPE) cell line that overexpresses and secretes human progranulin (PGRN), was used. The cells (passage number P3) were grown in T‐75 tissue culture flasks (Sarstedt) with Dulbecco ´s modified Eagle medium (DMEM, Glutamax, GibcoTM) supplemented with 10% fetal bovine serum (FBS), (GibcoTM) and 1% antibiotics (Antibiotic‐Antimycotic (100X), GibcoTM) and incubated at 37 °C with 5.0% CO2 under humidified atmosphere. At 90% confluency, the cells were passaged by detaching with 0.5% Trypsin‐ Ethylenediaminetetraacetic acid (EDTA), (Sigma). To encapsulate the cells within the gel, NT2RepCT protein solutions were mixed with the cells in media to achieve a final seeding density of 1000 cells mL−1. One hundred microliters of the protein‐cell mixture were added to the wells and allowed to gel at 37 °C with 5.0% CO2 in a cell culture incubator for 2 h. After gelation, 100 µL of medium was added on top of the gel. The medium was exchanged 3 times with fresh medium every 1 h for the first 3 h after gelation. As a control, ARPE‐19 cells were plated on standard wells of 96‐well tissue culture plates (Sarstedt) and cultured with 100 µL of medium. Thus, the same number of cells in each well was used, but it should be noted that proliferation rate of the cells growing in the gels and on plastic surfaces was not determined, respectively. The cells were incubated at 37 °C with 5.0% CO2 in a cell culture incubator with medium exchange every 2–3 days. On collection days 3, 7, 14, and 31 the old medium was removed, and fresh medium was added. After 4 h, aliquots of the fresh medium were collected and diluted 200 times with phosphate‐buffered saline containing 1% bovine serum albumin (BSA) and 0.1% Tween20. The diluted samples were used for enzyme linked immunosorbent assay (ELISA) to measure the amount of progranulin released by the cells, against standards provided by the manufacturer. ELISA was performed using Human Progranulin DuoSet ELISA kit (R&D Systems) following the manufacturer's protocol. The optical density of each well was measured using a microplate reader (SpectraMax) set to 450 nm and the amount of PGRN present in each sample was calculated from the standard curve. Each sample was done in triplicates and the experiment was repeated twice.
Cell Culture—Primary cell culture
Human fetal Mesenchymal stem cells (hfMSC), (from fetal liver obtained from Cecilia Götherström's group at Karolinska Institutet, ethical permits 428/01, 2006/308‐32 and 2013/566‐32) were grown in tissue culture treated standard flasks with complete DMEM – low glucose (DMEM – LG) supplemented with 10% FBS and 1% Antibiotic/Antimycotic solution (GibcoTM) at 37°C with 5.0% CO2 under humidified atmosphere. 0.5% Trypsin‐EDTA (Sigma) was used to detach the cells for passaging at 90% confluency.
Cell Culture—Cell proliferation assay
AlamarBlue assay was used to monitor the proliferation of the cells within the scaffolds during cell culturing. HfMSC, (P5) were cultured in T‐75 tissue culture flask (Sarstedt) and then plated on either 96‐well or 48‐well standard tissue culture plates (Sarstedt) in complete cell culture medium without phenol red. To encapsulate the cells within the gel, NT2RepCT protein solution was mixed with different number of cells in the medium to achieve a final protein concentration of 50 mg/ml and seeding densities of 1000, 10000, 25000, and 50000 cells per mL in the different samples. Either 100 µL of the protein and cell mixture was added to each well of a 96‐well plate to cover the whole well bottom with the gel or 10 µL of the mixture was carefully added at the center of either 96‐ or 48‐well plates to form an adherend drop. The solutions in the plates were allowed to gel at 37 °C with 5.0% CO2 in a cell culture incubator for 2 h. After gelation, 100 µL media was added to the gels in the 96‐well plate and 200 µL of media was added to the gels in the 48‐well plate. The medium was changed 3 times every hour for the first 3 h after gelation and then continued for further incubation at 37 °C with 5.0% CO2 in a cell culture incubator with medium exchange every third day.
To assay for viability and proliferation, the pre‐mixed AlamarBlue reagent (Invitrogen) was added (10% of the volume in the well) to the wells containing the gels with cells in complete cell culture medium without phenol red and incubated for 4 h. Following incubation, 70 or 100 µL of supernatant were collected (from 96‐ or 48‐well plates respectively) and transferred to a standard black opaque 96‐well plate (Nunc) and the fluorescence intensity was measured with a microplate reader (SpectraMax) using an excitation wavelength of 560 nm and an emission wavelength of 590 nm. Gels with cell culture medium but without cells were incubated with AlamarBlue as a control and subtracted from all measurements.
After collecting the supernatant any remaining medium was removed and the gels were washed 4 times for at least 1 h each time with medium to remove any traces of AlamarBlue in the samples and the culture was continued. This was performed on day 1, 3, 7, 14, and 21. Each sample was done in triplicates and the experiment was repeated thrice.
An AlamarBlue assay was also performed on 10 µl gel drops in which hfMSC (50000 cells mL−1) were grown for up to 7 days in hydrogels having final protein concentrations of 50, 100, and 150 mg mL−1.
Cell Culture—Cell viability and confocal laser scanning microscopy
24‐well imaging plates with 170 µm cover glass bottom (Zell‐Kontakt GmbH) were used for confocal imaging of the samples. NT2RepCT protein solutions were mixed with different number of cells (hfMSC, P5) in HEPES buffer (20 mM, pH 7.4) to achieve a final protein concentration of 50 mg mL−1 and cell seeding densities of 25000 and 50000 cells mL−1 respectively. Fifty microliters of the cell‐ protein mixture was carefully added to the center of the well to form an adherend drop. Imaging plates were placed in the CO2 incubator for 2 h for gelation after which 500 µL buffer was added. The medium was changed 3 times every 1 h for the first 3 h after gelation and then further incubated at 37 °C with 5.0% CO2 in a cell culture incubator with medium change every 2–3 day. On day 7, Calcein AM (Invitrogen) at a final concentration of 10 µM was added to the medium and incubated for 15 min followed by 3 washes (10 min each) with medium without phenol red. Five microliters of DRAQ7 (Invitrogen) was added to each well to achieve a final concentration of 3 µM and incubated for 10 min. The excess dye was washed with medium as before. The gel with encapsulated cells was removed from the complete culture medium without phenol red and imaged with an inverted Nikon Eclipse Ti confocal microscope equipped with a 10x and 20x objective. Fluorophores were imaged sequentially using a 488 nm laser and 515/30 filter and 640 nm and 700/75 filter for Calcein AM and DRAQ7 respectively. NIS‐Elements High Content Analysis software (Version 5.41.01) was used for imaging. Z‐scans were performed every 5 µm starting from the bottom of the sample to the top of the gel. For analysis ImageJ (Version 2.0.0‐rc‐69/1.52p) and NIS‐Elements Basic Research software (Version 5.30.03) were used for analysis including live/dead quantification. Each sample was done in duplicates and the experiment was repeated twice.
Statistics
Statistical analysis was performed with Prism 9.4.1 using one‐way ANOVA where appropriate. Statistical significance was indicated with asterisk *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Amino acid sequences of constructs tested for hydrogel formation.
| Name | Amino acid sequence |
|---|---|
| NT2RepCT | MGHHHHHHMSHTTPWTNPGLAENFMNSFMQGLSSMPGFTASQLDDMSTIAQSMVQSIQSLAAQGRTSPNKLQALNMAFASSMAEIAASEEGGGSLSTKTSSIASAMSNAFLQTTGVVNQPFINEITQLVSMFAQAGMNDVSAGNSGRGQGGYGQGSGGNAAAAAAAAAAAAAAAGQGGQGGYGRQSQGAGSAAAAAAAAAAAAAAGSGQGGYGGQGQGGYGQSGNSVTSGGYGYGTSAAAGAGVAAGSYAGAVNRLSSAEAASRVSSNIAAIASGGASALPSVISNIYSGVVASGVSSNEALIQALLELLSALVHVLSSASIGNVSSVGVDSTLNVVQDSVGQYVG* |
| His‐NT‐QC‐GFP | MGHHHHHHMSHTTPWTNPGLAENFMNSFMQGLSSMPGFTASQLDDMSTIAQSMVQSIQSLAAQGRTSPNKLQALNMAFASSMAEIAASEEGGGSLSTKTSSIASAMSNAFLQTTGVVNQPFINEITQLVSMFAQAGMNDVSAGNSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFICTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDDGNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITLGMDELYKLIN* |
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Supporting Information
Supplemental Movie 1
Supplemental Movie 2
Supplemental Movie 3
Supplemental Movie 4
Supplemental Movie 5
Supplemental Movie 6
Acknowledgements
T.A. and U.C. contributed equally to this work. This work was supported by European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement No 815357), the Center for Innovative Medicine (CIMED) at Karolinska Institutet and Stockholm City Council, Karolinska Institutet SFO Regen (FOR 4–1364/2019), and the Swedish Research Council (2019‐01257). G.G. is supported by Wenner‐Gren stiftelse (UPD2021‐0047). The authors would like to thank Annika Altskär, Niklas Lorén, and Nina Kronqvist for fruitful discussions. The study was partly performed at the Live Cell imaging Core facility/Nikon Center of Excellence, at the Karolinska Institute, supported by grants from the Swedish Research Council, KI infrastructure, and Centre for Innovative Medicine. All experiments involving human participants were carried out upon the approval from Regionala etikprövningsnämnden in Stockholm.
Arndt T., Chatterjee U., Shilkova O., Francis J., Lundkvist J., Johansson D., Schmuck B., Greco G., Nordberg Å. E., Li Y., Wahlberg L. U., Langton M., Johansson J., Götherström C., Rising A., Tuneable Recombinant Spider Silk Protein Hydrogels for Drug Release and 3D Cell Culture. Adv. Funct. Mater. 2024, 34, 2303622. 10.1002/adfm.202303622
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Conrad B., Han L.‐H., Yang F., Tissue Eng Part A 2018, 24, 1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guo J. L., Kim Y. S., Mikos A. G., Biomacromolecules 2019, 20, 2904. [DOI] [PubMed] [Google Scholar]
- 3. Klotz B. J., Oosterhoff L. A., Utomo L., Lim K. S., Vallmajo‐Martin Q., Clevers H., Woodfield T. B. F., Rosenberg A. J. W. P., Malda J., Ehrbar M., Spee B., Gawlitta D., Adv. Healthcare Mater. 2019, 8, 1900979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu M., Zeng X., Ma C., Yi H., Ali Z., Mou X., Li S., Deng Y., He N., Bone Res. 2017, 5, 17014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moxon S. R., Corbett N. J., Fisher K., Potjewyd G., Domingos M., Hooper N. M., Mater. Sci. Eng., C 2019, 104, 109904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geckil H., Xu F., Zhang X., Moon S., Demirci U., Nanomedicine 2010, 5, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guvendiren M., Burdick J. A., Curr. Opin. Biotechnol. 2013, 24, 841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Figueiredo L., Pace R., D'Arros C., Réthoré G., Guicheux J., Le Visage C., Weiss P., J Tissue Eng Regen Med 2018, 12, 1238. [DOI] [PubMed] [Google Scholar]
- 9. Ouyang L., Dan Y., Shao Z., Yang S., Yang C., Liu G., Duan D., Exp Ther Med 2019, 18, 2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pupkaite J., Rosenquist J., Hilborn J., Samanta A., Biomacromolecules 2019, 20, 3475. [DOI] [PubMed] [Google Scholar]
- 11. Roy T., James B. D., Allen J. B., Macromol. Biosci. 2021, 21, 2000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Samadian H., Farzamfar S., Vaez A., Ehterami A., Bit A., Alam M., Goodarzi A., Darya G., Salehi M., Sci. Rep. 2020, 10, 13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhai Z., Xu K., Mei L., Wu C., Liu J., Liu Z., Wan L., Zhong W., Soft Matter 2019, 15, 8603. [DOI] [PubMed] [Google Scholar]
- 14. Dorishetty P., Dutta N. K., Choudhury N. R., Adv. Colloid Interface Sci. 2020, 281, 102163. [DOI] [PubMed] [Google Scholar]
- 15. Hölzl K., Lin S., Tytgat L., Van Vlierberghe S., Gu L., Ovsianikov A., Biofabrication 2016, 8, 032002. [DOI] [PubMed] [Google Scholar]
- 16. Lee V. K., Dai G., Ann. Biomed. Eng. 2017, 45, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thiele J., Ma Y., Bruekers S. M. C., Ma S., Huck W. T. S., Adv. Mater. 2014, 26, 125. [DOI] [PubMed] [Google Scholar]
- 18. Yang J. M., Olanrele O. S., Zhang X., Hsu C. C., in Novel Biomaterials for Regenerative Medicine, Advances in Experimental Medicine and Biology, Vol. 1077 (Eds: Chun H., Park K., Kim C. H., Khang G.), Springer, Singapore: 2018, Ch. 19. [Google Scholar]
- 19. DeSimone E., Schacht K., Scheibel T., Mater. Lett. 2016, 183, 101. [Google Scholar]
- 20. Chai Q., Jiao Y., Yu X., Gels 2017, 3, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maitra J., Shukla V. K., Am. J. Polym. Sci. 2014, 4, 25. [Google Scholar]
- 22. Schacht K., Scheibel T., Biomacromolecules 2011, 12, 2488. [DOI] [PubMed] [Google Scholar]
- 23. Ahn T.‐K., Lee D. H., Kim T., chol Jang G., Choi S., Oh J. B., Ye G., Lee S., in Novel Biomaterials for Regenerative Medicine, Advances in Experimental Medicine and Biology, Vol. 1077 (Eds: Chun H., Park K., Kim C. H., Khang G.), Springer, Singapore: 2018, Ch. 19. [Google Scholar]
- 24. Prasad K., Bazaka O., Chua M., Rochford M., Fedrick L., Spoor J., Symes R., Tieppo M., Collins C., Cao A., Markwell D., (Ken) Ostrikov K., Bazaka K., Materials 2017, 10, 884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Żółtowska K., Piotrowska U., Oledzka E., Luchowska U., Sobczak M., Bocho‐Janiszewska A., Eur. J. Pharm. Sci. 2017, 96, 440. [DOI] [PubMed] [Google Scholar]
- 26. Dhandayuthapani B., Yoshida Y., Maekawa T., Kumar D. S., Int. J. Polym. Sci. 2011, 2011, 1. [Google Scholar]
- 27. Arndt T., Jaudzems K., Shilkova O., Francis J., Johansson M., Laity P. R., Sahin C., Chatterjee U., Kronqvist N., Barajas‐Ledesma E., Kumar R., Chen G., Strömberg R., Abelein A., Langton M., Landreh M., Barth A., Holland C., Johansson J., Rising A., Nat. Commun. 2022, 13, 4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neubauer V. J., Trossmann V. T., Jacobi S., Döbl A., Scheibel T., Angew. Chem. Int. Ed. 2021, 60, 11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schacht K., Jüngst T., Schweinlin M., Ewald A., Groll J., Scheibel T., Angew. Chem. Int. Ed. 2015, 54, 2816. [DOI] [PubMed] [Google Scholar]
- 30. Song W.‐W., Qian Z.‐G., Liu H., Chen H.‐F., Kaplan D. L., Xia X.‐X., ACS Macro Lett. 2021, 10, 395. [DOI] [PubMed] [Google Scholar]
- 31. Andersson M., Jia Q., Abella A., Lee X.‐Y., Landreh M., Purhonen P., Hebert H., Tenje M., Robinson C. V, Meng Q., Plaza G. R., Johansson J., Rising A., Nat. Chem. Biol. 2017, 13, 262. [DOI] [PubMed] [Google Scholar]
- 32. Arndt T., Laity P. R., Johansson J., Holland C., Rising A., ACS Biomater. Sci. Eng. 2021, 7, 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Landreh M., Andersson M., Marklund E. G., Jia Q., Meng Q., Johansson J., Robinson C. V., Rising A., Chem. Commun. 2017, 53, 3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Otikovs M., Andersson M., Jia Q., Nordling K., Meng Q., Andreas L. B., Pintacuda G., Johansson J., Rising A., Jaudzems K., Angew. Chem. Int. Ed. 2017, 56, 12571. [DOI] [PubMed] [Google Scholar]
- 35. Schmuck B., Greco G., Barth A., Pugno N. M., Johansson J., Rising A., Mater. Today 2021, 50, 16. [Google Scholar]
- 36. Dunn K. C., Aotaki‐Keen A. E., Putkey F. R., Hjelmeland L. M., Exp. Eye Res. 1996, 62, 155. [DOI] [PubMed] [Google Scholar]
- 37. Chaudhuri O., Gu L., Klumpers D., Darnell M., Bencherif S. A., Weaver J. C., Huebsch N., Lee H., Lippens E., Duda G. N., Mooney D. J., Nat. Mater. 2016, 15, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Engler A. J., Sen S., Sweeney H. L., Discher D. E., Cell 2006, 126, 677. [DOI] [PubMed] [Google Scholar]
- 39. Goldshmid R., Seliktar D., ACS Biomater. Sci. Eng. 2017, 3, 3433. [DOI] [PubMed] [Google Scholar]
- 40. Jeffords M. E., Wu J., Shah M., Hong Y., Zhang G., ACS Appl. Mater. Interfaces 2015, 7, 11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee J., Abdeen A. A., Huang T. H., Kilian K. A., J. Mech. Behav. Biomed. Mater. 2014, 38, 209. [DOI] [PubMed] [Google Scholar]
- 42. Leipzig N. D., Shoichet M. S., Biomaterials 2009, 30, 6867. [DOI] [PubMed] [Google Scholar]
- 43. Macrí‐Pellizzeri L., Pelacho B., Sancho A., Iglesias‐García O., Simón‐Yarza A. M., Soriano‐Navarro M., González‐Granero S., García‐Verdugo J. M., De‐Juan‐Pardo E. M., Prosper F., Tissue Eng. Part A 2015, 21, 1633. [DOI] [PubMed] [Google Scholar]
- 44. Park J. S., Chu J. S., Tsou A. D., Diop R., Tang Z., Wang A., Li S., Biomaterials 2011, 32, 3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tse J. R., Engler A. J., PLoS One 2011, 6, e15978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Young D. A., Choi Y. S., Engler A. J., Christman K. L., Biomaterials 2013, 34, 8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao W., Li X., Liu X., Zhang N., Wen X., Mater. Sci. Eng. C 2014, 40, 316. [DOI] [PubMed] [Google Scholar]
- 48. Lohrasbi S., Mirzaei E., Karimizade A., Takallu S., Rezaei A., Cellulose 2020, 27, 927. [Google Scholar]
- 49. Sun T.‐W., Zhu Y.‐J., Chen F., RSC Adv. 2018, 8, 26218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mirfakhrai T., Madden J. D. W., Baughman R. H., Mater. Today 2007, 10, 30. [Google Scholar]
- 51. Mathur A. B., Collinsworth A. M., Reichert W. M., Kraus W. E., Truskey G. A., J. Biomech. 2001, 34, 1545. [DOI] [PubMed] [Google Scholar]
- 52. Ogneva I. V., Lebedev D. V., Shenkman B. S., Biophys. J. 2010, 98, 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Little C. J., Bawolin N. K., Chen X., Tissue Eng. Part B Rev 2011, 17, 213. [DOI] [PubMed] [Google Scholar]
- 54. Buitrago J. O., Patel K. D., El‐Fiqi A., Lee J.‐H., Kundu B., Lee H.‐H., Kim H.‐W., Acta Biomater. 2018, 69, 218. [DOI] [PubMed] [Google Scholar]
- 55. Nada A. A., Ali E. A., Soliman A. A. F., Int. J. Biol. Macromol. 2019, 131, 624. [DOI] [PubMed] [Google Scholar]
- 56. Lorén N., Hagman J., Jonasson J. K., Deschout H., Bernin D., Cella‐Zanacchi F., Diaspro A., McNally J. G., Ameloot M., Smisdom N., Nydén M., Hermansson A.‐M., Rudemo M., Braeckmans K., Q Rev Biophys 2015, 48, 323. [DOI] [PubMed] [Google Scholar]
- 57. Branco M. C., Pochan D. J., Wagner N. J., Schneider J. P., Biomaterials 2009, 30, 1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brandl F., Kastner F., Gschwind R. M., Blunk T., Teßmar J., Göpferich A., J. Controlled Release 2010, 142, 221. [DOI] [PubMed] [Google Scholar]
- 59. Burke M. D., Park J. O., Srinivasarao M., Khan S. A., J. Controlled Release 2005, 104, 141. [DOI] [PubMed] [Google Scholar]
- 60. Censi R., Vermonden T., van Steenbergen M. J., Deschout H., Braeckmans K., De Smedt S. C., van Nostrum C. F., di Martino P., Hennink W. E., J. Controlled Release 2009, 140, 230. [DOI] [PubMed] [Google Scholar]
- 61. Henke M., Brandl F., Goepferich A. M., Tessmar J. K., Eur. J. Pharm. Biopharm. 2010, 74, 184. [DOI] [PubMed] [Google Scholar]
- 62. Kosto K. B., Deen W. M., AIChE J. 2004, 50, 2648. [Google Scholar]
- 63. Yodmuang S., McNamara S. L., Nover A. B., Mandal B. B., Agarwal M., Kelly T.‐A. N., Chao P. G., Hung C., Kaplan D. L., Vunjak‐Novakovic G., Acta Biomater. 2015, 11, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang Y., Yu T., Peng L., Sun Q., Wei Y., Han B., Front Pharmacol. 2020, 11, 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Merimi M., El‐Majzoub R., Lagneaux L., Moussa Agha D., Bouhtit F., Meuleman N., Fahmi H., Lewalle P., Fayyad‐Kazan M., Najar M., Front Cell Dev. Biol. 2021, 9, 661532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Götherström C., Best Pract. Res. Clin. Obstet. Gynaecol. 2016, 31, 82. [DOI] [PubMed] [Google Scholar]
- 67. Gillette B. M., Jensen J. A., Tang B., Yang G. J., Bazargan‐Lari A., Zhong M., Sia S. K., Nat. Mater. 2008, 7, 636. [DOI] [PubMed] [Google Scholar]
- 68. Mũnoz Z., Shih H., Lin C.‐C., Biomater. Sci. 2014, 2, 1063. [DOI] [PubMed] [Google Scholar]
- 69. Bidarra S. J., Barrias C. C., Barbosa M. A., Soares R., Granja P. L., Biomacromolecules 2010, 11, 1956. [DOI] [PubMed] [Google Scholar]
- 70. Hwang C. M., Ay B., Kaplan D. L., Rubin J. P., Marra K. G., Atala A., Yoo J. J., Lee S. J., Biomed. Mater. 2013, 8, 014105. [DOI] [PubMed] [Google Scholar]
- 71. Krishnamoorthy S., Noorani B., Xu C., Int. J. Mol. Sci. 2019, 20, 5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kuang L., Damayanti N. P., Jiang C., Fei X., Liu W., Narayanan N., Irudayaraj J., Campanella O., Deng M., J. Appl. Polym. Sci. 2019, 136, 47212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Trossmann V. T., Heltmann‐Meyer S., Amouei H., Wajant H., Horch R. E., Steiner D., Scheibel T., Biomacromolecules 2022, 23, 4427. [DOI] [PubMed] [Google Scholar]
- 74. Gaudet I. D., Shreiber D. I., Biointerphases 2012, 7, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bidarra S. J., Barrias C. C., Granja P. L., Acta Biomater. 2014, 10, 1646. [DOI] [PubMed] [Google Scholar]
- 76. Arndt T., Greco G., Schmuck B., Bunz J., Shilkova O., Francis J., Pugno N. M., Jaudzems K., Barth A., Johansson J., Rising A., Adv. Funct. Mater. 2022, 32, 2200986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bateman A., Bennett H., J. Endocrinol. 1998, 158, 145. [DOI] [PubMed] [Google Scholar]
- 78. Nguyen A. D., Nguyen T. A., Martens L. H., Mitic L. L., Farese R. V., Trends Endocrinol. Metab. 2013, 24, 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rhinn H., Tatton N., McCaughey S., Kurnellas M., Rosenthal A., Trends Pharmacol. Sci. 2022, 43, 641. [DOI] [PubMed] [Google Scholar]
- 80. Mitra S., Gera R., Linderoth B., Lind G., Wahlberg L., Almqvist P., Behbahani H., Eriksdotter M., Adv. Exp. Med. Biol. 2021, 1331, 167. [DOI] [PubMed] [Google Scholar]
- 81. da Silva A. J., Horta A. C. L., Velez A. M., Iemma M. R. C., Sargo C. R., Giordano R. L., Novo M. T. M., Giordano R. C., Zangirolami T. C., SpringerPlus 2013, 2, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Koulouras G., Panagopoulos A., Rapsomaniki M. A., Giakoumakis N. N., Taraviras S., Lygerou Z., Nucleic Acids Res. 2018, 46, W467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supplemental Movie 1
Supplemental Movie 2
Supplemental Movie 3
Supplemental Movie 4
Supplemental Movie 5
Supplemental Movie 6
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


