ABSTRACT
Viruses adapt and modulate cellular pathways to allow their replication in host cells. The catabolic pathway of macroautophagy, for simplicity referred to as autophagy, is no exception. In this review, we discuss anti-viral functions of both autophagy and select components of the autophagy machinery, and how viruses have evaded them. Some viruses use the membrane remodeling ability of the autophagy machinery to build their replication compartments in the cytosol or efficiently egress from cells in a non-lytic fashion. Some of the autophagy machinery components and their remodeled membranes can even be found in viral particles as envelopes or single membranes around virus packages that protect them during spreading and transmission. Therefore, studies on autophagy regulation by viral infections can reveal functions of the autophagy machinery beyond lysosomal degradation of cytosolic constituents. Furthermore, they can also pinpoint molecular interactions with which the autophagy machinery can most efficiently be manipulated, and this may be relevant to develop effective disease treatments based on autophagy modulation.
KEYWORDS: Endosomal damage, interferon, replication organelle, secretory autophagy, virophagy
1. Introduction on virophagy, the selective degradation of viruses by autophagy
Virus particles carry their blueprints as RNA or DNA genomes. They contain protein shells as capsids for blueprint protection. Additional proteins in the virus particles ensure host cell selection and immediate early host cell manipulation upon entry [1]. For all other aspects of their replication in host cells, viruses heavily depend on the host cell machinery which, at the same time, tries to restrict their replication, both by cell intrinsic mechanisms and communication with the host’s immune system. Therefore, successful viruses, whom we note as pathogens due to heterogenous outcomes of infections at least in immune compromised individuals, have developed strategies to evade cellular restriction mechanisms in their adaptation to at least one host species. Autophagy, a group of cellular degradation pathways for intracellular proteins, protein complexes and organelles, is no exception to this rule.
Macroautophagy, the most commonly studied of these pathways and hereafter referred to as autophagy, involves cytosolic membrane remodeling and formation of double-membrane vesicles termed autophagosomes to deliver macromolecules to lysosomal degradation [2-5]. Autophagosome biogenesis is mediated by the so-called ATG (autophagy related) proteins, which compose a protein kinase complex, a lipid kinase complex, ATG9A-positive vesicles, a lipid transfer complex, and two ubiquitin-like/Ubl conjugation systems [6]. The protein kinase complex consists of ULK1 (unc-51 like autophagy activating kinase 1) or ULK2, ATG13, RB1CC1/FIP200 (RB1 inducible coiled-coil 1) and ATG101. It is activated upon starvation, via inhibition of the MTOR (mechanistic target of rapamycin kinase) complex 1 (MTORC1) and activation of the AMP-activated protein kinase (AMPK) [6]. This complex then phosphorylates multiple targets [7], including the class III autophagy-specific phosphatidylinositol 3-kinase (PtdIns3K) complex I. This PtdIns3K complex I includes PIK3C3/VPS34 (phosphatidylinositol 3-kinase catalytic subunit type 3), PIK3R4/VPS15 (phosphoinositide-3-kinase regulatory subunit 4), BECN1 (beclin 1), ATG14, NRBF2 (nuclear receptor binding factor 2) and AMBRA1 (autophagy and beclin 1 regulator 1) and it is responsible for the synthesis of phosphatidylinositol-3-phosphate/PtdIns3P in the membranes of autophagosome precursors, known as phagophores, whose lipids are recruited from ATG9A-positive vesicles and an ATG2-containing lipid transfer complex that stabilizes channels with the ER. Phosphatidylinositol-3-phosphate formation leads to the recruitment of several effectors belonging to the ATG machinery, including WIPI1 (WD repeat domain, phosphoinositide interacting 1), WIPI2, WDR45B/WIPI3 (WD repeat domain 45B), WDR45/WIPI4 and ZFYVE1/DFCP1 (zinc finger FYVE-type containing 1). While WDR45/WIPI4 is important for the assembly of the ATG2 protein-WDR45/WIPI4 complexes, involved in the transfer of lipids from the endoplasmic reticulum (ER) to the phagophore for expansion, WIPI2 is crucial for the conjugation to phosphatidylethanolamine (PE) of the members of the Atg8-protein family, which contains six members in humans: MAP1LC3A/LC3A (microtubule associated protein 1 light chain 3 alpha), LC3B, LC3C, GABARAP (GABA type A receptor-associated protein), GABARAPL1 and GABARAPL2/GATE16. ATG8 proteins are first C-terminally processed by the ATG4 proteases (ATG4A to ATG4D), exposing a glycine that is covalently linked to PE or, in rare cases, to phosphatidylserine, through the action of the two Ubl conjugation systems [8]. For this purpose, ATG8 proteins are activated by ATG7, transferred to ATG3 and then finally conjugated to PE by the ATG12–ATG5-ATG16L1 complex. ATG12 is also an Ubl molecule that is activated by ATG7, transferred to ATG10 and then conjugated to ATG5 before associating with the ATG12–ATG5-ATG16L1 complex. ATG8 proteins which are located at the inner membrane of phagophores, mediate the recruitment of specific cargoes via the so-called selective autophagy receptors (SARs) that bridge phagophores to autophagy substrates via their LC3-interacting regions (LIRs) and ubiquitin-interacting motif (UIM)-like sequences. ATG8 proteins located at the outer membrane of phagophores mediate its elongation, autophagosome closure and recruit the machinery for fusion with late endosomes and/or lysosomes. They are finally recycled by ATG4-mediated cleavage from their lipid anchor. Autophagosomes fuse then with late endosomes and/or lysosomes with the help of specific RAB GTPases, tethering factors and SNARE proteins such as STX17 (syntaxin 17), YKT6 (YKT6 v-SNARE homolog), VAMP8 (vesicle associated membrane protein 8) and SNAP29 (synaptosome associated protein 29) [9,10], resulting in the degradation of their cargoes and of the inner autophagosomal membrane. Autophagy flux or autophagosome maturation refers to the completion of this pathway and to its cargo degradation. If this cargo is a pathogen, i.e. a bacterium or a virus, this selective type of autophagy is called xenophagy. In the case of viruses, the term virophagy is also used.
2. Anti-viral functions of autophagy
2.1. Restriction of viral entry by autophagy
Many viruses trigger autophagy early during infection. The same viruses often subsequently interfere with the late stages of autophagy, strongly suggesting that autophagy represents an innate and cell autonomous host defense. Early events associated with viral infection include virion attachment and internalization, and intracellular delivery of the viral genetic material and associated proteins. The foot-and-mouth disease virus/FMDV, a picornavirus, can interact with Arg-Gly-Asp (RGD)-binding integrins or with heparan sulfate to enter cells and both these virus binding partners appear to be able to activate an early autophagic response against the foot-and-mouth disease virus capsid proteins [11]. The complement regulating factor CD46 (CD46 molecule) mediates the attachment of vaccinal but not clinical strains of measles virus/MeV, a paramyxovirus, and rapidly induces autophagy by recruiting the PIK3C3-BECN1 complex via the CD46-Cyt1-GOPC pathway [12,13]. Similar to measles virus, the peste des petits ruminant’s virus/PPRV, another paramyxovirus, rapidly activates autophagy via the AKT1 (AKT serine/threonine kinase 1)-MTORC1 axis after binding to NECTIN4 (nectin cell adhesion molecule 4) [14]. The toll like receptor 2 (TLR2)-MYD88 axis can activate autophagy in response to the envelope gH/gL glycoproteins of herpes simplex virus 1/HSV-1 [15-17], and of human cytomegalovirus/HCMV, two orthoherpesviruses [18,19]. Upon infection of CD4+ T cells, the envelope glycoprotein Env of human immunodeficiency virus 1/HIV-1, a retrovirus, induces an early autophagy response that restricts viral production if not counteracted by the virus [20]. HIV-1 Env, expressed at the surface of infected cells, also induces autophagy in bystander uninfected CD4+ T cells, triggering their cell death [21,22]. In Drosophila, anti-viral autophagy has been observed in vivo upon recognition by Toll-7, the fly homolog of TLR7, of the glycoprotein of the vesicular stomatitis virus/VSV, a rhabdovirus [23]. Toll-7 is also important for Drosophila autophagic resistance to the infection by the Rift Valley fever virus/RVFV, a bunyavirus, but not to other viruses [24]. Remarkably, virion-containing endosomes can initiate antiviral mammalian autophagy through the endosomal protein SNX5 (sorting nexin 5), which activates the PtdIns3K complex I in cells infected with viruses, including the togaviruses Sindbis virus/SINV and chikungunya virus/CHIKV, West Nile virus/WNV, an orthoflavivirus, and a mutant autophagy-sensitive HSV-1. A curvature signature of virion-containing endosomes appears central for SNX5-mediated autophagy induction [25]. Activation of AMPK in response to hepatitis B virus/HBV-induced reactive oxygen species/ROS production stimulates autophagy initiation and HBV restriction [26]. Thus, virus entry or activation of pathogen-associated molecular pattern recognition receptors can stimulate autophagy during infection.
The detection of viral nucleic acids is another mechanism that rapidly mobilizes autophagy. Viral RNA sensing can involve TLRs [27-29], NOD-like receptors (NLRs), such as NOD2 (nucleotide binding oligomerization domain containing 2) [30,31] or, host enzymes such as EIF2AK2/PKR (eukaryotic translation initiation factor 2 alpha kinase 2) [32]. In the latter case, the autophagic degradation of HSV-1 virions is opposed by two viral proteins, Us11 and ICP34.5 [33,34]. The ICP34.5-inhibited autophagic restriction of HSV-1 appears particularly efficient in neurons [35]. Autophagy activation can also occur via the sensing of viral DNA of HSV-1 or HCMV, a mechanism that is connected with the CGAS (cyclic GMP-AMP synthase)-STING1 (stimulator of interferon response cGAMP interactor 1) pathway [18,36,37], potentially leading to viral DNA degradation (Figure 1).
Figure 1.
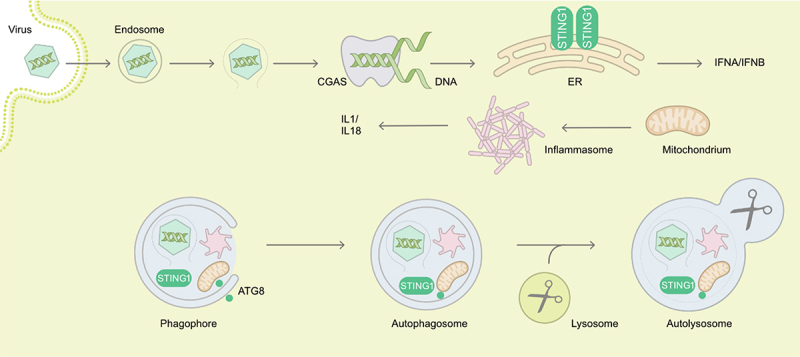
Examples of autophagy in targeting of viral entry, of sensors of viral genomes and of innate immune receptors of danger associated molecular patterns. During the cytoplasm entry by non-enveloped viruses, endosomal damage or virus capsids themselves can be recognized and targeted to degradation by autophagy. Viral DNA is detected by the CGAS-STING1 pathway, leading to type I interferon production. STING1 can also be degraded by autophagy. Damaged mitochondria activate inflammasomes triggering IL1 and IL18 production, and both are turned over by autophagy. Overall, autophagy restricts viruses during entry but at the same time dampens immune activation. ATG8, mammalian Atg8 homolog.
Host factors can play important roles in early antiviral autophagy. The E3 Ub ligase SMURF1 (SMAD specific E3 ubiquitin protein ligase 1) is instrumental for the degradative targeting of SINV capsid by the SAR SQSTM1/p62 (sequestosome 1) [38,39]. Fanconi anemia proteins, especially FANCC (FA complementation group C) serve also as SARs for virophagy [40]. Other central regulators are some members of the RING Ub ligases of the tripartite motif-containing (TRIM) protein family, such as TRIM5/TRIM5α which opposes retrovirus infection before reverse transcription takes place [41]. TRIM5 also acts as a receptor to target the HIV-1 capsid protein p24 for autophagic degradation [41]. In Langerhans cells, the C-type lectin CD207/Langerin is central for HIV-1 internalization and TRIM5-associated degradation [42]. Other TRIM proteins are involved against either a specific or several viruses. TRIM23, which is engaged in response to infections by influenza A virus/IAV, an orthomyxovirus, HSV-1 or encephalomyocarditis virus/EMCV, a picornavirus, can promote SQSTM1/p62-driven selective autophagy via TBK1 (TANK binding kinase 1) activation and contribute to adenovirus 5, HSV-1 and SINV antiviral autophagy [43]. The targeting of capsid proteins by selective autophagy is also observed in plants as the SAR NBR1 (NBR1 autophagy cargo receptor) drives autophagic degradation of the capsid protein or virions of cauliflower mosaic virus/CaMV, a caulimovirus, in a Ub-independent manner in Arabidopsis thaliana [44]. Host factors can also be recruited by viruses to counter the autophagic restriction that they face during cell invasion. A prominent example is adenovirus 5 whose capsid undergoes structural changes to expose the protein VI (PVI) and ruptures the endocytic vacuole to enter the cytosol. The detection of membrane damages by particular galectins (LGALS), primarily LGALS8, along with TBK1 activation and SAR engagement triggers the formation of autophagosomes. This is antagonized by the PVI-driven recruitment of the E3 Ub ligases NEDD4 and NEDD4L, leading to delayed autophagy until viral escape from endosomal remnants is achieved [45-47] (Figure 1). Kaposi sarcoma-associated herpesvirus/KSHV also causes endosomal damage during entry, recruiting LGALS8 and CALCOCO2/NDP52 (calcium binding and coiled-coil domain 2), resulting in autophagosomal degradation of incoming virions [48]. This, however, seems to be also rapidly inhibited by KSHV after 2 h but the underlying mechanism remains unclear. HBV envelope proteins are recognized and targeted by the SAR CALCOCO2/NDP52 to the lysosome for degradation, which is independent of LGALS8 [49]. In the case of endocytosed picornaviruses, conformational changes in the capsid proteins induce small pores into the endosomal membrane for translocation of viral RNA into the cytosol. This step requires the in-situ recruitment of host PLAAT3/PLA2G16 (phospholipase A and acyltransferase 3) by damaged membranes, causing an inhibition of the LGALS8-mediated autophagic response to viral entry [50]. Thus, ubiquitination of viral components and/or glycan exposure upon endosomal damage can recruit SARs and the ATG machinery to restrict viral entry.
Non-canonical LC3B conjugation to single endosomal membranes, called LC3-associated phagocytosis (LAP), LC3-associated endocytosis/LANDO or CASM (conjugation of ATG8/LC3 to single membranes) [51-54], can also delay virus entry, as has been shown for lung epithelial cells and IAV infection [55]. In mice expressing mutant ATG16L1 that lacks the WD40 domain that is required for LC3B conjugation to single membranes, IAV fusion with the limiting membrane of endosomes was accelerated, thereby increasing infection and associated pathology. Thus, both canonical autophagy and non-canonical functions of the autophagy machinery can compromise virus entry.
Overall, these findings highlight that autophagic responses opposing viral infections can be triggered by distinct early events. Those include virus receptor binding, sensing of infection-associated membrane remodeling/perturbation (fusion, curvature, rupture) or engagement of germline-encoded receptors able to detect conserved microbial molecular patterns (envelope/capsid proteins and nucleic acids).
2.2. Autosis of virus infected cells
Autosis, initially described by the laboratory of Beth Levine, is a unique form of non-apoptotic, non-necrotic cell death specifically relying on ATP1A1 (ATPase Na+/K+ transporting subunit alpha 1) and autophagy [56]. Autosis was characterized by increased numbers of autophagosomes, autolysosomes, and empty vacuoles, in combination with ER dilation and fragmentation. This is followed by nuclear membrane convolution, focal ballooning of the perinuclear space, depletion of ER, reduction of autophagosome and autolysosome numbers, mitochondria swelling, and focal rupture of the plasma membrane [56]. Autotic cells are resistant to apoptosis or necrosis inhibitors, but sensitive to autophagy machinery suppression. Inhibition of autophagosome fusion with lysosome does not affect autosis, indicating that excessive autophagosome formation, rather than degradation, is probably the trigger of this process [56]. Additionally, cardiac glycosides, e.g. digoxin, which inhibit Na+/K+-ATPases, suppress autosis [56]. The interaction between BECN1 and the α subunits of Na+/K+-ATPase, i.e., ATP1A1, ATP1A2 or ATP1A3, on both intracellular membranes and at the plasma membrane is crucial, though the precise molecular mechanisms underlying autosis remain unclear.
The potential for selective autosis is a promising area of research for cancer treatments. This is illustrated by oncolytic virotherapy with myxoma virus (MYXV), a poxvirus that is endemic to rabbits and hares [57]. Preclinical studies suggest that therapies combining oncolytic viruses with tumor-specific T cells expressing chimeric antigen receptor (CAR), could enhance efficacy of this latter system, particularly, via direct injection into tumors [58]. Tumor-specific T cells rely on direct contact to induce apoptosis and pyroptosis, but this often fails to elicit strong primary responses in solid tumors or prevent resistance due to tumor antigen escape. Research by Zheng and coworkers [59] demonstrated that MYXV-infected, CAR-expressing tumor-specific T cells (CAR-TMYXV) can overcome primary resistance by delivering MYXV systemically to solid tumors. These CAR-TMYXV cells induce not only apoptosis and pyroptosis but also ATP1A1-dependent autosis through a synergy of T cell-derived IFNG/IFNγ and AKT1 signaling with the MYXV M-T5 protein-triggered SKP1 (S-phase kinase associated protein 1) and PIK3C3 signaling, significantly assisting tumor eradication.
Another example is HIV-1 infection that increases ATP1A1 expression in macrophages and memory CD4+ T cells, making them more susceptible to autosis when treated with autophagy-inducing peptides or nanoparticles [60,61]. The autophagy-inducing peptide Tat-beclin 1, which inhibits HIV-1 replication at low doses [62], becomes a potent inducer of autosis at higher doses or upon prolonged exposure [56,59-61,63]. This peptide selectively triggers autosis in latent HIV-1-infected primary CD4+ T cells and macrophages [60,61]. Similarly, the anti-apoptotic α2-helix from the death effector domain 1 of the K13 protein of KSHV, i.e., v-FLIP-α2, which impairs FAS (Fas cell surface death receptor)-dependent cytotoxic T cell killing of HIV-1-infected cells and activates HIV-1 transcription [64], also selectively induces autosis in HIV-1-infected primary resting memory CD4+ T cells and macrophages [60,61]. These findings highlight the therapeutic potential of autosis in treating infectious diseases and cancers, by targeting infected or malignant cells while sparing normal tissues.
2.3. The ATG machinery as an effector or restriction factor of interferons
Interferons (IFNs) have been recognized since the mid-twentieth century as secreted host factors that interfere with productive viral infection [65,66]. Three IFN types (I, II, III), with multiple subtypes of type I and III and a single type II, i.e., IFNG, have been identified [67]. Both the IFN system and autophagy are evolutionarily ancient mediators of the cell autonomous host defense [67-70]. As key arms of the innate immune system, autophagy and IFN coordinate an effective antiviral response. As such, viruses have evolved mechanisms to subvert the signaling and effectors of these systems to evade or co-opt the host response (see sections 3 and 4 of this review). This section will focus on how canonical autophagy and in some cases selected ATG proteins outside their role in autophagy, mediate IFN-regulated host antiviral immunity.
Autophagy is an inducible process during various physiological cues and in response to environmental stressors, including immune responses [71]. The role of IFN in the regulation of autophagy has previously been reviewed [72]. Direct and indirect evidence of type I IFN involvement in autophagy regulation exists. Type I IFN treatment induces autophagy in various human cell lines [73,74], and primary cells [75]. Consistently, IFN-inducible antiviral and IFN-regulating proteins, called IFN-stimulated genes (ISGs), are required for the autophagic control of viruses, such as EIF2AK2 during HSV-1 infection [34], RNASE1/RNAse l in EMCV and VSV infection [76], OASL (2’-5’-oligoadenylate synthetase like) in the degradation of non-structural protein 2 (nsp2) of infectious bursal disease virus/IBDV, a birnavirus, [77], and BST2/tetherin 1 (bone marrow stromal cell antigen 2) in porcine epidemic diarrhea virus/PEDV, a coronavirus [78]. Additionally, the ISG SHISA5/SCOTIN (shisa family member 5) interferes with the replication of hepatitis C virus/HCV, a hepacivirus, through the autophagic degradation of its NS5A protein [79]. In murine RAW264.7 macrophage-like cells, however, the type I IFNs IFNA/IFNα and IFNB/IFNβ did not induce autophagy [80]. Therefore, type I IFN may play a role only in specific cell types, or potentially only in the context of viral infections. Extensive evidence for IFNG induction of autophagy exists for diverse cell types, including murine RAW264.7 cells [80-83], murine embryonic fibroblasts [84], various human cell lines [77,81,82,85,86], primary macrophages [82,87,88], and primary hepatocytes [85]. Type III IFN or IFNL/IFNλ, the most recently discovered type of IFN, is also implicated in regulating autophagy. Specifically, a short form of ATG10, ATG10S, can induce degradation of various viral proteins in the presence of IFNL in human cell lines [89].
The precise factors that transduce IFN signaling to increase autophagy activity are incompletely understood. For type I IFN, MTORC1 [72] and PtdIns3K [90] signaling have been implicated. The ISG RSAD2 (radical S-adenosyl methionine domain containing 2) mediates IFNB-induced autophagy in primary human cornea-associated trabecular mesh cells [75]. In the case of IFNG, an unconventional JAK2 (Janus kinase 2)-PtdIns3K-MAPK (mitogen-activated protein kinase)-dependent, STAT1-independent, pathway may be required in murine macrophages [83], while a role for IRF1 (interferon regulatory factor 1) [85] and MTORC1 [88] has been demonstrated in Huh7 hepatocytes and human macrophages, respectively. IFNG-induced autophagy may also be countered by other cytokines that induce STAT6 (signal transducer and activator of transcription 6) [87]. As autophagy regulates many facets of the antiviral sensing and IFN response, and viruses extensively target autophagy to suppress the IFN response, selective manipulation of these mechanisms may be leveraged for therapeutic induction of antiviral IFN responses via autophagy.
Beyond a regulatory role of IFN in autophagy induction, the role of the ATG machinery as an effector mechanism has also been described for diverse virus classes. As mentioned, the ISG EIF2AK2 is required for autophagic clearance of HSV-1 virions [34,91]. HSV-1 encoded neurovirulence protein ICP34.5 antagonizes BECN1 to evade antiviral autophagy and promote viral encephalitis [91]. However, the role of IFN in protection against the BECN1-binding mutant virus remains unknown. Autophagy is also activated by HSV-1 viral genomic DNA sensing by the CGAS-STING1 pathway to limit IFN production and exerts a negative feedback loop by directly targeting viral genomes, as well as CGAS and STING1 themselves to degradation [36] (Figure 1). In order to limit RNA sensing, Sendai virus/SeV and VSV infection activates CALCOCO2/NDP52-mediated selective autophagy that targets MAVS (mitochondrial antiviral signaling protein) into degradation by a mechanism involving the ISG BST2, limiting IFN signaling [92]. Similarly, autophagy of depolarized mitochondria removes endogenous danger associated molecular patterns for inflammasome activation and pro-inflammatory cytokine production, such as IL1 (interleukin 1) and IL18 [93]. Furthermore, components of inflammasomes are also directly targeted for autophagy (Figure 1). However, the relative contribution of viral genome targeting by autophagy versus inhibition by elevated IFN and pro-inflammatory cytokine production in restricting viral replication when autophagy is inhibited, has not been dissected [36]. Furthermore, in primary mouse neurons, ATG5 restricts HSV-1 replication via a type I IFN-independent mechanism [35]. Therefore, the role of IFN in regulating HSV-1 may be cell type-specific, and redundant IFN-mediated mechanisms may exist to control HSV-1.
A role for the ATG machinery in mediating IFNG-dependent restriction of murine norovirus (MNV), a calicivirus, in a non-canonical manner has been extensively studied. Initially, it was shown that type I IFN and IFNG exhibit non-redundant activity in limiting MNV, and IRF1 is required for cell intrinsic effects of IFNG [94]. Subsequently, a role for the Ubl conjugating machineries, in particular ATG5, ATG7, and ATG16L1, were demonstrated as the mediators of IFNG activity, while LC3B, ATG4B, and lysosomal fusion mediator RAB7A were dispensable [95]. In vivo, mice with myeloid-specific deletion of ATG5 infected with an acute strain of MNV exhibit increased susceptibility in the absence of type I IFN signaling, further underscoring both the non-redundant roles of these IFNs and the specificity of autophagy for IFNG [95]. The Ubl conjugation machinery required for MNV restriction also includes ATG3, and redundancy in the ATG8 homologs, while ATG14, ULK1 and ULK2 are dispensable [96]. In addition, the ISGs IGTP/IRGM3 (interferon gamma induced GTPase) and GBP2 (guanylate binding protein 2), but not IRGM1, GBP1, GBP3, GBP5 or GBP7, are required for IFNG activity, and are recruited to the MNV replication complexes to dismantle them [96]. Of note, murine GBP2 is orthologous to human GBP1.
A more complete list of ATG components required in IFNG-mediated restriction of viruses was elucidated recently. Using a candidate approach and CRISPR-Cas9-based screening of an autophagy-targeted library in the BV2 microglial cell line, ATG9A, WIPI2 and the ATG8 protein GABARAPL2/GATE16, but not LC3A, LC3B, GABARAP, or GABARAPL1, were identified as effectors of IFNG inhibition of MNV infection [97]. BECN1, ATG14, and UVRAG were dispensable [97]. Studying the role of human GBPs in HeLa cells treated with IFNG, the ATG16L1 interacting protein CAPRIN1 (cell cycle associated protein 1) was shown to mediate restriction of MNV in conjunction with WIPI2 and GBP1 [98]. IFNG also induces chicken GBP1 to restrict infectious bronchiolitis virus/IBV replication and to degrade nucleocapsid protein [89]. This activity is not pan-viral, as ECMV, murine hepatitis virus/MHV, and West Nile virus were equally well-controlled by IFNG in cells with or without ATG5 [95]. Collectively, these findings support a selective role for the Ubl conjugation machineries, together with the GBP1 system, in restricting different viruses in multiple species, without other ATG components.
The previous two decades have revealed multiple mechanisms for the regulation of both autophagy by IFNs and of viral infections by autophagy. As the evidence for IFN-regulation of viruses derives primarily from in vitro studies, the in vivo physiological relevance remains to be explored in many cases. Along these lines, the relative role of cell-autonomous IFN responses, for example type I IFN production, or IFN from immune cells, for example IFNG, in antiviral control remain to be determined. Moreover, the precise molecular mechanisms behind autophagy machinery-mediated control of viral replication, beyond degradation of virions and viral components, remains incompletely understood. Finally, the relative contribution of restricting viral infections via direct viral component degradation versus regulation of anti-viral cytokines remains poorly defined. Answers to these open questions will be important to translate our basic understanding into antiviral therapeutics.
3. Inhibition of the ATG machinery by viruses to prevent virophagy
Viruses benefit from inhibiting autophagy. The most obvious reason being to prevent their degradation since autophagy has been described as an antiviral mechanism. More generally, inhibition of autophagy enables viruses to create a favorable environment for their replication. This is also underscored by the fact that mutations in ATG genes have been associated with disease severity in patients infected by poliovirus, a picornavirus, herpes simplex virus 2/HSV-2, varicella zoster virus/VZV, another orthoherpesvirus, and possibly severe acute respiratory syndrome coronavirus 2/SARS-CoV-2 [99-102].
Many viral proteins are capable of interfering with the autophagy machinery. One of the first anti-autophagy viral protein to be identified was ICP34.5 encoded by HSV-1 [32]. ICP34.5 is a key protein for successful HSV-1 propagation in the central nervous system and multitasks in counteracting the host immune responses. Among those functions, ICP34.5 can interact with BECN1 to inhibit autophagy initiation [91]. Interestingly, the role of autophagy on the restriction of HSV-1 multiplication appears to be different depending on the cell type. The replication of HSV-1 is not modified in ATG5 deficient fibroblasts while ICP34.5 capacity to bind BECN1 is critical for neurovirulence [35,91]. HSV-1 encodes at least two other proteins that can inhibit autophagy initiation through different mechanisms: Us3, which directly phosphorylates BECN1, and Us11, which inhibits the PKR-EIF2A pathway to prevent virus-induced autophagy [33,103].
Targeting BECN1 is one of the main strategies developed by viruses to evade autophagy and several examples can be found in different viral families. Already among the Orthoherpesviridae family, HCMV, KSHV and murid herpesvirus 68/MHV-68, all express proteins that block BECN1 [104-106]. Additionally, HIV-1-encoded Nef protein impairs the formation of autophagosomes by interacting with BECN1 in T cells and might also inhibit the fusion of autophagosomes with lysosomes in macrophages [107,108].
Autophagy and innate immunity are closely interconnected, and autophagy can be both a modulator and an effector of antiviral immune responses. RVFV is an arbovirus from the Phenuiviridae family. In Drosophila, RVFV triggers antiviral autophagy via the Toll-7 receptor, a mechanism that seems to be conserved in mammalian cells [24]. A recent study showed that NSs, a virulence factor encoded by RVFV, can interact with ATG8 proteins and sequester them in the nucleus [109]. These interactions are detected at later stages of RVFV infection and lead to an inhibition of autophagy. Interestingly, although a virus expressing a variant of NSs unable to bind all six ATG8 orthologs, replicates less efficiently than the wild-type virus, this difference is no longer observed in IFN-deficient cells, confirming the important crosstalk between these pathways.
Activation of autophagy with drugs was shown to restrict replication of SARS-CoV-2 in different cell lines, giving hope for a potential antiviral treatment, although the interplay between autophagy and coronaviruses seems more complex [110]. Several proteins encoded by this virus seem to be able to inhibit autophagy at different steps of the pathway. For example, expression of the accessory factor ORF3a leads to an accumulation of autophagosomes by inhibiting their fusion with the lysosomes [111]. Specifically, ORF3a disrupts the interaction between the homotypic fusion and protein sorting (HOPS) tethering complex and RAB7, preventing the tethering of autophagosomes with lysosomes. Additionally, ORF7a inhibits the autophagic flux by inducing the cleavage of SNAP29 by the CASP3 (caspase 3) protease [112]. Another respiratory virus, IAV, is capable of blocking autophagosome-lysosome fusion by expression of its structural protein M2 [113,114]. Additionally, it has also been shown that M2 is able to hijack LC3B for stable IAV particle production. Finally, adenoviruses prevent autophagosome maturation via a conserved PPxY peptide motif in their capsid protein VI that recruits the ubiquitin ligase NEDD4L/NEDD4.2. Mutating the PPxY motif in turn renders adenoviruses susceptible to autophagic clearance and increases its antigen presentation [45,115].
Some of the latest publications on viruses and autophagy shed light on the modulation of selective autophagy. Viral proteins can be targeted for autophagic degradation by SARs. However, several members of the Picornaviridae family encode proteases that can cleave these receptors. Two recent publications on Seneca Valley virus/SVV, a picornavirus, showed that the SARs SQSTM1/p62 and OPTN (optineurin) can target the capsid protein VP1 for turnover by autophagy and OPTN is also involved in IFN signaling via the TBK1-IRF3 pathway [116,117]. SVV encodes a protease, 3Cpro, that cleaves SQSTM1/p62 and OPTN to counteract the antiviral activity of these SARs, preventing the degradation of capsid proteins and dampening IFN response. Other picornaviruses also evolved similar strategies to block selective autophagy, like coxsackievirus B3 (CVB3), which encodes two proteinases, 2A and 3C, that can cleave SQSTM1/p62, NBR1 and CALCOCO2/NDP52 [118,119]. In the case of CALCOCO2/NDP52, interestingly, the cleavage by 3Cpro allows the formation of a stable fragment that exerts proviral properties. Reticulophagy is a form of autophagy that selectively degrades the ER via specific ER-localized SARs, such as RETREG1/FAM134B, RTN3L, CCPG1 and ATL3 [120]. The NS3 protease of several orthoflaviviruses, including dengue virus/DENV and Zika virus/ZIKV, cleaves RETREG1, whereas SARS-CoV-2-encoded ORF8 sequesters FAM134B and ATL3 in SQSTM1/p62 condensates, inhibiting reticulophagy in both cases [121,122]. Inhibition of autophagy can also be a consequence of viral subversion of selective autophagy, as it has been described for a plant-infecting pathogen called Tomato bushy stunt virus/TBSV, a tombusvirus [123]. TBSV replication protein p33 diverts both ATG8 proteins and NBR1 into its replication organelles, resulting in the inhibition of autophagy in infected cells. These latest studies provide further demonstrations that manipulation of autophagy by viruses is a widely conserved mechanism.
4. Pro-viral functions of autophagy
4.1. Building viral replication compartments with the help of the autophagy machinery
Cellular stress caused by viral infection often triggers the activation of autophagy. Despite the antiviral nature of autophagy’s degradative ability, many viruses, particularly positive single-stranded RNA (+ssRNA) viruses, have evolved to resist and benefit from autophagy (Figure 2). The pro-viral nature of autophagy activation, or at least of components of the ATG machinery, is well documented for a wide variety of viruses [104,124-128]. This section focuses on how the autophagy machinery assists in the formation of virus-induced specialized organelles that harbor the replication of +ssRNA viruses.
Figure 2.
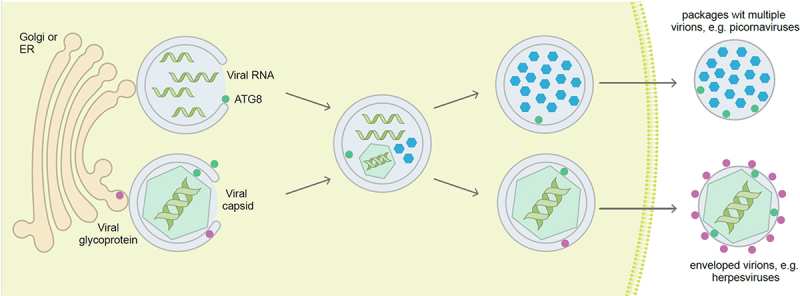
Viruses hijack autophagosomal membranes to establish or protect their replication in the cytosol, and for envelope acquisition during egress. Positive single-stranded RNA viruses often replicate in association with membrane compartments that can be assembled with the help of the ATG machinery. Once encapsulated, the viral particles can egress in virus packages within ATG8 protein-decorated membranes. Some DNA viruses also acquire their envelope from/with autophagosomal membranes and then carry parts of the ATG machinery in their virus particles. ATG8, mammalian Atg8 homolog.
The cytoplasmic replication of +ssRNA viruses requires membrane rearrangement to form replication factories, also called replication organelles/ROs, which concentrate and protect viral factors essential for efficient replication and transcription of the viral genomes. Pioneering ultrastructural studies of cells infected with viruses from the Picornaviridae, Flaviviridae, Caliciviridae, Coronaviridae, and Arteriviridae families by electron microscopy have revealed the cytoplasmic accumulation of double-membrane vesicles (DMVs) [129-134]. Because autophagosomes are also characterized by a double-membrane, it has been suggested that autophagy may contribute to the formation of these virus-induced DMVs [135,136], but it needs to be pointed out that the formation of DMVs might often utilize only individual components of the autophagy machinery. Notably, proteins involved in autophagy and related processes have been determined to be in close proximity to the coronavirus replication complex at DMVs [137]. Lately, 3D-reconstruction of replication organelles by electron tomography analysis confirms the presence of components of viral replicases on the inner membrane of DMVs, suggesting their involvement in the replication of at least coronaviruses [138] and hepaciviruses [139]. Importantly, the involvement of ATG proteins has been linked to DMVs formation by several +ssRNA viruses [140-144].
Coronaviruses and HCV are known to induce DMV accumulation in the cytoplasm of infected cells [139,145], to benefit from autophagy activation [144,146-148], and the direct involvement of the ATG machinery in DMV formation has been suggested [142,148-151]. However, due to conflicting data, it has remained unclear which part of the ATG machinery is essential for DMV formation. Recently, Twu and colleagues identified PtdIns3K as an essential factor for SARS-CoV-2 and HCV-induced DMV formation and their RNA replication [143], which in parallel requires the viral proteins SARS-CoV-2 nsp3 and nsp4 [150,152], or HCV NS5A and NS4B [139,153]. Interestingly, ATG5−/− Huh7-Lunet cells were still able to form virus-induced DMVs, suggesting that the autophagy elongation complex is dispensable [139,150,152,153]. Similarly, the replication of human astrovirus 1/HAstV-1, an astrovirus, localized at DMVs that required PtdIns3K for their formation [141], highlighting the importance of part of the ATG machinery in initiating DMV formation during infection. For coronaviruses, nsp6 is a multiple spanning membrane protein that localizes to the ER where it cooperates with nsp3 and nsp4 to generate DMVs as the replication organelle. Interactions between nsp3 and nsp4 extrude spherical replication organelles from the ER into the cytosol, where they house the viral RNA replicase proteins and at the same time 12 copies of nsp3 and nsp4 assemble into a pore complex to export viral RNA into the cytosol [154,155]. The replication organelles remain tethered to the ER by thin membrane connectors formed by nsp6, which induced ER zippering to collapse ER cisternae, assumed to exclude host proteins that might inhibit replication but to maintain membrane connections allowing entry of lipids [156]. A three amino acid deletion in nsp6 increased both ER zippering and virulence. Interestingly, when expressed alone in cells, nsp6 induces formation of LC3B puncta independently of activation of autophagy by starvation or inhibition of mTOR [149]. Formation of LC3B puncta by nsp6 required ATG5 suggesting conjugation of LC3 to membranes by the ATG12–ATG5-ATG16L1 complex. Nsp6 induced the formation of ZFYVE1/DFCP1-positive domains of the ER enriched for phosphatidylinositol-3-phosphate leading to recruitment of WIPI2 and the ATG8 conjugation machinery with ATG12–ATG5-ATG16L1. LC3B puncta are also induced by nsp6 of IBV, MHV and SARS-CoV-2 and the equivalent nsp5-nsp7 ortholog encoded by the arterivirus porcine reproductive and respiratory syndrome virus/PRRSV [149]. Interestingly, the LC3B puncta generated by nsp6 are small compared to LC3B puncta generated by canonical autophagy because nsp6 prevents autophagosome expansion normally seen in response to starvation [157]. Nsp6 proteins remain in the ER and do not travel with autophagosome membranes to lysosomes suggesting nsp6 disrupts early events in autophagosome formation. Affinity purification of tagged nsp6 [158] and proximity biotinylation [159] has shown that nsp6 interacts with proteins involved in membrane fusion such as SIGMAR1, VAMP7, ESYT2, ATP2A2/SERCA2 and TBK1 [159]. This complex of proteins regulates the formation of hybrid phagophore assembly sites/pre-autophagosomal structures/HyPAS generated early during autophagy by fusion of endosomal membranes containing ATG16L1 with cis-Golgi membranes enriched for RB1CC1 [159]. It is likely that nsp6 disrupts hybrid phagophore assembly sites formation to limit the maturation of autophagosomes induced during coronavirus infection.
Picornaviruses such as poliovirus and CVB3 also benefit from autophagy and induce the cytoplasmic accumulation of DMVs in infected cells [135,160,161]. Noticeably, CVB3-induced DMVs seem to rely on the autophagy machinery for their generation [140]. CVB3- and poliovirus-induced DMVs, however, appear somewhat later in the replication process [160,161]. In the case of poliovirus, it has been shown that poliovirus induced DMVs originate from cis-Golgi membranes appearing during the early stages of infection as single membrane structures that transform in a complex manner into double membrane structures [160]. Thus, additional studies on picornaviral infections are required to determine the interrelation between autophagy, DMV formation, and the viral life cycle.
HBV is a DNA virus that replicates both in the nucleus and cytoplasm. HBV infection stimulates autophagy through an increase in the PIK3C3/VPS34-BECN1 complex activity induced by the viral regulatory protein HBx [162,163]. HBV core particles are associated with the phagophore in the cytoplasm, which is required for the packaging of viral pregenomic RNA [164]. The precore protein derivatives, which form secreted viral antigens, are associated with autophagosomes. These findings suggest that differential autophagy membrane-related trafficking routes are involved in the secretion of virions and viral antigens [164].
In conclusion, the pro-viral roles of autophagy in viral replication have been well-documented for several viruses. Accumulating evidence demonstrates that virus-induced DMVs are the replication organelles of many +ssRNA viruses but the precise functions of ATG proteins in their formation still remain to be better understood.
4.2. Viral envelope acquisition from autophagosomal membranes
Enveloped viruses are generally composed of a capsid that houses the viral genome, enclosed in a lipid bilayer decorated with virus-encoded glycoproteins. For numerous virus families, non-glycosylated matrix or tegument proteins are found on the inner side of the envelope, ensuring the link between the capsid and the envelope. This latter is acquired by budding through either the host cell plasma membrane or organelles, such as the Golgi apparatus and the ER, but recent evidence also indicates that this can occur at autophagosome membranes (Table 1). For viruses that bud at the plasma membrane, such as IAV or rabies virus/RABV, a rhabdovius, this assembly stage also enables the release of the virion from the cell. Conversely, when the envelope originates from the endomembrane system, viral particles are transported luminally to the trans-Golgi network (TGN) and from there to the plasma membrane in vesicles to exit the cell by fusion.
Table 1.
Viruses that modulate autophagy to favor their envelopment or pseudoenvelopment and release.
| Virus | Structure/ family | Envelopment mode | Autophagy inhibition mechanism /Viral proteins | ATG proteins associated with virions | References |
|---|---|---|---|---|---|
| Budding at the plasma membrane | |||||
| RSV | ssRNA(-) linear genome Pneumoviridae |
|
No information | [165] | |
| RABV | ssRNA(-) linear genome Rhabdoviridae |
|
|
No information | [166] |
| LASV MOPV |
Segmented ssRNA(-) linear genome Arenaviridae |
|
|
No information | [168,169] |
| IAV | ssRNA(-) linear genome Orthomyxoviridae |
|
|
Absence of LC3B proteins in the virions | [113,114,171] |
| HBV | Partially dsDNA circular genome/ icosahedral capsid Hepadnaviridae |
|
|
ATG12 | [175,177] [125,164] |
| PIV3 | ssRNA(-) linear genome/helical capsid matrix Paramyxoviridae |
|
|
No information | [175] |
| SFTSV | Segmented ssRNA(-) linear genome Bunyaviridae |
|
|
PIK3C3, BECN1, ATG14, ATG7, ATG12–ATG5, ATG16L1, LC3B-II | [178] |
| HCMV | dsDNA genome/ icosahedral capsid/ tegument Orthoherpesviridae |
|
|
BECN1, ATG5, ATG12, LC3B-II, SQSTM1, GABARAP |
[104,179,180] |
| EBV | dsDNA genome/ icosahedral capsid/ tegument Orthoherpesviridae |
|
|
ATG3, ATG4C, ATG5, ATG7, ATG8 (LC3B and GABARAPL2), SARs (SQSTM1 NBR1 and TOLLIP), PIK3C3, PIK3R4, ATG14, BECN1 | [182,183] [186] |
| VZV | dsDNA genome/ icosahedral capsid/ tegument Orthoherpesviridae |
|
|
LC3B, RAB11 | [181,185,201] |
| DENV Zika virus |
ssRNA(+) linear genome/ icosahedral capsid Flaviviridae |
|
|
ATG proteins are associated with the infectious virion-containing extracellular vesicles | [187-189] [190,191] |
| HCV | ssRNA(+) linear genome Flaviviridae |
|
|
No information | [192,194] [193,195] |
| Picornaviruses (e.g., poliovirus, EMCV and CVB 3) | ssRNA(+) linear genome icosahedral capsid Picornaviridae |
|
|
LC3B-II | [135,196-198] |
dsDNA, double-stranded DNA; ssRNA(+), positive-sense single-stranded RNA; ssRNA(-), negative-sense single-stranded RNA.
Numerous studies have shown that the proviral effect of autophagy can be linked to the optimization of envelope acquisition and increased extracellular virus production. In this context, viral infections usually induce autophagosome accumulation, often associated with an inhibition of the autophagic flux, in particular by matrix proteins.
The autophagy machinery has been involved in the budding of several viruses at the plasma membrane [113,114,165-169]. For example, respiratory syncytial virus/RSV, a pneumovirus, induces the accumulation of immature autophagosomal structures by blocking autophagosome-lysosome fusion, to improve its viral production [165]. Knockdown of LC3B decreases RSV propagation and interestingly, treatment with exogenous IL22 restores the autophagic flux with concomitant reduction in RSV production. Both the mechanism of inhibition and the viral protein(s) involved in autophagy regulation are still unknown. RABV also induces incomplete autophagy in mouse neuroblastoma cells to favor its extracellular viral production [166]. At least two RABV proteins could be responsible for the accumulation of autophagosomes and the inhibition of autophagosome-lysosome fusion, i.e., the phosphoprotein RABV-P through interaction with BECN1 and the matrix protein RABV-M [166,167]. RABV-M interacts with NEDD4, an E3 Ub ligase and its ubiquitination is crucial for the accumulation of autophagosomes [166,167]. Inhibition of autophagy, by ATG5 knockdown or 3-methyladenine (3MA) treatment, decreases both RABV-M recruitment at the plasma membrane and generation of virus-like particles in the supernatant, revealing a possible role of autophagy in RABV budding [166,167]. The autophagy machinery is also hijacked by two viruses belonging to the Arenaviridae family, the Lassa virus/LASV, a rodent virus responsible for hemorrhagic fever in humans, and the Mopeia virus/MOPV, to favor their envelopment [168]. Silencing of ATG5 has no impact on viral DNA replication but decreases the generation of LASV and MOPV infectious particles. The matrix protein of LASV, i.e., LASV-Z, is able to impede autophagosome-lysosome fusion, lysosome acidification, and lysosomal enzyme transport by disrupting the cytoskeleton via its interaction with CCT2 (chaperonin containing TCP1 subunit 2) [169]. In the same way as for RABV-M, LASV-Z is decreased in plasma membrane-derived fractions and accumulates in the Golgi apparatus, when cells are silenced for ATG5 or ATG7, or treated with 3MA [169].
The proton-selective ion channel protein M2 of IAV is responsible for the stabilization of LC3B conjugated membranes during virus replication and egress, including SQSTM1/p62 containing vesicles with intraluminal membranes that resemble autophagosomes and amphisomes [113]. In particular, GFP-LC3B binds M2 via a highly conserved LC3-interacting region in M2, and this interaction localizes GFP-LC3B in two main pools, one in vesicles of the perinuclear region and one at the plasma membrane [114]. M2-LC3B interaction is not required for viral genome replication but is important for filamentous viral particle budding and stability of resulting virions. Indeed, knockdown of ATG16L1 decreased the proportion of filamentous particle budding. A small amount of membrane conjugated LC3B is found in extracellular viral particles and LC3B interaction with M2 that is disrupted by caspase cleavage seems required for optimal infectious particle production [170], but autophagy per se does not seem to directly contribute to IAV envelopment or egress. M2 induces an autophagy-related lipidation of LC3A/B, which nonetheless depends on ATG16L1 for the recruitment of the E3-like ATG12–ATG5-ATG16L1 complex, but it is independent of RB1CC1 and WIPI2 [171]. In fact, M2 induces the conjugation of LC3 to single membranes, in particular to the plasma membrane, through CASM. The proton-selective ion channel activity of M2 is responsible for this conjugation [172,173], via ATG16L1 recruitment by the ATP6V1H/V1H subunit of the vacuolar-type H+-translocating ATPase/V-ATPase [174]. Thus, IAV infection causes accumulation of LC3B conjugated membranes that are protected from lysosome fusion and required for stable virion production.
Manipulation of the early steps of autophagy has also been widely described for viruses acquiring their envelope through the endomembrane system, such as HBV, a hepadnavirus, parainfluenza virus 3/PIV3, a paramyxovirus, or Orthoflaviviridae family members. Both HBV and PIV3 block autophagosome-lysosome fusion to improve virion production. HBV decreases SNAP29 and RAB7 levels and impairs lysosomal acidification via its non-structural HBx protein [125,175]. PIV3 impairs SNARE complex formation via an interaction of its phosphoprotein P with SNAP29 [176]. Inhibition of autophagy markedly inhibited the production of extracellular virions of both HBV and PIV3, indicating a role of autophagy in viral envelopment and/or release. HBV seems to exploit autophagy for its envelopment and not for its egress, since the abundance of intracellular and extracellular enveloped HBV virions was similarly impacted by autophagy inhibition [177]. Moreover, HBV envelope proteins partially colocalize and interact with LC3B in the cytoplasm [125] and HBV core particles are associated with autophagosomes and phagophores, suggesting that these membranous structures are the site for the packaging of the viral pregenomic RNA [164]. Ding and co-workers observed that incomplete autophagy helps PIV3 extracellular viral production but does not influence viral protein synthesis or intracellular viral production, suggesting a role of autophagy in PIV3 extracellular release [176]. Severe fever with thrombocytopenia syndrome virus/SFTSV, a bunyavirus, exploits autophagy for both its envelopment, in the ER-Golgi intermediate compartment/ERGIC- and Golgi-originated autophagosomes, and its release by exocytosis [178]. Unlike HBV and PIV3, SFTSV triggers complete autophagy, via the interaction of its nucleoprotein NP with BECN1. Yan and co-workers proposed that mature SFTSV particles are released by fusion of autolysosomes with the plasma membrane.
Nucleocapsids of Herpesviridae family members exit the nucleus by transient envelopment across the intact nuclear membranes and acquire their final envelope in the cytoplasm before being released by exocytosis. It has been reported that several orthoherpesviruses use autophagy for their final envelopment in the cytoplasm (Figure 2). HCMV completely reorganizes pre-existing membranous compartments to create a unique compartment called viral assembly compartment (VAC), where SQSTM1/p62, LC3B and other ATG proteins are recruited [179,180]. VZV and Epstein–Barr virus (EBV), two orthoherpesviruses, generate a less-organized VAC but both exploit autophagic-derived vesicles, together with TGN- and endosome-derived vesicles for their final envelopment [181-183]. Autophagosomes accumulate in the cytoplasm of EBV, KSHV and HCMV-infected cells, due to an autophagic flux inhibition [104,182,184]. In contrast, VZV needs to induce a complete autophagy process [185]. Accordingly, the lysosomal inhibitor bafilomycin A1 treatment significantly decreases VZV secondary envelopment [185]. Finally, the process of orthoherpesvirus envelopment looks similar to phagophore expansion (Figure 2), and autophagosomes have been proposed as potential membrane donors for this viral envelope. Confirming this involvement, several ATG proteins are found in highly purified virions. In particular, different studies reported the presence of the lipidated form of LC3B and SQSTM1/p62 in EBV and HCMV particles and the association of LC3B and the recycling endosome protein RAB11 with VZV virions [179-182,186]. Recent mass spectrometry analysis of EBV particles identified numerous ATG proteins belonging to the PtdIns3K complex I and the Ubl conjugation systems, and SARs such as SQSTM1/p62, NBR1 and TOLLIP (toll interacting protein) [186]. Interestingly, ATG proteins are only present in a fraction of EBV virions, suggesting heterogeneity of virions with respect to their composition. The presence of ATG proteins in mature virions is not exclusive to orthoherpesviruses. It has been reported that HBV virions contain ATG12 [125] and SFSTV extracellular particles copurify with several ATG proteins, such as ATG5, BECN1 and lipidated LC3B, together with ER-Golgi intermediate compartment and TGN markers [178].
Although this is not strictly speaking for the envelope acquisition, DENV maturation depends on autophagy for the processing of its envelope glycoprotein prM and its infectivity [187]. DENV and ZIKV, two orthoflaviviruses, exit host cells in two distinct ways; one as free virions and the other enclosed within membranes. Whereas no data have connected autophagy in viral particle envelopment in the cytoplasm, the role of autophagy in the viral egress in secretory vesicles has been clearly demonstrated [188,189]. Unconventional secretory autophagy but not degradative autophagy allows the release of several infectious DENV and ZIKV particles within autophagosome-derived vesicles through a process that depends on the sequence of S-receptor kinases (Srk) [188]. A similar role of autophagy was observed for DENV in dendritic cells [190]. Moreover, hydrolysis of lipid droplets by a selective autophagy known as lipophagy plays a role in the assembly and secretion of DENV [191]. Autophagy is also involved in the maturation and release of HCV [192]. Trafficking of APOE (apolipoprotein E), a cellular apolipoprotein critical for viral morphogenesis, depends on autophagy and blocking autophagic flux improves APOE association with HCV and virion infectivity [193]. APOE is delivered by autophagosomes to the HCV assembly site where it interacts with the envelope protein complex E1/E2. HCV restricts the autophagic flux by increasing the expression of RUBCN (rubicon autophagy regulator), an inhibitor of autophagosome fusion with lysosomes [194,195].
Finally, picornaviruses, like poliovirus and CVB3, are non-enveloped viruses but they can also be non-lytically released from their host cells [196]. Stimulation of autophagy enhances non-lytic poliovirus spreading. This non-lytic spread of picornaviruses was found to occur by packages of multiple viruses, at least 20, within autophagosomal membranes [197,198] (Figure 2). These viruses utilize phosphatidylserine in the outer leaflet of autophagosomal membranes, which mostly originate from the ER and Golgi apparatus, to enter host cells via scavenger receptors such as TIMD4/TIM-4 (T cell immunoglobulin and mucin domain containing 4) and MERTK (T cell immunoglobulin and mucin domain containing 4) [197]. This mechanism increases viral spreading. In addition, membranes around the virus packages protect their content from environmental harm, such as antibodies. Along these lines, vesicle cloaked viruses have even been shown to survive fecal to oral transmission [199]. Therefore, both enveloped and non-enveloped viruses hijack the autophagy machinery for release in autophagosomal membranes and efficient spreading.
5. Conclusions and outlook
Some of the first electron microcopy pictures of double-membrane vesicles originated from virus infected cells [134], around the time when Christian de Duve had coined the term autophagosome [200]. The information gathered in this review suggests that nearly every virus has to cope with the anti-viral activity of autophagy and many modulate this pathway for their survival. Some viruses even utilize the machinery that generates autophagosomes for building their replication compartments and/or acquiring envelopes. Both of these strategies, inhibition of autophagosome formation and blocking autophagosome fusion with lysosomes, inhibit virophagy. Thus, autophagy stimulation might overcome these viral interference mechanisms and retore autophagic flux for virion or virus component degradation. A detailed understanding of how every virus interacts with autophagy might also enable us to more specifically tilt the balance of pro-viral and anti-viral functions of autophagy during viral infections for their treatment.
Acknowledgements
We thank Tara von Grebel of the Visual Design Department of the University of Zürich for the design of the figures.
F.R. is supported by a Novo Nordisk Foundation grant (0066384).
L.E. is supported by ANRS-MIE (MIE23159).
A.E. and M.L. are supported by Agence Nationale de la Recherche (ANR-22-CE15-0021-03).
H.W. is supported by the Agence Nationale de la Recherche, ANR-19-CE15-0013 Virmedaco.
M.F. and C.V. are supported by the Agence Nationale de la Recherche (ANR-23-CE15-0045).
P.L. was the recipient of a Discovery grant from the Natural Sciences and Engineering Research Council of Canada (NSERC), RGPIN06932
R.B. is supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Projektnummer 272983813 – TRR 179 and the Project “Virological and immunological determinants of COVID-19 pathogenesis – lessons to get prepared for future pandemics (KA1-Co-02 “COVIPA”)”, a grant from the Helmholtz Association’s Initiative and Networking Fund.
S.A.S. is supported by the National Institute of Neurological Disorders and Stroke of the NIH (www.ninds.nih.gov) (R01 NS084912 and R01NS104015) and the International Maternal Pediatric Adolescent AIDS Clinical Trials Network under awards UM1AI068632 (IMPAACT LOC), UM1AI068616 (IMPAACT SDMC), and UM1AI106716 (IMPAACT LC).
A.O. is supported by the National Institutes of Health (K08AI144033, DP2GM146457).
C.M. is supported by the Swiss National Science Foundation (310030_204470/1).
V.T. is supported by the Swiss National Science Foundation (310030B_201278).
Funding Statement
This work was supported by the Agence Nationale de Recherches sur le Sida et les Hépatites Virales [MIE23159]; Agence Nationale de la Recherche [ANR-22-CE15-0021-03]; Agence Nationale de la Recherche [ANR-19-CE15-0013]; Agence Nationale de la Recherche (ANR-23-CE15-0045); Deutsche Forschungsgemeinschaft [272983813]; Helmholtz-Gemeinschaft [KA1-Co-02]; International Maternal Pediatric Adolescent AIDS Clinical Trials Network [UM1AI068632, UM1AI068616, UM1AI106716]; National Institutes of Health [K08AI144033, DP2GM146457]; Natural Sciences and Engineering Research Council of Canada [RGPIN06932]; Novo Nordisk Foundation Center for Basic Metabolic Research [0066384]; National Institute of Neurological Disorders and Stroke [R01 NS084912, R01NS104015]; Swiss National Science Foundation [310030_204470/1]; Swiss National Science Foundation [310030B_201278].
Abbreviations
ATG, autophagy related; BECN1, beclin 1; CAR, chimeric antigen receptor; CASM, conjugation of ATG8/LC3 to single membranes; CVB3, coxsackie virus B3; DENV, dengue virus; DMVs, double-membrane vesicles; dsDNA, double-stranded DNA; EBV, Epstein–Barr virus; EMCV, encephalomyocarditis virus; ER, endoplasmic reticulum; HBV, hepatitis B virus; HCMV, human cytomegalovirus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; HSV, herpes simplex virus; IAV, influenza virus; IFN, interferon; ISG, interferon-stimulated gene; KSHV, Kaposi sarcoma associated herpesvirus; LASV, Lassa virus; MHV-68, murid herpesvirus 68; MNV, murine norovirus; MOPV, Mopeia virus; MYXV, myxoma virus; Nsp, non-structural protein; PE, phosphatidylethanolamine; PIV, parainfluenza virus; PRRSV, porcine reproductive and respiratory syndrome virus; PPRV, peste des petits ruminant’s virus; PtdIns3K, class 3 phosphatidylinositol 3-kinase; RABV, rabies virus; RSV, respiratory syncytial virus; RVFV, Rift Valley fever virus; SAR, selective autophagy receptor; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SFTSV, severe fever with thrombocytopenia syndrome virus; SINV, Sindbis virus; ssRNA, single-stranded RNA; TBK1, TANK binding kinase 1; TGN, trans-Golgi network; TRIM, tripartite motif containing; Ub, ubiquitin; Ubl, ubiquitin-like; VAC, viral assembly compartment; VSV, vesicular stomatitis virus; VZV, varicella zoster virus
References
- 1.Helenius A. Standing on the Shoulders of viruses. Annu Rev Biochem. 2020; 89:21–38. [DOI] [PubMed] [Google Scholar]
- 2.Münz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009; 27:423–429. [DOI] [PubMed] [Google Scholar]
- 3.Lamark T, Johansen T. Mechanisms of Selective Autophagy. Annu Rev Cell Dev Biol. 2021. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Yoshimori T, Ohsumi Y. The Role of Atg Proteins in Autophagosome Formation. Annu Rev Cell Dev Biol. 2011; 27:107–132. [DOI] [PubMed] [Google Scholar]
- 5.Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012; 30:611–646. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto H, Zhang S, Mizushima N. Autophagy genes in biology and disease. Nat Rev Genet. 2023; 24:382–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z, Sankar DS, Vu B, et al. ULK1 phosphorylation of striatin activates protein phosphatase 2A and autophagy. Cell Rep. 2021; 36:109762. [DOI] [PubMed] [Google Scholar]
- 8.Durgan J, Lystad AH, Sloan K, Carlsson SR, Wilson MI, Marcassa E, Ulferts R, Webster J, Lopez-Clavijo AF, Wakelam MJ, et al. Non-canonical autophagy drives alternative ATG8 conjugation to phosphatidylserine. Mol Cell. 2021; 81:2031–2040 e2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsui T, Jiang P, Nakano S, Sakamaki Y, Yamamoto H, Mizushima N. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J Cell Biol. 2018; 217:2633–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012; 151:1256–1269. [DOI] [PubMed] [Google Scholar]
- 11.Berryman S, Brooks E, Burman A, Hawes P, Roberts R, Netherton C, Monaghan P, Whelband M, Cottam E, Elazar Z, et al. Foot-and-mouth disease virus induces autophagosomes during cell entry via a class III phosphatidylinositol 3-kinase-independent pathway. J Virol. 2012; 86:12940–12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joubert PE, Meiffren G, Gregoire IP, Pontini G, Richetta C, Flacher M, Azocar O, Vidalain PO, Vidal M, Lotteau V, et al. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009; 6:354–366. [DOI] [PubMed] [Google Scholar]
- 13.Richetta C, Gregoire IP, Verlhac P, Azocar O, Baguet J, Flacher M, Tangy F, Rabourdin-Combe C, Faure M. Sustained autophagy contributes to measles virus infectivity. PLoS Pathog. 2013; 9:e1003599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang B, Xue Q, Guo J, Wang X, Zhang Y, Guo K, Li W, Chen S, Xue T, Qi X, et al.Autophagy induction by the pathogen receptor NECTIN4 and sustained autophagy contribute to peste des petits ruminants virus infectivity. Autophagy. 2020; 16:842–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siracusano G, Venuti A, Lombardo D, Mastino A, Esclatine A, Sciortino MT. Early activation of MyD88-mediated autophagy sustains HSV-1 replication in human monocytic THP-1 cells. Sci Rep. 2016; 6:31302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leoni V, Gianni T, Salvioli S, Campadelli-Fiume G. Herpes simplex virus glycoproteins gH/gL and gB bind Toll-like receptor 2, and soluble gH/gL is sufficient to activate NF-kappaB. J Virol. 2012; 86:6555–6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai M, Li M, Wang K, Wang S, Lu Q, Yan J, Mossman KL, Lin R, Zheng C. The herpes simplex virus 1-encoded envelope glycoprotein B activates NF-kappaB through the Toll-like receptor 2 and MyD88/TRAF6-dependent signaling pathway. PLoS One. 2013; 8:e54586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McFarlane S, Aitken J, Sutherland JS, Nicholl MJ, Preston VG, Preston CM. Early induction of autophagy in human fibroblasts after infection with human cytomegalovirus or herpes simplex virus 1. J Virol. 2011; 85:4212–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaumorcel M, Lussignol M, Mouna L, Cavignac Y, Fahie K, Cotte-Laffitte J, Geballe A, Brune W, Beau I, Codogno P, et al. The human cytomegalovirus protein TRS1 inhibits autophagy via its interaction with Beclin 1. J Virol. 2012; 86:2571–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sagnier S, Daussy CF, Borel S, Robert-Hebmann V, Faure M, Blanchet FP, Beaumelle B, Biard-Piechaczyk M, Espert L. Autophagy restricts HIV-1 infection by selectively degrading Tat in CD4+ T lymphocytes. J Virol. 2015; 89:615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, Codogno P, Biard-Piechaczyk M. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J Clin Invest. 2006; 116:2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denizot M, Varbanov M, Espert L, Robert-Hebmann V, Sagnier S, Garcia E, Curriu M, Mamoun R, Blanco J, Biard-Piechaczyk M. HIV-1 gp41 fusogenic function triggers autophagy in uninfected cells. Autophagy. 2008; 4:998–1008. [DOI] [PubMed] [Google Scholar]
- 23.Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, Shelly SS, Gold B, Cherry S. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. 2012; 36:658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moy RH, Gold B, Molleston JM, Schad V, Yanger K, Salzano MV, Yagi Y, Fitzgerald KA, Stanger BZ, Soldan SS, et al. Antiviral autophagy restricts Rift Valley fever virus infection and is conserved from flies to mammals. Immunity. 2014; 40:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong X, Yang Y, Zou Z, Zhao Y, Ci B, Zhong L, Bhave M, Wang L, Kuo YC, Zang X, et al. Sorting nexin 5 mediates virus-induced autophagy and immunity. Nature. 2021; 589:456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie N, Yuan K, Zhou L, Wang K, Chen HN, Lei Y, Lan J, Pu Q, Gao W, Zhang L, et al. PRKAA/AMPK restricts HBV replication through promotion of autophagic degradation. Autophagy. 2016; 12:1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008; 283:33175–33182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell GR, Spector SA. Toll-like receptor 8 ligands activate a vitamin D mediated autophagic response that inhibits human immunodeficiency virus type 1. PLoS Pathog 2012; 8:e1003017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell GR, Spector SA. Vitamin D inhibits human immunodeficiency virus type 1 and Mycobacterium tuberculosis infection in macrophages through the induction of autophagy. PLoS Pathog. 2012; 8:e1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010; 11:55–62. [DOI] [PubMed] [Google Scholar]
- 31.Lupfer C, Thomas PG, Anand PK, Vogel P, Milasta S, Martinez J, Huang G, Green M, Kundu M, Chi H, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol. 2013; 14:480–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talloczy Z, Jiang W, HWt Virgin, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S A. 2002; 99:190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lussignol M, Queval C, Bernet-Camard MF, Cotte-Laffitte J, Beau I, Codogno P, Esclatine A. The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J Virol. 2013; 87:859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talloczy Z, HWt Virgin, Levine B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006; 2:24–29. [DOI] [PubMed] [Google Scholar]
- 35.Yordy B, Iijima N, Huttner A, Leib D, Iwasaki A. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe. 2012; 12:334–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, Shi M, Leslie BJ, Hopfner KP, Ha T, et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe. 2014; 15:228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasmussen SB, Horan KA, Holm CK, Stranks AJ, Mettenleiter TC, Simon AK, Jensen SB, Rixon FJ, He B, Paludan SR. Activation of autophagy by alpha-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes. J Immunol. 2011; 187:5268–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orvedahl A, MacPherson S, Sumpter R, Jr. Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010; 7:115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orvedahl A, Sumpter R, Jr. Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011; 480:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sumpter R, Jr. Sirasanagandla S, Fernandez AF, Wei Y, Dong X, Franco L, Zou Z, Marchal C, Lee MY, Clapp DW, et al. Fanconi Anemia Proteins Function in Mitophagy and Immunity. Cell. 2016; 165:867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, Silvestri G, Munch J, Kirchhoff F, Simonsen A, et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev Cell. 2014; 30:394–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribeiro CM, Sarrami-Forooshani R, Setiawan LC, Zijlstra-Willems EM, van Hamme JL, Tigchelaar W, van der Wel NN, Kootstra NA, Gringhuis SI, Geijtenbeek TB. Receptor usage dictates HIV-1 restriction by human TRIM5alpha in dendritic cell subsets. Nature. 2016; 540:448–452. [DOI] [PubMed] [Google Scholar]
- 43.Sparrer KMJ, Gableske S, Zurenski MA, Parker ZM, Full F, Baumgart GJ, Kato J, Pacheco-Rodriguez G, Liang C, Pornillos O, et al. TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat Microbiol. 2017; 2:1543–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hafren A, Macia JL, Love AJ, Milner JJ, Drucker M, Hofius D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc Natl Acad Sci U S A. 2017; 114:E2026–E2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montespan C, Marvin SA, Austin S, Burrage AM, Roger B, Rayne F, Faure M, Campell EM, Schneider C, Reimer R, et al. Multi-layered control of Galectin-8 mediated autophagy during adenovirus cell entry through a conserved PPxY motif in the viral capsid. PLoS Pathog. 2017; 13:e1006217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luisoni S, Bauer M, Prasad V, Boucke K, Papadopoulos C, Meyer H, Hemmi S, Suomalainen M, Greber UF. Endosomophagy clears disrupted early endosomes but not virus particles during virus entry into cells. Matters. 2016; 6:13–22. [Google Scholar]
- 47.Pied N, Daussy CF, Denis Z, Ragues J, Faure M, Iggo R, Tschan MP, Roger B, Rayne F, Wodrich H. TBK1 is part of a galectin 8 dependent membrane damage recognition complex and drives autophagy upon Adenovirus endosomal escape. PLoS Pathog. 2022; 18:e1010736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt KW, Montespan C, Thompson D, Lucas MS, Ligeon LA, Wodrich H, Hahn AS, Greber UF, Münz C. Selective autophagy impedes KSHV entry after recruiting the membrane damage sensor galectin-8 to virus-containing endosomes. Cell Rep. 2024; 43:115019. [DOI] [PubMed] [Google Scholar]
- 49.Cui S, Xia T, Zhao J, Ren X, Wu T, Kameni M, Guo X, He L, Guo J, Duperray-Susini A, et al. NDP52 mediates an antiviral response to hepatitis B virus infection through Rab9-dependent lysosomal degradation pathway. Nat Commun. 2023; 14:8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staring J, von Castelmur E, Blomen VA, van den Hengel LG, Brockmann M, Baggen J, Thibaut HJ, Nieuwenhuis J, Janssen H, van Kuppeveld FJ, et al. PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature. 2017; 541:412–416. [DOI] [PubMed] [Google Scholar]
- 51.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007; 450:1253–1257. [DOI] [PubMed] [Google Scholar]
- 52.Heckmann BL, Teubner BJW, Tummers B, Boada-Romero E, Harris L, Yang M, Guy CS, Zakharenko SS, Green DR. LC3-Associated Endocytosis Facilitates beta-Amyloid Clearance and Mitigates Neurodegeneration in Murine Alzheimer’s Disease. Cell. 2019; 178:536–551 e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hooper KM, Jacquin E, Li T, Goodwin JM, Brumell JH, Durgan J, Florey O. V-ATPase is a universal regulator of LC3-associated phagocytosis and non-canonical autophagy. J Cell Biol. 2022; 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Durgan J, Florey O. Many roads lead to CASM: Diverse stimuli of noncanonical autophagy share a unifying molecular mechanism. Sci Adv. 2022; 8:eabo1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, Sharma P, Jefferson M, Zhang W, Bone B, Kipar A, Bitto D, Coombes JL, Pearson T, Man A, et al. Non-canonical autophagy functions of ATG16L1 in epithelial cells limit lethal infection by influenza A virus. EMBO J. 2021; 40:e105543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, Shoji-Kawata S, Sumpter RM, Jr. Wei Y, Ginet V, Zhang L, Posner B, Tran KA, Green DR, Xavier RJ, et al. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci U S A. 2013; 110:20364–20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mamola JA, Chen CY, Currier MA, Cassady K, Lee DA, Cripe TP. Opportunities and challenges of combining adoptive cellular therapy with oncolytic virotherapy. Mol Ther Oncolytics. 2023; 29:118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park AK, Fong Y, Kim SI, Yang J, Murad JP, Lu J, Jeang B, Chang WC, Chen NG, Thomas SH, et al. Effective combination immunotherapy using oncolytic viruses to deliver CAR targets to solid tumors. Sci Transl Med. 2020; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng N, Fang J, Xue G, Wang Z, Li X, Zhou M, Jin G, Rahman MM, McFadden G, Lu Y. Induction of tumor cell autosis by myxoma virus-infected CAR-T and TCR-T cells to overcome primary and acquired resistance. Cancer Cell 2022; 40:973–985 e977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang G, Luk BT, Hamidy M, Zhang L, Spector SA. Induction of a Na(+)/K(+)-ATPase-dependent form of autophagy triggers preferential cell death of human immunodeficiency virus type-1-infected macrophages. Autophagy. 2018; 14:1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang G, Luk BT, Wei X, Campbell GR, Fang RH, Zhang L, Spector SA. Selective cell death of latently HIV-infected CD4(+) T cells mediated by autosis inducing nanopeptides. Cell Death Dis. 2019; 10:419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013; 494:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature. 2019; 566:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun Q, Matta H, Chaudhary PM. Kaposi’s sarcoma associated herpes virus-encoded viral FLICE inhibitory protein activates transcription from HIV-1 Long Terminal Repeat via the classical NF-kappaB pathway and functionally cooperates with Tat. Retrovirology. 2005; 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Isaacs A, Lindenmann J, Valentine RC. Virus interference. II. Some properties of interferon. Proc R Soc Lond B Biol Sci. 1957; 147:268–273. [PubMed] [Google Scholar]
- 66.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957; 147:258–267. [PubMed] [Google Scholar]
- 67.Vilcek J. Fifty years of interferon research: aiming at a moving target. Immunity. 2006; 25:343–348. [DOI] [PubMed] [Google Scholar]
- 68.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005; 120:159–162. [DOI] [PubMed] [Google Scholar]
- 69.Paradkar PN, Trinidad L, Voysey R, Duchemin JB, Walker PJ. Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc Natl Acad Sci U S A. 2012; 109:18915–18920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qiao X, Wang L, Song L. The primitive interferon-like system and its antiviral function in molluscs. Dev Comp Immunol. 2021; 118:103997. [DOI] [PubMed] [Google Scholar]
- 71.Levine B, Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019; 176:11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmeisser H, Bekisz J, Zoon KC. New function of type I IFN: induction of autophagy. J Interferon Cytokine Res. 2014; 34:71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ambjorn M, Ejlerskov P, Liu Y, Lees M, Jaattela M, Issazadeh-Navikas S. IFNB1/interferon-beta-induced autophagy in MCF-7 breast cancer cells counteracts its proapoptotic function. Autophagy. 2013; 9:287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmeisser H, Fey SB, Horowitz J, Fischer ER, Balinsky CA, Miyake K, Bekisz J, Snow AL, Zoon KC. Type I interferons induce autophagy in certain human cancer cell lines. Autophagy. 2013; 9:683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang X, Zhou X, Zhang F, Wang X, Duan X, Liu K. DDX58 variant triggers IFN-beta-induced autophagy in trabecular meshwork and influences intraocular pressure. FASEB J. 2024; 38:e23651. [DOI] [PubMed] [Google Scholar]
- 76.Chakrabarti A, Ghosh PK, Banerjee S, Gaughan C, Silverman RH. RNase L triggers autophagy in response to viral infections. J Virol. 2012; 86:11311–11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang S, Xu Z, Liu Y, Yu M, Zhang T, Liu P, Qi X, Chen Y, Meng L, Guo R, et al. OASL suppresses infectious bursal disease virus replication by targeting VP2 for degrading through the autophagy pathway. J Virol. 2024; 98:e0018124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kong N, Shan T, Wang H, Jiao Y, Zuo Y, Li L, Tong W, Yu L, Jiang Y, Zhou Y, et al. BST2 suppresses porcine epidemic diarrhea virus replication by targeting and degrading virus nucleocapsid protein with selective autophagy. Autophagy. 2020; 16:1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim N, Kim MJ, Sung PS, Bae YC, Shin EC, Yoo JY. Interferon-inducible protein SCOTIN interferes with HCV replication through the autolysosomal degradation of NS5A. Nat Commun. 2016; 7:10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy Is a Defense Mechanism Inhibiting BCG and Mycobacterium tuberculosis Survival in Infected Macrophages. Cell. 2004; 119:753–766. [DOI] [PubMed] [Google Scholar]
- 81.Delgado MA, Deretic V: Toll-like receptors in control of immunological autophagy. Cell Death Differ. 2009; 16:976–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006; 313:1438–1441. [DOI] [PubMed] [Google Scholar]
- 83.Matsuzawa T, Kim BH, Shenoy AR, Kamitani S, Miyake M, Macmicking JD. IFN-gamma elicits macrophage autophagy via the p38 MAPK signaling pathway. J Immunol. 2012; 189:813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chang YP, Tsai CC, Huang WC, Wang CY, Chen CL, Lin YS, Kai JI, Hsieh CY, Cheng YL, Choi PC, et al. Autophagy facilitates IFN-gamma-induced Jak2-STAT1 activation and cellular inflammation. J Biol Chem. 2010; 285:28715–28722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li P, Du Q, Cao Z, Guo Z, Evankovich J, Yan W, Chang Y, Shao L, Stolz DB, Tsung A, et al. Interferon-gamma induces autophagy with growth inhibition and cell death in human hepatocellular carcinoma (HCC) cells through interferon-regulatory factor-1 (IRF-1). Cancer Lett. 2012; 314:213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol. 2002; 157:455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M, Deretic V. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007; 27:505–517. [DOI] [PubMed] [Google Scholar]
- 88.Su X, Yu Y, Zhong Y, Giannopoulou EG, Hu X, Liu H, Cross JR, Ratsch G, Rice CM, Ivashkiv LB. Interferon-gamma regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat Immunol. 2015; 16:838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang MQ, Li JR, Yang L, Peng ZG, Wu S, Zhang JP. ATG10S promotes IFNL1 expression and autophagic degradation of multiple viral proteins mediated by IFNL1. Autophagy. 2024:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Y, Zhu H, Zeng X, Fan J, Qian X, Wang S, Wang Z, Sun Y, Wang X, Wang W, et al. Suppression of autophagy enhanced growth inhibition and apoptosis of interferon-beta in human glioma cells. Mol Neurobiol. 2013; 47:1000–1010. [DOI] [PubMed] [Google Scholar]
- 91.Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib D, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host & Microbe. 2007; 1:23–35. [DOI] [PubMed] [Google Scholar]
- 92.Jin S, Tian S, Luo M, Xie W, Liu T, Duan T, Wu Y, Cui J. Tetherin Suppresses Type I Interferon Signaling by Targeting MAVS for NDP52-Mediated Selective Autophagic Degradation in Human Cells. Mol Cell. 2017; 68:308–322 e304. [DOI] [PubMed] [Google Scholar]
- 93.Chou WC, Jha S, Linhoff MW, Ting JP. The NLR gene family: from discovery to present day. Nat Rev Immunol. 2023; 23:635–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maloney NS, Thackray LB, Goel G, Hwang S, Duan E, Vachharajani P, Xavier R, Virgin HW. Essential cell-autonomous role for interferon (IFN) regulatory factor 1 in IFN-gamma-mediated inhibition of norovirus replication in macrophages. J Virol. 2012; 86:12655–12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, Zhang L, Shrestha B, Diamond MS, Dani A, Sosnovtsev SV, et al. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012; 11:397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Biering SB, Choi J, Halstrom RA, Brown HM, Beatty WL, Lee S, McCune BT, Dominici E, Williams LE, Orchard RC, et al. Viral Replication Complexes Are Targeted by LC3-Guided Interferon-Inducible GTPases. Cell Host Microbe. 2017; 22:74–85 e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McAllaster MR, Bhushan J, Balce DR, Orvedahl A, Park A, Hwang S, Sullender ME, Sibley LD, Virgin HW. Autophagy gene-dependent intracellular immunity triggered by interferon-gamma. mBio. 2023; 14:e0233223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kurhade C, Kang S, Biering SB, Hwang S, Randall G. CAPRIN1 Is Required for Control of Viral Replication Complexes by Interferon Gamma. mBio. 2023; 14:e0017223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hait AS, Olagnier D, Sancho-Shimizu V, Skipper KA, Helleberg M, Larsen SM, Bodda C, Moldovan LI, Ren F, Brinck Andersen NS, et al. Defects in LC3B2 and ATG4A underlie HSV2 meningitis and reveal a critical role for autophagy in antiviral defense in humans. Sci Immunol. 2020; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heinz JL, Hinke DM, Maimaitili M, Wang J, Sabli IKD, Thomsen M, Farahani E, Ren F, Hu L, Zillinger T, et al. Varicella zoster virus-induced autophagy in human neuronal and hematopoietic cells exerts antiviral activity. J Med Virol. 2024; 96:e29690. [DOI] [PubMed] [Google Scholar]
- 101.Ronit A, Jorgensen SE, Roed C, Eriksson R, Iepsen UW, Plovsing RR, Storgaard M, Gustafsson F, Hansen AE, Mogensen TH. Host Genetics and Antiviral Immune Responses in Adult Patients With Multisystem Inflammatory Syndrome. Front Immunol. 2021; 12:718744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thomsen MM, Tyrberg T, Skaalum K, Carter-Timofte M, Freytag MR, Norberg P, Helleberg M, Storgaard M, Nielsen H, Bodilsen J, et al. Genetic Variants and Immune Responses in a Cohort of Patients With Varicella Zoster Virus Encephalitis. J Infect Dis. 2021; 224:2122–2132. [DOI] [PubMed] [Google Scholar]
- 103.Rubio RM, Mohr I. Inhibition of ULK1 and Beclin1 by an alpha-herpesvirus Akt-like Ser/Thr kinase limits autophagy to stimulate virus replication. Proc Natl Acad Sci U S A. 2019; 116:26941–26950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mouna L, Hernandez E, Bonte D, Brost R, Amazit L, Delgui LR, Brune W, Geballe AP, Beau I, Esclatine A. Analysis of the role of autophagy inhibition by two complementary human cytomegalovirus BECN1/Beclin 1-binding proteins. Autophagy. 2015:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005; 122:927–939. [DOI] [PubMed] [Google Scholar]
- 106.X E, Hwang S, Oh S, Lee JS, Jeong JH, Gwack Y, Kowalik TF, Sun R, Jung JU, Liang C. Viral Bcl-2-mediated evasion of autophagy aids chronic infection of gammaherpesvirus 68. PLoS Pathog. 2009; 5:e1000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009; 186:255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Castro-Gonzalez S, Shi Y, Colomer-Lluch M, Song Y, Mowery K, Almodovar S, Bansal A, Kirchhoff F, Sparrer K, Liang C, et al. HIV-1 Nef counteracts autophagy restriction by enhancing the association between BECN1 and its inhibitor BCL2 in a PRKN-dependent manner. Autophagy. 2021; 17:553–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Petraccione K, Ali MGH, Cyr N, Wahba HM, Stocker T, Akhrymuk M, Akhrymuk I, Panny L, Bracci N, Cafaro R, et al. An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy. PLoS Pathog. 2024; 20:e1012093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gassen NC, Papies J, Bajaj T, Emanuel J, Dethloff F, Chua RL, Trimpert J, Heinemann N, Niemeyer C, Weege F, et al. SARS-CoV-2-mediated dysregulation of metabolism and autophagy uncovers host-targeting antivirals. Nat Commun. 2021; 12:3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang Y, Sun H, Pei R, Mao B, Zhao Z, Li H, Lin Y, Lu K. The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov. 2021; 7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hou P, Wang X, Wang H, Wang T, Yu Z, Xu C, Zhao Y, Wang W, Zhao Y, Chu F, et al. The ORF7a protein of SARS-CoV-2 initiates autophagy and limits autophagosome-lysosome fusion via degradation of SNAP29 to promote virus replication. Autophagy. 2023; 19:551–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gannage M, Dormann D, Albrecht R, Dengjel J, Torossi T, Ramer PC, Lee M, Strowig T, Arrey F, Conenello G, et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009; 6:367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Beale R, Wise H, Stuart A, Ravenhill BJ, Digard P, Randow F. A LIR motif in influenza A virus M2 is required for virion stability. Cell Host & Microbe. 2014; 5:239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wodrich H, Henaff D, Jammart B, Segura-Morales C, Seelmeir S, Coux O, Ruzsics Z, Wiethoff CM, Kremer EJ. A capsid-encoded PPxY-motif facilitates adenovirus entry. PLoS Pathog. 2010; 6:e1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wen W, Li X, Yin M, Wang H, Qin L, Li H, Liu W, Zhao Z, Zhao Q, Chen H, et al. Selective autophagy receptor SQSTM1/ p62 inhibits Seneca Valley virus replication by targeting viral VP1 and VP3. Autophagy. 2021; 17:3763–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Song J, Guo Y, Wang D, Quan R, Wang J, Liu J. Seneca Valley virus 3C protease cleaves OPTN (optineurin) to Impair selective autophagy and type I interferon signaling. Autophagy. 2024; 20:614–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mohamud Y, Qu J, Xue YC, Liu H, Deng H, Luo H. CALCOCO2/NDP52 and SQSTM1/p62 differentially regulate coxsackievirus B3 propagation. Cell Death Differ. 2019; 26:1062–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shi J, Fung G, Piesik P, Zhang J, Luo H. Dominant-negative function of the C-terminal fragments of NBR1 and SQSTM1 generated during enteroviral infection. Cell Death Differ. 2014; 21:1432–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reggiori F, Molinari M. ER-phagy: mechanisms, regulation, and diseases connected to the lysosomal clearance of the endoplasmic reticulum. Physiol Rev. 2022; 102:1393–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tan X, Cai K, Li J, Yuan Z, Chen R, Xiao H, Xu C, Hu B, Qin Y, Ding B. Coronavirus subverts ER-phagy by hijacking FAM134B and ATL3 into p62 condensates to facilitate viral replication. Cell Rep. 2023; 42:112286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lennemann NJ, Coyne CB. Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy. 2017; 13:322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kang Y, Lin W, Nagy PD. Subversion of selective autophagy for the biogenesis of tombusvirus replication organelles inhibits autophagy. PLoS Pathog. 2024; 20:e1012085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Delpeut S, Rudd PA, Labonte P, von Messling V. Membrane fusion-mediated autophagy induction enhances morbillivirus cell-to-cell spread. J Virol. 2012; 86:8527–8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Doring T, Zeyen L, Bartusch C, Prange R. Hepatitis B Virus Subverts the Autophagy Elongation Complex Atg5-12/16L1 and Does Not Require Atg8/LC3 Lipidation for Viral Maturation. J Virol. 2018; 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dreux M, Chisari FV. Autophagy proteins promote hepatitis C virus replication. Autophagy. 2009; 5:1224–1225. [DOI] [PubMed] [Google Scholar]
- 127.Jordan TX, Randall G. Dengue Virus Activates the AMP Kinase-mTOR Axis To Stimulate a Proviral Lipophagy. J Virol. 2017; 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Khabir M, Aliche AZ, Sureau C, Blanchet M, Labonte P. Hepatitis Delta Virus Alters the Autophagy Process To Promote Its Genome Replication. J Virol. 2020; 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Doerflinger SY, Cortese M, Romero-Brey I, Menne Z, Tubiana T, Schenk C, White PA, Bartenschlager R, Bressanelli S, Hansman GS, et al. Membrane alterations induced by nonstructural proteins of human norovirus. PLoS Pathog. 2017; 13:e1006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ferraris P, Blanchard E, Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol. 2010; 91:2230–2237. [DOI] [PubMed] [Google Scholar]
- 131.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008; 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pedersen KW, van der Meer Y, Roos N, Snijder EJ. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol. 1999; 73:2016–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schlegel A, Giddings TH, Jr. Ladinsky MS, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996; 70:6576–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dales S, Eggers HJ, Tamm I, Palade GE. Electron Microscopic Study of the Formation of Poliovirus. Virology. 1965; 26:379–389. [DOI] [PubMed] [Google Scholar]
- 135.Jackson WT, Giddings TH, Jr. Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005; 3:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wileman T. Aggresomes and autophagy generate sites for virus replication. Science. 2006; 312:875–878. [DOI] [PubMed] [Google Scholar]
- 137.V’Kovski P, Gerber M, Kelly J, Pfaender S, Ebert N, Braga Lagache S, Simillion C, Portmann J, Stalder H, Gaschen V, et al. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. Elife. 2019; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cortese M, Lee JY, Cerikan B, Neufeldt CJ, Oorschot VMJ, Kohrer S, Hennies J, Schieber NL, Ronchi P, Mizzon G, et al. Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies. Cell Host Microbe. 2020; 28:853–866 e855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Romero-Brey I, Berger C, Kallis S, Kolovou A, Paul D, Lohmann V, Bartenschlager R. NS5A Domain 1 and Polyprotein Cleavage Kinetics Are Critical for Induction of Double-Membrane Vesicles Associated with Hepatitis C Virus Replication. mBio. 2015; 6:e00759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Alirezaei M, Flynn CT, Whitton JL. Interactions between enteroviruses and autophagy in vivo. Autophagy. 2012; 8:973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bub T, Hargest V, Tan S, Smith M, Vazquez-Pagan A, Flerlage T, Brigleb P, Meliopoulos V, Lindenbach B, Ramanathan HN, et al. Astrovirus replication is dependent on induction of double-membrane vesicles through a PI3K-dependent, LC3-independent pathway. J Virol. 2023; 97:e0102523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fahmy AM, Labonte P. The autophagy elongation complex (ATG5-12/16L1) positively regulates HCV replication and is required for wild-type membranous web formation. Sci Rep. 2017; 7:40351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Twu WI, Lee JY, Kim H, Prasad V, Cerikan B, Haselmann U, Tabata K, Bartenschlager R. Contribution of autophagy machinery factors to HCV and SARS-CoV-2 replication organelle formation. Cell Rep. 2021; 37:110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Reggiori F, Monastyrska I, Verheije MH, Cali T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010; 7:500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wolff G, Limpens R, Zevenhoven-Dobbe JC, Laugks U, Zheng S, de Jong AWM, Koning RI, Agard DA, Grunewald K, Koster AJ, et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020; 369:1395–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009; 106:14046–14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guevin C, Manna D, Belanger C, Konan KV, Mak P, Labonte P. Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology. 2010; 405:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR. Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem. 2004; 279:10136–10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cottam EM, Maier HJ, Manifava M, Vaux LC, Chandra-Schoenfelder P, Gerner W, Britton P, Ktistakis NT, Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011; 7:1335–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ji M, Li M, Sun L, Zhao H, Li Y, Zhou L, Yang Z, Zhao X, Qu W, Xue H, et al. VMP1 and TMEM41B are essential for DMV formation during beta-coronavirus infection. J Cell Biol. 2022; 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sir D, Kuo CF, Tian Y, Liu HM, Huang EJ, Jung JU, Machida K, Ou JH. Replication of hepatitis C virus RNA on autophagosomal membranes. J Biol Chem. 2012; 287:18036–18043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Oudshoorn D, Rijs K, Limpens R, Groen K, Koster AJ, Snijder EJ, Kikkert M, Barcena M. Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication. mBio. 2017; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002; 76:5974–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zimmermann L, Zhao X, Makroczyova J, Wachsmuth-Melm M, Prasad V, Hensel Z, Bartenschlager R, Chlanda P. SARS-CoV-2 nsp3 and nsp4 are minimal constituents of a pore spanning replication organelle. Nat Commun. 2023; 14:7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Huang Y, Wang T, Zhong L, Zhang W, Zhang Y, Yu X, Yuan S, Ni T. Molecular architecture of coronavirus double-membrane vesicle pore complex. Nature. 2024; 633:224–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ricciardi S, Guarino AM, Giaquinto L, Polishchuk EV, Santoro M, Di Tullio G, Wilson C, Panariello F, Soares VC, Dias SSG, et al. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature. 2022; 606:761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cottam EM, Whelband MC, Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014; 10:1426–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, GuoJZ, Swaney DL, et al: A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020; 583:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kumar S, Javed R, Mudd M, Pallikkuth S, Lidke KA, Jain A, Tangavelou K, Gudmundsson SR, Ye C, Rusten TE, et al. Mammalian hybrid pre-autophagosomal structure HyPAS generates autophagosomes. Cell. 2021; 184:5950–5969 e5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Belov GA, Nair V, Hansen BT, Hoyt FH, Fischer ER, Ehrenfeld E. Complex dynamic development of poliovirus membranous replication complexes. J Virol. 2012; 86:302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Limpens RW, van der Schaar HM, Kumar D, Koster AJ, Snijder EJ, van Kuppeveld FJ, Barcena M. The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments. mBio. 2011; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, Li F, Wang Y, Tiollais P, Li T, et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009; 49:60–71. [DOI] [PubMed] [Google Scholar]
- 163.Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci U S A. 2010; 107:4383–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chu JYK, Chuang YC, Tsai KN, Pantuso J, Ishida Y, Saito T, Ou JJ. Autophagic membranes participate in hepatitis B virus nucleocapsid assembly, precore and core protein trafficking, and viral release. Proc Natl Acad Sci U S A. 2022; 119:e2201927119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Das S, St Croix C, Good M, Chen J, Zhao J, Hu S, Ross M, Myerburg MM, Pilewski JM, Williams J, et al. Interleukin-22 Inhibits Respiratory Syncytial Virus Production by Blocking Virus-Mediated Subversion of Cellular Autophagy. iScience. 2020; 23:101256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu J, Wang H, Gu J, Deng T, Yuan Z, Hu B, Xu Y, Yan Y, Zan J, Liao M, et al. BECN1-dependent CASP2 incomplete autophagy induction by binding to rabies virus phosphoprotein. Autophagy. 2017; 13:739–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yuan Y, Fang A, Wang Z, Chen H, Fu ZF, Zhou M, Zhao L. The matrix protein of lyssavirus hijacks autophagosome for efficient egress by recruiting NEDD4 through its PPxY motif. Autophagy. 2024; 20:1723–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Baillet N, Krieger S, Carnec X, Mateo M, Journeaux A, Merabet O, Caro V, Tangy F, Vidalain PO, Baize S. E3 Ligase ITCH Interacts with the Z Matrix Protein of Lassa and Mopeia Viruses and Is Required for the Release of Infectious Particles. Viruses. 2019; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yuan Y, Fang A, Zhang M, Zhou M, Fu ZF, Zhao L. Lassa virus Z protein hijacks the autophagy machinery for efficient transportation by interrupting CCT2-mediated cytoskeleton network formation. Autophagy. 2024:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Figueras-Novoa C, Akutsu M, Murata D, Jiang M, Montaner B, Dubois C, Shenoy A, Beale R. Caspase cleavage of Influenza A virus M2 disrupts M2-LC3 interaction and regulates virion production. BioRXiv. 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fletcher K, Ulferts R, Jacquin E, Veith T, Gammoh N, Arasteh JM, Mayer U, Carding SR, Wileman T, Beale R, et al. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ulferts R, Marcassa E, Timimi L, Lee LC, Daley A, Montaner B, Turner SD, Florey O, Baillie JK, Beale R. Subtractive CRISPR screen identifies the ATG16L1/vacuolar ATPase axis as required for non-canonical LC3 lipidation. Cell Rep. 2021; 37:109899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ren Y, Li C, Feng L, Pan W, Li L, Wang Q, Li J, Li N, Han L, Zheng X, et al. Proton Channel Activity of Influenza A Virus Matrix Protein 2 Contributes to Autophagy Arrest. J Virol. 2015; 90:591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Timimi L, Wrobel AG, Chiduza GN, Maslen SL, Torres-Mendez A, Montaner B, Davis C, Minckley T, Hole KL, Serio A, et al. The V-ATPase/ATG16L1 axis is controlled by the V(1)H subunit. Mol Cell. 2024; 84:2966–2983 e2969. [DOI] [PubMed] [Google Scholar]
- 175.Liu B, Fang M, Hu Y, Huang B, Li N, Chang C, Huang R, Xu X, Yang Z, Chen Z, et al. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy. 2014; 10:416–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ding B, Zhang G, Yang X, Zhang S, Chen L, Yan Q, Xu M, Banerjee AK, Chen M. Phosphoprotein of human parainfluenza virus type 3 blocks autophagosome-lysosome fusion to increase virus production. Cell Host Microbe. 2014; 15:564–577. [DOI] [PubMed] [Google Scholar]
- 177.Li J, Liu Y, Wang Z, Liu K, Wang Y, Liu J, Ding H, Yuan Z. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J Virol. 2011; 85:6319–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yan JM, Zhang WK, Yan LN, Jiao YJ, Zhou CM, Yu XJ. Bunyavirus SFTSV exploits autophagic flux for viral assembly and egress. Autophagy. 2022; 18:1599–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Taisne C, Lussignol M, Hernandez E, Moris A, Mouna L, Esclatine A. Human cytomegalovirus hijacks the autophagic machinery and LC3 homologs in order to optimize cytoplasmic envelopment of mature infectious particles. Sci Rep. 2019; 9:4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zimmermann C, Kramer N, Krauter S, Strand D, Sehn E, Wolfrum U, Freiwald A, Butter F, Plachter B. Autophagy interferes with human cytomegalovirus genome replication, morphogenesis, and progeny release. Autophagy. 2021; 17:779–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Buckingham EM, Jarosinski KW, Jackson W, Carpenter JE, Grose C. Exocytosis of varicella-zoster virions involves a convergence of endosomal and autophagy pathways. J Virol. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Nowag H, Guhl B, Thriene K, Romao S, Ziegler U, Dengjel J, Münz C. Macroautopphagy proteins assist Epstein Barr virus production and get incorporated into the virus particles. EBioMedicine. 2014; 1:116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Granato M, Santarelli R, Farina A, Gonnella R, Lotti LV, Faggioni A, Cirone M. EBV blocks the autophagic flux and appropriates the autophagic machinery to enhance viral replication. J Virol. 2014; 88:12715–12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Granato M, Santarelli R, Filardi M, Gonnella R, Farina A, Torrisi MR, Faggioni A, Cirone M. The activation of KSHV lytic cycle blocks autophagy in PEL cells. Autophagy. 2015; 11:1978–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Girsch JH, Walters K, Jackson W, Grose C. Progeny Varicella-Zoster Virus Capsids Exit the Nucleus but Never Undergo Secondary Envelopment during Autophagic Flux Inhibition by Bafilomycin A1. J Virol. 2019; 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pena-Francesch M, Vanoaica LD, Zhu GF, Stumpe M, Sankar DS, Nowag H, Valencia-Camargo AD, Hammerschmidt W, Dengjel J, Ligeon LA, et al. The autophagy machinery interacts with EBV capsids during viral envelope release. Proc Natl Acad Sci U S A. 2023; 120:e2211281120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mateo R, Nagamine CM, Spagnolo J, Mendez E, Rahe M, Gale M, Jr. Yuan J, Kirkegaard K. Inhibition of cellular autophagy deranges dengue virion maturation. J Virol. 2013; 87:1312–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Li MY, Naik TS, Siu LYL, Acuto O, Spooner E, Wang P, Yang X, Lin Y, Bruzzone R, Ashour J, et al. Lyn kinase regulates egress of flaviviruses in autophagosome-derived organelles. Nat Commun. 2020; 11:5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wu SY, Chen YL, Lee YR, Lin CF, Lan SH, Lan KY, Chu ML, Lin PW, Yang ZL, Chen YH, et al. The Autophagosomes Containing Dengue Virus Proteins and Full-Length Genomic RNA Are Infectious. Viruses. 2021; 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cloherty APM, Rader AG, Patel KS, Eisden T, van Piggelen S, Schreurs R, Ribeiro CMS. Dengue virus exploits autophagy vesicles and secretory pathways to promote transmission by human dendritic cells. Front Immunol. 2024; 15:1260439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lan Y, van Leur SW, Fernando JA, Wong HH, Kampmann M, Siu L, Zhang J, Li M, Nicholls JM, Sanyal S. Viral subversion of selective autophagy is critical for biogenesis of virus replication organelles. Nat Commun. 2023; 14:2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shrivastava S, Devhare P, Sujijantarat N, Steele R, Kwon YC, Ray R, Ray RB. Knockdown of Autophagy Inhibits Infectious Hepatitis C Virus Release by the Exosomal Pathway. J Virol. 2016; 90:1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kim JY, Ou JJ. Regulation of Apolipoprotein E Trafficking by Hepatitis C Virus-Induced Autophagy. J Virol. 2018; 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang L, Tian Y, Ou JH. HCV induces the expression of Rubicon and UVRAG to temporally regulate the maturation of autophagosomes and viral replication. PLoS Pathog. 2015; 11:e1004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Shiode Y, Hikita H, Tanaka S, Shirai K, Doi A, Sakane S, Kai Y, Nakabori T, Yamada R, Kodama T, et al. Hepatitis C virus enhances Rubicon expression, leading to autophagy inhibition and intracellular innate immune activation. Sci Rep. 2020; 10:15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bird SW, Maynard ND, Covert MW, Kirkegaard K. Nonlytic viral spread enhanced by autophagy components. Proc Natl Acad Sci U S A. 2014; 111:13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, Brantner CA, Stempinski ES, Connelly PS, Ma HC, et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015; 160:619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Dahmane S, Kerviel A, Morado DR, Shankar K, Ahlman B, Lazarou M, Altan-Bonnet N, Carlson LA. Membrane-assisted assembly and selective secretory autophagy of enteroviruses. Nat Commun. 2022; 13:5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Santiana M, Ghosh S, Ho BA, Rajasekaran V, Du WL, Mutsafi Y, De Jesus-Diaz DA, Sosnovtsev SV, Levenson EA, Parra GI, et al. Vesicle-Cloaked Virus Clusters Are Optimal Units for Inter-organismal Viral Transmission. Cell Host Microbe. 2018; 24:208–220 e208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966; 28:435–492. [DOI] [PubMed] [Google Scholar]
- 201.Girsch JH, Jackson W, Carpenter JE, Moninger TO, Jarosinski KW, Grose C. Exocytosis of Progeny Infectious Varicella-Zoster Virus Particles via a Mannose-6-Phosphate Receptor Pathway without Xenophagy following Secondary Envelopment. J Virol. 2020; 94. [DOI] [PMC free article] [PubMed] [Google Scholar]


