Summary
This study evaluates the efficacy of the postbiotic Probio-Eco in alleviating constipation in humans and mice. A randomized, double-blind, placebo-controlled crossover trial involving 110 adults with chronic constipation (Rome IV criteria) demonstrates that a 3-week Probio-Eco intervention significantly improves constipation symptoms, stool straining, and worry scores. Gut microbiota and metabolomic analyses reveal modulations in specific gut microbiota, succinate, tryptophan derivatives, deoxycholate, propionate, butyrate, and cortisol, correlating with symptom relief. A loperamide-induced mouse model confirms that Probio-Eco and its bioactive components (succinate, 3-indoleacrylic acid, and 5-hydroxytryptophan) alleviate constipation by stimulating mucin-2 secretion, regulating intestinal transport hormones, and promoting anti-inflammatory responses. Multi-omics integration identifies key pathways, including succinate-short-chain fatty acid, tryptophan-5-hydroxytryptophan-serotonin, and tryptophan-3-indoleacrylic acid, driving intestinal homeostasis and motility. These findings highlight the comprehensive efficacy of Probio-Eco and provide robust evidence for its clinical application in constipation management. This study was registered at Chinese Clinical Trial Registry (ChiCTR2100054376).
Keywords: gut microbiota, complete spontaneous bowel movements, succinate, 5-hydroxytryptophan, 3-indoleacrylic acid, loperamide, intestinal homeostasis, gastrointestinal motility
Graphical abstract

Highlights
-
•
Postbiotic Probio-Eco relieves chronic constipation in our human clinical trial
-
•
Modulates gut microbiota and metabolites to enhance intestinal motility and balance
-
•
Components activate mucin-2, gut hormones, and anti-inflammatory pathways synergistically
-
•
Mouse model confirms effects via succinate, 5-HTP, and 3-indoleacrylic acid pathways
Ma et al. demonstrate that the postbiotic Probio-Eco alleviates chronic constipation (Rome IV criteria) in a randomized clinical trial and mouse model. Multi-omics links symptom relief to succinate-SCFA and dual tryptophan pathways (5-HTP-serotonin and 3-indoleacrylic acid), enhancing intestinal motility and homeostasis. Findings support clinical application for constipation management.
Introduction
Chronic constipation, a prevalent gastrointestinal disorder associated with intestinal inflammation, significantly impacts patients’ quality of life. Management typically involves dietary adjustments, bowel habit education, and over-the-counter laxatives.1 While high-fiber diets are recommended, adequate fluid intake is essential to prevent side effects.2 Despite their accessibility, laxatives often cause gastrointestinal discomfort, with nearly half of patients expressing dissatisfaction.3 Multiple studies have demonstrated that the composition and stability of gut microbiota are disrupted in patients with constipation, leading to changes in intestinal metabolic environments, which in turn affect intestinal motility and secretory functions.4,5 Microbial metabolites, such as bile acid (BA), short-chain fatty acids (SCFAs), and tryptophan derivatives, influence gut motility by activating intestinal chemoreceptors.6 Furthermore, gut phageome alterations have been linked to several intestinal diseases, including inflammatory bowel disease and digestive tract tumors.7,8
Recently, probiotic supplementation has emerged as a promising approach for constipation management,9,10 though efficacy varies and the specific biomarkers related to constipation remain unclear. Advances in microbiome research highlight the potential of probiotics to alleviate constipation by modulating gut microbiota and metabolites.11 A large-scale, randomized, double-blind, placebo-controlled trial demonstrated that Lactiplantibacillus plantarum P9 improved the weekly mean frequency of complete spontaneous bowel movements (CSBMs) and alleviated constipation-related symptoms. The probiotic group exhibited increased levels of beneficial bacteria, amino acids, and SCFAs, while reducing Oscillospiraceae, Herelleviridae, p-cresol, and methylamine associated with intestinal barrier function and transit.12 Despite their potential benefits, probiotic activity is limited by host factors, formulation, and storage and transportation conditions.9,13 Moreover, there is growing recognition that non-biological materials, such as inactivated microorganisms and metabolites, can also influence physiological functions. In 2021, the International Scientific Association for Probiotics and Prebiotics defined these substances as “postbiotics,” referring to preparations of inanimate microorganisms and/or their components that confer health benefits to the host.14
Postbiotics offer a safe and stable alternative to probiotics, with advantages including defined chemical structures, resistance to antibiotics, and ease of storage.15,16 Several studies demonstrate their efficacy in constipation management: Lacticaseibacillus paracasei metabolites improved intestinal barrier function and water and sodium metabolism in constipated mice,17 while heat-inactivated Bifidobacterium bifidum MIMBb75 reduced irritable bowel syndrome (IBS)-related abdominal pain.18 A clinical trial further confirmed that heat-killed Bifidobacterium longum CLA8013 enhanced stool volume and improved defecation-related parameters.19 Despite promising results, postbiotic research remains hindered by a lack of high-quality clinical trials,16 highlighting the urgent need for large-scale studies to confirm their efficacy and safety in chronic constipation. Probio-Eco, a postbiotic product, is fermented by Lacticaseibacillus paracasei Zhang, Lactiplantibacillus plantarum P-8, and Bifidobacterium lactis V9 using soy protein, skimmed milk powder, and sodium citrate as substrates.20 These strains not only have well-documented origins but also exhibit synergistic interactions.21,22 Metabolomic analyses show that it contains bioactive compounds like organic acids, bacteriocins, and SCFAs. Animal studies indicated that Probio-Eco significantly reduced inflammation and improved symptoms related to IBS,20 while a population trial in dental caries patients showed that it suppressed pathogenic bacteria and fostered oxidative stress-tolerant oral bacteria.23
This study evaluated the efficacy of Probio-Eco in chronic constipation through a 9-week randomized, double-blind, placebo-controlled crossover clinical trial. The primary outcome was the weekly mean frequency of CSBMs, while secondary endpoints included stool consistency, stool straining, quality of life assessments, depression-anxiety-stress scores, and changes in gut microbiota and metabolomics. To validate clinical observations, a loperamide-induced constipation mouse model assessed Probio-Eco and its individual postbiotic components, namely succinate (Succ), 3-indoleacrylic acid (3-IA), and 5-hydroxytryptophan (5-HTP), on intestinal function, immune response, and gastrointestinal peptide levels. This study integrated pre-clinical, clinical, and multi-omics approaches to demonstrate how Probio-Eco and its metabolites improve gut function and alleviate constipation and provides a solid theoretical framework for the clinical application of postbiotics. Furthermore, it opens avenues for managing chronic constipation and other related digestive disorders.
Results
Characterization and quantification of Probio-Eco metabolites
A comprehensive metabolomic analysis identified various bioactive metabolites in Probio-Eco, including organic acids, SCFAs, and indole derivatives (Table S1). In addition to common organic acid, Probio-Eco contained notable levels of Succ (15.0 μg/g) and 4-hydroxyphenyllactic acid (24.2 μg/g), along with predominant SCFAs including acetic (65.46 μg/g), propionic (8.2 μg/g), and butyric acids (11.3 μg/g). Furthermore, several metabolites were identified associated with gastrointestinal health benefits, including 3-IA, 3-hydroxyflavone, and monobutyrin. The presence of these metabolites underscores the potential of Probio-Eco to promote gastrointestinal homeostasis and overall well-being.
Demographic data, compliance, and adverse effects
The trial enrolled 110 participants in the intention-to-treat (ITT) population, who were randomly assigned to either the postbiotic-placebo group (group A; n = 54) or the placebo-postbiotic group (group B; n = 56). After the 9-week intervention, three in group A and two in group B were excluded due to antibiotic use, withdrawal, or missing diary submissions (Figure S1A). Consequently, the per-protocol (PP) analysis included 105 participants (group A, n = 52; group B, n = 53; Figure 1A). Baseline characteristics of all 110 participants are summarized in Table S2. The mean (SD) age was 21.8 (2.1) years in group A and 21.8 (2.2) years in group B. Gender distribution was comparable between groups, with female accounting for 81.5% in group A and 80.4% in group B. Demographic and clinical characteristics (age, gender, BMI, ethnicity, drug allergies, smoking status, comorbidities, and medication history) were balanced between groups (p > 0.05; Table S2).
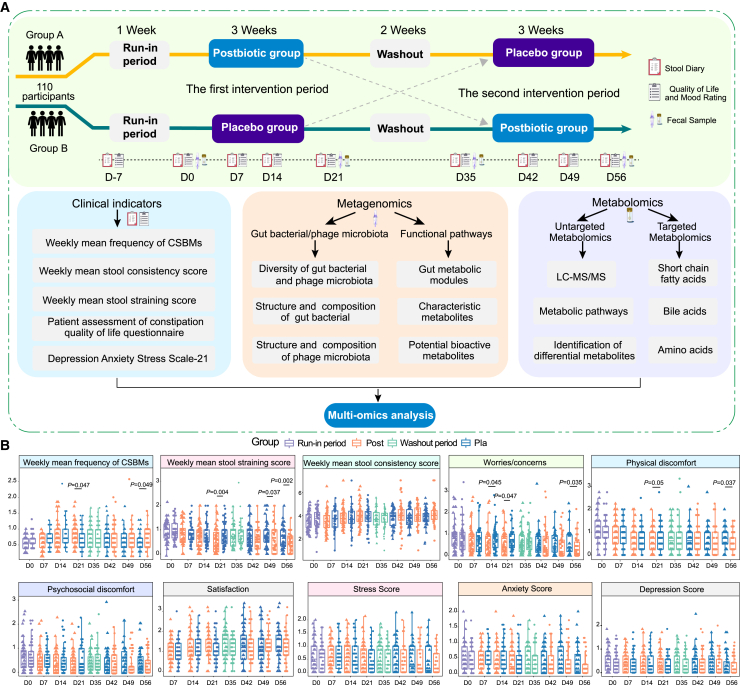
Figure 1.
Multi-omics analysis pipeline and changes in clinical indicators associated with constipation symptoms during the intervention trial
(A) Overview of the trial design, clinical indicators, fecal metagenomics, and metabolomics. The trial comprised two 3-week interventions, separated by a 2-week washout period. Data were collected using a stool diary and self-administered questionnaires at designated time points. Fecal samples were also collected for subsequent analyses.
(B) Boxplots showing the changes in complete spontaneous bowel movements (CSBMs); stool consistency score; stool straining score; patient assessment of constipation-quality of life; and the depression, anxiety, and stress scale-21 for group A (n = 52) and group B (n = 53) in the per-protocol population throughout the intervention trial. Data for group A (left) and group B (right) are presented side by side at each time point. Significant intergroup differences were evaluated by unpaired or two-sided paired Wilcoxon rank-sum test, with significant p values shown in the plots. In the boxplots, boxes represent the interquartile range of each group’s value distribution, lines within the boxes indicate median values, whiskers denote the lowest and highest values within 1.5 times the interquartile range, and dots above or below the whiskers represent outliers.
The postbiotic was well tolerated, with no serious adverse events. Mild gastrointestinal symptoms (bloating, diarrhea, and nausea) were most common; isolated cases of dental ulcer and anorexia also occurred. Fewer adverse events were reported during the postbiotic phase compared to the placebo phase (5 vs. 7 adverse events, respectively), but the difference was not statistically significant between groups (p = 0.811; Table S2). Both groups maintained high overall compliance, with no statistical difference between them (mean [SD] = 105.39 [7.7] in group A and 105.96 [8.9] in group B; p > 0.05; Table S2).
Postbiotic intervention improved constipation symptoms
The design included two 3-week intervention periods separated by a 2-week washout, with participants receiving Probio-Eco or placebo in reciprocal sequences (Figure 1A). The primary outcome was weekly CSBM frequency. In the ITT analysis, the mean (SD) weekly CSBMs at baseline were 0.60 (0.17) in group A and 0.64 (0.18) in group B. After the first 21-day intervention, the CSBMs demonstrated a significant increase in the postbiotic group, reaching 0.81 (0.35) compared to 0.68 (0.30) in the placebo group, resulting in a difference of 0.13 and a noteworthy 19.12% improvement (p = 0.047; Table S3). Similar results were observed in the PP analysis, which also indicated a significant improvement in the CSBMs, with a difference of 0.13 (0.81 [0.35] vs. 0.68 [0.31]; p = 0.047; Figure 1B; Table S4). After the second 21-day intervention period (day 56), both ITT and PP analyses revealed significant differences in mean weekly CSBM between the two groups (p = 0.048 and p = 0.049), with a difference of 0.11 (Tables S3 and S4). To further elucidate the differences, we performed a covariance analysis using baseline measurements as a covariate, revealing significant differences in mean weekly CSBMs between the two groups at days 21 and 56 (p = 0.010; p = 0.044). Additionally, a significant interaction in group and time analyses was observed (interaction p = 0.038), demonstrating sustained improvement in weekly CSBMs with postbiotic treatment compared to placebo group. This provides compelling evidence that the therapeutic effect of the postbiotic in alleviating constipation is genuine rather than coincidental.
In the ITT analysis, the postbiotic group showed significantly greater reductions in stool straining scores than placebo at all time points: 30.49% at day 21 (p = 0.006), 23.08% at day 49 (p = 0.046), and 34.67% at day 56 (p = 0.003, Table S3). The PP analysis corroborated these findings, showing significant reductions of 32.10%, 23.68%, and 36.99%, respectively, in stool straining scores at the same time points (p = 0.004, p = 0.037, and p = 0.002, respectively; Figure 1B; Table S4). Interestingly, the postbiotic also positively impacted quality of life measures. In the PP population, postbiotic supplementation significantly reduced worries (23.19%, p = 0.047) and physical discomfort (25.93%, p = 0.048) compared to placebo at day 21, with sustained improvements at day 56 (worries: p = 0.035; discomfort: p = 0.037). In the ITT analysis, postbiotics significantly reduced worries at both day 21 (22.06%, p = 0.049) and day 56 (27.45%, p = 0.046). For physical discomfort, improvement reached significance only at day 56 (p = 0.037), with a trend at day 21 (p = 0.055; Table S3). The other clinical parameters showed no significant between-group differences in ITT or PP analyses (Figure 1B; Tables S3 and S4). Collectively, these results indicate that the Probio-Eco postbiotic intervention significantly alleviated constipation symptoms, reduced stool straining, and improved patients’ worries and physical discomfort.
Postbiotic intervention modulated gut bacterial and bacteriophage composition
During the 21-day intervention, postbiotic supplementation significantly improved chronic constipation compared to the placebo group. However, by day 35, after a 14-day washout, no significant differences remained between groups; the positive effects were lost after the washout, indicating that sustained postbiotic delivery is essential to maintain therapeutic benefits. We integrated multi-omics data, with days 0 and 35 as baselines and days 21 and 56 as endpoints for the two intervention periods, to elucidate the taxonomic, functional, and metabolic changes driven by postbiotic administration in patients. We performed metagenomic analysis on fecal samples from 103 patients at four time points, with two subjects failing to provide samples (Figure S1A). The alpha diversity showed no significant differences (Figures 2A and S1B), and beta diversity analysis via non-metric multidimensional scaling (NMDS) revealed no significant restructuring of the bacterial community (Figure 2B). Species-level analysis revealed targeted modulations, with the postbiotic group showing significantly higher abundances of several beneficial species compared to the placebo group after intervention, including Lacticaseibacillus paracasei, Lactiplantibacillus plantarum, and Clostridium sp001916075 (p < 0.05; Figure 2C). In summary, postbiotic supplementation selectively enriched beneficial bacterial species without altering overall microbiota diversity, which could be a contributing factor to improved constipation symptoms among participants.
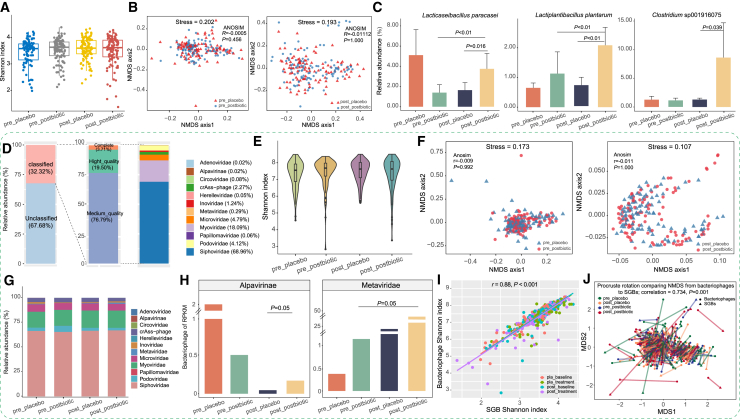
Figure 2.
Alterations in fecal bacteriome and phageome during the postbiotic and placebo interventions
Data presented in this figure were derived from the postbiotic (n = 205) and placebo (n = 203) groups. Sample codes use a prefix to indicate whether samples were collected pre-intervention (pre) or post-intervention (post) and a suffix to denote the type of intervention (postbiotic or placebo).
(A and B) (A) Shannon diversity index and (B) non-metric multidimensional scaling (NMDS) score plots of the gut bacteriome in both groups, before and after intervention.
(C) Differentially abundant species-level genome bins (SGBs) between the postbiotic and placebo groups, before and after intervention (p < 0.05, Wilcoxon rank-sum test). Error bars represent standard deviation.
(D) Family-level taxonomic annotation of the phage metagenome, comprising a total of 38,593 qualified non-redundant viral operational taxonomic units.
(E and F) (E) Shannon diversity index and (F) NMDS analysis of the gut phageome in both groups, before and after intervention.
(G and H) (G) Family-level distribution of phageome and (H) relative abundance of the Alpavirinae and Metaviridae in both groups, before and after intervention.
(I) Correlation between the Shannon diversity index of gut bacterial and phage, revealing a strong positive correlation.
(J) Procrustes analysis performed on fecal SGBs and bacteriophages in the two groups, before and after intervention, confirmed positive co-operativity.
In the boxplots and violin plots, boxes represent the interquartile range of each group’s value distribution, lines within boxes indicate median values, whiskers denote the lowest and highest values within 1.5 times the interquartile range, and dots above or below the whiskers represent outliers.
Recognizing the phageome’s role in host health, we investigated fecal phageome changes post-intervention. Metagenomic sequencing identified 12,475 viral operational taxonomic units (vOTUs) annotated against the Metagenomic Gut Virus catalog, and 32.32% belonged to 12 known bacteriophage families (Figure 2D). No significant differences in phage community alpha diversity or overall structure were observed between the groups (Figures 2E, 2F, and S1B). To explore bacteriophage composition, we analyzed longitudinal fecal metagenomes from both groups, identifying postbiotic-associated changes and revealing Siphoviridae, Myoviridae, and Microviridae as the dominant families (Figure 2G). Notably, Alpavirinae phages significantly increased in the postbiotic group compared to the placebo group (p = 0.05; Figure 2H). A strong positive correlation was observed between gut bacteriome and phageome Shannon indices (r = 0.88, p < 0.001; Figure 2I), further supported by Procrustes analysis showing significant concordance between bacterial and bacteriophage community structures (correlation = 0.734, p = 0.001; Figure 2J). These findings highlight strong co-operativity and interdependence between the gut bacterial and phageome. To investigate phage-bacterial interactions, we assessed phage sequences within bacterial genomes. Among 12,475 annotated vOTUs, 11,514 were linked to specific bacterial hosts, with Siphoviridae primarily infecting Faecalibacterium, Ruminococcus, and Clostridium. In contrast, Alpavirinae phages preferentially targeted Prevotella and Bacteroides. These findings underscore the intricate interdependence between the gut bacteriome and phageome, offering insights into the mechanisms of postbiotic health benefits.
Postbiotic intervention modulated gut metabolic pathways and metabolome
To identify intervention-related gut metabolic modules (GMMs), we conducted genome-centric metabolic reconstruction using the 539 identified species-level genome bins (SGBs), alongside the MetaCyc and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (Table S5). We identified 74 GMMs involved in tryptophan derivatives, SCFAs, amino acids, neurotransmitters, vitamins, BAs, and unsaturated fatty acids (Figure 3A). These GMMs were encoded by 13 phyla, with S-adenosylmethionine synthesis showing the highest sample coverage (Table S6). Four GMMs showed differential abundance between two groups: tryptophan synthesis, propionate synthesis, and arginine degradation pathways were elevated in postbiotic recipients, whereas p-cresol synthesis was reduced (p < 0.05; Figure S1C).
Figure 3.
Phylum-level distribution of predicted GMMs in the fecal metagenome dataset and metabolome changes during the intervention trial
Data presented in this figure were derived from the postbiotic (n = 205) and placebo (n = 203) groups. Sample codes use a prefix to indicate whether samples were collected pre-intervention (pre) or post-intervention (post) and a suffix to denote the type of intervention (postbiotic or placebo).
(A) Heatmap depicting the distribution of 74 identified GMMs, corresponding to 539 species-level genome bins (SGBs). These GMMs are associated with the development, pathophysiology, immunity, and quality of life aspects of constipation. The color scale represents the cumulative count of SGBs encoding GMMs, ranging from low (purple) to high (red).
(B and C) (B) Principal-component analysis and (C) partial least squares-discriminant analysis (PLS-DA) score scatterplots of the fecal metabolome of the postbiotic and placebo groups, before and after intervention. Adonis test results are shown in the lower right corner of the PLS-DA score plots.
(D) Significant differential fecal metabolites identified in the postbiotic group post-intervention. Variable importance in projection (VIP) scores generated by the PLS-DA were used to identify metabolites contributing to discrimination (VIP score > 2, p < 0.05).
(E) Boxplots showing relevant responsive fecal metabolites to the postbiotic intervention. Significant differences between groups are shown (evaluated by the Wilcoxon rank-sum test). In the boxplots, boxes represent the interquartile range of each group’s value distribution, lines within boxes indicate median values, whiskers denote the lowest and highest values within 1.5 times the interquartile range, and dots above or below the whiskers represent outliers.
To investigate the effects of the postbiotic intervention on the fecal metabolome, we conducted untargeted metabolomic analysis, demonstrating stable instrumental performance through close clustering of quality control samples (Figure 3B). Partial least squares-discriminant analysis (PLS-DA) revealed clear separation between postbiotic and placebo groups after the intervention (p = 0.038), but not at baseline (p = 0.064; Figure 3C). Significantly differential metabolites were identified based on a combined threshold from the PLS-DA model, which included variable importance in projection (VIP) scores (VIP score > 2) and Wilcoxon rank-sum test p values (p < 0.05), with 10 metabolites meeting both criteria (Figure 3D; Table S7). Notably, all identified differential metabolites were not significantly different at baseline. After the intervention, the postbiotic group exhibited significantly higher levels of Succ, cholate, 3-IA, carnitine, and 5-HTP than the placebo group, and 2-hydroxyethylamine, glycerol monostearate, and 17α-hydroxypregnenolone were significantly reduced.
Postbiotic intervention modulated metabolites associated with constipation symptoms
We conducted targeted metabolomics to characterize postbiotic-induced changes in key metabolites, including amino acids, organic acids, SCFAs, vitamins, BAs, 5-HTP, and cortisol (Table S8). After the intervention, the postbiotic group exhibited significantly higher levels of several metabolites compared to the placebo group, including glutamate, tryptophan, Succ, 4-hydroxyphenyllactic acid, propionate, butyrate, chenodeoxycholate, and deoxycholate, while methionine, oxalic acid, and glycoursodeoxycholate were markedly reduced (p < 0.05; Figure S2A). Furthermore, the postbiotic intervention significantly increased 5-HTP and 3-IA, while decreasing cortisol levels compared to the placebo group (p < 0.05; Figure S2B). These findings demonstrate that postbiotic intervention can modulate various functional intestinal metabolites related to intestinal transit and stress response.
To explore links between postbiotic metabolite changes and constipation symptoms, we conducted a multivariate analysis using a MaAsLin2 model and found that carnosine levels were positively correlated with stool consistency score (coefficient [coef] = 0.71, p = 0.03) and slightly with CSBMs (coef = 0.61, p = 0.06). Conversely, p-cresol synthesis module was positively associated with discomfort scores (coef = 0.16, p = 0.01), while propionate synthesis module was inversely associated with worries (coef = −0.25, p = 0.01). Furthermore, several amino acids and organic acids were associated with CSBMs and patients' quality of life (QoL). Valine and tyrosine were positively correlated with CSBMs (coef > 0.17, p < 0.05), while tryptophan showed an almost significant positive correlation (coef = 0.15, p = 0.06). Phenyllactic acid was inversely correlated with worries (coef = −0.73, p = 0.03). Additionally, the metabolite 3-IA was negatively associated with both worries (coef = −0.36, p = 0.05) and physical discomfort scores (coef = −0.39, p = 0.05; Figure S2C). In summary, these findings indicate that the postbiotic intervention may alleviate constipation symptoms and improve QoL by modulating bioactive metabolite levels.
Postbiotic and its metabolites mitigated loperamide-induced constipation in mice
To validate clinical observations, loperamide was administered for 7 days to induce a constipation model in mice. After acclimation, sixty mice were randomized into six groups: Con (control), Mod (model), Post (postbiotic), Succ, 3-IA, and 5-HTP groups (Figure 4A). On day 0, no significant differences were observed in body weight, food intake, water consumption, fecal weight, and water content among groups (Table S9). On day 7, the Mod and all treatment groups exhibited significantly reduced fecal weight and water content, along with delayed first black defecation compared to the Con group (p < 0.05; Figure 4B), confirming successful loperamide-induced constipation. On day 14, Succ, 3-IA, and 5-HTP treatment significantly increased fecal weight (p < 0.05), though only Succ and 3-IA restored output to near-normal levels when compared to the Con group (Figure S3A). On day 21, the Post and 3-IA groups showed significantly higher fecal weight than the Mod group (p < 0.05). Additionally, all intervention groups showed significantly increased fecal water content and small intestinal transit rate, with Post, 3-IA, and 5-HTP treatments reducing the time to first black defecation compared to the Mod group (p < 0.05; Figure 4C; Table S10). Collectively, the postbiotic and its metabolites effectively restored gastrointestinal function in loperamide-induced constipation.
Figure 4.
Postbiotic and its metabolite component’s effects on constipation alleviation in mice
(A) Schematic diagram illustrating the design of the intervention study. The trial involved 60 mice (n = 10 per group), which were randomly divided into six groups following a 1-week acclimatization period: control (Con), model (Mod), Probio-Eco (Post), succinate (Succ), 3-indoleacrylic acid (3-IA), and 5-hydroxytryptophan (5-HTP). The Mod, Post, Succ, 3-IA, and 5-HTP groups received loperamide (Lop) for the first week to induce constipation, while the Con group received normal saline (NS). From days 7–21, mice received the respective interventions.
(B) Effects of continuous intragastric loperamide administration on defecation at day 7.
(C) Effects of postbiotic and component interventions on defecation at day 21 (n = 10).
(D–F) (D) Colon histopathology and mucin expression (n = 5) and (E and F) intestinal levels of immune factors and gut hormones (n = 8) in constipated mice at day 21. Colon tissue sections were stained with hematoxylin and eosin (H&E), alcian blue-periodic acid-Schiff (AB-PAS), and wheat germ agglutinin-fluorescein isothiocyanate (WGA-FITC) (scale bar, 100 μm).
Error bars in the bar charts represent standard deviation. In the boxplots, boxes represent the interquartile range of each group’s value distribution, lines inside the boxes indicate median values, whiskers denote the lowest and highest values within 1.5 times the interquartile range, and dots above or below the whiskers represent outliers. Significance was determined using the Wilcoxon rank-sum test, with p < 0.05 considered statistically significant (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
Histological analysis of colon pathology revealed that the Mod group had thinner muscle layers than the Con group. Postbiotic and its metabolite treatments induced muscle layer thickening to varying degrees (Figure 4D). Alcian blue-periodic acid-Schiff and wheat germ agglutinin-fluorescein isothiocyanate staining revealed that the Mod group showed mucosal atrophy, marked by mucus layer damage and thinner surface neutral mucins. Postbiotic and its metabolite treatments variably increased colonic mucosa thickness and enhanced mucus secretion, particularly in the neutral mucus layer (Figure 4D). These findings indicate that the postbiotic and its metabolite interventions effectively restored the colonic mucus barrier and improved histological features of constipated mice. Serum analysis by ELISA revealed that the Mod group had elevated pro-inflammatory cytokines (interleukin [IL]-6 and IL-1β) and reduced IL-10 compared to the Con group, and postbiotic and 3-IA significantly reversed these effects (p < 0.05; Figure 4E). Furthermore, the Mod group showed elevated vasoactive intestinal peptide but reduced gastrin and motilin levels compared to the Con group (p < 0.05; Figure 4F). Specifically, all treatments restored gastrin levels, but only the postbiotic significantly increased motilin. Both postbiotic and 3-IA effectively reduced the vasoactive intestinal peptide level (p < 0.05; Figure 4F). These results suggest that the postbiotic alleviated constipation in mice by enhancing mucin secretion, regulating gut hormones, and modulating immunity. However, Succ, 3-IA, and 5-HTP exhibited distinct hormonal regulatory effects, warranting further investigation and analysis.
Postbiotic-derived metabolites restored gut microbiome in constipated mice
To evaluate the effects of postbiotic metabolites on gut microbiota in constipated mice, we analyzed microbial diversity over 21 days. Baseline showed no intergroup differences in alpha and beta diversity, providing a standardized foundation for subsequent interventions. By day 7, the Mod group showed reduced alpha diversity (p < 0.05; Figure 5A) and altered community structure compared to the Con group (R2 = 0.25, p = 0.003; Figure 5B), confirming loperamide-induced gut dysbiosis. After metabolite intervention (day 14), Succ and 3-IA groups showed recovery in alpha diversity, whereas 5-HTP remained significantly lower than controls (p < 0.05). Beta diversity analysis revealed significant structural shifts (R2 = 0.26, p = 0.001), with greater inter-/intra-group variation in treated groups compared to the Con group (R2 = 0.26, p = 0.001; Figures S3B–S3D).
Figure 5.
Effects of individual postbiotic components on gut microbiome, metabolites, and colonic transit-associated genes in constipated mice
(A and B) (A) Shannon index and (B) principal coordinates analysis (PCoA) score plots of the gut microbiome in the control (Con), model (Mod), succinate (Succ), 3-indoleacrylic acid (3-IA), and 5-hydroxytryptophan (5-HTP) groups (n = 6) at days 0, 7, and 21. Adonis test results are displayed in the upper left corner of the PCoA score plots.
(C) Expression of colonic transit-related genes in different groups at day 21 (n = 8). Significance was determined using the Wilcoxon rank-sum test, with p < 0.05 considered statistically significant (∗p < 0.05; ∗∗p < 0.01). In the boxplots, boxes represent the interquartile range of each group’s value distribution, lines inside the boxes indicate median values, whiskers denote the lowest and highest values within 1.5 times the interquartile range, and dots above or below the whiskers represent outliers.
(D) Correlation network analysis of metabolite levels, physiological factors, and defecation parameters following the Succ, 3-IA, and 5-HTP interventions. Spearman correlation coefficients were calculated between datasets, and features with |r| > 0.6 and p < 0.05 were selected to construct the correlation network plots. Blue lines represent negative correlations, and red lines indicate positive correlations between connected nodes.
AAAD, Aromatic amino acid decarboxylase; CLDN1, Claudin-1; AQP3, Aquaporin 3; AQP4, Aquaporin 4; GAS, Gastrin; GLP-1, Glucagon-like peptide-1; IL, interleukin; MTL, Motilin; MUC2, Mucin-2; PYY, Peptide YY,; SCFAs, short-chain fatty acids; SGBs, species-level genome bins; 5-HT, 5-hydroxytryptamine; VIP, Vasoactive intestinal polypeptide; ZO-1, Zonula occludens protein.
To identify treatment-induced differentially abundant taxa, we selected SGBs with no baseline differences among Con, Mod, and intervention groups but significant changes at later time points (relative abundance ≥ 0.05%, prevalence ≥ 5%). On day 7, the Mod group exhibited significantly lower relative abundances of Ligilactobacillus murinus, Alistipes timonensis, and Duncaniella freteri compared to the Con group, while Christensenella sp. and Erysipelotrichaceae sp. increased significantly, reflecting loperamide-induced alterations in bacterial composition. By day 21, treatments with Succ, 3-IA, and 5-HTP significantly increased Acetatifactor sp910586835, while decreasing Dubosiella sp004793885 (p < 0.05; Table S11). Additionally, Succ treatment significantly increased Kineothrix sp910587275, Prevotella sp., and Lachnospiraceae sp., while reducing Erysipelatoclostridium saccharogumia and Acetatifactor sp910578815. The 3-IA treatment increased Lachnospiraceae sp., Parasutterella sp., and Olsenella sp003150175, while significantly decreasing Erysipelotrichaceae sp. For the 5-HTP intervention, the relative abundances of Paramuribaculum sp001689535 and Parasutterella sp. increased significantly, whereas Erysipelatoclostridium sp. decreased (p < 0.05; Table S11). Collectively, these findings show that the implemented metabolites selectively restore key microbial species disrupted by loperamide-induced constipation.
Postbiotic metabolites modulated colon transit-related genes in constipated mice
We employed quantitative real-time PCR to evaluate postbiotic metabolites’ effects on genes regulating colonic mucosal barrier, water-electrolyte balance, gut hormones, and motility in constipated mice. Loperamide treatment reduced the gene expression of CLDN1, ZO-1, and MUC2, while elevating aquaporin (AQP3 and AQP4) compared to the Con group (p < 0.05). Succ and 3-IA treatments upregulated CLDN1 expression, with Succ significantly enhancing ZO-1 expression compared to the Mod group (p < 0.05). Although 5-HTP treatment also increased CLDN1 and ZO-1 expression, the changes were not statistically significant. Interestingly, all three treatments significantly elevated MUC2 expression while reducing AQP3 and AQP4 expression compared to the Mod group (p < 0.05). Furthermore, the Mod group showed elevated colonic PYY gene expression compared to the Con group, which was normalized by Succ, 3-IA, and 5-HTP treatments (p < 0.05; Figure 5C).
To explore the 5-HTP regulatory pathway on colon transit, we assessed the expression of aromatic amino acid decarboxylase (AAAD), the enzyme that converts amino acid precursors to serotonin (5-HT). Loperamide treatment significantly reduced AAAD expression, while 5-HTP supplementation significantly increased AAAD expression (p < 0.05; Figure 5C), an effect not observed for Succ and 3-IA. These findings indicate that loperamide disrupts AAAD and 5-HT regulation, while 5-HTP specifically enhances 5-HT biosynthesis, potentially contributing to its modulatory effects on colon transit genes. The data suggest that postbiotic metabolites, such as Succ, 3-IA, and 5-HTP, can counteract loperamide-induced dysregulation of genes involved in colonic mucosal barrier function, water-electrolyte balance, and gut peptide hormones. The ability of 5-HTP to upregulate AAAD highlights its role in enhancing colon transit via the serotonergic pathway.
Postbiotic metabolites modulated fecal SCFA and serum 5-HT in constipated mice
To further verify the effects of individual postbiotic metabolites on downstream signaling molecules, intestinal SCFA and serum 5-HT levels were enumerated using targeted metabolomics and ELISA (Figure S3E). Loperamide treatment significantly reduced intestinal propionate, butyrate, isovalerate, and serum 5-HT levels compared to the Con group (p < 0.05). Succ treatment elevated propionate, butyrate, and isovalerate, while 3-IA increased butyrate, isobutyrate, and isovalerate levels (p < 0.05). Interestingly, 5-HTP treatment significantly increased serum 5-HT compared to the Mod group (p < 0.05). Meanwhile, 3-IA supplementation specifically elevated aryl hydrocarbon receptor (AhR) and IL-22 levels (p < 0.05). Collectively, these findings demonstrate that loperamide disrupts SCFA and 5-HT homeostasis, while postbiotic metabolites selectively restore these pathways to varying degrees. Notably, 3-IA activates the AhR-IL-22 axis, highlighting potential immunomodulatory mechanisms for further investigation.
Correlation analysis links postbiotic interventions, gut physiology, and microbiome
We conducted correlation analyses to examine relationships between key markers, gut function, and microbial features after intervention with the three postbiotic metabolite components (Figure 5D). Succ intervention revealed a positive correlation between small intestinal transit rate and gastrin (r = 0.690, p = 0.05), while showing a negative correlation with Parabacteroides sp. (r = −0.639, p = 0.05). The expression of AQP3 was negatively correlated with fecal water content (r = −0.788, p = 0.02), and AQP4 showed a negative correlation with Prevotella sp. (r = −0.647, p = 0.05). Interestingly, propionate showed a significant negative correlation with IL-6 (r = −0.814, p = 0.01), which was inversely associated with CLDN1 (r = −0.755, p = 0.03). Butyrate negatively correlated with the mucin-degrading Acetatifactor sp910578815 (r = −0.711, p = 0.048), which was positively associated with PYY gene expression (r = 0.695, p = 0.05). Post 3-IA intervention, small intestinal transit rate positively correlated with IL-10 (r = 0.667, p = 0.05). Furthermore, Lachnospiraceae sp. positively correlated with AhR gene expression (r = 0.761, p = 0.03), which was positively correlated with IL-22 (r = 0.738, p = 0.04). The AQP3 gene expression positively correlated with black defecation time (r = 0.643, p = 0.05), but negatively with Acetatifactor sp910586835 (r = −0.695, p = 0.05). This Acetatifactor species positively correlated with motilin (r = 0.814, p = 0.01), and both AQP3 gene expression and vasoactive intestinal peptide negatively correlated with gastrin (r > 0.690, p < 0.05). In addition, CLDN1 gene expression positively correlated with butyrate (r = 0.762, p = 0.03).
In the 5-HTP intervention, small intestinal transit rate positively correlated with CLDN1 (r = 0.743, p = 0.03), but negatively with Erysipelatoclostridium sp. (r = −0.933, p < 0.001). Erysipelatoclostridium sp. negatively correlated with 5-HT (r = −0.707, p = 0.05), but positively with black defecation time (r = 0.687, p = 0.05). In addition, IL-1β negatively correlated with CLDN1 expression (r = −0.647, p = 0.05), but positively with vasoactive intestinal polypeptide (r = 0.695, p = 0.05). Notably, total fecal weight positively correlated with AAAD expression and Clostridia sp. (r > 0.762, p < 0.03), while Clostridia sp. was also positively associated with MUC2 expression (r = 0.764, p = 0.03). Collectively, these correlations reveal intricate links between postbiotic interventions, gut physiology, microbial ecology, and neuroimmune signaling, highlighting the systemic effects of Succ, 3-IA, and 5-HTP in regulating gut homeostasis.
Discussion
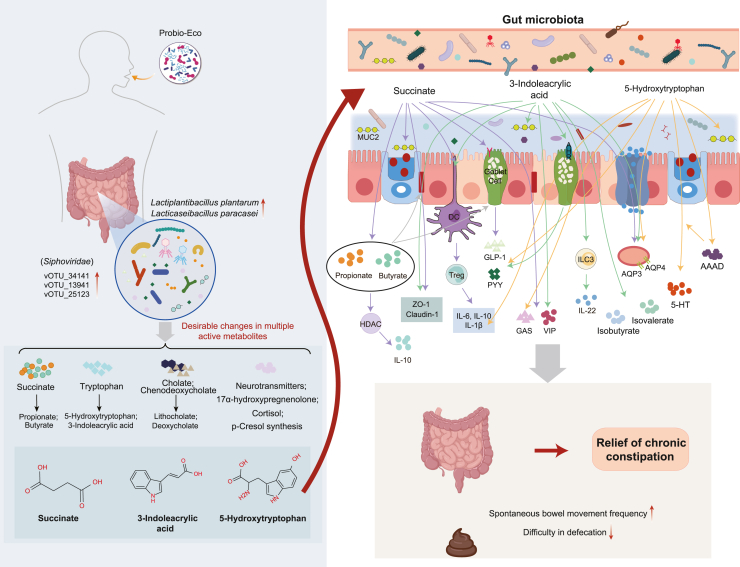
Chronic constipation, a prevalent gastrointestinal disorder, adversely affects patients’ quality of life and healthcare systems.1,24 Postbiotics have emerged as a promising strategy for managing gastrointestinal diseases, offering health benefits that may be similar to those of probiotics.16,25 However, the effects of postbiotics and the metabolites have not been fully elucidated. Building on previous work demonstrating constipation-alleviating effect of probiotic through regulation of host-microbiome interactions,12,26 this study comprehensively characterized the beneficial effects of the postbiotic Probio-Eco in both human and mouse constipation models. Our findings revealed that Probio-Eco activates the Succ and tryptophan metabolic pathways by modulating gut microbiota, resulting in the production of beneficial metabolites including SCFAs, 3-IA, and 5-HT that interact with several intestinal cells. This interaction enhances the secretion of intestinal hormones and mucin, modulates inflammation, and promotes mucosal homeostasis and intestinal transit, thus alleviating constipation symptoms (Figure 6). This study advances our understanding of postbiotics’ therapeutic potential for gastrointestinal disorders and supports developing more effective interventions.
Figure 6.
Schematic diagram illustrating the proposed mechanisms by which Probio-Eco modulates intestinal pathways to alleviate chronic constipation
Specifically, the postbiotic influences the succinate and tryptophan metabolic pathways, promoting intestinal homeostasis and motility. AQP3, Aquaporin 3; AQP4, Aquaporin 4; AAAD, Aromatic amino acid decarboxylase; GAS, Gastrin; GLP-1, Glucagon-like peptide-1; ILC3, Group 3 innate lymphoid cells; HDAC, Histone deacetylase; IL, interleukin; MUC2, Mucin-2; PYY, Peptide YY; Treg, regulatory T cells; 5-HT, 5-hydroxytryptamine; VIP, Vasoactive intestinal polypeptide; ZO-1, Zonula Occludens-1.
Our first finding is that Probio-Eco administration effectively improved constipation symptoms in both the ITT and PP analyses. Specifically, the postbiotic group demonstrated significantly improved weekly mean CSBMs and reduced stool straining scores compared to placebo at days 21 and 56, which is consistent with prior research.19 Moreover, chronic constipation can severely impact patients’ quality of life,27 and we found that the postbiotic intervention significantly reduced worries and physical discomfort on days 21 and 56. Notably, no significant differences were observed in the CSBMs; stool straining; stool consistency; Patient Assessment of Constipation Quality of Life (PAC-QoL); and depression, anxiety, and stress scale (DASS-21) scores between the two groups after the 14-day washout period (day 35). This suggests that the intervention effects were eliminated after the washout, providing a basis for the subsequent omics data analysis. Unlike probiotics, which can proliferate and expand in the colon, postbiotics do not have a sustained effect once the intervention is discontinued.28 Although postbiotics’ effects on the gut microbiome may be temporary, they significantly impact gastrointestinal health by inhibiting unfavorable microorganisms through antimicrobial components.14 Therefore, we evaluated gut microbiota changes during postbiotic intervention, and beta diversity analysis showed moderate gut bacteria and bacteriophage community shifts in patients. This mirrors previous observations of parallel shifts in gut bacteria, bacteriophages, and their interactions across health and disease states.29 At finer taxonomic resolution, the postbiotic group exhibited significant enrichment of Lacticaseibacillus paracasei, Lactiplantibacillus plantarum, and several vOTUs belonging to Siphoviridae. Notably, the fermentation strains used in Probio-Eco included two strains, and previous evidence demonstrated that Lactiplantibacillus plantarum-containing synbiotics improve bowel movement frequency in constipation patients,30 while Lacticaseibacillus paracasei metabolites alleviate constipation in aged mice by modulating gut microbiota.17 Furthermore, higher gut Siphoviridae levels were associated with improved brain executive function.31 In summary, these results suggest that the constipation-alleviating effects of Probio-Eco are accompanied by desirable changes in various health-related gut bacteria and bacteriophages.
We identified that Probio-Eco administration elevated tryptophan and its downstream metabolites (3-IA and 5-HTP) in fecal metabolomes. Gut bacteria convert tryptophan into various indole compounds that act as AhR ligands, and AhR activation can partially restore gut motility.32 Previous studies have found that elevated serum 3-IA levels were also linked to symptom improvement after fecal microbiota transplantation in constipation patients, suggesting a broader role in this metabolite in gut motility regulation.33 Another key tryptophan metabolic pathway involves 5-HT synthesis via 5-HTP as an intermediate, whose availability depends on tryptophan levels and enzyme activity.34 Constipation patients exhibit reduced colonic and serum levels of 5-HT and PYY, and a study found that serotonin can regulate gut motility, influencing gastrointestinal function.35 Moreover, the postbiotic group exhibited increased tryptophan synthesis pathway and reduced p-cresol synthesis pathway. p-cresol has been implicated in chronic constipation among children with autism,36 and our previous work identified a positive correlation between p-cresol and Oscillospiraceae, which can affect colonic transit.12 Taken together, these findings indicated that elevated tryptophan and its metabolites (3-IA and 5-HTP) and reduced p-cresol in postbiotic recipients may enhance colonic transit and relieve constipation.
Succ, a key microbial metabolite, is produced by Bacteroides and Prevotella through pentose and hexose metabolism,37 and our study revealed that postbiotic recipients showed elevated fecal fumarate and Succ levels. However, excessive Succ accumulation may be harmful.38 Its degradation by gut microbiota is thus essential, serving as the main pathway for intestinal propionate production. Additionally, some gut microbes can further convert Succ into acetate and butyrate.37 Consistent with this, our results showed that postbiotic intervention significantly increased fecal propionate and butyrate levels, which can improve constipation by stimulating intestinal smooth muscle contraction.39,40 Interestingly, we also observed that postbiotic intervention increased fecal chenodeoxycholate and deoxycholate levels, which are known to stimulate fluid secretion and trigger high-amplitude colon contractions.41,42 In addition, the postbiotic intervention was observed to alter neurotransmitter profiles, increasing propionate and glutamate while decreasing 17α-hydroxypregnenolone and cortisol. A recent trial showed that SCFA supplementation blunted cortisol responses to stress compared to the placebo group.43 Notably, 17-hydroxypregnenolone is a cortisol precursor,44 the key stress hormone associated with anxiety. Prior studies show that probiotic intake significantly reduces free cortisol in urine and feces, suggesting that probiotics may regulate cortisol levels and alleviate anxiety symptoms.45,46 Therefore, the observed improvement in worry and physical discomfort may result from postbiotic-modulated SCFAs and neurotransmitter regulation. In summary, the clinical results demonstrate that the Probio-Eco intervention effectively alleviates chronic constipation and enhances quality of life. Importantly, this alleviation effect was mediated not by broad gut microbiome shifts but by targeted modulation of beneficial microbes and their metabolites. Among these, the metabolic pathways involving tryptophan-3-IA, tryptophan-5-HT, and Succ-SCFAs mediated by Probio-Eco intervention are likely to play a pivotal role in these beneficial outcomes.
To validate clinical observations, we employed a loperamide-induced constipation mouse model that showed significantly decreased fecal water content, delayed gut transit, and prolonged defecation time. Notably, administration of the postbiotic and its components (Succ, 3-IA, and 5-HTP) reversed these effects, demonstrating efficacy against constipation. Given that impaired intestinal homeostasis is a hallmark of loperamide-induced constipation,47 we evaluated key regulatory aspects including intestinal hormones, mucosal barrier integrity, and immune responses. In the Mod group, we observed significantly reduced levels of excitatory intestinal hormones (gastrin and motilin) but elevated inhibitory hormones (vasoactive intestinal polypeptide) and pro-inflammatory cytokines (IL-6 and IL-1β). Additionally, intestinal mucosal epithelial cells exhibited atrophy, with both mucus layer damage and mucosal thinning. These physiological and pathological changes are consistent with previous reports, where Bifidobacterium longum supplementation alleviated constipation by increasing fecal water content, regulating intestinal hormones, and improving intestinal motility, corroborating our observations.48 Emerging evidence suggests that gut microbiota dysbiosis disrupts microbiome-gut-brain axis signaling, impairing intestinal motility.49 Our study found that loperamide-induced constipation in mice caused gut dysbiosis and dysregulated colonic transport gene expression, characterized by decreased CLDN1, ZO-1, and MUC2, alongside elevated PYY, AQP3, and AQP4 in the colon. Interestingly, Succ, 3-IA, and 5-HTP exhibited distinct efficacy pathways and hormonal regulation patterns, highlighting the nuanced mechanisms through which these metabolites alleviate constipation.
Treatment with the postbiotic component Succ significantly increased the abundance of SCFA-producing bacteria, while reducing potentially harmful taxa (Erysipelatoclostridium saccharogumia, Parabacteroides sp910577325, and Desulfovibrionaceae), collectively modulating intestinal motility.50,51 Previous studies found that Parabacteroides and Desulfovibrionaceae overgrowth are linked to the development of constipation.52,53 As downstream products of Succ, particularly propionate and butyrate can influence colon motility by regulating hormones release and stimulating MUC2 secretion from goblet cells.11,54 Our results show that Succ treatment increased gastrin levels and upregulated colonic expression of CLDN1, ZO-1, and MUC2 genes, which were associated with increased production of propionate and butyrate, leading to improved intestinal mechanical barrier function. Furthermore, SCFAs stimulate G protein-coupled receptors on enteroendocrine cells, mediating secretion of glucagon-like peptide-1 and PYY to regulate gastrointestinal motility.6 Interestingly, Succ treatment significantly reduced colonic PYY gene expression, a gut hormone that inhibits intestinal motility. This finding is consistent with studies showing that probiotics relieve constipation by decreasing PYY levels and promoting intestinal peristalsis.55,56 Additionally, we observed that Succ intervention reduced colonic expression of aquaporin genes AQP3 and AQP4, with AQP3 negatively correlating with fecal water content and AQP4 inversely associated with Prevotella sp. Colonic aquaporins promote water absorption, and their overexpression may cause excessive reabsorption and fecal dehydration.57 In summary, the postbiotic Succ activated the Succ-SCFA pathway, enriching Succ-utilizing bacteria and increasing propionate and butyrate production. These metabolites enhanced intestinal barrier function by modulating tight junction proteins and goblet cell-derived mucins, while also stimulating G protein-coupled receptors on enteroendocrine cells to improve intestinal transport, ultimately alleviating constipation.
Treatment with the postbiotic component 3-IA significantly increased the abundances of Roseburia sp., Eubacterium sp000435815, and Olsenella sp003150175, while Erysipelotrichaceae sp. and Mucispirillum schaedleri decreased significantly. Both Eubacterium and Roseburia are known butyrate producers, while probiotic supplementation has been shown to elevate Olsenella abundance, linked to reduced gut inflammation.58,59 Mucispirillum schaedleri has been identified as a major member of the mouse cecal crypt microbiome, which is linked to gut inflammation.60 Notably, indole derivatives can activate AhR in myenteric neurons to enhance colonic peristalsis.61,62 Our findings revealed that 3-IA treatment increased serum AhR levels while decreasing its downstream effector IL-22. Accumulating evidence indicates that AhR activation can modulate immune responses in a ligand-specific manner.63,64 Therefore, the 3-IA-mediated regulation of AHR and IL-22 may represent an additional pathway through which this postbiotic compound promotes intestinal health. We also found that 3-IA exerted stronger immune regulatory effects than other metabolites, significantly reducing IL-1β and IL-6 while increasing IL-10 in constipated mice. Furthermore, indole derivatives enhance intestinal barrier integrity by upregulating epithelial genes that promote mucus secretion,65,66 and we also observed that 3-IA treatment increased CLDN1 and MUC2 expression. A previous report found that Peptostreptococcus metabolizes tryptophan into 3-IA, which enhances intestinal barrier integrity in mice, potentially through AhR activation and regulation of goblet cell-related genes like MUC2.67 In summary, these findings suggest that the postbiotic 3-IA activated the indole-AhR axis by modulating gut microbiota. It maintains mucosal homeostasis by activating AhR in myenteric neurons, exerts ligand-specific anti-inflammatory immune modulation, and upregulates genes supporting epithelial function, thereby improving barrier integrity and alleviating constipation.
Treatment with 5-HTP markedly decreased the abundances of Oscillospiraceae sp. and Duncaniella dubosii, while significantly increasing Jeotgalicoccus sp., Ruminococcus sp., and Paramuribaculum sp001689535. Previous studies have linked a reduced abundance of Oscillospiraceae to looser stool consistency and implicated this family in altered colonic transit.68,69 Notably, a study found that the abundance of Jeotgalicoccus increased in constipated mice after prebiotic administration,70 and we observed that its negative correlation with AQP4 expression suggests a role in relieving constipation via colonic water transport regulation. Certain Ruminococcus species encode AAAD enzymes that convert aromatic amino acids into bioactive amines, stimulating colonic 5-HT production.71 Through serotonin signaling, the gut microbiome enhances gut motility by triggering enterochromaffin cell-derived 5-HT release.72,73,74 We also found that 5-HTP treatment upregulated AAAD gene expression, which encodes the enzyme that converts 5-HTP to 5-HT.75 This effect was unique to 5-HTP, as neither Succ nor 3-IA interventions altered these microbial pathways associated with 5-HT production. Furthermore, 5-HT can directly activate 5-HT receptor 4 located on goblet cells, stimulating MUC2 expression and secretion.76 We observed that 5-HTP treatment increased colonic MUC2 expression while reducing PYY levels in mice, which aligns with a previous animal study.77 In summary, the postbiotic 5-HTP treatment activates the 5-HTP-5-HP pathway by modulating gut microbiota-mediated AAAD. The activation stimulates enterochromaffin cells to release 5-HT, which regulates smooth muscle contraction and promotes MUC2 secretion in goblet cells, effectively relieving constipation symptoms.
In conclusion, this study provides compelling evidence for the efficacy of the postbiotic Probio-Eco in alleviating chronic constipation. The observed improvements in defecation were accompanied by the modulation of key microbial metabolic pathways, including the Succ-SCFA, tryptophan-5-HTP-5-HT, and tryptophan-3-IA pathways. By targeting specific microbial pathways and metabolites, postbiotic interventions may represent a promising, gut-centric therapeutic approach. These microbiome-mediated pathways provide a valuable framework for clinical application of postbiotics to treat chronic constipation and refractory gastrointestinal disorders.
Limitations of the study
This study has several notable limitations. Firstly, this study was conducted over a relatively short time frame, and, given that clinical evidence surrounding postbiotics is still in its early stages, there is a lack of comprehensive data regarding how factors such as intervention duration and dosage may influence their effectiveness in alleviating constipation. Therefore, whether long-term use enhances efficacy or induces dependency remains unclear, warranting future longitudinal studies. Secondly, the trial spanned from November 2021 to January 2022, during which the COVID-19 pandemic significantly impacted our ability to conduct social recruitment. This constraint yielded a predominantly young, university-based cohort, potentially limiting generalizability. While this demographic provides valuable insights, it may not fully represent the broader population of constipated patients. Additionally, to improve compliance and efficiency, colon transport tests were omitted due to participants’ concerns about tolerance and radiation exposure. Future studies should enhance participant engagement in assessment discussions to include colon transit time indicators. Furthermore, the trial design lacks detailed daily diet monitoring, potentially affecting intestinal motility and constipation outcomes, and future trials should include detailed dietary assessments to better understand these variables. Finally, we acknowledge that the loperamide-induced experimental constipation model, while widely used and validated for its reliability and similarity to human constipation, does not fully replicate the complexity of primary constipation in humans. Although this model offers valuable insights, developing more accurate animal models of human primary constipation is essential for improving intervention studies.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Heping Zhang (hepingdd@vip.sina.com).
Materials availability
This study did not generate any new unique reagents.
Data and code availability
-
•
The raw sequencing data are available in the CNGB Sequence Archive of China National GeneBank DataBase (CNGBdb; https://db.cngb.org/cnsa/) under accession numbers CNGBdb: CNP0005022 (human) and CNGBdb: CNP0005187 (mouse) and will be publicly accessible upon publication. The metabolomics data are available in MassIVE: MSV000096310 and are publicly available and accessible upon publication.
-
•
The original code has been deposited at GitHub and Zenodo and is available at https://github.com/TengMa-Cleap/Postbiotics-to-relieve-chronic-constipation (https://doi.org/10.5281/zenodo.14992672).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
This study was supported by the Research Fund for the National Key R&D Program of China (2022YFD2100700), the Inner Mongolia Autonomous Region Science and Technology Leading Talent Team Project (2022LJRC0003), and the National Natural Science Foundation of China (U22A20540). We thank Suzhou PANOMIX Biomedical Tech. Co., Ltd. for metabolite detection and Novogene Co., Ltd. for metagenomic sequencing.
Author contributions
H.Z. and Z.S. designed the trial. T.M. and Y. Li. collected clinical data. T.M., X.S., H.J., and Y. Liu. analyzed the data. H.W. and N.Y. accomplished the screening of enrolled patients. T.M. and Y. Li. wrote the manuscript. L.-Y.K. provided revisions to the manuscript.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Probio-Eco® | Jinhua Yinhe Biological Technology Co., Ltd. | N/A |
| Lacticaseibacillus paracasei Zhang (One of the main fermentation strains of Probio-Eco®) | Key Laboratory of Dairy Biotechnology and Engineering, Ministry of Education, Inner Mongolia Agricultural University | CGMCC5469 |
| Lactiplantibacillus plantarum P-8 (One of the main fermentation strains of Probio-Eco®) | Key Laboratory of Dairy Biotechnology and Engineering, Ministry of Education, Inner Mongolia Agricultural University | CGMCC6312 |
| Bifidobacterium animalis subsp. lactis V9 (One of the main fermentation strains of Probio-Eco®) | Key Laboratory of Dairy Biotechnology and Engineering, Ministry of Education, Inner Mongolia Agricultural University | CGMCC5470 |
| Biological samples | ||
| Human stool samples | This study | N/A |
| Mouse stool samples | This study | N/A |
| Mouse serum samples | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| TRIzol reagent | Thermo Fisher | Cat# 15596018 |
| Wheat germ agglutinin conjugated to fluorescein isothiocyanate | ZCIBIO Technology Co., Ltd. | Cat# 27072-45-3 |
| Loperamide hydrochloride | Xian Janssen Pharmaceutical Ltd | Cat# NAJ070H |
| 3-indoleacrylic acid | Shanghai yuanye Bio-Technology Co., Ltd | Cat# 1204-06-4 |
| 5-hydroxytryptophan | Shanghai yuanye Bio-Technology Co., Ltd | Cat# 314062-44-7 |
| Succinate | Shanghai yuanye Bio-Technology Co., Ltd | Cat# 150-90-3 |
| Methyl alcohol | Fisher Chemical | Cat# A452-4 |
| Aetonitrile | Merck | Cat# 1499230-935 |
| 2-Chloro-D-phenylalanine | Macklin | Cat# 14091-11-3 |
| Amonium acetate | Fluka | Cat# 17836-50G |
| Critical commercial assays | ||
| Magnetic Soil and Stool DNA Kit | TIANGEN Biotech Co., Ltd. | Cat# 51504 |
| Qubit™ dsDNA Quantification Assay Kits | Thermo Fisher | Cat# Q32854 |
| NEB Next Ultra™ RNA Library Prep Kit for Illumina | NEB | Cat# E7530S |
| cDNA first strand synthesis kit | Merck | Cat# F917866 |
| SYBR qPCR Master Mix | Tianya Biotech | Cat# P1503 |
| Hematoxylin and Eosin Staining Kit | Nanjing BioChannel Biotechnology Co., Ltd. | Cat# BP-DL001 |
| Alcian blue-periodic acid-Schiff staining kit | Nanjing BioChannel Biotechnology Co., Ltd. | Cat# BP-DL037 |
| Mouse motilin ELISA kit | Meimian Biotechnology | Cat# MM-0492M2 |
| Mouse gastrin ELISA kit | Meimian Biotechnology | Cat# MM-44405M2 |
| Mouse vasoactive intestinal peptide ELISA kit | Meimian Biotechnology | Cat# MM-0446M2 |
| Mouse 5-HT ELISA kit | Meimian Biotechnology | Cat# MM-0443M2 |
| Mouse IL-6 ELISA kit | Meimian Biotechnology | Cat# MM-0163M1 |
| Mouse IL-10 ELISA kit | Meimian Biotechnology | Cat# MM-0176M1 |
| Mouse IL-1β ELISA kit | Meimian Biotechnology | Cat# MM-0040M1 |
| Mouse IL-22 ELISA kit | Meimian Biotechnology | Cat# MM-0892M2 |
| Mouse AhR ELISA kit | Meimian Biotechnology | Cat# MM-46547M2 |
| Deposited data | ||
| Shotgun metagenomics data (Human) | This paper | CNGBdb: CNP0005022 |
| Untargeted Metagenomics data (Human) | This paper | MassIVEdb: MSV000096310 |
| Shotgun metagenomics data (Mice) | This paper | CNGBdb: CNP0005187 |
| Experimental models: Organisms/strains | ||
| Males C57BL/6 mice | Charles River Labs Co., Ltd | N/A |
| Software and algorithms | ||
| R (version 4.3.2) | R Project | https://www.r-project.org |
| MetaboAnalyst 5.0 | Ewald et al. | https://www.metaboanalyst.ca/MetaboAnalyst/home.xhtml |
| MEGAHIT V1.2.9 | Li et al.78 | https://github.com/voutcn/megahit |
| MetaBAT2 | Kang et al.79 | https://bitbucket.org/berkeleylab/metabat |
| VAMB | Johansen et al.80 | https://github.com/RasmussenLab/vamb. |
| DAS Tool 1.1.4 | Sharrar et al.81 | https://github.com/cmks/DAS_Tool |
| CheckM2 | Donovan H et al.82 | https://ecogenomics.github.io/CheckM/ |
| dRep | Matthew et al.83 | https://github.com/MrOlm/drep. |
| CoverM | Aroney et al. | https://github.com/wwood/CoverM |
| MetaCyc | Wikoff et al. | https://metacyc.org/ |
| KEGG | Minoru Kanehisa et al. | http://www.genome.ad.jp/kegg/ |
| Kraken2 | Wood et al. | https://ccb.jhu.edu/software/kraken2/ |
| VIBRANT | Kristopher et al.84 | https://github.com/AnantharamanLab/VIBRANT/ |
| CheckV | Pedro et al.85 | https://bitbucket.org/berkeleylab/CheckV |
| CD-HIT | Fu et al. | http://cd-hit.org |
| ProteoWizard | Darren et al. | http://proteowizard.sourceforge.net |
| Other | ||
| Placebo (consisted of soy protein powder, skim milk powder, and sodium citrate) | Jinhua Yinhe Biological Technology Co., Ltd. | N/A |
| Original code | This paper | https://doi.org/10.5281/zenodo.14992672 |
Experimental model and study participant details
Fermentation and production of the dry powder postbiotic product, Probio-Eco®
The postbiotic material was provided by Jinhua Yinhe Biological Technology Co., Ltd. (Zhejiang, China). The production process began with the thorough mixing of soybean powder, skim milk powder, and sodium citrate with water for 15 min, followed by preheating the mixture to a temperature range of 55-60°C. The mixture was then homogenized at a pressure of 18-20 MPa, sterilized at 95°C for 60 min, and subsequently cooled to 35°C. Next, three probiotic strains (Lacticaseibacillus paracasei Zhang, Lactiplantibacillus plantarum P-8, and Bifidobacterium animalis subsp. lactis V9) were inoculated into the mixture for fermentation. The fermentation was conducted with continuous stirring at 35°C until the pH of the broth reached a target range of 4.50 to 4.60. Following fermentation, the broth was pasteurized at 75°C for 15 min, followed by sterile homogenization at 25-30 MPa. The Probio-Eco® powder was obtained though spray-drying, with a feed temperature maintained at 60°C. The inlet and outlet air temperatures were set to 210°C and 88°C, respectively. Finally, the postbiotic powder was compressed into triangular tablets using a ZPW23 rotary tablet press (Shanghai Tianxiang & Chentai Pharmaceutical Machinery Co., Ltd., Shanghai, China). All procedures were performed in strict adherence to Good Manufacturing Practice.
Determination of major bioactive metabolites in Probio-Eco®
To determine the major bioactive metabolites in Probio-Eco®, a postbiotic sample of 0.1 g was accurately weighed and placed into a 2 mL centrifuge tube. To this sample, 600 μL of methanol, pre-cooled to -20°C, and 4 ppm of 2-amino-3-(2-chloro-phenyl)-propionic acid were added. The mixture was vortexed for 30 s and then subjected to ultrasonication at room temperature for 10 min. Following ultrasonication, the sample was centrifuged at 12,000 rpm for 10 min at 4°C. The resulting supernatant was filtered through a 0.22 μm membrane and transferred into an appropriate sample vial for subsequent liquid chromatograph-mass spectrometry analysis. Triplicate samples were prepared in parallel.
For the separation of metabolites, an Agilent 1290 Infinity LC Ultra-High-Performance Liquid Chromatography system (Agilent Technologies, Inc., Santa Clara, CA, USA) and a C18 chromatographic column (2.1 × 100 mm, 1.7 μm; Waters Corporation, Milford, MA, USA) were used. The AB 6500 + QTRAP mass spectrometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used for mass spectrometry analysis.
Human participants
We conducted a comprehensive randomized, double-blind, placebo-controlled crossover trial in collaboration with Inner Mongolia People’s Hospital from November 2021 to January 2022. The sample size calculation was based on the primary outcome variable, the weekly mean CSBMs. Drawing on insights from referenced studies and expert opinions from gastroenterologists,84,86 we anticipated a difference of 0.9 in CSBMs between the intervention and control groups, with a standard deviation of 1.3. To achieve a two-sided significance level of 5% and a power of 80%, we determined that a minimum sample size of 40 subjects per group was required, accounting for an expected dropout rate of less than 20%. Initially, 125 subjects were screened for eligibility to participate in this study. Of these, 15 were excluded due to not meeting the inclusion criteria, declining participation, or other reasons (Figure S1A). Consequently, a final cohort of 110 patients with chronic constipation was recruited from Inner Mongolia Agricultural University (Inner Mongolia, China), Nanchang University (Nanchang, China), and Jiangxi University of Chinese Medicine (Nanchang, China).
The inclusion criteria for the study were as follows: males and females aged 18-65 years who met the Rome IV criteria for chronic constipation, based on self-reported symptoms over the past three months,85 with symptom onset within the last six months. Participants had to experience two or more of the following symptoms: 1) straining during more than 25% of defecations; 2) lumpy or hard stools (Bristol Stool Form Scale type 1-2) during more than 25% of defecations; 3) sensation of incomplete evacuation during more than 25% of defecations; 4) sensation of anorectal obstruction or blockage during more than 25% of defecations; 5) at least 25% of defecations requiring manual assistance (such as finger assistance or pelvic floor support); 6) fewer than three spontaneous bowel movements per week. Exclusion criteria included: 1) individual or family history of colon cancer, celiac disease, or inflammatory bowel disease; 2) prior colonoscopy diagnosis of organic gastrointestinal diseases; 3) recent conception, pregnancy, or breastfeeding; 4) allergies to any study materials; 5) use of antibiotics, probiotic, or postbiotics within the past two weeks; 6) use of psychotropic medications within the past month; 7) need for long-term constipation medications; 8) serious medical conditions such as myocardial infarction, cerebral infarction, malignancy; 9) mental illnesses that could preclude study participation.
The trial comprised a 1-week run-in period, followed by two reciprocal 3-week intervention periods of either postbiotic or placebo, separated by a 2-week washout period. After the run-in phase, participants were randomized into two groups using a simple randomization method: Group A (received postbiotic first, followed by placebo; n = 54) and Group B (received placebo first, followed by postbiotic; n = 56). Randomization was executed by two independent project administrators using a computer-generated random number sequence to assign participants to their respective groups. All subjects, physicians, and investigators involved in the trial were blinded, and the group assignment information was concealed in opaque envelopes, which were opened only prior to the primary data analysis. Ultimately, 105 subjects successfully completed the trial and demonstrated good compliance with the protocol (Group A: n =52; Group B: n = 53; Figure S1A). An ITT analysis was also performed on all 110 subjects who participated in the study, revealing no significant differences in gender distribution between groups A and B.
The postbiotic intervention used was Probio-Eco®, which contained an inactivated bacterial count of 30 billion/g, as verified by flow cytometry. Participants received the intervention twice daily, with each dose consisting of six tablets (0.6 g per tablet). The placebo intervention consisted of soybean powder, skim milk powder, and sodium citrate. Both the postbiotic and placebo materials were provided by Jinhua Yinhe Biological Technology Co., Ltd. (Zhejiang, China) and were prepared in identical shape, texture, and appearance. For each 3-week intervention period, participants received a 21-day supply of either the postbiotic or placebo material, corresponding to their assigned group. The two spare doses were provided as a precaution against potential loss or damage. Compliance with the postbiotic intervention was assessed by counting the returned doses, with the compliance rate calculated as follows: [(Number of actual dispensed dose - Number of returned dose) / Number of prescribed doses] × 100%.
Mouse models
Sixty males C57BL/6 mice (6 weeks old; average body weight of 22.4 ± 0.8 g) were obtained from Charles River Labs Co., Ltd (Beijing, China) for this animal study. The mice were maintained in a specific pathogen-free animal facility, where they were maintained under controlled environmental conditions (temperature: 22 ± 2°C; humidity: 45% ± 5%) with a 12-h light-dark cycle. They were provided with a standard commercial diet and sterile water ad libitum throughout the study.
A mouse model of constipation was induced by administering loperamide. The effects of the postbiotic and other bioactive metabolites on constipation were assessed by their co-administering alongside loperamide. The metabolite intervention doses were determined based on previous literature.87,88,89 Mice were acclimatized for one week before being randomized into six groups (n = 10 per group), including: the Con group (received normal saline), the Mod group (received 15 mg/kg of loperamide), the Post group (received 15 mg/kg of loperamide and 500 mg/kg of Probio-Eco®), the Succ group (received 15 mg/kg of loperamide and 200 mg/kg of Succ), the 3-IA group (received 15 mg/kg of loperamide and 20 mg/kg of 3-IA), and the 5-HTP group (received 15 mg/kg of loperamide and 20 mg/kg of 5-HTP).
The intervention trial was conducted in two distinct stages: the loperamide-induced constipation modeling stage and the intervention stage. The modeling stage lasted 7 days, during which the Con group received daily intragastric gavage of 0.1 mL of saline, while the other groups received 0.1 mL of loperamide hydrochloride. The intervention stage lasted 14 days, during which the Con group continued to receive daily intragastric gavage of 0.1 mL of saline. In contrast, the remaining groups received 0.1 mL of loperamide hydrochloride. One hour later, the Con and Mod groups received an additional 0.1 mL of saline, while the other groups received 0.1 mL of postbiotic, Succ, 3-IA, and 5-HTP (dissolved in saline), respectively. Throughout the trial, food and water intake were recorded every other day at 9:00 am, and mouse body weights were measured weekly. Mouse fecal samples were collected at the beginning and end of both intervention periods.
At the end of the trial, after a 12-h fasting period, orbital blood samples were collected from all mice, followed by euthanasia via cervical dislocation. The blood samples were centrifuged (3,000 x g, 10 min) to obtain the sera, which were stored at -80°C for later use. The entire intestine, including the duodenum, ileum, colon, and cecum, was collected and stored in 0.5 mL sterile tubes. These tissue samples were quickly frozen in liquid nitrogen and immediately kept at -80°C for later use.
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Inner Mongolia Agricultural University (Permit No: SYXK-2020-0002) and were approved by the Animal Ethics Committee of Inner Mongolia Agricultural University (No: NND2023105).
Method details
Clinical endpoint definitions
The primary endpoint of this study was the weekly mean frequency of CSBMs, with a lower score indicating more severe constipation symptoms. Secondary endpoints included several parameters: the weekly mean stool consistency score, assessed using the Bristol Stool Form Scale, which categorizes stool types on an ordinal scale from the hardest (type 1) to the softest (type 7), scoring from 1 to 7, respectively; the weekly mean stool straining score, which was graded from 0 (not difficult), 1 (a little difficult, need some straining to defecate), 2 (difficult, need straining to defecate), 3 (very difficult, need hard straining to defecate); two comprehensive questionnaires, PAC-QoL and DASS-21, that evaluate the multi-dimensional impacts of constipation on patients’ quality of life and psychological well-being, respectively; and fecal metagenomics and metabolomics analyses. Data for the stool-related endpoints were collected using a daily electronic stool diary, with means calculated for each dimension at designated time points (days 7, 14, 21, 35, 42, 49, and 56). Adverse events, including systemic infections, deleterious metabolic activities, excessive immune stimulation, and gastrointestinal side effects, were recorded throughout the intervention period to ensure participant safety and assess the tolerability of the treatments.
Human fecal microbiome sample preparation
Fecal samples were self-collected by participants using sterile stool samplers, with a DNA protection solution (Guangdong Longsee Biomedical Co., Ltd., Guangzhou, China) added immediately after sampling. Samples were collected on days 0, 21, 35, and 56, and stored at -80°C until further metagenomics and metabolomics analyses. Fecal metagenomics analysis was conducted on samples from 103 subjects in the PP population. Metagenomic DNA was extracted from the collected stool samples using the Magnetic Soil and Stool DNA Kit (DP712; TIANGEN Biotech Co., Ltd., Beijing, China) following previously established procedures.12 Shotgun sequencing was performed on the extracted DNA samples using the Illumina Novaseq 6000 (Illumina Inc., San Diego, CA, USA). Notably, metagenomic sequencing failed for four samples from four different participants, resulting in a total of 408 successfully sequenced samples (Table S12).
Human fecal untargeted metabolomic sample preparation
Untargeted metabolomic analysis was performed on stool samples from the PP population. However, five samples were excluded due to insufficient stool volume, which posed challenges for fecal metabolomics analysis. A total of 407 samples were analyzed across all time points. Fecal samples were dried and then mixed with 600 μL of a methanol solution containing 2-chlorophenylalanine. The mixture was vortexed for 30 s, before grinding for 60 s at 55 Hz in the presence of 100 mg of glass beads. After ultrasonication and centrifugation at 12,000 rpm for 10 min, the supernatant was filtered through a 0.22 μm ultrafiltration membrane. Chromatographic separations were performed according to a previous work.12 Briefly, the separation was achieved using an ACQUITY UPLC BEH amide column (100 × 2.1 mm, 1.7 μm; Waters Corporation, Milford, MA, USA) at 25°C with a flow rate of 0.5 mL/min. The mobile phase consisted of a mixture of 25 mM ammonium acetate and 25 mM ammonia in water (A) and acetonitrile (B). Each sample was analyzed in both positive and negative ionization modes under electrospray ionization conditions.
Human fecal targeted metabolomics sample preparation
Fecal bioactive metabolites (SCFAs, BAs, organic acids, and amino acids) were isolated partly according to previously published methods.90,91,92 In brief, 100 mg of thawed feces was combined with 0.9 mL of a methanol-water solution (4:1, v/v), which was then homogenized using an OMNI Bead Ruptor at 55Hz for 1 min. The suspension was then vortexed and centrifuged at 12,000 × g for 10 min, and the supernatant was collected and stored at -20°C. Unlike other bioactive metabolites, the determination of SCFAs required a derivatization step. Specifically, 40 μL of the sample supernatant was mixed with 20 μL of 3-nitrophenylhydrazine (200 mM) and 20 μL of an N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide (EDC)-6% pyridine solution (120 mM). After incubation at 40°C for 30 min, 1.92 mL of a 10% acetonitrile-water solution was added to terminate the reaction. The sample was then centrifuged (12,000 × g, 4°C, 10 min), and 500 μL of the supernatant was filtered through a 0.22 μm membrane into a sample vial for subsequent analysis.
Record of constipation-related indicators in mice
Constipation-related indicators, including fecal weight, water content, first black stool defecation time, and small intestinal transit rate, were measured according to methods described in previous studies.93,94 Fecal water content was calculated on days 0, 7, 14, and 21. This was determined by the ratio of the difference between the wet and dry weights of the feces to the wet weight. The first black stool defecation time was determined on days 7 and 21, defined as the interval between the administration of activated carbon powder and the appearance of darkened feces. The small intestinal transit rate was evaluated on day 21 by measuring the distance traveled by the activated carbon solution relative to the total length of the small intestine.
Mice serum SCFAs sample preparation
The extraction and preparation of serum metabolites, along with quantification methods, were adapted from a prior study.95 Briefly, serum samples were centrifuged at 10,000 g for 10 min to remove particulates. Then, 100 μL of each serum sample was mixed with 400 μL of acetonitrile for 30 s to precipitate proteins. The mixtures were centrifuged again at 10,000 g for 10 min, and the supernatants were filtered through a 0.22 mm syringe filter. A 200 μL aliquot of each filtered supernatant was analyzed using Ultra-Performance Liquid Chromatography coupled with a Triple Quadrupole Mass Spectrometer (SCIEX QTRAP 6500+, CA, USA). Serum SCFA concentrations were quantified using the external standard method with analytical standards purchased from Sigma-Aldrich, Inc. (St. Louis, MO, USA).
Histological analysis of mice colon
Proximal colon tissue samples were fixed in 4% paraformaldehyde, embedded in paraffin wax, and sectioned to a thickness of 4 μm. The fixed tissue sections were deparaffinized with xylene, rehydrated through a graded series of ethanol, and rinsed with distilled water. Hematoxylin and eosin staining were performed to assess morphological changes in the colon tissue. Additionally, mucus-producing cells in the colon were stained using an alcian blue-periodic acid-Schiff staining kit (Nanjing BioChannel Biotechnology Co., Ltd., Nanjing, China). Wheat germ agglutinin conjugated to fluorescein isothiocyanate (ZCIBIO Technology Co., Ltd., Shanghai, China) was used to visualize glycosylated mucin expression in the colon sections. All staining procedures were performed according to the manufacturer’s instructions.
RNA extraction and real-time quantitative PCR of mice colon tissue
Total RNA was extracted from 30 mg of proximal colon tissue samples using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA). One microgram of the isolated RNA was reverse transcribed into complementary DNA using a cDNA first strand synthesis kit (Merck, Darmstadt, Germany). For real-time quantitative PCR analysis, 600 ng of complementary DNA was used as the amplification template, with reactions performed using SYBR qPCR Master Mix (Tianya Biotech, Guangzhou, China) on a BIO-RAD CFX Connect Real-Time PCR Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The primers sequences used to evaluate the expression of various genes encoding claudin-1, tight junction protein ZO-1, proto-oncogene c-Kit, aquaporin-3, aquaporin-4, peptide YY, mucin-2, aromatic L-amino acid decarboxylase, glucagon-like peptide-1 are provided in Table S13. The gene amplification protocol was as follows: initiation at 95°C for 10 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Target gene expression was normalized to the endogenous reference gene, β-actin.
Quantification and statistical analysis
Human fecal microbiome data analysis
A total of 2.52 Tbp of high-quality clean data were generated (mean = 6.32 Gbp/sample; Table S12). The clean data were assembled into contigs using MEGAHIT,78 and contigs larger than 2,000 bp were screened for binning using MetaBAT2,79 VAMB,80 and DAS Tool81 with default parameters. All resulting bins were then combined to generate metagenome-assembled genomes using custom scripts. The quality of these metagenome-assembled genomes was evaluated using CheckM,82 categorizing them into high-, medium-, and partial-quality based on completeness and contamination metrics. High-quality genomes were clustered, and the most representative genomes were selected to obtain SGBs using dRep (with parameters: -pa 0.95 and -sa 0.95).83 Taxonomic annotation of the SGBs was performed using Kraken2 and the NCBI Non-Redundant Protein Sequence Database. The relative abundance of each SGB was calculated using the CoverM tool (https://github.com/wwood/CoverM, with parameters set to --min-read-percent-identity 0.95 --min-covered-fraction 0.4.
To predict the GMMs encoded by the SGBs, relevant literature and the MetaCyc metabolic database were consulted.12,96 Open reading frames were annotated against the KEGG database to identify key metabolic modules associated with each SGB, and the distribution of synthesis or degradation modules within the SGBs was determined using Omixer-RPM, with the parameter set to -c 0.66.
Human fecal phageome identification and analysis
The gut DNA phageome was characterized through several steps. Firstly, potential bacteriophage features were identified from contigs greater than 1,000 bp using VIBRANT and CheckV.97,98 Secondly, viral contigs greater than 5,000 bp were selected and clustered into vOTUs based on a threshold of 95% nucleotide identity over 80% of the sequence length, using CD-HIT (https://github.com/weizhongli/cdhit).99 This process resulted in the identification of 158,640 vOTUs, which were then compared against a curated set of 189,680 viral genomes from the Metagenomic Gut Virus catalogue to assess the novelty of the identified vOTUs within our dataset.100 Finally, the average abundance of these vOTUs across the viral population was calculated using the CoverM contig pipeline (https://github.com/wwood/CoverM), applying parameters: --min-read-percent-identity 0.95, --min-read-aligned-percent 0.5, --proper-pairs-only, and –exclude-supplementary.
Human untargeted fecal metabolomics analysis
The raw data generated by ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry was processed using ProteoWizard software and the R package XCMS. Principal component analysis and partial least squares-discriminant analysis were applied to identify differential biomarker metabolites between groups (cut-off level: VIP value > 2 and P-value < 0.05). Further identification of selected biomarkers and potential metabolic pathways was achieved by searching the HMDB (http://www.hmdb.ca/), METLIN (http://metlin.scripps.edu/), Massbank (http://www.massbank.jp), and KEGG (http://www.kegg.com/) databases.
Human targeted fecal metabolomics analysis
Prepared samples were analyzed on a QTRAP 6500+ UPLC-MS system (AB Sciex LLC., Framingham, MA, USA) equipped with a Kinetex® EVO C18 100A column (2.6 μm, 2.1 × 100 mm; Phenomenex, Torrance, CA, USA). Standard curves were generated using concentration gradients and peak area ratios from constructed standards (Sigma-Aldrich, Inc., St. Louis, MO, USA), and the external standard method was employed to quantitatively analyze the concentrations of various bioactive metabolites of interest in the samples.
Mice fecal metagenomics analysis
Fecal DNA extraction from mouse samples, metagenomic sequencing, and subsequent data analysis, including sequence quality control, read assembly, binning, de-redundancy, species annotation, and abundance calculation, were achieved using the same methods and pipeline described in the human fecal metagenomic analysis.
Measurements of serum gastrointestinal regulatory peptides and immune factors
Serum levels of gastrointestinal regulatory peptides (motilin, gastrin, and vasoactive intestinal peptide), 5-HT, immune factors (IL-6, IL-10, IL-1β and IL-22, and AhR) were quantified using ELISA kits (Meimian Biotechnology, Jiangsu, China), following the instructions in the operation manuals.
Statistical analyses
All statistical analyses were conducted using R software (version 4.3.2), with data visualizations created in Adobe Illustrator. Results are presented as mean ± standard deviation. We evaluated differences in demographic and baseline characteristics, including gender, body mass index, allergy history, drug use, and adverse events, between Groups A and B using the chi-square test. Inter- and intragroup differences in primary and secondary outcomes were evaluated using either the unpaired or two-sided paired Wilcoxon rank-sum test. To address missing data in the ITT population analysis, the baseline-observation-carried-forward method was utilized for imputation, ensuring a comprehensive assessment of the entire population and a robust evaluation of the study findings. Additionally, sensitivity analyses for the primary outcome were conducted using a mixed-effects model, incorporating treatment group, time, and time × group interaction as fixed effects, along with a random intercept for subjects, using the nlme package in R. Non-parametric covariance analysis, with baseline data as covariates, was also performed using the sm package in R. The normality of the animal model data was evaluated by the Shapiro-Wilk test, while differences between skewed and normally distributed data across multiple groups were compared using the Kruskal-Wallis test and ANOVA, following by corresponding post hoc analyses. The Levene’s test was employed to evaluate the homogeneity of variances among groups, and the least significant difference (LDA) post hoc test was further applied. The Benjamini-Hochberg procedure was implemented to adjust P-values for multiple comparisons, with P < 0.05 considered statistically significant.
To calculate the GMM modules, we leveraged the dplyr R package, using the formula: cumulative abundance of GMM modules = number of metabolic modules encoded in the genomes × genome abundance. We calculated and visualized multivariable association using R packages including MaAsLin2, tidyverse, and ggplot2. Additionally, we conducted Spearman correlation analysis to investigate relationships between specific response markers and the defecation parameters in mice treated with Succ, 3-IA, and 5-HTP. This comprehensive analytical approach enabled us to identify potential links between gut microbiome, its metabolite profile, and clinical outcomes observed in our study. Other statistical analyses were performed using a suite of well-established R packages, including vegan, optparse, mixOmics, ggplot2, and ggpubr. We leveraged these tools to calculate Shannon and Simpson’s diversity indices, as well as to execute principal component analysis, principal coordinates analysis, partial least squares-discriminant analysis, adonis test, and Procrustes analysis.
Additional resources
The study was approved by the Ethics Committee of Inner Mongolia People’s Hospital (Approval number: 2021004). It was registered on the Chinese Clinical Trial Registry (http://www.chictr.org.cn; ChiCTR2100054376). The investigation conformed to the principles of the Declaration of Helsinki, with all subjects provided informed consent prior to participation in the study.
Published: April 25, 2025
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2025.102093.
Supplemental information
References
- 1.Bharucha A.E., Lacy B.E. Mechanisms, Evaluation, and Management of Chronic Constipation. Gastroenterology. 2020;158:1232–1249.e3. doi: 10.1053/j.gastro.2019.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rao S.S.C., Yu S., Fedewa A. Systematic review: dietary fibre and FODMAP-restricted diet in the management of constipation and irritable bowel syndrome. Aliment. Pharmacol. Ther. 2015;41:1256–1270. doi: 10.1111/apt.13167. [DOI] [PubMed] [Google Scholar]
- 3.Wlodarczyk J., Wasniewska A., Fichna J., Dziki A., Dziki L., Wlodarczyk M. Current Overview on Clinical Management of Chronic Constipation. J. Clin. Med. 2021;10 doi: 10.3390/jcm10081738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y., Yu Y.B. Intestinal microbiota and chronic constipation. SpringerPlus. 2016;5:1130. doi: 10.1186/s40064-016-2821-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prochazkova N., Falony G., Dragsted L.O., Licht T.R., Raes J., Roager H.M. Advancing human gut microbiota research by considering gut transit time. Gut. 2023;72:180–191. doi: 10.1136/gutjnl-2022-328166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Z., Tang J., Hu Y., Zhang W. Role of gut microbiota-derived signals in the regulation of gastrointestinal motility. Front. Med. 2022;9 doi: 10.3389/fmed.2022.961703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan Y., Young R., Schnabl B. Bacteriophages and their potential for treatment of gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2022;19:135–144. doi: 10.1038/s41575-021-00536-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mihindukulasuriya K.A., Mars R.A.T., Johnson A.J., Ward T., Priya S., Lekatz H.R., Kalari K.R., Droit L., Zheng T., Blekhman R., et al. Multi-omics analyses show disease, diet, and transcriptome interactions with the virome. Gastroenterology. 2021;161:1194–1207.e8. doi: 10.1053/j.gastro.2021.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang T., Liu W., Lu H., Cheng T., Wang L., Wang G., Zhang H., Chen W. Lactic acid bacteria in relieving constipation: Mechanism, clinical application, challenge, and opportunity. Crit. Rev. Food Sci. Nutr. 2025;65:551–574. doi: 10.1080/10408398.2023.2278155. [DOI] [PubMed] [Google Scholar]
- 10.Jin H., Ma T., Chen L., Kwok L.Y., Quan K., Li Y., Zhang Z., Chen T., Zhang J., Sun Z., Zhang H. The iLABdb: a web-based integrated lactic acid bacteria database. Sci. Bull. 2023;68:2527–2530. doi: 10.1016/j.scib.2023.09.016. [DOI] [PubMed] [Google Scholar]
- 11.Wang L., Wang L., Tian P., Wang B., Cui S., Zhao J., Zhang H., Qian L., Wang Q., Chen W., Wang G. A randomised, double-blind, placebo-controlled trial of Bifidobacterium bifidum CCFM16 for manipulation of the gut microbiota and relief from chronic constipation. Food Funct. 2022;13:1628–1640. doi: 10.1039/d1fo03896f. [DOI] [PubMed] [Google Scholar]
- 12.Ma T., Yang N., Xie Y., Li Y., Xiao Q., Li Q., Jin H., Zheng L., Sun Z., Zuo K., et al. Effect of the probiotic strain, Lactiplantibacillus plantarum P9, on chronic constipation: A randomized, double-blind, placebo-controlled study. Pharmacol. Res. 2023;191 doi: 10.1016/j.phrs.2023.106755. [DOI] [PubMed] [Google Scholar]
- 13.Ma T., Shen X., Shi X., Sakandar H.A., Quan K., Li Y., Jin H., Kwok L.-Y., Zhang H., Sun Z. Targeting gut microbiota and metabolism as the major probiotic mechanism-An evidence-based review. Trends Food Sci. Technol. 2023;138:178–198. doi: 10.1016/j.tifs.2023.06.013. [DOI] [Google Scholar]
- 14.Salminen S., Collado M.C., Endo A., Hill C., Lebeer S., Quigley E.M.M., Sanders M.E., Shamir R., Swann J.R., Szajewska H., Vinderola G. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021;18:649–667. doi: 10.1038/s41575-021-00440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nataraj B.H., Ali S.A., Behare P.V., Yadav H. Postbiotics-parabiotics: the new horizons in microbial biotherapy and functional foods. Microb. Cell Fact. 2020;19:168. doi: 10.1186/s12934-020-01426-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosca A., Abreu Y Abreu A.T., Gwee K.A., Ianiro G., Tack J., Nguyen T.V.H., Hill C. The clinical evidence for postbiotics as microbial therapeutics. Gut Microbes. 2022;14 doi: 10.1080/19490976.2022.2117508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei Y., Huang N., Ye X., Liu M., Wei M., Huang Y. The postbiotic of hawthorn-probiotic ameliorating constipation caused by loperamide in elderly mice by regulating intestinal microecology. Front. Nutr. 2023;10 doi: 10.3389/fnut.2023.1103463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andresen V., Gschossmann J., Layer P. Heat-inactivated Bifidobacterium bifidum MIMBb75 (SYN-HI-001) in the treatment of irritable bowel syndrome: a multicentre, randomised, double-blind, placebo-controlled clinical trial. Lancet Gastroenterol. Hepatol. 2020;5:658–666. doi: 10.1016/S2468-1253(20)30056-X. [DOI] [PubMed] [Google Scholar]
- 19.Okada K., Takami D., Makizaki Y., Tanaka Y., Nakajima S., Ohno H., Sagami T. Effects of Bifidobacterium longum CLA8013 on bowel movement improvement: a placebo-controlled, randomized, double-blind study. Biosci. Microbiota Food Health. 2023;42:213–221. doi: 10.12938/bmfh.2022-066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng C., Peng C., Zhang W., Zhang T., He Q., Kwok L.Y., Zhang H. Postbiotic Administration Ameliorates Colitis and Inflammation in Rats Possibly through Gut Microbiota Modulation. J. Agric. Food Chem. 2024;72:9054–9066. doi: 10.1021/acs.jafc.3c03901. [DOI] [PubMed] [Google Scholar]
- 21.Xu H., Ma C., Zhao F., Chen P., Liu Y., Sun Z., Cui L., Kwok L.Y., Zhang H. Adjunctive treatment with probiotics partially alleviates symptoms and reduces inflammation in patients with irritable bowel syndrome. Eur. J. Nutr. 2021;60:2553–2565. doi: 10.1007/s00394-020-02437-4. [DOI] [PubMed] [Google Scholar]
- 22.Chen P., Xu H., Tang H., Zhao F., Yang C., Kwok L.Y., Cong C., Wu Y., Zhang W., Zhou X., Zhang H. Modulation of gut mucosal microbiota as a mechanism of probiotics-based adjunctive therapy for ulcerative colitis. Microb. Biotechnol. 2020;13:2032–2043. doi: 10.1111/1751-7915.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q., Ma T., Feng C., Li Y., Jin H., Shi X., Kwok L.Y., Shi Y., Chen T., Zhang H. Adjuvant postbiotic administration improves dental caries prognosis by restoring the oral microbiota. Food Sci. Hum. Wellness. 2024;13:2690–2702. doi: 10.26599/FSHW.2022.9250217. [DOI] [Google Scholar]
- 24.Bharucha A.E., Wald A. Chronic constipation. Mayo Clin. Proc. 2019;94:2340–2357. doi: 10.1016/j.mayocp.2019.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbasi A., Rad A.H., Ghasempour Z., Sabahi S., Kafil H.S., Hasannezhad P., Rahbar Saadat Y., Shahbazi N. The biological activities of postbiotics in gastrointestinal disorders. Crit. Rev. Food Sci. Nutr. 2022;62:5983–6004. doi: 10.1080/10408398.2021.1895061. [DOI] [PubMed] [Google Scholar]
- 26.Ma T., Huang W., Li Y., Jin H., Kwok L.Y., Sun Z., Zhang H. Probiotics alleviate constipation and inflammation in late gestating and lactating sows. NPJ Biofilms Microbiomes. 2023;9:70. doi: 10.1038/s41522-023-00434-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng C., Chan A.O.O., Hui W.M., Lam S.K. Coping strategies, illness perception, anxiety and depression of patients with idiopathic constipation: a population-based study. Aliment. Pharmacol. Ther. 2003;18:319–326. doi: 10.1046/j.1365-2036.2003.01663.x. [DOI] [PubMed] [Google Scholar]
- 28.Rafique N., Jan S.Y., Dar A.H., Dash K.K., Sarkar A., Shams R., Pandey V.K., Khan S.A., Amin Q.A., Hussain S.Z. Promising bioactivities of postbiotics: A comprehensive review. J. Agric. Food Res. 2023;14 doi: 10.1016/j.jafr.2023.100708. [DOI] [Google Scholar]
- 29.Mukhopadhya I., Segal J.P., Carding S.R., Hart A.L., Hold G.L. The gut virome: the ‘missing link’between gut bacteria and host immunity? Ther. Adv. Gastroenterol. 2019;12 doi: 10.1177/1756284819836620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim Y.J., Jamaluddin R., Hazizi A.S., Chieng J.Y. Effects of synbiotics among constipated adults in serdang, selangor, malaysia—a randomised, double-blind, placebo-controlled trial. Nutrients. 2018;10:824. doi: 10.3390/nu10070824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiamani K., Luo S., Schulz S., Xue J., Costa R., Khan Mirzaei M., Deng L. The role of virome in the gastrointestinal tract and beyond. FEMS Microbiol. Rev. 2022;46 doi: 10.1093/femsre/fuac027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y., Pan R., Mei L., Tian P., Wang L., Zhao J., Chen W., Wang G. Colon-Targeted Delivery of Indole Acetic Acid Helps Regulate Gut Motility by Activating the AHR Signaling Pathway. Nutrients. 2023;15 doi: 10.3390/nu15194282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie L., Xu C., Fan Y., Li Y., Wang Y., Zhang X., Yu S., Wang J., Chai R., Zhao Z., et al. Effect of fecal microbiota transplantation in patients with slow transit constipation and the relative mechanisms based on the protein digestion and absorption pathway. J. Transl. Med. 2021;19:490. doi: 10.1186/s12967-021-03152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner E.H., Loftis J.M., Blackwell A.D. Serotonin a la carte: supplementation with the serotonin precursor 5-hydroxytryptophan. Pharmacol. Ther. 2006;109:325–338. doi: 10.1016/j.pharmthera.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Spiller R.C. Effects of serotonin on intestinal secretion and motility. Curr. Opin. Gastroenterol. 2001;17:99–103. doi: 10.1097/00001574-200103000-00001. [DOI] [PubMed] [Google Scholar]
- 36.Gabriele S., Sacco R., Altieri L., Neri C., Urbani A., Bravaccio C., Riccio M.P., Iovene M.R., Bombace F., De Magistris L., Persico A.M. Slow intestinal transit contributes to elevate urinary p-cresol level in Italian autistic children. Autism Res. 2016;9:752–759. doi: 10.1002/aur.1571. [DOI] [PubMed] [Google Scholar]
- 37.Wei Y.H., Ma X., Zhao J.C., Wang X.Q., Gao C.Q. Succinate metabolism and its regulation of host-microbe interactions. Gut Microbes. 2023;15 doi: 10.1080/19490976.2023.2190300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Veledo S., Vendrell J. Gut microbiota-derived succinate: Friend or foe in human metabolic diseases? Rev. Endocr. Metab. Disord. 2019;20:439–447. doi: 10.1007/s11154-019-09513-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian H., Chen Q., Yang B., Qin H., Li N. Analysis of Gut Microbiome and Metabolite Characteristics in Patients with Slow Transit Constipation. Dig. Dis. Sci. 2021;66:3026–3035. doi: 10.1007/s10620-020-06500-2. [DOI] [PubMed] [Google Scholar]
- 40.Song H., Guo R., Sun X., Kou Y., Ma X., Chen Y., Song L., Yuan C., Wu Y. Xylooligosaccharides from corn cobs alleviate loperamide-induced constipation in mice via modulation of gut microbiota and SCFA metabolism. Food Funct. 2023;14:8734–8746. doi: 10.1039/d3fo02688d. [DOI] [PubMed] [Google Scholar]
- 41.Camilleri M., Vijayvargiya P. The Role of Bile Acids in Chronic Diarrhea. Am. J. Gastroenterol. 2020;115:1596–1603. doi: 10.14309/ajg.0000000000000696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vijayvargiya P., Camilleri M. Current Practice in the Diagnosis of Bile Acid Diarrhea. Gastroenterology. 2019;156:1233–1238. doi: 10.1053/j.gastro.2018.11.069. [DOI] [PubMed] [Google Scholar]
- 43.Dalile B., Vervliet B., Bergonzelli G., Verbeke K., Van Oudenhove L. Colon-delivered short-chain fatty acids attenuate the cortisol response to psychosocial stress in healthy men: a randomized, placebo-controlled trial. Neuropsychopharmacology. 2020;45:2257–2266. doi: 10.1038/s41386-020-0732-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghayee H.K., Auchus R.J. Basic concepts and recent developments in human steroid hormone biosynthesis. Rev. Endocr. Metab. Disord. 2007;8:289–300. doi: 10.1007/s11154-007-9052-2. [DOI] [PubMed] [Google Scholar]
- 45.Evrensel A., Ceylan M.E. The Gut-Brain Axis: The Missing Link in Depression. Clin. Psychopharmacol. Neurosci. 2015;13:239–244. doi: 10.9758/cpn.2015.13.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma T., Jin H., Kwok L.Y., Sun Z., Liong M.T., Zhang H. Probiotic consumption relieved human stress and anxiety symptoms possibly via modulating the neuroactive potential of the gut microbiota. Neurobiol. Stress. 2021;14 doi: 10.1016/j.ynstr.2021.100294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin X., Liu Y., Ma L., Ma X., Shen L., Ma X., Chen Z., Chen H., Li D., Su Z., Chen X. Constipation induced gut microbiota dysbiosis exacerbates experimental autoimmune encephalomyelitis in C57BL/6 mice. J. Transl. Med. 2021;19:317. doi: 10.1186/s12967-021-02995-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou X., Mao B., Tang X., Zhang Q., Zhao J., Zhang H., Cui S. Exploring the Dose–Effect Relationship of Bifidobacterium longum in Relieving Loperamide Hydrochloride-Induced Constipation in Rats through Colon-Released Capsules. Int. J. Mol. Sci. 2023;24:6585. doi: 10.3390/ijms24076585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohkusa T., Koido S., Nishikawa Y., Sato N. Gut Microbiota and Chronic Constipation: A Review and Update. Front. Med. 2019;6:19. doi: 10.3389/fmed.2019.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reichardt N., Duncan S.H., Young P., Belenguer A., McWilliam Leitch C., Scott K.P., Flint H.J., Louis P. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Louis P., Flint H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 52.Zhuang M., Shang W., Ma Q., Strappe P., Zhou Z. Abundance of probiotics and butyrate-production microbiome manages constipation via short-chain fatty acids production and hormones secretion. Mol. Nutr. Food Res. 2019;63 doi: 10.1002/mnfr.201801187. [DOI] [PubMed] [Google Scholar]
- 53.de Meij T.G.J., de Groot E.F.J., Eck A., Budding A.E., Kneepkens C.M.F., Benninga M.A., van Bodegraven A.A., Savelkoul P.H.M. Characterization of Microbiota in Children with Chronic Functional Constipation. PLoS One. 2016;11 doi: 10.1371/journal.pone.0164731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan R., Wang L., Xu X., Chen Y., Wang H., Wang G., Zhao J., Chen W. Crosstalk between the gut microbiome and colonic motility in chronic constipation: potential mechanisms and microbiota modulation. Nutrients. 2022;14:3704. doi: 10.3390/nu14183704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang G., Yang S., Sun S., Si Q., Wang L., Zhang Q., Wu G., Zhao J., Zhang H., Chen W. Lactobacillus rhamnosus Strains Relieve Loperamide-Induced Constipation via Different Pathways Independent of Short-Chain Fatty Acids. Front. Cell. Infect. Microbiol. 2020;10:423. doi: 10.3389/fcimb.2020.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang C., Wang L., Liu X., Wang G., Guo X., Liu X., Zhao J., Chen W. The Different Ways Multi-Strain Probiotics with Different Ratios of Bifidobacterium and Lactobacillus Relieve Constipation Induced by Loperamide in Mice. Nutrients. 2023;15 doi: 10.3390/nu15194230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ikarashi N., Kon R., Sugiyama K. Aquaporins in the colon as a new therapeutic target in diarrhea and constipation. Int. J. Mol. Sci. 2016;17:1172. doi: 10.3390/ijms17071172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oh J.K., Vasquez R., Kim S.H., Hwang I.C., Song J.H., Park J.H., Kim I.H., Kang D.K. Multispecies probiotics alter fecal short-chain fatty acids and lactate levels in weaned pigs by modulating gut microbiota. J. Anim. Sci. Technol. 2021;63:1142–1158. doi: 10.5187/jast.2021.e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang C.Y., Chen J.S., Hsu B.M., Hussain B., Rathod J., Lee K.H. Colorectal Cancer Stage-Specific Fecal Bacterial Community Fingerprinting of the Taiwanese Population and Underpinning of Potential Taxonomic Biomarkers. Microorganisms. 2021;9 doi: 10.3390/microorganisms9081548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herp S., Durai Raj A.C., Salvado Silva M., Woelfel S., Stecher B. The human symbiont Mucispirillum schaedleri: causality in health and disease. Med. Microbiol. Immunol. 2021;210:173–179. doi: 10.1007/s00430-021-00702-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roager H.M., Licht T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018;9:3294. doi: 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Obata Y., Castaño Á., Boeing S., Bon-Frauches A.C., Fung C., Fallesen T., de Agüero M.G., Yilmaz B., Lopes R., Huseynova A., et al. Neuronal programming by microbiota regulates intestinal physiology. Nature. 2020;578:284–289. doi: 10.1038/s41586-020-1975-8. [DOI] [PubMed] [Google Scholar]
- 63.Bessede A., Gargaro M., Pallotta M.T., Matino D., Servillo G., Brunacci C., Bicciato S., Mazza E.M.C., Macchiarulo A., Vacca C., et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511:184–190. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gandhi R., Kumar D., Burns E.J., Nadeau M., Dake B., Laroni A., Kozoriz D., Weiner H.L., Quintana F.J. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell–like and Foxp3+ regulatory T cells. Nat. Immunol. 2010;11:846–853. doi: 10.1038/ni.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bansal T., Alaniz R.C., Wood T.K., Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. USA. 2010;107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li S., Qi C., Zhu H., Yu R., Xie C., Peng Y., Yin S.-W., Fan J., Zhao S., Sun J. Lactobacillus reuteri improves gut barrier function and affects diurnal variation of the gut microbiota in mice fed a high-fat diet. Food Funct. 2019;10:4705–4715. doi: 10.1039/C9FO00417C. [DOI] [PubMed] [Google Scholar]
- 67.Wlodarska M., Luo C., Kolde R., d'Hennezel E., Annand J.W., Heim C.E., Krastel P., Schmitt E.K., Omar A.S., Creasey E.A., et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe. 2017;22:25–37.e6. doi: 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mancabelli L., Milani C., Lugli G.A., Turroni F., Mangifesta M., Viappiani A., Ticinesi A., Nouvenne A., Meschi T., van Sinderen D., Ventura M. Unveiling the gut microbiota composition and functionality associated with constipation through metagenomic analyses. Sci. Rep. 2017;7:9879. doi: 10.1038/s41598-017-10663-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Su H., Chen J., Miao S., Deng K., Liu J., Zeng S., Zheng B., Lu X. Lotus seed oligosaccharides at various dosages with prebiotic activity regulate gut microbiota and relieve constipation in mice. Food Chem. Toxicol. 2019;134 doi: 10.1016/j.fct.2019.110838. [DOI] [PubMed] [Google Scholar]
- 70.Shen F., Wang Q., Zhong H., Pan Y., Zhang J., Wang J., Chen M., Feng F., Zhao M. Combination of galacto-oligosaccharides and wheat peptides synergistically relieved constipation via regulating water transport, gut barrier, inflammation and gut microbiota. Food Biosci. 2023;56 doi: 10.1016/j.fbio.2023.103208. [DOI] [Google Scholar]
- 71.Sugiyama Y., Mori Y., Nara M., Kotani Y., Nagai E., Kawada H., Kitamura M., Hirano R., Shimokawa H., Nakagawa A., et al. Gut bacterial aromatic amine production: aromatic amino acid decarboxylase and its effects on peripheral serotonin production. Gut Microbes. 2022;14 doi: 10.1080/19490976.2022.2128605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park J., Hosomi K., Kawashima H., Chen Y.A., Mohsen A., Ohno H., Konishi K., Tanisawa K., Kifushi M., Kogawa M., et al. Dietary Vitamin B1 Intake Influences Gut Microbial Community and the Consequent Production of Short-Chain Fatty Acids. Nutrients. 2022;14 doi: 10.3390/nu14102078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chidambaram S.B., Essa M.M., Rathipriya A.G., Bishir M., Ray B., Mahalakshmi A.M., Tousif A.H., Sakharkar M.K., Kashyap R.S., Friedland R.P., Monaghan T.M. Gut dysbiosis, defective autophagy and altered immune responses in neurodegenerative diseases: Tales of a vicious cycle. Pharmacol. Ther. 2022;231 doi: 10.1016/j.pharmthera.2021.107988. [DOI] [PubMed] [Google Scholar]
- 74.Mawe G.M., Hoffman J.M. Serotonin signalling in the gut—functions, dysfunctions and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 2013;10:473–486. doi: 10.1038/nrgastro.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Verbeek M.M., Geurtz P.B.H., Willemsen M.A.A.P., Wevers R.A. Aromatic L-amino acid decarboxylase enzyme activity in deficient patients and heterozygotes. Mol. Genet. Metab. 2007;90:363–369. doi: 10.1016/j.ymgme.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 76.Koopman N., Katsavelis D., Hove A.S.T., Brul S., Jonge W.J.d., Seppen J. The Multifaceted Role of Serotonin in Intestinal Homeostasis. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22179487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gu Y., Qin X., Zhou G., Wang C., Mu C., Liu X., Zhong W., Xu X., Wang B., Jiang K., et al. Lactobacillus rhamnosus GG supernatant promotes intestinal mucin production through regulating 5-HT4R and gut microbiota. Food Funct. 2022;13:12144–12155. doi: 10.1039/D2FO01900K. [DOI] [PubMed] [Google Scholar]
- 78.Li D., Liu C.M., Luo R., Sadakane K., Lam T.W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 79.Kang D.D., Li F., Kirton E., Thomas A., Egan R., An H., Wang Z. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7 doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nissen J.N., Johansen J., Allesøe R.L., Sønderby C.K., Armenteros J.J.A., Grønbech C.H., Jensen L.J., Nielsen H.B., Petersen T.N., Winther O., Rasmussen S. Improved metagenome binning and assembly using deep variational autoencoders. Nat. Biotechnol. 2021;39:555–560. doi: 10.1038/s41587-020-00777-4. [DOI] [PubMed] [Google Scholar]
- 81.Sieber C.M.K., Probst A.J., Sharrar A., Thomas B.C., Hess M., Tringe S.G., Banfield J.F. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 2018;3:836–843. doi: 10.1038/s41564-018-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olm M.R., Brown C.T., Brooks B., Banfield J.F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11:2864–2868. doi: 10.1038/ismej.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhong B. How to calculate sample size in randomized controlled trial? J. Thorac. Dis. 2009;1:51–54. [PMC free article] [PubMed] [Google Scholar]
- 85.Mearin F., Lacy B.E., Chang L., Chey W.D., Lembo A.J., Simren M., Spiller R. Bowel disorders. Gastroenterology. 2016;150:1393–1407.e5. doi: 10.1053/j.gastro.2016.02.031. [DOI] [PubMed] [Google Scholar]
- 86.Bang K.B., Choi J.H., Park J.H., Lee S., Rho M.-C., Lee S.W., Lee S., Shin J.E. Effect of Portulaca oleracea L. extract on functional constipation: A randomized, double-blind, placebo-controlled trial. Saudi J. Gastroenterol. 2022;28:296–303. doi: 10.4103/sjg.sjg_400_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang F.L., Chen X.W., Wang Y.F., Hu Z., Zhang W.J., Zhou B.W., Ci P.-F., Liu K.X. Microbiota-derived tryptophan metabolites indole-3-lactic acid is associated with intestinal ischemia/reperfusion injury via positive regulation of YAP and Nrf2. J. Transl. Med. 2023;21:264. doi: 10.1186/s12967-023-04109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yuan Y., Xu Y., Xu J., Liang B., Cai X., Zhu C., Wang L., Wang S., Zhu X., Gao P., et al. Succinate promotes skeletal muscle protein synthesis via Erk1/2 signaling pathway. Mol. Med. Rep. 2017;16:7361–7366. doi: 10.3892/mmr.2017.7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sonner J.K., Keil M., Falk-Paulsen M., Mishra N., Rehman A., Kramer M., Deumelandt K., Röwe J., Sanghvi K., Wolf L., et al. Dietary tryptophan links encephalogenicity of autoreactive T cells with gut microbial ecology. Nat. Commun. 2019;10:4877. doi: 10.1038/s41467-019-12776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Han J., Lin K., Sequeira C., Borchers C.H. An isotope-labeled chemical derivatization method for the quantitation of short-chain fatty acids in human feces by liquid chromatography–tandem mass spectrometry. Anal. Chim. Acta. 2015;854:86–94. doi: 10.1016/j.aca.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 91.Cumeras R., Shen T., Valdiviez L., Tippins Z., Haffner B.D., Fiehn O. Differences in the Stool Metabolome between Vegans and Omnivores: Analyzing the NIST Stool Reference Material. Metabolites. 2023;13:921. doi: 10.3390/metabo13080921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiang X., Chen J., Zhu M., Gao H., Liu X., Wang Q. Multiomics Revealed the Multi-Dimensional Effects of Late Sleep on Gut Microbiota and Metabolites in Children in Northwest China. Nutrients. 2023;15 doi: 10.3390/nu15204315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chai M., Wang L., Li X., Zhao J., Zhang H., Wang G., Chen W. Different Bifidobacterium bifidum strains change the intestinal flora composition of mice via different mechanisms to alleviate loperamide-induced constipation. Food Funct. 2021;12:6058–6069. doi: 10.1039/D1FO00559F. [DOI] [PubMed] [Google Scholar]
- 94.Huang J., Lin B., Zhang Y., Xie Z., Zheng Y., Wang Q., Xiao H. Bamboo shavings derived O-acetylated xylan alleviates loperamide-induced constipation in mice. Carbohydr. Polym. 2022;276 doi: 10.1016/j.carbpol.2021.118761. [DOI] [PubMed] [Google Scholar]
- 95.Sun B., Ma T., Li Y., Yang N., Li B., Zhou X., Guo S., Zhang S., Kwok L.Y., Sun Z., Zhang H. Bifidobacterium lactis Probio-M8 Adjuvant Treatment Confers Added Benefits to Patients with Coronary Artery Disease via Target Modulation of the Gut-Heart/-Brain Axes. mSystems. 2022;7 doi: 10.1128/msystems.00100-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sun H., Zhao F., Liu Y., Ma T., Jin H., Quan K., Leng B., Zhao J., Yuan X., Li Z., et al. Probiotics synergized with conventional regimen in managing Parkinson’s disease. NPJ Parkinsons Dis. 2022;8:62. doi: 10.1038/s41531-022-00327-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kieft K., Zhou Z., Anantharaman K. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome. 2020;8:90. doi: 10.1186/s40168-020-00867-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nayfach S., Camargo A.P., Schulz F., Eloe-Fadrosh E., Roux S., Kyrpides N.C. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 2021;39:578–585. doi: 10.1038/s41587-020-00774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nayfach S., Páez-Espino D., Call L., Low S.J., Sberro H., Ivanova N.N., Proal A.D., Fischbach M.A., Bhatt A.S., Hugenholtz P., Kyrpides N.C. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat. Microbiol. 2021;6:960–970. doi: 10.1038/s41564-021-00928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw sequencing data are available in the CNGB Sequence Archive of China National GeneBank DataBase (CNGBdb; https://db.cngb.org/cnsa/) under accession numbers CNGBdb: CNP0005022 (human) and CNGBdb: CNP0005187 (mouse) and will be publicly accessible upon publication. The metabolomics data are available in MassIVE: MSV000096310 and are publicly available and accessible upon publication.
-
•
The original code has been deposited at GitHub and Zenodo and is available at https://github.com/TengMa-Cleap/Postbiotics-to-relieve-chronic-constipation (https://doi.org/10.5281/zenodo.14992672).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.