Abstract
Background:
The DNA methylation statuses of PAX1 and ZNF582 show great promise as biomarkers for the detection of oral squamous cell carcinoma (OSCC). This study aims to investigate the distribution of PAX1 or ZNF582 methylation in the exfoliated oral epithelial cells (OECs) of OSCC.
Methods:
Methylation data from 528 tumors and 50 adjacent nontumor tissues were acquired from The Cancer Genome Atlas and analyzed using UALCAN database. Sixty-one OSCC cases from Peking University School and Hospital of Stomatology were included in this study and the exfoliated OECs collected by oral swabs were collected from the cancerous lesion (CL), adjacent normal (AN), and contralateral normal (CN) sites. The methylation levels of these 2 genes in different sites were evaluated.
Results:
PAX1 and ZNF582 were both hypermethylated in OSCC compared with nontumor sites but showed different methylation patterns within the oral environment. Generally, a CL-centric methylation pattern of PAX1 where methylation levels decrease gradually from CL through AN to CN was observed, suggesting a field cancerization effect. ZNF582 methylation levels are significantly higher at lesion sites compared with normal sites, but no significant difference is observed between AN and CN. Coexistence of ZNF582 methylation in CL and AN or CN sites was also observed in some patients with OSCC. Furthermore, ZNF582 methylation was more sensitive among patients with OSCC.
Conclusions:
DNA methylation detection of PAX1 and ZNF582 in the exfoliated OECs is helpful for OSCC diagnosis. Hypermethylated PAX1 and ZNF582 show different methylation patterns in the oral cavity of patients with OSCC.
Keywords: Oral squamous cell carcinoma, DNA methylation, oral swab, PAX1, ZNF582, field cancerization, tumor suppressor gene
Introduction
Oral squamous cell carcinoma (OSCC) is the most prevalent head and neck squamous cell carcinoma (HNSCC), 1 which is more prevalent in Asian countries than in Western countries. 2 OSCC can severely impact patients’ functions, appearance, quality of life, and survival outcomes. 3 It is characterized by a high rate of cervical lymph node metastasis and a tendency for recurrence. 4 The Asian region is a high-incidence and high-mortality area for HNSCC. 5 According to follow-up data of Chinese cancer patients from 2003 to 2021,6,7 the 5-year survival rate of patients with HNSCC increased from 42.2% to 52.1%. With the improvements in medical techniques and quality, early diagnosis and treatment are becoming increasingly effective in enhancing patient outcomes. 8 Thus, the development of biomarkers for OSCC as well as precancerous status has increased in recent decades.9-11
Tumor suppressor gene (TSG) methylation has been proven to be a promising marker for OSCC detection,9,12 a predictor of OSCC prognosis,13,14 and a predictor of malignant transformation of oral epithelial dysplasia. 11 Most studies focused on the methylation status in the lesion sites of OSCC,13,15-17 while a few mentioned about the overall oral environment, including adjacent normal sites 13 or contralateral normal sites. 18 In terms of the risk factors, areca nut chewing, 19 smoking, 20 and alcohol consumption 21 are considered to be associated with OSCC incidence. These factors might increase the hypermethylation of TSGs and induce carcinogenesis. Several studies have reported that diet, 22 inflammatory factors, 23 and other risk factors might also trigger carcinogenesis by increasing TSG methylation. However, some of the associated risk factors and the underlying mechanism are still unclear.
It has been proposed that DNA methylation pattern change start in the early stage of carcinogenic process. 24 Our previous study demonstrated that hypermethylated PAX1 and ZNF582 can be detected in tissue samples and the methylation levels are associated with the aggressive progression of OSCC. 14 Besides, we have confirmed that methylation levels of PAX1 and ZNF582 detected in oral swab samples from lesion sites accurately reflect those in tissue specimens and the status of OSCC carcinogenesis. 12 Thus, in this study, we aimed to investigate the hypermethylation of PAX1 and ZNF582 through collecting oral epithelial cells (OECs) by oral swabs from 3 different parts of patients with OSCC, to explore the hypermethylation pattern in the oral cavity of patients with OSCC. In addition, the associations between the coexistence of DNA methylation at cancer-adjacent and contralateral normal sites and risk factors or clinicopathological features among patients with OSCC with PAX1 and ZNF582 hypermethylation at cancerous lesion (CL) sites were further evaluated.
Methods
This study was performed at Peking University School and Hospital of Stomatology and the Chinese Clinical Trial Registry ID is ChiCTR1800015542. The reporting of this study conforms to the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement 25 (Supplementary Table).
DNA methylation analysis
The methylation and expression levels of PAX1 and ZNF582 in HNSCC tumors and adjacent nontumor tissues were obtained from The Cancer Genome Atlas (TCGA) and investigated in the UALCAN database (http://ualcan.path.uab.edu/analysis.html).
Study population
This study enrolled 61 patients with an OSCC diagnosis (ICD10-CM), who were older than 18 years old and had signed informed consent forms, as well as without a history of previous tumor-specific treatment, from April 2018 to November 2019 at the Department of Oral and Maxillofacial Surgery. Pregnant patients were excluded from this study. All patients with OSCC underwent lesion resection after recruitment. Two board-certified oral pathologists made all final histopathological diagnoses of the specimens.
Sample preparation
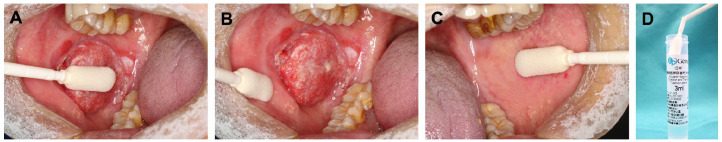
Oral epithelial cells were collected by the oral swabs from each patient with OSCC before surgery: one from the CL site (Figure 1A), one from the cancer-adjacent normal (AN) site (Figure 1B), one from the contralateral normal (CN) gross appearance mucosa or normal gross appearance mucosa in the non-contact-cancer area (Figure 1C). Before the collection procedure, patients rinsed with compound chlorhexidine mouthwash and then saline for 30 seconds each. The sampling area was cleaned with a sterile cotton ball until it was free of soft plaque and debris. Cells were collected from the target area using a noninvasive cell brush (iStat Biomedical Co., Ltd, New Taipei, Chinese Taiwan) by moving it up and down and rotating it approximately 10 times. Care was taken to ensure the cell brush did not touch the surrounding area during sampling. All collected OECs were stored in the independent phosphate-buffered saline solution (Figure 1D) at 4°C and sent to the laboratory for methylation detection within 1 week.
Figure 1.
Collect oral exfoliated cell samples from the cancerous lesion (A), cancer-adjacent normal area (B), contralateral normal area (C) sites, and break off the sampling brush head into a storage tube (D).
Genomic DNA extraction, bisulfite conversion, and methylation determination
Genomic DNA (gDNA) extraction, bisulfite conversion, and methylation determination were performed as previously described.12,14 Briefly, gDNA was extracted from OECs samples using the QIAamp DNA Mini Kit (Qiagen, Valencia, California) and then bisulfite conversion were performed using the “Epigene” Bisulfite Conversion Kit (iStat Biomedical Co., Ltd, New Taipei, Chinese Taiwan) according to the manufacturer’s instructions. Quantitative methylation-specific polymerase chain reaction was used to analyze the targeted bisulfite-converted gDNA with COL2A1 as the internal control. DNA methylation levels were estimated using ΔCp, defined as the difference between the Cp values of the target gene and COL2A1 (Cp target gene – Cp COL2A1 ). These levels are expressed as a Methylation Index (M-index), calculated using the formula: (2-ΔCp) × 10,000. The methylation result was judged as positive or negative at each site according to the manufacturer’s instructions.
Data analysis
The Wilcoxon signed-rank test was used for M-index comparison between each 2 paired groups, Kendall test was used for M-index comparison among 3 paired groups, the Mann-Whitney U test was used for M-index comparison between every 2 independent groups, and the chi-square test was used for comparison of positive rates by SPSS (version 26.0; IBM Corp., Armonk, New York) and GraphPad Prism (version 8.0; GraphPad Software, La Jolla, California). Statistical significance was set at P < .05.
Result
Analysis of the methylation and expression levels of PAX1 and ZNF582 in OSCC
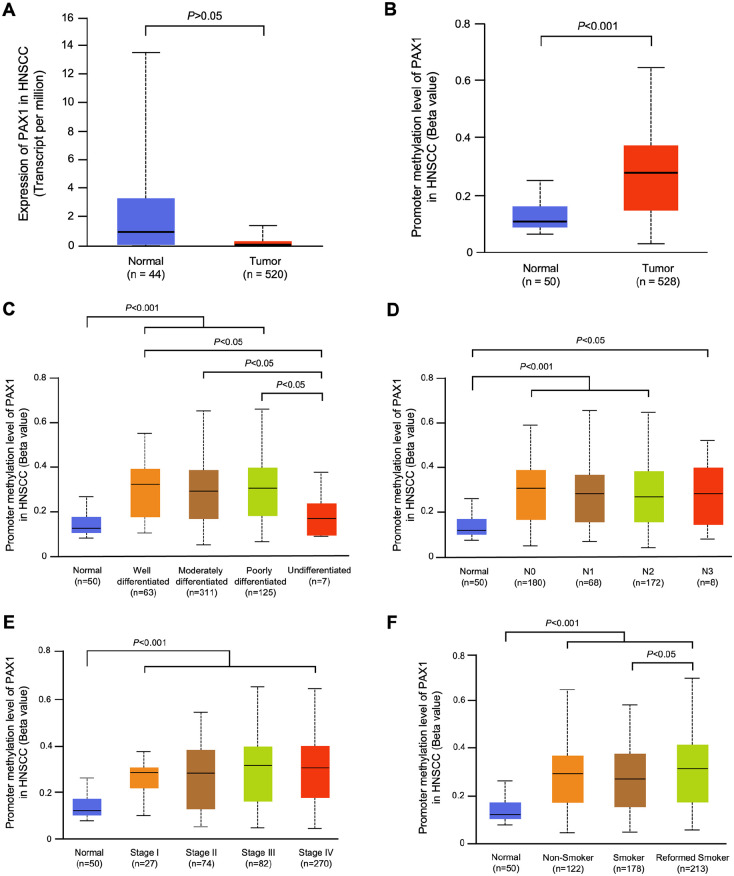
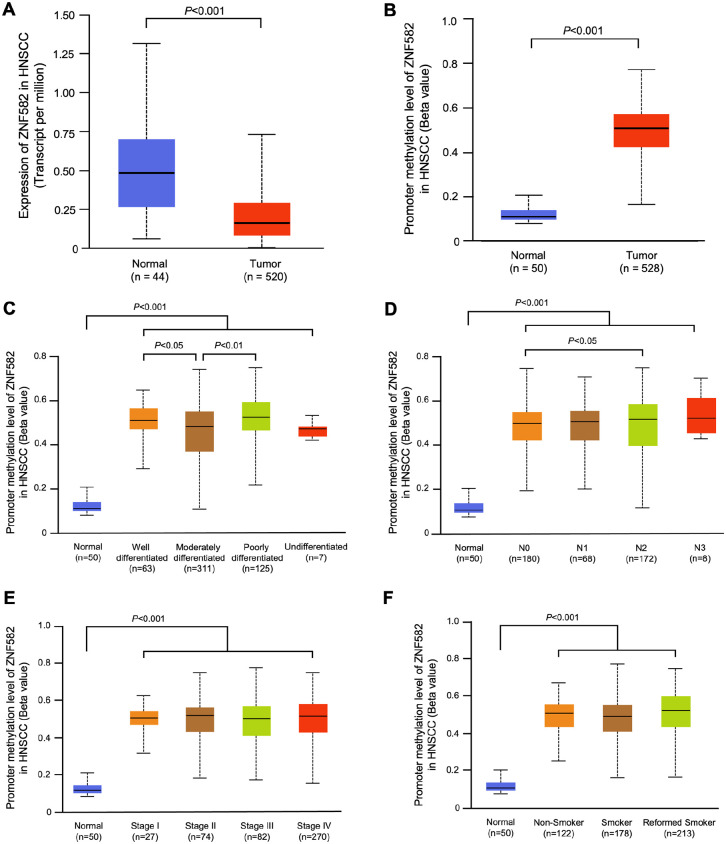
UALCAN is an integrated cancer data analysis platform which can perform in-depth analyses of TCGA gene expression data.26,27 Using this database, we compared the expression levels of PAX1 and ZNF582 in HNSCC between 520 HNSCC tumor tissues and 44 normal tissues, and the methylation levels of PAX1 and ZNF582 were compared between 528 HNSCC cancer tissues and 50 normal tissues. The expression levels of PAX1 and ZNF582 in HNSCC tissues were significantly lower compared with normal tissues (P < .05) (Figures 2A and 3A). The promoter methylation levels of PAX1 and ZNF582 in HNSCC were both significantly higher compared with normal tissues (n = 50) (Figures 2B and 3B). In the subgroup analysis, we analyzed the grade of differentiation, nodal metastasis status, TNM stage and smoking status with the promoter methylation levels of 2 genes. Higher methylation of PAX1 was observed in the reformed smoker patients with OSCC (Figure 2F), and higher methylation of ZNF582 was observed in patients with OSCC with poorly differentiated tumor (Figure 3C) and pN2 stage (Figure 3D).
Figure 2.
Use the UALCAN database to analyze the differences in PAX1 expression levels (A) in 520 HNSCC tumor tissues and 44 normal tissues and promoter methylation levels (B) in 528 HNSCC cancer tissues and 50 normal tissues between HNSCC and normal tissues sourced from The Cancer Genome Atlas. Compare the grade of differentiation (C), nodal metastasis status (D), TNM stage (E), and smoking status (F) with the PAX1 promoter methylation levels.
Figure 3.
Use the UALCAN database to analyze the differences in ZNF582 expression levels (A) in 520 HNSCC tumor tissues and 44 normal tissues and promoter methylation levels (B) in 528 HNSCC cancer tissues and 50 normal tissues between HNSCC and normal tissues sourced from The Cancer Genome Atlas. Compare the grade of differentiation (C), nodal metastasis status (D), TNM stage (E), and smoking status (F) with the ZNF582 promoter methylation levels.
Patient characteristics
From April 2018 to November 2019, 61 patients with OSCC at Peking University School and Hospital of Stomatology were enrolled in this study. Among these patients with OSCC, the ratio of males to females was 1.35, with a mean age of 59.6 ± 12.1 years. Table 1 shows the proportions of the location of the lesion, risk factors, tumor differentiation, and tumor stage. In general, nearly half of the patients have ever smoked (32/61, 52.46%) or drank alcohol (27/61, 44.26%). No patients have the habit of areca nut chewing. In addition, 73.8% (45/61) of patients had primary OSCC without lymph node metastasis.
Table 1.
Clinicopathological characteristics of 61 patients with OSCC.
| Total cases | ||
|---|---|---|
| Case number | % | |
| Gender | ||
| Male | 35 | 57.38 |
| Female | 26 | 42.62 |
| Age | ||
| <60 | 32 | 52.46 |
| ⩾60 | 29 | 47.54 |
| Location | ||
| Tongue | 25 | 40.98 |
| Cheek | 10 | 16.39 |
| Gum | 11 | 18.03 |
| Other | 15 | 24.59 |
| Risk factor (Ever) | ||
| Smoking | 32 | 52.46 |
| Alcohol consumption | 27 | 44.26 |
| T | ||
| Tis | 4 | 6.56 |
| T1 | 10 | 16.39 |
| T2 | 19 | 31.15 |
| T3 | 10 | 16.39 |
| T4 | 18 | 29.51 |
| N | ||
| pN0 | 45 | 73.77 |
| pN1 | 4 | 6.56 |
| pN2 | 5 | 8.20 |
| pN3 | 1 | 1.64 |
| missing | 6 | 9.84 |
| Stage | ||
| 0 | 4 | 6.56 |
| I | 8 | 13.11 |
| II | 17 | 27.87 |
| III | 10 | 16.39 |
| IV | 22 | 36.07 |
| Tumor differentiation | ||
| High | 28 | 45.90 |
| Moderate | 33 | 54.10 |
| Low | 0 | 0.00 |
Methylation levels of PAX1 and ZNF582 at the CL, AN, and CN sites
The methylation levels of PAX1 and ZNF582 at the CL, AN, and CN sites were examined and further evaluated according to patient characteristics at baseline (Tables 2 and 3). PAX1 methylation levels were significantly different among the CL, AN, and CN sites (PAX1: P < .001; P < .001, Figure 4A). Interestingly, PAX1 methylation was high at CL sites and gradually decreased at AN and CN sites. In other words, there is a CL-centric pattern of PAX1 methylation in patients with OSCC, implying a possible association between DNA methylation and field cancerization effects. In the subgroup analysis, higher methylation of PAX1 was observed in the CL site among patients with OSCC with T3-T4 tumor stage and bone invasion (P = .049 and P = .02, respectively, Table 2). A cancerous lesion-centric pattern of PAX1 methylation was also observed in subgroups including females, aged ⩾ 60 years, never smoked, never consumed alcohol, with 0-II stage, with moderate differentiation, and with bone invasion. Overall, the CL, AN, and CN sites in most of the subgroups, except for pN1-N3 (P = .08) showed significant differences in PAX1 methylation (Table 2).
Table 2.
PAX1 DNA methylation comparison in subgroups.
| No. | Cancerous lesion site (CL) | P 1 | Cancer-adjacent normal site (AN) | P 1 | Contralateral normal site (CN) | P 1 | P 2 | |
|---|---|---|---|---|---|---|---|---|
| Sex | ||||||||
| Male | 35 | 42.45 (0.17-476.96) | .39 | 0.02 (0.01-0.32) a | .21 | 0.02 (0.01-0.06) b | .27 | <.001 |
| Female | 26 | 150.8 (10-838.49) | 0.07 (0.02-6.62) a | 0.02 (0-0.07) bc | <.001 | |||
| Age | ||||||||
| <60 | 32 | 62.91 (0.44-700.35) | .95 | 0.03 (0.01-0.17) a | .21 | 0.03 (0.01-0.06) b | .30 | <.001 |
| ⩾60 | 29 | 70.9 (0.22-735.72) | 0.08 (0.01-8.31) a | 0.01 (0.01-0.07) bc | <.001 | |||
| Smoking | ||||||||
| Never | 29 | 139.85 (2.84-902.15) | .49 | 0.06 (0.02-4.19) a | .24 | 0.02 (0.01-0.08) bc | .27 | <.001 |
| Ever | 32 | 43.51 (0.22-465.18) | 0.02 (0.01-1.69) a | 0.02 (0.01-0.06) b | <.001 | |||
| Alcohol | ||||||||
| Never | 34 | 119.71 (10-1214.38) | .16 | 0.09 (0.01-6.62) a | .22 | 0.02 (0.01-0.07) bc | .27 | <.001 |
| Ever | 27 | 11.06 (0.17-429.86) | 0.02 (0.01-0.15) a | 0.02 (0.01-0.06) b | .001 | |||
| T | ||||||||
| Tis-T2 | 33 | 39.61 (0.04-303.12) | .049 | 0.08 (0.02-4.19) a | .35 | 0.02 (0.01-0.06) bc | .12 | .001 |
| T3 T4 | 28 | 204.96 (5.78-2451.51) | 0.03 (0.01-1.72) a | 0.03 (0.01-0.09) b | <.001 | |||
| pN | ||||||||
| pN0 | 45 | 66.61 (0.52-1132) | .33 | 0.05 (0.01-0.25) a | .32 | 0.02 (0.01-0.09) b | .54 | <.001 |
| pN1-pN3 | 10 | 43.58 (0.24-309.67) | 2.21 (0.02-34.68) | 0.03 (0.02-0.06) b | .08 | |||
| Stage | ||||||||
| 0+Ⅰ+Ⅱ | 29 | 42.45 (0.04-357.87) | .19 | 0.08 (0.02-1.82) a | .54 | 0.02 (0.01-0.06) bc | .16 | .001 |
| Ⅲ+Ⅳ | 32 | 86.74 (3.3-1161.4) | 0.03 (0.01-3.8) a | 0.03 (0.01-0.08) b | <.001 | |||
| Differentiate | ||||||||
| High | 28 | 114.19 (0.09-1074.34) | .84 | 0.06 (0.01-2.55) a | .70 | 0.02 (0.01-0.06) b | .25 | <.001 |
| Moderate | 33 | 44.56 (1.6-625.89) | 0.04 (0.01-3.26) a | 0.02 (0.01-0.08) b c | <.001 | |||
| Bone invasion | ||||||||
| No | 37 | 39.06 (0.16-257.04) | .02 | 0.03 (0.01-0.25) a | .67 | 0.02 (0.01-0.05) b | .40 | <.001 |
| Yes | 24 | 357.87 (12.85-1161.4) | 0.07 (0.01-4.43) a | 0.03 (0.01-0.11) bc | <.001 |
The significance values are conducted by Mann-Whitney U test. 2The significance values are conducted by Kendall test.
The significance values are conducted by Wilcoxon signed-rank test in subgroup comparison when P < .05, aCancerous lesion site (CL) versus Cancer-adjacent normal site (AN). bCancerous lesion site (CL) versus Contralateral normal site (CN). cCancer-adjacent normal site (AN) versus Contralateral normal site (CN).
Table 3.
ZNF582 DNA methylation comparison in subgroups.
| No. | Cancerous lesion site (CL) | P 1 | Cancer-adjacent normal site (AN) | P 1 | Contralateral normal site (CN) | P 1 | P 2 | |
|---|---|---|---|---|---|---|---|---|
| Sex | ||||||||
| Male | 35 | 149.89 (24.05-1103.38) | .52 | 2.48 (0.05-14.1) a | .15 | 5.06 (0.04-15.75) b | .61 | <.001 |
| Female | 26 | 190.15 (52.81-469.68) | 4.65 (0.41-34.33) a | 3.5 (0.88-32.81) b | <.001 | |||
| Age | ||||||||
| <60 | 32 | 102.46 (19.72-401.33) | .11 | 1.96 (0.1-14.47) a | .17 | 2.91 (0.09-13.11) b | .07 | <.001 |
| ⩾60 | 29 | 167.46 (50.38-1080.91) | 7.72 (0.3-34.68) a | 6.95 (1.24-51.45) b | <.001 | |||
| Smoking | ||||||||
| Never | 29 | 156.25 (47.06-475.45) | .64 | 2.75 (0.12-25.18) a | .51 | 2.84 (0.41-36.22) b | .99 | <.001 |
| Ever | 32 | 150.93 (26.65-1057.89) | 4.43 (0.09-14.47) a | 5.24 (0.18-15.49) b | <.001 | |||
| Alcohol | ||||||||
| Never | 34 | 154.11 (40.2-552.34) | .62 | 4.65 (0.33-44.64) a | .15 | 2.51 (0.07-19.17) b | .14 | <.001 |
| Ever | 27 | 149.89 (22.28-1103.38) | 2.48 (0.05-10.76) a | 7.72 (1.31-24.72) b | <.001 | |||
| T | ||||||||
| Tis-T2 | 33 | 126.04 (30.02-393.16) | .23 | 5.31 (0.16-33.24) a | .24 | 4.37 (0.78-15.23) b | .74 | <.001 |
| T3-T4 | 28 | 168.05 (49.07-1438.07) | 2.61 (0.08-9.27) a | 4.73 (0.07-29.81) b | <.001 | |||
| pN | ||||||||
| pN0 | 45 | 167.46 (35.83-989.92) | .57 | 4.62 (0.07-23.09) a | .77 | 4.4 (0.16-15.23) b | .38 | <.001 |
| pN1-pN3 | 10 | 116.99 (34.26-438.51) | 3.3 (1.64-14.59) a | 6.39 (2.02-39.84) b | .003 | |||
| Stage | ||||||||
| 0+Ⅰ+Ⅱ | 29 | 126.04 (26.93-420.36) | .22 | 5.31 (0.12-44.8) a | .39 | 4.37 (0.41-14.26) b | .82 | <.001 |
| Ⅲ+Ⅳ | 32 | 168.05 (49.07-1092.14) | 2.72 (0.09-10.36) a | 4.73 (0.18-34.1) b | <.001 | |||
| Differentiate | ||||||||
| High | 28 | 137.96 (33.43-683.06) | .71 | 2.33 (0.09-16.76) a | .39 | 1.71 (0.05-13.28) b | .03 | <.001 |
| Moderate | 33 | 156.25 (44.81-792) | 4.95 (0.12-24.22) a | 6.95 (1.27-41.69) b | <.001 | |||
| Bone invasion | ||||||||
| No | 37 | 60.45 (26.93-330.45) | .02 | 2.75 (0.14-16.04) a | .81 | 4.4 (0.21-15.23) b | .80 | <.001 |
| Yes | 24 | 431.9 (99.93-1672.89) | 4.3 (0.08-37.74) a | 3.85 (0.35-30.98) b | <.001 |
The significance values are conducted by Mann-Whitney U test. 2The significance values are conducted by Kendall test.
The significance values are conducted by Wilcoxon signed-rank test in subgroup comparison when P < .05. aCancerous lesion site (CL) versus Cancer-adjacent normal site (AN). bCancerous lesion site (CL) versus Contralateral normal site (CN).
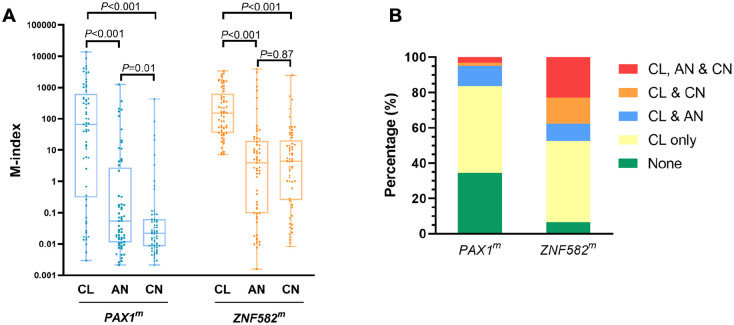
Figure 4.
The M-indexes (A) and coexistence (B) of PAX1 and ZNF582 at the cancerous lesion site (CL), cancer-adjacent normal site (AN), and contralateral normal site (CN).
Significant differences of ZNF582 methylation levels were also detected among different sites (ZNF582: P < .001, Figure 4A). ZNF582 methylation was significantly higher at CL sites than at AN or CN sites. No difference was detected between the AN and CN sites, which differs from the difference in PAX1 methylation, ZNF582 methylation did not occur in a cancer lesion-centric manner in patients with OSCC. According to our subgroup analysis, significantly higher methylation was detected at the CL site in patients with OSCC with bone invasion (P = .02, Table 3). For the CN site, ZNF582 methylation was higher in patients with moderate differentiation than in those with high differentiation (P = .03, Table 3).
DNA methylation coexists in CL, AN, and CN sites
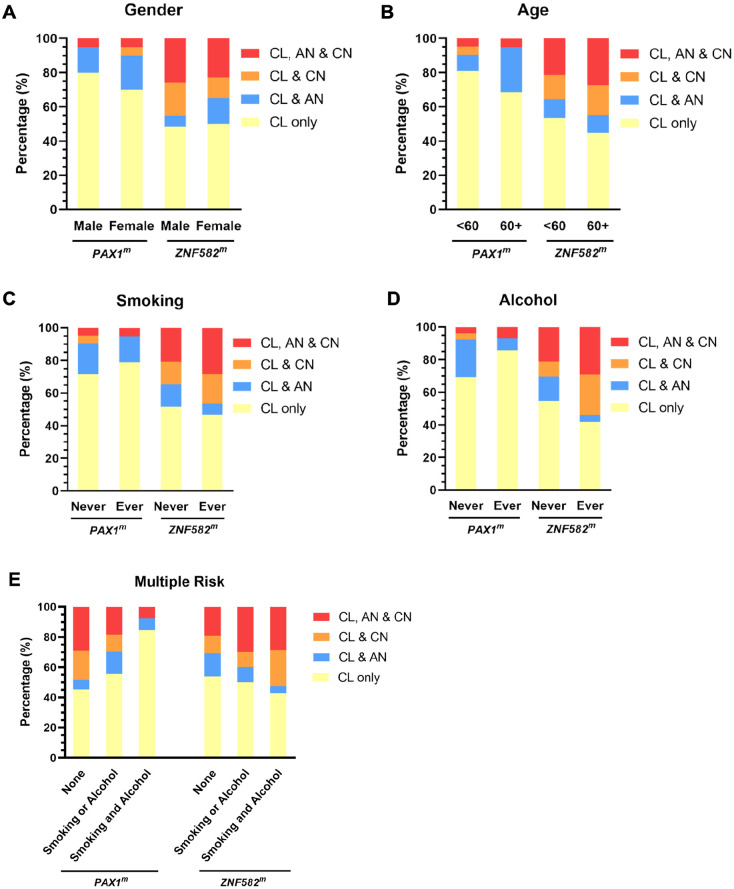
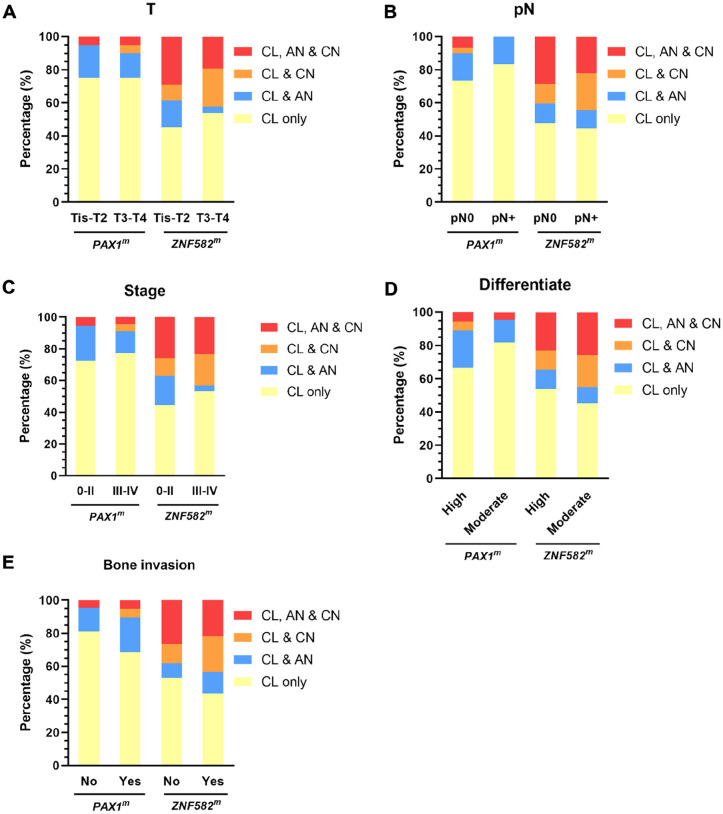
To explore the distribution of DNA methylation in the oral cavity, the patients were further classified into 5 groups according to their degree of hypermethylation at different sites. As shown in Figure 4B, there were no cases with only AN or only CN without concurrent CL positivity. In addition, 21 (34.4%) and 4 (6.6%) patients were negative for PAX1 and ZNF582 hypermethylation at all 3 sites, respectively. Furthermore, 40 (65.6%) and 57 (93.4%) patients were with PAX1 and ZNF582 hypermethylation in the CL site, respectively. Among them, 25% patients (10/40) had PAX1 methylation in AN or CN, whereas 50.9% patients (29/57) had ZNF582 methylation in AN or CN. In addition, 24.6% (14/57) patients with OSCC had ZNF582 methylation coexisted in AN and CN sites. The hypermethylation pattern was further stratified by patients’ characteristics. In female or elderly patients with OSCC, there is a slightly higher proportion of patients with hypermethylation in their AN or CN site (Figure 5A and B). Regarding the risk factors at baseline, a slight but not significant association between alcohol consumption and coexist of ZNF582 methylation in AN or CN site (Figure 5D). Comparing with patient with nonsmoking and alcohol consumption, coexistence of ZNF582 methylation in their AN and CN sites was observed the more proportion of patients with smoking or alcohol consumption (Figure 5E). No significant difference was found between DNA methylation coexistence in different sites in the settings of OECs and clinicopathologic characteristics of tumors among patients with OSCC (Figure 6), while a slight higher proportion of patients with OSCC with T3-T4 tumor stage (Figure 6A) or III-IV stage (Figure 6C) or bone invasion (Figure 6E) had concurrent CL and CN positivity for PAX1 and ZNF582 hypermethylation.
Figure 5.
Gender (A), age (B), smoking status (C), alcohol consumption (D), and multiple risk factors (E) effects on the coexistence of DNA methylation at the cancerous lesion site (CL), cancer-adjacent normal site (AN), and contralateral normal site (CN).
Figure 6.
The effect of tumor size (A), lymph node metastasis (B), tumor stage (C), differentiation (D), and bone invasion (E) on the coexistence of DNA methylation at the cancerous lesion site (CL), cancer-adjacent normal site (AN), and contralateral normal site (CN).
Discussion
Previous studies have suggested that TSG hypermethylation plays a vital role in OSCC carcinogenesis.28-32 Hypermethylation of PAX1 and ZNF582 have been demonstrated to be potential biomarkers for identifying OSCC using various specimens, including tissue, 16 OECs collected by oral scraping, or oral swabs,9,15 as well as mouth rinses. 17 Most studies focused on the epigenetic changes at lesion sites.9,15,16 In this study, oral swabs collected from exfoliate cells were used in this study to determine the epigenetic alteration pattern in the oral cavity of patients with OSCC. PAX1 methylation decreased in cancerous lesions, cancer-adjacent normal sites, and contralateral normal sites in a cancerous lesion-centric pattern. However, ZNF582 hypermethylation coexists in adjacent and contralateral normal sites in patients with OSCC. To our knowledge, this is the first study to reveal the general epigenetic changes in OECs, which were obtained from various sites in the oral cavity of patients with OSCC. These results provide information for developing epigenetic biomarkers with clinical implications or utility in the future.
DNA methylation changes in targeted TSGs were identified as the first epigenetic alterations in cancer and are related to the early stages of carcinogenesis.33,34 Previous studies have shown that PAX1 and ZNF582 hypermethylation in tissues can be used for OSCC detection and prognosis.13,24,35 DNA hypermethylation in OECs has also been shown, but studies have mostly focused on lesion sites. Collecting exfoliated cells via oral swab is notably less invasive than biopsy, which can enhance patient acceptance of oral cancer screening. Therefore, assessing PAX1 and ZNF582 methylation in OECs gathered by swab could be a noninvasive, viable alternative or supplementary diagnostic method for OSCC detection. 12 In the present study, PAX1 methylation in CL sites collected from oral swabs was higher in the T3-T4 tumor stage groups and bone invasion groups, whereas ZNF582 methylation in CL sites was higher in the bone invasion groups. This finding is consistent with our previous study using tissue samples. 13 Since bone invasion is considered as a significant independent factor that increases local recurrence of the tumor, metastasis, and median patient survival, our results provide evidence for the practical value of using swabbed oral cells for DNA methylation analysis in determining the prognosis of patients with OSCC.
The concept of “field cancerization” was proposed in 1953 based on the histopathological observations of multiple primary OSCC and their local recurrences. In conjunction with demonstrating of molecular abnormalities in tissues with a normal histological appearance, field cancerization may be used in the following areas of oncology, including risk assessment, early cancer detection, monitoring of tumor progression, and definition of tumor margins. 36 It has been revealed that histopathological and epigenetic alterations in both tumor and tumor-adjacent normal tissue from frozen tumor tissue, frozen tumor margin tissue and normal visual mucosa of patients with OSCC. 17 Notably, the 3-year overall survival rate demonstrated that PAX1 and ZNF582 hypermethylation in OECs collected from AN sites, rather than from CL sites, is associated with aggressive progression and poor prognosis of OSCC. 13 A previous study showed that approximately 51% and 73% of patients with OSCC had PAX1 and ZNF582 hypermethylation in their AN site, respectively. 8 Consistent with this pattern, this study revealed approximately 25% and 50.9% patients with OSCC with PAX1 and ZNF582 methylation, respectively, exhibited both in AN and CN sites. A cancerous lesion-centric pattern of PAX1 hypermethylation was observed in some subgroups of patients with OSCC in this study. ZNF582 hypermethylation coexists in AN and CN sites of patients with OSCC, which is widespread not just in lesion sites but also in surrounding normal tissues, pointing toward extensive epigenetic alterations in OSCC. Since field cancerization is known to be associated with primary cancer and recurrence, these results provide more information for understanding the role of DNA methylation in oncology. Our study further supported the widespread nature of epigenetic alterations in OSCC and further examination of potential oral cancer patients with precancerous lesions or those who are postoperative is also encouraged.
Smoking and alcohol consumption are common habits of patients with oral cancer in Western countries. Tobacco/cigarette use and alcohol have been reported to act in combination as primary carcinogens in the development of OSCC. 37 In this study, the PAX1 and ZNF582 hypermethylation was not significantly different between the AN and CN of patients with OSCC with risk factors of cigarette smoking or/and alcohol consumption (Figure 5C to E). Studies have shown that different environmental carcinogenic exposures in addition to genetic factors in global regions may contribute to distinct clinicopathological characteristics of oral cancer. 38 Areca nut chewing is also a common habit among oral cancer patients mainly in Southeast Asia. Poor oral hygiene, eating red meat or very spicy food, eating pickled food, and drinking boiling tea are reported to increase the risk of oral cancer.39,40 In mainland China, dietary and oral habits, as well as environmental factors varied across regions, and areca nut chewing is not the main habit in our study population. 41 This may explain why our currently enrolled patients with OSCC had never been exposed to areca nut chewing habit. However, the coexistence of DNA methylation of PAX1 and ZNF582 remains observable in nonsmoking and nondrinking patients with OSCC. Previous research 42 found that the major component of areca nut, arecoline, could induce hypermethylation of multiple genes in mouse OSCC tissues. A study based on a population in Chinese Taiwan 9 found that the positive rates for ZNF582/PAX1 methylation were significantly higher in areca-quid chewers (85%/63%) than in nonchewers (38%/26%, P < .001). Both ZNF582 and PAX1 hypermethylation showed significant associations with areca-quid chewing alone, areca-quid chewing combined with cigarette smoking, areca-quid chewing combined with alcohol drinking, and the combination of all 3 risk factors. Therefore, the association of diet, oral habits, areca nut chewing and environmental factors with the methylation levels of related genes in OSCC warrants further investigation.
Limitations of this study
There were still some limitations of this study. First, more detailed subgroup analysis including HPV status and macroscopic appearance are needed, which would provide deeper insights into the heterogeneity of OSCC. In addition, the sample size in certain subgroups (eg, pN+, poorly differentiated tumors) was relatively limited, which may reduce the statistical power to detect significant differences or draw definitive conclusions. Second, potential confounding factors, including the amount of smoking and alcohol consumption, as well as environmental factors and dietary habits, were not included in this study. Third, the molecular mechanism or factors contributing to the different patterns of PAX1 and ZNF582 hypermethylation are still unclear and warrant further investigation. Finally, due to the cross-sectional design of the study, data on long-term outcomes such as multiple primary OSCCs, recurrences, disease progression, and mortality were limited. Future longitudinal studies are needed to address these aspects comprehensively.
Conclusions
This study demonstrated that the hypermethylation of PAX1 and ZNF582 at lesion sites coupled with oral swab techniques provides a reliable diagnostic tool for OSCC detection. These 2 genes hypermethylation resulted in different epigenetic changes in the OECs of patients with OSCC. A cancerous lesion-centric pattern of PAX1 methylation was observed, suggesting a field cancerization in some patients with OSCC, while ZNF582 hypermethylation could coexist on adjacent or contralateral normal sides was observed.
Taken together, our study extends the investigation into the distribution of PAX1 and ZNF582 methylation within the oral cavity of patients with OSCC. The distinct patterns and coexistence phenomena of PAX1 and ZNF582 methylation provide novel insights into the epigenetic mechanisms of OSCC, particularly in relation to field cancerization and recurrence risk. These findings further highlight the potential clinical utility of PAX1 and ZNF582 methylation as biomarkers for OSCC prognosis and diagnosis and offer new perspectives on the understanding of the field cancerization effect in OSCC.
Supplemental Material
Supplemental material, sj-docx-1-onc-10.1177_11795549251335172 for Distribution of PAX1 and ZNF582 Hypermethylation in the Oral Exfoliated Cells of Oral Squamous Cell Carcinoma by Ya-Qing Mao, Rui Sun, Shuo Liu, Wen-Bo Zhang, Yao Yu, Ling-Fei Jia, Guang-Yan Yu and Xin Peng in Clinical Medicine Insights: Oncology
Acknowledgments
We would like to thank iStat Biomedical Co., Ltd for performing genomic DNA extraction, bisulfite conversion, and methylation determination. We would also like to thank the editors and anonymous reviewers who have helped us improve the manuscript.
Footnotes
ORCID iDs: Ya-Qing Mao  https://orcid.org/0009-0004-4537-7925
https://orcid.org/0009-0004-4537-7925
Ethical considerations: This study was approved by the Ethics Committee of Peking University School and Hospital of Stomatology (Beijing, China). The registration number of this study is PKUSSIRB-201525099a) and written informed consent form was obtained from each patient included in the study.
Consent for publication: Not applicable.
Author contributions: XP, LJ, and GY conceived the project. YM and RS designed and performed the study, and wrote the first draft. SL, WZ, and YY performed the data acquisition and analysis. XP supervised the project and guaranteed the integrity of the entire project. YM, RS, and XP participated in the manuscript revision. All authors carefully read and approved the final manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was financially supported by the Natural Science Foundation of Beijing Municipality (grant number 7222221), and the Research Foundation of Peking University School and Hospital of Stomatology (grant number PKUSS20220111).
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Data availability statement: The authors confirm that the data supporting the findings of this study are available within the article and can be requested from corresponding author by a reasonable necessity.
Supplemental material: Supplemental material for this article is available online.
References
- 1. Muller S, Tilakaratne WM. Update from the 5th edition of the World Health organization classification of head and neck tumors: tumours of the oral cavity and mobile tongue. Head Neck Pathol. 2022;16:54-62. doi: 10.1007/s12105-021-01402-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. doi: 10.3322/caac.21834 [DOI] [PubMed] [Google Scholar]
- 3. Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 2020;6:92. doi: 10.1038/s41572-020-00224-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology (NCCN Guidelines): head and neck cancers, 2025. [Google Scholar]
- 5. Chamoli A, Gosavi AS, Shirwadkar UP, et al. Overview of oral cavity squamous cell carcinoma: risk factors, mechanisms, and diagnostics. Oral Oncol. 2021;121:105451. doi: 10.1016/j.oraloncology.2021.105451 [DOI] [PubMed] [Google Scholar]
- 6. Zeng H, Chen W, Zheng R, et al. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018;6:e555-e567. doi: 10.1016/S2214-109X(18)30127-X [DOI] [PubMed] [Google Scholar]
- 7. Zeng H, Zheng R, Sun K, et al. Cancer survival statistics in China 2019-2021: a multicenter, population-based study. J Natl Cancer Cent. 2024;4:203-213. doi: 10.1016/j.jncc.2024.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wong AH-H. Pushing the boundary of cancer diagnostics through microfluidic technologies. Innov Med. 2023;1(1):100005. doi: 10.59717/j.xinn-med.2023.100005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng SJ, Chang CF, Lee JJ, et al. Hypermethylated ZNF582 and PAX1 are effective biomarkers for detection of oral dysplasia and oral cancer. Oral Oncol. 2016;62:34-43. doi: 10.1016/j.oraloncology.2016.09.007 [DOI] [PubMed] [Google Scholar]
- 10. Liu H, Liu XW, Dong G, et al. P16 methylation as an early predictor for cancer development from oral epithelial dysplasia: a double-blind multicentre prospective study. EBioMedicine. 2015;2:432-437. doi: 10.1016/j.ebiom.2015.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu H, Liu Z, Liu XW, et al. A similar effect of P16 hydroxymethylation and true-methylation on the prediction of malignant transformation of oral epithelial dysplasia: observation from a prospective study. BMC Cancer. 2018;18:918. doi: 10.1186/s12885-018-4787-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun R, Zhang WB, Yu Y, Yang HY, Yu GY, Peng X. Evaluation of DNA methylation in matched oral swab and tissue specimens from Chinese patients with oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2021;50:725-732. doi: 10.1016/j.ijom.2020.05.022 [DOI] [PubMed] [Google Scholar]
- 13. Cheng SJ, Chang CF, Ko HH, et al. Hypermethylated ZNF582 and PAX1 genes in oral scrapings collected from cancer-adjacent normal oral mucosal sites are associated with aggressive progression and poor prognosis of oral cancer. Oral Oncol. 2017;75:169-177. doi: 10.1016/j.oraloncology.2017.11.013 [DOI] [PubMed] [Google Scholar]
- 14. Sun R, Juan YC, Su YF, et al. Hypermethylated PAX1 and ZNF582 genes in the tissue sample are associated with aggressive progression of oral squamous cell carcinoma. J Oral Pathol Med. 2020;49:751-760. doi: 10.1111/jop.13035 [DOI] [PubMed] [Google Scholar]
- 15. Huang YK, Peng BY, Wu CY, Su CT, Wang HC, Lai HC. DNA methylation of PAX1 as a biomarker for oral squamous cell carcinoma. Clin Oral Investig. 2014;18:801-808. doi: 10.1007/s00784-013-1048-6 [DOI] [PubMed] [Google Scholar]
- 16. Yang CC, Wu CH, Chang CF, et al. DNA methylation confers clinical potential to predict the oral cancer prognosis. Clin Res Trials. 2018;4:1-8. doi: 10.15761/crt.1000232 [DOI] [Google Scholar]
- 17. Cheng SJ, Chang CF, Ko HH, et al. Hypermethylated ZNF582 and PAX1 genes in mouth rinse samples as biomarkers for oral dysplasia and oral cancer detection. Head Neck. 2018;40:355-368. doi: 10.1002/hed.24958 [DOI] [PubMed] [Google Scholar]
- 18. Eljabo N, Nikolic N, Carkic J, et al. Genetic and epigenetic alterations in the tumour, tumour margins, and normal buccal mucosa of patients with oral cancer. Int J Oral Maxillofac Surg. 2018;47:976-982. doi: 10.1016/j.ijom.2018.01.020 [DOI] [PubMed] [Google Scholar]
- 19. Islam S, Uehara O, Matsuoka H, et al. DNA hypermethylation of sirtuin 1 (SIRT1) caused by betel quid chewing-a possible predictive biomarker for malignant transformation. Clin Epigenetics. 2020;12:12. doi: 10.1186/s13148-019-0806-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joehanes R, Just AC, Marioni RE, et al. Epigenetic Signatures of Cigarette Smoking. Circ Cardiovasc Genet. 2016;9:436-447. doi: 10.1161/CIRCGENETICS.116.001506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Varela-Rey M, Woodhoo A, Martinez-Chantar ML, Mato JM, Lu SC. Alcohol, DNA methylation, and cancer. Alcohol Res. 2013;35:25-35 [PMC free article] [PubMed] [Google Scholar]
- 22. Mathers JC, Strathdee G, Relton CL. Induction of epigenetic alterations by dietary and other environmental factors. Adv Genet. 2010;71:3-39. doi: 10.1016/B978-0-12-380864-6.00001-8 [DOI] [PubMed] [Google Scholar]
- 23. Gasche JA, Hoffmann J, Boland CR, Goel A. Interleukin-6 promotes tumorigenesis by altering DNA methylation in oral cancer cells. Int J Cancer. 2011;129:1053-1063. doi: 10.1002/ijc.25764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rivera-Peña B, Folawiyo O, Turaga N, et al. Promoter DNA methylation patterns in oral, laryngeal and oropharyngeal anatomical regions are associated with tumor differentiation, nodal involvement and survival. Oncol Lett. 2024;27:89. doi: 10.3892/ol.2024.14223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med. 2007;147:573-577. doi: 10.7326/0003-4819-147-8-200710160-00010 [DOI] [PubMed] [Google Scholar]
- 26. Chandrashekar DS, Karthikeyan SK, Korla PK, et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18-27. doi: 10.1016/j.neo.2022.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649-658. doi: 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484-492. doi: 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- 29. Jones LK, Vaskar S. Chromatin modification, leukaemia and implications for therapy. Br J Haematol. 2002;118:714-727. [DOI] [PubMed] [Google Scholar]
- 30. Jones RS. Epigenetics: reversing the “irreversible.” Nature. 2007;450:357-359. [DOI] [PubMed] [Google Scholar]
- 31. Baylin SB, Ohm JE. Epigenetic gene silencing in cancer: a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107-116. [DOI] [PubMed] [Google Scholar]
- 32. Hellebrekers DM, Jair KW, Viré E, et al. Angiostatic activity of DNA methyltransferase inhibitors. Mol Cancer Ther. 2006;5:467-475. [DOI] [PubMed] [Google Scholar]
- 33. Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27-36. doi: 10.1093/carcin/bgp220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Khor GH, Froemming GR, Zain RB, et al. DNA methylation profiling revealed promoter hypermethylation-induced silencing of p16, DDAH2 and DUSP1 in primary oral squamous cell carcinoma. Int J Med Sci. 2013;10:1727-1739. doi: 10.7150/ijms.6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Prasad M, Veeraraghavan VP, Jayaraman S. Methylated ZNF582: a therapeutic target in oral cancer. Epigenomics. 2022;14:1389-1392. doi: 10.2217/epi-2022-0368 [DOI] [PubMed] [Google Scholar]
- 36. Dakubo GD, Jakupciak JP, Birch-Machin MA, Parr RL. Clinical implications and utility of field cancerization. Cancer Cell Int. 2007;7:2. doi: 10.1186/1475-2867-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mohan M, Jagannathan N. Oral field cancerization: an update on current concepts. Oncol Rev. 2014;8:244. doi: 10.4081/oncol.2014.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li YC, Cheng AJ, Lee LY, Huang YC, Chang JT. Multifaceted mechanisms of areca nuts in oral carcinogenesis: the molecular pathology from precancerous condition to malignant transformation. J Cancer. 2019;10:4054-4062. doi: 10.7150/jca.29765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gupta B, Bray F, Kumar N, Johnson NW. Associations between oral hygiene habits, diet, tobacco and alcohol and risk of oral cancer: a case-control study from India. Cancer Epidemiol. 2017;51:7-14. [DOI] [PubMed] [Google Scholar]
- 40. Huang JF, Qiu Y, Cai L, et al. Pickled food, fish, seafood intakes and oral squamous cell carcinoma: a case-control study. Zhonghua Yu Fang Yi Xue Za Zhi. 2017;51:680-685. doi: 10.3760/cma.j.issn.0253-9624.2017.08.005 [DOI] [PubMed] [Google Scholar]
- 41. Reichart PA, Zhang X. Misconceptions related to the areca nut chewing habits of Mainland China. Oral Oncol. 2007;43:958-959. doi: 10.1016/j.oraloncology.2007.01.020 [DOI] [PubMed] [Google Scholar]
- 42. Lai ZL, Tsou YA, Fan SR, et al. Methylation-associated gene silencing of RARB in areca carcinogens induced mouse oral squamous cell carcinoma. Biomed Res Int. 2014;2014:378358. doi: 10.1155/2014/378358 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-onc-10.1177_11795549251335172 for Distribution of PAX1 and ZNF582 Hypermethylation in the Oral Exfoliated Cells of Oral Squamous Cell Carcinoma by Ya-Qing Mao, Rui Sun, Shuo Liu, Wen-Bo Zhang, Yao Yu, Ling-Fei Jia, Guang-Yan Yu and Xin Peng in Clinical Medicine Insights: Oncology