Abstract
Background
Ammonia (NH3) is an environmental pollutant and a potent reproductive stressor widely found in rabbit houses. Exposure to ammonia can result in follicle atresia, affect oocyte maturation and cause cumulus cell apoptosis. Long non-coding RNA (lncRNA) is an important factor in the regulation of cumulus cell development and oocyte maturation. The potential molecular mechanism of NH3 in the induction of cumulus-oocyte complex (COCs) toxicity and the regulatory role of lncRNA in COCs are currently unclear.
Methods
A total of 150 female IRA rabbits (35 days old) were randomly divided into three groups, and kept in environmental control rooms for four weeks. The rabbits in the control group (CG) were kept under an NH3 concentration of < 3 ppm. The two treatment groups were kept under NH3 concentrations of 10 ppm (low ammonia concentration, LAC) and 30 ppm (high ammonia concentration, HAC). We used a combination of RNA deep sequencing, quantitative real-time polymerase chain reaction (qRT-PCR), and bioinformatic analysis to explore the regulatory mechanism of lncRNA and messenger RNA (mRNA) in COCs.
Results
We found that primordial follicles and primary follicles were significantly decreased while atretic follicles were significantly increased in the NH3-treated groups. The results from Gene Ontology (GO) items showed that female meiosis sister chromatid cohesion and the regulation of follicle-stimulating hormone secretion were involved in the mechanism of rabbits exposed to NH3. The results demonstrated that the mammalian target of the rapamycin (mTOR) signaling pathway and the transforming growth factor-beta (TGF-beta) signaling pathway inhibits germ cell development and follicular growth in the LAC versus the CG group. LncRNAs were involved in the apoptosis of female germ cells via the hypoxia-inducible factor (HIF-1) signaling pathway in the HAC versus the CG group. Co-expression analysis found that lncRNA MAPK3 and lncRNA SHC1 were correlated with changes in cumulus cell and oocyte function after NH3 exposure.
Conclusions
These results indicate that NH3 affected the development and function of COCs by influencing lncRNA expression.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12917-025-04806-9.
Keywords: Ammonia, Cumulus-oocyte complex, LncRNA, Rabbit, Reproductive dysfunction
Introduction
Ammonia (NH3), a strong irritant gas, is one of the major air pollutants in intensive animal production [1]. In China, the NH3 concentration in the rabbit houses has increased gradually with increasing breeding density. Relevant studies have shown that the NH3 concentration in most rabbit houses reaches 10 ppm before cleaning under normal ventilation conditions, while some rabbit houses reach or exceed 30 ppm in the winter under ventilation restrictions [2]. Previous studies found that 10ppm NH3 have affected the health of animal organs, such as the thymus, liver, kidneys, bursal lymphocytes and ovaries [3–5]. And our laboratory also showed that ovarian tissue structure damaged and hormone secretion disordered in NH3-treated rabbits, which indicated ovarian development disordered [2].
The follicle is the basic unit of the ovary [6], where follicular development is a process of mutual regulation between oocytes and the surrounding cumulus cells [7]. Cumulus cells can provide nutrients and maturation-enabling factors, secrete steroid hormones, and change the antioxidant system to protect oocytes [8]. In turn, oocytes can also regulate the function of cumulus cells by secreting certain paracrine factors [8]. Cumulus cells and oocytes form cumulus-oocyte complexes (COCs) through gap junctions [9] and the morphology of COCs is related to the atresia grade of the follicle that comprises it [10]. Yang et al. [11] found that NH3 treatment disrupted the oocyte maturation and caused apoptosis in cumulus cells. Nandi et al. [12] reported that the cell viability significantly decreased in cumulus cells after treated with NH3. These results suggested that the effect of ammonia on ovarian development may be caused by damaging COCs, but the mechanism is not clear.
Long non-coding RNA (lncRNA) are RNA transcripts with a length of more than 200 nucleotides which plays an important role in biological activities by regulating target genes [13–14]. They can participate in the regulation of the physiological and pathological functions of the ovary and affect follicle development at multiple levels [15]. For example, Non-ATPase 6 (PSMD6) and XLOC-011402 are related to oocyte maturation [16–17], H19 can regulate the number and quality of oocytes [18], and Hyaluronan Synthase-AS1 (HAS2-AS1) plays a role in regulating the expansion and migration of cumulus cells [19]. Nevertheless, the potential molecular mechanisms of NH3 in the induction of COCs toxicity and regulation of lncRNA in rabbits are currently unclear.
In this study, the potential molecular mechanisms of NH3 damage were discussed from the perspective of rabbit COCs. We identified differentially expressed lncRNA and mRNA through a combination of RNA deep sequencing, bioinformatic analysis, and quantitative real-time polymerase chain reaction (qRT-PCR) to explore the regulatory mechanisms of lncRNA and mRNA in the COCs of rabbits exposed to NH3.
Results
Ovarian structure and morphology
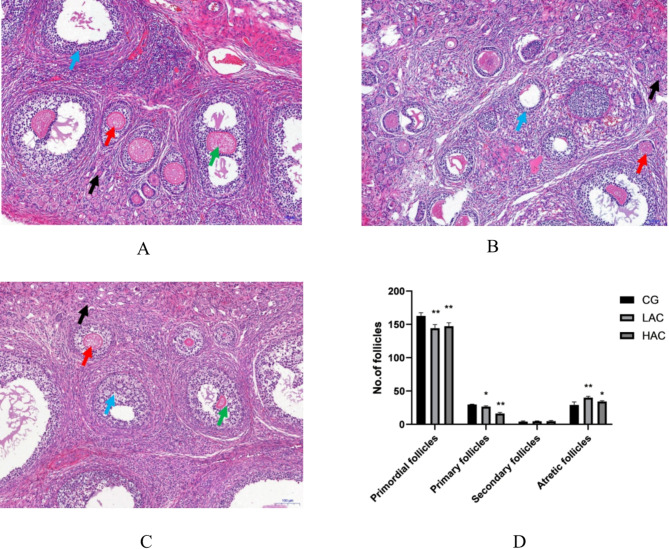
Histological changes in the ovaries were examined with H.E. staining (Fig. 1A, B and C). Compared with the CG, the ovarian tissue in the LAC and HAC groups showed pathological changes, collapse of the follicular cavity and increased atretic follicles. The amount of follicular cell necrosis was higher in the HAC group than in the LAC group. Follicle counts indicate that the number of primordial follicles and primary follicles was significantly lower (P < 0.05) in the NH3 treatments compared with the control group. While the number of atretic follicles was significantly more (P < 0.05) in the LAC group compared with the control group (Fig. 1D).
Fig. 1.
Effects of NH3 on ovarian structure and morphology (A-C) and the numbers of primordial, primary, secondary, and atretic follicles in the ovaries. (D) A: CG-< 3 ppm NH3, B: LAC-10 ppm NH3, C: HAC-30 ppm NH3; Key: Black arrows (↑) indicate primordial follicles. Red arrows (↑) indicate primary follicles. Green arrows (↑) indicate secondary follicles. Blue arrows (↑) indicate atretic follicles. Bars represent means ± SD, n = 4. * P < 0.05 and ** P < 0.01 indicate significantly different and highly significantly different values, respectively
Differentially expressed LncRNA and mRNA, and cluster analysis
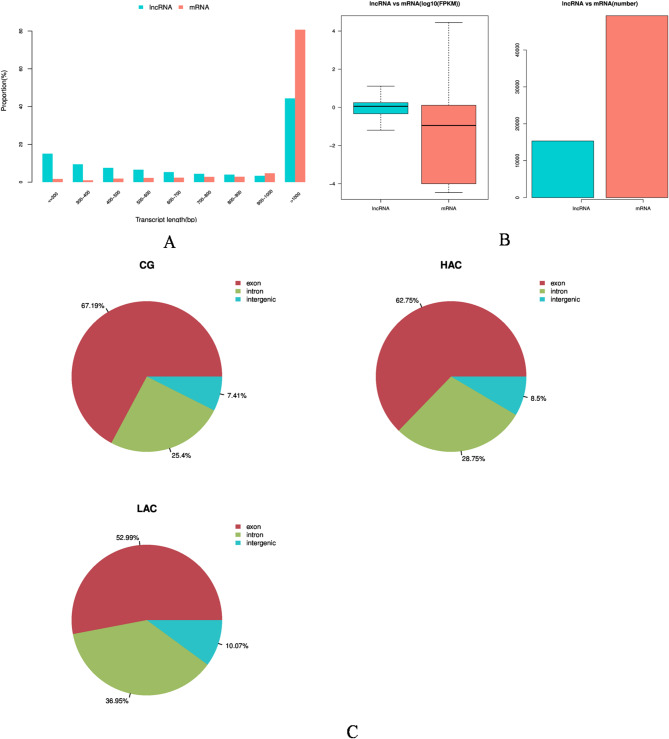
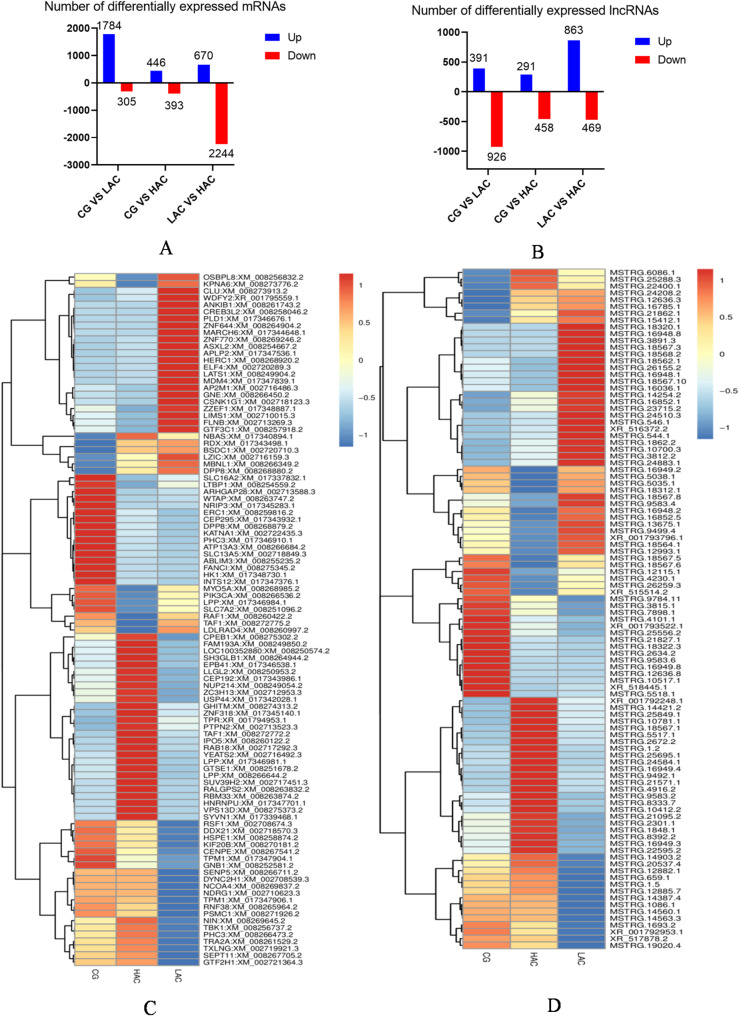
To investigate the effect of NH3 on lncRNAs in rabbit COCs, lncRNA sequencing was carried out. Statistical analysis showed that gene structure and expression levels were different among the three groups. The candidate lncRNAs were shorter than those of the mRNAs (Fig. 2A). The LncRNAs expression level was higher than the mRNAs (Fig. 2B), and the number of exons in the control group was higher than those in the treatment groups while the number of introns and intergenic regions was lower (Fig. 2C). Differentially expressed mRNAs (DEGs) for CG versus LAC and CG versus HAC identified 1784/305 and 446/393 up-regulated mRNAs, respectively (Fig. 3A). Differentially expressed lncRNAs (DELs) for CG versus LAC and CG versus HAC identified 391/926 and 291/458 up-regulated lncRNAs (Fig. 3B). Commonly expressed lncRNAs between the LAC and HAC groups identified 3 down-regulated lncRNAs and 7 up-regulated lncRNAs (Table 1). Hierarchical cluster analysis of the differentially expressed genes of mRNA and lncRNA are shown in Fig. 3C and D. Expression patterns of mRNA and lncRNA were different in the treatment groups compared with the control group.
Fig. 2.
Expression levels and genomic features of lncRNAs and mRNAs of rabbit cumulus-oocyte complexes under different levels of NH3 exposure. The length distribution of lncRNAs and mRNAs (A). Expression levels of mRNAs and lncRNAs in the CG, LAC and HAC groups were compared with the FPKM method (B). Boxplots of FPKM distributions were drawn using all the mRNAs and lncRNAs expression data, respectively. The lncRNAs from all samples generated exonic, intronic, and intergenic regions in the CG, LAC and HAC groups (C). CG: < 3 ppm NH3; LAC: 10 ppm NH3; HAC: 30 ppm NH3
Fig. 3.
Differentially expressed genes of rabbit cumulus-oocyte complexes under different levels of NH3 exposure (CG: < 3 ppm NH3; LAC: 10 ppm NH3; HAC: 30 ppm NH3). Differential expression levels of mRNAs (A) and lncRNAs (B). Hierarchical cluster analysis of significantly differentially expressed mRNAs (C) and lncRNAs (D). Red indicates up-regulated RNA and green indicates down-regulated RNA
Table 1.
Common up-regulated and down-regulated LncRNAs identified between the LAC and HAC groups of rabbits
| t_name | gene_id | gene_name | regulation |
|---|---|---|---|
| XR_001794626.1 | MSTRG.13,455 | ZBTB38 | down |
| XR_001792257.1 | MSTRG.21,901 | LOC108175509 | down |
| XR_517178.2 | 103,348,900 | LOC103348900 | down |
| XR_001792355.1 | MSTRG.22,541 | RPS28 | up |
| XR_001794087.1 | MSTRG.9827 | LOC103349551 | up |
| XR_001793515.1 | MSTRG.6208 | LOC108176812 | up |
| XR_001794723.1 | MSTRG.14,365 | LOC103350594 | up |
| XR_001793392.1 | MSTRG.5302 | LOC108176674 | up |
| XR_001795646.1 | MSTRG.20,881 | LOC108178897 | up |
| XR_001795241.1 | MSTRG.18,064 | LOC108178530 | up |
LAC: 10 ppm NH3; HAC: 30 ppm NH3.
GO and KEGG enrichment of differentially expressed LncRNA and mRNA
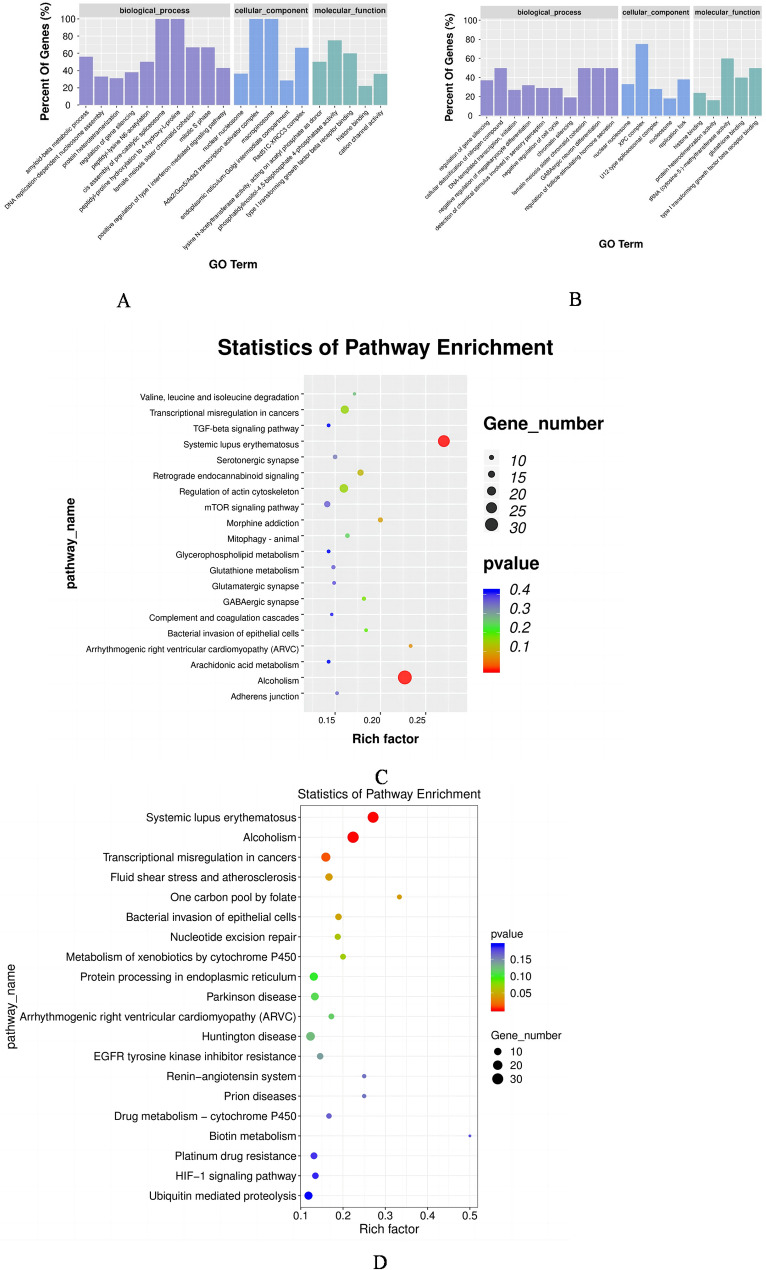
In this study, we performed GO analysis to gain functional categories of the DELs in the LAC versus the CG groups (Fig. 4A). Enriched biological processes (BP) included female meiosis sister chromatid cohesion and mitotic S phase. Enriched cellular components (CC) included nuclear nucleosome and Ada2/Gcn5/Ada3 transcription activator complex. Enriched molecular function (MF) included phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity and type I transforming growth factor beta receptor binding. The DELs were analyzed using GO analysis in the HAC versus CG groups (Fig. 4B). The most significantly enriched BPs included female meiosis sister chromatid cohesion and regulation of follicle-stimulating hormone secretion. Enriched CCs included nuclear nucleosome and XPC complex. Enriched MFs included histone binding and protein heterodimerization activity. The DELs were analyzed with the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and based on the first 20 pathway enrichments, the LAC versus CG group was mainly concentrated in Systemic lupus erythematosus, Alcoholism, Arrhythmogenic right ventricular cardiomyopathy (ARVC), Morphine addiction, Retrograde endocannabinoid signaling, the mammalian target of rapamycin (mTOR) signaling pathway and the transforming growth factor-beta (TGF-beta) signaling pathway (Fig. 4C). The HAC versus CG groups were mainly enriched in Systemic lupus erythematosus, Alcoholism, Transcriptional misregulation in cancers, Fluid shear stress and atherosclerosis, and the One carbon pool by folate and hypoxia-inducible factor-1α (HIF-1) signaling pathway (Fig. 4D).
Fig. 4.
Genes most enriched as determined by gene ontology (GO) and KEGG enrichment analysis of rabbit cumulus-oocyte complexes under different levels of NH3 exposure. The 20 most enriched differentially expressed genes (DEGs) by GO enrichment analysis in the LAC versus CG groups (A) and the HAC versus CG groups (B). The top 20 DEGs by KEGG enrichment analysis in the LAC versus CG groups (C) and the HAC versus CG groups (D). CG: < 3 ppm NH3; LAC: 10 ppm NH3; HAC: 30 ppm NH3
The construction of lncRNA-mRNA co-expression network
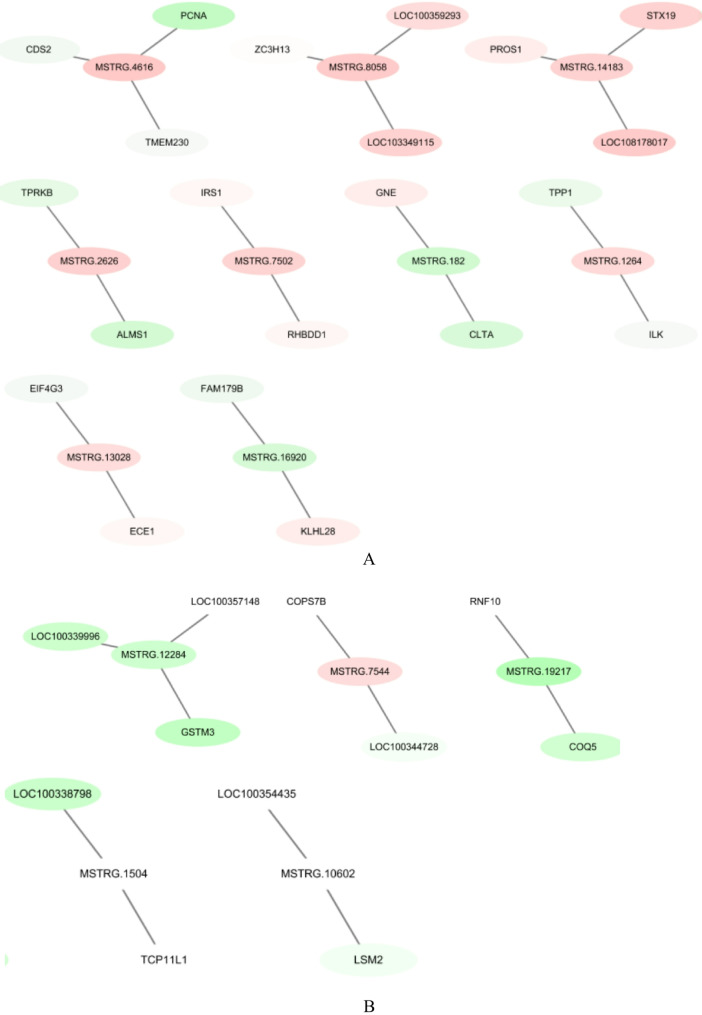
Co-expression networks were constructed between representative lncRNA and target mRNA to reveal the function of lncRNA (Fig. 5). The co-expression network map revealed that one lncRNA can correlate with several mRNAs. For example, LncRNA-MAPK3 was correlated with Yippee-like 3 (YPEL3) and Recombinant Human Fructose-Bisphosphate Aldolase A (ALDOA); and lncRNA-SHC1 was correlated with CDC28 protein kinase regulatory subunit 1B (CKS1B) and Homo sapiens zinc finger and BTB domain containing 7B (ZBTB7B).
Fig. 5.
Co-expression network of representative lncRNAs and their partial target mRNAs in the CG vs. LAC (A) and the CG vs. HAC groups (B). The relationships between lncRNA-mRNA were reconstructed based on expression correlation coefficients (Pearson correlation > 0.99 or < − 0.99) using Cytoscape software (v2.8.3). Red indicates up-regulated RNA and green indicates down-regulated RNA. CG: < 3 ppm NH3; LAC: 10 ppm NH3; HAC: 30 ppm NH3
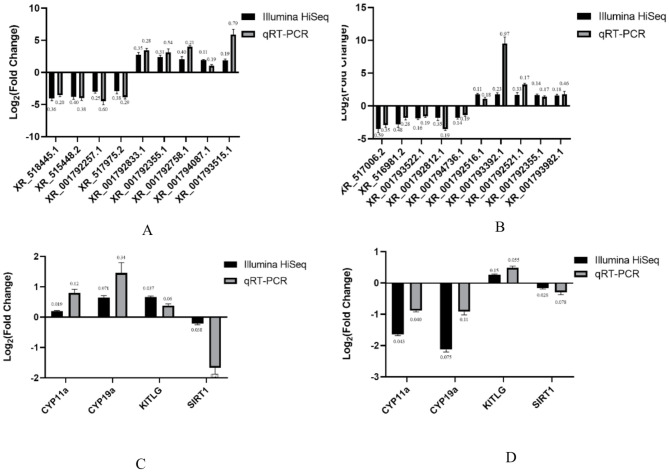
qRT-PCR analysis
Top nine differentially expressed lncRNAs in the sequencing results and four mRNAs related to reproduction were selected for the follow-up verification. The results demonstrated that the expression patterns of DELs and DEGs by qRT-PCR were consistent with those of RNA sequencing (Fig. 6A and B), verifying that the lncRNA expressions detected by RNA-seq analysis were reliable and accurate. Sirtuin 1 (Sirt1) mRNA expression increased significantly (P < 0.05) in the treatment groups compared with the control group (Fig. 6C). Luteinizing hormone receptor (LHR), Recombinant Cytochrome P450 11a (Cyp11a) and Recombinant Cytochrome P450 19a (Cyp19a) mRNA expression decreased significantly (P < 0.05) in the treatment groups compared with the control group (Fig. 6C).
Fig. 6.
The lncRNA and mRNA expressions were validated using qRT-PCR. Differentially expressed lncRNAs of LAC (A) and HAC (B) groups and genes of LAC (C) and HAC (D) groups were validated using qRT-PCR. Data of Illumina Hiseq and qRT-PCR were analyzed using SPSS version 21. Bars represent means ± SD, n = 4. CG: < 3 ppm NH3; LAC: 10 ppm NH3; HAC: 30 ppm NH3. CYP11a: recombinant Cytochrome P450 11a; CYP19a: recombinant Cytochrome P450 19a; KITLG: KIT Ligand; SIRT1: sirtuin 1
Discussion
Ammonia is toxic to animals and damages their reproductive performance [20–21]. Our laboratory found that exposure to NH3 destroyed the morphology, structure and reserve performance of rabbit ovaries [5]. In this experiment, histological examination of the ovary clearly showed abnormal morphological signs including collapsed follicular cavity and follicular cell necrosis, while primordial and primary follicular cells reduced and atretic follicular cells increased. These findings indicate that NH3 affects follicular development. Since rabbit COCs morphology is related to the atresia follicle [10], transcriptome was performed to investigate whether NH3 exposure impaired reproductive function in the COCs of rabbits. For the first time, we studied transcriptional alterations induced by NH3 in COCs and confirmed that lncRNA and mRNA genes may be involved in the progress of female reproductive function injury caused by NH3.
With the aid of high-throughput genomic analysis, the candidate genes related to the rabbit’s response to environmental stresses and reproductive traits were selected. We aimed to explore whether and how lncRNAs and mRNAs participated in rabbit COCs dysfunction caused by NH3. A total of 2066 lncRNAs, identified between the CG vs. LAC and CG vs. HAC groups, were associated with NH3 exposure. These dysregulated lncRNAs can be studied to investigate the damage and toxicity of NH3. In our experiment, GO analysis was performed to determine the functions of the identified differentially expressed genes. Two GO terms (female meiosis sister chromatid and cohesion and mitotic S phase) in the LAC versus CG groups and two GO terms (female meiosis sister chromatid cohesion and regulation of follicle-stimulating hormone secretion) in the HAC versus CG groups were involved in mammalian female fertility [22]. In the ovary, meiosis mainly produces ovum, and mitosis is used for the proliferation of granulosa cells and other auxiliary cells, which can produce and support follicle. Follicle-stimulating hormone plays a key role in regulating follicle development and producing mature ovum. GO analysis showed that NH3 affected the generation of germ cells and the development of follicles.
In this study, KEGG analysis revealed that lncRNAs were involved in reproductive function using the mTOR signaling pathway and TGF-beta signaling pathway in the LAC versus CG groups. The mTOR pathway plays an important role in maintaining follicle identity and preventing premature follicle exhaustion [23]. Previous studies showed that inhibition of the mTOR pathway could prolong ovarian life by inhibiting the activation of primordial follicle cells [24]. Both MSTRG.7502 and MSTRG.352 were significantly up-regulated in the COCs of the LAC group, and their target genes (Insulin receptor substrate 1 (IRS1) and ribosomal protein S6 (RPS6) were components of the mTOR signaling pathway. Here, IRS1 is associated with polycystic ovary syndrome and can produce or increase genetic variation, while RPS6 is a target of the mTOR pathway, and phosphorylation of RPS6 is a marker of cell senescence [25]. The TGF-beta pathway takes part in the growth of somatic cells and germ cells and is involved in the control of ovulation [26]. Additionally, MSTRG.9583, MSTRG.9580, MSTRG.9579 and MSTRG.9581 were significantly upregulated in the COCs of the LAC group, and their target gene, inhibin subunit beta A (INHBA), is involved in the TGF-beta signaling pathway. More specifically, INHBA can inhibit the synthesis and secretion of follicle-stimulating hormone in the pituitary gland and is associated with premature ovarian failure [27]. Furthermore, MSTRG.9058 was significantly upregulated in the COCs of the LAC group, and its target gene, transforming growth interacting factor (TGIF), is involved in the regulation of various cellular processes including development, proliferation and differentiation [28]. Three pathways (TGF-beta, Ras and MAPK signaling pathway) were oocyte maturation-related pathways in ochratoxin-induced oxidative stress 218 and apoptosis in porcine oocytes [29]. Two pathways (including mTOR and TGF-beta signaling pathways) regulated follicular development and atresia [30]. These findings suggest that 10 ppm NH3 affected the mTOR pathway and TGF-beta pathway by regulating lncRNA, which may inhibit germ cell development and follicular growth.
KEGG analysis revealed that LncRNAs were involved in the apoptosis of female germ cells via the HIF-1 signaling pathway in the HAC versus the CG groups. Ammonia can enter the blood through the alveoli, resulting in slow breathing and hypoxia in rabbits [31], which in turn harms ovarian follicular formation [32]. Additionally, follicles and oocytes are more prone to hypoxia than other cells [33–34]. Multiple target genes in the HIF-1 signaling pathway play an important role in metabolism, growth, proliferation, cytokines, mitogen secretion and cell death [35]. Hypoxic-specific genes are upregulated in the granulosa cells of elderly women, suggesting that hypoxia is the main mechanism of ovarian senescence and oocyte quality decline [36]. Also, HIF-1a plays a fundamental role in ovarian follicular development [37], while B-cell lymphoma/Leukemia-2 (Bcl-2), a specific target of HIF-1a, plays an important role in the apoptosis of female germ cells. The change expression of Bcl-2 can alter pro-apoptotic factors such as Bax, leading to the apoptosis of oocytes [38]. The results of this experiment showed that MSTRG.9345 was significantly up-regulated in the NH3-treated groups, and its target gene, Bcl-2, was enriched in the HIF-1 signaling pathway. Other researchers working on lncRNAs using transcriptome analysis have confirmed that the above mechanisms are stimulated by environmental factors. Consistent with the above studies, in this experiment, lncRNA was involved in reproductive toxicity via the HIF-1 signaling pathway in the HAC group, which resulted in germ cell apoptosis and follicle development inhibition.
Both lncRNAs and mRNAs share similar biogenesis pathways and are involved in multiple biological processes, but exert their functions in different ways [39–40]. Unlike other protein-coding mRNAs, lncRNAs exhibit functional uniqueness by participating in and modulating the various cellular processes such as histone modification, DNA methylation, and cellular transcription [41]. Changes in the amount of lncRNAs can induce aberrant gene expression that contributes to various disease states and biological dysfunctions [42]. Therefore, the co-expression of lncRNAs and mRNA should be analyzed to help predict their functional roles in COCs during NH3 exposure. Through co-expression network analysis, we identified differentially expressed lncRNAs strongly correlated with genes involved in reproductive cell development. For instance, INHBA is the target gene of MSTRG.9584. It is a gonadal glycoprotein hormone that can inhibit the synthesis and secretion of follicle-stimulating hormones from the pituitary gland and is strongly associated with premature ovarian failure [43–44]. Additionally, TGIF is involved in regulating various cellular processes, including development, proliferation and differentiation, which is correlated with MSTRG.9051 [45]. YPEL3 is associated with the induction of cell growth inhibition through apoptosis [46], while ALDOA is related to oocyte competence, glycolysis and developmental competence in COCs, which is the target gene of MSTRG.6032 [47]. CKS1B plays critical roles in cell cycle regulation and ovarian maturation and can regulate MSTRG.12,034 [48]. Therefore, lncRNAs can affect the development and function of follicles and adjust COCs biological functions through targeted regulation of mRNAs under an NH3-exposure environment.
Conclusions
Ammonia is one of the major air pollutants in intensive animal production. In the present study, we explore the potential mechanisms of ammonia toxicity at concentrations of 10 ppm and 30 ppm from the perspective of rabbit COCs. The number of atresia follicles increased in the NH3-treated rabbits. This is the first report of the correlation of lncRNA and COCs in rabbits after NH3 exposure. Co-expression analysis found that the lncRNAs, MAPK3 and SHC1 were correlated with a change in cumulus cell and oocyte function after NH3 exposure. The mTOR signaling pathway and the TGF-beta signaling pathway inhibited germ cell development and follicular growth in the NH3 exposure group. The results suggest that LncRNAs are involved in the apoptosis of female germ cells via the HIF-1 signaling pathway in NH3-exposed rabbits.
Materials and methods
Animal groups and treatments
A total of 150 female rabbits (35 days old) were purchased from Xingtai Kangming Breeding Co., LTD (Xingtai, China). The rabbits were randomly divided into three environmentally controlled compartments, and the concentration of NH3 was < 3 ppm (control group, CG), 10 ppm (low ammonia concentration, LAC) and 30 ppm (high ammonia concentration, HAC). In environmentally controlled compartments, the concentration of H2S was kept under 0.2 mg/m3 and the CO2 concentration was kept under 1500 mg/m3. All animals were placed in environmentally controlled compartments for four weeks and exogenous NH3 was added for 8 h per day for the NH3 groups. The rabbits were given ad libitum access to water and food throughout the study. The composition and nutrient levels of the basal diet are provided in Supplementary Table 1. We assessed the animals’ status four times a day referring to the guidelines of assessment for humane endpoints in animal experiments (RB/T173-2018) http://www.lascn.net/Item/75841.aspx.
On the 28th day of the experiment, 10 rabbits from each group were euthanized through intraperitoneal injection with sodium pentobarbital (100 mg/kg) by professional veterinarians with a Practicing Veterinary Certificate. The remaining rabbits were used for other studies under the guidance of the animal ethics regulations. Ovaries were harvested using aseptic techniques, stripped of surrounding fat tissue and washed three times in normal saline supplemented with two antibiotics. Ovary samples were kept in 10% formalin buffer (four rabbits in each group). The others were used to harvest COCs (six rabbits in each group). The follicles on the surface of the ovaries were extracted using a disposable syringe and COCs were selected from the follicles and immediately frozen in liquid nitrogen. Samples were stored at -80 ◦C until RNA extraction.
Sampling
Ovarian tissue morphology
Ovary samples were removed from 10% formalin buffer, dehydrated with a graded series of ethanol, cleared in xylene, and embedded in paraffin. Ovary tissue blocks were sliced into 4 mm thick slices and the sections were stained with hematoxylin and eosin (Saiweier Biotechnology Co., Ltd., Wuhan, China). Slices were then dehydrated and mounted for light microscopy examination.
Collection of COCs
Immediately after euthanasia, the ovaries were removed from the abdominal cavity of each rabbit and transported in normal saline (with added penicillin and streptomycin) at 37 °C. After two additional washes with phosphate-buffered saline (PBS), the ovaries were dipped into 70% ethanol for 30 s. Following several washes with warm PBS, the ovaries were transferred to a plastic Petri dish containing DMEM/F12 supplemented with 10% FBS and 0.1% PS. The follicular materials were aspirated from growing follicles (4–8 mm in diameter) by an 18-gauge needle attached to a 5 mL syringe. After this, Petri dishes containing COCs were analyzed using a stereomicroscope (Meiji EMZ 13TR, Meiji Techno Co, Saitama, Japan) for morphological selection. The COCs containing compact cumulus cells that demonstrated homogeneous ooplasm were removed (30 COCs were used per replicate), and placed in a dish containing a manipulation medium.
RNA library construction and sequencing
Total RNA was extracted using Trizol reagent (Invitrogen, CA, USA) following the manufacturer’s procedure. The total RNA quantity and purity were analyzed on a Bioanalyzer 2100 and an RNA 6000 Nano LabChip Kit (Agilent, CA, USA) with a RIN number > 7.0. Approximately 5 µg of total RNA was used to deplete ribosomal RNA according to the directions of the Epicentre Ribo-Zero Gold Kit (Illumina, San Diego, USA). Following purification, the poly(A)- or poly(A) + RNA fractions were fragmented into small pieces using divalent cations under elevated temperature. Then the cleaved RNA fragments were reverse-transcribed to create the final cDNA library following the protocol for the mRNA-Seq sample preparation kit (Illumina, San Diego, USA). The average insert size for the paired-end libraries was 300 bp (± 50 bp). Then, paired-end sequencing was performed on an Illumina Novaseq™ 6000 at the (lc-bio, China) following the vendor’s recommended protocol.
Transcript assembly
Firstly, Cutadapt [49] was used to remove the reads that contained adaptor contaminations, low-quality bases and undetermined bases. The sequence quality was verified using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) [50]. We used hisat2 [51] to map reads to the oryctolagus_cuniculus NCBI_2.0 version of the rabbit genome (Genome sequence file:
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/003/625/GCF_000003625.3_OryCun2.0/GCF_000003625.3_OryCun2.0_genomic.fna.gzoryctolagus_cuniculus; Comment file: http://www.ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/003/625/GCF_000003625.3_OryCun2.0/GCF_000003625.3_OryCun2.0_genomic.gff.gz ). The mapped reads of each sample were assembled using StringTie [52]. Then, all the transcriptomes from COCs samples were merged to reconstruct a comprehensive transcriptome using Perl scripts. After the final transcriptome was generated, StringTie and edgeR [53] were used to estimate the expression levels of all the transcripts.
LncRNA identification
First of all, transcripts that overlapped with known mRNAs and transcripts shorter than 200 bp were discarded. Then we utilized the Coding Potential Calculator (CPC) [54] and Coding-Non-Coding Index (CNCI) [55] to predict transcripts with coding potential. All transcripts with CPC score<-1 and CNCI score < 0 were removed. The remaining transcripts were considered lncRNAs.
Differential expression analysis of mRNAs and LncRNAs
StringTie [54] was used to determine the expression levels of mRNAs and lncRNAs by calculating FPKM [56]. The differentially expressed mRNAs and lncRNAs were selected with log2 (fold change) > 1 or log2 (fold change) <-1 and with statistical significance (P-value < 0.05) by the R package-edgeR [53].
Target gene prediction and functional analysis of LncRNAs
To explore the function of the lncRNAs, we predicted the cis-target genes of the lncRNAs as they may play a cis role, acting on neighboring target genes. In this study, coding genes in the 100,000 bp upstream and downstream were selected by a Python script. Then, we performed a functional analysis of the target genes for lncRNAs using the BLAST2GO program [57]. Significance was expressed as P < 0.05.
qRT-PCR
The differentially expressed lncRNAs and mRNAs related to reproduction were selected to verify the transcriptional expression profile by qRT-PCR. The primers were designed using Primer 5.0 and synthesized by the Beijing Genomics Institute (Table 2). Total RNA extraction from COCs was performed using Trizol (Takara, Dalian, China). The reverse transcription steps were carried out using the PrimerScript RT kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The PCR amplification was performed using the SYBR Premix Ex TagTM kit (TaKaRa, Dalian, China). The reaction mixtures were 20 µL, containing 12.5 µL SYBR premix, 1 µL of each sense and antisense primer (10 µmol/L), 8.5 µL water, and 2 µL complementary DNA (cDNA).
Table 2.
Sequence of primers for real-time PCR
| Type | Gene name | Gene ID | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|---|
| mRNA | GADPH | NM_080369 | GTGGCGCTGGACTTCGA | CGTTGCCGATGGTGATGA |
|
CYP11A KITLG SIRT CYP19A |
GCCGGGAACCTAGCTTCTTT AGTGGATGACCTTGTGGAGTG GGAACTGGATTTGGGGCTGAT TGGAATTACGAGGGCACGTC |
GATGCTCATCTCCACCTCGG GATGCTCATCTCCACCTCGG TTCTGTAGGCAAAACTGGACA CATCCACAGGAATCTGGCGT |
||
| lncRNA |
XR_518445. 1 XR_001792257. 1 XR_001792833. 1 XR_001792355. 1 XR_001792758. 1 XR_001794087. 1 |
GGGTCACTATCGGTTGCTTGTTTG GAGCGAGCATCTCATTACAAG GCCAGGTAAGGGTTTAGG AGGCTGAGTCAAAGAATGCTA ACCAGGTGAGGATGGACAA TTCATGGACGACACGAGCC GGTGAGCAGCCAAGAGCAATAC GGTCTCCCGATTCCAAAGT |
GGTATTCTGCTGACGGAGTGCTAA TCAGTTTGACGACGAAATAGC CAACCAGGAGAATCACAAAGTC GTGAGCAGCCAAGA GCAATA CACAGGCAGACGAGGACAG TCTGACTCCAGCAGGGTGA CATCGACTACAACGTGCCCT GGTCTGACATGAGCCTTCT |
Kitlg: KIT Ligand; Cyp11A: Recombinant Cytochrome P450 11a; Sirt1: sirtuin 1; Cyp19A: Recombinant Cytochrome P450 19a.
Statistical analysis
Data were analyzed using SPSS version 21 by performing one-way ANOVA, and differences between means were compared using Duncan’s multiple range test. Data are expressed as mean ± standard deviation (SD). Significant differences are indicated by P < 0.05 and P < 0.01.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We gratefully thank Dr. Fengyang Wu for his excellent technical assistance. We would like to thank MogoEdit (https://www.mogoedit.com) for its English editing during the preparation of this manuscript.
Abbreviations
- ALDOA
Recombinant Human Fructose-Bisphosphate Aldolase A
- ARVC
Arrhythmogenic right ventricular cardiomyopathy
- Bcl-2
B-cell lymphoma/Leukemia-2
- BP
Biological process
- CC
Cellular component
- cDNA
Complementary DNA
- CKS1B
CDC28 protein kinase regulatory subunit 1B
- CNCI
Coding-Non-Coding Index
- COCs
Cumulus oocyte complex
- CPC
Coding Potential Calculator
- Cyp11a
Recombinant Cytochrome P450 11a
- Cyp19a
Recombinant Cytochrome P450 19a
- DEGs
Differentially expressed mRNAs
- DELs
Differentially expressed lncRNAs
- FPKM
Fragments Per Kilobase of exon model per Million mapped fragments
- GO
Gene Ontology
- HAC
High ammonia concentration
- HAS2-AS1
Hyaluronan Synthase-AS1
- HIF-1
Hypoxia-inducible factor-1α
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LAC
Low ammonia concentration
- lncRNA
Long non-coding RNA
- MF
Molecular function
- mTOR
Mammalian target of rapamycin
- NH3
Ammonia
- PSMD6
Non ATPase 6
- qRT-PCR
Quantitative real-time polymerase chain reaction
- Sirt1
Sirtuin 1
- TGF-beta
Transforming growth factor-beta
- YPEL3
Yippee-like 3
- ZBTB7B
Homo sapiens zinc finger and BTB domain containing 7B
Author contributions
J and FY equally contributed to this work by performing the experiments and drafting the manuscript. J and JW conceived the study. XY and YH collected samples and assisted in data analysis. SD and SJ analyzed data. BJ edited and finalized the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This study was supported by the National Key R&D Program of China (2016YFD0500501). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
The datasets generated and analyzed during the current study are available in GEO (GSE282848).
Declarations
Ethics approval and consent to participate
All experiments were performed in accordance with the ARRIVE guidelines and were carried out in accordance with the U.K. Animals (Scientific Procedures) Act, 1986 and associated guidelines, EU Directive 2010/63/EU for animal experiments, and the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978), approved by the Ethics Committee of Animal Experimentation of Hebei Agricultural University (Protocol 2021083) and obtained informed consent from Xingtai Kangming Breeding Co., LTD (Xingtai, China) to use the animals.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jia Cui and Fengyang Wu contributed equally to this work.
Contributor Information
Jiawei Guo, Email: 1373528725@qq.com.
Baojiang Chen, Email: chenbaojiang@vip.sina.com.
References
- 1.David B, Mejdell C, Michel V, Lund V, Moe RO. Air Quality in Alternative Housing Systems may have an Impact on Laying Hen Welfare. Part II-Ammonia. Animals (Basel). (2015) Sep 3;5(3):886– 96. 10.3390/ani5030389 [DOI] [PMC free article] [PubMed]
- 2.Cui J, Wu F, Yang X, Liu S, Han S, Chen B. Effects of ammonia on hypothalamic-pituitary-ovarian axis in female rabbits. Ecotoxicol Environ Saf. (2021) Dec 20;227:112922. doi: 10. 1016/j.ecoenv.2021.112922. [DOI] [PubMed]
- 3.Xu Y, Li Z, Zhang S, Zhang H, Teng X. miR- 187-5p/apaf- 1 axis was involved in oxidative stress-mediated apoptosis caused by ammonia via mitochondrial pathway in chicken livers. Toxicol Appl Pharmacol. (2020)Feb 1;388:114869. doi: 10. 1016/j.taap.2019.114869. [DOI] [PubMed]
- 4.Han Q, Tong J, Sun Q, Teng X, Zhang H, Teng X. The involvement of miR-6615-5p/Smad7 axis and immune imbalance in ammonia-caused inflammatory injury via NF-κB pathway in broiler kidneys. Poult Sci. 2020;287(11):5378–88. 10. 1016/j.psj.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali Shah SW, Zhang S, Ishfaq M, Tang Y, Teng X. PTEN/AKT/mTOR pathway involvement in autophagy, mediated by miR-99a-3p and energy metabolism in ammonia-exposed chicken bursal lymphocytes. Poult Sci. (2021) Feb;100(2):553–564. doi: 10. 1016/j.psj.2020. 11.015. [DOI] [PMC free article] [PubMed]
- 6.Kidder GM, Vanderhyden BC. Bidirectional communication between oocytes and follicle cells: ensuring oocyte developmental competence. Can J Physiol Pharmacol. 2010;88(4):399–413. 10. 1139/y10-009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang LP, Jiang WX, Gao SX. Detection and evaluation of different types of rabbit houses in winter[J]. Shandong Agricultural Sci. (2009) (09): 91–3.
- 8.Shaeib F, Khan SN, Ali I, Thakur M, Saed MG, Dai J, Awonuga AO, Banerjee J, Abu-Soud HM. The Defensive Role of Cumulus Cells Against Reactive Oxygen Species Insult in Metaphase II Mouse Oocytes. Reprod Sci. (2016) Apr;23(4):498–507. doi: 10. 1177/1933719115607993. [DOI] [PMC free article] [PubMed]
- 9.Rouhollahi Varnosfaderani S, Hajian M, Jafarpour F, Ghazvini Zadegan F, Nasr-Esfahani MH. Granulosa secreted factors improve the developmental competence of cumulus oocyte complexes from small antral follicles in sheep. PLoS One. 2020 Mar 17;15(3):e0229043. doi: 10. 1371/journal.pone.0229043. [DOI] [PMC free article] [PubMed]
- 10.Boni R, Cuomo A, Tosti E. Developmental potential in bovine oocytes is related to cumulus-oocyte complex grade, calcium current activity, and calcium stores. Biol Reprod. 2002 Mar;66(3):836–42. 10. 1095/biolreprod66.3.836. [DOI] [PubMed]
- 11.Yang LL, Zhao Y, Luo SM, Ma JY, Ge ZJ, Shen W, Yin S. Toxic effects and possible mechanisms of hydrogen sulfide and/or ammonia on Porcine oocyte maturation in vitro. Toxicol Lett (2018) Mar 15;285:20–6. 10. 1016/j.toxlet.2017.12.019 [DOI] [PubMed]
- 12.Nandi S, Tripathi SK, Gupta PSP, Mondal S. Nutritional and metabolic stressors on ovine oocyte development and granulosa cell functions in vitro. Cell Stress Chaperones. 2018 May;23(3):357–71. 10. 1007/s12192-017-0846-1. [DOI] [PMC free article] [PubMed]
- 13.Gao X, Niu C, Wang Z, Jia S, Han M, Ma Y, Guan X, Wang L, Qiao X, Xu Y. Comprehensive analysis of LncRNA expression profiles in cytopathic biotype BVDV-infected MDBK cells provides an insight into biological contexts of host-BVDV interactions. Virulence. 2021;12(1):20–34. 10.1080/21505594.2020.1857572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sindhu P, Magotra A, Sindhu V, Chaudhary P. Unravelling the impact of epigenetic mechanisms on offspring growth, production, reproduction and disease susceptibility. Zygote. 2024;32(3):190–206. 10.1017/S0967199424000224. [DOI] [PubMed] [Google Scholar]
- 15.Sparber P, Filatova A, Khantemirova M, Skoblov M. The role of long non-coding RNAs in the pathogenesis of hereditary diseases. BMC Med Genomics. (2019) Mar 13;12(Suppl 2):42. 10. 1186/s12920-019-0487-6 [DOI] [PMC free article] [PubMed]
- 16.Kimura AP, Yoneda R, Kurihara M, Mayama S, Matsubara SA, Long Noncoding RNA. lncRNA-Amhr2, Plays a Role in Amhr2 Gene Activation in Mouse Ovarian Granulosa Cells. Endocrinology. (2017) Nov 1;158(11):4105–4121. doi: 10. 1210/en.2017– 00619. [DOI] [PubMed]
- 17.Yung Y, Ophir L, Yerushalmi GM, Baum M, Hourvitz A, Maman E. HAS2-AS1 is a novel LH/hCG target gene regulating HAS2 expression and enhancing cumulus cells migration. J Ovarian Res. (2019) Feb 28;12(1):21. 10. 1186/s13048-019-0495-3 [DOI] [PMC free article] [PubMed]
- 18.Xia X, Burn MS, Chen Y, Karakaya C, Kallen A. The relationship between H19 and parameters of ovarian reserve. Reprod BiolEndocrinol. (2020) May 13;18(1):46. doi: 10. 1186/s12958-020-00578-z. [DOI] [PMC free article] [PubMed]
- 19.Li L, Fu YC, Xu JJ, Lin XH, Chen XC, Zhang XM, Luo LL. Caloric restriction promotes the reserve of follicle pool in adult female rats by inhibiting the activation of mammalian target of Rapamycin signaling. Reprod Sci. 2015;22(1):60–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao Z, Xu W, Zhu C, Zhang S, Shi Z, Song W, Liu H, Li H. Effects of ammonia on intestinal microflora and productive performance of laying ducks. Poult Sci. (2019) May 1;98(5):1947–1959. 10.3382/ps/pey578 [DOI] [PubMed]
- 21.Li D, Tong Q, Shi Z, Li H, Wang Y, Li B, Yan G, Chen H, Zheng W. Effects of chronic heat stress and ammonia concentration on blood parameters of laying hens. Poult Sci. 2020;99(8):3784–92. 10.1016/j.psj.2020.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su YQ, Wigglesworth K, Pendola FL, O’Brien MJ, Eppig JJ. Mitogen-activated protein kinase activity in cumulus cells is essential for gonadotropin-induced oocyte meiotic resumption and cumulus expansion in the mouse. Endocrinology. 2002;143(6):2221–32. 10. 1210/endo.143.6.8845. [DOI] [PubMed] [Google Scholar]
- 23.Cui W. Oocyte Spontaneous Activation: An Overlooked Cellular Event That Impairs Female Fertility in Mammals. Front Cell Dev Biol. (2021) Mar 8;9:648057. 10.3389/fcell.2021.648057 [DOI] [PMC free article] [PubMed]
- 24.Correia B, Sousa MI, Ramalho-Santos J. The mTOR pathway in reproduction: from gonadal function to developmental coordination. Reproduction. 2020;159(4):R173–88. 1530/REP-. [DOI] [PubMed] [Google Scholar]
- 25.Albertini DF, Combelles CM, Benecchi E, Carabatsos MJ. Cellular basis for paracrine regulation of ovarian follicle development. Reproduction. 2001 May;121(5):647– 53. doi: 10. 1530/rep.0.1210647. [DOI] [PubMed]
- 26.Zheng GH, Liu CM, Sun JM, Feng ZJ, Cheng C. Nickel-induced oxidative stress and apoptosis in Carassius auratus liver by JNK pathway. Aquat Toxicol. 2014 Feb;147:105– 11. doi: 10. 1016/j.aquatox.2013. 12.015. [DOI] [PubMed]
- 27.Monsivais D, Matzuk MM, Pangas SA. The TGF- β Family in the Reproductive Tract. Cold Spring Harb Perspect Biol. (2017) Oct 3;9(10):a022251. doi: 10. 1101/cshperspect.a022251. [DOI] [PMC free article] [PubMed]
- 28.Mattar D, Samir M, Laird M, Knight PG. Modulatory effects of TGF- Β1 and BMP6 on the calangiogenesis and steroidogenesis in the bovine ovary. Reprod 2020 Apr;159(4):397–408. doi: 10. 1530/REP- 19–0311. [DOI] [PubMed]
- 29.Peng C. The TGF-beta superfamily and its roles in the human ovary and placenta. J Obstet Gynaecol Can. 2003 Oct;25(10):834–44. doi: 10. 1016/s1701-2163(16)30674-0. [DOI] [PubMed]
- 30.Lan M, Zhang Y, Wan X, Pan MH, Xu Y, Sun SC. Melatonin ameliorates Ochratoxin A-induced oxidative stress and apoptosis in Porcine oocytes. Environ Pollut. 2020;256:113374. 10.1016/j.envpol.2019.113374. [DOI] [PubMed] [Google Scholar]
- 31.Pan Y, Yang S, Cheng J, Lv Q, Xing Q, Zhang R, Liang J, Shi D, Deng Y. Whole-Transcriptome analysis of LncRNAs mediated CeRNA regulation in granulosa cells isolated from healthy and Atresia follicles of Chinese Buffalo. Front Vet Sci (2021) Jul 14;8:680182. 10.3389/fvets.2021.680182 [DOI] [PMC free article] [PubMed]
- 32.Stern RA, Mozdziak PE. Differential ammonia metabolism and toxicity between avian and mammalian species, and effect of ammonia on skeletal muscle: A comparative review. J Anim Physiol Anim Nutr (Berl). 2019 May;103(3):774–785. doi: 10. 1111/jpn.13080 [DOI] [PubMed]
- 33.Molinari E, Bar H, Pyle AM, Patrizio P. Transcriptome analysis of human cumulus cells reveals hypoxia as the main determinant of follicular senescence. Mol Hum Reprod. 2016 Aug;22(8):866–76. 10.1093/molehr/gaw038. [DOI] [PMC free article] [PubMed]
- 34.Van Blerkom J. Mitochondria in human oogenesis and preimplantation embryogenesis: engines of metabolism, ionic regulation and developmental competence. Reproduction. 2004 Sep;128(3):269–80. 10.1530/rep. [DOI] [PubMed]
- 35.Clark AR, Stokes YM. Follicle structure influences the availability of oxygen to the oocyte in antral follicles. Comput Math Methods Med. 2011;2011:287186. 1155/2011/287186. Epub 2011 Nov 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaluz S, Kaluzová M, Stanbridge EJ. Regulation of gene expression by hypoxia: integration of the HIF-transduced hypoxic signal at the hypoxia-responsive element. Clin Chim Acta. 2008 Sep;395(1–2):6–13. 10.1016/j.cca.2008.05.002. [DOI] [PMC free article] [PubMed]
- 37.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010 Oct 22;40(2):294–309. doi: 10. 1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed]
- 38.Wang F, Zhang Z, Wang Z, Xiao K, Wang Q, Su J, Wang Z. Expression and clinical significance of the HIF-1a/ET-2 signaling pathway during the development and treatment of polycystic ovary syndrome. J Mol Histol. 2015;46(2):173–81. 10.1007/s10735-015-9609-4. [DOI] [PubMed] [Google Scholar]
- 39.Dayal S, Chaubey D, Joshi DC, Ranmale S, Pillai B. Noncoding RNAs: emerging regulators of behavioral complexity. Wiley Interdiscip Rev RNA 2024 May-Jun;15(3):e1847. 10.1002/wrna.1847 [DOI] [PubMed]
- 40.Singh AK. Rules and impacts of nonsense-mediated mRNA decay in the degradation of long noncoding RNAs. Wiley Interdiscip Rev RNA. 2024 May-Jun;15(3):e1853. 10.1002/wrna.1853. [DOI] [PubMed]
- 41.Das S, Zea Rojas MP, Tran EJ. Novel insights on the positive correlation between sense and antisense pairs on gene expression. Wiley Interdiscip Rev RNA. 2024 Jul-Aug;15(4):e1864. 10.1002/wrna.1864. [DOI] [PMC free article] [PubMed]
- 42.Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC, Essig J, Otto GM, O’Sullivan MG, Largaespada DA, Schwertfeger KL, Marahrens Y, Kawakami Y, Bagchi A. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512(7512):82–6. 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao Xiaohan. Expression characteristics of Bcl-2 related apoptotic genes in sheep reproductive organs and the role of bad genes in sheep estrous cycle. Chin Acad ofAgricultural Sci (in chinese).
- 44.Mattar D, Samir M, Laird M, Knight PG. Modulatory effects ofTGF- Β1 and BMP6 on thecal angiogenesis and steroidogenesis in the bovine ovary. Reproduction. 2020;159(4):397–408. doi: 10. 1530/REP-. [DOI] [PubMed] [Google Scholar]
- 45.Peng C. The TGF-beta superfamily and its roles in the human ovary and placenta. J Obstet Gynaecol Can. 2003;25(10):834–44. 10. 1016/s1701-2163(16)30674-0. [DOI] [PubMed] [Google Scholar]
- 46.Dou X, Sun Y, Li J, Zhang J, Hao D, Liu W, Wu R, Kong F, Peng X, Li J. Short-term Rapamycin treatment increases ovarian lifespan in young and middle-aged female mice. Aging Cell. 2017;16(4):825–36. 10. 1111/acel.12617. Epub 2017 May 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan Y, Ida JM, Paczkowski M, Krisher RL. Identification of developmental competence-related genes in mature porcine oocytes. Mol Reprod Dev. (2011) Aug;78(8):565– 75. doi: 10. 1002/mrd.21351. [DOI] [PMC free article] [PubMed]
- 48.Yang Y, Zhang Y, Zhuang Y, Zhang C, Bao C, Cui Z. Identification of differentially abundant mRNA transcripts and autocrine/paracrine factors in oocytes and follicle cells of mud crabs. Anim Reprod Sci. (2021) Jul;230:106784. doi: 10. 1016/j.anireprosci.2021.106784. [DOI] [PubMed]
- 49.Kechin A, Boyarskikh U, Kel A, Filipenko M, cutPrimers:. A new tool for accurate cutting of primers from reads of targeted next generation sequencing. J Comput Biol. 2017;24(11):1138–43. 10.1089/cmb.2017.0096. Epub 2017 Jul 17. PMID: 28715235. [DOI] [PubMed] [Google Scholar]
- 50.FastQC. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 51.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–9. 10.1038/nmeth.1923. PMID: 22388286; PMCID: PMC3322381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–5. 10.1038/nbt.3122. Epub 2015 Feb 18. PMID: 25690850; PMCID: PMC4643835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ, Smyth GK. Bioinformatics. 2010;26(1):139–40. 10.1093/bioinformatics/btp616. Epub 2009 Nov 11. PMID: 19910308; PMCID: PMC2796818. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. [DOI] [PMC free article] [PubMed]
- 54.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35(Web Server issue):W345-9. 10.1093/nar/gkm391. PMID: 17631615; PMCID: PMC1933232. [DOI] [PMC free article] [PubMed]
- 55.Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41(17):e166. 10.1093/nar/gkt646. Epub 2013 Jul 27. PMID: 23892401; PMCID: PMC3783192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. 10.1038/nbt.1621. Epub 2010 May 2. PMID: 20436464; PMCID: PMC3146043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6. 10.1093/bioinformatics/bti610. Epub 2005 Aug 4. PMID: 16081474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Robinson MD, McCarthy DJ, Smyth GK. Bioinformatics. 2010;26(1):139–40. 10.1093/bioinformatics/btp616. Epub 2009 Nov 11. PMID: 19910308; PMCID: PMC2796818. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are available in GEO (GSE282848).