Abstract
Temozolomide (TMZ) remains the cornerstone chemotherapy for glioma, yet intrinsic and acquired resistance mechanisms significantly limit its clinical effectiveness. This review summarizes the multifaceted molecular pathways contributing to TMZ resistance, including enhanced DNA repair mechanisms such as O6-methylguanine-DNA methyltransferase (MGMT), mismatch repair (MMR), and base excision repair (BER). Additional resistance factors include genetic mutations that affect the drug response, dysregulated non-coding RNAs (miRNAs, lncRNAs, and circRNAs), glioma stem cells (GSCs), cytoprotective autophagy, an immunosuppressive tumor microenvironment (TME), altered signaling pathways, and active drug efflux transporters. Recent advancements to overcome these resistance mechanisms, including enhancing TMZ bioavailability through nanoparticle-based delivery systems and the inhibition of efflux transporters, have been explored. Novel therapeutic approaches that target DNA repair pathways and manipulate autophagy are highlighted. Immunotherapeutic interventions reversing immune suppression and metabolic strategies targeting tumor metabolism offer additional avenues. Emerging therapies such as CRISPR-based gene editing, phytochemical combinations, repurposed drugs, and novel TMZ analogs designed to bypass MGMT-mediated resistance are also discussed. This review highlights current developments and identifies emerging areas, with the goals of enhancing clinical outcomes and prolonging survival for glioma patients.
Keywords: Temozolomide, Glioma, Chemoresistance, Treatment strategies
Background
Glioma represent the most common primary malignant brain tumors in adults, constituting more than 80% of all central nervous system (CNS) malignancies. Among these, glioblastoma (GBM, isocitrate dehydrogenase [IDH]-wildtype, CNS WHO grade 4) is the most aggressive, with an annual incidence of 3.2 per 100,000 individuals [1]. Despite multimodal therapy involving maximal safe resection, radiotherapy (RT), and chemotherapy, GBM has a dismal prognosis: fewer than 12% of patients achieve 3-year survival (long-term survivors) [2, 3]. While surgical intervention may cure circumscribed glioma subtypes, such as pleomorphic xanthoastrocytoma, subependymal giant cell astrocytoma, pilocytic astrocytoma, chordoid glioma and so on, conventional therapies frequently fail in diffuse adult-type glioma because of intrinsic and acquired therapeutic resistance [4].
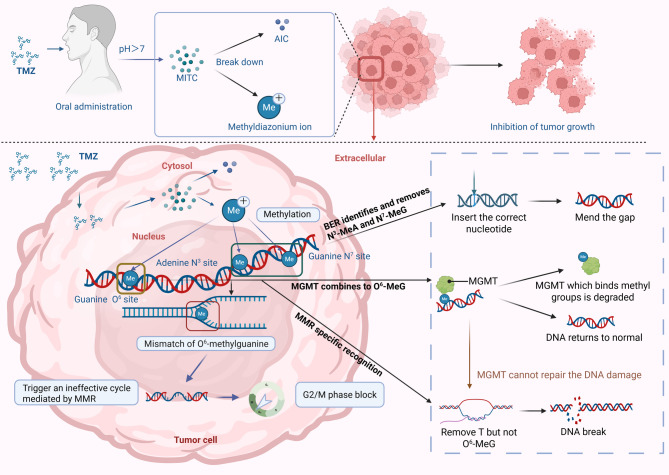
Temozolomide (TMZ), an orally administered imidazotetrazine prodrug, undergoes pH-dependent conversion to its active metabolite 5-(3-methyltriazen-1-yl) imidazole-4-carboxamide (MTIC) under physiological conditions [5–7]. Subsequent degradation releases the methyldiazonium cation, which preferentially methylates DNA at the guanine N7/O6 and adenine N3 positions [8]. The cytotoxic effect primarily arises from O6-methylguanine mispairing during replication, triggering mismatch repair (MMR)-mediated futile cycles and G2/M arrest [9]. Owing to its partial blood-brain barrier (BBB) penetration capability and oral bioavailability [10], TMZ has synergistic efficacy when combined with RT, establishing it as a first-line chemotherapeutic for GBM [11–15] (Fig. 1).
Fig. 1.
Mechanism of action of TMZ. TMZ is an orally administered imidazotetrazine prodrug that undergoes pH-dependent conversion under physiological conditions into its active metabolite MTIC. MTIC subsequently reacts with water, generating 5-aminoimidazole-4-carboxamide (AIC) and a highly reactive methyldiazonium cation. This methyldiazonium cation preferentially methylates DNA at the N7 position of guanine (N7-MeG; approximately 70%), predominantly in guanine-rich regions but also at adenine residues (N3-MeA; approximately 9%) and guanine residues at the O6 position (O6-MeG; approximately 6%). The cytotoxic effect of TMZ primarily results from the formation of O6-MeG lesions, which are carcinogenic, mutagenic, and toxic. These lesions are repaired directly by the suicide enzyme MGMT, which removes the methyl group from O6-MeG, restoring the original guanine residue. If left unrepaired, O6-MeG mispairs specifically with thymine during DNA replication, activating DNA MMR. MMR recognizes and excises the mispaired thymine on the daughter strand; however, the persistent O6-MeG lesion in the template strand results in futile cycles of thymine reinsertion and excision. These continuous futile repair cycles generate persistent DNA strand breaks, leading to G2/M cell cycle arrest and eventually cell death. The more abundant DNA adducts, N7-MeG and N3-MeA, are rapidly repaired via DNA BER. Therefore, the most important DNA repair systems affecting the mechanism of action and cytotoxicity of TMZ are MGMT, MMR, and BER
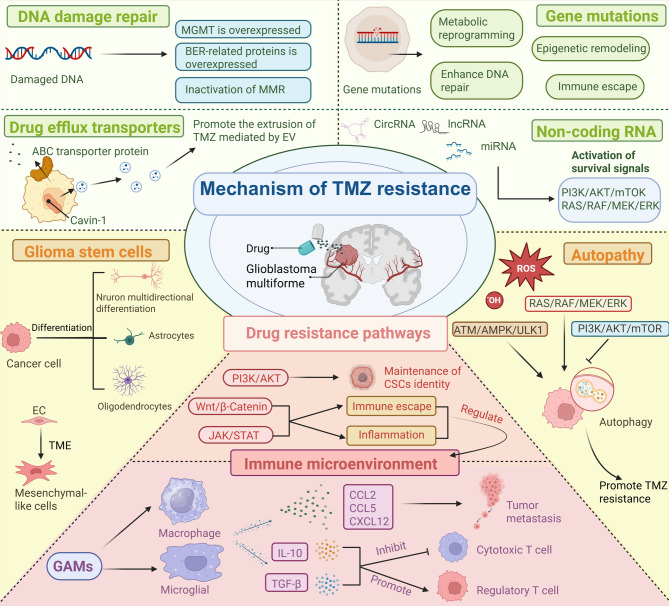
Despite its frontline status, the clinical utility of TMZ remains constrained by rapid pharmacokinetic clearance, suboptimal tumor accumulation [16], and multifaceted resistance mechanisms rooted in the molecular heterogeneity of GBM [17]. GBM resistance arises mainly from DNA repair mechanisms, cellular survival strategies, and factors within the tumor microenvironment. Major contributors include O6-methylguanine-DNA methyltransferase (MGMT) protein expression, MMR system dysfunction, and enhanced base-excision repair (BER). Additionally, glioma stem cells (GSCs), drug transporter proteins, autophagy, and non-coding RNA signaling further decrease TMZ effectiveness. Immunosuppression and abnormal signaling pathways within the tumor environment also contribute to resistance [18–21]. These mechanisms collectively enable tumor cells to repair or evade TMZ-induced DNA damage, emphasizing the need for improved treatment strategies (Fig. 2). The subsequent sections systematically analyze these mechanisms and evaluate emerging therapeutic approaches to restore TMZ sensitivity.
Fig. 2.
Mechanisms of TMZ resistance in GBM. Resistance arises through enhanced DNA damage repair pathways, including the overexpression of the MGMT and BER proteins and the inactivation of MMR. Drug efflux transporters promote TMZ extrusion, reducing intracellular drug levels. Genetic mutations and non-coding RNAs contribute to metabolic reprogramming, immune escape, and the activation of survival signaling pathways such as the PI3K/AKT/mTOR and RAS/RAF/MEK/ERK pathways. GSCs play crucial roles in tumor formation, treatment resistance, and recurrence, largely due to their self-renewal ability and adaptability. Autophagy is regulated through the RAS/RAF/MEK/ERK, ATM/AMPK/ULK1, and PI3K/AKT/mTOR pathways, further supporting cell survival under TMZ treatment. GSCs play a central role in maintaining therapeutic resistance via pathways such as the PI3K/AKT, Wnt/β-catenin, and JAK/STAT pathways, which sustain stemness, promote immune evasion, and modulate inflammation. Additionally, the tumor immune microenvironment, shaped by glioma-associated microglia and macrophages (GAMs), microglia, and secreted factors, inhibits cytotoxic T-cell activity and enhances regulatory T-cell function, facilitating tumor progression and metastasis
To ensure a comprehensive and systematic review of the current landscape of TMZ resistance mechanisms and therapeutic advancements in glioma, we conducted a rigorous literature search adhering to the following methodology: (1) Databases and Search Tools: The search was primarily performed using PubMed, supplemented by Web of Science and Scopus, to capture interdisciplinary insights. These platforms were chosen for their extensive coverage of biomedical literature and advanced filtering capabilities. (2) Time Frame: We focused on peer-reviewed articles published between 2019 and 2025 to emphasize recent breakthroughs while maintaining relevance to contemporary clinical and research contexts. (3) Keyword Strategy: Core search terms included “Temozolomide”, “glioma”, “chemoresistance”, “MGMT”, “DNA repair”, “autophagy”, “glioma stem cells”, and “immunotherapy”. Boolean operators (AND/OR) were employed to refine combinations, such as:“Temozolomide AND (glioma OR glioblastoma) AND (resistance mechanism OR DNA damage repair)”. Additional terms specific to subsections (e.g., “non-coding RNA”, “nanoparticles”, “CRISPR”) were integrated modularly. (4) Inclusion and Exclusion Criteria: Inclusion: Original research articles, meta-analyses, clinical trials, and authoritative reviews published in English. Priority was given to studies in JCR Zone 1/2 journals with high impact factors (> 5.0) and those validating mechanisms in in vivo models or patient-derived samples. Exclusion: Case reports, non-English publications, studies lacking mechanistic insights, and preclinical models without translational relevance. (5) Data Synthesis: Extracted data were categorized into thematic sections (e.g., DNA repair, autophagy, immunotherapy) to identify emerging trends and consensus findings. Discrepancies or contradictory results were critically analyzed to highlight unresolved questions. This structured approach ensured a balanced representation of foundational discoveries and cutting-edge innovations, enabling a cohesive narrative that bridges laboratory research and clinical translation. By focusing on high-impact studies with robust experimental designs, this review aims to serve as a reliable resource for both researchers and clinicians seeking to navigate the complexities of TMZ resistance in glioma.
Mechanisms of TMZ resistance
DNA damage repair (DDR)
MGMT
MGMT is a DNA repair protein that safeguards genomic integrity by transferring methyl groups from O6-benzylguanine (O6-BG) lesions to a cysteine residue (Cys145) at its active site, thereby reversing potentially carcinogenic DNA damage implicated in tumor initiation [22]. While MGMT protects normal cells from tumorigenesis, its overexpression in cancer cells confers resistance to O6-alkylating chemotherapeutics such as TMZ [23, 24]. The regulation of MGMT expression is predominantly governed by epigenetic modifications, with promoter CpG island methylation identified as the primary driver of transcriptional silencing [25]. In GBM and other infiltrating glioma, promoter methylation of MGMT correlated with reduced enzyme production, an enhanced TMZ response, and prolonged survival [26, 27]. Conversely, an unmethylated promoter status is associated with abundant MGMT protein, poor TMZ efficacy, and unfavorable clinical outcomes [28]. Notably, patients with methylated MGMT promoters exhibit 50–90% higher survival rates than those with unmethylated promoters [27], underscoring the therapeutic potential of epigenetic silencing to sensitize glioma to alkylating agents.
Interestingly, the impact of MGMT promoter methylation on prognosis varies depending on the molecular subtype of the tumor. For example, hypermethylation significantly benefits patients with certain subtypes, such as receptor tyrosine kinase II (RTK II) astrocytomas, but does not impact prognosis in other subtypes, such as RTK I or mesenchymal GBM [29]. Clinical studies suggest that methylated MGMT tumors may respond well to TMZ alone, allowing radiation therapy to be delayed. Additionally, recent research has shown that iron metabolism dysregulation, specifically increased ferritin expression, contributes significantly to TMZ resistance. High ferritin levels negate the survival benefit of MGMT methylation, highlighting the complexity of resistance mechanisms [30]. Clinically, testing for MGMT promoter methylation remains an important method to predict how GBM will respond to alkylating chemotherapy. However, the MGMT-STP27 method, which uses two specific CpG sites to determine methylation, has limitations. Although it works well for GBM without IDH mutations, it does not effectively predict outcomes in IDH-mutant astrocytomas. In the CATNON trial, researchers reported no difference in survival between patients with methylated and unmethylated IDH-mutant astrocytomas, even when the methylation cutoff levels were adjusted [31]. This indicates the need for better biomarkers that are specifically tailored to different tumor subtypes, and suggests the need to explore additional CpG sites or combine multiple biomarkers to improve predictions.
MGMT methylation status alone may not reliably predict the efficacy of TMZ treatment. Other factors, such as MGMT protein expression levels and the tumor’ s DNA repair capacity, significantly influence patient outcomes and thus should be considered during clinical decision-making [32]. Patient age further complicates this relationship: MGMT promoter methylation strongly predicts prognosis in younger patients, whereas, in elderly patients, the extent of surgical resection may have a more decisive impact on survival [28]. Additionally, treatment approaches for frail or elderly patients require careful consideration to balance therapeutic efficacy and potential toxicity. Although standard practice does not exclude elderly patients with unmethylated MGMT promoters from TMZ treatment, reanalysis of clinical trials indicates no survival advantage with TMZ in this subgroup, whereas RT demonstrates superior outcomes [33]. Therefore, comprehensive and precise assessment of MGMT status, alongside patient-specific factors such as age and general health, is essential for guiding optimal treatment decisions.
In addition to promoter methylation, several alternative mechanisms have been identified that regulate MGMT activity and contribute to TMZ resistance. Recent studies revealed that poly (ADP-ribose) polymerase (PARP) interacts directly with MGMT following TMZ treatment, enhancing MGMT’s DNA repair function through PARylation. This post-translational modification improves MGMT’s ability to bind DNA and repair TMZ-induced damage, thus promoting resistance.
Transcriptional and epigenetic factors also regulate MGMT expression. For example, NFAT5 leads to the upregulation of MGMT, a transcriptional target of NFAT5, which is responsible for unfavorable TMZ response. Inhibiting NFAT5K668 methylation significantly improves TMZ efficacy, especially in tumors with activated EGFR signaling [34]. Another factor influencing TMZ resistance involves TGF-β1 signaling. In GBM cells without MGMT promoter methylation, TGF-β1 activates certain long non-coding RNAs (lncRNAs). These lncRNAs prevent the maturation of miR-198, a small RNA molecule that normally reduces MGMT expression. Without miR-198, MGMT expression increases, causing TMZ resistance. Clinical data support this finding, showing that lower TGF-β1 and lncRNA levels correlate with better TMZ responses [35]. Decreasing the protein KSRP, which interacts with these lncRNAs, reversed resistance, highlighting another potential treatment approach.
Researchers have also identified genomic rearrangements as another pathway controlling MGMT expression independent of methylation status. Engineered structural changes via CRISPR/Cas9 technology led to increased MGMT production, directly causing TMZ resistance both in vitro and in animal models. Importantly, these genetic changes can be detected in exosomes from tumors, suggesting their potential as biomarkers for the early detection of treatment resistance or tumor recurrence [36]. Furthermore, resistance to TMZ can develop through pathways that do not involve MGMT at all. One such pathway involves RAD18 (an E3 ubiquitin-protein ligase)-mediated translesion synthesis, a mechanism allowing cancer cells to tolerate TMZ-induced DNA damage [37]. This pathway is critical for maintaining resistance in patient-derived GBM models that lack MGMT expression. This finding complements earlier observations showing TMZ resistance in certain glioma cell lines despite the absence of MGMT expression [38].
MMR
The DNA MMR system maintains genomic stability by correcting mismatched bases that occur during DNA replication [37]. When MGMT fails to repair DNA damage caused by TMZ, persistent mismatches, especially O6-methylguanine paired with thymine, are recognized by the MMR machinery [39]. This recognition initiates a futile repair cycle where repeated excision-repair attempts generate single- and double-strand DNA breaks [40, 41], ultimately triggering apoptotic cell death [32]. Dysfunction in the MMR pathway, such as the loss of key proteins such as MLH1, can lead to microsatellite instability, which contributes to cancer progression [42]. Importantly, alkylating agent-induced MMR protein inactivation has been identified as a resistance mechanism in GBM, suggesting that MMR deficiency can confer TMZ resistance even in MGMT-deficient tumors [21].
Analysis of tumor samples from The Cancer Genome Atlas (TCGA) has shown that recurrent GBM often develop mutations in MMR genes, especially in tumors with methylated MGMT promoters [43]. These findings suggest that tumors that are initially sensitive to TMZ may acquire resistance by developing secondary MMR defects. In support of this idea, recurrent GBM frequently exhibit reduced expression of several MMR proteins compared with initial GBM [44].
Emerging evidence indicates that epigenetic changes also influence MMR activity and TMZ resistance. For example, increased levels of histone lactylation (specifically H3K9 lactylation) observed in recurrent and TMZ-resistant GBM cells reduce MLH1 gene expression, weakening the MMR system. Interestingly, the anti-epileptic drug stiripentol reverses this resistance by inhibiting enzymes responsible for lactylation, restoring sensitivity to TMZ in experimental models [45]. This highlights the potential to target such modifications therapeutically.
Further complexity arises from RNA-binding proteins. MEX3A is upregulated in GBM tissues and cell lines following TMZ exposure, where it binds MSH2 mRNA to promote degradation. MEX3A knockdown restores MSH2 levels and chemosensitivity, with clinical correlations showing that high MEX3A expression predicts poor prognosis in MGMT-deficient patients [46]. These findings reveal a novel post-transcriptional regulatory axis involved in TMZ resistance.
Recent studies have also revealed adaptive interactions between the MMR system and other DNA repair pathways. Specifically, when the MMR system attempts to repair TMZ-induced damage, it creates secondary DNA damage that activates another repair mechanism called translesion synthesis (TLS). This TLS pathway, driven by RAD18 and DNA polymerase κ, allows cells to tolerate DNA damage caused by TMZ. In recurrent GBM, reduced RAD18 expression is linked to increased mutation, suggesting that dynamic interactions between these pathways help tumors adapt and survive despite treatment [37].
BER
While O6-MeG DNA damage is key to the therapeutic effects of TMZ, approximately 80–85% of TMZ-induced DNA modifications involve N7-MeG and N3-MeA [47]. These modifications result in the spontaneous loss of purine bases, creating toxic abasic (AP) sites. The enzyme APNG (also termed MPG) identifies and removes N3-MeA and N7-MeG damage, leading to AP sites that activate the BER pathway through another enzyme called apurinic/apyrimidinic endonuclease 1/redox effector factor-1 (APE1/Ref-1) [48]. BER is the main mechanism for repairing AP sites, thus playing a significant role in resistance to TMZ [49].
APNG expression in GBM is controlled by epigenetic mechanisms such as promoter methylation. Tumors with low APNG expression typically respond better to TMZ treatment, whereas tumors with high APNG levels, even those with methylated MGMT promoters, often show resistance [50]. This regulatory similarity to that of MGMT suggests that targeting the BER pathway might increase the effectiveness of TMZ.
Several studies have demonstrated that inhibiting BER pathway enzymes such as APE1/Ref-1 or DNA polymerase β can increase the effectiveness of TMZ in laboratory models [51, 52]. However, inhibiting APE1/Ref-1 presents challenges due to its involvement in critical cellular processes, such as DNA repair, regulation of transcription factors like NF-κB and p53, and its role in maintaining genomic stability [53]. These issues emphasize the need for selective inhibitors with fewer side effects.
Recently, alternative strategies focused on exploiting the BER pathway intermediates have emerged. TMZ treatment results in the accumulation of AP sites, which occur in both MMR-functional and MMR-deficient tumors [54]. A compound known as RA-1 selectively targets and cleaves these AP sites, increasing the effectiveness of TMZ regardless of the MMR status [54]. Rather than suppressing BER directly, this strategy utilizes the damage generated by BER, offering a promising new approach for overcoming TMZ resistance.
Gene mutations
Understanding genetic mutations and their relationship with TMZ resistance in glioma is essential, especially considering the latest 2021 WHO classification. The classification emphasizes genetic features such as IDH mutations, 1p/19q codeletion, H3F3A mutations, alterations of alpha thalassemia/mental retardation syndrome X-linked (ATRX) mutations, MGMT promoter methylation, CDKN2A loss, EGFR amplification, chromosomal imbalances (7+/10−), and mutations in PTEN, TP53, and the TERT promoter, as well as HFE polymorphisms [55–58].
IDH1 mutations, such as IDH1R132H, produce a molecule called D-2HG, which contributes to TMZ resistance in lower-grade glioma. Blocking D-2HG production enhances TMZ sensitivity and survival in preclinical studies [59–61]. On the other hand, the overexpression of normal IDH1 leads to TMZ resistance, whereas mutant IDH1 generally increases tumor sensitivity to TMZ [59, 60]. Additionally, targeting metabolic pathways such as NAD+ sequestration has been shown to effectively kill IDH-mutant cancer cells when combined with alkylating agents [62]. The TP53 tumor suppressor is frequently inactivated in GBM, promoting tumorigenesis and therapy resistance. Reactivating p53 or ATM-dependent HR and MMEJ pathways may counteract resistance in TP53-mutant tumors [63, 64]. ATRX mutations, which often co-occur with IDH1R132H and TP53 alterations, suppresses ATM dependent DNA damage repair by modulating H3K9me3 to enhance TMZ sensitivity in glioma [65]. However, ATRX loss in IDH1R132H/TP53mut gliomas activates a BRD4-dependent immune evasion mechanism. TMZ further exacerbates this mechanism, leading to reduced treatment efficacy and increased resistance, underscoring the complex and context-dependent role of ATRX in glioma progression [66].
TERT promoter mutations, which are frequently found in GBM, reactivate telomerase activity, resulting in cancer cell immortality. Blocking TERT can increase sensitivity to treatments that cause DNA damage [67–69]. EGFR amplification, particularly that of EGFRvIII, promotes cancer stem cell (CSC)-like behavior, tumor recurrence, and drug resistance. Combined inhibition of the EGFR/AKT and mevalonate pathways improves TMZ responses by disrupting membrane cholesterol dynamics and energy metabolism [70–72]. HFE polymorphisms (H63D/C282Y) correlate with TMZ resistance through p16INK4A upregulation [73, 74]. Additionally, selective pressure from TMZ therapy can lead to acquired MMR deficiencies in glioma, resulting in post-treatment hypermutation. This hypermutated phenotype enables tumor cells to withstand TMZ-induced DNA damage, fostering chemoresistance. Despite their high tumor mutational burden (TMB), these tumors respond poorly to immunotherapy due to limited immune cell infiltration, lack of clonal neoantigens, and significant intratumoral genetic diversity [75–78].
HRasV12 mutations drive epithelioid GBM (Ep-GBM)-like transformation by upregulating U3 small nucleolar RNAs (U3 snoRNAs) through activation of PHAX, which enhances ribosome biogenesis and malignant proliferation. Activated-PHAX also recruits TRIM24 to U3 snoRNAs and facilitates its phosphorylation via DNA-PKcs, linking RNA processing to epigenetic reprogramming [79]. Mutations in the H3F3A gene, particularly at positions K27 and G34, alter chromatin and DNA repair mechanisms, impacting TMZ sensitivity [80, 81]. The loss of CDKN2A enhances cell-cycle progression, helping tumor cells survive despite TMZ-induced DNA damage [82, 83]. PTEN mutations activate PI3K/AKT signaling, enhancing DNA repair and chemoresistance. Targeting PTEN C211 succination disrupts iron-sulfur cluster assembly, sensitizing GSCs to TMZ/radiation [84]. TMZ-resistant GBM exhibit guanine mutations destabilizing G-quadruplex (G4) structures and splice sites, creating vulnerabilities to G4-stabilizing agents such as TMPyP4 or splicing kinase inhibitors. Additionally, resistant GBM cells exhibit cytoplasmic aggregation of a protein EWSR1, which serves as a potential resistance biomarker [85].
Collectively, these genetic and adaptive mechanisms underscore the complexity of glioma resistance and highlight the necessity for innovative therapeutic strategies that anticipate and effectively target evolving tumor vulnerabilities.
Non-coding RNA
Emerging evidence highlights the critical role of non-coding RNAs (ncRNAs) in driving therapeutic resistance to TMZ through the modulation of key oncogenic pathways (Table 1). Dysregulated ncRNAs reinforce chemoresistance by aberrantly activating survival signaling cascades, including the RAS/RAF/MEK/ERK and PI3K/AKT/mTOR axes [86]. Therapeutic approaches targeting ncRNAs, including antisense oligonucleotides (ASOs) to inhibit harmful ncRNAs or mimics to enhance beneficial ones, offer potential for restoring TMZ sensitivity [87, 88]. Major classes of ncRNAs involved in glioma resistance include microRNAs (miRNAs), lncRNAs, and circular RNAs (circRNAs). The following Table 1 summarizes the expression patterns, signaling networks, and mechanisms of action of specific microRNAs, lncRNAs, and circRNAs that contribute to TMZ resistance, highlighting their potential roles as therapeutic targets.
Table 1.
Role of NcRNAs as regulators of the TMZ response in glioma
| ncRNA | Expression | Signaling network | Remark |
|---|---|---|---|
| miR-128-3p | Down | miR-128-3p/c-Met/epithelial-mesenchymal transition (EMT) | Overexpression of miR-128-3p downregulated the expression levels of EMT-transformed proteins (c-Met, PDGFRα, Notch1, and Slug) to enhance the effect of TMZ [89]. |
| miR-144 | Down | miR-144/CAV2 and FGF7 | MiR-144 repressed glioma progression and elevated susceptibility to TMZ by targeting CAV2 and FGF7 [90]. |
| miR-140 | Down | miR-140/CTSB/EMT | Overexpression of miR-140 reduced CTSB levels, enhanced TMZ cytotoxicity, suppressed the mesenchymal transition, and influenced CTSB-regulated tumor sphere formation and stemness marker expression [91]. |
| miR-517c | Down | miR-517c/KPNA2/P53/EMT, autophagy | MiR-517c inhibited autophagy and the epithelial-to-mesenchymal (-like) transition phenotype in human GBM through KPNA2-dependent disruption of TP53 nuclear translocation [92]. |
| miR-214-5p | Down | miR-214-5p/β-catenin/MGMT | Cyanidin-3-O-glucoside inhibited the β-catenin/MGMT pathway by upregulating miR-214-5p to reverse chemotherapy resistance in glioma cells [93]. |
| lncRNA HOXA-AS3 | up | HOXA-AS3/miR-455-5p/USP3 | LncRNA HOXA-AS3 promoted USP3 expression and EMT in vivo by negatively regulating miR-455-5p [94]. |
| lncRNA LINC00511 | Up | LINC00511/miR-524-5p/YB1/ZEB1 | LINC00511 indirectly promoted ZEB1 expression by sponging miR-524‐5p to target YB1, which promoted EMT and TMZ resistance of glioma cells [95]. |
| lncRNA LINC-PINT | Down | LINC-PINT/Wnt/β-catenin/EMT | LINC-PINT suppressed cell proliferation, invasion, and EMT by blocking Wnt/β-catenin signaling in GBM [96]. |
| lncRNA MEG3 | Down | Notch, TGF-β, Cell Adhesion Signaling Pathways | MEG3 acted as a tumor suppressor mainly regulating cell adhesion, EMT, and cell proliferation [97]. |
| lncRNA XLOC013218 | Up | XLOC/Sp1/PIK3R2/PI3K/AKT | XLOC recruited and promoted the binding of Sp1 to the promoters of PIK3R2 to elevate the expression of PIK3R2, then PIK3R2-mediated activation of the PI3K/AKT signaling pathway promoted TMZ resistance and cell proliferation [98]. |
| lncRNA HULC | Up | PI3K/AKT/mTOR | HULC may promote EMT by enhancing PI3K/AKT/mTOR signaling and upregulating TGF-β/Snail [99]. |
| lncRNA MSC-AS1 | Up | MSC-AS1/AKT/ miR-373-3p/CPEB4 | MSC-AS1 knockdown suppressed chemoresistance by regulating the miR-373-3p/CPEB4 axis in vitro and in vivo through activating the PI3K/AKT pathway [100]. |
| lncRNA RMRP | Up | RMRP/ZNRF3/Wnt/β-catenin | RMRP knockdown inhibited β-catenin expression by upregulating ZNRF3. RMRP/ZNRF3 axis and Wnt/β-catenin signaling formed a positive feedback loop to regulate TMZ resistance in glioma [101]. |
| lncRNA DLEU1 | Up | autophagy | Silencing DLEU1 suppressed TMZ-activated autophagy by regulating the expression of P62 and LC3, and promoted sensitivity of glioma cells to TMZ by triggering apoptosis [102]. |
| lncRNA SNHG12 | Up | SNHG12/miR-129-5p/MAPK1/E2F7 | In the cytoplasm, SNHG12 served as a sponge for miR-129-5p, leading to upregulation of MAPK1 and E2F7 and endowing the GBM cells with TMZ resistance [12]. |
| lncRNA MIR210HG | Up | hypoxia/MIR210HG/OCT1 | Hypoxia-induced MIR210HG interacted with OCT1 for modulating hypoxia‐promoted glioma stemness, TMZ resistance, and invasion [103]. |
| lncRNA TMEM161B-AS1 | Up | TMEM161B-AS1-has-miR-27a-3p-FANCD2/CD44 | Knockdown of TMEM161B-AS1 downregulated the expression of FANCD2 and CD44 by sponging hsa-miR-27a-3p, which can inhibit the proliferation, migration, invasion, and TMZ resistance of glioma [104]. |
| lnc-TALC | Up | p38/MAPK | Lnc-TALC regulated microglial M2 polarization and promoted TMZ resistance in GBM cells through C5a release induced by the p38 MAPK signaling pathway [105]. |
| lncRNA JPX | Up | JPX/FTO/PDK1 | JPX facilitated GBM progression and TMZ chemoresistance by modulating PDK1 [106]. |
| lncRNA OIP5-AS1 | Up | OIP5-AS1/miR-129-5p/ IGF2BP2 | OIP5-AS1 inhibition upregulated miR-129-5p to repress resistance to TMZ in GBM cells by downregulating IGF2BP2 [107]. |
| hsa_circ_0110757 | Up | hsa_circ_0110757/hsa-miR-1298-5p/ITGA | Hsa_circ_0110757 inhibited glioma cell apoptosis and promoted TMZ resistance by sponging hsa-miR-1298-5p to promote ITGA1 expression [108] |
| lncRNA LINC00470 | Up | LINC00470/miR-134/MYC/ABCC1 | LINC00470 promoted the expression of MYC and ABCC1 by suppressing miR-134, thus promoting glioma cell proliferation and invasion, and attenuating TMZ chemosensitivity [109]. |
| lncRNA LINC00473 | Up | CREB/LINC00473/CEBPα/MGMT | LINC00473, elevated in TMZ-resistant cells upon CREB activation, regulated the MGMT expression by binding to CEBPα [110]. |
| hsa_circ_0088732 | Up | hsa_circ_0088732/miR-661/RAB3D/EMT | Lcn2-derived Circular RNA (hsa_circ_0088732) inhibited cell apoptosis and promoted EMT in glioma via the miR-661/RAB3D Axis [111]. |
| circ_0059914 | Up | circ_0059914/miR-1249/VEGFA/EMT | EIF4A3induced circ_0059914 promoted angiogenesis and EMT of glioma via the miR-1249/VEGFA Pathway [112]. |
| hsa_circ_0067934 | Up | hsa_circ_0067934/PI3K/AKT/EMT | Upregulated circular RNA hsa_circ_0067934 contributed to GBM progression through activating PI3K/AKT pathway [113]. |
| circ_0003137 | Up | circ_0003137/PTBP1/PLOD3/EMT | Hypoxia-driven M2-polarized macrophages facilitated the epithelial-mesenchymal transition of GBM via extracellular vesicles [114]. |
| circZNF652 | Up | circZNF652/miR-486-5p/SERPINE1 | CircZNF652 regulated cancer aggressiveness through the miR-486-5p/SERPINE1 axis [115]. |
| circWDR62 | Up | circWDR62/miR-370-3p/MGMT | Exosomal circWDR62 promoted TMZ resistance and malignant progression through regulation of the miR-370-3p/MGMT axis [116]. |
MiRNAs
MiRNAs are short ncRNAs, typically approximately 24 nucleotides in length, that regulate gene expression by binding to mRNA. They influence both normal processes, such as development, and disease states, such as cancer [117–120]. In GBM, miRNAs function as either tumor suppressors or promoters, influencing chemotherapy resistance, CSC properties, and interactions within the tumor microenvironment [121, 122].
Specific miRNAs significantly impact TMZ resistance by controlling key signaling pathways [123, 124]. For example, miR-519a and miR-29b enhance TMZ sensitivity by suppressing STAT3 signaling to promote apoptosis [125, 126]. Conversely, miR-3129-5p and miR-199b-3p target the neural precursor cell expressed developmentally down-regulated NEDD4-1/PTEN/PI3K/AKT axis, where NEDD4-1 upregulation activates AKT/NRF2/HO-1 signaling to amplify reactive oxygen species (ROS) defense and TMZ resistance [127]. MiR-3116 sensitizes GBM to TMZ by downregulating FGFR1 and disrupting PI3K/AKT signaling, while the miR-223/PAX6 axis modulates PI3K/AKT to regulate stemness and chemoresistance [128, 129]. Oncogenic miRNAs such as miR-125b and miR-423-5p drive resistance via NF-κB activation and ING-4 suppression, respectively, whereas miR-221 reduces EGFR expression to impair TMZ efficacy [130–132]. Tumor-suppressive miRNAs, including miR-193a-5p and miR-23b-5p, increase TMZ sensitivity by inhibiting the mTOR and TLR4 pathways [133, 134].
MiRNAs also regulate cancer stemness, another key factor in TMZ resistance. GSC-derived extracellular vesicles (EVs) transfer miR-10b-5p to activate PI3K/AKT via PTEN suppression, fostering glycometabolic reprogramming and chemoresistance [135]. Similarly, miR-3065-5p in GSC exosomes transforms astrocytes into tumor-associated phenotypes via DLG2 downregulation, and hypoxic GSC-derived miR-30b-3p promotes resistance through intercellular transfer [136, 137]. The HIF1α/HIF2α-miR-210-3p axis sustains GSC proliferation and chemoresistance under hypoxia, while miR-146a suppresses stemness by targeting POU3F2/SMARCA5 [137, 138]. BMP-induced miR-199a-3p overexpression sensitizes GSCs to TMZ, whereas BC200 enhances stemness and resistance by inhibiting miR-218-5p [139, 140]. AP-2α/miR-26a interactions regulate the Nanog/Sox2/CD133 and IL6/STAT3 pathways, with miR-26a inhibition restoring the tumor-suppressive effects of AP-2α [141]. miR-132 drives stemness via TUSC3 suppression, and the PVT1/miR-365/ELF4/SOX2 axis maintains GSC self-renewal [142, 143]. miR-7-5p suppresses stemness by targeting YY1, resensitizing resistant cells to TMZ [144]. Beyond their role in signaling and stemness, miRNAs regulate EMT and DNA repair, further influencing chemoresistance. Collectively, miRNAs serve as pivotal regulators of TMZ response by intersecting with diverse molecular networks.
LncRNAs
LncRNAs, similar to miRNAs, are increasingly recognized as important regulators of chemoresistance in cancer. In glioma, lncRNAs significantly influence TMZ resistance by controlling DDR mechanisms, particularly through MGMT promoter methylation and alterations in the MMR and BER pathways. Recent studies have identified several lncRNAs that regulate TMZ resistance by modulating these DDR pathways, identifying new potential therapeutic targets.
For example, TMZ treatment activates CHK1, promoting structural changes in lnc01956. These changes allow lnc01956 to move from the nucleus to the cytoplasm, where it binds MGMT and induces chemoresistance. Blocking CHK1 with the inhibitor SRA737 reversed this effect, restoring TMZ sensitivity [145]. Another lncRNA, LIP, increases after TMZ treatment and enhances BER efficiency by directly interacting with PARP-1. Reducing LIP expression significantly improves glioma cell sensitivity to TMZ [146].
HOTAIR, an oncogenic lncRNA frequently overexpressed in GBM, also contributes significantly to TMZ resistance. HOTAIR interacts with PRC2/EZH2 to suppress tumor suppressor genes via H3K27 trimethylation, while also activating the miR-214/β-catenin/MGMT axis [147], the miR-125/Hexokinase 2 pathway [148], and the miR-526b-3p/EVA1 pathway [149]. The small molecule EPIC-0628 disrupts HOTAIR-EZH2 binding, enhancing TMZ efficacy by upregulating ATF3 and inhibiting the DDR [150]. CRISPR-mediated deletion of HOTAIR regulatory elements further highlights its transcriptional influence on chemoresistance [151]. Similarly, FoxD2-AS1 promotes TMZ resistance by reducing MGMT promoter methylation [152], recruiting EZH2 to silence tumor suppressors, and acting as a ceRNA for miR-98-5p/CPEB4 [153]. LINC00473, another resistance driver, amplifies MGMT expression via CREB/CCAAT/CEBPα signaling and transfers chemoresistance to neighboring cells via exosomal packaging [110, 154].
Beyond DDR, lncRNAs critically regulate glioma stemness to sustain TMZ resistance. SOX2OT, which is upregulated in recurrent GBM, promotes stemness and chemoresistance by activating the Wnt5a/β-catenin pathway via SOX2 [155]. GSCAR stabilizes SOX2 mRNA through DHX9-IGF2BP2 complex formation while acting as a ceRNA for miR-6760-5p/SRSF1 [156]. PVT1 enhances stemness via the miR-365/ELF4/SOX2 axis and JAK/STAT signaling [143, 157], whereas PDIA3P1 drives PMT through C/EBPβ stabilization [158]. Paradoxically, TUG1 downregulation in A172/TMZ cells enhances stemness and resistance by suppressing EZH2, while BC200 increases the expression of self-renewal markers (Oct4, SOX2) and ABC transporters (BCRP1, MDR1) via miR-218-5p inhibition [140]. The proto-oncogenic lncRNA NEAT1, which is elevated in recurrent glioma, promotes stemness through multiple mechanisms: downregulating connexin 43 via miR-454-3p [159], mediating HMGB1/TLR2/Wnt-driven GSC formation, and enhancing resistance via the let-7 g-5p/MAP3K1 axis [129].
LncRNAs further intersect with diverse signaling pathways to sustain TMZ resistance. The STAT pathway is activated by HOXD-AS2 [160], while PI3K/AKT signaling is modulated by XLOC013218 [98], LINC01410 [161], and MSC-AS1 [100]. Wnt/β-catenin signaling is reinforced by RMRP [101], LINC00511 [162], and MIR155HG [163], whereas autophagy-related chemoresistance is mediated by CRNDE and DLEU1 [102]. Notably, complex lncRNAs such as HOTAIR, FoxD2-AS1, and NEAT1 target multiple pathways simultaneously, emphasizing the need for comprehensive treatment strategies.
CircRNA
CircRNAs are a unique class of non-coding RNAs characterized by their closed-loop structure and play important roles in glioma biology, particularly in TMZ resistance. These molecules function by interacting with miRNAs, regulating cancer signaling pathways, and sometimes producing functional peptides, making them valuable targets for diagnosis and therapy [164].
Glioma actively secrete dysregulated circRNAs via exosomes to spread TMZ resistance. For example, exosomal circGLIS3 promotes resistance through interaction with miR-548 m and increased MED31 expression [165]. Similarly, circWDR62 enhances MGMT expression by binding to miR-370-3p, promoting resistance and aggressive tumor behavior. High circWDR62 levels are associated with poor patient outcomes [116]. The Warburg effect promotes the secretion of exosomal circ_0072083, which increases NANOG expression to reinforce resistance [166], and heparanase-mediated alterations in exosomal circRNA composition further exacerbate this phenotype [167]. Additionally, circHIPK3 and circCABIN1 promote tumor progression and resistance, respectively, via the miR-421/ZIC5 axis [168] and sustain ErbB signaling [169]. These findings underscore the critical role of exosomal circRNAs in the transmission of intercellular resistance.
In addition to their roles in exosomes, circRNAs directly activate cancer pathways to maintain resistance. CircTTLL13 activates Wnt/β-catenin signaling via OLR1 [170], while hsa_circ_0043949 and hsa_circ_0110757 amplify resistance through ITGA1-mediated PI3K/AKT pathway activation [108, 171]. CircASAP1, which is activated by EIF4A3, promotes tumor growth and resistance through NRAS/MEK/ERK signaling [172]. Conversely, reducing circHIPK3 levels improves sensitivity to TMZ by affecting the miR-524-5p/KIF2A/PI3K/AKT pathway [173]. Notably, circSPECC1 encodes the functional peptide SPECC1-415aa, which disrupts the ANXA2-EGFR interaction to inhibit EGFR/AKT phosphorylation, thereby restoring TMZ sensitivity in resistant cells [174, 175]. On the other hand, circRNAs such as circ_0005198 and circVPS18 increase chemoresistance and stem cell properties by sponging specific miRNAs and increasing TRIM14 and RUNX1 expression [176]. While most circRNAs contribute to resistance, some, such as hsa_circ_0072309, act as tumor suppressors by stabilizing the p53 protein, increasing TMZ sensitivity via autophagy. This protective effect is absent in tumors with mutated p53 [177].
CircRNAs exhibit complex functions that depend on the cellular context and their interactions with multiple targets. To fully understand their roles in resistance, detailed research into circRNA regulators (such as RNA-binding proteins) and downstream targets is necessary. Clinically, circRNAs are promising biomarkers because of their differential expression in resistant versus sensitive tumors and their ability to be detected in bodily fluids such as plasma and exosomes.
GSCs
GSCs, a unique subgroup within GBM, play crucial roles in tumor formation, treatment resistance, and recurrence, largely because of their self-renewal ability and adaptability [178]. A major mechanism of chemoresistance involves the transformation of endothelial cells (ECs) into mesenchymal-like cells within the TME. This process is mediated by c-Met-dependent activation of Wnt/β-catenin signaling, which upregulates multidrug resistance-associated protein-1 (MRP-1). Genetic ablation of β-catenin in ECs reverses TMZ resistance, while combining Wnt pathway inhibitors with TMZ reduces the number of tumor-associated ECs, suppresses tumor growth, and prolongs survival in preclinical models [179].
GSCs are organized in a hierarchy similar to neural development, with progenitor-like cells generating diverse tumor cell populations. This structure contributes to tumor heterogeneity and chemoresistance across different GBM subtypes [180]. Tumor recurrence involves shifts in cellular states, including increased EMT, stemness, and hypoxic signaling pathways. Single-cell studies have shown that recurrent tumors activate genes such as SOX4, SOX10, and HIF1A, shifting toward a therapy-resistant mesenchymal phenotype [181]. Quiescent GSC populations expressing specific receptors, such as F3, further increase resistance, becoming active and proliferating following chemotherapy [182].
Metabolic and epigenetic changes reinforce GSC chemoresistance. For example, cystathionine γ-lyase (CTH), which is overexpressed in resistant GSCs, increases stem cell properties, while its inhibition suppresses GSC renewal [183]. The activation of signaling pathways such as the Notch, Wnt/β-catenin, and PI3K-AKT pathways supports stemness, with markers such as CD133 indicating increased resistance and tumorigenic potential [184, 185]. Additionally, PDCD10, a regulator of stemness, is downregulated in resistant GBM, promoting dedifferentiation and TMZ resistance, mirroring its role in colon and breast cancers [186–188].
Ion channels and protein modifications also influence GSC behavior. Sodium channels (Nav) maintain GSC quiescence by regulating the resting membrane potential. Inhibiting these channels forces cells into active division, improving TMZ sensitivity [189]. FBXO7, which is stabilized by TMZ, promotes mesenchymal transformation in GSCs via Rbfox2 splicing regulation, while its depletion sensitizes tumors to chemotherapy [190]. Chaperone-mediated autophagy (CMA), which is mediated by LAMP2A, sustains GSC stemness and TMZ resistance. High LAMP2A levels correlated with poor survival [191]. Hypoxia-induced GLT8D1 stabilizes CD133 through glycosylation, activating Wnt/β-catenin signaling to drive tumorigenesis [185]. Furthermore, ubiquitin-specific peptidase USP36 stabilizes ALKBH5 to sustain GSC self-renewal and TMZ resistance, while MVP overexpression in resistant cells is correlated with multidrug resistance and poor prognosis [192, 193]. Interactions between proteins such as TRAF4 and CAV1 activate survival signaling pathways, which can be disrupted therapeutically to restore TMZ effectiveness [194].
Extracellular communication mediated by exosomes contributes to the dissemination of therapeutic resistance. For instance, CircCABIN1 packaged within exosomes derived from TMZ-resistant cells sponges miR-637 to upregulate OLFML3, activating ErbB signaling in recipient cells [169]. Glycosylation and RNA-binding proteins also regulate resistance: MAN1A1 deficiency in GSCs promotes CD133-DNMT1 interactions to maintain quiescence, whereas KHDRBS3 supports self-renewal and TMZ resistance [195–197]. Therapeutic strategies targeting GSC-specific pathways, such as MIDKINE/ALK blockade or HSP90 inhibition to impair HR, synergize with TMZ and prolong survival in models [198, 199]. Additionally, HDAC6 inhibitors disrupt Sp1-mediated stemness, inducing cell cycle arrest and senescence in resistant cells [200].
GSCs undergo metabolic reprogramming involving pathways such as oxidative phosphorylation and lipid synthesis, which are controlled by signaling pathways such as the PI3K/AKT and RAS/RAF/MEK/ERK pathways. Inhibiting enzymes critical for lipid synthesis, such as stearoyl-CoA desaturase (SCD), triggers cell death and enhances TMZ efficacy [201]. Epigenetic mechanisms, including MGMT methylation and histone modifications by KDM1A, protect GSCs from DNA damage [71, 178, 202]. Single-cell studies have shown that proneural-to-mesenchymal transition (PMT) plays a central role in cancer recurrence. It is driven by lncRNAs such as PDIA3P1 and regulated through proteins such as C/EBPβ. Targeting PMT pathways with specific inhibitors enhances TMZ effectiveness [158, 203–205]. Hypoxic GSC-derived EVs transfer miR-30b-3p to suppress RHOB, exacerbating chemoresistance, whereas elesclomol-induced ROS overcomes resistance by targeting mitochondrial metabolism [206, 207].
Autophagy
Autophagy plays a dual role in glioma progression and TMZ resistance, primarily by acting as a protective mechanism that helps tumor cells survive chemotherapy-induced stress. This protective form of autophagy enhances cell viability by alleviating cellular stress during TMZ-induced cell cycle arrest and removing damaged organelles and proteins, allowing glioma cells to resist TMZ treatment [208, 209].
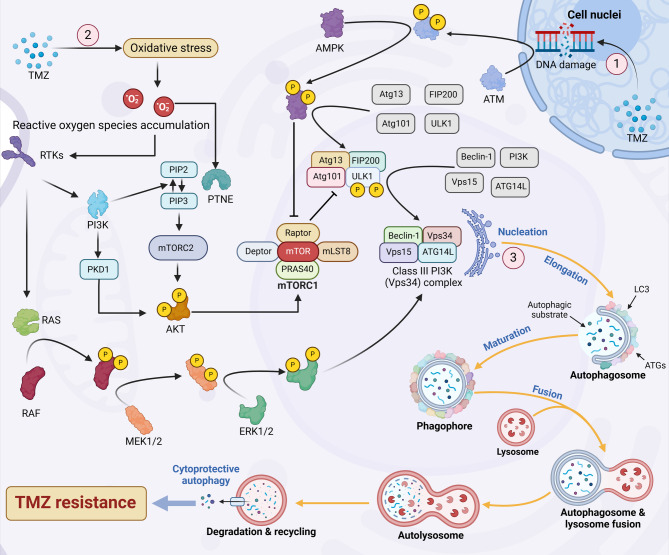
TMZ-induced damage in glioma cells triggers various stress responses, including DNA damage, oxidative stress, endoplasmic reticulum stress, and metabolic disruption. These stresses activate autophagy through multiple signaling pathways, such as the ATM/AMPK/ULK1 axis, the PI3K/AKT/mTOR pathway, and the RAS/RAF/MEK/ERK cascade driven by ROS. Collectively, these pathways stimulate autophagosome formation and enhance lysosomal degradation activity [210–212]. Autophagy then helps glioma cells degrade and recycle damaged cellular components, maintaining internal balance and promoting survival, particularly in GSCs [213] (Fig. 3).
Fig. 3.
Autophagy in TMZ-treated cells. Autophagy is a multistep process consisting of initiation, nucleation, elongation, maturation, and fusion. In glioma cells treated with TMZ, autophagy is activated through multiple signaling cascades. (1) TMZ induces DNA damage, which activates the ATM/AMPK/ULK1 signaling axis, subsequently promoting the assembly of the class III PI3K (Vps34) complex, which initiates autophagosome formation. (2) TMZ-induced oxidative stress results in the accumulation of ROS, which stimulates receptor tyrosine kinases (RTKs). Activated RTKs trigger the RAS/RAF/MEK/ERK and PI3K/AKT pathways, leading to the activation of downstream transcription factors that modulate autophagy. Specifically, ERK1/2 activation facilitates autophagy by enhancing the formation of the Vps34 complex, whereas AKT activation inhibits autophagy by promoting mTORC1 activity, which suppresses the ULK1 complex. Notably, elevated intracellular ROS levels also activate PTEN, a negative regulator of the PI3K/AKT pathway. This PTEN-mediated inhibition is more pronounced than the autophagy-promoting effect of RTKs, resulting in overall suppression of the PI3K/AKT pathway under TMZ treatment. (3) The Vps34 complex is essential for the nucleation of autophagic vesicles, whereas vesicle elongation and maturation into autophagosomes require additional autophagy-related proteins (ATG) and LC3. Mature autophagosomes subsequently fuse with lysosomes to form autolysosomes, where autophagic substrates are degraded. Cytoprotective autophagy supports protein synthesis, energy production, and cell survival, thereby contributing to TMZ resistance in glioma cells
SH3GLB1 (Bax-Interacting Factor 1 or endophilin B1) plays an essential role in initiating autophagy by recruiting Beclin-1 and activating PI3KC3, which is crucial for early autophagosome formation. Studies indicate that SH3GLB1 is regulated by the transcription factor Sp1 and contributes significantly to TMZ resistance by promoting autophagy and altering mitochondrial functions [214, 215]. In addition, DOC-2/DAB2IP suppresses TMZ-induced autophagy by downregulating ATG9B via inhibition of the Wnt/β-catenin pathway, thereby sensitizing GBM cells to TMZ [216]. Moreover, increased ADAR1 expression under TMZ treatment strengthens autophagy and enhances drug resistance via selective autophagy mediated by p62 [217]. These findings indicate a link between protective autophagy and TMZ resistance in GBM.
However, under certain genetic or molecular conditions, autophagy can also trigger cell death rather than survival. For example, p53 activation, increased Beclin-1 expression, or mTOR inhibition can result in hyperactive autophagy, inducing cell death and improving TMZ sensitivity [218, 219]. For example, hsa_circ_0072309 increases TMZ sensitivity in GBM with wild-type p53 by enhancing autophagy via the p53 signaling pathway [177]. Additionally, blocking the MIDKINE (MDK)/ALK pathway leads to the degradation of SOX9, a transcription factor involved in autophagy, thereby reducing glioma-initiating cell (GIC) self-renewal and enhancing TMZ effectiveness against this resistant cell population [198].
Immune microenvironment
The immune microenvironment of GBM significantly contributes to TMZ resistance through complex interactions involving multiple cell types and signaling molecules [220]. GAMs, which are primarily polarized to an immunosuppressive M2 phenotype, dominate this environment. These cells produce cytokines such as IL-10 and TGF-β, and chemokines such as CCL2, CCL5, and CXCL12, which attract regulatory T cells and reduce the effectiveness of cytotoxic T lymphocytes [221–225]. M2-type GAMs further promote resistance by activating survival pathways and altering tumor metabolism. Agents such as ginsenoside RK3, which shifts GAMs away from the M2 phenotype by targeting specific signaling axes (such as PPARG/CCL2), can enhance TMZ effectiveness [223, 226, 227].
Various molecular factors also support immune suppression and TMZ resistance. BCL7A contributes to immune exclusion by promoting EMT, creating physical barriers to immune cell infiltration [228]. Similarly, sodium-hydrogen exchanger 1 (NHE1) regulates immunosuppressive environments by altering glucose metabolism within GAMs [229]. Knocking out MXRA8 disrupts the recruitment of M2 macrophages and helps tumors regain sensitivity to TMZ, emphasizing the role of extracellular matrix-immune interactions in resistance [230].
Novel therapeutic strategies are being developed to reshape the immunosuppressive microenvironment. Targeted delivery of resiquimod to tumor-associated macrophages (TAMs) encourages their repolarization from the M2 phenotype to the M1 phenotype, enhancing antitumor immunity and overcoming TMZ resistance [231]. Additionally, modulating metabolism to increase nitric oxide production reactivates inflammatory pathways, improving chemotherapy responses [232]. Treatments such as piperlongumine, which increase CD8 + T-cell activity by increasing oxidative stress, also help reverse immune suppression [233]. Combination therapies, such as oxaliplatin/ferritin complexes, simultaneously trigger tumor cell death and reprogram immunosuppressive networks [234]. These multimodal interventions highlight the importance of simultaneously addressing intrinsic tumor resistance mechanisms and extrinsic immune barriers.
Drug efflux transporters
The overexpression of drug efflux transporters is a major cause of resistance to chemotherapeutic drugs such as TMZ in cancer cells. These transporters actively remove various anticancer drugs from tumor cells, reducing their accumulation and therapeutic effectiveness.
ATP-binding cassette (ABC) transporters are key players in TMZ resistance, actively removing TMZ and its active metabolites from cells via energy derived from ATP hydrolysis [235]. ABCB1(P-glycoprotein) specifically transports the methylated metabolite MTIC of TMZ, limiting its effects. High levels of ABCB1 in tumor cells and ECs of the blood-brain barrier reduce drug accumulation inside tumor cells and prevent TMZ from effectively penetrating brain tissue [73, 236]. Another important transporter, ABCC1 (MRP1), eliminates TMZ-induced DNA damage products by recognizing glutathione-bound methylated adducts, such as GS-MeG, formed by glutathione S-transferase. This action reduces oxidative stress signals and suppresses DNA damage response pathways, weakening the effectiveness of TMZ [237, 238]. ABCG2 (BCRP) also contributes to resistance by supporting the survival of GSC, particularly under low oxygen conditions. Under hypoxia, HIF-1α increases ABCG2 expression, and elevated LDHA activity lowers the intracellular pH, further activating ABCG2. While ABCG2 is not the primary transporter for TMZ, it removes molecules crucial for DNA repair and stress responses, indirectly enhancing resistance [239].
In addition to classical ABC transporters, polymerase I and transcript release factor (PTRF/cavin-1) also play a role in TMZ resistance. The overexpression of PTRF promotes the release of EVs by facilitating fusion between multivesicular bodies and the cell membrane [240]. PTRF overexpression in the glioma cell lines U87 and GL261 not only enhances EV production, uptake, and homing ability but also promotes EV-mediated proliferation of nearby glioma cells and the recruitment and activation of microglia/macrophages [241]. Recent studies have indicated that increased PTRF expression can induce intracellular TMZ efflux mediated by small EVs and large EVs, suggesting that PTRF can serve as an alternative drug target for which new therapies could be developed [242].
Drug resistance pathways
The molecular pathogenesis of GBM is characterized by the dysregulation of core signaling networks [243, 244]. More than 80% of GBM have alterations in the RTK/RAS/PI3K pathways, primarily through amplification of EGFR (including the oncogenic EGFRvIII variant) and loss of PTEN, leading to activation of survival signals such as AKT and MAPK pathways [245, 246]. The PI3K/AKT pathway promotes resistance through various mechanisms, including metabolic reprogramming via mTOR and the regulation of FOXO/GSK-3β signaling [247]. Similarly, excessive activation of RAS/RAF/MEK/ERK signaling, such as circASAP1, which promotes NRAS/MEK/ERK signaling via miR-502-5p, further reinforces malignant progression and TMZ unresponsiveness [172].
Hypoxia further increases resistance through activation of the Wnt/β-catenin pathway. Under low oxygen conditions, proteins such as FTL drive chemoresistance and EMT by activating AKT/GSK3β signaling [30, 248]. The canonical Wnt pathway stabilizes β-catenin, promoting stem cell characteristics and survival-related gene expression, whereas the non-canonical PCP-Wnt pathway during recurrence promotes neuronal transition via BRAF-mediated phosphorylation events [249–253]. ECs within tumors also enhance resistance through c-Met activation, increasing β-catenin levels, drug efflux proteins, and mesenchymal transformation. These effects can be reversed by EC-specific β-catenin ablation [179].
JAK/STAT signaling supports GBM resistance by activating the STAT3/STAT5 pathway, promoting survival signals and mesenchymal traits, and maintaining cancer stem cell populations [254, 255]. Chemoresistance is further enhanced through interactions in the TME, including activation of CCL5-CCR5 signaling in blood vessel regions [256], Notch-mediated stem cell maintenance [257, 258], and PTPN11 phosphorylation events identified through proteogenomic studies in recurrent tumors [181]. Metabolic changes, including altered lipid metabolism and responses to oxidative stress, add additional layers of resistance, highlighting the importance of comprehensive therapeutic approaches.
In conclusion, TMZ resistance in glioma arises from a complex interplay of molecular and cellular mechanisms (Table 2). Central to resistance is the enhanced DDR machinery, including the overexpression of MGMT, MMR deficiencies, and BER activation, which collectively mitigate TMZ-induced DNA lesions. Genetic mutations in IDH1/2, TP53, EGFR, and TERT further drive resistance by altering metabolic pathways, DNA repair fidelity, and stemness properties. Non-coding RNAs (miRNAs, lncRNAs, circRNAs) regulate chemoresistance by modulating survival signaling, EMT, and autophagy. GSCs contribute to therapeutic evasion through self-renewal, metabolic reprogramming, and interactions with the TME. Additionally, cytoprotective autophagy, drug efflux transporters (e.g., ABCB1, ABCC1), and dysregulated pathways such as PI3K/AKT, Wnt/β-catenin, and JAK/STAT amplify resistance by promoting survival and reducing drug accumulation. These multifaceted mechanisms underscore the need for multi-targeted strategies to overcome TMZ resistance in glioma.
Table 2.
Overview of major TMZ resistance mechanisms in glioma
| Resistance Mechanism | Key Factors/Pathways | Impact on TMZ |
|---|---|---|
| DNA damage repair | MGMT, MMR, BER | Repair the damage caused by TMZ, such as repairing O6-methylguanine lesions, N7-methylguanine/N3-methyladenine damage |
| Gene mutations | IDH1, TP53, ATRX, EGFR, PTEN, TERT, MMR, H3F3A | Reduce TMZ sensitivity by suppressing apoptosis, enhancing DNA repair, promoting CSC phenotypes, or inducing hypermutation |
| Non-coding RNA | miRNAs, lncRNAs, circRNAs | Reduce TMZ sensitivity by activating EMT, inhibiting apoptosis, enhancing DNA repair, driving resistance via exosomal transfer or regulating PI3K/AKT pathways |
| GSCs | Self-renewal, metabolic reprogramming, drug efflux, exosome signaling | Maintain tumor heterogeneity and resistance via stemness and signaling |
| Autophagy | PI3K/AKT/mTOR, ATM/AMPK/ULK1, MAPK/ERK, Beclin-1 | Promote survival under TMZ-induced stress. Hyperactive autophagy triggers cell death and improves TMZ sensitivity |
| Immune microenvironment | M2-polarized GAMs, MXRA8, IL-10, TGF-β, immune suppression, checkpoint resistance | Suppress anti-tumor immunity, reduce TMZ effectiveness |
| Drug efflux transporters | ABCB1 (P-gp), ABCC1 (MRP1), ABCG2 (BCRP), PTRF-mediated exosome efflux | Reduce TMZ cytotoxicity and drive tumor recurrence via enhanced DNA repair, stem cell survival, drug efflux, and immune suppression |
| Drug resistance pathways | PI3K/AKT RAS/RAF/MEK/ERK, JAK/STAT, Wnt/β-catenin | Promote stem cell characteristics and survival-related gene expression, leading to resistance |
This table provides an overview of major TMZ resistance mechanisms in glioma, summarizing key molecular pathways and their roles in reducing drug efficacy
Strategies for overcoming TMZ resistance in GBM
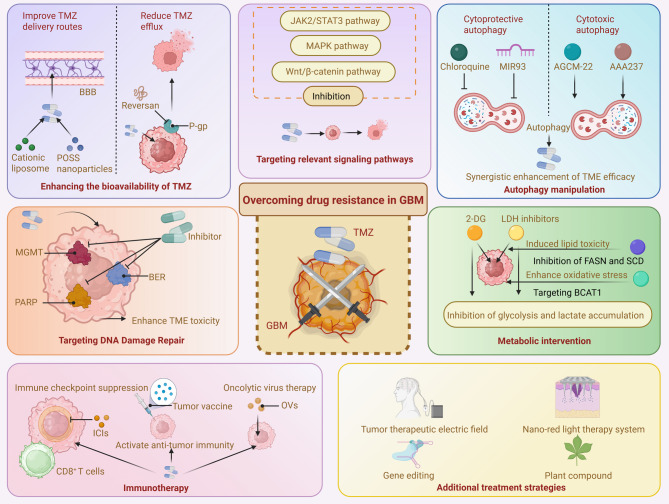
In recent years, considerable research has explored novel strategies to overcome TMZ resistance in GBM. In this section, We innovatively compile and discuss recent advancements, emphasizing cutting-edge areas such as nanodelivery systems, immunotherapy, metabolic interventions, and drug repurposing. By highlighting and integrating these emerging approaches, this work provides clear insights and directions for future clinical translation and personalized treatment strategies (Fig. 4).
Fig. 4.
Strategies for overcoming drug resistance in GBM. Strategies include improving the bioavailability of TMZ by enhancing its delivery across the blood-brain barrier and reducing efflux via P-glycoprotein inhibition. Targeting DNA damage repair pathways, such as the MGMT, PARP, and BER pathways, can increase TMZ-induced cytotoxicity. Modulation of key signaling pathways (JAK2/STAT3, MAPK, and Wnt/β-catenin) through targeted inhibitors offers another route to sensitize tumor cells. Autophagy manipulation, through the inhibition of cytoprotective autophagy or activation of cytotoxic autophagy, synergistically enhances the TMZ response. Metabolic interventions aim to disrupt glycolysis, lipid metabolism, and amino acid utilization by targeting enzymes such as LDH, FASN, and BCAT1. Immunotherapeutic strategies, including immune checkpoint inhibitors, tumor vaccines, and oncolytic viruses (OVs), are employed to boost anti-tumor immune responses. Additional treatments, such as tumor-treating fields, gene editing, nano-red light therapy, and plant-derived compounds, represent emerging modalities with the potential to overcome resistance and improve therapeutic outcomes in GBM
Enhancing the bioavailability of TMZ
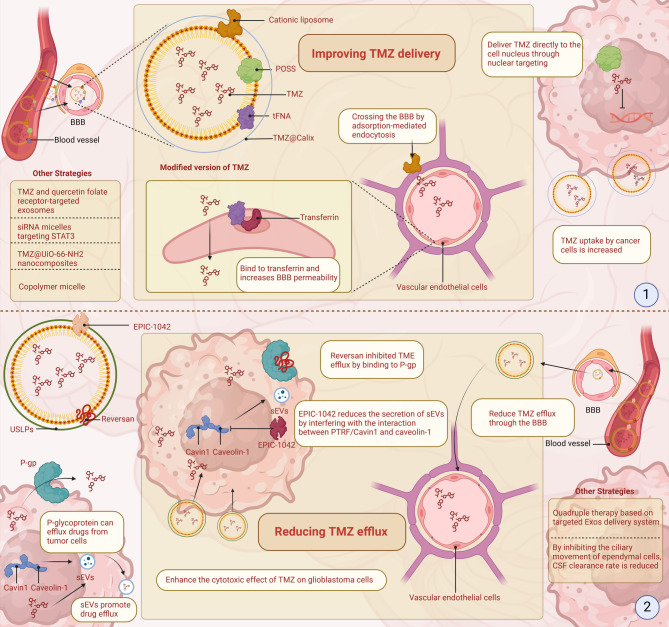
One significant challenge in TMZ treatment for GBM is its limited bioavailability. The BBB and active drug efflux mechanisms reduce drug accumulation within tumors, limiting therapeutic effectiveness [259]. Research has focused on two main strategies: enhancing drug delivery to bypass biological barriers and reducing drug efflux to maintain higher intracellular TMZ concentrations [260]. These approaches aim to increase TMZ levels in tumors while minimizing side effects.
Improving TMZ delivery
The effectiveness of TMZ is reduced by the rapid breakdown of its metabolite, MTIC, which poorly penetrates the BBB. This instability necessitates increased TMZ doses, increasing toxicity and promoting drug resistance [261–263]. Advances in drug delivery aim to address these issues by improving BBB permeability, tumor targeting, and retention within tumor cells.
Cationic liposomes bind to the negatively charged BBB, facilitating drug uptake through endocytosis and improving tumor-targeted delivery [264, 265]. Polyhedral oligomeric silsesquioxane (POSS) nanoparticles directly deliver TMZ to the nucleus, increasing DNA damage and suppressing tumor growth [266, 267]. Tetrahedral scaffold nucleic acid (tFNA) nanoparticles carrying TMZ also enhance BBB penetration and activate tumoricidal autophagy/apoptosis pathways [268]. TMZ encapsulated in calcium p-sulfonate [4]arene (Calix) nanocapsules results in faster cellular uptake than unbound TMZ [269].
Combination strategies with other drugs further enhance delivery. Folate receptor-targeted exosomes containing TMZ and quercetin improve drug delivery and block the PI3K/AKT/mTOR signaling pathway [270]. siRNA micelles targeting STAT3 effectively increase tumor sensitivity to TMZ, highlighting their strong drug loading capacity [271]. Modifying the metabolite MTIC of TMZ into a stable N-acylated prodrug combined with disulfide-linked copolymer micelles improved stability and targeted release [272]. TMZ@UiO-66-NH2 nanocomposites, delivered via ultrasound, increase the local TMZ concentration, effectively killing tumor cells while reducing toxicity to healthy tissues [273] (Fig. 5).
Fig. 5.
Strategies to improve TMZ delivery and reduce drug efflux in GBM. Research has focused on two main strategies: enhancing drug delivery to bypass biological barriers and reducing drug efflux to maintain higher intracellular TMZ concentrations. (1) Cationic liposomes and transferrin-modified nanoparticles facilitate BBB crossing via adsorption-mediated endocytosis and receptor targeting, respectively. POSS-based nanocarriers and tFNA nanoparticles enhance nuclear localization and tumor cell apoptosis. Encapsulation of TMZ in Calix nanocapsules increases early uptake and cytotoxicity. Additional strategies, including folate receptor-targeted exosomes co-loaded with quercetin, siRNA micelles targeting STAT3, MTIC prodrug micelles, and UiO-66-NH₂ nanocomposites activated by ultrasound, further improve BBB penetration and therapeutic efficiency while minimizing toxicity. (2) To counteract TMZ efflux, inhibitors such as Reversan block P-gp-mediated drug expulsion, and EPIC-1042 reduces the release of sEVs by disrupting PTRF/Cavin1-caveolin-1 interactions. Ultra-small, large-pore silica nanoparticles (USLPs) help evade efflux pump recognition and enhance cytotoxicity. Additional strategies include quadruple therapy using targeted exosome systems to downregulate TMZ-resistance genes such as RASGRP1 and VPS28, and approaches that reduce cerebrospinal fluid (CSF) clearance by modulating ependymal cilia activity, increasing TMZ accumulation at tumor sites
Reducing TMZ efflux
Reducing TMZ efflux from GBM cells is essential for enhancing its therapeutic effect. Efflux mechanisms, including cell membrane pumps and exosome-mediated drug release, significantly reduce the effectiveness of TMZ concentration in tumor cells [259].
P-glycoprotein (P-gp), a member of the ABC transporter family, actively removes chemotherapy drugs such as TMZ from cells, resulting in resistance. Inhibitors such as Reversan block P-gp, significantly increasing TMZ accumulation inside tumor cells and enhancing its efficacy [274]. Additionally, nanoparticle-based drug carriers avoid recognition by efflux pumps, reducing TMZ efflux. For example, USLPs, modified with PEG, reduce TMZ efflux at the BBB, increasing its toxicity toward GBM cells [275]. In addition to pump-driven resistance, the exosome-mediated efflux regulated by PTRF/Cavin-1 exacerbates chemoresistance. Targeting this axis with EPIC-1042 disrupts PTRF/Cavin-1-caveolin-1 interactions, suppresses exosome biogenesis, and induces PARP1 degradation via autophagy, thereby impairing DNA repair pathways and amplifying the antitumor effects of TMZ [242, 276].
Genetic factors can also affect TMZ efflux. RASGRP1 and VPS28 were identified as TMZ resistance genes that enhance the conversion of RAS-GDP to RAS-GTP and TMZ efflux. On this basis, a quadruple therapy based on a targeted Exos delivery system was constructed, which significantly reduced the tumor burden in vivo [277]. Additionally, fluid dynamics between the brain parenchyma and CSF play a role in drug distribution. By inhibiting ependymal cilia motility, CSF clearance is reduced, leading to increased TMZ concentrations at the glioblastoma site [278] (Fig. 5).
Targeting the DDR
Inhibiting MGMT
TMZ exerts its cytotoxic effects through DNA methylation at O6-MeG, N7-guanine, and N3-adenine, with O6-MeG being the most lethal lesion in glioma cells [279]. However, the DNA repair enzyme MGMT effectively removes these methyl groups from O6-MeG, significantly contributing to TMZ resistance in GBM. This resistance often increases in recurrent tumors due to genetic rearrangements and MGMT overexpression [36, 280].
Several approaches aim to overcome MGMT-driven resistance. Small molecules such as EPIC-0412 reduce MGMT levels through epigenetic mechanisms, involving UBXN1/ATF3-mediated recruitment of HDAC1 and subsequent removal of H3K27 acetylation, thereby decreasing MGMT expression [281]. EPIC-0628 prevents MGMT production by disrupting interactions between the RNA molecule HOTAIR and the EZH2 protein, whereas EPIC-0307 directly targets DNA repair proteins to increase TMZ effectiveness [150, 282]. These innovative methods help improve the ability of TMZ to kill resistant GBM cells.
Nanoparticle-based treatments further help overcome MGMT resistance. Examples include nucleic acid nanoparticles that deliver MGMT-targeting siRNA and LDL receptor-targeted nanoparticles that block Wnt/β-catenin signaling pathways involved in DNA repair [283, 284]. Natural compounds have also shown potential in combination with TMZ. Quercetin suppresses signaling pathways such as the Wnt3a/β-catenin and AKT/NF-κB pathways, effectively reducing MGMT expression [285]. Tubeimoside-I inhibits the EGFR-PI3K/AKT/mTOR pathway, further decreasing MGMT [286].
Additional therapeutic strategies include inhibition of bromodomain and extra-terminal (BET) proteins to suppress oncogenic transcriptional activity, the use of 2-deoxy-D-glucose-modified nanoparticles to enhance drug uptake and metabolic disruption, and the development of dual-target agents such as Compound 28a, which downregulates both Cyclin D1 and MGMT expression to restore chemosensitivity [287–289]. Furthermore, emerging molecular insights such as activation of the RIP2/NF-κB/MGMT axis have been shown to sustain MGMT expression, further reinforcing resistance to TMZ [34, 290]. These findings expand our understanding of resistance mechanisms and provide promising new targets for therapeutic intervention.
Inhibiting PARP
Targeting PARP, a key enzyme in DNA repair, has become a promising strategy to overcome TMZ resistance in GBM. PARP inhibitors (PARPis) increase tumor sensitivity to TMZ by blocking PARylation, which is essential for repairing TMZ-induced DNA damage [39, 291–293]. The dual inhibition of the PARP-1/2 catalytic domains with agents such as niraparib amplifies TMZ cytotoxicity, particularly in MGMT-deficient glioma, by suppressing telomerase activity and exacerbating DNA damage [265, 294]. Preclinical studies highlight synergistic efficacy: veliparib combined with TMZ suppresses MSH6-deficient xenograft growth, exploiting MMR deficiencies to re-sensitize resistant tumors [295]. Novel approaches include the use of KL-50, an MMR-independent DNA-damaging agent that is effective in MGMT-deficient models, with fractionated RT further increasing its anti-tumor activity [296].
BBB-penetrant PARPis, such as AZD9574, improve survival in GBM models when combined with TMZ [297]. Similarly, olaparib, combined with inhibitors of mitochondrial metabolism, effectively bypasses resistance mechanisms involving MGMT or PTEN [298, 299]. Recent research has identified resistance mechanisms involving proteins such as ATRX, which stabilizes PARP1 and enhances DNA repair [300]. Strategies that exploit synthetic lethality, such as combining Polθ inhibitors with PARP or RAD52 blockade, specifically target GBM cells, sparing normal cells and providing safer treatment options [301].
Inhibiting BER
APE1/Ref-1 is a pivotal enzyme in the BER pathway that is primarily responsible for cleaving AP sites in DNA to initiate the repair of alkylation-induced damage [302, 303]. Downregulation or pharmacological inhibition of APE1/Ref-1 markedly impairs the BER pathway’s capacity to resolve TMZ-induced DNA lesions, resulting in persistent DNA strand breaks, heightened cellular sensitivity to TMZ, and subsequent apoptosis. Preclinical research has demonstrated that silencing APE1/Ref-1 expression through RNA interference substantially reduces the survival of TMZ-resistant GBM cells. This is accompanied by increased levels of γ-H2AX, a marker of unresolved DNA damage, confirming that disrupting APE1/Ref-1 both prevents DNA repair and promotes apoptotic pathways [304]. Thus, APE1/Ref-1 inhibition represents a promising therapeutic strategy for enhancing TMZ sensitivity by disrupting essential DNA repair functions and overcoming chemoresistance.
Clinical investigations have further explored the ability of BER pathway inhibition to increase TMZ activity. For example, a phase I clinical trial involving methoxyamine (TRC102), a small-molecule inhibitor of the BER pathway, in combination with TMZ in patients with recurrent GBM (rGBM) showed promising safety and tolerability [305]. In addition to direct BER targeting, alternative approaches include EPIC-1042, which augments TMZ cytotoxicity by blocking drug efflux, degrading PARP1 via autolysosomal pathways, and inhibiting late-stage autophagy [306]. PARP1 itself is a crucial component of BER, as it coordinates DNA damage responses by facilitating the recruitment of other repair proteins via poly (ADP-ribose) (PAR) polymerization. Inhibition of PARP1 interrupts these processes, resulting in increased genomic instability and cell death [307]. Additionally, specific metabolic alterations such as the IDH1R132H mutation in GBM lead to excessive production of the metabolite 2-hydroxyglutarate, which disrupts BER activity by downregulating DNA polymerase β (Polβ). Tumors harboring such mutations are more vulnerable to alkylating agents and inhibitors that target poly (ADP-ribose) glycohydrolase (PARG) [308].
A novel therapeutic concept, known as the “repair accident model,” suggests that deliberately causing partial impairment in the repair synthesis or ligation steps shared between the BER and MMR pathways can generate lethal DNA double-strand breaks. This targeted disruption significantly amplifies TMZ-induced cell death and offers a potential route to overcoming chemoresistance [309].
Immunotherapy
Tumor immunotherapy represents a therapeutic approach designed to control and eliminate malignancies by reactivating and sustaining the tumor-immune cycle, thereby restoring the body’s intrinsic antitumor immune response [310]. This paradigm encompasses diverse strategies, including immune checkpoint blockade, tumor vaccines, and oncolytic virus therapy, which have demonstrated survival benefits in multiple cancer types and have emerged as transformative frontiers in oncology [311]. However, glioma exhibit diminished responsiveness to immunotherapy compared with other solid tumors, which is attributed to the BBB, brain tumor barrier (BTB), immunosuppressive TME, and low TMB [312]. Despite these challenges, immunotherapy offers advantages over conventional RT and chemotherapy, such as reduced off-target toxicity and durable therapeutic effects, positioning it as a compelling option for treatment-resistant glioma [313]. To increase immunotherapy efficacy in glioma, emerging strategies aim to optimize immune engagement through immune checkpoint inhibition, adoptive T-cell therapy, tumor antigen-targeted vaccination, and engineered OVs [314–316]. These approaches leverage the immune system’s precision to recognize and eliminate tumor cells while minimizing collateral damage to healthy tissues.
Immune checkpoint Inhibition
Immune checkpoint inhibitors (ICIs) significantly change the landscape of GBM treatment by blocking interactions between immune checkpoint receptors (such as CTLA-4, PD-1, TIM-3, and LAG-3) and their corresponding tumor ligands. This blockade restores the ability of CD8 + T-cells to recognize and eliminate tumor cells effectively [317]. Early clinical trials indicate variable outcomes for ICIs. For example, a randomized phase II clinical trial involving 35 patients demonstrated that, compared with postoperative pembrolizumab monotherapy, preoperative treatment with pembrolizumab combined with standard adjuvant therapy notably improved overall survival (OS) [318]. This finding underscores the potential advantages of early intervention with ICIs.
Further research has identified critical immunosuppressive mechanisms within GBM, particularly the soluble PD-L1 (sPD-L1) pathway. A phase II study involving 69 patients revealed that combining apatinib with TMZ therapy successfully reduced the levels of circulating sPD-1 and sPD-L1, potentially overcoming immune suppression and enhancing therapeutic efficacy in recurrent GBM [319]. Cytotoxic T-lymphocyte protein 4 (CTLA-4) is another checkpoint receptor associated with poorer outcomes in high-grade glioma (HGGs), as increased CTLA-4 expression is correlated with a worse prognosis. Early clinical trials combining CTLA-4 inhibitors such as ipilimumab with other ICIs such as nivolumab have demonstrated manageable side effects, although their clinical trial outcomes remain mixed [320, 321]. Nivolumab failed to outperform bevacizumab in rGBM or replace TMZ in unmethylated MGMT tumors, although subgroup analyses suggest a potential benefit in methylated MGMT patients without corticosteroid use [322, 323]. The lack of survival improvement with nivolumab-RT combinations in patients with unmethylated MGMT tumors underscores the need for biomarker-driven patient stratification [324]. Promisingly, the ipilimumab-nivolumab-TMZ regimens exhibit tolerable toxicity profiles that warrant phase II/III evaluation [325].
Emerging engineering strategies, including magnetic-driven photothermal nanorobots (BMPNs) that synergize with PD-L1 blockade [326]and RT-induced PD-L1/PD-L2 upregulation to potentiate ICIs, aim to overcome therapeutic barriers [327]. Additionally, biomimetic nanovesicles have been designed to improve BBB penetration, facilitating the targeted delivery of ICIs and chemotherapeutics [328]. Biopolymer-based implants allowing the controlled release of TMZ combined with immunomodulatory agents such as R848 and IOX1 also demonstrated potent anti-recurrence effects in preclinical models [329].
Mechanistic studies revealed that combination approaches involving D-2HG inhibition, RT, and anti-PD-L1 therapy achieve complete tumor regression in 60% of mIDH1 glioma by reducing T-cell exhaustion and promoting memory CD8 + T-cell formation [61]. Additionally, DDR score has emerged as an immunogenicity biomarker, with high DDR glioma showing elevated mutation burden and immune checkpoint expression [330]. Another promising biomarker, the gasdermin-related prognostic index (GPI), effectively predicts sensitivity to TMZ and ICIs, providing valuable guidance for patient selection [331].
Although combination therapies such as TMZ with ICIs have shown promise in preclinical models, clinical trial outcomes in glioma patients have been largely disappointing. This limited efficacy is attributed to multiple challenges, including the inherently immunosuppressive TME, poor T-cell infiltration, and the low tumor mutational burden typical of glioma. Moreover, the BBB continues to pose a substantial barrier to drug penetration. These limitations highlight the need for more refined combination strategies that incorporate immune modulation, enhanced delivery mechanisms, and biomarker-driven patient selection to improve clinical response rates.
Tumor vaccines
Tumor vaccines have emerged as a transformative strategy in GBM immunotherapy, leveraging tumor-specific antigens to activate antitumor immune responses [332]. These vaccines include various platforms such as peptide-based, dendritic cell (DC)-based, nucleic acid-based, and viral vector-based methods, enabling personalized treatments tailored to individual tumor characteristics [333].
Survivin is an anti-apoptotic protein that is highly expressed in GBM. It is an important target for vaccine development and is closely associated with treatment resistance and poor clinical outcomes [334, 335]. SurVaxM is a peptide vaccine conjugate that has been shown to activate the immune system against its target molecule, survivin. In a phase IIa open-label multicenter trial involving 64 newly diagnosed GBM (nGBM) patients treated with a combination of TMZ and SurVaxM, 95.2% of patients remained progression-free at 6 months after diagnosis. An apparent clinical benefit of SurVaxM was observed in both methylated and unmethylated patients [336, 337]. Similarly, the DCVax-L vaccine, which utilizes dendritic cells loaded with autologous tumor lysates, significantly extends survival when combined with standard treatments [338]. Additionally, vaccines targeting cytomegalovirus (CMV) antigens have shown beneficial outcomes in clinical studies with glioma patients [339].
mRNA vaccines are also gaining attention because of their rapid development, adaptability, and potential for personalized application [333, 340, 341]. However, one critical challenge is the expansion of immunosuppressive regulatory T cells (Tregs) induced by TMZ, potentially reducing vaccine efficacy [333]. Combining TMZ treatment with CMV pp65 RNA-loaded dendritic cells and lymphocyte transfer successfully reduced Treg numbers, amplified antigen-specific T-cell responses, and improved patient outcomes in terms of progression-free survival (PFS) [342]. Similarly, pp65 DC vaccines combined with dose-intensified TMZ (DI-TMZ) and GM-CSF enhanced survival in newly diagnosed GBM patients [343].
Novel delivery technologies have further optimized vaccine effectiveness. For example, synthetic high-density lipoprotein (sHDL) nanovaccines carrying CpG agonists and neoantigens, when combined with anti-PD-L1 therapy, successfully induced tumor regression in preclinical studies [344]. Autologous formalin-fixed GBM antigen vaccines have achieved impressive survival results, with an 80% three-year OS rate following tumor resection [345]. Additionally, allogeneic stem cell lysate-loaded dendritic cell vaccines have demonstrated safety in early-phase clinical trials [346].
Combination strategies are further reshaping GBM vaccine therapies. Peptide vaccines targeting VEGFRs potentially increase the efficacy of TMZ [347]. Tumor fusion DC (TFDC) vaccines are effective in treating chemotherapy-resistant GBM patients with unmethylated MGMT promoters [348]. Preclinical models in which Pmel-1 peptide vaccines are combined with Tc1/Tc17-1 T-cell therapy significantly prolong survival [348]. Personalized vaccines such as AV-GBM-1, consisting of dendritic cells loaded with irradiated tumor-initiating cell lysates, exhibit excellent tolerability and promising preliminary efficacy [349].
Despite these advancements, several challenges remain. While RT and chemotherapy enhance immunotherapeutic responses through antigen release and immunogenic cell death, they can also exhibit immunosuppressive effects that may limit vaccine efficacy, underscoring the need for optimized treatment sequencing and combinatorial strategies [350].
Oncolytic virus therapy
Oncolytic viruses (OVs) represent a promising therapeutic approach in cancer treatment, building upon the unique ability of certain viruses to selectively infect, replicate within, and destroy tumor cells [351]. First recognized in the mid-20th century, OVs utilize the host cell machinery to replicate, causing infected cancer cells to lyse (break open) and release viral progeny, which then infect neighboring tumor cells [352]. This self-amplifying cycle makes OVs particularly effective against invasive tumors such as GBM.
Herpes simplex virus (HSV) derivatives are among the most extensively developed and clinically tested OVs. One prominent example, G47Δ, is a third-generation HSV-1 variant engineered with multiple genetic modifications to increase safety and tumor specificity. Clinical trials have demonstrated significant survival benefits for GBM patients treated with G47Δ, leading to its approval in Japan as the first clinically available OV therapy for GBM [353]. Other modified HSV strains have been developed to further improve specificity and effectiveness. For example, the HSV variant rQNestin34.5 exploits the nestin promoter, which is highly active in glioma, resulting in significantly increased survival times in preclinical studies [354]. Another engineered virus, HSV-1 G207, which is enhanced with NKG2D ligands and bispecific T-cell engagers (BiTEs), shows promising synergy with RT and TMZ, effectively activating cytotoxic T cells and increasing immune-driven tumor cell death [355].
Adenovirus-based OVs also demonstrate significant potential for GBM therapy. The engineered adenovirus CRAd-S-pk7, which incorporates fiber modifications for enhanced targeting, has shown synergistic effects with TMZ in preclinical studies. In clinical trials, this combination led to improved PFS and OS, particularly benefiting patients with TMZ-resistant GBM characterized by unmethylated MGMT promoters, a challenging subset representing approximately 75% of the patient population [356]. Innovative delivery methods also increase the effectiveness of OV therapies. For example, combining Newcastle disease virus (NDV) with TMZ-loaded PLGA nanoparticles has been shown to increase tumor cell killing in GBM models, highlighting the valuable role of nanotechnology in optimizing OV-TMZ therapeutic combinations [357].
However, the interaction between TMZ and OVs is complex. Although laboratory studies initially suggested synergy between TMZ and OVs, further preclinical investigations revealed that TMZ administration can unintentionally impair the efficacy of HSV-based OVs. This discovery underscores the importance of carefully designing and sequencing treatment schedules in clinical settings to avoid negative interactions and maximize therapeutic benefits [358].
Targeting relevant signaling pathways
Resistance to TMZ in GBM is closely linked to disruptions in several critical cellular signaling pathways. Among these pathways, the JAK2/STAT3 pathway plays a central role. Persistent activation of STAT3 significantly increases MGMT expression and strengthens DNA damage response mechanisms, which in turn reduces TMZ effectiveness [359]. Notably, compared with initial tumors, rGBM tumors present elevated levels of MGMT and phosphorylated STAT3 (p-STAT3). Studies indicate that blocking STAT3 activity restores TMZ sensitivity in resistant GBM cell lines and decreases tumor growth in animal models [360]. GSC specifically utilize STAT3 to increase CYP3A5 expression, maintaining the NAD⁺/NADH balance to support mitochondrial function and chemotherapy resistance [361]. Thus, targeting CYP3A5 represents a potential therapeutic approach. Advanced delivery methods, such as bone marrow stem cell (BMSC)-derived exosomes decorated with HMOX1 peptides (HSSP-BMSCExos) and carrying TMZ alongside STAT3-targeting siRNA, have shown enhanced effectiveness in preclinical studies [362]. Similarly, biomimetic nanoparticles and cation-free siRNA micelles efficiently deliver STAT3 siRNA, successfully reversing drug resistance [271, 363].
The MAPK signaling pathway is another crucial mediator of GBM survival and TMZ resistance. The long non-coding RNA PDIA3P1 stabilizes the transcription factor C/EBP-β via p38α-MAPK signaling, promoting PMT and TMZ resistance [158, 364]. Targeting downstream MAPK effectors, such as MNK1/2 with osimertinib, suppresses eIF4E phosphorylation and tumor growth in xenografts [365]. Additionally, BRAF inhibitors such as vemurafenib reduce MAPK1/3 phosphorylation and synergize with TMZ to prolong survival in patient-derived models [253]. Targeting PDGF-Rα/β with CP-673,451 improved TMZ effectiveness by increasing DUSP1 expression and suppressing p38MAPK signaling [366]. Recent work identified ARNT as a p38α-MAPK activator, where disrupting the ARNT/p38α interaction restored TMZ sensitivity [367].
Inhibiting the Wnt/β-catenin pathway represents another effective approach. Natural compounds such as resveratrol have been demonstrated to downregulate Wnt signaling and MGMT expression, increasing TMZ-induced cell death [368]. Mannose also inhibits tumor cell proliferation by suppressing Wnt/β-catenin signaling [369]. Additionally, ApoE-functionalized liposomes delivering both artesunate and TMZ successfully disrupted Wnt/β-catenin activity, reversed MGMT-driven resistance, and improved survival in preclinical studies [284]. In tumors with MGMT deficiency but elevated PI3Kβ activity, inhibiting PI3Kβ synergizes effectively with TMZ, emphasizing the importance of the PI3K signaling axis [370].
Combining therapies that simultaneously target multiple pathways holds particular promise. Dual inhibition of p38MAPK and MEK/ERK (such as SB202190 and binimetinib) or PI3K and MAPK (such as dactolisib and trametinib) enhances TMZ sensitivity and overcomes kinome adaptation in preclinical models [371, 372]. While signaling pathway inhibitors demonstrate compelling preclinical potential to counteract TMZ resistance, translational success hinges on overcoming BBB penetration and bioavailability challenges.
Autophagy manipulation
Autophagy plays a dual and complex role in GBM resistance to TMZ therapy and represents a valuable therapeutic target for enhancing treatment effectiveness. Modulating autophagy can either sensitize tumors to TMZ or enhance chemoresistance, depending on the specific cellular context and signaling pathways involved.
Chloroquine (CQ), an inhibitor of late-stage autophagy, suppresses the fusion of autophagosomes and lysosomes, thus enhancing TMZ-induced apoptosis. In wild-type p53 GBM cells, CQ enhances TMZ efficacy by promoting the phosphorylation of p53. However, in cells with mutated p53, higher CQ concentrations or prolonged exposure are needed to effectively inhibit cell proliferation and induce G2-M cell-cycle arrest [373, 374]. Similarly, microRNA-93 (MIR93) can regulate autophagy by directly targeting critical autophagic genes such as BECN1, ATG5, ATG4B, and SQSTM1, thereby reducing cytoprotective autophagy and sensitizing GSC to both TMZ and RT [375]. Deubiquitinating enzymes (DUBs) increase TMZ resistance by promoting cytoprotective autophagy. Small-molecule inhibitors, such as compound G5, effectively inhibit DUB activity, thereby reversing this resistance mechanism [376]. Another promising approach involves NEO214, a conjugate of rolipram and perillyl alcohol, which blocks autophagosome-lysosome fusion. When combined with CQ and TMZ, NEO214 significantly overcomes TMZ resistance [377]. Genetic approaches, such as SH3GLB1 knockdown, suppress cytoprotective autophagy, disrupt mitochondrial metabolism, and restore TMZ sensitivity [215]. Additionally, Forkhead Box M1(FOXM1)-driven expression of NUF2 promotes resistance via activation of PI3K/AKT/mTOR-dependent autophagy. Inhibition of either FOXM1 or NUF2 reverses TMZ resistance, highlighting potential therapeutic targets [378]. Natural compounds such as daurisoline (DAS), a plant-derived alkaloid, similarly inhibit cytoprotective autophagy through PI3K/AKT/mTOR signaling, increasing TMZ sensitivity [379].
Interestingly, the activation of cytotoxic autophagy can also enhance the efficacy of chemotherapy. The antibody-drug conjugate AGCM-22, derived from cetuximab, enhances TMZ-induced cell death by simultaneously promoting apoptosis and autophagy-related processes [380]. In addition, the Skp2 inhibitor AAA237 induces cytotoxic autophagy through BNIP3-mediated mTOR inhibition, suppressing tumor cell proliferation and invasion [381]. Borneol stimulates the autophagic degradation of HIF-1α, thereby sensitizing glioma cells to radiation therapy by modulating the mTORC1/eIF4E axis [382, 383]. Cannabidiol (CBD) triggers ER stress and mitophagy via the TRPV4-ATF4-DDIT3-TRIB3-AKT-MTOR pathway, synergizing with TMZ in preclinical models [384].
Furthermore, targeting CMA by inhibiting LAMP2A disrupts GSC maintenance, reducing TMZ resistance. Clinically, higher levels of LAMP2 correlate with poorer patient outcomes, highlighting its potential as a prognostic biomarker [191]. Other emerging pathways illustrate the complex role of autophagy in GBM chemoresistance. TRIM7 silencing inhibits nuclear receptor coactivator 4 (NCOA4)-mediated ferritinophagy, inducing iron-dependent ferroptosis and sensitizing tumors to TMZ [385]. EPIC-1042, a novel PARP1 degrader, enhances TMZ efficacy by leveraging early-stage autophagy processes [306]. Dopamine D2 receptor (DRD2) antagonism, such as that caused by haloperidol, promotes autophagy and ferroptosis, countering adaptive resistance to TMZ [386]. Additionally, secretory autophagy induced by TMZ treatment releases HMGB1, driving macrophages toward a pro-inflammatory M1-like phenotype and improving chemotherapy responses, suggesting that HMGB1 is a potential therapeutic target [387].
Autophagy modulation in GBM is a double-edged sword in which both inhibition and strategic activation can enhance TMZ efficacy, depending on the molecular context. Although preclinical studies highlight promising strategies, clinical translation requires addressing challenges such as BBB penetration, tumor heterogeneity, and context-dependent roles of autophagy.
Metabolic intervention
GBM cells exhibit unique metabolic adaptations, notably increased reliance on aerobic glycolysis, known as the Warburg effect. This metabolic shift provides rapid energy production and supports aggressive tumor growth. Key enzymes involved in this process, such as hexokinase and phosphofructokinase, facilitate increased glucose uptake and glycolytic flux [388]. Targeting glycolysis with compounds such as 2-deoxyglucose (2-DG) effectively disrupts energy metabolism, suppressing tumor survival and proliferation [389]. Additionally, inhibiting the glucose transporter GLUT-3 further limits glycolysis, significantly reducing GBM cell proliferation [390]. Lactate dehydrogenase (LDH) also plays a critical role in converting pyruvate into lactate, acidifying the TME. LDH inhibition diminishes lactate accumulation, improving the therapeutic efficacy of TMZ by weakening tumor cell resilience [391, 392]. Furthermore, the circular RNA circKIF4A drives glycolysis by upregulating aldolase A (ALDOA), contributing to TMZ resistance. Targeting circKIF4A effectively reverses this glycolysis-mediated resistance [393].
Lipid metabolism is another key metabolic pathway exploited by GBM cells to support tumor growth and survival. Tumors heavily depend on fatty acids for essential membrane synthesis and energy storage. Inhibiting fatty acid synthase (FASN) and stearoyl-CoA desaturase (SCD) with agents such as orlistat induces lipotoxicity, disrupts DNA repair, and enhances TMZ efficacy [394]. Additionally, EGFR-driven lipid remodeling and cholesterol synthesis contribute significantly to chemoresistance. Preclinical studies highlight that lipid-lowering agents, such as atorvastatin, also demonstrate robust potential as additive therapies alongside TMZ [395].
Purine metabolism represents another therapeutic axis. Inhibition of inosine 5’-monophosphate dehydrogenase (IMPDH), a critical enzyme controlling de novo purine synthesis, significantly suppresses TERT activity, enhancing the impact of chemotherapy [396]. Gliocidin, an inhibitor that targets IMPDH2, induces nucleotide imbalance and effectively promotes tumor cell death [397]. Disruption of the interaction between ARL13B and IMPDH2 (for example, through mycophenolate mofetil) forces tumor cells to depend on nucleotide salvage pathways, thereby exacerbating TMZ-induced DNA damage [398]. Adenylosuccinate lyase (ADSL) inhibition further destabilizes GSC by affecting PTEN succinylation, underscoring its potential as a therapeutic target [84].
Tryptophan metabolism also contributes significantly to tumor immune evasion. Enzymes such as IDO1 and TDO2 produce immunosuppressive kynurenine, impairing effective immune surveillance [399–401]. Dual inhibitors such as AT-0174, which targets both IDO1 and TDO2, demonstrate significant synergy with TMZ treatment, restoring robust antitumor immunity by enhancing CD8 + T-cell responses and reducing immunosuppressive regulatory T-cell infiltration [402]. Additionally, branched-chain amino acid (BCAA) metabolism also supports tumor survival. Targeting the CHIP/BCAT1 axis leads to BCAT1 degradation, disrupts glutathione synthesis, increases oxidative stress, and significantly improves TMZ sensitivity [403].
However, metabolic intervention strategies face several critical limitations. Targeting essential metabolic pathways may inadvertently harm normal tissues, leading to significant toxicity and systemic side effects. Furthermore, the metabolic plasticity and heterogeneity of GBM cells allow tumors to adapt rapidly by activating alternative pathways, reducing the effectiveness of single-agent interventions.
Additional treatment strategies
Tumor treating field
TTFields therapy, a modality employing alternating electric fields to disrupt cancer cell division, received FDA approval in 2015 for nGBM on the basis of the pivotal EF-14 trial, marking a transformative milestone in neuro-oncology [404]. The arrays generate localized, low-intensity electric fields, selectively disrupting cancer cell division by interfering with critical processes such as microtubule assembly and organelle distribution during mitosis [405, 406].
Currently, the TTFields device is approved by the FDA for the treatment of recurrent and newly diagnosed GBM in adults aged 22 years and older. When TTFields therapy is combined with standard TMZ chemotherapy, it has demonstrated substantial improvements in patient outcomes, significantly extending PFS and OS [407–409]. Preclinical studies have revealed that TTFields disrupts tumor microtubule networks, inducing a “crooked microtubule” phenotype in 5–6% of treated GBM cells. This abnormality disrupts calcium signaling, impairs intercellular communication, and restricts tumor proliferation [410]. Clinically, TTFields therapy has a favorable safety profile, with mild-to-moderate skin irritation being the most common side effect, and importantly, it lacks significant systemic toxicity [411].
Emerging evidence supports the synergy between TTFields and other therapeutic modalities. Combining TTFields with chemoradiotherapy has enhanced local tumor control in nGBM patients [412, 413]. Additionally, preclinical studies combining TTFields with drug repurposing strategies, such as the CUSP9v3 regimen, have demonstrated increased anti-tumor efficacy [414]. Recent innovations in electrode array technology have further improved thermal management, patient comfort, and compliance, without compromising therapeutic effectiveness [415].
TTFields therapy represents a transformative advancement in GBM treatment, offering an effective, non-invasive modality capable of significantly improving patient survival outcomes [404, 405, 407, 408]. Its favorable safety profile, synergistic potential with existing therapies, and ongoing technological refinements highlight its growing role as a critical component of multimodal GBM treatment regimens [409, 412–415]. However, limitations such as high cost, patient compliance challenges, and restricted accessibility may hinder widespread adoption and long-term adherence.
Nanotherapy
Recent advances in nanomedicine, particularly nanodrug delivery systems (NDDSs), have shown substantial promise for enhancing therapeutic delivery to GBM tumors. However, only a few NDDSs have successfully overcome the BBB and the blood-brain tumor barrier (BBTB) [416–419]. Mesoporous silica nanoparticles (MSNs), known for their high surface area and pore volume, have attracted attention for their capacity for high drug loading and controlled drug release [420]. Surface-modified MSNs, particularly PEGylated MSNs with octyl groups, significantly enhance BBB penetration and tumor specificity. For example, PEGylated MSNs loaded with docetaxel (DTX) have shown superior penetration through the BBTB in animal models, resulting in reduced systemic toxicity and improved survival rates compared with free drug administration [421–424]. Additionally, advanced approaches such as Gint4. T-siHDGF chimera-capped MSN (TMSN@siHDGF-Gint4.T) enables the precise co-delivery of therapeutic agents, facilitating synergistic GBM suppression through the sequential release of siHDGF and TMZ [425]. Similar to MSNs, the development of USLPs loaded with TMZ and surface PEGylation has shown potential in enhancing BBB permeability, reducing TMZ efflux, and promoting GBM apoptosis [275].
Targeted nanocarrier systems have further improved treatment accuracy. Gold nanoparticles (Anti-EphA3-TMZ@GNPs) functionalized with EphA3 antibodies for intranasal administration bypass the BBB and directly target glioma cells, significantly enhancing TMZ efficacy while reducing systemic side effects. Moreover, gold nanoparticles modified with anti-EphA3 for chemical and auxiliary plasma photothermal treatment have demonstrated increased cellular uptake and induced apoptosis in glioma cells, overcoming drug resistance [426, 427]. Dual-targeting glutathione-responsive nanoparticles (T + A@Glu-NPs), which simultaneously deliver ARV-825 and TMZ, increase BBB penetration and tumor-specific uptake, markedly inhibiting tumor cell proliferation and promoting apoptosis [257]. Similarly, dual-functional nanoparticles (BIP-MPC-NP) concurrently inhibit EGFR/MET signaling pathways and DNA repair mechanisms, effectively resensitizing GBM to TMZ [428]. Furthermore, lipid-polymer nanoparticles modified with 2-deoxy-D-glucose (TMZ/siPD-L1@GLPN/dsb) simultaneously deliver TMZ and siPD-L1 to reverse chemoresistance and modulate immunosuppressive mechanisms [288].
Innovative nanotechnology-enabled physical and chemical therapies also enhance TMZ treatment. For example, magnetic carbon nanotubes (mCNTs) combined with precise magnetic field control have been utilized for mechanical disruption therapy against TMZ-resistant GBM. GBM cells can internalize mCNTs, and under the influence of a rotating magnetic field, cell death is induced, thereby inhibiting tumor growth in vivo. Additionally, functionalizing mCNTs with anti-CD44 antibodies, which recognize the CD44 antigen enriched on the surface of GBM cells, enhances the recognition of cancer cells, prolongs nanoparticle retention in tumors, and subsequently improves therapeutic efficacy [429]. Photodynamic therapy (PDT) using nanoparticle-based photosensitizers generates cytotoxic ROS, circumventing resistance mechanisms [430].
Multifunctional nanoplatforms that combine gene therapy and drug delivery have shown significant clinical promise. Cation-free siRNA micelles (siRNA-SS-PNIPAM) silence STAT3 in TMZ-resistant pathways, achieving synergistic effects [271]. ApoE-functionalized nanocapsules (ApoE-MT/siPKM2 NC) effectively co-deliver siPKM2 and TMZ, suppressing glycolysis and increasing cytotoxicity [431]. Iron oxide nanoparticles and framework nucleic acid nanoparticles (FNNs) effectively deliver siMGMT to inhibit DNA repair enzymes, significantly enhancing TMZ sensitivity [283, 432].
Gene editing
Recent advancements in CRISPR-Cas9 gene editing technology have shown significant potential for addressing TMZ resistance and enhancing therapeutic outcomes in GBM. CRISPR-Cas9, renowned for its precision and adaptability, employs guide RNA to facilitate targeted genetic modifications, allowing gene knockout, insertion, or regulation [433]. A notable application involves CRISPR-mediated targeting of the TIM3 gene, which has been shown to enhance natural killer (NK) cell-mediated glioma suppression, thereby amplifying antitumor immune responses [434]. Additionally, the silencing of microRNA-10b, a critical oncogenic driver in GBM, through CRISPR-Cas9 effectively inhibits tumor growth and progression [435]. Another prominent therapeutic target is the EGFR gene, which is frequently amplified or mutated in GBM. CRISPR-based disruption of the EGFR CE5B + 6B enhancer region effectively suppressed GBM cell proliferation and migration through apoptosis induction and metabolic reprogramming, highlighting the critical role of EGFR modulation in enhancing TMZ efficacy [436, 437].
CRISPR-Cas9 has also been effectively utilized to overcome resistance mechanisms associated with therapeutic stress. For example, knockout of GDNF or its receptor GFRA1 disrupts the GDNF/GFRA1 signaling pathway, sensitizing GBM cells to TMZ, lomustine, and RT [438]. Similarly, CRISPR-mediated ablation of the ABCB1 transporter significantly reduces tumor proliferation and restores sensitivity to TMZ [236]. Innovative strategies such as “cancer shredding,” which target non-coding sequences mutated during TMZ therapy, have enabled the precise removal of resistant GBM clones, effectively mitigating drug resistance [439].
To improve delivery efficiency, brain-targeted CRISPR-Cas9 nanomedicine systems have been developed. Polymer-locked fusosomes (plofsomes) designed by researchers facilitate crossing the BBB and effectively deliver CRISPR-Cas9 ribonucleoproteins into GBM cells, significantly suppressing MDK expression, reducing TMZ resistance, and inhibiting tumor growth [440]. Another promising approach employs glutathione-responsive nanocapsules encapsulating Cas9/sgRNA complexes, enabling precise BBB traversal, tumor-specific release, and efficient editing of the PLK1 gene while maintaining minimal off-target effects [441]. Despite these advances, challenges such as unintended off-target mutations and incomplete delivery specificity remain critical considerations for clinical translation [442].
Phytocompounds
Dietary nutrients and plant-derived compounds have become promising alternatives to standard glioma treatments because of their safety, affordability, and ability to target multiple resistance pathways (Table 3). Flavonoids, including quercetin, rutin, chrysin, apigenin, naringenin, silibinin, epigallocatechin gallate (EGCG), genistein, biochanin A, and cyanidin-3-glucoside (C3G), are commonly found in everyday diets and demonstrate therapeutic potential against HGGs. These compounds regulate autophagy-related proteins such as Beclin-1 and LC3B, reducing cytoprotective autophagy that can cause resistance to TMZ. Flavonoids also suppress key DNA repair enzymes such as MGMT and PARP in resistant glioma cells. Unlike small molecule inhibitors, flavonoids can simultaneously affect multiple resistance-related molecules, including caspase-3, thus minimizing side effects. Combining flavonoids with TMZ has shown stronger anticancer effects in preclinical studies, highlighting their promise in treating glioma [443–446].
Table 3.
Phytocompounds modulating GBM
| Substance | Primary Source | Mechanism of action |
|---|---|---|
| Alkaloids | Barberry | Induce DNA damage, cell cycle arrest, ER stress, apoptosis, and autophagy, inhibit angiogenesis and proliferation in tumor cells [457]. |
| Carboxylic Acid Derivatives | Cinnamon, Giant fennel | Regulate intracellular second messengers, inhibit DNA synthesis, transcriptional activity, and tumor cell proliferation [458]. |
| Carotenoids | Derivative of astaxanthin | Upregulate both extrinsic and intrinsic apoptosis pathways, impair migration and invasion of tumor cells [459]. |
| Coumarins | Celery, Carrot, Parsley family | Upregulate pro-apoptotic pathways, induce terminal differentiation, and reduce multi-drug resistance in cancer cells [460]. |
| Terpenes | Sunflower, White birch | Induce apoptosis through the ROS-JNK pathway, block cell cycle at G1/S phase, inhibit VEGF-mediated angiogenesis [450]. |
| Lignans | Greater burdock | Inhibit topoisomerase in tumor cells, disrupting DNA synthesis and proliferation [461]. |
| Steroids | Fenugreek, Ashwa-gandha | Induce apoptosis and cell cycle arrest in tumor cells [462] |
| Stilbenoids | Vitis vinifera | Activate apoptosis pathways, inhibit tumor cell proliferation, and reduce oxidative stress [463]. |
| Tannins | Oak | Inhibit angiogenesis via HIF-1α/VEGF signaling and induce protective autophagy through Beclin-1 [464]. |
| Triterpene | Tripterygium wilfordii | Modulate the PI3K/AKT/mTOR pathway to inhibit the formation of vasculogenic mimicry (VM) and angiogenesis [465]. |
This table summarizes natural substances, their primary sources, and the key molecular mechanisms by which they modulate glioblastoma progression, highlighting their potential therapeutic roles
Curcumin, a polyphenolic compound from the curcuminoid family, is highly permeable across the blood-brain barrier and accumulates effectively in the hippocampus. Its ability to dissolve in lipids helps its distribution and uptake in brain cells [447]. Studies have demonstrated that curcumin reduces glioma cell growth by triggering apoptosis, cell cycle arrest, and mitochondrial damage, ultimately suppressing tumor growth. Additionally, curcumin significantly inhibits angiogenesis and inflammation associated with tumor progression [448]. Clinical studies have confirmed the presence of curcumin in GBM tumor tissues following oral administration, increasing tumor sensitivity to RT and chemotherapy [449, 450]. Curcumin works by lowering the levels of protective proteins such as Bcl-2, blocking survival pathways such as the JNK/AKT signaling pathway, and reducing the activities of DNA repair enzymes, including MGMT, ERCC1, DNA-PK, and Ku70/80, suggesting potential synergy with standard treatments [451–453].
In addition to flavonoids and curcumin, other phytochemicals also have the potential to slow brain tumor progression and reduce treatment-related neurotoxicity [454]. A recent comprehensive review identified several medicinal plants, including Abutilon indicum, Anemone taipaiensis, Anisomeles indica, and Ardisia pusilla, whose bioactive compounds promote glioma cell death through DNA damage, mitochondrial disruption, and apoptosis by modulating critical signaling pathways [455]. However, challenges remain regarding the standardization of herbal treatments, including difficulties in purification, variable bioavailability, and unpredictable interactions with other medications. Improving phytochemical delivery across the BBB and developing combination therapies with conventional chemotherapeutics may increase treatment efficacy while limiting unwanted side effects [456].
Repurposing existing drugs
Drug repurposing involves the use of approved medications for new therapeutic purposes, providing faster development, lower costs, and established safety profiles than the development of new drugs from scratch [466, 467] (Table 4).
Table 4.
Repurposed drugs for glioma treatment
| Drug | Original Indication | Mechanism of action |
|---|---|---|
| Disulfiram | Alcohol dependence treatment | Downregulates Polo-like kinase 1 (PLK1) expression, inhibits MGMT activity, and activates NF-κB and proteasome [478]. |
| Chlorpromazine | Acute and chronic mental disorders treatment | Induces cytotoxic autophagy via endoplasmic reticulum stress and unfolded protein response, reduces Cx43, and inhibits DNA repair [479, 480]. |
| Gemcitabine | Lung cancer and breast cancer treatment | Inhibits DNA synthesis, induces cell cycle arrest and apoptosis [481, 482]. |
| Sildenafil | Erectile dysfunction | Modulates pro-apoptotic and anti-apoptotic signals via the cGMP/PKG pathway [483]. |
| BMS345541 | IKK-1/IKK-2 inhibition | Maintenance of the FOXG1 structure [484]. |
| Fluoxetine | Antidepressant | Reduces MGMT levels in GBM cells by disrupting NF-kB/p65 signaling [485]. |
| Iloperidone | atypical anti-psychotic | Shows synergistic effects with TMZ, potentially due to its inhibition of DRD2 and β-catenin expression [486]. |
| TmHg | Sensitizer | Inhibits the Trx system, leading to increased cancer cell death and reduced proliferation and angiogenesis [487]. |
| Acetazolamide | Altitude sickness treatment | Enhances GBM sensitivity to TMZ by inhibiting BCL-3-dependent carbonic anhydrase upregulation [488]. |
| Sunitinib | Gastrointestinal stromal tumors and metastatic renal cell carcinoma treatment | Inhibits alkyladenine DNA glycosylase (AAG), suppresses cell proliferation and stem-like characteristics, and induces cell cycle arrest [47]. |
| Sitagliptin | Type 2 diabetes treatment | Enhances TMZ cytotoxicity in glioma cells by inhibiting TMZ-induced protective autophagy [489]. |
| Hydroquinidine | Anti-arrhythmic agent | Modulates the gene expression profile in GBM cells, reducing viability, growth, and migration of TMZ-resistant GBM cells [220]. |
| Stiripentol | Antiepileptic agent | Augments TMZ cytotoxic activity in GBM cells by influencing the cell cycle, mirroring the effects of TMZ [490]. |
| Meclofenamate | Non-steroidal anti-inflammatory drug | Inhibits gap junction-mediated cytoplasmic transport and disrupts tumor microtubule network morphology [491]. |
| PCI-24,781/Abexinostat | Histone deacetylase inhibitor | Inhibits DDR and induces DNA damage [492]. |
| pimavanserin tartrate | Parkinson’s disease treatment | Suppresses the NFAT signaling pathway and inhibits the ATR/CDK2/E2F axis as well as MYC and Aurora A/B signaling pathways [493]. |
| Captopril | Hypertension and heart failure treatment | Modulates the MMP-2 pathway [494]. |
| Nicardipine | Hypertension and coronary artery disease treatment | Enhances TMZ cytotoxicity in GSCs by promoting apoptosis and sensitizing GSCs to TMZ via mTOR activation, inhibiting autophagy [495]. |
| Lovastatin | Prevention of atherosclerosis and coronary artery disease | Induces cell senescence by suppressing Skp2 expression, enhancing GBM sensitivity to TMZ both in vitro and in vivo [496]. |
| Roscovitine | Selective CDK inhibitor | Induces autophagy and caspase-3-dependent apoptosis [497]. |
| Ibudilast | MIF inhibitor | Increases GBM sensitivity to TMZ by downregulating macrophage migration inhibitory factor (MIF) expression [498]. |
| Pimozide | Antipsychotic drug | Inhibits EGFRvIII-Stat5-Fn14 signaling in GBM cells [499]. |
| Thioridazine | Antipsychotic drug | Sensitizes GBM to TMZ by impairing late-stage autophagy in GBM cells [500]. |
| Hydroxyurea | Myeloproliferative disorders and cancer treatment | Enhances TMZ sensitivity in GBM by inhibiting ribonucleotide reductase M2 [501]. |
| Dapagliflozin | Diabetes treatment | Inhibits cell cycle progression by interacting with CDK1/PBK/CHEK1 [502]. |
This table summarizes drugs originally developed for other medical conditions, highlighting their key mechanisms of action that confer potential therapeutic efficacy against glioma
Siramesine, initially created as an anxiety medication targeting the sigma-2 receptor, has shown promising effects against TMZ-resistant glioma by causing ferroptosis, a type of cell death involving lipid damage [468–470]. In TMZ-resistant glioma cells, siramesine decreases the activity of protective enzymes such as glutathione peroxidase 4 (GPX4) and HO-1, enhancing the effectiveness of TMZ in eliminating resistant cancer cells [471, 472].
Afatinib, an FDA-approved drug for non-small cell lung cancer (NSCLC) that targets EGFR/HER2/HER4, has shown limited results as a single treatment for recurrent GBM [473, 474]. However, combining afatinib with TMZ in laboratory studies significantly reduced the growth, survival, and invasion of glioma cells, including those expressing the aggressive EGFRvIII variant, highlighting its potential when used alongside standard treatments [72].
Metformin, which is commonly prescribed for diabetes, improves glioma cell sensitivity to TMZ [475]. It promotes cell death in TMZ-sensitive glioma cells by adjusting the Bax/Bcl-2 protein balance and reducing ROS. In TMZ-resistant glioma cells, metformin decreases resistance and suppresses markers of glioma stemness such as CD90. Animal studies have confirmed that combining metformin with TMZ specifically inhibits resistant tumor growth without negatively affecting TMZ-sensitive tumors [476].
Bortezomib, a proteasome inhibitor approved for multiple myeloma treatment, also acts as a sensitizer for glioma therapy. At non-toxic levels, bortezomib reduces glioma cell growth, spheroid formation, and stem-like characteristics, primarily by inducing apoptosis and cell cycle arrest. When combined with TMZ, bortezomib effectively targets the FOXM1-Survivin pathway, which is linked to poor glioma outcomes, resulting in improved treatment responses in both cell culture and animal models [477].
TMZ analogs
TMZ, an imidazotetrazine prodrug, faces significant clinical limitations because resistance is driven mainly by MGMT [36]. To overcome this resistance, novel TMZ analogs have been developed. For example, modified imidazotetrazine derivatives at the 3-methyl and 8-carboxamide positions, such as thiazole 13, show improved growth inhibition in glioma cells resistant to MGMT expression. These analogs cause cell cycle arrest, DNA damage, and cell death independently of the MGMT or MMR status, addressing key resistance pathways [503]. Other compounds, such as C8-imidazolyl (377) and C8-methylimidazole (465) tetrazines, also exhibit potent anticancer effects against resistant glioma and colorectal carcinoma cells. These analogs create DNA lesions similar to TMZ but avoid resistance linked to MGMT and MMR [6].
Imidazotetrazine 4a (KL-50), another novel analog, specifically targets drug-resistant glioma. KL-50 induces MMR-independent cell death by forming dynamic DNA lesions that evolve into interstrand cross-links in MGMT-silenced tumors, demonstrating efficacy in preclinical models with minimal toxicity [21]. Another novel boron-10 (10B)-boronated TMZ derivative, TMZB, combines the DNA-damaging effects of TMZ with those of boron neutron capture therapy (BNCT). TMZB efficiently delivers 10B across the BBB, enhancing tumor-specific radiation damage and outperforming conventional boron carriers such as boronophenylalanine (BPA) [504–506].
Lipophilic prodrug strategies further improve TMZ delivery. The hexadecyl ester TMZ16e, which is formulated into nanoparticles (TMZ16e-NPs) for intranasal administration, bypasses the BBB and prolongs survival in orthotopic glioma models by inducing G2/M arrest and downregulating Cyclin B1/CDK1 [263, 507]. DP68, another TMZ analog, inhibits GBM regrowth via interstrand DNA crosslinking and unique S-phase arrest, with enhanced efficacy upon FANCD2 or ATR suppression [508]. Additionally, the multifunctional analog NEO212 overcomes resistance driven by MGMT and MMR, enhances sensitivity to radiation, and exerts anti-angiogenic effects, significantly improving survival without severe side effects [509, 510].
Translational barriers and practical clinical guidance
Although recent advances in glioma treatment represent significant progress, translating these strategies into consistent clinical benefit remains a major challenge. Tumor heterogeneity and dynamic adaptation remain critical obstacles, limiting the effectiveness of single-agent therapies and highlighting the need for accurate biomarkers to stratify patients effectively [511, 512]. The inconsistent relationship between the MGMT promoter methylation status and actual protein expression complicates clinical decision-making, often resulting in unpredictable therapeutic outcomes [32]. Immunotherapy approaches encounter considerable hurdles, including restricted penetration across the BBB, inherently low tumor mutation burdens, and the profoundly immunosuppressive TME characteristic of gliomas [513]. Nanotherapy and gene-editing technologies face challenges related to precise delivery efficiency, specificity, potential off-target genetic effects, and issues with consistent scale-up for clinical application [514]. Additionally, strategies targeting autophagy require careful context-dependent optimization because of the dual role of autophagy in tumor cell survival and cell death mechanisms [515]. GSC plasticity and extensive metabolic adaptability further complicate sustained therapeutic efficacy, as these cells can quickly alter their phenotype to evade treatment pressures [516]. Translational efforts are frequently impeded by discrepancies between preclinical models, which often fail to recapitulate the complexity of human disease, and the clinical realities encountered in patient populations [517]. Furthermore, the potential for compounded toxicities with combination therapies poses significant risks, necessitating careful evaluation of dose, timing, and treatment sequence [518].
Addressing these limitations will require the integration of advanced multi-omics profiling techniques, including genomics, transcriptomics, proteomics, and metabolomics, to comprehensively decipher resistance mechanisms and reliably identify predictive biomarkers. Enhancing drug delivery systems through responsive nanoparticles, multifunctional nanocarriers, or sophisticated gene-editing platforms may significantly improve therapeutic specificity and effectiveness and overcome challenges associated with BBB penetration [519]. Personalized therapeutic regimens that simultaneously target DNA repair pathways, immune checkpoints, metabolic vulnerabilities, and stemness characteristics offer considerable promise for achieving durable and clinically meaningful responses [520, 521]. Further optimization and clinical validation of treatments such as TTFields and nanotechnology-based platforms are critical for broader clinical implementation, ensuring that these promising modalities are effectively integrated into treatment protocols. Future clinical trials should emphasize patient stratification based on molecular subtypes and resistance mechanisms to ensure more tailored interventions. Furthermore, leveraging artificial intelligence for drug repurposing, biomarker prediction, and regimen design may streamline decision-making and accelerate therapeutic breakthroughs [522].
While these forward-looking strategies evolve, TMZ remains a cornerstone of current glioma therapy [523]. Its widespread use, especially in combination with emerging agents, highlights the need for practical, evidence-based clinical guidance to maximize safety and therapeutic impact. Several critical considerations must be observed in daily practice. Older anti-epileptic drugs, such as phenobarbital, carbamazepine or phenytoin, stimulate the synthesis of hepatic cytochrome P450 enzymes and can affect the metabolism of TMZ. Non-enzyme inducing antiepileptic agents, such as levetiracetam, lacosamide, or clobazam, are preferred due to fewer drug-drug interactions and improved side effect profiles [524]. Due to the presence of compounded hematologic toxicities, patients receiving TMZ may experience myelosuppression, including persistent pancytopenias, which may lead to aplastic anemia [525]. Therefore, when combined with other myelotoxic agents (e.g., valproic acid), the potential for therapeutic synergy must be weighed against the increased risk of myelosuppression [526, 527]. Although glucocorticoids such as dexamethasone are commonly used to treat brain edema in patients with glioma, they may antagonize the effects of TMZ by inducing anti-apoptotic signals and altering tumor cell sensitivity. The use of dexamethasone during TMZ use may lead to adverse clinical outcomes [528]. Notably, recent evidence suggests that glucocorticoid receptor (GR) signaling in GBM follows a circadian rhythm, and fluctuations in daily glucocorticoid levels can regulate tumor growth in a time-dependent manner, further complicating its clinical impact [529]. Diet and supportive care likewise influence treatment success. TMZ should be taken on an empty stomach (≥ 2 h before or after meals), as high-fat meals can delay absorption and alter pharmacokinetics. Prophylactic antiemetics such as ondansetron are useful in managing nausea and vomiting [530]. Furthermore, proton pump inhibitors (e.g., omeprazole), which affect gastric acidity, may also impair TMZ absorption and should be timed carefully around dosing.
Ongoing monitoring and individualized assessment are essential throughout TMZ treatment [531]. Hematologic and hepatic function should be routinely evaluated, especially in patients with underlying liver dysfunction [532]. Clinicians should be vigilant in assessing new neurological symptoms, such as headaches or seizures, to distinguish between drug-related toxicity and tumor progression [533]. Patients should be encouraged to report infections, unexplained bleeding, severe fatigue, or the use of herbal/traditional supplements (e.g., curcumin, quercetin), which may interfere with the oxidative stress mechanisms central to TMZ’s action. Ultimately, integrating biomarker-informed decision-making (e.g., MGMT methylation status), personalized supportive care, and open patient communication is key to ensuring safe, effective, and context-sensitive use of TMZ in the clinic [534].
Conclusions
TMZ remains a cornerstone of glioma chemotherapy, but its long-term effectiveness is challenged by diverse resistance mechanisms, including MGMT-mediated repair, MMR deficiencies, enhanced BER activity, efflux transporter upregulation, and tumor mutations such as IDH1/2, TP53, EGFR, and ATRX. GSCs, non-coding RNAs, autophagy, and the TME further complicate resistance through metabolic plasticity, immune evasion, and therapeutic adaptation. Emerging treatment strategies targeting these mechanisms—such as PARP inhibition, advanced nanoparticle delivery systems, immunotherapies, autophagy modulators, TTFields, gene-editing technologies, and novel TMZ analogs—have shown significant promise. However, clinical translation remains constrained by tumor heterogeneity, biomarker inconsistency, drug delivery barriers, GSC plasticity, and discrepancies between preclinical and clinical outcomes.
Future progress will require comprehensive multi-omics approaches to identify reliable predictive biomarkers, precision-enhancing nanoparticle and gene-editing delivery systems, and personalized treatments addressing DNA repair, immune checkpoints, metabolic vulnerabilities, and stemness features. Advanced patient stratification in clinical trials, informed by detailed molecular profiling, alongside leveraging artificial intelligence for drug discovery and regimen optimization, will be essential to overcome these translational hurdles.
Ultimately, effective clinical care demands meticulous attention to practical aspects of TMZ administration. Careful management of drug interactions, dietary considerations, prophylactic and supportive care measures, vigilant monitoring for hematologic and neurologic toxicity, and individualized patient counseling remain crucial for maximizing treatment safety and efficacy. Through continued translational innovation and vigilant clinical practice, the therapeutic outcomes for glioma patients can be progressively improved.
Acknowledgements
Figures were created through BioRender.com.
Abbreviations
- ADSL
Adenylosuccinate lyase
- APE1/Ref-1
Apurinic/apyrimidinic endonuclease 1/Redox effector factor-1)
- ATRX
Alterations of alpha thalassemia/mental retardation syndrome X-linked
- BBB
Blood-brain barrier
- BBTB
Blood-brain tumor barrier
- BER
Base-excision repair
- BNCT
Boron neutron capture therapy
- BTB
Brain tumor barrier
- CHK1
Checkpoint kinase 1
- circRNAs
Circular RNAs
- CMA
Chaperone-mediated autophagy
- CNS
Central nervous system
- CSC
Cancer stem cell
- CSF
Cerebrospinal fluid
- CTLA-4
Cytotoxic T-lymphocyte protein 4
- D-2HG
D-2-hydroxyglutarate
- DC
Dendritic cell
- DDR
DNA damage repair
- DRD2
Dopamine D2 receptor
- DUBs
Deubiquitinating enzymes
- EC
Endothelial cell
- EGFR
Epidermal growth factor receptor
- EMT
Epithelial-mesenchymal transition
- EVs
Extracellular vesicles
- FGFR1
Fibroblast growth factor receptor 1
- FoxD2-AS1
FoxD2 adjacent opposite strand RNA 1
- FOXM1
Forkhead Box M1
- FTL
Ferritin light chain
- G4
G-quadruplex
- GAMs
Glioma-associated microglia and macrophages
- GBM
Glioblastoma
- GPI
Gasdermin-related prognostic index
- GSC
Glioma stem cell
- HGGs
High-grade glioma
- HOTAIR
HOX transcript antisense RNA
- HR
Homologous recombination
- HSV
Herpes simplex virus
- ICIs
Immune checkpoint inhibitors
- IDH
Isocitrate dehydrogenase
- IMPDH
Inosine 5’-monophosphate dehydrogenase
- LDH
Lactate dehydrogenase
- LGGs
Low-grade glioma
- lncRNAs
Long non-coding RNAs
- MCNTs
Magnetic carbon nanotubes
- MET
Metformin
- MEX3A
Mex-3 RNA binding family member A
- MGMT
O6-methylguanine-DNA methyltransferase
- miRNAs
MicroRNAs
- MMR
Mismatch repair
- MRP-1
Multidrug resistance-associated protein-1
- MSN
Mesoporous silica nanoparticles
- MTIC
5-(3-methyltriazen-1-yl) imidazole-4-carboxamide
- N3-MeA
N3-methyladenine
- N7-MeG
N7-methylguanine
- ncRNAs
Non-coding RNAs
- NDDS
Nanodrug delivery systems
- O6-BG
O6-benzylguanine
- O6-MeG
O6-methylguanine
- OS
Overall survival
- OVs
Oncolytic viruses
- P-gp
P-glycoprotein
- PAR
Poly (ADP-ribose)
- PARP
Poly (ADP-ribose) polymerase
- PARPis
PARP inhibitors
- PDT
Photodynamic therapy
- PFS
Progression-free survival
- PHAX
Phosphorylated adaptor for RNA export
- PMT
Proneural-to-mesenchymal transition
- POSS
Polyhedral oligomeric silsesquioxane
- PTRF
Transcript release factor
- rGBM
Recurrent glioblastoma
- ROS
Reactive oxygen species
- RT
Radiotherapy
- SCD
Stearoyl-CoA desaturase
- STAT3
Signal transducer and activator of transcription 3
- tFNA
Tetrahedral scaffold nucleic acid
- TGF-β1
Transforming growth factor-β1
- TLS
Translesion synthesis
- TMB
Tumor mutational burden
- TME
Tumor microenvironment
- TMZ
Temozolomide
- Tregs
Regulatory T cells
- TTFields
Tumor Treating Fields
- U3 snoRNAs
U3 small nucleolar RNAs
- USLPs
Ultra-small large-pore silica nanoparticles
Author contributions
H.L., Y.W., and Y.C. contributed equally to the conceptualization, literature review, and initial drafting of the manuscript. J.L., C.Q., Y.Z., C.Y., and F.L. assisted with figure preparation and manuscript editing. T.M. and S.G. provided critical revisions and contributed to refining the intellectual content. A.Y. and L.J. supervised the project, provided overall guidance throughout the writing process, and were responsible for funding acquisition. All authors contributed to the conception of the work and approved the final manuscript.
Funding
The present study was supported by the research project of 988th Hospital of Joint Logistic Support Force of PLA (grant no. YNZX2024005), the joint construction project of Henan Province (grant no. LHGJ20230703), and the opening project of State Key Laboratory of Explosion Science and Technology (Beijing Institute of Technology) (grant no. KFJJ23-09 M). The funding organizations played no role in the scientific conduct of this research, including but not limited to: study conceptualization, data acquisition/processing/interpretation, manuscript composition, or publication decision-making.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hengzeng Li, Yahui Wu, Yue Chen contributed equally to this work.
Contributor Information
Anhui Yao, Email: yaoanhui@aliyun.com.
Liyun Jia, Email: jialiyun0410@zzu.edu.cn.
References
- 1.Ostrom QT et al (2020) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the united States in 2013–2017. Neuro Oncol 22(12 Suppl 2):iv1–iv96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weller M et al (2024) Glioma. Nat Rev Dis Primers 10(1):33 [DOI] [PubMed] [Google Scholar]
- 3.Chang C et al (2024) Recurrent Glioblastoma-Molecular underpinnings and evolving treatment paradigms. Int J Mol Sci, 25(12) [DOI] [PMC free article] [PubMed]
- 4.Rong L, Li N, Zhang Z (2022) Emerging therapies for glioblastoma: current state and future directions. J Exp Clin Cancer Res 41(1):142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tonkin-Reeves A, Giuliani CM, Price JT (2023) Inhibition of autophagy; an opportunity for the treatment of cancer resistance. Front Cell Dev Biol 11:1177440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z et al (2019) C8-Substituted Imidazotetrazine analogs overcome Temozolomide resistance by inducing DNA adducts and DNA damage. Front Oncol 9:485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeon J et al (2021) Revisiting Platinum-Based anticancer drugs to overcome gliomas. Int J Mol Sci, 22(10):5111 [DOI] [PMC free article] [PubMed]
- 8.de Blank P, Fouladi M, Huse JT (2020) Molecular markers and targeted therapy in pediatric low-grade glioma. J Neurooncol 150(1):5–15 [DOI] [PubMed] [Google Scholar]
- 9.Shams B et al (2023) Improved prediction of glioma-related aphasia by diffusion MRI metrics, machine learning, and automated fiber bundle segmentation. Hum Brain Mapp 44(12):4480–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iturrioz-Rodríguez N, Sampron N, Matheu A (2023) Current advances in Temozolomide encapsulation for the enhancement of glioblastoma treatment. Theranostics 13(9):2734–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaff LR, Mellinghoff IK (2023) Glioblastoma and other primary brain malignancies in adults: A review. JAMA 329(7):574–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu C et al (2020) DNA-methylation-mediated activating of LncRNA SNHG12 promotes Temozolomide resistance in glioblastoma. Mol Cancer 19(1):28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J et al (2021) Regulation of tumor immune suppression and cancer cell survival by CXCL1/2 elevation in glioblastoma multiforme. Sci Adv, 7(5):eabc2511 [DOI] [PMC free article] [PubMed]
- 14.Collado J et al (2024) Understanding the glioblastoma tumor microenvironment: leveraging the extracellular matrix to increase immunotherapy efficacy. Front Immunol 15:1336476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moreno V et al (2023) Trotabresib, an oral potent bromodomain and extraterminal inhibitor, in patients with high-grade gliomas: A phase I, window-of-opportunity study. Neuro Oncol 25(6):1113–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SH et al (2020) Safety and feasibility of multiple blood-brain barrier disruptions for the treatment of glioblastoma in patients undergoing standard adjuvant chemotherapy. J Neurosurg 134(2):475–483 [DOI] [PubMed] [Google Scholar]
- 17.Oliver L et al (2020) Drug resistance in glioblastoma: are persisters the key to therapy? Cancer Drug Resist 3(3):287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janmey PA, Pogoda K (2019) Compressive tumours cause neuronal damage. Nat Biomed Eng 3(3):171–172 [DOI] [PubMed] [Google Scholar]
- 19.Takenaka MC et al (2019) Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat Neurosci 22(5):729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiao F et al (2020) RACK7 recognizes H3.3G34R mutation to suppress expression of MHC class II complex components and their delivery pathway in pediatric glioblastoma. Sci Adv 6(29):eaba2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin K et al (2022) Mechanism-based design of agents that selectively target drug-resistant glioma. Science 377(6605):502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bomsztyk K et al (2024) Analysis of DNA methylation in gliomas: assessment of preanalytical variables. Lab Invest 104(12):102160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansouri A et al (2019) MGMT promoter methylation status testing to guide therapy for glioblastoma: refining the approach based on emerging evidence and current challenges. Neuro Oncol 21(2):167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gramatzki D et al (2021) Telomerase reverse transcriptase promoter mutation- and O(6)-methylguanine DNA methyltransferase promoter methylation-mediated sensitivity to Temozolomide in isocitrate dehydrogenase-wild-type glioblastoma: is there a link? Eur J Cancer 147:84–94 [DOI] [PubMed] [Google Scholar]
- 25.Arcella A et al (2020) Dissecting molecular features of gliomas: genetic loci and validated biomarkers. Int J Mol Sci, 21(2):685 [DOI] [PMC free article] [PubMed]
- 26.Cruz JVR et al (2022) Obstacles to glioblastoma treatment two decades after Temozolomide. Cancers (Basel), 14(13):3203 [DOI] [PMC free article] [PubMed]
- 27.Papacocea SI et al (2024) Molecular profile as an outcome predictor in glioblastoma along with MRI features and surgical resection: A scoping review. Int J Mol Sci, 25(17):9714 [DOI] [PMC free article] [PubMed]
- 28.Brawanski KR et al (2023) Influence of MMR, MGMT promotor methylation and protein expression on overall and Progression-Free survival in primary glioblastoma patients treated with Temozolomide. Int J Mol Sci, 24(7):6184 [DOI] [PMC free article] [PubMed]
- 29.Wick A et al (2020) Superiority of Temozolomide over radiotherapy for elderly patients with RTK II methylation class, MGMT promoter methylated malignant Astrocytoma. Neuro Oncol 22(8):1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J et al (2020) Hypoxia induced ferritin light chain (FTL) promoted epithelia mesenchymal transition and chemoresistance of glioma. J Exp Clin Cancer Res 39(1):137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tesileanu CMS et al (2022) MGMT promoter methylation determined by the MGMT-STP27 algorithm is not predictive for outcome to Temozolomide in IDH-mutant anaplastic Astrocytomas. Neuro Oncol 24(4):665–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butler M et al (2020) MGMT status as a clinical biomarker in glioblastoma. Trends Cancer 6(5):380–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hegi ME et al (2024) No benefit from TMZ treatment in glioblastoma with truly unmethylated MGMT promoter: reanalysis of the CE.6 and the pooled Nordic/NOA-08 trials in elderly glioblastoma patients. Neuro Oncol 26(10):1867–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y et al (2023) Lysine methylation promotes NFAT5 activation and determines Temozolomide efficacy in glioblastoma. Nat Commun 14(1):4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nie E et al (2021) TGF-β1 modulates Temozolomide resistance in glioblastoma via altered MicroRNA processing and elevated MGMT. Neuro Oncol 23(3):435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oldrini B et al (2020) MGMT genomic rearrangements contribute to chemotherapy resistance in gliomas. Nat Commun 11(1):3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng X et al (2024) Trans-lesion synthesis and mismatch repair pathway crosstalk defines chemoresistance and hypermutation mechanisms in glioblastoma. Nat Commun, 15(1):1957 [DOI] [PMC free article] [PubMed]
- 38.Xu J et al (2024) MEN1 Deficiency-Driven activation of the β-Catenin-MGMT Axis promotes pancreatic neuroendocrine tumor growth and confers Temozolomide resistance. Adv Sci (Weinh) 11(35):e2308417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu S et al (2021) PARP-mediated parylation of MGMT is critical to promote repair of temozolomide-induced O6-methylguanine DNA damage in glioblastoma. Neuro Oncol 23(6):920–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu C et al (2021) DNA sensing in mismatch Repair-Deficient tumor cells is essential for Anti-tumor immunity. Cancer Cell 39(1):96–108.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang R, Zhou PK (2021) DNA damage repair: historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct Target Ther 6(1):254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He Y et al (2022) The role of DNA mismatch repair in immunotherapy of human cancer. Int J Biol Sci 18(7):2821–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El Touny LH et al (2021) ATR Inhibition reverses the resistance of homologous recombination deficient MGMT(low)/MMR(proficient) cancer cells to Temozolomide. Oncotarget 12(21):2114–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leong VWS et al (2024) MGMT function determines the differential response of ATR inhibitors with DNA-damaging agents in glioma stem cells for GBM therapy. Neurooncol Adv 6(1):vdad165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yue Q et al (2024) Histone H3K9 lactylation confers Temozolomide resistance in glioblastoma via LUC7L2-Mediated MLH1 intron retention. Adv Sci (Weinh) 11(19):e2309290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gan T et al (2022) MEX3A impairs DNA mismatch repair signaling and mediates acquired Temozolomide resistance in glioblastoma. Cancer Res 82(22):4234–4246 [DOI] [PubMed] [Google Scholar]
- 47.Song YQ et al (2023) A robust luminescent assay for screening alkyladenine DNA glycosylase inhibitors to overcome DNA repair and Temozolomide drug resistance. J Pharm Anal 13(5):514–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montaldo NP et al (2019) Alkyladenine DNA glycosylase associates with transcription elongation to coordinate DNA repair with gene expression. Nat Commun 10(1):5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caron C et al (2019) Interaction of functionalized naphthalenophanes with Abasic sites in DNA: DNA cleavage, DNA cleavage Inhibition, and formation of Ligand-DNA adducts. Chemistry 25(8):1949–1962 [DOI] [PubMed] [Google Scholar]
- 50.Golbourn B et al (2024) A Kinome drug screen identifies multi-TKI synergies and ERBB2 signaling as a therapeutic vulnerability in MYC/TYR subgroup atypical teratoid rhabdoid tumors. Neuro Oncol 26(10):1895–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng X et al (2024) Trans-Lesion Synthesis and Mismatch Repair Pathway Crosstalk Defines Chemoresistance and Hypermutation Mechanisms in Glioblastoma. Nat Commun 15(1):1957 [DOI] [PMC free article] [PubMed]
- 52.Kumar A et al (2022) Interlocking activities of DNA polymerase Β in the base excision repair pathway. Proc Natl Acad Sci U S A 119(10):e2118940119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tell G et al (2009) The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid Redox Signal 11(3):601–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bora A et al (2024) DNA Abasic sites act as rational therapeutic targets to synergize Temozolomide response in both MMR-proficient and deficient cancer. NAR Cancer 6(3):zcae034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruford EA et al (2020) Guidelines for human gene nomenclature. Nat Genet 52(8):754–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Z et al (2021) Chinese glioma genome atlas (CGGA): A comprehensive resource with functional genomic data from Chinese glioma patients. Genomics Proteom Bioinf 19(1):1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Louis DN et al (2021) The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 23(8):1231–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith HL, Wadhwani N, Horbinski C (2022) Major features of the 2021 WHO classification of CNS tumors. Neurotherapeutics 19(6):1691–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin L et al (2021) Mutant IDH1 enhances Temozolomide sensitivity via regulation of the ATM/CHK2 pathway in glioma. Cancer Res Treat 53(2):367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun X, Turcan S (2021) From laboratory studies to clinical trials: Temozolomide use in IDH-Mutant gliomas. Cells, 10(5):1225 [DOI] [PMC free article] [PubMed]
- 61.Kadiyala P et al (2021) Inhibition of 2-hydroxyglutarate elicits metabolic reprogramming and mutant IDH1 glioma immunity in mice. J Clin Invest, 131(4):e139542 [DOI] [PMC free article] [PubMed]
- 62.Nagashima H et al (2020) Poly(ADP-ribose) glycohydrolase Inhibition sequesters NAD(+) to potentiate the metabolic lethality of alkylating chemotherapy in IDH-Mutant tumor cells. Cancer Discov 10(11):1672–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forte IM et al (2019) Targeted therapy based on p53 reactivation reduces both glioblastoma cell growth and resistance to Temozolomide. Int J Oncol 54(6):2189–2199 [DOI] [PubMed] [Google Scholar]
- 64.Laverty DJ et al (2024) ATM Inhibition exploits checkpoint defects and ATM-dependent double strand break repair in TP53-mutant glioblastoma. Nat Commun 15(1):5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han B et al (2018) Loss of ATRX suppresses ATM dependent DNA damage repair by modulating H3K9me3 to enhance Temozolomide sensitivity in glioma. Cancer Lett 419:280–290 [DOI] [PubMed] [Google Scholar]
- 66.Hu C et al (2022) ATRX loss promotes immunosuppressive mechanisms in IDH1 mutant glioma. Neuro Oncol 24(6):888–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amen AM et al (2021) Cancer-specific loss of TERT activation sensitizes glioblastoma to DNA damage. Proc Natl Acad Sci U S A, 118(13):e2008772118 [DOI] [PMC free article] [PubMed]
- 68.Waitkus MS et al (2024) Mechanisms of telomere maintenance and associated therapeutic vulnerabilities in malignant gliomas. Neuro Oncol 26(6):1012–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McKinney AM et al (2022) GABP couples oncogene signaling to telomere regulation in TERT promoter mutant cancer. Cell Rep 40(12):111344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cui X et al (2023) Blockage of EGFR/AKT and mevalonate pathways synergize the antitumor effect of Temozolomide by reprogramming energy metabolism in glioblastoma. Cancer Commun (Lond) 43(12):1326–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tomar MS et al (2021) Elucidating the mechanisms of Temozolomide resistance in gliomas and the strategies to overcome the resistance. Biochim Biophys Acta Rev Cancer 1876(2):188616 [DOI] [PubMed] [Google Scholar]
- 72.Vengoji R et al (2019) Afatinib and Temozolomide combination inhibits tumorigenesis by targeting EGFRvIII-cMet signaling in glioblastoma cells. J Exp Clin Cancer Res 38(1):266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Souza I et al (2022) High levels of NRF2 sensitize temozolomide-resistant glioblastoma cells to ferroptosis via ABCC1/MRP1 upregulation. Cell Death Dis 13(7):591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nesterova DS et al (2020) Sexually dimorphic impact of the iron-regulating gene, HFE, on survival in glioblastoma. Neurooncol Adv 2(1):vdaa001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McAleenan A et al (2021) Prognostic value of test(s) for O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation for predicting overall survival in people with glioblastoma treated with Temozolomide. Cochrane Database Syst Rev, 3(3): p. Cd013316. [DOI] [PMC free article] [PubMed]
- 76.Haynes T et al (2024) Pathways to hypermutation in high-grade gliomas: mechanisms, syndromes, and opportunities for immunotherapy. Neurooncol Adv 6(1):vdae105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barthel FP et al (2019) Longitudinal molecular trajectories of diffuse glioma in adults. Nature 576(7785):112–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Touat M et al (2020) Mechanisms and therapeutic implications of hypermutation in gliomas. Nature 580(7804):517–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu C et al (2024) TRIM24 cooperates with Ras mutation to drive glioma progression through snorna recruitment of PHAX and DNA-PKcs. Adv Sci (Weinh) 11(29):e2400023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huchede P et al (2024) BMP2 and BMP7 cooperate with H3.3K27M to promote quiescence and invasiveness in pediatric diffuse midline gliomas. Elife, 12 [DOI] [PMC free article] [PubMed]
- 81.Cohen LRZ et al (2023) PRC2-independent actions of H3.3K27M in embryonic stem cell differentiation. Nucleic Acids Res 51(4):1662–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Appay R et al (2019) CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro Oncol 21(12):1519–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu VM et al (2020) The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: a systematic review of the contemporary literature. J Neurooncol 148(2):221–229 [DOI] [PubMed] [Google Scholar]
- 84.Yin J et al (2024) Reactivating PTEN to impair glioma stem cells by inhibiting cytosolic iron-sulfur assembly. Sci Transl Med 16(739):eadg5553 [DOI] [PubMed] [Google Scholar]
- 85.Tiek DM et al (2022) Temozolomide-induced guanine mutations create exploitable vulnerabilities of guanine-rich DNA and RNA regions in drug-resistant gliomas. Sci Adv 8(25):eabn3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uribe ML, Marrocco I, Yarden Y (2021) EGFR in cancer: signaling mechanisms, drugs, and acquired resistance. Cancers (Basel), 13(11):2748 [DOI] [PMC free article] [PubMed]
- 87.Lu H, Liu P, Pang Q (2021) MiR-27a-3p/miR-27b-3p promotes neurofibromatosis type 1 via targeting of NF1. J Mol Neurosci 71(11):2353–2363 [DOI] [PubMed] [Google Scholar]
- 88.Li X et al (2024) Biological characteristics of tissue engineered-nerve grafts enhancing peripheral nerve regeneration. Stem Cell Res Ther 15(1):215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao C et al (2020) MicroRNA-128-3p enhances the chemosensitivity of Temozolomide in glioblastoma by targeting c-Met and EMT. Sci Rep 10(1):9471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu ZQ et al (2020) MicroRNA-144 represses gliomas progression and elevates susceptibility to Temozolomide by targeting CAV2 and FGF7. Sci Rep 10(1):4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ho KH et al (2019) miR-140 targeting CTSB signaling suppresses the mesenchymal transition and enhances Temozolomide cytotoxicity in glioblastoma multiforme. Pharmacol Res 147:104390 [DOI] [PubMed] [Google Scholar]
- 92.Pourhanifeh MH et al (2020) Autophagy in cancers including brain tumors: role of MicroRNAs. Cell Commun Signal 18(1):88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou Y et al (2022) Cyanidin-3-O-glucoside inhibits the β-catenin/MGMT pathway by upregulating miR-214-5p to reverse chemotherapy resistance in glioma cells. Sci Rep 12(1):7773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen W et al (2020) LncRNA HOXA-AS3 promotes the malignancy of glioblastoma through regulating miR-455-5p/USP3 axis. J Cell Mol Med 24(20):11755–11767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Du X et al (2020) LINC00511 contributes to glioblastoma tumorigenesis and epithelial-mesenchymal transition via LINC00511/miR-524-5p/YB1/ZEB1 positive feedback loop. J Cell Mol Med 24(2):1474–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu H et al (2020) Long noncoding RNA LINC-PINT suppresses cell proliferation, invasion, and EMT by blocking Wnt/β-Catenin signaling in glioblastoma. Front Pharmacol 11:586653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Buccarelli M et al (2020) Deregulated expression of the imprinted DLK1-DIO3 region in glioblastoma stemlike cells: tumor suppressor role of LncRNA MEG3. Neuro Oncol 22(12):1771–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou J et al (2022) LncRNA XLOC013218 promotes cell proliferation and TMZ resistance by targeting the PIK3R2-mediated PI3K/AKT pathway in glioma. Cancer Sci 113(8):2681–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yin T et al (2021) Long non-coding RNA HULC stimulates the epithelial-mesenchymal transition process and vasculogenic mimicry in human glioblastoma. Cancer Med 10(15):5270–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li C, Feng S, Chen L (2021) MSC-AS1 knockdown inhibits cell growth and Temozolomide resistance by regulating miR-373-3p/CPEB4 axis in glioma through PI3K/Akt pathway. Mol Cell Biochem 476(2):699–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu T et al (2021) A positive feedback loop of lncRNA-RMRP/ZNRF3 axis and Wnt/β-catenin signaling regulates the progression and Temozolomide resistance in glioma. Cell Death Dis 12(11):952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lv QL et al (2020) Knockdown LncRNA DLEU1 inhibits gliomas progression and promotes Temozolomide chemosensitivity by regulating autophagy. Front Pharmacol 11:560543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ho KH et al (2022) Hypoxia-inducible LncRNA MIR210HG interacting with OCT1 is involved in glioblastoma multiforme malignancy. Cancer Sci 113(2):540–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen Q et al (2021) Over-expression of lncRNA TMEM161B-AS1 promotes the malignant biological behavior of glioma cells and the resistance to temozolomide via up-regulating the expression of multiple ferroptosis-related genes by sponging hsa-miR-27a-3p. Cell Death Discov, 7(1):311 [DOI] [PMC free article] [PubMed]
- 105.Li Z et al (2021) Glioblastoma Cell-Derived lncRNA-Containing exosomes induce microglia to produce complement C5, promoting chemotherapy resistance. Cancer Immunol Res 9(12):1383–1399 [DOI] [PubMed] [Google Scholar]
- 106.Li XD et al (2021) Long noncoding RNA just proximal to X-inactive specific transcript facilitates aerobic Glycolysis and Temozolomide chemoresistance by promoting stability of PDK1 mRNA in an m6A-dependent manner in glioblastoma multiforme cells. Cancer Sci 112(11):4543–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang X et al (2022) Long non-coding RNA OIP5-AS1 Inhibition upregulates microRNA-129-5p to repress resistance to Temozolomide in glioblastoma cells via downregulating IGF2BP2. Cell Biol Toxicol 38(6):963–977 [DOI] [PubMed] [Google Scholar]
- 108.Li H et al (2021) Hsa_circ_0110757 upregulates ITGA1 to facilitate temozolomide resistance in glioma by suppressing hsa-miR-1298-5p. Cell Death Dis, 12(3):252 [DOI] [PMC free article] [PubMed]
- 109.Wu C et al (2020) LINC00470 promotes tumour proliferation and invasion, and attenuates chemosensitivity through the LINC00470/miR-134/Myc/ABCC1 axis in glioma. J Cell Mol Med 24(20):12094–12106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jiang LY et al (2024) CREB-induced LINC00473 promotes chemoresistance to TMZ in glioblastoma by regulating O6-methylguanine-DNA-methyltransferase expression via CEBPα binding. Neuropharmacology 243:109790 [DOI] [PubMed] [Google Scholar]
- 111.Jin T et al (2020) Lcn2-derived circular RNA (hsa_circ_0088732) inhibits cell apoptosis and promotes EMT in glioma via the miR-661/RAB3D Axis. Front Oncol 10:170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu W et al (2025) EIF4A3-Induced circ_0059914 promoted angiogenesis and EMT of glioma via the miR-1249/VEGFA pathway. Mol Neurobiol 62(1):973–987 [DOI] [PubMed] [Google Scholar]
- 113.Qiu S et al (2024) Circular RNA PRKCI (hsa_circ_0067934): a potential target in the pathogenesis of human malignancies. Front Oncol 14:1365032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu L et al (2024) Hypoxia-driven M2-polarized macrophages facilitate the epithelial-mesenchymal transition of glioblastoma via extracellular vesicles. Theranostics 14(16):6392–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu L et al (2022) Identification of a novel circular RNA circZNF652/miR-486-5p/SERPINE1 signaling cascade that regulates cancer aggressiveness in glioblastoma (GBM). Bioengineered 13(1):1411–1423 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 116.Geng X et al (2022) Exosomal circWDR62 promotes Temozolomide resistance and malignant progression through regulation of the miR-370-3p/MGMT axis in glioma. Cell Death Dis 13(7):596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu W et al (2020) MiR-145 in cancer therapy resistance and sensitivity: A comprehensive review. Cancer Sci 111(9):3122–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim HS et al (2021) Exosomal miR-125b exerts Anti-Metastatic properties and predicts early metastasis of hepatocellular carcinoma. Front Oncol 11:637247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Riemann A et al (2023) The role of MicroRNAs in gene expression and signaling response of tumor cells to an acidic environment. Int J Mol Sci, 24(23):16919 [DOI] [PMC free article] [PubMed]
- 120.Abdul Manap AS et al (2024) Mapping the function of MicroRNAs as a critical regulator of tumor-immune cell communication in breast cancer and potential treatment strategies. Front Cell Dev Biol 12:1390704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Buruiană A et al (2020) The roles of MiRNA in glioblastoma tumor cell communication: diplomatic and aggressive negotiations. Int J Mol Sci, 21(6):1950 [DOI] [PMC free article] [PubMed]
- 122.Roscigno G et al (2020) miR-216a acts as a negative regulator of breast Cancer by modulating stemness properties and tumor microenvironment. Int J Mol Sci, 21(7):2313 [DOI] [PMC free article] [PubMed]
- 123.Cui P et al (2020) STAT3 Inhibition induced temozolomide-resistant glioblastoma apoptosis via triggering mitochondrial STAT3 translocation and respiratory chain dysfunction. Cell Signal 71:109598 [DOI] [PubMed] [Google Scholar]
- 124.Zhang ZX et al (2023) HOXD-AS2-STAT3 feedback loop attenuates sensitivity to Temozolomide in glioblastoma. CNS Neurosci Ther 29(11):3430–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Perrault EN et al (2023) Ribonucleotide reductase regulatory subunit M2 drives glioblastoma TMZ resistance through modulation of dNTP production. Sci Adv 9(20):eade7236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu JX et al (2020) MicroRNA-29b promotes cell sensitivity to Temozolomide by targeting STAT3 in glioma. Eur Rev Med Pharmacol Sci 24(4):1922–1931 [DOI] [PubMed] [Google Scholar]
- 127.Chuang HY et al (2021) The E3 ubiquitin ligase NEDD4-1 mediates Temozolomide-Resistant glioblastoma through PTEN Attenuation and redox imbalance in Nrf2-HO-1 Axis. Int J Mol Sci, 22(19):10247 [DOI] [PMC free article] [PubMed]
- 128.Kong S et al (2020) MiR-3116 sensitizes glioma cells to Temozolomide by targeting FGFR1 and regulating the FGFR1/PI3K/AKT pathway. J Cell Mol Med 24(8):4677–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zeng Z et al (2022) NcRNAs: Multi–angle participation in the regulation of glioma chemotherapy resistance (Review). Int J Oncol, 60(6):76 [DOI] [PMC free article] [PubMed]
- 130.Swellam M et al (2021) Emerging role of MiRNAs as liquid biopsy markers for prediction of glioblastoma multiforme prognosis. J Mol Neurosci 71(4):836–844 [DOI] [PubMed] [Google Scholar]
- 131.Li H et al (2024) Exosomal miR-423-5p Derived from Cerebrospinal Fluid Pulsation Stress-Stimulated Osteoblasts Improves Angiogenesis of Endothelial Cells via DUSP8/ERK1/2 Signaling Pathway. Stem Cells Int, 2024:5512423 [DOI] [PMC free article] [PubMed]
- 132.Areeb Z et al (2020) Reduced EGFR and increased miR-221 is associated with increased resistance to Temozolomide and radiotherapy in glioblastoma. Sci Rep 10(1):17768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yuan Y et al (2023) LncRNA CASC2 Regulate Cell Proliferation and Invasion by Targeting miR-155/SOCS1 Axis in Hepatocellular Carcinoma. J Oncol, 2023: p. 8457112 [DOI] [PMC free article] [PubMed]
- 134.Gao K et al (2021) miR-23b-5p promotes the chemosensitivity of Temozolomide via negatively regulating TLR4 in glioma. Acta Biochim Biophys Sin (Shanghai) 53(8):979–987 [DOI] [PubMed] [Google Scholar]
- 135.Li S et al (2024) Extracellular vesicles derived from glioma stem cells affect glycometabolic reprogramming of glioma cells through the miR-10b-5p/PTEN/PI3K/Akt pathway. Stem Cell Rev Rep 20(3):779–796 [DOI] [PubMed] [Google Scholar]
- 136.Li H et al (2024) Glioma stem cell-derived exosomes induce the transformation of astrocytes via the miR-3065-5p/DLG2 signaling axis. Glia 72(5):857–871 [DOI] [PubMed] [Google Scholar]
- 137.Wang P et al (2020) The HIF1α/HIF2α-miR210-3p network regulates glioblastoma cell proliferation, dedifferentiation and chemoresistance through EGF under hypoxic conditions. Cell Death Dis 11(11):992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cui T et al (2021) A novel miR-146a-POU3F2/SMARCA5 pathway regulates stemness and therapeutic response in glioblastoma. Mol Cancer Res 19(1):48–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Guo X et al (2020) Identification of MiRNA signature associated with BMP2 and chemosensitivity of TMZ in glioblastoma stem-like cells. Genes Dis 7(3):424–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Su YK et al (2020) Targeting BC200/miR218-5p signaling Axis for overcoming Temozolomide resistance and suppressing glioma stemness. Cells, 9(8):1859 [DOI] [PMC free article] [PubMed]
- 141.Huang W et al (2019) The miR-26a/AP-2α/Nanog signaling axis mediates stem cell self-renewal and Temozolomide resistance in glioma. Theranostics 9(19):5497–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen L et al (2020) MiR-132 inhibits migration and invasion and increases chemosensitivity of cisplatin-resistant oral squamous cell carcinoma cells via targeting TGF-β1. Bioengineered 11(1):91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gong R et al (2021) Long noncoding RNA PVT1 promotes stemness and Temozolomide resistance through miR-365/ELF4/SOX2 Axis in glioma. Exp Neurobiol 30(3):244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jia B et al (2019) MiR-7-5p suppresses stemness and enhances Temozolomide sensitivity of drug-resistant glioblastoma cells by targeting Yin Yang 1. Exp Cell Res 375(1):73–81 [DOI] [PubMed] [Google Scholar]
- 145.Liao X et al (2024) Dynamic structural remodeling of LINC01956 enhances Temozolomide resistance in MGMT-methylated glioblastoma. Sci Transl Med 16(767):eado1573 [DOI] [PubMed] [Google Scholar]
- 146.Zuo Y et al (2024) Long non-coding RNA LIP interacts with PARP-1 influencing the efficiency of base excision repair. Noncoding RNA Res 9(3):649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lan T et al (2024) High expression of LncRNA HOTAIR is a risk factor for Temozolomide resistance in glioblastoma via activation of the miR-214/β-catenin/MGMT pathway. Sci Rep 14(1):26224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang J et al (2020) HOTAIR/miR-125 axis-mediated hexokinase 2 expression promotes chemoresistance in human glioblastoma. J Cell Mol Med 24(10):5707–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang X et al (2022) Serum-derived extracellular vesicles facilitate Temozolomide resistance in glioblastoma through a HOTAIR-dependent mechanism. Cell Death Dis 13(4):344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yang E et al (2024) EPIC-0628 abrogates HOTAIR/EZH2 interaction and enhances the Temozolomide efficacy via promoting ATF3 expression and inhibiting DNA damage repair in glioblastoma. Cancer Lett 588:216812 [DOI] [PubMed] [Google Scholar]
- 151.Zhang L et al (2020) A HOTAIR regulatory element modulates glioma cell sensitivity to Temozolomide through long-range regulation of multiple target genes. Genome Res 30(2):155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shangguan W, Lv X, Tian N (2019) FoxD2-AS1 is a prognostic factor in glioma and promotes Temozolomide resistance in a O(6)-methylguanine-DNA methyltransferase-dependent manner. Korean J Physiol Pharmacol 23(6):475–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gu N et al (2019) Silencing LncRNA FOXD2-AS1 inhibits proliferation, migration, invasion and drug resistance of drug-resistant glioma cells and promotes their apoptosis via microRNA-98-5p/CPEB4 axis. Aging 11(22):10266–10283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang Q et al (2019) Long non-coding RNA LINC00473 promotes glioma cells proliferation and invasion by impairing miR-637/CDK6 axis. Artif Cells Nanomed Biotechnol 47(1):3896–3903 [DOI] [PubMed] [Google Scholar]
- 155.Liu B et al (2020) LncRNA SOX2OT promotes Temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis 11(5):384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jiang X et al (2023) LncRNA GSCAR promotes glioma stem cell maintenance via stabilizing SOX2 expression. Int J Biol Sci 19(6):1681–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen Y et al (2023) LncRNA-PVT1 was identified as a key regulator for TMZ resistance and STAT-related pathway in glioma. BMC Cancer 23(1):455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gao Z et al (2022) PDIA3P1 promotes Temozolomide resistance in glioblastoma by inhibiting C/EBPβ degradation to facilitate proneural-to-mesenchymal transition. J Exp Clin Cancer Res 41(1):223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.You X et al (2022) MiRNA let-7 family regulated by NEAT1 and ARID3A/NF-κB inhibits PRRSV-2 replication in vitro and in vivo. PLoS Pathog 18(10):e1010820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lin L et al (2022) LncRNA HOXA-AS2 promotes Temozolomide resistance in glioblastoma by regulated miR-302a-3p/IGF1 Axis. Genet Res (Camb) 2022:p3941952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fu T et al (2021) Silencing LncRNA LINC01410 suppresses cell viability yet promotes apoptosis and sensitivity to Temozolomide in glioblastoma cells by inactivating PTEN/AKT pathway via targeting miR-370-3p. Immunopharmacol Immunotoxicol 43(6):680–692 [DOI] [PubMed] [Google Scholar]
- 162.Lu Y et al (2021) LINC00511 facilitates Temozolomide resistance of glioblastoma cells via sponging miR-126-5p and activating Wnt/β-catenin signaling. J Biochem Mol Toxicol 35(9):e22848 [DOI] [PubMed] [Google Scholar]
- 163.He X et al (2021) LncRNA MIR155HG promotes Temozolomide resistance by activating the Wnt/β-Catenin pathway via binding to PTBP1 in glioma. Cell Mol Neurobiol 41(6):1271–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lei M et al (2020) Translation and functional roles of circular RNAs in human cancer. Mol Cancer 19(1):30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Li G, Lan Q (2023) Exosome-Mediated transfer of circ-GLIS3 enhances Temozolomide resistance in glioma cells through the miR-548m/MED31 Axis. Cancer Biother Radiopharm 38(1):62–73 [DOI] [PubMed] [Google Scholar]
- 166.Ding C et al (2021) Warburg effect-promoted Exosomal circ_0072083 releasing up-regulates NANGO expression through multiple pathways and enhances Temozolomide resistance in glioma. J Exp Clin Cancer Res 40(1):164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Si J et al (2021) Heparanase confers Temozolomide resistance by regulation of exosome secretion and circular RNA composition in glioma. Cancer Sci 112(9):3491–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Han C et al (2021) Exosomal circ-HIPK3 facilitates tumor progression and Temozolomide resistance by regulating miR-421/ZIC5 Axis in glioma. Cancer Biother Radiopharm 36(7):537–548 [DOI] [PubMed] [Google Scholar]
- 169.Liu X et al (2023) Exosome-transmitted circCABIN1 promotes Temozolomide resistance in glioblastoma via sustaining erbb downstream signaling. J Nanobiotechnol 21(1):45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li J et al (2023) CircTTLL13 promotes TMZ resistance in glioma via modulating OLR1-Mediated activation of the Wnt/β-Catenin pathway. Mol Cell Biol 43(7):354–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li X et al (2022) Hsa_circ_0043949 reinforces Temozolomide resistance via upregulating oncogene ITGA1 axis in glioblastoma. Metab Brain Dis 37(8):2979–2993 [DOI] [PubMed] [Google Scholar]
- 172.Wei Y et al (2021) EIF4A3-induced circular RNA ASAP1 promotes tumorigenesis and Temozolomide resistance of glioblastoma via NRAS/MEK1/ERK1-2 signaling. Neuro Oncol 23(4):611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang ZY et al (2022) Exosomal noncoding RNAs in central nervous system diseases: biological functions and potential clinical applications. Front Mol Neurosci 15:1004221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Woicke J et al (2021) International harmonization of nomenclature and diagnostic criteria (INHAND): nonproliferative and proliferative lesions of the dog. Toxicol Pathol 49(1):5–109 [DOI] [PubMed] [Google Scholar]
- 175.Goleij P et al (2025) Role of Non-coding RNAs in the response of glioblastoma to Temozolomide. Mol Neurobiol 62(2):1726–1755 [DOI] [PubMed] [Google Scholar]
- 176.Zhang B et al (2024) The regulatory role and clinical application prospects of circrna in the occurrence and development of CNS tumors. CNS Neurosci Ther 30(4):e14500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yuan F et al (2022) Hsa_circ_0072309 enhances autophagy and TMZ sensitivity in glioblastoma. CNS Neurosci Ther 28(6):897–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Biserova K et al (2021) Cancer stem cells: significance in origin, pathogenesis and treatment of glioblastoma. Cells, 10(3):621 [DOI] [PMC free article] [PubMed]
- 179.Huang M et al (2020) Wnt-mediated endothelial transformation into mesenchymal stem cell-like cells induces chemoresistance in glioblastoma. Sci Transl Med, 12(532):eaay7522 [DOI] [PMC free article] [PubMed]
- 180.Couturier CP et al (2020) Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat Commun 11(1):3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu J et al (2024) Multi-scale signaling and tumor evolution in high-grade gliomas. Cancer Cell 42(7):1217–1238e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Xie XP et al (2022) Quiescent human glioblastoma cancer stem cells drive tumor initiation, expansion, and recurrence following chemotherapy. Dev Cell 57(1):32–46e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Peleli M et al (2023) Cystathionine gamma-lyase (CTH) Inhibition attenuates glioblastoma formation. Redox Biol 64:102773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chehelgerdi M et al (2023) Exploring the promising potential of induced pluripotent stem cells in cancer research and therapy. Mol Cancer 22(1):189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu K et al (2022) Hypoxia-induced GLT8D1 promotes glioma stem cell maintenance by inhibiting CD133 degradation through N-linked glycosylation. Cell Death Differ 29(9):1834–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhu Y et al (2024) PDCD10 is a key player in TMZ-Resistance and tumor cell regrowth: insights into its underlying mechanism in glioblastoma cells. Cells, 13(17):1442 [DOI] [PMC free article] [PubMed]
- 187.Kuzmin E et al (2024) Evolution of chromosome-arm aberrations in breast cancer through genetic network rewiring. Cell Rep 43(4):113988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yang Q et al (2022) Extracellular vesicles in Cancer drug resistance: roles, mechanisms, and implications. Adv Sci (Weinh) 9(34):e2201609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Giammello F et al (2024) Modulating voltage-gated sodium channels to enhance differentiation and sensitize glioblastoma cells to chemotherapy. Cell Commun Signal 22(1):434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Li S et al (2023) FBXO7 confers mesenchymal properties and chemoresistance in glioblastoma by controlling Rbfox2-Mediated alternative splicing. Adv Sci (Weinh) 10(33):e2303561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Auzmendi-Iriarte J et al (2022) Chaperone-Mediated autophagy controls proteomic and transcriptomic pathways to maintain glioma stem cell activity. Cancer Res 82(7):1283–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Corrigendum (2023) to: USP36 promotes tumorigenesis and drug sensitivity of glioblastoma by deubiquitinating and stabilizing ALKBH5. Neuro Oncol, 25(10): p. 1913 [DOI] [PMC free article] [PubMed]
- 193.Noh KH et al (2022) Novel cancer stem cell marker MVP enhances temozolomide-resistance in glioblastoma. Transl Oncol 15(1):101255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Li Y et al (2022) TRAF4 maintains deubiquitination of Caveolin-1 to drive glioblastoma stemness and Temozolomide resistance. Cancer Res 82(19):3573–3587 [DOI] [PubMed] [Google Scholar]
- 195.Wei Y et al (2022) The interaction between DNMT1 and High-Mannose CD133 maintains the Slow-Cycling state and tumorigenic potential of glioma stem cell. Adv Sci (Weinh) 9(26):e2202216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Somrit K et al (2024) KHDRBS3 facilitates self-renewal and Temozolomide resistance of glioblastoma cell lines. Life Sci 358:123132 [DOI] [PubMed] [Google Scholar]
- 197.Moon BS et al (2020) Epigenetic modulator Inhibition overcomes Temozolomide chemoresistance and antagonizes tumor recurrence of glioblastoma. J Clin Invest 130(11):5782–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.López-Valero I et al (2020) Midkine signaling maintains the self-renewal and tumorigenic capacity of glioma initiating cells. Theranostics 10(11):5120–5136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Xu J et al (2022) Disruption of DNA repair and survival pathways through heat shock protein Inhibition by onalespib to sensitize malignant gliomas to chemoradiation therapy. Clin Cancer Res 28(9):1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yang WB et al (2020) Increased activation of HDAC1/2/6 and Sp1 underlies therapeutic resistance and tumor growth in glioblastoma. Neuro Oncol 22(10):1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Eyme KM et al (2023) Targeting de Novo lipid synthesis induces lipotoxicity and impairs DNA damage repair in glioblastoma mouse models. Sci Transl Med 15(679):eabq6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Alejo S et al (2023) Lysine-specific histone demethylase 1A (KDM1A/LSD1) Inhibition attenuates DNA double-strand break repair and augments the efficacy of Temozolomide in glioblastoma. Neuro Oncol 25(7):1249–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Fedele M et al (2019) Proneural-Mesenchymal transition: phenotypic plasticity to acquire multitherapy resistance in glioblastoma. Int J Mol Sci, 20(11):2746 [DOI] [PMC free article] [PubMed]
- 204.Luo Z et al (2024) Inhibition of iRhom1 by CD44-targeting nanocarrier for improved cancer immunochemotherapy. Nat Commun 15(1):255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Park JH et al (2024) Gene regulatory network topology governs resistance and treatment escape in glioma stem-like cells. Sci Adv 10(23):eadj7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yin J et al (2021) Extracellular vesicles derived from hypoxic glioma stem-like cells confer Temozolomide resistance on glioblastoma by delivering miR-30b-3p. Theranostics 11(4):1763–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Buccarelli M et al (2021) Elesclomol-induced increase of mitochondrial reactive oxygen species impairs glioblastoma stem-like cell survival and tumor growth. J Exp Clin Cancer Res 40(1):228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Amaravadi RK, Kimmelman AC, Debnath J (2019) Targeting autophagy in cancer: recent advances and future directions. Cancer Discov 9(9):1167–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wan S et al (2023) Pyroptosis, ferroptosis, and autophagy cross-talk in glioblastoma opens up new avenues for glioblastoma treatment. Cell Commun Signal 21(1):115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Xue YY et al (2022) CN-3 increases TMZ sensitivity and induces ROS-dependent apoptosis and autophagy in TMZ-resistance glioblastoma. J Biochem Mol Toxicol 36(3):e22973 [DOI] [PubMed] [Google Scholar]
- 211.Zou L et al (2022) Autophagy and beyond: unraveling the complexity of UNC-51-like kinase 1 (ULK1) from biological functions to therapeutic implications. Acta Pharm Sin B 12(10):3743–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Xu Z et al (2020) Targeting PI3K/AKT/mTOR-mediated autophagy for tumor therapy. Appl Microbiol Biotechnol 104(2):575–587 [DOI] [PubMed] [Google Scholar]
- 213.Hwang YK et al (2024) Importance of autophagy regulation in glioblastoma with Temozolomide resistance. Cells, 13(16):1332 [DOI] [PMC free article] [PubMed]
- 214.Yang LQ, Huang AF, Xu WD (2023) Biology of endophilin and it’s role in disease. Front Immunol 14:1297506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chien CH et al (2022) SH3GLB1-related autophagy mediates mitochondrial metabolism to acquire resistance against Temozolomide in glioblastoma. J Exp Clin Cancer Res 41(1):220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Yun EJ et al (2020) Wnt/β-catenin signaling pathway induces autophagy-mediated temozolomide-resistance in human glioblastoma. Cell Death Dis 11(9):771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhang Y et al (2025) ADAR1 promotes the progression and Temozolomide resistance of glioma through p62-Mediated selective autophagy. CNS Neurosci Ther 31(1):e70168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Luo M et al (2024) Revisiting the potential of regulated cell death in glioma treatment: a focus on autophagy-dependent cell death, Anoikis, ferroptosis, Cuproptosis, pyroptosis, Immunogenic cell death, and the crosstalk between them. Front Oncol 14:1397863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Li X, He S, Ma B (2020) Autophagy and autophagy-related proteins in cancer. Mol Cancer 19(1):12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Chiariello M et al (2023) Overcoming challenges in glioblastoma treatment: targeting infiltrating cancer cells and Harnessing the tumor microenvironment. Front Cell Neurosci 17:1327621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Eisenbarth D, Wang YA (2023) Glioblastoma heterogeneity at single cell resolution. Oncogene 42(27):2155–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhang L et al (2023) The diversity and dynamics of tumor-associated macrophages in recurrent glioblastoma. Front Immunol 14:1238233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhang H et al (2025) Ginsenoside RK3 inhibits glioblastoma by modulating macrophage M2 polarization via the PPARG/CCL2 axis. Phytomedicine 136:156271 [DOI] [PubMed] [Google Scholar]
- 224.Yang W et al (2024) T-cell infiltration and its regulatory mechanisms in cancers: insights at single-cell resolution. J Exp Clin Cancer Res 43(1):38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Bausart M, Préat V, Malfanti A (2022) Immunotherapy for glioblastoma: the promise of combination strategies. J Exp Clin Cancer Res 41(1):35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Liu M et al (2022) Targeting macrophages: a novel treatment strategy in solid tumors. J Transl Med 20(1):586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhu S et al (2021) Roles of tumor-associated macrophages in tumor progression: implications on therapeutic strategies. Exp Hematol Oncol 10(1):60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Liu J et al (2021) BCL7A as a novel prognostic biomarker for glioma patients. J Transl Med 19(1):335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hasan MN et al (2021) Blocking NHE1 stimulates glioma tumor immunity by restoring OXPHOS function of myeloid cells. Theranostics 11(3):1295–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Xu Z et al (2022) Matrix Remodeling-Associated protein 8 as a novel Indicator contributing to glioma immune response by regulating ferroptosis. Front Immunol 13:834595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Li J et al (2024) Targeted reprogramming of tumor-associated macrophages for overcoming glioblastoma resistance to chemotherapy and immunotherapy. Biomaterials 311:122708 [DOI] [PubMed] [Google Scholar]
- 232.Liu Y et al (2023) In situ nitric oxide gas nanogenerator reprograms glioma immunosuppressive microenvironment. Adv Sci (Weinh) 10(18):e2300679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Liu F et al (2023) Piperlongumine conquers Temozolomide chemoradiotherapy resistance to achieve immune cure in refractory glioblastoma via boosting oxidative stress-inflamation-CD8(+)-T cell immunity. J Exp Clin Cancer Res 42(1):118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Li X et al (2024) Glioma-targeted Oxaliplatin/ferritin clathrate reversing the immunosuppressive microenvironment through hijacking Fe(2+) and boosting Fenton reaction. J Nanobiotechnol 22(1):93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Phon BWS et al (2025) Revisiting ABC transporters and their clinical significance in glioblastoma. Pharmaceuticals (Basel), 18(1):102 [DOI] [PMC free article] [PubMed]
- 236.Radtke L et al (2022) CRISPR/Cas9-induced knockout reveals the role of ABCB1 in the response to temozolomide, carmustine and lomustine in glioblastoma multiforme. Pharmacol Res 185:106510 [DOI] [PubMed] [Google Scholar]
- 237.Chen Z et al (2022) Prrx1 promotes resistance to Temozolomide by upregulating ABCC1 and inducing vasculogenic mimicry in glioma. Am J Cancer Res 12(8):3892–3912 [PMC free article] [PubMed] [Google Scholar]
- 238.Khunweeraphong N, Kuchler K (2021) Multidrug resistance in mammals and Fungi-From MDR to PDR: A Rocky road from atomic structures to transport mechanisms. Int J Mol Sci, 22(9):4806 [DOI] [PMC free article] [PubMed]
- 239.Sim HW et al (2021) A randomized phase II trial of veliparib, radiotherapy, and Temozolomide in patients with unmethylated MGMT glioblastoma: the VERTU study. Neuro Oncol 23(10):1736–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Huang K et al (2019) Genome-Wide CRISPR-Cas9 screening identifies NF-κB/E2F6 responsible for EGFRvIII-Associated Temozolomide resistance in glioblastoma. Adv Sci (Weinh) 6(17):1900782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Wang L et al (2020) Homotrimer cavin1 interacts with caveolin1 to facilitate tumor growth and activate microglia through extracellular vesicles in glioma. Theranostics 10(15):6674–6694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yang E et al (2022) PTRF/Cavin-1 enhances chemo-resistance and promotes Temozolomide efflux through extracellular vesicles in glioblastoma. Theranostics 12(9):4330–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Sampson JH et al (2020) Brain immunology and immunotherapy in brain tumours. Nat Rev Cancer 20(1):12–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Dumas AA et al (2020) Microglia promote glioblastoma via mTOR-mediated immunosuppression of the tumour microenvironment. Embo J 39(15):e103790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Sabbah DA, Hajjo R, Sweidan K (2020) Review on epidermal growth factor receptor (EGFR) structure, signaling pathways, interactions, and recent updates of EGFR inhibitors. Curr Top Med Chem 20(10):815–834 [DOI] [PubMed] [Google Scholar]
- 246.Ezzati S et al (2024) Epidermal growth factor receptor inhibitors in glioblastoma: current status and future possibilities. Int J Mol Sci, 25(4):2316 [DOI] [PMC free article] [PubMed]
- 247.Yu X et al (2023) Integrating single-cell RNA-seq and Spatial transcriptomics reveals MDK-NCL dependent immunosuppressive environment in endometrial carcinoma. Front Immunol 14:1145300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Șovrea AS et al (2022) Multiple faces of the glioblastoma microenvironment. Int J Mol Sci, 23(2):595 [DOI] [PMC free article] [PubMed]
- 249.Zhang Y, Wang X (2020) Targeting the Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol 13(1):165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhang X, Yu X (2023) Crosstalk between Wnt/β-catenin signaling pathway and DNA damage response in cancer: a new direction for overcoming therapy resistance. Front Pharmacol 14:1230822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Tomar VS, Patil V, Somasundaram K (2020) Temozolomide induces activation of Wnt/β-catenin signaling in glioma cells via PI3K/Akt pathway: implications in glioma therapy. Cell Biol Toxicol 36(3):273–278 [DOI] [PubMed] [Google Scholar]
- 252.Xie Y et al (2023) Wnt signaling regulates MFSD2A-dependent drug delivery through endothelial transcytosis in glioma. Neuro Oncol 25(6):1073–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Kim KH et al (2024) Integrated proteogenomic characterization of glioblastoma evolution. Cancer Cell 42(3):358–377e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Xue C et al (2023) Evolving cognition of the JAK-STAT signaling pathway: autoimmune disorders and cancer. Signal Transduct Target Ther 8(1):204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hu Q et al (2023) JAK/STAT pathway: extracellular signals, diseases, immunity, and therapeutic regimens. Front Bioeng Biotechnol 11:1110765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Zhang XN et al (2021) Pericytes augment glioblastoma cell resistance to Temozolomide through CCL5-CCR5 paracrine signaling. Cell Res 31(10):1072–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Yi L et al (2024) BRD4 degradation enhanced glioma sensitivity to Temozolomide by regulating Notch1 via Glu-Modified GSH-Responsive nanoparticles. Adv Sci (Weinh) 11(48):e2409753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Alafate W et al (2020) Loss of PLK2 induces acquired resistance to Temozolomide in GBM via activation of Notch signaling. J Exp Clin Cancer Res 39(1):239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Noorani I, de la Rosa J (2023) Breaking barriers for glioblastoma with a path to enhanced drug delivery. Nat Commun 14(1):5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Narsinh KH et al (2024) Strategies to improve drug delivery across the Blood-Brain barrier for glioblastoma. Curr Neurol Neurosci Rep 24(5):123–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Yasaswi PS, Shetty K, Yadav KS (2021) Temozolomide nano enabled medicine: promises made by the nanocarriers in glioblastoma therapy. J Control Release 336:549–571 [DOI] [PubMed] [Google Scholar]
- 262.Amarandi RM et al (2022) Liposomal-Based formulations: A path from basic research to Temozolomide delivery inside glioblastoma tissue. Pharmaceutics, 14(2):308 [DOI] [PMC free article] [PubMed]
- 263.Wang S et al (2022) Temozolomide hexadecyl ester targeted Plga nanoparticles for drug-resistant glioblastoma therapy via intranasal administration. Front Pharmacol 13:965789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ziegler JN, Tian C (2023) Engineered extracellular vesicles: emerging therapeutic strategies for translational applications. Int J Mol Sci, 24(20):15206 [DOI] [PMC free article] [PubMed]
- 265.Tay MRJ, Seah JD, Chua KSG (2022) Long-Term outcomes of patients with primary brain tumors after acute rehabilitation: A retrospective analyses of factors. Life (Basel), 12(8):1208 [DOI] [PMC free article] [PubMed]
- 266.Soufi G et al (2022) Perylene diimide-POSS network for semi selective solid-phase Microextraction of lung cancer biomarkers in exhaled breath. Anal Chim Acta 1198:339550 [DOI] [PubMed] [Google Scholar]
- 267.Zhong X et al (2023) Polyhedral oligomeric Silsesquioxane-Based nanoparticles for efficient chemotherapy of glioblastoma. Small 19(18):e2207248 [DOI] [PubMed] [Google Scholar]
- 268.Fu W et al (2019) Enhanced efficacy of Temozolomide loaded by a tetrahedral framework DNA nanoparticle in the therapy for glioblastoma. ACS Appl Mater Interfaces 11(43):39525–39533 [DOI] [PubMed] [Google Scholar]
- 269.Renziehausen A et al (2019) Encapsulation of Temozolomide in a Calixarene nanocapsule improves its stability and enhances its therapeutic efficacy against glioblastoma. Mol Cancer Ther 18(9):1497–1505 [DOI] [PubMed] [Google Scholar]
- 270.Pourmasoumi P et al (2024) Co-delivery of Temozolomide and Quercetin with folic acid-conjugated exosomes in glioblastoma treatment. Nanomed (Lond) 19(27):2271–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Jiang T et al (2021) Cation-Free SiRNA micelles as effective drug delivery platform and potent RNAi nanomedicines for glioblastoma therapy. Adv Mater 33(45):e2104779 [DOI] [PubMed] [Google Scholar]
- 272.Du K et al (2021) Visible light and glutathione dually responsive delivery of a Polymer-Conjugated Temozolomide intermediate for glioblastoma chemotherapy. ACS Appl Mater Interfaces 13(47):55851–55861 [DOI] [PubMed] [Google Scholar]
- 273.Wan Z et al (2021) Accurately controlled delivery of Temozolomide by biocompatible UiO-66-NH(2) through ultrasound to enhance the antitumor efficacy and attenuate the toxicity for treatment of malignant glioma. Int J Nanomed 16:6905–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Hasan U, Rajakumara E, Giri J (2023) Reversal of multidrug resistance by the synergistic effect of reversan and hyperthermia to potentiate the chemotherapeutic response of doxorubicin in glioblastoma and glioblastoma stem cells. ACS Appl Bio Mater 6(12):5399–5413 [DOI] [PubMed] [Google Scholar]
- 275.Janjua TI et al (2023) Efficient delivery of Temozolomide using ultrasmall large-pore silica nanoparticles for glioblastoma. J Control Release 357:161–174 [DOI] [PubMed] [Google Scholar]
- 276.Yi K et al (2021) PTRF/cavin-1 remodels phospholipid metabolism to promote tumor proliferation and suppress immune responses in glioblastoma by stabilizing cPLA2. Neuro Oncol 23(3):387–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zhao J et al (2024) CRISPR-Cas9 library screening combined with an exosome-targeted delivery system addresses tumorigenesis/tmz resistance in the mesenchymal subtype of glioblastoma. Theranostics 14(7):2835–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Umlauf BJ et al (2023) A novel strategy to increase the therapeutic potency of GBM chemotherapy via altering parenchymal/cerebral spinal fluid clearance rate. J Control Release 364:195–205 [DOI] [PubMed] [Google Scholar]
- 279.Ortiz R et al (2021) Temozolomide: an updated overview of resistance mechanisms, nanotechnology advances and clinical applications. Curr Neuropharmacol 19(4):513–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Lang F et al (2021) Genotoxic therapy and resistance mechanism in gliomas. Pharmacol Ther 228:107922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Zhao J et al (2023) A novel compound EPIC-0412 reverses Temozolomide resistance via inhibiting DNA repair/mgmt in glioblastoma. Neuro Oncol 25(5):857–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Xin L et al (2023) EPIC-0307-mediated selective disruption of PRADX-EZH2 interaction and enhancement of Temozolomide sensitivity to glioblastoma via inhibiting DNA repair and MGMT. Neuro Oncol 25(11):1976–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Lan Y et al (2024) Framework nucleic acid-based nanoparticles enhance Temozolomide sensitivity in glioblastoma. Drug Resist Updat 76:101122 [DOI] [PubMed] [Google Scholar]
- 284.Ismail M et al (2022) Targeted liposomes for combined delivery of Artesunate and Temozolomide to resistant glioblastoma. Biomaterials 287:121608 [DOI] [PubMed] [Google Scholar]
- 285.Wang W et al (2023) Quercetin induces MGMT(+) glioblastoma cells apoptosis via dual Inhibition of Wnt3a/β-Catenin and Akt/NF-κB signaling pathways. Phytomedicine 118:154933 [DOI] [PubMed] [Google Scholar]
- 286.Tang Q et al (2022) Tubeimoside-I sensitizes temozolomide-resistant glioblastoma cells to chemotherapy by reducing MGMT expression and suppressing EGFR induced PI3K/Akt/mTOR/NF-κB-mediated signaling pathway. Phytomedicine 99:154016 [DOI] [PubMed] [Google Scholar]
- 287.Tancredi A et al (2022) BET protein Inhibition sensitizes glioblastoma cells to Temozolomide treatment by attenuating MGMT expression. Cell Death Dis 13(12):1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Liu D et al (2022) Nano-Codelivery of Temozolomide and siPD-L1 to reprogram the Drug-Resistant and immunosuppressive microenvironment in orthotopic glioblastoma. ACS Nano 16(5):7409–7427 [DOI] [PubMed] [Google Scholar]
- 289.Li Z et al (2024) Design and Synthesis of 7-Oxabicyclo[2.2.1]heptane-2,3-dicarboxylic Acid Derivatives as PP5 Inhibitors To Reverse Temozolomide Resistance in Glioblastoma Multiforme. J Med Chem, 67(17): pp. 15691–15710 [DOI] [PubMed]
- 290.Hu YH et al (2021) Regulation of Temozolomide resistance in glioma cells via the RIP2/NF-κB/MGMT pathway. CNS Neurosci Ther 27(5):552–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.How JA et al (2021) Modification of homologous recombination deficiency score threshold and association with Long-Term survival in epithelial ovarian Cancer. Cancers (Basel), 13(5):946 [DOI] [PMC free article] [PubMed]
- 292.Sim HW, Galanis E, Khasraw M (2022) PARP inhibitors in glioma: A review of therapeutic opportunities. Cancers (Basel), 14(4):1003 [DOI] [PMC free article] [PubMed]
- 293.Yuan AL et al (2020) PARP Inhibition suppresses the emergence of Temozolomide resistance in a model system. J Neurooncol 148(3):463–472 [DOI] [PubMed] [Google Scholar]
- 294.Kleinberg L et al (2023) A multi-site phase I trial of veliparib with standard radiation and Temozolomide in patients with newly diagnosed glioblastoma multiforme (GBM). J Neurooncol 165(3):499–507 [DOI] [PubMed] [Google Scholar]
- 295.Higuchi F et al (2020) Restoration of Temozolomide sensitivity by PARP inhibitors in mismatch repair deficient glioblastoma is independent of base excision repair. Clin Cancer Res 26(7):1690–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.McCord M et al (2024) The novel DNA cross-linking agent KL-50 is active against patient-derived models of new and recurrent post-temozolomide mismatch repair-deficient glioblastoma. Neuro Oncol 27(3):644–651 [DOI] [PMC free article] [PubMed]
- 297.Staniszewska AD et al (2024) Preclinical characterization of AZD9574, a Blood-Brain barrier penetrant inhibitor of PARP1. Clin Cancer Res 30(7):1338–1351 [DOI] [PubMed] [Google Scholar]
- 298.Zampieri LX et al (2021) Olaparib is a mitochondrial complex I inhibitor that kills Temozolomide-Resistant human glioblastoma cells. Int J Mol Sci, 22(21):11938 [DOI] [PMC free article] [PubMed]
- 299.Schnöller LE et al (2022) Integrative analysis of therapy resistance and transcriptomic profiling data in glioblastoma cells identifies sensitization vulnerabilities for combined modality radiochemotherapy. Radiat Oncol 17(1):79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Han B et al (2020) ATRX/EZH2 complex epigenetically regulates FADD/PARP1 axis, contributing to TMZ resistance in glioma. Theranostics 10(7):3351–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Barszczewska-Pietraszek G et al (2024) Polθ inhibitor (ART558) demonstrates a synthetic lethal effect with PARP and RAD52 inhibitors in glioblastoma cells. Int J Mol Sci, 25(17):9134 [DOI] [PMC free article] [PubMed]
- 302.Sahakian L et al (2021) Inhibition of APE1/Ref-1 redox signaling alleviates intestinal dysfunction and damage to myenteric neurons in a mouse model of spontaneous chronic colitis. Inflamm Bowel Dis 27(3):388–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Kumar S et al (2023) Elevated APE1 dysregulates homologous recombination and cell cycle driving genomic evolution, tumorigenesis, and chemoresistance in esophageal adenocarcinoma. Gastroenterology 165(2):357–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Zoi V et al (2023) Therapeutic potential of linearol in combination with radiotherapy for the treatment of glioblastoma in vitro. Int J Mol Sci, 24(4):3760 [DOI] [PMC free article] [PubMed]
- 305.Ahluwalia MS et al (2024) Evaluating the base excision repair inhibitor TRC102 and Temozolomide for patients with recurrent glioblastoma in the phase 2 adult brain tumor consortium trial BERT. Clin Cancer Res 30(15):3167–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Hong B et al (2024) EPIC-1042 as a potent PTRF/Cavin1-caveolin-1 interaction inhibitor to induce PARP1 autophagic degradation and suppress Temozolomide efflux for glioblastoma. Neuro Oncol 26(1):100–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.M, A., et al., Epigenetic basis for PARP mutagenesis in glioblastoma: A review. Eur J Pharmacol, (2023) 938: p. 175424 [DOI] [PubMed]
- 308.Saville KM et al (2024) Oncometabolite 2-hydroxyglutarate suppresses basal protein levels of DNA polymerase beta that enhances alkylating agent and PARG Inhibition induced cytotoxicity. DNA Repair (Amst) 140:103700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Fujii S, Fuchs RP (2024) Accidental encounter of repair intermediates in alkylated DNA May lead to Double-Strand breaks in resting cells. Int J Mol Sci, 25(15):8192 [DOI] [PMC free article] [PubMed]
- 310.Shin MH et al (2023) Recent advances in CAR-Based solid tumor immunotherapy. Cells, 12(12):1606 [DOI] [PMC free article] [PubMed]
- 311.Posey AD Jr., Young RM, June CH (2024) Future perspectives on engineered T cells for cancer. Trends Cancer 10(8):687–695 [DOI] [PubMed] [Google Scholar]
- 312.Lechpammer M et al (2022) Advances in immunotherapy for the treatment of adult glioblastoma: overcoming chemical and physical barriers. Cancers (Basel), 14(7):1627 [DOI] [PMC free article] [PubMed]
- 313.Jackson CM, Choi J, Lim M (2019) Mechanisms of immunotherapy resistance: lessons from glioblastoma. Nat Immunol 20(9):1100–1109 [DOI] [PubMed] [Google Scholar]
- 314.Borgeaud M et al (2023) Novel targets for immune-checkpoint Inhibition in cancer. Cancer Treat Rev 120:102614 [DOI] [PubMed] [Google Scholar]
- 315.Maggs L et al (2021) CAR T Cell-Based immunotherapy for the treatment of glioblastoma. Front Neurosci 15:662064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Keskin DB et al (2019) Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565(7738):234–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Ghouzlani A et al (2021) Immune checkpoint inhibitors in human glioma microenvironment. Front Immunol 12:679425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Cloughesy TF et al (2019) Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med 25(3):477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Kuang RZ et al (2024) Effects of apatinib combined with Temozolomide on levels of sPD-1 and sPD-L1 in patients with drug-resistant recurrent glioblastoma. Clin (Sao Paulo) 79:100376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Xu S et al (2020) Immunotherapy for glioma: current management and future application. Cancer Lett 476:1–12 [DOI] [PubMed] [Google Scholar]
- 321.VanderWalde A et al (2023) Ipilimumab with or without nivolumab in PD-1 or PD-L1 Blockade refractory metastatic melanoma: a randomized phase 2 trial. Nat Med 29(9):2278–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Omuro A (2022) Immune-checkpoint inhibitors for glioblastoma: what have we learned? Arq Neuropsiquiatr 80(5 Suppl 1):266–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Reardon DA et al (2020) Effect of nivolumab vs bevacizumab in patients with recurrent glioblastoma: the checkmate 143 phase 3 randomized clinical trial. JAMA Oncol 6(7):1003–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Rodriguez SMB et al (2024) Glioblastoma and immune checkpoint inhibitors: A glance at available treatment options and future directions. Int J Mol Sci, 25(19):10765 [DOI] [PMC free article] [PubMed]
- 325.Sloan AE et al (2024) NRG-BN002: phase I study of Ipilimumab, nivolumab, and the combination in patients with newly diagnosed glioblastoma. Neuro Oncol 26(9):1628–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Song X et al (2025) Hybrid membrane biomimetic photothermal nanorobots for enhanced Chemodynamic-Chemotherapy-Immunotherapy. ACS Appl Mater Interfaces 17(4):5784–5798 [DOI] [PubMed] [Google Scholar]
- 327.Sloan L et al (2024) Radiation immunodynamics in patients with glioblastoma receiving chemoradiation. Front Immunol 15:1438044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Meng L et al (2021) Targeted regulation of Blood-Brain barrier for enhanced therapeutic efficiency of Hypoxia-Modifier nanoparticles and immune checkpoint Blockade antibodies for glioblastoma. ACS Appl Mater Interfaces 13(10):11657–11671 [DOI] [PubMed] [Google Scholar]
- 329.Gao Q et al (2024) Versatile self-assembled near-infrared SERS nanoprobes for multidrug-resistant bacterial infection-specific surveillance and therapy. Acta Biomater 189:559–573 [DOI] [PubMed] [Google Scholar]
- 330.Chen M et al (2022) DNA damage response evaluation provides novel insights for personalized immunotherapy in glioma. Front Immunol 13:875648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Cai Y et al (2022) Lighting a fire: Gasdermin-Mediated pyroptosis remodels the glioma microenvironment and promotes immune checkpoint Blockade response. Front Immunol 13:910490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Yasinjan F et al (2023) Immunotherapy: a promising approach for glioma treatment. Front Immunol 14:1255611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Karimi-Sani I et al (2024) Personalized mRNA vaccines in glioblastoma therapy: from rational design to clinical trials. J Nanobiotechnol 22(1):601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Burkholz SR et al (2023) Survivin (BIRC5) peptide vaccine in the 4T1 murine mammary tumor model: A potential neoadjuvant T cell immunotherapy for triple negative breast cancer: A preliminary study. Vaccines (Basel), 11(3) [DOI] [PMC free article] [PubMed]
- 335.Miao H et al (2022) A nanobody-based molecular toolkit for ubiquitin-proteasome system explores the main role of survivin subcellular localization. Front Bioeng Biotechnol 10:952237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Wang X et al (2021) Immunotherapy for recurrent glioblastoma: practical insights and challenging prospects. Cell Death Dis 12(4):299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Ahluwalia MS et al (2023) Phase IIa study of survaxm plus adjuvant Temozolomide for newly diagnosed glioblastoma. J Clin Oncol 41(7):1453–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Liau LM et al (2023) Association of autologous tumor Lysate-Loaded dendritic cell vaccination with extension of survival among patients with newly diagnosed and recurrent glioblastoma: A phase 3 prospective externally controlled cohort trial. JAMA Oncol 9(1):112–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Ridolfi L et al (2024) First step results from a phase II study of a dendritic cell vaccine in glioblastoma patients (CombiG-vax). Front Immunol 15:1404861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Faghfuri E et al (2021) Recent developments of RNA-based vaccines in cancer immunotherapy. Expert Opin Biol Ther 21(2):201–218 [DOI] [PubMed] [Google Scholar]
- 341.Eralp Y (2022) Application of mRNA technology in Cancer therapeutics. Vaccines (Basel), 10(8) [DOI] [PMC free article] [PubMed]
- 342.Shan F et al (2022) Therapeutic targeting of regulatory T cells in cancer. Trends Cancer 8(11):944–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Alghamri MS et al (2021) Targeting neuroinflammation in brain cancer: Uncovering mechanisms, Pharmacological targets, and neuropharmaceutical developments. Front Pharmacol 12:680021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Scheetz L et al (2020) Synthetic High-density lipoprotein nanodiscs for personalized immunotherapy against gliomas. Clin Cancer Res 26(16):4369–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Muragaki Y et al (2023) A multicenter, randomized, placebo-controlled phase IIb trial of an autologous formalin-fixed tumor vaccine for newly diagnosed glioblastomas. J Neurosurg 139(2):344–354 [DOI] [PubMed] [Google Scholar]
- 346.Hu JL et al (2022) A phase I study of autologous dendritic cell vaccine pulsed with allogeneic Stem-like cell line lysate in patients with newly diagnosed or recurrent glioblastoma. Clin Cancer Res 28(4):689–696 [DOI] [PubMed] [Google Scholar]
- 347.Tamura R et al (2020) Clinical and histopathological analyses of VEGF receptors peptide vaccine in patients with primary glioblastoma - a case series. BMC Cancer 20(1):196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Takei J et al (2023) Prognostic survival biomarkers of tumor-fused dendritic cell vaccine therapy in patients with newly diagnosed glioblastoma. Cancer Immunol Immunother 72(10):3175–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Bota DA et al (2022) Phase 2 study of AV-GBM-1 (a tumor-initiating cell targeted dendritic cell vaccine) in newly diagnosed glioblastoma patients: safety and efficacy assessment. J Exp Clin Cancer Res 41(1):344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Dutoit V et al (2020) Impact of radiochemotherapy on immune cell subtypes in High-Grade glioma patients. Front Oncol 10:89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Hamad A et al (2023) Recent developments in glioblastoma therapy: oncolytic viruses and emerging future strategies. Viruses, 15(2):547 [DOI] [PMC free article] [PubMed]
- 352.Huang B et al (2020) Current immunotherapies for glioblastoma multiforme. Front Immunol 11:603911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Todo T et al (2022) Intratumoral oncolytic herpes virus G47∆ for residual or recurrent glioblastoma: a phase 2 trial. Nat Med 28(8):1630–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Sostoa J, Dutoit V, Migliorini D (2020) Oncolytic viruses as a platform for the treatment of malignant brain tumors. Int J Mol Sci, 21(20):7449 [DOI] [PMC free article] [PubMed]
- 355.Baugh R et al (2024) Targeting NKG2D ligands in glioblastoma with a bispecific T-cell engager is augmented with conventional therapy and enhances oncolytic virotherapy of glioma stem-like cells. J Immunother Cancer, 12(5):e008460 [DOI] [PMC free article] [PubMed]
- 356.Fares J et al (2021) Neural stem cell delivery of an oncolytic adenovirus in newly diagnosed malignant glioma: a first-in-human, phase 1, dose-escalation trial. Lancet Oncol 22(8):1103–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Kadhim ZA et al (2022) Oncolytic Newcastle disease virus Co-Delivered with modified PLGA nanoparticles encapsulating Temozolomide against glioblastoma cells: developing an effective treatment strategy. Molecules, 27(18):5757 [DOI] [PMC free article] [PubMed]
- 358.Saha D, Rabkin SD, Martuza RL (2020) Temozolomide antagonizes oncolytic immunovirotherapy in glioblastoma. J Immunother Cancer, 8(1):e000345 [DOI] [PMC free article] [PubMed]
- 359.Chen Y et al (2023) RPL22L1, a novel candidate oncogene promotes Temozolomide resistance by activating STAT3 in glioblastoma. Cell Death Dis 14(11):757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Tan MSY et al (2019) A STAT3-based gene signature stratifies glioma patients for targeted therapy. Nat Commun 10(1):3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Hu W et al (2025) CYP3A5 promotes glioblastoma stemness and chemoresistance through fine-tuning NAD(+)/NADH ratio. J Exp Clin Cancer Res 44(1):3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Rehman FU et al (2022) Heme Oxygenase-1 targeting exosomes for Temozolomide resistant glioblastoma synergistic therapy. J Control Release 345:696–708 [DOI] [PubMed] [Google Scholar]
- 363.Li S et al (2024) Brain targeted biomimetic SiRNA nanoparticles for drug resistance glioblastoma treatment. J Control Release 376:67–78 [DOI] [PubMed] [Google Scholar]
- 364.Yuan J et al (2020) The MAPK and AMPK signalings: interplay and implication in targeted cancer therapy. J Hematol Oncol 13(1):113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Chen C et al (2021) Osimertinib successfully combats EGFR-negative glioblastoma cells by inhibiting the MAPK pathway. Acta Pharmacol Sin 42(1):108–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Lane R et al (2022) PDGF-R Inhibition induces glioblastoma cell differentiation via DUSP1/p38(MAPK) signalling. Oncogene 41(19):2749–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Alafate W et al (2024) Targeting ARNT attenuates chemoresistance through destabilizing p38α-MAPK signaling in glioblastoma. Cell Death Dis 15(5):366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Yang HC et al (2019) Resveratrol restores sensitivity of glioma cells to Temozolamide through inhibiting the activation of Wnt signaling pathway. J Cell Physiol 234(5):6783–6800 [DOI] [PubMed] [Google Scholar]
- 369.Fei YQ et al (2022) Mannose inhibits proliferation and promotes apoptosis to enhance sensitivity of glioma cells to Temozolomide through Wnt/β-catenin signaling pathway. Neurochem Int 157:105348 [DOI] [PubMed] [Google Scholar]
- 370.Pridham KJ et al (2024) Selective regulation of chemosensitivity in glioblastoma by phosphatidylinositol 3-kinase beta. iScience, 27(6): p. 109921 [DOI] [PMC free article] [PubMed]
- 371.Cheng HS et al (2024) Dual p38MAPK and MEK Inhibition disrupts adaptive chemoresistance in mesenchymal glioblastoma to Temozolomide. Neuro Oncol 26(7):1247–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Lopez BGC et al (2022) Multimodal platform for assessing drug distribution and response in clinical trials. Neuro Oncol 24(1):64–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Wang CY et al (2020) Cordycepin inhibits human gestational choriocarcinoma cell growth by disrupting centrosome homeostasis. Drug Des Devel Ther 14:2987–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Lee SW et al (2015) The synergistic effect of combination Temozolomide and chloroquine treatment is dependent on autophagy formation and p53 status in glioma cells. Cancer Lett 360(2):195–204 [DOI] [PubMed] [Google Scholar]
- 375.Huang T et al (2019) MIR93 (microRNA– 93) regulates tumorigenicity and therapy response of glioblastoma by targeting autophagy. Autophagy 15(6):1100–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Zhao K et al (2021) Inhibition of carbonic anhydrase 2 overcomes Temozolomide resistance in glioblastoma cells. Int J Mol Sci, 23(1):157 [DOI] [PMC free article] [PubMed]
- 377.Ou M et al (2023) Inhibition of autophagy and induction of glioblastoma cell death by NEO214, a Perillyl alcohol-rolipram conjugate. Autophagy 19(12):3169–3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Tabnak P et al (2023) Forkhead box transcription factors (FOXOs and FOXM1) in glioma: from molecular mechanisms to therapeutics. Cancer Cell Int 23(1):238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Yin HT et al (2024) Daurisoline suppress glioma progression by inhibiting autophagy through PI3K/AKT/mTOR pathway and increases TMZ sensitivity. Biochem Pharmacol 223:116113 [DOI] [PubMed] [Google Scholar]
- 380.Li D et al (2024) AGCM-22, a novel cetuximab-based EGFR-targeting antibody-drug-conjugate with highly selective anti-glioblastoma efficacy. Bioorg Med Chem 102:117657 [DOI] [PubMed] [Google Scholar]
- 381.Zhang Y et al (2024) AAA237, an SKP2 inhibitor, suppresses glioblastoma by inducing BNIP3-dependent autophagy through the mTOR pathway. Cancer Cell Int 24(1):69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Lin L et al (2024) Borneol promotes autophagic degradation of HIF-1α and enhances chemotherapy sensitivity in malignant glioma. PeerJ 12:e16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Li Q et al (2021) Role of Borneol induced autophagy in enhancing radiosensitivity of malignant glioma. Front Oncol 11:749987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Huang T et al (2021) Cannabidiol inhibits human glioma by induction of lethal mitophagy through activating TRPV4. Autophagy 17(11):3592–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Li K et al (2022) TRIM7 modulates NCOA4-mediated ferritinophagy and ferroptosis in glioblastoma cells. Redox Biol 56:102451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Shi L et al (2023) The DRD2 antagonist haloperidol mediates Autophagy-Induced ferroptosis to increase Temozolomide sensitivity by promoting Endoplasmic reticulum stress in glioblastoma. Clin Cancer Res 29(16):3172–3188 [DOI] [PubMed] [Google Scholar]
- 387.Li Z et al (2022) Autophagy-based unconventional secretion of HMGB1 in glioblastoma promotes chemosensitivity to Temozolomide through macrophage M1-like polarization. J Exp Clin Cancer Res 41(1):74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Corrêa-Ferreira ML et al (2022) The mesoionic compound MI-D changes energy metabolism and induces apoptosis in T98G glioma cells. Mol Cell Biochem 477(8):2033–2045 [DOI] [PubMed] [Google Scholar]
- 389.Zhu JY et al (2022) SARS-CoV-2 Nsp6 damages Drosophila heart and mouse cardiomyocytes through MGA/MAX complex-mediated increased Glycolysis. Commun Biol 5(1):1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Pucci G et al (2024) Glut-3 gene knockdown as a potential strategy to overcome glioblastoma radioresistance. Int J Mol Sci, 25(4):2079 [DOI] [PMC free article] [PubMed]
- 391.Heuser C et al (2023) Targeting lactate metabolism for cancer immunotherapy - a matter of precision. Semin Cancer Biol 88:32–45 [DOI] [PubMed] [Google Scholar]
- 392.Sharma D, Singh M, Rani R (2022) Role of LDH in tumor glycolysis: regulation of LDHA by small molecules for cancer therapeutics. Semin Cancer Biol 87:184–195 [DOI] [PubMed] [Google Scholar]
- 393.Luo K et al (2022) CircKIF4A promotes glioma growth and Temozolomide resistance by accelerating Glycolysis. Cell Death Dis 13(8):740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Fhu CW, Ali A (2020) Fatty acid synthase: an emerging target in Cancer. Molecules, 25(17):3935 [DOI] [PMC free article] [PubMed]
- 395.Hao Z et al (2024) Identification of MGMT promoter methylation as a specific lipid metabolism biomarker, reveals the feasibility of Atorvastatin application in glioblastoma. Metabolism 153:155794 [DOI] [PubMed] [Google Scholar]
- 396.Liu X et al (2024) IMPDH Inhibition decreases TERT expression and synergizes the cytotoxic effect of chemotherapeutic agents in glioblastoma cells. Int J Mol Sci, 25(11):5992 [DOI] [PMC free article] [PubMed]
- 397.Chen YJ et al (2024) Gliocidin is a nicotinamide-mimetic prodrug that targets glioblastoma. Nature 636(8042):466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Shireman JM et al (2021) De Novo purine biosynthesis is a major driver of chemoresistance in glioblastoma. Brain 144(4):1230–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Platten M et al (2019) Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov 18(5):379–401 [DOI] [PubMed] [Google Scholar]
- 400.Du L et al (2020) Both IDO1 and TDO contribute to the malignancy of gliomas via the Kyn-AhR-AQP4 signaling pathway. Signal Transduct Target Ther 5(1):10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Winters M et al (2019) Diaryl hydroxylamines as pan or dual inhibitors of indoleamine 2,3-dioxygenase-1, indoleamine 2,3-dioxygenase-2 and Tryptophan dioxygenase. Eur J Med Chem 162:455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Bickerdike MJ et al (2024) AT-0174, a novel dual IDO1/TDO2 enzyme inhibitor, synergises with Temozolomide to improve survival in an orthotopic mouse model of glioblastoma. BMC Cancer 24(1):889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Lu Z et al (2024) CHIP-mediated ubiquitin degradation of BCAT1 regulates glioma cell proliferation and Temozolomide sensitivity. Cell Death Dis 15(7):538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Ballo MT et al (2019) Correlation of tumor treating fields dosimetry to survival outcomes in newly diagnosed glioblastoma: A Large-Scale numerical Simulation-Based analysis of data from the phase 3 EF-14 randomized trial. Int J Radiat Oncol Biol Phys 104(5):1106–1113 [DOI] [PubMed] [Google Scholar]
- 405.Liu S et al (2021) Progress and prospect in tumor treating fields treatment of glioblastoma. Biomed Pharmacother 141:111810 [DOI] [PubMed] [Google Scholar]
- 406.Moser JC et al (2022) The mechanisms of action of tumor treating fields. Cancer Res 82(20):3650–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Toms SA et al (2019) Increased compliance with tumor treating fields therapy is prognostic for improved survival in the treatment of glioblastoma: a subgroup analysis of the EF-14 phase III trial. J Neurooncol 141(2):467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Ram Z et al (2021) Efficacy and safety of tumor treating fields (TTFields) in elderly patients with newly diagnosed glioblastoma: subgroup analysis of the phase 3 EF-14 clinical trial. Front Oncol 11:671972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Chen C et al (2022) Tumor Treating Fields Combine with Temozolomide for Newly Diagnosed Glioblastoma: A Retrospective Analysis of Chinese Patients in a Single Center. J Clin Med, 11(19):5855 [DOI] [PMC free article] [PubMed]
- 410.Schlieper-Scherf S et al (2024) Disrupting glioblastoma networks with tumor treating fields (TTFields) in in vitro models. J Neurooncol 170(1):139–151 [DOI] [PMC free article] [PubMed]
- 411.Bokstein F et al (2020) Concurrent tumor treating fields (TTFields) and radiation therapy for newly diagnosed glioblastoma: A prospective safety and feasibility study. Front Oncol 10:411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Ali AS et al (2022) Concurrent chemoradiation and tumor treating fields (TTFields, 200 kHz) for patients with newly diagnosed glioblastoma: patterns of progression in a single institution pilot study. J Neurooncol 160(2):345–350 [DOI] [PubMed] [Google Scholar]
- 413.Song A et al (2020) Initial experience with scalp sparing radiation with concurrent Temozolomide and tumor treatment fields (SPARE) for patients with newly diagnosed glioblastoma. J Neurooncol 147(3):653–661 [DOI] [PubMed] [Google Scholar]
- 414.Cao Q et al (2024) Tumor treating fields (TTFields) combined with the drug repurposing approach CUSP9v3 induce metabolic reprogramming and synergistic anti-glioblastoma activity in vitro. Br J Cancer 130(8):1365–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Yang X, Hu C, Li L (2024) Technical note: computational study on thermal management schemes for tumor-treating fields therapy. Med Phys 51(10):7632–7644 [DOI] [PubMed]
- 416.Liu D et al (2023) Nanotechnology Meets glioblastoma multiforme: emerging therapeutic strategies. Wiley Interdiscip Rev Nanomed Nanobiotechnol 15(1):e1838 [DOI] [PubMed] [Google Scholar]
- 417.Khan I et al (2022) Nanomedicine for glioblastoma: progress and future prospects. Semin Cancer Biol 86(Pt 2):172–186 [DOI] [PubMed] [Google Scholar]
- 418.Fan Q et al (2021) Brain delivery of Plk1 inhibitor via chimaeric polypeptide polymersomes for safe and Superb treatment of orthotopic glioblastoma. J Control Release 329:1139–1149 [DOI] [PubMed] [Google Scholar]
- 419.Wei J et al (2022) Immunotherapy of malignant glioma by noninvasive administration of TLR9 agonist CpG Nano-Immunoadjuvant. Adv Sci (Weinh) 9(13):e2103689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Carvalho GC et al (2022) Cetyltrimethylammonium bromide in the synthesis of mesoporous silica nanoparticles: general aspects and in vitro toxicity. Adv Colloid Interface Sci 307:102746 [DOI] [PubMed] [Google Scholar]
- 421.Chen ZA et al (2024) Receptor Ligand-Free mesoporous silica nanoparticles: A streamlined strategy for targeted drug delivery across the Blood-Brain barrier. ACS Nano 18(20):12716–12736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Domb AJ et al (2021) Safety evaluation of nanotechnology products. Pharmaceutics, 13(10):1615 [DOI] [PMC free article] [PubMed]
- 423.Zeynalzadeh E et al (2024) Navigating the neurological frontier: macromolecular marvels in overcoming blood-brain barrier challenges for advanced drug delivery. Heliyon 10(15):e35562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Hsu TI et al (2024) Overcoming the Blood-Brain tumor barrier with Docetaxel-Loaded mesoporous silica nanoparticles for treatment of Temozolomide-Resistant glioblastoma. ACS Appl Mater Interfaces 16(17):21722–21735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Fei H et al (2024) Gint4.T-siHDGF chimera-capped mesoporous silica nanoparticles encapsulating Temozolomide for synergistic glioblastoma therapy. Biomaterials 306:122479 [DOI] [PubMed] [Google Scholar]
- 426.Wang L et al (2021) Intranasal delivery of Temozolomide-Conjugated gold nanoparticles functionalized with Anti-EphA3 for glioblastoma targeting. Mol Pharm 18(3):915–927 [DOI] [PubMed] [Google Scholar]
- 427.Yu Y et al (2022) Efficacy of Temozolomide-Conjugated gold nanoparticle photothermal therapy of Drug-Resistant glioblastoma and its mechanism study. Mol Pharm 19(4):1219–1229 [DOI] [PubMed] [Google Scholar]
- 428.Meng X et al (2020) Dual functionalized brain-targeting nanoinhibitors restrain temozolomide-resistant glioma via attenuating EGFR and MET signaling pathways. Nat Commun 11(1):594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Wang X et al (2023) Mechanical nanosurgery of chemoresistant glioblastoma using magnetically controlled carbon nanotubes. Sci Adv 9(13):eade5321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Liu Y et al (2021) Circumventing drug resistance pathways with a Nanoparticle-Based photodynamic method. Nano Lett 21(21):9115–9123 [DOI] [PubMed] [Google Scholar]
- 431.Zhang Y et al (2024) Dual-Targeted novel Temozolomide nanocapsules encapsulating siPKM2 inhibit aerobic Glycolysis to sensitize glioblastoma to chemotherapy. Adv Mater 36(29):e2400502 [DOI] [PubMed] [Google Scholar]
- 432.Wang K et al (2021) SiRNA nanoparticle suppresses drug-resistant gene and prolongs survival in an orthotopic glioblastoma xenograft mouse model. Adv Funct Mater, 31(6):2007166 [DOI] [PMC free article] [PubMed]
- 433.Yang Q et al (2021) Gene therapy for Drug-Resistant glioblastoma via Lipid-Polymer hybrid nanoparticles combined with focused ultrasound. Int J Nanomed 16:185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Morimoto T et al (2021) CRISPR-Cas9-Mediated TIM3 knockout in human natural killer cells enhances growth inhibitory effects on human glioma cells. Int J Mol Sci, 22(7):3489 [DOI] [PMC free article] [PubMed]
- 435.Nemeth K et al (2024) Non-coding RNAs in disease: from mechanisms to therapeutics. Nat Rev Genet 25(3):211–232 [DOI] [PubMed] [Google Scholar]
- 436.Luciani M et al (2024) Human iPSC-derived neural stem cells displaying radial glia signature exhibit long-term safety in mice. Nat Commun 15(1):9433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Vincent CA et al (2023) Epigenomic perturbation of novel EGFR enhancers reduces the proliferative and invasive capacity of glioblastoma and increases sensitivity to Temozolomide. BMC Cancer 23(1):945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Avenel ICN et al (2024) GDNF/GFRA1 signaling contributes to chemo- and radioresistance in glioblastoma. Sci Rep 14(1):17639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Tan IL et al (2023) Targeting the non-coding genome and Temozolomide signature enables CRISPR-mediated glioma Oncolysis. Cell Rep 42(11):113339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Zhao Y et al (2024) Polymer-locking fusogenic liposomes for glioblastoma-targeted SiRNA delivery and CRISPR-Cas gene editing. Nat Nanotechnol 19(12):1869–1879 [DOI] [PubMed] [Google Scholar]
- 441.Zou Y et al (2022) Blood-brain barrier-penetrating single CRISPR-Cas9 nanocapsules for effective and safe glioblastoma gene therapy. Sci Adv 8(16):eabm8011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Ruan W et al (2022) Brain-targeted CRISPR/Cas9 nanomedicine for effective glioblastoma therapy. J Control Release 351:739–751 [DOI] [PubMed] [Google Scholar]
- 443.Wong SC, Kamarudin MNA, Naidu R (2023) Anticancer mechanism of flavonoids on High-Grade Adult-Type diffuse gliomas. Nutrients, 15(4):797 [DOI] [PMC free article] [PubMed]
- 444.Zhai K et al (2021) Flavonoids synergistically enhance the Anti-Glioblastoma effects of chemotherapeutic drugs. Biomolecules, 11(12):1841 [DOI] [PMC free article] [PubMed]
- 445.Daisy Precilla S et al (2022) Integration of synthetic and natural derivatives revives the therapeutic potential of Temozolomide against glioma- an in vitro and in vivo perspective. Life Sci 301:120609 [DOI] [PubMed] [Google Scholar]
- 446.Dong Q et al (2022) Biochanin A sensitizes glioblastoma to Temozolomide by inhibiting autophagy. Mol Neurobiol 59(2):1262–1272 [DOI] [PubMed] [Google Scholar]
- 447.Wang L et al (2023) Therapeutic effects of Bombax ceiba flower aqueous extracts against loperamide-induced constipation in mice. Pharm Biol 61(1):125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Zhai K et al (2020) Curcumin’s Beneficial Effects on Neuroblastoma: Mechanisms, Challenges, and Potential Solutions. Biomolecules, 10(11):1469 [DOI] [PMC free article] [PubMed]
- 449.Kunnumakkara AB et al (2023) Role of turmeric and Curcumin in prevention and treatment of chronic diseases: lessons learned from clinical trials. ACS Pharmacol Transl Sci 6(4):447–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Zhai K et al (2021) Natural compounds in glioblastoma therapy: preclinical insights, mechanistic pathways, and outlook. Cancers (Basel), 13(10):2317 [DOI] [PMC free article] [PubMed]
- 451.Wong SC, Kamarudin MNA, Naidu R (2021) Anticancer mechanism of Curcumin on human glioblastoma. Nutrients, 13(3):950 [DOI] [PMC free article] [PubMed]
- 452.Beylerli O et al (2022) Therapeutic effect of natural polyphenols against glioblastoma. Front Cell Dev Biol 10:1036809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Mohamadian M et al (2022) Review on the therapeutic potential of Curcumin and its derivatives on glioma biology. Neurochem Res 47(10):2936–2953 [DOI] [PubMed] [Google Scholar]
- 454.Iegiani G, Di Cunto F, Pallavicini G (2021) Inhibiting microcephaly genes as alternative to microtubule targeting agents to treat brain tumors. Cell Death Dis 12(11):956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Yi L et al (2024) Chronic stress as an emerging risk factor for the development and progression of glioma. Chin Med J (Engl) 137(4):394–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Datta S, Luthra R, Bharadvaja N (2022) Medicinal plants for glioblastoma treatment. Anticancer Agents Med Chem 22(13):2367–2384 [DOI] [PubMed] [Google Scholar]
- 457.Mazumder K et al (2022) A review on mechanistic insight of plant derived anticancer bioactive phytocompounds and their structure activity relationship. Molecules, 27(9):3036 [DOI] [PMC free article] [PubMed]
- 458.Lipińska MM et al (2023) Active compounds with medicinal potential found in maxillariinae Benth. (Orchidaceae Juss.) Representatives-A review. Int J Mol Sci, 24(1):739 [DOI] [PMC free article] [PubMed]
- 459.Koklesova L et al (2020) Carotenoids in Cancer Metastasis-Status quo and outlook. Biomolecules, 10(12):1653 [DOI] [PMC free article] [PubMed]
- 460.Anywar G, Muhumuza E (2023) Bioactivity and toxicity of coumarins from African medicinal plants. Front Pharmacol 14:1231006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Jang WY, Kim MY, Cho JY (2022) Antioxidant, Anti-Inflammatory, Anti-Menopausal, and Anti-Cancer Effects of Lignans and Their Metabolites. Int J Mol Sci, 23(24):15482 [DOI] [PMC free article] [PubMed]
- 462.Sokolov MN et al (2024) The effects of the steroids 5-Androstenediol and dehydroepiandrosterone and their synthetic derivatives on the viability of K562, HeLa, and Wi-38 cells and the Luminol-Stimulated chemiluminescence of peripheral blood mononuclear cells from healthy volunteers. Biomolecules, 14(3):373 [DOI] [PMC free article] [PubMed]
- 463.Pucci S et al (2022) Evidence of a dual mechanism of action underlying the anti-proliferative and cytotoxic effects of ammonium-alkyloxy-stilbene-based α7- and α9-nicotinic ligands on glioblastoma cells. Pharmacol Res 175:105959 [DOI] [PubMed] [Google Scholar]
- 464.Chauhan SS et al (2020) Pectin-Tannic acid Nano-Complexes promote the delivery and bioactivity of drugs in pancreatic Cancer cells. Pharmaceutics, 12(3):285 [DOI] [PMC free article] [PubMed]
- 465.Zhu Y et al (2020) Celastrol suppresses glioma vasculogenic mimicry formation and angiogenesis by blocking the PI3K/Akt/mTOR signaling pathway. Front Pharmacol 11:25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Kim Y et al (2022) Drug-Disease association prediction using heterogeneous networks for computational drug repositioning. Biomolecules, 12(10):1497 [DOI] [PMC free article] [PubMed]
- 467.Pushpakom S et al (2019) Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov 18(1):41–58 [DOI] [PubMed] [Google Scholar]
- 468.Xu C et al (2023) Compounds targeting ferroptosis in breast cancer: progress and their therapeutic potential. Front Pharmacol 14:1243286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Chen X et al (2021) Ferroptosis: machinery and regulation. Autophagy 17(9):2054–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Jiang X, Stockwell BR, Conrad M (2021) Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol 22(4):266–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Guo J et al (2021) A combined model of human iPSC-Derived liver organoids and hepatocytes reveals ferroptosis in DGUOK mutant MtDNA depletion syndrome. Adv Sci (Weinh) 8(10):2004680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Nottingham E et al (2021) Synergistic effects of Methyl 2-cyano-3,11-dioxo-18beta-olean-1,-12-dien-30-oate and erlotinib on erlotinib-resistant non-small cell lung cancer cells. J Pharm Anal 11(6):799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Yang JC et al (2020) Afatinib for the treatment of NSCLC harboring uncommon EGFR mutations: A database of 693 cases. J Thorac Oncol 15(5):803–815 [DOI] [PubMed] [Google Scholar]
- 474.Sequist LV et al (2023) Phase III study of Afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 41(16):2869–2876 [DOI] [PubMed] [Google Scholar]
- 475.Foretz M, Guigas B, Viollet B (2023) Metformin: update on mechanisms of action and repurposing potential. Nat Rev Endocrinol 19(8):460–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Kuduvalli SS et al (2023) A combination of Metformin and Epigallocatechin gallate potentiates glioma chemotherapy in vivo. Front Pharmacol 14:1096614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Tang JH et al (2019) Bortezomib inhibits growth and sensitizes glioma to Temozolomide (TMZ) via down-regulating the FOXM1-Survivin axis. Cancer Commun (Lond) 39(1):81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Zhong S et al (2022) Disulfiram in glioma: literature review of drug repurposing. Front Pharmacol 13:933655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Matteoni S et al (2021) Chlorpromazine induces cytotoxic autophagy in glioblastoma cells via Endoplasmic reticulum stress and unfolded protein response. J Exp Clin Cancer Res 40(1):347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Matarrese P et al (2024) Chlorpromazine overcomes Temozolomide resistance in glioblastoma by inhibiting Cx43 and essential DNA repair pathways. J Transl Med 22(1):667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Ramalho MJ et al (2024) Chitosan-PLGA mucoadhesive nanoparticles for gemcitabine repurposing for glioblastoma therapy. Eur J Pharm Biopharm 200:114326 [DOI] [PubMed] [Google Scholar]
- 482.Liyanage PY et al (2020) Pediatric glioblastoma target-specific efficient delivery of gemcitabine across the blood-brain barrier via carbon nitride Dots. Nanoscale 12(14):7927–7938 [DOI] [PubMed] [Google Scholar]
- 483.Cruz-Burgos M et al (2021) New approaches in oncology for repositioning drugs: the case of PDE5 inhibitor sildenafil. Front Oncol 11:627229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Nayak R, Mallick B (2024) BMS345541 is predicted as a repurposed drug for the treatment of TMZ-resistant glioblastoma using target gene expression and virtual drug screening. Cancer Genet, 288–289:20–31 [DOI] [PubMed]
- 485.Kadasah SF et al (2024) Beyond psychotropic: potential repurposing of Fluoxetine toward Cancer therapy. Int J Mol Sci, 25(12):6314 [DOI] [PMC free article] [PubMed]
- 486.Mubeen S et al (2024) Iloperidone and Temozolomide synergistically inhibit growth, migration and enhance apoptosis in glioblastoma cells. Biomedicines, 12(6):1134 [DOI] [PMC free article] [PubMed]
- 487.Bramatti I et al (2024) Exposure of human glioblastoma cells to thimerosal inhibits the thioredoxin system and decreases tumor growth-related factors. Toxicol Appl Pharmacol 484:116844 [DOI] [PubMed] [Google Scholar]
- 488.Driscoll RK et al (2024) A multi-institutional phase I study of Acetazolamide with Temozolomide in adults with newly diagnosed MGMT-methylated malignant glioma. Neurooncol Adv 6(1):vdae014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.You F et al (2023) Sitagliptin inhibits the survival, stemness and autophagy of glioma cells, and enhances Temozolomide cytotoxicity. Biomed Pharmacother 162:114555 [DOI] [PubMed] [Google Scholar]
- 490.Yadav A et al (2022) Repurposing an antiepileptic drug for the treatment of glioblastoma. Pharm Res 39(11):2871–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Pepper NB et al (2024) ALA-RDT in GBM: protocol of the phase I/II dose escalation trial of radiodynamic therapy with 5-Aminolevulinic acid in patients with recurrent glioblastoma. Radiat Oncol 19(1):11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Vengoji R et al (2021) Differential gene expression-based connectivity mapping identified novel drug candidate and improved Temozolomide efficacy for glioblastoma. J Exp Clin Cancer Res 40(1):335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Liu ZZ et al (2021) Identification of Pimavanserin tartrate as a potent Ca(2+)-calcineurin-NFAT pathway inhibitor for glioblastoma therapy. Acta Pharmacol Sin 42(11):1860–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Hijazi MA, Gessner A, El-Najjar N (2023) Repurposing of chronically used drugs in Cancer therapy: A chance to Grasp. Cancers (Basel), 15(12):3199 [DOI] [PMC free article] [PubMed]
- 495.Shi J et al (2021) Nicardipine sensitizes Temozolomide by inhibiting autophagy and promoting cell apoptosis in glioma stem cells. Aging 13(5):6820–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Wu J et al (2020) Skp2 modulates proliferation, senescence and tumorigenesis of glioma. Cancer Cell Int 20:71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Pandey V et al (2019) Roscovitine effectively enhances antitumor activity of Temozolomide in vitro and in vivo mediated by increased autophagy and Caspase-3 dependent apoptosis. Sci Rep 9(1):5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Ha W et al (2019) Ibudilast sensitizes glioblastoma to Temozolomide by targeting macrophage migration inhibitory factor (MIF). Sci Rep 9(1):2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.van Kessel E et al (2022) Tumor-related molecular determinants of neurocognitive deficits in patients with diffuse glioma. Neuro Oncol 24(10):1660–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Johannessen TC et al (2019) Thioridazine inhibits autophagy and sensitizes glioblastoma cells to Temozolomide. Int J Cancer 144(7):1735–1745 [DOI] [PubMed] [Google Scholar]
- 501.Zhang Y et al (2021) Loss of COPZ1 induces NCOA4 mediated autophagy and ferroptosis in glioblastoma cell lines. Oncogene 40(8):1425–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Chinyama HA et al (2023) Identification of CDK1, PBK, and CHEK1 as an oncogenic signature in glioblastoma: A bioinformatics approach to repurpose Dapagliflozin as a therapeutic agent. Int J Mol Sci, 24(22):16396 [DOI] [PMC free article] [PubMed]
- 503.Summers HS et al (2023) Discovery of new imidazotetrazinones with potential to overcome tumor resistance. Eur J Med Chem 257:115507 [DOI] [PubMed] [Google Scholar]
- 504.Gazvoda M et al (2022) Palladium-Mediated incorporation of Carboranes into small molecules, peptides, and proteins. J Am Chem Soc 144(17):7852–7860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Coghi P et al (2023) Next generation of Boron neutron capture therapy (BNCT) agents for cancer treatment. Med Res Rev 43(5):1809–1830 [DOI] [PubMed] [Google Scholar]
- 506.Xiang J et al (2022) A boronated derivative of Temozolomide showing enhanced efficacy in Boron neutron capture therapy of glioblastoma. Cells, 11(7):1173 [DOI] [PMC free article] [PubMed]
- 507.Yu Y et al (2021) Synthesis and characterization of a series of Temozolomide esters and its Anti-glioma study. J Pharm Sci 110(10):3431–3438 [DOI] [PubMed] [Google Scholar]
- 508.Zhang X et al (2022) Acquired Temozolomide resistance in MGMT(low) gliomas is associated with regulation of homologous recombination repair by ROCK2. Cell Death Dis 13(2):138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Minea RO et al (2024) NEO212, Temozolomide conjugated to NEO100, exerts superior therapeutic activity over Temozolomide in preclinical chemoradiation models of glioblastoma. Neurooncol Adv 6(1):vdae095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Marín-Ramos NI et al (2019) NEO212, a conjugate of Temozolomide and Perillyl alcohol, blocks the endothelial-to-mesenchymal transition in tumor-associated brain endothelial cells in glioblastoma. Cancer Lett 442:170–180 [DOI] [PubMed] [Google Scholar]
- 511.Bae WH, Maraka S, Daher A (2024) Challenges and advances in glioblastoma targeted therapy: the promise of drug repurposing and biomarker exploration. Front Oncol 14:1441460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Nicholson JG, Fine HA (2021) Diffuse glioma heterogeneity and its therapeutic implications. Cancer Discov 11(3):575–590 [DOI] [PubMed] [Google Scholar]
- 513.Yu MW, Quail DF (2021) Immunotherapy for glioblastoma: current progress and challenges. Front Immunol 12:676301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Ganipineni LP, Danhier F, Préat V (2018) Drug delivery challenges and future of chemotherapeutic nanomedicine for glioblastoma treatment. J Control Release 281:42–57 [DOI] [PubMed] [Google Scholar]
- 515.Yun CW et al (2020) The dual role of autophagy in Cancer development and a therapeutic strategy for Cancer by targeting autophagy. Int J Mol Sci 22(1):179 [DOI] [PMC free article] [PubMed]
- 516.Badr CE et al (2020) Metabolic heterogeneity and adaptability in brain tumors. Cell Mol Life Sci 77(24):5101–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Dirnagl U et al (2022) Reproducibility, relevance and reliability as barriers to efficient and credible biomedical technology translation. Adv Drug Deliv Rev 182:114118 [DOI] [PubMed] [Google Scholar]
- 518.Ghosh D, Nandi S, Bhattacharjee S (2018) Combination therapy to checkmate glioblastoma: clinical challenges and advances. Clin Transl Med 7(1):33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Wu Y et al (2023) Functionalized nanoparticles crossing the brain-blood barrier to target glioma cells. PeerJ 11:e15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Meng W et al (2022) Overcoming radiation resistance in gliomas by targeting metabolism and DNA repair pathways. Int J Mol Sci 23(4):2246 [DOI] [PMC free article] [PubMed]
- 521.Mishchenko TA et al (2023) Glioma: bridging the tumor microenvironment, patient immune profiles and novel personalized immunotherapy. Front Immunol 14:1299064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Luo J et al (2023) Emerging role of artificial intelligence in diagnosis, classification and clinical management of glioma. Semin Cancer Biol 91:110–123 [DOI] [PubMed] [Google Scholar]
- 523.Mohile NA et al (2022) Therapy for diffuse astrocytic and oligodendroglial tumors in adults: ASCO-SNO guideline. J Clin Oncol 40(4):403–426 [DOI] [PubMed] [Google Scholar]
- 524.Walbert T et al (2021) SNO and EANO practice guideline update: anticonvulsant prophylaxis in patients with newly diagnosed brain tumors. Neuro Oncol 23(11):1835–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Park AK et al (2022) Characterization and prognosis of temozolomide-induced aplastic anemia in patients with central nervous system malignancies. Neuro Oncol 24(6):964–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Krauze AV et al (2015) A phase 2 study of concurrent radiation therapy, temozolomide, and the histone deacetylase inhibitor valproic acid for patients with glioblastoma. Int J Radiat Oncol Biol Phys 92(5):986–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Weller M et al (2011) Prolonged survival with valproic acid use in the EORTC/NCIC Temozolomide trial for glioblastoma. Neurology 77(12):1156–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Shields LB et al (2015) Dexamethasone administration during definitive radiation and Temozolomide renders a poor prognosis in a retrospective analysis of newly diagnosed glioblastoma patients. Radiat Oncol 10:222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Gonzalez-Aponte MF et al (2025) Daily glucocorticoids promote glioblastoma growth and circadian synchrony to the host. Cancer Cell 43(1):144–160 [DOI] [PMC free article] [PubMed]
- 530.Ha H, Lim JH (2022) Managing side effects of cytotoxic chemotherapy in patients with high grade gliomas. Brain Tumor Res Treat 10(3):158–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Stupp R et al (2017) Effect of Tumor-Treating fields plus maintenance Temozolomide vs maintenance Temozolomide alone on survival in patients with glioblastoma: A randomized clinical trial. JAMA 318(23):2306–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Weller M et al (2017) European association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol 18(6):e315–e329 [DOI] [PubMed] [Google Scholar]
- 533.van den Bent MJ et al (2017) Interim results from the CATNON trial (EORTC study 26053– 22054) of treatment with concurrent and adjuvant temozolomide for 1p/19q non-co-deleted anaplastic glioma: a phase 3, randomised, open-label intergroup study. Lancet, 390(10103):1645–1653 [DOI] [PMC free article] [PubMed]
- 534.Fanizzi C et al (2025) Optimal MGMT promoter methylation cut-off to predict better survival in glioblastoma patients undergoing gross-total resection. J Neurosurg Sci [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.