Abstract
The seed-setting rate has a significant effect on grain yield in rice (Oryza sativa L.). Embryo sac development is essential for seed setting; however, the molecular mechanism underlying this process remains unclear. Here, we isolated defective embryo sac1 (des1), a rice mutant with a low seed-setting rate. Cytological examination showed degenerated embryo sacs and reduced fertilization capacity in des1. Map-based cloning revealed a nonsense mutation in OsDES1, a gene that encodes a putative nuclear envelope membrane protein (NEMP)-domain-containing protein that is preferentially expressed in pistils. The OsDES1 mutation disrupts the normal formation of functional megaspores, which ultimately results in a degenerated embryo sac in des1. Reciprocal crosses showed that fertilization is abnormal and that the female reproductive organ is defective in des1. OsDES1 interacts with LONELY GUY (LOG), a cytokinin-activating enzyme that acts in the final step of cytokinin synthesis; mutation of LOG led to defective female reproductive organ development. These results demonstrate that OsDES1 functions in determining the rice seed-setting rate by regulating embryo sac development and fertilization. Our study sheds light on the function of NEMP-type proteins in rice reproductive development.
Keywords: Embryo sac, female sterile, fertilization, OsDES1, rice, seed-setting rate
OsDES1encodes a NEMP-domain protein that interacts with a cytokinin-activating enzyme, LOG, to determine the seed-setting rate in rice by regulating embryo sac development.
Introduction
In addition to its role as a primary staple for more than half the world’s population, rice (Oryza sativa L.) serves as a model species for studies of plant development and biology in monocotyledons. Rice yield is typically determined by the grain weight, panicle number, number of grains per panicle, and seed-setting rate. In the past few decades, significant advances have been made in understanding the molecular mechanisms that control the grain weight, panicle number, and number of grains per panicle (Zuo et al., 2014). Studies have shown that adverse environmental conditions can dramatically reduce the seed-setting rate in rice, resulting in serious yield reduction (Li et al., 2013). Recent findings have shed light on the molecular mechanisms that regulate the seed-setting rate in rice, and several genes related to this trait, such as PSS1, PTB1, OsSPX1, OsCNGC13, OsOAT, OsALMT7, DPS1, OsROS1, OsMLH3, and ESD1, have been reported (Zhou et al., 2011; Li et al., 2013; Zhang et al., 2016; Xu et al., 2017; Heng et al., 2018; Liu et al., 2018; Zafar et al., 2019; Xu et al., 2020; Mao et al., 2021; Wang et al., 2021). The low seed-setting rate in indica–japonica hybrids has become a major roadblock that restricts further improvements in grain yield (Li et al., 2016). Hence, more research is needed to explore the molecular mechanisms underlying seed setting in rice.
The seed-setting rate in rice is affected by many genetic and environmental factors, including defective embryo sac development, malformed floral organ morphology, disordered pollen grain formation, inadequate anther dehiscence, gametophytic incompatibility, and abnormal temperature (Xu et al., 2017). Embryo sac development is vital to the correct functioning of steps in the reproductive process such as pollen tube guidance, double fertilization, induced seed development, and maternal control of seed development after fertilization (Ray et al., 1997; Christensen et al., 1998; Drews et al., 1998; Yadegari and Drews, 2004). Embryo sac development is impacted by various physiological and environmental factors, such as plant hormones (Pischke et al., 2002; Bencivenga et al., 2012; Cheng et al., 2013) and climatic conditions (Li et al., 2013). Many genes controlling female reproductive organ development in plants have been studied. For example, Arabidopsis WUS and SPL and maize MAC1 regulate the differentiation of somatic cells into germ cells (Sheridan et al., 1996; Yang et al., 1999; Lieber et al., 2011). Several meiosis-related genes function in female fertility, including SWI1 (Boateng et al., 2008) and ARP6 (Qin et al., 2014) in Arabidopsis, and PAIR1 (Nonomura et al., 2004a), PAIR2 (Nonomura et al., 2004b), PAIR3 (Yuan et al., 2009), OsRPA1a (Chang et al., 2009), RAD51C (Kou et al., 2012), OsMSH4 (Wang et al., 2016), OsSHOC1 (Ren et al., 2019), OsMFS1 (Lu et al., 2020), and OsMLH3 (Mao et al., 2021) in rice. MYB64 and MYB119 (Rabiger and Drews, 2013) and BLH1 (Pagnussat et al., 2007) in Arabidopsis, and OsAPC6 (Kumar et al., 2010; Awasthi et al., 2012) and OsDEES1 (Wang et al., 2012) in rice, are associated with mitosis and regulate the development of the embryo sac and seed. The cytokinin-activating enzyme encoded by LONELY GUY (LOG) is essential for ovule and pistil formation (Kurakawa et al., 2007; Yamaki et al., 2011). Recently, OsROS1, ESD1, and OsMLH3 have been reported to function in rice embryo sac development (Xu et al., 2020; Wang et al., 2021; Mao et al., 2021).
In most angiosperms, embryo sac development generally includes megasporogenesis and megagametogenesis. The megasporocyte develops into a seven-celled structure through two meiotic divisions and three consecutive mitotic divisions (Yadegari and Drews, 2004; Drews and Koltunow, 2011; Nakajima, 2018). These cells make up four groups that function in fertilization, embryogenesis, and nutrition of the embryo sac and embryo (Reiser and Fischer, 1993; Chen et al., 2007; Li et al., 2015; Higashiyama and Yang, 2017; Meng et al., 2020; Sun et al., 2021). The male and female gametes undergo fusion and develop into the embryo and endosperm. The integuments go through structural and biochemical specialization as the ovule forms into a seed (Robinson-Beers et al., 1992; Li et al., 2013; Dresselhaus et al., 2016; Sankaranarayanan and Higashiyama, 2018). The embryo sac is embedded in sporophytic tissues of the ovule, making it difficult to directly isolate embryo sac tissue for research (Jones-Rhoades et al., 2007). In the past two decades, due to the difficulty of mutant acquisition and morphological identification and the complexity of genetic mechanisms, progress in research on female sterility has lagged. Therefore, we decided to evaluate female-sterility-related genes and apply them to target developmental mutants in rice.
In this study, we report on the role of a putative nuclear envelope membrane protein (NEMP) domain-containing protein, DEFECTIVE EMBRYO SAC1 (OsDES1), in seed setting by regulating embryo sac development. The loss-of-function mutant des1 displayed a significant reduction in seed-setting rate due to embryo sac degeneration and defective fertilization. We propose that the abnormal formation of functional megaspores in des1 causes embryo sac degeneration and reduced fertility. Our study provides insights into the roles of OsDES1 in female reproductive development and seed setting in rice.
Materials and methods
Plant materials and growth conditions
The rice plants used in this study were grown in paddy fields at the China National Rice Research Institute, Hangzhou, Zhejiang Province, and in Lingshui, Hainan Province, China. The des1 mutant, which shows an abnormal seed-setting rate, was isolated from a 60Co-γ-radiation-induced mutant library of the indica rice cv. ‘Zhonghui8015’ (ZH8015). An F2 mapping population was derived from a cross between the japonica rice cv. 02428 and the homozygous des1 mutant. The mutant plants showed genetic stability in both Zhejiang and Hainan.
Preparation of embryo sacs
Wild-type (WT) and mutant spikelets were collected at different stages of embryo sac development based on the length of the floret. The spikelets were fixed in ethanol:formaldehyde:glacial acetic acid (18:1:1) solution (FAA) and vacuum infiltrated. The samples were kept in 70% ethanol for embryo sac observation. The spikelets were dissected in 70% ethanol, hydrated sequentially in 50% ethanol, 30% ethanol, and distilled water, and then transferred to 2% aluminum potassium sulfate for 20 min. The samples were stained with 10 mg l–1 eosin B solution dissolved in 4% sucrose overnight at room temperature, pretreated with 2% aluminum potassium sulfate for 20 min, and then washed with distilled water followed by dehydration using ethanol solutions at concentrations of 30, 50, 70, 90, and 100%. The samples were transferred into a mixture of methylsalicylate and absolute ethanol (1:1) for 1 h and then moved to 100% methylsalicylate solution for 8 h (Zhao et al., 2007; Zeng et al., 2009). Finally, the ovaries were imaged with a Zeiss LSM710 laser scanning confocal microscope.
Histological analysis
The developmental stages of the rice anthers were identified as previously described (Zhang and Wilson, 2009; Zhang et al., 2011). Spikelets at different developmental stages were selected and fixed in FAA for semi-thin sectioning. Anthers at different developmental stages were selected and embedded in a standard resin for semi-thin sectioning according to a previously published protocol (Li et al., 2006). The samples were dehydrated in a graded ethanol series from 50% to 100% and embedded in Technovit 7100 resin (Heraeus, Kulzer, Germany), which was then allowed to solidify at 50 °C for 3–4 days. Transverse sections of 2 μm thickness were cut using a Leica RM2265 fully automated rotary microtome, stained with 0.25% toluidine blue O, and photographed under a Leica DM2000 light microscope. The observation of embryo sac development was performed as previously described (Yamaki et al., 2011). All samples were dehydrated in a graded ethanol series, substituted with xylene and embedded in paraffin, then cut at 8 μm thickness, and finally stained with hematoxylin, hyalinized, and sealed. The sealed sections were imaged with a Zeiss LSM710 laser scanning confocal microscope.
Acetocarmine and DAPI staining
For observations of microspore development, spikelet samples were chosen from the premeiotic to mature stages and fixed in FAA. The microspores from crushed anthers were stained with 1% (w/v) acetocarmine solution. After 2 min, microspores were observed under a Leica DM2000 light microscope. To observe the microspores of the mature stages better, DAPI (4ʹ,6-diamidino-2-phenylindole) staining was used on mature pollen grains, which were imaged using a Leica DM5000 B fluorescence microscope as previously described (Yu et al., 2018).
Evaluation of pollen fertility
To evaluate mature pollen fertility, WT and des1 mutant anthers were collected and stained with 1% (w/v) iodine–potassium iodide solution (I2-KI) and the accumulation of starch in pollen grains was observed using a Leica DM2000 microscope.
In vitro pollen germination assay
An in vitro pollen germination assay was performed as described previously (Zhou et al., 2011). Briefly, pollen grains were placed on Brewbaker and Kwack medium (10% sucrose, 200 mg l–1 magnesium sulfate, 300 mg l–1 calcium nitrate, 100 mg l–1 boric acid, and 100 mg l–1 potassium nitrate) for 1 h at 25 °C. The pollen grains were observed for germination using a Leica DM2000 light microscope. We defined successful germination as when the elongated length of the pollen tube exceeded the diameter of the pollen grain. The germination rate of the WT and the des1 mutant was calculated by examining at least 200 pollen grains per genotype.
Observation of pollen germination on the stigma
Observation of pollen germination on the stigma was performed as described previously (Xu et al., 2017). The pistils of WT and des1 mutants were fixed in FAA, dehydrated with an ethanol series, incubated in 10 M sodium hydroxide at 56 °C for 8 min, and then stained with 0.1% aniline blue solution. The pistils were imaged using a Zeiss LSM880 laser scanning confocal microscope. Pollen tube growth was defined as when at least one pollen tube in the ovule reached the micropyle.
Transmission and scanning electron microscopy
Mature anthers from WT and des1 plants were collected and fixed in 2.5% glutaraldehyde (pH 7.2) for 24 h, fixed in 1% OsO4 in phosphate buffer solution, and dehydrated with an ethanol series. Ultra-thin sections were stained with uranyl acetate and aqueous lead citrate solution, and then examined with a Hitachi H-7650 transmission electron microscope. For scanning electron microscopy, the mature anthers and pistils were fixed overnight with 2.5% glutaraldehyde (pH 7.2), rinsed three times using 0.1 M phosphate buffer solution, fixed in 1% OsO4 for 1.5 h, and dehydrated through an ethanol series. Subsequently, the samples were subjected to CO2 critical point drying, plated with gold by a sputter coater, and observed with a Hitachi TM-1000 scanning electron microscope.
Map-based cloning
To map the OsDES1 locus, eight individual plants with abnormal spikelets were chosen from the F2 population derived from the cross of 02428 and the des1 mutant for preliminary mapping using ~200 genome-wide insertion–deletion and simple sequence repeat markers. To fine map the OsDES1 locus, a total of 393 plants with the mutant phenotype were selected from the F2 population and 27 new molecular markers were designed by comparing the nucleotide polymorphisms in the reference sequences between cultivars 9311 and ‘Nipponbare’ (NIP). All primers used for mapping are listed in Supplementary Table S1.
RNA extraction and quantitative real-time reverse transcription–PCR
Total RNA was extracted from flag leaves at the heading stage and from pistils at different developmental stages using a RNAprep pure Plant kit (Tiangen Biotech Co. Ltd, Beijing, China) following the manufacturer’s instructions. First-strand cDNA was synthesized with a ReverTraAce® qPCR RT Master Mix with a gDNA Remover kit (Toyobo Co. Ltd, Osaka, Japan) using 1.5 μg of RNA. Quantitative real-time reverse transcription–PCR (qRT–PCR) assays were performed with a SYBR premix Ex Taq Kit (Takara Bio Inc., Kusatsu, Shiga, Japan). The relative mRNA levels of the investigated genes were normalized to Ubiquitin (Os03g0234350) and Actin (Os03g0718100) by the 2–ΔΔCT calculation method with three replicates, respectively. The primers used for qRT–PCR are shown in Supplementary Table S1.
Vector construction and plant transformation
For overexpression of OsDES1, the WT full-length cDNA was amplified and subcloned into the pCUbi1390 plasmid under the control of the maize Ubiquitin 1 promoter. The resulting construct was transformed into the des1 mutant by Agrobacterium-mediated transformation. The CRISPR/Cas9 system was used to knock out the OsDES1 gene as previously reported (Miao et al., 2013; Huang et al., 2017). The vector pBWA(V)HS_cas9i2 containing the target sequence was transformed into NIP callus tissue through Agrobacterium-mediated transformation. Individual plants carrying mutations were identified by sequencing before further analysis. For the promoter activity assay of OsDES1, a 2444 bp DNA fragment upstream of the OsDES1 start codon was amplified and ligated into the binary vector pCAMBIA1305 to serve as the OsDES1 promoter to drive the expression of the β-glucuronidase (GUS) reporter gene. The construct was transformed into the japonica variety NIP. The names and sequences of all primers used for vector construction and sequencing are listed in Supplementary Table S1.
β-Glucuronidase histochemical staining
Different tissues from OsDES1-promoter-GUS transgenic plants of NIP were collected at different developmental stages and stained as previously described (Jefferson, 1989). Images were obtained using a scanner (MRS-9600TFU2L) and a stereomicroscope (Leica MC120HD) with a digital camera.
RNA in situ hybridization
WT spikelets at different developmental stages were fixed overnight in an FAA (RNase-free) fixative solution at 4 °C. After being dehydrated in a graded ethanol series and xylene, pistils were embedded in paraffin. An OsDES1 cDNA fragment was amplified using primers (listed in Supplementary Table S1) and cloned into the pGEMT Easy vector. The antisense and sense probes were then transcribed in vitro using a DIG RNA Labeling Kit (SP6/T7) (Roche) according to the manufacturer’s instructions. RNA hybridization and immunological detection of the hybridized probes were performed as previously described (Kouchi and Hata, 1993).
Subcellular localization
To determine the subcellular localization of OsDES1, ΔOsDES1 (the mutant OsDES1 protein), and the NEMP domain, the coding sequence (CDS) of OsDES1, ΔOsDES1, and NEMP was amplified and inserted into the GFP vector pYBA1132. Rice leaf protoplasts were isolated from 10-day-old ZH8015 seedlings. The empty vector (as control) and the recombinant construct plasmids were transfected into protoplasts and incubated for 24 h in the dark (Yoo et al., 2007). The Ghd7-CFP construct was used as a nuclear marker. FM4-64 solution (8.2 μM; Molecular Probes) was added to protoplasts, which were incubated for 15 min to label the membranes and then observed immediately (Wang et al., 2018). The recombinant construct plasmids were co-expressed with Ghd7-CFP in Nicotiana benthamiana leaves. After 48 h, the fluorescent signal was detected with a Zeiss LSM710 confocal laser scanning microscope. All primers used are shown in Supplementary Table S1.
Yeast two-hybrid assay
The CDS of LOG was amplified and inserted into the prey vector pPR3-N. The CDS of OsDES1 was amplified and cloned into the bait vector pBT3-SUC. A yeast two-hybrid assay was performed according to the manufacturer’s instructions (Clontech). The DUALmembrane system was used for conducting the assays. All primers are listed in Supplementary Table S1.
Split luciferase complementation assay
The CDS of OsDES1 was cloned into the pCAMBIA-split_nLUC vector, and the CDS of LOG was cloned into pCAMBIA-split_cLUC. The constructs were transformed into Agrobacterium tumefaciens GV3101 and transiently expressed in N. benthamiana leaves. The primers used to construct nLUC-OsDES1 and cLUC-LOG are listed in Supplementary Table S1.
Co-immunoprecipitation assay
The full-length cDNA sequences of OsDES1 and LOG were amplified by PCR and fused with sequences encoding GFP and the Myc tag driven by the 35S promoter, respectively. The specific primer pairs used to amplify GFP-OsDES1 and Myc-LOG were GFP-OsDES1-F and GFP-OsDES1-R, and Myc-LOG-F and Myc-LOG-R, respectively. Leaves of N. benthamiana were transfected by injection with A. tumefaciens GV3101 containing the 35S:GFP-OsDES1 and 35S:Myc-LOG constructs as previously described (Voinnet et al., 2003). Total protein was extracted in extraction buffer [20 μg ml–1 MG132, 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 2% Triton X-100, 20% glycerol, 1× complete protease inhibitor cocktail (Roche), and 1 mM EDTA], and incubated with GFP-Trap A agarose beads for 60 min at 4 °C. The beads were rinsed three times with washing buffer [1× complete protease inhibitor cocktail (Roche), 150 mM NaCl, 50 mM Tris–HCl, pH 7.5, and 0.1% Triton X-100]. The immunoprecipitated proteins were separated by 10% SDS-PAGE and analyzed by immunoblot analysis with anti-Myc (LOG) and anti-GFP (OsDES1) antibodies. All primers used are shown in Supplementary Table S1.
Results
Identification of a low seed-setting rate rice mutant
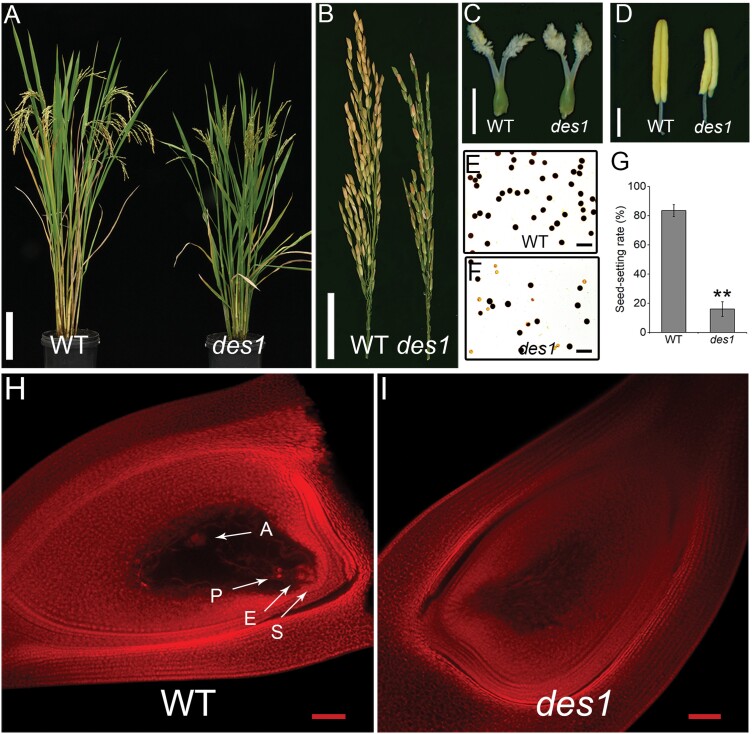
In an effort to characterize rice sterility phenotypes, we identified a low-seed-setting rice mutant, named defective embryo sac 1 (des1), from a 60Co-γ-irradiated library of the indica rice cv. ZH8015. Compared with the WT, des1 plants were slightly shorter in height and showed earlier heading but were otherwise normal in vegetative development (Supplementary Fig. S1A). The seed-setting rate of the WT was ~84%, whereas des1 showed a much lower seed-setting rate (~16%) under normal field conditions (Fig. 1A, B, G). In addition, des1 plants produced shorter brown panicles (Fig. 1B). However, no clear difference in pistil morphology was observed in des1 compared with the WT (Fig. 1C; Supplementary Fig. S2A, B). The WT embryo sacs contained egg cells, synergids, polar nuclei, and antipodal cells, whereas des1 embryo sacs degenerated and did not have these characteristic structures (Fig. 1H, I). In addition, the most obvious differences in the male reproductive organs between des1 and the WT were the number of stamens and pollen fertility. Seven different types of stamens were detected in des1 spikelets, and the numbers of stamens differed in each spikelet, whereas only six stamens were typically found in WT spikelets (Supplementary Fig. S1C, E). The stamen lengths were also shorter, whereas the development of other floral organs appeared normal, in des1 (Fig. 1D; Supplementary Fig. S2C, D). At anthesis, spikelet development in des1 was similar to that of the WT (Supplementary Fig. S1B). Pollen viability was approximately 98% in the WT but only 64% in des1 (Fig. 1E, F; Supplementary Fig. S1D). To determine the causes underlying the reduced fertility observed in des1, we performed reciprocal cross experiments. The results showed that the seed-setting rate of WT♀×WT♂ and des1♀×des1♂ crosses with full pollination were 74.4% and 29.3%, respectively. When the WT was pollinated with des1 pollen, the hybrid seed-setting rate was 57.3%. However, when des1 was used as the pollen recipient, the hybrid seed-setting rate was 35.4% (Supplementary Fig. S3). This finding suggests that the low seed-setting rate of des1 is mainly due to a maternal defect. The F2 population showed an approximate 3:1 segregation ratio of normal (108) and low (28) seed-setting (χ2=0.235<χ2 0.05=3.84, χ2 test). These results indicate that female sterility in des1 was inherited as a single recessive mutation.
Fig. 1.
Phenotypic characterization of des1 rice. (A) Comparison of WT and des1 plants at maturity. Scale bar=20 cm. (B) Mature panicles of WT and des1 plants. Scale bar=5 cm. (C) Pistils of WT and des1 at maturity. Scale bar=0.125 cm. (D) Anthers of WT and des1. Scale bar=1.25 mm. (E, F) I2-KI staining of pollen grains in WT (E) and des1 (F). Scale bars=25 μm. (G) Statistical analysis of the seed-setting rates in WT and des1 plants. Data are means ±SD (n=8 plants). Asterisks indicate significant differences (**P<0.01; Student’s t-test). (H, I) Microscopic observations of mature embryo sacs in WT (H) and des1 (I). Arrows indicate the eight-cell components in the embryo sac: A, antipodal cell; E, egg cell; P, polar nucleus; S, synergid cell. Scale bars=50 μm.
Disrupted embryo sac development in des1
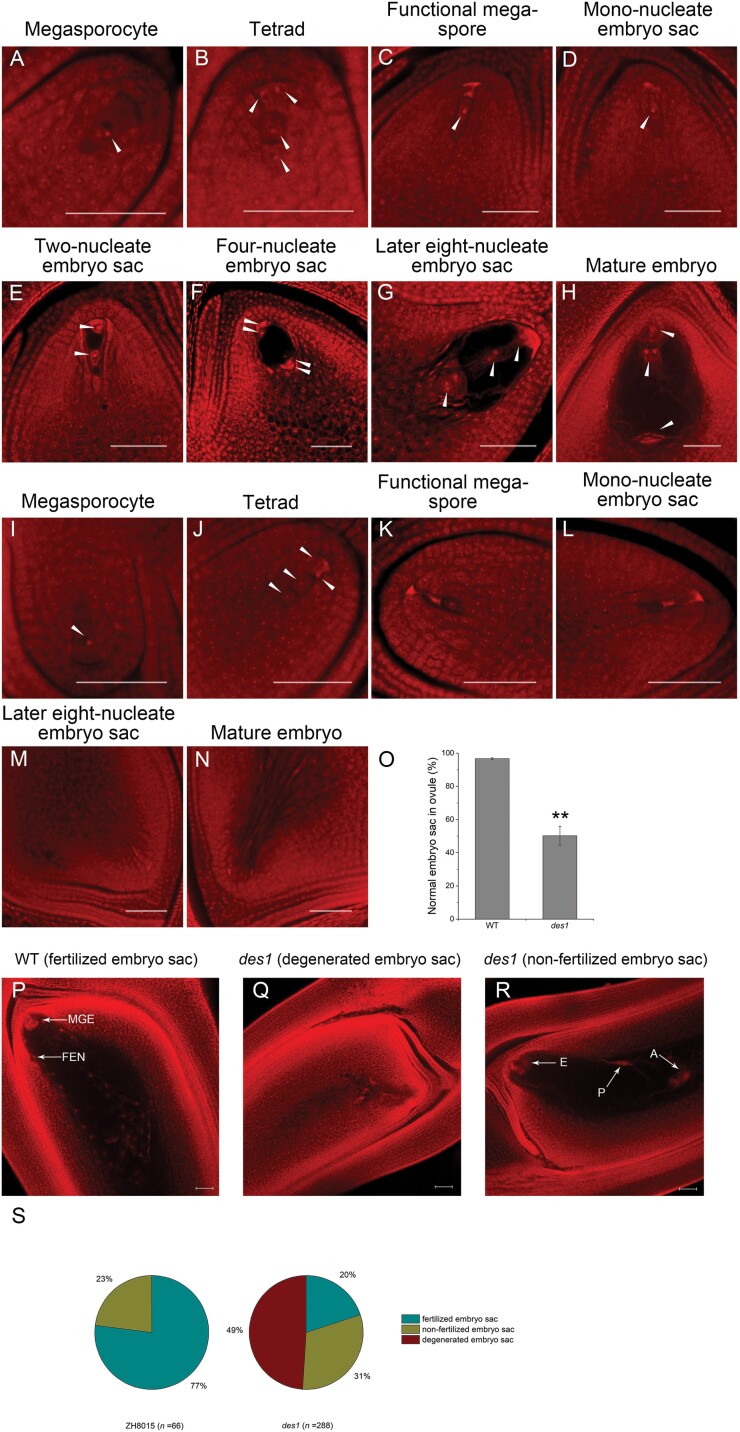
To uncover the cytological basis of female sterility, we compared the formation and development of the embryo sac of the WT and des1 using whole-mount stain-clearing laser scanning confocal microscopy. In the WT, megasporocytes undergo two meiotic divisions to produce a tetrad of megaspores (Fig. 2A, B). Subsequently, three megaspores situated at the micropylar end degenerate and the remaining megaspore remains functional and develops further by enlargement at the functional megaspore formation stage (Fig. 2C). The mono-nucleate embryo sac undergoes three rounds of mitotic division to form a two-nucleate, four-nucleate, and eight-nucleate embryo sac (Fig. 2D–G). Finally, the mature embryo sac forms, with polar nuclei, the egg cell, synergids, and antipodals (Fig. 2H). The megasporocyte and tetrads form normally in des1 (Fig. 2I, J), but some embryo sacs exhibit defects at the functional megaspore formation stage. The megaspore near the chalaza in des1 did not continue to grow into a functional megaspore. Instead, it gradually degenerated along with the other three megaspores near the micropyle (Fig. 2K). The aberrant nuclei then also began to degenerate along with the embryo sac in des1. Subsequently, only degenerated footprints of nuclei were observable, and these remained visible for a long time until embryo sac maturity. During the final developmental stage, the embryo sac ultimately developed into undifferentiated tissue, likely due to impaired mitotic processes (Fig. 2L–N). Additionally, ~97% of mature embryo sacs in the WT successfully formed eight-nucleate embryo sacs, whereas in des1, only ~50% of mature embryo sacs fully matured to this stage and the remaining embryo sacs degenerated (Fig. 2O). Furthermore, we observed the embryo sac in the WT and des1 by microscopic examination of paraffin sections. Consistent with the above results, des1 showed normal formation of the megasporocyte and tetrads, but no functional megaspores and undifferentiated embryo sacs were detected at maturity (Supplementary Fig. S4).
Fig. 2.
Development of the embryo sac in WT and des1 rice. (A–N) Different developmental stages of the embryo sac are shown in WT (A–H) and des1 (I–N). Arrows indicate nuclei during megasporogenesis and megagametogenesis. Scale bars=50 μm. (O) Statistical analysis of normal mature embryo sac formation in WT and des1. Data are means ±SD (n=3) (ovule number: WT 271, des1 607). (P–R) Morphology of embryo sacs 24 h after pollination in WT and des1: (P) Normal WT embryo sac with a multicellular globular embryo (MGE) and free endosperm nuclei (FEN); (Q, R) Abnormal des1 embryo sacs displaying a degenerated embryo sac (Q) and non-fertilized embryo sac (R). A, Antipodal cell; E, egg cell; P, polar nucleus. Scale bars=50 μm. (S) Statistical analysis of embryo sac morphology 24 h after pollination in WT (left) and des1 (right).
We also observed embryo sac development during the 24 h following pollination in both the WT and des1. Both multicellular globular embryos and free endosperm nuclei were observed in the WT, whereas fertilization did not occur in many of the des1 embryo sacs and most embryo sacs had degenerated or were not fertilized. Approximately 20% of embryo sacs developed normally and became fertilized in des1, whereas the proportion of successfully fertilized embryo sacs was 77% in the WT (Fig. 2P–S). This finding indicates that a reduced capacity for fertilization contributed to the low seed-setting rate observed in des1 (Fig. 1G). Together with our phenotypic observations, this suggests that the mutation in OsDES1 causes multiple defects in embryo sac formation and fertilization. In addition, we also observed the embryo sacs in des1/ZH8015 F1 plants. The results showed that normal embryo sacs were detected in 96.1% (n=179) of ovules and F1 plants were fertile (Supplementary Fig. S5), consistent with a des1 having a sporophytic effect on embryo sac development.
Abnormal stamen development and pollen tube growth in des1
We observed anther and pollen development in the WT and des1 and found that the number of aborted pollen grains identified by I2-KI staining was lower in the WT than in des1 (Fig. 1E, F; Supplementary Fig. S1D). We also performed in vitro pollen germination assays and found that, compared with WT pollen (~92% viable), only ~61% of des1 pollen grains successfully germinated, consistent with the results of I2-KI staining (Supplementary Fig. S6). Subsequently, we observed pollen germination on the stigma and pollen tube growth in the ovary in both the WT and des1. In the WT, 84% of pollen tubes in the ovule were able to grow and reach the micropyle, compared with 80% in des1 (Supplementary Fig. S7; Supplementary Table S2). To further characterize the differences in pollen development in the WT and des1, we examined microspores using acetocarmine and DAPI staining. Observations of microspores showed no clear differences between the WT and des1 until the mono-nucleate stage (Supplementary Fig. S8A–E, H–L). In des1, only a proportion of the pollen grains were able to undergo the first and second mitoses normally, while the remaining pollen grains maintained a single brightly staining nucleus at the bicellular stage (Supplementary Fig. S8F, M). The abnormal pollen grains in des1 also formed irregular shapes, and became abortive at maturity (Supplementary Fig. S8G, N–P).
No obvious differences between the WT and des1 were observed in semi-thin sections until the early microspore stage (Supplementary Fig. S9A–I, M). In des1, the pollen underwent vacuolization and the tapetum appeared much thicker and did not undergo complete degeneration (Supplementary Fig. S9J, K, N, O). Only a proportion of des1 microspores ultimately exhibited normal development, and the remaining microspores degenerated (Supplementary Fig. S9L, P). We also compared mature anther and pollen grain morphology between des1 and the WT by scanning and transmission electron microscopy. Compared with the WT, the des1 anther epidermis was more compact (Supplementary Fig. S2E, F) and the number of Ubisch bodies was higher (Supplementary Fig. S2G, H). A subset of des1 pollen grains exhibited a normal plump appearance (Supplementary Fig. S2I, J, L, M), while the remaining pollen grains were shrunken and had abnormal annular protrusions (Supplementary Fig. S2K, N). The pollen grains in the WT had a plump morphology and their internal structure was normal in appearance and contained starch granules (Supplementary Fig. S10A, B). By contrast, the shrunken pollen grains in des1 contained no starch granules (Supplementary Fig. S10D, E). The tectum and foot layer of des1 pollen grains were thicker than those in the WT, and the columella was degraded in des1 (Supplementary Fig. S10E) In addition, the tapetum in des1 was not completely degraded at maturity, as it was in the WT (Supplementary Fig. S10C, F). These results indicate that the des1 mutation affects anther and pollen development.
Map-based cloning of OsDES1
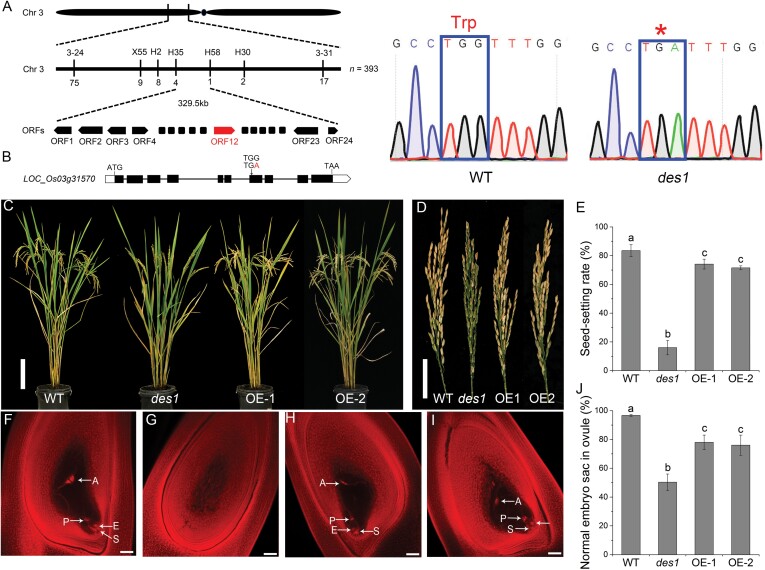
To identify the causal gene responsible for female sterility, we first crossed the des1 mutant with the japonica rice cultivar 02428 to generate an F2 mapping population. Using 393 recessive plants from the F2 population, we mapped OsDES1 to a 329.5 kb region on chromosome 3 located between marker loci H35 and H58, where a total of 24 open reading frames were predicted (Fig. 3A). Genomic sequence analysis revealed that LOC_Os03g31570 carried a nonsense mutation in the seventh exon in des1 (Fig. 3B). LOC_Os03g31570 was predicted to contain 10 exons and nine introns and encodes a 485 amino acid protein with a putative NEMP domain at amino acid residues 157–403 (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The mutation in des1 led to a truncated amino acid sequence lacking the NEMP domain.
Fig. 3.
Map-based cloning of OsDES1. (A) Mapping of the OsDES1 locus. The molecular marker loci and numbers of recombinant plants are shown above and below the line, respectively. The candidate gene OsDES1/LOC_Os03g31570 is shown in red. ORF, Open reading frame. (B) Schematic representation of the OsDES1 gene. White boxes, black boxes, and black lines indicate untranslated regions, exons, and introns, respectively. The single-base substitution of A for G in the seventh exon is shown. (C) Morphology of mature WT, des1, and OsDES1-overexpressing (OE-1 and OE-2) plants. Scale bar=20 cm. (D) Mature panicles of WT, des1, and OE plants. Scale bar=5 cm. (E) Seed-setting rates in WT, des1, and OsDES1-overexpressing plants. Data are means ±SD (n=8 plants). Different letters indicate significant differences (P<0.05; Duncan’s test). (F–I) Microscopic observations of mature embryo sacs in WT (F), des1 (G), and OE lines (H, I). A, antipodal cell; E, egg cell; P, polar nucleus; S, synergid cell. Scale bars=50 μm. (J) Statistical analysis of normal mature embryo sac formation in WT, des1, and OE plants. Data are means ±SD (n=3) (ovule number: WT 177; des1 162; OE 267). Different letters indicate significant differences (P<0.05; Duncan’s test).
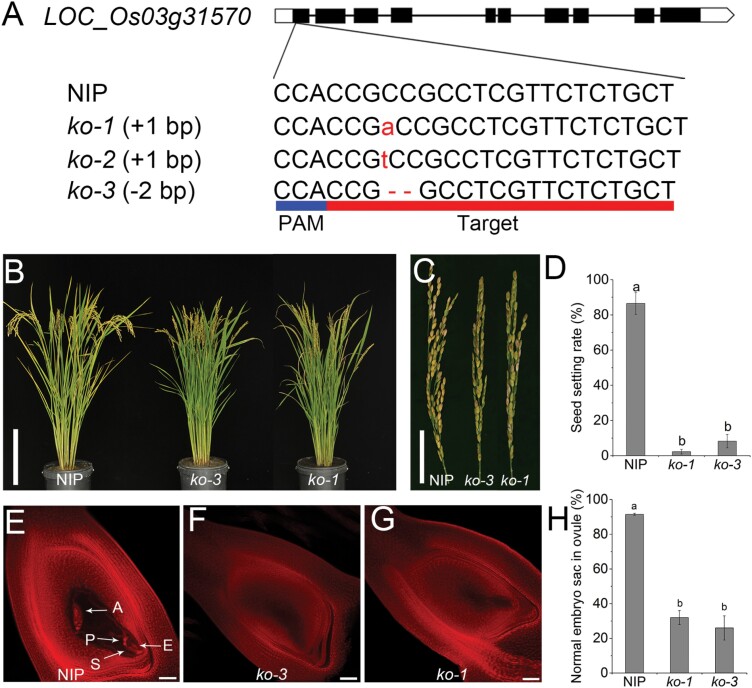
To verify whether the mutation in OsDES1 is responsible for the des1 mutant phenotype, we overexpressed OsDES1 under the control of the maize Ubiquitin1 promoter in the des1 background. As expected, the seed-setting rate in OsDES1-overexpressing plants was substantially increased compared with des1 (Fig. 3C–E). In addition, developmental defects observed in the spikelet, mature embryo sac, anther, and pollen grain were largely rescued in the transgenic plants grown under natural field conditions (Fig. 3F–J; Supplementary Fig. S11A–F). To further confirm that the mutation in OsDES1 is responsible for the mutant phenotype, we used CRISPR/Cas9 to generate five knockout mutant lines in the NIP background (Fig. 4A). Sequencing analysis revealed that these lines harbored three different types of independent homozygous mutations. All of these mutations resulted in predicted translational frame shifts (Fig. 4A). As expected, all knockout transgenic plants had the same phenotype as the des1 mutant, with the characteristic decreased seed-setting rate. The seed-setting rate of the WT (NIP) was ~87%, while both the ko-3 mutant (~8%) and ko-1 mutant (~2%) showed a significantly lower seed-setting rate (Fig. 4B–D). Similarly, ~92% of NIP mature embryo sacs successfully formed eight nuclei, whereas in the ko-3 and ko-1 lines only ~26% and ~32%, respectively, of mature embryo sacs had a normal appearance, and the rest developed into undifferentiated tissue (Fig. 4E–H). In addition, smaller brown panicles, shorter anthers, and abortive pollen grains were observed in the homozygous knockout lines (Supplementary Fig. S12). Together, these results confirmed that LOC_Os03g31570 corresponds to OsDES1, and mutation of this gene resulted in a low seed-setting rate.
Fig. 4.
CRISPR/Cas9 mutation of OsDES1. (A) Three types of mutations detected in the target site in the knockout (ko) lines. (B) Comparison of mature plants of NIP and the ko lines. Scale bar=20 cm. (C) Panicles of NIP and ko plants at maturity. Scale bar=5cm. (D) Seed-setting rate of NIP and ko plants. Data are means ±SD (n=10 plants). Different letters indicate significant differences (P<0.05; Duncan’s test). (E–G) Microscopic observations of mature embryo sacs in NIP (E) and ko (F, G) plants. A, antipodal cell; E, egg cell; P, polar nucleus; S, synergid cell. Scale bars=50 μm. (H) Statistical analysis of the numbers of normal mature embryo sacs in NIP and ko plants. Data are means ±SD (n=3) (ovule number: NIP 140; ko 292). Different letters indicate significant differences (P<0.05; Duncan’s test).
Subcellular localization of OsDES1 protein and expression pattern of the OsDES1 gene
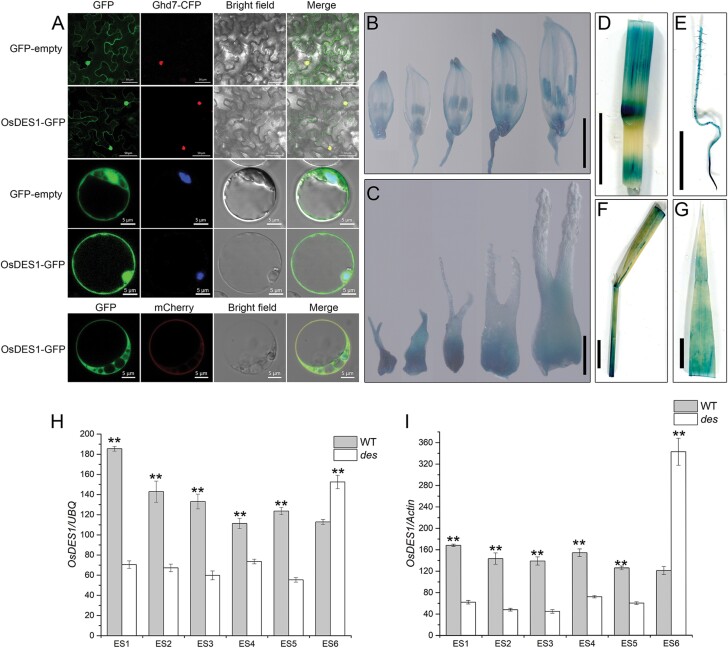
OsDES1 encodes a putative NEMP containing seven putative transmembrane regions. To determine the subcellular localization of OsDES1, we fused the full-length CDS of OsDES1 to the N-terminus of GFP driven by the CaMV35S promoter. In rice leaf protoplasts, GFP signals were clearly detected in the nuclear membrane, nucleus, plasma membrane, and cytoplasm (Fig. 5A). Notably, the ΔOsDES1 and NEMP domain showed a similar subcellular localization pattern to that of OsDES1 in rice leaf protoplasts (Supplementary Fig. S13). These results indicated that the mutation of OsDES1 did not change the subcellular localization of its protein product and the NEMP domain may play a crucial role for the function of OsDES1. The results of these experiments were further confirmed in N. benthamiana leaves, which displayed similar results to the rice leaf protoplasts.
Fig. 5.
Subcellular localization of OsDES1 and expression analysis of the OsDES1 gene in rice tissues. (A) Subcellular localization of OsDES1-GFP fusion protein in N. benthamiana leaf epidermal cells (top two rows) and rice leaf protoplasts (bottom three rows). Ghd7-CFP fusion protein was used as a nuclear marker. The plasma membrane was stained with FM4-64. (B–G) GUS staining of various tissues of proOsDES1-GUS transgenic plants: spikelets (B) and pistils (C) at different developmental stages, stem (D), primary root (E), leaf sheath (F), and leaf (G). The lengths of the spikelets from left to right in (B) and (C) are 2.5–3.0 mm, 3.0–3.4 mm, 3.4–4.5 mm, 4.5–5.6 mm, and 5.6–6.2 mm. Scale bars=50 μm and 5 μm in (A), 2 mm in (B), 200 μm in (C), and 1 cm in (D–G). (H, I) Expression levels of OsDES1 in WT and des1 during embryo sac development. ES1, spikelet lengths 5–5.9 mm; ES2, spikelet lengths 6–6.9 mm; ES3, spikelet lengths 7–7.9 mm; ES4, spikelet lengths 8–9.9 mm; ES5, spikelet lengths >10 mm; ES6, mature spikelets. The UBQ and Actin genes were used as internal controls for the data in (H) and (I), respectively. Data are the means ±SD of three independent biological replicates. Asterisks indicate significant differences (**P<0.01; Student’s t-test).
We then examined whether OsDES1 is transcriptionally expressed in specific tissues or during specific developmental stages in rice. To this end, we generated transgenic plants expressing the OsDES1pro::GUS reporter construct. Our results showed that OsDES1 is expressed in a range of rice tissues. Notably, the expression was strong in anthers (Fig. 5B) and pistils (Fig. 5C) from meiosis to maturity, whereas expression was found to be relatively weak in the culms (Fig. 5D), young roots (Fig. 5E), leaf sheaths (Fig. 5F), and leaves (Fig. 5G). To further elucidate the function of OsDES1 during embryo sac development, we analyzed the expression of OsDES1 in pistils of the WT and des1 using qRT–PCR at different developmental stages. In the megasporocyte (ES1), dyad (ES2), tetrad (ES3), functional megaspore formation (ES4), and mitosis (ES5) stages, the expression of OsDES1 in the WT was significantly greater than that in des1; however, the expression of OsDES1 was obviously lower in the WT at the mature stage (ES6) (Fig. 5H, I), which was consistent with the results of GUS staining (Fig. 5C).
To further elucidate the temporal and spatial expression patterns of OsDES1, we performed RNA in situ hybridization with WT pistil sections. The hybridization signals were detected in whole ovules, including embryo sacs, inner integuments, and outer integuments. As expected, strong signals were observed in the ovule at the megasporocyte stage, tetrad, functional megaspore formation stage, and mature embryo sac stage (Supplementary Fig. S14). The results of in situ hybridization are consistent with those of GUS staining and qRT–PCR, indicating that OsDES1 functions in ovules in reproductive development.
OsDES1 interacts with LOG
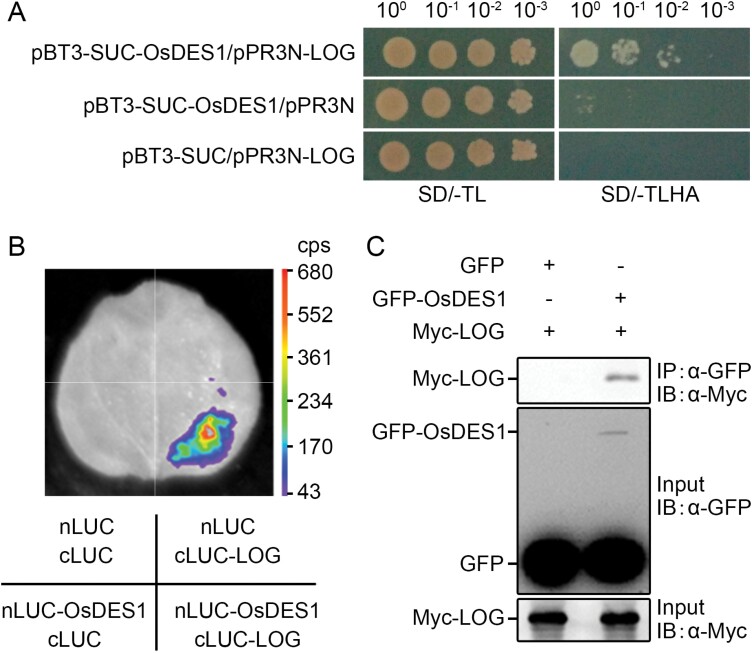
Previous studies showed that the mutation of LOG, a gene encoding a cytokinin-activating enzyme, resulted in similar phenotypic defects to des1 (Kurakawa et al., 2007; Yamaki et al., 2011). To identify potential interaction partners involved in OsDES1-mediated female organ development, we conducted yeast two-hybrid screening for OsDES1-interacting proteins. These assays revealed that LOG interacts with OsDES1 in yeast cells in vitro (Fig. 6A). To further verify this interaction, we carried out split luciferase complementation assays. Leaves of N. benthamiana co-transfected with nLUC-OsDES1 and cLUC-LOG constructs showed significant luciferase activity, whereas the negative control produced no luciferase signal, indicating that OsDES1 interacts with LOG in vivo (Fig. 6B). To further confirm the association between OsDES1 and LOG in planta, we used co-immunoprecipitation analysis to detect their interactions in vivo. We transiently co-expressed 35S:GFP-OsDES1 and 35S:Myc-LOG in N. benthamiana, with 35S:GFP and 35S:Myc-LOG serving as negative controls. As shown in Fig. 6C, we found that Myc-LOG interacts with GFP-OsDES1 but not with GFP in vivo. Thus, these results demonstrate that OsDES1 interacts with LOG both in vivo and in vitro.
Fig. 6.
Physical interaction of the OsDES1 and LOG proteins. (A) Yeast two-hybrid assay to detect the interaction between OsDES1 and LOG. The pPR3N/pBT3-SUC pair was used as the negative control. SD/-TL, Synthetic dropout medium lacking Trp and Leu; SD/-TLHA, synthetic dropout medium lacking Trp, Leu, His, and Ade. (B) Split luciferase complementation assay showing the interaction between OsDES1 and LOG in N. benthamiana. nLUC-OsDES1 and cLUC-LOG were co-expressed in N. benthamiana leaves. Luciferase activity was tested 24 h after infiltration. nLUC and cLUC were used as negative controls. (C) Co-immunoprecipitation assay confirming the interaction between OsDES1 and LOG. 35S:GFP-OsDES1 and 35S:Myc-LOG constructs were co-expressed in N. benthamiana. Proteins were immunoprecipitated (IP) using GFP beads and analyzed by immunoblotting (IB) with anti-Myc and anti-GFP antibodies.
LOG encodes a cytokinin-activating enzyme that acts in bioactive cytokinin synthesis (Kurakawa et al., 2007). Hence, we examined endogenous cytokinin levels in pistils of the WT and des1 at maturity by HPLC, and found that the levels of several cytokinins were significantly higher in the pistils of des1 compared with those of the WT (Supplementary Fig. S15A). We also assessed the expression levels of genes involved in the cytokinin signaling pathway, including cytokinin-response histidine protein kinases (OsHKs), histidine phosphotransfer proteins (OsHPs), and cytokinin response regulators (OsRRs). OsRR1, OsRR4, OsRR9, OsRR10, OsRR16, OsRR19, OsRR20, OsHK3, OsHK4, OsHP1, OsHP2, OsHP3, OsHP4, and OsHP5 were up-regulated in des1 (Supplementary Fig. S15D–G). The expression levels of LOG were significantly reduced in the pistils of des1 compared with the WT (Supplementary Fig. S15B, C). The expression levels of OsDES1 in des1 were significantly increased at the mature embryo stage, and were significantly higher than in the WT (Fig. 5H, I). Hence, we speculated that OsDES1 is involved in the regulation of cytokinin.
Discussion
The seed-setting rate is a major agronomic character that directly contributes to grain yield. Defective female reproductive organs lead to reduced fertility, which is one of the most common reasons for a reduction in the seed-setting rate in rice (Ren et al., 2019; Xu et al., 2020). In this study, we used map-based cloning to isolate a des1 rice mutant that exhibited an extremely low seed-setting rate. Cytological and genetic studies suggested that the low seed set in des1 is mainly caused by abnormal embryo sac development (Figs 2O, 3E, 4D). Similar cases have been reported previously. OsMLH3, which encodes a MutL-homolog 3 protein in rice, positively controls the panicle seed-setting rate by regulating embryo sac development (Mao et al., 2021). A reduced seed-setting rate can also result from different mechanisms. The female sterile variation 1 (fsv1) mutant shows low seed set, which is attributed to non-functional embryo sacs, while the low seed-setting rate of des1 is due to degeneration of the embryo sac and a reduced capacity for fertilization.
In OsDES1-overexpressing plants, the seed-setting rate and fertility were substantially increased compared with des1 (Fig. 3E, J; Supplementary Fig. S11F). However, the overexpression construct was driven by the maize Ubiquitin1 promoter instead of the endogenous promoter, and the relative transcription levels of OsDES1 were significantly higher than in the WT (Supplementary Fig. S11G, H), which may have interfered with seed setting, thus preventing a complete rescue. In our study, seed set was lower than embryo sac fertility in des1 (Figs 1G, 2O). When des1 was used as the maternal parent, varying degrees of reduced seed-setting rates (ranging from 29.3% to 35.4%) were identified. This finding suggests that low seed set in des1 is mainly due to a maternal effect. In addition, when the WT was used as the maternal parent in a cross with des1, we found a lower seed-setting rate than with WT self-crosses (Supplementary Fig. S3). These data suggest that fertilization is abnormal in both des1 and reciprocal crosses. Thus, we can hypothesize that there are other factors influencing seed setting in the des1 mutant besides embryo sac fertility.
Fertilization is a complex and robust process that is the core process in the reproductive development of angiosperms. The key to the success of fertilization is that two sperm cells can individually migrate to the egg cell and polar nuclei for karyogamy, ultimately forming viable seeds (Russell, 1996; Yadegari and Drews, 2004; Skinner and Sundaresan, 2018). Reduction in fertilization ability is another cause of the low seed-setting rate in des1. Approximately 51% of the embryo sacs were normal in des1. However, only ~20% of the normal embryo sacs in des1 were fertilized, compared with 77% in the WT. These findings suggest that both the egg cell and the polar nuclei do not undergo successful fertilization in some des1 normal embryo sacs at 24 h post fertilization and, as a result, seed set is reduced (Fig. 2P–S). Previous studies have shown that the fertilization process is affected by many factors, such as gametogenesis, pollen tube growth in the ovule, pollen tube reception, sperm cell release, and the recognition, activation, and fusion of male and female gametes (Berger, 2011; Dresselhaus et al., 2016; Sankaranarayanan and Higashiyama, 2018; Manrique et al., 2019; Sun et al., 2021). We observed pollen germination on the stigma and pollen tube growth in the ovule at 2 h after pollination in the WT and des1. The pollen tube could arrive at the micropyle at 2 h after pollination in des1, as in the WT (Supplementary Fig. S7; Supplementary Table S2). We speculate that there may be one or more points that could fail after the pollen tubes reach the micropyle, for example, in sperm cell delivery or the recognition, activation, or fusion of the gametes. Further studies are required to address this issue.
Through cytological and genetic studies, we found that sterility in des1 is mainly caused by abnormal development of the embryo sacs. Ultimately, des1 embryo sacs failed to divide or differentiate correctly, resulting in plant sterility (Fig 2A–N). Furthermore, reciprocal cross experiments also indicated that there are functional defects in the female reproductive organ of des1 (Supplementary Fig. S3). The expression of OsDES1 in the pistil was consistent with its functions (Fig. 5C, H, I; Supplementary Fig. S14). Therefore, OsDES1 plays a vital role in the regulation of embryo sac development in rice. Several studies have shown that defects in pollen and embryo sac development lead to sterility in indica–japonica hybrids (Song et al., 2005; Long et al., 2008; Zeng et al., 2009; Yang et al., 2012). Embryo sac fertility and pollen fertility are considered to be the most critical factors affecting spikelet fertility (Song et al., 2005; Zeng et al., 2009). The egg cell and central cell in the embryo sac develop into the embryo and endosperm by fertilization. Studies of the female gametophyte in flowering plants promote understanding of the molecular mechanism for cell specification, cell–cell interaction, and programmed cell death (Heydlauff and Groß-Hardt, 2014; Tekleyohans et al., 2017).
The process of embryo sac development involves the development of the archesporial cell and megaspore mother cell, functional megaspore formation, and gamete cell differentiation. Functional megaspore formation, which is known as the origin of the gametophytic lineage, is essential for embryo sac development (Demesa-Arévalo and Vielle-Calzada, 2013). Disruption of functional megaspore formation may cause female sterility. The osrpa1a, Osmsh4, and fsv1 mutants show defects in the megaspore at the tetrad stage, which lead to failures in functional megaspore formation (Chang et al., 2009; Wang et al., 2016; Mao et al., 2021). Cytological observation has shown that, unlike in the osrpa1a, Osmsh4, and fsv1 mutants, the megasporocyte can give rise to a tetrad megaspore in des1. However, the megaspore at the chalaza degenerated, together with the other three degenerating megaspores nearer the micropylar end, at the functional megaspore formation stage. Ultimately, the selected megaspore was unable to develop into a functional megaspore, which subsequently led to the formation of an undifferentiated embryo sac and female sterility (Fig. 2I–N). Intriguingly, female sterility in des1 is similar to that observed in plants in which DEFECT IN EARLY EMBRYO SAC1 (OsDEES1) has been silenced by RNAi, since both show normal tetrad megaspore and abnormal embryo sac formation (Wang et al., 2012). However, des1 shows defects during functional megaspore formation, whereas the OsDEES1 RNAi plants exhibit normal functional megaspore formation and severe defects in mitosis.
Previous results indicated that LOG regulates the development of the pistil and ovule. LOG activates cytokinin by catalyzing the conversion of inactive cytokinin species to active forms (Kurakawa et al., 2007; Yamaki et al., 2011). Therefore, the log-3 mutant is mainly considered to be a female-sterile mutant (Yamaki et al., 2011). Our results revealed a physical interaction between OsDES1 and LOG in vitro and in vivo (Fig. 6). Furthermore, LOG functions in the cytosol (Kurakawa et al., 2007), and OsDES1 was also detected in the cytoplasm (Fig. 5A). In addition, we found that the des1 and log mutants showed similar phenotypes in female gamete development. Therefore, we speculated that OsDES1 and LOG have a complex interaction in the regulation of cytokinin synthesis, and then both of them participate in the whole developmental process of embryo sacs from initiation to maturity. OsDES1 was also found to be localized to the nuclear membrane and plasma membrane in rice leaf protoplasts (Fig. 5A). This observation implies that OsDES1 might function in other biological pathways in rice reproductive development. Cytokinin is involved in diverse physiological functions, including cell proliferation, differentiation, shoot apical meristem function, seed germination, delayed leaf senescence, and plant immunity (Argueso et al., 2012; Hwang et al., 2012). In Arabidopsis, cytokinin signaling at the chalazal end of the developing embryo sac appears to be necessary for the selection of the functional megaspore. When cytokinin signaling in the sporophyte is disturbed, it leads to abnormalities in the functional megaspore. It has been shown that there is an uneven distribution of cytokinin signaling and biosynthesis in the ovule, particularly in the chalaza during megasporogenesis (Cheng et al., 2013). In maize, cytokinin signaling is not detected in the embryo sac, but in the outer periphery of the antipodal cells (Chettoor and Evans, 2015). Our results showed that the levels of several cytokinins were significantly higher in the mature pistils of des1 compared with the WT (Supplementary Fig. S15A). Hence, we speculate that cytokinin may participate in the regulation of female reproductive organ development. Future studies are needed to determine how cytokinin regulates female reproductive organ development in rice.
OsDES1 encodes a putative NEMP domain-containing protein. The nuclear envelope not only protects the genome from detrimental agents but also governs genome organization (Yang et al., 2017). The NEMP family includes a group of nuclear envelope integral membrane proteins in animals and plants. Nup154 is similar to known nucleoporins, and it is localized both at the nuclear envelope and in the nuclear interior, which further confirms its numerous roles in nuclear functions. In the ovary, Nup154 is required for egg chamber development and oocyte growth (Gigliotti et al., 1998). In Arabidopsis, loss of function of nucleoporin 1 (NUP1) causes defects in both female and male gametogenesis (Bao et al., 2019). Recently, two new homologs of rice OsDES1 (PNET1 and PNET2) have been characterized in Arabidopsis. PLANT NUCLEAR ENVELOPE TRANSMEMBRANE (PNET1) interacts with the nuclear pore complex outer ring complex nucleoporin Nup160 and is required for embryo development. nup160/pnet1 double mutants are embryo lethal and show undeveloped ovules and a reduction in seed set. However, the single pnet1 and nup160 mutants show normal development and seed set (Tang et al., 2020). PNET2 plays essential roles in establishing chromatin architecture and transcription programming. Both pnet2 single and triple mutants showed distinct defects in plant growth and development (Tang et al., 2022). In our study, des1 and the CRISPR/Cas9-based knockout lines had a low seed-setting rate. In addition, OsDES1 localized to the nuclear membrane, consistent with the previous localization of PNET1 and PNET2. The same-origin gene mutation causes diverse phenotypes. These results suggest that the NEMP protein OsDES1 in rice regulates embryo sac development and seed set. Our study provides new insight into the function of a NEMP protein in regulating reproductive development in a monocot, unlike other known and characterized NEMP proteins. In summary, our findings elucidate the essential regulatory mechanism of OsDES1 in embryo sac and pollen development, which may contribute to applications in rice production and the improvement of rice yield.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Phenotypic analysis in WT and des1.
Fig. S2. Scanning electron microscopy observation of the mature pistils, anthers, and pollen grains in WT and des1.
Fig. S3. Statistical data of the seed-setting rate of the reciprocal crosses.
Fig. S4. Paraffin section analysis of embryo sac development in WT and des1.
Fig. S5. Microscopic observations of mature embryo sacs in ZH8015 and F1 plants.
Fig. S6. In vitro pollen germination assay.
Fig. S7. Pollen germination on the stigma and pollen tube growth in WT and des1.
Fig. S8. Male gametogenesis in WT and des1 shown by acetocarmine and DAPI staining.
Fig. S9. Transverse sections of WT and des1 anthers at various developmental stages.
Fig. S10. Transmission electron microscopy observations of mature anthers in WT and des1.
Fig. S11. Anthers and pollen grains of WT, des1, and OsDES1-overexpressing plants.
Fig. S12. CRISPR/Cas9 characterization of OsDES1.
Fig. S13. Subcellular localization of the ΔOsDES1-GFP and NEMP-GFP fusion proteins.
Fig. S14. In situ analysis of OsDES1 expression in longitudinal sections of the embryo sacs.
Fig. S15. Cytokinin determination and cytokinin-related gene expression.
Table S1. Oligonucleotide primers used in this study.
Table S2. Statistical analysis of pollen tube growth observed in the ovule at 2 h after pollination in WT and des1.
Acknowledgements
We acknowledge Dr Zheng Wang at the Division of Agriculture and Natural Resources, University of California, for revising the manuscript. We thank Yunqin Li at the Analysis Center of Agrobiology and Environmental Sciences (ACAES), Zhejiang University, and the Public Laboratory of China National Rice Research Institute for helping with the whole-mount stain-clearing laser scanning confocal microscopy measurements. Finally, we thank Nianhang Rong, Junying Li, and Weilan Wang at ACAES, Zhejiang University, for helping with the scanning and transmission electron microscopy.
Contributor Information
Xia Hu, China National Rice Research Institute, Hangzhou, Zhejiang, 311400, China.
Ping Yu, China National Rice Research Institute, Hangzhou, Zhejiang, 311400, China.
Yingxin Zhang, China National Rice Research Institute, Hangzhou, Zhejiang, 311400, China.
Zhiqiang Gao, Gannan Normal University, Ganzhou, Jiangxi, 341000, China.
Bin Sun, Shanghai Academy of Agricultural Sciences, Shanghai, 201403, China.
Weixun Wu, China National Rice Research Institute, Hangzhou, Zhejiang, 311400, China.
Chenwei Deng, Zhoukou Academy of Agricultural Sciences, Zhoukou, Henan, 466001, China.
Adil Abbas, China National Rice Research Institute, Hangzhou, Zhejiang, 311400, China.
Yongbo Hong, China National Rice Research Institute, Hangzhou, Zhejiang, 311400, China.
Lianping Sun, China National Rice Research Institute, Hangzhou, Zhejiang, 311400, China.
Qunen Liu, China National Rice Research Institute, Hangzhou, Zhejiang, 311400, China.
Pao Xue, China National Rice Research Institute, Hangzhou, Zhejiang, 311400, China.
Beifang Wang, China National Rice Research Institute, Hangzhou, Zhejiang, 311400, China.
Xiaodeng Zhan, China National Rice Research Institute, Hangzhou, Zhejiang, 311400, China.
Liyong Cao, China National Rice Research Institute, Hangzhou, Zhejiang, 311400, China.
Shihua Cheng, China National Rice Research Institute, Hangzhou, Zhejiang, 311400, China.
Zoe Wilson, University of Nottingham, UK.
Author contributions
SC and LC conceived and supervised the project; XH designed the research, performed most of the experiments, and wrote the manuscript; XH and PY analyzed the data; YZ constructed the mutant plants; ZG, BS, WW, CD, AA, YH, LS, PX, and BW performed the experiments; PY, YZ, WW, QL, and XZ revised the manuscript and gave constructive comments on the experiments.
Conflict of interest
The authors declare no conflicts of interest.
Funding
This work was supported by grants from the Zhejiang Province Key Research and Development Program of China (2021C02056), Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Science (CAAS-ASTIP-2013-CNRRI), the earmarked fund for China Agriculture Research System (CARS-01), and Central Public Welfare Research Institutions (CPSIBRF-CNRRI-202102).
Data availability
All data supporting the findings of this study are available within the paper and within its supplementary data published online.
References
- Argueso CT, Ferreira FJ, Epple P, To JPC, Hutchison CE, Schaller GE, Dangl JL, Kieber JJ.. 2012. Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLoS Genetics 8, e1002448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi A, Paul P, Kumar S, Verma SK, Prasad R, Dhaliwal HS.. 2012. Abnormal endosperm development causes female sterility in rice insertional mutant OsAPC6. Plant Science 183, 167–174. [DOI] [PubMed] [Google Scholar]
- Bao SG, Shen GS, Li GC, Liu ZK, Arif M, Wei QQ, Men SZ.. 2019. The Arabidopsis nucleoporin NUP1 is essential for megasporogenesis and early stages of pollen development. Plant Cell Reports 38, 59–74. [DOI] [PubMed] [Google Scholar]
- Bencivenga S, Simonini S, Benkova E, Colombo L.. 2012. The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. The Plant Cell 24, 2886–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F. 2011. Imaging fertilization in flowering plants, not so abominable after all. Journal of Experimental Botany 62, 1651–1658. [DOI] [PubMed] [Google Scholar]
- Boateng KA, Yang X, Dong F, Owen HA, Makaroff CA.. 2008. SWI1 is required for meiotic chromosome remodeling events. Molecular Plant 1, 620–633. [DOI] [PubMed] [Google Scholar]
- Chang YX, Gong L, Yuan WY, Li XW, Chen GX, Li XH, Zhang QF, Wu CY.. 2009. Replication protein A (RPA1a) is required for meiotic and somatic DNA repair but is dispensable for DNA replication and homologous recombination in rice. Plant Physiology 151, 2162–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Li HJ, Shi DQ, Yuan L, Liu J, Sreenivasan R, Baskar R, Grossniklaus U, Yang WC.. 2007. The central cell plays a critical role in pollen tube guidance in Arabidopsis. The Plant Cell 19, 3563–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Mathews DE, Schaller GE, Kieber JJ.. 2013. Cytokinin-dependent specification of the functional megaspore in the Arabidopsis female gametophyte. The Plant Journal 73, 929–940. [DOI] [PubMed] [Google Scholar]
- Chettoor AM, Evans MMS.. 2015. Correlation between a loss of auxin signaling and a loss of proliferation in maize antipodal cells. Frontiers in Plant Science 6, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CA, Subramanian S, Drews GN.. 1998. Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Developmental Biology 202, 136–151. [DOI] [PubMed] [Google Scholar]
- Demesa-Arévalo E, Vielle-Calzada JP.. 2013. The classical arabinogalactan protein AGP18 mediates megaspore selection in Arabidopsis. The Plant Cell 25, 1274–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T, Sprunck S, Wessel GM.. 2016. Fertilization mechanisms in flowering plants. Current Biology 26, R125–R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Koltunow AM.. 2011. The female gametophyte. The Arabidopsis Book 9, e0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Lee D, Christensen CA.. 1998. Genetic analysis of female gametophyte development and function. The Plant Cell 10, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigliotti S, Callaini G, Andone S, Riparbelli MG, Pernas-Alonso R, Hoffmann G, Graziani F, Malva C.. 1998. Nup154, a new Drosophila gene essential for male and female gametogenesis is related to the Nup155 vertebrate nucleoporin gene. Journal of Cell Biology 142, 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng YQ, Wu CY, Long Y, et al. 2018. OsALMT7 maintains panicle size and grain yield in rice by mediating malate transport. The Plant Cell 30, 889–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydlauff J, Groß-Hardt R.. 2014. Love is a battlefield: programmed cell death during fertilization. Journal of Experimental Botany 65, 1323–1330. [DOI] [PubMed] [Google Scholar]
- Higashiyama T, Yang WC.. 2017. Gametophytic pollen tube guidance: attractant peptides, gametic controls, and receptors. Plant Physiology 173, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Peng X, Sun MX.. 2017. OsGCD1 is essential for rice fertility and required for embryo dorsal-ventral pattern formation and endosperm development. New Phytologist 215, 1039–1058. [DOI] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Muller B.. 2012. Cytokinin signaling setworks. Annual Review of Plant Biology 63, 353–380. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. 1989. The GUS reporter gene system. Nature 342, 837–838. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Borevitz JO, Preuss D.. 2007. Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genetics 3, 1848–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou YJ, Chang YX, Li H, Xiao JH, Wang SP.. 2012. The rice RAD51C gene is required for the meiosis of both female and male gametocytes and the DNA repair of somatic cells. Journal of Experimental Botany 63, 5323–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H, Hata S.. 1993. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Molecular and General Genetics 238, 106–119. [DOI] [PubMed] [Google Scholar]
- Kumar M, Basha PO, Puri A, Rajpurohit D, Randhawa GS, Sharma TR, Dhaliwal HS.. 2010. A candidate gene OsAPC6 of anaphase-promoting complex of rice identified through T-DNA insertion. Functional & Integrative Genomics 10, 349–358. [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J.. 2007. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. [DOI] [PubMed] [Google Scholar]
- Li DY, Huang ZY, Song SH, et al. 2016. Integrated analysis of phenome, genome, and transcriptome of hybrid rice uncovered multiple heterosis-related loci for yield increase. Proceedings of the National Academy of Sciences, USA 113, E6026–E6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SC, Li WB, Huang B, et al. 2013. Natural variation in PTB1 regulates rice seed setting rate by controlling pollen tube growth. Nature Communication 4, 2793. [DOI] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, et al. 2006. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. The Plant Cell 18, 2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HJ, Zhu SS, Zhang MX, Wang T, Liang L, Xue Y, Shi DQ, Liu J, Yang WC.. 2015. Arabidopsis CBP1 is a novel regulator of transcription initiation in central cell-mediated pollen tube guidance. The Plant Cell 27, 2880–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber D, Lora J, Schrempp S, Lenhard M, Laux T.. 2011. Arabidopsis WIH1 and WIH2 genes act in the transition from somatic to reproductive cell fate. Current Biology 21, 1009–1017. [DOI] [PubMed] [Google Scholar]
- Liu CZ, Xue ZH, Tang D, Shen Y, Shi WQ, Ren LJ, Du GJ, Li YF, Cheng ZK.. 2018. Ornithine δ-aminotransferase is critical for floret development and seed setting through mediating nitrogen reutilization in rice. The Plant Journal 96, 842–854. [DOI] [PubMed] [Google Scholar]
- Long YM, Zhao LF, Niu BX, et al. 2008. Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proceedings of the National Academy of Sciences, USA 105, 18871–18876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JY, Wang CL, Wang HY, et al. 2020. OsMFS1/OsHOP2 complex participates in rice male and female development. Frontiers in Plant Science 11, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrique S, Friel J, Gramazio P, Hasing T, Ezquer I, Bombarely A.. 2019. Genetic insights into the modification of the pre-fertilization mechanisms during plant domestication. Journal of Experimental Botany 70, 3007–3019. [DOI] [PubMed] [Google Scholar]
- Mao BG, Zheng WJ, Huang Z, et al. 2021. Rice MutLγ, the MLH1–LH3 heterodimer, participates in the formation of type I crossovers and regulation of embryo sac fertility. Plant Biotechnology Journal 19, 1443–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng JG, Liang L, Jia PF, Wang YC, Li HJ, Yang WC.. 2020. Integration of ovular signals and exocytosis of a Ca2+ channel by MLOs in pollen tube guidance. Nature Plants 6, 143–153. [DOI] [PubMed] [Google Scholar]
- Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ.. 2013. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Research 23, 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K. 2018. Be my baby: patterning toward plant germ cells. Current Opinion in Plant Biology 41, 110–115. [DOI] [PubMed] [Google Scholar]
- Nonomura KI, Nakano M, Fukuda T, Eiguchi M, Miyao A, Hirochika H, Kurata N.. 2004a. The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. The Plant Cell 16, 1008–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura KI, Nakano M, Murata K, Miyoshi K, Eiguchi M, Miyao A, Hirochika H, Kurata N.. 2004b. An insertional mutation in the rice PAIR2 gene, the ortholog of Arabidopsis ASY1, results in a defect in homologous chromosome pairing during meiosis. Molecular Genetics and Genomics 271, 121–129. [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Sundaresan V.. 2007. Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. The Plant Cell 19, 3578–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischke MS, Jones LG, Otsuga D, Fernandez DE, Drews GN, Sussman MR.. 2002. An Arabidopsis histidine kinase is essential for megagametogenesis. Proceedings of the National Academy of Sciences, USA 99, 15800–15805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Zhao LH, Skaggs MI, et al. 2014. ACTIN-RELATED PROTEIN6 regulates female meiosis by modulating meiotic gene expression in Arabidopsis. The Plant Cell 26, 1612–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiger DS, Drews GN.. 2013. MYB64 and MYB119 are required for cellularization and differentiation during female gametogenesis in Arabidopsis thaliana. PLoS Genetics 9, e1003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Park SS, Ray A.. 1997. Pollen tube guidance by the female gametophyte. Development 124, 2489–2498. [DOI] [PubMed] [Google Scholar]
- Reiser L, Fischer RL.. 1993. The ovule and the embryo sac. The Plant Cell 5, 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Chen D, Li WJ, et al. 2019. OsSHOC1 and OsPTD1 are essential for crossover formation during rice meiosis. The Plant Journal 98, 315–328. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers K, Pruitt RE, Gasser CS.. 1992. Ovule development in wild-type Arabidopsis and two female-sterile mutants. The Plant Cell 4, 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SD. 1996. Attraction and transport of male gametes for fertilization. Sexual Plant Reproduction 9, 337–342. [Google Scholar]
- Sankaranarayanan S, Higashiyama T.. 2018. Capacitation in plant and animal fertilization. Trends in Plant Science 23, 129–139. [DOI] [PubMed] [Google Scholar]
- Sheridan WF, Avalkina NA, Shamrov II, Batygina TB, Golubovskaya IN.. 1996. The mac1 gene: controlling the commitment to the meiotic pathway in maize. Genetics 142, 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner DJ, Sundaresan V.. 2018. Recent advances in understanding female gametophyte development [version 1; peer review: 2 approved]. F1000Research 7, 804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Qiu SQ, Xu CG, Li XH, Zhang QF.. 2005. Genetic dissection of embryo sac fertility, pollen fertility, and their contributions to spikelet fertility of intersubspecific hybrids in rice. Theoretical and Applied Genetics 110, 205–211. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wang X, Pan L, Xie F, Dai B, Sun MX, Peng XB.. 2021. Plant egg cell fate determination depends on its exact position in female gametophyte. Proceedings of the National Academy of Sciences, USA 118, e2017488118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Dong QL, Wang TY, Gong L, Gu YN.. 2022. PNET2 is a component of the plant nuclear lamina and is required for proper genome organization and activity. Developmental Cell 57, 19–31.e6. [DOI] [PubMed] [Google Scholar]
- Tang Y, Huang A, Gu YN.. 2020. Global profiling of plant nuclear membrane proteome in Arabidopsis. Nature Plants 6, 838–847. [DOI] [PubMed] [Google Scholar]
- Tekleyohans DG, Nakel T, Groß-Hardt R.. 2017. Patterning the female gametophyte of flowering plants. Plant Physiology 173, 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D.. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal 33, 949–956. [DOI] [PubMed] [Google Scholar]
- Wang N, Huang HJ, Ren ST, Li JJ, Sun Y, Sun DY, Zhang SQ.. 2012. The rice wall-associated receptor-like kinase gene OsDEES1 plays a role in female gametophyte development. Plant Physiology 160, 696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TK, Li YX, Song SF, Qiu MD, Zhang LC, Li CX, Dong H, Li L, Wang J, Li L.. 2021. EMBRYO SAC DEVELOPMENT 1 affects seed setting rate in rice by controlling embryo sac development. Plant Physiology 186, 1060–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CL, Wang Y, Cheng ZJ, et al. 2016. The role of OsMSH4 in male and female gamete development in rice meiosis. Journal of Experimental Botany 67, 1447–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Wang SZ, Hu K, et al. 2018. The kinase OsCPK4 regulates a buffering mechanism that fine-tunes innate immunity. Plant Physiology 176, 1835–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wang FQ, Chen ZH, Wang J, Li WQ, Fan FJ, Tao YJ, Jiang YJ, Zhu QH, Yang J.. 2020. CRISPR/Cas9-targeted mutagenesis of the OsROS1 gene induces pollen and embryo sac defects in rice. Plant Biotechnology Journal 18, 1999–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Yang J, Wang YH, et al. 2017. OsCNGC13 promotes seed-setting rate by facilitating pollen tube growth in stylar tissues. PLoS Genetics 13, e1006906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R, Drews GN.. 2004. Female gametophyte development. The Plant Cell 16, S133–S141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki S, Nagato Y, Kurata N, Nonomura KI.. 2011. Ovule is a lateral organ finally differentiated from the terminating floral meristem in rice. Developmental Biology 351, 208–216. [DOI] [PubMed] [Google Scholar]
- Yang HJ, Iwamoto M, Hiraoka Y, Haraguchi T.. 2017. Function of nuclear membrane proteins in shaping the nuclear envelope integrity during closed mitosis. Journal of Biochemistry 161, 471–477. [DOI] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V.. 1999. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes & Development 13, 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Zhao XB, Cheng K, et al. 2012. A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 337, 1336–1340. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J.. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu XW, Zhao ZG, Zheng XM, et al. 2018. A selfish genetic element confers non-Mendelian inheritance in rice. Science 360, 1130–1132. [DOI] [PubMed] [Google Scholar]
- Yuan WY, Li XW, Chang YX, Wen RY, Chen GX, Zhang QF, Wu CY.. 2009. Mutation of the rice gene PAIR3 results in lack of bivalent formation in meiosis. The Plant Journal 59, 303–315. [DOI] [PubMed] [Google Scholar]
- Zafar SA, Patil SB, Uzair M, et al. 2019. DEGENERATED PANICLE AND PARTIAL STERILITY 1 (DPS1) encodes a cystathionine β-synthase domain containing protein required for anther cuticle and panicle development in rice. New Phytologist 225, 356–375. [DOI] [PubMed] [Google Scholar]
- Zeng YX, Hu CY, Lu YG, Li JQ, Liu XD.. 2009. Abnormalities occurring during female gametophyte development result in the diversity of abnormal embryo sacs and leads to abnormal fertilization in indica/japonica hybrids in rice. Journal of Integrative Plant Biology 51, 3–12. [DOI] [PubMed] [Google Scholar]
- Zhang DB, Luo X, Zhu L.. 2011. Cytological analysis and genetic control of rice anther development. Journal of Genetics and Genomics 38, 379–390. [DOI] [PubMed] [Google Scholar]
- Zhang K, Song Q, Wei Q, Wang CC, Zhang LW, Xu WY, Su Z.. 2016. Down-regulation of OsSPX1 caused semi-male sterility, resulting in reduction of grain yield in rice. Plant Biotechnology Journal 14, 1661–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DB, Wilson ZA.. 2009. Stamen specification and anther development in rice. Chinese Science Bulletin 54, 2342–2353. [Google Scholar]
- Zhao ZG, Jiang L, Zhang WW, Yu CY, Zhu SS, Xie K, Tian H, Liu LL, Ikehashi H, Wan JM.. 2007. Fine mapping of S31, a gene responsible for hybrid embryo-sac abortion in rice (Oryza sativa L.). Planta 226, 1087–1096. [DOI] [PubMed] [Google Scholar]
- Zhou SR, Wang Y, Li WC, et al. 2011. Pollen semi-sterility1 encodes a kinesin-1-like protein important for male meiosis, anther dehiscence, and fertility in rice. The Plant Cell 23, 111–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo JR, Li JY.. 2014. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annual Review of Genetics 48, 99–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary data published online.