Summary
Fragile X syndrome (FXS), the most prevalent heritable form of intellectual disability, is caused by the transcriptional silencing of the FMR1 gene. While neuronal contribution to FXS has been extensively studied in both animal and human-based models of FXS, the roles of astrocytes, a type of glial cells in the brain, are largely unknown. Here, we generated a human-based FXS model via differentiation of astrocytes from human-induced pluripotent stem cells (hiPSCs) and human embryonic stem cells (hESCs) and characterized their development, function, and proteomic profiles. We identified shortened cell cycle, enhanced Ca2+ signaling, impaired sterol biosynthesis and pervasive alterations in the proteome of FXS astrocytes. Our work identified astrocytic impairments that could contribute to the pathogenesis of FXS and highlight astrocytes as a novel therapeutic target for FXS treatment.
Introduction
Fragile X syndrome (FXS), the most common form of inherited intellectual disability, and a condition often associated with autism spectrum disorder, results from mutations in the fragile X messenger ribonucleoprotein 1 (FMR1) gene (Pieretti et al. 1991, Wang et al. 2010). The main function of FMRP is regulation of mRNA translation (Corbin et al. 1997, Darnell et al. 2011, Richter and Zhao 2021). While the contribution of neuronal pathogenesis to FXS has been well studied, the role of astrocytes, a type of glial cells in the brain (Verkhratsky and Nedergaard 2018, Khakh and Deneen 2019, Bayraktar et al. 2020) that also express FMRP (Pacey and Doering 2007, Higashimori et al. 2013), is less well understood. Astrocytes, unlike neurons, are not electrically excitable and their participation in information transmission is an ongoing area of research. They regulate multiple aspects of brain function, including modulation of neuronal development and synaptic activity, regulation of ionic homeostasis, recycling of neurotransmitters, and maintenance of the blood-brain barrier (Verkhratsky and Nedergaard 2018). Thus, impairments in astrocyte function could exert a profound influence on the homeostasis of the brain, contributing to impairments in neurodevelopmental and neuropsychiatric disorders (Molofsky et al. 2012, Pekny et al. 2016, de Majo et al. 2020). Indeed, the absence of FMRP in astrocytes was found to contribute to the cellular and behavioral impairments in Fmr1 mutant animal models of FXS (Jacobs et al. 2010, Higashimori et al. 2013, Higashimori et al. 2016, Hodges et al. 2016, Reynolds et al. 2021). While these results based on the animal models suggest a critical contribution of astrocytes to the pathology of FXS, it remains unknown whether analogous astrocytic impairments are manifested in the FXS human brain.
The use of patient-derived induced pluripotent stem cells (iPSC) and FMR1 KO human embryonic stem cells (hESCs) provide an opportunity to understand the contribution of astrocytes to FXS (Thomson et al. 1998, Takahashi et al. 2007). While FXS and FMR1 KO human stem cell-derived neural progenitor cells (NPCs) and neurons have been increasingly described (Eiges et al. 2007, Urbach et al. 2010, Telias et al. 2013, Doers et al. 2014, Gerhardt et al. 2014, Telias et al. 2015, Achuta et al. 2018, Sunamura et al. 2018, Zhang et al. 2018, Brighi et al. 2021, Kang et al. 2021, Raj et al. 2021), the characterization of astrocytes derived from FXS human stem cells remains incomplete. Here, we differentiated FXS patient-derived iPSCs, FMR1 KO hESCs and isogenic control lines into astrocytes and characterized their properties in vitro. We found that patient-derived FXS astrocytes exhibit altered cell cycle dynamics with shortened S-phase length and enhanced expression of cyclin D1, a regulator in the G1/S checkpoint. Compared to typical controls, we found an increased number of Layer I cortical astrocytes in postmortem brain tissue from FXS patients. Functionally, we observed that while glutamate uptake was not impaired, patient-derived FXS astrocytes exhibited enhanced ATP-induced Ca2+ signaling. Proteomic profiling revealed pervasive alterations and identified multiple dysregulated pathways in FXS human astrocytes including impaired sterol biosynthesis. Our study, one of the first to model and characterize FXS astrocytes using human stem cells, demonstrated multiple intrinsic impairments in human FXS astrocytes which could be contributing to the pathology of FXS and highlights astrocytes and corresponding alterations as therapeutic targets in FXS treatment.
Experimental Procedures
Human Stem Cells
The FX11-9u (RRID: CVCL_EJ77), FX11-7 (RRID: CVCL_EJ76) hiPSC lines, and WA01 (RRID: CVCL_9771), H1-FMR1-KO hESC lines were obtained from WiCell. The FX11-9u and FX11-7 hiPSC lines were derived from the same FXS patient. The FX11-9u line retained the FMRP expression due to the mosaicism of CGG repeats during reprogramming (Doers et al. 2014). The H1-FMR1-KO (abbreviated as FMR1-KO) was engineered by CRISPR/Cas9 targeting exon 3 of the FMR1 gene in WA01 (Utami et al. 2020). The C1-2 and FXS hiPSC lines are in an isogenic pair from the same FXS patient. They were kindly provided by Dr. Stephen Warren at Emory University. The SC153 and SC135 hiPSC lines were generated by Dr. Philip Schwartz at Children’s Hospital of Orange County Research Institute and kindly provided by Dr. Lu Chen at Stanford University. The SC153 and SC153-N2P6 (abbreviated as N2P6) hiPSC lines were from the same FXS patient (Brick et al. 2014). The C1-2 and SC153 N2P6 hiPSC line were edited by the CRISPR/Cas9 system to restore the normal expression level of FMR1. For the proteomic and sterol analysis, we included the hESCs line WA01 and the isogenic line H1-FMR1-KO (Agulhon et al. 2010, Agarwal et al. 2017). All lines used in this study were derived from a single clone per line. Additional information about the cell lines is found in Table 1.
Table 1.
Information about hiPSC and hESC lines used in the study.
| ID | Diagnosis of source | Age of source | Gender | CGG repeats | Phenotype |
|---|---|---|---|---|---|
| FX11-7 | FXS | 7 years | male | > 435 | Full mutation |
| FX11-9u | FXS | 7 years | male | 114 | Pre-mutation (isogenic to FX11-7) |
| FXS | FXS | unknown | male | > 200 | Full mutation |
| C1-2 | FXS | unknown | male | < 40 | Control (isogenic to FXS) |
| SC135 | FXS and autism | 20 years | male | 450 | Full mutation |
| SC153 | FXS and autism | 23 years | male | > 350 | Full mutation |
| N2P6 | FXS and autism | 23 years | male | < 200 | Control (isogenic to SC153) |
| FMR1-KO | Normal | embryo | male | - | Full mutation |
| WA01 | Normal | embryo | male | < 200 | Control (isogenic to FMR1-KO) |
Human postmortem brain tissue
Human postmortem brain tissue of male FXS patients and healthy controls was acquired from the FXS/FXTAS Brain Repository (UC Davis). Tissue from Brodmann area 3 was embedded in optimal cutting temperature compound and sectioned at 14 μm. The donor IDs are as follows: UCD 14-15 11, UCD 18-08 16, UCD 14-12, UCD 15-07 18, UCD 14-01 20, UCD 18-07 17 from non-FXS individuals; 1061-09-JB 13, 1013-10-SK 18, 1031-08-GP 13, 1033-08-WS 10, 1007-18-RF 18, 1001-18-LO 13, 1031-09-LZ 9, 1005-19-JC 13 from FXS patients. The age of all donors is in the range of 50–70 years old.
Maintenance and expansion of stem cells
Human iPSCs and ESCs were cultured with feeder-free maintenance medium mTeSR™ Plus (STEMCELL Technologies, 100-0276) on petri dishes coated with Matrigel (Corning, 354277). ACCUTASE™ (STEMCELL Technologies, 07920) was used to detach stem cells during passaging. When plating the stem cells, Y-27632 dihydrochloride solution (10mM, Tocris, 1254), a selective p160 ROCK inhibitor, was added to the culture medium to optimize cell growth and viability (Ishizaki et al. 2000). mFreSR™ (STEMCELL Technologies, 05855) was used for the cryopreservation of human iPS and ES cells, which were then stored at −80 °C overnight and transferred to a liquid nitrogen tank for long-term storage. The age of human iPS and ES cells was recorded as the passage number.
Differentiation of stem cells to astrocytes
We modified a xeno-free, chemically defined astrocyte differentiation method that has been previously described (Chen et al., 2014). Each stem cell line, was independently differentiated into astroyctes 2–3 times. Briefly, colonies of hiPSCs and hESCs were split manually after treating with Dispase (1 U/ml, STEMCELL Technologies, 07923) to generate embryoid bodies. Embryoid bodies were grown in suspension culture in DME/F-12 (Hyclone, SH3002301), with 1× N2 supplement (Invitrogen, 17502048) and Noggin (40 ng/ml, PeproTech, 120-10C) for 3–5 days. Embryoid bodies were then plated on Petri dishes coated with growth factor reduced Matrigel (Corning, 354230) in DME/F-12, 1× N2 supplement, and laminin (1 μg/ml, Invitrogen, 2317-015). NPCs formed neural rosettes after growing for another 1 week. Manually isolated NPCs were plated on Petri dishes and expanded as neurospheres in suspension culture for 1 week in the presence of NPC medium, composed of DME/F-12 and Neurobasal medium (Invitrogen, 21103-049) in a 1:1 ratio, supplemented with 1× N2, 1× B27-RA (minus retinyl acetate, Invitrogen, 12587-010), and FGF2 (20 ng/ml, PeproTech, 100-18B). To differentiate NPCs to immature astrocytes, neurospheres were dissociated into single cells and were cultured on dishes coated with growth factor reduced Matrigel in astrocyte differentiation medium, consisting of DME/F-12, 1× N2 supplement, 1× B27-RA supplement, BMP4 (10 ng/ml, Peprotech, 120-05ET), and FGF2 (20 ng/ml). The age of immature astrocytes was calculated as days (D) in culture with the first day in astrocyte differentiation medium being “immature D0”.
To acquire mature astrocytes, immature D25 astrocytes were transferred to maturation medium consisting of DME/F-12, 1× N2 supplement, 1× B27-RA supplement, FGF1 (50 ng/ml, Peprotech, 100-17A), LIF (10 ng/ml, Peprotech, 300-05), CNTF (10 ng/ml, Peprotech, 450-13). After 3 weeks, mature astrocytes were used for Ca2+ imaging, glutamate uptake assay, and proteomic analysis. The age of mature astrocytes was recorded as days (D) in culture with the first day in maturation medium being “mature D0”. For hiPSCs, hESCs, embryoid bodies, and neurospheres, culture medium was changed every day. For neural rosettes and astrocytes, the medium was changed every other day.
Isolation and culture of mouse cortical astrocytes
Primary astrocyte cultures were prepared from cortices of Fmr1 KO and wild type (WT) mice on postnatal days 1–3. Each culture was prepared from the cortex of one mouse. Pups were euthanized with an overdose of isoflurane and buried in ice until dissection of the cortex. Following dissection, the cortex was placed in ice-cold Dulbecco’s phosphate buffered saline (DPBS, Gibco, 14190-144). Tissue was minced with sharp forceps, transferred to 0.05 % trypsin (Worthington, LS003707) in DPBS and incubated at 37 °C for 20 minutes with frequent agitation. Deoxyribonuclease (final concentration: 0.05 %, Sigma, DN25) was added to facilitate dissociation of tissue. Tissue was triturated twice, first with a 5 ¾ inch disposable Pasteur pipet (Fisherbrand, 13-678-20B) then with a 9 inch disposable Pasteur pipet (Fisherbrand, 13-678-20C). Complete astrocyte medium, equal to the volume of trypsin, was added to the cell suspension. Complete astrocyte medium consisted of Dulbecco’s Modified Eagle Medium (Gibco, 11965-092) with 10 % heat-inactivated fetal bovine serum (HI-FBS, Gibco, 16140-063), 1X penicillin/streptomycin (ThermoFisher Scientific, 15140-122), 1X GlutaMAX (Gibco, 35050-061), 1 mM oxaloacetate (Sigma, O7753), 0.45 mM pyruvate (Sigma, P2256), 0.2 U/mL insulin (Sigma, I6634), and 0.1 mM L-leucine methyl ester (Sigma, L1002) to induce microglial apoptosis. The suspension was filtered through a 70 μm cell strainer. Cells were collected by centrifuging at 1200 revolutions per minute (RPM) for 5 minutes at room temperature (RT), resuspended in complete astrocyte medium and plated in 25 cm2 flasks. Cells were passaged when they reached 80–90 % confluence. Before passaging, flasks were shaken at 150 RPM overnight to dislodge any remaining microglia. After the first passage, cells were maintained in 75 cm2 flasks until they were plated in dishes for experiments. After three passages, cells were plated at a density of 40,000 cells on 12 mm glass coverslips for cell cycle analysis.
Flow cytometry
Immature D16 hiPSC-astrocytes were plated on 10 cm dishes with a density of 500,000/dish. On the day of the experiment, immature D20 astrocytes were collected using TrypLE™ Express (Gibco, 12605-028) and prepared as a single cell suspension (1×107 cells/mL) with flow cytometry staining buffer (10 % bovine serum albumin (BSA) in PBS with 0.1 % sodium azide). The cell suspension was aliquoted to 100 μL per tube (Falcon, 352054) and incubated with a LIVE-DEAD™ Fixable Blue Dead Cell Stain Kit (Invitrogen, L34961) and 1 μL of mouse anti-CD44 antibody (Abcam, ab6124) per tube for 30 minutes on ice. Cells were washed with PBS by centrifuging at 500 × g for 5 minutes at 4°C. Cell pellets were resuspended in 100 μL of staining buffer containing 1 μL of goat anti-mouse Alexa Fluor 594 secondary antibody (Thermo Fisher Scientific, A-11005) and incubated for 30 minutes at RT, followed by three washes with PBS by centrifuging at 500 × g for 5 minutes at 4°C. Cells were resuspended in 1 % BSA in PBS before running flow cytometry. The flow cytometry BD LSR II Green (BD Biosciences) was used to detect CD44 labeling at 605 nm excitation wavelength and cell viability at 450 nm excitation wavelength. Forward scatter, side scatter and doublet discrimination were applied to screen for single astrocytes and assess the percentage of live and dead cells in the sample.
Immunocytochemistry
Immature D19 and mature D19 hiPSC- and hESC-astrocytes were plated on growth factor reduced Matrigel-coated 12 mm coverslips in 24-well plates with a density of 500,000 cells/coverslip. After 24 hours, cells were fixed with 4 % paraformaldehyde (PFA) in PBS, permeabilized with 0.1 % Triton X-100 (BP151-100) in PBS for 5 minutes and incubated with blocking buffer (5 % normal goat serum (NGS) and 0.1 % Triton X-100 in PBS) for 1 hour at RT. Coverslips were incubated with primary antibodies (in blocking buffer) overnight at 4 °C. Following three washes in PBS for 15 minutes, coverslips were incubated with secondary antibodies (in 1 % NGS in PBS) for 1 hour at RT. Coverslips were rinsed in PBS twice and deionized (D.I.) water once and mounted on slides using 4′,6-diamidino-2-phenylindole (DAPI)-containing mounting media (ThermoFisher Scientific, 00-4959-52). Slides were maintained at 4°C in the dark until confocal imaging. Information about antibodies and corresponding dilutions can be found in Table S2.
Immunohistochemistry of postmortem human tissue
Slide-mounted frozen tissue sections were immunostained with antibodies against glial fibrillary acidic protein (GFAP) or S100 calcium binding protein B (S100B). Briefly, sections were dehydrated for 1 hour at 60 °C. After cooling to RT, sections were cleaned twice with xylene for 10 minutes, followed by sequential rehydrating immersion in 100 %, 95 %, 75 %, and 50 % ethanol for 5 minutes each. Sections were then washed twice with PBS for 5 minutes. An antigen retrieval process was carried out by incubating the sections with pre-heated sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) for 40 minutes in a 95 °C water bath. Following cooling to RT, slides were washed 3 times with PBS and sections were dried and outlined with a PAP pen. Sections were incubated in blocking solution (10 % NGS and 0.3 % Triton-X in PBS) for 30 minutes at RT. Sections were then incubated with primary antibodies diluted in 2 % NGS overnight at 4 °C. After 3 washes with PBS, sections were incubated with secondary antibodies for 2 hours at RT, followed by 3 washes with PBS. Slides were then rinsed with tap water for 2 minutes. Finally, sections were dehydrated through sequential immersion in 50 %, 75 %, 95 %, and 100 % ethanol for 5 minutes each, then cleaned in xylene 3 times for 3 minutes each and cover slipped with Fluoro-Gel (with Tris Buffer, Electron Microscopy Science, 17985-11). Separate sections without GFAP and S100B staining were cover slipped with Mounting Medium with DAPI to estimate the thickness of cortical layer I. After mounting, sections were maintained at 4 °C in the dark until imaging.
Information about antibodies and corresponding dilutions can be found in Table S2.
EdU/BrdU pulse labeling and analysis
The stock solutions of 5-Bromo-2’-Deoxyuridine (BrdU, 10 mM, ThermoFisher Scientific, B23151) and 5-ethynyl-2’-deoxyuridine (EdU, 10 mM, Life Technologies, C10639) were prepared in dimethyl sulfoxide (DMSO, Life Technologies, C10639) and stored at −20 °C. These solutions were protected from light and were stable for at least 12 months. Immature D16 hiPSC-astrocytes or primary mouse astrocytes were plated at a concentration of 40,000 cells per coverslip in a 24-well plate and cultured for two additional days to reach confluence. After 48 hours, hiPSC-astrocytes or primary mouse astrocytes were incubated with EdU (10 μM) for four hours. BrdU (10 μM) was then added for another two hours.
Following fixation in 4 % PFA in PBS, astrocytes were washed with PBS 3 times and permeabilized with 0.1 % Triton X-100 (BP151-100) in PBS for 5 minutes. DNA was denatured with 2N HCl for 30 minutes at RT, followed by neutralization with PBS for 10 minutes at RT. After washing three times in PBS, coverslips were incubated at RT in blocking buffer (0.1 % Triton X-100, 5 % NGS in PBS) for 1 hour, then incubated overnight at 4 °C with Alexa Fluor 488-conjugated BrdU monoclonal antibody (MoBU-1, Invitrogen B35130) in blocking buffer with dilution ratio 1:100. The following day, coverslips were washed in PBS (3 times for 15 min each). The EdU staining solution was prepared with the Click-iT™ Plus EdU Alexa Fluor™ 594 Imaging Kit (Life Technologies, C10639). Coverslips were incubated with EdU staining solution for 30 minutes at RT. Next, coverslips were washed once with PBS and incubated with Hoechst 33342 (Life Technologies, C10639) diluted in PBS (1:1000) for 30 minutes at RT to label nuclei. Coverslips were rinsed again in PBS twice and D.I. water once, then mounted on slides using DAPI-containing mounting media (ThermoFisher Scientific, 00-4959-52). For each line of hiPSC-astrocytes, 3 independent experiments were performed, and data were collected from 3 coverslips per experiment. For primary mouse astrocytes, 3 Fmr1 KO mice and corresponding WT littermates were used in 3 independent experiments.
Cells were imaged with a Zeiss LSM 700 confocal microscope with 20x (0.5 NA) objective. Images were collected at 512 × 512 pixels (with a pixel size of 0.625 μm) with 405, 488, and 555 nm lasers. On each coverslip, 9 fields were imaged.
Image analysis was performed using ImageJ. The number of all cells (identified by Hoechst) and cells incorporated with EdU and BrdU were counted manually. Cells co-incorporated with EdU and BrdU were counted using the colocalization plugin in ImageJ. The calculation of cell cycle dynamics was adapted from (Martynoga et al. 2005, Harris et al. 2018). Briefly, the length of S-phase (Ts) and total cell cycle (Tc) were calculated using the following formulas:
Here, the incubation interval equals the time between the application of EdU and BrdU (4 hr). The (EdU+; BrdU+ cells) means the number of cells co-incorporated with both EdU and BrdU, and the (EdU+; BrdU− cells) means the number of cells that contained EdU. BrdU+/BrdU− represents the ratio of the number of cells with BrdU and without BrdU, regardless of the EdU incorporation.
Calcium imaging
Mature D20 astrocytes were plated on growth factor reduced Matrigel-coated 12 mm coverslips in a 24-well plate with a density of 40,000 cells/coverslip. Five days later, coverslips were incubated with Sulforhodamine 101 (SR101, Sigma Aldrich, 10 μM), Fluo-4 AM (5 μM, Molecular Probes), and 0.6 μL of Pluronic acid F-127 diluted in 1.5mL of astrocyte maturation medium for 40 minutes in a CO2 incubator. After incubating with culture medium 3 times for 10 minutes each, the coverslip was transferred into a submersion imaging chamber perfused with artificial cerebrospinal fluid (ACSF) saturated with 95 % O2/ 5 % CO2 at RT. ACSF was prepared with D.I. water containing NaCl (126 mM), KCl (3 mM), NaH2PO4 (1.25 mM), and NaHCO3 (26 mM), MgSO4 (1 mM), CaCl2 (2 mM) and dextrose (10 mM).
All imaging was performed with a two-photon microscope (Sutter instrument, Movable Objective Microscope®) using a Ti: sapphire laser (Chameleon Vision II, Coherent) tuned to 820 nm. Images were collected with a Nikon water-immersion objective (25X, 1.05 NA). Excitation power measured at the back aperture of the objective was typically around 5 mW. Pockels cell was used to adjust the power to achieve near identical levels of fluorescence for each imaged region. Two-channel imaging was achieved by using a 565 nm dichroic mirror and 2 external photomultiplier tubes. A 535/50 bandpass filter was used to detect Fluo-4 AM emission and a 610/75 bandpass filter was used to detect SR101. Scan-Image software written in MATLAB (Math-Works) was used to acquire the images (Pologruto et al. 2003). Upon selection of an imaging field, ATP (100 μM) was used to trigger Ca2+ activity (perfused for 5 minutes). Each time-lapse image was acquired at 512 × 512 pixels, 0.75 μm/pixel, every 2 seconds for 10 minutes. In total, 300 images were collected from each coverslip.
Calcium imaging analysis was performed in ImageJ software and Clampfit software. Briefly, the time lapse images were separated into two channels, one for Fluo-4 AM and one for SR101, and converted from signed 16-bit format to unsigned format. The GECIquant Ver 1.0 plugin was used to subtract background and detect regions of interest (ROIs) after manually delineating the astrocyte soma (Srinivasan et al. 2015). Time traces of fluorescence intensity were extracted from the ROIs and converted to 100 % ΔF/F. Ca2+ transients with at least two standard deviations above baseline fluorescence intensity were detected in Clampfit.
The following parameters were analyzed to assess the Ca2+ activity in the presence of ATP: proportion of responding astrocytes, average peak amplitude of the Ca2+ events, mean number of events per responding astrocyte, and average duration of the Ca2+ events. Analysis was performed on raw, unprocessed images. Images were despeckled for presentation purposes. Each cell line was analyzed in 3 independent experiments and for each experiment 3 coverslips were imaged.
Protein extraction
Protein was isolated using RIPA lysis buffer (Millipore, 20-188) containing 1X proteinase and phosphatase inhibitor (Thermo Scientific, 1861281). The concentration of protein was calculated using BCA protein assay kit (Thermo Scientific, 23228). An Albumin standard (Thermo Scientific, 23209) was used for generating the standard curve for the BCA assay. For western blot analysis, proteins were diluted with RIPA lysis buffer and 4X Laemmli sample buffer (BIO-RAD, 1610747) containing 10 % β-mercaptoethanol (Sigma, M7522). Then protein samples were heated at 95 °C for 5 minutes and stored at −80 °C for long-term storage.
Western blot analysis
Samples were briefly vortexed and centrifuged, then 20–30 ug were loaded into a 4–15 % polyacrylamide gel (BIO-RAD, 4568084). The gel was subjected to electrophoresis at 75V for 20 minutes, then at 150V for 30 minutes or until the loading dye reached the bottom. Proteins were then transferred onto a PVDF transfer membrane (Thermo Fisher Scientific, 88518) at 100V for 1 hour. In some cases, the membrane was cut into two sections to separately visualize proteins of different sizes. The membrane was washed with Tris-buffered saline with 0.1 % Tween® 20 (TBST, Fisher, BP2471-1, BP337-100) twice then blocked with 5 % BSA (Fisher, 196974) in TBST for 1 hour. The membrane was incubated with primary antibody diluted in blocking buffer overnight at 4 °C with shaking. The membrane was washed again with TBST and incubated with secondary antibody in TBST for 1.5 hours at RT with shaking. After washing with TBST, protein bands were detected with the chemiluminescence method using Pierce™ ECL Western Blotting Substrate (Thermo Scientific, 32106). Information about antibodies and corresponding dilutions can be found in Table S2.
Glutamate uptake
Mature D23 astrocytes were plated in a 24-well plate with a density of 40,000 cells per well. Forty-eight hours later, when cells reached confluency, astrocytes were equilibrated in Hank’s balanced salt solution (HBSS) without calcium and magnesium (Corning, 21-021-cv) for 30 minutes. Astrocytes were incubated with glutamate (100 μM) in HBSS buffer with calcium and magnesium (Hyclone, SH30268.01) for 3 hours. After 3 hours, the glutamate concentration remaining in the medium was measured using a glutamate assay kit (Sigma-Aldrich, MAK004) according to the manufacturer’s instructions. The change in glutamate concentration was normalized to total protein in each well, which was quantified using a BCA protein assay kit (Thermo Scientific, 23228).
Quantification of astrocytes in human post-mortem tissue
Post-mortem brain sections were imaged with a Zeiss LSM 700 confocal microscope with 20X (0.5 NA) objective. Images were collected at 512 × 512 pixels (with a pixel size of 0.625 μm) with a 488 nm laser. For each section, 2–3 fields containing the pial surface were imaged. Each image consisted of 3 × 3 tiled frames on the x-y axis (960.26 μm × 960.26 μm) and a 14–18 μm z-stack with a step size of 1 μm. Quantitative estimates of the number of astrocytes within cortical layer I were achieved using Optical Fractionator Workflow in the StereoInvestigator software (MBF Biosciences, Inc) as previously described (Gundersen and Jensen 1987, Shen et al. 2019). Briefly, layer I (250 microns from the pial surface) was traced as the ROI. The mounted thickness in the software was measured as 10 μm and the counting frame size was set as X: 90 μm and Y: 90 μm. The systematic random sampling grid layout was defined as 50 % of the region of interest. The dissector was centered with a distance of 10 μm between the guard zones and the manual focus method was selected. After defining the probe configurations above, astrocytes in each counting frame were labeled manually. The estimated cell numbers were calculated by the software as “Estimated Population using Number Weighted Section Thickness” in the probe run results. The density of astrocytes in the cortical layer I area was reported as number of astrocytes per 100 μm2.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted from hiPSC and hESCs growing in a culture dish to 80 % of confluence, using RNeasy Plus Mini Kit (QIAGEN, 74134). RNA purity and concentration were confirmed using NanoDrop 8000 (Thermo Scientific).
RT-qPCR was performed on a Mastercycler ep RealPlex2 Real Time PCR ThermoCycler (Eppendorf, 2894) using Verso SYBR Green 1-Step qRT-PCR Low ROX Kit (Thermo Scientific, AB-4106). Reactions were set up according to the manual using 1 ng RNA, 2.5 μL primers, 0.25 μL Verso Enzyme Mix, 1X SYBR mix, 1.25 μL RT Enhancer, and nuclease-free water for a total volume of 25 μL. The 1-Step RT-qPCR thermal cycling program was carried out at 50 °C for 15 min; 95 °C for 15 min; and 40 cycles of 95 °C for 15 s, 55 °C for 30 s and 72 °C for 30 s, with subsequent melting curve analysis. The primers used for RT-qPCR were Hs_FMR1_1_SG (QIAGEN QuantiTech® Primer Assay, QT00017479) and Hs_GAPDH_1_SG (QIAGEN QuantiTech® Primer Assay, QT00079247). Expression of transcripts of target genes was normalized to Gapdh.
Mass Spectrometry
Four biological replicates (100 μg of protein each) from each line were used for label-free mass spectrometry. Samples were subjected to reduction with 10 mM dithiothreitol at 56° C for 30 min and alkylation using 50 mM iodoacetamide at RT for 25 min. Detergent was removed using chloroform/methanol extraction. The protein pellet was resuspended in 100 mM ammonium bicarbonate and digested with Pierce™ Trypsin Protease, MS Grade (Thermo Scientific, 90057) overnight at 37°C. Tryptic peptides were cleaned from salts with PepClean C18 spin columns (Thermo Scientific, 89870) and were resuspended in 2 % acetonitrile (ACN) and 0.1 % formic acid (FA).
One μg of each sample was loaded onto Acclaim™ PepMap™ 100 75 μm × 2 cm C18 LC trap columns (Thermo Scientific, 164569) at a flow rate of 4 μL/min, then separated with a Thermo RSLC UltiMate™ 3000 (Thermo Scientific, ULTIM3000RSLCNANO) on a Thermo EASY-Spray™ PepMap RSLC C18 75 μm × 50 cm C-18 2 μm column (Thermo Scientific, 03-251-877) with a step gradient of 4–25 % solvent B (0.1 % FA in 80 % ACN) for 10–100 min and 25–45 % solvent B for 100–130 min at 300 nL/min and 50 °C with a 155 min total run time.
Eluted peptides were analyzed by a Thermo Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer (Thermo Scientific, IQLAAEGAAPFADBMBHQ) in a data-dependent acquisition mode. A full survey scan MS (from m/z 350–1800) was acquired in the Orbitrap with a resolution of 120,000. The automatic gain control (AGC) target for MS1 was set as 4 × 105 and ion filling time set as 100 ms. The most intense ions with charge state 2–6 were isolated in 3 s cycle and fragmented using HCD fragmentation with 35 % normalized collision energy and detected at a mass resolution of 30,000 at 200 m/z. The AGC target for MS/MS was set as 5 × 104 and ion filling time set as 60 ms. Dynamic exclusion was set for 30 s with a 10-ppm mass window. Each sample was run in duplicate.
Following conversion of Thermo raw format to mzML using msconvert with peakpicking (Chambers et al. 2012), protein identification was performed by searching DDA MS/MS data against the UniProt human protein database UP000005640 (20,601 searched entries) using the Fragpipe software suite (version 17.1) with built-in MSFragger (version 3.4) in closed search mode (Kong et al. 2017). The search was set up for fully tryptic peptides with a maximum of two missed cleavage sites. Oxidized methionine was included as a variable modification, carbamidomethylating of cysteine was set as a fixed modification, and the mass tolerance threshold was set to 20 ppm. The significance threshold of the ion score was calculated based on a false discovery rate of ≤ 1 % using the built-in Protein Prophet and Percolator algorithms. In total, 917 proteins were identified among all the samples.
Bioinformatics analysis
Post processing and data analysis were performed in R (version 4.1.1). Protein intensities wherein a single value was missing per group of bioreplicates (n=4) were defined as missing completely at random (MCAR), made up >2 % of all values, and were inputed using the missForest package (version 1.4)(Stekhoven 2022). Quantile normalization was performed using the preprocessCore package (version 1.54.0) followed by a Levene’s test for homogeneity of variance (F = 19.367, p < 2.2e-16) and a two sample equal variance t-test with Benjamini & Hochberg adjustment (Levene 1960, Benjamini and Hochberg 1995, Bolstad 2022). Differentially expressed proteins (DEPS) were defined as proteins with p.adj < 0.05 and log base 2 fold change ≥ 1 or ≤ −1. The volcano plot was generated using the ggplot package (version 3.3.5), the heatmap plots were generated using the pheatmap package (version 1.0.12), and the Venn diagrams were generated using the ggVennDiagram (version 1.2.0) and VennDiagram (version 1.7.3) packages (Wickham 2016, Kolde 2018, Gao et al. 2021, Chen 2022).
The 842 and 891 FMRP targets were identified in two mouse studies (Darnell et al. 2011, Maurin et al. 2018) and 3,718 FMRP targets were identified in human organoids (Kang et al. 2021). The 1,023 ASD-related human genes were collected from SFARI gene database (2021 Q3). The 459 and 325 genes enriched in fetal and mature human astrocytes were collected from transcriptome data in a previous study (Zhang et al. 2016). Genes expressed at fragments per kilobase of transcript sequence per million mapped fragments (FPKM) = 5 or higher and enriched by 4 folds or higher in fetal astrocytes over mature astrocytes were defined as fetal astrocyte-enriched genes. The same criteria were used for mature astrocyte-enriched genes. We also excluded genes with >2 fold higher expression in any cell type other than astrocytes to remove genes from contaminating cell types. The 9,996 and 8,634 genes expressed in fetal and mature human astrocytes were collected from transcriptome data in previous study (Zhang et al. 2016). Genes expressed at FPKM = 0.5 or higher in fetal or mature astrocytes were defined as fetal astrocyte or mature astrocyte expressed genes.
Global pathway analysis was conducted through Ingenuity Pathway Analysis (Kramer et al. 2014) using the log2 transformed mass spectrometry expression values for all quantified proteins between FXS and Ctr. Gene set enrichment analysis (GSEA) for CTR vs. FXS was performed by GSEA 4.1.0 (Mootha et al. 2003, Subramanian et al. 2005). Network analysis and Markov Clustering (MCL) were performed using the online STRING database (Szklarczyk et al. 2019) for the list of downregulated and upregulated DEPs with a minimum required interaction score of 0.700 (high confidence). Disconnected nodes in the network were hidden.
Astrocyte cultures for sterol extraction.
Mature human stem cell-induced astrocytes were plated in 96-well plates at 30,000 cells/well. After the incubation of cells at 37°C, in a 5 % CO2 cell culture incubator for 3 days, Hoechst dye was added to all wells in the 96-well plate and the total number of cells counted using an ImageXpress Pico and cell counting algorithm in CellReporterXpress. After removal of the medium, wells were rinsed twice with 1X PBS, treated with an antioxidant mixture (10 μl/well), and stored at −80°C for sterol analysis. The antioxidant mixture contained BHT (2.5 mg/ml) and PPh3 (1 mg/ml) in EtOH. All samples were analyzed within 1 week of freezing. For each condition there were 3–8 technical replicates.
HPLC grade solvents were purchased from Thermo Fisher Scientific Inc. (Waltham, MA). The derivatizing reagents 2-methyl-6-nitrobenzoic anhydride and N,N-dimethylglycine were purchased from Combi-Blocks (San Diego, CA). The synthesis of deuterated standards was described (Tallman et all 2021). All deuterated sterol standards are available from Kerafast, Inc. (Boston, MA).
Extraction of sterols.
A stock solution of deuterated standards was made containing 30 μM of d7-Chol, d6-Lan, d7-dHLan and 3 μM d7-7-DHC, d7-8-DHC, d6-Des, d6-7-DHD, d6-8-DHD, d7-Lath, d6-DHL, d7-Zyme, d6-Zym, d7-14d-Zyme, d6-14d-Zym in MeOH. The standards stock solution contained 1 % (v/v) Et3N and antioxidant mixture (BHT and PPh3) described above to prevent isomerization and/or oxidation. To each well of the 96-well plate was added the standards mixture (10 μL) and MeOH (100 μL). The plate was agitated on a shaker for 20 min and then allowed to rest to settle cell debris. The MeOH was transferred to an analysis plate, dried under vacuum, and derivatized as described below.
Derivatization and LC-MS/MS analysis of sterols.
Derivatizing reagent was freshly prepared with 2-methyl-6-nitrobenzoic anhydride (20 mg), N,N-dimethylglycine (14 mg), DMAP (6 mg), and Et3N (0.1 mL) in anhydrous CHCl3 (0.9 mL). To each sample was added derivatizing reagent (100 μL) and allowed to react at room temperature for 30 min. The samples were dried under vacuum and subsequently dissolved in MeOH (100 μL) for LC-MS/MS analysis. Samples were analyzed on an Acquity UPLC system equipped with ANSI-compliant well plate holder. The sterols (10 μL injection) were analyzed on an Agilent Poroshell EC-C18 (10 cm × 2.1 mm, 1.9 μm) with CH3CN:MeOH:H2O, 70:25:5 (0.01 % (v) formic acid, 1 mM NH4OAc) mobile phase at a column temperature of 40 °C. The flow rate was 400 μL/min for 11.5 min, then ramped to 600 μL/min at 11.6 min with a total run time of 16 min. A TSQ Quantum Ultra tandem mass spectrometer (ThermoFisher) was used for MS detections, and data were acquired with a Finnigan Xcalibur software package. Selected reaction monitoring (SRM) of the DMG derivatives was acquired in the positive ion mode using electrospray ionization (ESI). MS parameters were optimized using DMG-Chol and were as follows: spray voltage at 4500 V, capillary temperature at 300 °C, auxiliary nitrogen gas pressure at 55 psi, and sheath gas pressure at 60 psi. Collision energy (CE) was optimized for each sterol and oxysterol under a collision gas pressure of 1.5 mTorr. The monitored transitions were previously published (Tallman et al 2021). Endogenous sterol levels were quantified based on the known matched deuterated standard amount and then normalized to cell count. Oxysterol levels were quantified using d7-Chol as standard and a response factor that was determined for each compound, then also normalized to cell count.
Statistics
All analyses were performed while being blinded to the genotype of the cells and the tissue. Graphing and statistical analyses were conducted on GraphPad Prism 8.0 software (GraphPad Software, San Diego, CA). Before statistical analysis, outliers were identified and removed by robust regression and outlier removal (ROUT) with False Discovery Rate of 1 % (Motulsky and Brown 2006). To make comparison within isogenic pairs, relative values were calculated in each of three isogenic pairs. Statistical comparisons between two groups were made using unpaired Student’s t-test or Mann-Whitney test according to the normality and lognormality of data. All data are represented in scatter plot with a bar (displaying each N value) with the means ± SEM.
Results
Differentiation of astrocytes from hiPSCs and hESCs
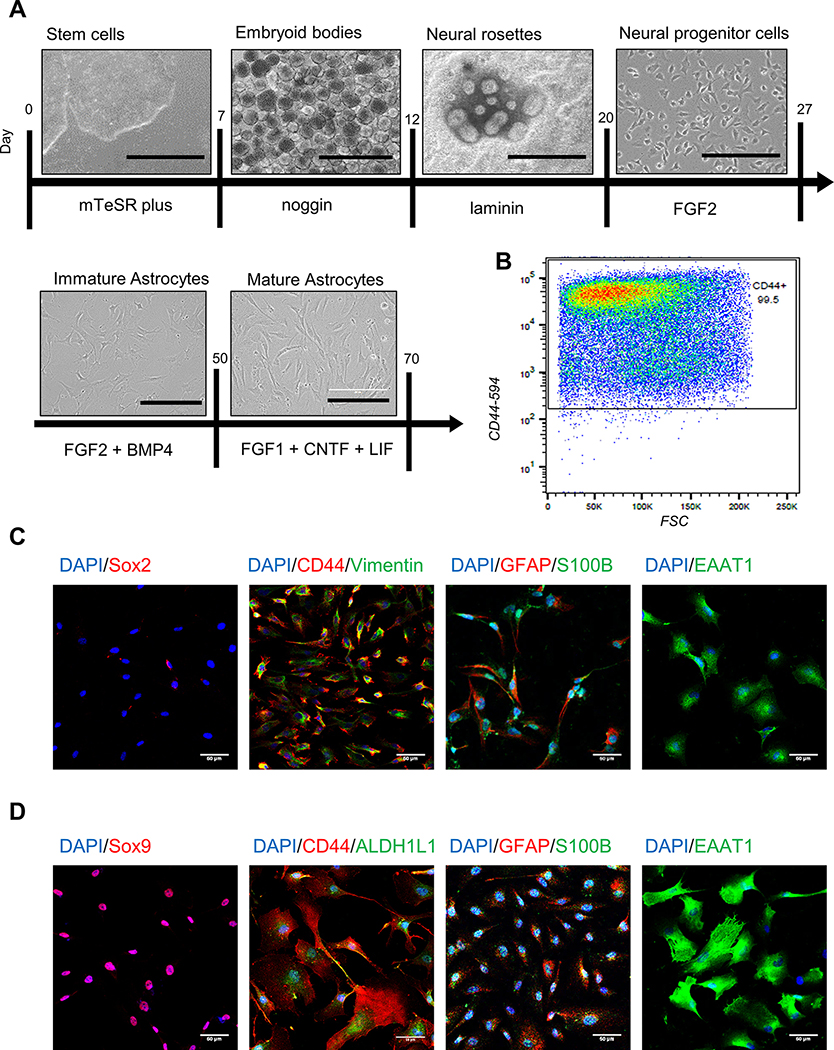
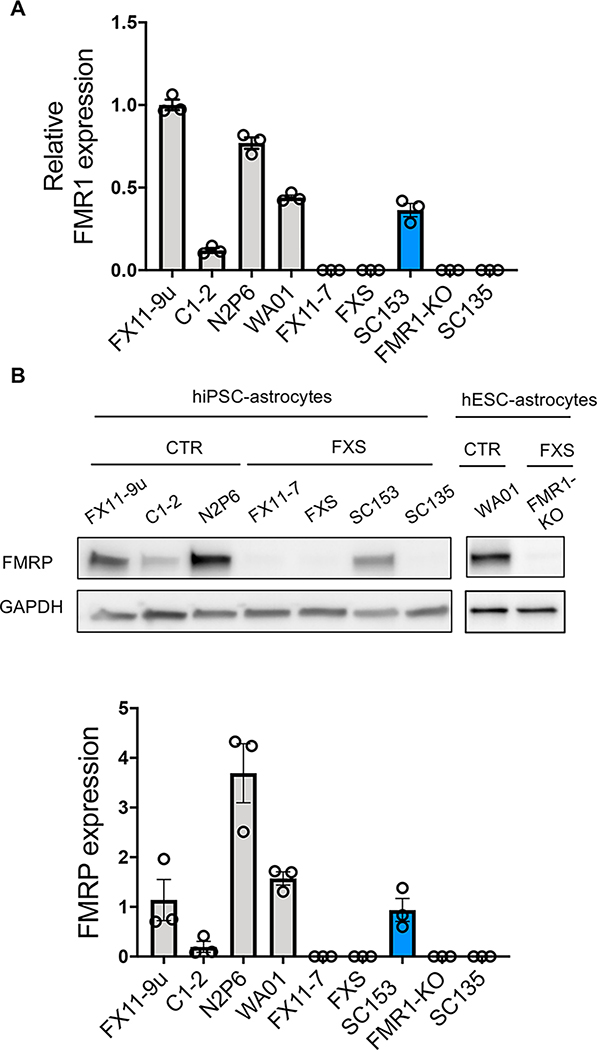
To model FXS and study the effects of FMRP deficiency in human astrocytes, we differentiated astrocytes from patient-derived FXS hiPSCs, FMR1-KO hESCs, and isogenic control (CTR) lines (Table 1) using a modified chemically defined and xeno-free method (Chen et al. 2014, Jiang et al. 2016) (Fig. 1). After 20 days, the vast majority of cells expressed cell surface glycoprotein CD44 as measured by FACS (Fig. 1B). Additionally, cells obtained expression of canonical astrocyte markers: SOX9, vimentin, GFAP, S-100B and ALDH1L1 and EAAT1 (Fig. 1C,D and Fig. S1, Table S2). All the stem cell lines were successfully differentiated into astrocytes, although some variability in morphology was observed between lines (Fig. S1). FXS hiPSCs, hESCs, and corresponding astrocytes had either no FMRP expression or greatly reduced FMRP expression as compared to their isogenic controls (Fig. 2). To study the properties of more mature astrocytes in vitro, immature astrocytes were exposed to additional growth factors for an additional 3–4 weeks. During the process of further maturation, astrocytes gradually exhibited decreased proliferation and finally became postmitotic. They also displayed enlarged soma and lengthened processes compared with immature astrocytes (Fig. 1D). We used immature astrocytes for cell cycle analysis and mature astrocytes for Ca2+ signaling, glutamate uptake and proteomic analysis (Table S1).
Figure 1. Generation of hiPSC- and hESC-astrocytes.
A. Schematic of hiPSC and hESC differentiation. Scale bar: 1000 μm for stem cells, embryoid bodies, neural rosettes; 200 μm for neural progenitor cells, immature astrocytes, mature astrocytes. B. CD44 expression in immature astrocytes by flow cytometry. C. Immature astrocytes do not express neural progenitor marker Sox2, but express canonical astrocyte markers. Scale bar: 50 μm. D. Mature astrocytes express canonical astrocyte markers and display enlarged soma and longer processes. Scale bar: 50 μm.
Figure 2. FMR1 and FMRP expression.
A. FMR1 expression in hiPSCs and hESCs by qRT-PCR. B. FMRP expression in hiPSC- and hESC-astrocytes by western blotting. N = 3 samples per line.
Altered cell cycle dynamics in FXS astrocytes
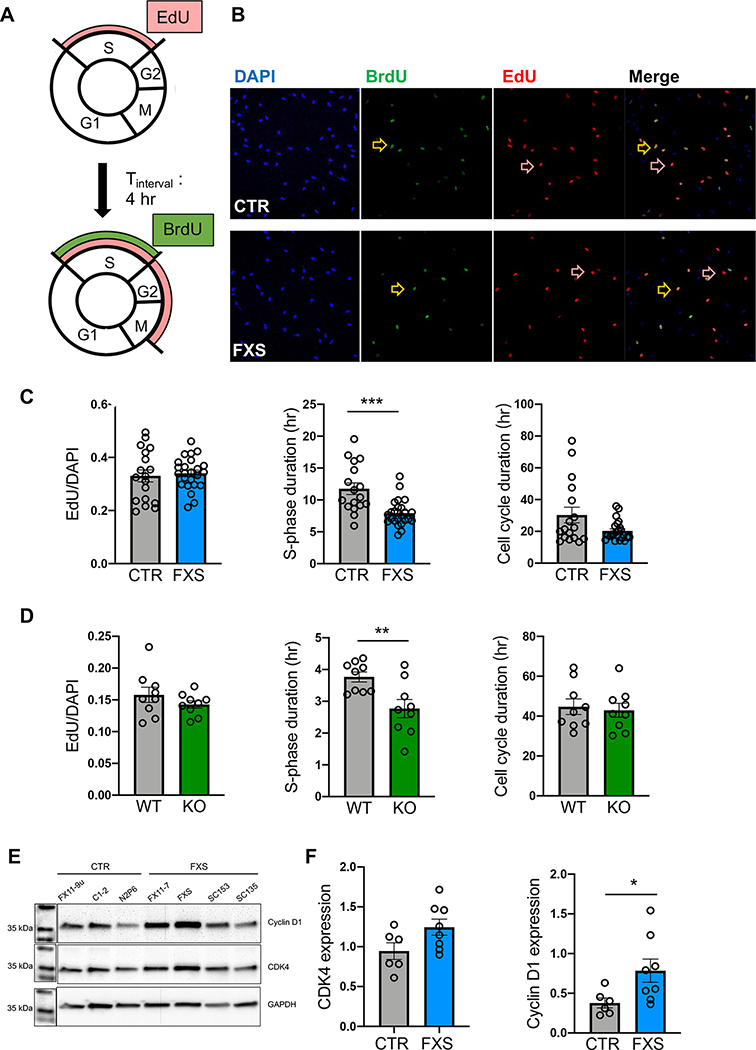
Previous studies found that FXS hESCs showed a bias towards glial lineage (Telias et al. 2013). Altered proliferation was also observed in FXS hiPSC-derived NPCs (Ge and Jia 2016, Kang et al. 2021, Raj et al. 2021). These results indicate dysregulated proliferation during stem cell differentiation in the absence of FMRP, however, whether similar dysregulation persists in FXS astrocytes has not been studied. To assess the cell cycle dynamics, two thymidine analogues, 5-bromo-2’-deoxyuridine (BrdU) and 5-ethynul-2’-deoxyuridine (EdU), which are incorporated into actively dividing cells during the S-phase (Golias et al. 2004, Chehrehasa et al. 2009), were sequentially applied to D18 immature astrocyte cultures. Actively proliferating astrocytes passing through S-phase were labeled with EdU. After four hours of incubation, astrocytes remaining in the S-phase were also labeled with BrdU (Fig. 3A). The proportion of EdU+ and EdU+/BrdU+ astrocytes was used to estimate the cell cycle dynamics (Fig. 3B) by calculating the length of the S phase and total cell cycle using formulas previously described (Harris et al. 2018).
Figure 3. Altered cell cycle dynamics of FXS astrocytes.
A. Schematic of EdU/BrdU pulse labeling with interval time of 4 hours. B. Representative images of CTR (top) and FXS (bottom) hiPSC-astrocytes labeled with BrdU (green) and EdU (red). Dual-labeled astrocytes are shown in orange. Yellow arrows point to cells that incorporated both BrdU and EdU. Red arrows point to the cells with only EdU. C. Proportion of EdU-labeled cells, S phase duration (Ts), of CTR and cell cycle duration (Tc) of CTR and FXS hiPSC-astrocytes. ***P < 0.001. N = 18–24 coverslips from 2 independent experiments. D. Proportion of EdU-labeled cells, S phase duration, and cell cycle duration of primary astrocytes from WT and Fmr1 KO mice **P < 0.01. N = 9 coverslips from 3 independent experiments. E. Representative western blot bands of cyclin D1 and CDK4 expression in hiPSC-astrocytes. F. Quantitative analysis of CDK4 and cyclin D1 expression in FXS hiPSC-astrocytes. *P < 0.05, N = 6–8 samples from 2 independent experiments, Mean ± SEM.
The proportion of astrocytes passing through S phase, as measured by the proportion of cells incorporating EdU, was similar in CTR and FXS groups (Fig. 3C). However, the estimated S phase duration (Ts) of FXS hiPSC-astrocytes was significantly shorter than that of CTR hiPSC-astrocytes. We did not observe a significant change in total cell cycle duration (Tc) of FXS hiPSC-astrocytes compared to CTR hiPSC-astrocytes (Fig. 3C). We next asked whether the cell cycle alteration in astrocytes is also present in an FXS mouse model. We measured cell cycle dynamics in primary astrocytes derived from Fmr1 KO mice. As with FXS-hiPSC astrocytes, the proportion of EdU+ astrocytes was similar between KO and WT littermates (Fig. 3D). Moreover, similar to hiPSC-astrocytes, the primary astrocytes from Fmr1 KO mice showed reduced Ts but similar Tc, compared to astrocytes from WT littermates. These results indicate that the cell cycle dynamics, in terms of the S phase progression, are altered in human FXS astrocytes and this impairment is conserved in an FXS mouse model.
As the cell cycle is strictly regulated by a series of checkpoints, we next examined whether the expression of cell cycle regulators is dysregulated in human FXS astrocytes. Given that FMRP interacts with mRNAs of cyclin-dependent kinase 4 (CDK4) and cyclin D1, and the loss of FMRP leads to increased protein expression of both regulators in Fmr1 KO mice (Luo et al. 2010), we asked whether dysregulated protein expression of these two factors is also present in human FXS astrocytes. We performed western blotting analysis using proteins from immature D18 astrocytes, the same age as the astrocytes used for cell cycle analysis (Fig. 3E). We found that, while the expression of CDK4 was not significantly different between CTR and FXS hiPSC-astrocytes (Fig. 3E), the expression of cyclin D1 showed a two-fold increase in FXS hiPSC-astrocytes (Fig. 3F). Taken together, these results indicate dysregulated cyclin D1 signaling in FXS hiPSC-astrocytes, potentially contributing to the observed altered cell cycle dynamics.
Increased number of astrocytes in the cortex of FXS patients
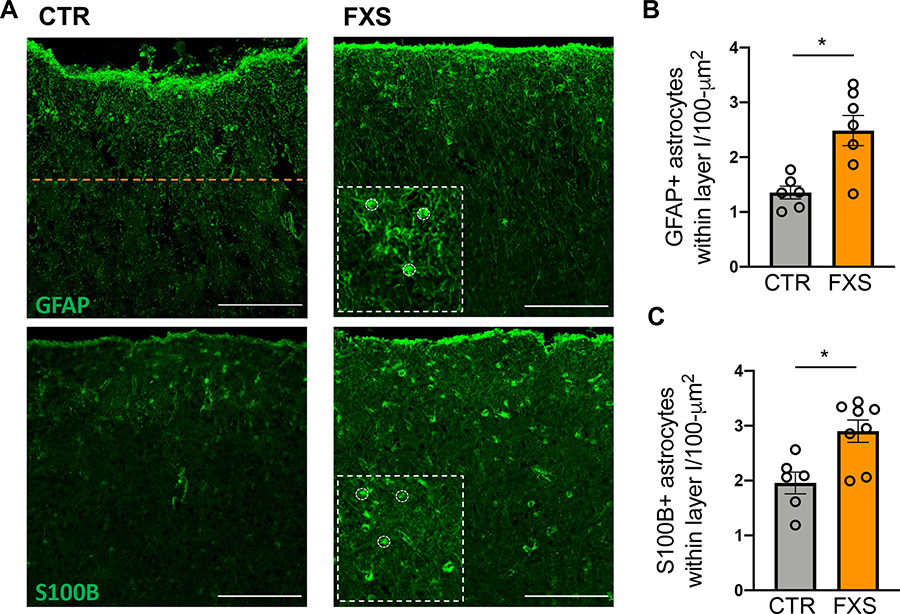
We reasoned that the enhanced proliferation of FXS astrocytes might influence the density of astrocytes in the cortex. We therefore used postmortem tissue to estimate the number of astrocytes in cortical layer I of Brodmann area 3 of FXS and control individuals. The number of astrocytes within cortical layer I was determined after immunostaining for the astrocyte markers GFAP and S100B (Fig. 4A). The depth of cortical layer I (~230 μm from pial surface) was determined by the density of cell nuclei stained with DAPI. Relative to control tissue, we observed a two-fold increase in the number of GFAP+ astrocytes in cortical layer I (Fig. 4B). Similarly, an increased number of S100B+ astrocytes was observed in FXS tissue compared to CTR (Fig. 4B). These results implicate a role for FMRP in the development of astrocytes in the human cortex and are consistent with increased proliferation of FXS hiPSC-astrocytes.
Figure 4. Astrocyte density is higher in postmortem tissue of FXS patients.
A. Representative images of astrocytes in human postmortem cortical sections stained with GFAP and S100B. The orange line indicates the boundary of cortical layer I from the pial surface. Scale bar: 200 μm. Insets: higher magnification with some of the counted astrocytes outlined. B. Density of GFAP+ astrocytes (per 100 μm2) within cortical layer I in postmortem tissue. *P < 0.05, N = 6–8 donors C. Density of S100B+ astrocytes (per 100 μm2) within cortical layer I in postmortem tissue. *P < 0.05, Mean ± SEM.
Altered Ca2+ signaling in FXS-astrocytes
Next, we examined functional properties of FXS hiPSC-astrocytes focusing on two canonical functions. Astrocytes respond to synaptic activity and environmental stimuli, such as glutamate and ATP, via elevation of intracellular Ca2+ concentration (Bazargani and Attwell 2016, Shigetomi and Koizumi 2016). A major source of [Ca2+]i elevation in the astrocyte soma is Ca2+ release from internal stores, triggered by Gq-coupled purinergic receptors (Bazargani and Attwell 2016). To investigate whether human astrocytes derived from FXS patients display altered Ca2+ signaling in response to extracellular cues, Ca2+ imaging was performed on mature hiPSC-astrocytes.
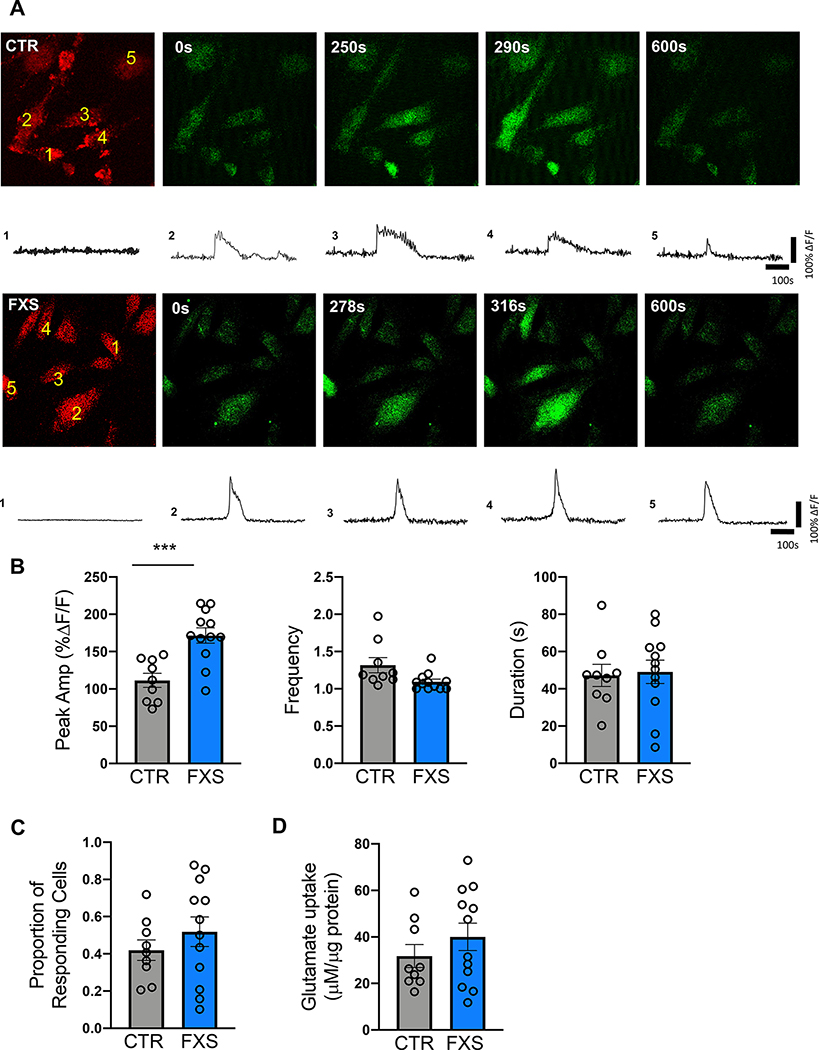
The calcium-sensitive dye Fluo-4 AM was used to capture the Ca2+ signaling dynamics and sulforhodamine 101 (SR101) was used as a structural marker for astrocytes (Fig. 5A). ATP (100 μM) was used to activate purinergic receptors and stimulate Ca2+ release from internal stores. Considering the lack of ramified and branched processes in our hiPSC-astrocytes in culture, we focused on Ca2+ transients in the soma. We did not observe a change in the proportion of FXS hiPSC-astrocytes responding to ATP compared to CTR hiPSC-astrocytes (Fig. 5C). The duration of the responses and the event frequency was not changed (Fig. 5B). However, the peak amplitude of the ATP response, shown as %ΔF/F, was significantly enhanced in FXS hiPSC-astrocytes compared to CTR hiPSC-astrocytes (Fig. 5B). These results indicate that human astrocytes derived from FXS patient cells have enhanced purinergic Ca2+ signaling properties.
Figure 5. Enhanced ATP-induced Ca2+ signaling of hiPSC-astrocytes.
A. Representative images and traces of ATP-induced Ca2+ signaling in numbered hiPSC-astrocytes (CTR: top, FXS: bottom) labeled with SR101 (red) and Fluo4-AM (green) obtained using two-photon microscopy. The amplitude of the Ca2+ signaling is shown as %ΔF/F. B. Peak amplitude (%ΔF/F), frequency (number of events per responding cell within 10-minute imaging period), and duration of ATP-induced Ca2+ signaling in control and FXS hiPSC-astrocytes. C. Proportion of hiPSC-astrocytes responding to ATP. D. Glutamate uptake analysis showing that both FXS and CTR hiPSC-astrocytes were capable of glutamate uptake. *** P < 0.001. N = 9–12 experiments (3 per line).
Glutamate uptake activity of FXS-astrocytes
Astrocytes can regulate synaptic transmission by rapidly removing neurotransmitters such as glutamate from the extracellular space on a millisecond timescale (Minelli et al. 1996, Bergles and Jahr 1997, Goubard et al. 2011). In both Fmr1 KO and astrocyte-specific Fmr1 KO mice, astrocytes lacking FMRP showed impaired glutamate uptake as well as reduced expression of the glutamate transporter GLT-1 (Higashimori et al. 2013, Higashimori et al. 2016). However, the glutamate uptake function of human astrocytes lacking FMRP has not been previously examined. In our study, a glutamate uptake assay demonstrated that differentiated astrocytes from FXS and CTR hiPSC were capable of glutamate uptake. However, we did not observe a difference in the glutamate uptake between CTR and FXS astrocytes (Fig. 5D). These results suggest that a degree of functional maturation of astrocytes differentiated from human stem cells in vitro is achieved in both FXS and control cells.
Altered proteome of FXS human astrocytes
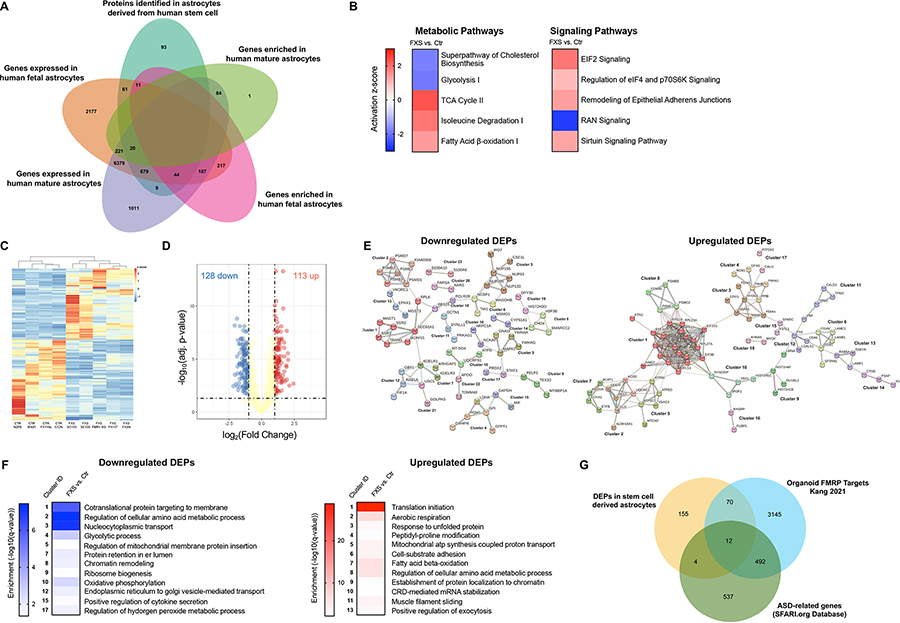
To better understand the observed cellular and functional alteration and to potentially identify novel therapeutic targets, we studied how the loss of FMRP affects protein expression in human stem cell-derived astrocytes. Given the role of FMRP as a regulator in translation processes, we examined the proteomic profiles of mature D20 FXS and CTR hiPSC-and hESC-astrocytes using liquid chromatography-mass spectrometry (LC-MS). In total, 917 proteins were identified in human stem cell-astrocytes (Table S3). Of 917 proteins identified in the CTR group, 743 overlapped with genes expressed in both fetal and mature human astrocytes (Zhang et al. 2016) (Fig. 6A, Table S4). Of all identified proteins in CTR group, 72 and 9 overlapped with genes specifically expressed in either fetal or mature astrocytes, respectively. Further, we found that 55 and 20 proteins overlapped with genes enriched in human fetal and mature astrocytes respectively (Zhang et al. 2016). Such results suggest that human stem cell-astrocytes in our study are more similar to fetal rather than mature human astrocytes in vivo.
Figure 6. Proteomic profiling of control and FXS stem cell derived astrocytes.
A. The number of proteins identified in human stem cell-astrocytes in our study (teal) and its overlaps with genes expressed in human fetal astrocytes (orange), genes expressed in human mature astrocytes (lilac), genes enriched in human fetal astrocytes (pink) and genes expressed in human mature astrocytes (green) identified by Zhang et al. B. Subset of pathways with specific alterations identified by ingenuity pathway analysis in FXS hiPSC- and hESC-astrocytes. C. Heatmap with hierarchical clustering of DEPs in CTR and FXS hiPSC- and hESC-astrocytes. Values are normalized as log2(protein expression+0.0001). Columns are averaged values of biological replicates of each line. Proteins with FDR < 0.05 were defined as DEPs. D. Volcano plot of protein expression change in hiPSC- and hESC-astrocytes upon loss of FMRP. Yellow dots are proteins with FDR>0.05 or log2(Fold Change) ≤1 or ≥−1; Red and blue dots are proteins with FDR < 0.05 and log2(Fold Change) ≥1 (up) or ≤−1 (down), respectively. E Bioinformatic analysis of the DEPs using the search tool for retrieval of interacting genes (STRING) revealed protein-protein interaction (PPI) networks. 23 downregulated and 18 upregulated significant clusters were found in the DEPs PPI network analysis using STRING MCL clustering. F. Heatmaps of significant enrichment for a GO term in the clusters.
We used different approaches to determine proteomic changes in FXS astrocytes. Assessment of the data by use of the bioinformatics software Ingenuity Pathway Analysis (IPA) revealed global proteomic changes induced by the loss of FMRP. Canonical pathways identified as significant (p-values < 0.05 determined in IPA using right-tailed Fisher’s Exact test) across the dataset included the metabolic pathways Superpathway of Cholesterol Biosynthesis and Glycolysis I which were predicted to be downregulated (negative activation z-score) and the TCA Cycle II, Isoleucine Degradation I, and Fatty Acid β-oxidation I which were predicted to be upregulated (positive activation z-score, Fig. 6B). Among the signaling pathways, EIF2 Signaling, Regulation of eIF4 and p70S6K Signaling, Remodeling of Epithelial Adherens Junctions, and Sirtuin Signaling Pathway were predicted to be upregulated (positive activation z-score) and RAN signaling was predicted to be downregulated (negative activation z-score, Fig. 6B). As an additional approach, we performed gene set enrichment analysis (GSEA) to identify dysregulated gene sets in FXS using the C2 collection of Molecular Signature Database (MSigDB) (Mootha et al. 2003, Subramanian et al. 2005). Using FDR < 0.05 we identified 36 gene sets that were enriched in FXS stem cell-derived astrocytes and none that were significantly enriched in the control cells (Table S5). Among the enriched gene sets several were associated with translation including initiation and elongation. Others included gene sets associated with metabolism of amino acids and derivatives and regulation of nervous system development (Fig. S2).
We next focused on differentially expressed proteins (DEPs) and identified 241 proteins with FDR < 0.05 (Table S6). The heatmap of DEPs with hierarchical clustering demonstrated the relative consistency across the human astrocytes within the same CTR or FXS group (Fig. 6C). We observed a similar number of DEPs with increased and decreased protein expression in FXS. Specifically, there were 113 upregulated DEPs and 128 downregulated DEPs (Fig. 6D, Table S6). Of the DEPs 4 overlapped with genes enriched in mature human astrocytes while 18 DEPs overlapped with genes enriched in fetal human astrocytes (Zhang et al. 2016) (Table S6). Surprisingly, very few DEPs overlapped with differentially expressed genes (DEGs) found in FXS human fetal brains and FXS organoids (Kang et al. 2021) (Table S6).
Bioinformatic analysis of the DEPs using the search tool for retrieval of interacting genes (STRING) revealed protein-protein interaction (PPI) networks. Fig. 6E shows the 23 significant clusters that were found in the downregulated DEPs PPI network analysis using STRING MCL clustering (Table S7). Functional annotation of the top PPI network cluster (Fig. 6F, cluster 1) revealed enrichment of the Gene Ontology (GO) Biological Process (BP) cotranslational protein targeting to membrane (GO-term: 0006613, FDR 1.08e-06) and the GO Cellular Component (CC) endoplasmic reticulum (GO-term: 0005783, FDR 2.00e-05). Fig. 6F also shows the 18 significant clusters that were found in the upregulated DEPs PPI network analysis using STRING MCL clustering (Table S7). Functional annotation of the top PPI network cluster (Fig. 6F, cluster 1) revealed enrichment of the GO BP translational initiation (GO-term: GO:0006410, FDR 1.54e-29) and the GO CC cytosolic ribosome (GO-term: 0022626, FDR 2.22e-27).
We next explored the potential link between the translational control function of FMRP and change in protein expression by comparing the overlap between the DEPs in our study and previously published FMRP binding targets. Of the 241 DEPs identified in our study, only a single protein overlapped with FMRP targets identified in mouse (Darnell et al. 2011, Maurin et al. 2018) brains. However, 82 DEPs overlapped with FMRP targets identified in FXS organoids (Kang et al. 2021) (Table S8, Fig. 6G). It is therefore likely that we are also detecting secondary effects of dysregulated primary mRNA targets. Finally, we found that 16 DEPs were also included in the Simons Foundation Autism Research Initiative (SFARI) genes, an ASD-related gene database and 12 of those genes were also identified as FMRP targets in FXS organoids (Abrahams et al. 2013, Kang et al. 2021) (Table S7, Fig. 6G). This analysis provides molecular candidates in astrocytes to study the mechanisms underlying the phenotypes shared in FXS and ASD.
Taken together, these data suggest that the loss of FMRP in human astrocytes causes an extensive protein expression alteration related to many signaling pathways and cellular activities, highlighting several molecular targets with therapeutic potential for FXS.
While some of the changes in the proteome were consistent with previous studies, such as the alteration in translational control, a few were surprising, and we aimed to validate them. The protein Drebrin (DBN1) has known roles in neuronal development but is also expressed at high levels in fetal astrocytes (Zhang et al. 2016). Indeed, proteomic analysis showed that Drebrin was expressed in in stem cell-derived astrocytes and was increased in the FXS lines (Table S3). Western blot analysis has confirmed the increase in Drebrin in three of the four FXS lines (Fig. S3). Drebrin with its actin binding activity is known to regulate process outgrowth in neurons and is likely to have a similar function in developing astrocytes.
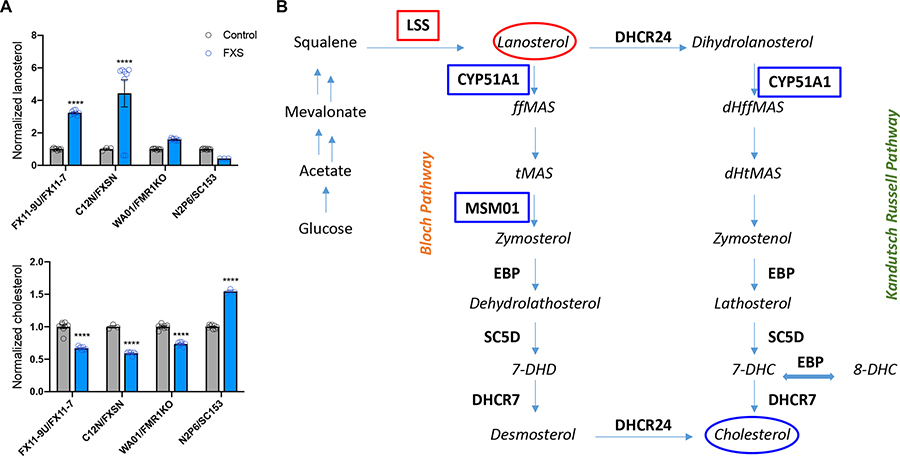
We next examined the altered cholesterol biosynthesis pathway that was identified by the proteomic analysis (Fig. 7, Table S3). We assayed 14 sterols and 7 oxysterols in stem cell-derived astrocytes by the N,N-dimethylglycyl (DMG) method (Tallman et al. 2021). We found that human stem cell-derived astrocytes express 13 sterols and 4 oxysterols. After cholesterol, the most abundant sterols were 8-DHC, 7-DHD, dihydrolanosterol and desmosterol (complete dataset is presented in Table S9). Oxysterols detected included 7-ketocholesterol, 25-hydroxycholesterol, 27-hydroxycholesterol and DHCAO. Consistent with the finding of increased expression of lanosterol synthase, an enzyme that promotes conversion of squalene into lanosterol, we observed increased levels of lanosterol (Fig. 7). In addition, consistent with reduced expression of the enzymes CYP51A1 and Sc4mol (MSM01), which target sterol intermediates in the pathway, we observed reduced levels of cholesterol in FXS astrocytes (Fig. 7). Thus, the proteomic analysis and the direct measurement of cholesterol and its precursors have identified previously unappreciated alteration of sterols in FXS astrocytes.
Figure 7. Altered cholesterol synthesis in FXS astrocytes.
A. Normalized levels of lanosterol and cholesterol in human stem cell-derived astrocytes. B. Simplified cholesterol synthesis pathway. Only a subset of intermediates is shown in italics. In bold are the enzymes involved in the pathways. Enzymes with increased or decreased levels in the proteomic analysis are marked with red and blue boxes, respectively. Sterols found to be increased or decreased are marked with red and blue ovals respectively. ****P<0.0001 N=3–8 replicates per line.
Discussion
The contribution of astrocytes to the development and function of the brain is increasingly recognized and astrocyte impairments have been implicated in neurodevelopmental disorders (de Majo et al. 2020). To better understand how FMRP deficiency affects astrocyte development and function, we generated a human-based astrocyte model using human stem cells and performed a series of characterization studies. Our study showed that human astrocytes with FMRP deficiency have altered proliferation patterns and Ca2+ signaling properties. We also identified altered proteomic profiles and dysregulated metabolic and signaling pathways in FXS human astrocytes, including altered sterol biosynthesis. These results provide insights into the potential roles of astrocytes in FXS. They also indicate that astrocytes are a potential therapeutic target for future FXS treatment.
Human stem cells are particularly suited for studying the role of astrocytes in FXS because the epigenetic silencing of the FMR1 gene only occurs in humans. In addition, there are considerable interspecies differences between human and rodent astrocytes, including vastly different morphologies, functions, and gene expression (Oberheim et al. 2009, Zhang et al. 2016) and therefore astrocytes could have unique and outsized contributions to FXS. Although multiple astrocyte differentiation methods have been generated (Jiang et al. 2016, Sloan et al. 2017, Tcw et al. 2017, Canals et al. 2018, Tchieu et al. 2019), here, we employed a two-dimensional differentiation process. This method resembles physiological developmental stages and lineage commitment progression by exposing cells to mitogens and morphogens, mimicking in vivo developmental cues (Krencik and Zhang 2011). However, it raises a common issue in this field, namely that we lack standards to define glial progenitors, immature astrocytes, and mature astrocytes in vitro. Although astrocytes develop simple morphologies with our approach, the cells do express canonical astrocytic markers and are functional, as indicated by glutamate uptake and calcium signaling assays. Nevertheless, the proteomic data indicate that even after 3 weeks in maturation medium, hiPSC-astrocytes share more similarities with fetal than mature human astrocytes.
Here we employed three isogenic pairs either naturally occurring due to mosaicism of CGG repeats or generated by CRISPR/Cas9 correction of the CGG repeat expansion (Doers et al. 2014, Zhang et al. 2018). While all isogenic control lines expressed FMRP, the FX11-9u line was previously reported to have 114 CGG repeats, which placed the FMR1 gene in the premutation range (Doers et al. 2014). Our analysis of FMR1 and FMRP expression in this line demonstrated expression levels that are similar to other control lines, which is consistent with previous work (Doers et al. 2014). In addition, the heatmap based on proteomic profiling demonstrated that FX11-9u clustered with the other 3 CTR lines, suggesting that it acts as a control line. However, we cannot rule out the possibility of additional differences in this line due to the premutation level expansion of the CGG repeats. In addition, the FXS line SC153, in contrast to a previous report (Zhang et al. 2018), showed expression of FMR1 in hiPSCs and FMRP in astrocytes, but in reduced level compared to its isogenic control pair. However, the SC153 line exhibited FXS phenotypes such as exaggerated purinergic signaling and shortened S-phase and clustered with the other FXS lines based on its proteome. This pair, however, stood out with its opposite alteration in cholesterol levels. Though evidence showed that restoration of greater than 5 % of overall FMRP expression in FXS neurons could rescue FXS deficits (Graef et al. 2020), and several treatments reactivating FMRP expression were generated as gene therapy (Shitik et al. 2020), the observations in our study suggest the existence of a threshold for the expression of FMRP, specifically in astrocytes, to restore normal activities.
Our results suggest that cell cycle progression of FXS astrocytes is altered. Specifically, the length of the S phase in the cell cycle is reduced in FXS astrocytes, suggesting increased proliferation rate driven by shortened S phase in FXS astrocytes. However, we also observed similar incorporation of EdU in CTR and FXS groups, which seems contradictory. Since we identified enhanced expression of cyclin D1 in FXS astrocytes, which is involved in the G1 phase progression, this discrepancy could be explained by the dysregulated progression to the G1 phase (Bertoli et al. 2013). If FXS cells both enter and exit S faster than CTR cells, the proportion of cells incorporating EdU could be similar between CTR and FXS astrocytes. These results are partially consistent with the previous study which showed adult NPCs from Fmr1 KO mice had increased protein expression of CDK4 and cyclin D1 compared with that of WT mice (Luo et al. 2010). However, a study using human stem cell-derived NPCs found age specific alteration in proliferation of FXS cells (Kang et al. 2021).
Neuroimaging studies of FXS patients revealed several alterations in brain morphology, including brain overgrowth and increased size of lateral ventricles (Hazlett et al. 2012, Hunter et al. 2014, Hustyi et al. 2014, Okazaki et al. 2021). Given the robust astrogenesis in early brain development, exaggerated astrocyte proliferation may contribute to macrocephaly in FXS patients. We found increased GFAP+ and S100B+ astrocytes in the FXS postmortem cortex. A previous study quantified the expression level of GFAP in Fmr1 KO mice and identified overexpression of GFAP in the cerebellum (Ellegood et al. 2015). Although we did not measure morphological changes, our result could also be associated with astrocyte activation, which is characterized by hypertrophy of astrocytes and notable upregulation of GFAP (Cai et al. 2000, Ma et al. 2016). While the increased number of astrocytes within the observed area is consistent with previously described enhanced proliferation, altered apoptosis which was not examined here, could also be a contributing factor. Nevertheless, an altered number of astrocytes might impact neuronal and circuit function in FXS.
Unlike neurons, astrocytes are not electrically excitable cells, instead, they communicate via intracellular Ca2+ elevation, which either occurs spontaneously or is evoked by environmental stimuli (Bazargani and Attwell 2016). The increased peak amplitude of ATP-induced Ca2+ signaling we observed in FXS human astrocytes is consistent with a recent study of primary astrocytes from Fmr1 KO mice that identified upregulated ATP-induced Ca2+ signaling mediated by increased expression of purinergic receptors (Reynolds et al. 2021). Enhanced excitability has been well documented in the FXS field with hyper-responsiveness and reduced adaptation to sensory stimulation and enhanced network synchronization (Castren et al. 2003, Van der Molen et al. 2012, Contractor et al. 2015, Ethridge et al. 2016, Lovelace et al. 2016, He et al. 2017). Astrocyte Ca2+ signaling has been shown to regulate neuronal activity as well as Up states in vivo (Poskanzer and Yuste 2011, Nagai et al. 2019). Therefore, enhanced astrocytic Ca2+ signaling could be an additional mechanism that contributes to enhanced neuronal excitability in FXS (Higashimori et al. 2016).
FMRP is a translational regulator that binds with mRNA in actively translating polyribosomes (Corbin et al. 1997). More than 3,700 mRNAs were identified as FMRP targets in human organoids (Kang et al. 2021), indicating the profound alteration in the transcriptomic profiles upon loss of FMRP. We examined the protein expression profiles of mature hiPSC- and hESC-astrocytes using LC-MS and identified several dysregulated pathways using IPA. Some of our findings, such as dysregulated ribosomes and altered translation while expected based on the literature (Li et al. 2020, Seo et al. 2022), are still novel as shown here for astrocytes specifically. However, the proteomic analysis also pointed towards novel, previously unappreciated changes in FXS astrocytes which we have validated. Specifically, we identified increased expression of drebrin, an actin-binding protein, in astrocytes derived from three of the four FXS lines. Drebrin has been mainly studied in neurons where it has well defined roles in developing neurons and synaptic plasticity (Borovac et al. 2018). However, drebrin is also enriched in human fetal astrocytes (Zhang et al. 2016) and recently has been shown to be upregulated in astrocytes following injury (Schiweck et al. 2021). The increased expression of drebrin suggests that FXS astrocytes are in a heightened reactive state and that astrocyte process outgrowth might be impaired in FXS.
A major finding of this study was the dysregulated cholesterol biosynthesis in astrocytes derived from human FXS stem cells. Reduced cholesterol in serum and platelets of FXS patients (Berry-Kravis et al. 2015, Caku et al. 2017) as well as in the serum of an FXS rat model (Parente et al. 2022) have previously been reported. Brain cholesterol metabolism is considered to be separated from the rest of the body because of the blood-brain barrier (Kong et al. 2017). Indeed, the report of cholesterol levels in the brain of a rat FXS model has shown increased levels of cholesterol and only in a single brain region (Parente et al. 2022). In the adult brain, astrocytes are considered the main producers of cholesterol (Ho et al. 2022). Our study was the first to measure cholesterol and multiple sterol precursors in astrocytes derived from FXS patient stem cells. The cholesterol biosynthesis pathway is complex with multiple feedback mechanisms maintaining cholesterol homeostasis (Mitsche et al. 2015). However, consistent with altered expression of several proteins involved in cholesterol biosynthesis pathway, we detected increased levels of lanosterol an early precursor while decreased levels of cholesterol. It will be important in future studies to measure levels of cholesterol released from astrocytes as well as to perform a comprehensive analysis of sterols in brains of Fmr1 KO mice.
In summary, we developed a human astrocyte model of FXS and showed that loss of FMRP could lead to alterations in development and function of astrocytes. Further, we identified alteration in proteomic profiles of human astrocytes associated with loss of FMRP. Among these alterations, some are consistent with previous results in Fmr1 KO mice, such as increased amplitude of Ca2+ signaling of FXS human astrocytes as well as dysregulated regulation of translation. Importantly, we discovered new features of FXS astrocytes that have not been described previously. An in-depth analysis on these DEPs could identify additional human-specific targets with therapeutic potentials. Taken together, our study sets a stage for understanding the role of astrocytes in the pathology of FXS and provides a human-based preclinical model for verifying and identifying molecular targets in FXS treatment.
Supplementary Material
Main points.
Astrocytes derived from FXS patient hiPSC display shortened cell cycle and enhanced calcium signaling.
Pervasively altered proteome identifies impaired cholesterol biosynthesis.
The study highlights astrocytes as a novel therapeutic target for FXS treatment.
Acknowledgments:
This research is supported by NIH funding R01NS109381 and R21NS122157 to A.D and Nebraska Stem Cell Grant to A.D. The University of Nebraska Medical Center Mass Spectrometry and Proteomics Core Facility is administrated through the Office of the Vice Chancellor for Research and supported by state funds from the Nebraska Research Initiative (NRI). We thank the Bioinformatics and Systems Biology Core at UNMC for providing data analysis services. The core receives support from Nebraska Research Initiative (NRI) and NIH (2P20GM103427, 5P30CA036727 and 2U54GM115458). We thank Dr. Thiago C Genaro Mattos for LS-MS/MS technical assistance, Dr. Terri Tallman for providing sterol standards and Kendall Panas for assistance with graphical design.
Footnotes
Data available on request from the authors
References
- Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, Menashe I, Wadkins T, Banerjee-Basu S and Packer A (2013). “SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs).” Mol Autism 4(1): 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achuta VS, Moykkynen T, Peteri UK, Turconi G, Rivera C, Keinanen K and Castren ML (2018). “Functional changes of AMPA responses in human induced pluripotent stem cell-derived neural progenitors in fragile X syndrome.” Sci Signal 11(513). [DOI] [PubMed] [Google Scholar]
- Agarwal A, Wu PH, Hughes EG, Fukaya M, Tischfield MA, Langseth AJ, Wirtz D and Bergles DE (2017). “Transient Opening of the Mitochondrial Permeability Transition Pore Induces Microdomain Calcium Transients in Astrocyte Processes.” Neuron 93(3): 587–605 e587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agulhon C, Fiacco TA and McCarthy KD (2010). “Hippocampal short- and long-term plasticity are not modulated by astrocyte Ca2+ signaling.” Science 327(5970): 1250–1254. [DOI] [PubMed] [Google Scholar]
- Bayraktar OA, Bartels T, Holmqvist S, Kleshchevnikov V, Martirosyan A, Polioudakis D, Ben Haim L, Young AMH, Batiuk MY, Prakash K, Brown A, Roberts K, Paredes MF, Kawaguchi R, Stockley JH, Sabeur K, Chang SM, Huang E, Hutchinson P, Ullian EM, Hemberg M, Coppola G, Holt MG, Geschwind DH and Rowitch DH (2020). “Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map.” Nat Neurosci 23(4): 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazargani N and Attwell D (2016). “Astrocyte calcium signaling: the third wave.” Nat Neurosci 19(2): 182–189. [DOI] [PubMed] [Google Scholar]
- Benjamini Y and Hochberg Y (1995). “Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing.” Journal of the Royal Statistical Society. 57: 289–300. [Google Scholar]
- Bergles DE and Jahr CE (1997). “Synaptic activation of glutamate transporters in hippocampal astrocytes.” Neuron 19(6): 1297–1308. [DOI] [PubMed] [Google Scholar]
- Berry-Kravis E, Levin R, Shah H, Mathur S, Darnell JC and Ouyang B (2015). “Cholesterol levels in fragile X syndrome.” Am J Med Genet A 167A(2): 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli C, Skotheim JM and de Bruin RA (2013). “Control of cell cycle transcription during G1 and S phases.” Nat Rev Mol Cell Biol 14(8): 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B (2022). “preprocessCore: A collection of pre-processing functions.” R package version 1.58.0,. [Google Scholar]
- Borovac J, Bosch M and Okamoto K (2018). “Regulation of actin dynamics during structural plasticity of dendritic spines: Signaling messengers and actin-binding proteins.” Mol Cell Neurosci 91: 122–130. [DOI] [PubMed] [Google Scholar]
- Brick DJ, Nethercott HE, Montesano S, Banuelos MG, Stover AE, Schutte SS, O’Dowd DK, Hagerman RJ, Ono M, Hessl DR, Tassone F and Schwartz PH (2014). “The Autism Spectrum Disorders Stem Cell Resource at Children’s Hospital of Orange County: Implications for Disease Modeling and Drug Discovery.” Stem Cells Transl Med 3(11): 1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brighi C, Salaris F, Soloperto A, Cordella F, Ghirga S, de Turris V, Rosito M, Porceddu PF, D’Antoni C, Reggiani A, Rosa A and Di Angelantonio S (2021). “Novel fragile X syndrome 2D and 3D brain models based on human isogenic FMRP-KO iPSCs.” Cell Death Dis 12(5): 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Pan ZL, Pang Y, Evans OB and Rhodes PG (2000). “Cytokine induction in fetal rat brains and brain injury in neonatal rats after maternal lipopolysaccharide administration.” Pediatr Res 47(1): 64–72. [DOI] [PubMed] [Google Scholar]
- Caku A, Seidah NG, Lortie A, Gagne N, Perron P, Dube J and Corbin F (2017). “New insights of altered lipid profile in Fragile X Syndrome.” PLoS One 12(3): e0174301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals I, Ginisty A, Quist E, Timmerman R, Fritze J, Miskinyte G, Monni E, Hansen MG, Hidalgo I, Bryder D, Bengzon J and Ahlenius H (2018). “Rapid and efficient induction of functional astrocytes from human pluripotent stem cells.” Nat Methods 15(9): 693–696. [DOI] [PubMed] [Google Scholar]
- Castren M, Paakkonen A, Tarkka IM, Ryynanen M and Partanen J (2003). “Augmentation of auditory N1 in children with fragile X syndrome.” Brain Topogr 15(3): 165–171. [DOI] [PubMed] [Google Scholar]
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak MY, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL and Mallick P (2012). “A cross-platform toolkit for mass spectrometry and proteomics.” Nat Biotechnol 30(10): 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehrehasa F, Meedeniya AC, Dwyer P, Abrahamsen G and Mackay-Sim A (2009). “EdU, a new thymidine analogue for labelling proliferating cells in the nervous system.” J Neurosci Methods 177(1): 122–130. [DOI] [PubMed] [Google Scholar]
- Chen C, Jiang P, Xue H, Peterson SE, Tran HT, McCann AE, Parast MM, Li S, Pleasure DE, Laurent LC, Loring JF, Liu Y and Deng W (2014). “Role of astroglia in Down’s syndrome revealed by patient-derived human-induced pluripotent stem cells.” Nat Commun 5: 4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H (2022). VennDiagram: Generate High-Resolution Venn and Euler Plots. R package version 1.7.3. [Google Scholar]
- Contractor A, Klyachko VA and Portera-Cailliau C (2015). “Altered Neuronal and Circuit Excitability in Fragile X Syndrome.” Neuron 87(4): 699–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin F, Bouillon M, Fortin A, Morin S, Rousseau F and Khandjian EW (1997). “The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes.” Hum Mol Genet 6(9): 1465–1472. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD and Darnell RB (2011). “FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism.” Cell 146(2): 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Majo M, Koontz M, Rowitch D and Ullian EM (2020). “An update on human astrocytes and their role in development and disease.” Glia 68(4): 685–704. [DOI] [PubMed] [Google Scholar]
- Doers ME, Musser MT, Nichol R, Berndt ER, Baker M, Gomez TM, Zhang SC, Abbeduto L and Bhattacharyya A (2014). “iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth.” Stem Cells Dev 23(15): 1777–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, Yaron Y, Eden A, Yanuka O, Benvenisty N and Ben-Yosef D (2007). “Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos.” Cell Stem Cell 1(5): 568–577. [DOI] [PubMed] [Google Scholar]
- Ellegood J, Anagnostou E, Babineau BA, Crawley JN, Lin L, Genestine M, DiCicco-Bloom E, Lai JK, Foster JA, Penagarikano O, Geschwind DH, Pacey LK, Hampson DR, Laliberte CL, Mills AA, Tam E, Osborne LR, Kouser M, Espinosa-Becerra F, Xuan Z, Powell CM, Raznahan A, Robins DM, Nakai N, Nakatani J, Takumi T, van Eede MC, Kerr TM, Muller C, Blakely RD, Veenstra-VanderWeele J, Henkelman RM and Lerch JP (2015). “Clustering autism: using neuroanatomical differences in 26 mouse models to gain insight into the heterogeneity.” Mol Psychiatry 20(1): 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethridge LE, White SP, Mosconi MW, Wang J, Byerly MJ and Sweeney JA (2016). “Reduced habituation of auditory evoked potentials indicate cortical hyper-excitability in Fragile X Syndrome.” Transl Psychiatry 6: e787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Yu G and Dusa A (2021). ggVennDiagram: A ‘ggplot2’ Implement of Venn Diagram. R package version 1.2.0. [Google Scholar]
- Ge WP and Jia JM (2016). “Local production of astrocytes in the cerebral cortex.” Neuroscience 323: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt J, Tomishima MJ, Zaninovic N, Colak D, Yan Z, Zhan Q, Rosenwaks Z, Jaffrey SR and Schildkraut CL (2014). “The DNA replication program is altered at the FMR1 locus in fragile X embryonic stem cells.” Mol Cell 53(1): 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golias CH, Charalabopoulos A and Charalabopoulos K (2004). “Cell proliferation and cell cycle control: a mini review.” Int J Clin Pract 58(12): 1134–1141. [DOI] [PubMed] [Google Scholar]
- Goubard V, Fino E and Venance L (2011). “Contribution of astrocytic glutamate and GABA uptake to corticostriatal information processing.” J Physiol 589(Pt 9): 2301–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef JD, Wu H, Ng C, Sun C, Villegas V, Qadir D, Jesseman K, Warren ST, Jaenisch R, Cacace A and Wallace O (2020). “Partial FMRP expression is sufficient to normalize neuronal hyperactivity in Fragile X neurons.” Eur J Neurosci 51(10): 2143–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen HJ and Jensen EB (1987). “The efficiency of systematic sampling in stereology and its prediction.” J Microsc 147(Pt 3): 229–263. [DOI] [PubMed] [Google Scholar]
- Harris L, Zalucki O and Piper M (2018). “BrdU/EdU dual labeling to determine the cell-cycle dynamics of defined cellular subpopulations.” J Mol Histol 49(3): 229–234. [DOI] [PubMed] [Google Scholar]
- Hazlett HC, Poe MD, Lightbody AA, Styner M, MacFall JR, Reiss AL and Piven J (2012). “Trajectories of early brain volume development in fragile X syndrome and autism.” J Am Acad Child Adolesc Psychiatry 51(9): 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CX, Cantu DA, Mantri SS, Zeiger WA, Goel A and Portera-Cailliau C (2017). “Tactile Defensiveness and Impaired Adaptation of Neuronal Activity in the Fmr1 Knock-Out Mouse Model of Autism.” J Neurosci 37(27): 6475–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashimori H, Morel L, Huth J, Lindemann L, Dulla C, Taylor A, Freeman M and Yang Y (2013). “Astroglial FMRP-dependent translational down-regulation of mGluR5 underlies glutamate transporter GLT1 dysregulation in the fragile X mouse.” Hum Mol Genet 22(10): 2041–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashimori H, Schin CS, Chiang MS, Morel L, Shoneye TA, Nelson DL and Yang Y (2016). “Selective Deletion of Astroglial FMRP Dysregulates Glutamate Transporter GLT1 and Contributes to Fragile X Syndrome Phenotypes In Vivo.” J Neurosci 36(27): 7079–7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho WY, Hartmann H and Ling SC (2022). “Central nervous system cholesterol metabolism in health and disease.” IUBMB Life 74(8): 826–841. [DOI] [PubMed] [Google Scholar]
- Hodges JL, Yu X, Gilmore A, Bennett H, Tjia M, Perna JF, Chen CC, Li X, Lu J and Zuo Y (2016). “Astrocytic Contributions to Synaptic and Learning Abnormalities in a Mouse Model of Fragile X Syndrome.” Biol Psychiatry s0006–3223 (16): 32779–32772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J, Rivero-Arias O, Angelov A, Kim E, Fotheringham I and Leal J (2014). “Epidemiology of fragile X syndrome: a systematic review and meta-analysis.” Am J Med Genet A 164A(7): 1648–1658. [DOI] [PubMed] [Google Scholar]
- Hustyi KM, Hall SS, Jo B, Lightbody AA and Reiss AL (2014). “Longitudinal trajectories of aberrant behavior in fragile X syndrome.” Res Dev Disabil 35(11): 2691–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki T, Uehata M, Tamechika I, Keel J, Nonomura K, Maekawa M and Narumiya S (2000). “Pharmacological properties of Y-27632, a specific inhibitor of rho-associated kinases.” Mol Pharmacol 57(5): 976–983. [PubMed] [Google Scholar]
- Jacobs S, Nathwani M and Doering LC (2010). “Fragile X astrocytes induce developmental delays in dendrite maturation and synaptic protein expression.” BMC Neurosci 11: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Chen C, Liu XB, Pleasure DE, Liu Y and Deng W (2016). “Human iPSC-Derived Immature Astroglia Promote Oligodendrogenesis by Increasing TIMP-1 Secretion.” Cell Rep 15(6): 1303–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Diaz-Castro B, Looger LL and Khakh BS (2016). “Dysfunctional Calcium and Glutamate Signaling in Striatal Astrocytes from Huntington’s Disease Model Mice.” J Neurosci 36(12): 3453–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Zhou Y, Li Y, Han Y, Xu J, Niu W, Li Z, Liu S, Feng H, Huang W, Duan R, Xu T, Raj N, Zhang F, Dou J, Xu C, Wu H, Bassell GJ, Warren ST, Allen EG, Jin P and Wen Z (2021). “A human forebrain organoid model of fragile X syndrome exhibits altered neurogenesis and highlights new treatment strategies.” Nat Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Zhou Y, Li Y, Han Y, Xu J, Niu W, Li Z, Liu S, Feng H, Huang W, Duan R, Xu T, Raj N, Zhang F, Dou J, Xu C, Wu H, Bassell GJ, Warren ST, Allen EG, Jin P and Wen Z (2021). “A human forebrain organoid model of fragile X syndrome exhibits altered neurogenesis and highlights new treatment strategies.” Nat Neurosci 24(10): 1377–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS and Deneen B (2019). “The Emerging Nature of Astrocyte Diversity.” Annu Rev Neurosci 42: 187–207. [DOI] [PubMed] [Google Scholar]
- Kolde R (2018). pheatmap : Implementation of heatmaps that offers more control over dimensions and appearance. R package version 1.0.12. [Google Scholar]
- Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D and Nesvizhskii AI (2017). “MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics.” Nat Methods 14(5): 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J Jr. and Tugendreich S (2014). “Causal analysis approaches in Ingenuity Pathway Analysis.” Bioinformatics 30(4): 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krencik R and Zhang SC (2011). “Directed differentiation of functional astroglial subtypes from human pluripotent stem cells.” Nat Protoc 6(11): 1710–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene H (1960). “Robust tests for equality of variances.” 278–292. [Google Scholar]
- Li M, Shin J, Risgaard RD, Parries MJ, Wang J, Chasman D, Liu S, Roy S, Bhattacharyya A and Zhao X (2020). “Identification of FMR1-regulated molecular networks in human neurodevelopment.” Genome Res 30(3): 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelace JW, Wen TH, Reinhard S, Hsu MS, Sidhu H, Ethell IM, Binder DK and Razak KA (2016). “Matrix metalloproteinase-9 deletion rescues auditory evoked potential habituation deficit in a mouse model of Fragile X Syndrome.” Neurobiol Dis 89: 126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Shan G, Guo W, Smrt RD, Johnson EB, Li X, Pfeiffer RL, Szulwach KE, Duan R, Barkho BZ, Li W, Liu C, Jin P and Zhao X (2010). “Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells.” PLoS Genet 6(4): e1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Qiao Q, Tsai JW, Yang G, Li W and Gan WB (2016). “Experience-dependent plasticity of dendritic spines of layer 2/3 pyramidal neurons in the mouse cortex.” Dev Neurobiol 76(3): 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Morrison H, Price DJ and Mason JO (2005). “Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis.” Dev Biol 283(1): 113–127. [DOI] [PubMed] [Google Scholar]
- Maurin T, Lebrigand K, Castagnola S, Paquet A, Jarjat M, Popa A, Grossi M, Rage F and Bardoni B (2018). “HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein.” Nucleic Acids Res 46(12): 6344–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minelli A, DeBiasi S, Brecha NC, Zuccarello LV and Conti F (1996). “GAT-3, a high-affinity GABA plasma membrane transporter, is localized to astrocytic processes, and it is not confined to the vicinity of GABAergic synapses in the cerebral cortex.” J Neurosci 16(19): 6255–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsche MA, McDonald JG, Hobbs HH and Cohen JC (2015). “Flux analysis of cholesterol biosynthesis in vivo reveals multiple tissue and cell-type specific pathways.” Elife 4: e07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Krencik R, Ullian EM, Tsai HH, Deneen B, Richardson WD, Barres BA and Rowitch DH (2012). “Astrocytes and disease: a neurodevelopmental perspective.” Genes Dev 26(9): 891–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D and Groop LC (2003). “PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes.” Nat Genet 34(3): 267–273. [DOI] [PubMed] [Google Scholar]
- Motulsky HJ and Brown RE (2006). “Detecting outliers when fitting data with nonlinear regression - a new method based on robust nonlinear regression and the false discovery rate.” BMC Bioinformatics 7: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai J, Rajbhandari AK, Gangwani MR, Hachisuka A, Coppola G, Masmanidis SC, Fanselow MS and Khakh BS (2019). “Hyperactivity with Disrupted Attention by Activation of an Astrocyte Synaptogenic Cue.” Cell 177(5): 1280–1292 e1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Takano T, Han X, He W, Lin JH, Wang F, Xu Q, Wyatt JD, Pilcher W, Ojemann JG, Ransom BR, Goldman SA and Nedergaard M (2009). “Uniquely hominid features of adult human astrocytes.” J Neurosci 29(10): 3276–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki T, Adachi K, Matsuura K, Oyama Y, Nose M, Shirahata E, Abe T, Hasegawa T, Maihara T, Maegaki Y and Nanba E (2021). “Clinical Characteristics of Fragile X Syndrome Patients in Japan.” Yonago Acta Med 64(1): 30–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacey LK and Doering LC (2007). “Developmental expression of FMRP in the astrocyte lineage: implications for fragile X syndrome.” Glia 55(15): 1601–1609. [DOI] [PubMed] [Google Scholar]
- Parente M, Tonini C, Buzzelli V, Carbone E, Trezza V and Pallottini V (2022). “Brain Cholesterol Biosynthetic Pathway Is Altered in a Preclinical Model of Fragile X Syndrome.” Int J Mol Sci 23(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekny M, Pekna M, Messing A, Steinhauser C, Lee JM, Parpura V, Hol EM, Sofroniew MV and Verkhratsky A (2016). “Astrocytes: a central element in neurological diseases.” Acta Neuropathol 131(3): 323–345. [DOI] [PubMed] [Google Scholar]
- Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT and Nelson DL (1991). “Absence of expression of the FMR-1 gene in fragile X syndrome.” Cell 66(4): 817–822. [DOI] [PubMed] [Google Scholar]
- Pologruto TA, Sabatini BL and Svoboda K (2003). “ScanImage: flexible software for operating laser scanning microscopes.” Biomed Eng Online 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poskanzer KE and Yuste R (2011). “Astrocytic regulation of cortical UP states.” Proc Natl Acad Sci U S A 108(45): 18453–18458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj N, McEachin ZT, Harousseau W, Zhou Y, Zhang F, Merritt-Garza ME, Taliaferro JM, Kalinowska M, Marro SG, Hales CM, Berry-Kravis E, Wolf-Ochoa MW, Martinez-Cerdeno V, Wernig M, Chen L, Klann E, Warren ST, Jin P, Wen Z and Bassell GJ (2021). “Cell-type-specific profiling of human cellular models of fragile X syndrome reveal PI3K-dependent defects in translation and neurogenesis.” Cell Rep 35(2): 108991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds KE, Krasovska V and Scott AL (2021). “Converging purinergic and immune signaling pathways drive IL-6 secretion by Fragile X cortical astrocytes via STAT3.” J Neuroimmunol 361: 577745. [DOI] [PubMed] [Google Scholar]
- Reynolds KE, Wong CR and Scott AL (2021). “Astrocyte-mediated purinergic signaling is upregulated in a mouse model of Fragile X syndrome.” Glia 69(7): 1816–1832. [DOI] [PubMed] [Google Scholar]
- Richter JD and Zhao X (2021). “The molecular biology of FMRP: new insights into fragile X syndrome.” Nat Rev Neurosci 22(4): 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiweck J, Murk K, Ledderose J, Munster-Wandowski A, Ornaghi M, Vida I and Eickholt BJ (2021). “Drebrin controls scar formation and astrocyte reactivity upon traumatic brain injury by regulating membrane trafficking.” Nat Commun 12(1): 1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo SS, Louros SR, Anstey N, Gonzalez-Lozano MA, Harper CB, Verity NC, Dando O, Thomson SR, Darnell JC, Kind PC, Li KW and Osterweil EK (2022). “Excess ribosomal protein production unbalances translation in a model of Fragile X Syndrome.” Nat Commun 13(1): 3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Wang F, Li M, Sah N, Stockton ME, Tidei JJ, Gao Y, Korabelnikov T, Kannan S, Vevea JD, Chapman ER, Bhattacharyya A, van Praag H and Zhao X (2019). “Reduced mitochondrial fusion and Huntingtin levels contribute to impaired dendritic maturation and behavioral deficits in Fmr1-mutant mice.” Nat Neurosci 22(3): 386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigetomi E and Koizumi S (2016). “Visualization of diversity of calcium signals in astrocytes.” Nihon Yakurigaku Zasshi 148(2): 75–80. [DOI] [PubMed] [Google Scholar]
- Shitik EM, Velmiskina AA, Dolskiy AA and Yudkin DV (2020). “Reactivation of FMR1 gene expression is a promising strategy for fragile X syndrome therapy.” Gene Ther 27(6): 247–253. [DOI] [PubMed] [Google Scholar]
- Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, Reimer R, Quake SR, Barres BA and Pasca SP (2017). “Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells.” Neuron 95(4): 779–790 e776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R, Huang BS, Venugopal S, Johnston AD, Chai H, Zeng H, Golshani P and Khakh BS (2015). “Ca(2+) signaling in astrocytes from Ip3r2(−/−) mice in brain slices and during startle responses in vivo.” Nat Neurosci 18(5): 708–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stekhoven D (2022). missForest: Nonparametric Missing Value Imputation using Random Forest. R package version 1.5. [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES and Mesirov JP (2005). “Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles.” Proc Natl Acad Sci U S A 102(43): 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunamura N, Iwashita S, Enomoto K, Kadoshima T and Isono F (2018). “Loss of the fragile X mental retardation protein causes aberrant differentiation in human neural progenitor cells.” Sci Rep 8(1): 11585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ and Mering CV (2019). “STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets.” Nucleic Acids Res 47(D1): D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K and Yamanaka S (2007). “Induction of pluripotent stem cells from adult human fibroblasts by defined factors.” Cell 131(5): 861–872. [DOI] [PubMed] [Google Scholar]
- Tallman KA, Allen LB, Klingelsmith KB, Anderson A, Genaro-Mattos TC, Mirnics K, Porter NA and Korade Z (2021). “Prescription Medications Alter Neuronal and Glial Cholesterol Synthesis.” ACS Chem Neurosci 12(4): 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchieu J, Calder EL, Guttikonda SR, Gutzwiller EM, Aromolaran KA, Steinbeck JA, Goldstein PA and Studer L (2019). “NFIA is a gliogenic switch enabling rapid derivation of functional human astrocytes from pluripotent stem cells.” Nat Biotechnol 37(3): 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcw J, Wang M, Pimenova AA, Bowles KR, Hartley BJ, Lacin E, Machlovi SI, Abdelaal R, Karch CM, Phatnani H, Slesinger PA, Zhang B, Goate AM and Brennand KJ (2017). “An Efficient Platform for Astrocyte Differentiation from Human Induced Pluripotent Stem Cells.” Stem Cell Reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telias M, Kuznitsov-Yanovsky L, Segal M and Ben-Yosef D (2015). “Functional Deficiencies in Fragile X Neurons Derived from Human Embryonic Stem Cells.” J Neurosci 35(46): 15295–15306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telias M, Segal M and Ben-Yosef D (2013). “Neural differentiation of Fragile X human Embryonic Stem Cells reveals abnormal patterns of development despite successful neurogenesis.” Dev Biol 374(1): 32–45. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS and Jones JM (1998). “Embryonic stem cell lines derived from human blastocysts.” Science 282(5391): 1145–1147. [DOI] [PubMed] [Google Scholar]
- Urbach A, Bar-Nur O, Daley GQ and Benvenisty N (2010). “Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells.” Cell Stem Cell 6(5): 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utami KH, Skotte NH, Colaco AR, Yusof N, Sim B, Yeo XY, Bae HG, Garcia-Miralles M, Radulescu CI, Chen Q, Chaldaiopoulou G, Liany H, Nama S, Peteri UA, Sampath P, Castren ML, Jung S, Mann M and Pouladi MA (2020). “Integrative Analysis Identifies Key Molecular Signatures Underlying Neurodevelopmental Deficits in Fragile X Syndrome.” Biol Psychiatry 88(6): 500–511. [DOI] [PubMed] [Google Scholar]
- Van der Molen MJ, Van der Molen MW, Ridderinkhof KR, Hamel BC, Curfs LM and Ramakers GJ (2012). “Auditory and visual cortical activity during selective attention in fragile X syndrome: a cascade of processing deficiencies.” Clin Neurophysiol 123(4): 720–729. [DOI] [PubMed] [Google Scholar]
- Verkhratsky A and Nedergaard M (2018). “Physiology of Astroglia.” Physiol Rev 98(1): 239–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LW, Berry-Kravis E and Hagerman RJ (2010). “Fragile X: leading the way for targeted treatments in autism.” Neurotherapeutics 7(3): 264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis, Springer-Verlag; New York. [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, Duncan JA 3rd, Cheshier SH, Shuer LM, Chang EF, Grant GA, Gephart MG and Barres BA (2016). “Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse.” Neuron 89(1): 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Marro SG, Zhang Y, Arendt KL, Patzke C, Zhou B, Fair T, Yang N, Sudhof TC, Wernig M and Chen L (2018). “The fragile X mutation impairs homeostatic plasticity in human neurons by blocking synaptic retinoic acid signaling.” Sci Transl Med 10(452). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.