Abstract
Background and Aims
Over the past decade, the DNA methylome has been increasingly studied in peripheral blood of inflammatory bowel disease [IBD] patients. However, a comprehensive summary and meta-analysis of peripheral blood leukocyte [PBL] DNA methylation studies has thus far not been conducted. Here, we systematically reviewed all available literature up to February 2022 and summarized the observations by means of meta-analysis.
Methods
We conducted a systematic search and critical appraisal of IBD-associated DNA methylation studies in PBL using the biomarker-based cross-sectional studies [BIOCROSS] tool. Subsequently, we performed meta-analyses on the summary statistics obtained from epigenome-wide association studies [EWAS] that included patients with Crohn’s disease [CD], ulcerative colitis [UC] and/or healthy controls [HC].
Results
Altogether, we included 15 studies for systematic review. Critical appraisal revealed large methodological and outcome heterogeneity between studies. Summary statistics were obtained from four studies based on a cumulative 552 samples [177 CD, 132 UC and 243 HC]. Consistent differential methylation was identified for 256 differentially methylated probes [DMPs; Bonferroni-adjusted p ≤ 0.05] when comparing CD with HC and 103 when comparing UC with HC. Comparing IBD [CD + UC] with HC resulted in 224 DMPs. Importantly, several of the previously identified DMPs, such as VMP1/TMEM49/MIR21 and RPS6KA2, were consistently differentially methylated across all studies.
Conclusion
Methodological homogenization of IBD epigenetic studies is needed to allow for easier aggregation and independent validation. Nonetheless, we were able to confirm previous observations. Our results can serve as the basis for future IBD epigenetic biomarker research in PBL.
Keywords: DNA methylation, epigenetics, inflammatory bowel diseases, biomarkers, systematic review and meta-analysis
1. Introduction
Inflammatory bowel diseases [IBD], including Crohn’s disease [CD] and ulcerative colitis [UC] are chronic, relapsing and remitting inflammatory disorders affecting an estimated 3 million patients in the USA and Europe alone.1,2 While the exact aetiopathogenesis of IBD remains unknown, current consensus suggests a dysregulated immune response to specific alterations in the microbial composition of genetically susceptible patients that either are triggered or caused by environmental factors.3 While numerous genome-wide association studies [GWAS] have been performed, the results thereof explain only part of the heritability observed in familial studies with high rates of discordance observed in monozygotic twins.4–10 Moreover, a rapid increase in the incidence of IBD has been observed in newly industrialized countries, which is thought to be the result of the increasing adoption of a western lifestyle.1 Together, these observations suggest an important role for gene–environment interactions in the aetiopathogenesis of IBD.
Epigenetics represent mitotically heritable mechanisms that affect the readability of the genome without changing the actual genetic sequence itself. Epigenetic modifications can regulate transcription and could therefore mediate gene–environment interactions. Epigenetic mechanisms are particularly interesting in complex multifactorial diseases, such as IBD, due to their dynamic interaction with the environment.11 One of the most widely studied epigenetic mechanisms is DNA methylation, representing a methyl group covalently bound to the cytosine of a cytosine–guanine dinucleotide [CpG]. The presence of DNA methylation in gene promoters is often inversely correlated with gene expression.12–14 Due to the growing popularity of epigenetics, an increasing number of studies have sought to understand the role of DNA methylation in IBD pathogenesis and aetiology, which was further strengthened by the discovery of an IBD-associated genetic variant associated with DNMT3A, a key de novo methylation enzyme.15 Accordingly, numerous differentially methylated positions [DMPs], regions [DMRs] or genes [DMGs] associated with various IBD phenotypes across different tissues have been identified in both paediatric and adult cohorts.16–24 Together, these studies make a case for using DNA methylation in classifying IBD phenotypes,20,25,26 thereby supplementing clinical care with potential novel drug and biomarker targets.
Despite the numerous studies available to date, translation to clinical practice has not occurred. All studies report different sets of biomarkers with different levels of statistical power. Moreover, many studies lack information on patient characteristics that are known to be associated with changes in methylation, such as smoking behaviour.27 To adequately understand the observations thus far, various attempts have been made to summarize the DNA methylation studies on IBD.
The most recent meta-analysis of IBD-associated epigenetic studies focused on DNA methylation differences in mucosal biopsies of both paediatric and adult IBD patients, including six separate datasets, yet a similar approach in peripheral blood was not performed.28 Additionally, four studies reviewed the available DNA methylation literature in paediatric and adult IBD patients,29–32 but did not perform a meta-analysis. Since 2014, the available literature on DNA methylation and IBD has further expanded as a result of the dropping costs of both microarray- and sequencing-based technologies, while the latter have become increasingly capable of covering larger numbers of cytosines. With the ever-increasing pool of data in peripheral blood, the opportunity to combine these data, thereby increasing statistical power, provides a potential to guide future methylation studies in IBD. We therefore sought to summarize the available peripheral blood DNA methylation data by systematically reviewing all currently available literature on peripheral blood leukocyte [PBL] DNA methylation in IBD and performing a meta-analysis on the available Illumina HumanMethylation BeadChip 450k data.
2. Methods
The methodological protocol implemented in this study was registered on PROSPERO [ID: CRD42020176655].
2.1. Systematic review
A literature search was performed on February 1, 2022 using the PubMed/MEDLINE, EMBASE [OVID], Cochrane library and CINAHL [ebsco] databases with the help of a scientific librarian at the Amsterdam University Medical Center. Main search criteria were ‘Crohn’s disease’, ‘Inflammatory Bowel Diseases’, ‘colitis’ and ‘DNA methylation’. In vitro and animal studies were excluded from this search. A detailed description of our search strategy can be found in the Supplementary information file. Screening and eligibility-based assessment was conducted separately by V.J. and I.L.H. adhering to pre-defined inclusion and exclusion criteria [Table 1] using the web application RAYYAN.33 Conflicts were resolved by A.Y.F.L.Y., whereupon eligible studies were read in full. A pre-specified data extraction form was then used to extract data from the included studies for assessment of study quality and a synthesis of evidence. Extracted information included author, year of publication, study population, sample size, patient demographics, type of assay, study design [targeted or whole-genome] and, if applicable, which CpGs of interest were analysed.
Table 1.
Inclusion and exclusion criteria
|
Inclusion criteria
1. Any prospective or retrospective study measuring peripheral blood leukocyte DNA methylation in both paediatric or adult IBD patients [cohort, case-control, RCT] 2. Epigenome-wide association studies [EWAS] and targeted DNA methylation study designs 3. DNA methylation analysis performed on peripheral whole blood or specific peripheral blood cell types 4. Studies comparing DNA methylation between Crohn’s disease and ulcerative colitis or comparing either disease with healthy controls Exclusion criteria 1. Studies using intestinal tissue samples 2. Studies investigating DNA methylation markers primarily in patients with cancer or pre-stages of cancer 3. Studies with fewer than five included patients per phenotype 4. Case reports, narrative reviews, in vitro studies [e.g. using cell lines], studies of genetic [rather than epigenetic] mutations or animal studies |
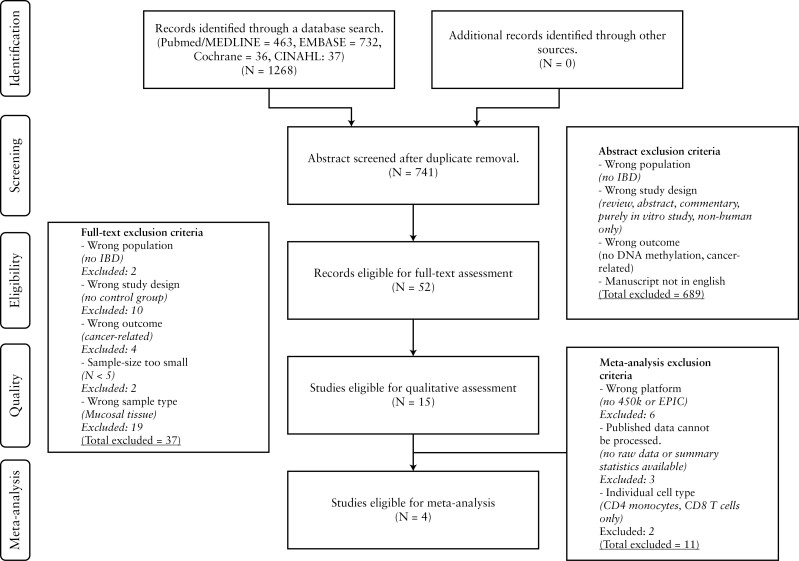
To assess the quality of the selected articles, we used a recently developed quality assessment tool specifically designed for biomarker-based cross-sectional studies [BIOCROSS].34 The BIOCROSS tool originally included ten items covering five domains: ‘Study rational’, ‘Design/Methods’, ‘Data analysis’, ‘Data interpretation’ and ‘Biomarker measurement’, aiming to assess different features of biomarker cross-sectional studies. For the purpose of this systematic review, we adapted the BIOCROSS tool for use in studies aimed at assessing DNA methylation [Supplementart information]. An overview of the entire process is presented in Figure 1.
Figure 1.
Study selection. Studies obtained through database searches were initially screened based on abstract exclusion criteria, followed by full-text exclusion criteria and final meta-analysis exclusion criteria.
2.2. Meta-analysis
For the meta-analysis, we acquired published data from studies that had been conducted in a genome-wide fashion using the Illumina HumanMethylation 450k or EPIC BeadChip array. As the analytical design could only utilize overlapping CpGs, including the Illumina HumanMethylation 27k BeadChip array would discard ~94% of the 450k probes, which we deemed excessive. Accordingly, we decided against including studies that had performed analyses using the Illumina HumanMethylation 27k BeadChip array. We specifically sought to acquire summary statistics to stay as close as possible to the interpretation of the original authors, where we sought to mitigate publication bias by acquiring the full summary statistics from both significant and non-significant CpGs. If the full summary statistics were unavailable, even after request, processed data was downloaded from the Gene Expression Omnibus [GEO], imported into the R statistical environment [v4.1] and processed per the methods outlined in the original publication. However, if the self-generated results were inconsistent with the results outlined in the original publication, the data were discarded. This restriction confined the analyses only to data obtained from the HumanMethylation 450k BeadChip array [Table 2]. Annotation of the CpGs to their location and genes was performed using the annotation file provided by Illumina [HumanMethylation450 v1.2 Manifest] as imported using minfi.35 If no annotated gene was found among the top 20 DMPs per comparison, a manual search was conducted using the UCSC genome browser [hg19] to identify the nearest gene. No specific selection for promoter-bound CpGs was applied. All genes have been annotated using the HUGO nomenclature.
Table 2.
Overview of the datasets included in the meta-analysis
| Samples | Array | Accession ID | |
|---|---|---|---|
| Harris et al. 201236 | 17 CD—PBL 11 UC—PBL 14 HC—PBL |
450k | GSE32148 |
| Adams et al. 201418 | 35 CD—PBL 36 HC—PBL |
450k | NA* |
| Li Yim et al. 201637 | 15 CD—PBL 25 HC—PBL |
450k | GSE81961 |
| Ventham et al. 201620 | 121 CD—PBL 119 UC—PBL 191 HC—PBL |
450k | GSE87650 |
CD, Crohn’s disease; UC, ulcerative colitis; HC, healthy controls; PBL, peripheral blood leukocytes.
*Data acquired following request.
The meta-analysis was subsequently performed by utilizing the framework outlined by Choi et al.38 and the GeneMeta package.39 In short, we calculated the standardized effect size [SES], as represented by Cohen’s d and Hedges’ g, and variance from the t-statistic per probe per study. Heterogeneity between the studies in effect size was quantified using the Cochran Q-test and corrected for by including the Cochran Q-derived moments estimator τ2 as a random effects in the subsequent random-effects model to combine the standardized effect into the unbiased estimator of effect size. Two-sided Z-tests were then performed to calculate the p-value per CpG, after which the resultant p-values were corrected using the Bonferroni correction where CpGs presenting a Bonferroni-adjusted p-value <0.05 were considered to be statistically significantly differentially methylated positions [DMPs].
For the differential methylated gene [DMG] analysis, we sought to investigate whether particular genes were enriched for low p-values. To this end, the meta-analysis p-values for CpGs annotated to a particular gene per the provided annotation obtained from Illumina’s annotation file were combined using Fisher’s method for combining p-values as implemented in the aggregation package.40 The resulting combined p-values were then corrected using the false discovery rate [FDR] where we defined FDR-adjusted p-values <0.05 as statistically significant. The resultant DMGs were then subjected to gene ontology [GO]41,42 overrepresentation analysis using the clusterProfiler package,43 where we considered an FDR-adjusted p-value <0.05 as a statistically significant difference.
3. Results
3.1. Study selection and quality appraisal
A total of 741 records were screened for eligibility according to our pre-defined inclusion criteria [Table 1], after which 15 studies were included for full text systematic review [Figure 1 and Supplementary Table 1] and qualitative assessment using our modified BIOCROSS tool for critical appraisal [Table 3].
Table 3.
Quality assessment
| Quality assessment | Study rationale | Design/Methods | Data analysis | Data interpretation | Biomarker measurement | Quality score | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1. Hypothesis/Objective | 2. Study population selection | 3. Study population representativeness | 4. Study population characteristics | 5. Statistical analysis | 6. Interpretation and evaluation of results | 7. Study limitations | 8. Specimen characteristics and assay methods | 9. Laboratory measurement | ||
| Balasa et al. 201044 | 3 | 2 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 8 |
| Harris et al. 201236 | 3 | 2 | 0 | 1 | 3 | 2 | 1 | 1 | 3 | 16 |
| Nimmo et al. 201222 | 3 | 2 | 1 | 1 | 2 | 2 | 0 | 1 | 2 | 12 |
| Kim et al. 201245 | 3 | 1 | 0 | 0 | 2 | 2 | 1 | 1 | 1 | 11 |
| Adams et al. 201418 | 3 | 3 | 1 | 1 | 4 | 2 | 2 | 2 | 3 | 21 |
| Karatzas et al. 201446 | 3 | 2 | 0 | 1 | 2 | 2 | 1 | 1 | 1 | 13 |
| Li Yim et al. 201637 | 3 | 2 | 0 | 0 | 3 | 2 | 2 | 2 | 2 | 16 |
| McDermott et al. 201623 | 3 | 3 | 0 | 1 | 4 | 2 | 2 | 1 | 3 | 19 |
| Ventham et al. 201620 | 2 | 3 | 1 | 1 | 4 | 2 | 3 | 1 | 3 | 20 |
| Klasic et al. 201847 | 3 | 3 | 1 | 1 | 3 | 2 | 0 | 2 | 1 | 16 |
| Moret-Tatay et al. 201948 | 3 | 2 | 0 | 1 | 1 | 2 | 1 | 2 | 3 | 15 |
| Somineni et al. 201926 | 3 | 3 | 1 | 1 | 3 | 1 | 2 | 1 | 3 | 18 |
| Li Yim et al. 202017 | 2 | 1 | 0 | 1 | 2 | 2 | 1 | 2 | 2 | 13 |
| Samarani et al. 202049 | 3 | 2 | 0 | 1 | 2 | 1 | 1 | 1 | 1 | 12 |
| Gasparetto et al. 202050 | 3 | 3 | 0 | 1 | 4 | 2 | 2 | 1 | 3 | 19 |
Quality items were scored as completely present [green], completely absent [red] or partly present [yellow]. The final quality score is an addition of all items present. All nine items were interpreted as defined by the modified version of the BIOCROSS tool [Supplementary Table 2].
Systematically reviewing all 15 studies indicated that 14 studies provided a clear rationale for studying DNA methylation in relation to IBD pathogenesis [domain 1]. Twelve studies presented a comparable case and control group [domain 2] and 13 provided a concise summary of the clinical characteristics [domain 4]. In addition, 11 studies described the laboratory procedures used in their study in detail [domain 8]. Finally, 12 contextualized the observed differences in DNA methylation with respect to existing literature, thereby providing insight into the functional implications [domain 6]. By contrast, we noted that many studies lacked important methodological aspects of a DNA methylation study design. First, key confounding factors [domain 5] were not reported on in five studies, with the most frequently missing factors being smoking behaviour [four studies] and whether or not the data were corrected for the cellular distribution [five studies]. Similarly, sample preservation, quality control and any potential batch effects were not reported on [domain 9]. Second, only five studies justified their sample size [domain 3]. Third, in six studies neither technical nor independent validation had been performed [domain 5]. Lastly, the associative nature of DNA methylation studies limits causal inference, which is only addressed in two studies [domain 7]. A summary of the quality appraisal for each of the nine domains per study is provided in Table 3.
3.2. Comparing Crohn’s disease with healthy controls
The CD with HC comparison represents the most investigated comparison in PBL [Nstudies = 10]18,20,22,26,36,37,46–49 and its constituents, such as peripheral blood mononuclear cells [PBMCs] [Nstudies = 3],23,44,45 CD14+ monocytes [Nstudies = 2],17,20 CD4+ T-cells [Nstudies = 1]20 and CD8+ T-cells [Nstudies = 2].20,50 Most studies sampled from an adult population,20,22,23,37,44,46–48 whereas a minority of studies sampled from a paediatric cohort.18,26,36,49,50 Overall, the studies can be categorized as either epigenome-wide [Nstudies = 11] or targeted [Nstudies = 4].
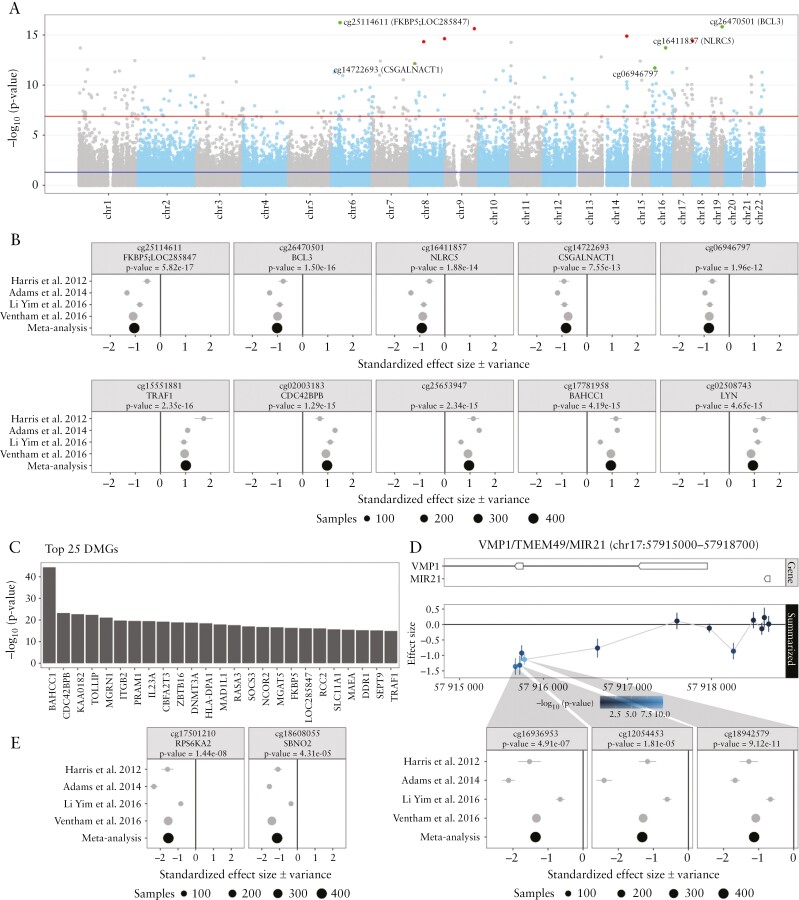
From four epigenome-wide association studies [EWAS] we were able to acquire full summary statistics, which was based on a combined 177 CD patients and 243 HC individuals.18,20,36,37 Meta-analysis thereon identified 256 consistently differentially methylated positions [DMPs] with a Bonferroni-adjusted p-value <0.05 that displayed a difference between CD and HC consistent across all studies [Figure 2a and Supplementary File a]. Among the top ten consistent DMPs, nine were annotated to known genes of which three [FKBP5, BCL3 and NLCR5] were hypomethylated, while the remaining six [TRAF1, CDC42BPB, BAHCC1, LYN, TOLLIP and KCNAB2] were hypermethylated among CD patients compared to healthy controls [Figure 2b and Supplementary Table 2]. Expectedly, two of the nine genes [CDC42BPB20 and TOLLIP18] were discussed in the included studies. While the genes FKBP5,51,52BCL3,53,54TRAF1,55BAHCC1,26NLRC556 and KCNAB220 were not specifically highlighted in the included studies, their association with inflammation or IBD has been described before. For example, DMPs cg25114611, cg26470501, cg15551881 and cg26599989 associate with FKBP5, BCL3, TRAF1 and TOLLIP, respectively, all of which are involved in NF-κB-signalling, underlining its importance in IBD pathogenesis.57–63 Furthermore, increased FKBP5 expression levels correlated with CD disease duration and endoscopic scoring [CDEIS] as well as were able to classify endoscopically active from non-active CD.51,52 Decreased expression of the transcriptional coactivator BCL3 has been identified as a novel risk factor for CD.53 In addition, Bcl3−/− deficient mice were found to be less sensitive to DSS-induced colitis,64 and mucosal tissue expression of BCL3 was strongly elevated in active UC as well as active/inactive CD.54 Significantly increased TRAF1 expression has been demonstrated in IBD tissue vs controls, as well as inflamed compared with non-inflamed tissue,55 while decreased levels of the TOLLIP protein have been found in mucosal tissue when comparing active UC and CD with HC,54 potentially resulting in a decreased inhibition of the NF-κB/JNK/MAPK signalling pathways. By contrast, LYN [cg02508743] has thus far not been associated with IBD. The encoded protein belongs to the family of tyrosine kinases known to function in cell-surface receptor signalling with particular importance in innate/adaptive immune responses, haematopoiesis, response to growth factors and cytokines, and integrin signalling.65Lyn−/− mice have been shown to be highly susceptible to T-cell (increased interferon-γ [IFNγ] production in CD4+ and CD8+ T-cells) dependent DSS-induced colitis which correlated with dysbiosis.66 Moreover, the reverse has been observed in Lynup mice, which presented a protective response during inflammation.67 Additional functional information for genes annotated to the top 20 DMPs can be found in Supplementary Table 2.
Figure 2.
Meta-analysis of the comparison Crohn’s disease [CD] with healthy controls [HC] using data from Harris et al. 2012, Adams et al. 2014, Li Yim et al. 2016 and Ventham et al. 2016. [A] A Manhattan plot representing the statistical significance depicted as −log10[p-value] on the y-axis relative to the location across the genome [hg19] on the x-axis. Red and green dots represent the five most significantly hyper- and hypomethylated differentially methylated probes [DMPs], respectively. [B] Forest plots representing the standardized effect size [SES] of interest per study of the five most significantly hyper- and hypomethylated DMPs annotated with the associated gene and p-value from the meta-analysis. [C] Barchart representing the top 25 most significant differentially methylated genes [DMGs]. The length of the bar is proportional to the −log10[p-value] obtained from the DMG analysis. [D] Genomic visualization and forest plots of the estimated difference in methylation for the DMPs cg16936953, cg12054453 and cg18942579, all of which have been associated with VMP1/TMEM49/MIR21. [E] Forest plots of DMPs cg17501210 and cg18608055, associated with RPS6KA2 and SBNO2, respectively, which had been reported on in multiple earlier studies.
Turning our attention to genes that were enriched for significant DMPs yielded 326 statistically significant DMGs. From the top 20, ten were previously identified within the context of CD [MGRN1,36ITGB2,20,23,68PRAM1,26,69IL23A,70CBFA2T3,71ZBTB16,72,73DNMT3A,15HLA-DPA1,18MAD1L1,74SOCS373,75] [Figure 2c, Supplementary Table 3 and File b]. Of particular interest is RASA3, a gene that to our knowledge has previously not been associated with IBD. RASA3 encodes RAS P21 Protein Activator 3, which is a negative regulator of the Ras signalling pathway through stimulation of GTPase.76In vitro RASA3 depletion of cultured endothelial cells was found to increase β1 integrin activation and cell adhesion to extracellular matrix components, decrease cell migration and block tubulogenesis.77 In addition, RASA-depleted cells showed reduced turnover of vascular endothelial cadherin-based adhesions, resulting in more stable endothelial cell–cell adhesion and decreased endothelial permeability.77 Differential methylation of RASA3 could thus potentially alter endothelial–leukocyte adhesions, known to be of major importance for gut homing of inflammatory cells in IBD, targeted by drugs such as vedolizumab.78 By performing overrepresentation analyses of the identified DMGs against the GO resource, we identified significant overrepresentation for 36 GO terms. Specifically, overrepresentation was observed for immune response [GO:0002683, GO:2000181, GO:0002683, GO:2000181, GO:0002366, GO:0050727, GO:0002861, GO:0006909, GO:0045638, GO:0002862, GO:0002263], cytokine production [GO:0001818, GO:0034341, GO:0032609, GO:0032649, GO:0001819] and cell–cell adhesion [GO:0045785, GO:1903037, GO:1903039, GO:0022409, GO:0022407, GO:0007156, GO:0007159] [Supplementary File h].
We next turned to DMPs that were reported as significant in multiple studies focusing on cg12054453, cg16936953 and cg18942579 [VMP1/TMEM49], cg17501210 [RPS6KA2] and cg18608055 [SBNO2].18,20,26 Meta-analysis statistics of these loci, expectedly, confirmed the direction of effect across all the individual studies included [Figure 2d and e].
3.3. Comparing ulcerative colitis with healthy controls
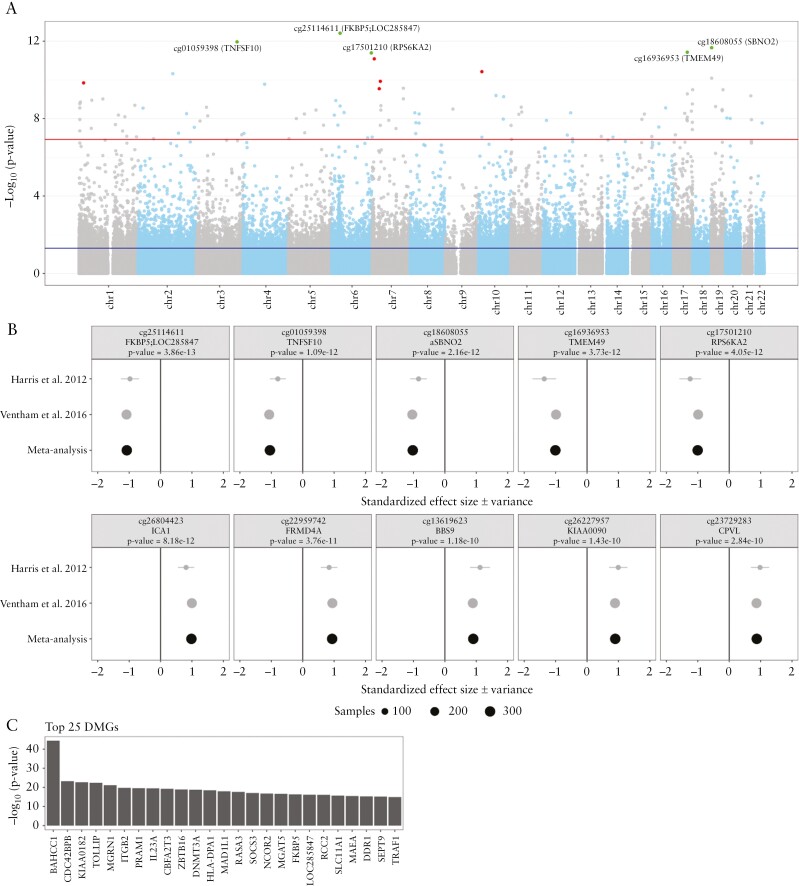
The comparison of UC with HC was performed in fewer studies, with studies interrogating the DNA methylome in PBL [Nstudies = 4]20,36,46,49 or its derivatives PBMCs [Nstudies = 2],23,44 CD4+ T-cells [Nstudies = 1]20 and CD8+ T-cells [Nstudies = 2].20,50 Notably, a more balanced distribution of paediatric [Nstudies = 3] and adult cohorts [Nstudies = 4] was observed. The majority of these studies adopted an EWAS approach [Nstudies = 5] using mostly the Illumina HumanMethylation BeadChip 450k array [Nstudies = 3]. Altogether, we obtained summary statistics from two studies totalling 132 UC patients and 217 HCs.20,36 We identified 104 DMPs with a Bonferroni-adjusted p-value <0.05 [Figure 3a and Supplementary File c]. All top ten DMPs annotated to known genes, of which seven showed consistent hypomethylation [FKBP5, TNFSF10, SBNO2, TMEM49/VMP1, RPS6KA2, ZEB2 and SBNO2] with the remaining three [ICA1, FRMD4A and BBS9] presenting consistent hypermethylation when comparing UC with HC [Figure 3b and Supplementary Table 4]. Expectedly, several DMPs among the top 20, namely cg01059398 [TNFSF10], cg18608055 [SBNO2], cg16936953 [TMEM49/VMP1/MIR21], cg17501210 [RPS6KA2], cg26804423 [ICA1], cg22959742 [FRMD4A] and cg20995564 [ZEB2], were reported as significant by Ventham et al.20 However, for six DMPs, namely cg02734358 [GPRIN3], cg20228731 [FLJ43663/LINC-PINT], cg23729283 [CPVL], cg16755922 [FOXK2], cg23761815 [SLC29A3] and cg27243685 [ABCG1], neither site nor gene has been associated with UC or IBD previously. For FLJ43663/LINC-PINT [Long Intergenic Non Coding RNA, P53 Induced Transcript] recent data in rheumatoid arthritis patients indicate that LINC-PINT affects the production of tumour necrosis factor α [TNFα].79 Moreover, the exact CpG [cg20228731] was found to be significantly hypomethylated in patients with Behçet’s disease compared to healthy controls,80 which was associated with an increased expression of the gene. Other DMP-associated genes, CPVL81 and ABCG1,82 have been associated with myeloid or macrophage function, with CPVL encoding a serine carboxypeptidase that is specifically expressed in macrophages and is proposed to function in proteolytic digestion of lysosomal components after phagocytosis.81ABCG1 encodes an ATP-binding cassette sub-family G member, which mediates cholesterol accumulation and efflux in macrophage foam cells and has therefore been associated with atherogenesis.82,83 Moreover, Abca1−/− mice demonstrated increased secretion of pro-inflammatory cytokines such as TNFα, IL-6, IL-1β and IL-12p7084 and increased neutrophil and monocyte populations in peripheral blood.85
Figure 3.
Meta-analysis of the comparison ulcerative colitis [UC] with healthy controls [HC] using data from Harris et al. 2012 and Ventham et al. 2016. [A] A Manhattan plot representing the statistical significance depicted as −log10[p-value] on the y-axis relative to the location across the genome [hg19] on the x-axis. Red and green dots represent the five most significantly hyper- and hypomethylated differentially methylated probes [DMPs], respectively. [B] Forest plots representing the standardized effect size [SES] of interest per study of the five most hyper- and hypomethylated DMPs annotated with the associated gene and p-value from the meta-analysis. [C] Barchart representing the top 25 most significant differentially methylated genes [DMGs]. The length of the bar is proportional to the −log10[p-value] obtained from the DMG analysis.
Aggregating the DMPs at the gene level yielded 45 statistically significant DMGs [Figure 3c and Supplementary File d]. Despite the considerable overlap with Ventham et al.,20 we observed novel DMGs of potential interest in UC pathogenesis, namely LOC285847, MZB1 and FOXK2. The long non-coding RNA gene LOC285847 was found to be differentially methylated in women with systemic lupus erythematosus.86MZB1 [Marginal Zone B And B1 Cell Specific Protein] is highly expressed in B-cell lineages and plasmacytoid dendritic cells [pDCs],87 where a knockout of Mzb1 in mice was associated with a reduction in immunoglobulin M [IgM]-88 and IFNα secretion.87FOXK2 is mostly described in cancer pathogenesis,89 where it is thought to be involved in autophagy,90 a process that has been associated extensively with IBD.91 Notably, functional overrepresentation analyses against the GO resource did not identify any significant GO terms that passed the multiple test correction.
3.4. Comparing Crohn’s disease with ulcerative colitis
In ~5–15% of all IBD patients, a definitive diagnosis of CD or UC is not always possible, leading to patients being diagnosed as either IBD-unclassified or indeterminate colitis.92 While the majority of these patients are re-diagnosed as either UC or CD at a later point in time, earlier classification might prevent the delay of an optimal treatment.93 Therefore, to explore the difference in DNA methylation between CD and UC we reviewed the available literature.
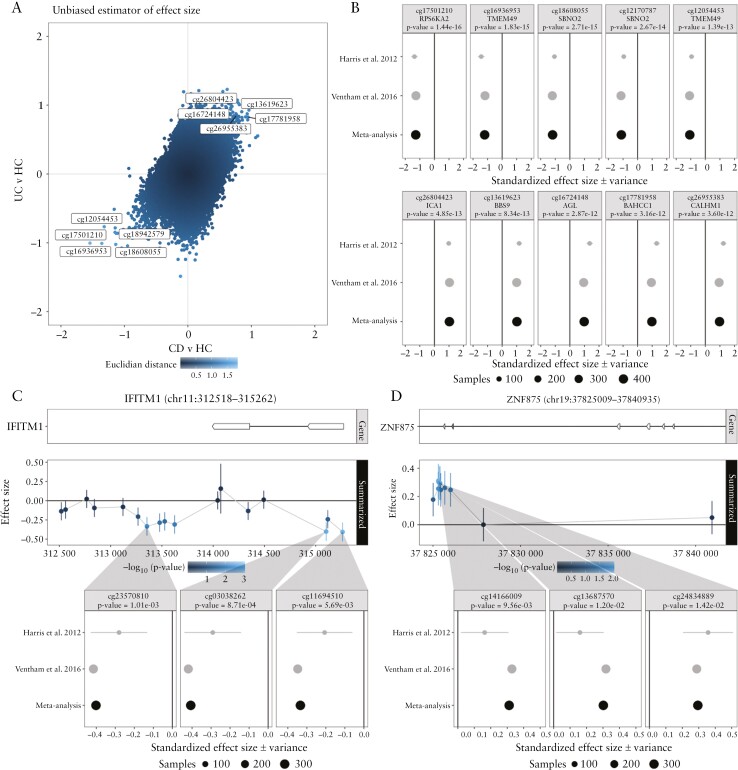
We included three studies for the systematic review that compared CD with UC in PBL20,46,49 and two studies in PBMCs.23,36 Three20,23,36 of these studies adopted an EWAS approach using the Illumina HumanMethylation BeadChip 450k array in an adult population, of which two provided summary statistics that enabled a meta-analysis. Both studies reported a considerable overlap between UC- and CD-associated DMPs with over 66–97% of the UC-associated DMPs overlapping with CD-associated DMPs, while 45% of the CD-associated DMPs overlapped with the UC-associated DMPs.20,23 Indeed, comparing the separate meta-analyses revealed a strong positive correlation [Figure 4a]. By combining both CD and UC and comparing that with HC, we identified DMPs annotated to FKBP5, TMFSF10, SBNO2, TMEM29/VMP1, RPS6KA2, ICA1, FRMD4A, BBS9, KIAA0090, CPVL, BCL3, BAHCC1 and CALHM1 and DMGs found in RPS6KA2, TMEM49/VMP1, TNFSF10/TRAIL, FKBP5, BAHCC1 and SOCS3 being identified as significantly different [Figure 4b, Supplementary Tables 7 and 8 and Files f and g]. GO term overrepresentation analysis between IBD patients and HC identified 224 significantly enriched GO terms. Among the top ten most enriched terms we observed significant overrepresentation for cell–cell adhesion [GO:0098742]. Notably, the majority of the remaining significantly associated GO terms were associated with developmental biology focusing on organogenesis [Supplementary File i].
Figure 4.
Meta-analysis of the comparisons Crohn’s disease [CD] with ulcerative colitis [UC] and inflammatory bowel disease [IBD] with healthy controls [HC] using data from Harris et al. 2012 and Ventham et al. 2016. [A] A scatter plot of the included CpGs representing aggregated effect size of CD with HC and UC with HC on the x- and y-axis, respectively. Colour intensity is proportional to the distance of each CpG to the origin [0,0]. Annotated are the CpGs that are concordantly hyper- and hypomethylated in both CD and UC, and hence IBD, relative to HC. [B] Forest plots representing the standardized effect size [SES] of interest per study of the five most hyper- and hypomethylated IBD-associated DMPs annotated with the associated gene and p-value from the meta-analysis. Genomic visualization and forest plots of the estimated difference in methylation for the CD/UC discordant genes [C] IFITM1 and [D] ZNF875, alongside for the most significant differentially methylated probes.
Comparing CD [N = 139] with UC [N = 133]20,36 did not yield any significant DMPs following Bonferroni correction, underlining the previously reported similarities in global DNA methylation between both diseases [Figure 4a]. Notably, a DMG analysis identified two genes enriched for low p-values, namely IFITM1 and ZNF875 [Figure 4c and d, Supplementary Table 6 and File e]. IFITM1 encodes Interferon Induced Transmembrane Protein 1, which is part of the interferon-stimulated gene families and is expressed in a variety of cells in response to interferon and viral immunity.94IFITM1 has shown to interact with IFNγ, a key interferon with anti-proliferative function.95 In IBD, IFNγ is known as a major cytokine involved in the pathogenesis and is found to be highly upregulated in both CD and UC patients.96 In addition, genetic polymorphisms of IFITM1 have been associated with UC patients, implicating IFITM1 as potential risk locus for UC.97
ZNF875 [Zinc Finger Protein 875] is part of the Krüppel-associated box zinc finger proteins [KRAB-ZFPs] which act as transcription factors thought to be involved in carcinogenesis98 and resistance to therapy in lung cancer.99 However, its relationship with IBD as yet remains unknown.
3.5. Classification of IBD using DNA methylation
While previous analyses were mostly geared at identifying and quantifying differences in methylation, an increasing number of studies have adopted a classification approach to distinguish CD, UC and HC focusing on the diagnostic potential of DNA methylation. Most of the studies that performed classification analyses were done so in PBL18,20,22,26,49 with a single study in mucosal tissue.21 Performance-wise, the resulting classification models indicated good distinctive capabilities in discriminating IBD from HC, reporting area under the curve (AUC) scores as high as 0.94 for CD and 0.91 for UC.49 Distinguishing CD from UC was notably less prominent, with an AUC score ranging between 0.71 and 0.81,18,20,49 corroborating the findings that both IBDs are similar.20,23,68 Most classification studies lacked details on the exact implementation of the model as well as the positions used as features for classification,19,20,26 thereby preventing independent validation. Only two previously reported paired-probe classification models, RPS6KA2/VMP1 [cg17501210/cg12054453] and RPS6KA2/TNFSF10 [cg17501210/cg01059398],18 were validated in a separate study.20 Besides the diagnostic capabilities of DNA methylation, several classification studies also investigated the use of DNA methylation in predicting disease progression, thereby distinguishing a different risk of treatment escalation over time100 or time to use of biologics.21
4. Discussion
Our study provides the most comprehensive summary of IBD-related DNA methylation studies in PBL to date. We initially performed a systematic review of all available literature using a pre-specified protocol according to the PRISMA guideline,101 whereupon we critically appraised both strengths and weaknesses using a modified version of the BIOCROSS tool.34 Despite finding multiple relevant studies, we noted substantial discrepancies in the way sample sizes were selected, clinical characteristics were documented and the level of data availability. Accordingly, we provide a methodological checklist that could aid future researchers with an interest in IBD epigenetics to increase the reproducibility of their work [Table 4].
Table 4.
Checklist of key components that should be described in DNA methylation studies
| Justify sample size: | • If previous literature has been published: estimate sample size102 • If no previous literature has been published: perform a pilot study in a small number of patients as an exploration. Report as explorative study design, a stepping-stone for future research |
| Limit confounding: | • Include detailed cohort description: ◦ Age, sex, smoking behaviour ◦ Disease duration, Montreal classification, surgical history ◦ Inflammatory status at time of sampling [CRP, faecal calprotectin, clinical and or endoscopy scores] ◦ Current and previous IBD-related medication, concomitant medication • If mixed tissue [whole blood, mucosal biopsy]: explore cellular heterogeneity • Describe sample quality control and describe actions taken to limit batch effects |
| Increase reproducibility: | •Report sample type [tube], sampling protocol, storage and isolation of DNA [kits] • When reporting differences in methylation, specify where the differences are found using genomic coordinates • Publish raw data in .idat or .fastq format in publicly available databases [Gene Expression Omnibus, ArrayExpress, European Genome-phenome Archive] |
| Increase interpretability: | • Include technical and/or independent validation in your design • Perform functional analysis of observed markers |
CRP, C-reactive protein; IBD, inflammatory bowel disease.
Through a meta-analysis, we identified consistent IBD-associated differences in DNA methylation between CD, UC and HC in PBL. The different comparisons in our meta-analyses show that there are certainly several notable markers that are indeed consistently differentially methylated across all studies. In addition, we corroborate the reported consistent differential methylation for previously identified probes: cg16936953, cg12054453, and cg18942579 [VMP1/TMEM49/MIR21] and cg17501210 [RPS6KA2], which demonstrated homogeneity across all studies despite the heterogeneity in clinical characteristics, such as age group, sex, smoking behaviour and inflammatory status. However, to understand the pathogenesis and aetiology of IBD, the results presented in this meta-analysis remain observational at best as we cannot ascertain the source of the signal, nor can we ascertain the causal relationship of the newly identified DMPs with the outcome of interest [IBD, CD or UC] due to reverse causation.103 That being said, the GO analyses of DMGs identified when comparing CD with HC returned multiple GO terms associated with inflammation, immune response and cell–cell adhesion, largely corroborating the GO analyses performed by the individual studies.18,20,37 By contrast, GO analyses of the DMGs obtained when comparing UC with HC yielded no significant GO terms. We are unsure why the UC vs HC DMGs present no statistically significant overrepresentations. We hypothesize that a possible cause may be the lack of included studies, with only two studies being eligible. Similarly, it is unclear why the DMGs obtained when comparing IBD with HC are overrepresented for GO terms associated with pathways related to developmental biology. Nonetheless, our observations open up novel routes for targeted functional studies to address the potential downstream role of the observed differences in methylation and its relationship to IBD pathogenesis.
Despite our best efforts at providing an overview of the existing literature on PBL-associated DNA methylation in IBD, we are aware that limitations exist due to the differences in sample sizes and clinical phenotypes between the included studies, and advances in analytical methods. We note that the majority of the reported DMPs are in line with the observations by Ventham et al.,20 which is the study with the largest sample size included in our meta-analysis, hence providing the largest weight. Nonetheless, the other studies provided similar direction of effect for the top DMPs, indicating that the observations were not solely restricted to Ventham et al.20 We utilized summary statistics from all the studies to remain as close as possible to the authors’ interpretations. This approach unfortunately excluded Somineni et al.,26 as their summary statistics were incomplete, nor were we able to reproduce their summary statistics due to the lack of patient metadata [GWAS-derived principal components and Houseman-estimated cell distribution]. That being said, the analytical approach of the included studies for calculating the summary statistics was not identical. In particular, correcting for the cellular composition was often inconsistent, with some not performing any,36 whereas others corrected for the measured leukocyte counts,18 remove unwanted variation-estimated surrogate variables,37 or used the Houseman algorithm to estimate the proportions.20 While an alternative would have been to redo the analyses per study using the raw data and a single analytical pipeline, the raw methylation data in .idat format were not provided by every study [Supplementary Table 9].
The studies included in our systematic review and meta-analysis have mostly been used for understanding the manifestations of IBD at the DNA methylation level. However, it remains to be seen whether such differences can be utilized for biomarker purposes. Indeed, the use of machine learning and high-dimensional statistics for classification analyses are becoming more prevalent,18,20,22,26,49 yet these models have to be translated to clinical practice before they can complement or even replace current diagnostic endoscopies. To this end, such models would need to be extensively validated, both in similar population cohorts as well as in randomized controlled trials. Unfortunately, many of the reported models lack implementation details, such as the weights or the actual predictive loci, complicating reproducibility. Moreover, the majority of the methylation data in IBD have been generated using a cross-sectional design. As such, no assurance on temporal stability of the identified biomarkers can be attributed. While independent validation is a key step towards clinical translation, focusing on markers that show limited intra-individual variability might increase the probability of replicating findings.104 Although studies in adult healthy individuals105,106 or patients with systemic lupus erythematosus107 have demonstrated time-stable methylation markers, no such studies in IBD have been performed. It is therefore questionable if the reported biomarkers in current IBD DNA methylation studies show similar patterns within the same individual across multiple time points, irrespective of inflammatory status. We expect future studies to further address this question.
Besides the diagnostic potential of DNA methylation, additional avenues are currently being investigated, such as risk of disease progression. Therapy response prediction remains an open target as well, as the use of DNA methylation has shown potential in cancer108,109 and rheumatology.110,111 To date, no such response-associated DNA methylation studies in IBD have been published. Taken together, we expect future DNA methylation studies in IBD to be more clinically relevant, where the focus will lie on the classification of specific IBD phenotypes, such as early disease progression, therapy response or post-operative recurrence.
5. Conclusion
Our findings demonstrate consistent differential methylation patterns discerning CD, UC and IBD from healthy controls across multiple studies, which provide a basis for future epigenetic biomarker research in IBD. While the utility of DNA methylation in prediction and classification is becoming more apparent, methodological homogenization is needed, allowing easier aggregation and validation of data, greatly enhancing the field.
Supplementary Material
Acknowledgments
We thank F. S. van Etten-Jamaludin, Amsterdam UMC, University of Amsterdam, Medical Library, Amsterdam, the Netherlands, for helping with the initial literature search.
Contributor Information
Vincent Joustra, Amsterdam UMC location University of Amsterdam, Department of Gastroenterology and Hepatology, Meibergdreef 9, Amsterdam, Netherlands; Amsterdam Gastroenterology Endocrinology Metabolism, Amsterdam, Netherlands.
Ishtu L Hageman, Amsterdam UMC location University of Amsterdam, Department of Gastroenterology and Hepatology, Meibergdreef 9, Amsterdam, Netherlands; Amsterdam Gastroenterology Endocrinology Metabolism, Amsterdam, Netherlands; Amsterdam UMC location University of Amsterdam, Tytgat Institute for Liver and Intestinal Research, Amsterdam, Netherlands.
Jack Satsangi, Translational Gastroenterology Unit, NIHR Oxford Biomedical Research Centre, Oxford University Hospitals NHS Foundation Trust, John Radcliffe Hospital, Oxford, UK.
Alex Adams, Translational Gastroenterology Unit, NIHR Oxford Biomedical Research Centre, Oxford University Hospitals NHS Foundation Trust, John Radcliffe Hospital, Oxford, UK.
Nicholas T Ventham, Institute of Genetics and Molecular Medicine, University of Edinburgh, UK.
Wouter J de Jonge, Amsterdam UMC location University of Amsterdam, Department of Gastroenterology and Hepatology, Meibergdreef 9, Amsterdam, Netherlands; Amsterdam Gastroenterology Endocrinology Metabolism, Amsterdam, Netherlands; Amsterdam UMC location University of Amsterdam, Tytgat Institute for Liver and Intestinal Research, Amsterdam, Netherlands.
Peter Henneman, Amsterdam UMC location University of Amsterdam, Department of Human Genetics, Genome Diagnostics Laboratory, Amsterdam, Netherlands; Amsterdam Reproduction & Development, Amsterdam, Netherlands.
Geert R D’Haens, Amsterdam UMC location University of Amsterdam, Department of Gastroenterology and Hepatology, Meibergdreef 9, Amsterdam, Netherlands; Amsterdam Gastroenterology Endocrinology Metabolism, Amsterdam, Netherlands.
Andrew Y F Li Yim, Amsterdam UMC location University of Amsterdam, Department of Gastroenterology and Hepatology, Meibergdreef 9, Amsterdam, Netherlands; Amsterdam Gastroenterology Endocrinology Metabolism, Amsterdam, Netherlands; Amsterdam UMC location University of Amsterdam, Tytgat Institute for Liver and Intestinal Research, Amsterdam, Netherlands; Amsterdam UMC location University of Amsterdam, Department of Human Genetics, Genome Diagnostics Laboratory, Amsterdam, Netherlands; Amsterdam Reproduction & Development, Amsterdam, Netherlands.
Funding
The authors received no financial support for research, authorship and/or publication of this article.
Conflicts of Interest
VJ, IH, JS, AA, NV, WdJ, PH, GD and ALY: none to declare.
Author Contributions
Conceptualization: VJ, IH, ALY, JS, AA, GD. Methodology: VJ, ALY, IH, JS, AA, GD. Formal analysis: ALY. Data acquisition: VJ, IH, ALY. Writing—original draft: VJ, IH, ALY. Writing—review and editing: VJ, IH, JS, AA, NV, WdJ, PH, GD and ALY. Approval of final manuscript: all authors.
Data Availability
The data that support the findings of this study are openly available in Gene Expression Omnibus [GEO] at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32148, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81961 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE87650 with reference numbers GSE32148, GSE81961 and GSE87650 respectively. Data from Adams et al [2014] were acquired after request to the corresponding author.
References
- 1. Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2017;390:2769–78. [DOI] [PubMed] [Google Scholar]
- 2. Collaborators GBDIBD. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol 2020;5:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Souza HS, Fiocchi C.. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol 2016;13:13–27. [DOI] [PubMed] [Google Scholar]
- 4. Moller FT, Andersen V, Wohlfahrt J, et al. Familial risk of inflammatory bowel disease: a population-based cohort study 1977–2011. Am J Gastroenterol 2015;110:564–71. [DOI] [PubMed] [Google Scholar]
- 5. Kim HJ, Shah SC, Hann HJ, et al. Familial risk of inflammatory bowel disease: a population-based cohort study in South Korea. Clin Gastroenterol Hepatol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Lange KM, Moutsianas L, Lee JC, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 2017;49:256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gordon H, Trier Moller F, Andersen V, et al. Heritability in inflammatory bowel disease: from the first twin study to genome-wide association studies. Inflamm Bowel Dis 2015;21:1428–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spehlmann ME, Begun AZ, Burghardt J, et al. Epidemiology of inflammatory bowel disease in a German twin cohort: results of a nationwide study. Inflamm Bowel Dis 2008;14:968–76. [DOI] [PubMed] [Google Scholar]
- 9. Orholm M, Binder V, Sorensen TI, et al. Concordance of inflammatory bowel disease among Danish twins. Results of a nationwide study. Scand J Gastroenterol 2000;35:1075–81. [DOI] [PubMed] [Google Scholar]
- 10. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature 2009;461:747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feil R, Fraga MF.. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 2012;13:97–109. [DOI] [PubMed] [Google Scholar]
- 12. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002;16:6–21. [DOI] [PubMed] [Google Scholar]
- 13. Bird AP. CpG-rich islands and the function of DNA methylation. Nature 1986;321:209–13. [DOI] [PubMed] [Google Scholar]
- 14. Deaton AM, Bird A.. CpG islands and the regulation of transcription. Genes Dev 2011;25:1010–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet 2010;42:1118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin Z, Hegarty JP, Cappel JJAJ, et al. Identification of disease-associated DNA methylation in intestinal tissues from patients with inflammatory bowel disease. Clin Genet 2011;80:59–67. [DOI] [PubMed] [Google Scholar]
- 17. Li Yim AYF, Duijvis NW, Ghiboub M, et al. Whole-genome DNA methylation profiling of CD14+ monocytes reveals disease status and activity differences in Crohn’s disease patients. J Clin Med 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adams AT, Kennedy NA, Hansen R, et al. Two-stage genome-wide methylation profiling in childhood-onset Crohnʼs disease implicates epigenetic alterations at the VMP1/MIR21 and HLA Loci. Inflamm Bowel Dis 2014;20:1784–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cooke J, Zhang H, Greger L, et al. Mucosal genome-wide methylation changes in inflammatory bowel disease. Inflamm Bowel Dis 2012;18:2128–37. [DOI] [PubMed] [Google Scholar]
- 20. Ventham NT, Kennedy NA, Adams AT, et al. Integrative epigenome-wide analysis demonstrates that DNA methylation may mediate genetic risk in inflammatory bowel disease. Nat Commun 2016;7:13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Howell KJ, Kraiczy J, Nayak KM, et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology 2018;154:585–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nimmo ER, Prendergast JG, Aldhous MC, et al. Genome-wide methylation profiling in Crohnʼs disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm Bowel Dis 2012;18:889–99. [DOI] [PubMed] [Google Scholar]
- 23. McDermott E, Ryan EJ, Tosetto M, et al. DNA methylation profiling in inflammatory bowel disease provides new insights into disease pathogenesis. J Crohns Colitis 2016;10:77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hasler R, Feng Z, Backdahl L, et al. A functional methylome map of ulcerative colitis. Genome Res 2012;22:2130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kang K, Bae JH, Han K, et al. A genome-wide methylation approach identifies a new hypermethylated gene panel in ulcerative colitis. Int J Mol Sci 2016;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Somineni HK, Venkateswaran S, Kilaru V, et al. Blood-derived DNA methylation signatures of Crohn disease and severity of intestinal inflammation. Gastroenterology 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsai PC, Glastonbury CA, Eliot MN, et al. Smoking induces coordinated DNA methylation and gene expression changes in adipose tissue with consequences for metabolic health. Clin Epigenetics 2018;10:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Agliata I, Fernandez-Jimenez N, Goldsmith C, et al. The DNA methylome of inflammatory bowel disease (IBD) reflects intrinsic and extrinsic factors in intestinal mucosal cells. Epigenetics 2020;15:1068–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ventham NT, Kennedy NA, Nimmo ER, et al. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology 2013;145:293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karatzas PS, Gazouli M, Safioleas M, et al. DNA methylation changes in inflammatory bowel disease. Ann Gastroenterol 2014;27:125–32. [PMC free article] [PubMed] [Google Scholar]
- 31. Li X, Song P, Timofeeva M, et al. Systematic meta-analyses and field synopsis of genetic and epigenetic studies in paediatric inflammatory bowel disease. Sci Rep 2016;6:34076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hornschuh M, Wirthgen E, Wolfien M, et al. The role of epigenetic modifications for the pathogenesis of Crohn’s disease. Clin Epigenetics 2021;13:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ouzzani M, Hammady H, Fedorowicz Z, et al. Rayyan – a web and mobile app for systematic reviews. Syst Rev 2016;5:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wirsching J, Grassmann S, Eichelmann F, et al. Development and reliability assessment of a new quality appraisal tool for cross-sectional studies using biomarker data (BIOCROSS). BMC Med Res Methodol 2018;18:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harris RA, Nagy-Szakal D, Pedersen N, et al. Genome-wide peripheral blood leukocyte DNA methylation microarrays identified a single association with inflammatory bowel diseases. Inflamm Bowel Dis 2012;18:2334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Yim AYF, Duijvis NW, Zhao J, et al. Peripheral blood methylation profiling of female Crohn’s disease patients. Clin Epigenetics 2016;8:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choi JK, Yu U, Kim S, et al. Combining multiple microarray studies and modeling interstudy variation. Bioinformatics 2003;19:i84–90. [DOI] [PubMed] [Google Scholar]
- 39. Lusa L, Gentlemen R, Ruschhaupt M.. GeneMeta: MetaAnalysis for high throughput experiments, R package version 1.68.0. 2022.
- 40. Yi L, Pimentel H, Bray NL, et al. Gene-level differential analysis at transcript-level resolution. Genome Biol 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gene Ontology C. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 2021;49:D325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Balasa A, Gathungu G, Kisfali P, et al. Assessment of DNA methylation at the interferon regulatory factor 5 (IRF5) promoter region in inflammatory bowel diseases. Int J Colorectal Dis 2010;25:553–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim SW, Kim ES, Moon CM, et al. Abnormal genetic and epigenetic changes in signal transducer and activator of transcription 4 in the pathogenesis of inflammatory bowel diseases. Dig Dis Sci 2012;57:2600–7. [DOI] [PubMed] [Google Scholar]
- 46. Karatzas PS, Mantzaris GJ, Safioleas M, et al. DNA methylation profile of genes involved in inflammation and autoimmunity in inflammatory bowel disease. Medicine 2014;93:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Klasic M, Markulin D, Vojta A, et al. Promoter methylation of the MGAT3 and BACH2 genes correlates with the composition of the immunoglobulin G glycome in inflammatory bowel disease. Clin Epigenetics 2018;10:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moret-Tatay I, Cerrillo E, Sáez-González E, et al. Identification of epigenetic methylation signatures with clinical value in Crohn’s disease. Clin Transl Gastroenterol 2019. doi: 10.14309/ctg.0000000000000083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Samarani S, Dupont-Lucas C, Marcil V, et al. CpG methylation in TGFbeta1 and IL-6 genes as surrogate biomarkers for diagnosis of IBD in children. Inflamm Bowel Dis 2020;26:1572–8. [DOI] [PubMed] [Google Scholar]
- 50. Gasparetto M, Payne F, Nayak K, et al. Transcription and DNA methylation patterns of blood-derived CD8(+) T cells are associated with age and inflammatory bowel disease but do not predict prognosis. Gastroenterology 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maltese P, Palma L, Sfara C, et al. Glucocorticoid resistance in Crohn’s disease and ulcerative colitis: an association study investigating GR and FKBP5 gene polymorphisms. Pharmacogenomics J 2012;12:432–8. [DOI] [PubMed] [Google Scholar]
- 52. Zhang C. Identification of hub genes and potential mechanisms in depressive disorder aggravated Crohn’s disease. In: Gastroenterology SU-Do, ed., Preprints with the lancet. 2021. https://ssrn.com/abstract=3773015 or 10.2139/ssrn.3773015. [DOI]
- 53. Fransen K, Visschedijk MC, van Sommeren S, et al. Analysis of SNPs with an effect on gene expression identifies UBE2L3 and BCL3 as potential new risk genes for Crohn’s disease. Hum Mol Genet 2010;19:3482–8. [DOI] [PubMed] [Google Scholar]
- 54. Fernandes P, MacSharry J, Darby T, et al. Differential expression of key regulators of Toll-like receptors in ulcerative colitis and Crohn’s disease: a role for Tollip and peroxisome proliferator-activated receptor gamma? Clin Exp Immunol 2016;183:358–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qiao YQ, Shen J, Gu Y, et al. Gene expression of tumor necrosis factor receptor associated-factor (TRAF)-1 and TRAF-2 in inflammatory bowel disease. J Dig Dis 2013;14:244–50. [DOI] [PubMed] [Google Scholar]
- 56. Wang JQ, Liu YR, Xia Q, et al. Emerging roles for NLRC5 in immune diseases. Front Pharmacol 2019;10:1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zannas AS, Jia M, Hafner K, et al. Epigenetic upregulation of FKBP5 by aging and stress contributes to NF-kappaB-driven inflammation and cardiovascular risk. Proc Natl Acad Sci USA 2019;116:11370–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu T, Zhang L, Joo D, et al. NF-kappaB signaling in inflammation. Signal Transduct Target Ther 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Comi M, Amodio G, Gregori S.. Interleukin-10-producing DC-10 is a unique tool to promote tolerance via antigen-specific T regulatory type 1 cells. Front Immunol 2018;9:682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lalani AI, Zhu S, Gokhale S, et al. TRAF molecules in inflammation and inflammatory diseases. Curr Pharmacol Rep 2018;4:64–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wicovsky A, Henkler F, Salzmann S, et al. Tumor necrosis factor receptor-associated factor-1 enhances proinflammatory TNF receptor-2 signaling and modifies TNFR1-TNFR2 cooperation. Oncogene 2009;28:1769–81. [DOI] [PubMed] [Google Scholar]
- 62. Didierlaurent A, Brissoni B, Velin D, et al. Tollip regulates proinflammatory responses to interleukin-1 and lipopolysaccharide. Mol Cell Biol 2006;26:735–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zaidi D, Wine E.. Regulation of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kappabeta) in inflammatory bowel diseases. Front Pediatr 2018;6:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. O’Carroll C, Moloney G, Hurley G, et al. Bcl-3 deficiency protects against dextran-sodium sulphate-induced colitis in the mouse. Clin Exp Immunol 2013;173:332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ingley E. Functions of the Lyn tyrosine kinase in health and disease. Cell Commun Signal 2012;10:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Roberts ME, Bishop JL, Fan X, et al. Lyn deficiency leads to increased microbiota-dependent intestinal inflammation and susceptibility to enteric pathogens. J Immunol 2014;193:5249–63. [DOI] [PubMed] [Google Scholar]
- 67. Bishop JL, Roberts ME, Beer JL, et al. Lyn activity protects mice from DSS colitis and regulates the production of IL-22 from innate lymphoid cells. Mucosal Immunol 2014;7:405–16. [DOI] [PubMed] [Google Scholar]
- 68. Harris RA, Nagy-Szakal D, Mir SA, et al. DNA methylation-associated colonic mucosal immune and defense responses in treatment-naive pediatric ulcerative colitis. Epigenetics 2014;9:1131–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wu YM, Gettler K, Giri M, et al. Identifying novel high-impact rare disease-causing mutations, genes and pathways in exomes of Ashkenazi Jewish inflammatory bowel disease patient. Gastroenterology 2021;160:S154–5. [Google Scholar]
- 70. Schmitt H, Neurath MF, Atreya R.. Role of the IL23/IL17 pathway in Crohn’s disease. Front Immunol 2021;12:622934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brown R. MTG16 (CBFA2T3) regulates colonic epithelial differentiation, colitis, and tumorigenesis by repressing E protein transcription factors. BioRxiv 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kitani T. In search of newer targets for inflammatory bowel disease: a systems and a network medicine approach. In: Network and Systems Medicine. Mary Ann Liebert, Inc.; 2021; 4. doi: 10.1089/nsm.2020.0012 [DOI] [Google Scholar]
- 73. Lin LJ, Zhang Y, Lin Y, et al. Identifying candidate genes for discrimination of ulcerative colitis and Crohn’s disease. Mol Biol Rep 2014;41:6349–55. [DOI] [PubMed] [Google Scholar]
- 74. Serena C, Millan M, Ejarque M, et al. Adipose stem cells from patients with Crohn’s disease show a distinctive DNA methylation pattern. Clin Epigenetics 2020;12:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lovato P, Brender C, Agnholt J, et al. Constitutive STAT3 activation in intestinal T cells from patients with Crohn’s disease. J Biol Chem 2003;278:16777–81. [DOI] [PubMed] [Google Scholar]
- 76. Schurmans S, Polizzi S, Scoumanne A, et al. The Ras/Rap GTPase activating protein RASA3: from gene structure to in vivo functions. Adv Biol Regul 2015;57:153–61. [DOI] [PubMed] [Google Scholar]
- 77. Molina-Ortiz P, Orban T, Martin M, et al. Rasa3 controls turnover of endothelial cell adhesion and vascular lumen integrity by a Rap1-dependent mechanism. PLoS Genet 2018;14:e1007195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Luzentales-Simpson M, Pang YCF, Zhang A, et al. Vedolizumab: potential mechanisms of action for reducing pathological inflammation in inflammatory bowel diseases. Front Cell Dev Biol 2021;9:612830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang J, Zhao Q.. LncRNA LINC-PINT increases SOCS1 expression by sponging miR-155-5p to inhibit the activation of ERK signaling pathway in rheumatoid arthritis synovial fibroblasts induced by TNF-alpha. Int Immunopharmacol 2020;84:106497. [DOI] [PubMed] [Google Scholar]
- 80. Yu HS, Du LP, Yi SL, et al. Epigenome-wide association study identifies Behcet’s disease-associated methylation loci in Han Chinese. Rheumatology 2019;58:1574–84. [DOI] [PubMed] [Google Scholar]
- 81. Mahoney JA, Ntolosi B, DaSilva RP, et al. Cloning and characterization of CPVL, a novel serine carboxypeptidase, from human macrophages. Genomics 2001;72:243–51. [DOI] [PubMed] [Google Scholar]
- 82. Baldan A, Gomes AV, Ping P, et al. Loss of ABCG1 results in chronic pulmonary inflammation. J Immunol 2008;180:3560–8. [DOI] [PubMed] [Google Scholar]
- 83. Yvan-Charvet L, Wang N, Tall AR.. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler Thromb Vasc Biol 2010;30:139–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yvan-Charvet L, Ranalletta M, Wang N, et al. Combined deficiency of ABCA1 and ABCG1 promotes foam cell accumulation and accelerates atherosclerosis in mice. J Clin Investig 2007;117:3900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Out R, Hoekstra M, Habets K, et al. Combined deletion of macrophage ABCA1 and ABCG1 leads to massive lipid accumulation in tissue macrophages and distinct atherosclerosis at relatively low plasma cholesterol levels. Arterioscler Thromb Vasc Biol 2008;28:258–64. [DOI] [PubMed] [Google Scholar]
- 86. Mok A, Solomon O, Nayak RR, et al. Genome-wide profiling identifies associations between lupus nephritis and differential methylation of genes regulating tissue hypoxia and type 1 interferon responses. Lupus Sci Med 2016;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kapoor T, Corrado M, Pearce EL, et al. MZB1 enables efficient interferon alpha secretion in stimulated plasmacytoid dendritic cells. Sci Rep 2020;10:21626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Flach H, Rosenbaum M, Duchniewicz M, et al. Mzb1 protein regulates calcium homeostasis, antibody secretion, and integrin activation in innate-like B cells. Immunity 2010;33:723–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nestal de Moraes G, Carneiro LDT, Maia RC, et al. FOXK2 transcription factor and its emerging roles in cancer. Cancers 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Li S, Wang P, Ju H, et al. FOXK2 promotes the proliferation of papillary thyroid cancer cell by down-regulating autophagy. J Cancer 2022;13:858–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shao BZ, Yao Y, Zhai JS, et al. The role of autophagy in inflammatory bowel disease. Front Physiol 2021;12:621132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Venkateswaran N, Weismiller S, Clarke K.. Indeterminate colitis – update on treatment options. J Inflamm Res 2021;14:6383–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Thurgate LE, Lemberg DA, Day AS, et al. An overview of inflammatory bowel disease unclassified in children. Inflamm Intest Dis 2019;4:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Siegrist F, Ebeling M, Certa U.. The small interferon-induced transmembrane genes and proteins. J Interferon Cytokine Res 2011;31:183–97. [DOI] [PubMed] [Google Scholar]
- 95. Yang G, Xu Y, Chen X, et al. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene 2007;26:594–603. [DOI] [PubMed] [Google Scholar]
- 96. Langer V, Vivi E, Regensburger D, et al. IFN-gamma drives inflammatory bowel disease pathogenesis through VE-cadherin-directed vascular barrier disruption. J Clin Invest 2019;129:4691–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Mo JS, Na KS, Yu JI, et al. Identification of the polymorphisms in IFITM1 gene and their association in a Korean population with ulcerative colitis. Immunol Lett 2013;156:118–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sobocinska J, Molenda S, Machnik M, et al. KRAB-ZFP transcriptional regulators acting as oncogenes and tumor suppressors: an overview. Int J Mol Sci 2021;22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Oguri T, Katoh O, Takahashi T, et al. The Kruppel-type zinc finger family gene, HKR1, is induced in lung cancer by exposure to platinum drugs. Gene 1998;222:61–7. [DOI] [PubMed] [Google Scholar]
- 100. Kalla R, Adams AT, Vatn S, et al. Epigenetic alterations at diagnosis predict susceptibility, prognosis and treatment escalation in inflammatory bowel disease-IBD character. Gut 2017;66:A24–5. [Google Scholar]
- 101. Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Syst Rev 2021;10:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tsai PC, Bell JT.. Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. Int J Epidemiol 2015;44:1429–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Birney E, Smith GD, Greally JM.. Epigenome-wide association studies and the interpretation of disease -omics. PLoS Genet 2016;12:e1006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sugden K, Hannon EJ, Arseneault L, et al. Patterns of reliability: assessing the reproducibility and integrity of DNA methylation measurement. Patterns 2020;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Li Y, Pan X, Roberts ML, et al. Stability of global methylation profiles of whole blood and extracted DNA under different storage durations and conditions. Epigenomics 2018;10:797–811. [DOI] [PubMed] [Google Scholar]
- 106. Gosselt HR, Griffioen PH, van Zelst BD, et al. Global DNA (hydroxy)methylation is stable over time under several storage conditions and temperatures. Epigenetics 2021;16:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Coit P, Ortiz-Fernandez L, Lewis EE, et al. A longitudinal and transancestral analysis of DNA methylation patterns and disease activity in lupus patients. JCI Insight 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Duruisseaux M, Martinez-Cardus A, Calleja-Cervantes ME, et al. Epigenetic prediction of response to anti-PD-1 treatment in non-small-cell lung cancer: a multicentre, retrospective analysis. Lancet Respir Med 2018;6:771–81. [DOI] [PubMed] [Google Scholar]
- 109. Kim JY, Choi JK, Jung H.. Genome-wide methylation patterns predict clinical benefit of immunotherapy in lung cancer. Clin Epigenetics 2020;12:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Nair N, Plant D, Verstappen SM, et al. Differential DNA methylation correlates with response to methotrexate in rheumatoid arthritis. Rheumatology 2020;59:1364–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Tao W, Concepcion AN, Vianen M, et al. Multi-omics and machine learning accurately predicts clinical response to Adalimumab and Etanercept therapy in patients with rheumatoid arthritis. Arthritis Rheumatol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in Gene Expression Omnibus [GEO] at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32148, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81961 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE87650 with reference numbers GSE32148, GSE81961 and GSE87650 respectively. Data from Adams et al [2014] were acquired after request to the corresponding author.