Abstract
Introduction:
The endocannabinoids (eCBs), 2-arachidonoylglycerol (2-AG) and arachidonoyl ethanolamine (AEA), are produced by separate enzymatic pathways, activate cannabinoid receptors with distinct pharmacology, and differentially regulate pathophysiological processes. The genetically encoded sensor, GRABeCB2.0, detects real-time changes in eCB levels in cells in culture and preclinical model systems; however, its activation by eCB analogues produced by cells and by phyto-cannabinoids remains uncharacterized, a current limitation when interpreting changes in its response. This information could provide additional utility for the tool in in vivo pharmacology studies of phyto-cannabinoid action.
Methods:
GRABeCB2.0 was expressed in cultured HEK293 cells. Live cell confocal microscopy and high-throughput fluorescent signal measurements.
Results:
2-AG increased GRABeCB2.0 fluorescent signal (EC50 = 85 nM), and the cannabinoid 1 receptor (CB1R) antagonist, SR141617, decreased GRABeCB2.0 signal (SR1, IC50 = 3.3 nM), responses that mirror their known potencies at cannabinoid 1 receptors (CB1R). GRABeCB2.0 fluorescent signal also increased in response to AEA (EC50 = 815 nM), the eCB analogues 2-linoleoylglycerol and 2-oleoylglycerol (2-LG and 2-OG, EC50s = 1.5 and 1.0 μM, respectively), Δ9-tetrahydrocannabinol (Δ9-THC) and Δ8-THC (EC50s = 1.6 and 2.0 μM, respectively), and the artificial CB1R agonist, CP55,940 (CP, EC50 = 82 nM); however their potencies were less than what has been described at CB1R. Cannabidiol (CBD) did not affect basal GRABeCB2.0 fluorescent signal and yet reduced the 2-AG stimulated GRABeCB2.0 responses (IC50 = 8.8 nM).
Conclusions:
2-AG and SR1 modulate the GRABeCB2.0 fluorescent signal with EC50s that mirror their potencies at CB1R whereas AEA, eCB analogues, THC and CP increase GRABeCB2.0 fluorescent signal with EC50s significantly lower than their potencies at CB1R. CBD reduces the 2-AG response without affecting basal signal, suggesting that GRABeCB2.0 retains the negative allosteric modulator (NAM) property of CBD at CB1R. This study describes the pharmacological profile of GRABeCB2.0 to improve interpretation of changes in fluorescent signal in response to a series of known eCBs and CB1R ligands.
Introduction
Many physiological functions and behaviors are differentially controlled by endogenously produced AEA and 2-AG that activate CB1R.1 Specifically, AEA or 2-AG production by select cells will partially or fully activate CB1Rs (EC50s: ≈10–100 nM and 30–300 nM, respectively) in an autocrine and paracrine fashion.2 The effects of CB1R activation are dependent on both cell type and coupling to intracellular signaling systems: CB1R are expressed at remarkably different levels by distinct excitatory and inhibitory neurons and by glial cells where they couple to specific signaling pathways. Thus, cell specific differential activation of CB1Rs by AEA and 2-AG in brain fine-tunes excitatory and inhibitory neurotransmission and neuromodulation, and regulates neuronal metabolism and phenotype.3;4 This fundamental signaling mechanism is modulated by Δ9-THC through its binding to the orthosteric binding site of CB1R where it acts as a partial agonist, and by cannabidiol (CBD) that interacts with a putative allosteric binding site on CB1R.5 For example, free CB1Rs expressed throughout the brain are partially activated by THC whereas localized activity-dependent increases in 2-AG and stimulation of CB1R signaling is reduced by THC.6;7 Multiple lines of evidence suggest that CBD acts as a NAM of CB1R, though direct CBD binding to CB1R has still not been demonstrated.8;9 Thus, CBD reduces 2-AG-stimulated CB1R signaling without influencing basal/tonic CB1R signaling. This premise emphasizes a need to better understand the role of endogenously produced AEA and 2-AG, their dynamics, and how the presence of THC and CBD affects their activation of the CB1R.
Mass spectrometry has demonstrated that 2-AG is 10–1000 times more abundant than AEA in select cell types and tissues; however, little is known about their activity-dependent and spatio-temporal changes that occur within seconds.1;10 Importantly, increased cellular activity not only enhances the production of AEA and 2-AG, but also enhances the production of lipid analogues synthesized by the same enzymatic pathways. For example, 2-LG and 2-OG activate CB1R yet with lower potency and efficacy than 2-AG.11–13 Thus, localized activity-dependent increases in eCBs and their analogues will differentially activate CB1R within seconds. The recent development of genetically encoded fluorescent sensors has enabled the real-time detection of changes in the levels of neurotransmitters and neuromodulators in live tissues.14 This technology leverages the selective binding of endogenous agonists to specific receptors that stabilize their conformation. For example, the GRABeCB2.0 sensor was recently engineered starting from CB1R by introducing a circularly permutated-green fluorescent protein (cpGFP) in its third intracellular loop.15;16 Thus, GRABeCB2.0 was developed by screening for constructs with functional insertion sites of cpGFP, followed by individual randomized mutations of amino acids that increased the fluorescent signal in response to 2-AG specifically.17 Several laboratories reported that GRABeCB2.0 signal increases within seconds when exogenously applying 2-AG or AEA to cells in culture, as well as when endogenously stimulating eCB production in cells in culture, mouse brain slices and behaving animals.15;18–20
In the current study, we measured GRABeCB2.0 signal in HEK293 cells in culture using live cell fluorescence microscopy and a high-throughput fluorescence plate reader assay. We found that 2-AG and SR1 formulated in buffer containing bovine serum albumin (BSA), a lipid binding protein known to assist eCB’s activation of CB1R, modulate GRABeCB2.0 fluorescent signal in HEK293 cells with potencies that closely mirror their reported activities at CB1R.15;21 Thus, we leveraged this experimental approach to characterize the pharmacological profile of eCB analogues and phyto-CBs at GRABeCB2.0.
Materials and Methods
Chemicals and Reagents:
2-AG (Cayman), 1-arachodonoylglycerol (Cayman), Δ9-THC (NIDA Drug Supply Program), CP55940 (Cayman), SR141716 (NIDA Drug Supply Program), arachidonoyl ethanolamine (Cayman), goat anti-CB1R antibody (1:1000 for ICC and 1:2,500 for immunoblotting; gift from Dr. Ken Mackie); AlexaFluor 647 conjugated donkey anti-goat (1:1000; Invitrogen); IRDye 800 CW conjugates donkey anti-goat (1:10,000; LI-COR).
Cloning:
GRABeCB2.0 and mut-GRABeCB2.0 DNA were subcloned into an AM/CBA-WPRE-bGH plasmid using the BamHI and EcoRI restriction sites. The plasmid was purified (Purelink HiPure Plasmid Maxiprep Kit, Invitrogen, CA) from transformed Stellar Competent Cells (Takara Bio Inc, Japan). The DNA was sequenced and verified (CLC Sequence Viewer 8) prior to use in transfection.
Cell Culture:
HEK293 cells were grown in DMEM (supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin) at 37°C and 5% CO2. To passage cells for experiments, a confluent 10 cm plate of cells was detached by incubating with 0.25% Trypsin-EDTA for 2–3 min at 37°C, adding 4–5 ml of supplemented DMEM and using gentle pipetting to remove any cells still attached, then added to a new plate with fresh supplemented DMEM. Cells were passaged every 3–4 days, and for no more than 25 passages.
Transfection:
All transfections were performed by incubating DNA with the transfection reagent polyethylenimine (PEI, 25K linear, Polysciences 23966) in a 1:3 ratio in serum free DMEM, incubating for 20–30 min. The DNA/PEI mixture was then added to cells in a dropwise fashion without changing the growth media. Cells were transfected when reaching >50% confluent and were incubated for 24 h after transfection before harvesting or using for GRABeCB2.0 assays.
Immunocytochemistry (ICC):
Glass coverslips (Fisher Scientific, 12-545-82) in a 6-well plate were coated with poly-D-lysine (50 ng/ml, Sigma, P6407) for 1–2 h at 37°C, after which the coverslips were washed 3X with sterile water and 1X with DMEM. HEK293 cells were detached and resuspended in supplemented DMEM as described above, plated at a density of 100,000 cells/well, and were transfected after 24 h with 0.75 g DNA. Twenty h after transfection, media was removed, and cells were fixed with 4% paraformaldehyde in PBS (Alfa Aeser) for 20 min at room temperature. Following fixation, cells were washed 5X with PBS and permeabilized and blocked with 0.1% saponin (Sigma-Aldrich) made and 1% BSA in PBS for 30 min at room temperature. Cells were incubated in goat anti-CB1R antibody (1:1000) overnight at 4°C. Cells were then washed with PBS 6X and incubated in AlexaFluor647 conjugated donkey anti-goat secondary antibody (Invitrogen, 1:1000) for 1 h at room temperature. Cells were washed with PBS 6X, air dried overnight, and mounted using ProLong Diamond Antifade Mountant with DAPI (ThermoFisher, P36966). Images were captured using a LeicaSP8X confocal microscope and a 40X oil objective.
Western Blotting:
HEK293 cells were plated as described above at a density of 500,000 cells/well in a 6-well plate 24 h after plating and were transfected the following day with 0.75 g DNA. Twenty h after transfection, cells were washed 3X with ice cold PBS, and in the last wash cells were harvested with a cell scraper and pelleted by centrifuging at 500 × g for 10 min. The supernatant was discarded, and cell pellets kept at −80°C until further use. Thus, pellets were thawed on ice, resuspended in lysis buffer (25 mM HEPES pH 7.4, 1 mM EDTA, 6 mM MgCl2, and 0.5% CHAPS), Dounce homogenized on ice (20–30 strokes), and incubated on a rotator at 4°C for 1 h. Homogenates were then centrifuged at 700 × g for 10 min at 4°C, supernatant collected, and protein concentration of supernatant determined using a DC Protein Assay. Samples were then mixed with 4X Laemmli Sample Buffer containing 10% β-mercaptoethanol and incubated at 65°C for 5 min. Twenty-five g of protein were loaded onto a 10% polyacrylamide gel and transferred to PVDF membrane. After transfer, membrane was washed once with tris-buffered saline (TBS) and incubated in blocking buffer (5% BSA in TBS) for 1 h at room temperature, followed by incubation in goat anti-CB1R antibody (1:1000) overnight at 4°C. After incubation in primary antibody, membranes were washed with TBS with 0.05% TWEEN-20 (TBST) 3 X, 10 min each. Membranes were then incubated in IRDye 800 CW conjugates donkey anti-goat (1:10,000) for 1 h at room temperature. Blots were then washed with TBST 3 X, 10 min each followed by 3 washes with TBS, 10 min each. Fluorescent signal was detected using a Chemidoc MP (Biorad). Primary antibodies were diluted in blocking buffer. Secondary antibodies were diluted in 1:1 TBS:Odyssey blocking buffer (LI-COR, 927–50000).
Live-cell imaging:
Glass bottom cell culture plates (MatTek, P35G-1.5-14-C) were coated with poly-D-lysine (50 ng/ml, Sigma, P6407) for 1–2 h at 37°C, after which the poly-D-lysine was removed, and coverslips were washed 3 X with sterile water and one time with DMEM. HEK293 cells were detached and resuspended in supplemented DMEM as described above, counted using a hemocytometer, plated (250,000 cells per well) and were transfected after 24 h with 0.75 g DNA. Twenty-four h after transfection, media was replaced by serum free DMEM and cells were incubated at 37°C and 5% CO2 for 1–2 h. To image, the serum free DMEM was replaced by phosphate-buffered saline (room temperature). The plates were transferred to the microscope (LeicaSP8X) and cells were imaged using a 40X oil objective with the following settings: 485 excitation and 525 emission wavelength, 5% laser power, HyD hybrid detector and a scan speed of 200 lines Hz (0.388 frames per second) with bidirectional scanning. All agents were prepared in PBS supplemented with 1 mg/ml BSA and spiked into buffer of cell on the microscope stage (Final concentration BSA = 0.1 mg/ml).
96-well Plate Reader:
Clear-bottom, black 96-well plates (USA Scientific 5665–5087) were coated with poly-D-lysine (50 ng/ml, Sigma, P6407) for 1–2 h at 37°C, and coverslips were washed 3 X with sterile water and 1 X with DMEM. HEK293 cells were detached using trypsin, resuspended in supplemented DMEM as described above, plated (20,000 cells per well) and transfected after 24 h with 0.1 g DNA and 0.3 g of PEI in 10 l of serum free DMEM. Twenty-four h after transfection, media was replaced with PBS (supplemented with BSA 0.1 mg/ml and at room temperature). Cells were incubated at room temperature for 20 min then a 1 min baseline fluorescent signal reading was obtained using a fluorescent plate reader (i.e., 485 emission and 525 emission filter settings with a 515 nm cutoff, and a speed of 1 reading every 20 sec). Immediately after baseline reading, agents in 1 mg/mL BSA and PBS were spiked into buffer in wells. Approximately 2 min after addition of treatment, the plate was reread for 5 min. For pre-treatment with SR1, this antagonist was prepped in PBS supplemented with BSA 0.1 mg/ml (room temperature) and spiked into the media.
Statistical Methods:
All GRABeCB2.0 fluorescent signals are expressed as ΔF/F0 as calculated by MATLAB for the high-throughput fluorescence assay or FIJI ImageJ for live cell confocal imaging. Data are shown as mean + s.e.m. and significance was determined by running a Two-Way ANOVA with Dunnett’s Multiple Comparison test using GraphPad Prism.
Results
Real time activation and antagonism of GRABeCB2.0 fluorescent signal in HEK293 cells in culture: Live cell microscopy.
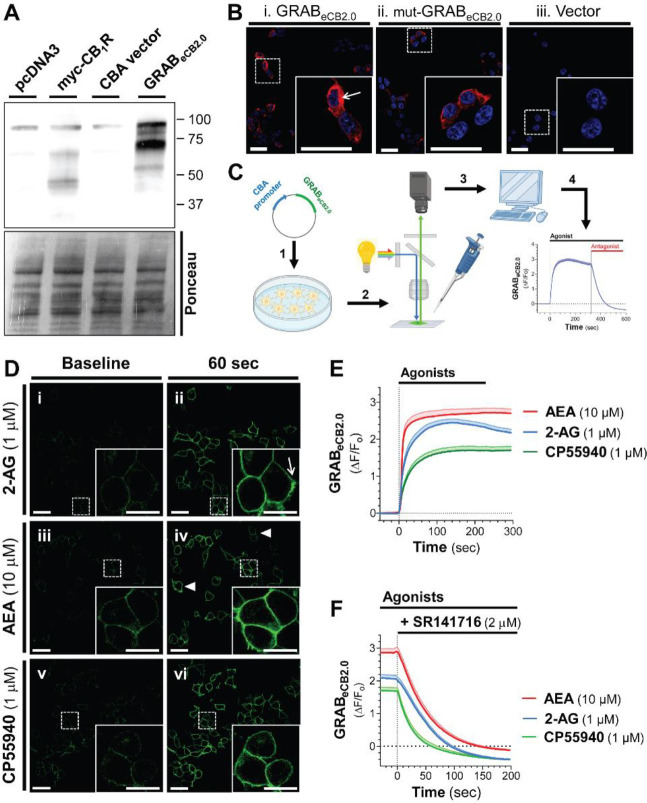
HEK293 cells in culture were transfected with eCB2.0 DNA plasmid constructs containing the chimeric cytomegalovirus-chicken β-actin promoter to drive expression.22–26 GRABeCB2.0 expression was confirmed by western blot using a rabbit polyclonal antibody developed against N-terminal amino acids of CB1R that remained unchanged in GRABeCB2.0 (Figure 1A and Supplementary Figure S1 for amino acid sequence alignment). Fluorescence confocal microscopy analysis of GRABeCB2.0 expression in fixed HEK293 cells using the CB1R antibody showed abundant expression in many cells, consistent with transient transfection approaches (Figure 1B).
Figure 1: Agonist triggered changes in GRABeCB2.0 fluorescent signal in HEK293 cells detected by live-cell confocal microscopy.
HEK293 cells were transfected with eCB2.0 DNA plasmid and GRABeCB2.0 expression was measured by western blot and ICC, and changes in fluorescent signal measured by live-cell confocal microscopy when treated with the CB1R agonists 2-arachidonoyl glycerol (2-AG), arachidonoyl ethanolamine (AEA), or CP55940 (CP). A] Detection of GRABeCB2.0 expression by western blot using lysates from HEK293 cells transfected with pcDNA3, myc-CB1R (positive control), CBA plasmid, or GRABeCB2.0. Both CB1R and GRABeCB2.0 were detected using an antibody against the CB1R. Loading control: Ponceau stain. B] Detection of GRABeCB2.0 (i) or mut-GRABeCB2.0 (ii) expression by ICC of fixed and permeabilized HEK293 cells transfected with eCB2.0 DNA plasmid (i), mut-eCB2.0 DNA plasmid (ii), or CBA plasmid (iii). Scale bar = 20 m (inset scale bar = 20 m). C] Schematic of live cell confocal imaging of HEK293 cells: cells were transfected with eCB2.0 DNA plasmid (1); 24 h later, cell growth media was exchanged for imaging PBS buffer and cells were placed on a confocal microscope (2); changes in GRABeCB2.0 fluorescent signal was determined by measuring fluorescent signal during baseline, spiking in treatments formulated with BSA (0.1%, 10X), and lastly spiking in SR141716 (SR1; 3 and 4). D] Live cell images of GRABeCB2.0-expressing HEK293 cells during baseline recording (i, iii, and v) and after 60 sec of treatment with 2-AG (1 μM, ii), AEA (10 μM, iv), or CP (1 μM, vi). Arrows indicate cells with different levels of fluorescent signal to show heterogeneous expression of GRABeCB2.0. Scale bars = 20 m. E] Time courses of GRABeCB2.0 activation (ΔF/F0) following treatment with 2-AG (1 μM), AEA (10 μM), and CP (1 μM) as measured by live-cell confocal microscopy. F] Effect of SR1 (2 μM) on agonist-stimulated increase in GRABeCB2.0 fluorescent signal. Shaded area represents s.e.m. n= 42–66 cells from 3–5 independent experiments.
We used live-cell confocal microscopy (line scanning frequency: 200 Hz) to establish the time-course of changes in GRABeCB2.0 fluorescent signal in response to 2-AG (the ligand used to develop this sensor) by spiking it directly into the BSA-containing PBS buffer inside the imaging chamber (Figure 1C). Figure 1Di-ii show that HEK293 cells exhibited low, yet clearly detectable basal fluorescent signal at the plasma membrane (treated with vehicle control, DMSO 0.1%); and that 2-AG (1 μM, 60 sec) increased this fluorescent signal. Of note, although the GRABeCB2.0 protein is also expressed in the intracellular compartment of HEK293 cells (see arrow in Figure 1B insert), 2-AG only increases GRABeCB2.0 fluorescent signal at the plasma membrane (see arrow in Figure 1Dii insert). Similarly, AEA (10 μM, 60 sec) and CP (1 μM, 60 sec) increased GRABeCB2.0 fluorescent signal at the plasma membrane (Figure 1Diii-iv). Basal fluorescent signal and all agonist-triggered increases in GRABeCB2.0 fluorescent signal reached different levels in HEK293 cells as expected by heterologous expression (for example, see Figure 1Div arrowheads).
Analysis of the time course of GRABeCB2.0 fluorescent signal (ΔF/F0) increase triggered by these agonists indicate that each agonist increased GRABeCB2.0 fluorescent signal within seconds and that these responses plateaued after approximately 60 sec (Figure 1E). Figure 1F shows that the CB1R antagonist, SR1 (2 μM), reduced these responses within 100 sec, and this reduction reached levels below basal. Calculation of the onset of activation (i.e., slope of the fluorescent signal increase within the first 20 sec after start of treatment), the magnitude of the response (i.e., peak fluorescent signal and area under the curve), and the decay following SR1 treatment (i.e., τ value following start of SR1 treatment) indicated that each agonist had significantly different pharmacological profiles at GRABeCB2.0 (Table 1). Specifically, 1] AEA triggered a 1.7- and 2.2-fold faster initial response compared to 2-AG and CP respectively, 2] 2-AG and AEA reached similar peak responses, and these were 47–58% greater than the CP maximal response, 3] 2-AG and AEA triggered a 1.4–1.6-fold overall greater response (area under the curve) compared to CP and 4] SR1 antagonized the CP and AEA responses with faster decay than the 2-AG response (24s < 37s < 43s, respectively). Together, these results indicate that GRABeCB2.0 expressed by HEK293 cells are activated by the CB1R agonists 2-AG ≈ AEA > CP, and these responses are differentially antagonized by SR1.
Table 1: Parameters of GRABeCB2.0 activation and antagonism.
Live cell images of GRABeCB2.0-expressing HEK293 cells treated with 2-AG (1 μM), AEA (10 μM), or CP (1 μM), followed by SR1 (2 μM) on agonist-stimulated increase in GRABeCB2.0 fluorescent signal. Calculation of the activation onset (i.e., the slope of the fluorescent signal increase within the first 20 sec after start of agonist treatment), the magnitude of the response (i.e., peak response and area under the curve), and the decay following SR1 treatment (i.e., τ value following treatment). Analysis of data presented in Figure 1, n= 42–66 cells from 3–5 independent experiments.
| CB1R agonists | ||||
|---|---|---|---|---|
| PD parameter | Units | 2-AG | CP | AEA |
| Initial response: slope | (× 10−2 DF/F0/sec) | 6.6 | 4.5 | 10.9 |
| Maximal response: peak | (DF/F0) | 2.5 | 1.7 | 2.7 |
| Overall response | (area under the curve) | 649 | 453 | 760 |
| Antagonism response: t | (sec) | 42.7 | 24.3 | 36.8 |
High-throughput measures of GRABeCB2.0 fluorescent signal in HEK293 cells in culture.
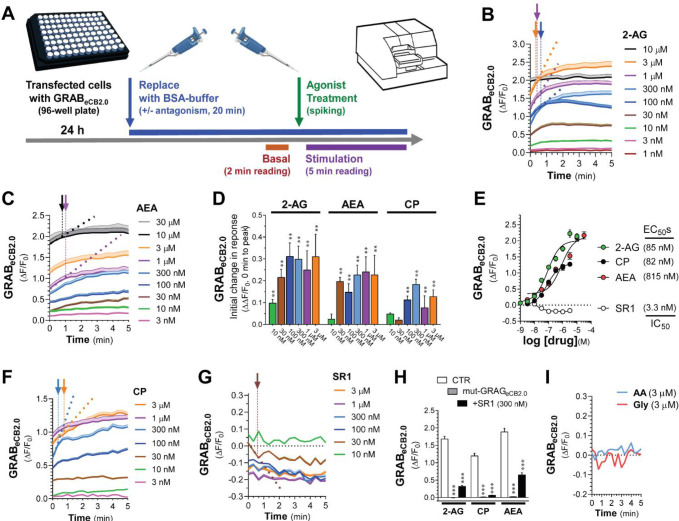
To further define the pharmacological profile and dynamics of changes in GRABeCB2.0 fluorescent signal, we developed and validated a fluorescent plate reader assay (96 wells, 3 Hz scanning frequency). Figure 2A outlines the experimental approach that consists of: 1] transfecting HEK293 cells in 96 well plates with eCB2.0 DNA plasmid constructs, 2] 24 h after transfection, replacing the cell culture media with buffer (20 min), inserting the plate into the reader and measuring GRABeCB2.0 fluorescent signal for the last 2 min of this preincubation period (i.e., Basal fluorescent signal), 3] promptly removing the plate from the reader, spiking agents (10X concentration) prepared in PBS supplemented with BSA (1 mg/ml) in media, and reinserting the plate in the reader, and finally 4] measuring fluorescence for 5 min (i.e., Stimulation fluorescent signal). To facilitate a data analysis pipeline, we developed a MATLAB R2021a algorithm that averages the fluorescent signal value of each well over time, for multiple experiments and at select timepoints (see the following link for the code: https://github.com/StellaLab/StellaLab.git). Figures 2B–C show that 2-AG and AEA induced concentration-dependent increases in GRABeCB2.0 fluorescent signals that were detected at the 0 sec timepoint (i.e., when the first Stimulation fluorescent signal was measured). For example, both the 10 nM and 100 nM 2-AG responses at 0 sec reached a GRABeCB2.0 fluorescent signals value of 0.19 and 0.96 ΔF/F0 over basal, respectively; and both these responses reached an initial inflection point at 0.66 and 0.33 min, respectively (Figures 2B, see color coded arrows). Of note, 2-AG at 3 μM induced the strongest response that reached an inflection point at ≈0.66 min, whereas 2-AG at 10 μM induced a response had already reached a maximum plateau response at the 0 min timepoint (Figures 2B). Thus, 2-AG rapidly activated GRABeCB2.0 in a concentration dependent manner as calculated by its initial response (i.e., slope: ΔΔF/F0 between 0 min and the initial inflection time point) (Figures 2D). To calculate EC50 values, we averaged the GRABeCB2.0 fluorescent signals between 4–5 min, and found 85 nM for 2-AG, a value that is consistent with its reported potency at CB1R (e.g., EC50 = 12–100 nM) (Figures 2E).27–30 AEA also induced a concentration-dependent and rapid increase in GRABeCB2.0 fluorescent signal that was detected at the 0 sec timepoint, but only reached a significant initial response starting at 30 nM, and a maximum plateau response at 10 μM (Figures 2C–D). Thus, AEA induced a concentration dependent increase in GRABeCB2.0 with an EC50 = 815 nM, an activity that is approximately 10-fold less potent than AEA’s potency at CB1R (e.g., EC50 = 69–276 nM) (Figures 2E).30–32 Of note, the EC50s values of 2-AG and AEA for GRABeCB2.0 measured here are lower than their previously reported values in cell culture models systems (i.e., 2-AG = 3.1–9.0 μM and AEA = 0.3–0.8 μM), most likely because we included BSA in the buffer to facilitate solubility and interaction with GRABeCB2.0.15;33
Figure 2: 2-AG, AEA, and CP differentially activate GRABeCB2.0: High-throughput fluorescence assay.
A] Schematic of high-throughput fluorescent plate reader assay: HEK293 cells were plated in a 96-well plate and transfected with eCB2.0 DNA plasmid; 24 h post transfection, growth media was replaced with imaging PBS buffer and incubated for 20 min. For the last 2 min of this incubation, the plate was placed in the fluorescent plate reader and a basal fluorescent signal was measured. Treatments were spiked in immediately after the basal reading and the pate was reinserted in the plate reader and fluorescent signal was recorded for 5 min. B, C, F, G and I] Kinetics of GRABeCB2.0 activation (ΔF/F0) following treatment with increasing concentrations of 2-AG (B), AEA (C), CP (F), SR1 (G) and arachidonic acid (AA) and glycerol (Gly) (I). Arrows represent inflection points and dotted lines represent initial change in response (slope between t=0 and time at inflection). D] Initial change in ΔF/F0 response elicited by increasing concentration of 2-AG, AEA, or CP shown in (B, C, and F). E] Concentration dependent responses and EC50s of 2-AG, AEA, and CP at inducing GRABeCB2.0 fluorescent signal and of SR1 at reducing GRABeCB2.0 fluorescent signal as determined by averaging ΔF/F0 between 4–5 min. H] 2-AG (1 μM), CP (1 μM), and AEA (10 μM)-induced increases in GRABeCB2.0 fluorescent signal was reduced by SR1 (300 nM) and absent in HEK293 cells expressing mut-GRABeCB2.0. Data represents mean ΔF/F0 between 4–5 min. Shaded areas on time-course plots and error bars on histograms represent s.e.m. Statistics: (D) **P<0.01 significantly different from basal (Two-Way ANOVA followed by Dunnett’s). (H) ***P<0.001 significantly different from corresponding CTR treatment (Two-way ANOVA followed by Dunnett’s). n= 9–50 independent experiments performed in triplicate.
To further characterize these high-throughput measures, we tested the effect of CP and SR1 on GRABeCB2.0 fluorescent signal. Figures 2F shows that CP induced a concentration-dependent increase in GRABeCB2.0 fluorescent signal that was detected at 0 sec and reached an initial inflection time point within ≈1.33 min and an EC50 = 82 nM, an activity that is at least 3-fold less potent than CP’s potency at CB1R (e.g., EC50 = 0.05–31 nM) (Figures 2D–E).34 As expected, SR1 induced a rapid and concentration-dependent decrease in GRABeCB2.0 fluorescent signal that was detected at 0 sec and plateaued below basal levels starting at 30 nM and for at least 5 min (Figures 2G). The IC50 for SR1 measured between 4–5 min was 3.3 nM, a value that mirrors SR1’s potency at CB1R (IC50 = 0.3–17 nM) (Figures 2E).34–37 Pre-treatment of HEK293 cells with SR1 (300 nM, 20 min) followed by treatment with 2-AG, AEA and CP significantly reduced GRABeCB2.0 activation (Figures 2H). Figure 2H also shows that 2-AG, AEA and CP failed to elicit increases in fluorescent signals in HEK293 cell expressing the mut-GRABeCB2.0, which has a similar expression profile as GRABeCB2.0 as determined by fluorescence confocal microscopy (Figure 1B). Thus, mut-GRABeCB2.0, which has a phenylalanine 177 to alanine mutation in the region within the orthosteric binding pocket to impair ligand binding, represents a valid negative control.38;39 As previously reported, the products of 2-AG hydrolysis, arachidonic acid, and glycerol, did not influence the GRABeCB2.0 fluorescent signal (Figure 2I).15 Together, these results provide strong support for use of this high throughput experimental approach to study the pharmacology and dynamics of changes in GRABeCB2.0 fluorescence when expressed by cells in culture, and show that: 1] Pronounced increases and decreases in GRABeCB2.0 fluorescent signal are reproducibly detected using a 96 well plate-reader format and promptly spiking agents in media, 2] 2-AG and SR1 modulate GRABeCB2.0 fluorescent signal with EC50s comparable to their activities at the CB1R, and 3] AEA and CP also increase GRABeCB2.0 fluorescent signal, although with 2–10-fold lesser activities than at CB1R.
Activity of 2-AG analogues at GRABeCB2.0.
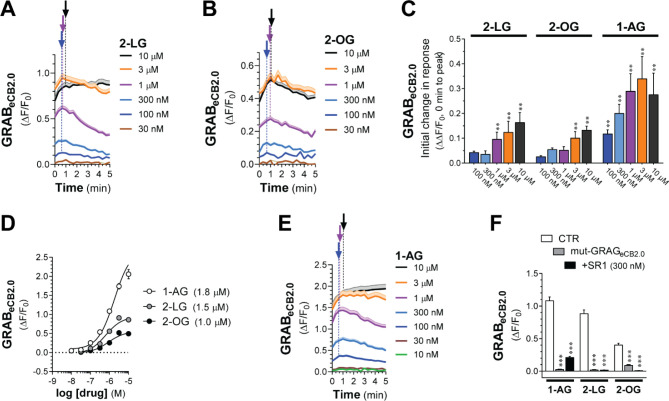
We leveraged the high-thought approach to determine whether 2-LG and 2-OG, which are produced by cells concomitantly to 2-AG, change GRABeCB2.0 fluorescent signals when expressed in HEK293 cells.11;12 Specifically, although 2-LG (18:2) and 2-OG (18:1) are lipid analogues of 2-AG (20:4) and are produced by the same biosynthetic and metabolic pathways as 2-AG, they are likely to activate CB1R signaling with much lower potency and efficacy than 2-AG. For example, 2-LG partially activates CB1R (EC50s = 16.6 μM) or antagonizes CB1R depending on the model system, while there is still no evidence that 2-OG might also activate CB1R.11;12 Figures 3A–B show that both 2-LG and 2-OG induced a concentration-dependent increase in GRABeCB2.0 fluorescent signal that was detected at 0 sec and that these responses rapidly reached their initial inflection time points within 1 min. Thus, 2-LG and 2-OG activated GRABeCB2.0 with increasingly rapid onset and EC50s = 1.0 and 1.5 μM, respectively, activities that are greater than their known potencies at CB1R (Figures 3C–D). We next tested 1-arachidonoylglycerol (1-AG, 20:4), which is a product of a non-enzymatic isomerization of 2-AG that is not endogenously produced by mammalian cells, but is commonly used to study the structure activity relationship of monoacylglycerols at CB1R as its acyl chain length and saturation match that of 2-AG (EC50s = 1–1.9 μM).29;40;41 We found that 1-AG induced a concentration-dependent increase in GRABeCB2.0 fluorescent signal that was detected at 0 sec, reached its initial inflection timepoint within 1 min and an EC50 = 1.8 μM, an activity that also mirrors its potency at CB1R (Figures 3D–E). The responses to these three monoacylglycerols were blocked by pretreatment with SR1 (300 nM) and absent in cells expressing the mut-GRABeCB2.0 (Figure 3F). Thus, 1-AG, 2-LG, and 2-OG increase GRABeCB2.0 fluorescent signal (rank order of potency: 2-OG > 2-LG > 1-AG; maximum efficacy: 1-AG >> 2-LG > 2-OG). Notably, all three lipid analogues had lower potencies than 2-AG and AEA, indicating that the GRABeCB2.0 has greater sensitivity for these two eCBs compared to similar monoacylglycerol lipids.
Figure 3: 2-LG, 2-OG, and 1-AG activate GRABeCB2.0.
HEK293 cells were transfected with eCB2.0 DNA plasmid and changes in fluorescent signal measured by high-throughput fluorescence assay. A, B, E] Kinetics of GRABeCB2.0 activation (ΔF/F0) following treatment with increasing concentrations of 2-LG (A), 2-OG (B), and 1-AG (E). Shaded areas in time courses and error bars represent s.e.m. Arrows represent inflection points and dotted lines represent initial change in response (slope between t=0 and time at inflection). C] Initial response (slope between time = 0 and inflection point) of ΔF/F0 response. D] Concentration dependent responses and EC50s of 2-LG, 2-OG, 1-AG at inducing GRABeCB2.0 fluorescent signal as determined by averaging ΔF/F0 between 4–5 min. F] 2-LG (1 μM), 2-OG (1 μM), and 1-AG (1 μM)-stimulated increases in GRABeCB2.0 fluorescent signal are reduced by SR1 (300 nM) and absent in HEK293 cells expressing mut-GRABeCB2.0. Statistics: (C) **P<0.01 significantly different from basal (Two-way ANOVA followed by Dunnett’s). (D) ***P<0.001 significantly different from corresponding CTR treatment (Two-Way ANOVA followed by Dunnett’s). n= 3–11 independent experiments performed in triplicate.
THC and CBD activity at GRABeCB2.0.
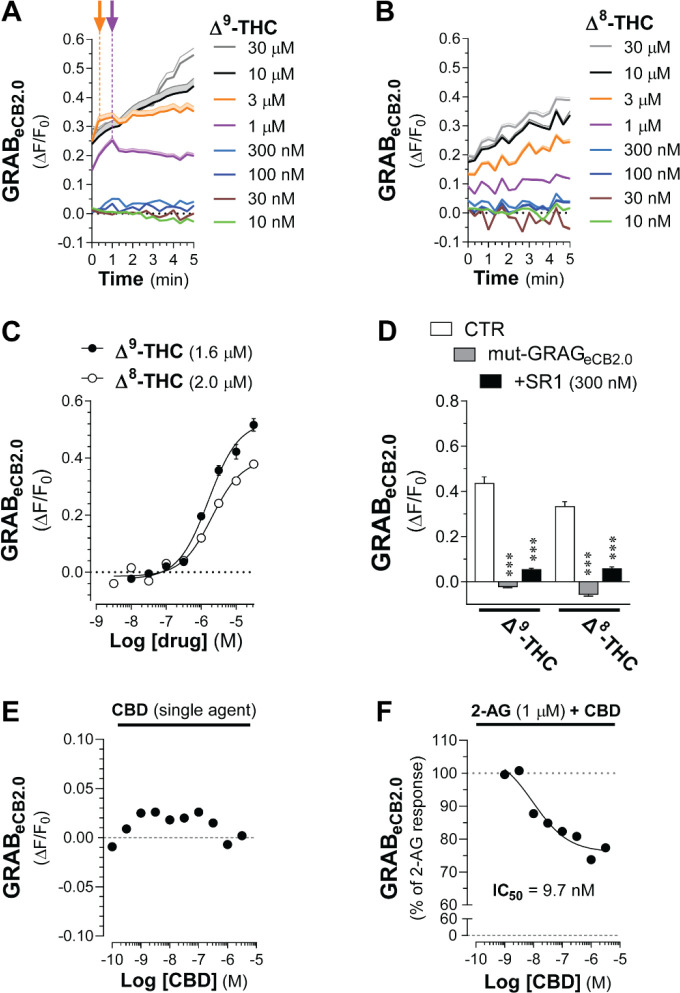
To extend the pharmacological characterization of GRABeCB2.0 using the high-throughput assay, we tested Δ9-THC, the principal psychoactive ingredient in Cannabis that activates CB1R as a high-affinity partial agonist,34 and Δ8-THC, which activates CB1R with a comparable pharmacology as Δ9-THC but represents a minor product of most Cannabis strains and remains poorly characterized.42 Δ9-THC increased GRABeCB2.0 fluorescent signal in a concentration-dependent manner that was detected at 0 sec starting at 1 and 3 μM, and these responses reached their initial inflection points within 1 min (Figure 4A). Of note, higher concentrations of Δ9-THC, i.e., 10 and 30 μM, triggered a response that continuously increased for at least 5 min without apparent inflection (Figure 4A). Figure 4B shows that Δ8-THC induced a concentration-dependent increase in GRABeCB2.0 fluorescent signal that was detected at 0 sec starting at 1 μM, and that all higher concentrations continuously increased for at least 5 min without inflection. Δ9-THC and Δ8-THC activate GRABeCB2.0 with comparable EC50s as calculated between 4–5 min (1.6 μM and 2 μM, respectively; Figure 4C). These 2 responses were blocked by pretreatment with SR1 (300 nM) and absent in cells expressing the mut-GRABeCB2.0 (Figure 4D). Thus, Δ9-THC and Δ8-THC similarly increase GRABeCB2.0 fluorescent signal, although with 10–100-fold lesser activities than their potencies at CB1R, and most of these responses at micromolar concentrations continuously increase for at least 5 min.
Figure 4: Δ9-THC and Δ8-THC activate GRABeCB2.0.
HEK293 cells were transfected with eCB2.0 DNA plasmid and changes in fluorescent signal measured by high-throughput fluorescence assay. A-B] Kinetics of GRABeCB2.0 activation (ΔF/F0) induced by increasing concentrations of Δ9-THC (A) or Δ8-THC (B). Shaded areas in time courses and error bars represent s.e.m. Arrows represent inflection points and dotted lines represent initial change in response (slope between t=0 and time at inflection). C] Concentration dependent responses and EC50s of Δ9-THC and Δ8-THC at inducing GRABeCB2.0 fluorescent signal as determined by averaging ΔF/F0 between 4–5 min. d] Δ9-THC (10 μM) and Δ8-THC (10 μM)-stimulated increases in GRABeCB2.0 fluorescent signal are reduced by SR1 (300 nM) and absent in HEK293 cells expressing mut-GRABeCB2.0. E-F] CBD (100 pM-10 μM) does not modulate GRABeCB2.0 fluorescent signal (E) and decreased the 2-AG (1 μM)-stimulated increase in GRABeCB2.0 fluorescent signal (IC50 of 9.7 nM). Statistics: (D) ***P<0.001 significantly different from corresponding CTR treatment (Two-way ANOVA followed by Dunnett’s). n= 3–7 independent experiments performed in triplicate.
CBD acts as a NAM of CB1R.8 We found that increasing concentrations of CBD (from 0.1 nM to 3 μM) did not significantly modulate basal GRABeCB2.0 fluorescent signal (Figure 4E and Supplementary Figure S4); however, it significantly reduced the 2-AG-induced increase in GRABeCB2.0 fluorescent signal by 26% at 1 μM and with an IC50 = 9.7 nM, values that are similar to CBD’s activity at CB1R (Figure 4E). Thus, our results suggests that GRABeCB2.0 retains the molecular mechanism that mediates the proposed NAM activity of CBD at CB1R.
Comparing changes in GRABeCB2.0 fluorescent signals.
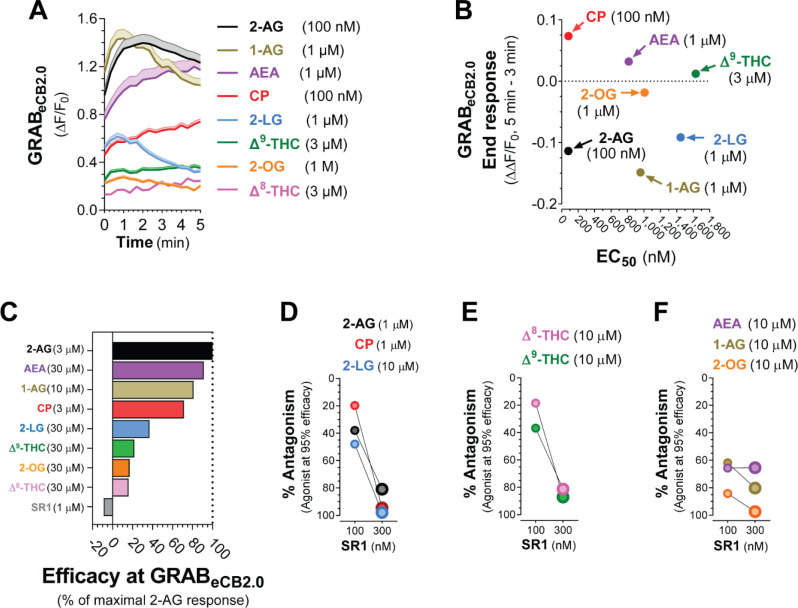
We sought to compare key pharmacological and dynamic parameters that characterize changes in GRABeCB2.0 fluorescent signal elicited by the agents tested in this study (chemical structures in Supplementary S5). Figure 5A shows GRABeCB2.0 fluorescent signal increased by each agonist applied at a concentration that approached their EC50s and the differences in their overall dynamics. Specifically, when analyzing the ΔΔF/F0 of these responses between 3–5 min, we found that the responses induced by monoacylglycerols were decaying between 3–5 min and thus had reached an earlier maximum response, whereas the GRABeCB2.0 fluorescent signals induced by AEA, CP and Δ9-THC continuously increased between 3–5 min and thus had not reached a maximum response within this time period (Figure 5B). Analysis of dose-responses indicated that 2-AG at 3 μM resulted in the most pronounced increase in GRABeCB2.0 fluorescent signal as measured by area under the curve between 0–5 min, and thus we calculated the relative efficacy of each agent at the of their maximal response concentration compared to the 2-AG response. Figure 5C shows that the maximal GRABeCB2.0 responses to AEA (30 μM), 1-AG (10 μM) and CP (3 μM) reached 91%, 81% and 71% of the 2-AG response, respectively. Figure 5C also shows that the maximal GRABeCB2.0 responses to 2-LG (10 μM) and 2-OG (10 μM) only reached 37% and 17% of the 2-AG response, respectively, and that Δ9-THC (30 μM) and Δ8-THC (30 μM) only reached 22% and 16% of the 2-AG response, respectively. Finally, we compared the antagonism of SR1 (100 and 300 nM) for each ligand. Figure 5D–E show that the 2-AG (1 μM), CP (1 μM), 2-LG (10 μM), as well as Δ9-THC (10 μM) and Δ8-THC (10 μM) we partial antagonized by SR1 100 nM and antagonized by more than 80% with SR1 300 nM. By contrast, 1-AG (10 μM) and 2-OG (10 μM) were strongly antagonized by SR1 at both 100 and 300 nM, and AEA (10 μM) was antagonized by only 65% by both 100 and 300 nM SR1. These results reveal differences in key pharmacological and dynamic parameters that describe changes in GRABeCB2.0 fluorescent signal elicited by eCBs and CB1R ligands.
Figure 5: Monoacylglycerols have distinct dynamics and pharmacological properties at GRABeCB2.0 compared to THC, CP, and AEA.
HEK293 cells were transfected with eCB2.0 DNA plasmid and changes in fluorescent signal measured by high-throughput fluorescence assay. A] Comparison of GRABeCB2.0 activation kinetics elicited by each ligand at a concentration that approaches their respective EC50s. Shaded areas in time courses and error bars represent s.e.m. B] Comparison of the EC50 of each ligand and their corresponding end response at a concentration that approached their respective EC50 and measured between 3–5 min. Y axis positive value represent increase in fluorescent signal (i.e., AEA, CP and D9-THC) and negative values represent decrease in fluorescent signal (i.e., 2-AG, 1-AG, 2-LG and 2-OG). C] Comparison of the maximum increase in GRABeCB2.0 fluorescent signal as expressed by area under the curve and as a percent of 3 μM 2-AG produced by each ligand. D-F] Antagonism of each response by SR1 (100 and 300 nM).
Discussion
Similarities and differences in GRABeCB2.0 and CB1R pharmacology.
GRABeCB2.0 was developed starting from CB1R and screening for genetic constructs and individual randomized mutations to improve the change in fluorescent signal in response to 2-AG.17 The initial pharmacological characterization of GRABeCB2.0 was performed in the absence of BSA in the buffer and resulted in EC50s values that are lower than their activities at CB1R: 2-AG = 3–9 μM, AEA = 0.3–0.8 μM, CP = 20 nM and THC = 2 μM).15 BSA is known to facilitate the solubility and interaction of CB agents with CB1R.33 We found that inclusion of BSA in the buffer improves 2-AG’s potency at GRABeCB2.0 to 82 nM compared to published values, but didn’t affect the activities of AEA, THC and CP at GRABeCB2.0 compared to published values. This result suggests that BSA might preferentially facilitate the solubility and interaction of select CB agents with CB1R and GRABeCB2.0, here 2-AG.
2-AG interacts with specific amino acids within the orthosteric binding site of CB1R that are likely conserved in GRABeCB2.0.5;38 By contrast, AEA, eCB analogues, phyto-CBs and artificial CBs bind to different amino acids within the orthosteric binding site that might have been mutated in GRABeCB2.0 and reduce their interaction with this sensor compared to CB1R. Thus, our study suggests that, compared to CB1R, GRABeCB2.0 expressed by cells in culture and mouse tissues reliably senses nanomolar changes in 2-AG levels, and micromolar amounts of AEA, eCB analogues, THC and CP.
GRABeCB2.0 pharmacology relative to abundance of eCB analogues and CB agents.
Diacylglycerol lipase (DAGL) produces several monoacylglycerols in addition to 2-AG that exhibit significant activity at CB1R, such as 2-LG.11;12 Furthermore, monoacylglycerol lipase (MAGL) and α/β domain containing 6 (ABHD6) hydrolyze several MAGs in addition to 2-AG, including 2-LG and 2-OG.43 However, 2-LG is 10 less abundant than 2-AG and requires 100-fold higher concentrations than 2-AG to activate CB1R (EC50 ≈ 25 μM). Considering these values, we conclude that the activity-dependent increases in GRABeCB2.0 fluorescent signal measured in cells in culture and mouse tissues that are blocked by DAGL inhibitors and increased by MAGL/ABHD6 inhibitors are much more likely to involve change in 2-AG levels than changes in 2-LG and 2-OG levels.
Δ9-THC increases GRABeCB2.0 fluorescent signal with an EC50 that is 100-fold higher than its activity at CB1R. Intraperitoneal injections of Δ9-THC (5 mg/kg) in adult mice results in ≈10 ng/ml of THC in mouse brain (31.8 nM).44;45 Accordingly, we conclude that GRABeCB2.0 expressed in mouse brain will therefore be able to sense i.p. injections of Δ9-THC and Δ8-THC and enable studying their PK profile and how this correlates with changes in behavior, or when either ligand arrives at a particular circuit or brain region in real time.
GRABeCB2.0 plateau versus decay dynamics.
When applied at a concentration that approaches their EC50 values, CB agents increased GRABeCB2.0 fluorescent signal with different kinetics: 2-AG, 1-AG, 2-LG, and 2-OG triggered rapid increases in fluorescent signal that were followed by slow decays, whereas AEA, CP, Δ9-THC and Δ8-THC triggered progressive increases in fluorescent signal that did not reach peak response within 5 min. HEK293 cells do not express MAGL, ABHD6 and fatty acid amid hydrolase (FAAH) but may express an unidentified hydrolase known to metabolize monoacylglycerols,46;47 an enzymatic activity that is likely involved in the slow decay response of 2-AG, 1-AG, 2-LG, and 2-OG. HEK293 cells also lack CYP2C9 and CYP3A4 enzymes that metabolize Δ9-THC and Δ8-THC. Thus, Δ9-THC and Δ8-THC are likely not enzymatically degraded by HEK293 cells in culture and will constantly increase GRABeCB2.0 fluorescent signal.
NAM activity of CBD at GRABeCB2.0.
Allosteric binding sites are distinct protein domains from orthosteric sites that bind small molecules and either increase or decrease orthosteric site-mediated changes in protein conformations and activities. Early in vitro and in vivo studies showed that CBD reduces CB1R signaling at concentrations well below its reported affinity (Ki) to the orthosteric agonist site of CB1R, providing the initial evidence that CBD acts as a NAM at this receptor.48–50 Accordingly, in neurons in cell culture, CBD reduces the efficacy and potency of 2-AG and THC at increasing CB1R signaling and inhibits eCB-mediated synaptic plasticity without influencing basal neurotransmission.51–53 Mutagenesis of CB1R indicated that several N-terminal residues of the CB1R, namely Cys98, Cys107, and Met1, interact with CBD when it occupies the putative allosteric site of the CB1R that CBD may target, and that this allosteric site overlaps with the orthosteric site which is near the second extracellular loop.8;54 In silico modeling of CB1R with an intact N-terminus revealed a potential binding pocket for NAMs of CB1R in close proximity its N terminus, one of the longest among class A GPCRs, and molecular docking studies suggest that binding to this site may results in a change in the 3 dimensional structure of the orthosteric binding site and thus in THC’s and 2-AG’s potencies.54;55 While these results suggest that CBD inhibits CB1R signaling by directly interacting with an allosteric binding site on this target, direct demonstration of CBD binding to CB1R is still needed. We found that CBD does not affect baseline GRABeCB2.0 fluorescent signal but reduces 2-AG’s activity at GRABeCB2.0, a result consistent with the premise that the molecular mechanism involved in mediating CBD’s allosteric modulation of CB1R remains functional in GRABeCB2.0. Accordingly, mutagenesis optimization of CB1R constructs to generate GRABeCB2.0 did not significantly affect its N-terminus (Supplementary Figure S1). Our results suggest that structural and mechanistic comparisons of CBD activity at CB1R and GRABeCB2.0 might help us better understand that molecular mechanism involved in the allosteric modulation of CB1R. That is- how allosteric ligands produce a distinctive receptor conformation with unique signaling and therapeutic value.
In conclusion:
We show that 2-AG increases GRABeCB2.0 fluorescent signal as a full agonist and with an EC50 similar to its activity at CB1R, and that GRABeCB2.0 responds to AEA, 2-LG and 2-OG with EC50s that are lower than their EC50s at CB1R. Considering the lower amount of AEA, 2-LG and 2-OG produced by cells, our results suggest that activity-dependent increases in GRABeCB2.0 fluorescent signal measured in cells in culture and mouse tissues will mainly reflect change in 2-AG levels, especially when this response is blocked by inhibitors of 2-AG production and inactivation. We also show that SR1 blocks GRABeCB2.0 fluorescent signal with an IC50 similar to its reported potency at CB1R, and that this leads to levels below baseline fluorescent signal, suggesting that GRABeCB2.0 exhibits basal fluorescence, and that SR1 may act as an inverse agonist and represents a useful pharmacological tool to validate GRABeCB2.0 functionality when expressed by various model systems. THC and CP increase GRABeCB2.0 fluorescent signal with EC50s lower than their activity at CB1R indicating that only high brain concentration of these agents will be detected in cell culture and mouse tissue model systems. CBD reduces the 2-AG-induced increase in GRABeCB2.0 fluorescent signal but not its basal fluorescent signal, suggesting that the molecular mechanism of CBD allosterism present in CB1R is maintained in GRABeCB2.0. Thus, GRABeCB2.0 provides an opportunity to study how changes in THC and CBD concentration and co-activity at CB1R might occur in cell culture and mouse tissue model systems. Our results outline the pharmacological profile and activation dynamics of GRABeCB2.0 to improve the interpretation of changes in its fluorescent signal when expressed in various model systems.
Supplementary Material
Funding Statement:
This work was supported by the National Institutes of Health (NS118130 and DA047626 to N.S., DA055448 to AE, DA033396 to MRB, and T32GM007750). We would also like to acknowledge support from the University of Washington Center of Excellence in Opioid Addiction Research/ Molecular Genetics Resource Core (P30DA048736). National Natural Science Foundation of China (31925017 and 31871087), the Beijing Municipal Science & Technology Commission (Z181100001318002 and Z181100001518004), the NIH BRAIN Initiative (1U01NS113358), the Shenzhen-Hong Kong Institute of Brain Science (NYKFKT2019013), the Science Fund for Creative Research Groups of the National Natural Science Foundation of China (81821092) and grants from the Peking-Tsinghua Center for Life Sciences and the State Key Laboratory of Membrane Biology at Peking University School of Life Sciences (to YL).
Footnotes
Conflict of Interest: The authors declare that they do not have any known competing financial interests or relationships that could have influenced the work in this paper.
References
- 1.Zou S, Kumar U. Cannabinoid Receptors and the Endocannabinoid System: Signaling and Function in the Central Nervous System. Int J Mol Sci 2018;19:833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu H-C, Mackie K. An Introduction to the Endogenous Cannabinoid System. Biol Psychiatry 2016;79:516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busquets-Garcia A, Bains J, Marsicano G. CB 1 receptor signaling in the brain: extracting specificity from ubiquity. Neuropsychopharmacology 2018;43:4–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lutz B. Neurobiology of cannabinoid receptor signaling. Dialogues in Clinical Neuroscience 2020;22:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shahbazi F, Grandi V, Banerjee A, et al. Cannabinoids and Cannabinoid Receptors: The Story so Far. iScience 2020;23:101301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen M, Thayer SA. Ð9-Tetrahydrocannabinol acts as a partial agonist to modulate glutamatergic synaptic transmission between rat hippocampal neurons in culture. Molecular Pharmacology 1999;55:8–13. [DOI] [PubMed] [Google Scholar]
- 7.Kelley BG, Thayer SA. Delta 9-tetrahydrocannabinol antagonizes endocannabinoid modulation of synaptic transmission between hippocampal neurons in culture. Neuropharmacology 2004;46:709–715. [DOI] [PubMed] [Google Scholar]
- 8.Laprairie RB, Bagher AM, Kelly ME, et al. Cannabidiol is a negative allosteric modulator of the cannabinoid CB1 receptor. Br J Pharmacol 2015;172:4790–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jakowiecki J, Abel R, Orzeł U, et al. Allosteric Modulation of the CB1 Cannabinoid Receptor by Cannabidiol-A Molecular Modeling Study of the N-Terminal Domain and the Allosteric-Orthosteric Coupling. Molecules 2021;26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zoerner AA, Gutzki F-M, Batkai S, et al. Quantification of endocannabinoids in biological systems by chromatography and mass spectrometry: A comprehensive review from an analytical and biological perspective. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2011;1811:706–723. [DOI] [PubMed] [Google Scholar]
- 11.Murataeva N, Straiker A, Mackie K. Parsing the players: 2-arachidonoylglycerol synthesis and degradation in the CNS. Br J Pharmacol 2014;171:1379–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu L, Williams G, Doherty P. 2-Linoleoylglycerol Is a Partial Agonist of the Human Cannabinoid Type 1 Receptor that Can Suppress 2-Arachidonolyglycerol and Anandamide Activity. Cannabis Cannabinoid Res 2019;4:231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tortoriello G, Beiersdorf J, Romani S, et al. Genetic Manipulation of sn-1-Diacylglycerol Lipase and CB(1) Cannabinoid Receptor Gain-of-Function Uncover Neuronal 2-Linoleoyl Glycerol Signaling in Drosophila melanogaster. Cannabis Cannabinoid Res 2021;6:119–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labouesse MA, Patriarchi T. A versatile GPCR toolkit to track in vivo neuromodulation: not a one-size-fits-all sensor. Neuropsychopharmacology 2021;46:2043–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong A, He K, Dudok B, et al. A fluorescent sensor for spatiotemporally resolved imaging of endocannabinoid dynamics in vivo. Nature biotechnology 2021:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patriarchi T, Cho JR, Merten K, et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science (New York, NY) 2018;360:eaat4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravotto L, Duffet L, Zhou X, et al. A bright and colorful future for G-protein coupled receptor sensors. Frontiers in Cellular Neuroscience 2020;14:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrell JS, Colangeli R, Dong A, et al. In vivo endocannabinoid dynamics at the timescale of physiological and pathological neural activity. Neuron 2021;109:2398–2403. e2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liput DJ, Puhl HL, Dong A, et al. 2-Arachidonoylglycerol mobilization following brief synaptic stimulation in the dorsal lateral striatum requires glutamatergic and cholinergic neurotransmission. Neuropharmacology 2022;205:108916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Yang N, Dong J, et al. Deficiency in endocannabinoid synthase DAGLB contributes to Parkinson’s disease and dopaminergic neuron dysfunction. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baggelaar MP, Chameau PJ, Kantae V, et al. Highly selective, reversible inhibitor identified by comparative chemoproteomics modulates diacylglycerol lipase activity in neurons. Journal of the American Chemical Society 2015;137:8851–8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozenfeld R. Type I Cannabinoid Receptor Trafficking: All Roads Lead to Lysosome. Traffic 2011;12:12–18. [DOI] [PubMed] [Google Scholar]
- 23.Rozenfeld R, Bushlin I, Gomes I, et al. Receptor Heteromerization Expands the Repertoire of Cannabinoid Signaling in Rodent Neurons. PLOS ONE 2012;7:e29239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He JC, Gomes I, Nguyen T, et al. The Gαo/i-coupled Cannabinoid Receptor-mediated Neurite Outgrowth Involves Rap Regulation of Src and Stat3*. Journal of Biological Chemistry 2005;280:33426–33434. [DOI] [PubMed] [Google Scholar]
- 25.Jordan JD, He JC, Eungdamrong NJ, et al. Cannabinoid Receptor-induced Neurite Outgrowth Is Mediated by Rap1 Activation through Gαo/i-triggered Proteasomal Degradation of Rap1GAPII*. Journal of Biological Chemistry 2005;280:11413–11421. [DOI] [PubMed] [Google Scholar]
- 26.Jung K-M, Astarita G, Thongkham D, et al. Diacylglycerol lipase-alpha and -beta control neurite outgrowth in neuro-2a cells through distinct molecular mechanisms. Mol Pharmacol 2011;80:60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stella N, Schweitzer P, Piomelli D. A second endogenous cannabinoid that modulates long-term potentiation. Nature 1997;388:773–778. [DOI] [PubMed] [Google Scholar]
- 28.Mechoulam R, Ben-Shabat S, Hanus L, et al. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochemical Pharmacology 1995;50:83–90. [DOI] [PubMed] [Google Scholar]
- 29.Sugiura T, Kodaka T, Nakane S, et al. Evidence That the Cannabinoid CB1 Receptor Is a 2-Arachidonoylglycerol Receptor: STRUCTURE-ACTIVITY RELATIONSHIP OF 2-ARACHIDONOYLGLYCEROL, ETHER-LINKED ANALOGUES, AND RELATED COMPOUNDS*. Journal of Biological Chemistry 1999;274:2794–2801. [DOI] [PubMed] [Google Scholar]
- 30.Steffens M, Zentner J, Honegger J, et al. Binding affinity and agonist activity of putative endogenous cannabinoids at the human neocortical CB1 receptor. Biochemical Pharmacology 2005;69:169–178. [DOI] [PubMed] [Google Scholar]
- 31.Kearn CS, Greenberg MJ, DiCamelli R, et al. Relationships between ligand affinities for the cerebellar cannabinoid receptor CB1 and the induction of GDP/GTP exchange. J Neurochem 1999;72:2379–2387. [DOI] [PubMed] [Google Scholar]
- 32.Shen M, Piser TM, Seybold VS, et al. Cannabinoid receptor agonists inhibit glutamatergic synaptic transmission in rat hippocampal cultures. J Neurosci 1996;16:4322–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makriyannis A, Guo J, Tian X. Albumin enhances the diffusion of lipophilic drugs into the membrane bilayer. Life sciences 2005;77:1605–1611. [DOI] [PubMed] [Google Scholar]
- 34.An D, Peigneur S, Hendrickx LA, et al. Targeting Cannabinoid Receptors: Current Status and Prospects of Natural Products. Int J Mol Sci 2020;21:5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinaldi-Carmona M, Barth F, Héaulme M, et al. SR141716A, a potent and selective antagonist of the brain cannabinoid receptor. FEBS letters 1994;350:240–244. [DOI] [PubMed] [Google Scholar]
- 36.Bauer M, Chicca A, Tamborrini M, et al. Identification and quantification of a new family of peptide endocannabinoids (Pepcans) showing negative allosteric modulation at CB1 receptors. J Biol Chem 2012;287:36944–36967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z, Iyer MR, Godlewski G, et al. Functional Selectivity of a Biased Cannabinoid-1 Receptor (CB1R) Antagonist. ACS Pharmacology & Translational Science 2021;4:1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hua T, Vemuri K, Nikas SP, et al. Crystal structures of agonist-bound human cannabinoid receptor CB(1). Nature 2017;547:468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shim J-Y, Bertalovitz AC, Kendall DA. Identification of essential cannabinoid-binding domains: structural insights into early dynamic events in receptor activation. J Biol Chem 2011;286:33422–33435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dócs K, Mészár Z, Gonda S, et al. The Ratio of 2-AG to Its Isomer 1-AG as an Intrinsic Fine Tuning Mechanism of CB1 Receptor Activation. Front Cell Neurosci 2017;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farah SI, Hilston S, Tran N, et al. 1-, 2- and 3-AG as substrates of the endocannabinoid enzymes and endogenous ligands of the cannabinoid receptor 1. Biochemical and Biophysical Research Communications 2022;591:31–36. [DOI] [PubMed] [Google Scholar]
- 42.Tagen M, Klumpers LE. Review of delta-8-tetrahydrocannabinol (Δ8-THC): Comparative pharmacology with Δ9-THC. British journal of pharmacology 2022;179:3915–3933. [DOI] [PubMed] [Google Scholar]
- 43.Navia-Paldanius D, Savinainen JR, Laitinen JT. Biochemical and pharmacological characterization of human α/β-hydrolase domain containing 6 (ABHD6) and 12 (ABHD12). J Lipid Res 2012;53:2413–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torrens A, Vozella V, Huff H, et al. Comparative Pharmacokinetics of Δ(9)-Tetrahydrocannabinol in Adolescent and Adult Male Mice. J Pharmacol Exp Ther 2020;374:151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vozella V, Zibardi C, Ahmed F, et al. Fast and Sensitive Quantification of Δ(9)-Tetrahydrocannabinol and Its Main Oxidative Metabolites by Liquid Chromatography/Tandem Mass Spectrometry. Cannabis Cannabinoid Res 2019;4:110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baggelaar MP, Maccarrone M, van der Stelt M. 2-Arachidonoylglycerol: A signaling lipid with manifold actions in the brain. Progress in Lipid Research 2018;71:1–17. [DOI] [PubMed] [Google Scholar]
- 47.Fezza F, Bari M, Florio R, et al. (2014) Endocannabinoids, Related Compounds and Their Metabolic Routes. In Molecules.) pp. 17078–17106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pertwee RG. Cannabinoids and multiple sclerosis. Pharmacol Ther 2002;95:165–174. [DOI] [PubMed] [Google Scholar]
- 49.Matsuda LA, Lolait SJ, Brownstein MJ, et al. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature 1990;346:561–564. [DOI] [PubMed] [Google Scholar]
- 50.Thomas BF, Gilliam AF, Burch DF, et al. Comparative receptor binding analyses of cannabinoid agonists and antagonists. Journal of Pharmacology and Experimental Therapeutics 1998;285:285–292. [PubMed] [Google Scholar]
- 51.Straiker A, Dvorakova M, Zimmowitch A, et al. Cannabidiol inhibits endocannabinoid signaling in autaptic hippocampal neurons. Molecular pharmacology 2018;94:743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laprairie RB, Bagher AM, Kelly ME, et al. Type 1 cannabinoid receptor ligands display functional selectivity in a cell culture model of striatal medium spiny projection neurons. J Biol Chem 2014;289:24845–24862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laprairie R, Bagher A, Kelly M, et al. Cannabidiol is a negative allosteric modulator of the cannabinoid CB1 receptor. British journal of pharmacology 2015;172:4790–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jakowiecki J, Abel R, Orzeł U, et al. Allosteric Modulation of the CB1 Cannabinoid Receptor by Cannabidiol—A Molecular Modeling Study of the N-Terminal Domain and the Allosteric-Orthosteric Coupling. Molecules 2021;26:2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shao Z, Yan W, Chapman K, et al. Structure of an allosteric modulator bound to the CB1 cannabinoid receptor. Nature chemical biology 2019;15:1199–1205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.