Abstract
Across biological systems, a number of genomic processes, including transcription, replication, DNA repair, and transcription factor binding, display intrinsic directionalities. These directionalities are reflected in the asymmetric distribution of nucleotides, motifs, genes, transposon integration sites, and other functional elements across the two complementary strands. Strand asymmetries, including GC skews and mutational biases, have shaped the nucleotide composition of diverse organisms. The investigation of strand asymmetries often serves as a method to understand underlying biological mechanisms, including protein binding preferences, transcription factor interactions, retrotransposition, DNA damage and repair preferences, transcription-replication collisions, and mutagenesis mechanisms. Research into this subject also enables the identification of functional genomic sites, such as replication origins and transcription start sites. Improvements in our ability to detect and quantify DNA strand asymmetries will provide insights into diverse functionalities of the genome, the contribution of different mutational mechanisms in germline and somatic mutagenesis, and our knowledge of genome instability and evolution, which all have significant clinical implications in human disease, including cancer. In this review, we describe key developments that have been made across the field of genomic strand asymmetries, as well as the discovery of associated mechanisms.
Keywords: Mutational strand asymmetries, Transcriptional strand asymmetries, Replicative strand asymmetries, Orientation
Graphical Abstract
1. Introduction
The DNA double helix shows rotational symmetry, whereas a number of biological processes such as transcription, replication, DNA repair, and transcription factor binding have intrinsic directionalities [1], [2]. Chargaff’s first parity rule, conceived over 70 years ago, states that the number of adenines (As) equals the number of thymines (Ts), while the number of guanines (Gs) equals the number of cytosines (Cs) [3]; this parity rule can be explained by base complementarity in double-stranded DNA. Chargaff’s second parity rule states that in long genomic windows, nucleotide sequences on the two complementary strands are found with approximately the same frequency [4], [5]. Although this rule is most accurate for long nucleotide sequences [4], it holds true for most double-stranded DNA organisms, with the notable exception of certain symbiotes [6].
In contrast to Chargaff’s first parity rule, which was explained by the elucidation of the double stranded DNA structure, a comprehensive explanation for the second rule has not yet been found. Although there are no clear evolutionary advantages associated with it, the second law has been observed across diverse organisms [7]. In addition, it is not attributed to a single biological mechanism, but is likely the result of multiple genomic processes [7]. Nevertheless, some research has pointed to inversions and inverted transposition events being major contributors to the validity of this rule [8], [9], while other models have proposed stem-loop structures [10] and duplication events [11] as potential explanations.
Importantly, when investigating particular genomic localities, there are clear deviations from the second parity rule, which can be attributed to specific functional elements. Biological processes, such as transcription and replication, possess intrinsic directionality, therefore resulting in the heterogeneous distribution of information. Identification of strand asymmetries can therefore enable the detection of biological mechanisms, the identification of novel genomic elements, and the characterization of selective environmental constraints. At the same time, strand asymmetry analyses can improve computational models across biological domains, such as in the estimation of the likelihood of mutagenesis, the identification of driver mutations, in cis-regulatory logic, in evolution, and in disease. In this review, we provide an overview of multiple biological processes that result in the asymmetric distribution of genomic information and demonstrate the utility of strand asymmetries as a tool to decipher new biological mechanisms.
2. Strand asymmetries shape the nucleotide composition of diverse genomes
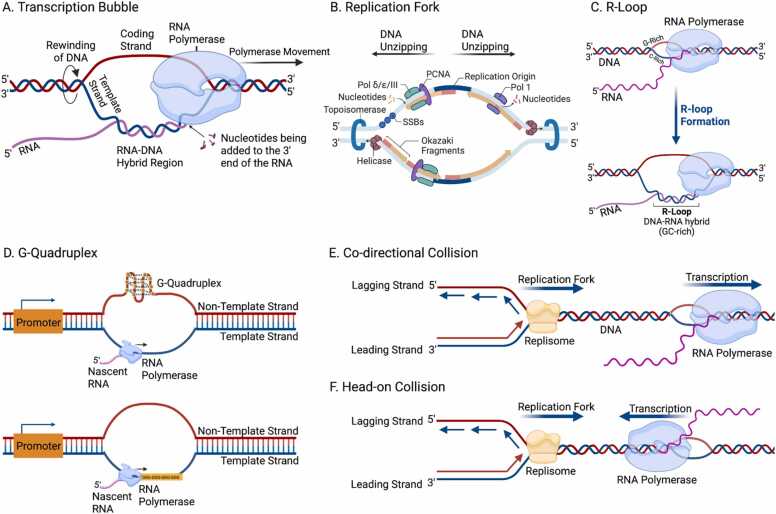
In transcription, which is a directional process, the elongating RNA polymerases synthesize nascent RNA complementary to the template strand (Fig. 1a). During replication, the leading strand is replicated continuously, whereas the lagging strand is replicated in short Okazaki fragments [12], [13] (Fig. 1b). Because DNA polymerases must add nucleotide monomers in the 5' and 3' directions, a discontinuous polymerization with Okazaki fragments on the lagging orientation is necessary.
Fig. 1.
Strand asymmetries associated with replication and transcription. A. Orientation of transcription fork, displaying the template and non-template strands and the generation of a nascent RNA. B. Replication fork schematic showing continuous replication in one orientation and discontinuous replication with Okazaki fragments in the opposite strand. Topoisomerase, helicase, and polymerases are crucial enzymes in this process. C. GC-skews favor formation of secondary structures such as R-loops. D. G-quadruplex at the template strand can impede RNA polymerase movement, whereas at the non-template strand, it can facilitate transcription. E-F. Replication-transcription collisions are a cause of instability and can result in replication fork arrest, premature transcription termination and genomic instability. Types include E. co-directional collisions and F. head-on collisions (Created with BioRender.com).
Transcriptional and replicative strand asymmetries refer to the asymmetric distribution of information such as nucleotides or motifs between the leading and lagging strands or between the template and non-template strands respectively. Both forms of asymmetry have been observed in the genomes of diverse organisms including prokaryotes, eukaryotes, and viruses [14], [15], [16], [17], [18], [19], [20], [21], [22]. These intrinsically asymmetric processes result in mutational asymmetries between the two DNA strands and have shaped the genomes of organisms across the tree of life [23]. For example, cytosine deamination occurs primarily at single-stranded DNA, resulting in C to T mutations. The likelihood of cytosine deamination is significantly higher on the leading-strand [22], and there is a higher repair rate on the lagging strand for C>T mutations [24]. As a result, in most studied bacteria, the leading strand has an excess of Gs and Ts relative to Cs and As [25]. Borrelia burgdorferi, a bacterium that causes Lyme disease, is one of the species with the most pronounced leading / lagging nucleotide asymmetries [26].
These asymmetries are frequently quantified with GC-skew and AT-skew, which measure statistical deviations of guanines or adenines between the two strands and which have been used to identify the location of replication origins, elucidate the direction of replication, and even to validate genome assemblies [27], [28], [29]. In the human genome, there is an enrichment of Gs and Ts relative to As and Cs on the non-template strand of genes [30]. Since the non-template DNA remains single-stranded for longer, while the template strand is used for the synthesis of the nascent RNA, cytosine deamination can explain the observed nucleotide asymmetries [31]. GC-skews favor the formation of non-canonical secondary structures including G-quadruplexes and R-loops, which are known to influence gene regulation and have also been associated with RNA polymerase pause sites in CpG island promoters [32], [33], [34], [35] (Fig. 1c-d).
In both prokaryotes and eukaryotes, a larger number of genes are usually found in the leading orientation [36], [37]. This phenomenon has been explained by a lower mutation rate, by competition between replication and gene expression [37], and as a way to limit collisions between the transcription and replication machineries [38] (Fig. 1e). A collision with the replication fork can halt transcription by the RNA polymerase in either orientation, and head-on collisions are the most common way replication is interrupted [39], [40] (Fig. 1e-f). Collisions can be a source of genomic instability, and prokaryotic genomes are therefore structured in ways that limit the number of collision events. Across 1552 studied bacterial and archaeal species, more than 90% of them subsequently display preference for their coding genes on the leading strand [41]. For instance, in the bacterium Bacillus subtilis, 75% of genes are transcribed in the same orientation as the direction of replication [42].
Further supporting this model, genes that are highly expressed and essential genes, such as ribosomal genes, which would experience more frequent collisions due to a higher density of elongating RNA polymerases, tend to be found on the leading strand [38], [43], [44], [45], [46]. For example, only 6% of essential genes are found on the lagging strand in Bacillus subtilis [47]; these essential genes found on the lagging strand in Bacillus subtilis have a higher rate of point mutations and non-synonymous mutations, indicating that they undergo faster adaptive evolution [48]. In addition to essential genes, longer operons are more likely to be found on the leading strand [48], [49]. As a result, head-on replication–transcription collisions result in a higher rate of mutagenesis than co-directional collisions, and there is a bias for co-orientation of transcription with replication that has been shaped by selection pressures [36]. In addition, essential genes tend to be at earlier positions in operon units in order to be more highly expressed [50], indicating how organismal genomes can be arranged to maximize protein efficacy.
In eukaryotic cells, multiple mechanisms are in place to limit collisions. These involve the separation of replication and transcription domains during S-phase [51], [52], replication fork barriers [53], coordinated changes between replication and transcription timing across different tissues or during differentiation [54], and a higher frequency of genes in early replicating domains [54], [55]. Nevertheless, replication-transcription collisions still occur in eukaryotic cells, particularly in the longer genes that require more time to be transcribed [56]. Collisions between the replication and transcription machineries are a cause of DNA damage, genomic instability, and recombination in eukaryotic cells [57].
Gene expression can be a mechanism that safeguards genome integrity. The testis is the tissue that expresses the highest number of genes in mammals; this results in a reduced mutation rate for the transcribed strand due to transcription-coupled repair, and in turn, leads to reduced population diversity across the expressed genes [58]. A study that investigated the contribution of transcriptional strand asymmetries in the usage of energetically cheaper nucleotides (“U”,”C”) in synonymous sites across 1550 prokaryotic genomes found substantial asymmetries resulting in strand-specific nucleotide usage [59]. The observed asymmetries were due to replication-related, transcriptional-related, and translational-related selection, and selection constraints were particularly amplified with higher expression levels [59].
3. Strand asymmetries in genes and gene features
The orientation of genes is often biased, and one extreme case of this is polycistronic gene expression, in which all genes have the same directionality. Prokaryotic operons are polycistronic, while the vast majority of eukaryotic mRNAs are monocistronic. However, it has been noted that polycistronic mRNAs can be rarely found in eukaryotic genomes [60], [61], [62]. Genes are heterogeneously distributed across the human genome. There are gene deserts, large genomic regions in which genes are largely absent, as well as gene clusters, in which gene density is significantly higher [63], [64]. This observation can be explained by common proximity-based regulation of multiple genes; genes are over-represented in early-replicating regions [65], [66].
In addition, gene pairing has been observed to be common across eukaryotic species, with genes being found in different orientations [67]. Gene pairs can be found in three orientations, which are tail to head, head to head, and tail to tail [66]. Genes in close proximity to each other are found more frequently in the head to head orientation, and this was observed for metabolism, DNA repair genes, housekeeping genes, and an unbiased set, while the expression of nearby genes has also been found to be correlated [68], [69].
Transcription in eukaryotes is inherently bidirectional, and antisense transcripts can arise from this process [70]. In contrast to mRNA transcripts, most of these antisense transcripts are unstable [71] and can be used for co-option and generation of new genes. However, it remains unclear what the exact mechanisms are that confer directional transcription. Long non-coding RNAs (lncRNAs) can be produced in the sense or antisense orientation of protein-coding genes [72]. For example, in yeast, the transcription factor Rap1 restricts transcription to the divergent orientation [73]; it remains unknown if additional transcription factors contribute to this effect.
Furthermore, key transcription initiation and termination signals, such as the TATA-box and the polyadenylation signal, display not only positional constraints but also intrinsic directionalities [74], [75], [76], and such directionalities have been used to identify genic regions [77], [78]. Nucleotide strand asymmetries have also been observed relative to splice sites [79], [80]. Strand asymmetries can be found in motifs associated with the splicing code, which are used for the recognition of core splicing signals, such as the 3’ and 5’ splice sites.
Exons and introns display opposite nucleotide strand asymmetries. In introns, Ts are more frequent than As and Gs are more abundant than Cs, a trend that is reversed in exons in both humans and mice [79]. This could serve as a mechanism to discriminate between exons and introns. Interestingly, intronless genes, in which splicing is absent, do not display these patterns [79]. Furthermore, the observed asymmetry trends do not translate to yeast. Zhang et al. found that exonic splicing enhancers and exonic splicing silencers display strand asymmetry patterns, and they utilized the observed strand asymmetry patterns to identify novel splicing regulatory elements. Another study found significant strand asymmetries in the distribution of G-quadruplexes between the template and non-template orientations relative to splice sites and provided evidence for their roles in the modulation of alternative splicing events [81]. As a result, a number of studies have used the inherent directionality in transcription initiation, splicing, and termination signals to identify mechanisms of gene regulatory control.
4. Mutational strand asymmetries and insights in operative biological processes
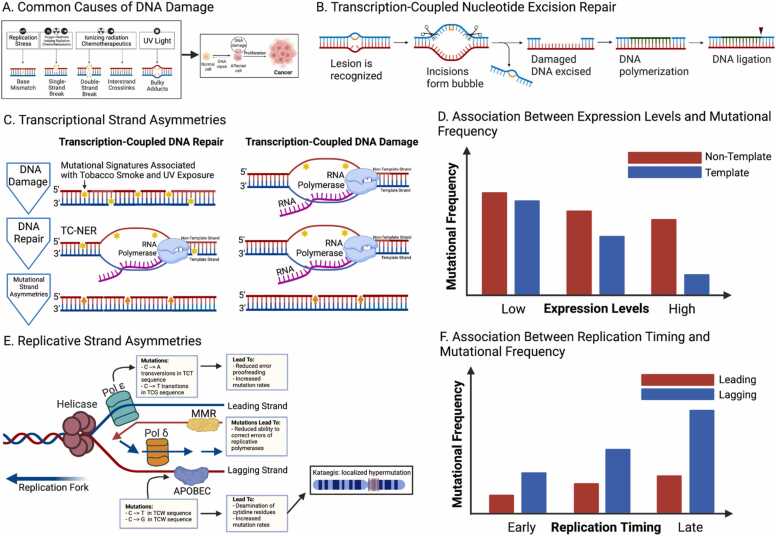
Throughout our lives, cells in the human body acquire and accumulate somatic mutations. Processes that cause the accumulation of somatic mutations can be divided into exogenous, such as UV light exposure, and endogenous, such as defects in DNA repair and oxidative damage (Fig. 2a). Therefore, mutational processes continuously shape the genome of somatic cells. Uncontrolled clonal expansion, usually through the accumulation of cancer driver mutations, can result in cancer development [82]. The vast majority of mutations in a cancer genome are passenger mutations, which have little to no effect on tumor progression. However, they can serve as signatures of operative mutational processes and also inform us about mutational strand asymmetries [83]; asymmetries can be inferred from the mutated nucleotides, depending on their frequency in leading versus lagging and template versus non-template orientations. DNA damage in either of the two complementary bases results in the same mutated site, and as a result, the base of the original DNA damage cannot be deduced with standard sequencing methods. However, substitution mutations at a reference nucleotide can be oriented on the template or the non-template strand relative to the transcriptional direction or on the leading or lagging strands relative to the directionality of the replication fork. Studies that have profiled the replicative and transcriptional strand biases relative to replication origins and transcription start sites have shown specific mutational patterns around those genomic sites [84], [85].
Fig. 2.
Mutational strand asymmetries. A. DNA damage from exogenous and endogenous processes can lead to the accumulation of somatic mutations and eventually can result in cancer development. B. Nucleotide excision repair schematic showing removal of transcription-blocking DNA lesions. C. In transcription-coupled repair, DNA damage at the non-template strand is more likely to escape repair from the TC-NER, leading to mutational strand asymmetries. With regards to transcription-coupled DNA damage, there is a lack of protection of the non-template strand during transcription, also leading to mutational strand asymmetry. D. Lowly expressed genes have higher mutation rates, while highly expressed genes have lower mutation rates. TC-NER activity is associated with expression levels and this results in an association between transcriptional strand asymmetry in mutations and expression levels. E. Deficiencies of proteins such as MMR or polymerase ε can lead to replicative strand asymmetries. F. Early replication timing is associated with lower mutation rates, while late replication timing is associated with higher mutation rates. For certain mutational processes, the replicative strand asymmetry aggravates with replication timing between early and late replicating regions. Schematics 2d and 2 f provide a model and do not include real data (Created with BioRender.com).
Strand asymmetric segregation of DNA lesions was observed in murine liver tumor genomes, resulting in chromosome-scale strand asymmetry of mutations [86]. Another study investigated how the orientation of the minor groove relative to histones influences germline and somatic mutation rate and found differences between sites with the DNA minor groove facing toward or away from the histones; this was observed across cancer types [87]. Moreover, the magnitude of the effect was higher for nucleosomes with strong rotational position, further supporting the model [87]. In a recent study, asymmetry in the distribution of structural population variants relative to the orientation of repeat elements was detected [88]. This likely reflects the jumping events of transposable elements in the population. Transposable element re-activation is frequently observed in cancer, and application of strand asymmetry analyses in structural variant datasets from cancer genomes could provide valuable mechanistic insights.
5. Transcriptional strand asymmetries in cancer genomes
Substitution mutations provide valuable information about underlying mutational processes. Previous research has used mutational classification of mutational signatures to further separate the standard 96 substitution classification system using the template and non-template orientation into 192 possible mutation classes [83]. The authors found strong transcriptional strand bias for mutational signatures associated with ultraviolet exposure and tobacco smoke among others [83]. In a recent study, a classification system for doublet-base substitutions and indel mutations was implemented across 4645 whole-genome and 19,184 whole-exome sequenced cancer tumors; the study identified additional mutational signatures with transcriptional strand asymmetries, which were also associated with tobacco smoke and ultraviolet exposure [89].
DNA damage is preferentially repaired at the template strand of expressed genes through transcription-coupled nucleotide excision repair (TC-NER), which removes transcription-blocking DNA lesions [90], [91] (Fig. 2b). In transcription-coupled repair, the recruitment of TC-NER correlates with expression levels, and highly transcribed genes have the most pronounced mutational strand asymmetries [92] (Fig. 2c-d). DNA damage at the non-template strand, however, is more likely to escape repair from TC-NER because it does not interfere with RNA polymerase progression and because it remains exposed as single stranded DNA, which is more likely to be mutated [93]. Therefore, transcription-associated mutations occur in part because the non-transcribed strand is single stranded and less protected from DNA damage and mutagens, which in turn can result in a higher rate of mutagenesis [94]. Recently, it was shown that transcription-associated mutagenesis is also observed in both germline and somatic mutations of higher eukaryotes at transcribed regions, a phenomenon that was previously seen primarily in microorganisms [95]. As a result, differences in DNA damage and repair between the template and non-template strands in transcribed regions are pervasive and can be reconstructed with mutational strand asymmetry analyses [96].
The accumulation of tobacco-related carcinogens at guanines in lung cancer results in the mutational imbalance of G>T site substitutions due to the preferential repair of these adducts at the template strands of expressed genes [97]. In liver cancer, a mutational signature that is correlated with alcohol consumption shows marked patterns associated with expression levels and transcription-coupled damage [98] (Fig. 2d). In bladder cancers, the mutational signature SBS92, which is enriched in smokers, has been shown to have a strong transcriptional strand asymmetry [99]. Another study oriented mononucleotide repeat tracts to observe transcriptional strand asymmetries in indel mutagenesis [100]. There is also evidence for significant differences in the strand asymmetries between introns and exons because exons are under stronger selection pressure and codon usage preference [101], [102]; there is also more efficient repair by mismatch repair (MMR) at exons [103]. However, transcription strand bias has been associated primarily with exogenous processes including tobacco smoking and UV light, which in turn are repaired by NER.
6. Replicative strand asymmetries in cancer genomes
Replicative strand biases are observed in cancer genomes, with one study showing significant replicative strand asymmetries across fourteen cancer types [96] (Fig. 2e). Systematic examination of mutational processes has indicated that replicative strand asymmetries are more common than transcriptional strand asymmetries across the mutational signatures examined [96]. In contrast with transcriptional strand asymmetries, replicative strand asymmetries are linked to endogenous processes; they are associated with repair enzyme deficiencies, such as MMR and polymerase ε deficiencies, as well as with the activity of the Apolipoprotein B mRNA editing catalytic polypeptide-like family (APOBEC) of cytidine deaminases [96]. Vöhringer et al. showed that out of twenty mutational signatures examined, nine exhibited significant replicative strand asymmetry, while only five showed significant transcriptional strand asymmetry [104]. Recently, replicative strand asymmetries have also been observed for specific mutational signatures in germline variants [105], [106].
In humans, leading and lagging strand DNA synthesis is performed primarily by polymerase ε and polymerase δ respectively [107], [108], [109]. MMR or polymerase ε deficiencies result in pronounced replicative strand asymmetries in the distribution of mutations, which indicates that these enzymes normally balance the likelihood of mutation during DNA replication [96]. It has also been observed that in certain cases, the magnitude of the replicative strand asymmetry can be associated with replication timing, with earlier replicating regions showing more pronounced replicative strand asymmetry in cancer genomes with polymerase ε deficiencies [96], [110] (Fig. 2f). Polymerase δ mutations in the exonuclease domain have also been reported; they are associated with increased mutability and show replicative strand asymmetries [111]. MMR also impacts the mutation rate between early and late replicating regions. Late replicating regions accumulate a higher number of mutations, while an MMR deficiency terminates this pattern [112]. Lujan et al. examined the contribution of MMR to replicative strand asymmetries with yeast as the model system and found that there is higher MMR efficiency for lagging-strand DNA polymerase α and DNA polymerase δ than for the leading-strand DNA polymerase ε [113]. Recent studies have also provided experimental proof for the roles of different repair enzymes in the observed mutational strand asymmetries. Zou et al. showed that the gene knockout of repair genes such as MSH6, MSH2 and MLH1 resulted in replication strand asymmetry effects in isogenic cell models [114], providing further experimental evidence regarding the contribution of the DNA mismatch repair system to mutational strand asymmetries. Knock outs of other DNA repair genes such as EXO1 and RNF168 showed specific transcription strand asymmetry effects [114].
Mutations associated with APOBEC, a cytidine deaminase with important roles in antiviral defense, cause off-target mutagenesis in the genome, especially at single-stranded DNA sites. There is evidence for episodic APOBEC mutagenesis across multiple cancer types [115], [116]. The APOBEC mutational signatures show a preference for early-replicating regions and highly expressed genes [117] with replicative strand asymmetry [22], [96] due to deamination of the lagging strand template during DNA replication [118]. APOBEC is also linked to kataegis, which is characterized by local strand-coordinated hypermutation [92] (Fig. 2e).
7. Orientation preferences in repeat elements
Transposable elements, originally discovered by McClintock [119], were initially thought of as junk DNA; however, this view has in many ways been disproven. Repeat elements represent a significant portion of the human genome and have contributed to its structure, functionalities, and evolution, while also contributing to genetic diversity between people. It is estimated that repetitive elements comprise two thirds of the human genome [120]. Some studies have suggested that transposable elements might offer an explanation for Chargaff's second parity rule [8] and account for the inversion events that could explain this rule [4], [9]. However, the integration of these elements is not random and exhibits biases in the sequence context and orientation preference [121], [122], [123], [124], as well as for preference for repeat pairs and clustering of repeat elements [125], [126], [127].
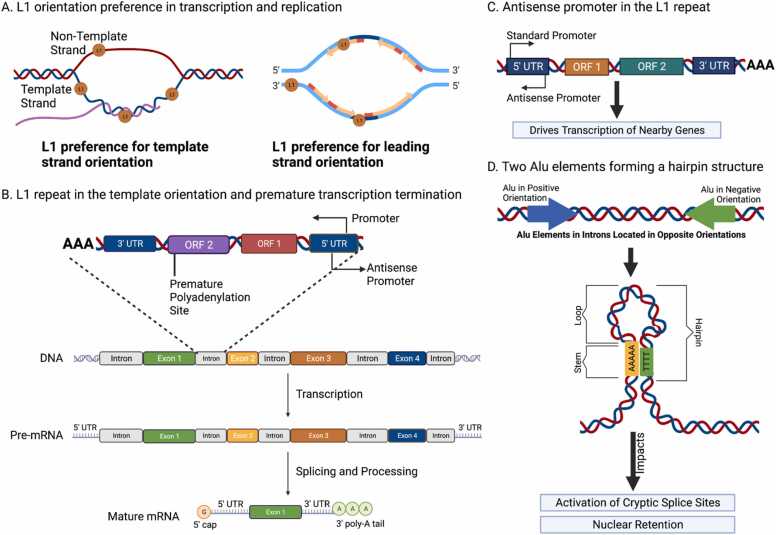
In the human genome, long interspersed nuclear elements (LINE) and short interspersed nuclear elements (SINE) show significant transcriptional and replicative strand asymmetries, while long terminal repeats (LTRs) exhibit pronounced transcriptional strand asymmetries [88]. LINE-1 (L1) elements are the most abundant subclass, comprising around 17% of the human genome [128]. Only approximately 100 L1 sites are still retrotransposition competent in the germline [129] and in disease [130]. The L1 distribution in the human genome shows a preference for the leading strand orientation relative to the replication direction [124] and for the template strand orientation in transcribed regions [123], [131] (Fig. 3a). Even though there is a higher density of L1 elements at late replicating regions, integration is more likely to occur at early-replicating sites, suggesting that evolutionary selection contributes to the observed patterns in the genome. Interestingly, the smaller subset of integrations at the non-template orientation are much more likely to be pathogenic or disease-causing [132]. However, when L1 repeats are present in introns in the template orientation, they can cause premature termination of transcription due to a polyadenylation signal within the L1 element [133], [134] (Fig. 3b). On the other hand, an antisense promoter in the L1 repeat, with opposite orientation than the open reading frames of the repeat, can drive transcription of nearby genes [135] (Fig. 3c); this has implications for both evolution and disease.
Fig. 3.
The effects of repeat element orientation. A. L1 shows an integration preference for the template strand and leading strand orientations. B. Polyadenylation signals within L1 repeats in introns in the template orientation can result in premature termination of transcription. C. Antisense promoters in L1 repeats can drive transcription of nearby genes. D. Two Alu repeats in opposite orientations can form hairpin structures (Created with BioRender.com).
Similarly, LTRs are more frequently found in the template orientation, and Alu repeats, which are a subset of SINE elements, also show a preference for the template orientation [136]. In lncRNAs, Alu repeats tend to be tolerated in the template strand across gene regions, whereas in the non-template strand, they tend to be found at the 3’ end [137]. Alu repeats are likely to be found clustered, closely positioned, and in direct orientation to one another [138], [139]. The orientation preference of multiple endogenous repeat elements for the template orientation in transcribed regions could be due to interference with transcription-associated signals in the non-template strand orientation, including splicing and polyadenylation motifs. Alu repeats in opposite orientations can form hairpin structures, in turn impacting biological processes such as alternative splicing and nuclear retention [125] (Fig. 3d). Overall, the orientation preference for the template strand across multiple endogenous repeat element categories could reflect the tendency to reduce the number of collisions between reverse transcription and gene transcription.
8. Orientation preferences in transcription factor binding
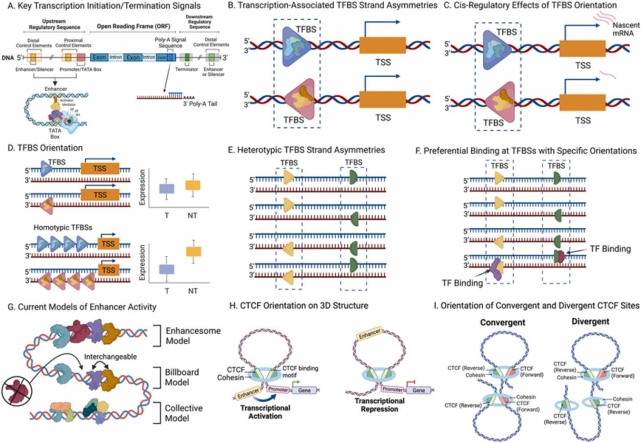
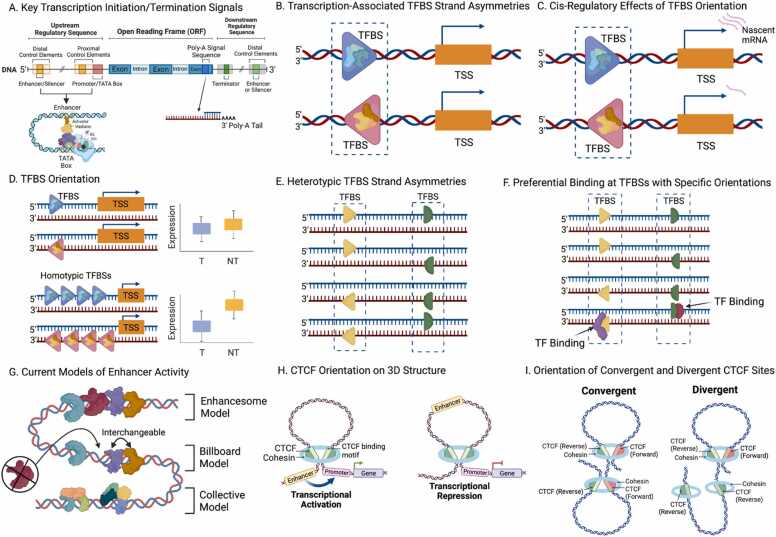
The orientation of DNA motifs in the genome impacts diverse biological processes, including gene regulation, through its effect on co-operative transcription factor binding at cis-regulatory elements (Fig. 4a). Combinatorial transcription factor binding is instrumental in organizing gene expression patterns across developmental time points and tissues [140], [141]. Even though only a limited number of studies have thoroughly investigated the impact of TFBS orientation, there is important evidence to suggest that TFBS orientation is a major factor in gene regulatory grammar [142], [143], [144]. TFBSs can be oriented relative to transcription direction and relative to one another (Fig. 4b-c). The orientation of homotypic or heterotypic transcription factor motif pairs is biased across the genome, and their relative orientation impacts homotypic and heterotypic transcription factor complex formation [143], [144], [145], [146], [147], [148] (Fig. 4d-e).
Fig. 4.
Transcription factor orientation and cis-regulatory grammar. A. Key transcription initiation and termination signals depicting the TATA-box and polyadenylation signal. B. and C. The orientation of a TFBS relative to the transcriptional direction in promoter upstream regions influences expression. D. and E. Orientation of TFBSs relative to each other. F. The orientation of TFBSs relative to each other influences transcription factor binding. G. Current models of enhancer activity include the enhancesome, billboard, and collective models. H. The orientation of CTCF is important for the formation of enhancer-promoter interactions and transcriptional activation. I. Convergent CTCF sites can create loops, but divergent CTCF sites disrupt the 3D structure (Created with BioRender.com).
At short inter-motif distances, the TFBS orientations impact protein-protein interactions (PPIs). In addition, even though the consensus TFBS motif of many TFs is palindromic, providing two templates for binding, there are significant binding biases depending on the orientation when considering flanking nucleotides [149]. There is also evidence to suggest that transcription factor pairs can bind to composite motifs with orientation and proximity preferences and that the composite motif sequences can differ from the constituent motif sequences of the individual transcription factors [142] (Fig. 4f). In the human transcriptome, the transcription factor binding sites for almost half of the transcription factors display strand asymmetry preference, which cannot be fully explained by nucleotide composition biases between the template and non-template strands [88]. The observed asymmetries could reflect binding preferences and not form impediments for RNA polymerase progression. Similarly, both at promoter upstream and downstream regions, there is orientation bias for a number of transcription factors [88]. In plants, orientation preference of TFBSs has been observed close to the transcription start site, which was attributed to background strand asymmetries in the dinucleotide composition of promoter upstream regions [150]. An association with expression levels was not identified.
At the core promoter, a number of motifs are positioned with respect to orientation, distance, and order preferences. For instance, transcription initiation in TATA-box-containing promoters requires the orientation and correct positioning of promoter-related motifs, including the initiation element, the TATA-box, and the upstream and downstream promoter elements, among others [74], [75]. Reversal of the TATA-box orientation can significantly reduce transcription levels [151]. In promoters with TATA and Inr motifs, correct spacing and orientation are important constituents for a synergistic effect [152]. At the 5’ end of the first intron in the non-template strand, G-quadruplexes and GrIn1 motifs have been shown to be associated with promoter-proximal pausing [153].
With regards to enhancers, studies that have investigated their mechanism of function have led to the proposition of two models, and there is currently evidence to support both of them. The “enhanceosome model” states that the function of the enhancer is dependent on the orientation, positioning, and order of TF binding sites, with changes in them resulting in significant changes in the enhancer’s activity [154] (Fig. 4g). The interferon-beta (IFN-beta) enhanceosome, which is highly conserved and for which an atomic model of cooperative TF binding has been produced, provided the first evidence to support the enhanceosome model [155], [156]. For example, within the IFN-beta enhanceosome, the ATF-2–c-jun heterodimer binds in a specific orientation which is necessary for the formation of the complex between ATF-2–c-jun and interferon regulatory factor 3 [157].
Second, the “billboard model”, which is also referred to as the information display model, proposes a more flexible structure for enhancer grammar in which the combination, orientation, order, and distance of cognate motifs are not fixed, but can instead vary without impacting enhancer function [157], [158] (Fig. 4g). In this model, only the binding sites themselves are critical. A number of studies have provided support for the billboard model [159], [160], indicating that both the enhanceosome and billboard models are likely to be true dependent on the specific enhancer.
Multiple studies have provided experimental evidence for the effect of orientation and spacing in cis-regulation. In a breakthrough study, researchers performed consecutive affinity-purification systematic evolution of ligands by exponential enrichment (CAP-SELEX) experiments, with which they examined 9400 TF–TF–DNA interactions. Interestingly, they were able to show that both the orientation and distance between the TF motifs determined heterodimer formation for a plethora of TF pairs [142]. Using massively parallel reporter assays (MRPAs), the orientation of enhancer tiles was found to have limited effects on expression levels [161]. However, this study did not capture orientation differences of individual TFs or of TF pairs within the enhancer tiles.
The transcription factor Yin-Yang can act as an activator or a repressor depending on motif orientation and positioning [162]. The orientation of the nuclear receptor for 1,25-dihydroxyvitamin D3 response elements in the basal promoter of the human calbindin D9k gene and the rat osteocalcin gene can change the expression 10-fold, and therefore, the orientation of the response elements dramatically influences the transcriptional response [163]. GABP–CREB1 motifs tend to be spaced with a one or two base pair gap with the two motifs in opposite orientations [164]. In the case of AP-1 transcription factor, the motif orientation, as well as its flanking base pairs at AP-1 binding sites, influence homo- and hetero-dimerization, and heterodimers of Fos and Jun bind in a preferred orientation [149], [165], [166]. In the IFN-β enhanceosome, the ATF-2–c-jun heterodimer does not show an orientation preference in the absence of IRF-1, whereas in its presence, it adopts an orientation-specific binding [157]. Therefore, in this particular case, the sequence orientation and the presence of specific proteins dictates the orientation of heterodimeric transcription factor binding. Another example of orientation preference has been observed in the NF-κB p50-p65 heterodimer, which is controlled by half-sites in the κB motif [167], [168].
The positioning of TFBSs within a nucleosome influences transcription factor binding, which can subsequently stabilize or destabilize a nucleosome [169], [170]. TFBSs can be found at different positions, such as near the edge or center of the nucleosome. Furthermore, studies have shown that TFs display directional binding to nucleosomes. TFBSs positioned along a nucleosome’s surface can face inward or outward. For the TFBSs of many transcription factors, especially of ETS and CREB bZIP factors, there is a preference for the end of the nucleosomal DNA or for periodic positions on the solvent-exposed side of the DNA [171]. This is likely due to steric hindrance and scaffolding by the nucleosome, resulting in specific positioning and orientation of TFBSs [171]. Furthermore, DNase I hypersensitivity analysis followed by sequencing (DNase-seq) experiments revealed unidirectional opening of chromatin relative to pioneer transcription factor motifs, with four out of the eight pioneer transcription factor families opening chromatin in a single orientation [172]. Nucleosome oriented binding has been observed for multiple pioneer transcription factors, including GATA3 and FOXA1 [173]; these TFs are able to bind to closed chromatin, recruit nucleosome remodelers, histone modification enzymes, and other transcription factors upon binding, and change the accessibility of a cis-regulatory region. However, additional research is required to examine the interplay between chromatin structure and the orientation of TFBSs and TF complexes.
9. CTCF motif orientation and genome organization
One of the most notable examples has been the CCCTC-binding factor (CTCF), which contributes to the formation of topologically-associating domains (TADs). Enhancer-promoter interactions are constrained within TADs, with the orientation of CTCF sites being important for their formation (Fig. 4h). The vast majority of CTCF sites are found to be bound by cohesin [174], which is associated with transcription factors and present in almost all active enhancer regions [175]. CTCF and the cohesin complex colocalize on chromatin, and their organization can help regulate three-dimensional genome structure through chromatin loop formation [176], [177]. These protein-mediated loops bring two loci that lie far apart along the chromosome into closer physical proximity; the CTCF binding sites stop loop extrusion with the ring-like cohesin complex [178]. The process of loop extrusion has been shown to link promoters and enhancers, be correlated with gene activation, and be conserved across both cell types and species [177], [179] (Fig. 4i). Interestingly, Rao et al. demonstrated that the deletion of CTCF sites interferes with loop formation and that after cohesin loss, loop domains disappear [177]. On the other hand, during cohesin recovery, the loop domains form again in minutes [177].
Loop extrusion can increase contact between loci that would typically lie in different sub-compartments [177]. The genome is separated into intervals based on distinctive histone marks, and these intervals are assigned to two compartments, A or B [177]. Intervals of the same type demonstrate increased contact frequency with one another, and loci in a compartment often form contact domains. When cohesin is lost, compartmentalization is preserved, demonstrating that it does not rely on cohesin, unlike the loop extrusion mechanism [177]. The loop extrusion mechanism interferes with compartmentalization by promoting the co-localization of loci not necessarily from the same compartment [177]. These loops are predominantly formed (greater than 90%) by convergent CTCF motif pairs that are asymmetric and face each other [180]. When their orientation is reversed, the 3D structure is disrupted (Fig. 4i).
Disruption of the loop extrusion mechanism has been associated with cancer due to alterations in enhancer-gene interactions [178]. This disruption is a result of the hypermutation of CTCF/cohesin binding sites, which are functional and alter CTCF binding, in almost all cancer types [175], [181]. Skin cancers specifically demonstrate distinct asymmetric mutations at CTCF-cohesin binding sites that form independently of replication timing; the specific mutations can be attributed to UV radiation and uneven nucleotide excision repair [181]. This mutation bias points towards cohesin being important for stabilization during CTCF-DNA binding and for impairing NER [181].
10. Conclusions
In this review, we have highlighted a number of genomic processes that are associated with strand asymmetries and have presented many of the underlying mechanisms that contribute to the asymmetric distribution of genomic features in organismal genomes. Strand asymmetries shape the nucleotide composition of viral, prokaryotic, and eukaryotic genomes and are genomic signatures of the biological processes that shape them. We have also highlighted the contribution of strand asymmetries in gene regulation, splicing, transcription factor binding, and retrotransposition. In addition, we summarize evidence regarding how mutational strand asymmetries reveal insights into DNA damage and repair in human health and disease. We argue that the implementation of sensitive methods to detect strand asymmetries in biological problems will enable breakthroughs in our understanding of genome biology.
The directionality of information in the DNA molecule is reflected in the orientation of motifs, genes, and other genomic elements. To conclude, an analogy can be drawn between genomic strand asymmetries and the road code, which dictates the rules by which vehicles have to move around in cities and with traffic signs that give instructions to road users. Similar to that, the orientation of motifs, genes, and other genomic elements in the genome provides instructions on how they should be interpreted.
CRediT authorship contribution statement
I.G.S. conceived and supervised the study. C.M., A.Z. and I.G.S. wrote the manuscript. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Acknowledgements
This study was funded by the startup funds of I.G.S. from the Penn State College of Medicine.
Contributions
I.G.S. conceived and supervised the study. C.M., A.Z. and I.G.S. wrote the manuscript.
Conflict of interest
No conflicts of interest.
Contributor Information
Apostolos Zaravinos, Email: A.Zaravinos@euc.ac.cy.
Ilias Georgakopoulos-Soares, Email: izg5139@psu.edu.
References
- 1.Smith D.J., Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belotserkovskii B.P., Tornaletti S., D’Souza A.D., Hanawalt P.C. R-loop generation during transcription: Formation, processing and cellular outcomes. DNA Repair. 2018;71:69–81. doi: 10.1016/j.dnarep.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chargaff E. Structure and function of nucleic acids as cell constituents. Fed Proc. 1951;10:654–659. [PubMed] [Google Scholar]
- 4.Baisnée P.-F., Hampson S., Baldi P. Why are complementary DNA strands symmetric? Bioinformatics. 2002;18:1021–1033. doi: 10.1093/bioinformatics/18.8.1021. [DOI] [PubMed] [Google Scholar]
- 5.Rudner R., Karkas J.D., Chargaff E. Separation of B. subtilis DNA into complementary strands, I. Biological properties. Proc Natl Acad Sci. 1968;60:630–635. doi: 10.1073/pnas.60.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nikolaou C., Almirantis Y. Deviations from Chargaff’s second parity rule in organellar DNA Insights into the evolution of organellar genomes. Gene. 2006;381:34–41. doi: 10.1016/j.gene.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell D., Bridge R. A test of Chargaff’s second rule. Biochem Biophys Res Commun. 2006;340:90–94. doi: 10.1016/j.bbrc.2005.11.160. [DOI] [PubMed] [Google Scholar]
- 8.Albrecht-Buehler G. Asymptotically increasing compliance of genomes with Chargaff’s second parity rules through inversions and inverted transpositions. Proc Natl Acad Sci. 2006;103:17828–17833. doi: 10.1073/pnas.0605553103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fickett J.W., Torney D.C., Wolf D.R. Base compositional structure of genomes. Genomics. 1992;13:1056–1064. doi: 10.1016/0888-7543(92)90019-o. [DOI] [PubMed] [Google Scholar]
- 10.Forsdyke D.R., Bell S.J. Purine loading, stem-loops and Chargaff’s second parity rule: a discussion of the application of elementary principles to early chemical observations. Appl Bioinforma. 2004;3:3–8. doi: 10.2165/00822942-200403010-00002. [DOI] [PubMed] [Google Scholar]
- 11.Jain S., Raviv N., Bruck J. Attaining the 2nd Chargaff Rule by Tandem Duplications. 2018 IEEE International Symposium on Information Theory (ISIT) 2018. https://doi.org/10.1109/isit.2018.8437526.
- 12.MacNeill S. The Eukaryotic Replisome: a Guide to Protein Structure and Function. Springer Science & Business Media; 2012.
- 13.Benkovic S.J., Valentine A.M., Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 14.Kano-Sueoka T., Lobry J.R., Sueoka N. Intra-strand biases in bacteriophage T4 genome. Gene. 1999;238:59–64. doi: 10.1016/s0378-1119(99)00296-6. [DOI] [PubMed] [Google Scholar]
- 15.Lobry J.R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 16.Touchon M., Nicolay S., Audit B., Brodie of Brodie E-B, d’Aubenton-Carafa Y, Arneodo A, et al. Replication-associated strand asymmetries in mammalian genomes: toward detection of replication origins. Proc Natl Acad Sci U S A 2005;102:9836–9841. [DOI] [PMC free article] [PubMed]
- 17.Pavlov Y.I., Newlon C.S., Kunkel T.A. Yeast origins establish a strand bias for replicational mutagenesis. Mol Cell. 2002;10:207–213. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- 18.Beletskii A., Bhagwat A.S. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia X. DNA replication and strand asymmetry in prokaryotic and mitochondrial genomes. Curr Genom. 2012;13:16–27. doi: 10.2174/138920212799034776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lind P.A., Andersson D.I. Whole-genome mutational biases in bacteria. Proc Natl Acad Sci USA. 2008;105:17878–17883. doi: 10.1073/pnas.0804445105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mrázek J., Karlin S. Strand compositional asymmetry in bacterial and large viral genomes. Proc Natl Acad Sci USA. 1998;95:3720–3725. doi: 10.1073/pnas.95.7.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhagwat A.S., Hao W., Townes J.P., Lee H., Tang H., Foster P.L. Strand-biased cytosine deamination at the replication fork causes cytosine to thymine mutations in Escherichia coli. Proc Natl Acad Sci USA. 2016;113:2176–2181. doi: 10.1073/pnas.1522325113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliverio A.M., Katz L.A. The dynamic nature of genomes across the tree of life. Genome Biol Evol. 2014;6:482–488. doi: 10.1093/gbe/evu024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank A.C., Lobry J.R. Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene. 1999;238:65–77. doi: 10.1016/s0378-1119(99)00297-8. [DOI] [PubMed] [Google Scholar]
- 25.McLean M.J., Wolfe K.H., Devine K.M. Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J Mol Evol. 1998;47:691–696. doi: 10.1007/pl00006428. [DOI] [PubMed] [Google Scholar]
- 26.Picardeau M., Lobry J.R., Hinnebusch B.J. Analyzing DNA strand compositional asymmetry to identify candidate replication origins of Borrelia burgdorferi linear and circular plasmids. Genome Res. 2000;10:1594–1604. doi: 10.1101/gr.124000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu J., Salzberg S.L. SkewIT: the skew index test for large-scale GC skew analysis of bacterial genomes. PLoS Comput Biol. 2020;16 doi: 10.1371/journal.pcbi.1008439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang G., Gao F. Quantitative analysis of correlation between AT and GC biases among bacterial genomes. PLoS One. 2017;12 doi: 10.1371/journal.pone.0171408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubert B., Skew D.B. A comprehensive database of GC and 10 other skews for over 30,000 chromosomes and plasmids. Sci Data. 2022;9:92. doi: 10.1038/s41597-022-01179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green P., Ewing B., Miller W., Thomas P.J. NISC Comparative Sequencing Program, Green ED. Transcription-associated mutational asymmetry in mammalian evolution. Nat Genet. 2003;33:514–517. doi: 10.1038/ng1103. [DOI] [PubMed] [Google Scholar]
- 31.Polak P., Arndt P.F. Transcription induces strand-specific mutations at the 5’ end of human genes. Genome Res. 2008;18:1216–1223. doi: 10.1101/gr.076570.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ginno P.A., Lott P.L., Christensen H.C., Korf I., Chédin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao S.-Q., Ghanbarian A.T., Spiegel J., Martínez Cuesta S., Beraldi D., Di Antonio M., et al. DNA G-quadruplex structures mold the DNA methylome. Nat Struct Mol Biol. 2018;25:951–957. doi: 10.1038/s41594-018-0131-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jara-Espejo M., Line S.R. DNA G-quadruplex stability, position and chromatin accessibility are associated with CpG island methylation. FEBS J. 2020;287 doi: 10.1111/febs.15065. 483–95. [DOI] [PubMed] [Google Scholar]
- 35.Georgakopoulos-Soares I., Victorino J., Parada G.E., Agarwal V., Zhao J., Wong H.Y., et al. High-throughput characterization of the role of non-B DNA motifs on promoter function. Cell Genom. 2022 doi: 10.1016/j.xgen.2022.100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merrikh H., Zhang Y., Grossman A.D., Wang J.D. Replication-transcription conflicts in bacteria. Nat Rev Microbiol. 2012;10:449–458. doi: 10.1038/nrmicro2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Million-Weaver S., Samadpour A.N., Moreno-Habel D.A., Nugent P., Brittnacher M.J., Weiss E., et al. An underlying mechanism for the increased mutagenesis of lagging-strand genes in Bacillus subtilis. Proc Natl Acad Sci USA. 2015;112:E1096–E1105. doi: 10.1073/pnas.1416651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brewer B.J. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53:679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- 39.French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992;258:1362–1365. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- 40.Bakthavachalam V., Baindur N., Madras B.K., Neumeyer J.L. Fluorescent probes for dopamine receptors: synthesis and characterization of fluorescein and 7-nitrobenz-2-oxa-1,3-diazol-4-yl conjugates of D-1 and D-2 receptor ligands. J Med Chem. 1991;34:3235–3241. doi: 10.1021/jm00115a012. [DOI] [PubMed] [Google Scholar]
- 41.Mao X., Zhang H., Yin Y., Xu Y. The percentage of bacterial genes on leading versus lagging strands is influenced by multiple balancing forces. Nucleic Acids Res. 2012;40:8210–8218. doi: 10.1093/nar/gks605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kunst F., Ogasawara N., Moszer I., Albertini A.M., Alloni G., Azevedo V., et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 43.Azvolinsky A., Giresi P.G., Lieb J.D., Zakian V.A. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell. 2009;34:722–734. doi: 10.1016/j.molcel.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srivatsan A., Tehranchi A., MacAlpine D.M., Wang J.D. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rocha E.P.C., Danchin A. Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat Genet. 2003;34:377–378. doi: 10.1038/ng1209. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi Y., Horiuchi T., Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rocha E.P.C., Danchin A. Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res. 2003;31:6570–6577. doi: 10.1093/nar/gkg859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paul S., Million-Weaver S., Chattopadhyay S., Sokurenko E., Merrikh H. Accelerated gene evolution through replication-transcription conflicts. Nature. 2013;495:512–515. doi: 10.1038/nature11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price M.N., Alm E.J., Arkin A.P. Interruptions in gene expression drive highly expressed operons to the leading strand of DNA replication. Nucleic Acids Res. 2005;33:3224–3234. doi: 10.1093/nar/gki638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu T., Luo H., Gao F. Position preference of essential genes in prokaryotic operons. PLoS One. 2021;16 doi: 10.1371/journal.pone.0250380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wansink D.G., Manders E.E., van der Kraan I., Aten J.A., van Driel R., de Jong L. RNA polymerase II transcription is concentrated outside replication domains throughout S-phase. J Cell Sci. 1994;107(Pt 6):1449–1456. doi: 10.1242/jcs.107.6.1449. [DOI] [PubMed] [Google Scholar]
- 52.Wei X., Samarabandu J., Devdhar R.S., Siegel A.J., Acharya R., Berezney R. Segregation of transcription and replication sites into higher order domains. Science. 1998;281:1502–1506. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- 53.López-estraño C., Schvartzman J.B., Krimer D.B., Hernández P. Co-localization of polar replication fork barriers and rRNA transcription terminators in mouse rDNA. J Mol Biol. 1998;277:249–256. doi: 10.1006/jmbi.1997.1607. [DOI] [PubMed] [Google Scholar]
- 54.Hiratani I., Takebayashi S.-I., Lu J., Gilbert D.M. Replication timing and transcriptional control: beyond cause and effect--part II. Curr Opin Genet Dev. 2009;19:142–149. doi: 10.1016/j.gde.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woodfine K., Fiegler H., Beare D.M., Collins J.E., McCann O.T., Young B.D., et al. Replication timing of the human genome. Hum Mol Genet. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- 56.Helmrich A., Ballarino M., Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 57.Vilette D., Ehrlich S.D., Michel B. Transcription-induced deletions in Escherichia coli plasmids. Mol Microbiol. 1995;17:493–504. doi: 10.1111/j.1365-2958.1995.mmi_17030493.x. [DOI] [PubMed] [Google Scholar]
- 58.Xia B., Yan Y., Baron M., Wagner F., Barkley D., Chiodin M., et al. Widespread transcriptional scanning in the testis modulates gene evolution rates. Cell. 2020;180:248–262. doi: 10.1016/j.cell.2019.12.015. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen W.-H., Lu G., Bork P., Hu S., Lercher M.J. Energy efficiency trade-offs drive nucleotide usage in transcribed regions. Nat Commun. 2016;7:11334. doi: 10.1038/ncomms11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gallaher S.D., Craig R.J., Ganesan I., Purvine S.O., McCorkle S.R., Grimwood J., et al. Widespread polycistronic gene expression in green algae. Proc Natl Acad Sci USA. 2021:118. doi: 10.1073/pnas.2017714118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.García-Ríos M., Fujita T., LaRosa P.C., Locy R.D., Clithero J.M., Bressan R.A., et al. Cloning of a polycistronic cDNA from tomato encoding gamma-glutamyl kinase and gamma-glutamyl phosphate reductase. Proc Natl Acad Sci USA. 1997;94:8249–8254. doi: 10.1073/pnas.94.15.8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gray T.A., Saitoh S., Nicholls R.D. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc Natl Acad Sci USA. 1999;96:5616–5621. doi: 10.1073/pnas.96.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zoubak S., Clay O., Bernardi G. The gene distribution of the human genome. Gene. 1996;174:95–102. doi: 10.1016/0378-1119(96)00393-9. [DOI] [PubMed] [Google Scholar]
- 64.Versteeg R., van Schaik B.D.C., van Batenburg M.F., Roos M., Monajemi R., Caron H., et al. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003;13:1998–2004. doi: 10.1101/gr.1649303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rhind N., Gilbert D.M. DNA replication timing. Cold Spring Harb Perspect Biol. 2013;5:a010132. doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rivera-Mulia J.C., Buckley Q., Sasaki T., Zimmerman J., Didier R.A., Nazor K., et al. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res. 2015;25:1091–1103. doi: 10.1101/gr.187989.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arnone J.T., Robbins-Pianka A., Arace J.R., Kass-Gergi S., McAlear M.A. The adjacent positioning of co-regulated gene pairs is widely conserved across eukaryotes. BMC Genom. 2012;13:546. doi: 10.1186/1471-2164-13-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adachi N., Lieber M.R. Bidirectional gene organization: a common architectural feature of the human genome. Cell. 2002;109:807–809. doi: 10.1016/s0092-8674(02)00758-4. [DOI] [PubMed] [Google Scholar]
- 69.Trinklein N.D., Aldred S.F., Hartman S.J., Schroeder D.I., Otillar R.P., Myers R.M. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jin Y., Eser U., Struhl K., Churchman L.S. The ground state and evolution of promoter region directionality. Cell. 2017;170:889–898. doi: 10.1016/j.cell.2017.07.006. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sigova A.A., Mullen A.C., Molinie B., Gupta S., Orlando D.A., Guenther M.G., et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci USA. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma L., Bajic V.B., Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu A.C.K., Van Werven F.J. Transcribe this way: Rap1 confers promoter directionality by repressing divergent transcription. Transcription. 2019;10:164–170. doi: 10.1080/21541264.2019.1608716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Butler J.E.F., Kadonaga J.T. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 75.Weingarten-Gabbay S., Nir R., Lubliner S., Sharon E., Kalma Y., Weinberger A., et al. Systematic interrogation of human promoters. Genome Res. 2019;29:171–183. doi: 10.1101/gr.236075.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tian B., Hu J., Zhang H., Lutz C.S. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y., Stumph W.E. RNA polymerase II/III transcription specificity determined by TATA box orientation. Proc Natl Acad Sci USA. 1995;92:8606–8610. doi: 10.1073/pnas.92.19.8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frith M.C., Valen E., Krogh A., Hayashizaki Y., Carninci P., Sandelin A. A code for transcription initiation in mammalian genomes. Genome Res. 2008;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang C., Li W.-H., Krainer A.R., Zhang M.Q. RNA landscape of evolution for optimal exon and intron discrimination. Proc Natl Acad Sci USA. 2008;105:5797–5802. doi: 10.1073/pnas.0801692105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Touchon M., Arneodo A., d’Aubenton-Carafa Y., Thermes C. Transcription-coupled and splicing-coupled strand asymmetries in eukaryotic genomes. Nucleic Acids Res. 2004;32:4969–4978. doi: 10.1093/nar/gkh823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Georgakopoulos-Soares I., Parada G.E., Wong H.Y., Miska E.A., Kwok C.K., Hemberg M. Alternative splicing modulation by G-quadruplexes n.d. https://doi.org/10.1101/700575. [DOI] [PMC free article] [PubMed]
- 82.Stratton M.R., Campbell P.J., Andrew Futreal P. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio SAJR, Behjati S., Biankin A.V., et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu H., Ness S., Li C.-I., Bai Y., Mao P., Guo Y. Surveying mutation density patterns around specific genomic features. Genome Res. 2022 doi: 10.1101/gr.276770.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tomkova M., Tomek J., Kriaucionis S., Schuster-Böckler B. Mutational signature distribution varies with DNA replication timing and strand asymmetry. Genome Biol. 2018;19:129. doi: 10.1186/s13059-018-1509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aitken S.J., Anderson C.J., Connor F., Pich O., Sundaram V., Feig C., et al. Pervasive lesion segregation shapes cancer genome evolution. Nature. 2020;583:265–270. doi: 10.1038/s41586-020-2435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pich O., Muiños F., Sabarinathan R., Reyes-Salazar I., Gonzalez-Perez A., Lopez-Bigas N. Somatic and germline mutation periodicity follow the orientation of the DNA minor groove around nucleosomes. Cell. 2018;175 doi: 10.1016/j.cell.2018.10.004. 1074–87.e18. [DOI] [PubMed] [Google Scholar]
- 88.Georgakopoulos-Soares I., Mouratidis I., Parada G.E., Matharu N., Hemberg M., Ahituv N. Asymmetron: a toolkit for the identification of strand asymmetry patterns in biological sequences. Nucleic Acids Res. 2021;49 doi: 10.1093/nar/gkaa1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alexandrov L.B., Kim J., Haradhvala N.J., Huang M.N., Tian Ng A.W., Wu Y., et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hanawalt P.C., Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 91.Mellon I., Spivak G., Hanawalt P.C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 92.Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jinks-Robertson S., Bhagwat A.S. Transcription-associated mutagenesis. Annu Rev Genet. 2014;48:341–359. doi: 10.1146/annurev-genet-120213-092015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Klapacz J., Bhagwat A.S. Transcription-dependent increase in multiple classes of base substitution mutations in Escherichia coli. J Bacteriol. 2002;184:6866–6872. doi: 10.1128/JB.184.24.6866-6872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reijns M.A.M., Parry D.A., Williams T.C., Nadeu F., Hindshaw R.L., Rios Szwed D.O., et al. Signatures of TOP1 transcription-associated mutagenesis in cancer and germline. Nature. 2022;602:623–631. doi: 10.1038/s41586-022-04403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Haradhvala N.J., Polak P., Stojanov P., Covington K.R., Shinbrot E., Hess J.M., et al. Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell. 2016;164:538–549. doi: 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kucab J.E., Zou X., Morganella S., Joel M., Nanda A.S., Nagy E., et al. A compendium of mutational signatures of environmental agents. Cell. 2019;177 doi: 10.1016/j.cell.2019.03.001. 821–36.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Letouzé E., Shinde J., Renault V., Couchy G., Blanc J.-F., Tubacher E., et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat Commun. 2017:8. doi: 10.1038/s41467-017-01358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Islam S.M.A., Ashiqul Islam S.M., Díaz-Gay M., Wu Y., Barnes M., Vangara R., et al. Uncovering novel mutational signatures by de novo extraction with SigProfilerExtractor. Cell Genom. 2022 doi: 10.1016/j.xgen.2022.100179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Georgakopoulos-Soares I., Koh G., Momen S.E., Jiricny J., Hemberg M., Nik-Zainal S. Transcription-coupled repair and mismatch repair contribute towards preserving genome integrity at mononucleotide repeat tracts. Nat Commun. 2020;11:1980. doi: 10.1038/s41467-020-15901-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heilbrun E.E., Merav M., Adar S. Exons and introns exhibit transcriptional strand asymmetry of dinucleotide distribution, damage formation and DNA repair. NAR Genom Bioinform. 2021;3:lqab020. doi: 10.1093/nargab/lqab020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vetsigian K., Goldenfeld N. Genome rhetoric and the emergence of compositional bias. Proc Natl Acad Sci USA. 2009;106:215–220. doi: 10.1073/pnas.0810122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frigola J., Sabarinathan R., Mularoni L., Muiños F., Gonzalez-Perez A., López-Bigas N. Reduced mutation rate in exons due to differential mismatch repair. Nat Genet. 2017;49:1684–1692. doi: 10.1038/ng.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vöhringer H., Van Hoeck A., Cuppen E., Gerstung M. Learning mutational signatures and their multidimensional genomic properties with TensorSignatures. Nat Commun. 2021;12:3628. doi: 10.1038/s41467-021-23551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Seplyarskiy V.B., Akkuratov E.E., Akkuratova N., Andrianova M.A., Nikolaev S.I., Bazykin G.A., et al. Error-prone bypass of DNA lesions during lagging-strand replication is a common source of germline and cancer mutations. Nat Genet. 2019;51:36–41. doi: 10.1038/s41588-018-0285-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Seplyarskiy V.B., Soldatov R.A., Koch E., McGinty R.J., Goldmann J.M., Hernandez R.D., et al. Population sequencing data reveal a compendium of mutational processes in the human germ line. Science. 2021;373:1030–1035. doi: 10.1126/science.aba7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pursell Z.F., Isoz I., Lundström E.-B., Johansson E., Kunkel T.A. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Morrison A., Araki H., Clark A.B., Hamatake R.K., Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990;62:1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- 109.McElhinny S.A.N., Nick McElhinny S.A., Gordenin D.A., Stith C.M., Burgers P.M.J., Kunkel T.A. Division of Labor at the Eukaryotic Replication Fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lujan S.A., Williams J.S., Pursell Z.F., Abdulovic-Cui A.A., Clark A.B., Nick McElhinny S.A., et al. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Robinson P.S., Coorens T.H.H., Palles C., Mitchell E., Abascal F., Olafsson S., et al. Increased somatic mutation burdens in normal human cells due to defective DNA polymerases. Nat Genet. 2021;53:1434–1442. doi: 10.1038/s41588-021-00930-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Supek F., Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. 2015;521:81–84. doi: 10.1038/nature14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lujan S.A., Clausen A.R., Clark A.B., MacAlpine H.K., MacAlpine D.M., Malc E.P., et al. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 2014;24:1751–1764. doi: 10.1101/gr.178335.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zou X., Koh G.C.C., Nanda A.S., Degasperi A., Urgo K., Roumeliotis T.I., et al. A systematic CRISPR screen defines mutational mechanisms underpinning signatures caused by replication errors and endogenous DNA damage. Nat Cancer. 2021;2:643–657. doi: 10.1038/s43018-021-00200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roberts S.A., Lawrence M.S., Klimczak L.J., Grimm S.A., Fargo D., Stojanov P., et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Petljak M., Alexandrov L.B., Brammeld J.S., Price S., Wedge D.C., Grossmann S., et al. Characterizing mutational signatures in human cancer cell lines reveals episodic APOBEC mutagenesis. Cell. 2019;176 doi: 10.1016/j.cell.2019.02.012. 1282–94.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kazanov M.D., Roberts S.A., Polak P., Stamatoyannopoulos J., Klimczak L.J., Gordenin D.A., et al. APOBEC-induced cancer mutations are uniquely enriched in early-replicating, gene-dense, and active chromatin regions. Cell Rep. 2015;13:1103–1109. doi: 10.1016/j.celrep.2015.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hoopes J.I., Cortez L.M., Mertz T.M., Malc E.P., Mieczkowski P.A., Roberts S.A. APOBEC3A and APOBEC3B preferentially deaminate the lagging strand template during DNA replication. Cell Rep. 2016;14:1273–1282. doi: 10.1016/j.celrep.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McCLINTOCK B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci USA. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.de Koning A.P.J., Gu W., Castoe T.A., Batzer M.A., Pollock D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cowperthwaite M., Park W., Xu Z., Yan X., Maurais S.C., Dooner H.K. Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell. 2002;14:713–726. doi: 10.1105/tpc.010468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Spradling A.C., Bellen H.J., Hoskins R.A. Drosophila P elements preferentially transpose to replication origins. Proc Natl Acad Sci USA. 2011;108:15948–15953. doi: 10.1073/pnas.1112960108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sultana T., van Essen D., Siol O., Bailly-Bechet M., Philippe C., Zine El Aabidine A., et al. The landscape of L1 retrotransposons in the human genome is shaped by pre-insertion sequence biases and post-insertion selection. Mol Cell. 2019;74 doi: 10.1016/j.molcel.2019.02.036. 555–70.e7. [DOI] [PubMed] [Google Scholar]
- 124.Flasch D.A., Macia Á., Sánchez L., Ljungman M., Heras S.R., García-Pérez J.L., et al. Genome-wide de novo L1 Retrotransposition Connects Endonuclease Activity with Replication. Cell. 2019;177 doi: 10.1016/j.cell.2019.02.050. 837–51.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Deininger P. Alu elements: know the SINEs. Genome Biol. 2011;12:236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lobachev K.S., Stenger J.E., Kozyreva O.G., Jurka J., Gordenin D.A., Resnick M.A. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lu J.Y., Chang L., Li T., Wang T., Yin Y., Zhan G., et al. Homotypic clustering of L1 and B1/Alu repeats compartmentalizes the 3D genome. Cell Res. 2021;31:613–630. doi: 10.1038/s41422-020-00466-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 129.Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rodriguez-Martin B., Alvarez E.G., Baez-Ortega A., Zamora J., Supek F., Demeulemeester J., et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat Genet. 2020;52:306–319. doi: 10.1038/s41588-019-0562-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Smit A.F. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 132.Hancks D.C., Kazazian H.H., Jr. Roles for retrotransposon insertions in human disease. Mob DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Han J.S., Szak S.T., Boeke J.D. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 134.Wheelan S.J., Aizawa Y., Han J.S., Boeke J.D. Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res. 2005;15:1073–1078. doi: 10.1101/gr.3688905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tsirigos A., Rigoutsos I. Alu and b1 repeats have been selectively retained in the upstream and intronic regions of genes of specific functional classes. PLoS Comput Biol. 2009;5 doi: 10.1371/journal.pcbi.1000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim E.Z., Wespiser A.R., Caffrey D.R. The domain structure and distribution of Alu elements in long noncoding RNAs and mRNAs. RNA. 2016;22:254–264. doi: 10.1261/rna.048280.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stenger J.E., Lobachev K.S., Gordenin D., Darden T.A., Jurka J., Resnick M.A. Biased distribution of inverted and direct Alus in the human genome: implications for insertion, exclusion, and genome stability. Genome Res. 2001;11:12–27. doi: 10.1101/gr.158801. [DOI] [PubMed] [Google Scholar]
- 139.Jurka J., Gentles A.J. Origin and diversification of minisatellites derived from human Alu sequences. Gene. 2006;365:21–26. doi: 10.1016/j.gene.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 140.Spitz F., Furlong E.E.M. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 141.Jindal G.A., Farley E.K. Enhancer grammar in development, evolution, and disease: dependencies and interplay. Dev Cell. 2021;56:575–587. doi: 10.1016/j.devcel.2021.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jolma A., Yin Y., Nitta K.R., Dave K., Popov A., Taipale M., et al. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015;527:384–388. doi: 10.1038/nature15518. [DOI] [PubMed] [Google Scholar]
- 143.Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 144.Bentsen M., Heger V., Schultheis H., Kuenne C., Looso M. TF-COMB - Discovering grammar of transcription factor binding sites. Comput Struct Biotechnol J. 2022;20:4040–4051. doi: 10.1016/j.csbj.2022.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McConkey G.A., Bogenhagen D.F. TFIIIA binds with equal affinity to somatic and major oocyte 5S RNA genes. Genes Dev. 1988;2:205–214. doi: 10.1101/gad.2.2.205. [DOI] [PubMed] [Google Scholar]
- 146.Kazemian M., Pham H., Wolfe S.A., Brodsky M.H., Sinha S. Widespread evidence of cooperative DNA binding by transcription factors in Drosophila development. Nucleic Acids Res. 2013;41:8237–8252. doi: 10.1093/nar/gkt598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lamber E.P., Vanhille L., Textor L.C., Kachalova G.S., Sieweke M.H., Wilmanns M. Regulation of the transcription factor Ets-1 by DNA-mediated homo-dimerization. EMBO J. 2008;27:2006–2017. doi: 10.1038/emboj.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jolma A., Kivioja T., Toivonen J., Cheng L., Wei G., Enge M., et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Leonard D.A., Kerppola T.K. DNA bending determines Fos-Jun heterodimer orientation. Nat Struct Biol. 1998;5:877–881. doi: 10.1038/2316. [DOI] [PubMed] [Google Scholar]
- 150.Lis M., Walther D. The orientation of transcription factor binding site motifs in gene promoter regions: does it matter? BMC Genom. 2016;17:185. doi: 10.1186/s12864-016-2549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nagawa F., Fink G.R. The relationship between the “TATA” sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1985;82:8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Emami K.H., Jain A., Smale S.T. Mechanism of synergy between TATA and initiator: synergistic binding of TFIID following a putative TFIIA-induced isomerization. Genes Dev. 1997;11:3007–3019. doi: 10.1101/gad.11.22.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Eddy J., Vallur A.C., Varma S., Liu H., Reinhold W.C., Pommier Y., et al. G4 motifs correlate with promoter-proximal transcriptional pausing in human genes. Nucleic Acids Res. 2011;39:4975–4983. doi: 10.1093/nar/gkr079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Panne D. The enhanceosome. Curr Opin Struct Biol. 2008;18:236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 155.Thanos D., Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 156.Panne D., Maniatis T., Harrison S.C. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Falvo J.V., Parekh B.S., Lin C.H., Fraenkel E., Maniatis T. Assembly of a functional beta interferon enhanceosome is dependent on ATF-2-c-jun heterodimer orientation. Mol Cell Biol. 2000;20:4814–4825. doi: 10.1128/mcb.20.13.4814-4825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Arnosti D.N., Kulkarni M.M. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- 159.Smith R.P., Taher L., Patwardhan R.P., Kim M.J., Inoue F., Shendure J., et al. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat Genet. 2013;45:1021–1028. doi: 10.1038/ng.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Patwardhan R.P., Hiatt J.B., Witten D.M., Kim M.J., Smith R.P., May D., et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Klein J.C., Agarwal V., Inoue F., Keith A., Martin B., Kircher M., et al. A systematic evaluation of the design and context dependencies of massively parallel reporter assays. Nat Methods. 2020;17:1083–1091. doi: 10.1038/s41592-020-0965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Natesan S., Gilman M.Z. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev. 1993;7:2497–2509. doi: 10.1101/gad.7.12b.2497. [DOI] [PubMed] [Google Scholar]
- 163.Schräder M., Nayeri S., Kahlen J.P., Müller K.M., Carlberg C. Natural vitamin D3 response elements formed by inverted palindromes: polarity-directed ligand sensitivity of vitamin D3 receptor-retinoid X receptor heterodimer-mediated transactivation. Mol Cell Biol. 1995;15:1154–1161. doi: 10.1128/mcb.15.3.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Whitington T., Frith M.C., Johnson J., Bailey T.L. Inferring transcription factor complexes from ChIP-seq data. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Leonard D.A., Rajaram N., Kerppola T.K. Structural basis of DNA bending and oriented heterodimer binding by the basic leucine zipper domains of Fos and Jun. Proc Natl Acad Sci USA. 1997;94:4913–4918. doi: 10.1073/pnas.94.10.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chytil M., Peterson B.R., Erlanson D.A., Verdine G.L. The orientation of the AP-1 heterodimer on DNA strongly affects transcriptional potency. Proc Natl Acad Sci USA. 1998;95:14076–14081. doi: 10.1073/pnas.95.24.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chen F.E., Huang D.B., Chen Y.Q., Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 168.Urban M.B., Schreck R., Baeuerle P.A. NF-kappa B contacts DNA by a heterodimer of the p50 and p65 subunit. EMBO J. 1991;10:1817–1825. doi: 10.1002/j.1460-2075.1991.tb07707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Morgunova E., Taipale J. Structural insights into the interaction between transcription factors and the nucleosome. Curr Opin Struct Biol. 2021;71:171–179. doi: 10.1016/j.sbi.2021.06.016. [DOI] [PubMed] [Google Scholar]
- 170.Grossman S.R., Engreitz J., Ray J.P., Nguyen T.H., Hacohen N., Lander E.S. Positional specificity of different transcription factor classes within enhancers. Proc Natl Acad Sci USA. 2018;115:E7222–E7230. doi: 10.1073/pnas.1804663115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhu F., Farnung L., Kaasinen E., Sahu B., Yin Y., Wei B., et al. The interaction landscape between transcription factors and the nucleosome. Nature. 2018;562:76–81. doi: 10.1038/s41586-018-0549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sherwood R.I., Hashimoto T., O’Donnell C.W., Lewis S., Barkal A.A., van Hoff J.P., et al. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–178. doi: 10.1038/nbt.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tanaka H., Takizawa Y., Takaku M., Kato D., Kumagawa Y., Grimm S.A., et al. Interaction of the pioneer transcription factor GATA3 with nucleosomes. Nat Commun. 2020;11:4136. doi: 10.1038/s41467-020-17959-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pugacheva E.M., Kubo N., Loukinov D., Tajmul M., Kang S., Kovalchuk A.L., et al. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc Natl Acad Sci USA. 2020;117:2020–2031. doi: 10.1073/pnas.1911708117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Katainen R., Dave K., Pitkänen E., Palin K., Kivioja T., Välimäki N., et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47:818–821. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- 176.Guo Y., Xu Q., Canzio D., Shou J., Li J., Gorkin D.U., et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rao S.S.P., Huang S.-C., Glenn St Hilaire B., Engreitz J.M., Perez E.M., Kieffer-Kwon K-R., et al. Cohesin loss eliminates all loop domains. Cell. 2017;171 doi: 10.1016/j.cell.2017.09.026. 305–20.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Grubert F., Srivas R., Spacek D.V., Kasowski M., Ruiz-Velasco M., Sinnott-Armstrong N., et al. Landscape of cohesin-mediated chromatin loops in the human genome. Nature. 2020;583:737–743. doi: 10.1038/s41586-020-2151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bauer B.W., Davidson I.F., Canena D., Wutz G., Tang W., Litos G., et al. Cohesin mediates DNA loop extrusion by a “swing and clamp” mechanism. Cell. 2021;184 doi: 10.1016/j.cell.2021.09.016. 5448–64.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Poulos R.C., Thoms J.A.I., Guan Y.F., Unnikrishnan A., Pimanda J.E., Wong J.W.H. Functional mutations form at CTCF-cohesin binding sites in melanoma due to uneven nucleotide excision repair across the motif. Cell Rep. 2016;17:2865–2872. doi: 10.1016/j.celrep.2016.11.055. [DOI] [PubMed] [Google Scholar]