Abstract
Tetronomycin (1), first isolated from a cultured broth of Streptomyces sp. by Juslen et al. in 1974, is a polycyclic polyether compound. However, the biological activity of 1 has not been thoroughly examined. In this study, we found that 1 exhibits more potent antibacterial activity than two well-known antibacterial drugs (vancomycin and linezolid) and is effective against several drug-resistant clinical isolates including methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococci. Furthermore, we reassigned the 13C NMR spectra of 1 and performed a preliminary structure–activity relationship study of 1 to synthesize a chemical probe for target identification, which implied different targets based on its ionophore activity.
Introduction
Antimicrobial resistance (AMR) is one of the major health threats to humankind, animals, and our shared environment. The number of AMR-related deaths in 2019 was estimated as 5 million annually.1 Vancomycin, which was first isolated in 1957 and approved by the Food and Drug Administration in 1958, is one of the oldest antibiotics in clinical use for ∼60 years and has long been considered a drug of the last resort, owing to its efficacy against Gram-positive bacteria as a broad-spectrum antibiotic.2 However, the increasing emergence of vancomycin-resistant bacteria such as vancomycin-intermediate Staphylococcus aureus (S. aureus) (VISA), which was initially discovered in methicillin-resistant S. aureus (MRSA), and vancomycin-resistant Enterococci (VRE) has been a global concern.3 The US Centers for Disease Control and Prevention projected that VRE caused 54,500 cases of infections and 5400 deaths in 2017.4 As alternatives to vancomycin, the development of new broad-spectrum antibiotics against Gram-positive bacteria is urgently required.
This study describes the identification of tetronomycin (1) as a broad-spectrum and potent antibiotic against Gram-positive bacteria including clinical isolates of VISA and VRE. Further, this study presents the reassignment of the NMR spectra of 1 and its preliminary structure–activity relationship (SAR).
Results and Discussion
Isolation of 1 and Reassignment of Its NMR Spectra
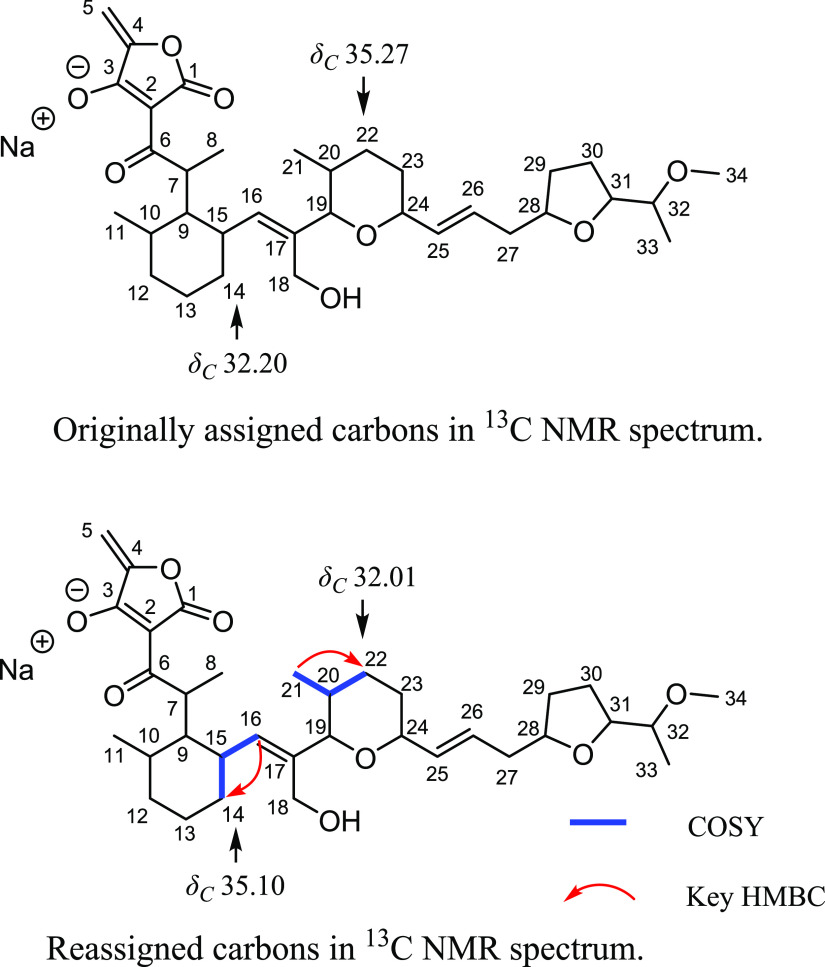
Compound 1 was isolated as a light-yellow oil from the culture broth of the Streptomyces sp. K20-0247 strain, which was isolated from the soil collected at Izu O̅shima, Tokyo, Japan (Figure 1). Originally, 1 was isolated by Juslen et al. in 1974, and its absolute structure was determined via the X-ray analysis of the mono-O-acetyltetronomycin silver salt.5 To assign the peaks in the 1H and 13C NMR spectra of 1, they derivatized 1 to its sodium salt 2, probably owing to the acidic nature of the tetronic acid moiety. Therefore, we also obtained 2; its 1H and 13C NMR spectra and optical rotation values are in good agreement with those described in the original report.5 However, as a result of our full reassignment of 2 by correlated spectroscopy (COSY), heteronuclear single quantum coherence (HSQC), and heteronuclear multiple bond correlation (HMBC) analyses (Figures S1–S7), we found that the chemical shifts of C-14 and C-22 in 13C NMR were found to be inconsistent with the original data; thus, we reassigned these shifts (Table 1 and Figure 2). The HSQC cross-peak from H2-14 (δH 1.34 and 1.01) to C-14 (δC 35.10), COSY cross-peaks of H-16 (δH 5.14)/H-15 (δH 2.49)/H2-14, and the HMBC cross-peak from H-16 to C-14 supported the reassignment of C-14 to δC 35.10. The HSQC cross-peaks from H2-22 (δH 1.78 and 1.17) to C-22 (δC 32.01), COSY cross-peaks of H3-21 (δH 0.52)/H-20 (δH 1.35)/H2-22, and the HMBC cross-peak from H-21 to C-22 supported the reassignment of C-22 to δC 32.01 (Table 1 and Figure 2). Therefore, based on the spectral data, the 13C NMR chemical shift values of C-4 and C-22 in 2 were reassigned.
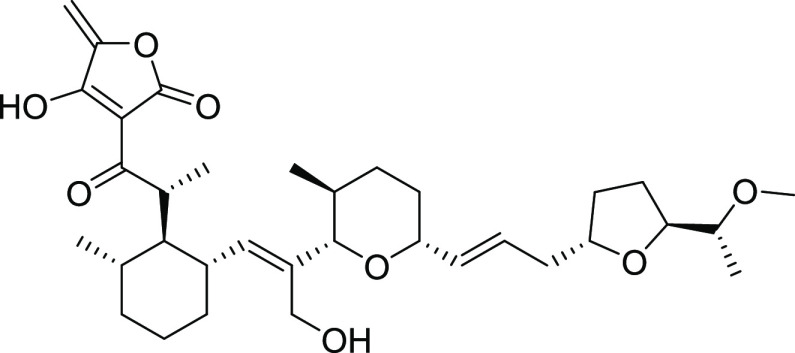
Figure 1.
Structure of tetronomycin (1).
Table 1. NMR Spectroscopy Data for Tetronomycin Sodium Salt (2)a.
| position | original δC, type | reassigned δC, type | original δH | reassigned δH (J in Hz) |
|---|---|---|---|---|
| 1 | 172.59, C | 172.32, C | ||
| 2 | 96.82, C | 96.74, C | ||
| 3 | 180.45, C | 180.40, C | ||
| 4 | 154.22, C | 153.87, C | ||
| 5 | 88.21, CH2 | 88.48, CH2 | 5.22 | 5.20, d (1.6) |
| 4.74 | 4.73, d (1.6) | |||
| 6 | 201.95, C | 202.00, C | ||
| 7 | 43.31, CH | 43.26, CH | 3.93 | 3.92, dq (4.0, 6.8) |
| 8 | 8.76, CH3 | 8.72, CH3 | 0.99 | 0.98c, d (6.8) |
| 9 | 47.32, CH | 47.08, CH | 1.89 | 1.87g, ddd (4.0, 10.8, 10.8) |
| 10 | 32.64b, CH | 33.92, CH | 1.32–1.42 | 1.37e, m |
| 11 | 19.84, CH3 | 19.76, CH3 | 1.21 | 1.20d, d (6.8) |
| 12 | 35.40a, CH2 | 35.22, CH2 | 1.52–1.67 | 1.61f, m |
| 1.1–1.3 | 1.14d, m | |||
| 13 | 25.76, CH2 | 25.62, CH2 | 1.52–1.67 | 1.57f, m |
| 1.1–1.3 | 1.22d, m | |||
| 14 | 32.20a, CH2 | 35.10, CH2 | 1.32–1.42 | 1.34e, m |
| 1 | 1.01c, m | |||
| 15 | 36.18, CH | 36.05, CH | 2.49 | 2.48, dddd (4.4, 10.2, 10.4, 10.4) |
| 16 | 141.94, CH | 141.68, CH | 5.15 | 5.14, d (10.2) |
| 17 | 130.72, C | 130.51, C | ||
| 18 | 56.29, CH2 | 56.15, CH2 | 4.29 | 4.27, d (11.0) |
| 3.78 | 3.79h, d (11.0) | |||
| 19 | 90.86, CH | 90.62, CH | 3.26 | 3.24, d (10.0) |
| 20 | 34.07b, CH | 32.47, CH | 1.32–1.42 | 1.39e, m |
| 21 | 18.16, CH3 | 18.15, CH3 | 0.52 | 0.52, d (6.8) |
| 22 | 35.27a, CH2 | 32.01, CH2 | 1.79 | 1.78, dddd (3.6, 3.6, 3.6, 13.4) |
| 1.1–1.3 | 1.17d, m | |||
| 23 | 31.58a, CH2 | 31.46, CH2 | 1.52–1.67 | 1.58f, m |
| 1.45 | 1.43e, dddd (3.7, 12.1, 12.1, 15.4) | |||
| 24 | 79.45, CH | 79.36, CH | 3.82 | 3.80h, ddd (1.8, 8.8, 8.8) |
| 25 | 132.62, CH | 132.92, CH | 5.48 | 5.47, ddd (1.6, 8.8, 15.2) |
| 26 | 135.23, CH | 135.10, CH | 6.16 | 6.13, ddd (3.8, 11.2, 15.2) |
| 27 | 39.87, CH2 | 39.77, CH2 | 2.33 | 2.34, dddd (1.6, 1.8, 3.8, 13.2) |
| 1.96 | 1.97g, ddd (10.8, 11.2, 13.2) | |||
| 28 | 78.05, CH | 77.97, CH | 4.1–4.2 | 4.13i, dddd (1.8, 6.2, 8.0, 10.8) |
| 29 | 31.93a, CH2 | 31.90, CH2 | 2.11 | 2.09, m |
| 1.52–1.67 | 1.56f, m | |||
| 30 | 27.73, CH2 | 27.74, CH2 | 1.85–1.95 | 1.90g, m |
| 1.52–1.67 | 1.55f, m | |||
| 31 | 80.17, CH | 80.08, CH | 4.1–4.2 | 4.16i, ddd (2.4, 6.0, 8.4) |
| 32 | 78.46, CH | 78.32, CH | 3.33 | 3.31, dq (2.4, 6.4) |
| 33 | 10.43, CH3 | 10.19, CH3 | 0.92 | 0.92, d (6.4) |
| 34 | 57.42, CH3 | 57.35, CH3 | 3.4 | 3.37, s |
| 18 | 4.36 | 4.41, br, s |
Original data were collected at 360 MHz for 1H NMR spectra and 90.5 MHz for 13C NMR spectra. Reassigned data were collected at 400 MHz for 1H NMR spectra and 100 MHz for 13C NMR spectra. a,bExchangable. c,d,e,f,g,h,iOverlapped.
Figure 2.
Reassignment of 13C NMR chemical shift values of 2.
Preliminary SAR Study of 1
As a first approach to identify the target molecules of 1 with broad antibacterial activity against Gram-positive bacteria, a preliminary SAR study was investigated (Scheme 1). Because the broad antibacterial activity spectrum of 1 is similar to that of related polyether tetronic compounds6,7 as well as based on the carrier-mediated transport studies of 1,8,9 it is considered as an ionophore compound. Furthermore, some studies have confirmed that polyether tetronic acid antibiotics accelerate the transport of ions across a model membrane.10,11 However, owing to insufficient evidence from these studies, a causal relationship between the ionophore and antibacterial activity could not be established. In addition, tetrodecamycin, one of the tetronate natural products, possesses an exo-methylene group on the tetronic acid moiety identical to 1, and the exo-methylene group is crucial for antibacterial activity.12,13 Therefore, we decided to chemically modify the exo-methylene group to determine its importance for antibiotic activity. Considering the electrophilicity of the exo-methylene group, thiophenol was used as a soft nucleophile, and a diastereomixture of the thioacetal derivative 3 was produced. However, as measured by the paper-disc method, the biological activity of 3 against S. aureus was slightly less than that of 1. We believed that the retro Michael reaction of 3 was possible to produce 1; therefore, we attempted the selective reduction of the exo-methylene group to prevent the retro Michael reaction. The use of H2 and 10% Pd/C afforded a diastereomixture of the target product 4. As anticipated, 4 lost its antibiological activity, indicating that the exo-methylene group on the tetronic acid moiety is crucial for antibacterial activity and that 1 may covalently bind to its target binding sites at the exo-methylene group by nucleophilic amino acid residues. This finding suggests that 1 has other targets of antibacterial activity in addition to ionophore activity. Subsequently, we derivatized the C-18 OH group using acid anhydrides to elucidate the role of the hydroxyl group in the antibiotic activity and provide insight into the chemical probe synthesis. The C-18 OH group barely tolerated the introduction of acyl groups, such as acetyl, propynoic, and pivaloyl, with reduced antibacterial activity slightly [the minimal inhibitory concentration (MIC) values of acyl derivatives (compounds 5–7) are shown in Table 2]. This result allowed us to use compound 6 as a chemical probe for target identification using the click-chemistry strategy.14
Scheme 1. Derivatization of 1.
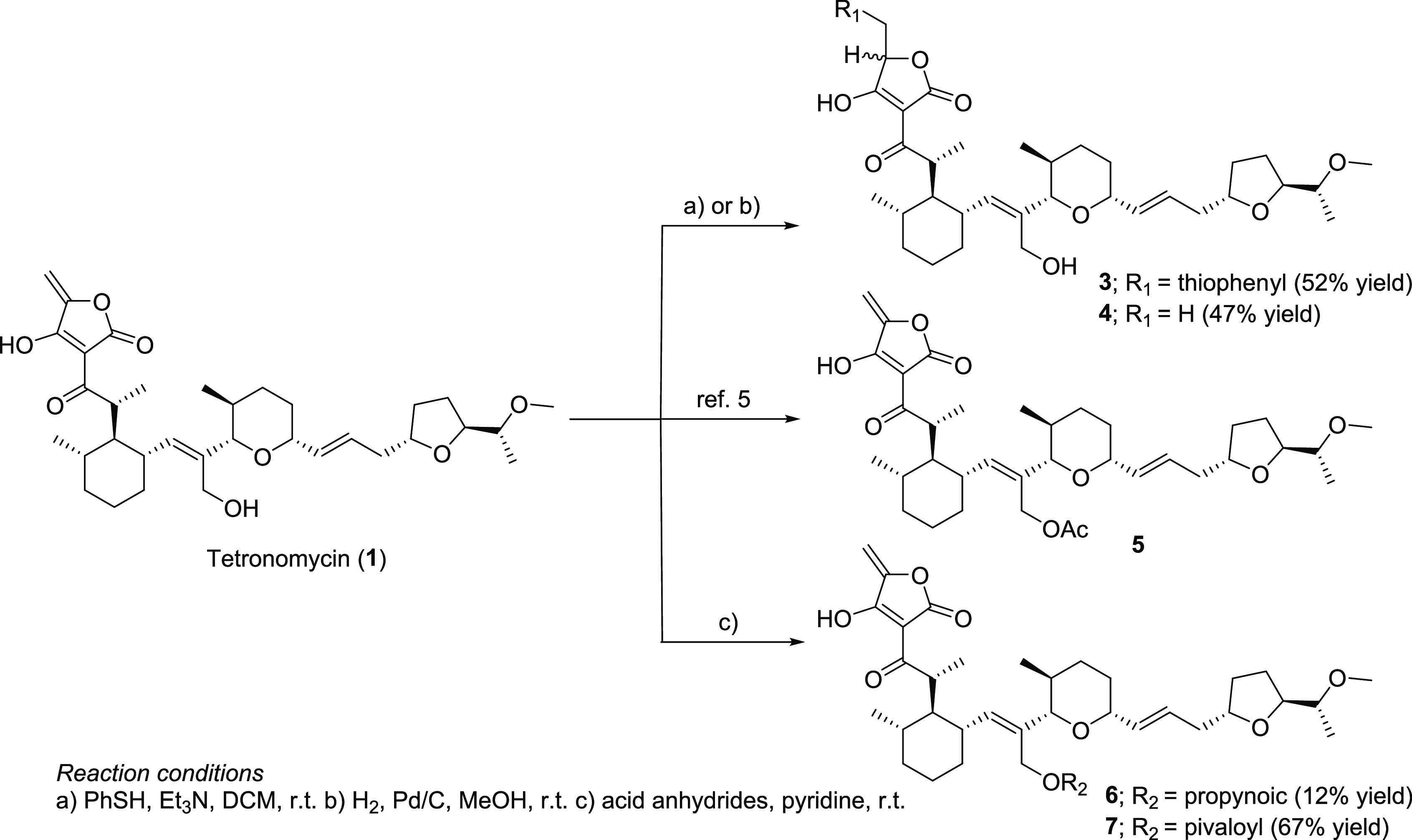
Table 2. Antibacterial Activity of 1 and Its Derivatives as well as the Known Drugsa.
| |
MIC (μg/mL) |
||||||
|---|---|---|---|---|---|---|---|
| strains | 1 | 5 | 6 | 7 | VCM | LZD | |
| Staphylococcus aureus | ATCC6538P | 0.125 | 0.5 | 1 | 2 | 1 | 1 |
| Staphylococcus aureus | ATCC 19636 | 0.063 | 0.25 | 1 | 2 | 1 | 1 |
| Staphylococcus aureus | ISP217 | 0.063 | 0.125 | 0.5 | 0.5 | 1 | 1 |
| Staphylococcus aureus | KUB854 | 0.125 | 0.25 | 1 | 2 | 1 | 2 |
| Staphylococcus aureus | KUB855 | 0.125 | 0.25 | 0.5 | 1 | 0.5 | 1 |
| Staphylococcus aureus | KUB856 | 0.125 | 0.5 | 1 | 2 | 1 | 2 |
| Staphylococcus aureus | ATCC700699 | 0.063 | 0.125 | 0.5 | 1 | 8 | 1 |
| Staphylococcus aureus | KUB877 | 0.125 | 0.25 | 1 | 2 | 1 | 16 |
| Staphylococcus epidermidis | KUB795 | 0.25 | 0.5 | 2 | 2 | 1 | 1 |
| Kocuria rhizophila | ATCC9341 | 0.063 | 0.125 | 0.5 | 0.125 | 0.5 | 1 |
| Enterococcus faecalis | ATCC29212 | 0.063 | 0.063 | 0.25 | 0.25 | 4 | 1 |
| Enterococcus faecalis | NCTC12201 | 0.063 | 0.125 | 0.25 | 0.25 | >64 | 1 |
| Enterococcus faecalis | KUB7012 | 0.016 | 0.063 | 0.25 | 0.016 | >64 | 1 |
| Enterococcus faecium | NCTC12204 | 0.063 | 0.125 | 0.5 | 0.25 | >64 | 1 |
| Enterococcus faecium | KUB7013 | 0.031 | 0.125 | 0.5 | 0.125 | >64 | 2 |
| Enterococcus gallinarum | KUB7014 | 0.063 | 0.031 | 0.25 | 0.25 | 8 | 1 |
| Escherichia coli | NIHJ JC-2 | >8 | >8 | >8 | >8 | >64 | >64 |
| Citrobacter freundii | ATCC8090 | >8 | >8 | >8 | >8 | >64 | >64 |
| Klebsiella pneumoniae | NCTC9632 | >8 | >8 | >8 | >8 | >64 | >64 |
| Klebsiella aerogenes | NCTC10006 | >8 | >8 | >8 | >8 | >64 | >64 |
| Proteus vulgaris | ATCC8427 | >8 | >8 | >8 | >8 | >64 | 16 |
| Morganella morganii | IID 602 | >8 | >8 | >8 | >8 | >64 | >64 |
| Serratia marcescens | IFO12648 | >8 | >8 | >8 | >8 | >64 | >64 |
| Enterobacter cloacae | IFO13535 | >8 | >8 | >8 | >8 | >64 | >64 |
| Pseudomonas aeruginosa | 46001 | >8 | >8 | >8 | >8 | >64 | >64 |
| Pseudomonas aeruginosa | E-2 | >8 | >8 | >8 | >8 | >64 | >64 |
| Acinetobacter calcoaceticus | IFO12552 | >8 | >8 | >8 | >8 | >64 | >64 |
Abbreviations: VCM, vancomycin; LZD, linezolid.
Evaluation of the Antibacterial Activity of 1 and Its Derivatives
According to the original study,5 the antibacterial activity evaluation of 1 against drug-resistant clinical isolates has not been performed yet. Therefore, using the paper-disc method and agar dilution method, we evaluated the antibacterial activity of 1 and its derivatives that maintained antibacterial activity against Gram-positive and Gram-negative bacteria (Table 2 and Table S2). The disc method is widely used in natural product chemistry to identify active compounds. Further, we compared the antibacterial potency of 1 to those of two well-known drugs (vancomycin and linezolid) (Table 2). Compound 1 showed a selective and more potent antibacterial activity against Gram-positive bacteria than the two known drugs. Compared with the MIC values of the known drugs toward S. aureus, Staphylococcus epidermidis, and Kocuria rhizophila, 1 exhibited antibacterial activity at a lower MIC. Furthermore, 1 was effective against S. aureus strains resistant to linezolid and macrolide. Notably, 1 exhibits remarkable antibacterial activity against VISA and other types of VRE. Thus, 1 is expected to be a new broad-spectrum antibiotic against Gram-positive bacteria and an effective alternative to vancomycin.
The antibacterial activity of acyl derivatives (compounds 5–7) slightly decreased but remained at a high level compared with the known drugs (Table 2). Strain information is provided in Table S3.
Conclusions
We reported the reassignment of the 13C NMR chemical shifts and showed that 1 is a broad-spectrum, potent antibiotic against Gram-positive bacteria and exhibits antibacterial activity against drug-resistant clinical isolates including VISA and VRE. As a preliminary attempt to identify the target molecules of 1 with broad antibacterial activity against Gram-positive bacteria, a preliminary SAR study was conducted. The exo-methylene group on the tetronic acid moiety is crucial for antibacterial activity, suggesting that in addition to its ionophore activity, 1 may covalently bind to its target binding sites. Overall, we expect that 1 will become an effective antibiotic against Gram-positive bacteria. While the MIC values of tetronomycin-nonproducing strains such as Streptomyces lividans TK24 and Staphylococcus albus J1074 were below 3 μg/mL, Streptomyces sp. K20-0247 was resistant to 1 (MIC >10 μg/mL). Thus, we speculated that Streptomyces sp. K20-0247 possesses resistance genes against 1 in the biosynthetic gene cluster of 1. The genome sequence of Streptomyces sp. K20-0247 was analyzed, and the biosynthetic gene cluster of 1 was discovered (Figure S18 and Table S1). This cluster was extremely similar to the previously reported tetronomycin biosynthetic gene cluster (Figure S18 and Table S1). In these clusters, resistance gene candidates such as genes encoding the DedA family and phosphatase were identified (Table S1). Thus, we hypothesized that these genes were responsible for Streptomyces sp. K20-0247 showing resistance against 1. Our next objective is to analyze the function of candidate resistance genes and the mode of action and the resistance mechanism using the propynoic derivative of 1.
Experimental Section
General Experimental Procedures
High- and low-resolution mass spectra were obtained using an AB Sciex QSTAR Hybrid LC/MS/MS system (AB Sciex, Framingham, MA, USA) and a JEOL JMS-T100LP (JEOL, Tokyo, Japan). NMR spectra were measured using a Varian XL-400 spectrometer (Agilent Technologies, CA, USA), with 1H NMR and 13C NMR at 400 and 100 MHz, respectively, and a JEOL JNM-ECA-500 (JEOL, Tokyo, Japan), with 1H NMR obtained at 500 Hz in CDCl3. The chemical shifts are reported in ppm and referenced to CDCl3 (7.26 ppm) in the 1H NMR spectra and CDCl3 (77.16 ppm) in the 13C NMR spectra. Optical rotation was measured using a JASCO P-2200 polarimeter (JASCO Corporation, Tokyo, Japan).
Isolation of Strain K20-0247
Strain K20-0247 was isolated from the soil sample collected from Izu O̅shima, Tokyo, Japan via incubation at 27 °C for two weeks on water-proline agar (proline 1%, agar 1.5%, and tap water) with 25 μg/mL benlate (DuPont de Nemours, Inc., Delaware, USA), 25 μg/mL nalidixic acid (TCI, Tokyo, Japan), and 1 μg/mL vancomycin (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan). After incubation, colonies were isolated and maintained on an agar medium (starch 1.0%, N-Z-amine 0.3%, yeast extract 0.1%, meat extract 0.1%, CaCO3 0.3%, agar 1.2%, and distilled water, with pH 7.0).
Genome Analysis
The genomic DNA of strain K20-0247 was obtained using the cetyltrimethylammonium bromide method.15 The genomes were sequenced using the Pacbio RSII technology (Macrogen, Inc., Seoul, Republic of Korea). The genome of Streptomyces sp. K20-0247 comprised a linear 8,931,955 bp genome and a circular 102,312 bp plasmid. The obtained genomic sequence was annotated using DFAST and analyzed via antiSMASH.16,17
The comparison of tetronomycin biosynthetic gene clusters generated from Streptomyces sp. K20-0247 and Streptomyces sp. NRRL 11266 was visualized using Clinker.18 BLAST search of the 16S rRNA gene sequence was performed using the EzBioCloud server (http://eztaxon-e.ezbiocloud.net) to determine the taxonomic position of the 16S rRNA gene sequence.
Fermentation
Strain K20-0247 was maintained on agar medium (glucose 1.0%, peptone 0.5%, 35% Ehrlich bonito extract 0.5%, NaCl 0.3%, and agar 1.0%). For seed culture, a loop of strain K20-0247 spores was inoculated into each 100 mL of seed medium (starch 2.4%, glucose 0.1%, peptone 0.3%, 35% Ehrlich bonito extract 0.3%, yeast extract 0.5%, and CaCO3 0.4%) (adjusted to pH 7.0 before sterilization) in a 500 mL Erlenmeyer flask (total = 2 flasks). The flask was incubated on a rotary shaker (210 rpm) at 27 °C for four days. A 1 mL portion of the seed culture was inoculated into each 100 mL of production medium (starch 2.0%, glycerol 0.5%, defatted wheat germ 1.0%, 35% Ehrlich bonito extract 0.3%, dry yeast 0.3%, and CaCO3 0.3%) in 500 mL Erlenmeyer flasks (total = 120 flasks). The flasks were incubated on a rotary shaker (210 rpm) at 27 °C for six days.
Isolation of Tetronomycin (1)
MeOH (12 L) was added to the culture, and the mixture was centrifuged at 3000 rpm for 10 min. The supernatant was collected and evaporated under reduced pressure to remove MeOH. The residual aqueous solution (12 L) was loaded onto an ODS column and eluted stepwise using a mixture of MeCN:H2O (0:100, 30:70, 60:40, and 100:0) and MeOH (with 0.1% TFA). The MeOH (0.1% TFA) fraction was concentrated in vacuo to remove MeOH. The residue was dissolved in 800 mL of EtOAc and rinsed three times using an equal volume of water. The organic layer was concentrated to dryness to afford a crude extract (477.3 mg). Finally, the crude extract was purified via preparative TLC (plate size = 200 × 200 mm and thickness = 1 mm) using two plates eluted with n-hexane:EtOAc (1:1, Rf = 0.35) to produce 129.1 mg of 1 as a light-yellow oil. 1 was isolated for its antimicrobial activity against S. aureus ATCC43300.
Preparation Procedure of the Tetronomycin Sodium Salt 2
To a solution of 1 (10.0 mg, 17.1 μmol) in dichloromethane (2.0 mL, 8.5 mM), 1.0 M NaHCO3 aq. (200.0 μL, 200.0 μmol) was added at room temperature. After stirring for 3 h, the reaction mixture was quenched using sat. NH4Cl aq., and the organic layer was separated. The organic layer was washed with brine, dried over Na2SO4, and concentrated under vacuum to afford 2 (7.0 mg, 11.4 μmol, 66.8%). [α]22D +80.5 (c 0.28, MeOH); 1H and 13C NMR are presented in Table 1; ESI-MS m/z 609.3401 [M + Na]+ (calcd. for C34H50O8Na, m/z 609.3403 [M + Na]+).
Preparation Procedure of the Tetronomycin Thioacetal Derivative 3
To a solution of 1 (10.0 mg, 17.1 μmol) in dichloromethane (0.3 mL, 56.8 mM), benzenethiol (8.0 μL, 78.4 μmol) and triethylamine (4.0 μL, 28.7 μmol) were added and stirred at room temperature for 5 h. The reaction mixture was purified via preparative TLC to afford 3 as a diastereomixture (6.2 mg, 8.9 μmol, 52.2%). 1H NMR (CDCl3, 500 MHz): δ 7.54–7.51 (2H, overlapped), 7.44–7.37 (2H, overlapped), 7.28–7.20 (4H, overlapped), 7.18–7.09 (2H, overlapped), 6.13 (2H, overlapped), 5.55–5.34 (2H, overlapped), 5.32 (1H, d, J = 10.3 Hz), 5.11 (1H, d, J = 10.3 Hz), 4.61 (1H, t, J = 3.7 Hz), 4.53 (1H, dd, J = 8.2, 2.9 Hz), 4.26 (1H, d, J = 11.1 Hz), 4.21 (1H, d, J = 11.1 Hz), 4.18–3.92 (4H, overlapped), 3.92–3.70 (6H, overlapped), 3.60–3.56 (4H, overlapped), 3.37–3.28 (6H, overlapped), 2.53–2.27 (4H, overlapped), 2.12–2.04 (2H, overlapped), 2.04–1.70 (8H, overlapped), 1.70–1.40 (8H, overlapped), 1.40–1.28 (6H, overlapped), 1.28–1.00 (4H, overlapped), 1.16 (6H, d, J = 6.4 Hz), 0.99–0.90 (12H, overlapped), 0.52 (3H, d, J = 6.5 Hz), 0.42 (3H, d, J = 6.5 Hz); 13C NMR (CDCl3, 125 MHz): δ 201.97, 201.57, 193.52, 193.23, 175.99, 175.80, 142.36, 141.60, 137.53, 136.36, 135.06, 134.98, 132.76, 132.67, 130.81, 130.00, 128.99, 128.93, 128.86, 126.15, 125.64, 90.71, 90.21, 82.11, 80.31, 80.23, 79.46, 79.13, 78.97, 78.60, 78.51, 78.47, 78.31, 78.21, 78.13, 57.48, 57.17, 56.33, 56.25, 47.62, 46.65, 43.34, 43.25, 39.92, 39.90, 38.74, 37.02, 36.36, 36.33, 36.10, 35.52, 35.35, 35.33, 35.00, 34.10, 33.96, 32.64, 32.52, 32.22, 32.08, 32.05, 31.99, 31.61, 31.57, 29.83, 27.84, 25.79, 25.74, 22.82, 19.98, 19.81, 18.32, 18.29, 15.72, 15.66, 14.25, 10.50, 10.48, 8.86, 8.80; ESI-MS m/z 714.4043 [M + NH4]+ (calcd. for C40H60NO8S, m/z 714.4040 [M + NH4]+).
Preparation Procedure of Reduced Tetronomycin 4
To a solution of 1 (11.7 mg, 20.0 μmol) in MeOH (0.5 mL, 40.0 mM), 10% Pd/C (1.5 mg, 1.4 mmol) was added and stirred at room temperature under a H2 atmosphere for 15 min. The reaction mixture was filtered and purified via preparative TLC to afford 4 as a diastereomixture (5.5 mg, 9.4 μmol, 46.9%). 1H NMR (CDCl3, 500 MHz): δ 6.15 (2H, overlapped), 5.48 (2H, overlapped), 5.14 (1H, d, J = 10.6 Hz), 5.08 (1H, d, J = 10.6 Hz), 4.58 (1H, br), 4.51 (1H, br), 4.44 (1H, q, J = 6.9 Hz), 4.34 (1H, q, J = 6.9 Hz), 4.27 (2H, d, J = 11.0 Hz), 4.20–4.09 (4H, overlapped), 3.91 (2H, dq, J = 7.0, 3.5 Hz), 3.89–3.76 (4H, overlapped), 3.36 (6H, s), 3.34–3.29 (2H, overlapped), 3.28 (1H, d, J = 9.9 Hz), 3.25 (1H, d, J = 9.9 Hz), 2.50 (2H, dddd, J = 10.8, 10.8, 10.6, 4.6 Hz), 2.32 (2H, m), 2.12–2.04 (2H, m), 2.01–1.72 (8H, overlapped), 1.67–1.50 (10H, overlapped), 1.48 (3H, d, J = 6.9 Hz), 1.43 (3H, d, J = 6.9 Hz), 1.50–1.27 (6H, overlapped), 1.27–1.06 (12H, overlapped), 1.05–0.95 (8H, overlapped), 0.95–0.91 (6H, overlapped), 0.57–0.49 (6H, overlapped); 13C NMR (CDCl3, 125 MHz): δ 201.96, 196.47, 196.21, 176.14, 141.66, 135.06, 134,89, 132.79, 132.68, 130.73, 95.61, 95.55, 90.87, 90.68, 80.23, 79.38, 78.37, 78.06, 57.44, 56.30, 47.52, 43.08, 41.05, 39.87, 36.27, 35.49, 35.47, 35.33, 34.08, 32.61, 32.17, 31.97, 31.94, 31.58, 31.56, 27.81, 27.74, 25.79, 19.94, 19.83, 18.31, 17.90, 17.78, 10.40, 8.89, 8.79; ESI-MS m/z 606.4006 [M + NH4]+ (calcd. for C34H56NO8, m/z 606.4006 [M + NH4]+).
Preparation Procedure of Acetyl Tetronomycin 5
According to ref (5), the acetyl tetronomycin Ag salt (15.8 mg, 26.0 μmol) was synthesized and purified via preparative TLC to afford 5 (6.2 mg, 9.9 μmol, 38.0%). [α]25D +94.0 (c 0.59, MeOH); 1H NMR (CDCl3, 500 MHz): δ 6.12 (1H, ddd, J = 15.1, 10.9, 4.1 Hz), 5.41 (1H, ddd, J = 15.1, 9.1, 1.4 Hz), 5.30 (1H, d, J = 10.2 Hz), 5.19 (1H, d, J = 1.7 Hz), 4.71 (1H, d, J = 1.7 Hz), 4.68 (1H, d, J = 12.1 Hz), 4.56 (1H, d, J = 12.1 Hz), 4.22–4.13 (1H, m), 4.13 (1H, ddd, J = 14.3, 7.3, 7.2 Hz), 4.10 (1H, ddd, J = 9.3, 6.5, 2.8 Hz), 3.86 (1H, ddd, J = 11.4, 9.1, 2.1 Hz), 3.78 (1H, dq, J = 6.7, 3.5 Hz), 3.44 (1H, s), 3.42 (1H, dq, J = 6.7, 2.8 Hz), 3.36 (1H, q, 3.78 J = 1.8 Hz), 3.35 (1H, s), 3.25 (1H, d, J = 10.1 Hz), 2.31 (2H, dddd, J = 10.9, 10.7, 10.7, 4.1 Hz), 2.39–2.25 (1H, m), 2.21–1.70 (8H, overlapped), 1.65–1.52 (5H, overlapped), 1.49–1.28 (3H, overlapped), 1.20 (3H, d, J = 6.4 Hz), 1.28–1.00 (4H, overlapped), 0.98 (3H, d, J = 6.7 Hz), 0.92 (3H, d, J = 6.7 Hz), 0.62 (3H, d, J = 6.6 Hz); 13C NMR (CDCl3, 125 MHz): δ 200.94, 180.04, 173.52, 171.56, 154.27, 144.50, 134.61, 133.02, 127.15, 96.72, 90.28, 88.38, 79.75, 79.14, 77.93, 59.62, 57.40, 57.20, 47.35, 43.92, 39.49, 37.16, 35.56, 35.00, 34.19, 32.84, 32.17, 31.67, 31.48, 29.85, 27.24, 25.93, 21.30, 19.85, 17.99, 10.41, 8.88; ESI-MS m/z 646.3956 [M + NH4]+ (calcd. for C36H56NO9, m/z 646.3955 [M + NH4]+, molecular formula).
Preparation Procedure of Propynoic Tetronomycin 6
To a solution of 1 (5.9 mg, 10.0 μmol) in propiolic acid (100.0 μL, 0.1 M), triethylamine (150.0 μL, 1.1 mmol) and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDCI) (19.0 mg, 99.1 μmol) were added at 0 °C. After stirring for 50 min, the reaction mixture was quenched using 1.0 M HCl aq. The organic layer was separated and washed with 1.0 M HCl aq., dried over Na2SO4, and concentrated under vacuum. The residue was purified via preparative TLC to afford 6 (0.8 mg, 1.2 μmol, 11.7%). [α]24D +95.9 (c 0.10, MeOH); 1H NMR (CDCl3, 500 MHz): δ 6.11 (1H, ddd, J = 15.3, 10.8, 4.3 Hz), 5.44 (1H, ddd, J = 15.3, 9.1, 1.7 Hz), 5.33 (1H, d, J = 10.3 Hz), 5.20 (1H, d, J = 1.9 Hz), 4.81 (1H, d, J = 12.1 Hz), 4.71 (1H, d, J = 1.9 Hz), 4.70 (1H, d, J = 12.1 Hz), 4.17 (1H, m), 4.11 (1H, m), 3.86 (1H, m), 3.78 (1H, m), 3.42 (3H, s), 3.27 (1H, d, J = 10.2 Hz), 2.87 (1H, s), 2.39–2.25 (2H, overlapped), 2.14–2.08 (1H, m), 2.06–1.72 (4H, overlapped), 1.68–1.54 (5H, overlapped), 1.20 (3H, d, J = 6.5 Hz), 1.48–0.98 (8H, overlapped), 0.97 (3H, d, J = 6.3 Hz), 0.92 (3H, d, J = 7.1 Hz), 0.64 (3H, d, J = 6.6 Hz); 13C NMR (CDCl3, 125 MHz): δ 201.00, 180.25, 173.48, 154.20, 153.32, 145.03, 134.59, 132.91, 126.28, 96.68, 90.00, 88.47, 79.75, 79.34, 77.93, 61.28, 57.43, 47.25, 43.91, 39.46, 37.26, 35.53, 34.80, 34.15, 32.86, 32.15, 32.06, 31.61, 31.50, 29.84, 27.26, 25.87, 22.83, 19.81, 17.97, 10.42, 8.86; ESI-MS m/z 656.3796 [M + NH4]+ (calcd. for C37H54NO9, m/z 656.3799 [M + NH4]+).
Preparation Procedure of Pivaloyl Tetronomycin 7
To a solution of 2 (4.1 mg, 6.7 μmol) in pivalic anhydride (0.1 mL, 0.1 mM), pyridine (0.1 mL, 1.2 mmol) was added and stirred at 80 °C for 8 h. The reaction mixture was quenched using sat. NaHCO3 aq. The organic layer was separated and washed with sat. NaHCO3 aq., dried over Na2SO4, and concentrated under vacuum. The residue was freeze-dried and purified via preparative TLC to afford 7 (3.0 mg, 4.5 μmol, 67.2%). [α]25D +125.4 (c 0.30, MeOH); 1H NMR (CDCl3, 500 MHz): δ 6.11 (1H, ddd, J = 15.3, 11.0, 4.1 Hz), 5.43 (1H, dd, J = 15.3, 9.1 Hz), 5.35 (1H, d, J = 10.2 Hz), 5.20 (1H, d, J = 1.7 Hz), 4.71 (1H, d, J = 1.7 Hz), 4.61 (1H, d, J = 11.6 Hz), 4.44 (1H, d, J = 11.6 Hz), 4.19 (1H, m), 4.06 (1H, ddd, J = 8.9, 6.2, 2.4 Hz), 3.88 (1H, ddd, J = 11.4, 9.1, 2.4 Hz), 3.76 (1H, dq, J = 6.7, 3.6 Hz), 3.48 (3H, s), 3.42 (1H, dq, J = 6.6, 2.4 Hz), 3.27 (1H, d, J = 10.0 Hz), 2.28 (1H, dddd, J = 10.8, 10.7, 10.5, 4.3 Hz), 2.32–2.23 (1H, m), 2.16–2.07 (1H, m), 2.07–1.97 (1H, overlapped), 1.96–1.82 (1H, overlapped), 1.86 (1H, ddd, J = 10.7, 10.6, 3.7 Hz), 1.82–1.74 (1H, m), 1.64–1.50 (5H, overlapped), 1.49–1.29 (4H, overlapped), 1.29–1.01 (4H, overlapped), 1.19 (3H, d, J = 6.5 Hz), 1.17 (9H, s), 0.98 (3H, d, J = 6.7 Hz), 0.92 (3H, d, J = 6.6 Hz), 0.67 (3H, d, J = 6.5 Hz); 13C NMR (CDCl3, 125 MHz): δ 201.14, 180.20, 179.05, 173.61, 154.23, 145.14, 134.51, 133.15, 127.34, 96.80, 89.93, 88.43, 79.27, 79.17, 77.85, 59.62, 57.48, 47.16, 44.00, 39.28, 38.86, 37.46, 35.55, 35.39, 34.20, 32.94, 32.12, 31.64, 31.22, 29.84, 27.25, 27.19, 25.82, 19.78, 18.15, 10.13, 8.85; ESI-MS m/z 688.4422 [M + NH4]+ (calcd. for C39H62NO9, m/z 688.4425 [M + NH4]+).
Antimicrobial Activity
The antimicrobial activity against S. aureus ATCC43300 was determined using the paper-disc method (6 mm discs from Advantec Co., Ltd., Tokyo, Japan). Sterile filter discs impregnated with compound solutions were placed on agar plates, and the plates were incubated. After incubation, inhibitory zones were determined. Culture conditions were as follows: 3.8% Mueller Hinton II agar (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan), 1.0% inoculation, 37 °C, 16 h.
The MICs of 1, its derivatives, vancomycin (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan; broad-spectrum activity against drug-resistant Gram-positive bacteria) and linezolid (Pfizer, Inc., NY, USA; used as a treatment for VRE and MRSA) were determined against 27 bacterial strains employing the agar dilution method using Mueller Hinton agar (Nippon Becton Dickinson Company, Ltd., Tokyo, Japan) according to the Clinical and Laboratory Standards Institute manuals.19
Acknowledgments
We are grateful to Distinguished Emeritus Professor S. O̅mura (Kitasato University, Tokyo, Japan) for his helpful support and valuable suggestions. We thank K. Shiraishi, S. Tanabe, and K. Abe (Kowa Company, Ltd., Aichi, Japan) for their valuable advice. Further, we are grateful to Dr. K. Nagai, R. Seki, and N. Sato, School of Pharmacy, Kitasato University for the measurements of mass and NMR spectra.
Glossary
Abbreviations
- AMR
antimicrobial resistance
- COSY
correlated spectroscopy
- HMBC
heteronuclear multiple bond correlation
- HSQC
heteronuclear single quantum coherence spectroscopy
- MIC
minimal inhibitory concentration
- MRSA
methicillin-resistant Staphylococcus aureus
- SAR
structure–activity relationship
- TFA
trifluoroacetic acid
- TLC
thin-layer chromatography
- VISA
vancomycin-intermediate Staphylococcus aureus
- VRE
vancomycin-resistant Enterococci
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c00651.
1H and 13C NMR spectra of compounds 2–7 and HSQC, COSY, and HMBC spectra of compound; comparison of biosynthetic gene clusters of tetronomycin between Streptomyces sp. K20-0247 and Streptomyces sp. NRRL 11266; annotation of biosynthetic gene clusters of tetronomycin in Streptomyces sp. K20-0247; results of determination of Ca2+ ionophobic activities of 1, 4, and EDTA and information on strains used in this study (PDF)
Author Contributions
∥ A.K. and I.T. contributed equally to this work.
This study was partially supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)), the Japan Agency for Medical Research & Development (AMED) under Grant Number JP21am0101096, and Grant-in-Aid for Scientific Research (KAKENHI C) 21K06616.
The authors declare no competing financial interest.
This paper originally published ASAP on March 14, 2023. An error in the abstract text was corrected, and a new version reposted on March 15, 2023.
Supplementary Material
References
- https://www.cdc.gov/drugresistance/biggest-threats.htmlMurray C. J. L.; Ikuta K. S.; Sharara F.; Swetschinski L.; Aguilar G. R.; Gray A.; Han C.; Bisignano C.; Rao P.; Wool E.; Johnson S. C.; Browne A. J.; Chipeta M. G.; Fell F.; Hackett S.; Woodhouse G. H.; Hamadani B. H. K.; Kumaran E. A. P.; McManigal B.; Agarwal R.; Akech S.; Albertson S.; Amuasi J.; Andrews J.; Aravkin A.; Ashley E.; Bailey F.; Baker S.; Basnyat B.; Bekker A.; Bender R.; Bethou A.; Bielicki J.; Boonkasidecha S.; Bukosia J.; Carvalheiro C.; Orjuela C. C.; Chansamouth V.; Chaurasia S.; Chiurchiù S.; Chowdhury F.; Cook A. J.; Cooper B.; Cressey T. R.; Criollo-Mora E.; Cunningham M.; Darboe S.; Day N. P. J.; Luca M. D.; Dokova K.; Dramowski A.; Eckmanns S. J. D. T.; Eibach D.; Emami A.; Feasey N.; Fisher-Pearson N.; Forrest K.; Garrett D.; Gastmeier P.; Giref A. Z.; Greer R. C.; Gupta V.; Haller S.; Haselbeck A.; Hay S. I.; Holm M.; Hopkins S.; Iregbu K. C.; Jacobs J.; Jarovsky D.; Javanmardi F.; Khorana M.; Kissoon N.; Kobeissi E.; Kostyanev T.; Krapp F.; Krumkamp R.; Kumar A.; Kyu H. H.; Lim C.; Limmathurotsakul D.; Loftus M. J.; Lunn M.; Ma J.; Mturi N.; Munera-Huertas T.; Musicha P.; Mussi-Pinhata M. M.; Nakamura T.; Nanavati R.; Nangia S.; Newton P.; Ngoun C.; Novotney A.; Nwakanma D.; Obiero C. W.; Olivas-Martinez A.; Olliaro P.; Ooko E.; Ortiz-Brizuela E.; Peleg A. Y.; Perrone C.; Plakkal N.; Ponce-de-Leon A.; Raad M.; Ramdin T.; Riddell A.; Roberts T.; Robotham J. V.; Roca A.; Rudd K. E.; Russell N.; Schnall J.; Scott J. A. G.; Shivamallappa M.; Sifuentes-Osornio J.; Steenkeste N.; Stewardson A. J.; Stoeva T.; Tasak N.; Thaiprakong A.; Thwaites G.; Turner C.; Turner P.; van Doorn H. R.; Velaphi S.; Vongpradith A.; Vu H.; Walsh T.; Waner S.; Wangrangsimakul T.; Wozniak T.; Zheng P.; Sartorius B.; Lopez A. D.; Stergachis A.; Moore C.; Dolecek C.; Naghavi M. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein E.; Keynan Y. Vancomycin Revisited - 60 Years Later. Front. Public Health 2014, 2, 217. 10.3389/fpubh.2014.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventola C. L. The Antibiotic Resistance Crisis: part 1: Causes and Threats. P & T 2015, 40, 277–283. [PMC free article] [PubMed] [Google Scholar]
- CDC’s Antibiotic Resistance Threats in the United States , 2019. (2019 AR Threats Report) Available at: https://www.cdc.gov/drugresistance/biggest-threats.html
- Keller-Juslen C.; King H. D.; Kuhn M.; Loosli H.-R.; Pache W.; Petcher T. J.; Weber H. P.; von Wartburg A. Tetronomycin, a Novel Polyether of Unusual Structure. J. Antibiot. (Tokyo) 1982, 35, 142–150. 10.7164/antibiotics.35.142. [DOI] [PubMed] [Google Scholar]
- Vieweg L.; Reichau S.; Schobert R.; Leadlay P. F.; Sussmuth R. D. Recent Advances in the Field of Bioactive Tetronates. Nat. Prod. Rep. 2014, 31, 1554–1584. 10.1039/C4NP00015C. [DOI] [PubMed] [Google Scholar]
- Gverzdys T.; Kramer G.; Nodwell J. R. Tetrodecamycin: An Unusual and Interesting Tetronate Antibiotic. Bioorg. Med. Chem. 2016, 24, 6269–6275. 10.1016/j.bmc.2016.05.028. [DOI] [PubMed] [Google Scholar]
- Grandjean J.; Laszlo P. Solution Structure and Cation-Binding Abilities of Two Quasi-Isomorphous Antibiotic Ionophores, M 139603 and Tetronomycin. Tetrahedron Lett. 1983, 24, 3319–3322. 10.1016/S0040-4039(00)86258-9. [DOI] [Google Scholar]
- Grandjean J.; Laszlo P. Synergistic Transport of Praseodymium(3+) Ion Across Lipid Bilayers in the Presence of Two Chemically Distinct Ionophores. J. Am. Chem. Soc. 1984, 106I, 1472–1476. [Google Scholar]
- Wyche T. P.; Alvarenga R. F. R.; Piotrowski J. S.; Duster M. N.; Warrack S. R.; Cornilescu G.; De Wolfe T. J. D.; Hou Y.; Braun D. R.; Ellis G. A.; Simpkins S. W.; Nelson J.; Myers C. L.; Steele J.; Mori H.; Safdar N.; Markley J. L.; Rajski S. R.; Bugni T. S. Chemical Genomics, Structure Elucidation, and In Vivo Studies of the Marine-Derived Anticlostridial Ecteinamycin. ACS Chem. Biol. 2017, 12, 2287–2295. 10.1021/acschembio.7b00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.; Liu H.; Svenningsen E. B.; Wollesen M.; Jacobsen K. M.; Andersen F. D.; Moyano-Villameriel J.; Pedersen C. N.; Norby P.; Torring T.; Poulsen T. B. Expanding the Antibacterial Selectivity of Polyether Ionophore Antibiotics Through Diversity-Focused Semisynthesis. Nat. Chem. 2021, 13, 47–55. 10.1038/s41557-020-00601-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T.; Iinuma H.; Sawa R.; Takahashi Y.; Nakamura H.; Nakamura K. T.; Sawa T.; Naganawa H.; Takeuchi T. Tetrodecamycin and Dihydrotetrodecamycin, New Antimicrobial Antibiotics Against Pasteurella piscicida Produced by Streptomyces nashvillensis MJ885-mF8 II. Structure Determination. J. Antibiot. (Tokyo) 1995, 48, 1110–1114. 10.7164/antibiotics.48.1110. [DOI] [PubMed] [Google Scholar]
- Tsuchida T.; Iinuma H.; Nakamura K. T.; Nakamura H.; Sawa T.; Hamada M.; Takeuchi T. Derivatives of Tetrodecamycin. J. Antibiot. 1995, 48, 1330–1335. 10.7164/antibiotics.48.1330. [DOI] [PubMed] [Google Scholar]
- Yang P.-Y.; Liu K.; Ngai M. H.; Lear M. J.; Wenk M. R.; Yao S. Q. Activity-Based Proteome Profiling of Potential Cellular Targets of Orlistat--An FDA-Approved Drug with Anti-tumor Activities. J. Am. Chem. Soc. 2010, 132, 656–666. 10.1021/ja907716f. [DOI] [PubMed] [Google Scholar]
- Park D. Genomic DNA Isolation from Different Biological Materials. Methods Mol. Biol. 2007, 353, 3–13. [DOI] [PubMed] [Google Scholar]
- Tanizawa Y.; Fujisawa T.; Nakamura Y. DFAST: A Flexible Prokaryotic Genome Annotation Pipeline for Faster Genome Publication. Bioinformatics 2018, 34, 1037–1039. 10.1093/bioinformatics/btx713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K.; Shaw S.; Kloosterman A. M.; Charlop-Powers Z.; van Wezel G. P.; Medema M. H.; Weber T. antiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist C. L. M.; Chooi Y.-H. C. Clustermap. js: Automatic Generation of Gene Cluster Comparison Figures. Bioinformatics 2021, 37, 2473–2475. 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing, 30th Ed.; Clinical and Laboratory Standards Institute, Wayne, PA; 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.