Abstract
The assembly of ribosomal subunits is a highly orchestrated process that involves a huge cohort of accessory factors. Most eukaryotic ribosome biogenesis factors were first identified by genetic screens and proteomic approaches of pre‐ribosomal particles in Saccharomyces cerevisiae. Later, research on human ribosome synthesis not only demonstrated that the requirement for many of these factors is conserved in evolution, but also revealed the involvement of additional players, reflecting a more complex assembly pathway in mammalian cells. Yet, it remained a challenge for the field to assign a function to many of the identified factors and to reveal their molecular mode of action. Over the past decade, structural, biochemical, and cellular studies have largely filled this gap in knowledge and led to a detailed understanding of the molecular role that many of the players have during the stepwise process of ribosome maturation. Such detailed knowledge of the function of ribosome biogenesis factors will be key to further understand and better treat diseases linked to disturbed ribosome assembly, including ribosomopathies, as well as different types of cancer.
Keywords: pre‐ribosomal particle, pre‐rRNA processing, ribosomal subunit, ribosome biogenesis factor, ribosome synthesis
Subject Categories: Translation & Protein Quality
As part of our 2023 RNA review series, Kutay and colleagues provide a comprehensive update on the diverse factors involved in ribosome biogenesis across different species.

Introduction
Ribosomes are at the heart of messenger (mRNA) translation, a key process of gene expression in all organisms. In eukaryotes, ribosomes consist of a small 40S and a large 60S subunit that together form the translation‐competent 80S ribosome. The 40S subunit comprises the 18S ribosomal RNA (rRNA) as well as 33 ribosomal proteins (RPs), whereas the 60S subunit contains three rRNAs, the 25S/28S, 5.8S, and 5S rRNAs, as well as 46 RPs in yeast and 47 RPs in humans (Table 1). During protein synthesis, mRNA is bound by the 40S subunit, which harbors the decoding center designed for pairing of an mRNA codon with a cognate tRNA. The peptidyl transferase center (PTC) in the 60S subunit catalyzes peptide bond formation in the emerging polypeptide chain, which leaves the ribosome through the peptide exit tunnel (PET) of the 60S subunit. As the catalytic PTC is formed by rRNA, the ribosome is classified as a ribozyme (Cech, 2000; Nissen et al, 2000).
Table 1.
Composition of ribosomes from different kingdoms.
| Bacteria | Archaea | Eukaryotes | ||
|---|---|---|---|---|
| E. coli | P. furiosus | S. cerevisiae | H. sapiens | |
| Molecular weight | ||||
| Ribosome | 2.3 MDa | 2.6 MDa | 3.3 MDa | 4.3 MDa |
| LSU | 1.45 MDa | 1.7 MDa | 2.1 MDa | 3.1 MDa |
| SSU | 0.8 MDa | 0.9 MDa | 1.2 MDa | 1.2 MDa |
| Sedimentation coefficients | ||||
| Ribosome | 70S | 70S | 80S | 80S |
| LSU | 50S | 50S | 60S | 60S |
| SSU | 30S | 30S | 40S | 40S |
| rRNAs | ||||
| LSU | 23S | 23S | 25S | 28S |
| 5S | 5S | 5S | 5S | |
| 5.8S | 5.8S | |||
| SSU | 16S | 16S | 18S | 18S |
| rRNA length (nucleotides) | ||||
| Total | 4,567 | 4,712 | 5,475 | 7,181 |
| LSU (23/25/28S +5S (+5.8S)) | 2,904 + 121 | 3,096 + 121 | 3,396 + 158 + 121 | 5,034 + 156 + 121 |
| SSU (18S) | 1,542 | 1,495 | 1,800 | 1,870 |
| Number of ribosomal proteins | ||||
| Ribosome | 54 | 69 | 79 | 80 |
| LSU | 33 | 42 | 46 | 47 |
| SSU | 21 | 27 | 33 | 33 |
Properties of ribosomes in bacteria (E. coli), archaea (P. furiosus), and lower (S. cerevisiae) and higher (H. sapiens) eukaryotes.
Due to their importance in mRNA translation, decoding center, PTC, PET, and translation factor binding sites are the evolutionarily most conserved regions of ribosomes (Klinge et al, 2012; Melnikov et al, 2012). In contrast to the conserved core structure, overall ribosomal composition, size, and complexity vary between the different kingdoms of life (Table 1). The number of rRNAs and their length increased significantly from bacteria and archaea to eukaryotes. Eukaryotic rRNAs display several large rRNA expansion segments with largely unexplored function. The accretion of these expansion segments bears the major contribution to the increase in rRNA length and ribosome size from yeast to vertebrates (Table 1) (Hariharan et al, 2022). Furthermore, eukaryotic ribosomes contain additional RPs, and most RPs of the conserved core harbor extensions and insertions (Spahn et al, 2001; Armache et al, 2010; Ben‐Shem et al, 2011; Klinge et al, 2011; Rabl et al, 2011; Melnikov et al, 2012; Khatter et al, 2015).
Assembly of both ribosomal subunits requires deposition of the numerous RPs on the pre‐rRNAs concomitant with rRNA transcription, modification, folding, and processing, which all occur in a hierarchical, highly orchestrated, and energetically expensive cellular process (Warner, 1999). Although bacterial ribosomal subunits can be reconstituted at elevated temperature in vitro by mixing mature rRNAs and ribosomal proteins, their assembly in vivo is supported by about 50 non‐ribosomal factors (Held et al, 1973; Nierhaus & Dohme, 1974; Kaczanowska & Rydén‐Aulin, 2007; Shajani et al, 2011; Gibbs & Fredrick, 2018). In contrast to prokaryotes, where the assembly process occurs in a single compartment, the cytoplasm, eukaryotic cells are highly compartmentalized and ribosomal subunit maturation starts in the nucleolus, continues in the nucleoplasm and is only finalized in the cytoplasm. The eukaryotic subunit assembly process involves several hundred non‐ribosomal factors, termed ribosome biogenesis factors (RBFs), which function as chaperones and as modification, processing, assembly, and remodeling factors. These RBFs transiently associate with pre‐ribosomal particles, but are not part of mature ribosomes. Along with the growing complexity of ribosome composition throughout evolution, the intricacy of the eukaryotic ribosome biogenesis pathway and the regulatory interplays with other processes also increased.
Eukaryotic ribosome assembly starts with the transcription of a polycistronic pre‐rRNA precursor by RNA polymerase I (RNAPI) in the nucleolus (Fig 1) (Turowski & Tollervey, 2015). Emerging rRNA stretches are quickly bound by some early assembling RPs as well as RBFs, giving rise to a large, 90S‐sized precursor particle in the nucleolus (Klinge & Woolford, 2019; vanden Broeck & Klinge, 2022). A critical endonucleolytic pre‐rRNA cleavage event then leads to the separation of the pre‐40S and pre‐60S particles (Baßler & Hurt, 2019; Bohnsack & Bohnsack, 2019). Both subunits subsequently undergo a number of nucle(ol)ar maturation steps, including the incorporation of the 5S rRNA, transcribed by RNA polymerase III (RNAPIII), into the pre‐60S subunit (Woolford & Baserga, 2013; de la Cruz et al, 2015; Kressler et al, 2017; Chaker‐Margot & Klinge, 2019; Frazier et al, 2021). After nuclear export, final assembly events take place in the cytoplasm, including the release of remaining RBFs and incorporation of missing RPs, giving rise to mature, translationally competent subunits (Kressler et al, 2017; Peña et al, 2017).
Figure 1. Overview of eukaryotic ribosome biogenesis.
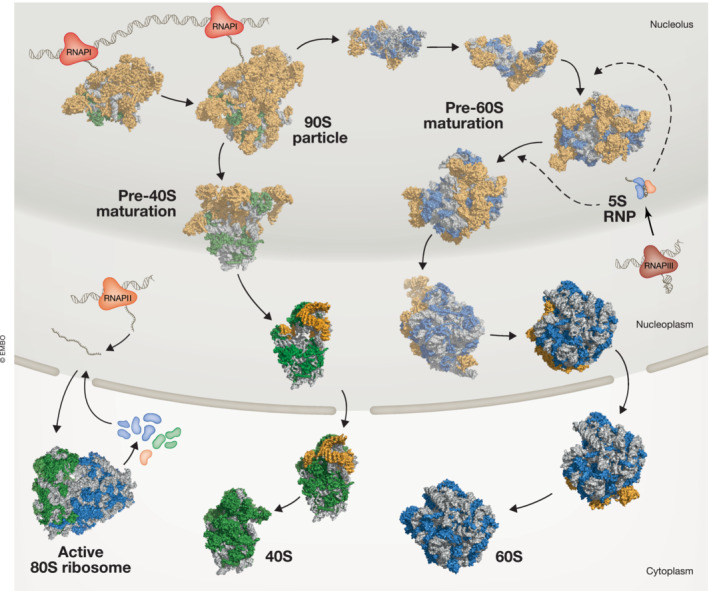
In the nucleolus, a rRNA precursor is transcribed by RNAPI and co‐transcriptionally joined by RPs and RBFs, giving rise to a 90S pre‐ribosomal particle. After pre‐rRNA cleavage (site 2 in human, A2 in yeast), the pre‐40S and pre‐60S particles further mature independently in the nucleolus and the nucleoplasm. After export through the nuclear pore complex, final maturation steps occur in the cytoplasm, yielding 40S and 60S subunits competent for mRNA translation. RNAPIII transcribes the 5S rRNA (nucleolar in human cells, nucleoplasmic in yeast), which joins pre‐60S particles in the nucleolus as part of the 5S RNP complex. RNAPII transcribes mRNAs of RP and RBF genes, which are translated by the 80S ribosome in the cytoplasm and then imported into the nucleus. Structural snapshots of maturing 90S (PDB ID: 6ZQA, 6ZQC), pre‐40S (PDB ID: 6G4W, 6G4S, 6ZQF), and pre‐60S particles (PDB ID: 6EM3, 6C0F, 6ELZ, 3JCT, 5JCS, 6LU8, 6LSR) as well as mature subunits (PDB ID: 6G5H, 3J7P) are shown. RBFs are displayed in orange, Rps in green, Rpl in blue, rRNA in gray. Structures solved in yeast are shown with reduced opacity, currently no structures of the corresponding human maturation stages are available.
Early work in the 1970s already indicated that yeast and human cells employ broadly similar principles for ribosome maturation, which differ substantially from prokaryotic ribosome synthesis (Darnell, 1968; Udem et al, 1971; Warner, 1971; Trapman et al, 1975). Many eukaryotic RBFs were first identified in genetic screens and proteomic approaches using budding yeast as a model organism, yet their function was often unraveled only later by biochemical studies (Hurt et al, 1999; Stage‐Zimmermann et al, 2000; Dragon et al, 2002; Nissan et al, 2002; Saveanu et al, 2003; Schäfer et al, 2003; Woolford & Baserga, 2013). Over time, an increasing number of structural snapshots of ribosomal pre‐particles from various stages of the assembly process has tremendously increased the understanding of RBF functionalities (Barrio‐Garcia et al, 2016; Kornprobst et al, 2016; Wu et al, 2016; Zhang et al, 2016b; Barandun et al, 2017; Cheng et al, 2017; Kater et al, 2017, 2020; Ma et al, 2017; Sun et al, 2017; Sanghai et al, 2018; Scaiola et al, 2018; Chaker‐Margot & Klinge, 2019; Kargas et al, 2019; Klinge & Woolford, 2019; Zhou et al, 2019a, 2019b). In the past 15 years, biochemical, cellular, and structural studies including large‐scale screening and proteomic approaches have revealed commonalities and differences in ribosome synthesis between lower and higher eukaryotes (Couté et al, 2008; Wild et al, 2010; Finkbeiner et al, 2011; Simabuco et al, 2012; Widmann et al, 2012; Tafforeau et al, 2013; Wyler et al, 2014; Zemp et al, 2014, 2009; Badertscher et al, 2015; Wandrey et al, 2015; Larburu et al, 2016; Raman et al, 2016; Fromm et al, 2017; Memet et al, 2017; Montellese et al, 2017, 2020; Ameismeier et al, 2018, 2020; Farley‐Barnes et al, 2018; Boneberg et al, 2019; Braun et al, 2020; Choudhury et al, 2020, 2019; Liang et al, 2020; Gerhardy et al, 2021; Ogawa et al, 2021; Pöll et al, 2021; Singh et al, 2021; Dörner et al, 2022; Sailer et al, 2022).
Underscoring the importance of ribosome maturation for cellular and organismal homeostasis, defects in ribosome biogenesis are associated with a variety of human diseases. Most RPs and RBFs are encoded by essential genes (Panić et al, 2006; Woolford & Baserga, 2013; Perucho et al, 2014). Genetic alterations in RPs and RBFs are frequently associated with haploinsufficiency and causative for a class of severe congenital diseases termed ribosomopathies (Bohnsack & Bohnsack, 2019; Farley‐Barnes et al, 2019; Kampen et al, 2020). A number of ribosomopathies are linked to increased cancer susceptibility and a growing body of evidence suggests that defects in ribosome biogenesis can drive tumorigenesis (Pelletier et al, 2018; Catez et al, 2019).
In this review, we present a current inventory of yeast and human ribosome biogenesis factors and their functions in eukaryotic ribosome maturation from rDNA organization and transcription, pre‐rRNA modification and processing, to subunit assembly and nuclear export.
rDNA organization in the nucleolus and pre‐rRNA transcription
Nucleoli are primarily dedicated to pre‐rRNA transcription and early steps of ribosomal subunit assembly. They are the most prominent nuclear, membrane‐less organelles. Nucleoli are built around rDNA loci which are organized in the form of nucleolar organizing regions (NORs) constituted by rDNA gene clusters (Ritossa & Spiegelman, 1965). While S. cerevisiae contains a single NOR on chromosome 12 with roughly 150 rDNA repeats, human cells possess NORs on the acrocentric human chromosomes 13, 14, 15, 21, and 22, harboring altogether roughly 400 rDNA repeats (Henderson et al, 1972; Stults et al, 2008; Lofgren et al, 2019). Not all NORs are active in a cell, and cell type‐specific differences in NOR activity exist (Roussel et al, 1996; Farley et al, 2015).
The organization of nucleoli differs between yeast and human. While yeast nucleoli consist of two subdomains, the fibrillar strands and granules, at least three subcompartments are found in human nucleoli, termed fibrillar center (FC), dense fibrillar center (DFC), and granular component (GC) (Fig 2A). rDNA loci are densely packed in the FC, whereas pre‐rRNA is transcribed at the boundary between FC and DFC (Koberna et al, 2002). Early processing steps including rRNA modification and cleavage also occur in the DFC, while pre‐ribosomal particles mature further in the surrounding GC. Nucleoli are considered to be multiphase liquid condensates (Lafontaine et al, 2021). However, their biogenesis and domain organization reflect the hierarchy in pre‐rRNA synthesis and ribosomal subunit assembly, highlighting the contribution of interaction specificity in shaping their molecular makeup and structure (Musacchio, 2022).
Figure 2. Organization of nucleoli, the rDNA locus and promoter architecture in yeast and human cells.
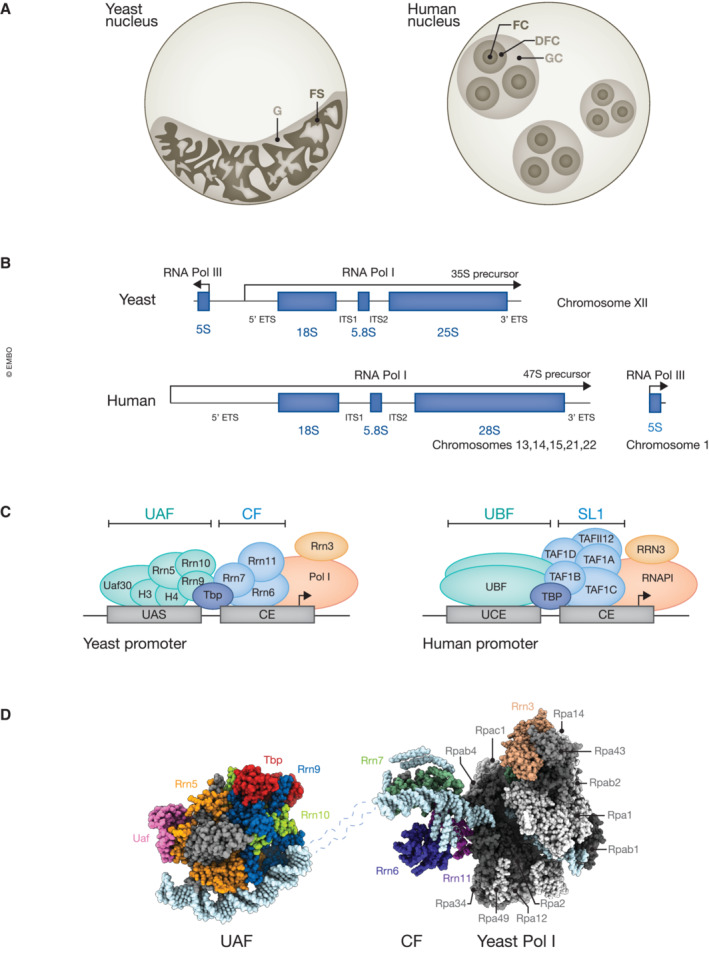
(A) Schematic representation of a yeast nucleolus composed of fibrillar strands (FS) and granules (G) and human nucleoli consisting of three subcompartments: fibrillar center (FC), dense fibrillar component (DFC), and granular component (GC). (B) Schematic representation of rDNA architecture in S. cerevisiae and human cells. (C) Comparison of 35S/47S rDNA promoter region with associated pre‐initiation complexes in yeast and human cells. Yeast promoters contain the upstream activation sequence (UAS) bound by the upstream activating factor (UAF) complex and the central element (CE) bound by the core factor (CF) complex. Human promoters also contain two elements; the upstream core element (UCE) bound by a UBF dimer and the central element (CE) bound by selectivity factor 1 (SL1) complex (Knutson & Hahn, 2013; Engel et al, 2018; Sadian et al, 2019; Pilsl & Engel, 2020; Baudin et al, 2022; Girbig et al, 2022). (D) Structural model of yeast RNAPI in complex with Rrn3, the CF (PDB ID: 7OBA), and UAF complexes, bound to Tbp and promoter DNA (PDB ID: 7Z0O). RNAPI subunits are in shadows of gray, factors are color‐coded, and DNA is shown in light blue.
Promoters of eukaryotic rDNA contain two regulatory elements: the core element (CE) and the upstream activating sequence or control element (UAS/UCE) (Fig 2C) (Knutson & Hahn, 2013). Although the general promoter architecture is conserved between yeast and mammals, regulation of RNAPI transcription likely differs between species, as there is little sequence similarity in the main promoter elements (Goodfellow & Zomerdijk, 2013). Several factors involved in yeast and human rDNA organization and transcription, including pre‐initiation complex (PIC) formation, elongation, and termination, have been identified (Table 2). In yeast, each rDNA repeat encodes two transcripts: the 5S rRNA, transcribed by RNAPIII, and the polycistronic 35S pre‐rRNA, produced by RNAPI. The 35S pre‐rRNA contains the 18S, 5.8S, and 25S rRNAs surrounded and separated by external and internal transcribed spacers (5′ ETS, ITS1, ITS2, 3′ ETS) (Fig 2B) (Woolford & Baserga, 2013). Recruitment of RNAPI to the rDNA promoter depends on the RNA polymerase I‐specific transcription factor Rrn3 and the heterotrimeric core factor (CF) complex, which recognizes the core promoter element upstream of the transcription start site (Fig 2C and D). The efficient initiation of pre‐rRNA transcription is further supported by the TATA‐binding protein (TBP) and the upstream activating factor (UAF) complex that binds an upstream activation sequence (UAS) (Russell & Zomerdijk, 2006; Girbig et al, 2022).
Table 2.
Factors involved in rDNA transcription.
| Yeast | 35S | Human | 47S | Function in rRNA transcription | Citation |
|---|---|---|---|---|---|
| RNA Polymerase I subunits | |||||
| Rpb5 | x | POLR2E | x | RNAPI subunits, shared with RNAPII and RNAPIII between polymerases | Russell and Zomerdijk (2006) |
| Rpb6 | x | POLR2F | x | ||
| Rpb8 | x | POLR2H | x | ||
| Rpb10 | x | POLR2L | x | ||
| Rpb12 | x | POLR2K | x | ||
| Rpa40 | x | POLR1C | x | RNAPI subunits, shared with RNAPIII | |
| Rpa19 | x | POLR1D | x | ||
| Rpa190 | x | POLR1A | x | RNAPI‐specific subunits | |
| Rpa135 | x | POLR1B | x | ||
| Rpa43 | x | POLR1F | x | ||
| Rpa14 | x | ||||
| Rpa12 | x | POLR1H | x | ||
| Rpa49 | x | POLR1E | x | ||
| Rpa34 | x | POLR1G | x | ||
| PIC formation/promoter escape | |||||
| Tbp | x | TBP | x | TATA box binding protein | Comai et al (1994) |
| UBF | x | Binds rDNA promoter as a dimer, has a role in promoter escape and regulation of elongation | Bell et al (1988) | ||
| Rrn3 | x | RRN3 | x | RNAPI‐specific initiation factor, stimulates RNAPI recruitment | Milkereit and Tschochner (1998) |
| Rrn6 | x | TAF1C | x | Involved in PIC formation. In yeast: part of CF Complex; in humans: part of selectivity factor SL1 | Comai et al (1994) |
| Rrn7 | x | TAF1B | x | ||
| Rrn11 | x | TAF1A | x | ||
| TAF1D | x | Involved in PIC formation: part of human Selectivity factor SL1 | Denissov et al (2007) and Gorski et al (2007) | ||
| TAF12 | x | ||||
| Rrn5 | x | Involved in PIC formation: part of yeast UAF complex | Keys et al (1996), Keener et al (1997) and Siddiqi et al (2001) | ||
| Rrn9 | x | ||||
| Rrn10 | x | ||||
| Uaf30 | x | ||||
| Hht1/H3 | x | H3C1/H3 | |||
| Hhf1/H4 | x | H4C1/H4 | |||
| Top1 | x | TOP1 | x | Facilitate PIC formation by removing super coils at rDNA promoters and promote elongation | Brill et al (1987) and Ray et al (2013) |
| Top2 | x | TOP2A | x | ||
| Cka1 | CK2A1 | x | Involved in PIC formation; part of tetrameric CK2 complex, regulate interaction between UBF and SL1 | Panova et al (2006) | |
| Cka2 | CK2A2 | x | |||
| Ckb1 | CK2N | x | |||
| Elongation | |||||
| Spt4 | x | SPT4H | x | DSIF complex, influences RNAPI activity; Regulates binding of UBF to rDNA | Schneider et al (2006) |
| Spt5 | x | SPT5H | x | ||
| Paf1 | x | PAF1 | x | Paf1C complex, influences RNAPI activity | Zhang et al (2009) |
| Ctr9 | x | CTR9 | x | ||
| Cdc73 | x | WDR61 | x | ||
| Rtf1 | x | RTF1 | x | ||
| Leo1 | x | LEO1 | x | ||
| Spt16 | x | SUPT16H | x | FACT complex, supports RNAPI transcription through nucleosomes | Birch et al (2009) |
| Pob3 | x | SSRP1 | x | ||
| Fcp1 | x | Dephosphorylates RNAPI for efficient RNA synthesis | Fath et al (2004) | ||
| Termination | |||||
| Rnt1 | x | Endonucleolytic cleavage at end of 25S transcript | el Hage et al (2008) | ||
| Rat1 | x | Release of RNAPI transcribing 3′ part of the transcript | el Hage et al (2008) | ||
| Reb1 | x | TTF1 | x | RNAPI transcription termination factors, bind to termination sequence T1, lead to RNAP I pausing | Reiter et al (2012) |
| Nsi1 | x | Reiter et al (2012) | |||
| Fob1 | x | Binds to replication fork barrier sequence, inhibits clashes with DNA replication machinery | el Hage et al (2008) | ||
| rDNA organization | |||||
| Hmo1 | x | Associates with active rDNA repeats, related to UBF | Gadal et al (2002) | ||
| CTCF | x | Regional organization of rDNA | van de Nobelen et al (2010) | ||
| MYC | x | Attachment of rDNA to nucleolar matrix; local histone acetylation | Grandori et al (2005) | ||
| MAX | x | Interacts with MYC | Nair and Burley (2003) | ||
| TCOF1 | x | Facilitates rDNA transcription, interacts with UBF and RNAPI, and is involved in rRNA methylation | Werner et al (2015) | ||
| Srp40 | NOLC1 | x | Interacts with TCOF1 | Valdez et al (2004) | |
| Bdf2 | BRD2 | x | Regulation of RNAPI activity by histone acetylation, recruited by LYAR | Izumikawa et al (2019) | |
| Bdf1 | BRD4 | x | |||
| KAT7 | x | ||||
| Nto1 | JADE3 | x | |||
| YCR087C | LYAR | x | Binds to UBF, recruits BRD2/4‐KAT7 | Izumikawa et al (2019) | |
| Factors acting both in rRNA transcription and early processing steps | |||||
| SIRT7 | x | Required for the activation of Pol I transcription at the exit from mitosis | Ford et al (2006) and Iyer‐Bierhoff et al (2018) | ||
| Nop1 | FBL | x | Methylates rRNA and histone H2AQ104 | Tessarz et al (2014) | |
| Utp4 | UTP4 | x | t‐UTPs, function in both rRNA transcription and the SSU processome | Gallagher et al (2004) and Prieto and McStay (2007) | |
| Utp5 | WDR43/UTP5 | x | |||
| Utp10 | HEATR1/UTP10 | x | |||
| Utp15 | UTP15 | x | |||
| Utp17 | UTP17/WDR75 | x | |||
Human rDNA repeats have by‐and‐large a similar architecture, although the 5S rRNA is transcribed from a distinct repeat region comprising roughly 100 loci on chromosome 1, located in the nucleoplasm in nucleolar proximity (Little & Braaten, 1989; Haeusler & Engelke, 2006; Stults et al, 2008). Also in human cells, several transcription factors and transcription factor complexes mediate RNAPI pre‐initiation complex assembly (Russell & Zomerdijk, 2006; Grummt, 2010). Initial binding of upstream binding factor (UBF) to the rDNA promoter allows for recruitment of the SL1 complex (containing TBP, factors analogous to CF in yeast (Fig 2C)), as well as metazoan‐specific factors (Table 2), before RNAPI is recruited via RRN3. Since pre‐rRNA transcription presents an initial rate‐limiting step of ribosome assembly, it is not surprising that many cellular signaling pathways target UBF, the SL1 complex, and RRN3 to regulate the production of ribosomes in human cells (Grummt, 2010; Bywater et al, 2013).
Pre‐rRNA modification
rRNA is heavily modified, in particular at functionally important regions such as the decoding center, the PTC, and the subunit interface (Decatur & Fournier, 2002; Polikanov et al, 2015; Sloan et al, 2017; Bailey et al, 2022). Many modifications are carried out co‐transcriptionally, and aid folding and compaction of the pre‐rRNA during assembly, but also support translation efficiency and accuracy (Liang et al, 2009; Sloan et al, 2017; Ojha et al, 2020; Khoshnevis et al, 2022). 2′‐O‐methylation of the ribose group in the rRNA backbone and pseudouridylation by isomerization of uridines are by far the most frequent modifications, introduced in a site‐specific manner by so‐called box C/D and box H/ACA small nucleolar ribonucleoproteins (snoRNPs) (see Table 3), respectively. Both types of RNPs contain four structural proteins and a dedicated modification enzyme, that is the methyltransferase fibrillarin (Nop1 in yeast) or the pseudouridine synthase dyskerin (Cbf5 in yeast). Each snoRNP also contains a 60–170 nt long snoRNA (with a few longer exceptions) (Marz et al, 2011; Jorjani et al, 2016), which guides the respective complex to its target site on the pre‐rRNA. Importantly, ribosome and snoRNA biogenesis are tightly interwoven in metazoans, since many snoRNAs are encoded within the introns of RP or RBF genes (Hirose & Steitz, 2001).
Table 3.
Factors involved in rRNA modification.
| Yeast | 40S | 60S | Human | 40S | 60S | Function in rRNA modification | Citation |
|---|---|---|---|---|---|---|---|
| snoRNP components | |||||||
| Nop1 | x | x | FBL | x | x | Box C/D snoRNP components, 2′O‐methylation of the ribose group in the RNA backbone | Grandi et al (2002) |
| Nop56 | x | x | NOP56 | x | x | ||
| Nop58 | x | x | NOP58 | x | x | ||
| Snu13 | x | x | SNU13 | x | x | ||
| Rrp9 | x | x | U3‐55K | x | x | ||
| Cbf5 | x | x | DKC | x | x | Box H/ACA snoRNP components, pseudouridylation of rRNA | Kiss‐László et al (1996) |
| Nhp2 | x | x | NHP2 | x | x | ||
| Nop10 | x | x | NOP10 | x | x | ||
| Gar1 | x | x | GAR1 | x | x | ||
| Stand‐alone enzymes | |||||||
| Tsr3 | x | TSR3 | x | Tsr3: aminocarboxypropyl transferase; Emg1: methyltransferase, 18S‐m1acp3Ψ1240 (y:1191) | Wurm et al (2010) and Meyer et al (2016) | ||
| Emg1 | x | EMG1 | x | ||||
| Kre33 | x | NAT10 | x | Acetyltransferase 18S‐ac4C1337,1842 (y:1280,1773) | Ito et al (2014) and Sharma et al (2017) | ||
| Bud23 | x | WBSCR22 | x | Methyltransferase 18S‐m7G1639 (y:1575) | White et al (2008) | ||
| Dim1 | x | DIMT1 | x | Methyltransferase 18S‐m2 6A1850/1 (y:1781/2) | Lafontaine et al (1998) | ||
| Rrp8 | x | NML/RRP8 | x | x | Methyltransferase 28S‐m1A1332 (y:645) | Peifer et al (2013) | |
| Bmt2 | x | Methyltransferase 25S‐m1A2142 | Sharma et al (2013) | ||||
| Rcm1 | x | NSUN5 | x | Methyltransferase 28S‐m5C3761 (y:2278) | Schosserer et al (2015) | ||
| Bmt5 | x | Methyltransferase 25S‐m3U2634 | Sharma et al (2014) | ||||
| Bmt6 | x | Methyltransferase 25S‐m3U2843 | Sharma et al (2014) | ||||
| Nop2 | x | NOP2 | x | Methyltransferase 28S‐m5C4414 (y:2870) | Sharma et al (2014) | ||
| Spb1 | x | x | Methyltransferase 28S‐Gm4469 (y:2922) | Lapeyre and Purushothaman (2004) | |||
| ZCCHC4 | x | Methyltransferase 28S‐m6A4220 | Ma et al (2019) and Pinto et al (2020) | ||||
| METTL5 | x | Methyltransferase 18S‐m6A1832 | van Tran et al (2019) | ||||
| Trm112 | TRMT112 | x | Activator of methyltransferases (Bud23/WBSCR22, METTL5) | Zorbas et al (2015) and van Tran et al (2019) | |||
Both the binding of snoRNAs to the pre‐rRNA and the resulting nucleotide modifications play important roles in pre‐rRNA folding (see recent reviews Mitterer & Pertschy, 2022; Oborská‐Oplová et al, 2022). Many snoRNA binding sites are found in pre‐rRNA regions that fold late during nucleolar assembly steps. By base‐pairing with these still unfolded pre‐rRNA elements, snoRNAs prevent premature or non‐productive formation of RNA helices. Furthermore, multivalent snoRNAs, exemplified by the U3 snoRNA that is involved in early pre‐rRNA folding and processing steps (see below), can force distant pre‐rRNA regions into a defined configuration while simultaneously inhibiting untimely RNA annealing events. Nucleotide modifications, in turn, affect base‐pairing preferences and can enhance the conformational rigidity of the RNA backbone (Sumita et al, 2005; Helm, 2006; Abou Assi et al, 2020).
In yeast, ~75 snoRNAs have been described, catalyzing the modification of 112 sites, while in humans 228 sites are targeted by more than 200 snoRNAs (Natchiar et al, 2017; Taoka et al, 2018, 2016). The lack of individual modifications is generally well tolerated in yeast cells, whereas their cumulative loss can cause defects in subunit assembly, translation, and cell growth (Liang et al, 2007, 2009). Notably, both in yeast and mammalian cells, there is some heterogeneity in the use of modification sites (Jaafar et al, 2021b), but evidence for a functional relevance of this variability remains scarce (Metge et al, 2021). More than 130 rRNA modifications have been visualized by single particle cryo‐EM in the human ribosome, of which 11 were found at universally conserved sites (Natchiar et al, 2017). As expected, the vast majority of modifications reside in the interior of the ribosome close to the functional centers. Interestingly, also a large number of novel sites were discovered, many of which contain base‐modified nucleotides.
Some rRNA modifications do not depend on snoRNPs, but are introduced by stand‐alone enzymes, many of which function in base methylation, but also support other modifications such as acetylation (listed in Table 3). For the majority of these enzymes only a single target nucleotide has been described, but the functions of these modifications remain largely enigmatic. The most complex ribosomal modification, a 1‐methyl‐3‐(3‐amino‐3‐carboxypropyl)‐pseudouridine (m1acp3Ψ) in helix 31 of the 18S rRNA (1248U), is established in a stepwise manner, starting with the formation of a pseudouridine catalyzed by the snR35 H/ACA snoRNP complex (Samarsky et al, 1995). After subsequent methylation by Emg1 (Wurm et al, 2010), the final modification is formed by the aminocarboxypropyl transferase Tsr3 during final cytoplasmic maturation (Meyer et al, 2016; Huang et al, 2022). Importantly, m1acp3Ψ, which is solvent exposed at the ribosomal P site, is frequently lost or hypomodified in cancer and has been suggested to lead to increased translation of RP mRNAs (Babaian et al, 2020). Recently, it was discovered that human rRNA also contains m6A modifications in the 18S and 28S rRNAs, mediated by the methyltransferases METTL5 and ZCCHC4 respectively, yet the molecular function of the two identified modification sites remains to be further explored (Ma et al, 2019; van Tran et al, 2019; Pinto et al, 2020).
Pre‐rRNA processing
A series of concerted pre‐rRNA cleavage and trimming reactions in the nucleus and cytoplasm leads to the excision of the mature 18S, 5.8S, and 25/28S rRNAs from the polycistronic rRNA precursor (Fig 3). The general hierarchical principle of sequential elimination of transcribed spacers and the function of several endo‐ and exonucleases (Table 4) in this process is largely conserved from yeast to human (for more details see Tomecki et al, 2017; Aubert et al, 2018; Bohnsack & Bohnsack, 2019). In both organisms, pre‐rRNA processing is tightly interconnected with other steps of ribosome biogenesis, such as rRNA modification, folding, and binding of RPs or RBFs. A critical step in the maturation pathway is the first cleavage event within the ITS1 (at site A2 in yeast, site 2 in humans), which separates the emerging pre‐40S and pre‐60S particles. In yeast, ~70% of nascent transcripts are cleaved co‐transcriptionally within ITS1 (Koš & Tollervey, 2010). Human pre‐rRNA processing also starts co‐transcriptionally, at least in the 5′ ETS region (Delannoy & Sollner‐Webb, 1997; Osheim et al, 2004). However, it remains to be defined whether the endonucleolytic cleavage within the ITS1 occurs only post‐transcriptionally or also co‐transcriptionally in mammalian cells, and how this critical event is coordinated with subunit assembly. The prominent presence of the long 47S/45S precursors in pulse‐labeling experiments and Northern blots indicates that at least a substantial fraction of ITS1 cleavage events may only occur after synthesis of the precursor has been completed (Bowman et al, 1981; Strezoska et al, 2000). Notably, in both yeast and mammals, alternative pre‐rRNA processing pathways exist, and their relative contribution to rRNA production is suggested to be governed by the respective kinetics of the processing reactions (Axt et al, 2014; Henras et al, 2015).
Figure 3. Pre‐rRNA maturation in yeast and human cells.
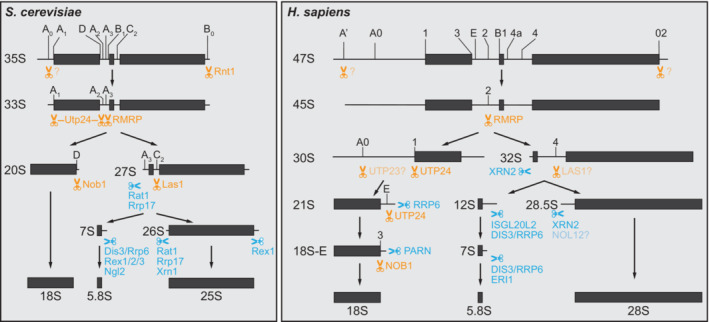
Simplified processing pathway of the 35S pre‐rRNA in yeast and 47S pre‐rRNA in human cells, indicating processing sites of endo‐ and exonucleases in orange and blue, respectively. Several alternative processing pathways exist, which are reviewed elsewhere (Tomecki et al, 2017; Aubert et al, 2018).
Table 4.
Factors involved in pre‐rRNA processing.
| Yeast | 40S | 60S | Human | 40S | 60S | Function in pre‐rRNA processing | Citation |
|---|---|---|---|---|---|---|---|
| Utp23 | UTP23 | x | Endoribonuclease, cleavage at site A0 (likely inactive) | Wells et al (2017) | |||
| Utp24/Fcf1 | x | UTP24 | x | Endoribonuclease, cleavage at site 1 (A1) | Bleichert et al (2006) | ||
| Nob1 | x | NOB1 | x | Endoribonuclease, cleavage at site 3 (D) | Fatica et al (2004) | ||
| RMRP | x | RMRP | x | Part of MRP complex, endoribonuclease with non‐canonical snoRNA, cleavage at site 2 (A3) | Perederina et al (2020), van Hoof et al (2000) and Goldfarb and Cech (2017) | ||
| Pop1 | x | POP1 | x | ||||
| Pop3 | x | RPP38 | x | ||||
| Pop4 | x | RPP29 | x | ||||
| Pop5 | x | POP5 | x | ||||
| Pop6 | x | RPP25 | x | ||||
| Pop7 | x | RPP20 | x | ||||
| Pop8 | x | RPP14 | x | ||||
| Rpp1 | x | RPP30 | x | ||||
| Rpr2 | x | RPP21 | x | ||||
| Snm1 | x | ||||||
| Rmp1 | x | ||||||
| RPP40 | x | ||||||
| Las1 | x | LAS1L | Endoribonuclease, maybe cleavage at site 4 (C2) | Schillewaert et al (2012) | |||
| Rnt1 | x | Endoribonuclease (B0) | Kufel et al (1999) | ||||
| Rat1 | x | XRN2 | x | x | Exoribonuclease, pre‐rRNA trimming | el Hage et al (2008) | |
| Rrp17 | x | NOL12 | Exoribonuclease, pre‐rRNA trimming | Oeffinger et al (2009) | |||
| Rrp44 | x | DIS3 | x | Nuclear exosome, exoribonuclease, pre‐rRNA Trimming 5.8S rRNA maturation | Briggs et al (1998) and Sloan et al (2012) | ||
| Rrp6 | x | EXOSC10 | x | ||||
| Csl4 | x | EXOSC1 | x | ||||
| Rrp4 | x | EXOSC2 | x | ||||
| Rrp40 | x | EXOSC3 | x | ||||
| Rrp41 | x | EXOSC4 | x | ||||
| Rrp46 | x | EXOSC5 | x | ||||
| Mtr3 | x | EXOSC6 | x | ||||
| Rrp42 | x | EXOSC7 | x | ||||
| Rrp43 | x | EXOSC8 | x | ||||
| Rrp45 | x | EXOSC9 | x | ||||
| Mtr4 | x | MTR4 | x | Cofactor of nuclear exosome | Briggs et al (1998) and Sloan et al (2012) | ||
| Trf4 | x | PAPD5 | x | ||||
| Trf5 | x | PAPD7 | x | ||||
| Air1 | x | ZCCHC7 | x | ||||
| Air2 | x | ZCCHC8 | x | ||||
| Mpp6 | x | MPP6 | x | ||||
| Lrp1/Rrp47 | x | C1D | x | ||||
| x | RBM7 | x | |||||
| Rex1 | x | REXO5 | Exoribonuclease, pre‐rRNA trimming | van Hoof et al (2000) | |||
| Rex2 | x | REXO2 | Exoribonuclease, pre‐rRNA trimming | van Hoof et al (2000) | |||
| Rex3 | x | REXO1 | Exoribonuclease, pre‐rRNA trimming | van Hoof et al (2000) | |||
| Ngl2 | x | Ccr4 | Exoribonuclease, pre‐rRNA trimming, 5.8S rRNA maturation | Faber et al (2002) | |||
| PARN | x | Exoribonuclease, pre‐rRNA trimming, 18S rRNA maturation | Montellese et al (2017) | ||||
| TUT4 | x | Uridylyltransferase, pre‐rRNA trimming, 18S rRNA maturation | Montellese et al (2017) | ||||
| TUT7 | x | Uridylyltransferase, pre‐rRNA trimming, 18S rRNA maturation | Montellese et al (2017) | ||||
| ISG20L2 | x | Exoribonuclease, pre‐rRNA trimming 5.8S rRNA maturation | Couté et al (2008) | ||||
| ERI1 | x | Exoribonuclease, pre‐rRNA trimming, 5.8S rRNA | Ansel et al (2008) |
The fourth rRNA, the 5S rRNA, is transcribed by RNAPIII. The immature 5S rRNA is initially bound by the La protein and TFIIIA in higher eukaryotes, protecting it from degradation (Ciganda & Williams, 2011; Layat et al, 2013). It needs to be trimmed at its 3′ end before getting incorporated into the maturing pre‐60S particle. This trimming is performed by the exonucleases Rex1, Rex2, and Rex3 in yeast, which act redundantly (Table 4) (van Hoof et al, 2000). Factors involved in human 5S rRNA maturation remain elusive. The Rex1 homolog REXO5 was shown to be functionally conserved in flies (Gerstberger et al, 2017), although mouse REXO5 is not essential for survival (Silva et al, 2017), pointing to potential redundancy. The processed 5S RNA associates with two newly synthesized RPs, RPL5/uL18, and RPL11/uL5, forming the 5S RNP which is then incorporated into nascent 60S subunits (see below).
Chaperones of ribosomal proteins
Ribosomal proteins are synthesized in the cytoplasm, but most are incorporated into pre‐ribosomal particles in the nucleolus, posing a logistic challenge to the cell. As RPs are enriched in basic amino acids and contain flexible tails as well as intrinsically disordered regions (Klinge et al, 2011; Rabl et al, 2011), they are prone to aggregation. Therefore, newly synthesized RPs need to be kept away from undesired interactions until they are incorporated into pre‐ribosomal particles. Several mechanisms contribute to avoiding adverse effects of unincorporated ribosomal proteins: dedicated chaperones for specific RPs (see below), the general chaperone network (Gong et al, 2009; Albanèse et al, 2010; Koplin et al, 2010; Leidig et al, 2013; Pillet et al, 2017) and association with nuclear transport receptors en route into the nucleus (Jäkel et al, 2002). In addition, excess, unincorporated RPs are degraded (Warner, 1977; Lam et al, 2007; Sung et al, 2016).
About a dozen dedicated chaperones for RPs have been described (Table 5). Some of them capture nascent RPs co‐translationally (e.g., Yar1 for Rps3/uS3, Rrb1 for Rpl3/uL3, Syo1 for Rpl5/uL18, Sqt1 for Rpl10/uL16, and Acl4 for Rpl4/uL4), whereas others bind their clients later (Pausch et al, 2015; Pillet et al, 2017). Interestingly, it has recently been revealed that the absence of the RP chaperones Acl4 or Rrb1 leads to a destabilization of the mRNAs encoding for their respective clients Rpl4 and Rpl3, revealing a novel regulatory mechanism of RP homeostasis (Pillet et al, 2022). RP chaperones not only shield their clients but can also aid their delivery into the nucleus. For those RPs that get incorporated into nascent subunits in the nuclear compartment, the nuclear localization signal can either be provided by the RP itself (e.g., Rps3/uS3, Rpl3/uL3, Rpl4/uL4), or by the dedicated chaperone (e.g., Syo1; Kressler et al, 2012; Bange et al, 2013; Calviño et al, 2015).
Table 5.
Chaperones of ribosomal proteins.
| Yeast | 40S | 60S | Human | 40S | 60S | Chaperoned RP | Citation |
|---|---|---|---|---|---|---|---|
| Rrb1 | x | GRWD1 | Rpl3/uL3 | Iouk et al (2001) | |||
| Acl4 | x | Rpl4/uL4 | Stelter et al (2015) | ||||
| Syo1 | x | HEATR3 | x | Rpl5/uL18 and Rpl11/uL5 | Kressler et al (2012), Hannan et al (2022) and O'Donohue et al (2022) | ||
| Sqt1 | x | AAMP | x | Rpl10/uL16 | Eisinger et al (1997) | ||
| Bcp1 | x | BCCIP | x | Rpl23/uL14 | Ting et al (2017) | ||
| Loc1 | x | Rpl43/eL43 | Liang et al (2019) | ||||
| Puf6 | x | Rpl43/eL43 | Liang et al (2019) | ||||
| Tsr4 | x | PDCD2 | Rps2/uS5 | Black et al (2019) and Rössler et al (2019) | |||
| Yar1 | x | Rps3/uS3 | Lindström and Zhang (2008) | ||||
| Nap1 | x | Rps6/eS6 | Rössler et al (2019) | ||||
| NPM | x | Rps9/uS4 | Lindström and Zhang (2008) | ||||
| Fap7 | x | AK/CINAP | x | Rps14/uS11, maybe also in complex with Rps26/eS26 | Hellmich et al (2013) and Peña et al (2016) | ||
| AROS | x | Rps19/eS19 | Singh et al (2021) | ||||
| Tsr2 | x | TSR2 | x | Rps26/eS26 | Schütz et al (2014) |
One peculiar case is yeast Tsr2, which serves as dedicated chaperone for Rps26/eS26. Tsr2 has been proposed to pick up newly synthesized Rps26/eS26 only after nuclear import by stimulating Rps26/eS26 release from importins in the nucleus, to then allow its nuclear association with pre‐40S subunits (Schütz et al, 2018). In the mammalian system, RPS26/eS26 is incorporated into the pre‐40S particle during its final cytoplasmic maturation (Ameismeier et al, 2018, 2020; Plassart et al, 2021). Here, TSR2 could help in preventing association of RPS26/eS26 with nuclear import receptors in the cytosol, but more work is needed to unravel the place of action of mammalian TSR2. Notably, also in yeast Tsr2 may harbor a cytosolic function, as it has recently been suggested to facilitate Rps26/eS26 release from and reincorporation into mature ribosomes in response to salt or pH stress (Yang & Karbstein, 2022).
The function of RP chaperones does not seem to be limited to factors acting in trans. In most eukaryotes, Rps31/RPS27A/eS31 and Rpl40/eL40 are initially synthesized as linear fusions with an N‐terminal ubiquitin, which facilitates their folding and enhances solubility of the respective RPs (Finley et al, 1989; Lacombe et al, 2009; Martín‐Villanueva et al, 2019, 2020, 2021). After synthesis, the ubiquitin moiety is rapidly released from these fusion proteins (Grou et al, 2015) and enters the cellular ubiquitin pool. Interestingly, in humans and other holozoan organisms, a second RP of the small subunit, RPS30/eS30, is synthesized as a fusion with a ubiquitin‐like protein called FUBI. Release of FUBI from the FUBI‐eS30 fusion protein is required for 40S subunit maturation, likely linked to its nuclear incorporation into pre‐40S subunits, and promoted by the deubiquitinase USP36 (van den Heuvel et al, 2021).
Formation and maturation of the SSU processome
When the pre‐rRNA emerges from the transcribing polymerase, it is soon bound by a subset of RPs of the small subunit, 40S RBFs as well as by small nucleolar ribonucleoproteins (snoRNPs), eventually giving rise to a large ribosomal pre‐particle that is referred to as the small subunit (SSU) processome or 90S pre‐ribosome (Fig 4) (Dragon et al, 2002; Grandi et al, 2002). Early pre‐rRNA processing events are executed during maturation of this complex, leading to the removal of the external transcribed spacer (5′ ETS) and cleavage within ITS1. Initially, high‐resolution structures of different fungal SSU processome particles were obtained, giving invaluable mechanistic insights into the function of a multitude of RBFs during early assembly, pre‐rRNA folding, and processing events (Kornprobst et al, 2016; Barandun et al, 2017; Chaker‐Margot et al, 2017; Cheng et al, 2017; Sun et al, 2017; Du et al, 2020; Lau et al, 2021). Then, in 2021, the first structures of the human SSU processome were published, providing visual evidence for the existence of SSU processome particles in higher eukaryotic cells and illustrating the vast conservation of its global architecture (Singh et al, 2021) (Fig 4, Table 6).
Figure 4. Structures of the yeast and human SSU processome.
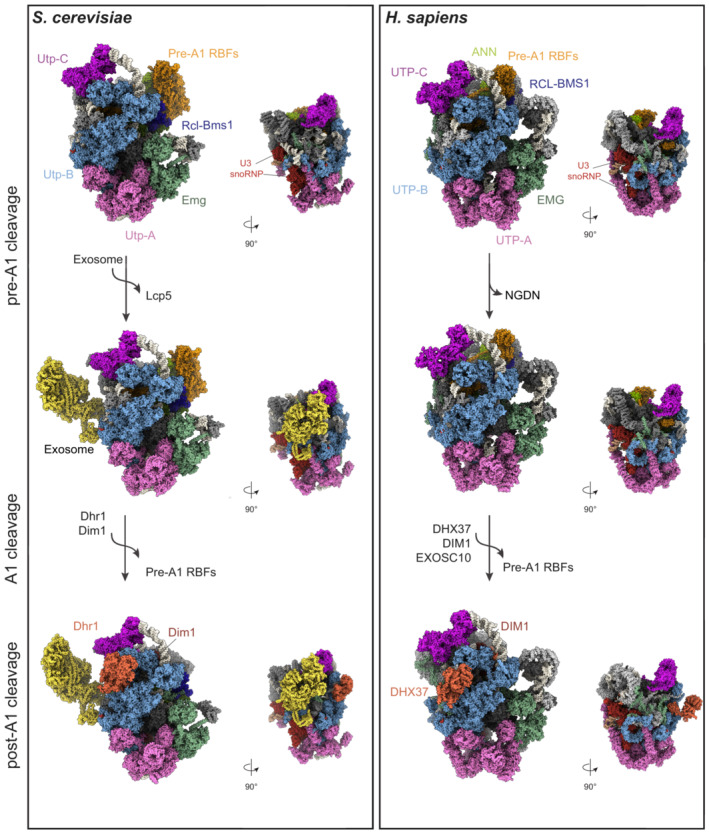
Cryo‐EM structures of the yeast (PDB ID: 6ZQB, 7AJT, and 7AJU) and human (PDB ID: 7MQ8, 7MQ9, 7MQA) SSU processome particles in the pre‐A1, pre‐A1* and post‐A1 cleavage states, showing a comparison of their overall architecture. The conserved RNA components and subcomplexes are color‐coded. The pre‐rRNA (white) and individual sub‐complexes such as UTP‐A (pink), UTP‐B (light blue), UTP‐C (violet), Emg1 (green), ANN (light green) complex, and additional RBFs, are shown as surfaces. KRR1/Krr1, C1orf131/Faf1, and the NAT10/Kre33 module are indicated as pre‐A1‐specific RBFs (sand). They are released after A1 cleavage. The exosome complex (yellow) is bound to the pre‐A1* yeast particle and it is associated only with the human post‐A1 structure. In the post‐A1 structures, DHX37/Dhr1 and DIM1/Dim1 are displayed in orange.
Table 6.
Small subunit processome subcomplexes and early assembly factors.
| Yeast | 40S | 60S | Human | 40S | 60S | Function in early nucleolar ribosome biogenesis | Citation |
|---|---|---|---|---|---|---|---|
| Utp4 | x | UTP4/CIRH1A | x | UTP‐A complex, interacts with the start of 5′ ETS, required for recruitment of UTP‐B and U3 snoRNP, essential for formation of 5′ ETS particle | Krogan et al (2004), Prieto and McStay (2007), Freed et al (2012), Chaker‐Margot et al (2017) and Singh et al (2021) | ||
| Utp5 | x | UTP5/WDR43 | x | ||||
| Utp8 | x | NOL11 | x | ||||
| Utp9 | x | ||||||
| Utp10 | x | UTP10/HEATR1 | x | ||||
| Utp15 | x | UTP15 | x | ||||
| Utp17 | x | UTP17/WDR75 | x | ||||
| Pol5 | x | x | MYBBP1A | x | |||
| Utp1/Pwp2 | x | PWP2 | x | UTP‐B complex, binds 5′ ETS and U3 snoRNA, supports structural pre‐rRNA remodeling, essential for formation of 5′ ETS particle, Utp18 contains exosome interaction motif | Krogan et al (2004), Sloan et al (2015), Barandun et al (2017), Barandun et al (2018) and Singh et al (2021) | ||
| Utp6 | x | UTP6 | x | ||||
| Utp12/Dip2 | x | UTP12/WDR3 | x | ||||
| Utp13 | x | TBL3 | x | ||||
| Utp18 | x | UTP18 | x | ||||
| Utp21 | x | WDR36 | x | ||||
| DDX21 | x | ||||||
| NOP2 | x | ||||||
| Nop1 | x | FBL | x | U3 snoRNP, U3 snoRNA base pairs with 5′ ETS, chaperones pre‐rRNA folding steps, essential for formation of 5′ ETS particle | Kiss‐László et al (1996), Grandi et al (2002), Barandun et al (2017) and Singh et al (2021) | ||
| Nop56 | x | NOP56 | x | ||||
| Nop58 | x | NOP58 | x | ||||
| Snu13 | x | SNU13/15.5 K | x | ||||
| Rrp9 | x | U3‐55K/U3IP2 | x | ||||
| Mpp10 | x | MPHOSPH10 | x | Mpp10‐Imp3‐Imp4 complex, interacts with U3 snoRNA, supports formation of 5′ ETS particle | Lee and Baserga (1999) and Granneman et al (2003) | ||
| Imp3 | x | IMP3 | x | ||||
| Imp4 | x | IMP4 | x | ||||
| Rrp7 | x | RRP7A | x | UTP‐C complex, chaperones 5′ domain of 18S rRNA | Baudin‐Baillieu et al (1997), Krogan et al (2004), Rudra et al (2007), Barandun et al (2017) and Singh et al (2021) | ||
| Utp22 | x | NOL6 | x | ||||
| Cka1 | x | CK2A1 | x | ||||
| Cka2 | x | CK2A2 | x | ||||
| Ckb1 | x | CK2N | x | ||||
| Ckb2 | x | CK2N | x | ||||
| Rrp36 | x | RRP36 | x | ||||
| Utp7 | x | WDR46 | x | Sof1‐Utp7 complex, aids organization of A1 cleavage site | Barandun et al (2017) | ||
| Utp14 | x | UTP14A | x | ||||
| Sof1 | x | WDSOF1 | x | ||||
| Rcl1 | x | RCL1 | x | Rcl1‐Bms1‐complex, GTPase activity (Bms1), mediates cleavage at A2 site, also required for A0 and A1 cleavage, interacts with U3 snoRNP | Karbstein and Doudna (2006) and Horn et al (2011) | ||
| Bms1 | x | BMS1 | x | ||||
| Enp1 | x | ENP1/BYSL | x | In yeast: Nop14‐Noc4‐Enp1 complex, in humans: NOP14‐NOC4L‐UTP14A‐EMG1, structural role: binds to 3′ domain of 18S pre‐RNA, Emg1: methyltransferase | Liu and Thiele (2001), Kühn et al (2009), Warda et al (2016) and Barandun et al (2017) | ||
| Utp2/Nop14 | x | NOP14 | x | ||||
| Noc4 | x | NOC4L | x | ||||
| Emg1 | x | EMG1 | x | ||||
| Bfr2 | x | AATF | x | in humans: ANN complex, required for cleavage at A0, 1 and in the ITS1, in yeast: Bfr2‐Enp2 recruit Dpb4 | Soltanieh et al (2014) and Bammert et al (2016) | ||
| Lcp5 | x | NGDN | x | ||||
| Enp2 | x | NOL10 | x | ||||
| XRN2 | x | XND complex, required for A′ cleavage, recruits XRN2 for degradation of excised spacer fragment | Dragon et al (2002) and Memet et al (2017) | ||||
| NKRF | x | ||||||
| Prp43 | DHX15 | x | |||||
| Utp30 | x | RSL1D1 | x | Utp30‐Rrt14 complex, binds 5′ ETS and pre‐18S rRNA, Rrt14 is non‐essential | Barandun et al (2017) | ||
| Rrt14 | x | ||||||
| Utp11 | x | UTP11 | x | ||||
| Bud21/Utp16 | x | NOL7 | x | Required for 18S pre‐rRNA processing | Dragon et al (2002) and Singh et al (2021) | ||
| Fcf2 | x | TDIF2 | x | Binds U3 snoRNP, TDIF2 contains AIM putative motif | Rempola et al (2006), Barandun et al (2017) and Singh et al (2021) | ||
| Sas10 | x | UTP3 | x | Stabilizes and chaperones Mpp10 complex to nucleolus, blocks Emg1 active site | Zhao et al (2019) | ||
| Utp24 | x | UTP24 | x | Endoribonuclease, couples pre‐rRNA cleavages in yeast at sites A1 and A2, in humans: site 1 and E | Bleichert et al (2006) and Wells et al (2016) | ||
| Esf1 | x | ESF1 | Involved in early pre‐rRNA processing | Peng et al (2004) | |||
| Esf2 | x | ABT1 | Stimulates Dbp8 | Granneman et al (2006b) | |||
| Dbp8 | x | DDX49 | x | DEAD box RNA helicase | Granneman et al (2006b) and Awasthi et al (2018) | ||
| Dbp4 | x | DDX10 | x | DEAD box RNA helicase | Granneman et al (2006a) and Turner et al (2009) | ||
| Rrp3 | x | DDX47 | x | DEAD box RNA helicase | Granneman et al (2006a) | ||
| Bud22 | x | SRFBP1 | Dakshinamurthy et al (2010) | ||||
| Efg1 | x | Required for 18S pre‐rRNA processing (A1, A2), initiates degradation of aberrant 23S pre‐rRNA | Choque et al (2018) | ||||
| Rrp5 | x | x | PDCD11 | x | x | Structural SSU component supporting pre‐rRNA, compaction, important for 18S maturation (site A0‐A2) and 5.8S processing (site A3) | Venema and Tollervey (1996) and Lebaron et al (2013) |
| Krr1 | x | KRR1 | x | Interacts with Faf1, important for 40S platform, assembly as it is replaced by Dim2 | Zheng et al (2014) and Sturm et al (2017) | ||
| Rok1 | x | DDX52 | x | DEAD box RNA helicase, releases Rrp5, releases snR30 | Khoshnevis et al (2016) | ||
| Utp25 | x | DEF/C1orf107 | x | Charette and Baserga (2010) and Tao et al (2017) | |||
| Kri1 | x | KRI1 | Interacts with Krr1 | Sasaki et al (2000) | |||
| Utp23 | x | UTP23 | x | Endoribonuclease, in yeast: likely inactive, in humans: cleavage at site A0 | Wells et al (2017) | ||
| Fyv7 | x | Peng et al (2003) | |||||
| Mrd1 | x | RBM19 | x | Aids formation of central pseudoknot, required for dynamic U3 snoRNA‐rRNA interaction (release) | Segerstolpe et al (2013) and Lackmann et al (2018) | ||
| Fal1 | x | DDX48/EIF4A3 | x | DEAD box RNA helicase | Kressler et al (1997) and Davila Gallesio et al (2020) | ||
| Sgd1 | x | NOM1 | x | Fal1 cofactor | Davila Gallesio et al (2020) | ||
| Cms1 | x | CMS1 | x | Grandi et al (2002) | |||
| Nop9 | x | NOP9 | Impedes Nob1 cleavage | García‐Gómez et al (2011) and Zhang et al (2016a) | |||
| Nop6 | x | Not essential for ribosome biogenesis | García‐Gómez et al (2011) | ||||
| Utp20 | x | UTP20 | x | Associates with faulty intermediates, might play a role in rRNA quality control | Dez et al (2007) | ||
| Kre33 | x | NAT10 | x | ATP‐dependent RNA acetyltransferase | Ito et al (2014) and Sharma et al (2017, 2015) | ||
| Faf1 | x | C1orf131 | Interacts with Krr1/KRR1 | Zheng et al (2014) and Singh et al (2021) | |||
| Dim2/Pno1 | x | PNO1/DIM2 | x | Interacts with Nob1 | Chaker‐Margot et al (2017) | ||
| Rrp12 | x | RRP12 | x | Part of the 3′ minor domain | Ameismeier et al (2018) | ||
| Nob1 | x | NOB1 | x | Endoribonuclease, associates with ITS1, catalyzes removal of final part of ITS1 (site D/3), might only associate in nucleoplasm | Fatica et al (2004) | ||
| Dhr1 | x | DHX37 | x | DEAH box RNA helicase, releases U3 snoRNA | Boneberg et al (2019) | ||
| Dhr2 | x | DEAH box RNA helicase, interacts with Utp25 and Nop19 | Granneman et al (2006a) | ||||
| Nop19 | x | Important for Utp25 incorporation | Choque et al (2011) | ||||
| Rrp8 | x | NML/RRP8 | x | Methyltransferase | Peifer et al (2013) | ||
| Slx9 | x | FAM207A/C21orf70 | Non‐essential, supports efficient ITS1 processing | Bax et al (2006) | |||
| Pol5 | x | x | MYBBP1A | x | Turnover of 5′ ETS fragment, required for recycling of SSU RBFs | Braun et al (2020) | |
| Bud23 | x | WBSCR22 | x | Methyltransferase, supports disassembly of SSU particle | Black et al (2020) | ||
| Nip7 | x | NIP7 | x | In contrast to yeast, human NIP7 and FTSJ3 were shown to function in 40S biogenesis | Morello et al (2011a) | ||
| Sbp1 | x | FTSJ3 | x |
Small subunit processome assembly is initiated when the 5′ ETS region of the nascent pre‐rRNA transcript is bound by the so‐called UTP‐A complex, a seven‐membered protein complex that provides binding interfaces for later joining SSU subcomplexes (Pérez‐Fernández et al, 2007; Zhang et al, 2016b; Barandun et al, 2017; Cheng et al, 2017; Hunziker et al, 2019). The UTP‐A complex is also needed for pre‐rRNA synthesis, thereby linking pre‐rRNA transcription and ribosome assembly (Gallagher et al, 2004). Following the UTP‐A complex, the six‐membered UTP‐B complex and the U3 snoRNP are co‐transcriptionally recruited to the 5′ ETS (Chaker‐Margot et al, 2015; Hunziker et al, 2016; Kornprobst et al, 2016). The UTP‐B complex acts as a chaperone for both the 5′ ETS and the U3 snoRNA, and upon incorporation of additional factors, including the Mpp10 complex (Granneman et al, 2003), a large 5′ ETS‐associated particle comprising more than 25 RBFs is formed (Chaker‐Margot et al, 2015; Zhang et al, 2016b; Barandun et al, 2017). Within the 5′ ETS‐associated particle, the U3 snoRNP serves as a key organizer orchestrating SSU processome formation. In both human and yeast cells, the U3 RNA base pairs with two regions each of the 5′ ETS and the 18S rRNA (Granneman et al, 2009; Dutca et al, 2011; Barandun et al, 2017; Singh et al, 2021), thereby functioning as a critical structural constraint for pre‐rRNA folding during SSU processome assembly (Fig 4). Importantly, this prevents the premature formation of the central pseudoknot, a universally conserved element of rRNA tertiary structure that is part of the small ribosomal subunit decoding center (Sardana et al, 2015). Notably, many RBFs within the SSU processome bind to more than one site of the pre‐rRNA, thereby reducing the conformational freedom of the rRNA during folding. In particular, SSU RBFs such as Faf1, Utp11, Mpp10, Sas10, and Nop14 exhibit long extensions that bridge distant rRNA regions within these precursors, thereby contributing to correct pre‐rRNA organization (Chaker‐Margot et al, 2017; Cheng et al, 2017).
A striking difference between yeast and mammalian cells is the length of the 5′ ETS that comprises roughly 700 nucleotides in yeast, but is extended to 3,600 nucleotides in humans. Only relatively short, structured parts of the 5′ ETS could be visualized in the structural models of the human SSU processome (Singh et al, 2021). Surprisingly, a minimal human 5′ ETS comprising these RNA segments (in total only ~25% of the entire 5′ ETS region) is sufficient for correct 40S and 60S assembly (Singh et al, 2021). In light of this data, the function of the other 5′ ETS regions is enigmatic. It has been proposed that they might contribute to nucleolar organization by supporting the formation of biomolecular condensates and thereby nucleolar phase separation (Yao et al, 2019). Along the same line of thought, it is conceivable that the increase in 5′ ETS length contributes to the observed differences in subnucleolar organization between mammals and yeast.
The 5′ ETS particle has originally been suggested to serve as a binding platform for the hierarchical recruitment of further protein complexes to sequentially promote the folding of spatially distant rRNA domains (Kornprobst et al, 2016; Cheng et al, 2017; Sun et al, 2017). However, since individual 18S rRNA domains can in principle recruit their respective biogenesis factors independently, it has also been proposed that the 5′ ETS and downstream 18S rRNA domains may function as independent units in the recruitment of their respective assembly factor complexes which may then support SSU processome assembly based on their mutual dependence (Hunziker et al, 2019). In either case, further assembly factors are recruited to the nascent rRNA concomitant with ongoing transcription in an rRNA‐subdomain dependent fashion (Pérez‐Fernández et al, 2007), among them the Bms1‐Rcl1 complex, the Nat10/Kre33 module (Kre33–Brf2–Lcp5–Enp2 in yeast/NAT10‐(AATF‐NGDN‐NOL10) (ANN) complex in humans) and the UTP‐C complex (Bammert et al, 2016; Barandun et al, 2018). Notably, also other snoRNAs including U14, snR30/U17, and snR10 transiently associate with the 5′ and central domains of the 18S rRNA (Zhang et al, 2016b) and aid 18S rRNA processing and folding. In the course of SSU processome assembly, cleavage at site A0 in the 5′ ETS occurs, eventually giving rise to a large, stable intermediate referred to as the “pre‐A1” particle in which site A1 is still uncut (Fig 4) (vanden Broeck & Klinge, 2022). The human pre‐A1 particle is 3.3 MDa in size and contains about 21 RPs and 50 RBFs (Singh et al, 2021).
To initiate cleavage at site A1, which generates the mature 5′ end of the 18S rRNA, the PIN domain endonuclease Utp24/UTP24 must gain access to the processing site (Tomecki et al, 2017; Barandun et al, 2018, 2017; Singh et al, 2021; vanden Broeck & Klinge, 2022). This involves a series of events, commencing with the release of Lcp5/NGDN (state pre‐A1*; Fig 4), followed by structural remodeling of the SSU particle that affects the positioning of the U3 snoRNA. Particle remodeling is associated with the release of the Utp24/UTP24 inhibitory factor Faf1/C1ORF131 and the residuals of the Kre33/NAT10 module (state post A1; Fig 4) (Cheng et al, 2020; Du et al, 2020; Singh et al, 2021). What drives these events and how the enzymatic activity of Kre33/NAT10 is coordinated with SSU processome maturation, potentially aided by the juxtaposed ANN complex or other factors, remains to be unraveled. Importantly, also the nuclear exosome may play a crucial role in the remodeling of the SSU processome (Du et al, 2020) by driving the 3′–5′ unwinding of the 5′ ETS after cleavage at site A0 (Du et al, 2020; Lau et al, 2021), contributing to structural remodeling of the SSU‐bound 5′ ETS particle and the SSU “head domain”. Recent structural and biochemical data obtained from yeast indeed revealed that the nuclear exosome already binds 90S particles prior to cleavage at site A1 as visualized in pre‐A1* particles (Fig 4) (Du et al, 2020; Lau et al, 2021). Yet, in humans, the exosome was only found to be associated with the post‐A1 structure and its binding sites are blocked in the described human pre‐A1 particles (Singh et al, 2021).
The disassembly of the SSU processome involves the activity of the conserved RNA helicase Dhr1/DHX37 (Sardana et al, 2015; Boneberg et al, 2019; Choudhury et al, 2019), which drives the release of the U3 snoRNP to allow the formation of the conserved pseudoknot in the 18S rRNA. Dhr1/DHX37 is recruited to the SSU processome already upon release of the ANN complex, and initially kept in an inhibited state. Activation is tightly linked to the choreography of SSU processome disassembly, when the RNA helicase gains spatial proximity to its activator UTP14 (Boneberg et al, 2019; Choudhury et al, 2019; Singh et al, 2021). Recent in vitro work reconstituted the Dhr1‐dependent release of the U3 snoRNA, allowing the visualization of these ATP‐dependent remodeling steps and details of central pseudoknot maturation (Cheng et al, 2022).
Cleavage in the ITS1 leads to the separation of pre‐40S and pre‐60S particles, which further mature independently in nucleoli and nucleoplasm. In both yeast and human, the ITS1 is cleaved at two sites (site A2 and A3, E and 2, respectively) by the conserved enzymes UTP24 (site A2, E) and RMRP (site A3, 2) (Udem et al, 1971; Lygerou et al, 1996; Rouquette et al, 2005; Tomecki et al, 2017; Aubert et al, 2018). The order of cleavages however differs: in yeast cleavage at site A2 by Utp24 is responsible for splitting the pre‐ribosomal particles, while in human cells the majority of pre‐rRNAs is first processed at site 2 by RMRP resulting in pre‐40S and pre‐60S separation (Allmang et al, 2000; Preti et al, 2013; Sloan et al, 2013b).
Nuclear maturation and export of the pre‐40S subunit
Recent structural analyses provided insights into the molecular organization of a series of nucleoplasmic 40S precursors from yeast and human cells (Cheng et al, 2022). Early nucleoplasmic 40S assembly intermediates already possess many features of mature 40S subunits, except that the head region remains largely delocalized and helix 18 of the 18S rRNA is kept in an immature conformation (Cheng et al, 2022). This is achieved by Bud23/WBRSC22, the binding of which has been implicated in the remodeling of the SSU processome into immature 40S particles (Black et al, 2020; Cheng et al, 2022). Bud23/WBRSC22 together with its adaptor Trm112/TRMT112 also acts as an RNA methyltransferase factor and is required for the methylation of a conserved guanosine in the P‐site of the 40S subunit (White et al, 2008; Figaro et al, 2012; Létoquart et al, 2014; Zorbas et al, 2015). Besides Bud23/WBRSC22, Tsr1/TSR1 and Slx9/C21orf70/FAM207A are also already bound to these early nucleoplasmic precursors (Table 7).
Table 7.
Factors involved in nucleoplasmic steps of pre‐40S maturation.
| Yeast | 40S | 60S | Human | 40S | 60S | Function in nucleoplasmic steps of pre‐40S maturation | Citation |
|---|---|---|---|---|---|---|---|
| Tsr1 | x | TSR1 | x | Associates | Gelperin et al (2001) | ||
| Nob1 | x | NOB1 | x | Endoribonuclease, associates with ITS1, catalyzes removal of final part of ITS1 (site D/3), might associate earlier | Fatica et al (2004), Zemp et al (2009) and Ameismeier et al (2018) | ||
| Hrr25 | x | CKI‐δ/CSNK1D | x | Associates | Schäfer et al (2006) and Zemp et al (2014) | ||
| Hrr25 | x | CKI‐ε/CSNK1E | x | Associates | Schäfer et al (2006) and Zemp et al (2014) | ||
| Rio1 | x | RIOK1/RIO1 | x | Associates | Rouquette et al (2005) and Widmann et al (2012) | ||
| Rio2 | x | RIOK2/RIO2 | x | Associates | Vanrobays et al (2003) and Zemp et al (2009) | ||
| Ltv1 | x | LTV1 | x | Associates | Seiser et al (2006) and Zemp et al (2014) | ||
| PARN | x | Processes 3′ end of the 18S pre‐rRNA | Montellese et al (2017) | ||||
| Slx9 | FAM207A/C21orf70 | x | Wyler et al (2011) |
The addition of further factors, including Hrr25/CKIδ/ε and Ltv1/LTV1, into the pre‐40S subunits then results in partial stabilization of the 40S head and neck region (Cheng et al, 2022). The large RBF RRP12, which embraces the immature head region, is central in further steps of head maturation. These entail the sequential joining of a set of ribosomal proteins, RPSA/uS2, RPS2/uS5, and RPS21/eS21, recruitment of Nob1/NOB1, and conformational changes in several rRNA helices (Ameismeier et al, 2018; Cheng et al, 2022). Incorporation of these RPs is followed by the release of Bud23, which occurs in coordination with the recruitment of Rio2 that associates with the decoding center between the platform and the head region, prior to nuclear export (Heuer et al, 2017; Ameismeier et al, 2018; Black & Johnson, 2022; Cheng et al, 2022). Stabilization of the head region thus precedes nuclear export of pre‐40S subunits, while beak formation is completed later in the cytoplasm.
Of the RBFs present in these 40S precursors, several are early‐associating RBFs that remain bound throughout maturation in the nucleoplasm and even accompany 40S precursors to the cytoplasm, namely Enp1/BYSL, Dim2/PNO1, and RRP12 (Table 6). One notable early‐binding RBF is the methyltransferase Dim1/DIM1, which remains particle‐bound until it methylates two adenosines in the 3′ region of the 18S rRNA (Lafontaine et al, 1994). While this occurs during cytoplasmic maturation steps in yeast, human DIM1 performs the corresponding modification earlier, in the nucleus, where it dissociates from maturing particles before they are eventually exported to the cytoplasm (Wyler et al, 2011; Zorbas et al, 2015).
Compared to yeast, the processing of the ITS1 is more complex in higher eukaryotes and involves an additional endonucleolytic cleavage step, generating the 21S pre‐rRNA (Fig 3). Moreover, 21S pre‐rRNA maturation also includes exonucleases, namely the exosome with its catalytic subunits DIS3 and EXOSC10, and the poly(A)‐specific ribonuclease PARN (Preti et al, 2013; Tafforeau et al, 2013; Sloan et al, 2013b; Montellese et al, 2017).
The association of several conserved RBFs, including Nob1/NOB1, Rio2/RIOK2, Ltv1/LTV1, Slx9/C21orf70/FAM207A, and Tsr1/TSR1 (Table 7) with 40S precursors during nuclear 40S maturation paves the way for nuclear export (Ferreira‐Cerca et al, 2005, 2007; Schäfer et al, 2006; Carron et al, 2011; Wyler et al, 2011; Zemp et al, 2014; Heuer et al, 2017; Ameismeier et al, 2018; Cheng et al, 2022). Pre‐ribosomal particles are among the largest transport cargos that pass through nuclear pore complexes (NPCs) and they need to be bound by multiple nuclear export receptors for NPC passage (Table 8). Binding of export factors to ribosomal pre‐particles is thought to function as a quality control step of subunit maturation (Johnson et al, 2002; Woolford & Baserga, 2013). In both yeast and mammalian cells, export of pre‐40S subunits depends on the RanGTP‐binding exportin Crm1/XPO1 (Hurt et al, 1999; Moy & Silver, 1999; Thomas & Kutay, 2003; Wild et al, 2010). The yeast 40S RBFs Dim2, Rio2, and Ltv1 contain nuclear export sequences (NES) and have been suggested to serve as redundant adaptors for Crm1 binding to the small subunit (Schäfer et al, 2003; Vanrobays et al, 2003, 2008; Seiser et al, 2006; Merwin et al, 2014). Further factors linked to pre‐40S export in yeast are the mRNA export factor Mex67/Mtr2 (Faza et al, 2012), Rrp12 (Oeffinger et al, 2004), and Slx9, the latter with a proposed role in mediating Crm1 binding to Rio2 (Fischer et al, 2015). Its human homolog SLX9/C21orf70/FAM207A is also associated with nucleoplasmic pre‐40S particles (Wyler et al, 2011), but is not known to accompany pre‐40S subunits into the cytoplasm. Like in yeast, pre‐40S export in human cells also exploits the atypical protein kinase RIOK2 as an adaptor for XPO1 (Zemp et al, 2009). Other NES‐containing RBFs are expected to contribute to the recruitment of XPO1 (Zemp et al, 2009), among them PDCD2L, which was suggested to support pre‐40S export in mammals (Landry‐Voyer et al, 2016).
Table 8.
Factors involved in nuclear export of ribosomal pre‐particles.
| Yeast | 40S | 60S | Human | 40S | 60S | Function in nuclear export | Citation |
|---|---|---|---|---|---|---|---|
| Crm1/Xpo1 | x | x | XPO1/CRM1 | x | x | Facilitates export in RanGTP‐dependent manner | Hurt et al (1999) and Thomas and Kutay (2003) |
| Dim2/Pno1 | x | PNO1/DIM2 | x | Possible adaptor for XPO1‐mediated export | Vanrobays et al (2008) | ||
| Rio2 | x | RIOK2 | x | Possible adaptor for XPO1‐mediated export | Vanrobays et al (2003) and Zemp et al (2014) | ||
| Ltv1 | x | LTV1 | x | Possible adaptor for XPO1‐mediated export | Seiser et al (2006) | ||
| PDCD2L | x | Possible adaptor for XPO1‐mediated export | Landry‐Voyer et al (2016) | ||||
| Yrb2 | x | RANBP3 | x | Stage‐Zimmermann et al (2000) and Badertscher et al (2015) | |||
| Slx9 | x | FAM207A/C21orf70 | Fischer et al (2015) | ||||
| Rrp12 | x | x | RRP12 | Potential function in export, interaction with FG repeat nucleoporins in vitro | Oeffinger et al (2004) | ||
| Mex67 | x | x | NXF1 |
Mex67‐Mtr2 complex Export receptor |
Yao et al (2008, 2007) | ||
| Mtr2 | x | x | NXT1 | ||||
| Nmd3 | x | NMD3 | x | Adaptor for XPO1‐mediated export | Ho et al (2000) and Thomas and Kutay (2003) | ||
| Arx1 | x | PA2G4/EBP1 | Induces structural changes allowing export | Bradatsch et al (2007) | |||
| Bud20 | x | ZNF593 | Binds FG repeats | Altvater et al (2012) | |||
| Ecm1 | x | Binds FG repeats | Yao et al (2010) | ||||
| Npl3/Nop3 | x | SRSF1 | Binds FG repeats | Hackmann et al (2011) | |||
| Gle2/Rae1 | x | RAE1 | x | x | Binds to Nup116 and recruits pre‐60S via second binding site | Wild et al (2010) and Occhipinti et al (2013) | |
| Msn5 | XPO5 | x | Facilitates export in RanGTP‐dependent manner | Wild et al (2010) |
Cytoplasmic steps of 40S subunit biogenesis
In the cytoplasm, several final maturation steps occur on both ribosomal pre‐particles, including incorporation of late‐assembling RPs, structural rearrangements, final pre‐rRNA processing steps, and the release of late‐acting RBFs (Zemp & Kutay, 2007; Nerurkar et al, 2015). Several RBFs accompany these particles from the nucleus to the cytoplasm and are thought to prevent premature 40S‐60S joining and translation initiation by keeping the particle in an inactive conformation and shielding functional sites on the subunit interface (Greber, 2016).
In yeast, exported pre‐40S particles are bound by the RBFs Dim1, Dim2, Enp1, Nob1, Hrr25, Rio2, Rrp12, and Tsr1 (Schäfer et al, 2006). The function of these factors is widely conserved in mammalian cells, although DIM1 acts earlier and is not part of human late 40S precursors (Table 9). Recently published cryo‐EM structures of cytoplasmic 40S pre‐particles from yeast and human cells have highlighted that cytoplasmic 40S maturation mostly involves structural changes in the head and beak region, formation of the decoding center by rearrangement of helix 44 (h44) and final processing of the 18S pre‐rRNA (Fatica et al, 2003; Lamanna & Karbstein, 2011; Larburu et al, 2016; Heuer et al, 2017; Scaiola et al, 2018; Ameismeier et al, 2020, 2018). These steps are coordinated by the conserved kinases Hrr25/CKIδ/ε, Rio2/RIOK2, and Rio1/RIOK1 (Vanrobays et al, 2003, 2001; Rouquette et al, 2005; Widmann et al, 2012; Ferreira‐Cerca et al, 2014, 2012; Zemp et al, 2014, 2009; Mitterer et al, 2019; Plassart et al, 2021), with an additional kinase, RIOK3, supporting 18S pre‐rRNA processing in mammalian cells (Baumas et al, 2012; Widmann et al, 2012). While Hrr25/CKIδ/ε is thought to phosphorylate and thereby trigger the release of Enp1/BYSL and Ltv1/LTV1 (Schäfer et al, 2006; Zemp et al, 2014), no substrates are known for the RIO kinases. Both Rio1 and Rio2 have been suggested to act as ATPases rather than kinases (Ferreira‐Cerca et al, 2012, 2014), with respective conformational changes regulating their association with 40S precursors. It is still unclear whether the kinase activity of Rio kinases is indeed exploited for structural remodeling of pre‐40S particles as originally suggested (Ferreira‐Cerca et al, 2012).
Table 9.
Factors involved in cytoplasmic pre‐40S maturation.
| Yeast | 40S | 60S | Human | 40S | 60S | Function in cytoplasmic pre‐40S maturation | Citation |
|---|---|---|---|---|---|---|---|
| Enp1 | x | ENP1/BYSL | x | Released by Hrr25/CKI phosphorylation, Release allows stable incorporation of Rps3/uS3, Rps10/eS10 and Rps20/uS10, Formation of the beak structure | Schäfer et al (2006), Zemp et al (2014) and Ameismeier et al (2018) | ||
| Ltv1 | x | LTV1 | x | ||||
| Tsr1 | x | TSR1 | x | Released, occludes binding sites for mRNA and translation initiation factors | Larburu et al (2016) and Scaiola et al (2018) | ||
| Dim1 | x | DIMT1 | x | Methyltransferase, yeast: modifies subsequent adenines near 3′ end of 18S rRNA; human: function occurs already in the nucleus | Strunk et al (2011) and Zorbas et al (2015) | ||
| Rrp12 | x | RRP12 | x | Released, which allows incorporation of Asc1 (RACK1 in humans) | Wyler et al (2011) and Larburu et al (2016) | ||
| Dim2/Pno1 | x | PNO1/DIM2 | x | Inhibits binding of eIF3; release contributes to Major structural rearrangements, allowing Nob1 activity | Ameismeier et al (2018) and Scaiola et al (2018) | ||
| Nob1 | x | NOB1 | x | Endonucleolytic cleavage of 20S pre‐rRNA (18S‐E pre‐rRNA in humans) to yield mature 18S rRNA | Fatica et al (2003) and Lamanna and Karbstein (2011) | ||
| Hrr25 | x | CKI‐δ/CSNK1D | x | Phosphorylation of Enp1‐Ltv1‐Rps3/uS3 complex, which triggers release of Ltv1 | Schäfer et al (2006) and Zemp et al (2014) | ||
| CKI‐ε/CSNK1E | x | ||||||
| Rio1 | x | RIOK1/RIO1 | x | Orchestrates structural changes, pre‐rRNA maturation and trans‐acting factor release | Vanrobays et al (2003) and Widmann et al (2012) | ||
| Rio2 | x | RIOK2/RIO2 | x | Kinase contributes to pre‐rRNA maturation and trans‐acting factor release | Geerlings et al (2003) and Zemp et al (2009) | ||
| RIOK3 | x | Promotes 18S‐E processing | Geerlings et al (2003) | ||||
| Prp43 | x | DHX15 | Conformational pre‐rRNA switch, which allows Nob1 activity | Pertschy et al (2009) | |||
| Pfa1/Sqs1 | x | SON | x | Cofactor for Prp43 | Sloan et al (2013b) | ||
| Pxr1/Gno1 | x | PINX1 | x | Cofactor for Prp43 | Guglielmi and Werner (2002) and Chen et al (2014) | ||
| Hbs1 | x | HBS1 | In yeast: facilitates formation of 80S like particle | Lebaron et al (2012), Strunk et al (2012) and Hector et al (2014) | |||
| Dom34 | x | PELO | |||||
| Rli1 | x | ABCE1 | |||||
| Fun12 | x | EIF5B | |||||
| USP16 | x | Deubiquitylates lysine 113 in RPS27A/eS31 | Montellese et al (2020) | ||||
| LRRC47 | x | Bound to late cytoplasmic pre‐40S particle | Ameismeier et al (2020) | ||||
| EIF1AD | x | Supports trans‐acting factor release and pre‐rRNA maturation | Ameismeier et al (2020) |
Intriguingly, the endonuclease Nob1/NOB1, which mediates the final cleavage of 20S pre‐rRNA (18S‐E in humans) to mature 18S rRNA, is already associated with nuclear pre‐40S particles. However, its access to the cleavage site at the 3′ end of 18S rRNA is restricted by Dim2/PNO1, thereby preventing premature removal of the remaining ITS1 fragment (Fig 5) (Turowski et al, 2014; Scaiola et al, 2018; Ameismeier et al, 2020). It has been suggested that in yeast, the formation of mature 18S rRNA is supported by the interaction of pre‐40S particles with mature 60S particles forming an 80S‐like complex, stimulated by the translation initiation factor Fun12/eIF5B (Lebaron et al, 2012; Strunk et al, 2012). The formation of an 80S‐like particle has not been described for human cells. However, two additional, human‐specific cytoplasmic RBFs were observed on late particles, namely EIF1AD and LRRC47 (Ameismeier et al, 2020; Montellese et al, 2020; Plassart et al, 2021). While LRRC47 associates with the subunit interface and might prevent premature 60S joining, binding of EIF1AD leads to a series of events, including repositioning of RIOK1 and the central helix h44, triggering PNO1 release and final pre‐rRNA processing by NOB1. These final maturation steps, entailing incorporation of RPS26/eS26, ATP hydrolysis on RIOK1, and dissociation of the few remaining RBFs, renders 40S subunit competent for 60S joining and mRNA translation (Plassart et al, 2021). The deubiquitinase USP16, which deubiquitylates RPS27A/eS31 in a translation‐dependent manner, supports these last maturation events in a not yet fully understood fashion, potentially linking surveillance of subunit maturation to translation initiation (Montellese et al, 2020).
Figure 5. Overview of late maturation steps of the small ribosomal subunit.
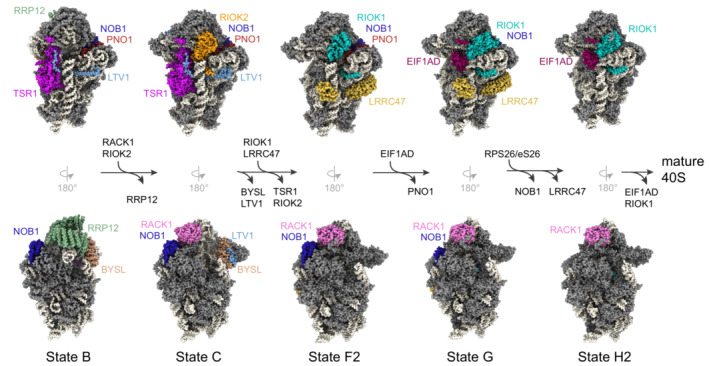
Front and back views of human pre‐40S particles at different stages of cytoplasmic maturation as derived from cryo‐EM analyses (PDB ID: 6G4S, 6G18, 6ZXE, 6ZXF, 6ZXH). Factors involved in these steps are color‐coded and pre‐rRNA is shown in white. After RRP12 release from the state B particle, RACK1 occupies its place and the pre‐rRNA is rearranged for head formation. PNO1 directly interacts with NOB1 and keeps it in an inactive state (from state B to F2). Association of EIF1AD, concomitant with rearrangements of RIOK1, triggers PNO1 dissociation, RPS26/eS26 incorporation, and final pre‐rRNA processing. LRRC47 association prevents 60S joining until the mature decoding region is formed.
Nucleolar pre‐60S biogenesis
After the subunit separating cleavage in the ITS1, the 60S precursor matures independently of the 40S subunit. At this point, the pre‐60S subunit contains the 27S (yeast) or 32S pre‐rRNA (human) that comprises both the 5.8S and 25S/28S rRNA portions (Fig 3) (Kater et al, 2017). Based on conserved secondary and tertiary structures, the 25/28S rRNA is subdivided into six domains, named I to VI from 5′ to 3′, which fold in a hierarchical process (Gamalinda et al, 2014; Klinge & Woolford, 2019). While the two most 5′ domains of the 25S rRNA were observed to fold first (Zhou et al, 2019a), later steps of 60S domain formation are more complex and intertwined as they do not follow the order of domain transcription, as illustrated by structural snapshots of several pre‐60S particles (Wu et al, 2016; Kater et al, 2017, 2020; Sanghai et al, 2018; Kargas et al, 2019; Zhou et al, 2019a, 2019b). Cryo‐EM structures of the first nucleolar pre‐60S particles could not be obtained so far, presumably due to the high flexibility and heterogeneity of these complexes, in which RNA–RNA and RNA‐protein interactions are gradually established (Burlacu et al, 2017; Pöll et al, 2017). In a recent study, an early pre‐60S was visualized in a bipartite structure with a 90S particle, but the existence of this intermediate needs to be confirmed in wild‐type cells (Ismail et al, 2022). The earliest visualized yeast particles reveal that 60S biogenesis first yields the solvent exposed subunit surface, initiated by folding of the 25S domains I and II (Zhou et al, 2019a). This pre‐particle is stabilized by several RBFs, including the Nsa1 module at the solvent‐exposed side. In intermediate pre‐60S particles, folding and positioning of domain VI toward the forming core can be observed, as well as folding of the pre‐5.8S rRNA, while domains III‐V remain too flexible to be resolved (Kater et al, 2017; Sanghai et al, 2018; Zhou et al, 2019a). In later stages of nuclear pre‐60S maturation (Fig 6), domains III, IV, and V, progressively fold and are positioned in the context of the maturing particle, leading to the gradual formation of PET, PTC, and the 60S/40S subunit interface (Barrio‐Garcia et al, 2016; Wu et al, 2016; Kater et al, 2017; Ma et al, 2017; Malyutin et al, 2017; Zhou et al, 2019b).
Figure 6. Overview of key nuclear maturation events of the large ribosomal subunit in yeast.
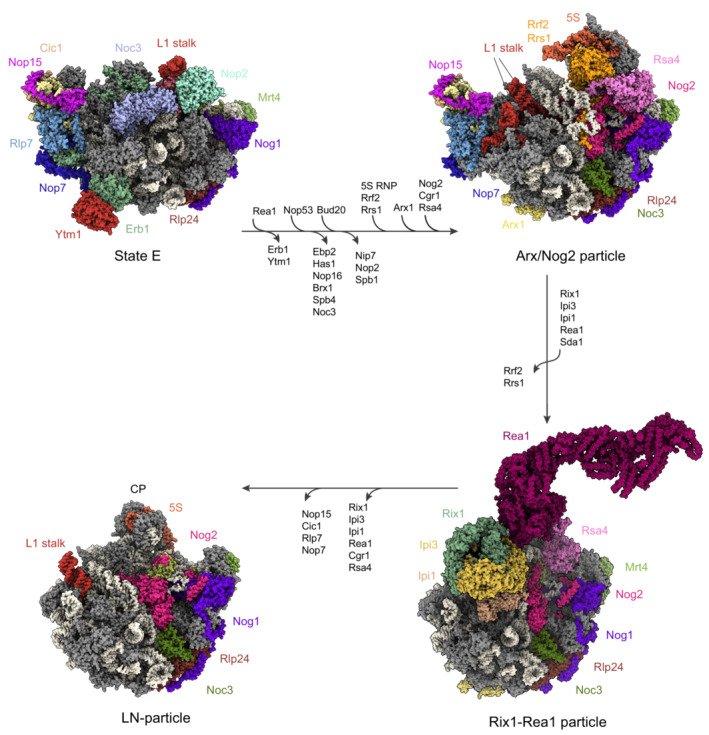
Cryo‐EM structures of yeast pre‐60S particles at different stages of maturation (PDB ID: 6ELZ, 3JCT, 6YLG, 6N8J). RBFs involved in these steps are color‐coded, as well as the pre‐rRNA (white), L1 stalk (red), and 5S rRNA (orange). The nucleolar pre‐60S in state E shows a displacement of the L1 stalk from its position in the mature subunit. The successive nucleoplasmic Arx/Nog2 particle is a result of a stepwise release and binding of the indicated RBFs and the 5S RNP. The Rix‐Rea1 remodeling machinery initiates the formation of the central protuberance (CP) and rotation of the 5S RNP to its mature conformation, visible in the successive late nuclear (LN) particle.
Pre‐rRNA folding, compaction, and cleavage steps, as well as incorporation of RPs, are supported by a number of 60S‐specific RBFs (Table 10). Many of these RBFs join the particle already early in nucleoli and the number of associated factors steadily decreases as 60S subunits mature on their way to the cytoplasm (Nerurkar et al, 2015). Importantly, directionality of the 60S assembly process is ensured by the activity of energy‐dependent RBFs, for example, ATP‐dependent RNA helicases (e.g., Has1/DDX18 and Dpb10/DDX54), AAA‐ATPases (e.g., Rix7/NVL2 and Rea1/MDN1) and GTPases (including Nug1/GNL3 and Nog1/GTPBP4) (Table 10) (Nissan et al, 2002; Bernstein et al, 2006; Ulbrich et al, 2009; Baßler et al, 2010; Kressler et al, 2010, 2008; Wild et al, 2010; Kappel et al, 2012; Dembowski et al, 2013; Matsuo et al, 2014; Manikas et al, 2016; Zhang et al, 2016b; Hiraishi et al, 2018; Klinge & Woolford, 2019). Several other RBFs involved in 60S biogenesis contain multiple RNA binding motifs, which likely provide structural support and reduce the conformational freedom of rRNA during folding and compaction. Importantly, correct binding and positioning of RPs also critically contribute to correct pre‐rRNA folding in the maturing 60S particle (de la Cruz et al, 2015; Pöll et al, 2021, 2009). This is exemplified by the largest Rpl, Rpl3/uL3, which binds very early during pre‐60S biogenesis and spans several rRNA domains (Ben‐Shem et al, 2011; de la Cruz et al, 2015). Rpl3 stabilizes the interaction between the 5′ and 3′ end of the 25S rRNA and its binding is prerequisite for the incorporation of most other Rpls (Pöll et al, 2009; Ohmayer et al, 2013; Gamalinda et al, 2014; de la Cruz et al, 2015).
Table 10.
Factors involved in nucleolar steps of 60S maturation.
| Yeast | 40S | 60S | Human | 40S | 60S | Function in nucleolar steps of pre‐60S maturation | Citation |
|---|---|---|---|---|---|---|---|
| Rrp5 | x | x | PDCD11 | x | Part of Rrp5‐Noc1‐Noc2 complex, supports early steps of pre‐rRNA Compaction, Rrp5 contains multiple RNA binding motifs | Hierlmeier et al (2013) | |
| Noc1 | x | CEBPZ | x | ||||
| Noc2/Rix3 | x | NOC2L/NIR | x | ||||
| Npa1/Urb1 | x | URB1 |
Npa1‐Npa2‐Nop8‐Rsa3‐Dbp6 complex, organization of early pre‐rRNA compaction steps, Dbp6/DDX51 is a DEAD box RNA helicase DDX51: 28S 3′ end maturation, release of U8 snoRNA (only in metazoans) |
Rosado et al (2007) and Srivastava et al (2010) | |||
| Npa2/Urb2 | x | URB2 | |||||
| Nop8 | x | ||||||
| Rsa3 | x | ||||||
| Dbp6 | x | DDX51 | x | ||||
| Dbp2 | x | DDX5 | x | DEAD box RNA helicases, involved in early pre‐rRNA remodeling steps, including release of snoRNPs (Dbp3) | Bond et al (2001) and Saporita et al (2011) | ||
| Dbp3 | x | Aquino et al (2021) | |||||
| Dbp7 | x | DDX31 | x | Bernstein et al (2006) | |||
| Dbp9 | x | Bernstein et al (2006) | |||||
| Mak5 | x | DDX24 | x | Bernstein et al (2006) | |||
| Nsa1 | x | WDR74 | x | Nsa1 module, stabilizes solvent exposed side, bridges 25S domains I and II, Rpf1 protrudes into PET | Kater et al (2017) and Lo et al (2017) | ||
| Rpf1 | x | RPF1 | |||||
| Mak16 | x | MAK16 | x | ||||
| Rrp1 | x | RRP1 | x | ||||
| Nop4 | x | RBM28 | Binds 5′ end of 5.8S rRNA | Granneman et al (2011) | |||
| Puf6 | x | PUM3 | Chaperone for Rpl43/eL43, aids 7S processing, interacts with H63 (25S rRNA), required for export of 60S at low temperature | Liang et al (2019) | |||
| Loc1 | x | Chaperone for Rpl43/eL43 | Liang et al (2019) | ||||
| Rrp15 | x | RRP15 | x | x | Rrp15‐Ssf1 complex, Ssf1 and Ssf2 are 94% identical | Fatica et al (2002) and de Marchis et al (2005) | |
| Ssf1 | x | PPAN | |||||
| Ssf2 | x | PPAN | |||||
| Rrp14 | x | SURF6 | Oeffinger et al (2007) | ||||
| METTL18 | x | Methylation of RPL3/uL3 His245 | Małecki et al (2021) | ||||
| “A3”‐factors, involved in A3 site processing and ITS1 trimming, incorporation of Rpl17/uL22, Rpl26/uL24, Rpl35/uL29, Rpl37/eL37, recruitment of Rrp17 | |||||||
| Nop7 | x | PES1 | x | Erb1‐Ytm1‐Nop7/PeBoW complex, Stabilizes early fold of 5.8S and domain I of 25S rRNA; important for structuring of PTC and PET, DDX27 also functions independently in 47S 3′ end formation, Drs1 is not associated with Erb1‐Ytm1‐Nop7 complex | Rohrmoser et al (2007), Kellner et al (2015) and Konikkat et al (2017) | ||
| Erb1 | x | BOP1 | x | ||||
| Ytm1 | x | WDR12 | x | ||||
| Drs1 | x | DDX27 | x | ||||
| Nop15 | x | MKI67IP | x | Nop15‐Rlp7‐Cic complex, Supports folding of ITS2, part of the foot structure of pre‐60S particles | Sahasranaman et al (2011) and Kater et al (2017) | ||
| Rlp7 | x | RLP7 | |||||
| Cic1 | x | ||||||
| Pwp1 | x | PWP1 | Important for 5.8S folding, ubiquitylated by CRL4VPRBP | Talkish et al (2014) and Han et al (2020) | |||
| Nop12 | x | RBM34 | Important for 5.8S folding | Talkish et al (2014) | |||
| Has1 | x | DDX18 | DEAD‐box RNA helicase, binding triggers trimming of 5′ end of 5.8S pre‐rRNA, important for 25S domain I folding, facilitates incorporation of Rpl17/uL22 and other PET forming proteins, also: linked to release of U14 snoRNA | Dembowski et al (2013) | |||
| Brx1 | x | BRIX1 | x |
Brx1‐Ebp2 complex, prevents premature RNA–RNA interactions of domains I and V, important for PET formation |
Sanghai et al (2018) | ||
| Ebp2 | x | EBNA1BP2 | x | ||||
| “B” factors, required for endonucleolytic cleavage at site C2 by Las1 | |||||||
| Nip7 | x | NIP7 | x | Nip7‐Nop2 complex, aids formation of PTC, recruits Rpf2‐Rrs1, Nop2 is a methyltransferase | Morello et al (2011b) and Kater et al (2017) | ||
| Nop2 | x | NOP2 | |||||
| Rrs1 | x | RRS1 | x | Rpf2‐Rrs1 complex, facilitates incorporation of 5S RNP | Wu et al (2016) | ||
| Rpf2 | x | BXDC1 | x | ||||
| Spb4 | x | DDX55 | x | Binds domain IV of 28S rRNA | Wu et al (2016) | ||
| Mak11 | x | PAK1IP1 | x | Manikas et al (2016) | |||
| Dbp10 | x | DDX54 | Binds h89, role in PTC formation | Bernstein et al (2006) and Manikas et al (2016) | |||
| Nug1 | x | GNL3 | x | GTPase, required for Dbp10 binding | Manikas et al (2016) | ||
| Rlp24 | x | RLP24/RSL24D1 | Placeholder for Rpl24/eL24, recruits and activates Drg1 | Kappel et al (2012) | |||
| Tif6 | x | EIF6 | x | Basu et al (2001) | |||
| Nog1 | x | GTPBP4 | x | GTPase, proofreading/maturation of PET | Wu et al (2016) and Liang et al (2020) | ||
| Nsa2 | x | NSA2 | x | Talkish et al (2012) | |||
| Rsa4 | x | NLE1 | de la Cruz et al (2005) | ||||
| Pol5 | x | x | MYBBP1A | x | Binds to domain III of 25S rRNA, important for PET formation | Braun et al (2020) | |
| Noc3 | x | NOC3L | Milkereit et al (2001) | ||||
| Other factors | |||||||
| Nog2 | x | GNL2 | x | Dembowski et al (2013) | |||
| Nop16 | x | NOP16 | x | Pratte et al (2013) | |||
| Nop53 | x | GLTSCR2/NOP53 | x | Binds to similar position as Erb1, after Erb1 is released, recruits nuclear exosome by Mtr4possibly structural role of Nop53 rearranging and stabilizing the foot interface | Falk et al (2017), Sanghai et al (2018) and Bagatelli et al (2021) | ||
| Spb1 | x | FTSJ3 | x | Methyltransferase, involved in PTC formation | Kater et al (2017) | ||
| Mrt4 | x | MRT4/MRTO4 | x | Structural placeholder for P stalk | Rodríguez‐Mateos et al (2009) | ||
| Mtq2 | x | N6AMT1 | Acts together with its cofactor Trm112 | Lacoux et al (2020) | |||
| YBL028C | x | LLPH | x | Kater et al (2017) and Liang et al (2020) | |||
| Bud20 | x | ZNF593 | Nuclear export factor | Altvater et al (2012) | |||
| Arx1 | x | PA2G4/EBP1 | x | Binds at PET exit, suggested to proofread PET exit region, supports structural changes allowing nuclear export | Greber et al (2012) | ||
| Rix7 | x | NVL2 | x | AAA‐ATPase, removes Nsa1 (+associated factors), which together with release of Erb1‐Ytm1 allows Las1 cleavage and transition of the particle to the nucleoplasm, NVL2 was shown to be associated with Mtr4 (exosome) and WDR74 | Kressler et al (2008) and Hiraishi et al (2018) | ||
| Rea1/Mdn1 | x | MDN1 | x | AAA‐ATPase, releases Erb1‐Ytm1 complex, binds again at a later stage of pre‐60S maturation | Baßler et al (2010) and Wild et al (2010) | ||
| Las1 | x | LAS1L | x | Endoribonuclease C2 cleavage | Schillewaert et al (2012) | ||
| Grc3 | x | NOL9 | Polynucleotide kinase, phosphorylation of ITS2 cleavage product, forms constitutive complex with Las1 in yeast | Pillon et al (2017) | |||
In general, DExD/H‐box ATPases facilitate ribosome biogenesis by unwinding snoRNA‐pre‐rRNA base pairs and remodeling of RNA–RNA and protein‐RNA interactions, thereby supporting major structural rearrangements in the forming subunits as well as pre‐rRNA folding (Dembowski et al, 2013; Rodríguez‐Galán et al, 2013; Martin et al, 2014; Khoshnevis et al, 2016; Brüning et al, 2018). Seven DExD/H‐box ATPases have been implicated in the first steps of 60S maturation (Table 10) (reviewed in Mitterer & Pertschy, 2022). Recently, it was shown that absence of yeast Dbp3 and Prp43 results in a drastic reduction of rRNA modifications (Aquino et al, 2021; Bailey et al, 2022), and several snoRNPs accumulate on early 60S pre‐particles (Leeds et al, 2006; Bohnsack et al, 2009; Aquino et al, 2021). Similarly, DDX51 facilitates release of U8 snoRNA (Srivastava et al, 2010), whereas Dbp7 was proposed to regulate the association of snR190, a snoRNA that structurally inhibits aberrant folding of 25S rRNA (Jaafar et al, 2021a).
Pre‐rRNA processing during 60S maturation commences with removal of the remaining ITS1 spacer at the 5′ end of 27S/32S pre‐rRNAs, before cleavage in the ITS2 (at site C2/4) separates the pre‐5.8S and pre‐25/28S rRNAs (Lygerou et al, 1996; Rouquette et al, 2005; Schillewaert et al, 2012; Gasse et al, 2015; Pillon et al, 2017). Removal of the ITS1 spacer is initiated by endonucleolytic cleavage at site A3 in yeast, while it is removed solely exonucleolytically in human cells (Tomecki et al, 2017). Depletion experiments revealed that a set of 12 yeast RBFs (Ytm1, Erb1, Nop7, Rlp7, Cic1, Nop15, Has1, Drs1, Rpf1, Pwp1, Nop12, and Rrp1) is required for A3 site processing, and they were consequently termed A3‐factors (Table 10) (Merl et al, 2010; Granneman et al, 2011; Sahasranaman et al, 2011; Shimoji et al, 2012; Dembowski et al, 2013; Woolford & Baserga, 2013; Talkish et al, 2014). Structural data showed that the binding sites of most of these factors are not located in proximity of the ITS1 fragment, indicating that they are rather needed as structural components for correct folding and configuration of a pre‐60S assembly intermediate to render it amenable to A3 cleavage (Sahasranaman et al, 2011; Woolford & Baserga, 2013; Kater et al, 2017; Konikkat & Woolford, 2017; Sanghai et al, 2018; Zhou et al, 2019a). Interestingly, A3 cleavage was found to be coupled with termination of RNAPI transcription and 3′ ETS processing, suggesting a link between 60S assembly and transcription, yet the underlying molecular mechanism remains to be deciphered (Allmang & Tollervey, 1998; Lebaron et al, 2012; Gamalinda et al, 2014; Chen et al, 2017).
After ITS1 removal, pre‐rRNA processing is focused on the separation of the 27SB/32S precursor into the 7/12S and 26/28.5S pre‐rRNAs by cleavage in the ITS2. More than a dozen RBFs, originally termed “B‐factors,” are required to prepare the particle for this processing step and many of them are bound to the subunit interface (Talkish et al, 2012; Woolford & Baserga, 2013). Interestingly, some B‐factors, including Nip7 and Nop2, are already part of the 90S particle, before endonucleolytic separation of the two subunits occurs (Kater et al, 2017). B‐factors contribute to the construction of the PET (e.g., Nog1; Fuentes et al, 2007; Wu et al, 2016) and the assembly of the PTC (e.g., Nsa2, Dpb10, Nug1, and Rsa4; Baßler et al, 2014; Matsuo et al, 2014; Barrio‐Garcia et al, 2016; Wu et al, 2016).
Before ITS2 cleavage can take place, pre‐60S subunits undergo major structural rearrangements driven by the AAA‐ATPases Rix7/NVL2 and Rea1/MDN1. First, Rix7 releases Nsa1, which leaves the particle together with Rpf1, Rrp1, and Mak16 (Saveanu et al, 2003; Kressler et al, 2008; Lo et al, 2017, 2019). This allows the formation of the outer part of the PET (Kater et al, 2017; Sanghai et al, 2018; Zhou et al, 2019a). Rix7 activity is then followed by ATP hydrolysis by Rea1. This giant 550 kDa protein removes the Erb1‐Ytm1 subcomplex (Fig 6), which interacts with many RBFs covering the yet to form intersubunit surface (Baßler et al, 2010, 2014; Thoms et al, 2016; Kater et al, 2017; Chen et al, 2018; Ahmed et al, 2019). The MIDAS domain of Rea1 was biochemically shown to bind the UBL domain of Ytm1, yet a structure illustrating the binding of Rea1 to Ytm1 is still missing. Erb1 likely stabilizes the premature architecture at this stage with its N‐terminus deeply embedded in the particle (Kater et al, 2017; Prattes et al, 2019). Its active removal by Rea1 presumably contributes to the major structural rearrangements and compositional changes observed during particle transition from the nucleolus to the nucleoplasm. Release of Erb1 also allows the recruitment of the GTPases Nog2 just before ITS2 cleavage and transition to the nucleoplasm (Talkish et al, 2012; Fromm et al, 2017; Biedka et al, 2018). Finally, cleavage at site C2/4 within ITS2 is mediated by the conserved endonuclease Las1/LAS1, which functions in a complex with the polynucleotide kinase Grc3, together called RNase PNK (Schillewaert et al, 2012; Castle et al, 2013; Gasse et al, 2015; Pillon et al, 2017; Frazier et al, 2021).
The release of pre‐particles from nucleoli could be governed by the state of pre‐rRNA compaction and processing. According to this model, pre‐ribosomal particles that still expose binding sites for general nucleolar RNA chaperones such as nucleophosmin (NPM) (Szebeni & Olson, 1999; Box et al, 2016) or nucleolin (NCL) (Mongelard & Bouvet, 2007) (Table 14) would remain partitioned in the nucleolar “phase” by low‐affinity interactions of exposed RNA segments with these multivalent RNA chaperones. Once the pre‐rRNAs in the emerging subunits have been sufficiently compacted, processed, and covered by RPs and RBFs, they would no longer be retained and released into the nucleoplasm. Along these lines, in vitro experiments showed a preferential partitioning of protein‐free bacterial rRNA but not mature ribosomal subunits into droplets formed by human NPM (Riback et al, 2020). Furthermore, a similar model has recently been proposed based on bioinformatic analyses of cryo‐EM structures, which revealed that earlier nucleolar ribosomal precursors contain more unstructured rRNA regions as well as RBFs with predicted intrinsically disordered regions compared to nucleoplasmic subunit assembly intermediates, and both these elements could contribute to nucleolar retention of subunits (LaPeruta et al, 2022).
Table 14.
Other factors involved in ribosome biogenesis.
| Yeast | 40S | 60S | Human | 40S | 60S | Function in rRNA transcription | Citation |
|---|---|---|---|---|---|---|---|
| NPM | x | x | Nuclear RNA‐binding protein and chaperone | Szebeni and Olson (1999) | |||
| Nsr1 | x | x | NCL | x | x | Nuclear RNA‐binding protein and chaperone | Mongelard and Bouvet (2007) |
| DDX1 | Suzuki et al (2021) | ||||||
| HECTD1 | Ubiquitylates ZNF622 | Lv et al (2021) | |||||
| USP36 | Promotes FAU processing and snoRNP maturation | Ryu et al (2021) and van den Heuvel et al (2021) | |||||
| UBR5 | x | Supports rRNA maturation by regulation of H/ACA RNPs | Saez et al (2020) | ||||
| VPRBP | x | Ubiquitylates PWP1 | Han et al (2020) |
Formation of the 5S RNP and its incorporation into pre‐60S subunits
During its maturation, the pre‐60S subunit must incorporate the mature 5S rRNA. The 5S rRNA first associates with its partner RPs Rpl5/uL18 and Rpl11/uL5, which are co‐imported into the nucleus with help of the RP chaperone Syo1/HEATR3 which also serves as a platform for 5S RNP assembly (Kressler et al, 2012; Calviño et al, 2015). The 5S RNP is then bound by pre‐60S particles in the nucleolus aided by the associated Rpf2‐Rrs1 complex (Table 11) (Wu et al, 2016). In mature 60S subunits, the 5S RNP forms the central protuberance, yet the 5S RNP is not immediately placed in its final position in the pre‐60S, but rotated by about 180°, as revealed by structures of Arx1/Nog2 pre‐60S particles from yeast (Leidig et al, 2014; Wu et al, 2016).
Table 11.
Factors involved in 5S RNP formation and incorporation.
| Yeast | 40S | 60S | Human | 40S | 60S | Function in 5S RNP maturation | Citation |
|---|---|---|---|---|---|---|---|
| Rex1 | x | REXO5 | Exoribonuclease, 5S pre‐rRNA trimming | van Hoof et al (2000) | |||
| Rex2 | x | REXO2 | Exoribonuclease, 5S pre‐rRNA trimming | van Hoof et al (2000) | |||
| Rex3 | x | REXO1 | Exoribonuclease, 5S pre‐rRNA trimming | van Hoof et al (2000) | |||
| La | x | Binds immature 5S pre‐rRNA | Madru et al (2015) | ||||
| TFIIIA | x | TFIIIA | x | Transcription factor for 5S DNA locus, but also binds 5S rRNA | Layat et al (2013) and Sloan et al (2013b) | ||
| Syo1 | x | HEATR3 | x | Chaperone for Rpl5/uL18 and Rpl11/uL5 | Kressler et al (2012), Hannan et al (2022) and O'Donohue et al (2022) | ||
| Rrs1 | x | RRS1 | x | Rpf2‐Rrs1 complex, aids incorporation of 5S RNP into mature ribosomes, in yeast also important for nucleolar localization of 5S RNP | Wu et al (2016) | ||
| Rpf2 | x | BXDC1 | x |
In mammalian cells, the incorporation of the 5S RNP into the nascent 60S subunit serves as an important checkpoint to relay defects in nucleolar ribosome synthesis into the p53 pathway, referred to as the “nucleolar stress response” (reviewed in Chakraborty et al, 2011; Bohnsack & Bohnsack, 2019). If nucleolar 60S maturation is perturbed, the unincorporated 5S RNP accumulates, to then bind and inhibit the p53 E3 ubiquitin ligase MDM2, leading to p53 stabilization and cell cycle arrest (Donati et al, 2013; Sloan et al, 2013a). This 5S RNP‐dependent mechanism has emerged as a key nuclear stress response pathway that reacts to a broad range of insults ranging from diverse DNA damaging insults, proteasome inactivation to nuclear export inhibition (Hannan et al, 2022), putting the 5S RNP and ribosome biogenesis defects into the center of nuclear stress sensing.
Nucleoplasmic assembly steps and export of pre‐60S subunits
The transition of pre‐60S particles from the nucleolus to the nucleoplasm is accompanied by the release and binding of a significant number of RBFs, resulting in substantial compositional and structural changes in the emerging subunits (Fig 6) (Kater et al, 2017, 2020; Sanghai et al, 2018). These include the repositioning of the L1 stalk, which is first delocalized and then accommodated into its mature position following the release of Spb1 (Kater et al, 2020). Furthermore, 2′O‐methylation of G2922 in the PTC A‐site loop by Spb1 was suggested to be prerequisite for stable binding of Nog2 to H92 (Kressler et al, 1999; Lapeyre & Purushothaman, 2004; preprint: Yelland et al, 2022). Finally, the association of Arx1, Cgr1, and Rsa4 to the forming subunit interface complete the well‐characterized Arx1/Nog2 particle (Fig 6) (Bradatsch et al, 2012; Leidig et al, 2014; Wu et al, 2016).
The L1 stalk together with the RBF Sda1 provides a binding platform for the Rix1 complex (Table 12) (Barrio‐Garcia et al, 2016; Wu et al, 2016; Kater et al, 2020). Association of Rix1‐Ipi3‐Ipi1 initiates the formation of the central protuberance as it recruits the ATPase Rea1 for a second round of action (Fig 6). Rea1 then catalyzes the release of the Rpf2‐Rrs1 complex powered by ATP hydrolysis (Baßler et al, 2010; Matsuo et al, 2014; Barrio‐Garcia et al, 2016). This leads to a 180° rotation of the 5S RNP and accommodation in its final position, coupled to maturation of PET and PTC (Micic et al, 2020). Moreover, the RBF Nop53 joins the pre‐60S subunits (Falk et al, 2017; Kater et al, 2020) and binds the RNA helicase Mtr4, thereby recruiting the nuclear exosome to trim the ITS2 part of the 7S pre‐rRNA (Michael et al, 2018). Elimination of this ITS2 fragment results in the removal of the so‐called foot region visible in cryo‐EM structures of Nog2 particles (Fromm et al, 2017; Zhou et al, 2019a). Coupled to its role in 5S RNP accommodation, Rea1 ATPase activity also triggers the release of Rsa4 (Baßler et al, 2014, 2010), and is prerequisite for GTP hydrolysis‐dependent release of the GTPase Nog2 (Matsuo et al, 2014). The precise molecular mechanism by which the mechano‐chemical force of ATP hydrolysis by Rea1 leads to these structural rearrangements needs to be further investigated, but the liberation of the Nog2 binding site on pre‐60S particles sets the stage for the recruitment of the export adaptor Nmd3. Nmd3 binding serves as a quality control checkpoint probing the correct assembly of the E‐ and P‐sites as well as of the L1 stalk (Sengupta et al, 2010; Matsuo et al, 2014; Malyutin et al, 2017).
Table 12.
Factors involved in nucleoplasmic steps of pre‐60S maturation.
| Yeast | 40S | 60S | Human | 40S | 60S | Function in nucleoplasmic steps of pre‐60S maturation | Citation |
|---|---|---|---|---|---|---|---|
| Alb1 | x | Arx1 binding partner | Greber et al (2012) | ||||
| Cgr1 | x | CCDC86 | Thoms et al (2018) | ||||
| Sda1 | x | SDAD1 | Sda1 suggested to initiate Rpf2‐Rrs1 release as it partially overlaps with Rpf2 binding site | Klinge and Woolford (2019) | |||
| Rix1 | x | PELP1 |
Rix1/PELP1 complex, important for Rea1/MDN1 positioning PELP1 is SUMOylated, which is required for its interaction with MDN1 |
Finkbeiner et al (2011), Barrio‐Garcia et al (2016) and Gordon et al (2022) | |||
| Ipi3 | x | TEX10 | |||||
| Ipi1 | x | WDR18 | |||||
| Rea1/Mdn1 | x | MDN1 | x | AAA‐ATPase, rebinds to 60S particles in nucleoplasm, releases Rsa4, triggers GTPase activity and release of Nog2 | Baßler et al (2010) | ||
| SENP3 | x | SUMO specific protease, activity results in disengagement of MDN1 and PELP1 deSUMOylates NPM | Finkbeiner et al (2011) and Raman et al (2016) | ||||
| NF45/ILF2 | x | NF45‐NF90 complex | Wandrey et al (2015) | ||||
| NF90/ILF3 | x | ||||||
| Nmd3 | x | NMD3 | x | Nuclear export adaptor | Ho et al (2000) and Thomas and Kutay (2003) |
Notably, the various RNA expansion segments, located at the solvent‐exposed surface of mature ribosomes (Yusupova & Yusupov, 2014), also play an important role in pre‐rRNA processing and subunit assembly. Studies in yeast have demonstrated that individual deletion of the majority of expansion segments in 25S rRNA leads to 60S biogenesis defects (Jeeninga et al, 1997; Ramesh & Woolford, 2016). Interestingly, certain eukaryotic RPs and their extensions (Ramesh & Woolford, 2016) as well as some RBFs such as Arx1 (Bradatsch et al, 2012), Nop7 (Granneman et al, 2011), Rlp7 (Dembowski et al, 2013), and Rrp5 (Lebaron et al, 2013) make contact to RNA expansion segments (Granneman et al, 2011; Bradatsch et al, 2012; Babiano et al, 2013; Dembowski et al, 2013; Lebaron et al, 2013), guiding models of coevolution of expansion segments with RPs and RBFs (Ramesh & Woolford, 2016).
While nucle(ol)ar pre‐60S maturation in yeast is relatively well understood, only a limited number of studies have addressed this process in human cells (Wild et al, 2010; Finkbeiner et al, 2011; Tafforeau et al, 2013; Wandrey et al, 2015; Dörner et al, 2022). Although the human homologs of many yeast RBFs have been identified (Wild et al, 2010; Tafforeau et al, 2013; Badertscher et al, 2015; Dörner et al, 2022), the functional conservation of most factors remains to be investigated (Table 12). Interestingly, SUMOylation of PELP1, a component of the PELP1‐TEX10‐WDR18 complex (Rix1 complex in yeast), was shown to be essential for recruitment of MDN1 (the human ortholog of yeast Rea1) to the pre‐60S particle (Finkbeiner et al, 2011; Raman et al, 2016). Recently, the structures of two late nuclear human pre‐60S particles already associated with the export factor NMD3 have been described (Liang et al, 2020). Overall, the structures showed similar architecture and composition as yeast particles at similar stages, indicating conservation of the function of bound RBFs. Yet, they also revealed mammalian‐specific features of 60S subunit maturation, for example, interaction of the N‐terminal domain of ZNF622 (Rei1 in yeast) with expansion segment ES27, which is much longer in human cells. Cryo‐EM structures of earlier nucle(ol)ar human pre‐60S particles remain to be solved and will provide additional insights into similarities and differences of the process between yeast and mammals.
Binding of the NES‐containing export adaptor Nmd3/NMD3 licenses pre‐60S subunits for Crm1/XPO1‐dependent nuclear export both in fungi and metazoan cells (Table 8) (Ho et al, 2000; Gadal et al, 2001; Thomas & Kutay, 2003; Trotta et al, 2003). In vertebrate cells, a second RanGTP‐binding exportin, XPO5, was shown to support pre‐60S export in addition (Moy & Silver, 1999; Wild et al, 2010). In yeast, a number of further auxiliary factors facilitate pre‐60S translocation through the NPC by directly interacting with FG‐repeats of nucleoporins, including Rrp12, Bud20, Ecm1, and Npl3 (Oeffinger et al, 2004; Yao et al, 2010; Hackmann et al, 2011; Altvater et al, 2012; Nerurkar et al, 2015) (Table 8).
In addition, the mRNA export receptor Mex67/Mtr2 aids export of both 40S and 60S pre‐particles in yeast, a function that is not conserved in human cells (Yao et al, 2008, 2007). Interestingly, recruitment of Mex67/Mtr2 has been linked to the assembly of the P stalk. Pre‐60S particles initially contain Mrt4, a structural homolog of the P‐stalk protein uL10/P0. As long as Mrt4 is bound, the recruitment of the nuclear export receptor Mex67/Mtr2 is inhibited (Sarkar et al, 2016). Yvh1/DUSP12 then dissociates Mrt4, thereby allowing P‐stalk formation by incorporation of the uL10/P0 (Kemmler et al, 2009; Rodríguez‐Mateos et al, 2009; Lo et al, 2010, 2009; Sarkar et al, 2016; Zhou et al, 2019b; Klingauf‐Nerurkar et al, 2020). It must be noted, however, that the exact timing of P‐stalk assembly is not fully resolved, since Mrt4 can be found on cytoplasmic particles upon expression of dominant‐negative Drg1, a cytoplasmic 60S‐RBF, indicating that exchange may occur later, in the cytoplasm (Klingauf‐Nerurkar et al, 2020).
Cytoplasmic 60S maturation steps
While cytoplasmic pre‐40S subunit maturation is primarily driven by kinases, cytoplasmic maturation of pre‐60S particles relies on the function of GTPases and AAA‐ATPases (Table 13) (Lo et al, 2010). These ensure the timely release of some remaining assembly factors (Fig 7), including the ribosomal‐like protein Rlp24/RSL24D1, the GTPase Nog1/GTPBP4, the export adaptor Nmd3/NMD3, Arx1/PA2G4, and Tif6/EIF6. At the same time, RBFs acting in the cytoplasm drive the incorporation of the last RPs and proofread the assembly state of the functional centers.
Table 13.
Factors involved in cytoplasmic pre‐60S maturation.
| Yeast | 40S | 60S | Human | 40S | 60S | Function in cytoplasmic pre‐60S maturation | Citation |
|---|---|---|---|---|---|---|---|
| Drg1/Afg2 | x | SPATA5 | x | ATPase activity releases Rlp24 and Nog1 | Pertschy et al (2007), Kappel et al (2012) and Ni et al (2022) | ||
| SPATA5L | x | ||||||
| CINP | x | ||||||
| C1orf109 | x | ||||||
| Rlp24 | x | RLP24/RSL24D1 | x | Released by Drg1, which allows incorporation of Rpl24/eL24 (placeholder) | Kappel et al (2012) | ||
| Bud20 | x | ZNF593 | x | Released | Srivastava et al (2010) | ||
| Nug1 | x | GNL3 | x | Released, timing of release not clear in humans | Altvater et al (2012) and Ma et al (2017) | ||
| YBL028C | x | LLPH | x | Released | Klingauf‐Nerurkar et al (2020) and Liang et al (2020) | ||
| TMA16 | x | TMA16 | x | Might be released before export | Liang et al (2020) | ||
| Nsa2 | x | NSA2 | Released | Altvater et al (2012) and Ma et al (2017) | |||
| Nog1 | x | GTPBP4/NOG1 | x | Inserts flexible helix into PET, which could function as proof reading or maturation step, released after/with Rlp24, coordinates bifurcation of pre‐60S maturation pathway | Pertschy et al (2007) and Kappel et al (2012) | ||
| Arx1 | x | PA2G4/EBP1 | x | Prevents binding of proteins and complexes typically engaging with the nascent peptide chain binding near the tunnel exit, released by Rei1 (with Ssa and Jjj), which frees peptide exit tunnel, function in human cells is less clear | Bradatsch et al (2012) and Greber et al (2012) | ||
| Alb1 | x | Binding partner of Arx1, released | Greber et al (2012) | ||||
| Rei1 | x | ZNF622 | x | Inserts flexible helix into PET, which could function as proof reading or maturation step; functions together with the ATPase Ssa and Jjj to release Arx1 | Meyer et al (2010), Bradatsch et al (2012) and Greber et al (2012) | ||
| Jjj1 | x | DNAJC21 | x | Cofactor for release of Arx1 by Rei1/Ssa | Demoinet et al (2007) and Greber et al (2012) | ||
| Ssa1 | x | HSPA1A | Binds together with Rei1 and Jjj1, ATPase Activity releases Arx1 | Pertschy et al (2007) and Ma et al (2017) | |||
| Ssa2 | x | HSPA1B | |||||
| Reh1 | x | Inserts C‐terminal helix into PET | Parnell and Bass (2009) | ||||
| Mrt4 | x | MRT4/MRTO4 | x | Placeholder for P stalk, released, which allows incorporation of ribosome stalk, initiated by binding of RplP0/uL10 | Kemmler et al (2009) and Lo et al (2009) | ||
| Yvh1 | x | DUSP12 | x | Phosphatase, releases Mrt4 | Kemmler et al (2009) and Lo et al (2009) | ||
| Efl1/Ria1 | x | EFL1/EFTUD1 | x | GTPase activity releases Tif6/EIF6, works together with Sdo1/SBDS | Basu et al (2001) and Finch et al (2011) | ||
| Sdo1 | x | SBDS | x | Works together with Efl1/EFL1 | Basu et al (2001) and Finch et al (2011) | ||
| Tif6 | x | EIF6 | x | Anti‐association factor, preventing premature association with 40S subunits, released by Efl1/EFL1 (with Sdo1/SDAD) | Gartmann et al (2010) | ||
| Lsg1/Kre35 | x | LSG1 | x | GTPase activity releases Nmd3/NMD3 | Kallstrom et al (2003) and Hedges et al (2005) | ||
| Sqt1 | x | AAMP | x | Chaperone of Rpl10/uL16 | Kallstrom et al (2003) and Hedges et al (2005) | ||
| Nmd3 | x | NMD3 | x | Released by Lsg1/LSG1, allows incorporation of Rpl10/uL16, before release: prevents joining of premature subunits | Kallstrom et al (2003) and Hedges et al (2005) |
Figure 7. Cytoplasmic maturation of 60S subunit in yeast.
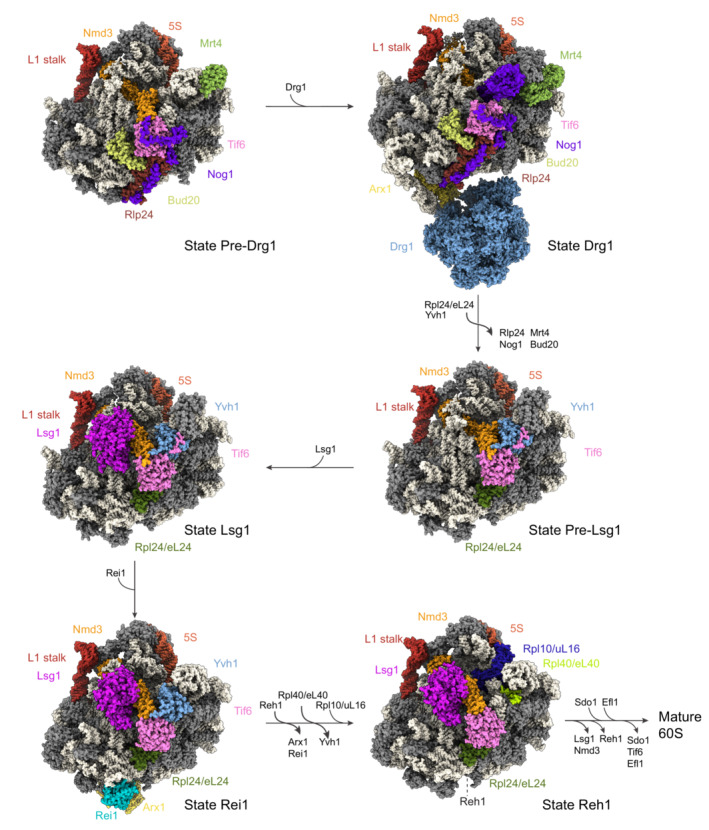
Cryo‐EM structures of pre‐60S particles illustrating snapshots of cytoplasmic maturation events (PDB ID: 6N8L, 7Z34, 6N8M, 6N8N, 6RZZ, 6QTZ). RBFs are color‐coded, as well as the pre‐rRNA (white), L1 stalk (red), and 5S rRNA (orange). After nuclear export, the ATPase Drg1 dissociates Rpl24 from early cytoplasmic pre‐60S particles. The release of multiple factors allows the association of the GTPase Lsg1, which in turn dissociates Nmd3 upon the incorporation of Rpl10/uL16. Following Reh1 dissociation, the GTPase Efl1 together with Sdo1 triggers the release of Tif6, resulting in mature 60S subunits.
In a first step, the AAA‐ATPase Drg1 releases Rlp24 (Fig 7), which acts as a placeholder for Rpl24/eL24 (Table 13) (Pertschy et al, 2007; Lo et al, 2010; Kappel et al, 2012). While Drg1 is the sole factor known to be responsible for Rlp24 dissociation in yeast, release of the human homolog of Rlp24, RLP24/RSL24D1, was recently reported to involve two Drg1‐related AAA‐ATPases, SPATA5, and SPATA5L1 (Ni et al, 2022). Interestingly, SPATA5 is localized in the cytosol, similar to yeast Drg1 (Puusepp et al, 2018), whereas SPATA5L1 is predominantly nuclear (Richard et al, 2021), suggesting that either RLP24 release can in principle occur in both compartments or that only one of these factors functions directly in RLP24 exchange. In addition to SPATA5 and SPATA5L, RLP24 release was suggested to depend on CINP and C1orf109 (Ni et al, 2022), two structurally related proteins. However, how these factors function together with SPATA5 and SPATA5L1 in RLP24/eL24 exchange remains to be mechanistically defined.
Drg1 not only acts on Rlp24, but also contributes to the dissociation of the GTPase Nog1 and additional factors that bind in close proximity to Rlp24. The GTPase Nog1 is deposited on the pre‐60S subunit already during nucleolar maturation steps and projects its long C‐terminal tail into the PET, almost reaching back to the PTC (Wu et al, 2016). Nog1 is liberated in a two‐step process. The first step exploits its own GTPase activity, driving the dissociation of both its N‐terminal and GTPase domains. In the second step, the C‐terminal part of Nog1 is dissociated by Drg1 (Pertschy et al, 2007; Kappel et al, 2012). After release of Nog1, the PET is further functionally probed and potentially matured by insertion of the C‐terminal domain of Rei1/ZNF622, which is recruited via Rpl24 (Greber et al, 2016, 2012; Kargas et al, 2019; Zhou et al, 2019b). Arx1 is bound close to the PET exit where it sterically impedes the premature loading of nascent chain binding factors. It is set free by Rei1, Jjj1 and the ATPase activity of Ssa1/Ssa2 (Hsp70) (Hung & Johnson, 2006; Lebreton et al, 2006; Meyer et al, 2010; Bradatsch et al, 2012; Greber et al, 2012). Then, the PET is again occluded by the C‐terminal α‐helix of Reh1, which binds inside the PET (Ma et al, 2017).
Cytoplasmic 60S maturation ends with the dissociation of Reh1, Nmd3, and Tif6, which are bound on the subunit interface. These factors are suggested to function as anti‐association factors, inhibiting premature interaction of cytoplasmic pre‐60S particles with mature 40S subunits (Gartmann et al, 2010; Weis et al, 2015; Ma et al, 2017). After incorporation of RPL40/eL40 and RPL10/uL16, Nmd3 is dissociated by the GTPase Lsg1/LSG1 (Fig 7) (Malyutin et al, 2017; Kargas et al, 2019; Zhou et al, 2019b). Finally, Tif6 removal is mediated by the GTPase Efl1/EFTUD1 and its guanine nucleotide exchange factor Sdo1/SBDS (Bécam et al, 2001; Senger et al, 2001; Menne et al, 2007), which have also been suggested to probe functionality of the P stalk, P‐site, and PTC (Ma et al, 2017; Zhou et al, 2019b).
Most factors involved in cytosolic pre‐60S maturation in yeast have human homologs, and recently solved structures of late human pre‐60S subunits revealed that the observed human homologs of yeast RBFs bind to similar positions and are likely functional homologs (Liang et al, 2020). One notable exception is PA2G4/EBP1, which is significantly smaller than its yeast homolog Arx1. PA2G4 has been implicated both in ribosome biogenesis and translation, as it was shown to bind to pre‐60S as well as mature 80S ribosomes in vivo (Liang et al, 2020; Bhaskar et al, 2021; Kraushar et al, 2021). Several structural features of yeast Arx1 that might play a role in 60S biogenesis are missing in PA2G4. Thus, further analysis is needed to address its functional conservation as an RBF.
Altogether, eukaryotic cells exploit several hundred RBFs along the intricate ribosome assembly line from the nucleolus to the cytoplasm (Tables 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14). These RBFs not only promote RP deposition, pre‐rRNA folding, processing, and maturation as well as remodeling of the emerging ribosomal particles, but also probe the correct configuration of the functional centers. Once all RBFs are released, both subunits are ready to fulfill their function in mRNA translation.
Concluding remarks
Over the past years, the field has made impressive progress in deciphering the molecular mechanisms of ribosomal subunit assembly and maturation. Today, we know for the vast majority of RBFs at which step of the complex pathway they function, how they associate with precursor particles, and which task they perform. What initially seemed an overwhelming list of accessory factors, first merely named as players in either 40S or 60S synthesis, has coalesced into an almost coherent molecular picture of the subunit assembly lines. Of course, some pieces of the puzzle are still missing. For instance, while we appreciate the impressive molecular snapshots of the SSU processome, we still do not fully understand how this gigantic RNP, similar in size to a mammalian ribosome, is assembled in the first place. How are structural and compositional remodeling steps of the SSU processome that are associated with pre‐rRNA folding and maturation driven? We also know little about whether and how early nucleolar steps of 40S and 60S biogenesis are coupled before RNA cleavage separates the precursors to both subunits. Likewise, we still miss a structural depiction of some key 60S subunit assembly intermediates.
But not only the assembly process itself still contains some uncharted territory, also other areas related to ribosome synthesis are expected to hide a number of exciting secrets. For instance, it is becoming increasingly clear that early errors in the assembly line affect nucleolar morphology and structure. But how is nucleolar organization governed in first place? And how are maturing particles expelled from nucleoli for further maturation into the nucleoplasm? Some first hints suggest, as discussed above, that pre‐rRNA length and compaction may govern this decisive step. Another almost unexplored area concerns quality control of ribosome synthesis. How are aberrant premature subunits recognized and eliminated? This question especially pertains to the fate of the proteinaceous parts of the precursor particles. Hence, which mechanisms counteract the potential proteotoxicity of unassembled RPs or aberrant precursors, especially in mammalian cells? Clearly, these and many other questions remain to be resolved. Thus, despite the impressive progress made, research on ribosome biogenesis will remain an active and rewarding field of biology in the future.
Author contributions
Kerstin Dörner: Conceptualization; data curation; formal analysis; writing – original draft; writing – review and editing. Chiara Ruggeri: Conceptualization; data curation; formal analysis; writing – review and editing. Ivo Zemp: Conceptualization; data curation; formal analysis; writing – original draft; writing – review and editing. Ulrike Kutay: Conceptualization; data curation; formal analysis; supervision; funding acquisition; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Acknowledgements
We thank Claudia Gafko, Annamaria Gamper, and Ivan Kisly for critical reading of the manuscript. We apologize for not being able to cite all original publications. Our work on ribosome synthesis is supported by the Swiss National Science Foundation (SNSF) in the framework of the NCCR “RNA and disease” (51NF40‐205601). Open access funding provided by Eidgenossische Technische Hochschule Zurich.
The EMBO Journal (2023) 42: e112699
References
- Abou Assi H, Rangadurai AK, Shi H, Liu B, Clay MC, Erharter K, Kreutz C, Holley CL, Al‐Hashimi HM (2020) 2′‐O‐Methylation can increase the abundance and lifetime of alternative RNA conformational states. Nucleic Acids Res 48: 12365–12379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed YL, Thoms M, Mitterer V, Sinning I, Hurt E (2019) Crystal structures of Rea1‐MIDAS bound to its ribosome assembly factor ligands resembling integrin–ligand‐type complexes. Nat Commun 10: 3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanèse V, Reissmann S, Frydman J (2010) A ribosome‐anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J Cell Biol 189: 69–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Tollervey D (1998) The role of the 3′ external transcribed spacer in yeast pre‐rRNA processing. J Mol Biol 278: 67–78 [DOI] [PubMed] [Google Scholar]
- Allmang C, Mitchell P, Petfalski E, Tollervey D (2000) Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res 28: 1684–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altvater M, Chang Y, Melnik A, Occhipinti L, Schütz S, Rothenbusch U, Picotti P, Panse VG (2012) Targeted proteomics reveals compositional dynamics of 60S pre‐ribosomes after nuclear export. Mol Syst Biol 8: 628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameismeier M, Cheng J, Berninghausen O, Beckmann R (2018) Visualizing late states of human 40S ribosomal subunit maturation. Nature 558: 249–253 [DOI] [PubMed] [Google Scholar]
- Ameismeier M, Zemp I, van den Heuvel J, Thoms M, Berninghausen O, Kutay U, Beckmann R (2020) Structural basis for the final steps of human 40S ribosome maturation. Nature 587: 683–687 [DOI] [PubMed] [Google Scholar]
- Ansel KM, Pastor WA, Rath N, Lapan AD, Glasmacher E, Wolf C, Smith LC, Papadopoulou N, Lamperti ED, Tahiliani M et al (2008) Mouse Eri1 interacts with the ribosome and catalyzes 5.8S rRNA processing. Nat Struct Mol Biol 15: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino GRR, Krogh N, Hackert P, Martin R, Gallesio JD, van Nues RW, Schneider C, Watkins NJ, Nielsen H, Bohnsack KE et al (2021) RNA helicase‐mediated regulation of snoRNP dynamics on pre‐ribosomes and rRNA 2′‐O‐methylation. Nucleic Acids Res 49: 4066–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S et al (2010) Cryo‐EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5‐Å resolution. Proc Natl Acad Sci USA 107: 19748–19753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert M, O'Donohue M‐F, Lebaron S, Gleizes P‐E (2018) Pre‐ribosomal RNA processing in human cells: from mechanisms to congenital diseases. Biomolecules 8: 123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi S, Verma M, Mahesh A, K Khan MI, Govindaraju G, Rajavelu A, Chavali PL, Chavali S, Dhayalan A (2018) DDX49 is an RNA helicase that affects translation by regulating mRNA export and the levels of pre‐ribosomal RNA. Nucleic Acids Res 46: 6304–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axt K, French SL, Beyer AL, Tollervey D (2014) Kinetic analysis demonstrates a requirement for the Rat1 exonuclease in cotranscriptional pre‐rRNA cleavage. PLoS One 9: e85703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaian A, Rothe K, Girodat D, Minia I, Djondovic S, Milek M, Spencer Miko SE, Wieden H‐J, Landthaler M, Morin GB et al (2020) Loss of m1acp3Ψ; ribosomal RNA modification is a major feature of cancer. Cell Rep 31: 107611 [DOI] [PubMed] [Google Scholar]
- Babiano R, Badis G, Saveanu C, Namane A, Doyen A, Díaz‐Quintana A, Jacquier A, Fromont‐Racine M, de la Cruz J (2013) Yeast ribosomal protein L7 and its homologue Rlp7 are simultaneously present at distinct sites on pre‐60S ribosomal particles. Nucleic Acids Res 41: 9461–9470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badertscher L, Wild T, Montellese C, Alexander LT, Bammert L, Sarazova M, Stebler M, Csucs G, Mayer TU, Zamboni N et al (2015) Genome‐wide RNAi screening identifies protein modules required for 40S subunit synthesis in human cells. Cell Rep 13: 2879–2891 [DOI] [PubMed] [Google Scholar]
- Bagatelli FFM, de Luna Vitorino FN, da Cunha JPC, Oliveira CC (2021) The ribosome assembly factor Nop53 has a structural role in the formation of nuclear pre‐60S intermediates, affecting late maturation events. Nucleic Acids Res 49: 7053–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AD, Talkish J, Ding H, Igel HA, Duran A, Mantripragada S, Paten B, Ares M Jr (2022) Concerted modification of nucleotides at functional centers of the ribosome revealed by single‐molecule RNA modification profiling. Elife 11: e76562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammert L, Jonas S, Ungricht R, Kutay U (2016) Human AATF/Che‐1 forms a nucleolar protein complex with NGDN and NOL10 required for 40S ribosomal subunit synthesis. Nucleic Acids Res 44: 9803–9820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bange G, Murat G, Sinning I, Hurt E, Kressler D (2013) New twist to nuclear import: when two travel together. Commun Integr Biol 6: e24792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barandun J, Chaker‐Margot M, Hunziker M, Molloy KR, Chait BT, Klinge S (2017) The complete structure of the small‐subunit processome. Nat Struct Mol Biol 24: 944–953 [DOI] [PubMed] [Google Scholar]
- Barandun J, Hunziker M, Klinge S (2018) Assembly and structure of the SSU processome—a nucleolar precursor of the small ribosomal subunit. Curr Opin Struct Biol 49: 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio‐Garcia C, Thoms M, Flemming D, Kater L, Berninghausen O, Baßler J, Beckmann R, Hurt E (2016) Architecture of the Rix1‐Rea1 checkpoint machinery during pre‐60S‐ribosome remodeling. Nat Struct Mol Biol 23: 37–44 [DOI] [PubMed] [Google Scholar]
- Baßler J, Hurt E (2019) Eukaryotic ribosome assembly. Annu Rev Biochem 88: 281–306 [DOI] [PubMed] [Google Scholar]
- Baßler J, Kallas M, Pertschy B, Ulbrich C, Thoms M, Hurt E (2010) The AAA‐ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol Cell 38: 712–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baßler J, Paternoga H, Holdermann I, Thoms M, Granneman S, Barrio‐Garcia C, Nyarko A, Stier G, Clark SA, Schraivogel D et al (2014) A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation. J Cell Biol 207: 481–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu U, Si K, Warner JR, Maitra U (2001) The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol Cell Biol 21: 1453–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin F, Murciano B, Fung HKH, Fromm SA, Mattei S, Mahamid J, Müller CW (2022) Mechanism of RNA polymerase I selection by transcription factor UAF. Sci Adv 8: eabn5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin‐Baillieu A, Tollervey D, Cullin C, Lacroute F (1997) Functional analysis of Rrp7p, an essential yeast protein involved in pre‐rRNA processing and ribosome assembly. Mol Cell Biol 17: 5023–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumas K, Soudet J, Caizergues‐Ferrer M, Faubladier M, Henry Y, Mougin A (2012) Human RioK3 is a novel component of cytoplasmic pre‐40S pre‐ribosomal particles. RNA Biol 9: 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax R, Raué HA, Vos JC (2006) Slx9p facilitates efficient ITS1 processing of pre‐rRNA in Saccharomyces cerevisiae . RNA 12: 2005–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécam AM, Nasr F, Racki W, Zagulski M, Herbert C (2001) Ria1p (Ynl163c), a protein similar to elongation factors 2, is involved in the biogenesis of the 60S subunit of the ribosome in Saccharomyces cerevisiae . Mol Genet Genomics 266: 454–462 [DOI] [PubMed] [Google Scholar]
- Bell SP, Learned RM, Jantzen H‐M, Tjian R (1988) Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241: 1192–1197 [DOI] [PubMed] [Google Scholar]
- Ben‐Shem A, de Loubresse NG, Melnikov S, Jenner L, Yusupova G, Yusupov M (2011) The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529 [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Granneman S, Lee AV, Manickam S, Baserga SJ (2006) Comprehensive mutational analysis of yeast DEXD/H Box RNA helicases involved in large ribosomal subunit biogenesis. Mol Cell Biol 26: 1195–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar V, Desogus J, Graff‐Meyer A, Schenk AD, Cavadini S, Chao JA (2021) Dynamic association of human Ebp1 with the ribosome. RNA 27: 411–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedka S, Micic J, Wilson D, Brown H, Diorio‐Toth L, Woolford JL Jr (2018) Hierarchical recruitment of ribosomal proteins and assembly factors remodels nucleolar pre‐60S ribosomes. J Cell Biol 217: 2503–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch JL, Tan BC‐M, Panov KI, Panova TB, Andersen JS, Owen‐Hughes TA, Russell J, Lee S‐C, Zomerdijk JCBM (2009) FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J 28: 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JJ, Johnson AW (2022) Release of the ribosome biogenesis factor Bud23 from small subunit precursors in yeast. RNA 28: 371–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JJ, Musalgaonkar S, Johnson AW (2019) Tsr4 is a cytoplasmic chaperone for the ribosomal protein Rps2 in Saccharomyces cerevisiae . Mol Cell Biol 39: e00094‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JJ, Sardana R, Elmir EW, Johnson AW (2020) Bud23 promotes the final disassembly of the small subunit processome in Saccharomyces cerevisiae . PLoS Genet 16: e1009215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Granneman S, Osheim YN, Beyer AL, Baserga SJ (2006) The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc Natl Acad Sci USA 103: 9464–9469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack KE, Bohnsack MT (2019) Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J 38: e100278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D (2009) Prp43 bound at different sites on the pre‐rRNA performs distinct functions in ribosome synthesis. Mol Cell 36: 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AT, Mangus DA, He F, Jacobson A (2001) Absence of Dbp2p alters both nonsense‐mediated mRNA decay and rRNA processing. Mol Cell Biol 21: 7366–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boneberg FM, Brandmann T, Kobel L, van den Heuvel J, Bargsten K, Bammert L, Kutay U, Jinek M (2019) Molecular mechanism of the RNA helicase DHX37 and its activation by UTP14A in ribosome biogenesis. RNA 25: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman LH, Rabin B, Schlessinger D (1981) Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res 9: 4951–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box JK, Paquet N, Adams MN, Boucher D, Bolderson E, O'Byrne KJ, Richard DJ (2016) Nucleophosmin: from structure and function to disease development. BMC Mol Biol 17: 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradatsch B, Katahira J, Kowalinski E, Bange G, Yao W, Sekimoto T, Baumgärtel V, Boese G, Bassler J, Wild K et al (2007) Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol Cell 27: 767–779 [DOI] [PubMed] [Google Scholar]
- Bradatsch B, Leidig C, Granneman S, Gnädig M, Tollervey D, Böttcher B, Beckmann R, Hurt E (2012) Structure of the pre‐60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat Struct Mol Biol 19: 1234–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun CM, Hackert P, Schmid CE, Bohnsack MT, Bohnsack KE, Perez‐Fernandez J (2020) Pol5 is required for recycling of small subunit biogenesis factors and for formation of the peptide exit tunnel of the large ribosomal subunit. Nucleic Acids Res 48: 405–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MW, Burkard KTD, Butler JS (1998) Rrp6p, the yeast homologue of the human PM‐Scl 100‐kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem 273: 13255–13263 [DOI] [PubMed] [Google Scholar]
- Brill SJ, DiNardo S, Voelkel‐Meiman K, Sternglanz R (1987) Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature 326: 414–416 [DOI] [PubMed] [Google Scholar]
- Brüning L, Hackert P, Martin R, Davila Gallesio J, Aquino GRR, Urlaub H, Sloan KE, Bohnsack MT (2018) RNA helicases mediate structural transitions and compositional changes in pre‐ribosomal complexes. Nat Commun 9: 5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlacu E, Lackmann F, Aguilar L‐C, Belikov S, van Nues R, Trahan C, Hector RD, Dominelli‐Whiteley N, Cockroft SL, Wieslander L et al (2017) High‐throughput RNA structure probing reveals critical folding events during early 60S ribosome assembly in yeast. Nat Commun 8: 714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater MJ, Pearson RB, McArthur GA, Hannan RD (2013) Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat Rev Cancer 13: 299–314 [DOI] [PubMed] [Google Scholar]
- Calviño FR, Kharde S, Ori A, Hendricks A, Wild K, Kressler D, Bange G, Hurt E, Beck M, Sinning I (2015) Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA‐binding site. Nat Commun 6: 6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carron C, O'Donohue MF, Choesmel V, Faubladier M, Gleizes PE (2011) Analysis of two human pre‐ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Res 39: 280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle CD, Sardana R, Dandekar V, Borgianini V, Johnson AW, Denicourt C (2013) Las1 interacts with Grc3 polynucleotide kinase and is required for ribosome synthesis in Saccharomyces cerevisiae . Nucleic Acids Res 41: 1135–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catez F, Dalla Venezia N, Marcel V, Zorbas C, Lafontaine DLJ, Diaz J‐J (2019) Ribosome biogenesis: An emerging druggable pathway for cancer therapeutics. Biochem Pharmacol 159: 74–81 [DOI] [PubMed] [Google Scholar]
- Cech TR (2000) The ribosome is a ribozyme. Science 289: 878–879 [DOI] [PubMed] [Google Scholar]
- Chaker‐Margot M, Klinge S (2019) Assembly and early maturation of large subunit precursors. RNA 25: 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker‐Margot M, Hunziker M, Barandun J, Dill BD, Klinge S (2015) Stage‐specific assembly events of the 6‐MDa small‐subunit processome initiate eukaryotic ribosome biogenesis. Nat Struct Mol Biol 22: 920–923 [DOI] [PubMed] [Google Scholar]
- Chaker‐Margot M, Barandun J, Hunziker M, Klinge S (2017) Architecture of the yeast small subunit processome. Science 355: eaal1880 [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Uechi T, Kenmochi N (2011) Guarding the ‘translation apparatus’: defective ribosome biogenesis and the p53 signaling pathway. Wiley Interdiscip Rev RNA 2: 507–522 [DOI] [PubMed] [Google Scholar]
- Charette JM, Baserga SJ (2010) The DEAD‐box RNA helicase‐like Utp25 is an SSU processome component. RNA 16: 2156–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y‐L, Capeyrou R, Humbert O, Mouffok S, Kadri YA, Lebaron S, Henras AK, Henry Y (2014) The telomerase inhibitor Gno1p/PINX1 activates the helicase Prp43p during ribosome biogenesis. Nucleic Acids Res 42: 7330–7345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Xie Z, Yang F, Ye K (2017) Stepwise assembly of the earliest precursors of large ribosomal subunits in yeast. Nucleic Acids Res 45: 6837–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Suzuki H, Kobayashi Y, Wang AC, DiMaio F, Kawashima SA, Walz T, Kapoor TM (2018) Structural insights into Mdn1, an essential AAA protein required for ribosome biogenesis. Cell 175: 822–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kellner N, Berninghausen O, Hurt E, Beckmann R (2017) 3.2‐Å‐resolution structure of the 90S preribosome before A1 pre‐rRNA cleavage. Nat Struct Mol Biol 24: 954–964 [DOI] [PubMed] [Google Scholar]
- Cheng J, Lau B, la Venuta G, Ameismeier M, Berninghausen O, Hurt E, Beckmann R (2020) 90S pre‐ribosome transformation into the primordial 40S subunit. Science 369: 1470–1476 [DOI] [PubMed] [Google Scholar]
- Cheng J, Lau B, Thoms M, Ameismeier M, Berninghausen O, Hurt E, Beckmann R (2022) The nucleoplasmic phase of pre‐40S formation prior to nuclear export. Nucleic Acids Res 50: 11924–11937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choque E, Marcellin M, Burlet‐Schiltz O, Gadal O, Dez C (2011) The nucleolar protein Nop19p interacts preferentially with Utp25p and Dhr2p and is essential for the production of the 40S ribosomal subunit in Saccharomyces cerevisiae . RNA Biol 8: 1158–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choque E, Schneider C, Gadal O, Dez C (2018) Turnover of aberrant pre‐40S pre‐ribosomal particles is initiated by a novel endonucleolytic decay pathway. Nucleic Acids Res 46: 4699–4714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury P, Hackert P, Memet I, Sloan KE, Bohnsack MT (2019) The human RNA helicase DHX37 is required for release of the U3 snoRNP from pre‐ribosomal particles. RNA Biol 16: 54–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury P, Kretschmer J, Hackert P, Bohnsack KE, Bohnsack MT (2020) The DExD box ATPase DDX55 is recruited to domain IV of the 28S ribosomal RNA by its C‐terminal region. RNA Biol 18: 1124–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciganda M, Williams N (2011) Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip Rev RNA 2: 523–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Zomerdijk JC, Beckmann H, Zhou S, Admon A, Tjian R (1994) Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science 266: 1966–1972 [DOI] [PubMed] [Google Scholar]
- Couté Y, Kindbeiter K, Belin S, Dieckmann R, Duret L, Bezin L, Sanchez J‐C, Diaz J‐J (2008) ISG20L2, a novel vertebrate nucleolar exoribonuclease involved in ribosome biogenesis. Mol Cell Proteomics 7: 546–559 [DOI] [PubMed] [Google Scholar]
- Dakshinamurthy A, Nyswaner KM, Farabaugh PJ, Garfinkel DJ (2010) BUD22 Affects Ty1 retrotransposition and ribosome biogenesis in Saccharomyces cerevisiae . Genetics 185: 1193–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE (1968) Ribonucleic acids from animal cells. Bacteriol Rev 32: 262–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila Gallesio J, Hackert P, Bohnsack KE, Bohnsack MT (2020) Sgd1 is an MIF4G domain‐containing cofactor of the RNA helicase Fal1 and associates with the 5′ domain of the 18S rRNA sequence. RNA Biol 17: 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Sanz‐Martínez E, Remacha M (2005) The essential WD‐repeat protein Rsa4p is required for rRNA processing and intra‐nuclear transport of 60S ribosomal subunits. Nucleic Acids Res 33: 5728–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Karbstein K, Woolford JL (2015) Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo . Annu Rev Biochem 84: 93–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Marchis ML, Giorgi A, Schinina ME, Bozzoni I, Fatica A (2005) Rrp15p, a novel component of pre‐ribosomal particles required for 60S ribosome subunit maturation. RNA 11: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ (2002) rRNA modifications and ribosome function. Trends Biochem Sci 27: 344–351 [DOI] [PubMed] [Google Scholar]
- Dembowski JA, Kuo B, Woolford JL Jr (2013) Has1 regulates consecutive maturation and processing steps for assembly of 60S ribosomal subunits. Nucleic Acids Res 41: 7889–7904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demoinet E, Jacquier A, Lutfalla G, Fromont‐Racine M (2007) The Hsp40 chaperone Jjj1 is required for the nucleo‐cytoplasmic recycling of preribosomal factors in Saccharomyces cerevisiae . RNA 13: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S, van Driel M, Voit R, Hekkelman M, Hulsen T, Hernandez N, Grummt I, Wehrens R, Stunnenberg H (2007) Identification of novel functional TBP‐binding sites and general factor repertoires. EMBO J 26: 944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Dlakić M, Tollervey D (2007) Roles of the HEAT repeat proteins Utp10 and Utp20 in 40S ribosome maturation. RNA 13: 1516–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G, Peddigari S, Mercer CA, Thomas G (2013) 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2‐p53 checkpoint. Cell Rep 4: 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörner K, Badertscher L, Horváth B, Hollandi R, Molnár C, Fuhrer T, Meier R, Sárazová M, van den Heuvel J, Zamboni N et al (2022) Genome‐wide RNAi screen identifies novel players in human 60S subunit biogenesis including key enzymes of polyamine metabolism. Nucleic Acids Res 50: 2872–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F, Compagnone‐Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, Beyer AL et al (2002) A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, An W, Zhu X, Sun Q, Qi J, Ye K (2020) Cryo‐EM structure of 90S small ribosomal subunit precursors in transition states. Science 369: 1477–1481 [DOI] [PubMed] [Google Scholar]
- Dutca LM, Gallagher JEG, Baserga SJ (2011) The initial U3 snoRNA:pre‐rRNA base pairing interaction required for pre‐18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res 39: 5164–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger DP, Dick FA, Denke E, Trumpower BL (1997) SQT1, which encodes an essential WD domain protein of Saccharomyces cerevisiae, suppresses dominant‐negative mutations of the ribosomal protein gene QSR1. Mol Cell Biol 17: 5146–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Hage A, Koper M, Kufel J, Tollervey D (2008) Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev 22: 1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel C, Neyer S, Cramer P (2018) Distinct mechanisms of transcription initiation by RNA polymerases I and II. Annu Rev Biophys 47: 425–446 [DOI] [PubMed] [Google Scholar]
- Faber AW, van Dijk M, Raué HA, Vos JC (2002) Ngl2p is a Ccr4p‐like RNA nuclease essential for the final step in 3′‐end processing of 5.8S rRNA in Saccharomyces cerevisiae . RNA 8: 1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S, Tants J‐N, Basquin J, Thoms M, Hurt E, Sattler M, Conti E (2017) Structural insights into the interaction of the nuclear exosome helicase Mtr4 with the preribosomal protein Nop53. RNA 23: 1780–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley KI, Surovtseva Y, Merkel J, Baserga SJ (2015) Determinants of mammalian nucleolar architecture. Chromosoma 124: 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley‐Barnes KI, McCann KL, Ogawa LM, Merkel J, Surovtseva YV, Baserga SJ (2018) Diverse regulators of human ribosome biogenesis discovered by changes in nucleolar number. Cell Rep 22: 1923–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley‐Barnes KI, Ogawa LM, Baserga SJ (2019) Ribosomopathies: old concepts, new controversies. Trends Genet 35: 754–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath S, Kobor MS, Philippi A, Greenblatt J, Tschochner H (2004) Dephosphorylation of RNA polymerase I by Fcp1p is required for efficient rRNA synthesis. J Biol Chem 279: 25251–25259 [DOI] [PubMed] [Google Scholar]
- Fatica A, Cronshaw AD, Dlaki M, Tollervey D (2002) Ssf1p prevents premature processing of an early pre‐60S ribosomal particle. Mol Cell 9: 341–351 [DOI] [PubMed] [Google Scholar]
- Fatica A, Oeffinger M, Dlakić M, Tollervey D (2003) Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol Cell Biol 23: 1798–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Tollervey D, Dlakić M (2004) PIN domain of Nob1p is required for D‐site cleavage in 20S pre‐rRNA. RNA 10: 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faza MB, Chang Y, Occhipinti L, Kemmler S, Panse VG (2012) Role of Mex67‐Mtr2 in the nuclear export of 40S pre‐ribosomes. PLoS Genet 8: e1002915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira‐Cerca S, Pöll G, Gleizes P‐E, Tschochner H, Milkereit P (2005) Roles of eukaryotic ribosomal proteins in maturation and transport of pre‐18S rRNA and ribosome function. Mol Cell 20: 263–275 [DOI] [PubMed] [Google Scholar]
- Ferreira‐Cerca S, Pöll G, Kühn H, Neueder A, Jakob S, Tschochner H, Milkereit P (2007) Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol Cell 28: 446–457 [DOI] [PubMed] [Google Scholar]
- Ferreira‐Cerca S, Sagar V, Schäfer T, Diop M, Wesseling AM, Lu H, Chai E, Hurt E, Laronde‐Leblanc N (2012) ATPase‐dependent role of the atypical kinase Rio2 on the evolving pre‐40S ribosomal subunit. Nat Struct Mol Biol 19: 1316–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira‐Cerca S, Kiburu I, Thomson E, LaRonde N, Hurt E (2014) Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre‐40S biogenesis factors in 80S‐like ribosomes. Nucleic Acids Res 42: 8635–8647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figaro S, Wacheul L, Schillewaert S, Graille M, Huvelle E, Mongeard R, Zorbas C, Lafontaine DLJ, Heurgué‐Hamard V (2012) Trm112 is required for Bud23‐mediated methylation of the 18S rRNA at position G1575. Mol Cell Biol 32: 2254–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch AJ, Hilcenko C, Basse N, Drynan LF, Goyenechea B, Menne TF, Fernández ÁG, Simpson P, D'Santos CS, Arends MJ et al (2011) Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman‐Diamond syndrome. Genes Dev 25: 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner E, Haindl M, Muller S (2011) The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J 30: 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Bartel B, Varshavsky A (1989) The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338: 394–401 [DOI] [PubMed] [Google Scholar]
- Fischer U, Schäuble N, Schütz S, Altvater M, Chang Y, Boulos Faza M, Panse VG (2015) A non‐canonical mechanism for Crm1‐export cargo complex assembly. Elife 4: e05745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L (2006) Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev 20: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier MN, Pillon MC, Kocaman S, Gordon J, Stanley RE (2021) Structural overview of macromolecular machines involved in ribosome biogenesis. Curr Opin Struct Biol 67: 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EF, Prieto J‐L, McCann KL, McStay B, Baserga SJ (2012) NOL11, implicated in the pathogenesis of north American Indian childhood cirrhosis, is required for pre‐rRNA transcription and processing. PLoS Genet 8: e1002892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm L, Falk S, Flemming D, Schuller JM, Thoms M, Conti E, Hurt E (2017) Reconstitution of the complete pathway of ITS2 processing at the pre‐ribosome. Nat Commun 8: 1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes JL, Datta K, Sullivan SM, Walker A, Maddock JR (2007) In vivo functional characterization of the Saccharomyces cerevisiae 60S biogenesis GTPase Nog1. Mol Genet Genomics 278: 105–123 [DOI] [PubMed] [Google Scholar]
- Gadal O, Strauß D, Kessl J, Trumpower B, Tollervey D, Hurt E (2001) Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence‐containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 21: 3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Labarre S, Boschiero C, Thuriaux P (2002) Hmo1, an HMG‐box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J 21: 5498–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher JEG, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ (2004) RNA polymerase I transcription and pre‐rRNA processing are linked by specific SSU processome components. Genes Dev 18: 2506–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalinda M, Ohmayer U, Jakovljevic J, Kumcuoglu B, Woolford J, Mbom B, Lin L, Woolford JL (2014) A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev 28: 198–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Gómez JJ, Babiano R, Lebaron S, Froment C, Monsarrat B, Henry Y, de la Cruz J (2011) Nop6, a component of 90S pre‐ribosomal particles, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae . RNA Biol 8: 112–124 [DOI] [PubMed] [Google Scholar]
- Gartmann M, Blau M, Armache J‐P, Mielke T, Topf M, Beckmann R (2010) Mechanism of eIF6‐mediated inhibition of ribosomal subunit joining. J Biol Chem 285: 14848–14851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasse L, Flemming D, Hurt E (2015) Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol Cell 60: 808–815 [DOI] [PubMed] [Google Scholar]
- Geerlings TH, Faber AW, Bister MD, Vos JC, Raué HA (2003) Rio2p, an evolutionarily conserved, low abundant protein kinase essential for processing of 20 S pre‐rRNA in Saccharomyces cerevisiae . J Biol Chem 278: 22537–22545 [DOI] [PubMed] [Google Scholar]
- Gelperin D, Horton L, Beckman J, Hensold J, Lemmon SK (2001) Bms1p, a novel GTP‐binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA 7: 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardy S, Oborská‐Oplová M, Gillet L, Börner R, van Nues R, Leitner A, Michel E, Petkowski JJ, Granneman S, Sigel RKO et al (2021) Puf6 primes 60S pre‐ribosome nuclear export at low temperature. Nat Commun 12: 4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Meyer C, Benjamin‐Hong S, Rodriguez J, Briskin D, Bognanni C, Bogardus K, Steller H, Tuschl T (2017) The conserved RNA exonuclease Rexo5 is required for 3′ end maturation of 28S rRNA, 5S rRNA, and snoRNAs. Cell Rep 21: 758–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs MR, Fredrick K (2018) Roles of elusive translational GTPases come to light and inform on the process of ribosome biogenesis in bacteria. Mol Microbiol 107: 445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girbig M, Misiaszek AD, Müller CW (2022) Structural insights into nuclear transcription by eukaryotic DNA‐dependent RNA polymerases. Nat Rev Mol Cell Biol 23: 603–622 [DOI] [PubMed] [Google Scholar]
- Goldfarb KC, Cech TR (2017) Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes Dev 31: 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Kakihara Y, Krogan N, Greenblatt J, Emili A, Zhang Z, Houry WA (2009) An atlas of chaperone–protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol Syst Biol 5: 275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow SJ, Zomerdijk JCBM (2013) Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcell Biochem 61: 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J, Chapus FL, Viverette EG, Williams JG, Deterding LJ, Krahn JM, Borgnia MJ, Rodriguez J, Warren AJ, Stanley RE (2022) Cryo‐EM reveals the architecture of the PELP1‐WDR18 molecular scaffold. Nat Commun 13: 6783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JJ, Pathak S, Panov K, Kasciukovic T, Panova T, Russell J, Zomerdijk JCBM (2007) A novel TBP‐associated factor of SL1 functions in RNA polymerase I transcription. EMBO J 26: 1560–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Rybin V, Baßler J, Petfalski E, Strauß D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D et al (2002) 90S pre‐ribosomes include the 35S pre‐rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 10: 105–115 [DOI] [PubMed] [Google Scholar]
- Grandori C, Gomez‐Roman N, Felton‐Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ (2005) c‐Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7: 311–318 [DOI] [PubMed] [Google Scholar]
- Granneman S, Gallagher JEG, Vogelzangs J, Horstman W, van Venrooij WJ, Baserga SJ, Pruijn GJM (2003) The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60–80S ribonucleoprotein complexes. Nucleic Acids Res 31: 1877–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Bernstein KA, Bleichert F, Baserga SJ (2006a) Comprehensive mutational analysis of yeast DEXD/H Box RNA helicases required for small ribosomal subunit synthesis. Mol Cell Biol 26: 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Lin C, Champion EA, Nandineni MR, Zorca C, Baserga SJ (2006b) The nucleolar protein Esf2 interacts directly with the DExD/H box RNA helicase, Dbp8, to stimulate ATP hydrolysis. Nucleic Acids Res 34: 3189–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Kudla G, Petfalski E, Tollervey D (2009) Identification of protein binding sites on U3 snoRNA and pre‐rRNA by UV cross‐linking and high‐throughput analysis of cDNAs. Proc Natl Acad Sci USA 106: 9613–9618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Petfalski E, Tollervey D (2011) A cluster of ribosome synthesis factors regulate pre‐rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J 30: 4006–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ (2016) Mechanistic insight into eukaryotic 60S ribosomal subunit biogenesis by cryo‐electron microscopy. RNA 22: 1643–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Montellese C, Ban N (2012) Cryo‐EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat Struct Mol Biol 19: 1228–1233 [DOI] [PubMed] [Google Scholar]
- Greber BJ, Gerhardy S, Leitner A, Leibundgut M, Salem M, Boehringer D, Leulliot N, Aebersold R, Panse VG, Ban N (2016) Insertion of the biogenesis factor Rei1 probes the ribosomal tunnel during 60S maturation. Cell 164: 91–102 [DOI] [PubMed] [Google Scholar]
- Grou CP, Pinto MP, Mendes AV, Domingues P, Azevedo JE (2015) The de novo synthesis of ubiquitin: identification of deubiquitinases acting on ubiquitin precursors. Sci Rep 5: 12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I (2010) Wisely chosen paths ‐ regulation of rRNA synthesis: delivered on 30 June 2010 at the 35th FEBS Congress in Gothenburg, Sweden. FEBS J 277: 4626–4639 [DOI] [PubMed] [Google Scholar]
- Guglielmi B, Werner M (2002) The yeast homolog of human PinX1 is involved in rRNA and small nucleolar RNA maturation, not in telomere elongation inhibition. J Biol Chem 277: 35712–35719 [DOI] [PubMed] [Google Scholar]
- Hackmann A, Gross T, Baierlein C, Krebber H (2011) The mRNA export factor Npl3 mediates the nuclear export of large ribosomal subunits. EMBO Rep 12: 1024–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler RA, Engelke DR (2006) Spatial organization of transcription by RNA polymerase III. Nucleic Acids Res 34: 4826–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X‐R, Sasaki N, Jackson SC, Wang P, Li Z, Smith MD, Xie L, Chen X, Zhang Y, Marzluff WF et al (2020) CRL4 DCAF1/VprBP E3 ubiquitin ligase controls ribosome biogenesis, cell proliferation, and development. Sci Adv 6: eabd6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan KM, Soo P, Wong MS, Lee JK, Hein N, Poh P, Wysoke KD, Williams TD, Montellese C, Smith LK et al (2022) Nuclear stabilization of p53 requires a functional nucleolar surveillance pathway. Cell Rep 41: 111571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan N, Ghosh S, Palakodeti D (2022) The story of rRNA expansion segments: finding functionality amidst diversity. Wiley Interdiscip Rev RNA 14: e1732 [DOI] [PubMed] [Google Scholar]
- Hector RD, Burlacu E, Aitken S, le Bihan T, Tuijtel M, Zaplatina A, Cook AG, Granneman S (2014) Snapshots of pre‐rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res 42: 12138–12154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges J, West M, Johnson AW (2005) Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J 24: 567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held WA, Mizushima S, Nomura M (1973) Reconstitution of Escherichia coli 30 S ribosomal subunits from purified molecular components. J Biol Chem 248: 5720–5730 [PubMed] [Google Scholar]
- Hellmich UA, Weis BL, Lioutikov A, Wurm JP, Kaiser M, Christ NA, Hantke K, Kötter P, Entian K‐D, Schleiff E et al (2013) Essential ribosome assembly factor Fap7 regulates a hierarchy of RNA–protein interactions during small ribosomal subunit biogenesis. Proc Natl Acad Sci USA 110: 15253–15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M (2006) Post‐transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res 34: 721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson AS, Warburton D, Atwood KC (1972) Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci USA 69: 3394–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Plisson‐Chastang C, O'Donohue M‐F, Chakraborty A, Gleizes P‐E (2015) An overview of pre‐ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6: 225–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer A, Thomson E, Schmidt C, Berninghausen O, Becker T, Hurt E, Beckmann R (2017) Cryo‐EM structure of a late pre‐40S ribosomal subunit from Saccharomyces cerevisiae . Elife 6: e30189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierlmeier T, Merl J, Sauert M, Perez‐Fernandez J, Schultz P, Bruckmann A, Hamperl S, Ohmayer U, Rachel R, Jacob A et al (2013) Rrp5p, Noc1p and Noc2p form a protein module which is part of early large ribosomal subunit precursors in S. cerevisiae . Nucleic Acids Res 41: 1191–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraishi N, Ishida Y, Sudo H, Nagahama M (2018) WDR74 participates in an early cleavage of the pre‐rRNA processing pathway in cooperation with the nucleolar AAA‐ATPase NVL2. Biochem Biophys Res Commun 495: 116–123 [DOI] [PubMed] [Google Scholar]
- Hirose T, Steitz JA (2001) Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proc Natl Acad Sci USA 98: 12914–12919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JH‐N, Kallstrom G, Johnson AW (2000) Nmd3p is a Crm1p‐dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol 151: 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn DM, Mason SL, Karbstein K (2011) Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production. J Biol Chem 286: 34082–34087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Parker M, Karbstein K (2022) The modifying enzyme Tsr3 establishes the hierarchy of Rio kinase binding in 40S ribosome assembly. RNA 28: 568–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung N‐J, Johnson AW (2006) Nuclear recycling of the pre‐60S ribosomal subunit‐associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae . Mol Cell Biol 26: 3718–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker M, Barandun J, Petfalski E, Tan D, Delan‐Forino C, Molloy KR, Kim KH, Dunn‐Davies H, Shi Y, Chaker‐Margot M et al (2016) UtpA and UtpB chaperone nascent pre‐ribosomal RNA and U3 snoRNA to initiate eukaryotic ribosome assembly. Nat Commun 7: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker M, Barandun J, Buzovetsky O, Steckler C, Molina H, Klinge S (2019) Conformational switches control early maturation of the eukaryotic small ribosomal subunit. Elife 8: e45185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E, Hannus S, Schmelzl B, Lau D, Tollervey D, Simos G (1999) A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran‐cycle and nucleoporin mutants. J Cell Biol 144: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iouk TL, Aitchison JD, Maguire S, Wozniak RW (2001) Rrb1p, a yeast nuclear WD‐repeat protein involved in the regulation of ribosome biosynthesis. Mol Cell Biol 21: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail S, Flemming D, Thoms M, Gomes‐Filho JV, Randau L, Beckmann R, Hurt E (2022) Emergence of the primordial pre‐60S from the 90S pre‐ribosome. Cell Rep 39: 110640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Akamatsu Y, Noma A, Kimura S, Miyauchi K, Ikeuchi Y, Suzuki T, Suzuki T (2014) A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae . J Biol Chem 289: 26201–26212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer‐Bierhoff A, Krogh N, Tessarz P, Ruppert T, Nielsen H, Grummt I (2018) SIRT7‐dependent deacetylation of fibrillarin controls histone H2A methylation and rRNA synthesis during the cell cycle. Cell Rep 25: 2946–2954 [DOI] [PubMed] [Google Scholar]
- Izumikawa K, Ishikawa H, Yoshikawa H, Fujiyama S, Watanabe A, Aburatani H, Tachikawa H, Hayano T, Miura Y, Isobe T et al (2019) LYAR potentiates rRNA synthesis by recruiting BRD2/4 and the MYST‐type acetyltransferase KAT7 to rDNA. Nucleic Acids Res 47: 10357–10372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafar M, Contreras J, Dominique C, Martín‐Villanueva S, Capeyrou R, Vitali P, Rodríguez‐Galán O, Velasco C, Humbert O, Watkins NJ et al (2021a) Association of snR190 snoRNA chaperone with early pre‐60S particles is regulated by the RNA helicase Dbp7 in yeast. Nat Commun 12: 6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafar M, Paraqindes H, Gabut M, Diaz J‐J, Marcel V, Durand S (2021b) 2′O‐ribose methylation of ribosomal RNAs: natural diversity in living organisms, biological processes, and diseases. Cell 10: 1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel S, Mingot J‐M, Schwarzmaier P, Hartmann E, Görlich D (2002) Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J 21: 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeeninga RE, van Delft Y, de Graaff‐Vincent M, Dirks‐Mulder A, Venema J, Raué HA (1997) Variable regions V13 and V3 of Saccharomyces cerevisiae contain structural features essential for normal biogenesis and stability of 5.8S and 25S rRNA. RNA 3: 476–488 [PMC free article] [PubMed] [Google Scholar]
- Johnson AW, Lund E, Dahlberg J (2002) Nuclear export of ribosomal subunits. Trends Biochem Sci 27: 580–585 [DOI] [PubMed] [Google Scholar]
- Jorjani H, Kehr S, Jedlinski DJ, Gumienny R, Hertel J, Stadler PF, Zavolan M, Gruber AR (2016) An updated human snoRNAome. Nucleic Acids Res 44: 5068–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczanowska M, Rydén‐Aulin M (2007) Ribosome biogenesis and the translation process in Escherichia coli . Microbiol Mol Biol Rev 71: 477–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallstrom G, Hedges J, Johnson A (2003) The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol Cell Biol 23: 4344–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampen KR, Sulima SO, Vereecke S, De Keersmaecker K (2020) Hallmarks of ribosomopathies. Nucleic Acids Res 48: 1013–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappel L, Loibl M, Zisser G, Klein I, Fruhmann G, Gruber C, Unterweger S, Rechberger G, Pertschy B, Bergler H (2012) Rlp24 activates the AAA‐ATPase Drg1 to initiate cytoplasmic pre‐60S maturation. J Cell Biol 199: 771–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K, Doudna JA (2006) GTP‐dependent formation of a ribonucleoprotein subcomplex required for ribosome biogenesis. J Mol Biol 356: 432–443 [DOI] [PubMed] [Google Scholar]
- Kargas V, Castro‐Hartmann P, Escudero‐Urquijo N, Dent K, Hilcenko C, Sailer C, Zisser G, Marques‐Carvalho MJ, Pellegrino S, Wawiórka L et al (2019) Mechanism of completion of peptidyltransferase centre assembly in eukaryotes. Elife 8: e44904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater L, Thoms M, Barrio‐Garcia C, Cheng J, Ismail S, Ahmed YL, Bange G, Kressler D, Berninghausen O, Sinning I et al (2017) Visualizing the assembly pathway of nucleolar pre‐60S ribosomes. Cell 171: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater L, Mitterer V, Thoms M, Cheng J, Berninghausen O, Beckmann R, Hurt E (2020) Construction of the central protuberance and L1 stalk during 60S subunit biogenesis. Mol Cell 79: 615–628 [DOI] [PubMed] [Google Scholar]
- Keener J, Dodd JA, Lalo D, Nomura M (1997) Histones H3 and H4 are components of upstream activation factor required for the high‐level transcription of yeast rDNA by RNA polymerase I. Proc Natl Acad Sci USA 94: 13458–13462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner M, Rohrmoser M, Forné I, Voss K, Burger K, Mühl B, Gruber‐Eber A, Kremmer E, Imhof A, Eick D (2015) DEAD‐box helicase DDX27 regulates 3′ end formation of ribosomal 47S RNA and stably associates with the PeBoW‐complex. Exp Cell Res 334: 146–159 [DOI] [PubMed] [Google Scholar]
- Kemmler S, Occhipinti L, Veisu M, Panse VG (2009) Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J Cell Biol 186: 863–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys DA, Lee BS, Dodd JA, Nguyen TT, Vu L, Fantino E, Burson LM, Nogi Y, Nomura M (1996) Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev 10: 887–903 [DOI] [PubMed] [Google Scholar]
- Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP (2015) Structure of the human 80S ribosome. Nature 520: 640–645 [DOI] [PubMed] [Google Scholar]
- Khoshnevis S, Askenasy I, Johnson MC, Dattolo MD, Young‐Erdos CL, Stroupe ME, Karbstein K (2016) The DEAD‐box protein Rok1 orchestrates 40S and 60S ribosome assembly by promoting the release of Rrp5 from pre‐40S ribosomes to allow for 60S maturation. PLoS Biol 14: e1002480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnevis S, Dreggors‐Walker RE, Marchand V, Motorin Y, Ghalei H (2022) Ribosomal RNA 2′‐O‐methylations regulate translation by impacting ribosome dynamics. Proc Natl Acad Sci USA 119: e2117334119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss‐László Z, Henry Y, Bachellerie JP, Caizergues‐Ferrer M, Kiss T (1996) Site‐specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Klingauf‐Nerurkar P, Gillet LC, Portugal‐Calisto D, Oborská‐Oplová M, Jäger M, Schubert OT, Pisano A, Peña C, Rao S, Altvater M et al (2020) The GTPase Nog1 co‐ordinates the assembly, maturation and quality control of distant ribosomal functional centers. Elife 9: e52474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Woolford JL (2019) Ribosome assembly coming into focus. Nat Rev Mol Cell Biol 20: 116–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S, Voigts‐Hoffmann F, Leibundgut M, Arpagaus S, Ban N (2011) Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334: 941–948 [DOI] [PubMed] [Google Scholar]
- Klinge S, Voigts‐Hoffmann F, Leibundgut M, Ban N (2012) Atomic structures of the eukaryotic ribosome. Trends Biochem Sci 37: 189–198 [DOI] [PubMed] [Google Scholar]
- Knutson BA, Hahn S (2013) TFIIB‐related factors in RNA polymerase I transcription. Biochim Biophys Acta 1829: 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberna K, Malínský J, Pliss A, Mašata M, Večeřová J, Fialová M, Bednár J, Raška I (2002) Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees” in situ . J Cell Biol 157: 743–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konikkat S, Woolford JL (2017) Principles of 60S ribosomal subunit assembly emerging from recent studies in yeast. Biochem J 474: 195–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konikkat S, Biedka S, Woolford JL (2017) The assembly factor Erb1 functions in multiple remodeling events during 60S ribosomal subunit assembly in S. cerevisiae . Nucleic Acids Res 45: 4853–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E (2010) A dual function for chaperones SSB–RAC and the NAC nascent polypeptide–associated complex on ribosomes. J Cell Biol 189: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornprobst M, Turk M, Kellner N, Cheng J, Flemming D, Koš‐Braun I, Koš M, Thoms M, Berninghausen O, Beckmann R et al (2016) Architecture of the 90S pre‐ribosome: a structural view on the birth of the eukaryotic ribosome. Cell 166: 380–393 [DOI] [PubMed] [Google Scholar]
- Koš M, Tollervey D (2010) Yeast pre‐rRNA processing and modification occur cotranscriptionally. Mol Cell 37: 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushar ML, Krupp F, Harnett D, Turko P, Ambrozkiewicz MC, Sprink T, Imami K, Günnigmann M, Zinnall U, Vieira‐Vieira CH et al (2021) Protein synthesis in the developing neocortex at near‐atomic resolution reveals Ebp1‐mediated neuronal proteostasis at the 60S tunnel exit. Mol Cell 81: 304–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, de la Cruz J, Rojo M, Linder P (1997) Fal1p is an essential DEAD‐box protein involved in 40S‐ribosomal‐subunit biogenesis in Saccharomyces cerevisiae . Mol Cell Biol 17: 7283–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Rojo M, Linder P, de la Cruz J (1999) Spb1p is a putative methyltransferase required for 60S ribosomal subunit biogenesis in Saccharomyces cerevisiae . Nucleic Acids Res 27: 4598–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Roser D, Pertschy B, Hurt E (2008) The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre‐60S particles. J Cell Biol 181: 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Baßler J (2010) Driving ribosome assembly. Biochim Biophys Acta 1803: 673–683 [DOI] [PubMed] [Google Scholar]
- Kressler D, Bange G, Ogawa Y, Stjepanovic G, Bradatsch B, Pratte D, Amlacher S, Strauß D, Yoneda Y, Katahira J et al (2012) Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science 338: 666–671 [DOI] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Baßler J (2017) A puzzle of life: crafting ribosomal subunits. Trends Biochem Sci 42: 640–654 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Peng W‐T, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP et al (2004) High‐definition macromolecular composition of yeast RNA‐processing complexes. Mol Cell 13: 225–239 [DOI] [PubMed] [Google Scholar]
- Kufel J, Dichtl B, Tollervey D (1999) Yeast Rnt1p is required for cleavage of the pre‐ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5: 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn H, Hierlmeier T, Merl J, Jakob S, Aguissa‐Touré A‐H, Milkereit P, Tschochner H (2009) The Noc‐domain containing C‐terminus of Noc4p mediates both formation of the Noc4p‐Nop14p submodule and its incorporation into the SSU processome. PLoS One 4: e8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackmann F, Belikov S, Burlacu E, Granneman S, Wieslander L (2018) Maturation of the 90S pre‐ribosome requires Mrd1 dependent U3 snoRNA and 35S pre‐rRNA structural rearrangements. Nucleic Acids Res 46: 3692–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe T, García‐Gómez JJ, de La Cruz J, Roser D, Hurt E, Linder P, Kressler D (2009) Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol Microbiol 72: 69–84 [DOI] [PubMed] [Google Scholar]
- Lacoux C, Wacheul L, Saraf K, Pythoud N, Huvelle E, Figaro S, Graille M, Carapito C, Lafontaine DLJ, Heurgué‐Hamard V (2020) The catalytic activity of the translation termination factor methyltransferase Mtq2‐Trm112 complex is required for large ribosomal subunit biogenesis. Nucleic Acids Res 48: 12310–12325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D, Delcour J, Glasser A‐L, Desgres J, Vandenhaute J (1994) The DIM1 gene responsible for the conserved m62Am62A dimethylation in the 3′‐terminal loop of 18 S rRNA is essential in yeast. J Mol Biol 241: 492–497 [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ, Preiss T, Tollervey D (1998) Yeast 18S rRNA Dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol Cell Biol 18: 2360–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP (2021) The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol 22: 165–182 [DOI] [PubMed] [Google Scholar]
- Lam YW, Lamond AI, Mann M, Andersen JS (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol 17: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamanna AC, Karbstein K (2011) An RNA conformational switch regulates pre‐18S rRNA cleavage. J Mol Biol 405: 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry‐Voyer A‐M, Bilodeau S, Bergeron D, Dionne KL, Port SA, Rouleau C, Boisvert F‐M, Kehlenbach RH, Bachand F (2016) Human PDCD2L is an export substrate of CRM1 that associates with 40S ribosomal subunit precursors. Mol Cell Biol 36: 3019–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPeruta AJ, Micic J, Woolford JL Jr (2022) Additional principles that govern the release of pre‐ribosomes from the nucleolus into the nucleoplasm in yeast. Nucleic Acids Res gkac430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre B, Purushothaman SK (2004) Spb1p‐directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing Stage. Mol Cell 16: 663–669 [DOI] [PubMed] [Google Scholar]
- Larburu N, Montellese C, O'Donohue MF, Kutay U, Gleizes PE, Plisson‐Chastang C (2016) Structure of a human pre‐40S particle points to a role for RACK1 in the final steps of 18S rRNA processing. Nucleic Acids Res 44: 8465–8478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau B, Cheng J, Flemming D, la Venuta G, Berninghausen O, Beckmann R, Hurt E (2021) Structure of the maturing 90S pre‐ribosome in association with the RNA exosome. Mol Cell 81: 293–303 [DOI] [PubMed] [Google Scholar]
- Layat E, Probst AV, Tourmente S (2013) Structure, function and regulation of transcription factor IIIA: from Xenopus to Arabidopsis. Biochim Biophys Acta 1829: 274–282 [DOI] [PubMed] [Google Scholar]
- Lazdins IB, Delannoy M, Sollner‐Webb B (1997) Analysis of nucleolar transcription and processing domains and pre‐rRNA movements by in situ hybridization. Chromosoma 105: 481–495 [DOI] [PubMed] [Google Scholar]
- Lebaron S, Schneider C, van Nues RW, Swiatkowska A, Walsh D, Böttcher B, Granneman S, Watkins NJ, Tollervey D (2012) Proofreading of pre‐40S ribosome maturation by a translation initiation factor and 60S subunits. Nat Struct Mol Biol 19: 744–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaron S, Segerstolpe Å, French SL, Dudnakova T, de Lima Alves F, Granneman S, Rappsilber J, Beyer AL, Wieslander L, Tollervey D (2013) Rrp5 binding at multiple sites coordinates pre‐rRNA processing and assembly. Mol Cell 52: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton A, Saveanu C, Decourty L, Rain JC, Jacquier A, Fromont‐Racine M (2006) A functional network involved in the recycling of nucleocytoplasmic pre‐60S factors. J Cell Biol 173: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Baserga SJ (1999) Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre‐18S rRNA processing. Mol Cell Biol 19: 5441–5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds NB, Small EC, Hiley S, Hughes TR, Staley JP (2006) The splicing factor Prp43p, a DEAH Box ATPase, functions in ribosome biogenesis. Mol Cell Biol 26: 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidig C, Bange G, Kopp J, Amlacher S, Aravind A, Wickles S, Witte G, Hurt E, Beckmann R, Sinning I (2013) Structural characterization of a eukaryotic chaperone—the ribosome‐associated complex. Nat Struct Mol Biol 20: 23–28 [DOI] [PubMed] [Google Scholar]
- Leidig C, Thoms M, Holdermann I, Bradatsch B, Berninghausen O, Bange G, Sinning I, Hurt E, Beckmann R (2014) 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat Commun 5: 3491 [DOI] [PubMed] [Google Scholar]
- Létoquart J, Huvelle E, Wacheul L, Bourgeois G, Zorbas C, Graille M, Heurgué‐Hamard V, Lafontaine DLJ (2014) Structural and functional studies of Bud23–Trm112 reveal 18S rRNA N7‐G1575 methylation occurs on late 40S precursor ribosomes. Proc Natl Acad Sci USA 111: E5518–E5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Liu Q, Fournier MJ (2007) rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell 28: 965–977 [DOI] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Fournier MJ (2009) Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre‐rRNA processing. RNA 15: 1716–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K‐J, Yueh L‐Y, Hsu N‐H, Lai J‐S, Lo K‐Y (2019) Puf6 and Loc1 are the dedicated chaperones of ribosomal protein Rpl43 in Saccharomyces cerevisiae . Int J Mol Sci 20: 5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Zuo M‐Q, Zhang Y, Li N, Ma C, Dong M‐Q, Gao N (2020) Structural snapshots of human pre‐60S ribosomal particles before and after nuclear export. Nat Commun 11: 3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström MS, Zhang Y (2008) Ribosomal protein S9 is a novel B23/NPM‐binding protein required for normal cell proliferation. J Biol Chem 283: 15568–15576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little RD, Braaten DC (1989) Genomic organization of human 5 S rDNA and sequence of one tandem repeat. Genomics 4: 376–383 [DOI] [PubMed] [Google Scholar]
- Liu PCC, Thiele DJ (2001) Novel stress‐responsive genes EMG1 and NOP14 encode conserved, interacting proteins required for 40S ribosome biogenesis. Mol Biol Cell 12: 3644–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KY, Li Z, Wang F, Marcotte EM, Johnson AW (2009) Ribosome stalk assembly requires the dual‐specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J Cell Biol 186: 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K‐Y, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW (2010) Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell 39: 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y‐H, Romes EM, Pillon MC, Sobhany M, Stanley RE (2017) Structural analysis reveals features of ribosome assembly factor Nsa1/WDR74 important for localization and interaction with Rix7/NVL2. Structure 25: 762–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y‐H, Sobhany M, Hsu AL, Ford BL, Krahn JM, Borgnia MJ, Stanley RE (2019) Cryo‐EM structure of the essential ribosome assembly AAA‐ATPase Rix7. Nat Commun 10: 513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofgren LA, Uehling JK, Branco S, Bruns TD, Martin F, Kennedy PG (2019) Genome‐based estimates of fungal rDNA copy number variation across phylogenetic scales and ecological lifestyles. Mol Ecol 28: 721–730 [DOI] [PubMed] [Google Scholar]
- Lv K, Gong C, Antony C, Han X, Ren J‐G, Donaghy R, Cheng Y, Pellegrino S, Warren AJ, Paralkar VR et al (2021) HectD1 controls hematopoietic stem cell regeneration by coordinating ribosome assembly and protein synthesis. Cell Stem Cell 28: 1275–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z, Allmang C, Tollervey D, Séraphin B (1996) Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro . Science 272: 268–270 [DOI] [PubMed] [Google Scholar]
- Ma C, Wu S, Li N, Chen Y, Yan K, Li Z, Zheng L, Lei J, Woolford JL, Gao N (2017) Structural snapshot of cytoplasmic pre‐60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1. Nat Struct Mol Biol 24: 214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R, Chen K, Lu Z, Chen H, Shi YG et al (2019) N6‐Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol 15: 88–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madru C, Lebaron S, Blaud M, Delbos L, Pipoli J, Pasmant E, Réty S, Leulliot N (2015) Chaperoning 5S RNA assembly. Genes Dev 29: 1432–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Małecki JM, Odonohue M‐F, Kim Y, Jakobsson ME, Gessa L, Pinto R, Wu J, Davydova E, Moen A, Olsen JV et al (2021) Human METTL18 is a histidine‐specific methyltransferase that targets RPL3 and affects ribosome biogenesis and function. Nucleic Acids Res 49: 3185–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyutin AG, Musalgaonkar S, Patchett S, Frank J, Johnson AW (2017) Nmd3 is a structural mimic of eIF 5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis. EMBO J 36: 854–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikas R‐G, Thomson E, Thoms M, Hurt E (2016) The K+‐dependent GTPase Nug1 is implicated in the association of the helicase Dbp10 to the immature peptidyl transferase centre during ribosome maturation. Nucleic Acids Res 44: 1800–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Hackert P, Ruprecht M, Simm S, Brüning L, Mirus O, Sloan KE, Kudla G, Schleiff E, Bohnsack MT (2014) A pre‐ribosomal RNA interaction network involving snoRNAs and the Rok1 helicase. RNA 20: 1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín‐Villanueva S, Fernández‐Pevida A, Kressler D, de la Cruz J (2019) The ubiquitin moiety of Ubi1 is required for productive expression of ribosomal protein eL40 in Saccharomyces cerevisiae . Cell 8: 850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín‐Villanueva S, Fernández‐Pevida A, Fernández‐Fernández J, Kressler D, de la Cruz J (2020) Ubiquitin release from eL40 is required for cytoplasmic maturation and function of 60S ribosomal subunits in Saccharomyces cerevisiae . FEBS J 287: 345–360 [DOI] [PubMed] [Google Scholar]
- Martín‐Villanueva S, Gutiérrez G, Kressler D, de la Cruz J (2021) Ubiquitin and ubiquitin‐like proteins and domains in ribosome production and function: chance or necessity? Int J Mol Sci 22: 4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marz M, Gruber AR, Höner zu Siederdissen C, Amman F, Badelt S, Bartschat S, Bernhart SH, Beyer W, Kehr S, Lorenz R et al (2011) Animal snoRNAs and scaRNAs with exceptional structures. RNA Biol 8: 938–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Granneman S, Thoms M, Manikas RG, Tollervey D, Hurt E (2014) Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 505: 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Ben‐Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M (2012) One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol 19: 560–567 [DOI] [PubMed] [Google Scholar]
- Memet I, Doebele C, Sloan KE, Bohnsack MT (2017) The G‐patch protein NF‐κB‐repressing factor mediates the recruitment of the exonuclease XRN2 and activation of the RNA helicase DHX15 in human ribosome biogenesis. Nucleic Acids Res 45: 5359–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menne TF, Goyenechea B, Sánchez‐Puig N, Wong CC, Tonkin LM, Ancliff PJ, Brost RL, Costanzo M, Boone C, Warren AJ (2007) The Shwachman‐Bodian‐Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet 39: 486–495 [DOI] [PubMed] [Google Scholar]
- Merl J, Jakob S, Ridinger K, Hierlmeier T, Deutzmann R, Milkereit P, Tschochner H (2010) Analysis of ribosome biogenesis factor‐modules in yeast cells depleted from pre‐ribosomes. Nucleic Acids Res 38: 3068–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merwin JR, Bogar LB, Poggi SB, Fitch RM, Johnson AW, Lycan DE (2014) Genetic analysis of the ribosome biogenesis factor Ltv1 of Saccharomyces cerevisiae . Genetics 198: 1071–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metge BJ, Kammerud SC, Pruitt HC, Shevde LA, Samant RS (2021) Hypoxia re‐programs 2′‐O‐Me modifications on ribosomal RNA. iScience 24: 102010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AE, Hoover LA, Craig EA (2010) The cytosolic J‐protein, Jjj1, and Rei1 function in the removal of the Pre‐60 S subunit factor Arx1. J Biol Chem 285: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B, Wurm JP, Sharma S, Immer C, Pogoryelov D, Kötter P, Lafontaine DLJ, Wöhnert J, Entian K‐D (2016) Ribosome biogenesis factor Tsr3 is the aminocarboxypropyl transferase responsible for 18S rRNA hypermodification in yeast and humans. Nucleic Acids Res 44: 4304–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael SJ, Sebastian F, Lisa F, Ed H, Elena C (2018) Structure of the nuclear exosome captured on a maturing preribosome. Science 360: 219–222 [DOI] [PubMed] [Google Scholar]
- Micic J, Li Y, Wu S, Wilson D, Tutuncuoglu B, Gao N, Woolford JL (2020) Coupling of 5S RNP rotation with maturation of functional centers during large ribosomal subunit assembly. Nat Commun 11: 3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P, Tschochner H (1998) A specialized form of RNA polymerase I, essential for initiation and growth‐dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J 17: 3692–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H (2001) Maturation and intranuclear transport of pre‐ribosomes requires Noc proteins. Cell 105: 499–509 [DOI] [PubMed] [Google Scholar]
- Mitterer V, Pertschy B (2022) RNA folding and functions of RNA helicases in ribosome biogenesis. RNA Biol 19: 781–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitterer V, Shayan R, Ferreira‐Cerca S, Murat G, Enne T, Rinaldi D, Weigl S, Omanic H, Gleizes P‐E, Kressler D et al (2019) Conformational proofreading of distant 40S ribosomal subunit maturation events by a long‐range communication mechanism. Nat Commun 10: 2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongelard F, Bouvet P (2007) Nucleolin: a multiFACeTed protein. Trends Cell Biol 17: 80–86 [DOI] [PubMed] [Google Scholar]
- Montellese C, Montel‐Lehry N, Henras AK, Kutay U, Gleizes P‐E, O'Donohue M‐F (2017) Poly(A)‐specific ribonuclease is a nuclear ribosome biogenesis factor involved in human 18S rRNA maturation. Nucleic Acids Res 45: 6822–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montellese C, van den Heuvel J, Ashiono C, Dörner K, Melnik A, Jonas S, Zemp I, Picotti P, Gillet LC, Kutay U (2020) USP16 counteracts mono‐ubiquitination of RPS27a and promotes maturation of the 40S ribosomal subunit. Elife 9: e54435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello LG, Coltri PP, Quaresma AJC, Simabuco FM, Silva TCL, Singh G, Nickerson JA, Oliveira CC, Moore MJ, Zanchin NIT (2011a) The human nucleolar protein FTSJ3 associates with NIP7 and functions in pre‐rRNA processing. PLoS One 6: e29174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello LG, Hesling C, Coltri PP, Castilho BA, Rimokh R, Zanchin NIT (2011b) The NIP7 protein is required for accurate pre‐rRNA processing in human cells. Nucleic Acids Res 39: 648–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy TI, Silver PA (1999) Nuclear export of the small ribosomal subunit requires the Ran‐GTPase cycle and certain nucleoporins. Genes Dev 13: 2118–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A (2022) On the role of phase separation in the biogenesis of membraneless compartments. EMBO J 41: e109952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SK, Burley SK (2003) X‐ray structures of Myc‐Max and Mad‐Max recognizing DNA: molecular bases of regulation by proto‐oncogenic transcription factors. Cell 112: 193–205 [DOI] [PubMed] [Google Scholar]
- Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I, Klaholz BP (2017) Visualization of chemical modifications in the human 80S ribosome structure. Nature 551: 472–477 [DOI] [PubMed] [Google Scholar]
- Nerurkar P, Altvater M, Gerhardy S, Schütz S, Fischer U, Weirich C, Panse VG (2015) Eukaryotic ribosome assembly and nuclear export. Int Rev Cell Mol Biol 319: 107–140 [DOI] [PubMed] [Google Scholar]
- Ni C, Schmitz DA, Lee J, Pawłowski K, Wu J, Buszczak M (2022) Labeling of heterochronic ribosomes reveals C1ORF109 and SPATA5 control a late step in human ribosome assembly. Cell Rep 38: 110597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus KH, Dohme F (1974) Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli . Proc Natl Acad Sci USA 71: 4713–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan TA, Baßler J, Petfalski E, Tollervey D, Hurt E (2002) 60S pre‐ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J 21: 5539–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA (2000) The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930 [DOI] [PubMed] [Google Scholar]
- Oborská‐Oplová M, Gerhardy S, Panse VG (2022) Orchestrating ribosomal RNA folding during ribosome assembly. Bioessays 44: e2200066 [DOI] [PubMed] [Google Scholar]
- Occhipinti L, Chang Y, Altvater M, Menet AM, Kemmler S, Panse VG (2013) Non‐FG mediated transport of the large pre‐ribosomal subunit through the nuclear pore complex by the mRNA export factor Gle2. Nucleic Acids Res 41: 8266–8279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donohue M‐F, da Costa L, Lezzerini M, Unal S, Joret C, Bartels M, Brilstra E, Scheijde‐Vermeulen M, Wacheul L, de Keersmaecker K et al (2022) HEATR3 variants impair nuclear import of uL18 (RPL5) and drive Diamond‐Blackfan anemia. Blood 139: 3111–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Dlakic M, Tollervey D (2004) A pre‐ribosome‐associated HEAT‐repeat protein is required for export of both ribosomal subunits. Genes Dev 18: 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Fatica A, Rout MP, Tollervey D (2007) Yeast Rrp14p is required for ribosomal subunit synthesis and for correct positioning of the mitotic spindle during mitosis. Nucleic Acids Res 35: 1354–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Zenklusen D, Ferguson A, Wei KE, el Hage A, Tollervey D, Chait BT, Singer RH, Rout MP (2009) Rrp17p is a eukaryotic exonuclease required for 5′ end processing of pre‐60S ribosomal RNA. Mol Cell 36: 768–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa LM, Buhagiar AF, Abriola L, Leland BA, Surovtseva YV, Baserga SJ (2021) Increased numbers of nucleoli in a genome‐wide RNAi screen reveal proteins that link the cell cycle to RNA polymerase I transcription. Mol Biol Cell 32: 956–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmayer U, Gamalinda M, Sauert M, Ossowski J, Pöll G, Linnemann J, Hierlmeier T, Perez‐Fernandez J, Kumcuoglu B, Leger‐Silvestre I et al (2013) Studies on the assembly characteristics of large subunit ribosomal proteins in S. cerevisae . PLoS One 8: e68412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha S, Malla S, Lyons SM (2020) snoRNPs: functions in ribosome biogenesis. Biomolecules 10: 783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL (2004) Pre‐18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae . Mol Cell 16: 943–954 [DOI] [PubMed] [Google Scholar]
- Panić L, Tamarut S, Sticker‐Jantscheff M, Barkić M, Solter D, Uzelac M, Grabušić K, Volarević S (2006) Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53‐dependent checkpoint during gastrulation. Mol Cell Biol 26: 8880–8891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panova TB, Panov KI, Russell J, Zomerdijk JCBM (2006) Casein kinase 2 associates with initiation‐competent RNA polymerase I and has multiple roles in ribosomal DNA transcription. Mol Cell Biol 26: 5957–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell KM, Bass BL (2009) Functional redundancy of yeast proteins Reh1 and Rei1 in cytoplasmic 60S subunit maturation. Mol Cell Biol 29: 4014–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pausch P, Singh U, Ahmed YL, Pillet B, Murat G, Altegoer F, Stier G, Thoms M, Hurt E, Sinning I et al (2015) Co‐translational capturing of nascent ribosomal proteins by their dedicated chaperones. Nat Commun 6: 7494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer C, Sharma S, Watzinger P, Lamberth S, Kötter P, Entian KD (2013) Yeast Rrp8p, a novel methyltransferase responsible for m1A 645 base modification of 25S rRNA. Nucleic Acids Res 41: 1151–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Thomas G, Volarević S (2018) Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer 18: 51–63 [DOI] [PubMed] [Google Scholar]
- Peña C, Schütz S, Fischer U, Chang Y, Panse VG (2016) Prefabrication of a ribosomal protein subcomplex essential for eukaryotic ribosome formation. Elife 5: e21755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña C, Hurt E, Panse VG (2017) Eukaryotic ribosome assembly, transport and quality control. Nat Struct Mol Biol 24: 689–699 [DOI] [PubMed] [Google Scholar]
- Peng W‐T, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W et al (2003) A panoramic view of yeast noncoding RNA processing. Cell 113: 919–933 [DOI] [PubMed] [Google Scholar]
- Peng W, Krogan NJ, Richards DP, Greenblatt JF, Hughes TR (2004) ESF1 is required for 18S rRNA synthesis in Saccharomyces cerevisiae . Nucleic Acids Res 32: 1993–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Li D, Lee H, Bator C, Berezin I, Hafenstein SL, Krasilnikov AS (2020) Cryo‐EM structure of catalytic ribonucleoprotein complex RNase MRP. Nat Commun 11: 3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Fernández J, Román A, de Las RJ, Bustelo XR, Dosil M (2007) The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol 27: 5414–5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertschy B, Saveanu C, Zisser G, Lebreton A, Tengg M, Jacquier A, Liebminger E, Nobis B, Kappel L, van der Klei I et al (2007) Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol Cell Biol 27: 6581–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertschy B, Schneider C, Gnädig M, Schäfer T, Tollervey D, Hurt E (2009) RNA helicase Prp43 and its co‐factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem 284: 35079–35091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perucho L, Artero‐Castro A, Guerrero S, Ramón y Cajal S, LLeonart ME, Wang Z‐Q (2014) RPLP1, a crucial ribosomal protein for embryonic development of the nervous system. PLoS One 9: e99956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillet B, Mitterer V, Kressler D, Pertschy B (2017) Hold on to your friends: dedicated chaperones of ribosomal proteins. Bioessays 39: e201600153 [DOI] [PubMed] [Google Scholar]
- Pillet B, Méndez‐Godoy A, Murat G, Favre S, Stumpe M, Falquet L, Kressler D (2022) Dedicated chaperones coordinate co‐translational regulation of ribosomal protein production with ribosome assembly to preserve proteostasis. Elife 11: e74255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon MC, Sobhany M, Borgnia MJ, Williams JG, Stanley RE (2017) Grc3 programs the essential endoribonuclease Las1 for specific RNA cleavage. Proc Natl Acad Sci USA 114: E5530–E5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsl M, Engel C (2020) Structural basis of RNA polymerase I pre‐initiation complex formation and promoter melting. Nat Commun 11: 1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto R, Vågbø CB, Jakobsson ME, Kim Y, Baltissen MP, O'Donohue M‐F, Guzmán UH, Małecki JM, Wu J, Kirpekar F et al (2020) The human methyltransferase ZCCHC4 catalyses N6‐methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res 48: 830–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plassart L, Shayan R, Montellese C, Rinaldi D, Larburu N, Pichereaux C, Froment C, Lebaron S, O'Donohue M‐F, Kutay U et al (2021) The final step of 40S ribosomal subunit maturation is controlled by a dual key lock. Elife 10: e61254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov YS, Melnikov S v, Söll D, Steitz TA (2015) Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat Struct Mol Biol 22: 342–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöll G, Braun T, Jakovljevic J, Neueder A, Jakob S, Woolford JL Jr, Tschochner H, Milkereit P (2009) rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS One 4: e8249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöll G, Müller C, Bodden M, Teubl F, Eichner N, Lehmann G, Griesenbeck J, Tschochner H, Milkereit P (2017) Structural transitions during large ribosomal subunit maturation analyzed by tethered nuclease structure probing in S. cerevisiae . PLoS One 12: e0179405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöll G, Pilsl M, Griesenbeck J, Tschochner H, Milkereit P (2021) Analysis of subunit folding contribution of three yeast large ribosomal subunit proteins required for stabilisation and processing of intermediate nuclear rRNA precursors. PLoS One 16: e0252497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratte D, Singh U, Murat G, Kressler D (2013) Mak5 and Ebp2 act together on early pre‐60S particles and their reduced functionality bypasses the requirement for the essential pre‐60S factor Nsa1. PLoS One 8: e82741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prattes M, Lo Y‐H, Bergler H, Stanley RE (2019) Shaping the nascent ribosome: AAA‐ATPases in eukaryotic ribosome biogenesis. Biomolecules 9: 715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti M, O'Donohue MF, Montel‐Lehry N, Bortolin‐Cavaillé ML, Choesmel V, Gleizes PE (2013) Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA. Nucleic Acids Res 41: 4709–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto J‐L, McStay B (2007) Recruitment of factors linking transcription and processing of pre‐rRNA to NOR chromatin is UBF‐dependent and occurs independent of transcription in human cells. Genes Dev 21: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puusepp S, Kovacs‐Nagy R, Alhaddad B, Braunisch M, Hoffmann GF, Kotzaeridou U, Lichvarova L, Liiv M, Makowski C, Mandel M et al (2018) Compound heterozygous SPATA5 variants in four families and functional studies of SPATA5 deficiency. Eur J Hum Genet 26: 407–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N (2011) Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331: 730–736 [DOI] [PubMed] [Google Scholar]
- Raman N, Weir E, Müller S (2016) The AAA ATPase MDN1 acts as a SUMO‐targeted regulator in mammalian pre‐ribosome remodeling. Mol Cell 64: 607–615 [DOI] [PubMed] [Google Scholar]
- Ramesh M, Woolford JL (2016) Eukaryote‐specific rRNA expansion segments function in ribosome biogenesis. RNA 22: 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Panova T, Miller G, Volkov A, Porter ACG, Russell J, Panov KI, Zomerdijk JCBM (2013) Topoisomerase IIα promotes activation of RNA polymerase I transcription by facilitating pre‐initiation complex formation. Nat Commun 4: 1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter A, Hamperl S, Seitz H, Merkl P, Perez‐Fernandez J, Williams L, Gerber J, Németh A, Léger I, Gadal O et al (2012) The Reb1‐homologue Ydr026c/Nsi1 is required for efficient RNA polymerase I termination in yeast. EMBO J 31: 3480–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempola B, Karkusiewicz I, Piekarska I, Rytka J (2006) Fcf1p and Fcf2p are novel nucleolar Saccharomyces cerevisiae proteins involved in pre‐rRNA processing. Biochem Biophys Res Commun 346: 546–554 [DOI] [PubMed] [Google Scholar]
- Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei M‐T, Kriwacki RW, Brangwynne CP (2020) Composition‐dependent thermodynamics of intracellular phase separation. Nature 581: 209–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard EM, Bakhtiari S, Marsh APL, Kaiyrzhanov R, Wagner M, Shetty S, Pagnozzi A, Nordlie SM, Guida BS, Cornejo P et al (2021) Bi‐allelic variants in SPATA5L lead to intellectual disability, spastic‐dystonic cerebral palsy, epilepsy, and hearing loss. Am J Hum Genet 108: 2006–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa FM, Spiegelman S (1965) Localization of DNA complementary to ribosomal RNA in the nucleolus. Proc Natl Acad Sci USA 53: 737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Galán O, García‐Gómez JJ, de la Cruz J (2013) Yeast and human RNA helicases involved in ribosome biogenesis: current status and perspectives. Biochim Biophys Acta 1829: 775–790 [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Mateos M, García‐Gómez JJ, Francisco‐Velilla R, Remacha M, de la Cruz J, Ballesta JPG (2009) Role and dynamics of the ribosomal protein P0 and its related trans ‐acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae . Nucleic Acids Res 37: 7519–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrmoser M, Hölzel M, Grimm T, Malamoussi A, Harasim T, Orban M, Pfisterer I, Gruber‐Eber A, Kremmer E, Eick D (2007) Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol Cell Biol 27: 3682–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado IV, Dez C, Lebaron S, Caizergues‐Ferrer M, Henry Y, de la Cruz J (2007) Characterization of Saccharomyces cerevisiae Npa2p (Urb2p) reveals a low‐molecular‐mass complex containing Dbp6p, Npa1p (Urb1p), Nop8p, and Rsa3p involved in early steps of 60S ribosomal subunit biogenesis. Mol Cell Biol 27: 1207–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rössler I, Embacher J, Pillet B, Murat G, Liesinger L, Hafner J, Unterluggauer JJ, Birner‐Gruenberger R, Kressler D, Pertschy B (2019) Tsr4 and Nap1, two novel members of the ribosomal protein chaperOME. Nucleic Acids Res 47: 6984–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette J, Choesmel V, Gleizes PE (2005) Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J 24: 2862–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel P, André C, Comai L, Hernandez‐Verdun D (1996) The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J Cell Biol 133: 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Mallick J, Zhao Y, Warner JR (2007) Potential interface between ribosomal protein production and pre‐rRNA processing. Mol Cell Biol 27: 4815–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J, Zomerdijk JCBM (2006) The RNA polymerase I transcription machinery. Biochem Soc Symp 73: 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Sun X‐X, Chen Y, Li Y, Wang X, Dai RS, Zhu H‐M, Klimek J, David L, Fedorov LM et al (2021) The deubiquitinase USP36 promotes snoRNP group SUMOylation and is essential for ribosome biogenesis. EMBO Rep 22: e50684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadian Y, Baudin F, Tafur L, Murciano B, Wetzel R, Weis F, Müller CW (2019) Molecular insight into RNA polymerase I promoter recognition and promoter melting. Nat Commun 10: 5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez I, Gerbracht JV, Koyuncu S, Lee HJ, Horn M, Kroef V, Denzel MS, Dieterich C, Gehring NH, Vilchez D (2020) The E3 ubiquitin ligase UBR5 interacts with the H/ACA ribonucleoprotein complex and regulates ribosomal RNA biogenesis in embryonic stem cells. FEBS Lett 594: 175–188 [DOI] [PubMed] [Google Scholar]
- Sahasranaman A, Dembowski J, Strahler J, Andrews P, Maddock J, Woolford JL Jr (2011) Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre‐rRNA processing. EMBO J 30: 4020–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailer C, Jansen J, Sekulski K, Cruz VE, Erzberger JP, Stengel F (2022) A comprehensive landscape of 60S ribosome biogenesis factors. Cell Rep 38: 110353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky DA, Balakin AG, Fournier MJ (1995) Characterization of three new snRNAs from Saccharomyces cerevisiae: snR34, snR35 and snR36. Nucleic Acids Res 23: 2548–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghai ZA, Miller L, Molloy KR, Barandun J, Hunziker M, Chaker‐Margot M, Wang J, Chait BT, Klinge S (2018) Modular assembly of the nucleolar pre‐60S ribosomal subunit. Nature 556: 126–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporita AJ, Chang H‐C, Winkeler CL, Apicelli AJ, Kladney RD, Wang J, Townsend RR, Michel LS, Weber JD (2011) RNA helicase DDX5 is a p53‐independent target of ARF that participates in ribosome biogenesis. Cancer Res 71: 6708–6717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardana R, Liu X, Granneman S, Zhu J, Gill M, Papoulas O, Marcotte EM, Tollervey D, Correll CC, Johnson AW (2015) The DEAH‐box helicase Dhr1 dissociates U3 from the pre‐rRNA to promote formation of the central pseudoknot. PLoS Biol 13: e1002083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Pech M, Thoms M, Beckmann R, Hurt E (2016) Ribosome‐stalk biogenesis is coupled with recruitment of nuclear‐export factor to the nascent 60S subunit. Nat Struct Mol Biol 23: 1074–1082 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Toh‐e A, Kikuchi Y (2000) Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol Cell Biol 20: 7971–7979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C, Namane A, Gleizes P‐E, Lebreton A, Rousselle J‐C, Noaillac‐Depeyre J, Gas N, Jacquier A, Fromont‐Racine M (2003) Sequential protein association with nascent 60S ribosomal particles. Mol Cell Biol 23: 4449–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaiola A, Peña C, Weisser M, Böhringer D, Leibundgut M, Klingauf‐Nerurkar P, Gerhardy S, Panse VG, Ban N (2018) Structure of a eukaryotic cytoplasmic pre‐40S ribosomal subunit. EMBO J 37: e98499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T, Strauss D, Petfalski E, Tollervey D, Hurt E (2003) The path from nucleolar 90S to cytoplasmic 40S pre‐ribosomes. EMBO J 22: 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T, Maco B, Petfalski E, Tollervey D, Böttcher B, Aebi U, Hurt E (2006) Hrr25‐dependent phosphorylation state regulates organization of the pre‐40S subunit. Nature 441: 651–655 [DOI] [PubMed] [Google Scholar]
- Schillewaert S, Wacheul L, Lhomme F, Lafontaine DLJ (2012) The evolutionarily conserved protein LAS1 is required for pre‐rRNA processing at both ends of ITS2. Mol Cell Biol 32: 430–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, French SL, Osheim YN, Bailey AO, Vu L, Dodd J, Yates JR, Beyer AL, Nomura M (2006) RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc Natl Acad Sci USA 103: 12707–12712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, Calle‐Perez A, Pircher A, Gerstl MP, Pfeifenberger S et al (2015) Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun 6: 6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz S, Fischer U, Altvater M, Nerurkar P, Peña C, Gerber M, Chang Y, Caesar S, Schubert OT, Schlenstedt G et al (2014) A RanGTP‐independent mechanism allows ribosomal protein nuclear import for ribosome assembly. Elife 3: e03473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz S, Michel E, Damberger FF, Oplová M, Peña C, Leitner A, Aebersold R, Allain FH‐T, Panse VG (2018) Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2. Nat Commun 9: 3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerstolpe Å, Granneman S, Björk P, de Lima AF, Rappsilber J, Andersson C, Högbom M, Tollervey D, Wieslander L (2013) Multiple RNA interactions position Mrd1 at the site of the small subunit pseudoknot within the 90S pre‐ribosome. Nucleic Acids Res 41: 1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiser RM, Sundberg AE, Wollam BJ, Zobel‐Thropp P, Baldwin K, Spector MD, Lycan DE (2006) Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae . Genetics 174: 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B, Lafontaine DLJ, Graindorge JS, Gadal O, Camasses A, Sanni A, Garnier JM, Breitenbach M, Hurt E, Fasiolo F (2001) The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol Cell 8: 1363–1373 [DOI] [PubMed] [Google Scholar]
- Sengupta J, Bussiere C, Pallesen J, West M, Johnson AW, Frank J (2010) Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J Cell Biol 189: 1079–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR (2011) Assembly of bacterial ribosomes. Annu Rev Biochem 80: 501–526 [DOI] [PubMed] [Google Scholar]
- Sharma S, Watzinger P, Kötter P, Entian KD (2013) Identification of a novel methyltransferase, Bmt2, responsible for the N‐1‐methyl‐adenosine base modification of 25S rRNA in Saccharomyces cerevisiae . Nucleic Acids Res 41: 5428–5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Yang J, Düttmann S, Watzinger P, Kötter P, Entian K‐D (2014) Identification of novel methyltransferases, Bmt5 and Bmt6, responsible for the m3U methylations of 25S rRNA in Saccharomyces cerevisiae . Nucleic Acids Res 42: 3246–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Langhendries JL, Watzinger P, Kotter P, Entian KD, Lafontaine DLJ (2015) Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res 43: 2242–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Yang J, van Nues R, Watzinger P, Kötter P, Lafontaine DLJ, Granneman S, Entian KD (2017) Specialized box C/D snoRNPs act as antisense guides to target RNA base acetylation. PLoS Genet 13: 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoji K, Jakovljevic J, Tsuchihashi K, Umeki Y, Wan K, Kawasaki S, Talkish J, Woolford JL Jr, Mizuta K (2012) Ebp2 and Brx1 function cooperatively in 60S ribosomal subunit assembly in Saccharomyces cerevisiae . Nucleic Acids Res 40: 4574–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi IN, Dodd JA, Vu L, Eliason K, Oakes ML, Keener J, Moore R, Young MK, Nomura M (2001) Transcription of chromosomal rRNA genes by both RNA polymerase I and II in yeast uaf30 mutants lacking the 30 kDa subunit of transcription factor UAF. EMBO J 20: 4512–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S, Homolka D, Pillai RS (2017) Characterization of the mammalian RNA exonuclease 5/NEF‐sp as a testis‐specific nuclear 3′ → 5′ exoribonuclease. RNA 23: 1385–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simabuco FM, Morello LG, Aragaão AZB, Leme AFP, Zanchin NIT (2012) Proteomic characterization of the human FTSJ3 preribosomal complexes. J Proteome Res 11: 3112–3126 [DOI] [PubMed] [Google Scholar]
- Singh S, vanden Broeck A, Miller L, Chaker‐Margot M, Klinge S (2021) Nucleolar maturation of the human small subunit processome. Science 373: eabj5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Schneider C, Watkins NJ (2012) Comparison of the yeast and human nuclear exosome complexes. Biochem Soc Trans 40: 850–855 [DOI] [PubMed] [Google Scholar]
- Sloan KE, Bohnsack MT, Watkins NJ (2013a) The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep 5: 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Mattijssen S, Lebaron S, Tollervey D, Pruijn GJM, Watkins NJ (2013b) Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J Cell Biol 200: 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Leisegang MS, Doebele C, Ramírez AS, Simm S, Safferthal C, Kretschmer J, Schorge T, Markoutsa S, Haag S et al (2015) The association of late‐acting snoRNPs with human pre‐ribosomal complexes requires the RNA helicase DDX21. Nucleic Acids Res 43: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Warda AS, Sharma S, Entian K‐D, Lafontaine DLJ, Bohnsack MT (2017) Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol 14: 1138–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltanieh S, Lapensée M, Dragon F (2014) Nucleolar proteins Bfr2 and Enp2 interact with DEAD‐box RNA helicase Dbp4 in two different complexes. Nucleic Acids Res 42: 3194–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CMT, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J (2001) Structure of the 80S ribosome from Saccharomyces cerevisiae–RNA‐ribosome and subunit‐subunit interactions. Cell 107: 373–386 [DOI] [PubMed] [Google Scholar]
- Srivastava L, Lapik YR, Wang M, Pestov DG (2010) Mammalian DEAD Box protein Ddx51 acts in 3′ end maturation of 28S rRNA by promoting the release of U8 snoRNA. Mol Cell Biol 30: 2947–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage‐Zimmermann T, Schmidt U, Silver PA (2000) Factors affecting nuclear export of the 60S ribosomal subunit in vivo . Mol Biol Cell 11: 3777–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Huber FM, Kunze R, Flemming D, Hoelz A, Hurt E (2015) Coordinated ribosomal L4 protein assembly into the pre‐ribosome is regulated by its eukaryote‐specific extension. Mol Cell 58: 854–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strezoska Ž, Pestov DG, Lau LF (2000) Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol Cell Biol 20: 5516–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, Karbstein K, Skiniotis G (2011) Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 333: 1449–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Novak MN, Young CL, Karbstein K (2012) A translation‐like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults DM, Killen MW, Pierce HH, Pierce AJ (2008) Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res 18: 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm M, Cheng J, Baßler J, Beckmann R, Hurt E (2017) Interdependent action of KH domain proteins Krr1 and Dim2 drive the 40S platform assembly. Nat Commun 8: 2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumita M, Desaulniers J‐P, Chang Y‐C, Chui HM‐P, Clos L, Chow CS (2005) Effects of nucleotide substitution and modification on the stability and structure of helix 69 from 28S rRNA. RNA 11: 1420–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhu X, Qi J, An W, Lan P, Tan D, Chen R, Wang B, Zheng S, Zhang C et al (2017) Molecular architecture of the 90S small subunit pre‐ribosome. Elife 6: e22086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M‐K, Reitsma JM, Sweredoski MJ, Hess S, Deshaies RJ (2016) Ribosomal proteins produced in excess are degraded by the ubiquitin–proteasome system. Mol Biol Cell 27: 2642–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Katada E, Mizuoka Y, Takagi S, Kazuki Y, Oshimura M, Shindo M, Hara T (2021) A novel all‐in‐one conditional knockout system uncovered an essential role of DDX1 in ribosomal RNA processing. Nucleic Acids Res 49: e40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebeni A, Olson MOJ (1999) Nucleolar protein B23 has molecular chaperone activities. Protein Sci 8: 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DLJ (2013) The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre‐rRNA processing factors. Mol Cell 51: 539–551 [DOI] [PubMed] [Google Scholar]
- Talkish J, Zhang J, Jakovljevic J, Horsey EW, Woolford JL Jr (2012) Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre‐rRNA processing in Saccharomyces cerevisiae . Nucleic Acids Res 40: 8646–8661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkish J, Campbell IW, Sahasranaman A, Jakovljevic J, Woolford JL (2014) Ribosome assembly factors Pwp1 and Nop12 are important for folding of 5.8S rRNA during ribosome biogenesis in Saccharomyces cerevisiae . Mol Cell Biol 34: 1863–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao T, Sondalle SB, Shi H, Zhu S, Perez‐Atayde AR, Peng J, Baserga SJ, Look AT (2017) The pre‐rRNA processing factor DEF is rate limiting for the pathogenesis of MYCN‐driven neuroblastoma. Oncogene 36: 3852–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka M, Nobe Y, Yamaki Y, Yamauchi Y, Ishikawa H, Takahashi N, Nakayama H, Isobe T (2016) The complete chemical structure of Saccharomyces cerevisiae rRNA: partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9. Nucleic Acids Res 44: 8951–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka M, Nobe Y, Yamaki Y, Sato K, Ishikawa H, Izumikawa K, Yamauchi Y, Hirota K, Nakayama H, Takahashi N et al (2018) Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res 46: 9289–9298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P, Santos‐Rosa H, Robson SC, Sylvestersen KB, Nelson CJ, Nielsen ML, Kouzarides T (2014) Glutamine methylation in histone H2A is an RNA‐polymerase‐I‐dedicated modification. Nature 505: 564–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas F, Kutay U (2003) Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci 116: 2409–2419 [DOI] [PubMed] [Google Scholar]
- Thoms M, Ahmed YL, Maddi K, Hurt E, Sinning I (2016) Concerted removal of the Erb1–Ytm1 complex in ribosome biogenesis relies on an elaborate interface. Nucleic Acids Res 44: 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M, Mitterer V, Kater L, Falquet L, Beckmann R, Kressler D, Hurt E (2018) Suppressor mutations in Rpf2–Rrs1 or Rpl5 bypass the Cgr1 function for pre‐ribosomal 5S RNP‐rotation. Nat Commun 9: 4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting Y‐H, Lu T‐J, Johnson AW, Shie J‐T, Chen B‐R, Kumar SS, Lo K‐Y (2017) Bcp1 is the nuclear chaperone of Rpl23 in Saccharomyces cerevisiae . J Biol Chem 292: 585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Sikorski PJ, Zakrzewska‐Placzek M (2017) Comparison of preribosomal RNA processing pathways in yeast, plant and human cells – focus on coordinated action of endo‐ and exoribonucleases. FEBS Lett 591: 1801–1850 [DOI] [PubMed] [Google Scholar]
- Trapman J, Retèl J, Planta RJ (1975) Ribosomal precursor particles from yeast. Exp Cell Res 90: 95–104 [DOI] [PubMed] [Google Scholar]
- Trotta CR, Lund E, Kahan L, Johnson AW, Dahlberg JE (2003) Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J 22: 2841–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AJ, Knox AA, Prieto J‐L, McStay B, Watkins NJ (2009) A novel small‐subunit processome assembly intermediate that contains the U3 snoRNP, nucleolin, RRP5, and DBP4. Mol Cell Biol 29: 3007–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowski TW, Tollervey D (2015) Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip Rev RNA 6: 129–139 [DOI] [PubMed] [Google Scholar]
- Turowski TW, Lebaron S, Zhang E, Peil L, Dudnakova T, Petfalski E, Granneman S, Rappsilber J, Tollervey D (2014) Rio1 mediates ATP‐dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res 42: 12189–12199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udem SA, Kaufman K, Warner JR (1971) Small ribosomal ribonucleic acid species of Saccharomyces cerevisiae . J Bacteriol 105: 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich C, Diepholz M, Baßler J, Kressler D, Pertschy B, Galani K, Böttcher B, Hurt E (2009) Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell 138: 911–922 [DOI] [PubMed] [Google Scholar]
- Valdez BC, Henning D, So RB, Dixon J, Dixon MJ (2004) The Treacher Collins syndrome TCOF1 gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci USA 101: 10709–10714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Nobelen S, Rosa‐Garrido M, Leers J, Heath H, Soochit W, Joosen L, Jonkers I, Demmers J, van der Reijden M, Torrano V et al (2010) CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenetics Chromatin 3: 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel J, Ashiono C, Gillet LC, Dörner K, Wyler E, Zemp I, Kutay U (2021) Processing of the ribosomal ubiquitin‐like fusion protein FUBI‐eS30/FAU is required for 40S maturation and depends on USP36. Elife 10: e70560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R (2000) Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J 19: 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, Bohnsack KE, Bohnsack MT, Jaffrey SR, Graille M et al (2019) The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res 47: 7719–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- vanden Broeck A, Klinge S (2022) An emerging mechanism for the maturation of the small subunit processome. Curr Opin Struct Biol 73: 102331 [DOI] [PubMed] [Google Scholar]
- Vanrobays E, Gleizes P‐E, Bousquet‐Antonelli C, Noaillac‐Depeyre J, Caizergues‐Ferrer M, Gélugne J‐P (2001) Processing of 20S pre‐rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non‐ribosomal cytoplasmic protein. EMBO J 20: 4204–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobays E, Gelugne J‐P, Gleizes P‐E, Caizergues‐Ferrer M (2003) Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae . Mol Cell Biol 23: 2083–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobays E, Leplus A, Osheim YN, Beyer AL, Wacheul L, Lafontaine DLJ (2008) TOR regulates the subcellular distribution of DIM2, a KH domain protein required for cotranscriptional ribosome assembly and pre‐40S ribosome export. RNA 14: 2061–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Tollervey D (1996) RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J 15: 5701–5714 [PMC free article] [PubMed] [Google Scholar]
- Wandrey F, Montellese C, Koos K, Badertscher L, Bammert L, Cook AG, Zemp I, Horvath P, Kutay U (2015) The NF45/NF90 heterodimer contributes to the biogenesis of 60S ribosomal subunits and influences nucleolar morphology. Mol Cell Biol 35: 3491–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warda AS, Freytag B, Haag S, Sloan KE, Görlich D, Bohnsack MT (2016) Effects of the Bowen‐Conradi syndrome mutation in EMG1 on its nuclear import, stability and nucleolar recruitment. Hum Mol Genet 25: 5353–5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR (1971) The assembly of ribosomes in yeast. J Biol Chem 246: 447–454 [PubMed] [Google Scholar]
- Warner JR (1977) In the absence of ribosomal RNA synthesis, the ribosomal proteins of HeLa cells are synthesized normally and degraded rapidly. J Mol Biol 115: 315–333 [DOI] [PubMed] [Google Scholar]
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Weis F, Giudice E, Churcher M, Jin L, Hilcenko C, Wong CC, Traynor D, Kay RR, Warren AJ (2015) Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat Struct Mol Biol 22: 914–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells GR, Weichmann F, Colvin D, Sloan KE, Kudla G, Tollervey D, Watkins NJ, Schneider C (2016) The PIN domain endonuclease Utp24 cleaves pre‐ribosomal RNA at two coupled sites in yeast and humans. Nucleic Acids Res 44: 5399–5409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells GR, Weichmann F, Sloan KE, Colvin D, Watkins NJ, Schneider C (2017) The ribosome biogenesis factor yUtp23/hUTP23 coordinates key interactions in the yeast and human pre‐40S particle and hUTP23 contains an essential PIN domain. Nucleic Acids Res 45: 4796–4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A, Iwasaki S, McGourty CA, Medina‐Ruiz S, Teerikorpi N, Fedrigo I, Ingolia NT, Rape M (2015) Cell‐fate determination by ubiquitin‐dependent regulation of translation. Nature 525: 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Li Z, Sardana R, Bujnicki JM, Marcotte EM, Johnson AW (2008) Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre‐40S subunits. Mol Cell Biol 28: 3151–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann B, Wandrey F, Badertscher L, Wyler E, Pfannstiel J, Zemp I, Kutay U (2012) The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol Biol Cell 23: 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E, Kutay U (2010) A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol 8: e1000522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL, Baserga SJ (2013) Ribosome biogenesis in the yeast Saccharomyces cerevisiae . Genetics 195: 643–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, Gamalinda M, Yuan Y, Li Z, Jakovljevic J et al (2016) Diverse roles of assembly factors revealed by structures of late nuclear pre‐60S ribosomes. Nature 534: 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm JP, Meyer B, Bahr U, Held M, Frolow O, Kötter P, Engels JW, Heckel A, Karas M, Entian K‐D et al (2010) The ribosome assembly factor Nep1 responsible for Bowen–Conradi syndrome is a pseudouridine‐N1‐specific methyltransferase. Nucleic Acids Res 38: 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyler E, Zimmermann M, Widmann B, Gstaiger M, Pfannstiel J, Kutay U, Zemp I (2011) Tandem affinity purification combined with inducible shRNA expression as a tool to study the maturation of macromolecular assemblies. RNA 17: 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyler E, Wandrey F, Badertscher L, Montellese C, Alper D, Kutay U (2014) The beta‐isoform of the BRCA2 and CDKN1A(p21)‐interacting protein (BCCIP) stabilizes nuclear RPL23/uL14. FEBS Lett 588: 3685–3691 [DOI] [PubMed] [Google Scholar]
- Yang Y‐M, Karbstein K (2022) The chaperone Tsr2 regulates Rps26 release and reincorporation from mature ribosomes to enable a reversible, ribosome‐mediated response to stress. Sci Adv 8: eabl4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W, Roser D, Köhler A, Bradatsch B, Baßler J, Hurt E (2007) Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67‐Mtr2. Mol Cell 26: 51–62 [DOI] [PubMed] [Google Scholar]
- Yao W, Lutzmann M, Hurt E (2008) A versatile interaction platform on the Mex67–Mtr2 receptor creates an overlap between mRNA and ribosome export. EMBO J 27: 6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Demoinet E, Saveanu C, Lenormand P, Jacquier A, Fromont‐Racine M (2010) Ecm1 is a new pre‐ribosomal factor involved in pre‐60S particle export. RNA 16: 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R‐W, Xu G, Wang Y, Shan L, Luan P‐F, Wang Y, Wu M, Yang L‐Z, Xing Y‐H, Yang L et al (2019) Nascent pre‐rRNA sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol Cell 76: 767–783 [DOI] [PubMed] [Google Scholar]
- Yelland JN, Bravo JPK, Black JJ, Taylor DW, Johnson AW (2022) A single 2′‐O‐methylation of ribosomal RNA gates assembly of a functional ribosome. bioRxiv 10.1101/2022.02.16.480697 [PREPRINT] [DOI] [PMC free article] [PubMed]
- Yusupova G, Yusupov M (2014) High‐resolution structure of the eukaryotic 80S ribosome. Annu Rev Biochem 83: 467–486 [DOI] [PubMed] [Google Scholar]
- Zemp I, Kutay U (2007) Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett 581: 2783–2793 [DOI] [PubMed] [Google Scholar]
- Zemp I, Wild T, O'Donohue MF, Wandrey F, Widmann B, Gleizes PE, Kutay U (2009) Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J Cell Biol 185: 1167–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I, Wandrey F, Rao S, Ashiono C, Wyler E, Montellese C, Kutay U (2014) CK1δ and CK1ε are components of human 40S subunit precursors required for cytoplasmic 40S maturation. J Cell Sci 127: 1242–1253 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sikes ML, Beyer AL, Schneider DA (2009) The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc Natl Acad Sci USA 106: 2153–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, McCann KL, Qiu C, Gonzalez LE, Baserga SJ, Hall TMT (2016a) Nop9 is a PUF‐like protein that prevents premature cleavage to correctly process pre‐18S rRNA. Nat Commun 7: 13085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wu C, Cai G, Chen S, Ye K (2016b) Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes Dev 30: 718–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Chen Y, Chen F, Huang D, Shi H, Lo LJ, Chen J, Peng J (2019) Sas10 controls ribosome biogenesis by stabilizing Mpp10 and delivering the Mpp10–Imp3–Imp4 complex to nucleolus. Nucleic Acids Res 47: 2996–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Lan P, Liu X, Ye K (2014) Interaction between ribosome assembly factors Krr1 and Faf1 is essential for formation of small ribosomal subunit in yeast. J Biol Chem 289: 22692–22703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Zhu X, Zheng S, Tan D, Dong M‐Q, Ye K (2019a) Cryo‐EM structure of an early precursor of large ribosomal subunit reveals a half‐assembled intermediate. Protein Cell 10: 120–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Musalgaonkar S, Johnson AW, Taylor DW (2019b) Tightly‐orchestrated rearrangements govern catalytic center assembly of the ribosome. Nat Commun 10: 958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorbas C, Nicolas E, Wacheul L, Huvelle E, Heurgué‐Hamard V, Lafontaine DLJ (2015) The human 18S rRNA base methyltransferases DIMT1L and WBSCR22‐TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol Biol Cell 26: 2080–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]


