Summary
Subtilase cytotoxin (SubAB) is mainly produced by locus of enterocyte effacement (LEE)-negative strains of Shiga-toxigenic Escherichia coli (STEC). SubAB cleaves an endoplasmic reticulum (ER) chaperone, BiP/Grp78, leading to induction of ER stress. This stress causes activation of ER stress sensor proteins and induction of caspase-dependent apoptosis. We found that SubAB induces stress granules (SG) in various cells. Aim of this study was to explore the mechanism by which SubAB induced SG formation. Here, we show that SubAB-induced SG formation is regulated by activation of double-stranded RNA-activated protein kinase (PKR)-like endoplasmic reticulum kinase (PERK). The culture supernatant of STEC O113:H21 dramatically induced SG in Caco2 cells, although subAB knockout STEC O113:H21 culture supernatant did not. Treatment with phorbol 12-myristate 13-acetate (PMA), a protein kinase C (PKC) activator, and lysosomal inhibitors, NH4Cl and chloroquine, suppressed SubAB-induced SG formation, which was enhanced by PKC and PKD inhibitors. SubAB attenuated the level of PKD1 phosphorylation. Depletion of PKCδ and PKD1 by siRNA promoted SG formation in response to SubAB. Furthermore, death-associated protein 1 (DAP1) knockdown increased basal phospho-PKD1(S916) and suppressed SG formation by SubAB. However, SG formation by an ER stress inducer, Thapsigargin, was not inhibited in PMA-treated cells. Our findings show that SubAB-induced SG formation is regulated by the PERK/DAP1 signalling pathway, which may be modulated by PKCδ/PKD1, and different from the signal transduction pathway that results in Thapsigargin-induced SG formation.
Introduction
Shiga-toxigenic Escherichia coli (STEC) produces Shiga toxin (Stx) 1 and 2, which are critical virulence factors (Karmali, 2004), resulting in hemorrhagic colitis and hemolytic uremic syndrome (Riley et al., 1983; Shiomi and Togawa, 1997; Latorre-Martinez et al., 2007). A new member of the AB5 toxin family from STEC, named subtilase cytotoxin (SubAB), was identified in E. coli O113: H21 strain 98NK2, which produces Stx2 and was responsible for an outbreak of hemolytic uremic syndrome (Paton et al., 2004). SubAB was mainly produced by locus of enterocyte effacement (LEE)-negative serotypes STEC (Velandia et al., 2011; Bentancor et al., 2012; Sanchez et al., 2012; Feng and Reddy, 2013). SubAB binds to receptors on the target cells (Yahiro et al., 2006; Byres et al., 2008; Yahiro et al., 2011) and then enters the cells via clathrin (Chong et al., 2008) or lipid rafts-dependent and an actin-dependent pathways (Nagasawa et al., 2014), followed by Golgi trafficking involving a COG/Rab6/COPI-dependent pathway (Chong et al., 2008; Smith et al., 2009). In the endoplasmic reticulum (ER), SubAB cleaved chaperone protein BiP/Grp78, which initiated an ER stress-induced unfolded protein response; this leads to activation of ER stress sensor proteins, which induce a variety of cellular events including cytotoxicity (Paton et al., 2006; Morinaga et al., 2007; Morinaga et al., 2008; Wolfson et al., 2008; Hu et al., 2009; Huang et al., 2009; Matsuura et al., 2009; May et al., 2010; Nakajima et al., 2010; Wang et al., 2010; Yahiro et al., 2010; Nakajima et al., 2011; Yahiro et al., 2012) and damage in mice (Wang et al., 2007; Furukawa et al., 2011; Wang et al., 2011; Amaral et al., 2013). In addition, we recently demonstrated in mouse macrophages that SubAB inhibited Lipopolysaccharide-stimulated nitric oxide (NO) production through inhibition of NF-κB nuclear translocation and iNOS expression (Tsutsuki et al., 2012). Death-associated protein 1 (DAP1) regulated SubAB-mediated apoptosis and autophagy in HeLa cells (Yahiro et al., 2014). These findings suggest that SubAB constitutes a novel bacterial strategy for resistance to host defence.
In eukaryotic cells, stress granule (SG) formation is induced by various environmental stresses (i.e. heat shock, oxidative stress, viral infection, UV irradiation, etc.) (Anderson and Kedersha, 2009). SG assembly is seen in non-membranous cytoplasmic foci, which typically contain poly(A) + mRNA, 40S ribosomal subunits, eIF4E, eIF4G, eIF4A, eIF4B, eIF3, eIF2, TIAR, G3BP1 and HuR (Kedersha et al., 1999; Kedersha and Anderson, 2002; Kimball et al., 2003; Anderson and Kedersha, 2006; Mazroui et al., 2006; White et al., 2007; Buchan and Parker, 2009). Recent studies have shown that SG contain various components that play an important role in mRNA translation and stability and the protein quality control (Kawaguchi et al., 2003; Mazroui et al., 2007; Anderson and Kedersha, 2008; Buchan and Parker, 2009; Athanasopoulos et al., 2010; Seguin et al., 2014). Previous studies showed that the pathological SG formation caused by mutations in RNA binding proteins might be involved in neurodegenerative disease (Wolozin, 2012; Vanderweyde et al., 2013; Wolozin, 2014).
Translational inhibition by phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) at Ser51 is a major trigger that induces SG formation (Kedersha et al., 1999). Thus, eIF2α phosphorylation in response to ER stress occurs through an ER stress transducer, protein kinase RNA (PKR)-like ER kinase (PERK), and induces preferential translation of activating transcription factor 4 (ATF4) (Ron and Walter, 2007). Recent reports indicate that the PERK-eIF2α-ATF4 pathway is associated with the maintenance of homeostasis (B’Chir et al., 2013; Matsumoto et al., 2013; Jiang et al., 2014), suggesting that eIF2α-related signalling controls a cross-talk between apoptosis, autophagy and SG formation.
Subtilase cytotoxin induces eIF2α phosphorylation through PERK activation (Morinaga et al., 2008; Wolfson et al., 2008; Yahiro et al., 2012). We hypothesized that SubAB-induced ER stress causes PERK/eIF2α-dependent SG formation. To our knowledge, this is the first report of a relationship between exotoxins and SG formation. In this study, we demonstrate that SG formation by SubAB is regulated by protein kinase C (PKC) isoforms. PKCs are ubiquitous serine/threonine kinases comprising a large superfamily composed of three classes: classical isoforms (α, β and γ); novel isoforms (δ, ε, η and θ); and atypical isoforms (λ and ζ) (Toker, 1998). Each isoform, exhibiting diversity in Ca2+ and phosphatidylserine sensitivity, is involved in multiple signal transduction actions through specific regulatory molecules (Mellor and Parker, 1998; Rosse et al., 2010). PKCμ/PKD1 is an atypical member of the PKC family (Johannes et al., 1994; Valverde et al., 1994), which participates in various cell signal processes (Sundram et al., 2011; Steinberg, 2012). The aim of this study was to investigate the mechanism of SubAB-induced SG formation. We show here that SubAB-induced SG formation is dependent on activation of the PERK/DAP1 signalling pathway with its modulation by PKCδ/PKD1.
Results
Subtilase cytotoxin-induced stress granule formation is dependent on PERK signalling pathway
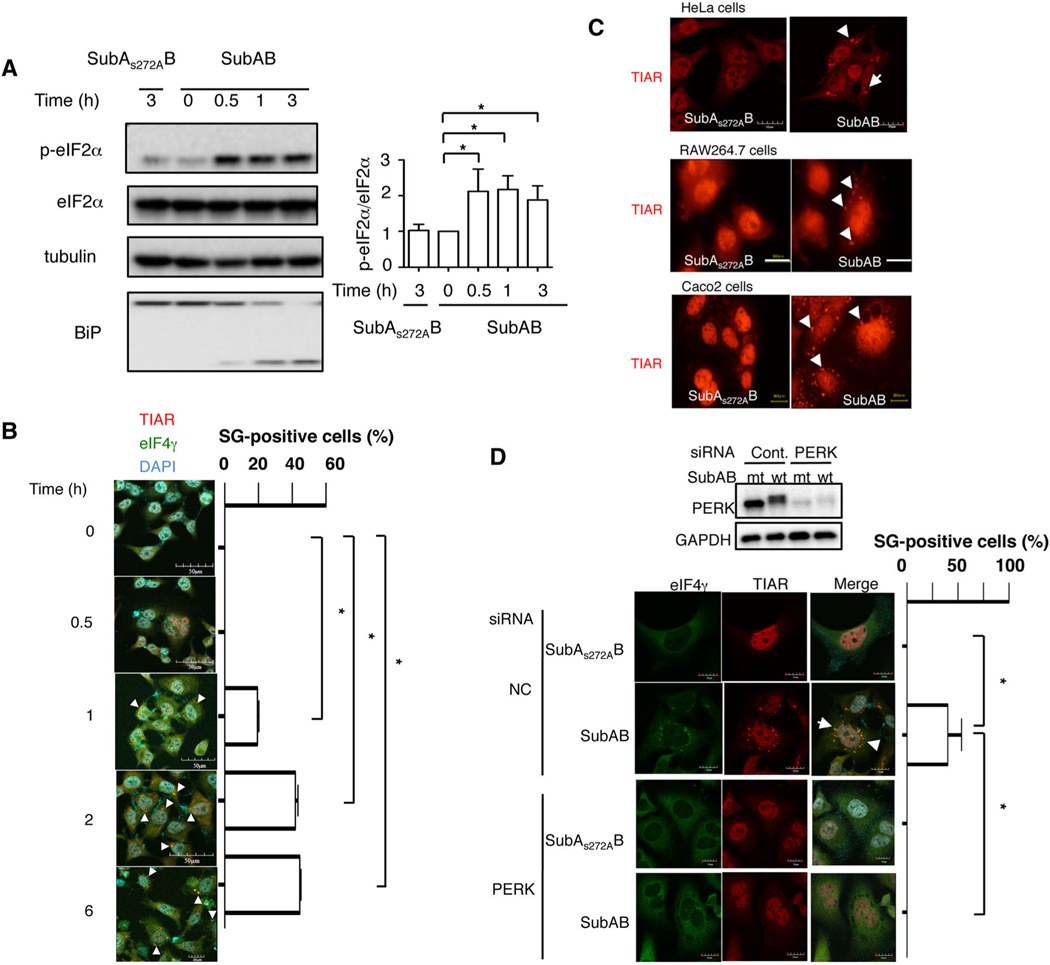
Our previous study demonstrated that SubAB-induced cell death was mediated by activation of PERK-eIF2α pathway (Yahiro et al., 2012). After 30 min incubation with SubAB, we observed eIF2α phosphorylation, which was not increased by catalytically inactive mutant SubAS272AB (Fig. 1A). A previous study showed that activation of the PERK-eIF2α pathway participates in SG formation (Kedersha et al., 1999). We next examined whether SubAB treatment induced SG. HeLa cells were incubated with SubAB for the indicated times, then fixed and immunostained for the SG marker proteins, TIAR and eIF4γ. As shown in Fig. 1B, TIAR was translocated from nucleus to dense cytoplasmic foci and colocalized with eIF4γ the cytoplasm in a time-dependent manner, consistent with the time-course of SG formation, SubAB-induced BiP cleavage and eIF2α phosphorylation (Fig. 1A). SubAB-induced TIAR translocation was observed in HeLa, RAW264.7 and Caco2 cells (Fig.1C).
Fig. 1.
Subtilase cytotoxin induces SG formation through a PERK-dependent pathway.
A. HeLa cells were incubated with catalytically inactive SubAS272AB or SubAB (400 ng ml−1) for the indicated times at 37°C, and cell lysates were subjected to immunoblotting with the indicated antibodies. α-tubulin served as a loading control.
B. After cells were treated with SubAB for the indicated times at 37°C, the fixed cells were reacted with anti-TIAR antibody (red) and anti-eIF4γ antibody (green) and observed by confocal microscopy. A merged picture shows colocalization in HeLa cells. Cell nuclei were stained by DAPI (cyan). The rate of SG formation is presented as mean ± standard deviation (SD) from four different fields, which included at least 20 cells/field (right panel). Bars represent 50 μm. Experiments were repeated two times with similar results, and significance is *P < 0.05. The white arrows indicate SG, including TIAR and eIF4γ.
C. The cells were incubated with 400 ng ml−1 of SubAB or SubAS272AB for 3 h at 37°C. The fixed cells were reacted with anti-TIAR antibody (red) observed by confocal microscopy. The arrows indicate SG formation, including TIAR. Bars represent 20 μm. Experiments were repeated three times with similar results.
D. Control (NC) and PERK siRNA-transfected cells were incubated with SubAS272AB (mt) or SubAB (wt) for 3 h at 37°C. Cell lysates were subjected to immunoblotting with the indicated antibodies. GAPDH served as a loading control. Cells were fixed, immunostained with the indicated antibodies and observed by confocal microscopy as described in Fig. 1. The rate of SG formation is presented as mean ± SD from four different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated three times with similar results, and significance is *P < 0.05. The arrows indicate SG.
Our previous study indicated that SubAB induced eIF2α-phosphorylation via a PERK-dependent pathway (Yahiro et al., 2012). PERK is a stress sensor in the ER and associated with the maintenance of homeostasis (B’Chir et al., 2013; Matsumoto et al., 2013; Jiang et al., 2014). We tested if activation of PERK was associated with SubAB-induced SG formation. Control or PERK siRNA-transfected HeLa cells were incubated with mt or wt SubAB for 2 ~ 3 h and reacted with antibodies against SG marker proteins. We found that SubAB-induced SG formation was not seen in PERK-knockdown cells (Fig. 1D). Further, a previous report showed that the activation of the PERK-eIF2α pathway induces preferential translation of activating transcription factor 4 (ATF4) (Ron and Walter, 2007). However, SubAB-induced SG formation was not altered in ATF4 knockdown cells compared with control cells (Fig. S1a). These findings suggest that SubAB-induced SG formation is a PERK-dependent and ATF4-independent pathway.
Depletion of G3BP1 inhibits subtilase cytotoxin-induced stress granule formation
Stress granule recruits RNA binding proteins such as Ras GTPase-activating protein-binding protein 1 (G3BP1) and Hu protein R (HuR) for assembly, and G3BP1 is essential for SG formation (Gallouzi et al., 2000; Tourriere et al., 2003). To investigate whether SG formation was associated with SubAB-induced apoptosis, we examined the effect of depletion of G3BP1 in HeLa cells. The expression level of G3BP1 was suppressed by the specific siRNA respectively (Fig S1b). G3BP1 depletion decreased SubAB-induced SG formation (Fig S1c). As shown in Fig S1d, knockdown of G3BP1 did not affect SubAB-induced PARP cleavage compared with control cells. These results indicate that SG formation is not essential for SubAB-induced apoptotic signalling.
Depletion of subAB gene in STEC O113:H21 impairs stress granule formation in Caco2 cells
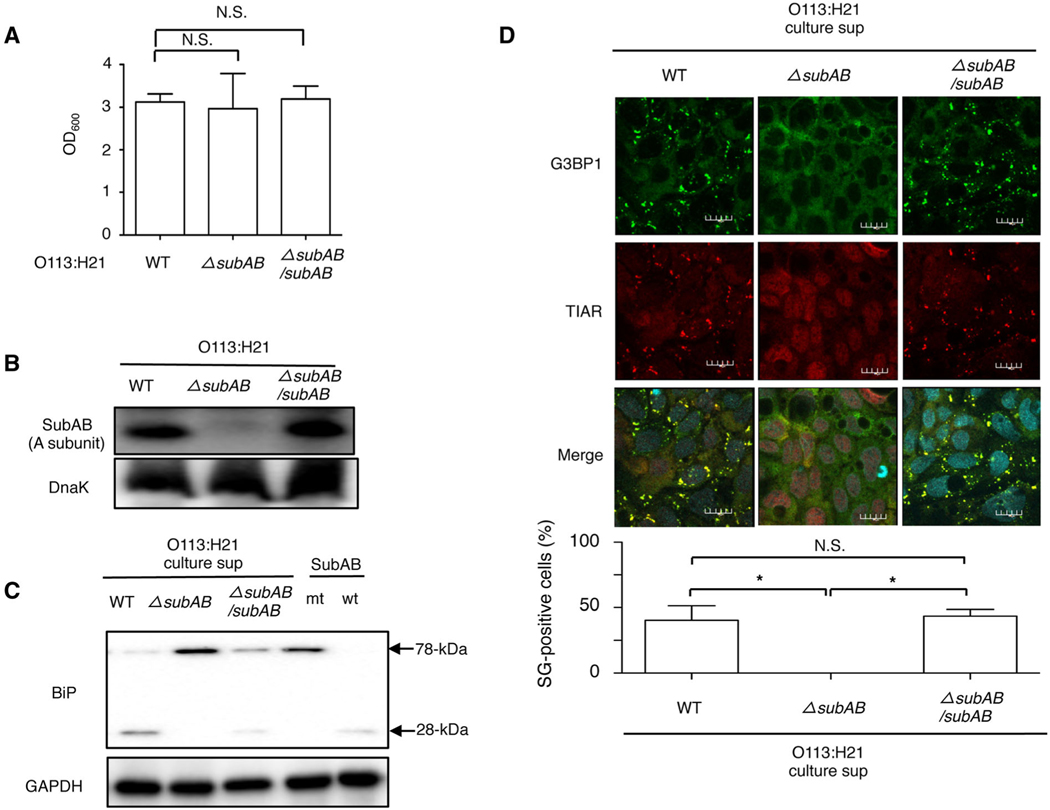
To determine whether STEC O113:H21-produced SubAB is an essential factor for SG formation, we used a wild-type strain, one lacking the subAB gene (ΔsubAB) strain and a ΔsubAB strain complemented with a wild-type SubAB expressing plasmid (ΔsubAB/subAB). These three strains exhibited similar growth, suggesting that deletion or complement of the subAB gene did not affect STEC growth (Fig. 2A). The expression of SubAB in STEC O113:H21 ΔsubAB strain was completely absent compared with the wild-type strain and the ΔsubAB/subAB strain (Fig. 2B). We next examined activity of SubAB-mediated BiP cleavage in the culture supernatant from each strain. Caco2 cells were incubated with the culture supernatants, with purified SubAB as a positive control and SubAS272AB as a negative control. Although the culture supernatant of wild-type strain, complement strain and purified SubAB induced BiP cleavage, ΔsubAB strain and purified SubAS272AB did not induce BiP cleavage, suggesting that the ΔsubAB strain completely lost SubAB activity (Fig. 2C). Consistent with the BiP cleavage patterns, we observed SG formation in Caco2 cells when incubated with the culture supernatant of the wild-type strain and complement strain but not with that of the ΔsubAB strain (Fig. 2D). These results suggest that STEC O113:H21-produced SubAB causes BiP cleavage, resulting in SG formation.
Fig. 2.
STEC O113:H21-produced SubAB induces SG formation.
A. Wild-type, ΔsubAB or ΔsubAB/subAB O113:H21 strains were cultured in BHI medium for 12 h at 37°C. Bacterial growth was measured by optical density at 600 nm (OD600).
B. The wild, ΔsubAB or ΔsubAB/subAB O113:H21 strains lysates were subjected to immunoblotting with anti-SubAB antibody or anti-DnaK antibody as a loading control.
C. Caco2 cells were incubated with purified SubAB (wt), SubAS272AB (mt), culture supernatant of wild-type, ΔsubAB or ΔsubAB/subAB O113:H21 strains for 1 h at 37°C. Cell lysates were subjected to immunoblotting with anti-BiP antibodies. GAPDH served as a loading control.
D. The culture supernatant of wild-type, ΔsubAB or ΔsubAB/subAB O113:H21 strains were added to Caco2 cells, which were incubated for 6 h, fixed with 4% PFA, reacted with anti-TIAR antibody (red) and anti-G3BP1 antibody (green) and observed by confocal microscopy. Cell nuclei were stained by DAPI (cyan). The rate of SG formation is presented as mean ± SD from four different fields, which included at least 20 cells/field (bottom panel). Bars represent 20 μm. All experiments were repeated two times with similar results, and significance is *P < 0.05.
Subtilase cytotoxin-induced stress granule formation is suppressed by lysosomal inhibition, not by caspase inhibition
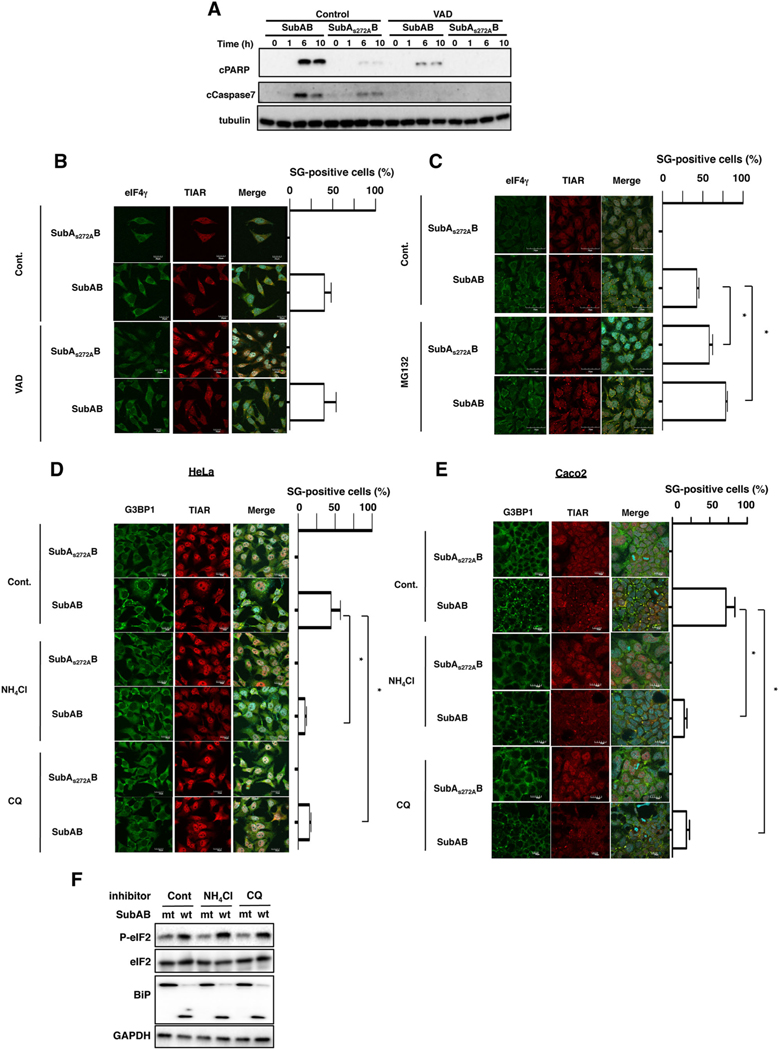
Next, we investigated whether SubAB-induced caspase activation was required for SG formation. As shown in Fig. 3A, a general caspase inhibitor, Z-VAD-FMK, suppressed SubAB-induced caspase-7 activation and PARP cleavage as previously reported (Yahiro et al., 2012). However, SG formation by SubAB was not inhibited in the presence of Z-VAD-FMK, suggesting that caspase activation by SubAB was not involved in SubAB-induced SG formation (Fig. 3B). Mazroui et al. (2007) have demonstrated that inhibition of ubiquitin-dependent proteasome system (UPS) activity by MG132 induced the formation of SG. We previously reported that SubAB-induced caspase activation was significantly inhibited by MG132 (Yahiro et al., 2012). We next monitored the effect of MG132 treatment on the SG formation by SubAB. Pretreatment cells with MG132 significantly increased SG formation, even in the absence of SubAB activity, and promoted SubAB-induced SG formation (Fig. 3C). These results support the conclusion that SubAB-induced SG fraction is independent of caspase activation.
Fig. 3.
Effect of caspase inhibitor and lysosomal inhibitor on SubAB-induced SG formation.
A. HeLa cells were pretreated with 10 μM of caspase inhibitor, Z-VAD-FMK (VAD), for 30 min and then incubated with SubAS272AB or SubAB for the indicated times at 37°C, and cell lysates were subjected to immunoblotting with the indicated antibodies. α-Tubulin served as a loading control.
B, C. HeLa cells were pretreated with control, 10 μM of VAD or 20 μM of MG132, incubated with SubAS272AB or SubAB for 3 h at 37°C, and then fixed cells were reacted with the anti-TIAR antibody (red) and anti-eIF4γ antibody (green) and observed by confocal microscopy. A merged picture shows colocalization in HeLa cells. Cell nuclei were stained by DAPI (cyan). The rate of SG formation is presented as mean ± SD from three different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated two times with similar results, and significance is *P < 0.05.
D, E. HeLa or Caco2 cells were pretreated with 10 mM of NH4Cl or 100 nM of CQ for 30 min and then incubated with SubAS272AB or SubAB for 3 h at 37°C, and the fixed cells were reacted with the anti-TIAR antibody (red) and anti-G3BP1 antibody (green) and observed by confocal microscopy. A merged picture shows colocalization in HeLa cells. Cell nuclei were stained by DAPI (cyan). The rate of SG formation is presented as mean ± SD from three different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated three times with similar results, and significance is *P < 0.05.
F. HeLa cells were pretreated with the indicated reagents as described earlier and then incubated with SubAS272AB (mt) or SubAB (wt) for 1 h at 37°C. Cell lysates were subjected to immunoblotting with the indicated antibodies. GAPDH served as a loading control. Experiments were repeated three times with similar results.
A previous study showed that co-incubation of the cells with MG132 and lysosomal inhibitors ammonium chloride (NH4Cl) or Chloroquine (CQ) reduced MG132-induced SG formation. They also suggested that NH4Cl suppressed MG132-induced SG by controlling steps downstream of polysome disassembly (Seguin et al., 2014). We next monitored whether NH4Cl and CQ inhibited SubAB-induced SG formation. In control cells, SubAB induced SG formation in approximately 40% of cells. Following pretreatment of cells with NH4Cl and CQ, SubAB-induced SG formation was suppressed throughout the cytoplasm (Fig. 3D). We also found that these inhibitors suppressed SubAB-induced SG formation in Caco2 cells (Fig. 3E). As shown in Fig. 3F, NH4Cl and CQ did not affect SubAB delivery and its activity. Thus, SubAB-induced SG does not result from a massive cell death by caspase activation, rather than ubiquitin proteasome system, and lysosomal activity may control SG formation.
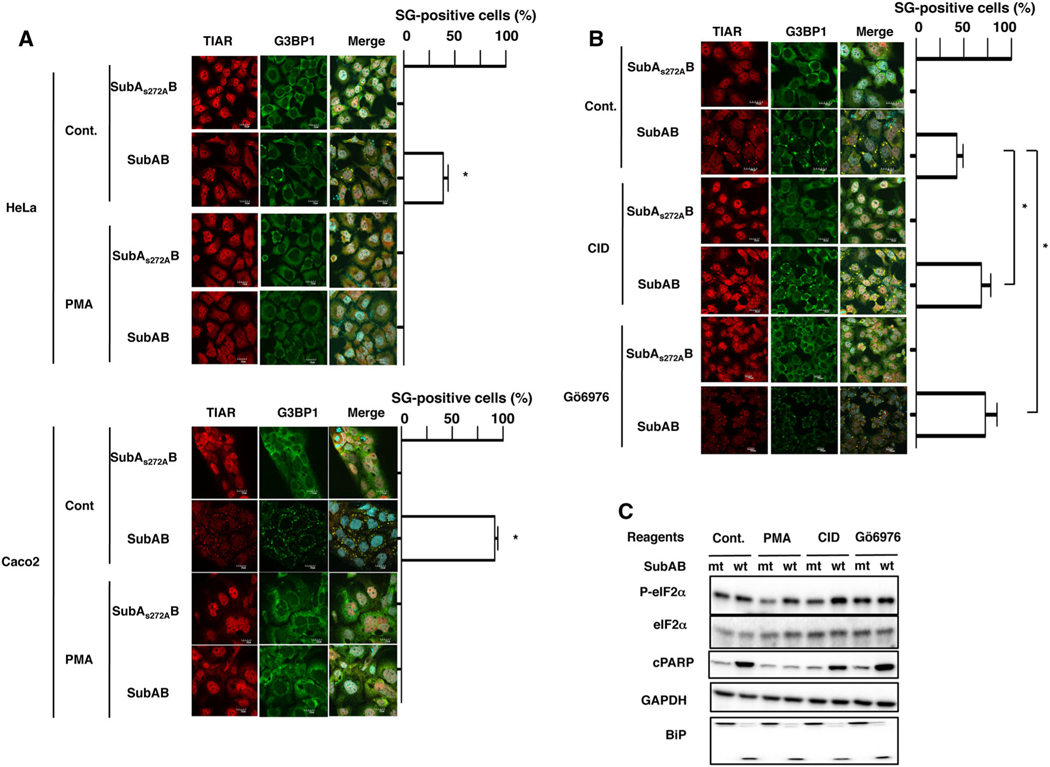
Subtilase cytotoxin-induced stress granule formation is inhibited by PMA treatment and enhanced by protein kinase C inhibition
Some protein kinases are associated with SG formation (Buchan and Parker, 2009; Shah et al., 2014). We screened effects of the kinase activators or inhibitors on SubAB-induced SG formation; effects of PKC activator, PMA; PKA activators, 8Br-cAMP and Forskolin; PKC inhibitors, Gö6976, Gö6983 and Bisindolylmaleimide II; PKD inhibitor, CID755673; PKA inhibitor, 14–22 Amide; ROCK II inhibitor, Y-27632; and CaM kinase II inhibitor, KN-93. In HeLa and Caco2 cells, we found that SubAB-induced SG formation was suppressed by PMA pretreatment (Fig. 4A). Further, we also found approximately twofold increase in SubAB-induced SG formation in the presence of Gö6976 and CID755673, although inhibitors with mutant SubAB did not affect SG formation (Fig. 4B). Moreover, other PKC inhibitors, Bisindolylmaleimide II and Gö6983, enhanced SubAB-induced SG formation (Fig S2). Other reagents (e.g. PKA inhibitor, PKA activator, ROCK II inhibitor and CaM kinase II inhibitor) did not affect SubAB-induced SG formation. Interestingly, PMA pretreatment did not suppress TG-induced SG formation in HeLa cells (Fig S4a). Although SubAB-induced eIF2α phosphorylation and BiP cleavage did not alter, SubAB-induced PARP cleavage was inhibited by PMA pretreatment (Fig. 4c). These results indicate that SubAB-induced SG formation was regulated by PKC activity, which acts downstream of eIF2α, and activation of PKC could suppress SubAB-induced activation of apoptosis.
Fig. 4.
Subtilase cytotoxin-induced SG formation is inhibited by PMA and enhanced by PKC inhibition.
A. HeLa and Caco2 cells were preincubated with or without 100 nM of PMA for 30 min and then incubated with SubAS272AB or SubAB for 3 h at 37°
C. Cells were fixed with 4% PFA and reacted with the anti-TIAR antibody (red) and anti-G3BP1 antibody (green), and then observed by confocal microscopy. Cell nuclei were stained by DAPI (cyan). The rate of SG formation is presented as mean ± SD from four different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated three times with similar results, and significance is *P < 0.05.
B. HeLa cells were preincubated with 20 μM of CID755673 (CID) or 4 μM of Gö6976 for 30 min and then incubated with SubAS272AB or SubAB for 3 h at 37°C. Cells were fixed with 4% PFA and reacted with the anti-TIAR antibody (red) and anti-G3BP1 antibody (green) and observed by confocal microscopy. Cell nuclei were stained by DAPI (cyan). The rate of SG formation is presented as mean ± SD from five different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated two times with similar results, and significance is *P < 0.05.
C. As described earlier, cells were treated with the indicated reagents and then incubated with SubAS272AB (mt) or SubAB (wt) for 3 h at 37°C. Cell lysates were subjected to immunoblotting with anti-eIF2α, anti-phospho-eIF2α, anti-cPARP and anti-BiP antibodies. GAPDH served as a loading control. Experiments were repeated three times with similar results.
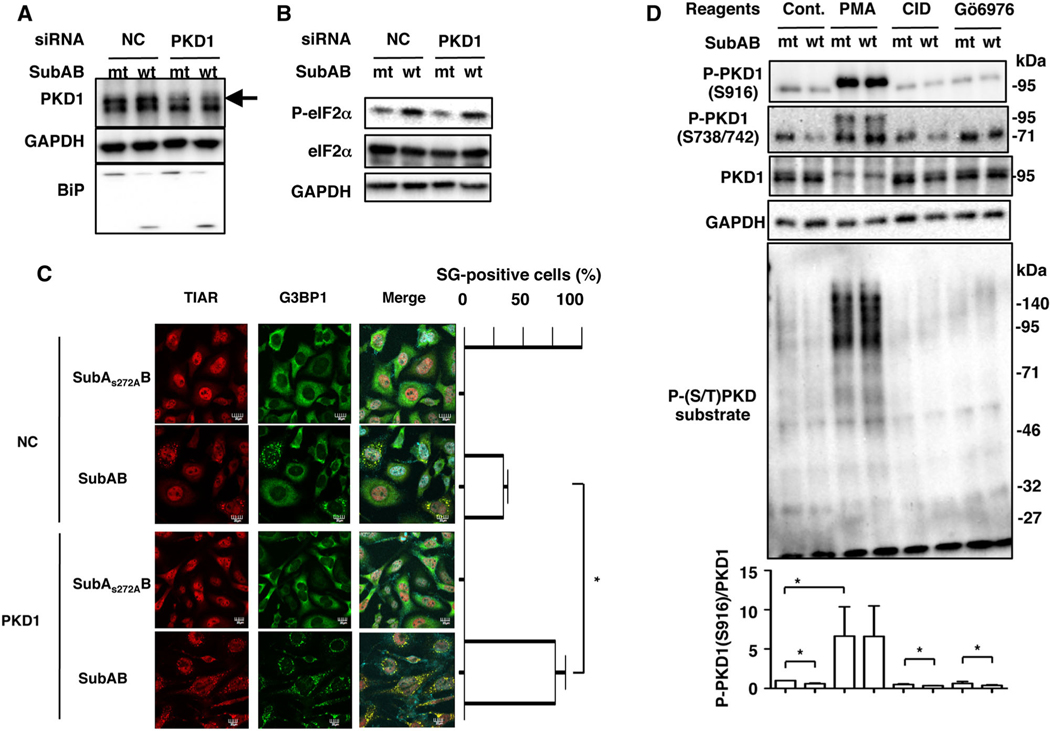
Next, we investigated whether PKD control SubAB-induced SG formation. We found that the amount of PKD1 protein was suppressed by PKD1 siRNA; however, that did not affect SubAB-mediated BiP cleavage and eIF2α phosphorylation (Fig. 5A and B). Consistent with these data with a PKD inhibitor CID755673, SubAB-induced SG formation in PKD1 knockdown cells approximately doubled that in control siRNA-transfected cells (Fig. 5C). These findings strongly suggest that PKD1 controls SubAB-induced SG formation.
Fig. 5.
Protein kinase D1-knockdown enhances SubAB-induced SG formation.
A, B. Control (NC) and PKD1 siRNA-transfected HeLa cells were incubated with 400 ng ml−1 of SubAS272AB (mt) or SubAB (wt) for 3 h at 37°C. Cell lysates were subjected to immunoblotting for PKD1 (arrow), phospho-eIF2α (P-eIF2α), eIF2α and BiP. GAPDH served as a loading control. Experiments were repeated three times with similar results.
C. As described earlier, NC-transfected and PKD1 siRNA-transfected cells were incubated with SubAS272AB (mt) or SubAB (wt) for 3–4 h at 37°C. Cells were fixed with 4% of PFA and reacted with the anti-TIAR antibody (red) and anti-G3BP1 antibody (green) and observed by confocal microscopy. Cell nuclei were stained by DAPI (cyan). The rate of SG formation is presented as mean ± SD from five different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated three times with similar results, and significance is *P < 0.01.
D. Cells were treated with the indicated reagents and then incubated with SubAS272AB (mt) or SubAB (wt) for 3 h at 37°C. Cell lysates were subjected to immunoblotting with anti-P-PKD1 (S916), anti-P-PKD1(S738/742), anti-P-(S/T) PKD substrates and anti-PKD1 antibodies. GAPDH served as a loading control. Experiments were repeated three times with similar results. Quantification of the level of P-PKD1 (S916)/PKD1 was performed by densitometry (lower panel). Data are means ± SD of values from three experiments, with n = 3 per experiment. Statistical significance is *P < 0.05.
Protein kinase D1 activity is regulated by PKC through phosphorylation at Ser738/742 in the activation loop, followed by autophosphorylation at Ser-916, which correlates with elevated PKD1 kinase activity (Matthews et al., 1999; Harrison et al., 2006). Next, we analysed if SubAB affects the level of phosho-PKD1. After 3 h of incubation, SubAB suppressed phospho-PKD1 (S916) in control cells. After treatment of cells with PMA, phospho-PKD1 (S738/742) and phospho-PKD1 (S916) were detected at increased molecular weight; these modifications were not suppressed by incubation with SubAB. Anti-phospho-PKD1 (S738/742) antibody recognized phospho-PKD1 (95 kDa) and unknown 70 kDa bands. Treatment with PKD inhibitors (CID755673, Gö6976) caused a reduction of the basal level of phospho-PKD1 (S916), which was accompanied by an additional decrease by SubAB. We also used anti-phospho (S/T) PKD substrates antibodies, which recognize phosphorylated PKD substrates, to investigate if SubAB causes downregulation of PKD1 activity. PMA treatment promoted the amount of phospho-(S/T) PKD substrate proteins, which were decreased by PKD inhibitors. SubAB suppressed phospho-PKD1 (S916) and phospho-(S/T) PKD substrate proteins (Fig. 5D).
Protein kinase Cδ is involved in subtilase cytotoxin-induced stress granule formation
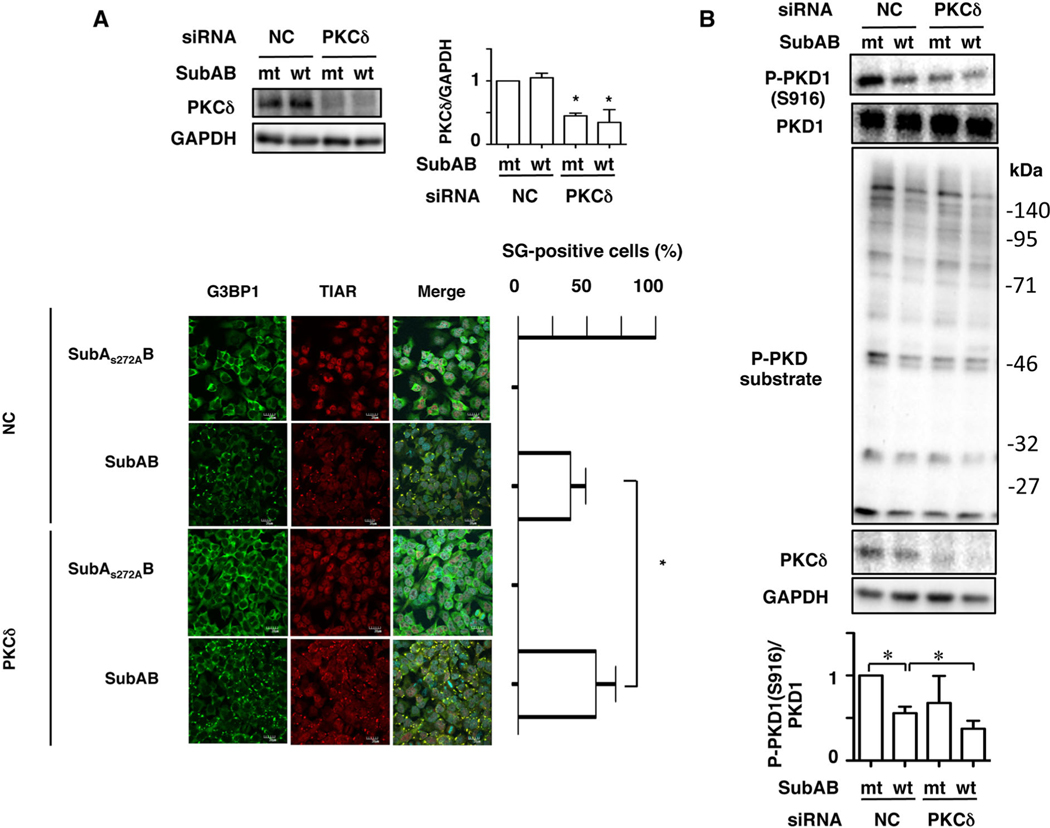
As shown earlier, broad-spectrum PKC inhibitors, Bisindolylmaleimide II and Gö6983, treatment enhanced SubAB-induced SG formation (Fig. S2). Both PKC inhibitors commonly suppress PKCα, PKCβ and PKCδ but not PKD1 (Sewald et al., 2011). Furthermore, it has been reported that PKD1 activation is PKC-dependent signalling (Rozengurt et al., 2005). In PKCδ siRNA-transfected cells, the amount of PKCδ was significantly suppressed. We found here that SubAB-induced SG formation was increased in PKCδ-knockdown cells compared with control cells (Fig. 6A). Next, we investigated effects of PKCδ-depletion on PKD1 phosphorylation with or without SubAB. PKCδ was suppressed by the specific siRNA. Depletion of PKCδ led to a reduction of basal phospho-PKD1 (S916) and phospho-(S/T) PKD substrate proteins, which were additionally suppressed by SubAB (Fig. 6B). These findings suggest that PKCδ/PKD1 signalling is involved in SubAB-induced SG formation.
Fig. 6.
Protein kinase Cδ-knockdown enhances SubAB-induced SG formation.
A. Control (NC) or PKCδ siRNA-transfected cells were incubated with SubAS272AB (mt) or SubAB (wt) for 3–4 h. Cell lysates were subjected to immunoblotting with anti-PKCδ and anti-GAPDH antibodies. Quantification of the level of PKCδ/GAPDH was performed by densitometry (right panel). Data are means ± SD of values from three experiments, with n = 3 per experiment. Statistical significance is *P < 0.05. The fixed cells were reacted with the indicated antibodies and observed by confocal microscopy (lower panel). The rate of SG formation is presented as mean ± SD from five different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated three times with similar results, and significance is *P < 0.05.
B. Control (NC) and PKCδ siRNA-transfected cells were incubated with SubAS272AB (mt) or SubAB (wt) for 3–4 h. Cell lysates were subjected to immunoblotting with the indicated antibodies. GAPDH served as a loading control. Experiments were repeated three times with similar results. Quantification of the level of P-PKD1 (S916)/PKD1 was performed by densitometry (bottom panel). Data are means ± SD of values from three experiments, with n =3 per experiment. Statistical significance is *P < 0.05.
Death-associated protein 1 controls subtilase cytotoxin-induced stress granule formation
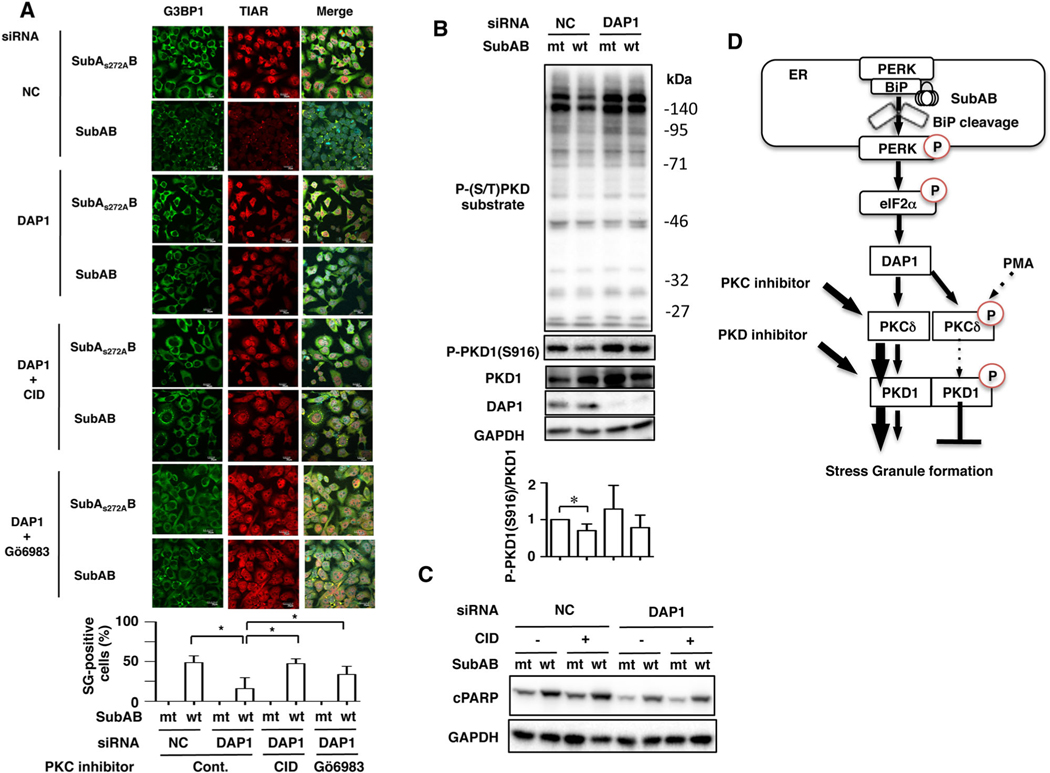
We previously demonstrated that DAP1 regulates SubAB-stimulated apoptotic pathway and acts downstream of PERK-eIF2α signalling (Yahiro et al., 2014). To examine if DAP1 involves in SubAB-induced SG formation, DAP1-knockdown cells were incubated with SubAB in the presence or absence of CID755673. SG formation by SubAB was dramatically decreased in DAP1-knockdown cells compared with control cells. Treatment of DAP1-knockdown cells with CID755673 and Gö6983 led to reappearance of SubAB-induced SG formation (Fig. 7A). Interestingly, TG-induced SG formation was slightly inhibited in DAP1-knockdown cells (Fig. S4b).
Fig. 7.
Death-associated protein 1 is associated with SubAB-induced SG formation.
A. Control (NC) or DAP1 siRNA-transfected cells were incubated with SubAS272AB (mt) or SubAB (wt) in the presence or absence of CID755673 (CID, 20 μM) or Gö6983 (4 μM) for 3 h. The fixed cells were reacted with the anti-TIAR antibody (red) and anti-G3BP1 antibody (green) and observed by confocal microscopy. The rate of SG formation is presented as mean ± SD from five different fields, which included at least 20 cells/field (bottom panel). Bars represent 20 μm. Experiments were repeated three times with similar results, and significance is *P < 0.05.
B. Control (NC) or DAP1 siRNA-transfected cells were incubated with SubAS272AB (mt) or SubAB (wt) for 3–4 h at 37°C, and cell lysates were subjected to immunoblotting with the indicated antibodies. GAPDH served as a loading control. Experiments were repeated three times with similar results. Quantification of the level of P-PKD1 (S916)/PKD1 was performed by densitometry (bottom panel). Data are means ± SD of values from three experiments, with an n = 3 per experiment. Statistical significance is *P < 0.05.
C. Cells were pretreated with or without 20 μM of CID755637 for 30 min and then incubated with SubAS272AB (mt) or SubAB (wt) for 3–4 h at 37°C. Cell lysates were subjected to immunoblotting with anti-cPARP antibodies. GAPDH served as a loading control. Experiments were repeated three times with similar results.
D. A possible model for SubAB-induced SG formation. BiP cleavage by SubAB induces activation of PERK-eIF2α, which affects downstream DAP1. DAP1 negatively regulates PKCδ/PKD1, resulting in SG formation. Treatment cells with PKD1 inhibitor CID755673 enhance SG formation by SubAB. In contrast, activation of PKC/PKD1 signalling by PMA or DAP1 depletion reduces SG formation.
We further examined if DAP1 controlled PKD1 phosphorylation in the presence or absence of SubAB. Depletion of DAP1 increased basal phospho-PKD1 (S916) and phospho-(S/T) PKD substrate proteins, which are slightly decreased by SubAB (Fig. 7B). These results indicate that DAP1 acts upstream of PKC/PKD1 and SubAB-induced SG formation. Furthermore, we next examined the effect of CID755673 on SubAB-induced PARP cleavage in DAP1-knockdown cells. As shown in Fig. 7C, depletion of DAP1 by siRNA suppressed SubAB-induced PARP cleavage as reported previously (Yahiro et al., 2014). Inhibition of PKD activity by CID755673 did not affect SubAB-induced PARP cleavage in DAP1-knockdown cells. Thus, PKD signalling pathway was not involved in SubAB-induced apoptotic signalling.
Discussion
Stress granules are known to aggregate in the cytoplasm when cells are exposed to stresses, e.g. heat shock, oxidative stress, viral infection and UV irradiation (Anderson and Kedersha, 2009). SG formation helps protect against stress-induced cell death (Arimoto et al., 2008; Tsai and Wei, 2010). Further, recent studies have shown that SG are associated with neurodegenerative diseases, e.g. Huntington’s disease, amyotrophic lateral sclerosis, frontotemporal lobar dementia and Alzheimer’s disease (Wolozin, 2012; Vanderweyde et al., 2013). In this study, we report that the bacterial toxin SubAB induced SG formation through BiP cleavage and PERK-eIF2α activation, followed by a DAP1-dependent and PKC-dependent pathway.
Knockdown of PERK by siRNA inhibited SubAB-induced SG formation. Activation of PERK by ER stress controls protein synthesis via eIF2α, this pathway is involved in autophagy and apoptosis (Yahiro et al., 2012; B’Chir et al., 2013; Matsumoto et al., 2013; Jiang et al., 2014). In agreement with our findings, recent studies demonstrated that cold shock or salubrinal, a PERK activator, caused activation of PERK-eIF2α pathway, which induces SG formation (Hofmann et al., 2012; Walker et al., 2013). Thus, these findings suggest that PERK plays an essential factor in SubAB-induced SG formation.
Treatment of HeLa and Caco2 cells with PMA completely suppressed SubAB-induced SG formation; hence, both PKD inhibitor CID755673 and PKD1 knockdown enhanced SG formation in HeLa cells. PKD1 is a serine/threonine kinase that is involved in crucial biological processes, including cell growth, apoptosis, adhesion and angiogenesis (Rozengurt et al., 2005; Sundram et al., 2011; Steinberg, 2012). In addition, subcellular localization of PKD1 is cell-specific (Van Lint et al., 2002). Although overexpressed PKD1 localizes in the trans-Golgi network and regulates anterograde membrane trafficking in HeLa cells (Prestle et al., 1996; Maeda et al., 2001), a previous study demonstrated that PKD1 interacts with transcription factor Snail1 in nuclei of HeLa cells and regulates cell proliferation (Eiseler et al., 2012). In addition, PKD1 phosphorylates Enabled/Vasodilator-stimulated phosphoprotein (Ena/VASP), leading to increased filopodia formation and length at focal adhesion contacts. These findings indicate that PKD1 acts not only in the trans-Golgi network but also in nuclei and at sites of actin remodelling to regulate biological processes. Hence, TG-induced SG formation was not inhibited by PMA, suggesting that TG-induced SG formation is independent of PKC activation. Our study now shows that PKD1 participates in SubAB-induced SG formation; PKD1 signalling regulates TIAR translocation from nuclei to cytoplasm and G3BP1 movement from cytosol to the SG compartment.
Because SubAB-induced SG formation was enhanced by depletion of PKD1 and its inhibitor CID755673, we focused on PKD1 in this study. However, PKD isoforms, PKD2 and PKD3, are also inhibited by CID755673 (Sharlow et al., 2008). SubAB-mediated SG was not enhanced in PKD2-knockdown and PKD3-knockdown cells (Fig. S3a). PKD1 knockdown did not affect the level of PKD2 and PKD3 (Fig. S3b). Thus, PKD1 is specifically involved in SubAB-induced SG formation.
Protein kinase D1 is activated by PMA or diacyl-glycerols (Rozengurt et al., 2005). Previous studies showed that activation of PKD1 by phosphorylation at Ser-738/742 causes autophosphorylation at other sites, including Ser910 in the C-terminal domain (Rybin et al., 2009; Steinberg, 2012). Treatment of the cells with PMA increased the molecular weight and basal phosphorylation of PKD1 and its substrates (Fig. 5D). In contrast, SubAB caused a slight reduction of phospho-PKD1 (S916 and S744/748) and PKD1 substrates. These findings raise the possibility that SubAB negatively regulates PKD1 function, which triggers SG formation, because PMA-activated PKD1 suppressed SubAB-induced SG formation. Seguin et al. (2014) also reported that both NH4Cl and chloroquine impaired SG formation induced by MG132. They suggested that interplay between proteasome, autophagy and lysosomes is needed to form optical SG assembly; NH4Cl suppressed MG132-induced SG by causing polysome disassembly. Our data demonstrated that inhibition of autophagy by Atg5 or Atg16L1 siRNA did not suppress SubAB-induced SG assembly (Fig. S5), suggesting that SubAB-induced SG formation is independent of autophagy and may occur by a different mechanism, as seen with MG132.
Although DAP1 is involved in negative regulation of autophagy and also in apoptosis (Koren et al., 2010; Yahiro et al., 2014), little is known of the biological process modulated by DAP1. We show here a novel function of DAP1; SubAB-induced SG formation was significantly inhibited in DAP1-knockdown cells; incubation with CID755673 and Gö6983 reversed the effect on SubAB-induced SG formation (Fig. 7A). In addition, the basal level of phospho-PKD1 (S916) increased in DAP1-knockdown cells. We found here that broad-spectrum PKC inhibitors Bisindolylmaleimide II and Gö6983, which do not affect PKD1 activity, significantly enhanced SubAB-induced SG (Fig. S2), suggesting that both PKC and PKD1 are downstream of DAP1 and regulate SubAB-induced SG formation. While SubAB-induced SG formation was still observed in DAP1-knockdown cells, these findings might reflect the fact that the siRNA transfection did not suppress the level of DAP1 in all cells or that a DAP1-independent pathway modulated SG formation.
Thapsigargin-induced SG formation was slightly inhibited in DAP1-knockdown cells, suggesting that TG-induced SG formation occurs predominantly by a DAP1-independent pathway. Thus, these data indicate that SubAB-induced SG formation pathway is different from that used by TG. However, we do not know how DAP1 is involved in PKCδ/PKD1 signal transduction. Immunoprecipitation by anti-FLAG antibodies using FLAG-tagged DAP1 overexpressed in cell lysates did not detect a direct interaction with PKCδ/PKD1 (data not shown). Thus, DAP1 might be indirectly involved with PKCδ/PKD1 to regulate SG formation by SubAB. Further experimentation is needed to clarify this point. Recent studies have shown that different PKC isoforms are acting upstream of PKD1 (Scheiter et al., 2013). For example, PKCδ is upstream of PKD1 in reactive oxygen species-mediated mitochondrial depolarization (Zhang et al., 2015), and PKCδ knockdown effectively attenuates PKD1 activation (Asaithambi et al., 2011). PKCε and PKCη interact and activate PKD1 (Waldron et al., 1999a,1999b; Brandlin et al., 2002a,2002b; Doppler and Storz, 2007). We provide evidence in this study that depletion of PKCδ suppressed basal phospho-PKD1 (S916) and phospho-(S/T) PKD substrate proteins, which were additionally decreased in the presence of SubAB, and enhanced SubAB-mediated SG formation. Thus, PKCδ is an important regulator involved in controlling PKD1 activity during SubAB-induced SG formation. These findings imply that, upon SubAB-induced ER stress, DAP1 may negatively regulate PKD1 activity through PKCδ.
Subtilase cytotoxin-induced ER stress caused a mitochondria-dependent apoptosis (Matsuura et al., 2009; May et al., 2010; Yahiro et al., 2010). Regarding the PKD1-associated cell death pathway, recent studies showed that PKD1 is a key mediator of necrosis in acute pancreatitis (Yuan et al., 2012), activated and downregulated by PMA through a PKC-dependent ubiquitin-proteasome pathway, which is also involved in induction of apoptosis in LNCaP prostate cancer cells (Chen et al., 2011); further, selenite, an anti-cancer reagent, induced apoptosis through suppression of PKD/CREB/Bcl2 pathway (Hui et al., 2014). Meanwhile, PKD1 inhibited H2O2-induced intestinal cell death via upregulation of NF-kB and downregulation of p38 MAPK (Song et al., 2009). Here, we show that inhibition of PKD activity by CID755673 (Figs 4C and 7C) or PKD1 knockdown did not affect SubAB-induced PARP cleavage (Fig. S3c); however, treatment of cells with PMA completely inhibited PARP cleavage. Our findings suggest that, upon stimulation by PMA, PKC activation has a protective role in SubAB-induced apoptotic pathway.
We found that SubAB-induced PARP cleavage was not suppressed in G3BP1-knockdown cells, suggesting that in HeLa cells, SG formation was not directly associated with apoptosis. On the other hand, a previous study indicated that G3BP1 mediates cross-talk between stress response and innate immune system (Reineke et al., 2015). Thus, this raised a possibility that SubAB-induced SG formation modifies host immune system by translational inhibition. In addition to their relevance in regulating translation by cellular stress, SG is induced during virus infection and countered by viruses to maximize replication efficiency (Raaben et al., 2007). Although SG are thought to be an anti-viral and host defence mechanism, pathogenic viruses such as herpes simplex virus (HSV) and influenza A virus (IAV) inhibit SG formation, resulting in suppression of the host immune system through varied mechanisms (Onomoto et al., 2014). These findings support the direct function of SG as a host defence system in viral infection. In the case of bacterial infection, the functional role of SG is still unclear. It was reported that translational arrest promotes host immune system through detection of pathogenic bacteria or an effector-triggered signalling pathway (McEwan et al., 2012; Stuart et al., 2013). SubAB also causes translational arrest through PERK-dependent phosphorylation of eIF2α. SubAB-induced SG formation may modify a host innate immune system and contribute to an anti-bacterial host defence system. Thus, determination if SG formation by SubAB affects the host immune system is critical.
In conclusion, we provide the proposed molecular mechanisms for SubAB-induced SG formation as shown in Fig. 7D. Stressed cells need to silence non-essential transcripts and produce cytoprotective proteins. The role of SubAB-induced SG formation in LEE-negative STEC infectious disease is unknown and is under investigation. Our novel findings suggest that SubAB induces translation arrest via PERK activation and phosphorylation of eIF2α. These signals negatively regulate PKCδ/PKD1 activity via DAP1, resulting in induction of SG formation, which is inhibited by PKC activation. Interestingly, PKC activation also inhibited SubAB-induced PARP cleavage. Thus, DAP1 is a key regulatory factor in SG formation and apoptosis.
Experimental procedures
Reagents
Anti-α-tubulin monoclonal antibody was purchased from Sigma-Aldrich. Anti-eIF4γ, anti-BiP/Grp78, anti-PKCδ and anti-G3BP1 monoclonal antibodies were from BD Bioscience; anti-Atg5, anti-Atg16L1, anti-eIF2α, anti-phospho-eIF2α, anti-cleaved caspase7 (cCas7), anti-PERK, anti-cleaved poly(ADP-ribose) polymerase (cPARP), anti-PKD1, anti-phospho-(Ser/Thr) PKD substrates and anti-TIAR antibodies were from Cell Signaling Technology; anti-DAP1, phospho-PKD1 (S738/742) and phospho-PKD1 (S916) antibodies were from Abcam; anti-ATF4 antibody was from Santa Cruz Biotechnology; and anti-GAPDH, anti-PKD2 and anti-PKD3 antibodies were from GeneTex. Anti-DnaK antibody was obtained from ENZO. Anti-SubAB antibody was prepared as previously described (Yahiro et al., 2006). PKC inhibitor Gö6976 was obtained from LC Laboratories; PKC activator PMA, PKC inhibitor Gö6983, PKD/PKCμ inhibitor CID755673, PKA activator 8Br-cAMP, Thapsigargin (TG), CaM kinase II inhibitor KN-93 were from Sigma Aldrich; and PKA inhibitor 14–22 Amide was from Calbiochem; PKC inhibitor Bisindolylmaleimide II was from ALEXIS Biochemicals; and ROCK II inhibitor Y-27632 was from Cayman Chemical.
Preparation of subtilase cytotoxin
Recombinant His-tagged SubAB and catalytically inactive mutant SubAS272AB were purified as reported previously (Morinaga et al., 2007).
Cell culture and gene silencing
HeLa and Caco2 cells were cultured at 37°C in a humidified 5% CO2 atmosphere in Eagle’s minimum essential medium (EMEM) (Sigma) containing 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin and 0.1 mg ml−1 streptomycin. RAW264.7 cells were cultured in RPMI-1640 medium (Sigma) containing 10% heat-inactivated FBS, 100 U/ml of penicillin and 0.1 mg ml−1 of streptomycin. All cells were incubated at 37°C in a humidified 5% CO2 atmosphere. RNA interference-mediated gene knockdown was performed using validated Qiagen HP small-interfering RNAs (siRNAs) for PERK as described previously (Yahiro et al., 2012). To suppress PKD1 expression, we transfected PKD1 siRNA mixture as follows: PKD1-a, 5′-GUCGAGAGAAGAGGUCAAATT-3′(Fuchs et al., 2009), PKD1-b, 5′-CAGGAAGAGAUGUAGCUAU-3′(Yin et al., 2008) and PKD1 siRNA pool (Santa Cruz Biotechnology, Inc.). As reported previously, we used specific siRNAs for PKD2 and PKD3 (Zou et al., 2012), which were synthesized by Sigma-Aldrich Japan. G3BP1 and DAP1 specific siRNAs were purchased from Dharmacon. Negative-control siRNAs were purchased from Sigma Aldrich or Dharmacon. HeLa cells were transfected with 100 nM of the indicated siRNAs for 48–72 h using Lipofectamine™ RNAiMax transfection reagent (Life Technologies) according to the manufacturer’s protocol. Transfection efficiency and effect were evaluated by Western blotting using the indicated antibodies.
Quantification of stress granule formation
To quantify SG formation in cells, 60× images of at least three to five random fields of view on the coverslip were used for analysis from at least three independent experiments. Colocalization of both TIAR-positive and G3BP1-positive or eIF4γ-positive puncta in cytoplasmic fractions was counted as SG-positive cells by two observers blinded to conditions using Image J software and then averaged.
Immunostaining
Cells were seeded in 12 well plates containing coverslips and incubated at 37°C overnight. After treatment with toxins for the indicated times at 37°C, the cells were fixed with 4% of formaldehyde in PBS at room temperature for 30 min and then washed three times with PBS. Cells were treated with PBS containing 5% of goat serum (Immuno BioScience) and 0.05% of Triton X-100 for 1 h. Cells were incubated with the indicated antibodies overnight at 4°C and washed three times with PBS, followed by incubation at room temperature for 1 h with Cy3-conjugated anti-rabbit IgG (Sigma Aldrich), Alexa 488-conjugated anti-rabbit IgG (Invitrogen) or Alexa 488-conjugated anti-mouse IgG (Invitrogen). Cells on the coverslips were then washed three times with PBS, once with water and then mounted on glass slides using ProLong Gold antifade reagent with DAPI (Invitrogen). The stained cells were visualized by FV10i-LIV confocal microscopy (Olympus). The images were arranged with Adobe Photoshop CS4.
Immunoblotting analysis
Cells lysed in SDS sample buffer were heated at 100°C for 10 min before proteins were analysed by SDS-PAGE. Separated proteins were transferred to polyvinylidene difluoride membranes (Millipore) at 100 V for 1 h, blocked with 5% of non-fat milk in TBS-T (20 mM Tris pH 7.6, 137 mM NaCl and 0.1% Tween 20) for 30 min and then incubated with the primary antibodies for 1 h at room temperature or overnight at 4°C. After washing with TBS-T, membranes were incubated with horseradish peroxidase-labelled secondary antibodies. Bands were detected using Las 1000 (Fuji film).
Knockout and complement of subAB gene in O113:H21 strain
The strain is an LEE-negative but stx2-positive, saa-positive and subAB-positive E. coli O113:H21. It was isolated from a patient with thrombotic thrombocytopenic purpura in Japan. To establish an O113:H21ΔsubAB strain, subAB gene in E. coli O113:H21 was disrupted by the insertion of a kanamycin-resistance gene (kan), as described previously (Datsenko and Wanner, 2000). The PCR primer sets for inserting Δ(subAB)::kan into the strain were subAB_F1 (5′-AGTCAATACGGCGCTCTGTTGACGCTTACATT TGTAACTAACTGGAGGAGCTTGTGTAGGCTGGAGCTGCTT-C-3′) and subAB_R1 (5′-GATCGGGACAGATCAGCGAGTCAGCGCCAGTGATATAAGACGATTATCACCATATGAATATCCTCCTTAG-3′). To complement wild-type subAB gene, SubAB expressing plasmid (pET-23b) was transferred into O113:H21 (ΔsubAB) strain by electroporation, and then we selected an ampicillin-resistant and kanamycin-resistant O113:H21(ΔsubAB/subAB) strain. These three strains (wild-type, ΔsubAB and ΔsubAB/subAB) were cultured in Brain Heart Infusion broth (BHI, Gibco) medium for 12 h at 37°C with shaking at 150 r.p.m. After centrifugation at 17 400 × g for 10 min, the culture supernatant was collected.
Statistical analysis
Student’s t-test was used to determine significant difference when only two treatment groups were being compared.
Supplementary Material
Fig. S1. G3BP1 knockdown suppresses SubAB-induced SG formation. (a) Control (NC) and ATF4 siRNA-transfected cells were incubated with of SubAS272AB (mt) or SubAB (wt) for 3 h at 37 °C. Cell lysates were subjected to immunoblotting with anti-ATF4 antibodies (arrow). GAPDH served as a loading control. Cells were fixed, immunostained with the indicated antibodies and observed by confocal microscopy as described in Fig. 1. The rate of SG formation is presented as mean ± SD from 4 different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated three times with similar results and significance is *P < 0.05. The arrows indicate SG. (b) Control (NC) and G3BP1 siRNA-transfected cells were incubated with SubAS272AB (mt) or SubAB (wt) for 3 h at 37 °C, and cell lysates were subjected to immunoblotting with the indicated antibodies. GAPDH served as a loading control. (c) The indicated siRNA-transfected cells were incubated with SubAS272AB or SubAB for 3 h at 37 °C. The fixed cells were reacted with the anti-TIAR antibody (red) and anti-eIF4γ antibody (green) and observed by confocal microscopy. Cell nuclei were stained by DAPI (cyan). The rate of SG formation is presented as mean ± SD from 5 different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated three times with similar results and significance is *P < 0.05. (d) The indicated siRNA-transfected cells were incubated with SubAS272AB (mt) or SubAB (wt) for 3 h and cell lysates were subjected to immunoblotting with the indicated antibodies. GAPDH served as a loading control. Experiments were repeated three times with similar results.
Fig. S2. PKC inhibitors enhance SubAB-induced SG formation. HeLa cells were incubated with SubAB or SubAS272AB in the presence or absence of Bisindolylmaleimide II (Bis II, 5 μM) or Gö6983 (4 μM) for 3 h. The fixed cells were reacted with the anti-TIAR antibody (red) and anti-G3BP1 antibody (green) and observed by confocal microscopy. The rate of SG formation is presented as mean ± SD from 5 different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated two times with similar results and significance is *P < 0.05.
Fig. S3. Effects of PKD isoforms depletion on SubAB-induced SG formation and apoptosis. (a) The indicated siRNA-transfected cell lysates were subjected to immunoblotting for PKD2 and PKD3. GAPDH served as a loading control. PKD2- and PKD3-knockdown cells were incubated with SubAB or SubAS272AB for 3 h. The fixed cells were reacted with the anti-TIAR antibody (red) and anti-G3BP1 antibody (green) and observed by confocal microscopy. The rate of SG formation is presented as mean ± SD from 5 different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated three times with similar results. (b) Control (NC)- and PKD1-transfcted cell lysates were subjected to immunoblotting for PKD2 and PKD3. GAPDH served as a loading control. Experiments were repeated three times with similar results. (c) Control (NC) and PKD1 siRNA-transfected HeLa cells were incubated with 400 ng ml-1 of SubAS272AB (mt) or SubAB (wt) for 3 h at 37 °C, and cell lysates were subjected to immunoblotting with the indicated antibodies. GAPDH served as a loading control. Experiments were repeated three times with similar results.
Fig. S4. Effect of Tapsigargin on SG formation in PMA-treated cells and DAP1-knockdown cells. (a) Control or PMA-treated HeLa cells were incubated with control or 20 μM Tapsigargin (TG) for 2 h. The fixed cells were reacted with anti-TIAR antibodies (red) and anti-G3BP1 antibodies (green) and observed by confocal microscopy. The rate of SG formation is presented as mean ± SD from 5 different fields, which included at least 20 cells/field. Bars represent 20 μm. Experiments were repeated two times with similar results. N.S.: not significant. (b) Control (NC) or DAP1 siRNA-transfected HeLa cells were incubated with control or 20 μM Tapsigargin (TG) for 2 h. The fixed cells were reacted with anti-TIAR antibodies (red) and anti-G3BP1 antibodies (green) and observed by confocal microscopy. The rate of SG formation is presented as mean ± SD from 5 different fields, which included at least 20 cells/field. Bars represent 20 μm. Experiments were repeated two times with similar results, significance is *P < 0.05.
Fig. S5. Effects of Atg5 and Atg16L1 depletion by siRNA on SubAB-induced SG formation. (a) Control (NC), Atg5 or tg16L1 knockdown cells were incubated with SubAS272AB (mt) or SubAB (wt) for 3 h at 37 °C, and cell lysates were subjected to immunoblotting for Atg5 and Atg16L1. GAPDH served as a loading control. Experiments were repeated three times with similar results. (b) The indicated siRNA-transfected cells were incubated with SubAB or SubAS272AB for 3 h. The fixed cells were reacted with anti-TIAR antibody (red) and anti-G3BP1 antibody (green) and observed by confocal microscopy. The rate of SG formation is presented as mean ± SD from 5 different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated tow times with similar results. N.S.: not significant.
Acknowledgements
This work was supported by grants-in-aid for Scientific Research from the Ministry of Education, Science and Culture of Japan, Improvement of Research Environment for Young Researchers from Japan Science and Technology Agency, Takeda Science Foundation, and Research Program on Emerging and Reemerging Infectious Diseases from Japan Agency for Medical Research and Development, AMED. Joel Moss was supported by the Intramural Research Program, National Institutes of Health, National Heart, Lung, and Blood Institute. We acknowledge the expert technical assistance of K. Hirano.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this articleat the publisher’s web-site:
References
- Amaral MM, Sacerdoti F, Jancic C, Repetto HA, Paton AW, Paton JC, and Ibarra C. (2013) Action of Shiga toxin type-2 and subtilase cytotoxin on human microvascular endothelial cells. PLoS One 8: e70431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, and Kedersha N. (2006) RNA granules. J Cell Biol 172: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, and Kedersha N. (2008) Stress granules: the Tao of RNA triage. Trends Biochem Sci 33: 141–150. [DOI] [PubMed] [Google Scholar]
- Anderson P, and Kedersha N. (2009) Stress granules. Curr Biol: CB 19: R397–398. [DOI] [PubMed] [Google Scholar]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, and Takekawa M. (2008) Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol 10: 1324–1332. [DOI] [PubMed] [Google Scholar]
- Asaithambi A, Kanthasamy A, Saminathan H, Anantharam V, and Kanthasamy AG (2011) Protein kinase D1 (PKD1) activation mediates a compensatory protective response during early stages of oxidative stress-induced neuronal degeneration. Mol Neurodegener 6: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasopoulos V, Barker A, Yu D, Tan AH, Srivastava M, Contreras N, et al. (2010) The ROQUIN family of proteins localizes to stress granules via the ROQ domain and binds target mRNAs. FEBS J 277: 2109–2127. [DOI] [PubMed] [Google Scholar]
- B’Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, et al. (2013) The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res 41: 7683–7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentancor A, Rumi MV, Carbonari C, Gerhardt E, Larzabal M, Vilte DA, et al. (2012) Profile of Shiga toxin-producing Escherichia coli strains isolated from dogs and cats and genetic relationships with isolates from cattle, meat and humans. Vet Microbiol 156: 336–342. [DOI] [PubMed] [Google Scholar]
- Brandlin I, Eiseler T, Salowsky R, and Johannes FJ (2002a) Protein kinase C(mu) regulation of the JNK pathway is triggered via phosphoinositide-dependent kinase 1 and protein kinase C(epsilon). J Biol Chem 277: 45451–45457. [DOI] [PubMed] [Google Scholar]
- Brandlin I, Hubner S, Eiseler T, Martinez-Moya M, Horschinek A, Hausser A, et al. (2002b) Protein kinase C (PKC)eta-mediated PKC mu activation modulates ERK and JNK signal pathways. J Biol Chem 277: 6490–6496. [DOI] [PubMed] [Google Scholar]
- Buchan JR, and Parker R. (2009) Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36: 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byres E, Paton AW, Paton JC, Lofling JC, Smith DF, Wilce MC, et al. (2008) Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature 456: 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Giridhar KV, Zhang L, Xu S, and Wang QJ (2011) A protein kinase C/protein kinase D pathway protects LNCaP prostate cancer cells from phorbol ester-induced apoptosis by promoting ERK1/2 and NF-{kappa}B activities. Carcinogenesis 32: 1198–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong DC, Paton JC, Thorpe CM, and Paton AW (2008) Clathrin-dependent trafficking of subtilase cytotoxin, a novel AB5 toxin that targets the endoplasmic reticulum chaperone BiP. Cell Microbiol 10: 795–806. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, and Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doppler H, and Storz P. (2007) A novel tyrosine phosphorylation site in protein kinase D contributes to oxidative stress-mediated activation. J Biol Chem 282: 31873–31881. [DOI] [PubMed] [Google Scholar]
- Eiseler T, Kohler C, Nimmagadda SC, Jamali A, Funk N, Joodi G, et al. (2012) Protein kinase D1 mediates anchorage-dependent and -independent growth of tumor cells via the zinc finger transcription factor Snail1. J Biol Chem 287: 32367–32380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng PC, and Reddy S. (2013) Prevalences of Shiga toxin subtypes and selected other virulence factors among Shiga-toxigenic Escherichia coli strains isolated from fresh produce. Appl Environ Microbiol 79: 6917–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs YF, Eisler SA, Link G, Schlicker O, Bunt G, Pfizenmaier K, and Hausser A. (2009) A Golgi PKD activity reporter reveals a crucial role of PKD in nocodazole-induced Golgi dispersal. Traffic 10: 858–867. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Yahiro K, Tsuji AB, Terasaki Y, Morinaga N, Miyazaki M, et al. (2011) Fatal hemorrhage induced by subtilase cytotoxin from Shiga-toxigenic Escherichia coli. Microb Pathog 50: 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, and Steitz JA (2000) HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci USA 97: 3073–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison BC, Kim MS, van Rooij E, Plato CF, Papst PJ, Vega RB, et al. (2006) Regulation of cardiac stress signaling by protein kinase D1. Mol Cell Biol 26: 3875–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Cherkasova V, Bankhead P, Bukau B, and Stoecklin G. (2012) Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol Biol Cell 23: 3786–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CC, Dougan SK, Winter SV, Paton AW, Paton JC, and Ploegh HL (2009) Subtilase cytotoxin cleaves newly synthesized BiP and blocks antibody secretion in B lymphocytes. J Exp Med 206: 2429–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Wan Y, Zhu Y, Fang X, Hiramatsu N, Hayakawa K, et al. (2009) Downregulation of gap junction expression and function by endoplasmic reticulum stress. J Cell Biochem 107: 973–983. [DOI] [PubMed] [Google Scholar]
- Hui K, Yang Y, Shi K, Luo H, Duan J, An J, et al. (2014) The p38 MAPK-regulated PKD1/CREB/Bcl-2 pathway contributes to selenite-induced colorectal cancer cell apoptosis in vitro and in vivo. Cancer Lett 354: 189–199. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Li F, Shi K, Wu P, An J, Yang Y, and Xu C. (2014) Involvement of p38 in signal switching from autophagy to apoptosis via the PERK/eIF2alpha/ATF4 axis in selenite-treated NB4 cells. Cell Death Dis 5: e1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes FJ, Prestle J, Eis S, Oberhagemann P, and Pfizenmaier K. (1994) PKCu is a novel, atypical member of the protein kinase C family. J Biol Chem 269: 6140–6148. [PubMed] [Google Scholar]
- Karmali MA (2004) Prospects for preventing serious systemic toxemic complications of Shiga toxin-producing Escherichia coli infections using Shiga toxin receptor analogues. J Infect Dis 189: 355–359. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, and Yao TP (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115: 727–738. [DOI] [PubMed] [Google Scholar]
- Kedersha N, and Anderson P. (2002) Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans 30: 963–969. [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, and Anderson P. (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Horetsky RL, Ron D, Jefferson LS, and Harding HP (2003) Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol 284: C273–284. [DOI] [PubMed] [Google Scholar]
- Koren I, Reem E, and Kimchi A. (2010) DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr Biol: CB 20: 1093–1098. [DOI] [PubMed] [Google Scholar]
- Latorre-Martinez JC, Garcia-Lozano T, Blanco J, and Buesa J. (2007) Characterization of Escherichia coli O157:H7 strains isolated from sporadic cases of hemolytic-uremic syndrome in children. Enferm Infecc Microbiol Clin 25: 603–604. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Beznoussenko GV, Van Lint J, Mironov AA, and Malhotra V. (2001) Recruitment of protein kinase D to the trans-Golgi network via the first cysteine-rich domain. EMBO J 20: 5982–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H, Miyazaki S, Matsuyama S, Takeda M, Kawano M, Nakagawa H, et al. (2013) Selection of autophagy or apoptosis in cells exposed to ER-stress depends on ATF4 expression pattern with or without CHOP expression. Biol Open 2: 1084–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura G, Morinaga N, Yahiro K, Komine R, Moss J, Yoshida H, and Noda M. (2009) Novel subtilase cytotoxin produced by Shiga-toxigenic Escherichia coli induces apoptosis in Vero cells via mitochondrial membrane damage. Infect Immun 77: 2919–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews SA, Rozengurt E, and Cantrell D. (1999) Characterization of serine 916 as an in vivo autophosphorylation site for protein kinase D/protein kinase Cmu. J Biol Chem 274: 26543–26549. [DOI] [PubMed] [Google Scholar]
- May KL, Paton JC, and Paton AW (2010) Escherichia coli subtilase cytotoxin induces apoptosis regulated by host Bcl-2 family proteins Bax/Bak. Infect Immun 78: 4691–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Di Marco S, Kaufman RJ, and Gallouzi IE (2007) Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol Biol Cell 18: 2603–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, et al. (2006) Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol Biol Cell 17: 4212–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DL, Kirienko NV, and Ausubel FM (2012) Host translational inhibition by Pseudomonas aeruginosa exotoxin A triggers an immune response in Caenorhabditis elegans. Cell Host Microbe 11: 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor H, and Parker PJ (1998) The extended protein kinase C superfamily. Biochem J 332(Part 2): 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaga N, Yahiro K, Matsuura G, Moss J, and Noda M. (2008) Subtilase cytotoxin, produced by Shiga-toxigenic Escherichia coli, transiently inhibits protein synthesis of Vero cells via degradation of BiP and induces cell cycle arrest at G1 by downregulation of cyclin D1. Cell Microbiol 10: 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaga N, Yahiro K, Matsuura G, Watanabe M, Nomura F, Moss J, and Noda M. (2007) Two distinct cytotoxic activities of subtilase cytotoxin produced by Shiga-toxigenic Escherichia coli. Infect Immun 75: 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa S, Ogura K, Tsutsuki H, Saitoh H, Moss J, Iwase H, et al. (2014) Uptake of Shiga-toxigenic Escherichia coli SubAB by HeLa cells requires an actin- and lipid raft-dependent pathway. Cell Microbiol 16: 1582–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima S, Hiramatsu N, Hayakawa K, Saito Y, Kato H, Huang T, et al. (2011) Selective abrogation of BiP/GRP78 blunts activation of NF-{kappa}B through the ATF6 branch of the UPR: involvement of C/EBP{beta} and mTOR-dependent dephosphorylation of Akt. Mol Cell Biol 31: 1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima S, Saito Y, Takahashi S, Hiramatsu N, Kato H, Johno H, et al. (2010) Anti-inflammatory subtilase cytotoxin up-regulates A20 through the unfolded protein response. Biochem Biophys Res Commun 397: 176–180. [DOI] [PubMed] [Google Scholar]
- Onomoto K, Yoneyama M, Fung G, Kato H, and Fujita T. (2014) Antiviral innate immunity and stress granule responses. Trends Immunol 35: 420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton AW, Beddoe T, Thorpe CM, Whisstock JC, Wilce MC, Rossjohn J, et al. (2006) AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature 443: 548–552. [DOI] [PubMed] [Google Scholar]
- Paton AW, Srimanote P, Talbot UM, Wang H, and Paton JC (2004) A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli. J Exp Med 200: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestle J, Pfizenmaier K, Brenner J, and Johannes FJ (1996) Protein kinase C mu is located at the Golgi compartment. J Cell Biol 134: 1401–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaben M, Groot Koerkamp MJ, Rottier PJ, and de Haan CA (2007) Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol 9: 2218–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke LC, Kedersha N, Langereis MA, van Kuppeveld FJ, and Lloyd RE (2015) Stress granules regulate double-stranded RNA-dependent protein kinase activation through a complex containing G3BP1 and Caprin1. mBio 6: e02486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, et al. (1983) Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med 308: 681–685. [DOI] [PubMed] [Google Scholar]
- Ron D, and Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529. [DOI] [PubMed] [Google Scholar]
- Rosse C, Linch M, Kermorgant S, Cameron AJ, Boeckeler K, and Parker PJ (2010) PKC and the control of localized signal dynamics. Nat Rev Mol Cell Biol 11: 103–112. [DOI] [PubMed] [Google Scholar]
- Rozengurt E, Rey O, and Waldron RT (2005) Protein kinase D signaling. J Biol Chem 280: 13205–13208. [DOI] [PubMed] [Google Scholar]
- Rybin VO, Guo J, and Steinberg SF (2009) Protein kinase D1 autophosphorylation via distinct mechanisms at Ser744/Ser748 and Ser916. J Biol Chem 284: 2332–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S, Beristain X, Martinez R, Garcia A, Martin C, Vidal D, et al. (2012) Subtilase cytotoxin encoding genes are present in human, sheep and deer intimin-negative, Shiga toxin-producing Escherichia coli O128:H2. Vet Microbiol 159: 531–535. [DOI] [PubMed] [Google Scholar]
- Scheiter M, Bulitta B, van Ham M, Klawonn F, Konig S, and Jansch L. (2013) Protein kinase inhibitors CK59 and CID755673 alter primary human NK cell effector functions. Front Immunol 4: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin SJ, Morelli FF, Vinet J, Amore D, De Biasi S, Poletti A, et al. (2014) Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ 21: 1838–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewald X, Jimenez-Soto L, and Haas R. (2011) PKC-dependent endocytosis of the Helicobacter pylori vacuolating cytotoxin in primary T lymphocytes. Cell Microbiol 13: 482–496. [DOI] [PubMed] [Google Scholar]
- Shah KH, Nostramo R, Zhang B, Varia SN, Klett BM, and Herman PK (2014) Protein kinases are associated with multiple, distinct cytoplasmic granules in quiescent yeast cells. Genetics 198: 1495–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharlow ER, Giridhar KV, LaValle CR, Chen J, Leimgruber S, Barrett R, et al. (2008) Potent and selective disruption of protein kinase D functionality by a benzoxoloazepinolone. J Biol Chem 283: 33516–33526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi M, and Togawa M. (1997) [Sporadic cases of hemolytic uremic syndrome and hemorrhagic colitis with serum IgM antibodies to lipopolysaccharides of enterohemorrhagic Escherichia coli O157]. Nihon rinsho. Jpn J Clin Med 55: 686–692. [PubMed] [Google Scholar]
- Smith RD, Willett R, Kudlyk T, Pokrovskaya I, Paton AW, Paton JC, and Lupashin VV (2009) The COG complex, Rab6 and COPI define a novel Golgi retrograde trafficking pathway that is exploited by SubAB toxin. Traffic 10: 1502–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Li J, Qiao J, Jain S, Mark Evers B, and Chung DH (2009) PKD prevents H2O2-induced apoptosis via NF-kappaB and p38 MAPK in RIE-1 cells. Biochem Biophys Res Commun 378: 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg SF (2012) Regulation of protein kinase D1 activity. Mol Pharmacol 81: 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart LM, Paquette N, and Boyer L. (2013) Effector-triggered versus pattern-triggered immunity: how animals sense pathogens. Nat Rev Immunol 13: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundram V, Chauhan SC, and Jaggi M. (2011) Emerging roles of protein kinase D1 in cancer. Mol Cancer Res: MCR 9: 985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A. (1998) Signaling through protein kinase C. Front Biosci: J Virtual Lib 3: D1134–1147. [DOI] [PubMed] [Google Scholar]
- Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, and Tazi J. (2003) The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160: 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tsai NP, and Wei LN (2010) RhoA/ROCK1 signaling regulates stress granule formation and apoptosis. Cell Signal 22: 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsuki H, Yahiro K, Suzuki K, Suto A, Ogura K, Nagasawa S, et al. (2012) Subtilase cytotoxin enhances Escherichia coli survival in macrophages by suppression of nitric oxide production through the inhibition of NF-kappaB activation. Infect Immun 80: 3939–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde AM, Sinnett-Smith J, Van Lint J, and Rozengurt E. (1994) Molecular cloning and characterization of protein kinase D: a target for diacylglycerol and phorbol esters with a distinctive catalytic domain. Proc Natl Acad Sci USA 91: 8572–8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint J, Rykx A, Maeda Y, Vantus T, Sturany S, Malhotra V, et al. (2002) Protein kinase D: an intracellular traffic regulator on the move. Trends Cell Biol 12: 193–200. [DOI] [PubMed] [Google Scholar]
- Vanderweyde T, Youmans K, Liu-Yesucevitz L, and Wolozin B. (2013) Role of stress granules and RNA-binding proteins in neurodegeneration: a mini-review. Gerontology 59: 524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velandia CV, Mariel Sanso A, Kruger A, Suarez LV, Lucchesi PM, and Parma AE (2011) Occurrence of subtilase cytotoxin and relation with other virulence factors in verocytotoxigenic Escherichia coli isolated from food and cattle in Argentina. Braz J Microbiol: Publ Braz Soc Microbiol 42: 711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron RT, Iglesias T, and Rozengurt E. (1999a) Phosphorylation-dependent protein kinase D activation. Electrophoresis 20: 382–390. [DOI] [PubMed] [Google Scholar]
- Waldron RT, Iglesias T, and Rozengurt E. (1999b) The pleckstrin homology domain of protein kinase D interacts preferentially with the eta isoform of protein kinase C. J Biol Chem 274: 9224–9230. [DOI] [PubMed] [Google Scholar]
- Walker AK, Soo KY, Sundaramoorthy V, Parakh S, Ma Y, Farg MA, et al. (2013) ALS-associated TDP-43 induces endoplasmic reticulum stress, which drives cytoplasmic TDP-43 accumulation and stress granule formation. PLoS One 8: e81170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Paton AW, McColl SR, and Paton JC (2011) In vivo leukocyte changes induced by Escherichia coli subtilase cytotoxin. Infect Immun 79: 1671–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Paton JC, and Paton AW (2007) Pathologic changes in mice induced by subtilase cytotoxin, a potent new Escherichia coli AB5 toxin that targets the endoplasmic reticulum. J Infect Dis 196: 1093–1101. [DOI] [PubMed] [Google Scholar]
- Wang H, Paton JC, Thorpe CM, Bonder CS, Sun WY, and Paton AW (2010) Tissue factor-dependent procoagulant activity of subtilase cytotoxin, a potent AB5 toxin produced by Shiga toxigenic Escherichia coli. J Infect Dis 202: 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Cardenas AM, Marissen WE, and Lloyd RE (2007) Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2: 295–305. [DOI] [PubMed] [Google Scholar]
- Wolfson JJ, May KL, Thorpe CM, Jandhyala DM, Paton JC, and Paton AW (2008) Subtilase cytotoxin activates PERK, IRE1 and ATF6 endoplasmic reticulum stress-signalling pathways. Cell Microbiol 10: 1775–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B. (2012) Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener 7: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B. (2014) Physiological protein aggregation run amuck: stress granules and the genesis of neurodegenerative disease. Discov Med 17: 47–52. [PMC free article] [PubMed] [Google Scholar]
- Yahiro K, Morinaga N, Moss J, and Noda M. (2010) Subtilase cytotoxin induces apoptosis in HeLa cells by mitochondrial permeabilization via activation of Bax/Bak, independent of C/EBF-homologue protein (CHOP), Ire1alpha or JNK signaling. Microb Pathog 49: 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahiro K, Morinaga N, Satoh M, Matsuura G, Tomonaga T, Nomura F, et al. (2006) Identification and characterization of receptors for vacuolating activity of subtilase cytotoxin. Mol Microbiol 62: 480–490. [DOI] [PubMed] [Google Scholar]
- Yahiro K, Satoh M, Morinaga N, Tsutsuki H, Ogura K, Nagasawa S, et al. (2011) Identification of subtilase cytotoxin (SubAB) receptors whose signaling, in association with SubAB-induced BiP cleavage, is responsible for apoptosis in HeLa cells. Infect Immun 79: 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahiro K, Tsutsuki H, Ogura K, Nagasawa S, Moss J, and Noda M. (2012) Regulation of subtilase cytotoxin-induced cell death by an RNA-dependent protein kinase-like endoplasmic reticulum kinase-dependent proteasome pathway in HeLa cells. Infect Immun 80: 1803–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahiro K., Tsutsuki H., Ogura K., Nagasawa S., Moss J., and Noda M. (2014) DAP1, a negative regulator of autophagy, controls SubAB-mediated apoptosis and autophagy. Infect Immun 82: 4899–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin DM, Huang YH, Zhu YB, and Wang Y. (2008) Both the establishment and maintenance of neuronal polarity require the activity of protein kinase D in the Golgi apparatus. J Neurosci: Off J Soc Neurosci 28: 8832–8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Liu Y, Tan T, Guha S, Gukovsky I, Gukovskaya A, and Pandol SJ (2012) Protein kinase D regulates cell death pathways in experimental pancreatitis. Front Physiol 3: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Sell P, Braun U, and Leitges M. (2015) PKD1 protein is involved in reactive oxygen species-mediated mitochondrial depolarization in cooperation with protein kinase Cdelta (PKCdelta). J Biol Chem 290: 10472–10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Zeng F, Xu W, Wang C, Ke Z, Wang QJ, and Deng F. (2012) PKD2 and PKD3 promote prostate cancer cell invasion by modulating NF-kappaB- and HDAC1-mediated expression and activation of uPA. J Cell Sci 125: 4800–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. G3BP1 knockdown suppresses SubAB-induced SG formation. (a) Control (NC) and ATF4 siRNA-transfected cells were incubated with of SubAS272AB (mt) or SubAB (wt) for 3 h at 37 °C. Cell lysates were subjected to immunoblotting with anti-ATF4 antibodies (arrow). GAPDH served as a loading control. Cells were fixed, immunostained with the indicated antibodies and observed by confocal microscopy as described in Fig. 1. The rate of SG formation is presented as mean ± SD from 4 different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated three times with similar results and significance is *P < 0.05. The arrows indicate SG. (b) Control (NC) and G3BP1 siRNA-transfected cells were incubated with SubAS272AB (mt) or SubAB (wt) for 3 h at 37 °C, and cell lysates were subjected to immunoblotting with the indicated antibodies. GAPDH served as a loading control. (c) The indicated siRNA-transfected cells were incubated with SubAS272AB or SubAB for 3 h at 37 °C. The fixed cells were reacted with the anti-TIAR antibody (red) and anti-eIF4γ antibody (green) and observed by confocal microscopy. Cell nuclei were stained by DAPI (cyan). The rate of SG formation is presented as mean ± SD from 5 different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated three times with similar results and significance is *P < 0.05. (d) The indicated siRNA-transfected cells were incubated with SubAS272AB (mt) or SubAB (wt) for 3 h and cell lysates were subjected to immunoblotting with the indicated antibodies. GAPDH served as a loading control. Experiments were repeated three times with similar results.
Fig. S2. PKC inhibitors enhance SubAB-induced SG formation. HeLa cells were incubated with SubAB or SubAS272AB in the presence or absence of Bisindolylmaleimide II (Bis II, 5 μM) or Gö6983 (4 μM) for 3 h. The fixed cells were reacted with the anti-TIAR antibody (red) and anti-G3BP1 antibody (green) and observed by confocal microscopy. The rate of SG formation is presented as mean ± SD from 5 different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated two times with similar results and significance is *P < 0.05.
Fig. S3. Effects of PKD isoforms depletion on SubAB-induced SG formation and apoptosis. (a) The indicated siRNA-transfected cell lysates were subjected to immunoblotting for PKD2 and PKD3. GAPDH served as a loading control. PKD2- and PKD3-knockdown cells were incubated with SubAB or SubAS272AB for 3 h. The fixed cells were reacted with the anti-TIAR antibody (red) and anti-G3BP1 antibody (green) and observed by confocal microscopy. The rate of SG formation is presented as mean ± SD from 5 different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated three times with similar results. (b) Control (NC)- and PKD1-transfcted cell lysates were subjected to immunoblotting for PKD2 and PKD3. GAPDH served as a loading control. Experiments were repeated three times with similar results. (c) Control (NC) and PKD1 siRNA-transfected HeLa cells were incubated with 400 ng ml-1 of SubAS272AB (mt) or SubAB (wt) for 3 h at 37 °C, and cell lysates were subjected to immunoblotting with the indicated antibodies. GAPDH served as a loading control. Experiments were repeated three times with similar results.
Fig. S4. Effect of Tapsigargin on SG formation in PMA-treated cells and DAP1-knockdown cells. (a) Control or PMA-treated HeLa cells were incubated with control or 20 μM Tapsigargin (TG) for 2 h. The fixed cells were reacted with anti-TIAR antibodies (red) and anti-G3BP1 antibodies (green) and observed by confocal microscopy. The rate of SG formation is presented as mean ± SD from 5 different fields, which included at least 20 cells/field. Bars represent 20 μm. Experiments were repeated two times with similar results. N.S.: not significant. (b) Control (NC) or DAP1 siRNA-transfected HeLa cells were incubated with control or 20 μM Tapsigargin (TG) for 2 h. The fixed cells were reacted with anti-TIAR antibodies (red) and anti-G3BP1 antibodies (green) and observed by confocal microscopy. The rate of SG formation is presented as mean ± SD from 5 different fields, which included at least 20 cells/field. Bars represent 20 μm. Experiments were repeated two times with similar results, significance is *P < 0.05.
Fig. S5. Effects of Atg5 and Atg16L1 depletion by siRNA on SubAB-induced SG formation. (a) Control (NC), Atg5 or tg16L1 knockdown cells were incubated with SubAS272AB (mt) or SubAB (wt) for 3 h at 37 °C, and cell lysates were subjected to immunoblotting for Atg5 and Atg16L1. GAPDH served as a loading control. Experiments were repeated three times with similar results. (b) The indicated siRNA-transfected cells were incubated with SubAB or SubAS272AB for 3 h. The fixed cells were reacted with anti-TIAR antibody (red) and anti-G3BP1 antibody (green) and observed by confocal microscopy. The rate of SG formation is presented as mean ± SD from 5 different fields, which included at least 20 cells/field (right panel). Bars represent 20 μm. Experiments were repeated tow times with similar results. N.S.: not significant.