Abstract
Background
Radiomics is the extraction of predefined mathematic features from medical images for the prediction of variables of clinical interest. While some studies report superlative accuracy of radiomic machine learning (ML) models, the published methodology is often incomplete, and the results are rarely validated in external testing data sets.
Purpose
To characterize the type, prevalence, and statistical impact of methodologic errors present in radiomic ML studies.
Materials and Methods
Radiomic ML publications were reviewed for the presence of performance-inflating methodologic flaws. Common flaws were subsequently reproduced with randomly generated features interpolated from publicly available radiomic data sets to demonstrate the precarious nature of reported findings.
Results
In an assessment of radiomic ML publications, the authors uncovered two general categories of data analysis errors: inconsistent partitioning and unproductive feature associations. In simulations, the authors demonstrated that inconsistent partitioning augments radiomic ML accuracy by 1.4 times from unbiased performance and that correcting for flawed methodologic results in areas under the receiver operating characteristic curve approaching a value of 0.5 (random chance). With use of randomly generated features, the authors illustrated that unproductive associations between radiomic features and gene sets can imply false causality for biologic phenomenon.
Conclusion
Radiomic machine learning studies may contain methodologic flaws that undermine their validity. This study provides a review template to avoid such flaws.
© RSNA, 2022
Supplemental material is available for this article.
See also the editorial by Jacobs in this issue.
Summary
By reproducing common methodologic flaws in radiomic machine learning publications, randomly generated radiomic features falsely inflated model performance by a factor of 1.4.
Key Results
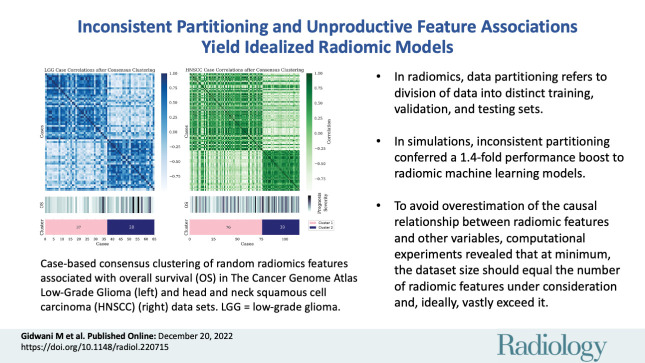
■ In radiomics research, data partitioning refers to the division of available data into distinct training, validation, and testing sets; in simulations, inconsistent partitioning conferred a 1.4-fold performance boost to radiomic machine learning models.
■ To avoid overestimation of the causal relationship between radiomic features and other variables, computational experiments revealed that at minimum, the data set size should equal the number of radiomic features under consideration and, ideally, vastly exceed it.
Introduction
Artificial intelligence and machine learning (ML) have received an outpouring of interest in the medical domain in recent years. In the area of image analysis, radiomic features, or hand-crafted numeric features calculated by applying mathematic operations to the intensity values within an image (1), have been used to characterize a region of interest in terms of intensity, size, shape, and texture. In the research setting, ML and handcrafted radiomic features have been combined for tasks including pathologic classification, survival prediction, and characterization of underlying biology (2–4) by taking advantage of the ability of ML models to model data sets with numerous covariates, such as radiomic features. Artificial intelligence models may also be used to fit radiomic features or may be trained directly with the source images (convolutional neural networks). Yet despite their widespread popularity and reported utility, published results of radiomic ML analysis are often overly optimistic due to methodologic errors.
This study evaluates the effect of two methodologic problem areas that may inflate the reported performance of radiomics models: inconsistent data partitioning and unproductive feature associations.
Data partitioning refers to the division of available data into distinct training, validation, and testing sets. Fundamentally, each partition is necessary for ML model development because it is used for learning parameters (training), assessing the impact of those parameters (validation), and evaluating the final model (testing) both internally and externally, respectively. Nevertheless, many radiomics studies eschew the split of primary data, partition inappropriately, or forego an external testing set, limiting the quality and scientific impact of the results. However, inconsistent partitioning allows inadvertent data leakage from the test set. “Information leak” is the unintentional incorporation of test set characteristics when training a prediction model, leading to artificial inflation of reported performance (5).
The second area of performance inflation is unproductive feature associations. This refers to the overestimation of the causal relationship between radiomics features and other variables. Radiomics features are high-dimensional data—some feature extraction packages can extract more than 1800 features by applying filters (eg, exponential, logarithm, wavelet) to image intensities before calculating feature values. Their large size makes radiomics data sets incredibly prone to overfitting by ML models, especially because most medical data sets contain few samples.
Compounding this problem, studies occasionally associate radiomics features with other high-dimensional data, such as gene ontology pathways, panels of genetic mutations, and blood metabolites, predisposing them to the discovery of spurious correlations (6,7). We hypothesize that these mistakes can artificially inflate performance accuracy. In this study, simulated features were randomly sampled by interpolation from publicly available radiomics data sets extracted from MRI scans of low-grade gliomas and from CT images of head and neck squamous cell carcinomas (HNSCCs) (8–13), uniquely enabling us to evaluate the impact of commonly observed methodologic errors on performance metrics.
Materials and Methods
Literature Review
To determine methodologic flaws that occurred frequently in the radiomics literature, we conducted a limited literature search with PubMed using the term “radiomics” and restricting the search years from 2017 to 2021 (Fig S1). We arbitrarily identified 10 journals with frequent publications on the topic of radiomics, eliminated duplicates, and randomly selected 50 articles for in-depth assessment. This literature search guided the subsequent experiments by highlighting which methodologic flaws occurred most frequently. For studies with multiple target tasks, the highest performing task as measured by using the area under the receiver operating characteristic curve (AUC) was evaluated. If a given flaw was not clearly attributable to the methodology, it was recorded as absent. The list of articles analyzed is available on request from the authors. Experimental code is available at: github.com/QTIM-lab/RandomRad.
Random Feature Generation
To create a realistic simulation of the radiomics pipeline, we aimed to generate a random radiomics data set representative of those seen in the medical imaging literature. Hence, the distribution of existing radiomics features served as the basis for feature generation with use of synthetic minority over-sampling technique (14,15).
Two original data sets from The Cancer Imaging Archive formed the basis of our random feature generation exercise. The first realistic random radiomics data set was derived from The Cancer Genome Atlas Low-Grade Glioma (TCGA-LGG) data set and comprises 65 samples with 220 features each (8–10). The second realistic random radiomics data set was derived from the HNSCC data set and comprises 125 samples with 9682 features each (10–13). These random features formed the basis of our analysis.
Inconsistent Partitioning Experiments
When implemented correctly, the training, validation, test, and external test data partitions are generated before further analyses. In the following experiments, we demonstrate the impact of disregarding partitioning at each step of the radiomic ML pipeline (Fig 1).
Figure 1:
Diagrams of inconsistent partitioning. Random features (R) based on published radiomics data form the basis of our experimentation (atypical from radiomics machine learning [ML] studies). (A) The upper level (blue and yellow) illustrates consistent partitioning that prevents information leak, while the lower level (green) demonstrates how the use of the entire data set for radiomics feature normalization, feature selection, hyperparameter selection, model selection, and performance reporting will result in an unrealistically optimistic assessment of the radiomics ML model. (B) Diagrams show normalization strategies. Data set normalization (green) is an example of inconsistent partitioning, with use of a mean and SD calculated with use of all samples, both the training and test sets, to scale. Train normalization (right) and split normalization (bottom) are different approaches to consistent partitioning (more details in Appendix S1).
Feature normalization and selection with use of the entire data set.—Because radiomics features are high dimensional and prone to overfitting, feature selection is used to reduce the number of features in the final model. However, feature selection strategies usually require correlation with the outcome label of interest. Hence, using the entire data set to select features causes an information leak by guaranteeing that features that are also associated with outcome labels from the test set are selected. This experiment measured the change in accuracy when normalizing and selecting radiomic features using the entire data set.
Hyperparameter tuning with use of the entire data set.—Hyperparameters are model components programmed before fitting to the data. They determine the behavior and therefore the performance of the model. In the radiomics literature, mistakenly tuning hyperparameters on the entire data set frequently materializes (Fig S2) in the form of cross validation (CV) without a held-out test set. CV creates temporary partitions within available data, iteratively fitting and evaluating models on each subsequent partition fold. It is often used in the medical domain because of the limited data availability for model development. This experiment measured the impact of extending hyperparameters chosen through CV to all data across all folds.
Model selection with use of the test set.—Even though the test set is designed to assess model generalizability on unseen data, our review revealed that model selection in the radiomics literature continues to be based on test set performance (Fig S2). In this experiment, eight ML models were fit to the training set of the TCGA-LGG random radiomics features and evaluated on the test set.
Previewing performance on the test set can also happen by partitioning the data multiple times to optimize the samples assorted to the training and test sets for the most favorable performance. Additionally, insufficiently small data sets can cause misestimation of the population accuracy. To demonstrate this, we partitioned increasingly large data sets and measured the range of AUC values on the test set in 10 replicates.
Selective class sampling or lack of external test set.—An external test set represents the ability of an ML model to generalize to unseen data from a different institution, which may have differences in patient demographics, data acquisition, diagnostic and treatment paradigms, and disease characteristics. To model the performance inflation gained by foregoing an external test set, we fit models to varying stratifications of random radiomic features from two institutions and measured the AUC.
Unproductive Feature Associations Experiments
Association of features with themselves.—In the medical radiomics literature, it is common to see samples clustered together to identify associations with known biologic groupings (Fig S2). In this experiment, random radiomics features correlated with overall survival (16–19) in the TCGA-LGG and HNSCC cohorts were selected, and clusters were formed based on selected features. Once each sample was assorted to a cluster, the difference in survival outcomes between clusters was measured (Mann-Whitney test).
Association of features with other high-dimensional variables (overfitting).—Given the important role radiomics features play in medical image analysis, creating avenues of explainability for the features is an area of great research interest. Some approaches to explainability found in our literature review (Fig S2) are the combination of radiomics features and clinically predictive variables and the correlation of radiomics features with quantitative biologic variables or primary outcomes.
Combination of random radiomics features with common clinical predictors.—Radiomics features reportedly augment clinical variables for the task of outcome prediction (2–4). In this experiment, models were trained on tumor volume, histologic grade (16–19), or random radiomics features of samples in the training set to predict overall survival. The added benefit of each variable is measured using the AUC.
Predictive power of a radiomics nomogram in Cox proportional hazards model.—The concordance index, or C index, is the ability of a variable to correctly rank survival outcomes (20). To determine if the authentic and random radiomics features are prognostic of overall survival, we integrated them into a radiomics nomogram, or RadScore, which is a linear combination of 10 radiomics features, and calculated the C index.
Random feature association with gene ontology pathways.—Forming post hoc correlations between radiomics features and gene ontology pathways represents the desire to ground medical descriptors in biology. Yet the association of two high-dimensional matrices is primed for overfitting. To demonstrate this, we correlated the TCGA-LGG random radiomics features with the corresponding single sample gene set enrichment scores for that cohort. Finally, we determined if the most frequently associated random feature could prognosticate overall survival.
Results
Literature Review
Upon surveying 50 radiomic ML publications, we observed that mistakes in inconsistent partitioning and unproductive feature associations rarely occur in isolation (Fig S2). In fact, the median number of methodologic mistakes observed in our literature review was six per publication (Fig S3). The average AUC for the training set reported was 0.84 and for the test set was 0.80. Encouragingly, there was high concordance between the frequency of the event of interest in the training set and the test set, which has been experimentally proven to be a source of performance manipulation (Fig S4).
Inconsistent Partitioning Experiments
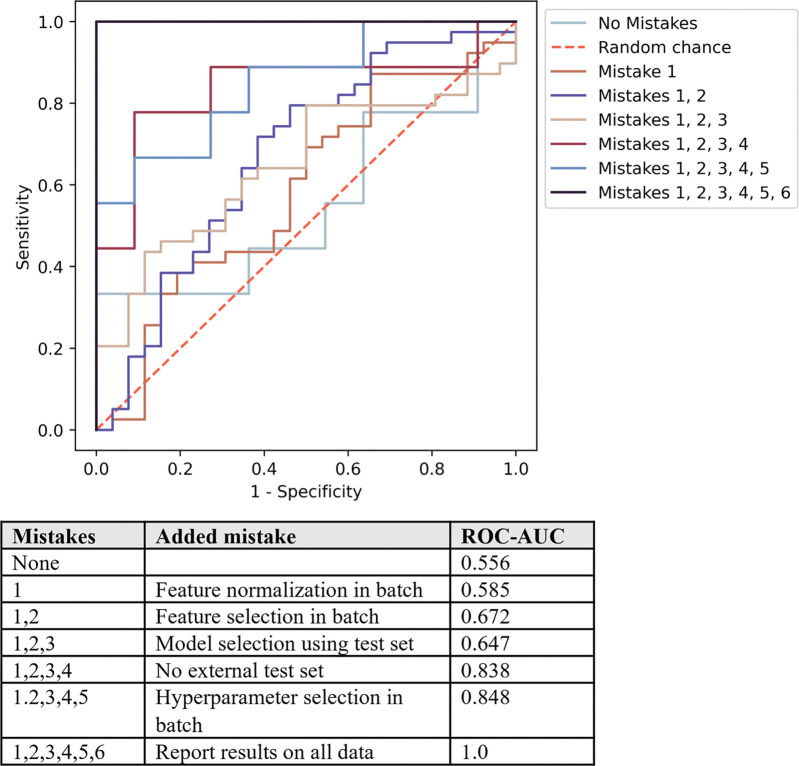
While each of the following methodologic flaws confers performance enhancement in isolation, when compounded they result in a 1.4 times magnification in reported AUC (Fig S5). Furthermore, if performance is not reported on an objective test set but rather on the data in totality, the model accuracy is wholly idealized (Fig 2).
Figure 2:
Receiver operating characteristic curves illustrate the performance inflation gained from each subsequent radiomics machine learning methodologic mistake as demonstrated on random radiomics features. Without mistakes, the area under the receiver operating characteristic curve (AUC) value (ROC-AUC) approximates 0.5 or random chance and compounding sufficient mistakes lead to idealized performance of a 1.0 AUC value.
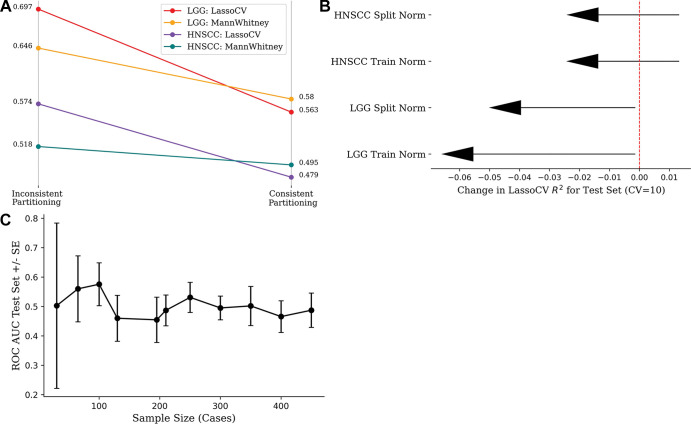
Feature selection with use of full data set.—When comparing models built with use of the entire data set (inconsistent partitioning) with those built with use of rigorous training and test set separation (consistent partitioning), a precipitous drop in accuracy was observed (Fig 3). The corrected performances after a thorough training and test split trend toward 0.5 because of the randomness of the used radiomics features and outcome labels. The performance of the HNSCC random features is consistently worse than the TCGA-LGG random features because of the greater number of samples (125 vs 65), resulting in a larger test set.
Figure 3:
(A) Strip chart shows mean accuracy loss from changing inconsistent partitioning (data set normalization and feature selection) to consistent partitioning (train normalization and feature selection) in 100 replicates. (B) Lollipop plot shows loss of mean model efficiency (LassoCV R2) over 100 iterations after changing from inconsistent to consistent partitioning. (C) Line chart shows effect of sample size on model performance, keeping number of radiomics features (10 features) and method of feature selection constant. Wide CIs are seen at low sample sizes because choice of data partition drastically alters the distribution of features in each partition. Performance plateaus at the area under the receiver operating characteristic curve (ROC AUC) value of 0.5 because the features and label are randomly generated. CV = cross validation, HNSCC = head and neck squamous cell carcinoma, LGG = low-grade glioma, SE = standard error.
Hyperparameter tuning with use of entire data set.—The loss of mean model efficiency (LassoCV R2) across data sets and normalization strategies when tuning hyperparameters after consistent partitioning (Fig 3) demonstrates that choosing hyperparameters using the full data set results in models with artificially increased performance.
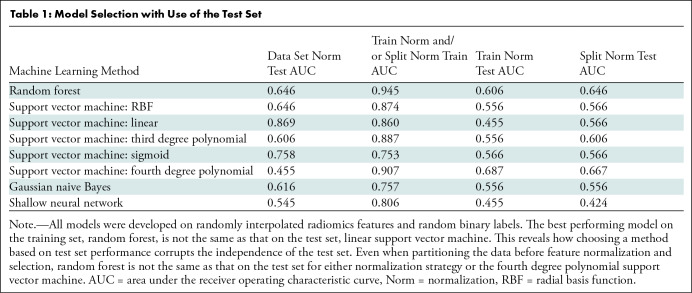
Method selection with use of test set.—Among the eight ML methods evaluated, the method that had the best performance on the training set was not the same as the one with the best performance on the test set (Table 1). Selecting an ML model and its parameters based on the test set negates the objective assessment of the generalizability of the model, necessitating more data to serve as a true test set. Cherry-picking the best performing CV folds and reporting their cumulative accuracy is yet another way this mistake manifests (Fig S5). CV methods, such as leave-one-out CV and the Jackknife procedure, are useful methods to ration data sets with few observations while quantifying the performance error of radiomics ML models (leave-one-out CV) or standard error (Jackknife) (21). Yet both these methods still require an independent test set so that a singular model is objectively evaluated for all data, rather than an average across CV folds.
Table 1:
Model Selection with Use of the Test Set
The range of AUC values generated from partitioning an increasing number of samples demonstrates that the minimum size of the data must approximate the feature dimension to confidently estimate the population accuracy (Fig 3). The widened uncertainty observed at lower sample sizes reflects that the training, validation, and test split can be manipulated to enhance statistical performance, taking advantage of the variability introduced by the partitioning procedure (Fig 3).
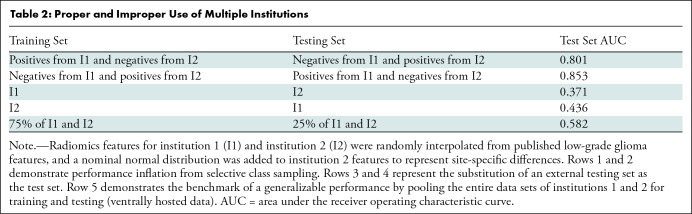
Selective class sampling or lack of external test set.—When selectively training on one pathologic label from each institution (eg, benign from one institution and malignant from another), the performance of the support vector machine model was enhanced (Table 2). When treating either institution as the test set (Table 2), the performance was below average because the support vector machine model learned the feature distributions of a single institution and was brittle to the nominal distribution, skewing the second institution. Because of the unique challenges of data privacy concerns when dealing with medical data, distributed learning strategies have been developed to expose an ML model to multiple institutions without requiring the data to be centrally hosted (Table 2) (22). Importantly, even when an external test set is included, it is often used for feature selection or model development, corrupting its independence.
Table 2:
Proper and Improper Use of Multiple Institutions
Unproductive Feature Associations
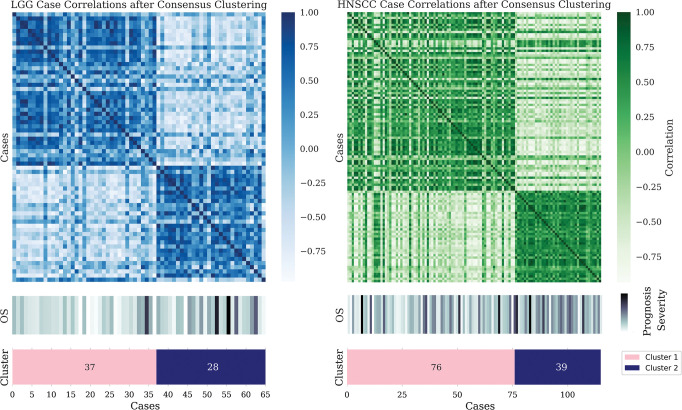
Association of features with themselves.—For the TCGA-LGG and HNSCC random radiomics features associated with overall survival, the number of clusters determined to be optimal is two, the same as the number of classes of the label (Fig 4). However, this does not imply a causal relationship between the prognosis and the clusters, most obviously because the features are random but also because the clusters have been formed based on features correlated with overall survival. The significant difference in overall survival between clusters in the TCGA-LGG (Mann-Whitney test, P = .044) and HNSCC (Mann-Whitney test, P = .041) cohorts demonstrates how overfitting to select features can render clustering a tautologic exercise. Variability between clustering algorithms and between clusters formed with use of varying initialization parameters emphasizes the limited utility in pairing clusters with biologic phenotypes without functional validation.
Figure 4:
Case-based consensus clustering of random radiomics features associated with overall survival (OS) in The Cancer Genome Atlas Low-Grade Glioma (left) and head and neck squamous cell carcinoma (HNSCC) (right) data sets. Despite sharp feature distribution differences, as seen in the heat maps, no statistically significant difference in outcome distribution exists between the assigned clusters. LGG = low-grade glioma.
Association of features with other high-dimensional variables (overfitting).—In the Cox proportional hazards model considering RadScore, tumor grade, and volume, the log of hazard ratios for both real and fake RadScore has a CI that intersects 0, implying that the hazard ratio may be greater than or less than 1 (Fig 5). Because a hazard ratio less than 1 implies a protective effect and a hazard ratio greater than 1 implies added risk, this uncertainty demonstrates the unreliable nature of the RadScore.
Figure 5:
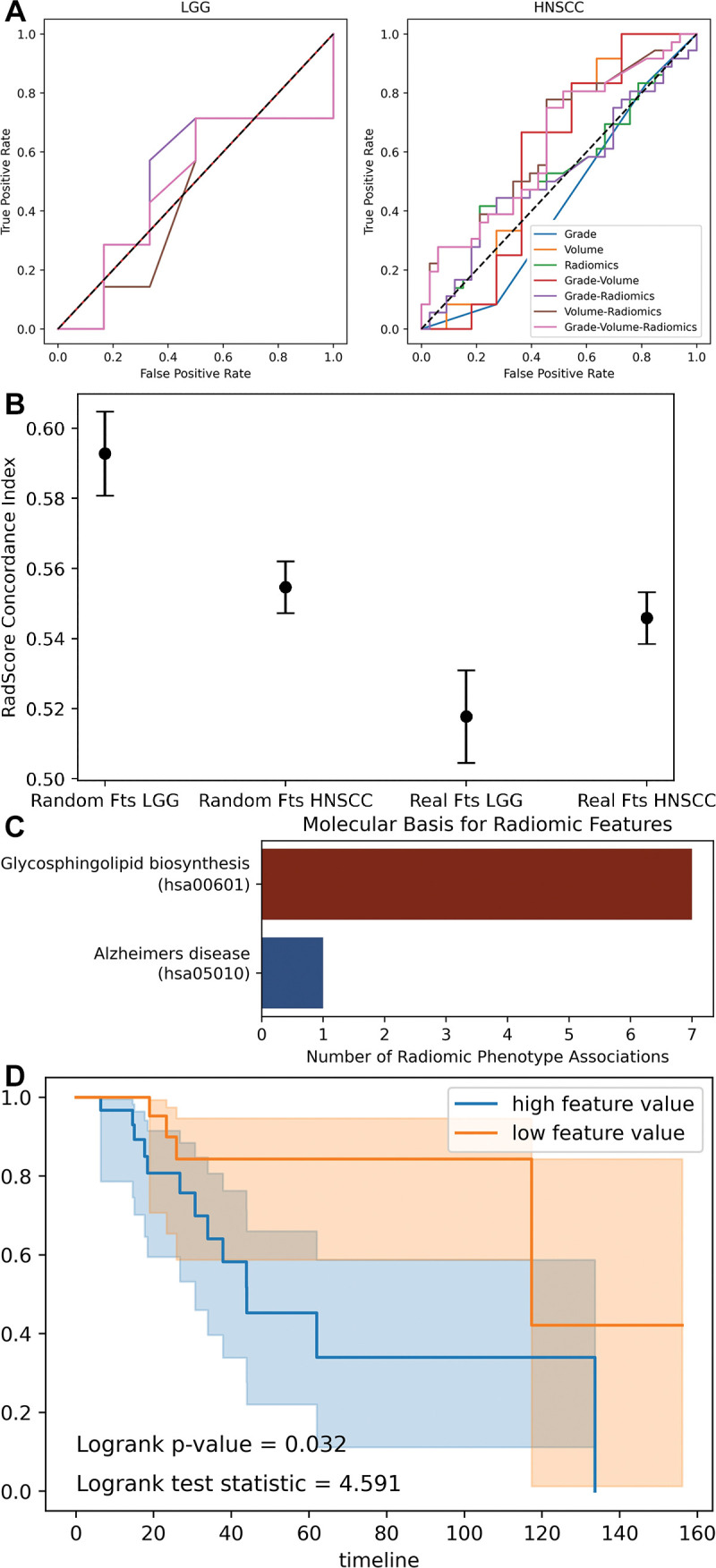
Combination of radiomics and biologic variables. (A) Receiver operating characteristic curves show support vector machine models fit to combinations of radiomics and biologic variables. (B) Dot plot with error bars show concordance index for radiomics score (RadScore) in Cox proportional hazards models. A concordance index of 0.5 represents random chance. The random radiomics features have higher concordance with true outcome (overall survival) than the authentic features. (C) Bar chart shows significant associations (Pearson) between random radiomics features and authentic gene ontology pathways in The Cancer Genome Atlas Low-Grade Glioma data set. (D) Kaplan-Meier curves show overall survival split by median feature value of a random feature observed to be spuriously yet significantly correlated with glycosphingolipid biosynthesis gene ontology pathway. Fts = features, HNSCC = head and neck squamous cell carcinoma, LGG = low-grade glioma.
As a succinct demonstration of overfitting, the 61160 correlations between fake radiomics features and gene ontology pathways resulted in eight significant associations, despite multiple hypothesis corrections (Fig 5). The high number of association of features with the glycosphingolipid biosynthesis pathway could otherwise suggest that imaging may reveal changes in this pathway, except that the radiomics features are randomly generated. Furthermore, a single simulated radiomics feature correlated with this gene ontology pathway results in significantly different survival functions when split across the median value. Thus, even random features can produce the supposedly meaningful survival predictions that pervade the radiomics literature.
Discussion
In this study we conducted a limited literature review of radiomics machine learning (ML) publications that identified two methodologic problem areas: inconsistent data partitioning and unproductive feature associations. We reproduced these flaws using randomly generated features based on authentic radiomics data sets. With facsimile features, we achieved state-of-the-art performance competitive with published studies (area under the receiver operating characteristic curve [AUC], 0.8–0.9). We experimentally demonstrated that correcting flawed methodology reduces AUCs on average by 1.4-fold to values approaching 0.5 or random chance. Inconsistent partitioning is hazardous to the objectivity of results because feature selection and normalization before partitioning, or selection of a ML model based on the test set, uses information about the entire feature distribution, inadvertently causing information leak. It is better to evaluate performance on a separate validation set as an early indication of the generalizability of the model, rather than extending all models to the test set before making a choice. Even with unbiased methodology, studies with too small a sample size can overestimate performance, and when a sufficiently large sample is taken, performance again approximates 0.5 (23). Our computational experiments revealed that at minimum, the data set size should equal the number of radiomics features under consideration and, ideally, vastly exceed it.
Previous studies also observed inconsistent partitioning: CV without a held-out test set or patient-level partitioning results in a 40%–55% boost to reported performance when classifying neurologic disorders from MRI (24). A meta-analysis of gut microbiome ML studies uncovered widespread test set omission (25). A seminal case study by Kapoor and Narayan (26) found that performance for ML models predicting civil war outbreak could not be reproduced when methodologic mistakes, specifically data leakage, were rectified. Their review is complementary to our study: imputation of data using the entire data set, information leakage from proxy variables, and CV using paired (nonindependent) samples are all manifestations of inconsistent partitioning.
Although the prevailing hope for ML in medicine is increased efficiency and precision in delivering care, many problems have been identified, including generalizability, explainability, and reinforcement of existing biases (27–29). While radiomics features also suffer from these limitations, they are often regarded as more explainable than convolutional neural networks because the mathematic basis of each feature is known, and they are associated either with the shape, intensity distribution, or texture of the region of interest. However, this is a fallacy of thought that contrasts engineering explainability, or the quantitative method in which a variable was derived, with biologic or human explainability (30). Even the most experienced radiologist does not understand the anatomic or molecular function driving “gray level co-occurrence matrix inverse difference moment,” just as they would not for a feature learned by a “black-box” neural network.
As with any disruptive innovation, investigation of radiomics ML was followed by guidelines and caution (31,32,33). The 2017 Radiomics Quality Score recommends best practices throughout the radiomics workflow but does not delve into granular details of model construction, such as consistent partitioning (34). It also encourages the combination of laboratory and biologic predictors with radiomics features. While these variables are paradigm shifting in medical evaluation, combining them with radiomics features presents the risk of overfitting (35). Similarly, the image biomarker standardization initiative that details best practices for radiomics feature extraction is upstream and complementary to our study, which begins with features already collected (36). Other guidelines provide useful considerations for data reporting, feature definition, extraction parameter standardization, the incremental value of radiomics features over common clinical variables, and broader application of ML algorithms (28,37–40). Of note, none of these works analyzed the vital importance of consistent data partitioning and tempered pairing of radiomics features with other high-dimensional variables as this study does. In addition, previous studies reported the influence of image acquisition protocols and instrumentation, image reconstruction and preprocessing, lesion segmentation, and feature extraction software on radiomics feature analysis. Here, we exclusively focus on methodologies downstream of feature extraction.
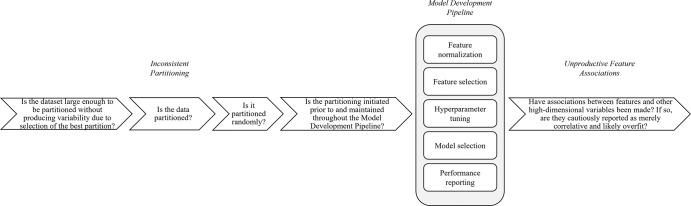
By deconstructing these two problem areas into their constitutive components, we suggest a template for independent researchers and reviewers alike (Fig 6). The following checklist summarizes the mistakes observed and modeled in this study: First, is the data set large enough to be partitioned in a way that does not produce high variability in results due to selection of the best partition? If so, is it partitioned? Is it partitioned randomly? Second, is the partitioning initiated before feature analysis and consistently observed throughout feature normalization, feature selection, hyperparameter tuning, model selection, and performance reporting? Third, has feature reproducibility been assessed? Fourth, have multiple hypotheses been tested and, if so, has a correction for multiple hypotheses been implemented? Fifth, is there an external test set and, if so, has model performance on an internal test set also been reported? Finally, is the importance of feature-feature, sample-sample, or feature-biologic variable correlations cautiously reported (multiple hypothesis correction) as merely correlative and likely overfit? If not, have the correlations been functionally validated on external test sets?
Figure 6:
Flow diagram shows reviewer questions when auditing radiomics machine learning studies for problem areas highlighted in this study: inconsistent partitioning and unproductive feature associations.
Random simulated features are uniquely illustrative for our purpose of estimating the impact of methodologic flaws on radiomic ML model performance. However, our study is limited in its narrow focus on model development and evaluation, when in fact performance inflation can occur further upstream of this step—for example, through homogeneous cohort selection and feature extraction parameter manipulation. Our simulation of a second institution is likewise limited by the naive addition of a nominal distribution rather than replicating true patient demographic or scanner differences. Finally, our study does not exhaustively consider the ways in which radiomics features can be overfit but rather provides an example in the form of gene ontology pathways. We encourage reviewers of radiomics studies to be open-minded when applying our recommendations to identify other sources of overfitting.
We conclude that radiomics machine learning studies require a rigorous analysis and review. Employing consistent data partitioning and appropriate feature associations will ensure the development of adaptive statistical models.
C.D.F. received/receives funding and salary support unrelated to this project during the period of study execution from the following: the National Institutes of Health (NIH) National Institute of Biomedical Imaging and Bioengineering (NIBIB) Research Education Programs for Residents and Clinical Fellows Grant (R25EB025787-01); the National Institute for Dental and Craniofacial Research Establishing Outcome Measures Award (1R01DE025248/R56DE025248) and Academic Industrial Partnership Grant (R01DE028290); NCI Early Phase Clinical Trials in Imaging and Image-Guided Interventions Program (1R01CA218148); an NIH/NCI Cancer Center Support Grant (CCSG) Pilot Research Program Award from the UT MD Anderson CCSG Radiation Oncology and Cancer Imaging Program (P30CA016672); an NIH/NCI Head and Neck Specialized Programs of Research Excellence (SPORE) Developmental Research Program Award (P50 CA097007); NIH Big Data to Knowledge (BD2K) Program of the National Cancer Institute (NCI) Early Stage Development of Technologies in Biomedical Computing, Informatics, and Big Data Science Award (1R01CA2148250); National Science Foundation (NSF), Division of Mathematical Sciences, Joint NIH/NSF Initiative on Quantitative Approaches to Biomedical Big Data (QuBBD) Grant (NSF 1557679); NSF Division of Civil, Mechanical, and Manufacturing Innovation (CMMI) grant (NSF 1933369); and Elekta.
Disclosures of conflicts of interest: M.G. No relevant relationships. K.C. No relevant relationships. J.B.P. No relevant relationships. K.V.H. No relevant relationships. S.R.A. No relevant relationships. P.S. No relevant relationships. C.D.F. Travel reimbursement and speaking honoraria from Elekta, National Institutes of Health, American Association of Physicists in Medicine, European Society for Therapeutic Radiation Oncology, American Society of Clinical Oncology, Varian Medical System, and Philips; unpaid service for advisory committee for the Dartmouth-Hitchcock Cancer Center Department of Radiation Oncology; serves in a committee or leadership service capacity for the National Institutes of Health, American Society of Clinical Oncology, Radiological Society of North America, American Association of Physicists in Medicine, NRG Oncology, American Cancer Society, Dutch Cancer Society, and Rice University; receives in-kind support from Elekta. J.K.C. Grants or contracts from the NIH; member of the Radiology-AI editorial board.
Abbreviations:
- AUC
- area under the receiver operating characteristic curve
- CV
- cross validation
- HNSCC
- head and neck squamous cell carcinoma
- ML
- machine learning
- TCGA-LGG
- The Cancer Genome Atlas Low-Grade Glioma
References
- 1. Gillies RJ , Kinahan PE , Hricak H . Radiomics: Images Are More than Pictures, They Are Data . Radiology 2016. ; 278 ( 2 ): 563 – 577 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang YQ , Liang CH , He L , et al . Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer . J Clin Oncol 2016. ; 34 ( 18 ): 2157 – 216 4 [Published correction appears in J Clin Oncol 2016;34(20):2436.]. [DOI] [PubMed] [Google Scholar]
- 3. Huang Y , Liu Z , He L , et al . Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer . Radiology 2016. ; 281 ( 3 ): 947 – 957 . [DOI] [PubMed] [Google Scholar]
- 4. Li H , Zhu Y , Burnside ES , et al . Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set . NPJ Breast Cancer 2016. ; 2 ( 1 ): 16012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaufman S , Rosset S , Perlich C , Stitelman O . Leakage in Data Mining: Formulation, Detection, and Avoidance . ACM Trans Knowl Discov Data 2012. ; 6 ( 4 ): 1 – 21 . [Google Scholar]
- 6. Diehn M , Nardini C , Wang DS , et al . Identification of noninvasive imaging surrogates for brain tumor gene-expression modules . Proc Natl Acad Sci U S A 2008. ; 105 ( 13 ): 5213 – 5218 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun R , Limkin EJ , Vakalopoulou M , et al . A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study . Lancet Oncol 2018. ; 19 ( 9 ): 1180 – 1191 . [DOI] [PubMed] [Google Scholar]
- 8. Bakas S , Akbari H , Sotiras A , et al . Segmentation Labels and Radiomic Features for the Pre-operative Scans of the TCGA-LGG collection [Data Set]. Cancer Imaging Archive . Published 2017. Accessed June 14, 2022 . [Google Scholar]
- 9. Bakas S , Akbari H , Sotiras A , et al . Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features . Sci Data 2017. ; 4 ( 1 ): 170117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clark K , Vendt B , Smith K , et al . The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository . J Digit Imaging 2013. ; 26 ( 6 ): 1045 – 1057 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grossberg AJ , Mohamed ASR , Elhalawani H , et al . Imaging and clinical data archive for head and neck squamous cell carcinoma patients treated with radiotherapy . Sci Data 2018. ; 5 ( 1 ): 18017 3 [Published correction appears in Sci Data 2018;5(1):1.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grossberg A , Elhalawani H , Mohamed A , et al . HNSCC [Dataset] . Cancer Imaging Archive . Published 2020. Accessed June 14, 2022 .
- 13. MICCAI/M.D. Anderson Cancer Center Head and Neck Quantitative Imaging Working Group . Matched computed tomography segmentation and demographic data for oropharyngeal cancer radiomics challenges . Sci Data 2017. ; 4 ( 1 ): 170077 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chawla NV , Bowyer KW , Hall LO , Kegelmeyer WP . SMOTE: synthetic minority over-sampling technique . J Artif Intell Res 2002. ; 16 : 321 – 357 . [Google Scholar]
- 15. Jeschkies K . NYAN . https://github.com/jeschkies/nyan. Published 2013. Accessed June 14, 2022 .
- 16. Cerami E , Gao J , Dogrusoz U , et al . The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data . Cancer Discov 2012. ; 2 ( 5 ): 401 – 404 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao J , Aksoy BA , Dogrusoz U , et al . Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal . Sci Signal 2013. ; 6 ( 269 ): pl1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoadley KA , Yau C , Hinoue T , et al . Cancer Genome Atlas Network. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer . Cell 2018. ; 173 ( 2 ): 291 – 304.e6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cancer Genome Atlas Network . Comprehensive genomic characterization of head and neck squamous cell carcinomas . Nature 2015. ; 517 ( 7536 ): 576 – 582 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cox DR . Regression Models and Life-Tables . J R Stat Soc Ser B Methodol 1972. ; 34 ( 2 ): 187 – 220 . [Google Scholar]
- 21. Lachenbruch PA . An almost unbiased method of obtaining confidence intervals for the probability of misclassification in discriminant analysis . Biometrics 1967. ; 23 ( 4 ): 639 – 645 . [PubMed] [Google Scholar]
- 22. Chang K , Balachandar N , Lam C , et al . Distributed deep learning networks among institutions for medical imaging . J Am Med Inform Assoc 2018. ; 25 ( 8 ): 945 – 954 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flint C , Cearns M , Opel N , et al . Systematic misestimation of machine learning performance in neuroimaging studies of depression . Neuropsychopharmacology 2021. ; 46 ( 8 ): 1510 – 1517 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yagis E , Atnafu SW , García Seco de Herrera A , et al . Effect of data leakage in brain MRI classification using 2D convolutional neural networks . Sci Rep 2021. ; 11 ( 1 ): 22544 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Quinn TP . Stool Studies Don’t Pass the Sniff Test: A Systematic Review of Human Gut Microbiome Research Suggests Widespread Misuse of Machine Learning . arXiv preprint arXiv:2107.03611. https://arxiv.org/abs/2107.03611. Posted July 8, 2021. Accessed June 14, 2022 .
- 26. Kapoor S , Narayanan A . (Ir)Reproducible Machine Learning: A Case Study . https://reproducible.cs.princeton.edu/. Published 2021. Accessed June 14, 2022 . [Google Scholar]
- 27. Zech JR , Badgeley MA , Liu M , Costa AB , Titano JJ , Oermann EK . Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study . PLoS Med 2018. ; 15 ( 11 ): e1002683 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghassemi M , Oakden-Rayner L , Beam AL . The false hope of current approaches to explainable artificial intelligence in health care . Lancet Digit Health 2021. ; 3 ( 11 ): e745 – e750 . [DOI] [PubMed] [Google Scholar]
- 29. Larrazabal AJ , Nieto N , Peterson V , Milone DH , Ferrante E . Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis . Proc Natl Acad Sci U S A 2020. ; 117 ( 23 ): 12592 – 12594 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller K . Should AI Models Be Explainable? That depends . Stanford HAI . https://hai.stanford.edu/news/should-ai-models-be-explainable-depends. Published 2021. Accessed June 14, 2022 .
- 31. Vollmer S , Mateen BA , Bohner G , et al . Machine learning and artificial intelligence research for patient benefit: 20 critical questions on transparency, replicability, ethics, and effectiveness . BMJ 2020. ; 368 : l692 7 [Published correction appears in BMJ 2020;369:m1312.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Collins GS , Reitsma JB , Altman DG , Moons KGM . Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement . BMJ 2015. ; 350 : g7594 . [DOI] [PubMed] [Google Scholar]
- 33. McKinlay JB . From “promising report” to “standard procedure”: seven stages in the career of a medical innovation . Milbank Mem Fund Q Health Soc 1981. ; 59 ( 3 ): 374 – 411 . [PubMed] [Google Scholar]
- 34. Lambin P , Leijenaar RTH , Deist TM , et al . Radiomics: the bridge between medical imaging and personalized medicine . Nat Rev Clin Oncol 2017. ; 14 ( 12 ): 749 – 762 . [DOI] [PubMed] [Google Scholar]
- 35. Welch ML , McIntosh C , Haibe-Kains B , et al . Vulnerabilities of radiomic signature development: The need for safeguards . Radiother Oncol 2019. ; 130 : 2 – 9 . [DOI] [PubMed] [Google Scholar]
- 36. Zwanenburg A , Vallières M , Abdalah MA , et al . The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping . Radiology 2020. ; 295 ( 2 ): 328 – 338 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hoebel KV , Patel JB , Beers AL , et al . Radiomics Repeatability Pitfalls in a Scan-Rescan MRI Study of Glioblastoma . Radiol Artif Intell 2020. ; 3 ( 1 ): e190199 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McNitt-Gray M , Napel S , Jaggi A , et al . Standardization in Quantitative Imaging: A Multicenter Comparison of Radiomic Features from Different Software Packages on Digital Reference Objects and Patient Data Sets . Tomography 2020. ; 6 ( 2 ): 118 – 128 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mali SA , Ibrahim A , Woodruff HC , et al . Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods . J Pers Med 2021. ; 11 ( 9 ): 842 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao B . Understanding Sources of Variation to Improve the Reproducibility of Radiomics . Front Oncol 2021. ; 11 : 633176 . [DOI] [PMC free article] [PubMed] [Google Scholar]



![Diagrams of inconsistent partitioning. Random features (R) based on published radiomics data form the basis of our experimentation (atypical from radiomics machine learning [ML] studies). (A) The upper level (blue and yellow) illustrates consistent partitioning that prevents information leak, while the lower level (green) demonstrates how the use of the entire data set for radiomics feature normalization, feature selection, hyperparameter selection, model selection, and performance reporting will result in an unrealistically optimistic assessment of the radiomics ML model. (B) Diagrams show normalization strategies. Data set normalization (green) is an example of inconsistent partitioning, with use of a mean and SD calculated with use of all samples, both the training and test sets, to scale. Train normalization (right) and split normalization (bottom) are different approaches to consistent partitioning (more details in Appendix S1).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/ab20/10068883/8c4c089be29a/radiol.220715.fig1.jpg)