SUMMARY PARAGRAPH
Treatment with BRAF/MEK-targeted therapy has revolutionized care in melanoma and other cancers, however therapeutic resistance is common and innovative treatment strategies are needed1,2. We studied a group of melanoma patients treated with neoadjuvant BRAF/MEK-targeted therapy (NCT02231775, n=51), and observed significantly higher rates of major pathologic response (MPR= <10% viable tumor at resection) and improved recurrence-free survival (RFS) in females versus males (MPR-66% versus 14%, p=0.001; RFS-64% versus 32% at 2 years, p=0.021). Findings were validated in a several additional cohorts2–4 patients with unresectable metastatic melanoma treated with BRAF and/or MEK-targeted therapy (n=664 patients in total), demonstrating improved progression-free survival (PFS) and overall survival (OS) in females versus males in several of these studies. Studies in pre-clinical models demonstrated significantly impaired anti-tumor activity in male versus female BRAF/MEK-treated mice (p=0.006), with significantly higher expression of androgen receptor (AR) in tumors of male and female BRAF/MEK-treated mice versus control (p=0.0006 and 0.0025). Pharmacologic inhibition of AR signaling improved responses to BRAF/MEK-targeted therapy in male and female mice (p=0.018 and p=0.003), whereas induction of AR signaling (via testosterone administration) was associated with significantly impaired response to BRAF/MEK-targeted therapy in males and females (p=0.021 and p<0.0001). Together, these results have important implications for therapy.
Keywords: cancer, targeted therapy, androgen, BRAF
To date, multiple studies have demonstrated that male sex is associated with worse outcomes in patients with melanoma, including patients with stage III and IV disease treated with BRAF targeted therapy1,2. However, the underlying biology is incompletely understood. In classical hormonally responsive malignancies such as prostate cancer, reciprocal interactions between androgen receptor (AR) signaling and MAPK and other signaling pathways are evident5 - though this has been less well-studied in melanoma and other cancers. Variations in the immune background6,7, tumor microenvironment8–11 and tumor cell susceptibility to targeted therapy6 have also been posited as driving factors of this sexual dimorphism in treatment outcomes. Additionally, innate hormonal differences in the estrogen and androgen pathways have been implicated as potential mechanisms driving observed disparities in melanoma preclinical models9,10,12–14 following immune checkpoint blockade15,16 and targeted therapy17. However, further insights from clinical and preclinical studies are needed to help derive innovative strategies to improve patient survival.
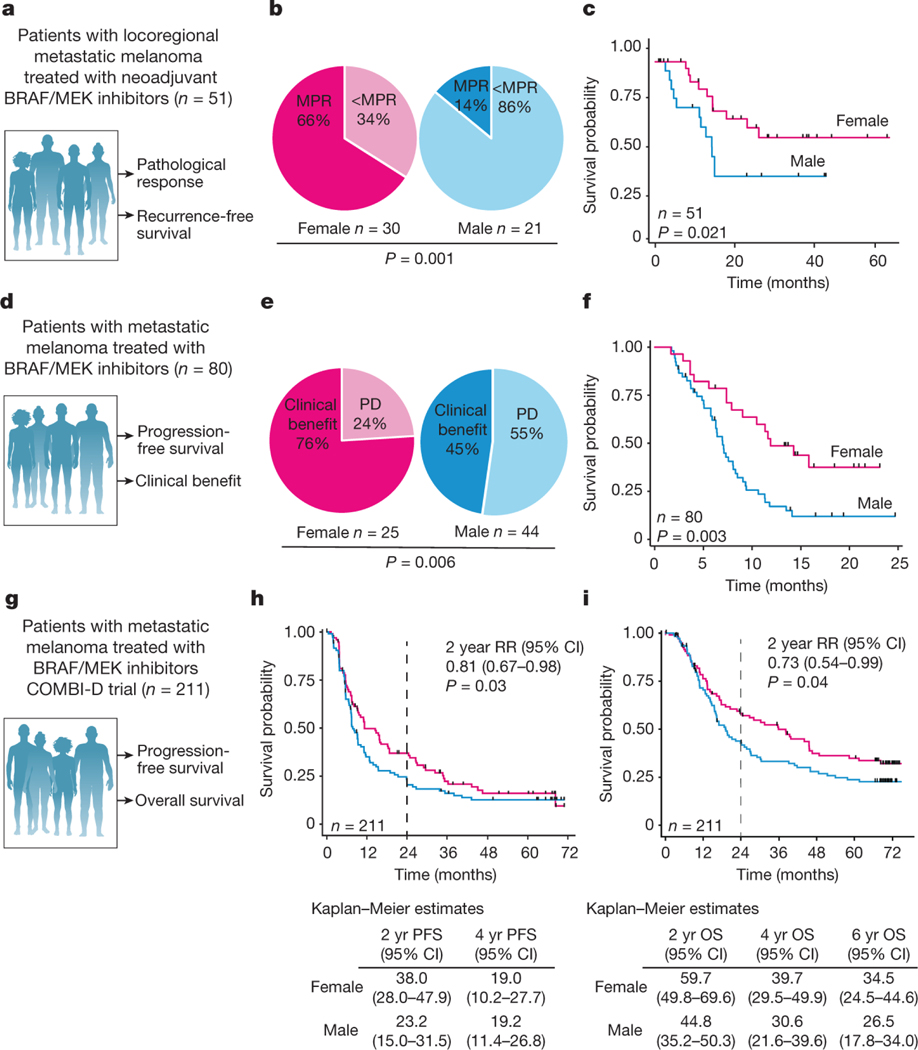
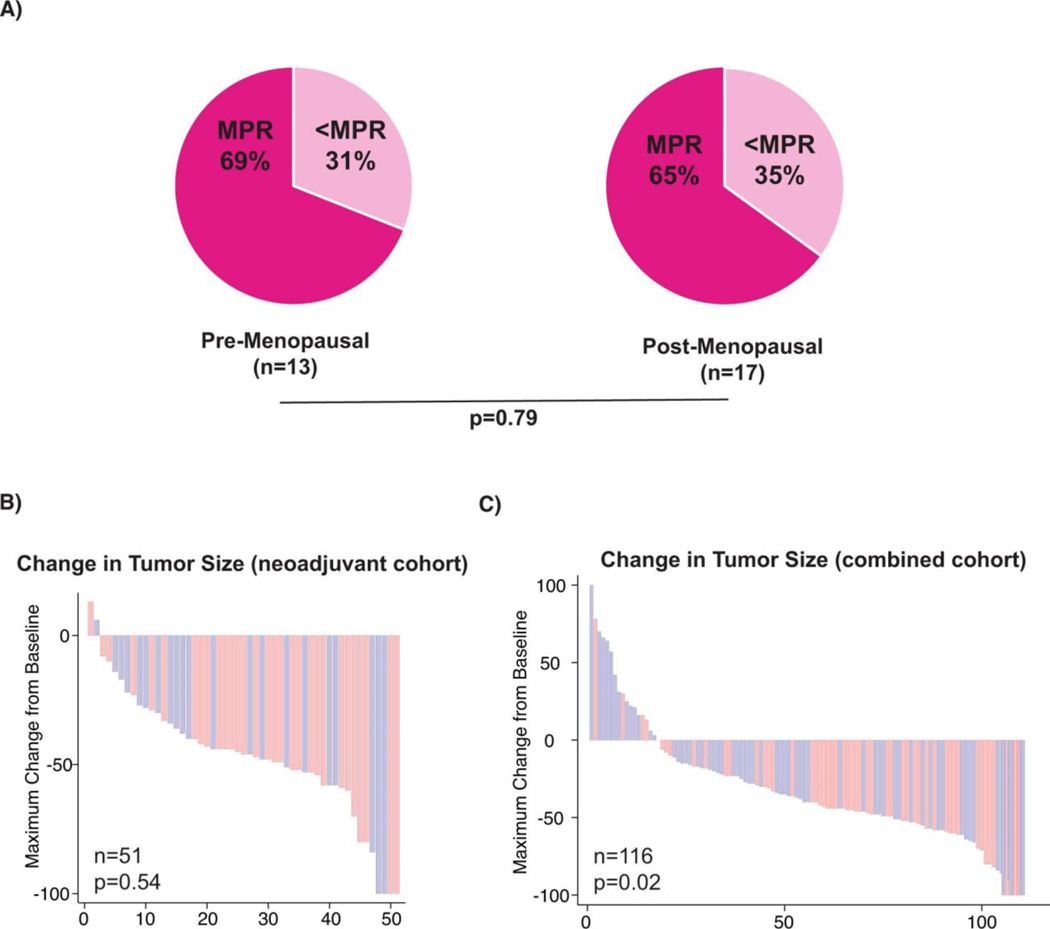
To help address this area of unmet need, we investigated the impact of biological sex on response to BRAF/MEK-targeted therapy in several independent cohorts of patients with metastatic melanoma. We first studied a cohort of patients with locoregional metastatic melanoma treated with 8–12 weeks of neoadjuvant BRAF/MEK-targeted therapy18 (n=51, Figure 1A). In this study, we observed a strong sexual dimorphism in response to treatment - with a significantly higher rate of major pathologic response (MPR; defined as < 10% viable tumor on pathologic evaluation of the surgically-resected tumor at time of operative intervention) in female versus males patients (66% versus 14% respectively, odds ratio of 12.0 95%CI [2.85–50.59] p=0.001) (Figure 1B). Importantly, relapse-free survival after neoadjuvant BRAF/MEK-targeted therapy was also noted to be significantly higher in female versus male patients (62% versus 34% at 2 years, p=0.021) (Figure 1C). No other clinical factors were significantly associated with achievement of MPR on logistic regression analysis, including age (p=0.40), stage IIIC or D vs. IIIA or B (p=0.45), stage IV vs. Stage IIIA or B (p=0.78), BRAF V600E versus V600 non-E mutation (p=0.81), Eastern Cooperative Oncology Group (ECOG) performance status (p=0.89), body mass index (BMI) (>30) (p=0.90), serum lactate dehydrogenase (LDH) (p=0.29), or recurrent vs. de novo disease (p=0.24) (Extended Data Table 1). There was no association between MPR and menopausal status in the female subset of the neoadjuvant cohort, where menopausal status was available, (MPR; 9/13 (69%) pre-menopausal women and 11/17 (65%) post-menopausal women (p=0.79) suggesting estrogen did not play a major role in our observations (Extended Data Figure 1A). RECIST response rates in the neoadjuvant cohort were not significantly different between females and males (Extended Data figure 1B, p=0.54), which is consistent with our previous observation of poor correlation of RECIST response rates and pathologic response in patients on neoadjuvant BRAF/MEK-targeted therapy18,19.
Figure 1. Female patients with melanoma have an improved response to BRAF/MEK inhibition compared with male patients.
a, Schematic of the neoadjuvant clinical cohort of patients treated with BRAF/MEK inhibition and their clinical outcomes studied. b, MPR (defined as ≤10% viable tumour) in female versus male patients (P = 0.001, χ 2 test). c, Recurrence-free survival by sex in the neoadjuvant patient cohort (n = 51, P = 0.021, log-normal Kaplan–Meier method). d, Schematic of the metastatic clinical cohort of patients treated with targeted therapy and their clinical outcomes studied. e, The clinical benefit of BRAF/MEK inhibition in the metastatic cohort (n = 69 evaluable) with clinical benefit defined as a complete response, partial response or stable disease on the basis of RECIST1.1 (P = 0.022, χ 2 test). f, PFS by sex in the metastatic cohort (n = 80, hazard ratio = 0.39, 95% CI = 0.24–0.62, P = 0.003, Kaplan–Meier method). g, Schematic of the COMBI-D trial clinical cohort of patients treated with BRAF/MEK inhibition and their clinical outcomes studied. h, PFS by sex in the COMBI-D cohort (2 year relative risk (RR) = 0.81, 95% CI = 0.67–0.98, P = 0.03, Kaplan–Meier method). i, Overall survival (OS) by sex in the COMBI-D cohort (2 year relative risk = 0.73, 95% CI = 0.54–0.99, P = 0.04, Kaplan–Meier method).
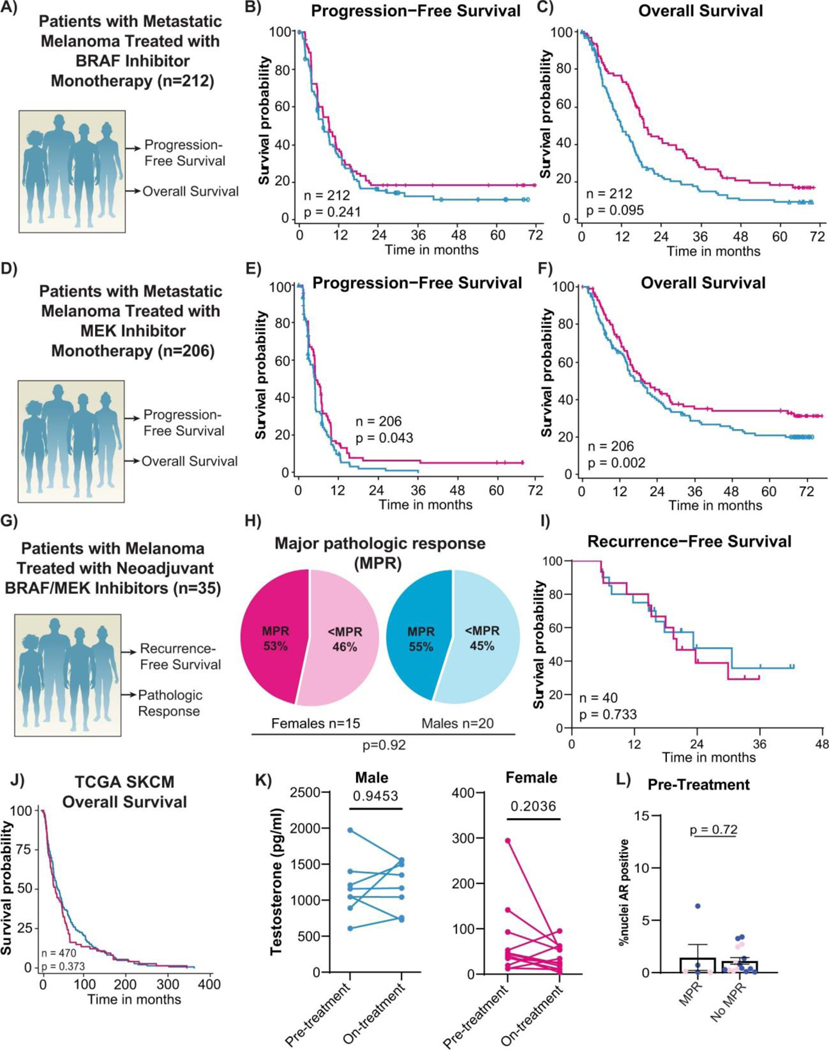
Intrigued by these findings, we sought to validate this in additional cohorts of patients. To do this, we next studied several cohorts of patients with unresectable metastatic disease treated with BRAF and/or MEK inhibitors until time of progression. This included a cohort of patients from our own institution treated with definitive BRAF/MEK-targeted therapy until time of progression (n=69) (Figure 1D, Extended Data Table 2). Clinical outcomes were assessed by both RECIST20 and progression-free survival. In this cohort a significantly higher rate of clinical benefit (defined as a complete response, partial response, or stable disease for 6 months or greater) was noted in female versus male patients on BRAF/MEK-targeted therapy (80% versus 68%; OR 3.65 95% CI [1.16–11.47], p=0.022 in 69 evaluable patients) (Figure 1E). Significantly improved progression-free survival was also noted in female versus male patients (median 12 months vs. 7 months; hazard ratio 0.42, 95% CI [0.23–0.75], p=0.003 in 80 evaluable patients) (Figure 1F). In addition, female patients exhibited a significantly larger reduction in overall tumor burden compared to males on combined analysis of both cohorts via cross sectional imaging (44% vs 27%, p=0.02, n=116 evaluable patients; Extended Data Figure 1C). We next validated these findings in several different cohorts of patients treated with BRAF and / or MEK inhibitor therapy for metastatic melanoma (n=664 patients total)2–4. Analysis of the COMBI-D study (NCT01584648) of patients treated with combined BRAF/MEK-targeted therapy (n=211) demonstrated that female patients had improved PFS/OS compared to male patients at 2 years (Figure 1G,H,I; relative risk 0.81, 95% CI [0.67–0.98], p=0.03 and relative risk 0.73, 95%CI [0.54–0.99], p=0.04 respectively). Differences were not as profound in the setting of treatment with BRAF inhibitor monotherapy (n=212 patients), with no significant differences observed in PFS / OS between male and female patients, though a trend toward improved OS was noted in female patients (Extended Data Figure 2A,B,C). Patients with metastatic melanoma treated with single agent MEK inhibition monotherapy were also studied (METRIC study, NCT01245062 - including 206 patients with 112 males and 94 females), demonstrating significantly improved PFS and OS in females versus males (Extended Data Figure 2D,E,F, p=0.043 and p=0.0021 respectively). Importantly, we also included an additional cohort of patients with locally advanced melanoma treated with neoadjuvant BRAF/MEK inhibition (NCT01972347)4. Analysis of this cohort did not reveal significant differences in MPR rates or RFS in females versus males - which is in contrast to our studies. This highlights heterogeneity in these clinical observations, though this cohort was substantially smaller than our own cohort, and was not balanced with regard to the number of enrolled female and male patients (n=35 patients total with the majority being male) (Extended Data Figure 2G,H,I).
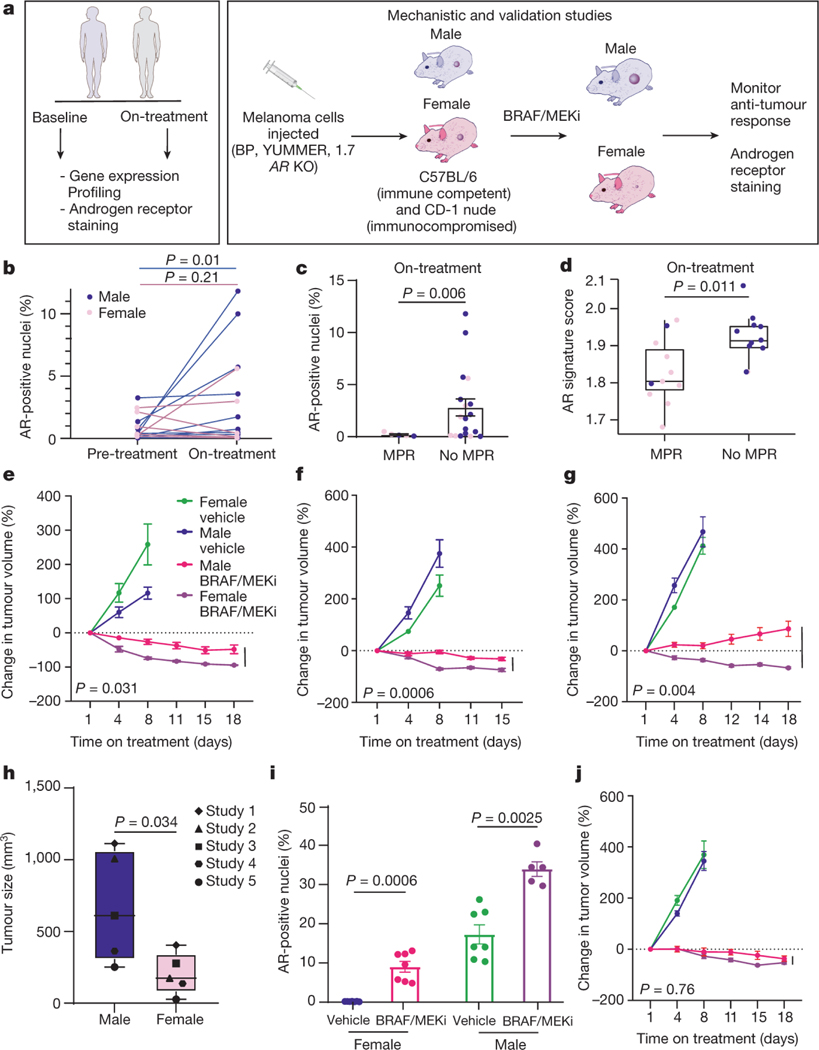
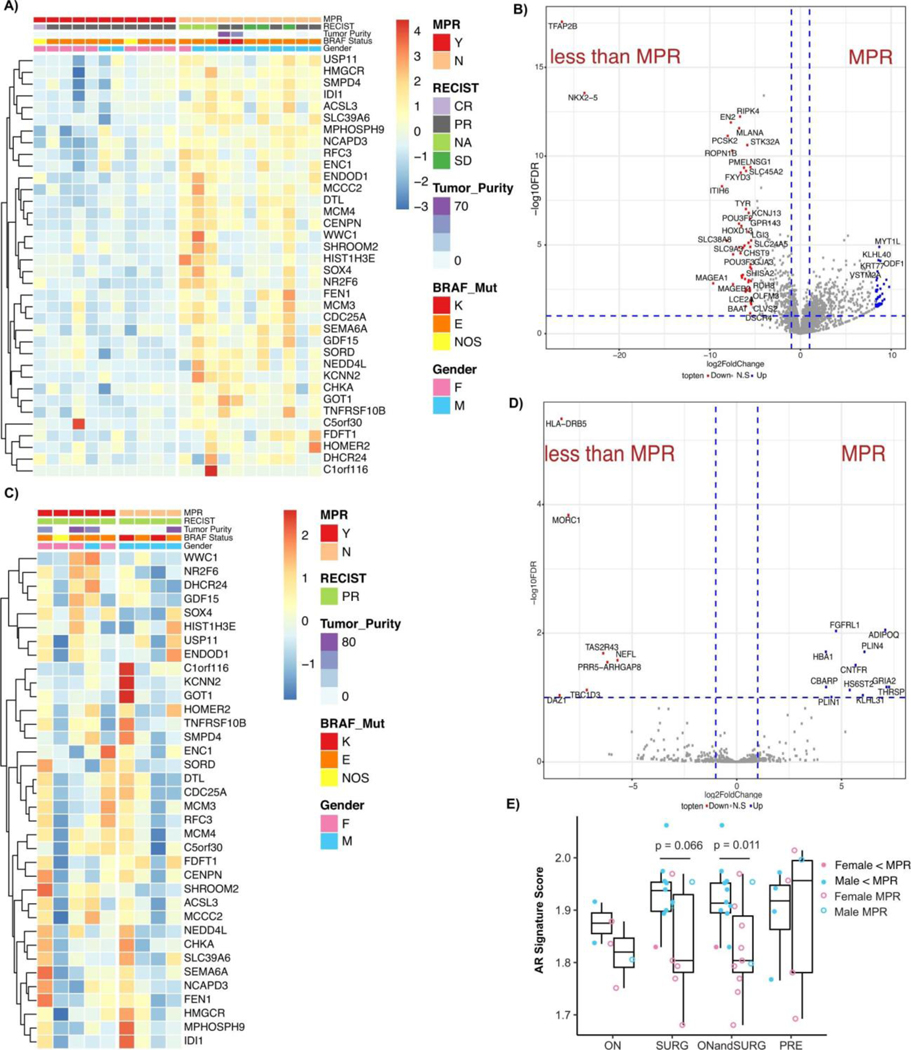
We next compared AR expression in matched pre- and on-treatment time points in patients receiving neoadjuvant BRAF/MEK-targeted therapy (Figure 2A), and observed significantly higher expression of AR in male patients on-treatment compared to pre-treatment time points (Figure 2B, p=0.01). In female patients, a trend toward increased AR expression was observed from pre-treatment to on-treatment samples but this did not reach statistical significance (Figure 2B, p=0.21), likely owing to lower baseline testosterone levels in female patients. Notably, circulating testosterone levels were not significantly impacted by treatment with BRAF/MEK-targeted therapy when assessing matched pre- and on-treatment blood samples of patients treated with neoadjuvant BRAF/MEK-targeted therapy (Extended Data Figure 2K). We specifically queried AR expression levels in pre- and on-treatment tumor samples of patients treated with neoadjuvant BRAF/MEK-targeted therapy, and observed significantly higher AR expression in on-treatment tumors of patients who failed to achieve an MPR compared to those who achieved an MPR (Figure 2C, p=0.006; Extended Data Table 3), intimating a possible association between AR expression post-treatment and response. However, this analysis was confounded by the lower proportion of viable tumor cells in tumors that had a significant response in on-treatment samples (Extended Data Figure 3, Extended Data Table 4). No significant differences in AR expression were observed in pre-treatment samples of patients achieving an MPR versus those who did not (Extended Data Figure 2L, p=0.72), suggesting that baseline AR expression is not predictive of resistance but that resistance is associated with upregulation of AR on treatment with BRAF/MEK-targeted therapy. AR signaling genes were assessed21 in bulk RNAseq data from available tumor biopsy samples of treated patients, again demonstrating a significantly higher AR signature score in on-treatment samples of patients who failed to achieve an MPR (Figure 2D, p=0.011), with no differences observed in pre-treatment samples (Extended Data Figure 3E, p=0.95). Additionally, no significant differences in AR related genes were observed when considering on-treatment and surgical samples independently rather than in aggregate (Extended Data Figure 3E, p=0.35, p=0.066, and p=0.011), though sample size was substantially lower limiting the power of such analyses.
Figure 2: Treatment with BRAF/MEK inhibition is associated with increased AR expression and phenotype is recapitulated in preclinical models.
b, AR staining in paired pre- and post-treatment male (blue) and female (pink) patient samples show increased AR expression in male samples (P = 0.01) but not in female patient samples (P = 0.21, two-sided t-test). c, AR staining post-treatment in male (n = 14, blue) and female (n = 9, pink) patients by MPR (P = 0.006, two-sided t-test). d, Androgen signalling score in patients achieving MPR (n = 11) versus <MPR (n = 11, P = 0.011, two-sided t-test). e, The percentage change in tumour volume in C57BL/6 mice that were implanted with YUMMER1.7 cells treated with vehicle or BRAF/MEK inhibition (BRAF/MEKi) (P = 0.031 male versus female BRAF/MEKi). n = 10 mice per group, aged 12–13 weeks. f, The percentage change in tumour volume in C57BL/6 mice implanted with Braf V600E Pten −/− mouse melanoma (BP) cells (P = 0.0006; male versus female BRAF/MEKi). Mice were treated as described in a. n = 10 mice per group, aged 12–13 weeks. The experiment was performed three times (Extended Data Fig. 3a,b). g, The percentage change in tumour volume in CD-1 nude mice that were implanted and treated as described in a (P = 0.004; male versus female BRAF/MEKi). n = 10 mice per group, aged 11 weeks. h, Aggregate end-point tumour volumes (n = 10 mice per study from five independent studies, P = 0.034, two-sided t-test). Individual points represent different studies. i, The percentage of cells with AR+ nuclei in YUMMER1.7 tumours in C57BL/6 mice that were treated with vehicle (n = 7 male and n = 7 female) or BRAF/MEKi (n = 7 female (P = 0.0006), n = 5 male (P = 0.0025), two-sided t-test) for 3 days. j, The percentage change in AR-KO BP tumour growth in CD-1 mice that were treated with vehicle or BRAF/MEKi (P = 0.76, male versus female BRAF/MEKi). n = 10 per group, aged 11 weeks. The tumour growth curves shown in e, f, g and j show mean ± s.e.m. and P values were computed using analysis of variance (ANOVA) with correction for multiple comparisons. The box plots in d and h show the median (centre line), interquartile range (IQR) (box limits) and the most extreme point within 1.5 × IQR (whiskers).
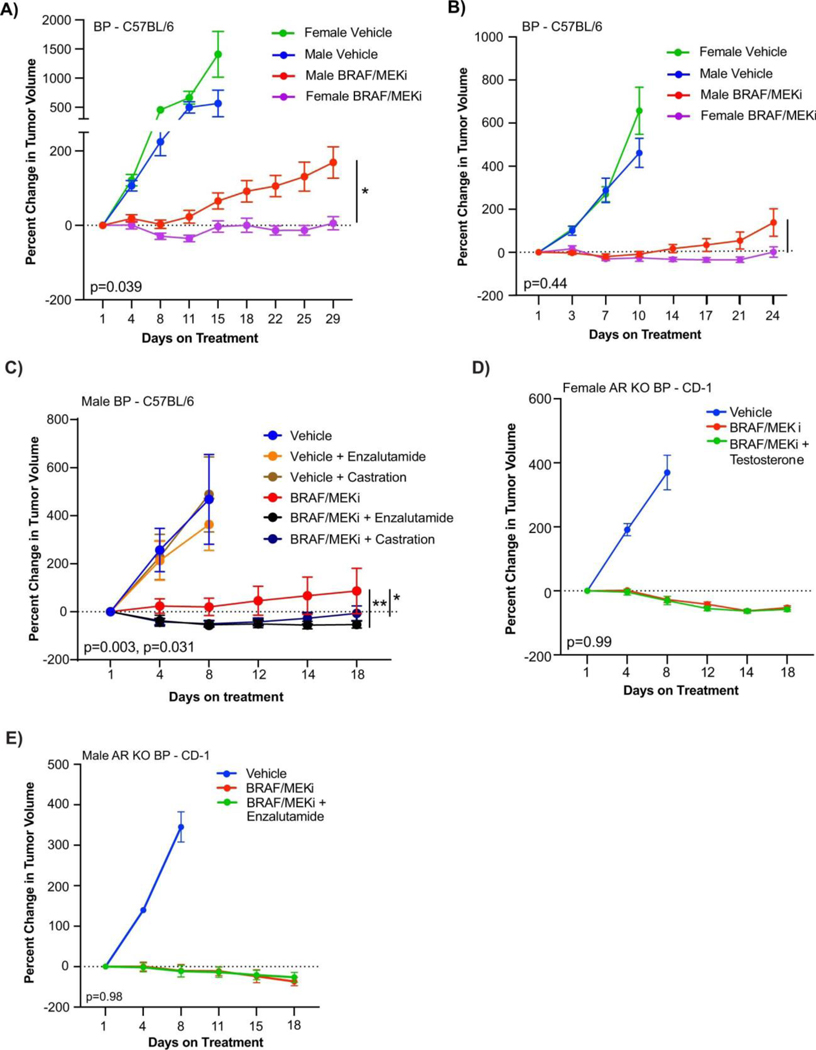
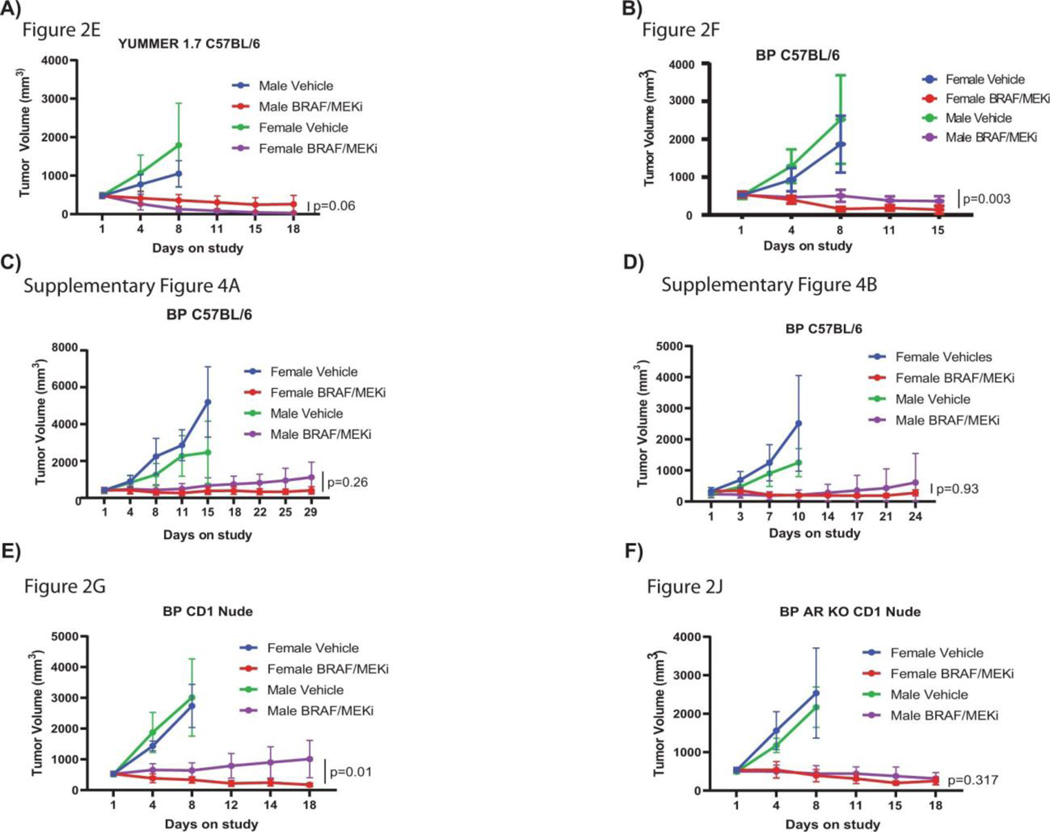
We next aimed to validate these findings in more carefully controlled pre-clinical models, with the goal of identifying potential therapeutic strategies to enhance response and survival rates to BRAF/MEK-targeted therapy. To do this, we studied the impact of biologic sex on response to BRAF/MEK targeted therapy in 3 independent pre-clinical models of BRAF-mutant melanoma (Figure 2A). We first assayed this in 2 immunocompetent models (C57BL/6 mice transplanted with 2 independently generated BRAF-mutant melanoma cell lines), demonstrating significantly impaired tumor control in male versus female mice treated with BRAF/MEK-targeted therapy (Figure 2E and 2F, p=0.031 and 0.0006 respectively, Extended Data Figure 4A and 4B, p=0.039 and p=0.45). No difference in tumor outgrowth was noted in female versus male mice treated with vehicle alone (control) (Figure 2E and 2F, p=0.13 and p=0.22, Extended Figure 4A and 4B, p=0.49 and p=0.47) or in male mice treated with vehicle and endocrine modulation via androgen blockade or castration (Extended Data Figure 4C, p=0.31 and 0.96, respectively) suggesting that the mechanism behind these findings was related to treatment with BRAF/MEK-targeted therapy and not in tumor onset in this particular model. We then assayed an immunocompromised model (CD-1 nude mice) and observed similar findings (Figure 2G, p=0.004), suggesting that the dominant mechanism behind this observation is not likely to be immune-mediated, though limitations exist with this particular model as B cells and macrophages are still present, with opportunities to validate this in an NOD SCID-gamma mouse model. Analysis of the three models confirmed larger tumor size in male mice across multiple studies (Figure 2H, p=0.034, tumor volume curves for associated experiments Extended Data Figure 5).
Following this, we sought to interrogate the mechanism through which male sex could be contributing to impaired tumor control in the setting of treatment with BRAF/MEK-targeted therapy. Given the observation that AR staining was increased in on-treatment models (Figure 2I, p=0.0006 and 0.0025), we hypothesized that increased AR signaling might be at play given our findings in patients and in pre-clinical models. This is supported by recent literature suggesting a role for androgen signaling in initiation and progression of melanoma14,22,23. To test whether AR signaling was driving resistance to BRAF/MEK-targeted therapy and whether this was tumor-cell intrinsic, we generated androgen receptor knock-out (AR KO) melanoma cell lines using CRISPR technology. AR KO cell lines were implanted into male and female mice and mice treated with BRAF/MEK-targeted therapy, demonstrating equivalent and effective tumor control in male and female mice compared to vehicle control. There was no evidence of sexual dimorphism in the setting of treatment with BRAF/MEK-targeted therapy alone (Figure 2J, p=0.76), or in the setting of modulation of androgen signaling via administration of supplementary testosterone or AR blockade with enzalutamide (Extended Data Figure 4D and 4E, p=0.99 and 0.98). This suggests that tumor-intrinsic AR expression is contributing to the therapeutic resistance to BRAF/MEK-targeted therapy.
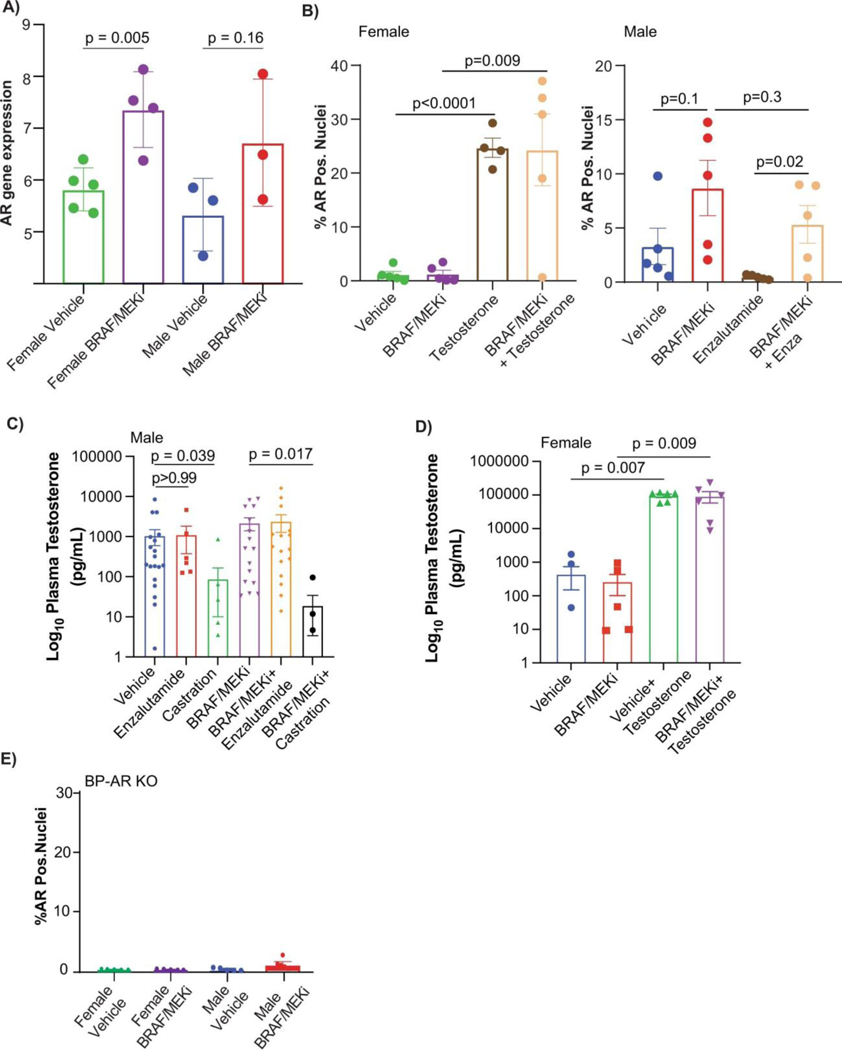
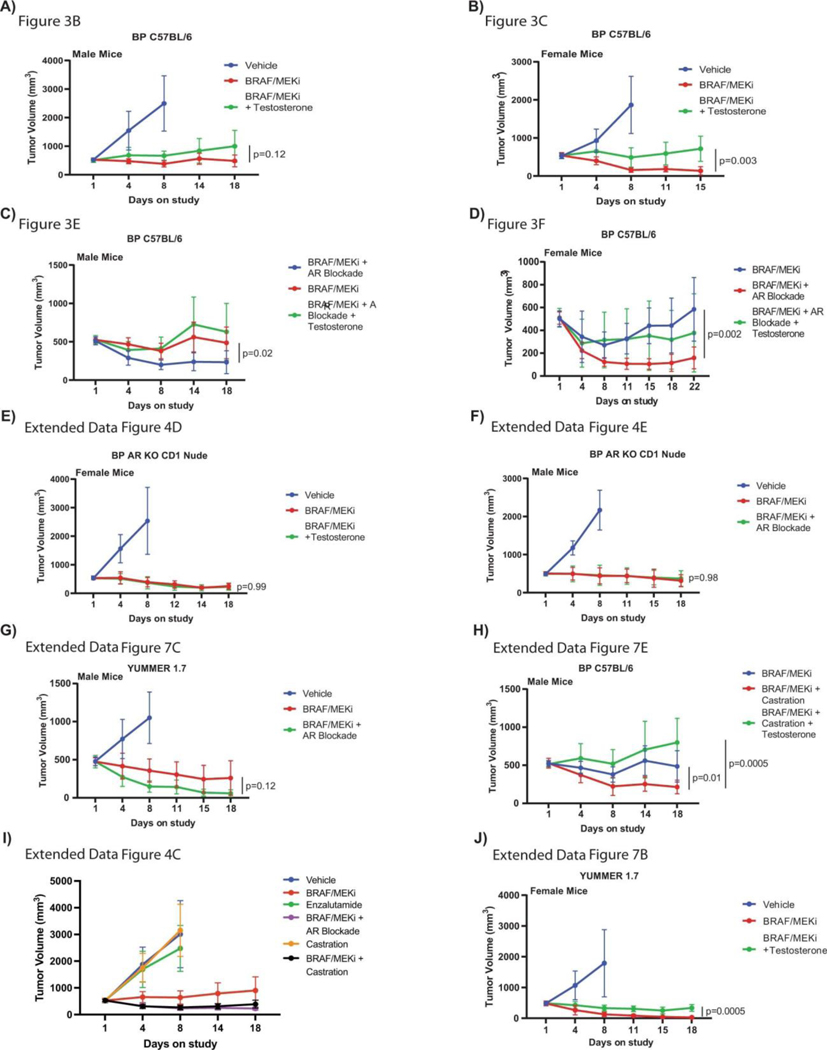
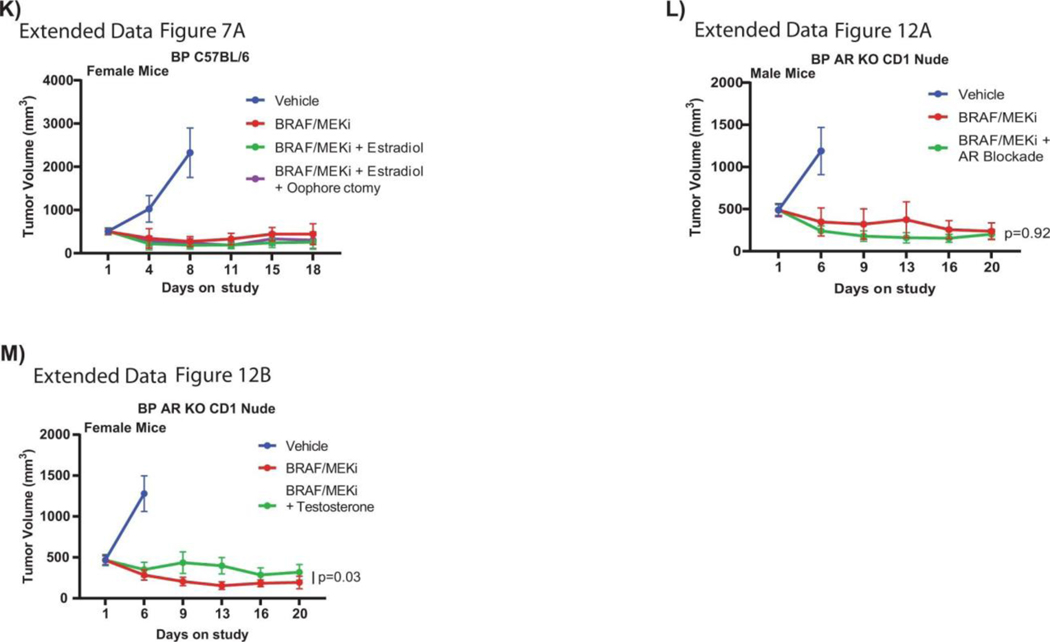
We next assessed AR staining and gene expression profiling in male and female mice treated with vehicle versus BRAF/MEK-targeted therapy, and in available pre- and on-treatment tumor samples from patients treated with neoadjuvant BRAF/MEK-targeted therapy. In these studies, we observed significantly higher expression of AR in both male and female mice following treatment with BRAF/MEK-targeted therapy versus vehicle control (Figure 2I, p=0.0006 and 0.0025, Extended Data Figures 6A and 6B), suggesting that treatment with BRAF/MEK-targeted therapy is associated with increased AR staining in both males and females. Notably, baseline and on-treatment levels of AR were significantly higher in males compared to females, likely related to higher levels of testosterone in males (Extended Data Figures 6C and 6D) - and might be attributable to the known role of testosterone in stabilizing AR protein as demonstrated in other malignancies24. There was not, however, a significant change in plasma testosterone comparing vehicle and BRAF/MEK-treated groups (Extended Data Figure 6C, p>0.99). Loss of AR staining was confirmed in AR KO tumors (Extended Data Figure 6E), which was accompanied by loss of sexual dimorphism in response to BRAF/MEK targeted therapy. This suggests that differences in response to BRAF/MEK targeted therapy is driven by tumor intrinsic AR activity. No differences in outcomes were noted in female mice treated with supplemental estrogen and oophorectomy in combination with BRAF/MEK-targeted therapy, compared to BRAF/MEK-targeted therapy alone (Extended Data Figure 7A, p=0.43), suggesting that estrogen signalling is not a dominant factor underlying the observed sexual dimorphism in this pre-clinical model, although additional study is needed. Together, these findings suggest that the sexually dimorphic phenotype is not primarily driven through the ovarian production of steroids.
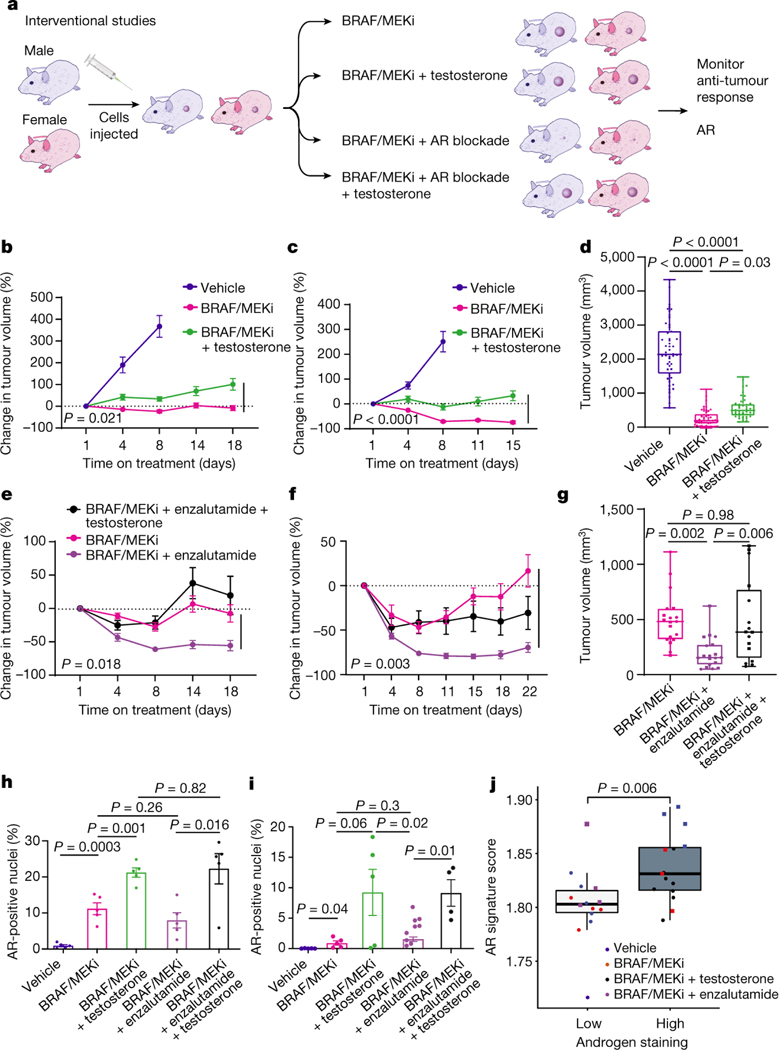
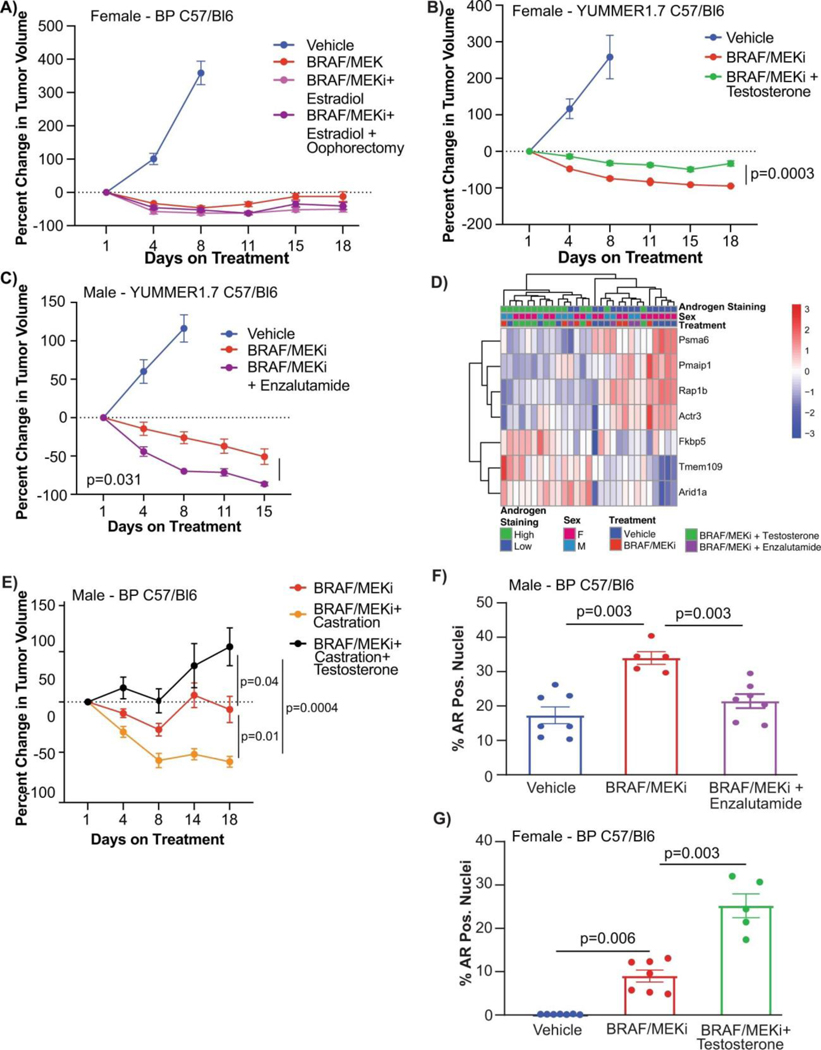
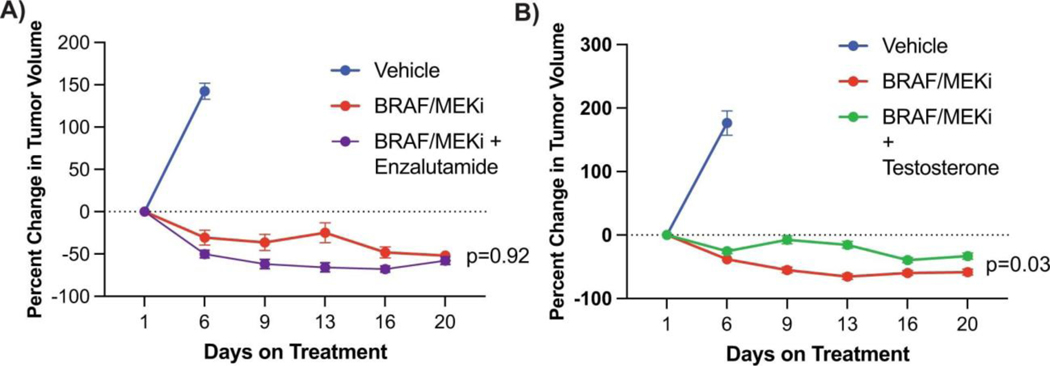
Based on these findings, we next conducted studies to test the hypothesis that pharmacologic manipulation of AR would be associated with differential responses to treatment with BRAF/MEK-targeted therapy. To do this, male and female mice were implanted with melanoma tumors and were treated with either vehicle or BRAF/MEK-targeted therapy, along with pharmacologic manipulation of AR activity (via systemic administration of testosterone or AR blockade) either alone or in combination (Figure 3A, Extended Data Figure 6C and 6D). We observed significantly impaired tumor control in male and female mice treated with testosterone and BRAF/MEK-targeted therapy compared to BRAF/MEK-targeted therapy alone (Figure 3B and 3C, p=0.021 male and p=<0.001 female, Extended Data Figure 7B, p=0.0003). With overall larger tumor volumes in testosterone treated mice (Figure 3D, p=0.031). AR blockade (via systemic administration of the anti-androgen enzalutamide) in combination with BRAF/MEK-targeted therapy demonstrated significantly improved tumor control compared to male and female mice treated with BRAF/MEK-targeted therapy alone (Figure 3E and 3F, p=0.018 and 0.003, Extended Data Figure 4C, p=0.003, Extended Data Figure 7C, p=0.031). Again, we saw improved tumor control across multiple studies and reduced tumor size in the AR blockade treated mice (Figure 3G, p=0.002, tumor volume curves for associated experiments Extended Figure 8). Importantly, these effects were abrogated by the addition of testosterone (Figure 3G, p=0.005).
Fig. 3: Modulation of AR activity is associated with differential response to BRAF/MEK-targeted therapy.
a, Schematic of mouse studies. b,c, The percentage change in BP tumour growth in male (b) or female (c) C57BL/6 mice treated with vehicle (n = 10 male and n = 10 female mice), BRAF/MEKi alone (n = 9 male and n = 10 female mice) or BRAF/MEKi + testosterone (n = 9 male and n = 10 female mice) (P = 0.021 (male) and P < 0.0001 (female)). d, Aggregate end-point tumour volumes showing larger volumes in mice treated with BRAF/MEKi + testosterone (n = 41) versus BRAF/MEKi (n = 49) (P = 0.031). n = 50 for vehicle. The squares represent male mice, and the dots represent female mice. e,f, The percentage change in BP tumour growth in male (n = 10 per group) (e) or female (n = 10 per group) (f) C57BL/6 mice treated with BRAF/MEKi alone or BRAF/MEKi + enzalutamide (P = 0.018 (male) and P < 0.003 (female)) or enzalutamide + testosterone (P < 0.0001 (male) and P < 0.0001 (female)). g, Aggregate end-point tumour volumes showing smaller volumes in mice treated with BRAF/MEKi + enzalutamide versus BRAF/MEKi (P = 0.002) and larger tumour volumes with the addition of testosterone (P = 0.005). The squares represent male mice and the dots represent female mice. Data from b and c are from the same experiment, and data from e and f are from separate experiments. h,i, Quantification of the percentage of AR+ nuclei by immunofluorescence analysis of BP tumour samples collected from male mice treated as indicated for 18 days (n = 5 per group; vehicle versus BRAF/MEKi (P = 0.0003), BRAF/MEKi versus BRAF/MEKi + testosterone (P = 0.001)) (h) or female mice treated for 15 days (i) (vehicle (n = 5) versus BRAF/MEKi (n = 5) (P = 0.04), BRAF/MEKi + testosterone (n = 5) versus BRAF/MEKi + enzalutamide (n = 9) (P = 0.02), BRAF/MEKi + enzalutamide versus BRAF/MEKi + enzalutamide + testosterone (n = 4) (P = 0.01)). P values were calculated using two-sided t-tests. Data are mean ± s.e.m. j, Androgen signalling score comparing mouse tumours with low (n = 14; female, vehicle; female, BRAF/MEKi; and male, BRAF/MEKi + enzalutamide) versus high (n = 15; male, vehicle; male, BRAF/MEKi; and female, BRAF/MEKi + testosterone) androgen staining (P = 0.0059). The squares represent male mice and the dots represent female mice. The box plots in d, g and j show the median (centre line), interquartile range (IQR) (box limits) and the most extreme point within 1.5 × IQR (whiskers). The tumour growth curves in b, c, e and f show mean ± s.e.m.
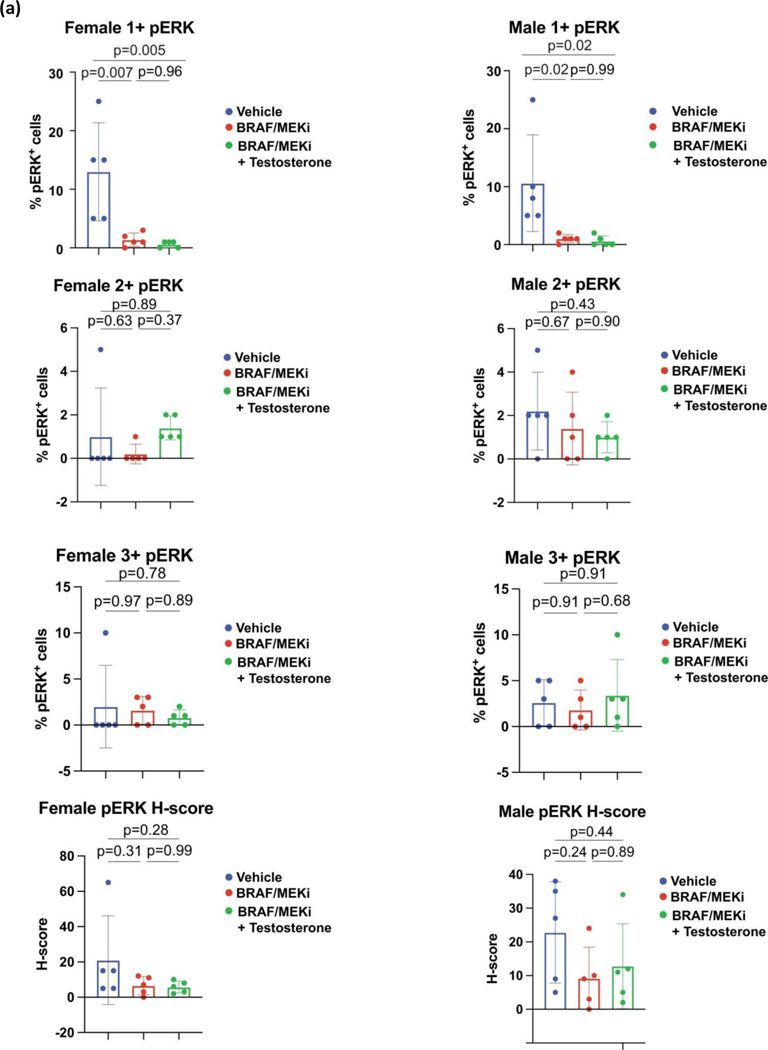
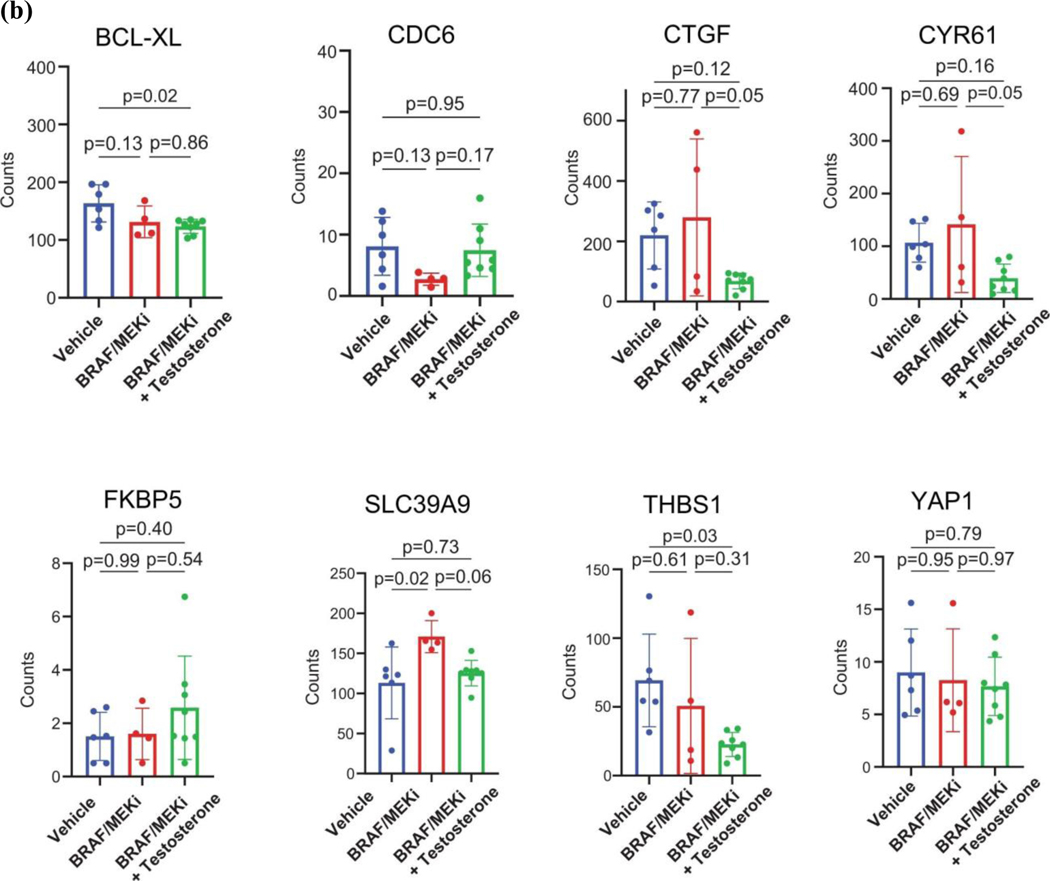
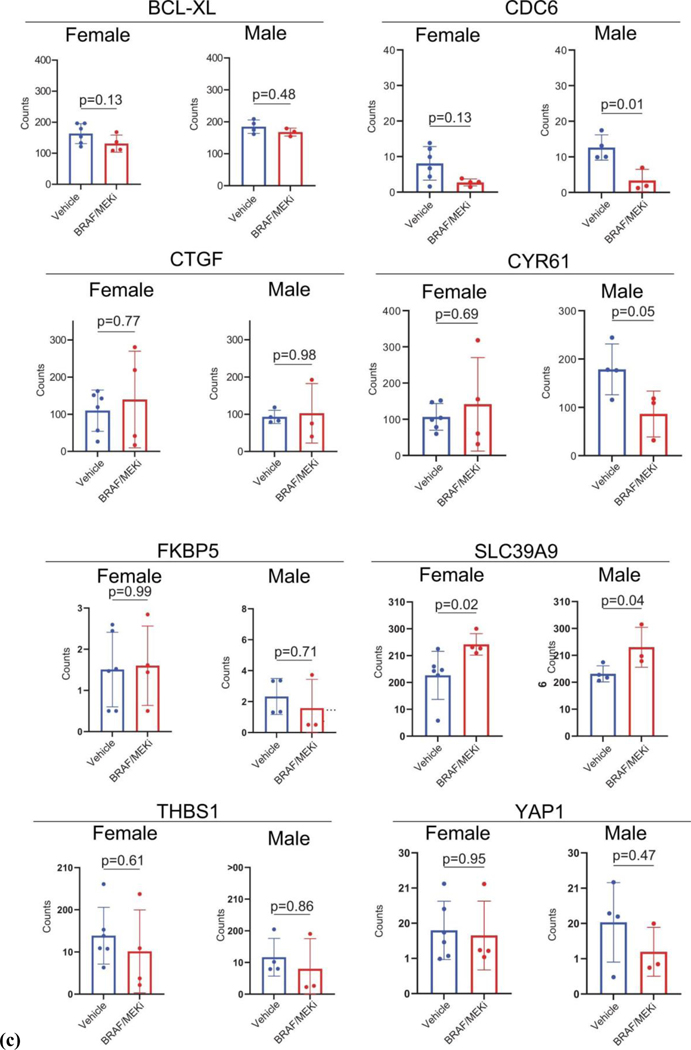
AR expression was assessed in male and female mice in each of these groups, demonstrating significantly increased AR expression in the setting of treatment with BRAF/MEK-targeted therapy (Figure 3H and 3I, p=0.0003 and p=0.04), as previously observed. Induction of AR expression via administration of testosterone along with BRAF/MEK-targeted therapy was also associated with significantly higher AR expression compared to BRAF/MEK alone in male and female mice (Figure 3H and 3I, p=0.001, p=0.06), which was corroborated by higher expression of androgen responsive genes in male mice and female mice with elevated AR activity (Figure 3J, p=0.0059, Extended Data Figure 7D)25. This suggests a potential feed forward loop whereby BRAF/MEK inhibition results in an increase in AR and AR-regulated targets. AR expression promotes proliferation of melanoma cells, and this response is magnified in testosterone rich environments24,26,27. Notably, treatment with enzalutamide was not associated with differences in the abundance of nuclear AR (Figure 3H and 3I), however, induction of AR activity upon exposure to exogenous testosterone together with BRAF/MEK-targeted therapy and enzalutamide increased AR expression compared to BRAF/MEK-targeted therapy and enzalutamide in male and female mice (Figure 3H and 3I, p=0.016, p=0.01, Extended Data Figure 7F and 7G). Consistent with this data, castration of tumor bearing mice significantly improved response to BRAF/MEK targeted therapy yet this benefit was lost upon testosterone administration (Extended Data Figure 7E and 8, p=0.0004). Notably, BRAF/MEK therapy resistance was independent of MAPK28–31 (Extended Data Figure 9), ZIP9/YAP1-MAPK32,33 (Extended Data Figures 9) or exogenous estrogen treatment (Extended Data Figure 7A)34,35.
Taken together, these data have important clinical implications. First, this data provides novel evidence that treatment with BRAF/MEK-targeted therapy is associated with an increase in AR expression in tumor cells promoting therapeutic resistance, and that AR blockade by anti-androgens such as enzalutamide may promote response to BRAF/MEK-targeted therapy in both males and females. Additionally, we provide provocative evidence that treatment with testosterone may promote resistance to BRAF/MEK-targeted therapy in males and females, which has important therapeutic implications as testosterone is widely used for multiple indications and perhaps its utilization needs to be carefully considered in patients with melanoma. These data have potential relevance in cancer beyond melanoma, as numerous malignancies are now being treated with BRAF/MEK-targeted therapy and other strategies targeting MAPK signaling pathways - warranting study of AR signaling in the setting of treatment with targeted therapy across cancer types and in the setting of other agents targeting MAPK and related pathways in cancer. Importantly, AR activity is also relevant in other forms of cancer treatment, as AR signaling has been shown to promote T cell exhaustion, and AR blockade may promote response to anti-PD-1 immune checkpoint blockade36. Findings from these studies have potential immediate clinical impact, though nuances exist, as treatment with AR blockade in the absence of hypothalamic suppression can result in increased AR signaling and higher testosterone levels37,38, and resistance to enzalutamide can occur through a number of different mechanisms including AR splice variants39,40 and glucocorticoid receptor expression41. Nonetheless, clinical trials investigating combination therapy strategies with AR blockade and BRAF/MEK-targeted therapy or immune checkpoint blockade are warranted with some trials currently underway (NCT02885649, NCT04926181, NCT01974765, NCT03207529).
Importantly, further insights are needed to better delineate the mechanism behind this, with opportunities to develop novel strategies to modulate AR signaling in a more targeted manner. This has the potential of abrogating side effects that are commonly experienced with androgen-deprivation therapy. There is also a critical need to better understand the impact of other hormones (estrogens, glucocorticoids) that are produced by the host or are modulated by gut microbiota in modulating responses to cancer treatment42–44. While the effect demonstrated here occurs on BRAF/MEK-targeted therapy and was not seen in mice treated with vehicle, sexual dimorphisms are noted across a variety of cancer therapies. Notably, these results overall have potential relevance beyond their impact on treatment with BRAF/MEK-targeted therapy or immune checkpoint blockade for patients with advanced cancer. AR signaling is implicated across numerous cancer types, and trials are underway incorporating AR blockade with conventional chemotherapy and other strategies in patients with advanced cancer (NCT02684227). Furthermore, AR signaling and targeting are also being studied in the context of carcinogenesis, with evidence that AR signaling may induce carcinogenesis and may promote increased invasiveness and metastasis45–49. Additional studies are needed to better understand the mechanism through which AR signaling promotes resistance to BRAF/MEK-targeted therapy and other cancer treatments, and should be interrogated in additional patient cohorts as well as in pre-clinical models - as important differences may exist and murine models are not a perfect surrogate to studying this in clinical cohorts. Further research is also needed to better understand the impact of androgens and other hormones on carcinogenesis, therapy response, and other disease conditions. Interrogation of clinical cohorts and studies in pre-clinical and other models will help us to better understand the relative contribution of sex hormones and related factors on cancer and other disease states, with opportunities to modulate these over a lifetime to promote overall precision health.
METHODS
Clinical cohorts:
Patients enrolled in the initial clinical trial cohort were previously described by Amaria et al.50 (NCT02231775). Briefly, patients ≥18 years old with histologically-proven clinical stage III or oligometastatic stage IV BRAFV600E/K melanoma deemed resectable by multidisciplinary consensus and measurable disease by RECIST 1.1 criteria were enrolled and those randomized to the experimental arm received 8 weeks of neoadjuvant dabrafenib (150mg PO BD) plus trametinib (2mg PO daily) prior to surgical resection, followed by up to 44 weeks of adjuvant dabrafenib and trametinib (neo-BRAF/MEKi)(n=12). Additional patients (n=20) from treated with neoadjuvant BRAF/MEKi along with an additional retrospective cohort of patients treated off protocol for logistic reasons with neoadjuvant BRAF/MEKi (n=16), dabrafenib (n=3), and encorafenib plus binimetinib (n=1) were included. Radiographic responses to neoadjuvant therapy were determined at week 8 prior to surgery and pathologic responses were determined by microscopic examination of the complete surgical specimen by a melanoma pathologist, including S0X10 immunostains when applicable to confirm presence or absence of viable melanoma cells. These patients were treated at the University of Texas MD Anderson Cancer Center and had tumor samples collected and analyzed under Institutional Review Board (IRB)-approved protocols. Of note, these studies were conducted in accordance with the Declaration of Helsinki and approved by the UT MD Anderson Cancer Center IRB.
The cohort of patients with metastatic disease treated at MD Anderson Cancer Center were collected and described in a retrospective analysis from a prospectively collected database51 with appropriate IRB approval. These patients were treated per under multidisciplinary consensus standard of care. Their measurable disease by RECIST 1.1 was measured on available CT scans and outcomes of progressive disease recorded. Of the 80 patients, 69 had measurable disease by RECIST 1.1 with 10 patients having had unmeasurable leptomeningeal or miliary lung recurrences, and another having his RECIST measured lesion resected per standard of care.
Patients enrolled in the COMBI-D trial were previously described by Robert et al. (NCT01584648)52. Patients with histologically-proven unresectable clinical stage III or stage IV BRAFV600E/K melanoma who met eligibility criteria were enrolled. Eligibility criteria included age greater than 18, measurable disease by RECIST 1.1 criteria, confirmed BRAF V600E/K mutation, ECOG performance status of 0 to 1, and no history of previous systemic treatment for advanced or metastatic cancer. Patients were randomized (1:1) to receive either oral dabrafenib (150mg twice daily) and oral trametinib (2mg one daily) or oral dabrafenib (150mg twice daily) and placebo. Disease baseline assessment was done by CT scan or MRI of the chest, abdomen and pelvis and skin lesions photographed. Follow-up was performed every 8 weeks for 52 weeks and every 12 weeks after until death, progression or withdrawal.
Patients enrolled in the METRIC trial53 (NCT01245062) had histologically-proven BRAF V600E/K mutant, unresectable clinical stage III or IV cutaneous melanoma and ECOG performance status of 0 or 1. Patients were randomized to receive either oral trametinib (2mg once daily) or IV chemotherapy consisting of either dacarbazine or paclitaxel every 3 weeks. For this study, data on only the patients who received trametinib were extrapolated and analyzed for sex-specific differences in clinical outcomes.
Patients enrolled in the NeoCombi trial (NCT01972347) were previously described by Long et al.54. Adults (age>18) with histologically-proven BRAF V600E/K mutant, resectable, clinical stage IIIB-C melanoma and ECOG performance status of 0 or 1 were enrolled for the study. Similar to the previous studies, the patient’s required RECIST 1.1 measurable disease. Patients received oral dabrafenib (150mg twice daily) and 2mg of trametinib (once daily) for 12 weeks prior to surgical resection and 40 weeks after surgery. CT and PET scans were performed prior to resection and pathologic responses were measured.
Overall survival and recurrence free survival within each clinical study were calculated using STATA (version 13.1) and R software (4.1.3). The study was conducted in accordance with the Declaration of Helsinki and approved by the medical ethics committee of MD Anderson Cancer Center. All subjects provided informed consent before their participation in the study.
RNA sequencing:
Tumor biopsies were obtained as feasible by punch or core biopsy prior to and during the neoadjuvant treatment period. Fresh-frozen tumor biopsy material was used for RNA sequencing library preparation. Total RNA was extracted from snap-frozen tumor specimens using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) following assessment of tumor content by a Pathologist, and macrodissection of tumor bed if required. RNA quality was assessed on an Agilent 2100 Bioanalyzer using the Agilent RNA 6000 Nano Chip with smear analysis to determine DV200 and original RNA concentration. Based on RNA quality, 40–80ng of total RNA from each sample then underwent library preparation using the Illumina TruSeq RNA Access Library Prep kit according to the manufacturer’s protocol. Barcoded libraries were pooled to produce final 10–12 plex pools prior to sequencing on an Illumina NextSeq 500 sequencer using one high-output run per pool of 76bp paired-end reads, generating 8 fastq files (4 lanes, paired reads) per sample.
RNA sequencing data processing:
RNA-seq FASTQ files were first processed through FastQC (v0.11.5)55, a quality control tool to evaluate the quality of sequencing reads at both the base and read levels. Reads having ≥15 contiguous low-quality bases (phred score <20) were removed from the FASTQ files prior to STAR 2-pass alignment (v2.5.3)56 with default parameters to generate one BAM file for each sequencing event. After that, RNA-SeQC (v1.1.8)57 was used to generate quality control metrics including read counts, coverage, and correlation. A matrix of Spearman correlation coefficients amongst all sequenced samples was subsequently generated by RNA-SeQC and after careful review the sequencing data generated from one library pool that showed poor correlation with other library pools from the same RNA sample was removed before sample-level merging of BAM files.
HTSeq-count (v0.9.1)58 tool was applied to aligned RNA-seq BAM files to count how many aligned reads overlapped with the exons of each gene. The raw read counts generated from HTSeq-count were normalized into fragments per kilobase of transcript per million mapped reads (FPKM) using the RNA-seq quantification approach suggested by the bioinformatics team of NCI Genomic Data Commons (GDC) [https://docs.gdc.cancer.gov/Data/Bioinformatics_Pipelines/Expression_mRNA_Pipeline/]. Briefly, FPKM normalizes read count by dividing it by the gene length and the total number of reads mapped to protein-coding genes using a calculation described below:
RCg, number of reads mapped to the gene; RCpc: number of reads mapped to all protein-coding genes; L, length of the gene in base pairs (calculated as the sum of all exons in a gene). The FPKM values were then log2-transformed for further downstream analyses.
Differential gene expression and pathway enrichment analysis
DESeq2 (v3.6) was identify differentially expressed genes between patients that achieved an MPR (defined as Y) and those that did not (defined as N). The Wilcoxon rank-sum test was used to identify significantly different differentially expressed genes between the MPR “Y” and “N” groups. A cutoff FDR q-value of <0.05 was applied to select the most significant DEGs. For pathway analysis, the curated AR gene sets59 was downloaded. Gene set enrichment analysis (GSEA) was applied and pathway scores were calculated for each sample using the fgsea software package60. The pathway scores were then compared between the MPR “Y” and “N” groups. For preclinical analyses there were 244 homologues within the 300 gene androgen signature utilized in analyses of clinical samples. Differential expression was studied between the “high testosterone” group comprised of male vehicle, male BRAF/MEKi, and female BRAF/MEKi with testosterone groups and the “low testosterone” group comprised of female vehicle, female BRAF/MEKi, and male BRAF/MEKi with enzalutamide.
AR signature score
Here, for sample s, ARS denotes its AR signature score; G is number of AR genes; EXPg is the Fragments Per Kilobase of transcript per Million mapped reads (FPKM) of AR gene g. Student t-test is applied, showing significant ARS different between sample group MPR Y and N.
Animals and xenograft studies:
Female or male C57BL/6 mice (strain code: 0000664, purchased from Jackson Lab), aged between 9 to 14 weeks, and weighing approximately 20 to 25 g were used for in vivo studies. Female or male CD-1 nude mice (strain code: 086, purchased from Charles River Laboratories), aged between 10 to 11 weeks, and weighing approximately 25–40 g were used for the immunodeficient model in vivo studies. Specific age of mice used in each experiment is indicated in the corresponding figure legends. Animal health was monitored daily by observation and sentinel blood sample analysis. Animal experiments were conducted in accordance with the Guideline of IACUC at MDACC.
BRAFV600E Ren−/− murine melanoma cell lines (BP) were previously generated by our group. YUMMER1.7 cells were a generous gift from Dr. Michael Davis from the University of Texas MD Anderson Cancer Center. Both tumor lines were periodically authenticated and tested for contamination by mycoplasma. BP cells were scaled up in DMEM culture media supplemented with 10% FBS, harvested, and prepared so that each mouse received 0.8×106 cells in 0.2 mL PBS. For YUMMER1.7 cells, mice received between 0.5×106 −2×106 cells. Cells were implanted subcutaneously in the right flank of each mouse. For some male mice, physical castration was required and performed two weeks before treatment or before cell implantation. For some female mice, physical oophorectomy was required and performed one week before treatment or before cell implantation. Testosterone pellets (5mg/day, Innovative Research of America) were implanted subcutaneously in the left flank one week before treatment with vehicle or BRAF/MEK-targeted therapy. Trametinib (Chemietek) at 1 mg/kg and dabrafenib (Chemietek) at 30 mg/kg were suspended at concentrations as needed in an aqueous vehicle containing 0.5% Hydroxypropylmethylcellulose and 0.2 % Tween 80 in distilled water and adjusted to pH 8.80 with diluted NaOH solution. Enzalutamide (Chemietek) at 10 mg/kg was formulated in 1% carboxymethyl cellulose (Sigma), 0.1% Tween-80, 5% DMSO.
BP and YUMMER1.7 tumors were monitored by calliper before randomly sorting and dividing into experimental groups (n=10 mice per group for efficacy, or n=5–7 for the acute pharmacodynamic (PD) 3-day study in the case of YUMMER1.7). Tumor dimensions for the in vivo experiments were performed by a dedicated team of technicians. As per animal welfare guidelines, mice were euthanized if tumors grew larger than 4000 mm3. Though these technicians were not blinded to the specific treatment groups, they did not have a working knowledge of the expected outcomes in these studies limiting but not eliminating potential bias. Treatment was started from day 14 to 17 days post-implantation depending on mouse strain and sex. Vehicle controls and compound treatments were given orally using a sterile 1- mL syringe and 18-gavage needle for the times noted for each study. Dosing was 5 hours apart between administration of trametinib + dabrafenib and enzalutamide for these specific treatment groups.
Tumor volume was calculated using the following formula: [L X (W2)/2] (in which L + length of tumors; W =width of tumor). Tumor and plasma were harvested 4 hours after the last dose. Tumors were snap of frozen and the plasma was divided for monitoring drug concentrations and hormone levels.
Tumor volumes are represented as raw volumes, percent change in tumor volume, as well as aggregate tumor volumes from independent studies. Percent change in tumor volume was calculated from study day 0 (Tumor day 14 to 17).
Quant-seq library construction and sequencing.
1000 ng or 500 ng of DNase-treated RNA samples were converted to cDNA using a QuantSeq 3’ mRNA according to the manufacturer’s protocol (Lexogen, Vienna, Austria). The libraries were amplified with 12 or 13 PCR Cycles and purified using the provided Lexogen. The purified libraries were quantified using a Kapa library quantification kit (KAPA biosystems) and loaded on NextSeq 500 Sequencer (Illumina, San Diego, CA) at a final concentration of 2.6 pM to perform cluster generation, followed by 1×76 bp sequencing on NextSeq 500 Sequencer (Illumina).
Generation of AR-KO CRISPR BP cell line
Three guide sequences were designed to target a 100 bp region of Exon 1 using HorizonDiscovery’s CRISPR Design Tool. Guide 1 (gacttgggtagtctacatgg AGG) was cloned into pLentiCRISPR.v2 according to addgene lentiCRISPRv2 and lentiGuide oligo cloning protocol for the purpose of pool selection. Guide 2 (gcttgatacgggcgtgtggat GGG) and Guide 3 (ctggagaacccattggacta CGG) were ordered as crRNA’s (IDT).
The Neon electroporation system was used to transfect ribonucleoprotein (RNP) complex + plasmid into the BP cells. The crRNAs were rehydrated to 200 nM and pooled in equal volume (3 μL each) for annealing with Alt-R CRISPR-Cas9 tracrRNA-ATTO 550 (IDT; 1075928); 95°C x 5 min, slowly cool to 10°C at 0.1 C/sec. 5 μL of crRNA:tracrRNA were combined with 5 μL of Alt-R S.p. HiFi Cas9 nuclease (IDT; 1081060) at room temperature for 10 min to generate the RNP complex. Electroporation Enhancer (2 μL) (IDT; 1075916) and 2 μg of plasmid were added to the RNP. During the annealing reaction, 1×106 BP cells were pelleted at 600xg for 3 min. The total volume of RNP + Plasmid + Electroporation Enhancer was transferred to an aspirated cell pellet. Next, 95 μL of R buffer was added to the cells and they were gently resuspended to single cells for an immediate 100 μL electroporation reaction. The Neon settings for the BP cells were two 30 ms pulses at 1150 volts. The cells were transferred to a single well of a 6 well plate and allowed to recover in growth media. After 24 hrs, the media was replaced and 1.5 μg/μL of Puromycin for a 48-hr selection. The cells were allowed to recover from selection for 48 hrs before single-cell clone selection. After clones were selected and expanded, AR protein was analyzed by Western Blot to confirm AR knockout. Briefly, cells were lysed for 30 min at 4 °C in 150 μL RIPA buffer with phosphatase and protease inhibitors. Protein (15 μg) was run on 10% SDS Page (Bio-Rad) and transferred overnight onto PDF membranes using TRIS Glycine Methanol buffer. After blocking for 1 hour at room temperature in 5% milk diluted in TSBS-T, AR antibody (Abcam) was incubated at 1:2000 dilution for 24 hrs. GAPDH was used as a loading control. The lentiviral expression vectors pLV-105 were purchased from Genecopeia to express either GFP as a transduction control. Lentivirus was generated using standard protocols and psPAX2 and pMD2.G as the packaging vectors. BP cells were transduced at 90% efficiency with viral supernatant and selected for 48 hours with 2 μg/mL puromycin.
Generation of AR-NT-KO BP Control Cell Line
The non-targeting control sequences targeted Luciferase and LacZ. The Luciferase specific target sequences (ACAACTTTACCGACCGCGCC) was cloned into pLentiCRISPR_v2. In addition, 2 crRNAs were designed and ordered Luciferase (ACAACTTTACCGACCGCGCC) and LacZ (CCCGAATCTCTATCGTGCGG). The transfection and selection of the sgRNA into the BP cells were the same as the KO cells. This quality metric is reported in Extended Figure Figure 10.
Measuring testosterone levels from plasma:
Testosterone quantification was determined using Agilent’s UHPLC Infinity II and 6495 triple quadrupole mass spectrometer, and MassHunter workstation software (8.0.8.23.5). Briefly, plasma samples containing 0.5 ng/mL 13C3-testosterone (Cerilliant, Round Rock, TX) were extracted with tert-butyl methyl ether (Sigma 34875), dried, and derivatized using hydroxylamine hydrochloride (Sigma 431362). The recovered ketoxime steroids were reconstituted in methanol/water (1:1 v/v) and injected into the Infinity II UHPLC. Ketoxime steroids were separated using a Chromolith reverse phase column (RP-18 endcapped 100–2mm, Sigma 152006) and introduced into a JetStream source (Agilent) for triple quadrupole analysis. Data were analyzed and quantified using MassHunter software (Agilent) and GraphPad PRISM (version 8.0) software was used to graph and perform statistics (two-sided Students t-test)61,62.
Immunofluorescence
FFPE blocks were sectioned (5 μm), mounted on charged microscope slides (Leica 38002092), and dried at 37°C overnight. Slides were then baked at 60°C for 1 hour in an oven (Biocare DRY2008US), deparaffinized in 3 changes of xylene, and then rehydrated in 3 changes of 100% ethanol followed by a series of 95%, 70%, and 50% ethanol and distilled water (5 min. each). Antigen retrieval (10 mM sodium citrate, pH 6.0 with 0.05% Tween 20) was performed by heating slides to 95°C for 15 minutes in a microwave (Biogenex EZ Retriever System v.3) followed by a 30 minute cool down at room temperature. Slides were then washed with TBST (Thermo TA-999-TT). The area around each section was traced with a PAP pen (Sigma Aldrich Z672548), blocked with Background Sniper (Biocare BS966) for 10 minutes, and then washed with TBST.
Primary antibody (rabbit anti-Androgen Receptor [EPR1535(2)], Abcam ab133273, 1:300, diluted in Dako S3022) was incubated overnight at 4°C, followed by a TBST wash. Secondary antibody (goat anti-rabbit Alexa-647 conjugate, Thermo A32733, 1:400, diluted in Fluorescence antibody Diluent, “FAD”, Biocare FAD901L) was added and allowed to incubate for 1 hour at room temperature, followed by a TBST wash. The sequence that the antibody was raised against was a human sequence and had been validated previously and used in numerous references13–16. This antibody was validated first in mouse by testing both AR-expressing (e.g. testis, prostate) and non-expressing tissues (e.g. surrounding tissue, fat) and then in human using KO cell lines (via western blot and immunofluorescence) as well as in AR-expressing and non-expressing human patient-derived xenograft (PDX) tumors. A sequential incubation with fluorophore-conjugated primary antibody was performed (rabbit anti-Sodium Potassium ATPase [EP1845Y] Alexa-488 conjugate, Abcam ab197713, 1:500, were diluted in FAD) for 1 hour at room temperature, followed by a TBST wash. Finally, slides were incubated with DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride (5 mg/mL stock in DMF, Thermo D1306) at 0.25 μg/mL (diluted in TBST) for 10 minutes at room temperature. Slides were then washed with a series of TBST, followed by TBS and then distilled H2O. Excess liquid was removed and slides were mounted with ProLong Diamond Gold (Life Tech P36930) and allowed to harden overnight. For patient samples, a TSA-amplification of AR signal was performed. Briefly, following antigen retrieval/tissue tracing with PAP pen, performed as above, slides were incubated with treated with Bloxall to remove endogenous peroxidase (Vector Labs SP-6000) for 10 minutes at RT. Slides were then rinsed in water (30 seconds) and TBST (30 seconds). Slides were blocked with 2.5% Normal Horse Serum (Vector Labs MP-7401 Component) for 25 minutes at RT and then block was tapped off. Slides were then blocked with Opal-specific PE/Diluent/Block (AKOYA ARD1001EA) for 10 minutes at RT and then block was tapped off. Primary antibody (rabbit anti-Androgen Receptor [EPR1535(2)], Abcam ab133273, 1:300, diluted in PE Diluent AKOYA ARD1001EA) was incubated overnight at 4°C, followed by a TBST wash for 3 minutes. Anti-Rabbit secondary HRP polymer was then added to slides (Impress MP-7401) for 25 minutes at RT after which slides were washed in TBST. Opal signal was generated by adding Opal 570 fluorophore (AKOYA FP1488001KT diluted 1:200 in 1x Plus Amplification Diluent AKOYA FP1609) for 10 minutes at RT and then washed in TBST. All following steps e.g. staining with the membrane marker (Abcam ab197713), DAPI, and coverslip mounting were identical to the steps described above.
IF Image acquisition and Analysis
Slides were imaged on a Vectra 3 or a Vectra Polaris using the A UPlanSApo 10x/0.40 air objective first. Images were acquired using all available channels with the Vectra software (3.0.5) and the raw data was saved as “.qptiff” files. Regions of interest (ROIs) were created using PhenoChart (1.0.10) and these areas were then imaged again at 20X magnification on the Vectra. 20x images were then spectrally unmixed using inForm software and saved as Component TIFFs. Files were opened in QuPath software, channels were split, and saved individually (or merged) as TIFFs. An APP was created in Visiopharm to segment cells and assess intensity of AR immunofluorescence. Percent positive cells was calculated for each sample. Raw analyzed data was exported as a.CSV file and graphed/statistics run using GraphPad PRISM 8 software. A t-test test was performed to test significance.
Extended Data
Extended Data Fig. 1: Expanded analysis of clinical cohorts.
A) Rates of major pathologic response (MPR) based on menopausal status of female patients within the neoadjuvant cohort (p = 0.79) by logistic regression. B) Waterfall plot of percent change in tumour size by cross-sectional imaging in the neoadjuvant cohort (p = 0.54). C) Waterfall plot of percent change in tumour size by cross-sectional imaging of the pooled neoadjuvant and metastatic cohorts (p = 0.02).
Extended Data Fig. 2: Analysis of published cohorts of BRAF/MEK inhibited patients.
A) Schema of metastatic BRAF inhibitor monotherapy clinical cohort of patients treated with targeted therapy and their clinical outcomes studied. B) Progression free survival by sex in the metastatic BRAF inhibitor monotherapy clinical cohort (n = 212, hazard ratio 1.2 CI [0.88–1.6] p = 0.241, by Kaplan-Meier method). C) Overall survival by sex in the metastatic BRAF inhibitor monotherapy clinical cohort (n = 212, hazard ratio 1.32 CI [0.95–1.82] p = 0.095, by Kaplan-Meier method). D) Schema of metastatic MEK inhibitor monotherapy clinical cohort of patients treated with targeted therapy and their clinical outcomes studied. E) Progression free survival by sex in the metastatic MEK inhibitor monotherapy clinical cohort (n = 206, hazard ratio 1.35 CI [1.01–1.81] p = 0.043, by Kaplan-Meier method). F) Overall survival by sex in the metastatic MEK inhibitor monotherapy clinical cohort (n = 206, hazard ratio 1.61 CI [1.19–2.18] p = 0.002, by Kaplan-Meier method). G) Schema of second neoadjuvant clinical cohort of patients treated with targeted therapy and their clinical outcomes studied. H) Major pathologic response defined as ≤ 10% viable tumour in females versus males (p = 0.92, by Chi-squared). I) Recurrence free survival by sex in the second neoadjuvant cohort (n = 40, hazard ratio 1.16, 95% CI [0.36–2.07], p = 0.733, by Kaplan-Meier method). J) Kaplan Meier survival curves of overall survival by sex of melanoma patients abstracted with The Cancer Genome Atlas (p = 0.373 by univariate analysis, p = 0.369 controlling for age at diagnosis and stage). K) Paired clinical samples of circulating testosterone prior to initiation and after treatment with BRAF/MEK targeted therapy (p = 0.9453 and 0.2036 respectively by two-sided Student’s t-test). L) AR staining pre-treatment in males (blue) and females (pink) by MPR (p = 0.72, by two-sided unpaired t-test).
Extended Data Fig. 3: Transcriptomic analysis of clinical specimens.
A) Heatmap of AR signature genes of on-treatment specimens demonstrating upregulation of AR signature pathways. B) Volcano plot of differentially expressed genes after whole transcriptomic analysis of on-treatment with BRAF/MEK inhibition clinical specimens. Vertical dotted blue lines represent log foldchange greater than 2. Horizontal dotted blue line represents q < 0.05. Genes with log fold change greater than 5 and q < 0.05 are labelled. C) Heatmap of AR signature genes of pre-treatment specimens of AR signature pathways. D) Volcano plot of differentially expressed genes after whole transcriptomic analysis of pre-treatment with BRAF/MEK inhibition clinical specimens. E) Androgen signalling score comparing clinical samples achieving an MPR and those not achieving an MPR for those on treatment (ON, <MPR n = 2, MPR n = 4), at the time of resection (SURG, <MPR n = 9, MPR = 7; p = 0.066), aggregate of those on treatment and surgically resected (ONandSURG, <MPR = 11, MPR = 11; p = 0.011), and samples collected prior to treatment (PRE, <MPR n = 4, MPR n = 5; p = 0.95); groups were compared using a two-sided Student’s t test. Box plot represents the median bar with the box bounding interquartile range (IQR) and whiskers to the most extreme point within 1.5 x IQR.
Extended Data Fig. 4: Murine model of melanoma validates a sexually dimorphic response and suggests AR activity as a mechanism of resistance.
A-B) Percent change in tumour volume for male and female C57BL/6 mice implanted subcutaneously with BP cells that were treated with Vehicle or BRAF/MEKi (n = 10 mice per group; A – mice aged 9 weeks, B - mice aged 12 weeks). Results from the second and third repeats of this experiment are shown in A and B, respectively (p = 0.039 and p = 0.45). C) Percent change in tumour volume in BP injected C57BL/6 male mice treated with vehicle in the presence or absence of BRAF/MEKi with endocrine modulation through androgen blockade with enzalutamide or castration (mice aged 14 weeks). All tumour growth curves were compared by ANOVA with multiple comparisons (n = 10/group except BRAF/MEKi + castration where n = 9; p = 0.003 BRAF/MEKi vs BRAF/MEKi + Enzalutamide; p = 0.031 BRAF/MEKi vs BRAF/MEKi + Castration). D-E) Percent change in tumour volume for AR-KO BP tumours in female (D) (p = 0.99) and male (E) (p = 0.98) CD-1 mice treated with vehicle or BRAF/MEKi in combination with either testosterone or enzalutamide, respectively (n = 10 mice/group; aged 11 weeks). All tumour growth represented as mean + SEM and p-values were calculated using ANOVA with multiple comparisons.
Extended Data Fig. 5: Mechanistic and Validations Murine Studies Tumour Volume Curves.
A) Tumour volumes for male and female C57BL/6 mice implanted subcutaneously with YUMMER 1.7 cells treated with Vehicle or BRAF/MEK inhibition (p = 0.06 between male and female BRAF/MEKi; 30 mpk dabrafenib and 1 mpk trametinib, PO, QD). n = 10 mice per group, aged 12–13 weeks. B-D) Percent change in tumour volume for male and female C57BL/6 mice implanted subcutaneously with BP (BRAFV600E, PTEN−/−) cells (p = 0.003, 0.26, and 0.93) between male and female BRAF/MEKi. Mice were treated as in A. n = 10 mice per group; aged 12–13 weeks. The experiment was performed in triplicate with each experiment represented above. E) Tumour volumes for male and female CD-1 nude mice implanted and treated as in A (p = 0.01 between male and female BRAF/MEKi). n = 10 mice per group; aged 11 weeks. F) Tumour volumes of male and female CD-1 mice with AR-KO BP tumours treated with vehicle or BRAF/MEKi (p = 0.317 between male and female BRAF/MEKi); n = 10/group; aged 11 weeks. All tumour growth represented as mean + SEM and p-values were calculated using ANOVA with multiple comparisons.
Extended Data Fig. 6: AR gene and protein expression analysis of preclinical models and serum testosterone measurements of clinical specimens.
A) AR gene expression of murine BP tumours from male and female mice treated with either Vehicle or BRAFi/MEKi (female Vehicle vs Female BRAF/MEKi treated, p = 0.005, male Vehicle vs male BRAF/MEKi treated, p = 0.16) p-values were calculated using two-sided Student’s t-test B) AR immunofluorescence staining of samples from female and male mice treated with Vehicle, BRAF/MEKi, BRAF/MEKI + Testosterone (females, vehicle vs BRAF/MEKi, p = 0.005) and BRAF/MEKi + enzalutamide (males, vehicle vs BRAF/MEKi, p = 0.16). p-values were calculated by two-sided Student’s t-test. C) Plasma testosterone levels for male mice across treatment groups. Decreased plasma testosterone was noted in both the castration group as compared to Vehicle (p = 0.039) as well as the BRAF/MEKi + castration group as compared to the BRAF/MEKi group (p = 0.017). p-values were calculated using a Kruskal Wallis test. D) Plasma testosterone levels for female mice across treatment groups. Increased plasma testosterone was noted in the Vehicle + testosterone group as compared to the BRAF/MEKi group (p = 0.007) and vehicle group (p = 0.007). Similarly increased testosterone was noted in the BRAF/MEKi + testosterone group as compared to the vehicle group (p = 0.009) and BRAF/MEKi group (p = 0.009). No other associations were significant p < 0.05 by Kruskal Wallis test. E) AR immunofluorescence staining of AR-KO BP tumour samples from female and male mice treated with either Vehicle or BRAFi/MEKi. p-values were calculated by Student’s t-test. Histograms in A and B represent mean + SD whereas C and D represent mean + SEM.
Extended Data Fig. 7: Modulation of AR activity is associated with differential response to BRAF/MEK inhibition.
A) Percent change in tumour volume in BP injected C57BL/6 female mice treated with vehicle in the presence or absence of BRAF/MEKi with endocrine modulation with either estradiol or estradiol and oophorectomy (n = 10/group; mice aged 14 weeks). No significant effects exist within either cohort as calculated by ANOVA. B) Percent change in tumour volume for female C57BL/6 mice implanted subcutaneously with YUMMER 1.7 cells treated with BRAF/MEKi or BRAF/MEKi + testosterone (n = 10/group; aged 13 weeks, p = 0.0003 between BRAF/MEKi and BRAF/MEKi + testosterone). C) Percent change in tumour volume in YUMMER1.7 injected into C57BL/6 male mice treated with vehicle in the presence or absence of BRAF/MEKi with endocrine modulation with enzalutamide (n = 10/group; mice aged 14 weeks, p = 0.031). D) Heatmap of differentially expressed androgen responsive genes between high versus low testosterone groups q < 0.05. Groups by androgen staining levels, sex, and treatment. E) Percent change in BP tumour volume in male C57BL/6 mice treated with vehicle in the presence or absence of BRAF/MEKi with physical castration or BRAF/MEKi with castration and exogenous testosterone (n = 10/group; aged 14 weeks, p = 0.01 for BRAF/MEKi + castration versus BRAF/MEKi, p = 0.04 for BRAF/MEKi versus BRAF/MEKi + castration + testosterone, and p = 0.0004 for BRAF/MEKi + castration vs BRAF/MEKi + castration + testosterone). F) Quantification of the percent of AR+ nuclei by immunofluorescence in BP tumours from male mice treated with vehicle (n = 7), BRAF/MEKi (n = 5), or BRAF/MEKi + enzalutamide (n = 7) (p = 0.003 between BRAF/MEKi and BRAF/MEKi + enzalutamide). p-values were calculated using two-sided Student’s t-test. G) Quantification of the percent of AR+ nuclei by immunofluorescence in BP tumours from female mice treated with vehicle (n = 7), BRAF/MEKi (n = 7), BRAF/MEKi + testosterone (n = 5) (p = 0.006 between vehicle and BRAF/MEKi and p = 0.003 between BRAF/MEKi and BRAF/MEKi + testosterone in female mice). p-values were calculated using two-sided Student’s t-test. Tumour growth curves in panels A-C and E represent mean + SEM. Histograms in panels F and G represent mean + SEM. All tumour growth curves were compared by ANOVA with multiple comparisons.
Extended Data Fig. 8: Interventional Murine Studies Tumour Volumes.
A) Tumour volumes curves of BP tumour growth in male mice treated with vehicle (n = 10), BRAF/MEKi alone (n = 9) or BRAF/MEKi in combination with testosterone (n = 9) (p = 0.12). B) Tumour volumes curves of BP tumour growth in female mice treated with vehicle, BRAF/MEKi alone or BRAF/MEKi in combination with testosterone (p = 0.003). (n = 10 mice/group). C) Tumour volume curves of BP tumour growth in male mice treated with BRAF/MEKi alone or BRAF/MEKi in combination with AR blockade (p = 0.02) or AR blockade with testosterone (p = 0.07) (n = 10 mice/group). D) Tumour volume curves of BP tumour growth in female mice treated with BRAF/MEKi alone or BRAF/MEKi in combination with AR blockade (p = 0.002) or AR blockade with testosterone (p = 0.34) (n = 10 mice/group). E) Tumour volume curves of BP AR KO tumour growth in CD-1 nude female mice treated with vehicle in the presence or absence of BRAF/MEKi with endocrine modulation (mice aged 14 weeks). BRAF/MEKi vs BRAF/MEKi + testosterone, p = 0.99; (n = 10 mice/group). F) Tumour volume curves of AR KO BP injected CD-1 nude male mice treated with vehicle in the presence or absence of BRAF/MEKi or BRAF/MEKi + enzalutamide (p = 0.98) (n = 10 mice/group). G) Tumour volume curves of YUMMER 1.7 tumour growth in male mice treated with vehicle, BRAF/MEKi or BRAF/MEKi + AR blockade (p = 0.12) (n = 10 mice/group) H) Tumour volume curves of BP tumour growth in C57BL/6 male mice treated with BRAF/MEKi, BRAF/MEKi + castration (p = 0.01) or BRAF/MEKi + castration + testosterone (p = 0.0005) (n = 10 mice/group). I) Change in tumour volume in BP injected C57BL/6 male mice treated with vehicle in the presence or absence of BRAF/MEKi with endocrine modulation through either androgen blockade, androgen blockade with testosterone, estradiol, or castration (mice aged 14 weeks) (n = 10 mice/group). J) Tumour volume curves of YUMMER 1.7 tumour growth in female mice treated with vehicle, BRAF/MEKi or BRAF/MEKi + testosterone (p = 0.0005) (n = 10 mice/group). K) Change in tumour volume in BP injected C57BL/6 female mice treated with vehicle in the presence or absence of BRAF/MEKi with endocrine modulation with either estradiol or estradiol and oophorectomy (n = 10 mice/group; mice aged 14 weeks). L) Change in tumour volume of BP tumours injected into CD-1 nude male mice and treated with vehicle, BRAF/MEKi, or BRAF/MEKi in combination with AR blockade (n = 10 mice/group, mice aged 11 weeks, BRAF/MEKi vs BRAF/MEKi + AR blockade p = 0.92). M) Change in tumour volume of BP tumours injected into CD-1 nude female mice and treated with vehicle, BRAF/MEKi, or BRAF/MEKi in combination with testosterone (n = 10 mice/group, mice aged 11 weeks, BRAF/MEKi vs BRAF/MEKi + testosterone p = 0.03). All tumour growth curves represent mean + SEM and were compared by ANOVA with multiple comparisons.
Extended Data Fig. 9: Effect of BRAF/MEKi on pERK, ZIP9/YAP1 associated transcripts and YAP1 associated transcripts in BP tumours.
A) Staining and quantification of phosphor-ERK in BP tumours of female and male mice on treated with vehicles, BRAF/MEK inhibition, or BRAF/MEK inhibition + testosterone. Histogram represent mean + SD. Differences were calculated using one-way ANOVA (n = 5/group). B) Staining and quantification of ZIP9/YAP1 associated transcripts in BP tumours of female and male mice on treatment with vehicles (n = 6), BRAF/MEK inhibition (n = 4), or BRAF/MEK inhibition + testosterone (n = 8). Histogram represent mean + SD. Differences were calculated using one-way ANOVA. C) Staining and quantification of YAP1 associated transcripts in BP tumours of female and male mice on treatment with vehicles (n = 6 females, 4 males) or BRAF/MEK inhibition (n = 4 females, 3 males. Histogram represent mean + SD. Differences were calculated using one-way ANOVA.
Extended Data Fig. 10: Non-targeting control line in BRAF/PTEN mice.
A) Volume of BP tumours established from the BP cell line engineered to express a non-targeting CRISPR control for male CD-1 nude mice treated with vehicle, BRAF/MEKi, or BRAF/MEKi in combination with AR blockade (n = 10 mice/group) B) Volume of BP tumours established from the BP cell line engineered to express a non-targeting CRISPR control for female CD-1 nude mice treated with vehicle, BRAF/MEKi, or BRAF/MEKi in combination with testosterone (n = 10 mice/group, p = 0.01).). All tumour growth curves represent mean + SEM and were compared by ANOVA with multiple comparisons.
Supplementary Material
ACKNOWLEDGEMENTS
JAW is supported by NIH (1 R01 CA219896-01A1), Melanoma Research Alliance (4022024), American Association for Cancer Research Stand Up To Cancer (SU2C-AACR-IRG-19-17), and MD Anderson Cancer Center’s Melanoma Moon Shots Program.
JLM is supported by the Melanoma Research Alliance, an American Society of Clinical Oncology and Conquer Cancer Foundation Career Development Award, the Elkins Foundation, Seerave Foundation, Rising Tide Foundation, the Mark Foundation, an MD Anderson Cancer Center (MDACC) Melanoma SPORE Developmental Research Program Award, and the MD Anderson Physician Scientist Program and acknowledges the Transdisciplinary Research in Energetics and Cancer Research Training Workshop R25CA203650 and the MDACC Center for Energy Balance in Cancer Prevention and Survivorship.
APC is supported by the CPRIT Research Training Program at MD Anderson Cancer Center (RP170067), The U.S. Department of State, Bureau of Educational and Cultural Affairs, and the Fulbright Franco-Américaine Commission.
MCA is supported by a National Health and Medical Research Council of Australia CJ Martin Early Career Fellowship (#1148680).
BH was supported by National Institutes of Health T32 CA 009599 and the MD Anderson Cancer Center support grant (P30 CA016672).
MGW is supported by National Institutes of Health T32 CA 009599 and the MD Anderson Cancer Center support grant (P30 CA016672).
AJL has served on advisory boards and/or consulted for: AbbVie, AstraZeneca, Bayer, BMS, Deciphera Pharmaceuticals, Foghorn Therapeutics, GSK, Invitae, Illumina, Iterion Therapeutics, Merck, Novartis, Nucleai, Paige.AI, Pfizer, Roche/Genentech, and ThermoFisher.
RGW is supported by National Institutes of Health T32 CA 009599 and the MD Anderson Cancer Center support grant (P30 CA016672).
GM is supported by the National Institute of Health (1F32CA260769-01).
MAD is supported by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the AIM at Melanoma Foundation, the NIH/NCI 1 P50 CA221703-02, Cancer Fighters of Houston, the Anne and John Mendelsohn Chair for Cancer Research, and philanthropic contributions to the Melanoma Moon Shots Program of MD Anderson
GOO is supported by National Institutes of Health T32 CA 009599 and the MD Anderson Cancer Center support grant (P30 CA016672).
GVL is consultant advisor for Agenus Inc, Amgen Inc, Array Biopharma Inc, Boehringer Ingelheim International GmbH, Bristol Myers Squibb, Evaxion Biotech A/S, Hexal AG (Sandoz Company), Highlight Therapeutics S.L., Merck Sharpe & Dohme (Australia) Pty Limited, Novartis Pharma AG, OncoSec Medical Australia, Pierre Fabre, Provectus, Qbiotics Group Limited, Regeneron Pharmaceuticals Inc,
SNW serves as a consultant for Agenus, AstraZeneca, Caris, Clovis Oncology, Eisai, EQRX, GSK, ImmunoGen, Lilly, Merck, Mersana, Roche/Genentech and Zentalis. She receives research support to her institution from AstraZeneca, Bayer, Bio-Path, Clovis Oncology, Cotinga Pharmaceuticals, GSK, Mereo, Novartis, OncXerna, Roche/Genentech, and Zentalis.
The University of Texas MD Anderson Cancer Center Science Park Next-Generation Sequencing (NGS) Facility is supported by CPRIT Core Facility Support Awards (CPRIT RP170002).
Footnotes
DISCLOSURE
J.A.W. is an inventor on a US patent application (PCT/US17/53.717); reports compensation for speaker’s bureau and honoraria from Imedex, Dava Oncology, Omniprex, Illumina, Gilead, PeerView, MedImmune, and Bristol-Myers Squibb (BMS); serves as a consultant/advisory board member for Roche/Genentech, Novartis, AstraZeneca, GlaxoSmithKline (GSK), BMS, Merck, Biothera Pharmaceuticals, and Micronoma. J.A.W. holds stock options from Micronoma.
VG is a co-inventor on a US patent application (PCT/US17/53 717) submitted by The University of Texas MD Anderson Cancer Center that covers methods to enhance checkpoint blockade therapy by the microbiome. V.G. has served as a consultant for Microbiome DX, and reports honoraria from ExpertConnect and Kansas Society of Clinical Oncology.
JLM serves as a consultant for Merck and in advisory board for Bristol-Myers Squibb.
ZAC is a current employee of and owns stock in AstraZeneca
MCA reports advisory board participation and honoraria from MSD Australia outside the current work; research funds to institution from MSD Australia and BMS Australia outside the current work; and is a named co-inventor on patent applications relating to Methods and compositions for treating cancer (WO2020106983A1) unrelated to the current work, and Methods for improving sex-dimorphic responses to targeted therapy in melanoma (US20200164034A1), related to the current work. RNA receives research support and advisory boards for Novartis, BMS, lovance. Research support from Genentech CL serves as a consultant/advisory board for Merck, Sharp & Dohme, Exelixis, Bayer, Amgen, reports honoraria from Merck, Sharp & Dohme, Bayer, Amgen and clinical grants from Janssen, ORIC Pharmaceuticals, Novartis, Aragon Pharmaceuticals. MAD is an advisory board member/consultant for BMS, Novartis, Array, Roche/Genentech, GSK, Sanofi-Aventis, and ABM Therapeutics. PI of research grant to institution from Astrazeneca, Roche/Genentech, GSK, Myriad, Oncothyreon, Sanofi-Aventis, and ABM Therapeutics.
PAF serves on the SAB for Scorpion Therapeutics
AEM has a sponsored research agreement with Medlmmune/AstraZeneca.
SNW serves as a consultant for Agenus, AstraZeneca, Clovis Oncology, Eisai, GSK/Tesaro, Merck, Pfizer, Roche/Genentech and Zentalis. She receives research support to her institution from AstraZeneca, Bayer, Clovis Oncology, Cotinga Pharmaceuticals, GSK/Tesaro, Mereo, Novartis and Roche/Genentech.
All other authors report no COI directly relevant to this work.
MATERIALS AND CORRESPONDENCE
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contacts, Timothy Heffernan (TPHeffernan@mdanderson.org), Joseph Marszalek (JRMarszalek@mdanderson.org), or Jennifer A. Wargo (jwargo@mdanderson.org).
DATA AVAILABILITY
The additional datasets generated during and/or analyzed during the current study of Clinical Trial NCT02231775 are now available in the European Genome-phenome Archive repository (EGAS00001006196). Other datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1.Long GV et al. Factors predictive of response, disease progression, and overall survival after dabrafenib and trametinib combination treatment: a pooled analysis of individual patient data from randomised trials. Lancet Oncol 17, 1743–1754, doi: 10.1016/S1470-2045(16)30578-2 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Robert C. et al. Five-Year Outcomes with Dabrafenib plus Trametinib in Metastatic Melanoma. The New England journal of medicine 381, 626–636, doi: 10.1056/NEJMoa1904059 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Flaherty KT et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 367, 107–114, doi: 10.1056/NEJMoa1203421 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Long GV et al. Neoadjuvant dabrafenib combined with trametinib for resectable, stage IIIB-C, BRAF(V600) mutation-positive melanoma (NeoCombi): a single-arm, open-label, single-centre, phase 2 trial. Lancet Oncol 20, 961–971, doi: 10.1016/S1470-2045(19)30331-6 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Leung JK & Sadar MD Non-Genomic Actions of the Androgen Receptor in Prostate Cancer. Front Endocrinol (Lausanne) 8, 2, doi: 10.3389/fendo.2017.00002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinto JA et al. Gender and outcomes in non-small cell lung cancer: an old prognostic variable comes back for targeted therapy and immunotherapy? ESMO Open 3, e000344, doi: 10.1136/esmoopen-2018-000344 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capone I, Marchetti P, Ascierto PA, Malorni W. & Gabriele L. Sexual Dimorphism of Immune Responses: A New Perspective in Cancer Immunotherapy. Front Immunol 9, 552, doi: 10.3389/fimmu.2018.00552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clocchiatti A. et al. Androgen receptor functions as transcriptional repressor of cancer-associated fibroblast activation. J Clin Invest 128, 5531–5548, doi: 10.1172/JCI99159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pequeux C. et al. Stromal estrogen receptor-alpha promotes tumor growth by normalizing an increased angiogenesis. Cancer Res 72, 3010–3019, doi: 10.1158/0008-5472.CAN-11-3768 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Zhao L. et al. Pharmacological activation of estrogen receptor beta augments innate immunity to suppress cancer metastasis. Proc Natl Acad Sci U S A 115, E3673–E3681, doi: 10.1073/pnas.1803291115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothenberger NJ, Somasundaram A. & Stabile LP The Role of the Estrogen Pathway in the Tumor Microenvironment. Int J Mol Sci 19, doi: 10.3390/ijms19020611 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribeiro MPC, Santos AE & Custodio JBA The activation of the G protein-coupled estrogen receptor (GPER) inhibits the proliferation of mouse melanoma K1735-M2 cells. Chem Biol Interact 277, 176–184, doi: 10.1016/j.cbi.2017.09.017 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Qi J. Therapy resistance by splicing: can the androgen receptor teach us about BRAF? Pigment Cell Melanoma Res 25, 293–294, doi: 10.1111/j.1755-148x.2012.01000.x (2012). [DOI] [PubMed] [Google Scholar]
- 14.Wang Y. et al. Androgen receptor promotes melanoma metastasis via altering the miRNA-539–3p/USP13/MITF/AXL signals. Oncogene 36, 1644–1654, doi: 10.1038/onc.2016.330 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Natale CA et al. Activation of G protein-coupled estrogen receptor signaling inhibits melanoma and improves response to immune checkpoint blockade. Elife 7, doi: 10.7554/eLife.31770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smalley KS Why do women with melanoma do better than men? Elife 7, doi: 10.7554/eLife.33511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marzagalli M. et al. Estrogen Receptor beta in Melanoma: From Molecular Insights to Potential Clinical Utility. Front Endocrinol (Lausanne) 7, 140, doi: 10.3389/fendo.2016.00140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amaria RN et al. Neoadjuvant plus adjuvant dabrafenib and trametinib versus standard of care in patients with high-risk, surgically resectable melanoma: a single-centre, open-label, randomised, phase 2 trial. Lancet Oncol 19, 181–193, doi: 10.1016/S1470-2045(18)30015-9 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Stacchiotti S. et al. High-grade soft-tissue sarcomas: tumor response assessment--pilot study to assess the correlation between radiologic and pathologic response by using RECIST and Choi criteria. Radiology 251, 447–456, doi: 10.1148/radiol.2512081403 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Eisenhauer EA New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). European Journal of Cancer 45, 228–247, doi: 10.1016/j.ejca.2008.10.026 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Mendiratta P. et al. Genomic strategy for targeting therapy in castration-resistant prostate cancer. J Clin Oncol 27, 2022–2029, doi: 10.1200/JC0.2008.17.2882 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Schmidt K. et al. The lncRNA SLNCR Recruits the Androgen Receptor to EGR1-Bound Genes in Melanoma and Inhibits Expression of Tumor Suppressor p21. Cell Rep 27, 2493–2507 e2494, doi: 10.1016/j.celrep.2019.04.101 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma M. et al. Sustained androgen receptor signaling is a determinant of melanoma cell growth potential and tumorigenesis. J Exp Med 218, doi: 10.1084/jem.20201137 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou ZX, Lane MV, Kemppainen JA, French FS & Wilson EM Specificity of ligand-dependent androgen receptor stabilization: receptor domain interactions influence ligand dissociation and receptor stability. Mol Endocrinol 9, 208–218, doi: 10.1210/mend.9.2.7776971 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Jin HJ, Kim J. & Yu J. Androgen receptor genomic regulation. Transl Androl Urol 2, 157–177, doi: 10.3978/j.issn.2223-4683.2013.09.01 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morvillo V. et al. Atypical androgen receptor in the human melanoma cell line IIB-MEL-J. Pigment Cell Res 8, 135–141, doi: 10.1111/j.1600-0749.1995.tb00654.x (1995). [DOI] [PubMed] [Google Scholar]
- 27.Aguirre-Portoles C. et al. ZIP9 Is a Druggable Determinant of Sex Differences in Melanoma. Cancer Res 81, 5991–6003, doi: 10.1158/0008-5472.CAN-21-0982 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang C. et al. TRIM36, a novel androgen-responsive gene, enhances anti-androgen efficacy against prostate cancer by inhibiting MAPK/ERK signaling pathways. Cell Death Dis 9, 155, doi: 10.1038/s41419-017-0197-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S. et al. Activation of MAPK Signaling by CXCR7 Leads to Enzalutamide Resistance in Prostate Cancer. Cancer Res 79, 2580–2592, doi: 10.1158/0008-5472.CAN-18-2812 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M. et al. Targeting AR-Beclin 1 complex-modulated growth factor signaling increases the antiandrogen Enzalutamide sensitivity to better suppress the castration-resistant prostate cancer growth. Cancer Lett 442, 483–490, doi: 10.1016/j.canlet.2018.11.008 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu H. et al. Combination of sorafenib and enzalutamide as a potential new approach for the treatment of castration-resistant prostate cancer. Cancer letters 385, 108–116, doi: 10.1016/j.canlet.2016.10.036 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Kuser-Abali G, Alptekin A, Lewis M, Garraway IP & Cinar B. YAP1 and AR interactions contribute to the switch from androgen-dependent to castration-resistant growth in prostate cancer. Nat Commun 6, 8126, doi: 10.1038/ncomms9126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin L. et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nature genetics 47, 250–256, doi: 10.1038/ng.3218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubin JB et al. Sex differences in cancer mechanisms. Biol Sex Differ 11, 17, doi: 10.1186/s13293-020-00291-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boese AC, Kim SC, Yin KJ, Lee JP & Hamblin MH Sex differences in vascular physiology and pathophysiology: estrogen and androgen signaling in health and disease. Am J Physiol Heart Circ Physiol 313, H524–H545, doi: 10.1152/ajpheart.00217.2016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan e. a. Suppressing androgen receptor activity in T cells promotes effective checkpoint blockade. (2021). [Google Scholar]
- 37.Balzano S. et al. The effect of androgen blockade on pulsatile gonadotrophin release and LH response to naloxone. Clin Endocrinol (Oxf) 27, 491–499, doi: 10.1111/j.1365-2265.1987.tb01178.x (1987). [DOI] [PubMed] [Google Scholar]
- 38.Kerrigan JR, Veldhuis JD & Rogol AD Androgen-receptor blockade enhances pulsatile luteinizing hormone production in late pubertal males: evidence for a hypothalamic site of physiologic androgen feedback action. Pediatr Res 35, 102–106, doi: 10.1203/00006450-199401000-00021 (1994). [DOI] [PubMed] [Google Scholar]
- 39.Maughan BL & Antonarakis ES Enzalutamide in chemo-naive castration-resistant prostate cancer: effective for most but not for all. Asian J Androl 16, 807–808, doi: 10.4103/1008-682X.137680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL & Tindall DJ Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res 68, 5469–5477, doi: 10.1158/0008-5472.CAN-08-0594 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arora VK et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 155, 1309–1322, doi: 10.1016/j.cell.2013.11.012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cross TL, Kasahara K. & Rey FE Sexual dimorphism of cardiometabolic dysfunction: Gut microbiome in the play? Mol Metab 15, 70–81, doi: 10.1016/j.molmet.2018.05.016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin AM, Sun EW, Rogers GB & Keating DJ The Influence of the Gut Microbiome on Host Metabolism Through the Regulation of Gut Hormone Release. Front Physiol 10, 428, doi: 10.3389/fphys.2019.00428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sudo N. Microbiome, HPA axis and production of endocrine hormones in the gut. Adv Exp Med Biol 817, 177–194, doi: 10.1007/978-1-4939-0897-4_8 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Gaballa R. et al. Exosomes-Mediated Transfer of Itga2 Promotes Migration and Invasion of Prostate Cancer Cells by Inducing Epithelial-Mesenchymal Transition. Cancers 12, doi: 10.3390/cancers12082300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ricke EA et al. Androgen hormone action in prostatic carcinogenesis: stromal androgen receptors mediate prostate cancer progression, malignant transformation and metastasis. Carcinogenesis 33, 1391–1398, doi: 10.1093/carcin/bgs153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scatena C. et al. Androgen receptor expression inversely correlates with histological grade and N stage in ER(+)/PgR(low) male breast cancer. Breast Cancer Res Treat 182, 55–65, doi: 10.1007/s10549-020-05682-7 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Wang D. & Tindall DJ Androgen action during prostate carcinogenesis. Methods Mol Biol 776, 25–44, doi: 10.1007/978-1-61779-243-4_2 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Xia N, Cui J, Zhu M, Xing R. & Lu Y. Androgen receptor variant 12 promotes migration and invasion by regulating MYLK in gastric cancer. J Pathol 248, 304–315, doi: 10.1002/path.5257 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Amaria RN et al. Neoadjuvant plus adjuvant dabrafenib and trametinib versus standard of care in patients with high-risk, surgically resectable melanoma: a single-centre, open-label, randomised, phase 2 trial. Lancet Oncol 19, 181–193, doi: 10.1016/S1470-2045(18)30015-9 (2018). [DOI] [PubMed] [Google Scholar]
- 51.McQuade JL et al. Association of body-mass index and outcomes in patients with metastatic melanoma treated with targeted therapy, immunotherapy, or chemotherapy: a retrospective, multicohort analysis. The Lancet Oncology 19, 310–322, doi: 10.1016/s1470-2045(18)30078-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robert C. et al. Five-Year Outcomes with Dabrafenib plus Trametinib in Metastatic Melanoma. The New England journal of medicine 381, 626–636, doi: 10.1056/NEJMoa1904059 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Robert C. et al. Five-year outcomes from a phase 3 METRIC study in patients with BRAF V600 E/K-mutant advanced or metastatic melanoma. Eur J Cancer 109, 61–69, doi: 10.1016/j.ejca.2018.12.015 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Long GV et al. Neoadjuvant dabrafenib combined with trametinib for resectable, stage IIIB-C, BRAF(V600) mutation-positive melanoma (NeoCombi): a single-arm, open-label, single-centre, phase 2 trial. Lancet Oncol 20, 961–971, doi: 10.1016/S1470-2045(19)30331-6 (2019). [DOI] [PubMed] [Google Scholar]
- 55.FastQC: A Quality Control Tool for High Throughput Sequence Data [Online] (2010).
- 56.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeLuca DS et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532, doi: 10.1093/bioinformatics/bts196 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anders S, Pyl PT & Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, doi: 10.1093/bioinformatics/btu638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mendiratta P. et al. Genomic strategy for targeting therapy in castration-resistant prostate cancer. J Clin Oncol 27, 2022–2029, doi: 10.1200/JC0.2008.17.2882 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Korotkevich G. et al. Fast gene set enrichment analysis. bioRxiv, 060012, doi: 10.1101/060012 (2021). [DOI] [Google Scholar]
- 61.Efstathiou E. et al. Enzalutamide in Combination with Abiraterone Acetate in Bone Metastatic Castration-resistant Prostate Cancer Patients. Eur Urol Oncol 3, 119–127, doi: 10.1016/j.euo.2019.01.008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maity SN et al. Targeting of CYP17A1 Lyase by VT-464 Inhibits Adrenal and Intratumoral Androgen Biosynthesis and Tumor Growth of Castration Resistant Prostate Cancer. Sci Rep 6, 35354, doi: 10.1038/srep35354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The additional datasets generated during and/or analyzed during the current study of Clinical Trial NCT02231775 are now available in the European Genome-phenome Archive repository (EGAS00001006196). Other datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.