Abstract
Background
Carbapenem-resistant Enterobacterales are among the most serious antimicrobial resistance (AMR) threats. Emerging resistance to polymyxins raises the specter of untreatable infections. These resistant organisms have spread globally but, as indicated in WHO reports, the surveillance needed to identify and track them is insufficient, particularly in less resourced countries. This study employs comprehensive search strategies with data extraction, meta-analysis and mapping to help address gaps in the understanding of the risks of carbapenem and polymyxin resistance in the nations of Africa.
Methods
Three comprehensive Boolean searches were constructed and utilized to query scientific and medical databases as well as grey literature sources through the end of 2019. Search results were screened to exclude irrelevant results and remaining studies were examined for relevant information regarding carbapenem and/or polymyxin(s) susceptibility and/or resistance amongst E. coli and Klebsiella isolates from humans. Such data and study characteristics were extracted and coded, and the resulting data was analyzed and geographically mapped.
Results
Our analysis yielded 1341 reports documenting carbapenem resistance in 40 of 54 nations. Resistance among E. coli was estimated as high (> 5%) in 3, moderate (1–5%) in 8 and low (< 1%) in 14 nations with at least 100 representative isolates from 2010 to 2019, while present in 9 others with insufficient isolates to support estimates. Carbapenem resistance was generally higher among Klebsiella: high in 10 nations, moderate in 6, low in 6, and present in 11 with insufficient isolates for estimates. While much less information was available concerning polymyxins, we found 341 reports from 33 of 54 nations, documenting resistance in 23. Resistance among E. coli was high in 2 nations, moderate in 1 and low in 6, while present in 10 with insufficient isolates for estimates. Among Klebsiella, resistance was low in 8 nations and present in 8 with insufficient isolates for estimates. The most widespread associated genotypes were, for carbapenems, blaOXA-48, blaNDM-1 and blaOXA-181 and, for polymyxins, mcr-1, mgrB, and phoPQ/pmrAB. Overlapping carbapenem and polymyxin resistance was documented in 23 nations.
Conclusions
While numerous data gaps remain, these data show that significant carbapenem resistance is widespread in Africa and polymyxin resistance is also widely distributed, indicating the need to support robust AMR surveillance, antimicrobial stewardship and infection control in a manner that also addresses broader animal and environmental health dimensions.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13756-023-01220-4.
Introduction
Antimicrobial resistance (AMR) is of growing concern as multidrug resistant organisms (MDRO) become more prevalent globally, undermining the efficacy of medicines needed for the treatment of infections and threatening patient safety and economic wellbeing [1]. Carbapenem-resistant Enterobacterales (CRE) infections are of particular concern as treatment options are highly limited [2] with carbapenems considered critical drugs for treatment of infections with documented or suspected resistance to alternative antimicrobials. Healthcare environments are the dominant source of human exposure to MDRO such as CRE [3] but exposure may also occur in the community, where organisms spread not only after transfer from patients exposed in healthcare settings, but also through contact with food, animals, and the environment [4–8].
Resistance to carbapenems arises through intrinsic or acquired mechanisms [3]. Acquired resistance [9–14] typically occurs due to carbapenemase enzymes encoded on plasmids or other genetic elements that are readily transferred among organisms [2, 15]. Major resistance determinants present worldwide include expression of Class A Klebsiella pneumoniae carbapenemases (KPC), Class B metallo-β-lactamases such as New Delhi metallo-β-lactamases (NDM), Verona integron-encoded metallo-β-lactamases (VIM), Imipenemase metallo-β-lactamases (IMP), and Class D oxacillinase β-lactamases (OXA), and alterations in outer membrane proteins (OMP) [15]. The polymyxin antibiotics, including polymyxin E (colistin) and polymyxin B, hereon in referred to as polymyxin(s), are polycationic peptides widely used until the 1970s, when largely abandoned as less toxic antibiotics became available [16, 17]. Currently, as one of few antimicrobial classes effective against CRE, polymyxins have regained importance. Determinants of acquired polymyxin resistance include transferable plasmid encoded mobile colistin resistance (mcr) genes as well as chromosomally encoded genes such as mgrB, phoP/phoQ, and pmrA/pmrB [16, 18]. The risk of organisms acquiring both carbapenem and polymyxin resistance is alarming as it severely limits treatment options. While rare to date, such dual resistance has been increasingly documented [19–22].
Despite the association of MDRO with excess morbidity, mortality and costs, major gaps exist in surveillance, particularly in under-resourced areas [23]. The WHO Global Action Plan to Tackle AMR (GAP-AMR) provides a roadmap for the treatment and prevention of resistant infections [24]. Since 2014, WHO has encouraged collection of data on carbapenem susceptibility and has published the limited available data in reports of the Global Antimicrobial Resistance Use and Surveillance System (GLASS) [1, 25–27]. In 2018, noting that only 7 of 47 WHO Africa nations had reported data on CRE to WHO [12, 28–30], we developed search and metanalytic approaches to utilize data from diverse sources to estimate and map carbapenem resistance and related genotypes in the WHO Africa region. We were able to identify and analyze data from 31 of 47 nations [2] documenting carbapenem-resistant Escherichia coli or Klebsiella spp. in 22, typically at low to moderate levels [2]. We subsequently refined these approaches to characterize carbapenem and polymyxin resistance and their concerning overlaps in Southeast Asia [31].
Since our initial study, reporting on carbapenem resistance in Africa has increased [32–34] but comprehensive analyses are not available. Information on polymyxin resistance is more limited but recent reviews document mcr plasmids as causes of resistance in several African nations [35, 36]. The 2020 WHO GLASS report included only 10 of 54 nations reporting data on carbapenems and just 4 on polymyxins. Given these persistent data gaps there is a major unmet need for information to inform medical and public health investments, strategies and practices. We applied our previously-developed approaches to locate available useful data on polymyxin/colistin resistance and related genes, as well as to broadly update analyses of carbapenem resistance to reflect emerging data and extend the scope of study to all continental Africa. The results provide a comprehensive database and maps of carbapenem and polymyxin resistance in Africa, documenting the significant ongoing spread of both throughout the continent.
Methods
Literature review and other data sources
Three comprehensive Boolean searches were constructed and utilized to query scientific and medical databases (Embase, Global Health, PubMed and Web of Science). Grey literature sources including ProMED-mail [37], ResistanceMap [38] and HealthMap [39] were also examined for data from African nations, as described [2, 31]. Data were further supplemented by review and, where meeting criteria, extraction of relevant primary data located based on citations identified through included studies or from other referenced reviews and meta-analyses, as well as directly utilizing data from World Health Organization GLASS reports [1, 25–27] and author correspondence. As detailed previously, for nations with fewer than 4 reports from these sources, manual Google Scholar searches were conducted and additional sources such as African Journals Online, Bioline International and Global Index Medicus were hand-searched for relevant documents [2].
Search strategy
As described [2, 31], search strategies were designed and executed to capture data describing susceptibility or resistance, and/or related genotypic findings, of Escherichia coli and Klebsiella isolates from humans. The searches (search operators capitalized) generally followed the structure of place (e.g. terms for Africa OR country names) AND terms for AMR (including general OR specific AMR terms OR synonym drug terms) AND species/mechanisms (including resistance enzymes and plasmid-mediated genotypes). As detailed (Additional file 1) the search strings also contained MeSH terms to optimize sensitivity while enhancing specificity. The first database search updated data from the WHO Africa Region nations (United Nations geoscheme) through 31 December 2019 [2]. The second search identified data published from 1 January 1996 to 31 December 2019 on carbapenem susceptibility or resistance for seven African countries not included in our original report (Djibouti, Egypt, Libya, Morocco, Tunisia, Somalia, and Sudan) [2]. The final search for 1 January 1996 to 31 December 2019 identified data for polymyxin susceptibility or resistance for all African nations.
Exclusion and inclusion criteria and data collection
Two authors (DMV and AYB) screened search result titles and excluded irrelevant materials. Remaining studies were examined for relevant information regarding carbapenem and/or polymyxin(s) susceptibility and/or resistance amongst E. coli and Klebsiella isolates from humans. Minimum criteria for inclusion in the study database were description of study design and sampling process, characteristics of participants, places and dates of data collection and use of recognized, standardized testing methods at the time of performance. Studies not including these data elements were excluded. Data were extracted and coded from studies meeting criteria and any coding questions resolved through mutual agreement amongst researchers.
Underlying data from 313 reports in our previous dataset [2] on carbapenem resistance in WHO Africa nations (from searches through 31 June 2017) were also incorporated into the current dataset. If a newly found study reported data duplicative of or overlapping with that included in earlier analyses, only the original report was included. We also examined the results of database searches for similar reports (e.g. in terms of country, dates and species) to detect potentially duplicative or overlapping reporting of the same data. In circumstances where searches yielded duplicative or overlapping data, the most complete study was utilized unless both included unique data, in which case any additional details from the second report were included on a separate line of the database without duplicate reporting. When a study provided potentially important findings, but substantive uncertainties were present, authors were contacted, when possible, for clarification. Outreach to authors was made for 167 studies and responses obtained for 85, of which 55 were included in the manuscript (see acknowledgements).
Database construction, definitions and data entry
A structured Microsoft Excel Version 1808 (Microsoft Corp., Redmond, WA, USA) template with predefined attributes was developed and utilized, as described [2, 31]. Data extracted included study characteristics, patient populations, and phenotypic and genotypic carbapenem and polymyxin resistance. Study type was classified as clinical laboratory-based, case series, outbreak, or surveillance, and populations were classified as from acute or chronic healthcare facilities, community-based, travelers or unknown [31]. Selected subpopulations, if studied, were defined by clinical attributes (e.g. pregnant, intensive care unit, clinical syndrome), travel status (e.g. immigrants, refugees) and/or occupation (e.g. farmers, students, healthcare workers). WHO age classification was utilized where applicable, unless age was otherwise classified by authors [31, 40].
Reports on subsets of laboratory isolates selected based on their resistance properties were coded noting the selection criteria utilized (e.g. ESBL or CRE). If results of susceptibility testing to multiple carbapenems were reported, all data were entered in the database with the value for the drug with the highest percentage resistance then used to represent overall carbapenem resistance, so long as the numbers of isolates tested for each drug were similar. On occasions where the differences in total numbers of isolates tested against different carbapenems were large (e.g. an order of magnitude), we used results from the drug with the most isolates tested to represent resistance. Isolates reported as having intermediate susceptibility were classified as resistant. For studies presenting disaggregated susceptibility results (e.g. by ESBL status), data were reaggregated to reflect resistance in the entire original set of isolates. Documentation of specific carbapenem or polymyxin resistance-associated genotypes was recorded whenever available. For quality control, all database entries were checked and confirmed by an additional reviewer.
Data analyses
Defining the presence of resistance and/or specific resistance genotypes
Any report of at least one carbapenem and/or polymyxin-resistant E. coli or Klebsiella isolate, or an isolate with a resistance-associated genotype, signified the presence of resistance in that nation. This included findings of phenotypic resistance or resistance inducing genotypes in any isolate, whether from population-based studies or narrower studies of outbreaks, case series, highly selected subpopulations, or from studies of isolates themselves selected for known resistance to any antibiotic(s) including carbapenem and polymyxin.
Crude national resistance proportion estimates
Analysis was conducted using R version 3.5.2 (R Core Team, 2014). To estimate crude national resistance proportions, data from studies with a minimum number of isolates tested (20 for carbapenems and 10 for polymyxin, given the paucity of available data) that were deemed to originate from reasonably ‘generalizable’ populations (i.e. representative of individuals in overall healthcare populations), were aggregated and analyzed across studies. These estimates excluded any data from outbreaks or from studies reporting resistance in certain highly selected subpopulations (burn injury, oncology or transplantation) that typically have levels of resistance significantly greater than general acute-care populations. Similarly, data reporting resistance among organisms selected based on their known resistance to any antibiotic were not considered generalizable and therefore not included in resistance estimates.
To better reflect recent resistance, crude resistance proportions were calculated using data available on isolates collected from 2010 onward. If the total of generalizable E. coli or Klebsiella isolates tested for susceptibility to carbapenems or polymyxin(s) from 2010 onward was at least 100, we calculated that nation’s mean and, across qualifying studies, median resistance proportions using R v.3.5.2. For nations with at least 100 generalizable isolates of E. coli or Klebsiella, a crude estimated median resistance category was assigned consistent with prior studies [2, 31] as follows: not detected, low (< 1%), moderate (1–5%) or high (> 5%). If the total of generalizable isolates for a nation was less than 100, a category of either ‘Insufficient isolates – Resistance detected’ or ‘Insufficient isolates – Resistance not detected’ was assigned.
Geocoding and mapping
ArcGIS Desktop 10.6 (ESRI, Redlands, CA, USA) was used to map median resistance proportions and genotypes at the national level. Sample origin was geocoded at facility level, or to the closest local administrative unit such as City or State/Province using Google Maps.
Data sharing
The supplementary material, including search strings (Additional file 1) and outputs (Additional file 2), explanation of data elements extracted for analyses (Additional file 3), and all study data (Additional file 4) are available through Mendeley.
Results
Data characteristics
The searches yielded 8631 studies of which 1191 passed initial screening and 749 then met inclusion criteria. Three were in French, all others were in English. Because a given study may contain data on more than one organism, sets of isolates, or populations, the 749 study documents yielded a total of 1479 unique data reports together providing data on carbapenem and/or polymyxin resistance from 48 of 54 African countries. Three nations (Egypt, Nigeria and South Africa) accounted for 647 (43.7%) of all reports in the database. In contrast, no relevant reports were identified from 6 nations and nearly 30 nations each accounted for less than 1% of reports.
Selected general attributes of the data reports are displayed in Table 1. Six hundred and ninety-two (46.8%) reported on E. coli, while 787 (53.2%) were on Klebsiella spp. More than half of the data reports (67.5%) were from clinical laboratory-based studies, while 22.6% were from case series, 8.2% from surveillance and 2% from outbreaks. Aside from 34.6% of reports of multiple sample sources, most reports were of isolates from urine (23.3%) or blood (20.6%). Subject ages were reported as all (30%), adults (20.2%) and children (13.1%) or as unknown (34.2%). The majority (83.4%) of reports included isolates collected in acute healthcare settings, others included community-based settings (29.0%), chronic health-care facilities (0.5%), unknown healthcare settings (4.2%), travelers (1.0%) and unknown sources (1.7%).
Table 1.
Key data attributes
| Age group | Number (%) | Population type | Number (%) | Study type | Number (%) | Specimen type | Number (%) | Specimen type | Number (%) | Species | Number (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Adolescent | 37 (2.5%) | Community | 429 (29.0%) | Case series | 334 (22.6%) | Ascitic fluid | 3 (0.2%) | Pus | 17 (1.2%) | E. coli a | 692 (46.8%) |
| Adult | 299 (20.2%) | HC-acute | 1234 (83.4%) | Clinical lab | 998 (67.5%) | Aspirate | 1 (0.1%) | Rectal | 1 (0.1%) | K. spp. b | 787 (53.2%) |
| All | 444 (30.0%) | HC-long | 8 (0.5%) | Outbreak | 29 (2.0%) | BAL | 1 (0.1%) | Respiratory | 6 (0.4%) | ||
| Child | 194 (13.1%) | HC-unknown | 62 (4.2%) | Surveillance | 121 (8.2%) | Bedsore | 1 (0.1%) | Sperm | 1 (0.1%) | ||
| Elderly | 16 (1.1%) | Travelers | 15 (1.0%) | Blood | 304 (20.6%) | Sputum | 9 (0.6%) | ||||
| Infant | 56 (3.8%) | Unknown | 25 (1.7%) | Catheter | 2 (0.1%) | Stool | 184 (12.4%) | ||||
| Neonate | 61 (4.1%) | Cervicovaginal | 2 (0.1%) | Tissue | 7 (0.5%) | ||||||
| Unknown | 506 (34.2%) | CSF | 8 (0.5%) | Umbilical | 1 (0.1%) | ||||||
| Ear | 6 (0.4%) | Unknown | 62 (4.2%) | ||||||||
| Endocervical | 2 (0.1%) | Urine | 345 (23.3%) | ||||||||
| Endotracheal | 3 (0.2%) | Vaginal | 7 (0.5%) | ||||||||
| ETA | 4 (0.3%) | Wound | 69 (4.7%) | ||||||||
| Gastric fluid | 2 (0.1%) | ||||||||||
| Hand | 3 (0.2%) | ||||||||||
| IV fluid | 1 (0.1%) | ||||||||||
| Multiple | 512 (34.6%) | ||||||||||
| Nasal | 7 (0.5%) | ||||||||||
| Otitis media | 2 (0.1%) | ||||||||||
| Peritoneal fluid | 11 (0.7%) | ||||||||||
| Peritoneum | 1 (0.1%) |
Numbers and % of 1479 unique data reports including the indicated subgroups. In some categories total is > 1479 as reports may contain multiple subgroups
aEscherichia coli
bKlebsiella spp.
Carbapenem resistance: overview of data from all years
There were a total of 1341 data reports, derived from 708 studies, providing data on carbapenem susceptibility from 48 of 54 nations (Table 2). These included 622 (46.4%) on E. coli isolates and 719 (53.6%) on Klebsiella from 48 and 42 nations, respectively. Of the total 1341 reports, 879 (65.5%) were from nations in WHO Africa (including 313 incorporated from the earlier analysis [2]) while 462 (34.5%) were from the other African nations. Phenotypic and or genotypic carbapenem resistance was reported among either species in 40 of 48 nations (83.3%) from which data were available. Specifically, resistance was detected among E. coli in 36 of 48 nations (75%) with available data and among Klebsiella in 35 of 42 (83.3%). There were no data available on E. coli or Klebsiella from 6 nations (Burundi, Comoros, Lesotho, Liberia, Seychelles and Swaziland) while data were available on E. coli but not Klebsiella from an additional 6 (Cape Verde, Djibouti, Eritrea, Guinea, Lesotho, Somalia and South Sudan). Tables 3 and 4 present national-level carbapenem resistance data for all years studied, including whether resistance was reported, specific genotypes detected and, for samples from generalizable studies, percent mean resistance.
Table 2.
Available reports on E. coli and Klebsiella carbapenem and polymyxin susceptibility, resistance, and related genes
| Nation | All reports on named species (reports identifying resistance or determinants related to resistance) | References | |||
|---|---|---|---|---|---|
| Carbapenem | Polymyxin (colistin and polymyxin B) | ||||
| E. coli | Klebsiella | E. coli | Klebsiella | ||
| Algeria | 33 (12) | 37(18) | 17(4) | 22(3) | [48–97] |
| Angola | 2(2) | 2(2) | 1(0) | 1(0) | [98, 99] |
| Benin | 6(4) | 2(1) | 2(0) | 2(0) | [100–105] |
| Botswana | 1(0) | 1(0) | 0 | 0 | [106] |
| Burkina Faso | 12(4) | 13(0) | 3(2) | 1(0) | [25, 107–121] |
| Cameroon | 11(3) | 7(3) | 3(2) | 1(0) | [116, 122–133] |
| Cape Verde | 1(0) | 0 | 0 | 0 | [116] |
| Central African Republic | 2(0) | 3(0) | 0 | 0 | [25, 134, 135] |
| Chad | 8(4) | 4(1) | 1(0) | 0 | [57, 116, 136–141] |
| Congo | 2 (0) | 1 (1) | 1(0) | 0 | [142, 143] |
| Cote d'lvoire | 4(1) | 5(1) | 1 (1) | 0 | [144–150] |
| Democratic Republic of the Congo | 3(0) | 2(1) | 0 | 0 | [151–153] |
| Djibouti | 2(1) | 0 | 1(0) | 0 | [57, 154] |
| Egypt | 106(66) | 125 (98) | 28 (14) | 34(15) | [1, 25–27, 57, 78, 116, 155–293] |
| Equatorial Guinea | 1(0) | 1 (1) | 0 | 0 | [294] |
| Eritrea | 1(0) | 0 | 1(0) | 0 | [295] |
| Ethiopia | 19(10) | 27(17) | 4(4) | 6(4) | [1, 112, 296–316] |
| Gabon | 4(0) | 5(1) | 0 | 0 | [317–321] |
| Gambia | 1 (1) | 1 (1) | 0 | 0 | [322] |
| Ghana | 15(5) | 15(8) | 1 (1) | 1 (1) | [116, 323–337] |
| Guinea | 1(0) | 0 | 0 | 0 | [116] |
| Guinea-Bissau | 1(0) | 1(0) | 0 | 0 | [338] |
| Kenva | 26(11) | 25 (21) | 2(1) | 2(0) | [116, 168, 268, 282, 339–362] |
| Libya | 17(10) | 22(20) | 3(0) | 8(4) | [57, 78, 363–385] |
| Madagascar | 14(3) | 12(6) | 0 | 0 | [1, 26, 27, 112, 116, 168, 386–395] |
| Malawi | 4(3) | 7(5) | 1(0) | 2(0) | [26, 27, 396–398] |
| Mali | 4(3) | 2(1) | 1(0) | 0 | [1, 57, 399, 400] |
| Mauritania | 1 (1) | 1(0) | 1 (1) | 1 (1) | [401] |
| Mauritius | 4(2) | 6(5) | 1(0) | 2(1) | [25, 57, 168, 183, 402–404] |
| Morocco | 24(13) | 39(24) | 6(2) | 10(2) | [25, 57, 78, 116, 183, 268, 282, 405–433] |
| Mozambique | 6(1) | 4(0) | 1(0) | 0 | [1, 116, 427, 434–438] |
| Namibia | 1(0) | 5(1) | 0 | 0 | [25, 183, 439] |
| Niger | 5(2) | 2(0) | 1 (1) | 1 (1) | [57, 440–443] |
| Nigeria | 82(53) | 80(46) | 3203) | 28(15) | [1, 27, 116, 444–561] |
| Rwanda | 7(3) | 6(2) | 1 (1) | 1 (1) | [562–568] |
| Sao Tome and Principe | 1 (1) | 1 (1) | 1 (1) | 0 | [569] |
| Senegal | 10(2) | 11(6) | 1 (1) | 3(1) | [56, 78, 112, 116, 145, 570–581] |
| Sierra Leone | 5(3) | 4(3) | 0 | 0 | [116, 582, 583] |
| Somalia | 1(0) | 0 | 1(0) | 0 | [295] |
| South Africa | 69(25) | 109(82) | 18(8) | 20(10) | [1, 25, 78, 168, 183, 268, 282, 519, 584–663] |
| South Sudan | 1(0) | 0 | 0 | 0 | [664] |
| Sudan | 12(7) | 10(5) | 1 (1) | 1 (1) | [1, 27, 112, 116, 665–673] |
| Tanzania | 29(6) | 26(7) | 2(1) | 2(1) | [56, 112, 116, 168, 674–700] |
| Togo | 6(4) | 4(3) | 3(1) | 3(1) | [116, 701–706] |
| Tunisia | 34(14) | 70(43) | 15(4) | 29(13) | [1, 26, 27, 78, 168, 183, 268, 282, 707–774] |
| Uganda | 17(10) | 16(11) | 2(1) | 2(1) | [1, 26, 27, 168, 775–788] |
| Zambia | 3(1) | 4(4) | 0 | 0 | [26, 27, 789, 790] |
| Zimbabwe | 3 (1) | 1 (1) | 0 | 0 | [116, 791, 792] |
| All reporting nations | 942(622) | 451 (19) | 75(158) | 76(183) | |
Reports on carbapenem or polymyxin susceptibility were not identified from the following searched nations: Burundi, Comoros, Lesotho, Liberia, Seychelles and Swaziland
Table 3.
Carbapenem resistance (R) and resistance determinants in Escherichia coli isolates: data from all years
| Findings in reports from all study years meeting criteria for generalizability | Identified resistance determinants | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Nations | Number of reports | Specimens in all reports | Any R | Reports meeting criteria | Total specimens meeting criteria | Range of specimens among studies | Resistant specimens (#) | Resistant specimens (%) | |
| Algeria | 33 | 4304 | Y | 15 | 4201 | 30—1184 | 13 | 0.3 | NDM-5, OXA-48, OXA-181, VIM-19 |
| Angola | 2 | 52 | Y | 0 | 0 | – | – | – | NDM-1, NDM-5, OXA-181 |
| Benin | 6 | 692 | Y | 5 | 687 | 84–221 | 18 | 2.6 | |
| Botswana | 1 | 27 | N | 0 | 0 | – | – | – | |
| Burkina Faso | 12 | 787 | Y | 6 | 743 | 26–296 | 5 | 0.7 | GES, OXA, OXA-181 |
| Cameroon | 11 | 330 | Y | 6 | 313 | 21–163 | 7 | 2.2 | NDM-4 |
| Cape Verde | 1 | 1 | N | 0 | 0 | – | – | – | |
| CAR | 2 | 84 | N | 2 | 84 | 33–51 | 0 | 0 | |
| Chad | 8 | 402 | Y | 5 | 382 | 31–128 | 6 | 1.6 | NDM-5, OXA, OXA-181 |
| Congo | 2 | 112 | Y | 2 | 112 | 23–89 | 4 | 3.6 | OXA-48 |
| Côte d'Ivoire | 4 | 145 | Y | 2 | 121 | 57–64 | 0 | 0 | |
| DRC | 3 | 451 | N | 3 | 451 | 21–376 | 0 | 0 | |
| Djibouti | 2 | 32 | Y | 1 | 31 | – | 0 | 0 | OXA-48 |
| Egypt | 106 | 8657 | Y | 56 | 7549 | 20–3177 | 425 | 5.6 | KPC, GES, IMP, NDM, NDM-1, NDM-5, OXA-48, OXA-181, VIM, VIM-1, VIM-2 |
| Equatorial Guinea | 1 | 39 | N | 1 | 39 | – | 0 | 0 | |
| Eritrea | 1 | 14 | N | 0 | 0 | – | – | – | |
| Ethiopia | 19 | 1794 | Y | 12 | 1729 | 31–235 | 54 | 3.1 | KPC |
| Gabon | 4 | 142 | N | 3 | 133 | 30–57 | 0 | 0 | |
| Gambia | 1 | 8 | Y | 0 | 0 | – | – | – | |
| Ghana | 15 | 621 | Y | 9 | 568 | 25–124 | 27 | 4.8 | NDM-1, OXA-48 |
| Guinea | 1 | 1 | N | 0 | 0 | – | – | – | |
| Guinea-Bissau | 1 | 83 | N | 1 | 83 | – | 0 | 0 | |
| Kenya | 26 | 10,654 | Y | 18 | 10,554 | 25–5165 | 57 | 0.5 | |
| Libya | 17 | 1387 | Y | 8 | 1154 | 75–346 | 62 | 5.4 | OXA-48 |
| Madagascar | 14 | 1381 | Y | 8 | 1355 | 31–672 | 7 | 0.5 | |
| Malawi | 4 | 2601 | Y | 2 | 2592 | 657–1935 | 54 | 2.1 | NDM-5, OXA-48 |
| Mali | 4 | 211 | Y | 3 | 210 | 31–132 | 25 | 11.9 | NDM-4, OXA-181 |
| Mauritania | 1 | 366 | Y | 1 | 366 | – | 4 | 1.1 | |
| Mauritius | 4 | 202 | Y | 1 | 183 | – | 5 | 2.7 | OXA-181 |
| Morocco | 24 | 3585 | Y | 10 | 3459 | 49–1174 | 41 | 1.2 | IMP-1, OXA-48 |
| Mozambique | 6 | 188 | Y | 3 | 161 | 35–75 | 0 | 0 | |
| Namibia | 1 | 23 | N | 1 | 23 | – | 0 | 0 | |
| Niger | 5 | 720 | Y | 3 | 502 | 27–434 | 0 | 0 | OXA-181 |
| Nigeria | 82 | 5072 | Y | 43 | 4161 | 21–400 | 319 | 7.7 | GES, NDM, OXA, OXA-48, OXA-181, VIM |
| Rwanda | 7 | 3009 | Y | 6 | 3002 | 55–2473 | 201 | 6.7 | |
| Sao Tome and Principe | 1 | 30 | Y | 0 | 0 | – | – | – | OXA-181 |
| Senegal | 10 | 581 | Y | 4 | 554 | 33–398 | 1 | 0.2 | OXA-48 |
| Sierra Leone | 5 | 14 | Y | 0 | 0 | – | – | – | DIM-1, OXA-58, VIM |
| Somalia | 1 | 27 | N | 1 | 27 | – | 0 | 0 | |
| South Africa | 69 | 36,224 | Y | 41 | 35,930 | 20–14,348 | 333 | 0.9 | NDM, NDM-1, NDM-5, OXA-48, VIM, VIM-1 |
| South Sudan | 1 | 65 | N | 0 | 0 | – | – | – | |
| Sudan | 12 | 1085 | Y | 4 | 978 | 71–458 | 72 | 7.4 | IMP, NDM |
| Tanzania | 29 | 1977 | Y | 16 | 1793 | 20–837 | 18 | 1 | KPC, IMP, NDM, OXA-48, VIM |
| Togo | 6 | 238 | Y | 2 | 109 | 35–74 | 1 | 0.9 | |
| Tunisia | 34 | 23,696 | Y | 17 | 23,619 | 31–9485 | 214 | 0.9 | KPC-2, NDM-1, OXA-48 |
| Uganda | 17 | 1532 | Y | 6 | 1302 | 22–930 | 167 | 12.8 | KPC, IMP, OXA-48, VIM |
| Zambia | 3 | 477 | Y | 3 | 477 | 56–343 | 341 | 71.5 | |
| Zimbabwe | 3 | 204 | Y | 2 | 203 | 23–180 | 27 | 13.3 | |
| All reporting countries | 622 | 114,327 | Y | 332 | 109,940 | 20–14,348 | 2508 | 2.3** | |
Y One or more resistant isolates identified phenotypically or genotypically
N No resistant isolates identified phenotypically or genotypically
**Calculation should not be considered an estimate of overall resistance due to varying totals of specimens meeting criteria across nations
–Data not available
Table 4.
Carbapenem resistance (R) and resistance determinants in Klebsiella spp. isolates: data from all years
| Findings in reports from all study years meeting criteria for generalizability | Identified resistance determinants | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Nations | Number of reports | Specimens in all reports | Any R | Reports meeting criteria | Total specimens meeting criteria | Range of specimens among studies | Resistant specimens (#) | Resistant specimens (%) | |
| Algeria | 37 | 2174 | Y | 12 | 1968 | 24–608 | 25 | 1.3 | KPC-3, NDM, NDM-1, OXA-48, VIM-19 |
| Angola | 2 | 49 | Y | 0 | 0 | – | – | – | NDM-1, NDM-5, OXA-181 |
| Benin | 2 | 51 | Y | 1 | 41 | – | 1 | 2.4 | |
| Botswana | 1 | 40 | N | 0 | 0 | – | – | – | |
| Burkina Faso | 13 | 297 | N | 4 | 242 | 20–109 | 0 | 0 | |
| Cameroon | 7 | 299 | Y | 4 | 276 | 28–99 | 5 | 1.8 | |
| CAR | 3 | 77 | N | 2 | 67 | 24–43 | 0 | 0 | |
| Chad | 4 | 87 | Y | 3 | 86 | 23–35 | 1 | 1.2 | OXA |
| Congo | 1 | 12 | Y | 0 | 0 | – | – | – | |
| Côte d'Ivoire | 5 | 237 | Y | 4 | 229 | 22–107 | 0 | 0 | |
| DRC | 2 | 167 | Y | 2 | 167 | 21–146 | 1 | 0.6 | |
| Egypt | 125 | 7320 | Y | 59 | 5501 | 20–594 | 1545 | 28.1 | KPC, KPC-2, IMP, IMP-1, NDM, NDM-1, OXA-48, VIM, VIM-1, VIM-2 |
| Equatorial Guinea | 1 | 30 | Y | 1 | 30 | – | 1 | 3.3 | |
| Ethiopia | 27 | 808 | Y | 9 | 675 | 30–154 | 78 | 11.6 | KPC, NDM-1 |
| Gabon | 5 | 161 | Y | 2 | 146 | 67–79 | 0 | 0 | NDM-7 |
| Gambia | 1 | 9 | Y | 0 | 0 | – | – | – | |
| Ghana | 15 | 537 | Y | 10 | 505 | 20–107 | 85 | 16.8 | NDM, OXA-48 |
| Guinea-Bissau | 1 | 91 | N | 1 | 91 | – | 0 | 0 | |
| Kenya | 25 | 1471 | Y | 15 | 1419 | 25–272 | 131 | 9.2 | KPC, NDM, NDM-1, NDM-5, OXA-48, OXA-58, VIM |
| Libya | 22 | 709 | Y | 8 | 514 | 24–158 | 202 | 39.3 | KPC, NDM, NDM-1, OXA-48 |
| Madagascar | 12 | 472 | Y | 6 | 418 | 22–122 | 13 | 3.1 | NDM-1 |
| Malawi | 7 | 1315 | Y | 2 | 1276 | 173–1103 | 60 | 4.7 | KPC-2, OXA-48 |
| Mali | 2 | 67 | Y | 2 | 67 | 26–41 | 7 | 10.4 | |
| Mauritania | 1 | 137 | N | 1 | 137 | – | 0 | 0 | |
| Mauritius | 6 | 235 | Y | 2 | 222 | 104–118 | 13 | 5.9 | NDM-1, OXA-181 |
| Morocco | 39 | 1784 | Y | 10 | 1380 | 24–389 | 69 | 5 | IMP-1, NDM-1, OXA-48, VIM-1 |
| Mozambique | 4 | 63 | N | 1 | 21 | – | 0 | 0 | |
| Namibia | 5 | 313 | Y | 2 | 303 | 23–280 | 1 | 0.3 | |
| Niger | 2 | 21 | N | 0 | 0 | – | – | – | |
| Nigeria | 80 | 4111 | Y | 42 | 3524 | 21–600 | 318 | 9 | GES, NDM, NDM-1, NDM-5, OXA, OXA-48, OXA-181, VIM |
| Rwanda | 6 | 1222 | Y | 5 | 1214 | 22–975 | 108 | 8.9 | |
| Sao Tome and Principe | 1 | 4 | Y | 0 | 0 | – | – | – | OXA-181 |
| Senegal | 11 | 249 | Y | 5 | 173 | 21–40 | 2 | 1.2 | OXA-48 |
| Sierra Leone | 4 | 15 | Y | 0 | 0 | – | – | – | DIM-1, OXA-58, VIM |
| South Africa | 109 | 45,588 | Y | 53 | 42,915 | 20–15,589 | 4214 | 9.8 | KPC, KPC-2, GES, IMP, NDM, NDM-1, OMP, OXA, OXA-48, OXA-181, OXA-232, VIM, VIM-1 |
| Sudan | 10 | 988 | Y | 5 | 940 | 21–404 | 98 | 10.4 | IMP, NDM |
| Tanzania | 26 | 947 | Y | 14 | 790 | 20–139 | 16 | 2 | KPC, IMP, NDM, OXA-48, VIM |
| Togo | 4 | 165 | Y | 1 | 86 | – | 3 | 3.5 | OXA-181 |
| Tunisia | 70 | 12,842 | Y | 26 | 12,117 | 21–2826 | 1417 | 11.7 | KPC, NDM, NDM-1, OMP, OXA-48, OXA-58, OXA-232, VIM, VIM-4 |
| Uganda | 16 | 319 | Y | 3 | 116 | 22–55 | 14 | 12.1 | KPC, IMP, NDM-1, OXA-48, VIM |
| Zambia | 4 | 683 | Y | 4 | 683 | 58–432 | 435 | 63.7 | |
| Zimbabwe | 1 | 130 | Y | 1 | 130 | – | 10 | 7.7 | |
| All reporting countries | 719 | 86,296 | Y | 322 | 78,469 | 20–15,589 | 8873 | 11.3** | |
Y One or more resistant isolates identified phenotypically or genotypically
N No resistant isolates identified phenotypically or genotypically
**Calculation should not be considered an estimate of overall resistance due to varying totals of specimens meeting criteria across nations
–Data not available
Carbapenem resistance among more recent E. coli isolates
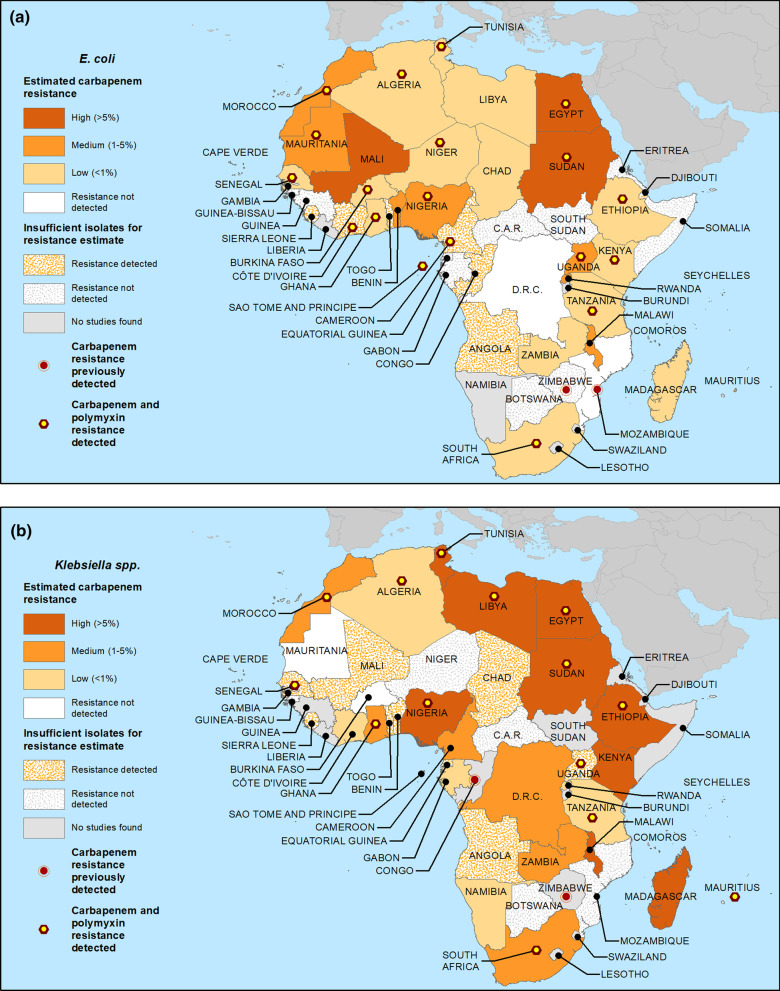
Table 5 displays carbapenem resistance data for E. coli based on samples collected in 2010 and later, including the mean and range of resistance percentages across studies, and, for nations with at least 100 generalizable isolates since 2010, crude estimated national resistance proportions (median across qualifying reports). Three nations (Egypt, Mali and Sudan) had high estimated resistance. Eight (Benin, Malawi, Mauritania, Mauritius, Morocco, Nigeria, Rwanda and Uganda) had moderate estimated resistance, and resistance in 14 nations (Algeria, Burkina Faso, Chad, Ethiopia, Ghana, Kenya, Libya, Madagascar, Niger, Senegal, South Africa, Tanzania, Tunisia and Zambia) was estimated as low. Resistance was not detected among ≥ 100 E. coli isolates from either the Democratic Republic of the Congo or Mozambique. Among nations with insufficient E. coli isolates to allow estimates, resistance was detected in nine (Angola, Cameroon, Congo, Côte d’Ivoire, Djibouti, Gambia, Sao Tome and Principe, Sierra Leone and Togo) and not detected in 11 (Botswana, Cape Verde, Central African Republic, Equatorial Guinea, Eritrea, Gabon, Guinea, Guinea-Bissau, Somalia, South Sudan and Zimbabwe). No relevant data were identified from Namibia. Resistance data for E. coli are mapped in Fig. 1a.
Table 5.
Carbapenem resistance (R) estimates and data for Escherichia coli isolates from studies including samples from 2010 and later
| Findings in reports from all study years meeting criteria for generalizability | Resistance estimate category | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nations | Number of reports | Specimens in all reports | Any R | Reports meeting criteria | Total specimens meeting criteria | Range of specimens among studies | Resistant specimens (#) | Resistant specimens (%) | Resistant range (%) | Median R | |
| Algeria | 21 | 2434 | Y | 10 | 2371 | 30–1184 | 13 | 0.5 | 0–12.7 | 0 | Low |
| Angola | 2 | 52 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected |
| Benin | 5 | 503 | Y | 4 | 498 | 84–221 | 11 | 2.2 | 0–8 | 2.3 | Moderate |
| Botswana | 1 | 27 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance not detected |
| Burkina Faso | 10 | 651 | Y | 5 | 611 | 26–296 | 5 | 0.8 | 0–16.1 | 0 | Low |
| Cameroon | 6 | 69 | Y | 2 | 54 | 24–30 | 5 | 9.3 | 0–16.7 | N/A* | Insufficient isolates—resistance detected |
| Cape Verde | 1 | 1 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance not detected |
| CAR | 2 | 84 | N | 2 | 84 | 33–51 | 0 | 0 | 0–0 | N/A* | Insufficient isolates—resistance not detected |
| Chad | 8 | 402 | Y | 5 | 382 | 31–128 | 6 | 1.6 | 0–4.7 | 0 | Low |
| Congo | 1 | 89 | Y | 1 | 89 | - | 3 | 3.4 | - | N/A* | Insufficient isolates—resistance detected |
| Côte d'Ivoire | 2 | 71 | Y | 1 | 57 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance detected |
| DRC | 3 | 451 | N | 3 | 451 | 21–376 | 0 | 0 | 0–0 | 0 | Resistance not detected |
| Djibouti | 2 | 32 | Y | 1 | 31 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance detected ^ |
| Egypt | 71 | 4094 | Y | 36 | 3274 | 21–486 | 377 | 11.5 | 0–83.3 | 7.9 | High |
| Equatorial Guinea | 1 | 39 | N | 1 | 39 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance not detected |
| Eritrea | 1 | 14 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—Resistance not detected |
| Ethiopia | 19 | 1794 | Y | 12 | 1729 | 31–235 | 54 | 3.1 | 0–41.8 | 0.9 | Low |
| Gabon | 3 | 85 | N | 2 | 76 | 30–46 | 0 | 0 | 0–0 | N/A* | Insufficient isolates—RESISTANCE not detected |
| Gambia | 1 | 8 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected |
| Ghana | 12 | 394 | Y | 6 | 341 | 25–118 | 27 | 7.9 | 0–40.6 | 0 | Low |
| Guinea | 1 | 1 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance not detected |
| Guinea-Bissau | 1 | 83 | N | 1 | 83 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance not detected |
| Kenya | 13 | 8603 | Y | 11 | 8595 | 25–5165 | 37 | 0.4 | 0–13 | 0 | Low |
| Libya | 14 | 1133 | Y | 7 | 1035 | 75–346 | 62 | 6 | 0–52 | 0.6 | Low |
| Madagascar | 9 | 1190 | Y | 5 | 1171 | 46–672 | 7 | 0.6 | 0–2 | 0.6 | Low |
| Malawi | 3 | 2600 | Y | 2 | 2592 | 657–1935 | 54 | 2.1 | 1.4–4.1 | 2.7 | Moderate |
| Mali | 3 | 164 | Y | 2 | 163 | 31–132 | 25 | 15.3 | 3.3–18.2 | 10.7 | High |
| Mauritania | 1 | 366 | Y | 1 | 366 | - | 4 | 1.1 | - | 1 | Moderate |
| Mauritius | 2 | 184 | Y | 1 | 183 | - | 5 | 2.7 | - | 3 | Moderate |
| Morocco | 15 | 3292 | Y | 7 | 3197 | 83–1174 | 41 | 1.3 | 0–5.7 | 1.1 | Moderate |
| Mozambique | 5 | 176 | N | 3 | 161 | 35–75 | 0 | 0 | 0–0 | 0 | Resistance not detected |
| Niger | 4 | 679 | Y | 2 | 461 | 27–434 | 0 | 0 | 0–0 | 0 | Low |
| Nigeria | 62 | 3095 | Y | 30 | 2567 | 21–278 | 265 | 10.3 | 0–63 | 2.7 | Moderate |
| Rwanda | 5 | 417 | Y | 4 | 410 | 55–139 | 8 | 2 | 0–8 | 1.7 | Moderate |
| Sao Tome and Principe | 1 | 30 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected |
| Senegal | 6 | 174 | Y | 3 | 156 | 33–74 | 1 | 0.6 | 0–3 | 0 | Low |
| Sierra Leone | 5 | 14 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected ^ |
| Somalia | 1 | 27 | N | 1 | 27 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance not detected |
| South Africa | 36 | 24,270 | Y | 22 | 24,135 | 20–14,348 | 264 | 1.1 | 0–82.6 | 0 | Low |
| South Sudan | 1 | 65 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—Resistance NOT detected |
| Sudan | 10 | 614 | Y | 3 | 520 | 71–326 | 72 | 13.8 | 9–36.6 | 10.7 | High |
| Tanzania | 22 | 912 | Y | 13 | 819 | 20–164 | 18 | 2.2 | 0–19.2 | 0 | Low |
| Togo | 5 | 164 | Y | 1 | 35 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance detected |
| Tunisia | 17 | 21,324 | Y | 10 | 21,299 | 48–9485 | 207 | 1 | 0–3.7 | 0.6 | Low |
| Uganda | 13 | 598 | Y | 5 | 372 | 22–181 | 18 | 4.8 | 0–19 | 4.5 | Moderate |
| Zambia | 3 | 477 | Y | 3 | 477 | 56–343 | 341 | 71.5 | 0–99.3 | 0 | Low |
| Zimbabwe | 2 | 24 | N | 1 | 23 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance not detected |
| All reporting countries | 432 | 81,970 | Y | 229 | 78,934 | 20–14,348 | 1930 | 2.4** | – | – | – |
Y one or more resistant isolates identified phenotypically or genotypically
N no resistant isolates identified phenotypically or genotypically
^Only genotypic resistance reported
*Insufficient isolates (< 100) for carbapenem resistance estimate
**Calculation should not be considered an estimate of overall resistance due to varying totals of specimens meeting criteria across nations
-Data not available
–Not calculated
Fig. 1.
Estimated crude median national carbapenem resistance proportions for a E. coli and b Klebsiella spp. for studies including samples from 2010 and later. For those nations with ≥ 100 isolates from qualifying studies (see Methods), median proportions across studies were calculated. Where < 100 isolates, data were deemed insufficient to estimate proportions and resistance is represented as either detected or not
Carbapenem resistance among more recent Klebsiella isolates
Median carbapenem resistance among recent Klebsiella isolates (Table 6) was estimated as high in 10 nations (Egypt, Ethiopia, Kenya, Libya, Madagascar, Malawi, Mauritius, Nigeria, Sudan and Tunisia). Six nations had moderate estimated resistance (Cameroon, Democratic Republic of the Congo, Ghana, Morocco, South Africa and Zambia), while resistance in 6 others (Algeria, Côte d’Ivoire, Gabon, Namibia, Rwanda and Tanzania) was estimated as low. Burkina and Mauritania had no resistance detected in ≥ 100 isolates. Among nations with insufficient Klebsiella isolates to allow estimates, resistance was detected in 11 (Angola, Benin, Chad, Equatorial Guinea, Gambia, Mali, Sao Tome and Principe, Senegal, Sierra Leone, Togo and Uganda) and not detected in 5 (Botswana, Central African Republic, Guinea-Bissau, Mozambique and Niger). No relevant data were identified from 8 nations (Cape Verde, Congo, Djibouti, Eritrea, Guinea, Somalia, South Sudan and Zimbabwe). Resistance data for Klebsiella are mapped in Fig. 1b.
Table 6.
Carbapenem resistance (R) estimates and data for Klebsiella spp. isolates from studies including samples from 2010 and later
| Findings in reports from all study years meeting criteria for generalizability | Resistance estimate category | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nations | Number of reports | Specimens in all reports | Any R | Reports meeting criteria | Total specimens meeting criteria | Range of specimens among studies | Resistant specimens (#) | Resistant specimens (%) | Resistant range (%) | Median R | |
| Algeria | 24 | 1205 | Y | 6 | 1029 | 24–608 | 25 | 2.4 | 0–20 | 0 | Low |
| Angola | 2 | 49 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected |
| Benin | 2 | 51 | Y | 1 | 41 | - | 1 | 2.4 | - | N/A* | Insufficient isolates—Resistance detected |
| Botswana | 1 | 40 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance not detected |
| Burkina Faso | 11 | 234 | N | 2 | 179 | 70–109 | 0 | 0 | 0–0 | 0 | Resistance not detected |
| Cameroon | 3 | 154 | Y | 2 | 151 | 52–99 | 4 | 2.6 | 0–4 | 2 | Moderate |
| CAR | 2 | 34 | N | 1 | 24 | - | 0 | 0 | - | N/A* | Insufficient isolates—Resistance not detected |
| Chad | 4 | 87 | Y | 3 | 86 | 23–35 | 1 | 1.2 | 0–2.9 | N/A* | Insufficient isolates—resistance detected |
| Côte d'Ivoire | 2 | 115 | Y | 1 | 107 | - | 0 | 0 | - | 0 | Low |
| DRC | 2 | 167 | Y | 2 | 167 | 21–146 | 1 | 0.6 | 0–4.8 | 2.4 | Moderate |
| Egypt | 94 | 4925 | Y | 45 | 3617 | 20–425 | 1321 | 36.5 | 0–86.4 | 26 | High |
| Equatorial Guinea | 1 | 30 | Y | 1 | 30 | - | 1 | 3.3 | - | N/A* | Insufficient isolates—resistance detected |
| Ethiopia | 27 | 808 | Y | 9 | 675 | 30–154 | 78 | 11.6 | 0–30 | 10.7 | High |
| Gabon | 5 | 161 | Y | 2 | 146 | 67–79 | 0 | 0 | 0–0 | 0 | Low |
| Gambia | 1 | 9 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—Resistance detected |
| Ghana | 12 | 366 | Y | 7 | 334 | 20–91 | 84 | 25.1 | 0–57.1 | 1.6 | Moderate |
| Guinea-Bissau | 1 | 91 | N | 1 | 91 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance not detected |
| Kenya | 17 | 964 | Y | 10 | 929 | 25–272 | 117 | 12.6 | 0–30 | 5.5 | High |
| Libya | 20 | 655 | Y | 7 | 464 | 24–158 | 202 | 43.5 | 0–92 | 26.9 | High |
| Madagascar | 8 | 306 | Y | 4 | 261 | 22–122 | 13 | 5 | 0–17 | 8.6 | High |
| Malawi | 5 | 1310 | Y | 2 | 1276 | 173–1103 | 60 | 4.7 | 2.7–17.3 | 10 | High |
| Mali | 2 | 67 | Y | 2 | 67 | 26–41 | 7 | 10.4 | 0–17.1 | N/A* | Insufficient isolates—resistance detected |
| Mauritania | 1 | 137 | N | 1 | 137 | - | 0 | 0 | - | 0 | Resistance not detected |
| Mauritius | 3 | 223 | Y | 2 | 222 | 104–118 | 13 | 5.9 | 1.9–9 | 5.4 | High |
| Morocco | 27 | 1671 | Y | 9 | 1348 | 24–389 | 69 | 5.1 | 0–22.5 | 3.1 | Moderate |
| Mozambique | 3 | 44 | N | 1 | 21 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance not detected |
| Namibia | 1 | 280 | Y | 1 | 280 | - | 1 | 0.4 | - | 0.4 | Low |
| Niger | 1 | 9 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance not detected |
| Nigeria | 58 | 2642 | Y | 28 | 2343 | 21–600 | 287 | 12.2 | 0–81 | 8.9 | High |
| Rwanda | 5 | 247 | Y | 4 | 239 | 22–91 | 4 | 1.7 | 0–4.6 | 0 | Low |
| Sao Tome and Principe | 1 | 4 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected |
| Senegal | 5 | 116 | Y | 2 | 55 | 21–34 | 2 | 3.6 | 2.9–5 | N/A* | Insufficient isolates—resistance detected |
| Sierra Leone | 4 | 15 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected ^ |
| South Africa | 62 | 37,049 | Y | 27 | 34,593 | 21–15,589 | 4051 | 11.7 | 0–90.1 | 3.5 | Moderate |
| Sudan | 8 | 576 | Y | 4 | 536 | 21–249 | 98 | 18.3 | 0–58 | 14.3 | High |
| Tanzania | 19 | 689 | Y | 11 | 618 | 20–139 | 16 | 2.6 | 0–13.6 | 0 | Low |
| Togo | 3 | 79 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected |
| Tunisia | 38 | 10,256 | Y | 12 | 9766 | 24–2826 | 1414 | 14.5 | 0–41.2 | 12.9 | High |
| Uganda | 14 | 262 | Y | 2 | 61 | 22–39 | 1 | 1.6 | 0–4.3 | N/A* | Insufficient isolates—resistance detected |
| Zambia | 4 | 683 | Y | 4 | 683 | 58–432 | 435 | 63.7 | 1–99.2 | 4.3 | Moderate |
| All reporting countries | 503 | 66,810 | Y | 216 | 60,576 | 20–15,589 | 8306 | 13.7** | – | – | – |
Y one or more resistant isolates identified phenotypically or genotypically
N no resistant isolates identified phenotypically or genotypically
^Only genotypic resistance reported
*Insufficient isolates (< 100) for carbapenem resistance estimate
**Calculation should not be considered an estimate of overall resistance due to varying totals of specimens meeting criteria across nations
-Data not available
–Not calculated
Carbapenem resistance genotypes
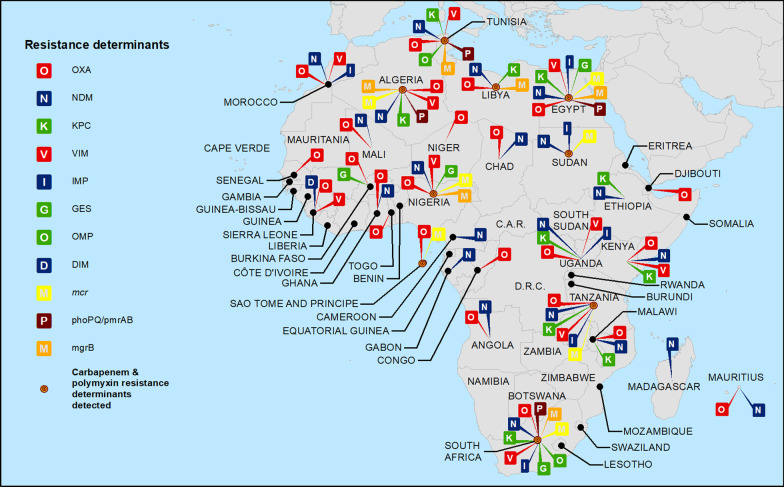
There were 94 data reports from 25 nations identifying at least one carbapenem resistance associated genotype among E. coli isolates (Table 3 and Fig. 2). The most common were blaOXA-48 and blaOXA-181, detected in 14 and 10 nations respectively. blaVIM was identified in 6 nations and blaNDM, blaNDM-1 and blaNDM-5 each reported in 5. blaGES was identified in 3 nations and blaNDM-4, blaOXA, and blaVIM-1 each identified in 2. blaDIM-1, blaIMP, blaIMP-1, blaKPC, blaKPC-2, blaOXA-58, blaVIM-2 and blaVIM-19 were each noted in one nation.
Fig. 2.
Carbapenem and polymyxin(s) resistance determinants reported from African nations
For Klebsiella spp., there were 187 reports from 24 nations identifying at least one carbapenem resistance genotype (Table 4 and Fig. 2). As also noted for E. coli, blaOXA-48 and blaOXA-181 were most common, detected in 14 and 10 nations, respectively. blaKPC was identified in 8 nations, blaNDM-5 and blaVIM in 6, with blaIMP, blaNDM and blaNDM-1 each found in 5. blaKPC-2 was identified in 3 nations and blaIMP-1, blaNDM-4, blaOXA and blaVIM-1 were each identified in 2. blaDIM-1, blaGES, blaKPC-3, blaVIM-2 and blaVIM-19 were each identified in 1 nation.
Polymyxin resistance: overview of data from all years
We found 341 unique data reports, derived from 208 studies, reporting data on polymyxin susceptibility from 33 of 54 African nations (Table 2). These reports included 158 (46.3%) on E. coli and 183 (53.7%) on Klebsiella, originating from 33 and 24 nations, respectively. Resistance was phenotypically or genotypically identified in 23 of the 33 nations (69.6%) from which any data were available. Tables 7 and 8 present national-level polymyxin resistance data for all years studied, including whether resistance was reported, specific genotypes detected and, for samples from generalizable studies, percent mean resistance.
Table 7.
Polymyxin (colistin and polymyxin B) resistance (R) and resistance determinants in Escherichia coli isolates: data from all years
| Findings in reports from all study years meeting criteria for generalizability | Identified resistance determinants | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Nations | Number of reports | Specimens in all reports | Any R | Reports meeting criteria | Total specimens meeting criteria | Range of specimens among studies | Resistant specimens (#) | Resistant specimens (%) | |
| Algeria | 17 | 2249 | Y | 11 | 2235 | 13–1184 | 1 | < 0.1 | mcr-1 |
| Angola | 1 | 23 | N | 0 | 0 | – | – | – | |
| Benin | 2 | 97 | N | 1 | 92 | – | 0 | 0 | |
| Burkina Faso | 3 | 262 | Y | 3 | 262 | 26–205 | 40 | 15.3 | |
| Cameroon | 3 | 41 | Y | 1 | 30 | – | 30 | 100 | |
| Chad | 1 | 18 | N | 1 | 18 | – | 0 | 0 | |
| Congo | 1 | 89 | N | 1 | 89 | – | 0 | 0 | |
| Côte d'Ivoire | 1 | 177 | Y | 1 | 177 | – | 14 | 7.9 | |
| Djibouti | 1 | 31 | N | 1 | 31 | – | 0 | 0 | |
| Egypt | 28 | 1276 | Y | 14 | 678 | 11–212 | 32 | 4.7 | mcr-1, mgrB, phoPQ/pmrAB |
| Eritrea | 1 | 14 | N | 0 | 0 | – | – | – | |
| Ethiopia | 4 | 163 | Y | 3 | 150 | 17–78 | 76 | 50.7 | |
| Ghana | 1 | 49 | Y | 1 | 49 | – | 3 | 6.1 | |
| Kenya | 2 | 7 | Y | 0 | 0 | – | – | – | |
| Libya | 3 | 127 | N | 2 | 126 | 51–75 | 0 | 0 | |
| Malawi | 1 | 8 | N | 0 | 0 | – | – | – | |
| Mali | 1 | 47 | N | 1 | 47 | – | 0 | 0 | |
| Mauritania | 1 | 366 | Y | 1 | 366 | – | 6 | 1.6 | |
| Mauritius | 1 | 183 | N | 1 | 183 | – | 0 | 0 | |
| Morocco | 6 | 896 | Y | 4 | 890 | 51–398 | 47 | 5.3 | |
| Mozambique | 1 | 33 | N | 1 | 33 | – | 0 | 0 | |
| Niger | 1 | 21 | Y | 1 | 21 | – | 4 | 19 | |
| Nigeria | 32 | 1757 | Y | 21 | 1607 | 12–568 | 674 | 41.9 | mcr-1 |
| Rwanda | 1 | 2473 | Y | 1 | 2473 | – | 35 | 1.4 | |
| Sao Tome and Principe | 1 | 1 | Y | 0 | 0 | – | – | – | mcr-1 |
| Senegal | 1 | 33 | Y | 1 | 33 | – | 1 | 3 | |
| Somalia | 1 | 27 | N | 0 | 0 | – | – | – | |
| South Africa | 18 | 2665 | Y | 10 | 2605 | 16–683 | 98 | 3.8 | mcr-1, mgrB, phoPQ/pmrAB |
| Sudan | 1 | 71 | Y | 0 | 0 | – | – | – | mcr-1 |
| Tanzania | 2 | 99 | Y | 1 | 30 | – | 0 | 0 | mcr-1 |
| Togo | 3 | 80 | Y | 1 | 74 | – | 1 | 1.4 | |
| Tunisia | 15 | 15,852 | Y | 10 | 15,839 | 26–12,574 | 24 | 0.2 | |
| Uganda | 2 | 66 | Y | 1 | 61 | – | 10 | 16.4 | |
| All reporting countries | 158 | 29,301 | Y | 95 | 28,199 | 11–12,574 | 1096 | 3.9** | |
Y one or more resistant isolates identified phenotypically or genotypically
N No resistant isolates identified phenotypically or genotypically
**Calculation should not be considered an estimate of overall resistance due to varying totals of specimens meeting criteria across nations
–Data not available
Table 8.
Polymyxin (colistin and polymyxin B) resistance (R) and resistance determinants in Klebsiella spp. isolates: data from all years
| Findings in reports from all study years meeting criteria for generalizability | Identified resistance determinants | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Nations | Number of reports | Specimens in all reports | Any R | Reports meeting criteria | Total specimens meeting criteria | Range of specimens among studies | Resistant specimens (#) | Resistant specimens (%) | |
| Algeria | 17 | 2249 | Y | 11 | 2235 | 13–1184 | 1 | < 0.1 | mcr-1 |
| Angola | 1 | 23 | N | 0 | 0 | – | – | – | |
| Benin | 2 | 97 | N | 1 | 92 | – | 0 | 0 | |
| Burkina Faso | 3 | 262 | Y | 3 | 262 | 26–205 | 40 | 15.3 | |
| Cameroon | 3 | 41 | Y | 1 | 30 | – | 30 | 100 | |
| Chad | 1 | 18 | N | 1 | 18 | – | 0 | 0 | |
| Congo | 1 | 89 | N | 1 | 89 | – | 0 | 0 | |
| Côte d'Ivoire | 1 | 177 | Y | 1 | 177 | – | 14 | 7.9 | |
| Djibouti | 1 | 31 | N | 1 | 31 | – | 0 | 0 | |
| Egypt | 28 | 1276 | Y | 14 | 678 | 11–212 | 32 | 4.7 | mcr-1, mgrB, phoPQ/pmrAB |
| Eritrea | 1 | 14 | N | 0 | 0 | – | – | – | |
| Ethiopia | 4 | 163 | Y | 3 | 150 | 17–78 | 76 | 50.7 | |
| Ghana | 1 | 49 | Y | 1 | 49 | – | 3 | 6.1 | |
| Kenya | 2 | 7 | Y | 0 | 0 | – | – | – | |
| Libya | 3 | 127 | N | 2 | 126 | 51–75 | 0 | 0 | |
| Malawi | 1 | 8 | N | 0 | 0 | – | – | – | |
| Mali | 1 | 47 | N | 1 | 47 | – | 0 | 0 | |
| Mauritania | 1 | 366 | Y | 1 | 366 | – | 6 | 1.6 | |
| Mauritius | 1 | 183 | N | 1 | 183 | – | 0 | 0 | |
| Morocco | 6 | 896 | Y | 4 | 890 | 51–398 | 47 | 5.3 | |
| Mozambique | 1 | 33 | N | 1 | 33 | – | 0 | 0 | |
| Niger | 1 | 21 | Y | 1 | 21 | – | 4 | 19 | |
| Nigeria | 32 | 1757 | Y | 21 | 1607 | 12–568 | 674 | 41.9 | mcr-1 |
| Rwanda | 1 | 2473 | Y | 1 | 2473 | – | 35 | 1.4 | |
| Sao Tome and Principe | 1 | 1 | Y | 0 | 0 | – | – | – | mcr-1 |
| Senegal | 1 | 33 | Y | 1 | 33 | – | 1 | 3 | |
| Somalia | 1 | 27 | N | 0 | 0 | – | – | – | |
| South Africa | 18 | 2665 | Y | 10 | 2605 | 16–683 | 98 | 3.8 | mcr-1, mgrB, phoPQ/pmrAB |
| Sudan | 1 | 71 | Y | 0 | 0 | – | – | – | mcr-1 |
| Tanzania | 2 | 99 | Y | 1 | 30 | – | 0 | 0 | mcr-1 |
| Togo | 3 | 80 | Y | 1 | 74 | – | 1 | 1.4 | |
| Tunisia | 15 | 15,852 | Y | 10 | 15,839 | 26–12,574 | 24 | 0.2 | |
| Uganda | 2 | 66 | Y | 1 | 61 | – | 10 | 16.4 | |
| All reporting countries | 158 | 29,301 | Y | 95 | 28,199 | 11–12,574 | 1096 | 3.9** | |
Y one or more resistant isolates identified phenotypically or genotypically
N No resistant isolates identified phenotypically or genotypically
**Calculation should not be considered an estimate of overall resistance due to varying totals of specimens meeting criteria across nations
–Data not available
Polymyxin resistance among more recent E. coli isolates
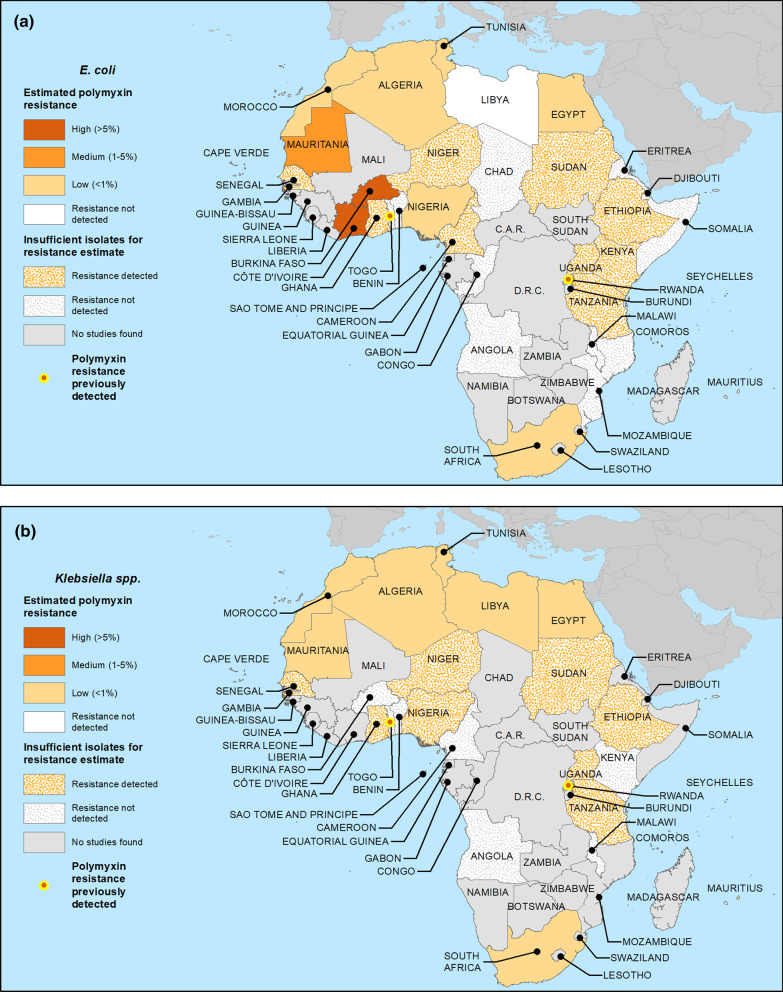
Polymyxin resistance was identified among more recent E. coli isolates from 21 of 33 nations where either phenotypic or genotypic testing was performed (Table 9). Among 11 nations where at least 100 relevant E. coli isolates from 2010 onwards were tested, median polymyxin resistance was estimated as high in Burkina Faso and Côte d’Ivoire, moderate in Mauritania, low in Algeria, Egypt, Morocco, Nigeria, South Africa and Tunisia, and was not detected in Libya and Mauritius. Although resistance was detected, there were insufficient isolates to support estimates for 10 nations (Cameroon, Ethiopia, Ghana, Kenya, Niger, Sao Tome and Principe, Senegal, Sudan, Tanzania and Uganda). Similarly, there were 10 nations with insufficient E. coli isolates to support estimates where resistance was not detected (Angola, Benin, Chad, Congo, Djibouti, Eritrea, Malawi, Mozambique, Somalia and Togo). No relevant data were found from 18 nations (Botswana, Cape Verde, Central African Republic, Democratic Republic of the Congo, Equatorial Guinea, Gabon, Gambia, Guinea, Guinea-Bissau, Madagascar, Mali, Namibia, Rwanda, Sierra Leone, South Sudan, Zambia and Zimbabwe). Resistance data for E. coli are mapped in Fig. 3a.
Table 9.
Polymyxin (colistin and polymyxin B) resistance (R) estimates and data for Escherichia coli isolates from studies including samples from 2010 and later
| Findings in reports from all study years meeting criteria for generalizability | Resistance estimate category | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nations | Number of reports | Specimens in all reports | Any R | Reports meeting criteria | Total specimens meeting criteria | Range of specimens among studies | Resistant specimens (#) | Resistant specimens (%) | Resistant range (%) | Median R | |
| Algeria | 14 | 2168 | Y | 9 | 2155 | 13–1184 | 1 | < 0.1 | 0–0.4 | 0 | Low |
| Angola | 1 | 23 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance not detected |
| Benin | 2 | 97 | N | 1 | 92 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance not detected |
| Burkina Faso | 3 | 262 | Y | 3 | 262 | 26–205 | 40 | 15.3 | 0–61.3 | 10 | High |
| Cameroon | 3 | 41 | Y | 1 | 30 | - | 30 | 100 | - | N/A* | Insufficient isolates—resistance detected |
| Chad | 1 | 18 | N | 1 | 18 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance not detected |
| Congo | 1 | 89 | N | 1 | 89 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance not detected |
| Côte d'Ivoire | 1 | 177 | Y | 1 | 177 | - | 14 | 7.9 | - | 7.9 | High |
| Djibouti | 1 | 31 | N | 1 | 31 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance not detected |
| Egypt | 20 | 1015 | Y | 9 | 431 | 11–212 | 17 | 3.9 | 0–17.4 | 0.9 | Low |
| Eritrea | 1 | 14 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance not detected |
| Ethiopia | 2 | 68 | Y | 1 | 55 | - | 50 | 90.9 | - | N/A* | Insufficient isolates—resistance detected |
| Ghana | 1 | 49 | Y | 1 | 49 | - | 3 | 6.1 | - | N/A* | Insufficient isolates—resistance detected |
| Kenya | 2 | 7 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected |
| Libya | 3 | 127 | N | 2 | 126 | 51–75 | 0 | 0 | 0–0 | 0 | Resistance not detected |
| Malawi | 1 | 8 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance not detected |
| Mauritania | 1 | 366 | Y | 1 | 366 | - | 6 | 1.6 | - | 1.7 | Moderate |
| Mauritius | 1 | 183 | N | 1 | 183 | - | 0 | 0 | - | 0 | Resistance not detected |
| Morocco | 5 | 893 | Y | 4 | 890 | 51–398 | 47 | 5.3 | 0–11.3 | 0.3 | Low |
| Mozambique | 1 | 33 | N | 1 | 33 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance not detected |
| Niger | 1 | 21 | Y | 1 | 21 | - | 4 | 19 | - | N/A* | Insufficient isolates—Resistance detected |
| Nigeria | 8 | 125 | Y | 3 | 111 | 18–50 | 1 | 0.9 | 0–2.3 | 0 | Low |
| Sao Tome and Principe | 1 | 1 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected |
| Senegal | 1 | 33 | Y | 1 | 33 | - | 1 | 3 | - | N/A* | Insufficient isolates—Resistance detected |
| Somalia | 1 | 27 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance not detected |
| South Africa | 14 | 2005 | Y | 6 | 1945 | 16–683 | 12 | 0.6 | 0–0.9 | 0.15 | Low |
| Sudan | 1 | 71 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected ^ |
| Tanzania | 2 | 99 | Y | 1 | 30 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance detected |
| Togo | 2 | 6 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—Resistance not detected |
| Tunisia | 11 | 3014 | Y | 7 | 3002 | 26–1075 | 13 | 0.4 | 0–1.3 | 0 | Low |
| Uganda | 2 | 66 | Y | 1 | 61 | - | 10 | 16.4 | - | N/A* | Insufficient isolates—resistance detected |
| All reporting countries | 109 | 11,137 | Y | 58 | 10,190 | 11–1184 | 249 | 2.4** | – | – | – |
Y one or more resistant isolates identified phenotypically or genotypically
N no resistant isolates identified phenotypically or genotypically
^Only genotypic resistance reported
*Insufficient isolates (< 100) for polymyxin resistance estimate
**Calculation should not be considered an estimate of overall resistance due to varying totals of specimens meeting criteria across nations
-Data not available
–Not calculated
Fig. 3.
Estimated crude median national polymyxin(s) resistance proportions for a E. coli and b Klebsiella spp. for studies including samples from 2010 and later. For those nations with ≥ 100 isolates from qualifying studies (see Methods), median proportions across studies were calculated. Where < 100 isolates, data were deemed insufficient to estimate proportions and resistance is represented as either detected or not
Polymyxin resistance among more recent Klebsiella isolates
Polymyxin resistance was identified among more recent Klebsiella isolates from 18 of 24 nations where either phenotypic or genotypic testing was performed (Table 10). Resistance was detected in all 8 nations with at least 100 generalizable Klebsiella isolates studied (Algeria, Egypt, Libya, Mauritania, Mauritius, Morocco, South Africa and Tunisia), and was estimated as low in each. Among nations with insufficient isolates to support a resistance estimate, resistance was detected in 8 (Ethiopia, Ghana, Niger, Nigeria, Senegal, Sudan, Tanzania and Uganda) and not detected in 7 (Angola, Benin, Burkina Faso, Cameroon, Kenya, Malawi and Togo). No studies were identified from 25 nations (Botswana, Cape Verde, Central African Republic, Chad, Congo, Côte d’Ivoire, Democratic Republic of the Congo, Djibouti, Equatorial Guinea, Eritrea, Gabon, Gambia, Guinea, Guinea-Bissau, Madagascar, Mali, Mozambique, Namibia, Rwanda, Sao Tome and Principe, Sierra Leone, Somalia, South Sudan, Zambia and Zimbabwe). Resistance data for Klebsiella are mapped in Fig. 3b.
Table 10.
Polymyxin (colistin and polymyxin B) resistance (R) estimates and data for Klebsiella spp. isolates from studies including samples from 2010 and later
| Findings in reports from all study years meeting criteria for generalizability | Resistance estimate category | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nations | Number of reports | Specimens in all reports | Any R | Reports meeting criteria | Total specimens meeting criteria | Range of specimens among studies | Resistant specimens (#) | Resistant specimens (%) | Resistant range (%) | Median R | |
| Algeria | 17 | 1056 | Y | 6 | 1015 | 13–608 | 2 | 0.2 | 0–4.3 | 0 | Low |
| Angola | 1 | 24 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance not detected |
| Benin | 2 | 51 | N | 2 | 51 | 10–41 | 0 | 0 | 0–0 | N/A* | Insufficient isolates—resistance not detected |
| Burkina Faso | 1 | 5 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance not detected |
| Cameroon | 1 | 3 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance not detected |
| Egypt | 27 | 1084 | Y | 10 | 605 | 14–183 | 11 | 1.8 | 0–50 | 0 | Low |
| Ethiopia | 4 | 53 | Y | 3 | 51 | 10–30 | 17 | 33.3 | 0–90 | N/A* | Insufficient isolates—resistance detected |
| Ghana | 1 | 38 | Y | 1 | 38 | - | 5 | 13.2 | - | N/A* | Insufficient isolates—Resistance detected |
| Kenya | 1 | 5 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—Resistance not detected |
| Libya | 8 | 179 | Y | 3 | 136 | 24–76 | 6 | 4.4 | 0–25 | 0 | Low |
| Malawi | 2 | 8 | N | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance not detected |
| Mauritania | 1 | 137 | Y | 1 | 137 | - | 1 | 0.7 | - | 0.8 | Low |
| Mauritius | 2 | 119 | Y | 1 | 118 | - | 0 | 0 | - | 0 | Low |
| Morocco | 7 | 245 | Y | 4 | 232 | 10–118 | 28 | 12.1 | 0–22.9 | 0.6 | Low |
| Niger | 1 | 4 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected |
| Nigeria | 9 | 95 | Y | 4 | 80 | 10–32 | 0 | 0 | 0–0 | N/A * | Insufficient isolates—Resistance detected |
| Senegal | 1 | 34 | Y | 1 | 34 | - | 3 | 8.8 | - | N/A* | Insufficient isolates—resistance detected |
| South Africa | 15 | 2286 | Y | 7 | 1682 | 10–839 | 18 | 1.1 | 0–2.9 | 0 | Low |
| Sudan | 1 | 50 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected^ |
| Tanzania | 2 | 59 | Y | 0 | 0 | - | - | - | - | - | Insufficient isolates—resistance detected |
| Togo | 2 | 31 | N | 1 | 30 | - | 0 | 0 | - | N/A* | Insufficient isolates—resistance not detected |
| Tunisia | 20 | 4250 | Y | 11 | 4148 | 11–2826 | 60 | 1.4 | 0–6.2 | 0.5 | Low |
| Uganda | 2 | 20 | Y | 1 | 12 | - | 3 | 25 | - | N/A* | Insufficient isolates—resistance detected |
| All reporting countries | 128 | 9836 | Y | 56 | 8369 | 10–2826 | 154 | 1.8** | – | – | – |
Y one or more resistant isolates identified phenotypically or genotypically
N no resistant isolates identified phenotypically or genotypically
^Only genotypic resistance reported
*Insufficient isolates (< 100) for polymyxin resistance estimate
**Calculation should not be considered an estimate of overall resistance due to varying totals of specimens meeting criteria across nations
-Data not available
–Not calculated
Polymyxin resistance genotypes
Genotypic determinants of polymyxin resistance in E. coli were characterized in 15 data reports on isolates from 7 nations (Table 7 and Fig. 2), with mcr-1 found in all (Algeria, Egypt, Nigeria, Sao Tome and Principe, South Africa, Sudan and Tanzania). phoPQ/pmrAB and mgrB were identified in E. coli from Egypt and South Africa. Among Klebsiella, genotypic polymyxin resistance determinants were identified in 12 reports on isolates from 7 nations (Table 8 and Fig. 2). mcr-1 was identified in Egypt, South Africa and Sudan, and mcr-8 in Algeria. mgrb was reported from six nations (Algeria, Egypt, Libya, Nigeria, South Africa and Tunisia), and phoPQ/pmrAB identified from 4 (Algeria, Egypt, South Africa and Tunisia).
Documented geographic overlaps of carbapenem and polymyxin resistance
Overlapping resistance to carbapenems and polymyxin(s) among E. coli or Klebsiella, whether phenotypic and/or genotypic, was documented in 23 nations with overlapping genotypic resistance present in 9 (Fig. 2). Specific geographic overlaps between NDM carbapenemases and mcr genetic determinants were identified in 6 nations (Algeria, Egypt, Nigeria, South Africa, Sudan and Tanzania).
Discussion
We searched for and conducted meta-analyses and mapping of available data on carbapenem and polymyxin resistance in E. coli and Klebsiella isolates from humans in Africa. These analyses, which included 1479 unique data reports through the end of 2019, show that resistance to each of these important antibiotic classes has become increasingly widespread on the continent.
The availability of a large amount of additional data since our prior report on WHO Africa nations [2] provided substantive new insights into the distribution of carbapenem resistance and its genotypic determinants, with resistance documented in approximately ¾ of African nations (compared to less than half previously for WHO Africa [2]). Carbapenem resistance among Klebsiella was significant in most countries with sufficient isolates to support a resistance estimate and categorized as high in 10, and moderate and low in 6 nations respectively. Among E. coli, estimated resistance was generally somewhat lower: high in 3, moderate in 7, and low in 14 nations with sufficient isolates. Levels of carbapenem resistance appeared high in contiguous areas of Northern and Eastern Africa (e.g. for Klebsiella in Libya, Egypt, Sudan, Ethiopia and Kenya, Fig. 1b). The most widespread genes conferring carbapenem resistance in both species, including in that area, were blaOXA-48, blaNDM-1 and blaOXA-181. Taken together, the analyses document continuing continent-wide spread of carbapenem resistance and of a broad variety of transferrable resistance plasmids, raising concerns about the future reliability of carbapenems.
Given their importance in treating resistant infections, and the paucity of available data, we also searched for and analyzed available information on polymyxin susceptibility. We located data on polymyxin susceptibility for E. coli and/or Klebsiella spp. isolates from 33 of 54 African nations, with resistance identified in 23 of those 33 nations (69.7%) from which any data were available. For the small minority of nations with ≥ 100 isolates studied from 2010 and later, estimated resistance among E. coli to polymyxins was high in 2, moderate in 1 and low in 6. Although resistance was estimated as high in two nations, estimates were based on relatively limited isolate and study numbers, and, in many cases, older methods of susceptibility testing, and should be interpreted with caution. Estimated resistance to polymyxins was low among Klebsiella in all 8 nations with sufficient isolates to support an estimate. Polymyxin resistance genetic determinants were evaluated among E. coli and Klebsiella in 7 nations each, with the mobile mcr-1 determinant shown to be predominant, consistent with recent reviews of the genetics of colistin resistance in E. coli both globally [35] and in Africa [36].
Our analyses also show, even based on limited information available from many areas (particularly with respect to polymyxins), that geographic overlapping of carbapenem and polymyxin resistance has become common and widespread, with 23 nations having documented phenotypic and/or genotypic resistance for both. Furthermore, overlapping plasmid mediated resistance to the two drug classes was documented in 9 nations, including the presence of both NDM carbapenemases and mcr genetic determinants in 6. These findings document highly concerning ongoing risks from transferrable resistance, including, were blaNDM and mcr to be acquired by the same organism(s), the risk of infections not susceptible to currently available antibiotics.
Despite efforts to enhance surveillance, major information gaps remain. For example, searches yielded no data on polymyxin resistance from 21 nations, and 6 nations with no available data on carbapenem resistance. Furthermore, even from countries where data were available, there were often less than 100 recent isolates studied, not meeting minimal pre-specified criteria to support crude estimation of resistance proportions.
It is important to note a number of limitations of these analyses, discussed in detail previously [2, 31]. Despite use of predefined study inclusion criteria and employment of common data elements, the inherently diverse data sources, time periods and locations, as well as study designs and methods, mean that inferences must be made with caution and the data should be interpreted in the context of the timing, location and populations studied. Interested readers can access further details, including the primary data from individual reports on specific nations, in the supplemental material (Additional file 4). In addition, susceptibility testing methods and standards for breakpoints to interpret their results have evolved considerably over time and often differ among laboratories. Therefore, comparability of results across laboratories, nations and time periods may be affected by such differences. For carbapenems, minimum inhibitory concentrations considered susceptible have decreased over time, meaning that some decrease in the proportion of isolates susceptible may be expected due to changing standards. There are also major caveats with respect to the interpretation of reported polymyxin susceptibility testing results. Rather than utilizing currently recommended broth microdilution methods, most studies were performed using previously employed disk diffusion methods which may be inconsistent and may overestimate susceptibility. Therefore, while the presence and spread of resistance to polymyxins is well documented, often at both phenotypic and genotypic levels, rate estimates must be interpreted with caution.
Looking at the totality of the data, despite well over a thousand data reports from hundreds of studies, the available information from many countries was limited or, in some cases, absent. Additionally, lag periods between data acquisition and reporting, along with the analysis time since the searches included in the current study, which utilized data available through December 31, 2019, mean that the continued documentation and spread of resistance to new areas is fully expected. Thus, the non-detection of resistance in a nation should not be considered as evidence that resistance was or is absent. Ensuring a more complete picture of resistance distribution and rates will require both ongoing surveillance and continued updating of data and analyses. As also noted, where resistance proportions have been estimated, these should generally be considered to be crude approximations based on non-random reporting and samples, although in our prior study of Southeast Asia [31] the results from similarly performed meta-analyses generally tracked with national surveillance where available. Similarly, available genotypic data are even more limited, with laboratories often assaying for a limited number of specific genotype(s) rather than broadly characterizing isolates with multiplex or sequence-based methods, likely leading to under-detection of less recognized or uncommon genotypes. Other potential factors may also affect the representativeness of the data, including the tendency toward publication of positive results and the likelihood that laboratories performing susceptibility testing may be located in more urban and regional centers, typically associated with more complex care and drug resistance. We attempted to address such issues by searching not only for positive but also for negative results such as in publications where susceptibility testing was reported but not as the focus of the studies.
Despite such limitations, the findings show the widespread and overlapping presence of carbapenem and polymyxin resistance among E. coli and Klebsiella isolates from humans in Africa and highlight the urgent need to better address remaining gaps in surveillance, including to systematically determine and track rates of carbapenem and polymyxin resistance, and to monitor for the emergence of dually resistant organisms. To do so will require adequate support for sustainable laboratory and epidemiologic capacity, as stressed by both WHO [41] and the African Union and Africa CDC [42]. Robust ongoing longitudinal AMR surveillance is also critical to inform antibiotic stewardship initiatives [41, 43]. Furthermore, the widespread nature of the CRE and polymyxin resistance threats reinforces the importance of strong infection prevention and control in healthcare facilities [41, 44]. Beyond enhanced stewardship of antimicrobials and measures to contain the spread of MDRO in healthcare, the continuing use of important antimicrobials, including colistin, in animal production remains a problem that must be fully addressed [45]. Resistant organisms may also be present in and spread through waste water, including from healthcare facilities [46], agriculture, and aquaculture [46].
Conclusions
Carbapenem resistance among E. coli and Klebsiella is widely distributed in Africa, and documented in 40 of 54 nations. Although resistance rates for nations with sufficient isolates to support estimates were typically low to moderate, high rates (> 5%) were found in several nations, including 10 nations with high rates among Klebsiella. Although far less data are available concerning polymyxins, resistance was documented in 23 of 33 nations with available data. The most widespread resistance associated genotypes were, for carbapenems, blaOXA-48, blaNDM-1 and blaOXA-181 and, for polymyxins, mcr-1, mgrB, and phoPQ/pmrAB. Overlapping phenotypic and/or genotypic resistance to both carbapenems and polymyxins was documented in 23 nations, including the presence of both transferrable NDM carbapenemases and mcr determinants of polymyxin resistance in 6. These findings point to ongoing and significant risks to patient safety and public health from carbapenem and polymyxin resistance. Despite progress in recent years, resistance appears to be spreading and numerous data gaps remain, indicating the need to fully support robust AMR surveillance, antimicrobial stewardship and infection control in a manner that also addresses animal and environmental health dimensions. A One Health approach that enhances surveillance and reduces both the inappropriate use of critical antibiotics and the spread of resistant organisms in all relevant settings is essential [47].
Supplementary Information
Additional file 1: Boolean search strings constructed for searches of scientific databases.
Additional file 3: Annotation on data entry columns and abbreviations.
Acknowledgements
The authors thank the following individuals who kindly discussed their study findings with us: Dr. Nadjat Aggoune [81], Dr. Carolyn S. Reid [109], Dr. Landry Beyala Bita’a [121], Dr. M. Y. Dehayem [126], Dr. Eugene Vernyuy Yeika [124], Dr. Oumar Ouchar Mahamat [137], Dr. Hisham A. Abbas [271], Dr. Mohamed A. El-Mokhtar [237], Dr. Rasha H. Hassan and Dr. Elham A. Hassan [265], Dr. Noha G. Khalaf [275], Dr. Reham Osama [164], Dr. Dina E. Rizk [173], Dr. Howayda E. Gomaa [184], Dr. Noha A. Hassuna [197], Dr. Anita Hallgren [167], Dr. Stephen Hawser [267], Dr. Harald Seifert [182], Dr. Philipp Zanger [294], Dr. Beza Eshetu [315], Dr. Alex Owusu-Ofori [329], Dr. Jibril Mohammed [324], Dr. Adelaide Ogutu Ayoyi [358], Dr. Tarig M.S. Alnour [371], Dr. Zoly Nantenaina Ranosiarisoa [388], Dr. Anthony G. Charles [395], Dr. Touria Essayagh [416], Dr. Adil Maleb [420], Dr. Anthony Ayodeji Adegoke [506, 517], Dr. Paul Akinniyi Akinduti [468], Dr. Charles J. Elikwu [490], Dr.Yusuf Ibrahim [497], Dr. Gbolahan O Babalola [498], Dr. Christiana Jesumirhewe [514], Dr. Ikechukwu Benjamin Moses [460], Dr. Mamadou Saidou Barry [576], Dr. Lo Seynabou [572], Dr. A. Dramowski [597], Dr. Brian Godman [602], Dr. Chetna Govind [603], Dr. Laurent Poirel [613], Dr. Johann D. D. Pitout [621], Dr. Jesús Rodríguez-Baño [643], Dr. Amidou Samie [654], Dr. John Osei Sekyere [656], Dr. A. Singh-Moodley [658], Dr. Sandeep Vasaikar [661], Dr. Jason S. Biswas [663], Dr. Malik I. A. [671], Dr. Mokline Amel [707], Dr. Carmen Torres [713], Dr. C. Chouchani [727], Dr. Ramzi Jeddi [748], Dr. Elaa Maamar [756], and Dr. Josephine Tumuhamye [781]. The authors also thank C. Scott Dorris of the Dahlgren Memorial Library at Georgetown University School of Medicine for advice on search strategies.
Author contributions
All authors made substantial contributions to the conceptualization and design of the study, acquisition of data, analysis of data, and/or drafting of the manuscript. DMV, DMH, MDM, MK and JLG participated in the conceptual design and development of the original and/or current study. DMH, MDM, MK and JLG assisted in the design and analysis of search strategies. DMV also designed, tested and conducted searches, screened and reviewed literature, performed data extraction, spreadsheet production and geographic mapping, with the assistance of AYB who reviewed and screened studies and performed data extraction. DMV, DMH and JLG shared in data analysis, while DMV constructed tables, DMH created the R code used to analyze extracted data, and JLG lead in development of analytic strategies and methods. DMV, DMH and JLG played a major role in drafting and reviewing the manuscript with the assistance of MDM, MK and AYB. All authors agreed to submit it to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This study was financially supported by the Armed Forces Health Surveillance Branch and its Global Emerging Infections Surveillance (GEIS) Section, Silver Spring, MD 20904, 2016-17 (ProMIS ID PO151_15_UN) and by the Georgetown University Medical Center. The funding sources played no role in study design, data collection, analysis, or interpretation, or in writing the report or the decision to submit for publication.
Availability of data and materials
The dataset supporting the conclusions of this article is available in the Harvard Dataverse repository, https://doi.org/10.7910/DVN/JIJH3W. The dataset(s) supporting the conclusions of this article is also included within the article as Additional file 4.
Declarations
Ethics approval and consent to participate
Not required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/12/2024
A Correction to this paper has been published: 10.1186/s13756-024-01403-7
References
- 1.World Health Organization (WHO). Global antimicrobial resistance surveillance system (GLASS) report: early implementation 2020 [Internet]. Geneva, Switzerland; 2020. Available from: https://apps.who.int/iris/bitstream/handle/10665/332081/9789240005587-eng.pdf?ua=1%0Ahttp://www.who.int/glass/resources/publications/early-implementation-report-2020/en/%0Ahttp://apps.who.int/iris/bitstream/10665/188783/1/9789241549400_eng.pdf?ua=1
- 2.Mitgang EA, Hartley DM, Malchione MD, Koch M, Goodman JL. Review and mapping of carbapenem-resistant Enterobacteriaceae in Africa: using diverse data to inform surveillance gaps. Int J Antimicrob Agents. 2018;52(3):372–384. doi: 10.1016/j.ijantimicag.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 3.Codjoe F, Donkor E. Carbapenem resistance: a review. Med Sci. 2017;6(1):1. doi: 10.3390/medsci6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein EY, Van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, et al. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci USA. 2018;115(15):E3463–E3470. doi: 10.1073/pnas.1717295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torres NF, Chibi B, Kuupiel D, Solomon VP, Mashamba-Thompson TP, Middleton LE. The use of non-prescribed antibiotics; prevalence estimates in low-and-middle-income countries. A systematic review and meta-analysis. Arch Public Heal. 2021;79(1):1–15. doi: 10.1186/s13690-020-00517-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Köck R, Daniels-Haardt I, Becker K, Mellmann A, Friedrich AW, Mevius D, et al. Carbapenem-resistant enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin Microbiol Infec. 2018;24:1241–1250. doi: 10.1016/j.cmi.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Kelly AM, Mathema B, Larson EL. Carbapenem-resistant Enterobacteriaceae in the community: a scoping review. Int J Antimicrob Agents. 2017;50(2):127–134. doi: 10.1016/j.ijantimicag.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katale BZ, Misinzo G, Mshana SE, Chiyangi H, Campino S, Clark TG, et al. Genetic diversity and risk factors for the transmission of antimicrobial resistance across human, animals and environmental compartments in East Africa: a review. Antimicrob Resist Infect Control. 2020 doi: 10.1186/s13756-020-00786-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, et al. Molecular characterization of an enterobacterial metallo β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother. 1994;38(1):71–78. doi: 10.1128/AAC.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh TR. Emerging carbapenemases: a global perspective. Int J Antimicrob Agents. 2010;36(SUPPL. 3):S8. doi: 10.1016/S0924-8579(10)70004-2. [DOI] [PubMed] [Google Scholar]
- 12.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:1–30. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos G, Cormican M, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Logan LK, Weinstein RA. The epidemiology of Carbapenem-resistant enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(Suppl 1):S28–36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diene SM, Rolain JM. Carbapenemase genes and genetic platforms in gram-negative bacilli: enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2014;20(9):831–838. doi: 10.1111/1469-0691.12655. [DOI] [PubMed] [Google Scholar]
- 16.Poirel L, Jayol A, Nordmanna P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, et al. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 2006;6(9):589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 18.Aghapour Z, Gholizadeh P, Ganbarov K, Bialvaei AZ, Mahmood SS, Tanomand A, et al. Molecular mechanisms related to colistin resistance in enterobacteriaceae. Infect Drug Resist. 2019;12:965–975. doi: 10.2147/IDR.S199844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Z, Hu Y, Sun Q, Hu F, Zhou H, Shu L, et al. Emerging carriage of NDM-5 and MCR-1 in Escherichia coli from healthy people in multiple regions in China: a cross sectional observational study. EClinicalMedicine. 2018;6:11–20. doi: 10.1016/j.eclinm.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng B, Dong H, Xu H, Lv J, Zhang J, Jiang X, et al. Coexistence of MCR-1 and NDM-1 in clinical Escherichia coli Isolates. Clin Infect Dis. 2016;63(10):1393–1395. doi: 10.1093/cid/ciw553. [DOI] [PubMed] [Google Scholar]
- 21.Huang H, Dong N, Shu L, Lu J, Sun, Qiaoling, Waichi Chan E, Chen S, et al. Colistin-resistance gene mcr in clinical carbapenem-resistant Enterobacteriaceae strains in China, 2014–2019. Emerg Microbes Infect. 2020;9(1):237–45. [DOI] [PMC free article] [PubMed]
- 22.Mediavilla JR, Patrawalla A, Chen L, Chavda KD, Mathema B, Vinnard C, et al. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. MBio. 2016;7(4):1–4. doi: 10.1128/mBio.01191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartsch SM, Mckinnell JA, Mueller LE, Miller LG, Gohil SK, Huang SS, et al. Potential economic burden of carbapenem-resistant Enterobacteriaceae (CRE) in the United States. Clin Microbiol Infect. 2017; [DOI] [PMC free article] [PubMed]
- 24.World Health Organization. Global Antimicriobial Resistance Surveillance System (GLASS) [Internet]. 2021 [cited 2022 Oct 4]. Available from: https://www.who.int/initiatives/glass
- 25.World Health Organization (WHO). Antimicrobial resistance: global report on surveillance [Internet]. 2014. Available from: https://www.who.int/antimicrobial-resistance/publications/surveillancereport/en/
- 26.World Health Organization (WHO). Global antimicrobial resistance surveillance system (GLASS) report: early implementation 2016–2017 [Internet]. Geneva, Switzerland; 2017. Available from: https://apps.who.int/iris/bitstream/handle/10665/259744/9789241513449-eng.pdf
- 27.World Health Organization (WHO). Global antimicrobial resistance surveillance system (GLASS) report: early implementation 2017–2018 [Internet]. Geneva, Switzerland; 2018. Available from: https://apps.who.int/iris/bitstream/handle/10665/279656/9789241515061-eng.pdf?ua=1
- 28.Berrazeg M, Diene SM, Medjahed L, Parola P, Drissi M, Raoult D, et al. New Delhi metallo-beta-lactamase around the world: an eReview using google maps. Eurosurveillance. 2014;19(20):1–14. doi: 10.2807/1560-7917.ES2014.19.20.20809. [DOI] [PubMed] [Google Scholar]
- 29.Manenzhe RI, Zar HJ, Nicol MP, Kaba M. The spread of carbapenemase-producing bacteria in Africa: a systematic review. J Antimicrob Chemother. 2015;70(1):23–40. doi: 10.1093/jac/dku356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tadesse BT, Ashley EA, Ongarello S, Havumaki J, Wijegoonewardena M, González IJ, et al. Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis. 2017;17(1):1–17. doi: 10.1186/s12879-017-2713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malchione MD, Torres LM, Hartley DM, Koch M, Goodman JL. Carbapenem and colistin resistance in Enterobacteriaceae in Southeast Asia: review and mapping of emerging and overlapping challenges. Int J Antimicrob Agents. 2019;54(4):381–399. doi: 10.1016/j.ijantimicag.2019.07.019. [DOI] [PubMed] [Google Scholar]
- 32.Ssekatawa K, Byarugaba DK, Wampande E, Ejobi F. A systematic review: the current status of carbapenem resistance in East Africa. BMC Res Notes. 2018;11(1):1–9. doi: 10.1186/s13104-018-3738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irek EO, Amupitan AA, Obadare TO, Aboderin AO. A systematic review of healthcare-associated infections in Africa: an antimicrobial resistance perspective. Afr J Lab Med. 2018 doi: 10.4102/ajlm.v7i2.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okomo U, Akpalu ENK, Le Doare K, Roca A, Cousens S, Jarde A, et al. Aetiology of invasive bacterial infection and antimicrobial resistance in neonates in sub-Saharan Africa: a systematic review and meta-analysis in line with the STROBE-NI reporting guidelines. Lancet Infect Dis. 2019;19(11):1219–1234. doi: 10.1016/S1473-3099(19)30414-1. [DOI] [PubMed] [Google Scholar]
- 35.Dadashi M, Sameni F, Bostanshirin N, Yaslianifard S, Khosravi-Dehaghi N, Nasiri MJ, et al. Global prevalence and molecular epidemiology of mcr-mediated Colistin resistance in Escherichia coli clinical isolates: a systematic review. J Glob Antimicrob Resist. 2021 doi: 10.1016/j.jgar.2021.10.022. [DOI] [PubMed] [Google Scholar]
- 36.Olowo-Okere A, Yacouba A. Molecular mechanisms of colistin resistance in africa: a systematic review of literature. Germs. 2020;10(4):367–379. doi: 10.18683/germs.2020.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.ProMED-Mail. International Society for Infectious Diseases [Internet]. [cited 2022 Oct 4]. Available from: https://www.promedmail.org/
- 38.Center for Disease Dynamics Economics and Policy. Resistance Map. [Internet]. [cited 2022 Oct 4]. Available from: https://resistancemap.cddep.org/
- 39.HealthMap. HealthMap - virus and contagious disease surveillance [Internet]. [cited 2022 Oct 4]. Available from: https://healthmap.org/en/
- 40.World Health Organization (WHO). The use of antiretroviral drugs for treating and preventing HIV infection Recommendations for a public health approach [Internet]. 2nd ed. World Health Organisation (WHO). Geneva; 2016. Definition of key terms. Available from: https://www.ncbi.nlm.nih.gov/books/NBK374295/
- 41.World Health Organization (WHO). Antimicrobial stewardship programmes in health-care facilities in low- and middle-income countries: a WHO practical toolkit [Internet]. Vol. 1, JAC-Antimicrobial Resistance. 2019. Available from: https://www.who.int/publications/i/item/9789241515481 [DOI] [PMC free article] [PubMed]
- 42.Africa Centres for Disease Control and Prevention. African Union Framework for Antimicrobial Resistance Control 2020–2025 [Internet]. 2020. Available from: https://africacdc.org/download/african-union-framework-for-antimicrobial-resistance-control-2020-2025/
- 43.Africa Centres for Disease Control and Prevention. African antibiotic treatment guidelines for common bacterial infections and syndromes. 2021. Available from: https://africacdc.org/download/african-antibiotic-treatment-guidelines-for-common-bacterial-infections-and-syndromes-2/
- 44.World Health Organization (WHO). Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in health care facilities [Internet]. Geneva; 2017. Available from: https://www.who.int/publications/i/item/9789241550178 [PubMed]
- 45.Dhaouadi S, Soufi L, Hamza A, Fedida D, Zied C, Awadhi E, et al. Co-occurrence of mcr-1 mediated colistin resistance and β-lactamase-encoding genes in multidrug-resistant Escherichia coli from broiler chickens with colibacillosis in Tunisia. J Glob Antimicrob Resist. 2020;22:538–545. doi: 10.1016/j.jgar.2020.03.017. [DOI] [PubMed] [Google Scholar]
- 46.WHO, FAO, OIE. Technical brief on water, sanitation, hygiene and wastewater management to prevent infections and reduce the spread of antimicrobial resistance [Internet]. 2020. Available from: https://www.who.int/water_sanitation_health/publications/wash-wastewater-management-to-prevent-infections-and-reduce-amr/en/
- 47.Mendelson M, Brink A, Gouws J, Mbelle N, Naidoo V, Pople T, et al. The One Health stewardship of colistin as an antibiotic of last resort for human health in South Africa. Lancet Infect Dis. 2018;18(9):e288–e294. doi: 10.1016/S1473-3099(18)30119-1. [DOI] [PubMed] [Google Scholar]
- 48.Abderrahim A, Djahmi N, Pujol C, Nedjai S, Bentakouk MC, Kirane-Gacemi D, et al. First case of NDM-1-producing Klebsiella pneumoniae in Annaba University Hospital, Algeria. Microb Drug Resist. 2017;23(7):895–900. doi: 10.1089/mdr.2016.0213. [DOI] [PubMed] [Google Scholar]
- 49.Agabou A, Lezzar N, Ouchenane Z, Khemissi S, Satta D, Sotto A, et al. Clonal relationship between human and avian ciprofloxacin-resistant Escherichia coli isolates in North-Eastern Algeria. Eur J Clin Microbiol Infect Dis. 2016. [DOI] [PubMed]
- 50.Belbel Z, Lalaoui R, Bakour S, Nedjai S, Djahmi N, Rolain JM. First report of colistin resistance in an OXA-48- and a CTX-M-15 producing Klebsiella pneumoniae clinical isolate in Algeria due to PmrB protein modification and mgrB inactivation. J Glob Antimicrob Resist. 2018;14:158–160. doi: 10.1016/j.jgar.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 51.Bentroki AA, Gouri A, Yakhlef A, Touaref A, Gueroudj A, Bensouilah T. Antibiotic resistance of strains isolated from community acquired urinary tract infections between 2007 and 2011 in Guelma (Algeria) Ann Biol Clin (Paris) 2012;70(6):666–668. doi: 10.1684/abc.2012.0760. [DOI] [PubMed] [Google Scholar]
- 52.Berrazeg M, Drissi M, Medjahed L, Rolain JM. Hierarchical clustering as a rapid tool for surveillance of emerging antibiotic-resistance phenotypes in Klebsiella pneumoniae strains. J Med Microbiol. 2013 doi: 10.1099/jmm.0.049437-0. [DOI] [PubMed] [Google Scholar]
- 53.Berrazeg M, Hadjadj L, Ayad A, Drissi M, Rolain JM. First detected human case in Algeria of mcr-1 plasmid-mediated colistin resistance in a 2011 Escherichia coli isolate. Antimicrob Agents Chemother. 2016;60(11):6996–6997. doi: 10.1128/AAC.01117-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Betitra Y, Teresa V, Miguel V, Abdelaziz T. Determinants of quinolone resistance in Escherichia coli causing community-acquired urinary tract infection in Bejaia, Algeria. Asian Pac J Trop Med. 2014. [DOI] [PubMed]
- 55.Cuzon G, Bentchouala C, Vogel A, Héry M, Lezzar A, Smati F, et al. First outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Constantine, Algeria. Int J Antimicrob Agents. 2015 doi: 10.1016/j.ijantimicag.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 56.Epelboin L, Robert J, Tsyrina-Kouyoumdjian E, Laouira S, Meyssonnier V, Caumes E. High rate of multidrug-resistant gram-negative bacilli carriage and infection in hospitalized returning travelers: a cross-sectional cohort study. J Travel Med. 2015;22(5):292–299. doi: 10.1111/jtm.12211. [DOI] [PubMed] [Google Scholar]
- 57.Gauthier L, Dortet L, Cotellon G, Creton E, Cuzon G, Ponties V, et al. Diversity of carbapenemase-producing Escherichia coli isolates in France in 2012–2013. Antimicrob Agents Chemother. 2018 doi: 10.1128/AAC.00266-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gharout-Sait A, Touati A, Benallaoua S, Guillard T, Brasme L, de Champs C. CTX-M from community-acquired urinary tract infections in Algeria. Afr J Microbiol Res. 2012;6(25). Available from: http://www.academicjournals.org/ajmr/abstracts/abstracts/abstract2012/5July/Gharout-Sait et al.htm
- 59.Gharout-Sait A, Touati A, Guillard T, Brasme L, de Champs C. Molecular characterization and epidemiology of cefoxitin resistance among Enterobacteriaceae lacking inducible chromosomal ampC genes from hospitalized and non-hospitalized patients in Algeria: description of new sequence type in Klebsiella pneumoniae iso. Brazil J Infect Dis. 2015 doi: 10.1016/j.bjid.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agabou A, Pantel A, Ouchenane Z, Lezzar N, Khemissi S, Satta D, et al. First description of OXA-48-producing Escherichia coli and the pandemic clone ST131 from patients hospitalised at a military hospital in Algeria. Eur J Clin Microbiol Infect Dis. 2014. [DOI] [PubMed]
- 61.Hecini-Hannachi A, Bentchouala C, Lezzar A, Laouar H, Benlabed K, Smati F. Multidrug-resistant bacteria isolated from patients hospitalized in intensive care unit in University Hospital of Constantine, Algeria (2011–2015) Afr J Microbiol Res. 2016;10(33):1328–1336. doi: 10.5897/AJMR2016.8257. [DOI] [Google Scholar]
- 62.Iabadene H, Messai Y, Ammari H, Alouache S, Verdet C, Bakour R, et al. Prevalence of plasmid-mediated AmpC β-lactamases among Enterobacteriaceae in Algiers hospitals. Int J Antimicrob Agents. 2009;34(4):340–342. doi: 10.1016/j.ijantimicag.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 63.Labid A, Gacemi-Kirane D, Timinouni M, Amoura K, Rolain J-M. High prevalence of extended spectrum beta-lactamase (ESBL) producers in fatal cases of pediatric septicemia among the Enterobacteriaceae in the pediatric hospital of Annaba, Algeria. Afr J Microbiol Res. 2014;8(9):947–954. doi: 10.5897/AJMR2013.6291. [DOI] [Google Scholar]
- 64.Lagha N, Abdelouahid D-E, Hassaine H, Robin F, Bonnet R. First characterization of CTX-M-15 and DHA-1 -lactamases among clinical isolates of Klebsiella pneumoniae in Laghouat Hospital, Algeria. Afr J Microbiol Res. 2014;8(11):1221–1227. doi: 10.5897/AJMR2013.6229. [DOI] [Google Scholar]
- 65.Lagha N, Hassaine H, Robin F, Bonnet R, Abdelouahid D-E. Prevalence and molecular typing of extended-spectrum -lactamases in Escherichia coli, Enterobacter cloacae and Citrobacter freundii isolates from Laghouat Hospital, Algeria. Afr J Microbiol Res. 2016;10(35):1430–1438. doi: 10.5897/AJMR2016.8263. [DOI] [Google Scholar]
- 66.Loucif L, Chelaghma W, Helis Y, Sebaa F, Douniazed Baoune R, Zaatout W, et al. First detection of OXA-48-producing Klebsiella pneumoniae in community-acquired urinary tract infection in Algeria. J Glob Antimicrob Resist. 2018;12:115–116. doi: 10.1016/j.jgar.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 67.Loucif L, Kassah-Laouar A, Saidi M, Messala A, Chelaghma W, Rolain JM. Outbreak of OXA-48-producing Klebsiella pneumoniae involving a sequence type 101 clone in Batna University Hospital, Algeria. Antimicrob Agents Chemother. 2016. [DOI] [PMC free article] [PubMed]
- 68.Mairi A, Pantel A, Sotto A, Lavigne JP, Touati A. OXA-48-like carbapenemases producing Enterobacteriaceae in different niches in Algeria: clonal expansion, plasmid characteristics and virulence traits. Eur J Clin Microbiol Infect Dis. 2018. [DOI] [PubMed]
- 69.Mairi A, Touati A, Ait Bessai S, Boutabtoub Y, Khelifi F, Sotto A, et al. Carbapenemase-producing Enterobacteriaceae among pregnant women and newborns in Algeria: prevalence, molecular characterization, maternal-neonatal transmission, and risk factors for carriage. Am J Infect Control. 2019;47(1):105–108. doi: 10.1016/j.ajic.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 70.Medboua-Benbalagh C, Touati A, Kermas R, Gharout-Sait A, Brasme L, Mezhoud H, et al. Fecal carriage of extended-spectrum beta-lactamase-producing enterobacteriaceae strains is associated with worse outcome in patients hospitalized in the pediatric oncology unit of Beni-Messous Hospital in Algiers. Algeria Microb Drug Resist. 2017;23(6):757–763. doi: 10.1089/mdr.2016.0153. [DOI] [PubMed] [Google Scholar]
- 71.Aggoune N, Tali-Maamar H, Assaous F, Benamrouche N, Naim M, Rahal K. Emergence of plasmid mediated carbapenemase OXA-48 in a Klebsiella pneumoniae strain in Algeria. J Glob Antimicrob Resist. 2014 doi: 10.1016/j.jgar.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 72.Mellouk FZ, Bakour S, Meradji S, Al-Bayssari C, Bentakouk MC, Zouyed F, et al. First detection of VIM-4-producing Pseudomonas aeruginosa and OXA-48-producing Klebsiella pneumoniae in Northeastern (Annaba, Skikda) Algeria. Microb Drug Resist. 2017;23(3):335–344. doi: 10.1089/mdr.2016.0032. [DOI] [PubMed] [Google Scholar]
- 73.Messai Y, Benhassine T, Naim M, Paul G, Bakour R. Prevalence of β-lactams resistance among Escherichia coli clinical isolates from a hospital in Algiers. Rev Esp Quimioter. 2006. [PubMed]
- 74.Messai Y, Iabadene H, Benhassine T, Alouache S, Tazir M, Gautier V, et al. Prevalence and characterization of extended-spectrum β-lactamases in Klebsiella pneumoniae in Algiers hospitals (Algeria) Pathol Biol. 2008 doi: 10.1016/j.patbio.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 75.Nabti LZ, Sahli F, Hadjadj L, Ngaignam EP, Lupande-Mwenebitu D, Rolain J-M, et al. Autochthonous case of mobile colistin resistance gene mcr-1 from a uropathogenic Escherichia coli isolate in Sétif Hospital Algeria. J Antimicrob Resist. 2019;19:356–357. doi: 10.1016/j.jgar.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 76.Nabti LZ, Sahli F, Ngaiganam EP, Radji N, Mezaghcha W, Lupande-Mwenebitu D, et al. Development of real-time PCR assay allowed describing the first clinical Klebsiella pneumoniae isolate harboring plasmid-mediated colistin resistance mcr-8 gene in Algeria. J Glob Antimicrob Resist. 2020;20:266–271. doi: 10.1016/j.jgar.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 77.Nedjai S, Barguigua A, Djahmi N, Jamali L, Zerouali K, Dekhil M, et al. Prevalence and characterization of extended spectrum β-lactamases in Klebsiella-Enterobacter-Serratia group bacteria. Algeria. Med Mal Infect. 2012 doi: 10.1016/j.medmal.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 78.Potron A, Rondinaud E, Nordmann P, Poirel L, Rondinaud E, Nordmann P. Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Surveill Outbreak Reports. 2013;18(31). Available from: www.eurosurveillance.org [DOI] [PubMed]
- 79.Ramdani-Bouguessa N, Manageiro V, Jones-Dias D, Ferreira E, Tazir M, Caniça M. Role of SHV β-lactamase variants in resistance of clinical Klebsiella pneumoniae strains to β-lactams in an Algerian hospital. J Med Microbiol. 2011 doi: 10.1099/jmm.0.030577-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramdani-Bouguessa N, Mendonça N, Leitão J, Ferreira E, Tazir M, Caniça M. CTX-M-3 and CTX-M-15 extended-spectrum β-lactamases in isolates of Escherichia coli from a hospital in Algiers. Algeria. J Clin Microbiol. 2006 doi: 10.1128/JCM.01445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robin F, Aggoune-Khinache N, Delmas J, Naim M, Bonnet R. Novel VIM metallo-β-lactamase variant from clinical isolates of Enterobacteriaceae from Algeria. Antimicrob Agents Chemother. 2010 doi: 10.1128/AAC.00017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aggoune N, Tali-Maamar H, Assaous F, Guettou B, Laliam R, Benamrouche N, et al. Wide spread of oxa-48-producing enterobacteriaceae in algerian hospitals: a four years’ study. J Infect Dev Ctries. 2018;12(11):1039–1044. doi: 10.3855/jidc.9692. [DOI] [PubMed] [Google Scholar]
- 83.Rodriguez-Martinez JM, Nordmann P, Fortineau N, Poirel L. VIM-19, a metallo-β-lactamase with increased carbapenemase activity from Escherichia coli and Klebsiella pneumoniae. Antimicrob Agents Chemother. 2010;54(1):471–476. doi: 10.1128/AAC.00458-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sassi A, Loucif L, Gupta SK, Dekhil M, Chettibi H, Rolain JM. NDM-5 carbapenemase-encoding gene in multidrug-resistant clinical isolates of Escherichia coli from Algeria. Antimicrob Agents Chemother. 2014 doi: 10.1128/AAC.02818-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Touati A, Benallaoua S, Forte D, Madoux J, Brasme L, de Champs C. First report of CTX-M-15 and CTX-M-3 β-lactamases among clinical isolates of Enterobacteriaceae in Béjaia, Algeria. Int J Antimicrob Agents. 2006. [DOI] [PubMed]
- 86.Toumi S, Meliani S, Amoura K, Rachereche A, Djebien M, Djahoudi A. Multidrug-resistant Gram-negative bacilli producing oxacillinases and Metallo-β-lactamases isolated from patients in intensive care unit - Annaba hospital - Algeria (2014–2016) J Appl Pharm Sci. 2018;8(7):107–113. doi: 10.7324/JAPS.2018.8717. [DOI] [Google Scholar]
- 87.Yagoubat M, Ould El-Hadj-Khelil A, Malki A, Bakour S, Touati A, Rolain JM. Genetic characterisation of carbapenem-resistant Gram-negative bacteria isolated from the University Hospital Mohamed Boudiaf in Ouargla, southern Algeria. J Glob Antimicrob Resist. 2017;8:55–59. doi: 10.1016/j.jgar.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 88.Yahiaoui M, Robin F, Bakour R, Hamidi M, Bonnet R, Messai Y. Antibiotic resistance, virulence, and genetic background of community-acquired uropathogenic Escherichia coli from Algeria. Microb Drug Resist. 2015;21(5):516–526. doi: 10.1089/mdr.2015.0045. [DOI] [PubMed] [Google Scholar]
- 89.Yanat B, Machuca J, Díaz-De-Alba P, Mezhoud H, Touati A, Pascual Á, et al. Characterization of plasmid-mediated quinolone resistance determinants in high-level quinolone-resistant enterobacteriaceae isolates from the community: first Report of qnrD gene in Algeria. Microb Drug Resist. 2017;23(1):90–97. doi: 10.1089/mdr.2016.0031. [DOI] [PubMed] [Google Scholar]
- 90.Yanat B, Machuca J, Yahia RD, Touati A, Pascual Á, Rodriguez-Martinez JM, et al. First report of the plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate in Algeria. Int J Antimicrob Agents. 2016;48(6):760–761. doi: 10.1016/j.ijantimicag.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 91.Yousfi H, Hadjadj L, Dandachi I, Lalaoui R, Merah A, Amoura K, et al. Colistin- and carbapenem-resistant Klebsiella pneumoniae clinical isolates: Algeria. Microb Drug Resist. 2019;25(2):258–263. doi: 10.1089/mdr.2018.0147. [DOI] [PubMed] [Google Scholar]
- 92.Zenati F, Barguigua A, Nayme K, Benbelaïd F, Khadir A, Bellahsene C, et al. Characterization of uropathogenic ESBL-producing Escherichia coli isolated from hospitalized patients in western Algeria. J Infect Dev Ctries. 2019;13(4):291–302. doi: 10.3855/jidc.10702. [DOI] [PubMed] [Google Scholar]
- 93.Ahmed ZB, Ayad A, Mesli E, Messai Y, Bakour R, Drissi M. CTX-M-15 extended-spectrum β-lactamases in Enterobacteriaceae in the intensive care unit of Tlemcen Hospital, Algeria. East Mediterr Heal J. 2012; [DOI] [PubMed]
- 94.Aouf A, Gueddi T, Djeghout B, Ammari H. Frequency and susceptibility pattern of uropathogenic enterobacteriaceae isolated from patients in Algiers. Algeria J Infect Dev Ctries. 2018;12(4):244–249. doi: 10.3855/jidc.10017. [DOI] [PubMed] [Google Scholar]
- 95.Ayad A, Drissi M, de Curraize C, Dupont C, Hartmann A, Solanas S, et al. Occurence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in extended-spectrum β-lactamases producing Escherichia coli in Algerian hospitals. Front Microbiol. 2016;7(SEP). [DOI] [PMC free article] [PubMed]
- 96.Bakour S, Sahli F, Touati A, Rolain JM. Emergence of KPC-producing Klebsiella pneumoniae ST512 isolated from cerebrospinal fluid of a child in Algeria. New Microbes New Infect. 2015;3(C):34–6. [DOI] [PMC free article] [PubMed]
- 97.Belbel Z, Chettibi H, Dekhil M, Ladjama A, Nedjai S, Rolain JM. Outbreak of an armA Methyltransferase-producing ST39 Klebsiella pneumoniae clone in a pediatric Algerian hospital. Microb Drug Resist. 2014. [DOI] [PubMed]
- 98.Kieffer N, Nordmann P, Aires-De-Sousa M, Poirel L. High prevalence of carbapenemase-producing Enterobacteriaceae among hospitalized children in Luanda, Angola. Antimicrob Agents Chemother. 2016. [DOI] [PMC free article] [PubMed]
- 99.Poirel L, Goutines J, Aires-De-Sousa M, Nordmann P. High rate of association of 16S rRNA methylases and carbapenemases in enterobacteriaceae recovered from Hospitalized Children in Angola. Antimicrob Agents Chemother. 2018;62(4):1–7. doi: 10.1128/AAC.00021-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ahoyo AT, Baba-Moussa L, Anago AE, Avogbe P, Missihoun TD, Loko F, et al. Incidence of infections due to Escherichia coli strains producing extended spectrum betalactamase, in the Zou/Collines Hospital Centre (CHDZ/C) in Benin. Med Mal Infect. 2007. [DOI] [PubMed]
- 101.Ahoyo TA, Bankolé HS, Adéoti FM, Gbohoun AA, Assavèdo S, Amoussou-Guénou M, et al. Prevalence of nosocomial infections and anti-infective therapy in Benin: Results of the first nationwide survey in 2012. Antimicrob Resist Infect Control. 2014 doi: 10.1186/2047-2994-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Anago E, Ayi-Fanou L, Akpovi CD, Hounkpe WB, Agassounon-Djikpo Tchibozo M, Bankole HS, et al. Antibiotic resistance and genotype of beta-lactamase produci.g Escherichia coli in nosocomial infections in Cotonou, Benin. Ann Clin Microbiol Antimicrob. 2015. [DOI] [PMC free article] [PubMed]
- 103.Dougnon V, Koudokpon H, Hounmanou YMG, Azonbakin S, Fabiyi K, Oussou A, et al. High prevalence of multidrug-resistant bacteria in the centre hospitalier et Universitaire de la Mère et de l’Enfant Lagune (CHU-MEL) reveals implications of poor hygiene practices in healthcare. SN Compr Clin Med. 2019;1(12):1029–1037. doi: 10.1007/s42399-019-00149-3. [DOI] [Google Scholar]
- 104.Koudokon H, Dougnon V, Hadjadj L, Kissira I, Fanou B, Loko F, et al. First Sequence Analysis of genes mediating extended-spectrum beta-lactamase (ESBL) bla-TEM, SHV-and CTX-M production in isolates of enterobacteriaceae in Southern Benin. Int J Infect. 2018 doi: 10.5812/iji.83194. [DOI] [Google Scholar]
- 105.Mousse W, Sina H, Wele M, Chabi N, Nouvlessounon DD, Bade FT, et al. Molecular characterization and Antibiotic resistance profiles of Escherichia coli extended-spectrum β-lactamases producer strains isolated from urine samples in Benin. Eur Sci J. 2018;14(30):323–337. [Google Scholar]
- 106.Mpinda-Joseph P, Anand Paramadhas BD, Reyes G, Maruatona MB, Chise M, Monokwane-Thupiso BB, et al. Healthcare-associated infections including neonatal bloodstream infections in a leading tertiary hospital in Botswana. Hosp Pract. 2019;47(4):203–210. doi: 10.1080/21548331.2019.1650608. [DOI] [PubMed] [Google Scholar]
- 107.Amana MD, Wend-Kuni TRY, Aminata BY, Mahoukede ZT, Serge S, Koudbi ZJ, et al. Detection of multidrug-resistant enterobacteria simultaneously producing extended-spectrum β- lactamases of the PER and GES types isolated at Saint Camille Hospital Center, Ouagadougou, Burkina Faso. Afr J Microbiol Res. 2019;13(26):414–420. doi: 10.5897/AJMR2019.9147. [DOI] [Google Scholar]
- 108.Ouédraogo AS, Sanou S, Kissou A, Poda A, Aberkane S, Bouzinbi N, et al. Fecal carriage of enterobacteriaceae producing extended-spectrum beta-lactamases in hospitalized patients and healthy community volunteers in Burkina Faso. Microb Drug Resist. 2017;23(1):63–70. doi: 10.1089/mdr.2015.0356. [DOI] [PubMed] [Google Scholar]
- 109.Ouedraogo A-S, Sanou M, Kissou A, Sanou S, Solaré H, Kaboré F, et al. High prevalence of extended-spectrum β-lactamase producing enterobacteriaceae among clinical isolates in Burkina Faso. BMC Infect Dis. 2016 doi: 10.1186/s12879-016-1655-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Reid CS, Bonkoungou K. Vesico-umbilical fistula in a child with severe vesico-ureteral reflux and bladder diverticulum. Trop Doct. 2017;47(3):271–273. doi: 10.1177/0049475516687433. [DOI] [PubMed] [Google Scholar]
- 111.Sanou M, Ky A, Ouangre E, Bisseye C, Sanou A, Nagalo BM, et al. Characterization of bacterial flora in community peritonitis carried out in Burkina Faso. Pan Afr Med J. 2014 doi: 10.11604/pamj.2014.18.17.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Toy T, Pak GD, Duc TP, Campbell JI, El Tayeb MA, Von Kalckreuth V, et al. Multicountry distribution and characterization of extended-spectrum beta-lactamase-associated gram-negative bacteria from bloodstream infections in Sub-Saharan Africa. Clin Infect Dis. 2019;69(S6):449–458. doi: 10.1093/cid/ciz450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zongo KJ, Dabire AM, Compaore LG, Sanou I, Sangare L, Simpore J, et al. First detection of bla TEM, SHV and CTX-M among Gram negative bacilli exhibiting extended spectrum -lactamase phenotype isolated at University Hospital Center, Yalgado Ouedraogo, Ouagadougou, Burkina Faso. Afr J Biotechnol. 2015;14(14):1174–1180. doi: 10.5897/AJB2014.13908. [DOI] [Google Scholar]
- 114.Frida ST, Karim OA, Theodora ZM, Dorcas O-Y, Theophane YA, Florencia DW, et al. Prevalence of lower genital tract infections in women: case of Saint Camille Hospital of Ouagadougou from 2015 to 2018. Int J Curr Res. 2019;11(10):7721–7727. [Google Scholar]
- 115.Guira O, Tiéno H, Sagna Y, Yaméogo TM, Zoungrana L, Traoré S, et al. Antibiotic susceptibility of bacteria isolated from diabetic foot infections and prospects for empiric antibiotic therapy in Ouagadougou (Burkina Faso) Med Sante Trop. 2015;25(3):291–295. doi: 10.1684/mst.2015.0493. [DOI] [PubMed] [Google Scholar]
- 116.Guiral E, Gonçalves Quiles M, Munoz L, Moreno-Morales J, Aejo-Cancho I, Salvador P, et al. Emergence of resistance to quinolones and B-lactam antibiotics in enteroaggregative and enterotoxigenic Escherichia coli causing traveler’s diarrhea. Antimicrob Agents Chemother. 2019;63(2):e01745–e1818. doi: 10.1128/AAC.01745-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Konaté A, Dembélé R, Guessennd NK, Kouadio FK, Kouadio IK, Ouattara MB, et al. Epidemiology and antibiotic resistance phenotypes of diarrheagenic Escherichia coli responsible for infantile gastroenteritis in Ouagadougou, Burkina Faso. Eur J Microbiol Immunol. 2017;7(3):168–175. doi: 10.1556/1886.2017.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kpoda DSS, Guessennd N, Bonkoungou JI, Ouattara MB, Konan F, Ajayi A, et al. Prevalence and resistance profile of extended-spectrum beta -lactamases-producing Enterobacteriaceae in Ouagadougou, Burkina Faso. Afr J Microbiol Res. 2017;11(27):1120–1126. doi: 10.5897/AJMR2017.8598. [DOI] [Google Scholar]
- 119.Maltha J, Guiraud I, Kaboré B, Lompo P, Ley B, Bottieau E, et al. Frequency of severe malaria and invasive bacterial infections among children admitted to a rural hospital in Burkina Faso. PLoS ONE. 2014 doi: 10.1371/journal.pone.0089103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Metuor-Dabire A, Zongo JK, Zeba B, Ouédraogo RT, Moussawi J, Baucher M, et al. First detection of SHV-type extended spectrum B-Lactamases in the University Hospital Complex Paediatric Charles de Gaulle (CUP-CDG) of Ouagadougou in Burkina Faso. J Asian Sci Res. 2014;4(5):214–221. [Google Scholar]
- 121.Ouédraogo AS, Compain F, Sanou M, Aberkane S, Bouzinbi N, Hide M, et al. First description of IncX3 plasmids carrying blaOXA-181 in Escherichia coli clinical isolates in Burkina Faso. Antimicrob Agents Chemother. 2016;60(5):3240–3242. doi: 10.1128/AAC.00147-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ateudjieu J, Bita’a LB, Guenou E, Chebe AN, Chukuwchindun BA, Goura AP, et al. Profile and antibiotic susceptibility pattern of bacterial pathogens associated with diarrheas in patients presenting at the Kousseri regional hospital Anne, Far North, Cameroon. Pan Afr Med J. 2018 doi: 10.11604/pamj.2018.29.170.14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lonchel CM, Melin P, Gangoué-Piéboji J, Assoumou MCO, Boreux R, De Mol P. Extended-spectrum β-lactamase-producing Enterobacteriaceae in Cameroonian hospitals. Eur J Clin Microbiol Infect Dis. 2013;32(1):79–87. doi: 10.1007/s10096-012-1717-4. [DOI] [PubMed] [Google Scholar]
- 124.Ngalani OJT, Mbaveng AT, Marbou WJT, Ngai RY, Kuete V. Antibiotic resistance of enteric bacteria in HIV-infected patients at the Banka Ad-Lucem Hospital, West Region of Cameroon. Can J Infect Dis Med Microbiol. 2019. [DOI] [PMC free article] [PubMed]
- 125.Yeika EV, Foryoung JB, Efie DT, Nkwetateba EA, Tolefac PN, Ngowe MN. Multidrug resistant Proteus mirabilis and Escherichia coli causing fulminant necrotising fasciitis: a case report. BMC Res Notes. 2018 doi: 10.1186/s13104-018-3413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Betbeui A, Kamga H, Toukam M, Mbakop C, Lyonga E, Bilong S, et al. Phenotypic Detection of Extended Spectrum Beta-Lactamase and Carbapenemases Produced by Klebsiella spp Isolated from Three Referrals Hospitals in Yaounde, Cameroon. Br Microbiol Res J. 2015.
- 127.Dehayem M, Ngassam E, Mendane F, Balla V, Saji J, Sobngwi E, et al. OP67 Bacteriology of diabetic foot infections and susceptibility to antimicrobial agents in Cameroon. Diabetes Res Clin Pract. 2014;103:S27. doi: 10.1016/S0168-8227(14)70090-7. [DOI] [Google Scholar]
- 128.Dortet L, Poirel L, Anguel N, Nordmann P. New Delhi metallo-β-lactamase 4-producing Escherichia coli in Cameroon. Emerg Infect Dis. 2012. [DOI] [PMC free article] [PubMed]
- 129.Founou LL, Founou RC, Allam M, Ismail A, Essack SY. Draft genome sequence of an extended-spectrum β-lactamase (CTX-M-15)-producing Escherichia coli ST10 isolated from a nasal sample of an abattoir worker in Cameroon. J Glob Antimicrob Resist. 2018;14:68–69. doi: 10.1016/j.jgar.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 130.Founou LL, Founou RC, Ntshobeni N, Govinden U, Bester LA, Chenia HY, et al. Emergence and spread of extended spectrum β-lactamase producing enterobacteriaceae (ESBL-PE) in pigs and exposed workers: a multicentre comparative study between Cameroon and South Africa. Pathogens. 2019;8(1). [DOI] [PMC free article] [PubMed]
- 131.Gangoue-Pieboji J, Koulla-Shiro S, Ngassam P, Adiogo D, Ndumbe P. Antimicrobial activity against gram negative bacilli from Yaounde Central Hospital, Cameroon. Afr Health Sci. 2006; [DOI] [PMC free article] [PubMed]
- 132.Gangoué-Piéboji J, Miriagou V, Vourli S, Tzelepi E, Ngassam P, Tzouvelekis LS. Emergence of CTX-M-15-producing enterobacteria in Cameroon and characterization of a blaCTX-M-15-carrying element. Antimicrob Agents Chemother. 2005;49(1):441–443. doi: 10.1128/AAC.49.1.441-443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lonchel CM, Meex C, Gangoue-Piebogji J, Boreux R, Assoumou M-CO, Melin P, et al. Proportion of extended-spectrum ß-lactamase-producing Enterobacteriaceae in community setting in Ngaoundere, Cameroon. BMC Infect Dis. 2012; [DOI] [PMC free article] [PubMed]
- 134.Farra A, Frank T, Tondeur L, Bata P, Gody JC, Onambele M, et al. High rate of faecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae in healthy children in Bangui, Central African Republic. Clin Microbiol Infect. 2016; [DOI] [PubMed]
- 135.Rafai C, Frank T, Manirakiza A, Gaudeuille A, Mbecko J-R, Nghario L, et al. Dissemination of IncF-type plasmids in multiresistant CTX-M-15-producing Enterobacteriaceae isolates from surgical-site infections in Bangui, Central African Republic Clotaire. BMC Microbiol. 2015;15(15). [DOI] [PMC free article] [PubMed]
- 136.Kengne M, Dounia AT, Nwobegahay JM. Bacteriological profile and antimicrobial susceptibility patterns of urine culture isolates from patients in Ndjamena. Chad. Pan Afr Med J. 2017 doi: 10.11604/pamj.2017.28.258.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mahamat OO, Lounnas M, Hide M, Dumont Y, Tidjani A, Kamougam K, et al. High prevalence and characterization of extended-spectrum ß-lactamase producing Enterobacteriaceae in Chadian hospitals. BMC Infect Dis. 2019 doi: 10.1186/s12879-019-3838-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mahamat OO, Lounnas M, Hide M, Tidjani A, Benavides J, Diack A, et al. Spread of NDM-5 and OXA-181 Carbapenemase-Producing Escherichia coli in Chad. Antimicrob Agents Chemother. 2019;63(11):1–5. doi: 10.1128/AAC.00646-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ndoutamia G, Yandai FH, Nadlaou B. Antimicrobial resistance in extended spectrum β-lactamases (ESBL)-producing Escherichia coli isolated from human urinary tract infections in Ndjamena, Chad. African J Microbiol Res. 2015.
- 140.Yandaïab F, Zongoa C, Moussac A, Bessimbayea N, Tapsobaa F, Savadogoa A, et al. Prevalence and antimicrobial susceptibility of faecal carriage of Extended-Spectrum β -lactamase (ESBL) producing Escherichia coli at the “ Hôpital de la M ère et de l’Enfant ” in N’ Djamena. Chad Sci J Microbiol. 2014;3:25–31. [Google Scholar]
- 141.Yandai FH, Ndoutamia G, Nadlaou B, Barro N. Prevalence and resistance profile of Escherichia coli and Klebsiella pneumoniae isolated from urinary tract infections in N’Djamena. Tchad Int J Biol Chem Sci. 2019;13(4):2065–2073. doi: 10.4314/ijbcs.v13i4.13. [DOI] [Google Scholar]
- 142.Moyen R, Ahombo G, Nguimbi E, Ontsira NE, Niama RF, Yala GC, et al. Activity of beta-lactam antibiotics and production of beta-lactamases in bacteria isolated from wound infections in Brazzaville, Congo. African J Microbiol Res. 2014;
- 143.Mpelle FL, Ngoyi ENO, Kayath CA, Nguimbi E, Moyen R, Kobawila SC. First report of the types TEM, CTX-M, SHV and OXA-48 of beta-lactamases in Escherichia coli, from Brazzaville, Congo. Afr J Microbiol Res. 2019;13(8):158–167. doi: 10.5897/AJMR2018.9042. [DOI] [Google Scholar]
- 144.Abe IA, Koffi M, Sokouri PD, Ahouty BA, N’djetchi MK, Simaro S, et al. Assessment of drugs pressure on Escherichia coli and Klebsiella spp. uropathogens in patients attending Abobo-Avocatier Hospital, North of Abidjan (Côte d’Ivoire) African J Microbiol Res. 2019;13(29):658–666. doi: 10.5897/AJMR2019.9166. [DOI] [Google Scholar]
- 145.Breurec S, Guessennd N, Timinouni M, Le TAH, Cao V, Ngandjio A, et al. Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: Multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin Microbiol Infect. 2013. [DOI] [PubMed]
- 146.Guessend KN, Toty AA, Gbonon MC, Dondelinger M, Toe E, Ouattara MB, et al. CTX-M-15 extended-spectrum-Β-lactamase among clinical isolates of enterobacteriaceae in Abidjan. Côte d’Ivoire Int J Biol Res. 2017;2(3):5–8. [Google Scholar]
- 147.Guessennd N, Bremont S, Gbonon V, Kacou-NDouba A, Ekaza E, Lambert T, et al. Qnr-type quinolone resistance in extended-spectrum beta-lactamase producing enterobacteria in Abidjan, Ivory Coast. Pathol Biol. 2008. [DOI] [PubMed]
- 148.Maataoui N, Mayet A, Duron S, Delacour H, Mentre F, Laouenan C, et al. High acquisition rate of extended-spectrum beta-lactamase-producing Enterobacteriaceae among French military personnel on mission abroad, without evidence of inter-individual transmission. Clin Microbiol Infect. 2019;25(5):631.e1–631.e9. doi: 10.1016/j.cmi.2018.07.030. [DOI] [PubMed] [Google Scholar]
- 149.Moroh JLA, Fleury Y, Tia H, Bahi C, Lietard C, Coroller L, et al. Diversity and antibiotic resistance of uropathogenic bacteria from Abidjan. Afr J Urol. 2014;20(1):18–24. doi: 10.1016/j.afju.2013.11.005. [DOI] [Google Scholar]
- 150.Müller-Schulte E, Tuo MN, Akoua-Koffi C, Schaumburg F, Becker SL. High prevalence of ESBL-producing Klebsiella pneumoniae in clinical samples from central Côte d’Ivoire. Int J Infect Dis. 2020;91:207–209. doi: 10.1016/j.ijid.2019.11.024. [DOI] [PubMed] [Google Scholar]
- 151.Irenge LM, Ambroise J, Bearzatto B, Durant JF, Chirimwami RB, Gala JL. Whole-genome sequences of multidrug-resistant Escherichia coli in South-Kivu Province, Democratic Republic of Congo: characterization of phylogenomic changes, virulence and resistance genes. BMC Infect Dis. 2019;19(1). [DOI] [PMC free article] [PubMed]
- 152.Irenge LM, Kabego L, Kinunu FB, Itongwa M, Mitangala PN, Gala JL, et al. Antimicrobial resistance of bacteria isolated from patients with bloodstream infections at a tertiary care hospital in the Democratic Republic of the Congo. S Afr Med J. 2015;105(9):752–755. doi: 10.7196/SAMJnew.7937. [DOI] [PubMed] [Google Scholar]
- 153.Irenge LM, Kabego L, Vandenberg O, Chirimwami RB, Gala JL. Antimicrobial resistance in urinary isolates from inpatients and outpatients at a tertiary care hospital in South-Kivu Province (Democratic Republic of Congo). BMC Res Notes. 2014. [DOI] [PMC free article] [PubMed]
- 154.Plantamura J, Bousquet A, Védy S, Larréché S, Bigaillon C, Delacour H, et al. Molecular epidemiological of extended-spectrum β-lactamase producing Escherichia coli isolated in Djibouti. J Infect Dev Ctries. 2019;13(8):753–758. doi: 10.3855/jidc.11283. [DOI] [PubMed] [Google Scholar]
- 155.Abdallah HM, Wintermans BB, Reuland EA, Koek A, Naiemi N Al, Ammar AM, et al. Extended-spectrum β-lactamase- and carbapenemase-producing enterobacteriaceae isolated from Egyptian patients with suspected blood stream infection. PLoS ONE. 2015;10(5). [DOI] [PMC free article] [PubMed]
- 156.Mohamed NM, Youssef AAF. In vitro activity of tigecycline and comparators against gram-negative bacteria isolated from a tertiary hospital in Alexandria, Egypt. Microb Drug Resist. 2011;17(4):489–495. doi: 10.1089/mdr.2010.0195. [DOI] [PubMed] [Google Scholar]
- 157.Mohamed T, Yousef LM, Darweesh EI, Khalil AH, Meghezel EM. Detection and characterization of carbapenem resistant enterobacteriacea in Sohag University Hospitals, Egypt. J Med Microbiol. 2018;27(4):61–69. [Google Scholar]
- 158.Mohammed ESH, Fakhr AE, El Sayed HM, Al Johery SAE, Hassanein WAG. Spread of TEM, VIM, SHV, and CTX-M β -Lactamases in Imipenem-Resistant Gram-Negative Bacilli Isolated from Egyptian Hospitals. Int J Microbiol. 2016;2016. [DOI] [PMC free article] [PubMed]
- 159.Mohsen L, Ramy N, Saied D, Akmal D, Salama N, Abdel Haleim MM, et al. Emerging antimicrobial resistance in early and late-onset neonatal sepsis. Antimicrob Resist Infect Control. 2017;6(1). [DOI] [PMC free article] [PubMed]
- 160.Moore KL, Kainer MA, Badrawi N, Afifi S, Wasfy M, Bashir M, et al. Neonatal sepsis in Egypt associated with bacterial contamination of glucose-containing intravenous fluids. Pediatr Infect Dis J. 2005;24(7):590–594. doi: 10.1097/01.inf.0000168804.09875.95. [DOI] [PubMed] [Google Scholar]
- 161.Mukhtar A, Abdelaal A, Hussein M, Dabous H, Fawzy I, Obayah G, et al. Infection complications and pattern of bacterial resistance in living-donor liver transplantation: a multicenter epidemiologic study in Egypt. Transplant Proc. 2014;46(5):1444–1447. doi: 10.1016/j.transproceed.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 162.Nazeih S, Serry F, Abbas H. Study on increased antimicrobial resistance among bacteria isolated from ICUs Zagazig University Hospitals. Zagazig J Pharm Sci. 2019;28(1):13–25. doi: 10.21608/zjps.2019.13544.1001. [DOI] [Google Scholar]
- 163.Newire EA, Ahmed SF, House B, Valiente E, Pimentel G. Detection of new SHV-12, SHV-5 and SHV-2a variants of extended spectrum beta-lactamase in Klebsiella pneumoniae in Egypt. Ann Clin Microbiol Antimicrob. 2013 doi: 10.1186/1476-0711-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nour I, Eldegla HE, Nasef N, Shouman B, Abdel-Hady H, Shabaan AE. Risk factors and clinical outcomes for carbapenem-resistant Gram-negative late-onset sepsis in a neonatal intensive care unit. J Hosp Infect. 2017;97:52–58. doi: 10.1016/j.jhin.2017.05.025. [DOI] [PubMed] [Google Scholar]
- 165.Osama R, Bakeer W, Fadel S, Amin M. Association of carbapenem and colistin resistance in pathogenic Gram negative bacteria. J Pure Appl Microbiol. 2019;13(2):733–739. doi: 10.22207/JPAM.13.2.09. [DOI] [Google Scholar]
- 166.Abdelaziz MO, Bonura C, Aleo A, El-Domany RA, Fasciana T, Mammina C. OXA-163-producing Klebsiella pneumoniae in Cairo, Egypt, in 2009 and 2010. J Clin Microbiol. 2012;50(7):2489–2491. doi: 10.1128/JCM.06710-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Osman KM, Kappell AD, Elhofy F, Orabi A, Mubarak AS, Dawoud TM, et al. Urinary tract infection attributed to Escherichia coli isolated from participants attending an unorganized gathering. Future Microbiol. 2018;13(7):745–756. doi: 10.2217/fmb-2017-0304. [DOI] [PubMed] [Google Scholar]
- 168.Östholm-Balkhed Å, Tärnberg M, Nilsson M, Nilsson LE, Hanberger H, Hällgren A. Travel-associated faecal colonization with esbl-producing enterobacteriaceae: Incidence and risk factors. J Antimicrob Chemother. 2013;68(9):2144–2153. doi: 10.1093/jac/dkt167. [DOI] [PubMed] [Google Scholar]
- 169.Poirel L, Abdelaziz MO, Bernabeu S, Nordmann P. Occurrence of OXA-48 and VIM-1 carbapenemase-producing Enterobacteriaceae in Egypt. Int J Antimicrob Agents. 2013;41(1):90–91. doi: 10.1016/j.ijantimicag.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 170.Principe L, Mauri C, Conte V, Pini B, Giani T, Rossolini GM, et al. First report of NDM-1-producing Klebsiella pneumoniae imported from Africa to Italy: evidence of the need for continuous surveillance. J Glob Antimicrob Resist. 2017;8:23–27. doi: 10.1016/j.jgar.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 171.Putnam SD, Riddle MS, Wierzba TF, Pittner BT, Elyazeed RA, El-Gendy A, et al. Antimicrobial susceptibility trends among Escherichia coli and Shigella spp. isolated from rural Egyptian paediatric populations with diarrhoea between 1995 and 2000. Clin Microbiol Infect. 2004;10(9):804–810. doi: 10.1111/j.1469-0691.2004.00927.x. [DOI] [PubMed] [Google Scholar]
- 172.Ramadan H, Gupta SK, Sharma P, Ahmed M, Hiott LM, Barrett JB, et al. Circulation of emerging NDM-5-producing Escherichia coli among humans and dogs in Egypt. Zoonoses Public Health. 2020;67(3):324–329. doi: 10.1111/zph.12676. [DOI] [PubMed] [Google Scholar]
- 173.Ramadan H, Rasha B, Mona IS, Lamiaa A. Random amplified DNA polymorphism of Klebsiella pneumoniae isolates from Mansoura University Hospitals, Egypt. Afr J Microbiol Res. 2015;9(9):621–630. doi: 10.5897/AJMR2014.7256. [DOI] [Google Scholar]
- 174.Rizk DE, El-Mahdy AM. Emergence of class 1 to 3 integrons among members of Enterobacteriaceae in Egypt. Microb Pathog. 2017;112:50–56. doi: 10.1016/j.micpath.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 175.Saied GM. Microbial pattern and antimicrobial resistance, a surgeon’s perspective: retrospective study in surgical wards and seven intensive-care units in two university hospitals in Cairo. Egypt Dermatol. 2006;212(SUPPL. 1):8–14. doi: 10.1159/000089193. [DOI] [PubMed] [Google Scholar]
- 176.Saied T, Elkholy A, Hafez SF, Basim H, Wasfy MO, El-Shoubary W, et al. Antimicrobial resistance in pathogens causing nosocomial bloodstream infections in university hospitals in Egypt. Am J Infect Control. 2011;39(9):e61–e65. doi: 10.1016/j.ajic.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 177.Abdelaziz MO, Bonura C, Aleo A, Fasciana T, Calà C, Mammina C. Cephalosporin resistant Escherichia coli from cancer patients in Cairo. Egypt Microbiol Immunol. 2013;57(5):391–395. doi: 10.1111/1348-0421.12046. [DOI] [PubMed] [Google Scholar]
- 178.Salem MM, Muharram M, Alhosiny IM. Distribution of classes 1 and 2 Integrons among Multi Drug Resistant E. coli Isolated from Hospitalized Patients with Urinary Tract Infection in Cairo, Egypt. Aust J Basic Appl Sci. 2010;4(3):398–407.
- 179.Sallam SA, Arafa MA, Razek AA, Naga M, Hamid MA. Device-related nosocomial infection in intensive care units of Alexandria University Students Hospital. East Mediterr Heal J. 2005;11(1/2):52–61. [PubMed] [Google Scholar]
- 180.Samah SE-K, Ghada E-SM, Amr ME-S, Dina SAE. Resistance genes to sulphonamide in commensal Escherichia coli isolated from stool of patients in Mansoura University Children Hospital. African J Microbiol Res. 2016;10(33):1363–70.
- 181.Samra MA-A, Ali NK, El-Madboly AAE. Detection of Multi-Drug Resistant Klebsiella pneumoniae in Al-Zahraa University Hospital. Egypt J Hosp Med. 2019;75(6):3006–12.
- 182.See I, Lessa FC, ElAta OA, Hafez S, Samy K, El-Kholy A, et al. Incidence and Pathogen Distribution of Healthcare-Associated Infections in Pilot Hospitals in Egypt. Infect Control Hosp Epidemiol. 2013;34(12):1281–1288. doi: 10.1086/673985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Seifert H, Blondeau J, Dowzicky MJ. In vitro activity of tigecycline and comparators (2014–2016) among key WHO ‘priority pathogens’ and longitudinal assessment (2004–2016) of antimicrobial resistance: a report from the T.E.S.T. study. Int J Antimicrob Agents. 2018;52(4):474–84. [DOI] [PubMed]
- 184.Selim S, Aziz MA, El-Alfay S, Zakaria H. Incidence and Antibiotics Resistance of Staphylococci and Escherichia coli Isolated from Diabetic Urinary Tract Infection Patients in Egypt. J Pure Appl Microbiol. 2019;13(3):1697–1702. doi: 10.22207/JPAM.13.3.44. [DOI] [Google Scholar]
- 185.Shaker OA, Gomaa HE, Elmasry SA, Abdel Halim RM, Abdelrahman AH, Kamal JS. Evaluation of combined use of temocillin disk and mastdisks inhibitor combination set against polymerase chain reaction for detection of carbapenem-resistant enterobacteriaceae. Open Access Maced J Med Sci. 2018;6(2):242–247. doi: 10.3889/oamjms.2018.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shash RY, Elshimy AA, Soliman MY, Mosharafa AA. Molecular characterization of extended-spectrum β-lactamase enterobacteriaceae isolated from egyptian patients with community- and hospital-acquired urinary tract infection. Am J Trop Med Hyg. 2019;100(3):522–528. doi: 10.4269/ajtmh.18-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shehab El-Din EMR, El-Sokkary MMA, Bassiouny MR, Hassan R. Epidemiology of neonatal sepsis and implicated pathogens: A Study from Egypt. Biomed Res Int. 2015;2015. [DOI] [PMC free article] [PubMed]
- 188.Abdelaziz MO, Bonura C, Aleo A, Fasciana T, Mammina C. NDM-1- and OXA-163-producing Klebsiella pneumoniae isolates in Cairo, Egypt, 2012. J Glob Antimicrob Resist. 2013;1(4):213–215. doi: 10.1016/j.jgar.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 189.Soliman AM, Khalifa HO, Ahmed AM, Shimamoto T, Shimamoto T. Emergence of an NDM-5-producing clinical Escherichia coli isolate in Egypt. Int J Infect Dis. 2016;48:46–48. doi: 10.1016/j.ijid.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 190.Soliman AM, Zarad HO, Nariya H, Shimamoto T, Shimamoto T. Genetic analysis of carbapenemase-producing Gram-negative bacteria isolated from a university teaching hospital in Egypt. Infect Genet Evol. 2020;77. [DOI] [PubMed]
- 191.Talaat M, El-Shokry M, El-Kholy J, Ismail G, Kotb S, Hafez S, et al. National surveillance of health care–associated infections in Egypt: developing a sustainable program in a resource-limited country. Am J Infect Control. 2016;44(11):1296–1301. doi: 10.1016/j.ajic.2016.04.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tohamy EY, Abo-Zeid AM, Shaheen AA, El-Awadi SF. Nosocomial Infection in Surgical Hospital in Zagazig University. J Agric Sci Ain Shams Univ. 2006;14(1):133–145. [Google Scholar]
- 193.Tohamy ST, Aboshanab KM, El-Mahallawy HA, El-Ansary MR, Afifi SS. Prevalence of multidrug-resistant Gram-negative pathogens isolated from febrile neutropenic cancer patients with bloodstream infections in Egypt and new synergistic antibiotic combinations. Infect Drug Resist. 2018;11:791–803. doi: 10.2147/IDR.S163293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wasfi R, Elkhatib WF, Ashour HM. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci Rep. 2016;6. [DOI] [PMC free article] [PubMed]
- 195.Wassef M, Abdelhaleim M, Abdulrahman E, Ghaith D. The role of OmpK35, OmpK36 porins, and production of β-lactamases on imipenem susceptibility in Klebsiella pneumoniae clinical isolates, Cairo. Egypt Microb Drug Resist. 2015;21(6):577–580. doi: 10.1089/mdr.2014.0226. [DOI] [PubMed] [Google Scholar]
- 196.Wassef M, Abdelhaleim M, Ghaith D, El-Mahdy Y. Emerging New Delhi metallo-β-lactamase-1-type-producing Gram-negative bacteria isolated from Cairo University Pediatric Hospital, Cairo. Egypt J Glob Antimicrob Resist. 2016;7:84–87. doi: 10.1016/j.jgar.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 197.Younus H-EMA, Jiman-Fatani AAM. Spontaneous bacterial peritonitis in Egyptian and Saudi patients with liver cirrhosis. J King Abdulaziz Univ Med Sci. 2011;18(3):29–46.
- 198.Youssef MM, Rizk HA, Hassuna NA. Phenotypic and genotypic characterization of extended-spectrum β-lactamase-producing enterobacteriaceae in asymptomatic bacteriuria in pregnancy. Microb Drug Resist. 2019;25(5):731–738. doi: 10.1089/mdr.2018.0088. [DOI] [PubMed] [Google Scholar]
- 199.Abdel-Hady H, Hawas S, El-Daker M, El-Kady R. Extended-spectrum β-lactamase producing Klebsiella pneumoniae in neonatal intensive care unit. J Perinatol. 2008;28(10):685–690. doi: 10.1038/jp.2008.73. [DOI] [PubMed] [Google Scholar]
- 200.Yousseff AS, El Feky SAM, El-Asser SA, Allah RAMA. Microorganisms isolated from surgical wounds infection and treatment with different natural products and antibiotics. Int J Med Heal Sci. 2013;7(6):236–239. [Google Scholar]
- 201.Zafer MM, El-Mahallawy HA, Abdulhak A, Amin MA, Al-Agamy MH, Radwan HH. Emergence of colistin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli strains isolated from cancer patients. Ann Clin Microbiol Antimicrob. 2019;18(1):1–8. doi: 10.1186/s12941-019-0339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zaki AE, Amer WH, Elezz AAA, Mohamed WM. Study of some enteropathogens causing acute diarrhea in infants and children less than 5 years old. Egypt J Med Microbiol. 2019;28(2):145–151. doi: 10.21608/ejmm.2019.282758. [DOI] [Google Scholar]
- 203.Zaki MES. Extended spectrum β-lactamases among gram-negative bacteria from an Egyptian pediatric hospital: a two-year experience. J Infect Dev Ctries. 2007;1(3):269–274. doi: 10.3855/jidc.363. [DOI] [PubMed] [Google Scholar]
- 204.Zayed ME, Alharbi SA, Masoud IM, Ammar RA. Utilization of bacteria as virulence agents for urinary tract infectionin Egyptian patients. Biosci Biotechnol Res Asia. 2012;9(2):521–530. doi: 10.13005/bbra/1029. [DOI] [Google Scholar]
- 205.Zowawi H, Thomas M, Abdelrahman S, Shabban M, Harris P, Paterson D. Molecular characterization of multidrug-resistant gram-negative bacilli in Egypt: a snapshot study. J Infect Public Health. 2019;12(1):130. doi: 10.1016/j.jiph.2018.10.081. [DOI] [Google Scholar]
- 206.Abdelhamid SM, Abozahra, Rania R. Expression of the fluoroquinolones efflux pump genes acrA and mdfA in Urinary Escherichia coli isolates. Polish J Microbiol. 2017;66(1):25–30. [DOI] [PubMed]
- 207.Abdelkader MM, Aboshanab KM, El-Ashry MA, Aboulwafa MM. Prevalence of MDR pathogens of bacterial meningitis in Egypt and new synergistic antibiotic combinations. PLoS ONE. 2017;12(2). [DOI] [PMC free article] [PubMed]
- 208.Abdel-Moaty MM, Mohamed WS, Abdel-All SM, El-Hendawy HH. Prevalence and molecular epidemiology of extended spectrum beta-lactamase producing Escherichia coli from hospital and community settings in Egypt. J Appl Pharm Sci. 2016;6(01):042–47. doi: 10.7324/JAPS.2016.600107. [DOI] [Google Scholar]
- 209.Abd-Elmonsef MME, Elsharawy D, Abd-Elsalam AS. Mechanical ventilator as a major cause of infection and drug resistance in intensive care unit. Environ Sci Pollut Res. 2018;25:30787–30792. doi: 10.1007/s11356-017-8613-5. [DOI] [PubMed] [Google Scholar]
- 210.Abdelsalam MFA, Abdalla MS, El-Abhar HSED. Prospective, comparative clinical study between high-dose colistin monotherapy and colistin–meropenem combination therapy for treatment of hospital-acquired pneumonia and ventilator-associated pneumonia caused by multidrug-resistant Klebsiella pneumoniae. J Glob Antimicrob Resist. 2018;15:127–135. doi: 10.1016/j.jgar.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 211.Abdulall AK, Tawfick MM, El Manakhly AR, El Kholy A. Carbapenem-resistant Gram-negative bacteria associated with catheter-related bloodstream infections in three intensive care units in Egypt. Eur J Clin Microbiol Infect Dis. 2018;37(9):1647–1652. doi: 10.1007/s10096-018-3294-7. [DOI] [PubMed] [Google Scholar]
- 212.Abduo EM, El-Kholy J, Abdou S, Hafez S, Omar N, Talaat M. Incidence and Microbial Etiology of Surgical Site Infections at Select Hospitals in Egypt. Am J Infect Control. 2016;44(6):S52–S53. doi: 10.1016/j.ajic.2016.04.049. [DOI] [Google Scholar]
- 213.Abou-Dobara MI, Deyab MA, Elsawy EM, Mohamed HH. Antibiotic susceptibility and genotype patterns of Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa Isolated from urinary tract infected patients. Polish J Microbiol. 2010;59(3):207–212. doi: 10.33073/pjm-2010-032. [DOI] [PubMed] [Google Scholar]
- 214.Alabsi MS, Ghazal A, Sabry SA, Alasaly MM. Association of some virulence genes with antibiotic resistance among uropathogenic Escherichia coli isolated from urinary tract infection patients in Alexandria, Egypt: a hospital-based study. J Glob Antimicrob Resist. 2014;2(2):83–86. doi: 10.1016/j.jgar.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 215.Al-Agamy MHM, Ashour MSE-D, Wiegand I, Mohamed Al-Agamy MH, Ashour MSED, Wiegand I. First description of CTX-M beta-lactamase-producing clinical Escherichia coli isolates from Egypt. Int J Antimicrob Agents. 2006;27(27):545–8. [DOI] [PubMed]
- 216.Ali MMM, Ahmed SF, Klena JD, Mohamed ZK, Moussa TAA, Ghenghesh KS. Enteroaggregative Escherichia coli in diarrheic children in Egypt: molecular characterization and antimicrobial susceptibility. J Infect Dev Ctries. 2014;8(5):589–596. doi: 10.3855/jidc.4077. [DOI] [PubMed] [Google Scholar]
- 217.Aly MEA, Essam TM, Amin MA. Antibiotic resistance profile of E. coli strains isolated from clinical specimens and food samples in Egypt. Int J Microbiol Res. 2012;3(3):176–82.
- 218.Amer WH, Elsweikh SAR, Hablas NM. Comparative study between beta-lactam/beta-lactamase inhibitors as alternatives for carbapenems in the treatment of extended-spectrum beta-lactamase-producing Enterobacteriaceae. Infect Dis Clin Pract. 2019;27(3).
- 219.Amira MEG, Areej MEM, Hemat KAEL, Ramadan HI, Heba IA. Phenotypic and genotypic detection of β-lactams resistance in Klebsiella species from Egyptian hospitals revealed carbapenem resistance by OXA and NDM genes. Afr J Microbiol Res. 2016;10(10):339–347. doi: 10.5897/AJMR2015.7871. [DOI] [Google Scholar]
- 220.Ashour HM, El-Sharif A. Species distribution and antimicrobial susceptibility of gram-negative aerobic bacteria in hospitalized cancer patients. J Transl Med. 2009;19:7. doi: 10.1186/1479-5876-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Attia H, Szubin R, Yassin AS, Monk JM, Aziz RK. Draft genome sequences of four metallo-beta-lactamase-producing multidrug-resistant Klebsiella pneumoniae clinical isolates, including two colistin-resistant strains, from Cairo, Egypt. Am Soc Mircobiol. 2019;8(7). Available from: 10.1128/MRA [DOI] [PMC free article] [PubMed]
- 222.Aziz MA, El-Kholy I, Abdo A, Selim S. Influence of multi drug resistance Gram negative bacteria in liver transplant recipient. African J Microbiol Res. 2013;7(41):4857–4861. doi: 10.5897/AJMR2013.5959. [DOI] [Google Scholar]
- 223.Azzab MM, El-Sokkary RH, Tawfeek MM, Gebriel MG. Multidrug-resistant bacteria among patients with ventilator-associated pneumonia in an emergency intensive care unit, Egypt. East Mediterr Heal J. 2016;22(12). [DOI] [PubMed]
- 224.Bassyouni RH, Gaber SN, Wegdan AA. Fecal carriage of extended-spectrum β-lactamase- and AmpC- producing Escherichia coli among healthcare workers. J Infect Dev Ctries. 2015;9(3):304–308. doi: 10.3855/jidc.5633. [DOI] [PubMed] [Google Scholar]
- 225.Bathoorn E, Friedrich AW, Zhou K, Arends JP, Borst DM, Grundmann H, et al. Latent introduction to the Netherlands of multiple antibiotic resistance including NDM-1 after hospitalisation in Egypt, August 2013. Eurosurveillance. 2013;18(42). [DOI] [PubMed]
- 226.EL Bedewy RMS. Multi drug resistant bacteria and its antibiotic susceptibility at percutanous endoscopic gastrostomy (PEG) tube site of long term care facility elderly residents. Egypt J Hosp Med. 2017;68(2):1094–1100. doi: 10.12816/0039035. [DOI] [Google Scholar]
- 227.Behiry IK, Abada EA, Ahmed EA, Labeeb RS. Enteropathogenic Escherichia coli associated with diarrhea in children in Cairo. Egypt Sci World J. 2011;11:2613–2619. doi: 10.1100/2011/485381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Biedenbach D, Bouchillon S, Hackel M, Hoban D, Kazmierczak K, Hawser S, et al. Dissemination of NDM metallo-beta-lactamase genes among clinical isolates of Enterobacteriaceae collected during the SMART global surveillance study from 2008 to 2012. Antimicrob Agents Chemother. 2015;59(59):826–830. doi: 10.1128/AAC.03938-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Eida M, Nasser M, El-Maraghy N, Azab K. Pattern of hospital-acquired pneumonia in Intensive Care Unit of Suez Canal University Hospital. Egypt J Chest Dis Tuberc. 2015;64(3):625–631. doi: 10.1016/j.ejcdt.2015.03.028. [DOI] [Google Scholar]
- 230.El Awady BA, Anan MG, Gohar HA, Saleh MH. Detection of carbapenemase-producing enterobacteriaceae using chromogenic medium, ChromidID OXA-48, in critical care patients of kasr Al-Ainy hospital in Egypt. J Pure Appl Microbiol. 2017;11(4):1655–1664. doi: 10.22207/JPAM.11.4.03. [DOI] [Google Scholar]
- 231.El Kholy A, Baseem H, Hall GS, Procop GW, Longworth DL. Antimicrobial resistance in Cairo, Egypt 1999–2000: a survey of five hospitals. J Antimicrob Chemother. 2003;51(3):625–630. doi: 10.1093/jac/dkg101. [DOI] [PubMed] [Google Scholar]
- 232.El Metwally HAR, Ibrahim HAH, El-Athamna MN, Amer MA, El MHAR, Ibrahim HAH, et al. Multiplex PCR for detection of diarrheagenic Escherichia coli in Egyptian children. J Med Sci. 2007;7(2):255–262. doi: 10.3923/jms.2007.255.262. [DOI] [Google Scholar]
- 233.Elawady B, Ghobashy M, Balbaa A. Rapidec carba NP for detection of carbapenemase-producing enterobacteriaceae in clinical isolates: a cross-sectional study. Surg Infect (Larchmt) 2019;20(8):672–676. doi: 10.1089/sur.2019.084. [DOI] [PubMed] [Google Scholar]
- 234.El-Badawy MF, Tawakol WM, El-Far SW, Maghrabi IA, Al-Ghamdi SA, Mansy MS, et al. Molecular identification of aminoglycoside-modifying enzymes and plasmid-mediated quinolone resistance genes among Klebsiella pneumoniae clinical isolates recovered from Egyptian patients. Int J Microbiol. 2017;2017. [DOI] [PMC free article] [PubMed]
- 235.El-Badawy MF, Tawakol WM, Maghrabi IA, Mansy MS, Shohayeb MM, Ashour MS. Iodometric and Molecular Detection of ESBL Production among Clinical Isolates of E. coli Fingerprinted by ERIC-PCR: The First Egyptian Report Declares the Emergence of E. coli O25b-ST131clone Harboring blaGES. Microb Drug Resist. 2017;23(6):703–17. [DOI] [PubMed]
- 236.El-Din R, Elbaset A, Naim A. Epidemiology, Phenotyping and Antimicrobial Susceptibility Profile of Enterohaemrrhagic Escherichia coli Strains Isolated from Cases of Diarrhea. Br Microbiol Res J. 2015;8(4):546–553. doi: 10.9734/BMRJ/2015/18252. [DOI] [Google Scholar]
- 237.El-Din RAE-HA, El-Sanosy MG. Phenotypic Study on Some Virulence Factors and Molecular Screening of Aminoglycoside Resistance among Klebsiella pneumoniae Strains Isolated from Urinary Tract Infections in Pediatric Cases in Egypt. Microbiol Res J Int. 2018;26(2):1–11.
- 238.Elgendy SG, Abdel Hameed MR, El-Mokhtar MA. Tigecycline resistance among Klebsiella pneumoniae isolated from febrile neutropenic patients. J Med Microbiol. 2018;67(7):972–975. doi: 10.1099/jmm.0.000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.El-Kazzaz SS, El-khier NTA. AmpC and metallo beta-lactamases producing Gram negative bacteria in patients with hematological malignancy. Afr J Microbiol Res. 2015;9(18):1247–1254. doi: 10.5897/AJMR2015.7435. [DOI] [Google Scholar]
- 240.El-latif RSA, Elbadawy NE, El-Hady HA. Checkboard antimicrobial susceptibility testing of multidrug resistant Klebsiella pneumoniae isolated from patients with ventilator associated pneumonia. Egypt J Med Microbiol. 2012;21(4):89–98. doi: 10.12816/0004911. [DOI] [Google Scholar]
- 241.ElMahallawy HA, Zafer MM, Amin MA, Ragab MM, Al-Agamy MH. Spread of carbapenem resistant Enterobacteriaceae at tertiary care cancer hospital in Egypt. Infect Dis (Auckl) 2018;50(7):560–564. doi: 10.1080/23744235.2018.1428824. [DOI] [PubMed] [Google Scholar]
- 242.El-Mahdy R, El-Kannishy G, Salama H. Hypervirulent Klebsiella pneumoniae as a hospital-acquired pathogen in the intensive care unit in Mansoura, Egypt. GERMS [Internet]. 2018;8(3):140–6. Available from: www.germs.ro [DOI] [PMC free article] [PubMed]
- 243.El-Masry EA, Melake NA, Taher IA. Phenotypic and Molecular Characterization of Extended-Spectrum P-Lactamase Producing Klebsiella spp. from Nosocomial Infections in Egypt. Int Med J. 2019;26(5):376–80.
- 244.El-Moghazy A-N, Tawfick MM, El-Habibi MM. Prevalence, antimicrobial susceptibilities and molecular characterization of enteric bacterial pathogens isolated from patients with infectious diarrhoea in Cairo. Int J Curr Microbiol Appl Sci. 2016;5(4):553–564. doi: 10.20546/ijcmas.2016.504.063. [DOI] [Google Scholar]
- 245.Elnahriry SS, Khalifa HO, Soliman AM, Ahmed AM, Hussein AM, Shimamoto T, et al. Emergence of plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate from Egypt. Antimicrob Agents Chemother. 2016;60(5):3249–3250. doi: 10.1128/AAC.00269-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Elraghy NA, Zahran WA, Makled AF, El-Sebaey HM, El-Hendawy GR, Melake NA, et al. Enterobacteriaceae nosocomial uropathogens at Menoufia University Hospitals: phenotypic characterization and detection of resistance genes using real-time PCR. Menoufia Med J. 2016;29(4):855–861. [Google Scholar]
- 247.El-Sahrigy SAF, Shouman MG, Ibrahim HM, Rahman AMOA, Habib SA, Khattab AA, et al. Prevalence and anti-microbial susceptibility of hospital acquired infections in two pediatric intensive care units in Egypt. Open Access Maced J Med Sci. 2019;7(11):1744–1749. doi: 10.3889/oamjms.2019.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Elsherif RH, Ismail DK, El-Kholy YS, Gohar NM, Elnagdy SM, Elkraly OA. Integron-mediated multidrug resistance in extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolated from fecal specimens in Egypt. J Egypt Public Health Assoc. 2016;91(2):73–79. doi: 10.1097/01.EPX.0000483165.56114.d8. [DOI] [PubMed] [Google Scholar]
- 249.El-Sweify MA, Gomaa NI, El-Maraghy NN, Mohamed HA. Phenotypic detection of carbapenem resistance among Klebsiella pneumoniae in Suez Canal University Hospitals, Ismailiya, Egypt. Int J Curr Microbiol Appl Sci. 2015;4(2):10–18. [Google Scholar]
- 250.Elzayat MAA, Barnett-Vanes A, Dabour MFE, Cheng F, Abdel-Aziz Elzayat M, Barnett-Vanes A, et al. Prevalence of undiagnosed asymptomatic bacteriuria and associated risk factors during pregnancy: a cross-sectional study at two tertiary centres in Cairo, Egypt. BMJ Open [Internet]. 2017;7. Available from: http://bmjopen.bmj.com/ [DOI] [PMC free article] [PubMed]
- 251.Esmat MM, Saif A, Islam A. Diabetic foot infection: bacteriological causes and antimicrobial therapy. J Am Sci. 2012;8(10):389–393. [Google Scholar]
- 252.Fahmey SS. Early-onset sepsis in a neonatal intensive care unit in Beni Suef, Egypt: bacterial isolates and antibiotic resistance pattern. Korean J Pediatr. 2013;56(8):332–337. doi: 10.3345/kjp.2013.56.8.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Fam N, Leflon-Guibout V, Fouad S, Aboul-Fadl L, Marcon E, Desouky D, et al. CTX-M-15-producing Escherichia coli clinical isolates in Cairo (Egypt), including isolates of clonal complex ST10 and clones ST131, ST73, and ST405 in both community and hospital settings. Microb Drug Resist. 2011;17(1):67–73. doi: 10.1089/mdr.2010.0063. [DOI] [PubMed] [Google Scholar]
- 254.Fattouh M, Nasr El-Din A, Abdelgalil W. Bacteriologic and immunologic profile of blood stream infected patients in intensive care unit of Sohag University Hospital, Egypt. Int J Curr Microbiol Appl Sci. 2014;3(8):265–281. [Google Scholar]
- 255.Fouda R, Soliman MS, ElAnany MG, Abadeer M, Soliman G. Prevalence and risk factors of MRSA, ESBL and MDR bacterial colonization upon admission to an Egyptian medical ICU. J Infect Dev Ctries. 2016;10(4):329–336. doi: 10.3855/jidc.6798. [DOI] [PubMed] [Google Scholar]
- 256.Gamal D, Fernández-Martínez M, El-Defrawy I, Ocampo-Sosa AA, Martínez-Martínez L. First identification of NDM-5 associated with OXA-181 in Escherichia coli from Egypt. Emerg Microbes Infect. 2016;5. [DOI] [PMC free article] [PubMed]
- 257.Gamal D, Fernández-Martínez M, Salem D, El-Defrawy I, Montes LÁ, Ocampo-Sosa AA, et al. Carbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM-1 on IncR plasmids and its association with rmtF. Int J Infect Dis. 2016;43:17–20. doi: 10.1016/j.ijid.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 258.Gawad WE, Helmy OM, Tawakkol WM, Hashem AM. Antimicrobial resistance, biofilm formation, and phylogenetic grouping of uropathogenic Escherichia coli isolates in Egypt: the role of efflux pump-mediated resistance. Jundishapur J Microbiol. 2018;11(2).
- 259.Ghaith DM, Mohamed ZK, Farahat MG, Aboulkasem Shahin W, Mohamed HO. Colonization of intestinal microbiota with carbapenemase-producing Enterobacteriaceae in paediatric intensive care units in Cairo. Egypt Arab J Gastroenterol. 2019;20(1):19–22. doi: 10.1016/j.ajg.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 260.Ghaith DM, Zafer MM, Said HM, Elanwary S, Elsaban S, Al-Agamy MH, et al. Genetic diversity of carbapenem-resistant Klebsiella pneumoniae causing neonatal sepsis in intensive care unit, Cairo. Egypt Eur J Clin Microbiol Infect Dis. 2020;39(3):583–591. doi: 10.1007/s10096-019-03761-2. [DOI] [PubMed] [Google Scholar]
- 261.Aamir MM, Abu El-Wafa WM, Ali AE, Hamouda, Hayam M, Mourad FE. Prevalence of Multidrug Resistant Bacteria Causing Late-Onset Neonatal Sepsis. Int J Curr Microbiol Appl Sci. 2015;4(5):172–90
- 262.Grisold AJ, Hoenigl M, Ovcina I, Valentin T, Fruhwald S. Ventilator-associated pneumonia caused by OXA-48-producing Escherichia coli complicated by ciprofloxacin-associated rhabdomyolysis. J Infect Chemother. 2013;19(6):1214–1217. doi: 10.1007/s10156-013-0628-3. [DOI] [PubMed] [Google Scholar]
- 263.Hasanin A, Eladawy A, Mohamed H, Salah Y, Lotfy A, Mostafa H, et al. Prevalence of extensively drug-resistant gram negative bacilli in surgical intensive care in Egypt. Pan Afr Med J. 2014;19. [DOI] [PMC free article] [PubMed]
- 264.Hashem AA, Taha SA, Anani MM. Antibiotic Susceptibility Pattern and Biofilm Production of Multidrug-Resistant Organisms (MDROs) Isolated from Suez-Canal University Hospitals. Egypt J Med. 2018;27(4):113–121. [Google Scholar]
- 265.Hassan A, Mohamed S, Mohamed M, El-Mokhtar M. Acute exacerbations of chronic obstructive pulmonary disease: etiological bacterial pathogens and antibiotic resistance in Upper Egypt. Egypt J Bronchol. 2016;10(3):283–290. doi: 10.4103/1687-8426.193640. [DOI] [Google Scholar]
- 266.Hassan EA, Elsherbiny NM, Abd El-Rehim AS, Soliman AMA, Ahmed AO. Health care-associated infections in pre-transplant liver intensive care unit: Perspectives and challenges. J Infect Public Health. 2018;11(3):398–404. doi: 10.1016/j.jiph.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 267.Hassan MA, Tamer TM, Rageh AA, Abou-Zeid AM, Abd El-Zaher EHF, Kenawy ER. Insight into multidrug-resistant microorganisms from microbial infected diabetic foot ulcers. Diabetes Metab Syndr Clin Res Rev. 2019;13(2):1261–1270. doi: 10.1016/j.dsx.2019.01.044. [DOI] [PubMed] [Google Scholar]
- 268.Hawser SP, Badal RE, Bouchillon SK, Hoban DJ, Biedenbach DJ, Cantón R, et al. Monitoring the global in vitro activity of ertapenem against Escherichia coli from intra-abdominal infections: SMART 2002–2010. Int J Antimicrob Agents. 2013;41(3):224–228. doi: 10.1016/j.ijantimicag.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 269.Hefzy EM, Hassuna NA. Fluoroquinolone-resistant sequence type 131 subgroups O25b and O16 among extraintestinal Escherichia coli Isolates from community-acquired urinary tract infections. Microb Drug Resist. 2017;23(2):224–229. doi: 10.1089/mdr.2016.0040. [DOI] [PubMed] [Google Scholar]
- 270.Helal SF, El-Rachidi NGE, AbdulRahman EM, Hassan DMA. The presence of blaKPC-mediated resistance in Enterobacteriaceae in Cairo University hospitals in Egypt and its correlation with in vitro carbapenem susceptibility. J Chemother. 2014;26(2):125–128. doi: 10.1179/1973947813Y.0000000099. [DOI] [PubMed] [Google Scholar]
- 271.Henderson J, Ciesielczuk H, Nelson SM, Wilks M. Community prevalence of carbapenemase-producing organisms in East London. J Hosp Infect. 2019;103(2):142–146. doi: 10.1016/j.jhin.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 272.Abbas HA, Kadry AA, Shaker GH, Goda RM. Impact of specific inhibitors on metallo-β-carbapenemases detected in Escherichia coli and Klebsiella pneumoniae isolates. Microb Pathog. 2019;132:266–274. doi: 10.1016/j.micpath.2019.05.022. [DOI] [PubMed] [Google Scholar]
- 273.Iman FEG, Marwa AM, Doaa AY. Phenotypic and genotypic methods for detection of metallo beta lactamases among carbapenem resistant Enterobacteriaceae clinical isolates in Alexandria Main University Hospital. Afr J Microbiol Res. 2016;10(1):32–40. doi: 10.5897/AJMR2015.7821. [DOI] [Google Scholar]
- 274.Kamel NA, Abouelwafa MM, El-tayeb WN, Aboshanab KM. Antibacterial resistance pattern of aerobic bacteria isolated from patients with diabetic foot ulcers in Egypt. Afr J Microbiol Res. 2014;8(31):2947–2954. doi: 10.5897/AJMR2014.6909. [DOI] [Google Scholar]
- 275.Kamel NA, El-tayeb WN, El-Ansary MR, Mansour MT, Aboshanab KM. Phenotypic screening and molecular characterization of carbapenemase-producing Gram-negative bacilli recovered from febrile neutropenic pediatric cancer patients in Egypt. PLoS ONE. 2018;13(8). [DOI] [PMC free article] [PubMed]
- 276.Khalaf NG, Eletreby MM, Hanson ND. Characterization of CTX-M ESBLs in Enterobacter cloacae, Escherichia coli and Klebsiella pneumoniae clinical isolates from Cairo, Egypt. BMC Infect Dis. 2009;9. [DOI] [PMC free article] [PubMed]
- 277.Khalifa HO, Soliman AM, Ahmed AM, Shimamoto T, Hara T, Ikeda M, et al. High carbapenem resistance in clinical gram-negative pathogens isolated in Egypt. Microb Drug Resist. 2017;23(7):838–844. doi: 10.1089/mdr.2015.0339. [DOI] [PubMed] [Google Scholar]
- 278.Khalil MAF, Elgaml A, El-Mowafy M. Emergence of multidrug-resistant New Delhi metallo-β-lactamase-1-producing Klebsiella pneumoniae in Egypt. Microb Drug Resist. 2017;23(4):480–487. doi: 10.1089/mdr.2016.0003. [DOI] [PubMed] [Google Scholar]
- 279.Khalil MAF, Hager R, Abd-El Reheem F, Mahmoud EE, Samir T, Moawad SS, et al. A study of the virulence traits of carbapenem-resistant Klebsiella pneumoniae Isolates in a Galleria mellonella model. Microb Drug Resist. 2019;25(7):1063–1071. doi: 10.1089/mdr.2018.0270. [DOI] [PubMed] [Google Scholar]
- 280.Labib JR, Ibrahim SK, Salem MR, Youssef MRL, Meligy B. Infection with gram-negative bacteria among children in a tertiary pediatric hospital in Egypt. Am J Infect Control. 2018;46(7):798–801. doi: 10.1016/j.ajic.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 281.Lashin GMA, Tohamy EY, Askora AA, Mahmoud FE-Z. Use of probiotic acid bacteria for the control of multidrug resistant bacteria isolated from clinical infections. Bull Fac Sci Zagazig Univ. 2017;39:61–81. doi: 10.21608/bfszu.2017.31043. [DOI] [Google Scholar]
- 282.Lob SH, Hoban DJ, Young K, Motyl MR, Sahm DF. Activity of imipenem/relebactam against Gram-negative bacilli from global ICU and non-ICU wards: SMART 2015–2016. J Glob Antimicrob Resist. 2018;15:12–19. doi: 10.1016/j.jgar.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 283.Abdallah HM, Alnaiemi N, Reuland EA, Wintermans BB, Koek A, Abdelwahab AM, et al. Fecal carriage of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae in Egyptian patients with community-onset gastrointestinal complaints: a hospital-based cross-sectional study. Antimicrob Resist Infect Control. 2017 doi: 10.1186/s13756-017-0219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Mahdi WKM, Abd H, Ahmed A, Abo M, Euoon E, Mohamed MH. Extended Spectrum-lactamase producing Klebsiella pneumoniae in Neonatal Units of Minya Governorate. Int J Curr Microbiol App Sci. 2014;3(12):787–800. [Google Scholar]
- 285.Malek MM, Amer FA, Allam AA, El-Sokkary RH, Gheith T, Arafa MA. Occurrence of classes I and II integrons in Enterobacteriaceae collected from Zagazig University Hospitals, Egypt. Front Microbiol. 2015;6. [DOI] [PMC free article] [PubMed]
- 286.Metwally L, Gomaa N, Attallah M, Kamel N. High prevalence of Klebsiella pneumoniae carbapenemase-mediated resistance in K. pneumoniae isolates from Egypt. East Mediterr Heal J. 2013;19(11):947–952. doi: 10.26719/2013.19.11.947. [DOI] [PubMed] [Google Scholar]
- 287.Mohamad EA, El Shalakany AH. Detection of biofilm formation in uropathogenic bacteria. Egypt J Med Microbiol. 2015;24(1):49–458. doi: 10.12816/0024809. [DOI] [Google Scholar]
- 288.Mohamed DS, Ahmed EF, Mahmoud AM, El-Baky RMA, John J. Isolation and evaluation of cocktail phages for the control of multidrug-resistant Escherichia coli serotype O104: H4 and E. coli O157: H7 isolates causing diarrhea. FEMS Microbiol Lett. 2018;365(2). [DOI] [PubMed]
- 289.Mohamed ER, Ali MY, Waly NGFM, Halby HM, El-baky RMA. The inc FII plasmid and its contribution in the transmission of blaNDM-1 and blaKPC-2 in Klebsiella pneumoniae in Egypt. Antibiotics. 2019;8(4):1–12. doi: 10.3390/antibiotics8040266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Mohamed ER, Aly SA, Halby HM, Ahmed SH, Zakaria AM, El-Asheer OM. Epidemiological typing of multidrug-resistant Klebsiella pneumoniae, which causes paediatric ventilator-associated pneumonia in Egypt. J Med Microbiol. 2017;66(5):628–634. doi: 10.1099/jmm.0.000473. [DOI] [PubMed] [Google Scholar]
- 291.Al-Agamy MH. Genetic basis of cefotaxime resistant isolates of Klebsiella pneumoniae from Cairo. Afr J Microbiol Res. 2012 doi: 10.5897/AJMRX11.022. [DOI] [Google Scholar]
- 292.Mohamed M, El-Mokhtar M, Hassan A. Bacterial profile and antibiotic susceptibility patterns of acute exacerbation of chronic obstructive pulmonary disease in Assiut University Hospitals, upper Egypt; a one-year prospective study. Br Microbiol Res J. 2015;7(6):288–305. doi: 10.9734/BMRJ/2015/16317. [DOI] [Google Scholar]
- 293.Mohamed MAES, Eman SA. Antibacterial resistance pattern among Escherichia coli strains isolated from Mansoura hospitals in Egypt with a special reference to quinolones. Afr J Microbiol Res. 2015;9(9):662–670. doi: 10.5897/AJMR2014.7351. [DOI] [Google Scholar]
- 294.Shatalov A. Prevalence and antibiotic resistance pattern of Escherichia coli and Klebsiella pneumoniae in Urine Tract Infections at the La Paz Medical Center, Malabo, Equatorial Guinea. Open J Med Microbiol. 2015;5:177–183. doi: 10.4236/ojmm.2015.54022. [DOI] [Google Scholar]
- 295.Ehlkes L, Pfeifer Y, Werner G, Ignatius R, Vogt M, Eckmanns T, et al. No evidence of carbapenemase-producing Enterobacteriaceae in stool samples of 1,544 asylum seekers arriving in Rhineland-Palatinate, Germany, April 2016 to March, 2017. Eurosurveillance. 2019;24(8). [DOI] [PMC free article] [PubMed]
- 296.Gashaw M, Berhane M, Bekele S, Kibru G, Teshager L, Yilma Y, et al. Emergence of high drug resistant bacterial isolates from patients with health care associated infections at Jimma University medical center: A cross sectional study. Antimicrob Resist Infect Control. 2018;7(1). [DOI] [PMC free article] [PubMed]
- 297.Gebre-Sealssie S. Antimicrobial resistance patterns of clinical bacterial isolates in Southwestern Ethiopia. Ethiop Med J. 2007;45(4):363–370. [PubMed] [Google Scholar]
- 298.Gizachew Z, Kassa T, Beyene G, Howe R, Yeshitila B. Multi-drug resistant bacteria and associated factors among reproductive age women with significant bacteriuria. Ethiop Med J. 2019;1:31–43. [Google Scholar]
- 299.Kalayu AA, Diriba K, Girma C, Abdella E. Incidence and Bacterial etiologies of surgical site infections in a public hospital, Addis Ababa, Ethiopia. Open Microbiol J. 2020;13(1):301–307. doi: 10.2174/1874285801913010301. [DOI] [Google Scholar]
- 300.Legese MH, Weldearegay GM, Asrat D. Extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae among Ethiopian children. Infect Drug Resist. 2017;10:27–34. doi: 10.2147/IDR.S127177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Moges F, Eshetie S, Abebe W, Mekonnen F, Dagnew M, Endale A, et al. High prevalence of extended-spectrum beta-lactamase-producing Gram-negative pathogens from patients attending Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Amhara region. PLoS ONE. 2019;14(4). [DOI] [PMC free article] [PubMed]
- 302.Moges F, Mengistu G, Genetu A. Multiple drug resistance in urinary pathogens at Gondar College of Medical Sciences Hospital, Ethiopia. East Afr Med J. 2002;8(415–419):415–419. doi: 10.4314/eamj.v79i8.8827. [DOI] [PubMed] [Google Scholar]
- 303.Saba MG. Magnitude of Extended-spectrum Beta-lactamase, AmpC Beta-lactamase and Carbapenemase producing gram negative bacilli isolated from clinical specimens at International Clinical Laboratories, Addis Ababa, Ethiopia. [Addis Ababa, Ethiopia]: Addis Ababa University; 2018. [DOI] [PMC free article] [PubMed]
- 304.Teklu DS, Negeri AA, Legese MH, Bedada TL, Woldemariam HK, Tullu KD. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob Resist Infect Control. 2019;8(1). [DOI] [PMC free article] [PubMed]
- 305.Ten Hove RJ, Tesfaye M, ten Hove WF, Nigussie M. Profiling of antibiotic resistance of bacterial species recovered from routine clinical isolates in Ethiopia. Ann Clin Microbiol Antimicrob. 2017; [DOI] [PMC free article] [PubMed]
- 306.Abayneh M, Tesfaw G, Abdissa A. Isolation of Extended-Spectrum beta-lactamase-(ESBL-) Producing Escherichia coli and Klebsiella pneumoniae from Patients with Community-Onset Urinary Tract Infections in Jimma University Specialized Hospital, Southwest Ethiopia. Can J Infect Dis Med Microbiol. 2018;2018. [DOI] [PMC free article] [PubMed]
- 307.Tuem KB, Desta R, Bitew H, Ibrahim S, Hishe HZ. Antimicrobial resistance patterns of uropathogens isolated between 2012 and 2017 from a tertiary hospital in Northern Ethiopia. J Glob Antimicrob Resist. 2019;18:109–114. doi: 10.1016/j.jgar.2019.01.022. [DOI] [PubMed] [Google Scholar]
- 308.Tufa TB, Andre F, Abdissa S, Achim K, Colin M, Klaus P, et al. Resistance to third generation cephalosporin due to tem and CTX-M-1 type extended-spectrum beta-lactamase genes among clinical isolates of gram-negative bacilli in Asella, Central Ethiopia. Antimicrob Resist Infect Control. 2019;8((Suppl 1)P51):40–1.
- 309.Zeynudin A, Pritsch M, Schubert S, Messerer M, Liegl G, Hoelscher M, et al. Prevalence and antibiotic susceptibility pattern of CTX-M type extended-spectrum β-lactamases among clinical isolates of gram-negative bacilli in Jimma, Ethiopia. BMC Infect Dis. 2018;18(1). [DOI] [PMC free article] [PubMed]
- 310.Alemayehu T, Ali M, Mitiku E, Hailemariam M. The burden of antimicrobial resistance at tertiary care hospital, southern Ethiopia: A three years’ retrospective study. BMC Infect Dis. 2019;19(1). [DOI] [PMC free article] [PubMed]
- 311.Alemu M. Extended Spectrum Beta-lactamase producing E. coli and K. pneumoniae carriage among under five years children in Addis Raey public health center, Addis Ababa, Ethiopia. [Addis Ababa, Ethiopia]: Addis Ababa University; 2018.
- 312.Beyene D, Bitew A, Fantew S, Mihret A, Evans M. Multidrug-resistant profile and prevalence of extended spectrum β-lactamase and carbapenemase production in fermentative Gram-negative bacilli recovered from patients and specimens referred to National Reference Laboratory, Addis Ababa, Ethiopia. PLoS One. 2019;14(9). [DOI] [PMC free article] [PubMed]
- 313.Dadi BR, Abebe T, Zhang L, Mihret A, Abebe W, Amogne W. Drug resistance and plasmid profile of uropathogenic Escherichia coli among urinary tract infection patients in Addis Abeba. J Infect Dev Ctries. 2018;12(8):608–615. doi: 10.3855/jidc.9916. [DOI] [PubMed] [Google Scholar]
- 314.Desta K, Woldeamanuel Y, Azazh A, Mohammod H, Desalegn D, Shimelis D, et al. High gastrointestinal colonization rate with extended-spectrum β-lactamase-producing Enterobacteriaceae in hospitalized patients: Emergence of carbapenemase-producing K. pneumoniae in Ethiopia. PLoS ONE. 2016;11(8). [DOI] [PMC free article] [PubMed]
- 315.Eshetie S, Unakal C, Gelaw A, Ayelign B, Endris M, Moges F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral Hospital, Northwest Ethiopia. Antimicrob Resist Infect Control. 2015;4(1). [DOI] [PMC free article] [PubMed]
- 316.Eshetu B, Gashaw M, Berhane M, Abdissa A, McClure EM, Goldenberg RL, et al. Intravenous fluid contaminated with Klebsiella oxytoca as a source of sepsis in a preterm newborn: case report. Am J Infect Control. 2019;47(7):840–842. doi: 10.1016/j.ajic.2018.12.025. [DOI] [PubMed] [Google Scholar]
- 317.Rerambiah LK, Ndong J-C, Massoua PMM, Medzegue S, Elisee-Ndam M, Mintsa-Ndong A, et al. Antimicrobial profiles of bacterial clinical isolates from the Gabonese National Laboratory of Public Health: data from routine activity. Int J Infect Dis. 2014;29:48–53. doi: 10.1016/j.ijid.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 318.Moussounda M, Diene SM, Dos Santos S, Goudeau A, François P, Mee-Marquet N van der A. Emergence of bla NDM-7 producing enterobacteriaceae in Gabon 2016. Emerg Infect Dis. 2017;23(2):2–4. [DOI] [PMC free article] [PubMed]
- 319.Presterl E, Zwick RH, Reichmann S, Aichelburg A, Winkler S, Kremsner PG, et al. Frequency and virulence properties of diarrheagenic Escherichia coli in children with diarrhea in Gabon. Am J Trop Med Hyg. 2003;69(4):406–410. doi: 10.4269/ajtmh.2003.69.406. [DOI] [PubMed] [Google Scholar]
- 320.Rogombe SM, Jean K, Mimbila M, Kamgaing EK, M’ella RM, Pambou RKM ep. N, et al. The epidemiological aspects and evolution of nosocomial infection in Hospital, neonatology unit of Angondje Teaching. Neonatal Pediatr Med. 2018;4(2).
- 321.Schaumburg F, Alabi A, Kokou C, Grobusch MP, Köck R, Kaba H, et al. High burden of extended-spectrum β-lactamase-producing enterobacteriaceae in Gabon. J Antimicrob Chemother. 2013;68(9):2140–2143. doi: 10.1093/jac/dkt164. [DOI] [PubMed] [Google Scholar]
- 322.Sanneh B, Kebbeh A, Jallow HS, Camara Y, Mwamakamba LW, Ceesay IF, et al. Prevalence and risk factors for faecal carriage of Extended Spectrum Î2-lactamase producing Enterobacteriaceae among food handlers in lower basic schools in West Coast Region of The Gambia. PLoS One. 2018;13(8). [DOI] [PMC free article] [PubMed]
- 323.Agyekum A, Fajardo-Lubián A, Ansong D, Partridge SR, Agbenyega T, Iredell JR. blaCTX-M-15 carried by IncF-type plasmids is the dominant ESBL gene in Escherichia coli and Klebsiella pneumoniae at a hospital in Ghana. Diagn Microbiol Infect Dis. 2016; [DOI] [PubMed]
- 324.Labi AK, Obeng-Nkrumah N, Bjerrum S, Enweronu-Laryea C, Newman MJ. Neonatal bloodstream infections in a Ghanaian Tertiary Hospital: Are the current antibiotic recommendations adequate? BMC Infect Dis. 2016;16(1). [DOI] [PMC free article] [PubMed]
- 325.Mohammed J, Hounmanou YMG, Thomsen LE. Antimicrobial resistance among clinically relevant bacterial isolates in Accra: a retrospective study. BMC Res Notes. 2018;11(1):254. doi: 10.1186/s13104-018-3377-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Obeng-Nkrumah N, Labi AK, Addison NO, Labi JEM, Awuah-Mensah G. Trends in paediatric and adult bloodstream infections at a Ghanaian referral hospital: a retrospective study. Ann Clin Microbiol Antimicrob. 2016. [DOI] [PMC free article] [PubMed]
- 327.Obeng-Nkrumah N, Twum-Danso K, Krogfelt KA, Newman MJ. High levels of extended-spectrum beta-lactamases in a major teaching hospital in Ghana: The need for regular monitoring and evaluation of antibiotic resistance. Am J Trop Med Hyg. 2013; [DOI] [PMC free article] [PubMed]
- 328.Opintan JA, Newman MJ. Prevalence of antimicrobial resistant pathogens from blood cultures: results from a laboratory based nationwide surveillance in Ghana. Antimicrob Resist Infect Control. 2017;6(1). [DOI] [PMC free article] [PubMed]
- 329.Quansah E, Barnie PA, Acheampong DO, Obiri-Yeboah D, Mills RO, Asmah E, et al. Geographical distribution of β-lactam resistance among Klebsiella spp. From selected health facilities in Ghana. Trop Med Infect Dis. 2019;4(3). [DOI] [PMC free article] [PubMed]
- 330.Agyepong N, Govinden U, Owusu-Ofori A, Essack SY. Multidrug-resistant gram-negative bacterial infections in a teaching hospital in Ghana. Antimicrob Resist Infect Control. 2018;7(1). [DOI] [PMC free article] [PubMed]
- 331.Amankwa R, Tay SC, Agbenorku P, Frimpong E, Gyampomah TK, Osei Sampene PP. Bacteriological profile of burn wound isolates in a burns center of a tertiary hospital. J Acute Dis. 2017;6(4):181–6.
- 332.Ayibieke A, Sato W, Mahazu S, Prah I, Addow-Thompson J, Ohashi M, et al. Molecular characterisation of the NDM-1encoding plasmid p2189-NDM in an Escherichia coli ST410 clinical isolate from Ghana. PLoS ONE. 2018;13(12). [DOI] [PMC free article] [PubMed]
- 333.Bourafa N, Chaalal W, Bakour S, Lalaoui R, Boutefnouchet N, Diene SM, et al. Molecular characterization of carbapenem-resistant Gram-negative bacilli clinical isolates in Algeria. Infect Drug Resist. 2018;11:735–742. doi: 10.2147/IDR.S150005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Codjoe FS, Donkor ES, Smith TJ, Miller K. Phenotypic and genotypic characterization of carbapenem-resistant gram-negative bacilli pathogens from hospitals in Ghana. Microb Drug Resist. 2019;25(10):1449–1457. doi: 10.1089/mdr.2018.0278. [DOI] [PubMed] [Google Scholar]
- 335.Eibach D, Campos CB, Krumkamp R, Al-Emran HM, Dekker D, Boahen KG, et al. Extended spectrum beta-lactamase producing Enterobacteriaceae causing bloodstream infections in rural Ghana, 2007–2012. Int J Med Microbiol. 2016. [DOI] [PubMed]
- 336.Janssen H, Janssen I, Cooper P, Kainyah C, Pellio T, Quintel M, et al. Antimicrobial-resistant bacteria in infected wounds, Ghana, 2014. Emerg Infect Dis. 2018;24(5):916–919. doi: 10.3201/eid2405.171506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Christian KG-S, Bernard N, Edwin MTY, Arhin AA, Ken A, Roland A, et al. Resistance pattern of uropathogenic bacteria in males with lower urinary tract obstruction in Kumasi, Ghana. Afr J Microbiol Res. 2014;
- 338.Isendahl J, Turlej-Rogacka A, Manjuba C, Rodrigues A, Giske CG, Nauclér P. Fecal carriage of ESBL-producing E. coli and K. pneumoniae in Children in Guinea-Bissau: a hospital-based cross-sectional study. PLoS ONE. 2012. [DOI] [PMC free article] [PubMed]
- 339.Kagia N, Kosgei P, Ooko M, Wafula L, Mturi N, Anampiu K, et al. Carriage and acquisition of extended-spectrum β-lactamase-producing Enterobacterales among Neonates Admitted to Hospital in Kilifi, Kenya. Clin Infect Dis. 2019;69(5):751–759. doi: 10.1093/cid/ciy976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Kariuki S, Corkill JE, Revathi G, Musoke R, Hart CA. Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob Agents Chemother. 2001;45(7):2141–2143. doi: 10.1128/AAC.45.7.2141-2143.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Kariuki S, Revathi G, Corkill J, Kiiru J, Mwituria J, Mirza N, et al. Escherichia coli from community-acquired urinary tract infections resistant to fluoroquinolones and extended-spectrum beta-lactams. J Infect Dev Ctries. 2007;1(3):257–262. doi: 10.3855/jidc.361. [DOI] [PubMed] [Google Scholar]
- 342.Kassim A, Omuse G, Premji Z, Revathi G. Comparison of Clinical Laboratory Standards Institute and European Committee on Antimicrobial Susceptibility Testing guidelines for the interpretation of antibiotic susceptibility at a University teaching hospital in Nairobi, Kenya: A cross-sectional stud. Ann Clin Microbiol Antimicrob. 2016; [DOI] [PMC free article] [PubMed]
- 343.Kiiru J, Kariuki S, Godderis BM, Butaye P. Analysis of β-lactamase phenotypes and carriage of selected β-lactamase genes among Escherichia coli strains obtained from Kenyan patients during an 18-year period. BMC Microbiol. 2012; [DOI] [PMC free article] [PubMed]
- 344.Kiiru J, Kariuki S, Goddeeris BM, Revathi G, Maina TW, Ndegwa DW, et al. Escherichia coli strains from Kenyan patients carrying conjugatively transferable broad-spectrum β-lactamase, qnr, aac(6’)-ib-cr and 16s rRNA methyltransferase genes. J Antimicrob Chemother. 2011;66(7):1639–1642. doi: 10.1093/jac/dkr149. [DOI] [PubMed] [Google Scholar]
- 345.Kohli R, Omuse G, Revathi G. Antibacterial susceptibility patterns of blood stream isolates in patients investigated at the Aga Khan university Hospital, Nairobi. East Afr Med J. 2010; [DOI] [PubMed]
- 346.Maina D, Omuse G, Revathi G, Adam RD. Spectrum of microbial diseases and resistance patterns at a private teaching hospital in Kenya: Implications for clinical practice. PLoS One. 2016; [DOI] [PMC free article] [PubMed]
- 347.Ndung’u C, Muigai AWT, Kenyatta J, Kariuki S. Prevalence and antibiotic resistance patterns of Escherichia coli among hospitalised patents at Thika District Hospital. East Afr Med J. 2014;91(6):185–90.
- 348.Nibogora C, Nyerere AK, Nguigi CW, Makau P. Phenotypic and Genotypic Characterization of Antibiotics Resistance Klebsiella pneumoniae Isolated from Clinical Samples at The Nairobi Hospital, Kenya. J Biol Agric Healthc. 2018;8(19–29).
- 349.Njagi LN, Odera S, Mutua F. Antimicrobial susceptibility patterns of urinary bacteria amongst paediatric patients at the Nairobi Hospital, Kenya. East Afr Med J. 2015;92(3).
- 350.Oneko M, Kariuki S, Muturi-Kioi V, Otieno K, Otieno VO, Williamson JM, et al. Emergence of community-acquired, multidrug-resistant invasive nontyphoidal Salmonella disease in Rural Western Kenya, 2009–2013. Clin Infect Dis. 2015;61:S310–S316. doi: 10.1093/cid/civ674. [DOI] [PubMed] [Google Scholar]
- 351.Poirel L, Revathi G, Bernabeu S, Nordmann P. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob Agents Chemother. 2011; [DOI] [PMC free article] [PubMed]
- 352.Shah M, Kathiiko C, Wada A, Odoyo E, Bundi M, Miringu G, et al. Prevalence, seasonal variation, and antibiotic resistance pattern of enteric bacterial pathogens among hospitalized diarrheic children in suburban regions of central Kenya. Trop Med Health. 2016; [DOI] [PMC free article] [PubMed]
- 353.Swierczewski BE, Odundo EA, Koech MC, Ndonye JN, Kirera RK, Kirera CP, et al. Surveillance for enteric pathogens in a case-control study of acute diarrhea in Western Kenya. Trans R Soc Trop Med Hyg. 2013; [DOI] [PubMed]
- 354.Taitt CR, Leski TA, Erwin DP, Odundo EA, Kipkemoi NC, Ndonye JN, et al. Antimicrobial resistance of Klebsiella pneumoniae stool isolates circulating in Kenya. PLoS One. 2017;12(6). [DOI] [PMC free article] [PubMed]
- 355.Tornberg-Belanger SN, Rwigi D, Brander RL, Tickell KD, McGrath CJ, Muraya M, et al. Prevalence of antimicrobial resistance in commensal E. coli from children dischrarged from hospital in western Kenya. Am Soc Trop Med Hyg. 2019;101(S5):553.
- 356.Wangai FK, Masika MM, Lule GN, Karari EM, Maritim MC, Jaoko WG, et al. Bridging antimicrobial resistance knowledge gaps: The East African perspective on a global problem. PLoS One. 2019;14(2). [DOI] [PMC free article] [PubMed]
- 357.Aduda DSO, Macharia IM, Mugwe P, Oburra H, Farragher B, Brabin B, et al. Bacteriology of chronic suppurative otitis media (CSOM) in children in Garissa district, Kenya: A point prevalence study. Int J Pediatr Otorhinolaryngol. 2013;77(7):1107–1111. doi: 10.1016/j.ijporl.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 358.Apondi OE, Oduor OC, Gye BK, Kipkoech MK. High prevalence of multi-drug resistant Klebsiella pneumoniae in a tertiary teaching hospital in Western Kenya. African J Infect Dis. 2016; [DOI] [PMC free article] [PubMed]
- 359.Ayoyi AO, Kikuvi G, Bii C, Kariuki S. Prevalence, aetiology and antibiotic sensitivity profile of asymptomatic bacteriuria isolates from pregnant women in selected antenatal clinic from Nairobi, Kenya. Pan Afr Med J. 2017;26. [DOI] [PMC free article] [PubMed]
- 360.Carattoli A, Villa L, Poirel L, Bonnin RA, Nordmann P. Evolution of IncA/C blaCMY-2-carrying plasmids by acquisition of the blaNDM-1 carbapenemase gene. Antimicrob Agents Chemother. 2012;56(2):783–786. doi: 10.1128/AAC.05116-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Henson SP, Boinett CJ, Ellington MJ, Kagia N, Mwarumba S, Nyongesa S, et al. Molecular epidemiology of Klebsiella pneumoniae invasive infections over a decade at Kilifi County Hospital in Kenya. Int J Med Microbiol. 2017;307(7):422–429. doi: 10.1016/j.ijmm.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Juma A, Caroline N. Antimicrobial susceptibility profiles and prevalence of ESBLs among E. coli isolates recovered from people working in hospitality industry within Nairobi, Kenya. East Afr Med J [Internet]. 2017;94(6):445–58. Available from: https://www.researchgate.net/publication/328334021
- 363.Frickmann H, Köller T, Hagen RM, Ebert K-P, Müller M, Wenzel W, et al. Molecular epidemiology of multidrug-resistant bacteria isolated from Libyan and Syrian patients with war injuries in two Bundeswehr hospitals in Germany. Eur J Microbiol Immunol. 2018;8(1):1–11. doi: 10.1556/1886.2018.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Hammerum AM, Larsen AR, Hansen F, Justesen US, Friis-Møller A, Lemming LE, et al. Patients transferred from Libya to Denmark carried OXA-48-producing Klebsiella pneumoniae, NDM-1-producing Acinetobacter baumannii and meticillin-resistant Staphylococcus aureus. Int J Antimicrob Agents. 2012;40(2):191–192. doi: 10.1016/j.ijantimicag.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 365.Kaase M, Eckmanns T, Pfennigwerth N, Szabados F, Gatermann S. PRP08 Carbapenemases found in patients from Libyia. Int J Med Microbiol. 2012;302:91. [Google Scholar]
- 366.Kieffer N, Ahmed MO, Elramalli AK, Daw MA, Poirel L, Álvarez R, et al. Colistin-resistant carbapenemase-producing isolates among Klebsiella spp. and Acinetobacter baumannii in Tripoli, Libya. J Glob Antimicrob Resist. 2018;13:37–9. [DOI] [PubMed]
- 367.Kocsis E, Savio C, Piccoli M, Cornaglia G, Mazzariol A. Klebsiella pneumoniae harbouring OXA-48 carbapenemase in a Libyan refugee in Italy. Clin Microbiol Infect. 2013;19(9). [DOI] [PubMed]
- 368.Kraiem AG, Zorgani A, Elahmer O, Hammami A, Chaaben BM, Ghenghesh KS. New Delhi metallo-β-lactamase and OXA-48 carbapenemases in gram-negative bacilli isolates in Libya. Libyan J Med. 2015;10. [DOI] [PMC free article] [PubMed]
- 369.Lafeuille E, Decré D, Mahjoub-Messai F, Bidet P, Arlet G, Bingen E. OXA-48 carbapenemase-producing Klebsiella pneumoniae isolated from Libyan patients. Microb Drug Resist. 2013;19(6):491–497. doi: 10.1089/mdr.2012.0219. [DOI] [PubMed] [Google Scholar]
- 370.Leistner R, Denkel LA, Gastmeier P, Werner G, Layer F, Pfeifer Y. Prevalence of MRSA and Gram-negative bacteria with ESBLs and carbapenemases in patients from Northern Africa at a German hospital. J Antimicrob Chemother. 2015;70(11):3161–3164. doi: 10.1093/jac/dkv219. [DOI] [PubMed] [Google Scholar]
- 371.Mathlouthi N, Al-Bayssari C, El Salabi A, Bakour S, Ben Gwierif S, Zorgani AA, et al. Carbapenemases and extended-spectrum β-lactamases producing enterobacteriaceae isolated from Tunisian and Libyan hospitals. J Infect Dev Ctries. 2016;10(7):718–727. doi: 10.3855/jidc.7426. [DOI] [PubMed] [Google Scholar]
- 372.Mohammed MA, Alnour TMS, Shakurfo OM, Aburass MM. Prevalence and antimicrobial resistance pattern of bacterial strains isolated from patients with urinary tract infection in Messalata Central Hospital. Libya Asian Pac J Trop Med. 2016;9(8):771–776. doi: 10.1016/j.apjtm.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 373.Abujnah AA, Zorgani A, Sabri MAM, El-Mohammady H, Khalek RA, Ghenghesh KS. Multidrug resistance and extended-spectrum beta-lactamases genes among Escherichia coli from patients with urinary tract infections in Northwestern Libya. Libyan J Med. 2015;10(1). [DOI] [PMC free article] [PubMed]
- 374.Pirš M, Andlovic A, Cerar T, Žohar-Čretnik T, Kobola L, Kolman J, et al. A case of OXA-48 carbapenemase-producing Klebsiella pneumoniae in a patient transferred to Slovenia from Libya, November 2011. Eurosurveillance [Internet]. 2011;16(50):pii=20042. Available from: www.eurosurveillance.orghttp://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20042 [PubMed]
- 375.Salem MA, Ahmed FA. Bacterial Profile of Urinary Tract Infection and Antimicrobial Susceptibility Pattern Among Patients Attending at Bushra Medical Laboratory, Tripoli. Libya J Gastroenterol Hepatol Res. 2018;7(4):2671–2675. doi: 10.17554/j.issn.2224-3992.2018.07.788. [DOI] [Google Scholar]
- 376.Zorgani A, Abofayed A, Glia A, Albarbar A, Hanish S. Prevalence of device-associated nosocomial infections caused by gram-negative bacteria in a trauma intensive care unit in Libya. Oman Med J. 2015;30(4):270–275. doi: 10.5001/omj.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Zorgani A, Almagatef A, Sufya N, Bashein A, Tubbal A. Detection of CTX-M-15 among uropathogenic Escherichia coli isolated from five major hospitals in Tripoli, Libya. Oman Med J. 2017;32(4):322–327. doi: 10.5001/omj.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Zorgani A, Daw H, Sufya N, Bashein A, Elahmer O, Chouchani C. Co-occurrence of plasmid-mediated AmpC β-lactamase activity among Klebsiella pneumoniae and Escherichia coli. Open Microbiol J. 2017;11(1):195–202. doi: 10.2174/1874285801711010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Ahmed SF, Ali MMM, Mohamed ZK, Moussa TA, Klena JD. Fecal carriage of extended-spectrum beta-lactamases and AmpC-producing Escherichia coli in a Libyan community. Ann Clin Microbiol Antimicrob. 2014;13(22). [DOI] [PMC free article] [PubMed]
- 380.Ali IM, Amirthalingam R. Clinical isolates of MEC A, methicillin, vancomycin resistance S. aureus; ESBLs producing K. pneumoniae, E. coli, P. auregenosa from various clinical source and its antimicrobial resistance patterns. Int J Med Res Heal Sci. 2015;4(1):123.
- 381.Buzayan MM, El-Garbulli FR. Detection of ESBL and AmpC-lactamases producing in uropathogen Escherichia coli isolates at Benghazi Center of Infectious Diseases and Immunity. Int J Curr Microbiol Appl Sci [Internet]. 2014;3(2):145–53. Available from: http://www.ijcmas.com
- 382.Buzayan MM, Tobgi RS, Ibrahim AAT. Detection of extended spectrum β-lactamases among urinary Escherichia coli and Klebsiella pneumoniae from two centres. Jamahiriya Med J. 2010;10(1):10–16. [Google Scholar]
- 383.Carannante N, Pallotto C, Bernardo M, Di Caprio G, Tascini C. Treatment of a Klebsiella pneumoniae KPC cellulitis and gut decolonization with ceftazidime/avibactam in a migrant from Libya. J Chemother. 2018;30(3):183–184. doi: 10.1080/1120009X.2018.1424504. [DOI] [PubMed] [Google Scholar]
- 384.Dau AA, Tloba S, Daw MA. Characterization of wound infections among patients injured during the 2011 Libyan conflict. East Mediterr Heal J. 2013;19(4):356–361. doi: 10.26719/2013.19.4.356. [DOI] [PubMed] [Google Scholar]
- 385.Elramalli A, Almshawt N, Ahmed MO. Current problematic and emergence of carbapenemase-producing bacteria: A brief report from a libyan hospital. Pan Afr Med J. 2017;26. [DOI] [PMC free article] [PubMed]
- 386.Rakotovao-Ravahatra ZD, Randriatsarafara FM, Rasoanandrasana S, Raverohanta L, Rakotovao AL. Resistant phenotypes of Escherichia coli strains responsible for urinary tract infection in the laboratory of the University Hospital Joseph Raseta Befelatanana, Antananarivo. Pan Afr Med J. 2017;26. [DOI] [PMC free article] [PubMed]
- 387.Randrianirina F, Vaillant L, Ramarokoto CE, Rakotoarijaona A, Andriamanarivo ML, Razafimahandry HC, et al. Antimicrobial resistance in pathogens causing nosocomial infections in surgery and intensive care units of two hospitals in Antananarivo, Madagascar. J Infect Dev Ctries. 2010; [DOI] [PubMed]
- 388.Randrianirina F, Vedy S, Rakotovao D, Ramarokoto CE, Ratsitohaina H, Carod JF, et al. Role of contaminated aspiration tubes in nosocomial outbreak of Klebsiella pneumoniae producing SHV-2 and CTX-M-15 extended-spectrum β-lactamases. J Hosp Infect. 2009;72(1):23–29. doi: 10.1016/j.jhin.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 389.Ranosiarisoa ZN, El Harrif S, Andrianirina AZ, Duron S, Simon-Ghediri MJ, Ramparany L, et al. Epidemiology of Early-onset Bacterial Neonatal Infections in Madagascar. Pediatr Infect Dis J. 2019;38(1):76–81. doi: 10.1097/INF.0000000000001993. [DOI] [PubMed] [Google Scholar]
- 390.Rasamiravaka T, Batavisoa E, Ranaivosoa MK, Rasamindrakotroka A. Profile and antimicrobial resistance to newly available drugs of urinary tract pathogens among Malagasy pregnant women. Trop Biomed. 2016; [PubMed]
- 391.Rasamiravaka T, Shaista Sheila HSL, Rakotomavojaona T, Rakoto-Alson AO, Rasamindrakotroka A. Changing profile and increasing antimicrobial resistance of uropathogenic bacteria in Madagascar. Med Mal Infect. 2015; [DOI] [PubMed]
- 392.Andriatahina T, Randrianirina F, Hariniana ER, Talarmin A, Raobijaona H, Buisson Y, et al. High prevalence of fecal carriage of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a pediatric unit in Madagascar. BMC Infect Dis. 2010;10. [DOI] [PMC free article] [PubMed]
- 393.Chereau F, Herindrainy P, Garin B, Huynh BT, Randrianirina F, Padget M, et al. Colonization of extended-spectrum-β-lactamase- and NDM-1-producing Enterobacteriaceae among pregnant women in the community in a low-income country: A potential reservoir for transmission of multiresistant Enterobacteriaceae to neonates. Antimicrob Agents Chemother. 2015;59(6):3652–3655. doi: 10.1128/AAC.00029-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Herindrainy P, Randrianirina F, Ratovoson R, Hariniana E, Buisson Y, Genel N, et al. Rectal carriage of extended-spectrum beta-lactamase-producing Gram-negative bacilli in community settings in Madagascar. PLoS One. 2011; [DOI] [PMC free article] [PubMed]
- 395.Naas T, Cuzon G, Robinson AL, Andrianirina Z, Imbert P, Ratsima E, et al. Neonatal infections with multidrug-resistant ESBL-producing E. cloacae and K. pneumoniae in Neonatal Units of two different Hospitals in Antananarivo, Madagascar. BMC Infect Dis. 2016;16(1). [DOI] [PMC free article] [PubMed]
- 396.Gallaher JR, Banda W, Lachiewicz AM, Krysiak R, Cairns BA, Charles AG. Colonization with Multidrug-Resistant Enterobacteriaceae is Associated with Increased Mortality Following Burn Injury in Sub-Saharan Africa. World J Surg. 2018;42(10):3089–3096. doi: 10.1007/s00268-018-4633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Gray KJ, Wilson LK, Phiri A, Corkill JE, French N, Hart CA. Identification and characterization of ceftriaxone resistance and extended-spectrum β-lactamases in Malawian bacteraemic Enterobacteriaceae. J Antimicrob Chemother. 2006;57(4):661–665. doi: 10.1093/jac/dkl037. [DOI] [PubMed] [Google Scholar]
- 398.Kumwenda GP, Sugawara Y, Abe R, Akeda Y, Kasambara W, Chizani K, et al. First Identification and genomic characterization of multidrug-resistant carbapenemase-producing Enterobacteriaceae clinical isolates in Malawi. Africa J Med Microbiol. 2019;68(12):1707–1715. doi: 10.1099/jmm.0.001087. [DOI] [PubMed] [Google Scholar]
- 399.Sangare SA, Rondinaud E, Maataoui N, Maiga AI, Guindo I, Maiga A, et al. Very high prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in bacteriemic patients hospitalized in teaching hospitals in Bamako, Mali. PLoS One. 2017; [DOI] [PMC free article] [PubMed]
- 400.Tandé D, Boisramé-Gastrin S, Münck MR, Héry-Arnaud G, Gouriou S, Jallot N, et al. Intrafamilial transmission of extended-spectrum-β-lactamase-producing Escherichia coli and Salmonella enterica Babelsberg among the families of internationally adopted children. J Antimicrob Chemother. 2010;65(5):859–865. doi: 10.1093/jac/dkq068. [DOI] [PubMed] [Google Scholar]
- 401.Hailaji NSM, Ould Salem ML, Ghaber SM. La sensibilité aux antibiotiques des bactéries uropathogènes dans la ville de Nouakchott – Mauritanie. Prog en Urol. 2016;26(6):346–352. doi: 10.1016/j.purol.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 402.Allyn J, Coolen-Allou N, De Parseval B, Galas T, Belmonte O, Allou N, et al. Medical evacuation from abroad of critically ill patients: A case report and ethical issues. Med (United States). 2018;97(38). [DOI] [PMC free article] [PubMed]
- 403.Issack M. Antibiotic resistance among hospitalized patients in Mauritius in 2014. Int J Infect Dis. 2016;45:94. doi: 10.1016/j.ijid.2016.02.250. [DOI] [Google Scholar]
- 404.Poirel L, Lascols C, Bernabeu S, Nordmann P. NDM-1-producing Klebsiella pneumoniae in Mauritius. Antimicrobial Agents and Chemotherapy. 2012. [DOI] [PMC free article] [PubMed]
- 405.Barguigua A, El otmani F, Lakbakbi FEY, Talmi M, Zerouali K, Timinouni M. First report of a Klebsiella pneumoniae strain coproducing NDM-1, VIM-1 and OXA-48 carbapenemases isolated in Morocco. J Pathol Microbiol Immunol. 2012;121(7):675–7. [DOI] [PubMed]
- 406.Barguigua A, El Otmani F, Talmi M, Zerouali K, Timinouni M. Emergence of carbapenem-resistant Enterobacteriaceae isolates in the Moroccan community. Diagn Microbiol Infect Dis [Internet]. 2012;73(3):290–1. Available from: http://dx.doi.org/10.1016/j.diagmicrobio.2012.03.011 [DOI] [PubMed]
- 407.Barguigua A, El Otmani F, Talmi M, Zerouali K, Timinouni M. Prevalence and types of extended spectrum β-lactamases among urinary Escherichia coli isolates in Moroccan community. Microb Pathog. 2013;61–62:16–22. doi: 10.1016/j.micpath.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 408.Barguigua A, El OF, Talmi M, Bourjilat F, Haouzane F, Zerouali K, et al. Characterization of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates from the community in Morocco. J Med Microbiol. 2011;60(9):1344–1352. doi: 10.1099/jmm.0.032482-0. [DOI] [PubMed] [Google Scholar]
- 409.Barguigua A, Zerouali K, Katfy K, El Otmani F, Timinouni M, Elmdaghri N. Occurrence of OXA-48 and NDM-1 carbapenemase-producing Klebsiella pneumoniae in a Moroccan university hospital in Casablanca. Morocco Infect Genet Evol. 2015;31:142–148. doi: 10.1016/j.meegid.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 410.Benaicha H, Barrijal S, Ezzakkioui F, Elmalki F. Prevalence of PMQR genes in E. coli and Klebsiella spp. isolated from North-West of Morocco. J Glob Antimicrob Resist. 2017;10:321–5. [DOI] [PubMed]
- 411.Benouda A, Touzani O, Khairallah MT, Araj GF, Matar GM. First detection of oxacillinase-mediated resistance to carbapenems in Klebsiella pneumoniae from Morocco. Ann Trop Med Parasitol. 2010;104(4):327–330. doi: 10.1179/136485910X12743554760108. [DOI] [PubMed] [Google Scholar]
- 412.Bourjilat F, Bouchrif B, Dersi N, Claude JDPG, Amarouch H, Timinouni M. Emergence of extended-spectrum beta-lactamase-producing Escherichia coli in community-acquired urinary infections in Casablanca. Morocco J Infect Dev Ctries. 2011;5(12):850–855. doi: 10.3855/jidc.1490. [DOI] [PubMed] [Google Scholar]
- 413.Chabah M, Chemsi M, Zerouali K, Alloula O, Lehlimi M, Habzi A, et al. Healthcare-associated infections due to carbapenemase-producing Enterobacteriaceae: Bacteriological profile and risk factors. Med Mal Infect. 2016;46(3):157–162. doi: 10.1016/j.medmal.2015.12.015. [DOI] [PubMed] [Google Scholar]
- 414.Cheikh A, Belefquih B, Chajai Y, Cheikhaoui Y, El Hassani A, Benouda A. Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) colonization as a risk factor for developing ESBL infections in pediatric cardiac surgery patients: “retrospective cohort study.” BMC Infect Dis. 2017;17(237). [DOI] [PMC free article] [PubMed]
- 415.El Bouamri MC, Arsalane L, El Kamouni Y, Zouhair S. Antimicrobial susceptibility of urinary Klebsiella pneumoniae and the emergence of carbapenem-resistant strains: A retrospective study from a university hospital in Morocco. North Africa African J Urol. 2015;21(1):36–40. doi: 10.1016/j.afju.2014.10.004. [DOI] [Google Scholar]
- 416.El Bouamri MC, Arsalane L, Zerouali K, Katfy K, El kamouni Y, Zouhair S. Molecular characterization of extended spectrum β-lactamase-producing Escherichia coli in a university hospital in Morocco, North Africa. African J Urol. 2015;21(3):161–6.
- 417.Essayagh T, Zohoun A, Tourabi K, Ennouhi MA, Boumaarouf A, Ihrai H, et al. Burn unit: Colonization of burn wounds and local environment. Ulus Travma ve Acil Cerrahi Derg. 2012;18(4):296–300. doi: 10.5505/tjtes.2012.26928. [DOI] [PubMed] [Google Scholar]
- 418.Girlich D, Bouihat N, Poirel L, Benouda A, Nordmann P. High rate of faecal carriage of extended-spectrum β-lactamase and OXA-48 carbapenemase-producing Enterobacteriaceae at a University hospital in Morocco. Clin Microbiol Infect. 2014;20(4):350–354. doi: 10.1111/1469-0691.12325. [DOI] [PubMed] [Google Scholar]
- 419.Hays C, Benouda A, Poirel L, Elouennas M, Nordmann P. Nosocomial occurrence of OXA-48-producing enterobacterial isolates in a Moroccan hospital. Int J Antimicrob Agents. 2012;39(6):545–547. doi: 10.1016/j.ijantimicag.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 420.El KA, Zerouali K, Diawara I, Ouhadous M, Harrar N, Belabbes H, et al. Healthcare-associated bacteraemia in intensive care units of Ibn Rochd University Hospital. Casablanca Sante publizue. 2017;29(2):209–213. [PubMed] [Google Scholar]
- 421.Lachhab Z, Frikh M, Maleb A, Kasouati J, Doghmi N, Ben Lahlou Y, et al. Bacteraemia in Intensive Care Unit: Clinical, Bacteriological, and Prognostic Prospective Study. Can J Infect Dis Med Microbiol. 2017;2017. [DOI] [PMC free article] [PubMed]
- 422.Mouaffak Y, Boutbaoucht M, Soraa N, Chabaa L, Salama T, Oulad Saiad M, et al. Bacteriology of community-acquired peritonitis in children treated in the University Hospital of Marrakech. Ann Fr Anesth Reanim. 2013;32:60–62. doi: 10.1016/j.annfar.2012.10.031. [DOI] [PubMed] [Google Scholar]
- 423.Nadmi H, Elotmani F, Talmi M, Zerouali K, Perrier-Gros-Claude JD, Timinouni M. Antibiotic resistance profile ofcommunity acquired uropathogenic enterobacteria in El Jadida (Morocco) Med Mal Infect. 2010;40(5):303–305. doi: 10.1016/j.medmal.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 424.Natoubi S, Barguigua A, Baghdad N, Nayme K, Timinouni M, Hilali A, et al. Occurrence of carbapenemases and extended-spectrum beta-lactamases in uropathogenic Enterobacteriaceae isolated from a community setting, Settat. Morocco Asian J Pharm Clin Res. 2017;10(1):211–215. doi: 10.22159/ajpcr.2017.v10i1.14924. [DOI] [Google Scholar]
- 425.Nejma H, Laghla B, Boutbaoucht M, Samkaou MA. Evolution of Escherichia coli resistance in community acquired peritonitis. Med Mal Infect. 2011;41:218–220. doi: 10.1016/j.medmal.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 426.Poirel L, Benouda A, Hays C, Nordmann P. Emergence of NDM-1-producing Klebsiella pneumoniae in Morocco. J Antimicrob Chemother. 2011;66(12):2781–2783. doi: 10.1093/jac/dkr384. [DOI] [PubMed] [Google Scholar]
- 427.Sáez-López E, Cossa A, Benmessaoud R, Madrid L, Moraleda C, Villanueva S, et al. Characterization of vaginal Escherichia coli isolated from pregnant women in two different African sites. PLoS One. 2016;11(7). [DOI] [PMC free article] [PubMed]
- 428.Shimi A, Touzani S, Elbakouri N, Bechri B, Derkaoui A, Khatouf M. Nosocomial pneumonia in ICU CHU Hassan II of Fez. Pan Afr Med J. 2015;22:285. doi: 10.11604/pamj.2015.22.285.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Tali A, Zahlane K, Chabaa L, Soraa N, Tayeb Z, El Bakkouri J, et al. Klebsiella pneumoniae infections in children: Epidemiological patterns and antibiotics susceptbility in Mother and Child Hospital of the Unviersity Hospital Mohammed VI in Marrakech. 13th Arab Congr Clin Biol 12th Moroccan Congr CCLMP. 2002;A131.
- 430.Taoufik L, Amrani Hanchi A, Fatiha B, Nissrine S, Mrabih Rabou MF, Nabila S. Emergence of OXA-48 Carbapenemase Producing Klebsiella pneumoniae in a Neonatal Intensive Care Unit in Marrakech. Morocco Clin Med Insights Pediatr. 2019;13:1–5. doi: 10.1177/1179556519834524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Amiyare R, Ouhssine M. Bacteriological profile of infection nosocomial urinary in intensive care unit of hospital El idrissiKenitra in Morocco. Sch Res Libr. 2015;7(10):53–56. [Google Scholar]
- 432.Arhoune B, Oumokhtar B, Hmami F, Barguigua A, Timinouni M, el Fakir S, et al. Rectal carriage of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae among hospitalised neonates in a neonatal intensive care unit in Fez. Morocco J Glob Antimicrob Resist. 2017;8:90–96. doi: 10.1016/j.jgar.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 433.Arsalane L, Zouhair S, Lahlou Amine I, Louzi L, Bouskraoui M. Urinary tract infection in infants (376 cases) in a Moroccan hospital (2009–2010)—frequency and prevalence of resistance. Pathol Biol. 2012;60(6):e90–e91. doi: 10.1016/j.patbio.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 434.Chirindze LM, Zimba TF, Sekyere JO, Govinden U, Chenia HY, Sundsfjord A, et al. Faecal colonization of E. coli and Klebsiella spp. producing extended-spectrum beta-lactamases and plasmid-mediated AmpC in Mozambican university students. BMC Infect Dis. 2018;18(1). [DOI] [PMC free article] [PubMed]
- 435.Estaleva CEL. Extended spectrum B-lactamase and plasmid mediated AmpC resistance in clinical isolates of Escherichia coli from the central hospital of Maputo, Mozambique. [Internet]. 2016. Available from: https://www.infodesign.org.br/infodesign/article/view/355%0Ahttp://www.abergo.org.br/revista/index.php/ae/article/view/731%0Ahttp://www.abergo.org.br/revista/index.php/ae/article/view/269%0Ahttp://www.abergo.org.br/revista/index.php/ae/article/view/106
- 436.Guiral E, Pons MJ, Vubil D, Marí-Almirall M, Sigaúque B, Soto SM, et al. Epidemiology and molecular characterization of multidrug-resistant Escherichia coli isolates harboring blaCTX-M group 1 extended-spectrum β-lactamases causing bacteremia and urinary tract infection in Manhiça. Mozambique Infect Drug Resist. 2018;11:927–936. doi: 10.2147/IDR.S153601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Pons MJ, Vubil D, Guiral E, Jaintilal D, Fraile O, Soto SM, et al. Characterisation of extended-spectrum β-lactamases among Klebsiella pneumoniae isolates causing bacteraemia and urinary tract infection in Mozambique. J Glob Antimicrob Resist. 2015;3(1):19–25. doi: 10.1016/j.jgar.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 438.Preziosi M, Zimba TF, Lee K, Tomas M, Kinlin S, Nhatave-Paiva C, et al. A prospective observational study of bacteraemia in adults admitted to an urban Mozambican hospital. South African Med J. 2017;105(5):370–374. doi: 10.7196/SAMJ.8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Mengistu A, Gaeseb J, Uaaka G, Ndjavera C, Kambyambya K, Indongo L, et al. Antimicrobial sensitivity patterns of cerebrospinal fluid (CSF) isolates in Namibia: Implications for empirical antibiotic treatment of meningitis. J Pharm Policy Pract. 2013;6(1). [DOI] [PMC free article] [PubMed]
- 440.Abdoulaye O, Amadou MLH, Amadou O, Adakal O, Larwanou HM, Boubou L, et al. Epidemiological and bacteriological features of surgical site infections (ISO) in the Division of Surgery at the Niamey National Hospital (HNN). Pan Afr Med J. 2018;31. [DOI] [PMC free article] [PubMed]
- 441.Alio MF, Laouali B, Ali M, Hadiza IB, Ali K, Chaibou Y, et al. Phenotypic detection of extended spectrum beta-lactamase in multidrug-resistant Escherichia coli from clinical isolates in Niamey. Niger African J Microbiol Res. 2017;11(18):712–717. doi: 10.5897/AJMR2017.8535. [DOI] [Google Scholar]
- 442.Langendorf C, Le Hello S, Moumouni A, Gouali M, Mamaty AA, Grais RF, et al. Enteric bacterial pathogens in children with diarrhea in Niger: Diversity and antimicrobial resistance. PLoS One. 2015; [DOI] [PMC free article] [PubMed]
- 443.Page AL, de Rekeneire N, Sayadi S, Aberrane S, Janssens AC, Rieux C, et al. Infections in Children Admitted with Complicated Severe Acute Malnutrition in Niger. PLoS One. 2013; [DOI] [PMC free article] [PubMed]
- 444.Aibinu I, Odugbemi T, Koenig W, Ghebremedhin B. Sequence Type ST131 and ST10 Complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin Microbiol Infect. 2012; [DOI] [PubMed]
- 445.Oshun P, Ogunsola F, Lagos NG. Carbapenem resistant Klebsiella pneumoniae at the Lagos University teaching hospital, Lagos, Nigeria. Clin Microbiol Infect. 2012;18(3):765. [Google Scholar]
- 446.Osundiya O, Oladele R, Oduyebo O. Multiple Antibiotic Resistance (MAR) indices of Pseudomonas and Klebsiella species isolates in Lagos University Teaching Hospital. African J Clin Exp Microbiol. 2013;
- 447.Otokunefor K, Tamunokuro E, Amadi A. Molecular detection of mobilized colistin resistance (mcr-1) gene in Escherichia coli isolates from Port Harcourt, Nigeria. J Appl Sci Environ Manag. 2019;23(3):401. [Google Scholar]
- 448.Ozumba UC, Jiburum BC. Bacteriology of burn wounds in Enugu. Nigeria Burns. 2000;26:178–180. doi: 10.1016/S0305-4179(99)00075-3. [DOI] [PubMed] [Google Scholar]
- 449.Raji MA, Jamal W, Ojemeh O, Rotimi VO. Sequence analysis of genes mediating extended-spectrum beta-lactamase (ESBL) production in isolates of Enterobacteriaceae in a Lagos Teaching Hospital, Nigeria. BMC Infect Dis. 2015;15(1). [DOI] [PMC free article] [PubMed]
- 450.Raji MA, Jamal W, Ojemhen O, Rotimi VO. Point-surveillance of antibiotic resistance in Enterobacteriaceae isolates from patients in a Lagos Teaching Hospital. Nigeria: J Infect Public Health; 2013. [DOI] [PubMed] [Google Scholar]
- 451.Iroha IR, Egwu OA, Ngozi AT, Chidiebube NA, Chika EP. Extended spectrum beta - Lactamase (ESBL) mediated resistance to antibiotics among Klebsiella pneumoniae in Enugu metropolis. Maced J Med Sci. 2009;
- 452.Romanus II, Eze AT. Antibiotics susceptibility patterns and clonal relatedness of uropathogenic Escherichia coli in Abakaliki, Ebonyi State. Can J Pure Appl Sci. 2009;5(2):1475. [Google Scholar]
- 453.Seni J, Peirano G, Okon KO, Jibrin YB, Mohammed A, Mshana SE, et al. The population structure of clinical extra-intestinal Escherichia coli in a teaching hospital from Nigeria. Diagn Microbiol Infect Dis. 2018;92(1):46–49. doi: 10.1016/j.diagmicrobio.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 454.Shettima SA, Tickler IA, dela Cruz CM, Tenover FC. Characterisation of carbapenem-resistant Gram-negative organisms from clinical specimens in Yola. Nigeria J Glob Antimicrob Resist. 2020;21:42–45. doi: 10.1016/j.jgar.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 455.Aiyegoro OA, Igbinosa OO, Ogunmwonyi IN, Odjadjare EE, Igbinosa OE, Okoh AI. Incidence of urinary tract infection (UTI) among children and adolescents of Ile-Ife. African J Microbiol Res [Internet]. 2007;013–9. Available from: https://www.researchgate.net/publication/258027967
- 456.Shu’aibu SS, Arzai AH, Nura S, Shaaibu AS. Antimicrobial susceptibility profile of class D Oxa B-lactamases producing bacteria in Kano state, Nigeria. Bayero J Pure Appl Sci. 2019;11(1):471–6.
- 457.Soge OO, Queenan AM, Ojo KK, Adeniyi BA, Roberts MC. CTX-M-15 extended-spectrum β-lactamase from Nigerian Klebsiella pneumoniae. J Antimicrob Chemother. 2006;57(1):24–30. doi: 10.1093/jac/dki429. [DOI] [PubMed] [Google Scholar]
- 458.State E, Moses U. Prevalence and antibiotic resistance profiles of extended spectrum β -lactamase-producing Escherichia coli among paediatric patients with urinary tract infection in St. Patricks’ Hospital, Mile Four, Abstract: Profils. Africal J Clin Exp Microbiol. 2019;20(4):332–6.
- 459.Sule H, Kumurya AS. The Prevalence of Klebsiella Species Causing Urinary Tract Infections in Murtala Muhammad Specialist Hospital, Kano. Nigeria Am J Biomed Life Sci. 2016;4(2):11–15. doi: 10.11648/j.ajbls.20160402.11. [DOI] [Google Scholar]
- 460.Taiwo S, Aderounmu A. Catheter associated urinary tract infection: Aetiologic agents and antimicrobial susceptibility pattern in Ladoke Akintola University Teaching Hospital, Osogbo. Nigeria African J Biomed Res. 2009;9(3):141–148. [Google Scholar]
- 461.Ugbo EN, Moses IB, Orji JO, Ukpai EG, Eluu SC, Egbule CU, et al. Antimicrobial susceptibility patterns of uropathogenic microorganisms associated with vesico-vaginal fistula (VVF) patients in Abakaliki, South eastern Nigeria. African J Microbiol Res. 2018;12(46):1039–1043. doi: 10.5897/AJMR2018.8984. [DOI] [Google Scholar]
- 462.Uwaezuoke NS, Kieffer N, Iregbu KC, Nordmann P. First report of OXA-181 and NDM-1 from a clinical Klebsiella pneumoniae isolate from Nigeria. Int J Infect Dis. 2017;61:1–2. doi: 10.1016/j.ijid.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 463.Uzoamaka M, Ngozi O, Johnbull OS, Martin O. Bacterial Etiology of Lower Respiratory Tract Infections and Their Antimicrobial Susceptibility. Am J Med Sci [Internet]. 2017;354(5):471–5. Available from: www.amjmedsci.com [DOI] [PubMed]
- 464.Walkty A, Gilmour M, Simner P, Embil JM, Boyd D, Mulvey M, et al. Isolation of multiple carbapenemase-producing Gram-negative bacilli from a patient recently hospitalized in Nigeria. Diagn Microbiol Infect Dis. 2015;81(4):296–298. doi: 10.1016/j.diagmicrobio.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 465.Yunusa T, Adeoye AM, Akitoye OA. Profile of septic work up among patients admitted into the intensive care unit in University of Abuja teaching hospital Gwagwalada, Abuja. African J Clin Exp Microbiol. 2018;20(1):9. doi: 10.4314/ajcem.v20i1.2. [DOI] [Google Scholar]
- 466.Ajayi AO, Osanyinlusi SA, Ogeneh B, Ojerinde OA, Oladeji SJ. Antibiotic Resistance Patterns among Gram-negative Bacteria from Patients with Urinary Tract Infection at a Healthcare Center in Ekiti-State. Nigeria Am J Microbiol Res. 2019;7(2):37–44. doi: 10.12691/ajmr-7-2-1. [DOI] [Google Scholar]
- 467.Yusuf I, Arzai AH, Haruna M, Sharif AA, Getso MI. Detection of multi drug resistant bacteria in major hospitals in Kano, North-West, Nigeria. Brazilian J Microbiol. 2014; [DOI] [PMC free article] [PubMed]
- 468.Akinduti P, Oluwadun A, Iwalokun B, Onagbesan O, Ejilude O. Community-Acquire CTX-M Beta-Lactamase Enteric Isolates in Abeokuta. Nigeria Br Microbiol Res J. 2015;5(4):351–358. doi: 10.9734/BMRJ/2015/5622. [DOI] [Google Scholar]
- 469.Akinduti PA, Olasehinde GI, Ejilude O, Taiwo OS, Obafemi YD. Fecal carriage and phylodiversity of community-acquired blaTEM enteric bacilli in southwest Nigeria. Infect Drug Resist. 2018;11:2425–2433. doi: 10.2147/IDR.S178243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Akingbade OA, Olalekan AO, Okerentugba PO, Innocent-Adiele HC, Ohoh CC, Nwanze JC, et al. Multi-Drug Resistant (MDR) Urinopathogens among Patients Attending a Tertiary Hospital in Lagos, Nigeria. Researcher [Internet]. 2012;4(5):35–9. Available from: http://www.researchgate.net/publication/258340422_Multi-Drug_Resistant_(MDR)_Urinopathogens_among_Patients_Attending_a_Tertiary_Hospital_in_Lagos_Nigeria/file/60b7d52aac3a07fb7c.pdf
- 471.Eghafona NO, Enabulele IO. Aetiologic Agents of Acute Otitis Media (AOM): Prevalence, Antibiotic Susceptibility, Β-Lactamase (Βl) and Extended Spectrum Β-Lactamase (ESBL) Production. J Microbiol Biotechnol Food Sci. 2019;10:333–353. [Google Scholar]
- 472.Akinyemi KO, Iwalokun B, Ayoola R, Alafua, Elsse O. Abegunrin R, Fakorede CO, Adunmo M, et al. Extended spectrum beta-lactamase producing Klebsiella pneumoniae bacteraemia and reduced susceptibility to carbapenem in Lagos, Nigeria. Am Soc Trop Med Hyg. 2018;5(1):146–7.
- 473.Akujobi CN, Ezeanya CC, Emeka-Okafor KM, Ebenebe JC. A Study on Significant Bacteriuria among Children Attendint the Outpatient Clinic of a University Teaching Hospital. Nigeria Int J Microbiol Res. 2013;5(4):448–451. doi: 10.9735/0975-5276.5.4.448-451. [DOI] [Google Scholar]
- 474.Akujobi CN, Ezeanya CC. Emergence of Carbapenem Resistance among Extended Spectrum Beta-lactamase Isolate of Escherichia coli from Clinical Specimens in a Tertiary Hospital. Nigeria Int J Microbiol Res. 2013;5(2):367–370. doi: 10.9735/0975-5276.5.2.367-370. [DOI] [Google Scholar]
- 475.A AI, Katsa M, Yakubu H, Habibu T, Daniel A, Solomon AJ. Non-Salmonella Bacteremia Among Seropositive Hiv Patients Attending Three Tertiary Hospital In Nasarawa State , Nigeria . J Nat Sci Res. 2013;3(5):60–7.
- 476.Alabi OS, Obisesan AO, Ola AA. Prevalence of methicillin-resistant Staphylococcus aureus and extended spectrum β–lactamase producers among bacteria isolated from infected wounds in a tertiary hospital in Ibadan City. African J Clin Exp Microbiol. 2016;17(4):235. doi: 10.4314/ajcem.v17i4.3. [DOI] [Google Scholar]
- 477.Anibijuwon II, Gbala ID, Adebisi OO. Carbapenem-Resistant Enterobacteriaceae among In-Patients of Tertiary Hospitals in Southwest. Nigeria Not Sci Biol. 2018;10(3):310–317. doi: 10.15835/nsb10310300. [DOI] [Google Scholar]
- 478.Awopeju A, Ide L, Obunge O. Antibiotic Susceptibilities and Plasmid Profile of Extended Spectrum Beta Lactamase- producing Escherichia coli from Community Acquired Urinary Tract Infection at the University of Port Harcourt Teaching Hospital. Nigeria Br Microbiol Res J. 2015;9(6):1–9. doi: 10.9734/BMRJ/2015/19045. [DOI] [Google Scholar]
- 479.Bashir A, Garba I, Aliero AA, Kibiya A, Abubakar MH, Ntulume I, et al. Superbugs-related prolonged admissions in three tertiary hospitals, Kano State. Nigeria Pan Afr Med J. 2019;32:166. doi: 10.11604/pamj.2019.32.166.18481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Bigwan EI, David E. Prevalence of Escherichia coli among uropathogens in asymptomatic bacteriuria in a Nigerian Tertiary school in Jos. Nigeria Int J Biomed Adv Res. 2013;4(3):198–202. doi: 10.7439/ijbar.v4i3.250. [DOI] [Google Scholar]
- 481.Brinkac LM, White R, D’Souza R, Nguyen K, Obaro SK, Fouts DE, et al. Emergence of New Delhi Metallo-Lactamase (NDM-5) in Klebsiella quasipneumoniae from Neonates in a Nigerian Hospital. Clin Sci Epidemiol [Internet]. 2019;4(2):1–10. Available from: http://msphere.asm.org/ [DOI] [PMC free article] [PubMed]
- 482.Brown B, Dada-Adegbola H, Trippe C, Olopade O. Prevalence and etiology of bacteremia in febrile children with sickle cell disease at a Nigeria tertiary hospital. Mediterr J Hematol Infect Dis. 2017;9(1). [DOI] [PMC free article] [PubMed]
- 483.Chika E, Ifeanyichukwu I, Michael A, Charles E. Susceptibility and detection of extended spectrum β-Lactamase enzymes from Otitis media pathogens. Am J Infect Dis. 2013;9(1):24–29. doi: 10.3844/ajidsp.2013.24.29. [DOI] [Google Scholar]
- 484.Chinedu AE. Occurence of Beta-Lactamases in Escherichia coli and Klebsiella species isolated from environmental sources and hospital patients in Nsukka. Enugu State: University of Nigeria Nsukka; 2014. [Google Scholar]
- 485.Abba PO, Umeh EU, Gberikon GM, Agbo EB. Emergence of bla TEM resistance gene in ESBL-producing Escherichia coli clinical isolates from Health facilities in Makurdi, Benue State Nigeria. Int J Adv Sci Res Eng. 2019;5(8):96–101. [Google Scholar]
- 486.Chukwu BF, Okafor HU, Ikefuna AN. Asymptomatic bacteriuria in children with sickle cell anemia at the University of Nigeria teaching hospital, Enugu, South East, Nigeria. Vol. 37, Italian Journal of Pediatrics. 2011. [DOI] [PMC free article] [PubMed]
- 487.Dayyab FM, Iliyasu G, Aminu A, Habib ZG, Tiamiyu AB, Tambuwal SH, et al. A prospective study of hospital-acquired infections among adults in a tertiary hospital in north-western Nigeria. Trans R Soc Trop Med Hyg. 2018;112(1):36–42. doi: 10.1093/trstmh/try020. [DOI] [PubMed] [Google Scholar]
- 488.Ebenebe J. Plasmid Profile of Uropathogens among Children. Br J Med Med Res. 2014;4(5):1195–1203. doi: 10.9734/BJMMR/2014/6243. [DOI] [Google Scholar]
- 489.Egbe CA, Omoregie R, Igbarumah IO, Onemu S. Microbiology of wound infections and its associated risk factors among patients of a Tertiary hospital in Benin City, Nigeria. J Res Health Sci. 2011; [PubMed]
- 490.Ejiofor OS, Ajunwa OM, Ezeudu CE, Emechebe GO, Okeke KN, Ifezulike CC, et al. The Bacteriology and Its Virulence Factors in Neonatal Infections: Threats to Child Survival Strategies. J Pathog. 2018;2018:1–11. doi: 10.1155/2018/4801247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Elikwu CJ, Shobowale EO, Oluyemi OY, Afolabi DO, Aderinto DA, Onyedibe KI, et al. The etiology and antimicrobial susceptibility patterns of urinary tract infections at a private Nigerian teaching hospital in South West Nigeria. African J Clin Exp Microbiol. 2016;18(1):21. doi: 10.4314/ajcem.v18i1.3. [DOI] [Google Scholar]
- 492.Ella E, Ahmad AA, Ogala WN, Umoh VJ, Balogun TB. Characterization and antimicrobial sensitivity assay of gram negative rods isolated from neonates with septicaemia in Zaria. Artic J Pure Appl Microbiol [Internet]. 2007; Available from: https://www.researchgate.net/publication/288740308
- 493.Faari B, Akanbi A, Fadeyi A, Wahab K, Nwabuisi C, Faari B: B. Prevalence of extended spectrum beta-lactamase-producing Klebsiella species at the University of Ilorin Teaching Hospital. J Med Investig Pract [Internet]. 2015;10(1). Available from: https://go.gale.com/ps/anonymous?id=GALE%7CA435519035&sid=googleScholar&v=2.1&it=r&linkaccess=abs&issn=97831230&p=HRCA&sw=w
- 494.Fadeyi A, Zumuk CP, Raheem RA, Nwabuisi C, Desalu OO. Prevalence and antibiotic susceptibility pattern of ESBL producing Klebsiellae isolated from clinical specimens in a Nigerian tertiary hospital. African J Infect Dis. 2016;
- 495.Giwa F, Ige O, Haruna D, Yaqub Y, Lamido T, Usman S. Extended-Spectrum beta-lactamase production and antimicrobial susceptibility pattern of uropathogens in a Tertiary Hospital in Northwestern Nigeria. Ann Trop Pathol. 2018;9(1):11. doi: 10.4103/atp.atp_39_17. [DOI] [Google Scholar]
- 496.Abubakar E-MM. Antimicrobial susceptibility pattern of pathogenic bacteria causing urinary tract infections at the Specialist Hospital, Yola, Adamawa state, Nigeria. J Clin Med Res [Internet]. 2009;1(1):001–8. Available from: http://www.academicjournals.org/JCMR
- 497.Ibadin EE, Omoregie R, Igbarumah IO, Anogie NA, Ogefere HO. Prevalence of Extended Spectrum β-Lactamase, AmpC β-Lactamase and Metallo-β-Lactamase Among Gram Negative Bacilli Recovered From Clinical Specimens in Benin City. Nigeria Int J Enteric Pathog. 2017;5(3):85–91. doi: 10.15171/ijep.2017.20. [DOI] [Google Scholar]
- 498.Ibrahim Y, Sani Y, Saleh Q, Saleh A, Hakeem G. Phenotypic Detection of Extended Spectrum Beta lactamase and Carbapenemase Co-producing Clinical Isolates from Two Tertiary Hospitals in Kano, North West Nigeria. Ethiop J Health Sci. 2017;27(1):3–10. doi: 10.4314/ejhs.v27i1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Idowu OJ, Onipede AO, Orimolade AE, Akinyoola LA, Babalola GO. Extended-spectrum beta-lactamase orthopedic wound infections in Nigeria. J Glob Infect Dis. 2011;3(3):211–215. doi: 10.4103/0974-777X.83524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Igwe JC, Olayinka BO, Ehnimidu JO, Onanolapo JA. Virulent Characteristics of Multidrug Resistant E. coli from Zaria, Nigeria. Clin Microbiol. 2016;5(6).
- 501.Iliyasu G, Dayyab FM, Abubakar S, Inuwa S, Tambuwal SH, Tiamiyu AB, et al. Laboratory-confirmed hospital-acquired infections: An analysis of a hospital’s surveillance data in Nigeria. Heliyon [Internet]. 2018;4:720. Available from: 10.1016/j.heliyon.2018.e00720 [DOI] [PMC free article] [PubMed]
- 502.Inwezerua C, Mendonça N, Calhau V, Domingues S, Adeleke OE, Da Silva GJ. Occurrence of extended-spectrum beta-lactamases in human and bovine isolates of Escherichia colifrom Oyo state. Nigeria J Infect Dev Ctries. 2014;8(6):774–779. doi: 10.3855/jidc.3430. [DOI] [PubMed] [Google Scholar]
- 503.Iregbu K, Abdullahi N. Profiles of acute bacterial meningitis isolates in children in National Hospital, Abuja. Niger Med J. 2015;56(4):297. doi: 10.4103/0300-1652.169749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Iregbu K, Nwajiobi-Princewill P. Urinary tract infections in a Tertiary Hospital in Abuja, Nigeria. African J Clin Exp Microbiol. 2013;
- 505.Iregbu K, Sonibare S. Profile of Infections in Intensive Care Unit (Icu) in a Central Nigeria Tertiary Hospital. African J Clin Exp Microbiol. 2014;
- 506.Iregbu K, Uwaezuoke N, Nwajiobi-Princewill I, Eze S, Medugu N, Shettima S, et al. A profile of wound infections in National Hospital Abuja. African J Clin Exp Microbiol. 2013;
- 507.Adegoke AA, Okoh AI, Adegoke AA, Mvuyo T, Steve J. Studies on multiple antibiotic resistant bacteria isolated from surgical site infection. Sci Res Essays [Internet]. 2010;5(24):3876–81. Available from: http://www.academicjournals.org/SRE
- 508.Iregbu K, Zubair K, Modibbo I, Aigbe A, Sonibare S, Ayoola O. Neonatal infections caused by Escherichia coli at the National Hospital, Abuja: a three-year retrospective study. African J Clin Exp Microbiol. 2013;
- 509.Iroha IR, Afiuka FN, Oji AE, Ejikeugwu PC, Nwakeze EA. Occurrence of extended spectrum beta lactamase producing Escherichia coli from human clinical and wild birds (pigeons, bats, parrots and ducks)samples from Ebonyi State, Nigeria. World J Pharm Pharm Sci [Internet]. 2015;4(7):20–9. Available from: www.wjpps.com
- 510.Iroha IR, Oji AE, Ayogu, Oji. Analysis of antibiotic susceptibility of Klebsiella pneumoniae strains isolated from different clinical specimens in Enugu State. Int J Curr Res [Internet]. 2011;2(1):8–14. Available from: http://www.journalcra.comhttp//www.journalcra.com
- 511.Iroha IR, Okafor-Alu FN, Ugbo EN, Ejikeugwu CP, Nwuzo AC, Nwakeze AE, et al. Antibiogram of pathogenic bacteria isolated from pre- and post-surgery vesicovaginal fistula (VVF) patients in Abakaliki, Ebonyi State. Int J Pharm Sci Res. 2016;
- 512.Iroha IR, Ukwauani EO, Moses IB, Ajah MI, Iroha CS, Ajah LO. Prevalence and Characterization of Multi-drug resistant Uropathogens from Children with Urinary Tract Infections in Children Emergency Unit of Federal Teaching Hospital, Abakaliki (FETHA), Nigeria. Int J Med Heal Sci [Internet]. 2016;5(4). Available from: https://www.researchgate.net/publication/309040027
- 513.Isaiah IN, Nche BT, Nwagu IG, Nwagu II. Incidence of temonera, sulphuhydryl variables and cefotaximase genes associated with β-lactamase producing Escherichia coli in clinical isolates. N Am J Med Sci. 2011; [DOI] [PMC free article] [PubMed]
- 514.Iwuafor AA, Ogunsola FT, Oladele RO, Oduyebo OO, Desalu I, Egwuatu CC, et al. Incidence, clinical outcome and risk factors of intensive care unit infections in the Lagos University Teaching Hospital (LUTH), Lagos, Nigeria. PLoS One. 2016;11(10). [DOI] [PMC free article] [PubMed]
- 515.Jesumirhewe C, Springer B, Lepuschitz S, Allerberger F, Ruppitsch W. Carbapenemase-producing Enterobacteriaceae isolates from Edo State, Nigeria. Antimicrob Agents Chemother. 2017;61(8). [DOI] [PMC free article] [PubMed]
- 516.Jombo G, Emanghe U, Amefule E, Dahmen J. Urinary tract infections at a Nigerian university hospital: Causes, patterns and antimicrobial susceptibility profile. J Microbiol Antimicrob [Internet]. 2011;3(6):153–9. Available from: http://www.academicjournals.org/JMA
- 517.Kandakai-Olukemi Y, Mawak J, Onojo M. Isolation of Enteropathogenic Escherichia coli from Children with Diarrhoea Attending the National Hospital in Abuja, Nigeria. Shiraz E-Medical J [Internet]. 2009;10(3):99–106. Available from: http://semj.sums.ac.ir/vol10/jul2009/87022.htm
- 518.Adegoke AA, Okoh AI. Prevalence, antibiotic susceptibility profile and extended spectrum β-lactamase production among Escherichia coli from high vaginal swab (HVS) African J Pharm Pharmacol. 2011;5(10):1287–1291. doi: 10.5897/AJPP11.146. [DOI] [Google Scholar]
- 519.Kazmierczak KM, Rabine S, Hackel M, McLaughlin RE, Biedenbach DJ, Bouchillon SK, et al. Multiyear, multinational survey of the incidence and global distribution of metallo-β-lactamase-producing enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60(2):1067–1078. doi: 10.1128/AAC.02379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Chiedozie Kingsley O, Onwuezobe Ifeanyi A, Emmanuel Edet A, Chukwuemeka Smart O. Bacteriologic profile and antibiotics susceptibility pattern of suspected septicaemic patients in Uyo, Nigeria. Res J Med Sci. 2013;
- 521.Li X, Goodman D, Mihalov SA, Oswald M. Poster 118 New Delhi metallo-B-lactamase (NDM-1) producing Klebsiella pneumoniae urinary tract infection (UTI) in acute inpatient rehabilitation after right hip open reduction internal fixation revision (ORIF): a case report. PM&R. 2014;6(9):S224–S225. [Google Scholar]
- 522.Makanjuola OB, Fayemiwo SA, Gbaja A, Ogunleye VA, Kehinde AO, Bakare RA. Pattern of Multidrug Resistant Bacteria Associated with Intensive Care Unit Infections in Ibadan. Nigeria Ann Ibadan Postgrad Med. 2018;16(2):162–169. [PMC free article] [PubMed] [Google Scholar]
- 523.Medugu N, Iregbu KC. Trends in profiles of bacteria causing neonatal sepsis in Central Nigeria Hospital. African J Clin Exp Microbiol. 2016;18(1):49. doi: 10.4314/ajcem.v18i1.7. [DOI] [Google Scholar]
- 524.Mohammed Y, Gadzama G, Zailani S, Abubakar A, Dalhat M, Ibrahim B, et al. Determination of the antimicrobial susceptibility pattern of extended spectrum beta lactamase (esbl) producing and the non-esbl producing strains of Escherichia coli. African J Clin Exp Microbiol. 2016;
- 525.Mordi RM, Erah PO. Susceptibility of common urinary isolates to the commonly used antibiotics in a tertiary hospital in southern Nigeria. African J Biotechnol. 2006;5(11):1067–1071. [Google Scholar]
- 526.Motayo BO, Akinduti PA, Adeyakinu FA, Okerentugba PO, Nwanze JC, Onoh CC, et al. Antibiogram and plasmid profiling of carbapenemase and extended spectrum beta-lactamase (ESBL) producing Escherichia coli and Klebsiella pneumoniae in Abeokuta, South Western, Nigeria. Afr Health Sci. 2013; [DOI] [PMC free article] [PubMed]
- 527.Murphy RA, Okoli O, Essien I, Teicher C, Elder G, Pena J, et al. Multidrug-resistant surgical site infections in a humanitarian surgery project. Epidemiol Infect. 2016; [DOI] [PMC free article] [PubMed]
- 528.Ngwai Y, Akpotu MO, Obidake RE, Sounyo AA, Onanuga A, Origbo SO. Antimicrobial susceptibility of Escherichia coli and other coliforms isolated from urine of asymptomatic students in Bayelsa State, Nigeria. African J Microbiol Res [Internet]. 2010;5(3):184–91. Available from: https://www.researchgate.net/publication/225071308
- 529.Adekunle OC, Falade-Fatila AJ, Odewale G. Molecular Detection ctx-M, TEM and VIM in ESBL-Producing E. coli Strains Isolated from Pregnant Women in Osogbo. Microbiol Res J Int. 2019;28(2):1–8.
- 530.Njoku CO, Njoku AN. Microbiological pattern of surgical site infection following caesarean section at the University of Calabar Teaching Hospital. Open Access Maced J Med Sci. 2019;7(9):1430–1435. doi: 10.3889/oamjms.2019.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Nsofor CA, Anyanwu NC, Ogbulie TE. High Antibiotic Resistance Pattern Observed in Bacterial Isolates from a Tertiary Hospital in South East Nigeria. Int J Res Pharm Biosci. 2016;3(1):1–6. [Google Scholar]
- 532.Nwabuisi C, Ologe FE. Pathogenic Agents of Chronic Suppurative Otitis Media in Ilorin. Nigeria East African J. 2002;79(1):202–205. doi: 10.4314/eamj.v79i4.8879. [DOI] [PubMed] [Google Scholar]
- 533.Nwafia IN, Ohanu ME, Ebede SO, Ozumba UC. Molecular detection and antibiotic resistance pattern of extended-spectrum beta-lactamase producing Escherichia coli in a Tertiary Hospital in Enugu, Nigeria. Ann Clin Microbiol Antimicrob [Internet]. 2019;18(1):1–7. Available from: 10.1186/s12941-019-0342-9 [DOI] [PMC free article] [PubMed]
- 534.Nwankwo EO, Magaji NS, Tijjani J. Antibiotic susceptibility pattern of extended spectrum betalactamase (ESBL) producers and other bacterial pathogens in Kano. Nigeria Trop J Pharm Res. 2015;14(7):1273–1278. doi: 10.4314/tjpr.v14i7.21. [DOI] [Google Scholar]
- 535.Obi CL, Coker AO, Epoke J. Distributional patterns of bacterial diarrhoeagenic agents and antibiograms of isolates from diarrhoeaic and non-diarrhoeaic patients in urban and rural areas of Nigeria. Cent Afr J Med. 1998;44(9):223–229. [PubMed] [Google Scholar]
- 536.Ochada N, Nasiru I, Thairu Y, Okanlowan M, Abdulakeem Y. Antimicrobial Susceptibility Pattern of Urinary Pathogens Isolated from Two Tertiary Hospitals in Southwestern Nigeria. African J Clin Exp Microbiol. 2014;
- 537.Odedina EA, Eletta EA, Balogun RA, Idowu O. Isolates from wound infections at Federal Medical Centre. BIDA African J Clin Exp Microbiol. 2008;8(2):26–32. doi: 10.4314/ajcem.v9i1.7479. [DOI] [Google Scholar]
- 538.Ogbolu DO, Piddock LJV, Webber MA. Opening Pandora’s box: high level resistance to antibiotics of last resort in Gram negative bacteria from Nigeria. J Glob Antimicrob Resist. 2019 [DOI] [PubMed]
- 539.Ogbolu DO, Terry Alli OA, Webber MA, Oluremi AS, Oloyede OM. CTX-M-15 is established in most multidrug-resistant uropathogenic Enterobacteriaceae and Pseudomonaceae from hospitals in Nigeria. Eur J Microbiol Immunol. 2018;8(1):20–24. doi: 10.1556/1886.2017.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Adenipekun EO, Jackson CR, Ramadan H, Iwalokun BA, Oyedeji KS, Frye JG, et al. Prevalence and multidrug resistance of Escherichia coli from community-acquired infections in Lagos, Nigeria. J Infect Dev Ctries. 2016; [DOI] [PubMed]
- 541.Ogbolu DO, Webber MA. Carbapenem resistance in gram-negative bacteria in south-western Nigeria: The role of extended-spectrum β-lactamase CTX-M-15. West Indian Med J. 2018;67(4):344–349. doi: 10.7727/wimj.2017.122. [DOI] [Google Scholar]
- 542.Ogbolu DO, Webber MA. High-level and novel mechanisms of carbapenem resistance in Gram-negative bacteria from tertiary hospitals in Nigeria. Int J Antimicrob Agents. 2014; [DOI] [PubMed]
- 543.Ogefere HO, Aigbiremwen PA, Omoregie R. Extended-spectrum beta-lactamase (esbl)–producing gram-negative isolates from urine and wound specimens in a tertiary health facility in southern nigeria. Trop J Pharm Res. 2015;
- 544.Ogunshe AAO, Gbadamosi ME. Pediatric health implication of ògì and omi’dùn as potential complementary therapy for infantile teething-diarrheal control. Rawal Med J. 2011;36(1):45–49. [Google Scholar]
- 545.Okafor UE, Ogunsola FT, Osinupebi OA. Aetiology of Catheter-Associated bacteriuria in Lagos University Teaching Hospital. Niger Postgrad Med J. 2005;12(2):89–92. doi: 10.4103/1117-1936.175258. [DOI] [PubMed] [Google Scholar]
- 546.Oladipo AO, Olowe OA, Olafimihan KF, Udoh SJ. Ctx-m, Tem, and Shv Beta- lactamases in clinical isolates of Klebsiella species in Ile-Ife, Nigeria. Int J Infect Dis. 2014;
- 547.Olaitan AO, Diene SM, Kempf M, Berrazeg M, Bakour S, Gupta SK, et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: An epidemiological and molecular study. Int J Antimicrob Agents. 2014;44(6):500–507. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 548.Oli AN, Akabueze VB, Ezeudu CE, Eleje GU, Ejiofor OS, Ezebialu IU, et al. Bacteriology and Antibiogram of Urinary Tract Infection Among Female Patients in a Tertiary Health Facility in South Eastern Nigeria. Open Microbiol J. 2017;11(1):292–300. doi: 10.2174/1874285801711010292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Oli AN, Itumo CJ, Okam PC, Ezebialu IU, Okeke KN, Ifezulike CC, et al. Carbapenem-resistant enterobacteriaceae posing a dilemma in effective healthcare delivery. Antibiotics. 2019;8(4):1–11. doi: 10.3390/antibiotics8040156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Oli AN, Ogbuagu VI, Ejikeugwu CP, Iroha IR, Ugwu MC, Ofomata CM, et al. Multi-Antibiotic Resistance and Factors Affecting Carriage of Extended Spectrum β-Lactamase-Producing Enterobacteriaceae in Pediatric Population of Enugu Metropolis. Nigeria Med Sci. 2019;7(11):104–116. doi: 10.3390/medsci7110104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Adeyankinnu FA, Motayo BO, Akinduti A, Akinbo J, Ogiogwa JI, Aboderin BW, et al. A multicenter study of beta-lactamase resistant Escherichia coli and Klebsiella pneumoniae reveals high level chromosome mediated extended spectrum β lactamase resistance in ogun state, Nigeria. Interdiscip Perspect Infect Dis. 2014; [DOI] [PMC free article] [PubMed]
- 552.Olowe OA, Oladipo GO, Makanjuola OA, Olaitan JO. Prevalence of extended spectrum beta-lactamases (ESBLs) carrying genes in Klebsiella spp. from Clinical Samples at Ile-Ife, South Western Nigeria. Int J Pharma Med Biol Serv. 2012;1(2):129–38.
- 553.Olowe OA, Adefioye OJ, Ajayeoba TA, Schiebel J, Weinreich J, Ali A, et al. Phylogenetic grouping and biofilm formation of multidrug resistant Escherichia coli isolates from humans, animals and food products in South-West Nigeria. Sci African. 2019;6.
- 554.Olowe OA, Ayilara OA, Oladipo GO, Makanjuola OA, Olaitan JO. Multidrug resistance Escherichia coli carrying extended-spectrum β-lactamases enzymes in a tertiary care hospital in Osogbo, South Western Nigeria. Int J Pharma Med Biol Sci. 2012;
- 555.Olowe OA, Grobbel M, Büchter B, Lübke-Becker A, Fruth A, Wieler LH. Detection of blaCTX-M-15 extended-spectrum beta-lactamase genes in Escherichia coli from hospital patients in Nigeria. Int J Antimicrob Agents. 2010;35(2):206–207. doi: 10.1016/j.ijantimicag.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 556.Omoyibo EE, Oladele AO, Ibrahim MH, Adekunle OT. Antibiotic susceptibility of wound swab isolates in a tertiary hospital in Southwest Nigeria. Ann Afr Med. 2018;17(3):110–116. doi: 10.4103/aam.aam_22_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Onanuga A, Mahindroo J, Singh S, Taneja N. Phenotypic and molecular characterization of antimicrobial resistant Escherichia coli from urinary tract infections in port-harcourt, nigeria. Pan Afr Med J. 2019;34(144):1–14. doi: 10.11604/pamj.2019.34.144.18182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Onanuga A, Vincent CH, Eboh DD. Carbapenem Resistance among Extended Spectrum Beta-Lactamases Producing Escherichia coli and Klebsiella pneumoniae isolates from Patients with Urinary Tract Infections in Port-Harcourt, Nigeria. Niger J Pharm Appl Sci Res. 2019;8(1):16–23. [Google Scholar]
- 559.Onifade J, Oladipo A. Prevalence of Urinary Tract Infections (UTIs) Among Pregnant Women Attending Antenatal Clinic at Ile-Ife. Southwestern Nigeria Am Soc Clin Pathol. 2019;152:128–129. [Google Scholar]
- 560.Onwuezobe A, Orok F. Extended spectrum beta-lactamase producing Uropathogens in Asymptomaticpregnant women attending antenatal care in an urban community secondary health facility. African J Clin Exp Microbiol. 2015;
- 561.Onyedibe K, Bode-Thomas F, Afolaranmi T, Okolo M, Banwat E, Egah D. Bacteriologic Profile, Antibiotic Regimen and Clinical Outcome of Neonatal Sepsis in a University Teaching Hospital in North Central Nigeria. Br J Med Med Res. 2015;
- 562.Bayingana C, Kayitare E, Nteziyaremye J, Sendegeya A, Ndoli J, Busumbigabo A. Update on Antibiotic Activity on Bacterial Strains Isolated from Urine Samples at Butare University Teaching Hospital (BUTH) Laboratory in Rwanda. Online Int Interdiscip Res J. 2016;6(2):6–16. [Google Scholar]
- 563.Carroll M, Rangaiahagari A, Musabeyezu E, Singer D, Ogbuagu O. Five-year antimicrobial susceptibility trends among bacterial isolates from a tertiary health-care facility in Kigali. Rwanda Am J Trop Med Hyg. 2016;95(6):1277–1283. doi: 10.4269/ajtmh.16-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Halfon J, Thielman N, Hill W, Rulisa S. Alarming Rates of Antibiotic Resistance Among Organisms Causing Post-Cesarean Section Peritonitis in Rwanda. Obstet Gynecol [Internet]. 2016;127:86S. Available from: https://journals.lww.com/greenjournal
- 565.Kurz MSE, Bayingana C, Ndoli JM, Sendegeya A, Durst A, Pfüller R, et al. Intense pre-admission carriage and further acquisition of ESBL-producing Enterobacteriaceae among patients and their caregivers in a tertiary hospital in Rwanda. Trop Med Int Heal. 2017; [DOI] [PubMed]
- 566.Muvunyi CM, Masaisa F, Bayingana C, Mutesa L, Musemakweri A, Muhirwa G, et al. Decreased susceptibility to commonly used antimicrobial agents in bacterial pathogens isolated from urinary tract infections in Rwanda: Need for new antimicrobial guidelines. Am J Trop Med Hyg. 2011; [DOI] [PMC free article] [PubMed]
- 567.Ntirenganya C, Manzi O, Muvunyi CM, Ogbuagu O. High prevalence of antimicrobial resistance among common bacterial isolates in a tertiary healthcare facility in Rwanda. Am J Trop Med Hyg. 2015; [DOI] [PMC free article] [PubMed]
- 568.Sutherland T, Mpirimbanyi C, Nziyomaze E, Niyomugabo JP, Niyonsenga Z, Muvunyi CM, et al. Widespread antimicrobial resistance among bacterial infections in a Rwandan referral hospital. PLoS ONE. 2019;14(8):1–15. doi: 10.1371/journal.pone.0221121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Poirel L, Aires-De-Sousa M, Kudyba P, Kieffer N, Nordmann P. Screening and characterization of multidrug-resistant gram-negative bacteria from a remote African Area, São Tomé and Príncipe. Antimicrob Agents Chemother. 2018;62(9):1–7. doi: 10.1128/AAC.01021-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Dromigny JA, Nabeth P, Juergens-Behr A, Perrier-Gros-Claude JD. Risk factors for antibiotic-resistant Escherichia coli isolated from community-acquired urinary tract infections in Dakar, Senegal. J Antimicrob Chemother. 2005; [DOI] [PubMed]
- 571.Lakhe NA, Diallo Mbaye K, Sylla K, Dia Badaine NM, Ndiaye R, Cisse Diallo VMP, et al. Urinary Tract Infection Profile at the Clinic of Infectious and Tropical Diseases at Fann University Hospital, Dakar. Senegal J Antimicrob Agents. 2018;4(3):1–8. [Google Scholar]
- 572.Lo S, Robin F, Ba-Diallo A, Diallo OF, Dia ML, Beyrouthy R, et al. Fortuitous Detection of cmy-2 and dha-1 from ESBL-producing Escherichia coli in Senegal. Bull la Soc Pathol Exot. 2017;110(4):221–223. doi: 10.1007/s13149-017-0573-y. [DOI] [PubMed] [Google Scholar]
- 573.Lo S, Robin F, Beyrouthy R, Ba-Diallo A, Niang AA, Diagne R, et al. OXA-48 type carbapenemase in Klebsiella pneumoniae producing extended spectrum B-lactamases (ESBL) in Senegal. African J Microbiol Res. 2018;12(18):413–418. doi: 10.5897/AJMR2018.8830. [DOI] [Google Scholar]
- 574.Moquet O, Bouchiat C, Kinana A, Seck A, Arouna O, Bercion R, et al. Class D OXA-48 Carbapenemase in Multidrug-Resistant Enterobacteria, Senegal. Emerg Infect Dis. 2011; [DOI] [PMC free article] [PubMed]
- 575.Ndoye B, Konate NN. National prevalence survey of healthcare associated infections: Lessons learned in Senegal in 2017. Antimicrob Resist Infect Control. 2019;8(S1).
- 576.Ruppé E, Woerther PL, Diop A, Sene AM, Da Costa A, Arlet G, et al. Carriage of CTX-M-15-producing Escherichia coli isolates among children living in a remote village in Senegal. Antimicrob Agents Chemother. 2009;53(7):3135–3137. doi: 10.1128/AAC.00139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Barry MS, Diallo BA, Kanté D, Diallo IS. Antimicrobial susceptibility profile of community-acquired urinary tract infection in adults: A seven months prospective cross-sectional study in Dakar Town. Senegal African J Urol. 2017;23(2):166–171. doi: 10.1016/j.afju.2016.11.005. [DOI] [Google Scholar]
- 578.Breurec S, Bouchiat C, Sire JM, Moquet O, Bercion R, Cisse MF, et al. High third-generation cephalosporin resistant Enterobacteriaceae prevalence rate among neonatal infections in Dakar, Senegal. BMC Infect Dis. 2016; [DOI] [PMC free article] [PubMed]
- 579.Cisse CT, Mbengue-Diop R, Moubarek M, Ndiaye O, Dotou CR, Boye CS, et al. Neonatal bacterial infections in UTH in Dakar. Gynécologie Obs Fertil. 2001;29:433–439. doi: 10.1016/S1297-9589(01)00157-6. [DOI] [PubMed] [Google Scholar]
- 580.Dia M, Ndour C, Ka R, Diagne R, Diop A, Sow A, et al. P087: Multiresistant bacteria bacteremia cases in a Dakar University Hospital (Senegal) Antimicrob Resist Infect Control. 2013;2(S1):2013. doi: 10.1186/2047-2994-2-S1-P87. [DOI] [Google Scholar]
- 581.Dromigny J. A, Nabeth P, Perrier JD, Claude PG. Distribution and susceptibility of bacterial urinary tract infections in Dakar, Senegal. Int J Antimicrob Agents [Internet]. 2002;20:339–47. Available from: www.isochem.org [DOI] [PubMed]
- 582.Leski TA, Bangura U, Jimmy DH, Ansumana R, Lizewski SE, Li RW, et al. Identification of blaOXA-51-like, blaOXA-58, bla DIM-1, and blaVIM carbapenemase genes in hospital Enterobacteriaceae isolates from Sierra Leone. J Clin Microbiol. 2013; [DOI] [PMC free article] [PubMed]
- 583.Leski TA, Taitt CR, Bangura U, Stockelman MG, Ansumana R, Cooper WH, et al. High prevalence of multidrug resistant Enterobacteriaceae isolated from outpatient urine samples but not the hospital environment in Bo, Sierra Leone. BMC Infect Dis. 2016; [DOI] [PMC free article] [PubMed]
- 584.Ballot DE, Bandini R, Nana T, Bosman N, Thomas T, Davies VA, et al. A review of multidrug-resistant Enterobacteriaceae in a neonatal unit in Johannesburg, South Africa. BMC Pediatr. 2019;19(320). [DOI] [PMC free article] [PubMed]
- 585.Bamford C, Bonorchis K, Ryan A, Simpson J, Elliott E, Hoffmann R, et al. Antimicrobial Susceptibility Patterns of Selected Bacteraemic Isolates from South African Public Sector Hospitals, 2010. South African J Epidemiol Infect. 2011;
- 586.Bhat VG, Vasaikar SD. Bacteriological profile and antibiogram of aerobic burn wound isolates in Mthatha, Eastern Cape, South Africa. South African J Epidemiol Infect. 2010;
- 587.Bolukaoto JY, Kock MM, Strydom KA, Mbelle NM, Ehlers MM. Molecular characteristics and genotypic diversity of enterohaemorrhagic Escherichia coli O157:H7 isolates in Gauteng region. South Africa Sci Total Environ. 2019;692:297–304. doi: 10.1016/j.scitotenv.2019.07.119. [DOI] [PubMed] [Google Scholar]
- 588.Brink A, Feldman C, Richards G, Moolman J, Senekal M. Emergence of extensive drug resistance (XDR) among Gram-negative bacilli in South Africa looms nearer. South African Med J. 2008; [PubMed]
- 589.Brink A, Moolman GJJ, Cruz da Silva M, Botha M, Badenhorst L, Botha F, et al. Antimicrobial susceptibility profile of selected bacteraemic pathogens from private institutions in South Africa. South African Med J. 2007; [PubMed]
- 590.Brink AJ, Coetzee J, Clay CG, Sithol S, Richards GA, Poirel L, et al. Emergence of New Delhi metallo-beta-lactamase (NDM-1) and Klebsiella pneumoniae carbapenemase (KPC-2) in South Africa. J Clin Microbiol. 2012; [DOI] [PMC free article] [PubMed]
- 591.Brink AJ, Coetzee J, Corcoran C, Clay CG, Hari-Makkan D, Jacobson RK, et al. Emergence of OXA-48 and OXA-181 carbapenemases among Enterobacteriaceae in South Africa and evidence of in vivo selection of colistin resistance as a consequence of selective decontamination of the gastrointestinal tract. J Clin Microbiol. 2013;51(1):369–372. doi: 10.1128/JCM.02234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Buys H, Muloiwa R, Bamford C, Eley B. Klebsiella pneumoniae bloodstream infections at a South African children’s hospital 2006–2011, a cross-sectional study. BMC Infect Dis. 2016;16(1):1–10. doi: 10.1186/s12879-016-1919-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593.Chibabhai V, Perovic O. Epidemiology of carbapenem resistant Enterobacteriaceae at Charlotte Maxeke Johannesburg Academic Hospital. Int J Infect Dis. 2014;21:410. doi: 10.1016/j.ijid.2014.03.1265. [DOI] [Google Scholar]
- 594.Chukwu MO, Abia ALK, Ubomba-Jaswa E, Obi LC, Dewar JB. Antibiotic resistance profile and clonality of E. coli isolated from water and paediatric stool samples in the north-west, province South Africa. J Pure Appl Microbiol. 2019;13(1):517–30.
- 595.Crowther-Gibson P, Govender N, Lewis DA, Bamford C, Brink A, von Gottberg A, et al. Part IV. Human infections and antibiotic resistance. South African Med J. 2011; [PubMed]
- 596.De Jager P, Chirwa T, Naidoo S, Perovic O, Thomas J. Nosocomial outbreak of New Delhi metalloβ-lactamase-1-producing Gram-negative bacteria in South Africa: A case-control study. PLoS One. 2015; [DOI] [PMC free article] [PubMed]
- 597.Defrancesco AS, Tanih NF, Samie A, Guerrant RL, Bessong PO. Antibiotic resistance patterns and beta-lactamase identification in Escherichia coli isolated from young children in rural Limpopo Province, South Africa: The MAL-ED cohort. South African Med J. 2017;107(3):205–214. doi: 10.7196/SAMJ.2017.v107i3.12111. [DOI] [PubMed] [Google Scholar]
- 598.Dramowski A, Whitelaw A, Cotton MF. Burden, spectrum, and impact of healthcare-associated infection at a South African children’s hospital. J Hosp Infect. 2016;94(4):364–372. doi: 10.1016/j.jhin.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Elliott E, Brink AJ, van Greune J, Els Z, Woodford N, Turton J, et al. In vivo development of ertapenem resistance in a patient with pneumonia caused by Klebsiella pneumoniae with an extended-spectrum beta-lactamase. Clin Infect Dis. 2006; [DOI] [PubMed]
- 600.Essack, S Y, Connolly, C. Treatment guidelines and nosocomial infections: The South African experience. African J Microbiol Res. 2011;
- 601.Founou RC, Founou LL, Allam M, Ismail A, Essack SY. Whole Genome Sequencing of Extended Spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae Isolated from Hospitalized Patients in KwaZulu-Natal, South Africa. Sci Rep. 2019;9(6266). [DOI] [PMC free article] [PubMed]
- 602.Founou RC, Founou LL, Arshad Ismail MA, Essack SY. Genomic characterisation of Klebsiella michiganensis co-producing OXA-181 and NDM-1 carbapenemases isolated from a cancer patient in uMgungundlovu District, KwaZulu-Natal Province. South Africa SAMJ. 2019;109(1):7–8. doi: 10.7196/SAMJ.2018.v109i1.13696. [DOI] [PubMed] [Google Scholar]
- 603.Fourie T, Schellack N, Bronkhorst E, Coetzee J, Godman B. Antibiotic prescribing practices in the presence of extended-spectrum β-lactamase (ESBL) positive organisms in an adult intensive care unit in South Africa – A pilot study. Alexandria J Med. 2018;54(4):541–547. doi: 10.1016/j.ajme.2018.09.001. [DOI] [Google Scholar]
- 604.Govind C, Moodley K. The Epidemiology of Carbapenem Resistant Enterobacteriaceae in Kwa-Zulu Natal. South Africa FIDSSA Congr. 2015;2015:143. [Google Scholar]
- 605.Gqunta K, Govender S. Characterization of ESBL-producing Escherichia coli ST131 isolates from Port Elizabeth. Diagn Microbiol Infect Dis. 2015; [DOI] [PubMed]
- 606.Greatorex B, Oosthuizen G. Organisms cultured and resistance patterns seen in a secondary referral centre ICU and burns unit. South African J Crit Care. 2015;
- 607.Han KS, Gustavo L, Rajkumar VC, Swe S-H. Antimicrobial stewardship approach: Prevalence of antimicrobial resistant bacteria at a regional hospital in South Africa. J Infect Dev Ctries. 2019;13(8):748–752. doi: 10.3855/jidc.10685. [DOI] [PubMed] [Google Scholar]
- 608.Hirakata Y, Matsuda J, Miyazaki Y, Kamihira S, Kawakami S, Miyazawa Y, et al. Regional variation in the prevalence of extended-spectrum β-lactamase-producing clinical isolates in the Asia-Pacific region (SENTRY 1998–2002) Diagn Microbiol Infect Dis. 2005;52(4):323–329. doi: 10.1016/j.diagmicrobio.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 609.International Society for Infectious Diseases. Antibiotic-resistant Klebsiella - South Africa. Neonatal Intensive Care Unit [Internet]. ProMED-mail. 2016 [cited 2018 May 22]. Available from: http://www.promedmail.org/
- 610.Ismail H, Lowman W, Govind CN, Swe-Han KS, Maloba MRB, Bamford C, et al. Surveillance and comparison of antimicrobial susceptibility patterns of ESKAPE organisms isolated from patients with bacteraemia in South Africa, 2016–2017. South African Med J. 2019;109(12):934–940. doi: 10.7196/SAMJ.2019.v109i12.14079. [DOI] [PubMed] [Google Scholar]
- 611.Iweriebor BC, Obi CL, Akinyemi O, Ramalivhana NJ, Hattori T, Okoh AI. Uropathogens isolated from HIV-infected patients from Limpopo Province. South Africa African J Biotechnol. 2012;11(46):10598–10604. [Google Scholar]
- 612.Jacobson RK, Manesen MR, Moodley C, Smith M, Williams S, Nicol M, et al. Molecular characterisation and epidemiological investigation of an outbreak of blaOXA-181 carbapenemaseproducing isolates of Klebsiella pneumoniae in South Africa. South African Med J. 2015;105(12):1030–1035. doi: 10.7196/SAMJ.2015.v105i12.9926. [DOI] [PubMed] [Google Scholar]
- 613.Jaspan HB, Huang LC, Cotton MF, Whitelaw A, Myer L. Bacterial disease and antimicrobial susceptibility patterns in HIV-infected, hospitalized children: A retrospective cohort study. PLoS One. 2008; [DOI] [PMC free article] [PubMed]
- 614.Jayol A, Nordmann P, Brink A, Villegas M-V, Dubois V, Poirel L. High-Level Resistance to Colistin Mediated by Various Mutations in the crrB Gene among Carbapenemase-Producing Klebsiella pneumoniae. Antimicrob Agents Chemother. 2017. Available from: 10.1128/AAC [DOI] [PMC free article] [PubMed]
- 615.Kalule JB, Fortuin S, Calder B, Robberts L, Keddy KH, Nel AJM, et al. Proteomic comparison of three clinical diarrhoeagenic drug-resistant Escherichia coli isolates grown on CHROMagarTMSTEC media. J Proteomics. 2018;180:25–35. doi: 10.1016/j.jprot.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 616.Kalule JB, Keddy KH, Nicol MP. Characterisation of STEC and other diarrheic E. coli isolated on CHROMagarTMSTEC at a tertiary referral hospital, Cape Town. BMC Microbiol. 2018;18(1). [DOI] [PMC free article] [PubMed]
- 617.Karama M, Cenci-Goga BT, Malahlela M, Smith AM, Keddy KH, El-Ashram S, et al. Virulence Characteristics and Antimicrobial Resistance Profiles of Shiga Toxin-Producing Escherichia coli Isolates from Humans in South Africa: 2006–2013. Toxins (Basel). 2019;11(7). [DOI] [PMC free article] [PubMed]
- 618.Lebea MM, Davies V. Evaluation of culture-proven neonatal sepsis at a tertiary care hospital in Johannesburg, South Africa. SAJCH South African J Child Heal. 2017;11(4):170–173. [Google Scholar]
- 619.Liebowitz LD, Klugman KP. Comparative in vitro activity of piperacillin/taxobactam against Gram-negative bacilli. South African Med J. 1996;86:1276–1280. [PubMed] [Google Scholar]
- 620.Lochan H, Pillay V, Bamford C, Nuttall J, Eley B. Bloodstream infections at a tertiary level paediatric hospital in South Africa. BMC Infect Dis. 2017;17(1). [DOI] [PMC free article] [PubMed]
- 621.Louw V, van der Westhuizen J, Rautenbach W, van der Berg E, Wamelink M, Joubert G. The antibiotic susceptibility of bacteria isolated from blood cultures during episodes of neutropenic fever in patients with acute myeloid leukaemia. South African J Epidemiol Infect. 2010;25(2):9–11. doi: 10.1080/10158782.2010.11441380. [DOI] [Google Scholar]
- 622.Lowe M, Kock MM, Coetzee J, Hoosien E, Peirano G, Strydom KA, et al. Klebsiella pneumoniae ST307 with blaoxa-181, South Africa, 2014–2016. Emerg Infect Dis. 2019;25(4):739–747. doi: 10.3201/eid2504.181482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Lowman W, Marais M, Ahmed K, Marcus L. Routine active surveillance for carbapenemase-producing enterobacteriaceae from rectal swabs: Diagnostic implications of multiplex polymerase chain reaction. J Hosp Infect. 2014;88(2):66–71. doi: 10.1016/j.jhin.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 624.Lowman W, Schleicher G. Antimicrobial treatment and outcomes of critically ill patients with OXA-48like carbapenemase-producing Enterobacteriaceae infections. Diagn Microbiol Infect Dis. 2015;81(2):138–140. doi: 10.1016/j.diagmicrobio.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 625.Magazi B, Holl R. P075 Colistin ‘MIC’ creep, a harbinger for resistance? Study for monitoring antimicrobial resistance trends (SMART) - South Africa. Crit Care. 2018;22(S1):30. [Google Scholar]
- 626.Magazi BT, Khan S, Dlamini S, Pasipanodya J. Molecular characterization and antimicrobial susceptibility of extended-spectrum b-lactamases (ESBL) producing enterobacteriaceae (ESBL-PE) causing urinary tract infections (UTI): Results from the Study for Monitoring Antimicrobial Resistance Trends (SMAR. OFID. 2017;4(Suppl 1):S590–S591. doi: 10.1093/ofid/ofx163.1549. [DOI] [Google Scholar]
- 627.Malande OO, du Plessis A, Rip D, Bamford C, Eley B. Invasive carbapenem-resistant Enterobacteriaceae infection at a paediatric hospital: A case series. South African Med J. 2016;106(9):877–882. doi: 10.7196/SAMJ.2016.v106i9.11028. [DOI] [PubMed] [Google Scholar]
- 628.Malande OO, Nuttall J, Pillay V, Bamford C, Eley B. A ten-year review of ESBL and non-ESBL Escherichia coli bloodstream infections among children at a tertiary referral hospital in South Africa. PLoS One. 2019;14(9). [DOI] [PMC free article] [PubMed]
- 629.Maweya S. Microbiological profile of organisms causing bloodstream infections between 2004 and 2016 in a tertiary hospital, Limpopo province, South Africa. [Pretoria, South Africa]: University of Pretoria; 2017.
- 630.Mbelle N, Mogoloane M, Mthombeni R, Le Roux M, De Villiers B, Fernandes L. The distribution of blaSHV, blaTEM and blaCTX-M genes in Klebsiella pneumoniae and E. coli clinical isolates at a tertiary hospital in South Africa. Int J Infect Dis. 2012;
- 631.Mbelle NM, Feldman C, Osei Sekyere J, Maningi NE, Modipane L, Essack SY. The Resistome, Mobilome, Virulome and Phylogenomics of Multidrug-Resistant Escherichia coli Clinical Isolates from Pretoria, South Africa. Sci Rep. 2019 Dec 1;9(1). [DOI] [PMC free article] [PubMed]
- 632.Mocktar C, Govinden U, Sturm AW, Essack S. Complexity and diversity of betβ-lactamase expression in inhibitor-resistant Escherichia coli from public hospitals in KwaZulu-Natal, South Africa. South African J Epidemiol Infect. 2009;24(4):29–33. doi: 10.1080/10158782.2009.11441359. [DOI] [Google Scholar]
- 633.Mohlabeng RM, Singh-Moodley A, Iyaloo S, Perovic O. Molecular characterization of carbapenems-producing Klebsiella pneumoniae at the National Antimicrobial Resistance Reference Laboratory. South Africa FIDSSA Congr. 2015;2015:133. [Google Scholar]
- 634.Morkel G, Bekker A, Marais BJ, Kirsten G, van Wyk J, Dramowski A. Bloodstream infections and antimicrobial resistance patterns in a South African neonatal intensive care unit. Paediatr Int Child Health. 2014; [DOI] [PubMed]
- 635.Mthembu W, Penduka D, Mosa R, Zobolo A, Opoku A. Antibiotic susceptibility patterns of bacteria recovered from wounds of diabetic patients in some Northern Kwazulu-Natal Hospitals. South Africa J Biol Sci. 2017;18(1):13–20. [Google Scholar]
- 636.National Institute for Communicable Diseases. Communicable Diseases Surveillance Bulletin [Internet]. Vol. 10. 2012 [cited 2018 May 22]. Available from: http://www.nicd.ac.za/index.php/publications/nicd-nhls-communicable-diseases-communique/archives/
- 637.National Institute for Communicable Diseases. Communicable Diseases Surveillance Bulletin [Internet]. Vol. 11. 2013 [cited 2018 May 22]. Available from: http://www.nicd.ac.za/index.php/publications/nicd-nhls-communicable-diseases-communique/archives/
- 638.National Institute for Communicable Diseases. Communicable Diseases Surveillance Bulletin [Internet]. Vol. 12. 2014 [cited 2018 May 22]. Available from: http://www.nicd.ac.za/index.php/publications/nicd-nhls-communicable-diseases-communique/archives/
- 639.National Institute for Communicable Diseases. Monthly Surveillance Report [Internet]. 2015 [cited 2018 May 22]. Available from: http://www.nicd.ac.za/index.php/publications/nicd-nhls-communicable-diseases-communique/archives/
- 640.National Institute for Communicable Diseases. Monthly Surveillance Report [Internet]. 2016 [cited 2018 May 22]. Available from: http://www.nicd.ac.za/index.php/publications/nicd-nhls-communicable-diseases-communique/archives/
- 641.Nel P, Roberts LA, Hoffmann R. Carbapenemase-producing Enterobacteriaceae colonisation in adult inpatients: A point prevalence study. South African J Infect Dis. 2019;34(1):1–5. doi: 10.4102/sajid.v34i1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642.Newton-Foot M, Snyman Y, Maloba MRB, Whitelaw AC. Plasmid-mediated mcr-1 colistin resistance in Escherichia coli and Klebsiella spp. clinical isolates from the Western Cape region of South Africa. Antimicrob Resist Infect Control. 2017;6(1). [DOI] [PMC free article] [PubMed]
- 643.Nyasulu PS, Murray J, Perovic O, Koornhof H. Laboratory information system for reporting antimicrobial resistant isolates from academic hospitals. South Africa J Infect Dev Ctries. 2017;11(9):705–718. doi: 10.3855/jidc.7159. [DOI] [PubMed] [Google Scholar]
- 644.Palacios-Baena ZR, Gutiérrez-Gutiérrez B, Calbo E, Almirante B, Viale P, Oliver A, et al. Empiric Therapy with Carbapenem-Sparing Regimens for Bloodstream Infections due to Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: Results from the INCREMENT Cohort. Clin Infect Dis. 2017;65(10):1615–1623. doi: 10.1093/cid/cix606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 645.Peirano G, Moolman J, Pitondo-Silva A, Pitout JDD. The characteristics of VIM-1-producing Klebsiella pneumoniae from South Africa. Scand J Infect Dis. 2012;44(1):74–78. doi: 10.3109/00365548.2011.614276. [DOI] [PubMed] [Google Scholar]
- 646.Peirano G, Van Der Bij AK, Freeman JL, Poirel L, Nordmann P, Costello M, et al. Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum β-lactamases: Global distribution of the H30-Rx sublineage. Antimicrob Agents Chemother. 2014;58(7):3762–3767. doi: 10.1128/AAC.02428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 647.Pepper DJ, Rebe K, Morroni C, Wilkinson RJ, Meintjes G. Clinical deterioration during antitubercular treatment at a district hospital in South Africa: The importance of drug resistance and AIDS defining illnesses. PLoS One. 2009;4(2). [DOI] [PMC free article] [PubMed]
- 648.Perovic O, Britz E, Chetty V, Singh-Moodley A. Molecular detection of carbapenemase-producing genes in referral enterobacteriaceae in South Africa: A short report. South African Med J. 2016; [DOI] [PubMed]
- 649.Pillay T, Pillay DG, Adhikari M, Sturm AW. Piperacillin/Tazobactam in the treatment of Klebsiella pneumoniae infections in neonates. Am J Perinatol. 1998;15(1):47–51. doi: 10.1055/s-2007-993898. [DOI] [PubMed] [Google Scholar]
- 650.Pitout JDD, Thomson KS, Hanson ND, Ehrhardt AF, Moland ES, Sanders CC. β-lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabills isolates recovered in South Africa. Antimicrob Agents Chemother. 1998;42(6):1350–1354. doi: 10.1128/AAC.42.6.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 651.Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob Agents Chemother. 2016;60(7):4394–4397. doi: 10.1128/AAC.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 652.Ramsamy Y, Milsana KP, Allam M, Amoako D, Ismail A, Akebe A, et al. P54 Molecular characterization of carbapenem resistant Klebsiella pneumoniae isolates collected from a central public hospital in Durban, South Africa between 2016 and 2017. Antimicrob Resist Infect Control. 2019;8(S1):41–42. [Google Scholar]
- 653.Ramsamy Y, Muckart DJJ, Han KSS. Microbiological surveillance and antimicrobial stewardship minimise the need for ultrabroad-spectrum combination therapy for treatment of nosocomial infections in a trauma intensive care unit: An audit of an evidence-based empiric antimicrobial policy. South African Med J. 2013;103(6):371–376. doi: 10.7196/SAMJ.6459. [DOI] [PubMed] [Google Scholar]
- 654.Alliance for Prudent Use of Antibiotics. NASF private sector antibiotic susceptibility data: Jan–June 2005 [Internet]. [cited 2018 May 22]. Available from: http://emerald.tufts.edu/med/apua/intl_chapters/private_sector.pdf
- 655.Samie A, Nkgau TF, Bessong PO, Obi CL, Dillingham R, Guerrant RL. Escherichia coli pathotypes among human immunodeficiency virus infected patients in the Limpopo Province. African J Microbiol Res. 2012;6(32):6022–6030. [Google Scholar]
- 656.Segal H, Gay EB. Resistance to β-lactams, and reduced susceptibility to carbapenems, in clinical isolates of Klebsiella pneumoniae due to interplay between CTX-M-15 and altered outer membrane permeability. South African J Epidemiol Infect. 2006;21(2):41–44. doi: 10.1080/10158782.2006.11441262. [DOI] [Google Scholar]
- 657.Sekyere JO, Amoako DG. Carbonyl cyanide m-chlorophenylhydrazine (CCCP) reverses resistance to colistin, but not to Carbapenems and tigecycline in multidrug-resistant Enterobacteriaceae. Front Microbiol. 2017;8. [DOI] [PMC free article] [PubMed]
- 658.Shipton SE, Cotton MF, Wessels G. Nosocomial endocarditis due to Extended-Spectrum Beta Lactamase producing Klebisiella pneumoniae in a child. South African Med J. 2001;91(4):321–322. [PubMed] [Google Scholar]
- 659.Singh-Moodley A, Perovic O. Phenotypic and genotypic correlation of carbapenememase-producing Enterobacteriaceae and problems experienced in routine screening. South African Med J. 2018;108(6):495–501. doi: 10.7196/SAMJ.2018.v108i6.12878. [DOI] [PubMed] [Google Scholar]
- 660.Smith AM, Tau NP, Sooka A, Keddy KH. Surveillance for enterohaemorrhagic Escherichia coli associated with human diarrhoea in South Africa, 2006–2009. J Med Microbiol. 2011;60(5):681–683. doi: 10.1099/jmm.0.022947-0. [DOI] [PubMed] [Google Scholar]
- 661.Tau NP, Meidany P, Smith AM, Sooka A, Keddy KH. Escherichia coli o104 associated with human diarrhea, South Africa, 2004–2011. Emerg Infect Dis. 2012;18(8):1314–1317. doi: 10.3201/eid1808.111616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 662.Vasaikar S, Obi L, Morobe I, Bisi-Johnson M. Molecular characteristics and antibiotic resistance profiles of Klebsiella isolates in Mthatha, Eastern Cape province, South Africa. Int J Microbiol. 2017; [DOI] [PMC free article] [PubMed]
- 663.Archary M, Adler H, La Russa P, Mahabeer P, Bobat RA. Bacterial infections in HIV-infected children admitted with severe acute malnutrition in Durban. South Africa Paediatr Int Child Health. 2017;37(1):6–13. doi: 10.1080/20469047.2016.1198561. [DOI] [PubMed] [Google Scholar]
- 664.Biswas JS, Lentaigne J, Hill NE, Harrison JJ, Mackenzie H, Akorli E, et al. Epidemiology and etiology of diarrhea in UK military personnel serving on the United Nations Mission in South Sudan in 2017: A prospective cohort study. Travel Med Infect Dis. 2019;28:34–40. doi: 10.1016/j.tmaid.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 665.Nurain AM, Bilal NE, Ibrahim ME. The frequency and antimicrobial resistance patterns of nosocomial pathogens recovered from cancer patients and hospital environments. Asian Pac J Trop Biomed. 2015;5(12):1055–1059. doi: 10.1016/j.apjtb.2015.09.015. [DOI] [Google Scholar]
- 666.Saad ESA, Fahal AH. Broncho-pleuro-cutaneous fistula and pneumothorax: Rare challenging complications of chest wall eumycetoma. PLoS Negl Trop Dis. 2017;11(9). [DOI] [PMC free article] [PubMed]
- 667.Shingeray OH, Abd Elrhaman MA, Musa AM, Mohamid N. Prevalence and Antimicrobial Susceptibility Pattern of Bacteria Causing Postoperative Wound Infections in Port-Sudan. J Biomed Pharm Res [Internet]. 2013;2(6):82–5. Available from: www.jbpr.in
- 668.Adam MA, Elhag WI. Prevalence of metallo-β-lactamase acquired genes among carbapenems susceptible and resistant Gram-negative clinical isolates using multiplex PCR, Khartoum hospitals, Khartoum Sudan. BMC Infect Dis. 2018;18(1). [DOI] [PMC free article] [PubMed]
- 669.Elbadawi HS, Elhag KM, Mahgoub E, Altayb HN, Abdel Hamid MM. Antimicrobial resistance surveillance among gram negative bacterial isolates from patients in khartoum state hospitals. F1000Research. 2019;8(156).
- 670.Gafar F, Ekarim A, Hassan AN. Proportion of bacteria causing healthcare associated infection in Khartoum North Teaching Hospital. Sudan J Med Sci. 2009;4(4):351–355. [Google Scholar]
- 671.Ibrahim AH, Abdelhalim KA. Detection of extended specgtrum beta-lactamase in Klebsiella pneumoniae isolated form sputum in Khartoum, Sudan. World J Pharm Res [Internet]. 2015;4(3):131–40. Available from: www.wjpr.net
- 672.Malik IA, Elhag KM. Characterisation of extended-spectrum b-lactamases among multidrug resistant Enterobacteriaceae from Sudan. J Pure Appl Microbiol. 2019;13(1):61–68. doi: 10.22207/JPAM.13.1.06. [DOI] [Google Scholar]
- 673.Mohammed I, Abass E. Phenotypic detection of Extended Spectrum β-Lactamases (ESBL) among gram negative uropathogens reveals highly susceptibility to imipenem. Pakistan J Med Sci. 2019;35(4):1104–1109. doi: 10.12669/pjms.35.4.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 674.Fleece ME, Nshama R, Walongo T, Kimathi C, Gratz J, Rogawski McQuade ET, et al. Longitudinal assessment of antibiotic resistance in fecal Escherichia coli in Tanzanian Children. Am J Trop Med Hyg. 2019;100(5):1110–1114. doi: 10.4269/ajtmh.18-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 675.Kaduma J, Seni J, Chuma C, Kirita R, Mujuni F, Mushi MF, et al. Urinary tract infections and preeclampsia among pregnant women attending two hospitals in Mwanza City, Tanzania: A 1:2 Matched case-control study. Biomed Res Int. 2019;2019. [DOI] [PMC free article] [PubMed]
- 676.Kiponza R, Balandya B, Majigo M V., Matee M. Laboratory confirmed puerperal sepsis in a national referral hospital in Tanzania: Etiological agents and their susceptibility to commonly prescribed antibiotics. BMC Infect Dis. 2019;19(1). [DOI] [PMC free article] [PubMed]
- 677.Manyahi J, Matee MI, Majigo M, Moyo S, Mshana SE, Lyamuya EF. Predominance of multi-drug resistant bacterial pathogens causing surgical site infections in Muhimbili national hospital, Tanzania. BMC Res Notes. 2014;7(1). [DOI] [PMC free article] [PubMed]
- 678.Manyahi J, Moyo SJ, Tellevik MG, Ndugulile F, Urassa W, Blomberg B, et al. Detection of CTX-M-15 beta-lactamases in Enterobacteriaceae causing hospital- and community-acquired urinary tract infections as early as 2004, in Dar es Salaam, Tanzania. BMC Infect Dis. 2017; [DOI] [PMC free article] [PubMed]
- 679.Masinde A, Gumodoka B, Kilonzo A, Mshana SE. Prevalence of urinary tract infection among pregnant women at Bugando Medical Centre, Mwanza. Tanzania Tanzan J Health Res. 2009;11(3):154–159. doi: 10.4314/thrb.v11i3.47704. [DOI] [PubMed] [Google Scholar]
- 680.Moremi N, Claus H, Mshana SE. Antimicrobial resistance pattern: A report of microbiological cultures at a tertiary hospital in Tanzania. BMC Infectious Diseases. 2016. [DOI] [PMC free article] [PubMed]
- 681.Moremi N, Claus H, Vogel U, Mshana SE. Faecal carriage of CTX-M extended-spectrum beta-lactamase-producing Enterobacteriaceae among street children dwelling in Mwanza city, Tanzania. PLoS One. 2017;12(9). [DOI] [PMC free article] [PubMed]
- 682.Moremi N, Mushi MF, Fidelis M, Chalya P, Mirambo M, Mshana SE. Predominance of multi-resistant gram-negative bacteria colonizing chronic lower limb ulcers (CLLUs) at Bugando Medical Center. BMC Res Notes. 2014; [DOI] [PMC free article] [PubMed]
- 683.Moyo SJ, Aboud S, Kasubi M, Lyamuya EF, Maselle SY. Antimicrobial resistance among producers and non-producers of extended spectrum beta-lactamases in urinary isolates at a tertiary Hospital in Tanzania. BMC Res Notes. 2010;3(348):1–5. doi: 10.1186/1756-0500-3-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 684.Msaki BP, Mshana SE, Hokororo A, Mazigo HD, Morona D. Prevalence and predictors of urinary tract infection and severe malaria among febrile children attending Makongoro health centre in Mwanza city, North-Western Tanzania. Arch Public Heal. 2012;70(1). [DOI] [PMC free article] [PubMed]
- 685.Mshana SE, Hain T, Domann E, Lyamuya EF, Chakraborty T, Imirzalioglu C. Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect Dis. 2013; [DOI] [PMC free article] [PubMed]
- 686.Mushi MF, Alex VG, Seugendo M, Silago V, Mshana SE. C-reactive protein and urinary tract infection due to gram-negative bacteria in a pediatric population at a tertiary hospital, Mwanza. Tanzania Afr Health Sci. 2019;19(4):3217–3224. doi: 10.4314/ahs.v19i4.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 687.Mushi MF, Mshana SE, Imirzalioglu C, Bwanga F. Carbapenemase genes among multidrug resistant gram negative clinical isolates from a tertiary hospital in Mwanza, Tanzania. Biomed Res Int. 2014; [DOI] [PMC free article] [PubMed]
- 688.Mushi MF, Mwalutende AE, Gilyoma JM, Chalya PL, Seni J, Mirambo MM, et al. Predictors of disease complications and treatment outcome among patients with chronic suppurative otitis media attending a tertiary hospital, Mwanza Tanzania. BMC Ear, Nose Throat Disord. 2016; [DOI] [PMC free article] [PubMed]
- 689.Nelson E, Kayega J, Seni J, Mushi MF, Kidenya BR, Hokororo A, et al. Evaluation of existence and transmission of extended spectrum beta lactamase producing bacteria from post-delivery women to neonates at Bugando Medical Center, Mwanza-Tanzania. BMC Res Notes. 2014; [DOI] [PMC free article] [PubMed]
- 690.Onken A, Said AK, Jørstad M, Jenum PA, Blomberg B. Prevalence and antimicrobial resistance of microbes causing bloodstream infections in Unguja, Zanzibar. PLoS One. 2015 Dec 1;10(12). [DOI] [PMC free article] [PubMed]
- 691.Seni J, Mwakyoma AA, Mashuda F, Marando R, Ahmed M, Devinney R, et al. Deciphering risk factors for blood stream infections, bacteria species and antimicrobial resistance profiles among children under five years of age in North-Western Tanzania: A multicentre study in a cascade of referral health care system. BMC Pediatr. 2019;19(1). [DOI] [PMC free article] [PubMed]
- 692.Seni J, Sweya E, Mabewa A, Mshana SE, Gilyoma JM. Comparison of antimicrobial resistance patterns of ESBL and non ESBL bacterial isolates among patients with secondary peritonitis at Bugando Medical Centre, Mwanza - Tanzania. BMC Emerg Med. 2016;16(1). [DOI] [PMC free article] [PubMed]
- 693.Seni J, Tito JN, Makoye SJ, Mbena H, Alfred HS, van der Meer F, et al. Multicentre evaluation of significant bacteriuria among pregnant women in the cascade of referral healthcare system in North-western Tanzania: Bacterial pathogens, antimicrobial resistance profiles and predictors. J Glob Antimicrob Resist. 2019;17:173–179. doi: 10.1016/j.jgar.2018.12.024. [DOI] [PubMed] [Google Scholar]
- 694.Tellevik MG, Blomberg B, Kommedal Ø, Maselle SY, Langeland N, Moyo SJ. High prevalence of faecal carriage of esbl-producing enterobacteriaceae among children in Dar es Salaam, Tanzania. PLoS One. 2016; [DOI] [PMC free article] [PubMed]
- 695.Ahmed M, Moremi N, Mirambo MM, Hokororo A, Mushi MF, Seni J, et al. Multi-resistant gram negative enteric bacteria causing urinary tract infection among malnourished underfives admitted at a tertiary hospital, northwestern, Tanzania. Ital J Pediatr. 2015; [DOI] [PMC free article] [PubMed]
- 696.Blomberg B, Manji KP, Urassa WK, Tamim BS, Mwakagile DSM, Jureen R, et al. Antimicrobial resistance predicts death in Tanzanian children with bloodstream infections: A prospective cohort study. BMC Infect Dis. 2007; [DOI] [PMC free article] [PubMed]
- 697.Büdel T, Kuenzli E, Clément M, Bernasconi OJ, Fehr J, Mohammed AH, et al. Polyclonal gut colonization with extended-spectrum cephalosporin- and/or colistin-resistant Enterobacteriaceae: a normal status for hotel employees on the island of Zanzibar. Tanzania J Antimicrob Chemother. 2019;74(10):2880–2890. doi: 10.1093/jac/dkz296. [DOI] [PubMed] [Google Scholar]
- 698.Chalya PL, Igenge JZ, Mabula JB, Simbila S. Fournier’s gangrene at a tertiary health facility in northwestern Tanzania: A single centre experiences with 84 patients. BMC Res Notes. 2015;8(1). [DOI] [PMC free article] [PubMed]
- 699.Chaula T, Seni J, Ng’walida N, Kajura A, Mirambo MM, Devinney R, et al. Urinary tract infections among HIV-positive pregnant women in Mwanza city, Tanzania, are high and predicted by low CD4+ Count. Int J Microbiol. 2017; [DOI] [PMC free article] [PubMed]
- 700.Christopher A, Mshana SE, Kidenya BR, Hokororo A, Morona D. Bacteremia and resistant gram-negative pathogens among under-fives in Tanzania. Ital J Pediatr [Internet]. 2013;39(27):1–8. Available from: http://www.ijponline.net/content/39/1/27 [DOI] [PMC free article] [PubMed]
- 701.Dossim S, Bonnin RA, Salou M, Tanga K, Godonou V, Dagnra AY, et al. Occurrence of carbapenemase-producing Enterobacteriaceae in Togo. West Africa Int J Antimicrob Agents. 2019;53(4):530–532. doi: 10.1016/j.ijantimicag.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 702.Douti NK, Fiawoo M, Salou M, Senagbe KM, Hemou M, Sanni EE, et al. Klebsiella pneumoniae Nosocomial Infection in an African Pediatrics Health Center: Case of Campus-Teaching Hospital in Togo. Int J Clin Pediatr. 2018;7(4):51–54. doi: 10.14740/ijcp313w. [DOI] [Google Scholar]
- 703.Gambogou B, Anani K, Karou SD, Ameyapoh YA, Simpore J. Effect of Aqueous garlic extract on biofilm formation and antibiotic susceptibility of multidrug-resistant uropathogenic Escherichia coli clinical isolates in Togo. Int J Adv Multidiscip Res [Internet]. 2018;5(7):23–33. Available from: http://dx.doi.org/10.22192/ijamr.2018.05.06.006
- 704.Pessinaba NC, Landoh DE, Dossim S, Bidjada B, Kere-Banla A, Tamekloe TA, et al. Screening for extended-spectrum beta-lactamase-producing Enterobacteriaceae intestinal carriage among children aged under five in Lome. Togo Med Mal Infect. 2018;48:551–554. doi: 10.1016/j.medmal.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 705.Salah FD, Soubeiga ST, Ouattara AK, Sadji AY, Metuor-Dabire A, Obiri-Yeboah D, et al. Distribution of quinolone resistance gene (qnr) in ESBL-producing Escherichia coli and Klebsiella spp. in Lomé, Togo. Antimicrob Resist Infect Control. 2019;8(1). [DOI] [PMC free article] [PubMed]
- 706.Toudji AG, Djeri B, Karou SD, Tigossou S, Ameyapoh Y, Souza C De. Prevalence of Extended Spectrum Beta Lactamases Producing Enterobacteriaceae and the Antibiotic Susceptibility in Lome, Togo. Asian J Life Sci. 2017;
- 707.Alibi S, Ferjani A, Boukadida J. Molecular characterization of extended spectrum beta-lactamases produced by Klebsiella pneumoniae clinical strains from a Tunisian hospital. Med Mal Infect. 2015;45(4):139–143. doi: 10.1016/j.medmal.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 708.Amel M, Ameni K, Lazheri G, Nada BS, Imen R, Lamia T, et al. Role of carbapenemase detection in optimization antimicrobial therapy in burns. Ann Intensive Care. 2018;8(S1):101. [Google Scholar]
- 709.Ayari K, Bourouis A, Chihi H, Mahrouki S, Naas T, Belhadj O. Dissemination and genetic support of broad-spectrum beta-lactam-resistant Escherichia coli strain isolated from two Tunisian hospitals during 2004–2012. Afr Health Sci. 2017;17(2):346–355. doi: 10.4314/ahs.v17i2.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 710.Battikh H, Harchay C, Dekhili A, Khazar K, Kechrid F, Zribi M, et al. Clonal Spread of Colistin-Resistant Klebsiella pneumoniae Coproducing KPC and VIM Carbapenemases in Neonates at a Tunisian University Hospital. Microb Drug Resist. 2017;23(4):468–472. doi: 10.1089/mdr.2016.0175. [DOI] [PubMed] [Google Scholar]
- 711.Bauernfeind A, Hohl P, Schneider I, Jungwirth R, Frei R. Escherichia coli producing a cephamycinase (CMY-2) from a patient from the Libyan-Tunisian border region. Clin Microbiol Infect. 1998;4(3):168–170. doi: 10.1111/j.1469-0691.1998.tb00384.x. [DOI] [PubMed] [Google Scholar]
- 712.Ben Achour N, Mercuri PS, Belhadj C, Moussa M Ben, Galleni M, Belhadj O. Cefotaxime and ceftriaxon resistant Klebsiella pneumoniae associated with SHV-11 hyperproduction. Ann Microbiol. 2008;58(4):727–30.
- 713.Ben Jemaa Z, Mahjoubi F, Ben Haj H’mida Y, Hammami N, Ben Ayed M, Hammami A. Antimicrobial susceptibility and frequency of occurrence of clinical blood isolates in Sfax-Tunisia (1993–1998). Pathol Biol. 2004;52(2):82–8. [DOI] [PubMed]
- 714.Ben Sallem R, Ben Slama K, Estepa V, Jouini A, Gharsa H, Klibi N, et al. Prevalence and characterisation of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolates in healthy volunteers in Tunisia. Eur J Clin Microbiol Infect Dis. 2012;31(7):1511–1516. doi: 10.1007/s10096-011-1471-z. [DOI] [PubMed] [Google Scholar]
- 715.Ben Tanfous F, Achour W, Raddaoui A, Ben HA. Molecular characterisation and epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates from immunocompromised patients in Tunisia. J Glob Antimicrob Resist. 2018;13:154–160. doi: 10.1016/j.jgar.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 716.Ben Tanfous F, Alonso CA, Achour W, Ruiz-Ripa L, Torres C, Ben HA. First description of KPC-2-producing Escherichia coli and ST15 OXA-48-positive Klebsiella pneumoniae in Tunisia. Microb Drug Resist. 2017;23(3):365–375. doi: 10.1089/mdr.2016.0090. [DOI] [PubMed] [Google Scholar]
- 717.Ben-Hamouda T, Foulon T, Ben-Cheikh-Masmoudi A, Fendri C, Belhadj O, Ben-Mahrez K. Molecular epidemiology of an outbreak of multiresistant Klebsiella pneumoniae in a Tunisian neonatal ward. J Med Microbiol. 2003;52(5):427–433. doi: 10.1099/jmm.0.04981-0. [DOI] [PubMed] [Google Scholar]
- 718.Bouallègue-Godet O, Grimont F, Salem Y Ben, Saidani M, Mzoughi R, Sboui H, et al. Investigation of the clonal dissemination of Klebsiella pneumoniae isolates producing extended-spectrum beta-lactamases in a neonatal ward, Sousse, Tunisia. Pathol Biol. 2005;53(2):75–80. [DOI] [PubMed]
- 719.Boutiba Ben Boubaker I, Ghozzi R, Abdallah H Ben, Mamlouk K, Kamoun A, Redjeb S Ben. Evolution of acquired resistance to third-generation cephalosporins in Enterobacteriaceae in a Tunisian hospital 1993–2001. Clin Microbiol Infect. 2004;10(7):665–7. [DOI] [PubMed]
- 720.Boukadida J, Boukadida N, Elraii S. Profile and sensitivity to antibiotics of 2063 uropathogenic bacteria in midTunisia. Bull Soc Exot Pathol. 2002;95(1):8–10. [PubMed] [Google Scholar]
- 721.Boukadida J, Monastiri K, Lamouri N, Bouallegue O, Snoussi N, Essoussi AS, et al. Epidemiological aspects of the resistanc eof enterobacteriaceae to cephalosporins of third generation in Central Tunisia. Med Infect Dis. 1996;26:1155–1158. [Google Scholar]
- 722.Boukadida J, Salem N, Hannachi N, Monastiri K, Snoussi N. Genotypic exploration of a hospital neonatal outbreak due to Klebsiella pneumoniae producing extended-spectrum-betalactamase. Arch Pediatr. 2002;9:463–468. doi: 10.1016/S0929-693X(01)00827-2. [DOI] [PubMed] [Google Scholar]
- 723.Boutiba-Ben Boubaker I, Ben Salah D, Besbes M, Mahjoubi F, Ghozzi F, Ben Redjeb S, et al. Multidrug Resistant Klebsiella pneumoniae: A Multicentrique Study. Tunis Med. 2002;80(1):26–28. [PubMed] [Google Scholar]
- 724.Charfi K, Grami R, Ben Jeddou A, Messaoudi A, Mani Y, Bouallegue O, et al. Extended-spectrum β-lactamases and plasmid-mediated quinolone resistance in enterobacterial clinical isolates from neonates in Tunisia. Microb Pathog. 2017;110:184–188. doi: 10.1016/j.micpath.2017.06.030. [DOI] [PubMed] [Google Scholar]
- 725.Charfi K, Mansour W, Ben Haj Khalifa A, Mastouri M, Aouni M, Mammeri H. Emergence of OXA-204 β-lactamase in Tunisia. Diagn Microbiol Infect Dis. 2015;82(4):314–7. [DOI] [PubMed]
- 726.Chérif T, Saidani M, Decré D, Boutiba-Ben Boubaker I, Arlet G. Cooccurrence of multiple AmpC β-lactamases in Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis in Tunisia. Antimicrob Agents Chemother. 2016;60(1):44–51. doi: 10.1128/AAC.00828-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 727.Chouchani C, Ben-Achour N, M’charek A, Belhadj O. Cefotaxime and ceftazidime-resistant Escherichia coli isolate producing TEM-15 β-lactamase from a Tunisian hospital. Comptes Rendus - Biol. 2007;330(8):565–570. doi: 10.1016/j.crvi.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 728.Chouchani C, El Salabi A, Marrakchi R, Ferchichi L, Walsh TR. Characterization of IncA/C conjugative plasmid harboring bla TEM-52 and bla CTX-M-15 extended-spectrum β-lactamases in clinical isolates of Escherichia coli in Tunisia. Eur J Clin Microbiol Infect Dis. 2012;31(6):1081–1087. doi: 10.1007/s10096-011-1410-z. [DOI] [PubMed] [Google Scholar]
- 729.Cuzon G, Naas T, Lesenne A, Benhamou M, Nordmann P. Plasmid-mediated carbapenem-hydrolysing OXA-48 beta-lactamase in Klebsiella pneumoniae from Tunisia. Int J Antimicrob Agents. 2010;36(1):91–93. doi: 10.1016/j.ijantimicag.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 730.Dahmen S, Bettaieb D, Mansour W, Boujaafar N, Bouallè Gue O, Arlet G. Characterization and Molecular Epidemiology of Extended-Spectrum b-Lactamases in Clinical Isolates of Enterobacteriaceae in a Tunisian University Hospital. Microb Drug Resist [Internet]. 2010;16(2):163–70. Available from: www.liebertpub.com [DOI] [PubMed]
- 731.Dahmen S, Mansour W, Charfi K, Boujaafar N, Arlet G, Bouallègue O. Imipenem resistance in Klebsiella pneumoniae is associated to the combination of plasmid-mediated CMY-4 AmpC β-Lactamase and loss of an outer membrane protein. Microb Drug Resist. 2012;18(5):479–483. doi: 10.1089/mdr.2011.0214. [DOI] [PubMed] [Google Scholar]
- 732.Dziri O, Alonso CA, Dziri R, Gharsa H, Maraoub A, Torres C, et al. Metallo-β-lactamases and class D carbapenemases in south-east Tunisia: Implication of mobile genetic elements in their dissemination. Int J Antimicrob Agents. 2018;52(6):871–877. doi: 10.1016/j.ijantimicag.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 733.Dziri O, Dziri R, Maraoub A, Chouchani C. First report of SHV-148-Type ESBL and CMY-42-type AmpC β-lactamase in Klebsiella pneumoniae clinical isolates in Tunisia. Microb Drug Resist. 2018;24(10):1483–1488. doi: 10.1089/mdr.2018.0073. [DOI] [PubMed] [Google Scholar]
- 734.Elhani D, Bakir L, Aouni M, Passet V, Arlet G, Brisse S, et al. Molecular epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae strains in a university hospital in Tunis, Tunisia, 1999–2005. Clin Microbiol Infect. 2010;16(2):157–164. doi: 10.1111/j.1469-0691.2009.03057.x. [DOI] [PubMed] [Google Scholar]
- 735.Ferjani S, Saidani M, Amine FS, Boutiba Ben Boubaker I. A comparative study of antimicrobial resistance rates and phylogenetic groups of community-acquired versus hospital-acquired invasive Escherichia coli. Med Mal Infect. 2015;45(4):133–8. [DOI] [PubMed]
- 736.Ferjani S, Saidani M, Ennigrou S, Hsairi M, Slim AF aouz., Ben Boubaker IB outib. Multidrug resistance and high virulence genotype in uropathogenic Escherichia coli due to diffusion of ST131 clonal group producing CTX-M-15: an emerging problem in a Tunisian hospital. Folia Microbiol (Praha). 2014;59(3):257–62. [DOI] [PubMed]
- 737.Ferjani S, Saidani M, Maamar E, Harbaoui S, Hamzaoui Z, Hosni H, et al. Escherichia coli colonizing healthy children in Tunisia: High prevalence of extra-intestinal pathovar and occurrence of non-extended-spectrum-β-lactamase-producing ST131 clone. Int J Antimicrob Agents. 2018;52(6):878–885. doi: 10.1016/j.ijantimicag.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 738.Ghali H, Ben Cheikh A, Hannachi H, Khefacha-Aissa S, Ben Rejeb M, Said-Latiri H. Screening for MRSA, VRE and carbapenemase producing enterobacteriaceae in Tunisian intensive care units. Antimicrob Resist Infect Control. 2019;8:2–3. [Google Scholar]
- 739.Girlich D, Karim A, Spicq C, Nordmann P. Plasmid-mediated cephalosporinase ACC-1 in clinical isolates of Proteus mirabilis and Escherichia coli. Eur J Clin Microbiol Infect Dis. 2000;19(11):893–895. doi: 10.1007/s100960000386. [DOI] [PubMed] [Google Scholar]
- 740.Grami R, Mansour W, Khalifa ABH, Dahmen S, Chatre P, Haenni M, et al. Emergence of ST147 Klebsiella pneumoniae producing OXA-204 carbapenemase in a University Hospital, Tunisia. Microb Drug Resist. 2016;22(2):137–140. doi: 10.1089/mdr.2014.0278. [DOI] [PubMed] [Google Scholar]
- 741.Guermazi-Toumi S, Boujlel S, Assoudi M, Issaoui R, Tlili S, Hlaiem ME. Susceptibility profiles of bacteria causing urinary tract infections in Southern Tunisia. J Glob Antimicrob Resist. 2018;12:48–52. doi: 10.1016/j.jgar.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 742.Hammami S, Dahdeh C, Mamlouk K, Ferjeni S, Maamar E, Hamzaoui Z, et al. Rectal carriage of extended-spectrum beta-lactamase and carbapenemase producing gram-negative bacilli in intensive care units in Tunisia. Microb Drug Resist. 2017;23(6):695–702. doi: 10.1089/mdr.2016.0205. [DOI] [PubMed] [Google Scholar]
- 743.Hammami S, Saidani M, Ferjeni S, Aissa I, Slim A, Boutiba-Ben BI. Characterization of extended spectrum β-lactamase-producing Escherichia coli in community-acquired urinary tract infections in Tunisia. Microb Drug Resist. 2013;19(3):231–236. doi: 10.1089/mdr.2012.0172. [DOI] [PubMed] [Google Scholar]
- 744.Hamzaoui Z, Ocampo-Sosa A, Fernandez Martinez M, Landolsi S, Ferjani S, Maamar E, et al. Role of association of OmpK35 and OmpK36 alteration and bla ESBL and/or bla AmpC genes in conferring carbapenem resistance among non-carbapenemase-producing Klebsiella pneumoniae. Int J Antimicrob Agents. 2018;52(6):898–905. doi: 10.1016/j.ijantimicag.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 745.Harchay C, Rachid S, Ben OA, et al. Epidemic diffusion of Klebsiella pneumoniae isolates producing extended-spectrum beta-lactamases in neonatal and pediatric wards in Rabta hospital of Tunisia. Afr J Microbiol Res. 2013;7(21):2497–2504. doi: 10.5897/AJMR12.941. [DOI] [Google Scholar]
- 746.Izdebski R, Bojarska K, Baraniak A, Literacka E, Herda M, Zabicka D, et al. NDM-1- or OXA-48-producing enterobacteriaceae colonising polish tourists following a terrorist attack in Tunis, March 2015. Euro Surveill. 2015;20(23):21150. doi: 10.2807/1560-7917.ES2015.20.23.21150. [DOI] [PubMed] [Google Scholar]
- 747.Jaballah NB, Bouziri A, Mnif K, Hamdi A, Khaldi A, Kchaou W. Epidemiology of hospital-acquired bloodstream infections in a Tunisian pediatric intensive care unit: a 2-year prospective study. Am J Infect Control. 2007;35(9):613–618. doi: 10.1016/j.ajic.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 748.Jaidane N, Bonnin RA, Mansour W, Girlich D, Creton E, Cotellon G, et al. Genomic insights into Colistin-resistant Klebsiella pneumoniae from a Tunisian teaching hospital. Antimicrob Agents Chemother. 2017;62:E01601–E1617. doi: 10.1128/AAC.01601-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 749.Jeddi R, Achour M, Amor R Ben, Aissaoui L, Bouterĝa W, Kacem K, et al. Factors associated with severe sepsis: Prospective study of 94 neutropenic febrile episodes. Hematology. 2010;15(1):28–32. [DOI] [PubMed]
- 750.Khalifa ABH, Khedher M. Epidemiological study of Klebsiella spp. uropathogenic strains producing extended-spectrum b-lactamase in a Tunisian university hospital, 2009. Pathol Biol. 2012 Apr;60(2). [DOI] [PubMed]
- 751.Kollenda H, Frickmann H, Helal R Ben, Wiemer DF, Naija H, El Asli MS, et al. Screening for carbapenemases in ertapenem-resistant Enterobacteriaceae collected at a Tunisian hospital between 2014 and 2018. Eur J Microbiol Immunol. 2019;9(1):9–13. [DOI] [PMC free article] [PubMed]
- 752.Ktari S, Arlet G, Mnif B, Gautier V, Mahjoubi F, Jmeaa M Ben, et al. Emergence of multidrug-resistant Klebsiella pneumoniae isolates producing VIM-4 metallo-β-lactamase, CTX-M-15 extended-spectrum β-lactamase, and CMY-4 AmpC β-lactamase in a Tunisian University Hospital. Antimicrob Agents Chemother. 2006;50(12):4198–201. [DOI] [PMC free article] [PubMed]
- 753.Ktari S, Mnif B, Louati F, Rekik S, Mezghani S, Mahjoubi F, et al. Spread of Klebsiella pneumoniae isolates producing OXA-48 β-lactamase in a Tunisian University Hospital. J Antimicrob Chemother. 2011;66(7):1644–1646. doi: 10.1093/jac/dkr181. [DOI] [PubMed] [Google Scholar]
- 754.Lahlaoui H, Bonnin RA, Moussa MB, Khelifa ABH, Naas T. First report of OXA-232-producing Klebsiella pneumoniae strains in Tunisia. Diagn Microbiol Infect Dis. 2017;88(2):195–197. doi: 10.1016/j.diagmicrobio.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 755.Lahlaoui H, Poirel L, Barguellil F, Moussa MB, Nordmann P. Carbapenem-hydrolyzing class D β-lactamase OXA-48 in Klebsiella pneumoniae isolates from Tunisia. Eur J Clin Microbiol Infect Dis. 2012;31(6):937–939. doi: 10.1007/s10096-011-1389-5. [DOI] [PubMed] [Google Scholar]
- 756.Lamia T, Zoubeir C, Beya M, Yosra B, Allah MA. Molecular characterization of carbapenemase-producing enterobacteriaceae in burn patients. Ann Intensive Care. 2018;8(S1):101–102. [Google Scholar]
- 757.Maamar E, Ferjani S, Jendoubi A, Hammami S, Hamzaoui Z, Mayonnove-Coulange L, et al. High prevalence of gut microbiota colonization with broad-spectrum cephalosporin resistant Enterobacteriaceae in a Tunisian intensive care unit. Front Microbiol. 1859;2016(7):1–10. doi: 10.3389/fmicb.2016.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 758.Maamar E, Hammami S, Ferjani S, Hamzaoui Z, Jlizi A, Saidani M, et al. Molecular characterization of extended spectrum β-lactamases, ampccephalosporinases and carbapenemases in Klebsiella pneumoniae causing bacteremia at Charles Nicolle Hospital of Tunisia. Acta Medica Int. 2016;3(2):40. doi: 10.5530/ami.2016.2.10. [DOI] [Google Scholar]
- 759.Mamlouk K, Boubaker IB, Gautier V, Vimont S, Picard B, Ben Redjeb S, et al. Emergence and outbreaks of CTX-M β-lactamase-producing Escherichia coli and Klebsiella pneumoniae strains in a Tunisian hospital. J Clin Microbiol. 2006;44(11):4049–4056. doi: 10.1128/JCM.01076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 760.Mansour W, Grami R, Jaidane N, Messaoudi A, Charfi K, Ben Romdhane L, et al. Epidemiology and whole-genome analysis of NDM-1-producing Klebsiella pneumoniae KP3771 from Tunisia. Microb Drug Resist. 2019;25(5):644–651. doi: 10.1089/mdr.2018.0204. [DOI] [PubMed] [Google Scholar]
- 761.Mansour W, Haenni M, Saras E, Grami R, Mani Y, Khalifa ABH, et al. Outbreak of Colistin-resistant carbapenemase-producing Klebsiella pneumoniae in Tunisia. J Glob Antimicrob Resist. 2017;10:88–94. doi: 10.1016/j.jgar.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 762.Messaoudi A, Haenni M, Mansour W, Saras E, Khalifa ABH, Chaouch C, et al. ST147 NDM-1-producing Klebsiella pneumoniae spread in two Tunisian hospitals. J Antimicrob Chemother. 2017;72(1):315–316. doi: 10.1093/jac/dkw401. [DOI] [PubMed] [Google Scholar]
- 763.Messous S, Grissa MH, Beltaief K, Boukef R, Nouira S, Mastouri M. Bacteriology of acute exacerbations of chronic obstructive pulmonary disease in Tunisia. Rev Mal Respir. 2018;35(1):36–47. doi: 10.1016/j.rmr.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 764.Mezghani Maalej S, Rekik Meziou M, Mahjoubi F, Hammami A. Epidemiological study of enterobacteriaceae resistance to colistin in Sfax (Tunisia) Med Mal Infect. 2012;42(6):256–263. doi: 10.1016/j.medmal.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 765.Najwa D, Salah AM, Yolanda S, Monia K, Dorsaf M, Chiheb BR, et al. Low antibiotic resistance rates and high genetic heterogeneity of Escherichia coli isolates from urinary tract infections of diabetic patients in Tunisia. J Chemother. 2016;28(2):89–94. doi: 10.1179/1973947814Y.0000000229. [DOI] [PubMed] [Google Scholar]
- 766.Nasr AB, Decré D, Compain F, Genel N, Barguellil F, Arlet G. Emergence of NDM-1 in association with OXA-48 in Klebsiella pneumoniae from Tunisia. Antimicrob Agents Chemother. 2013;57(8):4089–4090. doi: 10.1128/AAC.00536-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 767.Ouertani R, Ben Jomàa-Jemili M, Gharsa H, Limelette A, Guillard T, Brasme L, et al. Prevalence of a new variant OXA-204 and OXA-48 carbapenemases plasmids encoded in Klebsiella pneumoniae clinical isolates in Tunisia. Microb Drug Resist. 2018;24(2):142–149. doi: 10.1089/mdr.2016.0236. [DOI] [PubMed] [Google Scholar]
- 768.Ouertani R, Limelette A, Guillard T, Brasme L, Jridi Y, Barguellil F, et al. First report of nosocomial infection caused by Klebsiella pneumoniae ST147 producing OXA-48 and VEB-8 β-lactamases in Tunisia. J Glob Antimicrob Resist. 2016;4:53–56. doi: 10.1016/j.jgar.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 769.Potron A, Nordmann P, Poirel L. Characterization of OXA-204, a carbapenem-hydrolyzing class D β-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2013;57(1):633–636. doi: 10.1128/AAC.01034-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 770.Saïdani M, Hammami S, Kammoun A, Slim A, Boutiba-Ben BI. Emergence of carbapenem-resistant OXA-48 carbapenemase-producing enterobacteriaceae in Tunisia. J Med Microbiol. 2012;61(PART12):1746–1749. doi: 10.1099/jmm.0.045229-0. [DOI] [PubMed] [Google Scholar]
- 771.Tanfous FB, Raddaoui A, Chebbi Y, Achour W. Epidemiology and molecular characterisation of colistin-resistant Klebsiella pneumoniae isolates from immunocompromised patients in Tunisia. Int J Antimicrob Agents. 2018;52(6):861–865. doi: 10.1016/j.ijantimicag.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 772.Toumi A, Kadri Y, Abdallah HB, Noomen S, Chakroun M, Mastouri M. P659 Clinical, epidemiological and microbiological features of urinary tract infections caused by ESBL-producing Enterobacteriaceae in hospitalised patients. Clin Microbiol Infect: S142.
- 773.Abbassi MS, Torres C, Achour W, Vinué L, Sáenz Y, Costa D, et al. Genetic characterisation of CTX-M-15-producing Klebsiella pneumoniae and Escherichia coli strains isolated from stem cell transplant patients in Tunisia. Int J Antimicrob Agents. 2008;32(4):308–314. doi: 10.1016/j.ijantimicag.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 774.Achour NB, Mercuri PS, Moussa MB, Galleni M, Belhadj O. Characterization of a novel extended-spectrum TEM-type b-lactamase, TEM-164, in a clinical strain of Klebsiella pneumoniae in Tunisia. Microb Drug Resist. 2009;15(3):195–199. doi: 10.1089/mdr.2009.0900. [DOI] [PubMed] [Google Scholar]
- 775.Lamorde M, Mpimbaza A, Walwema R, Kamya M, Kapisi J, Kajumbula H, et al. A cross-cutting approach to surveillance and laboratory capacity as a platform to improve health security in Uganda. Heal Secur. 2018;16:S76–86. doi: 10.1089/hs.2018.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 776.Lubwama M, Phipps W, Najjuka CF, Kajumbula H, Ddungu H, Kambugu JB, et al. Bacteremia in febrile cancer patients in Uganda. BMC Res Notes. 2019 doi: 10.1186/s13104-019-4520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 777.Najjuka CF, Kateete DP, Kajumbula HM, Joloba ML, Essack SY. Antimicrobial susceptibility profiles of Escherichia coli and Klebsiella pneumoniae isolated from outpatients in urban and rural districts of Uganda. BMC Res Notes. 2016; [DOI] [PMC free article] [PubMed]
- 778.Okoche D, Asiimwe BB, Katabazi FA, Kato L, Najjuka CF. Prevalence and characterization of carbapenem-resistant enterobacteriaceae isolated from Mulago National Referral Hospital, Uganda. PLoS One. 2015; [DOI] [PMC free article] [PubMed]
- 779.Seni J, Najjuka CF, Kateete DP, Makobore P, Joloba ML, Kajumbula H, et al. Antimicrobial resistance in hospitalized surgical patients: A silently emerging public health concern in Uganda. BMC Res Notes. 2013; [DOI] [PMC free article] [PubMed]
- 780.Ssemogerere L, Sendagire C, Mbabazi C, Namungoma Y, Oketayot AN, Namuyonga J, et al. Hand Colonization with Gram-Negative Organisms of Healthcare Workers Accessing the Cardiac Intensive Care Unit: A Cross-Sectional Study at the Uganda Heart Institute. Crit Care Res Pract. 2019;2019. [DOI] [PMC free article] [PubMed]
- 781.Stanley IJ, Kajumbula H, Bazira J, Kansiime C, Rwego IB, Asiimwe BB. Multidrug resistance among Escherichia coli and Klebsiella pneumoniae carried in the gut of out-patients from pastoralist communities of Kasese district, Uganda. PLoS One. 2018;13(7). [DOI] [PMC free article] [PubMed]
- 782.Tumuhamye J, Sommerfelt H, Bwanga F, Tumwine JK, Mukunya D, Nankabirwa V. Etiology and antimicrobial resistance patterns of neonatal sepsis at Mulago National Referral Hospital, Uganda. Intensive Care Med Exp. 2019;7(S2). [DOI] [PMC free article] [PubMed]
- 783.Agaba P, Tumukunde J, Tindimwebwa JVB, Kwizera A. Nosocomial bacterial infections and their antimicrobial susceptibility patterns among patients in Ugandan intensive care units: a cross sectional study. BMC Res Notes. 2017 doi: 10.1186/s13104-017-2695-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 784.Ampaire L, Katawera V, Nyehangane D, Boum Y, Bazira J. Epidemiology of carbapenem resistance among multi-drug resistant enterobacteriaceae in Uganda. Br Microbiol Res J. 2015. [DOI] [PMC free article] [PubMed]
- 785.Bebell LM, Ngonzi J, Bazira J, Fajardo Y, Boatin AA, Siedner MJ, et al. Antimicrobial-resistant infections among postpartum women at a Ugandan referral hospital. PLoS One. 2017. [DOI] [PMC free article] [PubMed]
- 786.George M, Iramiot J, Muhindo R, Olupot-Olupot P, Nanteza A. Bacterial aetiology and antibiotic susceptibility profile of post-operative sepsis among surgical patients in a tertiary hospital in rural eastern Uganda. Microbiol Res J Int. 2018;24(2):1–8. doi: 10.9734/MRJI/2018/41690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 787.Kajumbula H, Fujita AW, Mbabazi O, Najjuka C, Izale C, Akampurira A, et al. Antimicrobial drug resistance in blood culture isolates at a tertiary hospital. Uganda Emerg Infect Dis. 2018;24(1):174–175. doi: 10.3201/eid2401.171112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 788.Kemigisha E, Nanjebe D, Ii YB, Langendorf C, Aberrane S, Nyehangane D, et al. Antimicrobial treatment practices among Ugandan children with suspicion of central nervous system infection. PLoS ONE. 2018 doi: 10.1371/journal.pone.0205316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 789.Kabwe M, Tembo J, Chilukutu L, Chilufya M, Ngulube F, Lukwesa C, et al. Etiology, antibiotic resistance and risk factors for neonatal sepsis in a large referral center in Zambia. Pediatr Infect Dis J. 2016. [DOI] [PubMed]
- 790.Roth B, Laps A, Stafford K, Heil E, Hachaambwa L, Yamba K, et al. High-frequency of multi-drug-resistant organisms (MDRO) at university teaching hospital (UTH), Lusaka, Zambia. OFID. 2018;5(Suppl 1):S169–S170. doi: 10.1093/ofid/ofy210.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 791.Obi CL, Makandiramba SA, Twsana SA, Robertson V, Moyo SR, Nziramasanga P. In-vitro disk diffusion sensitivitiy of meropenem against bacterial pathogens in Harare. East Afr Med J. 1999;76(7):365–369. [PubMed] [Google Scholar]
- 792.Wilmore SMS, Kranzer K, Williams A, Makamure B, Nhidza AF, Mayini J, et al. Carriage of extended-spectrum beta-lactamase-producing enterobacteriaceae in HIV-infected children in Zimbabwe. J Med Microbiol. 2017. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Boolean search strings constructed for searches of scientific databases.
Additional file 3: Annotation on data entry columns and abbreviations.
Data Availability Statement
The dataset supporting the conclusions of this article is available in the Harvard Dataverse repository, https://doi.org/10.7910/DVN/JIJH3W. The dataset(s) supporting the conclusions of this article is also included within the article as Additional file 4.