Summary
l-Alanine is an important amino acid widely used in food, medicine, materials, and other fields. Here, we develop Bacillus licheniformis as an efficient l-alanine microbial cell factory capable of realizing high-temperature fermentation. By enhancing the glycolytic pathway, knocking out the by-product pathways and overexpressing the thermostable alanine dehydrogenase, the engineered B. licheniformis strain BLA3 produced 93.7 g/L optically pure l-alanine at 50°C. Subsequently, d-alanine dependence of an alanine racemase-deficient strain is relieved by adaptive laboratory evolution, implying that a dormant alternative pathway for d-alanine synthesis is activated in the evolved strain. The d-amino acid aminotransferase Dat1 is shown to be a key enzyme in the dormant alternative pathway. Molecular mechanism of the d-alanine dependence is revealed via mutational analysis. This study demonstrates a novel technology for high-temperature l-alanine production and shows that activating dormant metabolic pathway(s) is an effective strategy of metabolic engineering.
Subject areas: Microbiology, Applied microbiology, Microbial biotechnology
Graphical abstract
Highlights
-
•
Efficient high-temperature l-alanine production is achieved by Bacillus licheniformis
-
•
d-Alanine dependence of the alanine racemase-deficient B. licheniformis is relieved
-
•
d-Amino acid aminotransferase Dat1 is a key enzyme in the dormant alternative pathway
-
•
Molecular mechanism of the d-alanine dependence is revealed via mutational analysis
Microbiology; Applied microbiology; Microbial biotechnology
Introduction
l-Alanine is an important industrial chemical with a global annual demand around 50,000 tons.1 It is not only an important sweetener and seasoning,2,3 but also the raw material for a novel green chelating agent, methylglycinediacetic acid (MGDA).4 In the field of medicine, l-alanine is an important stimulator of glucagon; therefore, it plays an important role in the research and treatment of diabetes.5 In addition, incorporation of l-alanine into polyester amides can significantly improve their biodegradability and biocompatibility for drug delivery applications.6 Currently, l-alanine is mainly produced from l-aspartate by enzymatic methods, but l-alanine production via microbial fermentation is attractive due to the use of renewable and cheap raw materials.1 NADH-dependent alanine dehydrogenase that can reduce pyruvate to l-alanine is the most important enzyme involved in l-alanine biosynthesis (Figure 1A).7 By introducing heterologous alanine dehydrogenase, l-alanine fermentation production processes have been successfully established in Escherichia coli, and the final titer and yield can reach about 120.0 g/L and 0.9 g/g, respectively.8,9 E. coli is a common microbial chassis for biomanufacturing and has significant advantages in growth performance. However, the optimum growth temperature for E. coli is only 37°C, which results in high cooling costs in large-scale fermentation processes. This increases the economic cost of biomanufacturing and has become a new focus for industrial upgrading.
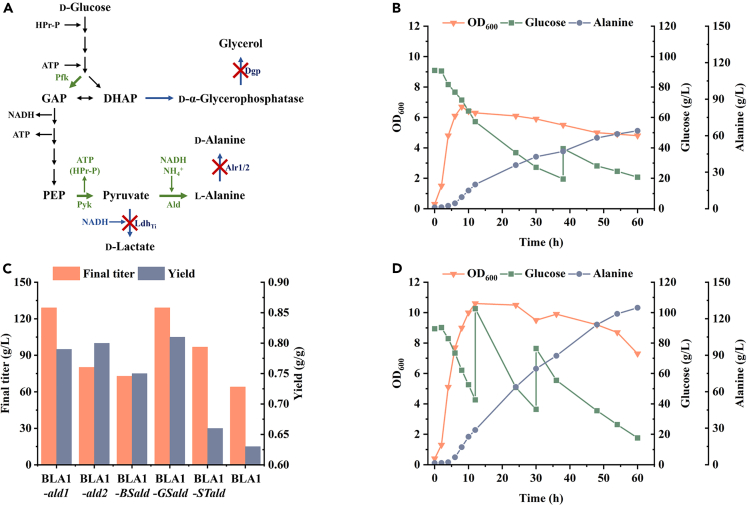
Figure 1.
Redirection of carbon metabolism in B. licheniformis to obtain efficient alanine producer
(A) Alanine synthesis pathway and main by-product pathways in B. licheniformis. The overexpressed enzymes were marked in green, while the knockout enzymes were marked in blue.
(B) Fed-batch fermentation of B. licheniformis BLA1.
(C) The final titers and yields of fed-batch fermentations of B. licheniformis overexpressing alanine dehydrogenases from different sources.
(D) Fed-batch fermentation of B. licheniformis BLA2.
Industrially, the fermentation process using thermophiles (> 45°C) is defined as high-temperature fermentation,10 which is considered a potential next-generation biotechnology with multiple advantages, including improved biochemical reaction efficiency, enhanced resistance to bacterial contamination, and reduced heat exchange costs associated with cooling.11 For a 30,000-kL scale ethanol plant, a 5°C increase in fermentation temperature has been reported to save about $390,000 per year.12 To reduce production costs, simultaneous saccharification and fermentation (SSF) processes that can efficiently utilize renewable lignocellulosic biomass have received increasing attention.13 Since the optimum temperature of cellulase and hemicellulase is usually over 50°C,14 high-temperature fermentation can improve the SSF process by increasing saccharification efficiency.15,16 The currently developed microbial fermentation processes for l-alanine production mainly focus on mesophilic bacteria, such as E. coli, which cannot meet the requirements of high-temperature fermentation. Bacillus licheniformis is an excellent high-temperature platform strain that is capable of rapid growth at 50°C. As a generally regarded as safe (GRAS) strain,17 B. licheniformis has been successfully used for the biosynthesis of d-lactic acid,11 2,3-butanediol,18 bacitracin,19 and other compounds. On the other hand, Bacillus is an important source of alanine dehydrogenase7; therefore, B. licheniformis may have a genetic potential for naturally accumulating l-alanine.
Optical purity is an important parameter for l-alanine production; however, it is challenging to achieve high optical purity of l-alanine without affecting bacterial growth, as d-alanine is an essential component of the bacterial cell wall.20 Alanine is a kind of chiral molecule and is divided into d and l configurations.21 d-Alanine is mainly converted from l-alanine by alanine racemase in bacterial cells.20 As an essential pathway for bacterial growth, the d-alanine synthesis pathway is widely present in bacteria; therefore, it is inevitable to accumulate d-alanine when l-alanine is produced. On the one hand, the accumulation of d-alanine requires l-alanine as a precursor that significantly reduces the productivity and yield of l-alanine. However, the accumulation of d-alanine reduces the optical purity of l-alanine products, and it is difficult to separate from l-alanine, thus affecting the application value of l-alanine products. In microbial fermentation for l-alanine production, it is necessary to provide sufficient d-alanine to maintain cell growth and prevent excessive accumulation of d-alanine to ensure the optical purity of l-alanine. This requires that the biosynthesis of d-alanine in microbial cells is perfectly balanced, which poses a new challenge to the construction of l-alanine producers.
Bacteria typically contain genomes of several thousand kilobases in size and thousands of genes.22 The products encoded by these genes constitute complex metabolic networks in cells, which can serve as a library of components for synthetic biology. However, most genes are expressed at very low levels, leaving many potentially dormant pathways.23 Several studies have attempted to activate these dormant pathways by various means, thereby endowing microorganisms with new functions to synthesize new products or to utilize new substrates.24,25 Therefore, the activation of intracellular dormant metabolic pathways can significantly rewire the metabolism of microbial cells, which may be a new solution for solving complex intracellular metabolic problems.
In this study, we engineered B. licheniformis as a high-temperature microbial cell factory capable of efficiently producing l-alanine. To achieve efficient production of alanine, the carbon metabolic flux of B. licheniformis was redirected to the alanine biosynthesis pathway by eliminating competing pathways, enhancing the glycolytic pathway, and overexpressing alanine dehydrogenase. To reconstitute the balance of intracellular d-alanine synthesis to prevent excessive accumulation of d-alanine, we activated the dormant d-alanine synthesis pathway in alanine racemase-deficient strains by adaptive laboratory evolution. Simultaneously, we attempted to explain the fine-tuning of d-alanine metabolic patterns in mutant strains by the cell wall synthesis characterization based on fluorescently labeled d-alanine and genome resequencing analysis.
Results
Redirection of carbon flux for alanine biosynthesis
NADH-dependent alanine dehydrogenase catalyzes the synthesis of l-alanine from pyruvate, the final product of the EMP pathway. Two alanine dehydrogenases (encoded by ald1 and ald2) were found in the genome of our previously reported efficient d-lactic acid producer, B. licheniformis BN11.11 Considering the similar substrates and cofactors of alanine dehydrogenase and lactate dehydrogenase, strain BN11 was used as the parent strain for strain engineering. To further enhance the flux of the EMP pathway, the pfkA and pyk genes (encoding 6-phosphofructokinase and pyruvate kinase, respectively) from Bacillus coagulans were introduced into strain BN11 with the strong promoter Pals, resulting in the recombinant strain B. licheniformis BN11-PFYAK. To prevent carbon flux from flowing into d-lactic acid, the ldhTi gene encoding d-lactate dehydrogenase was deleted, resulting in the recombinant strain B. licheniformis BLA1. After 60 h of fed-batch fermentation, a total of 64.0 g/L alanine was produced by B. licheniformis BLA1 (Figure 1B). The yield of alanine reached 0.63 g/g, indicating that the carbon flux was successfully redirected into the alanine biosynthesis pathway. The fermentation process was divided into two stages: aerobic growth and microaerobic production. During the aerobic growth phase, the growth of BLA1 was not so fast, and the maximum OD600 (optical density at a wavelength of 600 nm) was 6.7. During the microaerobic production phase, the productivity of alanine was also slow. The average productivity of BLA1 to produce alanine was only 1.1 g/(L·h), which might be limited by the natural activity and expression of Ald1 and Ald2 in B. licheniformis.
Improving alanine production by overexpressing efficient thermostable alanine dehydrogenase
Excessive accumulation of alanine is toxic to cells,26 so the natural expression of ald1 and ald2 may not be sufficient for efficient alanine production. To increase the expression of alanine dehydrogenase, a second copy of ald1 or ald2 was introduced into B. licheniformis BLA1 with the strong promoter Pals. The gene fragment was integrated at the same position as the ldhTi gene in B. licheniformis BN11. The final titers of alanine increased to 129.0 g/L and 80.1 g/L using recombinant strains BLA1-ald1 and BLA1-ald2, respectively (Figures 1C and S1A–S1B). The fermentation results showed that increasing the expression of alanine dehydrogenase in B. licheniformis BLA1 can improve alanine production and that Ald1 seems to have higher activity than Ald2. To further improve alanine production, several thermostable alanine dehydrogenases were mined using a co-evolution-based mining method for thermostable enzymes.27 This method adopts the concept of co-evolution for mining thermostable enzymes from thermophilic microbial genomes. Three alanine dehydrogenase genes from Geobacillus stearothermophilus (GSald), Bacillus smithii (BSald), and Streptomyces thermoautotrophicus (STald) were selected for further testing. Heterologous alanine dehydrogenase genes were introduced into B. licheniformis BLA1 in the same way as genes ald1 and ald2. The final titers of alanine increased to 129.0 g/L, 72.8 g/L, and 96.7 g/L, respectively using recombinant strains BLA1-GSald, BLA1-BSald, and BLA1-STald (Figures 1C–1D and S1C–S1D). Both BLA1-ald1 and BLA1-GSald exhibited efficient alanine production, but the yield of BLA1-GSald was higher than that of BLA1-ald1. Therefore, BLA1-GSald was selected for subsequent genetic manipulation and was named BLA2. In fed-batch fermentation, strain BLA2 showed advantages over BLA1 in terms of both aerobic growth and microaerobic production. The maximum OD600 reached 10.6, and the average productivity was 2.2 g/(L·h) (Figure 1D). The final titer and yield of l-alanine were increased by 101.6% and 28.6%, respectively.
Deleting alanine racemase genes to increase the optical purity
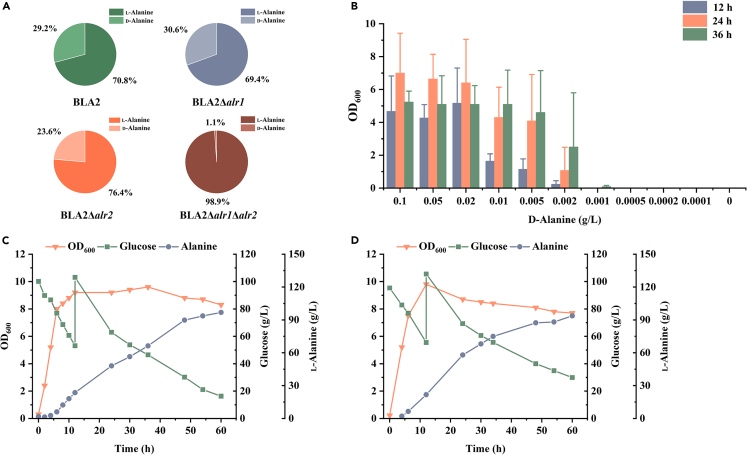
Our experiment up to this point has led to the development of an efficient alanine producer, B. licheniformis BLA2. However, the alanine product was a mixture of l-alanine and d-alanine with an optical purity of only 70.8% (Figure 2A). d-Alanine was converted from l-alanine catalyzed by alanine racemase, which is encoded by genes alr1 and alr2 in B. licheniformis BLA2. To improve optical purity, the alr1 and alr2 genes were knocked out separately to obtain the recombinant strains BLA2Δalr1 and BLA2Δalr2. Unfortunately, deleting gene alr1 or alr2 alone did not effectively improve the optical purity of l-alanine, and the optical purities were maintained at 69.4% and 76.4%, respectively (Figure 2A). To further inhibit the synthesis of d-alanine, both genes alr1 and alr2 were knocked out resulting in the recombinant strain BLA3. Strain BLA3 exhibited severe defect in d-alanine synthesis and could not grow in a medium without the addition of d-alanine. To characterize the d-alanine dependence of strain BLA3, it was cultured in seed media containing various concentrations of d-alanine (Figure 2B). Strain BLA3 was able to maintain normal growth with 0.02 g/L d-alanine and hardly grow when the concentration of d-alanine was lower than 0.001 g/L. The alr1/alr2 double-knockout strain successfully produced l-alanine with an optical purity of 98.9% in a 5-L fermenter by adding 1 g/L d-alanine (Figure 2A). Affected by the defect of d-alanine synthesis, the growth rate of BLA3 in the aerobic growth phase was slightly reduced, and the maximum OD600 was reduced to 9.6. The final titer and yield of l-alanine were 97.0 g/L and 0.77 g/g, respectively (Figure 2C).
Figure 2.
Efficient production of optically pure l-alanine achieved by knocking out alanine racemase and glycerol by-product biosynthesis pathway
(A) Effects of individual and simultaneous knockout of two alanine racemase alr1 and alr2 on the optical purity of l-alanine produced by fed-batch fermentation of B. licheniformis.
(B) Characterization of the d-alanine dependence of B. licheniformis BLA3 by testing the growth performance in media containing various concentrations of d-alanine. Triplicate experiments were carried out for physiological measurements, and error bars represent standard deviations.
(C) Fed-batch fermentation of B. licheniformis BLA3.
(D) Fed-batch fermentation of B. licheniformis BLA4.
Eliminating glycerol synthesis pathway to increase the yield
In the commercial production of l-alanine, yield is an important production indicator that can significantly affect the cost of raw materials. To further improve the yield of l-alanine, organic acid, alcohol, and amino acid by-products of the strain BLA3 fermentation samples were identified. Glycerol was identified as the predominant by-product, although formic acid, acetic acid, and succinic acid also accumulated to some extent (Table S1). In addition to alanine, only lysine, valine, glutamic acid, γ-aminobutyric acid, and glycine were detected at concentrations below 0.3 g/L (Figure S2). This may be because most of the ammonium ions in the cell are used to synthesize alanine.
Although strain BLA3 accumulated up to 6.6 g/L glycerol, no gene responsible for glycerol synthesis was annotated in the genome. A recent study has reported the role of d-α-glycerophosphatase encoded by the dgp gene in glycerol synthesis in B. licheniformis 4071.28 After amino acid sequence alignment, the mtnX gene in B. licheniformis BLA3 genome was confirmed as the dgp gene (100% amino acid identity). To eliminate the glycerol synthesis pathway, gene dgp was deleted, resulting in the recombinant strain B. licheniformis BLA4. After 60 h of fed-batch fermentation, strain BLA4 was able to produce 93.7 g/L of l-alanine with an average productivity of 1.6 g/(L·h). Knockout of gene dgp resulted in a slight decrease in the final titer of l-alanine, but the yield increased to 0.81 g/g (Figure 2D). Glycerol was completely undetectable after 60 h of fermentation, and the concentration of acetic acid decreased slightly (Table S1). The maximum OD600 of strain BLA4 was comparable to that of strain BLA3, but it entered the death phase earlier in the fermentation process, which may explain the decreased final titer.
Removing growth dependence on exogenous d-alanine through adaptive evolution
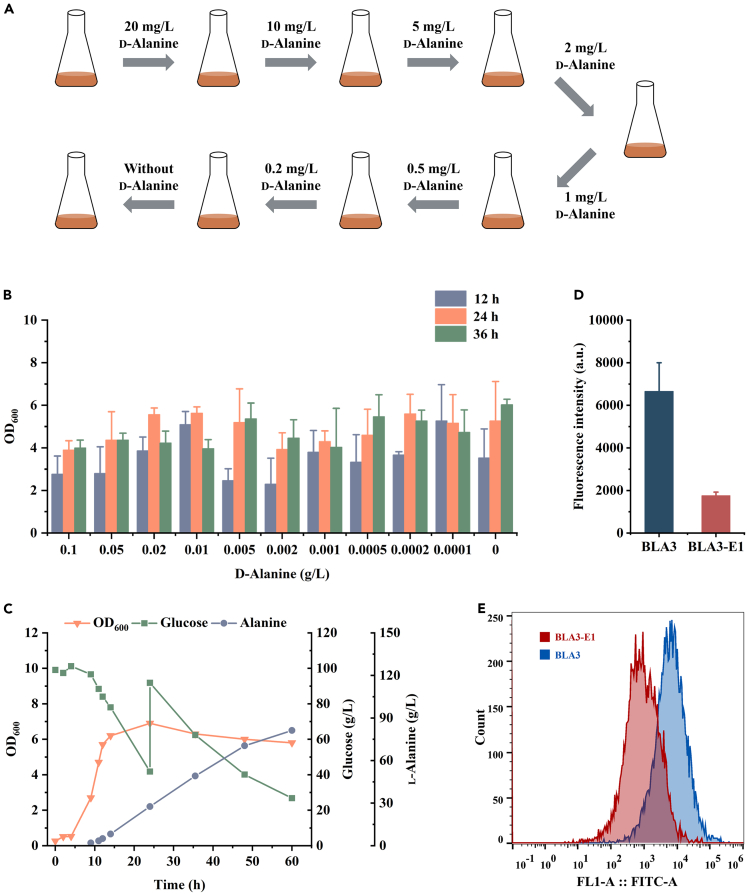
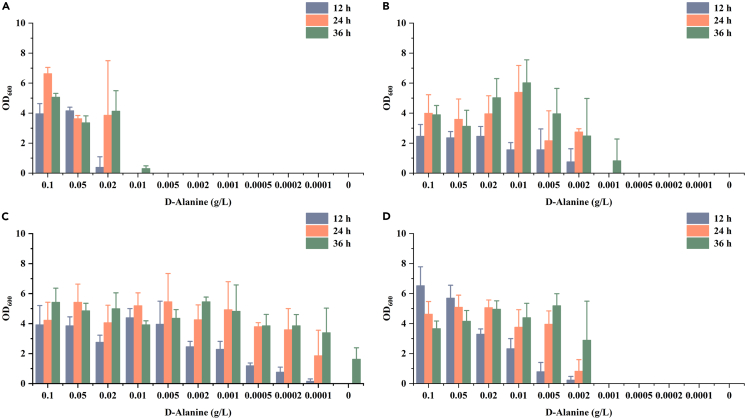
d-alanine is an essential component of the bacterial cell wall peptidoglycan and is widely involved in the growth and division of bacterial cells. Therefore, it is plausible that B. licheniformis deficient in alanine racemase exhibited severe growth defects without the addition of d-alanine. To eliminate the growth dependence of strain BLA3 on exogenous d-alanine, adaptive laboratory evolution experiments with gradually decreasing concentrations of d-alanine were carried out. Strain BLA3 was first transferred to seed medium supplemented with d-alanine at a concentration of 20 mg/L. Subsequently, the strain was continuously transferred to medium containing a lower concentration of d-alanine (Figure 3A). When the concentration of d-alanine was reduced to 0.2 mg/L, we found that the resulting strain, which was named BLA3-E1, was able to grow in the seed medium without d-alanine. The d-alanine dependence of the evolved B. licheniformis BLA3-E1 strain was identified using the same method as for strain BLA3 (Figure 3B). Although the evolved strain grew worse than strain BLA3 at a higher d-alanine concentration, it was able to maintain normal growth even without the addition of d-alanine. These results indicated that there was a novel d-alanine biosynthesis pathway in strain BLA3 that does not depend on racemase Alr1 or Alr2. In fed-batch fermentation without d-alanine, strain BLA3-E1 was able to produce 81.2 g/L L-alanine with a yield of 0.71 g/g and an optical purity of 99.5% (Figure 3C). The evolved strain showed growth defects with a longer lag phase and a lower maximum OD600, which may be an important reason for the lower final titer and yield of l-alanine.
Figure 3.
Removing the growth dependence of B. licheniformis BLA3 on d-alanine through adaptive laboratory evolution
(A) Scheme of the laboratory evolutionary approach to remove the growth dependence of mutant strains on d-alanine.
(B) Characterization of the d-alanine dependence on evolved B. licheniformis BLA3-E1 by testing the growth performance in media containing various concentrations of d-alanine. Triplicate experiments were carried out for physiological measurements, and error bars represent standard deviations.
(C) Fed-batch fermentation of evolved B. licheniformis BLA3-E1.
(D) Mean fluorescence intensity of B. licheniformis BLA3 and evolved B. licheniformis BLA3-E1 incubated with 5-FAM labeled d-alanine. Fluorescence intensity represented the requirement of extracellular d-alanine for the biological process of cell wall synthesis. Triplicate experiments were carried out for physiological measurements, and error bars represent standard deviations.
(E) Flow cytometry analysis of B. licheniformis BLA3 and evolved B. licheniformis BLA3-E1 incubated with 5-FAM labeled d-alanine. The X axis is fluorescence intensity, and the Y axis is cell number. B. licheniformis BLA3 and BLA3-E1 are shown in blue and red, respectively.
d-alanine supplemented in the medium was mainly used for the synthesis of the cell wall of B. licheniformis. Here, the fluorescent labeling reagent 5-FAM was used to label d-alanine to characterize the biological process of cell wall synthesis involving extracellular d-alanine.29 After incubation with 5-FAM labeled d-alanine for 2 h, the mean fluorescence intensity of the B. licheniformis cells reflected the rate of extracellular d-alanine-involved cell wall synthesis. Compared with strain BLA3, the fluorescence intensity of strain BLA3-E1 was significantly reduced by 73.7% (Figures 3D and 3E). This suggests that the evolved strain reduced its reliance on extracellular d-alanine for cell wall biosynthesis, which may be either by reducing the content of peptidoglycan in the cell wall, thereby reducing the need for d-alanine, or by providing additional d-alanine intracellularly through other pathways.
Identification of the alternate d-alanine synthetic pathway
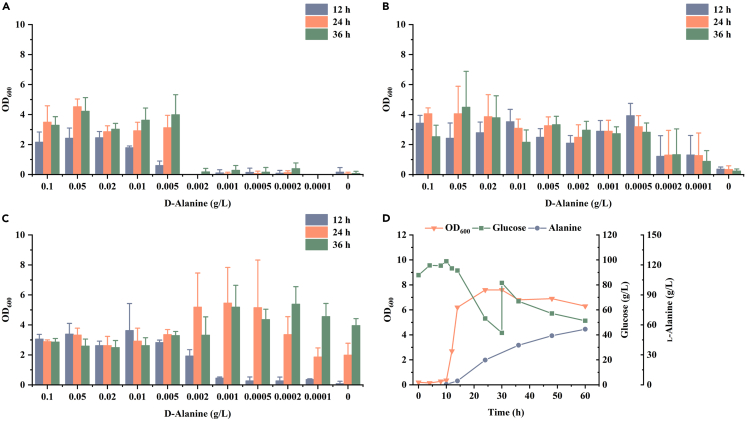
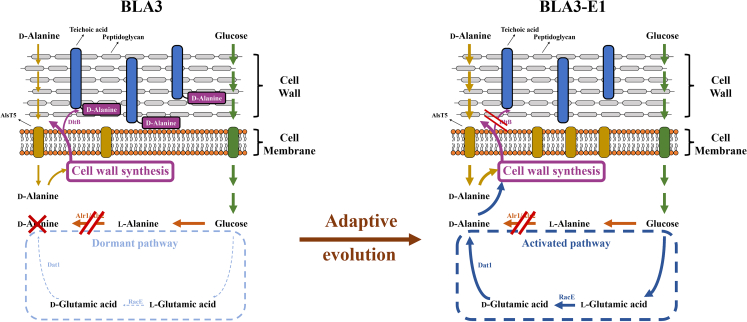
The evolved strain BLA3-E1 removed growth dependence on exogenous d-alanine, suggesting that a dormant d-alanine biosynthesis pathway was activated. Through analysis of the B. licheniformis genome, dat1, which encodes a d-amino acid aminotransferase, was suggested to be involved in the alternative d-alanine synthesis pathway. The d-amino acid aminotransferase Dat1 can achieve the mutual conversion of different d-amino acids, and this gene was also found to synthesize d-alanine in Bacillus subtilis.30 In fact, the B. licheniformis genome also contains a glutamate racemase encoded by racE, which is capable of converting intracellular l-glutamate to d-glutamate, and subsequently converting d-glutamate to d-alanine through Dat1 (Figure S3). To verify the function of dat1, the dat1 genes of strains BLA3 and BLA3-E1 were knocked out to obtain recombinant strains BLA3Δdat1 and BLA3-E1Δdat1. Consistent with our speculation, knockout of gene dat1 resulted in limited growth of strain BLA3 at all concentrations of d-alanine, and in particular, almost complete loss of growth at 0.002 g/L of d-alanine (Figure 4A). In contrast, knockout of dat1 in the evolved strain BLA3-E1 resulted in restricted growth of the strain at low concentrations of d-alanine and an OD600 of less than 1.0 in medium without d-alanine (Figure 4B). In addition, a second copy of dat1 was introduced into strain BLA3 with strong promoter Pals to obtain the recombinant strain BLA3-Pdat1. The recombinant strain exhibited better growth performance than strain BLA3 with a low concentration of d-alanine and was able to grow in medium without d-alanine (Figure 4C). However, when the concentration of d-alanine was lower than 0.001 g/L, the strain showed a significant lag phase and growth was restricted in the early stage. In fed-batch fermentation without d-alanine, strain BLA3-Pdat1 was able to produce 55.7 g/L L-alanine (Figure 4D). Compared with strain BLA3-E1, strain BLA3-Pdat1 exhibited a longer lag phase. Although the maximum OD600 of strain BLA3-Pdat1 was higher than that of strain BLA3-E1, l-alanine production by the strain was significantly reduced in the middle and late stages of fermentation. These results demonstrated that the evolved strains indeed supplied d-alanine necessary for growth through the d-alanine synthesis pathway involving Dat1.
Figure 4.
Identification of the alternate d-alanine synthetic pathway involving d-amino acid aminotransferase Dat1
(A–C) Characterization of the d-alanine dependence on B. licheniformis BLA3Δdat1, BLA3-E1Δdat1, and BLA3-Pdat1 by testing the growth performance in media containing various concentrations of d-alanine. Triplicate experiments were carried out for physiological measurements, and error bars represent standard deviations.
(D) Fed-batch fermentation of B. licheniformis BLA3-Pdat1.
Genome resequencing analysis of the evolved strain
To explore the mechanism, by which the evolved strain could maintain growth without exogenous d-alanine, genome resequencing of strain BLA3-E1 was performed. Similar to our hypothesis, evolved strain BLA3-E1 was obtained with fewer mutations. Compared with strain BLA3, strain BLA3-E1 had a total of nine nucleotide polymorphisms (SNPs) and two short insertion-deletions (InDels) (Table S2). In the genome of strain BLA3-E1, only alsT5, encoding a sodium:alanine symporter family protein, appeared to have two related mutations. The G at position 493 in alsT5 was replaced with A, resulting in the substitution of V165I in AlsT5. In addition, there was a 16 bp deletion at the 143 bp position upstream of alsT5, which may affect the expression of this gene. Interestingly, ald1 is not located far upstream of alsT5 implying that this gene is related to alanine transport (Figure S4). Therefore, we speculated that this transporter was responsible for the uptake d-alanine with sodium ions. The evolved strain BLA3-E1 improved the activity and expression of AlsT5 through an SNP and a short indel and could make better use of d-alanine released by other dead cells.
Another interesting mutation occurred in the dltB gene, which encodes teichoic acid d-alanyltransferase. Deletion of C at position 409 of dltB triggered a frameshift mutation and inactivated the gene. As part of the dlt operon, the dltB gene participates in the incorporation of d-alanine into teichoic acid and affects the net charge of the cell wall.31 Another study reported that knocking out gene dltB could reduce the production of poly-γ-glutamic acid in B. licheniformis.32 Inactivation of the dltB gene prevented the flow of d-alanine to pathways other than the cell wall synthesis pathway, which may be an important factor in improving the growth of strain BLA3-E1 in the seed medium without d-alanine.
In addition to the aforementioned mutations, SNP that occur in the mtnW gene also received our attention. As a RuBisCO-like protein, MtnW is considered a key enzyme in the methionine salvage pathway. Although MtnW function is difficult to relate to cellular d-alanine dependence, an SNP mutation in MtnW results in a G352V substitution at the substrate-binding site of the protein product (Figure S5), which may alter the enzyme activity or specificity.33 This may imply that MtnW affected the d-alanine dependence of strain BLA3 in an unexpected manner.
To further verify these hypotheses, genes alsT5, dltB, and mtnW were individually knocked out in strain BLA3 to obtain the recombinant strains BLA3ΔalsT5, BLA3ΔdltB, and BLA3ΔmtnW, respectively. The d-alanine dependence of the recombinant strains was characterized using the same method as strain BLA3. Consistent with our conjecture, knockout of alsT5 resulted in the weakened growth ability of the strain at low concentrations of d-alanine, and it was unable to grow in the medium with 0.01 g/L d-alanine (Figure 5A). This may be because the deletion of alsT5 restricted the transport of extracellular d-alanine into the cell. To further confirm the effect of this gene, alsT5 of strain BLA3-E1 was also knocked out. The obtained recombinant strain exhibited obvious growth inhibition when the d-alanine concentration was lower than 0.002 g/L and completely lost the ability to grow in the medium with d-alanine concentrations lower than 0.001 g/L (Figure 5B). This result further demonstrates the effect of AlsT5 on d-alanine dependence of strain BLA3. Compared with strain BLA3, strain BLA3ΔdltB significantly reduced d-alanine dependence (Figure 5C). Although the growth was not as efficient as that of strain BLA3 at a higher concentration of d-alanine, strain BLA3ΔdltB showed an obvious growth advantage when the concentration of d-alanine was lower than 0.01 g/L. In contrast, strain BLA3ΔdltB was able to grow in seed medium without d-alanine, but when the concentration of d-alanine was lower than 0.001 g/L, the growth rate was significantly lower than that of strain BLA3-E1. These results demonstrated the contribution of the dltB inactivation to the evolved strain BLA3-E1. Knockout of mtnW alone did not cause any change in strain BLA3 for d-alanine dependence, unexpectedly demonstrating that MtnW did not directly affect d-alanine dependence (Figure 5D). However, knockout of gene mtnW in strain BLA3ΔdltB improved the growth of the strain in 0.0001 g/L d-alanine or absence of d-alanine (Figure S6). Therefore, the effect of MtnW on d-alanine dependence of strain BLA3 may be indirect.
Figure 5.
Identification of the effects of alsT5, dltB, and mtnW on the exogenous d-alanine dependence of the alr1/alr2 double-knockout B. licheniformis
(A–D) Characterization of the d-alanine dependence on B. licheniformis BLA3ΔalsT5, BLA3-E1ΔalsT5, BLA3ΔdltB, and BLA3ΔmtnW by testing the growth performance in media containing various concentrations of d-alanine. Triplicate experiments were carried out for physiological measurements, and error bars represent standard deviations.
Discussion
As one of the 20 basic amino acids, l-alanine is an important industrial chemical with important applications in food, medicine, materials, and other fields. At present, E. coli is the most important l-alanine producer in the industry, with a high potential final titer and yield. In this study, we developed an efficient high-temperature l-alanine production microbial cell factory, and the final titer and yield of l-alanine reached 93.7 g/L and 0.81 g/g at 50°C, respectively. The level of l-alanine production achieved by our process was close to that has been reported in E. coli.8,9 In addition, our process can support high-temperature fermentation at 50°C, which has the dual advantages of reducing energy consumption and resisting bacterial contamination. Compared with the 37°C production process of E. coli, our technology increases the fermentation temperature by 13°C, thereby significantly reducing the cooling cost of the production process.12 At the same time, the fermentation condition of 50°C also effectively eliminated the contamination of mesophilic bacteria that could not tolerate high temperatures, thereby improving the robustness of the entire process. Finally, the strains used in the fermentation processes reported in this study were biosafe B. licheniformis,17 which is more easily recognized by society in terms of safety than E. coli. Therefore, our process offers additional industrial production advantages with the potential for large-scale industrialization.
The optical purity of l-alanine, a chiral molecule, is an important parameter for microbial fermentation. Since d-alanine is an essential component of bacterial cell walls,20 L-alanine production requires the prevention of d-alanine accumulation and secretion at the same time as providing sufficient d-alanine to support cell growth. This did not seem to be a difficult problem to solve in E. coli, since the remaining alanine racemase, Alr, after knocking out one alanine racemase, DadX, was sufficient to maintain cell growth without compromising the optical purity.8 In this study, we found that although B. licheniformis has two alanine racemases, this strain cannot obtain optically pure l-alanine by knocking out only one racemase like E. coli, but requires simultaneous inactivation of both enzymes. The obtained alr1/2 double-knockout strain had growth defects, and additional d-alanine was needed to restore growth. Other gram-positive bacteria face similar problem when engineered to produce l-alanine. Some studies solved this problem by supplementing additional d-alanine,34 others used resting cells to separate bacterial growth and alanine production.2,35 Compared to E. coli, the gram-positive bacteria have thicker cell wall and higher peptidoglycan content,36 and require a stronger d-alanine synthesis pathway for cell growth. Therefore, it is challenging for the gram-positive bacteria to achieve a balanced state of d-alanine biosynthesis to meet the needs of cell growth and prevent d-alanine accumulation.
The gram-positive bacterial growth usually depends on the d-alanine biosynthesis pathway catalyzed by alanine racemase, which leads to excessive accumulation of d-alanine. To solve this problem, the d-alanine dependence of the alanine racemase-deficient strain was relieved by adaptive laboratory evolution. Through a series of mutational analysis and verification experiments, we found the mechanism of the evolved strain BLA3-E1 removing the growth dependence on exogenous d-alanine (Figure 6). Activation of dormant alternative d-alanine synthesis pathway, which produces additional endogenous d-alanine, is an important strategy for BLA3-E1 to grow without exogenous d-alanine. B. licheniformis BLA3-E1 utilizes glucose and ammonium ions in the medium to synthesize l-glutamic acid, which can be converted to d-glutamic acid by glutamate racemase RacE. By activating the d-amino acid aminotransferase Dat1, d-glutamic acid in bacterial cells can be converted into d-alanine required for cell wall synthesis. In addition, the sodium:alanine symporter AlsT5, which was considered to be the predominant transporter responsible for d-alanine uptake, also plays an important role in improving the utilization of exogenous d-alanine derived from dead bacterial cells. Inactivation of teichoic acid d-alanyltransferase DltB in evolved strain reduces consumption of d-alanine by blocking the incorporation of d-alanine into teichoic acid. d-Alanine, which was originally used for the d-alanylation of teichoic acid can be used for the synthesis of cell wall peptidoglycan and improve the growth of BLA3-E1 without exogenous d-alanine. d-Alanylation of teichoic acid also have multiple effects on microbial cells, such as changing the cell surface charge to affect the recruitment of signaling molecules,32 increasing cell wall thickness,37 affecting enzyme secretion,38 membrane protein activity,32 and γ-polyglutamic acid production.32 This may explain the difference in growth between BLA3-E1 and BLA3. Finally, our study also found that RuBisCO-like 2,3-diketo-5-methylthiopentyl-1-phosphate enolase MtnW plays a role in d-alanine metabolism, which may help restore the adverse effects of DltB deletion on bacterial growth. However, the growth-improving mechanism of MtnW is unknown.
Figure 6.
Schematic of evolved strain BLA3-E1 removing the growth dependence on exogenous d-alanine
The elimination of alanine racemase in strain BLA3 blocks the endogenous d-alanine synthesis pathway, which results in the growth dependence on exogenous d-alanine. The evolved strain BLA3-E1 produces additional endogenous d-alanine by activating dormant alternative d-alanine synthesis pathway, improves the utilization of exogenous d-alanine by enhancing the expression of d-alanine transporter and reduces the consumption of d-alanine by blocking the incorporation of d-alanine into teichoic acid.
Many dormant metabolic pathways exist in microbial cells that are untapped treasure troves in synthetic biology. Activation of these dormant metabolic pathways can endow microbial cells with new functions and may serve as a novel strategy for solving complex metabolic problems. In this study, we activated the dormant d-alanine alternative synthesis pathway through laboratory adaptive evolution, thereby successfully releasing the growth dependence of the alanine racemase-deficient strains on exogenous d-alanine. The d-amino acid aminotransferase Dat1 was identified as a key enzyme in the d-alanine alternative synthesis pathway and direct overexpression of dat1 was able to activate this pathway, thereby relieving the growth dependence of alanine racemase-deficient strains on exogenous d-alanine. A similar alternative d-alanine synthesis pathway also exists in B. subtilis. It was reported that the d-amino acid aminotransferase encoded by the dat gene in alanine racemase-deficient B. subtilis was capable of converting other d-amino acids to d-alanine.30 This suggests that the alternative d-alanine synthesis pathway may be widespread in Bacillus spp. Our study showed that the paradoxical problem of d-alanine synthesis in l-alanine production can be solved by activating the dormant d-alanine alternative synthesis pathway. These results demonstrate the value of this strategy.
In this study, we have successfully constructed a high-temperature microbial cell factory capable of efficiently producing l-alanine by metabolic engineering. Subsequently, d-alanine dependence of the alanine racemase-deficient strain is also relieved by activating a dormant alternative pathway for d-alanine synthesis; d-amino acid aminotransferase Dat1 is involved in the d-alanine alternative synthesis pathway. In comparison to previously reported processes, our strategy realizes a high-temperature production of l-alanine for the first time. Importantly, we suggest that activating dormant metabolic pathway(s) may be an effective strategy for solving complex metabolic problems.
Limitation of the study
The process of high-temperature l-alanine developed in this study has strong potentials of high robustness and low energy consumption, being ready for industrialization. However, the fed-batch fermentation experiments were carried out in a 5-L bioreactor, and the performance on a larger scale needs to be further tested. In addition, the knockout of alanine racemase gene leads to decreased l-alanine production, which needs to be solved through subsequent studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| All strains used in this study were listed in Table S3 | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 5-FAM labeled d-alanine | Chinese Peptide | Cat#997300 |
| Clone-express Vltra One Step Cloning kit | Vazyme | Cat#C115-01 |
| Deposited data | ||
| Resequencing data | This study, NCBI Sequence Read Archive (SRA) database | NCBI SRA accession number: PRJNA893198 |
| Oligonucleotides | ||
| All primers used in this study were listed in Table S4 | Shanghai Generay Biotech | N/A |
| Recombinant DNA | ||
| All plasmids used in this study were listed in Table S3 | This study | N/A |
| The heterologous gene sequences used in this study were listed in Table S5 | BGI Geneland Scientific | N/A |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Ping Xu (pingxu@sjtu.edu.cn).
Materials availability
All the requests for the generated plasmids and strains should be directed to the lead contact and will be made available on request after completion of a Materials Transfer Agreement.
Experimental model and subject details
Strains, plasmids, and growth conditions
All strains and plasmids used in this study were listed in Table S3. B. licheniformis BN1111 was used as the starting strain for metabolic engineering. E. coli S17-1 was used as the conjugative donor strain. The temperature-sensitive shuttle plasmid pKVM139 was used for genetic manipulation in B. licheniformis. All of the E. coli strains were grown at 37°C in Luria-Bertani (LB) broth (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl). The LB broth was also regarded as the seed medium for B. licheniformis. The fermentation medium of the following composition was used in this study: 100 g/L glucose, 5 g/L yeast extract, 5 g/L (NH4)2SO4, 1.3 g/L K2HPO4·3H2O, 0.5 g/L KH2PO4, 0.5 g/L MgSO4·7H2O, 0.02 g/L FeSO4·7H2O, 0.02 g/L MnSO4·4H2O. Appropriate concentrations of d-alanine (1 g/L), ampicillin (100 mg/L), erythromycin (5 mg/L), and polymyxin B (40 mg/L) were also added to the medium if necessary.
Method details
DNA manipulation and strain construction
All primers used in this study were listed in Table S4. The heterologous gene sequences used in this study were listed in Table S5. To introduce the pfkA and pyk genes from B. coagulans with the strong promoter Pals, the DNA fragment PFYAK containing these two genes and Pals were amplified using the primer pairs Pals-PFYAK-F/BLAldh-PFYAK-R and BLAldh-Pals-F/PFYAK-Pals-R. To insert the PFYAK fragment into the site where the ldh gene was knockout, the upstream and downstream of original ldh gene were also amplified using the primer pairs PKVM-BLAldh-UpF/Pals-BLAldh-UpR and PFYAK-BLAldh-DnF/PKVM-BLAldh-DnR. These DNA fragments were ligated together by recombinant PCR and integrated into the EcoRI/BamHI site of pKVM1, resulting in pKVM1-PFYAK. To overexpress the ald1, ald2, GSald, STald, BSald, and dat1 genes, the plasmids pKVM1-ald1, pKVM1-ald2, pKVM1-GSald, pKVM1-STald, pKVM1-BSald, and pKVM1-dat1 were constructed using the similar method to the PFYAK fragment. To delete the ldhTi gene, the upstream and downstream of ldhTi gene were amplified using the primer pairs BLAldhTi-UpF/BLAldhTi-UpR and BLAldhTi-DnF/PKVM-BLAldhTi-DnR. These DNA fragments were ligated together by recombinant PCR and integrated into the EcoRI/BamHI site of pKVM1, resulting in pKVM1ΔldhTi. To delete the alr1, alr2, dltB, mtnW, alsT5, and dat1 genes, the plasmids pKVM1Δalr1, pKVM1Δalr2, pKVM1ΔdltB, pKVM1ΔmtnW, pKVM1ΔalsT5, and pKVM1Δdat1 were constructed using the similar method to the ldhTi gene. All of the plasmids were subsequently transformed directly into E. coli S17-1.
E. coli S17-1 harboring plasmids and B. licheniformis were cultured to an OD600 of 1.2, centrifuged at 6,000 rpm for 5 min and washed twice with PBS solution, mixed and added dropwise to LB plates at a ratio of 7:1. After overnight culture at 30°C, the strains were screened on LB plates with 5 g/mL erythromycin and 40 g/mL polymyxin B at 30°C. The screened transformants were cultured at 37°C in LB medium containing erythromycin, and then cultured at 50°C in LB medium. The single-crossover transformants were obtained after screening on LB plates containing erythromycin. Single-crossover transformants were continuously cultured in LB medium at 30°C for two generations, diluted and spread on LB plates, cultured at 37°C overnight. Transformants that lost erythromycin resistance were selected for PCR verification, and the successfully constructed B. licheniformis strains were obtained.
Fed-batch fermentation
The fed-batch fermentation experiments were carried out in a 5-L bioreactor containing 3 L fermentation medium. The fermentation temperature was controlled at 50°C and the inoculum volume was 5% (v/v). The culture pH was maintained at 7.0 by adding 25% (v/v) NH3·H2O. The aeration rate was set to 3 L/min (1.0 vvm) and the initial speed of bioreactor was set to 300 rpm. When the OD600 value reached 6.0–8.0, stop aeration and reduce the speed of the bioreactor to 80 rpm. The glucose concentration was maintained between 30 g/L and 100 g/L by adding glucose powder. The final and intermediate samples were collected to determine the OD600 value, optimal purity and concentrations of glucose, l-alanine, and other by-products.
Adaptive laboratory evolution
The adaptive laboratory evolution was aimed to improve the growth defect of B. licheniformis BLA3 without d-alanine. B. licheniformis BLA3 was first inoculated into LB medium without d-alanine, demonstrating that the strain was unable to grow without d-alanine. Subsequently, B. licheniformis BLA3 was cultured at 50°C in LB medium containing 0.02 g/L d-alanine. When the OD600 value of B. licheniformis BLA3 exceeded 3.0, the concentration of d-alanine in the medium was reduced to 0.01 g/L. By the same method, the concentration of d-alanine in the medium was sequentially decreased to 0.005 g/L, 0.002 g/L, 0.001 g/L, 0.0005 g/L, and 0.0002 g/L. Evolved B. licheniformis were simultaneously inoculated into LB medium without d-alanine at each time the concentration of d-alanine was decreased. When the OD600 value of B. licheniformis could exceed 3.0 without d-alanine, the adaptive laboratory evolution experiment was stopped, and the evolved strain BLA-E1 was obtained.
d-Alanine dependence of alr1/alr2 double-knockout strains
To test the d-Alanine dependence, the alr1/alr2 double-knockout strains were first cultured at 50°C in LB medium containing 1 g/L of d-Alanine. The activated strains were centrifuged at 6,000 rpm for 5 min and washed three times with fresh LB medium. Then appropriate volumes of fresh LB mediums were added to obtain the seed culture solutions, so that the OD600 value of the culture solutions were 5.0. The seed culture solutions were inoculated into LB medium containing 0.1 g/L, 0.05 g/L, 0.02 g/L, 0.01 g/L, 0.005 g/L, 0.002 g/L, 0.001 g/L, 0.0005 g/L, 0.0002 g/L, 0.0001 g/L, and 0 g/L according to the inoculation amount of 1%, respectively, and cultured at 50°C. Each concentration contained three biological replicates. The 12 h, 24 h, and 36 h samples were collected to determine the OD600 value.
Genome annotation, mining of thermostable alanine dehydrogenase, and genome resequencing analysis
The B. licheniformis ATCC 14580 genome sequence was download from the NCBI database (GenBank assembly accession number: GCA_000011645.1). The B. licheniformis ATCC 14580 genome was annotated by Prokka.40 The thermostable alanine dehydrogenases were mined by the co-evolution mining approach previously developed by our group. The Genbank protein IDs of alanine dehydrogenase Ald1, alanine dehydrogenase Ald2, d-lactate dehydrogenase LdhTi, alanine dehydrogenase GSAld, alanine dehydrogenase BSAld, alanine dehydrogenase STAld, alanine racemase Alr1, alanine racemase Alr2, d-α-glycerophosphatase Dgp, d-amino acid aminotransferase Dat1, glutamate racemase RacE, sodium:alanine symporter AlsT5, teichoic acid d-alanyltransferase DltB, and 2,3-diketo-5-methylthiopentyl-1-phosphate enolase MtnW are AAU24844.1, AAU25710.1, WP_013906894.1, RLP89197.1, AKP47217.1, KWX03169.1, AAU22152.2, AAU24747.1, AAU25694.1, AAU22605.1, AAU24493.1, AAU24842.2, AAU25521.1, and AAU23062.1, respectively.
B. licheniformis cells at the stationary phases were collected for DNA isolation. DNA isolation, DNA library construction, high-throughput sequencing (Illumina), genome assembly and subsequent bioinformatics analysis were performed at Guangdong Magigene Biotechnology Co., Ltd. (Guangdong, China). To construct DNA libraries, the extracted DNA samples were randomly interrupted to generate DNA fragments of desired length. The sticky ends formed by the break were repaired into blunt ends and the base A was added at the 3' end, so that the DNA fragments were able to be ligated with a special adaptor with a T base at the 3' end. Finally, PCR technology was used to amplify the DNA fragments with adapters at both ends to obtain the DNA libraries. Afterwards, the constructed qualified libraries were clustered and sequenced on the machine. All of the raw sequencing reads were filtered to remove low quality reads, resulting in clean data. The Prokka-annotated B. licheniformis ATCC 14580 genome was used as the reference genome. Resequencing data have been deposited in the NCBI SRA database (NCBI SRA accession number: PRJNA893198). The SNPs and InDels were obtained by aligning clean data with reference genome sequence by SAMTOOLS.41 The genomic structural variants (SVs) were detected by BreakDancer.42
Characterization of cell wall synthesis involving extracellular d-alanine based on 5-FAM labeled d-alanine
5-FAM labeled d-alanine was used to characterize the biological process of cell wall synthesis involving extracellular d-alanine.29 All of the B. licheniformis were cultured in LB medium with 1 g/L d-alanine till the OD600 value reached 1.2. The cultures were centrifuged at 6,000 rpm for 5 min, washed three times with fresh LB medium, resuspended in fresh LB medium, and separated into three aliquots (1 mL each). 5-FAM labeled d-alanine was also added to the aliquots to a final concentration of 300 μM. The aliquots were cultured at 50°C for 2 h, washed five times with phosphate buffered saline (PBS) and resuspended in PBS. After being diluted to appropriate concentrations, the fluorescence intensities of FAM-labeled samples were detected by CytoFLex (Beckman Coulter Co., Ltd) excited with 488 nm laser.
Analytical methods
The cell density was determined by measuring the optical density at a wavelength of 600 nm (OD600). The concentration of glucose was estimated by an SBA-40D biosensor analyzer. The fermentation samples were heated at 100°C, diluted to an appropriate multiple, and centrifuged at 8,000 rpm for 10 min. Filtered supernatant can be used for subsequent detection. The concentration of alanine was determined by a high-performance liquid chromatography (HPLC) equipped with a ZORBAX Eclipse XDB-C18 column and a diode array detector (DAD) at 210 nm. A solution containing 0.54 g/L of sodium 1-octanesulfonate and 10% methanol was used as the mobile phase, and the pH value was adjusted to 2.1 with phosphoric acid. The column temperature and flow rate were set at 30°C and 0.8 ml/min, respectively. The optical purity of l-alanine was assayed by HPLC using a SUMICHIRAL OA-5000 column and a DAD at 254 nm. Analysis was performed with a mobile phase of 2 mM CuSO4 at a flow rate of 0.5 mL/min at 30°C. Other organic acid by-products were detected by HPLC using a Bio-Rad Aminex HPX-87H column and a DAD at 210 nm or a refractive index detector (RID). Analysis was performed with a mobile phase of 5 mM H2SO4 at a flow rate of 0.5 mL/min at 55°C. The concentrations of free amino acids were detected by L-8900 automatic amino acid analyzer.
Quantification and statistical analysis
The flask experiments to test the d-Alanine dependence contained three biological replicates. The characterization experiments of cell wall synthesis involving extracellular d-alanine based on 5-FAM labeled d-alanine also contained three biological replicates. The fed-batch fermentations of B. licheniformis BLA3 and BLA4 were validated in the laboratory in multiple replicates. All of the results reported in the article are reproducible. Data are given as the means ± SD.
Acknowledgments
This study was supported by the grants (22138007, 32170105, and 31870088) from National Natural Science Foundation of China.
Author contributions
F.T., X.H., and P.X. designed the experiments. X.H., J.L., Y.W., and Y.Y. performed the experiments. X.H. wrote the manuscript. X.H., F.T., and P.X. revised the manuscript. P.X. and F.T. conceived the projects.
Declaration of interests
The authors declare no competing interests.
Published: March 15, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106397.
Contributor Information
Fei Tao, Email: taofei@sjtu.edu.cn.
Ping Xu, Email: pingxu@sjtu.edu.cn.
Supplemental information
Data and code availability
The resequencing data have been deposited at the NCBI Sequence Read Archive (SRA) database and are publicly available as of the data of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Liu P., Xu H., Zhang X. Metabolic engineering of microorganisms for l-alanine production. J. Ind. Microbiol. Biotechnol. 2022;49 doi: 10.1093/jimb/kuab057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hols P., Kleerebezem M., Schanck A.N., Ferain T., Hugenholtz J., Delcour J., de Vos W.M. Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering. Nat. Biotechnol. 1999;17:588–592. doi: 10.1038/9902. [DOI] [PubMed] [Google Scholar]
- 3.Khattab S.N., Massoud M.I., Jad Y.E.S., Bekhit A.A., El-Faham A. Production and physicochemical assessment of new stevia amino acid sweeteners from the natural stevioside. Food Chem. 2015;173:979–985. doi: 10.1016/j.foodchem.2014.10.093. [DOI] [PubMed] [Google Scholar]
- 4.Li T., Cui X., Cui Y., Sun J., Chen Y., Zhu T., Li C., Li R., Wu B. Exploration of transaminase diversity for the oxidative conversion of natural amino acids into 2-ketoacids and high-value chemicals. ACS Catal. 2020;10:7950–7957. doi: 10.1021/acscatal.0c01895. [DOI] [Google Scholar]
- 5.Porcellati F., Pampanelli S., Rossetti P., Busciantella Ricci N., Marzotti S., Lucidi P., Santeusanio F., Bolli G.B., Fanelli C.G. Effect of the amino acid alanine on glucagon secretion in non-diabetic and type 1 diabetic subjects during hyperinsulinaemic euglycaemia, hypoglycaemia and post-hypoglycaemic hyperglycaemia. Diabetologia. 2007;50:422–430. doi: 10.1007/s00125-006-0519-6. [DOI] [PubMed] [Google Scholar]
- 6.Bonillo Martínez A.D., Galán I.C.R., Bellver M.V.M. Application of a biodegradable polyesteramide derived from l-alanine as novel excipient for controlled release matrix tablets. AAPS PharmSciTech. 2017;18:3286–3295. doi: 10.1208/s12249-017-0809-y. [DOI] [PubMed] [Google Scholar]
- 7.Dave U.C., Kadeppagari R.K. Alanine dehydrogenase and its applications - a review. Crit. Rev. Biotechnol. 2019;39:648–664. doi: 10.1080/07388551.2019.1594153. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X., Jantama K., Moore J.C., Shanmugam K.T., Ingram L.O. Production of l-alanine by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2007;77:355–366. doi: 10.1007/s00253-007-1170-y. [DOI] [PubMed] [Google Scholar]
- 9.Zhou L., Deng C., Cui W.J., Liu Z.M., Zhou Z.M. Efficient l-alanine production by a thermo-regulated switch in Escherichia coli. Appl. Biochem. Biotechnol. 2016;178:324–337. doi: 10.1007/s12010-015-1874-x. [DOI] [PubMed] [Google Scholar]
- 10.Arbab S., Ullah H., Khan M.I.U., Khattak M.N.K., Zhang J., Li K., Hassan I.U. Diversity and distribution of thermophilic microorganisms and their applications in biotechnology. J. Basic Microbiol. 2022;62:95–108. doi: 10.1002/jobm.202100529. [DOI] [PubMed] [Google Scholar]
- 11.Li C., Tao F., Xu P. Carbon flux trapping: highly efficient production of polymer-grade d-lactic acid with a thermophilic d-lactate dehydrogenase. Chembiochem. 2016;17:1491–1494. doi: 10.1002/cbic.201600288. [DOI] [PubMed] [Google Scholar]
- 12.Abdel-Banat B.M.A., Hoshida H., Ano A., Nonklang S., Akada R. High-temperature fermentation: how can processes for ethanol production at high temperatures become superior to the traditional process using mesophilic yeast? Appl. Microbiol. Biotechnol. 2010;85:861–867. doi: 10.1007/s00253-009-2248-5. [DOI] [PubMed] [Google Scholar]
- 13.Han X., Hong F., Liu G., Bao J. An approach of utilizing water-soluble carbohydrates in lignocellulose feedstock for promotion of cellulosic l-lactic acid production. J. Agric. Food Chem. 2018;66:10225–10232. doi: 10.1021/acs.jafc.8b03592. [DOI] [PubMed] [Google Scholar]
- 14.Prajapati B.P., Kumar Suryawanshi R., Agrawal S., Ghosh M., Kango N. Characterization of cellulase from Aspergillus tubingensis NKBP-55 for generation of fermentable sugars from agricultural residues. Bioresour. Technol. 2018;250:733–740. doi: 10.1016/j.biortech.2017.11.099. [DOI] [PubMed] [Google Scholar]
- 15.Jiang S., Xu P., Tao F. l-Lactic acid production by Bacillus coagulans through simultaneous saccharification and fermentation of lignocellulosic corncob residue. Bioresour. Technol. Rep. 2019;6:131–137. doi: 10.1016/j.biteb.2019.02.005. [DOI] [Google Scholar]
- 16.Kong X., Zhang B., Hua Y., Zhu Y., Li W., Wang D., Hong J. Efficient l-lactic acid production from corncob residue using metabolically engineered thermo-tolerant yeast. Bioresour. Technol. 2019;273:220–230. doi: 10.1016/j.biortech.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 17.Li L., Zhang L., Li K., Wang Y., Gao C., Han B., Ma C., Xu P. A newly isolated Bacillus licheniformis strain thermophilically produces 2,3-butanediol, a platform and fuel bio-chemical. Biotechnol. Biofuels. 2013;6:123. doi: 10.1186/1754-6834-6-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu Y., Zhang J., Li L., Wen Z., Nomura C.T., Wu S., Chen S. Engineering Bacillus licheniformis for the production of meso-2,3-butanediol. Biotechnol. Biofuels. 2016;9:117. doi: 10.1186/s13068-016-0522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai D., Zhu J., Zhu S., Lu Y., Zhang B., Lu K., Li J., Ma X., Chen S. Metabolic engineering of main transcription factors in carbon, nitrogen, and phosphorus metabolisms for enhanced production of bacitracin in Bacillus licheniformis. ACS Synth. Biol. 2019;8:866–875. doi: 10.1021/acssynbio.9b00005. [DOI] [PubMed] [Google Scholar]
- 20.Milligan D.L., Tran S.L., Strych U., Cook G.M., Krause K.L. The alanine racemase of Mycobacterium smegmatis is essential for growth in the absence of d-alanine. J. Bacteriol. 2007;189:8381–8386. doi: 10.1128/JB.01201-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Souza J.M., Freire P.T.C., Bordallo H.N., Argyriou D.N. Structural isotopic effects in the smallest chiral amino acid: observation of a structural phase transition in fully deuterated alanine. J. Phys. Chem. B. 2007;111:5034–5039. doi: 10.1021/jp070366z. [DOI] [PubMed] [Google Scholar]
- 22.Trevors J.T. Genome size in bacteria. Antonie Leeuwenhoek. 1996;69:293–303. doi: 10.1007/BF00399618. [DOI] [PubMed] [Google Scholar]
- 23.DeRisi J.L., Iyer V.R., Brown P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 24.Dong Y., Cui C.B., Li C.W., Hua W., Wu C.J., Zhu T.J., Gu Q.Q. Activation of dormant secondary metabolite production by introducing neomycin resistance into the deep-sea fungus, Aspergillus versicolor ZBY-3. Mar. Drugs. 2014;12:4326–4352. doi: 10.3390/md12084326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryu S., Trinh C.T. Understanding functional roles of native pentose-specific transporters for activating dormant pentose metabolism in Yarrowia lipolytica. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.02146-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S., Ihara K., Katsube S., Hori H., Ando T., Isogai E., Yoneyama H. Characterization of the l-alanine exporter AlaE of Escherichia coli and its potential role in protecting cells from a toxic-level accumulation of l-alanine and its derivatives. Microbiologyopen. 2015;4:632–643. doi: 10.1002/mbo3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y., Liu J., Han X., Meng X., Li M., Wang J., Xue H., Yang Y., Xu P., Tao F. Eliminating host-guest incompatibility via enzyme mining enables the high-temperature production of N-acetylglucosamine. iScience. 2023;26 doi: 10.1016/j.isci.2022.105774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song C.W., Rathnasingh C., Song H. CRISPR-Cas9 mediated metabolic engineering of a mucoid Bacillus licheniformis isolate for mass production of 2, 3-butanediol. Biochem. Eng. J. 2021;175 doi: 10.1016/j.bej.2021.108141. [DOI] [Google Scholar]
- 29.Lin L., Song J., Du Y., Wu Q., Gao J., Song Y., Yang C., Wang W. Quantification of bacterial metabolic activities in the gut by d-amino acid-based in vivo labeling. Angew. Chem. Int. Ed. Engl. 2020;59:11923–11926. doi: 10.1002/anie.202004703. [DOI] [PubMed] [Google Scholar]
- 30.Sidiq K.R., Chow M.W., Zhao Z., Daniel R.A. Alanine metabolism in Bacillus subtilis. Mol. Microbiol. 2021;115:739–757. doi: 10.1111/mmi.14640. [DOI] [PubMed] [Google Scholar]
- 31.Abi Khattar Z., Rejasse A., Destoumieux-Garzón D., Escoubas J.M., Sanchis V., Lereclus D., Givaudan A., Kallassy M., Nielsen-Leroux C., Gaudriault S. The dlt operon of Bacillus cereus is required for resistance to cationic antimicrobial peptides and for virulence in insects. J. Bacteriol. 2009;191:7063–7073. doi: 10.1128/JB.00892-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He P., Wan N., Cai D., Hu S., Chen Y., Li S., Chen S. 13C-Metabolic flux analysis reveals the metabolic flux redistribution for enhanced production of poly-γ-glutamic acid in dlt over-expressed Bacillus licheniformis. Front. Microbiol. 2019;10:105. doi: 10.3389/fmicb.2019.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura H., Saito Y., Ashida H., Kai Y., Inoue T., Yokota A., Matsumura H. Structure of the apo decarbamylated form of 2,3-diketo-5-methylthiopentyl-1-phosphate enolase from Bacillus subtilis. Acta Crystallogr. D Biol. Crystallogr. 2009;65:942–951. doi: 10.1107/S0907444909022422. [DOI] [PubMed] [Google Scholar]
- 34.Hashimoto S.I., Katsumata R. l-Alanine fermentation by an alanine racemase-deficient mutant of the dl-alanine hyperproducing bacterium Arthrobacter oxydans HAP-1. J. Ferment. Bioeng. 1998;86:385–390. doi: 10.1016/S0922-338X(99)89009-6. [DOI] [Google Scholar]
- 35.Yamamoto S., Gunji W., Suzuki H., Toda H., Suda M., Jojima T., Inui M., Yukawa H. Overexpression of genes encoding glycolytic enzymes in Corynebacterium glutamicum enhances glucose metabolism and alanine production under oxygen deprivation conditions. Appl. Environ. Microbiol. 2012;78:4447–4457. doi: 10.1128/AEM.07998-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohde M. The Gram-positive bacterial cell wall. Microbiol. Spectr. 2019;7 doi: 10.1128/microbiolspec.GPP3-0044-2018. GPP3–0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin J., Meng Q., Cheng D., Fu J., Luo Q., Liu Y., Yu Z. Mechanisms of bactericidal action and resistance of polymyxins for Gram-positive bacteria. Appl. Microbiol. Biotechnol. 2020;104:3771–3780. doi: 10.1007/s00253-020-10525-y. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y., Cai D., He P., Mo F., Zhang Q., Ma X., Chen S. Enhanced production of heterologous proteins by Bacillus licheniformis with defective d-alanylation of lipoteichoic acid. World J. Microbiol. Biotechnol. 2018;34:135. doi: 10.1007/s11274-018-2520-x. [DOI] [PubMed] [Google Scholar]
- 39.Rachinger M., Bauch M., Strittmatter A., Bongaerts J., Evers S., Maurer K.H., Daniel R., Liebl W., Liesegang H., Ehrenreich A. Size unlimited markerless deletions by a transconjugative plasmid-system in Bacillus licheniformis. J. Biotechnol. 2013;167:365–369. doi: 10.1016/j.jbiotec.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 40.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 41.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen K., Wallis J.W., McLellan M.D., Larson D.E., Kalicki J.M., Pohl C.S., McGrath S.D., Wendl M.C., Zhang Q., Locke D.P., et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The resequencing data have been deposited at the NCBI Sequence Read Archive (SRA) database and are publicly available as of the data of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.