Abstract
Photosynthetic algae have evolved mechanisms to cope with suboptimal light and CO2 conditions. When light energy exceeds CO2 fixation capacity, Chlamydomonas reinhardtii activates photoprotection, mediated by LHCSR1/3 and PSBS, and the CO2 Concentrating Mechanism (CCM). How light and CO2 signals converge to regulate these processes remains unclear. Here, we show that excess light activates photoprotection- and CCM-related genes by altering intracellular CO2 concentrations and that depletion of CO2 drives these responses, even in total darkness. High CO2 levels, derived from respiration or impaired photosynthetic fixation, repress LHCSR3/CCM genes while stabilizing the LHCSR1 protein. Finally, we show that the CCM regulator CIA5 also regulates photoprotection, controlling LHCSR3 and PSBS transcript accumulation while inhibiting LHCSR1 protein accumulation. This work has allowed us to dissect the effect of CO2 and light on CCM and photoprotection, demonstrating that light often indirectly affects these processes by impacting intracellular CO2 levels.
Subject terms: Non-photochemical quenching, Plant signalling, Light responses
Photosynthetic algae have evolved to survive in suboptimal light and CO2 conditions. Here, the authors show that depletion of CO2 can drive photoprotection and carbon acquisition even in the absence of light, that was previously believed to be indispensable for the activation of these processes.
Introduction
A major challenge for photosynthetic organisms is to efficiently acclimate to highly dynamic light and nutrient conditions that occur in natural environments. While light provides the energy that fuels photosynthetic CO2 fixation, excess light can cause oxidative damage and ultimately result in cell death. Therefore, light absorption must be precisely managed via photoprotective mechanisms that help integrate the use of light energy with CO2 availability and the potential of the organism to grow and store fixed carbon. A dominant photoprotective mechanism, called qE (energy-dependent quenching), results in the harmless dissipation of excess absorbed light energy as heat1,2. Triggering qE requires the synthesis of specific proteins and pigments that are controlled both transcriptionally and post-transcriptionally.
In the green microalga Chlamydomonas reinhardtii (hereafter Chlamydomonas), qE depends on the nucleus-encoded, chloroplast-localized Light Harvesting Complex-Stress Related (LHCSR) proteins LHCSR1, LHCSR3 and Photosystem II Subunit S, PSBS, which are present in many algae and lower plants3 and belong to the Light Harvesting Complex protein superfamily4. The LHCSR3.1 and LHCSR3.2 genes in Chlamydomonas encode identical LHCSR3 proteins5, while PSBS1 and PSBS2 encode proteins that differ by only one amino acid of the chloroplast transit peptide6. While LHCSR1 and LHCSR3 are present in algae but not in vascular plants, PSBS is present in both4. PSBS in Chlamydomonas is transiently expressed in cells exposed to high light (HL)6,7 and accumulates in cells exposed to UV-B irradiation8. LHCSR3 is the main qE effector protein in HL5, although LHCSR1 can significantly contribute to qE under certain conditions9. In Chlamydomonas, expression of LHCSR3 has been reported to increase upon absorption of blue-light by the photoreceptor phototropin (PHOT1)10 and involves calcium ion signaling11, active photosynthetic electron transport (PET)10,11 and the transcriptional factor CONSTANS, which is also required for activation of the LHCSR1 and PSBS genes12,13.
Similar to the dynamic light cue, the concentration of inorganic carbon (HCO3−, CO2 and CO32−, together designated Ci) in aquatic environments varies spatially and temporally; aquatic CO2 levels can also fluctuate from extremely high (hyper-saturated) to extremely low14. Because low CO2 levels limit photoautotrophic growth, microalgae have evolved a CO2 Concentrating Mechanism (CCM) that elevates the level of CO2 at the site of fixation by Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO). Major components of the CCM are carbonic anhydrases (CAH), which facilitate interconversions among the different Ci species, and Ci transporters. The genes encoding many Ci transporters and CAHs are under the control of the zinc-finger type potential transcription regulator CIA5 (also CCM1)15,16, which is localized in the nucleus17 and controls expression of low-CO2 responsive genes.
In addition to the use of CO2 to support phototrophic growth in the light, Chlamydomonas can also use the two-carbon molecule acetate either in the dark to support heterotrophic growth, or in the light, to support photoheterotrophic or mixotrophic growth18. Acetate is incorporated into acetyl-CoA either in a one-step reaction catalyzed by acetyl-CoA synthetase (ACS), or in two steps that use acetate kinase (ACK) and phosphate acetyltransferase (PAT), which sequentially catalyze the formation of acetyl-phosphate and acetyl-CoA19. Acetyl-CoA can then enter the glyoxylate cycle, a shunt of the tricarboxylic acid (TCA) cycle20, recently characterized in Chlamydomonas21, where it can be converted to metabolites that are used for anabolic metabolism. Alternatively, acetyl-CoA enters the TCA cycle to feed the respiratory chain with reducing equivalents. Both, the glyoxylate cycle and respiration are essential for growth in the dark since Chlamydomonas mutants affected in either of these processes are unable to grow heterotrophically21,22.
Despite the evident connection between light and CO2 levels, the physiological responses to different light and CO2 availabilities have been traditionally studied separately. However, several lines of evidence indicate that both acetate and Ci abundance impact not only qE but also the establishment of the CCM in Chlamydomonas23–26, while LHCSR3 transcripts accumulation have been reported to be CIA5-dependent26–28. Yet, the mechanism(s) associated with carbon-dependent regulation of qE and CCM induction and the intimate link between the two processes have still not been clearly defined.
Here, using genetic and mathematical modelling approaches, we demonstrate that inhibition of LHCSR3 accumulation and CCM activity by acetate is at the level of transcription and a consequence of metabolically produced CO2. We also show that exposure of Chlamydomonas to HL triggers not only HL responses, but also low-CO2 responses, and we report the discovery of a novel CO2- and CIA5-dependent mechanism that activates LHCSR3 gene expression even in complete darkness. Finally, we propose that PET is critical for the activation of LHCSR3 transcription because it sustains CO2 fixation, consuming intracellular CO2 and thereby relieving its inhibitory effect. This work emphasizes the importance of CO2 in regulating photoprotection and the CCM, and demonstrates that light often indirectly affects these processes by altering intracellular CO2 levels.
Results
CO2 generated from acetate metabolism inhibits LHCSR3
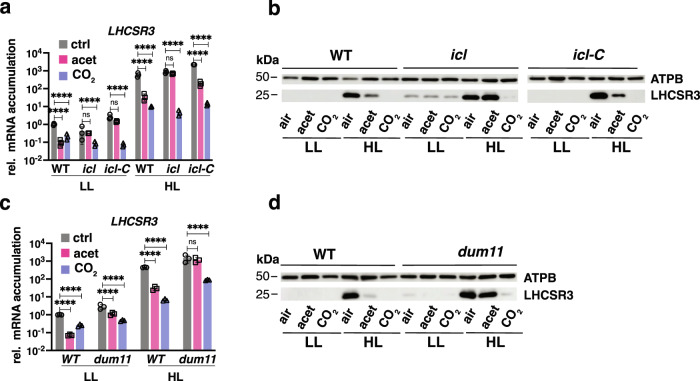
To gain insights into the effect of carbon metabolism on photoprotection, we explored the impact of acetate and high CO2 on LHCSR3 mRNA and protein levels in wild-type (WT) cells and in two mutants impaired in acetate metabolism; the icl mutant, which lacks isocitrate lyase, a key enzyme of the glyoxylate cycle21, and the dum11 mutant, which is defective in the ubiquinol cytochrome c oxidoreductase (respiratory complex III)29. The presence of acetate in the medium of WT cells inhibited the accumulation of the LHCSR3 transcript (Fig. 1a, note the logarithmic scale) in both low light (LL) and high light (HL) conditions. No protein was detected in WT under any condition in LL, but inhibition by acetate was apparent in HL (Fig. 1b), as previously reported25. However, in the icl mutant, acetate had no inhibitory effect on the accumulation of LHCSR3 mRNA (Fig. 1a) or protein (Fig. 1b) in either HL or LL, while the icl::ICL-complemented line, designated icl-C, exhibited similar behavior to that of WT cells (Fig. 1a, b). Additionally, acetate did not alter LHCSR3 transcript or protein accumulation in HL-treated dum11 mutant cells (Fig. 1c, d), while under LL, acetate inhibited LHCSR3 transcript in the dum11 mutant but to a much smaller extent than in WT (Fig. 1c). Together, these results suggest that the acetate administered to the cells must be metabolized for it to have a suppressive effect on the accumulation of LHCSR3 transcript and protein in HL. We also sparged WT, icl, icl-C and dum11 cells with 5% CO2 both in LL and HL. CO2 strongly repressed the accumulation of LHCSR3 mRNA and protein in all genotypes, including the metabolic mutants icl and dum11 for which expression of LHCSR3 was unaffected by acetate (Fig. 1a-d).
Fig. 1. Acetate needs to be metabolized to inhibit LHCSR3 accumulation.
WT, icl, icl-C and dum11 strains were acclimated for 16 h in LL (15 µmol photons m−2 s−1) in HSM; sparged with air (labelled as “air”); sparged with air and supplemented with 10 mM sodium acetate (labelled as “acet”); sparged with air enriched with 5% CO2 (labelled as “CO2”). After sampling for the LL conditions, light intensity was increased to 600 µmol photons m−2 s−1 (HL); samples were taken 1 h (RNA) or 4 h (protein) after exposure to HL. a, c. Accumulation of LHCSR3 mRNA at the indicated conditions normalized to WT LL ctrl (n = 3 biological samples, mean ± s.d.). The p-values for the comparisons of acetate and CO2 conditions to air are based on ANOVA Dunnett’s multiple comparisons test of log10 transformed mRNA data as indicated in the graphs (*P < 0.005, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant). Exact p-values can be found at the Source Data file. b, d. Immunoblot analyses of LHCSR3 and ATPB (loading control) under the indicated conditions. Representative datasets of experiments repeated three times.
We evaluated the impact of carbon availability on the photosynthetic properties of cells. The presence of acetate in the medium of WT cells enhanced photosynthetic electron transport (rETR) and strongly suppressed qE (Supplementary Fig. 1). In the icl mutant, acetate enhanced the extent of rETR only by ~10% compared with 60% for WT cells. Additionally, acetate caused less pronounced suppression of qE in the icl mutant (by 40%) compared to the level of suppression in WT cells (by 95%); icl-C, behaved similarly to WT cells. As expected, CO2 enhanced rETR and suppressed qE in WT, icl and icl-C (Supplementary Fig. 1).
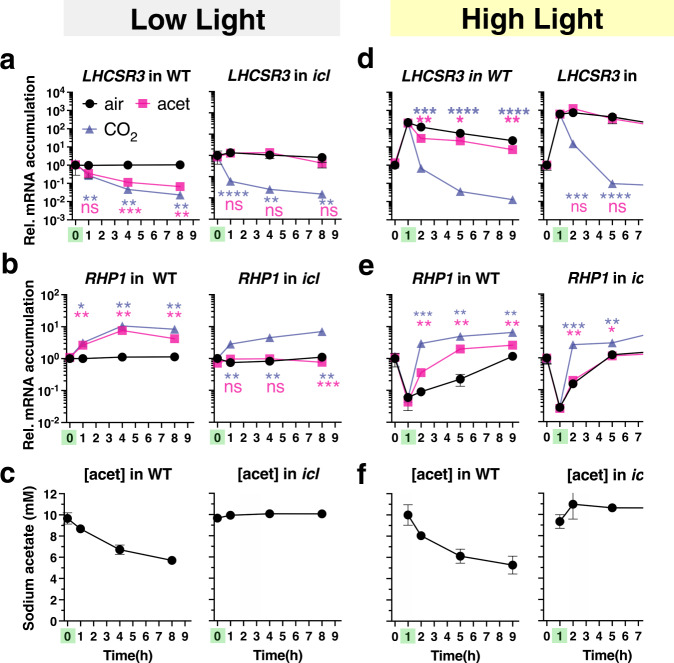
The similarity between the impact of acetate and 5% CO2 on LHCSR3 expression in WT and icl-C cells (Fig. 1a) as well as on their photosynthetic properties (Supplementary Fig. 1) raised the possibility that both treatments elicited a common mechanism of LHCSR3 control, possibly reflecting a change in the CO2 concentration within the cell or growth medium. This possibility is plausible based on the finding that acetate metabolism leads to the generation of CO230. To investigate whether the generation of CO2 via acetate metabolism can explain the repression of LHCSR3 transcript and protein levels, we monitored the levels of transcripts from the RHP1 gene in the mutant and WT cells; RHP1 (aka RH1) encodes a CO2 channel shown to be CO2 responsive and to accumulate in cells grown in a high CO2 atmosphere31. Acetate or 5% CO2 were introduced to WT and icl mutant cells acclimated in LL and air and the levels of the LHCSR3 and RHP1 transcripts were assayed over a period of 8 h in LL (Fig. 2a, b). The LHCSR3 transcript accumulation patterns observed agreed with the findings presented in Fig. 1a (LL panel). In WT cells, acetate and CO2 caused a reduction in LHCSR3 mRNA accumulation over the LL period relative to the control (no acetate, air). Additionally, in the icl mutant, acetate did not affect the accumulation of this transcript while CO2 efficiently repressed the LHCSR3 transcript level (Fig. 2a). Under these experimental conditions, acetate levels in the medium decreased in WT cultures but remained unchanged in cultures of the icl mutant (Fig. 2c). Lastly, RHP1expression increased in WT and icl mutant cells when the culture was sparged with CO2, but only in the WT cells when the cultures were not sparged with CO2 and only supplemented with acetate, suggesting that acetate metabolism resulted in higher intracellular CO2 levels (Fig. 2b).
Fig. 2. LHCSR3 inhibition is driven by CO2 derived from the metabolism of acetate.
Experiment at LL: a, b mRNA accumulation of LHCSR3 and RHP1 and c concentration of sodium acetate in the growth medium in WT and icl strains. Cells were acclimated overnight at LL (15 µmol photons m−2 s−1) in HSM sparged with air. At t = 0 cells either continued being sparged with air (labelled “air”); or sparged with air and supplemented with 10 mM sodium acetate (labelled “acet”); or sparged with air enriched with 5% CO2 (labelled “CO2”). The addition of acetate or CO2 is indicated with a green mark on the x-axis. Samples were taken at t = 0, 1 h, 4 h and 8 h. Experiment at HL: d, e mRNA accumulation of LHCSR3 and RHP1 and f concentration of sodium acetate in the growth medium in WT and icl strains. Cells were acclimated overnight at LL (15 µmol photons m−2 s−1) in HSM sparged with air; at t = 0 light intensity was increased to 600 µmol photons m−2 s−1. At t = 1 h cells either continued being sparged with air (labelled “air”); or sparged with air and supplemented with 10 mM sodium acetate (labelled “acet”); or bubbled with air enriched with 5% CO2 (labelled “CO2”), always at 600 µmol photons m−2 s−1. The time of addition of acetate or CO2 is highlighted in green on the x-axis. Samples were taken at t = 0, 1 h, 2 h, 5 h and 9 h. (n = 3 biological samples, mean ± s.d.). The p values for the comparisons of acetate and CO2 conditions to air (LL; t = 1, 4, 8 h, HL; t = 2, 5, 9 h) are based on ANOVA Dunnett’s multiple comparisons test of log10 transformed mRNA data as indicated in the graphs (*P < 0.005, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant), following the color-code of the datasets. Exact p-values can be found at the Source Data file. Please note that in some cases the error bars are smaller than the data point symbols.
In another experiment, LL acclimated cells were shifted to HL (t = 0) and LHCSR3 and RHP1 transcript levels were assayed over a period of 9 h; acetate or high CO2 were introduced 1 h after the shift to HL (Fig. 2d, e, note the 1 h time point highlighted in green on the x-axis). In agreement with Fig. 1 (HL panel), LHCSR3 transcript accumulation increased by two orders of magnitude after 1 h exposure to HL in both WT and the icl mutant (Fig. 2d), while RHP1 transcripts rapidly decreased (Fig. 2e), which likely resulted from a reduction in the concentration of intracellular CO2 as a consequence of enhanced photosynthetic CO2 fixation in the HL. Introduction of acetate or CO2 to the cultures caused a rapid reduction in the level of LHCSR3 expression in WT (Fig. 2d), with the decline much more pronounced with CO2 supplementation. Supplementation with CO2 or acetate also caused an increase of RHP1 transcript relative to the control. In contrast, in the icl mutant, the decline in the level of the LHCSR3 transcript and the increase in the level of the RHP1 transcript was the same in cells with and without acetate supplementation, while the effect of CO2 was similar to that of WT cells (Fig. 2d, e). Furthermore, WT cells consumed about half of the acetate in the medium over the course of the experiment, while none of the acetate was consumed by the icl mutant (Fig. 2f). These results strongly suggest that CO2 inhibits the accumulation of the LHCSR3 transcript and that the decline of LHCSR3 mRNA in WT cells supplemented with acetate is a consequence of the CO2 released as the acetate is metabolized. The extent of this inhibition by acetate-derived CO2 appears to depend mostly on the rate of photosynthetic CO2 fixation (consumption of CO2) because acetate was taken up by WT cells at similar rates under both LL and HL conditions (Fig. 2c, f). Indeed, under LL conditions, where CO2 fixation is slow, acetate and CO2 repressed LHCSR3 to the same extent (Fig. 2a); under HL conditions, where CO2 fixation is much faster, the effect of acetate on the LHCSR3 transcript level was much smaller than that of CO2, which was continuously provided in excess (5%) via sparging (Fig. 2d).
We also employed constraint-based metabolic modelling to assess in silico whether acetate metabolism in Chlamydomonas leads to an increase in the concentration of intracellular CO2 under different growth conditions (Supplementary Note 1, Supplementary Fig. 2, Supplementary Tables 1–3, Supplementary Data 1–3). The findings from this approach support the hypothesis that there are changes in the internal CO2 concentration under autotrophic and mixotrophic growth conditions at different light intensities. These predicted changes in internal CO2 levels under the different conditions for the WT and mutant cells are congruent with the levels of accumulation of LHCSR3 transcripts that were measured.
CIA5 links HL and low CO2 responses
The responses to HL and low CO2 have been traditionally studied separately, despite several lines of evidence suggesting that they are integrated26,27,32. To elucidate the molecular connection between photoprotection and CCM, we analyzed mRNA accumulation of twelve genes implicated as functionally involved in the CCM, previously shown to be strongly expressed under low CO2 conditions33,34 and/or to be under the control of CIA526. Specifically, we analysed twelve CCM-related genes encoding LOW-CO2-INDUCIBLE PROTEIN B (LCIB) and E (LCIE), involved in CO2 uptake; HIGH-LIGHT ACTIVATED 3 (HLA3), LOW CO2-INDUCED 1 (LCI1), CHLOROPLAST CARRIER PROTEIN 1 (CCP1), CCP2, LCIA, BESTROPHINE-LIKE PROTEIN 1 (BST1), acting as Ci transporters; carbonic anhydrases CAH1, CAH3, CAH4; the nuclear regulator LOW-CO2 -STRESS RESPONSE 1 (LCR1).
When LL-acclimated, air-sparged WT, icl and icl-C strains were exposed to HL (experiment described in Fig. 1) a marked increase (5 to 600-fold) in CCM transcript levels was observed in WT cells (Supplementary Fig. 3), in accordance with recent studies;28,32 this increase was strongly suppressed by CO2 and to a lesser extent by acetate, which did not affect CCM gene expression in the icl mutant (Supplementary Fig. 3). This pattern of mRNA accumulation was essentially identical to that of LHCSR3 (Fig. 1a), highlighting the tight connection between HL and low-CO2 responses in Chlamydomonas. The CO2-mediated repression was more pronounced for most of the CCM genes relative to LHCSR3 (Fig. 1a, c, Supplementary Fig. 3).
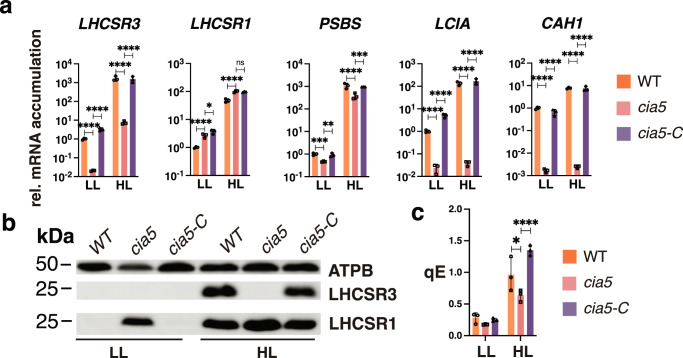
CIA5 has been shown to regulate the accumulation of transcripts from both the CCM genes26,27 and LHCSR328. To obtain a comprehensive view of the photoprotection capacity of the cia5 mutant, air-sparged WT and cia5 cells grown in LL were shifted to HL, and the transcript and protein levels from the qE effector genes were monitored. Remarkably, a lack of CIA5 resulted in much lower accumulation of LHCSR3 mRNA than in WT cells; 50 times lower at LL and over 200 times lower at HL. This phenotype was fully reversed by ectopic expression of the WT CIA5 gene (Fig. 3a). PSBS also showed a significant CIA5-dependent control at the mRNA level, although at a smaller extent (Fig. 3a). The cia5 mutant accumulated slightly more LHCSR1 mRNA in both LL and HL (~2 fold), however, this phenotype was not restored in the complemented cia5-C strain (Fig. 3a); we conclude that LHCSR1 mRNA accumulation is CIA5-independent. We also quantified the accumulation of mRNAs of CAH1 and LCIA, which are known to be strongly dependent on CIA527,35. As expected, the cia5 mutant cells failed to activate either of those genes in HL while their activation was fully restored in the complemented cia5-C strain (Fig. 3a).
Fig. 3. Cross-talk of responses to HL and low-CO2.
a CC-125 WT, cia5 and cia5-c strains were acclimated for 16 h in LL (15 µmol photons m−2 s−1) in HSM bubbled with air (labelled as “LL”); after sampling for the LL conditions, light intensity was increased to 600 µmol photons m−2 s−1 (HL); samples were taken after 1 h (RNA) and 4 h (protein and photosynthesis measurements). Accumulation of mRNA of genes at the indicated conditions were normalized to WT LL ctrl. (n = 3 biological samples, mean ± s.d.). The p-values for the comparisons of WT with cia5 and cia5 with cia5-C are based on ANOVA Dunnett’s multiple comparisons test of log10 transformed mRNA data as indicated in the graphs. b Immunoblot analyses of LHCSR3, LHCSR1 and ATPB (loading control) under the indicated conditions; PSBS was non-detectable at these experimental conditions. Representative dataset of experiment repeated three times. c qE of WT, cia5 and cia5-C under LL and HL conditions (n = 3 biological samples, mean ± s.d.). The statistical analyses (two-way ANOVA Tukey’s multiple comparison test) are shown in the graph. Exact p-values can be found at the Source Data file. Raw fluorescence and NPQ curves can be seen in Supplementary Fig. 4.
At the protein level, no LHCSR3 protein was detected in the cia5 mutant in either LL or HL (Fig. 3b). We were unable to immunologically detect the PSBS protein under these experimental conditions, in agreement with previous findings showing that PSBS protein accumulation is highly transient in cell cultures bubbled with air6. Importantly, the LHCSR1 protein accumulated to high levels in the mutant under both LL (conditions in which no protein is apparent in the WT) and HL conditions; this phenotype was fully reversed by ectopic expression of the WT CIA5 gene (Fig. 3b). This result suggests that CIA5 acts as a suppressor of LHCSR1 translation (and/or decreases protein stability) in both LL and HL. Our data additionally suggest that accumulation of LHCSR1 protein occurs through a compensatory, CIA5-controlled posttranscriptional mechanism that provides photoprotection under conditions in which the cells have almost no LHCSR3 protein (compare LHCSR1 and LHCSR3 immunoblots in Fig. 3b). Supporting this idea, the qE levels in cia5, although lower than WT and cia5-C (Fig. 3c and Supplementary Fig. 4), they were unexpectedly high considering the absence of LHCSR3 protein (Fig. 3b); we attribute this result to overaccumulation of LHCSR1 in this mutant (Fig. 3b). Together, our results demonstrate a key role of CIA5 in regulating photoprotection, activating LHCSR3 and to a lesser extent PSBS transcription and suppressing LHCSR1 protein accumulation.
CIA5 and CO2 availability regulate LHCSR1 protein stability
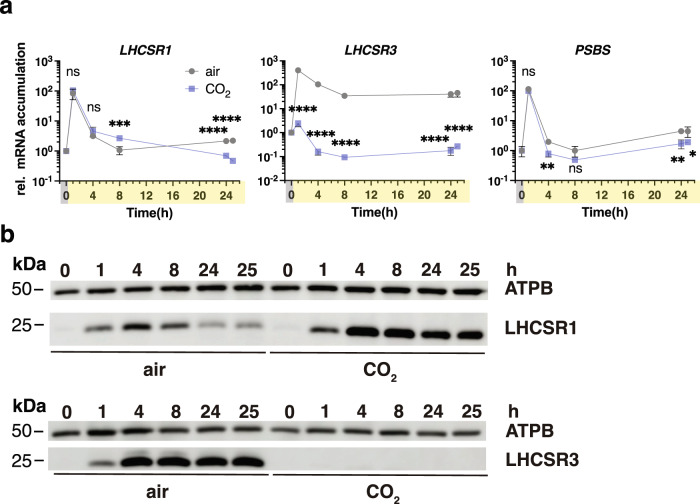
The high levels of accumulation of LHCSR1 protein in the cia5 mutant (Fig. 3b) suggest that CO2 availability could be the key determinant for LHCSR1 protein accumulation, as CIA5 is not functional under high CO2 levels15–17. Given the novelty of this finding, we decided to perform additional experiments to provide more details concerning LHCSR1 regulation. LL-acclimated WT cells sparged with air were exposed to HL sparged with air or 5% CO2 and the mRNA and protein levels were quantified over a 25-h period. Upon initial exposure to HL, LHCSR1 mRNA rapidly increased (2 orders of magnitude in 1 h) and then decreased to the initial level (between 4 and 8 h), in agreement with a previous report36, in the presence or absence of high CO2 (Fig. 4a). In contrast, the presence of high CO2 sustained high levels of LHCSR1 protein over the 25-h incubation period relative to cultures sparged with air (Fig. 4b). These results suggest that elevated CO2 either promotes translation of LHCSR1 mRNA or is involved in stabilizing the protein once it is synthesized. This contrasts with the behaviour of LHCSR3 for which there was a strong correlation between the level of mRNA and protein (the RNA was 3 orders of magnitude lower in 5% CO2 and the protein was no longer detected) (Fig. 4). The kinetics of PSBS transcript accumulation in HL very much resembled those of LHCSR1, with CO2 not having a strong impact on transcript accumulation (Fig. 4a). PSBS protein accumulation was not detectable under the experimental conditions used. Taken together, our data demonstrate the critical importance of CIA5 and CO2 in regulating the different qE effectors, mainly LHCSR3 and less strongly PSBS at the transcript level, and LHCSR1 at the protein level.
Fig. 4. Kinetic resolution of photoprotective gene and protein expression at different light and CO2 availabilities.
Cells were acclimated overnight at LL (15 µmol photons m−2 s−1) bubbled with air (labelled “air”). At t = 0 the light intensity was raised to 600 µmol photons m−2 s−1 under air bubbling or bubbling with 5% CO2 and mRNA and protein were followed for 25 h. a LHCSR1, LHCSR3 and PSBS mRNA accumulation. (n = 3 biological samples, mean ± s.d.). The p-values for the comparisons of CO2 conditions to air for t = 1, 4, 8, 24 and 25 h are based on two-way ANOVA Šídák’s multiple comparisons test of log10 transformed mRNA data as indicated in the graphs (*P < 0.005, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant). Exact p-values can be found at the Source Data file. b Immunoblot analyses of LHCSR1, LHCSR3 and ATPB (loading control). Representative dataset of experiment repeated three times.
Intracellular CO2 levels regulate photoprotective and CCM gene expression in the absence of light
To de-convolute the light and CO2 signals regulating LHCSR3, we exposed the cells to different light intensities and CO2 concentrations (Supplementary Fig. 5). High CO2 levels completely abolished the accumulation of LHCSR3 protein at all light intensities, in accord with the results of Fig. 1b, d and Fig. 4b. On the contrary, low CO2 levels led to very high accumulation of LHCSR3 protein at 150 and 300 µmol photons m−2 s−1. Under low CO2, LHCSR3 protein was also detectable even at the very low light intensity of 10 µmol photons m−2 s−1 (Supplementary Fig. 5), as previously demonstrated6.
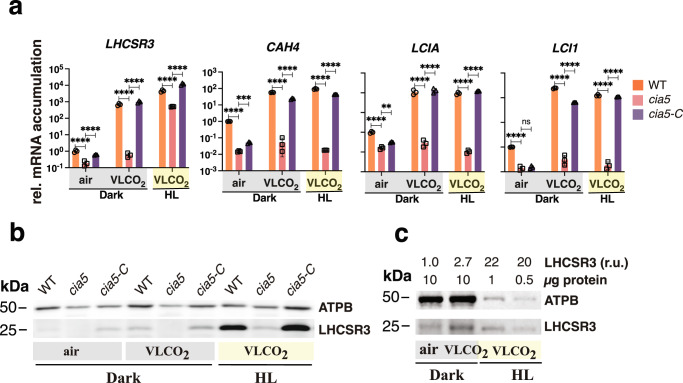
Prompted by this result, we tested whether changes in CO2 levels could activate transcription of LHCSR3 in complete darkness. We shifted air-sparged cells to sparging with CO2-free air (Very low CO2; VLCO2) in complete darkness and to our surprise, we observed that despite the absence of light, that a drop in CO2 availability was sufficient to trigger LHCSR3 mRNA accumulation by ~ 700-fold (Fig. 5a), with an increase in accumulation of the protein by 3-fold (Fig. 5b, c; compare WT air with WT VLCO2). In addition, when HL was combined with VLCO2, which is expected to result in an even greater reduction in the intracellular CO2 concentration, the levels of LHCSR3 mRNA and protein further increased, reaching levels of ~4500-fold (mRNA) and 21-fold (protein) compared to air dark conditions (Fig. 5a-c). Interestingly, this light-independent regulation of mRNA accumulation was under the control of CIA5 as the accumulation of LHCSR3 transcripts was abolished in the cia5 mutant (Fig. 5a) and a full reversal of these phenotypes (gene expression and protein levels) was observed in the cia5-C strain (Fig. 5a, b). We also observed significant LHCSR3 transcript accumulation in the cia5 mutant when cells were shifted from dark-air to HL-VLCO2, which was, however, 9-fold lower compared to the WT (Fig. 5a), and that was rescued to WT-levels in the cia5-C complemented line. This CIA5-independent regulation of mRNA in the presence of light could account for the contribution of light signaling in LHCSR3 gene expression, possibly via phototropin10 or via the generation of reactive oxygen species28.
Fig. 5. Low CO2 levels can trigger qE and CCM genes in the absence of light.
WT, cia5 and cia5-C cells were bubbled with air overnight in darkness; next day air bubbling was either maintained or replaced by CO2-limited-air bubbling in the darkness or in the presence of 600 µmol photons m−2 s−1 light. Sampling was performed after 1 h (RNA) or 4 h (protein). a mRNA accumulation of LHCSR3.1 (qE gene) and CAH4, LCIA, LCI1 (CCM genes) in WT, cia5 and cia5-C. Data were normalized to WT air dark; (n = 3 biological samples, mean ± s.d.). The p-values for the comparisons of WT with cia5 and cia5 with cia5-C are based on ANOVA Dunnett’s multiple comparisons test of log10 transformed mRNA data as indicated in the graphs (*P < 0.005, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant). Exact p-values can be found at the Source Data file. b Immunoblot analyses of LHCSR3 and ATPB (loading control) under the indicated conditions. Representative dataset of experiment repeated three times. c Immunoblot analyses of LHCSR3 and ATPB (loading control) of WT samples presented in b. Above the immunoblot shown are the amount of protein loaded per lane and the quantification of LHCSR3 protein accumulation (calculated as LHCSR3 /ATPB ratio) normalized to the air dark conditions. Representative dataset of experiment repeated three times.
We could observe that LHCSR1 transcripts were also induced in the dark (shift from dark-air to dark-VLCO2), but this induction was very low (7-fold) and appeared to be CIA5 independent (Supplementary Fig. 6a). At the protein level however, LHCSR1 over-accumulated in the cia5 mutant under all conditions tested (Supplementary Fig. 6b), confirming our previous findings (Fig. 3b). PSBS also showed a CIA5-dependent dark induction of transcripts (shift from dark-air to dark-VLCO2), although this induction was low (5-fold); complementation with the CIA5 gene (cia5-C strain) did not rescue the phenotype in dark-air conditions and only partially rescued it under dark-VLCO2 (Supplementary Fig. 6a, b). Both mRNA and protein accumulation of PSBS accumulated in a CIA5-dependent manner when cells were shifted from dark-air to HL-VLCO2 (Supplementary Fig. 6a, b); under these conditions the phenotypes were fully reversed in the cia5-C strain. This CIA5-dependent regulation of PSBS can most likely explain previously reported findings that PSBS protein accumulation was responsive to CO2 abundance, with its accumulation reaching maximum levels under low CO2 and HL conditions6.
We also measured CCM-related gene expression in the dark. As shown in Fig. 5a and Supplementary Fig. 6a, high levels of CCM-related transcripts were observed in the dark when the cells experienced VLCO2 conditions (compare “dark air” with “dark VLCO2”). The combination of HL and VLCO2 conditions, either elicited very small (less than two-fold) or no additional increase (compare “HL VLCO2” to “dark VLCO2”) in their level of the mRNA accumulation (Fig. 5a and Supplementary Fig. 6a). As expected CIA5 was critical for expression of the CCM genes under all conditions tested (Fig. 5a and Supplementary Fig. 6a).
Our data points out that the LHCSR1 protein overaccumulation in cia5 was fully reversed only when cia5-C cells were pre-acclimated in the light (Fig. 3b); when pre-acclimation took place in the dark the phenotype was only partially rescued (Supplementary Fig. 6b). The same is true for the mRNA accumulation of PSBS (compare LL; Fig. 3a with air-dark; Supplementary Fig. 6a), CAH1 (compare LL; Fig. 3a with air-dark; Supplementary Fig. 6a), LCIA (compare LL; Fig. 3a with air-dark; Fig. 5a), while in the case of LHCSR3 a full reversal of the CIA5-dependent phenotype was seen no matter what pre-acclimation strategy was followed (Figs. 3a, b and 5a, b). A plausible explanation for these results is the differential accumulation of CIA5 protein in the different acclimation regimes due to the promoter used. In line with this explanation, CIA5 expression in cia5-C is driven by the light-inducible promoter of the PSAD gene, and, as a result, less CIA5 protein accumulated in the dark-acclimated cia5-C compared to the LL-acclimated (Supplementary Fig. 7a). This in turn affects the relative abundance of CIA5 available for binding with its target molecules (DNA binding sites or CIA5-interacting proteins), ultimately affecting the reversal of the CIA5-related phenotypes.
Overall, these data challenge the view concerning the regulation of photoprotection and CCM and bring CO2 to the forefront as a crucial signal controlling LHCSR3 and CCM-related genes induction in the absence of light.
Link between photosynthetic electron transfer and CO2 intracellular concentration
Our finding that LHCSR3 is regulated by light-independent CO2 availability has guided us in revising the way in which we view the impact of photosynthetic electron transfer (PET) on LHCSR3 accumulation; i.e. inhibition of LHCSR3 accumulation in photosynthetic mutants or WT cells treated with photosynthetic inhibitors10,11.
We propose that enhanced PET, occurring under HL conditions, facilitates CO2 fixation, draws down the intracellular CO2 concentration and induces LHCSR3 transcription. In contrast, when PET is impaired, intracellular CO2 levels increase, thus promoting LHCSR3 transcript inhibition. In accordance, there is a marked increase of CO2 in cultures treated with DCMU, an inhibitor of photosystem II37, measured either as dissolved CO2 in the culture medium30, or as CO2 in the air stream coming from the headspace of the column bioreactor (Fig. 6a). In order to test our hypothesis, we analyzed the combined effect of DCMU and CO2 on the accumulation of mRNA from the LHCSR3 and two CCM genes in WT cultures shaken without or with VLCO2 sparging. In accord with previous reports10,38, DCMU completely blocked the HL elicited accumulation of LHCSR3 mRNA; LHCSR3 mRNA after 1 h exposure to HL diminished to ten times lower levels than the initial LL levels (shown as dotted line in graph) (Fig. 6b), which most likely reflects the degradation of the transcripts following inactivation of the gene after the addition of DCMU. Previous work has shown that LHCSR3 transcripts are rapidly lost once the gene becomes inactive10 which has also been observed for the CAH4 transcript39. However, when the cultures were sparged with VLCO2 air, which would result in the maintenance of a continuous VLCO2 concentration in the cultures, a large part of the DCMU elicited inhibition was relieved (Fig. 6b), supporting the idea that light primarily impacts LHCSR3 transcript levels by altering CO2 consumption and the intracellular (and/or extracellular) CO2 concentration. In contrast to LHCSR3, sparging with VLCO2 only partly relieved the suppression of transcript accumulation for the CCM genes in the presence of DCMU (Fig. 6b). This difference may reflect the fact that CCM gene expression is solely regulated by CO2 via CIA5 (Fig. 5a) and that sparging with VLCO2 in the presence of DCMU does not reduce the CO2 levels enough to attain full gene activation. It is also possible that longer incubation time with VLCO2 would have relieved a larger part of the DCMU-elicited inhibition of CCM genes (Fig. 6b) as implied by the slow kinetics of CAH4/5 mRNA accumulation when cells are shifted from 5% CO2 to air39.
Fig. 6. Photosynthetic electron transfer draws down the intracellular CO2 concentration, relieving inhibition of LHCSR3 transcription.
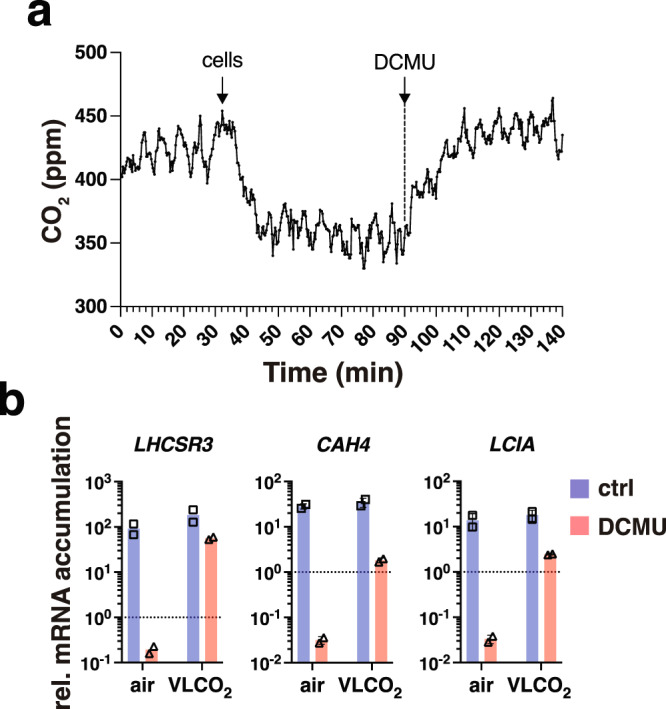
a CO2 concentration measured in the air stream coming out of the headspace of a column filled with 50 mL of HSM, sparged with air under HL. The two arrows in the graph indicate the addition of cells or DCMU. b WT cells were acclimated to LL HSM overnight shaken in flasks; the next day they were exposed to 300 µmol photons m−2 s−1 light in the presence or absence of 40 µM DCMU, shaken in flasks without or with sparging with VLCO2. Samples were taken after 1 h. Presented are mRNA accumulation of LHCSR3, CAH4, LCIA. Data were normalized to LL (shown as dotted line in graph); n = 2 biological samples.
Discussion
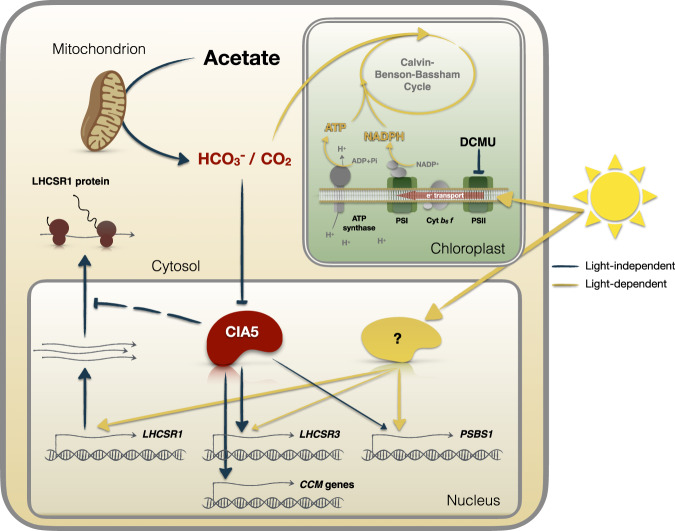
In this work, we presented findings that advance our understanding of integration between CO2- and light-dependent signaling in Chlamydomonas. We propose that the intracellular level of CO2, defined by the equilibrium between light-driven CO2 fixation in chloroplasts and the generation of CO2 by mitochondrial metabolism (e.g. acetate assimilation), is a key regulator of two major processes in photosynthetic organisms: the CCM and photoprotection (Fig. 7).
Fig. 7. CO2- and light-dependent signals converge to regulate photoprotection and CCM in Chlamydomonas.
The intracellular levels of CO2, defined by the equilibrium between CO2 fixation in chloroplasts and the generation of CO2 by mitochondrial metabolism (e.g. acetate assimilation) is the key determinant of the regulation of gene expression controlling two major processes of photosynthetic organisms: CCM and photoprotection. Changes in light availability have a direct impact on intracellular CO2 levels; exposure to HL increases CO2 fixation rates leading to depletion of CO2 and to activation of not only photoprotection- but also CCM-related genes. Conversely, depletion of CO2 is sufficient to drive high expression levels of CCM genes and LHCSR3 even in complete darkness (indicated by the black arrows). High CO2 levels, either exogenously supplied by sparging or metabolically produced via acetate metabolism or by inhibiting photosynthetic electron flow using DCMU, repress LHCSR3 and CCM genes while at the same time they stabilize LHCSR1 protein levels. The close interconnection of photoprotection and CCM is further corroborated by the fact that CIA5, the regulator of expression of genes associated with the CCM, also exerts control over LHCSR3 and to a lesser extent over PSBS mRNA levels and acts as repressor of LHCSR1 protein accumulation. Independent of CIA5, light strongly impacts expression of all of these photoprotective genes (yellow arrows). This impact can be the consequence of both photoperception (e.g. phototropin) and the production of reactive oxygen species.
To better understand the role of CO2 in regulating photoprotection and its integration with light, we designed experiments to separate the effects of the two signals (Figs. 5–7); we reduced the concentration of CO2 in the microalgae medium by sparging it with VLCO2 in complete darkness. This abrupt change in CO2 levels experienced by the cultures in the dark may be considered a condition only encountered in the laboratory. However, in certain ecological niches, such as soil or catchments with elevated levels of organic matter33, Chlamydomonas would encounter changes in the levels of CO2 that would be dependent on the microbes and the ratio between respiration and photosynthesis in the environment. Our experimental setup allowed us to observe a strong increase of LHCSR3 transcript levels when cells were shifted from air-CO2 to VLCO2 levels in the dark (Fig. 5), a very surprising result as the accumulation of LHCSR3 mRNA was considered so far to be strictly light-dependent5,11,38. Moreover, with this strategy we can disentangle light from CO2 signalling effects; while dark induction of LHCSR3 under CO2-depletion was completely dependent on CIA5, light could still strongly impact expression of all photoprotective genes in the cia5 mutant, which was not the case for CCM gene expression that was completely abolished in the light or dark in the absence of CIA5 (Fig. 5 and Supplementary Fig. 6). This impact of light on qE gene expression may be the consequence of photoperception (e.g. PHOT1)10, but also the generation of light-dependent signals such as reactive oxygen species28. Furthermore, a CIA5-independent regulation (also observed in Fig. 3a) explains LHCSR3 induction in high CO2-acclimated WT cells (cells in which CIA5 is not functional15–17) as they transition from LL to HL (Fig. 1a), which was not observed for CCM genes tested under identical conditions (Supplementary Fig. 3); it also explains why the CO2-mediated repression was more pronounced for most of the CCM genes relative to LHCSR3 (Fig. 1a, c, Supplementary Fig. 3). CO2 and CIA5 appear to be of paramount importance in signal integration and transduction, regulating expression of both photoprotection and CCM genes. For instance, CO2 represses the UV-B elicited, UVR8-mediated expression of LHCSR3, and CIA5 is absolutely required for this expression28. Moreover, our results have shown that high CO2 levels or the absence of CIA5 have a severe impact on LHCSR3 gene expression and, although HL can still induce LHCSR3 transcription, no protein is detected (Figs. 1, 3 and 5).
Besides transcriptionally controlling LHCSR3, CIA5 post-transcriptionally controls LHCSR1. Our view on LHCSR1 regulation by light and CIA5 is as follows: under LL conditions, LHCSR1 protein accumulates in cia5 while it is non-detectable in WT and cia5-C (Fig. 3b), suggesting that CIA5 suppresses LHCSR1 protein accumulation. Exposure to HL triggers a CIA5-independent LHCSR1 mRNA accumulation (Fig. 3a), possibly driven by reactive species, previously shown to favor LHCSR1 mRNA accumulation40. As a result, LHCSR1 protein accumulates in WT and cia5-C in HL, despite the fact that suppression of LHCSR1 protein by CIA5 still occurs; indeed, LHCSR1 accumulates to higher levels in the cia5 mutant as compared to WT and cia5-C under HL conditions (Fig. 3b). In line with the above observations in the cia5 mutant, high levels of LHCSR1 protein accumulate in WT under high CO2, conditions that inactivate CIA5 (Fig. 4b). Put together, our findings unveil a multilevel role of CIA5 in regulating qE; inactivation of CIA5 in high CO2 or by eliminating the CIA5 gene blocks LHCSR3 transcript accumulation, while it promotes LHCSR1 protein accumulation (Figs. 3, 4). Further investigation will be required to explain how a single nuclear factor, CIA5, can control cellular processes happening in different cellular compartments; transcription in the nucleus and translation in the cytosol.
Our results provide an interpretation of the findings that PET is required for LHCSR3 accumulation11, activation of the CCM and expression of CCM genes41. We propose that CO2, either provided directly or indirectly through metabolic generation, represents a critical link between PET and transcriptional regulation of LHCSR3 and the CCM genes (Fig. 6). Photosynthesis draws down cellular CO2 levels, and therefore, blocking photosynthesis with DCMU leads to the accumulation of CO2 (Fig. 6a) which elicits LHCSR3 repression, while sparging DCMU-treated cells with VLCO2 almost fully derepresses LHCSR3 (and partially CCM) expression (Fig. 6b). DCMU also upregulates genes of the leucine degradation pathway42 leading to the generation of acetoacetate and acetyl-CoA, which can lead to oxidative CO2 production. Whether leucine itself has a regulatory role or CO2 is the key regulator deserves further attention. It is tempting to propose that CO2 is a retrograde signal that readily diffuses through the cell and impacts nuclear gene expression, which would integrate both mitochondrial and chloroplastic metabolic activities.
The way in which Chlamydomonas senses CO2 is not clear. Our data, i.e. accumulation of LHCSR3 and CCM genes in the dark, exclude the possibility that a metabolite produced by photorespiration plays a major signalling role, as previously proposed43. CO2 itself might also serve as the metabolite being recognized by a putative sensor that could be controlled by carbamylation, a CO2-mediated post-translational modification that regulates, among others, the activation of Rubisco44. Furthermore, the large number of adenosine and guanylyl cyclases in Chlamydomonas45 suggests that cyclic nucleotides play an important role in controlling various processes in this alga; these metabolites have been shown to be involved in mating46, regulation of flagellar beating and phototaxis47–49, in regulating inorganic nitrogen assimilation50 and in restoring LHCSR3 accumulation in the absence of phototropin10. Cyclases have been shown to act as CO2 sensors (as bicarbonate) in mammalian cells51, making it plausible that they can also serve as sensors in Chlamydomonas. As cyclic nucleotide signalling and calcium are tightly linked51, we anticipate an important role for calcium in CO2 sensing; calcium signalling has already been shown to be involved in the regulation of both LHCSR3 and CCM genes11,52.
Overall, our work shows that the intracellular CO2 level is the main factor in regulating CCM genes and LHCSR3 in Chlamydomonas (Fig. 7). Exposure to HL increases the CO2 fixation rate which causes a drop in intracellular CO2 which, in turn, actives both photoprotection- and CCM-related genes. Depletion of CO2 is sufficient to drive high expression levels of CCM genes and LHCSR3 even in complete darkness. On the other hand, high CO2 levels, either generated through enhanced respiratory activity or impaired photosynthetic electron transport, repress LHCSR3 and CCM genes while at the same time stabilizing the LHCSR1 protein, which likely acts as a backup photoprotection protein under conditions where LHCSR3 is not expressed. Furthermore, our data reveals a closer interconnection of photoprotection and CCM as CIA5, the CCM master regulator, also exerts control over LHCSR3 and to a lesser extent over PSBS mRNA levels, while repressing LHCSR1 protein accumulation. Our findings highlight the need to develop an integrated approach that examines the role of CO2 and light, not only as substrates of photosynthetic CO2 fixation, but also as signals regulating photoprotection, CCM, and at a wider context genome-wide gene expression.
Methods
Chemicals
DCMU (3-(3,4-dichlorophenyl)−1,1-dimethylurea) was purchased from Sigma. Stock solutions of DCMU were prepared in ethanol (40 mM).
Strains and conditions
Chlamydomonas strains were grown under 20 µmol photons m−2 s−1 in Tris-acetate-phosphate (TAP) medium53 at 23 °C in Erlenmeyer flasks shaken at 125 rpm. For all experiments cells were transferred to Sueoka’s High Salt medium54 supplemented when needed with 10 mM sodium acetate, at 2 million cells mL−1 in 80 mL capacity columns, unless otherwise stated, sparged with air, air enriched with 5% CO2, or very low CO2 air (VLCO2; generated by passing the air through soda lime) and exposed to light intensities as described in the text and figure legends. Chlamydomonas strain 137c mt+ was used as WT. The icl (defective in ICL1; gene ID: Cre06.g282800), icl-C (icl strain complemented with the WT ICL gene), dum11 (defective in defective in ubiquinol cytochrome c oxidoreductase of the respiratory complex III; geneID: CreMt.g000300) and cia5 (defective in CIA5, aka CCM1; geneID: Cre02.g096300; Chlamydomonas Resource Centre strain CC-2702) mutants were previously generated15,21,29. For complementation of cia5, a 3.5-kbp genomic DNA fragment from CC-125 containing the CIA5 coding region was amplified by PCR using Platinum™ SuperFi™ DNA Polymerase (Thermo Fisher Scientific) and primers gib-cia5-fw and gib-cia5-rev (Supplementary Table 4), gel purified and cloned into pLM00555 by Gibson assembly56 for expression under control of the PSAD promoter. Junctions and insertion were sequenced, and constructs were linearized by EcoRV before transformation into cia5. Eleven ng/kb of linearized plasmid55 mixed with 400 μL of 1.0 × 107 cells mL−1 were electroporated in a volume of 120 mL in a 2-mm-gap electro cuvette using a NEPA21 square-pulse electroporator (NEPAGENE, Japan). The electroporation parameters were set as follows: Poring Pulse (300 V; 8 ms length; 50 ms interval; one pulse; 40% decay rate; + Polarity), Transfer Pulse (20 V; 50 ms length; 50 ms interval; five pulses; 40% decay rate; +/- Polarity). Transformants were plated onto solid agar medium containing 10 μg/ml paromomycin and screened for fluorescence using a Tecan fluorescence microplate reader (Tecan Group Ltd., Switzerland). Parameters used were as follows: YFP (excitation 515/12 nm and emission 550/12 nm) and chlorophyll (excitation 440/9 nm and 680/20 nm). Transformants showing a high YFP/chlorophyll ratio were further analyzed by immunoblotting using anti-FLAG antibodies (Supplementary Fig. 7b). Among the transformants analyzed the cia5-C-a1 (cia5-C throughout the text) was retained for further analyses in the present study, after verifying that it grows similarly with the WT under phototrophic conditions on agar (Supplementary Fig. 7c). Unless otherwise stated, LL conditions corresponded to 20 µmol photons m−2 s−1 while HL conditions corresponded to 600 µmol photons m−2 s−1 of white light (Neptune L.E.D., France; see Supplementary Fig. 8 for light spectrum). All experiments were repeated three times to examine reproducibility, unless otherwise stated.
Light acclimation experiments
Cells were acclimated overnight in High Salt Medium (HSM) in LL sparged with air, in the presence or absence of acetate, or sparged with 5% CO2. Following this acclimation period, cells were transferred from LL to HL, with all other conditions identical to those of the acclimation period. Samples were collected after 1 h for RNA analyses and after 4 h for protein analysis and measurements of photosynthetic activity.
Fluorescence-based measurements
Fluorescence-based photosynthetic parameters were measured with a pulse modulated amplitude fluorimeter (MAXI-IMAGING-PAM, HeinzWaltz GmbH, Germany). Prior to the onset of the measurements, cells were acclimated to darkness for 15 min. Chlorophyll fluorescence was recorded under different intensities of actinic light; starting with measurements in the dark (indicated as D below the x-axis of the graphs), followed by measurements at 21 μmol photons m−2 s−1 (indicated as L1 below the x-axis of the graphs) and 336 μmol photons m−2 s−1 (indicated as L2 below the x-axis of the graphs) and finishing with measurements of fluorescence relaxation in the dark. The calculations of the different photosynthetic parameter was performed based on57 as follows: The relative photosynthetic electron transfer rate (rETR) was calculated as (Fm′ − F)/Fm ′ × I; F and Fm′ are the fluorescence yield in steady state light and after a saturating pulse in the actinic light, respectively; I is the light irradiance in μmol photons m−2 s−1; NPQ was calculated as (Fm − Fm′)/Fm′; Fm is the maximal fluorescence yield in dark-adapted cells; the effective photochemical quantum yield of photosystem II was calculated as Y(II) = (Fm’-F)/Fm’; qE was estimated as the fraction of NPQ that is rapidly inducible in the light and reversible in the dark.
mRNA quantification
Total RNA was extracted using the RNeasy Mini Kit (Qiagen) and treated with the RNase-Free DNase Set (Qiagen). 1 µg total RNA was reverse transcribed with oligo dT using Sensifast cDNA Synthesis kit (Meridian Bioscience, USA). qPCR reactions were performed and quantitated in a Bio-Rad CFX96 system using SsoAdvanced Universal SYBR Green Supermix (BioRad). The primers (0.3 µM) used for qPCR are listed in Supplementary Table 4. A gene encoding G protein subunit-like protein (GBLP)58 was used as the endogenous control, and relative expression values relative to GBLP were calculated from three biological replicates, each of which contained three technical replicates.
CO2 measurements
CO2 concentration was measured in the air stream coming from the headspace of a HSM or culture-containing column using the CO2 Probe GMP251 connected to the MI70 data logger from Vaisala (Vantaa, Finland).
Immunoblotting
Protein samples of whole cell extracts (0.5 µg chlorophyll or 10 µg protein) were loaded on 4-20% SDS-PAGE gels (Mini-PROTEAN TGX Precast Protein Gels, Bio-Rad) and blotted onto nitrocellulose membranes. Antisera against LHCSR1 (AS14 2819, 1:15000 dilution), LHCSR3 (AS14 2766, 1:15000 dilution), ATPB (AS05 085, 1:15000 dilution) were from Agrisera (Vännäs, Sweden); previously described was antisera against C. reinhardtii PSBS6 (used at a dilution of 1:1000). ATPB was used as a loading control. An antirabbit horseradish peroxidase-conjugated antiserum was used for detection at 1:10000 dilution. Mouse monoclonal antibody against FLAG was purchased from Sigma-Aldrich (F3165, St. Louis, MO, USA) and was used at a dilution of 1:15000. An anti-mouse horseradish peroxidase-conjugated antiserum (Jackson Immuno Research Europe LTD) was used as a secondary antibody for 3xFLAG immunoblotting (1:10000 dilution). The blots were developed with ECL detection reagent, and images of the blots were obtained using a CCD imager (ChemiDoc MP System, Bio-Rad). For the densitometric quantification, data were normalized with ATPB.
Statistical analyses
Statistical methods were not used to predetermine the sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experimental procedures and data assessment. All statistical tests were performed using the computing environment Prism 9 (Graphpad Software, LLC), at a significance level of 0.05. In order to conform mRNA accumulation data to the distributional assumptions of Analysis of Variance (ANOVA), i.e. the residuals should be normally distributed and variances should be equal among groups, two-way ANOVA was performed with log-transformed data Y = logX where X is mRNA accumulation59.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We are grateful to Dr. Konomi Fujimura-Kamada for performing experiments to validate the dark induction of LHCSR3 in the Minagawa lab; to Claire Remacle for sharing the icl and icl-C strains and Pierre Cardol for the dum11 strain. We thank Eric Soupene for valuable insights in the RHP1 induction conditions. We thank Dimitra Karageorgou for performing preliminary experiments in the project and Gilles Curien for fruitful discussions on aspects of microalgae metabolism. We would like to thank the following agencies for funding: The Human Frontiers Science Program through the funding of the project RGP0046/2018 (DP, ARG, PR, ES-L, ZN, AK); the French National Research Agency in the framework of the Young Investigators program ANR-18-CE20-0006 through the funding of the project MetaboLight (DP); the French National Research Agency through the funding of the Grenoble Alliance for Integrated Structural & Cell Biology GRAL project ANR-17-EURE-0003 (DP, MAR-S, GV, YY); the French National Research Agency in the framework of the Investissements d’Avenir program ANR-15-IDEX-02, through the funding of the “Origin of Life” project of the Univ. Grenoble-Alpes (DP, YY); the Prestige Marie-Curie co-financing grant PRESTIGE-2017-1-0028 (MAR-S); the International Max Planck Research School “Primary Metabolism and Plant Growth” at the Max Planck Institute of Molecular Plant Physiology (MA, ZN); the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement no. 751039 (ES-L); the program ‘Plan Propio UCO’ from University of Cordoba, Spain for postdoctoral Support (ES-L); the Carnegie Institution for Science and the Department of Energy, DE-SC0019417 (ARG); the Marie Curie Initial Training Network Accliphot FP7-PEPOPLE-2012-ITN; 316427 (SF, GF, DP); the Japan Society for the Promotion of Science, JSPS, for the grants-in-Aid for Scientific Research, KAKENHI, 21H04778 and 21H05040 (JM) and the German Research Foundation DFG HI 739/9.2 (MH).
Source data
Author contributions
Conceptualization: M.A.R.-S., ES.-L., P.R., M.H., Z.N., J.M., A.R.G., D.P.; Methodology: M.A.R.-S., Y.Y., G.V., P.R., E.S.-L., Z.N., D.P.; Investigation: M.A.R.-S., S.F., Y.Y., G.V., R.T., A.K., A.T., G.K., G.A., M.A., F.I.; Supervision: G.F., Z.N., J.M., A.R.G., D.P.; Writing—original draft: M.A.R.-S., D.P.; Writing—review & editing: all authors
Peer review
Peer review information
Nature Communications thanks Marco Fondi, Congming Lu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The source data underlying Figs. 1–6 and Supplementary Figures 1, 3, 4, 6, 7 are provided as a Source Data file. The Source Data file also includes the exact p-values for Figs. 1a, c, 2a, b, d, e, 3a, c, 4a, 5a, and Supplementary figures 1a, b, 3, 6a. All biological material described in this study is available upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: M. Águila Ruiz-Sola, Serena Flori, Yizhong Yuan.
Change history
7/24/2023
In the original version of this article, the given and family names of M. Águila Ruiz-Sola were incorrectly structured. The name was displayed correctly in all versions at the time of publication. The original article has been corrected.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-023-37800-6.
References
- 1.Li Z, Wakao S, Fischer BB, Niyogi KK. Sensing and responding to excess light. Annu. Rev. Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- 2.Bennett DIG, et al. Models and mechanisms of the rapidly reversible regulation of photosynthetic light harvesting. Open Biol. 2019;9:190043. doi: 10.1098/rsob.190043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alboresi A, Gerotto C, Giacometti GM, Bassi R, Morosinotto T. Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc. Natl Acad. Sci. USA. 2010;107:11128–11133. doi: 10.1073/pnas.1002873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niyogi KK, Truong TB. Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis. Curr. Opin. Plant Biol. 2013;16:307–314. doi: 10.1016/j.pbi.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Peers G, et al. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462:518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- 6.Correa-Galvis V, et al. Photosystem II Subunit PsbS Is Involved in the Induction of LHCSR Protein-dependent Energy Dissipation in Chlamydomonas reinhardtii. J. Biol. Chem. 2016;291:17478–17487. doi: 10.1074/jbc.M116.737312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tibiletti T, Auroy P, Peltier G, Caffarri S. Chlamydomonas reinhardtii PsbS Protein Is Functional and Accumulates Rapidly and Transiently under High Light. Plant Physiol. 2016;171:2717–2730. doi: 10.1104/pp.16.00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allorent G, et al. UV-B photoreceptor-mediated protection of the photosynthetic machinery in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA. 2016;113:14864–14869. doi: 10.1073/pnas.1607695114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinc E, et al. LHCSR1 induces a fast and reversible pH-dependent fluorescence quenching in LHCII in Chlamydomonas reinhardtii cells. Proc. Natl Acad. Sci. USA. 2016;113:7673–7678. doi: 10.1073/pnas.1605380113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petroutsos D, et al. A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature. 2016;537:563–566. doi: 10.1038/nature19358. [DOI] [PubMed] [Google Scholar]
- 11.Petroutsos D, et al. The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. Plant Cell. 2011;23:2950–2963. doi: 10.1105/tpc.111.087973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokutsu R, Fujimura-Kamada K, Matsuo T, Yamasaki T, Minagawa J. The CONSTANS flowering complex controls the protective response of photosynthesis in the green alga Chlamydomonas. Nat. Commun. 2019;10:655–10. doi: 10.1038/s41467-019-11989-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabilly ST, et al. Regulation of photoprotection gene expression in Chlamydomonas by a putative E3 ubiquitin ligase complex and a homolog of CONSTANS. Proc. Natl Acad. Sci. USA. 2019;116:17556–17562. doi: 10.1073/pnas.1821689116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maberly SC, Gontero B. Ecological imperatives for aquatic CO2-concentrating mechanisms. J. Exp. Bot. 2017;68:3797–3814. doi: 10.1093/jxb/erx201. [DOI] [PubMed] [Google Scholar]
- 15.Moroney JV, et al. Isolation and Characterization of a Mutant of Chlamydomonas reinhardtii Deficient in the CO2 Concentrating Mechanism. Plant Physiol. 1989;89:897–903. doi: 10.1104/pp.89.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuzawa H, et al. Ccm1, a regulatory gene controlling the induction of a carbon-concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proc. Natl Acad. Sci. USA. 2001;98:5347–5352. doi: 10.1073/pnas.081593498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang Y, Zhang J, Weeks DP. The Cia5 gene controls formation of the carbon concentrating mechanism in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA. 2001;98:5341–5346. doi: 10.1073/pnas.101534498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris, E. H. Chlamydomonas as a Model Organism. Annu. Rev. Plant. Physiol.10.1146/annurev.arplant.52.1.363 (2001). [DOI] [PubMed]
- 19.Wolfe AJ. The acetate switch. Microbiol. Mol. Biol. Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornberg HL, Krebs HA. Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle . Nature. 1957;179:988–991. doi: 10.1038/179988a0. [DOI] [PubMed] [Google Scholar]
- 21.Plancke C, et al. Lack of isocitrate lyase in Chlamydomonas leads to changes in carbon metabolism and in the response to oxidative stress under mixotrophic growth. Plant J. 2014;77:404–417. doi: 10.1111/tpj.12392. [DOI] [PubMed] [Google Scholar]
- 22.Salinas, T., Larosa, V., Cardol, P., Marechal-Drouard, L. & Remacle, C. Respiratory-deficient mutants of the unicellular green alga Chlamydomonas: A review. Biochimie.10.1016/j.biochi.2013.10.006 (2013). [DOI] [PubMed]
- 23.Fett JP, Coleman JR. Regulation of Periplasmic Carbonic Anhydrase Expression in Chlamydomonas reinhardtii by Acetate and pH. Plant Physiol. 1994;106:103–108. doi: 10.1104/pp.106.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moroney JV, Kitayama M, Togasaki RK, Tolbert NE. Evidence for Inorganic Carbon Transport by Intact Chloroplasts of Chlamydomonas reinhardtii. Plant Physiol. 1987;83:460–463. doi: 10.1104/pp.83.3.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polukhina, I., Fristedt, R., Dinc, E., Cardol, P. & Croce, R. Carbon supply and photoacclimation crosstalk in the green alga Chlamydomonas reinhardtii. Plant Physiol. 1494–1505. 10.1104/pp.16.01310 (2016) [DOI] [PMC free article] [PubMed]
- 26.Fang W, et al. Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. Plant Cell. 2012;24:1876–1893. doi: 10.1105/tpc.112.097949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miura K, et al. Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol. 2004;135:1595–1607. doi: 10.1104/pp.104.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redekop P, et al. Transcriptional regulation of photoprotection in dark-to-light transition- more than just a matter of excess light energy. Sci. Adv. 2022;8:eabn1832. doi: 10.1126/sciadv.abn1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colin M, et al. Mutations affecting the mitochondrial genes encoding the cytochrome oxidase subunit I and apocytochrome b of Chlamydomonas reinhardtii. Mol. Gen. Genet. 1995;249:179–184. doi: 10.1007/BF00290364. [DOI] [PubMed] [Google Scholar]
- 30.Hanawa Y, Watanabe M, Karatsu Y, Fukuzawa H, Shiraiwa Y. Induction of a high-CO2-inducible, periplasmic protein, H43, and its application as a high-CO2-responsive marker for study of the high-CO2-sensing mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol. 2007;48:299–309. doi: 10.1093/pcp/pcl066. [DOI] [PubMed] [Google Scholar]
- 31.Soupene E, et al. Rhesus expression in a green alga is regulated by CO(2) Proc. Natl Acad. Sci. USA. 2002;99:7769–7773. doi: 10.1073/pnas.112225599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scholz M, et al. Light-dependent N-terminal phosphorylation of LHCSR3 and LHCB4 are interlinked in Chlamydomonas reinhardtii. Plant J. 2019;99:877–894. doi: 10.1111/tpj.14368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Stessman DJ, Spalding MH. The CO2 concentrating mechanism and photosynthetic carbon assimilation in limiting CO2: how Chlamydomonas works against the gradient. Plant J. 2015;82:429–448. doi: 10.1111/tpj.12829. [DOI] [PubMed] [Google Scholar]
- 34.Mukherjee A, et al. Thylakoid localized bestrophin-like proteins are essential for the CO2 concentrating mechanism of Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA. 2019;116:16915–16920. doi: 10.1073/pnas.1909706116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshioka S, et al. The novel Myb transcription factor LCR1 regulates the CO2-responsive gene Cah1, encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Cell. 2004;16:1466–1477. doi: 10.1105/tpc.021162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aihara Y, Fujimura-Kamada K, Yamasaki T, Minagawa J. Algal photoprotection is regulated by the E3 ligase CUL4-DDB1DET1. Nat. Plants. 2019;5:34–40. doi: 10.1038/s41477-018-0332-5. [DOI] [PubMed] [Google Scholar]
- 37.Trebst A. Inhibitors in the functional dissection of the photosynthetic electron transport system. Photosyn. Res. 2007;92:217–224. doi: 10.1007/s11120-007-9213-x. [DOI] [PubMed] [Google Scholar]
- 38.Maruyama S, Tokutsu R, Minagawa J. Transcriptional regulation of the stress-responsive light harvesting complex genes in Chlamydomonas reinhardtii. Plant Cell Physiol. 2014;55:1304–1310. doi: 10.1093/pcp/pcu068. [DOI] [PubMed] [Google Scholar]
- 39.Eriksson M, Villand P, Gardeström P, Samuelsson G. Induction and Regulation of Expression of a Low-CO2-Induced Mitochondrial Carbonic Anhydrase inChlamydomonas reinhardtii. Plant Physiol. 1998;116:637–641. doi: 10.1104/pp.116.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roach T, Na CS, Stöggl W, Krieger-Liszkay A. The non-photochemical quenching protein LHCSR3 prevents oxygen-dependent photoinhibition in Chlamydomonas reinhardtii. J. Exp. Bot. 2020;71:2650–2660. doi: 10.1093/jxb/eraa022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spalding, M. H. in The molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas (eds. Rochaix, J. D., Goldschmidt-Clermont, M. & Merchant, S. S.) (1998).
- 42.Hemschemeier A, et al. COPPER RESPONSE REGULATOR1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia. Plant Cell. 2013;25:3186–3211. doi: 10.1105/tpc.113.115741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santhanagopalan I, Wong R, Mathur T, Griffiths H. Orchestral manoeuvres in the light: crosstalk needed for regulation of the Chlamydomonas carbon concentration mechanism. J. Exp. Bot. 2021;72:4604–4624. doi: 10.1093/jxb/erab169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linthwaite VL, et al. The identification of carbon dioxide mediated protein post-translational modifications. Nat. Commun. 2018;9:3092. doi: 10.1038/s41467-018-05475-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hasegawa E, Hayashi H, Asakura S, Kamiya R. Stimulation of in vitro motility of Chlamydomonas axonemes by inhibition of cAMP-dependent phosphorylation. Cell Motil. Cytoskel. 1987;8:302–311. doi: 10.1002/cm.970080403. [DOI] [PubMed] [Google Scholar]
- 47.Pasquale SM, Goodenough UW. Cyclic AMP functions as a primary sexual signal in gametes of Chlamydomonas reinhardtii. J. Cell Biol. 1987;105:2279–2292. doi: 10.1083/jcb.105.5.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaillard AR, Fox LA, Rhea JM, Craige B, Sale WS. Disruption of the A-kinase anchoring domain in flagellar radial spoke protein 3 results in unregulated axonemal cAMP-dependent protein kinase activity and abnormal flagellar motility. Mol. Biol. Cell. 2006;17:2626–2635. doi: 10.1091/mbc.E06-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boonyareth M, Saranak J, Pinthong D, Sanvarinda Y, Foster KW. Roles of cyclic AMP in regulation of phototaxis in Chlamydomonas reinhardtii. Biologia. 2009;64:1058–1065. [Google Scholar]
- 50.de Montaigu A, Sanz-Luque E, Galván A, Fernández E. A soluble guanylate cyclase mediates negative signaling by ammonium on expression of nitrate reductase in Chlamydomonas. Plant Cell. 2010;22:1532–1548. doi: 10.1105/tpc.108.062380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beavo JA, Brunton LL. Cyclic nucleotide research - still expanding after half a century. Nature. 2002;3:710–718. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, et al. Chloroplast-mediated regulation of CO2-concentrating mechanism by Ca2+-binding protein CAS in the green alga Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA. 2016;113:12586–12591. doi: 10.1073/pnas.1606519113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardii. Proc. Natl Acad. Sci. USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sueoka N. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardii. Proc. Natl Acad. Sci. USA. 1960;46:83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mackinder LCM, et al. A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proc. Natl Acad. Sci. USA. 2016;113:5958–5963. doi: 10.1073/pnas.1522866113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 57.Genty B, Briantais J-M, Baker NR. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. et. Biophys. Acta (BBA) 1989;990:87–92. [Google Scholar]
- 58.Schloss JA. A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Mol. Gen. Genet. 1990;221:443–452. doi: 10.1007/BF00259410. [DOI] [PubMed] [Google Scholar]
- 59.Schlesselman, J. J. Data Transformation in Two-Way Analysis of Variance. J. Am. Stat. Assoc.10.1080/01621459.1973.10482435 (2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The source data underlying Figs. 1–6 and Supplementary Figures 1, 3, 4, 6, 7 are provided as a Source Data file. The Source Data file also includes the exact p-values for Figs. 1a, c, 2a, b, d, e, 3a, c, 4a, 5a, and Supplementary figures 1a, b, 3, 6a. All biological material described in this study is available upon request. Source data are provided with this paper.