Summary
Background
Age-related changes in immune cell composition and functionality are associated with multimorbidity and mortality. However, many centenarians delay the onset of aging-related disease suggesting the presence of elite immunity that remains highly functional at extreme old age.
Methods
To identify immune-specific patterns of aging and extreme human longevity, we analyzed novel single cell profiles from the peripheral blood mononuclear cells (PBMCs) of a random sample of 7 centenarians (mean age 106) and publicly available single cell RNA-sequencing (scRNA-seq) datasets that included an additional 7 centenarians as well as 52 people at younger ages (20–89 years).
Findings
The analysis confirmed known shifts in the ratio of lymphocytes to myeloid cells, and noncytotoxic to cytotoxic cell distributions with aging, but also identified significant shifts from CD4+ T cell to B cell populations in centenarians suggesting a history of exposure to natural and environmental immunogens. We validated several of these findings using flow cytometry analysis of the same samples. Our transcriptional analysis identified cell type signatures specific to exceptional longevity that included genes with age-related changes (e.g., increased expression of STK17A, a gene known to be involved in DNA damage response) as well as genes expressed uniquely in centenarians’ PBMCs (e.g., S100A4, part of the S100 protein family studied in age-related disease and connected to longevity and metabolic regulation).
Interpretation
Collectively, these data suggest that centenarians harbor unique, highly functional immune systems that have successfully adapted to a history of insults allowing for the achievement of exceptional longevity.
Funding
TK, SM, PS, GM, SA, TP are supported by NIH-NIAUH2AG064704 and U19AG023122. MM and PS are supported by NIHNIA Pepper center: P30 AG031679-10. This project is supported by the Flow Cytometry Core Facility at BUSM. FCCF is funded by the NIH Instrumentation grant: S10 OD021587.
Keywords: Aging, Extreme human longevity, Elite immunity, Single cell RNA sequencing
Research in context.
Evidence before this study
Aging is associated with multi-morbidity and mortality. Transcriptional studies of blood have previously identified global immune cell dysfunction with changes in the composition and gene expression profiles of lymphocytes and myeloid cells with age (Geiger et al., 2013, Petersone et al., 2015, Alpert et al., 2019). Centenarians, a rare population of individuals that reach 100 years, experience delays in aging-related diseases and mortality suggesting their immune systems remain functional at extreme old age. A recent study of peripheral blood mononuclear cells (PBMC) from Japanese centenarians observed changes in composition of immune cells and increased cytotoxic lymphocytes compared to younger ages (Hashimoto et al., 2019). However, the generalizability of these results to other ethnicities and most importantly the transcriptional changes that occur in peripheral immune cell types of centenarians compared to younger individuals are still unclear. Further investigation to characterize the repertoire of immune cells of individuals who reach Extreme Longevity (EL) may indicate important mechanisms that promote EL.
Added value of this study
We performed CITE-seq on PBMCs from 7 centenarians to provide transcriptome-wide expression data in addition to a 10 cell-surface protein marker panel to increase the specificity of the identification of major immune cell types. We then integrated this dataset with two publicly available scRNA-seq datasets of PBMCs to investigate compositional and transcriptional changes in circulating immune profiles across the human lifespan and extreme old age. We found three patterns of changes in the immune cell type composition and gene expression profiles across the human lifespan that include aging related changes and changes unique to centenarians. We also validated these patterns using an orthogonal methodology (flow cytometry). The unique transcriptional changes we observed in centenarians compared to younger ages point to changes in metabolic regulation. In addition, we observed compositional changes within myeloid and lymphocyte lineages that reflect a greater exposure to infections and a shift in the immune resiliency strategy in EL compared to younger ages, which could be used for further investigation into immune resilience of EL and healthy aging.
Implications of all the available evidence
Our single cell analysis of peripheral blood immune cell populations across the human lifespan confirms observations made in previous studies of aging and identifies novel cell type specific compositional and transcriptional changes that are specific to centenarians and reflect immunocompetent profiles. These findings provide a foundation to investigate immune resilience mechanisms of extreme longevity as a target for healthy aging therapeutics.
Introduction
While a decline in cellular, organismic, and overall functionality is an inexorable outcome of aging, the rate and impact of aging is increasingly recognized to be affected by multiple factors including environment, genetics, and immune history.1, 2, 3, 4 At a phenotypic level, aging leads to functional abnormalities and alterations in hematopoietic cell populations that prevent a proper immune response and lead to increased susceptibility to infections, cancers, and auto-immune diseases.1, 2, 3 Driven by transcriptional changes and alterations in gene expression, the global immune cell dysfunction generally observed with aging results in distinct shifts in the composition of peripheral immune cell types characterized by a loss of naive B and T cells and an accumulation of memory effector T and B cells,3,5,6 and more recently the age-associated expansion of granzyme K (GZMK)-expressing CD8+ T cells.7 In addition, an increase of inflammation-promoting cell populations, such as Natural Killer and myeloid cells (e.g., monocytes) are observed with aging,3,5,6 in parallel with gene expression changes in these populations.6,8,9
At the extreme of the human aging process is extreme longevity (EL), characterized by survival beyond an age reached by less than 1% of a cohort.10 EL is often, but not always, associated with a marked delay of disability and in majority (about 60%), common aging-related diseases.11, 12, 13 Changes in immune cells are considered one of the hallmarks of aging, with growing recognition that the loss of immune competence to control inflammation and rebound from immune stressors is central to the progression of age-limiting morbidities.14 Since centenarians—individuals who live to at least 100 years—appear to experience a slower pace of aging,13,15 characterizing the repertoire of immune cells of these elite individuals may point to important mechanisms that promote EL.
A recent study using single cell RNA sequencing (scRNA-seq) of peripheral blood mononuclear cells (PBMCs) displayed changes in the distribution of lymphocytes and myeloid cells, and a significant expansion of cytotoxic CD4+ T cells in individuals who live to at least 105 years compared to younger individuals.16 Hashimoto's study focused on a cohort of Japanese individuals,16 thus it is not clear whether those results generalize to other ethnicities. Furthermore, the study did not perform an extensive characterization of the transcriptional changes—i.e., changes in expression levels as a function of age and EL—that occur in these cell types.
In this study, we employ a multi-modal approach that combines single-cell transcriptomics with cell-surface protein profiling to characterize both the composition and transcriptional profiles of the peripheral immune system of centenarians. We perform a harmonization of a novel single cell dataset of centenarians with diverse, publicly available peripheral blood scRNA-seq datasets of aging and longevity in an effort to understand the dynamics of circulating immune cell populations throughout the human lifespan, and in particular, in EL.
Methods
Experimental procedure
Centenarians were enrolled in North America in 2019. The study was approved by the Boston Medical Center and Boston University Medical Campus IRB and all participants provided written informed consent. We provide a brief overview of the overall approach, and detailed information and methods for the recruitment for human subjects, blood sample collection and processing, CITE-seq and flow cytometry analysis of PBMCs, and all statistical methods in Supplementary Materials.
Single cell analysis
CITE-seq data of centenarians
Cellular Indexing and epitopes sequencing (CITE-seq) was performed on the 7 centenarians and 2 younger age individuals using a commercial droplet-based platform (10x Chromium). Detailed methods for preprocessing data, filtering, PCA analysis, batch correction, clustering, and cell type identification are provided in the Supplementary Materials.
Harmonization of datasets of aging and EL
We harmonized the CITE-seq data of centenarians from NECS with publicly available scRNA-seq data sets using the Harmony algorithm17 to perform integrative analyses across four age groups of the human lifespan (Supplementary Table S3). To avoid assumptions on the age patterns (e.g. linearity) we determined four age groups in the integrated datasets by grouping subjects into four approximate, quantile-based groups of age (Supplementary Table S3).
Heterogeneity of the overall cell type distribution
We calculated a normalized entropy-based metric18 to describe the heterogeneity of the proportions of all cell types of each sample. This metric is higher in samples with more heterogenous (uniform) proportions of cell types and lower in samples that are more enriched in specific cell types.
Effect of age and sex on cell type distribution
We analyzed the effect of age and sex on the proportions of all the 13 cell types at subject level, using a Bayesian multinomial regression model.
Analysis of the hierarchy of peripheral immune compartments
We used K2Taxonomer (v1.0.5)19 to perform top-down partitioning of cell types based on the relative similarity of their transcriptomic profiles and generate a data-driven hierarchy of peripheral immune compartments. Within each level of the hierarchy, we tested the significance of the differences in the cell type diversity statistics between age groups using ANOVA with p-value threshold of 0.05.
Validation of cell type patterns
We calculated cell type proportions of the NECS samples using a spectral flow cytometry panel that included specific markers for all identified populations. We integrated these new data with a mass cytometry dataset previously published as a part of the immune monitoring study across multiple age groups.5
Cell type specific differential gene expression analysis
To investigate the cell type specific differences across age groups, we analyzed the log-normalized gene expression abundances using a Bayesian mixed effects regression model of age adjusted by sex, ethnicity, batches of data. We included a random effect to account for the grouping of cells into subjects. We calculated the FDR for each age group comparison based on the Benjamin and Hochberg correction for multiple testing across all genes tested. We selected significantly different genes based on FDR < 0.05 and fold change (FC) of at least 10 percent.
Bulk level differential gene expression analysis
We performed differential gene expression analysis at the bulk level between age groups using DESeq220 of genes expressed in at least 50% of the smallest cell type population. Significant genes at the bulk level were evaluated according to a fold change of at least 50% and FDR < 0.05.
Role of funding source
No funding sources had any role in the study design, data collection, data analyses, interpretation, writing of report, and decision to submit the paper for publication.
Results
The peripheral immune landscape of centenarians at single cell resolution
Using the 10X Genomics platform for droplet-based Cellular Indexing of Transcriptomes and Epitopes sequencing (CITE-seq),21 we simultaneously profiled the transcriptome-wide expression and the 10 marker cell surface-level protein expression of 16,082 PBMCs from seven centenarians (100–119 years of age) and two younger individuals with no known history of familial longevity (20–59 years of age). All nine subjects were of European descent and from the New England Centenarian Study (NECS), with an average sample capture of 1833 cells (Table 1). We accounted for technical differences and integrated multiple samples using Harmony17 (Supplementary Figure S1). We then performed Louvain graph-based clustering to group cells into populations of similar expression profiles, and used Uniform Manifold Approximation and Projection (UMAP)22 of cell expression profiles to visualize the single cells in a 2-dimensional space.
Table 1.
Table of demographic characteristics of the 7 centenarians included in this study.
| Sample ID | Age range | Sex |
|---|---|---|
| EL1 | 100–105 | Male |
| EL2 | 105+ | Female |
| EL3 | 105+ | Female |
| EL4 | 100–105 | Female |
| EL5 | 105+ | Male |
| EL6 | 105+ | Male |
| EL7 | 100–105 | Female |
| YA1 | 44 | Male |
| YA2 | 34 | Female |
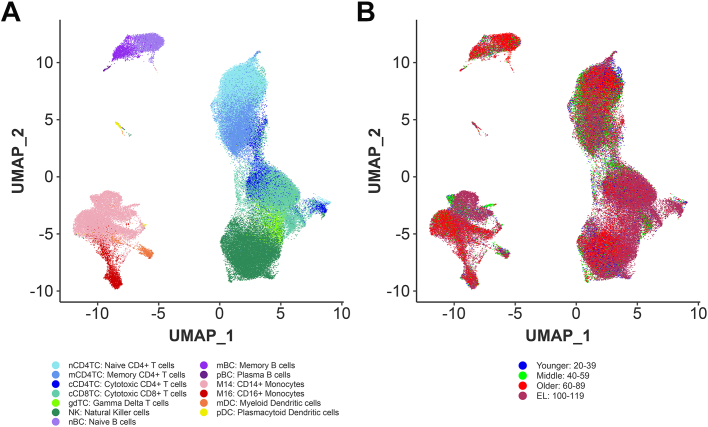
We leveraged the 10 immune cell-surface protein expression markers to identify the major lymphocyte and myeloid cell types (Supplementary Figure S2, Supplementary Table S1), with subtypes subsequently characterized by transcriptional immune cell signatures previously characterized in human peripheral blood23 and fetal liver24 (Supplementary Figures S3–S7, Supplementary Table S2). This approach identified 11 immune cell types that included major lymphocyte populations: CD4+ T cells (CD4TC) with noncytotoxic naive and memory subtypes (nCD4TC, mCD4TC) and cytotoxic subtype (cCD4TC), CD8+ T cells (CD8TC) with cytotoxic subtype (cCD8TC), B cells (BC) with naive and memory subtypes (nBC and mBC), and Natural Killer cells (NK) (Supplementary Figures S2 and S3). In addition, we identified major myeloid populations: monocytes with CD14+ and CD16+ subtypes (M14 and M16) and dendritic cells (DC) with myeloid and plasmacytoid subtypes (mDC and pDC) (Supplementary Figures S2 and S3).
Centenarians display alterations in immune cell repertoire in comparison to younger age groups
To characterize peripheral immune cell type composition and gene expression profiles across the human lifespan, we integrated our data with two publicly available PBMC datasets of aging and longevity that include subjects of European and Japanese descent16,25 (Fig. 1A and B, Supplementary Figure S8). The integration of these datasets with our novel NECs dataset produced a total of 102,284 cells from 66 individuals across four age groups: 12 subjects of younger age (20–39 years), 26 subjects of middle age (40–59 years), 14 subjects of older age (60–89 years), and 14 EL subjects (100–119 years) (Supplementary Tables S3 and S4). Technical differences between datasets were accounted for using Harmony.17 This analysis identified two additional cell types: plasma B cells (pBC) and gamma delta T cells (gdTC) for a total of 13 cell types across the datasets (Fig. 1B).
Fig. 1.
The immune landscape of peripheral blood cells from subjects with extreme longevity at single cell resolution.A. UMAP embeddings of PBMCs from all 66 subjects representative of the human lifespan from the integrated scRNA-seq datasets (NECS, PNAS, and NATGEN, labelled by the identified immune cell subtypes. B. UMAP embeddings of PBMCs from all 66 subjects representative of the human lifespan from the integrated scRNA-seq datasets (NECS, PNAS, and NATGEN, labelled by the four age groups.
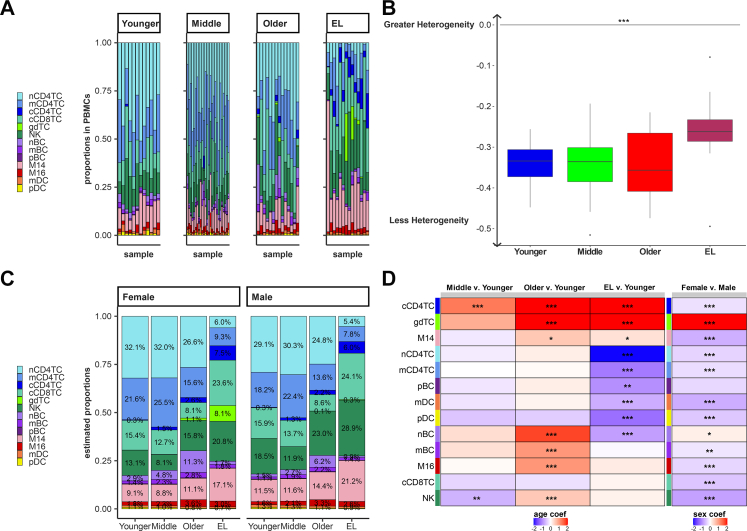
Fig. 2A displays the observed proportions of the 13 immune cell types in the 66 subjects, stratified by age. Notably, the EL group was characterized by an increase in the proportion of myeloid cells and a reduction in the proportion of lymphocytes: The ratio of myeloid cells to lymphocytes was approximately 13.8/86.2% across the three younger age groups (20–89 years) and shifted to 25.2/74.8% in the EL group (Fig. 2A, Supplementary Table S5). This shift in myeloid cells and lymphocytes is an expected trend in aging.26 The barplot in Fig. 2A suggests that the distribution of proportions of the 13 immune cell types becomes more uniform in the EL group. To formalize this observation, we next calculated the cell type diversity statistic of each sample.18 This statistic is essentially an entropy-based score that we introduced to summarize the vector of proportions of cell types in a sample. The score is normalized between −1 and 0, with −1 corresponding to the case of a single cell type being present and 0 corresponding to the case when all cell types are present in the exact same proportion and therefore more uniform. The analysis of the 13 immune populations showed a trend towards an increase of the cell type diversity statistic in EL compared to younger age groups, although this difference was not statistically significant (F-test, p-value = 0.7231) (Supplementary Figure S9). When nCD4TC and mCD4TC were combined as noncytotoxic CD4+ T cells, the increase of the cell type diversity statistic in EL compared to younger ages was statistically significant (F-test, p-value = 0.0001875) (Fig. 2B, Supplementary Table S6). This analysis formalized the observation that the composition of PBMCs in EL subjects becomes more heterogeneous.
Fig. 2.
Extreme longevity demonstrates shifts in immune cell repertoire compared to younger age groups.A. Bar chart of the relative proportions of the lymphocyte and myeloid immune cell subtypes for each sample across the integrated scRNA-seq datasets: Younger Age, Middle age, Older age, and EL. B. Boxplot of subject specific cell type diversity statistic of 12 immune cell subtypes, grouped by age groups (F-test p-value = 0.0001875). C. Bar chart of the estimated proportions of the lymphocyte and myeloid immune cell subtypes in each age group, grouped separately for males and females. The proportions were estimated using a Bayesian multinomial regression model, and the average 1833 cells per sample allowed us to estimate the smallest proportion 0.3% with 95% confidence. D. Heatmap of the age coefficient comparing Middle, Older, and EL age groups to the Younger age group (right) and heatmap of the sex coefficient for each cell type comparing Females compared to Males. We calculated the Z-statistic and p-value of significance for each coefficient, represented with: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To estimate the proportions of cell types by age and sex, we next analyzed the observed cell type proportions from all 66 subjects using a Bayesian multinomial regression (See Methods, Supplementary Tables S7–S10). This analysis produced age and sex specific estimates of each of the 13 cell type proportions across the four age groups (Fig. 2C and D). The estimates suggest that there are three main groups of immune cells based on their distributions at different ages and EL: 1) cell types whose proportions increase or decrease monotonically with age and EL (Aging-Related), 2) cell types whose proportions increase or decrease only in the EL group (EL-Specific), 3) cell types whose proportions increase or decrease with age, but these changes do not continue in the EL group (Aging-Specific).
We observed Aging-Related changes (i.e., change in both aging and EL) in cCD4TC, gdTC, and M14 populations. The estimated proportion of cCD4TC increased steadily with increasing age, representing 7.5% of PBMCs in males with EL and 6.0% of PBMCs in females with EL compared to less than 1% of the PBMCs in the younger age groups (Fig. 2C and D). This significant change in cCD4TC in centenarians compared to younger individuals is consistent with previous findings.16 We observed a similar aging-related change in the estimated proportions of gdTC and M14 (Fig. 2C and D).
Five lymphocyte and myeloid populations were observed to have EL-Specific changes (i.e., change only in EL) and include nCD4TC, mCD4TC, pBC, mDC, and pDC. The lower frequency of nCD4TC and mCD4TC in EL is known.16 However, the estimated lower proportion of mDC that decreased to 0.9% of PBMCs in males with EL and 0.7% of PBMCs in females with EL compared to 1.3% in males and 1.1% in females in the younger age group has not been reported (Fig. 2C and D). Furthermore, we observed Aging-Specific changes (i.e., change in aging but not EL) in nBC, mBC, and M16 populations. The estimated proportion of nBC significantly increased to 6.2% of PBMCs in males and 11.3% of PBMCs in females in the older age group compared to 1.5% of PBMCs in males and 2.9% of PBMCs in females in the younger age group (Fig. 2C and D). However, the estimated proportions of PBMCs that were nBC in EL were 0.9% in males and 1.7% in females (Fig. 2C and D).
In addition, we identified significant changes in composition between older age compared to EL in nCD4TC, cCD4TC, cCD8TC, gdTC, nBC, NK, and M14 (Supplementary Table S11). For example, we found a significant decrease in proportion of cCD4TC in older age compared to EL, which is consistent with the increase in proportion of cCD4TC we observed in the older and EL age groups compared to younger age (Supplementary Table S11).
By examining the proportion of all 13 cell types together, we highlighted major differences in the overall makeup of PBMCs of centenarians and showed a major shift from innate to adaptive cell types with older age.
Extreme longevity displays a shift in immune resilience strategy within lymphocyte and myeloid populations compared to younger age groups
To investigate whether the makeup within immune compartments also changed with age and EL and with possible effects on their biological functions and immune resiliency strategies, we next examined the make-up of various immune cell types within their myeloid and lymphocyte lineages. To this end, we generated a hierarchy of peripheral immune compartments, partitioned cell types, based on the gene expression profiles of the immune cell types using K2Taxonomer,19 and then calculated the average proportions of cell types within each level of the hierarchy (see Methods) (Supplementary Tables S12 and S13). Fig. 3 displays the results of this analysis and reveals specific changes of the composition of myeloid and lymphocyte compartments in the EL group as compared to younger age groups. In addition, we generated independent estimates of a large portion of the same cell counts in the NECS subjects using flow cytometry, and integrated the data with publicly available mass cytometry data from Alpert et al.5 that included 39 younger individuals (mean age = 29) and 50 older individuals (mean age = 80). Supplementary Figure S11 contrasts the age trend of cell types based on the cytometry dataset to the original analysis based on scRNA-seq data. We discuss selected results next, with the complete analysis available in Supplementary Tables S14–S26.
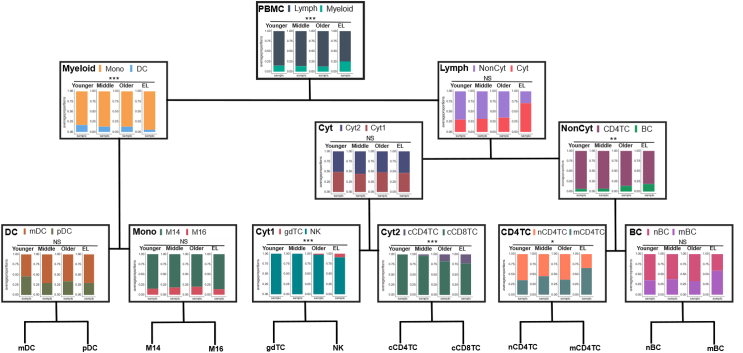
Fig. 3.
Shift in the immune resilience strategy within lymphocyte and myeloid populations in centenarians. The average immune cell type proportions across the four age groups of the human lifespan along the hierarchy of peripheral immune compartments: PBMC (Myeloid v. Lymph), Myeloid (Mono v. DC), Lymph (NonCyt v. Cyt), DC (mDC v. pDC), Mono (M14 v. M16), NonCyt (CD4TC v. BC), Cyt (Cyt1 v. Cyt2), Cyt1 (gdTC v. NK), Cyt2 (cCD4TC v. cCD8TC), and CD4TC (nCD4TC v. mCD4TC) and BC (nBC v. mBC).
The top of the hierarchy recapitulates the significantly larger proportion of myeloid cells (Myeloid) and smaller proportion of lymphocytes (Lymph) observed in centenarians' PBMCs discussed earlier. The reduction of lymphocytes was validated by flow cytometry analysis of 7 centenarians and 3 younger age individuals from NECS, and 39 younger age and 50 older age individuals from Alpert et al.5 (Supplementary Figure S11, Supplementary Table S26). The analysis of the Myeloid compartment (left branch of the tree in Fig. 3) showed that centenarians’ Myeloid cells were mainly monocytes (Mono) rather than dendritic cells (DC) (EL Mono to DC ratio 94.58%/5.42%) compared to a lower fraction of Mono and higher fraction of DC observed in all other age groups (Mono to DC ratio 85.59%/14.41%). The difference in the two distributions was statistically significant, (F-test of cell type diversity statistic, p-value = 0.0001315) (Fig. 3, Supplementary Figure S10). We observed the increase in Mono compared to DC in EL based on the proportions measured by cytometric of EL and younger individuals from NECS (Supplementary Figure S11, Supplementary Table S26). This change in composition of DC has not been reported before.6,27 Although no additional changes were detected within Mono and DC, we observed similar trends in cell proportions measured by flow cytometry across age within Mono and DC (Supplementary Figure S11, Supplementary Table S26).
The analysis of the lymphocyte compartments (right branch of the tree in Fig. 3) showed that more than 70% of centenarians' lymphocytes were Cytotoxic (Cyt: 70.94% and NonCyt: 29.06%) compared to the younger age group (Cyt: 30.30% and NonCyt: 69.70%). This difference in distributions was only borderline statistically significant in our analysis (F-test of cell type diversity statistic, p-value = 0.05032) but consistent with results reported by other investigators.2 In addition, the hierarchy showed further sub-types of Cyt and NonCyt that had unique composition in centenarians. For example, centenarians’ NonCyt had a significant lower proportion of CD4TC (81.55%) and larger proportion of BC (18.45%) compared to all younger age groups (CD4TC: 90.86% and BC: 9.41%, F-test, p-value = 0.003313) (Fig. 3, Supplementary Figure S10). We observed a similar shift between CD4TC and BC in EL compared to younger individuals from NECS based on proportions measured by flow cytometry, but was not observed in the younger and older individuals from Alpert et al.5 (Supplementary Figure S11, Supplementary Table S26). In previous studies, both noncytotoxic CD4TC and BC have been reported to decrease in PBMCs of long-lived individuals,16 but this shift between the two cell types has not been previously observed.
The composition of CD4TC in centenarians was characterized by an expansion of mCD4TC (65.72%) and a reduction of nCD4TC (34.28%) compared to all younger age groups (mCD4TC: 39.46% and nCD4TC: 60.54%, F-test, p-value = 0.02852) (Fig. 3, Supplementary Figure S10). The composition of BC in the EL group had a similar shift from naive (nBC: 41.06%) to memory cells (mBC: 58.94%) but the change did not reach statistical significance (F-test, p-value = 0.7168) (Fig. 3, Supplementary Figure S10). In addition, we observed similar shifts between nBC and mBC between EL and younger individuals of NECS based on flow cytometry analysis (Supplementary Figure S11, Supplementary Table S26).
In summary, using the hierarchy of peripheral immune compartments, we identified major shifts in the makeup of centenarian PBMCs compared to those of younger ages within the myeloid and lymphocyte lineages that were obscured globally in terms of all 13 cell types together.
Centenarians display unique transcriptional profiles associated with extreme longevity
The previous two analyses characterized the makeup of centenarian PBMCs globally and within their myeloid and lymphocyte lineages in terms of proportions of the various cell types. We next examined their expression profiles relative to younger age groups. For each cell type, we performed an analysis to discover genes with differential expression as a function of age and/or EL, which identified 151 genes with age- or EL-associated differential expression in at least one cell type (Fig. 4, Supplementary Table S27). The number of significantly differentially expressed genes varied by cell types and comparison groups (Fig. 4A): on average, the comparison of expression profiles of cell types in the middle vs. the younger age group produced a smaller number of differentially expressed genes (i.e., fewer differences) than the comparisons of the older vs. younger age and the EL vs. younger age group. Fig. 4B shows clear differential gene expression patterns for all cell type-specific signatures across each age group comparison. Of these cell type-specific signatures, we identified 20 differential genes that have been previously identified to change with age in transcriptional studies of aging in PBMCs,6,8,28 including leucine-rich repeat neuronal protein 3 (LRRN3), lymphoid enhancer-binding factor-1 (LEF1), and Cathepsin H (CTSH).
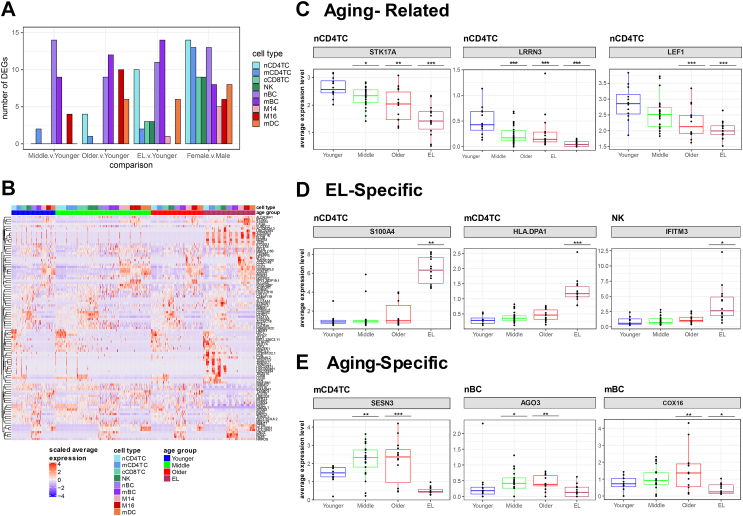
Fig. 4.
Cell type gene expression changes demonstrate three patterns across the human lifespan.A. Table of the number of significant differentially expressed genes across aging comparisons: Middle v. Younger age, Older v. Younger age, EL v. Younger age based on fold change threshold of minimum 10% change and FDR less than 0.05. B. Heatmap of scaled average expression per sample of all significant genes across all cell types grouped by age group. C. Boxplots of expression levels of specific significant genes in particular cell types demonstrating changes in aging and EL (Aging-Related) with at least a nominal significance, D. changes only in EL (EL-Specific), and E. changes in aging not in EL (Aging-Specific). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Further examination of the age-specific expression changes identified three main patterns that are summarized in Fig. 4C–E, and closely mirror the changes in cell type composition described above. These patterns include 1) genes whose expression increase or decrease monotonically with age and EL (Aging-Related) with at least a nominal significance, 2) genes whose expression increase or decrease only in the EL group (EL-Specific), 3) genes whose expression increase or decrease with age, but these changes do not continue in the EL group (Aging-Specific).
We identified 35 genes with the Aging-Related pattern (i.e., change in both aging and EL) of differential expression across the various immune cell types. Fig. 4C shows selected examples of genes in the first group and Supplementary Figures S12–S20 include more examples. The set includes genes involved in DNA damage response such as serine/threonine-protein kinase 17A (STK17A) in nCD4TC (Fig. 4C). STK17A is a positive regulator of apoptosis and a variant near STK17A was previously reported to be associated with longevity in WGS analysis.29 In addition, STK17A has been previously identified to change in age in T cells of PBMCs.6 Furthermore, two genes previously reported to increase in expression with age in peripheral blood29,30 including in T cells6 were in group 1: LRRN3 and LEF1 in nCD4TC (Fig. 4C).
We identified 25 genes with an EL-Specific pattern (i.e., change only in EL) in several cell types (Fig. 4D, Supplementary Figures S12–S20). Fig. 4D shows examples of genes that appear to be only expressed in immune cells from the EL group. An example of a gene only expressed in nCD4TC of the EL group is S100 protein-coding gene, S100A4 (Fig. 4D). S100 proteins have been implicated in aging-related diseases such as Alzheimer's disease as well as longevity.31,32 In addition, S100A4 has been previously implicated in age-related changes in T cells of PBMCs.6 Group 2 also included the HLA class II histocompatibility antigen gene HLA-DPA1 in mCD4TC (Fig. 4D) and cCD8TC (Supplementary Figure S14), and interferon-induced transmembrane proteins IFITM3 in NK (Fig. 4D) and in mDC (Supplementary Figure S18), and IFITM2 in cCD8TC (Supplementary Figure S12). These major histocompatibility and interferon-related genes are involved in antigen presentation and activation of immune response pathways.
We identified 26 genes with the Aging-Specific pattern (i.e., change in aging but not in EL), with expression levels that change with age but not in EL across various cell populations. Fig. 4E shows examples of 3 genes and additional examples are in Supplementary Figures S12–S20. This set includes genes that respond to oxidative stress including sestrin 3 (SESN3) in nCD4TC (Fig. 4E) and mCD4TC (Supplementary Figure S13), argonaute RISC catalytic component 3 (AGO3) in nBC, and cytochrome C oxidase assembly factor (COX16) in mBC (Fig. 4E). SESN3 is part of the sestrin family of stress-induced metabolic proteins and is stimulated in response to oxidative stress/damage by FOXO3, a transcription factor associated with longevity,33,34 while AGO3 is part of the argonaute (AGO) family of proteins that have been previously implicated in aging and oxidative stress-induced senescence.35
In addition, we found 62 significant cell type specific differential genes between older and EL age groups, including S100A4 which is significantly lower in older age compared to EL in nCD4TC (Supplementary Table S28). This is consistent with the significant increase observed in EL compared to younger age (Fig. 4D). Furthermore, we examined the gene expression changes between females and males. We detected 28 significant differential genes between females compared to males across cell types (Fig. 4A, Supplementary Table S27). Of the 28 genes, six genes were differentially expressed across most or all cell types. This set of genes included X-chromosome and Y-chromosome linked genes such as ribosomal protein S4 X-linked (RPS4X), X inactive specific transcript (XIST), and eukaryotic translation initiation factor 1A X-linked (EIF1AX), ribosomal protein S4 Y-linked (RPS4Y1), DEAD-box helicase 3 Y-linked (DDX3Y), and eukaryotic translation initiation factor 1A Y-linked (EIF1AY) (Supplementary Table S27). With the exception of EIF1AX, these genes have been previously reported to have gene expression differences based on sex across immune cell types in PBMCs.9 In addition, CD99, involved in T cell activation and pro-inflammatory mechanisms,36 had significantly lower expression in females nCD4TC than males.
In addition to cell type specific expression profiles, we analyzed gene expression aggregated over different cell types (see Methods). This analysis identified a greater number of genes with significantly different expression in the EL v. younger age group comparison (387 genes) compared to middle v. younger age group (0 genes) and older v. younger age group (3 genes) (Supplementary Figure S21, Supplementary Tables S29). Among the 387 significant differential genes identified in EL v. younger age, we found 136 genes over-expressed and 251 under-expressed in the EL group compared to the younger group (Supplementary Figure S21, Supplementary Tables S29). In addition, of the 387 significant genes identified in EL v. younger age, we identified 164 genes previously identified to change in expression with age8,28 including LEF1 and LRRN38,28 that we also identified at the single cell level as well as CD28 antigen molecule.28
Discussion
Overview of main results
Using a multi-modal, single cell approach, we generated cell composition and transcriptional profiles from the PBMCs of 7 centenarians using CITE-seq. We integrated this novel data set with publicly available scRNA-seq datasets of aging and longevity across the human lifespan to characterize cell type composition and gene expression profiles unique to centenarians. We observed substantial changes in the composition of immune cells with age, including novel changes in myeloid cell types: M14, M16, mDC, and pDC. We also conducted a novel analysis of a data-driven hierarchy of peripheral immune compartments, which revealed previously undetected changes in the composition of T cells and B cells in centenarians and we validated several of these changes using orthogonal approaches (flow cytometry). Based on gene expression changes, we identified cell type-specific transcriptional signatures of extreme longevity that include aging-related changes as well as unique gene changes in the immune profiles of centenarians.
Cell type composition profiles based on the total number of PBMC populations
The peripheral blood immune cell repertoire of individuals is known to change with age.5,6 Previous transcriptional studies have shown decreases in lymphocytes and increases in myeloid cells with age,26 which we also observed in the peripheral blood of centenarians (Fig. 2). However, in addition to these common changes across aging, our analysis identified patterns of immune cell profiles and compositional alterations that are unique to centenarians. We observed expected shifts in the composition of centenarians’ PBMCs from non-cytotoxic (e.g., nCD4TC and mCD4TC) to cytotoxic lymphocytes (e.g., cCD4TC) that have been observed previously in studies of human longevity.16 Similarly, the decrease of nBC with aging and longevity has also been reported previously.5,6 However, we also discovered novel compositional patterns of extreme old age including aging-related changes (e.g. a significant increase of M14 in older age that continues in the EL group), EL-specific changes (e.g. mDC and pDC display no significant change among the three younger age groups but a unique, significant decrease occurs in EL), and aging-specific changes independent of EL (e.g. a significant increase of M16 in older age that then decreases in the EL age group) (Fig. 2). The extent to which these unique patterns in centenarians are the drivers of extreme longevity or just the consequence of having reached an extreme old age remains an open question, since not everything we see in centenarians is necessarily important to reach extreme old ages. Additional data are needed to understand the effect of these patterns on human longevity.
Cell type composition profiles within peripheral cell compartments
Utilizing the traditional method for characterizing cell type composition profiles based on the total number of PBMC populations, we identified novel compositional changes of extreme old age (Fig. 2). However, this analysis provides limited insight into immune cell type change within the lymphocyte and myeloid compartments. We used a novel data-driven approach to create a hierarchy of peripheral immune compartments. The analysis summarized in Fig. 3 shows, for example, that the proportion of lymphocytes in the PBMCs of centenarians decreases compared to younger age groups, but a significant change in composition also occurs. Specifically, centenarians' lymphocytes are characterized by an almost 50% decrease of NonCyt, which become enriched for BC that are themselves enriched for mBC. Notably, CD4TC and BC have a significant role in the immune system's response to infection.37 BC are associated with the antibody-mediated immune response that triggers a quick response against pathogens, while T lymphocytes such as CD4TC are associated with cell-mediated immunity that develops at a slower rate. Studies have also found crosstalk between BC and CD4TC, showing their co-dependence in protective immune response.37 The shift from CD4TC to BC suggests that centenarians develop a more immediate immune response to infections. We also observed a significant shift from naive to memory subtypes within CD4TC and, to an extent, within BC suggesting that centenarians did not escape infection but experienced a greater exposure to infections and were able to develop robust responses to them. Previous studies have also shown associations between the capacity to control inflammation and preserving immunocompetence with longevity as an immune resilience phenotype.38, 39, 40 It is possible that the unique make up of immune cells we observed in centenarians may represent an adaptation of their immune system or a compensatory mechanism to the loss of key immune cell types. Interestingly, in almost all compartments displayed in Fig. 3, the EL group was characterized by a lesser skewed distribution of cell types compared to younger age groups. A more heterogeneous distribution of immune cells may be the driver of their immune resiliency. Compositional heterogeneity may reflect immunocompetence as a dynamic balance or homeostasis.40 Reciprocally, immune imbalance is often characteristic of suboptimal responses to infections (e.g., COVID-1941) or compensation by less effective or exhausted cell mediators (e.g., NK cells42) that compromise health span.
Gene expression profiles
Our analysis identified three patterns of age-related changes: monotonic changes across the lifespan, age-related changes that are absent in the EL group, and changes that are unique to centenarians. Interestingly, we noticed similar patterns in the serum proteome of centenarians.43 By comparing the serum proteome of centenarians to septuagenarians, we discovered aging-related protein signatures as well as a protein signature that was unique to centenarians. We also discovered an age-specific protein signature that was not extended as expected in centenarians. Without the additional data of blood expression profiling of centenarians when they were of younger age, we cannot determine whether these EL specific patterns are the drivers to extreme human longevity or the effect of extreme old age. However, some of the genes with differential expression in the EL group have been linked to aging and longevity studies. For example, the expression of a variant of STK17A that we found associated with age in nCD4TC (Fig. 4C) was higher in centenarians.29 STK17A is involved in DNA damage response, positive regulation of apoptosis, and mitochondrial and metabolic regulation of reactive oxygen species (ROS). This association is consistent with results from previous studies that correlated DNA damage repair mechanisms to aging and longevity.25,40,41 S100A4, part of the S100 family of calcium-binding proteins, showed an EL specific change in nCD4TC (Fig. 4D). S100 proteins such as S100A13 have been implicated in their role in longevity, including an association with APOE genotypes in centenarians.28 In addition, the S100 family of proteins are associated with inflammatory pathways in the brain connected to aging-related diseases such as Alzheimer's disease.27 In CD4+ T cells, S100A4 was found to be higher expressed in older mice compared to younger mice, and involved in inflammation and activation.44,45 A recent study discovered an S100 protein to be a critical regulator of hematopoietic stem cell renewal through mitochondrial metabolic regulation and function.46 In rats, recombinant S100A4 demonstrates an anti-apoptotic function in response to oxidative stress injury.47 Other transcriptional signatures that we identified in our analysis, such as SESN3 and AGO3 (Fig. 4E), are involved in DNA damage response and mitochondrial and metabolic regulation activated in response to oxidative stress.33,34 Sestrins such as SESN3 are highly conserved stress inducible proteins that protect the immune system in response to DNA damage and oxidative stress.34 More specifically, the induction of SESN3 in response to oxidative damage is activated by FOXO3,34 a transcription factor associated with longevity.33 AGO3 is part of the AGO family of proteins involved in miRNA association and stability that has been implicated in oxidative stress-induced senescence and aging.35 In the immune system, mitochondrial regulation plays a role in immune cell transcription and activation48,49 and may promote longevity.50 In addition, the decline of mitochondrial quality and activity is associated with aging and senescence.48,49 The connection to mitochondrial and metabolic regulation suggests that centenarians may have changes that occur in mitochondrial and metabolic regulation and function and should be further investigated. We note that our signatures of aging derived from “pseudo-bulk” data closely match previously published signatures of aging derived from bulk data8,28 and included a much larger number of differentially expressed genes compared to the analysis at single cell level. The difference between the bulk and single-cell signatures could point to the fact that much of the observed differential expression in bulk RNA is driven by differences in cell composition rather than by differences in within-cell type expression.
Caveats and limitations and future directions
This study has several limitations, particularly the cross-sectional nature of the data and the small sample size. We integrated our data with multiple single cell datasets to increase sample size, but we intentionally adopted a conservative approach to identify cell type specific signatures across age groups. Larger studies of centenarians will be needed to detect robust transcriptional changes that characterize EL. Although we identified a small set of transcriptional signatures, we were still able to identify patterns of EL that have been discovered in previous studies including EL specific changes. In addition, the compositional and gene expression changes that we observed in centenarians displayed not only EL specific changes but also age-related changes. How EL differs from regular aging remains unclear, and more investigation and future studies will be required to elucidate this difference and investigate the mechanisms behind the patterns observed in extreme old age. Access to the peripheral blood of centenarian offspring and studying longitudinal changes in PBMC populations may help to better define immunocompetence causal drivers of the beneficial health outcome observed in EL.
Conclusion
Overall, these findings display age-related changes in composition and transcription in both lymphocyte and myeloid cell types that collectively reflect immunocompetent profiles that may in part account for centenarians' ability to reach extreme ages. The extent to which some of the unique compositional and transcriptional patterns we identified in centenarians’ PBMCs are the drivers or markers of extreme old age remains an open question. To our knowledge, this is the first study to define cell compositional and transcriptional signatures of EL across immune cell types in peripheral blood. This study provides a foundation and resource to explore immune resilience mechanisms engaged in exceptional longevity.
Contributors
PS, SM, and GM conceptualized the study. TD prepared all scRNA-seq libraries. CVM performed processing and generation of scRNA-seq count data. ER performed K2Taxonomer and the generation of the peripheral immune hierarchy. ACB generated flow cytometry data and analyses. TK performed all other computational analyses. TP and GM organized collection of human samples. MM, TP, and SA provided insights and advice. TK, TD, ACB, SM, GM, and PS interpreted the data and wrote the manuscript with the feedback of all the authors. All authors read and approved the final version of the manuscript. TK, ER, ACB, SM, and PS have accessed and verified the underlying data.
Data sharing statement
The data that support these findings are publicly available and were accessed from several repositories. NATGEN single cell expression data and subject level data were publicly available as referenced in25: https://molgenis58.target.rug.nl/scrna-seq/. PNAS single cell expression data and subject level data was available as referenced in16: http://gerg.gsc.riken.jp/SC2018/. NECS will be available from Synapse (https://adknowledgeportal.synapse.org/Explore/Projects/DetailsPage?Grant%20Number=UH2AG064704). All scripts to reproduce analyses and figures reported in this paper are available on github (https://github.com/Integrative-Longevity-Omics/sc_pbmc_centenarians).
Declaration of interests
TK, TD, MM, SM, PS, GM, SA, and TP report grants from National Institute on Aging, during the conduct of the study. ACB reports grants from NIH Instrumentation grant: S10 OD021587, during the conduct of the study. CVM and ER declare no competing interests.
Acknowledgements
TK, SM, PS, GM, SA, TP are supported by NIH-NIA UH2AG064704 and U19AG023122. MM and PS are supported by NIH NIA Pepper center: P30 AG031679-10. This project is supported by the Flow Cytometry Core Facility at BUSM. FCCF is funded by the NIH Instrumentation grant: S10 OD021587.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104514.
Appendix A. Supplementary data
References
- 1.Aw D., Silva A.B., Palmer D.B. Immunosenescence: emerging challenges for an ageing population. Immunology. 2007;120:435–446. doi: 10.1111/j.1365-2567.2007.02555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deleidi M., Jäggle M., Rubino G. Immune aging, dysmetabolism, and inflammation in neurological diseases. Front Neurosci. 2015;9 doi: 10.3389/fnins.2015.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiello A., Farzaneh F., Candore G., et al. Immunosenescence and its hallmarks: how to oppose aging strategically? A review of potential options for therapeutic intervention. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.02247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franceschi C., Garagnani P., Parini P., Giuliani C., Santoro A. Inflammaging: a new immune–metabolic viewpoint for age-related diseases. Nat Rev Endocrinol. 2018;14:576–590. doi: 10.1038/s41574-018-0059-4. [DOI] [PubMed] [Google Scholar]
- 5.Alpert A., Pickman Y., Leipold M., et al. A clinically meaningful metric of immune age derived from high-dimensional longitudinal monitoring. Nat Med. 2019;25:487–495. doi: 10.1038/s41591-019-0381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Y., Liu X., Le W., et al. A human circulating immune cell landscape in aging and COVID-19. Protein Cell. 2020;11:740–770. doi: 10.1007/s13238-020-00762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mogilenko D.A., Shpynov O., Andhey P.S., et al. Comprehensive profiling of an aging immune system reveals clonal GZMK+ CD8+ T cells as conserved hallmark of inflammaging. Immunity. 2021;54:99–115.e12. doi: 10.1016/j.immuni.2020.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Peters M.J., Joehanes R., Pilling L.C., et al. The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. doi: 10.1038/ncomms9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Z., Chen B., Liu X., et al. Effects of sex and aging on the immune cell landscape as assessed by single-cell transcriptomic analysis. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2023216118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebastiani P., Perls T.T. The genetics of extreme longevity: lessons from the new England centenarian study. Front Genet. 2012;3 doi: 10.3389/fgene.2012.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hitt R., Young-Xu Y., Silver M., Perls T. Centenarians: the older you get, the healthier you have been. Lancet. 1999;354:652. doi: 10.1016/S0140-6736(99)01987-X. [DOI] [PubMed] [Google Scholar]
- 12.Terry D.F., Sebastiani P., Andersen S.L., Perls T.T. Disentangling the roles of disability and morbidity in survival to exceptional old age. Arch Intern Med. 2008;168:277–283. doi: 10.1001/archinternmed.2007.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen S.L., Sebastiani P., Dworkis D.A., Feldman L., Perls T.T. Health span approximates life span among many supercentenarians: compression of morbidity at the approximate limit of life span. J Gerontol A Biol Sci Med Sci. 2012;67:395–405. doi: 10.1093/gerona/glr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayed N., Huang Y., Nguyen K., et al. An inflammatory aging clock (iAge) based on deep learning tracks multimorbidity, immunosenescence, frailty and cardiovascular aging. Nat Aging. 2021;1:598–615. doi: 10.1038/s43587-021-00082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrucci L., Fabbri E. Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat Rev Cardiol. 2018;15:505–522. doi: 10.1038/s41569-018-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto K., Kouno T., Ikawa T., et al. Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proc Natl Acad Sci U S A. 2019;116:24242–24251. doi: 10.1073/pnas.1907883116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korsunsky I., Millard N., Fan J., et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karagiannis T.T., Monti S., Sebastiani P. Cell type diversity statistic: an entropy-based metric to compare overall cell type composition across samples. Front Genet. 2022;13 doi: 10.3389/fgene.2022.855076. https://www.frontiersin.org/article/10.3389/fgene.2022.855076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reed E.R., Monti S. Multi-resolution characterization of molecular taxonomies in bulk and single-cell transcriptomics data. Nucleic Acids Res. 2021;49:e98. doi: 10.1093/nar/gkab552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoeckius M., Hafemeister C., Stephenson W., et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McInnes L., Healy J., Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. arXiv. 2020 http://arxiv.org/abs/1802.03426 180203426 [cs, stat]; published online Sept 17. [Google Scholar]
- 23.Chen B., Khodadoust M.S., Liu C.L., Newman A.M., Alizadeh A.A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popescu D.-M., Botting R.A., Stephenson E., et al. Decoding human fetal liver haematopoiesis. Nature. 2019;574:365–371. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Wijst M.G.P., Brugge H., de Vries D.H., Deelen P., Swertz M.A., Franke L. Single-cell RNA sequencing identifies celltype-specific cis-eQTLs and co-expression QTLs. Nat Genet. 2018;50:493–497. doi: 10.1038/s41588-018-0089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiger H., de Haan G., Florian M.C. The ageing haematopoietic stem cell compartment. Nat Rev Immunol. 2013;13:376–389. doi: 10.1038/nri3433. [DOI] [PubMed] [Google Scholar]
- 27.Agrawal A., Gupta S. Impact of aging on dendritic cell functions in humans. Ageing Res Rev. 2011;10:336–345. doi: 10.1016/j.arr.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vo T.K.D., Godard P., de Saint-Hubert M., et al. Transcriptomic biomarkers of human ageing in peripheral blood mononuclear cell total RNA. Exp Gerontol. 2010;45:188–194. doi: 10.1016/j.exger.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Garagnani P., Marquis J., Delledonne M., et al. Whole-genome sequencing analysis of semi-supercentenarians. Elife. 2021;10 doi: 10.7554/eLife.57849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harries L.W., Hernandez D., Henley W., et al. Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell. 2011;10:868–878. doi: 10.1111/j.1474-9726.2011.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cristóvão J.S., Gomes C.M. S100 proteins in Alzheimer's disease. Front Neurosci. 2019 doi: 10.3389/fnins.2019.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sebastiani P., Monti S., Morris M., et al. A serum protein signature of APOE genotypes in centenarians. Aging Cell. 2019;18 doi: 10.1111/acel.13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris B.J., Willcox D.C., Donlon T.A., Willcox B.J. FOXO3 – a major gene for human longevity. Gerontology. 2015;61:515. doi: 10.1159/000375235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y., Huang T., Yu Z., et al. The functions and roles of sestrins in regulating human diseases. Cell Mol Biol Lett. 2022;27 doi: 10.1186/s11658-021-00302-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bu H., Wedel S., Cavinato M., Jansen-Dürr P. MicroRNA regulation of oxidative stress-induced cellular senescence. Oxid Med Cell Longev. 2017;2017 doi: 10.1155/2017/2398696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takheaw N., Earwong P., Laopajon W., Pata S., Kasinrerk W. Interaction of CD99 and its ligand upregulates IL-6 and TNF-α upon T cell activation. PLoS One. 2019;14 doi: 10.1371/journal.pone.0217393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petersone L., Edner N.M., Ovcinnikovs V., et al. T cell/B cell collaboration and autoimmunity: an intimate relationship. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.01941. https://www.frontiersin.org/article/10.3389/fimmu.2018.01941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fulop T., Larbi A., Dupuis G., et al. Immunosenescence and inflamm-aging as two sides of the same coin: friends or foes? Front Immunol. 2018;8 doi: 10.3389/fimmu.2017.01960. https://www.frontiersin.org/article/10.3389/fimmu.2017.01960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee G.C., Restrepo M.I., Harper N., et al. Immunologic resilience and COVID-19 survival advantage. J Allergy Clin Immunol. 2021;148:1176–1191. doi: 10.1016/j.jaci.2021.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marconi V.C., Krishnan V., Ely E.W., Montano M. Immune health grades: finding resilience in the COVID-19 pandemic and beyond. J Allergy Clin Immunol. 2022;149:565–568. doi: 10.1016/j.jaci.2021.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schafer J.L., Li H., Evans T.I., Estes J.D., Reeves R.K. Accumulation of cytotoxic CD16+ NK cells in simian immunodeficiency virus-infected lymph nodes associated with in situ differentiation and functional anergy. J Virol. 2015;89:6887–6894. doi: 10.1128/JVI.00660-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sebastiani P., Federico A., Morris M., et al. Protein signatures of centenarians and their offspring suggest centenarians age slower than other humans. Aging Cell. 2021;20 doi: 10.1111/acel.13290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zemmour D., Zilionis R., Kiner E., Klein A.M., Mathis D., Benoist C. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat Immunol. 2018;19:291–301. doi: 10.1038/s41590-018-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elyahu Y., Hekselman I., Eizenberg-Magar I., et al. Aging promotes reorganization of the CD4 T cell landscape toward extreme regulatory and effector phenotypes. Sci Adv. 2019;5 doi: 10.1126/sciadv.aaw8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grahn T.H.M., Niroula A., Á Végvári, et al. S100A6 is a critical regulator of hematopoietic stem cells. Leukemia. 2020;34:3323–3337. doi: 10.1038/s41375-020-0901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng X., Gao X., Zhang Z., et al. Protective effect and mechanism of rat recombinant S100 calcium-binding protein A4 on oxidative stress injury of rat vascular endothelial cells. Oncol Lett. 2018;16:3614–3622. doi: 10.3892/ol.2018.9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun N., Youle R.J., Finkel T. The mitochondrial basis of aging. Mol Cell. 2016;61:654–666. doi: 10.1016/j.molcel.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Angajala A., Lim S., Phillips J.B., et al. Diverse roles of mitochondria in immune responses: novel insights into immuno-metabolism. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.01605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gureev A.P., Shaforostova E.A., Popov V.N. Regulation of mitochondrial biogenesis as a way for active longevity: interaction between the Nrf2 and PGC-1α signaling pathways. Front Genet. 2019;10 doi: 10.3389/fgene.2019.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.