Abstract
The ongoing SARS-CoV-2 pandemic is controlled but not halted by public health measures and mass vaccination strategies which have exclusively relied on intramuscular vaccines. Intranasal vaccines can prime or recruit to the respiratory epithelium mucosal immune cells capable of preventing infection. Here we report a comprehensive series of studies on this concept using various mouse models, including HLA class II-humanized transgenic strains. We found that a single intranasal (i.n.) dose of serotype-5 adenoviral vectors expressing either the receptor binding domain (Ad5-RBD) or the complete ectodomain (Ad5-S) of the SARS-CoV-2 spike protein was effective in inducing i) serum and bronchoalveolar lavage (BAL) anti-spike IgA and IgG, ii) robust SARS-CoV-2-neutralizing activity in the serum and BAL, iii) rigorous spike-directed T helper 1 cell/cytotoxic T cell immunity, and iv) protection of mice from a challenge with the SARS-CoV-2 beta variant. Intramuscular (i.m.) Ad5-RBD or Ad5-S administration did not induce serum or BAL IgA, and resulted in lower neutralizing titers in the serum. Moreover, prior immunity induced by an intramuscular mRNA vaccine could be potently enhanced and modulated towards a mucosal IgA response by an i.n. Ad5-S booster. Notably, Ad5 DNA was found in the liver or spleen after i.m. but not i.n. administration, indicating a lack of systemic spread of the vaccine vector, which has been associated with a risk of thrombotic thrombocytopenia. Unlike in otherwise genetically identical HLA-DQ6 mice, in HLA-DQ8 mice Ad5-RBD vaccine was inferior to Ad5-S, suggesting that the RBD fragment does not contain a sufficient collection of helper-T cell epitopes to constitute an optimal vaccine antigen. Our data add to previous promising preclinical results on intranasal SARS-CoV-2 vaccination and support the potential of this approach to elicit mucosal immunity for preventing transmission of SARS-CoV-2.
Keywords: SARS-CoV-2, COVID-19, Intranasal vaccination, Mucosal immunity, Adenoviral vector
1. Introduction
Since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late 2019 [1] the coronavirus disease 2019 (COVID-19) pandemic has spread around the globe [2].
The development and approval of SARS-CoV-2 vaccines with an unprecedented speed has been a remarkable success story. However, whereas the currently available vaccines offer good protection against severe outcomes of COVID-19 they fail to prevent SARS-CoV-2 reinfection, especially by viral variants that have emerged more recently [3], [4], [5]. Also, for economic reasons and due to logistic challenges associated with mRNA vaccine technology, the SARS-CoV-2 vaccines currently in clinical use have mainly been distributed in developed countries, leaving many people in the developing world unprotected from COVID-19. This failure is permissive to excess mortality, as well as the emergence and rapid dissemination of new SARS-CoV-2 variants that evade acquired SARS-CoV-2 immunity [6], [7].
All currently approved vaccines are based on the SARS-CoV-2 spike glycoprotein as the vaccine antigen, but many vaccine designs under development or in clinical trials include only the highly immunogenic receptor binding domain (RBD) of spike [8], which is the target of the large majority of neutralizing antibodies in convalescent sera of COVID-19 patients [9].
Of note, all SARS-COV-2 vaccines currently in clinical use are delivered into skeletal muscle. This is suboptimal, given the fact that vaccines administered via this route induce only weak mucosal IgA and memory T cell responses [10]. Consequently, they fail to prevent infection and transmission of SARS-CoV-2. Nasal vaccines against SARS-CoV-2 mimic natural respiratory infection, induce SARS-CoV-2-directed secretory IgA and mucosal T cells and reduce viral shedding in pre-clinical infectious challenges in rodents and non-human primates [11], [12], [13], [14].
To improve SARS-CoV-2 vaccine efficacy, and to better address the global demand for safe and affordable vaccines against COVID-19, we have developed two human adenovirus 5-vectored vaccines, one expressing an RBD fragment (Ad5-RBD) and the other a long version of the spike protein (Ad5-S). Here, we report promising data on pre-clinical efficacy testing of these vectored vaccines using different mouse models. These models include two commonly used inbred mouse strains that show opposing patterns of T helper cell functional responses (C57BL/6, Balb/c), and include two HLA-transgenic strains carrying common class II alleles, mimicking SARS-CoV-2 spike peptide presentation and CD4 + T cell restriction in humans (HLA-DQ6 or HLA-DQ8, huCD4 transgenic Ab0 NOD).
Our findings agree with and extend results from previous studies [11], [12], [13], [14], and suggest that the use of adeno-vectored intranasal vaccines could result in significant reductions of respiratory tract viral loads following natural infection, thereby potentially preventing not only severe COVID-19, but also transmission and spreading of SARS-CoV-2.
2. Results
2.1. Ad5-RBD administered intranasally to mice induces serum anti-spike IgA, serum SARS-CoV-2 neutralizing activity, and mucosal anti-spike IgA and IgG in the respiratory tract (single dose testing)
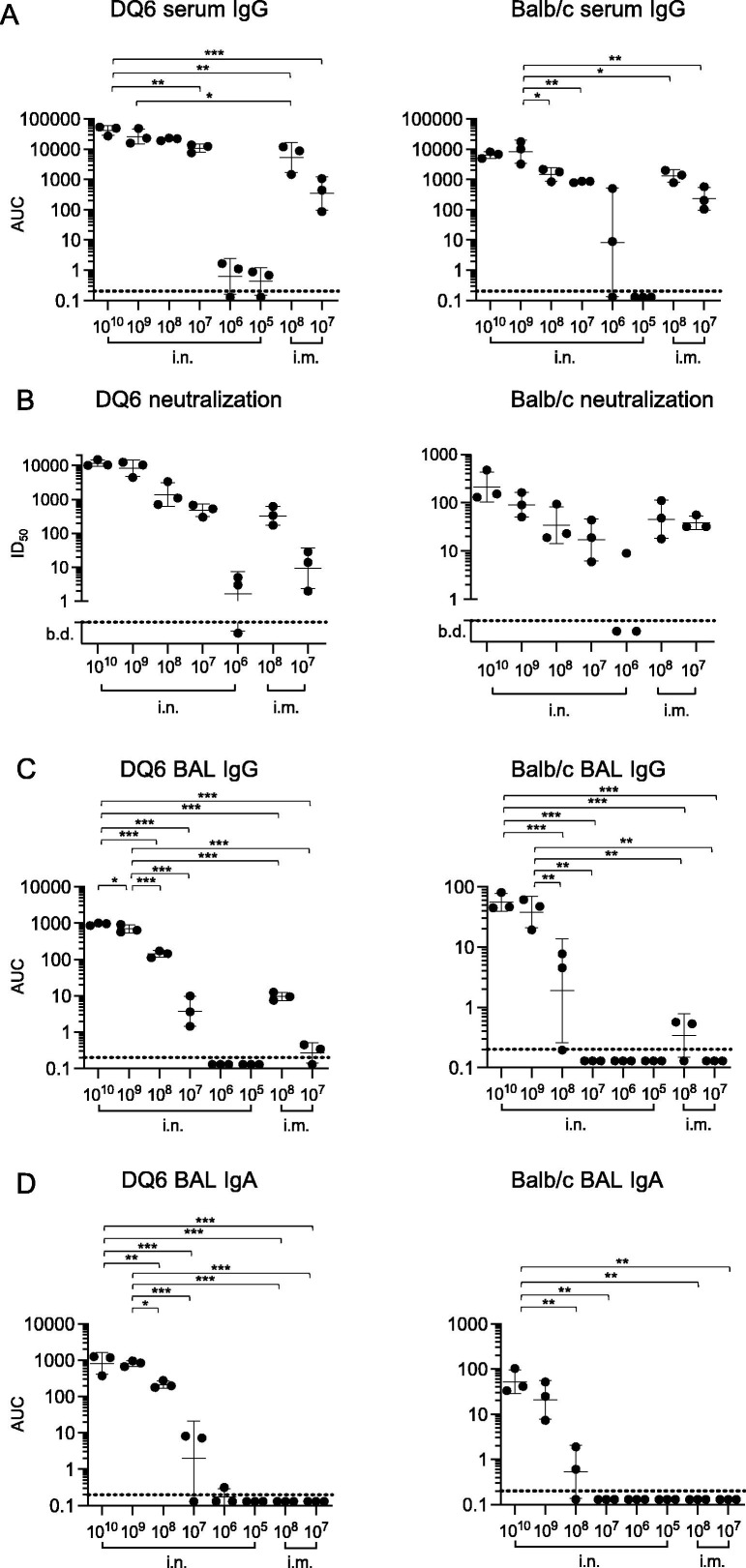
To test the efficacy of an Ad5-vectored, codon-optimized RBD vaccine (Ad5-RBD), we performed dose testing by single (1x) treatment via the nose (intranasal, i.n.) or muscle (intramuscular, i.m.) of wildtype Balb/c and HLA-DQB1*06:02 transgenic Ab0 NOD (HLA-DQ6) mice [15]. HLA-DQ6 mice were used as a humanized vaccination model, because in these mice all CD4 + helper T cells that aid in the production of virus-directed, neutralizing antibodies are restricted by the HLA class II molecule DQ6.2, expressed in up to 35% of European or North African populations, and 15% worldwide [16]. On day 25 after vaccination, sera, bronchial lavage fluid and spleens were collected. Intranasal administration of the vaccine induced higher serum anti-spike antibody titers in HLA-DQ6 (NOD) mice than in Balb/c mice, with evidence of a dose dependent response in both strains. Intranasal administration of Ad5-RBD produced marginally higher serum anti-spike IgG titers in comparison to intramuscular administration in HLA-DQ6, and similar titers in Balb/c mice (Fig. 1 a,Fig. S1). The results for serum neutralization of a pseudovirus expressing SARS-CoV-2 spike protein correlated with the findings for anti-spike IgG in ELISA, demonstrating higher efficacy for intranasal compared to intramuscular administration in DQ6 mice (non-significant trends), but similar serum neutralization efficacy in Balb/c mice, with evidence of a dose effect in both strains (Fig. 1b). Finally, intranasal administration of Ad5-RBD appeared to produce higher anti-spike IgG titers in comparison to intramuscular in bronchoalveolar lavage fluid from HLA-DQ6 and Balb/c mice (non-significant trends; Fig. 1c Fig. S1). No anti-spike IgA was detected in any group receiving vaccine intramuscularly (Fig. 1d Fig. S1). In contrast, intranasal administration induced mucosal anti-spike IgA, with higher titers in HLA-DQ6 than in Balb/c mice, and evidence of a dose effect in both strains. These results demonstrated the induction of robust humoral immunity in both wild type and humanized strains treated with a single dose of Ad5-RBD.
Fig. 1.
Humoral responses to single treatment with Ad5-RBD administered intranasally (i.n.) or intramuscularly (I.m.) in HLA-DQ6 vs. Balb/c mice. Groups of adult female mice (n = 3) were inoculated with Ad5-RBD at doses ranging from 105-1010 viral particles/mouse. Anti-spike IgG serum antibodies (a; area under the curve (AUC)), SARS-CoV-2 pseudovirus neutralization (b; calculated serum dilution producing 50% inhibition (ID50)), and anti-spike IgG and IgA bronchoalveolar lavage fluid antibodies (c, d; AUC) were measured after 25 days. For statistical comparisons, one-way ANOVA and Tukey’s test for multiple comparisons (a, c, d) or Kruskal-Wallis test and Dunn’s test for multiple comparisons (b) were used. Adjusted p-values are displayed as * <0.05, ** <0.01 and *** <0.001. To improve readability, adjusted p-values calculated in comparisons involving groups treated with doses 105 or 106 are not shown. Lines and bars represent geometric means and standard deviations. Dotted line represents the limit of detection of the assay.
2.2. Ad5-RBD administered intranasally does not lead to systemic spread of the adenoviral vector
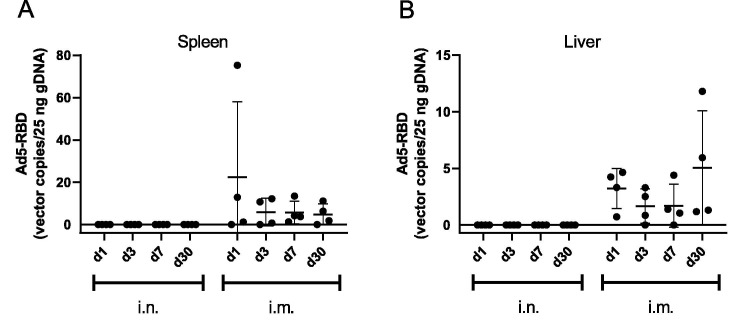
To examine the potential for systemic spread of the Ad5 vector following different administration routes, mouse liver and spleen samples were collected following delivery of Ad5-RBD (109 vp) either i.n. or i.m. Samples were collected on days 1, 3, 7 and 30 following vector delivery. Whereas the viral vector was readily detected in the spleen and liver at all the tested timepoints following i.m. delivery, none of the spleen or liver samples from the i.n. groups had detectable Ad5-RBD viral vector at the different timepoints (Fig. 2 a and b). These results demonstrate that the nasally delivered Ad5 vector does not enter the blood circulation and is hence not carried to these organs in measurable quantities, thereby suggesting an improved safety profile compared to intramuscular administration.
Fig. 2.
Distribution of Ad5 vector DNA to the spleen and liver. Spleen (A) and liver (B) samples were collected from C57BL/6 mice (n = 4) at 1, 3, 7 or 30 days following either intranasal (i.n.) or intramuscular (i.m.) administration of Ad-RBD (109 virus particles). Ad5-RBD vector was quantified from genomic DNA samples (25 ng) by real-time PCR using a specific probe-based assay to detect the RBD. Vector copy numbers were determined by comparison to plasmid standard (pAdapt-RBD). Individual values and means ± SEM are plotted.
2.3. A single administration of Ad5-RBD intranasally is sufficient to induce high serum and mucosal anti-spike antibodies and SARS-CoV-2 neutralizing activity (single vs. repeated dose testing)
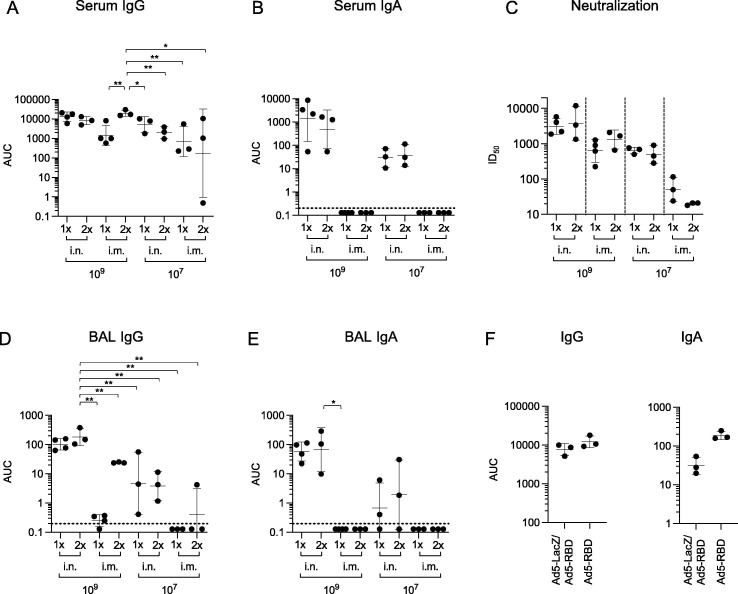
To test whether the efficacy of Ad5-RBD could be improved by the administration of a second vaccine dose, we treated HLA-DQ6 mice either once (at day 1) or twice (at days 1 and 14) via the nose or muscle. Based on the results for intranasal treatment above, a suboptimal (107 virus particles) or a fully sufficient dose (109) were chosen for comparisons. On day 28, sera, bronchial lavage fluid and spleens were collected. The results confirmed that intranasal or intramuscular administration of the vaccine both induced serum anti-spike IgG, with evidence of a dose effect. However, intramuscular administration did not produce specific serum IgA antibodies, in contrast to intranasal administration (Fig. 3 a, b, Fig. S1). Interestingly, specific serum IgG and IgA titers did not show significant differences between groups vaccinated either once or twice intranasally, although higher anti-spike IgG levels were seen in mice vaccinated intramuscularly. In line with the findings in ELISA, the neutralization efficacy was similar in groups that were treated either once or twice (Fig. 3c). After intramuscular administration, no anti-spike IgA was detected in bronchoalveolar lavage (BAL) fluid. Mucosal anti-spike IgG was detected at relatively low levels, and mainly after the second intramuscular dose. In contrast, intranasal administration induced mucosal anti-spike IgG and IgA, with evidence of a dose effect, but without an apparent effect of boosting (Fig. 3d and e, Fig. S1).
Fig. 3.
Humoral responses to one vs. two treatments with Ad5-RBD administered intranasally (i.n.) or intramuscularly (i.m.) in HLA-DQ6 mice, and lack of effect of pre-treatment with Ad5 vector on priming with Ad5-RBD. Groups of adult male and female mice (n = 3) were inoculated either once or twice (14 days apart) with Ad5-RBD at doses 107 or 109 viral particles/mouse (a-e). Anti-spike IgG or IgA serum antibodies (a, b; area under the curve (AUC)), SARS-CoV-2 pseudovirus neutralization (c; calculated serum dilution producing 50% inhibition (ID50)), and anti-spike IgG and IgA bronchoalveolar lavage fluid antibodies (d, e; AUC) were measured after 28 days. In a different experiment, one group of HLA-DQ6 mice was pre-treated i.n. with Ad5-vector encoding β-galactosidase (Ad5-LacZ; 109 vp/mouse), while a second group remained untreated (n = 3; 3f). After 21 days, a priming dose of Ad5-RBD i.n. was administered to both groups (109 vp/mouse). Serum was collected after 42 days, and anti-spike IgG or IgA serum antibodies were measured (f; AUC). For statistical comparisons, one-way ANOVA and Tukey’s test for multiple comparisons (a, b, d, e, f) or Kruskal-Wallis test and Dunn’s test for multiple comparisons (c) were used. Adjusted p-values are displayed as * <0.05, ** <0.01 and *** <0.001. Lines and bars represent geometric means and standard deviations. Dotted line represents the limit of detection of the assay.
To address the possibility that vector-directed immunity induced by the first dose would render boosting with a second dose of Ad5-RBD vaccine inefficient, we conducted an additional experiment in HLA-DQ6 mice. One group was pre-treated intranasally with a similar Ad5-vector encoding β-galactosidase (Ad5-LacZ; 109 vp/mouse), while a second group remained untreated. After three weeks, a priming dose of Ad5-RBD was administered to both groups via the same route (109 vp/mouse). Serum was collected after six weeks. The results demonstrated that exposure of mice to the Ad5-vector three weeks before Ad5-RBD priming did not lead to a significant reduction of serum anti-spike IgG or IgA (Fig. 3f, Fig. S1), arguing against the possibility that vector-directed immunity prevented effective boosting with a second dose of Ad5-RBD.
Taken together, these results confirmed the induction of humoral immunity, including secretion of specific IgA and IgG antibody in the respiratory epithelium in mice treated with a single dose of Ad5-RBD via the nose, without an apparent benefit of a second dose.
2.4. Ad5-RBD administered intranasally to HLA-DQ6 mice induces a spike-directed systemic T helper 1/cytotoxic T cell response pattern
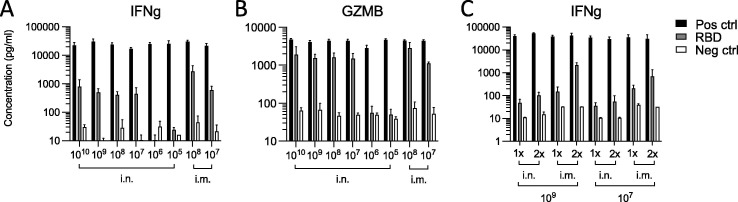
To test induction of cellular immunity against the spike protein by Ad5-RBD, we stimulated ex vivo the spleen cells of vaccinated HLA-DQ6 mice from the above experiments with recombinant RBD. Spleen cells responded to stimulation with the secretion of IFN γ and granzyme B into cell culture supernatants. High concentrations of both lymphocyte effector molecules were reached with a single dose as low as 107 virus particles (vp), matching the effective dose level determined for humoral vaccine responses (see above). Higher vaccine doses (>107 vp), repeated treatment or intramuscular route of administration did not increase secretion significantly (Fig. 4 a-c). These findings confirmed that a single dose of Ad5-RBD administered via the nose induced systemic cellular immunity against the spike protein in mice.
Fig. 4.
Cellular responses to treatment with Ad5-RBD administered intranasally (i.n.) or intramuscularly (i.m.) in HLA-DQ6 mice. Groups of female HLA-DQ6 mice (n = 3) were inoculated once with Ad5-RBD at doses ranging from 105-1010 viral particles/mouse (a, b). Groups of male or female HLA-DQ6 mice were inoculated once or twice (14 days apart) with Ad5-RBD at doses 107 or 109 viral particles/mouse (c). After 25–28 days, spleen cells were restimulated in vitro with recombinant SARS-CoV-2 spike protein receptor binding domain (RBD). Supernatants were harvested after 3 days, and the effector proteins IFNg and granzyme B were measured by ELISA. Results expressed as concentrations (pg/ml). Statistical comparisons were performed using Kruskal-Wallis test and Dunn’s test for multiple comparisons. Group differences were non-significant (adjusted p-values > 0.05).
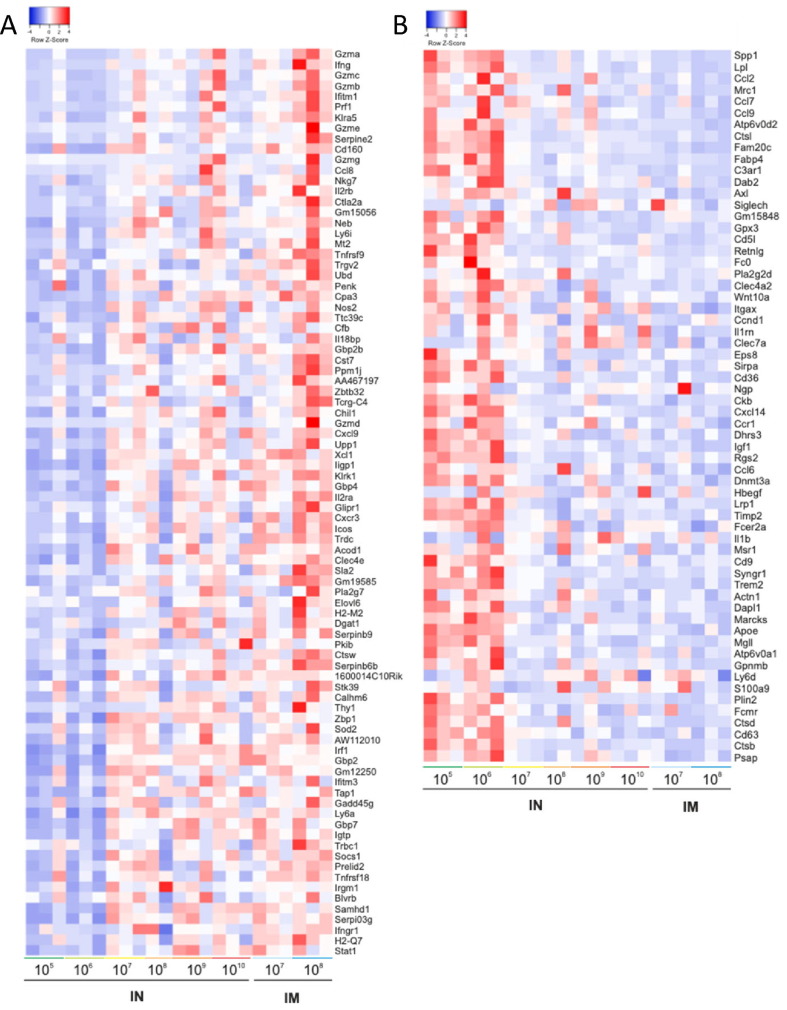
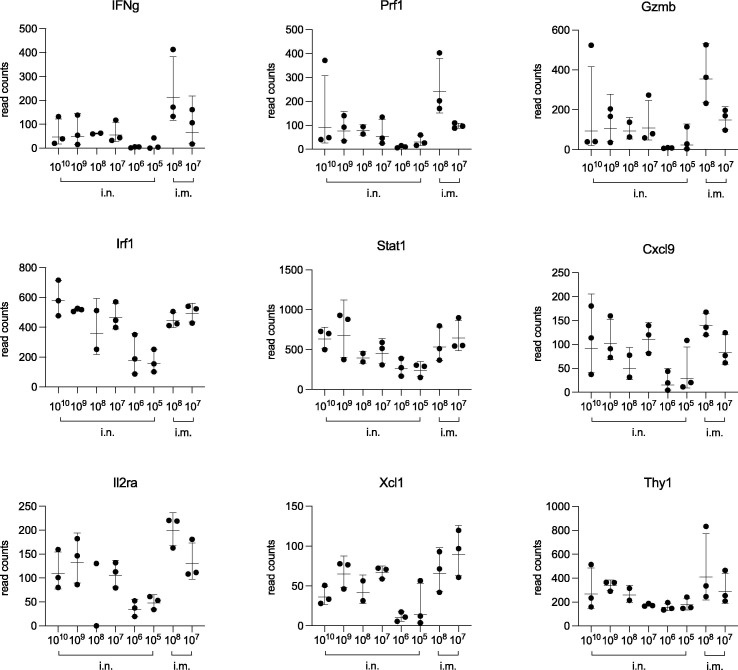
To characterize more extensively the systemic cellular immune response primed by treatment with the Ad5-RBD vaccine, we studied gene expression levels in the spleen cells of HLA-DQ6 mice stimulated with recombinant RBD ex vivo by total RNA sequencing. In two comparisons between the highest doses administered (either 1010/109 vp intranasally or 108/107 vp intramuscularly) vs. the lowest, ineffective doses (negative control; 106/105 vp intranasally), 148 genes were differentially expressed (Fig. 5 a, b). These included IFN γ, granzyme B and perforin 1, effector molecules characteristic of a cytotoxic T cell response. Other immune genes differentially expressed included the IFN γ -related genes interferon regulatory factor 1 (Irf1), signal transducer and activator of transcription 1 (Stat1) and chemokine (C-X-C motif) ligand 9 (Cxcl9), as well as the lymphocyte activation-related genes interleukin 2-receptor alpha (Il2ra), chemokine (C motif) ligand (Xcl1) and thymocyte differentiation antigen 1 (Thy1; Fig. 6 ). In summary, the results were consistent with a Th1-mediated/cytotoxic T cell response induced by treatment with a single dose of Ad5-RBD as low as 107 vp. A principal component analysis demonstrated distancing of samples from mice that received effective vs. ineffective vaccine doses (1010-107 vs. 106-105 vp), and co-clustering of samples from mice treated with the same dose either intranasally or intramuscularly (Fig. S2).
Fig. 5.
Transcriptomic profiles of RBD-stimulated spleen cells from mice treated with Ad5-RBD administered intranasally (i.n.) or intramuscularly (i.m.). Groups of female HLA-DQ6 mice (n = 3) were inoculated once with Ad5-RBD at doses ranging from 105-1010 viral particles/mouse. After 25 days, spleen cells were restimulated in vitro with recombinant SARS-CoV-2 spike protein receptor binding domain (RBD). Cells were harvested after 3 days of culture, and bulk RNA was isolated. Heatmaps depicting the up- (a; red color, 86 genes) or down-regulation (b; blue color, 62 genes) of altogether 148 genes differentially expressed after treatment with Ad5-RBD in at least one of two separate comparisons, and are ordered to show the genes with the highest log2 fold changes from the top [Ad5-RBD at doses 105/106 viral particles i.n. (ineffective treatment) vs. doses 107/108 viral particles i.m. or doses 109/1010 viral particles i.n. (n = 6; adjusted P value, P ≤ 0.001; RNA sequencing)]. Statistical analyses were performed using edgeR. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
Upregulation of selected immune genes, differentially expressed in RBD-stimulated spleen cells from mice treated with Ad5-RBD administered intranasally (i.n.) or intramuscularly (i.m.). Groups of female HLA-DQ6 mice (n = 3) were inoculated once with Ad5-RBD at doses ranging from 105-1010 viral particles/mouse. After 25 days, spleen cells were restimulated in vitro with recombinant SARS-CoV-2 spike protein receptor binding domain (RBD). Cells were harvested after 3 days of culture, and bulk RNA was isolated for transcriptomic analyses (RNA sequencing; compare Fig. 5). Panels depict the results for 9 selected immune genes [T helper 1/cytotoxic T cell markers: Interferon gamma (Ifng), perforin 1 (Prf1), granzyme B (Gzmb); Interferon gamma-related genes: Interferon regulatory factor 1 (Irf1), signal transducer and activator of transcription 1 (Stat1), chemokine (C-X-C motif) ligand 9 (CXCL9); Lymphocyte activation-related genes: Interleukin 2-receptor alpha (Il2ra), chemokine (C motif) ligand (XCL1), thymocyte differentiation antigen 1 (Thy1)]. Results expressed as read counts.
2.5. Ad5-S, Ad5-RBD and ChAdOx1 nCoV-19 are all effective in eliciting serum anti-spike IgA and SARS-CoV-2 neutralizing antibodies in C57BL/6 mice
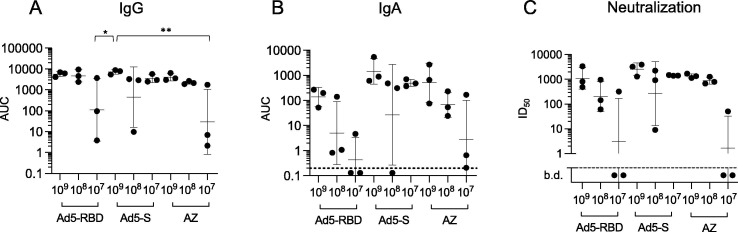
Next we compared the efficacy of Ad5-RBD, coding for a relatively short region of the spike protein, with an Ad5-vectored, codon-optimized complete spike ectodomain protein vaccine (Ad5-S). The chimpanzee adenovirus-vectored spike protein vaccine ChAdOx1 nCoV-19 (Vaxzevria, AstraZeneca), currently in clinical use by intramuscular administration, was also included in this comparison. All three vaccines were administered by single treatment intranasally in C57BL/6 mice. C57BL/6 mice were chosen to gain more information on vaccine performance in another healthy wildtype strain. In this limited comparison involving relatively few mice Ad5-S appeared more effective than Ad5-RBD or ChAdOx1 nCoV-19 in inducing serum anti-spike IgG and IgA (Fig. 7 a and b, Fig. S1), and in eliciting SARS-CoV-2 neutralizing antibodies (Fig. 7c), particularly at the lowest dose levels, but these trends were non-significant.
Fig. 7.
Humoral responses to a single treatment with either Ad5-RBD, Ad5-S or ChAdOx1 nCoV-19 (AstraZeneca, “AZ”) administered intranasally in C57BL/6 mice. Groups of adult male or female mice (n = 3) were inoculated with 3 different vaccines, at doses ranging from 107-109 viral particles/mouse, as indicated. Anti-spike IgG serum antibodies (a; area under the curve (AUC)), anti-spike IgA serum antibodies (b; AUC) and SARS-CoV-2 pseudovirus neutralization (c; calculated serum dilution producing 50% inhibition (ID50)) were measured after 25 days. For statistical comparisons, one-way ANOVA and Tukey’s test for multiple comparisons (a, b) or Kruskal-Wallis test and Dunn’s test for multiple comparisons (c) were used. Adjusted p-values are displayed as * <0.05, ** <0.01 and *** <0.001. Lines and bars represent geometric means and standard deviations. Dotted line represents the limit of detection of the assay.
2.6. Ad5-S is similarly effective in HLA-DQ6 and HLA-DQ8 mice, but HLA-DQ8 mice respond poorly to priming with Ad5-RBD
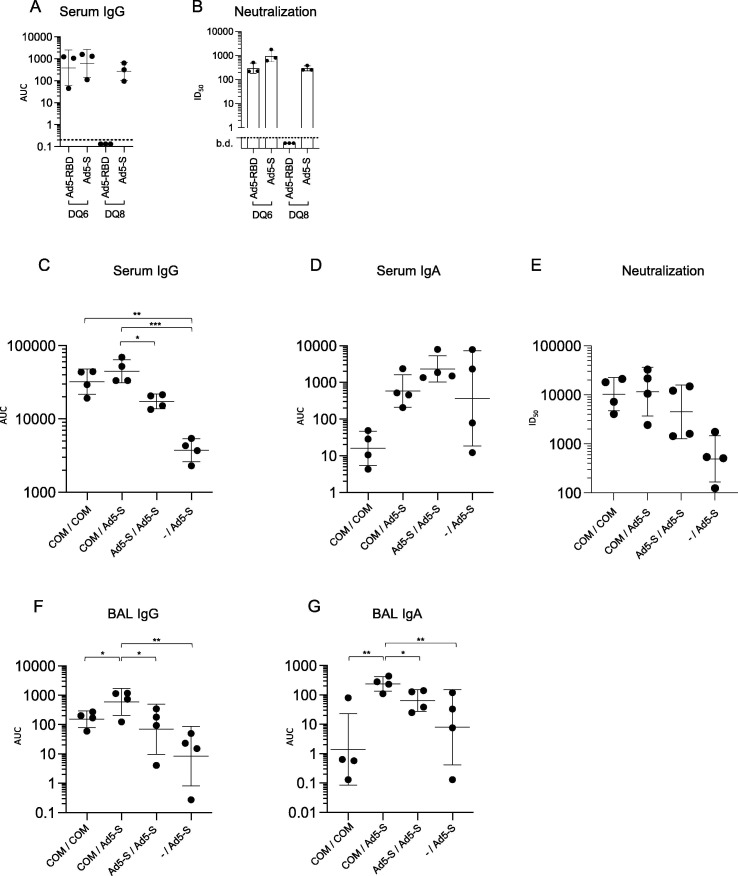
To mimic vaccination in heterogeneous human populations, and to further compare the efficacy of Ad5-RBD and Ad5-S vaccines in priming humoral responses against the spike protein, we immunized HLA-DQ8 transgenic Ab0 NOD (HLA-DQ8) mice in addition to HLA-DQ6 transgenic mice [15], as they carry another common HLA class II allele, found in up to 40% of Europeans or North Africans, and 19% worldwide [16]. Mice were vaccinated by single treatment with 107 vp of either Ad5-RBD or Ad5-S intranasally, shown to be effective in the experiments with HLA-DQ6 mice above. The results demonstrated that whereas Ad5-S was effective at inducing serum anti-spike IgG and pseudovirus-neutralizing activity both in HLA-DQ6 and HLA-DQ8 mice, Ad5-RBD failed to induce either of them in HLA-DQ8 mice (Fig. 8 a and b, Fig. S1).
Fig. 8.
Humoral responses to single treatment with Ad5-RBD or Ad5-S administered intranasally in HLA-DQ6 vs. HLA-DQ8 mice, and responses to Ad5-S administered intranasally after priming with mRNA vaccine Comirnaty (Pfizer-BioNTech, “COM”) intramuscularly. Groups of adult female HLA-DQ6 or -DQ8 mice (n = 3) were inoculated with 107 viral particles (vp)/mouse of either Ad5-RBD or Ad5-S (a, b), as indicated. Anti-spike IgG serum antibodies (a; AUC) and SARS-CoV-2 pseudovirus neutralization (b; pooled sera, dots representing the values of triplicates; calculated serum dilution producing 50% inhibition (ID50)) were measured after three weeks. Groups of adult male and female HLA-DQ8 mice (n = 4) were inoculated either with Comirnaty (2 µg/dose, i.m.), Ad5-S (109 vp/dose, i.n.) or were not pre-treated (c-g). Three weeks later, they received either Comirnaty i.m. or Ad5-S i.n. at the same doses, as indicated. The Ad5-S vector used in this experiment encoded for the beta variant strain of SARS-CoV-2, containing three major receptor-binding domain mutations. Anti-spike IgG and IgA serum or bronchoalveolar lavage (BAL) antibodies (c, d, f, g; AUC) and pseudovirus neutralization (e; individual sera; ID50) were measured after six weeks. For statistical comparisons, one-way ANOVA and Tukey’s test for multiple comparisons (a, c, d, f, g) or Kruskal-Wallis test and Dunn’s test for multiple comparisons (b, e) were used. Adjusted p-values are displayed as * <0.05, ** <0.01 and *** <0.001. Lines and bars represent geometric means and standard deviations. Dotted line represents the limit of detection of the assay.
These results combined demonstrated high efficacy of Ad5-S in priming humoral responses against the spike protein in different wildtype and humanized mouse strains, and suggested that the immunogenicity of the RBD fragment of the spike protein in human vaccinees could be limited by the HLA class II genetic background.
2.7. Ad5-S administered intranasally enhances and modulates the immune response induced by a prior mRNA SARS-CoV-2 vaccine administered intramuscularly
To address whether Ad5-S (i.n.) could be used for heterologous boosting after priming with other vaccines (i.m.), we compared four groups of HLA-DQ8 mice that received either the mRNA spike vaccine Comirnaty (2 μg/mouse; Pfizer-BioNTech) or Ad5-S (109 vp), as prime/boost in three different combinations, or as a control received only a single dose of Ad5-S at the same time the other groups received their second dose. As shown in Fig. 8 (c, e, and f, Fig. S1), serum and BAL anti-spike IgG levels and neutralizing activity were comparable after Comirnaty priming followed by the use of either Ad5-S (i.n.) or Comirnaty (i.m.) as a booster. In fact, in this comparison intranasal Ad5-S seemed to be a somewhat more potent booster, with a significant statistical difference reached for BAL IgG. By contrast, serum and BAL anti-spike IgA levels were effectively increased only in mice boosted with Ad5-S (Fig. 8d and g, Fig. S1). However, it is noteworthy that when comparing to the sera and BAL samples of mice that received only a single dose of Ad5-S at the 3-week time point, it was clear that a prior dose of Comirnaty could enhance serum and mucosal antibody responses elicited by a subsequent Ad5-S vaccination. This indicated that the immune response induced by an intramuscular vaccine was not only increased but also qualitatively modulated and directed towards the respiratory mucosa by boosting with Ad5-S intranasally (Fig. 8d and g, Fig. S1).
2.8. Ad5-S and Ad5-RBD administered intranasally protect Balb/c mice from infectious challenge with SARS-CoV-2 beta variant
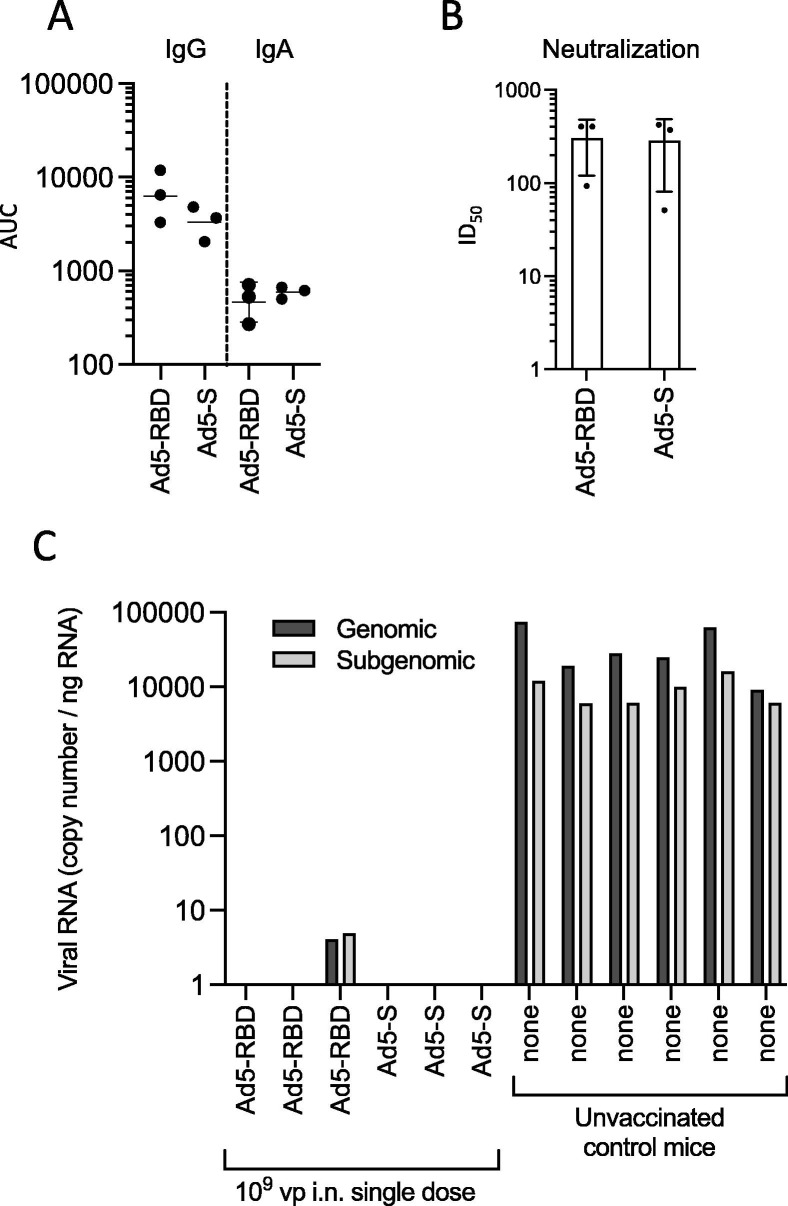
To test the efficacy of Ad5-RBD and Ad5-S vaccines in providing protection from infection we used the SARS-CoV-2 beta variant nasal challenge model recently described in Balb/c mice [17]. Two groups of female Balb/c mice (n = 3) were vaccinated by administration of a single dose of 109 vp of either Ad5-RBD or Ad5-S intranasally. Another group (n = 6) remained without pre-treatment. Three weeks later, the vaccinated mice were bled, and five weeks after the vaccination, all mice were inoculated intranasally with 2 × 105 plaque-forming units of the beta variant of SARS-CoV-2. Three days post-inoculation, the mice were euthanized, and the lungs were collected for virological examinations (RT-PCR of the right lung) as well as histological and immunohistochemical examinations (left lung). The results showed that the sera from vaccinated mice had anti-spike IgG and IgA titers and SARS-CoV-2 neutralizing activity (Fig. 9 a and b, Fig. S1). Three days after the infectious challenge abundant viral RNA was found in the lungs of all unvaccinated mice, whereas no viral RNA could be detected in five of the six vaccinated mice. Only in one mouse that had received the Ad5-RBD vaccine, low amounts of viral RNA were found but at levels that were strongly reduced in comparison to unvaccinated mice (Fig. 9c).
Fig. 9.
Infectious challenge of Balb/c mice with SARS-CoV-2 after single treatment with Ad5-RBD or Ad5-S administered intranasally (I). Groups of adult female mice (n = 3) were inoculated with 109 viral particles/mouse of either Ad5-RBD or Ad5-S, or received no pre-treatment (n = 6). Anti-spike IgG or IgA serum antibodies (a; AUC) and SARS-CoV-2 pseudovirus neutralization (b; pooled sera, dots representing the values of triplicates; calculated serum dilution producing 50% inhibition (ID50)) were measured after three weeks. Lines and bars represent geometric means and standard deviations. After 5 weeks, all mice were inoculated intranasally with 2 × 105 plaque-forming units of the beta variant of SARS-CoV-2. Three days later, viral RNA transcripts for (genomic) RNA-dependent RNA polymerase or (subgenomic) E were quantified in tissue of the right lung (c; RNA copies per ng of total lung RNA, based on SARS-CoV-2 genomic RNA standard included in the RT-qPCR run).
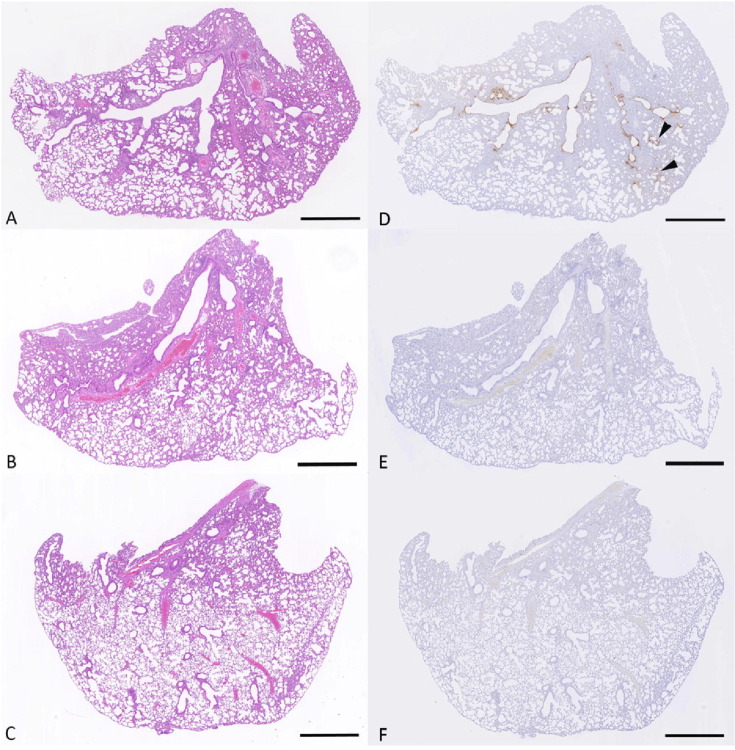
The lungs of all unvaccinated animals showed the histological changes previously described at day 3 post SARS-CoV-2 beta variant infection in wildtype mice [17] represented by degeneration and loss of low numbers of epithelial cells in bronchi and bronchioles, and rare small groups of alveoli with degenerate epithelial cells and desquamated alveolar macrophages (Fig. 10 a, d). In contrast, the vaccinated animals generally exhibited neither histological changes (Fig. 10b, c) nor viral antigen expression (Fig. 10e, f). No viral antigen was detected in the lungs of animals vaccinated with Ad5-S, whereas in two mice vaccinated with Ad5-RBD, some viral antigen could be detected, but it was restricted to a few epithelial cells in individual bronchioles and alveoli. In stark contrast to unvaccinated mice, bronchial lymph nodes were clear of viral antigen in all but one vaccinated animal that showed rare antigen-positive macrophages/dendritic cells. Detailed information on the results in individual animals is provided in Supplementary Table 1. These results show that both vaccines block or at least strongly suppress lung infection.
Fig. 10.
Infectious challenge of Balb/c mice with SARS-CoV-2 after single treatment with Ad5-RBD or Ad5-S administered intranasally (II). The left lung was subjected to histological (H/E; a-c) and immunohistochemical (SARS-CoV-2 nucleoprotein, hematoxylin counterstain; d-f) examination on day 3 post intranasal infection with 2 × 105 plaque-forming units of the beta variant of SARS-CoV-2. Unvaccinated animal no. 3 (a, d): There is extensive SARS-CoV-2 nucleoprotein expression in bronchial and bronchiolar epithelial cells and, focally (arrow heads) in the parenchyma. Ad5-RBD vaccinated mouse no. 1 (b, e): The lung is widely unaltered and there is no evidence of viral antigen expression. Ad5-S vaccinated animal no. 1 (c, f): The lung is widely unaltered and there is no evidence of viral antigen expression. Bars = 1 mm.
3. Discussion
This study reports a comprehensive series of experiments to explore the concept of an Ad5-vectored intranasal vaccine against SARS-CoV-2. Our studies utilized the spike protein sequence of the original Wuhan strain of SARS-CoV-2 for vaccine antigen design and likewise for construction of the pseudoviruses to measure vaccination-induced neutralizing antibodies. Apart from using the beta variant to evaluate the protection provided by these Wuhan-based vaccines in challenged mice, in this work we have not addressed the challenge for vaccine development posed by the constantly emerging antigenically divergent SARS-CoV-2 variants. However, while the optimal spike protein sequences for future vaccines remains to be determined, the data reported here provide valuable insights for intranasal adeno-vectored SARS-CoV-2 vaccine development that are relevant also to newer variants.
We found that a single intranasal dose of serotype 5-based adenoviral vectors expressing either the receptor binding domain (Ad5-RBD) or the entire SARS-CoV-2 spike protein ectodomain (Ad5-S) was effective in inducing I) mucosal and serum anti-spike IgA and IgG, II) robust SARS-CoV-2-neutralizing activity in the serum and in respiratory secretions, III) rigorous spike-directed T helper 1 cell/cytotoxic T cell immunity, and IV) protection from a challenge with the SARS-CoV-2 beta variant in mice. In addition to eliciting circulating spike-directed IgA antibodies and mucosal responses not observed after intramuscular immunization, the intranasal administration led to higher SARS-CoV-2 neutralizing titers in the serum. Moreover, intranasal Ad5-S could serve as an effective booster in mice that had received a prior intramuscular vaccination with the mRNA vaccine Comirnaty. Interestingly, when compared to a single dose of Ad5-S in unprimed mice, it was evident that prior mRNA vaccination administered i.m. strongly enhanced the mucosal IgA response elicited by a subsequent i.n. dose of Ad5-S, although two doses of Comirnaty were equally unable to induce IgA as were two doses of Ad5-RBD or Ad5-S when injected intramuscularly. These results provide further support for the concept of developing an intranasal adenovirus-vectored vaccine against SARS-CoV-2 for use in humans.
Our study confirms that mucosa-associated lymphoid tissue (MALT) can protect against respiratory pathogens such as SARS-CoV-2 [11], [12], [13], [14]. Due to the unique properties of MALT, and due to a general inefficacy of intramuscular vaccines to prevent mucosal infection [10], [18], intranasal vaccines represent an attractive alternative to the intramuscular SARS-CoV-2 vaccines currently in clinical use. Mucosal IgA, induced only by mucosal vaccines, plays a key role in viral neutralization on epithelial surfaces and can block viral transmission, reducing the risk of infection for those in close contact with infected individuals. Several SARS-CoV-2 vaccine candidates designed for intranasal delivery have already entered clinical trials, some of them based on adenovirus vectors [19].
We found that a single intranasal dose of serotype 5-based adenovirus-vectored vaccines was effective in inducing humoral and cellular immunity in mice, without a benefit of a second dose. This lack of an apparent booster effect by a second intranasal dose, was not due to an anti-Ad5 vector response caused by the first dose, because mice that received the same i.n. dose of a mock Ad5-vaccine (encoding β-galactosidase) could be efficiently immunized by Ad5-RBD administered intranasally 3 weeks later. These results confirm previous reports for human or chimpanzee adenovirus-vectored vaccines in rodents and non-human primates [11], [12], [13], [14] and point to strong adjuvant properties provided by adenovirus vector-based vaccines [20], [21]. However, administration of a second vaccine dose at a later timepoint is likely to provide a boosting effect, particularly in maintaining SARS-CoV-2-directed antibodies titers, and may improve vaccine efficacy when used in humans. Duration of protection was not assessed in our experiments.
To mimic vaccination in heterogeneous human populations, we compared the Ad5-RBD and Ad5-S vaccine responses in HLA-DQ6 and HLA-DQ8 transgenic mice [15]. HLA class II molecules restrict CD4 + T cells that provide help both to CD8 + cytotoxic T cells and antibody-producing B cells, and thereby control virus-directed cellular and humoral immune responses. The class II alleles DQB1*0602 and DQB1*0302 that give rise to the molecules HLA-DQ6.2 and -DQ8, respectively, represent two different class II supertypes (of seven identified in total) and are carried by 15 and 19 percent of the human population, respectively [16]. HLA transgenic mice have been used previously to test and predict immunogenicity of vaccines developed against tumors and infectious diseases in humans [22]. Interestingly, priming with the Ad5-S vaccine was similarly effective in HLA-DQ6 and HLA-DQ8 mice, whereas HLA-DQ8 mice responded poorly to the Ad5-RBD vaccine. Since these mouse strains differ only in the HLA class II transgene they carry, our findings suggest that the number of HLA-DQ8-binding CD4 + T cell peptides derived from the relatively short RBD protein sequence is suboptimal, while the longer S protein sequence harbors a broader repertoire of T cell epitopes for recognition in the context of various HLA class II (and class I) alleles. In this regard, the low immunogenicity of a live-attenuated influenza vector RBD-based intranasal vaccine against SARS-CoV-2 reported in clinical phase 1 and 2 trials may be related to the size and protein sequence characteristics of RBD [23]. The entire ectodomain of the spike protein therefore appears to be more suitable than RBD for use as a vaccine antigen, capable of inducing SARS-CoV-2-directed immune responses in vaccinees of diverse genetic backgrounds.
In this study, intranasal Ad5-S appeared (without reaching statistical significance) more effective than intranasal ChAdOx1 nCoV-19 (AstraZeneca) in inducing serum anti-spike IgA and SARS-CoV-2 neutralizing antibodies. ChAdOx1 nCoV-19 was approved for emergency use as an intramuscular vaccine against SARS-CoV-2 in January 2021, and its immunogenicity when used as an intranasal vaccine is being explored in a clinical trial (NCT04816019). In a previous study the human Ad5 vector was found to elicit higher antigen-specific immune responses than the chimpanzee ChAdOx1 vector in vaccinations in mice, due to higher levels of transgene expression induced by Ad5, and/or more effective transduction of antigen-presenting cells [24]. The results of our direct comparison in C57BL/6 mice might reflect these findings, but could also be due to other differences in the design of these spike protein-encoding vectors. For example, the ectodomain fragment of Ad5-S is designed to be expressed as a soluble and predominantly monomeric protein in contrast to the membrane anchored and mainly trimeric Spike produced by ChAdOx1 that also contains the polybasic S1/S2 cleavage site mutated in Ad5-S.
The intramuscular use of adenovirus-vectored vaccines ChadOx1 nCoV-19 (AstraZeneca) and Ad26.COV2.S (Janssen) has been associated with a rare syndrome of vaccine-induced thrombotic thrombocytopenia (VITT) [25], [26]. This syndrome is thought to be triggered by interactions of circulating adenoviral vectors and their components with platelets, followed by platelet activation, platelet factor 4 (PF4) release, and PF4-directed autoimmunity. In this regard it is of importance that our sensitive PCR assay detected no evidence of adenoviral DNA in spleen or liver of mice that had been intranasally immunized with an Ad5 vaccine vector. In contrast, Ad5 DNA could be readily detected in these organs in mice that had received the same vaccine dose intramuscularly, an indication of systemic spread of the adenoviral vector. It has also been suggested that microvascular damage due intramuscular vaccine injection would contribute to VITT [27]. In summary, intranasal vaccination with adenoviral vector most likely carries a reduced risk for the development of VITT in comparison to intramuscular vaccination.
The continuous rise over the course of the pandemic of new SARS-CoV-2 variants capable of evading vaccine-induced immunity, establishing mucosal infection, and transmitting to new contacts is challenging current vaccine strategies. Adenovirus-vectors allow for rapid adaptation to new variants that could be used alone or in combination with vectors based on the Wuhan strain for priming and/or boosting immune responses against SARS-CoV-2. At the same time, vaccines based on the Wuhan strain continue to provide strong protection from severe outcomes in COVID-19. The introduction of intranasal adeno-vectored vaccines such as Ad5-RBD or Ad5-S could reinforce the mucosal immunity to SARS-CoV-2 when administered as a booster in persons previously vaccinated intramuscularly, and could play a role in reducing person-to-person transmission. Intranasal adeno-vectored vaccines could also provide affordable and easily administered alternatives to intramuscular vaccines currently of limited reach in developing countries. The results presented in this study strongly encourage further efforts in this area, to attain the outlined goals as soon as possible.
4. Methods
4.1. Ad5-RBD and Ad5-S vector vaccine development
To obtain vaccine vectors, we cloned RBD (Ad5-RBD; residues 331–529) and the soluble spike protein (Ad5-S; residues 14–1214) with a human Interleukin 3 (IL3) signal peptide into the pAdApt adenovirus vector. The furin cleavage site in S is blocked by Arg682Gly, Arg683Ser and Arg685Ser mutations. The pAdApt adenoviral vectors were verified by DNA sequencing and by transient protein expression in HEK293 cells. Protein expression was analyzed by direct western blotting of the conditioned supernatants. Production of the Ad5-viral vaccines from pAdApt took place in the National Virus Vector Laboratories (Kuopio, Finland). Vector plasmid DNA was transfected into HEK-293 cells and the resulting recombinant vectors were colony-purified for three rounds prior to cesium chloride gradient purification. The vector titers were quantified spectrophotometrically and tested for sterility and mycoplasma contamination [28]. Expression of the Ad5-RBD and –S vaccine antigens was confirmed in HeLa and A549 cells upon adenoviral infection.
4.2. Animals
HLA-DQ6 or HLA-DQ8, huCD4 transgenic Ab0 NOD mice [15] and C57BL/6 mice were originally purchased from The Jackson Laboratory (Bar Harbor, ME, USA), and then bred at the University of Helsinki Laboratory Animal Centre. Balb/c mice were purchased from Scanbur (Sollentuna, Sweden), and used for experiments after an acclimatization phase of 7 days. C57BL/6 mice were also purchased from Charles River Laboratories (Wilmington, MA, USA), and bred at the University of Eastern Finland Laboratory Animal Center. All mice were maintained under specific pathogen–free conditions, on normal chow. All groups were sex-matched if male and female mice were used in the same experiment.
4.3. Vaccinations
Mice were assigned to sex-matched groups, sedated with isoflurane, and placed in a supine position for intranasal inoculation during normal inhalation using a pipette, or onto the left side, for intramuscular injection into the right thigh using a 27G needle [11]. Mice were vaccinated with different doses of Ad5-RBD, Ad5-S or ChAdOx1-S, administered in 25–30 µl of PBS either once or twice (dose range 106-1010 vp; 2–3 weeks apart, using the same route), and were allowed to recover naturally. In one experiment, and Ad5-vector [29] with a backbone identical to Ad5-RBD and Ad5-S but encoding β-galactosidase (Ad5-LacZ) was used as control. ChAdOx1-S vaccine (Vaxzevria, AstraZeneca; lot ABV7764) was obtained from the Finnish National Public Health Institute, stored at 4 °C and used before the expiry date. The mRNA SARS-CoV-2 vaccine Comirnaty (Pfizer-BioNTech; lot EY3014) was also obtained from the Finnish National Public Health Institute, stored at −80 °C, and used at 2 µg/dose.
4.4. Sampling of serum, bronchoalveolar lavage fluid, liver and spleen from mice
Mice were anesthesized with ketamine/xylazine intraperitoneally, and exsanguinated by retroorbital bleeding. Sera were separated after 2 h at RT. After cervical dislocation, spleens were removed from the abdomen aseptically, and bronchoalveolar lavage was performed by instillation of 800 µl of PBS containing Roche complete protease inhibitor (Roche Diagnostics, Mannheim, Germany) via the exposed trachea into the lungs, followed by aspiration after 30 s, using a 18G IV cannula (B. Braun, Melsungen, Germany). For the biodistribution studies, mice were put down with CO2 and perfused with PBS at the given timepoints. Spleen and liver samples were collected, snap-frozen in liquid nitrogen and stored at −70 °C.
4.5. SARS-CoV-2 infectious challenge
Female Balb/c mice, vaccinated or not by administration of Ad5-RBD or Ad5-S via the nose, were intranasally inoculated in a biosafety level 3 facility at the University of Helsinki with 2 × 105 plaque-forming units of the SARS-CoV-2 beta variant in 20 µl PBS under isoflurane anesthesia, as described previously [17]. The strain sequence has been deposited in the NCBI GenBank database under accession number MW717678 [30]. Immediately after the inoculation, the mice were held in an upright position for 15 s. All mice were monitored for signs of illness and were weighed on a daily basis. Three days after infection, the mice were euthanized under terminal isoflurane anesthesia with cervical dislocation, and lungs were collected for virological examinations (RT-PCR of the right lung; frozen tissue) as well as histological and immunohistochemical examinations (left lung; fixed in 10% buffered formalin for 48 h, and then stored in 70% ethanol until processing).
4.6. Ethical permissions
All animal procedures were approved by the Board of Animal Research (Ella), Southern Finnish State Administrative Agency (license numbers ESAVI/11326/2020, ESAVI/28687/2020 and ESAVI/21694/2020).
4.7. Recombinant spike and spike RBD protein
The expression and purification of SARS-CoV-2 spike protein has been previously described [31]. Recombinant spike RBD was produced in HEK293 cells. Cells were seeded at 0.5 × 106 cells/ml into 100 ml SFM II medium (Gibco, ThermoFisher Scientific) supplemented with penicillin, streptomycin, and glutamine and incubated in an orbital shaker incubator at 37 °C, 200 rpm, and 5% CO2 until cells reached a density of 1.0 × 106 cells/ml. Cells were pelleted, washed with 40 ml culture grade phosphate-buffered saline (PBS) and resuspended in 100 ml FreeStyle™ 293 Expression medium (Gibco, ThermoFisher Scientific) supplemented with 1% fetal bovine serum (Gibco, ThermoFisher Scientific). Transfection mixture was made by combining 0.2 ml filter-sterilized (1 mg/ml) polyethylenimine (Sigma-Aldrich) with 100 µg of filter-sterilized plasmid DNA (pCAGGS Containing Wuhan-Hu-1 spike glycoprotein gene RBD with C-Terminal Hexa-Histidine Tag, Beiresources, NR-52309) diluted in 10 ml of PBS. Transfection mixture preincubated at room temperature for 20 min was added to the cell suspension and incubated in an orbital shaker incubator for a further 72 hr at 37 °C, 200 rpm, and 5% CO2. Culture supernatant was collected and filtered through a 0.22 µm Stericup vacuum filtration system (Sigma-Aldrich) and NaCl concentration was adjusted to 150 mM and further supplemented with imidazole to a final concentration of 10 mM, pH was also adjusted to 8. Culture supernatant was then passed through His GraviTrap column (GE Healthcare) pre-equilibrated with PBS/10 mM imidazole, column was then washed with 20 ml PBS/20 mM imidazole and bound RBD protein was eluted with PBS/250 mM imidazole solution. RBD-containing fractions were verified by SDS-PAGE, dialyzed against PBS, and concentrated using Amicon™ Ultra Centrifugal Filter Units, 10 kDA (Sigma-Aldrich). Concentrated RBD protein was filter-sterilized, and the concentration was estimated by the NanoDrop ND-1000 Spectrophotometer (Thermo Scientific). RBD protein expression and purification yield was estimated to be about 8 mg/l culture.
4.8. Anti-spike IgG and IgA ELISA
ELISA was performed by coating 96-well microtiter plates (Nunc Polysorb; Thermo Fischer Scientific) with spike protein (1 µg/ml in PBS, 50 µl volume/well) at 4 °C for 16 h. The plates were washed with PBS/Tween-20 0.05% (PBS-T) and blocked with 3% skim milk in PBS-T at RT for 1 h before addition of 50 µl of serum or bronchoalveolar lavage (BAL) dilutions. After incubation at RT for 2 h, the plates were washed three times with PBS-T followed by the addition of HRP-conjugated anti-mouse IgG or IgA antibodies in 1 % skim milk in PBS-T (IgG: 1:5000 dilution, Sigma Aldrich, cat# A4416; IgA: 1:600 dilution, Abcam, cat# ab7235). After incubation at RT for 1 h, plates were washed three times with PBS-T before addition of substrate (1-Step™ Ultra TMB-ELISA Substrate Solution, Thermo Fisher Scientific). After incubation at room temperature for 20 min, the reaction was stopped by adding 2 M H2SO4. Absorbance (OD450) was measured with a Hidex Sense microplate reader (Hidex Oy, Finland). At least two independently mixed replicates were measured for each experiment. Experiments were repeated twice and yielded similar results. Antibody levels are presented as the area under the curve (AUC), calculated for each sample using the trapezoidal rule from graphs showing optical density (y-axis) plotted against serial dilutions (x-axis). ELISA data were analyzed using Prism 9 (GraphPad).
4.9. Neutralization of pseudovirus expressing SARS-CoV-2 spike protein
HEK293T cells were maintained in DMEM supplemented with 10% fetal bovine serum, 2% L-Glutamine, and 1% penicillin/streptomycin (complete medium). Angiotensin-converting enzyme 2 (ACE2) expressing HEK293T cells were generated by lentivirus-mediated gene transduction. Briefly, pWPI-puro plasmid containing ACE2 cDNA (AB046569.1) was co-transfected with p8.9NdSB and vesicular stomatitis virus G protein (VSV-G) expressing envelope plasmids into HEK293T cells in complete medium using polyethylenimine. The recombinant lentivirus containing supernatant was collected two days after transfection, filtered and used to infect wild-type HEK293T cells. Transduced cells were selected with puromycin. Luciferase encoding SARS-CoV-2 pseudotyped reporter virus was generated by transfecting HEK293T cells with p8.9NdSB, pWPI-GFP expressing Renilla luciferase, and pCAGGS, an expression vector containing the SARS-CoV-2 S protein cDNA of the Wuhan-Hu-1 reference strain (NC_045512.2). The last 18 amino acids containing an endoplasmic reticulum (ER)-retention signal of the spike protein was removed to enhance transport to the plasma membrane. D614G mutation was introduced with site directed mutagenesis. Pseudovirus stock was harvested 48 h post-transfection, filtered and stored at −80 °C. SARS-CoV-2 pseudovirus-mediated expression of luciferase is directly proportional to the quantity of internalized virus. 10 μl of serum or BAL dilutions were mixed with 40 μl of luciferase encoding SARS- CoV-2 pseudotyped reporter virus in 96-well cell culture plates and incubated at 37 °C for 1 h. After incubation, 2x104 HEK293T-ACE2 cells (in 50 μl) were added on the wells and the plates were further incubated at 37 °C for 48 h. The amount of internalized pseudovirus in infected cells was quantified by measuring luciferase activity using Renilla-GLO assay (Promega). Half maximal inhibitory dilutions (ID50) were determined using Prism9 software (GraphPad), expressed as calculated serum dilution. At least two independently mixed replicates were measured for each experiment.
4.10. Genomic DNA extraction
Genomic DNA was extracted from mouse tissues (stored at −70 °C) using NucleoSpin DNA RapidLyse (Macherey-Nagel) according to the manufacturer’s instructions. DNA was quantified using the Nanodrop-1000 spectrophotometer and diluted to 25 ng/µl prior to assaying.
4.11. Quantitative real-time PCR for Ad5 vector
Quantitative real-time PCR (RT-qPCR) was performed using an RBD-specific probe-primers assay (IDTDNA) and TaqMan™Fast Advanced Master Mix (Applied Biosystems) according to the manufacturer’s recommendations. Assays were performed using a QuantStudio™ 3 Real-Time PCR System (Applied Biosystems). All samples were assayed in duplicate wells and all assays included plasmid standards, that were generated from a 1:10 serial dilution of pAdapt-RBD (1 ng/µl). The vector copy numbers in the samples were determined from comparison to the plasmid standard.
4.12. Spleen cell stimulation
Spleen cells from individual mice were seeded in 48 well plates at 5x106 cells/ml in RPMI 1640 medium (750 μl volume) containing heat-inactivated fetal calf serum (10%), penicillin/ streptomycin, glutamine and HEPES (25 mM), at 37 °C and 5% CO2 (all from Sigma-Aldrich). Cells were stimulated for 3 days with a combination of anti-CD3 and anti-CD28 antibodies (positive control, 3 μg and 2 μg/ml; clones 145–2C11 and 37.51; eBioscience, San Diego, CA), medium (negative control) or recombinant spike RBD protein, produced in HEK-293 cells (10 μg/ml).
4.13. IFN gamma and granzyme B ELISA
IFN gamma and granzyme B were measured in supernatants from mouse spleen cell stimulations using ELISA kits, according to manufacturer instructions (Duoset ELISA kits, cat#s DY485 and DY1865; Bio-techne, UK).
Mouse spleen RNA extraction. RNA was extracted from mouse spleen cells cultured and stimulated for 3 days at 37 °C in RPMI 1640, and then stored at −20 °C in RNAlater (ThermoFisher Scientific), using the RNeasy Mini Kit (Qiagen) and following manufacturer’s guidelines. Cells were homogenized with QIAshredder columns (Qiagen). After extraction, RNA was quantified using a NanoDrop spectrophotometer (ThermoFisher Scientific). RNA samples were stored at −70 °C.
RNA-sequencing, read alignment, generation of digital expression data and principal component analysis. RNA sequencing was performed at the Functional Genomics Unit of the University of Helsinki, Finland, as described previously [32]. The method of read alignment and generation of digital expression data were described in the same publication [32]. Differentially expressed genes were identified using edgeR [33]. Principal component analysis was performed, using the DESeq2 Bioconductor package [34].
4.14. Viral RNA measurement by RT-qPCR from lung tissue
The right lungs were dissected, immediately transferred to tubes containing Trizol reagent (Thermo Fisher) and frozen at – 80 °C for later use. Total RNA was extracted following recommendations by the manufacturer (Trizol, ThermoFisher) and 1/100 of the total RNA was used for multiplex one-step reverse transcriptase (RT)- real time PCR using 1–step fast virus master mix (ThermoFisher) to detect one viral RNA target and housekeeping gene actin mRNA levels in the same reaction with AriaMx instrumentation (Agilent). The primers and probes for detection of genomic RNA-dependent RNA polymerase (RdRp) and subgenomic E (subE) with either FAM- or HEX-fluoroconjugates, are described elsewhere [35], [36] and were synthesized by Metabion, Planegg, Germany. Viral RdRp and subE RNA copy numbers were calculated based on a standard curve generated using Ct values obtained by running the PCR assay with serially diluted in vitro transcribed RdRp RNA.
4.15. Histological and immunochemical examinations
The left lung and remaining thoracic organs were fixed in 10% buffered formalin for 48 h and stored in 70% ethanol until they were trimmed for histological examination and routinely paraffin wax embedded. Consecutive sections (3–5 µm) were prepared from lungs and routinely stained with hematoxylin-eosin (HE) or subjected to immunohistochemistry (IHC) for the detection of SARS-CoV-2 antigen, as previously described [17]. IHC was performed in an autostainer (Agilent) using a rabbit polyclonal anti-SARS-CoV nucleoprotein antibody (Rockland, 200–402-A50) and the horseradish peroxidase (HRP) method. Briefly, sections were deparaffinized and rehydrated through graded alcohol. Antigen retrieval was achieved by 20 min incubation in citrate buffer (pH 6.0) at 98 °C in a pressure cooker. This was followed by incubation with the primary antibody (diluted 1:3,000 in dilution buffer; Dako) overnight at 4 °C, a 10 min incubation at room temperature (RT) with peroxidase blocking buffer (Agilent) and a 30 min incubation at RT with Envision + System HRP Rabbit (Agilent). The reaction was visualized with diaminobenzidin (DAB; Dako) for 10 min at RT. After counterstaining with hematoxylin for 2 s, sections were dehydrated and placed on a coverslip with Tissue-Tek Film (Sysmex, Kobe, Japan).
4.16. Statistical comparisons
For serum and bronchoalveolar antibody ELISA, groups were compared using one-way ANOVA and Tukey's test for multiple comparisons. Results of serum neutralization of pseudovirus and cytokine ELISA were analyzed using Kruskal-Wallis and Dunn’s tests for multiple comparisons. Statistical analyses of results from RNA sequencing were performed using edgeR.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Tobias Freitag reports a relationship with Rokote Laboratories Finland Ltd that includes: employment. Kalle Saksela reports a relationship with Rokote Laboratories Finland Ltd that includes: board membership and equity or stocks. Seppo Yla-Herttuala reports a relationship with Rokote Laboratories Finland Ltd that includes: board membership and equity or stocks. Kari Alitalo reports a relationship with Rokote Laboratories Finland Ltd that includes: equity or stocks.
Acknowledgments
Acknowledgments
We thank Virpi Syvälahti, Sanna Mäki, Marjo Rissanen and Marcel Messing (University of Helsinki) and Sari Järveläinen and Tiina Koponen (Biocenter Kuopio National Virus Vector Laboratory, Finland) for expert technical assistance, and the laboratory technicians of the Histology Laboratory, Institute of Veterinary Pathology, Vetsuisse Faculty, University of Zurich, for performing the histological and immunohistochemical stainings. This study was supported primarily by funding from the Academy of Finland to K.A., K.S., and S.Y.-H., and from the Sakari Alhopuro Foundation to KS. The funders had no role in the study design, the, data collection, analysis, and interpretation, writing of the report, or the decision to submit the article for publication.
The following reagent was obtained through BEI Resources, NIAID, NIH: Vector pCAGGS, containing the SARS-Related Coronavirus 2, Wuhan-Hu-1 Spike Glycoprotein Gene RBD with C-Terminal Hexa-Histidine Tag, NR-52309.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2023.04.020.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ioannou P., Karakonstantis S., Astrinaki E., Saplamidou S., Vitsaxaki E., Hamilos G., et al. Transmission of SARS-CoV-2 variant B.1.1.7 among vaccinated health care workers. Infect Dis (Lond) 2021;53:876–879. doi: 10.1080/23744235.2021.1945139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine-Tiefenbrun M., Yelin I., Katz R., Herzel E., Golan Z., Schreiber L., et al. Initial report of decreased SARS-CoV-2 viral load after inoculation with the BNT162b2 vaccine. Nat Med. 2021;27:790–792. doi: 10.1038/s41591-021-01316-7. [DOI] [PubMed] [Google Scholar]
- 5.Nanduri S., Pilishvili T., Derado G., Soe M.M., Dollard P., Wu H., et al. Effectiveness of Pfizer-BioNTech and Moderna Vaccines in preventing SARS-CoV-2 infection among nursing home residents before and during widespread circulation of the SARS-CoV-2 B.1.617.2 (Delta) variant - National Healthcare Safety Network, March 1-August 1, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1163–1166. doi: 10.15585/mmwr.mm7034e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hotez P.J. SARS-CoV-2 variants offer a second chance to fix vaccine inequities. Nat Rev Microbiol. 2022:1–2. doi: 10.1038/s41579-022-00824-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savinkina A., Bilinski A., Fitzpatrick M., Paltiel A.D., Rizvi Z., Salomon J., et al. Estimating deaths averted and cost per life saved by scaling up mRNA COVID-19 vaccination in low-income and lower-middle-income countries in the COVID-19 Omicron variant era: a modelling study. BMJ Open. 2022;12:e061752. doi: 10.1136/bmjopen-2022-061752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z., Shen Q., Chang H. Vaccines for COVID-19: a systematic review of immunogenicity, current development, and future prospects. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.843928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lund F.E., Randall T.D. Scent of a vaccine. Science. 2021;373:397–399. doi: 10.1126/science.abg9857. [DOI] [PubMed] [Google Scholar]
- 11.Hassan A.O., Kafai N.M., Dmitriev I.P., Fox J.M., Smith B.K., Harvey I.B., et al. A single-dose Intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183:169–184.e13. doi: 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King R.G., Silva-Sanchez A., Peel J.N., Botta D., Dickson A.M., Pinto A.K., et al. Single-Dose Intranasal Administration of AdCOVID Elicits Systemic and Mucosal Immunity against SARS-CoV-2 and Fully Protects Mice from Lethal Challenge. Vaccines (Basel) 2021:9. doi: 10.3390/vaccines9080881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langel S.N., Johnson S., Martinez C.I., Tedjakusuma S.N., Peinovich N., Dora E.G., et al. Adenovirus type 5 SARS-CoV-2 vaccines delivered orally or intranasally reduced disease severity and transmission in a hamster model. Sci Transl Med. 2022;14:eabn6868. doi: 10.1126/scitranslmed.abn6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Doremalen N., Purushotham J.N., Schulz J.E., Holbrook M.G., Bushmaker T., Carmody A., et al. Intranasal ChAdOx1 nCoV-19/AZD1222 vaccination reduces viral shedding after SARS-CoV-2 D614G challenge in preclinical models. Sci Transl Med. 2021:13. doi: 10.1126/scitranslmed.abh0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayward S.L., Bautista-Lopez N., Suzuki K., Atrazhev A., Dickie P., Elliott J.F. CD4 T cells play major effector role and CD8 T cells initiating role in spontaneous autoimmune myocarditis of HLA-DQ8 transgenic IAb knockout nonobese diabetic mice. J Immunol. 2006;176:7715–7725. doi: 10.4049/jimmunol.176.12.7715. [DOI] [PubMed] [Google Scholar]
- 16.Greenbaum J., Sidney J., Chung J., Brander C., Peters B., Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kant R., Kareinen L., Smura T., Freitag T.L., Jha S.K., Alitalo K., et al. Common laboratory mice are susceptible to infection with the sars-cov-2 beta variant. Viruses. 2021:13. doi: 10.3390/v13112263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamichhane A., Azegamia T., Kiyonoa H. The mucosal immune system for vaccine development. Vaccine. 2014;32:6711–6723. doi: 10.1016/j.vaccine.2014.08.089. [DOI] [PubMed] [Google Scholar]
- 19.Kar S., Devnath P., Emran T.B., Tallei T.E., Mitra S., Dhama K. Oral and intranasal vaccines against SARS-CoV-2: Current progress, prospects, advantages, and challenges. Immun Inflamm Dis. 2022;10:e604. doi: 10.1002/iid3.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coughlan L. Factors Which Contribute to the Immunogenicity of Non-replicating Adenoviral Vectored Vaccines. Front Immunol. 2020;11:909. doi: 10.3389/fimmu.2020.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee E.G., Blattman J.N., Kasturi S.P., Kelley R.P., Kaufman D.R., Lynch D.M., et al. Multiple innate immune pathways contribute to the immunogenicity of recombinant adenovirus vaccine vectors. J Virol. 2011;85:315–323. doi: 10.1128/JVI.01597-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao L., Zhang M., Cong H. Advances in the study of HLA-restricted epitope vaccines. Hum Vaccin Immunother. 2013;9:2566–2577. doi: 10.4161/hv.26088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu F., Zhuang C., Chu K., Zhang L., Zhao H., Huang S., et al. Safety and immunogenicity of a live-attenuated influenza virus vector-based intranasal SARS-CoV-2 vaccine in adults: randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Respir Med. 2022;10:749–760. doi: 10.1016/S2213-2600(22)00131-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dicks M.D., Spencer A.J., Coughlan L., Bauza K., Gilbert S.C., Hill A.V., et al. Differential immunogenicity between HAdV-5 and chimpanzee adenovirus vector ChAdOx1 is independent of fiber and penton RGD loop sequences in mice. Sci Rep. 2015;5:16756. doi: 10.1038/srep16756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greinacher A., Schönborn L., Siegerist F., Steil L., Palankar R., Handtke S., et al. Pathogenesis of vaccine-induced immune thrombotic thrombocytopenia (VITT) Semin Hematol. 2022;59:97–107. doi: 10.1053/j.seminhematol.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klok F.A., Pai M., Huisman M.V., Makris M. Vaccine-induced immune thrombotic thrombocytopenia. Lancet Haematol. 2022;9:e73–e80. doi: 10.1016/S2352-3026(21)00306-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGonagle D., De Marco G., Bridgewood C. Mechanisms of Immunothrombosis in Vaccine-Induced Thrombotic Thrombocytopenia (VITT) Compared to Natural SARS-CoV-2 Infection. J Autoimmun. 2021;121 doi: 10.1016/j.jaut.2021.102662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leikas A.J., Laham-Karam N., Agtereek E., Peltonen H.M., Selander T., Korpisalo P., et al. Efficacy and safety of clinical-grade human vascular endothelial growth factor-D(ΔNΔC) gene therapy containing residual replication-competent adenoviruses. Hum Gene Ther. 2021;32:761–770. doi: 10.1089/hum.2020.299. [DOI] [PubMed] [Google Scholar]
- 29.Puumalainen A.M., Vapalahti M., Agrawal R.S., Kossila M., Laukkanen J., Lehtolainen P., et al. Beta-galactosidase gene transfer to human malignant glioma in vivo using replication-deficient retroviruses and adenoviruses. Hum Gene Ther. 1998;9:1769–1774. doi: 10.1089/hum.1998.9.12-1769. [DOI] [PubMed] [Google Scholar]
- 30.Virtanen J., Uusitalo R., Korhonen E.M., Aaltonen K., Smura T., Kuivanen S., et al. Kinetics of Neutralizing Antibodies of COVID-19 Patients Tested Using Clinical D614G, B.1.1.7, and B 1.351 Isolates in Microneutralization Assays. Viruses. 2021:13. doi: 10.3390/v13060996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusanen J., Kareinen L., Levanov L., Mero S., Pakkanen S.H., Kantele A., et al. A 10-minute ”mix and read“ antibody assay for SARS-CoV-2. Viruses. 2021;13 doi: 10.3390/v13020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freitag T.L., Podojil J.R., Pearson R.M., Fokta F.J., Sahl C., Messing M., et al. Gliadin nanoparticles induce immune tolerance to gliadin in mouse models of celiac disease. Gastroenterology. 2020;158:1667–1681.e12. doi: 10.1053/j.gastro.2020.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2019;2020:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dagotto G., Mercado N.B., Martinez D.R., Hou Y.J., Nkolola J.P., Carnahan R.H., et al. Comparison of subgenomic and total RNA in SARS-CoV-2 challenged Rhesus macaques. J Virol. 2021;95 doi: 10.1128/JVI.02370-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.