Abstract
The past three decades have yielded a wealth of information regarding the chromatin regulatory mechanisms that control transcription. The “histone code” hypothesis—which posits that distinct combinations of posttranslational histone modifications are “read” by downstream effector proteins to regulate gene expression—has guided chromatin research to uncover fundamental mechanisms relevant to many aspects of biology. However, recent molecular and genetic studies revealed that the function of many histone-modifying enzymes extends independently and beyond their catalytic activities. In this review, we highlight original and recent advances in the understanding of noncatalytic functions of histone modifiers. Many of the histone modifications deposited by these enzymes—previously considered to be required for transcriptional activation—have been demonstrated to be dispensable for gene expression in living organisms. This perspective aims to prompt further examination of these enigmatic chromatin modifications by inspiring studies to define the noncatalytic “epigenetic moonlighting” functions of chromatin-modifying enzymes.
Chromatin modifiers have “moonlighting” functions in transcriptional regulation that go beyond their enzymatic activities.
INTRODUCTION
It is a long-appreciated fact that the posttranslational modifications on diverse histone residues, which are catalyzed by histone-modifying enzymes, are linked to transcriptional regulation. Early biochemical studies correlatively associated histone acetylation with increased RNA synthesis, suggesting a role in transcriptional activation (1, 2), and subsequent genetic loss-of-function experiments revealed that many histone acetyltransferases (HATs) and methyltransferases play crucial roles in transcriptional regulation (3, 4). The catalytic activities of these enzymes have widely been assumed to directly instruct gene expression via distinct combinations of interpretable histone marks, in accordance with what is referred to as the “histone code” hypothesis (5, 6). This hypothesis holds that it is the “written” combination of histone modifications (on the same histone, on different histones within the same nucleosome, or even on adjacent nucleosomes within a local chromatin environment) that drive downstream transcriptional responses by recruiting “reader” proteins to bind. Because many histone-modifying enzymes contain one or more reader domains themselves, these combinatorial histone modifications are proposed to create feedback circuits to ensure robust responses (6–11). Over the past two decades, studies guided by the histone code hypothesis have revolutionized our understanding of the implementation and function of chromatin modifications and have unequivocally established chromatin-modifying enzymes as central regulators of many biological processes (7). However, recent studies have revealed that the cellular functions of many chromatin-modifying enzymes extend beyond their catalytic activities and, in several cases, have been shown to be catalytic independent. Our initial studies in generating a comprehensive library of histone mutants in yeast suggested that no lysine residues are essential for viability (12). This initial observation suggested that either lysine modifications are not required for specific regulation of gene expression or that the enzymes have multiple lysine substrates (12). In addition, some histone modifications that were previously assumed to directly affect transcription have recently been shown to have negligible effects on gene expression. For instance, we recently demonstrated that H3K4 monomethylation (H3K4me1) and H3K27 acetylation (H3K27ac) marks at enhancers are not required for transcriptional regulation, let alone for the development of metazoan life (13, 14). We were able to generate viable Drosophila that express SET domainless Trr/COMPASS (complex of proteins associated with Set1) H3K4me1 methyltransferase but still develop normally, despite the resultant loss of both H3K4me1 and H3K27ac at enhancers (13, 14). Several recent studies by our group and others have effectively shifted the paradigm in which histone methylation was held to be essential (15–18), and this shift has now arrived for histone acetylation by CBP/p300 and Gcn5 as well (19). Facilitated by modern gene editing approaches, direct interrogation of histone modifier catalytic activity in transcriptional regulation has revealed that catalytic-independent functions overshadow catalytic activities in the case of many enzymes, suggesting that their histone marks may not necessarily be instructive for transcriptional regulation. Rather, for these enzymes, it may be their protein complex context and not their catalytic activity that primarily contributes to their biological role in the regulation of gene expression. In this review, we highlight recent studies of catalysis-independent functions that illuminate intriguing aspects of these multifaceted protein/enzyme complexes.
CATALYTIC INDEPENDENT FUNCTIONS OF HATS
HATs off to ZGA: Transcription in the absence of two major HAT activities
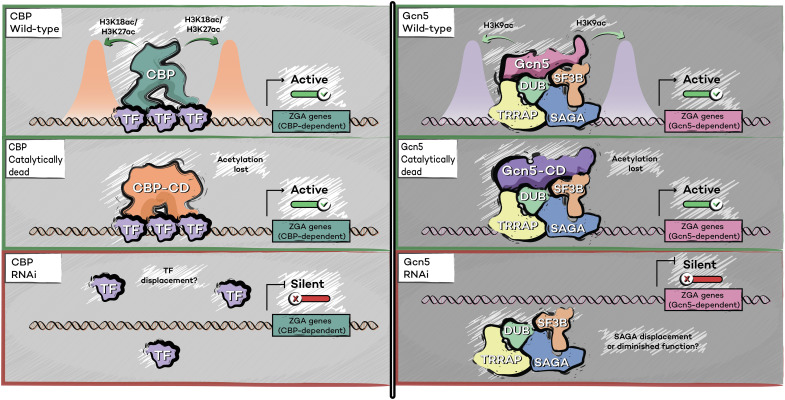
The earliest phase of metazoan development is supported by protein and mRNA that was maternally deposited into the oocyte during female gametogenesis. A crucial transition occurs when the zygotic genome begins to transcribe RNA, an event termed zygotic genome activation (ZGA) (20). In this issue of Science Advances, Iovino’s laboratory examined the requirement for catalytic and catalytic-independent activities of HATs in Drosophila ZGA (Fig. 1) (19). Through an in vivo RNA interference (RNAi) screen of maternally deposited chromatin regulatory factors, they identified a number of HATs and histone deacetylases that are essential for ZGA. They focus on Nejire (a homolog of mammalian CBP and p300) and Gcn5 (a homolog of mammalian GCN5 and PCAF), two HATs that have been studied previously in several model systems and have HAT activities considered to be instructive for transcriptional activation (21–26). Acetylation of histone lysine residues directly affects the structure and biophysical properties of nucleosomes: Acetylation neutralizes the positive charge of lysine side chains, thereby weakening their interaction with DNA (27). In vitro studies demonstrated that H4K16ac is sufficient both to inhibit formation of the compacted 30-nm chromatin fiber and to alter interactions with chromatin remodeling proteins (28–30). These observations led to the notion that the general function of histone acetylation is alteration of histone charge to trigger chromatin decompaction and increase accessibility for activating transcription factors (27). Although CBP/Nejire and Gcn5 are both involved in transcriptional activation, they acetylate distinct histone residues: CBP/Nejire acetylates histone H3 at lysine-18 (H3K18ac) and the enhancer-associated H3K27ac, whereas Gcn5 acetylates histone H3 lysine-9 (H3K9ac) (31). Protein depletion of either CBP/Nejire or Gcn5 by RNAi results in a failure to initiate ZGA and in defective embryogenesis, suggesting that their HAT activities may be essential for ZGA (19). However, ZGA can be rescued by expression of catalytically dead Gcn5 or CBP/Nejire in their respective knockdown Drosophila lines, despite failure of these transgenes to rescue the loss of histone acetylation in these animals (Fig. 1) (19). This observation suggests that the HAT activities of these enzymes may not play an instructive role in the regulation of gene expression during ZGA.
Fig. 1. Catalytic-independent functions of CBP/p300 (Nejire) and Gcn5 in Drosophila ZGA.
In wild-type Drosophila (top), CBP/p300 (Nejire) and Gcn5 regulate the expression of distinct gene sets and catalyze H3K18ac/H3K27ac and H3K9ac, respectively. When wild-type Nejire and Gcn5 are replaced with catalytically dead (CD) mutants, histone acetylation is lost from their target genes; however, these genes still become activated during ZGA. By contrast, when Nejire and Gcn5 protein is eliminated by RNAi (bottom), target genes fail to activate. DUB, deubiquitinase module; SF3B, splicing factor 3B module; TF, transcription factor; TRRAP, transformation/transcription domain–associated protein module.
These results demonstrate that catalytic-independent mechanisms are central to the function of Gcn5 and CBP/p300 homologs in Drosophila ZGA and imply that features such as interactions with cofactors may play a more critical role than histone acetylation for this process (Fig. 1). Because Nejire and its homologs CBP and p300 are thought to function by forming interactions with various activating transcription factors (32), it will be interesting to determine whether the absence of CBP/Nejire protein disrupts transcription factor recruitment to chromatin. Perhaps, protein-protein interactions formed between CBP/Nejire, and multiple transcription factors facilitate cooperative binding to DNA and increase avidity (Fig. 1). Gcn5 participates in a large complex called Spt-Ada-Gcn5 acetyltransferase (SAGA) (33). In addition to its Gcn5-containing HAT module, SAGA also contains additional functional modules involved in processes including histone H2B deubiquitination, mRNA splicing, and transcription factor interactions (Fig. 1) (33). How catalytic-independent functions of Gcn5 may participate in SAGA function will be an important area for future studies to examine. Although Iovino and colleagues (19) clearly demonstrate that the catalytic activity of Nejire and Gcn5 is dispensable for Drosophila ZGA and early embryonic development, it will be interesting to further examine the phenotype of animals in which both maternal and embryonic Nejire or Gcn5 have been substituted with a catalytically dead form, to determine the extent to which Drosophila can develop in the total absence of these HAT activities. It will also be important to examine the mammalian homologs of these proteins to determine whether HAT catalytic-independent activity is a conserved feature of mammalian ZGA.
Catalytic-independent functions of Nejire/CBP/p300 in transcriptional regulation
Another recent study identified an unexpected role for Nejire/CBP/p300 catalytic-independent activity in the promotion of gene repression by Polycomb repressive complex 2 (PRC2) (34). The HAT activity of Nejire/CBP/p300 is impeded by the histone methyltransferase activity of PRC2 (35, 36), which catalyzes methylation of H3K27, a histone mark that exists in obligate mutual exclusivity with H3K27ac. Loss-of-function mutations of PRC2 and inhibition of PRC2 catalytic activity both result in increased H3K27ac levels in several model systems (35–38). In continuation of previous work that observed binding of CBP/Nejire at Drosophila Polycomb response elements (PREs) (39), Hunt and coworkers (34) examined whether Nejire might play a functional role in Polycomb-mediated silencing. Treatment of Drosophila cells with C646, a catalytic inhibitor of CBP/p300 (40), unexpectedly resulted in the depletion of Nejire from chromatin. Both C646 treatment and Nejire RNAi resulted in Nejire depletion and reduced Polycomb complex subunit recruitment at PREs (34). In contrast, treatment of cells with the structurally distinct CBP/p300 catalytic inhibitor A-485 (41) did not displace Nejire from chromatin and had no effect on Polycomb occupancy at PREs. Hunt and coworkers went on to show that Nejire and CBP/p300 promote RNA polymerase II pausing through a catalytic-independent mechanism (34). These findings implicate the CBP/p300 homolog Nejire, a protein that has predominantly been thought to function as a transcriptional activator, in the facilitation of transcriptional repression by an antagonistic complex. The ability of the CBP/p300 inhibitor C646 to induce depletion of Nejire from chromatin is an unexpected finding that merits further examination of the underlying molecular mechanism. It will also be informative to determine the effects of CBP/p300 bromodomain inhibitors such as SGC-CBP30, which displace the protein from chromatin while leaving its HAT activity unperturbed (42).
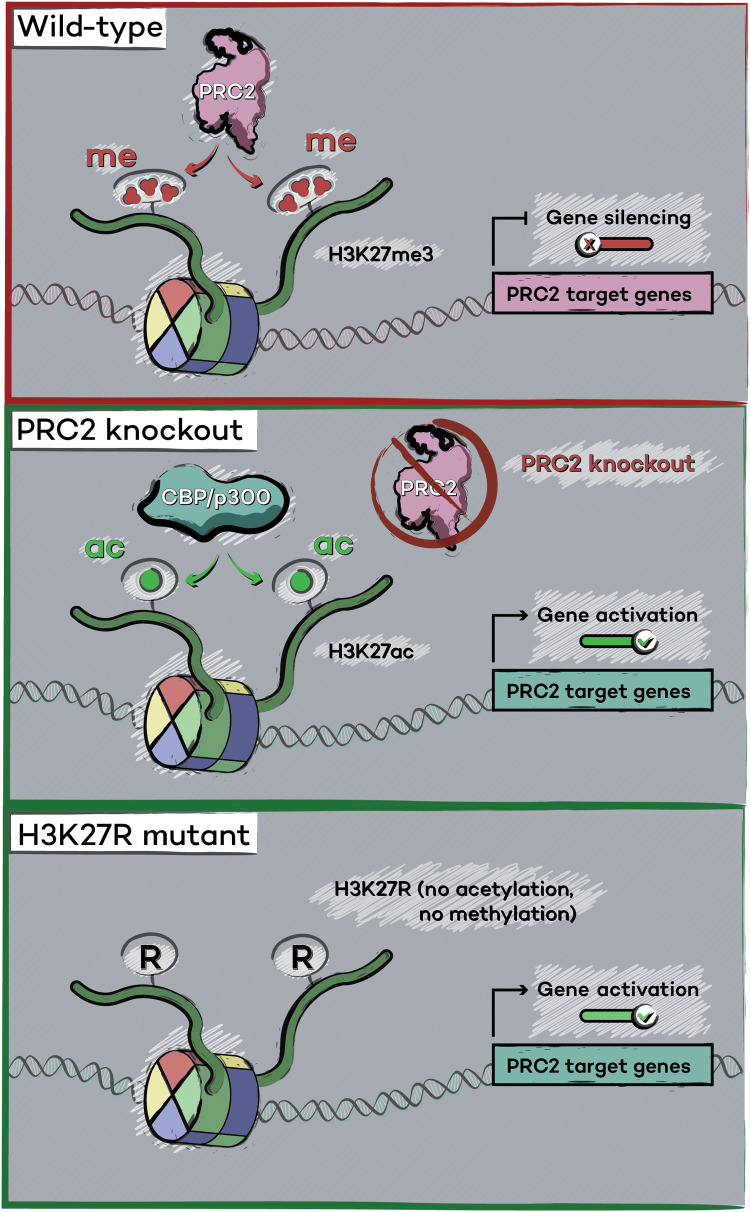
Histone gene mutagenesis experiments also support a model in which H3K27ac is generally dispensable for transcriptional activation (Fig. 2). In Drosophila, replacement of histone H3.1 or H3.3 with a lysine-27–to–arginine (K27R) point mutant, which can no longer be acetylated by Nejire or methylated by PRC2, results in the aberrant activation of PRC2-repressed genes and a developmental phenotype that mirrors PRC2 loss of function (43–46). This result strongly suggests that while H3K27me3 mediated by PRC2 is critical for repression of PRC2 targets, H3K27ac is not required for their activation. Because of the multiple histone clusters in mammalian genomes, studies of H3K27 in mammals were until recently limited to the H3.3 variant, which is encoded by only two genes (47). These studies revealed that modifications of H3.3K27 are not essential for gene expression or for embryonic stem cell viability (47). A recent study accomplished the monumental task of mutagenizing all copies of mammalian H3.1 and H3.3 (48). Sankar and coworkers (48) developed a CRISPR-guided DNA base-editing strategy to create mouse embryonic stem cells (mESCs) in which each copy of H3.1 and H3.3 contains the K27R point mutation. Analysis of these cells revealed that, akin to Drosophila (43–46), genes normally repressed by PRC2 displayed spurious activation (48). Moreover, these cells activate lineage-specific genes upon in vitro differentiation, demonstrating that H3K27ac is not required for activation of PRC2 target genes in the absence of H3K27me3 nor is it required for the activation of genes involved in specific developmental programs. Instead, these studies strongly suggest that H3K27 methylation by PRC2, which is crucial to PRC2-mediated repression, is the major biologically relevant activity of this complex (48).
Fig. 2. Gene activation in the absence of H3K27 modifications.
In wild-type mESCs and Drosophila, PRC2 maintains target gene silencing by catalyzing methylation of the H3K27 residue. Upon genetic elimination of PRC2 (middle), its targets acquire CBP/p300-catalyzed H3K27ac and become transcriptionally activated, suggesting that the possibility that H3K27ac could instruct activation. However, replacement of H3K27 with an arginine residue (bottom) revealed that PRC2 repressed genes are activated even in the absence of H3K27ac.
It should be noted that although H3K27ac is not required for transcriptional activation in general, this does not exclude context-dependent requirements for the HAT activity of CBP/p300. The role of H3K18ac, which is also catalyzed by CBP/p300, is yet to be investigated by histone gene mutagenesis in mammals. Multiple studies have observed altered gene expression upon CBP/p300 catalytic inhibition, suggesting that blocking CBP/p300 substrate acetylation alters transcription, whether directly or indirectly (49–51). However, an important caveat when evaluating experiments involving CBP/p300 catalytic inhibitors is that CBP/p300 also acetylates a wide range of nonhistone substrates (52). Therefore, it cannot necessarily be concluded that alterations in gene expression due to CBP/p300 catalytic inhibition are caused by loss of H3K18ac/H3K27ac, as they could alternatively result from the loss of acetylation of other CBP/p300 targets including transcription factors, chromatin modifiers, and cohesin subunits (52). Now that methods for generating mammalian histone gene point mutations have been developed (48), it will be important to compare and contrast the cellular phenotypes resulting from H3K27R and H3K18R mutation with the effects of CBP/p300 catalytic inhibition or total CBP/p300 protein depletion. In addition, the mammalian experiments performed thus far have been executed in mESCs, and further experiments in mouse embryos carrying the H3K27R mutant are needed to evaluate the developmental requirement for modifications at this site.
Essential role of H4K16ac in Drosophila dosage compensation
Although there is strong evidence that H3K27ac does not generally play a direct role in transcriptional activation, this is not the case for all histone acetylation marks. For instance, the H4K16ac modification has a well-defined role in the regulation of developmental gene expression in Drosophila. In organisms that use XY sex determination, various systems exist to equalize the gene dosage of X-linked genes between XX and XY individuals (53–55). In Drosophila, this is accomplished by a twofold up-regulation of genes on the single male X chromosome to match the expression levels in XX females—an effect termed dosage compensation (56–58). The transcriptional up-regulation of X-linked genes in male Drosophila is dependent on the multiprotein male-specific lethal (MSL) complex that contains MOF, a H4K16 acetyltransferase (59–61). Histone H4K16ac is enriched on the male X chromosome, implicating this mark in the up-regulation of gene expression during dosage compensation (56, 58), and recent histone gene mutagenesis experiments have also examined this phenomenon. Copur and coworkers (62) engineered Drosophila lines in which all histone H4 genes (including the replication-independent His4r genes) were deleted and rescued using a transgene containing either wild-type H4 or H4 in which K16 was substituted with arginine (H4K16R), glutamine (H4K16Q, an acetyl-mimic), or alanine (H4K16A). The resultant phenotypes support the model that dosage compensation is the primary biological function of MSL-MOF H4K16ac HAT activity during Drosophila development. Drosophila with an H4K16R mutation display a phenotype akin to MOF loss-of-function mutants, with males dying at the end of larval development, whereas females undergo metamorphosis and die within 7 days of adult life. This later requirement in females is likely due to MOF participating in a second HAT complex called the nonspecific lethal complex (NSL) (56, 58). The H4K16Q mutant allows a very small number of males to survive to adulthood, suggesting that an acetyl mimic can support dosage compensation, albeit with very low penetrance (62). Intriguingly, the H4K16A mutant displays a more severe phenotype than H4K16R, with both males and females dying before the end of embryogenesis, implying that the presence of a longer amino acid side chain may be essential for interactions with critical nucleosome binding proteins. It should also be noted that the number of female Drosophila surviving until adulthood is also reduced for both the H4K16R and H4K16Q mutants relative to the wild-type histone H4 rescue transgene. This raises several possibilities: (i) The point mutants disrupt the dynamic acetylation and deacetylation of H4K16, which may play a role in dosage compensation, (ii) the unmodified H4K16 residue is important for protein-protein interactions, or (iii) additional unidentified modifications regulate the H4K16 site.
Another group performed similar studies with H4K16A mutant Drosophila, and although they observed distinct developmental effects, they reached similar overall conclusions regarding the critical role of H4K16ac in dosage compensation (46). When Zhang and coworkers (46) analyzed Drosophila expressing H4K16A, they observed that males exhibit lethality at the larval stage due to defective dosage compensation, whereas females develop into adults but have fertility defects (46). This stands in stark contrast to the findings of Copur and coworkers (62), who observed that both male and female H4K16A Drosophila die much earlier during embryogenesis. There are significant technical differences between the two studies that may explain these differences. Copur and coworkers (62) deleted all copies of histone H4 including the His4r variant and rescued this with transgenes containing a total 12 histone gene units (GUs). It should be noted that only approximately 30% of Drosophila with 12 wild-type H4 GUs survive to adulthood, so the gene dosage from this transgene is clearly insufficient to match that of the endogenous 23 histone GUs, resulting in partially penetrant lethality (62). In contrast, Zhang and colleagues (46) deleted the canonical histone H4 genes but did not disrupt the His4r variant, so the animals that they analyzed did have residual expression of wild-type H4. However, Zhang and coworkers (46) performed their transgenic rescue experiments using 20 histone GUs, a number that they observed to be sufficient to support normal development in close to 100% of offspring. Thus, while the function of H4K16ac in dosage compensation in male Drosophila is clear, the requirements for H4K16ac and the H4K16 residue itself outside of dosage compensation have not yet been fully resolved. Copur and coworkers (62) may have observed a more severe developmental phenotype because their rescue system used an insufficient number of histone GUs. Alternatively, because Zhang and coworkers (46) did not disrupt the His4r histone H4 variant, it is also possible that the residual levels of wild-type H4 expressed in their system is capable of rescuing the mutant phenotype. It will be important for future studies to combine the strengths of these two genetic systems to resolve the intricacies of H4K16 regulation.
Similar to its Drosophila counterpart, mammalian MOF (also known as KAT8) is responsible for implementing the majority of H4K16ac (63, 64). However, in mammals, sex-chromosome dosage compensation is not achieved by up-regulating the male X chromosome; therefore, a role for MSL-MOF is unlikely to be conserved. In vitro biochemical experiments suggest that H4K16ac impairs chromatin compaction both by weakening internucleosomal interactions and by altering interactions with chromatin remodeling proteins (28–30), providing a possible biophysical mechanism by which H4K16ac could function in transcriptional activation. However, experiments performed in mammalian cells found that although H4K16ac is associated with active genes, it does not appear to be essential for either large-scale chromatin decompaction or local chromatin accessibility (65, 66). Radzisheuskaya and coworkers (65) recently performed a genetic analysis of the requirements for multiple subunits of the mammalian MSL and NSL complexes (65). The authors found that the MSL complex and the H4K16ac mark were dispensable, both for cell proliferation and for chromatin accessibility. In contrast, the NSL complex promotes the expression of genes that are essential for critical cellular processes and is itself essential for cell viability. Intriguingly, when MOF participates in the NSL complex, it acetylates H4K5 and H4K8 in lieu of H4K16. The roles of H4K5 and H4K8 in transcriptional activation and the mechanism by which the context of the NSL complex switches MOF substrate specificity together comprise an important area for future work.
CATALYTIC INDEPENDENT FUNCTIONS OF HISTONE METHYLTRANSFERASES
Life without H3K4me1 at enhancers by Trr-MLL3-MLL4-COMPASS
In addition to histone acetylation, H3K4 methylation is another histone posttranslational modification that is evolutionarily conserved from yeast to human and has long been implicated in transcriptional regulation (67, 68). Trimethylation of H3K4 (H3K4me3) correlates with active promoters, whereas H3K4me1 is associated with distal cis-regulatory enhancer sequences (67, 69–73). The multiprotein complexes that catalyze H3K4 methylation belong to the evolutionarily conserved COMPASS family of enzymes (67, 74). Many single-celled eukaryotes including Saccharomyces cerevisiae have a single COMPASS enzyme, Set1, which is responsible for all H3K4 methylation states (67, 75–78). In contrast, metazoans contain three versions of COMPASS: SET1-COMPASS, which is structurally similar to yeast COMPASS; Trx-MLL1-MLL2-COMPASS, which mediates H3K4me2 and H3K4me3 at the promoters of developmentally regulated genes; and Trr-MLL3-MLL4-COMPASS, which mediates H3K4me1 at intergenic enhancer sequences (67, 74).These divergent COMPASS family display unique subunit compositions and are thought to play distinct roles in transcriptional regulation. However, despite the strong correlation of H3K4 methylation with active transcription, there is little evidence that this histone modification itself is directly involved in transcriptional activation at most genes (16–18, 79). Studies of S. cerevisiae Set1 mutants, which entirely lack H3K4 methylation, revealed that cells can survive in the absence of this modification. While displaying only modest alterations in gene expression, these yeast do exhibit growth defects and, paradoxically, display impairments in gene silencing rather than activation (75, 78, 80–82). Mosaic clonal analysis of histone H3K4R and H3K4A gene replacement in Drosophila also revealed that, although cells lacking H3K4 methylation display pronounced proliferation defects, they nevertheless retain normal expression of many developmentally regulated genes (83). While these experiments do not rule out gene-specific transcriptional regulatory requirements for H3K4 methylation, they clearly demonstrated that this mark is not an absolute requirement for gene expression. A more recent study using a global H3K4A gene replacement strategy observed early embryonic lethality (46), suggesting that H3K4 methylation may play an important role in development; however, it is also possible that the unmodified H3K4 residue itself could play a critical role through recruitment of H3K4me0 reader proteins (84). Moreover, none of these experiments distinguish which H3K4 methylation state (H3K4me1/2/3) might be important. Thus, it has until recently remained unclear whether and in what contexts the catalytic activity of the three distinct metazoan COMPASS complexes might play a crucial role in animal development.
Catalytic-independent activities of the COMPASS family of H3K4 methyltransferases
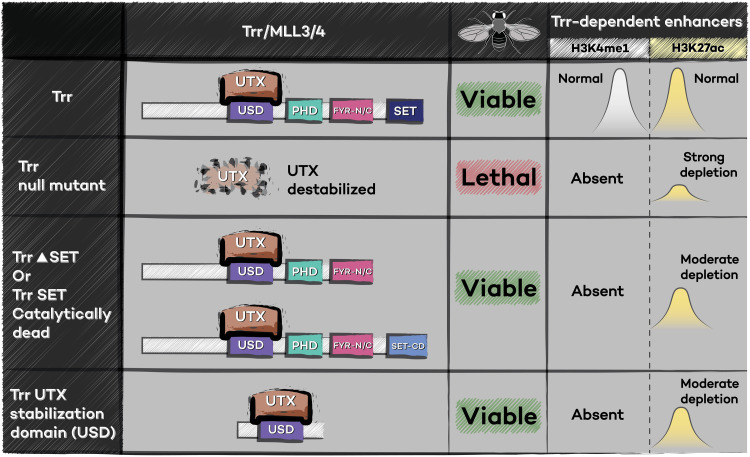
Recent studies have examined catalytic-independent functions for the multiple metazoan COMPASS complexes. Gene enhancer regulation has been the subject of intense interest for several decades, as enhancer sequences drive accurate developmental gene expression patterns even when separated from their cognate promoters by significant distances (85, 86). The discovery that enhancers often contain a combination of the H3K4me1 and H3K27ac histone modifications led to speculation that these chromatin marks might be important for enhancer function (70, 87). It was subsequently found by our laboratory that Drosophila Trr and its homologous mammalian MLL3-MLL4-COMPASS counterparts are responsible for implementing H3K4me1 specifically at enhancers (88, 89). A major breakthrough in our understanding of the role for Trr-MLL3-MLL4-COMPASS in enhancer regulation came when ours and the Wysocka laboratories found that the H3K4me1 methyltransferase proteins, but not their methyltransferase activities, are essential for enhancer activation (13, 14, 69, 90). Drosophila carrying a catalytically dead Trr allele or Trr lacking a SET domain develop normally, despite an absence of H3K4me1 and reduced H3K27ac at enhancers (13, 14). Similarly, studies in mESCs revealed that complete deletion of MLL3-MLL4 protein eliminates H3K4me1 and H3K27ac from enhancer sequences, alters mESC gene expression, and disrupts differentiation (13, 14, 90, 91). MLL3-MLL4 catalytically dead mESCs also lack H3K4me1 at enhancers but display only modestly reduced H3K27ac, minimal changes in gene expression, and, unlike MLL3-MLL4–deficient mESCs, are capable of the naïve to primed pluripotency transition (13, 90, 91). These results demonstrate that it is the Trr-MLL3-MLL4 proteins themselves, not their H3K4me1 methyltransferase activities, that are crucial for regulating enhancer function. Our laboratory then undertook a genetic domain-mapping screen to determine which regions of the Trr protein can rescue development in Trr-deficient Drosophila (Fig. 3) (14). Consistent with our previous study, we found that the SET methyltransferase domain is dispensable for Trr function; however, we also identified a small, central region of Trr that can rescue a Trr null condition (14). This region is required both for binding of Trr-MLL3-MLL4 to the coregulator UTX (also known as KDM6A) and for stabilization of the UTX protein (14). Intriguingly, mutations of MLL4 or UTX are responsible for two closely related genetic syndromes called Kabuki syndrome 1 and Kabuki syndrome 2 (92). UTX itself is a demethylase that removes the repressive, Polycomb-deposited H3K27me3 mark (92), although this catalytic activity has been demonstrated to be nonessential in some contexts (93–96). Our results demonstrate that in the context of Drosophila development, the major biologically relevant function of Trr is to stabilize UTX protein and that H3K4me1 methyltransferase activity is dispensable for viability. Although in vitro experiments in mESCs also demonstrate differences between null and catalytically dead MLL3-MLL4 mutants (13, 90, 91), the requirement for their noncatalytic activity in organismal development has not been resolved in vivo because the catalytically dead alleles analyzed also result in destabilization of the MLL3 and MLL4 proteins (97). Thus, further experiments are required to determine whether the major function of MLL3-MLL4 in mammals is to stabilize the UTX protein. Similarly, underscoring the importance of noncatalytic stabilizing functions, mutations that disrupt the interaction between MLL3-COMPASS and BAP1, another negative regulator of Polycomb repression, are associated with human cancer (98, 99).
Fig. 3. Life and transcription in the absence of H3K4me1 at enhancers.
In wild-type Drosophila, Trr catalyzes H3K4me1 at enhancers, coincident with the presence of H3K27ac. In Trr protein null mutants, which are lethal, H3K4me1 is completely lost from enhancers and H3K27ac is strongly reduced. In addition, the coactivator protein UTX becomes destabilized in the absence of Trr. When Trr null mutants are complemented with either Trr lacking the SET methyltransferase domain (Trr∆SET) or Trr with a catalytically dead SET mutation (SET-CD), viability is rescued; however, H3K4me1 remains absent from enhancers. Expression of these mutant proteins also rescues stability of the UTX protein and partially restores H3K27ac at enhancers. Remarkably, when Trr null mutants are complemented with a small region of Trr that lacks all other known functional domains but is sufficient to stabilize UTX, this is sufficient to rescue viability.
Since this discovery, catalytic-independent functions have also been described for other COMPASS family methyltransferases, including SET1A (100, 101), MLL1 (102, 103), and the MLL1-associated proteins and Dot1 (91). Complete deletion of Set1a in mice and mESCs results in a cell lethal phenotype (104). However, when our laboratory deleted the Set1 SET methyltransferase domain (Set1a-∆SET), we found that, although differentiated Set1a-∆SET cells do display substantial transcriptional alterations and impaired survival, the mutant protein causes only minimal alterations to gene expression and is sufficient to support normal survival in undifferentiated mESCs (101). Consistent with this, our analysis of mouse embryos revealed that Set1a-∆SET mutant embryos are still present at embryonic day 10.5 (E10.5), and although they display significantly impaired growth (105), their phenotype is distinct from that of Set1a null embryos, which are absent after E7.5 (104). This suggests that catalytic-independent functions are essential for Set1a’s ability to support general cellular survival, whereas catalytic-dependent functions are required specifically during cellular differentiation and embryonic development. MLL1 similarly exhibits both catalytic-dependent and -independent functions (102, 103). MLL1 null mice exhibit embryonic lethality due to a failure in hematopoiesis and altered expression of Hox genes that results in skeletal segmental identity defects (106–108). Although they display skeletal defects similar to these null mutants, mice homozygous for a catalytically dead MLL1 allele survive until adulthood and exhibit normal hematopoiesis (102, 103). Thus, MLL1 catalytic activity appears to be required for control of skeletal segmentation but dispensable for hematopoiesis.
Recent work from our laboratory also suggests that H3K4 methylation may be a by-product of active transcription rather than a signal directly instructing transcriptional activation (109, 110). The COMPASS family enzyme MLL2 (also known as Kmt2B) controls the expression of a subset of genes on mESCs. Our laboratory performed a genome-wide CRISPR-Cas9 screen for regulators of MLL2-dependent genes and identified the PRC2 complex and DNA methyltransferases (DNMTs) functioning in antagonism of MLL2 (109). Genes that are down-regulated upon genetic deletion of MLL2 display a loss of H3K4me3 at their promoters. Although the expression levels of these genes can be rescued by genetic or chemical disruption of either PRC2 or DNMTs (109), they do not reacquire H3K4me3 at their promoters when these genes are derepressed; rather, H3K4me3 remains at similar levels to those observed in MLL2 null mESCs. These findings have three main implications: (i) H3K4me3 is not required for gene expression at MLL2 targets; (ii) other COMPASS HMTs are unable to compensate for H3K4me3 loss when MLL2 is deleted, even in genes whose expression is rescued by loss of PRC2 or DNMT activity; and (iii) the main function of trithorax/MLL2 is to repel the PRC2 and DNMT binding that would lead to transcriptional inhibition at these genomic loci (109).
Using an orthogonal strategy, the Lis and Danko laboratories used molecular biological approaches in combination with computational modeling to correlate gene expression with histone modifications and investigate their causal relationship (110). Similar to our study, they also observed that H3K4me3 levels were dependent on transcription but not vice versa (110). By blocking transcription initiation using the chemical triptolide, Wang and colleagues (110) observed that H3K4me3 and H3K27ac are rapidly depleted from promoters within 1 hour after treatment, whereas other histone modifications assayed, such as H3K4me1 and H3K36me3, remained stable. Thus, it appears that while gene activation can trigger the accumulation of H3K4me3 at promoters in the presence of the appropriate methyltransferase enzyme, the levels of H3K4me3 are tightly coupled specifically to the active transcriptional state and, thus, do not play an instructive role in this process.
CONCLUSION
Despite decades of intense interest, the scientific community still has a great deal to learn about the intricate functions of histone modifying enzyme and their complexes in transcriptional regulation and other DNA-templated processes that use chromatin as a substrate. From the initial discovery/observation that generating a comprehensive library of histone mutants in yeast demonstrated that no lysine residues are essential for viability (12) to today, we have learned that histone-modifying enzymes have “moonlighting” activities in transcriptional regulation independent of their catalytic activities. The development of CRISPR-Cas9 technology now allows for the rapid mutagenesis of multiple genes, a crucial advantage when examining the many potentially redundant, closely related modifiers in mammalian cells. Perhaps, the most outstanding recent technical advance in the study of chromatin is the mutagenesis of all mammalian histone H3 genes, which was achieved through a dCas9 base-editor strategy (48, 111). If this approach can be successfully applied to additional sites of histone posttranslational modifications, then it will likely lead to a new wave in the understanding of histone modifier function. Although some histone modifications that correlate with active transcription (H3K4me1, H3K4me3 H3K9ac, H3K16ac, and H3K27ac) do not appear to generally instruct the activation of gene expression, we must be cautious not to rule out context- or gene-specific instructive functions for these modifications that may more subtly modulate gene expression or perhaps contribute to robustness of gene expression programs during development or stress responses. In addition, it is possible that certain histone modifications have important roles outside of transcriptional regulation, as it has been demonstrated that several histone modifiers and histone modification “readers” also moonlight in other DNA-based processes, such as DNA damage repair (17). Thus, emerging evidence that histone modifications have only minor or context-specific roles in transcriptional regulation should not diminish their potential biological importance but, instead, should guide chromatin research into new areas such as cell cycle control, DNA damage responses, and genome replication or other chromatin tempated processes. Further advances may result from the use of specific modifier-targeting small-molecule inhibitors, powerful tools that can also be leveraged to create PROTACs capable of triggering target degradation. An expanded slate of molecules targeting chromatin regulators may eventually furnish researchers with tools that can be used to inhibit catalytic activity, inhibit chromatin binding, or completely ablate the target protein. The combination of state-of-the-art genetics and chemical biology approaches promises unprecedented insight into the regulation and function of chromatin, which we hope will answer not only why many histone marks that are highly conserved from yeast to human do not appear to directly instruct gene expression per se but also how the protein complexes implementing these marks can have "moonlighting" activities to regulate gene expression independently of their catalytic functions.
Acknowledgments
We thank B. M. Monroe for scientific illustrations and S. Gold and A. Piunti for critical reading and editing of this manuscript.
Funding: Research in the A.S. laboratory is supported by an NIH Outstanding Investigator Award R35CA197569.
Author contributions: M.A.J.M. and A.S. wrote and edited the manuscript.
Competing interests: The authors declare that they have no competing interests
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the materials cited herein.
REFERENCES AND NOTES
- 1.V. G. Allfrey, R. Faulkner, A. E. Mirsky, Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 51, 786–794 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.B. G. Pogo, V. G. Allfrey, A. E. Mirsky, RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 55, 805–812 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.K. K. Lee, J. L. Workman, Histone acetyltransferase complexes: One size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295 (2007). [DOI] [PubMed] [Google Scholar]
- 4.H. M. Herz, A. Garruss, A. Shilatifard, SET for life: Biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem. Sci. 38, 621–639 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.B. D. Strahl, C. D. Allis, The language of covalent histone modifications. Nature 403, 41–45 (2000). [DOI] [PubMed] [Google Scholar]
- 6.T. Jenuwein, C. D. Allis, Translating the histone code. Science 293, 1074–1080 (2001). [DOI] [PubMed] [Google Scholar]
- 7.C. D. Allis, T. Jenuwein, The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016). [DOI] [PubMed] [Google Scholar]
- 8.T. Kouzarides, Chromatin modifications and their function. Cell 128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 9.S. Maurer-Stroh, N. J. Dickens, L. Hughes-Davies, T. Kouzarides, F. Eisenhaber, C. P. Ponting, The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28, 69–74 (2003). [DOI] [PubMed] [Google Scholar]
- 10.C. A. Musselman, M. E. Lalonde, J. Côté, T. G. Kutateladze, Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19, 1218–1227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. J. Ruthenburg, H. Li, D. J. Patel, C. D. Allis, Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S. Nakanishi, B. W. Sanderson, K. M. Delventhal, W. D. Bradford, K. Staehling-Hampton, A. Shilatifard, A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 15, 881–888 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.R. Rickels, H. M. Herz, C. C. Sze, K. Cao, M. A. Morgan, C. K. Collings, M. Gause, Y. H. Takahashi, L. Wang, E. J. Rendleman, S. A. Marshall, A. Krueger, E. T. Bartom, A. Piunti, E. R. Smith, N. A. Abshiru, N. L. Kelleher, D. Dorsett, A. Shilatifard, Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat. Genet. 49, 1647–1653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R. Rickels, L. Wang, M. Iwanaszko, P. A. Ozark, M. A. Morgan, A. Piunti, N. Khalatyan, S. H. A. Soliman, E. J. Rendleman, J. N. Savas, E. R. Smith, A. Shilatifard, A small UTX stabilization domain of Trr is conserved within mammalian MLL3-4/COMPASS and is sufficient to rescue loss of viability in null animals. Genes Dev. 34, 1493–1502 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Y. Aubert, S. Egolf, B. C. Capell, The unexpected noncatalytic roles of histone modifiers in development and disease. Trends Genet. 35, 645–657 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.S. Henikoff, A. Shilatifard, Histone modification: Cause or cog? Trends Genet. 27, 389–396 (2011). [DOI] [PubMed] [Google Scholar]
- 17.M. A. J. Morgan, A. Shilatifard, Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 52, 1271–1281 (2020). [DOI] [PubMed] [Google Scholar]
- 18.T. Pollex, E. E. M. Furlong, Correlation does not imply causation: Histone methyltransferases, but not histone methylation, SET the stage for enhancer activation. Mol. Cell 66, 439–441 (2017). [DOI] [PubMed] [Google Scholar]
- 19.F. Ciabrelli, L. Rabbani, F. Cardamone, F. Zenk, E. Löser, M. A. Schächtle, M. Mazina, V. Loubiere, N. Iovino, CBP and Gcn5 drive zygotic genome activation independently of their catalytic activity. Sci. Adv. 9, eadf2687 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.N. L. Vastenhouw, W. X. Cao, H. D. Lipshitz, The maternal-to-zygotic transition revisited. Development 146, dev161471 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Z. Arany, W. R. Sellers, D. M. Livingston, R. Eckner, E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell 77, 799–800 (1994). [DOI] [PubMed] [Google Scholar]
- 22.J. E. Brownell, J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, C. D. Allis, Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84, 843–851 (1996). [DOI] [PubMed] [Google Scholar]
- 23.V. V. Ogryzko, R. L. Schiltz, V. Russanova, B. H. Howard, Y. Nakatani, The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959 (1996). [DOI] [PubMed] [Google Scholar]
- 24.P. A. Grant, L. Duggan, J. Côté, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, J. L. Workman, Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11, 1640–1650 (1997). [DOI] [PubMed] [Google Scholar]
- 25.R. C. Trievel, J. R. Rojas, D. E. Sterner, R. N. Venkataramani, L. Wang, J. Zhou, C. D. Allis, S. L. Berger, R. Marmorstein, Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc. Natl. Acad. Sci. U.S.A. 96, 8931–8936 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.W. Zhang, J. R. Bone, D. G. Edmondson, B. M. Turner, S. Y. Roth, Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17, 3155–3167 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.A. J. Bannister, T. Kouzarides, Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Y. Liu, C. Lu, Y. Yang, Y. Fan, R. Yang, C. F. Liu, N. Korolev, L. Nordenskiöld, Influence of histone tails and H4 tail acetylations on nucleosome-nucleosome interactions. J. Mol. Biol. 414, 749–764 (2011). [DOI] [PubMed] [Google Scholar]
- 29.P. J. J. Robinson, W. An, A. Routh, F. Martino, L. Chapman, R. G. Roeder, D. Rhodes, 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J. Mol. Biol. 381, 816–825 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. Shogren-Knaak, H. Ishii, J. M. Sun, M. J. Pazin, J. R. Davie, C. L. Peterson, Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Q. Jin, L. R. Yu, L. Wang, Z. Zhang, L. H. Kasper, J. E. Lee, C. Wang, P. K. Brindle, S. Y. R. Dent, K. Ge, Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.N. Vo, R. H. Goodman, CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 276, 13505–13508 (2001). [DOI] [PubMed] [Google Scholar]
- 33.D. Helmlinger, L. Tora, Sharing the SAGA. Trends Biochem. Sci. 42, 850–861 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.G. Hunt, A. Boija, M. Mannervik, p300/CBP sustains Polycomb silencing by non-enzymatic functions. Mol. Cell 82, 3580–3597.e9 (2022). [DOI] [PubMed] [Google Scholar]
- 35.D. Pasini, M. Malatesta, H. R. Jung, J. Walfridsson, A. Willer, L. Olsson, J. Skotte, A. Wutz, B. Porse, O. N. Jensen, K. Helin, Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 38, 4958–4969 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.F. Tie, R. Banerjee, C. A. Stratton, J. Prasad-Sinha, V. Stepanik, A. Zlobin, M. O. Diaz, P. C. Scacheri, P. J. Harte, CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136, 3131–3141 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.H. M. Herz, M. Morgan, X. Gao, J. Jackson, R. Rickels, S. K. Swanson, L. Florens, M. P. Washburn, J. C. Eissenberg, A. Shilatifard, Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science 345, 1065–1070 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A. Piunti, E. R. Smith, M. A. J. Morgan, M. Ugarenko, N. Khaltyan, K. A. Helmin, C. A. Ryan, D. C. Murray, R. A. Rickels, B. D. Yilmaz, E. J. Rendleman, J. N. Savas, B. D. Singer, S. E. Bulun, A. Shilatifard, CATACOMB: An endogenous inducible gene that antagonizes H3K27 methylation activity of Polycomb repressive complex 2 via an H3K27M-like mechanism. Sci. Adv. 5, eaax2887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.P. Philip, A. Boija, R. Vaid, A. M. Churcher, D. J. Meyers, P. A. Cole, M. Mannervik, P. Stenberg, CBP binding outside of promoters and enhancers in Drosophila melanogaster. Epigenetics Chromatin 8, 48 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.E. M. Bowers, G. Yan, C. Mukherjee, A. Orry, L. Wang, M. A. Holbert, N. T. Crump, C. A. Hazzalin, G. Liszczak, H. Yuan, C. Larocca, S. A. Saldanha, R. Abagyan, Y. Sun, D. J. Meyers, R. Marmorstein, L. C. Mahadevan, R. M. Alani, P. A. Cole, Virtual ligand screening of the p300/CBP histone acetyltransferase: Identification of a selective small molecule inhibitor. Chem. Biol. 17, 471–482 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.L. M. Lasko, C. G. Jakob, R. P. Edalji, W. Qiu, D. Montgomery, E. L. Digiammarino, T. M. Hansen, R. M. Risi, R. Frey, V. Manaves, B. Shaw, M. Algire, P. Hessler, L. T. Lam, T. Uziel, E. Faivre, D. Ferguson, F. G. Buchanan, R. L. Martin, M. Torrent, G. G. Chiang, K. Karukurichi, J. W. Langston, B. T. Weinert, C. Choudhary, P. de Vries, A. F. Kluge, M. A. Patane, J. H. van Drie, C. Wang, D. McElligott, E. Kesicki, R. Marmorstein, C. Sun, P. A. Cole, S. H. Rosenberg, M. R. Michaelides, A. Lai, K. D. Bromberg, Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550, 128–132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.A. Hammitzsch, C. Tallant, O. Fedorov, A. O’Mahony, P. E. Brennan, D. A. Hay, F. O. Martinez, M. H. al-Mossawi, J. de Wit, M. Vecellio, C. Wells, P. Wordsworth, S. Müller, S. Knapp, P. Bowness, CBP30, a selective CBP/p300 bromodomain inhibitor, suppresses human Th17 responses. Proc. Natl. Acad. Sci. U.S.A. 112, 10768–10773 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.M. Leatham-Jensen, C. M. Uyehara, B. D. Strahl, A. G. Matera, R. J. Duronio, D. J. McKay, Lysine 27 of replication-independent histone H3.3 is required for Polycomb target gene silencing but not for gene activation. PLOS Genet. 15, e1007932 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D. J. McKay, S. Klusza, T. J. R. Penke, M. P. Meers, K. P. Curry, S. L. McDaniel, P. Y. Malek, S. W. Cooper, D. C. Tatomer, J. D. Lieb, B. D. Strahl, R. J. Duronio, A. G. Matera, Interrogating the function of metazoan histones using engineered gene clusters. Dev. Cell 32, 373–386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.A. R. Pengelly, Ö. Copur, H. Jäckle, A. Herzig, J. Müller, A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339, 698–699 (2013). [DOI] [PubMed] [Google Scholar]
- 46.W. Zhang, X. Zhang, Z. Xue, Y. Li, Q. Ma, X. Ren, J. Zhang, S. Yang, L. Yang, M. Wu, M. Ren, R. Xi, Z. Wu, J. L. Liu, E. Matunis, J. Dai, G. Gao, Probing the function of metazoan histones with a systematic library of H3 and H4 mutants. Dev. Cell 48, 406–419.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.T. Zhang, Z. Zhang, Q. Dong, J. Xiong, B. Zhu, Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells. Genome Biol. 21, 45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. Sankar, F. Mohammad, A. K. Sundaramurthy, H. Wang, M. Lerdrup, T. Tatar, K. Helin, Histone editing elucidates the functional roles of H3K27 methylation and acetylation in mammals. Nat. Genet. 54, 754–760 (2022). [DOI] [PubMed] [Google Scholar]
- 49.E. Hsu, N. R. Zemke, A. J. Berk, Promoter-specific changes in initiation, elongation, and homeostasis of histone H3 acetylation during CBP/p300 inhibition. eLife 10, e63512 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.T. Narita, S. Ito, Y. Higashijima, W. K. Chu, K. Neumann, J. Walter, S. Satpathy, T. Liebner, W. B. Hamilton, E. Maskey, G. Prus, M. Shibata, V. Iesmantavicius, J. M. Brickman, K. Anastassiadis, H. Koseki, C. Choudhary, Enhancers are activated by p300/CBP activity-dependent PIC assembly, RNAPII recruitment, and pause release. Mol. Cell 81, 2166–2182.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 51.S. J. Hogg, O. Motorna, L. A. Cluse, T. M. Johanson, H. D. Coughlan, R. Raviram, R. M. Myers, M. Costacurta, I. Todorovski, L. Pijpers, S. Bjelosevic, T. Williams, S. N. Huskins, C. J. Kearney, J. R. Devlin, Z. Fan, J. S. Jabbari, B. P. Martin, M. Fareh, M. J. Kelly, D. Dupéré-Richer, J. J. Sandow, B. Feran, D. Knight, T. Khong, A. Spencer, S. J. Harrison, G. Gregory, V. O. Wickramasinghe, A. I. Webb, P. C. Taberlay, K. D. Bromberg, A. Lai, A. T. Papenfuss, G. K. Smyth, R. S. Allan, J. D. Licht, D. A. Landau, O. Abdel-Wahab, J. Shortt, S. J. Vervoort, R. W. Johnstone, Targeting histone acetylation dynamics and oncogenic transcription by catalytic P300/CBP inhibition. Mol. Cell 81, 2183–2200.e13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.B. T. Weinert, T. Narita, S. Satpathy, B. Srinivasan, B. K. Hansen, C. Schölz, W. B. Hamilton, B. E. Zucconi, W. W. Wang, W. R. Liu, J. M. Brickman, E. A. Kesicki, A. Lai, K. D. Bromberg, P. A. Cole, C. Choudhary, Time-resolved analysis reveals rapid dynamics and broad scope of the CBP/p300 Acetylome. Cell 174, 231–244.e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.J. C. Lucchesi, W. G. Kelly, B. Panning, Chromatin remodeling in dosage compensation. Annu. Rev. Genet. 39, 615–651 (2005). [DOI] [PubMed] [Google Scholar]
- 54.M. Samata, A. Akhtar, Dosage compensation of the X chromosome: A complex epigenetic assignment involving chromatin regulators and long noncoding RNAs. Annu. Rev. Biochem. 87, 323–350 (2018). [DOI] [PubMed] [Google Scholar]
- 55.T. Straub, P. B. Becker, Dosage compensation: The beginning and end of generalization. Nat. Rev. Genet. 8, 47–57 (2007). [DOI] [PubMed] [Google Scholar]
- 56.T. Conrad, A. Akhtar, Dosage compensation in Drosophila melanogaster: Epigenetic fine-tuning of chromosome-wide transcription. Nat. Rev. Genet. 13, 123–134 (2012). [DOI] [PubMed] [Google Scholar]
- 57.A. S. Mukherjee, W. Beermann, Synthesis of ribonucleic acid by the X-chromosomes of Drosophila melanogaster and the problem of dosage compensation. Nature 207, 785–786 (1965). [DOI] [PubMed] [Google Scholar]
- 58.J. C. Lucchesi, M. I. Kuroda, Dosage compensation in Drosophila. Cold Spring Harb. Perspect. Biol. 7, 123–134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.A. Akhtar, P. B. Becker, Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5, 367–375 (2000). [DOI] [PubMed] [Google Scholar]
- 60.A. Hilfiker, D. Hilfiker-Kleiner, A. Pannuti, J. C. Lucchesi, mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 16, 2054–2060 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.E. R. Smith, A. Pannuti, W. Gu, A. Steurnagel, R. G. Cook, C. D. Allis, J. C. Lucchesi, The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20, 312–318 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ö. Copur, A. Gorchakov, K. Finkl, M. I. Kuroda, J. Müller, Sex-specific phenotypes of histone H4 point mutants establish dosage compensation as the critical function of H4K16 acetylation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 115, 13336–13341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.E. R. Smith, C. Cayrou, R. Huang, W. S. Lane, J. Côté, J. C. Lucchesi, A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol. Cell. Biol. 25, 9175–9188 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.M. Taipale, S. Rea, K. Richter, A. Vilar, P. Lichter, A. Imhof, A. Akhtar, hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol. Cell. Biol. 25, 6798–6810 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.A. Radzisheuskaya, P. V. Shliaha, V. V. Grinev, D. Shlyueva, H. Damhofer, R. Koche, V. Gorshkov, S. Kovalchuk, Y. Zhan, K. L. Rodriguez, A. L. Johnstone, M. C. Keogh, R. C. Hendrickson, O. N. Jensen, K. Helin, Complex-dependent histone acetyltransferase activity of KAT8 determines its role in transcription and cellular homeostasis. Mol. Cell 81, 1749–1765.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.G. C. Taylor, R. Eskeland, B. Hekimoglu-Balkan, M. M. Pradeepa, W. A. Bickmore, H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction. Genome Res. 23, 2053–2065 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.A. Shilatifard, The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81, 65–95 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.B. D. Strahl, R. Ohba, R. G. Cook, C. D. Allis, Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. U.S.A. 96, 14967–14972 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.E. Calo, J. Wysocka, Modification of enhancer chromatin: What, how, and why? Mol. Cell 49, 825–837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.M. P. Creyghton, A. W. Cheng, G. G. Welstead, T. Kooistra, B. W. Carey, E. J. Steine, J. Hanna, M. A. Lodato, G. M. Frampton, P. A. Sharp, L. A. Boyer, R. A. Young, R. Jaenisch, Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.M. G. Guenther, S. S. Levine, L. A. Boyer, R. Jaenisch, R. A. Young, A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.B. E. Bernstein, E. L. Humphrey, R. L. Erlich, R. Schneider, P. Bouman, J. S. Liu, T. Kouzarides, S. L. Schreiber, Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. U.S.A. 99, 8695–8700 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.H. Santos-Rosa, R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. T. Emre, S. L. Schreiber, J. Mellor, T. Kouzarides, Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411 (2002). [DOI] [PubMed] [Google Scholar]
- 74.B. K. Cenik, A. Shilatifard, COMPASS and SWI/SNF complexes in development and disease. Nat. Rev. Genet. 22, 38–58 (2021). [DOI] [PubMed] [Google Scholar]
- 75.T. Miller, N. J. Krogan, J. Dover, H. Erdjument-Bromage, P. Tempst, M. Johnston, J. F. Greenblatt, A. Shilatifard, COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. U.S.A. 98, 12902–12907 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.P. L. Nagy, J. Griesenbeck, R. D. Kornberg, M. L. Cleary, A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. U.S.A. 99, 90–94 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.A. Roguev, D. Schaft, A. Shevchenko, W. W. Pijnappel, M. Wilm, R. Aasland, A. F. Stewart, The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20, 7137–7148 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.N. J. Krogan, J. Dover, S. Khorrami, J. F. Greenblatt, J. Schneider, M. Johnston, A. Shilatifard, COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277, 10753–10755 (2002). [DOI] [PubMed] [Google Scholar]
- 79.F. S. Howe, H. Fischl, S. C. Murray, J. Mellor, Is H3K4me3 instructive for transcription activation? Bioessays 39, 1–12 (2017). [DOI] [PubMed] [Google Scholar]
- 80.S. D. Briggs, M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie, S. Y. R. Dent, F. Winston, C. D. Allis, Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15, 3286–3295 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.M. Bryk, S. D. Briggs, B. D. Strahl, M. J. Curcio, C. D. Allis, F. Winston, Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12, 165–170 (2002). [DOI] [PubMed] [Google Scholar]
- 82.C. Nislow, E. Ray, L. Pillus, SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell 8, 2421–2436 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.M. Hödl, K. Basler, Transcription in the absence of histone H3.2 and H3K4 methylation. Curr. Biol. 22, 2253–2257 (2012). [DOI] [PubMed] [Google Scholar]
- 84.K. Jain, C. S. Fraser, M. R. Marunde, M. M. Parker, C. Sagum, J. M. Burg, N. Hall, I. K. Popova, K. L. Rodriguez, A. Vaidya, K. Krajewski, M. C. Keogh, M. T. Bedford, B. D. Strahl, Characterization of the plant homeodomain (PHD) reader family for their histone tail interactions. Epigenetics Chromatin 13, 3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.E. M. Blackwood, J. T. Kadonaga, Going the distance: A current view of enhancer action. Science 281, 60–63 (1998). [DOI] [PubMed] [Google Scholar]
- 86.M. Bulger, M. Groudine, Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.N. D. Heintzman, G. C. Hon, R. D. Hawkins, P. Kheradpour, A. Stark, L. F. Harp, Z. Ye, L. K. Lee, R. K. Stuart, C. W. Ching, K. A. Ching, J. E. Antosiewicz-Bourget, H. Liu, X. Zhang, R. D. Green, V. V. Lobanenkov, R. Stewart, J. A. Thomson, G. E. Crawford, M. Kellis, B. Ren, Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.H. M. Herz, M. Mohan, A. S. Garruss, K. Liang, Y. H. Takahashi, K. Mickey, O. Voets, C. P. Verrijzer, A. Shilatifard, Enhancer-associated H3K4 monomethylation by trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 26, 2604–2620 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.D. Hu, X. Gao, M. A. Morgan, H. M. Herz, E. R. Smith, A. Shilatifard, The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell. Biol. 33, 4745–4754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.K. M. Dorighi, T. Swigut, T. Henriques, N. V. Bhanu, B. S. Scruggs, N. Nady, C. D. Still II, B. A. Garcia, K. Adelman, J. Wysocka, Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol. Cell 66, 568–576.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.K. Cao, C. K. Collings, M. A. Morgan, S. A. Marshall, E. J. Rendleman, P. A. Ozark, E. R. Smith, A. Shilatifard, An Mll4/COMPASS-Lsd1 epigenetic axis governs enhancer function and pluripotency transition in embryonic stem cells. Sci. Adv. 4, eaap8747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.H. M. Herz, Enhancer deregulation in cancer and other diseases. Bioessays 38, 1003–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.M. Gozdecka, E. Meduri, M. Mazan, K. Tzelepis, M. Dudek, A. J. Knights, M. Pardo, L. Yu, J. S. Choudhary, E. Metzakopian, V. Iyer, H. Yun, N. Park, I. Varela, R. Bautista, G. Collord, O. Dovey, D. A. Garyfallos, E. de Braekeleer, S. Kondo, J. Cooper, B. Göttgens, L. Bullinger, P. A. Northcott, D. Adams, G. S. Vassiliou, B. J. P. Huntly, UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat. Genet. 50, 883–894 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.K. B. Shpargel, T. Sengoku, S. Yokoyama, T. Magnuson, UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLOS Genet. 8, e1002964 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.J. Vandamme, G. Lettier, S. Sidoli, E. di Schiavi, O. Nørregaard Jensen, A. E. Salcini, The C. elegans H3K27 demethylase UTX-1 is essential for normal development, independent of its enzymatic activity. PLOS Genet. 8, e1002647 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.C. Wang, J. E. Lee, Y. W. Cho, Y. Xiao, Q. Jin, C. Liu, K. Ge, UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc. Natl. Acad. Sci. U.S.A. 109, 15324–15329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Y. Jang, C. Wang, L. Zhuang, C. Liu, K. Ge, H3K4 methyltransferase activity is required for MLL4 protein stability. J. Mol. Biol. 429, 2046–2054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.L. Wang, A. Shilatifard, UTX mutations in human cancer. Cancer Cell 35, 168–176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.L. Wang, Z. Zhao, P. A. Ozark, D. Fantini, S. A. Marshall, E. J. Rendleman, K. A. Cozzolino, N. Louis, X. He, M. A. Morgan, Y. H. Takahashi, C. K. Collings, E. R. Smith, P. Ntziachristos, J. N. Savas, L. Zou, R. Hashizume, J. J. Meeks, A. Shilatifard, Resetting the epigenetic balance of Polycomb and COMPASS function at enhancers for cancer therapy. Nat. Med. 24, 758–769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.T. Hoshii, P. Cifani, Z. Feng, C. H. Huang, R. Koche, C. W. Chen, C. D. Delaney, S. W. Lowe, A. Kentsis, S. A. Armstrong, A non-catalytic function of SETD1A regulates cyclin K and the DNA damage response. Cell 172, 1007–1021.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.C. C. Sze, K. Cao, C. K. Collings, S. A. Marshall, E. J. Rendleman, P. A. Ozark, F. X. Chen, M. A. Morgan, L. Wang, A. Shilatifard, Histone H3K4 methylation-dependent and -independent functions of Set1A/COMPASS in embryonic stem cell self-renewal and differentiation. Genes Dev. 31, 1732–1737 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.B. P. Mishra, K. M. Zaffuto, E. L. Artinger, T. Org, H. K. A. Mikkola, C. Cheng, M. Djabali, P. Ernst, The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell Rep. 7, 1239–1247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.R. Terranova, H. Agherbi, A. Boned, S. Meresse, M. Djabali, Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc. Natl. Acad. Sci. U.S.A. 103, 6629–6634 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.A. S. Bledau, K. Schmidt, K. Neumann, U. Hill, G. Ciotta, A. Gupta, D. C. Torres, J. Fu, A. Kranz, A. F. Stewart, K. Anastassiadis, The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development 141, 1022–1035 (2014). [DOI] [PubMed] [Google Scholar]
- 105.B. K. Cenik, C. C. Sze, C. A. Ryan, S. das, K. Cao, D. Douillet, E. J. Rendleman, D. Zha, N. H. Khan, E. Bartom, A. Shilatifard, A synthetic lethality screen reveals ING5 as a genetic dependency of catalytically dead Set1A/COMPASS in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 119, e2118385119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.J. L. Hess, B. D. Yu, B. Li, R. Hanson, S. J. Korsmeyer, Defects in yolk sac hematopoiesis in Mll-null embryos. Blood 90, 1799–1806 (1997). [PubMed] [Google Scholar]
- 107.H. Yagi, K. Deguchi, A. Aono, Y. Tani, T. Kishimoto, T. Komori, Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood 92, 108–117 (1998). [PubMed] [Google Scholar]
- 108.B. D. Yu, J. L. Hess, S. E. Horning, G. A. Brown, S. J. Korsmeyer, Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378, 505–508 (1995). [DOI] [PubMed] [Google Scholar]
- 109.D. Douillet, C. C. Sze, C. Ryan, A. Piunti, A. P. Shah, M. Ugarenko, S. A. Marshall, E. J. Rendleman, D. Zha, K. A. Helmin, Z. Zhao, K. Cao, M. A. Morgan, B. D. Singer, E. T. Bartom, E. R. Smith, A. Shilatifard, Uncoupling histone H3K4 trimethylation from developmental gene expression via an equilibrium of COMPASS, Polycomb and DNA methylation. Nat. Genet. 52, 615–625 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Z. Wang, A. G. Chivu, L. A. Choate, E. J. Rice, D. C. Miller, T. Chu, S. P. Chou, N. B. Kingsley, J. L. Petersen, C. J. Finno, R. R. Bellone, D. F. Antczak, J. T. Lis, C. G. Danko, Prediction of histone post-translational modification patterns based on nascent transcription data. Nat. Genet. 54, 295–305 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.N. M. Gaudelli, A. C. Komor, H. A. Rees, M. S. Packer, A. H. Badran, D. I. Bryson, D. R. Liu, Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]