Abstract
Proteinase 3 (PR3) is the main target antigen of antineutrophil cytoplasmic antibodies (ANCAs) in PR3-ANCA–associated vasculitis. A small fraction of PR3 is constitutively exposed on the surface of quiescent blood neutrophils in a proteolytically inactive form. When activated, neutrophils expose an induced form of membrane-bound PR3 (PR3mb) on their surface as well, which is enzymatically less active than unbound PR3 in solution due to its altered conformation. In this work, our objective was to understand the respective role of constitutive and induced PR3mb in the immune activation of neutrophils triggered by murine anti-PR3 mAbs and human PR3-ANCA. We quantified immune activation of neutrophils by the measurement of the production of superoxide anions and secreted protease activity in the cell supernatant before and after treatment of the cells by alpha-1 protease inhibitor that clears induced PR3mb from the cell surface. Incubation of TNFα-primed neutrophils with anti-PR3 antibodies resulted in a significant increase in superoxide anion production, membrane activation marker exposition, and secreted protease activity. When primed neutrophils were first treated with alpha-1 protease inhibitor, we observed a partial reduction in antibody-induced neutrophil activation, suggesting that constitutive PR3mb is sufficient to activate neutrophils. The pretreatment of primed neutrophils with purified antigen-binding fragments used as competitor significantly reduced cell activation by whole antibodies. This led us to the conclusion that PR3mb promoted immune activation of neutrophils. We propose that blocking and/or elimination of PR3mb offers a new therapeutic strategy to attenuate neutrophil activation in patients with PR3-ANCA–associated vasculitis.
Keywords: alpha-1 protease inhibitor, anti-PR3 mAbs, autoantibody, immune activation, inflammation, neutrophil, PR3-ANCA, proteinase 3, therapeutic strategy, vasculitis
Granulomatosis with polyangiitis (GPA) is one of the antineutrophil cytoplasmic antibodies (ANCAs)-associated systemic vasculitis syndromes characterized by inflammation of blood vessels (1). GPA is characterized by the presence of ANCA which are mainly directed against proteinase 3 (PR3, EC 3.4.21.76), stored in cytoplasmic granules of neutrophilic granulocytes and monocytes. This uncommon chronic inflammatory disease is characterized by necrotizing granulomatous inflammation and vasculitis of small vessels which can affect any organ. But the most common sites of involvement are the upper and lower respiratory tract and the kidneys. The most devastating disease manifestations, such as progressive glomerulonephritis and lung hemorrhage are caused by capillaritis resulting from the activation of innate immune responses driven by the interaction of pathogenic PR3-ANCA with membrane-bound PR3 (PR3mb) on primed neutrophils (2). Depending on which conformational epitope of PR3 is engaged by PR3-ANCA, they can be pathogenic and able to fully activate neutrophils or they can be nonpathogenic (2). Once primed neutrophils are fully activated by PR3-ANCA, the alternative complement pathway is activated, and the complement activation product C5a and its interaction with the C5a receptor (C5aR) on neutrophils enhances the inflammation (3). Despite recent advances in therapy, the relapsing nature of PR3-ANCA–associated vasculitis continues to expose these patients to substantial risk of cumulative tissue damage from active disease and drug toxicities (1, 4). Elucidation of the exact nature of the interaction of pathogenic PR3-ANCA with a specific epitope on PR3mb is fundamental for the design of novel therapeutic approaches.
PR3 displaying a chymotrypsin/trypsin fold is one of the four neutrophil serine proteases (NSPs: elastase, PR3, cathepsin G, and NSP-4) (5), which are finally maturated by cathepsin C (CatC) during neutrophil differentiation in the bone marrow (6). By contrast, to these other homologs, PR3 appears to be constitutively exposed on the surface of quiescent neutrophils in a variable genetically controlled manner (7, 8). Individuals carry two subsets of neutrophils with low (PR3mb(low)) or high (PR3mb(high)) amounts of “constitutive” form of PR3mb on their surfaces (9) without proteolytic activity (10). The induced form of PR3mb reveals enzymatic activity in the presence of high substrate and suicide inhibitor concentrations (11). In other words, the neutrophil membrane functions like an allosteric inhibitor, which affects the conformational state of the induced PR3mb and thereby impairs substrate or inhibitor binding. Alpha-1 protease inhibitor (α1PI), the major natural plasma inhibitor of PR3, also hardly interacts with induced PR3mb but removes the protease from the surface when used in excess (10, 11). Covalent complexation to α1PI changes the structure of PR3mb and its hydrophobic interaction with the membrane (12). The unique conformational transition of PR3 on the cell surface most probably makes it an easy target for autoantibodies in PR3-ANCA–associated vasculitis (10, 11). Consistent with this view are clinical findings in patients with PR3-ANCA–associated vasculitis, which show an increased expression of PR3mb on resting neutrophils during remissions (13, 14). PR3-ANCA activates primed neutrophils by binding to PR3mb, whereas their Fc portion interacts with FcγR (15). This results in an intensified neutrophil activation with the release of proteolytic enzymes including elastase, reactive oxygen species (ROS), and cytokines that significantly contribute to the vessel wall damage observed in patients (2).
A collection of mouse anti-PR3 mAbs were characterized and grouped into four major subsets that recognize different surface areas of PR3 (5, 16). The mAbs (CLB12.8, 6A6, PR3G-2) of group 1 bind to epitope 1 which is altered by a conformational change in the PR3–α1PI complex (17). The group 2 mAbs (MCPR3-1, MCPR3-2, 4A3) bind to a region located to N-terminal subdomain involving loops composed by the 60 loop and the beginning of the long 99 loop, designated as epitope 4 (17). MAbs of group 3 defining epitope 3 (4A5, WGM2, 1B10) recognize an antigenic region located on the backside of PR3 (16, 17, 18). The mAbs named MCPR3-7 and MCPR3-11 binding to epitope 5 are unique in that they much better bind to the PR3 zymogen than to mature PR3 (16, 19). A vast majority of PR3-ANCA share the binding specificity with the group 1 mAbs and do not bind to PR3 in a complex with α1PI (17). In addition, several studies reported that purified PR3-ANCA were able to interfere with the proteolytic activity of soluble PR3 (20, 21). The investigation of longitudinally collected samples from participants in a large randomized controlled trial provided evidence for PR3 activity-modulating PR3-ANCA (22). The inhibitory type of PR3-ANCA was most prevalent, but autoantibodies with PR3 activity-enhancing effects were also identified (22). Inhibitory PR3-ANCA partially blocks PR3 activity by an allosteric mechanism of inhibition (19, 22).
Autoimmune activation of neutrophils can be reproduced using monoclonal antibodies directed to PR3mb. TNF-α–primed purified blood neutrophils are activated by mouse monoclonal anti-PR3 antibodies and autoantibodies from PR3-ANCA–associated vasculitis patients (23, 24). Mouse monoclonal antiPR3 antibodies and autoantibodies from patients recognize conformational epitopes on PR3 that may vary during the course of the disease. Notably, the intensity of PR3-ANCA–induced oxidative burst is correlated with the percentage of neutrophil subsets expressing PR3mb (25). Epitope of PR3 recognized by PR3-ANCA, isotype, and glycosylation of PR3-ANCA are additional factors influencing autoantibody pathogenicity (2). Different therapeutic strategies targeting PR3mb could be considered to block autoimmune activation of neutrophils by PR3-ANCA (26). Strong reduction of PR3mb has been shown using a CatC inhibitor in a CD34+ hematopoietic stem cell model (27). Recently, the first nonactivating human IgG1κ anti-PR3 monoclonal antibody named 4C3 was identified using immortalizing B cells from a patient with PR3-ANCA–associated vasculitis (28).
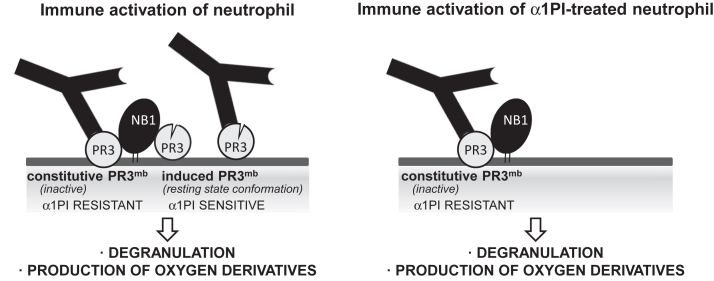
The observation that genetic α1PI deficiency (Z-variant) is a risk factor for PR3-ANCA–associated vasculitis argues for a potential role of induced proteolytically active PR3mb in neutrophil activation (29). However, this does not exclude that inactive constitutive PR3mb is indeed the unique target of autoantibodies which triggers the immunopathologic process. Indeed, higher levels of constitutive PR3mb are generally observed on purified quiescent neutrophils from PR3-ANCA–associated vasculitis patients (14) and are positively correlated with ANCA-induced neutrophil activation and clinical relapses of disease (13). Whether constitutive or induced PR3mb or both forms are the preferential targets of PR3-ANCA in the course of the disease remains unclear. Therefore, we conducted the present study to better understand the molecular details of PR3-ANCA–mediated effects on neutrophils with the goal to inform the design of new specific targeted therapies for the disease. Induced active PR3mb exposed at the surface of calcium ionophore A23187-activated neutrophils is inhibited and cleared by α1PI (10). But the exposure of induced PR3mb at the neutrophil surface depends on the nature of the stimulus. Consequently, here, we first studied the exposure and amount of PR3mb on quiescent and activated neutrophil surfaces using a variety of activators and characterized constitutive and induced PR3mb before and after incubation with α1PI. We then investigated the respective role of constitutive and induced PR3mb pools for neutrophil activation by murine anti-PR3 antibodies and PR3-ANCA from patients (Fig. 1).
Figure 1.
Exploration of α1PI for investigation of the respective role of constitutive and inducible PR3mbin neutrophils activation by anti-PR3 antibodies. The diagram illustrates two distinct pool of PR3mb colocalized with NB1 (CD177) (34) on activated neutrophils: one catalytically inactive, which is α1PI-resistant (constitutive PR3mb), and the other in an activatable conformation that is α1PI-sensitive (induced PR3mb) (10, 11). The property of α1PI to discriminate between constitutive and induced PR3mb was explored to assess their relative role in immune activation of neutrophils by anti-PR3 mAbs and PR3-ANCA. α1PI, alpha-1 protease inhibitor; ANCA, antineutrophil cytoplasmic antibody; PR3, proteinase 3.
Results
Load in PR3mb on activated healthy and patient neutrophils after incubation with α1PI: assessment of constitutive PR3mb(α1PI resistant) and induced PR3mb(α1PI sensitive)
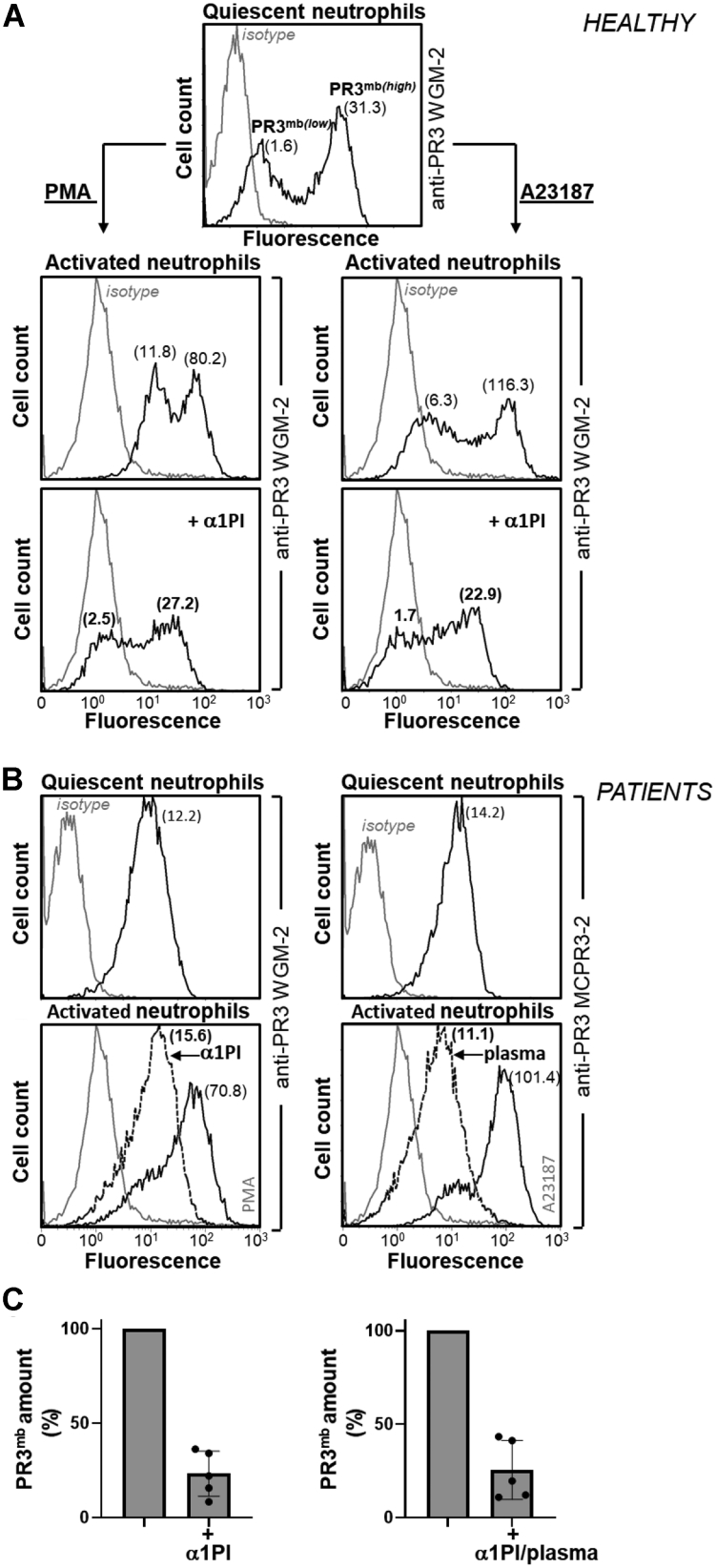
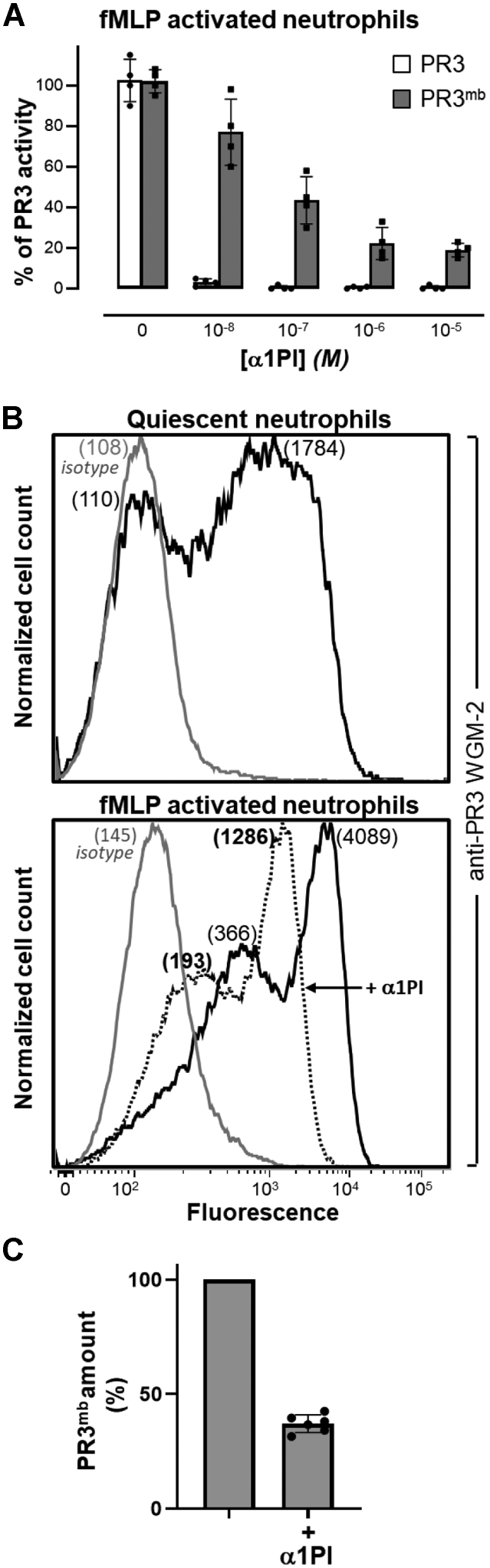
We first looked at whether two strong neutrophil activators, phorbol myristate acetate (PMA) PKC agonist and formyl-methionyl-leucyl-phenylalanine (fMLP) FPR agonist also induced the surface expression of α1PI-sensitive PR3mb compared to the calcium ionophore A23187. The load of PR3mb on activated cells before and after treatment with purified human α1PI (10 and 50 μM) was quantified by flow cytometry. We used for this purpose the anti-PR3 WGM-2 mAb that recognizes epitope 3 both on the free and the α1PI-bound protease. Results show that resting quiescent neutrophils expose a low level and a high level of PR3 at the cell surface, and PMA, A23187, and fMLP dramatically increased these PR3 expositions (Figs. 2, 3, S1 and S2). Interestingly, whatever activator used, we observed a large decrease in the PR3mb load at the membrane surface after α1PI treatment. We observed that the percentage of PR3mb cleared by α1PI increased proportionally with the total amount of PR3 exposed at the cell surface. This means that most of the PR3mb exposed at the cell surface in response to a chemical or a biological stimulus is a proteolytically active protease, whereas the part which is not removed by α1PI remains almost constant. Similar results were obtained using purified blood neutrophils from patients after incubation with human α1PI or human plasma. Results however depended on the variable, sample-dependent level of constitutive PR3mb exposed on the neutrophil surface. Based on the mean of the results, we found that ∼30% of PR3mb exposed at the neutrophils membrane upon stimulation with a chemical or physiologic activator are in an inactive α1PI-resistant conformation (PMA-activated cells: 23% ± 12 (n = 5); A23187-activated cells: 25.5% ± 15.5 (n = 5); fMLP-activated cells: 37.1% ± 4 (n = 5)), whereas ∼70% are in an activable, α1PI-sensitive, conformation (Figs. 2C and 3C).
Figure 2.
Surface exposition of PR3 on activated healthy and patient neutrophils before and after treatment with α1PI.A, PR3mb on purified quiescent blood neutrophils from a healthy volunteer was stained using murine anti-PR3 WGM-2 (25 μg/ml). Flow cytometry analysis showed two populations of neutrophils with low and high amounts of constitutive PR3mb. Quiescent cells were then stimulated using PMA (100 ng/μl) or calcium ionophore A23187 (1 μM) for 15 min at 37 °C and the load of PR3mb before and after treatment with α1PI (50 μM) was quantified by flow cytometry using anti-PR3 WGM-2 (25 μg/ml). Flow cytometry studies showed that after neutrophil stimulation, PR3mb load was increased and as expected, the proportion of low and high PR3-exposing cells remained stable (14, 27). The displacement of fluorescence of the histogram plots to the left shows that induced but not constitutive PR3mb was cleared from the surface of triggered neutrophils by α1PI. Two populations of neutrophils with low and high amounts of constitutive PR3mb remained unchanged. Similar results were obtained in n ≥ 10 independents experiments using 10 or 50 μM of α1PI. B, purified blood neutrophils from two patients were stimulated using PMA or calcium ionophore A23187. The load of PR3mb before and after treatment with α1PI (50 μM) or plasma (dilution 1/3) was quantified by flow cytometry using murine anti-PR3 WGM-2 (25 μg/ml, epitope 3) or MCPR3-2 (40 μg/ml, epitope 4). A monomodal exposition of PR3mb on GPA patient neutrophils was observed as documented in (13, 14, 33). The gray lines represent nonspecific binding of mouse isotype-specific IgG control antibody used at the same concentration. C, PR3mb amount on PMA- or A23187-activated neutrophils after α1PI treatment by flow cytometry (PMA-activated cells, n = 5 independent experiments; A23187-activated cells, n = 5 independent experiments; See Fig. S1). Individuals results and the means ± SD are given. The results showed that the induced PR3mb was removed from the cell surface after α1PI treatment of activated healthy and patient neutrophils. α1PI, alpha-1 protease inhibitor; GPA, granulomatosis with polyangiitis; PMA, phorbol myristate acetate; PR3, proteinase 3.
Figure 3.
Inhibition and surface exposition of PR3 on fMLP-activated neutrophils treated with α1PI.A, residual activity of soluble PR3 (gray bars) and PR3mb (black bars) after incubation with increasing amounts of α1PI (0.01–10 μM). Neutrophils were primed with CB (10 μg/ml) for 10 min and stimulated with chemotactic N-formyl-methionyl-leucyl-phenylalanine (fMLP, 1 μM) for 20 min at 37 °C and the activity of PR3mb was measured using FRET substrate ABZ-VAD(nor)VADYQ-EDDnp (20 μM) (43, 57) after incubation with increasing amounts of α1PI in comparison with that of soluble purified PR3. The starting rates of hydrolysis were adjusted on that of a 1 nM PR3 solution. Individuals results and the means ± SD are given (n = 4 independents experiments). B, representative flow cytometry analysis of fMLP-activated neutrophils as analyzed using anti-PR3 WGM-2 (25 μg/ml) showing a partial decrease of the load in PR3mb after incubation with α1PI (50 μM). The proportion of PR3mb over the total neutrophil population remained unchanged. The gray line represents nonspecific binding of mouse isotype-specific IgG control antibody used at the same concentration. C, PR3mb amount on fMLP-activated neutrophils after α1PI treatment by flow cytometry (n = 5 independents experiments, See Fig. S2). Individuals results and the means ± SD are given. α1PI, alpha-1 protease inhibitor; fMLP, formyl-methionyl-leucyl-phenylalanine; PR3, proteinase 3.
Inhibition of α1PI-sensitive–induced PR3mb by purified inhibitory PR3-ANCA
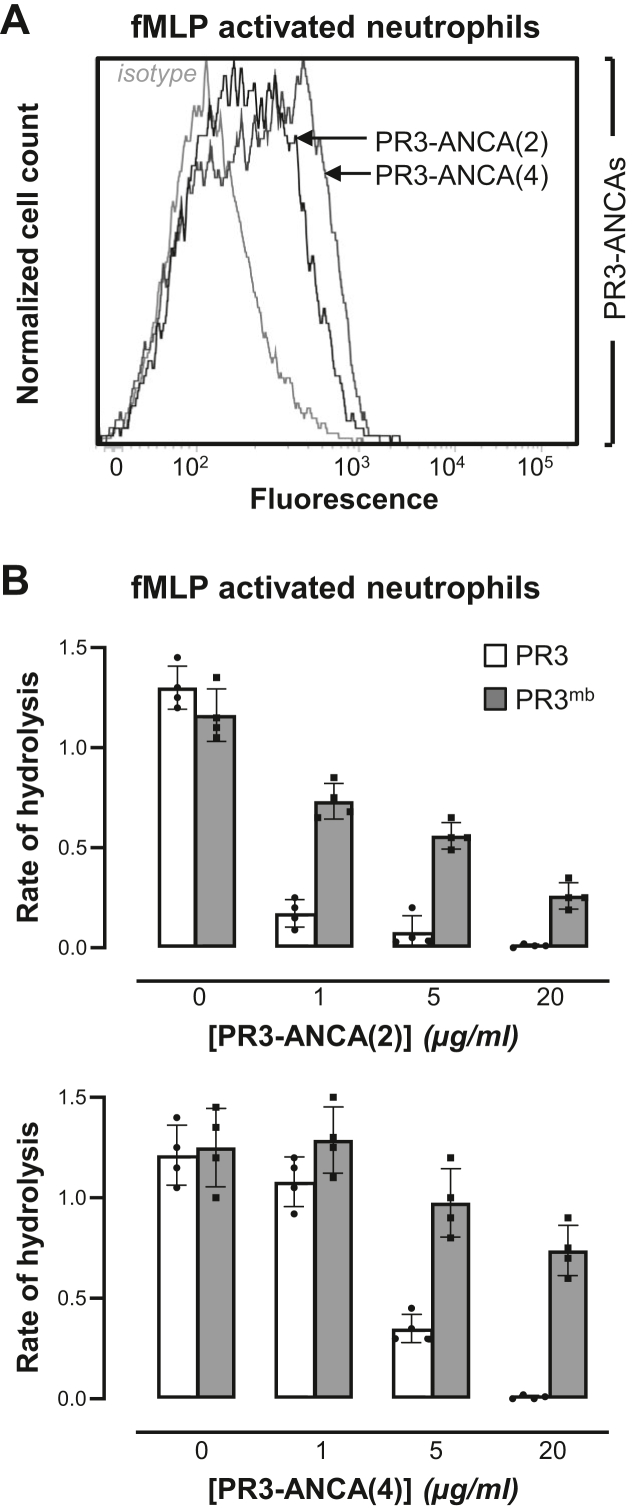
The majority of PR3-ANCA from patients display inhibitory properties towards soluble purified PR3 (22). Inhibition is substrate-dependent and occurs through an allosteric mechanism with an antibody-binding site located on the active site surface of PR3. We therefore wondered whether PR3-ANCA interacted with PR3mb and were able to inhibit its activable form (22). PR3-ANCA were purified from patients’ plasma removed by therapeutic plasma exchange using immobilized human PR3. After purification steps, the interaction of purified PR3-ANCA with PR3 was checked by ELISA. Flow cytometry studies showed that PR3-ANCA recognized both constitutive and induced PR3mb on quiescent and fMLP-activated purified neutrophils (Figs. 4A and S3). Furthermore, purified PR3-ANCA were able to induce autoimmune activation of neutrophils (not shown). After the functional assays, we compared the inhibition of soluble PR3 and PR3mb by five purified PR3-ANCA from patients. We used for this purpose a low concentration of the PR3 substrate ABZ Tyr-Tyr-Abu-Asn-Glu-Pro-Tyr(3-NO2)-NH2 (7 μM final) to avoid competition with the antibody. Four of the five PR3-ANCA were able to inhibit the activity of soluble PR3 almost completely. However, inhibition of PR3mb by these purified PR3-ANCA never exceeded 80% of the total PR3mb activity in the same conditions (Fig. 4B). Thus, induced PR3mb differs from soluble PR3 in terms of sensitivity to PR3-ANCA and interaction/inhibition.
Figure 4.
Inhibition of induced PR3mbby purified PR3-ANCA from patients.A, representative flow cytometry pattern of PR3mb on fMLP-activated neutrophils using purified PR3-ANCA. Neutrophils were labeled with purified PR3-ANCA (5 μg/ml) from two patients (sample (2, 4)) and revealed by FITC-conjugated antihuman IgG to visualize cell surface PR3 (black lines). The gray line represents nonspecific binding of human isotype-specific control IgG (5 μg/ml). B, residual activity of soluble (gray bars) and PR3mb (black bars) in the presence of increased concentrations of purified PR3-ANCA (1–20 μg/ml). The activities of soluble PR3 and PR3mb on the surface of fMLP-stimulated neutrophils were measured using the FRET substrate ABZ Tyr-Tyr-Abu-Asn-Glu-Pro-Tyr(3-NO2)-NH2 (58) (7 μM final) after a 30 min incubation time at 37 °C. The starting rates of hydrolysis were adjusted on that of a 1 nM PR3 solution. The rates of hydrolysis are expressed as the relative fluorescence unit per second (FU/s). Individuals results and the means ± SD are given (n = 4 independents experiments). Similar results were obtained using three other purified PR3-ANCA from patients (samples [(1), (3), (5)]). ANCA, antineutrophil cytoplasmic antibody; fMLP, formyl-methionyl-leucyl-phenylalanine; PR3, proteinase 3.
Immune activation of neutrophils in response to mouse anti-PR3 mAbs or human PR3-ANCA before and after incubation with α1PI
Superoxide anions production
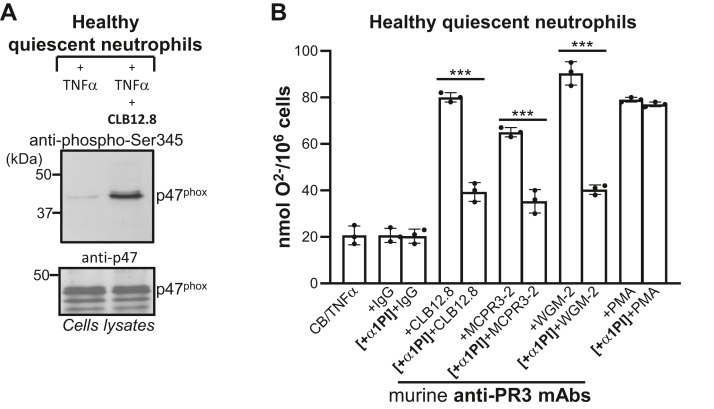
The NADPH oxidase is composed of two membrane proteins (gp91phox/NOX2 and p22phox) and four cytosolic proteins (p47phox, p67phox, p40phox, and Rac1/2). Superoxide anion (O2-) is generated by the NADPH oxidase complex after its cytosolic components have been phosphorylated and assembled at the cell membrane. We first examined whether anti-PR3 mAbs were able to induce p47phox phosphorylation. Neutrophils primed with TNF-α were incubated for 90 min with anti-PR3 mAbs and cell lysates were analyzed by Western blotting using an anti-phospho-Ser345 that specifically recognizes the phosphorylated sequence on p47phox (30). A band of 45 kDa corresponding to phosphorylated p47phox was detected (Fig. 5A). We ensured that this incubation did not affect the intracellular level of p47phox by Western blot using an anti-p47phox antibody. Next, we quantified O2- production before and after pretreatment of primed neutrophils with α1PI. Cells were first incubated with 50 μM α1PI, then stimulated with anti-PR3 mAbs antibodies that recognize epitope 1 (CLB12.8), epitope 4 (MCPR3-2), or epitope 3 (WGM-2). Cytochrome c reduction increased considerably when cells were stimulated with anti-PR3 mAbs compared to primed cells. The superoxide anion production by α1PI-treated cells was reduced by only 30 to 50% (CLB12.8: 40.7% ± 2.3; MCPR3-2: 29.7% ± 4.3; WGM-2: 50% ± 2.5) regardless of the antibody used, which means that cell activation occurs even when active PR3mb has been cleared from the cell surface (Fig. 5B). However, this decrease depended on the cell preparation since the total amount of PR3mb at the cell surface and the proportion between constitutive and inducible PR3mb may differ significantly.
Figure 5.
Effects of α1PI on superoxide anion production by anti-PR3–stimulated neutrophils.A, anti-PR3 CLB12.8–induced phosphorylation of p47phox from freshly prepared neutrophils from healthy volunteers pretreated by cytochalasin B (CB) and then primed by TNF-α. Phosphorylated p47phox was revealed using an anti-phospho-Ser345 antibody (p-Ser345). The intracellular level of p47phox was checked using an anti p47phox Ab. B, production of superoxide anion as quantified by the measurement at 550 nm of the SOD-inhibitable reduction of cytochrome c. Neutrophils were treated as in (A) and then incubated or not with α1PI (10 μM) before adding the anti-PR3 CLB12.8 antibody (40 μg/ml), an IgG mouse isotype (negative control, 40 μg/ml), or PMA (positive control). The same experiment was repeated using the MCPR3-2 antibody (40 μg/ml) and the WGM-2 antibody (40 μg/ml). The 30 to 40% decrease in superoxide anions generated by α1PI-treated cells indicates that activation occurs even when induced PR3 has been cleared from the membrane. The histogram shows the mean ± SD of three independent experiments, each done in duplicates. Statistical analysis of the data was performed using two-way ANOVA (Bonferroni’s multiple comparison tests), ∗∗∗p < 0.001. α1PI, alpha-1 protease inhibitor; PMA, phorbol myristate acetate; PR3, proteinase 3.
Neutrophil degranulation
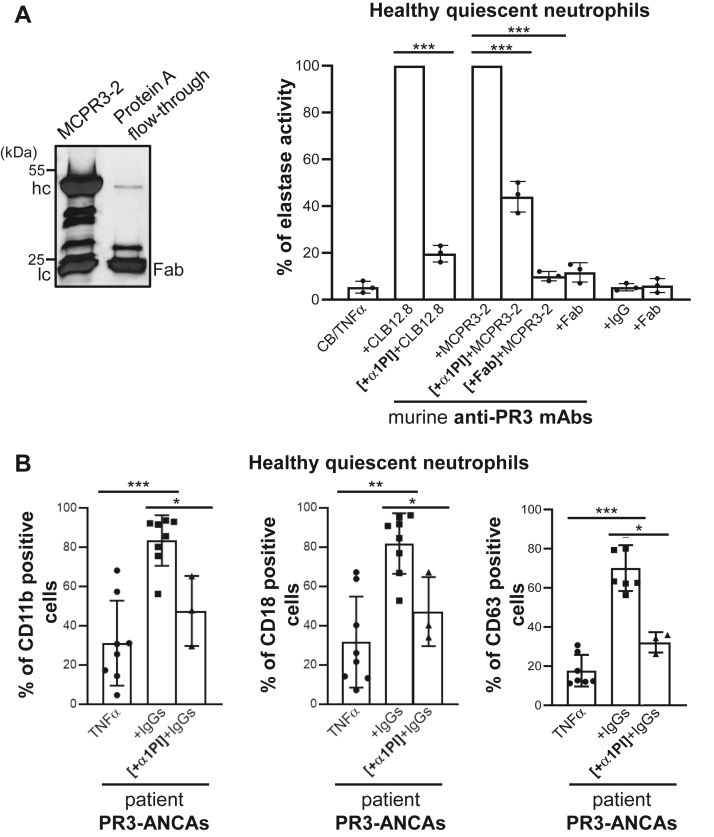
Degranulation of activated neutrophils is reflected in the extracellular release of elastase-like proteases. We quantified the neutrophil degranulation induced by anti-PR3 antibodies by measuring neutrophil elastase activity, an accepted marker of neutrophil activation (31), kinetically in cell-free supernatants after the cells were incubated with antibodies recognizing three different epitopes. We measured a significant increase in elastase activity in the supernatants of neutrophils stimulated by anti-PR3 CLB12.8 (epitope 1), MCPR3-2 (epitope 4) (Fig. 6A), and to a lesser extent by anti-PR3 WGM-2 (epitope 3) (not shown). This probably depends on a less favorable orientation of epitope 3 on PR3mb though this had no influence on the secretion of superoxide anions. When primed cells were first treated with an excess of α1PI, the incubation with anti-PR3 MCPR3-2 resulted only in a partial reduction of the elastase activity (56% ± 4.6 less elastase activity) (Fig. 6A). Anti-PR3 mAbs trigger the immune response of neutrophils by interacting with PR3mb via their Fab fragment and with the Fcγ receptors via their Fc fragment. We used here Fab fragments of MCPR3-2 mAbs as competitors of whole mAbs to tentatively prevent a neutrophil immune response (ratio Fab fragments/MCPR3-2 = 3). Fabs were incubated with primed neutrophils before the cells were treated with MCPR3-2 mAbs and the activity of elastase measured in supernatants as reported above. Incubation of primed neutrophils with purified MCPR3-2 Fab fragments induced no increase of the elastase activity. Following incubation with MCPR3-2, Fab pretreated cells released 90% ± 1.4 less elastase activity in the cell supernatants compared to controls (Fig. 6A). The treatment of cells with CLB12.8 Fab fragments before the incubation with CLB12.8 (ratio Fab fragments/CLB12.8 = 3) resulted also in significant reduction of the elastase release in the supernatant (80.3% ± 2.5 less elastase activity). The upregulation of adhesion molecules on α1PI-treated cells was also partially reduced (35%-40%) using PR3-ANCA (Fig. 6B). Taken together, these results indicate that both constitutive and induced PR3mb participated in the immune activation of neutrophils.
Figure 6.
Effects of α1PI and purified Fab fragments on degranulation by anti-PR3–stimulated neutrophils.A, effects of α1PI and purified Fab fragments on secretion of elastase activity from neutrophils stimulated by anti-PR3 antibodies. (Left) SDS-PAGE (reducing conditions) of Fab fragments prepared from anti-PR3 MCPR3-2 mAbs after digestion with papain. Contaminating intact immunoglobulins and intact Fc parts were removed by the protein A affinity column. IgG heavy and light chains migrate respectively with an apparent molecular weight near 50 and 25 kDa, while Fab fragments display as a single band of 25 kDa. (Right) Purified neutrophils from healthy volunteers were primed, then incubated with anti-PR3 CLB12.8 (40 μg/ml), anti-PR3 WGM-2 (40 μg/ml), or anti-PR3 MCPR3-2 (40 μg/ml) as reported in Figure 5. The residual proteolytic activity of elastase in cell-free neutrophil supernatants was measured as an indicator of cell degranulation induced by anti-PR3 mAbs. Preincubation of the primed-cells with α1PI (50 μM) caused only a partial (50–60%) reduction in anti-PR3 mAbs-induced degranulation. Purified Fab fragments of anti-PR3 CLB12.8 or anti-PR3 MCPR3-2 mAbs were incubated with primed-neutrophils before the cells were treated with the entire antibody in a molar ratio of 4 and the cell degranulation measured as reported above. Results show that Fab fragments compete with the whole antibody for binding to both constitutive and inducible PR3mb, which results in a significant decrease in cell activation. The histogram shows the mean ± SD of three independent experiments, each done in duplicates. Statistical analysis of the data was performed using two-way ANOVA (Bonferroni’s multiple comparison tests), ∗∗∗p < 0.001. B, surface markers were analyzed using TNFα-primed neutrophils stimulated with IgGs from patients (200 μg/ml) before (eight independent experiments for CD11b and CD11b; seven independent experiments for CD63) and after incubation with α1PI (three independent experiments) as in A. After stimulation, cells were washed and stained with CD63-FITC or double stained with CD11b VioBlue/CD18 FITC before flow cytometry analysis. Results show that preincubation of the primed cells with α1PI caused only a partial reduction in anti-PR3–induced degranulation. Histogram shows the mean ± SD of independent experiments. Statistical analysis of the data was performed using a nonparametric test (Mann-Whitney test), ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. α1PI, alpha-1 protease inhibitor; PR3, proteinase 3.
Discussion
In this study, we show (i) that both “constitutive” and “induced” PR3mb, which exhibit conformational differences from soluble active PR3, are recognized by murine anti-PR3 mAbs (epitope 1, epitope 3, or epitope 4) and pathogenic human PR3-ANCAs from patients resulting in immune activation of neutrophils, (ii) that the temporary increase of PR3mb exposure is required for full immune activation of neutrophils by anti-PR3 mAbs and PR3-ANCA, and (iii) that Fab fragments compete with PR3-ANCA without inducing neutrophil activation. Like no other previous study, our study describes the details of important biochemical and molecular mechanisms of PR3-ANCA–induced neutrophil activation resulting in important novel insights necessary for the rational design of novel treatment strategies for PR3-ANCA–associated vasculitis.
The vast majority of PR3 is expressed by neutrophils and to a much lesser degree by certain populations of monocytes (32). PR3 is constitutively expressed to a variable, genetically determined extent on the surface of quiescent isolated blood neutrophils (8). There is a high interindividual variability in the percentage of PR3mb-exposing neutrophils ranging from 0 to 100% of total neutrophils. Bimodal exposition of PR3mb was detected in ∼30% of healthy individuals and GPA patients. Approximately, 70% of healthy controls and GPA patients have one uniform population of neutrophils with an interindividual variable level of PR3mb exposition. The constitutive PR3mb exposed on freshly isolated neutrophils is not eliminated from the cell surface in the presence of high concentrations of serpins such as α1PI (10) and is not labeled using selective PR3 activity–based probe (11). PR3-FRET substrates are not significantly cleaved by purified quiescent blood neutrophils. Furthermore, CD34+ hematopoietic stem cells from the human umbilical cord, differentiated into functional neutrophils in vitro in a medium containing fetal bovine serum rich in α1PI, exposed PR3 on their cell surface progressively (33). All data suggest that constitutive PR3mb is proteolytically inactive but the events leading to an inactive PR3 fraction on the quiescent neutrophil surface are still unknown.
PR3 is also expressed as a proteolytically latent, inducible, form at the cell surface of primed and activated neutrophils (11). CD177 (also known as NB1 receptor) functions as a high-affinity PR3 receptor on neutrophil (34). CD177-transfected Chinese hamster ovary (CHO) cells expressing the entire human CD177 showed positive staining after human PR3 incubations. Elafin (6 kDa), a canonical low molecular weight inhibitor, abolished the activity of PR3 on CD177-positive CHO cells, but in contrast to α1PI, this interaction did not result in a dissociation of the hPR3–CD177 complex on CHO membranes (12). In a recent work, Ebert et al. showed using surface plasmon resonance measurement that PR3 preincubation with native α1PI, but not inactive mutated α1PI, prevented PR3 binding to immobilized CD177 (35). Moreover, the authors performed a PR3mb shedding assay after incubation of surface-biotinylated neutrophils with wt α1PI and inactive mutated α1PI. Significantly, more biotinylated α1PI was found in the supernatants from wt α1PI–treated neutrophils, indicating that PR3 was indeed released form the neutrophil surface (35). The weak activity of induced PR3mb and its hard inhibition with α1PI might be due to its altered conformation on the cell surface. An excess of substrate or inhibitor would convert the latent conformation to more active form (11). The monoclonal Ab MCPR3-7 inhibits PR3 activity by an allosteric mechanism and competes with the neutrophil membrane for the binding of PR3 (19). Interaction of the hydrophobic patch of PR3mb with the neutrophil surface might well employ the same mechanism to affect the substrate and inhibitor binding subsites. The allosteric inhibition of the induced PR3mb by the membrane means that it shows activity at high substrate concentrations and is fully inhibited at high levels of activity-dependent suicide inhibitors (11). Furthermore, we observed that serpins such as α1PI (10) or serpin B1(S/DAR) (36) at high concentration inhibited and almost completely removed induced PR3mb from the membrane of calcium ionophore A23187-activated neutrophils. The solubilization of PR3mb might be due to its hydrophobic patch being distorted in the complex with α1PI (12). These results were in good agreement with Rooney et al. who reported that PR3mb detection on the surface was reduced by α1PI (1 mg/ml) when it was added to TNFα-primed neutrophils before the addition of the anti-PR3 mAb CLB12.8 antibody (29). Since the mobilization of intracellular granules and the subsequent exposition of induced PR3mb at the neutrophil surface depends on the nature of the stimulus, in this current work, we investigated the load in PR3mb on activated neutrophils using a variety of activator including calcium ionophore A23187, PMA, fMLP before and after treatment with α1PI at plasma concentrations. The levels used in our experiments do not modulate immune cell functions instantaneously and differentially from the endogenous α1PI in vivo. Our results revealed that ∼30% of PR3mb on the neutrophil membrane upon stimulation with a chemical or physiologic activator are in an inactive α1PI-resistant conformation, whereas ∼70% are in an activatable, α1PI-sensitive, conformation. This is true for purified blood neutrophils from both healthy donors and patients. In the current study, we found that murine anti-PR3 mAbs binding to three different epitopes recognized both mature PR3 and its zymogen, which is consistent with our hypothesis that constitutive PR3mb could be the zymogen form of PR3 (11, 27). Furthermore, PR3 zymogens do not form irreversible complexes with α1PI unlike neutrophil elastase (Korkmaz and Jenne, unpublished results), which can explain the α1PI-resistant trait of constitutive PR3mb.
A large collection of longitudinally collected samples from the Wegener Granulomatosis Etanercept Trial allowed us to determine the functional effects of PR3-ANCA in comparison to clinical disease manifestations (22). PR3-ANCA may become undetectable after patients go into remission, and subsequently rising titers are associated with an increased risk of relapse (37, 38, 39). However, contradictory observations have been reported about the significance of inhibitory PR3-ANCA and their impact on disease activity, clinical course, and prognosis (20, 21, 40, 41). In patient samples from the Wegener Granulomatosis Etanercept Trial study, PR3-ANCA with inhibitory capacity but also PR3-ANCA with enhancing effects on soluble PR3 activity were noticed (22). Epitope mapping of the activity-modulating PR3-ANCA revealed a binding on the active site surface of soluble PR3 and an allosteric mechanism of inhibition. Since a particular subset of PR3-ANCA interferes with the enzymatic activity of soluble PR3 and impairs its inhibition by α1PI (22), one may wonder how PR3-ANCA interact with constitutive and induced PR3mb at the cellular surface. We first showed the ability of inhibitory PR3-ANCA to bind PR3mb using flow cytometry. A significant decrease in the mean fluorescence signal was observed when neutrophils were preincubated with α1PI, suggesting the interaction of PR3-ANCA with constitutive and induced PR3mb. PR3-ANCA were also able to induce immune activation of neutrophils. We then investigated whether activity modulating purified PR3-ANCA from patient samples could interfere with induced PR3mb activity. Unlike soluble PR3, which almost completely inhibited, induced PR3mb activity was only partially inhibited by inhibitory PR3-ANCA. These results were consistent with the resistance of induced PR3mb to inhibition by α1PI (10, 11) and confirmed that PR3mb differs in conformation from its active soluble form.
We used the property of α1PI to clear induced PR3mb from the membrane surface (10) and were thereby able to discriminate between constitutive and inducible PR3mb. We investigated whether immune activation of neutrophil was induced by only one or both forms of PR3mb. This approach was made possible thanks to the design and use of highly selective and sensitive FRET substrates of NSPs (42, 43). Primed neutrophils in a suspension medium were triggered by murine anti-mouse PR3 mAbs or human anti-PR3 ANCA to release superoxide ions and elastase in the supernatant even in the presence of a large excess of α1PI with near-total inhibition of PR3mb activity. This means that constitutive PR3mb alone can interact with antibodies and contributes to cell activation. A large drop in superoxide ion production and elastase activity was, however, observed after α1PI treatment, indicating that inducible PR3mb also participated with 30 to 60% in cell activation. Clearing PR3mb from the cell surface, however, requires a large excess of inhibitor because of the allosteric inhibitory effect of the membrane. Poor inhibition and clearance of induced PR3mb by its main plasma inhibitor could favor immune activation of neutrophils by PR3-ANCA, competing with circulating α1PI in spite of its important plasma concentration. Since both inducible and constitutive PR3mb appear to be the target of PR3-ANCA during disease, a serpin would be of limited value in controlling immune activation of neutrophils and subsequent inflammation. The results of the current study are in agreement with those of Schreiber et al. who explored the consequences of PR3mb(high) and PR3mb(low) neutrophil phenotypes in autoimmune activation. PR3-ANCA–induced generation of ROS and release of granule proteins were significantly higher in the PR3mb(high) neutrophil subset than in the PR3mb(low) subset (25). There are auxiliary mechanisms in neutrophil activation by PR3-ANCA and anti-PR3 mAbs, which most likely involves Fc-receptor interactions (2). Mechanistically, anti-PR3 mAbs binding to PR3mb, however, initiate immune activation of neutrophils and is widely regarded as the primary interaction with neutrophils (Fig. 7). Furthermore, therapeutic elimination of PR3 together with PR3mb by inhibiting CatC, an enzyme involved in its maturation, reduced immune activation of neutrophils in response to PR3-ANCA and endothelial cell injury (32). The pharmacological inhibition of CatC using a cell-permeable inhibitor reduced PR3mb by ∼85% (32). Consequently, superoxide release diminished approximately 50% (32). There is little doubt that the degree of neutrophil PR3mb exposure has functional consequences on immune activation of neutrophil.
Figure 7.
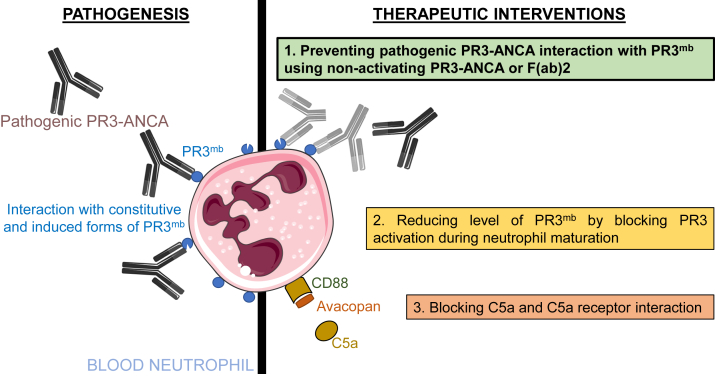
Targeting neutrophil activation in PR3-ANCA vasculitis. Our study focusing on the neutrophil-activating effects of pathogenic PR3-ANCA shows that both enzymatically inactive constitutive PR3mb as well as enzymatically active PR3mb contribute to the activation of neutrophils when interacting with anti-PR3 mAbs and pathogenic PR3-ANCA; this can be prevented completely by targeting both constitutive and induced PR3mb using nonactivating PR3-ANCA or F(ab)2, but not α1PI. This novel information is crucial for the design of a novel therapeutic approach (shown in green). Other therapeutic approaches are also depicted: the prevention of intracellular maturation of pro-PR3 to the active enzyme using CatC inhibition (shown in beige) is under clinical investigation, and inhibition of further neutrophil activation by blocking the interaction of C5a with its receptor (shown in salmon) has become an approved therapy for ANCA-associated vasculitis. ANCA, antineutrophil cytoplasmic antibody; CatC, cathepsin C; PR3, proteinase 3.
The cellular response to ANCA requires two steps: first, ANCA must bind to PR3mb through their F(ab)2 fragment, and second, they interact with FcγRs on the surface of neutrophils and other cells via their Fc domain. This cross-linking is a signal to activate neutrophils and induces an inflammatory response by increasing neutrophil adherence to endothelial cells (44), triggering the secretion of proinflammatory cytokines and ROS (45), inducing the release of proteolytic enzymes (23), and triggering NETosis (46). Such an inflammatory response around the vessel walls and in the surrounding tissue will result in the development of necrotizing small-vessel vasculitis (47). Fab fragments of anti-PR3 mAbs could possibly compete with whole antibodies to bind to constitutive and inducible PR3mb, which would impair the interaction of Fc fragments with FcγR on the cell surface. Indeed, pretreatment of primed neutrophils with purified Fab fragments from the anti-PR3 mAb MCPR3-2 resulted in a lower release of proteolytic activity into the cell supernatant. We anticipate that intravenous administration of anti-PR3 Fab or F(ab)2 fragment with extended plasma half-life will significantly reduce immune activation in PR3-ANCA–associated vasculitis patients. This, however, must be regarded with certain constraints, as the true role of F(ab)2 binding to the antigen and Fc binding to receptors in neutrophil activation is unclear. F(ab)2 fragments of PR3-ANCA have been reported to mediate a neutrophil respiratory burst mechanism (48) but these results were not confirmed by others (45, 49). We recently obtained a nonactivating human IgG1κ anti-PR3 mAb, named 4C3, that binds with high affinity soluble PR3 and PR3mb close to the PR3 hydrophobic patch (28). 4C3 did not induce immune activation of neutrophils and is therefore nonpathogenic (28). Our data support the view that the interaction of PR3mb with Fab or F(ab)2 ANCA fragments is not sufficient to fully activate neutrophils (Fig. 7). Mechanistic studies showed the alternative complement pathway, specifically C5a receptor activation, in the pathogenesis of ANCA-associated vasculitis. Results from a recent phase 3 trial suggest that C5a receptor blockade could enable the reduced use or complete withdrawal of steroids from induction protocols (50). Experimental data support that inhibition of neutrophil activation in ANCA-associated vasculitis by blocking PR3mb or C5a receptor could be an effective treatment during the active phase of the disease (Fig. 7).
The therapeutic targeting of soluble and PR3mb is not expected to increase the patients’ risk for bacterial or viral infections considering the following clinical observations. Loss-of-function mutation in the CatC gene result in the so-called Papillon-Lefèvre syndrome characterized by severe prepubertal periodontitis and palmoplantar keratoderma without marked immunodeficiency (51, 52). The lack of CatC involved in the maturation of PR3 and related NSP zymogens reduces both the activity (active NE/PR3 < 5%; active CatG < 1%) (27) and the protein amount of these proteases in blood neutrophils from Papillon-Lefèvre syndrome patients (6, 26). The pharmacological inhibition of CatC using a nitrile inhibitor termed brensocatib (53) in noncystic fibrosis bronchiectasis (Phase 3, NCT04594369) and in patients with severe acute respiratory syndrome coronavirus 2 (NCT04817332) are underway (54). CatC-dependent proteolytic mechanisms are also therapeutic targets in cancer (55). Pharmacological inhibition of CatC could be a strategy to help fight cancer (56).
In conclusion, both constitutive and induced PR3mb, which exhibit conformational differences from soluble active PR3, are recognized by murine anti-PR3 mAbs and pathogenic human PR3-ANCA resulting in immune activation of neutrophils. Blocking both constitutive and induced PR3mb with Fab, F(ab)2 fragments or whole nonactivating PR3-ANCA prevents neutrophil activation by pathogenic PR3-ANCA offering a novel therapeutic approach targeting neutrophil activation in PR3-ANCA–associated vasculitis. This direct approach would be disease-specific and different from alternative approaches, such as preventing the expression of PR3 on the neutrophil surface with a CatC inhibitor or blocking further neutrophil activation with a C5a receptor antagonist. Effective prevention of neutrophil activation by pathogenic PR3-ANCA by blocking their binding to both constitutive and induced PR3mb offers a promising new targeted therapeutic perspective for both remission induction and maintenance therapy of PR3-ANCA–associated vasculitis.
Experimental procedures
Isolation of human neutrophils
Blood samples were obtained from normal healthy volunteers and PR3-ANCA–associated vasculitis patients after informed consent. Neutrophils were purified using negative magnetic selection with the commercial kit "EasySep Direct Human Neutrophil Isolation Kit" (StemCells) following the manufacturer’s instructions. At the end of isolation, neutrophils were suspended in Hanks’ Balanced Salt Solution (HBSS) solution without calcium and magnesium. Neutrophils were examined for viability using trypan blue dye exclusion, which were around 98% immediately after isolation. The purity of isolated neutrophils was around >95%, as assessed by flow cytometry (CD15-PE and Live dead, Miltenyi Biotech).
Neutrophils priming and activation
Freshly purified neutrophils were studied as resting (quiescent) cells or after activation using different stimuli. Neutrophil activation by PMA (Sigma Chemical Co) (100 ng/ml) was achieved at 37 °C for 15 min. Stimulation with calcium ionophore A23187 (1 μM) was performed as described in (57). Activation with formylated peptide fMLP (Sigma Chemical Co) was preceded by a priming step with cytochalasin B (CB, Sigma Chemical Co.) as a cytoskeleton-disrupting substance. Firstly, cells (10 × 106/ml) were primed with CB (10 μg/ml) for 10 min and then were treated with fMLP (1 μM) for 20 min; both steps have been performed at 37 °C.
Antibody-triggered activation was performed for 90 min at room temperature (RT) after pretreatment of neutrophils with the priming agents CB (10 μg/ml) and TNF-α (2 or 10 ng/ml): the priming step was performed by pretreating cells (10 × 106/ml) with CB for 5 min and then by directly adding TNF-α for 20 min. In all instances, cell activation was stopped by centrifuging the samples at 500g for 5 min at 4 °C.
Purification of IgGs and PR3-ANCA from patients
For the functional tests, we selected sera of patients with active disease and positive PR3-ANCA. PR3-ANCA–associated vasculitis patients with active disease were consecutively diagnosed at the University Hospital Centre of Tours between 2017 and 2019. We purified IgGs from five independent patient sera obtained during the active phase of the disease after diagnosis with a protein G SepharoseTM 4 fast flow kit (GE HealthCare). Briefly, nonpooled sera were incubated with protein G for 1 hour at RT before elution. Filtration of samples was conducted using a Spin-X UF Concentrator ultrafiltration system. Purified IgG from nonpooled sera underwent electrophoresis on a 4 to 12% Bis-tris Gel before staining with Coomassie Blue. The presence of PR3-ANCA in separate IgG preparations from active vasculitis patients was confirmed by PR3-ANCA ELISA (EuroImmun kit).
Total IgGs from plasmapheresis material of patients, kindly provided by the Klinikum Großhadern, were precipitated. PR3-ANCA were affinity-purified from the total IgG solution. To this end, biotinylated recombinant PR3 (12) was bound to streptavidin columns, and the PR3-specific ANCA were isolated. The elution fractions were tested for the presence of PR3-ANCA by ELISA. Recombinant PR3 with a poly-His tag (12) was immobilized on nickel-plates, and the ANCA in the elution fractions were bound. The fractions, which were positively tested for the presence of PR3-ANCA, were then analyzed in an activity assay for their inhibitory capacity.
Activity and inhibition of PR3mb
PR3mb activity was quantified by comparing the rates of hydrolysis of its specific FRET substrates (ABZ-VAD(nor)VADRQ-EDDnp (57) (Genecust) or ABZ Tyr-Tyr-Abu-Asn-Glu-Pro-Tyr(3-NO2)-NH2 (58) supplied by Dr Adam Lesner (University of Gdansk)) with that of titrated of purified PR3 under the same experimental conditions. Activated neutrophils (0.5–2 × 106 cells) and purified PR3 used as control (1 × 10–9 M) were incubated with FRET substrates in wells of polypropylene microplates selected for their low binding properties (Hard-Shell Thin-Wall Microplates; MJ Research) at 37 °C in PBS. Their residual activity in the presence of increased concentrations of α1PI (0.01–10 μM) and purified human PR3-ANCA (1–20 μg/ml) were also monitored. The fluorescence was recorded at λex = 320 nm and λem = 420 nm using a microplate fluorescence reader (Spectra Max Gemini; Molecular Devices) under continuous stirring.
Flow cytometry
PR3mb was analyzed on resting and activated purified neutrophils (1 × 106). Cells were resuspended in PBS and a blocking step has been performed with 5% bovine serum albumin, 2.5 mM EDTA in PBS for 15 min at 4 °C. When required, cells were preincubated with α1PI (10 or 50 μM) or plasma (dilution 1/3) for 60 min at RT and then washed three times in PBS to remove inhibitors. The chosen amount of α1PI was selected to mimic optimal plasma levels. Cells were treated with mouse anti-PR3 mAbs (CLB12.8 (Sanquin), MCPR3-2 (Thermo Fisher Scientific), WGM-2 (GeneTex Inc) and with purified PR3-ANCA in a total assay volume of 100 μl for 1 h at RT. They were than incubated with the respective secondary antibody (goat anti-mouse IgG-FITC 1:100 dilution, goat anti-human IgG-FITC 1:100 dilution) for 30 min at RT in the dark. Neutrophils were then washed twice in PBS 5% bovine serum albumin, 2.5 mM EDTA, and the analysis was performed immediately after this last wash in a final volume of 200 μl. The samples were examined by flow cytometry: (i) Beckman Coulter XL flow cytometer equipped with a 488 nm argon laser (Beckman Coulter), (ii) MACSQuant, Miltenyi Biotec, (Bergisch-Gladbach), or (iii) BD FACS Melody (BD Biosciences). Results were analyzed with i) the Expo32 software (Beckman Coulter), (ii) VenturiOne software (Applied Cytometry), or (iii) FlowJo software (BD Biosciences).
Surface markers were analyzed using TNF-α–primed neutrophils stimulated with IgGs from patients (200 μg/ml) for 45 min at 37 °C before and after incubation with α1PI. After activation, cells were washed and stained with CD63-FITC (degranulation) or double stained with CD11b VioBlue/CD18 FITC (BD Biosciences) (adhesion phenotype) for 20 min at 4 °C before flow cytometry analysis. The percentage of positive cells was determined using FlowJo software.
p47phox immunodetection
TNF-α–primed purified neutrophils (3 × 106) were analyzed for their respiratory burst response in the presence of anti-PR3 mAbs. Treated cells were lysed by resuspension in NP-40 lysis buffer (1% v/v Nonidet P-40, 150 mM NaCl, 10 mM Tris–Cl, pH 7.5, 0.4 mM EDTA) containing a phosphatase inhibitor cocktail (P0044, Sigma). The lysates were centrifuged at 10, 000g for 30 min at 4 °C, and the cleared supernatants were separated by SDS-PAGE, 12% NaDodSO4-PAGE under denaturing conditions, and transferred to a nitrocellulose (Hybond)-ECL membrane. Free sites on the membranes were blocked by incubation with 5% non-fat dried milk in PBS-0.1% Tween for 90 min at RT. Membranes were washed twice with PBS-Tween 0.1% and incubated overnight with anti-p47phox (1:500) or with anti-phospho-Ser345 antibody (p-Ser345) (1:8000), followed by a goat anti-rabbit IgG secondary antibody (1:5000). These membranes were then washed, and reactive bands were identified by chemiluminescence (ECL Kit).
Superoxide generation assay
Measurement of superoxide was performed following the SOD-inhibitable reduction of cytochrome c (59) (Merck Millipore). Freshly purified neutrophils (1 × 106) were primed and/or activated as described earlier using murine anti-PR3 mAbs (20–90 μg/ml) and PMA (100 ng/ml). When needed, cells were preincubated with α1PI (10 μM or 50 μM) as reported above. They were added to a reaction mixture containing stimulating antibodies (or PMA) and cytochrome c (100 μM) under gentle agitation for 90 min at RT. After centrifugation (400g, 5 min), absorbance of the cell-free supernatant was measured at 550 nm with a Cary 100 UV-Visible Spectrophotometer, and the absorption of samples with and without superoxide dismutase (1.5 μg/ml) was also measured. The amount of O2- produced was calculated using an extinction coefficient of 21.1 nmol/cm-1 as molar extinction coefficient of cytochrome c.
Degranulation assay
Cells (1 × 106) were primed and/or activated, treated with α1PI (1, 10 or 50 μM) when required, as described previously. The cell-free supernatant of each tested condition was recovered to assess the residual activity of neutrophil elastase with ABZ-HPVPVYAFSPQ-YNO2 (60).
Digestion and purification of Fab Fragments from mouse IgG
Fab fragments were generated using Pierce Fab Micro Preparation (Thermo Fisher Scientific) which is based on immobilized papain. First, mouse anti-PR3 MCPR3-2 antibody produced as in (61) and CLB12.8 were concentrated by using Vivaspin 2 centrifugation tubes (Hydrosart Membrane, 5000 MWCO, Sartorius Stedim Biotech GmbH), and then papain digestion was performed at 37 °C for 5 h. The obtained Fab fragments were separated from undigested IgGS and Fc fragments with protein A column and were eluted in sterile-filtered PBS. They were concentrated by Vivaspin 2 centrifugation tubes, and their concentration was estimated by NanoDrop 2000c (Thermo Fisher Scientific). The purity of Fab fragments was analyzed by SDS-PAGE under reducing and denaturing conditions.
Data availability
All data are contained within the manuscript.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank Dr Lisa Hinkofer (Comprehensive Pneumology Center) for PR3-ANCA purification, Lise Vanderlynden (INSERM UMR-1100) for technical assistance, and Dr Elodie Kara for statistical analyses. This work was supported by “Labex MabImprove” and “Ministère de l'Enseignement Supérieur”.
Author contributions
B. K. supervision; C. G., S. S., R. L., A. M. H., and J.-E. M. investigation; C. G., S. S., R. L., A. M. H., J.-E. M., J. E.-B., C. H., U. S., D. E. J., and B. K. formal analysis; B. K. writing–original draft; C. G., S. S., R. L., A. M. H., J.-E. M., J. E.-B., C. H., U. S., D. E. J., and B. K. writing–review and editing.
Funding and additional information
B. K. acknowledges the “Alexandre von Humboldt Foundation”.
Reviewed by members of the JBC Editorial Board. Edited by George DeMartino
Supporting information
References
- 1.Kitching A.R., Anders H.J., Basu N., Brouwer E., Gordon J., Jayne D.R., et al. ANCA-associated vasculitis. Nat. Rev. Dis. Primers. 2020;6:71. doi: 10.1038/s41572-020-0204-y. [DOI] [PubMed] [Google Scholar]
- 2.Granel J., Korkmaz B., Nouar D., Weiss S.A.I., Jenne D.E., Lemoine R., et al. Pathogenicity of proteinase 3-anti-neutrophil cytoplasmic antibody in granulomatosis with polyangiitis: implications as Biomarker and future therapies. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.571933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kettritz R. Vasculitis: a clear argument for targeting complement in ANCA vasculitis. Nat. Rev. Nephrol. 2017;13:448–450. doi: 10.1038/nrneph.2017.69. [DOI] [PubMed] [Google Scholar]
- 4.Specks U., Merkel P.A., Seo P., Spiera R., Langford C.A., Hoffman G.S., et al. Efficacy of remission-induction regimens for ANCA-associated vasculitis. N. Engl. J. Med. 2013;369:417–427. doi: 10.1056/NEJMoa1213277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korkmaz B., Lesner A., Guarino C., Wysocka M., Kellenberger C., Watier H., et al. Inhibitors and antibody fragments as potential anti-inflammatory therapeutics targeting neutrophil proteinase 3 in human disease. Pharmacol. Rev. 2016;68:603–630. doi: 10.1124/pr.115.012104. [DOI] [PubMed] [Google Scholar]
- 6.Korkmaz B., Caughey G.H., Chapple I., Gauthier F., Hirschfeld J., Jenne D.E., et al. Therapeutic targeting of cathepsin C: from pathophysiology to treatment. Pharmacol. Ther. 2018;190:202–236. doi: 10.1016/j.pharmthera.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Korkmaz B., Lesner A., Letast S., Mahdi Y.K., Jourdan M.L., Dallet-Choisy S., et al. Neutrophil proteinase 3 and dipeptidyl peptidase I (cathepsin C) as pharmacological targets in granulomatosis with polyangiitis (Wegener granulomatosis) Semin. Immunopathol. 2013;35:411–421. doi: 10.1007/s00281-013-0362-z. [DOI] [PubMed] [Google Scholar]
- 8.Schreiber A., Busjahn A., Luft F.C., Kettritz R. Membrane expression of proteinase 3 is genetically determined. J. Am. Soc. Nephrol. 2003;14:68–75. doi: 10.1097/01.asn.0000040751.83734.d1. [DOI] [PubMed] [Google Scholar]
- 9.Halbwachs-Mecarelli L., Bessou G., Lesavre P., Lopez S., Witko-Sarsat V. Bimodal distribution of proteinase 3 (PR3) surface expression reflects a constitutive heterogeneity in the polymorphonuclear neutrophil pool. FEBS Lett. 1995;374:29–33. doi: 10.1016/0014-5793(95)01073-n. [DOI] [PubMed] [Google Scholar]
- 10.Korkmaz B., Jaillet J., Jourdan M.L., Gauthier A., Gauthier F., Attucci S. Catalytic activity and inhibition of wegener antigen proteinase 3 on the cell surface of human polymorphonuclear neutrophils. J. Biol. Chem. 2009;284:19896–19902. doi: 10.1074/jbc.M901471200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guarino C., Legowska M., Epinette C., Kellenberger C., Dallet-Choisy S., Sienczyk M., et al. New selective peptidyl di(chlorophenyl) phosphonate esters for visualizing and blocking neutrophil proteinase 3 in human diseases. J. Biol. Chem. 2014;289:31777–31791. doi: 10.1074/jbc.M114.591339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korkmaz B., Kuhl A., Bayat B., Santoso S., Jenne D.E. A hydrophobic patch on proteinase 3, the target of autoantibodies in Wegener granulomatosis, mediates membrane binding via NB1 receptors. J. Biol. Chem. 2008;283:35976–35982. doi: 10.1074/jbc.M806754200. [DOI] [PubMed] [Google Scholar]
- 13.Rarok A.A., Stegeman C.A., Limburg P.C., Kallenberg C.G. Neutrophil membrane expression of proteinase 3 (PR3) is related to relapse in PR3-ANCA-associated vasculitis. J. Am. Soc. Nephrol. 2002;13:2232–2238. doi: 10.1097/01.asn.0000028642.26222.00. [DOI] [PubMed] [Google Scholar]
- 14.Witko-Sarsat V., Lesavre P., Lopez S., Bessou G., Hieblot C., Prum B., et al. A large subset of neutrophils expressing membrane proteinase 3 is a risk factor for vasculitis and rheumatoid arthritis. J. Am. Soc. Nephrol. 1999;10:1224–1233. doi: 10.1681/ASN.V1061224. [DOI] [PubMed] [Google Scholar]
- 15.Hu N., Westra J., Kallenberg C.G. Membrane-bound proteinase 3 and its receptors: relevance for the pathogenesis of wegener's granulomatosis. Autoimmun. Rev. 2009;8:510–514. doi: 10.1016/j.autrev.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Silva F., Hummel A.M., Jenne D.E., Specks U. Discrimination and variable impact of ANCA binding to different surface epitopes on proteinase 3, the Wegener's autoantigen. J. Autoimmun. 2010;35:299–308. doi: 10.1016/j.jaut.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhl A., Korkmaz B., Utecht B., Kniepert A., Schonermarck U., Specks U., et al. Mapping of conformational epitopes on human proteinase 3, the autoantigen of Wegener's granulomatosis. J. Immunol. 2010;185:387–399. doi: 10.4049/jimmunol.0903887. [DOI] [PubMed] [Google Scholar]
- 18.Pang Y.P., Casal Moura M., Thompson G.E., Nelson D.R., Hummel A.M., Jenne D.E., et al. Remote activation of a latent epitope in an autoantigen decoded with simulated B-factors. Front. Immunol. 2019;10:2467. doi: 10.3389/fimmu.2019.02467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinkofer L.C., Seidel S.A., Korkmaz B., Silva F., Hummel A.M., Braun D., et al. A monoclonal antibody (MCPR3-7) interfering with the activity of proteinase 3 by an allosteric mechanism. J. Biol. Chem. 2013;288:26635–26648. doi: 10.1074/jbc.M113.495770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolman K.M., van de Wiel B.A., Kam C.M., Kerrigan J.E., Hack C.E., von dem Borne A.E., et al. Proteinase 3: substrate specificity and possible pathogenetic effect of wegener's granulomatosis autoantibodies (c-ANCA) by dysregulation of the enzyme. Adv. Exp. Med. Biol. 1993;336:55–60. doi: 10.1007/978-1-4757-9182-2_7. [DOI] [PubMed] [Google Scholar]
- 21.van der Geld Y.M., Tool A.T., Videler J., de Haas M., Cohen Tervaert J.W., Stegeman C.A., et al. Interference of PR3-ANCA with the enzymatic activity of PR3: differences in patients during active disease or remission of wegener's granulomatosis. Clin. Exp. Immunol. 2002;129:562–570. doi: 10.1046/j.1365-2249.2002.01926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinkofer L.C., Hummel A.M., Stone J.H., Hoffman G.S., Merkel P.A., Spiera E.R., et al. Allosteric modulation of proteinase 3 activity by anti-neutrophil cytoplasmic antibodies in granulomatosis with polyangiitis. J. Autoimmun. 2015;59:43–52. doi: 10.1016/j.jaut.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Falk R.J., Terrell R.S., Charles L.A., Jennette J.C. Anti-neutrophil cytoplasmic autoantibodies induce neutrophils to degranulate and produce oxygen radicals in vitro. Proc. Natl. Acad. Sci. U. S. A. 1990;87:4115–4119. doi: 10.1073/pnas.87.11.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerke U., Rolle S., Dittmar G., Bayat B., Santoso S., Sporbert A., et al. Complement receptor Mac-1 is an adaptor for NB1 (CD177)-mediated PR3-ANCA neutrophil activation. J. Biol. Chem. 2011;286:7070–7081. doi: 10.1074/jbc.M110.171256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schreiber A., Luft F.C., Kettritz R. Membrane proteinase 3 expression and ANCA-induced neutrophil activation. Kidney Int. 2004;65:2172–2183. doi: 10.1111/j.1523-1755.2004.00640.x. [DOI] [PubMed] [Google Scholar]
- 26.Korkmaz B., Horwitz M., Jenne D.E., Gauthier F. Neutrophil elastase, proteinase 3 and cathepsin G as therapeutic targets in human diseases. Pharmacol. Rev. 2010;62:726–759. doi: 10.1124/pr.110.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seren S., Rashed Abouzaid M., Eulenberg-Gustavus C., Hirschfeld J., Nasr Soliman H., Jerke U., et al. Consequences of cathepsin C inactivation for membrane exposure of proteinase 3, the target antigen in autoimmune vasculitis. J. Biol. Chem. 2018;293:12415–12428. doi: 10.1074/jbc.RA118.001922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granel J., Lemoine R., Morello E., Gallais Y., Mariot J., Drapeau M., et al. 4C3 human monoclonal antibody: a proof of concept for non-pathogenic proteinase 3 anti-neutrophil cytoplasmic antibodies in granulomatosis with polyangiitis. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.573040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rooney C.P., Taggart C., Coakley R., McElvaney N.G., O'Neill S.J. Anti-proteinase 3 antibody activation of neutrophils can be inhibited by alpha1-antitrypsin. Am. J. Respir. Cell Mol. Biol. 2001;24:747–754. doi: 10.1165/ajrcmb.24.6.4147. [DOI] [PubMed] [Google Scholar]
- 30.Dang P.M., Stensballe A., Boussetta T., Raad H., Dewas C., Kroviarski Y., et al. A specific p47phox -serine phosphorylated by convergent MAPKs mediates neutrophil NADPH oxidase priming at inflammatory sites. J. Clin. Invest. 2006;116:2033–2043. doi: 10.1172/JCI27544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faurschou M., Borregaard N. Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. 2003;5:1317–1327. doi: 10.1016/j.micinf.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Jerke U., Eulenberg-Gustavus C., Rousselle A., Nicklin P., Kreideweiss S., Grundl M.A., et al. Targeting cathepsin C in PR3-ANCA vasculitis. J. Am. Soc. Nephrol. 2022;33:936–947. doi: 10.1681/ASN.2021081112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schreiber A., Otto B., Ju X., Zenke M., Goebel U., Luft F.C., et al. Membrane proteinase 3 expression in patients with wegener's granulomatosis and in human hematopoietic stem cell-derived neutrophils. J. Am. Soc. Nephrol. 2005;16:2216–2224. doi: 10.1681/ASN.2004070609. [DOI] [PubMed] [Google Scholar]
- 34.von Vietinghoff S., Tunnemann G., Eulenberg C., Wellner M., Cristina Cardoso M., Luft F.C., et al. NB1 mediates surface expression of the ANCA antigen proteinase 3 on human neutrophils. Blood. 2007;109:4487–4493. doi: 10.1182/blood-2006-10-055327. [DOI] [PubMed] [Google Scholar]
- 35.Ebert M., Jerke U., Eulenberg-Gustavus C., Kling L., Jenne D., Kirchner M., et al. Protective alpha1-antitrypsin effects in autoimmune vasculitis are compromised by methionine oxidation. J. Clin. Invest. 2022;132 doi: 10.1172/JCI160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jegot G., Derache C., Castella S., Lahouassa H., Pitois E., Jourdan M.L., et al. A substrate-based approach to convert SerpinB1 into a specific inhibitor of proteinase 3, the Wegener's granulomatosis autoantigen. FASEB J. 2011;25:3019–3031. doi: 10.1096/fj.10-176552. [DOI] [PubMed] [Google Scholar]
- 37.Sanders J.S., Stassen P.M., van Rossum A.P., Kallenberg C.G., Stegeman C.A. Risk factors for relapse in anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis: tools for treatment decisions? Clin. Exp. Rheumatol. 2004;22:S94–101. [PubMed] [Google Scholar]
- 38.Fussner L.A., Hummel A.M., Schroeder D.R., Silva F., Cartin-Ceba R., Snyder M.R., et al. Factors determining the clinical utility of serial measurements of antineutrophil cytoplasmic antibodies targeting proteinase 3. Arthritis Rheumatol. 2016;68:1700–1710. doi: 10.1002/art.39637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kemna M.J., Damoiseaux J., Austen J., Winkens B., Peters J., van Paassen P., et al. ANCA as a predictor of relapse: useful in patients with renal involvement but not in patients with nonrenal disease. J. Am. Soc. Nephrol. 2015;26:537–542. doi: 10.1681/ASN.2013111233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finkielman J.D., Merkel P.A., Schroeder D., Hoffman G.S., Spiera R., St Clair E.W., et al. Antiproteinase 3 antineutrophil cytoplasmic antibodies and disease activity in Wegener granulomatosis. Ann. Intern. Med. 2007;147:611–619. doi: 10.7326/0003-4819-147-9-200711060-00005. [DOI] [PubMed] [Google Scholar]
- 41.Daouk G.H., Palsson R., Arnaout M.A. Inhibition of proteinase 3 by ANCA and its correlation with disease activity in Wegener's granulomatosis. Kidney Int. 1995;47:1528–1536. doi: 10.1038/ki.1995.216. [DOI] [PubMed] [Google Scholar]
- 42.Korkmaz B., Attucci S., Moreau T., Godat E., Juliano L., Gauthier F. Design and use of highly specific substrates of neutrophil elastase and proteinase 3. Am. J. Respir. Cell Mol. Biol. 2004;30:801–807. doi: 10.1165/rcmb.2003-0139OC. [DOI] [PubMed] [Google Scholar]
- 43.Korkmaz B., Hajjar E., Kalupov T., Reuter N., Brillard-Bourdet M., Moreau T., et al. Influence of charge distribution at the active site surface on the substrate specificity of human neutrophil protease 3 and elastase. A kinetic and molecular modeling analysis. J. Biol. Chem. 2007;282:1989–1997. doi: 10.1074/jbc.M608700200. [DOI] [PubMed] [Google Scholar]
- 44.Ewert B.H., Jennette J.C., Falk R.J. Anti-myeloperoxidase antibodies stimulate neutrophils to damage human endothelial cells. Kidney Int. 1992;41:375–383. doi: 10.1038/ki.1992.52. [DOI] [PubMed] [Google Scholar]
- 45.Reumaux D., Vossebeld P.J., Roos D., Verhoeven A.J. Effect of tumor necrosis factor-induced integrin activation on Fc gamma receptor II-mediated signal transduction: relevance for activation of neutrophils by anti-proteinase 3 or anti-myeloperoxidase antibodies. Blood. 1995;86:3189–3195. [PubMed] [Google Scholar]
- 46.Kessenbrock K., Krumbholz M., Schonermarck U., Back W., Gross W.L., Werb Z., et al. Netting neutrophils in autoimmune small-vessel vasculitis. Nat. Med. 2009;15:623–625. doi: 10.1038/nm.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kallenberg C.G., Heeringa P., Stegeman C.A. Mechanisms of disease: pathogenesis and treatment of ANCA-associated vasculitides. Nat. Clin. Pract. Rheumatol. 2006;2:661–670. doi: 10.1038/ncprheum0355. [DOI] [PubMed] [Google Scholar]
- 48.Keogan M.T., Esnault V.L., Green A.J., Lockwood C.M., Brown D.L. Activation of normal neutrophils by anti-neutrophil cytoplasm antibodies. Clin. Exp. Immunol. 1992;90:228–234. doi: 10.1111/j.1365-2249.1992.tb07934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porges A.J., Redecha P.B., Kimberly W.T., Csernok E., Gross W.L., Kimberly R.P. Anti-neutrophil cytoplasmic antibodies engage and activate human neutrophils via Fc gamma RIIa. J. Immunol. 1994;153:1271–1280. [PubMed] [Google Scholar]
- 50.Jayne D.R.W., Merkel P.A., Schall T.J., Bekker P., Group A.S. Avacopan for the treatment of ANCA-associated vasculitis. N. Engl. J. Med. 2021;384:599–609. doi: 10.1056/NEJMoa2023386. [DOI] [PubMed] [Google Scholar]
- 51.Toomes C., James J., Wood A.J., Wu C.L., McCormick D., Lench N., et al. Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat. Genet. 1999;23:421–424. doi: 10.1038/70525. [DOI] [PubMed] [Google Scholar]
- 52.Pham C.T., Ivanovich J.L., Raptis S.Z., Zehnbauer B., Ley T.J. Papillon-lefevre syndrome: correlating the molecular, cellular, and clinical consequences of cathepsin C/dipeptidyl peptidase I deficiency in humans. J. Immunol. 2004;173:7277–7281. doi: 10.4049/jimmunol.173.12.7277. [DOI] [PubMed] [Google Scholar]
- 53.Chalmers J.D., Haworth C.S., Metersky M.L., Loebinger M.R., Blasi F., Sibila O., et al. Phase 2 trial of the DPP-1 inhibitor brensocatib in bronchiectasis. New Engl. J. Med. 2020;383:2127–2137. doi: 10.1056/NEJMoa2021713. [DOI] [PubMed] [Google Scholar]
- 54.Korkmaz B., Lesner A., Marchand-Adam S., Moss C., Jenne D.E. Lung protection by cathepsin C inhibition: a new hope for COVID-19 and ARDS? J. Med. Chem. 2020;63:13258–13265. doi: 10.1021/acs.jmedchem.0c00776. [DOI] [PubMed] [Google Scholar]
- 55.Korkmaz B., Lamort A.S., Domain R., Beauvillain C., Gieldon A., Yildirim A.O., et al. Cathepsin C inhibition as a potential treatment strategy in cancer. Biochem. Pharmacol. 2021;194 doi: 10.1016/j.bcp.2021.114803. [DOI] [PubMed] [Google Scholar]
- 56.Xiao Y., Cong M., Li J., He D., Wu Q., Tian P., et al. Cathepsin C promotes breast cancer lung metastasis by modulating neutrophil infiltration and neutrophil extracellular trap formation. Cancer Cell. 2021;39:423–437 e427. doi: 10.1016/j.ccell.2020.12.012. [DOI] [PubMed] [Google Scholar]
- 57.Korkmaz B., Attucci S., Juliano M.A., Kalupov T., Jourdan M.L., Juliano L., et al. Measuring elastase, proteinase 3 and cathepsin G activities at the surface of human neutrophils with fluorescence resonance energy transfer substrates. Nat. Protoc. 2008;3:991–1000. doi: 10.1038/nprot.2008.63. [DOI] [PubMed] [Google Scholar]
- 58.Popow-Stellmaszyk J., Wysocka M., Lesner A., Korkmaz B., Rolka K. A new proteinase 3 substrate with improved selectivity over human neutrophil elastase. Anal. Biochem. 2013;442:75–82. doi: 10.1016/j.ab.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 59.Pick E., Mizel D. Rapid microassays for the measurement of superoxide and hydrogen peroxide production by macrophages in culture using an automatic enzyme immunoassay reader. J. Immunol. Met. 1981;46:211–226. doi: 10.1016/0022-1759(81)90138-1. [DOI] [PubMed] [Google Scholar]
- 60.Kalupov T., Brillard-Bourdet M., Dade S., Serrano H., Wartelle J., Guyot N., et al. Structural characterization of mouse neutrophil serine proteases and identification of their substrate specificities: relevance to mouse models of human inflammatory diseases. J .Biol. Chem. 2009;284:34084–34091. doi: 10.1074/jbc.M109.042903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun J., Fass D.N., Viss M.A., Hummel A.M., Tang H., Homburger H.A., et al. A proportion of proteinase 3 (PR3)-specific anti-neutrophil cytoplasmic antibodies (ANCA) only react with PR3 after cleavage of its N-terminal activation dipeptide. Clin. Exp. Immunol. 1998;114:320–326. doi: 10.1046/j.1365-2249.1998.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript.