ABSTRACT
Microbes possess conserved microbe-associated molecular patterns (MAMPs) that are recognized by plant receptors to induce pattern-triggered immunity (PTI). Despite containing the same MAMPs as pathogens, commensals thrive in the plant rhizosphere microbiome, indicating they must suppress or evade host immunity. Previous work found that bacterial-secreted gluconic acid is sufficient to suppress PTI. Here, we show that gluconic acid biosynthesis is not necessary for immunity suppression by the beneficial bacterial strain Pseudomonas simiae WCS417. We performed a forward genetic screen with EMS-mutagenized P. simiae WCS417 and a flagellin-inducible CYP71A12pro:GUS reporter as a PTI readout. We identified a loss of function mutant in ornithine carbamoyltransferase argF, which is required for ornithine conversion to arginine, that cannot suppress PTI or acidify the rhizosphere. Fungal pathogens use alkalization through production of ammonia and glutamate, and arginine biosynthetic precursors, to promote their own growth and virulence. While a ΔargF mutant has a growth defect in the rhizosphere, we found that restoring growth with exogenous arginine resulted in rhizosphere alkalization in a mutant that cannot make gluconic acid, indicating that arginine biosynthesis is required for both growth and acidification. Furthermore, blocking bacterial arginine, glutamine, or proline biosynthesis through genetic mutations or feedback inhibition by adding corresponding amino acids, resulted in rhizosphere alkalization. Untargeted metabolomics determined that ornithine, an alkaline molecule, accumulates under conditions associated with rhizosphere alkalization. Our findings show that bacterial amino acid biosynthesis contributes to acidification by preventing accumulation of ornithine and the resulting alkalization.
KEYWORDS: PTI, amino acid biosynthesis, innate immunity, pH, plant-microbe interactions, rhizosphere-inhabiting microbes
INTRODUCTION
A myriad of microorganisms, including pathogens and mutualists, live in the plant rhizosphere and actively influence plant fitness (1). To protect themselves from pathogens, plants use pattern recognition receptors (PRRs) that can specifically sense microbe-associated molecular patterns (MAMPs), which are evolutionarily conserved across diverse microbes. Perception of MAMPs results in pattern-triggered immunity (PTI), which includes a reactive oxygen species burst, calcium influx, and defense gene expression (2). As both pathogens and commensals contain MAMPs that can be recognized by the plant innate immune system, both must suppress or evade immunity in order to successfully colonize. While mechanisms of immunity suppression by pathogenic bacteria are well-established, how commensal microbes evade or suppress plant immunity to promote their own fitness is poorly understood.
Some successful pathogens can suppress PTI by injecting effector proteins into the plant cytosol via the type III secretion systems (T3SS) (3). In addition to injecting effectors, pathogenic bacteria can manipulate phytohormones to suppress host immunity. The pathogenic bacterium Pseudomonas syringae pv tomato DC3000 (Pto) can secrete the phytotoxin coronatine (COR) that mimics the active form of jasmonic acid (JA), JA-lIe, to promote JA-dependent defense. Since the JA and salicylic acid (SA) pathways antagonize each other, inducing JA signaling by COR suppresses SA signaling, which is critical for resistance against Pto (4). Lastly, instead of suppressing host immunity, pathogenic microbes can degrade their MAMPs to prevent recognition by PRRs. For instance, P. syringae can secrete AprA, an extracellular alkaline protease that can degrade flagellin monomers, thereby avoiding immune recognition (5).
Although commensals also have MAMPs that have the potential to induce PTI, many do not induce immune responses, suggesting that commensals can suppress or evade the plant immune system (6). A growth promoting and biocontrol bacterial strain, Pseudomonas sp. WCS365, can evade host immunity by fine tuning biofilm formation (7). Pseudomonas capeferrum WCS358 can suppress root local immunity by secreting organic acids to lower the pH of the rhizosphere (8). Dyella japonica suppresses root immunity through a Type II secretion-dependent mechanism without affecting rhizosphere pH (9). Collectively, these findings indicate that rhizosphere commensals possess diverse mechanisms to modulate host immunity.
The beneficial root-associated bacterial strain, P. simiae WCS417, was previously shown to suppress root immunity (8, 10). It lowers the pH of the rhizosphere to a greater extent than P. capeferrum WCS358, and produces more gluconic acid (8); however, we found that deletion of pqqF, which results in loss of gluconic acid biosynthesis and immunity suppression in P. capeferrum WCS358 (8), does not impair the ability of WCS417 to suppress rhizosphere immunity. Here, we describe a forward genetic screen that identified a novel mechanism of root immunity suppression in P. simiae WCS417, where amino acid biosynthesis prevents rhizosphere alkalization and suppresses immunity.
RESULTS
Acidification is sufficient, but not necessary, for Arabidopsis immunity suppression by P. simiae WCS417.
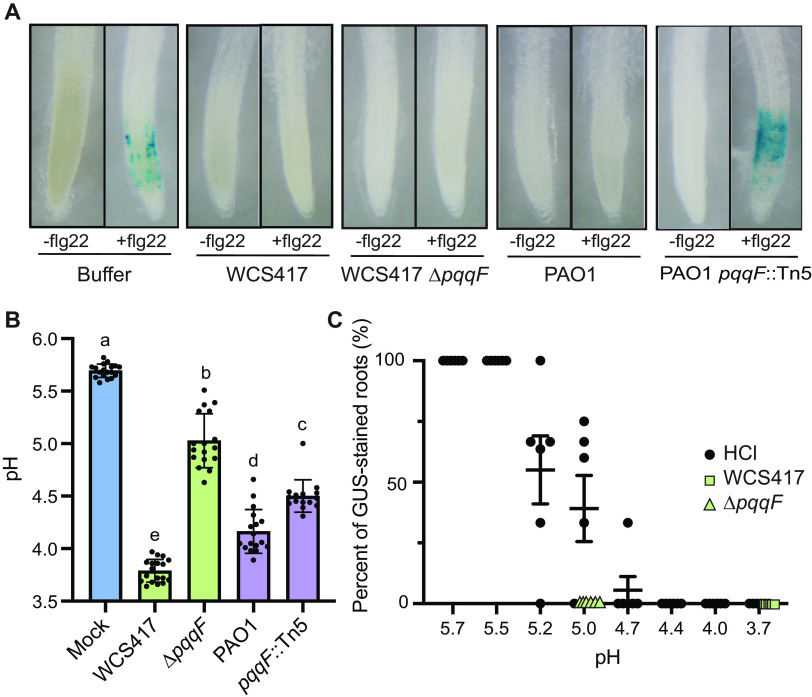
Gluconic acid biosynthesis via pqqF is necessary for Arabidopsis rhizosphere immunity suppression in P. capeferrum WCS358 and Pseudomonas aeruginosa PAO1 as measured by expression of a PTI-inducible reporter CYP71A12pro:GUS expression (8). CYP71A12 is involved in biosynthesis of the antimicrobial camalexin and is induced in the root elongation zone or maturation zone upon sensing MAMPs such as flg22 or chitin (10). As a result, induction of CYP71A12pro:GUS reports the activation of PTI. P. simiae WCS417 produces more gluconic acid and lowers the pH of the rhizosphere to a greater extent than WCS358 suggesting WCS417 pqqF is also required for rhizosphere immunity suppression (8). Surprisingly, we found that the WCS417 ΔpqqF mutant can still completely suppress flg22-triggered expression of CYP71A12pro:GUS (Fig. 1A). We found that while P. simiae WCS417 acidifies seedling exudates to a pH of 3.7, a clean deletion of pqqF in P. simiae WCS417 resulted in a significant increase in the pH of seedling exudates to pH 5.0 (Fig. 1B). In contrast, while wildtype P. aeruginosa PAO1 lowers the pH of the seedling exudates to pH 4.0 and supresses CYP71A12pro:GUS, disruption of PAO1 pqqF resulted in a less dramatic increase in the pH of seedling exudates to pH 4.5 (Fig. 1B) but results in a complete loss of suppression in host immunity (Fig. 1A; (8)). Collectively these data suggest a gluconic acid independent mechanism of immunity suppression by P. simiae WCS417.
FIG 1.
pqqF is not required for host immunity suppression in P. simiae WCS417. (A) While loss of function of P. aeruginosa PAO1 pqqF results in inability to suppress expression of CYP71A12pro:GUS expression, the P. simiae WCS417 pqqF-deficient mutant retains the ability to suppress flg22-induced CYP71A12pro:GUS expression. (B) Deletion or disruption of pqqF results in a pH increase in WCS417 and P. aeruginosa PAO1. Individual data are the result of a single well of seedling exudates. Statistics were determined by using one-way ANOVA and Tukey’s HSD. Error bars represent mean ± SD, and letters indicate differences at P < 0.05. (C) Percent of GUS-stained CYP71A12pro:GUS roots in the presence of flg22 under a pH gradient (black dots). pH of WCS417 (green squares) and the pqqF mutant fully suppress flg22-triggered CYP71A12pro:GUS expression. Each dot represents the percent of blue roots from 1 to 3 wells of a temporal experiment containing 3 to 4 roots per well. All experiments were independently repeated at least 3 times.
Previous work showed that lowering the rhizosphere pH to 3.7 with hydrochloric acid (HCl) resulted in complete inhibition of CYP71A12pro:GUS expression (8). Lowering the rhizosphere pH to between 5.5 to 4.6 resulted in partial immunity suppression, indicating that we should expect about 50% of roots to retain CYP71A12pro:GUS expression at pH 5.0, the pH of the P. simiae WCS417 ΔpqqF mutant growing in seedling exudates. We tested the effect of a pH gradient on suppression of CYP71A12pro:GUS and indeed found that modifying the pH of the rhizosphere to 4.7–5.0 in the absence of bacteria results in intermediate suppression of plant immunity, with about 50% of roots retaining CYP71A12pro:GUS expression (Fig. 1C). The pH of the WCS417 ΔpqqF mutant is around 5.0, but results in complete inhibition of CYP71A12pro:GUS expression (Fig. 1C). These data suggest that WCS417 possesses additional mechanisms to suppress host immunity.
P. simiae WCS417 argF is required for rhizosphere acidification and immunity suppression.
To identify additional genes that are necessary for host immunity suppression, we generated an EMS-mutagenized library of P. simiae WCS417, and screened for mutants that were unable to suppress flg22-mediated induction of the CYP71A12pro:GUS reporter. We screened 960 EMS-mutagenized colonies of WCS417 in duplicate for their ability to suppress flg22-induced expression of the CYP71A12pro:GUS reporter. A single mutant, named 10E10, was found incapable of suppressing flg22-induced immunity (Fig. 2A). We found that 10E10 completely failed to reduce the pH of seedling exudates, suggesting that it might contribute to immunity suppression through rhizosphere acidification (Fig. 2B).
FIG 2.
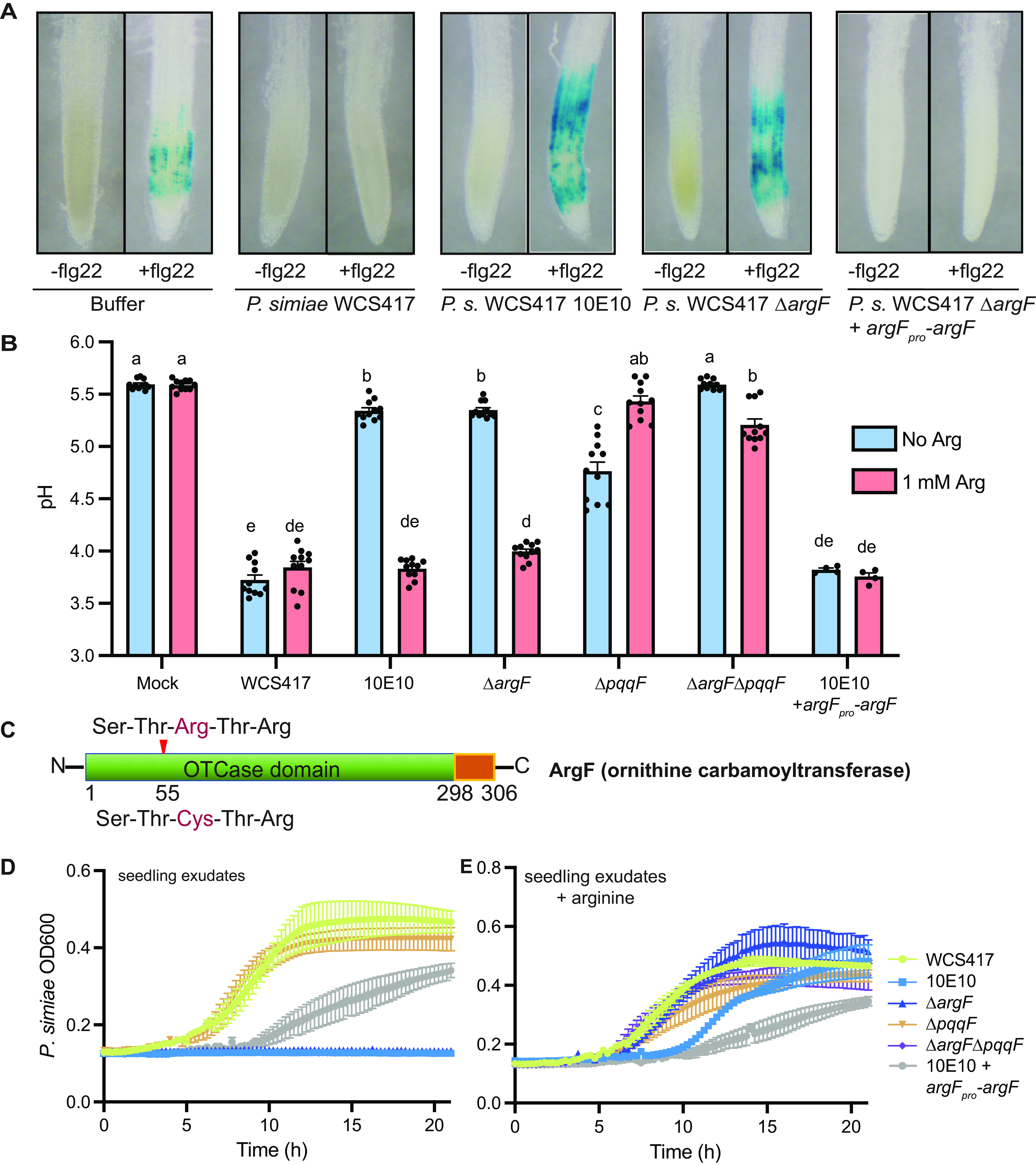
An EMS screen identified a mutant in argF that is required for both Pseudomonas growth and rhizosphere acidification. (A) A mutant, 10E10, was identified from an EMS screen for P. simiae mutants that cannot supress flg22-triggered induction of CYP71A12pro:GUS expression or (B) acidify the rhizosphere. An ΔargF mutant phenocopied the inability of the 10E10 mutant to suppress immunity suppression and acidification, and expression of argFpro-argF on a plasmid restored the immunity suppression and acidification of the 10E10 mutant. Addition of exogenous arginine lowered the pH of the ΔargF and 10E10 mutant, but resulted in alkalization of a ΔpqqF or a ΔpqqFΔargF double mutant growing in seedling exudates. All the experiments were independently repeated at least 3 times. Statistics were calculated by using one-way ANOVA and Tukey’s HSD. Error bars represent mean ± SD, and letters indicate differences at P < 0.05. (C) The mutation in the 10E10 mutant is within the catalytic site of ΔargF, and results in a predicted loss of function mutation. (D) The 10E10, the ΔargF, and the ΔargFΔpqqF mutants had a growth defect in the seedling exudates. (E) Growth of ΔargF and the ΔargFΔpqqF mutants were rescued by the addition of exogenous arginine.
Because we only identified one mutant from the screen, we wondered if mutations in WCS417 that result in a loss of immunity suppression are rare, or if our screen was not saturating. To test this, we determined whether we could identify an allele of pqqF in our screen. Gluconic acid has previously been shown to be required for zinc solubilization, and so we tested whether we could identify a mutant unable to solubilize zinc. By growing bacteria on zinc phosphate media (11), only the strains that can solubilize zinc phosphate will produce a clear halo on the plate. We found a single mutant, 4E4, that cannot solubilize zinc phosphate (Fig. S1). The genome of 4E4 was sequenced, and we identified mutations in both pqqF and pqqB (Data Set S1, Table S1). However, 4E4 can still suppress flg22-triggered CYP71A12pro:GUS expression, which is consistent with our finding that the WCS417 ΔpqqF mutant can still suppress plant immunity. As our screen successfully identified a mutation in pqqF, which suggests that mutants that fail to suppress root immunity might be rare in P. simiae WCS417.
P. simiae WCS417 can solubilize ZnSO4, while the mutant 4E4 cannot. Rescreening the P. simiae WCS417 EMS library on media containing ZnSO4 and glucose identified a single mutant that could not solubilize zinc. Sequencing the 4E4 mutant identified point mutations in both pqqF and pqqB. The 7A1 strain can solubilize ZnSO4, and is shown as a positive control. Download FIG S1, PDF file, 0.5 MB (528.7KB, pdf) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The process of HILIC+/− shows results from metabolomics analysis. Data Set S1, Table S1, A total of 35 non-synonymous candidate genes from genome mapping of the 10E10 WCS417 mutant. Data Set S1, Table S2, total of 48 non-synonymous candidate genes from genome mapping of the 4E4 WCS417 mutant. Data Set S1, Table S3 Stains used in this study. Download Data Set S1, XLSX file, 0.5 MB (546.8KB, xlsx) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We sequenced the genome of the P. simiae WCS417 10E10 mutant, and identified 35 non-synonymous mutations with respect to the parental WCS417 strain (Data Set S1, Table S2). To narrow down candidate genes, we made use of a PAO1 transposon insertion library (12), and tested transposon insertion mutants in PAO1 orthologs of genes from the mapping list that we hypothesized were most likely to contribute to immunity suppression (Fig. S2). We found that an insertion in argF uniquely impaired the ability of PAO1 to acidify seedling exudates (Fig. S2). ArgF encodes ornithine carbamoyltransferase, which is involved in arginine biosynthesis, by converting l-ornithine to L-citrulline (13, 14). The mutation in argF in P. simiae WCS417 is predicted to affect its catalytic site Ser-Thr-Arg-Thr-Arg, where a C to T change is predicted to convert the third arginine to cysteine (Fig. 2C). Thus, we hypothesized that a loss of function of argF likely underlines the lack of immunity suppression by 10E10.
A screen of P. aeruginosa PAO1 transposon insertion mutants in candidate genes with point mutations in P. simiae WCS417 10E10 identified argF, which cannot acidify seedling exudates. Genome sequencing of 10E10 identified 35 non-synonymous candidate genes (Data Set S1, Table S1). Orthologs were identified in PAO1. argF::Tn5-1/-2: PS417_05595, ornithine carbamoyltransferase; maeB: PS417_01950, malate dehydrogenase; PA4465: PS417_04330, glmZ(sRNA)-inactivating NTPase; PA0214: PS417_26590, malonate decarboxylase subunit epsilon; PA0575: PS417_25885, diguanylate cyclase; and PA0148: PS417_03205, adenine deaminase. Statistics were calculated by using two-way ANOVA and Tukey’s HSD. Error bars represent mean +/− SD, and * indicate differences at P < 0.05. Download FIG S2, PDF file, 0.2 MB (208.5KB, pdf) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We tested whether a loss of P. simiae WCS417 argF could explain the inability of the 10E10 mutant to suppress immunity. First, we confirmed the C to T mutation in the argF catalytic site in the 10E10 mutant by PCR. We then complemented the 10E10 mutant with argF expressed by its native promoter in the pBBR1MCS-5 plasmid, and found that expression of argF under its native promoter (argFpro-argF) rescued the 10E10 mutant, and the complemented strain suppressed flg22-triggered CYP71A12pro:GUS expression, resulting in acidification of seedling exudates to a similar degree as wildtype WCS417 (Fig. 2A and B). We made a clean deletion of argF in WCS417, and found that the ΔargF mutant phenocopied the inability of the 10E10 mutant to suppress flg22-triggered CYP71A12pro:GUS expression, and could not acidify seedling exudates (Fig. 2A and B). These results illustrate that a loss of function of argF underlies the inability to suppress immunity by the 10E10 mutant.
pqqF and argF pathways synergistically regulate rhizosphere pH.
Fungal pathogens can produce glutamate or glutamine to increase the local concentration of ammonia, and raise host pH to promote their own virulence (15, 16). As a result, it is an intriguing possibility that a loss of function mutation in argF may result in accumulation of alkaline arginine precursors such as ornithine, polyamines, and ammonia (Fig. 3A). A second, potentially confounding possibility is that arginine is limiting for growth in the rhizosphere, and so the ΔargF mutant may not be able to acidify because a lack of arginine is insufficient to support growth. If the former is true, then conditions resulting in accumulation of arginine precursors, such as addition of exogenous arginine, should result in accumulation of alkaline arginine precursors and rhizosphere alkalization. If the latter is true, and arginine is only required for growth, then addition of arginine should always result in increased growth and acidification.
FIG 3.
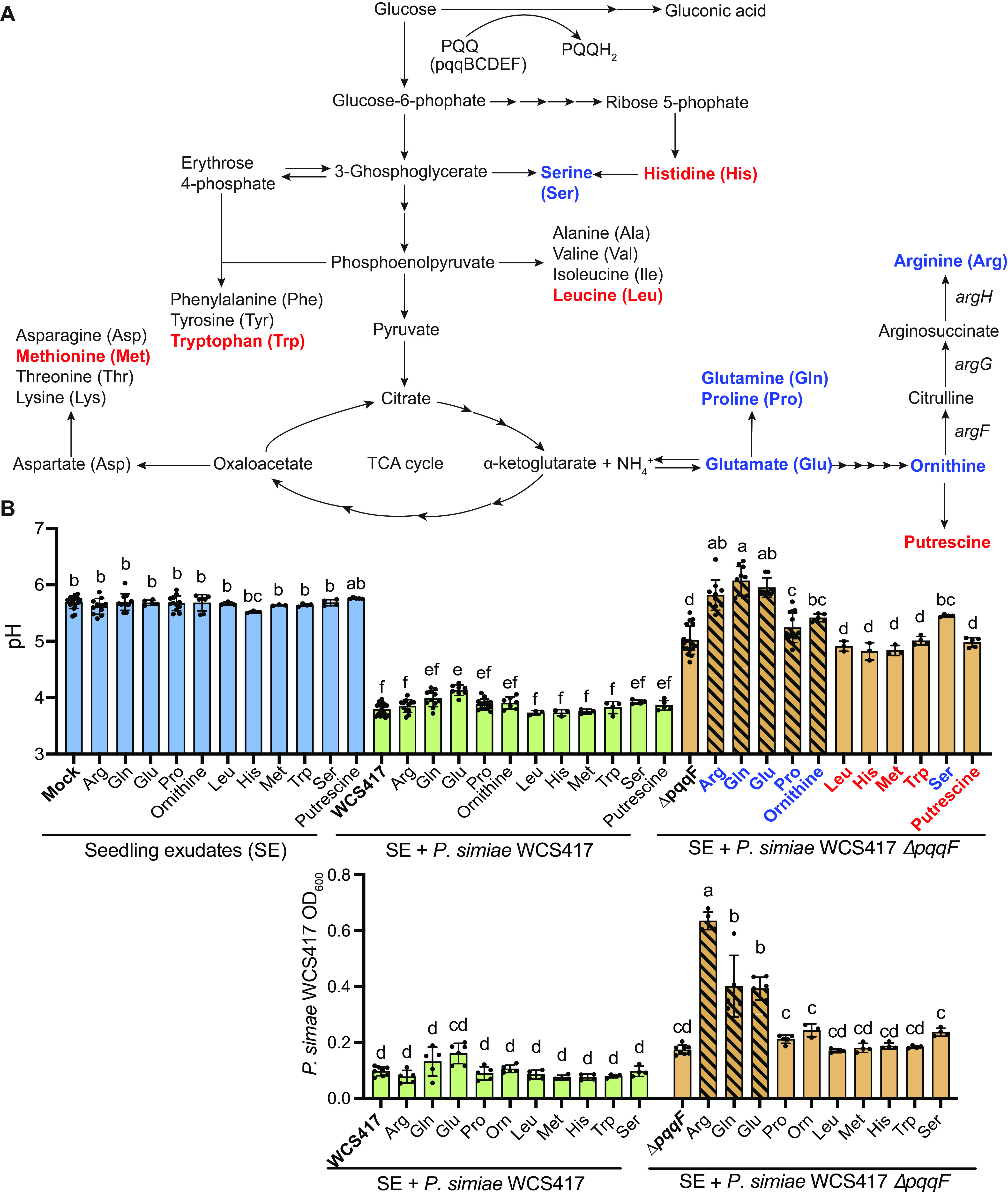
Accumulation of intermediates in the glutamate biosynthesis pathway contributes to rhizosphere alkalization. (A) Glutamate and gluconic acid biosynthesis pathways in Pseudomonas. (B) Inhibiting glutamate biosynthesis by providing arginine, proline, glutamine, and glutamate (striped bars) significantly raised the pH of WCS417 growing in seedling exudates. (C) Inhibiting glutamate biosynthesis by addition of exogenous arginine, glutamine, and glutamate resulted in overgrowth of the WCS417 ΔpqqF mutant. All the experiments were independently repeated at least 3 times. Statistics were calculated by using one-way ANOVA and Tukey’s HSD. Error bars represent mean ± SD, and letters indicate differences at P < 0.05.
We tested whether exogenous arginine would result in an increase or decrease in rhizosphere pH in P. simiae WCS417, the ΔargF mutant, and the ΔpqqF mutant. As exogenous arginine should suppress argF expression, it should mimic the argF mutant, and result in accumulation of alkaline precursors. We found that there was no effect on pH following the addition of 1 mM exogenous arginine in wildtype P. simiae WCS417 growing in seedling exudates (Fig. 2B). However, exogenous arginine fully restored acidification by both the 10E10 and ΔargF mutants, indicating that their inability to acidify is at least partly due to limited availability of arginine (Fig. 2B). Indeed, we confirmed that both mutants have growth defects in seedling exudates, which can be rescued by addition of exogenous arginine (Fig. 2D and E). Intriguingly, we found that arginine resulted in a significant increase in the pH of a ΔpqqF mutant growing in seedling exudates (Fig. 2B). This suggests that inhibition of argF may indeed result in increased pH through accumulation of arginine precursors, but that this effect may be masked by the large amount of gluconic acid produced by P. simiae WCS417.
P. simiae WCS417 produces large amounts of gluconic acid (8), and we only observed an increase in pH with application of exogenous amino acids in the ΔpqqF mutant (Fig. 2B). As a result, we hypothesized that argF might be necessary for lowering pH, but that gluconic acid might mask the effect of argF. If this is the case, we would expect that exogenous arginine would restore growth, but not lower the pH of a double ΔpqqFΔargF mutant growing in seedling exudates. In contrast, if argF only contributes to growth, addition of exogenous arginine should restore pH of the ΔpqqFΔargF mutant to the level of the single ΔpqqF mutant. Consistent with a role of ArgF in rhizosphere acidification, we found that addition of exogenous arginine fully restored growth of a ΔpqqFΔargF or ΔargF mutants to wildtype levels (Fig. 2E). However, arginine addition to the ΔpqqFΔargF double mutant resulted in significantly higher rhizosphere pH than the ΔpqqF mutant alone (Fig. 2B). These data indicate that argF is required for regulation of pH in the rhizosphere independent of growth.
We initially observed that the P. simiae WCS417 ΔpqqF mutant could not acidify the rhizosphere to a level that should result in full suppression of CYP71A12pro:GUS expression. However, the mutant can still fully suppress CYP71A12pro:GUS expression, suggesting a pH-independent mechanism of immunity suppression. While exogenous arginine resulted in a rhizosphere pH of 5.8, at which we would predict no suppression of the CYP71A12pro:GUS reporter (Fig. 1C), we found that arginine treatment of the ΔpqqFΔargF double mutant fully restored immunity suppression (Fig. S3). These data indicate that, while low pH is sufficient to suppress immunity, it is not necessary in WCS417, again supporting that there is a second, pH-independent mechanism of immunity suppression by P. simiae WCS417.
Addition of arginine to the P. simiae WCS417 ΔargF ΔpqqF mutant restores immunity suppression, but not acidification. Exogenous arginine had no effect on the ability of wild type P. simiae WCS417 to suppress expression of the CYP71A12pro:GUS reporter in the presence of flg22, but restored the immunity suppression by the 10E10 mutant, the ΔargF mutant, and the ΔpqqFΔargF double mutant. The addition of arginine to the ΔpqqF and pqqFΔargF double mutant fails to restore acidification (Fig. 2B). Download FIG S3, JPG file, 0.3 MB (330.5KB, jpg) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ornithine accumulation contributes to rhizosphere alkalization.
To test the hypothesis that accumulation of alkaline precursors of arginine, such as glutamate and ammonia could result in an increase in rhizosphere pH (Fig. 3A), we added arginine, proline, or glutamine to seedling exudates containing bacteria, which should result in an accumulation of their precursors, glutamate, and ammonia (17). We also tested the addition of exogenous methionine, leucine, tryptophan, serine, and histidine (which should not affect glutamate catabolism), as controls. To avoid confounding our findings with acidification through gluconic acid biosynthesis, we added amino acids to the P. simiae WCS417 ΔpqqF mutant. We found that exogenous arginine, proline, glutamine, serine and glutamate, but not methionine, leucine, tryptophan, or histidine, significantly raised the pH of the ΔpqqF mutant close to the level of mock seedling exudates (Fig. 3B). Moreover, we found this alkalization phenotype is even more dramatic in PAO1 as arginine, glutamine, and glutamate raised the pH of seedling exudates inoculated with PAO1 pqqF::Tn5 to around 8.0 (Fig. S4). These data indicate that, similar to pathogenic fungi, exogenous arginine, proline, glutamine, or glutamate likely result in rhizosphere alkalization through ammonia accumulation.
Addition of amino acids downstream of glutamine biosynthesis raised the pH of P. aeruginosa PAO1 growing in seedling exudates. Seedling exudates were treated with 1 mM amino acids and mock treated, or treated with PAO1 or the pqqF::Tn5 mutant. This showed that arginine, proline, glutamine, and glutamate significantly raised the pH of the pqqF mutant growing in seedling exudates. All the experiments were independently repeated at least 3 times. Statistics were calculated by using one-way ANOVA and Tukey’s HSD. Error bars represent mean +/− SD, and letters indicate differences at P < 0.05. Download FIG S4, PDF file, 0.4 MB (471.9KB, pdf) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To genetically test whether a loss of arginine, proline, glutamine, or glutamate could specifically result in increased pH of seedling exudates, we selected 6 mutants that have insertions in the genes that are required for amino acid biosynthesis from the P. aeruginosa PAO1 transposon insertion library (Data Set S1, Table S3). We found that proA::Tn5 (deficient in proline biosynthesis) and gltB::Tn5 (deficient in glutamine biosynthesis) neither acidify seedling exudates, nor suppress PTI (Fig. S5), which is consistent with the glutamate biosynthetic pathway being required for rhizosphere acidification. Two additional amino acid insertions in metZ::Tn5 (deficient in methionine biosynthesis) and leuC::Tn5 (deficient in leucine biosynthesis) also resulted in a loss of acidification of seedling exudates and immunity suppression of seedlings, suggesting that leucine and methionine may be limiting for bacterial growth in the rhizosphere (Fig. S5). In addition, serA:: Tn5 (deficient in serine biosynthesis) suppressed host immunity and had decreased pH of seedling exudates, indicating that the rhizosphere may contain enough serine to support bacterial growth (Fig. S5). Interestingly, a hisB::Tn5 mutant (deficient in histidine biosynthesis) can also acidify seedling exudates but induced immunity on its own, further indicating that low pH alone is not sufficient for immunity suppression (Fig. S5). Collectively, these data confirm that the rhizosphere is deficient in glutamate and downstream amino acids, and so bacteria must actively synthesize these in the rhizosphere.
Bacterial auxotrophs cannot suppress plant immunity or acidify the rhizosphere. (A) P. aeruginosa PAO1 amino acid auxotrophic mutants fail to acidify the seedling exudates with the exception of hisB::Tn5 and serA::Tn5 in PAO1. (B) Amino acid auxotrophic mutants fail to suppress flg22-induced CYP71A12pro:GUS expression, with the exception of serA::Tn5. The hisB::Tn5 PAO1 mutant induces CYP71A12pro:GUS expression in the absence of exogenous flg22 treatment. All the experiments were independently repeated 3 times. Statistics were calculated by using one-way ANOVA. Error bar represents mean +/− SD. Download FIG S5, JPG file, 0.4 MB (464.3KB, jpg) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
As fungal alkalization through glutamate secretion is accompanied by increased fungal growth and virulence (18), we tested whether an increase in pH might result in bacterial overgrowth. We observed that arginine, glutamine, and glutamate-mediated alkalization were accompanied by dramatic bacteria overgrowth of both the WCS417 and PAO1 ΔpqqF mutants (Fig. 3C and Fig. S6). To determine if immunity was suppressed at high pH, we tested whether flg22 could still trigger CYP71A12pro:GUS expression at pH 8, and found that expression still occurred (Fig. S7), indicating that overgrowth is occurring at a pH that is not sufficient to suppress immunity. These data indicate that pH correlates with bacteria growth, and that by limiting certain amino acids, it is possible that plants can control the rhizosphere pH and bacterial growth via arginine biosynthesis.
The addition of arginine, glutamine, or glutamate resulted in overgrowth of the P. aeruginosa pqqF mutant. P. aeruginosa PAO1 or the pqqF:Tn5 mutant were grown in seedling exudates, and inoculated with 1 mM of the indicated amino acid. Statistics were calculated by using one-way ANOVA and Tukey’s HSD. Error bars represent mean +/− SD, and letters indicate differences at P < 0.05. Download FIG S6, PDF file, 0.4 MB (430.9KB, pdf) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Flg22-triggered immunity is intact at pH 8. We found, upon raising the pH to 8.0 (the pH associated with bacterial overgrowth) with KOH, that flg22-triggered expression of the CYP71A12pro:GUS promoter, meaning it is not suppressed. Download FIG S7, PDF file, 0.9 MB (900.2KB, pdf) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To test our prediction that rhizosphere alkalization is due to accumulation of ammonia from inhibiting the arginine biosynthesis pathway, we measured the ammonium concentration in seedling exudates. In contrast to our prediction that addition of arginine, glutamine, or glutamate would increase ammonium, we found that the ΔpqqF mutant consumes more ammonium than wild type bacteria in the presence of these amino acids (Fig. S8). This suggests that, when arginine biosynthesis is inhibited, ammonia is converted to a distinct compound that contributes to rhizosphere alkalization.
Ammonium concentration does explain alkalinization in the P. simiae WCS417 ΔpqqF mutant growing in seedling exudates treated with arginine. Ammonium was quantified in seedling exudates (mock), or seedling exudates grown with P. simiae WCS417 or the ΔpqqF mutant. Seedling exudates were treated with 1 mM of the indicated amino acid. Statistics were calculated by using one-way ANOVA and Tukey’s HSD. Error bars represent mean +/− SD, * indicate differences at P < 0.05, and *** indicate differences at P < 0.01. Download FIG S8, PDF file, 0.4 MB (442.7KB, pdf) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
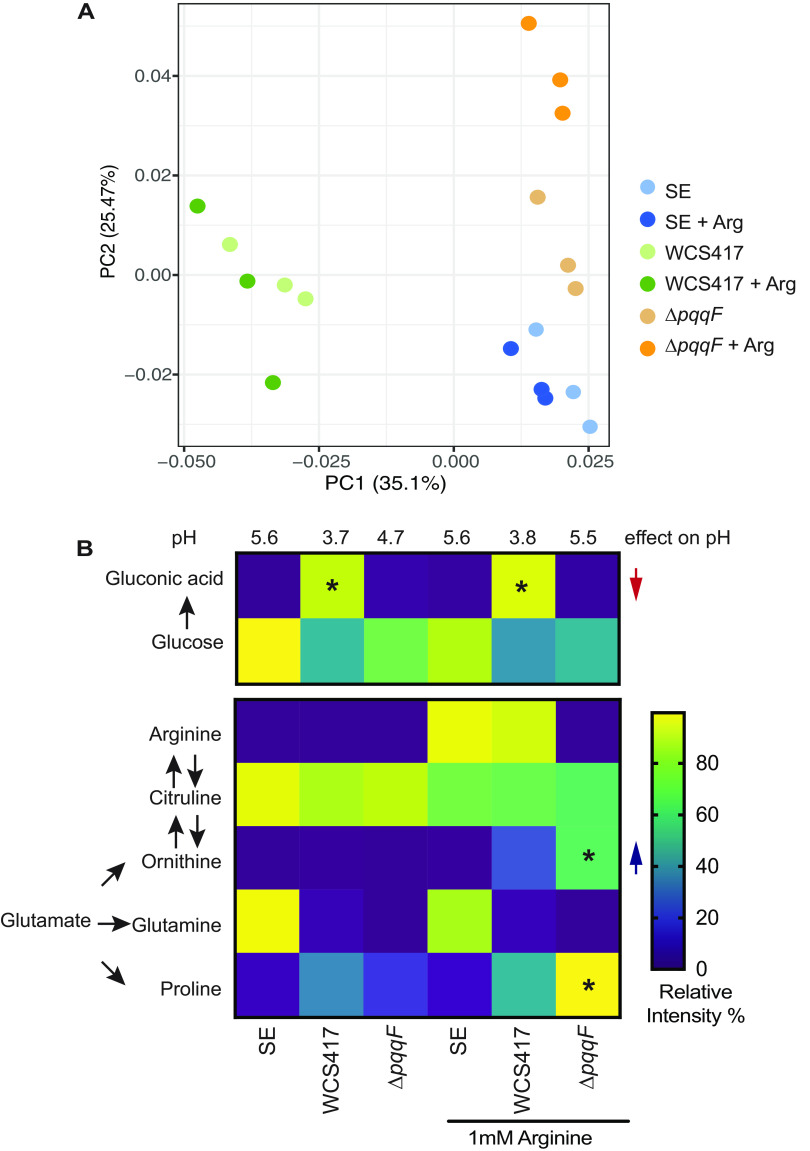
To uncover the compound that caused the rhizosphere alkalization, we performed untargeted metabolomics of seedling exudates, or seedling exudates containing P. simiae WCS417 or the ΔpqqF mutant with or without arginine. We found that mock and bacterial treatments were clearly separated in the pooled principal coordinates analysis (PCoA), as shown in PC1 (P < 0.001) (Fig. 4A). We found that the wild type WCS417 and the ΔpqqF mutant also have distinct metabolite profiles, regardless of the presence of exogenous arginine, as shown in PC2 (P < 0.002), indicating that loss of pqqF has a significant impact on the bacterial metabolism (Fig. 4A). While arginine did not cause a significant global change across all conditions (P < 0.1), we saw that arginine affected the metabolite profile of the ΔpqqF mutant to a greater degree than it did when added to wildtype P. simiae WCS417, as shown in PC2 (Fig. 4B).
FIG 4.
Untargeted metabolomics identified ornithine accumulation is associated with rhizosphere alkalization. (A) Untargeted metabolomics revealed that genotype has the largest impact on the metabolite profiles as seedling exudates, or seedling exudates treated with P. simiae WCS417 or the P. simiae WCS417. The ΔpqqF mutant showed distinct metabolite profile, regardless of addition of arginine. PCoA calculated by Bray-Curtis distances of the pooled samples that appeared in both HILIC positive mode and negative mode. Statistical significance was calculated by permutational multivariate ANOVA (n = 3 for each treatment). (B) Ornithine accumulates in the presence of arginine uniquely in the ΔpqqF mutant, indicating that ΔpqqF mutant converted into exogenous arginine to ornithine, an alkaline molecule. All the experiments were independently repeated at least 3 times. Statistics were calculated by using one-way ANOVA and Tukey’s HSD. Error bars represent mean ± SD, * indicate differences at P < 0.05, and *** indicate differences at P < 0.01.
When we observed individual metabolites, we found that, while wildtype bacteria produced a large amount of gluconic acid, the ΔpqqF mutant produced no detectable gluconic acid (Fig. 4B). We also found that, while seedling exudates with wild type WCS417 contain similar amount of arginine as seedling exudates with no bacteria, the ΔpqqF mutant completely depleted the exogenous arginine, indicating that the ΔpqqF mutant converted the exogenous arginine to a distinct compound (Fig. 4B). Although we did not observe significant differences of citrulline between the WCS417 and ΔpqqF mutant, addition of exogenous arginine resulted in a reduction in the overall amount of citrulline, indicating feedback inhibition of arginine on argF (13) (Fig. 4B).
We queried our untargeted metabolomics data for compounds that uniquely accumulated in the P. simiae WCS417 ΔpqqF mutant in the presence of arginine (Data Set S1). We found that the ΔpqqF mutant uniquely accumulates significantly higher ornithine and proline than the wild type in the presence of exogenous arginine (Fig. 4B). While proline has a neutral pKa, ornithine is an alkaline non-proteogenic amino acid with pKa of 10.29, and it is a substrate of ArgF in the arginine biosynthesis pathway. Although spermidine and putrescine are also alkaline polyamine compounds derived from ornithine, we only detected trace amount of putrescine, spermidine, and spermine that were too low to be quantified. Additionally, exogenous arginine did not significantly change the amount of malic acid in the ΔpqqF mutant relative to wildtype WCS417 (Data Set S1), suggesting that addition of exogenous arginine unlikely affected the TCA cycle. These data suggest that, upon arginine addition to the ΔpqqF mutant, the accumulation of ornithine underlies the rhizosphere alkalization.
To test the possibility that ornithine, but not polyamines, resulted in increase in rhizosphere pH, we added exogenous polyamines or ornithine to WCS417 or the ΔpqqF mutant. We found that the addition of exogenous ornithine or putrescine had no effect on the pH of WCS417 in seedling exudates, and that ornithine, but not putrescine, resulted in a significant increase in the pH of the ΔpqqF mutant (Fig. 3B). However, we observed no effect of ornithine addition to the wildtype P. aeruginosa PAO1 or the PAO1 ΔpqqF mutant (Fig. S4), indicating that the compound that results in alkalization might differ in PAO1 from WCS417.
Acidification-mediated immunity suppression is peptide-specific in roots.
Our data show that Pseudomonas have multiple, partially redundant mechanisms to acidify the rhizosphere, indicating that maintenance of correct pH may be critical to establishing plant immune homeostasis. Previously, we found that acidification to pH 3.7 partially blocked flg22-mediated induction of the MYB51pro:GUS reporter gene, but had no effect on the expression of SA-triggered NPR1pro:GUS and MYB72pro:GUS triggered by WCS417 or WCS358 at pH 3.7 in roots, indicating that acidification can only impair a section of plant innate immunity (8). Thus, we wondered whether acidification suppression of immunity affects MAMP binding to receptors, or affects downstream signaling.
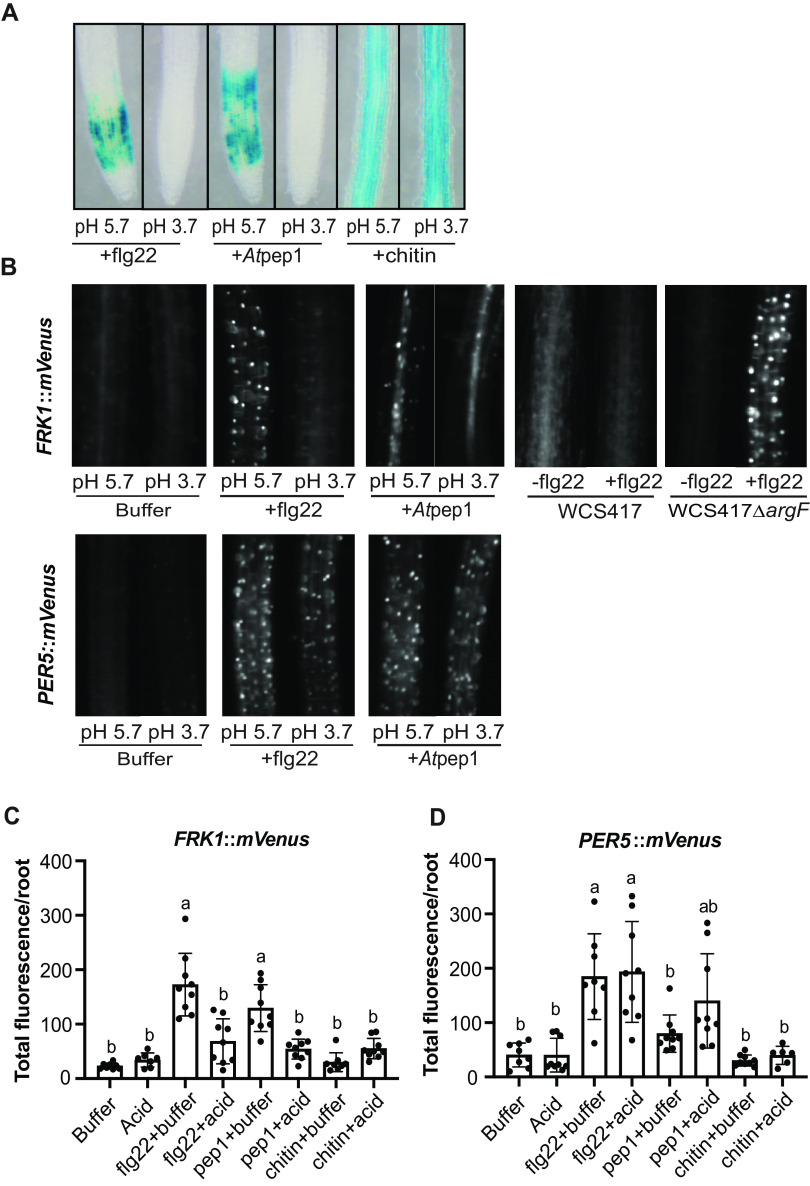
If acidification specifically blocks flg22-induced immunity, then low pH should not affect other MAMP-triggered immunity pathways. We tested the effect of acidification on the damage-associated molecular pattern Atpep1, which is BAK1-dependent (19, 20), and chitin, a sugar polymer from fungal cell walls, which is BAK1-independent in Arabidopsis (21, 22). All 3 MAMPs (flg22, Atpep1, and chitin) share a MAPK cascade (23–25). We tested whether acidification can also suppress chitin- and Atpep1-triggered immunity using the MAMPs marker gene reporters CYP71A12pro:GUS. We found that pH 3.7 abolished Atpep1-, but not chitin-triggered, expression of the CYP71A12pro:GUS reporter (Fig. 5A). These data indicate that acidification likely interferes with the peptide-triggered immunity.
FIG 5.
Acidification only suppresses a sector of plant innate immunity. (A) Acidification blocks flg22- but not Atpep1- or chitin-triggered CYP71A12pro:GUS expression. (B) Acidification and WCS417, but not the WCS417 ΔargF mutant can block flg22-induced FRK1::mVenus fluorescent reporter expression. (C-D) Acidification blocks flg22-induced FRK1::mVenus expression, but not PER5::mVenus expression. All the experiments were independently repeated 3 times. Statistics were calculated by using one-way ANOVA and Tukey’s HSD. Error bars represent mean ± SD, and letters indicate differences at P < 0.05.
To test if low pH affects specific defense responses, we tested whether expression of multiple PTI-induced genes was blocked by using additional PTI-inducible reporters. We used PER5 (AT1G14550), which is a peroxidase (26), and FRK1 (AT2G19190), which encodes a LRR receptor kinase (27). Both genes are flg22 and Atpep1-inducible in the root (28–30). We found that both flg22- and Atpep1-induced FRK1::mVenus expression, which was significantly suppressed by low pH (Fig. 5B, C). In contrast, lower pH did not affect flg22-induced PER5::mVenus expression or Atpep1-induced PER5::mVenus expression (Fig. 5D). These results suggest that acidification can only affect a sector of PTI, and rhizosphere pH may be critical to determine the baseline setting on the plant immune thermostat.
DISCUSSION
Here we report a forward genetic screen that identified a bacterial gene ornithine carbamoyltransferase argF from P. simiae WCS417 that is required for host immunity suppression, colonization, and acidification. The ΔargF mutant is auxotrophic, and exogenous arginine restored ΔargF-mediated host immunity suppression, colonization, and acidification to wildtype levels. This indicates that amino acid biosynthesis plays an important role in rhizosphere colonization. This is not the first time that amino acid biosynthesis has been shown to be necessary for root colonization (7, 31). Interestingly, a previous TnSeq screen found that amino acid auxotrophs, including insertions in argF in P. simiae WCS417, exhibited enhanced fitness in the Arabidopsis rhizosphere (32). We suspect the difference between these findings may be because of a community of transposon insertion mutants in a TnSeq screen, where the presence of other mutants could potentially provide amino acids in trans to auxotrophs. In fact, metabolic exchange, including amino acid cross-feeding among microbes, is characteristic and reciprocal in a microbial community (33–35). Our data suggest that the rhizosphere may be limiting in many amino acids, and that by synthesizing certain amino acids, bacteria will alter the rhizosphere pH and affect plant immune homeostasis.
To disentangle the role of amino acid biosynthesis in rhizosphere acidification from their role in growth, we supplied different amino acids to Pseudomonas pqqF mutants, which cannot produce gluconic acid but retain some rhizosphere acidification (Fig. 2B). We found that only arginine, proline, ornithine, glutamine, and glutamate caused rhizosphere alkalization, and arginine, glutamine, and glutamate caused bacterial overgrowth in the pqqF-deficient mutants. Metabolic profiling suggests that alkalization by Pseudomonas was likely the consequence of ornithine accumulation, an alkaline non-proteogenic amino acid, and the substrate of ArgF. Interestingly, alkalization is associated with overgrowth of pqqF-deficient mutants, which is reminiscent of alkalization-mediated invasive growth in fungi (18). However, Pseudomonas does not utilize ammonia for alkalization, as ammonia-driven alkalization requires carbon deprivation (15, 16). We found alkalization in the presence of glucose, as the ΔpqqF mutant retains more glucose than the wild type because it does not convert glucose to gluconic acid. We found that the overgrowth of the ΔpqqF mutant in the presence of arginine was accompanied by significantly more glucose consumption than the ΔpqqF mutant without exogenous arginine. These data suggest that maintaining a balance of carbon and nitrogen containing compounds is essential for pH homeostasis and bacterial growth regulation.
We found that, while P. simiae WCS417 produces gluconic acid and significantly acidifies the rhizosphere, it is not necessary for immunity suppression in P. simiae WCS417, indicating there is a distinct mechanism of immunity suppression. The ΔpqqF mutant and the ΔpqqFΔargF mutant supplemented with exogenous arginine caused an increased in rhizosphere pH, but still suppressed immunity. This indicates that there are additional pH-independent mechanisms for host immunity manipulation in P. simiae WCS417.
Rhizosphere acidification seems to be a general characteristic of many root-associated microbes (8, 9). Thus, rhizosphere acidification could be a conserved evolutionary trait of root-associated microbes. However, suppression of host immunity may also open the window for pathogens. In this study, we found that acidification can only dampen a sector of immunity, indicating that plant must also evolve novel mechanisms to counteract acidification-mediated immunity suppression, which may act as a selection force for microbial colonization. Recently, roots were shown to contain both acidic (early elongation zone and root tip) and alkaline domains (late elongation zone/root hair zone) (36). Interestingly, we found that acidic pH does not suppress chitin-triggered immunity in the maturation zone, suggesting that acidification-mediated immunity suppression might be zone-dependent (Fig. 5) rather than ligand dependent.
Collectively, host immunity suppression is crucial for host colonization of both commensals and pathogens. Crosstalk between the microbes and the plant immune system is an ongoing process. Our results highlight that, apart from serving as colonization factors and nutrients, bacterial amino acid biosynthesis plays a novel dual role in the rhizosphere acidification and host immunity suppression (Fig. 6). As acidification quenches only a sector of host immunity, it is clear that more mechanisms of host immunity suppression are still to be discovered.
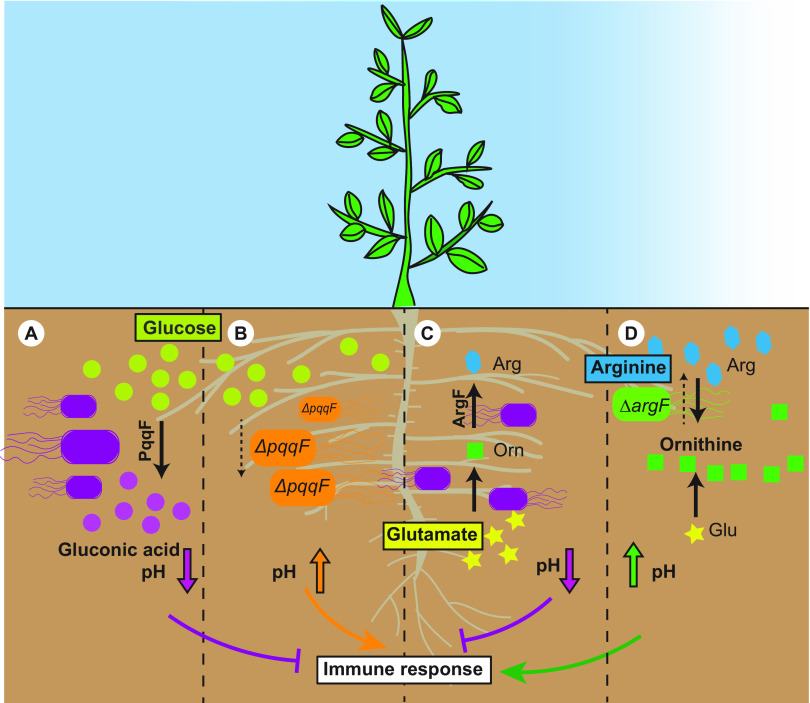
FIG 6.
Model of bacteria-mediated rhizosphere acidification and acidification-mediated plant innate immunity suppression. (A) Bacteria convert plant-derived glucose to gluconic acid in order to acidify the rhizosphere and suppress plant innate immunity. (B) Loss of pqqF in bacteria (orange bacteria) results in a loss of gluconic acid biosynthesis, loss in acidification, and, in some cases, the inability to suppress plant immunity. (C) Bacteria actively synthesize certain amino acids (i.e., arginine) to avoid buildup of alkaline precursors. (D) When plants provide sufficient arginine, ornithine builds up, resulting in rhizosphere alkalization and significant bacterial overgrowth. Overall, this work suggests that maintaining an amino acid-glucose balance is crucial for regulating plant rhizosphere microbiome assembly.
MATERIALS AND METHODS
Plant materials and growth conditions.
We used Arabidopsis thaliana wild type Col-0, CYP71A12pro:GUS reporter (10) and FRK1::mVenus, PER5::mVenus (29) in our study. All the plant material used in our study were grown in the same condition, which was in a climate-controlled growth room at 21°C, 16 h light/8 h dark cycle with light intensity of 100 μM. Plants were grown in the ½ X Murashige and Skoog (MS) media with 1% MES [2-(N-morpholino) ethanesulfonic acid] with 0.5% sucrose, adjusted by KOH to a pH of 5.7 (10).
Bacterial strains and growth condition.
Strains that were used in this study are listed in Data Set S1, Table S2. P. simiae WCS417 and mutants were cultured overnight in LB or King’s B at 28°C with shaking at 180 rpm. P. aeruginosa PAO1 was cultured overnight in LB at 37°C with shaking at 180 rpm. PAO1 transposon insertion mutants were obtained from the 2-allele PAO1 transposon insertion library (12). Wildtype PAO1 and the transposon insertion mutants used in this study were cultured in LB with 25 μg/mL tetracycline at 37°C. Escherichia coli were cultured in 37°C with 15 μg/mL or 100 μg/mL gentamicin, depending on the experiment.
ß-glucosidase histochemical assays.
The reporter lines CYP71A12pro:GUS in the Arabidopsis Col-0 genetic background contains the CYP71A12 promoter driving the expression of the ß-glucosidase (GUS) reporter gene (10). Seeds were grown in 48-well plates for 1 week after surface sterilization, and were grown in the condition described above. Each well contained 300 μL ½x MS media and 0.5% sucrose. On day 8, the media was replaced by fresh ½x MS with 0.5% sucrose media. Bacteria were grown overnight in LB, washed in 10 mM MgSO4, and serially diluted to an OD600 of 0.02 in 10 mM MgSO4. On day 9, 30 μL bacteria were added to each well (final OD600 of 0.002), and the plates were returned to the growth room for at least 18 h before adding flg22. On day 10, 500 μM flg22 was added to a final concentration of 500 nm, and the media was replaced with GUS staining solution after 4.5 h of incubation. The GUS staining solution was made fresh at a final concentration of 0.5 M sodium phosphate buffer (pH 7), 0.5M EDTA, 50 mM potassium ferricyanide, 50 mM potassium ferrocyanide, 50 mM X-Gluc (5-bromo-4-chloro-3-indolyl-beta-d-glucuronic acid), and 10 μL Triton X-100. Plates were then incubated at 37°C without light until the control roots treated with flg22 developed a visible blue color (approximately 3 to 4 h). Finally, to clear the tissue, the GUS stain was replaced with 95% ethanol, and washed with water afterwards. Images were taken with a Macro Zoom Fluorescence Microscope MVX10.
Bacterial growth curves.
Overnight cultures of bacteria were grown in LB, then were serially diluted to an OD600 of 0.2 in 10 mM MgSO4 for growth curves. Growth curves were performed by adding 10 μL of the diluted culture to 90 μL rich media (LB), minimal media (M9 salts supplemented with 30 mM succinate), or seedling exudates (M9 salts supplemented with 30 mM succinate, with or without 1 mM arginine). Bacteria growth was quantified by measuring OD600 on a Versamax plate reader (Molecular Devices). Data presented in this study represent the average of 3 biological replicates.
WCS417 EMS mutant library construction and screening.
P. simiae WCS417r (rif resistant variant) was mutagenized by spinning down and washing an overnight culture, and exposing it to 1, 2, or 4% EMS for 1 h. Mutagenized cells were plated on King’s B with 50 μg/mL streptomycin, and it was found that after treatment with 4% EMS there was ~ 100-fold increase in the number of resistant cells relative to the parental strain, so these cells were used for library construction. For library construction, mutagenized cells were plated on LB + rifampicin 50 μg/mL, and individual colonies were placed into wells of 96-well deep well plates in LB media. Each plate contained 92 EMS mutants, and 4 wells contained the parental strain as positive controls. After overnight growth, 75 μL of LB containing library bacteria were pipetted into a fresh 96-well plate, and 25 μL of 80% glycerol was added. The library was stored at −80°C.
To screen the library, seedlings were grown in 96-well plates in MS media as described above. Three seedlings were grown in each well containing 100 μL MS media. The library was stamped onto rectangular plates containing solid LB media, and then subcultured into 96-well deep bottom plates containing LB. After overnight growth, the OD600 of 4 independent wells was measured, and the average was taken. This was used to calculate an approximate dilution factor to dilute all 96 wells to a final OD600 = 0.05. A total of 10 μL of the diluted culture was added to each well, containing 90 μL MS, for a final average bacterial concentration of 0.005. The screen was repeated in duplicate, and candidates that failed to suppress immunity in both replicates were retested.
Mapping WCS417 EMS mutations.
To map WCS417 EMS mutations, we sequenced the genomes of the parental line used to make the library, as well as the genomes of each individual mutant. Genomic DNA was extracted from WCS417 and the 10E10 EMS mutant with the Puregene Core Kit (Qiagen). A PE150 short insert library was prepared and sequenced on an Illumina HiSeq 2500 (Novogene). After adapter trimming with Cutadapt (37), reads were aligned to the WCS417 genome using the Bowtie2 (38) aligner with default parameters. Variant calling was performed with BCFtools (39). Low quality SNPs with a quality score under 20 were filtered out, and SNPs found in the parent strain were discarded from consideration.
Bacteria mutant complementation.
Complementation of the 10E10 mutant was performed by PCR amplifying the coding sequence and native promoter of argF in WCS417. The PCR product containing HindIII and BamHI restriction sites was ligated to the plasmid pBBR1MSC5, and the ligation product was transformed into the competent cell E.coli DH5α, and plated on gentamicin 100 μg/mL plates for selection of positive colonies. Positive colonies containing the proargF:pBBR1MSC5 construct were confirmed by colony PCR, and further confirmed by Sanger sequencing. The confirmed pBBR1MCS-5::ProargF-argF construct was then transformed into the 10E10 mutant.
Generation of deletion mutants in WCS417.
Clean deletions of argF or pqqF in WCS417 were made using a double-recombination method in Gram-negative bacteria using counter selection with sacB (40). Two sets of primers were designed to amplify the 500 bp flanking region upstream and downstream of argF or pqqF. Primer 1 with the restriction enzymes site HinIII, and primer 4 with the restriction enzyme site BamHI are the left and right primers of the upstream and downstream flanking region of the target gene, respectively. Primer 2 and primer 3 are the right and left primers of the upstream and downstream flanking region of the target gene, respectively. Both primer 2 and primer 3 consist of 15 bp of region upstream and 15 bp of region downstream of the target gene. Thus, primer pairs primer 1 and primer 2, and primer 3 and primer 4 were used to amplify the 500 bp regions upstream and downstream of the target gene, respectively. Overlap PCR was performed with the upstream and downstream PCR products, which were digested with HindIII and BamHI, and ligated to the pEXG2 suicide vector containing sacB (41), and then transformed into E.coli DH5α. The positive colonies were selected for plating on LB plates with gentamicin 15 μg/mL, and then confirmed by colony PCR. The deletion constructs for argF or pqqF were further confirmed by Sanger sequencing. The confirmed argF or pqqF deletion constructs were then transformed into the competent SM10λ cells, and were selected on LB plates with gentamicin 15 μg/mL. Conjugation of the SM10λ containing the deletion construct and WCS417 was performed, and the transconjugants were selected on plates containing nalidixic acid 15 μg/mL and gentamicin 100 μg/mL. Positive colonies were re-streaked, and cultured overnight in plain LB. Cell pellets were diluted to 10X and 100X, and were each plated onto 10% sucrose plates and gentamicin 100 μg/mL plates to select for the second recombination. The ΔargFΔpqqF double mutant was made by conjugating the ΔpqqF mutant with the SM10λ strain containing the argF-pPEXG2 deletion construct, and the selection was performed as described above.
Seedling exudates.
To generate seedling exudates, A. thaliana Col-0 seeds were grown in half strength MS media containing 0.5% sucrose for 7 days, as described above. The seedling exudates were collected from all the wells, and immediately syringe filtered with a 0.22 μm filter and frozen at −20°C.
Amino acid solutions.
A total of 100 mM (100X) stock solutions of l-arginine, L-proline, l-glutamine, l-glutamate, l-ornithine, l-leucine, l-methionine, l-histidine, L-tryptophan, and l-serine were made in water and filter sterilized using a 0.22 μm filter, before storing at 4°C.
pH assay in seeding exudates.
Bacteria were grown in LB overnight and were serially diluted to OD600 0.02 in 10 mM MgSO4. Bacteria were inoculated into 24-well plates containing seedling exudates to a final OD of 0.002, and the plates were incubated in a 28°C or 37°C incubator for 18 h. Final concentration of 1 mM amino acids (100X dilution of stocks) were added when indicated. Then, 1 mL of culture was directly taken from each well, and the OD was measured by a spectrophotometer. Each experiment included 3 technical replicates, and was independently repeated at least three times.
Ammonium quantification.
Ammonium concentration in the pH assay was measured by an Ammonia assay kit (Sigma). The kit provides reagents that reacts with ammonia/ammonium ions, which produces fluorescence signals that are proportional to the ammonia concentration in the sample. A total of 1 mL of each sample was taken from the pH assay and centrifuged for 5 min at 14,000 rpm. Then, 10 μL of the supernatant was used for ammonia quantification following the manufacturer’s instructions. Plates were read by a Spectramax plate reader (λex = 360/λem = 450 nm).
Metabolomic profiling.
The samples described above in the “pH assays in seedling exudates” were used for untargeted metabolomics analysis. A 60 μL aliquot of each media sample was diluted with 60 μL of acetonitrile (ACN), and vortexed for 10 sec. The mixture was then centrifuged at 14000 rpm for 15 min at 4°C. The supernatant was transferred to glass inserts in 2 mL autosampler vials for liquid chromatography-mass spectrometry (LC-MS) analysis. The analysis was performed using a Bruker Impact II Ultra-High Resolution Qq-Time-of-Flight mass spectrometer coupled to an Agilent 1290 Infinity Liquid Chromatography system. For each sample, 2 μL was injected onto an EMD Millipore SeQuant ZIC-pHILIC column (200 Å, 5 μm, 2.1 × 150 mm) for hydrophilic interaction chromatography (HILIC) separation. For negative ionization mode, the mobile phases (MPs) were 10 mM ammonium acetate in 95/5 water/ACN at pH 9.8 (MP A), and 95/5 ACN/water (MP B). The MPs for positive ionization were the same, except MP A was pH 4.8. The same LC gradient was used for both ionization modes at a flow rate of 0.150 mL/min. The separation gradient, described in percentage of MP B, started at 95%, dropped to 5% over 20 min, and then increased to 95% over 1 min; the column was equilibrated for 14 min between injections. A pooled quality control sample was injected at 5 different volumes for metabolic signal correction, and high-quality feature selection (42). Sodium formate was injected for mass calibration. The mass spectrometer was operated in Auto MS/MS mode. The ionization capillary voltage was 3.6 kV for negative mode, and 4.5 kV for positive mode. The nebulizer gas was 1.6 bar. The dry gas was 7 L/min, and the dry temperature was 220°C. The mass range collected was from 70 to 1500 m/z at 8 Hz. The collision energy was 20 to 50 eV. The acquired data were calibrated and processed with MS-DIAL (ver. 4.80). The resulting metabolite intensity tables were exported for statistical analysis.
Fluorescence reporter imaging and quantification.
FRK1::mVenus and PER5::mVenus seedlings were grown in 600 μL of ½x MS media with 0.5% sucrose and a pH of 5.7 in 24-well plates. On day 8, the media was replaced with 540 μL of fresh ½x MS with 0.5% sucrose at a pH 5.7. On day 9, MAMPs were added to a final concentration of 500 μM flg22, 100 nM Atpep1, or 0.1 mg/mL chitin were added. For low pH conditions, ½x MS with 0.5% sucrose with pH 3.7 were added, along with the elicitors described above, and incubated for 4.5 h. Images were taken with a Macro Zoom Fluorescence Microscope MVX10 microscope.
ACKNOWLEDGMENTS
This work was supported by NSERC Discovery Grants NSERC-RGPIN-2016-04121 and NSERC-RGPIN-2021-03587, and Weston Seeding Food Innovation grants to C.H.H. Y.L. was supported by a Chinese Graduate Scholarship Council award and an NSERC CREATE-PRoTECT award. Early stages of this work were supported by NIH grant R37 GM48707, and NSF grants MCB-0519898 and IOS-0929226, which were awarded to Frederick M. Ausubel.
We thank Niko Geldner for providing the fluorescent reporter lines FRK1::mVenus and PER5::mVenus. We thank Yi Song and David Thoms training in the gnotobiotic plate assay and microscopy experiment, respectively. We thank Sarzana Hossain and Nicole R. Wang for insights and critical reading of the manuscript.
Contributor Information
Cara H. Haney, Email: cara.haney@ubc.ca.
David S. Guttman, University of Toronto
REFERENCES
- 1.Berendsen RL, Pieterse CMJ, Bakker PAHM. 2012. The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Jones JDG, Dangl JL. 2006. The plant immune system. Nature 444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Macho AP, Zipfel C. 2015. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr Opin Microbiol 23:14–22. doi: 10.1016/j.mib.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Brooks DM, Bender CL, Kunkel BN. 2005. The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol Plant Pathol 6:629–639. doi: 10.1111/j.1364-3703.2005.00311.x. [DOI] [PubMed] [Google Scholar]
- 5.Pel MJC, Van Dijken AJH, Bardoel BW, Seidl MF, Van Der Ent S, Van Strijp JAG, Pieterse CMJ. 2014. Pseudomonas syringae evades host immunity by degrading flagellin monomers with alkaline protease AprA. Mol Plant Microbe Interact 27:603–610. doi: 10.1094/MPMI-02-14-0032-R. [DOI] [PubMed] [Google Scholar]
- 6.Colaianni NR, Parys K, Lee H-S, Conway JM, Kim NH, Edelbacher N, Mucyn TS, Madalinski M, Law TF, Jones CD, Belkhadir Y, Dangl JL. 2021. A complex immune response to flagellin epitope variation in commensal communities. Cell Host Microbe 29:635–649. doi: 10.1016/j.chom.2021.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Beskrovnaya P, Melnyk RA, Hossain SS, Khorasani S, O’sullivan LR, Wiesmann CL, Bush J, Richard JD, Haney CH. 2018. A genome-wide screen identifies genes in rhizosphere-associated Pseudomonas required to evade plant defenses. mBio 9:1–17. doi: 10.1128/mBio.00433-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu K, Liu Y, Tichelaar R, Savant N, Lagendijk E, van Kuijk SJL, Stringlis IA, van Dijken AJH, Pieterse CMJ, Bakker PAHM, Haney CH, Berendsen RL. 2019. Rhizosphere-associated Pseudomonas suppress local root immune responses by gluconic acid-mediated lowering of environmental pH. Curr Biol 29:3913–3920.e4. doi: 10.1016/j.cub.2019.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Teixeira PJPL, Colaianni NR, Law TF, Conway JM, Gilbert S, Li H, Salas-González I, Panda D, Del Risco NM, Finkel OM, Castrillo G, Mieczkowski P, Jones CD, Dangl JL. 2021. Specific modulation of the root immune system by a community of commensal bacteria. Proc Natl Acad Sci USA 118:e2100678118. doi: 10.1073/pnas.2100678118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel FM. 2010. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22:973–990. doi: 10.1105/tpc.109.069658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Simine CD, Sayer JA, Gadd GM. 1998. Solubilization of zinc phosphate by a strain of Pseudomonas fluorescens isolated from a forest soil. Biol Fertil Soils 28:87–94. doi: 10.1007/s003740050467. [DOI] [Google Scholar]
- 12.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu CD, Yang Z, Li W. 2004. Transcriptome analysis of the ArgR regulon in Pseudomonas aeruginosa. J Bacteriol 186:3855–3861. doi: 10.1128/JB.186.12.3855-3861.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh Y, Soldati L, Stalon V, Falmagne P, Terawaki Y, Leisinger T, Haas D. 1988. Anabolic ornithine carbamoyltransferase of Pseudomonas aeruginosa: nucleotide sequence and transcriptional control of the argF structural gene. J Bacteriol 170:2725–2734. doi: 10.1128/jb.170.6.2725-2734.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes TR, Segorbe D, Prusky D, Di Pietro A. 2017. How alkalinization drives fungal pathogenicity. PLoS Pathog 13:e1006621. doi: 10.1371/journal.ppat.1006621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vylkova S. 2017. Environmental pH modulation by pathogenic fungi as a strategy to conquer the host. PLoS Pathog 13:e1006149. doi: 10.1371/journal.ppat.1006149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikami Y, Yoneda H, Tatsukami Y, Aoki W, Ueda M. 2017. Ammonia production from amino acid-based biomass-like sources by engineered Escherichia coli. AMB Express 7:83. doi: 10.1186/s13568-017-0385-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masachis S, Segorbe D, Turrà D, Leon-Ruiz M, Fürst U, El Ghalid M, Leonard G, López-Berges MS, Richards TA, Felix G, Di Pietro A. 2016. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat Microbiol 1:1–9. doi: 10.1038/nmicrobiol.2016.73. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA. 2010. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22:508–522. doi: 10.1105/tpc.109.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaguchi Y, Pearce G, Ryan CA. 2006. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc Natl Acad Sci USA 103:10104–10109. doi: 10.1073/pnas.0603729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, Stacey G. 2014. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 3:e03766. doi: 10.7554/eLife.03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan J, Zhang X-C, Neece D, Ramonell KM, Clough S, Kim S, Stacey MG, Stacey G. 2008. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rich-Griffin C, Eichmann R, Reitz MU, Hermann S, Woolley-Allen K, Brown PE, Wiwatdirekkul K, Esteban E, Pasha A, Kogel KH, Provart NJ, Ott S, Schäfer P. 2020. Regulation of cell type-specific immunity networks in Arabidopsis roots. Plant Cell 32:2742–2762. doi: 10.1105/tpc.20.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada K, Yamashita-Yamada M, Hirase T, Fujiwara T, Tsuda K, Hiruma K, Saijo Y. 2016. Danger peptide receptor signaling in plants ensures basal immunity upon pathogen-induced depletion of BAK 1. EMBO J 35:46–61. doi: 10.15252/embj.201591807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bi G, Zhou Z, Wang W, Li L, Rao S, Wu Y, Zhang X, Menke FLH, Chen S, Zhou J-M. 2018. Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell 30:1543–1561. doi: 10.1105/tpc.17.00981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tognolli M, Penel C, Greppin H, Simon P. 2002. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288:129–138. doi: 10.1016/s0378-1119(02)00465-1. [DOI] [PubMed] [Google Scholar]
- 27.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. 2002. Map kinase signalling cascade in Arabidopsis innate immunity. Nature 415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 28.Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou J-M, Chai J. 2013. Structural basis for flg22-Induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342:624–628. doi: 10.1126/science.1243825. [DOI] [PubMed] [Google Scholar]
- 29.Poncini L, Wyrsch I, Tendon VD, Vorley T, Boller T, Geldner N, Métraux JP, Lehmann S. 2017. In roots of Arabidopsis thaliana, the damage-associated molecular pattern AtPep1 is a stronger elicitor of immune signalling than flg22 or the chitin heptamer. PLoS One 12:e0185808. doi: 10.1371/journal.pone.0185808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou J-M. 2013. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci USA 110:6205–6210. doi: 10.1073/pnas.1215543110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons M, Permentier HP, De Weger LA, Wijffelman CA, Lugtenberg BJJ. 1997. Amino acid synthesis is necessary for tomato root colonization by Pseudomonas fluorescens strain WCS365. Mol Plant Microbe Interact 10:102–106. doi: 10.1094/MPMI.1997.10.1.102. [DOI] [Google Scholar]
- 32.Cole BJ, Feltcher ME, Waters RJ, Wetmore KM, Mucyn TS, Ryan EM, Wang G, Ul-Hasan S, McDonald M, Yoshikuni Y, Malmstrom RR, Deutschbauer AM, Dangl JL, Visel A. 2017. Genome-wide identification of bacterial plant colonization genes. PLoS Biol 15:e2002860. doi: 10.1371/journal.pbio.2002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mee MT, Collins JJ, Church GM, Wang HH. 2014. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci USA 111:E2149–E2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zengler K, Zaramela LS. 2018. The social network of microorganisms — how auxotrophies shape complex communities. Nat Rev Microbiol 16:383–390. doi: 10.1038/s41579-018-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelly EE, Fischer AM, Collins CH. 2021. Drawing up a collaborative contract: Amino acid cross-feeding between interspecies bacterial pairs. Biotechnol Bioeng 118:3138–3149. doi: 10.1002/bit.27837. [DOI] [PubMed] [Google Scholar]
- 36.Bc Serre N, Wernerová D, Vittal P, Dubey SM, Medvecká E, Jelínková A, Petrášek J, Grossmann G, Fendrych M. 2022. The AUX1-AFB1-CNGC14 module establishes longitudinal root surface pH profile. BioRxiv. doi: 10.1101/2022.11.23.517700. [DOI] [PMC free article] [PubMed]
- 37.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 38.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. 2015. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu H, Huan T. 2021. Patterned signal ratio biases in mass spectrometry-based quantitative metabolomics. Anal Chem 93:2254–2262. doi: 10.1021/acs.analchem.0c04113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P. simiae WCS417 can solubilize ZnSO4, while the mutant 4E4 cannot. Rescreening the P. simiae WCS417 EMS library on media containing ZnSO4 and glucose identified a single mutant that could not solubilize zinc. Sequencing the 4E4 mutant identified point mutations in both pqqF and pqqB. The 7A1 strain can solubilize ZnSO4, and is shown as a positive control. Download FIG S1, PDF file, 0.5 MB (528.7KB, pdf) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The process of HILIC+/− shows results from metabolomics analysis. Data Set S1, Table S1, A total of 35 non-synonymous candidate genes from genome mapping of the 10E10 WCS417 mutant. Data Set S1, Table S2, total of 48 non-synonymous candidate genes from genome mapping of the 4E4 WCS417 mutant. Data Set S1, Table S3 Stains used in this study. Download Data Set S1, XLSX file, 0.5 MB (546.8KB, xlsx) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A screen of P. aeruginosa PAO1 transposon insertion mutants in candidate genes with point mutations in P. simiae WCS417 10E10 identified argF, which cannot acidify seedling exudates. Genome sequencing of 10E10 identified 35 non-synonymous candidate genes (Data Set S1, Table S1). Orthologs were identified in PAO1. argF::Tn5-1/-2: PS417_05595, ornithine carbamoyltransferase; maeB: PS417_01950, malate dehydrogenase; PA4465: PS417_04330, glmZ(sRNA)-inactivating NTPase; PA0214: PS417_26590, malonate decarboxylase subunit epsilon; PA0575: PS417_25885, diguanylate cyclase; and PA0148: PS417_03205, adenine deaminase. Statistics were calculated by using two-way ANOVA and Tukey’s HSD. Error bars represent mean +/− SD, and * indicate differences at P < 0.05. Download FIG S2, PDF file, 0.2 MB (208.5KB, pdf) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Addition of arginine to the P. simiae WCS417 ΔargF ΔpqqF mutant restores immunity suppression, but not acidification. Exogenous arginine had no effect on the ability of wild type P. simiae WCS417 to suppress expression of the CYP71A12pro:GUS reporter in the presence of flg22, but restored the immunity suppression by the 10E10 mutant, the ΔargF mutant, and the ΔpqqFΔargF double mutant. The addition of arginine to the ΔpqqF and pqqFΔargF double mutant fails to restore acidification (Fig. 2B). Download FIG S3, JPG file, 0.3 MB (330.5KB, jpg) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Addition of amino acids downstream of glutamine biosynthesis raised the pH of P. aeruginosa PAO1 growing in seedling exudates. Seedling exudates were treated with 1 mM amino acids and mock treated, or treated with PAO1 or the pqqF::Tn5 mutant. This showed that arginine, proline, glutamine, and glutamate significantly raised the pH of the pqqF mutant growing in seedling exudates. All the experiments were independently repeated at least 3 times. Statistics were calculated by using one-way ANOVA and Tukey’s HSD. Error bars represent mean +/− SD, and letters indicate differences at P < 0.05. Download FIG S4, PDF file, 0.4 MB (471.9KB, pdf) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial auxotrophs cannot suppress plant immunity or acidify the rhizosphere. (A) P. aeruginosa PAO1 amino acid auxotrophic mutants fail to acidify the seedling exudates with the exception of hisB::Tn5 and serA::Tn5 in PAO1. (B) Amino acid auxotrophic mutants fail to suppress flg22-induced CYP71A12pro:GUS expression, with the exception of serA::Tn5. The hisB::Tn5 PAO1 mutant induces CYP71A12pro:GUS expression in the absence of exogenous flg22 treatment. All the experiments were independently repeated 3 times. Statistics were calculated by using one-way ANOVA. Error bar represents mean +/− SD. Download FIG S5, JPG file, 0.4 MB (464.3KB, jpg) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The addition of arginine, glutamine, or glutamate resulted in overgrowth of the P. aeruginosa pqqF mutant. P. aeruginosa PAO1 or the pqqF:Tn5 mutant were grown in seedling exudates, and inoculated with 1 mM of the indicated amino acid. Statistics were calculated by using one-way ANOVA and Tukey’s HSD. Error bars represent mean +/− SD, and letters indicate differences at P < 0.05. Download FIG S6, PDF file, 0.4 MB (430.9KB, pdf) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Flg22-triggered immunity is intact at pH 8. We found, upon raising the pH to 8.0 (the pH associated with bacterial overgrowth) with KOH, that flg22-triggered expression of the CYP71A12pro:GUS promoter, meaning it is not suppressed. Download FIG S7, PDF file, 0.9 MB (900.2KB, pdf) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ammonium concentration does explain alkalinization in the P. simiae WCS417 ΔpqqF mutant growing in seedling exudates treated with arginine. Ammonium was quantified in seedling exudates (mock), or seedling exudates grown with P. simiae WCS417 or the ΔpqqF mutant. Seedling exudates were treated with 1 mM of the indicated amino acid. Statistics were calculated by using one-way ANOVA and Tukey’s HSD. Error bars represent mean +/− SD, * indicate differences at P < 0.05, and *** indicate differences at P < 0.01. Download FIG S8, PDF file, 0.4 MB (442.7KB, pdf) .
Copyright © 2023 Liu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.