Abstract
Few studies have demonstrated reproducible gene–diet interactions (GDIs) impacting metabolic disease risk factors, likely due in part to measurement error in dietary intake estimation and insufficient capture of rare genetic variation. We aimed to identify GDIs across the genetic frequency spectrum impacting the macronutrient–glycemia relationship in genetically and culturally diverse cohorts. We analyzed 33,187 participants free of diabetes from 10 National Heart, Lung, and Blood Institute Trans-Omics for Precision Medicine program cohorts with whole-genome sequencing, self-reported diet, and glycemic trait data. We fit cohort-specific, multivariable-adjusted linear mixed models for the effect of diet, modeled as an isocaloric substitution of carbohydrate for fat, and its interactions with common and rare variants genome-wide. In main effect meta-analyses, participants consuming more carbohydrate had modestly lower glycemic trait values (e.g., for glycated hemoglobin [HbA1c], −0.013% HbA1c/250 kcal substitution). In GDI meta-analyses, a common African ancestry–enriched variant (rs79762542) reached study-wide significance and replicated in the UK Biobank cohort, indicating a negative carbohydrate–HbA1c association among major allele homozygotes only. Simulations revealed that >150,000 samples may be necessary to identify similar macronutrient GDIs under realistic assumptions about effect size and measurement error. These results generate hypotheses for further exploration of modifiable metabolic disease risk in additional cohorts with African ancestry.
Article Highlights
We aimed to identify genetic modifiers of the dietary macronutrient–glycemia relationship using whole-genome sequence data from 10 Trans-Omics for Precision Medicine program cohorts.
Substitution models indicated a modest reduction in glycemia associated with an increase in dietary carbohydrate at the expense of fat.
Genome-wide interaction analysis identified one African ancestry–enriched variant near the FRAS1 gene that may interact with macronutrient intake to influence hemoglobin A1c.
Simulation-based power calculations accounting for measurement error suggested that substantially larger sample sizes may be necessary to discover further gene–macronutrient interactions.
Introduction
Diet is an established modifiable factor associated with risk of type 2 diabetes (T2D) and related cardiometabolic diseases (1). However, evidence is mixed regarding the ideal dietary macronutrient composition for risk reduction. Dietary interventions with differing proportions of energy from carbohydrates versus fat have shown varied efficacy for T2D risk reduction and substantial between-person heterogeneity in effects on cardiometabolic risk factors (2–4). Further, acute glycemic responses to meals with specific macronutrient composition are reproducible within individuals (5,6). Genetically different mouse strains have varying sensitivity of glycemic biomarkers to a high-fat diet (7) and to human-relevant dietary patterns (8). Retrospective analysis of human trials manipulating macronutrient intake has found genetic modifiers of glycemic response (9). Taken together, such studies suggest that genetics could be a key contributor to variability in the association between dietary macronutrient composition and glycemic health.
Gene–diet interaction (GDI) studies aim to identify genetic variants that modify the association between dietary behaviors and health. Furthermore, GDI studies support differential associations of dietary factors with glycemic traits according to genotypes, using both hypothesis-driven (10) and hypothesis-free (11,12) strategies. However, in general, discovery and replication of GDI with T2D risk has been poor, possibly due to measurement error in assessing habitual diet, low statistical power for interaction analysis, and biological and behavioral heterogeneity across populations (13). Additionally, to date, there has been little exploration of GDIs involving rare genetic variants, which affect a smaller proportion of the population but may have larger effect sizes (14).
Our primary aim was to discover novel putative genetic modifiers for the association between dietary macronutrient composition and glycemic traits. To this end, we performed a GDI analysis using common and rare genetic variants in >30,000 individuals with diverse ancestral backgrounds from the National Heart, Lung, and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) program. We focused on modeling a dietary carbohydrate–fat exchange, which can be reasonably assessed via self-reported diet questionnaires and can be straightforwardly modified in the context of a healthful diet. Furthermore, the use of whole-genome sequencing (WGS) permitted the analysis of rare variants using set-based association tests. As a secondary aim, we sought to inform subsequent GDI research by exploring the impact of dietary exposure measurement error on statistical power in the context of realistic effect size estimates.
Research Design and Methods
WGS
WGS was conducted through the NHLBI TOPMed program (Freeze 8 data release). Sequencing and alignment to the GRCh38 reference genome was performed at seven centers across the U.S.: Broad Institute of MIT and Harvard, Northwest Genomics Center, New York Genome Center, Illumina Genomic Services, Psomagen (formerly Macrogen), Baylor College of Medicine Human Genome Sequencing Center, and McDonnell Genome Institute at Washington University. Data harmonization and joint variant discovery and genotype calling were performed within the TOPMed Informatics Research Center at the University of Michigan. Sequence quality control filters were as follows: estimated DNA sample contamination <10% and at least 95% of the genome having coverage of at least 10 times. After genotyping, variants were further filtered for Mendelian inconsistency (based on a support vector machine classifier) and excess heterozygosity. Additional sample quality control was performed within the Data Coordinating Center at the University of Washington, including: matching sex as annotated and inferred from WGS, concordance of WGS genotypes with prior array-based “fingerprints,” and agreement of inferred relatedness with expectations based on pedigrees. Additional details on the processing steps are available at: https://www.nhlbiwgs.org/topmed-whole-genome-sequencing-methods-freeze-8.
Global measures of ancestry and relatedness were calculated on the entire TOPMed Freeze 8 sample by the TOPMed Data Coordinating Center. Genetic principal components reflecting ancestry were calculated using the PC-AiR method (allowing for related individuals) (15), and kinship matrices were calculated using the PC-Relate method (accounting for principal components) (16), both from the GENESIS R package. A sparse genetic relationship matrix containing only relationships of degree four or closer was extracted for analysis. Samples were grouped into race/ethnicity categories based on cohort-reported values. Some individuals are represented in multiple studies in the TOPMed program (e.g., Jackson Heart Study and Atherosclerosis Risk in Communities [ARIC] study); in such cases, one individual from each duplicate pair was removed prior to analysis.
Harmonization of Glycemic Traits
Phenotypes were harmonized across the 10 studies based on a protocol developed within the TOPMed Diabetes Working Group. Glycemic traits, including fasting glucose (FG; millimoles per liter), fasting insulin (FI; picomoles per liter), and glycated hemoglobin (HbA1c; percentage), were collected where available. Fasting (for FG and FI) was defined as at least 8 h without food or drink. FG measurements made in blood rather than plasma were adjusted by multiplying by a correction factor of 1.13. When multiple values were available for a given participant, blood draws were chosen to favor measurements made at study baselines and to maximize overlap with time points at which dietary data were collected. Participants were excluded if their glycemic trait blood draw was >1 year before or after diet measurement or if they had diabetes (defined as any of: taking antidiabetic medication, FG ≥7 mmol/L, or HbA1c ≥6.5%). See Supplementary Fig. 1 for a participant inclusion flowchart. Further study-specific details are available in the Supplementary Methods. Phenotype data harmonization and all other post–genome-wide analyses and visualizations were conducted using R version 4.1.1 (17). Unless otherwise noted, all analyses including harmonization were performed on the NHLBI BioData Catalyst cloud computing platform (18).
Harmonization of Dietary Data
Estimates of dietary intake were derived from self-reported diet questionnaires, either food frequency questionnaires, diet history, or 24-h recalls. Reported quantities of food and beverage consumption were converted into daily nutrient intake estimates via standard nutrient databases (see Supplementary Methods for study-specific details), with energy and macronutrients (carbohydrate, protein, and total fat) expressed in kilocalories per day. Participants were excluded if responses were deemed implausible, based on having total caloric intake <600 kcal/d or >4,800 kcal/d. Nutrient intake values were analyzed in units of kilocalories per day and Winsorized at 3 SDs from the mean within each cohort. Dietary fiber was represented in grams per day, and alcohol intake was reported as number of drinks per day. Diet questionnaires were completed at the same time point as blood draws for glycemic trait measurement, with the exception of a 3-year gap between diet and HbA1c measurement in the ARIC study (see Supplementary Methods for details).
Genome-wide GDI Scans
For each cohort and glycemic trait, four genome-wide GDI scans were performed to identify diet-interacting loci: one for common variants and three gene-based aggregate tests for rare variants using different variant masks (described below). Mixed linear models were used to allow for random effects of kinship capturing close family relationships (degree four relatives or closer). The linear model setup was as follows:
where is the genotype at the variant of interest, is dietary carbohydrate intake (kilocalories per day), and is a random effect governed by a sparse kinship matrix. General covariates included sex, age, age2, five genetic principal components to capture genetic ancestry, cohort-reported race/ethnicity to capture potential confounding by ethnicity-related dietary behavior, and additional study-specific covariates (Supplementary Table 2). Dietary protein intake and total energy (also expressed in kilocalories per day) were included as covariates to set up an isocaloric substitution model in which increases in carbohydrate were implicitly exchanged for decreases in dietary fat. Dietary fiber (grams per day), alcohol intake (standard drinks per day), and BMI (kilograms per meter squared) were included as additional covariates to account for further lifestyle-related confounding. Though the inclusion of dietary fiber as a covariate impacts the interpretation of the carbohydrate term of interest, we found in preliminary analyses that its inclusion substantially decreased cross-study heterogeneity in parameter estimates, possibly due to a reduction in the confounding mentioned above. During null model fitting, heterogeneous variances were allowed within each cohort-reported race/ethnicity group (equivalent to including a random effect for this grouping variable). For variants on the X chromosome, male genotypes were coded as (0, 2).
Genome-wide interaction analysis was performed using the MAGEE package (19). Single-variant analysis (glmm.gei function) was conducted for variants with minor allele frequency (MAF) >1%. METAL (20) was used to perform fixed-effects meta-analysis across cohorts. Specifically, the 2-df joint meta-analysis patch was used (21), with genetic main effect and interaction P values derived downstream based on the resulting effect and SE estimates.
Gene-centric, set-based rare-variant analysis (MAGEE function) was conducted for variants with MAF <1%. Variant annotations derived from the WGSA v0.8 and WGSAParsr v6.3.8 were retrieved from the National Center for Biotechnology Information Database of Genotypes and Phenotypes (dbGaP). A genome-wide interaction meta-analysis was conducted for each of three variant masks: loss of function variants (VEP_ensembl_Consequence has terms transcript_ablation, splice_acceptor_variant, splice_donor_variant, stop_gained, frameshift_variant, stop_lost, start_lost or transcript_amplification), missense variants (VEP_ensembl_Consequence has the term missense_variant), and a broad coding and noncoding filter (containing high-confidence loss-of-function variants, missense variants, protein-altering variants, synonymous variants, variants overlapping enhancers, and variants overlapping promoters). MAGEE calculates three interaction P values: an adjusted variance component-like test, a burden test (assuming a consistent direction of effect for all variants), and a hybrid test (which combines the first two P values using the Fisher method). P values from the hybrid test were used in this study to balance the increased power of the burden test with the possibility that its assumption of homogeneous effect directions is violated. Meta-analysis was then performed using a fixed-effects strategy.
Linear mixed models without genotype terms, meant to understand the marginal dietary effects prior to considering genetic effects, were fit in R using analogous models to those with diet–genotype interaction terms. Diet–genetic principal component interaction terms were excluded from these models, and individuals in cohort-reported race/ethnicity groups with less than five members were excluded. Fixed-effect meta-analysis of the carbohydrate association (implicitly modeling an exchange with fat due to the additional dietary covariates) was conducted using the meta package.
Variant Follow-up
Sensitivity analysis was conducted to understand the impact of modeling choices on the interaction effect estimates derived in the genome-wide analysis. These linear mixed models were fit in R, with G × CHO interaction terms subject to fixed-effects meta-analysis using the meta package as with the models without genotype effects. Some of these involved subsets of the population: male and female subsets were tested separately, as well as subsets without obesity (BMI <30 kg/m2) and with and without prediabetes (defined as FG >5.6 mmol/L or HbA1c >5.7% [39 mmol/mol]). Additional models included adjustment for smoking status (never/former/current, coded as 0/1/2 and analyzed as a continuous variable), the Alternative Healthy Eating Index 2010 (22) (a diet quality score), or a categorical coding of alcohol intake (none, modest [less than 1 drink per day for females or less than two drinks per day for males], or high), where available. These models with additional covariate adjustments also included adjustment for their interactions with genotype. Finally, a model including genotype interaction terms for other main dietary components and lifestyle confounders (total energy, protein, fiber, and alcohol) was included. This type of residual confounding by genotype–covariate interaction terms has been previously documented (23), but would have decreased statistical power if included in the genome-wide analysis, especially for lower-frequency variants.
Variant rs79762542 was investigated in greater depth as the only variant reaching study-wide significance. Based on its expression quantitative trait locus (eQTL) relationship impacting FRAS1 gene expression in thyroid from the Genotype-Tissue Expression (GTEx) v8 data set (https://gtexportal.org/), we tested for colocalization of this signal with the carbohydrate interaction signal impacting HbA1c. Interaction summary statistics were retrieved in a window of 1 Mb around the index variant rs79762542, and all thyroid-specific cis-eQTL summary statistics related to FRAS1 were retrieved from GTEx. Colocalization was tested using the coloc package for R, assuming a single causal variant (coloc.abf function). Visualizations used the carbohydrate-to-fat ratio (simple ratio of kcalories from carbohydrate to kcalories from fat) as a summary variable to capture the modeled carbohydrate–fat exchange in a single variable for stratification. Tertiles of this ratio were defined in the entire pooled study cohort (with nonmissing HbA1c values).
Replication Analysis in the UK Biobank
UK Biobank (UKB) is a large prospective cohort with both deep phenotyping and molecular data, including genome-wide genotyping, on >500,000 individuals aged 40–69 years living throughout the U.K. between 2006 and 2010 (24). Genotyping, imputation, and initial quality control on the genetic data set have been described previously (25). Analyses were conducted on genetic data release version 3, with imputation to a joint reference panel including the Haplotype Reference Consortium and the 1000 Genomes Project under UKB application 27892. This work was conducted under a Not Human Subjects Research determination (NHSR-4298 at the Broad Institute of MIT and Harvard).
Ancestry group labels, genetic principal components, and labels defining an unrelated subset of individuals were retrieved from the Pan-UKB project (https://pan.ukbb.broadinstitute.org/; data retrieved from UKB return of results number 2442). Only unrelated individuals were used for analysis, with additional removal of individuals who were pregnant or had diabetes at the study center visit. Two glycemic traits were available for testing in UKB: HbA1c (provided in units of millimoles per mole and transformed after regression to units of HbA1c percentage by dividing by 10.929) and glucose (collected as a random glucose measurement with subsequent removal of nonfasting individuals). Outliers for both traits (defined as more than 5 SDs from the mean) were removed. Dietary data came from one or more Oxford WebQ 24-h dietary assessments (26) completed at the study center or during online follow-up over the course of 2 years. Daily nutrient intake estimates (calculated centrally by the UKB) were averaged across all questionnaires for each individual and Winsorized at 3 SDs from the mean. After all exclusions, 178,352 individuals without diabetes had available genotype, biomarker, and dietary data.
Regression analysis in the UKB mirrored that of the primary analysis, replacing cohort-reported race/ethnicity with genetically defined ancestry groups as defined by the Pan-UKB project. Given the larger sample size available, gene–covariate interactions were included for dietary covariates (total energy, protein, fiber, and alcohol). When analyzing glucose, only the subset of individuals with reported fasting times of at least 8 h were included, reducing the sample size to 5,183. Due to the African-ancestry specificity of some of the top variants, a second replication analysis was performed in the African-ancestry subset of UKB.
Power Calculations
Interaction test power calculations were performed using the ESPRESSO.GxE R package, which uses a simulation-based approach to calculate empirical power estimates (given some sample size) and sample size requirements (to achieve 80% power). The following parameters were fixed for this analysis, chosen to mimic an analysis of HbA1c: random seed = 1; significance threshold = 5 × 10−8; phenotype mean = 5.5; phenotype SD = 0.5; phenotype reliability = 1; genetic main effect = 0.1; exposure mean = 0; exposure SD = 1; and exposure main effect = 0.2. The following parameters were varied: interaction effect {0.025, 0.0375, 0.05, 0.0625, 0.075, 0.0875, 0.1}, MAF {0.01, 0.05, 0.1, 0.5}, and exposure reliability {0.25, 0.5, 0.75, 1}. In this study, reliability is used to quantify the simulated measurement error of the phenotype and exposure and is equivalent to an intraclass correlation coefficient (ratio of between-subject variance to total [between-subject plus measurement error] variance).
To enable simulation-based power calculations for aggregate tests of rare variants while accounting for exposure measurement error, we developed an extension of the ESPRESSO.GxE package, called ESPRESSO.GxE.RV. In this extension, the basic structure of the simulations remains the same, but an additional parameter allows the user to specify a number of variants (M) to test in aggregate. Within each simulation run, M variants are simulated, with some portion having equal interaction and main effects on the outcome (according to a user-specified causal variant fraction) and the rest generated randomly. The final P value from that simulation is calculated using the Fisher method on the full set of M P values. The following parameters were given different values for this set of simulations: interaction effect {0.025, 0.05, 0.075, 0.1, 0.125, 0.15, 0.175, 0.2} and MAF {0.0025, 0.005, 0.0075, 0.01}. Other parameters were specific to rare-variant tests: number of variants per aggregate test {1, 5, 10, 20} and causal fraction {0.1, 0.25, 0.5, 1}. Code for this extension of the package can be found on GitHub: https://github.com/kwesterman/ESPRESSO.GxE.RV.
To provide context for realistic GDI effect sizes despite few well-replicated examples of such interactions for glycemic traits in the literature, we retrieved results from variants reaching significance in a recent trans-ancestry genome-wide association studies for HbA1c (27) and an estimated effect for the carbohydrate–HbA1c relationship from a recent nutritional epidemiological analysis (28).
Data and Resource Availability
The TOPMed study data that support the findings of this study are available from the National Center for Biotechnology Information dbGaP, which were used under license for the current study and, therefore, are not publicly available. UKB data are available through a process described at: https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access. No applicable resources were generated or analyzed during the current study.
Results
We analyzed data from 33,178 individuals without diabetes (based on FG, HbA1c, or medication use) from 10 TOPMed program cohorts. Participants had diverse cohort-reported race/ethnicities, including: African American (N = 6,158), American Indian (N = 35), Asian (N = 124), White (N = 19,721), and Hispanic/Latino (N = 7,114). Dietary carbohydrate and fat as a percentage of total energy intake on average were 50.5% (SD 8.5%) and 32.2% (6.9%), respectively, in the full pooled sample, estimated using validated food frequency questionnaires or 24-h dietary recalls. Cohort-specific carbohydrate intake estimates (as percent of total energy [percent kilocalories]), glycemic trait values (FG, FI [or log-transformed (lnFI)], and HbA1c), and additional population characteristics are presented in Supplementary Table 1 and Supplementary Fig. 2.
We first modeled the main association of macronutrient compositions with each of the glycemic traits. By adjusting for total energy and energy from protein, resulting regression estimates for carbohydrate represented a macronutrient exchange (increased 250 kcal from carbohydrate replacing an equivalent 250 kcal from fat; see Research Design and Methods). Meta-analysis of the individual cohorts indicated that a higher proportion of kcal from carbohydrate at the expense of fat was associated with lower FG (−0.030 mmol/L/250 kcal; P = 2.2 × 10−6), lnFI (−0.008 log[pmol/L]/250 kcal; P = 0.15), and HbA1c (−0.012% [−0.13 mmol/mol] HbA1c/250 kcal; P = 0.029). Forest plots of these results are shown in Supplementary Fig. 3.
Common Variant Interactions
We sought to identify macronutrient GDIs with the maximal sample available in TOPMed program cohorts to provide a baseline for discovery and evaluate our assumptions about expected effect sizes. Common variants (MAF >1%) were analyzed in a primary, single-variant analysis of gene–carbohydrate interactions, with the same regression adjustments as above. This GDI analysis produces interaction estimates for the difference in the macronutrient–glycemic trait association per alternate allele at the variant of interest. After genome-wide, cohort-specific analysis and cross-cohort meta-analysis, one variant reached a study-wide significance threshold of 1.67 × 10−8 (5 × 10−8/3 glycemic traits). Two additional variants passed a standard genome-wide threshold of 5 × 10−8 (Table 1). We note that this threshold is liberal given the greater testing burden involved in the analysis of multiple ancestry groups (29). Of these three, none had evidence of a genetic main effect on the associated trait. Results are visualized in Supplementary Fig. 4 for all variants and shown in Supplementary Table 3 for variants with interaction P < 10−5.
Table 1.
Top variants interacting with carbohydrate intake from the common-variant genome-wide interaction study
| Trait | rsID | Chromosome | Position | Effect allele | Average EAF | Main effect estimate | Interaction estimate | P interaction |
|---|---|---|---|---|---|---|---|---|
| HbA1c | rs79762542 | 4 | 77979164 | G | 0.03 | −0.013 (−0.038 to 0.012) | 0.048 (0.031–0.064) | 1.1 × 10−8 |
| FG | rs1288694 | 3 | 71275429 | C | 0.61 | −0.003 (−0.011 to 0.004) | 0.016 (0.01–0.022) | 1.9 × 10−8 |
| lnFI | rs782681704 | X | 155084576 | G | 0.01 | −0.049 (−0.209 to 0.11) | 0.284 (0.182–0.385) | 4.6 × 10−8 |
Interaction estimates with 95% CIs are given in units of (trait units/allele/250 kcal carbohydrate). All variants passed a significance threshold of Pinteraction < 5 × 10−8.
EAF, effect allele frequency.
As the only variant reaching study-wide significance in the primary analysis, we looked deeper into the biological function of rs79762542 and the functional form of its interaction. Variant rs79762542 is observed on African-ancestry haplotypes and was discovered with respect to HbA1c. The variant does not have known regulatory activity based on epigenomic assays in RegulomeDB, but there is evidence for a role in regulating expression of the nearby gene FRAS1, especially in thyroid, where this gene is most strongly expressed (GTEx project). Colocalization analysis did not support a shared causal signal between our interaction results and thyroid-specific eQTL signal (posterior probability of shared causal variant = 0.003%).
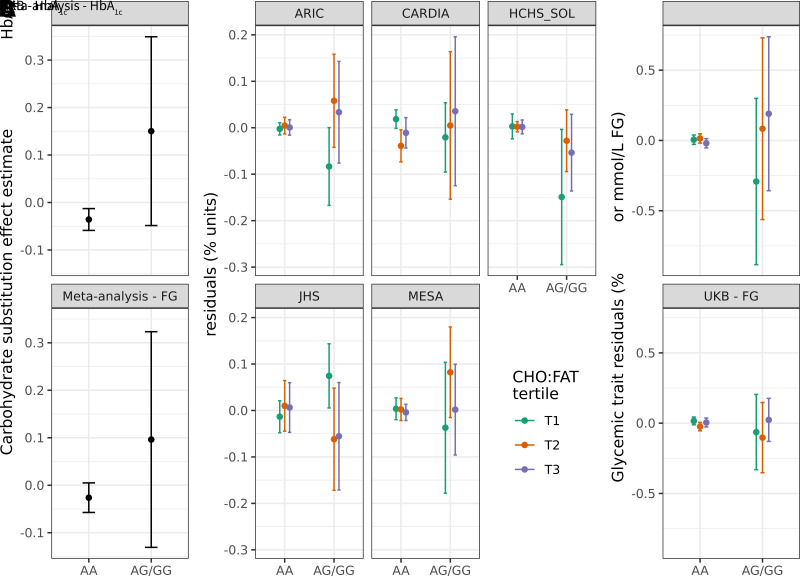
In genotype-stratified meta-analysis, HbA1c showed a modest negative association with increasing carbohydrate relative to fat intake in major allele homozygotes (−0.033% [−0.36 mmol/mol] HbA1c/250 kcal; P = 0.004) versus a positive association in minor allele carriers (0.10% [1.1 mmol/mol] HbA1c/250 kcal; P = 0.42) that may not have reached significance due to the much lower sample size in this group (N = 1,055 across all studies) (Fig. 1A). This genetic effect modification was moderately consistent across cohorts, as visualized through stratification by genotype and carbohydrate/fat ratio (Fig. 1B; identical visualization in the African American race/ethnicity subset in Supplementary Fig. 5). Finally, with respect to the other glycemic traits in our analysis, interaction effects were directionally consistent but did not reach nominal significance (0.02 mmol/L/allele/250 kcal; P = 0.07 for FG and 0.003 log[pmol/L]/allele/250 kcal; P = 0.74 for lnFI).
Figure 1.
Exploration of the rs79762542 interaction and replication. A: Genotype-stratified dietary main effect estimates. B: Stratified plots (one for each cohort with HbA1c available) display residualized HbA1c within strata defined by both genotype at rs79762542 (none vs. any minor alleles) and tertile of carbohydrate/fat ratio. This ratio was defined in the pooled data set on a caloric basis and is used to provide a visual representation of the modeled macronutrient exchange. C: Similar stratified plots for the UKB replication cohort. For B and C, the y-axis displays residuals after regressing the relevant trait (HbA1c or FG) on the set of covariates used in the replication analysis. Error bars indicate 95% CIs for the effect estimates (A) or mean residual values after stratification (B and C).
Lookups for the other two variants passing P < 5 × 10−8 revealed potential functional roles for these variants. Variant rs1288694 (common in multiple ancestries) impacted FG in our analysis. The variant is intronic to the FOXP1 gene and may regulate splicing of the same gene (GTEx project). FOXP1 has a demonstrated role in hepatic gluconeogenesis (30). Variant rs782681704 is observed on African-ancestry haplotypes and was discovered with respect to FI in our analysis. The variant is intronic to BRCC3 and has likely regulatory activity (RegulomeDB score of 0.59), but does not have clear evidence as an eQTL for BRCC3.
We explored these three prioritized single-variant loci through sensitivity analysis (Supplementary Figs. 6 [rs79762542] and 7 [all three variants]). Interaction effects were robust in population subsets: only males, only females, and individuals without obesity. Exclusion of individuals either with or without prediabetes (beyond the predefined exclusion of individuals with diabetes) partially attenuated the interaction signal; this might be expected due to the removal of a substantial portion of the glycemic trait spectrum. Further, adjustment for either a diet quality score (Alternative Health Eating Index 2010) or smoking status (along with their genotype interactions) did not meaningfully impact estimates. Interaction estimates were also generally consistent in the African American race/ethnicity subset, indicating that the interactions for African ancestry–specific variants do not solely reflect population stratification.
Common Variant Replication
For the three prioritized single-variant loci, we tested for replication of these signals in the UKB [N = 178,352 with 24-h dietary assessment data (26) and glycemic biomarkers; see Research Design and Methods]. Of these, 5,183 individuals were included in FG analyses (based on fasting for at least 8 h prior to the associated blood draw). In the full multiancestry group (Supplementary Table 5), we saw nominal replication of the interaction at rs79762542 with respect to both HbA1c (the discovery trait; P = 0.025) and FG (P = 0.013) (Fig. 1C). The interaction effect size with respect to HbA1c (0.07% [0.77 mmol/mol] HbA1c/allele/250 kcal) was of a similar magnitude to that from the primary meta-analysis (0.048% [0.52 mmol/mol] HbA1c/allele/250 kcal). Because most of the prioritized variants were specific to African-ancestry individuals, we conducted a similar replication in just this subgroup of the UKB (Supplementary Table 6). This analysis revealed an even closer HbA1c effect size to that of the meta-analysis despite a lack of significance (0.05% [0.55 mmol/mol] HbA1c/allele/250 kcal; P = 0.29) and supported the rs79762542 interaction influencing FG (P = 0.046).
Rare Variant Interactions
Rare variants (MAF <1%) were analyzed in gene-centric, set-based tests, which help to overcome power limitations for low-MAF variants by aggregating signal across multiple variants annotated to the same gene. We used three variant aggregation strategies to define sets: selecting missense variants, loss-of-function variants, or a broader coding plus noncoding variant set annotated to each gene (see Research Design and Methods). No rare-variant interaction signals showed genome-wide significance (P < 0.05/28,111 total genes = 1.78 × 10−6) (Supplementary Table 4 and Supplementary Fig. 8).
Since the set of rare variants used does not overlap with those from the common-variant tests, these gene-based tests can provide orthogonal evidence supporting common-variant signals while further clarifying potential effector genes. Each of the three prioritized single-variant findings were annotated to one or more genes based on proximity and/or eQTL data. None of these pairings showed supporting gene-based signals for the corresponding glycemic trait, though the single study-wide significant variant (rs79762542, discovered in relation to HbA1c) showed a nominal corresponding signal from the gene-based test of FRAS1 impacting FG (P = 0.028).
Power Calculations Incorporating Measurement Error
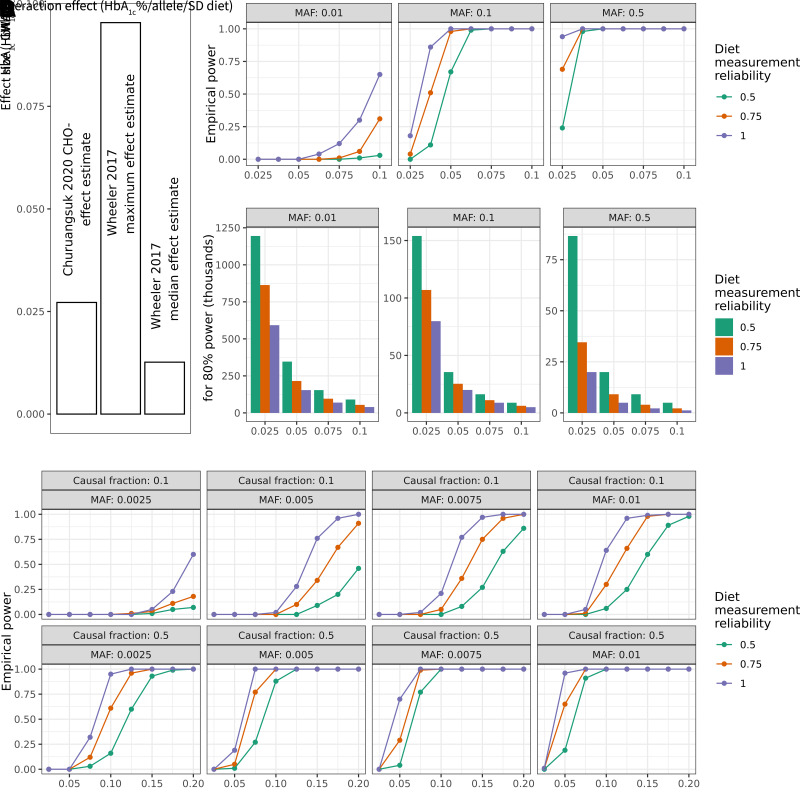
Given the modest discovery of GDIs despite the use of the maximal sample available within TOPMed cohorts and substantial harmonization effort, we sought to better understand the necessary power to detect expected GDI effects using literature-based anchors for context-specific expected effect sizes (Fig. 2A). Simulation-based power calculations for single-variant tests were conducted with added noise to account for the known random measurement error in dietary data. Assuming a conservative but realistic dietary measurement reliability of 0.5 (see Research Design and Methods for details), we established that a sample size of >150,000 would be required to detect a GDI effect of 0.025% (0.27 mmol/mol) HbA1c/allele/SD carbohydrate at genome-wide significance for a variant with an MAF of 0.1 (Fig. 2B and C). As previously explored in the literature (31,32), power scaled approximately linearly with exposure measurement fidelity. If we alternatively assume perfect dietary exposure measurement, the associated sample size to detect the same effect was reduced to 80,000, indicating the importance of accounting for this measurement error. The necessary sample size, given realistic measurement reliabilities, increased even further for lower-frequency variants (e.g., 1.2 million for MAF = 1%).
Figure 2.
Power calculations for gene–environment interaction incorporating exposure measurement error. For all plots above, HbA1c is used as a basis for parameter choices. A: Genetic and dietary effect sizes on HbA1c for reference for potential interaction effects. Bars are annotated with the source study, either Churuangsuk et al. (28) or Wheeler et al. (27). B: Simulation-based empirical power estimates are shown as a function of the interaction effect (x-axis), MAF (panels left to right), and diet measurement reliability (colors). C: Bar plots show the estimated sample size needed to achieve 80% statistical power. Panels and colors are as in B. D: As in A, but modeling empirical power for simulated aggregate tests of 20 rare variants with a causal fraction of 0.1 or 0.5 (indicated in panel labels). Additional assumptions for these simulations (full details in Research Design and Methods): N = 35,000; phenotype mean of 5.5; phenotype SD of 0.5; exposure mean of 0; exposure SD of 1; genetic main effect of 0.01; and environmental main effect of 0.2.
We extended this simulation-based power calculation approach to test multiple variants jointly, mimicking the variant set-based test implemented for rare variants. Assuming similar measurement fidelity and effect sizes as for single variants and fixing the sample size to match the full sample used in this study (∼35,000), an aggregate test of 20 rare variants with a causal fraction of 50% and MAF of 0.25% had negligible power (Fig. 2D). Power increased somewhat but remained low when incorporating larger effect sizes (as are known to be present for rare-variant main effects on cardiometabolic traits) (33). For example, using an effect size of 0.1, approximately equal to the largest genetic main effect on HbA1c reported by Wheeler et al. (27), power increased to 0.16. The full set of power simulation results is provided in Supplementary Tables 7 and 8 for single variants and set-based rare variants, respectively.
Discussion
Our goal was to investigate genotype-related variability in the association of dietary macronutrient composition with glycemic traits. Importantly, this was based on a regression strategy modeling an isocaloric increase in dietary carbohydrate at the expense of fat (34). We conducted our comprehensive analyses in cohorts with racial/ethnic diversity with data for both common and rare variants from WGS. We examined multiple single variants with potential modifying roles for the relationship of carbohydrate intake with glycemic traits but did not find substantial evidence from gene-based tests for a role of rare variants in modifying this diet–glycemia relationship. Furthermore, our simulation-based power analysis highlighted the impact of dietary measurement error on statistical power for the GDI tests.
Dietary main effect models indicated that an increase in carbohydrate at the expense of dietary fat was associated with lower FG and HbA1c. The impact of this macronutrient exchange on glucose homeostasis and diabetes risk is complex and likely depends on the respective macronutrient quality. Prior studies suggest null associations of total carbohydrate to total fat exchange on diabetes risk (35,36). However, an exchange of animal-sourced fat for carbohydrate or vegetable fat appears to have favorable associations with HbA1c (37,38).
Our primary genome-wide common-variant interaction analysis yielded an interaction between a 250-kcal carbohydrate–fat substitution and HbA1c with the African-ancestry rs79762542 variant, which was validated in the multiancestry UKB. Genotype-stratified analyses suggested that minor allele carriers generally had a small negative association between carbohydrate and HbA1c (−0.033% [−0.36 mmol/mol] HbA1c/250 kcal; P = 0.004) versus a larger but nonsignificant association in minor allele carriers, where the sample size was much lower (−0.10% [−1.1 mmol/mol] HbA1c/250 kcal; P = 0.42). However, this precise pattern was not observed in all cohorts, possibly due to the low sample size of minor allele carriers in the populations studied. These results warrant further exploration in additional cohorts with African-ancestry individuals and dietary intervention studies to examine whether glycemic traits among minor allele carriers may benefit from higher-fat and lower-carbohydrate diet composition. This primary discovery was made with HbA1c as an outcome, but our results from set-based rare variant analysis and the UKB replication suggest similar patterns with respect to other traits, such as FG.
Beyond GDI discovery, the genome-wide interaction study results provided an opportunity to inform and evaluate the effect size assumptions used in the power calculations. For example, the rs79762542 interaction had an effect size of 0.048% (0.52 mmol/mol) HbA1c/allele/250 kcal carbohydrate, or 0.068% (0.74 mmol/mol) HbA1c/allele/SD carbohydrate. This effect size is comparable to the relevant anchor for the power analysis [the referenced main effect association of carbohydrate with HbA1c (28)].
This analysis leveraged WGS data along with multivariant set-based tests to better incorporate rare variants (MAF <0.01). While these variants do not contribute substantially to the overall population variance of glycemic or other traits, they tend to have larger effect sizes and thus may be important for the specific individuals carrying them (14). For example, phenylketonuria, a well-known inborn error of metabolism, acts through a rare-variant GDI in which severe adverse effects of phenylalanine intake are seen only in individuals with a particular genotype (39). In our study, despite helping to reinforce common-variant signals, the rare-variant analysis did not contribute additional findings after aggregation at the gene level. Substantially larger sample sizes will likely be necessary to uncover macronutrient GDIs involving rare variants.
We explored the statistical power for these interaction tests through simulations incorporating random dietary measurement error using available simulation-based power calculation software [ESPRESSO.GxE (40) for single variants] with additional extensions to allow for aggregate rare-variant tests. We estimated that substantially higher sample sizes (almost five times that used in this study) are required for sufficient power to detect macronutrient–gene interactions at expected effect sizes obtained from genetics and nutrition literature. This prompts two directions of further inquiry. First, it suggests the importance of complementary approaches that assess where there is any whole-genome contribution to the diet–glycemia association, at least in observational data sets. These whole-genome analyses trade resolution for statistical power (41) and have a precedent for GDIs in smaller study samples (42). Second, it reinforces the importance of collecting dietary intake data in the growing group of large-scale biobanks and cohorts. Improvements in study design, data collection methods, and analysis that can improve quality of dietary assessments are also warranted. For example, conducting rigorous validation studies of the data collection tools and approaches and ascertaining repeated dietary data can greatly improve the precision of these measurements on a population level. Advancements in objectively quantifying habitual diet from biospecimen samples are also underway and have potential to improve discovery for genetic analyses.
An important strength of this study is the breadth of ethnic and cultural diversity of the sample (increasing the likelihood that findings are robust) and of genetic variation (with WGS data enabling exploration of ancestry-specific genetic variation across the frequency spectrum). We also conducted a systematic investigation into the available statistical power while incorporating both realistic degrees of measurement error and evidence-based estimations of realistic effect sizes for gene–macronutrient interactions. However, the diversity of the included study sample also introduces heterogeneity that may be problematic. For example, the cohorts used different dietary assessment tools to capture habitual intake, leading to differences in the degree and direction of random and systematic measurement error. This is compounded by general, culturally driven differences in food intake across race/ethnicity groups. Furthermore, heterogeneity arises from the time of data collection; the perceptions of carbohydrate intake have trended as more and less healthful in recent decades, potentially resulting in differential confounding between diet and other health-related behaviors depending on the time of data collection (43). Future work can step beyond broad macronutrient categories by harmonizing intakes of specific foods or dietary patterns and analyzing macronutrient subtypes (e.g., added sugar and specific fatty acids). These approaches, combined with improved methods for detecting rare-variant gene–environment interactions, will help use the increasing volume of WGS data to discover new GDIs relevant for metabolic disease risk.
Article Information
Funding. K.E.W., H.C., and A.K.M. were supported by NIH grant R01 HL145025. K.E.W. was also supported by NIH grants K01DK133637 and T32DK007028. L.M.R. was supported by the National Center for Advancing Translational Sciences, NIH, through grant KL2TR002490. V.S.R. is supported in part by the Evans Medical Foundation and the Jay and Louis Coffman Endowment from the Department of Medicine, Boston University School of Medicine. M.E.W. is supported in part by the American Heart Association (20CDA35310237), the Doris Duke Charitable Foundation (2021261), and the National Center for Advancing Translational Sciences, NIH, through BU-CTSI (1UL1TR001430). The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health (NIH).
Duality of Interest. K.E.W. has provided consulting services for FOXO Bioscience. L.M.R. is a consultant for the TOPMed Administrative Coordinating Center (through Westat). No other potential conflicts of interest relevant to this article were reported.
Author Contributions. K.E.W., M.E.W., J.M., and A.K.M. designed the research. K.E.W., S.M.G., J.W., D.K.T., and A.K.M. contributed to the harmonization of glycemic trait variables across studies. K.E.W. conducted the research and performed the primary data analysis. J.C.F., J.B.M., D.K.T., H.C., and A.K.M. supervised the research. K.E.W. and M.E.W. wrote the manuscript. All additional authors contributed to the collection and curation of the study-specific or TOPMed-wide data sets. K.E.W. and A.K.M. had primary responsibility for the final content. All authors read and approved the final manuscript. K.E.W. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
GTEx. The GTEx project was supported by the Common Fund of the Office of the Director of the NIH and the National Cancer Institute, National Human Genome Research Institute (NHGRI), NHLBI, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke. The data used for the analyses described in this study were obtained from the GTEx Portal on 28 March 2022. Support for this work was provided by the NIH, NHLBI, through the BioData Catalyst program (award 1OT3HL142479-01, 1OT3HL142478-01, 1OT3HL142481-01, 1OT3HL142480-01, and 1OT3HL147154-01). Any opinions expressed in this document are those of the authors and do not necessarily reflect the views of NHLBI, individual BioData Catalyst team members, or affiliated organizations and institutions. Furthermore, the authors acknowledge the contributions of the consortium working on the development of the NHLBI BioData Catalyst ecosystem. The authors also thank L. Adrienne Cupples (deceased) for the contributions to the Framingham Heart Study (FHS) and the TOPMed program.
TOPMed. Molecular data for the TOPMed program were supported by the NHLBI. Study-specific omics support information is detailed below. Core support, including centralized genomic read mapping and genotype calling, along with variant quality metrics and filtering, was provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1, contract HHSN268201800002I). Core support, including phenotype harmonization, data management, sample-identity quality control, and general program coordination, was provided by the TOPMed Data Coordinating Center (R01HL-120393, U01HL-120393, contract HHSN268201800001I). The authors gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed.
ARIC. The ARIC study has been funded in whole or in part with federal funds from the NHLBI, NIH, Department of Health and Human Services, under contract numbers (75N92022D00001, 75N92022D00002, 75N92022D00003, 75N92022D00004, 75N92022D00005). The authors thank the staff and participants of the ARIC study for the important contributions. WGS for “NHLBI TOPMed: Atherosclerosis Risk in Communities” (phs001211) was performed at the Baylor College of Medicine Human Genome Sequencing Center (HHSN268201500015C and 3U54HG003273–12S2) and the Broad Institute for MIT and Harvard (3R01HL092577–06S1). The Genome Sequencing Program (GSP) was funded by the NHGRI, NHLBI, and National Eye Institute. The GSP Coordinating Center (U24 HG008956) contributed to cross program scientific initiatives and provided logistical and general study coordination. The Centers for Common Disease Genomics program was supported by NHGRI and NHLBI, and WGS was performed at the Baylor College of Medicine Human Genome Sequencing Center (UM1 HG008898).
CARDIA. The Coronary Artery Risk Development in Young Adults Study (CARDIA) is conducted and supported by the NHLBI in collaboration with the University of Alabama at Birmingham (HHSN268201800005I and HHSN268201800007I), Northwestern University (HHSN268201800003I), University of Minnesota (HHSN268201800006I), and Kaiser Foundation Research Institute (HHSN268201800004I). CARDIA was also partially supported by the Intramural Research Program of the National Institute on Aging (NIA) and an intra-agency agreement between the NIA and NHLBI (AG0005).
CHS. The Cardiovascular Health Study (CHS) was supported by contracts 75N92021D00006, HHSN268201200036C, HHSN268200800007C, HHSN268201800001C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, and grants R01HL120393, U01HL080295 and U01HL130114 from the NHLBI, with additional contribution from the National Institute of Neurological Disorders and Stroke. Additional support was provided by R01AG023629 from the NIA. A full list of principal CHS investigators and institutions can be found at https://CHS-NHLBI.org. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
FHS. WGS for “NHLBI TOPMed: Whole-Genome Sequencing and Related Phenotypes in the Framingham Heart Study” (phs000974.v1.p1) was performed at the Broad Institute of MIT and Harvard (3R01HL092577-06S1 [AFGen]). This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the NIH and Boston University School of Medicine. The FHS acknowledges the support of contracts NO1-HC-25195, HHSN268201500001I, and 75N92019D00031 from the NHLBI and grant supplement R01 HL092577-06S1 for this research. The authors also acknowledge the dedication of the FHS study participants, without whom this research would not be possible.
GeneSTAR. GeneSTAR was supported by grants from the NIH/NHLBI (U01 HL72518, HL087698, HL49762, HL59684, HL58625, HL071025, HL092165, HL099747, K23HL105897, and HL112064), grants from the NIH/National Institute of Nursing Research (NR0224103 and NR008153), a grant from the NIH/National Institute of Neurological Disorders and Stroke (NS062059), a grant from the NIH/National Center for Research Resources (M01-RR000052) to the Johns Hopkins General Clinical Research Center, and a grant from the National Center for Advancing Translational Sciences, NIH (UL1TR001079) to the Johns Hopkins Institute for Clinical and Translational Research.
GOLDN. GOLDN biospecimens, baseline phenotype data, and intervention phenotype data were collected with funding from NHLBI grant U01 HL072524. WGS in GOLDN was funded by NHLBI grant R01 HL104135–04S1. The authors thank GOLDN participants and investigators for the significant contributions.
HCHS/SOL. The Hispanic Community Health Study/Study of Latinos (HCHS/SOL) is a collaborative study supported by contracts from the NHLBI to the University of North Carolina (HHSN268201300001I/N01-HC-65233), University of Miami (HHSN268201300004I/N01-HC-65234), Albert Einstein College of Medicine (HHSN268201300002I/N01-HC-65235), University of Illinois at Chicago (HHSN268201300003I/N01-HC-65236 Northwestern University), and San Diego State University (HHSN268201300005I/N01-HC-65237). The following institutes/centers/offices have contributed to the HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, and NIH Institution-Office of Dietary Supplements.
JHS. The Jackson Heart Study (JHS) is supported and conducted in collaboration with Jackson State University (HHSN268201800013I), Tougaloo College (HHSN268201800014I), Mississippi State Department of Health (HHSN268201800015I), and University of Mississippi Medical Center (HHSN268201800010I, HHSN268201800011I, and HHSN268201800012I) contracts from the NHLBI and the National Institute on Minority Health and Health Disparities. The authors thank the participants and data collection staff of the JHS. The views expressed in this study are those of the authors and do not necessarily represent the views of the NHLBI, NIH, or the U.S. Department of Health and Human Services.
MESA. WGS for the TOPMed program was supported by the NHLBI. WGS for “NHLBI TOPMed: Multi-Ethnic Study of Atherosclerosis (MESA)” (phs001416.v1.p1) was performed at the Broad Institute of MIT and Harvard (3U54HG003067–13S1). Centralized read mapping and genotype calling, along with variant quality metrics and filtering, were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1). Phenotype harmonization, data management, sample-identity quality control, and general study coordination were provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1) and TOPMed MESA Multi-Omics (HHSN2682015000031/HSN26800004). The MESA projects are conducted and supported by the NHLBI in collaboration with MESA investigators. The MESA projects are conducted and supported by the NHLBI in collaboration with MESA investigators. Support for MESA is provided by contracts 75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1TR001881, DK063491, and R01HL105756 as well as in part by NHLBI contracts R01HL151855 and R01HL146860. The authors thank the other investigators, the staff, and the participants of MESA for the valuable contributions. A full list of participating MESA investigators and institutes can be found at https://www.mesa-nhlbi.org.
WHI. The Women’s Health Initiative program is funded by the NHLBI, NIH, U.S. Department of Health and Human Services through contracts 75N92021D00001, 75N92021D00002, 75N92021D00003, 75N92021D00004, and 75N92021D00005.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.22080683.
References
- 1. Ajala O, English P, Pinkney J. Systematic review and meta-analysis of different dietary approaches to the management of type 2 diabetes. Am J Clin Nutr 2013;97:505–516 [DOI] [PubMed] [Google Scholar]
- 2. Salas-Salvadó J, Bulló M, Babio N, et al.; PREDIMED Study Investigators . Reduction in the incidence of type 2 diabetes with the Mediterranean diet: results of the PREDIMED-Reus nutrition intervention randomized trial. Diabetes Care 2011;34:14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Diabetes Prevention Program (DPP) Research Group . The Diabetes Prevention Program (DPP): description of lifestyle intervention. Diabetes Care 2002;25:2165–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davis NJ, Tomuta N, Schechter C, et al. Comparative study of the effects of a 1-year dietary intervention of a low-carbohydrate diet versus a low-fat diet on weight and glycemic control in type 2 diabetes. Diabetes Care 2009;32:1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Korem T, Zeevi D, Zmora N, et al. Bread affects clinical parameters and induces gut microbiome-associated personal glycemic responses. Cell Metab 2017;25:1243–1253.e5 [DOI] [PubMed] [Google Scholar]
- 6. Berry SE, Valdes AM, Drew DA, et al. Human postprandial responses to food and potential for precision nutrition. Nat Med 2020;26:964–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bachmann AM, Morel JD, El Alam G, et al. Genetic background and sex control the outcome of high-fat diet feeding in mice. iScience 2022;25:104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barrington WT, Wulfridge P, Wells AE, et al. Improving metabolic health through precision dietetics in mice. Genetics 2018;208:399–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qi Q, Bray GA, Hu FB, Sacks FM, Qi L. Weight-loss diets modify glucose-dependent insulinotropic polypeptide receptor rs2287019 genotype effects on changes in body weight, fasting glucose, and insulin resistance: the Preventing Overweight Using Novel Dietary Strategies trial. Am J Clin Nutr 2012;95:506–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corella D, Carrasco P, Sorlí JV, et al. Mediterranean diet reduces the adverse effect of the TCF7L2-rs7903146 polymorphism on cardiovascular risk factors and stroke incidence: a randomized controlled trial in a high-cardiovascular-risk population. Diabetes Care 2013;36:3803–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Westerman KE, Miao J, Chasman DI, et al. Genome-wide gene-diet interaction analysis in the UK Biobank identifies novel effects on hemoglobin A1c. Hum Mol Genet 2021;30:1773–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Francis M, Li C, Sun Y, et al. Genome-wide association study of fish oil supplementation on lipid traits in 81,246 individuals reveals new gene-diet interaction loci. PLoS Genet 2021;17:e1009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Franks PW, Merino J. Gene-lifestyle interplay in type 2 diabetes. Curr Opin Genet Dev 2018;50:35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 2018;50:1505–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Conomos MP, Miller MB, Thornton TA. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol 2015;39:276–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Conomos MP, Reiner AP, Weir BS, Thornton TA. Model-free estimation of recent genetic relatedness. Am J Hum Genet 2016;98:127–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria, R Foundation for Statistical Computing, 2021 [Google Scholar]
- 18. National Heart, Lung, and Blood Institute, National Institutes of Health . The NHLBI BioData Catalyst. Washington, DC, U.S. Department of Health and Human Services, 2020. Accessed 5 January 2023. Available from https://biodatacatalyst.nhlbi.nih.gov/ [Google Scholar]
- 19. Wang X, Lim E, Liu CT, et al. Efficient gene-environment interaction tests for large biobank-scale sequencing studies. Genet Epidemiol 2020;44:908–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Manning AK, LaValley M, Liu CT, et al. Meta-analysis of gene-environment interaction: joint estimation of SNP and SNP × environment regression coefficients. Genet Epidemiol 2011;35:11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chiuve SE, Fung TT, Rimm EB, et al. Alternative dietary indices both strongly predict risk of chronic disease. J Nutr 2012;142:1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Keller MC. Gene × environment interaction studies have not properly controlled for potential confounders: the problem and the (simple) solution. Biol Psychiatry 2014;75:18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu B, Young H, Crowe FL, et al. Development and evaluation of the Oxford WebQ, a low-cost, web-based method for assessment of previous 24 h dietary intakes in large-scale prospective studies. Public Health Nutr 2011;14:1998–2005 [DOI] [PubMed] [Google Scholar]
- 27. Wheeler E, Leong A, Liu CT, et al. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: a transethnic genome-wide meta-analysis. PLoS Med 2017;14:e1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Churuangsuk C, Lean MEJ, Combet E. Lower carbohydrate and higher fat intakes are associated with higher hemoglobin A1c: findings from the UK National Diet and Nutrition Survey 2008-2016. Eur J Nutr 2020;59:2771–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fadista J, Manning AK, Florez JC, Groop L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur J Hum Genet 2016;24:1202–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zou Y, Gong N, Cui Y, et al. Forkhead box P1 (FOXP1) transcription factor regulates hepatic glucose homeostasis. J Biol Chem 2015;290:30607–30615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wong MY, Day NE, Luan JA, Chan KP, Wareham NJ. The detection of gene-environment interaction for continuous traits: should we deal with measurement error by bigger studies or better measurement? Int J Epidemiol 2003;32:51–57 [DOI] [PubMed] [Google Scholar]
- 32. Osazuwa-Peters OL, Schwander K, Waken RJ, et al. The promise of selecting individuals from the extremes of exposure in the analysis of gene-physical activity interactions. Hum Hered 2018;83:315–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jurgens SJ, Choi SH, Morrill VN, et al.; Regeneron Genetics Center . Analysis of rare genetic variation underlying cardiometabolic diseases and traits among 200,000 individuals in the UK Biobank. Nat Genet 2022;54:240–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arnold KF, Berrie L, Tennant PWG, Gilthorpe MS. A causal inference perspective on the analysis of compositional data. Int J Epidemiol 2020;49:1307–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salmerón J, Hu FB, Manson JE, et al. Dietary fat intake and risk of type 2 diabetes in women. Am J Clin Nutr 2001;73:1019–1026 [DOI] [PubMed] [Google Scholar]
- 36. Merino J, Guasch-Ferré M, Ellervik C, et al. Quality of dietary fat and genetic risk of type 2 diabetes: individual participant data meta-analysis. BMJ 2019;366:l4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harding AH, Sargeant LA, Welch A, et al.; EPIC-Norfolk Study . Fat consumption and HbA(1c) levels: the EPIC-Norfolk study. Diabetes Care 2001;24:1911–1916 [DOI] [PubMed] [Google Scholar]
- 38. Imamura F, Micha R, Wu JHY, et al. Effects of saturated fat, polyunsaturated fat, monounsaturated fat, and carbohydrate on glucose-insulin homeostasis: a systematic review and meta-analysis of randomised controlled feeding trials. PLoS Med 2016;13:e1002087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pérusse L, Bouchard C. Gene-diet interactions in obesity. Am J Clin Nutr 2000;72(Suppl. 5):1285S–1290S [DOI] [PubMed] [Google Scholar]
- 40. Gaye A, Burton TWY, Burton PR. ESPRESSO: taking into account assessment errors on outcome and exposures in power analysis for association studies. Bioinformatics 2015;31:2691–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dahl A, Nguyen K, Cai N, Gandal MJ, Flint J, Zaitlen N. A robust method uncovers significant context-specific heritability in diverse complex traits. Am J Hum Genet 2020;106:71–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zheng JS, Arnett DK, Lee YC, et al. Genome-wide contribution of genotype by environment interaction to variation of diabetes-related traits. PLoS One 2013;8:e77442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oster E. Health recommendations and selection in health behaviors. Am Econ Rev Insights 2020;2:143–160 [Google Scholar]