Abstract
Purpose
Age is the main risk factor for age-related macular degeneration (AMD), a leading cause of blindness in the elderly, with limited therapeutic options.
Methods
Here, we analyze the transcriptomic characteristics and cellular landscape of the aging retinas from controls and patients with AMD.
Results
We identify the aging genes in the neural retina, which are associated with innate immune response and inflammation. Deconvolution analysis reveals that the estimated proportions of M2 macrophages are significantly increased with both age and AMD severity. Moreover, we find that proportions of Müller glia are significantly increased only with age but not with AMD severity. Several genes associated with both age and AMD severity, particularly C1s and MR1, are strong positively correlated with the proportions of Müller glia.
Conclusions
Our studies expand the genetic and cellular landscape of AMD and provide avenues for further studies on the relationship between age and AMD.
Keywords: age-related macular degeneration (AMD), transcriptome, aging, RNA-seq, retina
Aging is one of the most important demographic risk factors for age-related macular degeneration (AMD),1,2 the leading causes of blindness among people aged over 55-year-old in the developed world.3 A review summarizing the prevalence of AMD over age from various studies suggested a clear trend that the prevalence of AMD regardless of mild or severe clinical features is gradually increased with age.2 Advances in sequencing technologies, genetic analyses, such as genomewide association studies (GWAS), using large numbers of samples from donors with AMD, enables identification of common and rare variations that are associated with AMD.4 Moreover, two expression quantitative trait loci (eQTL) analyses using transcriptional profiles of postmortem retinas from 453 and 129 donors, respectively, expanded the genetic landscape of AMD.5–7 Such studies have also implicated roles for the immune system, in particular the dysregulation of the complement system, in the pathogenesis and severity of AMD.8 Whereas these studies provided useful information on possible causal genes or immune cell types associated with AMD, there is little information on how age affects the progression of AMD.
In this study, we assess the transcriptomic profiles, and the immune and retinal cellular landscapes of the aging retinas in association with the progression of AMD. We systematically define the aging genes in the retina and show that the aging retina is transcriptionally featured by innate immune response and inflammation. Deconvolution analysis reveals increased proportion of M2 macrophages with both age and disease severity during the progression of AMD. We further identify specific genes that altered with age and disease severity, which are relevant to regulation of immune response, lymphocyte activation, and angiogenesis. Our results suggest that proportions of Müller glia in the retina are increased with age but not with disease severity, further indicating that age is the principal risk factor that uniquely contributes to the progression of AMD. Our study pinpoints detailed genetic and cellular characteristics of the aging retina that contribute to the progression of AMD.
Materials and Methods
Data Accession
RNA-Seq raw counts of two transcriptomic datasets (GSE115828 and GSE135092) were obtained from Gene Expression Omnibus (accessed on June 28, 2022).5–7 Raw counts were combined and converted to transcripts per million (TPM) in R (version 4.1.0) for downstream analyses after removing batch effect. Quality-controlled samples (199 Minnesota Grading System [MGS]1, 175 MGS2, 112 MGS3, and 61 MGS4 samples) were selected from the raw dataset using the subset function from the R package Seurat (version 4.0.5).9 Detailed clinical information, including patient age and MGS level, were accessed from both datasets. Single-cell transcriptomes of human neural retina were directly downloaded from Human Cell Atlas (https://www.humancellatlas.org/, E-MTAB-7316, accessed on January 15, 2022).10
Identification of Age-Associated Genes in Human Neural Retina From Donors With AMD
To identify age-associated genes, the RNA-Seq raw count from quality-controlled samples was analyzed by DESeq2 (version 1.32.0),11 using the likelihood-ratio test (LRT). According to the design of batch correction in the EyeGEx database, gene expression was determined by three variables, including batch effect, age, and MGS level.6,7 Thus, batch effect, age, and MGS level were included in the LRT model for controlling these factors. Age was categorized into decades, including 50 to 59, 60 to 69, 70 to 79, 80 to 89, 90 to 99, and 100 to 109 years, for downstream analyses. Significant age-associated genes were determined by adjusted P < 0.05. The resulting genes were then scaled to z-score and clustered using the degPatterns function from the R package DEGreport (version 1.28.0). Gene clusters with consistent and progressive changes (increasing or decreasing with age) were selected for downstream analyses.
Enrichment analyses for selected genes were performed using Metascape (https://metascape.org/gp/index.html#/main/step1), a web-based tool that involves a variety of terms, including Gene Ontology (GO) Biological Processes.12 Importantly, Metascape provides more frequently updated bioinformatics analyses than DAVID.13
Identification of Age-MGS Genes in Human Neural Retina From Donors With AMD
To identify age-MGS associated genes, the TPM values of age-associated genes from both clusters (increasing or decreasing with age) were analyzed by R package MASS (version 7.3-54) and ordinal (version 2019.12-10) using the ordinal logistic regression (OLR) model.14 Using the Cumulative Link Model function in ordinal,14 we controlled age to screen input genes that are associated with the MGS level. Age-MGS genes were determined at a significance threshold of P < 0.05. The resulting genes were represented in a forest plot and ranked according to their beta coefficient.
For gene-gene correlation of age-MGS genes, the raw counts were processed by the Variance Stabilizing Transformation function in DESeq2,11 followed by statistical adjustment for age and visualization using the removeBatchEffect function in limma (version 3.48.3).15 The correlation heatmap was plotted using the R package corrplot (version 0.92).16
RNA-Seq Gene Expression Visualization and Statistical Analyses
For visualization of adjusted gene expression levels, the raw counts were processed by the Variance Stabilizing Transformation function in DESeq2,11 followed by statistical adjustment for the MGS level or age using the Remove Batch Effect function in limma (version 3.48.3).15 Comparison of gene expression among different age groups or MGS groups are presented as Tukey boxplots, with interquartile range boxes and 1.5 times interquartile range whiskers. Two-tailed Mann-Whitney test was used to assess pairwise comparisons in the plots, whereas the Kruskal-Wallis test was used to perform statistical comparison across all the age groups or MGS groups. Such statistics were performed to confirm the results of age-associated genes generated by the two-sided DESeq2 LRT.11
Normal Human Neural Retina Single Cell RNA-Seq Data Analysis
The single cell RNA (scRNA)-seq raw counts from the Human Cell Atlas (HCA) were analyzed using Seurat (version 4.0.5).9 Cells with >200 detected genes and expressing <10% mitochondrial genes were retained, resulting in 19,950 cells. Genes were retained if they were expressed in more than or equal to 3 cells. The LogNormalize method was used to normalize the dataset, followed by removing inherent variation caused by mitochondrial gene expression and the number of unique molecular identifiers per cell.10 Using the graph-based shared nearest neighbor method, clustering at a resolution of 0.9 was performed on principal component analysis PCA)-reduced expression data because it yields high maximum cluster assignment probability for most of the cells. Clusters were visualized using uniform manifold approximation and projection (UMAP). Retinal cell markers were referred to studies analyzing retina scRNA-Seq data.10,17
Of the 50 age-MGS-associated genes identified from the EyeGEx transcriptome, 43 genes were matched in the HCA dataset. To calculate the proportions of cells expressing a given gene, the expression matrices were converted to binary matrices using the threshold of expression >0.18 Expression frequencies of a given gene in different cell types were determined as per the cell type annotation resulting from the above analyses. We further scaled the expression frequencies in cell types in z-score using the NMF package (version 0.23.0).
Deconvolution Analyses of the Immune Cells and Retinal Cells
The CIBERSORTx, a machine learning deconvolution algorithm that enables inference of cell-type-specific gene expression profiles,19 was applied to infer the estimated proportions of immune cells or retinal cells of each bulk retinal transcriptome. We used the LM22 signature matrix to define 22 infiltrating immune cells.20 Normalized data were uploaded to the CIBERSORTx web portal (https://cibersortx.stanford.edu/), with the algorithm run using LM22 signature matrix at 100 permutations with B-mode batch correction.
To infer retinal cellular profile, we applied a previously derived retinal signature matrix using the HCA scRNA-Seq dataset of the human retina.10 Briefly, the retinal signature matrix contains rod, Müller glia, bipolar, cone, amacrine, microglia, ganglion, astrocytes, and horizontal cells, which were used to impute retinal cellular fraction from the bulk transcriptome at 100 permutations with S-mode batch correction. Statistical significance of cellular numbers associated with age or MGS level was assessed by an OLR model using the nonparametric Kruskal-Wallis test that allows for multifactorial designs. The estimated coefficients and P values were presented as a forest plot with 95% confidence intervals.
Statistical Analyses
Detailed information of statistical analyses was described in different places, including results, Figures, and Figure legends, in which the test methods, significance, P values, error bars, and coefficients were included.
Results
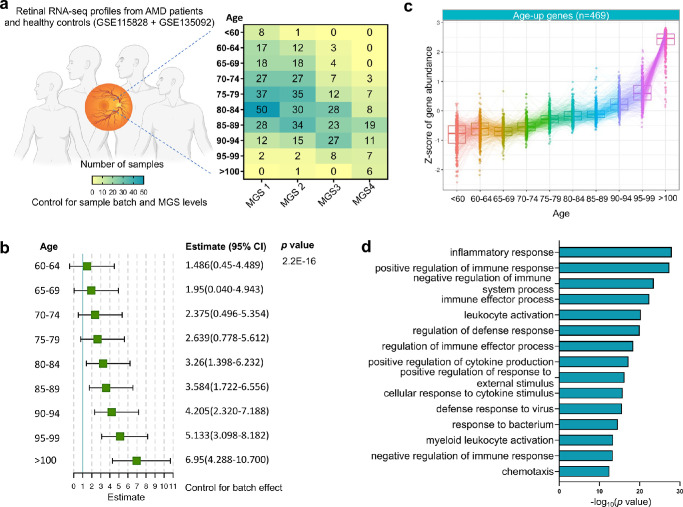
We analyzed two neural retinal transcriptomic datasets (the Eye Genotype Expression [EyeGEx] database GSE115828 and GSE135092) from patients with AMD and healthy controls.5–7 A total of 453 previously quality-controlled6,7 retinal RNA-seq profiles from controls and cases at distinct stages of AMD incorporated with 94 retinal RNA-seq profiles from controls5 were analyzed, following the batch correction (Fig. 1a, Supplementary Fig. S1). The MGS was used in this database to classify the disease severity. MGS1 categorizes donor retinas without AMD symptoms and serves as controls, whereas MGS2 to MGS4 categorize donor retinas at the progressively advanced AMD stages.
Figure 1.
Identification of age-associated genes in the human retina from donors with AMD. (a) Demographics of the human retina RNA-Seq profiles in the EyeGEx dataset (n = 453, previously quality-controlled dataset), detailed by age group and AMD severity (MGS level). (b) Forest plot of the ordinal logistic regression with MGS level as dependent variable. The green box and black line represent the estimate and the corresponding 95% confidence interval (CI), respectively. (c) Tukey boxplots (interquartile range [IQR] boxes with 1.5 × IQR whiskers) of age-associated genes in the human retina. Expression of age-up genes increase with age (left, n = 469). Age-associated genes were defined with DESeq2 two-sided likelihood-ratio test (adjusted p [adj P] < 0.05) by controlling for MGS level. Adjusted gene expressions were shown as z-score. (d) Functional enrichment analyses (biological processes) of age-up genes by Metascape.
Identification of Age-Associated Genes in the Human Retina
Gene expression levels in both of the databases are previously defined by a number of clinical variables, including MGS level and age.5–7 We revealed that there is a significant correlation between age and MGS level using the OLR (Fig. 1b), suggesting the chance of developing advanced stage of AMD is significantly higher in older people comparing to younger ones. By controlling for MGS level, therefore, we identified a cluster of genes associated with progressive aging (adjusted P < 0.05; Supplementary Data S1) using a LRT.11 Specifically, we found that expression levels of 469 genes were increased over age (Fig. 1c; hereafter referred to age-up genes). The Gene Ontology (GO biological process) analysis of age-up genes by Metascape, a comprehensive and combinational knowledgebase,12 indicates that inflammatory response, regulation of immune responses, and leukocyte activation are among the top GO terms (Fig. 1d, Supplementary Fig. S2, Supplementary Data S2). Our results were consistent with previous reports that retinal aging is a major risk factor of developing various retinal degenerative diseases mainly via dysregulation of innate immune systems, including AMD, diabetic retinopathy, and glaucomatous retinopathy.21,22 Studies on the retinal immune cells in particular macrophages suggested its activation in the retina is reported in both patients with AMD23 and old mice.24
The Immune Cellular Landscape in the Retina of Patients With AMD
In order to understand different immune cell types and to assess their relationship with age or MGS level in the retinas of patients with AMD, we deconvoluted bulk transcriptional data from the combined database by CIBERSORTx, a machine learning method that enables inference of cell-type-specific gene expression profiles without physical cell isolation.19 LM22, a signature matrix that enables distinction of 22 immune cell types, was used as a reference for deconvolution analysis.20 We specifically looked at how age or disease severity (MGS level) as an individual factor impact the cellular proportions of immune cells using an OLR model. By controlling for batch effect and MGS level, we found that proportions of M2 macrophages and gamma delta T cells were significantly increased with age, whereas proportions of M0 macrophages were decreased (Supplementary Figs. S3, S4). By controlling for batch effect and age, we only observed that proportions of activated M2 macrophages were significantly increased with MGS level (Supplementary Figs. S5, S6). It is known that human peripheral monocytes were differentiated into inactivated M0 macrophages and then polarized to M1 and M2 phenotypes under various stress or stimuli.25 Previous studies have shown that numbers of M2 macrophages were increased in the retina23 or aqueous humors26 of a small number of patients with neovascular AMD or laser-induced murine model with choroidal neovascularization.26,27 In addition, activated macrophages were observed in the peripheral blood from patients with neovascular AMD.28 Gamma delta T cells were identified as a major source of interleukin-17 production, a crucial inflammatory cytokine, involved in age-dependent retinal pigment epithelium (RPE) degeneration, a typical clinical feature during AMD.29 In line with these reports, our results further revealed that age alone as a predominant factor that triggers the polarization of M0 macrophages toward pro-angiogenic M2 phenotypes,23 whereas disease severity alone seems to be majorly affecting the numbers of M2 macrophages that feature in the pro-angiogenic process. Collectively, these results together showed that activated M2 macrophages caused by age and disease severity is the major immune cell type involved in the immune response in the pathogenesis of AMD.
Identification of Age- and Disease-Severity-Associated Genes in Relation to Progression of AMD
We next investigated whether retinal aging is related to disease-severity (ranked by MGS level) in patients with AMD. Using the OLR model that controls for age, we correlated age-up genes to MGS level, resulting in 34 genes that their expression levels were significantly altered with the change of MGS level (Fig. 2, Supplementary Data S3). Notably, the expression level of some age-up genes is decreased with the MGS level after controlling for age, suggesting either age or MGS level alone affects gene expression. Therefore, we specifically focused on those genes that are expressed at the same pattern with both age and MGS level, resulting in 26 age-up genes with increased expression level over disease severity (hereafter referred to age-MGS genes, adjusted P [adj P] < 0.05). Interestingly, many of the identified age-MGS genes have not been reported as AMD-associated genes in GWAS studies. We then explored the biological processes of each age-MGS gene involved by analyzing their corresponding GO annotations (Supplementary Data S4). We found that half of the age-MGS genes are involved in the activation of immune cells or inflammatory pathways (see Supplementary Data S4), indicating a strong immune response and inflammation occurred during the development of AMD over increasing age. Other age-MGS were clustered to groups with other biological processes, such as retinal endothelium-mediated angiogenesis.
Figure 2.
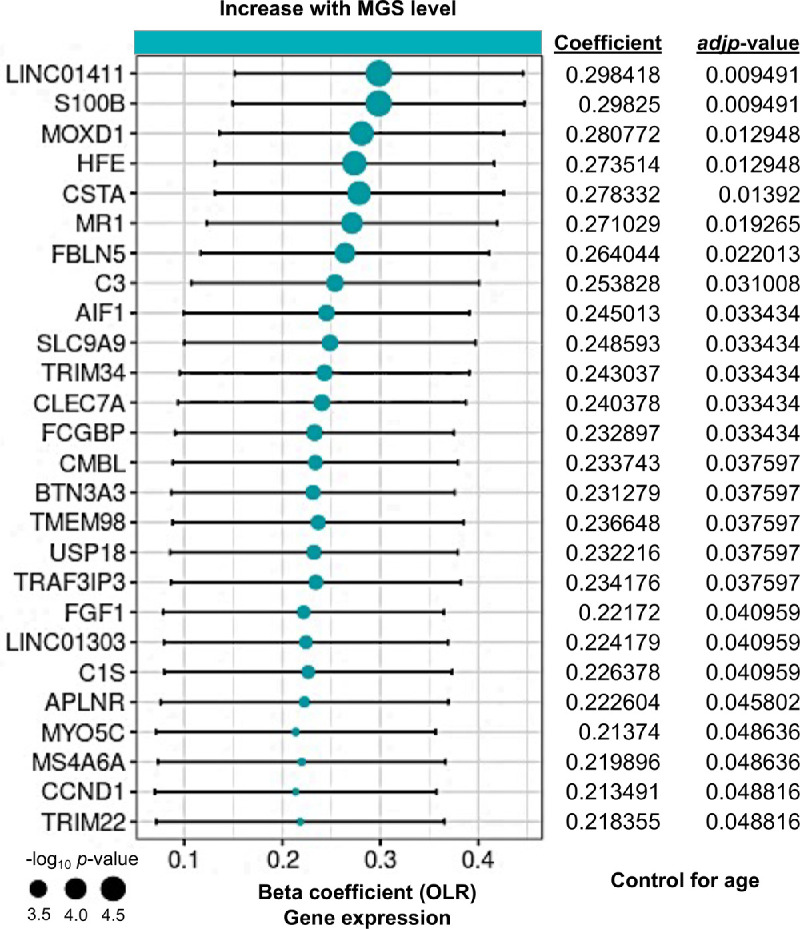
Identification of age-associated genes progressively increased with AMD severity (MGS level). Forest plot of age-associated gene having significant correlation with MGS level (age-MGS genes, n = 26, adjusted P < 0.05). Positive coefficients indicate age-associated genes increase in expression with MGS level. We focused the genes with increased expression over MGS level and defined them. Statistical analyses of MGS association for age-up genes were assessed by a nonparametric ordinal logistic regression model that controls for age. Point sizes are scaled by statistical significance. Error bars represent 95% confidence intervals.
Immune dysregulation is an underling alteration in AMD as in other inflammatory diseases thought to be degenerative due to aging.30 Many age-MGS genes play critical roles in immune responses and inflammation in the retina. For example, S100 calcium binding protein B (S100B), a well-known biomarker of active neural distress that is involved in many neurodegenerative diseases,31 are pro-inflammation and can induce retinal degeneration in animal models.32–34 Homeostatic iron regulator (HFE), an RPE-enriched protein, regulates iron absorption by decreasing the affinity of the transferrin receptor for transferrin.35 Mutation of this gene may lead to retinal iron overload predisposing to AMD.36 Membrane spanning 4 domains A6A (MS4A6A) has been identified as an age-related gene and been indicated in associated with Alzheimer's disease.37 It is expressed in monocytes, macrophages, and microglia; thus, its expression level is likely reflective of expression in innate immune cells. However, its role in retina-related disease has never been explored.
Retinal endothelium plays important roles in maintenance of retinal statics particularly in the development of retinal vasculature. Among the 26 age-MGS genes, a cluster of genes are related to endothelium or angiogenesis (see Supplementary Data S4), including C3, apelin receptor (APLNR), fibroblast growth factor 1 (FGF1), and fibulin 5 (FBLN5). C3 is a well-reported risky gene implicated in AMD in several GWAS studies,4,38,39 which appear to be involved in immune modulation and the complement system. C3 also plays a role in the imbalance between pro-angiogenic stimulation by vascular endothelial growth factor (VEGF) and anti-angiogenic activation by pigment epithelial derived factor (PEDF) in human retinal RPEs,40 suggesting its potential contribution in angiogenesis in AMD. APLNR expression is limited in the veins and proliferative endothelial cells during physiological retinal angiogenesis,41 suggesting endothelial cells might be active at the early stage of AMD. Indeed, using APLNR inhibitors can selectively prevent pathological retinal angiogenesis in an oxygen-induced retinopathy mouse model.41 Studies have shown FGF1 plays a role in retinal neovascularization through organization of various angiogenic pathways and coordination of cell-cell interactions.42,43 Several studies reported that missense mutations of FBLN5, a calcium-binding epidermal growth factor-rich extracellular matrix protein, cause structural changes that has been associated with AMD.44,45 Although these genes appear to be involved in the angiogenic activities at different stage of AMD, their role in contributing to the severity of AMD remains unstudied.
Some age-MGS genes were not clustered into the two groups mentioned above (see Supplementary Data S4) but their significance in relation to disease severity remained high (see Fig. 2). For example, long non-coding RNA genes (lncRNAs) are widely expressed and are distinct from mRNA, which are important regulators of the epigenetic status of the human genome, leading to modulation of chromatin function, regulation of the stability of mRNA, and ultimately alteration of gene expression.46 We found that long intergenic non-protein coding RNA 1411 (LINC01411) and 1303 (LINC01303) are among the top ranked age-MGS genes, suggesting these lncRNAs are directly involved in the progression of AMD, although its function remains unclear. Few studies investigated the function of monooxygenase DBH like 1 (MOXD1), however, GO annotation of MOXD1 revealed its involvement in octopamine biosynthetic process, dopamine catabolic process, and norepinephrine biosynthetic process (see Supplementary Data S4). Our results showed an increased expression level of MOXD1 with disease severity, implying a progressive elevation of those neurotransmitters in the retinas of patients with AMD with age and disease severity. Increasing evidence showed that neural inputs, such as chronic stress or inflammation, resulted in the release of neurotransmitters (e.g. norepinephrine) at specific vessels in the blood-brain barrier, that in turn augment the expression of chemokines in the endothelium to establish gateways through which immune cells can reach the central nervous system.47–50 Given that the retina displays similarities to the brain and spinal cord in terms of functional, anatomic, and immunological properties,51 it is possible that neurotransmitters in the retina might also be increased to stimulate immune responses under chronic stress resulting from AMD.
Correlation of Age-MGS Genes and Their Enrichment in Retinal Cell Types
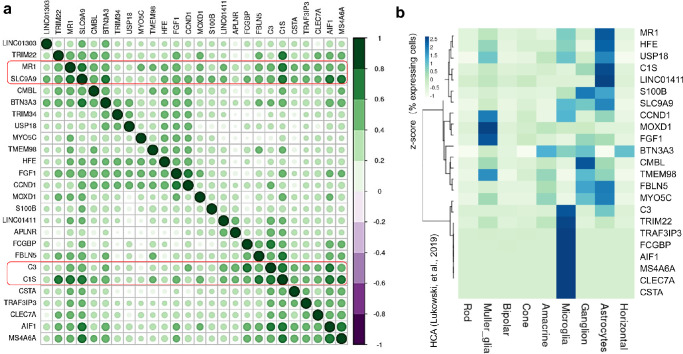
We next sought to understand the correlation between the expression of 26 age-MGS genes. Using the combined transcriptomic data with age controlled, correlation analysis revealed several age-MGS genes have stronger correlation (|R| ≥ 0.4) with other genes, including C3, complement C1s (C1s), major histocompatibility complex (MHC) class I-related (MR1) and solute carrier family 9 member A9 (SLC9A9; Fig. 3a), suggesting their crucial roles in coordinating other genes for multiple biological processes in AMD. The C3 gene variants are strongly in relation to AMD development.4,38,39 Increasing evidence also pinpoints the potential of C3 as a therapeutic target for AMD preceding geographic atrophy.52 Indeed, C3 inhibitor pegcetacoplan has been clinically trialed to treat the geographic atrophy of AMD, currently in the phase II and III stages.53 Results from the phase II trial indicated local C3 inhibition with pegcetacoplan resulted in significant decrease of the growth of geographic atrophy compared with the sham treatment.53 C1s is a single chain glycoprotein and cleaves the components C4 and C2 to initiate the complement cascade when it becomes activated.54 Transcriptome profiling of retina tissue from AMD donor eyes (31 healthy and 26 eyes with AMD) suggested a direct role of dysregulation of C1s among other complement pathways in retinal degeneration.55 Moreover, upregulation of C1s along with C4a and C2 were found in retinas of rats subjected to photo-oxidative damage.56 However, the functional role of C1s in retinal degeneration remains unclear. The MR1 molecule is distinct from MHC class I molecules that can present microbial riboflavin metabolites to a specialized subset of T cells known as mucosal-associated invariant T (MAIT) cells.57 Interestingly, a study showed that MR1 was upregulated at early stages of retinal degeneration in DBA/2J mice featured an immune response in the iris and death of retinal ganglion cells,58 suggesting its potential role in the progression of immune-relevant retinal diseases. SLC9A9 is highly expressed in the brain and is implicated with immune regulation and endocytosis via mTOR signaling and cell survival.59 However, little is known about its role in the retina.
Figure 3.
Correlation of age-MGS genes and their enrichment in retinal cell types. (a) Heatmap of the correlation matrix across 26 age-MGS genes identified in Figure 2. Pearson's correlation was calculated among 26 age-MGS genes to show the co-expression patterns of genes adjusted by age in the heatmap. Color key denotes the Pearson's correlation higher than absolute value 0.3 between genes. Genes highlighted in red indicates their stronger correlation with many other genes. (b) Heatmap showing the percentage of retina cells expressing each of age-MGS genes, scaled by gene across the different cell types. Data are from HCA.
To assess the cell type-specific expression pattern of the age-associated genes and age-MGS genes, we further analyzed the neural retina scRNA-seq data from the HCA (Supplementary Fig. S7).10 We found that the expression pattern of the age-associated genes is distinctive among nine retinal cell types, and a higher percentage of cells expressing these genes are Müller glia, microglia, ganglion, and astrocytes (Supplementary Fig. S8). Age-MGS genes are more enriched in Müller glia, microglia and astrocytes (Fig. 3b). Müller glia and astrocytes express a few similar age-MGS genes but their percentage of expressing cells are clearly different (see Fig. 3b). Consistent with our finding, a previous study reported that Müller glia and astrocytes among other major retinal cell types express the highest number of AMD-associated genes identified by a GWAS study.17 Müller glia and astrocytes assist metabolism of retinal neurons through releasing trophic factors, recycling neurotransmitter glutamate, and controlling extracellular ion homeostasis,60,61 whereas astrocytes are resident macrophages and are implicated in neuroinflammatory changes during age-dependent retinal diseases, including AMD.62 Together, these analyses highlight the cellular landscape of age-MGS genes in the context of retinal cell types in which these genes are normally expressed. Of note, many age-MGS genes were not expressed in any of the neural retina cell types, and their expression in retinal cell types in the condition of AMD is not clear. We also found that correlation between AMD-associated genes identified through different methodologies, such as LRT combined with OLR (present study), the transcriptome-wide association study (TWAS; using EyeGEx)6,7 and GWAS,4 is clearly distinct (Supplementary Fig. S9a), indicating that these are limited interactions between these genes. However, genes identified by TWAS (9 genes) and GWAS (25 leading genes from 34 risk loci associated with AMD, sample size >15,000) are closer to each other, as analyses of TWAS are based on GWAS result. Using HCA data, we validated leading AMD-associated genes previously identified GWAS4 and showed similar results that higher percentage of associated cells expressing AMD-associated genes are Müller glia and astrocytes (Supplementary Fig. S9b).
Functional Roles of Age-MGS-Associated Genes in AMD
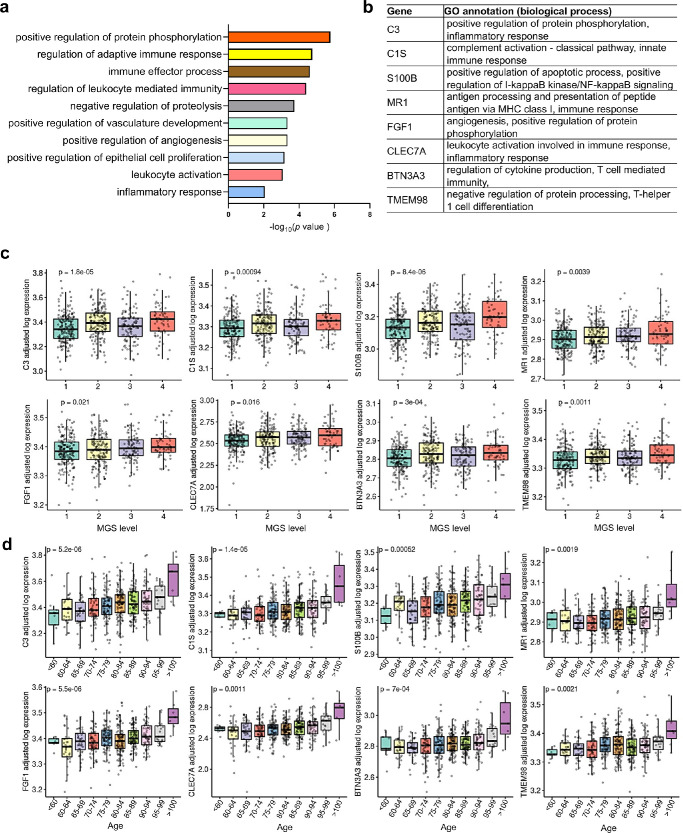
To determine the functional roles of these age-MGS genes involved in the pathogenesis of AMD, we applied GO enrichment analysis (biological process) by Metascape. The results revealed top significant enrichment most related to regulation of immune response and vasculature development (Fig. 4a, Supplementary Data S5). We combined genes involved in these biological processes (see Supplementary Data S5) and focused on the eight age-MGS genes (Fig. 4b) with significant changes with both MGS levels (Fig. 4c) and age (Fig. 4d), including C3, C1s, S100B, MR1, FGF1, C-type lectin domain family 7 member A (CLEC7A), butyrophilin subfamily 3 member A3 (BTN3A3), and transmembrane protein 98 (TMEM98).
Figure 4.
Functional roles of age-MGS genes in AMD. (a) Functional enrichment analyses (gene ontology annotations of biological processes) of 50 age-MGS genes by Metascape. (b) Eight selected age-MGS genes based on the GO annotations in Figure 4a. (c) Tukey boxplots (interquartile range [IQR] boxes with 1.5 × IQR whiskers) showing the expression of C3, C1s, S100B, MR1, FGF1, CLEC7A, BTN3A3, and TMEM98. Gene expression value are shown as log-transformed, controlled for age (Fig. 4c) or MGS level (Fig. 4d). Statistical significance of age-associated or MGS-associated difference was assessed by two-sided Kruskal-Wallis test on the adjusted expression values.
C3, C1s, and MR1 may play a central role in the regulation of innate immune responses because these genes have higher correlation scores with more age-MGS genes that we aforementioned (see Fig. 3a) and are reported to be related to retinal degeneration.56,58 However, their individual roles in regulating immune response in the retina remains unclear. CLEC7A is a pattern recognition receptor that recognizes β-1,3 glucans, and its stimulation initiates inflammatory signaling events from human dendritic cells.63 Knockout of CLEC7A in mice induces autoimmune pathologies and generates stronger cytotoxic T cell-mediated immune responses against apoptotic cells.64 BTN3A3 has been suggested to contribute to the maintenance of the immune system through inhibition of excessive cellular immune response.65 BTN3A3 is also indicated in the modulation of gamma delta T cells that participate in tissue surveillance and infection.66 It is reasonable to speculate that BTN3A3 is a potentially important regulator of increased gamma delta T cells during the progression of AMD that our deconvolution analyses identified. These results collectively suggested that CLEC7A may take part in the regulation of immune response in AMD. TMEM98 is involved in biological processes, such as immune response or inflammation (see Supplementary Data S5), whereas their function in the stressed retina is not clear. Together, our results suggested that these eight age-MGS genes collectively regulate innate immune response, promote leukocyte activation and endothelial cell proliferation, and ultimately retinal angiogenesis. As little is known about the role of the eight gene in the degenerated retina, functional studies in the future would be important to explore how they interact and lead to retinal lesions.
The Cellular Landscape of the Retina of Patients With AMD and its Relation to Age-MGS Genes
Previous studies have shown cellular changes occurred in the retinas of patients with AMD, including Müller glia, astrocytes, microglia, and RPE.67,68 However, detailed quantification of these cellular changes remains a challenge. Here, we deconvoluted the combined bulk retina transcriptomes with CIBERSORTx to quantify cellular changes, using the scRNA-seq data from HCA as a reference (see Supplementary Fig. S7, Supplementary Data S6). To increase the confidence of down-stream analyses, the cell types were retained if their estimated proportions were >0 in all samples (Supplementary Fig. S10).
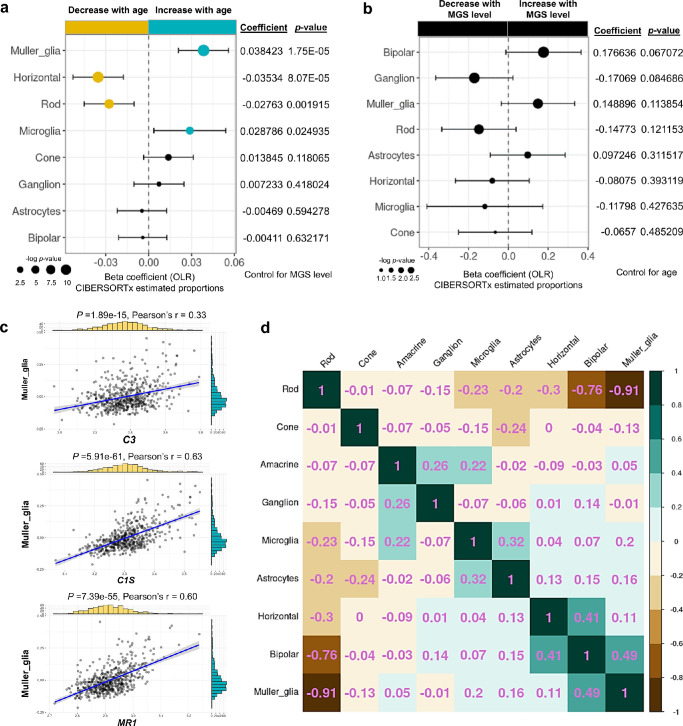
To assess whether age as a risk factor affects cell type proportions, we used an OLR model to control for MGS level and identified age-associated alteration in the proportions of retinal cell types. We observed that the proportions of Müller glia and microglia were significantly increased with age, whereas the proportions of horizontal and rod cells were significantly decreased with age (see Fig. 5a, Supplementary Fig. S11). Next, we focused on the risk factor of MGS level to assess its effect on cell proportions by controlling for age using an OLR model. Although the proportions of Müller glia and rod were increased and decreased, respectively, with MGS levels, the changes were not significant (Fig. 5b, Supplementary Fig. S12), suggesting that disease severity has limited effects to alter cell proportions in the retinas of patients with AMD. Of note, amacrine cells were removed in Figure 5b due to its zero estimated expression profile across all samples after adjustment for age. Previous study using immunolabeling of glial fibrillary acidic protein (GFAP), a marker of retinal glial cells, observed morphological changes and up-modulation of astrocytes and Müller glia in the inner and outer retina, respectively, in a small number of human donor retinas (normal = 6, AMD = 11).67 Retina microglia are primary resident immune cells in the retina, which are believed to be stimulated to infiltrate into the outer retina under the condition of advanced age and during the progression of AMD.69 Rods loss in aging and age-related maculopathy has been widely reported.70,71 Moreover, rods die in older eyes without evidence of overt retinal pigment epithelial degeneration,70 indicating that age is a main factor to cause rods loss. Horizontal cells form triad synapses with photoreceptors and bipolar cells to module signal transmission in between these cells.72 The scRNA-Seq profiling of the aging human retina revealed a horizontal cell subtype (ISL1+ CALB1+) were greatly reduced particularly in the fovea, suggesting that age may affect different regions in the retina.73 Together, these results suggested that age is a central risk factor in AMD that enables alterations in the proportions of cell types in the retina (i.e. Müller glia, microglia, horizontal cells, and rod photoreceptors).
Figure 5.
The cellular landscape of the retina of patients with AMD and its relation to Age-MGS genes. (a, b) Forest plot of estimated proportions of retinal cells having significant correlation with age or MGS level, respectively (P < 0.05). Positive coefficients indicate the proportion of cells increases with age or MGS level, while negative coefficients indicate the proportion of cells decrease with age or MGS level. Statistical analyses of age association or MGS association for the proportions of cells were assessed by a nonparametric ordinal logistic regression model, adjusted for MGS level (a) or age (b), respectively. Point sizes are scaled by statistical significance. Error bars represent 95% confidence intervals. (c) Analyses of the correlation between expression level of C3, C1s, or MR1 and the estimated proportion of Müller glia, adjusted for age. Pearson's correlation coefficient was used to test the strength of linear relationships between gene expression and the estimated proportion of Müller glia. Black dots denote individual samples (n = 547). Blue lines denote regression lines. (d) Heatmap of the correlation matrix among retinal cell types, based on the proportions of retina cells estimated by CIBERSORTx and adjusted for age. Pearson's correlation was calculated among retinal cells to show the co-expression patterns of cells in the heatmap. Values in the heatmap indicates the Pearson correlation coefficient between two cell types.
As most of age-MGS genes are enriched in retinal glial cells, including Müller glia and the increased proportions of Müller glia with age in the retina, we specifically investigated the relationship between Müller glia and three core age-MGS genes we identified (Fig. 5c). We showed that expression of C1s and MR1 or C3 have a strong (Pearson correlation coefficient, 0.6 < r < 1) or weak (0.2 < r < 0.4) positive relationship with the estimated proportions of Müller glia. We also observed moderate negative (−0.4 < r < −0.6) correlation among the expression of C1s, MR1, and the estimated proportions of rod photoreceptors (Supplementary Fig. S13a), whereas weak positive correlation (0.2 < r < 0.4) between these two genes and the estimated proportions of microglia (Supplementary Figs. S13b, S13c). Moreover, there was weak correlation between expression of C3 and the estimated proportions of rod photoreceptors or microglia. Notably, no correlations (0 < r < 0.2) were observed among the expression of three age-MGS genes and the estimated proportions of horizontal cells. These results suggested that C1s and MR1 are potential core age-MGS genes to regulate Müller glia and rod photoreceptors but not horizontal cells and microglia that are involved in retina aging. Photoceptor degeneration and Müller glia activation are indicative of structure and molecular abnormalities in the development of AMD,74 suggesting their possible complementary relationship in this form of retinal degeneration. Indeed, our results revealed that there is a strong negative correlation (r = −0.91) between rod photoreceptors and Müller glia based on its estimated proportions after controlling for age (Fig. 5d).
Discussion
In the present study, we systematically analyzed the retinal aging transcriptome and cellular landscape in association with the progression of AMD. We revealed that the aging retina is characterized by a range of biological processes, in particular inflammatory response and immune response, that could lead to the progressively severe AMD. We specifically studied the immune cell profiles of the combined retinal transcriptomic database and identified the upregulation of M2 macrophages with both age and disease severity in patients with AMD. We subsequently pinpointed a group of retinal aging genes that change with disease severity (age-MGS genes) by controlling for age. Many of the age-MGS genes have function mainly in relation to immune response, endothelium-mediated angiogenesis, and retinoic acid metabolic process. Through gene correlation analysis, we demonstrated that C3, C1s, MR1, and SLC9A9 have stronger correlation with other age-MGS genes, and all are involved in regulation of immune response. By integrating these data with the single-cell transcriptome of the human neural retina, we further identified specific cell types that normally express these age-MGS genes. GO enrichment analysis of the age-MGS genes further suggested the aging and AMD retina is characterized by adaptive immune response, regulation of angiogenesis, and leukocyte activation. We further identified eight age-MGS genes in these biological processes that potentially play critical roles in the progressive AMD. Through bulk deconvolution analyses, we showed that the proportions of Müller glia and microglia are increased with age, whereas the proportions of rod photoreceptors and horizontal cells are decreased with age. Moreover, the proportions of all retinal cell types do not have significant changes with disease severity after factoring out age, suggesting that age is potentially the central risk factor that uniquely contributes to the severity of AMD. Correlation analysis among eight age-MGS genes and the proportions of cells with significant alteration revealed that C1s and MR1 have a strong relationship with Müller glia and rod photoreceptors, both of which are negatively correlated. Future research to investigate the function of these age-MGS genes in the retina would be important to understand the mechanism underly AMD pathology.
Because the transcriptome data from human donors are complexed with a variety of latent factors, such as sex, age, and clinical diagnosis, it is important to use batch correction to adjust those factors for the downstream analysis. Throughout our analysis, all the raw data were subjected to the batch correction before relevant analysis. We noted a study using various deconvolution algorithms to analyze involvement of retinal cell types in patients with AMD with different databases, including EyeGEx.75 However, all the raw data from this study were not subjected to batch correction, thus the conclusion resulted from was less stringent compared to the present study. A clear example of difference between adjusted and unadjusted data analysis can be referred to the cell type proportion estimated by CIBERSORTx (see Supplementary Figs. S4, S6, S11, S12). There were several limitations of this study. For example, the sample size in each of age group is relatively small particularly the group of the youngest (<60) and the oldest (100–109), which likely affects the number of aging genes defined by the LRT model. Moreover, the HCA dataset does not include RPE cells for which we might miss information on how the age-MGS genes involving in the degeneration of RPE cells in AMD.76 Whereas the MGS grading system, a classification system for the postmortem human eye bank tissue, was used to classify the samples in the present study, such grading system corresponds with the classification of AMD described in the Age-related Eye Disease Study (AREDS) that features clear symptoms of dry and wet AMD.77 In addition, we used the human hematopoietic immune cell matrix, LM22, to deconvolute immune cell profiles in the retinas, potentially missing information on resident immune cell types, such as microglia, in neural tissues, including the brain and retinas. Future studies to determine the change of the expression of the age-MGS gene in RPE cells and to generate transcriptome dataset for resident immune cells in neural tissues would be helpful. Furthermore, whether any of these age-MGS-associated changes causally contribute to the progression of AMD in older populations remains to be experimentally tested. It is worth noting that the cellular proportions are different between fovea and peripheral retinal under the normal condition.78 Our datasets were largely acquired from whole retina samples so that our prediction of cellular proportion in AMD might be confounded by cells of the fovea and peripheral retinas. Besides, given that the transcriptomic dataset required here were from neural retina, caution should be taken when apply our findings to outer retinas in patients with AMD, a disease primarily affecting the outer retina.79 Our study defined the aging genes in the retina using the LRT model by controlling for disease severity of AMD and identified novel aging genes associated with AMD progression. We also highlighted the potential for using deconvolution analysis to understand cellular and transcriptomic changes in AMD. The functions of most of the age-MGS genes remain unclear in the retina and more research is needed to understand their role in progressive development of AMD. Our analyses provide potential directions for subsequent research on the aging retina for which AMD progressively gets worse over age.
Supplementary Material
Acknowledgments
The authors thank Ryan D. Chow for sharing codes for data analyses at Github.
Supported by grants from the National Health and Medical Research Council of Australia (GNT1185600). The Centre for Eye Research Australia receives Operational Infrastructure Support from the Victorian Government.
Author Contribution: J.-H.W. and G.-S.L. conceived and designed the study. J.-H.W. developed the analysis approach, performed all data analyses, and generated the figures. J.-H.W., R.C.B.W., and G.-S.L. prepared the manuscript. G.-S.L. supervised the work.
Data Availability: All processed data files generated for this study are included in this article and its supplementary information files. Raw data files from two sources are available for download through GEO under the accession numbers GSE115828 and GSE135092 and Human Cell Atlas under accession number E-MTAB-7316.
Code Availability: Analysis scripts will be made available upon request.
Disclosure: J.-H. Wang, None; R.C.B. Wong, None; G.-S. Liu, None
References
- 1. Evans JR. Risk factors for age-related macular degeneration. Prog Retin Eye Res. 2001; 20: 227–253. [DOI] [PubMed] [Google Scholar]
- 2. Lambert NG, ElShelmani H, Singh MK, et al.. Risk factors and biomarkers of age-related macular degeneration. Prog Retin Eye Res. 2016; 54: 64–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bressler NM. Age-related macular degeneration is the leading cause of blindness. JAMA. 2004; 291: 1900–1901. [DOI] [PubMed] [Google Scholar]
- 4. Fritsche LG, Igl W, Bailey JN, et al.. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016; 48: 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Orozco LD, Chen HH, Cox C, et al.. Integration of eQTL and a single-cell atlas in the human eye identifies causal genes for age-related macular degeneration. Cell Rep. 2020; 30: 1246–1259.e1246. [DOI] [PubMed] [Google Scholar]
- 6. Ratnapriya R, Sosina OA, Starostik MR, et al.. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat Genet. 2019; 51: 606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ratnapriya R, Sosina OA, Starostik MR, et al.. Author Correction: Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat Genet. 2019; 51: 1067. [DOI] [PubMed] [Google Scholar]
- 8. Whitcup SM, Sodhi A, Atkinson JP, et al.. The role of the immune response in age-related macular degeneration. Int J Inflam. 2013; 2013: 348092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stuart T, Butler A, Hoffman P, et al.. Comprehensive integration of single-cell data. Cell. 2019; 177: 1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lukowski SW, Lo CY, Sharov AA, et al.. A single-cell transcriptome atlas of the adult human retina. EMBO J. 2019; 38: e100811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou Y, Zhou B, Pache L, et al.. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019; 10: 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wadi L, Meyer M, Weiser J, Stein LD, Reimand J.. Impact of outdated gene annotations on pathway enrichment analysis. Nat Methods. 2016; 13: 705–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Q, Shepherd BE, Li C, Harrell FE Jr. Modeling continuous response variables using ordinal regression. Stat Med. 2017; 36: 4316–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ritchie ME, Phipson B, Wu D, et al.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei T, Simko V, Levy M, Xie Y, Jin Y, Zemla J. Corrplot: Visualization of a correlation matrix. R package version 073 2013;230.
- 17. Menon M, Mohammadi S, Davila-Velderrain J, et al.. Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat Commun. 2019; 10: 4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chow RD, Majety M, Chen S.. The aging transcriptome and cellular landscape of the human lung in relation to SARS-CoV-2. Nat Commun. 2021; 12: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Newman AM, Steen CB, Liu CL, et al.. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019; 37: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA.. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018; 1711: 243–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen M, Muckersie E, Forrester JV, Xu H.. Immune activation in retinal aging: A gene expression study. Invest Ophthalmol Vis Sci. 2010; 51: 5888–5896. [DOI] [PubMed] [Google Scholar]
- 22. Chen M, Luo C, Zhao J, Devarajan G, Xu H.. Immune regulation in the aging retina. Prog Retin Eye Res. 2019; 69: 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cao X, Shen D, Patel MM, et al.. Macrophage polarization in the maculae of age-related macular degeneration: A pilot study. Pathol Int. 2011; 61: 528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kelly J, Ali Khan A, Yin J, Ferguson TA, Apte RS. Senescence regulates macrophage activation and angiogenic fate at sites of tissue injury in mice. J Clin Invest. 2007; 117: 3421–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tarique AA, Logan J, Thomas E, Holt PG, Sly PD, Fantino E.. Phenotypic, functional, and plasticity features of classical and alternatively activated human macrophages. Am J Respir Cell Mol Biol. 2015; 53: 676–688. [DOI] [PubMed] [Google Scholar]
- 26. Yang Y, Liu F, Tang M, et al.. Macrophage polarization in experimental and clinical choroidal neovascularization. Sci Rep. 2016; 6: 30933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou Y, Yoshida S, Kubo Y, et al.. Different distributions of M1 and M2 macrophages in a mouse model of laser-induced choroidal neovascularization. Mol Med Rep. 2017; 15: 3949–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hagbi-Levi S, Grunin M, Jaouni T, et al.. Proangiogenic characteristics of activated macrophages from patients with age-related macular degeneration. Neurobiol Aging. 2017; 51: 71–82. [DOI] [PubMed] [Google Scholar]
- 29. Zhao Z, Xu P, Jie Z, et al.. gammadelta T cells as a major source of IL-17 production during age-dependent RPE degeneration. Invest Ophthalmol Vis Sci. 2014; 55: 6580–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nussenblatt RB, Lee RW, Chew E, et al.. Immune responses in age-related macular degeneration and a possible long-term therapeutic strategy for prevention. Am J Ophthalmol. 2014; 158: 5–11.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Michetti F, D'Ambrosi N, Toesca A, et al.. The S100B story: From biomarker to active factor in neural injury. J Neurochem. 2019; 148: 168–187. [DOI] [PubMed] [Google Scholar]
- 32. Reinehr S, Reinhard J, Gandej M, et al.. S100B immunization triggers NFkappaB and complement activation in an autoimmune glaucoma model. Sci Rep. 2018; 8: 9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grotegut P, Perumal N, Kuehn S, et al.. Minocycline reduces inflammatory response and cell death in a S100B retina degeneration model. J Neuroinflammation. 2020; 17: 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Niven J, Hoare J, McGowan D, et al.. S100B up-regulates macrophage production of IL1beta and CCL22 and influences severity of retinal inflammation. PLoS One. 2015; 10: e0132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Feder JN, Penny DM, Irrinki A, et al.. The hemochromatosis gene product complexes with the transferrin receptor and lowers its affinity for ligand binding. Proc Natl Acad Sci USA. 1998; 95: 1472–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dunaief JL. Iron induced oxidative damage as a potential factor in age-related macular degeneration: The Cogan Lecture. Invest Ophthalmol Vis Sci. 2006; 47: 4660–4664. [DOI] [PubMed] [Google Scholar]
- 37. Hollingworth P, Harold D, Sims R, et al.. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011; 43: 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maller J, George S, Purcell S, et al.. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006; 38: 1055–1059. [DOI] [PubMed] [Google Scholar]
- 39. Yates JR, Sepp T, Matharu BK, et al.. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007; 357: 553–561. [DOI] [PubMed] [Google Scholar]
- 40. Long Q, Cao X, Bian A, Li Y.. C3a increases VEGF and decreases PEDF mRNA levels in human retinal pigment epithelial cells. Biomed Res Int. 2016; 2016: 6958752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ishimaru Y, Shibagaki F, Yamamuro A, Yoshioka Y, Maeda S.. An apelin receptor antagonist prevents pathological retinal angiogenesis with ischemic retinopathy in mice. Sci Rep. 2017; 7: 15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Murakami M, Simons M.. Fibroblast growth factor regulation of neovascularization. Curr Opin Hematol. 2008; 15: 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guillonneau X, Regnier-Ricard F, Dupuis C, Courtois Y, Mascarelli F.. FGF2-stimulated release of endogenous FGF1 is associated with reduced apoptosis in retinal pigmented epithelial cells. Exp Cell Res. 1997; 233: 198–206. [DOI] [PubMed] [Google Scholar]
- 44. Jones RP, Ridley C, Jowitt TA, et al.. Structural effects of fibulin 5 missense mutations associated with age-related macular degeneration and cutis laxa. Invest Ophthalmol Vis Sci. 2010; 51: 2356–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schneider R, Jensen SA, Whiteman P, McCullagh JS, Redfield C, Handford PA.. Biophysical characterisation of fibulin-5 proteins associated with disease. J Mol Biol. 2010; 401: 605–617. [DOI] [PubMed] [Google Scholar]
- 46. Statello L, Guo CJ, Chen LL, Huarte M.. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021; 22: 96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deutschman CS, Tracey KJ.. Sepsis: Current dogma and new perspectives. Immunity. 2014; 40: 463–475. [DOI] [PubMed] [Google Scholar]
- 48. Tracey KJ. Reflexes in immunity. Cell. 2016; 164: 343–344. [DOI] [PubMed] [Google Scholar]
- 49. Pavlov VA, Tracey KJ.. Neural regulation of immunity: Molecular mechanisms and clinical translation. Nat Neurosci. 2017; 20: 156–166. [DOI] [PubMed] [Google Scholar]
- 50. Stofkova A, Kamimura D, Ohki T, Ota M, Arima Y, Murakami M.. Photopic light-mediated down-regulation of local alpha1A-adrenergic signaling protects blood-retina barrier in experimental autoimmune uveoretinitis. Sci Rep. 2019; 9: 2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. London A, Benhar I, Schwartz M.. The retina as a window to the brain-from eye research to CNS disorders. Nat Rev Neurol. 2013; 9: 44–53. [DOI] [PubMed] [Google Scholar]
- 52. Kim BJ, Liu T, Mastellos DC, Lambris JD.. Emerging opportunities for C3 inhibition in the eye. Semin Immunol. 2022;101633. [DOI] [PubMed] [Google Scholar]
- 53. Liao DS, Grossi FV, El Mehdi D, et al.. Complement C3 inhibitor pegcetacoplan for geographic atrophy secondary to age-related macular degeneration: A randomized phase 2 trial. Ophthalmology. 2020; 127: 186–195. [DOI] [PubMed] [Google Scholar]
- 54. Daugan MV, Revel M, Russick J, et al.. Complement C1s and C4d as prognostic biomarkers in renal cancer: Emergence of noncanonical functions of C1s. Cancer Immunol Res. 2021; 9: 891–908. [DOI] [PubMed] [Google Scholar]
- 55. Newman AM, Gallo NB, Hancox LS, et al.. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med. 2012; 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Natoli R, Fernando N, Jiao H, et al.. Retinal macrophages synthesize C3 and activate complement in AMD and in models of focal retinal degeneration. Invest Ophthalmol Vis Sci. 2017; 58: 2977–2990. [DOI] [PubMed] [Google Scholar]
- 57. Gold MC, Napier RJ, Lewinsohn DM.. MR1-restricted mucosal associated invariant T (MAIT) cells in the immune response to mycobacterium tuberculosis. Immunol Rev. 2015; 264: 154–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fan W, Li X, Wang W, Mo JS, Kaplan H, Cooper NG.. Early involvement of immune/inflammatory response genes in retinal degeneration in DBA/2J mice. Ophthalmol Eye Dis. 2010; 1: 23–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Patak J, Hess JL, Zhang-James Y, Glatt SJ, Faraone SV.. SLC9A9 co-expression modules in autism-associated brain regions. Autism Res. 2017; 10: 414–429. [DOI] [PubMed] [Google Scholar]
- 60. Vecino E, Rodriguez FD, Ruzafa N, Pereiro X, Sharma SC.. Glia-neuron interactions in the mammalian retina. Prog Retin Eye Res. 2016; 51: 1–40. [DOI] [PubMed] [Google Scholar]
- 61. Reichenbach A, Bringmann A.. New functions of Muller cells. Glia. 2013; 61: 651–678. [DOI] [PubMed] [Google Scholar]
- 62. Ma W, Wong WT.. Aging changes in retinal microglia and their relevance to age-related retinal disease. Adv Exp Med Biol. 2016; 854: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Goodridge HS, Reyes CN, Becker CA, et al.. Activation of the innate immune receptor Dectin-1 upon formation of a 'phagocytic synapse'. Nature. 2011; 472: 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bode K, Bujupi F, Link C, et al.. Dectin-1 binding to annexins on apoptotic cells induces peripheral immune tolerance via NADPH oxidase-2. Cell Rep. 2019; 29: 4435–4446.e4439. [DOI] [PubMed] [Google Scholar]
- 65. Yamashiro H, Yoshizaki S, Tadaki T, Egawa K, Seo N.. Stimulation of human butyrophilin 3 molecules results in negative regulation of cellular immunity. J Leukoc Biol. 2010; 88: 757–767. [DOI] [PubMed] [Google Scholar]
- 66. Rhodes DA, Chen HC, Price AJ, et al.. Activation of human gammadelta T cells by cytosolic interactions of BTN3A1 with soluble phosphoantigens and the cytoskeletal adaptor periplakin. J Immunol. 2015; 194: 2390–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wu KH, Madigan MC, Billson FA, Penfold PL.. Differential expression of GFAP in early v late AMD: A quantitative analysis. Br J Ophthalmol. 2003; 87: 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jones BW, Pfeiffer RL, Ferrell WD, Watt CB, Tucker J, Marc RE.. Retinal remodeling and metabolic alterations in human AMD. Front Cell Neurosci. 2016; 10: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ma W, Zhao L, Wong WT.. Microglia in the outer retina and their relevance to pathogenesis of age-related macular degeneration. Adv Exp Med Biol. 2012; 723: 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Curcio CA, Medeiros NE, Millican CL.. Photoreceptor loss in age-related macular degeneration. Invest Ophthalmol Vis Sci. 1996; 37: 1236–1249. [PubMed] [Google Scholar]
- 71. Curcio CA. Photoreceptor topography in ageing and age-related maculopathy. Eye (Lond). 2001; 15: 376–383. [DOI] [PubMed] [Google Scholar]
- 72. Sonntag S, Dedek K, Dorgau B, et al.. Ablation of retinal horizontal cells from adult mice leads to rod degeneration and remodeling in the outer retina. J Neurosci. 2012; 32: 10713–10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yi W, Lu Y, Zhong S, et al.. A single-cell transcriptome atlas of the aging human and macaque retina. Natl Sci Rev. 2021; 8: nwaa179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Johnson PT, Lewis GP, Talaga KC, et al.. Drusen-associated degeneration in the retina. Invest Ophthalmol Vis Sci. 2003; 44: 4481–4488. [DOI] [PubMed] [Google Scholar]
- 75. Lyu Y, Zauhar R, Dana N, et al.. Implication of specific retinal cell-type involvement and gene expression changes in AMD progression using integrative analysis of single-cell and bulk RNA-seq profiling. Sci Rep. 2021; 11: 15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. McLeod DS, Grebe R, Bhutto I, Merges C, Baba T, Lutty GA.. Relationship between RPE and choriocapillaris in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009; 50: 4982–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Olsen TW, Feng X.. The Minnesota Grading System of eye bank eyes for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2004; 45: 4484–4490. [DOI] [PubMed] [Google Scholar]
- 78. Voigt AP, Whitmore SS, Flamme-Wiese MJ, et al.. Molecular characterization of foveal versus peripheral human retina by single-cell RNA sequencing. Exp Eye Res. 2019; 184: 234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Acton JH, Smith RT, Hood DC, Greenstein VC.. Relationship between retinal layer thickness and the visual field in early age-related macular degeneration. Invest Ophthalmol Vis Sci. 2012; 53: 7618–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.