Abstract
Flowering transition is tightly coordinated by complex gene regulatory networks, in which AGAMOUS-LIKE 16 (AGL16) plays important roles. Here, we identified the molecular function and binding properties of AGL16 and demonstrated its partial dependency on the SUPPRESSOR OF CONSTANS 1 (SOC1) function in regulating flowering. AGL16 bound to promoters of more than 2,000 genes via CArG-box motifs with high similarity to that of SOC1 in Arabidopsis (Arabidopsis thaliana). Approximately 70 flowering genes involved in multiple pathways were potential targets of AGL16. AGL16 formed a protein complex with SOC1 and shared a common set of targets. Intriguingly, only a limited number of genes were differentially expressed in the agl16-1 loss-of-function mutant. However, in the soc1-2 knockout background, AGL16 repressed and activated the expression of 375 and 182 genes, respectively, with more than a quarter bound by AGL16. Corroborating these findings, AGL16 repressed the flowering time more strongly in soc1-2 than in the Col-0 background. These data identify a partial inter-dependency between AGL16 and SOC1 in regulating genome-wide gene expression and flowering time, while AGL16 provides a feedback regulation on SOC1 expression. Our study sheds light on the complex background dependency of AGL16 in flowering regulation, thus providing additional insights into the molecular coordination of development and environmental adaptation.
Introduction
Timely transitions from vegetative to reproductive growth (floral transition) and from dormant to germinating seeds determine the capacity of plant adaptation to changing environments, thus are under tight control by complex interactions between endogenous signals and exogenous environmental factors (Michaels 2009; Andres and Coupland 2012; Nee et al. 2017). The gene regulatory network (GRN) controlling floral transition converges at several floral integrator genes like SUPPRESSOR OF CONSTANS1 (SOC1) and FLOWERING LOCUS T (FT). These genes often encode transcription regulators controlling the transcription of their downstream targets by binding to specific cis-motifs, for example, CArG-boxes (Michaels 2009; Fornara et al. 2010; Andres and Coupland 2012). CArG-box motifs are binding sites specific for MADS-box transcription factors (TFs) like SOC1, FLOWERING LOCUS C (FLC), SHORT VEGETATIVE PHASE (SVP) and SEPALLATA 3 (SEP3) (Immink et al. 2009; Kaufmann et al. 2009; Kaufmann et al. 2010; Deng et al. 2011; Immink et al. 2012; Tao et al. 2012; Gregis et al. 2013; Mateos et al. 2015; Mateos et al. 2017; Aerts et al. 2018). These MADS-box TFs often form homo- and/or hetero-protein complexes that act in concert and bind to the CArG-box motifs in promoters of more than hundreds of downstream genes to regulate flowering time and other developmental processes in Arabidopsis (Arabidopsis thaliana).
SOC1 is one key flowering promoter integrating signals from photoperiod, temperature, hormones, and age-related pathways (Lee and Lee 2010). SOC1 forms a protein complex with AGL24 to activate LFY and AP1 to initiate and maintain flower meristem identity but represses SEP3 to prevent premature differentiation of floral meristem (Lee et al. 2008). SOC1 activates the expression of TARGET OF FLC AND SVP1 (TFS1) via recruiting histone demethylase RELATED TO EARLY FLOWERING 6 (REF6) and chromatin remodeler BRAHMA (BRM), and cooperates with SQUAMOSAL PROMOTER BINDING PROTEIN-LIKE 15 (SPL15) to modulate their targets’ expression thereby regulating flowering time (Hyun et al. 2016; Richter et al. 2019). SOC1 forms a set of heterologous complexes with other MADS-box TFs (de Folter et al. 2005; Immink et al. 2009). Furthermore, SOC1 times flowering downstream of several hormone signaling pathways including gibberellin acid (GA), ABA, and BRs (Jung et al. 2012; Li et al. 2017; Hwang et al. 2019) and of nutrient status (Liu et al. 2013; Olas et al. 2019; Yan et al. 2021). Interestingly, profiling of SOC1 targets also identifies genes involved in the signaling processes of these hormones and nutrients (Immink et al. 2012; Tao et al. 2012). Via protein complexing with many TFs and binding to its own promoter, SOC1 regulates its own expression with auto-regulatory feedback loops (Immink et al. 2012). However, the biological importance of these molecular interactions remains to be explored further.
AGL16 is a floral repressor with dependency on genetic background, photoperiod of growth conditions, and gene dosages (Hu et al. 2014). Only under the inductive long-day conditions loss-of-function mutants for AGL16 show early flowering especially in the functional FRI-FLC background (Johanson et al. 2000; Michaels and Amasino 2001; Hu et al. 2014). AGL16 expression can be modulated by the level of the Brassicaceae-specific miR824, for which natural variation has been identified (Rajagopalan et al. 2006; Fahlgren et al. 2007; Kutter et al. 2007; de Meaux et al. 2008; Hu et al. 2014). Change in miR824 expression results in a significant modification of the plant flowering (Hu et al. 2014). AGL16 acts in flowering time regulation via transcriptional regulation of FT, whose expression is also regulated by TFs like SVP and FLC and many others (Aukerman and Sakai 2003; Searle et al. 2006; Jung et al. 2007; Castillejo and Pelaz 2008; Li et al. 2008; Mathieu et al. 2009). AGL16 forms complexes with SVP and FLC, and mildly represses their expression (Hu et al. 2014). AGL16 is a direct downstream target of both FLC and SVP, but the expression of AGL16 changes only weakly in loss-of-function mutants of both genes (Deng et al. 2011; Gregis et al. 2013; Mateos et al. 2015). Yeast two-hybrid assays suggest that AGL16 interacts with SOC1 and other MADS-box TFs and is hypothesized to modulate the SOC1 expression (de Folter et al. 2005; Immink et al. 2009; Immink et al. 2012). Besides its roles in development (Kutter et al. 2007; Hu et al. 2014), AGL16 represses plant responses to salt stress and drought resistance via binding to a specific set of downstream genes (Zhao et al. 2020; Zhao et al. 2021). The miR824-AGL16 module participates also in heat stress adaptation (Szaker et al. 2019). However, the exact AGL16 target spectra and the impacts of interactions between AGL16 and its partners remain under-explored.
In this study, we profiled the target spectra of AGL16 and demonstrated the molecular and genetic links between AGL16 and SOC1 played important roles in flowering time regulation. We found that, in contrast to its weak effects in flowering time regulation in Col-0 background, AGL16 could bind to more than 2,000 target genes that were involved in regulation of flowering time and many other biological processes. We showed that the regulatory roles of AGL16 on genome-wide gene expression and flowering time depended partially on the SOC1 activity.
Results
AGL16 binds to a large set of genomic segments with CArG-boxes
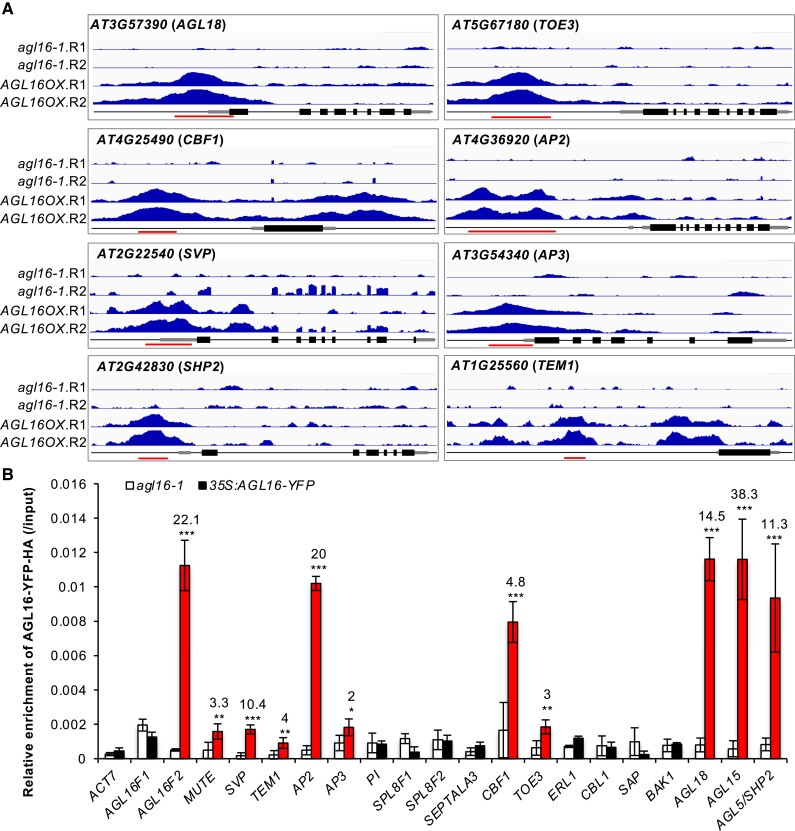
We profiled AGL16 binding sites by a ChIP-seq approach (chromatin immunoprecipitation followed by sequencing). We used a line expressing AGL16 fused to a combined Yellow Fluorescent Protein (YFP)-HA epitope tag under the control of the Cauliflower Mosaic Virus 35S promoter (AGL16OX), which restores the early flowering of agl16-1 to wild type Col-0 level (Supplemental Fig. S1) (Hu et al. 2014). In two independent trials, we identified, respectively, 5,463 and 3,294 DNA segments statistically enriched for AGL16 binding, of which 3,086 were shared (Supplemental Tables S1 and S2). Most of the peaks were around 150–500 bp in both trials (Supplemental Fig. S2). To test whether these segments were real binding sites for AGL16, we carried out ChIP-qPCR assays with two independent chromatin preparations for 20 peaks identified by ChIP-seq. These efforts confirmed 12 regions bound by AGL16-YFP-HA with a minimum 2-fold enrichment in the AGL16OX line compared with agl16-1 background (Fig. 1). Besides these, four known targets including HKT1; 1 (AT4G10310), HsfA6a (AT5G43840), MYB102 (AT4G21440), and CYP707A3 (AT5G45340) were also among the list (Zhao et al. 2020, 2021). Hence, a majority proportion of peaks detected via the ChIP-seq method were reproducibly enriched.
Figure 1.
Validation of the AGL16 binding on target DNA fragments. A) Binding profiles for selected target genes. The TAIR10 annotation of the genomic locus was shown at the bottom of each box. For each panel, the profiles for two trials (R1 and R2) in agl16-1 background line were shown in the upper panel, while the profiles for agl16-1 35S:AGL16-YFP-HA (AGL16OX; two trials) were shown in the middle panel of each box. All the genes were from 5′-end to 3′-end with scale bars indicating sequence lengths of 500 bp. Note that data range for each gene in agl16-1 and AGL16OX was the same scale, but different genes could have different scales. Red lines marked the binding regions tested via ChIP-qPCR assays (B). B) ChIP-qPCR validation of AGL16 binding on 20 DNA segments. Significant enrichment (red bars) was defined with the following criteria: mean enrichment must be at least 2-fold higher than negative control ACT7, the enrichment for AGL16OX (in agl16-1 background) than agl16-1 must be higher than 2-fold change, and the amplification CT number of IP samples must be at least two cycles less than no-antibody controls. This experiment was repeated with another independent trial, which gave similar pattern. Statistics was carried out with Student's t-test with Bonferroni correction. ***P < 0.001; **P < 0.01; *P < 0.05.
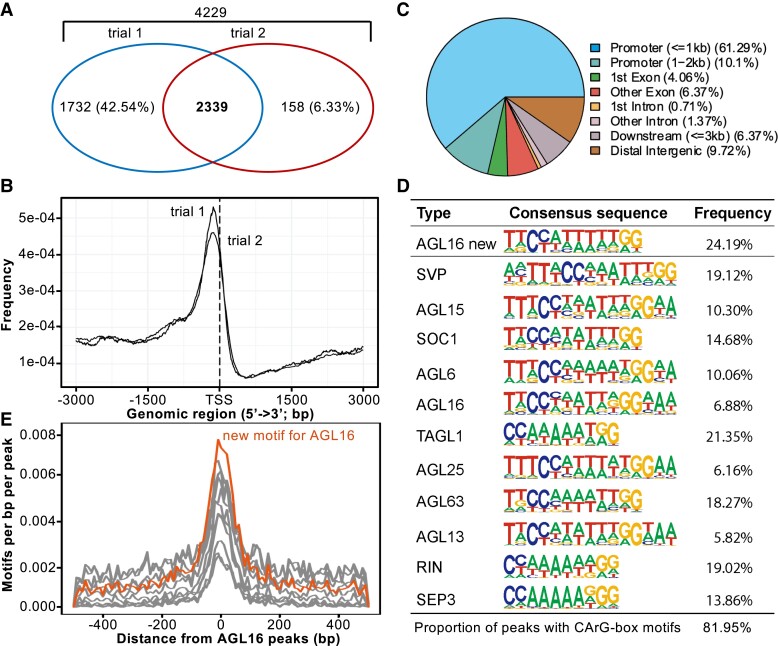
Peaks bound by AGL16 were annotated using Arabidopsis TAIR10 data to identify their distribution and genomic features (Fig. 2). The peaks from both trials were centered to the 3-kb regions around transcriptional start sites (TSS; Fig. 2B). Around 60% of peaks located in the 1-kb regions surrounding TSS (Fig. 2C and Supplemental Table S2). About 10% of peaks were in the 1- to 2-kb promoter regions upstream of TSS, while 10–12% of peaks were in exons/introns. Thus, AGL16 bound to DNA fragments close to TSS of a large set of genes. The 2,339 genes with peaks mapped to gene body or up to 2-kb upstream of their TSS were taken as AGL16 targets (Supplemental Table S2).
Figure 2.
Genome-wide identification of AGL16 target genes via ChIP-seq. A) Venn diagram of AGL16 targets identified in two independent trials. B) Distribution of AGL16 binding sites for two trials surrounding the transcriptional starting site (TSS). C) Location distribution in relative to nearby genes for AGL16 binding sites of trial 1. Peaks within the 3-kb promoter region were taken as AGL16 targets. D) CArG type of motifs over-represented in the AGL16 binding peaks. AGL16 new, which was highly similar to known SOC1 type, showed the de novo motif predicted for AGL16. Frequency gave the percentage for each motif presented in the binding peaks. E) Distribution of new (orange) and known (gray; shown in (D) CArG type of motifs around AGL16 peaks center.
We next searched for potential cis-motifs in the common peaks bound by AGL16 using HOMER, which could predict new motifs and identify known motifs (Heinz et al. 2010). This analysis reported a de novo CArG-box motif CCATTTTTGG for AGL16 in 707 peaks (24.2% of all common peaks; Fisher P = 1e-340, in comparison with 3.8% at genome level; Fig. 2D and Supplemental Table S2). Ten other CArG-box motifs were also significantly enriched, and matched to the known motifs of SVP, SOC1, SEP3, TAGL1, AGL63, and other MADS-box TFs, most of which could potentially interact with AGL16 (Fig. 2D; Supplemental Fig. S3 and Table S2). The de novo and the 10 significantly enriched CArG-box motifs were all distributed around the peak center, indicating that AGL16 bound to its targets via the cluster of CArG-box motifs, just like SOC1 and other MADS-box proteins (Deng et al. 2011; Immink et al. 2012; Tao et al. 2012). There were also other motifs significantly enriched in the AGL16-bound peaks, such as those bound by TCPs (321 peaks), bHLHs (1,131), C2C2 DOFs (2,524), WRKYs (1,039). However, these motifs were not in the peaks center. Since AGL16 modulated significantly the flowering time in Arabidopsis (Hu et al. 2014), we next asked which flowering time genes could be targeted by AGL16.
AGL16 targets flowering time genes in multiple pathways
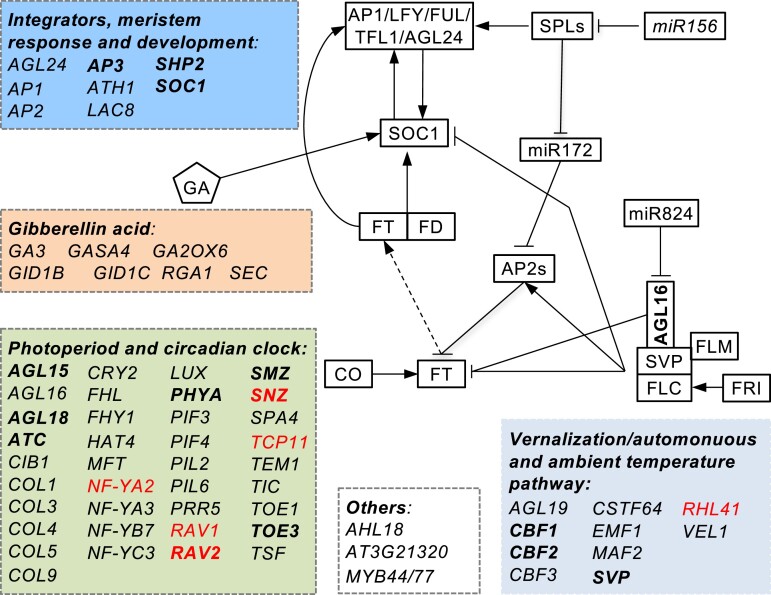
The Arabidopsis genome contains ∼400 flowering time genes, among which around 70 were targeted by AGL16 (Fig. 3 and Supplemental Table S2). This number was significantly larger than randomly expected (Yates’ χ2test, P < 0.0001). Consistent with the described photoperiod dependency for AGL16-mediated flowering regulation (Hu et al. 2014), 37 genes (for example, AGAMOUS-LIKE 15/16/18 (AGL15/AGL16/AGL18), CONSTANS LIKE 1/3/4/5 (COL1/3/4/5), TWIN SISTER OF FT (TSF) and MOTHER OF FT (MFT), etc.) were related to photoperiod and circadian clock pathways (Bouche et al. 2016). Ten genes (like AGL19 and SVP, etc.) were in the vernalization and ambient temperature pathway, seven genes were involved in the GA pathway, and nine genes are integrators or related to meristem response and developmental process. Four genes bound by AGL16 were not clearly defined for the flowering pathways (Boxall et al. 2005; Xiao et al. 2009; Zhao et al. 2011). Taken together, AGL16 might impact several flowering pathways, and the alteration of flowering time in mutants of AGL16 could be a net effect of multiple flowering pathways.
Figure 3.
Molecular pathways (indicated with different color boxes) targeted by AGL16. Genes with names in bold were common targets for AGL16 and SOC1, while those in red were differentially expressed between the agl16 soc1 and soc1-2 mutants.
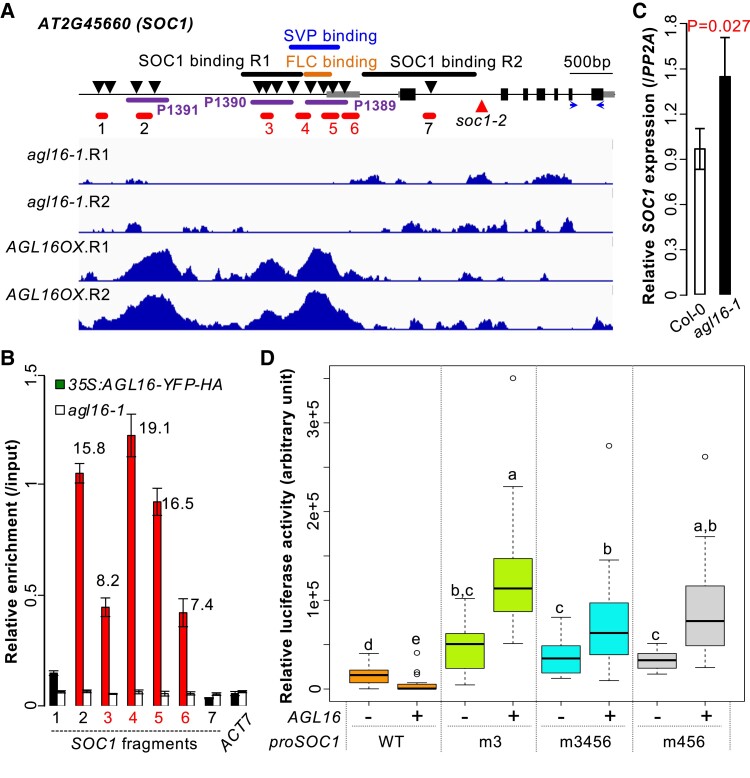
AGL16 binds to SOC1 promoter and modifies its expression
The floral integrator gene SOC1 was one of the targets bound by AGL16 (Fig. 4A and Supplemental Table S2). AGL16 interacted with three DNA segments (peaks 1,389; 1,390; and 1,391) in the promoter region of SOC1 that harbored several CArG-motifs, consistent with a previous observation that the whole intergenic region is required for proper SOC1 expression (Hepworth et al. 2002). Peak 1,390 overlapped with a region bound by SOC1 itself (SOC1 binding region 1) (Tao et al. 2012), while peak 1,389 overlapped with regions previously shown to be targeted by SVP (Tao et al. 2012; Mateos et al. 2015) or FLC (Deng et al. 2011; Mateos et al. 2015). An independent ChIP-qPCR assay confirmed AGL16 binding on all three peaks with the binding on peaks 1,389 and 1,391 relatively stronger than on peak 1,390 (Fig. 4B). The second segment bound by SOC1 itself (SOC1 binding region 2 or fragment 7) was not targeted by AGL16. As AGL16 forms protein complexes with SVP and FLC (Hu et al. 2014), it is likely that AGL16 binds target regions together with these MADS-box TFs. However, SOC1 transcription was weakly up-regulated by loss-of-function of AGL16 in both Col-0 (Fig. 4C) and Col-FRI backgrounds (Supplemental Fig. S4), a pattern likely caused by the very complex regulation of SOC1 expression (Hepworth et al. 2002; Immink et al. 2012). A transient luciferase assay with the 1.7-kb SOC1 promoter in Nicotiana benthamiana leaves demonstrated that the co-transfection of AGL16 significantly repressed the pSOC1 expression compared with the co-transfection control with an empty vector (Fig. 4D and Supplemental Fig. S5). In contrast, when the CArG-box(es) bound by MADS-box TFs were mutated (m3, m456, and m3456), pSOC1 expression increased remarkedly, and this increase became even more prominent when AGL16 was co-transfected additionally. These data together suggest that AGL16 represses directly the SOC1 expression.
Figure 4.
AGL16 targets SOC1 and represses its expression. A) Schematic representation of the SOC1 locus. Filled bars indicated exons and gray bars marked the 5′- and 3′-UTR regions while the line indicated the non-coding region of SOC1. Arrows downward labeled the putative CArG-boxes potentially bound by MADS-box proteins. The dark purple lines indicated the three peaks (P1389, P1390, and P1391) bound by AGL16. Orange, blue and black thick lines marked the known regions targeted by FLC, SVP and SOC1, respectively. Note that two sites in the regulatory region of SOC1 were bound by itself (SOC1 binding R1 and R2; see ref. Tao et al. 2012). Red lines (1–7) showed the regions tested for AGL16-YFP-HA binding on SOC1 chromatin. Horizontal arrows marked the position of primers used for quantification of CDS regions. The lower panel showed the ChIP-seq profile at SOC1. B) Relative enrichment of AGL16 on SOC1 chromatin tested with ChIP-qPCR. Mean fold change values with significant enrichment was labelled above bars together with standard deviation. ACT7 was taken as a negative enrichment control. C) Relative expression of SOC1 against PP2A in Col-0 and agl16-1 plants. Mean relative expression was given with standard deviation and the significant difference was examined using Student's t-test. D) Quantitative luciferase assay showing that AGL16 regulated the expression of 1.7-kb promoter of SOC1 via cis-motifs 3–6 described in (A). Box plots mark the 25–75% quartiles with the line in box representing the median. The lines extending from each box marked the minimum (5%) and maximum (95%) values of the dataset. Circles showed the outliers. WT marked the 1.7-kb promoter without any sequence modification, while the m3 indicated the mutation of a CCW6GG-box in WT background. The m3456 and m456 showed the relative expression level for the WT promoter with cis-motifs 3, 4, 5, 6 and 4, 5, 6 mutated, respectively. At least 15 randomly selected fields each from one individual N. benthamiana leaf per treatment were used for measuring with + and – labeling the presence and absence of 35S:AGL16, respectively. Different letters above the boxes represented the signficant differences among treatments using one-way ANOVA Dunnett's test (P < 0.05). This experiment was triplicated and each trial gave similar results.
AGL16 forms protein complex with SOC1
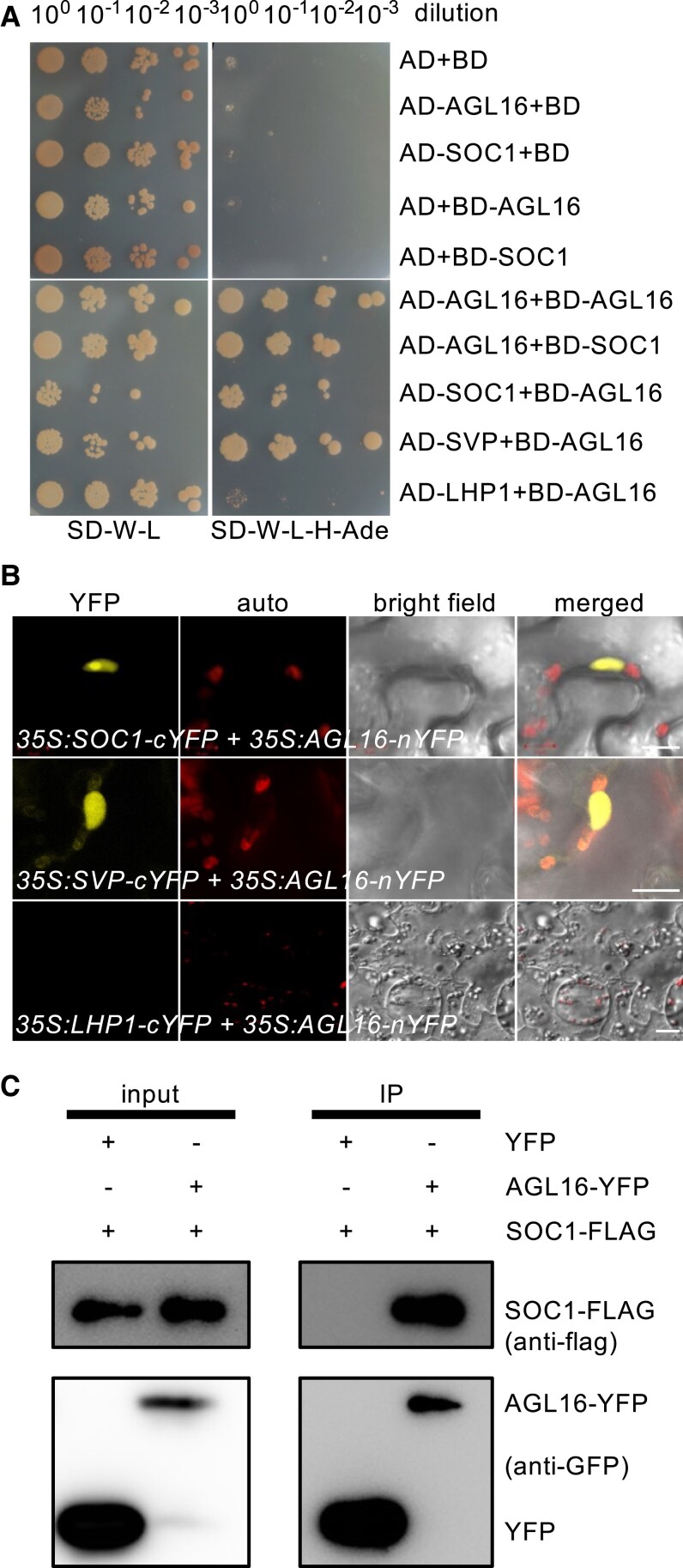
AGL16 dimers likely with SOC1 (Supplemental Fig. S3) (de Folter et al. 2005; Immink et al. 2009). We verified this interaction with Yeast-2-Hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) techniques. Y2H assays confirmed interactions between SOC1 and AGL16 (Fig. 5A), which was as strong as the previously reported direct interaction between AGL16 and SVP, in clear contrast to the negative interaction between AGL16 and LHP1 (Hu et al. 2014). BiFC assays by fusing the N-terminal half of yellow fluorescent protein (nYFP) with AGL16 (35S:AGL16-nYFP) and the C-terminal of YFP with SOC1 (35S:SOC1-cYFP) detected an interaction of AGL16 with SOC1 in the nuclei of Agrobacterium-infiltrated N. benthamiana leaves (Fig. 5B). A further co-immunoprecipitation (co-IP) assay with AGL16 fused to a YFP tag and SOC1 fused with a Flag tag in Arabidopsis cells also confirmed the physical interaction between these two TFs (Fig. 5C). Hence, AGL16 and SOC1 form hetero-protein complexes.
Figure 5.
AGL16 forms protein complex with SOC1. A) Yeast two-hybrid assay revealed a direct interaction between AGL16 and SOC1. Each protein was fused to either the activation domain (AD) as prey or the DNA-binding domain (BD) as bait. Serial dilutions (100x to 10−3x) of J69-4A cells containing different construct combinations indicated on the left were grown on control (left) and selective (right) medium. The AGL16-SVP and the AGL16-LHP1/empty vector combinations provided positive and negative controls, respectively. B) BiFC assay evidenced the formation of AGL16–SOC1 complex in nucleus of N. benthamiana leaf epidermis. The interaction was tested with constructs 35S:SOC1-cYFP and 35S:AGL16-nYFP. A negative interaction between AGL16 and LHP1 and a positive interaction between AGL16 and SVP were tested as well. Bars = 10 µm. C) Co-immunoprecipitation (co-IP) assay confirmed the AGL16–SOC1 interaction in Arabidopsis protoplast. SOC1 was FLAG-tagged while the AGL16 was fused with an YFP tag. Total protein of the transfected wild type protoplasts was immuno-precipitated with antibody against GFP (anti-GFP) first, and further analyzed by Western blot using antibody against FLAG (anti-FLAG).
AGL16 and SOC1 co-target a common set of genes
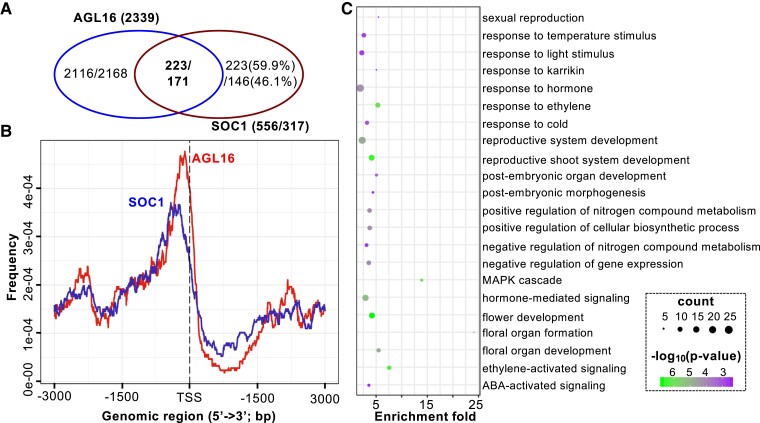
We next examined whether AGL16 and SOC1 had common targets. For this aim, the previously generated binding profiles for SOC1 were used to identify shared targets with AGL16 (Immink et al. 2012; Tao et al. 2012). We applied the same annotation procedure for both AGL16 and SOC1 binding profiles in order to identify common genes. There were 193 AGL16-bound segments that overlapped with 240 SOC1 peaks (Supplemental Table S3). These peaks were in the ±2-kb vicinity of 223 genes (five without annotation information), which were then taken as AGL16 and SOC1 common targets (Fig. 6A). Most of these common peaks were in the 1-kb region surrounding TSS with AGL16 peaks a bit more proximal (Fig. 6B). We further identified 211 CArG-box motifs in 144 common peaks (400 bp surrounding peak centers; 74.6% of all overlapped peaks) with MEME-ChIP. Eighty-seven peaks harbored one CArG-box (DCCAAAAAWGGAAAR; 60.4%), while the rest featured two (49 or 34%) or three (6 peaks or 4.2%) or more (2 peaks; Supplemental Fig. S6A). The distances between the CArG-box motifs were spaced with 20–40 bases (Supplemental Fig. S6B). Among these common targets, genes involved in floral organ development (or reproductive growth) and responses to hormone stimulus including ethylene and ABA were significantly enriched (Fig. 6C and Supplemental Table S3). Eight genes of the photoperiod and circadian clock related pathways (AGL15, AGL18, ATC, PHYA, RAV2, SMZ, SNZ, and TOE3), three genes of the temperature-related pathways (CBF1, CBF2, and SVP), and SOC1 itself were involved in flowering (Fig. 3), suggesting that AGL16 and SOC1 act likely together to time floral transition in Arabidopsis.
Figure 6.
AGL16 and SOC1 share a common set of target genes involved in multiple functions. A) Venn diagram showing that 223/171 genes (Immink et al. 2012; Tao et al. 2012) were co-bound potentially by both AGL16 and SOC1. B) Binding intensities for AGL16 (red) and SOC1 (blue) peaks surrounding transcription starting sites (TSS). Regions 3-kb upstream and downstream of TSS were plotted. C) Selected significantly enriched GO terms for the common targets.
AGL16 regulates genome-wide gene expression depending partially on SOC1 function
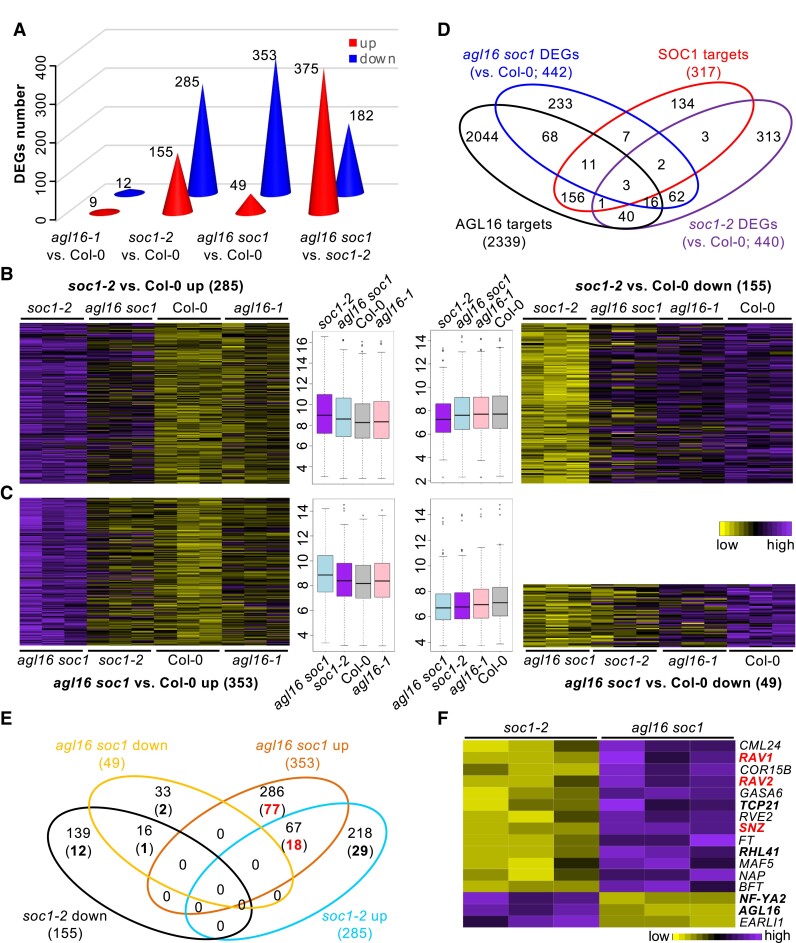
We next determined to what extent the gene expression at the genome-wide level could be affected by the AGL16–SOC1 module (Supplemental Table S1). For this, we carried out a comparative transcriptomics analysis using the single and double mutants between the agl16-1 and soc1-2 lines. In contrast to the very broad binding spectrum of AGL16, we only detected very small number of genes showing differential expression (differentially expressed genes [DEGs]) in agl16-1 single mutant (9 up and 12 down) compared with Col-0 (Fig. 7A and Supplemental Table S4). The soc1-2 single (155 up and 285 down) and the agl16 soc1 double (49 up and 353 down) mutants had similar number of DEGs but soc1-2 featured more up and less down DEGs (Yate's χ2test, P < 0.001; Fig. 7A), indicating that AGL16 either countered SOC1's repressive or inductive role on gene expression. A heatmap analysis of DEGs in the soc1-2 vs Col-0 revealed that absence of agl16 mostly reverted the differential gene expression observed in soc1-2 to wild type levels (Fig. 7B). Genes down-regulated in the agl16 soc1 mutants showed also down-regulation in soc1-2 (Fig. 7C). In contrast, genes up-regulated in agl16 soc1 were barely affected by either single mutation, suggesting that for these genes, AGL16 and SOC1 contribute redundantly to the repression. Accordingly, only 83 soc1-2 DEGs (in total 155 up and 285 down; ∼18.9%) overlapped with the agl16 soc1 DEGs (375 up and 182 down; ∼14.9%; Fig. 7D). Therefore, AGL16 has potential in regulating gene expression at the genome-wide level, but apparently depends on its genetic background, i.e. here the SOC1 activity.
Figure 7.
The AGL16–SOC1 module collaborates on regulation of genome-wide gene expression. A) The number of differentially expressed genes (DEGs) in three mutants. The exact number of up (red) or down (blue) regulated DEGs were given on each cone. B and C) Heatmaps showing the normalized relative expression of soc1-2 (B) and agl16 soc1 (C) DEGs in all four lines. The boxplots in the middle gave the data distribution pattern for each cluster. Box plots mark the 25–75% quartiles with the line in box representing the median. The lines extending from each box marked the minimum (5%) and maximum (95%) values of the dataset. Circles showed the outliers. D) Venn diagram demonstrating the overlap between DEGs and the AGL16 targets profile. E) A detailed comparison between the DEGs in soc1-2 and agl16 soc1 mutants with the AGL16 binding profile. Bold numbers in brackets showed the number of DEGs bound by AGL16. F) A heatmap showing the normalized relative expression of the DEGs related to flowering time regulation in the soc1-2 and agl16 soc1 mutants.
We next examined to what extent these DEGs associated with AGL16 targeting. Among the 557 agl16 soc1 DEGs, AGL16 bound to 98 genes (∼22.2%), in which only 23 (∼4.1%) were also targeted by SOC1 (Fig. 7D). About 13.6% or 60 soc1-2 DEGs were likely the AGL16 targets (Yate's χ2test, P = 2e-8, in comparison with genome-wide level of AGL16 binding). However, we noticed that only nine soc1-2 DEGs (∼2% among 440) were potential targets of SOC1, a pattern similar to a previous report, in which 52 SOC1 targets were among the 1,186 DEGs (Tao et al. 2012). There were six targets (∼28.6%) showing differential expression in the 21 agl16-1 DEGs. Moreover, we identified more than a quarter of up-regulated DEGs in the agl16 soc1 line (77 among 286) being AGL16 targets in contrast to about 13.3% of up-regulated DEGs in the soc1-2 mutant (29 among 218; Yate's χ2 test, P = 0.0035; Fig. 7E). Among the 67 up-regulated DEGs shared between the soc1-2 and agl16 soc1 mutants, 18 (26.9%) were potentially AGL16 targets. In contrast, less than 8% of down-regulated DEGs in both mutants were targeted by AGL16. Together, these data suggest that AGL16 may act mainly as a transcriptional repressor and exert an antagonistic role against SOC1 regulation on target gene expression.
AGL16–SOC1 module is important for flowering time regulation
Among the DEGs between agl16 soc1 and soc1-2 plants, we identified 17 known genes involved in floral regulation with seven being targeted by AGL16 (NF-YA2, TCP21, RHL41, AGL16, three AP2-like genes RAV1, RAV2/TEM2, and SNZ; Fig. 7F and Supplemental Table S4). Expression of FT was significantly enhanced in the agl16 soc1 double mutant. In line with this, the double mutant agl16 soc1 flowered significantly earlier (∼20 rosette leaves) than the soc1-2 single mutant (∼25.6 rosette leaves; about 21.6% reduction in rosette leaf number) but still later than both agl16-1 (∼11.1 rosette leaves; ∼13.6% reduction) and wild type Col-0 (∼12.9 rosette leaves) plants (Fig. 8). This indicated that AGL16 could counteract the SOC1 regulation on flowering, while the regulatory role of AGL16 in floral transition depends on SOC1, a pattern like the genetic dependency of AGL16 on FLC (Hu et al. 2014).
Figure 8.
AGL16 and SOC1 regulate additively flowering time. A) Overall flowering behaviors of LD-growing wild type Col-0, agl16-1, soc1-2, and agl16 soc1 mutants. B) Leaf number production upon flowering under LD conditions. Mean rosette (filled bars, RLN) and cauline (open bars, CLN) leaves were shown with standard deviation. Numbers in percentage showed the earlier flowering level of agl16-1 and agl16 soc1 comparing with Col-0 and soc1-2, respectively. Analyses were triplicated and all had similar patterns. Statistical comparisons were performed with the Wilcoxon rank sum test in R.
Discussion
AGL16 acts in the hubs of GRN related to various biological processes
The MADS-box TF AGL16 is an important regulator in flowering time (Hu et al. 2014), stomata development (Kutter et al. 2007), heat stress adaptation (Szaker et al. 2019), drought resistance (Zhao et al. 2020) and salt stress adaptation (Zhao et al. 2021), suggesting that it might have very broad spectra of downstream targets. In this study, indeed, our ChIP-seq assay demonstrated that AGL16 could target more than 2,000 genes featuring characteristic CArG-box motifs (Figs. 1–3). These genes were involved not only plant development but also various hormone signaling processes (Supplemental Table S2) including some targets in the ABA signaling pathway that previously identified (Zhao et al. 2020, 2021). These broad spectra are not rare, however, especially for MADS-box TFs. Two such examples would be SVP and SEP3, both of which can bind to thousands of downstream targets involved in a very broad set of biological processes (Kaufmann et al. 2009; Gregis et al. 2013; Mateos et al. 2015). Intriguingly, both SVP and SEP3 can or potentially form hetero-protein complexes with AGL16 (de Folter et al. 2005; Hu et al. 2014), indicating that they may work together to fine-tune plant developmental programs in responding to ever-changing environments, a hypothesis awaits for further investigation.
Interestingly, expression of AGL16 responds to ABA treatment as well as multiple stresses (Szaker et al. 2019; Zhao et al. 2021), thus revealing a very complex role of AGL16 (and its potential partners) in abiotic adaptation. Since both AGL16 and SOC1 play important roles in stomata development and movement (Kutter et al. 2007; Zhao et al. 2020), it is likely that the hetero-protein complexes formed between AGL16 and its partners may be the key molecule that functions in these abiotic adaptations. Indeed, both AGL16 and SOC1 can directly bind and regulate the expression of a shared set of genes involved in ABA signaling and abiotic stresses (Fig. 6) (Immink et al. 2012; Tao et al. 2012). Considering the essential roles of ABA in seed dormancy and germination regulation, the fact that several AGL16 targets encode for ABA receptors may invoke us to further examine the regulatory roles of AGL16 and its related protein complexes in seed dormancy and germination (Supplemental Tables S2 and S3). Corroborating with this, AGL16 expression drops substantially during seed germination (Das et al. 2018).
AGL16 regulates multiple floral pathways
AGL16 might exert its regulation potential in several pathways controlling flowering time (Fig. 3). Being congruent with its photoperiod dependency in regulation of flowering time, AGL16 targets 37 genes (including AGL16 itself) related to photoperiod and circadian clock pathways. Though under the tested environmental conditions agl16-1 still shows a normal vernalization response (Hu et al. 2014), several genes related to temperature responses are directly targeted by AGL16. FLC, SVP, and SOC1 might be partners of AGL16 in this respect as all three proteins target also directly on some of these temperature-related genes (Deng et al. 2011; Immink et al. 2012; Tao et al. 2012; Mateos et al. 2015). The binding of AGL16 may cause both positive and negative influences on the transcription of these targets (Fig. 7), which encompass both repressors and promoters of the floral transition. Indeed, several of the flowering time genes targeted by AGL16 show an enhanced or decreased expression when AGL16 activity is modified in the soc1-2 background (Figs. 3 and 7and Supplemental Tables S3 and S4). Therefore, the early flowering phenotypes present in, AGL16 loss-of-function mutants might be a net effect or balanced regulation on different pathways (Fig. 8) (Hu et al. 2014).
It should be noted that AGL16 also targets and represses the expression of MYC2, which is previously claimed to modulate flowering time (Kazan and Manners 2013; Zhai et al. 2015; Wang et al. 2017; Bao et al. 2019). However, our recent efforts have demonstrated, partially based on the data generated from this study, that the MYC2-family TFs only play very limited roles in timing floral transition, because it's the hidden mutation of COP1, not the MYC mutations, causing early flowering observed in the original jin1-2 mutant (Yu et al. 2023). Whether the AGL16-MYC2 interaction regulates flowering time upon different stress conditions needs to be tested later.
AGL16 and SOC1 collaborate in regulation of genome-wide gene expression
The formation of AGL16–SOC1 complex identifies the collaborative potential in targeting and regulation of genome-wide gene expression like other MADS-box TFs (Fig. 5) (de Folter et al. 2005; Lee et al. 2008; Immink et al. 2009; Kaufmann et al. 2009; Kaufmann et al. 2010; Deng et al. 2011; Immink et al. 2012; Tao et al. 2012; Mateos et al. 2015). AGL16 binds more than 2,000 genes, which is in line with its very broad expression in many tissues and organs (Alvarez-Buylla et al. 2000), but affects the expression of a very limited number of genes in the background of Col-0 (Fig. 7). When SOC1 becomes non-functional (soc1-2), AGL16 modulates the expression of more than 550 genes and acts both as a transcriptional repressor and activator. In the soc1-2 background, AGL16 seems mainly act as a transcriptional repressor as more than a quarter of the up-regulated DEGs, in contrast to the less than 8.5% of the down-regulated DEGs, are potential targets of AGL16. Hence AGL16's activity in gene expression regulation requires partially SOC1, and corroborating with this, both AGL16 and SOC1 expression can be detected in the shoot apex (Corbesier et al. 2007; Immink et al. 2012; Hu et al. 2014). On the other hand, SOC1 also needs AGL16 as SOC1's repressive activity substantially drops (from 155 to 49 genes) but the promoting activity increases (from 285 to 353 genes) when AGL16 has no function. Many soc1-2 DEGs are not differentially expressed any more in agl16 soc1 mutant (Fig. 7). Indeed, AGL16 and SOC1 co-bind a common set of targets and regulate the expression of many known flowering time genes (Figs. 6 and 7). As expected, these two TFs collaborate in regulation of flowering time (Fig. 8). The agl16 soc1 double mutant flowered significantly earlier than the soc1-2 single mutant, on the other hand, still later than both agl16-1 and wild type plants. AGL16 could counteract SOC1 effects in flowering time regulation, and vice versa, similar to the genetic dependency of AGL16 on FLC (Hu et al. 2014). It's possible that these TFs may form higher-order protein complexes to regulate downstream genes, for example FT expression, which should be tested further.
The identification of three DNA fragments bound by AGL16 in the upstream ∼4-kb intergenic region raises a possibility that the SOC1 expression regulation might be more complicated than we have expected (Fig. 4) (Hepworth et al. 2002; Immink et al. 2012; Jung et al. 2012; Liu et al. 2013; Li et al. 2017; Hwang et al. 2019; Olas et al. 2019; Yan et al. 2021). Though AGL16 can repress the SOC1 expression in planta and when the frequently used ∼1.7-kb promoter was included in transient assays (Fig. 4), this short fragment may not be enough for full mechanistic understanding the regulation of SOC1 expression. Whether higher order 3D chromatin loop presents for SOC1, like the ones for FT and FLC (Crevillen et al. 2013; Liu et al. 2014), and whether AGL16 has a role in the loop formation need further investigation.
Together, as a master regulator in GRNs connecting multiple pathways, AGL16's function has a partial inter-dependency with SOC1. AGL16 might act as a glue molecule, like other MADS-box TFs do, to micro-tune the expression of downstream genes at proper stages and environmental conditions (Immink et al. 2009; Kaufmann et al. 2010; Pajoro et al. 2014; Richter et al. 2019). It will be important to address these further to understand their precise roles and mechanisms in balancing development and environmental adaptation.
Materials and methods
Plant materials, growth conditions, and phenotype assays
A. thaliana plants including wild-type Col-0, agl16-1, 35S:AGL16-YFP-HA in agl16-1 background, Col-FRI, agl16-1 Col-FRI, and m3 have been described previously (Kutter et al. 2007; Hu et al. 2014). The soc1-2 mutant in Col-0 background (Torti et al. 2012) was kindly provided by Prof. George Coupland. To test the genetic interactions between AGL16 and SOC1, agl16-1 and soc1-2 were crossed and double mutant was screened with gene-specific primers (Supplemental Table S5) (Kutter et al. 2007; Torti et al. 2012; Hu et al. 2014).
Arabidopsis seeds were stratified in distilled water at 4°C for 72 h and sown in soil and grown under LD conditions (16-h light at 21°C and 8-h night at 18°C). Seedlings for phenotyping were planted either in growth rooms or chambers, while materials for gene expression analysis and ChIP assays were sown on Murashige and Skoog medium plates (Hu et al. 2014).
Flowering time assays were carried out according to previous report (Hu et al. 2014). Four independent trials were applied and each gave similar pattern. Phenotype comparisons were performed with Student's t-test with Bonferroni-correction.
RNA isolation, Rt-qPCR, and RNA-seq assays
Total RNA was extracted with TRI Reagent® (Molecular Research Center, Inc. Cincinati, USA). Ten-day-old seedlings were used for quantification of relative expression of selected genes with PP2A as reference (Hu et al. 2014). Reverse transcription was carried out with the HiScript® II Q RT SuperMix for RT-qPCR (+gDNA wiper) and quantification PCRs were performed with ChamQ™ SYBR qPCR Master Mix (both from Vazyme Biotech co. Ltd, Nanjing) on QuantStudio™ 7 Flex Real-Time PCR System (ThermoFisher). Three to four biological replicates from each of two to three independent trials were applied for each experiment. A similar protocol was developed for monitoring relative enrichment of DNA fragments in ChIP-qPCR experiments. All the primers used in this study are included in Supplemental Table S1.
For RNA-seq, materials were collected from three independent biological replicates for each genotype, and DNA-free total RNA was generated as described above. Illumina True-seq library preparation was performed from 3 μg DNA-free total RNA and sequenced by the Biomarker Technologies Corporation, Beijing, China. Quality trimmed pair-end RNA-seq reads were mapped to the Arabidopsis TAIR10 annotation using the HISAT2 v2.1.0 (Kim et al. 2019). The featureCounts included in subread v1.6.4 package was applied to calculate reads counts on each gene (Liao et al. 2013, 2014). DESeq2 v1.14.1 was used to detect differentially expressed genes (DEGs; fold change above 1.5 and Padj < 0.1). Only uniquely mapped reads were used for downstream analysis. Transcriptional clustering analysis was performed using the heatmap.2 function in R. GO analysis was performed with PANTHER in TAIR web-tool (https://www.arabidopsis.org/tools/go_term_enrichment.jsp) or agriGO pipeline (Mi et al. 2017; Tian et al. 2017).
ChIP-seq, ChIP-qPCR assays, and data analysis
ChIP experiments were carried out following protocols described (Reimer and Turck 2010; Zhou et al. 2016). Chromatin for both agl16-1 and agl16-1 AGL16OX plants was extracted from ten-day-old seedlings grown under LD conditions at ZT14, and precipitated with antibody against GFP (Abcam, Ab290). For ChIP-seq, the immuno-precipitations from two independent trials were used for NGS library preparation with NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® (E7645, New England BioLabs Inc.) and high-throughput sequencing with HiSeq2000 platform. ChIP-seq reads were mapped to the TAIR10 assembly of A. thaliana using BWA-MEM (v0.7.17-r1188) (Li 2013). Reads with mapping quality below 30 were discarded with SAMtools v1.7 (Li et al. 2009). Duplicated reads were removed using Picard MarkDuplicates v1.119. The resulted .bam file was used as input to call AGL16 enriched regions with MACS v2.2.7.1 (Zhang et al. 2008). Enriched regions were generated by the comparison of immune-precipitated products to input for AGL16OX and then compared against agl16-1. For annotation of AGL16 targets, the R package ChIPseeker was used (Yu et al. 2015). The position and strand information of nearest genes were reported with the distance from peak to the TSS of its closest gene identified. As annotations might overlap, we use “promoter” definition in ChIPseeker as the highest priority for annotation. Each binding site was assigned to only one gene. IGV was used for data visualization of the binding profiles for targets (Thorvaldsdottir et al. 2013). Enriched motifs in AGL16 binding peaks were identified using Homer suite with findMotifsGenome.pl function (Heinz et al. 2010). Motifs in AGL16–SOC1 co-targeted regions were analyzed with MEME-ChIP tools (Machanick and Bailey 2011), and the spacing between primary and secondary motifs was analyzed with SpaMo (spamo -dumpseqs -bin 20 -verbosity 1 -oc spamo_out_1 -bgfile./background -keepprimary -primary DCCAAAAAWGGAAAR). We compared the AGL16 targets with SOC1 targets from both Immink (2012) and Tao (2012) with the same annotation procedures for AGL16 (Immink et al. 2012; Tao et al. 2012). In an earlier independent trial, we pooled the immuno-precipitations from two biological replicates and sequenced the products. This pooled sequencing results gave similar pattern of AGL16 targets profile but with a lower coverage. Yate's χ2tests were performed online (http://www.quantpsy.org/chisq/chisq.htm). The ∼400 flowering time genes were downloaded from https://www.mpipz.mpg.de (Bouche et al. 2016) with self-curations. Reads data for RNA-seq and ChIP-seq experiments were accessible at NCBI under accession code SUB5067038.
Yeast two-hybrid and biomolecular fluorescence complementation (BiFC) experiments
Yeast two-hybrid and the BiFC assays were carried out to test the physical interaction between AGL16 and SOC1 proteins according to previous report (Hu et al. 2014). In yeast two-hybrid assay, interactions between AGL16-SVP and AGL16-AGL16 were applied as positive controls while the AGL16-LHP1, AGL16-BD, SOC1-BD, AD-AGL16, and AD–SOC1 were applied as negative controls together with empty vectors. For BiFC assay in N. benthamiana plants, 35S:SOC1-cYFP construct was built by cloning the full-length encoding-region without stop codon of SOC1 (from Col-0) into pDONR221 entry vector first and later transferred into RfA-sYFPc-pBatTL-B vector. The interactions between AGL16 and SVP, between AGL16 and LHP1, were used as positive and negative controls, respectively.
Co-IP assay
To test the AGL16 and SOC1 interactions, coding sequences of AGL16 and SOC1 were amplified from the wild type cDNA with Phanta Max Super-Fidelity DNA Polymerase (P505, Vazyme). All sequences were cloned into the pDONR201 entry vector and verified via Sanger sequencing. The resulting destination vectors containing N-terminal tagged pENSG-YFP-AGL16 and pICH47811–SOC1 were used to transfect protoplasts prepared from leaves of wild type seedlings (Yoo et al. 2007). The transfected protoplasts were incubated at room temperature for 16 h and used for co-IP assays as described previously (Cui et al. 2018). In brief, the protoplasts were lysed in immunoprecipitation buffer (50 mM Tris pH7.5, 150 mM NaCl, 10% [v/v] glycerol, 2 mM EDTA, 5 mM DTT, protease inhibitor, 0.1% Triton). Lysates were centrifuged at 14,000 × g for 15 min at 4°C with aliquots of supernatants as input controls. Immunoprecipitations (Ips) were performed by incubating the supernatants with 15 μl GFP-Trap beads (gta-10, ChromoTek) for 2 h at 4°C. After centrifugation at 1,000 × g and washing four times with extraction buffer, beads were eluted with 2× Laemmli loading buffer. The proteins were then separated with SDS-PAGE and analyzed by immuno-blotting with antibodies against GFP (ab290, Abcam) and FLAG (ab49763, Abcam).
Transient transactivation assay
To test the regulatory effects of AGL16 on SOC1 expression, the coding region of AGL16 was inserted into the pOCA30 vector to generate the effector, while the 1.7-kb promoter and its mutated versions of SOC1 was fused with a pZP vector to generate the reporter constructs (Chen et al. 2021). Equal amounts of the effector and reporter constructs in Agrobacterium tumefaciens strain GV3101 were used to co-infiltrate N. benthamiana leaves with at least 15 biological replicates that were randomly distributed. After 2 days of infiltration, the luciferase intensity was collected and quantified with a low-light cooled CCD imaging apparatus. Experiments were triplicated with each containing at least 15 replicates. Relative expression was examined for statistical significance using ANOVA followed by Dunnett's test.
Accession numbers
Sequence data from this article can be found in the CNSA database (https://db.cngb.org/) under project number CNP0003940.
Supplementary Material
Acknowledgments
We thank Franziska Turck, Ligang Chen, Liangyu Liu, Feihong Yan, Fei He, Yibo Sun, Shulan Chen, and Zhijia Gu for insightful discussion and assistance in experiments.
Contributor Information
Xue Dong, CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan Province, China; Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Li-Ping Zhang, CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan Province, China.
Yin-Hua Tang, CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan Province, China; Kunming College of Life Sciences, University of Chinese Academy of Sciences, Kunming 650201, Yunnan Province, China.
Dongmei Yu, CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan Province, China.
Fang Cheng, CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan Province, China.
Yin-Xin Dong, CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan Province, China.
Xiao-Dong Jiang, CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan Province, China.
Fu-Ming Qian, CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan Province, China.
Zhen-Hua Guo, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Jin-Yong Hu, CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan Province, China.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Overexpression of AGL16 cDNA restored the early flowering of agl16-1 to a wild-type level.
Supplemental Figure S2 . The length distribution of AGL16 peaks in two trials and the overlapped peaks.
Supplemental Figure S3 . Potential protein partners interacting with AGL16 in Arabidopsis thaliana.
Supplemental Figure S4 . Relative expression of SOC1 CDS against PP2A in Col-FRI and Col-FRI-agl16-1 plants.
Supplemental Figure S5 . AGL16 repressed proSOC1 expression (1.7-kb promoter) with a dependency on cis-motifs.
Supplemental Figure S6 . Characters of secondary CArG-box motifs in the common targets between AGL16 and SOC1.
Supplemental Table S1 . Statistics of ChIP-seq and RNA-seq results.
Supplemental Table S2 . The results of AGL16OX ChIP-seqs.
Supplemental Table S3 . AGL16 and SOC1 share common targets.
Supplemental Table S4 . RNA-seq results of agl16-1, soc1-2, agl16 soc1 in comparison with Col-0.
Supplemental Table S5 . Primers used in this study.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31570311 to J.-Y.H., 31501034 to X.-D.J., 31700275 to Y.-X.D., 31800261 to F.C.), the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) to J.-Y.H. (XDB31000000), the China Postdoctoral Science Foundation (2017M613023 to Y.-X.D.), the Postdoctoral targeted funding from Yunnan Province to F.C. and Y.-X.D., the CAS Pioneer Hundred Talents Program (292015312D11035 to J.-Y.H), and the Yunnan basic and applied research funding to F.C. This work is partially facilitated by the Germplasm Bank of Wild Species of China and the Platform for Plant Multi-dimensional Imaging and Diversity Analysis.
Data availability
All data and materials needed to evaluate the conclusions in the paper are present in the paper and the Supplemental materials.
References
- Aerts N, de Bruijn S, van Mourik H, Angenent GC, van Dijk ADJ. Comparative analysis of binding patterns of MADS-domain proteins in Arabidopsis thaliana. BMC Plant Biol. 2018:18(1): 131. 10.1186/s12870-018-1348-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, Vergara-Silva F, Yanofsky MF. MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 2000:24(4): 457–466. 10.1046/j.1365-313x.2000.00891.x [DOI] [PubMed] [Google Scholar]
- Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012:13(9): 627–639. 10.1038/nrg3291 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003:15(11): 2730–2741. 10.1105/tpc.016238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Hua C, Huang G, Cheng P, Gong X, Shen L, Yu H. Molecular basis of natural variation in photoperiodic flowering responses. Dev Cell. 2019:50(1): 90–101.e3. 10.1016/j.devcel.2019.05.018 [DOI] [PubMed] [Google Scholar]
- Bouche F, Lobet G, Tocquin P, Perilleux C. FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2016:44(D1): D1167–D1171. 10.1093/nar/gkv1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxall SF, Foster JM, Bohnert HJ, Cushman JC, Nimmo HG, Hartwell J. Conservation and divergence of circadian clock operation in a stress-inducible crassulacean acid metabolism species reveals clock compensation against stress. Plant Physiol. 2005:137(3): 969–982. 10.1104/pp.104.054577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol. 2008:18(17): 1338–1343. 10.1016/j.cub.2008.07.075 [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhang L, Zhang H, Chen L, Yu D. ERF1 Delays flowering through direct inhibition of FLOWERING LOCUS T expression in Arabidopsis. J Integr Plant Biol. 2021:63(10): 1712–1723. 10.1111/jipb.13144 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. FT Protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007:316(5827): 1030–1033. 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- Crevillen P, Sonmez C, Wu Z, Dean C. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J. 2013:32(1): 140–148. 10.1038/emboj.2012.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui HT, Qiu JD, Zhou Y, Bhandari DD, Zhao CH, Bautor J, Parker JE. Antagonism of transcription factor MYC2 by EDS1/PAD4 complexes bolsters salicylic acid defense in Arabidopsis effector-triggered immunity. Mol Plant. 2018:11(8): 1053–1066. 10.1016/j.molp.2018.05.007 [DOI] [PubMed] [Google Scholar]
- Das SS, Yadav S, Singh A, Gautam V, Sarkar AK, Nandi AK, Karmakar P, Majee M, Sanan-Mishra N. Expression dynamics of miRNAs and their targets in seed germination conditions reveals miRNA-ta-siRNA crosstalk as regulator of seed germination. Sci Rep. 2018:8(1): 13. 10.1038/s41598-017-18823-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S, Immink RG, Kieffer M, Parenicova L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell. 2005:17(5): 1424–1433. 10.1105/tpc.105.031831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meaux J, Hu JY, Tartler U, Goebel U. Structurally different alleles of the ath-MIR824 microRNA precursor are maintained at high frequency in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008:105(26): 8994–8999. 10.1073/pnas.0803218105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc Natl Acad Sci U S A. 2011:108(16): 6680–6685. 10.1073/pnas.1103175108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One. 2007:2(2): e219. 10.1371/journal.pone.0000219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Coupland G. Snapshot: control of flowering in Arabidopsis. Cell. 2010:141(3): 550–550.e2. 10.1016/j.cell.2010.04.024 [DOI] [PubMed] [Google Scholar]
- Gregis V, Andres F, Sessa A, Guerra R, Simonini S, Mateos J, Torti S, Zambelli F, Prazzoli G, Bjerkan K, et al. Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genom Biol. 2013:14(6): R56. 10.1186/gb-2013-14-6-r56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010:38(4): 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 2002:21(16): 4327–4337. 10.1093/emboj/cdf432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JY, Zhou Y, He F, Dong X, Liu LY, Coupland G, Turck F, de Meaux J. miR824-Regulated AGAMOUS-LIKE16 contributes to flowering time repression in Arabidopsis. Plant Cell. 2014:26(5): 2024–2037. 10.1105/tpc.114.124685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang K, Susila H, Nasim Z, Jung J-Y, Ahn JH. Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC Complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol Plant. 2019:12(4): 489–505. 10.1016/j.molp.2019.01.002 [DOI] [PubMed] [Google Scholar]
- Hyun Y, Richter R, Vincent C, Martinez-Gallegos R, Porri A, Coupland G. Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev Cell. 2016:37(3): 254–266. 10.1016/j.devcel.2016.04.001 [DOI] [PubMed] [Google Scholar]
- Immink RGH, Posé D, Ferrario S, Ott F, Kaufmann K, Valentim FL, de Folter S, van der Wal F, van Dijk ADJ, Schmid M, et al. Characterization of SOC1's central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol. 2012:160(1): 433–449. 10.1104/pp.112.202614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RG, Tonaco IA, de Folter S, Shchennikova A, van Dijk AD, Busscher-Lange J, Borst JW, Angenent GC. SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biol. 2009:10(2): R24. 10.1186/gb-2009-10-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000:290(5490): 344–347. 10.1126/science.290.5490.344 [DOI] [PubMed] [Google Scholar]
- Jung JH, Ju Y, Seo PJ, Lee JH, Park CM. The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 2012:69(4): 577–588. 10.1111/j.1365-313X.2011.04813.x [DOI] [PubMed] [Google Scholar]
- Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. 2007:19(9): 2736–2748. 10.1105/tpc.107.054528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Muino JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 2009:7(4): e1000090. 10.1371/journal.pbio.1000090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muino JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueno F, Krajewski P, Meyerowitz EM, et al. Orchestration of floral initiation by APETALA1. Science. 2010:328(5974): 85–89. 10.1126/science.1185244 [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. MYC2: the master in action. Mol Plant. 2013:6(3): 686–703. 10.1093/mp/sss128 [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019:37(8): 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter C, Schob H, Stadler M, Meins F Jr, Si-Ammour A. MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell. 2007:19(8): 2417–2429. 10.1105/tpc.107.050377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee I. Regulation and function of SOC1, a flowering pathway integrator. J Exp Bot. 2010:61(9): 2247–2254. 10.1093/jxb/erq098 [DOI] [PubMed] [Google Scholar]
- Lee J, Oh M, Park H, Lee I. SOC1 Translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J. 2008:55(5): 832–843. 10.1111/j.1365-313X.2008.03552.x [DOI] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1303.3997. 10.48550/arXiv.1303.3997 [DOI]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup . The sequence alignment/map format and SAMtools. Bioinformatics. 2009:25(16): 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, Wu Y, Chen H, Robertson M, Helliwell CA, Ito T, Meyerowitz E, Yu H. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev Cell. 2008:15(1): 110–120. 10.1016/j.devcel.2008.05.002 [DOI] [PubMed] [Google Scholar]
- Li H, Ye K, Shi Y, Cheng J, Zhang X, Yang S. BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol Plant. 2017:10(4): 545–559. 10.1016/j.molp.2017.01.004 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013:41(10):e108. 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014:30(7): 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu L, Adrian J, Pankin A, Hu J, Dong X, von Korff M, Turck F. Induced and natural variation of promoter length modulates the photoperiodic response of FLOWERING LOCUS T. Nat Commun. 2014:5: 4558. 10.1038/ncomms5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Li Y, Ren J, Qian Y, Yang X, Duan W, Hou X. Nitrate or NaCl regulates floral induction in Arabidopsis thaliana. Biologia. 2013:68(2): 215–222. 10.2478/s11756-013-0004-x [DOI] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011:27(12): 1696–1697. 10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos J, Madrigal P, Tsuda K, Rawat V, Richter R, Romera-Branchat M, Fornara F, Schneeberger K, Krajewski P, Coupland G. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 2015:16(1): 31. 10.1186/s13059-015-0597-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos JL, Tilmes V, Madrigal P, Severing E, Richter R, Rijkenberg CWM, Krajewski P, Coupland G. Divergence of regulatory networks governed by the orthologous transcription factors FLC and PEP1 in Brassicaceae species. Proc Natl Acad Sci U S A. 2017:114(51): E11037–E11046. 10.1073/pnas.1618075114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Murdter F, Kuttner F, Schmid M. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009:7(7): e1000148. 10.1371/journal.pbio.1000148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi HY, Huang XS, Muruganujan A, Tang HM, Mills C, Kang D, Thomas PD. PANTHER Version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017:45(D1): D183–D189. 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD. Flowering time regulation produces much fruit. Curr Opin Plant Biol. 2009:12(1): 75–80. 10.1016/j.pbi.2008.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001:13(4): 935–941. 10.1105/tpc.13.4.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nee G, Xiang Y, Soppe WJJ. The release of dormancy, a wake-up call for seeds to germinate. Curr Opin Plant Biol. 2017:35: 8–14. 10.1016/j.pbi.2016.09.002 [DOI] [PubMed] [Google Scholar]
- Olas JJ, Van Dingenen J, Abel C, Dzialo MA, Feil R, Krapp A, Schlereth A, Wahl V. Nitrate acts at the Arabidopsis thaliana shoot apical meristem to regulate flowering time. New Phytol. 2019:223(2): 814–827. 10.1111/nph.15812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajoro A, Madrigal P, Muiño JM, Matus JT, Jin J, Mecchia MA, Debernardi JM, Palatnik JF, Balazadeh S, Arif M, et al. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014:15(3): R41. 10.1186/gb-2014-15-3-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006:20(24): 3407–3425. 10.1101/gad.1476406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer JJ, Turck F. Genome-wide mapping of protein-DNA interaction by chromatin immunoprecipitation and DNA microarray hybridization (ChIP-chip). part A: ChIP-chip molecular methods. Methods Mol Biol. 2010:631: 139–160. 10.1007/978-1-60761-646-7_12 [DOI] [PubMed] [Google Scholar]
- Richter R, Kinoshita A, Vincent C, Martinez-Gallegos R, Gao H, van Driel AD, Hyun Y, Mateos JL, Coupland G. Floral regulators FLC and SOC1 directly regulate expression of the B3-type transcription factor TARGET OF FLC AND SVP 1 at the Arabidopsis shoot apex via antagonistic chromatin modifications. PLoS Genet. 2019:15(4): e1008065. 10.1371/journal.pgen.1008065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006:20(7): 898–912. 10.1101/gad.373506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaker HM, Darko E, Medzihradszky A, Janda T, Liu H-C, Charng Y-Y, Csorba T. Mir824/AGAMOUS-LIKE16 module integrates recurring environmental heat stress changes to fine-tune poststress development. Front Plant Sci. 2019:10: 1454. 10.3389/fpls.2019.01454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Shen LS, Liu C, Liu L, Yan YY, Yu H. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 2012:70(4): 549–561. 10.1111/j.1365-313X.2012.04919.x [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinformatics. 2013:14(2): 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan HY, You Q, Yi X, Du Z, Xu WY, Su Z. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017:45(W1): W122–W129. 10.1093/nar/gkx382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti S, Fornara F, Vincent C, Andrés F, Nordström K, Göbel U, Knoll D, Schoof H, Coupland G. Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a Leucine-Rich repeat protein that promotes flowering. Plant Cell. 2012:24(2): 444–462. 10.1105/tpc.111.092791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li Y, Pan J, Lou D, Hu Y, Yu D. The bHLH transcription factors MYC2, MYC3, and MYC4 are required for jasmonate-mediated inhibition of flowering in Arabidopsis. Mol Plant. 2017:10(11): 1461–1464. 10.1016/j.molp.2017.08.007 [DOI] [PubMed] [Google Scholar]
- Xiao CW, Chen FL, Yu XH, Lin CT, Fu YF. Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol Biol. 2009:71(1-2): 39–50. 10.1007/s11103-009-9507-9 [DOI] [PubMed] [Google Scholar]
- Yan F-H, Zhang L-P, Cheng F, Yu D-M, Hu J-Y. Accession-specific flowering time variation in response to nitrate fluctuation in Arabidopsis thaliana. Plant Diversity. 2021:43(1): 78–85. 10.1016/j.pld.2020.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007:2(7): 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Yu D, Dong X, Zou K, Jiang X-D, Sun Y-B, Min Z, Zhang L-P, Cui H, Hu J-Y. A hidden mutation in the seventh WD40-repeat of COP1 determines the early flowering trait in a set of Arabidopsis myc mutants. Plant Cell. 2023:35(1):345–350. 10.1093/plcell/koac319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GC, Wang LG, He QY. ChIPseeker: an R/bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015:31(14): 2382–2383. 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- Zhai Q, Zhang X, Wu F, Feng H, Deng L, Xu L, Zhang M, Wang Q, Li C. Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell. 2015:27(10): 2814–2828. 10.1105/tpc.15.00619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008:9(9): 9. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Hanada A, Yamaguchi S, Kamiya Y, Beers EP. The Arabidopsis Myb genes MYR1 and MYR2 are redundant negative regulators of flowering time under decreased light intensity. Plant J. 2011:66(3): 502–515. 10.1111/j.1365-313X.2011.04508.x [DOI] [PubMed] [Google Scholar]
- Zhao P-X, Miao Z-Q, Zhang J, Chen S-Y, Liu Q-Q, Xiang C-B. MADS-box factor AGL16 negatively regulates drought resistance via stomatal density and stomatal movement. J Exp Bot. 2020:71(19): 6092–6610. 10.1093/jxb/eraa303 [DOI] [PubMed] [Google Scholar]
- Zhao PX, Zhang J, Chen SY, Wu J, Xia JQ, Sun LQ, Ma SS, Xiang CB. Arabidopsis MADS-box factor AGL16 is a negative regulator of plant response to salt stress by downregulating salt-responsive genes. New Phytologist. 2021:232(6): 2418–2439. 10.1111/nph.17760 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Hartwig B, James GV, Schneeberger K, Turck F. Complementary activities of TELOMERE REPEAT BINDING proteins and polycomb group complexes in transcriptional regulation of target genes. Plant Cell. 2016:28(1): 87–101. 10.1105/tpc.15.00787 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and materials needed to evaluate the conclusions in the paper are present in the paper and the Supplemental materials.