Abstract
Polygenic inheritance plays a pivotal role in driving multiple sclerosis susceptibility, an inflammatory demyelinating disease of the CNS. We developed polygenic risk scores (PRS) of multiple sclerosis and assessed associations with both disease status and severity in cohorts of European descent.
The largest genome-wide association dataset for multiple sclerosis to date (n = 41 505) was leveraged to generate PRS scores, serving as an informative susceptibility marker, tested in two independent datasets, UK Biobank [area under the curve (AUC) = 0.73, 95% confidence interval (CI): 0.72–0.74, P = 6.41 × 10−146] and Kaiser Permanente in Northern California (KPNC, AUC = 0.8, 95% CI: 0.76–0.82, P = 1.5 × 10−53).
Individuals within the top 10% of PRS were at higher than 5-fold increased risk in UK Biobank (95% CI: 4.7–6, P = 2.8 × 10−45) and 15-fold higher risk in KPNC (95% CI: 10.4–24, P = 3.7 × 10−11), relative to the median decile. The cumulative absolute risk of developing multiple sclerosis from age 20 onwards was significantly higher in genetically predisposed individuals according to PRS. Furthermore, inclusion of PRS in clinical risk models increased the risk discrimination by 13% to 26% over models based only on conventional risk factors in UK Biobank and KPNC, respectively. Stratifying disease risk by gene sets representative of curated cellular signalling cascades, nominated promising genetic candidate programmes for functional characterization. These pathways include inflammatory signalling mediation, response to viral infection, oxidative damage, RNA polymerase transcription, and epigenetic regulation of gene expression to be among significant contributors to multiple sclerosis susceptibility. This study also indicates that PRS is a useful measure for estimating susceptibility within related individuals in multicase families. We show a significant association of genetic predisposition with thalamic atrophy within 10 years of disease progression in the UCSF-EPIC cohort (P < 0.001), consistent with a partial overlap between the genetics of susceptibility and end-organ tissue injury. Mendelian randomization analysis suggested an effect of multiple sclerosis susceptibility on thalamic volume, which was further indicated to be through horizontal pleiotropy rather than a causal effect.
In summary, this study indicates important, replicable associations of PRS with enhanced risk assessment and radiographic outcomes of tissue injury, potentially informing targeted screening and prevention strategies.
Keywords: polygenic risk score, multiple sclerosis, pathway-specific risk score, phenotype association
Shams et al. show that polygenic risk scores identify individuals at high risk of developing multiple sclerosis and who may benefit from preventive interventions. A strong genetic predisposition also associates with hallmarks of disease progression, advancing understanding of biochemical pathways related to disease inheritance.
Introduction
Multiple sclerosis is a chronic disease of the CNS with established genetic susceptibility footprints. Leveraging genome-wide genotype data from 47 429 multiple sclerosis cases and 68 374 controls, the International Multiple Sclerosis Genetics Consortium (IMSGC) developed a dataset that yielded statistical evidence for the association of 200 autosomal susceptibility variants outside the major histocompatibility complex (MHC), 32 within the extended MHC region, and one in chromosome X.1 These associations together with an additional 416 highly suggestive, albeit not genome-wide significant variants explain 48% of multiple sclerosis heritability, and collectively highlight gene networks operating in the adaptive and innate arms of immune response, as well as enrichment of genes expressed in microglia.1
The polygenic mode of multiple sclerosis inheritance provided the rationale for developing aggregated genetic burden scores including all identified genome-wide significant susceptibility variants, in an attempt to better predict the cumulative effects of genetic liability.2,3 Polygenic risk scores (PRS) combine all genetic effects into a single metric of inherited susceptibility with multiple important potential applications: they can be used to assess genetic heritability, measure genetic overlap between different traits, improve screening, assist in identifying biomarkers of complex diseases, stratify patients in clinical trials, and adjust treatment strategies.4–9 However, the early application of this tool in multiple sclerosis was compromised by gaps in the number of known risk loci and imperfect estimation of allelic weights, leading to poor sensitivity.10–12 Methodological and analytical advances, and increasing sample sizes in genome-wide association studies (GWAS) have facilitated genetic discoveries and provided adequate power for detecting variations with small effects on the target phenotype, thus enabling more compelling studies of the risk distribution in a given population. Specifically, the LDPred algorithm considerably enhanced the prediction accuracy of the genetic load for autoimmune diseases including type 1 diabetes, Crohn’s disease, and rheumatoid arthritis.13 Recent updates of the program (LDPred2) provide even higher performance and more accurate effect size adjustments compared to both the old version and other competitive methods, particularly for causal variants in complex long-range linkage disequilibrium (LD) regions such as the MHC.14 LDPred2 also identifies variants with true zero effect size, significantly reducing the number of variants used in the PRS calculations without affecting its predictive performance. Here, we developed both genome-wide and pathway-specific PRS for well-characterized, independent multiple sclerosis datasets to assess the aggregated genetic burden effects on disease risk and activity.
Materials and methods
Polygenic risk score derivation
The post-quality control (QC) IMSGC Discovery summary statistics based on 14 802 multiple sclerosis cases and 26 703 controls including 8 589 719 variants was used for PRS derivation to predict multiple sclerosis risk in people of European ancestry. Please refer to IMSGC for details of multisite cohorts, inclusion criteria, demographics and analytical methods.1 Datasets used in this study are summarized in Supplementary Table 1. All duplicate single nucleotide polymorphisms (SNPs), multi-allelic markers, variants with low imputation quality (INFO < 0.6), rare variants with minor allele frequencies of <1%, variants with genotype missingness >10%, and variants deviating from Hardy–Weinberg equilibrium (P < 1 × 10−6) were excluded. For specific details about each dataset please refer to the Supplementary material.
The LDPred2 algorithm was used to obtain posterior mean effect sizes by adjusting a prior probability and accounting for LD.14 As recommended by recent studies,14,15 only HapMap3 variants that passed rigorous QC in our validation and replication datasets, namely UK Biobank (UKBB), Kaiser Permanente in Northern California (KPNC), and UCSF-EPIC, were included in PRS development (n = 800 702). This set contained established disease-associated variants and other loci. The advantage of using HapMap3 variants is that they have passed extensive QC and have a good coverage of the whole genome.
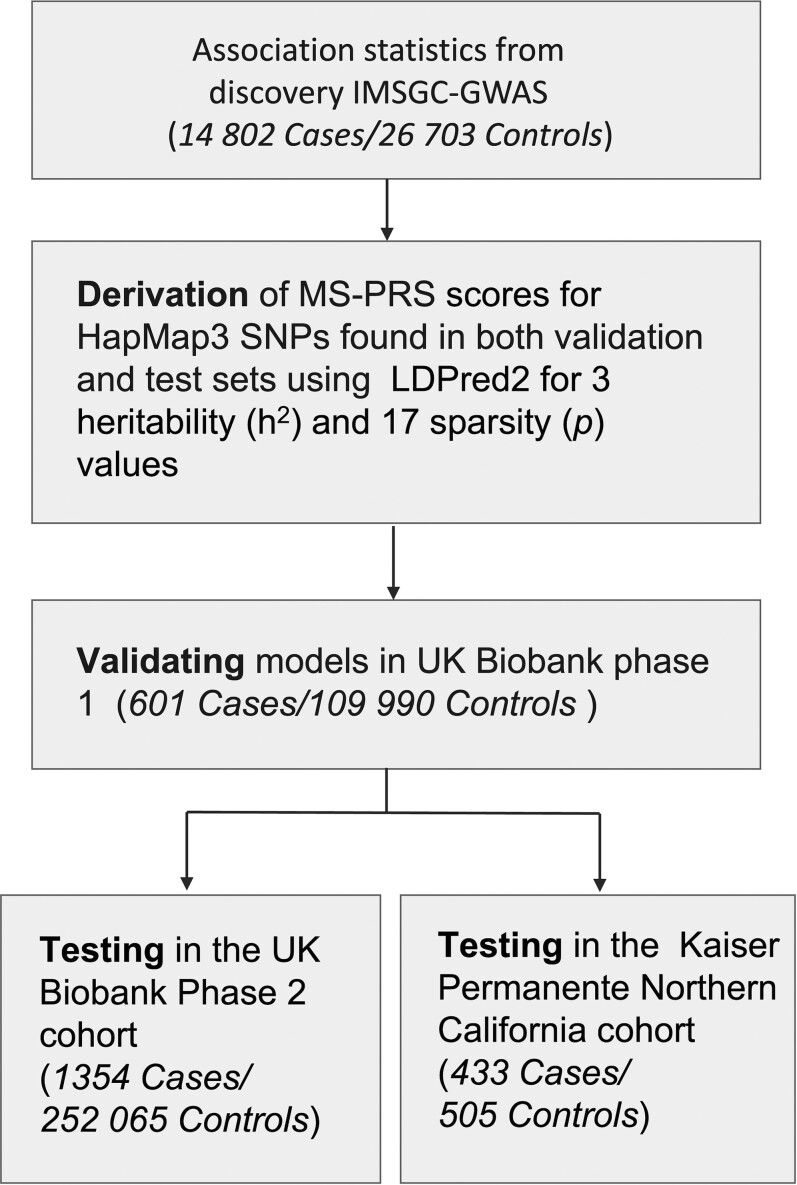
The underlying population structure of both UKBB and KPNC datasets is homogeneous and strictly European, while 6.6% of UCSF-EPIC subjects were identified as Admixed Americans (AMR), which was demonstrated by performing principal component analysis (PCA) of each dataset with 1000 Genomes Phase 3 populations as a reference (Supplementary Fig. 1A–H). The LD reference used was based on 362 320 European individuals in UKBB. The tuning hyperparameters, sparsity (p) and heritability (h2), were optimized in UKBB Phase 1 (UKBB1) (validation set) comprising 601 multiple sclerosis cases and 109 990 unaffected subjects. Heritability values tested in the model included the one estimated by LD score regression (h2 = 0.38) as well as that multiplied by 0.7, and 1.4. A total of 17 P-values between 1 × 10−4 and 1 evenly spaced on a logarithmic scale were examined. Discriminative capacity of PRS was assessed as the maximum area under the receiver-operating curve (AUC) and 95% confidence intervals (CI) obtained from 10 000 non-parametric bootstrap replicates. The sparse option was enabled in LDPred2 to allow for computing true zero effect sizes. A total of 455 902 (57% of included variants) had non-zero effect sizes β = {β1, β2, …, βn} and PRS of the j-th individual was computed by taking the weighted sum of risk alleles, . All PRS scores are standardized to have zero mean and unit variance. The best performing model out of 61 in the validation dataset (P = 0.018, h2 = 0.38, AUC = 70%, 95% CI: 68% to 72%) was then tested in UKBB Phase 2 (UKBB2), consisting of 1354 multiple sclerosis cases and 252 065 unaffected subjects after QC. The top model was then tested in the KPNC dataset. Scores were adjusted for the first 20 principal components of ancestry. The flowchart of PRS development is shown in Fig. 1.
Figure 1.
Flow diagram of the MS-PRS derivation. The post-QC discovery IMSGC summary statistics were used to construct a predictive model of multiple sclerosis risk. Multiple genetic models comprising significant variants and their associated effect sizes were optimized over a range of parameters using LDpred2. Performance of genetic models was validated in the UKBB1 and independently tested in UKBB2 and KPNC datasets.
Since the UCSF-EPIC cohort was part of the IMSGC meta-analysis, two sets of PRS scores were developed for this dataset. In the first set, PRS is computed using the same effect sizes as described above, which are based on the summary statistics incorporating all IMSGC-GWAS datasets. These scores are referred to as MS-PRS for all cohorts used in this study. Additionally, a second set of scores for UCSF-EPIC was developed for which a summary statistics based on only SNPs contributed by all the IMSGC sites except UCSF (n = 14) was used (PRSLOO). The PRSLOO also included SNPs that passed QC in all datasets, as described above. However, before computing the effect sizes in LDPred2, variants with SDss < 0.5.SDval or SDss > 0.1 + SDval or SDss < 0.1 or SDval < 0.05, in which SDss is the standard deviations from the summary statistics and SDval is the standard deviations of genotypes of subjects in the validation set, are removed as suggested by Privé et al.14 Applying this condition using the leave-one-out summary statistics resulted in removing 5320 SNPs from PRSLOO.
To develop familial PRS, DNA samples of 135 individuals from 35 families with an unaffected parent, a co-affected parent-child pair, and a discordant sib-pair were genotyped using a custom Illumina Chip.16 Due to the limited overlap between variants in the familial dataset and the HapMap3 markers (∼18%), the familial PRS was computed based on the common variants present in this dataset, the IMSGC summary statistics, and UKBB1, which was used as the validation set (n = 921 610). Chromosome-specific scores were computed by summing over variants within each chromosome multiplied by their corresponding effect sizes, which were estimated genome-wide, as described above.
Gene set selection and derivation of pathway-specific risk scores
Pathway-specific risk scores were developed for the 2852 gene sets identified in the Canonical Pathway subset of Molecular Signatures Database (MSigDB version 7.2) consisting of curated subsets representing biological pathways using PRSice2.17 The LD pruning was carried out using r2 of 0.1 and a 1 mb window. To generate competitive P-value estimates for each pathway, 10 000 permutations of sample labels were implemented. Nagelkerke’s pseudo R2 value was adjusted for an estimated prevalence of 0.00127,18 gender, age and the first 20 principal components. The gene sets surviving the Bonferroni correction for multiple testing (corrected P = 1.75 × 10−5) in UKBB1 were further replicated in UKBB2. Cumulative scores of 132 gene sets were significantly associated with multiple sclerosis risk in the UKBB1 (Bonferroni corrected P < 1.75 × 10−5). A total of 85 associations were replicated in UKBB2 (Supplementary Table 2).
Polygenic risk score association with neuroimaging phenotypes
Calibrated volumetric measurements of the total brain (BV), white matter (WMV), peripheral grey matter (pGMV), CSF, and three compartments of the deep grey matter, namely thalamus, caudate and putamen in 467 UCSF-EPIC participants with 10 years of annual follow-up were investigated as hallmarks of disease progression.19 First, the association of both PRSLOO and MS-PRS with each metric at baseline was examined in a linear model including sex, age and disease duration as covariates. To replicate associations in UKBB, only cases from both phases with available neuroimaging data were included in the analysis (n = 132).20
Longitudinal associations were then examined by computing percentage change in neuroimaging phenotypes with respect to the baseline at each annual visit within a 10-year follow-up period. Using linear mixed modelling for repeated measurements (MMRM), the associations between polygenic risk scores and longitudinal per cent change of volumetric measurements of BV, WMV, pGMV, thalamus, caudate and putamen were investigated. To avoid potential biases imposed by inflated MS-PRS in UCSF-EPIC, analyses were only performed with PRSLOO. Years six and seven were excluded due to the high proportion of missing data. Specifically, fixed effects in the model included PRSLOO, baseline phenotype values, age at baseline, disease duration at baseline, sex and visit as nominal variables, interaction terms between visit and PRSLOO (visit × PRSLOO), visit and age at baseline (visit × age), and visit and sex (visit × sex). PRSLOO was treated as a continuous variable. Antedependent covariance structure was used in all models.
For details on datasets used in this study, the conventional risk factors, coefficient of determination, absolute risk stratification, and Mendelian randomization, please refer to the online Supplementary material.
Data availability
This study includes no data deposited in external repositories. All the data supporting the findings of this study are available through application to UK Biobank or request from the corresponding author.
Results
Polygenic susceptibility of multiple sclerosis
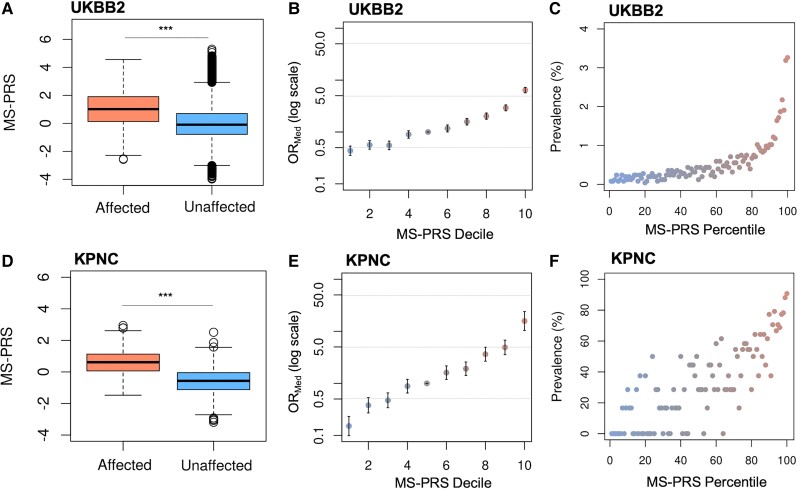
The MS-PRS performance in UKBB2 evaluated by the AUC (AUC = 0.73, 95% CI: 0.72–0.74; Supplementary Fig. 2A) was higher than that in UKBB1 (AUC = 0.7, 95% CI: 0.68–0.72). Increased MS-PRS of multiple sclerosis cases compared to unaffected subjects in UKBB2 was confirmed by unpaired two-sample Wilcoxon test (P = 1.29 × 10−190) shown in Fig. 2A and Supplementary Fig. 2B. The risk of multiple sclerosis across equal strata of increasing MS-PRS was estimated by odds ratios relative to the median decile (ORMed) shown in Fig. 2B as well as relative to the remainder of the population, corrected for the first 20 principal components (PCs), age and sex. Both measures indicated that individuals at the tail of the MS-PRS distribution were at markedly higher risk of developing the disease. Specifically, individuals in the top 5% and 10% of MS-PRS in UKBB2 were at more than 6- (95% CI: 5.7–6.5, P = 9.9 × 10−134) and 5-fold (95% CI: 4.8–5.4, P = 6.41 × 10−146) increased risk, respectively, compared to the rest of the population (Supplementary Fig. 2C). Similarly, individuals at the top MS-PRS decile were at greater risk relative to the median decile (ORMed > 5.3, 95% CI: 4.7–6, P = 2.8 × 10−45), shown in Fig. 2B. Prevalence of multiple sclerosis notably increased according to the MS-PRS percentile (Fig. 2C). These results confirm that genetic predisposition is an important component of multiple sclerosis risk in a population-level cohort.
Figure 2.
Discriminative power of MS-PRS in the UKBB2 and KPNC datasets. (A) The MS-PRS of subjects affected by multiple sclerosis (mean = 1, IQR = 1.770, min = −2.575, max = 4.561) is greater than unaffected individuals (mean = −0.005, IQR = 1.484, min = −3.979, max = 5.3) in UKBB2 (***P = 1.29 × 10−190). (B) The ORMed at higher MS-PRS deciles relative to the median decile is increased more than 5-fold in UKBB2. The y-axis is on a log scale. Bars are standard errors. (C) Prevalence of multiple sclerosis in UKBB2 increased according to the MS-PRS percentile. (D) Consistent with UKBB2, the MS-PRS of cases (mean = 0.585, IQR = 1.064, min = −1.474, max = 2.945) is higher than controls (mean = −0.579, IQR = 1.076, min = −3.193, max = 2.514) in KPNC (***P = 1.5 × 10−53). (E) The ORMed in the top MS-PRS decile of KPNC is higher than 15-fold relative to the median decile. (F) A linear increase of the multiple sclerosis prevalence with respect to MS-PRS is observed in the KPNC cohort. IQR = interquartile range; min = minimum; max = maximum.
The discriminative power of MS-PRS in the well-curated, case-control KPNC cohort was enhanced (Supplementary Fig. 2D; AUC = 0.8, 95% CI: 0.76–0.82) compared to UKBB2. The scores of KPNC cases were greater than controls (P = 1.5 × 10−53), as shown in Fig. 2D. Similar to UKBB2, the proportion of KPNC subjects with increased MS-PRS had higher OR values relative to the remainder of this dataset (Supplementary Fig. 2E and F). The top MS-PRS decile relative to the median decile indicated a 15-fold higher OR (95% CI: 10.4–24, P = 3.7 × 10−11), as shown in Fig. 2E. The multiple sclerosis prevalence in the KPNC dataset according to MS-PRS percentile linearly increased, reflecting the size and higher proportion of multiple sclerosis cases in this dataset (Fig. 2F).
Splitting MS-PRS by chromosome demonstrated the higher statistical significance of chromosome 6, containing the extended MHC region, in predicting the multiple sclerosis disease status in both UKBB2 and KPNC datasets (Supplementary Fig. 3A and B). The predictive power of collective scores based on all autosomes excluding chromosome 6, marked by ‘A-6’ in Supplementary Fig. 3A and B, was almost equal to that of the scores based on chromosome 6 alone, showing that excluding chromosome 6 in both datasets significantly affected P-values of cumulative MS-PRS. The MS-PRS distributions in males and females were not significantly different in KPNC, while the unaffected female MS-PRS was slightly higher than that of males (P < 0.01) in UKBB2 (Supplementary Fig. 3C and D). Such difference was driven by the outliers and became insignificant when those were excluded from the UKBB2 analysis.
To assess the prediction accuracy of MS-PRS, we used R2 on the liability scale , which was suggested to be directly comparable to heritability by Lee et al.21 Please refer to the Supplementary material for prevalence and derivation details. Assuming that multiple sclerosis liability has a normal distribution, coefficients of determination on the liability scale in the UKBB2 were , 7.7% (95% CI: 6.8–8.7%), , 8.3% (95% CI: 7.3–9.2%), and , 10.7% (95% CI: 9.4–11.8%), while R2 values corrected for the ascertainment bias in KPNC were increased to , 12.5% (95% CI: 11.5–13.5%), , 13.3% (95% CI: 12.3–14.4%), and , 16.9% (95% CI: 15.7–18.2%) shown in Table 1. Under the logistic distribution assumption, in UKBB2 and KPNC were 16.23% (95% CI: 16.2–16.3%) and 30% (95% CI: 28.1–31.9%), respectively. The UKBB is a population-level cohort, whereas KPNC is a case-control study in which cases are identified and matched to controls arising in the same population. Nonetheless, was comparable between these datasets, particularly under the normal distribution assumption, in agreement with Lee et al.21 Limiting the analysis to only first, fifth, and 10th MS-PRS deciles combined, increased the prediction accuracy by 30% (P = 6.97 × 10−145) and 60% (P = 1.01 × 10−38) in UKBB2 and KPNC datasets, repectively.
Table 1.
Prediction accuracy of MS-PRS in the test datasets
| MS prevalence | (95% CI) | |
|---|---|---|
| UKBB2 | KPNC | |
| 0.00127 | 7.7 (6.8–8.7) | 12.5 (11.5–13.5) |
| 0.0019 | 8.3 (7.3–9.2) | 13.3 (12.3–14.4) |
| 0.0069 | 10.6 (9.4–11.8) | 16.9 (15.7–18.2) |
The regression P-values were 3.13 × 10−170 and 1.56 × 10−57 for UKBB2 and KPNC datasets, respectively. = coefficient of determination on the liability scale.
We also computed Nagelkerke’s pseudo-R2 on the observed scale in order to compare the MS-PRS performance with a previous case-control study by IMSGC.2 Genetic burden scores in this study showed 3% association with susceptibility estimated by . The case-control ratio in this study (∼0.38) was significantly higher than that in UKBB2 (∼0.005), but lower than KPNC (∼0.85), while having a substantially sparser genotyping density than both UKBB and KPNC. The was 8% (P = 7.26 × 10−224) and 33% (P = 9.4 × 10−60) in UKBB2 and KPNC, respectively. Although captured the improved predictive power of MS-PRS in independent test sets, it is relatively sensitive to dataset composition and thus may not serve as an accurate measure for comparing PRS models between population-level and case-control cohorts.
The cumulative incidence of multiple sclerosis as a function of age was used to assess the ability of PRS to refine risk estimates in the UK population. Risk stratification was primarily driven by PRS as indicated by significantly diverging risk trajectories after age 20 (Supplementary Fig. 4A). Multiple sclerosis clinical onset age typically ranges between 20 and 40. The cumulative risk of individuals in the UKBB2 within the top 5% of MS-PRS for developing multiple sclerosis up to age 40 was more than 8- and 30-fold higher than those within the 30–60% of MS-PRS (P < 0.0001; Supplementary Table 2) and the bottom 5% of MS-PRS percentile (P < 0.0001), respectively. Coarser MS-PRS strata were used for this analysis in the KPNC dataset to account for the smaller sample size. Significant divergence of risk trajectories after the age of 20 and higher cumulative incidence for KPNC subjects in the top 20% of MS-PRS up to age 40 (fold change compared to the lowest 20% MS-PRS, 6.5; P < 1 × 10−4) is shown in Supplementary Fig. 4B and Supplementary Table 3.
Combining polygenic risk score of multiple sclerosis with conventional risk factors
Classification accuracies of basic models, consisting of age, sex, and established conventional risk factors (CRF) of multiple sclerosis were compared against models in which MS-PRS was also included (Supplementary Table 4). An increase in the AUC, net reclassification index (NRI), and integrated discrimination index (IDI) are reported as measures of risk discrimination improvement for MS-PRS-included versus basic models (Table 2). For more details, please refer to the Supplementary material.
Table 2.
Risk model improvement upon PRS addition in UKBB2 and KPNC datasets
| Prediction model | AUC (95% CI) | NRI (95% CI) | IDI (95% CI) | P a |
|---|---|---|---|---|
| UKBB2 | ||||
| ȃsex + age | 0.64 (0.62–0.65) | NA | NA | NA |
| ȃsex + age + PRS | 0.77 (0.75–0.78) | 0.729 (0.67–0.788) | 0.007 (0.006–0.008) | <0.001 |
| ȃsex + age + HLA.DRB1*15:01 (rs3135391) | 0.69 (0.68–0.71) | 0.47 (0.407–0.533) | 0.002 (0.002–0.003) | <0.001 |
| ȃsex + age + mono | 0.64 (0.63–0.66) | NA | NA | NA |
| ȃsex + age + mono + PRS | 0.77 (0.75–0.78) | 0.727 (0.668–0.786) | 0.007 (0.006–0.008) | <0.001 |
| ȃsex + age + mono + smoking | 0.65 (0.64–0.67) | NA | NA | NA |
| ȃsex + age + mono + smoking + PRS | 0.78 (0.76–0.79) | 0.726 (0.667–0.785) | 0.007 (0.007–0.008) | <0.001 |
| ȃsex + age + mono + smoking + BMI | 0.66 (0.64–0.67) | NA | NA | NA |
| ȃsex + age + mono + smoking + BMI + PRS | 0.77 (0.76–0.79) | 0.719 (0.658–0.781) | 0.007 (0.006–0.008) | <0.001 |
| KPNC | ||||
| ȃsex + age | 0.62 (0.58–0.65) | NA | NA | NA |
| ȃsex + age + PRS | 0.80 (0.77–0.83) | 0.838 (0.718–0.958) | 0.238 (0.21–0.266) | <0.001 |
| ȃsex + age + HLA.DRB1*15:01 (rs3135391) | 0.72 (0.68–0.75) | 0.64 (0.517–0.762) | 0.104 (0.083–0.124) | <0.001 |
| ȃsex + age + mono | 0.62 (0.58–0.65) | NA | NA | NA |
| ȃsex + age + mono + PRS | 0.80 (0.77–0.83) | 0.859 (0.74–0.978) | 0.238 (0.21–0.266) | <0.001 |
| ȃsex + age + mono + smoking | 0.64 (0.60–0.67) | NA | NA | NA |
| ȃsex + age + mono + smoking + PRS | 0.81 (0.78–0.84) | 0.874 (0.756–0.993) | 0.234 (0.206–0.262) | <0.001 |
| ȃsex + age + mono + smoking + family history | 0.67 (0.63–0.70) | NA | NA | NA |
| ȃsex + age + mono + smoking + family history + PRS | 0.82 (0.79–0.85) | 0.844 (0.725–0.964) | 0.214 (0.187–0.241) | <0.001 |
| ȃsex + age + mono + smoking + family history + PRS + PRS*family history | 0.82 (0.80–0.85) | 0.813 (0.692–0.933) | 0.222 (0.195–0.25) | 0.001 |
| ȃsex + age + mono + smoking + family history + overweight as a child | 0.67 (0.64–0.71) | NA | NA | NA |
| ȃsex + age + mono + smoking + family history + overweight as a child + PRS | 0.82 (0.80–0.85) | 0.844 (0.725–0.964) | 0.212 (0.185–0.239) | <0.001 |
| ȃsex + age + mono + smoking + family history + overweight as a child + BMI | 0.71 (0.68–0.75) | NA | NA | NA |
| ȃsex + age + mono + smoking + family history + overweight as a child + BMI + PRS | 0.83 (0.80–0.86) | 0.814 (0.687–0.941) | 0.184 (0.157–0.211) | 0.001 |
| ȃsex + age + mono + smoking + family history + overweight as a child + BMI20 | 0.68 (0.60–0.75) | NA | NA | NA |
| ȃsex + age + mono + smoking + family history + overweight as a child + BMI20 + PRS | 0.85 (0.79–0.92) | 1.051 (0.805–1.297) | 0.202 (0.142–0.262) | <0.001 |
Basic models are shown in bold. NA = not applicable; BMI = body mass index; BMI20 = BMI at 20; mono = mononucleosis infection.
Significance of AUC difference, NRI and IDI.
Inclusion of PRS in all basic models consistently enhanced model performance in UKBB2 (Table 2). An alternative genetic model consisting of only a single SNP tagging HLA-DRB1*15:01, considered as the major genetic contributor to multiple sclerosis susceptibility,22 achieved an AUC of 0.69 (95% CI: 0.68–0.71), lagging the PRS-included model by 8% (P < 0.001). The NRI of the PRS-included model was 0.73 (95% CI: 0.67–0.79, P < 0.001), higher than that of a single SNP model (0.47, 95% CI: 0.41–0.53, P < 0.001). The best-performing model in this dataset included PRS, mono, smoking history, sex, and age (AUC = 0.78, 95% CI: 0.76–0.79, P < 0.001).
Other established risk factors, such as family history and being overweight as a child are available in KPNC, thus additional models are presented for this dataset. Consistent with the UKBB2 results, the single SNP tagging HLA-DRB1*15:01 achieved an AUC of 0.69 (95% CI: 0.65–0.72), while the AUC of PRS-included model reached 0.8 (95% CI: 0.77–0.83). Adding PRS to the model including mono, smoking history, family history, current body mass index (BMI), being overweight as a child, sex, and age increased the AUC by 12% to 0.83 (95% CI: 0.80–0.85), with an NRI of 0.818 (95% CI: 0.698–0.938, P < 0.001), IDI of 0.207 (95% CI: 0.146–0.268), and a false positive rate of <1%. Replacing the current body mass index (BMI) by BMI at 20s, which was only available in the KPNC dataset increased the AUC to 86% (95% CI: 0.80–0.85). Nevertheless, due to the high proportion of missing data (Supplementary Table 4), this result is likely biased. Altogether, addition of PRS to CRF notably improved model performance in both UKBB2 and KPNC.
This outcome was further tested in the UCSF-EPIC cohort.23 The prediction accuracy of PRSLOO, described in the ‘Materials and methods’ section, is negatively impacted by both re-adjusted effect sizes and the number of included SNPs. Nevertheless, addition of PRSLOO to CRFs significantly enhanced model performance, increasing the AUC of the basic model including mononucleosis infection, smoking, family history, current BMI, being overweight as a child, sex, and age by 16% (NRI = 0.792, 95% CI: 0.662–0.922, Supplementary Table 5) and decreasing the false positive rate by 6%. The interaction term between PRS and family history did not affect model performance in either KPNC or EPIC. Repeating this analysis excluding the AMR subjects from UCSF-EPIC (6.6%) increased the AUC by 1% (Supplementary Table 5). Including MS-PRS in EPIC overestimated the prediction accuracy of the full model (AUC > 0.9), as expected.
Pathway-specific polygenic risk scores
To investigate whether PRS can identify genetic circuits underlying multiple sclerosis risk, pathway-specific risk scores were computed. Out of 85 risk-associated pathway-based scores replicated in UKBB2, several were related to adaptive immune response, such as IL-5 (R2 = 1.8%, P = 2.03 × 10−64) and IL-12 signalling (R2 = 2.1%, P = 7.89 × 10−77), T cell receptor (TCR) signalling (R2 = 2.1%, P = 5 × 10−86), MHC class II antigen presentation (R2 = 2%, P = 9.7 × 10−78), interferon gamma signalling (R2 = 2%, P = 2.7 × 10−76), and complement cascade (R2 = 1.4%, P = 3 × 10−54). Viral and parasite infection response pathways also emerged as significantly associated with multiple sclerosis risk (P < 1 × 10−58). Signature pathways for other autoimmune chronic conditions such as lupus (R2 = 2.2%, 'P = 9.84 × 10−86), Hashimoto’s thyroiditis (R2 = 2%, P = 1.78 × 10−78), and diabetes type I (R2 = 2%, P = 3.39 × 10−81) appeared common with multiple sclerosis susceptibility pathways. Gene sets involved in cell adhesion (R2 = 2%, P = 1.54 × 10−77) and extracellular matrix (ECM) organization (R2 = 1.6%, P = 1.16 × 10−56) and protein glycosylation (R2 = 1%, P = 6.95 × 10−38) were also among the top pathway scores. Other signalling cascades such as the VEGF (R2 = 2%, P = 4.83 × 10−75) and several NOTCH pathways (R2 < 1.3%, P < 1 × 10−36) also contribute to multiple sclerosis susceptibility. The risk of the top 10% relative to the remainder of the population ranged between 2 (95% CI: 0.92–1.06) and 3 (95% CI: 2.8–3.2), according to the pathway-specific scores.
Multiple sclerosis risk and parental genetic load
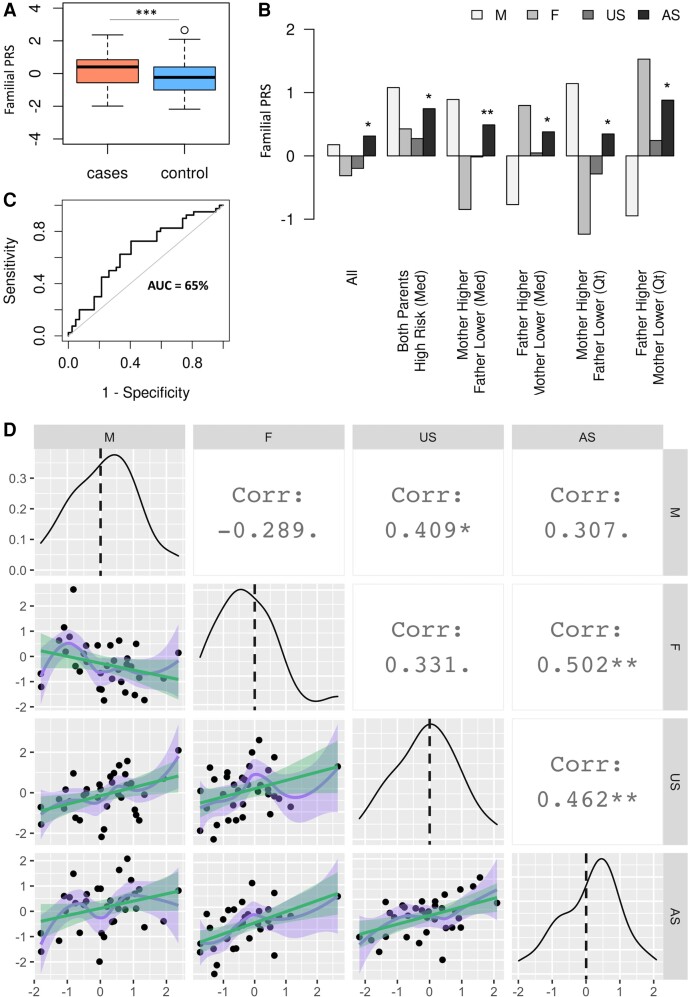
The best-performing PRS for the UCSF Multi-case Quartets dataset, validated in UKBB1, reached an AUC of 0.66. For details on the dataset and PRS derivation, please refer to the Supplementary material. The PRS distribution of cases was significantly higher than controls (P = 0.0004), as shown in Fig. 3A and Supplementary Fig. 5A. The statistical significance and the predictive power of the familial PRS compared to that of MS-PRS in UKBB2 and KPNC datasets was negatively impacted by several factors, including the limited number of subjects, the kinship among individuals, and the limited overlap of variants with those included in the HapMap3-based MS-PRS. Consistently, the chromosome-based PRS highlighted the importance of chromosome 6 in risk discrimination in affected families (Supplementary Fig. 5B). Moreover, the PRS distribution of siblings was not impacted by sex stratification, as indicated in Supplementary Fig. 5C. Families were then classified according to parents’ risk scores, i.e. lower or higher than the median PRS (PRSMedian), and lower or higher than 25% and 75% quantiles of PRS of all subjects, to further elucidate patterns of inheritance. The average PRS of mothers (M), fathers (F), affected (AS) and unaffected siblings (US) in each subgroup are shown in Fig. 3B. For comparisons across subgroups, we included 12 additional siblings data available for nine families to enhance statistical power. The PRS scores of all affected siblings (n = 40) were higher than all unaffected siblings (n = 42, P < 0.05), shown in the ‘All’ subgroup of Fig. 3B. In only four families, both parents were at higher risk than PRSMedian. The most common pattern in the dataset, according to the subgroups defined in Fig. 3B, was mothers’ scores higher and fathers’ scores lower than PRSMedian (n = 15). Therefore, the affected sibling scores in this subgroup compared to all unaffected siblings reached the highest statistical significance across all subgroups (P = 0.001). Although the statistical power was reduced in other subgroups due to the small number of families that met the criteria, the mean PRS of affected siblings was consistently higher (P < 0.05) than all unaffected siblings in families of one high and one low parental risk (Fig. 3B). The AUC for predicting disease status among siblings was 0.65 (Fig. 3C), which is comparable to overall AUC in this dataset.
Figure 3.
The PRS analysis of families with multiple sclerosis. (A) The PRS distribution of multiple sclerosis cases (mean = 0.265, IQR = 1.364, min = −1.990, max = 2.362) in families with co-affected parent-child pairs is higher than that of controls (mean = −0.27, IQR = 1.414, min = −2.180, max = 2.655; ***P = 0.0004). (B) The discriminative power of PRS among siblings (AUC = 0.65) is equal to the overall AUC of 0.66 in this dataset. (C) Families are stratified according to parents’ risk scores and average PRS of affected siblings in each subgroup is compared to that of all unaffected siblings (US in the ‘All’ subgroup). The median PRS and the 25% and 75% PRS quantiles are used for grouping families. For families in which one parent is at high and one at low risk relative to both median and top and bottom 25% PRS quantiles, affected siblings’ PRS scores in subgroups are consistently higher than all unaffected siblings. Subgroups: ‘All’ = all mothers, fathers, affected and unaffected siblings in the dataset; ‘Mother Higher Father Lower (Med)’ = mothers and fathers having higher and lower PRS scores relative to the median score, respectively; ‘Father Higher Mather Lower (Med)’ = fathers and mothers having higher and lower PRS scores relative to the median score, respectively; ‘Mother Higher Father Lower (Qt)’ = mothers and fathers having higher and lower PRS scores relative to the top and bottom 25% PRS quantiles, respectively; ‘Father Higher Mother Lower (Qt)’ = fathers and mothers having higher and lower PRS scores relative to the top and bottom 25% PRS quantiles, respectively. (D) Genetic correlations between family members depicted in the matrix scatter plot. Density plots across the diagonal are PRS distributions of M, F, US and AS. Linear (green) and loess (purple) regressions of pairwise PRS graphs are shown in the lower half of the matrix plot, shadings reflect data spread. The highest positive correlation is between PRS of fathers and that of AS (P < 0.001) followed by the correlation between US and AS (P < 0.001). AS = affected siblings; Corr = correlation; F = fathers; M = mothers; Med = median; Qt = quantile; US = unaffected siblings.
Pairwise correlations of MS-PRS between family members are depicted in Fig. 3D. Diagonal panels are PRS distributions of parents and siblings. Mothers constituted 65% of affected parents in this dataset, and the mother and affected siblings' distributions were skewed towards higher PRS values, while fathers’ distribution was shifted towards lower scores. The unaffected siblings' distribution was relatively symmetric. The correlation indices shown in the upper-half panels were obtained from the data-points in the lower-half of the matrix plot. The negative correlation between mothers and fathers reflected that only one spouse was affected in each family. The strongest correlation was observed among fathers and affected children (0.5, P < 0.005), while PRS of mothers better correlated with unaffected children (0.41, P < 0.05), suggesting that genetic predisposition of fathers is an important risk factor for children. The correlation among siblings was the second highest (0.46, P < 0.005).
Polygenic risk score association with disease progression and activity
Changes in CNS volumes represent a quantifiable surrogate of tissue loss and long-term disease progression in multiple sclerosis.24–26 Assessing the proportion of phenotypic variations at baseline explained by MS-PRS and PRSLOO showed modest associations of BV (βLOO = −0.09, ; βMS-PRS = −0.11, ), WMV (βLOO = −0.10, ; βMS-PRS = −0.13, ), thalamus (βMS-PRS, −0.11, ), putamen (βLOO = −0.10, ; βMS-PRS = −0.11, ), and CSF (βLOO = 0.12, ; βMS-PRS = 0.11, ) with at least one of the risk scores (Supplementary Table 6). Replicating these observations in UKBB showed consistency in terms of directionality of associations with EPIC as shown in Supplementary Table 6, but only associations with thalamic (β = −0.15, R2 = 3.2%) and putamen (β = −0.15, R2 = 3%) volumes remained significant in UKBB (P < 0.05). Furthermore, only CSF volume association with MS-PRS emerged as nominally significant (β = 0.1, R2 = 0.3%) in high-risk unaffected subjects in UKBB (top 5% MS-PRS, n = 1495) as shown in Supplementary Table 7.
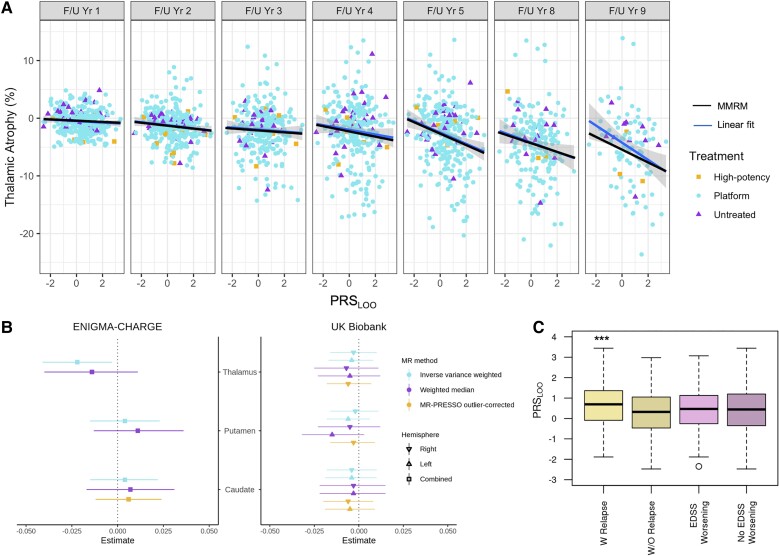
Next, we investigated the association of PRS with longitudinal per cent change of regional brain volumes at each annual visit relative to the baseline in the UCSF-EPIC dataset over a 10-year follow-up period. Association of PRSLOO with peripheral grey (β = −0.26) and thalamic atrophy (β = −0.53) remained significant upon Bonferroni correction for multiple testing across phenotypes (P < 0.007, Supplementary Table 8). Model predictions showed the highest atrophy rate in thalamus (Supplementary Table 9). An elevated PRSLOO was associated with increased thalamic volume loss at each visit (Fig. 4A). Of note, fluctuations in thalamic volume varied over time mainly due to the measurement noise, while group means decreased monotonically (Supplementary Fig. 6). Adjusting for treatment did not affect model predictions since most individuals (>55%) were on ‘platform therapy’ or ‘other’, grouped as one, in each follow-up year (Supplementary Table 10). However, associations may have been partially masked by longer periods of treatment and/or increased frequency of high potency therapy among patients throughout the 10-year course.
Figure 4.
Genetic risk association with clinical and radiographic phenotypes. (A) Thalamic atrophy at each visit against PRSLOO in the 10-year follow-up period. The blue line is a fit through the raw data at each visit and the shading shows the CI. The black line is the MMRM predictions including measurements from all visits. Different shapes for data-points are used to mark treatment. (B) The forest plot shows Mendelian randomization estimates (standard deviation change in imaging phenotype per doubling in odds of multiple sclerosis) in ENIGMA-CHARGE and UKBB and 95%CI. Inverse variance weighted Mendelian randomization analysis shows a weak effect of multiple sclerosis susceptibility on thalamic volume only in the ENIGMA-CHARGE cohort (P = 0.02). (C) Association of PRSLOO with disease activity represented by presence of relapses is significant (***P = 0.0002; W Relapse, mean = 0.651, IQR = 1.450, min = −1.886, max = 3.442; W/O Relapse, mean = 0.278, IQR = 1.519, min = −2.474, max = 2.978), but not with EDSS worsening (EDSS Worsening, mean = 0.429, IQR = 1.360, min = −2.348, max = 3.069; No EDSS Worsening, mean = 0.410, IQR = 1.545, min = −2.474, max = 3.442) from baseline to Year 5. EDSS = Expanded Disability Status Scale; IQR = interquartile range; min = minimum; max = maximum; PRESSO = Pleiotropy RESidual Sum and Outlier.
We additionally tested the existence of a putative causal effect of liability to multiple sclerosis on the same baseline imaging phenotypes, except CSF volume, in the ENIGMA-CHARGE cohort (n = 37 741) and broad unaffected UKBB population (n = 31 968) within a two-sample Mendelian randomization framework.27,28 Summary statistics for left and right parts of thalamus, caudate and putamen volumes as well as BV and WMV were available in UKBB, while only total thalamus, caudate and putamen were available in ENIGMA-CHARGE. For these analyses, we only used genome-wide significant non-MHC disease-associated SNPs (P < 5 × 10−8) as instrumental variables, to reduce the likelihood of weak instrument bias and distortion from horizontal pleiotropy.29,30 We found weak evidence of an effect of liability to multiple sclerosis (scaled per doubling in odds) and thalamic volume in the main inverse variance weighted Mendelian randomization analysis in the ENIGMA-CHARGE cohort (β = −0.22, P = 0.02), but not in the UKBB (Fig. 4B and Supplementary Tables 11 and 12). Sensitivity analyses revealed that the association in ENIGMA-CHARGE was likely driven by horizontal pleiotropy (Mendelian randomization-Egger intercept = −0.009, 95% CI: −0.016 to −0.001, P = 0.04) and the effect did not persist using a pleiotropy-robust method (Supplementary Table 11). Results for BV and WMV were not significant in UKBB (Supplementary Table 12). Despite substantial heterogeneity (Q statistic 144 to 207), sensitivity analyses were consistent with a null causal effect.
To further assess the association of polygenic scores with disease activity, UCSF-EPIC participants (n = 464) were divided into two groups according to whether they had experienced one or more relapses within a 5-year interval from the baseline visit, regardless of disease worsening within the same timeframe based on Expanded Disability Status Scale (EDSS) scores. Since no phenotypic information was utilized for the risk score calculations, we examined phenotypic associations with both PRSLOO and MS-PRS. An increase in both risk scores was associated with relapse activity, which remained significant after correcting for age, sex, and disease duration (βLOO = 0.34, P = 0.002; βMS-PRS = 0.26, P = 0.002; Fig. 4C). On the other hand, when EPIC patients were stratified according to EDSS worsening regardless of relapse co-occurrence (Supplementary Table 13), no significant difference was observed between the two groups (Fig. 4C). Multiple sclerosis age of onset is negatively associated with PRS in all datasets, but only significant in UKBB2 (β = −0.006, P = 0.01).
Discussion
Understanding the genetic architecture of polygenic diseases like multiple sclerosis requires accommodating significant variability in the number, relative weight, and ontological type of risk variants each individual carries. Polygenic risk scores developed and tested in different cohorts substantially outperformed previous susceptibility scores, including conventional genetic burdens based on a subset of 233 bona fide multiple sclerosis susceptibility variants.12,31,32 The ability of PRS to serve as a predictive biomarker for high-risk individuals in population-based cohorts has been demonstrated for multiple diseases,33–35 and was suggested to facilitate early diagnosis and implementation of preventive or therapeutic interventions.6,36,37 Interestingly, a recent study showed that polygenic risk profiling can assist in prioritizing individuals with low PRS for identification of rare pathogenic variant heterozygotes.38
Our results demonstrated that including PRS improves risk stratification of basic models including age, sex and established conventional multiple sclerosis risk factors, increasing AUC by up to 0.25 (Table 2 and Supplementary Table 4). Furthermore, comparing similar metrics between population-level UKBB cohort and the KPNC case-control study suggested that despite fundamental differences in the dataset design and compositions, individuals at significantly increased risk could be identified via MS-PRS in both cohorts. The MS-PRS scores explained 7–11% and 12–16% of multiple sclerosis liability, and even higher in extreme PRS deciles, in UKBB2 and KPNC, respectively. UKBB is a remarkable resource, steadily making progress in linking the diagnostic data with other health records in the UK. New disease cases are periodically added to this dataset. Nonetheless, lack of hospitalization records for all multiple sclerosis subjects and possibility of inaccurate self-reports may have resulted in the presence of false negatives in this dataset, affecting modestly the overall precision of MS-PRS. On the other hand, multiple sclerosis cases in the KPNC case-control study are neurologist-diagnosed and the controls are sex, age and locality matched, resulting in the absence of false negatives and improved performance of MS-PRS. Despite these differences affecting the overall AUC of the top MS-PRS, results were consistent between these datasets. Thus, although genetic prediction of future disease status in the general population is not sensitive enough due to the low prior probability to multiple sclerosis, if the target sample is at higher risk according to conventional risk factors and/or those experiencing suggestive symptoms, PRS can assist the diagnosis and the choice of management strategy. Of note, in the EPIC dataset, clinically isolated syndrome (CIS) patients' PRS scores were similar to multiple sclerosis patients but significantly different from healthy controls (Supplementary Fig. 7).
Stratifying disease risk by cellular pathways may provide insights into pathological mechanisms and unravel important biological overlap between different disorders. Pathway-based scores related to regulation of immune response showed the most significant association with disease status, confirming the central role of adaptive immunity in driving multiple sclerosis risk. TCR signalling and MHC class II antigen presentation are among the top disease-related pathways, highlighting the role of canonical antigen presentation processes in multiple sclerosis pathogenesis.39 Dysregulation in specific interleukin-mediated pathways may also contribute to possible imbalance toward pro-inflammatory signals in subjects at risk of multiple sclerosis. For example, our analysis pinpoints IL-12, a master regulator of Th1 responses, and IL-5, a potent chemoattractant and differentiation factor for eosinophils and basophils40,41 as key underlying processes of risk. In this context, the importance of cell adhesion and ECM organization for lymphocyte extravasation and CNS infiltration is also highlighted.42 Regulatory pathways underlying gene expression such as epigenetic processes are also emerging as essential.43 Our data suggest that post-translational regulatory processes, such as defective protein glycosylation, might be equally involved in multiple sclerosis pathogenesis. Notably, aberrant glycosylation patterns can modulate the self/non-self identification of multiple cellular proteins as well as switching antibodies from protective to autoreactive.44 The significance of the NOTCH signalling cascade in our analysis strengthens the interface between genetic predisposition and both neurodegeneration and immune response.45–47 Another significant pathway, the VEGF cascade, similarly modulates both CNS inflammation and neuronal survival in autoimmune demyelination.48 In addition, the association of oxidative stress pathways and multiple sclerosis susceptibility may support the role of biological ageing in multiple sclerosis pathology.49–52 Finally, a robust body of data supports an aetiological and pathological contribution of viral infection, mainly the Epstein–Barr virus, to multiple sclerosis,53–55 and the results presented here are consistent with an association of viral infection pathway with multiple sclerosis risk.
A higher aggregation of susceptibility variants in multi-case compared to single-case multiple sclerosis families has been previously reported.10 Yet, these studies showed a limited power in predicting the case-control status.12 To further our understanding of the heritability patterns within families, we studied a multi-case familial dataset in which one parent and at least one child were diagnosed with multiple sclerosis and incorporated all variants identified by a custom genotyping array for multiple sclerosis.16 We confirmed that a greater PRS in families of disease-discordant parents is associated with an increased risk of multiple sclerosis among all subjects (AUC = 66%), as well as just among the siblings (AUC = 65%). In this study, the PRS of the affected siblings were significantly higher if either or both parents were at high risk compared to the rest of the cohort. Unaffected siblings at high risk may especially benefit from this knowledge. These results suggest that polygenic profiling provides a compelling opportunity to forecast multiple sclerosis within sibships but needs to be further tested in larger familial cohorts.
Associations between MS-PRS and relapses and regional brain volumes were modest, yet important. Our results suggest a robust association of longitudinal peripheral and deep grey matter atrophy with high genetic predisposition, the strongest association being with thalamic atrophy. Thalamic atrophy is an important marker of multiple sclerosis progression occurring early and declining consistently throughout the course of multiple sclerosis and across clinical subtypes.56 Thalamic volume loss in multiple sclerosis patients is associated with decreased neuroperformance in all scales.57 Evidence on genetic correlation does not necessarily imply direct genetic modulation of the CNS tissue. Indeed, ours and prior studies in multiple sclerosis described heritability enrichment mainly in immune-related tissues.58 The Mendelian randomization analysis in ENIGMA-CHARGE replicated an effect of multiple sclerosis liability on thalamic volume, although the results indicated that this was through horizontal pleiotropy rather than a causal effect. Conversely, in UKBB where there was little evidence of pleiotropy, no Mendelian randomization association was observed. Differences between the two cohorts could be due to averaging bilateral structures in ENIMGA-CHARGE, and its inclusion of case-control studies with psychiatric diagnoses. Taken together, our results indicate that genetic liability to multiple sclerosis is unlikely to cause global or regional subcortical volume changes in the general adult population; rather, the association between MS-PRS and peripheral and deep grey matter atrophy is specific to those with multiple sclerosis. Lastly, we observed a negative association between MS-PRS and age of onset, consistent with a recent report on the genetic underpinning of early disease onset.59
In summary, PRS has the advantage of being accessible at any time and incorporating it in the current clinical risk models can be a promising basis for intensive monitoring, reducing modifiable risk factors that may delay the disease onset, and promoting early diagnosis or inform treatment options in cohorts at a higher prior probability, e.g. individuals with suggestive symptoms or those with family history asshown for other diseases.60,61 Also, considering that multiple sclerosis clinical onset typically occurs between 20 and 40 years of age, implementing preventive strategies for those at higher risk in early adolescence could be an effective strategy to control the rising global incidence of multiple sclerosis and its detrimental consequences. It is noteworthy that diverse population-level GWAS screening, for example in African Americans and Hispanic Americans, is a pressing need in multiple sclerosis genetics and essential for utilizing polygenic profiling in non-European populations. Given the increasing incidence rate of multiple sclerosis, PRS can play an important role in future public health as a part of multifactorial predictive models along with modifiable lifestyle factors, family history, and rare variations. Therefore, this study is an important step towards translating GWAS studies into relevant biology and clinically meaningful outcomes.
Supplementary Material
Acknowledgements
The resources and collaborative efforts by the IMSGC Consortium resulted in the summary statistics used in this work. This research has been conducted using the UK Biobank Resource (Project ID: 59309), UCSF-EPIC, Kaiser Permanente in Northern California, and the GWAS meta-analysis of ENIGMA and CHARGE consortia. The authors acknowledge the contributions of Stacy Caillier, Nicholas Lee, and Rosa Guerrero for sample processing and management. The authors appreciate Cameron Adams at UC Berkeley for his help with KPNC data acquisition and QC. The authors thank the staff at the John P. Hussman Institute for Human Genomics, University of Miami for their assistance in data acquisition and quality control. We thank Dr Wallace Wang for sharing his expertise in PRS calculation methods and Deborah Gordon for language editing.
Appendix 1
UCSF-EPIC Team in alphabetical order: Jessa Alexander, Riley Bove, Sergio Baranzini, Bruce A. C. Cree, Eduardo Caverzasi, Richard Cuneo, Stacy J. Caillier, Tiffany Cooper, Ari J. Green, Chu-Yueh Guo, Jeffrey M. Gelfand, Refujia Gomez-O’shea, Sasha Gupta, Jill Hollenbach, Meagan Harms, Roland G. Henry, Stephen L. Hauser, Myra Mendoza, Jorge R. Oksenberg, Nico Papinutto, Sam Pleasure, Kyra Powers, Adam Renschen, Adam Santaniello, Joseph J. Sabatino Jr., William A. Stern, Michael R. Wilson, Scott S. Zamvil.
Contributor Information
Hengameh Shams, Weill Institute for Neurosciences, Department of Neurology, University of California San Francisco, San Francisco, CA 94158, USA; Division of Epidemiology and Biostatistics, School of Public Health, University of California Berkeley, Berkeley, CA 94720, USA.
Xiaorong Shao, Division of Epidemiology and Biostatistics, School of Public Health, University of California Berkeley, Berkeley, CA 94720, USA.
Adam Santaniello, Weill Institute for Neurosciences, Department of Neurology, University of California San Francisco, San Francisco, CA 94158, USA.
Gina Kirkish, Weill Institute for Neurosciences, Department of Neurology, University of California San Francisco, San Francisco, CA 94158, USA.
Adil Harroud, Weill Institute for Neurosciences, Department of Neurology, University of California San Francisco, San Francisco, CA 94158, USA.
Qin Ma, Weill Institute for Neurosciences, Department of Neurology, University of California San Francisco, San Francisco, CA 94158, USA.
Noriko Isobe, Department of Neurology, Graduate School of medical Sciences, Kyushu University, Fukuoka, 812-8582, Japan.
Catherine A Schaefer, Kaiser Permanente Division of Research, Oakland, CA 94612, USA.
Jacob L McCauley, John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami, FL, USA; Dr. John T. Macdonald Department of Human Genetics, Miller School of Medicine, University of Miami, Miami, FL, USA.
Bruce A C Cree, Weill Institute for Neurosciences, Department of Neurology, University of California San Francisco, San Francisco, CA 94158, USA.
Alessandro Didonna, Weill Institute for Neurosciences, Department of Neurology, University of California San Francisco, San Francisco, CA 94158, USA; Department of Anatomy and Cell Biology, East Carolina University, Greenville, NC 27834, USA.
Sergio E Baranzini, Weill Institute for Neurosciences, Department of Neurology, University of California San Francisco, San Francisco, CA 94158, USA.
Nikolaos A Patsopoulos, Systems Biology and Computer Science Program, Ann Romney Center for Neurological Diseases, Department of Neurology, Brigham and Women’s Hospital, Boston, 02115 MA, USA; Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA; Harvard Medical School, Boston, MA 02115, USA; Broad Institute of Harvard and Massachusetts Institute of Technology, Cambridge, MA, USA.
Stephen L Hauser, Weill Institute for Neurosciences, Department of Neurology, University of California San Francisco, San Francisco, CA 94158, USA.
Lisa F Barcellos, Division of Epidemiology and Biostatistics, School of Public Health, University of California Berkeley, Berkeley, CA 94720, USA.
Roland G Henry, Weill Institute for Neurosciences, Department of Neurology, University of California San Francisco, San Francisco, CA 94158, USA.
Jorge R Oksenberg, Weill Institute for Neurosciences, Department of Neurology, University of California San Francisco, San Francisco, CA 94158, USA.
University of California San Francisco MS-EPIC Team:
Jessa Alexander, Riley Bove, Sergio Baranzini, Bruce A C Cree, Eduardo Caverzasi, Richard Cuneo, Stacy J Caillier, Tiffany Cooper, Ari J Green, Chu-Yueh Guo, Jeffrey M Gelfand, Refujia Gomez-O’shea, Sasha Gupta, Jill Hollenbach, Meagan Harms, Roland G Henry, Stephen L Hauser, Myra Mendoza, Jorge R Oksenberg, Nico Papinutto, Sam Pleasure, Kyra Powers, Adam Renschen, Adam Santaniello, Joseph J Sabatino, Jr, William A Stern, Michael R Wilson, and Scott S Zamvil
Funding
This study was supported primarily by grants from the National Multiple Sclerosis Society RG-1707-28775 to R.G.H. and J.R.O., National Multiple Sclerosis Society RFA-2104-37474 to J.R.O., and National Institutes of Health R35NS111644 and the Valhalla Foundation to S.L.H., as well as National Institutes of Health R01NS099240 to S.E.B. and National Institutes of Health R01ES017080, R01AI076544, R01NS049510 to L.B. Also, N.A.P. is supported by RG-1707-28657 and JF-1808-32223, Harry Weaver Award from the National Multiple Sclerosis Society. H.S. is supported by a postdoctoral fellowship from National Multiple Sclerosis Society (FG-1807-31603). This publication is also supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSF-CTSI Grant Number TL1 TR001871 to H.S. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Competing interests
The authors have no conflicts of interest to declare.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. International Multiple Sclerosis Genetics Consortium . Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science. 2019;365(6460):eaav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. International Multiple Sclerosis Genetics Consortium . Evidence for polygenic susceptibility to multiple sclerosis—The shape of things to come. Am J Hum Genet. 2010;86(4):621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17(10):1520–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andersen MS, Bandres-Ciga S, Reynolds RH, et al. . Heritability enrichment implicates microglia in Parkinson’s disease pathogenesis. Ann Neurol. 2021;89(5):942–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lobo JJ, McLean SA, Tungate AS, et al. . Polygenic risk scoring to assess genetic overlap and protective factors influencing posttraumatic stress, depression, and chronic pain after motor vehicle collision trauma. Transl Psychiatry. 2021;11(1):359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lu T, Forgetta V, Keller-Baruch J, et al. . Improved prediction of fracture risk leveraging a genome-wide polygenic risk score. Genome Med. 2021;13(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li QS, Wajs E, Ochs-Ross R, Singh J, Drevets WC. Genome-wide association study and polygenic risk score analysis of esketamine treatment response. Sci Rep. 2020;10(1):12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharp SA, Rich SS, Wood AR, et al. . Development and standardization of an improved type 1 diabetes genetic risk score for use in newborn screening and incident diagnosis. Diabetes Care. 2019;42(2):200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schumacher FR, Al Olama AA, Berndt SI, et al. . Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50(7):928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gourraud PA, McElroy JP, Caillier SJ, et al. . Aggregation of multiple sclerosis genetic risk variants in multiple and single case families. Ann Neurol. 2011;69(1):65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Jager PL, Chibnik LB, Cui J, et al. . Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8(12):1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Isobe N, Damotte V, Lo RV, et al. . Genetic burden in multiple sclerosis families. Genes Immun. 2013;14(7):434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vilhjálmsson BJ, Yang J, Finucane HK, et al. . Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015;97(4):576–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Privé F, Arbel J, Vilhjálmsson BJ. LDpred2: Better, faster, stronger. Bioinformatics. 2021;36(22–23):5424–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ge T, Chen CY, Ni Y, Feng YCA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10(1):1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. International Multiple Sclerosis Genetics Consortium (IMSGC) . Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Choi SW, Mak TSH, O’Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc. 2020;15(September):2759–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. GBD 2016 Multiple Sclerosis Collaborators . Global, regional, and national burden of multiple sclerosis 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18(3):269–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bischof A, Papinutto N, Keshavan A, et al. . Spinal cord atrophy predicts progressive disease in relapsing multiple sclerosis. Ann Neurol. 2022;91(2):268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bycroft C, Freeman C, Petkova D, et al. . The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee SH, Goddard ME, Wray NR, Visscher PM. A better coefficient of determination for genetic profile analysis. Genet Epidemiol. 2012;36(3):214–224. [DOI] [PubMed] [Google Scholar]
- 22. Lincoln MR, Montpetit A, Cader MZ, et al. . A predominant role for the HLA class II region in the association of the MHC region with multiple sclerosis. Nat Genet. 2005;37(10):1108–1112. [DOI] [PubMed] [Google Scholar]
- 23. University of California San Francisco MS-EPIC Team . Long-term evolution of multiple sclerosis disability in the treatment era. Ann Neurol. 2016;80(4):499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oh J, Sicotte NL. New imaging approaches for precision diagnosis and disease staging of MS? Mult Scler J. 2020;25(5):568–575. [DOI] [PubMed] [Google Scholar]
- 25. Bakshi R, Healy BC, Dupuy SL, et al. . Brain MRI predicts worsening multiple sclerosis disability over 5 years in the SUMMIT study. J Neuroimaging. 2020;30(2):212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barnett Y, Garber JY, Barnett MH. MRI biomarkers of disease progression in multiple sclerosis: old dog, new tricks? Quant Imaging Med Surg. 2020;10(2):527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith SM, Douaud G, Chen W, et al. . An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat Neurosci. 2021;24(May):737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Satizabal CL, Adams HHH, Hibar DP, et al. . Genetic architecture of subcortical brain structures in 38,851 individuals. Nat Genet. 2019;51(11):1624–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burgess S, Thompson SG, CRP CHD Genetics Collaboration . Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–764. [DOI] [PubMed] [Google Scholar]
- 30. Richardson TG, Harrison S, Hemani G, Smith GD. An atlas of polygenic risk score associations to highlight putative causal relationships across the human phenome. Elife. 2019;8:e43657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Isobe N, Keshavan A, Gourraud PA, et al. . Association of HLA genetic risk burden with disease phenotypes in multiple sclerosis. JAMA Neurol. 2016;73(7):795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harbo HF, Isobe N, Berg-Hansen P, et al. . Oligoclonal bands and age at onset correlate with genetic risk score in multiple sclerosis. Mult Scler. 2014;20(6):660–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khera A V, Chaffin M, Aragam KG, et al. . Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Khera A V, Chaffin M, Wade KH, et al. . Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell. 2019;177(12):587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rammos A, Gonzalez LAN, Weinberger DR, Mitchell KJ, Nicodemus KK. The role of polygenic risk score gene-set analysis in the context of the omnigenic model of schizophrenia. Neuropsychopharmacology. 2019;44(9):1562–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Natarajan P, Young R, Stitziel NO, et al. . Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135:2091–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wolfson M, Gribble S, Pashayan N, et al. . Potential of polygenic risk scores for improving population estimates of women’s breast cancer genetic risks. Genet Med. 2021;23(11):2114–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu T, Zhou S, Wu H, Forgetta V, Greenwood CMT, Richards JB. Individuals with common diseases but with a low polygenic risk score could be prioritized for rare variant screening. Genet Med. 2021;23:508–515. [DOI] [PubMed] [Google Scholar]
- 39. Yuseff MI, Pierobon P, Reversat A. How B cells capture, process and present antigens: a crucial role for cell polarity. Nat Rev Immunol. 2013;13(7):475–486. [DOI] [PubMed] [Google Scholar]
- 40. Athie-Morales V, Smits HH, Cantrell DA, Hilkens CMU. Sustained IL-12 signaling is required for Th1 development. J Immunol. 2004;172(1):61–69. [DOI] [PubMed] [Google Scholar]
- 41. Collins PD, Marleau S, Griffiths-Johnson DA, Jose PJ, Williams TJ. Cooperation between interleukin-5 and the chemokine eotaxin to induce eosinophil accumulation in vivo. J Exp Med. 1995;182(4):1169–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Damotte V, Guillot-Noel L, Patsopoulos NA, et al. . A gene pathway analysis highlights the role of cellular adhesion molecules in multiple sclerosis susceptibility. Genes Immun. 2014;15:126–132. [DOI] [PubMed] [Google Scholar]
- 43. Vakhitov VA, Kuzmina US, Bakhtiyarova KZ, et al. . Epigenetic mechanisms of the pathogenesis of multiple sclerosis. Hum Physiol. 2020;46(1):104–112. [Google Scholar]
- 44. Maverakis E, Kim K, Shimoda M, et al. . Glycans in the immune system and the altered glycan theory of autoimmunity: a critical review. J Autoimmun. 2015;57:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Garis M, Garrett-sinha LA. Notch signaling in B Cell immune responses. Front Immunol. 2021;11:609324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sega FVD, Fortini F, Aquila G, Campo G, Vaccarezza M, Rizzo P. Notch signaling regulates immune responses in atherosclerosis. Front Immunol. 2019;10:1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ho DM, Artavanis-Tsakonas S, Louvi A. The Notch pathway in CNS homeostasis and neurodegeneration. WIREs Dev Biol. 2020;9(1):e358. [DOI] [PubMed] [Google Scholar]
- 48. Lin W. Neuroprotective effects of vascular endothelial growth factor A in the experimental autoimmune encephalomyelitis model of multiple sclerosis. Neural Regen Res. 2017;12(1):70–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Satoh JI, Nakanishi M, Koike F, et al. . Microarray analysis identifies an aberrant expression of apoptosis and DNA damage-regulatory genes in multiple sclerosis. Neurobiol Dis. 2005;18(3):537–550. [DOI] [PubMed] [Google Scholar]
- 50. Briggs FBS, Goldstein BA, McCauley JL, et al. . Variation within DNA repair pathway genes and risk of multiple sclerosis. Am J Epidemiol. 2010;172:217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Papadopoulos D, Magliozzi R, Mitsikostas DD, Gorgoulis VG, Nicholas RS. Aging, cellular senescence, and progressive multiple sclerosis. Front Cell Neurosci. 2020;14:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Krysko KM, Henry RG, Cree BAC, et al. . Telomere length is associated with disability progression in multiple sclerosis. Ann Neurol. 2019;86(5):671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Donati D. Viral infections and multiple sclerosis. Drug Discov Today Dis Model. 2020;32:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bar-or A, Pender MP, Khanna R, et al. . Epstein–Barr virus in multiple sclerosis: Theory and emerging immunotherapies. Trends Mol Med. 2020;26(3):296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bjornevik K, Cortese M, Healy BC, et al. . Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science. 2022;375(January):296–301. [DOI] [PubMed] [Google Scholar]
- 56. Azevedo CJ, Cen SY, Khadka S, et al. . Thalamic atrophy in multiple sclerosis: a magnetic resonance imaging marker of neurodegeneration throughout disease. Ann Neurol. 2018;83(2):223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bergsland N, Benedict RHB, Dwyer MG, et al. . Thalamic nuclei volumes and their relationships to neuroperformance in multiple sclerosis: a cross-sectional structural MRI study. J Magn Reson Imaging. 2021;53(3):731–739. [DOI] [PubMed] [Google Scholar]
- 58. Finucane HK, Reshef YA, Anttila V, et al. . Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. 2018;50(4):621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Misicka E, Davis MF, Kim W, et al. . A higher burden of multiple sclerosis genetic risk confers an earlier onset. Mult Scler J. 2022;28(8):1189–1197. [DOI] [PubMed] [Google Scholar]
- 60. Hadley TD, Agha AM, Ballantyne CM. How do we incorporate polygenic risk scores in cardiovascular disease risk assessment and management? Curr Atheroscler Rep. 2021;23(6):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yanes T, Young MA, Meiser B, James PA. Clinical applications of polygenic breast cancer risk: a critical review and perspectives of an emerging field. Breast Cancer Res. 2020;22(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study includes no data deposited in external repositories. All the data supporting the findings of this study are available through application to UK Biobank or request from the corresponding author.