Abstract
The main carcinogen for keratinocyte skin cancers (KCs) such as basal and squamous cell carcinomas is ultraviolet (UV) radiation. There is growing evidence that accumulation of mutations and clonal expansion play a key role in KC development. The relationship between UV exposure, epidermal mutation load, and KCs remains unclear. Here, we examined the mutation load in both murine (n = 23) and human (n = 37) epidermal samples. Epidermal mutations accumulated in a UV dose–dependent manner, and this mutation load correlated with the KC burden. Epidermal ablation (either mechanical or laser induced), followed by spontaneous healing from underlying epithelial adnexae reduced the mutation load markedly in both mouse (n = 8) and human (n = 6) clinical trials. In a model of UV-induced basal cell carcinoma, epidermal ablation reduced incident lesions by >80% (n = 5). Overall, our findings suggest that mutation burden is strongly associated with KC burden and represents a target to prevent subsequent KCs.
The role of epidermal mutation load in skin carcinogenesis is explored as a target and indicator of burden.
INTRODUCTION
Keratinocyte skin cancers (KCs) are the most common malignancy (1). The two major types of KCs, basal cell carcinomas (BCCs) and squamous cell carcinomas (SCCs), originate from the epidermis and represent a major global health burden. In Australia alone, there are more than a million interventions annually for KCs through mostly surgical excision (2, 3). The main environmental factor driving KC formation is solar ultraviolet (UV) irradiation. This is also reflected in several key patient-associated risk factors of KCs such as skin type and age. Red hair, inability to tan, and propensity to sunburn are key elements determining the risk of skin cancer at the individual level (4). Similarly, advancing age is another important factor associated with KC formation (5). The cumulative effect of sun damage is classically thought to result in field cancerization, where the entire area of the skin exposed to the sun is susceptible to cancer development. The high burden of the disease is due to multiplicity of new KCs—more than 50 to 80% of patients with a history of KCs will develop subsequent KCs (2, 5).
UV exposure is thought to contribute to carcinogenesis through a variety of mechanisms. It induces mutations directly (6, 7), allows the expansion of clones through a proliferative stimulus (8), and acts as an immunosuppressant (9). Recently, multiple studies reported that normal skin exposed to the sun could harbor mutations commonly found in KCs (10–13). This suggests that carcinogenesis might be a more complex and intricate process, and simple disruption of genes that are commonly considered as cancer causing may not be enough to drive KCs. This was replicated in murine studies where single oncogenic events were not enough to drive KCs (14). The accumulation of mutations in the same cell or the occurrence of the mutation in a specific cell of origin were prerequisites, suggesting that similar mechanisms might be at play in humans (15, 16).
Previous studies have found that mutations accumulate as we age (17, 18). To date, the link between accumulated mutations and KC burden has not been explored. It is also unclear whether targeting epidermal mutational burden is an effective strategy to lower the risk of KCs. Thus, better understanding of the relationship between sun exposure, epidermal mutation accumulation, and cancer risk is essential in an attempt at lowering the KC burden.
In this study, we explored the relationship between mutation load and clinical characteristics in a cohort of patients and in mouse models. We also explored an intervention that reduced mutation load in the prevention of skin cancers. We showed that continuous chronic suberythermal ultraviolet B (UVB) exposure resulted in increased mutation load. Furthermore, KC burden in patients was associated with increased epidermal mutation load. Last, we showed that epidermal ablation resulted in a major reduction in mutation load with a notable reduction in cancer occurrence and size.
RESULTS
Chronic suberythemal UV exposure promotes mutation accumulation over time
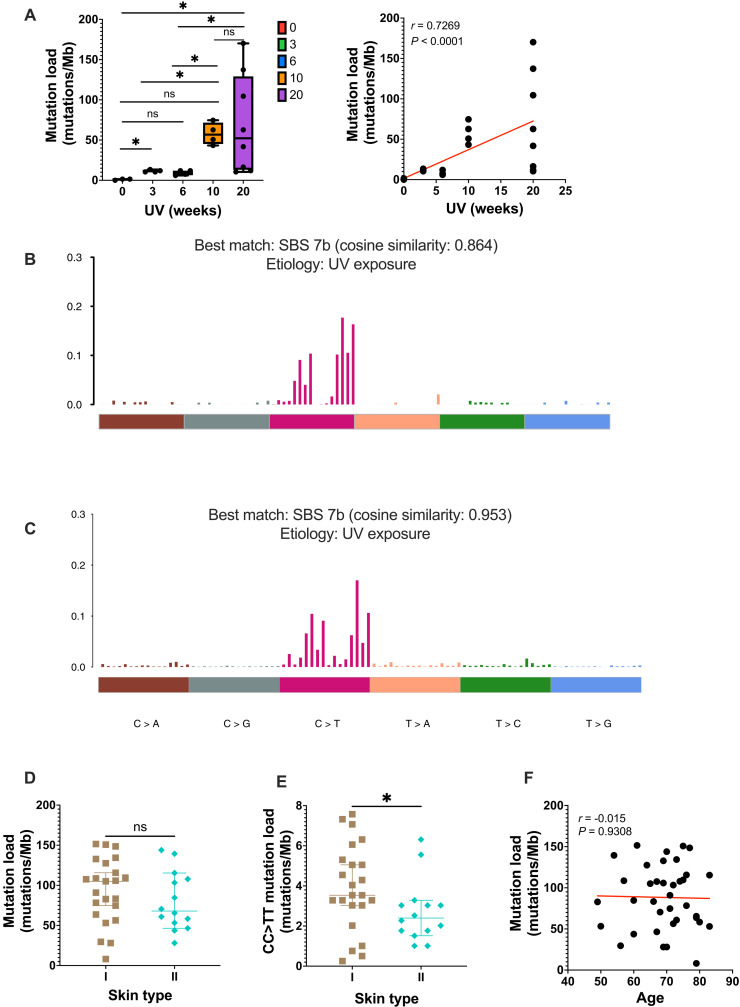
UVB is a direct mutagen of DNA, and accumulation of mutation is thought to be one of the key mechanisms in KC formation. To investigate whether skin mutations accumulate over time during chronic UV exposure, C57BL/6 mice were exposed to different duration/doses of suberythermal chronic broadband UVB irradiation (see irradiation spectrum in Materials and Methods) from baseline (n = 3), 3 (n = 4), 6 (n = 4), 10 (n = 4), and 20 (n = 8) weeks. Two-millimeter-diameter portions of the dorsal epidermis separated from the dermis were subjected to deep sequencing targeting 152 genes. We took extreme care to ensure that the panel of 152 genes was sequenced to saturation. This corresponded to an average coverage of 2625× (421 to 28,680) per gene (fig. S1, A and B). Somatic mutations were called and filtered using six syngeneic liver DNA. A total of 585 somatic mutations were identified in 23 samples with most of them being single-nucleotide variants (SNVs) (fig. S1C). We ensured that sequencing was performed at a depth to detect many mutations with a variant allele frequency (VAF) of >0.001 (fig. S1D). There was an average of 0.99 mutations/Mb in the unexposed group, 11.6 mutations/Mb in the 3-week samples, and 9.3 mutations/Mb in the 6-week samples. Epidermal mutation load increased substantially to 57.8 mutations/Mb at 10 weeks and 69.4 mutations/Mb at 20 weeks of UV exposure (Fig. 1A, left). This supports a near linear (r = 0.73, P < 0.0001, Spearman correlation) relationship between cumulative exposure time and mutation load in murine dorsal skin (Fig. 1A, right). We examined the mutational signature in the 23 samples combined and identified UV exposure signature single base substitution (SBS7b) (Fig. 1B) (19). Overall, these findings highlighted the accumulation of mutations in the epidermis secondary to cumulative UV exposure.
Fig. 1. Skin mutation load reflects ultraviolet (UV) exposure.
(A) Left: Mutation load of chronic ultraviolet B (UVB)–irradiated mice. Right: Spearman correlation of mutation load and cumulative UV exposure. Red line indicates relationship of the correlation. Baseline (n = 3), 3 (n = 4), 6 (n = 4), 10 (n = 4), and 20 (n = 8) weeks. (B) Trinucleotide spectrum for single base substitution (SBS) of all murine samples. (C) Trinucleotide spectrum for SBS of patient cohort. (D) Mutation load in patients with skin phototype I (n = 23) and II (n = 14). (E) CC > TT mutation load in patients with skin phototype I (n = 23) and II (n = 14). Each data point represents each individual. Mann-Whitney test. (F) Correlation between patient age and mutation load (Spearman correlation). Mb, megabase; ns, not significant. ns P > 0.05 and *P ≤ 0.05.
Skin mutational load in patients is not reflected by age
Previous studies have reported the presence of mutations in normal-looking skin, and some association with age of participants has been established (13). To assess the link between exposure, risk of skin cancer, and epidermal mutation load, we conducted an analysis on 37 individual participants. Each participant was assessed for their demographic data and their Fitzpatrick skin phototype (combining their skin, hair and eye color, and ability to tan). For consistency in a limited sample of the population, only patients with skin phototypes I and II were recruited. This corresponded to Caucasians with pale skin, green or blue eyes, red or blonde hair, freckles, and no ability (type I) or minimal ability (type II) to tan. A 2-mm punch biopsy was collected from the left dorsal forearm, a chronically sun-exposed area. Matching saliva samples of each individual were used as controls to filter out germline variants. The mean age of the 37 individuals was 69 (range, 49 to 83). Twenty-five were male, and 23 had skin phototype I. Similar to the murine samples, targeted sequencing of 352 genes was performed on the epidermal preparation of each sample. On average, the skin samples coverage reached 1261× for each participant, and each gene was sequenced to a depth of 1718× (fig. S2, A and B). Most variants were SNVs (fig. S2C). We ensured that sequencing was performed at a depth to detect many mutations with VAF of >0.001 (fig. S2D). This ensured that all epidermal clones within the 2-mm samples were sequenced to allow the identification of most of the mutations.
A total of 12,939 mutations were identified across the 37 individual samples, upon comparison to germline sequences of the same participant. Overall, across the 37 samples, a mutational burden of 88.3 mutations/Mb (8 to 151.5 mutations/Mb) was uncovered on the normal-looking (defined as skin with some degree of sun damage but no sign of tumor or precancerous lesion) dorsal forearm of these individuals living in a high-incidence region of the world. Expectedly, the specific mutational spectrum closely resembled COSMIC mutational signature SBS7b (cosine similarity 0.953) (19), which is linked to UV exposure (Fig. 1C). This clearly suggested the role of UV exposure in driving mutation accumulation in normal-looking skin. In sensitivity analysis, we also calculated the mutation burden per cell (10) to provide a more stringent method of accounting for mutations with low allele frequency. Of importance, mutation burden and mutation burden per cell were strongly correlated (r = 0.65, P < 0.0001, Spearman correlation) (fig. S3). When grouping participants by skin type, no difference was observed in mutation load between participants with a type I versus a type II skin phototype (Fig. 1D). The groups differed significantly when CC > TT mutations that are highly specific consequences of UV exposure were examined (3.5 versus 2.4, P = 0.046, Mann-Whitney test) (Fig. 1E). However, despite a trend, this was not reproduced when examining the average mutation per cell in both groups (fig. S4, A and B). We next considered age as a proxy for duration of sun exposure. There was no correlation between age and the mutation load (r = −0.015, P = 0.9308, Spearman correlation) (Fig. 1F and fig. S4C). Overall, our murine findings suggest that there is a linear accumulation of UV-induced mutations with time; however, in patients, lighter skin types were more prone to display CC > TT UV-induced mutations.
Mutant clones of keratinocyte increase in size over time
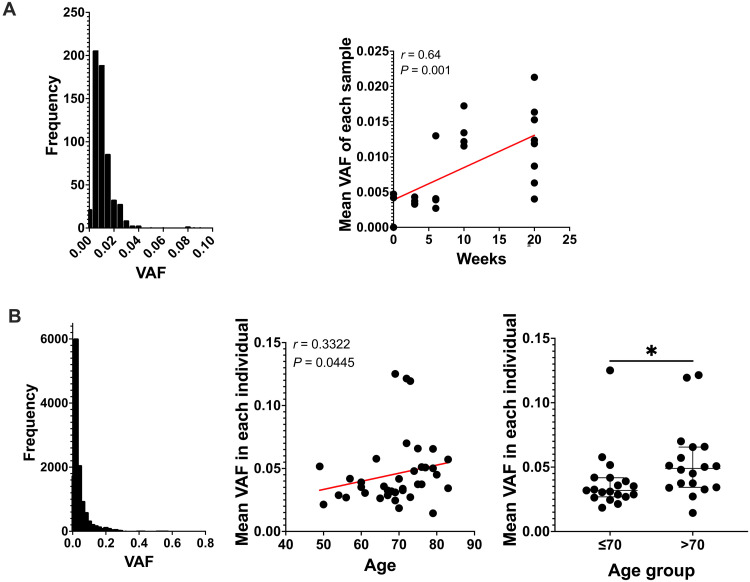
Previous studies have reported that clones of mutant cells and normal epithelial cells tend to increase in size over time (20, 21). Using the VAF in a standard 2-mm sample of the epidermis as a proxy for clone size, we examined the evolution of clone size over time in both murine and human samples. On the basis of previous reports of the number of basal keratinocytes in humans in the basal layer of the forearm skin (22) in a 2-mm punch biopsy, between 38,079 and 47,100 basal keratinocytes could be expected. Assuming that the distribution of clones resembles epidermal proliferative units of around 10 basal keratinocytes, a maximum of 3808 to 4710 clones were subjected to sequencing. Many groups have shown that epidermal clones are larger than what epidermal proliferation units predicted (23–25). Therefore, a mutation with a neutral effect would be expected at a minimum VAF of 0.0001 to 0.00013 [10/(38079 × 2)]. Given the smaller size of murine keratinocytes, one would expect a smaller VAF in individual clones in the absence of any other interfering factors. In our murine samples, the distribution of VAF for each mutation ranged from 0.0017 to 0.092 with an average of 0.011 (Fig. 2A). The average VAF of all mutations per sample increased over time (P = 0.001, Spearman correlation) (Fig. 2A). The average VAF in the nonexposed group was 0.0030. At 3 weeks, mutations had a mean VAF of 0.0037; at 6 weeks, 0.0059; and reached 0.014 and 0.012 at 10 and 20 weeks, corresponding to a fourfold increase compared to the unexposed group (Fig. 2A). Similarly, in human samples, the average VAF increased with age. The average VAF for all detected mutations was 0.044 and ranged from 0.0016 to 0.63 (Fig. 2B). When comparing individuals older than 70 to those who were younger, there was a significant increase in the VAF with age (0.032 versus 0.049, respectively, P = 0.014, Mann-Whitney test) (Fig. 2B). This was due in large part to more mutations with VAF > 0.1 in those older than 70 (fig. S5). These results showed that the clonal representation was substantially larger than predicted by the theoretical number of clones. This prompted us to examine the potential role of specific variants that would drive the observed increase in clone size.
Fig. 2. Clone size expands over time.
(A) Chronic exposed murine samples. Distribution of variant allele frequency (VAF) size (left). Spearman correlation of the ultraviolet (UV) dose/duration and mean VAF of each sample (right). Red line represents trend line of the correlation. (B) Patient cohort. Distribution of VAF size (left). Spearman correlation of age and mean VAF of each individual (middle). Red line represents trend line of the correlation. Distribution of mean VAF when grouping patient younger (n = 19) or older (n = 18) than 70 years old (right). Each dot represents each patient. Mann-Whitney test. *P ≤ 0.05.
Mutations in specific genes allow clone size expansion
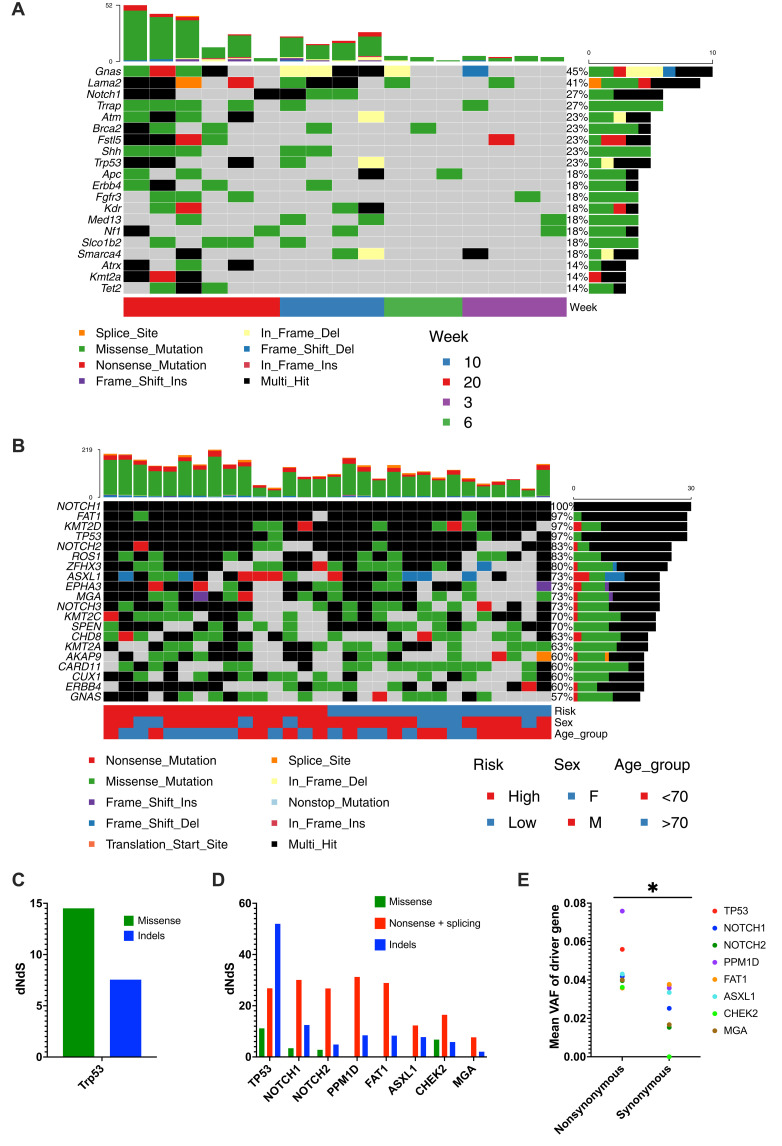
We examined murine and human samples for recurrent mutations (Fig. 3, A and B). Across human samples, NOTCH1, FAT1, TP53, KMT2D, and NOTCH2 were the most frequently mutated genes (Fig. 3B). Similarly, in murine samples, Trp53 and Notch1 could be identified most frequently among a more diverse group of mutated genes (Fig. 3A). To evaluate the impact of each gene in driving clone size more directly, we examined the ratio between silent (synonymous) and nonsynonymous mutations of the gene that would potentially affect its product function ratio of nonsynonymous to synonymous substitutions (dNdS). In murine samples, the only gene with a significant alteration between the synonymous and nonsynonymous variants was Trp53 (qglobal_cv = 0.0076; Fig. 3C). In human samples, eight driver genes (TP53, NOTCH1, NOTCH2, PPM1D, FAT1, ASXL1, CHEK2, and MGA) were identified using the dNdS analysis (Fig. 3D). TP53, NOTCH1, NOTCH2, PPMID, and FAT1 were the leading candidates. Each of these genes harbored a variety of mutation types of nonsynonymous nature with indels and nonsense mutations being the most frequent types. We further showed a significant increase in the average VAF of nonsynonymous versus synonymous mutations for these genes suggesting their role as drivers of clone size (P = 0.0156, Wilcoxon; Fig. 3E).
Fig. 3. Driver mutations in sun exposed skin.
(A and B) Waterfall plot of chronic ultraviolet (UV)–exposed murine samples (A) and patient cohort (B). Top shows mutation count of each sample. Right shows the percentage of samples with mutation in specific gene. (C and D) dNdS for missense, nonsense, and splicing as well as indels for genes under significant positive selection (global q < 0.01) in the chronic UV-exposed murine samples (C) and patient cohort (D). (E) Mean variant allele frequency (VAF) of nonsynonymous and synonymous mutations in each of the eight positive selected genes. Each dot represents each gene. Wilcoxon test. *P ≤ 0.05.
Mutation load is associated with risk of keratinocyte cancer
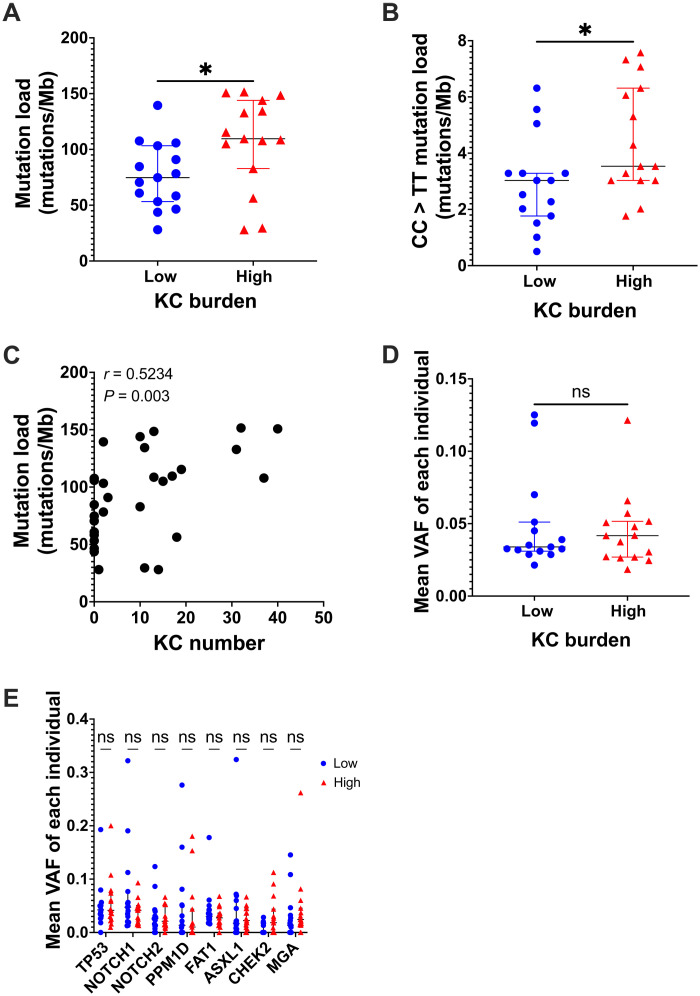
Despite the widespread presence of mutations in normal-looking skin, there has been uncertainty whether their presence or their load is meaningful in the development of KCs. To address this question, we conducted a case-control study among the 37 participants described above. We matched patients with a high burden of KCs (>10 KCs in the past 5 years) to those with a low burden (<3 KCs in the past 5 years). The 15 pairs of cases and controls were matched for age (±5 years) and sex. Comparing the mutation load in the normal-looking dorsal forearm skin of these individuals, we showed that those with a high KC burden had an elevated mutation load (109.6 versus 74.75, P = 0.0148, Mann-Whitney test) (Fig. 4A). This difference held true when only CC > TT mutations were considered (P = 0.0281, Mann-Whitney test) (Fig. 4B). Using mutational burden per cell, we observed similar trends for both comparisons (fig. S6, A and B). We then wondered whether mutation load was associated with cancer burden. The number of KCs in the past 5 years in each patient was counted. There was a positive correlation between the mutation load and the number of KCs (P = 0.003, r = 0.5234, Spearman correlation) (Fig. 4C). This was further validated by examining mutational burden per cell (P = 0.031, r = 0.40; fig. S6C).
Fig. 4. Mutation load is associated with risk of keratinocyte cancer.
(A and B) Mutation load (A) and CC > TT mutation load (B) in patient with low (n = 15) or high (n = 15) keratinocyte cancer burden. Mann-Whitney test. (C) Spearman correlation of mutation load and the number of keratinocyte cancers in the past 5 years. (D) Mean variant allele frequency (VAF) in patient with low or high keratinocyte cancer burden. Mann-Whitney test. (E) Mean VAF of each of the eight positive selected genes in low or high keratinocyte cancer burden patients. Each dot represents each individual. Mann-Whitney test. ns P > 0.05 and *P ≤ 0.05.
Furthermore, we asked whether VAF reflecting mutant clone size was associated with the KC risk. The mean VAF of all variants in each sample was compared between both groups. No difference was identified between the VAF as a proxy for clone size in the low- and high-risk group (P = 0.87, Mann-Whitney test; Fig. 4D). Last, when looking at the specific driver mutations described above, there was no difference in the VAF of the nonsynonymous mutations between the cases or controls (Fig. 4E).
Overall, our findings established an association between mutation load and KC burden. We therefore examined whether reducing this mutation load would reduce the burden of KCs.
Epidermal abrasion recruits cells from hair follicles replicating wound healing
As KC risk is associated with the skin mutation load, we reasoned that a procedure that lowers the epidermal mutation burden may be able to reduce KC occurrence. During wound healing, the hair follicle stem cells’ progeny migrates to the surface epidermis to repair the interfollicular epidermis (IFE) (26). Furthermore, it has been shown that mutation load gradually decreases from epidermis down to the bottom of the hair follicles (11). Therefore, we wondered whether dermabrasion, a technique that ablates the IFE, can induce a wound healing response to recruit cells from the deeper part of the hair follicles without resulting in substantial scarring. Hence, the procedure would allow replacement of the ablated, mutation-bearing epidermis with less UV-damaged hair-derived epidermal cells.
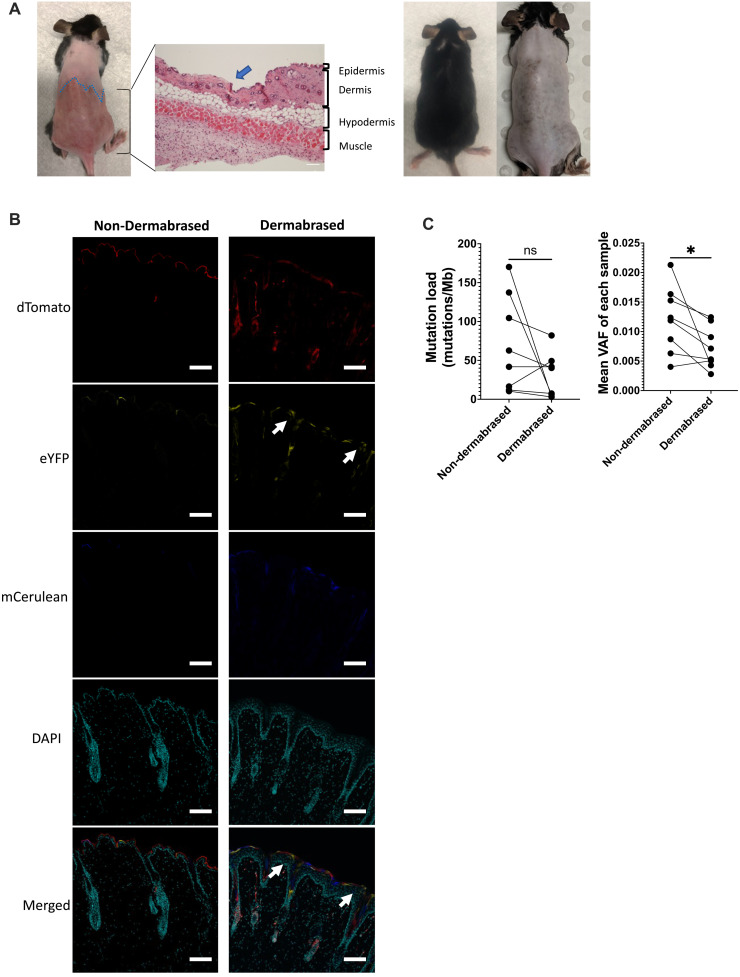
To test this idea, a multicolor lineage tracing Skinbow murine model (K14ERTCre x CAGSBOW) was used (23). Upon tamoxifen-induced Cre recombinase activation, keratin 14–expressing cells will adopt one of the six possible colors (23). Keratin 14 is expressed in the basal layer of epidermis and the hair follicles. Skinbow mice were injected with tamoxifen, and half of their dorsal epidermis was ablated on the next day using dermabrasion (Fig. 5A). We ensured that the dermabrasion technique removed the entire epidermis but did not affect the hair follicles nor induce fibrosis (Fig. 5A and fig. S7). Upon healing, dermabrased skin recovered all structure and components of normal skin. Immunofluorescence for keratin 14 and keratin 10 showed a similar pattern suggesting full reconstitution of the epidermis with proper differentiation (fig. S7). Last, trichrome or Sirius red staining did not demonstrate any increased level of fibrosis in the dermabrased area (fig. S7). Moreover, after dermabrasion, the entire treated area did not display any scars and was still harboring hair follicles suggesting their integrity (Fig. 5A and fig. S7). Skin samples from both dermabrased and non-dermabrased areas of Skinbow mice were collected 7 days after the procedure for confocal imaging. As expected, in the non-dermabrased skin area, groups of epidermal cells displaying the same color were either in the hair follicle or in the epidermis, suggesting that there were no clones from the hair follicles overlapping an area of the IFE. In contrast, in the dermabrased area, we found multiple clones overlapping the hair follicles and the skin surface, suggesting that hair follicle cells had in part restored the IFE that was initially ablated (Fig. 5B).
Fig. 5. Dermabrasion lowers mutation load and variant allele frequency (VAF).
(A) From left to right: Dorsal skin right after dermabrasion of the bottom half of the dorsal skin. Hematoxylin and eosin staining of the dorsal skin (blue arrow shows the junction of nondermabased and dermabrased skin). Picture showing dorsal skin was fully healed at 4 weeks after dermabrasion without scarring or damage to the hair follicles. (B) Skinbow mice (K14ERTCre × CAGSBOW) were injected with tamoxifen, and half of their dorsal epidermis was dermabrased the next day. One week after dermabrasion, skin from non-dermabrased and dermabrased areas were collected. Confocal images of skin sections from non-dermabrased and dermabrased areas were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). White arrows indicate the clones overlapping between the hair follicle and skin surface (n = 3). Scale bars, 50 μm. (C) Mutation load (left) and VAF (right) in non-dermabrased and dermabrased skin (n = 8). Wilcoxon test. ns P > 0.05 and *P ≤ 0.05.
Epidermal ablation reduces mutation load and clone size
We next investigated whether epidermal dermabrasion could reduce skin mutation load. C57BL/6 wild-type mice were exposed to a total of 20 weeks of chronic suberythermal UVB irradiation. The lower half of the dorsal epidermis was dermabrased after the first 10 weeks of UVB irradiation, whereas the upper half of the dorsal epidermis was left intact. At 20 weeks, animals were sacrificed, and the upper and lower portions of the back were sampled for targeted deep sequencing as described above. Notably, the average mutation load markedly reduced from 69.4 mutations/Mb in the non-dermabrased control area to 29.1 mutations/Mb in the dermabrased area (P = 0.1562, Wilcoxon) (Fig. 5C). This reduction was observed in six of eight mice, and only in two mice an increase in mutation load was observed. This was further confirmed when examining mutation burden per cell (P = 0.12; fig. S8). Furthermore, the average estimated VAF reflecting clone size reduced from 0.012 in the control non-dermabrased area to 0.0072 in the dermabrased area (P = 0.023, Wilcoxon) (Fig. 5C). More specifically, upon dermabrasion, there was also a reduction in oncogenic mutations in genes such as Trp53, Notch1, Kras, or Patch1 (fig. S9). Fewer oncogenic mutations in these genes could be identified, and when they were present, the VAF of these mutations tended to be in average lower (fig. S10, A to D).
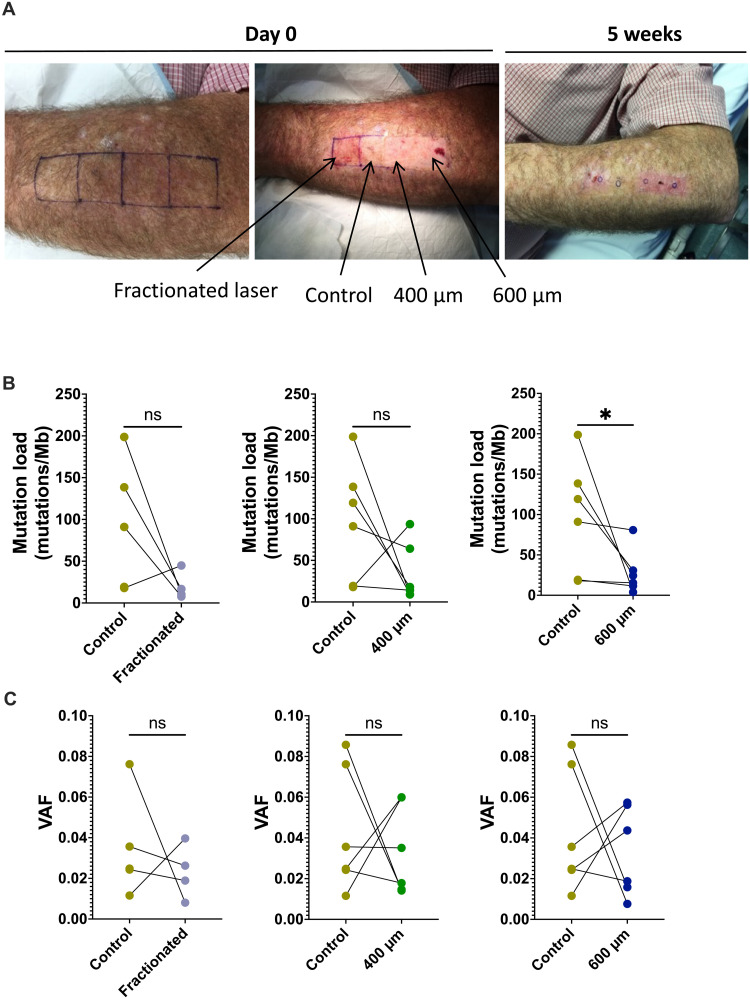
Laser ablation in patients reduces mutation load
To test the translational value of this procedure in reducing skin mutations in a clinical setting, we conducted a pilot clinical trial comparing the effectiveness of different epidermal ablative procedures versus control in reducing mutation load. Patients with a history of multiple epidermal cancers were subjected to epidermal ablation on 2 cm–by–2 cm areas of their dorsal forearms with three different modalities of Erbium Yag laser (600-μm-depth full ablation, 400-μm full ablation, and fractional 15% 200 J/cm2) versus a control area that was left untouched (Fig. 6A). Five weeks after the procedure, the participant’s skin was biopsied (2-mm punch), and the epidermis was recovered for sequencing. Deep targeted DNA sequencing of 152 cancer-related genes revealed 97.4mutations/Mb in the nonablated control epidermis compared to 27.7 mutations/Mb in 600 μm (n = 6), 34.6 mutation/Mb in 400 μm (n = 6), and 19.8 mutations/Mb in fractional laser (n = 4) ablated and regenerated epidermis (Fig. 6B). The difference between the control area was significant for the 600-μm laser ablation group (P = 0.0312, Wilcoxon), suggesting that all modalities of ablation resulted in a lower epidermal mutation load with the deeper module 600 μm reaching significance (similar trend with mutational burden per cell; fig. S11). There was no difference in the VAF in each treatment group compared to control (Fig. 6C). Together, these results suggest that epidermal ablation can effectively reduce skin mutational burden. Similar to our observation in mice, the reduction in mutational burden also affected our ability to detect oncogenic mutations in TP53, PTCH1, and SUFU, especially at 600 μm, and in fractional laser modalities (fig. S12). NOTCH1 mutations did not seem to reduce in numbers. However, all oncogenic mutations, even when identified, tended to have lower VAFs after 600-μm ablative laser treatment (nonsignificant; fig. S13, A to D).
Fig. 6. Laser ablation reduces mutation load in patients with high keratinocyte cancer burden.
(A) Representative images of the forearm that was treated with three different laser setting or modalities. (B and C) Mutation load (B) and variant allele frequency (VAF) (C) of control skin and the skin 5 weeks after either fractionated (n = 4), 400-μm (n = 6), or 600-μm laser treatment (n = 6). Wilcoxon test. ns P > 0.05 and *P ≤ 0.05.
Epidermal ablation lowers the number and size of cancers in a murine model of UV-induced BCC
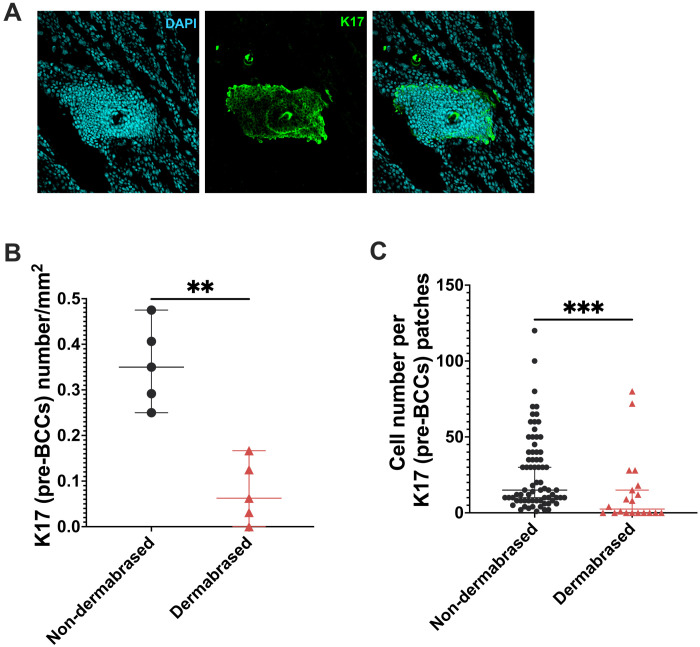
As skin mutation load was reduced after epidermal ablation and in murine models there seemed to be a reduction in clone size as reflected by VAF, we next wondered whether this procedure could reduce cancer burden. We used a mouse model of UV-induced BCCs (K14ERTCre × Ptch+/lox injected with tamoxifen) where a single copy of the Ptch1 gene is genetically ablated before UV irradiation to trigger the loss of the second copy. This is a well-established model of carcinogenesis based on a single pathway offering more consistent time courses than other models of UV-induced SCCs. Using this model, we performed the same dermabrasion experiment to examine the effect. The dorsal skin was UVB-irradiated for 10 weeks. The lower half of the dorsal epidermis was dermabrased and allowed to heal before further irradiation for another 10 weeks. At the term of the experiment, the dermabrased and control areas were recovered, and whole-mount immunostaining of the IFE with keratin 17 (K17) was used as an indicator of hedgehog activation in the IFE for early developing BCC lesions (called here pre-BCC lesions) (Fig. 7A) (8). Compared to the non-dermabrased site, dermabrasion reduced the number of K17+ pre-BCC lesions by fivefold (0.35 versus 0.06 pre-BCCs per mm2, P = 0.0079 Mann-Whitney) (Fig. 7B). There was also a significant reduction in the size of K17+ patches in the dermabrased site (15 versus 2.5 cells per BCC, P = 0.0006, Mann-Whitney) (Fig. 7C). These results suggest that epidermal ablation results in a reduction of cancer burden and size by lowering the skin mutation load.
Fig. 7. Epidermal abrasion lowers the numbers and size of basal cell carcinoma (BCC) precursors.
(A) Representative images of keratin 17 (K17) staining on whole-mount dorsal skin. Counterstained with 4′,6-diamidino-2-phenylindole (DAPI). (B) Quantification of the number of K17 patches in non-dermabrased and dermabrased skin (n = 5). Each dot represents an average number of patches from three images from each mouse. Mann-Whitney test. (C) Quantification of the size of K17 patches in non-dermabrased and dermabrased skin. Each dot represents a K17 patch. Mann-Whitney test. **P ≤ 0.01 and ***P ≤ 0.001.
DISCUSSION
It has become evident that normal skin that has been exposed to the sun carries mutations. This implies that accumulation of mutations might play an important role in skin field cancerization. By sequencing chronically sun-exposed epidermis from normal-looking forearm skin, we showed that both mutation load and allele frequency of individual mutations reflecting clone size increased in a near linear manner with cumulative UV exposure. We also showed that skin mutation load was associated with KC burden and correlated with the number of KCs, possibly reflecting one’s sun damage and KC risk. Epidermal mutation load could be reset to levels observed with lower UV exposure through ablation techniques such as dermabrasion or laser ablation and significantly reduced K17+ pre-BCC lesions in a mouse model of UV induced BCCs, which further reaffirmed the link between epidermal mutation load and KC burden.
Multiple studies have consistently found mutations in normal-looking skin, with a special focus on the same cancer-related genes (TP53, NOTCH1, and FAT1) that was previously identified by Martincorena et al. (10) with various mutation burden. The normal skin mutation load estimated by the current literature ranges from 3 to 132 mutations/Mb (10–13). The variation may be due to the different body site, punch size, the targeted panel of genes, and the choice of variant caller. Fowler et al. (11) showed that mutation load varied across five different body sites tested. The highest mutation load ~20 mutations/Mb was found on the forearm skin compared to that from the abdomen, head, leg, or trunk (11). In our study, we also focused on the forearm as a commonly sun-exposed site. There is no standardized panel for mutation load estimation. The panel in previous studies ranged between 0.0055 and 0.67 Mb and targeted 12 to 74 of the most commonly mutated genes in the skin (10–13). However, it has been shown that a panel of at least 1.1 Mb is required to estimate mutation load accurately especially for samples with 0 to 30 mutations/Mb (27, 28). Furthermore, the choice of variant calling tool and the use of blood, saliva, or dermal controls to filter germline variants also varied between studies. Although these studies gave important insight into skin mutation burden, they focused on smaller cohorts to demonstrate variability of mutation load within an individual, using panel size of up to 0.67 Mb that may affect reproducibility of mutation load estimation. To overcome some of these challenges, we used a panel of 3.96 Mb targeting 352 genes in the present study. A cohort of 37 individuals including a case-control series with matched age and sex was recruited in an attempt to eliminate the potential contributing factors to mutation burden. Saliva was also collected from everyone to filter germline variants to identify somatic variants more accurately.
Consistent with previous studies, TP53, NOTCH1, NOTCH2, and FAT1 were among the most mutated genes in our samples. In our cohort, an average of 88.3 mutations/Mb was identified in the normal skin compared to 20 mutations/Mb previously found in the forearm skin (11). The higher mutation load found in our study is likely due to the fact that we examined patients with skin cancers and that Australia, particularly Brisbane, has a two to four times higher level of UV irradiation compared to European countries and the United Kingdom where most of the aforementioned studies were carried out (29). In agreement with previous studies, we found that the VAF reflecting clone size in a standardized sample increased with the amount of UV exposure in mice or with age in patients. This may reflect the promotional effect of UV irradiation. It has been shown that some normal and mutated epidermal clones increase in size upon chronic UVB irradiation, and once the irradiation stops, these clones can regress in size (8, 30). In addition to the already identified driver genes (TP53, NOTCH1, NOTCH2, and FAT1), we found four driver genes (PPM1D, ASXL1, CHEK2, and MGA) that have not been reported before in this context. Notably, PPM1D and CHEK2 are involved in cell cycle regulation upon environmental stress through TP53 (31, 32). Of interest, the number of oncogenic driver genes and their VAF tended to be lower after epidermal ablative procedures.
Individuals with previous KCs are at high risk of developing multiple subsequent cancers in the field of sun exposure (2, 5). To reduce cancer burden, we tested a minimally invasive technique that removed the surface epidermis to reduce mutation load and cancer occurrence. We also showed that epidermal ablation promoted the contribution from deeper hair follicle cells to the surface epidermis. The lower mutation load after ablation supports the previous finding that cells deeper in the skin are better protected from UV irradiation and harbor fewer mutations (11). However, the reduced mutation burden and mutation burden per cell shown in our study could result also from a strong reduction in the size of mutant clones, making them undetectable within the sequencing depth of our study. Alternative possibilities also exist to explain the reduction in mutation load. It has been shown that fractional laser resurfacing reduced the size of p53 clones (33). Consistent with our finding of fewer BCCs after dermabrasion, a previous study has found that the number of KCs reduced from 24 on the control arm to 2 on the single laser–treated arm (34). Although our model addressed BCC carcinogenesis for simplicity, it is expected that addressing chronic sun damage may have more profound effects on SCCs and actinic keratoses given the stronger epidemiological link. In xeroderma pigmentosum, a genetic disease related to defective DNA repair in response to UV exposure, dermabrasion of the epidermis has been reported to protect from SCCs (35, 36). Overall, these findings strongly support the reduction of the mutation load as a potential mechanism for the effect of epidermal ablation on skin cancer occurrence.
In summary, our study sheds light on the association between mutation load and field cancerization and establishes epidermal mutation load as a potential target for therapy and indicator of risk. The findings in this study give insights into establishing a threshold in particular CC > TT mutation load to estimate cancer risk more objectively, opening avenues to guide and better evaluate future treatments of field cancerization.
MATERIALS AND METHODS
Study design
The objective of this study was to investigate the association of epidermal mutation load that can be used with KC burden and if it can be a target. For risk prediction cohort, patients aged 45 years or older with skin phototypes I and II were recruited during scheduled visits to the Dermatology Outpatients Department of the Princess Alexandra Hospital (PAH), Brisbane. Informed consent was obtained after the nature and possible consequences of the studies were explained. This project was approved by the Metro South Human Research Ethics Committee (HREC/17/QPAH/823). High-risk patients defined as >10 KCs in the past 5 years and low risk as <3 KCs in the past 5 years. Patients were excluded if they were on immunosuppressive therapy; if they had used field therapy such as topical fluorouracil, photodynamic therapy, ingenol or imiquimod on the areas considered for biopsy in the past 6 months; if they had an active bloodborne disease (e.g., HIV); or if they had a hereditary genetic condition favoring skin cancer formation. The use of nicotinamide or oral retinoids was permitted. A 2-mm-diameter biopsy was collected from an area devoid of skin cancer on the left dorsal forearms. Saliva samples were collected from everyone to remove germline mutations in the downstream analysis. The primary outcome was the difference in mutation load between high- and low-risk patients. A higher mutation burden was expected in high-risk patients.
For the epidermal ablation trial (a single-arm, open-label, pilot trial, registered as ACTRN12618001513202), high-risk patients were recruited during scheduled visits to the Dermatology Outpatients Department of the PAH, Brisbane. Inclusion criteria for the definition of high risk were the same as that in the risk prediction cohort. Eligible individuals were provided with project details as well as the potential risks and benefits of the procedure. Upon written informed consent, high-risk patients were subjected to a single session of epidermal ablation on 2 cm–by–2 cm areas of their dorsal forearms with three different modalities of Erbium Yag laser (600-μm depth full ablation, 400-μm full ablation, and fractional 15% 200 J/cm2) versus a control area that was left untreated. Five weeks after the procedure, a 2-mm-diameter biopsy was collected from each of the four areas on the dorsal forearms and matching saliva samples used to remove germline mutations in the downstream analysis. This project was approved by the Metro South Human Research Ethics Committee (HREC/17/QPAH/823).
Tamoxifen induction and UVB irradiation
For K14CreERT × Ptch+/lox mice, P24 mice were injected with 1 mg of tamoxifen intraperitoneally for five consecutive days to allow the activation of the Cre recombinase and the deletion of one allele of the Ptch1 gene (8, 37). Care of experimental animals was in accordance with institutional guidelines.
For UV treatment, six UVB broadband lamps (referred as UVB lamps) were used (TL 40W/12 RS SLV, Philips) as described previously by Roy et al. (8). The UV spectral emission of the setup can be found in fig. S14. The dorsal hair was shaved using a hair clipper. Mice were exposed to suberythemal dose of UVB (0.8 kJ/m2/min) three times a week for 0.5 min initially and then increased by 0.5 min every 2 weeks. Mice were either not exposed to UVB or they were exposed to 3, 6, 10, or 20 weeks of UVB irradiation. For K14CreERT × Ptch+/lox mice, triweekly UVB irradiation commenced 2 days after the last dose of tamoxifen injection as described above (8, 37).
Dermabrasion on murine dorsal skin
Dermabrasion was performed 2 days after the last UVB irradiation at 10 weeks. The day before dermabrasion, the shaved dorsal area was treated with Veet cream to remove the remaining hair from the skin surface. On the next day, the lower half of the back was dermabrased using a Scholl Velvet Smooth Electronic Foot Care System until the skin appeared shinny and red. Mice were then allowed to heal. After healing (~4 weeks), mice were subjected to 10 more weeks of UVB irradiation. Samples were collected 48 hours after the last UVB exposure. Tissue was fixed in 4% paraformaldehyde/phosphate-buffered saline (PBS) solution for 2 hours and transferred to 30% sucrose solution for storage.
Whole-mount immunofluorescence staining of K17
Whole-mount staining of K17 was completed as previously described (8). Briefly, whole-mount back-skin samples were permeabilized and blocked at 4°C overnight for 24 hours with 1× (PBS) + 0.5% Triton X-100, 2% bovine serum albumin and 20% normal goat serum (NGS), 1% dimethyl sulfoxide, and 100 mM maleic acid (pH at 7.5) before incubation with 1:50 (stock: 0.059 mg/ml) rabbit anti-K17 (ab109725, Abcam, Cambridge, UK) diluted in the blocking solution without NGS and left at 4°C for 24 hours. A secondary antibody conjugated with Alexa Fluor 647 (Invitrogen) was incubated at a 1:500 (stock: 2 μg/ml) dilution in blocking solution without serum, for 24 hours at 4°C. Nuclei were revealed using a 4′,6-diamidino-2-phenylindole (DAPI) counterstain at 0.2 μg/ml overnight at 4°C. Tissue was cleared in RapiClear for 3 to 6 hours (SunJin Lab Co., Taiwan). Samples were then mounted with fluorescent mounting media (Dako, Glostrup, Denmark) on single concave microscope slides (Sail Brand, China). All experiments were approved by the Animal Ethics Committee at the University of Queensland.
Epidermal DNA extraction for sequencing
Samples were snap-frozen for DNA sequencing. To extract epidermal DNA, the hypodermis was scrapped off using a scalpel. Skin was placed floating on deoxyribonuclease- and ribonuclease-free 5 mM EDTA solution with only the dermis in contact with the solution. After overnight incubation at 4°C, the epidermis was peeled from the dermis using sterile forceps. DNA was extracted from a 2-mm (diameter) epidermis using the Qiagen QIAamp DNA Micro Kit (560354) according to the manufacturer’s protocol. DNA was stored at −80°C until sequencing.
Library preparation and variant calling
The Sureselect ClearSeq Cancer Comprehensive panel (0.78 Mb) that targets 152 genes was used for the laser ablation trial. The same 152 genes were targeted in the mouse samples (tables S1 and S2). For the samples in the risk prediction trial, the Agilent SureSelect Community Design Glasglow Cancer plus panel (3.96 Mb) targeting 352 genes was used (tables S1 and S3). Input DNA was between 50 and 100 ng. Libraries were generated as per the manufacturer’s protocol and were sequenced in NovaSeq 6000 2 × 100 bp. The raw reads were trimmed using Agilent AGeNT trimmer v2.0.3 and fastp v0.23 (38). The resulting reads were aligned to GRCm38p6 (mm10) or GRCh38 (hg19) reference genome with BWA v0.7.17 (39). Duplicates were marked using Agilent Locatit. After removing the duplicates, indel realignment and base quality score recalibration were performed. Mutect2 GATK 4.1.9 was used to call variants (40). Variants were filtered using six liver samples for mouse samples, or they were filtered using matching saliva samples for human samples. Variants were annotated with Snpeff v5.0 (41). Dbsnp was annotated using Snpsift (41). Maftools was used for mutation signature analysis (42). Driver gene analysis was performed using dNdScv (43).
Statistical analysis
All analysis and graph construction were performed in Prism version 9. Briefly, the Mann-Whitney test was used to compare two independent groups, and the Wilcoxon test was used to compare two dependent groups. Spearman correlation was used to indicate the relationships between groups. Details of the statistical tests used are stated in the figure legends and in-text. A P value of <0.05 was considered significant.
Down-sampling analysis
Saturation analysis was performed by subsampling the reads with Picard DownsampleSam (40) to 0.1, 0.2, 0.5, 0.7, and 0.9 of the original number of reads, and the variant calling analysis was performed as described above.
Estimation of mutation load and mutation burden per cell
Mutation load was calculated by the number of mutations divided by the panel size. The average mutation burden per cell for a given sample was estimated using the following equation as previously described (10). Where LMb is the number of megabases sequenced with good coverage (>1000×). Per-base coverage was estimated using mosdepth (44). Samples with a good coverage region less than 1 Mb were excluded in the mutation burden per cell analysis for dermabrasion and laser ablation group.
Acknowledgments
We would like to thank the patients and their families for agreeing to be a part of this study. We wish to acknowledge the staff at the University of Queensland Institute of Molecular Biology sequencing facility for support on library preparation and sequencing generation. We would also like to thank the staff at the University of Queensland PACE animal house who helped facilitate the animal work in this study.
Funding: This work was supported by the National Health and Medical Research Council of Australia project grant 1145350 (to H.P.S., E.R., and K.K.), Advancing Queensland Clinical Research Fellowship (to K.K.), University of Queensland Graduate School Scholarship (to H.Y.W. and R.C.L.). Author contributions: Conceptualization: K.K. and E.R. Methodology: H.Y.W., R.C.L., S.C., S.K., E.R., M.F., K.K., and S.B. Investigation: H.Y.W., R.C.L., S.K., E.R., S.C. and M.F. Visualization: H.Y.W., R.C.L., V.M., S.K., and E.R. Supervision: E.R. and K.K. Writing—original draft: H.Y.W. and K.K. Writing—review and editing: R.C.L., V.M., H.P.S., and S.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Data were generated by the authors and deposited in European Nucleotide Archive PRJEB55817. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
Table S1 to S3
REFERENCES AND NOTES
- 1.T. W. Ridky, Nonmelanoma skin cancer. J. Am. Acad. Dermatol. 57, 484–501 (2007). [DOI] [PubMed] [Google Scholar]
- 2.L. G. Gordon, T. M. Elliott, C. M. Olsen, N. Pandeya, D. C. Whiteman, Multiplicity of skin cancers in Queensland and their cost burden to government and patients. Aust. N. Z. J. Public Health 42, 86–91 (2018). [DOI] [PubMed] [Google Scholar]
- 3.L. G. Gordon, D. Rowell, Health system costs of skin cancer and cost-effectiveness of skin cancer prevention and screening: A systematic review. Eur. J. Cancer Prev. 24, 141–149 (2015). [DOI] [PubMed] [Google Scholar]
- 4.D. Mitra, X. Luo, A. Morgan, J. Wang, M. P. Hoang, J. Lo, C. R. Guerrero, J. K. Lennerz, M. C. Mihm, J. A. Wargo, K. C. Robinson, S. P. Devi, J. C. Vanover, J. A. D'Orazio, M. McMahon, M. W. Bosenberg, K. M. Haigis, D. A. Haber, Y. Wang, D. E. Fisher, An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in the red hair/fair skin background. Nature 491, 449–453 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.N. Pandeya, C. M. Olsen, D. C. Whiteman, The incidence and multiplicity rates of keratinocyte cancers in Australia. Med. J. Aust. 207, 339–343 (2017). [DOI] [PubMed] [Google Scholar]
- 6.W. A. Bruls, H. Slaper, J. C. van der Leun, L. Berrens, Transmission of human epidermis and stratum corneum as a function of thickness in the ultraviolet and visible wavelengths. Photochem. Photobiol. 40, 485–494 (1984). [DOI] [PubMed] [Google Scholar]
- 7.D. E. Brash, UV signature mutations. Photochem. Photobiol. 91, 15–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.E. Roy, H. Y. Wong, R. Villani, T. Rouille, B. Salik, S. L. Sim, V. Murigneux, M. S. Stark, J. L. Fink, H. P. Soyer, G. Walker, J. G. Lyons, N. Saunders, K. Khosrotehrani, Regional variation in epidermal susceptibility to UV-induced carcinogenesis reflects proliferative activity of epidermal progenitors. Cell Rep. 31, 107702 (2020). [DOI] [PubMed] [Google Scholar]
- 9.P. H. Hart, M. A. Grimbaldeston, J. J. Finlay-Jones, Sunlight, immunosuppression and skin cancer: Role of histamine and mast cells. Clin. Exp. Pharmacol. Physiol. 28, 1–8 (2001). [DOI] [PubMed] [Google Scholar]
- 10.I. Martincorena, A. Roshan, M. Gerstung, P. Ellis, P. Van Loo, S. McLaren, D. C. Wedge, A. Fullam, L. B. Alexandrov, J. M. Tubio, L. Stebbings, A. Menzies, S. Widaa, M. R. Stratton, P. H. Jones, P. J. Campbell, Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.J. C. Fowler, C. King, C. Bryant, M. W. J. Hall, R. Sood, S. H. Ong, E. Earp, D. Fernandez-Antoran, J. Koeppel, S. C. Dentro, D. Shorthouse, A. Durrani, K. Fife, E. Rytina, D. Milne, A. Roshan, K. Mahububani, K. Saeb-Parsy, B. A. Hall, M. Gerstung, P. H. Jones, Selection of oncogenic mutant clones in normal human skin varies with body site. Cancer Discov. 11, 340–361 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.L. Wei, S. R. Christensen, M. E. Fitzgerald, J. Graham, N. D. Hutson, C. Zhang, Z. Huang, Q. Hu, F. Zhan, J. Xie, J. Zhang, S. Liu, E. Remenyik, E. Gellen, O. R. Colegio, M. Bax, J. Xu, H. Lin, W. J. Huss, B. A. Foster, G. Paragh, Ultradeep sequencing differentiates patterns of skin clonal mutations associated with sun-exposure status and skin cancer burden. Sci. Adv. 7, eabd7703 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.B. Hernando, M. Dietzen, G. Parra, M. Gil-Barrachina, G. Pitarch, L. Mahiques, F. Valcuende-Cavero, N. McGranahan, C. Martinez-Cadenas, The effect of age on the acquisition and selection of cancer driver mutations in sun-exposed normal skin. Ann. Oncol. 32, 412–421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A. C. White, K. Tran, J. Khuu, C. Dang, Y. Cui, S. W. Binder, W. E. Lowry, Defining the origins of Ras/p53-mediated squamous cell carcinoma. Proc. Natl. Acad. Sci. U.S.A. 108, 7425–7430 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.B. L. Van Duuren, A. Sivak, C. Katz, I. Seidman, S. Melchionne, The effect of aging and interval between primary and secondary treatment in two-stage carcinogenesis on mouse skin. Cancer Res. 35, 502–505 (1975). [PubMed] [Google Scholar]
- 16.G. Lapouge, K. K. Youssef, B. Vokaer, Y. Achouri, C. Michaux, P. A. Sotiropoulou, C. Blanpain, Identifying the cellular origin of squamous skin tumors. Proc. Natl. Acad. Sci. U.S.A. 108, 7431–7436 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.F. Blokzijl, J. de Ligt, M. Jager, V. Sasselli, S. Roerink, N. Sasaki, M. Huch, S. Boymans, E. Kuijk, P. Prins, I. J. Nijman, I. Martincorena, M. Mokry, C. L. Wiegerinck, S. Middendorp, T. Sato, G. Schwank, E. E. Nieuwenhuis, M. M. Verstegen, L. J. van der Laan, J. de Jonge, I. J. JN, R. G. Vries, M. van de Wetering, M. R. Stratton, H. Clevers, E. Cuppen, R. van Boxtel, Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.C. C. Laurie, C. A. Laurie, K. Rice, K. F. Doheny, L. R. Zelnick, C. P. McHugh, H. Ling, K. N. Hetrick, E. W. Pugh, C. Amos, Q. Wei, L. E. Wang, J. E. Lee, K. C. Barnes, N. N. Hansel, R. Mathias, D. Daley, T. H. Beaty, A. F. Scott, I. Ruczinski, R. B. Scharpf, L. J. Bierut, S. M. Hartz, M. T. Landi, N. D. Freedman, L. R. Goldin, D. Ginsburg, J. Li, K. C. Desch, S. S. Strom, W. J. Blot, L. B. Signorello, S. A. Ingles, S. J. Chanock, S. I. Berndt, L. Le Marchand, B. E. Henderson, K. R. Monroe, J. A. Heit, M. de Andrade, S. M. Armasu, C. Regnier, W. L. Lowe, M. G. Hayes, M. L. Marazita, E. Feingold, J. C. Murray, M. Melbye, B. Feenstra, J. H. Kang, J. L. Wiggs, G. P. Jarvik, A. N. McDavid, V. E. Seshan, D. B. Mirel, A. Crenshaw, N. Sharopova, A. Wise, J. Shen, D. R. Crosslin, D. M. Levine, X. Zheng, J. I. Udren, S. Bennett, S. C. Nelson, S. M. Gogarten, M. P. Conomos, P. Heagerty, T. Manolio, L. R. Pasquale, C. A. Haiman, N. Caporaso, B. S. Weir, Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat. Genet. 44, 642–650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.J. G. Tate, S. Bamford, H. C. Jubb, Z. Sondka, D. M. Beare, N. Bindal, H. Boutselakis, C. G. Cole, C. Creatore, E. Dawson, P. Fish, B. Harsha, C. Hathaway, S. C. Jupe, C. Y. Kok, K. Noble, L. Ponting, C. C. Ramshaw, C. E. Rye, H. E. Speedy, R. Stefancsik, S. L. Thompson, S. Wang, S. Ward, P. J. Campbell, S. A. Forbes, COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A. M. Klein, D. E. Brash, P. H. Jones, B. D. Simons, Stochastic fate of p53-mutant epidermal progenitor cells is tilted toward proliferation by UV B during preneoplasia. Proc. Natl. Acad. Sci. U.S.A. 107, 270–275 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.B. Colom, M. P. Alcolea, G. Piedrafita, M. W. J. Hall, A. Wabik, S. C. Dentro, J. C. Fowler, A. Herms, C. King, S. H. Ong, R. K. Sood, M. Gerstung, I. Martincorena, B. A. Hall, P. H. Jones, Spatial competition shapes the dynamic mutational landscape of normal esophageal epithelium. Nat. Genet. 52, 604–614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.P. R. Bergstresser, R. J. Pariser, J. R. Taylor, Counting and sizing of epidermal cells in normal human skin. J. Invest. Dermatol. 70, 280–284 (1978). [DOI] [PubMed] [Google Scholar]
- 23.E. Roy, Z. Neufeld, L. Cerone, H. Y. Wong, S. Hodgson, J. Livet, K. Khosrotehrani, Bimodal behaviour of interfollicular epidermal progenitors regulated by hair follicle position and cycling. EMBO J. 35, 2658–2670 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.G. Mascre, S. Dekoninck, B. Drogat, K. K. Youssef, S. Brohee, P. A. Sotiropoulou, B. D. Simons, C. Blanpain, Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 489, 257–262 (2012). [DOI] [PubMed] [Google Scholar]
- 25.E. Clayton, D. P. Doupe, A. M. Klein, D. J. Winton, B. D. Simons, P. H. Jones, A single type of progenitor cell maintains normal epidermis. Nature 446, 185–189 (2007). [DOI] [PubMed] [Google Scholar]
- 26.M. Ito, Y. Liu, Z. Yang, J. Nguyen, F. Liang, R. J. Morris, G. Cotsarelis, Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 11, 1351–1354 (2005). [DOI] [PubMed] [Google Scholar]
- 27.I. Buchhalter, E. Rempel, V. Endris, M. Allgauer, O. Neumann, A. L. Volckmar, M. Kirchner, J. Leichsenring, A. Lier, M. von Winterfeld, R. Penzel, P. Christopoulos, M. Thomas, S. Frohling, P. Schirmacher, J. Budczies, A. Stenzinger, Size matters: Dissecting key parameters for panel-based tumor mutational burden analysis. Int. J. Cancer 144, 848–858 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Z. R. Chalmers, C. F. Connelly, D. Fabrizio, L. Gay, S. M. Ali, R. Ennis, A. Schrock, B. Campbell, A. Shlien, J. Chmielecki, F. Huang, Y. He, J. Sun, U. Tabori, M. Kennedy, D. S. Lieber, S. Roels, J. White, G. A. Otto, J. S. Ross, L. Garraway, V. A. Miller, P. J. Stephens, G. M. Frampton, Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 9, 34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.P. Gies, C. Roy, J. Javorniczky, S. Henderson, L. Lemus-Deschamps, C. Driscoll, Global Solar UV Index: Australian measurements, forecasts and comparison with the UK. Photochem. Photobiol. 79, 32–39 (2004). [PubMed] [Google Scholar]
- 30.W. Zhang, E. Remenyik, D. Zelterman, D. E. Brash, N. M. Wikonkal, Escaping the stem cell compartment: Sustained UVB exposure allows p53-mutant keratinocytes to colonize adjacent epidermal proliferating units without incurring additional mutations. Proc. Natl. Acad. Sci. U.S.A. 98, 13948–13953 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.M. Magni, V. Ruscica, G. Buscemi, J. E. Kim, B. T. Nachimuthu, E. Fontanella, D. Delia, L. Zannini, Chk2 and REGγ-dependent DBC1 regulation in DNA damage induced apoptosis. Nucleic Acids Res. 42, 13150–13160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.M. Fiscella, H. Zhang, S. Fan, K. Sakaguchi, S. Shen, W. E. Mercer, G. F. Vande Woude, P. M. O'Connor, E. Appella, Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 94, 6048–6053 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.J. S. Orringer, T. M. Johnson, S. Kang, D. J. Karimipour, C. Hammerberg, T. Hamilton, J. J. Voorhees, G. J. Fisher, Effect of carbon dioxide laser resurfacing on epidermal p53 immunostaining in photodamaged skin. Arch. Dermatol. 140, 1073–1077 (2004). [DOI] [PubMed] [Google Scholar]
- 34.D. F. Spandau, R. Chen, J. J. Wargo, C. A. Rohan, D. Southern, A. Zhang, M. Loesch, J. Weyerbacher, S. S. Tholpady, D. A. Lewis, M. Kuhar, K. Y. Tsai, A. J. Castellanos, M. G. Kemp, M. Markey, E. Cates, A. R. Williams, C. Knisely, S. Bashir, R. Gabbard, R. Hoopes, J. B. Travers, Randomized controlled trial of fractionated laser resurfacing on aged skin as prophylaxis against actinic neoplasia. J. Clin. Investig. 131, e150972 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.J. Ocampo-Candiani, G. Silva-Siwady, L. Fernandez-Gutierrez, L. M. Field, Dermabrasion in xeroderma pigmentosum. Dermatol. Surg. 22, 575–577 (1996). [DOI] [PubMed] [Google Scholar]
- 36.A. Konig, H. C. Friederich, R. Hoffmann, R. Happle, Dermabrasion for the treatment of xeroderma pigmentosum. Arch. Dermatol. 134, 241–242 (1998). [DOI] [PubMed] [Google Scholar]
- 37.H. Y. Wong, K. Khosrotehrani, E. Roy, Whole-mount staining coupled to a UV-inducible basal cell carcinoma murine model. STAR Protoc. 2, 100329 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.S. Chen, Y. Zhou, Y. Chen, J. Gu, fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.H. Li, R. Durbin, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. A. DePristo, E. Banks, R. Poplin, K. V. Garimella, J. R. Maguire, C. Hartl, A. A. Philippakis, G. del Angel, M. A. Rivas, M. Hanna, A. McKenna, T. J. Fennell, A. M. Kernytsky, A. Y. Sivachenko, K. Cibulskis, S. B. Gabriel, D. Altshuler, M. J. Daly, A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.P. Cingolani, A. Platts, L. Wang le, M. Coon, T. Nguyen, L. Wang, S. J. Land, X. Lu, D. M. Ruden, A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.A. Mayakonda, D. C. Lin, Y. Assenov, C. Plass, H. P. Koeffler, Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 28, 1747–1756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.I. Martincorena, K. M. Raine, M. Gerstung, K. J. Dawson, K. Haase, P. Van Loo, H. Davies, M. R. Stratton, P. J. Campbell, Universal patterns of selection in cancer and somatic tissues. Cell 173, 1823 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.B. S. Pedersen, A. R. Quinlan, Mosdepth: Quick coverage calculation for genomes and exomes. Bioinformatics 34, 867–868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14
Table S1 to S3