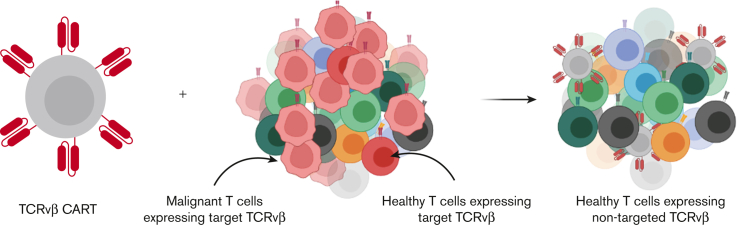
Key Points
-
•
TCRvβ-CARTs deplete antigen-expressing populations, including malignant clones, and spare nontargeted T cells in vitro and in vivo.
-
•
TCRvβ-CARTs exhibit limited fratricide and retain immune reactivity.
Visual Abstract
Abstract
Peripheral T-cell lymphomas (PTCLs) are a heterogeneous group of lymphoid malignancies associated with poor prognosis due to ineffective treatment options and high rates of relapse. The success of chimeric antigen receptor T-cell (CART) therapy for certain hematologic malignancies makes it an attractive treatment option for PTCLs. However, shared expression of potential target antigens by both malignant and healthy T cells poses a challenge. Current prospective CART approaches cause a high degree of on-target, off-tumor activity, resulting in fratricide during CART expansion, depletion of healthy T cells in vivo, and immune compromise in the patient. To limit off-tumor targeting, we sought to develop a CART platform specific for a given T-cell receptor vβ (TCRvβ) family that would endow CAR-modified T cells with the ability to mediate lysis of the clonal malignant population while preserving the majority of healthy T cells. Here, CAR constructs specific for multiple TCRvβ family members were designed and validated. Our results demonstrate that TCRvβ-family–specific CARTs (TCRvβ-CARTs) recognize and kill TCRvβ-expressing target cells. This includes specific self-depletion of the targeted cell subpopulation in the CART product and lysis of cell lines engineered to express a target TCRvβ family. Furthermore, TCRvβ-CARTs eliminated the dominant malignant TCRvβ clone in 2 patient samples. Finally, in immunodeficient mice, TCRvβ-CARTs eradicated malignant cells in a TCRvβ-dependent manner. Importantly, the nontargeted TCRvβ families were spared in all cases. Thus, TCRvβ-CART therapy provides a potential option for high-precision treatment of PTCL with limited healthy T-cell depletion.
Introduction
Peripheral T-cell lymphomas (PTCLs) are a heterogeneous group of aggressive cancers that account for 10% to 15% of non-Hodgkin lymphomas.1 Patients with PTCLs are primarily treated with chemotherapy regimens designed for treatment of B-cell lymphomas, with limited efficacy.1,2 By 7 months after diagnosis, half of the patients relapse or experience disease progression, and the median overall survival for these patients is 5.5 months.3 By 5 years after diagnosis, only 30% of the patients are progression free,1 highlighting the need for novel and improved therapies.
Cellular immunotherapies, particularly those involving chimeric antigen receptor T cells (CARTs), represent a promising treatment option for select blood cancers.4,5 CART therapies targeting CD19 or B-cell maturation antigen have demonstrated successful treatment of B-cell malignancies and myeloma, respectively, leading to US Food and Drug Administration approvals for multiple indications.6, 7, 8, 9, 10, 11, 12, 13 Based on these encouraging results, the opportunity to extend CART therapy for treatment of PTCLs is being explored.
Developing a T-cell platform to treat T-cell malignancies is challenging due to shared expression of lineage antigens on cancerous and healthy T cells, which comprise the immune repertoire of the patient and the CART product. One approach is to target surface markers with restricted expression, such as CD4, CD30, or CCR4.14, 15, 16 However, only certain patients with PTCL will benefit from this option, and targeting these molecules can deplete T-cell subsets that serve important functions (eg, CD4 helper T cells), compromising immune function. Other prospective CART platforms aim to treat most PTCL subsets by targeting common T-cell proteins, such as CD3, CD5, and CD7.17, 18, 19 Such approaches can cause substantial self-depletion, or fratricide, during ex vivo expansion and limit the ability to produce a viable product without further genetic modifications or using another immune cell type.19, 20, 21 Pan–T-cell–targeting CARTs can also mediate on-target, off-tumor killing of healthy T cells in patients, resulting in T-cell aplasia. Patients treated with CARTs targeting the pan–B-cell protein CD19 can experience profound B-cell aplasia, but this is considered a well-tolerated side effect resolved by steroid treatment and immunoglobulin replacement.22 However, there is no established equivalent treatment of T-cell aplasia, so these platforms only act as a bridge to stem cell transplantation.23 Current CART approaches are summarized in Table 1. To avoid complications while maintaining sufficient antitumor response, a more precise target limited to malignant T cells is needed.
Table 1.
CART approaches for T-cell malignancies
| Target antigen | Expression in malignant T cells, %∗ | Expression in healthy T cells, % | Predicted level of healthy T-cell depletion (%) | Notes | Representative reference |
|---|---|---|---|---|---|
| CD3 | ∼100 | 100 | Pan-depleting (100) | Pan–T-cell depleting fratricide as an issue; as a bridge to HSCT only | Chen et al40 |
| CD5 | ∼100 | 100 | Pan-depleting (100) | Pan–T-cell depleting fratricide as an issue; as a bridge to HSCT only | Mamonkin et al18 |
| CD7 | ∼100 | 100 | Pan-depleting (100) | Pan–T-cell depleting fratricide as an issue; as a bridge to HSCT only | Gomes-Silva et al19 |
| CD4 | Only in CD4+ T-ALL | 65 | Partial depletion (65) | Utility in CD4+ PTCLs only, elimination of all CD4+ cells; preservation of CD8+ T cells | Pinz et al14 |
| CCR4 | ∼50 | 50 | Partial depletion (50) | Incomplete cancer cell killing; partial depletion of healthy T cells | Perera et al16 |
| CD30 | 100 in the anaplastic large-cell lymphoma subtype; lower in other subtypes |
Expression in various leukocyte subsets | Partial depletion; eosinopenia noted in trial | Expressed in activated B and T cells and natural killer cells; low levels in activated monocytes and eosinophils; activity largely limited to anaplastic large-cell lymphoma subtype (∼13%) | Ramos et al15 |
| TRBC1 | 100 in ∼50 of cases; 0 in the other half | 50 | Partial depletion (50) | Utility in only ∼50% of PTCL cases; depletion of ∼50% of healthy T cells | Maciocia et al31 |
| T- cell receptor vβ family | ∼100 | ∼3-5 | Minimal depletion (∼3-5) | Minimal impact on healthy T cells; only limited fratricide; not an issue; no need for HSCT | This report |
HCST, hematopoietic stem cell transplant; T-ALL, T-cell acute lymphoblastic leukemia; TCR, T-cell receptor; TRBC1; TCRβ chain 1.
Represents expression in T-cell lymphomas bearing αβ TCRs (does not include less prevalent γδ or natural killer subtypes).
The TCR has become an attractive target for PTCLs, which, by nature of their clonal expansion from mature T cells, express a conserved TCR.24 In all, 95% of the TCRs comprise an α- and β-chain heterodimer (TCRα/β). The TCRβ chain is derived from a unique combination of variable, diverse, and joining segments assembled through DNA recombination and irreversibly selected during early T-cell maturation.25 The possible variable regions of the TCRβ chain are diversified into families referred to as TCRvβs.26, 27, 28 Malignant T-cell population in a patient with PTCL can be identified by a dominant TCR clone belonging to one of these TCRvβ families.24,29,30 Others have explored the possibility of targeting 1 of 2 constant regions of the TRBC1,31 but this approach leads to the elimination of half of the healthy T-cell repertoire. Because TRBC1 and TRBC2 T-cell populations differ in their differentiation status,32 this approach may also impede sufficient immune health. Alternatively, another group reported the ability to develop personalized CARs against the TCR complementarity-determining region 3.33 However, this is a resource- and time-intensive approach that may not be feasible for routine clinical care. Ultimately, the most successful approach for treating PTCLs will be specific enough to preserve most of the healthy T-cell repertoire but broad enough to make production practical. We therefore sought to determine whether CARTs specific for a TCRvβ family member will enable efficient killing of malignant T cells while preserving most of the healthy T-cell population as a high-precision treatment option for patients with PTCL.
Materials and methods
Design of TCRvβ-CAR constructs
Briefly, to generate TCRvβ-CARs, mouse antihuman TCRvβ antibody sequences were obtained from hybridoma cell lines (Table 2). Codon optimization of variable heavy- and light-chain antigen–binding elements was performed, and the single-chain variable fragments (scFvs) were designed in both heavy- and light-chain orientations, connected by a GGGS repeat domain. Each scFv was linked via CD8a hinge and transmembrane domains to intracellular activation domains derived from either CD28 or 4-1BB and CD3z. In some cases, constructs contained enhanced green fluorescent protein (eGFP) upstream of a T2A self-cleaving peptide as a marker for transduction. CAR constructs were cloned into a lentiviral vector via Nhe1 and BamH1 cut sites under the control of the EF-1α promoter and confirmed by Sanger sequencing.
Table 2.
Nomenclature of TCRvβ targets and antibodies used
| TCRvβ antibody | Gene name(s) | Clone | Tube; fluorochrome | Targets used in this report |
|---|---|---|---|---|
| TCRvβ 1 | TRBV9 | F; PE + FITC | ||
| TCRvβ 2 | TRBV20-1 | E; PE + FITC | ||
| TCRvβ 3 | TRBV28 | A; FITC | ||
| TCRvβ 4 | TRBV29-1 | P-LS-01 | H; PE + FITC | Healthy T cells (fratricide) |
| TCRvβ 5.1 | TRBV5-1 | P-LS-02 | C; PE + FITC |
Healthy T cells (fratricide); patient cells |
| TCRvβ 5.2 | TRBV5-6 | E; PE | ||
| TCRvβ 5.3 | TRBV5-5 | A; PE | ||
| TCRvβ 7.1 | TRBV4-1, 4-2, 4-3 | A; PE + FITC | ||
| TCRvβ 7.2 | TRBV4-3 | P-LS-03 | H; FITC | Healthy T cells (fratricide) |
| TCRvβ 8 | TRBV12-3, 12-4 | D; FITC | ||
| TCRvβ 9 | TRBV3-1 | P-LS-04 | B; PE |
Healthy T cells (fratricide); engineered cell line |
| TCRvβ 11 | TRBV25-1 | G; PE | ||
| TCRvβ 12 | TRBV10-3 | P-LS-05 | E; FITC |
Healthy T cells (fratricide); engineered cell line; patient cells |
| TCRvβ 13.1 | TRBV6-5, 6-6, 6-9 | D; PE | ||
| TCRvβ 13.2 | TRBV6-2 | P-LS-06 | H; PE | Healthy T cells (fratricide) |
| TCRvβ 13.6 | TRBV6-6 | D; PE + FITC | ||
| TCRvβ 14 | TRBV27 | G; FITC | ||
| TCRvβ 16 | TRBV14 | B; FITC | ||
| TCRvβ 17 | TRBV19 | B; PE + FITC | ||
| TCRvβ 18 | TRBV18 | C; PE | ||
| TCRvβ 20 | TRBV30 | C; FITC | ||
| TCRvβ 21.3 | TRBV11-2 | F; FITC | ||
| TCRvβ 22 | TRBV2 | P-LS-07 | G; PE + FITC | Healthy T cells |
| TCRvβ 23 | TRBV13 | F; PE |
Sequences derived from bolded antibodies were used in CAR design.
FITC, fluorescein isothiocyanate; PE, phycoerythrin; TRBV, T-cell receptor beta variable.
Generation of CAR-modified T cells
Healthy donor human T cells were purchased from the Human Immunology Core (University of Pennsylvania, Philadelphia, PA) and cultured at a density of 1 × 106 cells per mL in RPMI 1640 supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL of streptomycin at 37°C and 5% CO2. Unless specified, CD4 and CD8 T cells were cultured at a 1:1 ratio. The T cells were activated using anti-CD3/CD28 beads (Invitrogen) at a 3:1 bead-to-cell ratio. After 24 hours, the lentivirus was added at a multiplicity of infection of 5. Seven days after activation, beads were removed from culture via magnetic separation. T cells were maintained at a density of 0.5 × 106 to 1 × 106 cells per mL in RPMI 1640 supplemented with 100 IU/mL interleukin 2 (Prometheus Therapeutics and Diagnostics) for 5 days and then rested without additional interleukin 2 for 2 days before use in functional assays. T-cell count and volume were tracked using a Multisizer 3 (Beckman Coulter).
CART phenotype analysis
Expanded T cells were washed in phosphate-buffered saline (PBS) and resuspended in 100 μL staining master mix (1% fetal bovine serum and fluorescently labeled antibodies in PBS) for 30 minutes at 4°C in the dark. Cells were then washed 3 times and analyzed on a BD LSRFortessa cytometer. Compensation was performed on FlowJo version 10.8 using OneComp eBeads. Antibodies included CD3-BV605, protein L-biotin, streptavidin-APC, vβ12-PE, vβ5.1-PE, and vβ9-APC. LIVE/DEAD Fixable Aqua was used to determine live-cell populations. The Beckman Coulter IO Test Beta Mark kit was used to stain for 24 TCRvβ family members, according to the manufacturer’s protocol (Table 2). Fratricide was determined by comparing expression of an individual vβ family to that of nontransduced (NTD) T cells. Relative expression = 100 × (expressionCARTs − expressionNTD)/expressionNTD. For TCR repertoire sequencing, on day 14 (end of expansion) 0.2 × 106 TCRvβ-CART or NTD T cells were pelleted, resuspended in lysis buffer, and then bulk DNA sequencing was performed by the Human Immunology Core (University of Pennsylvania). Weighted, productive reads of TRBV genes were obtained and analyzed for relative change compared with NTD cells, using the aforementioned equation.
Patient cell preparation and phenotyping
Peripheral blood mononuclear cells from patients with T-cell malignancies were obtained from whole blood by Ficoll-Paque (GE Healthcare) gradient centrifugation. Cells were phenotyped using CD3-BV605, CD4-FITC, TCRαβ-APC, and CD8-APC/H7 antibodies using the aforementioned staining protocol. Dominant clones were identified using the Beckman Coulter IO Test Beta Mark kit. A sample of 200,000 cells of the patient sample with a Vb12-dominant clone was pelleted, placed in lysis buffer, and sent for bulk DNA TCR sequencing (Human Immunology Core, University of Pennsylvania) using a TCR sequencing kit from Adaptive Biotech.
Cell lines
The SupT1 cell line was maintained in culture using the same conditions as the aforementioned T cells. To generate TCR-expressing SupT1 cell lines, SupT1s were transduced with a retrovirus to express either MART-1-DMF4-TCR (vβ12) or HER2-TCR (vβ9) as previously described.34,35 Cells were transduced with lentivirus to express GFP/firefly luciferase. Cells were propagated and sorted by fluorescence-activated cell sorting (FACS) for those expressing both GFP and the designated TCRvβ. All cell lines were routinely tested for mycoplasma and surface expression of target antigens.
In vitro cytotoxicity assays
For cytotoxicity assays, CARTs were cocultured with target cells at various effector-to-target (E:T) ratios, defined as the ratio of CAR-expressing T cells to target T cells and time points as indicated in the text. Lysis of SupT1s was measured via bioluminescence, and lysis of patient tumor cells was measured by flow cytometry. Further details are provided in supplemental Methods.
Xenograft models and in vivo antitumor activity
NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice were engrafted IV with 3 × 106 SupT1s (CD4+CD8+)–expressing GFP/firefly luciferase and a TCRvβ family. After 3 days, mice were injected with 5 × 106 CARTs. Total T cells injected were equilibrated to account for differences in transduction efficiency. Tumors were monitored with bioluminescence imaging after intraperitoneal injection of 150 mg/kg D-luciferin in PBS. Living Image software (PerkinElmer) was used to visualize and calculate average radiance. At termination, mice were euthanized, and cells were collected and prepared for FACS analysis using antibodies CD3-BV605, CD4-FITC, CD8-APC-H7, TCRvβ12-PE, and TCRvβ9-APC. The brain contained SupT1s most consistently in preliminary results and was chosen for collection in final experiments (supplemental Figure 7A). Mice were bred, treated, and maintained in-house in accordance with the University of Pennsylvania Institutional Animal Care and Use Committee–approved protocols.
Statistical analysis
Data are reported as mean ± standard deviation unless specified. When more than 2 groups were compared, one- or two-way analysis of variance (ANOVA) tests were used, as appropriate, with Dunnett or Sídák test for multiple comparisons, respectively. Kaplan-Meier survival curves were generated. GraphPad Prism version 9.2.0 was used for statistical analysis. P < 0.05 was considered significant.
Results
Generation of TCRvβ-CARTs to detect PTCL malignant clones
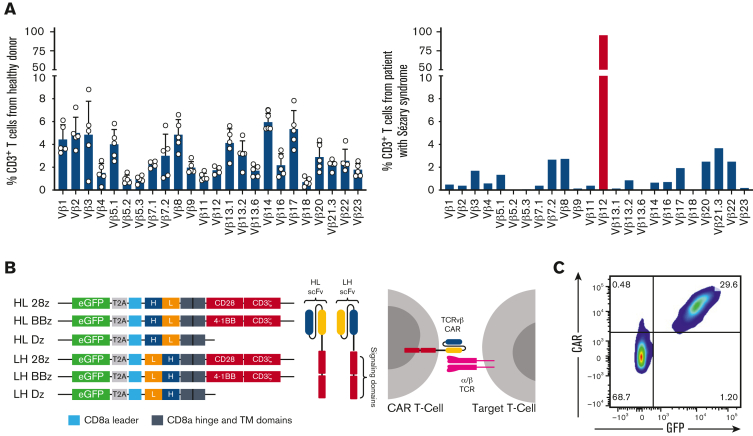
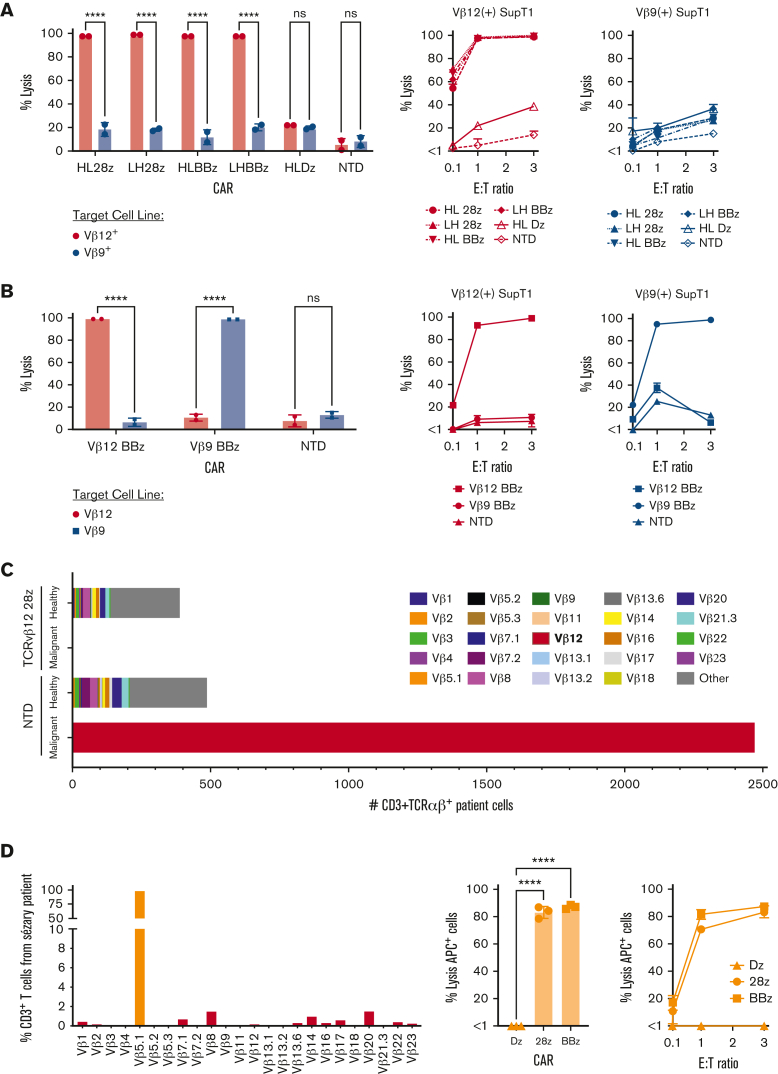
PTCLs are defined by the clonal expansion of malignant T cells and express a conserved TCR.24,29,30 As such, malignant cells can be identified as a dominant clone in 1 TCRvβ family. Peripheral blood lymphocytes from healthy donors and patients with Sézary syndrome were analyzed for expression of 24 TCRvβ families (supplemental Figure 1A). Although 1% to 10% of the total T cells from healthy donors expressed each TCRvβ family, the patient sample had a dominant TCRvβ12 clone representing 98% of the total T cells (Figure 1A). This was confirmed at the genomic level via bulk DNA sequencing (supplemental Figure 1B). Targeting this malignant clone would allow the healthy T-cell population, represented by all other TCRvβ families, to be spared and avoid immunosuppression in the clinical setting.
Figure 1.
Generation of TCRvβ-CARTs to detect PTCL malignant clones. (A) TCRvβ repertoire of CD3+ T cells from healthy donors (left) or a patient with Sézary syndrome (right), determined by flow cytometry. Each dot represents an individual healthy donor (n = 5). Malignant cells (red) can be identified as a dominant clone in 1 TCRvβ family. (B) Design of second-generation CARs including scFvs derived from the sequences of heavy (H) and light (L) chains of antibodies against TCRvβ families and intracellular signaling domains (Dz) (left). Expression of the CAR on the T-cell surface allows targeting of malignant cells expressing the TCRvβ family (right). (C) Representative plot of expanded normal donor T cells that were transduced with a TCRvβ12-targeting CAR. CAR expression was confirmed by staining with protein L and observing coexpression with the transduction marker, GFP, by flow cytometry.
To design CARs against dominant clones, we identified sequences for binding elements of antibodies specific for 7 distinct TCRvβ families, including TCRvβ4, 5.1, 7.2, 9, 12, 13.2, and 22, and constructed second-generation TCRvβ-targeting CARs (Figure 1B; Table 2). CARs devoid of intracellular signaling domains were designed as controls. T cells from multiple healthy donors were transduced with TCRvβ-CARs, expanded, and evaluated for CAR expression (Figure 1C; supplemental Figure 1C).
Engineering TCRvβ-CARs into healthy T cells results in target elimination
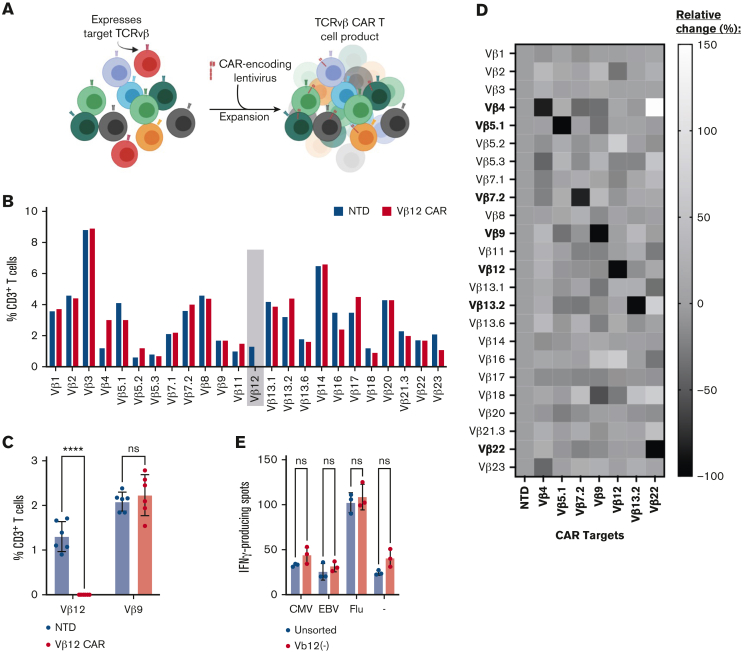
We produced CARTs using healthy donor cells containing 1% to 10% of each TCRvβ family and hypothesized that TCRvβ-targeting T cells would specifically deplete the target population via fratricide without impairing expansion kinetics (Figure 2A). Indeed, cell count and size were similar between TCRvβ-CAR and NTD T cells throughout expansion (supplemental Figure 2B-C). Following expansion, T cells were examined for surface expression of 24 distinct TCRvβs. As expected, TCRvβ12 was no longer detected in TCRvβ12-CARTs, but the 23 other TCRvβs were maintained, showing specific depletion of target cells (Figure 2B; supplemental Figure 2A). TCRvβ12-CARTs from 6 healthy donors were similarly analyzed and showed significant depletion of the target TCRvβ12 population while sparing the nontargeted TCRvβ9 population (Figure 2C). The data for all TCRvβ families were further represented as a relative change compared with NTD T cells and repeated for CARTs specific for TCRvβ4, 5.1, 7.2, 9, 13.2, and 22 (Figure 2D; supplemental Figure 2B-C). Each of these CART products exhibited specific depletion of their target TCRvβ family and, importantly, preservation of nontargeted populations. These data demonstrate the feasibility of creating a cadre of CARs that can be selected for tumor targeting depending on each patient’s malignant clone.
Figure 2.
Engineering TCRvβ CARs into healthy T cells results in target elimination. (A) Schematic demonstrating the elimination of a target TCRvβ family (red) within the bulk of healthy donor T cells after transduction with a TCRvβ-targeting CAR and expansion. (B) Representative TCRvβ repertoire of TCRvβ12-CARTs or NTD T cells after expansion determined by flow cytometry. Target population identified with gray box. (C) Targeted (TCRvβ12) and nontargeted (TCRvβ9) surface expression in TCRvβ12-CARTs and NTD T cells after expansion across multiple healthy donors determined by flow cytometry (n = 6) (∗∗∗∗P < .0001 by 2-way ANOVA with Sídák multiple comparison test). (D) Relative surface expression of TCRvβ family members compared with NTD control T cells after transduction with CARs targeting various TCRvβ families. (E) Representative interferon γ (IFN-γ) production by T cells with their full repertoire or sorted to remove the TCRvβ12 population, stimulated with peptides of common viruses, cytomegalovirus (CMV), Epstein-Barr virus (EBV), and influenza virus, and analyzed for production of IFN-γ by ELISpot (not significant [ns] by 2-way ANOVA with Sídák multiple comparison test).
T cells depleted of 1 TCRvβ family retain reactivity against common pathogens
Some CART approaches for PTCLs target pan–T-cell markers, which may cause T-cell aplasia, rendering the patient immunosuppressed. Other approaches, such as the TRBC1-CAR, are more selective; however, they still deplete a large component of the T-cell repertoire.32,36 We sought to determine whether redundancy in the T-cell repertoire would maintain an immune response against common viruses (CMV, EBV, and influenza virus) after a single TCRvβ family was depleted. T cells from 3 healthy donors depleted of populations belonging to TCRvβ5.1, 9, or 12 were exposed to viral peptides and analyzed for IFN-γ production (Figure 2E; supplemental Figure 3). Across all donors, there was no loss of reactivity or consistent difference in level of response against the viral peptides when compared with unsorted cells. Our results indicate that elimination of a single TCRvβ family had little or no impact on the virus-specific response mounted by the remaining T-cell repertoire.
“Epitope masking” occurs in healthy T cells expressing signaling-deficient CARs
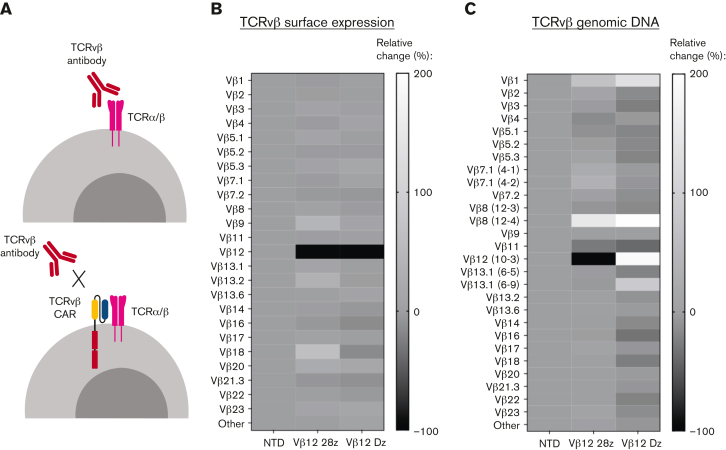
Thus far, our analysis of fratricide relies on surface detection of TCRvβs, which could potentially be affected by epitope masking by the CAR. Epitope masking may occur if a target cell is transduced with the CAR, which can bind to the TCR target on its own surface and block the epitope from detection antibodies (Figure 3A). Masking may be an issue in settings with patients with PTCL, where autologous T cells collected for CART production are contaminated by malignant cells that are not sufficiently separated. A single cell can cause relapse in a patient if masking occurs during the manufacturing process.37 In our CART products, we did not remove the target population before transduction, allowing us to explore the potential for epitope masking. We became especially interested in masking after we observed that T cells expressing the TCRvβ12-Dz CAR, despite lacking intracellular stimulatory domains, had no detectable TCRvβ12 population by flow cytometry (Figure 3B; supplemental Figure 4A). To investigate this phenomenon, we performed TCR sequencing of the TCRvβ12-CARTs to identify TCRvβ populations at the genetic level. Analysis of the TCR repertoire showed specific depletion of the TCRvβ12 (TRBV10-3) population in the TCRvβ12-28z CARTs but not in T cells expressing the truncated TCRvβ12-Dz CAR (Figure 3C; supplemental Figure 4B). Thus, substantial masking appears to occur only in the signaling-deficient CARTs. This indicates that our TCRvβ-CARs bind to the same epitope as the detection antibodies and that although masking is possible with TCRvβ scFvs, it is less likely an issue with functional CARs to be used in a clinical setting. Importantly, fratricide mediated by CARTs may in fact aid in the manufacturing process by removing any malignant cells remaining in the starting product. Similar studies on TCRvβ CARTs and other T-cell–target CARTs should be performed as they are further developed for clinical use. Other approaches to avoid potential contamination include using natural killer cells or allogeneic T cells as starting material, approaches that are currently under development by multiple groups.20,21,38,39
Figure 3.
“Epitope masking” occurs in healthy T cells expressing signaling-deficient CARs. (A) Schematic representing the proposed mechanism of “epitope masking” by the CAR on antigen-expressing cells. Relative change of TCRvβ surface expression (B) or genomic DNA (C) in healthy donor T cells engineered to express TCRvβ12-CARs with or without stimulatory domains compared with NTD T cells. All experiments were run with 2 donors. Data shown are from 1 representative donor.
TCRvβ-CARTs effectively and specifically lyse antigen-expressing cells in vitro
To test the functionality of TCRvβ-CARTs in vitro, we transduced SupT1s, which lack an endogenous TCR, to express previously validated TCRs that included either TCRvβ9 or TCRvβ12 (supplemental Figure 5A).34,35 Lysis of these cell lines was analyzed following overnight incubation with TCRvβ12-CARTs. TCRvβ12-CARs were able to mediate specific lysis of TCRvβ12-expressing SupT1s but not TCRvβ9-expressing SupT1s, achieving ∼100% lysis at 1:1 E:T ratios (Figure 4A). NTD and TCRvβ12-Dz–CARTs did not significantly lyse target cells. No significant difference was detected between costimulatory domains or orientation of the scFv. Analogous cocultures with TCRvβ12-BBz and TCRvβ9-BBz CARTs led to TCRvβ-specific lysis of their respective target cell lines with similar efficacy (Figure 4B).
Figure 4.
TCRvβ-CARTs effectively and specifically lyse target cells in vitro. (A) Cytotoxicity against SupT1s engineered to express a TCR containing either TCRvβ12 or TCRvβ9 by TCRvβ12-CARTs. Lysis was determined using a bioluminescence-based assay at various E:T ratios. Representative plots from 1 donor at 1:1 E:T ratio (left) or E:T ratios ranging from 0.1:1 to 3:1 (right) (∗∗∗∗P < .0001 by 2-way ANOVA with Sídák multiple comparison test). (B) Killing of engineered SupT1s by either TCRvβ12-BBz or TCRvβ9-BBz CARTs. Representative plots from 1 donor at 3:1 E:T ratio (left) or E:T ratios ranging from 0.1:1 to 3:1 (right) (∗∗∗∗P < .0001 by 2-way ANOVA with Sídák multiple comparison test). (C) Quantification of the number of patient cells expressing TCRvβ families remaining after coculture with either NTD T cells or TCRvβ12-28z–CARTs as identified by flow cytometry. TCRvβ12-expressing cells (red) represent the malignant clone. (D) Lysis against a patient sample identified to have a TCRvβ5.1 dominant clone (left) by T cells engineered to express CAR constructs targeting TCRvβ5.1. Representative plots at 3:1 E:T ratio (middle) or various E:T ratios (right) (∗∗∗∗P < .0001 by one-way ANOVA with Dunnett multiple comparison test). Data for all figures are representative of experiments repeated 3 times with CARTs derived from different healthy donors.
TCRvβ-CARTs lyse target patient cells and spare nontargeted T-cell populations
To further demonstrate the efficacy and specificity of TCRvβ12-CARTs, a sample from a patient with Sézary syndrome, containing a TCRvβ12 dominant clone was cocultured with TCRvβ12-CARTs, and cells expressing the 24 TCRvβ families were identified (supplemental Figure 5B). After 48 hours, the TCRvβ12-28z–CARTs had eliminated the malignant clone while sparing the other TCRvβ families. This reduction was not seen when patient samples were cultured with NTD T cells (Figure 4C). These findings validate the lytic function of TCRvβ12-CARTs against a patient sample and, importantly, support that this approach will spare nontargeted T cells.
For further validation of this approach, efficacy of TCRvβ-CARTs was tested against another sample from a patient with Sézary syndrome, identified by its TCRvβ5.1 dominant clone. This patient sample was obtained before treatment and contained an overwhelming TCRvβ5.1 fraction (Figure 4D; supplemental Figure 5C). This patient’s cells were cocultured with TCRvβ5.1-CARTs and analyzed for TCRvβ5.1 expression and cell count (supplemental Figure 5D). TCRvβ5.1-28z and -BBz CARTs significantly lysed target cells when compared with the truncated CARTs at 1:1 and 3:1 E:T ratios (Figure 4D).
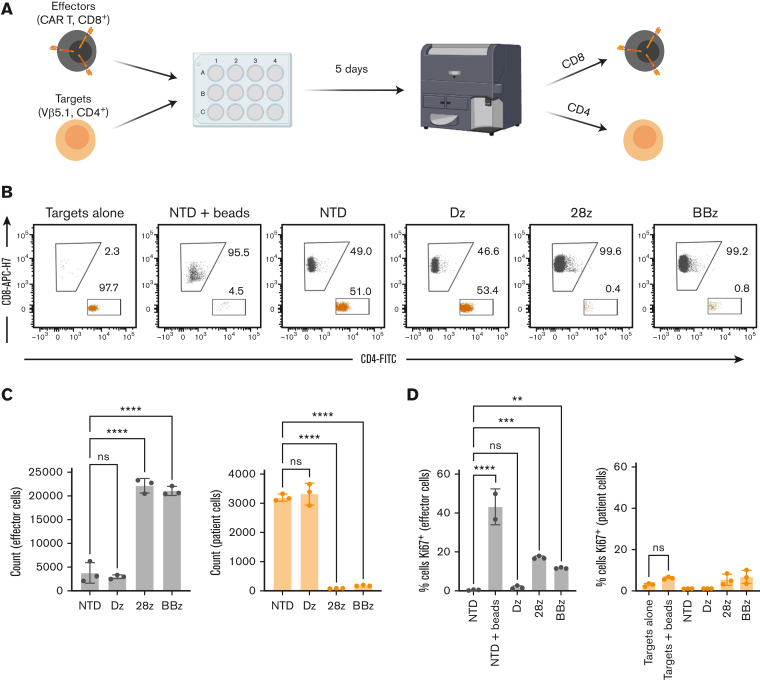
Long-term exposure to TCRvβ-CARTs does not result in bidirectional killing or proliferation of patient cancer cells
Although others have proposed targeting a component of the TCR to eliminate malignant T cells,31,33,40 there has been limited investigation into the potential complications arising from engaging their TCR. A group using bispecific antibodies to target TCRvβ families saw significant bidirectional killing between antigen(−) and antigen(+) T cells from healthy donors, which reduced the specificity of the treatment and caused elimination of effector cells.41 We observed an increase in the target population within expanded healthy T cells expressing the signaling-deficient CAR, which may be due to TCR engagement (Figure 3C; supplemental Figure 4B). However, this was not observed with the functional CAR. We hypothesized that the fast kinetics of CART killing along with the potential abnormality of TCR signaling in malignant cells would limit any unintended activity by malignant cells upon TCRvβ-CAR engagement.42
To investigate this, we performed long-term coculture experiments with the sample from the patient with Sézary syndrome, containing a TCRvβ5.1-dominant clone and TCRvβ5.1-CARTs (Figure 5A). After 5 days, there was significant lysis of target cells with both 28z and BBz CARTs, but target cell count was not impacted by, and was similar between, signaling-deficient CAR or NTD T cells, indicating no proliferation. No reduction of effector cells was observed in any condition when compared with the NTD control, indicating there was no bidirectional killing (Figure 5B-C). Moreover, no significant upregulation of Ki67 was observed in target cells after the coculture (Figure 5D; supplemental Figure 6A). This is particularly relevant in the signaling-deficient CAR condition, where the target cells’ TCR is engaged, but target cells are not killed. Notably, Ki67 was not upregulated in target cells stimulated with CD3/CD28 beads, indicating abnormal TCR signaling. Ki67 expression was upregulated in effector cells that were involved in killing as well as in T cells stimulated with CD3/CD28 beads. Similar results were seen with TCRvβ12-expressing patient cells (supplemental Figure 6B). In addition, when TCRvB12-CARTs were cocultured with healthy, sorted autologous TCRvB12+ T cells at low (1:10) E:T ratios, the number of effector cells was not significantly reduced when compared with untransduced cells, indicating that reciprocal lysis did not occur (supplemental Figure 6C). Together, these results suggest that TCR engagement by TCRvβ-CARTs does not lead to substantial bidirectional killing by or proliferation of patient malignant cells.
Figure 5.
Long-term exposure to TCRvβ-CARTs does not result in bidirectional killing or proliferation of patient cancer cells. (A) Schematic of the experimental setup. CD8+ effector cells were produced to be distinguished from the CD4+ target patient cells, and both populations were analyzed for cell count and Ki67 expression at the end of a 5-day coculture. (B) Dot plots of the target patient (orange) and effector CART (gray) cell populations after 5 days of coculture. (C) Quantified cell counts and (D) Ki67 staining of identified cells at the end of the coculture. Data are representative of experiments repeated 3 times with CARTs derived from different healthy donors (∗P < .0332, ∗∗P < .0021, ∗∗∗P < .0002, ∗∗∗∗P < .0001 by one-way ANOVA with Dunnett multiple comparison test). Data for all figures are representative of experiments repeated 3 times with CARTs derived from different healthy donors.
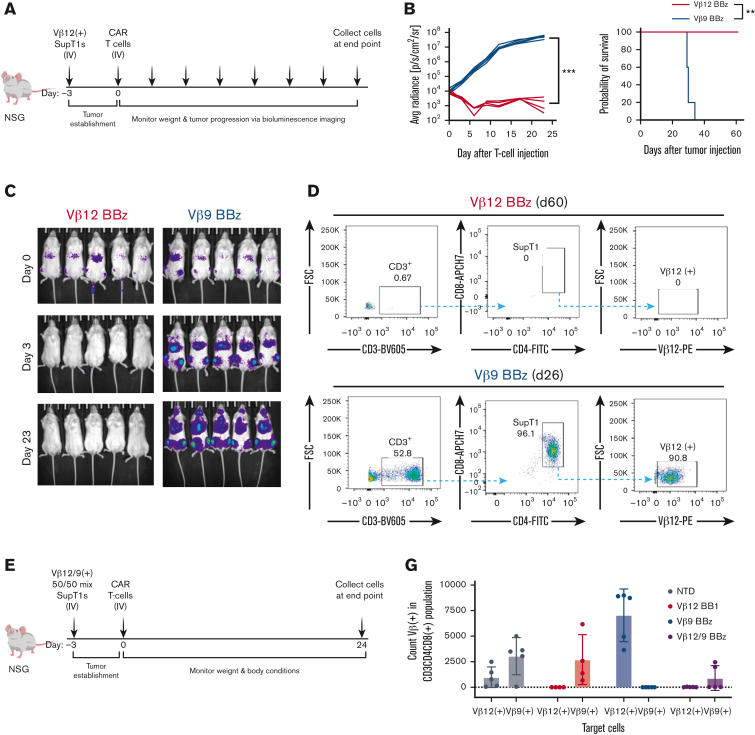
TCRvβ-CARTs specifically reduce tumor burden in vivo
After validating TCRvβ-CARTs function in vitro, we tested their abilities in a disseminated cancer mouse model. NSG mice were engrafted with TCRvβ12-expressing SupT1s and treated with either TCRvβ12- or TCRvβ9-CARTs (Figure 6A). Because 28z and BBz CARs had similar efficacy in vitro, BBz CARs were selected for in vivo studies owing to growing evidence that BBz CARTs have better persistence and lower risk of cytokine release syndrome and neurotoxicity in patients with hematologic malignancies.43,44 Three days after injection of the CARTs, tumor burden was significantly decreased in the treatment group. This reduction continued throughout the experiment and led to a significant survival benefit for the mice that received TCRvβ12-CARTs (Figure 6B-C). Organs collected from mice were processed and evaluated for TCRvβ12-expressing SupT1s. Target cells were found in the control group but were undetectable in the treatment group even after 60 days (Figure 6D; supplemental Figure 7B).
Figure 6.
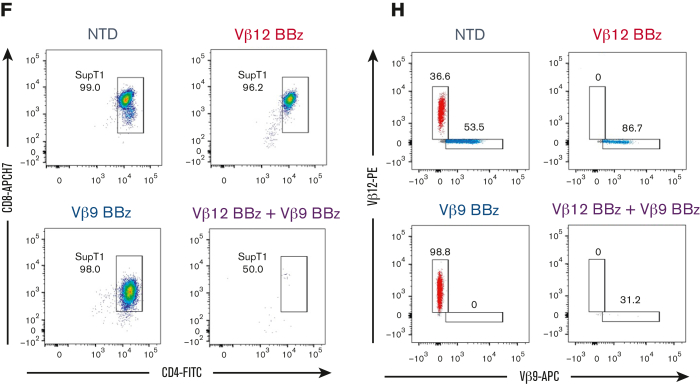
TCRvβ-CARTs specifically reduce tumor burden in vivo. (A) Experimental setup of the in vivo experiment with a homogenous target population. (B) Quantification of luminescence and survival of mice (n = 5 per group) (∗∗∗P = .0008 by mixed-effects analysis, ∗∗P = .0039 by Gehan-Breslow-Wilcoxon test). (C) In vivo imaging system imaging of tumor cells after treatment with TCRvβ12-CARTs (left) or TCRvβ9-CARTs (right) over the course of the experiment. (D) Representative dot plots of CD3+CD4+CD8+TCRvβ12+ SupT1s identified in brain tissue collected at the respective end points. (E) Experimental setup of the in vivo experiment with a heterogeneous target population. (F) Representative dot plots of CD8+CD4+ SupT1s identified in brain tissue collected 24 days after T-cell infusion. Each plot represents an individual mouse. (G) Quantified TCRvβ12- and TCRvβ9-expressing cells within the identified SupT1s in all mice measured by flow cytometry. (H) Representative plots of TCRvβ12 and TCRvβ9 expression of the identified SupT1s (n = 5 per group). APC, activated protein C; FSC, forward scatter.
To recapitulate a heterogeneous patient T-cell environment more closely, NSG mice were engrafted with a 50:50 mix of SupT1s expressing either TCRvβ12 or TCRvβ9 and then treated with CAR or NTD T cells (Figure 6E). Twenty-four days after T-cell infusion, organs were collected, and cells were analyzed by flow cytometry. CD4+CD8+ SupT1s were readily detected in mice receiving NTD, TCRvβ12-CARTs, and TCRvβ9-CARTs but were greatly reduced in the group that received both CARTs (Figure 6F), indicating their clearance. Within the bulk SupT1s, both TCRvβ12- and TCRvβ9-expressing populations were detected in the NTD group. By contrast, the TCRvβ12 or TCRvβ9 populations were specifically and deeply eliminated by their respective CARTs (Figure 6G-H; supplemental Figure 7C), leaving the nontargeted cells intact. Few to no TCRvβ12 or TCRvβ9 populations were detectable in mice treated with both CARTs. These data corroborate our in vitro data and establish that TCRvβ-CARTs can mediate TCRvβ-specific lysis of malignant T cells while sparing nontargeted T cells in vivo.
Finally, NSG mice engrafted with TCRvβ5.1-expressing patient cells were treated with TCRvβ-CARTs and evaluated for patient cell elimination. The results trended toward a decrease in the treatment group, but this was not statistically significant, likely due to the low number and variability of cells across groups (supplemental figure 8).
Discussion
CART therapy is a promising treatment option for hematologic malignancies, but there are significant difficulties adapting it for the treatment of PTCLs. Here, we show that selective targeting of dominant TCRvβ clones permits production of CART products without other genetic modifications that eliminate the malignant cells and preserve most of the healthy T-cell repertoire. The main limitation of this approach is the practicality of developing a complete library of TCRvβ CARs to cover all potential dominant clones that arise in patients. However, our work demonstrates the feasibility of this approach, as we were able to develop CARs against 7 TCRvβ families in this study. Continued expansion of this library is achievable owing to the availability of antibodies specific for 24 TCRvβ families. In addition, there is evidence of overrepresentation of certain TCRvβs in PTCL cases, including TCRvβ5.1, which made up almost half of the cases tested in 1 study.29 In another study of samples from a patient with Sézary syndrome, 11 families encompassed 82% of cases.45 Notably, our most deeply interrogated targets (TCRvβ5.1, TCRvβ9, and TCRvβ12) were among the highest represented. and CARs specific for these subfamilies are being prioritized for a planned umbrella clinical trial for patients with PTCL.
As an alternative approach to increase accessibility, universal immune receptors or CARs designed to bind to a tag conjugated to antigen-specific antibodies,46 can be investigated. In this way, only 1 CAR needs to be validated, and the existing TCRvβ antibodies could be repurposed into targeting moieties delivered on demand. A similar approach was adopted in a previous report that introduced the concept of targeting TCRvβ families with monoclonal antibodies that bound to a CD64 immune receptor on T cells.47 However, this approach requires repeat dosing of targeted antibodies as opposed to a single CART infusion. Similarly, TCRvβ bispecific antibodies were proposed in a recent study.41 These caused significant bidirectional killing that can be avoided by CARTs but may offer an additional “off-the-shelf” approach.
Manufacturing of autologous CARTs from patients with PTCL comes with considerable challenges. Specifically, with any CART approach there will be a scarcity of healthy T cells isolated among the dominant malignant population. This could be improved by frontline therapy that reduces the dominance of malignant cells before cell collection and CART therapy. Alternatively, allogeneic “off-the-shelf” CARTs can be created and implemented.48 If this method is standardized for the clinic, the TCRvβ-CAR approach would be attractive because manufactured products could be selected at will after identification of the dominant clone. Given the frequent urgency for effective therapy for patients with PTCL, off-the-shelf CARTs could be a preferable approach over the time needed to manufacture CARTs from autologous cells. Allogeneic T-cell products would also mitigate the need for cancer cell separation and risk of masking in CART products for T-cell malignancies. Still, our results indicate that fratricide may aid in the autologous setting, leading to selective depletion of any target cells in the starting T-cell material.
Based on the long-term B-cell aplasia often observed in patients treated with currently approved CD19-directed CARTs, it is possible that CARTs against a T-cell antigen could similarly prevent recovery of the targeted healthy T-cell population. Current proposed CAR approaches are prone to pan–T-cell aplasia or elimination of important T-cell subsets, leaving the patient susceptible to potentially life-threatening infections. One group aimed to circumvent T-cell aplasia by knocking out CD7 before transduction with a CD7-CAR for the treatment of T-ALL, reducing fratricide and allowing outgrowth of healthy CD7 T cells.19 This approach increases manufacturing obstacles, and the consequences of CD7 loss are not yet fully understood, but it offers a potential solution for patients with T-ALL. In this study, by targeting a single TCRvβ family, only a small portion of the TCR repertoire is eliminated, and aplasia is limited without further genetic modifications. Across all experiments, we observed that nontargeted T-cell populations were spared. Furthermore, we found that healthy T cells depleted of 1 TCRvβ family are still able to mount responses against common pathogens. Therefore, although TCRvβ-CARTs may persist and prevent recovery of the targeted TCRvβ family, severe immunosuppression necessitating stem cell transplantation is unlikely.
Although others have begun to investigate TCR-related targets, there is a lack of information on the risks of engaging the TCR on malignant T cells. Our study shows that when using TCRvβ-CARTs, there is no evidence of bidirectional killing or proliferation of malignant cells following TCR engagement. This may be due to disruption of TCR signaling during malignant transformation. In 1 report, 46% of the PTCL cases studied showed at least 1 mutation in the TCR or its downstream mediators, often leading to a constitutively activated phenotype.42 In these cases, engagement of the TCR may not result in TCR-mediated activity. However, further studies into the pathogenesis of PTCLs and their relationship to TCR signaling are needed.
In conclusion, targeting TCRvβ families by CARTs represents a promising approach to a precision treatment of T-cell malignancies that spares the T-cell repertoire. We demonstrated the ability to manufacture TCRvβ-specific CARTs and use them against malignant T cells in vitro and in vivo. In all cases, nontargeted, healthy T cells were spared. Therefore, this approach has the potential to overcome the current barriers to safe and effective CART therapy for T-cell malignancies.
Conflict-of-interest disclosure: L.C.S., S.J.S., and D.J.P. are listed as inventors on a patent related to this work and held by the University of Pennsylvania. The remaining authors declare no competing financial interests.
Acknowledgments
The authors thank the University of Pennsylvania Stem Cell and Xenograft Core and Human Immunology Core for their resources and expertise. The authors also thank Saar Gill for providing samples from patients with Sézary syndrome, Wenzhao Meng and Eline Luning-Prak for assistance with genomic sequencing, and Tatiana Blanchard for assistance with the ELISpot assay.
This work was supported by generous gifts from the Berman Fund and Laffery McHugh, as well as a grant from the National Institutes of Health (R01-EB-026892).
BioRender was used for schematic design in the figures of this report.
Authorship
Contribution: L.C.S. performed the experiments, analyzed the results, and created the figures; M.P. assisted with in vivo experiments; A.R.-G. created and maintained engineered SupT1 cells; N.G.M. provided assistance in experimental design and CAR construction; A.H.R. provided crucial samples from patients with Sézary syndrome; J.E. provided expertise in FACS; L.C.S., S.J.S., and D.J.P. conceived and designed the research; J.W., S.J.S., A.H.R., and D.J.P. provided clinical expertise and aided in obtaining patient samples; L.C.S. wrote the manuscript; and all authors read, edited, and approved the manuscript.
Footnotes
The sequencing data reported in this article have been deposited in the BioProject database (accession number PRJNA865733).
Data are available on request from the corresponding author, Daniel J. Powell Jr. (poda@pennmedicine.upenn.edu).
The full-text version of this article contains a data supplement.
Supplementary Material
References
- 1.Vose J, Armitage J, Weisenburger D, International T-Cell Lymphoma Project International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008;26(25):4124–4130. doi: 10.1200/JCO.2008.16.4558. [DOI] [PubMed] [Google Scholar]
- 2.Casulo C, O’Connor O, Shustov A, et al. T-Cell lymphoma: recent advances in characterization and new opportunities for treatment. J Natl Cancer Inst. 2016;109(2):djw248. doi: 10.1093/jnci/djw248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mak V, Hamm J, Chhanabhai M, et al. Survival of patients with peripheral T-cell lymphoma after first relapse or progression: spectrum of disease and rare long-term survivors. J Clin Oncol. 2013;31(16):1970–1976. doi: 10.1200/JCO.2012.44.7524. [DOI] [PubMed] [Google Scholar]
- 4.Sadelain M, Brentjens R, Rivière I. The promise and potential pitfalls of chimeric antigen receptors. Curr Opin Immunol. 2009;21(2):215–223. doi: 10.1016/j.coi.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.June CH, Blazar BR, Riley JL. Engineering lymphocyte subsets: tools, trials and tribulations. Nat Rev Immunol. 2009;9(10):704–716. doi: 10.1038/nri2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porter DL, Bagg A. Chimeric antigen receptor–modified T cells in chronic lymphoid leukemia. N Engl J Med. 2011;365(8):725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalos M, Levine BL, Porter DL, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3(95) doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grupp SA, Kalos M, Barrett D, et al. Chimeric antigen receptor–modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368(16):1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maude SL, Frey N, Shaw PA, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. 2014;371(16):1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuster SJ, Svoboda J, Chong EA, et al. Chimeric antigen receptor T cells in refractory B-cell lymphomas. N Engl J Med. 2017;377(26):2545–2554. doi: 10.1056/NEJMoa1708566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locke FL, Neelapu SS, Bartlett NL, et al. Phase 1 results of ZUMA-1: a multicenter study of KTE-C19 anti-CD19 CAR T cell therapy in refractory aggressive lymphoma. Mol Ther. 2017;25(1):285–295. doi: 10.1016/j.ymthe.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao W-H, Liu J, Wang B-Y, et al. A phase 1, open-label study of LCAR-B38M, a chimeric antigen receptor T cell therapy directed against B cell maturation antigen, in patients with relapsed or refractory multiple myeloma. J Hematol Oncol. 2018;11(1):141. doi: 10.1186/s13045-018-0681-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B-Y, Zhao W-H, Liu J, et al. Long-term follow-up of a phase 1, first-in-human open-label study of LCAR-B38M, a structurally differentiated chimeric antigen receptor T (CAR-T) cell therapy targeting B-cell maturation antigen (BCMA), in patients (pts) with relapsed/refractory multiple myeloma (RRMM) Blood. 2019;134(suppl 1):579. [Google Scholar]
- 14.Pinz K, Liu H, Golightly M, et al. Preclinical targeting of human T-cell malignancies using CD4-specific chimeric antigen receptor (CAR)-engineered T cells. Leukemia. 2016;30(3):701–707. doi: 10.1038/leu.2015.311. [DOI] [PubMed] [Google Scholar]
- 15.Ramos CA, Ballard B, Zhang H, et al. Clinical and immunological responses after CD30-specific chimeric antigen receptor-redirected lymphocytes. J Clin Invest. 2017;127(9):3462–3471. doi: 10.1172/JCI94306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perera LP, Zhang M, Nakagawa M, et al. Chimeric antigen receptor modified T cells that target chemokine receptor CCR4 as a therapeutic modality for T-cell malignancies. Am J Hematol. 2017;92(9):892–901. doi: 10.1002/ajh.24794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen KH, Wada M, Pinz KG, et al. Preclinical targeting of aggressive T-cell malignancies using anti-CD5 chimeric antigen receptor. Leukemia. 2017;31(10):2151–2160. doi: 10.1038/leu.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mamonkin M, Rouce RH, Tashiro H, Brenner MK. A T-cell-directed chimeric antigen receptor for the selective treatment of T-cell malignancies. Blood. 2015;126(8):983–992. doi: 10.1182/blood-2015-02-629527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomes-Silva D, Srinivasan M, Sharma S, et al. CD7-edited T cells expressing a CD7-specific CAR for the therapy of T-cell malignancies. Blood. 2017;130(3):285–296. doi: 10.1182/blood-2017-01-761320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper ML, Choi J, Staser K, et al. An ‘off-the-shelf’ fratricide-resistant CAR-T for the treatment of T cell hematologic malignancies. Leukemia. 2018;32(9):1970–1983. doi: 10.1038/s41375-018-0065-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinz KG, Yakaboski E, Jares A, et al. Targeting T-cell malignancies using anti-CD4 CAR NK-92 cells. Oncotarget. 2017;8(68):112783–112796. doi: 10.18632/oncotarget.22626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamers CH, Sleijfer S, van Steenbergen S, et al. Treatment of metastatic renal cell carcinoma with CAIX CAR-engineered T cells: clinical evaluation and management of on-target toxicity. Mol Ther. 2013;21(4):904–912. doi: 10.1038/mt.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scherer LD, Brenner MK, Mamonkin M. Chimeric antigen receptors for T-cell malignancies. Front Oncol. 2019;9:126. doi: 10.3389/fonc.2019.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown SD, Hapgood G, Steidl C, et al. Defining the clonality of peripheral T cell lymphomas using RNA-seq. Bioinformatics. 2017;33(8):1111–1115. doi: 10.1093/bioinformatics/btw810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis MM. Molecular genetics of the T cell-receptor beta chain. Annu Rev Immunol. 1985;3(1):537–560. doi: 10.1146/annurev.iy.03.040185.002541. [DOI] [PubMed] [Google Scholar]
- 26.Robinson MA. The human T cell receptor beta-chain gene complex contains at least 57 variable gene segments. Identification of six V beta genes in four new gene families. J Immunol. 1991;146(12):4392–4397. [PubMed] [Google Scholar]
- 27.Robinson MA, Mitchell MP, Wei S, et al. Organization of human T-cell receptor beta-chain genes: clusters of V beta genes are present on chromosomes 7 and 9. Proc Natl Acad Sci. 1993;90(6):2433–2437. doi: 10.1073/pnas.90.6.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.IMGT Repertoire (IG and TR) Immunogenetics database search engine. https://imgt.org/IMGTrepertoire/ Deposited 17 October 1997. Available at:
- 29.Vonderheid EC, Boselli CM, Conroy M, et al. Evidence for restricted Vbeta usage in the leukemic phase of cutaneous T cell lymphoma. J Invest Dermatol. 2005;124(3):651–661. doi: 10.1111/j.0022-202X.2004.23586.x. [DOI] [PubMed] [Google Scholar]
- 30.Potoczna N, Boehncke WH, Nestle FO, et al. T-cell receptor beta variable region (V beta) usage in cutaneous T-cell lymphomas (CTCL) in comparison to normal and eczematous skin. J Cutan Pathol. 1996;23(4):298–305. doi: 10.1111/j.1600-0560.1996.tb01301.x. [DOI] [PubMed] [Google Scholar]
- 31.Maciocia PM, Wawrzyniecka PA, Philip B, et al. Targeting the T cell receptor β-chain constant region for immunotherapy of T cell malignancies. Nat Med. 2017;23(12):1416–1423. doi: 10.1038/nm.4444. [DOI] [PubMed] [Google Scholar]
- 32.Kaewpreedee P, Boonrat P, Tansiri Y, Rowland-Jones SL, Hansasuta P. Dimorphism in the T-cell receptor constant region affects T-cell function, phenotype and HIV outcome. AIDS. 2019;33(9):1421–1429. doi: 10.1097/QAD.0000000000002187. [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Alexey S, Li J, et al. Unique CDR3 epitope targeting by CAR-T cells is a viable approach for treating T-cell malignancies. Leukemia. 2019;33(9):2315–2319. doi: 10.1038/s41375-019-0455-3. [DOI] [PubMed] [Google Scholar]
- 34.Johnson LA, Heemskerk B, Powell DJ, et al. Gene transfer of tumor-reactive TCR confers both high avidity and tumor reactivity to nonreactive peripheral blood mononuclear cells and tumor-infiltrating lymphocytes. J Immunol. 2006;177(9):6548–6559. doi: 10.4049/jimmunol.177.9.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanitis E, Smith JB, Dangaj D, et al. A human ErbB2-specific T-cell receptor confers potent antitumor effector functions in genetically engineered primary cytotoxic lymphocytes. Hum Gene Ther. 2014;25(8):730–739. doi: 10.1089/hum.2014.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikolich-Žugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. 2004;4(2):123–132. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- 37.Ruella M, Xu J, Barrett DM, et al. Induction of resistance to chimeric antigen receptor T cell therapy by transduction of a single leukemic B cell. Nat Med. 2018;24(10):1499–1503. doi: 10.1038/s41591-018-0201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salman H, Pinz KG, Wada M, et al. Preclinical targeting of human acute myeloid leukemia using CD4-specific chimeric antigen receptor (CAR) T cells and NK cells. J Cancer. 2019;10(18):4408–4419. doi: 10.7150/jca.28952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qasim W. Allogeneic CAR T cell therapies for leukemia. Am J Hematol. 2019;94(S1):S50–S54. doi: 10.1002/ajh.25399. [DOI] [PubMed] [Google Scholar]
- 40.Chen KH, Wada M, Firor AE, et al. Novel anti-CD3 chimeric antigen receptor targeting of aggressive T cell malignancies. Oncotarget. 2016;7(35):56219–56232. doi: 10.18632/oncotarget.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul S, Pearlman AH, Douglass J, et al. TCR β chain–directed bispecific antibodies for the treatment of T cell cancers. Sci Transl Med. 2021;13(584):eabd3595. doi: 10.1126/scitranslmed.abd3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schatz JH, Horwitz SM, Teruya-Feldstein J, et al. Targeted mutational profiling of peripheral T-cell lymphoma not otherwise specified highlights new mechanisms in a heterogeneous pathogenesis. Leukemia. 2015;29(1):237–241. doi: 10.1038/leu.2014.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Tao Z, Xu Y, et al. CD33-specific chimeric antigen receptor T cells with different co-stimulators showed potent anti-leukemia efficacy and different phenotype. Hum Gene Ther. 2018;29(5):626–639. doi: 10.1089/hum.2017.241. [DOI] [PubMed] [Google Scholar]
- 44.Ying Z, He T, Wang X, et al. Parallel Comparison of 4-1BB or CD28 Co-stimulated CD19-Targeted CAR-T Cells for B Cell Non-Hodgkin’s Lymphoma. Mol Ther Oncolytics. 2019;15:60–68. doi: 10.1016/j.omto.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Masson A, O’Malley JT, Elco CP, et al. High-throughput sequencing of the T cell receptor β gene identifies aggressive early-stage mycosis fungoides. Sci Transl Med. 2018;10(440) doi: 10.1126/scitranslmed.aar5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minutolo NG, Hollander EE, Powell DJ. The emergence of universal immune receptor T cell therapy for cancer. Front Oncol. 2019;9:176. doi: 10.3389/fonc.2019.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Urbanska K, Sharma P, et al. A novel approach for the treatment of T cell malignancies: targeting T cell receptor Vβ families. Vaccines. 2020;8(4):631. doi: 10.3390/vaccines8040631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poirot L, Philip B, Schiffer-Mannioui C, et al. Multiplex genome-edited T-cell manufacturing platform for “off-the-shelf” adoptive T-cell immunotherapies. Cancer Res. 2015;75(18):3853–3864. doi: 10.1158/0008-5472.CAN-14-3321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.