Abstract
RNA metabolism, broadly defined as the compendium of all processes that involve RNA, including transcription, processing and modification of transcripts, translation, RNA degradation and its regulation, is the central and most evolutionarily conserved part of cell physiology. A comprehensive, genome-wide census of all enzymatic and non-enzymatic protein domains involved in RNA metabolism was conducted by using sequence profile analysis and structural comparisons. Proteins related to RNA metabolism comprise from 3 to 11% of the complete protein repertoire in bacteria, archaea and eukaryotes, with the greatest fraction seen in parasitic bacteria with small genomes. Approximately one-half of protein domains involved in RNA metabolism are present in most, if not all, species from all three primary kingdoms and are traceable to the last universal common ancestor (LUCA). The principal features of LUCA’s RNA metabolism system were reconstructed by parsimony-based evolutionary analysis of all relevant groups of orthologous proteins. This reconstruction shows that LUCA possessed not only the basal translation system, but also the principal forms of RNA modification, such as methylation, pseudouridylation and thiouridylation, as well as simple mechanisms for polyadenylation and RNA degradation. Some of these ancient domains form paralogous groups whose evolution can be traced back in time beyond LUCA, towards low-specificity proteins, which probably functioned as cofactors for ribozymes within the RNA world framework. The main lineage-specific innovations of RNA metabolism systems were identified. The most notable phase of innovation in RNA metabolism coincides with the advent of eukaryotes and was brought about by the merge of the archaeal and bacterial systems via mitochondrial endosymbiosis, but also involved emergence of several new, eukaryote-specific RNA-binding domains. Subsequent, vast expansions of these domains mark the origin of alternative splicing in animals and probably in plants. In addition to the reconstruction of the evolutionary history of RNA metabolism, this analysis produced numerous functional predictions, e.g. of previously undetected enzymes of RNA modification.
INTRODUCTION
All cells synthesize a vast array of RNAs, using DNA or RNA templates, through a nucleoside polymerization reaction catalyzed by RNA polymerases (1). The mRNAs are templates for the ribosomal synthesis of proteins. Ribosomal RNAs are central to the functions of the ribosome, such as recognition and positioning of the mRNA and formation of the peptide bond during protein synthesis, whereas tRNAs are adaptors that deliver aminoacyl units to the site of protein synthesis and read the genetic code during translation through complementary pairing with codons in mRNA. In addition to these ubiquitous RNAs that are embedded in the Central Dogma of molecular biology (2), there is a plethora of other RNAs whose occurrence ranges from universality to a presence in only one of the terminal lineages of life. These include, among others, the ubiquitous signal recognition particle RNA involved in secretion, the nearly universal RNase P ribozyme, the small guide RNAs of eukaryotes and archaea that participate in processing and modification events to produce mature mRNAs, rRNAs and tRNAs, the bacterial tmRNA involved in protein degradation, the telomerase RNA from eukaryotes that acts as the template for the synthesis of chromosomal termini, the guide RNAs of trypanosomes involved in RNA editing, the small temporal (st) RNA, such as Lin-4, implicated in post-transcriptional regulation in animals, and the animal RoX1/2 and XIST RNAs, which contribute to chromosomal organization (1,3–8). From the time a RNA chain is elongated by the RNA polymerase to its ultimate destruction by ribonucleases, it undergoes interactions with numerous proteins that either form a variety of ribonucleoprotein (RNP) complexes or catalyze various reactions that modify the RNA’s composition or structure. This complex set of processes centered around RNA–protein and RNA–RNA interactions constitutes what can be termed ‘RNA metabolism’.
Thus defined, RNA metabolism is an integral part of the basal processes of molecular biology, namely transcription, translation and secretion, as well as numerous other cellular systems that employ RNAs in various capacities. The diversity of functional contexts notwithstanding, a number of computational analyses of proteins involved in RNA metabolism have brought out several unifying themes in the form of domains that bind RNA molecules and/or catalyze reactions of RNA processing and modification across these different contexts. This justifies the above definition of RNA metabolism and calls for a synthetic treatment of the cellular processes that involve RNA. Several previous computational analyses have considered specific aspects of RNA metabolism and concentrated on the identification of previously undetected domains in proteins involved in these processes (9–23). The results from such studies cast light on the early evolution of life, the last universal common ancestor (LUCA), the events surrounding the divergence of the major lineages of life, and potentially even the transition from the ancient RNA world to the modern-type, protein-dominated cellular systems.
In order to obtain a comprehensive view of the evolution of RNA metabolism, we conducted a large-scale computational analysis of the proteins involved in RNA metabolism. This analysis was chiefly based on detection of statistically significant similarities through sequence and structure comparisons, determination of orthologous and paralogous relationships between proteins, and utilization of contextual information derived from domain fusions, operon organization and phyletic patterns. This allowed us to define the major transitions and relative temporal order in the evolution of the principal branches of RNA metabolism and to gain some insights into the earliest phases of life’s evolution. Using the parsimony principle, we reconstructed the probable repertoire of genes and functions related to RNA metabolism that were present in LUCA. The analysis also enabled systematization of the vast amounts of information on RNA metabolism that have become available through genome sequencing, and produced structural and functional predictions that might facilitate further experimental studies on RNA metabolism.
MATERIALS AND METHODS
Eighteen complete bacterial genomes [Aquifex aeolicus (Aae), Bacillus subtilis (Bs), Borrelia burgdorferi (Bb), Campylobacter jejuni (Cj), Chlamydia trachomatis (Ct), Deinococcus radiodurans (Dr), Escherichia coli (Ec), Haemophilus influenzae (Hi), Helicobacter pylori (Hp), Mycobacterium tuberculosis (Mt), Neisseria meningitides (Nm), Pseudomonas aeruginosa (Pa), Rickettsia prowazekii (Rp), Synechocystis PCC6803 (Ssp), Thermotoga maritima (Tm), Treponema pallidum (Tp), Ureaplasma urealyticum (Uu) and Xylella fastidiosa (Xf)], seven complete archaeal genomes [Aeropyrum pernix (Ap), Archaeoglobus fulgidus (Af), Halobacterium sp. NRC-1 (Hsp), Methanobacterium thermoautotrophicum (Mta), Methanococcus jannashii (Mj), Pyrococcus horikoshii (Ph) and Thermoplasma acidophilum (Ta)] and six (nearly) complete eukaryotic genomes [Arabidopsis thaliana (At), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Homo sapiens (Hs), Saccharomyces cerevisiae (Sc) and Schizosaccharomyces pombe (Sp)] were investigated. The sequence data for all genomes were obtained using the Genome Division of the Entrez system at the National Center for Biotechnology (NCBI) (http://www.ncbi.nlm.nih.gov/Entrez/Genome/main_genomes.html).
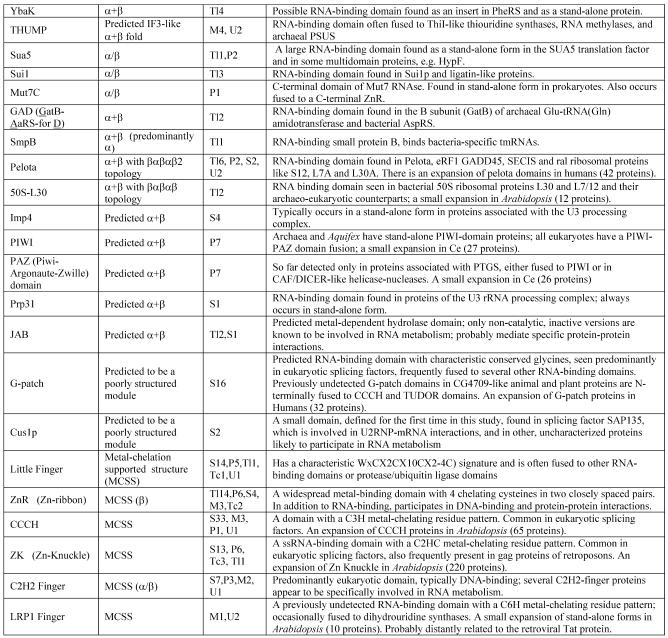
The domains listed in Table 1 were included in this study. A set of representative sequences was chosen for each domain, and position-specific scoring matrices (PSSMs or profiles) were generated by running the PSI-BLAST program (24–26) against the non-redundant protein sequence database at the NCBI, with the expectation (E) value of 0.01 typically used as the cut-off for including sequences into the profile. PSI-BLAST searches (E value = 0.01) using the constructed profiles were run against the protein sets from each of the genomes included in the study, and lists of all proteins containing the given domain were compiled. After verifying the presence of the target domain through examination of the conservation of the salient amino acid sequence and structural motifs, other domains present in the respective proteins were identified by running PSI-BLAST searches with these sequences as queries and by comparing them with libraries of domain-specific profiles using the NCBI CD-search (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) and an additional profile collection (L.Aravind, unpublished data) or by using hidden Markov models (HMMs) for conserved domains (27). The proteins were then clustered by sequence similarity using the BLASTCLUST program (I.Dondoshansky, Y.I.Wolf and E.V.Koonin, unpublished data). Multiple alignments, whenever deemed necessary, were constructed using the T_coffee program (28) and phylogenetic trees were constructed using the PHYLIP and MOLPHY packages (29–31). Protein secondary structure was predicted using the PHD program (32) and structure coordinate files were handled using the Swiss-PDB Viewer program (33).
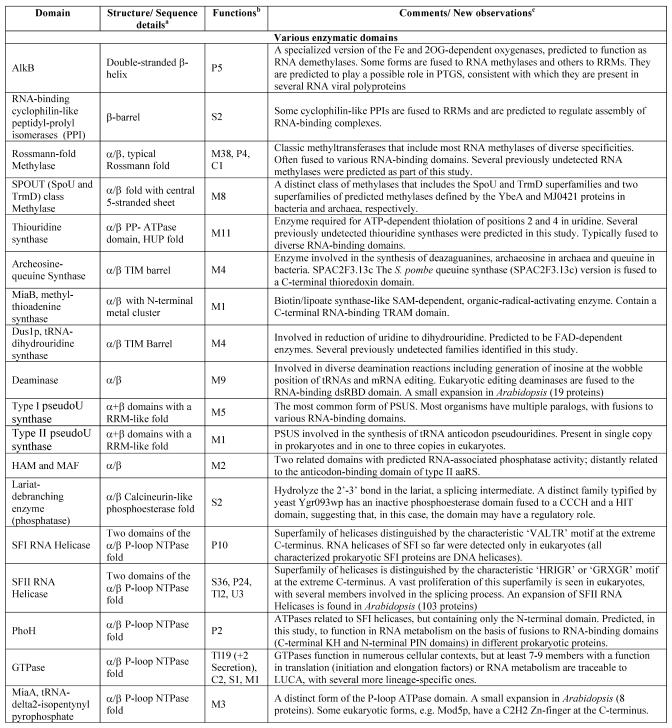
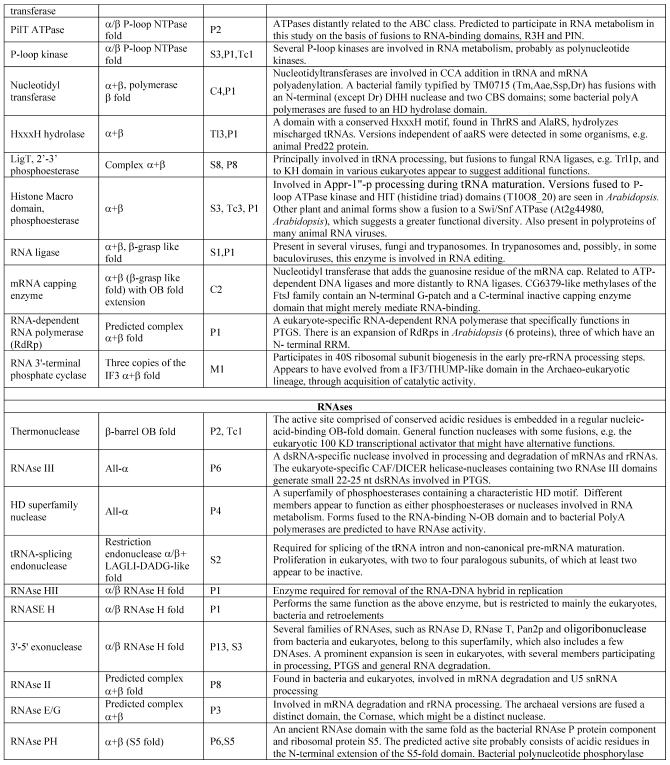
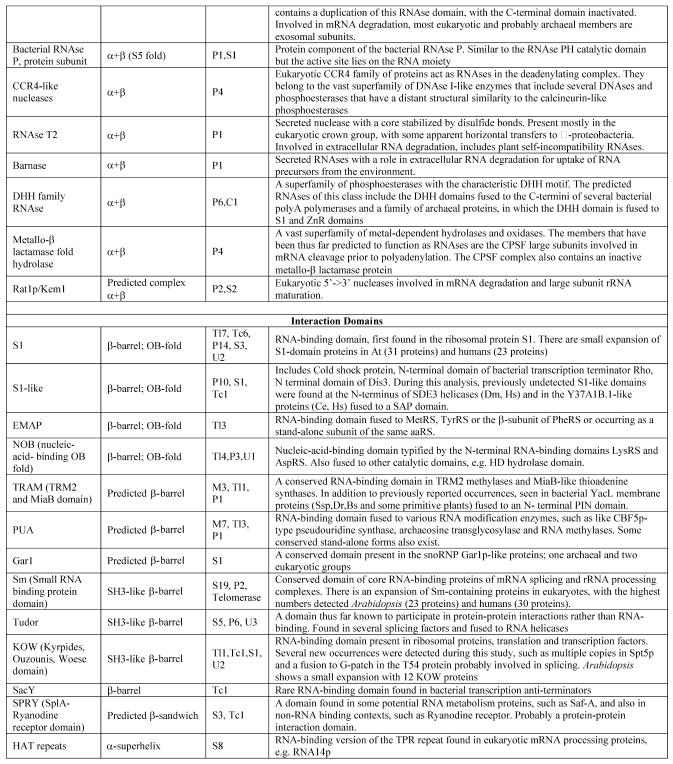
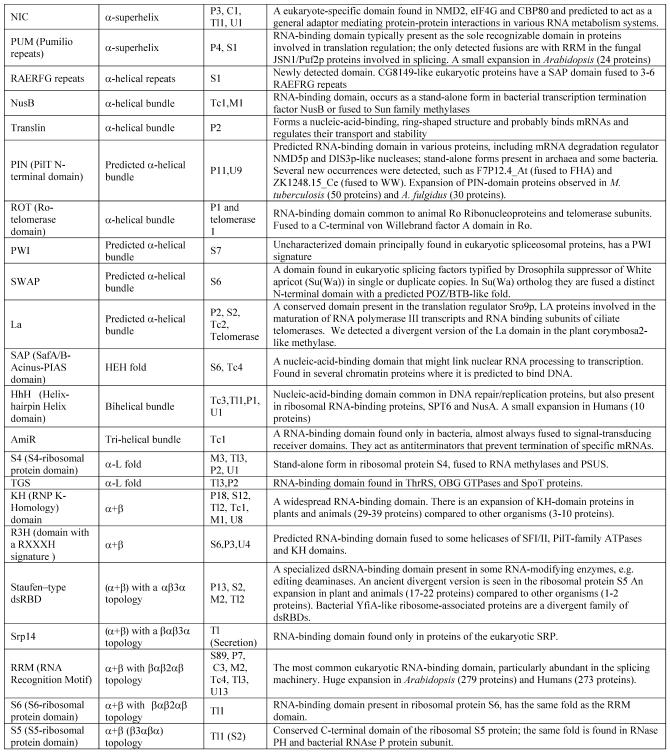
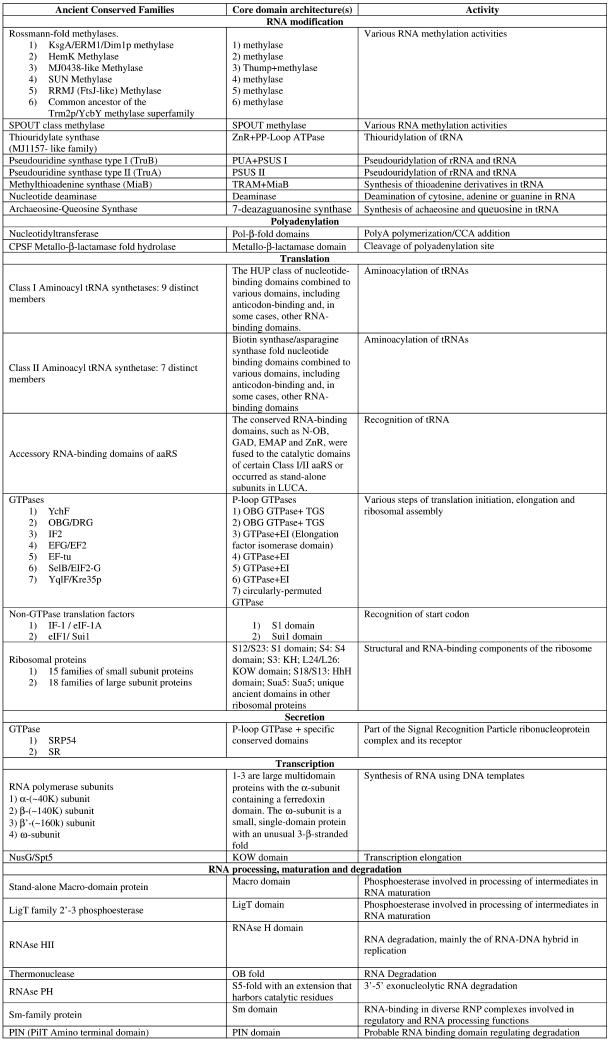
Table 1. Protein domains involved in RNA metabolism.
aα/β, regular alternating αβ units with a typically parallel β sheet; α + β, domains isolated α and β elements with a typically antiparallell β sheet.
bClassification of protein functions involved in RNA metabolism. S, splicing and processing; P, PTGR; C, capping and polyadenylation; Tl, translation; Tc, transcription; M, modification; U, miscellaneous. The numbers after the function designations indicate the number of orthologous groups of proteins containing the given domain for each function.
cThe number of paralogs is indicated in parentheses whenever a lineage-specific expansion of a protein family is mentioned.
RESULTS AND DISCUSSION
Scope and approach
A meaningful computational analysis of proteins involved in RNA metabolism requires a clear definition of the components under investigation so that it does not draw in every other cellular protein based on an indirect connection with such a pivotal class of molecules as RNA. We restrict our scope essentially to those proteins that more or less directly interact with every known type of RNA from the time it is synthesized by the RNA polymerase until its ultimate degradation by nucleases. Briefly, this includes (i) certain components of the transcription machinery itself that directly interact with the transcript in the process of elongation and termination; (ii) proteins involved in the processes that, at least in eukaryotes, occur shortly after transcription, namely polyadenylation and capping; (iii) various complexes and enzymes involved in the maturation of RNAs, including numerous enzymes that catalyze covalent modification of RNA (e.g. methylation) and the complex splicing machinery involved in pre-mRNA processing in eukaryotes; (iv) translation and its regulation; (v) post-transcriptional gene regulation (PTGR), which, in its simplest form, involves various RNAses that catalyze mRNA degradation [a more complex form of such regulation in eukaryotes is post-transcriptional gene silencing (PTGS)]; (vi) proteins that interact with diverse RNAs during maturation of functional complexes, such as the ribosome, the signal recognition particle, RNase P, SnuRPs, SnoRPs, telomerase, the SsrA particle and the Ro-yRNA particles. Not included in this analysis are proteins that regulate transcription through interaction with DNA and generic structural proteins of various complexes, such as those containing WD40 or tetratricopeptide (TPR) repeats, which have similar roles in protein–protein interaction in both RNA metabolism and other systems.
Proteins involved in RNA metabolism were collected through a systematic survey of the literature. This was followed by profile sequence analysis using PSI-BLAST to identify the domains present in these proteins. Once identified, these domains were categorized into two principal classes: (i) enzymatic domains and (ii) interaction domains. The latter class mainly consists of non-catalytic, RNA-binding domains (RBDs) and some protein–protein interaction domains that are predominantly associated with the formation of multisubunit complexes involved in RNA metabolism. Table 1 shows the list of the principal domains included in this analysis. One or more PSI-BLAST PSSMs that ensured complete coverage without inclusion of false positives were prepared for each of the domains and a representative set of complete proteomes (see Materials and Methods) sampled across the three primary kingdoms (bacteria, archaea and eukaryotes) was searched with these profiles to detect all occurrences of each domain. The proteins recovered from all the proteomes were then pooled together and potential orthologous sets were delineated by clustering with BLASTCLUST. These groups of orthologs were corrected and optimized using the symmetry of recovery in single-pass BLAST searches (34) and phylogenetic tree construction and analysis using the minimum evolution (least squares) and maximum likelihood methods (29–31). The domain architecture of each individual protein was then determined by using libraries of PSSMs and HMMs. Finally, we attempted to reconstruct the conservation patterns of functional complexes and pathways across the entire phyletic range of analyzed genomes by combining the results of protein domain analysis with experimental evidence extracted from the literature.
The most likely points of origin of domains and individual protein families involved in RNA metabolism were inferred from the patterns of phyletic distribution and phylogenetic tree topology and on the basis of the parsimony principle. If a particular domain or protein family is widely represented in all three primary kingdoms, bacteria, archaea and eukaryotes, the most parsimonious scenario of evolution points to its presence in LUCA. This conclusion is reinforced when the phylogenetic tree for the family in question family conforms to the ‘standard model’ topology, with a bacterial and archaeo-eukaryotic primary clades (35,36). Conversely, the derivation of a family in LUCA or earlier was considered less likely when a fundamentally different topology was observed, such as grouping of bacteria with eukaryotes. In such a case, a (pre)LUCA origin of the given family would require the extra assumption of displacement of the ancestral form with the bacterial one in eukaryotes, which makes a bacterial or archaeal origin with subsequent dissemination by horizontal gene transfer a viable alternative. Along similar lines, the parsimony principle dictates that, for example, when a domain or a family is widely represented in bacteria and eukaryotes, but is only sporadically encountered in archaea, the most likely scenario involves derivation within the bacterial kingdom and independent acquisitions by eukaryotes and archaea via horizontal gene transfer. Below, in the discussion of the evolution of domains and protein families, we follow these principles of phylogenetic inference, not necessarily referring to them explicitly. All conclusions arrived at with this approach are necessarily probabilistic, but this appears to be the best we can do when reconstruction of ancient evolutionary events is concerned.
In a number of cases, detection of homologs of proteins involved in RNA metabolism required additional correction because a subset of RNA-interacting domains are also involved in DNA binding. We utilized a variety of inputs from experimental studies, phylogenetic relationships and contextual information to exclude those domains and proteins that are primarily involved in DNA binding and metabolism. Nevertheless, a relatively small fraction of the detected domains and proteins either are indeed bifunctional, being involved in both RNA and DNA metabolism, or cannot be assigned specific function with confidence due to insufficient information; such proteins were included in the present analysis for the sake of completeness.
Phyletic patterns and genome-wide demography of protein domains involved in RNA metabolism
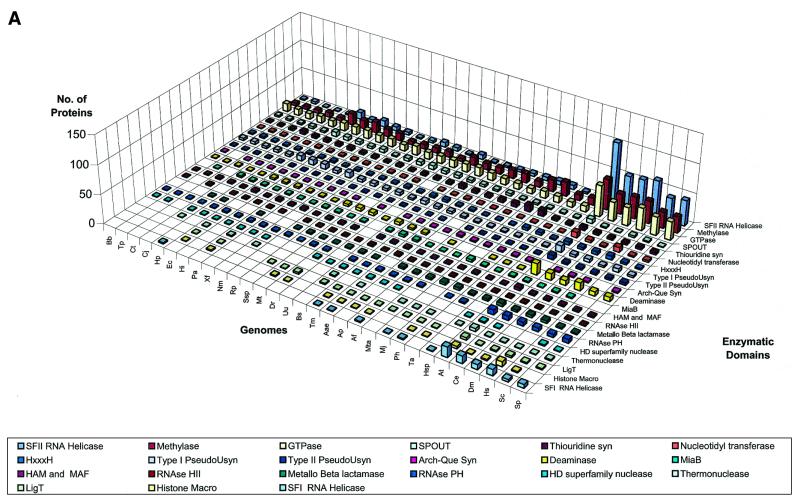
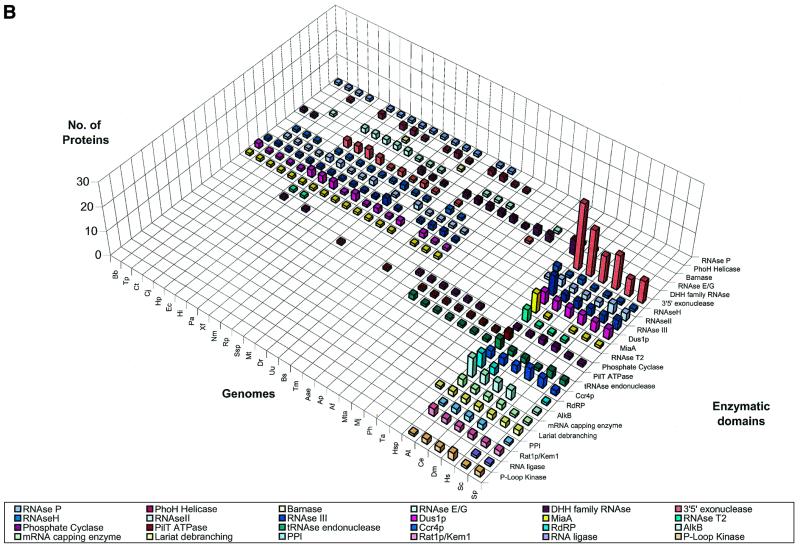
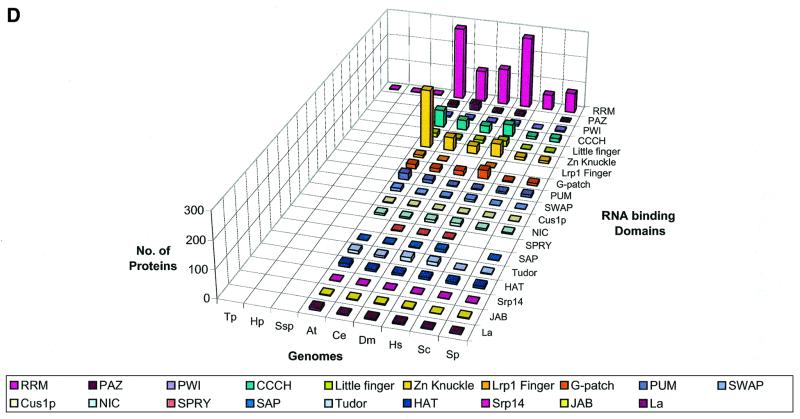
We delineated domains involved in RNA metabolism as described above and conducted a survey of their demography across the genomes of representative organisms considered in this study. This overall demographic survey revealed a number of general trends in the evolution of these domains (Fig. 1A–D). The most notable, if not unexpected, feature was the separation of the three primary kingdoms by specific phyletic patterns of many domains. A large set of enzymes and interaction domains are present universally and, in all likelihood, are part of the LUCA inheritance (Fig. 1A and C). However, another substantial fraction of the domains involved in RNA metabolism appear to have evolved in a particular superkingdom or lineage, with the greatest number of lineage-specific inventions found in eukaryotes (Fig. 1D). Many eukaryote-specific domains belong to ancient folds, but acquired their RNA-related function only in the eukaryotic lineage. Examples of such exaptation of ancient domains for functions in RNA metabolism include the mRNA-capping enzyme that was derived, at the onset of eukaryotic evolution, from the more ancient DNA ligases (37–39), and the lariat-debranching enzyme that was derived from the ubiquitous calcineurin-like phosphoesterases (40,41). Similarly, superfamily (SF)-I helicases were recruited for important RNA-related functions, such as nonsense-mediated decay, only in eukaryotes, although several such helicases function in bacterial DNA recombination and repair. Some eukaryote-specific enzymes, such as the RNA-dependent RNA polymerase involved in PTGS and the Kem1/Rat1 family of 5′→3′ nucleases, have large, complex catalytic domains that so far could not be traced to any ancient enzymatic fold. Although structural innovation is less common in prokaryotes than it is in eukaryotes, there are a few enzymes, for example, the RNase domains of the RNaseE/G superfamily, that appear to be innovations of the bacterial lineage.
Figure 1.
Absolute counts of proteins containing domains involved in RNA metabolism in completely sequenced genomes. (A) RNA metabolism enzymes found in all three superkingdoms. (B) Enzymes of RNA metabolism restricted to one or two superkingdoms. (C) RNA-binding and interaction domains found two or three superkingdoms. (D) RBDs restricted to eukaryotes. For species abbreviations, see Materials and Methods. The domain names/acronyms are as in Table 1.
The interaction domains also show a strong trend of eukaryote-specific innovation, the most prominent one being the RNA recognition motif (RRM), which apparently was derived from a more ancient nucleic acid-binding fold with a characteristic four-stranded core found in diverse DNA- and RNA-binding domains (Table 1). Another theme seen in eukaryotes is the recruitment of α-helical superstructures, such as the TPR-like fold (the HAT repeat module found in RNA processing proteins), the pumilio (PUM) repeat (42,43), and the NIC domains (16) for functions in RNA metabolism. This parallels the widespread utilization of these α-helical repeat modules in a number of other contexts in eukaryotes. Many of the distinct, small RBDs that evolved in eukaryotes, such as CCCH, Zn knuckle, C2H2-, LRP1- and C4-Little fingers utilize the common theme of stabilization through metal chelating cysteines and histidines (Fig. 1D). This type of structure is ancient, with numerous Zn-ribbon modules found in archaea (44), but many of these metal- and RNA-binding domains seem to have evolved de novo in eukaryotes, given that utilization of metal coordination to stabilize the core of a domain requires relatively few evolutionary changes, namely the emergence of a strategically placed set of metal-chelating residues.
Another major pattern in the phyletic distribution is the presence of numerous catalytic and interaction domains that are shared by eukaryotes and bacteria, to the exclusion of archaea (Fig. 1A–D). Another distinct set of domains is solely shared by archaea and eukaryotes, which supports the chimeric origin of the eukaryotic systems of RNA metabolism. A subset of proteins containing domains shared by eukaryotes and bacteria function in the mitochondria and chloroplasts that have descended from endosymbiotic bacteria. This is reflected in the larger average number of proteins with such a phyletic pattern in plants that have two distinct endosymbiont organelles, mitochondria and chloroplasts. However, several domains with a bacterio-eukaryotic distribution pattern function in non-organellar contexts, such as cytoplasmic RNA degradation. Enzymes of apparent bacterial origin recruited for cytoplasmic functions include several superfamilies of RNases, such as the 3′→5′ exonucleases (45). Of the domains with an archaeo-eukaryotic phyletic pattern, several are involved in core processes, such as RNA maturation, e.g. the tRNA endonucleases, and translation, e.g. PIWI (14), pelota and SUI1 domains (9).
Most of the domains involved in ancient functions, such as RNA modification enzymes and RBDs associated with RNA modification, translation and transcription (Table 1 and Fig. 1), are present in nearly constant numbers in all life forms, except that eukaryotes often have more paralogs, partly owing to the presence of organelles derived from bacteria. Eukaryotes show a striking expansion of ancient SFII RNA helicases and, to a lesser extent, of other ancient catalytic domains, such as SFI helicases, GTPases, Rossmann-fold methylases, 3′→5′ exonucleases, RNase III and deaminases. A corresponding expansion of non-catalytic domains is mainly restricted to those newly invented or recruited in eukaryotes, including RRM, CCCH, Zn-Knuckle and G-patch. The advent of these RBDs correlates with the emergence of eukaryote-specific functional systems, such as pre-mRNA splicing, PTGR, and mRNA editing and modification (Fig. 1).
These observations indicate that 40–45 of the approximately 100 principal domains associated with RNA metabolism originated at early stages of evolution, prior to LUCA. These domains were associated with the most ancient and conserved cellular functions, such as translation, transcription and some forms of RNA modification. The next phase of innovation marked the separation of the bacterial and archaeo-eukaryotic lineages and saw the origin of some proteins, which are involved in basic cellular functions, but are specific to one of these lineages. Finally, with the emergence of the chimeric eukaryotic lineage, domains from both the bacterial and the archaeo-eukaryotic precursor were incorporated into the eukaryotic RNA metabolism pathways. In addition, eukaryotes also ‘invented’ several new domains and recruited or expanded preexisting ones, concomitant with the origin of new RNA processing systems that were largely absent in prokaryotes. No archaea-specific domains involved in RNA metabolism were identified. This might reflect the retention of most core archaeal systems in eukaryotes, which makes the corresponding domains archaeo-eukaryotic in distribution. In addition, archaea could possess some distinct domains that were not detectable through homology and remain unknown due to the paucity of experimental studies in archaeal systems.
The surveyed organisms dedicate, approximately, between 3 and 11% of their proteomes to RNA metabolism, with the highest fraction, predictably, seen in parasitic bacteria with small genomes and the lowest fraction in multicellular eukaryotes and complex bacteria. Generally, this seems to reflect (i) the central place that RNA metabolism systems occupy in all cells, compared with the substantially more variable systems of transcription, replication or DNA repair, and (ii) a more or less linear growth of the number of proteins involved in RNA metabolism with the increase of the total number of encoded proteins in free-living organisms. Below we discuss in detail specific trends in evolution of catalytic and interaction domains involved in RNA metabolism.
Evolutionary histories of catalytic domains involved in RNA metabolism
RNA modifying enzymes. Cellular RNAs are subject to a number of post-transcriptional modifications that involve modification of the bases and sugars or synthesis of non-canonical bases or nucleotides (46–48). The direct nucleotide modifications include methylation of bases and sugars on N, C or O atoms, deamination and demethylation, whereas formation of non-canonical bases includes thiouridylation, pseudouridylation, thioadenylation, dihydrouridylation, and synthesis of archaeosine and queuine.
Methylases. The most common among RNA modifications are the numerous methylations of all types of RNA molecules (46). The RNA methylases come in two major classes (Table 1): (i) the Rossmann-fold methylases, which include the majority of N-, C- and O-methylases that modify both sugars and bases in RNA, and (ii) the recently described SPOUT (49) superfamily, which consists of the m1G-specific methylase TrmD (50,51), the 2′-O-methylguanosine-specific methylase SpoU (52–54), and several other poorly characterized predicted RNA. The SPOUT superfamily is traceable to LUCA, but the evolution of these methylases is not considered here in detail because it has been recently described in detail elsewhere (49).
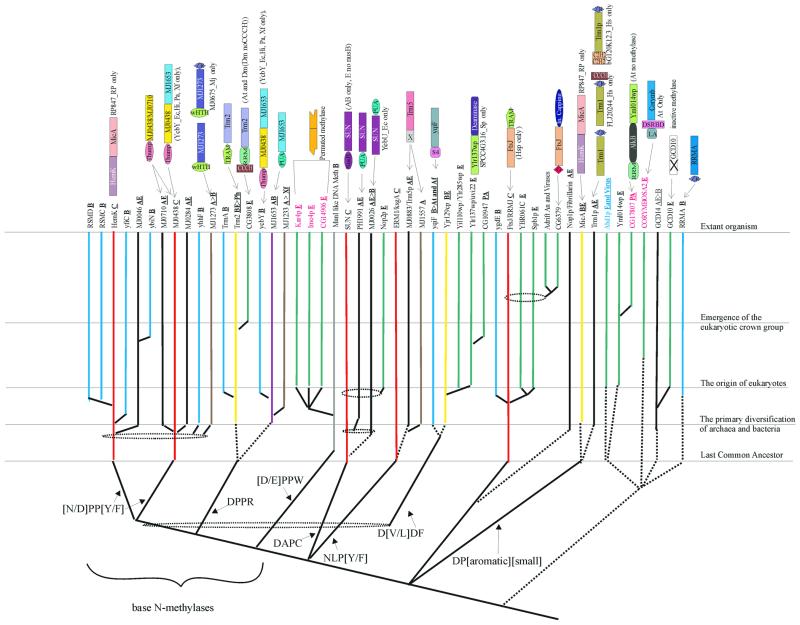
The methylases of the Rossmann-fold class share a six-stranded Rossmann-fold core with the dinucleotide-binding dehydrogenases and are distinguished from them by a methylase-specific 7th strand (20,55). This class contains the great majority of the known methylases that participate in almost every conceivable methylation reaction in biological systems, and RNA specificity appears to have emerged on multiple occasions among them. We sought to resolve the evolutionary relationships among Rossmann-fold RNA methylases using a combination of conventional phylogenetic trees and cladistic analysis based on specific shared sequence motifs (Fig. 2). Several distinct lineages of dedicated RNA methylases can be detected; some of the corresponding protein families also include related DNA methylases. The RNA methylases, typically, are highly conserved and are often associated with specific RBDs, which distinguish them from the DNA methylases; many of the latter are large proteins occurring in restriction-modification operons with a sporadic phyletic distribution. The largest monophyletic superfamily of nucleic acid methylases are the base N-methylases (the BNM superfamily). These methylases are characterized by a shared derived character, the [N/D]PP[Y/F] motif at the end of strand 4, which is associated with base specificity (Fig. 2). Phylogenetic analysis helped in identifying several distinct families within the BNM superfamily, and most of these families can be distinguished by specific derived characters in the above motif. Within the BNM superfamily, two families, namely the HemK family (19) and the MJ0438 family of predicted methylases containing the RNA-binding THUMP domain (12), are represented in all three primary kingdoms and are thus traceable to LUCA. Along with several other related families with more restricted phyletic patterns, these families form a large assemblage of (predicted) purine N-methylases with the NPP[Y/F] motif associated with strand 4. Some of the smaller families appear to be more closely related to either the HemK or the MJ0438 family and might have emerged from them through duplications much later in evolution. The RsmC family methylases that methylate G1207 in 16S rRNA (56) and RsmD,YfiC and YbiN families are bacteria-specific elaborations that are related to the HemK family, whereas the MJ0046 family apparently was derived from the HemK family in the archaeo-eukaryotic lineage. The MJ0438-related elaborations, namely the MJ0710 and MJ0284 lineages, are present in archaea and eukaryotes. The YhhF and MJ1273 families, which are restricted in their distribution to bacteria and archaea, respectively, also belong to this assemblage, but do not show a specific relationship with either the HemK or the MJ0438 family. The functions of the HemK and MJ0438 families are poorly characterized, but their nearly universal conservation pattern suggests a role in purine methylation in rRNA. In Rickettsia, the HemK methylase is fused with another methyltransferase of a different family, MicA (Fig. 2). This suggests that these two methylases coordinately function in rRNA methylation.
Figure 2.
An evolutionary scheme for Rossmann-fold RNA methylases. The conserved families and their probable temporal points of origin are shown for each of the major lineages. Dotted ellipses indicate the general phylogenetic affinities whose branching points could not be more precisely assigned, and the dotted lines indicate temporal uncertainty regarding the point of emergence. The gray line indicates related DNA methylase lineages, which are not shown in detail. Any special domain architecture present in a given lineage is shown on the top. The motif at the end of strand 4 is shown at the first branchpoint of each major family. The methylase domains in the domain architectures are given respective family names. The lines are colored to indicate the phyletic pattern of the corresponding families: A, archaea (brown); B, bacteria (blue); E, PA, An or Ver, eukaryotes, plants and animals, animals, or vertebrates (green); AB, archaeo-bacterial (purple); AE, archaeao-eukaryotic (black); BE, bacterio-eukaryotic (yellow); and C, conserved (universal) (red). These phyletic pattern abbreviations are additionally shown next to the family names and are underlined. The family names are colored according to the function: black, modification; red, PTGR; blue, capping. The domain names/acronyms are as explained in Table 1; additionally, X is a possible OB fold domain specific to Trm5; G is G-Patch domain and ‘In. capping’ is an inactive mRNA capping enzyme.
The next major assemblage within the BNM superfamily is distinguished by the motif DPP followed by a polar residue (typically R) after strand 4. One of the main families within this assemblage is the Trm2 family, which is involved in methylation of U54 in tRNA at the 5 position (57). This family with its pan-bacterial distribution appears to have emerged early in bacterial evolution and apparently was subsequently transferred to the eukaryotic lineage through the mitochondrial symbiosis. Certain bacteria encode an additional methylase family of this assemblage, TrmA, which has the same specificity (58), and appears to have branched off the more widespread Trm2 family. Similarly, eukaryotes have their own, specific methylase family related to the Trm2 family proteins and typified by CG3808 from Drosophila. Another prominent group within this assemblage is the MJ1653 family that shows a fusion to the RNA-binding PUA domain and is widespread in both archaea and bacteria. Families with a more restricted distribution, which are probably more recent offshoots of this lineage, include the YcbY family seen only in some bacteria and the archaeal MJ1233 family (Fig. 2).
The last major group of the BNM assemblage are the methylases with a circularly permuted methylase domain. All members of this group that are widespread in prokaryotes are DNA adenine methylases associated with restriction–modification systems. In eukaryotes, this group diversified into three distinct families of adenine mRNA methylases (59) typified by the yeast proteins Kar4p and Ime4p, and Drosophila CG14906 (lost in S.cerevisiae), respectively (Fig. 2). In these families, the motif associated with strand 4 assumes the form [D/E]PPW, which is shared with DNA adenine methylases, such as MunI.
The SUN superfamily is the next major assemblage of Rossmann-like fold RNA methylases, which is the sister group of the BNM superfamily (Fig. 2) and has the diagnostic motif DAPC associated with strand 4. The Sun family enzymes, which methylate rRNA at the cytosine 5 position (23), are represented in all three primary kingdoms, consistent with their presence in LUCA. The SUN superfamily has undergone extensive radiation in archaea and eukaryotes, giving rise to two distinct families prior to the separation of eukaryotes and archaea and the eukaryote-specific Nop1 family involved in rRNA and snU RNA methylation (60).
The Erm1/KsgA family that has the motif NLP[Y/F] associated with strand 4 is another close sister group of the BNM superfamily (Fig. 2). These methylases are conserved in all life forms and are responsible for diadenine 2-methylation in rRNA (61), which suggests the presence of this modification in LUCA. The archaeo-eukaryotic Trm5 tRNA methylase family and the archaea-specific MJ1557 family also have a similar form of the strand-4 motif, suggesting that these families form a monophyletic superfamily with the KsgA family (Fig. 2).
Generically related to the BNM, SUN and KsgA-Trm5-like superfamilies are two methylase groups with a more restricted distribution. One of these is the bacterial YqlF family, which has an N-terminal S4 domain and a strand-4 motif of the form D[V/L]DF. Thus, this family shares the conserved D or N followed by two small residues and the predicted base-interacting aromatic or hydrophobic residue with the former superfamilies. The second group, the Uvi22 superfamily, also has a similar strand-4 motif, but has a unique, two small amino acid insert prior to the conserved D at the end of strand 4. While none of the members of this superfamily has been experimentally characterized as RNA methylases, the presence of the characteristic form of the above mentioned strand-4 motif supports this function. Additionally, one of the yeast members of this family is fused to a RNA deaminase (see below), suggesting a role in RNA modification (Fig. 2). This superfamily is restricted to the proteobacteria (conserved in all α-proteobacteria) amidst the bacteria, while it vastly expanded into several distinct families in eukaryotes. This pattern, taken together with phylogenetic analysis results (data not shown), suggests an origin from the mitochondrial endosymbiont. Members of this superfamily might represent a major, as yet unexplored group of eukaryotic nucleic acid methylases.
Sequence evidence and the distinct form of the strand-4 motif suggest that all methylase superfamilies described above descended from a common RNA-methylating ancestor well before the emergence of LUCA. Structural comparisons reveal even deeper links, suggesting that these methylases, in turn, form a higher-order monophyletic group with the FtsJ superfamily of methylases involved in 2′-O-methylation of uridine in LSU rRNA (62) (Fig. 2). The FtsJ/RrmJ family proper is represented in all three primary kingdoms, which points to its presence in LUCA. Several other related families, such as YgdE in bacteria and at least four distinct eukaryotic families, including two animal-specific ones, were derived at various later points in evolution, probably from a FtsJ-like precursor. Some of these, e.g. the Spb1 family, might methylate Sno RNAs (63), suggesting that other, unexplored specificities exist within this family of methylases. Structural comparisons indicate that the group of RNA methylases closest to the FtsJ superfamily is the Fibrillarin/Nop1 family, which is involved in snoRNA methylation (64). This family is restricted to the archaeo-eukaryotic lineage and might have been derived from the FtsJ superfamily through extreme divergent evolution. The archaeo-eukaryotic Trm1 methylase family and the MicA family shared by bacteria and eukaryotes appear to comprise another monophyletic group, which appears to be a sister group of all of the rRNA methylases described above (Fig. 2). Both these families share a similar form of the strand-4 motif with the signature DP followed by an aromatic and then by a small residue. Trm1 functions as a tRNA N2,N2-dimethylguanosine-26 methyltransferase (65,66) and MicA probably performs a similar, although not identical, role in bacteria and eukaryotic mitochondria. These two families might represent the archaeo-eukaryotic and bacterial branches, respectively, of an ancestral methylase that was represented in LUCA.
All the other groups of RNA methylases appear to have been derived, independently, on more than one occasion in evolution, from within the vast assemblage of small molecule and protein methylases. None of these families is traceable to LUCA; instead, they are restricted in their distribution to only one or two of the primary kingdoms. Two of these families, the Abd1p family that methylates the eukaryotic mRNA cap, and Yml014w family that is fused, in some cases, to the AlkB domain (see below), have a dyad of aromatic residues in the 4th position after the end of strand 4. This feature suggests their derivation from within the vast class of small molecule methylases. The Yml014w family has additionally lost the polar residue (D/N) at the end of strand 4. Also derived from within this small molecule methylase assemblage is the family typified by the plant Corymbosa2/Hen-1 protein. Predicted methylases of this family are present in the crown group eukaryotes and in some bacteria, such as Streptomyces and Nostoc, and retain a single aromatic residue in the 4th position after the end of strand 4. The plant representatives of this family are fused to an N-terminal RNA-binding LA domain and a double-stranded RNA-binding domain (dsRBD) (Table 1 and Fig. 2), which suggests that these proteins are RNA methylases that probably methylate substrates containing double-stranded regions (see below). The GCD14 family of methylases (67,68), which methylate A58 of tRNAs in position 1, was derived in the archaeo-eukaryotic lineage and is more closely related to protein arginine and carboxyl group methylases than to other RNA methylases. These methylases have been sporadically transferred to bacteria, such as M.tuberculosis and A.aeolicus. They are distinguished by the presence of a distinct C-terminal domain similar to the transcript cleavage factor GreA (69). This family appears to have undergone a duplication in eukaryotes, giving rise to a paralog, GCD10, whose methylase domain shows a disruption of the Rossmann-fold loop and the strand-4 region. The RrmA family that methylates G745 in position 1 in LSU rRNA (70) is another family that appears to have been derived from the small molecule methylases late in bacterial evolution, followed by inter-bacterial dispersion via horizontal transfer.
Thus, Rossmann-fold methylase appear to have been recruited for RNA methylation at an early stage of evolution, well before LUCA. From this ancient, ancestral methylase, the significant majority of the RNA methylases, including the five to six aforementioned methylase families that were probably already present in LUCA, were derived. Extensive duplication, later in evolution, particularly in eukaryotes, resulted in the formation of several more families within this large, monophyletic assembly of RNA methylases. Additionally, lineage-specific RNA methylases were apparently derived independently, on multiple occasions, from within the small molecule and protein methylase clade. At early stages of their evolution, RNA methylases formed stable fusions with several distinct RBDs, such as the S4, PUA (9), TRAM (11), THUMP (12), NusB and a potential OB-fold domain (in Trm5) (71) (Fig. 2). In addition, in eukaryotes, fusions of RNA methylases to eukaryotic-specific RBDs, including RRM and CCCH domains in the TrmA-family methylases and a G-patch domain (18) in the FtsJ family, were detected. These fusions appear to have emerged relatively late in eukaryotic evolution and probably participate in the methylation of eukaryote-specific snRNAs. Most of these pan-bacterial families of methylases appear to have been horizontally transferred to the eukaryotic genomes as a consequence of organellar endosymbiosis, resulting in a bacterial–eukaryotic distribution pattern. The identification of several uncharacterized RNA methylase groups in this analysis (Table 1) may help in further investigations of the diversity of this crucial RNA modification.
Pseudouridine synthases. The modified base pseudouridine is synthesized by pseudouridine synthases via in situ isomerization of uridines in tRNAs, rRNAs and eukaryotic snRNAs, such as U5 and U3 (46,72). Pseudouridine synthases belong to two apparently unrelated superfamilies, one of which (Type I PSUS) includes the four principal ancient families, RluD, RsuA, TruB and MJ0041, whereas the other superfamily (Type II PSUS) consists of a single ancient lineage typified by TruA (22,73,74) (Fig. 3). Type II PSUS are present in a single copy in all proteomes, except for eukaryotes that have at least three enzymes of this superfamily. Within the Type I PSUS superfamily, the TruB family is traceable to LUCA; several members of this family are fused to a PUA domain, suggesting that this was the ancestral PSUS Type I domain architecture. The RluD and RsuA families originated in bacteria; each family includes several members containing the S4 RBD (9), which was probably present in the ancestor of these families, but was subsequently lost on multiple secondary occasions. Conversely, the THUMP-domain-containing MJ0041 family of PSUS appears to be an innovation specific to the archaeal lineage. The RluD family has been secondarily transferred to the eukaryotes, probably via the pro-mitochondrial endosymbiont. Type I PSUS are predicted to adopt an α+β fold; the crystal structure of the Type II PSUS shows the presence of a core RRM-like fold common to several ancient nucleic acid-binding domains (75). This, taken together with the use of guide RNAs by the eukaryotic PSUS, suggests that Type II PSUS might have evolved from an ancient RBD that functioned in conjunction with a ribozyme, with a gradual shift of the active site from the RNA to the protein component.
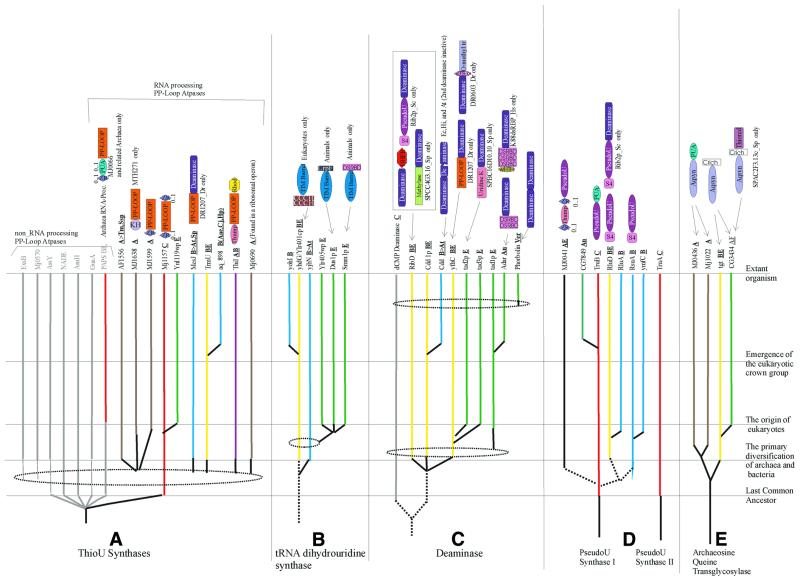
Figure 3.
An evolutionary scheme for other RNA modification enzymes. (A) Thiouridine synthases; the PP-loop ATPases not involved in RNA processing are shown in gray. (B) tRNA-dihydrouridine synthases. (C) Deaminases. (D) Pseudouridine synthases I and II. (E) Archaeosine/queuine synthases (transglycosylases). The conventions for color-coding and line patterns are the same as in Figure 2. The domain names/acronyms are: Rhod, rhodanese domain; DHFR, dihydrofolate reductase; Uridine K, uridine kinase; wHTH, winged helix–turn–helix domain; ZR, zinc ribbon; and Crich, cysteine rich. The remaining designations are as in Table 1.
Enzymes involved in base thiolation. A variety of thio-bases are represented in cellular RNAs, the most common ones being 2- or 4-thiouridines and their derivatives, and 2-methylthioadenine derivatives. The methylthioadenines are typically additionally modified with bulky adducts, such as threonine or 4-hydroxyisopentene in the N6 position. Recently, the enzyme responsible for adenine thiolation in E.coli, MiaB (76), has been identified and shown to consist of a C-terminal RNA-binding TRAM domain and an N-terminal biotin synthase-like, metal cluster-containing catalytic domain that is predicted to catalyze sulfur insertion via SAM-dependent organic radical generation (11,77,78). MiaB-like proteins are universally present in all life forms, indicating their origin prior to LUCA. Several organisms encode more than one version of this enzyme, which appear to have diversified through early duplications; these multiple forms might differentially function in the synthesis of different 2-methylthioadenine derivatives, such as 2-methylthio-N6-threonyl carbamoyladenosine and 2-methylthio-N6-methyladenosine (46).
Thiouridine synthase (ThioUS; ThiI protein in E.coli) is involved in the synthesis of 4-thiouridine in tRNAs and has a core PP-ATPase domain (79), which catalyzes adenylylation of the 4-carbonyl group of uridine, followed by sulfur insertion catalyzed by a rhodanese-like enzyme (80,81). This rhodanese-like enzyme either comprises a distinct domain of the ThiI protein or functions as a stand-alone protein. 2-Thiouridine is universally present in tRNA, and 2-thiouridine derivatives, typically containing an additional modification of a methyl or aminomethyl group in position 5, are also common. One of the enzymes involved in 2-thiouridine synthesis, TrmU, has been identified (82). This protein contains a PP-ATPase domain with an unusual conserved cysteine dyad inserted after strand 3 in the PP-loop domain. This suggests that syntheses of 2-thiouridine and 4-thiouridine follow similar biochemistry, which involves activation of the carbonyl group by adenylylation. In TrmU-like enzymes, the internal conserved cysteines might directly participate in sulfur insertion as a functional counterpart of the separate rhodanese-like domain, which is required for 4-thiouridine formation.
Previously, we predicted that the MJ0066 family represents a novel family of archaeal ThioUS, on the basis of the fusion of a PP-ATPase domain with a PUA domain (9). Here, we systemically investigated other PP-ATPase families that potentially could be involved in thiouridine or thiocytidine synthesis by examining fusions with RBDs, association with the ribosomal super-operon and conserved phyletic patterns typical of RNA metabolism proteins. As a result, the MTH271-MJ1157 family, which showed fusions with the KH and Zn-ribbon domains, and the MJ0690 family, which is associated with ribosomal super-operon in different archaeal genomes, emerged as candidates for these functions (Fig. 3). Furthermore, the MesJ family, which is closely related to the TrmU family, is universally conserved in all bacteria and potentially also could be involved in base thiolation.
The ThiI-family proteins contain a N-terminal THUMP domain and are bifunctional proteins that additionally participate in thiamin biosynthesis (80,81,83). These proteins are ubiquitous in archaea, but sporadic in bacteria, suggesting that they originated in archaea, with several subsequent horizontal transfers to bacteria. In contrast, the TrmU family is absent in archaea, but is nearly universal in bacteria and eukaryotes, suggesting origin in the bacterial lineage, followed by an early transfer to the eukaryotes, probably via the pro-mitochondrial route. These phyletic patterns do not seem to be consistent with the universal distribution of the simple 2-thiouridine modification in tRNAs (46,84). The only predicted universal ThioUS are the members of the MJ1157 subfamily (Fig. 3) of the MTH271-MJ1157 family containing N- and C-terminal Zn-ribbon domains. The universal distribution, taken together with the distinct bacterial and archaeo-eukaryotic clades detected during the phylogenetic analysis of this family (data not shown), suggests that these enzymes are the 2-thiouridine synthases, whereas TrmU is likely to be specifically involved in 5-methyl-2-thiouridine biosynthesis. The presence of a conserved cysteine dyad insert in the PP-ATPase domain of the MTH271-MJ1157 family, similar to TrmU, might indicate an analogous catalytic mechanism. Archaea and eukaryotes have additional subfamilies of the MTH271-MJ1157 family (Fig. 3) that, along with the MJ0066 family, could compensate for the absence of TrmU or ThiI in some of these lineages. The sporadic presence of ThiI in bacteria suggests that it might be substituted by a more widespread, but so far unidentified 4-thiouridine synthase specific to bacteria; the conserved MesJ protein could be one candidate for this function.
Queuosine and archaeosine synthases. The 7-deazaguanosines, queuosine and archaeosine, found, respectively, in bacteria and eukaryotes, and in archaea, are incorporated into tRNA through transglycosylation (46,84). The queuosine transglycosylase, Tgt (85,86), is present in a single copy in most bacteria, whereas eukaryotes, with the exception of yeast, encode two forms of this enzyme. Archaea have two distinct proteins of the archaeosine transglycosylase family, which are distantly related to Tgt (87). The MJ1022 subfamily so far has been found only in Euryarchaea; A.fulgidus, in addition, has a single copy of queuosine transglycosylase, apparently a lineage-specific acquisition from bacteria (Fig. 3). The complementary distribution of queuosine and archaeosine transglycosylases suggests that they originally diverged from a common ancestor with a TIM barrel fold (88), concomitantly with the split of the bacterial and archaeo-eukaryotic lineages. In archaea, the catalytic domain was fused with the RNA-binding PUA domain and this form of archaeosine transglycosylase underwent a duplication in Euryarchaea (Fig. 3). In eukaryotes, acquisition of the bacterial queuosine synthase through horizontal transfer from the pro-mitochondrion probably resulted in displacement of the ancestral archaeo-eukaryotic archaeosine synthase, with a further duplication leading to the forms involved in modification of organellar and cytoplasmic tRNAs.
RNA deaminases. RNA deaminases are responsible for the synthesis of certain modified nucleosides, such as inosine, and for base conversions during various RNA editing reactions. The cytidine deaminase family includes generic enzymes that catalyze generation of uridine from cytidine. In yeast, these enzymes are responsible for C→U editing (89), suggesting that they might perform a similar function in many, if not all, eukaryotes. Plants show an expansion of a specialized form of this family, with an N-terminal inactive deaminase domain, in addition to the C-terminal active one; conceivably, these proteins might be involved in a plant-specific form of regulated RNA editing. Deaminases of the Tad2p family, which generate uracil from cytosine and inosine from adenosine in the wobble position of tRNAs (90,91), are present in most bacteria and all eukaryotes, but not in archaea (Fig. 3). The Tad3p family, which comprises the second subunit of the inosine-generating deaminase, is eukaryote specific. The combination of Tad2p and Tad3p probably confers the specificity that differentiates this enzyme from generic cytosine deaminases. The eukaryote-specific Tad1p family of deaminases (92) is involved in inosine generation at A37 of tRNAAla and in adenine editing of mRNAs in animals (93,94). The animal versions typically have the characteristic dsRBD fused to the catalytic domain, whereas one of the vertebrate paralogs contains a winged helix–turn–helix domain (Fig. 3). Cytosine deaminases of the vertebrate-specific APOBEC family are involved in C→U editing and are represented by at least eight paralogs in mammals (95). These enzymes appear to have been recently derived from the cytidine deaminases through rapid divergent evolution. The deaminases related to the RibD protein, which is involved in riboflavin biosynthesis, are fused to a Type I PSUS in S.cerevisiae and to a potential RNA methylase in S.pombe, suggesting that, similarly to cytidine deaminases, they might be involved in specific editing processes (Figs 2 and 3).
Specific RNA deaminases of known families are nearly absent in archaea. The corresponding functions might have been taken over by unrelated, still unknown enzymes or, at least in some cases, could be provided by related enzymes of the deoxycytidine deaminase family that are present in some archaea. This phyletic pattern suggests a bacterial origin for at least two of the major deaminase lineages, cytidine deaminases and cytosine deaminases. Following their acquisition by eukaryotes from the bacterial endosymbiont, cytosine deaminase underwent duplication to give rise to the two A→I deaminases involved in wobble-specific inosine synthesis. Additionally, members of both the cytidine and cytosine deaminase lineages were independently recruited for mRNA editing in vertebrates and possibly in other eukaryotic lineages (Fig. 3).
Dihydrouridine synthases. Dihydrouridine synthases are poorly characterized enzymes that synthesize dihydrouridine through the reduction of the aromatic ring of uracil. This base is widely found in tRNAs from all three primary kingdoms and in LSU rRNA from prokaryotes (96,97). The yeast dihydrouridine synthase Dus1p belongs to the superfamily of FAD-binding TIM barrel oxidoreductases typified by dihydroorotate dehydrogenase (98). This enzyme is universally represented in eukaryotes and bacteria, but completely missing in archaea. Eukaryotes have four main lineages within this family, which are typified by the yeast proteins Dus1p, Smm1p, Ylr405wp and Ylr401cp. The members of the first three families typically show fusions with the LRP1 Zn-finger, dsRBD and CCCH RBDs, respectively (Fig. 3); these RBDs probably target dihydrouridine synthases to specific sites in the substrate RNAs. Bacteria have at least three principal lineages of dihydrouridine synthases typified by the YhdG, YohI and YjbN proteins from E.coli (Fig. 3). The phyletic pattern of dihydrouridine synthases suggests that this enzyme emerged early in bacterial evolution and was transferred to eukaryotes, probably via the endosymbiotic route. The diversification of dihydrouridine synthases into multiple forms apparently occurred independently in bacteria and eukaryotes. Dihydrouridine has been detected in tRNAs of T.acidophilum and M.thermoautotrophicum, but appears to be missing in other archaea studied to date (99,100). Hence, at least in those archaea that appear to contain this modification, an alternative as yet undiscovered enzyme is likely to be present.
NTP-dependent enzymes involved in RNA metabolism
In addition to the PP-loop ATPases discussed above in the context of base modification, a variety of ATP- and GTP-utilizing enzymes of the P-loop NTPase fold are involved in RNA modification, processing and splicing and especially in translation itself. In addition, aminoacyl-tRNA synthetases (aaRS), which belong to two other distinct, ancient classes of ATP-utilizing enzymes, are central to the translation process. Evolutionary relationships of aminoacyl-tRNA synthetases have been examined in detail in several recent studies (10,36,101,102). Here, we briefly summarize the evolutionary history of the vast class of P-loop NTPases in the context of their repeated utilization in RNA metabolism.
GTPases. P-loop GTPases are among the central, most ancient components of RNP complexes and at least nine distinct GTPases associated with different aspects of translation are traceable to LUCA. These include the four translation factors involved in initiation and elongation, two distinct versions of the OBG family of GTPases containing the RNA-binding TGS domain, the circularly permuted YlqF-like GTPases, and two GTPases associated with the signal recognition particle and its receptor. The first seven of these families belong to a large assemblage of GTPases related to the translation factors (the TRAFAC class), whereas the remaining two are members of the signal recognition/MinD/BioD (SIMIBI) class of GTPases and related ATPases (103). These two classes correspond to the first fundamental split in the evolution of GTPases and, because both classes include proteins involved in translation, it appears likely that the primordial GTPase was a component of an ancient RNP complex that functioned as a generic regulator of translation. Even prior to LUCA, the GTPases have diversified through several duplications to perform more specific, essential functions in translation and secretion. After the radiation of the major lineages of life, many GTPases were recruited for specific functions within the translation system, such as translation-termination and RNA modification and processing. The Era family GTPases (104), which contain a C-terminal domain that is a topologically rearranged version of the KH domain, the PseudoKH domain (105), and the TrmE (ThdF) family were derived in bacteria within the TRAFAC class of GTPases and participate in rRNA and tRNA modification. TrmE is involved in the synthesis of the modified nucleotide 5-methylaminomethyl-2-thiouridine in tRNAs (106). The archaeo- eukaryotic Clp1 GTPase family of the SIMIBI class was recruited to participate in polyadenylation site selection (107). In eukaryotes, a distinct paralogous derivative of the universal translation factor EF-2, typified by Snu114p, acquired a new function in splicing as a component of the U5 RNP (103). Further details of GTPase evolution are presented elsewhere (108).
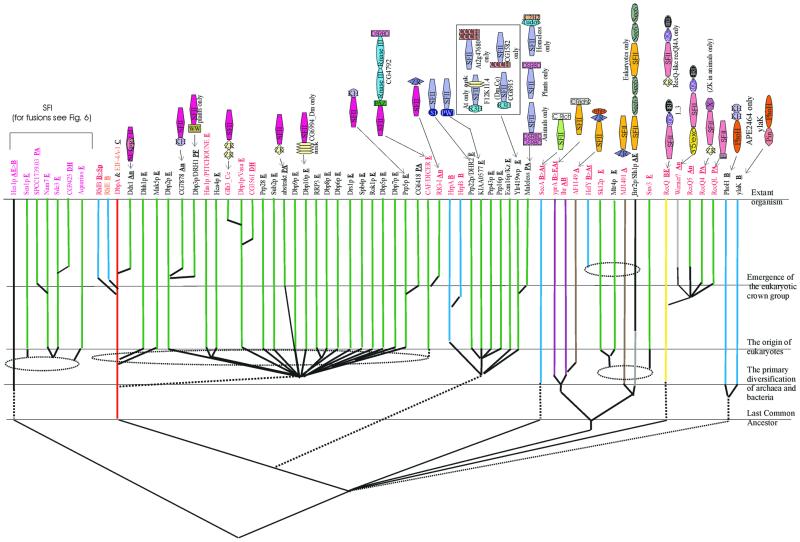
RNA helicases. The next major class of P-loop NTPases that are associated with RNA metabolism are RNA helicases and related ATPases. The known RNA helicases of cellular life forms belong to two major superfamilies, SFI and SFII, that descend from an ancient common ancestor antedating LUCA. This ancestral helicase contained two distinct α/β domains that are present in both SFI and SFII (109). The N-terminal domain is a classic P-loop ATPase domain that belongs to the RecA-like subclass of P-loop domains (110,111). The C-terminal domain appears to represent an extremely divergent P-loop domain that might have evolved through an ancient duplication of the N-terminal domain, followed by extreme sequence divergence, which probably accompanied a functional shift to single-strand nucleic acid binding. The extant lineages of SFI and SFII helicases include both DNA and RNA helicases, and other nucleic acid-dependent ATPases. Among the helicases involved in RNA metabolism, SFII occupies a more prominent position than SFI; SFII helicases are much more prevalent in eukaryotes than in bacteria (Fig. 4). Seven major families of SFII helicases have experimentally characterized or clearly predicted roles in RNA metabolism. Two of these, namely the eIF4A-DeaD family (with the classic DEAD motif in the Walker B site) and the Ski2p-Lhr family, are widespread in all three primary kingdoms, which points to their presence in LUCA. Within the eIF4A-DeaD family, the orthologous group typified by the bacterial DeaD protein, which is involved in translation regulation (112), is widely represented in bacteria and archaea and might be the form closest to the ancestor of this family. In eukaryotes, this family has vastly expanded to include at least 30 distinct lineages, with almost 25 of them traceable to the common ancestor of the crown group (Fig. 4). Most members of this expanded helicase subfamily are subunits of pre-mRNA splicing complexes, whereas some others, such as Rrp3p (113), function in other RNA processing pathways, and Upf1p is involved in mRNA degradation (114). The pan-eukaryotic translation initiation factor eIF4A appears to be the direct equivalent of the prokaryotic DeaD-like helicases, and its function in eukaryotes might be an extension of the ancient role of these helicases in regulatory unwinding of mRNA secondary structure. Proteobacteria have a lineage-specific expansion of the DeaD lineage, with additional orthologous groups, such as RhlE and RhlB (115), whereas most of the other bacteria have only a single member.
Figure 4.
An evolutionary scheme for RNA helicases and related ATPases. The family names are colored according to their function: black, splicing and processing; red, PTGR; orange, translation. The lineages containing DNA helicases are shown in gray. The conventions for color-coding and line patterns are the same as in Figure 2. The domain names/acronyms are as explained in Table 1. Additionally: ank, ankyrin repeats; Crich and Crich2, different lineage-specific cysteine-rich domains; RQC, C-terminal domain of RecQ family helicases; Zr, zinc ribbon; and HRD, the C-terminal domain common to RecQ and RNase D.
The Ski2p-LHR family is a much smaller family whose ancestral form probably was involved in RNA degradation and processing (116,117). Archaea typically have three distinct helicases of this family, whereas eukaryotes have four members of the Ski2p-Mtr4p-like subfamily, all of which apparently function in conjunction with the exosomal nucleases in RNA degradation (Fig. 4). Another eukaryote-specific orthologous group within this family includes Brr2p-like proteins, which contain two helicase and sec63 domains and are involved in both cytoplasmic RNA processing and splicing as a component of U5 snRNP (118). One orthologous group within the Ski2p-LHR family, which is typified by mus308 of D.melanogaster and MJ1401 of M.jannaschii, appears to have been recruited for DNA-related functions in the archaeo-eukaryotic lineage and, in eukaryotes, shows a fusion to a DNA polymerase domain (119,120).
The remaining families of SFII helicases involved in RNA metabolism show purely eukaryotic, bacterial or bacterio-eukaryotic distribution. The Suv3 family involved in mitochondrial RNA degradation (121) and the CAF family involved in PTGS are small groups that are restricted to eukaryotes and appear to function in eukaryote-specific regulatory processes (see below). The Prp2p-Mle subfamily is found in both bacteria and eukaryotes. Eight distinct orthologous groups can be delineated within this family in eukaryotes, with the majority involved in splicing, including Prp2p, Prp16p, Prp43p, Prp22p and Mle (122). The HrpA/B proteins are bacterial representatives of the Prp2p-Mle family that are present only in proteobacteria, spirochetes and Deinococcus, which suggests dissemination via horizontal transfer among bacteria, although the initial direction of horizontal transfer responsible for the bacterio-eukaryotic distribution remains uncertain. The SecA family proteins are ubiquitous in bacteria and plants and have been shown to possess RNA helicase activity (123–125). However, the role of this activity in vivo remains unclear because SecA also has a well characterized function as an ATP-dependent translocase involved in protein secretion. The RecQ family of SFII helicases is unusual in that these proteins have functions in both DNA repair and RNA metabolism. This family is represented only in bacteria and eukaryotes, with a single horizontal transfer into the crenarchaeon A.pernix. This distribution suggests that the RecQ family originally evolved in bacteria and was subsequently acquired by eukaryotes from the pro-mitochondrial endosymbiont, which was followed by extensive diversification into at least five distinct orthologous, eukaryote-specific groups. Many members of this family share a predicted RBD, the HRDC domain (126), with the RNase D family of nucleases, suggesting that the ancestral function of the RecQ family helicases might have been in RNA metabolism, with a subsequent shift to DNA-related functions. A member of this family from Neurospora has been shown to have a role in RNA metabolism, in particular PTGS (127). Orthologs of this protein are present in other eukaryotes; furthermore, fusion of the RecQ family helicases with the Zn-knuckle and the F-box domains in plants and animals (see Figs 4 and 6) indicate that this family might have more extensive RNA-related functions than presently conceived.
Figure 6.
An evolutionary scenario for the PTGR systems. The architectures and names of proteins involved in PTGR are shown attached to the nodes, at which they are inferred to have been derived. Architectures with stand-alone domains and repeats of a single domain are shown by names; architectures are separated by semicolons. In some cases, the details of phyletic patterns are indicated as: PA, plant and animals; AF, animals and fungi; CH, nematodes and vertebrates. When a given architecture is represented in more than one orthologous group, the number of groups is given alongside the name (bold and underlined numbers denote distinct, distantly related groups; plain numbers denote recently diverged paralogous groups). The range of numbers under a repeated domain (example 7..10) denotes the range seen in individual proteins of the corresponding group. The domain names/acronyms are as given in Table 1. Additionally: S1L, S1-like; MI, MA-3 and eIF4G; Cr3, cysteine-rich domain with three cysteines; Crich, Crich1, Crich2, different lineage-specific cysteine-rich domains; RQC, C-terminal domain of RecQ family helicases; ank, ankyrin repeats; SAM, S-adenosylmethionine-binding domain; TPR, tetratricopeptide repeats; FHA, fork head-associated domain; RNA Lig, RNA ligase; F, F box; M-bet-lac, metallo-β-lactamase; DHH, hydrolase of the DHH family; Corn, cornase domain; HD, hydrolase of the HD superfamily; ZR, zinc ribbon; 2C, an uncharacterized domain with two conserved cysteines; X, X1, different lineage-specific uncharacterized domains. The boxes are marked: C, ancient conserved (universal); A, archaea; B, bacteria; AE, archaeo-eukaryotic; E, eukaryotes; P, plants; CDH, animals; F, fungi; Ce, nematodes; D, arthropods; H, vertebrates; and DH, arthropods and vertebrates.
Several SFI helicases are implicated in RNA-related functions in eukaryotes; they all belong to the Smubp-Sen1p family, which is conserved throughout the archaeo-eukaryotic lineage and in a few bacteria (128). This family includes both DNA and RNA helicases and probably emerged early during the evolution of the archaeo-eukaryotic clade, rather than in LUCA. All archaeal members of the Smubp-Sen1p family are orthologs of the eukaryotic Smubp, which is a DNA-binding protein (129). However, the presence of the single-stranded nucleic acid-binding R3H domain (15) in some of the eukaryotic members of this family might point to an undiscovered role in RNA metabolism (see Figs 4 and 6). All known eukaryotic SFI RNA helicases of the Smubp-Sen1p family were derived after the divergence of eukaryotes from the common ancestor with archaea (Fig. 4). Five distinct lineages of RNA helicases of this family emerged prior to the divergence of the crown group eukaryotes and include proteins involved in a variety of functions, such as snoRNA maturation [Sen1p (130)], mRNA degradation [Nam7p (131)] and PTGS [Sde3 (132)] (Fig. 4). One of the eukaryotic SFI lineages, represented by the S.pombe SPCC1739.03 and its orthologs, is closely related to the NAM7p subfamily and is an uncharacterized group of predicted RNA helicases, which, on the basis of their phyletic pattern (133), are likely to participate in PTGS (Fig. 4). Another distinct pan-eukaryotic family, typified by the Aquarius protein (134), is predicted to include inactive helicases as indicated by the disruption of the P-loop and Walker B motifs; these proteins probably function as RNA-binding regulators, rather than as enzymes. Two small, lineage-specific expansions of these helicases were detected in Arabidopsis and C.elegans, typified by the F1E22.14 (eight members) and K08D10.5 (six members) proteins, respectively; these might represent specific adaptations for antiviral response or related processes. Most of the other SFI families, such as the RecD family, appear to have evolved in bacteria and are known to be involved only in DNA repair and recombination (119).
The PhoH family of ATPases (135) evolved in bacteria, apparently through the loss of the C-terminal α/β domain that was present in the common ancestor of SFI and SFII helicases. A role in RNA metabolism is strongly suggested by the presence of RNA-binding PIN and KH domains in different members of the PhoH family (Fig. 4). There are two orthologous groups of PhoH-like ATPases, typified by PhoH and YlaK, respectively, that evolved as a result of an early duplication in the bacterial lineage. The PhoH proteins could either function as helicases or could be involved in ATP-dependent dynamics of as yet uncharacterized RNP complexes in bacteria.
Miscellaneous P-loop NTPases involved in RNA metabolism. In addition to the above, well characterized classes of P-loop NTPases involved in RNA metabolism, several others have less common and less thoroughly understood RNA-related functions. The most notable of such groups includes the PilT ATPases, which form a distinct class within the P-loop fold and appear to be a sister group to the ABC class (D.D.Leipe, E.V.Koonin and L.Aravind, unpublished data). The PilT ATPases implicated in RNA metabolism appear to be predominantly an archaeal innovation and are typified by MJ1533 and its orthologs that are highly conserved in archaea (136). These proteins combine the PilT ATPase domain with RNA-binding PIN and KH domains. In bacteria, a group of PilT ATPases is present sporadically in Bacillus and Synechocystis and form fusions with the RNA-binding R3H domain. These ATPases might represent a novel class of RNA helicases or could participate in other ATP-dependent reactions of RNA metabolism.
Some kinases of the P-loop fold, such as polynucleotide kinases, also participate in RNA metabolism. A generic polynucleotide kinase that probably acts on both DNA and RNA seems to be conserved in all eukaryotes except for S.cerevisiae (137–139). Additionally, some lineage-specific P-loop kinases are implicated in RNA metabolism on the basis of suggestive domain fusions, including the kinase fused to yeast RNA ligase (140,141) and the animal-specific hnRNP-U (SAF-A) proteins, which contain a SAP domain, and might function as chromatin-bound polynucleotide kinases in pre-mRNA splicing (142,143). P-loop kinase domains are also fused to the ligase-related nucleotidyltransferase domains of the capping enzyme in trypanosomes (144).
The P-loop proteins of the MiaA family modify adenines, chiefly in tRNAs, through the addition of bulky adducts, such as isopentene, in position 6, using organic phosphates, e.g. dimethylallyl diphosphate, as donors of the modifying groups (145,146). These enzymes are distantly related to the AAA+ class of P-loop ATPases and are nearly ubiquitous in bacteria and eukaryotes, which is consistent with the phyletic pattern of 6-isopentenyl adenines in tRNA. MiaA probably evolved in the common ancestor of bacteria and was acquired by eukaryotes from the promitochondrial endosymbiont. On the basis of operon organization, it can be predicted that, at least in certain bacteria, such as proteobacteria, Aquifex and Synechocystis, MiaA utilizes the Hfq protein (the bacterial homolog of the eukaryotic SM proteins) as an RNA-binding subunit.
Other enzymes of RNA metabolism
At least 15 superfamilies of RNases are involved in a variety of processes, such as maturation of tRNAs and rRNAs, polyadenylation site-specific cleavage of mRNAs, and RNA degradation in various contexts and cellular compartments. A detailed evolutionary classification of RNases has been published recently (45), and therefore individual groups of these enzymes are not discussed here in detail. However, we cover some specific aspects of their evolution when reconstructing the evolution of individual functional systems in RNA metabolism (see below).
In addition, a number of other enzymes that form relatively small families, sometimes with restricted phyletic distribution, are involved in RNA metabolism. One such group is the RNA ligases that are related to the DNA ligases and appear sporadically in cellular life forms. The fungi possess a RNA ligase, which is required for the maturation of tRNAs and non-spliceosomal mRNA maturation (147), whereas in trypanosomes RNA ligases participate in mRNA editing (38,148). Homologs of these RNA ligases are encoded by several DNA viruses, including phage T4, baculoviruses and entomopox viruses (38). This observation, together with the sporadic distribution of RNA ligases, might suggest that cellular organisms acquired these enzymes independently from DNA viral sources. Additionally, a variety of other nucleotidyltransferases are involved in non-templated polymerization of ribonucleotides during polyadenylation of mRNAs, CCA addition in tRNAs and RNA editing. All these enzymes have the DNA polymerase β-fold (149) and are considered in greater detail below in the context of evolution of the capping and polyadenylation systems.
Cyclic phosphodiesterases of the LigT superfamily hydrolyze 2′-5′ phosphoesters in various contexts in RNA metabolism. The most conserved of these enzymes form the core LigT family, which apparently evolved in the archaeo-eukaryotic lineage, with a few transfers into bacteria; the animal members of this family have a fusion with the RNA-binding KH domain. They apparently catalyze hydrolysis of ADP-ribose 1″,2″-cyclic phosphate that is formed as an intermediate in tRNA processing (150,151). Additional members of this superfamily, which are not orthologs of LigT, were identified as fusions with RNA ligases in yeast, in RNA viral polyproteins, and as stand-alone proteins in Arabidopsis (L.Aravind and E.V.Koonin, unpublished observations); these proteins might have related phosphodiesterase activities in RNA metabolism.
The Macro domain (first detected in vertebrate macrohistone 2) is another highly conserved phosphoesterase that is involved in Appr-1″-p-processing (152), as part of tRNA maturation. Macro domain phosphoesterases are conserved across the three superkingdoms of life, which is compatible with the presence of such an enzyme in LUCA. Finally, several families of enzymes, such as the enigmatic RNA-dependent RNA polymerases (153–156), and AlkB-like oxoglutarate-dependent dioxygenases (157), show a limited phyletic distribution. Most of these are known or predicted components of the eukaryotic post-transcription regulatory systems and are further explored below in the context of evolution of these functional systems.
Evolutionary history and trends of non-catalytic domains involved in RNA metabolism
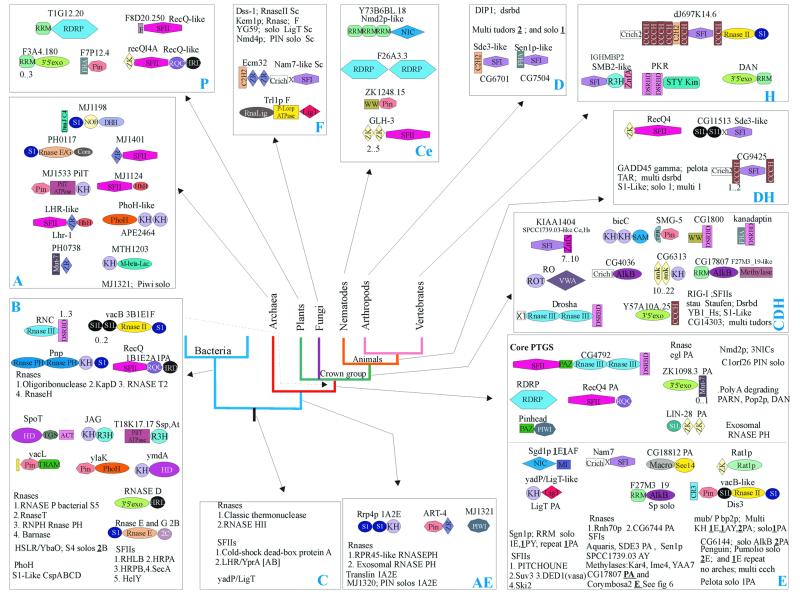
Approximately 50 major superfamilies of non-catalytic domains, primarily RNA-binding ones, are implicated in RNA metabolism (Fig. 1A and B and Table 1). In addition, several conserved domains are found exclusively in ribosomal proteins. Below we consider some of the general and specific features of the natural history of these domains that emerge from a detailed analysis of their phyletic patterns combined with attempts on evolutionary classification.
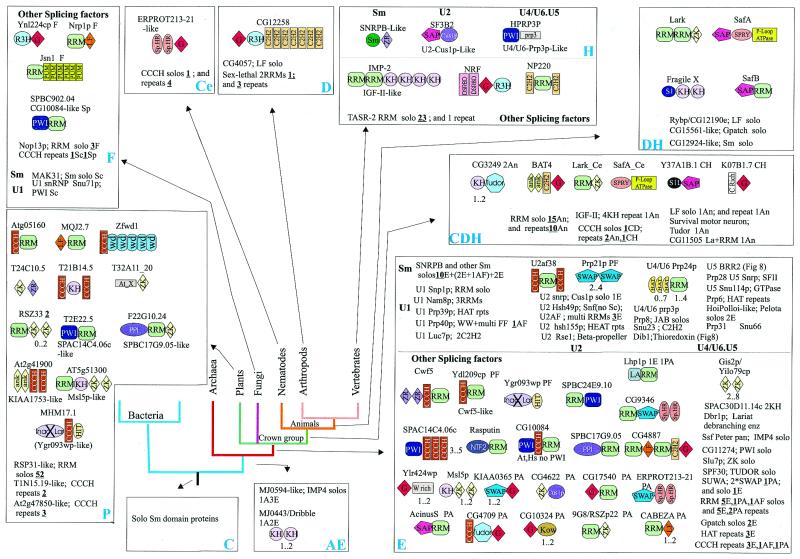
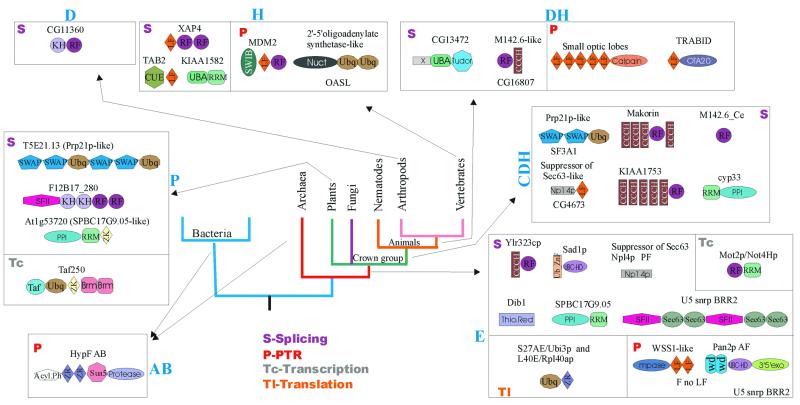
Evolutionary mobility of domains. RBDs show remarkable diversity in terms of domain architectures. Several RBDs, such as ribosomal protein L30 and the SRP14-domain, typically occur as stand-alone proteins and in a single copy per genome. At the other end of the spectrum are ‘promiscuous’ domains, such as RRM, which display over 35 distinct multidomain architectures and are found in combination with up to 20 different domains (Figs 5–7). These observations suggests major differences in evolutionary mobility among RBDs. Certain highly conserved, ancient RBDs, such as L30, S6 and SmpB, appear to have largely stabilized in specific functional niches in the ribosome or in lineage-specific RNP complexes and are not typically recruited to roles in more general contexts related to RNA metabolism. In contrast, some other conserved domains found in ribosomal proteins, such as S1 (158), KOW (13) and S4 domains (9), have been recruited for a variety of other functions which involve RNA binding. Some of these domains (KOW, S4), along with other mobile RBDs, such as EMAP, PUA, PIN, TRAM, THUMP, TGS, N-OB, NusB (9–12,71,136) and several conserved domains found in aaRS (10), form a group of moderately mobile, ancient domains. The majority of the fusions that involved these domains appear to have evolved close to the origin of one of the superkingdoms or, in some cases, even in or prior to LUCA. Most of these architectures show remarkable parallelism of fusions of different RBDs to various RNA modification and processing enzymes. It appears that these RBDs emerged at early stages of evolution and, shortly after their origin, formed fusions that facilitated the delivery of diverse catalytic activities to RNA and hence were maintained in most lineages. These moderately mobile domains formed lineage-specific fusions on relatively rare occasions, such as those of N-OB and EMAP to the C-termini of plant and vertebrate TyrRS, respectively (10), or the fusion of TRAM to a FtsJ-like methylase in Thermoplasma (11).
Figure 5.
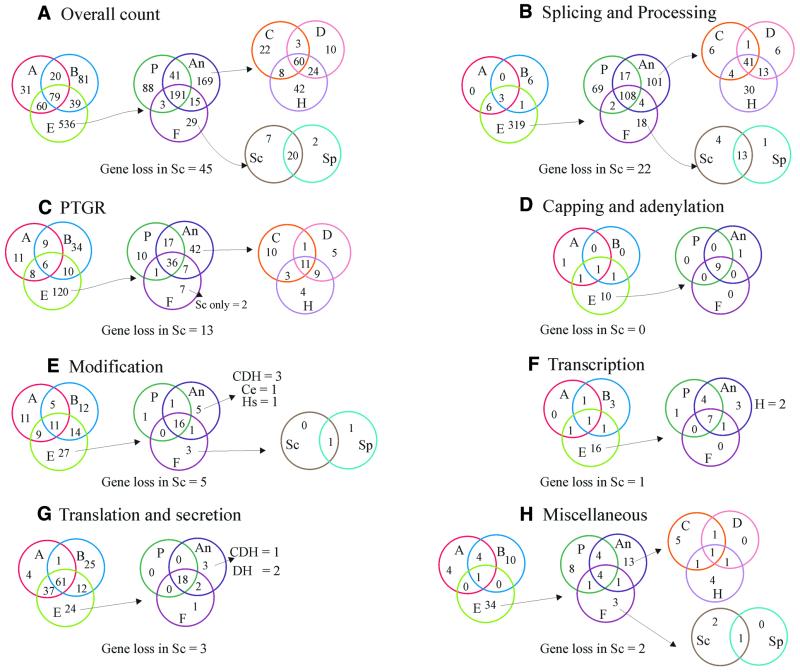
A Venn diagram of phyletic patterns of RNA metabolism systems. The number of orthologous groups of proteins detected in each lineage is shown according to their functions. Each number in a given compartment of a Venn diagram is exclusive of the numbers in the other compartments. The number in the intersection of two circles is the number of orthologous groups shared by the two lineages (e.g. 20 groups in the AB compartment of the overall counts represents the 20 orthologous groups shared by the archaeal and bacterial lineages), whereas the intersection of three circles shows the number of orthologous groups shared by three lineages. A, archaea; B, bacteria; E, eukaryotes; P, plants; An, animals; F, fungi; C, C.elegans; D, D.melanogaster; H, H.sapiens.
Figure 7.
Architectural diversity and points of origin of the splicing machinery components. The conventions for the representation of architecture, points of derivation and lineage abbreviations are as in Figure 6. The domain names/acronyms are as given in Table 1. Additionally: G, G-Patch domain; WW, conserved domain with two characteristic tryptophans; Sp.Ht., specialized HEAT repeats; WD, WD40 repeats; Crich, lineage-specific cysteine-rich domain; At_X, an uncharacterized domain expanded in Arabidopsis; ‘Inac. Lar.’, inactive lariat-debranching enzyme; ZR, zinc ribbon; HIT, histidine triad domain.
The next major phase of domain mobility coincided with the emergence of eukaryotes and continued through the divergence of the major eukaryotic lineages. This burst of mobility correlates with the origin of splicing and other post-transcriptional regulatory mechanisms in eukaryotes. Some of these domains, such as S1, dsRBD (159) and KH (160,161), were already present in LUCA as parts of ubiquitous ribosomal proteins or enzymes. These domains went through an initial phase of moderate evolutionary mobility, but experienced a new spurt of mobility in eukaryotes, each giving rise to several new architectures associated with splicing and other post-transcriptional regulatory processes. However, most of the domain shuffling events in eukaryotes involve relatively new, eukaryote-specific domains, such as RRM, Zn-Knuckle, CCCH, Little Finger, G-patch, SWAP and PWI.
Differential utilization of some ancient RBDs and high mobility of the eukaryote-specific domains point to two distinct evolutionary forces involved in the emergence of the complexity of eukaryotic RNA metabolism. First, it appears that most of the ancient, moderately mobile RBDs were not sufficiently versatile to occupy the new functional niches, such as splicing and PTGR. Exceptions include several ancient mobile domains, such as S1 and KH; proteins containing these domains in eukaryote-specific architectures have undergone lineage-specific expansions, which indicates greater functional versatility and adaptation to some of the new functional niches. These domains, however, largely formed combinations amidst themselves or with catalytic domains, akin to their more ancient versions, rather than with more recently invented domains. Secondly, the newly invented domains appear to have been recruited en masse to the new, eukaryote-specific functions close to the points of origin of these functions. Thus, through an evolutionary feedback process driven by duplication and repeated selection for the same set of newly derived domains, they started rapidly colonizing the new functional niches to the exclusion of the older, moderately mobile RBDs. This strong selection favoring the proliferation of the recently evolved, mobile domains also appears to have resulted in architectures that most frequently involved combinations among themselves rather than with the less common, ancient RBDs.
A brief history of major families of RBDs. The specific evolutionary histories of the common RBDs are important for understanding the emergence of the functional systems that comprise cellular RNA metabolism. Below we briefly consider the main events in the diversification of major RBD families.
OB-fold and other all-β strand domains. The OB-fold is a six-stranded β-barrel, which is common to several superfamilies of nucleic acid-binding domains. Among the domains involved in RNA metabolism, the S1, S1-like, EMAP, N-OB and thermonuclease domains adopt the OB fold (55,71,158). Most of these domains were already represented in LUCA, which indicates that a major phase of divergent evolution of OB-fold domains took place at even earlier stages of evolution. Several of the OB-fold domains are seen in proteins that have been conserved throughout evolution as central components of the translation system. Ribosomal protein S12 and initiation factor IF1/eIF1A are the most conserved orthologous groups of S1-domain proteins, each traceable to LUCA. In addition, several conserved versions of the S1 domain are present in ribosomal protein S1, RNase E, RNase II, polynucleotide phosphorylase, the circularly permuted GTPases of the YjeQ family, Tex and NusA, all of which are (nearly) ubiquitous in bacteria and probably evolved at the onset of bacterial evolution. Conversely, the forms of the S1 domain present in eIF2-α, RpoE and Rrp4p/Rrp40p exosomal subunits go back to the base of the archaeo-eukaryotic clade. The Rrp5p and Prp22 lineages of S1 domains evolved in eukaryotes, whereas the SPT5p family appears to have evolved in eukaryotes, from a Tex-like ancestor that was acquired from bacteria. ‘S1-like’ domains belong to a lineage that is of bacterial origin and is represented by orthologous groups, such as the major cold shock protein (CspA), RNase II and transcription terminator Rho. Another OB-fold domain related to the S1 domains is the C-terminal domain of the universal translation factor EF-P/eIF5A,which appears to have branched off from all the other S1 domains prior to LUCA and has not shown any evolutionary mobility ever since.
The most ancient form of the EMAP domain seems to be the one in methionyl-tRNA synthetase (10), which is widely distributed throughout all three primary kingdoms. Additionally, a duplication at the base of the bacterial clade gave rise to the EMAP domain in the β-subunit of PheRS. Similarly, the most ancient lineage of N-OB domains (162) is the one that is present in AspRS; this domain underwent duplications to give rise to the forms present in LysRS and AsnRS in bacteria and eukaryotes, respectively. Other N-OB domains appear to have been recruited widely in various DNA metabolism enzymes, which suggests exaptation of an ancient RBD for DNA binding (162).
The SH3-like barrel (163) is another all-β fold, which is present in several non-catalytic domains involved in RNA metabolism, such as the KOW, SM, L21E, L2 and tudor domains. The KOW domain present in the ribosomal protein L24, NusG/Spt6 and EF-P/eIF5A evolved prior to LUCA and the KOW-domain-containing proteins have largely retained their architectures ever since. The eukaryotic ortholog of NusG, Spt6, contains four or five divergent copies of the KOW domain, apparently resulting from a previously undetected amplification. The SM domain (164–166) also appears to have been present in LUCA, although it seems to have been subsequently lost in several bacterial lineages. This domain is unusual in that it always occurs as a stand-alone protein, suggesting selection against the formation of multidomain architectures, the underlying cause of which remains unclear. Prokaryotes encode one or two SM-domain proteins, whereas, in eukaryotes, 16 distinct orthologous groups of SM proteins already evolved prior to the radiation of the crown group, which is consistent with large-scale recruitment of this domain to snRNP complexes involved in splicing. The L2 domain seen in the universal ribosomal protein L2 is an orphan version of the SH3-like barrel fold that might have been derived from the ancient KOW superfamily, with subsequent extreme sequence divergence. Similarly, the L21E domain of the archaeo-eukaryotic lineage might be a divergent derivative of the more universal superfamilies of the SH3-like fold. The TUDOR domain (167) is also related to the SM and L2 domains and appears to have been derived from one of them in eukaryotes. Several members of the tudor superfamily appear to have lost the RNA-binding function and participate in protein–protein interactions in the splicing snRNP complexes (168); some divergent versions even function in chromatin structure maintenance (L.Aravind, unpublished observations). At least four distinct orthologous groups of proteins containing TUDOR domains with functions in RNA, such as Drosophila TUDOR itself and its orthologs and the splicing factor SPF30, had evolved prior to the radiation of the eukaryotic crown group. A few additional TUDOR-containing proteins emerged in animals, such as the Drosophila RNA helicase Homeless and the SMN-like proteins.
The PUA domain and the TRAM domain are two other RBDs that are confidently predicted to have an all-β fold (9,11); however, they cannot be classified with any of the folds discussed above in the absence of a 3D structure. Both PUA and TRAM are ancient, moderately mobile domains that typically are associated with various RNA-modifying enzymes (Table 1 and Figs 2 and 3). Eukaryotes and bacteria have some lineage-specific architectures of PUA domain-containing proteins, such as the fusion with another RBD, SUI1, in eukaryotes and an unexpected, conserved fusion with glutamate kinase in bacteria. The TRAM domain is principally associated with the ancient MiaB-like enzymes (see above) and is also fused to the predominantly archaeo-eukaryotic TRM2-like methylases.
Major RBDs with α/β and α + β folds. The RRM, which is the most prevalent RBD in eukaryotes and is involved in all aspects of RNA metabolism, is a eukaryotic invention. At least 40 distinct orthologous groups of RRM-containing proteins appear to have emerged prior to the radiation of the eukaryotic crown group and, additionally, several more orthologous groups are confined to animals, plants or fungi. This explosive diversification of the RRM domain is surprising given the absence of this domain in archaea and bacteria, except for a few occurrences, which probably are horizontal transfers from eukaryotes. The RRM domain belongs to an ancient fold of nucleic acid-binding domains, which is present, for example, in ribosomal protein S6 (75) and also in the catalytic domains of a variety of enzymes, including RNA and DNA polymerases and Type II PSUS (55) (L.Aravind, unpublished data). It appears most likely that the RRM domain proper has been derived from a S6-like ancestor at an early stage of eukaryotic evolution.
Several other α/β and α + β domains, such as KH, dsRBD and THUMP (Table 1), have ancient representatives among ribosomal proteins or RBDs of conserved RNA-modifying enzymes. The lineage-specific orthologous groups of proteins containing these domains appear to have evolved through duplication and diversification of these ancient lineages. The TGS domain and the S4 domain that have a distinct α + β fold, called the α-L fold (169), appear to have diverged from a common ancestor and become distinct lineages prior to LUCA.
All α-helical domains. A distinct version of the helix–hairpin–helix (HhH) domain, which is typified by the RBD of ribosomal proteins S13/S18, is ubiquitous in all three primary kingdoms and may represent one of the most ancient lineages of the HhH domains (170). This domain was subsequently sporadically recruited to RNA metabolism, e.g. in the NusA and Tex-Spt6 families, but is far more prevalent in DNA-binding contexts. Thus, this might be another case of an ancient RBD, which diversified extensively only after recruitment for DNA binding.
The PIN domain is another predominantly α-helical domain found in proteins, which, in eukaryotes, are associated with PTGR and RNA degradation (136,171). Stand-alone PIN-domain proteins are widespread across all three primary kingdoms, with distinct architectures in the form of fusions with PilT and PhoH ATPases conserved, respectively, in archaea and in bacteria. A protein containing a PIN protein and a Zn-ribbon domain (human ART-4 orthologs) is conserved in the archaeo-eukaryotic lineage, whereas eukaryotes additionally have a unique architecture of PIN fused to RNase II and TPR repeats. These domain fusions suggest that PIN domains perform a wide range of functions and experimental analysis of PIN-domain proteins might unravel new facets of RNA metabolism. An enigmatic aspect of the evolution of the PIN domains is the expansion of the stand-alone versions of these domains in archaea, such as Archaeoglobus and Aeropyrum, and bacteria, such as Mycobacterium and Synechocystis. These PIN domains potentially might be involved in some unusual regulatory mechanism or in defense against RNA viruses. It has been hypothesized, on the basis of limited similarity to 3′-5′ exonucleases of the RNase H fold, that PIN domains, particularly those involved in RNA degradation in eukaryotes, might have exonuclease activity (171). However, the proposed catalytic residues are not conserved in all PIN domains and a nuclease activity appears unlikely at least for the expanded prokaryotic forms.
The translin domain is an α-helical RBD that is found in a single copy in archaea and in two copies in eukaryotes. The eukaryotic translin protein might be part of a cytoplasmic RNP complex that mediates localization or tethering of mRNAs (172). Given the conservation of this protein in archaea, it seems that these RNP complexes have an ancient function in maintaining RNA stability. As discussed above, several α-helical superstructure-forming domains, such as PUM, HAT [a specific version of the TPR repeat (173)] and NIC, have been recruited for functions related to RNA metabolism in eukaryotes.
Metal-chelating domains. Of the large number of mobile metal-chelating domains that are utilized in RNA metabolism, only the Zn-ribbon (44) (ZNR) is of ancient provenance. The ZNR is a four-stranded domain stabilized by a metal atom typically chelated by four cysteine side chains (sometimes replaced by histidines). The ZNRs function as RBDs and DNA-binding domains and as cofactors in redox reactions, and are also involved in structural stabilization of various proteins (44). The ZNRs in MetRS, IleRS and ribosomal protein S14 are traceable to LUCA. Several ZNRs in translation-associated proteins, such as L40A, L36AE, S27, eIF5 and eIF5β, are conserved throughout the archaeo-eukaryotic lineage, whereas many others are specific to archaea and some to bacteria. This is indicative of massive recruitment of ZNRs during the emergence of the archaeal clade, which might correlate with the iron respiration typical of archaea (174).
The Zn-chelating RBDs that evolved in eukaryotes include the Zn-knuckle with a C2HC pattern of metal ligands, the CCCH domain (named after its conserved chelating cysteines), the little finger with a C4 metal-binding pattern and characteristic conserved tryptophan, the LRP1 finger and the classic C2H2 Zn-finger. There are approximately 12 orthologous groups of proteins containing Zn-knuckles, 13 groups of proteins with CCCH domains and three groups with Little Fingers that are conserved throughout the eukaryotic crown group. All these domains are highly mobile and several lineage-specific fusions of ancient or recently derived proteins to these domains were detected. This suggests a burst of proliferation in early eukaryotes resulting in the establishment of the major orthologous groups, followed by sporadic duplications in individual lineages.
The LRP1 finger is a previously undetected domain that we identified as part of this study. LRP1 has a C6H ligand pattern, which suggests chelation of two metal ions. In animals, this domain is fused to the dihydrouridine synthase Dus1p (Fig. 3), whereas in plants, it has undergone a lineage-specific expansion, with at least 10 stand-alone members, including the namesake LRP1 protein (175). The classic C2H2 Zn-finger is typically associated with DNA binding in eukaryotes and is part of numerous transcription factors and chromatin-associated proteins. However, several members of this family are associated with known or predicted RBDs, e.g. in the experimentally confirmed RNA-binding proteins TFIIIA and dsRBP-Zfa (JAZ) (176–178). However, no distinct sequence features or specific phylogenetic relationships of the RNA-binding versions of this domain were detected so far, making it impossible to predict the fraction of C2H2 fingers in eukaryotic proteomes that have RNA-related functions. We only documented those occurrences where the evidence was sufficiently clear from either experimental data or association with other specific RBDs. This is likely to represent the lower boundary of the C2H2 fingers involved in RNA metabolism.
Evolutionary history of RNA metabolism systems and reconstruction of their ancestral states
Analysis of evolution of individual domain families, a summary of which is presented above, provided a means of reconstructing the evolutionary history and probable ancestral states of the numerous functional systems, pathways and protein complexes that comprise RNA metabolism. We summarize below the results of this reconstruction, which is based on the data gathered for principal conserved domains involved in RNA metabolism. Figure 5 is a Venn diagram that shows the numbers of conserved orthologous groups of proteins shared by various lineages across the entire phylogenetic spectrum that we sampled, for various functional systems.
The ancient core: translation, transcription and RNA modification. Comparative genomics showed that the basic translation apparatus contains the largest number of (nearly) universally conserved proteins. The set of translation-associated proteins whose origin is traceable to LUCA and possibly beyond includes 15 proteins associated with the small subunit of the ribosome, 18 proteins associated with the large subunit, nine class I aaRS, seven class II aaRS, seven GTPases associated with various aspects of translation, and at least two other translation factors. Other ancient proteins associated with translation are the glutamate (aspartate) amidating enzyme subunits, which are necessary for glutamine (and in some cases, asparagine) incorporation into proteins in most bacteria and archaea (179), the signal recognition particle GTPases that form the link between translation and secretion, possibly a SFII helicase associated with translation regulation or initiation, and a variety of RNA-modifying enzymes (Table 2). The modification enzymes that could be confidently traced back to LUCA include two distinct classes of methyltransferases with six to seven representatives altogether, two classes of pseudouridine synthases, and enzymes involved in the synthesis of thiouridine and thioadenine derivatives and 7-deazaguanosines. Thus, LUCA possessed an abbreviated protein core of the modern ribosome and the basic repertoire of accessory proteins required for translation. From this pivotal point, it is possible both to track back the early, pre-LUCA stages in the evolution of RNA metabolism and to examine its elaborations in the major clades of life.
Table 2. Proteins involved in RNA metabolism that are traceable to the LUCA of the extant life forms.
As pointed out above for individual domains, many components of the ribosome, translation factors and RBDs of RNA-modifying enzymes, which are traceable to LUCA, descended from even more ancient common ancestors. Numerous ribosomal proteins and other translation/modification-associated RBDs in the ancestral set belong to a small number of folds, such as OB-fold, SH3-like barrel and the α-L fold. Thus, prior to the divergence of the S1, N-OB and EMAP domains, or the KOW, SM and L2 domains, or the TGS and S4 domains, their respective ancestors probably functioned as RBDs with generic properties. The same logic applies to enzymes of RNA metabolism. The case is particularly clear for aaRS, which are indispensable components of the modern translation machinery responsible for the specificity and efficacy of amino acid incorporation into protein. Since most of class I and class II aaRS were already present in LUCA, there is obviously a history of pre-LUCA duplications in each of the classes (102). The ancestral aaRS of each class, which functioned in the primitive translation system, most likely was a non-specific amino acid-activating enzyme, with the specificity determined by tRNAs themselves. This type of translation system appears to be a transition state between a primordial machinery based entirely on RNA catalysts and the modern, largely protein-based system. Furthermore, the catalytic domains of both classes of aaRS are homologous to certain other NTPases and nucleotidyl transferases, whose functions are unrelated to translation; some of these, for example, are enzymes of coenzyme biosynthesis, such as NAD synthase in the case of class I (102) and biotin synthase for class II (180). Thus, the progenitors of the two classes of aaRS, which evolved from within the primitive RNA world, probably were non-specific nucleotidyl transferases, which combined functions in translation with those in other branches of metabolism. Similarly, at this stage of evolution, the individual translation factors and RNA-modifying enzymes, such as methyltransferases, had probably not yet differentiated into their specific versions, but were represented by the corresponding ancestral forms, which functioned in multiple contexts with a low specificity.
Looking forward from LUCA, it is immediately apparent that several major additions to the translation apparatus and its accessories map to the point of divergence of the two principal branches of life, the bacterial and the archaeo-eukaryotic clades. Approximately 28 proteins were added to the ancestral ribosomal core in the archaeo-eukaryotic lineage and, conversely, 21 ribosomal proteins are specific to the bacterial lineage, which results in the profound differences in the ribosomal superstructure between the two clades. The translation termination factors and several initiation factors also were added to the conserved set as these major lineages diverged. Eukaryotes showed a further development in the complexity of the translation initiation system: several new translation regulators emerged in the eukaryotic lineage, some of which consist of the RRM domain or newly derived α-helical domains, such as NIC, MI and W2 (16), whereas others have new combinations of ancient RNA-binding and enzymatic domains, such as PUA, SUI1 and SFII helicases. The complexity of RNA modification also increased during the post-LUCA phase of evolution as a result of several duplications within various enzyme families and the origin of several new enzymes, such as dihydrouridine synthetase and MiaA (Figs 2 and 3). Most of the RNA modification enzyme superfamilies, in addition to the highly conserved groups of orthologs, include many smaller groups, which are restricted to a specific lineage or show a sporadic distribution (Figs 2 and 3). Thus, a subset of RNA modifications, while not universally essential, are likely to have specific adaptive value for particular organisms in their ecological niches. These adaptations might include tolerance to extreme environmental conditions, such as high temperature or osmolarity, or resistance to anti-translation antibiotics or particular xenobiotics. The relatively late emergence of many RNA modifications suggests that the RNA modification state in LUCA and especially at earlier stages of evolution was relatively simple and therefore these modifications might not have been a major factor in modulation of the catalytic activities of primordial ribozymes.
Several RNA-binding proteins contribute to transcription. The best-studied proteins in this category are the transcription elongation/antitermination factors that include the universally conserved NusG-Spt5p family of KOW-domain proteins (181). Bacteria additionally possess several distinct subunits of the transcription antitermination complex, including NusB, which contains the prototype of the α-helical NusB domain, ribosomal protein S4 and the S1 and KH domain-containing protein NusA (182–184). The functionally equivalent eukaryotic transcription elongation complex contains Spt6 (185,186), which is the ortholog of the bacterial Tex protein (187). Similarly to NusA, this protein contains an S1 domain and is likely to be the functional counterpart of NusA. In animals, this complex additionally contains the RRM-containing RD protein (188). The ancestral form of the transcription elongation/antitermination complex, which was present in LUCA, might have consisted of a single KOW-domain protein and perhaps the ribosomal protein S4. This was followed by accretion of additional subunits, at least in bacteria. Bacteria also evolved transcription antiterminators containing the α-helical AmiR domain that relieve specific mRNAs from termination in response to stimulation of specific signaling pathways that lie upstream of them (Table 1) (189). The corresponding additions in archaea, if any, remain unknown, but in eukaryotes, SPT6, apparently acquired from bacteria via horizontal transfer, was recruited to this complex, followed by other lineage-specific additions.
The archaeo-eukaryotic RNA polymerase E1 subunit containing the S1 domain and eukaryotic transcription factors EWS/TAF68 and TAFII250 containing the Zn-knuckle domain are other transcription-related RNA-binding proteins. Fusion of the SAP domain with RBDs (143) suggests that eukaryotes might have still uncharacterized RNP complexes, which could couple nuclear RNA processing with transcription. Finally, in animals, several chromosomal RNAs, such as RoX1/2 and XIST, have been described that have a role in regulating chromosomal structure, and thereby transcription, on a global scale. A specific class of Chromodomains typified by the MSL proteins (190) and other proteins, such as the SFII helicase Mle (122), interact with these RNA molecules.
Polyadenylation and capping. Polyadenylation occurs in all three primary kingdoms. Prokaryotic poly(A) tails are short (~30 nt) compared with the eukaryotic ones, which extend to several hundred nucleotides (191). Bacterial poly(A) polymerases also have CCA-adding activity and are often fused to HD or DHH phosphohydrolase domains (149). The eukaryotic Poly(A) polymerases are only distantly related to the bacterial versions and, instead, are more closely related to the Trf4/5 family of eukaryotic DNA polymerases and archaeal CCA-adding enzymes (149), suggesting that these archaeal enzymes probably have a second function as Poly(A) polymerases. In eukaryotes, the free 3′ end for the Poly(A) polymerase is generated by a predicted nuclease of the metallo-β-lactamase fold, CPSF-I (192–194). This enzyme is conserved throughout the archaeo-eukaryotic lineage and is also present in many bacteria. Thus, LUCA probably had a polyadenylation system that consisted, at least, of a CPSF-I-like enzyme that cleaved the transcript and a polymerase β family nucleotidyltransferase that added the adenylates. The reasons for the rapid evolution of the poly(A) polymerases in each of the three primary kingdoms are unclear. It seems plausible that, in eukaryotes, the displacement of the CCA-adding function by a horizontally transferred bacterial enzyme resulted in the divergence of the poly(A) polymerase from the ancestral, bifunctional form seen in the archaea. Eukaryotes additionally recruited to the CSPF complex several new RNA-binding proteins containing eukaryote-specific domains, such as RRM, CCCH and Zn-knuckle. Furthermore, RRM and NIC-domain-containing proteins were recruited to form a eukaryote-specific poly(A) tail-binding complex.
The cap is a unique structure present in eukaryotic mRNAs; the minimal form of the cap is synthesized through the following steps: (i) removal of the terminal phosphate of the triphosphate at the 5′ end of mRNA, (ii) guanylylation of the 5′ diphosphate and (iii) methylation of the guanine at the N-7 position (195). The first two steps are catalyzed by the capping enzyme, which consists of a triphosphatase and a nucleotidyltransferase, whereas the N-7 methylation is catalyzed by methylases of the Abd1p family (196). The enzymes that catalyze the latter two capping reactions appear to be conserved throughout the eukaryotes. The capping guanylyl transferase apparently was derived from the more ancient ATP-dependent DNA ligase (38,39), whereas the capping methylase probably evolved from within the vast small-molecule methylase class, rather than from the regular, monophyletic RNA N-methylases (see above). The capping triphosphatase, however, shows great variability among eukaryotes. Animals and plants share a triphosphatase of the tyrosine phosphatase superfamily that is fused to the N-terminus of the guanylyl transferase (197,198). The fungi and Plasmodium falciparum contain a distinct phosphoesterase of an all-β fold, which occurs as a stand-alone subunit and is also present in large DNA viruses, such as PBCV (199). The earlier branching trypanosomes have a phosphoesterase domain of the P-loop-containing adenylate kinase family fused to the N-terminus of the guanylyl transferase (144). This unusual diversification of the triphosphatase domain suggests that, whereas the capping methylase and guanylyl transferase were derived early in eukaryotic evolution, there was no specific triphosphatase at the corresponding stage of evolution. Instead, the triphosphatase reaction might have been performed by a non-specific phosphatase. Subsequently, in each lineage, an independent triphosphatase appears to have been recruited for this function. We found that the animal-specific CG6379 family of methylases of the FtsJ-like superfamily have a divergent, catalytically inactive version of the capping enzyme nucleotidyltransferase domain fused to the methylase domain. These RNA methylases might function as regulators of the capping process that bind cap through the inactive capping enzyme domain.
The principal proteins of the nuclear and cytoplasmic cap-binding complexes, CBP80 and eIF4G, respectively, appear to have diverged from an NIC-domain-containing ancestor, which was probably the core subunit of the ancestral cap-binding complex (16,17). After the divergence of these central components, new subunits, such as CBP20 (200), a RRM domain protein and eIF4E (201), appear to have been independently recruited to the respective complexes, at least prior to the divergence of the eukaryotic crown group. EIF4E also has a core RRM-like fold, although no sequence similarity to RRM domains is detectable; this domain might have been derived from a common precursor with the RRM.
Post-transcriptional regulatory mechanisms. Mechanisms of PTGR that act directly on the transcript and affect its stability or association with the ribosome are common in both bacteria and eukaryotes. At the core of these mechanisms are the ribonucleases that mediate RNA degradation; these enzymes are conserved in all three primary kingdoms (45). Eukaryotes evolved a specific elaboration of this system whereby a whole class of dedicated proteins and RNAs lend specificity to the degradation system with respect to the transcripts that are regulated (202–205). This phenomenon has been termed PTGS and, in many eukaryotes, depends on the amplification of small regulatory RNAs by an RNA-dependent RNA polymerase (153–156). Additionally, while distinct from the chromatin-level transcriptional silencing, the PTGS system appears to interact with it (133,206).
The most ancient PTGR systems are comprised of RNases and helicases that unwind RNA secondary structures to aid degradation or regulate translation (Fig. 6). Many, if not all, of the nucleases implicated in PTGR appear to be involved also in the processing of RNA precursors. The RNA degradation enzymes that can be traced back to LUCA are RNase HII and RNase PH, of which the former is responsible for the removal of the RNA primer during DNA replication and apparently has no direct role in PTGR. In contrast, RNase PH is one of the principal RNA degradation enzymes, along with RNase P. RNase P is present in all extant organisms, but its protein subunits are not homologous in bacteria and archaea-eukaryotes, which suggests that, in LUCA, RNase P existed as pure ribozyme. RNase PH and the bacterial RNase P protein subunit have a common nucleic acid-binding domain of the S5 fold (207,208). This suggests an evolutionary scenario whereby the S5 domain was recruited by a common ribozyme ancestor of RNases PH and P and, during the subsequent evolution, the ribozyme was gradually replaced entirely by a protein catalytic scaffold in RNase PH-like enzymes, whereas RNase P retained the ribozyme and the RNA-binding subunit. This scenario implies that the protein subunit of the bacterial RNase P retains the ancestral state and probably has been displaced by unrelated proteins in the archaeo-eukaryotic lineage. The primitive RNA degradation system of LUCA might also have included a LHR-Ski2p family helicase and, possibly, a generic thermonuclease-like protein of the OB fold and RNA-binding PIN domains. Another component that might have been represented in LUCA is the SM domain. In prokaryotes, SM domain-containing proteins bind numerous specialized small RNAs, such as the DsrA/RprA RNA, and regulate mRNA stability and association with the ribosome (209). It remains to be seen if any of the small RNAs bound by the SM proteins possess ribozyme activities.
With the separation of the archaeo-eukaryotic and bacterial lineages, several distinct superfamilies of nucleases were independently recruited in each of them for RNA degradation and processing [see the recent detailed evolutionary classification of RNases (45)]. The most important innovations in bacteria included 3′→5′ exoRNases, RNase E/G, RNase II and RNase III. In the archaeo-eukaryotic lineage, a 3′→5′ RNA degradation and processing complex, the exosome, has evolved. The eukaryotic exosome has been extensively characterized experimentally (116,210,211), whereas the existence of the archaeal counterpart and, by inference, the presence of the exosome in the common ancestor of archaea and eukaryotes, have been postulated through comparative analysis of archaeal genomes. Genes for predicted exosomal components form some of the most conspicuously conserved gene strings (probable operons) in archaea (212). The exosome consists of Rrp41p- and Rrp42p-like RNase PH family nucleases, RNA-binding proteins containing S1 domains combined with KH or Zn-ribbon domains, such as Rrp4p and Csl4p, PIN domain proteins, a LHR/Ski2p-like helicase and, possibly, also RNase P as predicted during archaeal genome analysis.
The archaea also evolved a distinct RNase of the DHH hydrolase family, which contains S1 and ZnR domains and, as suggested by the comparative genome analysis, might interact with the exosome (45). In addition to these conserved complexes involved in RNA degradation, other RNA-binding complexes, which might contribute to PTGR by affecting mRNA stability and association with the ribosome, evolved after the split of the primary lineages. Cold shock proteins (CspA) containing S1-like domains are among such bacterial regulatory RNA-binding protein (213). Additionally, proteins such as Hsp15, with a stand-alone S4 domain, which bind RNA and regulate translation, point to the existence of diverse PTGR systems in bacteria (214). Some of the RNA-binding proteins predicted during this study, e.g. a protein that combines a PIN and a TRAM domain, could provide leads for discovery and investigation of poorly understood PTGR systems in prokaryotes (Fig. 6).
The emergence of eukaryotes was accompanied by several major elaborations of the PTGR systems, which involved several types of evolutionary processes. One of the major factors was the collusion of the archaeal and bacterial inheritances that gave rise to more complex forms of ancient PTGR systems. A case in point are nucleases, such as 3′→5′ exoRNases (e.g. Rrp6p) and RNase II (e.g. Rrp44p), which apparently were acquired by eukaryotes from bacteria, probably via the pro-mitochondrial endosymbiont, and added to the exosome whose core was inherited from the archaeo-eukaryotic ancestor. The large-scale, intra-familial duplication, e.g. among helicases such as Mtr4p and Ski2p (Fig. 4), was the second major evolutionary phenomenon that contributed to the elaboration of the eukaryotic exosome complex. The third trend in the ontology of these complexes was the recruitment of pan-eukaryotic, superstructure-forming domains, such as WD40 and TPR, which probably provided scaffolding for the enlarged eukaryotic complexes.
The eukaryote-specific mRNA degradation system, which destroys both nonsense codon-containing (nonsense-mediated decay or NMD) and normal mRNAs, appears to have been assembled, in part, from various translation-related components. Among these components, NMD3p appears to have emerged in the archaeo-eukaryotic lineage and functions in ribosomal assembly (215,216). The other components of this system are eukaryote-specific innovations that mimic the set of similar components that have been added to the exosome. NMD2p (217) contains an NIC domain and shares a common ancestor with the translation factor eIF4G. NMD4p and its metazoan equivalents, such as SMG6 (171,218), contain PIN domains and might ultimately have descended from the stand-alone PIN-domain proteins detected in archaea. NMD5p is a HEAT repeat protein and UPF1p is a SFII RNA helicase (217). The poly(A)-degrading complex also appears to have emerged prior to the divergence of the major eukaryotic lineages and contains at least three conserved nuclease components, namely Pan1p, Pop2p and DAN-like nucleases, which belong to the 3′→5′ exonuclease family, and CCR4, which is a derivative of the DNase I superfamily (45,219,220).
The eukaryote-specific PTGS system is present throughout the crown group, at least. Recent experimental results combined with computational predictions based on phyletic patterns resulted in the identification of a complex PTGS apparatus that can be traced back to the common ancestor of the eukaryotic crown group. The core of this system includes a SFII helicase–RNaseIII fusion protein of the carpel factory (CAF, also called DICER) family, which generates small, 21–25 nt RNAs [small interfering RNAs (siRNAs)] used as guides to promote degradation of specific RNAs by a nuclease complex (133,221–223). Additionally, the DICER helicase–nuclease appears to be involved in the processing of numerous other small regulatory RNAs, including the stRNAs, such as Lin-4 and Let-7, which regulate specific transcripts through antisense interactions (224). A LIN-28-like RNA-binding protein containing an S1-like domain and homologous to bacterial Csp (225), which binds these small RNAs, probably is another ancestral component of the PTGS system. The siRNAs function as primers in an amplificatory degradative PCR-like reaction that generates dsRNA and is catalyzed by a specialized RNA-dependent RNA polymerase that is thus far traceable to the base of the eukaryotic crown group (153–156). Proteins of the PIWI-argonaute family, which combine PIWI and PAZ domains (14), also probably participated in the ancestral PTGS as siRNA-binding components (226). The actual RNA destruction apparently depends on several other components, including a RecQ-like helicase (127) and RNase D family 3′→5′ nucleases, such as Mut-7 and Egl (227). From the time of its emergence, the PTGS system probably closely interacted with the more generic RNA degradation systems, including the exosome, NMD and the poly(A)-tail degradation system.
A substantial part of the PTGS system, including the progenitor of most of the 3′→5′ exonucleases, RNase III, the RecQ-like helicase and the RNA-binding CSP proteins are part of the bacterial inheritance of the eukaryotes. The 3′→5′ exonucleases and RNase III, after their acquisition by eukaryotes, each underwent series of duplications to give rise to several distinct groups of orthologs and also formed new architectures through domain fusions. The Mut-7 proteins contain a module, C-terminal to the 3′→5′ exonuclease domain, which consists of a unique α/β domain fused to a Zn-ribbon, which might bind RNA (45). This Mut-7C module appears as a stand-alone protein in archaea and bacteria and potentially might interact with a 3′→5′ nuclease already in prokaryotes, followed by the fusion in eukaryotes. The Argonaute-like proteins are represented in archaea and Aquifex; one of the eukaryotic members of this family has been described as translation initiation factor eIF2C (228). These ancient versions contain only a PIWI domain and their phyletic pattern is typical of translation machinery components, suggesting that their original function was related to translation. Prior to the divergence of the eukaryotic crown group, the PIWI domain combined with a predicted RBD, PAZ, which is also fused to the helicase and nuclease domains in the CAF family proteins (Fig. 6). The PAZ domain, which might bind the small RNAs that are generated as part of PTGS, evolved in eukaryotes with the emergence of this system.
Within the crown group, PTGS shows considerable variability, with extensive gene loss completely or partially eliminating the system in various lineages. In yeast S.cerevisiae, the entire system appears to have been lost (133), whereas in Drosophila and humans, the apparent loss is restricted to the RdRp and the Mut-7 nuclease. However, the detection of a functional PTGS system in Drosophila (229) suggests that the role of the RNA polymerase may have been taken over by other enzymes, such as the DNA-dependent RNA polymerase or a reverse transcriptase-like enzyme, which are known to possess similar activities in vitro. In contrast, plants and Dictyostelium show expansions of the RdRp family, with at least six and four distinct members, respectively. Furthermore, the architectures of the proteins involved in PTGS show lineage-specific variability, e.g. fusion of RRM domains to the RdRp in plants and a duplication of the RdRp within a single protein in C.elegans. Several eukaryotic proteins were identified that, on the basis of their domain architectures, seem to be likely candidates for participation in PTGS. Examples include a nuclease of the RNase II family that is fused to a Sen1p-like SFI helicase in humans and a family of plant 3′→5′ exonucleases fused to the RRM domain (45). Analysis of phyletic patterns and domain architectures also resulted in the identification of several novel candidates, which could be parts of a more extended PTGS network (133) (Fig. 6). The most notable of these include an orthologous group of predicted adenine methylases (the CG14906 group) related to the Kar4-Ime-4 family of mRNA methylases (Fig. 2). Another group of predicted RNA methylases with a similar phyletic pattern are the Corymbosa2/Hen1 family of methylases that are predicted to be dsRNA methylases (see above). These enzymes could specifically regulate the stability of dsRNA regions formed by pairing of mRNAs with anti-sense RNAs (Figs 2 and 6). Homologs of the DNA repair protein AlkB fused to the RRM domain might be involved in RNA modification (Fig. 6). It has been predicted that this subfamily of AlkB proteins, similarly to their homologs involved in DNA repair, possess iron- and 2-oxoglutarate-dependent oxidative demethylating activity (157). Consistent with this prediction, these AlkB homologs, in addition to the RRM domain fusion, also show fusions to a distinct family of methylases. Taken together with the widespread distribution of these enzymes in the crown group, with the exception of S.cerevisiae [a phyletic pattern typical of other PTGS components (133)], these observations suggest that a mRNA methylation–demethylation circuit might be another component of PTGS.
Finally, numerous other uncharacterized eukaryotic RNA-binding proteins were predicted, which could point to still unknown PTGR systems and complexes. For example, Ro protein, which shares the RNA-binding ROT domain with telomerase subunits (230), binds small RNAs called Y RNAs in animals and the resulting RNPs might be involved in several poorly characterized regulatory functions, such as RNA quality control (231). Ro protein homologs are also present in certain bacteria, such as Deinococcus and Streptomyces, probably as a result of horizontal gene transfer from eukaryotes and it has been shown that, in Deinococcus, the Ro homolog binds several small RNAs and belongs to a PTGR system that regulates radiation resistance (232).
RNA processing and splicing. In both eukaryotes and prokaryotes, rRNAs and tRNAs are released from larger precursors through RNA processing events mediated by the same nucleases that are involved in RNA degradation, such as RNase PH and RNase P. As discussed above, the presence of distinct nuclease families in the archaeo-eukaryotic and bacterial lineages suggests that many of these processing systems evolved only after the separation of these primary lineages, with the eukaryotes processing machinery combining the archaeal and bacterial inheritances. Archaea-eukaryotes evolved a specific system of tRNA processing, which removes an intron present in the middle of the tRNA precursor (233). The tRNA splicing endonuclease is a distinct member of the restriction endonuclease fold (234), which might have been derived from an ancient, restriction enzyme-like genomic parasite. This is consistent with the mobile parasitic behavior of several members of the restriction endonuclease superfamily (235,236). In eukaryotes, this enzyme underwent a tetraplication followed by inactivation of two of the copies and resulting in a heterotetrameric functional complex (21,45). The U3 RNP complex is involved in rRNA processing, which involves chiefly rRNA modifications guided by the associated small RNAs (237). This complex consists of, at least, Imp4p, Prp31p and the methylase fibrillarin and evolved in the common ancestor of archaea and eukaryotes; archaeal genome comparisons suggest that it might functionally interact with the exosome (212). In eukaryotes, some of the components of this complex, e.g. PRP31p (238), appear to have been additionally recruited for pre-mRNA splicing.
The most distinctive RNA-processing pathway is mRNA splicing, which, in its entirety, is seen only in eukaryotes (Fig. 7). Eukaryotic spliceosomal mRNA introns share with Type II self-splicing introns the intermediate step of lariat formation. This observation prompted the hypothesis that Type II introns, which existed as parasitic retroelements in the genomes of the organellar precursors, invaded the eukaryotic nucleus, giving rise to the spliceosomal introns (239–241). The analysis of the spliceosomal components that we present here suggests that a version of this hypothesis is plausible and argues against the competing ‘introns early’ hypothesis, which postulates extensive presence of introns in LUCA (242–244).
The eukaryotic splicing apparatus consists of five principal snRNP particles, U1, U2, U4, U5 and U6 (245–248), which contain their namesake small RNAs (Fig. 7). Many specialized spliceosomal particles, especially in multicellular eukaryotes, contain alternative counterparts of these main U RNAs and are dedicated to the processing of special (non-canonical) splice junctions (249). The components that are common to all five spliceosomal U RNP particles can be traced back to the common ancestor of the eukaryotic crown group, suggesting that the core of the spliceosomal machinery was firmly established by the time the crown-group eukaryotes radiated. Examination of the inferred domain composition of the ancestral spliceosomal machinery shows marked enrichment of several conserved domains (Fig. 7). These include SFII helicases and RBDs, namely RRM, SM, Zn-knuckle, CCCH, G-patch, SWAP and PWI. Thus, the spliceosomal particles are largely made up of paralogous forms of a relatively small set of domains. It appears that the ancestral spliceosome was assembled mainly from eukaryote-specific domains and its elaboration resulting in the origin of the five principal spliceosomal particles had occurred largely through the proliferation and shuffling of just these few domains that, in the early spliceosome, were represented by their common ancestors. Common to all these U snRNPs are small, stand-alone SM proteins, which belong to a class of RNA-binding SH3-fold β-barrel domains (see above); this RBD probably bound small RNAs already at a pre-LUCA stage of evolution.
The expansion of the SM family from a single ancestral form found in archaea to the numerous lineages seen in eukaryotes suggests that the SM protein formed the ancestral core of the splicing complex by acting as a protein cofactor for the self-splicing Type II introns that invaded eukaryotes. This could have increased the efficiency of splicing of the Type II introns and diminished their deleterious effects, thereby contributing to their spread. At this point, proteins containing some of the newly emerged eukaryote-specific domains, such as RRM, Zn-knuckle and CCCH, and RNA helicases of the eIF4A-DEAD and Maleless families, might have been added to the set of protein cofactors of the Type II introns. Additionally, some proteins that were initially associated with exosomal function, such as helicases of the Ski2p-Lhr family, also might have been recruited to the emerging spliceosome. The next stage of evolution probably involved partial degeneration of the introns themselves and the emergence of distinct intron fragments as precursors of the U RNAs, which possess ribozyme activity and appear to be the primary catalysts of splicing (250). Simultaneous evolution of eukaryotic chromatin allowed the major increase in genome size in eukaryotes and thus provided the niche for selectively neutral or advantageous (although the nature of these potential advantages is not clear) expansion of the introns throughout eukaryotic evolution. This expansion probably was accompanied by a feedback loop that selected for the proliferation and diversification of the original protein cofactors recruited for splicing, causing an explosive expansion of RRM, SFII helicase and other eukaryote-specific domains involved in splicing.
Genome sequences of early-branching eukaryotes might provide the details of the actual temporal order of the duplications in the evolution of the splicing system, but some inferences on the relative branching pattern already can be drawn from the currently available eukaryotic genome sequences. At least 70–80 orthologous lineages of proteins containing one or more of the common RBDs mentioned above, 15 or more lineages of SFII helicases (249), and several other single-copy proteins with no mobile domains, such as PRP38 or Snu66, are traceable to the ancestor of the crown group. Among the RRM-domain proteins, the most common architectures include the single- and multi-RRM proteins, followed by fusions to the G-patch, CCCH and Zn-knuckle domains (Fig. 7). From this ancestral state that existed prior to the radiation of the crown group, several lineage-specific developments ensued, which correlate with the origin of alternative splicing in multicellular eukaryotes. The common ancestor of animals apparently had approximately 40 orthologous groups of splicing-related proteins that evolved after the divergence of the major crown group lineages (Figs 5 and 7). However, the most striking development is seen in vertebrates, which have at least 30 distinct RRM-domain proteins with no orthologs in arthropods or nematodes and several vertebrate-specific expansions within other ancient ortholog groups of RRM proteins. This diversity of RRM proteins correlates with and is probably functionally linked to the extensive utilization of alternative splicing as a means of generating protein diversity (251,252). A similar situation seems to exist in plants because over 50 plant-specific RRM proteins were detected in Arabidopsis; however, the exact point of origin of this diversity is currently unclear, given the absence of other plant genomes. In contrast, in yeast, the U2 and, to a lesser extent, U5 snRNPs show extensive degeneration, which correlates with the near-complete elimination of spliceosomal introns (133,253).
Links between molecular chaperones, protein degradation, the ubiquitin system and RNA metabolism. Several deep evolutionary links seem to exist between RNA metabolism, protein degradation and ubiquitin signaling pathways, suggesting that these cellular systems have a long history of interactions. The earliest of these links appears to be the potential functional coupling of the RNA-degrading exosome, the protein-degrading proteasome and co-translational protein folding facilitated by prefoldins, as indicated by the juxtaposition of the corresponding genes within a superoperon, which is conserved in most archaeal genomes (212). Such functional coupling can be rationalized in terms of coupled pre- and post-translational regulation of the protein level through mRNA and protein stability, respectively. This type of interaction appears to have extended into eukaryotes as suggested, in particular, by the presence of the shared Sec63 domain in chaperones involved in endoplasmic protein translocation and degradation and the exosome/splicing-related helicase Brr2p (118), and by the presence of the Little Finger domain in the animal versions of the Npl4p (suppressor of Sec63p) protein (Fig. 8). A pan-eukaryotic SPBC17G9.05-like protein containing a cyclophilin-like PPIase fused to a RRM domain (with an additional Zn-knuckle in plants) might be another component of such a system, through coupling protein unfolding to RNA metabolism. Furthermore, animals possess another distinct cyclophilin–RRM fusion (Fig. 8) that might also perform a similar function.
Figure 8.
Architectural diversity of proteins that link RNA metabolism with protein degradation and folding–unfolding. The conventions for the representation of architecture, points of derivation and lineage abbreviations are as in Figure 6. Protein functions are shown in bold type and explained in the key. The domain names/acronyms are as given in Table 1. Additionally: Ubq, ubiquitin; mpase, metalloprotease; OTA20, OTU-A20-like protease; WD, WD40 repeats; UBC-HD, ubiquitin C-terminal hydrolase; RF, RING finger; Ub-Znf, ubiquitin-specific Zn-finger; UBA, ubiquitin-associated domain; ‘acyl. ph.’, acyl phosphatase; ZR, zinc ribbon; X, a lineage-specific uncharacterized domain.
Another ancient link between RNA metabolism and protein degradation is suggested by the domain architecture of the prokaryotic protease HypF, which is involved in hydrogenase maturation and assembly. The HypF protein consists of a dsRNA-binding Sua5 domain (254), an OSGP metallo-protease domain of the Hsp70 fold (38) and an acyl phosphatase domain; this domain architecture is suggestive of complex regulation of specific protein processing events through interaction with RNA (Fig. 8).
In eukaryotes, the elaborate ubiquitin signaling system has a central role in targeting proteins for degradation (255,256). Ubiquitin also acts as a signaling moiety to direct specific protein–protein interactions. A number of domain architectures seen among eukaryotic proteins involved in RNA metabolism suggest close interactions with the ubiquitin system. These include numerous fusions of RING-finger domains, which function as ubiquitin E3 ligases, with RBDs, such as KH, Little Finger and CCCH; these domain combinations are present in MDM2, Makorin and several other proteins (Fig. 8). These proteins might function as E3 ligases that specifically tag certain splicing or other RNP complexes with ubiquitin or ubiquitin-like molecules and thereby target them for degradation or regulate their assembly. Furthermore, fusions of other domains involved in ubiquitin signaling, such as ubiquitin itself, UBA, F-box and CUE with various domains involved in RNA metabolism are also seen in several proteins, such as PRP21, TAFII250 and TAB2 (Fig. 8). These architectures are again suggestive of a role in bringing the ubiqutination machinery to the RNA-binding complexes, in which these proteins reside.
A protease of the ubiquitin C-terminal hydrolase family, Sad1p, is required for the assembly of the U4/U6.U5 tri-snRNP and might act a protease in processing of some of the subunits of this complex (257,258). Eukaryotes have re-used inactive versions of a predicted ancient hydrolytic enzyme, the JAB domain, which is also found in several components of the proteasome/signalosome, in two distinct RNA-associated complexes (259,260). Specifically, the JAB-domain protein PRP8 is a subunit of the U5 and U6 snRNP complexes, and the translation-related eIF3 complex also contains several JAB-domain subunits. In the context of potential links between RNA and protein degradation, among the most interesting architectures are the fusions of the Little Finger domain with three distinct protease domains (Fig. 8), namely an inactive version of the Otu-A20 family protease (261) in TRABID, a calpain protease in Small optic lobes (262), and a metalloprotease domain of the WSS1p family in Arabidopsis and Trypanosoma F14J16.17. This, taken together with the fusion of the Little Finger with E3 ubiquitin ligases in certain proteins, such as MDM2 (263), suggests that this domain might provide a specific link between RNA metabolism and protein degradation. The exact nature of this connection is unclear, but it seems plausible that still uncharacterized, small RNAs regulate the function of the protein degradation complexes. Alternatively or additionally, the Little Finger might function as a tether to target proteolytic machinery to proteins associated with specific cellular RNAs. Most of these architectures are restricted to a few eukaryotic lineages, suggesting the existence of numerous lineage-specific mechanisms for modulation of RNA metabolism.
The significance of the phyletic patterns of conserved proteins in RNA metabolism systems for inferring evolutionary relationships between major taxa. Examination of the phyletic patterns of the conserved proteins in the RNA metabolism systems potentially could help in testing phylogenetic hypotheses regarding the relationships between major lineages. At the deepest level, the presence of a distinct archaeo-eukaryotic lineage is supported by approximately 60 conserved orthologous groups that are shared exclusively by archaea and eukaryotes. This is contrasted by a mere 20 or so orthologous groups common exclusively to archaea and bacteria and approximately 39 bacterial–eukaryotic groups. This pattern is consistent with the domain distribution data and supports a model whereby eukaryotes are a chimeric lineage, which combines archaeal and bacterial inheritances. This massive chimerism in the eukaryotic inheritance most likely reflects the endosymbiotic interaction between the pro-mitochondrial α-proteobacterium and an archaeon. The evidence for the presence of an ancestral mitochondrion in all, including the earliest branching eukaryotes (264–267), and the extensive bacterial contribution that can be seen in the available genomic data from early-branching eukaryotes (268–270) supports this model. There has been a smaller, but noticeable gene flow between the two prokaryotic superkingdoms, apparently driven by the regular process of horizontal gene transfer rather than large-scale chimerism; however, in some cases, such as the bacterial hyperthermophiles, this gene transfer probably made a much greater contribution (271,272).
Within the eukaryotes, the observed phyletic distribution of domains and proteins involved in RNA metabolism seems to conflict with two well known phylogenetic hypotheses. The number of orthologous groups of proteins shared exclusively by animals and plants is approximately 41, in contrast to just 15 that are exclusively shared by fungi and animals. At face value, this contradicts the currently accepted phylogeny, in which fungi and animals are sister groups (273). A possible explanation for this pattern, however, is a massive loss of ancestral genes in the currently available fungal genomes, those of two yeasts. Comparative genomics indeed provides support for large-scale gene loss in the yeasts (133,274). However, in some cases, such as the capping enzyme, TAFii250, eIF4G and Whi3p, the yeast versions have domain architectures distinct from those that are shared by their orthologs in animals and plants. Thus, the topology of the primary branches within the eukaryotic crown group probably should be considered unresolved, emphasizing the need for further investigation from the comparative genomics angle, in addition to individual phylogenies of multiple proteins.
The second piece of evidence that contradicts a popular phylogenetic hypothesis is the presence of 24 exclusive orthologous groups shared by arthropods and vertebrates as opposed to only three that are shared by arthropods and nematodes. A similar phyletic pattern has been reported in the case of orthologous groups shared by nematodes and vertebrates in other functional systems, such as chromatin structure and organization and the apoptosis apparatus (275,276). These observations are not consistent with the existence of a nematode–arthropod clade, which is favored by the ecdysozoan model of eukaryotic evolution (277). Although some gene loss in C.elegans is a possibility, the minimal animal proteomes are of approximately the same size (once lineage-specific family expansions are factored in), and therefore it appears less likely that the specific link between vertebrates and arthropods can be attributed to massive gene loss in nematodes. This suggests that the traditional model of a coelomate clade (278), as opposed to an ecdysozoan clade (277), could be a more accurate representation of animal phylogeny.
CONCLUSION
The RNA metabolism system includes approximately 80 orthologous groups of proteins traceable to LUCA, which makes it the most evolutionarily conserved system among all cellular functional systems. This simple observation is consistent with the idea of a primordial ‘RNA world’ wherein RNA-related functions had a dominant role. Even before the radiation of the bacterial and archaeo-eukaryotic clades from LUCA, RNA metabolism had already differentiated into several distinct functional complexes: the ribosome involved in protein synthesis, the accessory apparatus of protein synthesis, which includes aaRS and translation factors, a battery of RNA-modifying enzymes involved in production of functional RNAs, a RNA degradation system with nucleases involved in both recycling and maturation of RNAs, and complexes with more specialized functions, such as transcription elongation and polyadenylation (Table 2). The majority of these proteins can be dissected into a limited set of about 40–50 principal domains, including several paralogous versions, which were present already in LUCA. This observation points to a pre-LUCA phase of evolution, with an even more limited set of RNA-associated proteins. More specifically, comparisons of the paralogous domains/proteins traceable to LUCA indicate that, at this early stage of evolution, the primitive organisms had single, ancestral GTPases, methylase, helicase (the ancestor of both SFI and SFII) and several other enzymes, as well as single versions of proteins containing RBDs, such as the progenitors of the α-L domains, the OB fold, the SH3-like barrels, KH, dsRBD and ZnR. Each of these ancestral proteins probably performed a wide range of functions, albeit with low specificity. The inevitable corollary of this notion is that, unlike the modern systems of RNA metabolism, the primitive system relied primarily on RNA for specificity of interactions and even catalysis, with proteins functioning largely as co-factors. Thus, these reconstructions seem to provide support for an ancient RNA world in which simple proteins with generic functions facilitated catalysis and specific interactions that were primarily mediated by RNAs. With the gradual increase in the number of proteins interacting with RNAs, as a result of multiple duplications, proteins gradually evolved greater diversity to occupy most functional niches that, in the primordial organisms, belonged to RNAs. This led to the gradual displacement of the ribozymes, while leaving behind remnants, such as RNase P, the guide RNAs involved in RNA modifications, the spliceosomal U RNAs and, most prominently, the 23S rRNA. Most but not all of these displacements appear to have already taken place prior to the LUCA. Previous studies on the evolution of DNA replication systems suggested that LUCA most likely did not possess a modern-type DNA genome, but instead had a mixed RNA–DNA genetic system (279). Thus, as long as the nature of the genetic material can be considered a criterion, LUCA itself probably still was one of the terminal stages of the evolution of the RNA world.
As discussed above with regard to numerous protein families, evolution of the RNA metabolism system involved multiple horizontal gene transfers which, in principle, could jeopardize the use of the parsimony principle for evolutionary reconstructions (see Materials and Methods). Furthermore, backward extrapolation suggests that horizontal transfer was ever more rampant early during evolution, which could potentially refute the very concept of a single LUCA (280). However, the (near) omnipresence of numerous translation components and a substantial set of RNA modification enzymes, together with the fact that most of them conform with the standard model of evolution, indicate that reconstruction of LUCA, although necessarily probabilistic, is feasible (Table 2). These reconstructions indicate that LUCA probably was an organism or, more precisely, a population of organisms, certain major characteristics of which were very different from those of modern organisms. In particular, LUCA’s genome probably consisted of multiple RNA and DNA segments (279), which led to extreme genome fluidity (279,280). Nevertheless, the previous and present evolutionary reconstructions show that many functional systems, including, above all, the RNA metabolism system, have already ‘crystallized’ in this organism (281).
During the post-LUCA phase of evolution, the ontology of RNA metabolism followed an evolutionary course essentially similar to other biological systems, but showed a strong tendency toward conservation of its ancient components. While some novelties evolved in both archaeal and bacterial lineages, the emergence of eukaryotes was marked with the most remarkable burst of innovation. Many of these can be traced to the ‘cross-fertilization’ between the archaeal and bacterial inheritances of the eukaryotic protein complement, whereas others involve new, eukaryote-specific domains. These innovations led to the origin and development of new functional systems, such as splicing, PTGS and other forms of post-transcriptional regulation; via a feedback loop, the evolution of these systems apparently stimulated lineage-specific expansion of numerous domains, particularly eukaryote-specific ones, through multiple rounds of duplication. This phase of eukaryotic evolution culminated in the extensive expansion of RBDs in the vertebrate and plant lineages, which seems to correlate with the advent of alternative splicing as a major force in the diversification of the functional potential of an organism.
Availability of complete results
An annotated list of all detected proteins from completely sequenced genomes that are known or predicted to be involved in RNA metabolism is available at ftp://ncbi.nlm.nih.gov/pub/aravind/RNA
NOTE ADDED IN PROOF
After this paper was submitted, the crystal structure of Type I pseudouridine synthase TruB was published (282). Like Type II pseudouridine synthases, TruB has an RRM-like fold, which indicates that all known pseudouridine synthases have evolved from a common ancestor, which originally probably was derived from a primitive RNA-binding protein. Two members of the HemK family of predicted methylases, HemK itself and YfcB, have been shown to methylate a specific glutamine residue in bacterial class 1 peptide release factors and in ribosomal protein L3, respectively (283). The role of HemK in release factor methylation was also demonstrated in an independent study (284). Thus, although the HemK family belongs to the BNM superfamily, which consists predominantly of RNA- and DNA-methylases, these proteins turned out to be protein N-methylases specific for protein with fundamental roles in translation. This specificity is compatible with the universal presence of the HemK family in all forms of life. It appears likely that these protein methylases evolved from RNA methylases at an early, pre-LUCA stage of evolution, in a fundamental switch of specificity, which resembles similar transitions in other methylases, e.g. the origin of cap methylases from small-molecule methylases. Finally, it has been shown that yeast Mrm2, a member of the FtsJ family of RNA methylases, is responsible for the 2’-O-ribose methylation of two nucleotides in the peptidyltransferase center of yeast mitochondrial 21S rNA (285).
Acknowledgments
ACKNOWLEDGEMENTS
We thank J. Bujnicki for helpful discussions on RNA methylases. We gratefully acknowledge all researchers who contributed to the current understanding of diverse aspects of RNA metabolism and apologize for inevitable omissions in citation of their work due to space considerations.
REFERENCES
- 1.Alberts B., Bray,D., Lewis,J., Raff,M., Roberts,K. and Watson,J.D. (1999) Molecular Biology of the Cell. Garland Publishing, New York, NY.
- 2.Crick F.H.C. (1958) Symp. Soc. Exp. Biol. XII, 139–163. [Google Scholar]
- 3.Reinhart B.J., Slack,F.J., Basson,M., Pasquinelli,A.E., Bettinger,J.C., Rougvie,A.E., Horvitz,H.R. and Ruvkun,G. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature, 403, 901–906. [DOI] [PubMed] [Google Scholar]
- 4.Erdmann V.A., Szymanski,M., Hochberg,A., de Groot,N. and Barciszewski,J. (1999) Collection of mRNA-like non-coding RNAs. Nucleic Acids Res., 27, 192–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franke A. and Baker,B.S. (1999) The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol. Cell, 4, 117–122. [DOI] [PubMed] [Google Scholar]
- 6.Kelley R.L., Meller,V.H., Gordadze,P.R., Roman,G., Davis,R.L. and Kuroda,M.I. (1999) Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell, 98, 513–522. [DOI] [PubMed] [Google Scholar]
- 7.Meller V.H., Wu,K.H., Roman,G., Kuroda,M.I. and Davis,R.L. (1997) roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell, 88, 445–457. [DOI] [PubMed] [Google Scholar]
- 8.Keiler K.C., Waller,P.R. and Sauer,R.T. (1996) Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science, 271, 990–993. [DOI] [PubMed] [Google Scholar]
- 9.Aravind L. and Koonin,E.V. (1999) Novel predicted RNA-binding domains associated with the translation machinery. J. Mol. Evol., 48, 291–302. [DOI] [PubMed] [Google Scholar]
- 10.Wolf Y.I., Aravind,L., Grishin,N.V. and Koonin,E.V. (1999) Evolution of aminoacyl-tRNA synthetases—analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res., 9, 689–710. [PubMed] [Google Scholar]
- 11.Anantharaman V., Koonin,E.V. and Aravind,L. (2001) TRAM, a predicted RNA-binding domain, common to tRNA uracil methylation and adenine thiolation enzymes. FEMS Microbiol. Lett., 197, 215–221. [DOI] [PubMed] [Google Scholar]
- 12.Aravind L. and Koonin,E.V. (2001) THUMP—a predicted RNA-binding domain shared by 4-thiouridine and pseudouridine synthases and RNA methylases. Trends Biochem. Sci., 26, 215–217. [DOI] [PubMed] [Google Scholar]
- 13.Kyrpides N., Woese,C. and Ouzounis,C. (1996) KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci., 21, 425–426. [DOI] [PubMed] [Google Scholar]
- 14.Cerutti L., Mian,N. and Bateman,A. (2000) Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci., 25, 481–482. [DOI] [PubMed] [Google Scholar]
- 15.Grishin N.V. (1998) The R3H motif: a domain that binds single-stranded nucleic acids. Trends Biochem. Sci., 23, 329–330. [DOI] [PubMed] [Google Scholar]
- 16.Aravind L. and Koonin,E.V. (2000) Eukaryote-specific domains in translation initiation factors: implications for translation regulation and evolution of the translation system. Genome Res., 10, 1172–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponting C.P. (2000) Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem. Sci., 25, 423–426. [DOI] [PubMed] [Google Scholar]
- 18.Aravind L. and Koonin,E.V. (1999) G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci., 24, 342–344. [DOI] [PubMed] [Google Scholar]
- 19.Bujnicki J.M. and Radlinska,M. (1999) Is the HemK family of putative S-adenosylmethionine-dependent methyltransferases a ‘missing’ zeta subfamily of adenine methyltransferases? A hypothesis. IUBMB Life, 48, 247–249. [DOI] [PubMed] [Google Scholar]
- 20.Bujnicki J.M. (1999) Comparison of protein structures reveals monophyletic origin of the AdoMet-dependent methyltransferase family and mechanistic convergence rather than recent differentiation of N4-cytosine and N6-adenine DNA methylation. In Silico Biol., 1, 175–182. [PubMed] [Google Scholar]
- 21.Bujnicki J.M. and Rychlewski,L. (2000) Prediction of a common fold for all four subunits of the yeast tRNA splicing endonuclease: implications for the evolution of the EndA/Sen family. FEBS Lett., 486, 328–329. [DOI] [PubMed] [Google Scholar]
- 22.Gustafsson C., Reid,R., Greene,P.J. and Santi,D.V. (1996) Identification of new RNA modifying enzymes by iterative genome search using known modifying enzymes as probes. Nucleic Acids Res., 24, 3756–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reid R., Greene,P.J. and Santi,D.V. (1999) Exposition of a family of RNA m(5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res., 27, 3138–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaffer A.A., Aravind,L., Madden,T.L., Shavirin,S., Spouge,J.L., Wolf,Y.I., Koonin,E.V. and Altschul,S.F. (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res., 29, 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaffer A.A., Wolf,Y.I., Ponting,C.P., Koonin,E.V., Aravind,L. and Altschul,S.F. (1999) IMPALA: matching a protein sequence against a collection of PSI-BLAST-constructed position-specific score matrices. Bioinformatics, 15, 1000–1011. [DOI] [PubMed] [Google Scholar]
- 27.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 28.Notredame C., Higgins,D.G. and Heringa,J. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol., 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 29.Adachi J. and Hasegawa,M. (1994) Institute of Statistical Mathematics, Tokyo.
- 30.Felsenstein J. (1989) PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics, 5, 164–166. [Google Scholar]
- 31.Felsenstein J. (1996) Inferring phylogenies from protein sequences by parsimony, distance and likelihood methods. Methods Enzymol., 266, 418–427. [DOI] [PubMed] [Google Scholar]
- 32.Rost B. and Sander,C. (1993) Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol., 232, 584–599. [DOI] [PubMed] [Google Scholar]
- 33.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 34.Tatusov R.L., Koonin,E.V. and Lipman,D.J. (1997) A genomic perspective on protein families. Science, 278, 631–637. [DOI] [PubMed] [Google Scholar]
- 35.Woese C.R., Kandler,O. and Wheelis,M.L. (1990) Towards a natural system of organisms: proposal for the domains Archaea, Bacteria and Eucarya. Proc. Natl Acad. Sci. USA, 87, 4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doolittle R.F. and Handy,J. (1998) Evolutionary anomalies among the aminoacyl-tRNA synthetases. Curr. Opin. Genet. Dev., 8, 630–636. [DOI] [PubMed] [Google Scholar]
- 37.Cong P. and Shuman,S. (1993) Covalent catalysis in nucleotidyl transfer. A KTDG motif essential for enzyme–GMP complex formation by mRNA capping enzyme is conserved at the active sites of RNA and DNA ligases. J. Biol. Chem., 268, 7256–7260. [PubMed] [Google Scholar]
- 38.Aravind L. and Koonin,E.V. (1999) Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol., 287, 1023–1040. [DOI] [PubMed] [Google Scholar]
- 39.Shuman S. and Schwer,B. (1995) RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol. Microbiol., 17, 405–410. [DOI] [PubMed] [Google Scholar]
- 40.Aravind L. and Koonin,E.V. (1998) Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res., 26, 3746–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koonin E.V. (1994) Conserved sequence pattern in a wide variety of phosphoesterases. Protein Sci., 3, 356–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X., Zamore,P.D. and Hall,T.M. (2001) Crystal structure of a Pumilio homology domain. Mol. Cell, 7, 855–865. [DOI] [PubMed] [Google Scholar]
- 43.Edwards T.A., Pyle,S.E., Wharton,R.P. and Aggarwal,A.K. (2001) Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell, 105, 281–289. [DOI] [PubMed] [Google Scholar]
- 44.Aravind L. and Koonin,E.V. (1999) DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res., 27, 4658–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aravind L. and Koonin,E.V. (2001) A natural classification of ribonucleases. Methods Enzymol., 341, 3–28. [DOI] [PubMed] [Google Scholar]
- 46.Limbach P.A., Crain,P.F. and McCloskey,J.A. (1994) Summary: the modified nucleosides of RNA. Nucleic Acids Res., 22, 2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ofengand J., Bakin,A., Wrzesinski,J., Nurse,K. and Lane,B.G. (1995) The pseudouridine residues of ribosomal RNA. Biochem. Cell Biol., 73, 915–924. [DOI] [PubMed] [Google Scholar]
- 48.Maden B.E.H. and Hughes,J.M.X. (1997) Eukaryotic ribosomal RNA: the recent excitement in the nucleotide modification problem. Chromosoma, 105, 391–400. [DOI] [PubMed] [Google Scholar]
- 49.Anantharaman V., Koonin,E.V. and Aravind,L. (2002) SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol., 4, 71–75. [PubMed] [Google Scholar]
- 50.Bjork G.R., Wikstrom,P.M. and Bystrom,A.S. (1989) Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science, 244, 986–989. [DOI] [PubMed] [Google Scholar]
- 51.Li J.N. and Bjork,G.R. (1999) Structural alterations of the tRNA(m1G37)methyltransferase from Salmonella typhimurium affect tRNA substrate specificity. RNA, 5, 395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cavaille J., Chetouani,F. and Bachellerie,J.P. (1999) The yeast Saccharomyces cerevisiae YDL112w ORF encodes the putative 2′-O-ribose methyltransferase catalyzing the formation of Gm18 in tRNAs. RNA, 5, 66–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Persson B.C., Jager,G. and Gustafsson,C. (1997) The spoU gene of Escherichia coli, the fourth gene of the spoT operon, is essential for tRNA (Gm18) 2′-O-methyltransferase activity. Nucleic Acids Res., 25, 4093–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koonin E.V. and Rudd,K.E. (1993) SpoU protein of Escherichia coli belongs to a new family of putative rRNA methylases. Nucleic Acids Res., 21, 5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lo Conte L., Ailey,B., Hubbard,T.J., Brenner,S.E., Murzin,A.G. and Chothia,C. (2000) SCOP: a structural classification of proteins database. Nucleic Acids Res., 28, 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tscherne J.S., Nurse,K., Popienick,P. and Ofengand,J. (1999) Purification, cloning and characterization of the 16 S RNA m2G1207 methyltransferase from Escherichia coli. J. Biol. Chem., 274, 924–929. [DOI] [PubMed] [Google Scholar]
- 57.Nordlund M.E., Johansson,J.O., von Pawel-Rammingen,U. and Bystrom,A.S. (2000) Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae. RNA, 6, 844–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Persson B.C., Gustafsson,C., Berg,D.E. and Bjork,G.R. (1992) The gene for a tRNA modifying enzyme, m5U54-methyltransferase, is essential for viability in Escherichia coli. Proc. Natl Acad. Sci. USA, 89, 3995–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bokar J.A., Shambaugh,M.E., Polayes,D., Matera,A.G. and Rottman,F.M. (1997) Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA, 3, 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 60.Hong B., Wu,K., Brockenbrough,J.S., Wu,P. and Aris,J.P. (2001) Temperature sensitive nop2 alleles defective in synthesis of 25S rRNA and large ribosomal subunits in Saccharomyces cerevisiae. Nucleic Acids Res., 29, 2927–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lafontaine D., Delcour,J., Glasser,A.L., Desgres,J. and Vandenhaute,J. (1994) The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3′-terminal loop of 18 S rRNA is essential in yeast. J. Mol. Biol., 241, 492–497. [DOI] [PubMed] [Google Scholar]
- 62.Caldas T., Binet,E., Bouloc,P., Costa,A., Desgres,J. and Richarme,G. (2000) The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem., 275, 16414–16419. [DOI] [PubMed] [Google Scholar]
- 63.Pintard L., Kressler,D. and Lapeyre,B. (2000) Spb1p is a yeast nucleolar protein associated with Nop1p and Nop58p that is able to bind S-adenosyl-l-methionine in vitro. Mol. Cell. Biol., 20, 1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lafontaine D.L. and Tollervey,D. (2000) Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol., 20, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ellis S.R., Morales,M.J., Li,J.M., Hopper,A.K. and Martin,N.C. (1986) Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J. Biol. Chem., 261, 9703–9709. [PubMed] [Google Scholar]
- 66.Edqvist J., Straby,K.B. and Grosjean,H. (1995) Enzymatic formation of N2,N2-dimethylguanosine in eukaryotic tRNA: importance of the tRNA architecture. Biochimie, 77, 54–61. [DOI] [PubMed] [Google Scholar]
- 67.Anderson J., Phan,L., Cuesta,R., Carlson,B., Pak,M., Asano,K., Bjork,G., Tamame,M. and Hinnebusch,A.G. (1998) The essential Gcd10p–Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev., 12, 3650–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson J., Phan,L. and Hinnebusch,A.G. (2000) The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 5173–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gupta A., Kumar,P.H., Dineshkumar,T.K., Varshney,U. and Subramanya,H.S. (2001) Crystal structure of Rv2118c: an AdoMet-dependent methyltransferase from Mycobacterium tuberculosis H37Rv. J. Mol. Biol., 312, 381–391. [DOI] [PubMed] [Google Scholar]
- 70.Gustafsson C. and Persson,B.C. (1998) Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J. Bacteriol., 180, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murzin A.G. (1993) OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J., 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ofengand J. (1967) The function of pseudouridylic acid in transfer ribonucleic acid. I. The specific cyanoethylation of pseudouridine, inosine and 4-thiouridine by acrylonitrile. J. Biol. Chem., 242, 5034–5045. [PubMed] [Google Scholar]
- 73.Koonin E.V. (1996) Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res., 24, 2411–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang L., Pookanjanatavip,M., Gu,X. and Santi,D.V. (1998) A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry, 37, 344–351. [DOI] [PubMed] [Google Scholar]
- 75.Foster P.G., Huang,L., Santi,D.V. and Stroud,R.M. (2000) The structural basis for tRNA recognition and pseudouridine formation by pseudouridine synthase I. Nature Struct. Biol., 7, 23–27. [DOI] [PubMed] [Google Scholar]
- 76.Esberg B., Leung,H.C., Tsui,H.C., Bjork,G.R. and Winkler,M.E. (1999) Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol., 181, 7256–7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guianvarc’h D., Florentin,D., Tse Sum Bui,B., Nunzi,F. and Marquet,A. (1997) Biotin synthase, a new member of the family of enzymes which uses S-adenosylmethionine as a source of deoxyadenosyl radical [published erratum appears in Biochem. Biophys. Res. Commun. (1997) 240, 246]. Biochem. Biophys. Res. Commun., 236, 402–406. [DOI] [PubMed] [Google Scholar]
- 78.McIver L., Baxter,R.L. and Campopiano,D.J. (2000) Identification of the [Fe-S] cluster-binding residues of Escherichia coli biotin synthase. J. Biol. Chem., 275, 13888–13894. [DOI] [PubMed] [Google Scholar]
- 79.Bork P. and Koonin,E.V. (1994) A P-loop-like motif in a widespread ATP pyrophosphatase domain: implications for the evolution of sequence motifs and enzyme activity. Proteins, 20, 347–355. [DOI] [PubMed] [Google Scholar]
- 80.Mueller E.G., Buck,C.J., Palenchar,P.M., Barnhart,L.E. and Paulson,J.L. (1998) Identification of a gene involved in the generation of 4-thiouridine in tRNA. Nucleic Acids Res., 26, 2606–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mueller E.G. and Palenchar,P.M. (1999) Using genomic information to investigate the function of ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis. Protein Sci., 8, 2424–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hagervall T.G., Pomerantz,S.C. and McCloskey,J.A. (1998) Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol., 284, 33–42. [DOI] [PubMed] [Google Scholar]
- 83.Webb E., Claas,K. and Downs,D. (1997) Characterization of thiI, a new gene involved in thiazole biosynthesis in Salmonella typhimurium. J. Bacteriol., 179, 4399–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCloskey J.A. and Crain,P.F. (1998) The RNA modification database—1998. Nucleic Acids Res., 26, 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Okada N., Noguchi,S., Kasai,H., Shindo-Okada,N., Ohgi,T., Goto,T. and Nishimura,S. (1979) Novel mechanism of post-transcriptional modification of tRNA. Insertion of bases of Q precursors into tRNA by a specific tRNA transglycosylase reaction. J. Biol. Chem., 254, 3067-3073. [PubMed] [Google Scholar]
- 86.Slany R.K. and Kersten,H. (1994) Genes, enzymes and coenzymes of queuosine biosynthesis in procaryotes. Biochimie, 76, 1178–1182. [DOI] [PubMed] [Google Scholar]
- 87.Watanabe M., Matsuo,M., Tanaka,S., Akimoto,H., Asahi,S., Nishimura,S., Katze,J.R., Hashizume,T., Crain,P.F., McCloskey,J.A. et al. (1997) Biosynthesis of archaeosine, a novel derivative of 7-deazaguanosine specific to archaeal tRNA, proceeds via a pathway involving base replacement on the tRNA polynucleotide chain. J. Biol. Chem., 272, 20146–20151. [DOI] [PubMed] [Google Scholar]
- 88.Romier C., Reuter,K., Suck,D. and Ficner,R. (1996) Crystal structure of tRNA-guanine transglycosylase: RNA modification by base exchange. EMBO J., 15, 2850–2857. [PMC free article] [PubMed] [Google Scholar]
- 89.Dance G., Beemiller,P., Yang,Y., Mater,D., Mian,I. and Smith,H. (2001) Identification of the yeast cytidine deaminase CDD1 as an orphan C→U RNA editase. Nucleic Acids Res., 29, 1772–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gerber A.P. and Keller,W. (1999) An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science, 286, 1146–1149. [DOI] [PubMed] [Google Scholar]
- 91.Keller W., Wolf,J. and Gerber,A. (1999) Editing of messenger RNA precursors and of tRNAs by adenosine to inosine conversion. FEBS Lett., 452, 71–76. [DOI] [PubMed] [Google Scholar]
- 92.Gerber A., Grosjean,H., Melcher,T. and Keller,W. (1998) Tad1p, a yeast tRNA-specific adenosine deaminase, is related to the mammalian pre-mRNA editing enzymes ADAR1 and ADAR2. EMBO J., 17, 4780–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Keegan L.P., Gerber,A.P., Brindle,J., Leemans,R., Gallo,A., Keller,W. and O’Connell,M.A. (2000) The properties of a tRNA-specific adenosine deaminase from Drosophila melanogaster support an evolutionary link between pre-mRNA editing and tRNA modification. Mol. Cell. Biol., 20, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maas S., Gerber,A.P. and Rich,A. (1999) Identification and characterization of a human tRNA-specific adenosine deaminase related to the ADAR family of pre-mRNA editing enzymes. Proc. Natl Acad. Sci. USA, 96, 8895–8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Anant S., MacGinnitie,A.J. and Davidson,N.O. (1995) apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, is a novel RNA-binding protein. J. Biol. Chem., 270, 14762–14767. [PubMed] [Google Scholar]
- 96.Naora H. (1977) In Stewart,P.R. and Letham,D.S. (eds), The Ribonucleic Acids. Springer-Verlag, Berlin, pp. 43–80.
- 97.Kowalak J.A., Bruenger,E. and McCloskey,J.A. (1995) Posttranscriptional modification of the central loop of domain V in Escherichia coli 23 S ribosomal RNA. J. Biol. Chem., 270, 17758–17764. [DOI] [PubMed] [Google Scholar]
- 98.Copley R.R. and Bork,P. (2000) Homology among (betaalpha)(8) barrels: implications for the evolution of metabolic pathways. J. Mol. Biol., 303, 627–641. [DOI] [PubMed] [Google Scholar]
- 99.Edmonds C.G., Crain,P.F., Gupta,R., Hashizume,T., Hocart,C.H., Kowalak,J.A., Pomerantz,S.C., Stetter,K.O. and McCloskey,J.A. (1991) Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria). J. Bacteriol., 173, 3138–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.House C.H. and Miller,S.L. (1996) Hydrolysis of dihydrouridine and related compounds. Biochemistry, 35, 315–320. [DOI] [PubMed] [Google Scholar]
- 101.Woese C.R., Olsen,G.J., Ibba,M. and Soll,D. (2000) Aminoacyl-tRNA synthetases, the genetic code and the evolutionary process. Microbiol. Mol. Biol. Rev., 64, 202–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aravind L., Anantharaman,V. and Koonin,E.V. (2002) Monophyly of Class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA world. Proteins, in press. [DOI] [PubMed] [Google Scholar]
- 103.Fabrizio P., Laggerbauer,B., Lauber,J., Lane,W.S. and Luhrmann,R. (1997) An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J., 16, 4092–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen X., Court,D.L. and Ji,X. (1999) Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif. Proc. Natl Acad. Sci. USA, 96, 8396–8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grishin N.V. (2001) KH domain: one motif, two folds. Nucleic Acids Res., 29, 638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cabedo H., Macian,F., Villarroya,M., Escudero,J.C., Martinez-Vicente,M., Knecht,E. and Armengod,M.E. (1999) The Escherichia coli trmE (mnmE) gene, involved in tRNA modification, codes for an evolutionarily conserved GTPase with unusual biochemical properties. EMBO J., 18, 7063–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.de Vries H., Ruegsegger,U., Hubner,W., Friedlein,A., Langen,H. and Keller,W. (2000) Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J., 19, 5895–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Leipe D.D., Koonin,E.V. and Aravind,L. (2002) Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol., 317, 41–72. [DOI] [PubMed] [Google Scholar]
- 109.Gorbalenya A.E. and Koonin,E.V. (1993) Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol., 3, 419–429. [Google Scholar]
- 110.Korolev S., Yao,N., Lohman,T.M., Weber,P.C. and Waksman,G. (1998) Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci., 7, 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bird L.E., Subramanya,H.S. and Wigley,D.B. (1998) Helicases: a unifying structural theme? Curr. Opin. Struct. Biol., 8, 14–18. [DOI] [PubMed] [Google Scholar]
- 112.Jones P.G., Mitta,M., Kim,Y., Jiang,W. and Inouye,M. (1996) Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc. Natl Acad. Sci. USA, 93, 76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.O’Day C., Chavanikamannil,F. and Abelson,J. (1996) 18S rRNA processing requires the RNA helicase-like protein Rrp3. Nucleic Acids Res., 24, 3201–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.He F. and Jacobson,A. (1995) Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev., 9, 437–454. [DOI] [PubMed] [Google Scholar]
- 115.Coburn G.A., Miao,X., Briant,D.J. and Mackie,G.A. (1999) Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes Dev., 13, 2594–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mitchell P., Petfalski,E., Shevchenko,A., Mann,M. and Tollervey,D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- 117.van Hoof A. and Parker,R. (1999) The exosome: a proteasome for RNA? Cell, 99, 347–350. [DOI] [PubMed] [Google Scholar]
- 118.Ponting C.P. (2000) Proteins of the endoplasmic-reticulum-associated degradation pathway: domain detection and function prediction. Biochem. J., 351, 527–535. [PMC free article] [PubMed] [Google Scholar]
- 119.Aravind L., Walker,D.R. and Koonin,E.V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Burtis K.C. and Harris,P.V. (1997) A possible functional role for a new class of eukaryotic DNA polymerases. Curr. Biol., 7, R743–R744. [DOI] [PubMed] [Google Scholar]
- 121.Margossian S., Li,H., Zassenhaus,H. and Butow,R. (1996) The DExH box protein Suv3p is a component of a yeast mitochondrial 3′-to-5′ exoribonuclease that suppresses group I intron toxicity. Cell, 84, 199–209. [DOI] [PubMed] [Google Scholar]
- 122.Sanjuan R. and Marin,I. (2001) Tracing the origin of the compensasome: evolutionary history of DEAH helicase and MYST acetyltransferase gene families. Mol. Biol. Evol., 18, 330–343. [DOI] [PubMed] [Google Scholar]
- 123.Koonin E. and Gorbalenya,A. (1992) Autogenous translation regulation by Escherichia coli ATPase SecA may be mediated by an intrinsic RNA helicase activity of this protein. FEBS Lett., 298, 6–8. [DOI] [PubMed] [Google Scholar]
- 124.Park S., Kim,D., Choe,J. and Kim,H. (1997) RNA helicase activity of Escherichia coli SecA protein. Biochem. Biophys. Res. Commun., 235, 593–597. [DOI] [PubMed] [Google Scholar]
- 125.Schmidt M.O., Brosh,R.M.,Jr and Oliver,D.B. (2001) Escherichia coli SecA helicase activity is not required in vivo for efficient protein translocation or autogenous regulation. J. Biol. Chem., 276, 37076–37085. [DOI] [PubMed] [Google Scholar]
- 126.Morozov V., Mushegian,A.R., Koonin,E.V. and Bork,P. (1997) A putative nucleic acid-binding domain in Bloom’s and Werner’s syndrome helicases. Trends Biochem. Sci., 22, 417–418. [DOI] [PubMed] [Google Scholar]
- 127.Cogoni C. and Macino,G. (1999) Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science, 286, 2342–2344. [DOI] [PubMed] [Google Scholar]
- 128.Koonin E.V. (1992) A new group of putative RNA helicases. Trends Biochem. Sci., 17, 495–497. [DOI] [PubMed] [Google Scholar]
- 129.Fukita Y., Mizuta,T.R., Shirozu,M., Ozawa,K., Shimizu,A. and Honjo,T. (1993) The human S mu bp-2, a DNA-binding protein specific to the single-stranded guanine-rich sequence related to the immunoglobulin mu chain switch region. J. Biol. Chem., 268, 17463–17470. [PubMed] [Google Scholar]
- 130.Ursic D., Himmel,K.L., Gurley,K.A., Webb,F. and Culbertson,M.R. (1997) The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res., 25, 4778–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Altamura N., Groudinsky,O., Dujardin,G. and Slonimski,P.P. (1992) NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J. Mol. Biol., 224, 575–587. [DOI] [PubMed] [Google Scholar]
- 132.Dalmay T., Horsefield,R., Braunstein,T. and Baulcombe,D. (2001) SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J., 20, 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Aravind L., Watanabe,H., Lipman,D.J. and Koonin,E.V. (2000) Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl Acad. Sci. USA, 97, 11319–11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sam M., Wurst,W., Kluppel,M., Jin,O., Heng,H. and Bernstein,A. (1998) Aquarius, a novel gene isolated by gene trapping with an RNA-dependent RNA polymerase motif. Dev. Dyn., 212, 304–317. [DOI] [PubMed] [Google Scholar]
- 135.Koonin E.V. and Rudd,K.E. (1996) Two domains of superfamily I helicases may exist as separate proteins. Protein Sci., 5, 178–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Makarova K.S., Aravind,L., Galperin,M.Y., Grishin,N.V., Tatusov,R.L., Wolf,Y.I. and Koonin,E.V. (1999) Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core and the variable shell. Genome Res., 9, 608–628. [PubMed] [Google Scholar]
- 137.Wang L.K. and Shuman,S. (2001) Domain structure and mutational analysis of T4 polynucleotide kinase. J. Biol. Chem., 276, 26868–26874. [DOI] [PubMed] [Google Scholar]
- 138.Jilani A., Ramotar,D., Slack,C., Ong,C., Yang,X.M., Scherer,S.W. and Lasko,D.D. (1999) Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J. Biol. Chem., 274, 24176–24186. [DOI] [PubMed] [Google Scholar]
- 139.Karimi-Busheri F., Daly,G., Robins,P., Canas,B., Pappin,D.J., Sgouros,J., Miller,G.G., Fakhrai,H., Davis,E.M., Le Beau,M.M. et al. (1999) Molecular characterization of a human DNA kinase. J. Biol. Chem., 274, 24187–24194. [DOI] [PubMed] [Google Scholar]
- 140.Koonin E.V. and Gorbalenya,A.E. (1990) Related domains in yeast tRNA ligase, bacteriophage T4 polynucleotide kinase and RNA ligase and mammalian myelin 2′,3′-cyclic nucleotide phosphohydrolase revealed by amino acid sequence comparison. FEBS Lett., 268, 231–234. [DOI] [PubMed] [Google Scholar]
- 141.Xu Q., Teplow,D., Lee,T.D. and Abelson,J. (1990) Domain structure in yeast tRNA ligase. Biochemistry, 29, 6132–6138. [DOI] [PubMed] [Google Scholar]
- 142.Romig H., Fackelmayer,F.O., Renz,A., Ramsperger,U. and Richter,A. (1992) Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J., 11, 3431–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Aravind L. and Koonin,E.V. (2000) SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci., 25, 112–114. [DOI] [PubMed] [Google Scholar]
- 144.Silva E., Ullu,E., Kobayashi,R. and Tschudi,C. (1998) Trypanosome capping enzymes display a novel two-domain structure. Mol. Cell. Biol., 18, 4612–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moore J.A. and Poulter,C.D. (1997) Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: a binding mechanism for recombinant enzyme. Biochemistry, 36, 604–614. [DOI] [PubMed] [Google Scholar]
- 146.Leung H.C., Chen,Y. and Winkler,M.E. (1997) Regulation of substrate recognition by the MiaA tRNA prenyltransferase modification enzyme of Escherichia coli K-12. J. Biol. Chem., 272, 13073–13083. [DOI] [PubMed] [Google Scholar]
- 147.Sidrauski C., Cox,J.S. and Walter,P. (1996) tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell, 87, 405–413. [DOI] [PubMed] [Google Scholar]
- 148.McManus M.T., Shimamura,M., Grams,J. and Hajduk,S.L. (2001) Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA, 7, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Aravind L. and Koonin,E. (1999) DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res., 27, 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hofmann A., Zdanov,A., Genschik,P., Ruvinov,S., Filipowicz,W. and Wlodawer,A. (2000) Structure and mechanism of activity of the cyclic phosphodiesterase of Appr>p, a product of the tRNA splicing reaction. EMBO J., 19, 6207–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nasr F. and Filipowicz,W. (2000) Characterization of the Saccharomyces cerevisiae cyclic nucleotide phosphodiesterase involved in the metabolism of ADP-ribose 1″,2″-cyclic phosphate. Nucleic Acids Res., 28, 1676–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Martzen M.R., McCraith,S.M., Spinelli,S.L., Torres,F.M., Fields,S., Grayhack,E.J. and Phizicky,E.M. (1999) A biochemical genomics approach for identifying genes by the activity of their products. Science, 286, 1153–1155. [DOI] [PubMed] [Google Scholar]
- 153.Dalmay T., Hamilton,A., Rudd,S., Angell,S. and Baulcombe,D.C. (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- 154.Schiebel W., Pelissier,T., Riedel,L., Thalmeir,S., Schiebel,R., Kempe,D., Lottspeich,F., Sanger,H.L. and Wassenegger,M. (1998) Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell, 10, 2087–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cogoni C. and Macino,G. (1999) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature, 399, 166–169. [DOI] [PubMed] [Google Scholar]
- 156.Smardon A., Spoerke,J.M., Stacey,S.C., Klein,M.E., Mackin,N. and Maine,E.M. (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol., 10, 169–178. [DOI] [PubMed] [Google Scholar]
- 157.Aravind L. and Koonin,E.V. (2001) The DNA-repair protein AlkB, EGL-9 and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol., 2, RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bycroft M., Hubbard,T.J., Proctor,M., Freund,S.M. and Murzin,A.G. (1997) The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell, 88, 235–242. [DOI] [PubMed] [Google Scholar]
- 159.Bycroft M., Grunert,S., Murzin,A.G., Proctor,M. and St Johnston,D. (1995) NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J., 14, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Siomi H., Matunis,M.J., Michael,W.M. and Dreyfuss,G. (1993) The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res., 21, 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gibson T.J., Thompson,J.D. and Heringa,J. (1993) The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett., 324, 361–366. [DOI] [PubMed] [Google Scholar]
- 162.Koonin E.V., Wolf,Y.I. and Aravind,L. (2000) Protein fold recognition using sequence profiles and its application in structural genomics. Adv. Protein Chem., 54, 245–275. [DOI] [PubMed] [Google Scholar]
- 163.Murzin A.G. (1992) Familiar strangers. Nature, 360, 635. [DOI] [PubMed] [Google Scholar]
- 164.Neuwald A.F. and Koonin,E.V. (1998) Ataxin-2, global regulators of bacterial gene expression and spliceosomal snRNP proteins share a conserved domain. J. Mol. Med., 76, 3–5. [DOI] [PubMed] [Google Scholar]
- 165.Collins B.M., Harrop,S.J., Kornfeld,G.D., Dawes,I.W., Curmi,P.M. and Mabbutt,B.C. (2001) Crystal structure of a heptameric Sm-like protein complex from archaea: implications for the structure and evolution of snRNPs. J. Mol. Biol., 309, 915–923. [DOI] [PubMed] [Google Scholar]
- 166.Toro I., Thore,S., Mayer,C., Basquin,J., Seraphin,B. and Suck,D. (2001) RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J., 20, 2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ponting C.P. (1997) Tudor domains in proteins that interact with RNA. Trends Biochem. Sci., 22, 51–52. [DOI] [PubMed] [Google Scholar]
- 168.Selenko P., Sprangers,R., Stier,G., Buhler,D., Fischer,U. and Sattler,M. (2001) SMN tudor domain structure and its interaction with the Sm proteins. Nature Struct. Biol., 8, 27–31. [DOI] [PubMed] [Google Scholar]
- 169.Staker B.L., Korber,P., Bardwell,J.C. and Saper,M.A. (2000) Structure of Hsp15 reveals a novel RNA-binding motif. EMBO J., 19, 749–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Doherty A.J., Serpell,L.C. and Ponting,C.P. (1996) The helix–hairpin–helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res., 24, 2488–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Clissold P.M. and Ponting,C.P. (2000) PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol., 10, R888–R890. [DOI] [PubMed] [Google Scholar]
- 172.Wu X.Q., Gu,W., Meng,X. and Hecht,N.B. (1997) The RNA-binding protein, TB-RBP, is the mouse homologue of translin, a recombination protein associated with chromosomal translocations. Proc. Natl Acad. Sci. USA, 94, 5640–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Preker P.J. and Keller,W. (1998) The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem. Sci., 23, 15–16. [DOI] [PubMed] [Google Scholar]
- 174.Huber R., Huber,H. and Stetter,K.O. (2000) Towards the ecology of hyperthermophiles: biotopes, new isolation strategies and novel metabolic properties. FEMS Microbiol. Rev., 24, 615–623. [DOI] [PubMed] [Google Scholar]
- 175.Smith D.L. and Fedoroff,N.V. (1995) LRP1, a gene expressed in lateral and adventitious root primordia of arabidopsis. Plant Cell, 7, 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Finerty P.J. Jr and Bass,B.L. (1997) A Xenopus zinc finger protein that specifically binds dsRNA and RNA-DNA hybrids. J. Mol. Biol., 271, 195–208. [DOI] [PubMed] [Google Scholar]
- 177.Friesen W.J. and Darby,M.K. (2001) Specific RNA binding by a single C2H2 zinc finger. J. Biol. Chem., 276, 1968–1973. [DOI] [PubMed] [Google Scholar]
- 178.Finerty P.J. Jr and Bass,B.L. (1999) Subsets of the zinc finger motifs in dsRBP-ZFa can bind double-stranded RNA. Biochemistry, 38, 4001–4007. [DOI] [PubMed] [Google Scholar]
- 179.Curnow A.W., Hong,K., Yuan,R., Kim,S., Martins,O., Winkler,W., Henkin,T.M. and Soll,D. (1997) Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl Acad. Sci. USA, 94, 11819–11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Artymiuk P.J., Rice,D.W., Poirrette,A.R. and Willet,P. (1994) A tale of two synthetases. Nature Struct. Biol., 1, 758–760. [DOI] [PubMed] [Google Scholar]
- 181.Sullivan S.L. and Gottesman,M.E. (1992) Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell, 68, 989–994. [DOI] [PubMed] [Google Scholar]
- 182.Squires C.L., Greenblatt,J., Li,J. and Condon,C. (1993) Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl Acad. Sci. USA, 90, 970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mason S.W. and Greenblatt,J. (1991) Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG and S10. Genes Dev., 5, 1504–1512. [DOI] [PubMed] [Google Scholar]
- 184.Torres M., Condon,C., Balada,J.M., Squires,C. and Squires,C.L. (2001) Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J., 20, 3811–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hartzog G.A., Wada,T., Handa,H. and Winston,F. (1998) Evidence that Spt4, Spt5 and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev., 12, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wada T., Takagi,T., Yamaguchi,Y., Ferdous,A., Imai,T., Hirose,S., Sugimoto,S., Yano,K., Hartzog,G.A., Winston,F. et al. (1998) DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev., 12, 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Fuchs T.M., Deppisch,H., Scarlato,V. and Gross,R. (1996) A new gene locus of Bordetella pertussis defines a novel family of prokaryotic transcriptional accessory proteins. J. Bacteriol., 178, 4445–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yamaguchi Y., Takagi,T., Wada ,T., Yano,K., Furuya,A., Sugimoto,S., Hasegawa,J. and Handa,H. (1999) NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell, 97, 41–51. [DOI] [PubMed] [Google Scholar]
- 189.O’Hara B.P., Norman,R.A., Wan,P.T., Roe,S.M., Barrett,T.E., Drew,R.E. and Pearl,L.H. (1999) Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. EMBO J., 18, 5175–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Akhtar A., Zink,D. and Becker,P.B. (2000) Chromodomains are protein-RNA interaction modules. Nature, 407, 405–409. [DOI] [PubMed] [Google Scholar]
- 191.Sarkar N. (1997) Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem., 66, 173–197. [DOI] [PubMed] [Google Scholar]
- 192.Aravind L. (1999) An evolutionary classification of the metallo-beta-lactamase fold proteins. In Silico Biol., 1, 69–91. [PubMed] [Google Scholar]
- 193.Jenny A., Minvielle-Sebastia,L., Preker,P.J. and Keller,W. (1996) Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science, 274, 1514–1517. [DOI] [PubMed] [Google Scholar]
- 194.Chanfreau G., Noble,S.M. and Guthrie,C. (1996) Essential yeast protein with unexpected similarity to subunits of mammalian cleavage and polyadenylation specificity factor (CPSF). Science, 274, 1511–1514. [DOI] [PubMed] [Google Scholar]
- 195.Shuman S. (2000) Structure, mechanism and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol., 66, 1–40. [DOI] [PubMed] [Google Scholar]
- 196.Mao X., Schwer,B. and Shuman,S. (1995) Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol. Cell. Biol., 15, 4167–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Takagi T., Moore,C.R., Diehn,F. and Buratowski,S. (1997) An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell, 89, 867–873. [DOI] [PubMed] [Google Scholar]
- 198.Hakansson K., Doherty,A.J., Shuman,S. and Wigley,D.B. (1997) X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell, 89, 545–553. [DOI] [PubMed] [Google Scholar]
- 199.Lima C.D., Wang,L.K. and Shuman,S. (1999) Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell, 99, 533–543. [DOI] [PubMed] [Google Scholar]
- 200.Izaurralde E., Lewis,J., McGuigan,C., Jankowska,M., Darzynkiewicz,E. and Mattaj,I.W. (1994) A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell, 78, 657–668. [DOI] [PubMed] [Google Scholar]
- 201.Marcotrigiano J., Gingras,A.C., Sonenberg,N. and Burley,S.K. (1999) Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell, 3, 707–716. [DOI] [PubMed] [Google Scholar]
- 202.Cogoni C. and Macino,G. (2000) Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev., 10, 638–643. [DOI] [PubMed] [Google Scholar]
- 203.Iyer L.M., Kumpatla,S.P., Chandrasekharan,M.B. and Hall,T.C. (2000) Transgene silencing in monocots. Plant Mol. Biol., 43, 323–346. [DOI] [PubMed] [Google Scholar]
- 204.Vance V. and Vaucheret,H. (2001) RNA silencing in plants—defense and counterdefense. Science, 292, 2277–2280. [DOI] [PubMed] [Google Scholar]
- 205.Waterhouse P.M., Wang,M.B. and Lough,T. (2001) Gene silencing as an adaptive defence against viruses. Nature, 411, 834–842. [DOI] [PubMed] [Google Scholar]
- 206.Jones L., Ratcliff,F. and Baulcombe,D.C. (2001) RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol., 11, 747–757. [DOI] [PubMed] [Google Scholar]
- 207.Stams T., Niranjanakumari,S., Fierke,C.A. and Christianson,D.W. (1998) Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science, 280, 752–755. [DOI] [PubMed] [Google Scholar]
- 208.Symmons M.F., Jones,G.H. and Luisi,B.F. (2000) A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity and regulation. Struct. Fold Des., 8, 1215–1226. [DOI] [PubMed] [Google Scholar]
- 209.Wassarman K., Repoila,F., Rosenow,C., Storz,G. and Gottesman,S. (2001) Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev., 15, 1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Brouwer R., Allmang,C., Raijmakers,R., van Aarssen,Y., Egberts,W.V., Petfalski,E., van Venrooij,W.J., Tollervey,D. and Pruijn,G.J. (2001) Three novel components of the human exosome. J. Biol. Chem., 276, 6177–6184. [DOI] [PubMed] [Google Scholar]
- 211.Mitchell P. and Tollervey,D. (2000) Musing on the structural organization of the exosome complex. Nature Struct. Biol., 7, 843–846. [DOI] [PubMed] [Google Scholar]
- 212.Koonin E.V., Wolf,Y.I. and Aravind,L. (2001) Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res., 11, 240–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jiang W., Hou,Y. and Inouye,M. (1997) CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem., 272, 196–202. [DOI] [PubMed] [Google Scholar]
- 214.Korber P., Stahl,J.M., Nierhaus,K.H. and Bardwell,J.C. (2000) Hsp15: a ribosome-associated heat shock protein. EMBO J., 19, 741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ho J.H. and Johnson,A.W. (1999) NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 2389–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ho J.H., Kallstrom,G. and Johnson,A.W. (2000) Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p. RNA, 6, 1625–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.He F. and Jacobson,A. (2001) Upf1p, Nmd2p and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol. Cell. Biol., 21, 1515–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Domeier M.E., Morse,D.P., Knight,S.W., Portereiko,M., Bass,B.L. and Mango,S.E. (2000) A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science, 289, 1928–1931. [DOI] [PubMed] [Google Scholar]
- 219.Tucker M., Valencia-Sanchez,M.A., Staples,R.R., Chen,J., Denis,C.L. and Parker,R. (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell, 104, 377–386. [DOI] [PubMed] [Google Scholar]
- 220.Korner C.G., Wormington,M., Muckenthaler,M., Schneider,S., Dehlin,E. and Wahle,E. (1998) The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J., 17, 5427–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hutvagner G., McLachlan,J., Pasquinelli,A.E., Balint,E., Tuschl,T. and Zamore,P.D. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science, 293, 834–838. [DOI] [PubMed] [Google Scholar]
- 222.Knight S.W. and Bass,B.L. (2001) A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in C. elegans. Science, 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 224.Grishok A., Pasquinelli,A.E., Conte,D., Li,N., Parrish,S., Ha,I., Baillie,D.L., Fire,A., Ruvkun,G. and Mello,C.C. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell, 106, 23–34. [DOI] [PubMed] [Google Scholar]
- 225.Moss E.G., Lee,R.C. and Ambros,V. (1997) The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell, 88, 637–646. [DOI] [PubMed] [Google Scholar]
- 226.Hammond S.M., Boettcher,S., Caudy,A.A., Kobayashi,R. and Hannon,G.J. (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- 227.Ketting R.F., Haverkamp,T.H., van Luenen,H.G. and Plasterk,R.H. (1999) Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell, 99, 133–141. [DOI] [PubMed] [Google Scholar]
- 228.Zou C., Zhang,Z., Wu,S. and Osterman,J.C. (1998) Molecular cloning and characterization of a rabbit eIF2C protein. Gene, 211, 187–194. [DOI] [PubMed] [Google Scholar]
- 229.Lipardi C., Wei,Q. and Paterson,B.M. (2001) RNAi as random degradative PCR. siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell, 107, 297–307. [DOI] [PubMed] [Google Scholar]
- 230.Harrington L., McPhail,T., Mar,V., Zhou,W., Oulton,R., Bass,M.B., Arruda,I. and Robinson,M.O. (1997) A mammalian telomerase-associated protein. Science, 275, 973–977. [DOI] [PubMed] [Google Scholar]
- 231.O’Brien C.A. and Wolin,S.L. (1994) A possible role for the 60-kD Ro autoantigen in a discard pathway for defective 5S rRNA precursors. Genes Dev., 8, 2891–2903. [DOI] [PubMed] [Google Scholar]
- 232.Chen X., Quinn,A.M. and Wolin,S.L. (2000) Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev., 14, 777–782. [PMC free article] [PubMed] [Google Scholar]
- 233.Li H., Trotta,C.R. and Abelson,J. (1998) Crystal structure and evolution of a transfer RNA splicing enzyme. Science, 280, 279–284. [DOI] [PubMed] [Google Scholar]
- 234.Li H. and Abelson,J. (2000) Crystal structure of a dimeric archaeal splicing endonuclease. J. Mol. Biol., 302, 639–648. [DOI] [PubMed] [Google Scholar]
- 235.Kobayashi I., Nobusato,A., Kobayashi-Takahashi,N. and Uchiyama,I. (1999) Shaping the genome-restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev., 9, 649–656. [DOI] [PubMed] [Google Scholar]
- 236.Aravind L., Makarova,K.S. and Koonin,E.V. (2000) Survey and summary: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res., 28, 3417–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Omer A.D., Lowe,T.M., Russell,A.G., Ebhardt,H., Eddy,S.R. and Dennis,P.P. (2000) Homologs of small nucleolar RNAs in Archaea. Science, 288, 517–522. [DOI] [PubMed] [Google Scholar]
- 238.Newman D.R., Kuhn,J.F., Shanab,G.M. and Maxwell,E.S. (2000) Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA, 6, 861–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Cavalier-Smith T. (1985) Selfish DNA and the origin of introns. Nature, 315, 283–284. [DOI] [PubMed] [Google Scholar]
- 240.Cavalier-Smith T. (1991) Intron phylogeny: a new hypothesis. Trends Genet., 7, 145–148. [PubMed] [Google Scholar]
- 241.Logsdon J.M. Jr (1998) The recent origins of spliceosomal introns revisited. Curr. Opin. Genet. Dev., 8, 637–648. [DOI] [PubMed] [Google Scholar]
- 242.Gilbert W. and Glynias,M. (1993) On the ancient nature of introns. Gene, 135, 137–144. [DOI] [PubMed] [Google Scholar]
- 243.de Souza S.J., Long,M., Schoenbach,L., Roy,S.W. and Gilbert,W. (1996) Intron positions correlate with module boundaries in ancient proteins. Proc. Natl Acad. Sci. USA, 93, 14632–14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.de Souza S.J., Long,M., Klein,R.J., Roy,S., Lin,S. and Gilbert,W. (1998) Toward a resolution of the introns early/late debate: only phase zero introns are correlated with the structure of ancient proteins. Proc. Natl Acad. Sci. USA, 95, 5094–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hastings M.L. and Krainer,A.R. (2001) Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol., 13, 302–309. [DOI] [PubMed] [Google Scholar]
- 246.Kambach C., Walke,S. and Nagai,K. (1999) Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol., 9, 222–230. [DOI] [PubMed] [Google Scholar]
- 247.Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- 248.Will C.L. and Luhrmann,R. (1997) Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol., 9, 320–328. [DOI] [PubMed] [Google Scholar]
- 249.Will C.L. and Luhrmann,R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol., 13, 290–301. [DOI] [PubMed] [Google Scholar]
- 250.Valadkhan S. and Manley,J.L. (2001) Splicing-related catalysis by protein-free snRNAs. Nature, 413, 701–707. [DOI] [PubMed] [Google Scholar]
- 251.Mironov A.A., Fickett,J.W. and Gelfand,M.S. (1999) Frequent alternative splicing of human genes. Genome Res., 9, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Brett D., Hanke,J., Lehmann,G., Haase,S., Delbruck,S., Krueger,S., Reich,J. and Bork,P. (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett., 474, 83–86. [DOI] [PubMed] [Google Scholar]
- 253.Kaufer N.F. and Potashkin,J. (2000) Analysis of the splicing machinery in fission yeast: a comparison with budding yeast and mammals. Nucleic Acids Res., 28, 3003–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Teplova M., Tereshko,V., Sanishvili,R., Joachimiak,A., Bushueva,T., Anderson,W.F. and Egli,M. (2000) The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding. Protein Sci., 9, 2557–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- 256.Mayer R.J. (2000) The meteoric rise of regulated intracellular proteolysis. Nature Rev. Mol. Cell. Biol., 1, 145–148. [DOI] [PubMed] [Google Scholar]
- 257.Makarova O.V., Makarov,E.M. and Luhrmann,R. (2001) The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J., 20, 2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lygerou Z., Christophides,G. and Seraphin,B. (1999) A novel genetic screen for snRNP assembly factors in yeast identifies a conserved protein, Sad1p, also required for pre-mRNA splicing. Mol. Cell. Biol., 19, 2008–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Aravind L. and Ponting,C.P. (1998) Homologues of 26S proteasome subunits are regulators of transcription and translation. Protein Sci., 7, 1250–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Hofmann K. and Bucher,P. (1998) The PCI domain: a common theme in three multiprotein complexes. Trends Biochem. Sci., 23, 204–205. [DOI] [PubMed] [Google Scholar]
- 261.Makarova K.S., Aravind,L. and Koonin,E.V. (2000) A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci., 25, 50–52. [DOI] [PubMed] [Google Scholar]
- 262.Delaney S.J., Hayward,D.C., Barleben,F., Fischbach,K.F. and Miklos,G.L. (1991) Molecular cloning and analysis of small optic lobes, a structural brain gene of Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 88, 7214–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Fang S., Jensen,J.P., Ludwig,R.L., Vousden,K.H. and Weissman,A.M. (2000) Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem., 275, 8945–8951. [DOI] [PubMed] [Google Scholar]
- 264.Brown J.R. and Doolittle,W.F. (1997) Archaea and the prokaryote-to-eukaryote transition. Microbiol. Mol. Biol. Rev., 61, 456–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Sogin M.L. (1991) Early evolution and the origin of eukaryotes. Curr. Opin. Genet. Dev., 1, 457–463. [DOI] [PubMed] [Google Scholar]
- 266.Akhmanova A., Voncken,F., van Alen,T., van Hoek,A., Boxma,B., Vogels,G., Veenhuis,M. and Hackstein,J.H. (1998) A hydrogenosome with a genome. Nature, 396, 527–528. [DOI] [PubMed] [Google Scholar]
- 267.Dyall S.D. and Johnson,P.J. (2000) Origins of hydrogenosomes and mitochondria: evolution and organelle biogenesis. Curr. Opin. Microbiol., 3, 404–411. [DOI] [PubMed] [Google Scholar]
- 268.Myler P.J. and Stuart,K.D. (2000) Recent developments from the Leishmania genome project. Curr. Opin. Microbiol., 3, 412–416. [DOI] [PubMed] [Google Scholar]
- 269.Smith M.W., Aley,S.B., Sogin,M., Gillin,F.D. and Evans,G.A. (1998) Sequence survey of the Giardia lamblia genome. Mol. Biochem. Parasitol., 95, 267–280. [DOI] [PubMed] [Google Scholar]
- 270.Gardner M.J., Tettelin,H., Carucci,D.J., Cummings,L.M., Aravind,L., Koonin,E.V., Shallom,S., Mason,T., Yu,K., Fujii,C. et al. (1998) Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science, 282, 1126–1132. [DOI] [PubMed] [Google Scholar]
- 271.Aravind L., Tatusov,R.L., Wolf,Y.I., Walker,D.R. and Koonin,E.V. (1998) Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet., 14, 442–444. [DOI] [PubMed] [Google Scholar]
- 272.Nelson K.E., Clayton,R.A., Gill,S.R., Gwinn,M.L., Dodson,R.J., Haft,D.H., Hickey,E.K., Peterson,J.D., Nelson,W.C., Ketchum,K.A. et al. (1999) Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature, 399, 323–329. [DOI] [PubMed] [Google Scholar]
- 273.Wainright P.O., Hinkle,G., Sogin,M.L. and Stickel,S.K. (1993) Monophyletic origins of the metazoa: an evolutionary link with fungi. Science, 260, 340–342. [DOI] [PubMed] [Google Scholar]
- 274.Braun E.L., Halpern,A.L., Nelson,M.A. and Natvig,D.O. (2000) Large-scale comparison of fungal sequence information: mechanisms of innovation in Neurospora crassa and gene loss in Saccharomyces cerevisiae. Genome Res., 10, 416–430. [DOI] [PubMed] [Google Scholar]
- 275.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 276.Aravind L., Dixit,V.M. and Koonin,E.V. (2001) Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science, 291, 1279–1284. [DOI] [PubMed] [Google Scholar]
- 277.Aguinaldo A.M., Turbeville,J.M., Linford,L.S., Rivera,M.C., Garey,J.R., Raff,R.A. and Lake,J.A. (1997) Evidence for a clade of nematodes, arthropods and other moulting animals. Nature, 387, 489–493. [DOI] [PubMed] [Google Scholar]
- 278.Ruppert E.E. and Barnes,R.D. (1994) Invertebrate Zoology. Harcourt Brace College Publishers, Orlando, FL.
- 279.Leipe D.D., Aravind,L. and Koonin,E.V. (1999) Did DNA replication evolve twice independently? Nucleic Acids Res., 27, 3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Woese C. (1998) The universal ancestor. Proc. Natl Acad. Sci. USA, 95, 6854–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Woese C.R. (2000) Interpreting the universal phylogenetic tree. Proc. Natl Acad. Sci. USA, 97, 8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Hoang C. and Ferre-D’Amare,A.R. (2001) Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell, 28, 929–939. [DOI] [PubMed] [Google Scholar]
- 283.Heurgue-Hamard V., Champ,S., Engstrom,A., Ehrenberg,M. and Buckingham,R.H. (2002) The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J., 21, 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Nakahigashi K., Kubo,N., Narita,S., Shimaoka,T., Goto,S., Oshima,T., Mori,H., Maeda,M., Wada,C. and Inokuchi,H. (2002) HemK, a class of protein methyl transferase with similarity to methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc. Natl Acad. Sci. USA, 99, 1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Pintard L., Bujnicki,J.M., Lapeyre,B. and Bonnerot,C. (2002) MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. EMBO J., 21, 1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]