Abstract
The diagnosis of minimal prostatic adenocarcinoma can be challenging on prostate needle biopsy, and immunohistochemistry may be used to support the diagnosis of cancer. The International Society of Urologic Pathology currently recommends the use of the basal cell markers high–molecular-weight cytokeraratin and p63, and α-methylacyl-coenzyme-A racemase. However, there are caveats associated with the interpretation of these markers, particularly with benign mimickers. Another issue is that of early detection of presence and progression of disease and prediction of recurrence after Clinical intervention. There remains a lack of reliable biomarkers to accurately predict low-risk cancer and avoid over treatment. As such, aggressive forms of prostate cancer may be missed and indolent disease may be subjected to unnecessary radical therapy. New biomarker discovery promises to improve early detection and prognosis and to provide targets for therapeutic interventions. In this review, we present the emerging immunohistochemical biomarkers of prostate cancer PTEN, ERG, FASN, MAGI-2, and SPINK1, and address their diagnostic and prognostic advantages and limitations.
Keywords: prostate cancer, biomarkers, diagnosis, prognosis, PTEN, ERG, MAGI-2, SPINK1, FASN
The diagnosis of prostatic adenocarcinoma is based on the evaluation of a combination of cytologic and architectural features. However, evaluation based solely on morphology is often insufficient, particularly in cases with minimal carcinoma. Immunohistochemistry is commonly used to support the morphologic impression of prostate cancer. The most commonly used markers include those specific for basal cells, high–molecular-weight cytokeratin (HMWCK) and p63, and α-methylacyl-CoA racemase (AMACR). Although typically adenocarcinoma lacks expression of basal cell markers, benign lesions such as adenosis, atrophy, or benign glands may also demonstrate similar basal cell loss.1–4 Conversely, HMWCK staining in a nonbasal distribution and aberrant diffuse expression of p63 may occasionally be observed in prostate cancer.5–7 In addition, AMACR also stains 5% to 21% of benign prostatic glands,1,3,8,9 and up to 18% of cases of adenosis,10 which limits its specificity for the diagnosis of adenocarcinoma. The current International Society of Urologic Pathology recommendations suggest using “HMWCK, or p63 or a combination of the two with AMACR either in a double or triple cocktail” for the workup of “small foci of atypical glands suspicious for prostatic adenocarcinoma.”11 However, successful treatment of prostatic adenocarcinoma relies not only on sensitive and specific methods used for diagnosis, but also on prognostication of progressive versus indolent disease and detection of recurrence. Current advances in molecular techniques have provided new methods for biomarker discovery. This review will focus on discussing new concepts and existing challenges of emerging prognostic markers of prostatic adenocarcinoma, and address their utility in the immunohistochemical diagnosis of prostate cancer.
ETS-RELATED GENE (ERG)
Biology and Role in Tumorigenesis
ERG, located on chromosome 21q22.2 encodes a member of the erythroblast transformation-specific (ETS) family of transcriptions factors, which includes 30 ETS family genes.12 The proteins encoded by this gene are mainly expressed in the nucleus and act as activators or repressors of transcription, affecting development and differentiation of multiple cell types. Initially reported by Tomlins et al,13 ERG gene fusion with the androgen-driven promoter of the TMPRSS2 gene is the most common recurrent genetic alteration in prostate cancer, accounting for 50% of Clinically localized prostate cancers in prostatespecific antigen-screened patient cohorts,14 and with a reported frequency of 15% in a population-based cohort.15 Gene fusions involving other ETS family members, primarily ETV1 but also ETV4 and ETV5, together constitute <10% of prostate cancer genetic abnormalities.16,17 ERG translocation has been detected in 10% to 20% of highgrade prostatic intraepithelial neoplasia (HGPIN) adjacent to prostate cancer harboring the ERG fusion, but it is otherwise infrequent in isolated HGPIN. This suggests a possible early role for ERG in the progression from HGPIN to cancer.18 In prostate cancer, the result of the TMPRSS2:ERG translocation induces the promoter region containing androgen-sensitive elements of TMPRSS2 to fuse with the coding region of ERG, leading to androgen-induced ERG overexpression.13
Prognostic Value of ETS-related Gene
The prognostic value of ERG is a matter of debate. Several studies have described an association between ERG fusion and poor prognostic indicators, including higher Gleason grade, pathologic stage, biochemical recurrence, metastases, and cancer-specific mortality.15,19–24 However, other studies have shown no correlation with outcome and biochemical recurrence.25–27
Clinical Applications
The diagnostic utility of ERG has been evaluated in several studies. ERG expression by immunohistochemistry was detected in 9% of prostatic “atypical glands suspicious for carcinoma.”28 When using a combined ERG/AMACR/HMWCK/p63 immunohistochemistry, ERG and AMACR were positive in 45% and 94% of prostate cancers, while ERG modified an initial diagnosis of atypical glands to prostate cancer in 28% cases.29 In a study of limited prostatic adenocarcinoma, combined p63 and ERG staining detected 42% of carcinoma, and ERG expression was identified in 5% of HGPIN cases and in rare cases of benign glands adjacent to cancer.30 In another study, 36% of prostatic adenocarcinomas, 27% of HGPIN, 13% of HGPIN with adjacent atypical glands, and none of benign mimickers were positive for ERG. Furthermore, ERG demonstrated higher specificity for cancer than AMACR (0.87 vs. 0.23), but lower sensitivity (0.36 vs. 0.95).31 ERG expression was also detected in 58% of intraductal carcinomas and 27% of borderline intraductal proliferations,32 and ERG rearrangements were identified in 45% of prostatic small cell carcinomas, but not in lung small cell carcinomas.33 Intertumoral and intratumoral heterogenous ERG expression by immunohistochemistry has been described.30,34,35 Figure 1 illustrates examples of the utility and role of ERG in routine Clinical applications.
FIGURE 1.
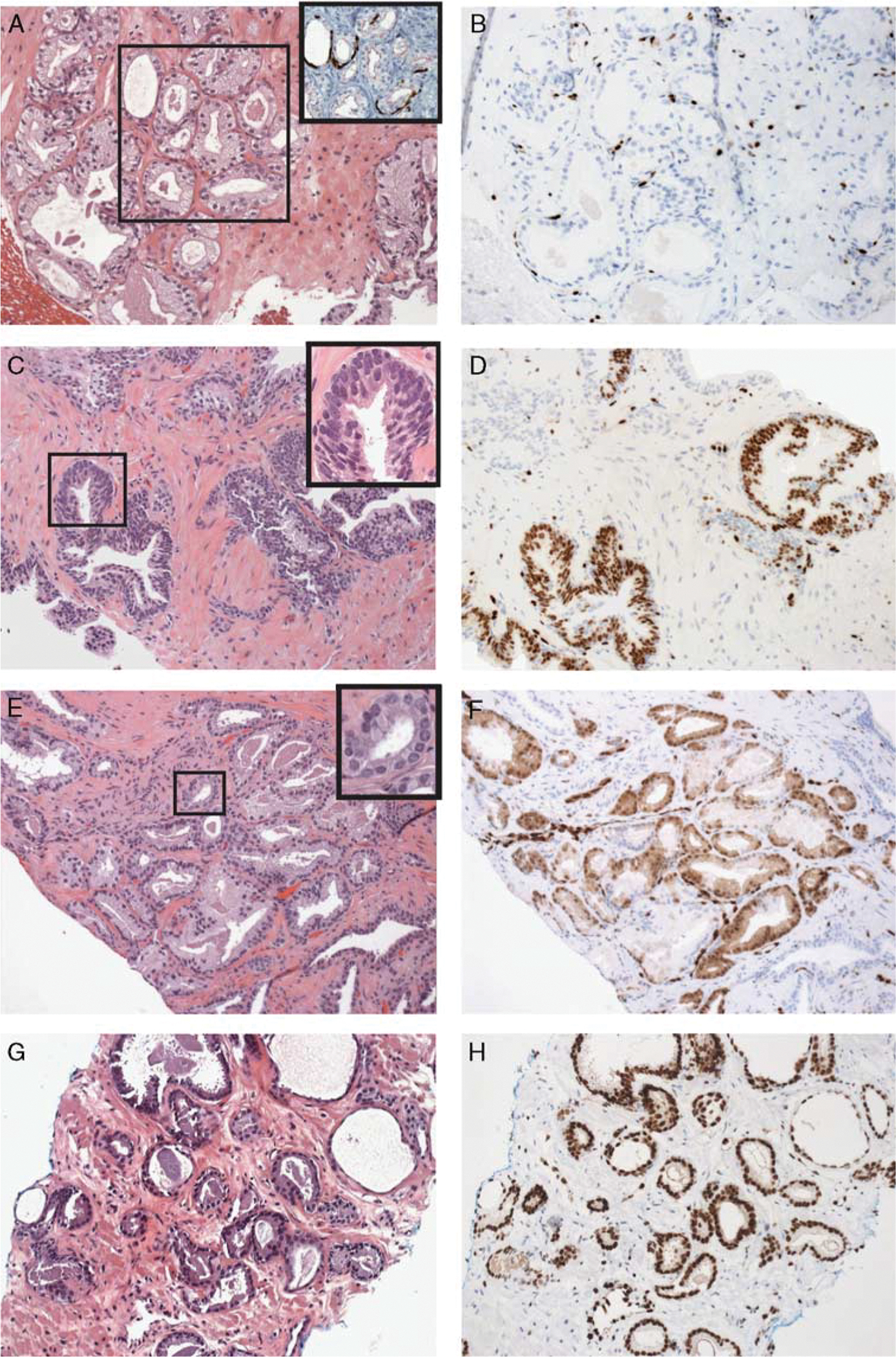
A and B, Adenosis (inset of A: prostate cocktail immuno staining) with negative staining for ETS-related gene (ERG). C and D, High-grade prostatic intraepithelial neoplasia with positive ERG staining. Insect of C shows nuclear atypia and prominent nucleoli. E and F, Small focus of atypical glands with positive ERG staining. Insect of E shows focal nuclear atypia. G and H, Gleason 6 adenocarcinoma with positive ERG staining (original magnification, _20). Reprinted from Tomlins et al36 with permission from Archives of Pathology & Laboratory Medicine. Copyright 2012 College of American Pathologists. All permission requests for this image should be made to the copyright holder.
A strong correlation between ERG gene rearrangements by fluorescence in situ hybridization (FISH) and immunohistochemistry has been reported with sensitivity and specificity ranging from 95.7% to 100% and 96.5% to 100%, respectively. However, despite high specificity, the potential utility of ERG in biopsy specimens is limited in view of the relatively frequent expression in foci of HGPIN adjacent to adenocarcinoma, low sensitivity, and intratumoral heterogeneity.11
PHOSPHATASE AND TENSIN HOMOLOG (PTEN)
Biology and Role in Tumorigenesis
PTEN, localized on chromosome 10 encodes a phosphatase that dephosphorylates phosphatidylinositol-3,4,5-trisphosphate, a second messenger in the phosphatidylinositide 3-kinase (PI3K)-protein kinase B (PKB) signaling pathway. By negatively regulating the (PI3K)/PKB signaling pathway it functions as a tumor suppressor. As a result, mutation of PTEN is a key step in disinhibition of the PI3K-PKB pathway, which in turn becomes over-active in prostate cancer. PTEN alterations consist of genomic deletions with heterozygous and, less likely homozygous mutations,37–42 epigenetic silencing,43,44 posttranscriptional regulation,45 and proto-oncogenic miRNA-dependent regulation.46 Haploinsufficiency may also contribute to genomic instability.47–49
Prognostic Value of Phosphatase and Tensin Homolog
Frequent PTEN alterations are associated with Clinically significant prostate cancer, high Gleason score, and biochemical recurrence. PTEN loss is infrequent in Gleason score 6 biopsies,49 and has not been detected in HGPIN, suggesting a later event in carcinogenesis.50,51 Genomic deletion of PTEN and loss of PTEN expression in Gleason score 6 biopsies have been associated with increased risk of upgrading at radical prostatectomy,52 and unfavorable Clinical outcome.38,53–55 PTEN inactivation may also be implicated in the mechanisms of progression to androgen independence.56,57 PTEN deletion or mutation has been reported in 20% to 40% of localized cancers,53,57,58 and up to 60% of metastases.38
Methods of Measurement
Although FISH is the gold standard for detection of genomic alterations of PTEN, recently immunohistochemistry has been validated as an alternative assay in prostate cancer59 (Figs. 2A, B). This may provide additional benefits to PTEN detection as a screening methodology, in view of the relatively inexpensive and less cumbersome immunohistochemistry-based methods, as well as the potential to detect PTEN loss by alternative mechanisms that would not be otherwise detected by FISH, such as posttranscriptional, epigenetic, and proto-oncogenic miRNA-dependent regulation. In a manual and Clinical-grade automated platform, sensitivity of PTEN immunohistochemistry for hemizygous and homozygous deletion was 87% and 86% versus 65% and 97%, respectively.59,60
FIGURE 2.
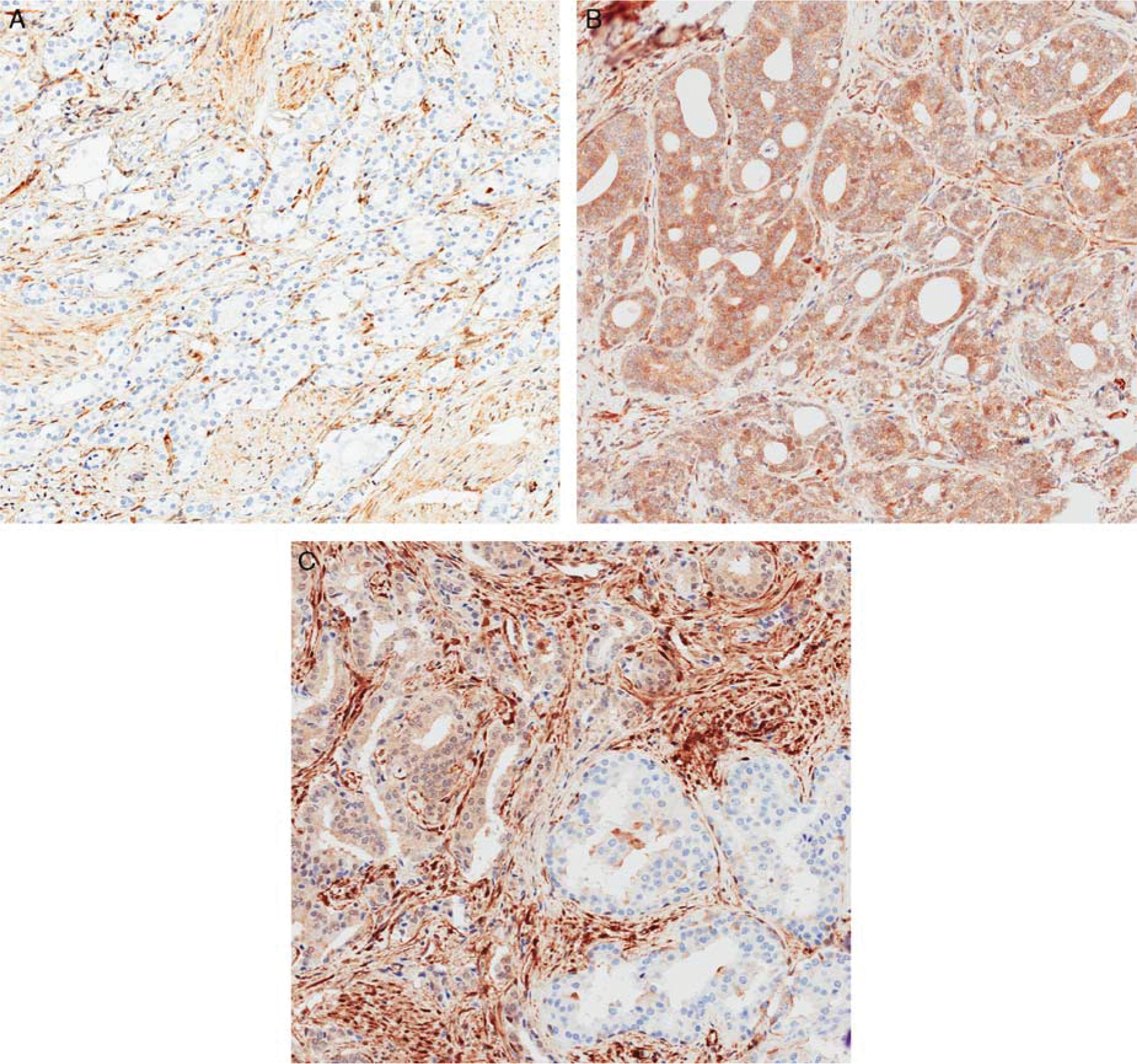
Phosphatase and tensin homolog (PTEN) expression by immunohistochemistry in prostate tissue: A, cancer with PTEN loss. The intervening stroma shows retained PTEN expression. B, Cribriform adenocarcinoma with retained PTEN expression. C, Heterogenous PTEN expression in malignant glands (original magnification, × 200).
Intratumoral and intertumoral PTEN heterogeneity has been demonstrated by immunohistochemistry and FISH, likely reflecting the presence of an “index tumor” or different tumor clones, and reflecting different biology underlying different morphologic forms of prostate cancer49,61,62 (Fig. 2C). Although heterogenous expression has also been observed for ERG, the pattern of PTEN loss is even more diverse, with 68% of cases demonstrating partial loss and 32% demonstrating complete loss in tumor cells by immunohistochemistry, versus 42% and 5% of cases with intertumoral and intratumoral staining heterogeneity for ERG.63
Clinical Applications
Although PTEN has been predominantly identified as a prognostic rather than diagnostic marker for prostate cancer, recent studies have shown cytoplasmic PTEN loss in the majority of intraductal carcinoma and atypical intraductal cribriform proliferation cases, and intact PTEN expression in HGPIN, suggesting a potential role for this marker in distinguishing intraductal carcinoma from PIN and in the differential diagnosis of atypical intraductal cribriform proliferations of the prostate.32,50
FATTY ACID SYNTHASE (FASN)
Biology and Role in Tumorigenesis
FASN is a 250 to 270kD multifunctional, homodimeric enzyme that synthesizes long-chain fatty acids. The main product of FASN is a 16-carbon fatty acid, palmitate. In normal conditions FASN converts excess carbohydrate into fatty acids, which are esterified to storage triacylglycerols and represent a source of energy obtained through β-oxidation. In a well-nourishment state with sufficient levels of dietary fat, FASN has a limited role, as fatty acids are normally obtained through the diet.64 The enzyme is expressed minimally in normal cells, but highly in liver and adipose tissue.
A significant role of FASN in cancer biology has been documented in several studies. FASN upregulation could be induced by an increased use of the glycolytic pathway for energy production, leading, in turn to an increase in the substrates for de novo fatty acid synthesis (Warburg effect).65,66 Alternatively, actively proliferating tumor cells could activate mechanisms to supply the increased demand of structural components of the cell membrane.66 In the redox balance despite surrounding conditions of extreme highly hypoxic and acidotic environment of tumors, FASN hypoxia, contributing to develop alternative survival overexpression could result in a significant improvement in mechanism.65,67,68 Finally, FASN can act as a prostate cancer oncogene by inhibiting the intrinsic pathway of apoptosis in mouse models.69
Prognostic Value of Fatty Acid Synthase
FASN expression has been detected in multiple cancer types, including kidney, pancreas, lung, colorectal, ovarian, breast, stomach, prostate, retinoblastoma, and soft tissue sarcomas,70–77 among others. Higher levels of FASN correlate with increasing tumor burden, later stages of disease, and poor prognosis.78–83
FASN overexpression has been described in prostate cancer in several studies.67,69,80,84–86 FASN germline polymorphisms have been significantly associated with risk of lethal cancer,87 while correlation with higher Gleason grade has been identified with nuclear FASN expression by immunoblot analysis of cell lysates, immunohistochemistry, and confocal microscopy.79 Further, immunohistochemical expression of FASN was a significant predictor of cancer progression,80 as well as pathologic stage.88
Clinical Applications
FASN expression has been detected by immunohistochemistry in the cytoplasm of normal prostatic epithelium and HGPIN with significantly higher intensity in neoplastic tissue (Fig. 3) and with a pattern of staining that was sufficiently distinct in prostatic adenocarcinoma compared to benign glands.89 In a comparison study with AMACR, 91% of AMACR-negative neoplastic glands demonstrated cytoplasmic FASN expression,90 and both markers showed comparable areas under the curve in receiver operating characteristic analysis, and comparable sensitivity, specificity, and accuracy rates at optimal cutoff intensities.89 Thus, FASN could represent a potential marker to complement but not substitute AMACR in the diagnosis of prostatic adenocarcinoma.
FIGURE 3.
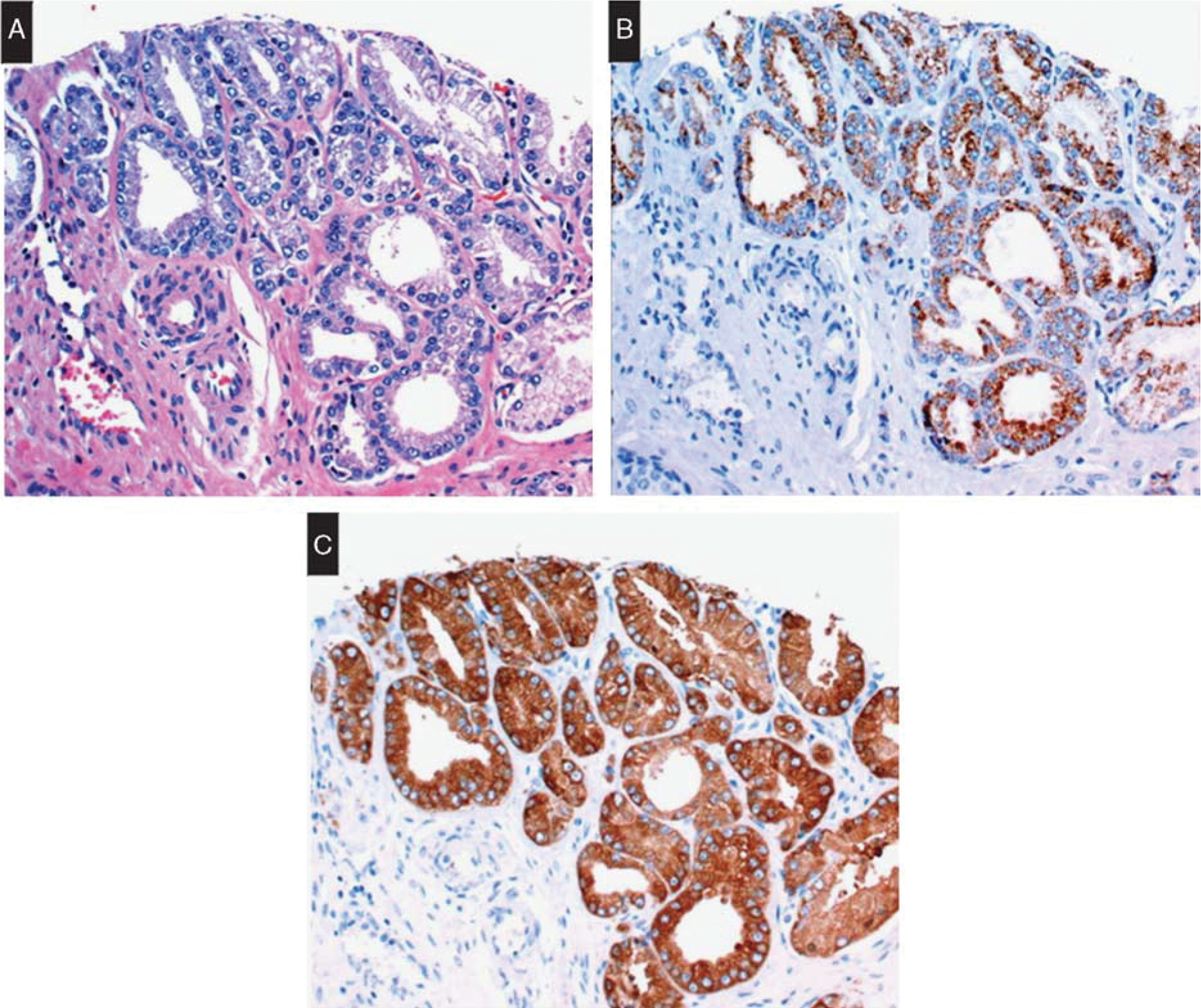
Fatty acid synthase in prostatic adenocarcinoma. A, Focus of Gleason 6 adenocarcinoma. B, Cytoplasmic AMACR expression. C, Cytoplasmic FASN expression. Reprinted from Wu et al89 with permission from Oxford University Press. Copyright 2016 Copyright Clearance Center Inc. All permission requests for this image should be made to the copyright holder.
MAGI-2
Biology and Role in Tumorigenesis
Membrane-associated guanylate kinase, WW and PDZ domain containing 2 (MAGI-2), also known as S-SCAM, GKAP91,92 or atrophin interacting protein-193 is a member of the membrane-associated guanylate kinase with an inverted arrangement of protein-protein interaction domains family, which also includes MAGI-1 and MAGI-3. There are 3 isoforms of MAGI-2-α, MAGI-2-β, and MAGI-2-γ, which are generated by differential translational initiations from multiple sites.94 Membrane-associated guanylate kinases are highly expressed in mouse brain at synaptic junctions,92,93 and MAGI-2 gene mutations have been implicated in neurological diseases such as schizophrenia,95 and infantile spasms.96 MAGI-2 is also expressed in kidney podocytes, where it interacts with the slit diaphragm protein nephrin. Studies in MAGI-2 knock-out mice have demonstrated that loss of MAGI-2 expression leads to slit diaphragm disruption, podocyte foot process effacement, and severe podocyte loss. MAGI-2-null mice develop rapidly progressive glomerular disease and renal failure.97 In the intestine, MAGI-2 binds to G-protein-coupled receptor involved in fluid and electrolyte secretion98 and has been implicated in the pathogenesis of celiac disease99 and inflammatory bowel disease.100
In cancer biology, MAGI-2 acts as a scaffolding protein that interacts with signaling proteins in multiple pathways, including PTEN.101,102 MAGI-2 binds to the C-terminus of PTEN through its PDZ domain in a yeast 2-hybrid assay, resulting in increased PTEN stability and phosphatase activity.101 MAGI-2 rearrangements with frame shift mutation have been demonstrated in a melanoma Cancer Cell line.103 In hepatocellular carcinoma, MAGI-2 upregulates PTEN expression by decreasing protein degradation and inhibits cell migration and proliferation,104 whereas epigenetic silencing has been shown in cervical cancer.105 In lung cancer, miR-134, miR-487b, and miR-655 target MAGI-2 and promote epithelial-mesenchymal transition, a mechanism involved in the process of metastasis.106
Role of MAGI-2 in Prostate Cancer
Previous studies have proposed that rearrangements of the MAGI-2 gene could alter the interaction of MAGI-2 with PTEN or other scaffolding proteins, including SMAD3, β-catenin, or the activin type 2 receptor.107 In cell culture, MAGI-2 inhibited epithelial-mesenchymal transition, a key event in the mechanism of invasion and metastasis, while miRNA-induced downregulation of MAGI-2 was permissive for transforming growth factor β−1–induced resistance to the tyrosine kinase inhibitor gefinitib.106 MAGI-2 gene rearrangement, including two independent but closely aligned inversions and two long-range intrachromosomal inversions have been recently demonstrated.108 Mutation of MAGI-2 is theorized to contribute to prostate carcinogenesis by driving PKB phosphorylation.109 A recent study has shown decreased expression of MAGI-2 mRNA in prostate cancer and prostate Cancer Cell lines, with no difference in expression between benign prostatic hyperplasia and normal tissue. Genetic alterations of PTEN result in loss of MAGI-2 mRNA, but not always PTEN mRNA.110
Clinical Applications
We have previously demonstrated that MAGI-2 staining by immunohistochemistry is expressed in 96% of adenocarcinomas with different Gleason grades and 21% of adjacent benign tissue with a trend in decreased staining intensity in higher Gleason grades (Fig. 4). MAGI-2 was positive in secretory cells and focally in basal cells with predominantly cytoplasmic and focally nuclear expression. Intensity of cytoplasmic staining of MAGI-2 was significantly higher in HGPIN and cancer compared with benign glands and benign prostatic hyperplasia by both visual and image analysis.111 There was no significant difference in MAGI-2 expression in HGPIN and adenocarcinoma, suggesting that this marker may not be useful in differentiating these lesions. In another study, MAGI-2 and AMACR were similarly significantly overexpressed in cancer in all ranges of Gleason grades compared with benign glands. At all H-score cutoffs, MAGI-2 was more sensitive but less specific in distinguishing benign from malignant glands by receiver operating characteristic analysis. Using digital analysis, the area under the curve for AMACR was higher than that for MAGI-2. However, visual evaluation showed that MAGI-2 had slightly higher accuracy than AMACR in distinguishing between benign glands and adenocarcinoma with a greater discriminatory power, especially evident in foci of adenocarcinoma that lacked AMACR expression. Thus, MAGI-2 could be of Clinical utility in the evaluation of prostate needle biopsy material, especially when other markers are not discriminatory.112
FIGURE 4.
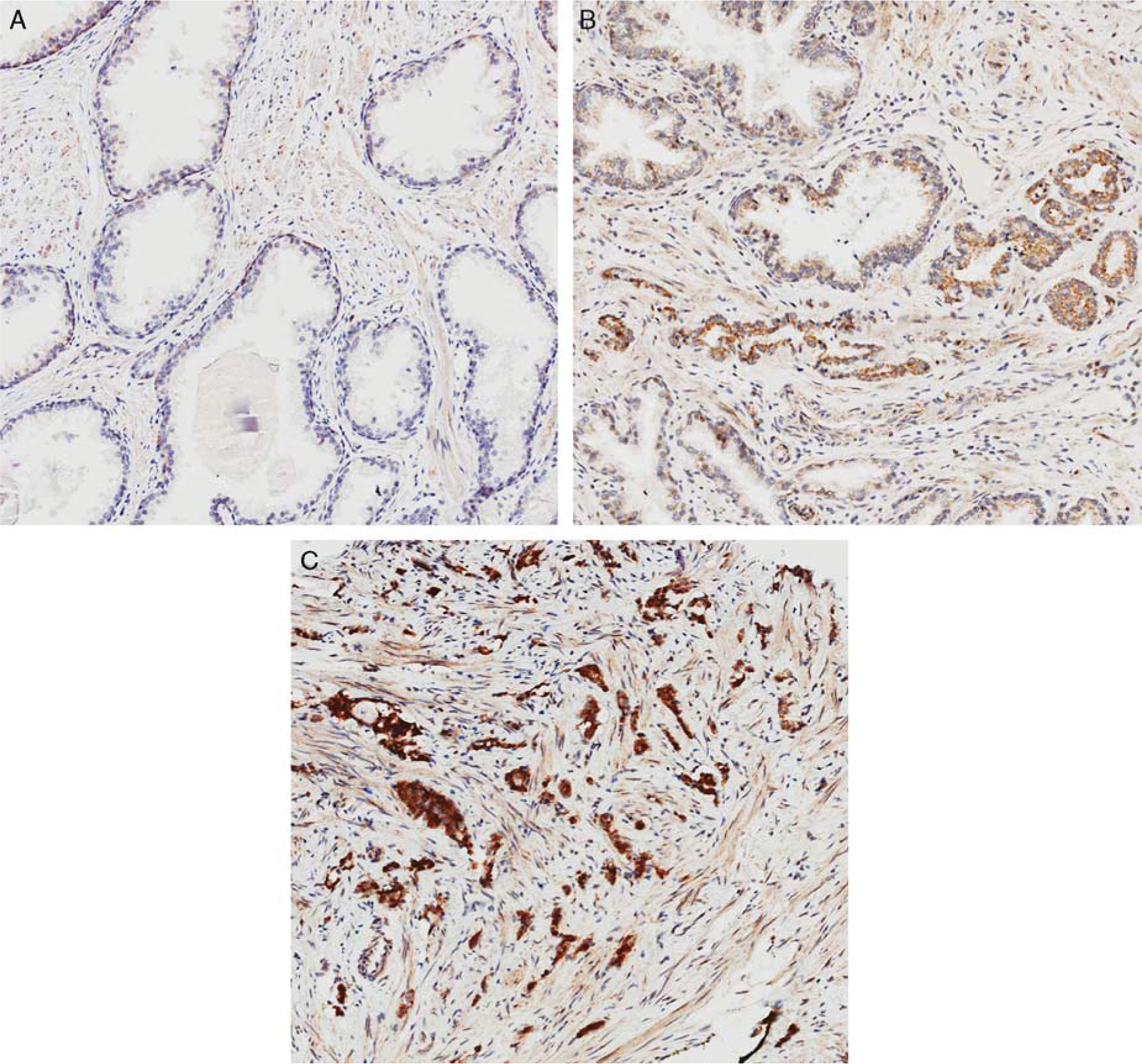
MAGI-2 expression by immunohistochemistry in prostate tissue: A, benign glands with focal basal cell staining for MAGI-2 and lack of expression in secretory cells. B, A small focus of Glason pattern 3 adenocarcinoma infiltrating between benign glands. Focally benign glands show weak cytoplasmic MAGI-2 expression. C, Gleason pattern 5 adenocarcinoma with strong MAGI-2 expression (original magnification, × 200).
SERINE PEPTIDASE INHIBITOR, KAZAL TYPE 1 (SPINK1)
Biology and Role in Tumorigenesis
SPINK1 is a trypsin inhibitor secreted from acinar cells of the exocrine pancreas, and prevents trypsin-catalyzed intra-acinar premature activation of zymogens. Overexpression of SPINK1 protein has been associated with oncogenic properties in several studies. SPINK1 has been found to be the most overexpressed gene in hepatitis C virus–induced hepatocellular carcinoma by gene expression profiling.113 Overexpression of SPINK1 protein has also been detected in carcinoma of the colon,114,115 lung,116 and pancreas.117
In prostate cancer, SPINK1 has been identified as the second-ranked candidate oncogene from transcriptomic data in a “cancer profile outlier analysis” detecting “oncogene outliers,” genes with a systematic increase in expression in a small number of cancer samples.118 SPINK1 is overexpressed in about 5% to 10% of prostate cancers (Fig. 5).118 Although the majority of SPINK1-positive cancers have been observed in ETS rearrangement–negative prostate cancers, recent studies have detected SPINK1 overexpression in 4% of ERG-positive cases.119 SPINK1 overexpression has been associated with an increased risk of disease progression and biochemical recurrence in hormonally and surgically treated prostate cancer cohorts,118,120,121 higher Gleason scores and African American population.27 However, other studies have shown no significant association between SPINK1 and Gleason grade, tumor stage and biochemical recurrence, as well as prostate cancer mortality.119
FIGURE 5.
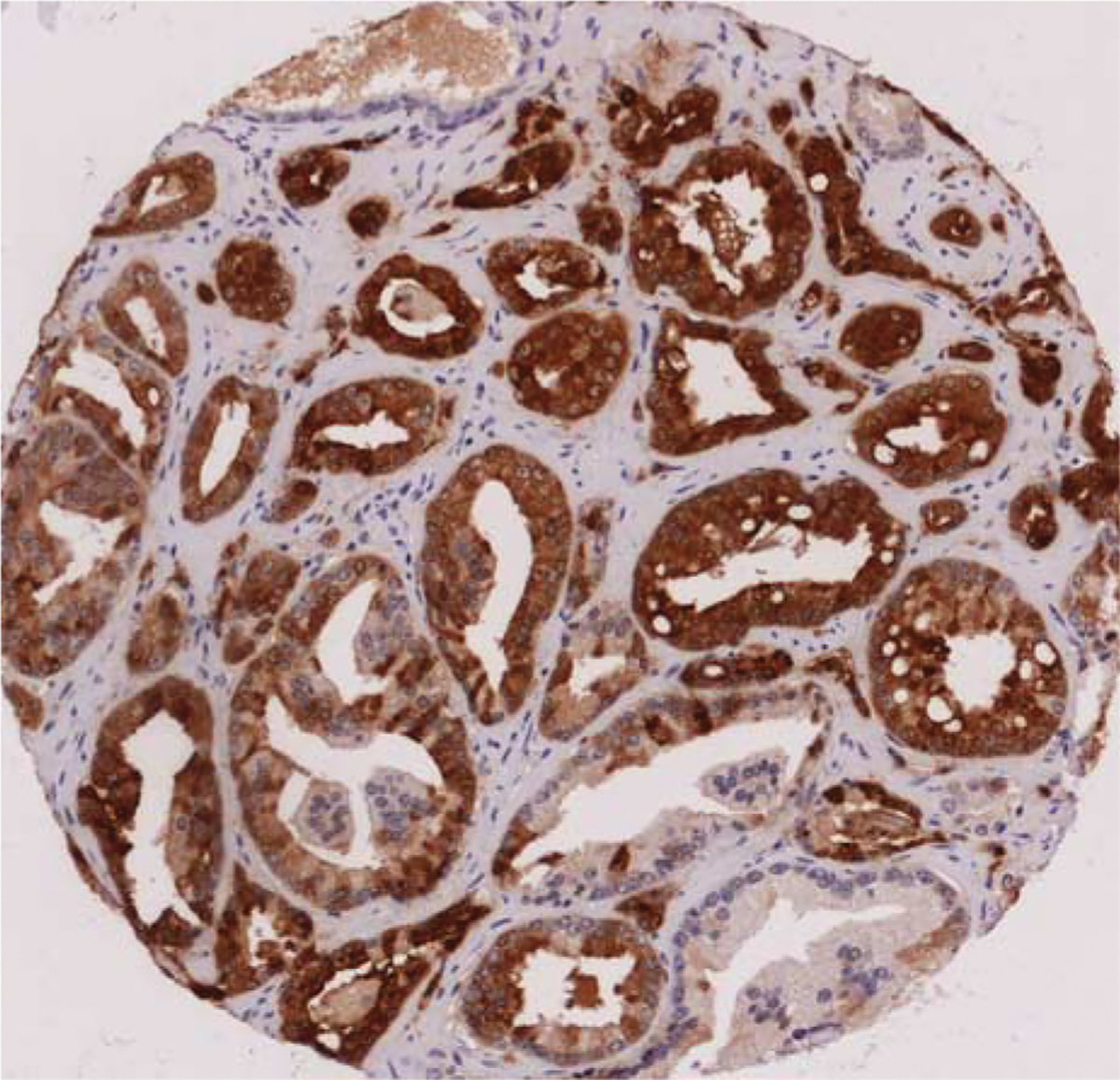
SPINK1 expression in Gleason 6 prostatic adenocarcinoma (original magnification, × 100). Image courtesy of Dr Richard Flavin, Center for Molecular Oncologic Pathology; Department of Histopathology, St. James’s Hospital and Trinity College Dublin Medical School, Dublin, Ireland (rflavin@stjames.ie).
Clinical Applications
The role of SPINK1 in dual immunohistochemistry with ERG has been explored to assess tumor clonality in prostate biopsies with discontinuous involvement by prostate cancer. In this setting, 25% of biopsies with discontinuous tumor involvement had discordant ERG/SPINK1 cancer foci, consistent with molecularly distinct cancer in the same core. This finding may have significant impact in the evaluation of percent of core involved by prostate cancer and eligibility for active surveillance protocols.
CONCLUSIONS
Immunohistochemistry plays a very important role in diagnostic surgical pathology of the prostate, especially in the setting of minimal prostatic adenocarcinoma. However, caveats in the interpretation of currently recommended immunohistochemical markers, particularly with benign mimickers may limit the usefulness of these studies. With recent advances in understanding of molecular pathways associated with prostate cancer, new biomarkers with prognostic and diagnostic potential have been developed. However, the Clinical value of the proposed biomarkers awaits definitive confirmation as Clinically reliable indicators with high specificity for the diagnosis and prognosis of prostate cancer.
ACKNOWLEDGMENT
The authors would like to thank Dr Richard Flavin for kindly supplying the SPINK1 image.
S.A.A. was supported by the Department of Veterans Affairs (VA CDA IK2BX002498). The remaining authors have no funding or conflicts of interest to disclose.
REFERENCES
- 1.Jiang Z, Woda BA, Rock KL, et al. P504S: a new molecular marker for the detection of prostate carcinoma. Am J Surg Pathol. 2001;25:1397–1404. [DOI] [PubMed] [Google Scholar]
- 2.Gaudin PB, Epstein JI. Adenosis of the prostate. Histologic features in needle biopsy specimens. Am J Surg Pathol. 1995;19:737–747. [DOI] [PubMed] [Google Scholar]
- 3.Beach R, Gown AM, De Peralta-Venturina MN, et al. P504Simmunohistochemical detection in 405 prostatic specimens including 376 18-gauge needle biopsies. Am J Surg Pathol. 2002;26:1588–1596. [DOI] [PubMed] [Google Scholar]
- 4.Amin MB, Tamboli P, Varma M, et al. Postatrophic hyperplasia of the prostate gland: a detailed analysis of its morphology in needle biopsy specimens. Am J Surg Pathol. 1999;23:925–931. [DOI] [PubMed] [Google Scholar]
- 5.Brimo F, Epstein JI. Immunohistochemical pitfalls in prostate pathology. Hum Pathol. 2012;43:313–324. [DOI] [PubMed] [Google Scholar]
- 6.Osunkoya AO, Hansel DE, Sun X, et al. Aberrant diffuse expression of p63 in adenocarcinoma of the prostate on needle biopsy and radical prostatectomy: report of 21 cases. Am J Surg Pathol. 2008;32:461–467. [DOI] [PubMed] [Google Scholar]
- 7.Giannico GA, Ross HM, Lotan T, et al. Aberrant expression of p63 in adenocarcinoma of the prostate: a radical prostatectomy study. Am J Surg Pathol. 2013;37:1401–1406. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Z, Wu CL, Woda BA, et al. Alpha-methylacyl-CoA racemase: a multi-institutional study of a new prostate cancer marker. Histopathology. 2004;45:218–225. [DOI] [PubMed] [Google Scholar]
- 9.Zhou M, Chinnaiyan AM, Kleer CG, et al. Alpha-methylacyl-CoA racemase: a novel tumor marker over-expressed in several human cancers and their precursor lesions. Am J Surg Pathol. 2002;26:926–931. [DOI] [PubMed] [Google Scholar]
- 10.Yang XJ, Tretiakova MS, Sengupta E, et al. Florid basal cell hyperplasia of the prostate: a histological, ultrastructural, and immunohistochemical analysis. Hum Pathol. 2003;34:462–470. [DOI] [PubMed] [Google Scholar]
- 11.Epstein JI, Egevad L, Humphrey PA, et al. Best practicesrecommendations in the application of immunohistochemistry in the prostate: report from the International Society of Urologic Pathology consensus conference. Am J Surg Pathol. 2014;38:e6–e19. [DOI] [PubMed] [Google Scholar]
- 12.Rao VN, Modi WS, Drabkin HD, et al. The human erg gene maps to chromosome 21, band q22: relationship to the 8; 21 translocation of acute myelogenous leukemia. Oncogene. 1988;3:497–500. [PubMed] [Google Scholar]
- 13.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion ofTMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. [DOI] [PubMed] [Google Scholar]
- 14.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. [DOI] [PubMed] [Google Scholar]
- 16.Tomlins SA, Mehra R, Rhodes DR, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. [DOI] [PubMed] [Google Scholar]
- 17.Helgeson BE, Tomlins SA, Shah N, et al. Characterization ofTMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. [DOI] [PubMed] [Google Scholar]
- 18.Carver BS, Tran J, Chen Z, et al. ETS rearrangements and prostate cancer initiation. Nature. 2009;457:E1. Discussion E2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehra R, Tomlins SA, Shen R, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in Clinically localized prostate cancer. Mod Pathol. 2007;20:538–544. [DOI] [PubMed] [Google Scholar]
- 20.Nam RK, Sugar L, Wang Z, et al. Expression of TMPRSS2:ERG gene fusion in prostate Cancer Cells is an important prognostic factor for cancer progression. Cancer Biol Ther. 2007;6:40–45. [DOI] [PubMed] [Google Scholar]
- 21.Nam RK, Sugar L, Yang W, et al. Expression of theTMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007;97:1690–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng LH, Wang C, Dolph M, et al. ERG protein expression is of limited prognostic value in men with localized prostate cancer. ISRN Urol. 2013;2013:786545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajput AB, Miller MA, De Luca A, et al. Frequency of theTMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60:1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang KC, Dolph M, Donnelly B, et al. ERG expression is associated with increased risk of biochemical relapse following radical prostatectomy in early onset prostate cancer. Clin Transl Oncol. 2014;16:973–979. [DOI] [PubMed] [Google Scholar]
- 25.Saramaki OR, Harjula AE, Martikainen PM, et al. TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin Cancer Res. 2008;14: 3395–3400. [DOI] [PubMed] [Google Scholar]
- 26.Gopalan A, Leversha MA, Satagopan JM, et al. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomlins SA, Alshalalfa M, Davicioni E, et al. Characterization of 1577 primary prostate cancers reveals novel biological and Clinicopathologic insights into molecular subtypes. Eur Urol. 2015;68:555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He H, Magi-Galluzzi C, Li J, et al. The diagnostic utility of novel immunohistochemical marker ERG in the workup of prostate biopsies with “atypical glands suspicious for cancer”. Am J Surg Pathol. 2011;35:608–614. [DOI] [PubMed] [Google Scholar]
- 29.Shah RB, Tadros Y, Brummell B, et al. The diagnostic use ofERG in resolving an “atypical glands suspicious for cancer” diagnosis in prostate biopsies beyond that provided by basal cell and alphα-methylacyl-CoA-racemase markers. Hum Pathol. 2013;44:786–794. [DOI] [PubMed] [Google Scholar]
- 30.Yaskiv O, Zhang X, Simmerman K, et al. The utility of ERG/P63 double immunohistochemical staining in the diagnosis of limited cancer in prostate needle biopsies. Am J Surg Pathol. 2011;35:1062–1068. [DOI] [PubMed] [Google Scholar]
- 31.Lee SL, Yu D, Wang C, et al. ERG expression in prostate needle biopsy: potential diagnostic and prognostic implications. Appl Immunohistochem Mol Morphol. 2015;23:499–505. [DOI] [PubMed] [Google Scholar]
- 32.Lotan TL, Gumuskaya B, Rahimi H, et al. Cytoplasmic PTEN protein loss distinguishes intraductal carcinoma of the prostate from high-grade prostatic intraepithelial neoplasia. Mod Pathol. 2013;26:587–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lotan TL, Gupta NS, Wang W, et al. ERG gene rearrangements are common in prostatic small cell carcinomas. Mod Pathol. 2011;24:820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minner S, Gartner M, Freudenthaler F, et al. Marked heterogeneity of ERG expression in large primary prostate cancers. Mod Pathol. 2013;26:106–116. [DOI] [PubMed] [Google Scholar]
- 35.Mertz KD, Horcic M, Hailemariam S, et al. Heterogeneity of ERG expression in core needle biopsies of patients with early prostate cancer. Hum Pathol. 2013;44:2727–2735. [DOI] [PubMed] [Google Scholar]
- 36.Tomlins SA, Palanisamy N, Siddiqui J, et al. Antibody-based detection of ERG rearrangements in prostate core biopsies, including diagnostically challenging cases: ERG staining in prostate core biopsies. Arch Pathol Lab Med. 2012;136:935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang SI, Parsons R, Ittmann M. Homozygous deletion of the PTEN tumor suppressor gene in a subset of prostate adenocarcinomas. Clin Cancer Res. 1998;4:811–815. [PubMed] [Google Scholar]
- 38.Suzuki H, Freije D, Nusskern DR, et al. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer Res. 1998;58: 204–209. [PubMed] [Google Scholar]
- 39.Yoshimoto M, Joshua AM, Cunha IW, et al. Absence ofTMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21:1451–1460. [DOI] [PubMed] [Google Scholar]
- 40.Reid AH, Attard G, Ambroisine L, et al. Molecular characterisation of ERG, ETV1 and PTEN gene loci identifies patients at low and high risk of death from prostate cancer. Br J Cancer. 2010;102:678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verhagen PC, van Duijn PW, Hermans KG, et al. The PTEN gene in locally progressive prostate cancer is preferentially inactivated by bi-allelic gene deletion. J Pathol. 2006; 208:699–707. [DOI] [PubMed] [Google Scholar]
- 42.Bismar TA, Yoshimoto M, Vollmer RT, et al. PTEN genomic deletion is an early event associated with ERG gene rearrangements in prostate cancer. BJU Int. 2011;107:477–485. [DOI] [PubMed] [Google Scholar]
- 43.Whang YE, Wu X, Suzuki H, et al. Inactivation of the tumor suppressor PTEN/MMAC1 in advanced human prostate cancer through loss of expression. Proc Natl Acad Sci U S A. 1998;95:5246–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konishi N, Nakamura M, Kishi M, et al. Heterogeneous methylation and deletion patterns of the INK4a/ARF locus within prostate carcinomas. Am J Pathol. 2002;160: 1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Jiang X. Post-translational regulation of PTEN.Oncogene. 2008;27:5454–5463. [DOI] [PubMed] [Google Scholar]
- 46.Poliseno L, Salmena L, Riccardi L, et al. Identification of themiR-106b 25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen MM, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24: 1967–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alimonti A, Carracedo A, Clohessy JG, et al. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet. 2010;42:454–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshimoto M, Ding K, Sweet JM, et al. PTEN losses exhibit heterogeneity in multifocal prostatic adenocarcinoma and are associated with higher Gleason grade. Mod Pathol. 2013;26:435–447. [DOI] [PubMed] [Google Scholar]
- 50.Morais CL, Han JS, Gordetsky J, et al. Utility of PTEN and ERG immunostaining for distinguishing high-grade PIN from intraductal carcinoma of the prostate on needle biopsy. Am J Surg Pathol. 2015;39:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy SJ, Karnes RJ, Kosari F, et al. Integrated analysis of the genomic instability of PTEN in Clinically insignificant and significant prostate cancer. Mod Pathol. 2016;29:143–156. [DOI] [PubMed] [Google Scholar]
- 52.Lotan TL, Carvalho FL, Peskoe SB, et al. PTEN loss is associated with upgrading of prostate cancer from biopsy to radical prostatectomy. Mod Pathol. 2015;28:128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshimoto M, Cunha IW, Coudry RA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor Clinical outcome. Br J Cancer. 2007;97:678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McMenamin ME, Soung P, Perera S, et al. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res. 1999;59:4291–4296. [PubMed] [Google Scholar]
- 55.Mithal P, Allott E, Gerber L, et al. PTEN loss in biopsy tissue predicts poor Clinical outcomes in prostate cancer. Int J Urol. 2014;21:1209–1214. [DOI] [PubMed] [Google Scholar]
- 56.Bertram J, Peacock JW, Fazli L, et al. Loss of PTEN is associated with progression to androgen independence. Prostate. 2006;66:895–902. [DOI] [PubMed] [Google Scholar]
- 57.Jiao J, Wang S, Qiao R, et al. Murine cell lines derived from Pten null prostate cancer show the critical role of PTEN in hormone refractory prostate cancer development. Cancer Res. 2007;67:6083–6091. [DOI] [PubMed] [Google Scholar]
- 58.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. [DOI] [PubMed] [Google Scholar]
- 59.Lotan TL, Gurel B, Sutcliffe S, et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res. 2011;17:6563–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lotan TL, Wei W, Ludkovski O, et al. Analytic validation of a Clinical-grade PTEN immunohistochemistry assay in prostate cancer by comparison with PTEN FISH. Mod Pathol. 2016;29:904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gumuskaya B, Gurel B, Fedor H, et al. Assessing the order of critical alterations in prostate cancer development and progression by IHC: further evidence that PTEN loss occurs subsequent to ERG gene fusion. Prostate Cancer Prostatic Dis. 2013;16:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krohn A, Freudenthaler F, Harasimowicz S, et al. Heterogeneity and chronology of PTEN deletion and ERG fusion in prostate cancer. Mod Pathol. 2014;27:1612–1620. [DOI] [PubMed] [Google Scholar]
- 63.Shah RB, Bentley J, Jeffery Z, et al. Heterogeneity of PTENand ERG expression in prostate cancer on core needle biopsies: implications for cancer risk stratification and biomarker sampling. Hum Pathol. 2015;46:698–706. [DOI] [PubMed] [Google Scholar]
- 64.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7: 763–777. [DOI] [PubMed] [Google Scholar]
- 65.Menendez JA, Lupu R. Oncogenic properties of the endogenous fatty acid metabolism: molecular pathology of fatty acid synthase in Cancer Cells. Curr Opin Clin Nutr Metab Care. 2006;9:346–357. [DOI] [PubMed] [Google Scholar]
- 66.Flavin R, Zadra G, Loda M. Metabolic alterations and targeted therapies in prostate cancer. J Pathol. 2011;223:283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baron A, Migita T, Tang D, et al. Fatty acid synthase: a metabolic oncogene in prostate cancer? J Cell Biochem. 2004;91:47–53. [DOI] [PubMed] [Google Scholar]
- 68.Hochachka PW, Rupert JL, Goldenberg L, et al. Going malignant: the hypoxia-cancer connection in the prostate. BioEssays. 2002;24:749–757. [DOI] [PubMed] [Google Scholar]
- 69.Migita T, Ruiz S, Fornari A, et al. Fatty acid synthase: a metabolic enzyme and candidate oncogene in prostate cancer. J Natl Cancer Inst. 2009;101:519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horiguchi A, Asano T, Asano T, et al. Fatty acid synthase over expression is an indicator of tumor aggressiveness and poor prognosis in renal cell carcinoma. J Urol. 2008;180: 1137–1140. [DOI] [PubMed] [Google Scholar]
- 71.Rashid A, Pizer ES, Moga M, et al. Elevated expression of fatty acid synthase and fatty acid synthetic activity in colorectal neoplasia. Am J Pathol. 1997;150:201–208. [PMC free article] [PubMed] [Google Scholar]
- 72.Walter K, Hong SM, Nyhan S, et al. Serum fatty acid synthase as a marker of pancreatic neoplasia. Cancer Epidemiol Biomarkers Prev. 2009;18:2380–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Witkiewicz AK, Nguyen KH, Dasgupta A, et al. Co-expression of fatty acid synthase and caveolin-1 in pancreatic ductal adenocarcinoma: implications for tumor progression and Clinical outcome. Cell cycle. 2008;7:3021–3025. [DOI] [PubMed] [Google Scholar]
- 74.Cai Y, Wang J, Zhang L, et al. Expressions of fatty acid synthase and HER2 are correlated with poor prognosis of ovarian cancer. Med Oncol. 2015;32:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vazquez-Martin A, Colomer R, Brunet J, et al. Overexpression of fatty acid synthase gene activates HER1/HER2 tyrosine kinase receptors in human breast epithelial cells. Cell Prolif. 2008;41:59–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sangeetha M, Deepa PR, Rishi P, et al. Global gene deregulations in FASN silenced retinoblastoma Cancer Cells: molecular and Clinico-pathological correlations. J Cell Biochem. 2015;116:2676–2694. [DOI] [PubMed] [Google Scholar]
- 77.Patel AV, Johansson G, Colbert MC, et al. Fatty acid synthase is a metabolic oncogene targetable in malignant peripheral nerve sheath tumors. Neuro Oncol. 2015;17:1599–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duan J, Sun L, Huang H, et al. Overexpression of fatty acid synthase predicts a poor prognosis for human gastric cancer. Mol Med Rep. 2016;13:3027–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Madigan AA, Rycyna KJ, Parwani AV, et al. Novel nuclear localization of fatty acid synthase correlates with prostate cancer aggressiveness. Am J Pathol. 2014;184:2156–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shurbaji MS, Kalbfleisch JH, Thurmond TS. Immunohistochemical detection of a fatty acid synthase (OA-519) as a predictor of progression of prostate cancer. Hum Pathol. 1996;27:917–921. [DOI] [PubMed] [Google Scholar]
- 81.Takahiro T, Shinichi K, Toshimitsu S. Expression of fatty acid synthase as a prognostic indicator in soft tissue sarcomas. Clin Cancer Res. 2003;9:2204–2212. [PubMed] [Google Scholar]
- 82.Visca P, Sebastiani V, Botti C, et al. Fatty acid synthase(FAS) is a marker of increased risk of recurrence in lung carcinoma. AntiCancer Res. 2004;24:4169–4173. [PubMed] [Google Scholar]
- 83.Ogino S, Nosho K, Meyerhardt JA, et al. Cohort study of fatty acid synthase expression and patient survival in colon cancer. J Clin Oncol. 2008;26:5713–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shah US, Dhir R, Gollin SM, et al. Fatty acid synthase gene overexpression and copy number gain in prostate adenocarcinoma. Hum Pathol. 2006;37:401–409. [DOI] [PubMed] [Google Scholar]
- 85.Dhanasekaran SM, Dash A, Yu J, et al. Molecular profiling of human prostate tissues: insights into gene expression patterns of prostate development during puberty. FASEB J. 2005;19:243–245. [DOI] [PubMed] [Google Scholar]
- 86.Ashida S, Nakagawa H, Katagiri T, et al. Molecular features of the transition from prostatic intraepithelial neoplasia (PIN) to prostate cancer: genome-wide gene-expression profiles of prostate cancers and PINs. Cancer Res. 2004;64:5963–5972. [DOI] [PubMed] [Google Scholar]
- 87.Nguyen PL, Ma J, Chavarro JE, et al. Fatty acid synthase polymorphisms, tumor expression, body mass index, prostate cancer risk, and survival. J Clin Oncol. 2010;28:3958–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Epstein JI, Carmichael M, Partin AW. OA-519 (fatty acid synthase) as an independent predictor of pathologic state in adenocarcinoma of the prostate. Urology. 1995;45:81–86. [DOI] [PubMed] [Google Scholar]
- 89.Wu X, Zayzafoon M, Zhang X, et al. Is there a role for fatty acid synthase in the diagnosis of prostatic adenocarcinoma?: a comparison with AMACR. Am J Clin Pathol. 2011;136:239–246. [DOI] [PubMed] [Google Scholar]
- 90.Tischler V, Fritzsche FR, Gerhardt J, et al. Comparison of the diagnostic value of fatty acid synthase (FASN) with alpha-methylacyl-CoA racemase (AMACR) as prostatic cancer tissue marker. Histopathology. 2010;56:811–815. [DOI] [PubMed] [Google Scholar]
- 91.Kim E, Naisbitt S, Hsueh YP, et al. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol. 1997;136:669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hirao K, Hata Y, Ide N, et al. A novel multiple PDZ domain containing molecule interacting with N-methyl-D-aspartate receptors and neuronal cell adhesion proteins. J Biol Chem. 1998;273:21105–21110. [DOI] [PubMed] [Google Scholar]
- 93.Wood JD, Yuan J, Margolis RL, et al. Atrophin-1, the DRPLA gene product, interacts with two families of WW domain-containing proteins. Mol Cell Neurosci. 1998;11: 149–160. [DOI] [PubMed] [Google Scholar]
- 94.Hirao K, Hata Y, Yao I, et al. Three isoforms of synaptic scaffolding molecule and their characterization. Multimerization between the isoforms and their interaction with N-methyl-D-aspartate receptors and SAP90/PSD-95-associated protein. J Biol Chem. 2000;275:2966–2972. [DOI] [PubMed] [Google Scholar]
- 95.Walsh T, McClellan JM, McCarthy SE, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. [DOI] [PubMed] [Google Scholar]
- 96.Marshall CR, Young EJ, Pani AM, et al. Infantile spasms is associated with deletion of the MAGI2 gene on chromosome 7q11.23-q21.11. Am J Hum Genet. 2008;83:106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Balbas MD, Burgess MR, Murali R, et al. MAGI-2 scaffold protein is critical for kidney barrier function. Proc Natl Acad Sci U S A. 2014;111:14876–14881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gee HY, Kim YW, Jo MJ, et al. Synaptic scaffolding molecule binds to and regulates vasoactive intestinal polypeptide type-1 receptor in epithelial cells. Gastroenterology. 2009;137:607–617. 617.e1–4. [DOI] [PubMed] [Google Scholar]
- 99.Wapenaar MC, Monsuur AJ, van Bodegraven AA, et al. Associations with tight junction genes PARD3 and MAGI2 in Dutch patients point to a common barrier defect for coeliac disease and ulcerative colitis. Gut. 2008;57:463–467. [DOI] [PubMed] [Google Scholar]
- 100.McGovern DP, Taylor KD, Landers C, et al. MAGI2 genetic variation and inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu X, Hepner K, Castelino-Prabhu S, et al. Evidence for regulation of the PTEN tumor suppressor by a membrane localized multi-PDZ domain containing scaffold protein MAGI-2. Proc Natl Acad Sci U S A. 2000;97:4233–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vazquez F, Grossman SR, Takahashi Y, et al. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J Biol Chem. 2001;276:48627–48630. [DOI] [PubMed] [Google Scholar]
- 103.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hu Y, Li Z, Guo L, et al. MAGI-2 Inhibits cell migration and proliferation via PTEN in human hepatocarcinoma cells. Arch Biochem Biophys. 2007;467:1–9. [DOI] [PubMed] [Google Scholar]
- 105.Chen YC, Huang RL, Huang YK, et al. Methylomics analysis identifies epigenetically silenced genes and implies an activation of beta-catenin signaling in cervical cancer. Int J Cancer. 2014;135:117–127. [DOI] [PubMed] [Google Scholar]
- 106.Kitamura K, Seike M, Okano T, et al. MiR-134/487b/655 cluster regulates TGF-beta-induced epithelial-mesenchymal transition and drug resistance to gefitinib by targeting MAGI2 in lung adenocarcinoma cells. Mol Cancer Ther. 2014;13:444–453. [DOI] [PubMed] [Google Scholar]
- 107.Nagashima S, Kodaka M, Iwasa H, et al. MAGI2/S-SCAM outside brain. J Biochem. 2015;157:177–184. [DOI] [PubMed] [Google Scholar]
- 108.Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brenner JC, Chinnaiyan AM. Disruptive events in the life of prostate cancer. Cancer Cell. 2011;19:301–303. [DOI] [PubMed] [Google Scholar]
- 110.Mahdian R, Nodouzi V, Asgari M, et al. Expression profile of MAGI2 gene as a novel biomarker in combination with major deregulated genes in prostate cancer. Mol Biol Rep. 2014;41:6125–6131. [DOI] [PubMed] [Google Scholar]
- 111.Goldstein J, Borowsky AD, Goyal R, et al. MAGI-2 in prostate cancer: an immunohistochemical study. Hum Pathol. 2016;52:83–91. [DOI] [PubMed] [Google Scholar]
- 112.Goldstein J, Roland JT, Gellert LL, et al. MAGI-2 is a sensitive and specific marker of prostatic adenocarcinoma: a comparison with AMACR. Am J Clin Pathol. 2016;146:294–302. [DOI] [PubMed] [Google Scholar]
- 113.Ohmachi Y, Murata A, Matsuura N, et al. Specific expression of the pancreatic-secretory-trypsin-inhibitor (PSTI) gene in hepatocellular carcinoma. Int J Cancer. 1993;55:728–734. [DOI] [PubMed] [Google Scholar]
- 114.Higashiyama M, Monden T, Tomita N, et al. Expression of pancreatic secretory trypsin inhibitor (PSTI) in colorectal cancer. Br J Cancer. 1990;62:954–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tomita N, Doi S, Higashiyama M, et al. Expression of pancreatic secretory trypsin inhibitor gene in human colorectal tumor. Cancer. 1990;66:2144–2149. [DOI] [PubMed] [Google Scholar]
- 116.Higashiyama M, Doi O, Kodama K, et al. Immunohistochemical analysis of pancreatic secretory trypsin inhibitor expression in pulmonary adenocarcinoma: its possible participation in scar formation of the tumor tissues. Tumour Biol. 1992;13:299–307. [DOI] [PubMed] [Google Scholar]
- 117.Ozaki N, Ohmuraya M, Hirota M, et al. Serine protease inhibitor Kazal type 1 promotes proliferation of pancreatic Cancer Cells through the epidermal growth factor receptor. Mol Cancer Res. 2009;7:1572–1581. [DOI] [PubMed] [Google Scholar]
- 118.Tomlins SA, Rhodes DR, Yu J, et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell. 2008;13:519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Flavin R, Pettersson A, Hendrickson WK, et al. SPINK1 protein expression and prostate cancer progression. Clin Cancer Res. 2014;20:4904–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Leinonen KA, Tolonen TT, Bracken H, et al. Association ofSPINK1 expression and TMPRSS2:ERG fusion with prognosis in endocrine-treated prostate cancer. Clin Cancer Res. 2010;16:2845–2851. [DOI] [PubMed] [Google Scholar]
- 121.Terry S, Nicolaiew N, Basset V, et al. Clinical value of ERG, TFF3, and SPINK1 for molecular subtyping of prostate cancer. Cancer. 2015;121:1422–1430. [DOI] [PubMed] [Google Scholar]


