Abstract
Bacterial autotrophs often rely on CO2 concentrating mechanisms (CCMs) to assimilate carbon. Although many CCM proteins have been identified, a systematic screen of CCM components has not been carried out. Here, we performed a genome-wide barcoded transposon screen to identify essential and CCM-related genes in the ɣ-proteobacterium H. neapolitanus. Screening revealed that the CCM comprises at least 17 and likely no more than 25 genes most of which are encoded in 3 operons. Two of these operons contain a two-gene locus encoding a domain of unknown function (PFAM:PF10070) and a putative cation transporter (PFAM:PF00361). Physiological and biochemical assays demonstrate that these proteins, which we name DabA and DabB for “DABs accumulate bicarbonate,” assemble into a heterodimeric complex, contain a putative β-carbonic anhydrase-like active site, and function as an energy-coupled inorganic carbon (Ci) pump. Surprisingly, DabAB operons are found in diverse bacteria and archaea. We demonstrate that functional DABs are present in the human pathogens B. anthracis and V. cholerae. Based on these results, we propose that DABs constitute a class of energized Ci pump and play a critical role in Ci metabolism throughout prokaryotic phyla.
Introduction
Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase (Rubisco) is the primary carboxylase of the Calvin-Benson-Bassham (CBB) cycle and the major entry point of Ci into the biosphere. Rubisco activity is critical to agriculture and a major flux removing anthropogenic CO2 from the atmosphere. Despite its centrality and abundance, Rubisco is not a fast enzyme1,2. Nor is Rubisco specific - all known Rubiscos can use molecular oxygen (O2) as a substrate in place of CO2 3. Oxygenation does not fix carbon and produces a product that must be recycled through metabolically-expensive photorespiratory pathways4. Many studies support the hypothesis that improvements to Rubisco could improve crop yields, but Rubisco has proven recalcitrant to protein engineering. It remains unclear whether or how Rubisco can be improved2,5,6.
Organisms that depend on Rubisco for growth often employ CO2 concentrating mechanisms (CCMs) that concentrate CO2 near Rubisco so that carboxylation proceeds at high rate and specificity7,8. All cyanobacteria and many chemolithoautotrophic proteobacteria have a CCM9. The bacterial CCM has garnered particular interest among bioengineers because it is well-understood, thought to consist of relatively few genes and operates inside single cells10. Detailed modeling suggests that transplantation of the bacterial CCM into crops might improve yields11,12 and efforts towards transplantation are already underway13.
Diverse experimental studies make it clear that the bacterial CCM requires two components to function: active Ci transport driving accumulation of HCO3− in the cytosol and organization of Rubisco with carbonic anhydrase (CA) in the lumen of a protein organelle called the carboxysome7,14,15. Energy-coupled Ci pumps keep the cytosolic HCO3− concentration high (> 10 mM) and, crucially, out-of-equilibrium with CO214,16,17. CA activity interconverts HCO3− + H+ with CO2 + H2O, and thus, the carboxysomal CA converts a high cytosolic HCO3− concentration into a high carboxysomal CO2 concentration, promoting faster carboxylation by Rubisco and competitively inhibiting oxygenation7. Genetic lesions to either component - Ci uptake systems or carboxysomes - disrupt the CCM and mutants have growth defects unless CO2 is supplemented18,19. This high-CO2 requiring (HCR) mutant phenotype is commonly-used to identify CCM components in screens18,19.
Despite early screens, a comprehensive list of bacterial CCM components remains unknown, leaving the possibility that additional activities are required for CCM function. Although well-assembled carboxysome structures can be produced in bacteria and plants13,20, the functionality of these carboxysomes in a heterologous CCM has not been demonstrated. Bioinformatic studies show that several non-carboxysomal genes are associated with carboxysome operons21,22. Further, experimental14,23 and modeling studies7,15 make it clear that energy-coupled Ci uptake systems are required for CCM function. Several different Ci pump families, including transporters and facilitated uptake systems are known for cyanobacterial lineages, but mechanistic understanding of Ci uptake is limited24.
Here we use a genome-wide barcoded transposon mutagenesis screen (RB-TnSeq) to interrogate the CCM of Halothiobacillus neapolitanus (henceforth Hnea). Hnea is a sulfur oxidizing Ɣ-proteobacterial chemolithoautotroph and a model system for studying ɑ-carboxysomes25–27. Older physiological measurements suggest that Hnea possesses an energized Ci uptake system, but the molecular identity of this activity is unknown17. In addition to producing the essential gene set for a bacterial chemolithoautotroph, we leverage our pooled mutant library to comprehensively screen for knockouts that produce an HCR phenotype. This screen identified all known CCM components and confirmed that a two-gene operon containing a large, conserved, poorly-characterized protein (PFAM:PF10070, hereafter DabA) and a cation transporter (PFAM:PF00361, hereafter DabB) is required for CCM function. Scott and colleagues have recently identified and validated homologs of these genes as a Ci import system in hydrothermal vent chemolithoautotrophs28–30. Based on this work and results further described below, we propose naming this locus the DAB operon for “DABs Accumulate Bicarbonate.”
Here we show that the products of the DAB operon form a protein complex that is capable of energetically-coupled Ci uptake. Both proteins are necessary for activity and treatment with an ionophore abrogates DAB-mediated Ci uptake. Structural homology modeling suggests that DabA contains a domain distantly homologous to a type II β-CA. Indeed, DabA binds zinc, likely in a manner similar to β-CAs. These results are consistent with a model of activity dependant on unidirectional hydration of CO2 to HCO3− in the cytosol via a CA-like mechanism and energized by coupling to a cation gradient. Phylogenomic analysis demonstrates that DAB operons are widespread throughout prokaryotes including carbon-fixing bacteria and archaea. Surprisingly, DAB operons are also found in many heterotrophic bacteria. We demonstrate that functional operons are present in the notable pathogens V. cholera and B. anthracis. We therefore propose that DABs constitute a class of Ci uptake pump whose biochemical tractability facilitates mechanistic analyses and whose widespread occurrence merits further investigation.
Results
Transposon mutagenesis and gene essentiality
We constructed a randomly-barcoded genome-wide pooled knockout library of Hnea by conjugation (Figure 1a). The conjugated vector contained a barcoded Tn5-derived transposon encoding a kanamycin resistance marker. The library was produced in 5% CO2 enabling isolation of CCM gene knockouts.
Figure 1. Transposon mutagenesis reveals the essential gene set of a chemolithoautotrophic organism.
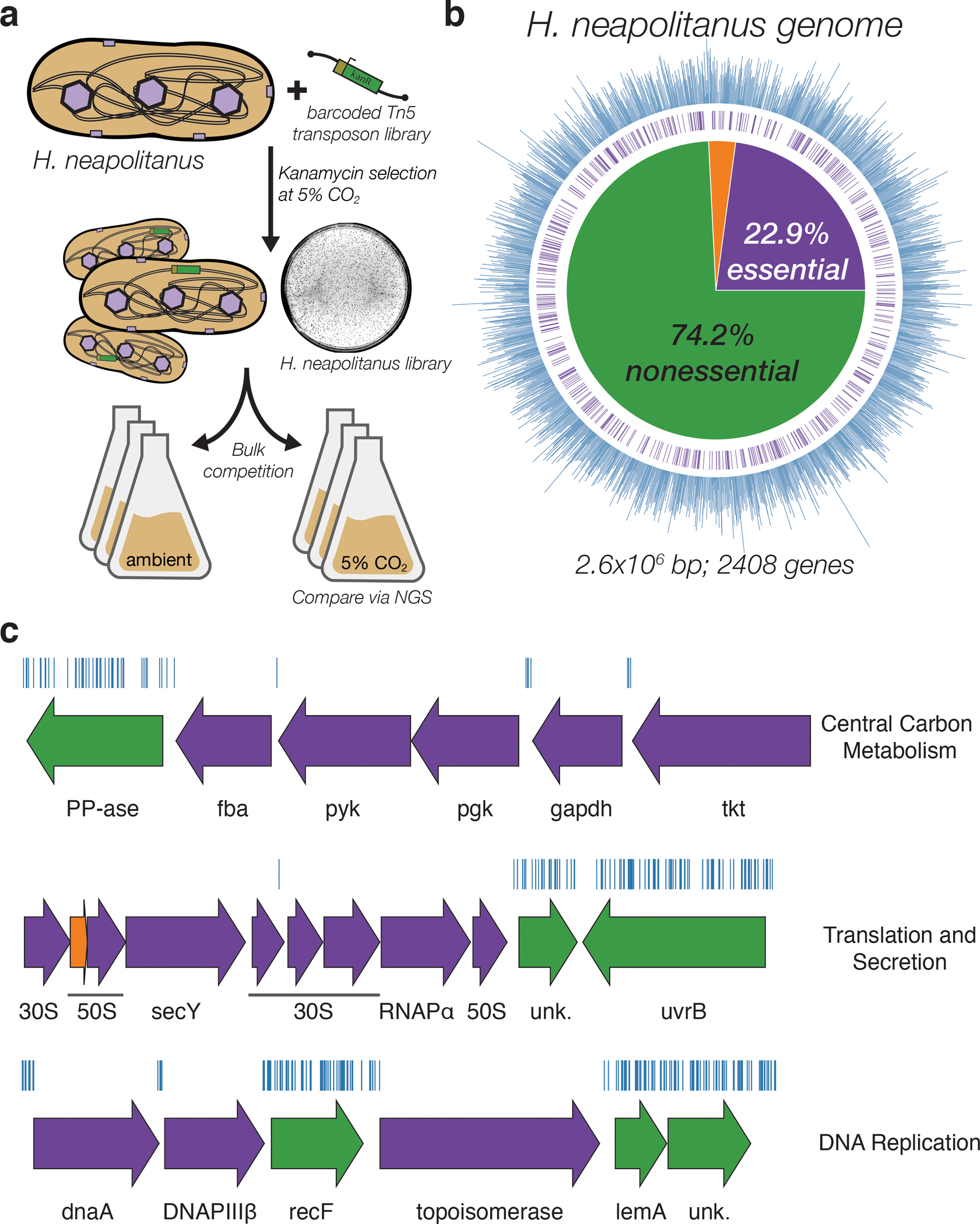
a. Schematic depicting the generation and screening of the RB-TnSeq library. Transposons were inserted into the Hnea genome by conjugation with an E. coli donor strain. The transposon contains a random 20 base pair barcode (yellow) and a kanamycin selection marker (green). Selection for colonies containing insertions was performed in the presence of kanamycin at 5% CO2 and insertions were mapped by sequencing as described in the Methods. Subsequent screens were carried out as bulk competition assays and quantified by BarSeq. b. Insertions and essential genes are well-distributed throughout the Hnea genome. The outer track (blue) is a histogram of the number of barcodes that were mapped to a 1 kb window. The inner track annotates essential genes in purple. The pie chart shows the percentages of the genome called essential (purple), ambiguous (orange), and nonessential (green). c. Representative essential genes and nonessential genes in the Hnea genome. The blue track indicates the presence of an insertion. Genes in purple were called essential and genes in green are nonessential. Genes labeled “unk.” are hypothetical proteins. The first genomic locus contains 5 essential genes involved in glycolysis or the CBB cycle including pyruvate kinase (pyk) and transketolase (tkt). The 8 essential genes in the second locus encoding 30S and 50S subunits of the ribosome, the secY secretory channel, and an RNA polymerase subunit. Essential genes in the third example locus include topoisomerase and DNA polymerase III β. A full analysis with gene names is in Supplemental Figure 1 and essentiality information for every gene can be found in supplemental file 2.
Transposon barcodes simplify the use of the library for pooled screens using the ‘barseq’ approach (Methods)31. Transposon insertion sites and cognate barcodes were mapped using standard TnSeq methods (Methods)31. The library was found to contain ~105 insertions, or one insertion for every ≈25 base pairs in the Hnea genome. Since the average gene contains ≈35 insertions, genes with no insertions are very likely essential for growth. A simple statistical model identified 551 essential genes and 1787 nonessential genes out of 2408 genes in the Hnea genome (Methods, Figure 1a–b, Supplemental File 2). The remaining 70 genes were classified as “ambiguous” due either to their short length or because replicate mapping experiments were discordant (Methods). Genes associated with known crucial functions including central carbon metabolism, ribosome production, and DNA replication were found to be essential (Figure 1c and Supplemental Figure 1). Importantly, known CCM genes, including carboxysome components, were not essential for growth at 5% CO2 (Figure 2).
Figure 2. A systematic screen for high CO2-requiring mutants identifies genes putatively associated with the CCM.
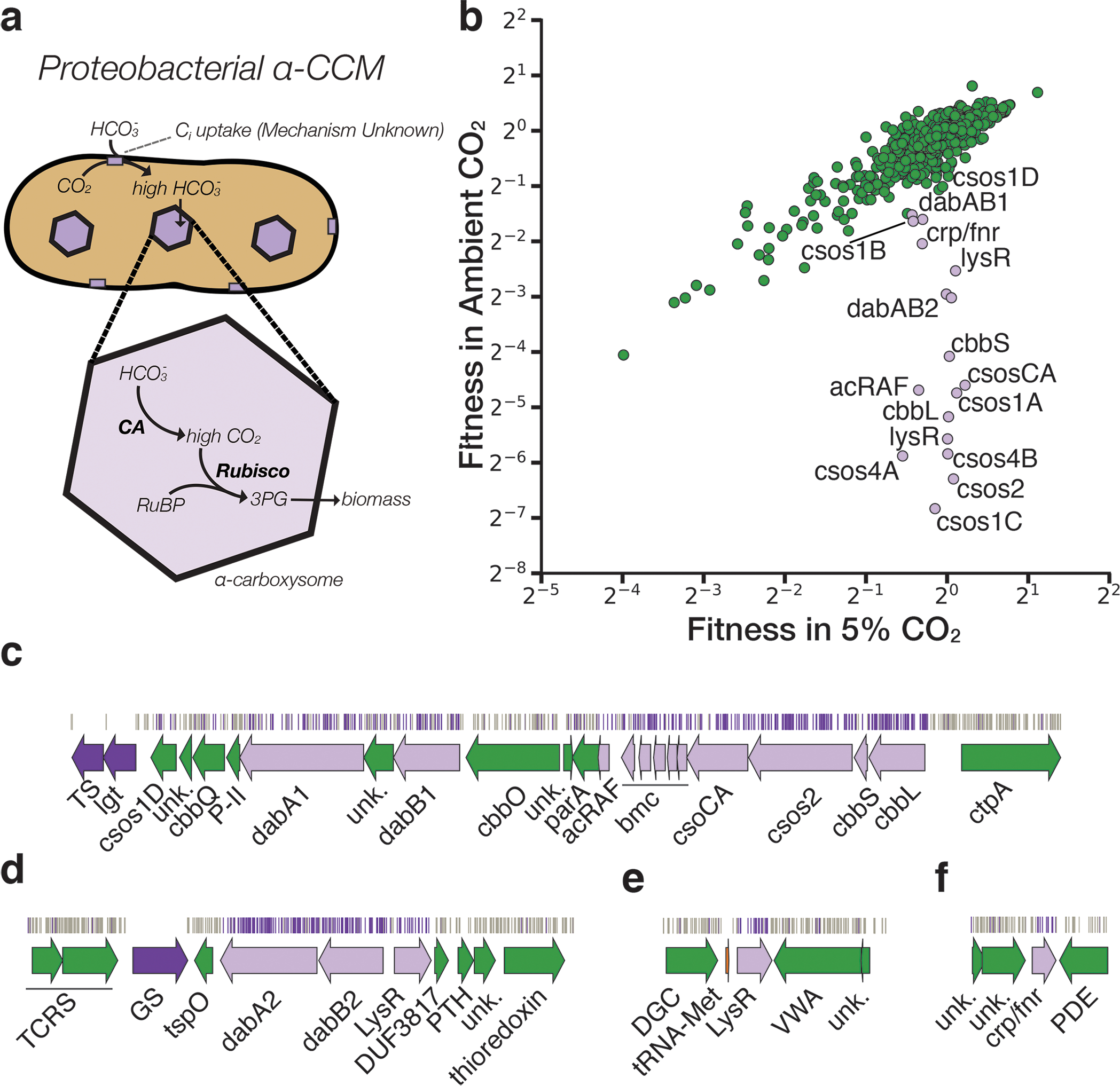
a. Simplified model of the ɑ-CCM of chemolithoautotrophic proteobacteria. Inorganic carbon is concentrated via an unknown mechanism, producing a high cytosolic HCO3− concentration. High cytosolic HCO3− is converted into high carboxysomal CO2 by CA, which is localized only to the carboxysome. b. Fitness effects of gene knockouts in 5% CO2 as compared to ambient CO2. Data is from one of two replicates of BarSeq. The effects of single transposon insertions into a gene are averaged to produce the gene-level fitness value plotted. We define HCR mutants as those displaying a twofold fitness defect in ambient CO2 relative to 5% CO2 in both replicates. HCR genes are colored light purple. Data from both replicates and the associated standard errors are shown in Supplemental Figure 2 and in supplemental file 3. Panels c-f show regions of the Hnea genome containing genes annotated as HCR in panel A. Essential genes are in dark purple, HCR genes are in light purple, and other genes are in green. The top tracks show the presence of an insertion in that location. Insertions are colored grey unless they display a twofold or greater fitness defect in ambient CO2, in which case they are colored light purple. c. The gene cluster containing the carboxysome operon and a second CCM-associated operon. This second operon contains acRAF, a Form IC associated cbbOQ-type Rubisco activase and dabAB1. d. The DAB2 operon and surrounding genomic context. e. The genomic context of a lysR-type transcriptional regulator that shows an HCR phenotype. f Genomic context of a crp/fnr-type transcriptional regulator that displays an HCR phenotype. Genes labeled “unk.” are hypothetical proteins. Full gene names are given in Supplemental Figure 3. Accession numbers and gi numbers for selected genes can be found in Supplemental Table 1.
Comprehensive screen for Hnea CCM components
Based on the current model of the bacterial CCM (Figure 2a), knockouts of CCM genes are expected to have reduced fitness in atmospheric CO2 conditions18,19. As our pooled library contains ~70,000 barcodes that map to exactly one position in the Hnea genome, we were able to use barseq to quantify the fitness effects of single gene knockouts for all nonessential Hnea genes in a pooled competition experiment (Methods, Figure 2b)31. Since the library contains roughly 20 uniquely-mapped knockouts per gene, this screen contains multiple internal biological replicates testing the effect of gene knockouts. Mutants in a particular gene were designated as HCR if the average effect of a knockout in that gene was a twofold (or greater) growth defect in ambient CO2 as compared to 5% in two replicate experiments.
As expected, knockouts of carboxysome genes consistently produced large and specific fitness defects in ambient CO2 (Figures 2b–c)27. These genes include cbbLS - the large and small subunits of the ɑ-carboxysomal Rubisco9; csoS2 - an intrinsically disordered protein required for ɑ-carboxysome assembly32; csoSCA - the carboxysomal carbonic anhydrase9; csoS4AB - the pentameric proteins thought to form vertices of the ɑ-carboxysome33; and csoS1CAB - the hexamers that form the faces of the ɑ-carboxysome shell9,25. Knockouts of csoS1D, a shell hexamer with a large central pore20,34, had too weak a phenotype to be considered HCR (Figures 2b–c). The Hnea genome also contains a non-carboxysomal Form II Rubisco that is likely not involved in CCM activity as its disruption confers no fitness defect. A number of genes that are not associated with the carboxysome structure also exhibited HCR phenotypes. These include two LysR transcriptional regulators, a Crp/Fnr type transcriptional regulator, a protein called acRAF that is involved in Rubisco assembly35,36, and two paralogous loci encoding DAB genes (hereafter DAB1 and DAB2, Figure 2b–f).
DAB operon composition
DAB1 is a cluster of 2 genes found in an operon directly downstream of the carboxysome operon (Figure 2c). Though DAB1 is part of an 11-gene operon containing several genes associated with Rubisco proteostasis, including acRAF35,36 and a cbbOQ-type Rubisco activase37, we refer to DAB1 as an “operon” for simplicity. DAB2 is a true operon and is not proximal to the carboxysome operon in the Hnea genome. These “operons” are unified in that they both display HCR phenotypes and possess similar genes (Figures 2b–d).
Both operons contain a conserved protein of unknown function (PFAM:PF10070) that we term DabA. DabAs have no predicted transmembrane helices or signal peptides and appear to be large (DabA1: 118.5 kDa, DabA2: 91.7 kDa), soluble, cytoplasmic proteins (Methods, Figure 3a). Both DAB operons also contain a member of the cation transporter family (PFAM:PF00361) that includes H+-pumping subunits of respiratory complex I and Mrp Na+:H+ antiporters38. This protein, which we call DabB (DabB1: 62.2 kDa, DabB2: 59.3 kDa), is predicted to have 12–13 transmembrane helices (Figure 3a). The complex I subunits in PF00361 are H+-pumping proteins and do not contain redox active groups, e.g. iron-sulfur clusters or quinone binding sites. Phylogenetic analysis suggests DabB proteins form a clade among PF00361 members (Supplemental Figure 4a) distinct from complex I subunits. Therefore, homology between DabB and complex I subunits (e.g. NuoL) suggests cation transport but does not imply redox activity. Importantly, operons of this type were recently demonstrated to be capable of Ci uptake in the hydrothermal vent chemolithoautotroph Hydrogenovibrio crunogenus28-30.
Figure 3. The DABs catalyze active transport of Ci and are energized by a cation gradient.
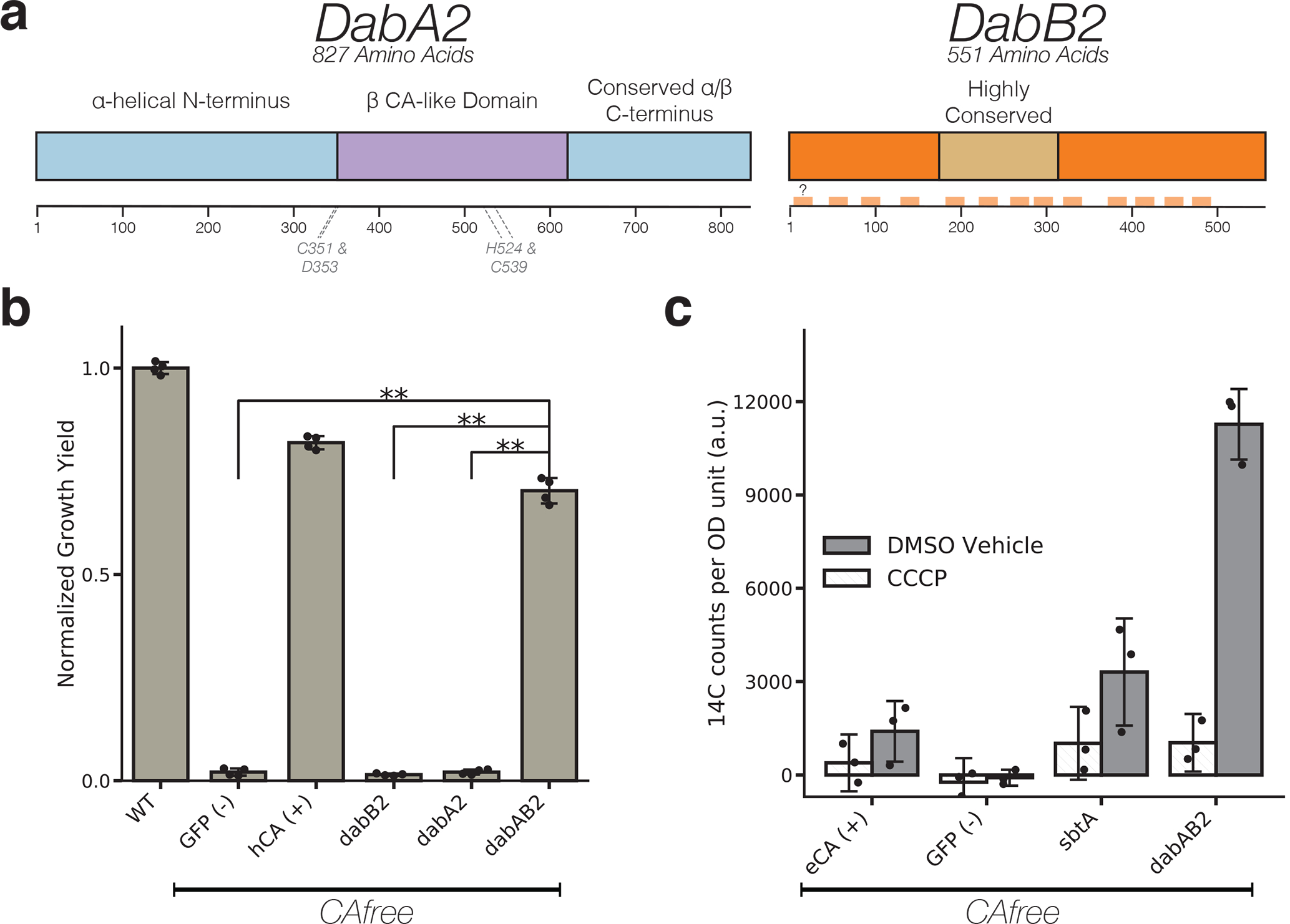
a. Diagrammatic representation of DabA2 and DabB2 based on bioinformatic annotation. The four predicted active site residues (C351, D353, H524, C539) are marked on the primary amino acid sequence. Amino acid numbers are marked below each gene and predicted transmembrane helices are marked in light orange. b. DAB2 was tested for ability to rescue growth of CAfree E. coli in ambient CO2 conditions. Expression of the full operon (DabAB2) rescues growth, as does the positive control, and human carbonic anhydrase II (hCA). dabAB2 has a larger rescue than GFP (t=42.6, corrected p=3.4*10−8), dabA2 (t=43.4, corrected p=3*10−8), and dabB2 (t=44.5, corrected p=2.6*10−8). “**” denotes that means are significantly different with Bonferroni corrected p < 5X10−4 according to a two-tailed t-test. Bar heights represent means and error bars represent standard deviations of 4 biological replicates. Consistent results were seen after 2 independent experiments. c. CAfree E. coli were tested for Ci uptake using the silicone-oil centrifugation method. Expression of DabAB2 produced a large increase in 14C uptake as compared to all controls. Moreover, treatment with the ionophore CCCP greatly reduces DabAB2-mediated 14C uptake, suggesting that DabAB2 is coupled to a cation gradient. E. coli CA (eCA) was used as a control for a non-vectorial CA. Synechococcus elongatus PCC 7942 sbtA was used as a known Ci transporter. GFP was used as a vector control. Bar heights represent the mean and error bars represent standard deviations of 3 technical replicates. Consistent results were seen with 3 independent experiments.
dabA2 and dabB2 are necessary and sufficient for energy-coupled Ci accumulation in E. coli
In order to facilitate testing for Ci transport activity, we generated an E. coli strain, CAfree, that has knockouts of both CA genes (Methods). It was previously shown that deletion of the constitutive CA, can, gene produces an HCR phenotype in E. coli39 that is complemented by expression of cyanobacterial bicarbonate transporters40. Deleting both CA genes replicates this phenotype and greatly reduces the likelihood of escape mutants. Since DAB2 disruption is associated with a larger fitness defect than DAB1 (Figure 2b), we used CAfree to test DAB2 for Ci uptake activity. DAB2 expression enables growth of CAfree in ambient CO2 while expression of either gene alone is not sufficient (Figure 3b and Supplemental Figure 5). 14Ci uptake assays demonstrate that DAB2 facilitates import of extracellular Ci to levels above that of the appropriate control (Figure 3c). Moreover, DAB2-associated Ci uptake is strongly inhibited by the ionophore CCCP (white bars in Figure 3c), indicating that DAB2 is energetically-coupled, either directly or indirectly, to a cation gradient (e.g. H+ or Na+). This is consistent with previous observations that Ci uptake in Hnea is powered by a membrane gradient17.
DabA2 and DabB2 interact to form a complex
In order to determine if the genetic interaction between dabA2 and dabB2 reflects a physical interaction, we attempted to co-purify the two proteins. DabA2 was fused to a C-terminal Strep-tag, DabB2 was fused to a C-terminal GFP with 6xHis-tag, and the genes were co-expressed in E. coli (Methods). Tandem-affinity purification following detergent solubilization revealed that DabA2 and DabB2 form a complex in E. coli (Figure 4a). The complex runs as a single major peak on size exclusion chromatography and has a retention volume consistent with a heterodimer of DabA2 and DabB2 (Figure 4b). We did not observe co-purification of any E. coli proteins suggesting that DAB2 operates as an independent complex within the membrane (Figure 4a). Moreover, DAB2 expression rescues CAfree growth even when complex I is knocked out (Δ(nuoA-nuoN)) (Supplemental Figure 6), providing further evidence that DAB function is independent of complex I.
Figure 4. DabA contains a β-CA-like active site but is not active outside of the membrane.
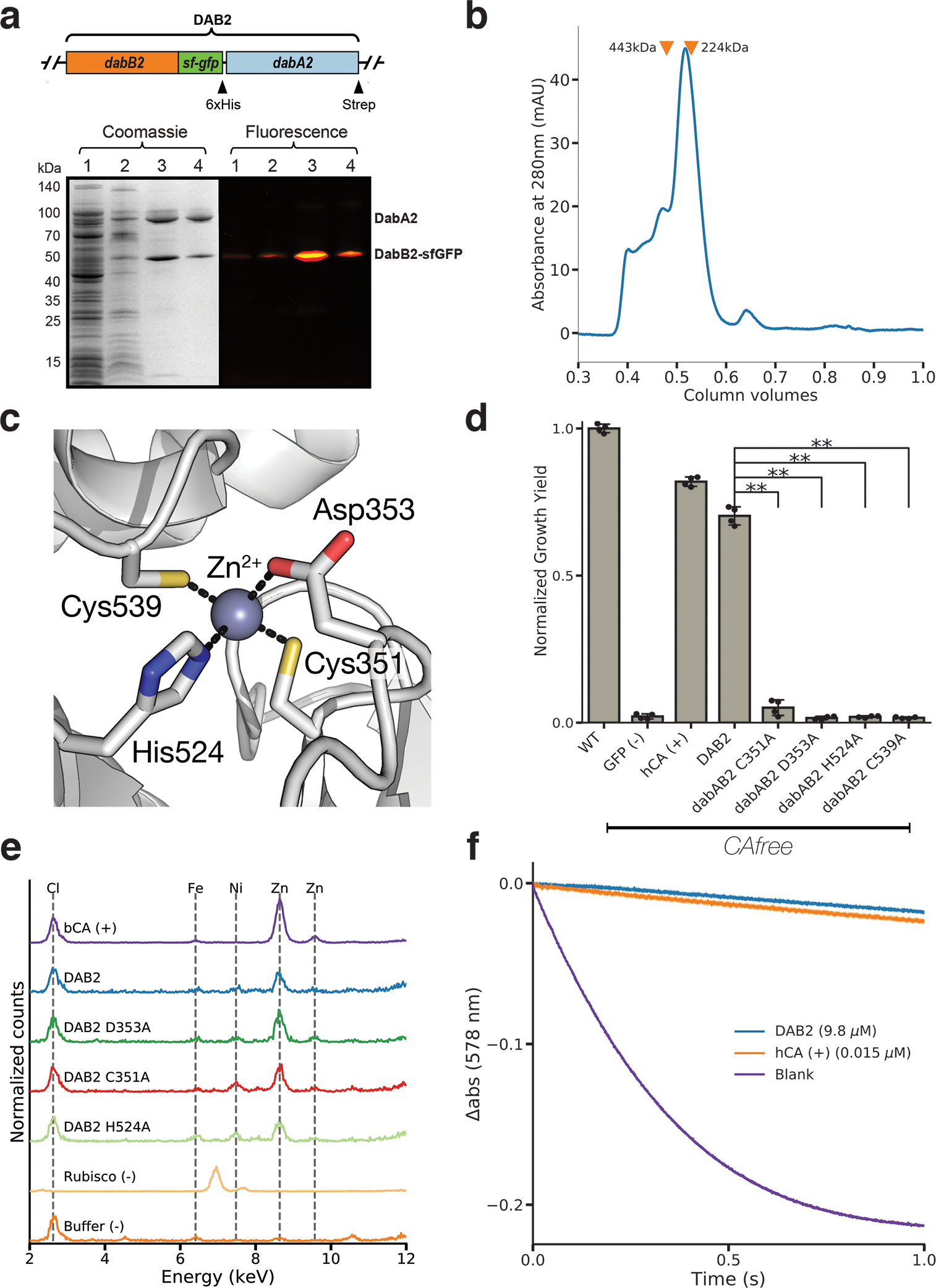
a. Purification of DabAB2 complex from E. coli. DabA2 was C-terminally tagged with a Strep-tag and DabB2 was C-terminally tagged with sf-GFP and a 6xHis-tag. Purification was monitored using SDS-PAGE imaged with fluorescence (right view) before coomassie staining (left view). Lane 1: clarified lysate; 2: solubilized membranes; 3: Ni-NTA resin eluent; 4: strep-tactin resin eluent. DabA2 and DabB2 co-purify as a single complex without any obvious interactors. Similar results were observed after 3 independent purifications. b. Size-exclusion chromatogram of His/Strep purified DabAB2 with retention volumes (orange arrows) and molecular weights (kDa) indicated for standard samples (apoferritin, 443 kDa; β-amylase, 224 kDa). DabAB2 runs with an effective mass of ~270 kDa, which likely reflects an oligomer of DabA and DabB. Similar results were observed after 3 independent purifications. c. Structural model of the DabA2 active site based on a β-CA from E. coli (PDB 1I6P). Typical β-CAs rely on two cysteines and one histidine to bind Zn2+. The aspartic acid coordinates Zn2+ but is likely displaced during catalysis43. d. Alanine mutants of the putative DabA2 active site residues abrogate rescue of CAfree E. coli compared to wild-type dabAB2 (C351A, t=54.3, p=1.1*10−8; D353A, t=144, p=3.1*10−11; H524A, t=44, p=3.7*10−8; C539A, t=44.3, p=3.5*10−8; all p values listed here are Bonferroni corrected). “**” denotes that means are significantly different with Bonferroni corrected p < 5X10−4 according to a two-tailed t-test. Bar heights indicate means and error bars give standard deviations of four biological replicate cultures. e. X-ray fluorescence data indicate that DabAB2 binds zinc like all known β-CAs. Single mutations to the active site do not abrogate zinc binding. Curves are from representative samples. Technical replicate traces were concordant (WT: 9, D353A:5, H524A:4, C351A:3, Rubisco: 2, Blank: 4, BCA: 3). Replicate traces for DAB2 H524A include samples from two independent purifications. f. Purified DabAB2 does not display any obvious CA activity despite being present in 650-fold excess over the positive control (Human carbonic anhydrase II, hCA) in our assays. Curves display averages of 7 experimental traces +/− standard error of the mean. Similar results were observed in two independent purifications.
pH independence of DAB2 rescue suggests that CO2 is likely the true substrate
Aqueous CO2 spontaneously interconverts with the gas phase as well as hydrated Ci species (H2CO3, HCO3−, CO3−2). The equilibrium of CO2(aq) and CO2(gas) is not affected by pH, but the conversion from CO2 to hydrated Ci is pH dependent. Thus, the equilibrium concentration of HCO3− increases 100 fold between pH 5 and 7 without an accompanying change in CO2 concentration (Supplemental Figure 7a)7. Expression of SbtA, a known HCO3− transporter, rescues CAfree growth at pH 7 but not at pH 5, while DabAB2 rescues growth at both pHs (Supplemental Figure 8). Since DabAB2 rescue is pH-independent in this range, its substrate is likely CO2 and not H2CO3, HCO3−, or CO3-2. This is consistent with previous observations that CO2 is the likely substrate of Hnea Ci uptake17.
Requirement of putative Zn-binding residues for DAB function
Structural homology modeling software predicts that the middle of DabA2 has sequence elements related to a β-CA (Figure 3a). Phyre2 predictions identify C539 and H524 as part of a potential Zn2+ binding site distantly homologous to a bacterial type II β-CA (10% coverage of DabA, 90.8% confidence). I-TASSER predicts a Zn2+ binding site including the same residues along with an additional cysteine (C351), and aspartic acid (D353). As shown in Figure 4c, these residues could make up the active site of a type II β-CA41. We generated individual alanine mutants for each of these putative active site residues (C351A, D353A, H524A and C539A) and tested them in CAfree. All mutants failed to rescue CAfree growth in ambient CO2 (Figure 4d). We proceeded to assay zinc binding of purified DabAB2 complex using X-ray fluorescence spectroscopy and found that wild-type DabAB2 and three of the single mutants (C351A, D353A, and H524A) bind zinc (Figure 4e). Single mutants retain three of four zinc-coordinating residues41, which could explain why the mutants bind zinc. Indeed, mutational studies of the human CA II show that single mutations to Zn2+-binding residues reduce but do not abrogate zinc binding42.
Purified DabAB2 complex does not have conspicuous CA activity.
The assay of detergent solubilized, purified DabAB2 did not show carbonic anhydrase activity over controls (Figure 4f). DabAB2 was assayed at high protein concentrations (> 650-fold more protein than the positive control) and under CO2 concentrations that are typically saturating for CAs, but displayed no activity (Figure 4f). Absence of activity in vitro argues either that DabAB2 has extremely low CA activity or, perhaps, that DabAB2 must reside in a cell membrane holding a cation gradient to function as an energetically-activated carbonic anhydrase.
DAB operons are widespread in prokaryotes and present in human pathogens
A query of the Uniprot database with the DabA PFAM (PF10070) yielded 878 putative DabA sequences. DabAs were found in a wide variety of prokaryotes including bacteria and archaea (Figure 5a and Supplemental Figure 9), as is consistent with previous work28. Represented clades include not only Proteobacteria, but also Euryarchaeota, Firmicutes, Planctomycetes, and Bacterioides. However, we were surprised to observe many dabA sequences were found in genomes of organisms that cannot fix CO2 including the heterotrophic human pathogens V. cholera, B. anthracis, and L. pneumophila (Figure 5a). Notably, 843 (96%) of the identified dabA sequences were either within three genes of, or fused to, a dabB.
Figure 5. DAB operons are widespread among prokaryotes.
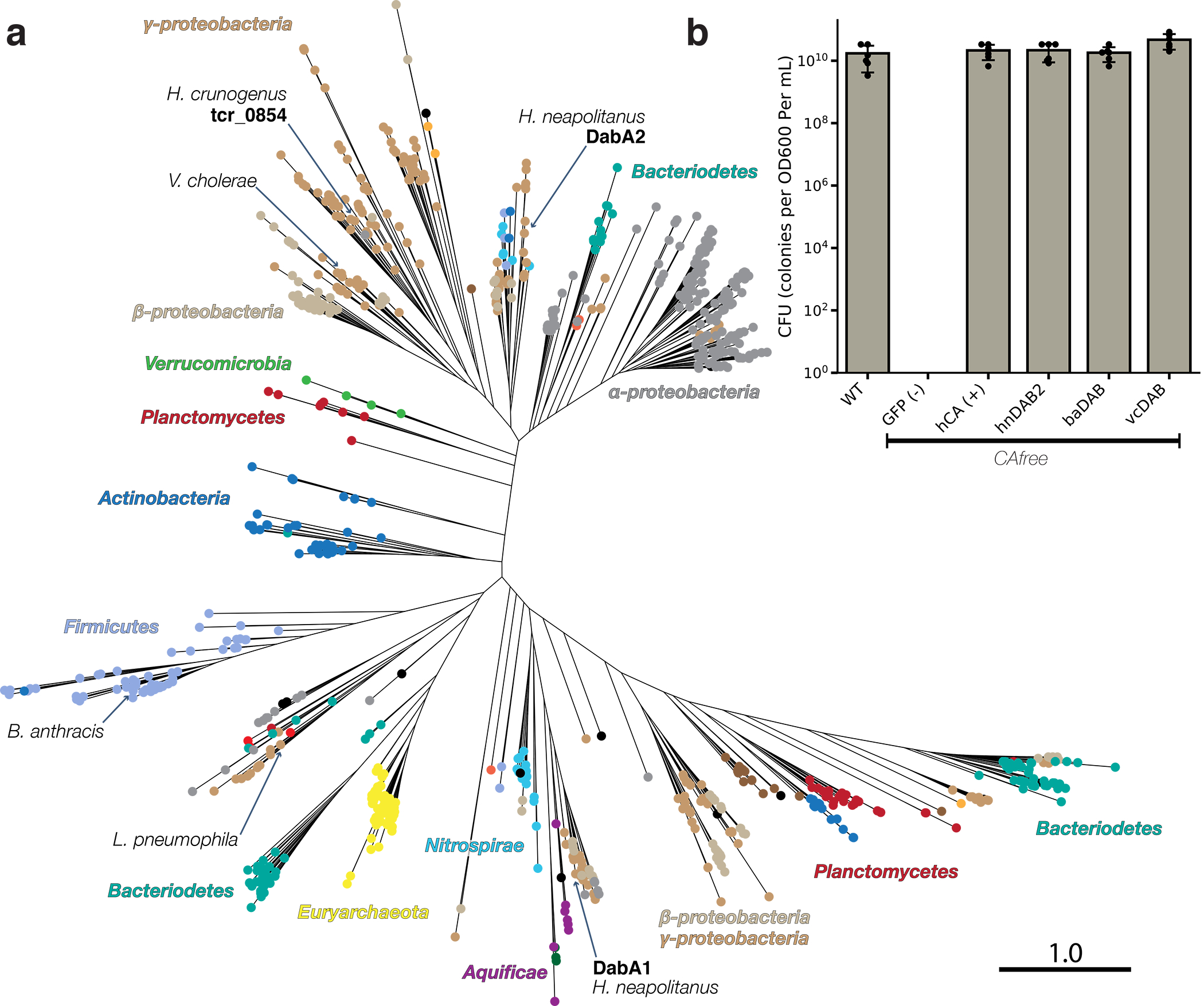
a. Approximate maximum likelihood phylogenetic tree of 878 DabA homologs associated with PF10070.9 (Methods). DabA homologs are found in > 15 prokaryotic clades, including archaea. Hnea DabA1 and DabA2 represent two different groupings that are commonly found in proteobacteria. Inspecting the tree reveals several likely incidents of horizontal transfer, e.g. between Proteobacteria and Firmicutes, Nitrospirae and Actinobacteria. Moreover, the genomes of several known pathogens contain a high-confidence DabA homolog, including B. anthracis, V. cholerae, and L. pneumophila. Detailed annotations are given in Supplemental Figure 9. Scale bar indicates one substitution per site. Sequences used to generate the tree can be found in Supplemental File 5. b. Functional DABs are found in human pathogens. Colony forming units per OD600 per ml were measured on LB plates with induction in air. DAB operons from B. anthracis (baDAB) and V. cholerae (vcDAB) rescued growth of CAfree cells. The Hnea operon DAB2 is abbreviated as hnDAB2. Bars represent means. Error bars represent the standard deviation of 6 technical replicate platings. Consistent results were achieved in biologically independent platings of baDAB and vcDAB.
Finally, we assayed whether the DAB homologs from heterotrophic pathogens are functional Ci pumps. Operons from V. cholera E7946 El Tor Ogawa and B. anthracis Sterne were cloned and expressed in CAfree. Both DAB operons rescued CAfree growth in ambient CO2 (Fig. 5B and Supplemental Figure 10). Thus, DAB operons from heterotrophic, human pathogens are functional.
Discussion
Here, we generated a knockout library containing ≈35 individual knockouts for every gene in the genome of the proteobacterial chemolithoautotroph H. neapolitanus. Using these data, we compiled the essential gene set of a chemolithoautotroph (Figure 1) and were able to confidently identify 551 essential genes and 1787 nonessential genes. Mapping essential genes will provide insight into the metabolism and growth physiology of sulfur-oxidizing chemolithoautotrophs.
In addition to mapping essential genes, this library would be used to measure conditional phenotypes for nonessential genes. These mutants were isolated in high CO2 and so we were able to disrupt all known components of the bacterial CCM (Figure 2). The resulting genome-wide knockout library was used to perform a comprehensive screen for bacterial CCM genes. This screen highlighted a small number of genes (17) as having the HCR phenotype associated with the CCM (Figure 2b–f), nearly all of these genes are known to be associated with the ɑ-carboxysome. Though it is possible that genetic redundancy, conditional phenotypes, or impairment only at sub-ambient CO2 permit some genes to escape notice, these data suggest that the proteobacterial CCM is composed of < 30 functionally distinct components. Moreover, none of the genes identified have unexpected functions, suggesting that current models of bacterial CCMs incorporate all necessary functions.
Our screen identified 3 transcriptional regulators as well as 3 distinct CCM operons (Figures 2b–f). Identification of transcriptional regulators with HCR phenotypes (Figures 2d–f) may inform the study of CCM regulation. The first operon contains nearly all known components of the ɑ-carboxysome, all of which confer HCR phenotypes upon disruption (Figure 2c). The second operon is adjacent to the carboxysome operon and contains 11 genes. Only 3 of these genes - the Rubisco chaperone acRAF and dabAB1 - displayed HCR phenotypes (Figure 2c). The remaining 8 genes had no associated phenotype but might nonetheless have roles in the CCM. These genes include cbbOQ, csos1D, p-II, and a parA homolog (Figure 2c). The third operon contains two genes, dabAB2, both with HCR phenotypes (Figure 2d).
A previous physiological study suggested that Hnea Ci uptake is coupled to the membrane electrochemical potential and uses CO2 as a substrate, but the protein(s) responsible for this activity were unknown17. DAB1 and DAB2 are homologous to Ci pumps from hydrothermal vent chemolithoautotrophs recently discovered by Scott and colleagues28,30 and our screen suggests that DAB1 and DAB2 are likely the Ci pumps in Hnea. These observations raise many mechanistic questions as to how DABs function, and we therefore sought to establish a biochemical system to investigate DAB structure-function.
We showed that the DAB2 operon encodes a two-component protein complex that has Ci uptake activity when heterologously expressed in E. coli (Figures 3b–c & 4a). This complex is likely a heterodimer as suggested by size-exclusion chromatography (Figure 4b). As Ci uptake is inhibited by the ionophore CCCP (Figure 3c), we suspect that DAB2 activity is energetically-coupled to a cation gradient (Figure 5a). Moreover, since DabAB2 shows pH-independent rescue of CAfree E. coli, CO2 is likely the transported substrate (Figure 4c). This is further supported by the fact that DabA has distant homology to a type II β-CA and binds a zinc (Figures 3–4), suggesting that a CA active site hydrates transported CO2. Finally, mutations to the putative zinc-binding residues (C351A, D353A, H524A, and C539A) ablate function in vivo (Figure 4d). We therefore propose a speculative model of DAB activity wherein CO2 is passively taken into the cell and then unidirectionally hydrated to HCO3− by energy-coupled CA activity of DabA (Figure 6).
Figure 6. A speculative model of the unidirectional energy-coupled CA activity of DAB complexes.
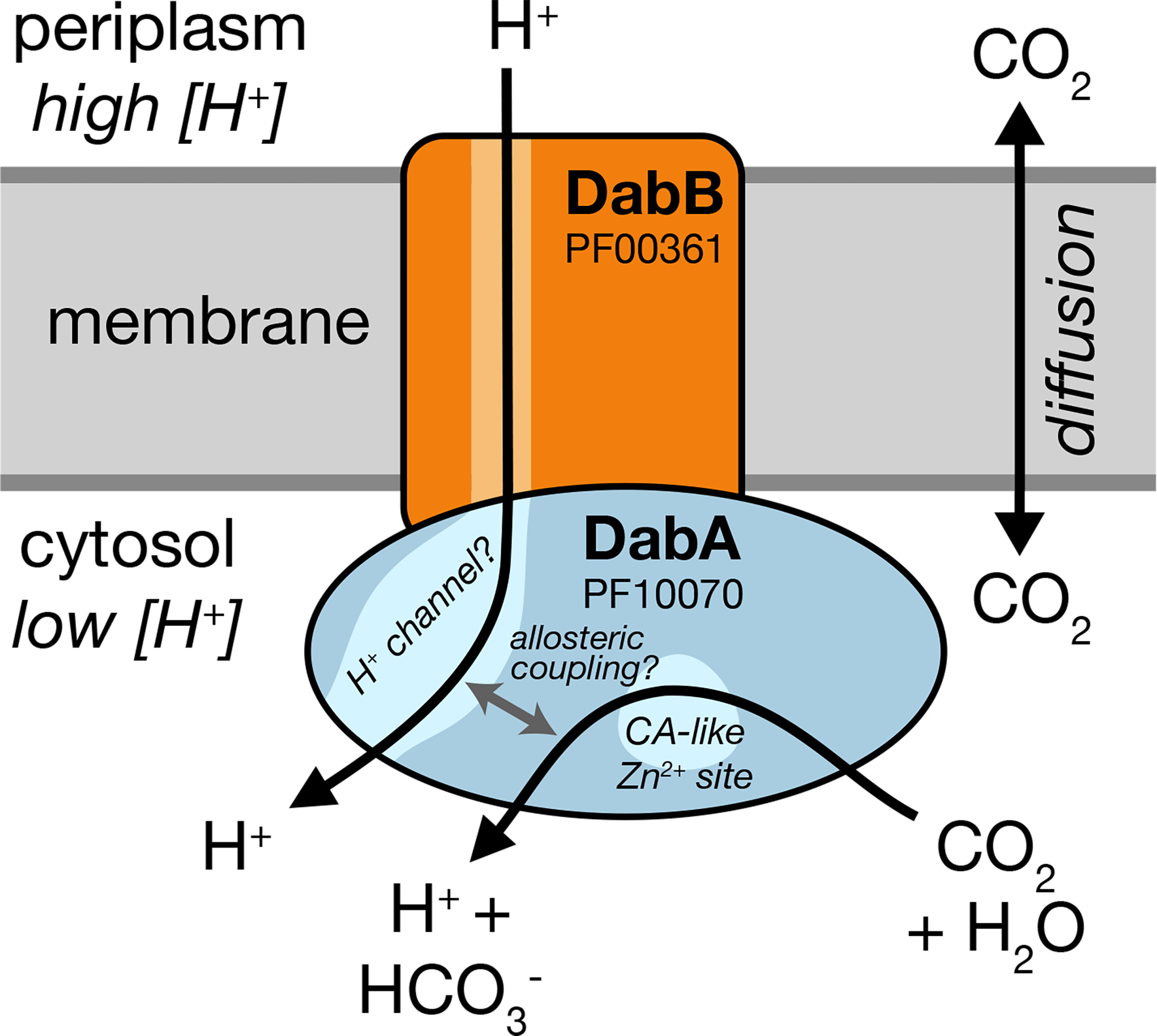
We propose that DabAB complexes couple CA activity of DabA to a cation gradient across the cell membrane, producing unidirectional hydration of CO2 to HCO3−. The cation gradient could be H+ or Na+. Energy-coupled CA activity is required for the DABs role as a Ci uptake system in the proteobacterial CCM, as discussed in the text. Because it appears that DabAB2 is not active as a purified complex outside of the membrane, it is assumed protein tightly couples the inflow of cations with CO2 hydration so that there is no “slippage.” Indeed, slippage - i.e., uncoupled CA activity - would be counterproductive for CCM function7,14. Notably, Zn2+ binding by the active site aspartic acid of type II β-CAs (D353 in DabA2, Figure 4a) is thought to allosterically regulate activity43. This Asp-mediated activity switch could, therefore, provide a means for allosteric coupling of a β-CA active site to distal ion transport.
Model carbonic anhydrases are not coupled to any energy source (e.g., ATP, cation gradient). Rather, they equilibrate CO2 and HCO3- 42. However, energy-coupled CA activity could favor CO2 hydration, allowing the DAB system to actively accumulate HCO3− in the cytosol and power the CCM (Figure 2a). Given the similarity of DabB to H+-pumping proteins, we propose that DABs use the H+ gradient, though our results are equally consistent with other cation gradients, e.g. Na+. This mechanism would require tight coupling of cation flow to CA activity by DabA, consistent with our observation that purified DabAB2 displays no measurable CA activity. Interestingly, type II β-CAs are the only CAs known to display allosteric regulation43. Allosteric control is thought to be mediated by Zn2+ capping and uncapping by the active site aspartic acid (D353 in DabA2)43. A similar mechanism might couple cation flow through DabB to the active site of DabA.
Cyanobacteria possess vectorial CAs called CUPs, which may provide clues to the DAB mechanism24,44,45. Indeed, both DAB and CUP systems contain subunits in the Mrp protein family (DabB and NdhD/F are in PF00361) that also contains the H+-pumping subunits of complex I. This commonality might suggest similar mechanisms. CO2 hydration by CUPs is thought to be coupled to energetically-favorable electron flow because CUPs associate with complex I46 (Supplemental Figure 9b). However, the Mrp protein family (PF00361) is very diverse and contains many cation transporters that do not associate with complex I or any other redox-coupled complex38. Moreover, DabB sequences are only distantly related to complex I and CUP subunits (Supplemental Figure 4a), DAB2 subunits do not co-purify with E. coli complex I (Figure 4a) and DAB2 rescues CAfree growth in a complex I knockout (Supplemental Figure 6). We therefore propose that DAB activity is not coupled to electron flow through complex I but, rather, to a cation gradient across the membrane (Figure 6).
DabAB2 functions robustly, as demonstrated by complementation of CAfree (Figure 3b) and 14C uptake measurements (Figure 3c). Indeed, we observed that DabAB2 functions substantially better in E. coli than SbtA, a Ci transporter from cyanobacteria9,40 (Figure 3c). As E. coli and Hnea are proteobacteria, this observation could result from greater “compatibility” of proteobacterial proteins with E. coli expression. It may also be the case, though, that the ɑ-CCM of proteobacteria is more “portable” than the β-CCM of freshwater cyanobacteria. Indeed, ɑ-CCM genes are typically found in a single gene cluster in chemolithoautotrophs throughout ɑ- β- and Ɣ-proteobacteria and the ɑ-CCM was clearly horizontally transferred from proteobacteria to marine cyanobacteria9. DabA homologs are widespread in prokaryotes and were likely horizontally transferred multiple times (Figure 5a). Since DAB complexes are prevalent among prokaryotes and have superlative activity, DAB-family transporters are an attractive target for protein engineering and heterologous expression in plants and industrial microbes, where elevated intracellular Ci could be useful47.
Finally, DABs are present in a wide variety of bacteria and archaea28. High-confidence DabA homologs are found not only in large numbers of autotrophs but also in heterotrophs (Figure 5a & Supplemental Figure 9). Moreover, homologs are present in the notable heterotrophic pathogens V. cholerae, B. anthracis, and L. pneumophila (Figure 5a). We showed that DABs from V. cholerae and B. anthracis are active in E. coli (Figure 5b). This leads us to wonder: what do heterotrophic pathogens use Ci uptake systems for? Carbonic anhydrase activity is essential for growth of the heterotrophs E. coli and S. cerevisiae in ambient CO239,48. In the heterotrophic context, CA activity is thought to supply bicarbonate for biotin-dependent carboxylases in central metabolism, for which HCO3− is the substrate39,48. Additionally, bicarbonate levels have been linked to virulence in both V. cholera and B. anthracis49,50. Perhaps DAB-family Ci uptake systems play roles in the growth or virulence of these important pathogens? We hope that future research will delineate the role of energetically-activated Ci uptake in heterotrophic and pathogenic organisms.
Materials and Methods
Important strains and reagents
A detailed listing of key strains and reagents is given in Supplemental File 1.
Bacterial strains and growth conditions
E. coli strain APA766 was used as the conjugation donor to transfer the Tn5 transposon to Halothiobacillus neapolitanus C2 (Hnea) via conjugation31. The E. coli double CA deletion strain “CAfree” (BW25113 Δcan ΔcynT) was generated by curing the KEIO collection cynT knockout (BW25113 ΔcynT, KEIO strain JW0330) of kanamycin resistance via pCP20-mediated FLP recombination and subsequent P1 transduction (and curing) of kanamycin resistance from the can knockout strain EDCM636 (MG1655 Δcan, Yale Coli Genomic Stock Center, 39,51). Complex I knockout strains (Δ(nuoA-nuoN)) were generated in the BW25113 and CAfree backgrounds. These strains were generated by lambda red mediated recombination of a KanR resistance cassette flanked by FRT sites into the nuo locus such that the entire operon was removed. The pSIM5 plasmid carrying the lambda red recombinase was heat cured at 42 °C. Lysogeny broth (LB) and LB agar were used as E. coli growth media unless otherwise specified. E. coli strains were grown at 37 °C in the presence of 0.1 mg/ml carbenicillin, 0.06 mg/ml kanamycin, or 0.025 mg/ml chloramphenicol as appropriate. Hnea was grown in DSMZ-68 media at 30 °C and in the presence of 0.03 mg/ml kanamycin when appropriate.
Transposon mutagenesis and RB-TnSeq library production
A barcoded library of Hnea transposon mutants was generated by adapting the methods of Wetmore et al.31. Conjugations were performed as follows. Hnea and APA766 were cultured and harvested by centrifugation. Both cultures were washed once in 10 mL antibiotic-free growth media per conjugation reaction and resuspended in 100 μl. 5 OD600 units of Hnea were mixed with 20 OD600 units of APA766 on a 0.45 μM Millipore MCE membrane filter and cultured overnight at 30 °C in 5% CO2 on an antibiotic-free LB agar plate containing 0.06 mg/ml diaminopimelic acid. Cells were scraped from the filter into 2 mL DSMZ-68 and collected in a 2 mL microcentrifuge tube. Recovered cells were pelleted by centrifugation at 16000 × g for 1 minute, washed in 2 mL DSMZ-68, pelleted again at 9000 × g for 1 minute, and resuspended in 2 ml DSMZ-68 before 200 μl was plated onto 10 separate DSMZ-68 kanamycin plates (per conjugation). Plates were incubated at 30 °C under 5% CO2 until colonies formed (~ 7 days). Colonies were counted and scraped into 55 mL DSMZ-68. Two 1.4 OD600 unit samples were taken and used to prepare genomic DNA (Qiagen DNeasy blood and tissue kit). Transposon insertions were amplified from gDNA and transposons were mapped after Illumina sequencing using protocols and software from Wetmore et al.31 1.6 OD600 unit aliquots were then flash frozen in 50% glycerol for subsequent BarSeq experiments.
Essential gene assignment
Following the logic of Wetmore et al. and Rubin et al.31,52, we categorized genes as essential if we observed significantly fewer transposon insertions than would be expected by chance. If insertion occurred uniformly at random, the number of insertions per gene would be expected to follow a binomial distribution. The probability of observing at most insertions into a gene of length is therefore expressed as:
Here, is the average rate of transposon insertion per base pair genome-wide. Genes were determined to be essential if they received a lower-than-expected number of insertions in both replicates of the library mapping, i.e. if the probability of observing or fewer insertions was beneath 0.05 after Bonferroni correction. Genes were called “ambiguously essential” in two cases: (i) replicates were discordant or (ii) zero insertions were observed but the gene was short enough that the formula could not yield a Bonferroni-corrected p-value below a 0.05 threshold even in the case of zero insertions.
Gene fitness experiments
Fitness experiments were performed according to a modification of the protocol in Wetmore et al.31. This method allows pooled library fitness experiments to be performed comparing different growth conditions by comparing barcode abundance changes in order to track changes in the abundance of the transposon mutants. In short, a library aliquot was thawed and used to inoculate three 33 mL cultures. Cultures were grown to OD600 ~0.08 in 5% CO2. At this point, 20 mL were removed and harvested by centrifugation as two t0 (input) samples. Cultures were back-diluted 1:64 into 128 mL and incubated for 6.5–7.5 doublings under 5% CO2 or ambient conditions. 50 mL of culture was harvested by centrifugation. gDNA was prepared and barcodes were amplified for fitness determination via Illumina sequencing as described previously31. Fitness values were calculated using existing software31. Genes were assigned an HCR phenotype if they had a fitness defect of two fold or greater in ambient CO2 compared to 5% CO2 in two replicate experiments.
CAfree rescue experiments
Electrocompetent CAfree cells were prepared using standard protocols53 and transformed with pFE plasmids expressing genes of interest by electroporation. CAfree pre-cultures were grown overnight in 10% CO2 and diluted into 96 well plates (3 μl cells in 250 μl media). Growth curves were measured by culturing cells in a Tecan M1000 microplate reader under ambient conditions with continuous shaking, and measuring OD600 every 15 minutes. When samples are marked “induced,” 200 nM anhydrotetracycline (aTc) was added to the media. Growth yields are calculated as the maximum OD600 achieved after 24 hours of growth and normalized to the yield of a wild type control. CFU experiments were performed by back diluting cultures to OD600 0.2 before performing 10X serial dilutions. 3 μl of the OD600 0.2 sample and each of the serial dilutions were then spotted on plates with 200 nM aTc and grown overnight in ambient conditions (400 ppm CO2). The spot with the highest dilution that yielded more than one colony was counted and a minimum of six replicates were averaged for each strain.
Silicone oil centrifugation measurement of Ci uptake
The silicone oil filtration method was modified from Dobrinski et al.54 and used to measure uptake of radiolabeled inorganic carbon. Assay tubes were generated using 0.6 ml microcentrifuge tubes containing 20 μl of dense kill solution (66.7% v/v 1 M glycine pH 10, 33.3% v/v triton X-100) covered by 260 μl of silicone oil (4 parts AR20:3.5 parts AR200). Electrocompetent CAfree cells were prepared using standard protocols and transformed with pFA-based plasmids containing genes of interest by electroporation. CAfree cultures were grown overnight in 10% CO2, back diluted to an OD600 of 0.1 and allowed to grow to mid-log phase in 10% CO2 in the presence of 200 nM aTc for induction. Cells were then harvested by centrifugation, washed once in PBS (pH 7.55) and resuspended to OD600 0.6 in PBS + 0.4% glucose. 14C-labeled sodium bicarbonate (PerkinElmer) was added to a final concentration of 4.1 nM and an activity of 0.23 μCi. Cells were incubated with 14C for 4 minutes before centrifugation at 17,000 × g for 4 minutes to separate cells from buffer. Pellets were clipped into scintillation vials containing 5 ml Ultima Gold scintillation fluid and 300 μl 3M NaOH using microcentrifuge tube clippers or medium dog toenail clippers. Counts were measured on a PerkinElmer scintillation counter. 14C counts are normalized to 1 OD600 unit of cells added. During inhibition assays, cells were incubated in PBS pH 7.55 with 0.4% glucose + 0.4% DMSO and the inhibitor (100 μM CCCP) for 10 minutes before assay.
Generation of DabA phylogenetic tree
We searched the Uniprot reference proteome database using the Pfam Hidden Markov Model PF10070.9 with a cutoff e-value of 10-4. Our search recovered 941 candidate DabA proteins. These sequences were aligned using MAFFT and manually pruned to remove fragments and poorly aligning sequences. The remaining 878 candidate DabA sequences were re-aligned with MAFFT and an approximate maximum likelihood phylogenetic tree was constructed using FastTree. Taxonomy was assigned to nodes in the tree based on NCBI taxonomy information for the genomes harboring each sequence. Genomic neighborhoods for each gene in the tree were determined using the EFIGNT online server55 and genomes with a dabB gene within 3 genes of dabA and oriented in the same direction were considered to have full DAB operons. dabAB fusions were found by visual inspection of genomic neighborhoods from those genomes that did not have separate dabB genes located close to dabA.
Generation of DabB phylogenetic tree
DabB homologs were collected manually by searching MicrobesOnline for close homologs of four PF00361 members in the Hnea genome (dabB1, dabB2, Hneap_1953, Hneap_1130) and other characterized PF00361 members including Synechococcus elongatus ndhF1, Synechococcus elongatus ndhF3, and Synechococcus elongatus ndhF4. Genes were clustered to 95% similarity and genes with divergent operon structure were removed manually using MicrobesOnline treeview56. nuoL from Escherichia coli, nqo12 from Thermus thermophilus, and ndhF1/3/4 from Thermosynechococcus elongatus BP-1 were added as markers. ClustalOmega was used to construct a multiple sequence alignment and an approximate maximum likelihood phylogenetic tree was constructed using FastTree57,58. The tree was visualized using the Interactive Tree of Life59.
Protein annotation and structural homology modeling
Secondary structural annotations for DabA and DabB were generated using XtalPred60. Structural Homology modeling of DabA was performed using Phyre2 and I-TASSER web servers with default parameters61,62. A list of close DabB homologs was assembled by searching MicrobesOnline for PF00361 members with similar operon structure. A ClustalOmega alignment was used to calculate residue-level conservation of DabB proteins while the MAFFT alignment generated during the creation of the DabA tree was used to calculate residue level conservation of DabA proteins (Supplemental Figure 4b).
Purification of DAB2
Chemically competent BL21-AI E. coli were transformed with a pET14b-based vector containing the dabAB genes. 1 liter of 2xYT media was inoculated with 20 ml of an overnight culture of BL21-AI E. coli in LB+CARB and allowed to grow to mid log at 37 °C. When midlog was reached, cells were induced with 20 ml of 50 mg/ml arabinose and transitioned to 20 °C for overnight growth. Cultures were pelleted and resuspended in 10 ml TBS (50 mM Tris, 150 mM NaCl, pH 7.5) supplemented with 1.2 mM phenylmethylsulfonyl fluoride, 0.075 mg/ml lysozyme and 0.8 ug/ml DNAse I per liter of starting culture and then incubated at room temperature on a rocker for 20 minutes. Cells were lysed with four passes through a homogenizer (Avestin). Lysate was clarified at 15,000 × g for 30 minutes. Membranes were pelleted at 140,000 × g for 90 minutes. Membrane pellets were resuspended overnight in 25 ml TBS supplemented with 1 mM phenylmethylsulfonyl fluoride and 1% β-dodecyl-maltoside (DDM, Anatrace) per liter of culture following63. Membranes were then re-pelleted at 140,000 – 200,000 × g for 60 minutes and the supernatant was incubated with Ni-NTA beads (Thermo Fisher) for 90 min at 4 °C. The resin was washed with “Ni buffer” (20 mM Tris + 300 mM NaCl + 0.03% DDM, pH 7.5) supplemented with 30 mM imidazole and eluted with Ni buffer supplemented with 300 mM imidazole. Eluent was then incubated with Strep-Tactin (Millipore) resin for 90 min at 4 °C. Resin was washed with “strep buffer” (TBS + 0.03% DDM) and eluted with strep buffer supplemented with 2.5 mM desthiobiotin. Eluent was concentrated using Vivaspin 6 100 kDa spin concentrators and buffer exchanged into strep buffer by either spin concentration or using Econo-Pac 10DG (Biorad) desalting columns. For analytical purposes, 300 μg of strep-purified protein was injected onto a Superdex 200 Increase 3.2/300 size-exclusion column pre-equilibrated in strep buffer and eluted isocratically in the same buffer.
Carbonic anhydrase assays
CA-catalyzed CO2 hydration of purified DAB2 complex and human carbonic anhydrase (hCA) was measured using the buffer/indicator assay of Khalifah64 on a KinTek AutoSF-120 stopped-flow spectrophotometer at 25 °C. The buffer/indicator pair used was TAPS/m-cresol purple measured at a wavelength of 578 nm using a pathlength of 0.5 cm. Final buffer concentration after mixing was 50 mM TAPS, pH 8.0 with the ionic strength adjusted to 50 mM with Na2SO4, and 50 μM of pH-indicator. Final protein concentration used was: 9.8 μM DAB2 (His-elution) and 0.015 μM hCA (positive control; Sigma Aldrich C6624). Saturated solution of CO2 (32.9 mM) was prepared by bubbling CO2 gas into milli-Q water at 25 °C. The saturated solution was injected into the stopped-flow using a gas-tight Hamilton syringe, and measurements were performed in a final CO2 concentration of 16.5 mM. Progression curves were measured in 7 replicates.
X-ray fluorescence spectroscopy for metal analysis
50–100 μg of protein in 20–200 μl of TBS + 0.03% DDM was precipitated by addition of 4 volumes of acetone and incubation at −20 °C for 1 hour. Samples were centrifuged at 21,130 × g for 15 minutes in a benchtop centrifuge and the supernatant was removed. Pellets were stored at 4 °C until analysis. Fluorescence analysis was performed by breaking up the pellet into 5 μl of TBS + 0.03% DDM with a pipette tip. Small pieces of the pellet were looped with a nylon loop and flash frozen at the beamline under a nitrogen stream. The sample was excited with a 14 keV X-ray beam and a fluorescence spectrum was collected. Sample emission spectra were then used to identify metals. Metal analysis was performed on wild-type DAB2, Zn-binding mutants C351A, D353A, and H524A, bovine CA (positive control; Sigma Aldrich C7025), and a buffer blank was used as a negative control. A Rubisco crystal containing cobalt salts was also used as a zinc free control. Experiments were performed at the Lawrence Berkeley National Laboratory Advanced Light Source Beamline 8.3.1.
Supplementary Material
Acknowledgements
We thank Adam Deutschbauer and Morgan Price for assistance with RB-TnSeq experiments and analysis, respectively. Genomic DNA samples were kindly provided by Zoe Netter and Kimberly Seed (V. cholera) and Dan Portnoy and Richard Calendar (B. anthracis Sterne). We thank Andreas Martin and Jared Bard for assistance with stopped flow experiments. Thanks to Emeric Charles, Woodward Fischer, Britta Forster, Ben Long, Robert Nichols, Dean Price and Patrick Shih for useful conversations and comments on the manuscript. X-ray-based experiments were performed at the Lawrence Berkeley National Laboratory Advanced Light Source Beamline 8.3.1. J.J.D. was supported by National Institute of General Medical Sciences grant-T32GM066698. A.F. and T.G.L. were supported by a National Science Foundation Graduate Research Fellowship. C.B. was supported by an International Postdoctoral grant from the Swedish Research Council 637-2014-6914. D.F.S was supported by National Science Foundation grant MCB-1818377 (for genetic screen) and by US Department of Energy Grant DE-SC00016240 (for DAB characterization).
Footnotes
Competing Interests
UC Regents have filed a patent related to this work on which J.J.D., A.F., and D.F.S. are inventors. D.F.S. is a co-founder of Scribe Therapeutics and a scientific advisory board member of Scribe Therapeutics and Mammoth Biosciences. All other authors declare no competing interests.
Code availability
All custom code is available on github at: https://github.com/jackdesmarais/DabTransporterPaper.
Data availability
All illumina sequencing data is accessible as an NCBI SRA (BioProject accession number: PRJNA546024). All other data is available on github at: https://github.com/jackdesmarais/DabTransporterPaper.
References
- 1.Bathellier C, Tcherkez G, Lorimer GH & Farquhar GD Rubisco is not really so bad. Plant Cell Environ. 41, 705–716 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Flamholz A et al. Revisiting tradeoffs in Rubisco kinetic parameters. bioRxiv 470021 (2018). doi: 10.1101/470021 [DOI] [Google Scholar]
- 3.Tcherkez G The mechanism of Rubisco-catalysed oxygenation. Plant Cell Environ. 39, 983–997 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Bauwe H, Hagemann M & Fernie AR Photorespiration: players, partners and origin. Trends Plant Sci. 15, 330–336 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Tcherkez GGB, Farquhar GD & Andrews TJ Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. U. S. A. 103, 7246–7251 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savir Y, Noor E, Milo R & Tlusty T Cross-species analysis traces adaptation of Rubisco toward optimality in a low-dimensional landscape. Proc. Natl. Acad. Sci. U. S. A. 107, 3475–3480 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangan NM, Flamholz A, Hood RD, Milo R & Savage DF pH determines the energetic efficiency of the cyanobacterial CO2 concentrating mechanism. Proc. Natl. Acad. Sci. U. S. A. 113, E5354–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raven JA, Beardall J & Sánchez-Baracaldo P The possible evolution and future of CO2-concentrating mechanisms. J. Exp. Bot. 68, 3701–3716 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Rae BD, Long BM, Badger MR & Price GD Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol. Mol. Biol. Rev. 77, 357–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long BM, Rae BD, Rolland V, Förster B & Price GD Cyanobacterial CO2-concentrating mechanism components: function and prospects for plant metabolic engineering. Curr. Opin. Plant Biol. 31, 1–8 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Price GD, Badger MR & von Caemmerer S The prospect of using cyanobacterial bicarbonate transporters to improve leaf photosynthesis in C3 crop plants. Plant Physiol. 155, 20–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGrath JM & Long SP Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol. 164, 2247–2261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long BM et al. Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat. Commun. 9, 3570 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price GD & Badger MR Expression of Human Carbonic Anhydrase in the Cyanobacterium Synechococcus PCC7942 Creates a High CO2-Requiring Phenotype Evidence for a Central Role for Carboxysomes in the CO2 Concentrating Mechanism. Plant Physiol. 91, 505–513 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopkinson BM, Young JN, Tansik a. L. & Binder BJ The Minimal CO2-Concentrating Mechanism of Prochlorococcus spp. MED4 Is Effective and Efficient. Plant Physiol. 166, 2205–2217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitehead L, Long BM, Price GD & Badger MR Comparing the in Vivo Function of α-Carboxysomes and β-Carboxysomes in Two Model Cyanobacteria. Plant Physiol. 165, 398–411 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holthuijzen YA, van Dissel-Emiliani FFM, Kuenen JG & Konings WN Energetic aspects of CO2 uptake in Thiobacillus neapolitanus. Arch. Microbiol. 147, 285–290 (1987). [Google Scholar]
- 18.Price GD & Badger MR Isolation and characterization of high CO2-requiring-mutants of the cyanobacterium Synechococcus PCC7942: two phenotypes that accumulate inorganic carbon but are apparently unable to generate CO2 within the carboxysome. Plant Physiol. 91, 514–525 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcus Y, Schwarz R, Friedberg D & Kaplan A High CO(2) Requiring Mutant of Anacystis nidulans R(2). Plant Physiol. 82, 610–612 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonacci W et al. Modularity of a carbon-fixing protein organelle. Proc. Natl. Acad. Sci. U. S. A. 109, 478–483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jorda J, Lopez D, Wheatley NM & Yeates TO Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Sci. 22, 179–195 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Axen SD, Erbilgin O & Kerfeld CA A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput. Biol. 10, e1003898 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibata M, Ohkawa H, Katoh H, Shimoyama M & Ogawa T Two CO2 uptake systems in cyanobacteria: four systems for inorganic carbon acquisition in Synechocystis sp. strain PCC6803. Funct. Plant Biol. 29, 123–129 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Price GD Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism. Photosynth. Res. 109, 47–57 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Heinhorst S, Cannon GC & Shively JM Carboxysomes and Carboxysome-like Inclusions. in Complex Intracellular Structures in Prokaryotes (ed. Shively JM) 141–165 (Springer Berlin; Heidelberg, 2006). [Google Scholar]
- 26.Shively JM, Ball F, Brown DH & Saunders RE Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182, 584–586 (1973). [DOI] [PubMed] [Google Scholar]
- 27.Cannon GC et al. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl. Environ. Microbiol. 67, 5351–5361 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangiapia M et al. Proteomic and mutant analysis of the CO2 concentrating mechanism of hydrothermal vent chemolithoautotroph Thiomicrospira crunogena. J. Bacteriol. (2017). doi: 10.1128/JB.00871-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott KM et al. Genomes of ubiquitous marine and hypersaline Hydrogenovibrio, Thiomicrorhabdus and Thiomicrospira spp. encode a diversity of mechanisms to sustain chemolithoautotrophy in heterogeneous environments. Environ. Microbiol. 20, 2686–2708 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Scott KM et al. Diversity in CO2-Concentrating Mechanisms among Chemolithoautotrophs from the Genera Hydrogenovibrio, Thiomicrorhabdus, and Thiomicrospira, Ubiquitous in Sulfidic Habitats Worldwide. Appl. Environ. Microbiol. 85, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wetmore KM et al. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. MBio 6, e00306–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaijarasphong T et al. Programmed Ribosomal Frameshifting Mediates Expression of the α-Carboxysome. J. Mol. Biol. 428, 153–164 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Cai F et al. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One 4, e7521 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts EW, Cai F, Kerfeld CA, Cannon GC & Heinhorst S Isolation and characterization of the Prochlorococcus carboxysome reveal the presence of the novel shell protein CsoS1D. J. Bacteriol. 194, 787–795 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheatley NM, Sundberg CD, Gidaniyan SD, Cascio D & Yeates TO Structure and Identification of a Pterin Dehydratase-like Protein as a Ribulose-bisphosphate Carboxylase/Oxygenase (RuBisCO) Assembly Factor in the α-Carboxysome. J. Biol. Chem. 289, 7973–7981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aigner H et al. Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2. Science 358, 1272–1278 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Mueller-Cajar O The Diverse AAA+ Machines that Repair Inhibited Rubisco Active Sites. Front Mol Biosci 4, 31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krulwich TA, Hicks DB & Ito M Cation/proton antiporter complements of bacteria: why so large and diverse? Mol. Microbiol. 74, 257–260 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merlin C & Masters M Why is carbonic anhydrase essential to Escherichia coli? J. Bacteriol. 185, (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du J, Förster B, Rourke L, Howitt SM & Price GD Characterisation of Cyanobacterial Bicarbonate Transporters in E. coli Shows that SbtA Homologs Are Functional in This Heterologous Expression System. PLoS One 9, e115905 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cronk JD, Endrizzi J. a., Cronk MR, O’neill JW & Zhang KY Crystal structure of E. coli beta-carbonic anhydrase, an enzyme with an unusual pH-dependent activity. Protein Sci. 10, 911–922 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnamurthy VM et al. Carbonic anhydrase as a model for biophysical and physical-organic studies of proteins and protein- ligand binding. Chem. Rev. 108, 946–1051 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cronk JD et al. Identification of a novel noncatalytic bicarbonate binding site in eubacterial beta-carbonic anhydrase. Biochemistry 45, 4351–4361 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Shibata M et al. Distinct constitutive and low-CO2-induced CO2 uptake systems in cyanobacteria: genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc. Natl. Acad. Sci. U. S. A. 98, 11789–11794 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maeda S-I, Badger MR & Price GD Novel gene products associated with NdhD3/D4-containing NDH-1 complexes are involved in photosynthetic CO2 hydration in the cyanobacterium, Synechococcus sp. PCC7942. Mol. Microbiol. 43, 425–435 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Battchikova N, Eisenhut M & Aro EM Cyanobacterial NDH-1 complexes: Novel insights and remaining puzzles. Biochimica et Biophysica Acta - Bioenergetics 1807, 935–944 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Antonovsky N et al. Sugar Synthesis from CO2 in Escherichia coli. Cell 166, 115–125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aguilera J, Van Dijken JP, De Winde JH & Pronk JT Carbonic anhydrase (Nce103p): an essential biosynthetic enzyme for growth of Saccharomyces cerevisiae at atmospheric carbon dioxide pressure. Biochem. J. 391, 311–316 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sirard JC, Mock M & Fouet A The three Bacillus anthracis toxin genes are coordinately regulated by bicarbonate and temperature. J. Bacteriol. 176, 5188–5192 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abuaita BH & Withey JH Bicarbonate Induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect. Immun. 77, 4111–4120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baba T et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rubin BE et al. The essential gene set of a photosynthetic organism. Proceedings of the National Academy of Sciences 201519220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oakes BL, Nadler DC & Savage DF Chapter Twenty-Three - Protein Engineering of Cas9 for Enhanced Function. in Methods in Enzymology (ed. Sontheimer JADAEJ) 546, 491–511 (Academic Press, 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobrinski KP, Longo DL & Scott KM The Carbon-Concentrating Mechanism of the Hydrothermal Vent Chemolithoautotroph Thiomicrospira crunogena. J. Bacteriol. 187, 5761–5766 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zallot R, Oberg NO & Gerlt JA ‘Democratized’ genomic enzymology web tools for functional assignment. Curr. Opin. Chem. Biol. 47, 77–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dehal PS et al. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 38, D396–400 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sievers F & Higgins DG Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price MN, Dehal PS & Arkin AP FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Letunic I & Bork P Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slabinski L et al. XtalPred: a web server for prediction of protein crystallizability. Bioinformatics 23, 3403–3405 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Kelley LA, Mezulis S, Yates CM, Wass MN & Sternberg MJE The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roy A, Kucukural A & Zhang Y I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newby ZER et al. A general protocol for the crystallization of membrane proteins for X-ray structural investigation. Nat. Protoc. 4, 619–637 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khalifah RG The Carbon Dioxide Hydration Activity of Carbonic Anhydrase. J. Biol. Chem. 246, 2561–2573 (1971). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All illumina sequencing data is accessible as an NCBI SRA (BioProject accession number: PRJNA546024). All other data is available on github at: https://github.com/jackdesmarais/DabTransporterPaper.


