Abstract
Tumor metabolism has emerged as a hallmark of cancer and is involved in carcinogenesis and tumor growth. Reprogramming of tumor metabolism is necessary for cancer cells to sustain high proliferation rates and enhanced demands for nutrients. Recent studies suggest that metabolic plasticity in cancer cells can decrease the efficacy of anticancer therapies by enhancing antioxidant defenses and DNA repair mechanisms. However, there are few robust studies characterizing the metabolic changes induced by radiation therapy in cancer. Extensive study of radiation-induced metabolic changes will lead to a better understanding of radiation response mechanisms as well as the identification of new therapeutic targets. In this review, we will highlight studies that provide information on the metabolic changes induced by radiation and oxidative stress in cancer cells and the underlying mechanisms.
Keywords: Cancer, Radiation therapy, Metabolism, Radiosensitivity, Oxidative stress
Introduction
Cancer is the second leading cause of death in the United States, with approximately 1.8 million cases recorded domestically per year1,2 and over 19 million cases worldwide3. About 50% of cancer patients receive radiation therapy during the course of their treatment, often in combination with surgery4,5, chemotherapeutics5–8, or immunotherapy9,10. Recent efforts have focused on the unique role of cancer metabolism as a diagnostic tool and therapeutic target11–14.
Over a century after the discovery of a highly glycolytic metabolism in cancer cells by Otto Warburg, now known as the Warburg effect15,16, the study of cancer metabolism has seen a marked resurgence13,14,17,18. Relative to their non-malignant counterparts, cancer cells are in general characterized by an enhanced catabolism of glucose19, glutamine20, lactate21 and fatty acids22–24, accompanied by pronounced changes in glycolytic flux19, increased uptake of amino acids, especially glutamine14,25, fatty acid synthesis26–28, and redox metabolism14,29. Moreover, cancer cell metabolism is highly dynamic in response to various environmental stresses, such as hypoxia30,31 or cytotoxic therapies such as chemotherapy or radiation therapy32,33. Ionizing radiation typically induces cell death via indirect ionization of oxide species that form free radical species and attack the nearby DNA backbone, generating double strand breaks34,35. Metabolic radiosensitizers generally operate by depleting reducing equivalents such as the reduced form of nicotinamide adenine dinucleotide phosphate (NADPH), in affected cells36, inhibiting tumor-specific metabolic programs37, or by decreasing the rate of oxygen consumption in the tumor, resulting in a less hypoxic tumor microenvironment38–40.
Cancer metabolism has considerable utility both as a diagnostic41,42 and as a predictor of the overall progression43–47, chemotherapeutic response41,48–50 and radiosensitivity51,52 of tumors, and several chemotherapeutics targeting cancer metabolism have been demonstrated as radiosensitizers in recent trials53. Despite the growing interest in cancer metabolism and its extension into the field of radiation therapy, comparably little attention has been paid to the effects of ionizing radiation on cancer cell metabolism, which may be a useful predictor of the tumor response to radiation therapy and offer novel radiotherapeutic targets54,55.
Here, we will first review methods commonly used in cancer metabolism studies and their relative utility in assessing metabolic changes after radiation therapy. We will further expand on the metabolic alterations that have been reported to be induced by ionizing radiation in cancer cells, and the underlying potential molecular mechanisms. Lastly, we will comment on future directions that could further our understanding of the mechanistic and clinical implications of cancer cell metabolism following radiation therapy, including their potential as prognostic or therapeutic markers.
Section 1: Methods for tracking metabolic changes
Several methods have been developed to investigate the concentration and flux of various cellular metabolites and have proven useful both in basic science and in clinical applications. Here, we will discuss five important methods utilized in cancer metabolism studies, as well as which metabolites each method allows to be studied and the temporal dynamics of each. These methods are summarized in Figure 1.
Figure 1. A summary of methods for the study of cancer cell metabolism and its alterations after radiation.
Methods are grouped by their utility in vivo or in vitro, with suggested applications listed in blue boxes. Created with BioRender.com.
A. Extracellular flux analysis
Extracellular flux analysis studies the oxygen consumption rate and extracellular acidification rate of cells cultured in proprietary multi-well plates, allowing for quantification of the rate of ATP production through mitochondrial and glycolytic sources56. Sequential treatment with inhibitors of proteins involved in oxidative phosphorylation (i.e. oligomycin, FCCP, and rotenone) determines a cell’s basal respiration rate, ATP-linked respiration, proton leak and maximum respiratory capacity, in addition to oxygen consumption due to non-mitochondrial respiration. Extracellular flux assays are typically performed in vitro and, since inhibitors of mitochondrial respiration are added sequentially, do not typically permit multiple analyses of the same population in the same well, but readily permit analysis of irradiated cell cultures. The analysis is high-throughput, easily multiplexed, and provides useful information not just on the baseline activity of mitochondrial and glycolytic respiration, but also on their response to stimulus.
B. Mass spectrometry-based metabolomics
Mass spectrometry-based metabolomics have proven useful for analyzing the absolute levels of multiple metabolites as well as metabolic flux, with applications both in vivo and in vitro. Among the different types of mass spectrometry, metabolomics uses either separation-based techniques, such as liquid chromatography57 and gas chromatography (GC), or separation-free techniques, such as Matrix-Assisted Laser Desorption Ionization (MALDI). These techniques each have their own advantages and limitations, which have been discussed extensively elsewhere58. What is particularly interesting about metabolomics studies is the possibility of studying treatment-induced changes in pathway flux by performing targeted metabolomics in the presence of stable isotopic tracers59. Tracers are chosen based on the pathway studied60 and may be incubated for different periods of time based on the pathway of interest, and at different timepoints after interventions such as radiation therapy. As an example, the use of uniformly-labeled 13C-glucose allows for carbon tracing through glucose metabolism and downstream pathways, but specific pathways might not be distinguishable. To specifically quantify the glycolytic flux relative to the pentose phosphate pathway flux, 1,2-13C-glucose may be used as it results in metabolites with a different labeling pattern compared to uniformly-labeled 13C-glucose59. Among the metabolites that can be quantitatively analyzed are amino acids, lipids, nucleotides, carbohydrates and organic acids, making mass spectrometry-based metabolomics a broad approach to study metabolism. While this technique is widely used in vitro, it can also be applied in vivo. The latter has been made possible thanks to the development of minimally invasive surgical techniques for isotope infusion61. Finally, spatial (or imaging) mass spectrometry that detects the spatial distribution of metabolites in tissues and cells62 has also been developed and can be applied in vitro and in vivo. By ionizing select regions of a sample at a time, the identities and positions of metabolites within the sample can be detected with63 or without64 metabolic tracers. This technique holds great promise for studying in situ metabolism65. This is of particular importance in the context of cancer and tumor heterogeneity. The metabolic response to radiation is likely to vary within a tumor, for example based on oxygen availability.
C. Nuclear magnetic resonance spectroscopy
Hyperpolarized nuclear magnetic resonance spectroscopy, or similarly, hyperpolarized magnetic resonance spectroscopic imaging (HP-MRSI) typically uses metabolic probes designed with a single carbon-13 (13C) at a known location, which is then subjected to extremely low temperatures and a strong magnetic field66,67. This results in a single, highly polarized, 13C-labeled metabolite, whose concentration and conversion to other metabolites can be measured in real-time using conventional MRSI methods68,69. Notably, this allows not only for localization and quantification of chosen metabolites, but, owing to the different chemical shifts of downstream metabolites, also allows for determination of the downstream metabolites to which the injected probe was converted over time. Several 13C metabolites have been developed, including labelled pyruvate70, alpha-ketoglutarate71,72, acetyl-CoA73, and arginine74, to name a few75,76, allowing for the study of various metabolic pathways. Selective labeling of particular carbons can be used to specify unique downstream pathways for further study. However, given the rapid decay of spin-polarization experienced by hyperpolarized samples, detectable signal tends to decay quickly after application of 13C probes. This presents logistical challenges, since probes are typically administered concomitantly with imaging, and limits the duration of HP-MRSI experiments. Accordingly, experimental design using hyperpolarized metabolites should take into account the time scale at which metabolic changes are expected, since continuous imaging over a prolonged time course is difficult with hyperpolarized metabolites.
HP-NMR has been successfully used to track metabolite conversion over time in cells cultured in NMR-compatible bioreactors, providing a useful system for studying cancer cell organoids77, tissue slices78 or alginate bead cultures of cancer cell lines after ionizing radiation79. This method has successfully tracked the concentration and conversion of multiple metabolites in cancer cells, including the conversion of pyruvate to lactate, alanine and bicarbonate66,80 and the conversion of alpha-ketoglutarate to dextro-2-hydroxyglutarate71 in cells with mutations in isocitrate dehydrogenase 1 (IDH1)81. Similarly, HP-MRSI was used to track metabolism in mouse xenografts and in pilot studies of human tumors, and is the subject of ongoing clinical trials (Table 1). Nuclear magnetic resonance without hyperpolarization of naturally-occurring 31P or 1H has also been used clinically for decades, and is capable of identifying changes in the levels of multiple metabolites after radiation therapy in patient tumors82–84 or in plasma, urine, and stool85.
Table 1. A summary of clinical trials involving HP-MRSI.
Status and descriptions of clinical trials were taken from clinicaltrials.gov on 2 August 2021.
| Trial ID | Status | Phase | Etiology | Purpose | Metabolites studied |
|---|---|---|---|---|---|
| NCT04540107 | Active | Phase II | Glioma | Investigate whether changes in levels of HP-pyruvate, lactate, and bicarbonate predict tumor progression in low-grade glioma patients | Pyruvate, lactate, bicarbonate |
| NCT04258462 | Active | Phase II | Renal | Investigate association between HP-Pyruvate-to-lactate conversion and renal tumor histoloy. Predict benign renal tumors versus renal cell carcinoma and grading | Pyruvate, lactate |
| NCT04565327 | Active | Phase II | Pancreatic | Evaluation of early treatment response | Pyruvate, lactate |
| NCT04019002 | Active | Phase I | Glioblastoma | Detection of early response to standard therapy | Pyruvate, lactate |
| NCT03933670 | Active | Phase II | Prostate | Associate intratumoral pyruvate-to-lactate and pyruvate-to-glutamate conversion with Gleason grade | Pyruvate, lactate, glutamate |
| NCT03739411 | Active | Phase I | Glioma | Assess safety of hyperpolarized metabolites, evaluate lactate-to-pyruvate response to standard RT/temozolomide | Pyruvate, lactate |
| NCT02526368 | Active | Early Phase I | Prostate | Determine metabolite levels that accurately detect Gleason 4 component cancer | Pyruvate, lactate, urea |
| NCT04589624 | Active | Phase I | Thryoid | Assess early metabolic changes in response to radiation and/or systemic therapy | Pyruvate, lactate |
| NCT04346225 | Active | Phase II | Prostate | Assess rates of conversion and intratumoral heterogeneity of metabolites, determine if changes from baseline conversion predict clinical outcomes | Pyruvate, lactate, glutamate |
| NCT04044872 | Active | Phase IV | Cardiotoxicity in breast or thoracic tumors | Determine if radiation-induced cardiac injury disrupts mitochondrial metabolism, determine prognostic value of mitochondrial pyruvate flux on cardiotoxicity | Pyruvate, lactate, bicarbonate, glutamate |
| NCT03830151 | Active | Phase I | Glioma | Assess correlation between pyruvate-to-lactate conversion and Ki-67 levels in tumor | Pyruvate, lactate |
| NCT03581500 | Active | Phase II | Prostate | Assess sensitivity and specificity of pre-therapy HP-MRSI for detecting localized prostate cancer | Pyruvate, lactate |
| NCT02421380 | Active | Phase II | Sarcoma, Prostate, Breast, Brain | Test reproducibility of HP-MRSI in vivo dynamics | Pyruvate, lactate |
| NCT04656431 | Closed due to accrual | Phase I | Primary central nervous system lymphoma | Investigate connections between pre and post-treatment lactate signal and NF-kB | Pyruvate, lactate |
| NCT03685175 | Active | Phase I | Cardiotoxicity in breast tumors | Test correlation between doxorubicin-induced cardiotoxicity and aerobic cardiac metabolism | Pyruvate |
| NCT03759704 | Active | Phase 0 | Musculoskeletal sarcoma | Determine if pyruvate-to-lactate conversion correlates with malignancy or tumor grade | Pyruvate, lactate, bicarbonate, glutamate |
| NCT04286386 | Active | N/A | Prostate | Assess reproducibility of pyruvate-to-lactate conversion, assess correlation between conversion and tumor grade | Pyruvate, lactate |
| NCT03067467 | Active | N/A | Glioma, meningioma, brain metastases | Determine whether ratio of metabolites indexes balance between glycolytic and mitochondrial metabolism | Pyruvate, lactate, bicarbonate |
| NCT01229618 | Completed | Phase I | Prostate | Determine time course and signal-to-noise ratio of hyperpolarized pyruvate in cancer and benign tissue | Pyruvate, lactate |
| NCT02450201 | Terminated | Early Phase I | Prostate Cancer | Evaluate reproducibility of pyruvate signal in patients treated with androgen depletion | Pyruvate, lactate |
| NCT02647983 | Withdrawn (Lack of participants) | Phase I | Prostatic Neoplasms | Ability to predict Gleason grade using metabolic profile | Pyruvate, lactate, alanine, bicarbonate |
| NCT02844647 | Recruiting | Phase I | Prostate Cancer | Evaluate pyruvate conversion in bone-metastatic castration-resistant prostate cancer | Pyruvate, lactate, bicarbonate |
| NCT02911467 | Terminated | Phase I | Prostate Cancer | Investigate baseline intratumoral peak lactate-to-pyruvate ratio in androgen signaling inhibitor refractory and responsive tumors | Pyruvate, lactate |
| NCT02913131 | Terminated | Phase I/II | Prostate Cancer | Measure percent change from baseline lactate-to-pyruvate ratio in patients treated with PI3K/mTOR inhibitors | Pyruvate, lactate |
| NCT02947373 | Completed | Phase I | Pediatric Brain Tumors | Categorize adverse events, describe imaging quality in HP-MRSI of pediatric tumors | Pyruvate, lactate |
| NCT03121989 | Recruiting | Phase I | Breast Cancer | Determine diagnostic accuracy of 13C pyruvate, distinguish tumor necroses from viable tumor cells in patients given neoadjuvant chemotherapy | Pyruvate, lactate |
| NCT03129776 | Recruiting | Phase I | Uterine Cervical Neoplasms | Correlate MRSI and 18FDG-PET images in cervical cancer | Pyruvate, lactate, fluorodeoxyglucose |
| NCT03279250 | Completed | Phase II | Prostate | Investigate proportion of patients with pathological complete response via HP-pyruvate imaging | Pyruvate |
| NCT03324360 | Recruiting | Phase I | Brain Metastases | Characterize metabolic features of intracranial metastasis, predict tumor aggressiveness | Pyruvate |
| NCT03526809 | Unknown status | N/A | Ovarian, breast, prostate, pancreatic, hepatic, renal, brain tumors | Correlate hyperpolarization images with tumor histology, biochemistry and genetics. Detect change in metabolism after therapy. Determine correlation between HP and FDG-PET images | Pyruvate, fluorodeoxyglucose |
| NCT03565367 | Suspended (Logistics) | Phase I | Central nervous system, including metastases | Assess frequency and sensitivity of lactate and bicarbonate signal in malignant brain tumors after HP-pyruvate injection | Pyruvate, lactate, bicarbonate |
| NCT03687645 | Unknown status | N/A | Prostate, renal, breast cancer, lymphoma | Determine rate of pyruvate-to-lactate conversion, correlation of rate with genetic markers and to predict response | Pyruvate, lactate |
| NCT03849963 | Recruiting | Early Phase I | Brain Cancer | Evaluate sensitivity and feasibility of HP-MRSI for imaging oxidative metabolism and neurotransmitter synthesis | 1–13C and 2–13C pyruvate, bicarbonate, lactate, glutamate |
| NCT04698564 | Not yet recruiting | Phase II | Prostate Cancer | Determine diagnostic accuracy and utility of 13C pyruvate, compare metabolic MRI to standard MRI imaging | 1–13C, 2–13C, 1,2–13C pyruvate “and its metabolites” |
| NCT04772456 | Recruiting | Phase I | Glioma | Determine utility of metabolic MRI over standard MRI in diagnosis of glioma | 1–13C, 2–13C, 1,2–13C pyruvate “and its metabolites” |
D. Measuring oxygenation
Multiple methods exist for imaging oxygenation of tumor tissues, including blood oxygen level-dependent (BOLD) MRI86, positron emission tomography (PET) with 18F-imidazoles87 and electron paramagnetic resonance oximetry88, all of which can provide useful insight into the hypoxic status of heterogenous solid tumors. Considering the frequency with which metabolic radiosensitizers alter the oxygenation status of tumors, imaging of tumor oxygenation89 can enhance effective studies of changes in tumor metabolism in vivo.
E. Positron emission tomography
PET is a non-invasive imaging technique commonly used to visualize patient tumors, in particular with the use of 18F-fluoreodeoxyglucose (FDG). This technique is based on the high glucose uptake by cancer cells relative to the surrounding normal cells. Other PET tracers have also been developed in the context of tumor detection90. In addition to its role in clinical oncology, PET has been used in pre-clinical research to measure metabolite uptake in vivo and study pathways such as glutamine metabolism91 (11C-L-glutamine) and fatty acid synthesis92 (11C-acetate). PET can also be useful for evaluating treatment response in vivo. If normalized to tumor size instead of being used to define it, this technique can also be used to determine the uptake of a given tracer per cancer cell or tumor unit93,94. However, it provides no indication on the metabolic flux downstream of the tracer. Studies using FDG-PET early in the course of combination therapy have shown prognostic value in several tumor types, implying utility of these imaging methods both before and after radiation therapy95–98.
Section 2: Changes to major metabolic pathways induced by ionizing radiation
The effects of ionizing radiation on cancer cell metabolism are potentially a reflection of the indirect damages that radiation-induced reactive oxygen species (ROS) cause on macromolecules, leading to disruption of redox homeostasis and ‘oxidative stress’. Oxidative stress can be defined as an imbalance between ROS generation and ROS scavenging by cellular antioxidant defenses99. If not successfully repaired, the oxidative damage can be lethal. The effectiveness of radiation therapy is in part due to the therapeutic index created by the superior repair capacity of normal tissues relative to cancer cells, with radiation sensitivity in cancer thought to be a function of DNA repair capacities and antioxidant defenses52. Rewiring of metabolic pathways so as to provide cells with metabolites for antioxidant defenses and DNA repair is necessary for both these mechanisms. Cancer cells with high metabolic plasticity may therefore be able to better respond to oxidative stress and mitigate the cytotoxic effects of ionizing radiation.
It is important to point out that many studies aimed at investigating the effects of ionizing radiation on cells use oxidizing agents in lieu of radiation to mimic its effects. As an example, although hydrogen peroxide (H2O2) does induce oxidative stress100, its effects are not equivalent to those of ionizing radiation101,102. H2O2 is converted to water and dioxygen within minutes by the catalase enzyme and studies therefore require high concentrations of H2O2 to induce potentially lethal oxidative stress103. A recent study using various types of cancer cells showed that while radiosensitivity predicted sensitivity to peroxide, the converse was not always true, as H2O2-resistant cancer cells were sensitive to X-ray-induced cell death101. Such studies caution against equating observations made under oxidative stress conditions induced by means other than ionizing radiation to those of radiation-induced effects. In this section, we will specifically focus on metabolic changes after oxidative stress induced by ionizing radiation.
A. Metabolic response to oxidative stress induced by radiation
Robust antioxidant defenses play a major role in radiation sensitivity. For example, cancer stem cells, notorious for their resistance to therapies, have higher basal activation of ROS scavenging systems, which is correlated to radiation sensitivity104. It is reasonable to postulate that cancers with poor responses to radiotherapy may be more likely to have either strong basal antioxidant defenses or a great deal of metabolic plasticity that facilitates rewiring of metabolic fluxes to generate a powerful antioxidant response after radiation. In agreement with this idea, two very recent studies using personalized genome-scale metabolic flux models identified tumor redox metabolism as a major predictor for radiation sensitivity105,106. It seems that tumors with poor radiation response reroute metabolism to boost the levels of reducing factors of the cell, such as NADPH and glutathione, thus enhancing clearance of ROS105. This implies a dependency on rerouting metabolic fluxes for maintaining antioxidant defenses and survival after lethal oxidative damage, ultimately pointing to metabolic plasticity as a driver of radiosensitivity53,107,108. This mechanism of radiation sensitivity is distinct from enhanced DNA repair capacity, although successful DNA repair ultimately depends on enhancing flux through metabolic pathways that generate precursors for DNA building blocks (more below). Considering radiation sensitivity from this angle opens up the possibility that metabolically targeted interventions might improve radiotherapeutic outcomes. Multiple metabolic pathways feed into antioxidant defenses and DNA repair, including glycolysis, the pentose phosphate pathway (PPP), glutaminolysis, and 1-carbon metabolism (1CM), all of which will be discussed more in detail below.
B. bGlycolysis and the pentose phosphate pathway
Glucose is one of the major nutrients (the other being glutamine) which supports the growth and survival of cancer cells. Irradiated breast cancer cells have been shown to increase glucose consumption, accompanied by increased extracellular lactate109,110. While the Warburg effect has been well characterized to increase glycolysis in cancer cells at baseline16,111, changes to glycolytic flux after ionizing radiation may be a function of cellular capacity to transport metabolites112,113. Glucose transporters (GLUTs) are responsible for glucose import and GLUT1 expression in particular has been linked to radiosensitivity112. Studies have shown HIF-1-dependent upregulation of GLUT1 after ionizing radiation in cancerous110,114 and normal tissue115, including under normoxic conditions115, thus explaining in part the radiation-induced increase in glucose consumption, although the exact mechanisms remain to be elucidated.
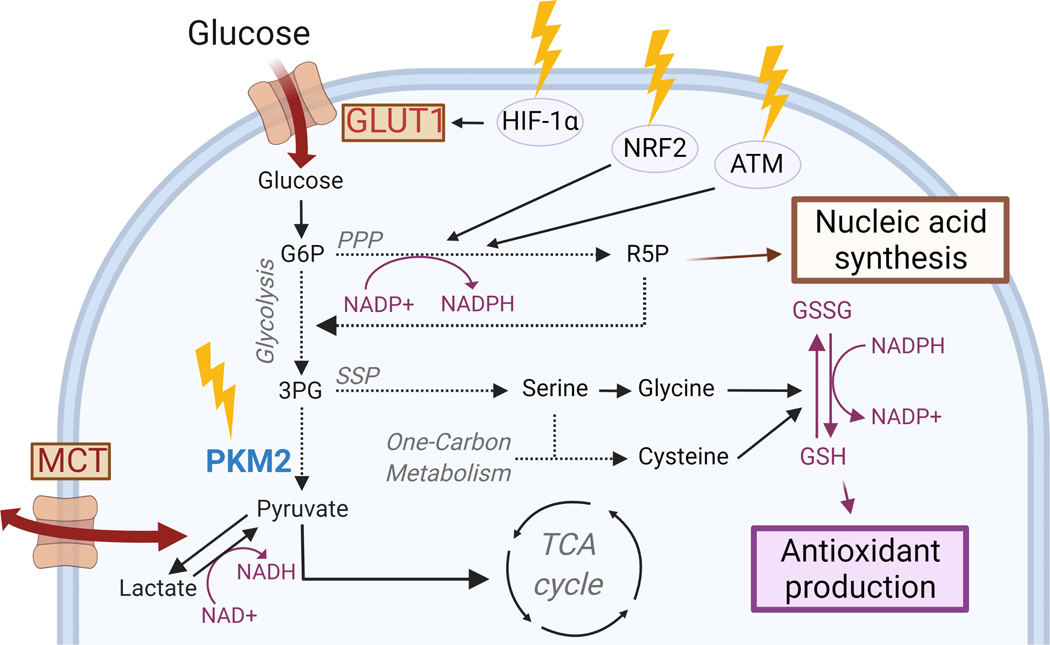
The altered glycolytic flux that seems to be induced by radiation is accompanied by a suppression in the enzymatic activity of the M2 isoform of pyruvate kinase (PKM2), which converts phosphoenolpyruvate to pyruvate immediately before pyruvate enters the TCA cycle109. Others have shown similar effects on PKM2 with chemically-induced oxidative stress116, suggesting that inactivation of this redox-sensitive glycolytic enzyme may be a shared response to oxidative stress. The role of PKM2 in reprogramming cancer cell metabolism to promote tumor growth is well established117,118 and it has provided an explanation for the preferential expression of the M2 isoform in cancer cells. While all other isoforms of PK exist as constitutively active tetramers, PKM2 can shift between enzymatically active tetramers and inactive dimers; tetrameric PKM2 efficiently converts phosphoenolpyruvate to pyruvate119. In contrast, kinase-inactive PKM2 blocks pyruvate production, creating a bottleneck in glycolysis that makes upstream glycolytic intermediates available for auxiliary glycolytic pathways, such as the pentose phosphate pathway (PPP) and the serine synthesis pathway (SSP). In proliferating cells, the PPP and SSP provide building blocks necessary for cellular anabolism29,120,121. These same pathways also provide the two main reducing equivalents in cells, NAPDH and glutathione (Figure 2). NADPH fuels antioxidant systems and recycles oxidized glutathione to combat oxidative stress so as to restore cellular redox homeostasis122. It therefore appears that one advantage of the seemingly paradoxical radiation-induced increase in glucose consumption in breast cancer cells while PKM2 activity is suppressed may be the availability of glycolytic intermediates for funneling into antioxidant pathways subsidiary to glycolysis that can support survival during oxidative stress conditions. In this manner, PKM2 may act as a ‘metabolic switch’ that provides cells with metabolic plasticity in a context-dependent manner123,124. A study by Tuttle and colleagues shows that cells with high PPP activity are less sensitive to radiation therapy and our own work (unpublished data) suggests that forced activation of PKM2 can sensitize breast cancer cells109 as well as glioblastoma tumors to radiation.
Figure 2. Radiation increases glucose metabolism to enhance antioxidant production and nucleic acid synthesis.
Ionizing radiation (yellow arrows) decreases PKM2 activity (in green) while increasing GLUT1-mediated glucose uptake and MCT-mediated exchange of pyruvate and lactate (in red), providing intermediates for nucleic acid synthesis and antioxidant production. Created with BioRender.com.
Enhanced activity of the PPP after ionizing radiation is possible via the activation of diverse molecular pathways that are responsive to oxidative stress, for example, the activation of the transcription factor nuclear factor erythroid 2-related factor 2 (NRF2), a master regulator of oxidative stress. Under normal conditions, NRF2 is targeted for degradation by its negative regulator Kelch-like ECH-associated protein 1 (KEAP1)125, but oxidative stress inactivates KEAP1, resulting in accumulation of newly translated NRF2, which can then translocate to the nucleus and initiate transcription of its target genes that contain ‘antioxidant response elements’ (ARE) largely involved in maintaining cellular redox homeostasis125,126. Among NRF2’s target genes are a number of metabolic genes that drive the PPP, which generates NADPH (Figure 2). Ionizing radiation activates the NRF2 pathway and protects normal cells (mouse embryonic fibroblasts) from ROS and radiation-induced toxicity. Our own data also shows that cancer cells activate the NRF2 pathway after radiation127 (Figure 2), and that NRF2 activation is involved in radiation-induced metabolic reprogramming (unpublished data). An additional way of upregulating the PPP is via the redox-sensitive, protein kinase ataxia telangiectasia mutated (ATM). ATM is activated in response to radiation-induced DNA damage128,129 and prevents radiation-induced cell death by promoting homologous recombination and DNA repair130. Evidence exists that ATM can induce the activity of G6PDH, the first rate-limiting, and NADPH-producing enzyme in the PPP (Figure 2). Others have shown that in addition to promoting an antioxidant response, ATM-induced PPP activation also leads to increased nucleotide synthesis via the generation of the sugar backbone for dNTPs, the ribose-5-phosphate (R5P)131 (Figure 2).
C. Pyruvate and lactate metabolism
In addition to its stereotypical role as a waste product of anaerobic glycolysis, lactate serves several important roles in cancer cell metabolism. Given the high demand for NAD+ imposed by a highly glycolytic phenotype14, the conversion of pyruvate to lactate serves as a critical mechanism for replenishing NAD+ levels from the cellular NADH pool132 (Figure 3). On the other hand, lactate also serves as an important source for gluconeogenesis through the hepatic Cori cycle, and can serve as an important extracellular signaling molecule133,134, altering the activity of several key metabolic enzymes at high concentrations135–138. Moreover, high lactate concentration in the tumor microenvironment has been implicated in alterations of local immune cell populations139–141, decreased radiosensitivity142,143, higher cancer cell proliferation144, metastasis145–147 and worsened prognosis145,148–150 in a variety of cancer etiologies.
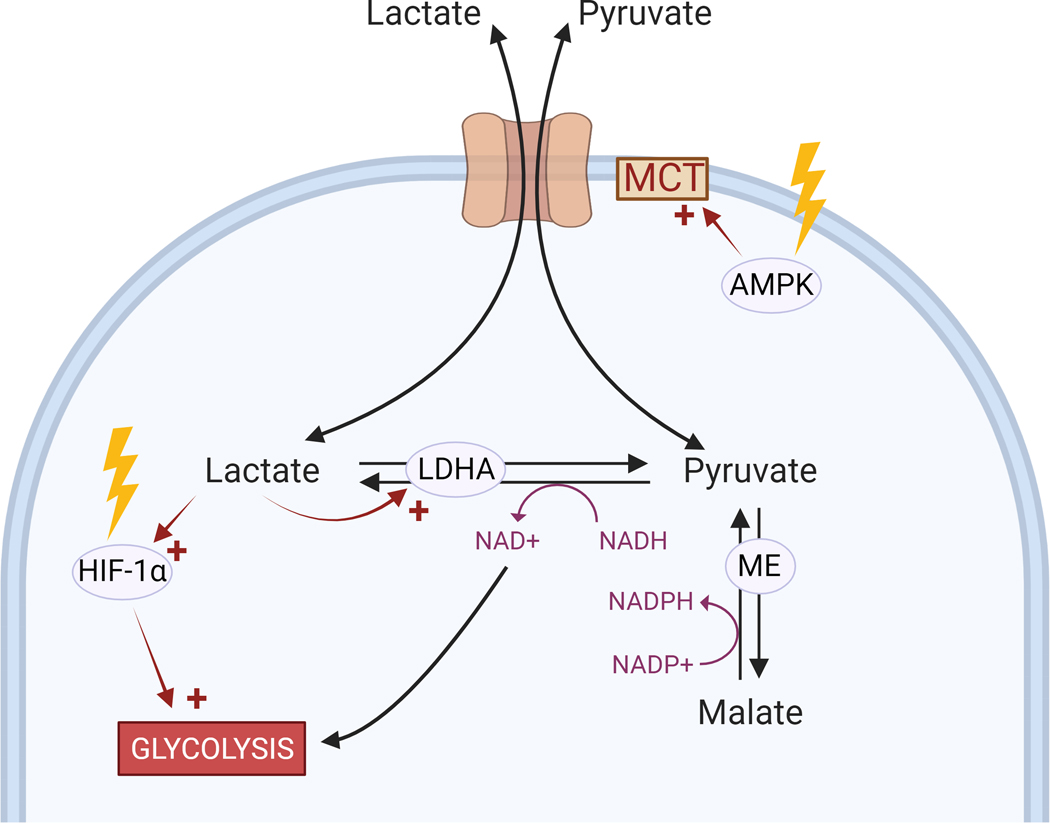
Figure 3. Radiation affects the lactate to pyruvate ratio via MCT transporters.
Ionizing radiation (yellow arrows) activates several pathways (indicated by red arrows) implicated in lactate production and transport. Created with BioRender.com.
The radiation-induced increase in extracellular lactate concentration despite the suppressed enzymatic activity of PKM2 in breast cancer cells is unexpected109, as low PKM2 activity should decrease pyruvate production from glycolysis. One possible explanation is the generation of pyruvate via the oxidative decarboxylation of malate to pyruvate (Figure 3). This reaction is catalyzed by the cytosolic malic enzyme 1 (ME1), a target gene of NRF2151, or the mitochondrial isoforms (ME2/ME3). It is possible therefore that the activation of the NRF2 pathway by radiation127 contributes to increased lactate production by upregulating the malic enzyme levels. The conversion of malate to pyruvate by malic enzyme generates NADPH, thus further contributing to the antioxidant defenses, as well as to lipid and cholesterol biosynthesis152.
The source and rate of lactate usage in cancer cells has been the subject of a number of studies19,21. Notably, even cells with net lactate production import exogenous lactate in in vivo rat glioma models, using it both as an energy source and to generate alanine and glutamate153,154. Production of lactate through lactate dehydrogenase A (LDHA)148 and transport of lactate via monocarboxylate transporter (MCT) families have been correlated with cancer prognosis150,155,156. Several key cancer-associated signaling pathways also influence lactate production, including Ras, Akt, Myc, and the Dek oncogene157. The Myc signaling pathway upregulates LDHA149,158 and cellular MCTs159. Lactate itself has an effect on several key signaling pathways, upregulating expression of HIF-1α, an upstream regulator of GLUT1160,161, and many other glycolytic enzymes, including LDHA162 (Figure 3).
Several studies have found that the interconversion between lactate and pyruvate is highly responsive to chemotherapeutics113,163–165 and radiotherapy79,163,164 in both three-dimensional alginate cultures79,113 and mouse xenografts163–165. The lactate-to-pyruvate ratio after radiation shows significant temporal dynamics, with a marked decrease in lactate levels and lactate-to-pyruvate ratio shortly after irradiation79,166,167 and increased lactate levels over time166,168, which may imply differences in tumor reducing potential over time after ionizing radiation167,169,170. Radiation can upregulate expression of MCT1 after 8 hours in an AMPK/NFkB-dependent manner, providing a mechanism for the radiation-induced decrease in lactate-to-pyruvate ratio171. MCT inhibition radiosensitizes small cell lung cancer cells in vitro and decreases tumor growth172. Interestingly, studies of tumor oxygenation following MCT inhibition found a notable increase in tumor oxygenation, arguing that radiosensitization of MCT-inhibited tumors may stem from loss of the hypoxic core173. Similar mechanisms targeting inhibition of the mitochondrial pyruvate carrier (MPC) found that inhibition of extracellular lactate uptake and accumulation of intracellular pyruvate radiosensitized spheroid cultures by decreasing oxygen consumption and decreasing the size of the hypoxic core independent of extracellular lactate or glucose concentrations40.
Pyruvate and lactate levels are strongly influenced by the expression of the bidirectional cell membrane monocarboxylate transporter family, including MCT1174,175 (Figure 3). Studies of siRNA-mediated MCT knockouts in three-dimensional alginate cultures found that the lactate-to-pyruvate ratio was predominantly a function of MCT expression regulating pyruvate influx, rather than LDHA expression or glycolytic flux113. This implied that the lactate-to-pyruvate ratio in cancer cells may be a function of transport rather than synthesis. MCT1 expression was found to rate-limit the conversion of pyruvate to lactate in non-irradiated cells113. Together with observations that ionizing radiation alters expression of MCT1, this provides an independent mechanism by which lactate production can increase while the lactate-to-pyruvate ratio decreases in irradiated cells. Given the importance of both lactate synthesis and transport following ionizing radiation, further studies are necessary to gain more insight into this seemingly competitive utilization of lactate by irradiated cancer cells, paying particular attention to alterations of signaling pathways and the temporal dynamics of post-irradiation metabolic responses.
D. Mitochondrial metabolism
Despite the heavy reliance on Warburg metabolism in many cancer cells, mitochondrial metabolism remains a significant source of energy production and oxygen consumption, which may have significant effects on radiation response176,177. Interestingly, ionizing radiation upregulates mitochondrial oxygen consumption, ATP production and ROS generation within 24 hours after irradiation178–182, possibly due to mTOR-dependent upregulation of oxidative phosphorylation183. Inhibition of this mitochondrial increase in ATP production after ionizing radiation has been demonstrated to radiosensitize lung cancer cells181, in addition to other mitochondrial radiosensitizers that may also work to decrease oxygen consumption.
Mitochondria are also responsible for approximately 90% of intracellular ROS generation184, implying that mitochondrial ROS generation could play a significant role in radiosensitivity. Several key antioxidant proteins with important roles in radiation response, including SOD2, thioredoxin reductase (TXNRD) and glutathione peroxidase, are frequently upregulated in tumors and are downstream targets of the mitochondrial KEAP1-NRF2 pathway185,186. Several mitochondrial genes are upregulated in response to ionizing radiation in both directly irradiated and bystander cells, including ATP synthases MT-ATP6 and MT-ATP8, and NADH dehydrogenases MT-ND1, MT-ND5 and MT-ND6187,188. Murine cancer models also show upregulation of superoxide dismutase after radiation188, consistent with upregulation of genes involved in radiation-induced oxidative stress response189.
Ionizing radiation has also been shown to affect mitochondrial dynamics, inducing mitochondrial fission in normal fibroblasts in a DRP1-dependent manner190,191, consistent with upregulation by ionizing radiation of the upstream regulators of DRP1, AKT and mTOR192,193 and increased mitochondrial localization of DRP1190. In cancer cells, metabolic reprogramming typically leads to higher cytoplasmic expression of DRP1 at baseline, paired with accumulation of both functional and defective mitochondria 194–197. Accordingly, mitochondrial autophagy to remove defective mitochondria is correlated with tumor malignancy198 and strongly tied to loss of mitochondrial membrane polarization, which is induced by mitochondrial ROS. Inhibition of mitophagy after ionizing radiation increased rates of cellular autophagy and slowed growth in a HeLa cell culture model199, consistent with similar reports connecting inhibition of mitophagy to increased radiosensitivity in in vitro and in vivo nasopharyngeal carcinoma models200. Mitochondrial fission is frequently implicated in other stress responses201, including ultraviolet radiation, genotoxic stress, and nutrient deprivation202. Interestingly, oxidative stress following brief administration of hydrogen peroxide stimulated mitochondrial fusion in human umbilical vein epithelial cells203, implying different mitochondrial dynamics in response to the duration and severity of stress presented204. After ionizing radiation, increased mitochondrial fission has been implicated in calcium release, and mitotic catastrophe via dysregulation of centromeres independent of apoptotic pathways190,205, with inhibition of DRP1 increasing the rate of mitotic catastrophe190.
E. Amino acids and one-carbon metabolism
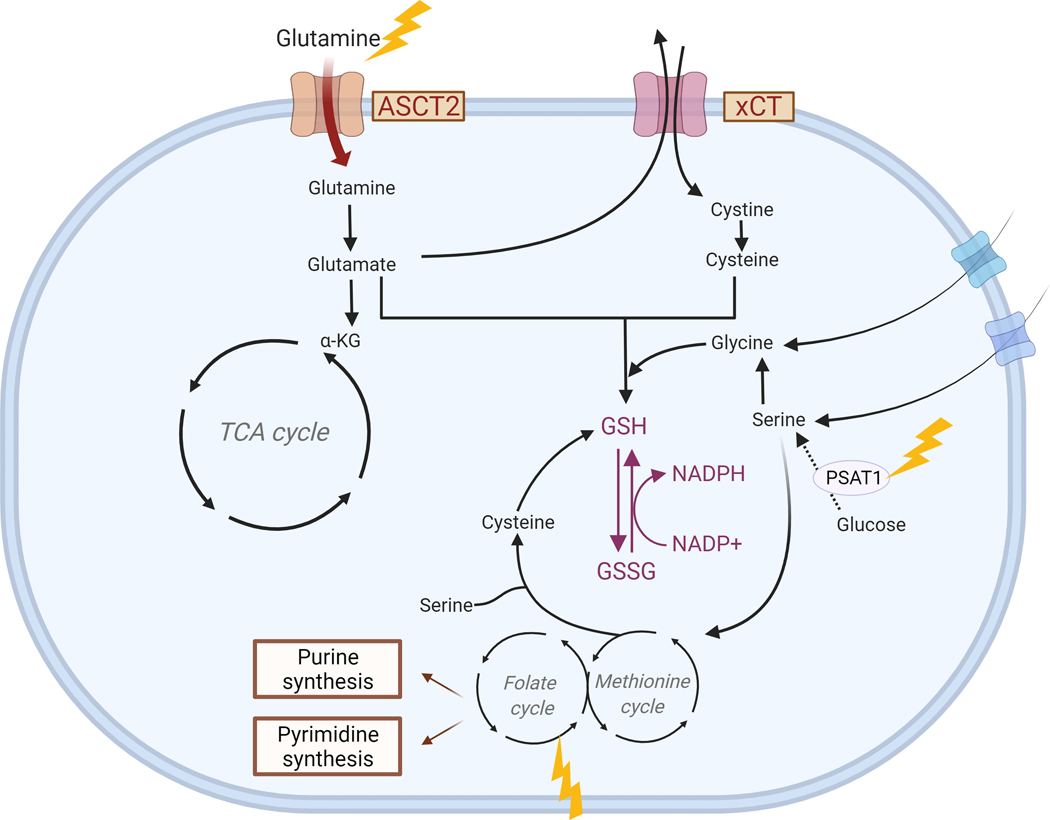
Amino acids constitute a necessary fuel for tumor growth. Glutamine, the second major nutrient for cancer cells after glucose, plays an important role in the cancer cell biology. Upon entry in the cell via ASCT2 transporter, glutamine is converted to glutamate and serves as a nitrogen source for amino acid and nucleotide biosynthesis. Glutamate is particularly important as it can either be converted to α-ketoglutarate (αKG), and enter the tricarboxylic acid (TCA) cycle, or serve as one of the main building blocks in glutathione synthesis (Figure 4). Cancer cells from various origins are highly dependent on glutamine206–208 and therapeutic targeting of glutamine metabolism is an area of significant interest.
Figure 4. Effects of radiation on amino acids and one-carbon metabolism.
Ionizing radiation (yellow arrows) affects expression of several pathways (in red text) associated with amino acid synthesis and redox metabolism. Other important signaling pathways that provide intermediates for these reactions are also shown. Created with BioRender.com.
Multiple targets associated with glutamine metabolism have also been associated with radiosensitivity209–211. However, very few studies have investigated the effects of ionizing radiation on glutamine metabolism in cancer. Previous work has demonstrated that matched cancer cells selected for radiation resistance upregulated glutamine synthetase relative to parental lines211. The authors noted an increase in glutamine anabolism while glycolysis, OCR, and ATP production were reduced, but did not study the direct effect of ionizing radiation on glutamine metabolism. Our own data also suggest that ionizing radiation leads to an enhanced consumption of glutamine 24 hours after radiation109, most likely as a response to oxidative stress to generate more glutathione210 (Figure 4). There is also evidence that glutathione levels are increased by radiation in the context of cancer57,212. In addition to glutamate, glutathione is made from cysteine and glycine, the latter of which can either be imported or synthesized from serine. While cells can take up free serine from their environment or synthesize it de novo from glucose, some cancer subtypes become addicted to de novo production of serine213–215. The serine synthesis pathway in combination with one-carbon metabolism generates both NADPH and the precursors for reduced glutathione synthesis. Interestingly, levels of phosphoserine aminotransferase 1 (PSAT1), which converts 3-phospho-hydroxypyruvate and glutamate to 3-phospho-serine and αKG, have been shown to increase after ionizing radiation (Figure 4), and targeting PSAT1 leads to radiation- and glutamine-deprivation sensitivity in lung cancer cells216. Although there is a lack of studies on serine metabolism after radiation, the field is starting to recognize the role of SSP in redox metabolism217–219.
One-carbon metabolism, responsible for maintenance of methylation reactions in the cell, is heavily implicated in cancer and frequently altered by ionizing radiation (Figure 4). It is well established that cancer cells maintain significant alterations in both DNA220,221 and histone methylation222,223, implicated in carcinogenesis224, proliferation225,226 and drug resistance227 of several cancers. Importantly, one-carbon metabolism is also closely tied to synthesis of purines and pyrimidines via the folate cycle. While a recent review has already covered radiation-induced alterations to methylation and one-carbon metabolism in depth228, we will summarize important points here.
One-carbon metabolism involves the conversion of folate from circulating monoglutamates229 or the import of extracellular folate by reduced folate carrier (RFC) transporters230, with outputs capable of generating purines, thymidylates, and the S-adenosylmethionine and S-adenosylhomocysteine responsible for DNA and histone methylation. Importantly, homocysteine is also a precursor for glutathione synthesis, implicating one-carbon metabolism in glutathione-mediated oxidative responses. Mechanistically, antifolate chemotherapeutics have been shown to be effective via inhibition of nucleotide synthesis231,232.
Total body irradiation of normal mice resulted in an increase in expression of several key enzymes in the folate cycle, but a decrease in expression of the rate-limiting enzyme methylenetetrahydrofolate reductase 1 (MTHFR1), resulting in a net decrease in total folate levels in liver233, plasma and bone marrow samples234. Direct oxidation of folate by ionizing radiation has also been observed, potentially contributing to a general decrease in folate levels in irradiated animals235. Cancer cells also seem to be vulnerable to depletion of methionine, the precursor to S-adenosylmethionine, a key player in both the folate and methionine cycles236. Despite their influence on several key signaling pathways and the longstanding use of chemotherapeutic antifolates, little literature exists detailing changes to folate metabolism after ionizing radiation in cancer cell models. Future studies investigating changes in folate metabolism after ionizing radiation, especially with respect to nucleotide synthesis and methionine metabolism, may be beneficial.
F. Nucleotide metabolism
Since ionizing radiation primarily affects cells through oxidative damage to DNA, the demand for de novo nucleotide synthesis to aid in DNA repair is high in irradiated cells. Independently of ionizing radiation, proteins involved in the metabolism of nucleic acids are frequently altered in cancer cells237,238, and are often predictive of cancer prognosis239–241. Ionizing radiation has been shown to differentially upregulate enzymes involved in nucleotide synthesis in some cell lines but not others242–244, with a greater change in expression observed in less radiosensitive cell lines.
De novo nucleotide synthesis is a 10-step process catalyzed by six enzymes245, taking inputs from ribose 5-phosphate produced by the pentose phosphate pathway (Figure 2), glycine, glutamine, and aspartate, with a rate-limiting step catalyzed by ribonucleotide reductase246, a homologue of which is a downstream target of p53247,248. Likewise, contributions from other metabolic pathways to nucleotide synthesis are altered by ionizing radiation: the rate-limiting enzyme of the pentose phosphate pathway, glucose-6 phosphate dehydrogenase (G6PD), is a downstream target of the key DNA repair kinase ATM249,250, and alterations to glutamine metabolism are discussed elsewhere in this review. Interestingly, radiosensitization of nasopharyngeal carcinoma cells by inhibition of glutamine synthetase showed that this radiosensitization was dependent on the activity of de novo pyrimidine and purine synthesis, with glutamine synthetase-knockdown cells demonstrating significantly lower levels of homologous recombination, implying that a significant portion of nucleotide synthesis in irradiated cells is dependent on glutamine metabolism211.
Similarly, nucleotides can be generated via salvage pathways, which add activated ribose-5 phosphate to bases to form nucleoside monophosphates. In particular, given the high need for NAD+ in cancer cells251, the key salvaging enzyme nicotinate phosphoribosyltransferase (NAPRT) is widely upregulated in cancer cells252. While nucleic acid salvage pathways also require many of the same intermediates as the synthesis pathway discussed above, membrane transporters can import hypoxanthine, guanine, and adenine, which are capable of being converted to inosine-, guanosine-, and adenosine 5’-monophosphate. Such nucleotide salvage pathways are likely upregulated by ionizing radiation; for example, deoxycytidine kinase, which is responsible for phosphorylation of deoxyadenosine, deoxyguanosine, and deoxycytidine, is upregulated by the master DNA damage regulator ATM253,254. Inhibition of de novo nucleotide synthesis forces cancer cells to rely on these nucleotide salvage pathways, increasing the efficiency of radiolabeled nucleotide analogues255,256, implying that both de novo nucleotide synthesis and scavenging may play an important role in cancer cell nucleotide metabolism after radiation-induced DNA damage. Recent studies have demonstrated that purine, but not pyrimidine levels correlate inversely with radiosensitivity, as inhibition of guanylate synthesis radiosensitized glioblastoma cells and addition of exogenous purines rescued this effect257. While little is known about the difference between purine and pyrimidine metabolism after irradiation, prior studies have shown that decreased proliferation of nucleotide-depleted cells can be rescued by addition of exogenous thymine258, implying that differences in purine and pyrimidine metabolism may be contextually relevant. Additionally, investigation of expression of enzymes involved in nucleotide synthesis after ionizing radiation showed increases in pyrimidine synthetic pathways, but not purine pathways242. While concentrations of nucleotides broadly affect the synthesis of the other nucleobases259, a specific need for purine synthesis is consistent with observations that purine and GTP levels predict aggressiveness of glioblastoma260,261. As DNA repair, requires contributions from both purine and pyrimidine pools, especially through homologous recombination, the disparate need for purine synthesis may point at a significant need for GTP in irradiated cells, though this mechanism requires further study257.
G. Fatty acids/lipids
While the prominence of the Warburg effect has led to considerable study of sugar metabolism in cancer cells, lipid metabolism is also significantly altered262, with upregulation of lipogenic pathways predictive of increased cancer incidence263–265 and increased proliferation265,266, seemingly in an AKT-mTOR-dependent manner267. In addition to rewiring their own lipid metabolism, cancer cells can also scavenge lipids from surrounding cells268,269. Accordingly, irradiation of prostate cancer induces detectable, significant, and durable changes in several fatty acids in the plasma and stool of patients given pelvic radiation therapy, with further studies planned to investigate the prognostic potential of changes to short-chain fatty acids in these samples85.
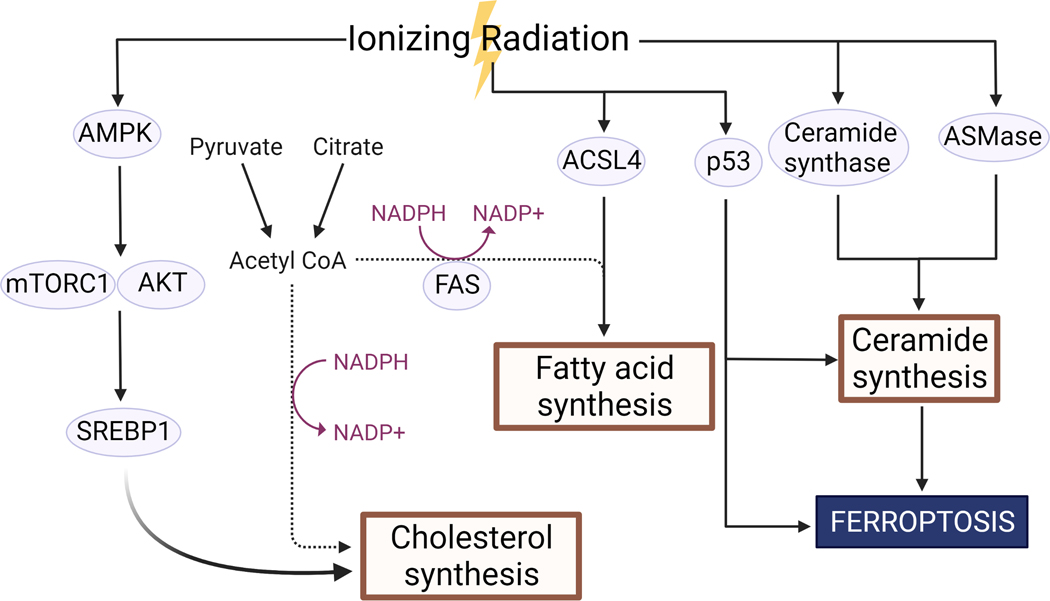
Inhibition of stearoyl-CoA desaturase 1 (SCD1), the rate-limiting enzyme in the formation of monounsaturated fatty acids, decreased clonogenic survival after ionizing radiation in hepatocellular carcinoma cell culture, while overexpression decreased genomic instability as measured by comet assay270. Additionally, radiosensitivity of hepatocellular carcinoma can be decreased by addition of monounsaturated fatty acids270, and radiosensitivity of colorectal cancer cell culture and xenografts can be diminished by exogenous addition of cholesterol271. Ionizing radiation was found to alter expression of several key regulators of lipogenesis, upregulating AMPK, an upstream regulator of the key mediator of cholesterol synthesis SREBP1 for 6–24h after irradiation (Figure 5). This alteration was accompanied by an increase in cholesterol and decrease in triacylglycerol levels271. Interestingly, inhibition of stress-responsive cholesterol synthesis in glioblastoma after combination therapy with ionizing radiation and dopamine receptor antagonists did not significantly increase radiosensitivity in cell culture, but did significantly improve median survival and reduce proliferation in murine xenograft models272. Ionizing radiation has also been shown to upregulate the long-chain-fatty-acid CoA ligase ACSL4 (Figure 5), which is responsible for esterification of CoA onto fatty acids273, with ASCL4 inhibition decreasing cancer cell ferroptosis after radiation274.
Figure 5. Ionizing radiation affects fatty acid and lipid metabolism.
Ionizing radiation affects several pathways associated with the synthesis of cholesterol, fatty acids, and ceramides. Created with BioRender.com.
While the molecular mechanisms governing alterations to lipid metabolism after ionizing radiation are unclear, possible mechanisms include interplay between monounsaturated fatty acid levels, AKT phosphorylation275, and p53 activity276,277. Moreover, despite the utility of lipids as a reservoir for the production of ATP, CoA, and NADPH in cancer metabolism, comparably little is known about the alterations to redox metabolism following radiation, especially in light of Akt-dependent acetyl-CoA production being utilized in de novo production of fatty acids in cancer262,278,279. Accordingly, inhibition of fatty acid synthesis in matched lines of different radiosensitivity in vivo280 showed that the less radiosensitive cells had an increased sensitivity to PPP inhibitors, and elevated NADPH levels, and displayed greater radiosensitivity following inhibition of fatty acid synthase (FASN)281.
In addition to its well-documented effects on nucleotide backbones, ionizing radiation has pronounced effects on both lipid backbones282,283 and on lipogenesis in normal cells284,285, while studies in cancer cells are lacking. Typically measured using proton magnetic resonance spectroscopy (1H-MRS), studies of lipogenesis in irradiated cancer cells have found an increase in production of both saturated and unsaturated lipid levels after ionizing radiation286,287. Cervical cancer biopsies from patients undergoing radiotherapy showed increases in lipid and fatty acid -CH2 levels predictive of apoptosis in irradiated cells, while changes to creatine, taurine, glucose, or lactate metabolism were not individually associated with apoptosis286, consistent with prior observations of changes in lipid levels in necrotic cancer tissue287.
Specifically, considerable research has focused on sphingomyelinase (ASMase)-mediated hydrolysis of sphingomyelin to form ceramide after ionizing radiation288, which induces a translocation of ASMase from the lysosomes to the plasma membrane289,290, followed by increased activity of ceramide synthase291 (Figure 5), resulting in significant increases to ceramide concentrations after ionizing radiation and a consequent increase in ceramide-derived lipid rafts290. Accordingly, ceramide has emerged as a useful biomarker of irradiated cells288,292,293, with ceramide accumulation shown to result in G0/G1 cell cycle arrest294,295 and apoptosis296–298 via permeabilization of the mitochondrial outer membrane299 after ionizing radiation. Interestingly, p53 appears necessary for ceramide accumulation after ionizing radiation in cells with intact p53 independent of ASMase activity300. Both mitochondrial apoptosis and de novo synthesis of ceramide are dependent on Bcl-2 in p53-positive cells301, but ceramide-derived growth suppression appears to be p53-independent302.
Recent studies have also investigated the potential for radiation-induced oxidation of lipids to stimulate ferroptosis (Figure 5), an erastin and RSL3-dependent form of cell death independent of canonical apoptotic or necrotic pathways triggered by lipid peroxidation274,303,304. Accordingly, a decrease in available redox scavengers such as glutathione radiosensitize cells in a ferroptosis-dependent manner305,306, as does inhibition of SLC7A11 (xCT), a glutamate-cystine antiporter, in line with cysteine’s role as a scavenger and necessity for glutathione synthesis274,307–309. Similarly, de novo lipogenesis suppresses ferroptosis310.
H. Radiation dose fractionation and metabolic response
Clinical radiotherapy is usually delivered across fractionated doses as opposed to a single high dose. This approach provides several clinical benefits, including improved radiation response of formerly hypoxic and reoxygenated tumor regions and facilitated repair of sublethal DNA damage in surrounding normal tissues35,311. While many different fractionation plans exist clinically, the metabolic response of tumor cells irradiated in fractionated doses may be significantly different from the metabolic responses of cells given single high doses, which may therefore offer new opportunities for interventions directed at tumor metabolism during a course of radiation therapy. Numerous studies have documented the effects of fractionated radiotherapy on hypoxia and repopulation in the tumor microenvironment312–315, and the metabolic effects of those stimuli have been thoroughly discussed elsewhere316,317.
Notably, gene expression patterns in cells exposed to fractionated radiation can differ significantly from responses in cells given single equivalent doses, with significant changes in timing and magnitude of responses. For example, irradiated 3-dimensional prostate cancer cultures upregulated AKT within 2 hours of a multifractionated dose regimen, but did not show the same response following an equivalent single dose, resulting in enhanced sensitivity to an AKT inhibitor only when administered after fractionated radiation318. These changes occur rapidly after fractionated radiotherapy, and altered signaling in integrin expression has been shown to continue for up to a month after fractionated radiotherapy, implying sustained changes in signaling after fractionated radiotherapy319,320. Biological context also informs response to fractionated radiotherapy, with prostate cancer xenografts showing significantly different activation of several important signaling pathways (e.g. interferon-related genes, STAT1) to fractionated radiation than the same cell line cultured in vitro321.
Since radiotherapy is typically delievered in fractionated regimens, it is also important to note that the size and frequency of fractions can be informed by metabolic response as measured by the metabolic imaging methods discussed above. For example, recent clinical trials (NCT01507428, NCT01576796, NCT02473133) track the effects of modifying fractionation patterns to introduce more fractions to NSCLC patients whose tumors do not show metabolic improvement as measured by mid-treatment PET imaging322–324. Given the clinical benefits of fractionated radiotherapy and the differential dynamics of metabolic response over time, future radiation therapy studies of cancer cell metabolism and stress responses after ionizing radiation should investigate the responses to fractionated radiation.
I. Effects on normal cells
Despite recent technical and technological advances in clinical radiation therapy, normal tissue toxicity remains a significant dose-limiting factor325. To this end, several studies have aimed to identify easily-assessable metabolic markers of normal tissue toxicity after ionizing radiation, either at a systemic level by measuring changes in blood326,327 or urine328 metabolomes, or in specific organs of interest329. These studies in both animal models328,330 and humans327, aside from their value in predicting normal tissue toxicity, have revealed interesting differences in metabolic response to radiation between normal and cancerous tissue, resulting in a series of metabolite panels useful for uniquely predicting radiation-induced normal tissue damage331–333.
Normal tissue irradiation produces characteristic metabolite profiles that can serve as useful predictors of damage to normal tissue or of associated inflammation in multiple models326, including total body irradiation334,335, lung cancer336, head and neck cancer337, and glioblastoma338. Studies of metabolic changes induced in normal tissues have typically focused on some of the metabolites also upregulated in cancer cells after ionizing radiation, including phospholipids and triglycerides339,340, nucleic acids341,342, and precursors to nucleotide synthesis333. Modification of metabolic pathways in normal cells can be highly dependent on radiation quality343, dose rate344,345, and timing346,347, with significant alterations in pyruvate metabolism and nucleic acid synthesis precursors detected in cells exposed to doses as low as 10 cGy347 and increasing in proportion to registered dose344,348.
Additionally, several studies have investigated radiotoxicity to tissues in the field of irradiated tumors. For example, cardiomyocyte metabolism is primarily mitochondrial349 and cardiac tissue is often damaged by thoracic radiation therapy350. Rat hearts treated with a single dose of 2 Gy showed dysregulation of several metabolic proteins isolated from mitochondrial fractions, including proteins involved in the electron transport chain, glycolysis, and lipid metabolism351. Interestingly, murine cardiac models of high-dose radiation have showed downregulation and decreased activity of multiple parts of the electron transport chain, paired with increased activity of pyruvate dehydrogenase E1α352,353. These data contrast with observations from irradiated cancer cells, which show a HIF-1α-dependent upregulation of mitochondrial complex proteins after comparable doses of X-rays or gamma irradiation182,354, highlighting the differential impact of metabolic reprogramming in cancerous tissue.
Section 3: Changes to the tumor environment
A. Tumor microenvironment
Irradiation of a solid tumor inevitably affects all the cells in the tumor microenvironment (TME)355, but the metabolic response of TME cells to radiation is largely unknown. Some evidence suggest that radiation therapy will affect stromal cells, such as cancer-associated fibroblasts (CAFs), so as to promote a metabolic re-wiring and increased glutamine consumption in cancer cells in a paracrine manner356. Radiation could also potentially have direct effects on the metabolism of cells in the tumor microenvironment, or their metabolic phenotype could be indirectly affected by the metabolic changes of the cancer cells induced by ionizing radiation. For example, as discussed above, ionizing radiation can induce an increase in lactate production and secretion109,110,357,358. The sudden increase in lactate concentration following ionizing radiation in the microenvironment is likely to impact the surrounding cells. Apart from serving as a fuel source for cancer cells, lactate participates in tumor acidification, which plays a role in local invasion359, metastasis360, therapy resistance361, immune escape362, and angiogenesis363. Intratumoral lactate levels are capable of modulating the count and activity of several immune cell populations in the TME. For example, Treg cells, which suppress effector T cells in the TME139,364,365, preferentially suppress antitumor immunity in low glucose, high lactate conditions139. Interestingly, inhibition of lactate dehydrogenase prevented lactate-mediated decreases in T-cell cytotoxic activity, implying that T-cell lactate metabolism within the tumor microenvironment has a strong effect on antitumor cytotoxicity. High lactate can decrease antitumor T-cell cytotoxic activity up to 50% in a spheroid model, with cytotoxic activity restored 24 hours after removal of the exogenous lactate140. Similarly, high lactate decreases rates of monocyte migration and macrophages polarize towards an immunosuppressive M2-like phenotype in tumors with high lactate141. More recent evidence has suggested that MCT1-mediated lactate export is required for Treg function intratumorally, but not in peripheral Treg cells139. Similarly, inhibition of lactate transport in intratumoral cytotoxic T lymphocytes decreased T-cell cytotoxicity in a tumor spheroid model140, in agreement with the above observations that high concentrations of exogenous lactate are sufficient to enhance the immunosuppressive activity of the Treg population. Increased lactate secretion after radiation therapy can also affect myeloid-derived suppressor cells (MDSCs) in pancreatic cancer358. In this study, the authors demonstrate that radiation-induced lactate secretion in pancreatic cancer cells promotes the activation of immunosuppressive, pro-tumoral MDSCs via induction of HIF1358.
Cancer-associated fibroblasts (CAFs) are another important cell population found in the TME, and are known to contribute to tumor growth and metastasis366, and radiotherapy is one of many factors shown to activate CAFs356,367–369. Interestingly, radiation-activated CAFs have been shown to induce metabolic reprogramming in colorectal cancer cells356. By using conditioned media from irradiated CAFs, the authors demonstrated that radiation-activated CAFs induce an increase in glucose metabolism, lactate release, and glutamine consumption, while the expression of many metabolic genes was enhanced, including xCT responsible for glutamine transport. These changes happened in a paracrine manner through IGF1 secretion by CAFs356. Although this study did not investigate radiation-induced metabolic changes in the CAFs themselves, there is evidence that CAFs also undergo metabolic reprogramming to provide the cancer cells with additional sources of carbon370, such as glutamine371 or alanine372. As radiation therapy activates CAFs356,367–369, it is reasonable to postulate that ionizing radiation can induce metabolic reprogramming in activated fibroblasts that will further sustain cancer growth. Another study demonstrated that radiation therapy induces high inducible nitric oxide synthase139 expression and increased levels of nitric oxide (NO) secretion in CAFs, which in turn enhanced iNOS/NO signaling in pancreatic cancer cells, contributing to an acidic microenvironment368. Interestingly, NO is involved in metabolic reprogramming by promoting nuclear translocation of PKM2 and macromolecules biosynthesis through accumulation of glycolytic intermediates in ovarian cancer373. Epigenetic changes can also be responsible for metabolic reprogramming in CAFs, via induction of HIF1 and the subsequent expression of glycolytic target genes374. This is of particular interest because, as we will discuss below, radiation can induce HIF1 expression in cancer cells375,376.
B. Role of hypoxia
Hypoxia is an indisputable player in radiation response377, due to the direct effects of molecular oxygen in facilitating oxidative damage after ionizing radiation34 and the hypoxia-induced shift in cell metabolism, resulting in suppression of the oxygen-consuming oxidative phosphorylation while enhancing glycolysis378. Under hypoxic conditions, hypoxia-inducible factor (HIF) is activated secondary to both lower oxygen availability379 and mitochondria-generated ROS380. Of note, while HIF-1β is constitutively expressed, the activity of the HIF-1 pathway is largely dependent on the oxygen-reliant modification and stability of HIF-1α, the main effector of cellular response to hypoxia381. Surprisingly, given the importance of hypoxia on radiation sensitivity, studies investigating the effect of ionizing radiation on the HIF1 pathway are sparse, but there is some evidence that ionizing radiation activates the HIF-1 pathway in cancer cells375,376. Part of the mechanism of HIF1 activation by ionizing radiation has been attributed to the ROS generated from tumor re-oxygenation after radiation357. HIF1 is a positive regulator of a number of glycolytic enzymes, including lactate dehydrogenase A (LDHA) and pyruvate dehydrogenase kinase 1 (PDK1)162,382. While PDK1 inhibits pyruvate dehydrogenase (PDH), which forms acetyl-CoA from pyruvate, LDHA generates lactate from pyruvate. Therefore, following HIF1 activation, pyruvate is routed away from the TCA to form lactate. Another important HIF1 target is GLUT1383, through which HIF1 can regulate glucose import. This could explain in part the enhanced glucose consumption observed in irradiated cancer cells109,110,357, although activation of HIF1 pathway by ionizing radiation seems to only occur under hypoxic conditions in vivo357 while the radiation-induced increase in glucose consumption is observed in cancer cells propagated under normoxia in vitro109. Only a few studies have investigated the effect of ionizing radiation on the HIF1 pathway in cancer cells and these are limited to specific cancer cell lines358,375,376. It is likely that the pre-irradiation metabolic state of a cancer cell will dictate its metabolic response after radiation. From the limited studies available it seems that ionizing radiation induced metabolic changes may resemble those of hypoxia109,110,357. It remains to be determined whether the radiation-induced metabolic changes on cancer cells under hypoxic conditions are different from those of normoxic cells.
Section 4: Metabolic radiosensitizers
A recent review has summarized radiosensitizing drugs with potential metabolic mechanisms53 and several radiosensitizers associated with rewired glucose metabolism have been documented. Most notably, 2-deoxyglucose (2-DG) has been shown to radiosensitize prostate, pancreatic, and cultured HeLa cells384,385 via multiple proposed mechanisms, including decreased glutathione levels384 or autophagy induction via AMPK stimulation386. 2-DG treatment in combination with radiation therapy in glioma patients was well tolerated and increased patient survival in a Phase I/II clinical trial387,388, though the relatively short biological half-life and high drug concentrations involved in such studies may be problematic without development of improved therapeutics with similar mechanisms389. Inhibitors of the glucose transporter GLUT1 have also been demonstrated to have potential as radiosensitizers37,390. Other metabolic pathways, including amino acid and fatty acid metabolism, have been targeted for radiosensitization. Notably, the conversion of arginine to citrulline by arginine deiminase-polyethylene glycol (ADI-PEG 20) has been shown to increase radiosensitivity via arginine depletion, especially in cells lacking expression of argininosuccinate synthetase391. Given the heightened requirement for fatty acid synthesis in irradiated cells, inhibition of fatty acid synthase (FASN) has potential as a radiosensitizer. While several FASN inhibitors exist392–394 and have radiosensitized diverse cancer types in preclinical studies281,395–397, none have shown efficacy in vivo, likely due to the ability of cancer cells to scavenge fatty acids from the tumor microenvironment after ionizing radiation397. This suggests that the efficacy of treatments involving FASN inhibition may be significantly improved by co-targeting of fatty acid scavenging. These studies demonstrate the potential of therapeutics aimed at targeting metabolites especially significant to cancer metabolism, especially in the context of metabolic alterations following radiation therapy.
Metformin, frequently implicated in several relevant metabolic pathways in cancer, including AMPK, mTOR, MAPK, and PI3K signaling, has been the subject of considerable recent study in combination with chemo- and radiation therapy. Interestingly, a recent Phase II clinical trial of metformin combined with paclitaxel and carboplatin in non-diabetic NSCLC patients398 found no significant increase in progression-free survival, despite models suggesting metabolic response in the majority of similar tumors399. Patients in this study demonstrated increased glucose uptake in the majority of tumors, despite preclinical evidence suggesting that metformin decreases glucose consumption via inhibition of PKM2400 and demonstrated utility of metformin as a monotherapy both in vitro and in vivo401. Several mechanistic explanations for metformin’s inability to increase overall survival in this trial are plausible. Metformin’s antineoplastic activity may be more important than its effects on cytotoxicity, or the cytotoxic effects of metformin may be mitigated by altered local immune function after ionizing radiation402. Further study of metabolic flux alterations after metformin treatment, especially in in vivo models, may help to elucidate the components of the whole-organism response. Such understanding may enable combination therapies directed at the metabolic context of chemotherapy-treated tumors and tumor-infiltrating lymphocytes after radiation.
Since cellular metabolism is a significant consumer of oxygen and local oxygen is required for the cytotoxic effects of radiation, several metabolic radiosensitizers have functioned by decreasing the size of the hypoxic core of in vivo tumors38,39,403. Radiosensitization brought on by these radiosensitizers in three-dimensional cultures with differential oxygen tension likely stems in large part from an increased capacity for oxygen-mediated DNA damage in newly-oxygenated tissues. Consistent with the importance of redox biology in cancer cell radiosensitivity, pharmacological alteration of cellular redox pools can significantly alter radiosensitivity. For example, decreased glutathione levels, either through inhibition of glutaminase209,404 or modification of the glutamine-cysteine antiporter xCT309, have been demonstrated to alter radiosensitivity inversely with glutathione concentration. Similarly, thioredoxin reductase inhibitors radiosensitize via ROS accumulation405. While drugs targeting glutaminase have been shown to increase radiosensitivity in vitro, in vivo application has suffered from either poor potency or poor bioavailability406. The glutaminase inhibitor CB-839 increased radiosensitivity in head and neck squamous cell carcinoma xenograft models407 and has been tested in early phase clinical trials of renal cancer alongside the VEGFR inhibitor cabozantinib408 (NCT02071862), demonstrating potential for clinical applications of metabolic radiosensitizers aimed at altering metabolism.
Conclusions
In light of the centrality of of radiation therapy to multidisciplinary cancer care and the rapidly expanding body of work surrounding cancer metabolism, it is critical to gain additional insight both into the capacity for radiotherapy to alter cancer cell metabolism and the ways in which metabolism impacts the efficacy of radiotherapy. In this review, we outline methods capable of tracking relevant alterations to cancer cell metabolism that are compatible with radiobiological studies, including in vitro models and metabolism-directed imaging modalities transferrable to clinical studies. These methods informed numerous studies into radiation-induced alterations to signaling pathways and metabolite concentrations in cancer cells, as well as studies investigating alterations to the tumor microenvironment after ionizing radiation. Studies of cancer metabolism before and after ionizing radiation offer significant prognostic and therapeutic potential.
Acknowledgements
This study was supported by National Cancer Institute CA251872 (to E.V) and CA222493 (to A.H.K). The authors report no conflicts of interest.
Works Cited
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA: A Cancer Journal for Clinicians. 2021;71(1):7–33. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention DoCPaC. An Update on Cancer Deaths in the United States. 2021. [Google Scholar]
- 3.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians. 2021;71(3):209–249. [DOI] [PubMed] [Google Scholar]
- 4.Jang JY, Han Y, Lee H, et al. Oncological Benefits of Neoadjuvant Chemoradiation With Gemcitabine Versus Upfront Surgery in Patients With Borderline Resectable Pancreatic Cancer: A Prospective, Randomized, Open-label, Multicenter Phase 2/3 Trial. Annals of surgery. 2018;268(2):215–222. [DOI] [PubMed] [Google Scholar]
- 5.Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA: A Cancer Journal for Clinicians. 2019;69(5):363–385. [DOI] [PubMed] [Google Scholar]
- 6.Membreno PV, Luttrell JB, Mamidala MP, et al. Outcomes of primary radiotherapy with or without chemotherapy for advanced oral cavity squamous cell carcinoma: A systematic review. Head & neck. 2021. [DOI] [PubMed] [Google Scholar]
- 7.Grinnell M, Appiah AK, Baine M, et al. Adjuvant chemotherapy following SBRT for early stage non-small cell lung cancer (NSCLC) in older patients. Journal of Geriatric Oncology. 2020;11(7):1145–1153. [DOI] [PubMed] [Google Scholar]
- 8.Shipley WU, Seiferheld W, Lukka HR, et al. Radiation with or without Antiandrogen Therapy in Recurrent Prostate Cancer. New England Journal of Medicine. 2017;376(5):417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demaria S, Coleman CN, Formenti SC. Radiotherapy: Changing the Game in Immunotherapy. Trends Cancer. 2016;2(6):286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vatner RE, Cooper BT, Vanpouille-Box C, Demaria S, Formenti SC. Combinations of immunotherapy and radiation in cancer therapy. Frontiers in oncology. 2014;4:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asai A, Konno M, Koseki J, Taniguchi M, Vecchione A, Ishii H. One-carbon metabolism for cancer diagnostic and therapeutic approaches. Cancer letters. 2020;470:141–148. [DOI] [PubMed] [Google Scholar]
- 12.Phan LM, Yeung S-CJ, Lee M-H. Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol Med. 2014;11(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luengo A, Gui DY, Vander Heiden MG. Targeting Metabolism for Cancer Therapy. Cell chemical biology. 2017;24(9):1161–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23(1):27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warburg O. The Metabolism of Carcinoma Cells. The Journal of Cancer Research. 1925;9(1):148. [Google Scholar]
- 16.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. The Journal of general physiology. 1927;8(6):519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 18.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324(5930):1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaupel P, Schmidberger H, Mayer A. The Warburg effect: essential part of metabolic reprogramming and central contributor to cancer progression. International journal of radiation biology. 2019;95(7):912–919. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Le A. Glutamine Metabolism in Cancer. Advances in experimental medicine and biology. 2018;1063:13–32. [DOI] [PubMed] [Google Scholar]
- 21.San-Millán I, Brooks GA. Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis. 2017;38(2):119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Outschoorn UE, Peiris-Pagés M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nature reviews Clinical oncologyHead & neck 2017;14(1):11–31. [DOI] [PubMed] [Google Scholar]
- 23.Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nature reviews Cancer. 2013;13(4):227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koundouros N, Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. British journal of cancer. 2020;122(1):4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauer LA, Stayman JW 3rd, Dauchy RT. Amino acid, glucose, and lactic acid utilization in vivo by rat tumors. Cancer research. 1982;42(10):4090–4097. [PubMed] [Google Scholar]
- 26.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nature reviews Cancer. 2007;7(10):763–777. [DOI] [PubMed] [Google Scholar]
- 27.Swierczynski J, Hebanowska A, Sledzinski T. Role of abnormal lipid metabolism in development, progression, diagnosis and therapy of pancreatic cancer. World journal of gastroenterology. 2014;20(9):2279–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cellular and Molecular Life Sciences. 2016;73(2):377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2(5):e1600200-e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao X, Ren Y, Feng M, Wang Q, Wang Y. Metabolic reprogramming due to hypoxia in pancreatic cancer: Implications for tumor formation, immunity, and more. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2021;141:111798. [DOI] [PubMed] [Google Scholar]
- 31.Payen VL, Brisson L, Dewhirst MW, Sonveaux P. Common responses of tumors and wounds to hypoxia. Cancer journal (Sudbury, Mass). 2015;21(2):75–87. [DOI] [PubMed] [Google Scholar]
- 32.Vidal RS, Quarti J, Rodrigues MF, Rumjanek FD, Rumjanek VM. Metabolic Reprogramming During Multidrug Resistance in Leukemias. Frontiers in oncology. 2018;8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gadéa E, Thivat E, Planchat E, Morio B, Durando X. Importance of metabolic changes induced by chemotherapy on prognosis of early-stage breast cancer patients: a review of potential mechanisms. Obesity reviews : an official journal of the International Association for the Study of Obesity. 2012;13(4):368–380. [DOI] [PubMed] [Google Scholar]
- 34.O’Neill P, Wardman P. Radiation chemistry comes before radiation biology. International journal of radiation biology. 2009;85(1):9–25. [DOI] [PubMed] [Google Scholar]
- 35.Hall EJ, Giaccia AJ. Radiobiology for the Radiologist. Lippincott Williams & Wilkins; 2006. [Google Scholar]
- 36.Gerber DE, Beg MS, Fattah F, et al. Phase 1 study of ARQ 761, a β-lapachone analogue that promotes NQO1-mediated programmed cancer cell necrosis. British journal of cancer. 2018;119(8):928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao F, Ming J, Zhou Y, Fan L. Inhibition of Glut1 by WZB117 sensitizes radioresistant breast cancer cells to irradiation. Cancer chemotherapy and pharmacology. 2016;77(5):963–972. [DOI] [PubMed] [Google Scholar]
- 38.Benej M, Hong X, Vibhute S, et al. Papaverine and its derivatives radiosensitize solid tumors by inhibiting mitochondrial metabolism. Proceedings of the National Academy of Sciences. 2018;115(42):10756–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zannella VE, Dal Pra A, Muaddi H, et al. Reprogramming Metabolism with Metformin Improves Tumor Oxygenation and Radiotherapy Response. Clinical Cancer Research. 2013;19(24):6741. [DOI] [PubMed] [Google Scholar]
- 40.Corbet C, Bastien E, Draoui N, et al. Interruption of lactate uptake by inhibiting mitochondrial pyruvate transport unravels direct antitumor and radiosensitizing effects. Nature Communications. 2018;9(1):1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin X, Xu R, Mao S, et al. Metabolic biomarker signature for predicting the effect of neoadjuvant chemotherapy of breast cancer. Ann Transl Med. 2019;7(22):670–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Issaq HJ, Fox SD, Chan KC, Veenstra TD. Global proteomics and metabolomics in cancer biomarker discovery. Journal of separation science. 2011;34(24):3484–3492. [DOI] [PubMed] [Google Scholar]
- 43.Zuo D, Li C, Liu T, Yue M, Zhang J, Ning G. Construction and validation of a metabolic risk model predicting prognosis of colon cancer. Scientific Reports. 2021;11(1):6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sreekumar A, Poisson LM, Rajendiran TM, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457(7231):910–914. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 45.Hakimi AA, Reznik E, Lee CH, et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer cell. 2016;29(1):104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng X, Chen Z, Farshidfar F, et al. Molecular Characterization and Clinical Relevance of Metabolic Expression Subtypes in Human Cancers. Cell Rep. 2018;23(1):255–269.e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai D, Duan X, Wang W, et al. A Metabolism-Related Radiomics Signature for Predicting the Prognosis of Colorectal Cancer. Frontiers in Molecular Biosciences. 2021;7(458). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cardoso MR, Santos JC, Ribeiro ML, Talarico MCR, Viana LR, Derchain SFM. A Metabolomic Approach to Predict Breast Cancer Behavior and Chemotherapy Response. Int J Mol Sci. 2018;19(2):617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lau LF, Williams DS, Lee ST, Scott AM, Christophi C, Muralidharan V. Metabolic response to preoperative chemotherapy predicts prognosis for patients undergoing surgical resection of colorectal cancer metastatic to the liver. Annals of surgical oncology. 2014;21(7):2420–2428. [DOI] [PubMed] [Google Scholar]
- 50.Day SE, Kettunen MI, Gallagher FA, et al. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nature medicine. 2007;13(11):1382–1387. [DOI] [PubMed] [Google Scholar]
- 51.Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nature chemical biology. 2015;11(1):9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang L, Wei F, Wu Y, et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. Journal of Experimental & Clinical Cancer Research. 2018;37(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Gisbergen MW, Zwilling E, Dubois LJ. Metabolic Rewiring in Radiation Oncology Toward Improving the Therapeutic Ratio. Frontiers in oncology. 2021;11:653621–653621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai SY, Fuller CD, Bhattacharya PK, Frank SJ. Metabolic Imaging as a Biomarker of Early Radiation Response in Tumors. Clinical Cancer Research. 2015;21(22):4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saito K, Matsumoto S, Takakusagi Y, et al. 13C-MR Spectroscopic Imaging with Hyperpolarized [1–13C]pyruvate Detects Early Response to Radiotherapy in SCC Tumors and HT-29 Tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21(22):5073–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu M, Neilson A, Swift AL, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. American journal of physiology Cell physiology. 2007;292(1):C125–136. [DOI] [PubMed] [Google Scholar]
- 57.Olivares A, Alcaraz-Saura M, Achel DG, Alcaraz M. Effect of Rosmarinic Acid and Ionizing Radiation on Glutathione in Melanoma B16F10 Cells: A Translational Opportunity. Antioxidants (Basel). 2020;9(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ren J-L, Zhang A-H, Kong L, Wang X-J. Advances in mass spectrometry-based metabolomics for investigation of metabolites. RSC Advances. 2018;8(40):22335–22350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zamboni N, Saghatelian A, Patti GJ. Defining the metabolome: size, flux, and regulation. Mol Cell. 2015;58(4):699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jang C, Chen L, Rabinowitz JD. Metabolomics and Isotope Tracing. Cell. 2018;173(4):822–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bednarski TK, Rahim M, Young JD. In vivo(2)H/(13)C flux analysis in metabolism research. Curr Opin Biotechnol. 2021;71:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X-w Li Q-h, Xu Z-d Dou J-j. Mass spectrometry-based metabolomics in health and medical science: a systematic review. RSC Advances. 2020;10(6):3092–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao J, Balluff B, Arts M, et al. Mass spectrometry imaging of L-[ring-(13)C(6)]-labeled phenylalanine and tyrosine kinetics in non-small cell lung carcinoma. Cancer Metab. 2021;9(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aichler M, Walch A . MALDI Imaging mass spectrometry: current frontiers and perspectives in pathology research and practice. Laboratory investigation; a journal of technical methods and pathology. 2015;95(4):422–431. [DOI] [PubMed] [Google Scholar]
- 65.Buchberger AR, DeLaney K, Johnson J, Li L. Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Anal Chem. 2018;90(1):240–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kohler SJ, Yen Y, Wolber J, et al. In vivo 13 carbon metabolic imaging at 3T with hyperpolarized 13C-1-pyruvate. Magnetic resonance in medicine. 2007;58(1):65–69. [DOI] [PubMed] [Google Scholar]
- 67.Abragam A, Goldman M. Principles of dynamic nuclear polarisation. Reports on Progress in Physics. 1978;41(3):395–467. [Google Scholar]
- 68.Ardenkjær-Larsen JH, Fridlund B, Gram A, et al. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proceedings of the National Academy of Sciences. 2003;100(18):10158–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Golman K, Zandt Ri, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic Imaging by Hyperpolarized <sup>13</sup>C Magnetic Resonance Imaging for <em>In vivo</em> Tumor Diagnosis. Cancer research. 2006;66(22):10855. [DOI] [PubMed] [Google Scholar]
- 70.Nelson SJ, Kurhanewicz J, Vigneron DB, et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-13C]pyruvate. Science translational medicine. 2013;5(198):198ra108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miura N, Mushti C, Sail D, et al. Synthesis of [1–13C-5–12C]-alpha-ketoglutarate enables noninvasive detection of 2-hydroxyglutarate. NMR in Biomedicine. 2021;n/a(n/a):e4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chaumeil MM, Larson PEZ, Yoshihara HAI, et al. Non-invasive in vivo assessment of IDH1 mutational status in glioma. Nature communications. 2013;4:2429–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bastiaansen JA, Cheng T, Mishkovsky M, Duarte JM, Comment A, Gruetter R. In vivo enzymatic activity of acetylCoA synthetase in skeletal muscle revealed by (13)C turnover from hyperpolarized [1-(13)C]acetate to [1-(13)C]acetylcarnitine. Biochimica et biophysica acta. 2013;1830(8):4171–4178. [DOI] [PubMed] [Google Scholar]
- 74.Najac C, Chaumeil MM, Kohanbash G, et al. Detection of inflammatory cell function using (13)C magnetic resonance spectroscopy of hyperpolarized [6-(13)C]-arginine. Sci Rep. 2016;6:31397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kurhanewicz J, Vigneron DB, Ardenkjaer-Larsen JH, et al. Hyperpolarized (13)C MRI: Path to Clinical Translation in Oncology. Neoplasia. 2019;21(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salamanca-Cardona L, Keshari KR. (13)C-labeled biochemical probes for the study of cancer metabolism with dynamic nuclear polarization-enhanced magnetic resonance imaging. Cancer Metab. 2015;3:9–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keshari KR, Kurhanewicz J, Jeffries RE, et al. Hyperpolarized (13)C spectroscopy and an NMR-compatible bioreactor system for the investigation of real-time cellular metabolism. Magnetic resonance in medicine. 2010;63(2):322–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sriram R, Van Criekinge M, DeLos Santos J, et al. Elevated Tumor Lactate and Efflux in High-grade Prostate Cancer demonstrated by Hyperpolarized (13)C Magnetic Resonance Spectroscopy of Prostate Tissue Slice Cultures. Cancers. 2020;12(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Read GH, Miura N, Carter JL, et al. Three-dimensional alginate hydrogels for radiobiological and metabolic studies of cancer cells. Colloids and surfaces B, Biointerfaces. 2018;171:197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen AP, Chu W, Gu Y-P, Cunningham CH. Probing early tumor response to radiation therapy using hyperpolarized [1-13C]pyruvate in MDA-MB-231 xenografts. PLoS One. 2013;8(2):e56551-e56551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chaumeil MM, Larson PE, Yoshihara HA, et al. Non-invasive in vivo assessment of IDH1 mutational status in glioma. Nat Commun. 2013;4:2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cai K, Tain R-W, Zhou XJ, et al. Creatine CEST MRI for Differentiating Gliomas with Different Degrees of Aggressiveness. Mol Imaging Biol. 2017;19(2):225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Monteiro MS, Barros AS, Pinto J, et al. Nuclear Magnetic Resonance metabolomics reveals an excretory metabolic signature of renal cell carcinoma. Scientific Reports. 2016;6(1):37275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Emwas A-H, Roy R, McKay RT, et al. NMR Spectroscopy for Metabolomics Research. Metabolites. 2019;9(7):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ferreira MR, Sands CJ, Li JV, et al. Impact of pelvic radiotherapy for prostate cancer on global metabolic profiles and microbiota-driven gastrointestinal late side-effects: a longitudinal observational study. International Journal of Radiation Oncology, Biology, Physics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ogawa S, Lee TM, Kay AR, Tank DW. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc Natl Acad Sci U S A. 1990;87(24):9868–9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Read SJ, Hirano T, Abbott DF, et al. Identifying hypoxic tissue after acute ischemic stroke using PET and 18F-fluoromisonidazole. Neurology. 1998;51(6):1617–1621. [DOI] [PubMed] [Google Scholar]
- 88.Ahmad R, Kuppusamy P. Theory, instrumentation, and applications of electron paramagnetic resonance oximetry. Chemical reviews. 2010;110(5):3212–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Matsuo M, Matsumoto S, Mitchell JB, Krishna MC, Camphausen K. Magnetic resonance imaging of the tumor microenvironment in radiotherapy: perfusion, hypoxia, and metabolism. Semin Radiat Oncol. 2014;24(3):210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oriuchi N, Higuchi T, Ishikita T, et al. Present role and future prospects of positron emission tomography in clinical oncology. Cancer Sci. 2006;97(12):1291–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu L, Ploessl K, Zhou R, Mankoff D, Kung HF. Metabolic Imaging of Glutamine in Cancer. J Nucl Med. 2017;58(4):533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deford-Watts LM, Mintz A, Kridel SJ. The potential of 11C-acetate PET for monitoring the Fatty acid synthesis pathway in Tumors. Curr Pharm Biotechnol. 2013;14(3):300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sertic M, Kilcoyne A, Catalano OA, Lee SI. Quantitative imaging of uterine cancers with diffusion-weighted MRI and 18-fluorodeoxyglucose PET/CT. Abdominal radiology (New York). 2021. [DOI] [PubMed] [Google Scholar]
- 94.Hirata K, Tamaki N. Quantitative FDG PET Assessment for Oncology Therapy. Cancers. 2021;13(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Keller H, Goda JS, Vines DC, Lockwood G, Tsang R. Quantification of Local Tumor Response to Fractionated Radiation Therapy for Non-Hodgkin Lymphoma Using Weekly 18F-FDG PET/CT Imaging. International Journal of Radiation Oncology*Biology*Physics. 2010;76(3):850–858. [DOI] [PubMed] [Google Scholar]
- 96.Phillips EH, Iype R, Wirth A. PET-guided treatment for personalised therapy of Hodgkin lymphoma and aggressive non-Hodgkin lymphoma. The British journal of radiology. 2021:20210576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sachpekidis C, Kopp-Schneider A, Hassel JC, Dimitrakopoulou-Strauss A. Assessment of early metabolic progression in melanoma patients under immunotherapy: an (18)F-FDG PET/CT study. EJNMMI research. 2021;11(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang D, Liu X, Wang W, et al. The Role of the Metabolic Parameters of (18)F-FDG PET/CT in Patients With Locally Advanced Cervical Cancer. Frontiers in oncology. 2021;11:698744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12(12):931–947. [DOI] [PubMed] [Google Scholar]
- 100.Miguel F, Augusto AC, Gurgueira SA. Effect of acute vs chronic H2O2-induced oxidative stress on antioxidant enzyme activities. Free Radic Res. 2009;43(4):340–347. [DOI] [PubMed] [Google Scholar]
- 101.Kuwahara Y, Tomita K, Roudkenar MH, et al. The Effects of Hydrogen Peroxide and/or Radiation on the Survival of Clinically Relevant Radioresistant Cells. Technol Cancer Res Treat. 2020;19:1533033820980077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dahm-Daphi J, Sass C, Alberti W. Comparison of biological effects of DNA damage induced by ionizing radiation and hydrogen peroxide in CHO cells. International journal of radiation biology. 2000;76(1):67–75. [DOI] [PubMed] [Google Scholar]
- 103.Ransy C, Vaz C, Lombes A, Bouillaud F. Use of H2O2 to Cause Oxidative Stress, the Catalase Issue. Int J Mol Sci. 2020;21(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Diehn M, Cho RW, Lobo NA, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458(7239):780–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lewis JE, Forshaw TE, Boothman DA, Furdui CM, Kemp ML. Personalized Genome-Scale Metabolic Models Identify Targets of Redox Metabolism in Radiation-Resistant Tumors. Cell Syst. 2021;12(1):68–81.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lewis JE, Kemp ML. Integration of machine learning and genome-scale metabolic modeling identifies multi-omics biomarkers for radiation resistance. Nat Commun. 2021;12(1):2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lyssiotis CA, Kimmelman AC. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017;27(11):863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhou W, Wahl DR. Metabolic Abnormalities in Glioblastoma and Metabolic Strategies to Overcome Treatment Resistance. Cancers. 2019;11(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang L, Bailleul J, Yazal T, et al. PK-M2-mediated metabolic changes in breast cancer cells induced by ionizing radiation. Breast Cancer Res Treat. 2019;178(1):75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao CB, Shi L, Pu HH, Zhang QY. The Promoting Effect of Radiation on Glucose Metabolism in Breast Cancer Cells under the Treatment of Cobalt Chloride. Pathol Oncol Res. 2017;23(1):47–53. [DOI] [PubMed] [Google Scholar]
- 111.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annual review of cell and developmental biology. 2011;27:441–464. [DOI] [PubMed] [Google Scholar]
- 112.Huang XQ, Chen X, Xie XX, et al. Co-expression of CD147 and GLUT-1 indicates radiation resistance and poor prognosis in cervical squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7(4):1651–1666. [PMC free article] [PubMed] [Google Scholar]
- 113.Rao Y, Gammon S, Zacharias NM, et al. Hyperpolarized [1-(13)C]pyruvate-to-[1-(13)C]lactate conversion is rate-limited by monocarboxylate transporter-1 in the plasma membrane. Proc Natl Acad Sci U S A. 2020;117(36):22378–22389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang J, Zhang Y, Mo F, et al. The Roles of HIF-1alpha in Radiosensitivity and Radiation-Induced Bystander Effects Under Hypoxia. Front Cell Dev Biol. 2021;9:637454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lall R, Ganapathy S, Yang M, et al. Low-dose radiation exposure induces a HIF-1-mediated adaptive and protective metabolic response. Cell Death & Differentiation. 2014;21(5):836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Anastasiou D, Poulogiannis G, Asara JM, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334(6060):1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Anastasiou D, Yu Y, Israelsen WJ, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nature chemical biology. 2012;8(10):839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vander Heiden MG, Locasale JW, Swanson KD, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329(5998):1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moore HM, Bai B, Boisvert FM, et al. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Molecular & cellular proteomics : MCP. 2011;10(10):M111.009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.De Santis MC, Porporato PE, Martini M, Morandi A. Signaling Pathways Regulating Redox Balance in Cancer Metabolism. Frontiers in oncology. 2018;8:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tedeschi PM, Markert EK, Gounder M, et al. Contribution of serine, folate and glycine metabolism to the ATP, NADPH and purine requirements of cancer cells. Cell Death Dis. 2013;4:e877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Murray DM R; McBride WH Defenses against Pro-oxidant Forces - Maintenance of Cellular and Genomic Integrity and Longevity. Radiation Research. 2018;190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chaneton B, Gottlieb E. Rocking cell metabolism: revised functions of the key glycolytic regulator PKM2 in cancer. Trends in biochemical sciences. 2012;37(8):309–316. [DOI] [PubMed] [Google Scholar]
- 124.Tamada M, Suematsu M, Saya H. Pyruvate kinase M2: multiple faces for conferring benefits on cancer cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(20):5554–5561. [DOI] [PubMed] [Google Scholar]
- 125.Kobayashi M, Yamamoto M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxidants & redox signaling. 2005;7(3–4):385–394. [DOI] [PubMed] [Google Scholar]
- 126.Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nature reviews Cancer. 2012;12(8):564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McDonald JT, Kim K, Norris AJ, et al. Ionizing radiation activates the Nrf2 antioxidant response. Cancer research. 2010;70(21):8886–8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mirzayans R, Severin D, Murray D. Relationship between DNA double-strand break rejoining and cell survival after exposure to ionizing radiation in human fibroblast strains with differing ATM/p53 status: implications for evaluation of clinical radiosensitivity. Int J Radiat Oncol Biol Phys. 2006;66(5):1498–1505. [DOI] [PubMed] [Google Scholar]
- 129.Pandita TK, Lieberman HB, Lim DS, et al. Ionizing radiation activates the ATM kinase throughout the cell cycle. Oncogene. 2000;19(11):1386–1391. [DOI] [PubMed] [Google Scholar]
- 130.Golding SE, Rosenberg E, Khalil A, et al. Double strand break repair by homologous recombination is regulated by cell cycle-independent signaling via ATM in human glioma cells. J Biol Chem. 2004;279(15):15402–15410. [DOI] [PubMed] [Google Scholar]
- 131.Dahl ES, Aird KM. Ataxia-Telangiectasia Mutated Modulation of Carbon Metabolism in Cancer. Frontiers in oncology. 2017;7:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Feron O. Pyruvate into lactate and back: from the Warburg effect to symbiotic energy fuel exchange in cancer cells. Radiother Oncol. 2009;92(3):329–333. [DOI] [PubMed] [Google Scholar]
- 133.Philp A, Macdonald AL, Watt PW. Lactate--a signal coordinating cell and systemic function. The Journal of experimental biology. 2005;208(Pt 24):4561–4575. [DOI] [PubMed] [Google Scholar]
- 134.Sola-Penna M. Metabolic regulation by lactate. IUBMB Life. 2008;60(9):605–608. [DOI] [PubMed] [Google Scholar]
- 135.Costa Leite T, Da Silva D, Guimarães Coelho R, Zancan P, Sola-Penna M. Lactate favours the dissociation of skeletal muscle 6-phosphofructo-1-kinase tetramers down-regulating the enzyme and muscle glycolysis. Biochem J. 2007;408(1):123–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dietl K, Renner K, Dettmer K, et al. Lactic acid and acidification inhibit TNF secretion and glycolysis of human monocytes. Journal of immunology (Baltimore, Md : 1950). 2010;184(3):1200–1209. [DOI] [PubMed] [Google Scholar]
- 137.Liu C, Wu J, Zhu J, et al. Lactate inhibits lipolysis in fat cells through activation of an orphan G-protein-coupled receptor, GPR81. J Biol Chem. 2009;284(5):2811–2822. [DOI] [PubMed] [Google Scholar]
- 138.Roland CL, Arumugam T, Deng D, et al. Cell surface lactate receptor GPR81 is crucial for cancer cell survival. Cancer research. 2014;74(18):5301–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Watson MJ, Vignali PDA, Mullett SJ, et al. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature. 2021;591(7851):645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fischer K, Hoffmann P, Voelkl S, et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. 2007;109(9):3812–3819. [DOI] [PubMed] [Google Scholar]
- 141.Ohashi T, Aoki M, Tomita H, et al. M2-like macrophage polarization in high lactic acid-producing head and neck cancer. Cancer science. 2017;108(6):1128–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sattler UG, Meyer SS, Quennet V, et al. Glycolytic metabolism and tumour response to fractionated irradiation. Radiother Oncol. 2010;94(1):102–109. [DOI] [PubMed] [Google Scholar]
- 143.Grotius J, Dittfeld C, Huether M, Mueller-Klieser W, Baumann M, Kunz-Schughart LA. Impact of exogenous lactate on survival and radioresponse of carcinoma cells in vitro. International journal of radiation biology. 2009;85(11):989–1001. [DOI] [PubMed] [Google Scholar]
- 144.Farabegoli F, Vettraino M, Manerba M, Fiume L, Roberti M, Di Stefano G. Galloflavin, a new lactate dehydrogenase inhibitor, induces the death of human breast cancer cells with different glycolytic attitude by affecting distinct signaling pathways. European Journal of Pharmaceutical Sciences. 2012;47(4):729–738. [DOI] [PubMed] [Google Scholar]
- 145.Brizel DM, Schroeder T, Scher RL, et al. Elevated tumor lactate concentrations predict for an increased risk of metastases in head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2001;51(2):349–353. [DOI] [PubMed] [Google Scholar]
- 146.Walenta S, Wetterling M, Lehrke M, et al. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer research. 2000;60(4):916–921. [PubMed] [Google Scholar]
- 147.Walenta S, Mueller-Klieser WF. Lactate: mirror and motor of tumor malignancy. Semin Radiat Oncol. 2004;14(3):267–274. [DOI] [PubMed] [Google Scholar]
- 148.Koukourakis MI, Giatromanolaki A, Winter S, Leek R, Sivridis E, Harris AL. Lactate Dehydrogenase 5 Expression in Squamous Cell Head and Neck Cancer Relates to Prognosis following Radical or Postoperative Radiotherapy. Oncology. 2009;77(5):285–292. [DOI] [PubMed] [Google Scholar]
- 149.He T-L, Zhang Y-J, Jiang H, Li X-H, Zhu H, Zheng K-L. The c-Myc-LDHA axis positively regulates aerobic glycolysis and promotes tumor progression in pancreatic cancer. Med Oncol. 2015;32(7):187–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kennedy KM, Dewhirst MW. Tumor metabolism of lactate: the influence and therapeutic potential for MCT and CD147 regulation. Future Oncol. 2010;6(1):127–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mitsuishi Y, Motohashi H, Yamamoto M. The Keap1-Nrf2 system in cancers: stress response and anabolic metabolism. Frontiers in oncology. 2012;2:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Simmen FA, Alhallak I, Simmen RCM. Malic enzyme 1 (ME1) in the biology of cancer: it is not just intermediary metabolism. J Mol Endocrinol. 2020;65(4):R77–R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bouzier AK, Voisin P, Goodwin R, Canioni P, Merle M. Glucose and lactate metabolism in C6 glioma cells: evidence for the preferential utilization of lactate for cell oxidative metabolism. Developmental neuroscience. 1998;20(4–5):331–338. [DOI] [PubMed] [Google Scholar]
- 154.Bouzier-Sore AK, Canioni P, Merle M. Effect of exogenous lactate on rat glioma metabolism. Journal of neuroscience research. 2001;65(6):543–548. [DOI] [PubMed] [Google Scholar]
- 155.Latif A, Chadwick AL, Kitson SJ, et al. Monocarboxylate Transporter 1 (MCT1) is an independent prognostic biomarker in endometrial cancer. BMC clinical pathology. 2017;17:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Leu M, Kitz J, Pilavakis Y, et al. Monocarboxylate transporter-1 (MCT1) protein expression in head and neck cancer affects clinical outcome. Sci Rep. 2021;11(1):4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sertorio M, Perentesis JP, Vatner RE, Mascia AE, Zheng Y, Wells SI. Cancer Cell Metabolism: Implications for X-ray and Particle Radiation Therapy. Int J Part Ther. 2018;5(1):40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wahlström T, Henriksson MA. Impact of MYC in regulation of tumor cell metabolism. Biochimica et biophysica acta. 2015;1849(5):563–569. [DOI] [PubMed] [Google Scholar]
- 159.Doherty JR, Yang C, Scott KE, et al. Blocking lactate export by inhibiting the Myc target MCT1 Disables glycolysis and glutathione synthesis. Cancer research. 2014;74(3):908–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.De Saedeleer CJ, Copetti T, Porporato PE, Verrax J, Feron O, Sonveaux P. Lactate activates HIF-1 in oxidative but not in Warburg-phenotype human tumor cells. PLoS One. 2012;7(10):e46571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kozlov AM, Lone A, Betts DH, Cumming RC. Lactate preconditioning promotes a HIF-1α-mediated metabolic shift from OXPHOS to glycolysis in normal human diploid fibroblasts. Scientific reports. 2020;10(1):8388–8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cui XG, Han ZT, He SH, et al. HIF1/2α mediates hypoxia-induced LDHA expression in human pancreatic cancer cells. Oncotarget. 2017;8(15):24840–24852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Day SE, Kettunen MI, Cherukuri MK, et al. Detecting response of rat C6 glioma tumors to radiotherapy using hyperpolarized [1– 13C]pyruvate and 13C magnetic resonance spectroscopic imaging. Magnetic resonance in medicine. 2011;65(2):557–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lim H, Martínez-Santiesteban F, Jensen MD, Chen A, Wong E, Scholl TJ. Monitoring Early Changes in Tumor Metabolism in Response to Therapy Using Hyperpolarized (13)C MRSI in a Preclinical Model of Glioma. Tomography (Ann Arbor, Mich). 2020;6(3):290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chaumeil MM, Ozawa T, Park I, et al. Hyperpolarized 13C MR spectroscopic imaging can be used to monitor Everolimus treatment in vivo in an orthotopic rodent model of glioblastoma. Neuroimage. 2012;59(1):193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Matsuo M, Kawai T, Kishimoto S, et al. Co-imaging of the tumor oxygenation and metabolism using electron paramagnetic resonance imaging and 13-C hyperpolarized magnetic resonance imaging before and after irradiation. Oncotarget. 2018;9(38):25089–25100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sandulache VC, Chen Y, Lee J, et al. Evaluation of hyperpolarized [1-13C]-pyruvate by magnetic resonance to detect ionizing radiation effects in real time. PLoS One. 2014;9(1):e87031-e87031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kawai T, Brender JR, Lee JA, et al. Detection of metabolic change in glioblastoma cells after radiotherapy using hyperpolarized (13) C-MRI. NMR Biomed. 2021;34(7):e4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Albers MJ, Bok R, Chen AP, et al. Hyperpolarized 13C lactate, pyruvate, and alanine: noninvasive biomarkers for prostate cancer detection and grading. Cancer research. 2008;68(20):8607–8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sandulache VC, Skinner HD, Wang Y, et al. Glycolytic inhibition alters anaplastic thyroid carcinoma tumor metabolism and improves response to conventional chemotherapy and radiation. Molecular cancer therapeutics. 2012;11(6):1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liao EC, Hsu YT, Chuah QY, et al. Radiation induces senescence and a bystander effect through metabolic alterations. Cell Death Dis. 2014;5(5):e1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bola BM, Chadwick AL, Michopoulos F, et al. Inhibition of Monocarboxylate Transporter-1 (MCT1) by AZD3965 Enhances Radiosensitivity by Reducing Lactate Transport. Molecular cancer therapeutics. 2014;13(12):2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sonveaux P, Végran F, Schroeder T, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. The Journal of Clinical Investigation. 2008;118(12):3930–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Halestrap AP, Wilson MC. The monocarboxylate transporter family--role and regulation. IUBMB Life. 2012;64(2):109–119. [DOI] [PubMed] [Google Scholar]
- 175.Halestrap AP. The monocarboxylate transporter family--Structure and functional characterization. IUBMB Life. 2012;64(1):1–9. [DOI] [PubMed] [Google Scholar]
- 176.Porporato PE, Filigheddu N, Pedro JMB-S, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Research. 2018;28(3):265–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vyas S, Zaganjor E, Haigis MC. Mitochondria and Cancer. Cell. 2016;166(3):555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Leach JK, Van Tuyle G, Lin PS, Schmidt-Ullrich R, Mikkelsen RB. Ionizing radiation-induced, mitochondria-dependent generation of reactive oxygen/nitrogen. Cancer research. 2001;61(10):3894–3901. [PubMed] [Google Scholar]
- 179.Nugent SM, Mothersill CE, Seymour C, McClean B, Lyng FM, Murphy JE. Increased mitochondrial mass in cells with functionally compromised mitochondria after exposure to both direct gamma radiation and bystander factors. Radiat Res. 2007;168(1):134–142. [DOI] [PubMed] [Google Scholar]
- 180.Ogura A, Oowada S, Kon Y, et al. Redox regulation in radiation-induced cytochrome c release from mitochondria of human lung carcinoma A549 cells. Cancer letters. 2009;277(1):64–71. [DOI] [PubMed] [Google Scholar]
- 181.Yasui H, Yamamoto K, Suzuki M, et al. Lipophilic triphenylphosphonium derivatives enhance radiation-induced cell killing via inhibition of mitochondrial energy metabolism in tumor cells. Cancer letters. 2017;390:160–167. [DOI] [PubMed] [Google Scholar]
- 182.Yamamori T, Yasui H, Yamazumi M, et al. Ionizing radiation induces mitochondrial reactive oxygen species production accompanied by upregulation of mitochondrial electron transport chain function and mitochondrial content under control of the cell cycle checkpoint. Free Radic Biol Med. 2012;53(2):260–270. [DOI] [PubMed] [Google Scholar]
- 183.Lu CL, Qin L, Liu HC, Candas D, Fan M, Li JJ. Tumor cells switch to mitochondrial oxidative phosphorylation under radiation via mTOR-mediated hexokinase II inhibition--a Warburg-reversing effect. PLoS One. 2015;10(3):e0121046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nissanka N, Moraes CT. Mitochondrial DNA damage and reactive oxygen species in neurodegenerative disease. FEBS letters. 2018;592(5):728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Cebula M, Schmidt EE, Arnér ES. TrxR1 as a potent regulator of the Nrf2-Keap1 response system. Antioxidants & redox signaling. 2015;23(10):823–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dinkova-Kostova AT, Abramov AY. The emerging role of Nrf2 in mitochondrial function. Free radical biology & medicine. 2015;88(Pt B):179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chaudhry MA, Omaruddin RA. Mitochondrial gene expression in directly irradiated and nonirradiated bystander cells. Cancer biotherapy & radiopharmaceuticals. 2011;26(5):657–663. [DOI] [PubMed] [Google Scholar]
- 188.Lin RX, Zhao HB, Li CR, Sun YN, Qian XH, Wang SQ. Proteomic analysis of ionizing radiation-induced proteins at the subcellular level. Journal of proteome research. 2009;8(1):390–399. [DOI] [PubMed] [Google Scholar]
- 189.Patwardhan RS, Sharma D, Checker R, Thoh M, Sandur SK. Spatio-temporal changes in glutathione and thioredoxin redox couples during ionizing radiation-induced oxidative stress regulate tumor radio-resistance. Free Radic Res. 2015;49(10):1218–1232. [DOI] [PubMed] [Google Scholar]
- 190.Yamamori T, Ike S, Bo T, et al. Inhibition of the mitochondrial fission protein dynamin-related protein 1 (Drp1) impairs mitochondrial fission and mitotic catastrophe after x-irradiation. Mol Biol Cell. 2015;26(25):4607–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kobashigawa S, Suzuki K, Yamashita S. Ionizing radiation accelerates Drp1-dependent mitochondrial fission, which involves delayed mitochondrial reactive oxygen species production in normal human fibroblast-like cells. Biochemical and Biophysical Research Communications. 2011;414(4):795–800. [DOI] [PubMed] [Google Scholar]
- 192.Toulany M, Rodemann HP. Phosphatidylinositol 3-kinase/Akt signaling as a key mediator of tumor cell responsiveness to radiation. Seminars in cancer biology. 2015;35:180–190. [DOI] [PubMed] [Google Scholar]
- 193.Morita M, Prudent J, Basu K, et al. mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Mol Cell. 2017;67(6):922–935.e925. [DOI] [PubMed] [Google Scholar]
- 194.Rehman J, Zhang HJ, Toth PT, et al. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26(5):2175–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Inoue-Yamauchi A, Oda H. Depletion of mitochondrial fission factor DRP1 causes increased apoptosis in human colon cancer cells. Biochemical and Biophysical Research Communications. 2012;421(1):81–85. [DOI] [PubMed] [Google Scholar]
- 196.Rodrigues T, Ferraz LS. Therapeutic potential of targeting mitochondrial dynamics in cancer. Biochemical pharmacology. 2020;182:114282. [DOI] [PubMed] [Google Scholar]
- 197.Ferreira-da-Silva A, Valacca C, Rios E, et al. Mitochondrial dynamics protein Drp1 is overexpressed in oncocytic thyroid tumors and regulates cancer cell migration. PLoS One. 2015;10(3):e0122308-e0122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zong W-X, Rabinowitz JD, White E. Mitochondria and Cancer. Molecular Cell. 2016;61(5):667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yu L, Yang X, Li X, et al. Pink1/PARK2/mROS-Dependent Mitophagy Initiates the Sensitization of Cancer Cells to Radiation. Oxid Med Cell Longev. 2021;2021:5595652–5595652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chen Q, Zheng W, Zhu L, et al. LACTB2 renders radioresistance by activating PINK1/Parkin-dependent mitophagy in nasopharyngeal carcinoma. Cancer letters. 2021;518:127–139. [DOI] [PubMed] [Google Scholar]
- 201.Meyer JN, Leuthner TC, Luz AL. Mitochondrial fusion, fission, and mitochondrial toxicity. Toxicology. 2017;391:42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tondera D, Grandemange S, Jourdain A, et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. The EMBO journal. 2009;28(11):1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Jendrach M, Mai S, Pohl S, Vöth M, Bereiter-Hahn J. Short- and long-term alterations of mitochondrial morphology, dynamics and mtDNA after transient oxidative stress. Mitochondrion. 2008;8(4):293–304. [DOI] [PubMed] [Google Scholar]
- 204.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337(6098):1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bo T, Yamamori T, Yamamoto K, Fujimoto M, Yasui H, Inanami O. Mitochondrial fission promotes radiation-induced increase in intracellular Ca(2+) level leading to mitotic catastrophe in mouse breast cancer EMT6 cells. Biochem Biophys Res Commun. 2020;522(1):144–150. [DOI] [PubMed] [Google Scholar]
- 206.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends in biochemical sciences. 2010;35(8):427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Marquez J, Alonso FJ, Mates JM, Segura JA, Martin-Rufian M, Campos-Sandoval JA. Glutamine Addiction In Gliomas. Neurochem Res. 2017;42(6):1735–1746. [DOI] [PubMed] [Google Scholar]
- 208.Vanhove K, Derveaux E, Graulus GJ, et al. Glutamine Addiction and Therapeutic Strategies in Lung Cancer. Int J Mol Sci. 2019;20(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Boysen G, Jamshidi-Parsian A, Davis MA, et al. Glutaminase inhibitor CB-839 increases radiation sensitivity of lung tumor cells and human lung tumor xenografts in mice. International journal of radiation biology. 2019;95(4):436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sappington DR, Siegel ER, Hiatt G, et al. Glutamine drives glutathione synthesis and contributes to radiation sensitivity of A549 and H460 lung cancer cell lines. Biochimica et biophysica acta. 2016;1860(4):836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Fu S, Li Z, Xiao L, et al. Glutamine Synthetase Promotes Radiation Resistance via Facilitating Nucleotide Metabolism and Subsequent DNA Damage Repair. Cell Rep. 2019;28(5):1136–1143.e1134. [DOI] [PubMed] [Google Scholar]
- 212.Daukantienė L, Kazbarienė B, Valuckas KP, Didžiapetrienė J, Krikštaponienė A, Aleknavičius E. The significance of reduced glutathione and glutathione S-transferase during chemoradiotherapy of locally advanced cervical cancer. Medicina. 2014;50(4):222–229. [DOI] [PubMed] [Google Scholar]
- 213.Possemato R, Marks KM, Shaul YD, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Locasale JW, Grassian AR, Melman T, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43(9):869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sullivan MR, Vander Heiden MG. When cancer needs what’s non-essential. Nat Cell Biol. 2017;19(5):418–420. [DOI] [PubMed] [Google Scholar]
- 216.Jin HO, Hong SE, Kim JY, et al. Knock-down of PSAT1 Enhances Sensitivity of NSCLC Cells to Glutamine-limiting Conditions. Anticancer Res. 2019;39(12):6723–6730. [DOI] [PubMed] [Google Scholar]
- 217.Ye J, Fan J, Venneti S, et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 2014;4(12):1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yang M, Vousden KH. Serine and one-carbon metabolism in cancer. Nature reviews Cancer. 2016;16(10):650–662. [DOI] [PubMed] [Google Scholar]
- 219.Sánchez-Castillo A, Vooijs M, Kampen KR. Linking Serine/Glycine Metabolism to Radiotherapy Resistance. Cancers. 2021;13(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Koch A, Joosten SC, Feng Z, et al. Analysis of DNA methylation in cancer: location revisited. Nature reviews Clinical oncology. 2018;15(7):459–466. [DOI] [PubMed] [Google Scholar]
- 221.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. [DOI] [PubMed] [Google Scholar]
- 222.Audia JE, Campbell RM. Histone Modifications and Cancer. Cold Spring Harb Perspect Biol. 2016;8(4):a019521-a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.McCabe MT, Mohammad HP, Barbash O, Kruger RG. Targeting Histone Methylation in Cancer. The Cancer Journal. 2017;23(5). [DOI] [PubMed] [Google Scholar]
- 224.Harris WJ, Huang X, Lynch JT, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer cell. 2012;21(4):473–487. [DOI] [PubMed] [Google Scholar]
- 225.Mitra D, Das PM, Huynh FC, Jones FE. Jumonji/ARID1 B (JARID1B) protein promotes breast tumor cell cycle progression through epigenetic repression of microRNA let-7e. J Biol Chem. 2011;286(47):40531–40535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. The EMBO journal. 2003;22(20):5323–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Sharma SV, Lee DY, Li B, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Miousse IR, Tobacyk J, Melnyk S, et al. One-carbon metabolism and ionizing radiation: a multifaceted interaction. Biomol Concepts. 2017;8(2):83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Fowler B. The folate cycle and disease in humans. Kidney International. 2001;59:S221–S229. [DOI] [PubMed] [Google Scholar]
- 230.Matherly LH, Goldman DI. Membrane transport of folates. Vitamins and hormones. 2003;66:403–456. [DOI] [PubMed] [Google Scholar]
- 231.Hitchings GH, Burchall JJ. Inhibition of folate biosynthesis and function as a basis for chemotherapy. Advances in enzymology and related areas of molecular biology. 1965;27:417–468. [DOI] [PubMed] [Google Scholar]
- 232.Kanarek N, Keys HR, Cantor JR, et al. Histidine catabolism is a major determinant of methotrexate sensitivity. Nature. 2018;559(7715):632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Batra V, Kesavan V, Mishra KP. Modulation of Enzymes Involved in Folate Dependent One-carbon Metabolism by γ-radiation Stress in Mice. Journal of Radiation Research. 2004;45(4):527–533. [DOI] [PubMed] [Google Scholar]
- 234.Endoh K, Murakami M, Umegaki K. Vulnerability of folate in plasma and bone marrow to total body irradiation in mice. International journal of radiation biology. 2007;83(1):65–71. [DOI] [PubMed] [Google Scholar]
- 235.Kesavan V, Pote MS, Batra V, Viswanathan G. Increased Folate Catabolism Following Total Body γ-Irradiation in Mice. Journal of Radiation Research. 2003;44(2):141–144. [DOI] [PubMed] [Google Scholar]
- 236.Hoffman RM. Development of recombinant methioninase to target the general cancer-specific metabolic defect of methionine dependence: a 40-year odyssey. Expert Opinion on Biological Therapy. 2015;15(1):21–31. [DOI] [PubMed] [Google Scholar]
- 237.Camici M, Garcia-Gil M, Pesi R, Allegrini S, Tozzi MG. Purine-Metabolising Enzymes and Apoptosis in Cancer. Cancers. 2019;11(9):1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Zhang B. CD73 promotes tumor growth and metastasis. Oncoimmunology. 2012;1(1):67–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Galmarini CM, Graham K, Thomas X, et al. Expression of high Km 5’-nucleotidase in leukemic blasts is an independent prognostic factor in adults with acute myeloid leukemia. Blood. 2001;98(6):1922–1926. [DOI] [PubMed] [Google Scholar]
- 240.Lv Y, Wang X, Li X, et al. Nucleotide de novo synthesis increases breast cancer stemness and metastasis via cGMP-PKG-MAPK signaling pathway. PLoS Biol. 2020;18(11):e3000872-e3000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Gao ZW, Dong K, Zhang HZ. The roles of CD73 in cancer. BioMed research international. 2014;2014:460654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wei AA-BFPJBSBCLS. Radiation-induced changes in nucleotide metabolism of two colon cancer cell lines with different radiosensitivities. International journal of radiation biology. 1999;75(8):1005–1013. [DOI] [PubMed] [Google Scholar]
- 243.Boothman DA, Davis TW, Sahijdak WM. Enhanced expression of thymidine kinase in human cells following ionizing radiation. Int J Radiat Oncol Biol Phys. 1994;30(2):391–398. [DOI] [PubMed] [Google Scholar]
- 244.Boothman DA, Meyers M, Fukunaga N, Lee SW. Isolation of x-ray-inducible transcripts from radioresistant human melanoma cells. Proc Natl Acad Sci U S A. 1993;90(15):7200–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ipata PL, Balestri F, Camici M, Tozzi MG. Molecular mechanisms of nucleoside recycling in the brain. The international journal of biochemistry & cell biology. 2011;43(1):140–145. [DOI] [PubMed] [Google Scholar]
- 246.Elledge SJ, Zhou Z, Allen JB. Ribonucleotide reductase: regulation, regulation, regulation. Trends in biochemical sciences. 1992;17(3):119–123. [DOI] [PubMed] [Google Scholar]
- 247.Guittet O, Håkansson P, Voevodskaya N, et al. Mammalian p53R2 protein forms an active ribonucleotide reductase in vitro with the R1 protein, which is expressed both in resting cells in response to DNA damage and in proliferating cells. J Biol Chem. 2001;276(44):40647–40651. [DOI] [PubMed] [Google Scholar]
- 248.Pontarin G, Ferraro P, Håkansson P, Thelander L, Reichard P, Bianchi V. p53R2-dependent ribonucleotide reduction provides deoxyribonucleotides in quiescent human fibroblasts in the absence of induced DNA damage. J Biol Chem. 2007;282(23):16820–16828. [DOI] [PubMed] [Google Scholar]
- 249.Cosentino C, Grieco D, Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. The EMBO journal. 2011;30(3):546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Bravard A, Luccioni C, Moustacchi E, Rigaud O. Contribution of antioxidant enzymes to the adaptive response to ionizing radiation of human lymphoblasts. International journal of radiation biology. 1999;75(5):639–645. [DOI] [PubMed] [Google Scholar]
- 251.Chiarugi A, Dölle C, Felici R, Ziegler M. The NAD metabolome--a key determinant of cancer cell biology. Nature reviews Cancer. 2012;12(11):741–752. [DOI] [PubMed] [Google Scholar]
- 252.Duarte-Pereira S, Pereira-Castro I, Silva SS, et al. Extensive regulation of nicotinate phosphoribosyltransferase (NAPRT) expression in human tissues and tumors. Oncotarget. 2016;7(2):1973–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Bunimovich YL, Nair-Gill E, Riedinger M, et al. Deoxycytidine Kinase Augments ATM-Mediated DNA Repair and Contributes to Radiation Resistance. PLoS One. 2014;9(8):e104125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kunos CA, Ferris G, Pyatka N, Pink J, Radivoyevitch T. Deoxynucleoside salvage facilitates DNA repair during ribonucleotide reductase blockade in human cervical cancers. Radiat Res. 2011;176(4):425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Miran T, Vogg ATJ, El Moussaoui L, et al. Dual addressing of thymidine synthesis pathways for effective targeting of proliferating melanoma. Cancer Med. 2017;6(7):1639–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Morgenroth A, Vogg AT, Mottaghy FM, Schmaljohann J. Targeted endoradiotherapy using nucleotides. Methods (San Diego, Calif). 2011;55(3):203–214. [DOI] [PubMed] [Google Scholar]
- 257.Zhou W, Yao Y, Scott AJ, et al. Purine metabolism regulates DNA repair and therapy resistance in glioblastoma. Nat Commun. 2020;11(1):3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lunt SY, Muralidhar V, Hosios AM, et al. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Molecular cell. 2015;57(1):95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Reaves ML, Young BD, Hosios AM, Xu YF, Rabinowitz JD. Pyrimidine homeostasis is accomplished by directed overflow metabolism. Nature. 2013;500(7461):237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Hu J, Locasale JW, Bielas JH, et al. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nature biotechnology. 2013;31(6):522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Kofuji S, Hirayama A, Eberhardt AO, et al. IMP dehydrogenase-2 drives aberrant nucleolar activity and promotes tumorigenesis in glioblastoma. Nat Cell Biol. 2019;21(8):1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Li J, Cheng JX. Direct visualization of de novo lipogenesis in single living cells. Sci Rep. 2014;4:6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Mounier C, Bouraoui L, Rassart E . Lipogenesis in cancer progression (review). International journal of oncology. 2014;45(2):485–492. [DOI] [PubMed] [Google Scholar]
- 264.Igal RA. Roles of StearoylCoA Desaturase-1 in the Regulation of Cancer Cell Growth, Survival and Tumorigenesis. Cancers. 2011;3(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Chen L, Ren J, Yang L, et al. Stearoyl-CoA desaturase-1 mediated cell apoptosis in colorectal cancer by promoting ceramide synthesis. Sci Rep. 2016;6:19665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Scaglia N, Chisholm JW, Igal RA. Inhibition of stearoylCoA desaturase-1 inactivates acetyl-CoA carboxylase and impairs proliferation in cancer cells: role of AMPK. PLoS One. 2009;4(8):e6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Calvisi DF, Wang C, Ho C, et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology. 2011;140(3):1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140(1):49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Nieman KM, Kenny HA, Penicka CV, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nature medicine. 2011;17(11):1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.de Lima Luna AC, Forti FL. Modulation of SCD1 activity in hepatocyte cell lines: evaluation of genomic stability and proliferation. Molecular and Cellular Biochemistry. 2021. [DOI] [PubMed] [Google Scholar]
- 271.Jin Y, Chen Z, Dong J, et al. SREBP1/FASN/cholesterol axis facilitates radioresistance in colorectal cancer. FEBS Open Bio. 2021;11(5):1343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Bhat K, Saki M, Cheng F, et al. Dopamine Receptor Antagonists, Radiation, and Cholesterol Biosynthesis in Mouse Models of Glioblastoma. JNCI: Journal of the National Cancer Institute. 2021;113(8):1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Küch E-M, Vellaramkalayil R, Zhang I, et al. Differentially localized acyl-CoA synthetase 4 isoenzymes mediate the metabolic channeling of fatty acids towards phosphatidylinositol. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2014;1841(2):227–239. [DOI] [PubMed] [Google Scholar]
- 274.Lei G, Zhang Y, Koppula P, et al. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell research. 2020;30(2):146–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Hirsch HA, Iliopoulos D, Joshi A, et al. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer cell. 2010;17(4):348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Jeffries KA, Krupenko NI. Chapter Seven - Ceramide Signaling and p53 Pathways. In: Chalfant CE, Fisher PB, eds. Advances in Cancer Research. Vol 140. Academic Press; 2018:191–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Mirza A, Wu Q, Wang L, et al. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene. 2003;22(23):3645–3654. [DOI] [PubMed] [Google Scholar]
- 278.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24(41):6314–6322. [DOI] [PubMed] [Google Scholar]
- 279.Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J Biol Chem. 2002;277(37):33895–33900. [DOI] [PubMed] [Google Scholar]
- 280.Bansal N, Mims J, Kuremsky JG, et al. Broad phenotypic changes associated with gain of radiation resistance in head and neck squamous cell cancer. Antioxidants & redox signaling. 2014;21(2):221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Mims J, Bansal N, Bharadwaj MS, et al. Energy Metabolism in a Matched Model of Radiation Resistance for Head and Neck Squamous Cell Cancer. Radiation Research. 2015;183(3):291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Walden T, Hughes HN. Prostaglandin and Lipid Metabolism in Radiation Injury. Paper presented at: Springer US1987. [Google Scholar]
- 283.Reisz JA, Bansal N, Qian J, Zhao W, Furdui CM. Effects of ionizing radiation on biological molecules--mechanisms of damage and emerging methods of detection. Antioxidants & redox signaling. 2014;21(2):260–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Kolomiytseva IK, Novoselova EG, Kulagina TP, Kuzin AM. The effect of ionizing radiation on lipid metabolism in lymphoid cells. International journal of radiation biology and related studies in physics, chemistry, and medicine. 1987;51(1):53–58. [DOI] [PubMed] [Google Scholar]
- 285.Jo SK, Seol MA, Park HR, Jung U, Roh C. Ionising radiation triggers fat accumulation in white adipose tissue. International journal of radiation biology. 2011;87(3):302–310. [DOI] [PubMed] [Google Scholar]
- 286.Lyng H, Sitter B, Bathen TF, et al. Metabolic mapping by use of high-resolution magic angle spinning 1H MR spectroscopy for assessment of apoptosis in cervical carcinomas. BMC Cancer. 2007;7(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Kuesel AC, Sutherland GR, Halliday W, Smith ICP. 1H MRS of high grade astrocytomas: Mobile lipid accumulation in necrotic tissue. NMR in Biomedicine. 1994;7(3):149–155. [DOI] [PubMed] [Google Scholar]
- 288.Shaw J, Costa-Pinheiro P, Patterson L, Drews K, Spiegel S, Kester M. Novel Sphingolipid-Based Cancer Therapeutics in the Personalized Medicine Era. Adv Cancer Res. 2018;140:327–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Haimovitz-Friedman A, Kan CC, Ehleiter D, et al. Ionizing radiation acts on cellular membranes to generate ceramide and initiate apoptosis. The Journal of experimental medicine. 1994;180(2):525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Carroll B, Donaldson JC, Obeid L. Sphingolipids in the DNA damage response. Advances in biological regulation. 2015;58:38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Vit JP, Rosselli F. Role of the ceramide-signaling pathways in ionizing radiation-induced apoptosis. Oncogene. 2003;22(54):8645–8652. [DOI] [PubMed] [Google Scholar]
- 292.Modrak DE, Gold DV, Goldenberg DM. Sphingolipid targets in cancer therapy. Molecular cancer therapeutics. 2006;5(2):200–208. [DOI] [PubMed] [Google Scholar]
- 293.Vicente E, Vujaskovic Z, Jackson IL. A Systematic Review of Metabolomic and Lipidomic Candidates for Biomarkers in Radiation Injury. Metabolites. 2020;10(6):259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Wang J, Lv X-W, Shi J-P, Hu X-S. Mechanisms involved in ceramide-induced cell cycle arrest in human hepatocarcinoma cells. World journal of gastroenterology. 2007;13(7):1129–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Kim WH, Kang KH, Kim MY, Choi KH. Induction of p53-independent p21 during ceramide-induced G1 arrest in human hepatocarcinoma cells. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2000;78(2):127–135. [PubMed] [Google Scholar]
- 296.Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nature reviews Cancer. 2004;4(8):604–616. [DOI] [PubMed] [Google Scholar]
- 297.Aureli M, Murdica V, Loberto N, et al. Exploring the link between ceramide and ionizing radiation. Glycoconjugate journal. 2014;31(6–7):449–459. [DOI] [PubMed] [Google Scholar]
- 298.Hara S, Nakashima S, Kiyono T, et al. p53-Independent ceramide formation in human glioma cells during gamma-radiation-induced apoptosis. Cell death and differentiation. 2004;11(8):853–861. [DOI] [PubMed] [Google Scholar]
- 299.Colombini M. Ceramide channels and mitochondrial outer membrane permeability. Journal of Bioenergetics and Biomembranes. 2017;49(1):57–64. [DOI] [PubMed] [Google Scholar]
- 300.Santana P, Peña LA, Haimovitz-Friedman A, et al. Acid sphingomyelinase-deficient human lymphoblasts and mice are defective in radiation-induced apoptosis. Cell. 1996;86(2):189–199. [DOI] [PubMed] [Google Scholar]
- 301.Kobeissy H, Hage-Sleiman R, Dakdouk Z, Kozhaya L, Dbaibo G. Crosstalk between Noxa, Bcl-2, and ceramide in mediating p53-dependent apoptosis in Molt-4 human T-cell leukemia. Mol Cell Biochem. 2020;475(1–2):215–226. [DOI] [PubMed] [Google Scholar]
- 302.Dbaibo GS, Pushkareva MY, Rachid RA, et al. p53-dependent ceramide response to genotoxic stress. J Clin Invest. 1998;102(2):329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer cell. 2003;3(3):285–296. [DOI] [PubMed] [Google Scholar]
- 304.Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nature Reviews Clinical Oncology. 2021;18(5):280–296. [DOI] [PubMed] [Google Scholar]
- 305.Yang WS, SriRamaratnam R, Welsch ME, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Yang WS, Stockwell BR. Ferroptosis: Death by Lipid Peroxidation. Trends in cell biology. 2016;26(3):165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Koppula P, Zhang Y, Zhuang L, Gan B. Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer communications (London, England). 2018;38(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Patt HM, Tyree EB, Straube RL, Smith DE. Cysteine Protection against X Irradiation. Science. 1949;110(2852):213–214. [DOI] [PubMed] [Google Scholar]
- 309.Cobler L, Zhang H, Suri P, Park C, Timmerman LA. xCT inhibition sensitizes tumors to γ-radiation via glutathione reduction. Oncotarget. 2018;9(64):32280–32297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Yi J, Zhu J, Wu J, Thompson CB, Jiang X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proceedings of the National Academy of Sciences. 2020;117(49):31189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Steel GG, McMillan TJ, Peacock JH. The 5Rs of radiobiology. International journal of radiation biology. 1989;56(6):1045–1048. [DOI] [PubMed] [Google Scholar]
- 312.Bollineni VR, Koole MJ, Pruim J, et al. Dynamics of tumor hypoxia assessed by 18F-FAZA PET/CT in head and neck and lung cancer patients during chemoradiation: possible implications for radiotherapy treatment planning strategies. Radiother Oncol. 2014;113(2):198–203. [DOI] [PubMed] [Google Scholar]
- 313.Bollineni VR, Kerner GS, Pruim J, et al. PET imaging of tumor hypoxia using 18F-fluoroazomycin arabinoside in stage III-IV non-small cell lung cancer patients. J Nucl Med. 2013;54(8):1175–1180. [DOI] [PubMed] [Google Scholar]
- 314.Guerrero M, Carlson DJ. A radiobiological model of reoxygenation and fractionation effects. Medical Physics. 2017;44(5):2002–2010. [DOI] [PubMed] [Google Scholar]
- 315.Kocher M, Treuer H, Müller RP. Quantification of tumor reoxygenation during accelerated radiation therapy. Radiology. 1997;205(1):263–268. [DOI] [PubMed] [Google Scholar]
- 316.Rakotomalala A, Escande A, Furlan A, Meignan S, Lartigau E. Hypoxia in Solid Tumors: How Low Oxygenation Impacts the “Six Rs” of Radiotherapy. Frontiers in endocrinology. 2021;12:742215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Al Tameemi W, Dale TP, Al-Jumaily RMK, Forsyth NR. Hypoxia-Modified Cancer Cell Metabolism. Front Cell Dev Biol. 2019;7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Coleman CN, Eke I, Makinde AY, et al. Radiation-induced Adaptive Response: New Potential for Cancer Treatment. Clinical cancer research : an official journal of the American Association for Cancer Research. 2020;26(22):5781–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Eke I, Makinde AY, Aryankalayil MJ, et al. Exploiting Radiation-Induced Signaling to Increase the Susceptibility of Resistant Cancer Cells to Targeted Drugs: AKT and mTOR Inhibitors as an Example. Molecular cancer therapeutics. 2018;17(2):355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Eke I, Makinde AY, Aryankalayil MJ, et al. Long-term Tumor Adaptation after Radiotherapy: Therapeutic Implications for Targeting Integrins in Prostate Cancer. Mol Cancer Res. 2018;16(12):1855–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Tsai M-H, Cook JA, Chandramouli GVR, et al. Gene Expression Profiling of Breast, Prostate, and Glioma Cells following Single versus Fractionated Doses of Radiation. Cancer research. 2007;67(8):3845. [DOI] [PubMed] [Google Scholar]
- 322.Kong FM, Ten Haken RK, Schipper M, et al. Effect of Midtreatment PET/CT-Adapted Radiation Therapy With Concurrent Chemotherapy in Patients With Locally Advanced Non-Small-Cell Lung Cancer: A Phase 2 Clinical Trial. JAMA Oncol. 2017;3(10):1358–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Thureau S, Dubray B, Modzelewski R, et al. FDG and FMISO PET-guided dose escalation with intensity-modulated radiotherapy in lung cancer. Radiation Oncology. 2018;13(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Ganem J, Thureau S, Gardin I, Modzelewski R, Hapdey S, Vera P. Delineation of lung cancer with FDG PET/CT during radiation therapy. Radiat Oncol. 2018;13(1):219–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Prasanna PG, Rawojc K, Guha C, Buchsbaum JC, Miszczyk JU, Coleman CN. Normal Tissue Injury Induced by Photon and Proton Therapies: Gaps and Opportunities. International Journal of Radiation Oncology*Biology*Physics. 2021;110(5):1325–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Jelonek K, Pietrowska M, Widlak P. Systemic effects of ionizing radiation at the proteome and metabolome levels in the blood of cancer patients treated with radiotherapy: the influence of inflammation and radiation toxicity. International journal of radiation biology. 2017;93(7):683–696. [DOI] [PubMed] [Google Scholar]
- 327.Laiakis EC, Pannkuk EL, Chauthe SK, et al. A Serum Small Molecule Biosignature of Radiation Exposure from Total Body Irradiated Patients. Journal of proteome research. 2017;16(10):3805–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Chen C, Brenner DJ, Brown TR. Identification of urinary biomarkers from X-irradiated mice using NMR spectroscopy. Radiat Res. 2011;175(5):622–630. [DOI] [PubMed] [Google Scholar]
- 329.Gramatyka M, Sokół M. Radiation metabolomics in the quest of cardiotoxicity biomarkers: the review. International journal of radiation biology. 2020;96(3):349–359. [DOI] [PubMed] [Google Scholar]
- 330.Pannkuk EL, Laiakis EC, Girgis M, et al. Temporal Effects on Radiation Responses in Nonhuman Primates: Identification of Biofluid Small Molecule Signatures by Gas Chromatography⁻Mass Spectrometry Metabolomics. Metabolites. 2019;9(5):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Menon SS, Uppal M, Randhawa S, et al. Radiation Metabolomics: Current Status and Future Directions. Frontiers in oncology. 2016;6:20–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Johnson CH, Patterson AD, Krausz KW, et al. Radiation metabolomics. 5. Identification of urinary biomarkers of ionizing radiation exposure in nonhuman primates by mass spectrometry-based metabolomics. Radiat Res. 2012;178(4):328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Laiakis EC, Mak TD, Anizan S, et al. Development of a metabolomic radiation signature in urine from patients undergoing total body irradiation. Radiat Res. 2014;181(4):350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Sproull M, Kramp T, Tandle A, Shankavaram U, Camphausen K. Multivariate Analysis of Radiation Responsive Proteins to Predict Radiation Exposure in Total-Body Irradiation and Partial-Body Irradiation Models. Radiat Res. 2017;187(2):251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Ossetrova NI, Condliffe DP, Ney PH, et al. Early-response biomarkers for assessment of radiation exposure in a mouse total-body irradiation model. Health physics. 2014;106(6):772–786. [DOI] [PubMed] [Google Scholar]
- 336.Hao D, Sarfaraz MO, Farshidfar F, et al. Temporal characterization of serum metabolite signatures in lung cancer patients undergoing treatment. Metabolomics : Official journal of the Metabolomic Society. 2016;12:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Roś-Mazurczyk M, Wojakowska A, Marczak Ł, et al. Ionizing radiation affects profile of serum metabolites: increased level of 3-hydroxybutyric acid in serum of cancer patients treated with radiotherapy. Acta biochimica Polonica. 2017;64(1):189–193. [DOI] [PubMed] [Google Scholar]
- 338.Mörén L, Wibom C, Bergström P, Johansson M, Antti H, Bergenheim AT. Characterization of the serum metabolome following radiation treatment in patients with high-grade gliomas. Radiat Oncol. 2016;11:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Ahlers I. Changes in whole-body metabolic parameters associated with radiation. Advances in space research : the official journal of the Committee on Space Research (COSPAR). 1994;14(10):531–539. [DOI] [PubMed] [Google Scholar]
- 340.Laiakis EC, Canadell MP, Grilj V, et al. Serum lipidomic analysis from mixed neutron/X-ray radiation fields reveals a hyperlipidemic and pro-inflammatory phenotype. Scientific Reports. 2019;9(1):4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Tyburski JB, Patterson AD, Krausz KW, et al. Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal gamma-radiation exposure in mice. Radiat Res. 2009;172(1):42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Manna SK, Krausz KW, Bonzo JA, Idle JR, Gonzalez FJ. Metabolomics reveals aging-associated attenuation of noninvasive radiation biomarkers in mice: potential role of polyamine catabolism and incoherent DNA damage-repair. Journal of proteome research. 2013;12(5):2269–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Laiakis EC, Canadell MP, Grilj V, et al. Small Molecule Responses to Sequential Irradiation with Neutrons and Photons for Biodosimetry Applications: An Initial Assessment. Radiat Res. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Pannkuk EL, Fornace AJ, Jr., Laiakis EC. Metabolomic applications in radiation biodosimetry: exploring radiation effects through small molecules. International journal of radiation biology. 2017;93(10):1151–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Pannkuk EL, Laiakis EC, Girgis M, et al. Biofluid Metabolomics of Mice Exposed to External Low-Dose Rate Radiation in a Novel Irradiation System, the Variable Dose-Rate External 137Cs Irradiator. Journal of proteome research. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Patterson AD, Li H, Eichler GS, et al. UPLC-ESI-TOFMS-based metabolomics and gene expression dynamics inspector self-organizing metabolomic maps as tools for understanding the cellular response to ionizing radiation. Anal Chem. 2008;80(3):665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Hu ZP, Kim YM, Sowa MB, et al. Metabolomic response of human skin tissue to low dose ionizing radiation. Molecular bioSystems. 2012;8(7):1979–1986. [DOI] [PubMed] [Google Scholar]
- 348.Taraboletti A, Goudarzi M, Kabir A, et al. Fabric Phase Sorptive Extraction-A Metabolomic Preprocessing Approach for Ionizing Radiation Exposure Assessment. Journal of proteome research. 2019;18(8):3020–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.White MY, Edwards AVG, Cordwell SJ, Van Eyk JE. Mitochondria: A mirror into cellular dysfunction in heart disease. PROTEOMICS – Clinical Applications. 2008;2(6):845–861. [DOI] [PubMed] [Google Scholar]
- 350.Livingston K, Schlaak RA, Puckett LL, Bergom C. The Role of Mitochondrial Dysfunction in Radiation-Induced Heart Disease: From Bench to Bedside. Frontiers in cardiovascular medicine. 2020;7:20–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Barjaktarovic Z, Schmaltz D, Shyla A, et al. Radiation-induced signaling results in mitochondrial impairment in mouse heart at 4 weeks after exposure to X-rays. PLoS One. 2011;6(12):e27811-e27811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Barjaktarovic Z, Schmaltz D, Shyla A, et al. Radiation-induced signaling results in mitochondrial impairment in mouse heart at 4 weeks after exposure to X-rays. PLoS One. 2011;6(12):e27811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Schlaak RA, Frei A, SenthilKumar G, et al. Differences in Expression of Mitochondrial Complexes Due to Genetic Variants May Alter Sensitivity to Radiation-Induced Cardiac Dysfunction. Frontiers in cardiovascular medicine. 2020;7(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Bartoletti-Stella A, Mariani E, Kurelac I, et al. Gamma rays induce a p53-independent mitochondrial biogenesis that is counter-regulated by HIF1α. Cell Death & Disease. 2013;4(6):e663–e663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Barker HE, Paget JT, Khan AA, Harrington KJ. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nature reviews Cancer. 2015;15(7):409–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Tommelein J, De Vlieghere E, Verset L, et al. Radiotherapy-Activated Cancer-Associated Fibroblasts Promote Tumor Progression through Paracrine IGF1R Activation. Cancer research. 2018;78(3):659–670. [DOI] [PubMed] [Google Scholar]
- 357.Zhong J, Rajaram N, Brizel DM, et al. Radiation induces aerobic glycolysis through reactive oxygen species. Radiother Oncol. 2013;106(3):390–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Yang X, Lu Y, Hang J, et al. Lactate-Modulated Immunosuppression of Myeloid-Derived Suppressor Cells Contributes to the Radioresistance of Pancreatic Cancer. Cancer Immunol Res. 2020;8(11):1440–1451. [DOI] [PubMed] [Google Scholar]
- 359.Estrella V, Chen T, Lloyd M, et al. Acidity generated by the tumor microenvironment drives local invasion. Cancer research. 2013;73(5):1524–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Boedtkjer E, Pedersen SF. The Acidic Tumor Microenvironment as a Driver of Cancer. Annu Rev Physiol. 2020;82:103–126. [DOI] [PubMed] [Google Scholar]
- 361.Kolosenko I, Avnet S, Baldini N, Viklund J, De Milito A. Therapeutic implications of tumor interstitial acidification. Seminars in cancer biology. 2017;43:119–133. [DOI] [PubMed] [Google Scholar]
- 362.Renner K, Singer K, Koehl GE, et al. Metabolic Hallmarks of Tumor and Immune Cells in the Tumor Microenvironment. Front Immunol. 2017;8:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Vegran F, Boidot R, Michiels C, Sonveaux P, Feron O. Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-kappaB/IL-8 pathway that drives tumor angiogenesis. Cancer research. 2011;71(7):2550–2560. [DOI] [PubMed] [Google Scholar]
- 364.Wang D, Quiros J, Mahuron K, et al. Targeting EZH2 Reprograms Intratumoral Regulatory T Cells to Enhance Cancer Immunity. Cell Rep. 2018;23(11):3262–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Delgoffe GM, Woo SR, Turnis ME, et al. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature. 2013;501(7466):252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Liao Z, Tan ZW, Zhu P, Tan NS. Cancer-associated fibroblasts in tumor microenvironment - Accomplices in tumor malignancy. Cell Immunol. 2019;343:103729. [DOI] [PubMed] [Google Scholar]
- 367.Ohuchida K, Mizumoto K, Murakami M, et al. Radiation to stromal fibroblasts increases invasiveness of pancreatic cancer cells through tumor-stromal interactions. Cancer research. 2004;64(9):3215–3222. [DOI] [PubMed] [Google Scholar]
- 368.Pereira PMR, Edwards KJ, Mandleywala K, et al. iNOS Regulates the Therapeutic Response of Pancreatic Cancer Cells to Radiotherapy. Cancer research. 2020;80(8):1681–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Shimura T, Sasatani M, Kawai H, et al. Radiation-Induced Myofibroblasts Promote Tumor Growth via Mitochondrial ROS-Activated TGFbeta Signaling. Mol Cancer Res. 2018;16(11):1676–1686. [DOI] [PubMed] [Google Scholar]
- 370.Reina-Campos M, Moscat J, Diaz-Meco M. Metabolism shapes the tumor microenvironment. Curr Opin Cell Biol. 2017;48:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Yang L, Achreja A, Yeung TL, et al. Targeting Stromal Glutamine Synthetase in Tumors Disrupts Tumor Microenvironment-Regulated Cancer Cell Growth. Cell Metab. 2016;24(5):685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Sousa CM, Biancur DE, Wang X, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536(7617):479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Li L, Zhu L, Hao B, et al. iNOS-derived nitric oxide promotes glycolysis by inducing pyruvate kinase M2 nuclear translocation in ovarian cancer. Oncotarget. 2017;8(20):33047–33063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Becker LM, O’Connell JT, Vo AP, et al. Epigenetic Reprogramming of Cancer-Associated Fibroblasts Deregulates Glucose Metabolism and Facilitates Progression of Breast Cancer. Cell Rep. 2020;31(9):107701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Harada H, Inoue M, Itasaka S, et al. Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate towards tumour blood vessels. Nat Commun. 2012;3:783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer cell. 2004;5(5):429–441. [DOI] [PubMed] [Google Scholar]
- 377.Huang R, Zhou P-K. HIF-1 signaling: A key orchestrator of cancer radioresistance. Radiation Medicine and Protection. 2020;1(1):7–14. [Google Scholar]
- 378.Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123(9):3664–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Lee G, Won HS, Lee YM, et al. Oxidative Dimerization of PHD2 is Responsible for its Inactivation and Contributes to Metabolic Reprogramming via HIF-1α Activation. Sci Rep. 2016;6:18928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci U S A. 1998;95(20):11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3(3):177–185. [DOI] [PubMed] [Google Scholar]
- 383.Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J Biol Chem. 2001;276(12):9519–9525. [DOI] [PubMed] [Google Scholar]
- 384.Lin X, Zhang F, Bradbury CM, et al. 2-Deoxy-D-glucose-induced cytotoxicity and radiosensitization in tumor cells is mediated via disruptions in thiol metabolism. Cancer research. 2003;63(12):3413–3417. [PubMed] [Google Scholar]
- 385.Coleman MC, Asbury CR, Daniels D, et al. 2-deoxy-D-glucose causes cytotoxicity, oxidative stress, and radiosensitization in pancreatic cancer. Free Radic Biol Med. 2008;44(3):322–331. [DOI] [PubMed] [Google Scholar]
- 386.Rashmi R, Huang X, Floberg JM, et al. Radioresistant Cervical Cancers Are Sensitive to Inhibition of Glycolysis and Redox Metabolism. Cancer research. 2018;78(6):1392–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Dwarakanath BS, Singh D, Banerji AK, et al. Clinical studies for improving radiotherapy with 2-deoxy-D-glucose: present status and future prospects. Journal of cancer research and therapeutics. 2009;5 Suppl 1:S21–26. [DOI] [PubMed] [Google Scholar]
- 388.Singh D, Banerji AK, Dwarakanath BS, et al. Optimizing cancer radiotherapy with 2-deoxy-d-glucose dose escalation studies in patients with glioblastoma multiforme. Strahlentherapie und Onkologie : Organ der Deutschen Rontgengesellschaft [et al]. 2005;181(8):507–514. [DOI] [PubMed] [Google Scholar]
- 389.Pajak B, Siwiak E, Sołtyka M, et al. 2-Deoxy-d-Glucose and Its Analogs: From Diagnostic to Therapeutic Agents. Int J Mol Sci. 2019;21(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Peng Y, Xing SN, Tang HY, et al. Influence of glucose transporter 1 activity inhibition on neuroblastoma in vitro. Gene. 2019;689:11–17. [DOI] [PubMed] [Google Scholar]
- 391.Singh PK, Deorukhkar AA, Venkatesulu BP, et al. Exploiting Arginine Auxotrophy with Pegylated Arginine Deiminase (ADI-PEG20) to Sensitize Pancreatic Cancer to Radiotherapy via Metabolic Dysregulation. Molecular cancer therapeutics. 2019;18(12):2381–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Oslob JD, Johnson RJ, Cai H, et al. Imidazopyridine-Based Fatty Acid Synthase Inhibitors That Show Anti-HCV Activity and in Vivo Target Modulation. ACS medicinal chemistry letters. 2013;4(1):113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Fhu CW, Ali A. Fatty Acid Synthase: An Emerging Target in Cancer. Molecules. 2020;25(17):3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Qu H, Shan K, Tang C, et al. A novel small-molecule fatty acid synthase inhibitor with antitumor activity by cell cycle arrest and cell division inhibition. European journal of medicinal chemistry. 2021;219:113407. [DOI] [PubMed] [Google Scholar]
- 395.Chuang HY, Lee YP, Lin WC, Lin YH, Hwang JJ. Fatty Acid Inhibition Sensitizes Androgen-Dependent and -Independent Prostate Cancer to Radiotherapy via FASN/NF-κB Pathway. Sci Rep. 2019;9(1):13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Rae C, Haberkorn U, Babich JW, Mairs RJ. Inhibition of Fatty Acid Synthase Sensitizes Prostate Cancer Cells to Radiotherapy. Radiat Res. 2015;184(5):482–493. [DOI] [PubMed] [Google Scholar]
- 397.Rae C, Fragkoulis GI, Chalmers AJ. Cytotoxicity and Radiosensitizing Activity of the Fatty Acid Synthase Inhibitor C75 Is Enhanced by Blocking Fatty Acid Uptake in Prostate Cancer Cells. Advances in Radiation Oncology. 2020;5(5):994–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Skinner H, Hu C, Tsakiridis T, et al. Addition of Metformin to Concurrent Chemoradiation in Patients With Locally Advanced Non–Small Cell Lung Cancer: The NRG-LU001 Phase 2 Randomized Clinical Trial. JAMA Oncology. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Chun SG, Liao Z, Jeter MD, et al. Metabolic Responses to Metformin in Inoperable Early-stage Non–Small Cell Lung Cancer Treated With Stereotactic Radiotherapy: Results of a Randomized Phase II Clinical Trial. American Journal of Clinical Oncology. 2020;43(4). [DOI] [PubMed] [Google Scholar]
- 400.Liu Y, He C, Huang X. Metformin partially reverses the carboplatin-resistance in NSCLC by inhibiting glucose metabolism. Oncotarget. 2017;8(43):75206–75216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Ashinuma H, Takiguchi Y, Kitazono S, et al. Antiproliferative action of metformin in human lung cancer cell lines. Oncol Rep. 2012;28(1):8–14. [DOI] [PubMed] [Google Scholar]
- 402.Eze C, Belka C, Manapov F. Forging a Path for Metformin Use in Inoperable Locally Advanced Non–Small Cell Lung Cancer. JAMA Oncology. 2021. [DOI] [PubMed] [Google Scholar]
- 403.Buckley AM, Dunne MR, Lynam-Lennon N, et al. Pyrazinib (P3), [(E)-2-(2-Pyrazin-2-yl-vinyl)-phenol], a small molecule pyrazine compound enhances radiosensitivity in oesophageal adenocarcinoma. Cancer letters. 2019;447:115–129. [DOI] [PubMed] [Google Scholar]
- 404.Gross MI, Demo SD, Dennison JB, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Molecular cancer therapeutics. 2014;13(4):890–901. [DOI] [PubMed] [Google Scholar]
- 405.Wang H, Bouzakoura S, de Mey S, et al. Auranofin radiosensitizes tumor cells through targeting thioredoxin reductase and resulting overproduction of reactive oxygen species. Oncotarget. 2017;8(22):35728–35742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Shukla K, Ferraris DV, Thomas AG, et al. Design, synthesis, and pharmacological evaluation of bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide 3 (BPTES) analogs as glutaminase inhibitors. Journal of medicinal chemistry. 2012;55(23):10551–10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Wicker CA, Hunt BG, Krishnan S, et al. Glutaminase inhibition with telaglenastat (CB-839) improves treatment response in combination with ionizing radiation in head and neck squamous cell carcinoma models. Cancer letters. 2021;502:180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Meric-Bernstam F, Lee RJ, Carthon BC, et al. CB-839, a glutaminase inhibitor, in combination with cabozantinib in patients with clear cell and papillary metastatic renal cell cancer (mRCC): Results of a phase I study. Journal of Clinical Oncology. 2019;37(7_suppl):549–549. [Google Scholar]