Abstract
The large diversity of cell types in nervous systems presents a challenge in identifying the genetic mechanisms that encode it. Here, we report that nearly 200 distinct neurons in the Drosophila visual system can each be defined by unique combinations of ~10 continuously expressed transcription factors. We show that targeted modifications of this terminal selector code induce predictable conversions of neuronal fates that appear morphologically and transcriptionally complete. Cis-regulatory analysis of open chromatin links one of these genes to an upstream patterning factor that specifies neuronal fates in stem cells. Experimentally validated network models describe the synergistic regulation of downstream effectors by terminal selectors and ecdysone signaling during brain wiring. Our results provide a generalizable framework of how specific fates are implemented in postmitotic neurons.
One sentence summary:
Neuronal diversity in the Drosophila brain is defined by a relatively simple code of continuously expressed transcriptions factors.
Introduction
Neurons are by far the most diverse of all cell types in animals. Understanding the molecular mechanisms that produce this diversity is a central goal of neurobiology. The Drosophila brain provides a tractable system to approach this challenge due to its manageable size and genetically hardwired development. The optic lobes constitute two thirds of the fly brain and each of their neuropils: lamina, medulla, lobula and lobula plate (Fig. 1A) is divided into ~800 columns, corresponding to the same number of ommatidia (unit eyes) in the retina. Because of this retinotopic organization with multiple repeats of the same circuits, most neuronal types are present in high number of cells per brain. We previously completed a large single-cell RNA sequencing (scRNA-seq) atlas of the optic lobes, resolving around 200 cell types that we consistently tracked across 6 timepoints from early pupal stages to adult (1). Almost all annotated clusters in this atlas correspond to a distinct neuronal type with unique morphology (2). This strongly suggests that most of our clusters represent biologically homogeneous groups, giving us access to the cell-type specific transcriptome of every neuron throughout its development.
Figure 1: Pdm3 instructs complete switches of neuronal fates.
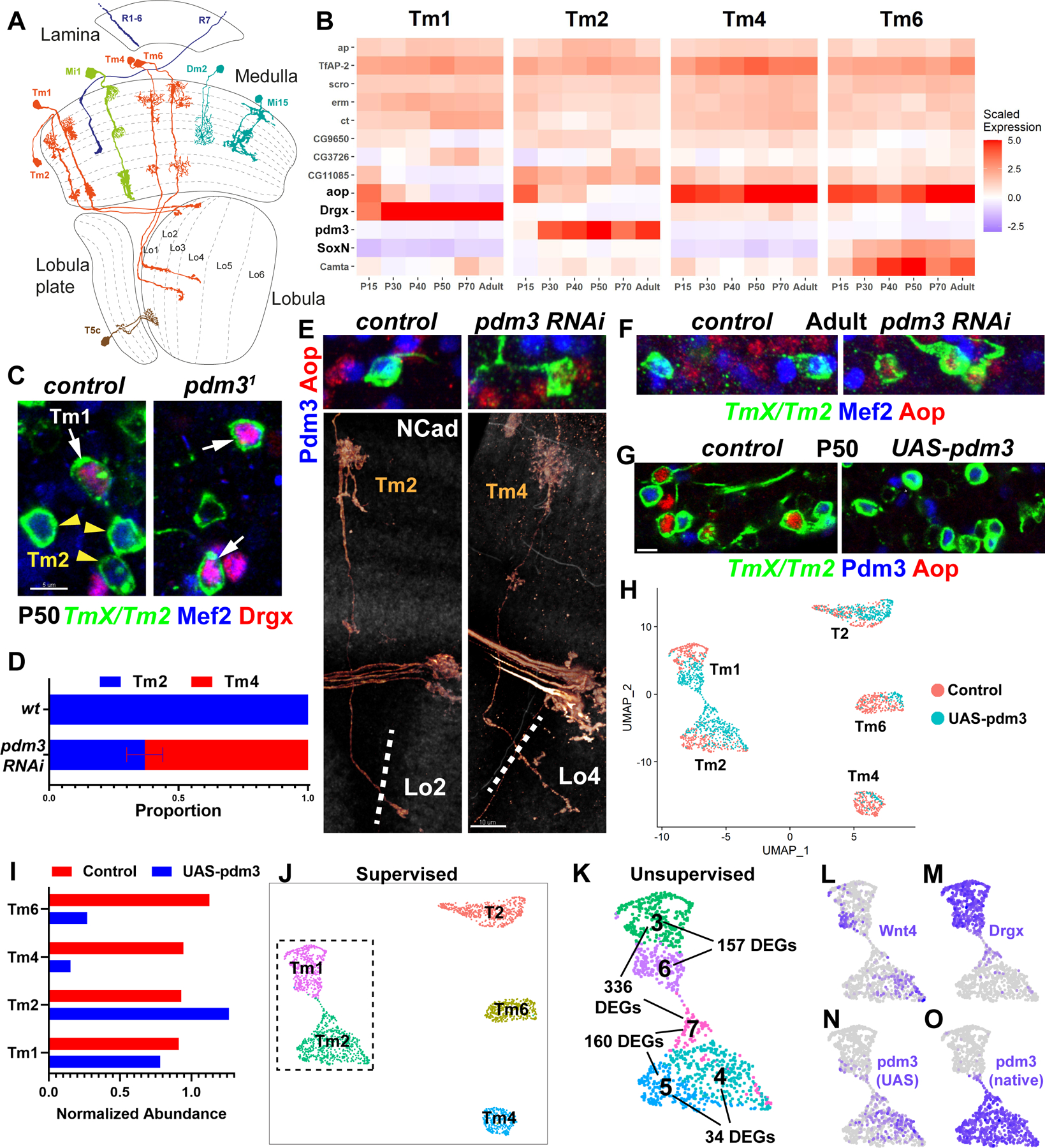
A, D. melanogaster optic lobe in cross-section, with drawings of select cell types. Partially adapted from (2). B, Developmental (scaled) expression patterns (1) of all genes that are candidate selectors in any of the displayed cell types. C, FRT40A and pdm31 MARCM clones labeled with TmX/Tm2-Gal4 and CD4-tdGFP in P50 brains (maximum projection), with anti-Mef2 (blue) and anti-Drgx (red). n= 8 (control) and 4 (mutant) brains. D-F, TmX/Tm2-Gal4 driving pdm3 RNAi and CD4-tdGFP (flip-out). D, Quantification of E-F. n= 54/6 (control), 96/8 (RNAi) neurons/brains, p<0.0001. Error bar denotes SEM. E, 3D reconstructions of GFP (bottom) or max projections (top) for the same representative adult neurons in each condition, with anti-NCad (white), anti-Pdm3 (blue) and anti-Aop (red). The dashed lines indicate the border of the lobula neuropil based on NCad staining. F, Same as (E, top) with anti-Mef2 instead of Pdm3. G, TmX/Tm2-Gal4 driving UAS-pdm3 and CD4-tdGFP (flip-out). Max. projections of somas in P50 medulla cortex, with anti-Pdm3 (blue) and anti-Aop (red). n=129 (control), 118 (pdm3) neurons. Scale bars: 5 μm (C, E-top, F, G) and 10 μm (E-bottom). H-O, scRNA-seq of FACSed neurons, same experiment as (G). UMAP visualizations were calculated using top 6 principal components. Cells are colored according to library (condition) of origin (H), supervised classifications (J), unsupervised clustering (K, inset only, see also Fig. S4A, DEGs: differentially expressed genes) and the log-normalized expression of indicated genes (L-O). I, Numbers of Tm neurons in each library, divided by the number of T2 neurons.
The identity of optic lobe neurons is specified deterministically by their progenitors during neurogenesis that occurs from late larval stages (L3) until about 20% of pupal development (P20) (3). Neurons from the medulla neuropil are produced from a neuroepithelium called the outer proliferation center, which is progressively converted into neuroblasts that asymmetrically divide multiple times, each time self-renewing and producing an intermediate progenitor that divides once to generate two different neurons (4). Neurons are diversified by the intersection of three patterning mechanisms: Compartmentalization of the neuroepithelium into at least 8 spatial regions by transcription factors (TFs) and signaling molecules (5), sequential expression of at least 11 temporal TFs (tTFs) in neuroblasts (6), and Notch signaling between sister neurons (7). Similar patterning mechanisms are also utilized in other parts of the fly brain, as well as mammalian neural stem cells to generate diversity (reviewed in 8). However, most spatial and tTFs are not maintained in neurons (6), and thus it is not clear how these cell fate decisions are implemented and maintained in postmitotic neurons.
Much of our knowledge about neuronal identity control originates from C. elegans. The terminal selector hypothesis (9) posits that type-specific gene expression in neurons is controlled by combinations of TFs that are continuously maintained in each neuron throughout its life. Terminal selectors control both the developmental features such as synaptic connectivity (10) and the functional features such as neurotransmitter identity (11), but they are largely not required for the pan-neuronal gene expression programs (12). This model also implies that individual selectors do not specialize in distinct phenotypic features of a neuron. Although a few TFs that could function as terminal selectors have been identified in mammalian neurons (13–15), it remains unclear how generally applicable this regulatory logic is beyond the relatively simple nervous system of worms. Moreover, the ultimate test of this model, i.e. the predictive and complete transformation of one neuronal type into another through targeted modification of its selector code, has been difficult to assess, even in C. elegans (16).
Results
Terminal selectors of optic lobe neurons
To investigate if a sustained code of TFs maintains the identity of each neuron throughout development, we sought to identify the combinations of candidate terminal selectors expressed in each of the 174 neuronal clusters in our scRNA-seq atlas (1). We determined the sets of TFs continuously expressed in each cluster throughout all six stages of development (P15 to Adult), excluding those expressed in all clusters (pan-neuronal or ubiquitous genes, see Methods). We found on average unique combinations of 10 such genes per cluster, representing 95 TFs in total (Fig. S1A, Table S1); 72 of these were expressed in fewer than 25 clusters (Fig. S1B). Homeobox genes were enriched in this list (Fig. S1C), but unlike in the C. elegans nervous system (17), they were not sufficient to uniquely define every neuron. Furthermore, while the selectors could delineate developmentally related lineages e.g., from lamina or inner proliferation center (Fig. S2A), homeobox genes alone could not (Fig. S2B).
Terminal selector hypothesis predicts that, if continuously maintained TFs are primarily responsible for cell type-specific neuronal differentiation, it should be possible to engineer complete switches of identity between different neurons by modifying these TFs alone. All genes that were previously reported to interfere with neuronal type identity in the optic lobe, including bsh, hth, drifter (vvl), Lim1, erm, SoxN and Sox102F (18–22), were indeed candidate selectors for the respective neurons (Fig. S1A). However, these studies generally reported disruptions rather than switches of morphological identity: e.g. loss of hth/bsh in Mi1 results in an incomplete conversion to “Tm1-like” neurons (19), likely because Tm1 differs from Mi1 by the expression of the additional selectors Drgx and TfAP-2. It remains challenging to simultaneously perturb more than one or two genes at once using classical genetic methods. In order to provide definitive evidence for the sufficiency of terminal selectors in determining neuronal type identity, we thereby looked for groups of closely related neurons whose selector codes differ only by one or two genes, where complete conversions from one cell-type to another may be feasible.
Transmedullary (Tm) neurons 1, 2, 4 and an unidentified cluster (#62) have nearly indistinguishable transcriptomes shortly after their terminal division at P15 (1), suggesting a very close developmental relationship. We annotated cluster 62 as Tm6 neurons, based on its expression of the unique combination of aop, SoxN and Wnt10 (Fig. S3A–C). Although these Tms share similar overall morphology, adult neurons are readily distinguishable from one another by their distinct dendritic shapes as well as the different target layers of their axons in the lobula (Fig. 1A). Analysis of candidate selector expression in these clusters across development (Fig. 1B) revealed that the 4 neurons indeed share a similar code: ap, TfAP-2, scro, erm and ct are continuously expressed in all four clusters. CG9650, CG3726, CG11085 and aop could also be found in all 4 Tm neurons at some point during development, though they are only transiently expressed in some of them. Camta is expressed at much higher levels in Tm6 but is also detected in the others. Among these 4 neurons, Drgx is specific to Tm1, pdm3 to Tm2 and SoxN to Tm6, while there are no candidate selectors exclusive to Tm4. Therefore, these TFs that are each continuously and specifically expressed in one of these types are strong candidates to differentially specify their fates.
Pdm3 instructs transcriptionally complete neuronal fate conversions
As pdm3 is the only TF that continuously distinguishes Tm2 from Tm4 (Fig. 1B), the terminal selector code predicts that its loss should reprogram Tm2 neurons to Tm4 fate. R71F05-Gal4 is expressed in all 4 Tm neurons (1/2/4/6) until P50; however, it is only maintained in Tm2 in adults (Fig. S3D, TmX/Tm2-Gal4). Using this driver, we generated MARCM (23) clones of a pdm3 null allele (24). No mutant Tm neurons were recovered in adult brains, suggesting that Tm2 were not specified properly (Fig. S3E). Mef2 is an effector (downstream) TF normally expressed specifically in both Tm1 and Tm2 after P40 (Figs. 1C, S7J). At P50, we observed that the only remaining Mef2+ Tm neurons in pdm31 clones were Tm1 that expressed Drgx (Fig. 1C), indicating that Tm2 were either lost or converted to another fate. Unlike the mutant, upon RNAi knock-down of pdm3 using TmX/Tm2-Gal4, 65% of Tm2s that retained the expression of the driver in adult brains were converted to neurons with Tm4 morphology, characterized by wider dendritic arbors that are symmetrical around the main fiber of the neuron as well as axons targeting the deeper lobula layer 4 (Fig. 1D–E, compare to 1A). It is likely that the knock-down retains low levels of Pdm3 in Tm2 that are sufficient to maintain expression of TmX/Tm2-Gal4, but are insufficient for instructing the Tm2 fate. These Tm4-looking neurons did not express Mef2 and instead expressed the putative Tm4 selector Aop (Fig. 1E–F).
We then asked whether ectopic expression of pdm3 in Tm4 and Tm6 could be sufficient to convert them to Tm2 fate. We used TmX/Tm2-Gal4 to express UAS-pdm3.short (25) and found that more than 90% of Aop+ neurons (Tm4 and Tm6) were eliminated at P50 (Fig. 1G), suggesting that they had been lost or converted. To address the completeness of these conversions at P50, when the neurons have not fully acquired their adult morphology but display the greatest transcriptomic diversity (1), we analyzed their gene expression with scRNA-seq. As the driver weakly labels several other cell-types (Fig. S4A, see Methods), we only retained the cells classified as Tm1/2/4/6 by a neural-network trained on our reference atlas (1), in addition to those classified as T2 that are also strongly labeled by TmX/Tm2-Gal4 (Fig. 1H,J). T2 neurons, like Tm2, natively express pdm3, and thus they should not be affected by this perturbation and serve as an internal control. We observed a depletion of Tm4 and Tm6 in the UAS-pdm3 library compared to control and an increase in the number of Tm2s (Fig. 1I), indicating that ectopic pdm3 converts Tm4 and Tm6 to Tm2. We noted that the increased number of Tm2s upon pdm3 overexpression was not sufficient to fully account for the lost Tm4s and Tm6s. Some optic lobe neurons are known to be generated in excess, followed by widespread apoptosis in the first half of pupal development (26). Staining against cleaved Dcp-1, an activated caspase that marks dying cells (27), indeed showed significantly increased rate of apoptosis in brains overexpressing pdm3 at P25 (Fig. S4B–C), while no Tm2 neurons (GFP+Pdm3+) were stained with Dcp-1 in the control brains. Together, these results suggest that when excess Tm2 neurons are produced through conversions from Tm4 and Tm6, this is compensated by increased cell death. This mechanism potentially helps ensure that only one Tm neuron of each type is present per column in wild-type brains.
To distinguish the wild-type Tm2 neurons from those converted from another cell type, we performed unsupervised clustering on the dataset. This revealed heterogeneous populations among the cells classified as Tm1 and Tm2 (Figs. S4D, 1K). Tm2 subclusters 4 and 5 were extremely similar, with only 34 significant differentially expressed genes (DEGs). Most of these differences were consistent with markers of the control Tm4/6 clusters (Fig. S4E), including the strongest one, Wnt4, found in cluster 4 (Fig. 1L). We recently showed that Wnt4 is expressed in ventral Tm4 and Tm6, but not in Tm2 (1), suggesting that cells in cluster 4 were converted neurons that had retained these markers from their initial specification as Tm4/6. Nevertheless, these differences between the converted and ‘original’ Tm2s were minimal, as compared to more than 700 DEGs observed between wild-type Tm2 and Tm4 at this stage (Table S2). We therefore conclude that conversion from Tm4/6 to Tm2 induced by ectopic pdm3 appears complete.
The third subgroup of the cells classified as Tm2, cluster 7, consisted entirely of cells from the UAS-pdm3 library and expressed the Tm1 selector Drgx (Fig. 1M), suggesting that they were originally Tm1s converted to a Tm2-like state. These were still significantly different from cluster 5 (wt Tm2) with 160 DEGs (Fig. 1K). The cells classified as Tm1 were clustered into two groups: cluster 6, made entirely of cells from the UAS-pdm3 library, and was significantly different from cluster 3 that consisted essentially of wt Tm1s (Fig. 1H,K). Thus, both clusters 6 and 7 contained Tm1s with ectopic pdm3. UMAP visualization showed a thin stripe of cells bridging the Tm1-like (cluster 6) and Tm2-like (cluster 7) states. We observed that while both clusters 6 and 7 displayed reads coming from the UAS-pdm3 construct as expected (Fig. 1N), cluster 7 also expressed pdm3 from the native locus (Fig. 1O). We thereby conclude that the amount of protein produced from the UAS construct is insufficient for conversion into Tm2, but instead Pdm3 must autoactivate above a certain threshold. Once this threshold is reached, Pdm3 quickly drives Tm1 and Tm4/6 to a Tm2-like state; however, this conversion is incomplete in Tm1 (cluster 7) since Drgx remains expressed. Morphologically, Tm1 neurons overexpressing pdm3 appeared normal in adults (Fig. S3F), suggesting that the 157 DEGs between clusters 3 and 6 are not important for morphology.
In summary, pdm3 is necessary and sufficient to instruct the fate choice between Tm2 and Tm4 neurons, as predicted by the terminal selector code. Its loss results in morphological conversion of Tm2 into Tm4, and its ectopic expression can induce essentially complete transcriptomic conversions of Tm4 and Tm6 to Tm2 fate. It is also an upstream repressor of the Tm4/6 selector aop (Fig. 1E,G).
Tm1 selector Drgx is regulated by Klumpfuss
Similar to pdm3 in Tm2, Drgx is the only TF that continuously distinguishes Tm1 from Tm4 (Fig. 1B). We ectopically expressed Drgx using R35H01-Gal4, which is expressed in all 4 Tm neurons until P50 but is only maintained in Tm4 and Tm6 in adults (Fig. S5A–B, TmX/Tm4,6-Gal4). In these adult brains, the proportion of Tm6 remained unchanged, but most Tm4s were converted into Tm1s, characterized by much narrower dendritic arbors and axons terminating in the first layer of the lobula (Fig. 2A–B, see Fig. 1A). The converted neurons also lost Aop expression and instead expressed Mef2 (Tm1/2 marker) (Fig. 2C). Some of the converted neurons displayed morphological features atypical of Tm1, such as targeting to the Lo2 layer instead of Lo1 (Fig. S5C). We suspect that this partial expressivity is due to low Gal4 expression from TmX/Tm4,6-Gal4. Moreover, the fact that this driver is expressed at even weaker levels in Tm6 compared to Tm4 (Fig. S5B) might explain our failure to affect Tm6 fate. Loss of Drgx (described below) resulted in conversion of Tm1 neurons into Tm4 (Fig. 2G–H). Thus, Drgx specifies the Tm1 fate; it can repress aop and mediate the conversion of Tm4 into Tm1 as predicted by the selector code.
Figure 2. Tm1 selector Drgx is regulated by Klumpfuss.
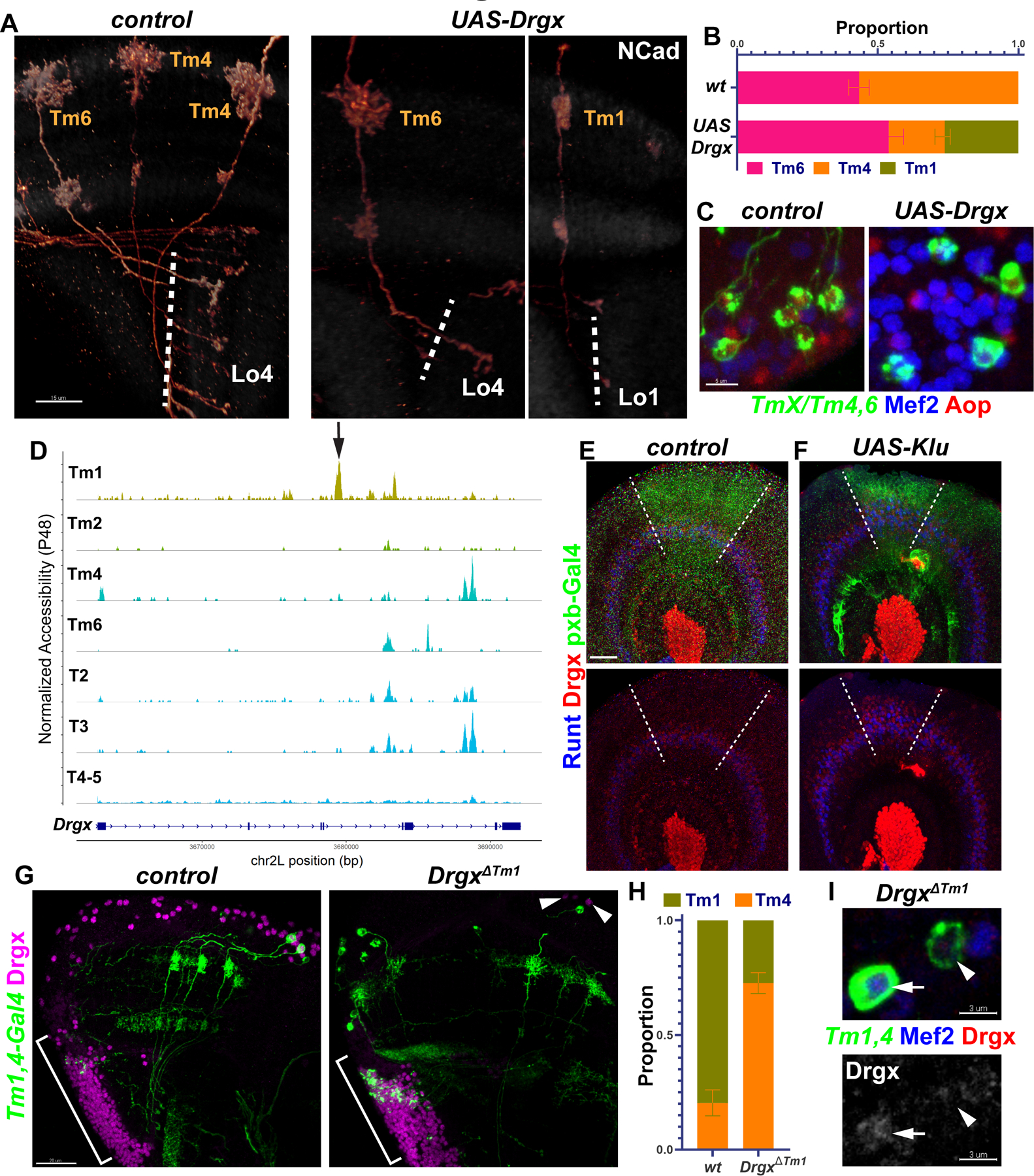
A-C, TmX/Tm4,6-Gal4 driving UAS-Drgx and CD4-tdGFP (flip-out). A, 3D reconstructions of GFP for representative adult neurons, with anti-NCad (white). Dashed lines mark the border of lobula neuropil. B, Quantification of A. n= 92/4 (control), 45/6 (Drgx) neurons/brain, p=0.0003. C, Same as (A) with max. projections of somas with anti-Mef2 (blue) and anti-Aop (red). D, Aggregated accessibility tracks of Drgx locus from the TF-IDF normalized snATAC-seq data at P48 (28). Arrow: Tm1-specific enhancer deleted in (G-I). E-F, pxb-Gal4 driving CD8-GFP and UAS-Klu (F, n=5 brains) in L3 optic lobes, with anti-Runt (blue) and anti-Drgx (red). Dashed lines mark the borders of driver expression. G-I, Tm1,4-Gal4 driving CD4-tdGFP (flip-out) in heterozygous (control) or homozygous DrgxΔTm1 mutants. G, Max projections of adult optic lobes with anti-Drgx (magenta). Brackets mark the location of the lobula plate cortex (T2–5 neurons). Arrowheads: glia (see Fig. S5E) that maintain Drgx expression in the mutants. H, Quantification of G (see also Fig. S5D). Tm1 were normally observed more frequently than Tm4 as the driver expression is much lower in Tm4. n = 57/6 (control) and 181/10 (DrgxΔTm1) neurons/brains, p<0.0001. I, Same as (G), displaying instead somas with anti-Mef2 (blue) and anti-Drgx (red), or only anti-Drgx (bottom). Arrow: Tm1, Arrowhead: Tm4. Scale bars: 15 μm (A,E,F), 5 μm (C), 20 μm (G), 3 μm (I). Error bars denote SEM.
Neither Drgx nor pdm3 is expressed in the progenitors (neuroblasts) of Tm neurons (6), implying that their postmitotic expression in specific neurons is instructed by tTFs in the neuroblasts. To investigate how this is controlled, we used a single-nucleus ATAC-seq (chromatin accessibility) dataset of the developing Drosophila brain (28). We identified the cells belonging to optic lobe neurons at adult, P48 and P24 stages, re-clustered and annotated them using our scRNA-seq atlas (1) as reference (Fig. S6A–D, Methods). We found a putative enhancer in the 4th intron of Drgx that was specifically accessible in Tm1 throughout development (Fig. S6E) but was not accessible in the other Tm neurons, or in T2, T3 and T4/5, which also express Drgx (Fig. 2D). We found that the only enriched binding motifs for any of the TFs expressed in the optic lobe (E-value<100, see Methods) within this 700bp region belonged to the tTF Klumpfuss (Klu). Klu is expressed at higher levels in neuroblasts during early temporal windows, when Tm1 is generated, and its overexpression in neuroblasts can expand Runt+ neurons (29) that are likely born in the same temporal window as Tm1 (Fig. 2E) (6). Thus, Klu might also regulate Drgx expression. Indeed, Klu overexpression using pxb-Gal4, which is expressed in the central region of the neuroepithelium (Fig. 2E–F, dashed lines), resulted in the expansion of Drgx+ neurons (i.e. Tm1) in this region, similar to Runt (Fig. 2F). We therefore conclude that Klu expression in neuroblasts helps specify Tm1 from an early temporal window by activating the selector Drgx in their neuronal progeny.
Next, we asked whether Drgx expression is regulated by this enhancer element by engineering a CRISPR deletion (DrgxΔTm1, Methods), which should function as a conditional mutant specifically in Tm1 neurons. 27b-Gal4 (30) is expressed in Tm1, and much more weakly in Tm4 (Fig. S5D, Tm1,4-Gal4) throughout development. In DrgxΔTm1 mutant adults, Drgx expression in medulla cortex (where all Tm somas are located) was almost completely lost (Fig. 2G) but was still normally present in Repo+ perineurial glia (Fig. S5E) at the surface of the brain (Fig. 2G, arrowheads) and in T-neurons originating from the lobula plate (Fig. 2G, brackets). The observed ratio of Tm1/Tm4 labeled by Tm1,4-Gal4 decreased significantly in the mutant brains (Fig. 2H). This was not due to loss of Tm1s as we could observe no apoptotic Tm1 neurons at P25 in either condition (Fig. S5F–G), suggesting instead that most Tm1s were converted to Tm4. Furthermore, 69% of the few remaining Tm1s displayed abnormal morphological features such as disrupted dendritic arbors, and/or axons reaching to deeper layers in the lobula (Fig. S5D). Close examination of somas revealed that these neurons that maintained Mef2 (Tm1/2 marker) still expressed Drgx at very low levels (Fig. 2I, arrow). In addition, Drgx expression was normal in DrgxΔTm1 mutants at L3 stage (Fig. S5H). These results suggest that there are other, partially redundant enhancers regulating Drgx that control its initial activation in newly born neurons, while the robust maintenance of expression in Tm1s requires this specific enhancer. Even though it is not maintained later, Klu could be priming this enhancer in newborn Tm1s to ensure sustained expression of Drgx.
Selectors jointly control developmental and functional features
Similar to Drgx in Tm1 and pdm3 in Tm2, SoxN is the sole candidate selector distinguishing Tm6 neurons from Tm4 (Fig. 1B). In control MARCM clones marked with TmX/Tm4,6-Gal4, we observed roughly equal numbers of adult Tm4 and Tm6. In contrast, only Tm4s could be observed in SoxN null mutant (31) clones (Fig. 3A). All four Tms are unicolumnar neurons produced by all neuroblasts; therefore, a given column should contain one Tm4 and one Tm6 generated from the same neuroblast (5). We consistently observed Tm4 and Tm6 neurons occupying the same column in sparse control MARCM neuroblast clones (Fig. 3B). However, these clones consisted of two Tm4s in SoxN mutants (Fig. 3B), indicating that loss of SoxN converted Tm6 neurons to Tm4, rather than eliminating them. However, columns with these pairs were rare, suggesting that the extra Tm4s often undergo apoptosis as shown above for Tm2s. Overexpressing SoxN using TmX/Tm4,6-Gal4 did not convert Tm4s to Tm6 (Fig. S7A). This is again likely due to the weak Gal4 driver, as the amount of SoxN protein detected in these Tm4s was an order of magnitude lower than in wild-type Tm6 (Fig. S7A, insets). Thus, we could engineer predictable switches of type identity between all 4 Tm neurons guided solely by a code of sustained transcription factors.
Figure 3: Selectors jointly control developmental and functional features.
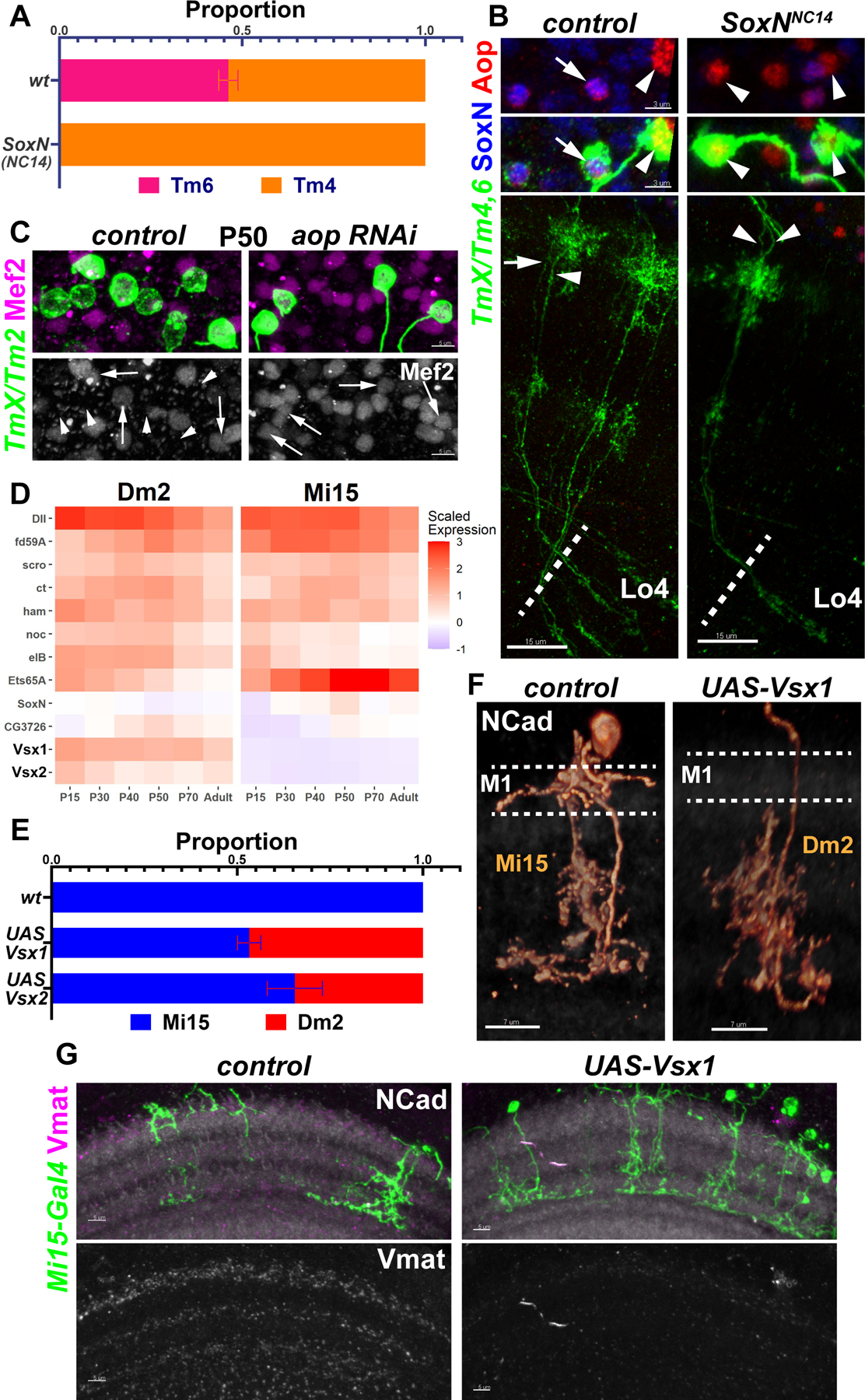
A-B, FRT40A and SoxNNC14 MARCM clones labeled with TmX/Tm4,6-Gal4 and CD4-tdGFP in adult brains. A, Quantification of B, based on Aop-only (Tm4) and Aop+SoxN (Tm6) neurons. n=225/9 (control) and 258/10 (SoxNNC14) neurons/brains, p<0.0001. No neurons with Tm6 morphology were observed in the mutant clones. B, Max projections with anti-SoxN (blue) and anti-Aop (red), displaying the neurites (bottom) and the somas (top) of the same two neurons. Arrow: Tm6, Arrowheads: Tm4. C, TmX/Tm2-Gal4 driving aop RNAi (n=6 brains) and CD4-tdGFP (flip-out). Maximum projections of somas at P50 with anti-Mef2 (top: magenta, bottom: white). Arrows: GFP+Mef2+, arrowheads: GFP+Mef2− Tm neurons. D, Developmental (scaled) expression patterns (1) of all genes that are candidate selectors in either of the displayed cell types. E, Quantification of (F) and Fig. S7D. Cells labeled by Mi15(R76F01)-Gal4 were identified based on their morphology in each condition. n=104/11 (wt), 108/11 (UAS-Vsx1) and 24/4 (UAS-Vsx2) neurons/brains, p<0.0001 for change in Dm2 proportions in both conditions. Error bars denote SEM. F-G, Mi15-Gal4 driving UAS-Vsx1 and CD4-tdGFP (flip-out). F, 3D reconstructions of GFP for representative adult neurons in each condition (see Fig. 1A), with anti-NCad (white). Dashed lines mark the M1 layer where Mi15 arborizes but Dm2 does not. Also note that Mi15 has two descending branches while Dm2 has one. G, Max projections of adult optic lobes with anti-NCad (white) and anti-Vmat (top: magenta, bottom: white). n= 4 (control) and 7 (UAS-Vsx1) brains. Scale bars: 15 μm (B), 5 μm (C,G), 7 μm (F).
Combined, our results suggest that Tm4 is the default fate among these Tm neurons, which is overridden by Drgx in Tm1, pdm3 in Tm2 and SoxN in Tm6. aop is expressed in both Tm4 and Tm6, but it is repressed by Drgx in Tm1 and by pdm3 in Tm2. To address whether Aop also functions as a selector, we generated aop null MARCM clones (32) and also performed RNAi knock-down using TmX/Tm4,6-Gal4; in both cases, the driver was turned off (Fig. S7B–C). Instead using TmX/Tm2-Gal4 to express aop RNAi, we observed that all Tm neurons at P50 (when this driver normally labels all 4 Tms) expressed the Tm1/2 marker Mef2 (Fig. 3C), indicating that aop is necessary for Tm4 and Tm6 identity. However, we could not determine the exact fate of these neurons i.e., eliminated or transformed to Tm1 or Tm2, as both were labeled by the driver.
To further validate the terminal selector concept for functional features of neurons, we sought other neuronal types whose selector codes differ only by a few genes, and in addition use different neurotransmitters. Dm2 and Mi15 are both cholinergic, but Mi15 are also the only aminergic neurons in the optic lobe (33), expressing the vesicular monoamine transporter (Vmat). They both express the candidate selectors Dll, fd59A, scro, ct, ham, noc, eIB and Ets65A, but Vsx1 and Vsx2 are specific to Dm2 and are the only TFs that continuously distinguish the two cell types (Fig. 3D). Ectopic expression of either Vsx1 or Vsx2 using an Mi15-specific (early) driver was sufficient to convert them to Dm2 morphology (Figs. 3E–F, S7D, see 1A). Vsx1 and Vsx2 could function redundantly due to their sequence similarity, or they could cross-activate each other’s expression. However, we did not observe Vsx2 protein in Mi15s ectopically expressing Vsx1 (Fig. S7E), suggesting redundancy.
Next, we evaluated these conversions for more terminal features that are likely to be important for neurotransmission. We observed a drastic reduction in Vmat protein levels in the medulla upon Vsx1 overexpression in Mi15 neurons (Fig. 3G). In addition, we checked 4 neurotransmitter/modulator receptor genes that are differentially expressed between Mi15 and Dm2: 5-HT7, Octβ1R, Or63a and Dh44-R1 (Fig. S7I). Using in situ hybridizations, we observed that Mi15s overexpressing Vsx1 downregulated Or63a and Dh44-R1, and they upregulated 5-HT7 and Octβ1R (Fig. S7F–H), as expected. Taken together, our results show that Vsx genes function as terminal selectors in Dm2, controlling both morphological and functional features.
RTK signaling stabilizes the Tm selector network
Even though the mRNA of the Tm4/6-specific selector aop could be found in all 4 Tm neurons up to P40 (Fig. S7J), Aop protein was no longer localized to Tm1 nuclei already by P25 (Fig. S5F). This could be explained by a well-known post-translational regulatory mechanism: Aop is exported from the nucleus and degraded after phosphorylation by MAPK (34). This regulation is essential for specification of R7 photoreceptors in the developing eye through receptor tyrosine kinase (RTK) signaling (35). We could not detect phosphorylated MAPK in Tm4/6 that strongly express Aop, but P-MAPK was present in Tm1/2 where Aop protein could sometimes be observed as a ‘ring’ outside the nucleus at P30 (Fig. S7K, arrows), suggestive of nuclear export. This suggests that Drgx and Pdm3 initially repress Aop protein indirectly in Tm1 and Tm2, respectively, by rendering them sensitive to RTK signaling.
After P40, aop mRNA is also downregulated in Tm1 and Tm2; this coincides with Mef2 upregulation in these cell types (Fig. S7J) downstream of Pdm3 and Drgx (Figs. 1F, 2C). Knocking-down Mef2 using TmX/Tm2-Gal4 resulted in a very rare (1/38 neurons observed) conversion of Tm2 into Tm4 in adults, and 18% of Tm neurons observed were morphologically unrecognizable (Fig. 4A,D). Aop could be detected in some of these cells, but only outside the nucleus (Fig. 4E), similar to wild-type Tm1/2 at P30 (Fig. S7K). This indicates that aop was transcriptionally de-repressed without Mef2, but its post-translational repression through MAPK remained intact, even in adult brains. Therefore, aop appears to be downregulated in Tm1/2 through two independent mechanisms (Fig. 4H): Degradation of the protein through MAPK at all stages, and transcriptional suppression by Mef2 after P40. Drgx and Pdm3 could control the first mechanism by regulating the expression of RTK genes such as InR, Ror and Alk that are differentially expressed between Tm1/2 and Tm4/6 clusters (Fig. S7L).
Figure 4: RTK signaling stabilizes the Tm selector network.
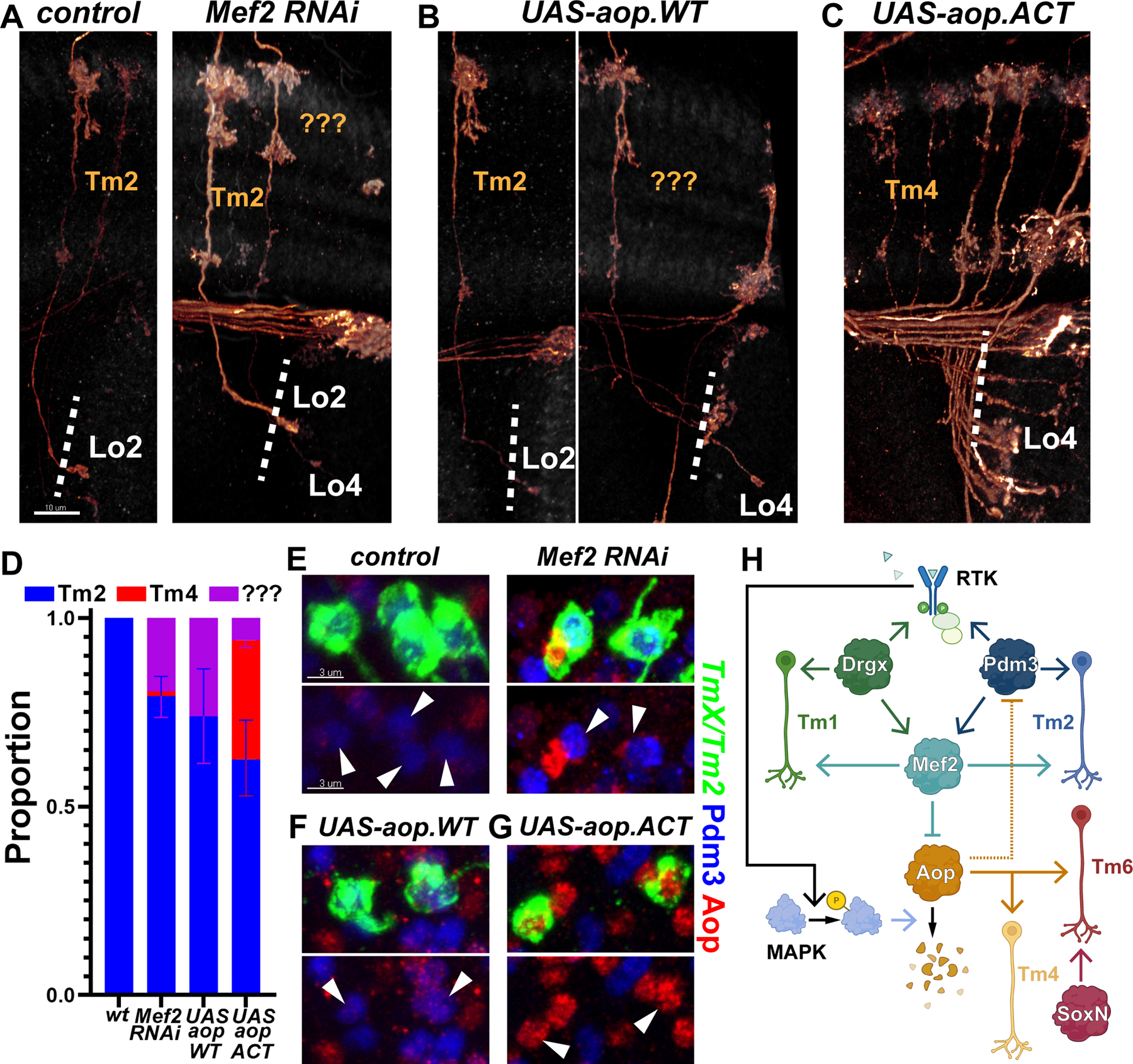
A-G, TmX/Tm2-Gal4 driving CD4-tdGFP (flip-out) and Mef2 RNAi (A,E), UAS-aop.WT (B,F) or UAS-aop.ACT (C,G). A-C, 3D reconstructions of GFP for the representative adult neurons in each condition, with anti-NCad. “???” marks neurons that typically target to Lo4 but could not be recognized as any known optic lobe neuron based on their morphology. Dashed lines mark the border of lobula neuropil. D, Quantification of A-C. n= 54/6 (control), 38/8 (Mef2 RNAi), 16/6 (aop.WT) and 75/5 (aop.ACT) neurons/brains. p=0.03 (Mef2 RNAi), p=0.02 (aop.WT), p=0.005 (aop.ACT) for change in cell-type proportions. Error bars denote SEM. E-G, Same as (A-C) with max. projections of somas with anti-Pdm3 (blue) and anti-Aop (red). Arrowheads: GFP+ neurons. Scale bars: 10 μm (A-C) and 3 μm (E-G). H, Summary of the experimentally validated regulatory interactions between Drgx, Pdm3, Mef2 and Aop in Tm neurons. Negative regulation of Pdm3 by Aop (dashed line) is only applicable when Aop cannot be degraded through the MAPK pathway. RTK: receptor tyrosine kinase.
As pdm3 and Drgx both negatively regulate Aop expression (Figs. 1G, 2C), we asked if the opposite was also true. Overexpression of wild-type aop with TmX/Tm2-Gal4 did not convert Tm2 to either Tm4 or Tm6, but 25% of Tm neurons were morphologically unrecognizable (Fig. 4A,D), similar to those observed with Mef2 RNAi where aop transcription was derepressed. These neurons still maintained Pdm3 (Fig. 4F) whose co-expression with Aop might create a confused state. The signal for the ectopic Aop protein was weak (Fig. 4F), suggesting that it was being degraded by the active MAPK pathway in Tm2. We therefore overexpressed a constitutively active form of Aop (aop.ACT) that cannot be phosphorylated and degraded (34). In these brains, 40% of Tm2s were converted to Tm4s (Fig. 4C–D) that had lost Pdm3 expression (Fig. 4G). This indicates that while Aop can suppress pdm3 and promote Tm4 fate (Fig. 4H, dashed arrow), this regulation is not relevant with wild-type aop due to RTK signaling which ensures that pdm3 acts upstream.
The apparent destabilization of the fate choice between Tm2 and Tm4 with the postmitotic expression of aop.ACT (but not with aop.WT) implies that neuronal identity remains dependent on the signaling conditions even after the initial specification events. Similar mechanisms in organisms with larger and more complex brains could be exploited to further diversify the neurons generated from the same stem cell pool with a common identity, but then migrate to distinct brain regions where different signals might be available (36).
Decoding neuronal gene regulation through network inference
Cell type identity encoded by terminal selectors represents only one aspect of neuronal gene regulation. Neuronal transcriptomes are dynamic throughout differentiation, typically in response to external signals such as the steroid hormone ecdysone (37–39). It remains unclear how these two top-level regulatory programs, i.e. the identity and developmental state, interact to combinatorially determine the expression of downstream genes in each neuron.
We implemented the Inferelator 3.0 framework (40) to build gene regulatory network (GRN) models with the goal of gaining a more complete understanding of the regulatory programs employed by developing optic lobe neurons. A key feature of this method is its use of Transcription Factor Activity (TFA) that allows the Inferelator to estimate the underlying activity of each TF using the expression levels of its known targets from prior information (‘priors’) (Fig. 5A). Calculating TFA before fitting a linear regression model to infer a GRN circumvents the issue that mRNA levels are often not a good substitute for a TF’s latent activity (41), which may vary with post-transcriptional modifications or the presence of cofactors. We modeled GRNs in Tm1/2/4/6 neurons, as well as the five types of lamina monopolar neurons (L1–5), which provide a useful benchmark for our models. For inference, we used single-cell transcriptomes from both our atlas (1), as well as one generated by another group (42) between stages P24–50, when optic lobe neurons acquire most of their morphological features and begin to form synapses. We constructed priors for each network using the corresponding P48 clusters (Fig. S6C) in the snATAC-seq dataset described above (28). Figure 5C displays the entire GRN inferred from the Tm neurons, highlighting the top 10 TFs with the highest number of targets predicted, which includes Mef2, the selectors pdm3, cut and ap, as well as the ecdysone-responsive TFs Hr3, Hr39, Eip74EF and Eip93F.
Figure 5: Computational inference of gene regulatory networks.
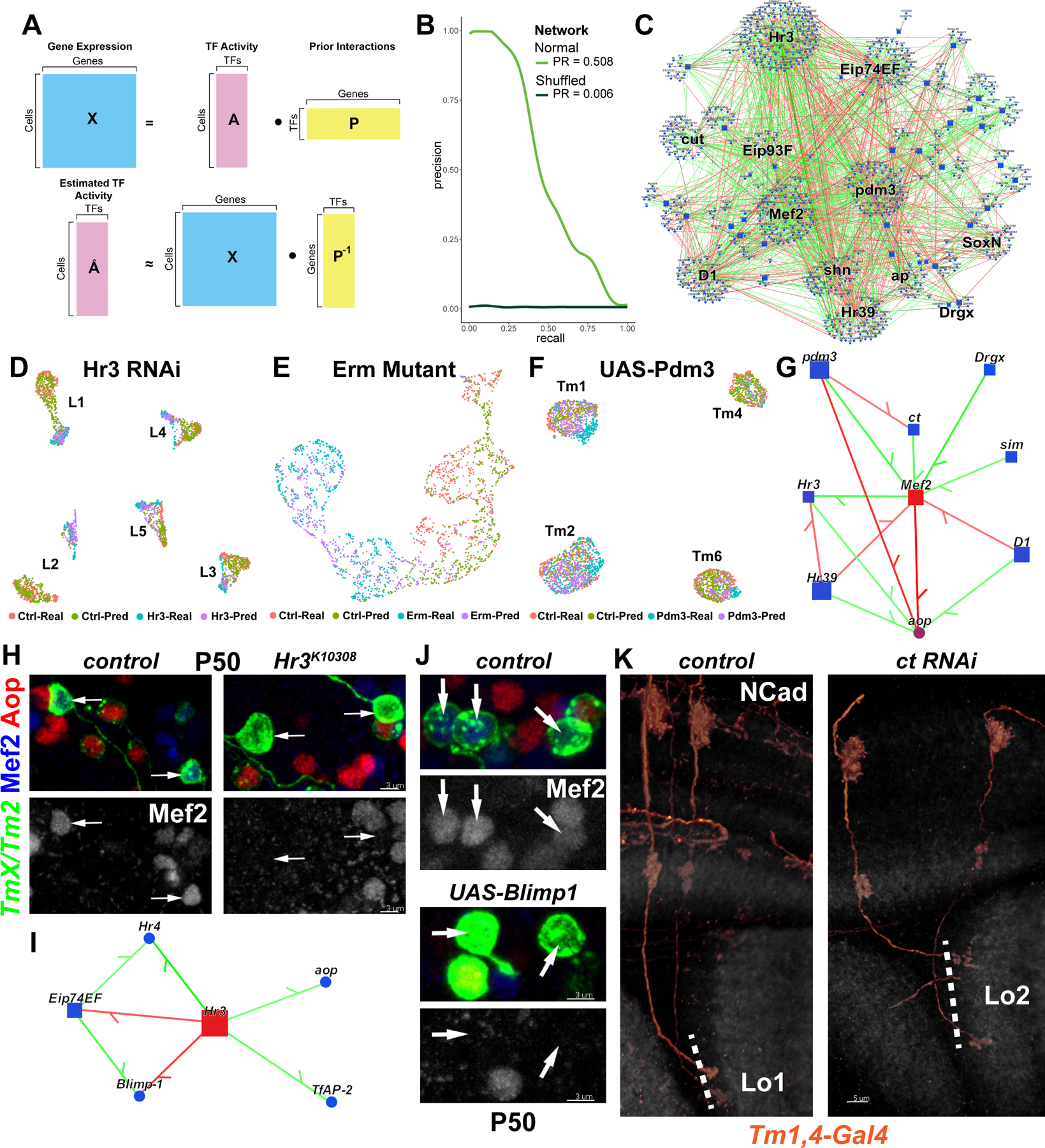
A, Gene expression in each cell is assumed to be a linear product of latent TF activities and the connectivity matrix (prior) between TFs and their targets (top). TFA is estimated as the dot product of the expression matrix and the pseudoinverse of the prior matrix (bottom). B, AUPR curves for the Tm network built using the MergedDA prior (see also Fig. S6A). C, Visualization of the network in (B), displaying all interactions with a minimum of 80% confidence (combined) and variance explained of 1%. Top 10 TFs that had the highest number of target genes in the network were highlighted, in addition to SoxN and Drgx. D-F, Single-cell transcriptomes of (D) L1–5 neurons at P48 expressing Hr3 RNAi (39), (E) simulated single L3 neurons (Fig. S8D) at P40 mutant for erm (21), (F) Tm1/2/4/6 neurons at P50 overexpressing pdm3 (Fig. 1). UMAPs were calculated using 30 PCs (D,F) or 3 PCs (E) on the integrated gene expression. (see also Fig. S8E–G). G, Network visualization displaying all TFs predicted to regulate Mef2 or aop with confidence >95%, and all inferred interactions between the displayed genes in the Tm network. Green: positive, red: negative correlation between mRNA levels of the gene pairs. H, FRT42D and Hr3K10308 MARCM clones labeled with TmX/Tm2-Gal4 and CD4-tdGFP in P50 medulla cortex (maximum projection), with anti-Mef2 (blue) and anti-Aop (red). Arrows: Aop− Tm neurons that should normally be Mef2+ (n=6 brains). I, Same as (G), displaying all TFs predicted to be regulated by Hr3 (confidence>95%). J, TmX/Tm2-Gal4 driving CD4-tdGFP (flip-out) and UAS-Blimp1 (n=5 brains). Max. projection same as (H). K, Tm1,4-Gal4 driving CD4-tdGFP (flip-out) and ct RNAi (n=40/6 neurons/brains). 3D reconstructions of GFP for the representative adult neurons in each condition with anti-NCad (white). Dashed lines mark the border of lobula neuropil. Scale bars: 3 μm (H-J) and 5 μm (K).
The scarcity of ground-truth networks presents a challenge when benchmarking inferred GRNs in complex multicellular organisms. To assess the predictive power of our models, we exploited available RNA-seq datasets collected from perturbed neurons. For lamina neurons, we used two relevant datasets: knock-down of Hr3 in all 5 lamina neurons at P48 (39), and L3 neurons at P40 mutant for erm (21). For Tm neurons, we used the UAS-pdm3 scRNA-seq experiment we performed at P50 (Fig. 1H). For each experiment, TFA was calculated using the same corresponding priors used for inference. We then applied matrix multiplication (dot product) between the estimated TFA and the learned weights between TFs and targets (betas) to generate a predicted expression matrix (Fig. S8C). Even though these betas were determined from completely independent, wild-type datasets discussed above, the real and predicted transcriptomes aligned nearly perfectly according to their cell-type and condition of origin (Fig. 5D–F), after Seurat integration (43). However, we also observed that before integration (Fig. S8E–G) the differences between control and perturbed conditions were much smaller in predicted clusters. Consistently, we found that the predicted transcriptomes recapitulated only a small proportion (10–30%) of the real DEGs between the control and perturbed conditions (low recall), but the predicted DEGs were mostly (>50%) correct (high precision) (Fig. S8H–J). Thus, our benchmarks suggest that the interactions learned by our models are largely accurate, though they represent only a snippet of the true underlying GRNs.
Selectors and ecdysone signaling regulate downstream targets
We found that nearly all the regulatory relationships we experimentally validated in the previous sections (Fig. 4H) were also captured by our GRN model (Fig. 5G), such as the regulation of Mef2 by Drgx and pdm3. Drgx and Pdm3, while required, are not sufficient to activate Mef2 expression, which does not occur until P40 despite the continuous expression of the selectors. Ecdysone-responsive Hr3 emerged as a candidate for this temporal trigger, as its activation around P30 precedes Mef2 (Fig. S7J). We tested this prediction by generating Hr3 mutant MARCM clones using TmX/Tm2-Gal4. In adult brains, Mef2 expression was not affected and Tm2 neurons appeared morphologically normal (Fig. S9A). However, at P50, Mef2 could not be detected in mutant clones (Fig. 5H), indicating that Mef2 expression was delayed, but not abolished in Hr3 mutants. This implies that there are redundant temporal mechanisms regulating Mef2 expression and/or that Hr3 acts indirectly to control Mef2. Among the predicted downstream targets of Hr3 (Fig. 5I), Hr4, Eip74EF and Blimp-1 were all shown to be regulated by Hr3 in lamina neurons, whose knock-down delays downregulation of Blimp-1 (39) that normally occurs around P40 (Fig. S7J). Indeed, overexpression of Blimp-1 using TmX/Tm2-Gal4 also repressed Mef2 at P50 (Fig. 5J), suggesting that Hr3 acts through Blimp-1 for this function. Altogether, our results show that combinatorial action of the selectors Drgx/Pdm3 and the ecdysone-responsive TFs Hr3/Blimp-1 enables Mef2 to be expressed specifically in Tm1 and Tm2 neurons, and only after P40.
Another inferred edge in this sub-network (Fig. 5G), the negative regulation of cut (ct) by pdm3, was consistent with the lower levels of ct expression in Tm2s compared to other Tm neurons (Fig. S9B). Since different Cut expression levels in larval ‘da’ sensory neurons regulate the size of their dendritic arborizations (44), this difference could be functionally significant. Indeed, we found that 73% of Tm1s targeted to Lo2 layer instead of Lo1 upon ct knock-down (Fig. 5K). Thus, the level of ct expression controls a specific subroutine during brain wiring downstream of pdm3. However, overexpression of ct in Tm2 neurons did not lead to their axons projecting to Lo1 instead of Lo2 (Fig. S9C), suggesting that there are redundant mechanisms allowing Tm2 to arborize in this layer.
Lastly, we inspected the GRN model of the Tm neurons (Fig. 5C) to assess if different types of TFs specialize on different types of targets. We previously reported that TFs and cell-surface proteins (CSPs) are overrepresented in DEGs between optic lobe neurons; CSPs are particularly upregulated at P40–50 (1) during synaptogenesis. Accordingly, CSPs that may be involved in cell-cell recognition (45) were strongly enriched among the network targets (Fig. S9D). However, we did not see a clear bias for any TF class to regulate more or fewer CSPs than others. We also performed GO-term enrichment analysis on the predicted targets of the top 3 regulators in the network: ecdysone TF Hr3, terminal selector Pdm3 and effector TF Mef2. The same general terms were enriched for all of them: ion channels, cell adhesion and signaling molecules (Fig. S9E). These results further highlight the combinatorial nature of neuronal gene regulation and they are consistent with other findings that most targets of the ecdysone-responsive TFs are cell-type specific (39), despite the uniform expression of these TFs in all neurons.
Discussion
We set out to test whether neuronal type identity is primarily encoded by unique and sustained combinations of TFs in each cell type. The terminal selector hypothesis has been extensively supported in C. elegans, and a few selectors have also been described in mice, such as Fezf2 (homolog of Erm) whose expression in cortical progenitors induces corticospinal motor neuron-like fates (15). However, the previous studies focused on the roles of individual selector genes and did not systematically test the sufficiency of a continuously expressed TF code in instructing all type-specific gene expression in neurons. We explicitly addressed this prediction by determining the selector codes of every neuron in the fly visual system using a developmental scRNA-seq atlas and by engineering predictable transformations between different neurons using only these TFs. This has so far been difficult to demonstrate, even in C. elegans, likely because neurons typically diverge by multiple selectors (46). Our results suggest that effector genes are controlled by different permutations of available selectors in each neuron, which implies that every effector might not be regulated by all selectors. Similar selector combinations generally resulted in similar transcriptomes and vice versa (Fig. S1D), but this relationship was not strict, reflecting the combinatorial nature of TF action. For instance, during brain wiring (P50), we observed a distinct branch (Fig. S2C, red circle) in which all annotated clusters corresponded to neurons that connect to the central brain, irrespective of their developmental origin and the similarity of selector expression.
We defined terminal selectors broadly to satisfy two key criteria: continuous expression in postmitotic neurons and involvement in the control of neuronal type identity. It has also been proposed that selectors directly regulate most effector genes by binding to their cis-regulatory enhancers (47). In our GRN models (Table S3), the interactions that were present in our prior network (‘gold_standard=1’) represent those that were directly supported by cis-regulatory evidence, i.e. enrichment of motifs in differentially accessible regions near the target gene, and thereby indicate direct regulation. For instance, 27 of the 120 high-confidence targets of Pdm3 (Fig. S9D–E) are supported to be direct interactions by scATAC-seq. This does not imply that all other interactions are indirect, since the Inferelator-Prior pipeline was designed to favor accuracy by only retaining the highest confidence targets for TFA calculation (40). These direct targets include several wiring molecules like Dscam4 and the Netrin receptor frazzled, as well as effector TFs NK7.1 and CG9932. Consistently, overexpression of fra in Tm4s led to 54% of these neurons forming dendritic branches that split into a ‘fork’ in the M2 layer, which is normally characteristic of Pdm3-expressing Tm2 neurons (Fig. S9F, arrowhead, compare to S9C). Nevertheless, extensive utilization of effector TFs (such as Mef2 in Tm1/2 neurons) makes it likely that many targets are also regulated indirectly. Such transcriptional ‘subroutines’ may be more prevalent in flies and higher organisms where neuronal differentiation occurs over several days or weeks, in contrast to worm neurons that are typically functional within a few hours after their terminal division. Another feature that has been commonly, but not universally, associated with terminal selectors is auto-regulation (47). Our results support that Pdm3 indeed auto-activates (Fig. 1N–O), but this is unlikely to be the case for Drgx, SoxN and Vsx1/2, given the lower efficiency of their overexpression (Figs. 2B, S7A, 3E).
The apparent conservation of this regulatory logic in both C. elegans and Drosophila, whose last common ancestor lived over 600 million years ago (48), makes it likely that the terminal selector concept will also be useful to understand and manipulate the neuronal diversity of mammalian brains. This could have large implications for the emerging field of cell replacement therapy. The usage of lineage-specific TFs for the generation of specific neuronal types in vitro have significantly improved the efficiency of these protocols (49). Some of these TFs, such as Pet1 and Lmx1b for serotonergic neurons (50) or Lmx1a and Nurr1 for dopaminergic neurons (51) are also likely to be terminal selectors. However, the protocols used still tend to produce heterogenous populations of related cell types (52). We propose that more specific combinations of such re-programming TFs could be identified in specific cell types of interest by virtue of their sustained postmitotic expression.
We described that tTF Klu from neuroblasts activates the selector Drgx in newly born Tm1 neurons. But we still know little about how the combined action of temporal, spatial and Notch patterning (5) activates a unique set of selectors in every neuronal type, and subsequently how the selector combination enacts precise gene batteries over the course of development. There are also some limitations of this framework: We and others previously reported that a few neuronal types have distinct transcriptomes during development but then converge to a common state in adult brains (1, 53). These are generally very similar (sub)types that only differ in their connectivity, thus the TFs that encode their differences do not need to be maintained after their wiring is complete. In addition, TFs can be post-transcriptionally regulated by, for instance, RNA binding proteins Imp and Syp that are widely utilized in Drosophila nervous system to generate neuronal diversity (54). This could complicate the identification of correct selector combinations from RNA-seq data alone.
Both this work and other previous efforts to decipher gene regulation in the fly brain (28) have now made it possible to study the molecular mechanisms of synaptic specificity within the framework of gene regulatory mechanisms that encode neuronal type identity. We propose a “top-down” approach where terminal selectors that cause broad changes in neuronal fates are identified first, followed by the dissection of downstream mechanisms aided by GRN modeling. Perhaps the most promising targets are effector TFs (like cut), that still regulate many other genes but have more limited functions (sub-routines) than the selectors. Nevertheless, the current models still have limitations, imposed mainly by the quality of snATAC-seq priors. Single-nucleus multiome studies that simultaneously profile gene expression and chromatin accessibility could remove many of these limitations in the near future.
Methods Summary
Candidate terminal selectors:
We selected for each neuronal cluster in our developmental scRNA-seq atlas (1) the transcription factors that were i) found as a consistent marker of that cluster, or ii) continuously expressed at all stages according to binarized expression. We discarded all TFs that were expressed in more than 150 clusters (likely pan-neuronal).
Genetics:
The precise genotypes and temperatures used for experiments in each figure panel are detailed in Table S4. Source details for all fly strains are specified in Table S5. Full names of all genes references in the manuscript are listed in Table S6.
Immunohistochemistry and RNA-FISH experiments were performed according to previously described protocols. New polyclonal antibodies were generated against Drgx, Pdm3, SoxN, Mef2, Vsx2, Brp and Repo by Genscript. Source details for all other antibodies are specified in Table S5. Custom FISH probes were designed by Molecular Instruments against the transcripts of 5-HT7, Octβ1R, Or63a and Dh44-R1. All samples were imaged using a Leica SP8 confocal microscope with a 63x (NA=1.3) glycerol objective. Images were analyzed using Imaris (details in the supplement). Parametric, two-sided t-tests were used for all pairwise comparisons.
DrgxΔTm1 deletion was produced by WellGenetics through CRISPR-mediated mutagenesis.
Single-cell RNA sequencing:
We labeled all 4 Tm neurons (TmX/Tm2-Gal4) with nuclear GFP, crossed to either UAS-pdm3 or yw (control), isolated the labeled cells using FACS, and prepared libraries using 10x Genomics 3’ kit (v3.1) that were sequenced on Illumina NovaSeq 6000 platform. The data were analyzed with CellRanger v5 and Seurat v4.
Network Inference:
We used a published scATAC-seq dataset (28) to identify differentially accessible regions between Tm1/2/4/6 clusters (Tm network) and L1–5 clusters (Lamina network) at P48. We then used Inferelator-Prior (v0.2.3 faf5e47) package to scan and score these regions within 10kb of every gene for TF binding motifs. The scores were then clustered to retain only the highest confidence targets of each TF for TFA calculation. For inference, we used single-cell transcriptomes from (1), at stages P30, P40 and P50, also from (42), at stages P24, P36 and P48. The networks were modeled only on the genes that displayed differential expression between either the cell-types or the time-points analyzed, using Inferelator 3 (v0.5.6 dd532f4). The network performances were evaluated against the respective priors using 4 different metrics (Fig. S8, see also supplemental Methods). We additionally compared the performance of each network to negative control networks that were built with shuffled priors.
Supplementary Material
Acknowledgements
We would like to thank all members of the Desplan and Bonneau Labs, Oliver Hobert, Richard Mann and Denis Jabaudon for helpful discussions. We thank Chris Doe, Stein Aerts, Justin Blau, Esteban Mazzoni, Robin Hiesinger, Nikos Konstantinides, David Chen, Ryan Loker and Sromana Mukherjee for critical reading of the manuscript. We further thank David Krantz, Ilaria Rebay, Cheng-Ting Chien, Wesley Grueber and Jens Rister for reagents, and Yen-Chung Chen for help with plotting. We would like to extend special thanks to Jasper Janssens and Stein Aerts for sharing the snATAC-seq data ahead of publication, and to Giuseppe-Antonio Saldi for help with network inference.
Funding:
This work was supported by NIH grants EY13010 and EY017916 to C.D. and by R01HD096770, R01CA229235 and RM1HG011014 to R.B and by Simons Foundation. M.N.O. was a Leon Levy Neuroscience Fellow and is supported by NINDS K99NS125117. C.S.G. is supported by the NSF Award 1922658 to NYU CDS. I.H. was supported by an HFSP postdoctoral fellowship (LT000757/2017-L) and by the Kimmel Center for Stem Cell Biology Senior Postdoctoral Fellowship.
Footnotes
Data Availability:
All data that support the conclusions in this manuscript can be found in the main text or in the supplemental materials. Raw and processed scRNA-seq data are publicly accessible on GEO: GSE199734). Newly created materials from this study may be requested from the corresponding authors.
References
- 1.Özel MN et al. , Neuronal diversity and convergence in a visual system developmental atlas. Nature 589, 88–95 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fischbach KF, Dittrich APM, The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res 258, 441–475 (1989). [Google Scholar]
- 3.Ngo KT, Andrade I, Hartenstein V, Spatio-temporal pattern of neuronal differentiation in the Drosophila visual system: A user’s guide to the dynamic morphology of the developing optic lobe. Developmental Biology 428, 1–24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu F, Kuo CT, Jan YN, Drosophila neuroblast asymmetric cell division: recent advances and implications for stem cell biology. Neuron 51, 13–20 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Erclik T et al. , Integration of temporal and spatial patterning generates neural diversity. Nature 541, 365–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konstantinides N et al. , A complete temporal transcription factor series in the fly visual system. Nature 604, 316–322 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X et al. , Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature 498, 456–462 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holguera I, Desplan C, Neuronal specification in space and time. Science 362, 176–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobert O, in Current topics in developmental biology, Wassarman PM, Ed. (Academic Press, 2016), vol. 116, pp. 455–475. [DOI] [PubMed] [Google Scholar]
- 10.Berghoff EG et al. , The Prop1-like homeobox gene unc-42 specifies the identity of synaptically connected neurons. eLife 10, e64903 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira L et al. , A cellular and regulatory map of the cholinergic nervous system of C. elegans. eLife 4, e12432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefanakis N, Carrera I, Hobert O, Regulatory Logic of Pan-Neuronal Gene Expression in C. elegans. Neuron 87, 733–750 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serrano-Saiz E, Leyva-Díaz E, De La Cruz E, Hobert O, BRN3-type POU Homeobox Genes Maintain the Identity of Mature Postmitotic Neurons in Nematodes and Mice. Current Biology 28, 2813–2823.e2812 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Remesal L et al. , PBX1 acts as terminal selector for olfactory bulb dopaminergic neurons. Development (Cambridge, England) 147, (2020). [DOI] [PubMed] [Google Scholar]
- 15.Lodato S et al. , Gene co-regulation by Fezf2 selects neurotransmitter identity and connectivity of corticospinal neurons. Nature Neuroscience 17, 1046–1054 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arlotta P, Hobert O, Homeotic Transformations of Neuronal Cell Identities. Trends in neurosciences 38, 751–762 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Reilly MB, Cros C, Varol E, Yemini E, Hobert O, Unique homeobox codes delineate all the neuron classes of C. elegans. Nature 584, 595–601 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa E, Kaido M, Takayama R, Sato M, Brain-specific-homeobox is required for the specification of neuronal types in the Drosophila optic lobe. Developmental Biology 377, 90–99 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Hasegawa E et al. , Concentric zones, cell migration and neuronal circuits in the Drosophila visual center. Development (Cambridge, England) 138, 983–993 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Suzuki T et al. , Formation of Neuronal Circuits by Interactions between Neuronal Populations Derived from Different Origins in the Drosophila Visual Center. Cell reports 15, 499–509 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Peng J et al. , Drosophila Fezf coordinates laminar-specific connectivity through cell-intrinsic and cell-extrinsic mechanisms. eLife 7, e33962 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schilling T, Ali AH, Leonhardt A, Borst A, Pujol-Martí J, Transcriptional control of morphological properties of direction-selective T4/T5 neurons in Drosophila. Development (Cambridge, England) 146, dev169763 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee T, Luo L, Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends in neurosciences 24, 251–254 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Chen C-K, Chen W-Y, Chien C-T, The POU-domain protein Pdm3 regulates axonal targeting of R neurons in the Drosophila ellipsoid body. Developmental neurobiology 72, 1422–1432 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Tichy AL, Ray A, Carlson JR, A New Drosophila POU Gene, pdm3, Acts in Odor Receptor Expression and Axon Targeting of Olfactory Neurons. The Journal of Neuroscience 28, 7121–7129 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Togane Y et al. , Spatio-temporal pattern of programmed cell death in the developing Drosophila optic lobe. Dev Growth Differ 54, 503–518 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Song Z, McCall K, Steller H, DCP-1, a Drosophila Cell Death Protease Essential for Development. Science 275, 536–540 (1997). [DOI] [PubMed] [Google Scholar]
- 28.Janssens J et al. , Decoding gene regulation in the fly brain. Nature, (2022). [DOI] [PubMed]
- 29.Suzuki T, Kaido M, Takayama R, Sato M, A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Dev Biol 380, 12–24 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Morante J, Desplan C, The Color-Vision Circuit in the Medulla of Drosophila. Current Biology 18, 553–565 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chao AT, Jones WM, Bejsovec A, The HMG-box transcription factor SoxNeuro acts with Tcf to control Wg/Wnt signaling activity. Development (Cambridge, England) 134, 989–997 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Schober M, Rebay I, Perrimon N, Function of the ETS transcription factor Yan in border cell migration. Development (Cambridge, England) 132, 3493–3504 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Davis FP et al. , A genetic, genomic, and computational resource for exploring neural circuit function. eLife 9, e50901 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rebay I, Rubin GM, Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell 81, 857–866 (1995). [DOI] [PubMed] [Google Scholar]
- 35.O’Neill EM, Rebay I, Tjian R, Rubin GM, The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78, 137–147 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Mayer C et al. , Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White KP, Hurban P, Watanabe T, Hogness DS, Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science 276, 114–117 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Riddiford LM, Truman JW, Nern A, Juvenile hormone reveals mosaic developmental programs in the metamorphosing optic lobe of Drosophila melanogaster. Biology Open 7, bio034025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain S et al. , A global timing mechanism regulates cell-type-specific wiring programmes. Nature, (2022). [DOI] [PubMed]
- 40.Skok Gibbs C et al. , High-performance single-cell gene regulatory network inference at scale: the Inferelator 3.0. Bioinformatics, btac117 (2022). [DOI] [PMC free article] [PubMed]
- 41.Schacht T, Oswald M, Eils R, Eichmüller SB, König R, Estimating the activity of transcription factors by the effect on their target genes. Bioinformatics 30, i401–i407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurmangaliyev YZ, Yoo J, Valdes-Aleman J, Sanfilippo P, Zipursky SL, Transcriptional Programs of Circuit Assembly in the Drosophila Visual System. Neuron 108, 1045–1057.e1046 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Stuart T et al. , Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grueber WB, Jan LY, Jan YN, Different Levels of the Homeodomain Protein Cut Regulate Distinct Dendrite Branching Patterns of Drosophila Multidendritic Neurons. Cell 112, 805–818 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Kurusu M et al. , A Screen of Cell-Surface Molecules Identifies Leucine-Rich Repeat Proteins as Key Mediators of Synaptic Target Selection. Neuron 59, 972–985 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kratsios P et al. , An intersectional gene regulatory strategy defines subclass diversity of C. elegans motor neurons. eLife 6, e25751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hobert O, Kratsios P, Neuronal identity control by terminal selectors in worms, flies, and chordates. Current Opinion in Neurobiology 56, 97–105 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Holton TA, Pisani D, Deep Genomic-Scale Analyses of the Metazoa Reject Coelomata: Evidence from Single- and Multigene Families Analyzed Under a Supertree and Supermatrix Paradigm. Genome Biology and Evolution 2, 310–324 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fitzgerald M, Sotuyo N, Tischfield DJ, Anderson SA, Generation of cerebral cortical GABAergic interneurons from pluripotent stem cells. STEM CELLS 38, 1375–1386 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Xu Z et al. , Direct conversion of human fibroblasts to induced serotonergic neurons. Molecular Psychiatry 21, 62–70 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caiazzo M et al. , Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Konstantinides N, Desplan C, Neuronal differentiation strategies: insights from single-cell sequencing and machine learning. Development (Cambridge, England) 147, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H et al. , Classifying Drosophila Olfactory Projection Neuron Subtypes by Single-Cell RNA Sequencing. Cell 171, 1206–1220.e1222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossi AM, Desplan C, Extrinsic activin signaling cooperates with an intrinsic temporal program to increase mushroom body neuronal diversity. eLife 9, e58880 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferrero E, Fischer B, Russell S, SoxNeuro orchestrates central nervous system specification and differentiation in Drosophila and is only partially redundant with Dichaete. Genome Biology 15, R74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romero-Calderón R et al. , A Glial Variant of the Vesicular Monoamine Transporter Is Required To Store Histamine in the Drosophila Visual System. PLOS Genetics 4, e1000245 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferreira AAG, Sieriebriennikov B, Whitbeck H (ZappyLab, Inc., 2021). [Google Scholar]
- 58.Hao Y et al. , Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stuart T, Srivastava A, Madad S, Lareau CA, Satija R, Single-cell chromatin state analysis with Signac. Nature methods 18, 1333–1341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McLeay RC, Bailey TL, Motif Enrichment Analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics 11, 165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grant CE, Bailey TL, Noble WS, FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gabitto MI et al. , Characterizing chromatin landscape from aggregate and single-cell genomic assays using flexible duration modeling. Nat Commun 11, 747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castro DM, de Veaux NR, Miraldi ER, Bonneau R, Multi-study inference of regulatory networks for more accurate models of gene regulation. PLoS Comput Biol 15, e1006591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Supek F, Bošnjak M, Škunca N, Šmuc T, REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLOS ONE 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuznetsova I, Lugmayr A, Siira SJ, Rackham O, Filipovska A, CirGO: an alternative circular way of visualising gene ontology terms. BMC Bioinformatics 20, 84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nern A, Pfeiffer BD, Rubin GM, Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proceedings of the National Academy of Sciences 112, E2967–E2976 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the conclusions in this manuscript can be found in the main text or in the supplemental materials. Raw and processed scRNA-seq data are publicly accessible on GEO: GSE199734). Newly created materials from this study may be requested from the corresponding authors.


