Abstract
The mevalonate pathway and the glyceraldehyde 3-phosphate (GAP)–pyruvate pathway are alternative routes for the biosynthesis of the central isoprenoid precursor, isopentenyl diphosphate. Genomic analysis revealed that the staphylococci, streptococci, and enterococci possess genes predicted to encode all of the enzymes of the mevalonate pathway and not the GAP-pyruvate pathway, unlike Bacillus subtilis and most gram-negative bacteria studied, which possess only components of the latter pathway. Phylogenetic and comparative genome analyses suggest that the genes for mevalonate biosynthesis in gram-positive cocci, which are highly divergent from those of mammals, were horizontally transferred from a primitive eukaryotic cell. Enterococci uniquely encode a bifunctional protein predicted to possess both 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase and acetyl-CoA acetyltransferase activities. Genetic disruption experiments have shown that five genes encoding proteins involved in this pathway (HMG-CoA synthase, HMG-CoA reductase, mevalonate kinase, phosphomevalonate kinase, and mevalonate diphosphate decarboxylase) are essential for the in vitro growth of Streptococcus pneumoniae under standard conditions. Allelic replacement of the HMG-CoA synthase gene rendered the organism auxotrophic for mevalonate and severely attenuated in a murine respiratory tract infection model. The mevalonate pathway thus represents a potential antibacterial target in the low-G+C gram-positive cocci.
Isoprenoids, which are ubiquitous in nature, comprise a family of over 23,000 products, each composed of repeating five carbon isopentenyl diphosphate (IPP) subunits. Among the isoprenoids are sterols, which contribute to eukaryotic membrane architecture, steroid hormones, chlorophylls, and the acyclic polyprenoid plastoquinone (42). In bacteria, the principal products of IPP include the lipid carrier undecaprenol which is involved in cell wall biosynthesis (36), menaquinones and ubiquinones involved in electron transport (32), and carotenoids (23).
Two pathways for the biosynthesis of IPP have been described, the classical mevalonate pathway and the more recently identified glyceraldehyde 3-phosphate (GAP)–pyruvate pathway. Rohmer et al. (38) demonstrated the presence of the non-mevalonate pathway, originating from pyruvate and GAP, in several gram-negative bacteria, including Escherichia coli. The GAP-pyruvate pathway is also present in Chlamydia trachomatis (44) and Aquifex aeolicus (13), the gram-positive bacteria Bacillus subtilis (52) and Mycobacterium tuberculosis (35), the cyanobacterium Synechocystis (14), chloroplasts (29), and some unicellular algae, including Scenedesmus oliquus (40).
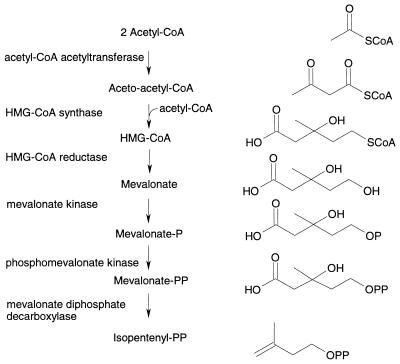
The mevalonate pathway for IPP biosynthesis was first identified in eukaryotic cells (17). In this pathway (Fig. 1), three acetyl coenzyme A (acetyl-CoA) units are joined successively to form 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA). HMG-CoA is then reduced to mevalonate, which is subsequently phosphorylated, decarboxylated, and dehydrated to form IPP. The enzymes of the mevalonate pathway from a number of organisms, including humans, have been studied. HMG-CoA reductase, the best-characterized enzyme in the pathway, is the target of the statin class of cholesterol-lowering drugs (1). Particularly well characterized is the biodegradative HMG-CoA reductase from Pseudomonas mevalonii, which catalyzes the conversion of mevalonate to HMG-CoA, enabling the organism to use mevalonate as its sole source of carbon (4). P. mevalonii, however, does not appear to possess the other enzymes of the mevalonate pathway and hence probably synthesizes IPP via the GAP-pyruvate pathway.
FIG. 1.
Biosynthesis of IPP via the mevalonate pathway.
While the genome of B. subtilis revealed genes encoding enzymes of the GAP-pyruvate pathway for IPP biosynthesis, other gram-positive bacteria, including Lactobacillus plantarum (49), Staphylococcus aureus (18, 33), Staphylococcus carnosus (21), and Streptococcus mutans (50), incorporate radiolabeled mevalonate into isoprenoids, suggesting that these bacteria use the mevalonate pathway for the synthesis of IPP. Furthermore, Doolittle and Lodgson (15) and Bochar et al. (7) reported the presence of the HMG-CoA reductase gene in the genomes of Streptococcus pneumoniae and Streptococcus pyogenes. The majority of Streptomyces species are reported to possess only the GAP-pyruvate pathway, but some species, such as Streptomyces aeriouvifer, possess both pathways for the synthesis of isoprenoids (41), perhaps accounting for the abundance of isoprenoid compounds produced as low-molecular-weight secondary metabolites. Among microorganisms there is genomic and/or biochemical evidence to suggest the presence of the mevalonate pathway in the spirochete Borrelia burgdorferi (AE001169), the gram-negative bacteria Chloropseudomonas ethylica and Myxococcus fulvus (21), the archaea (5, 6), fungi (28), and species of algae such as Euglena gracilis (14, 43).
We report here that, unlike most gram-negative bacteria and B. subtilis, species of enterococci, staphylococci, and streptococci possess homologs of five genes predicted to encode enzymes involved in the mevalonate route to IPP but not homologs of the genes involved in the GAP-pyruvate pathway. Phylogenetic analysis suggests that gram-positive cocci obtained the genes for the mevalonate pathway through one or more horizontal gene transfer events involving a primitive eukaryote. The genes are arranged in one or two clusters. In all species, the HMG-CoA synthase and HMG-CoA reductase genes are closely linked but are divergently transcribed in the enterococci and staphylococci. The mevalonate kinase, mevalonate decarboxylase, and phosphomevalonate kinase genes constitute an apparent single operon, whose structure is highly conserved in the species examined. The enterococci appear to encode a bifunctional protein predicted to possess both HMG-CoA reductase and acetyl-CoA acetyltransferase (thiolase) activities. Using allelic replacement techniques, we have demonstrated that each of the five genes of the mevalonate pathway is essential for the growth of Streptococcus pneumoniae in vitro without added mevalonate.
MATERIALS AND METHODS
Identification and phylogenetic analysis of mevalonate pathway gene sequences.
Separate database searches and phylogenetic analyses were performed for the five proteins MvaS, MvaA, MvaK1, MvaK2, and MvaD. Homologous protein sequences were retrieved from public and proprietary genomic sequence databases including the Incyte Pharmaceutical Inc. PathoSeq sequence database by using BLASTP and TBLASTN software (2). Preliminary sequence data were also obtained from The Institute for Genomic Research website at http://www.tigr.org. The proteins were aligned using the program CLUSTALW version 1.7 (48) with the BLOSUM62 (20) similarity matrix and gap opening and extension penalties of 10.0 and 0.05, respectively. The multiple-sequence alignments were refined manually using the program SEQLAB of the GCG version 9.0 software package (Genetics Computer Group, Madison, Wis.).
Phylogenetic trees were constructed by maximum-parsimony (MP) and neighbor-joining (NJ) methods for each set of alignments. NJ trees were based on pairwise distances between amino acid sequences using the programs NEIGHBOR and PROTDIST of the PHYLIP 3.57c package (http://evolution.genetics.washington.edu/phylip.html) (16). The Dayhoff program option was invoked in the latter program, which estimates based on the Dayhoff 120 matrix. The programs SEQBOOT and CONSENSE were used to estimate the confidence limits of branching points from 1,000 bootstrap replications. MP analysis was done using the software package PAUP* (45). Given the large size of the data set, it was not possible to exhaustively search for the total number of minimal-length trees. Instead, the numbers and lengths of minimal trees were estimated from 100 replicate random heuristic searches while confidence limits of branch points were estimated by 1,000 bootstrap replications.
Bacterial strains, media, plasmid, enzymes and chemicals.
S. pneumoniae R6 was as reported by Avery et al. (3); S. pneumoniae 0100993 was a clinical isolate kindly provided by D. Felmington (University College Hospital, London, United Kingdom); Enterococcus faecium 2, Staphylococcus haemolyticus 225, and Staphylococcus epidermidis CL-7 were clinical isolates from the SmithKline Beecham culture collection. E. faecium was grown in Luria-Bertani broth (39). S. haemolyticus and S. epidermidis were grown on tryptic soy broth (TSB) and agar (TSA) (Oxoid). pAMβ1 was as reported by Martin et al. (31). Chromosomal DNA was prepared using published procedures (30), except that 0.1 μg of lysostaphin per ml (Applied Microbiology Inc.) was included for the lysis of S. epidermidis and S. haemolyticus. Chromosomal DNA was amplified using PfuTurbo DNA polymerase (Stratagene) under conditions recommended by the manufacturer. Restriction enzymes were obtained from GibcoBRL and used as specified by the manufacturer. All chemicals were from Sigma Chemical Co., unless otherwise specified.
Completion of nucleotide sequences.
Analysis of sequence databases revealed gaps in the nucleotide sequences of mvaA, mvaK1, mvaK2, and mvaD from E. faecium and mvaK2 from S. epidermidis and S. haemolyticus. Gaps were completed by comparing the operon structure from other members of the genus, designing primers, and amplifying the missing sequence. PCR products were sequenced directly on both strands. For mvaK1 of E. faecium, chromosomal DNA was sequenced directly to complete the 5′ end of the gene. Nucleotide and amino acid sequences were analyzed with the GCG version 9.0 and Lasergene (DNASTAR, Madison, Wis.) software packages.
Generation of S. pneumoniae allelic replacement mutants.
Chromosomal DNA fragments (500 bp long) flanking the genes of interest were PCR amplified from S. pneumoniae 0100993 chromosomal DNA. Primers were designed so that flanking genes and potential promoters would remain intact in the deletion mutant to minimize polar effects. The fragments were used to make constructs in which they flanked the erythromycin resistance gene ermAM from pAMβ1. A total of 106 S. pneumoniae R6 competent cells prepared by published methods were incubated with 500 ng of allelic replacement construct at 30°C for 30 min and transferred to 37°C for 90 min to allow expression of antibiotic resistance. The transformation mixes were plated in AGCH agar (24) containing 1 μg of erythromycin per ml and incubated at 37°C for 36 h under 5% CO2. Where appropriate, transformation and growth media were supplemented with 10 mM mevalonate. If no transformants were obtained in three separate transformation experiments with positive allelic replacement and transformation controls, the target gene was considered to be essential in vitro under the conditions chosen.
Erythromycin-resistant S. pneumoniae R6 colonies were picked and grown overnight in Todd-Hewitt broth (Difco) supplemented with 5% (wt/vol) yeast extract. Chromosomal DNA was prepared and used to transform S. pneumoniae 0100993 in the presence of 1 μg of competence-stimulating heptadecapeptide per ml (19). Chromosomal DNA from S. pneumoniae 0100993 Ermr clones was examined using Southern blot analysis and diagnostic PCR to verify that the appropriate chromosomal DNA rearrangement had occurred. In the former, flanking DNA fragments labeled using the ECL random-prime labeling kit (Amersham Pharmacia Biotech) were used as probes to chromosomal DNA restricted with appropriate enzymes and blotted using standard methods (39). In the latter, DNA primers designed to hybridize within ermAM were paired with primers hybridizing to distal chromosomal sequences to generate DNA amplification products of characteristic sizes.
Mouse respiratory tract infection model using S. pneumoniae.
S. pneumoniae 0100993 and the mvaS null mutant were grown on TSA plates containing 5% (vol/vol) sheep blood (BBL) (supplemented with 10 mM mevalonate as appropriate) at 37°C in 5% CO2 overnight. Bacteria were recovered from the plates and resuspended in phosphate-buffered saline to an absorbance at 600 nm of ∼0.9 (∼107 CFU ml−1). For each strain used, five male CBA/J mice (14 to 16 g) were anesthetized with 3% isoflurane and 50 μl of inoculum was administered by intranasal instillation. The mice were given food and water ad libitum. Mice were sacrificed at 12 or 48 h postinoculation by CO2 overdose, their lungs were aseptically removed and homogenized in 1 ml of phosphate-buffered saline, and viable bacteria were enumerated by serial dilution and viable counting on blood agar plates supplemented with 10 mM mevalonate for the mvaS null mutant.
RESULTS
Identification of mevalonate pathway genes in gram-positive cocci.
Through genomic analysis, the five genes encoding enzymes involved in the mevalonate pathway (Fig. 1) were identified in the low-G+C gram-positive cocci S. aureus, S. epidermidis, S. haemolyticus, S. pyogenes, S. pneumoniae, E. faecalis, and E. faecium. Primers for amplification of missing gene sequences were designed by comparing the gene arrangements in these related species, and additional sequencing resulted in the completion of the full-length gene sequences. An extensive homology search of sequence databases comprising over 60 partial and complete bacterial genomes did not reveal the presence of the complete set of mevalonate pathway genes in bacteria other than the low-G+C gram-positive cocci and B. burgdorferi. With the exception of the recently discovered ygbP gene, which encodes 4-diphosphocytidyl-2-C-methylerythritol synthase (37), genomes of the gram-positive cocci do not possess genes predicted to encode enzymes involved in the GAP-pyruvate pathway, although the genomes are incomplete. Only a single copy of each gene was identified in each species, except for the first enzyme in the pathway (acetyl-CoA acetyltransferase). Although three homologs were found in S. pyogenes, no putative acetyl-CoA acetyltransferase homologs were identified in the partial genomes of four strains of S. pneumoniae. A single open reading frame (mvaE) predicted to encode a single polypeptide with both acetyl-CoA acetyltransferase and HMG-CoA reductase activities was identified in E. faecalis and E. faecium. The amino-terminal region of the E. faecalis enzyme was 48% identical to the full length acetyl-CoA acetyltransferase enzyme from Thermoanaerobacterium thermosaccharolyticum (Z82038.1), and the carboxy-terminal end was 42% identical to the full length HMG-CoA reductase enzyme from Archaeoglobus fulgidus (AE000983.1). Sequence alignment programs and visual inspection were employed to compare the derived amino acid sequences from the five enzymes involved in the mevalonate pathway. Amino acid residues identified as important in catalysis in homologous eukaryotic, eubacterial, and archaeal enzymes are conserved in the enzymes of the gram-positive bacteria (Fig. 2). Multiple sequence alignments are available upon request from the authors (James_R_Brown@sbphrd.com).
FIG. 2.
Alignment of the conserved regions of the mevalonate pathway enzymes from gram-positive bacteria, B. burgdorferi, P. mevalonii, and eukaryotes. Bold type indicates that amino acids are identical in all proteins. The asterisks indicate amino acids proposed to function in catalysis and substrate binding. Dashes indicate gaps introduced into the sequence to optimize the alignment. Numbers refer to amino acid positions. In all alignments, abbreviations are as follows: EFA, E. faecalis; EFM, E. faecium; SEP, S. epidermidis; SHA, S. haemolyticus; SAU, S. aureus; SPN, S. pneumoniae; SPY, S. pyogenes; BBO, B. burgdorferi (AE001169). In HMG-CoA synthase, abbreviations are as follows: SCA, Staphylococcus carnosus (U450157); HUM, human cytoplasmic enzyme (Q01581); MIT, human mitochondrial enzyme (NP005509); SAC, S. cerevisiae (P54839). In HMG-CoA reductase, abbreviations are as follows: PME, P. mevalonii (P13702). In mevalonate kinase, abbreviations are as follows: HUM, human (NP000422); SAC, S. cerevisiae (NP000422). In phosphomevalonate kinase, abbreviations are as follows: SAC, S. cerevisiae (P24521). In mevalonate diphosphate decarboxylase, abbreviations are as follows: HUM, human (AAC50440); SAC, S. cerevisiae (AAC49252).
Phylogenetic analysis.
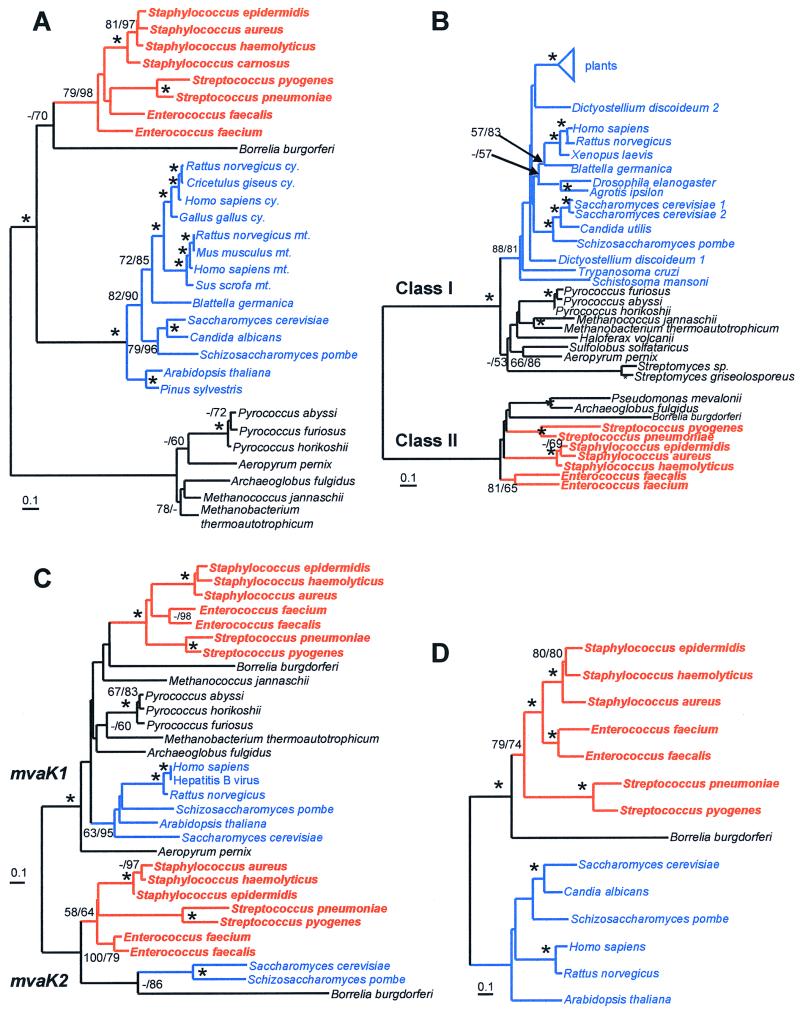
MP and NJ methods showed similar overall tree topologies for the four enzyme families (Fig. 3). Each of the phylogenetic analyses showed strong statistical support, in terms of bootstrap values and minimal-length trees, for clustering the enzymes from gram-positive bacteria together. Phylogenetic analyses were based on the total number of sites in edited multiple-sequence alignments for the proteins HMG-CoA synthase (275 sites), HMG-CoA reductase (331 sites), mevalonate and phosphomevalonate kinase (184 sites), and mevalonate diphosphate decarboxylase (292 sites). The clusters of gram-positive bacterial enzymes showed high levels of sequence identity (based on the length of the shorter sequence without gaps from the edited alignment) for each of the five enzymes HMG-CoA synthase (48 to 90%), HMG-CoA reductase (45 to 90%), mevalonate kinase (39 to 81%), phosphomevalonate kinase (34 to 87%), and mevalonate diphosphate decarboxylase (39 to 80%). Single minimal-length MP trees were determined for HMG-CoA synthase (minimal length [ml] = 1,617 steps) and mevalonate diphosphate decarboxylase (ml = 1,177 steps). For the mevalonate kinase family, two MP trees were found (ml = 1,812 steps), one of which clustered eukaryotes and gram-positive cocci together and one of which grouped the eukaryotes with the archaea. Sixteen MP trees were found for HMG-CoA reductase (ml = 3,430 steps) which differed only in the arrangements of the terminal class I taxa.
FIG. 3.
Phylogenetic trees of HMG-CoA synthase (A), HMG-CoA reductase (B), mevalonate and phosphomevalonate kinase (C), and mevalonate diphosphate decarboxylase (D). Trees were constructed using the NJ method as implemented by the program NEIGHBOR of the PHYLIP 3.57c package (16). The scale bar represents 0.1 expected amino acid replacements per site as estimated by the program PROTDIST using the Dayhoff PAM-120 substitution matrix. Branches containing or linking low-G+C gram-positive coccal species are colored red, those of eukaryotes are blue, while those of archaea and other bacteria are black. Numbers at the branching points represent the percent occurrence in 1,000 random bootstrap replications of MP and NJ analyses. Values less than 50% are not shown or are indicated by a dash (–), while nodes supported in more than 90% of bootstrap replications for both methods are marked with an asterisk (∗). Nodes separating classes I and II of HMG-CoA reductase as well as the mevalonate (mvaK1) and phosphomevalonate (mvaK2) kinases are indicated.
Although all protein trees are unrooted, the gram-positive bacterial mevalonate pathway enzymes are generally most similar to eukaryotic versions with the exception of HMG-CoA reductase, where the bacteria have enzymes of the class II rather than class I type (7). In the HMG-CoA synthase tree, there are three major clusters, the archaea, bacteria, and eukaryotes, and the tree could be rooted in any branch, although overall sequence similarities are slightly greater between eukaryotes and bacteria. Two distinct types of kinase were found. The mevalonate kinase gene (mvaK1) occurs in the archaea, bacteria, and eukaryotes, while the phosphomevalonate kinase gene (mvaK2) is restricted to gram-positive cocci, B. burgdorferi, and Saccharomyces cerevisiae. Phylogenetic analysis suggests that phosphomevalonate kinases are an ancient group of proteins and did not arise from gene duplications within the gram-positive cocci. The branching order of archaeal, eukaryotic, and bacterial mvaK1 could not be resolved, mainly because the archaea were not monophyletic. Extensive homology searches failed to detect the mvaD gene, encoding mevalonate diphosphate decarboxylase, in any archaeal genomes, although this enzyme is present in eukaryotes, gram-positive cocci, and B. burgdorferi.
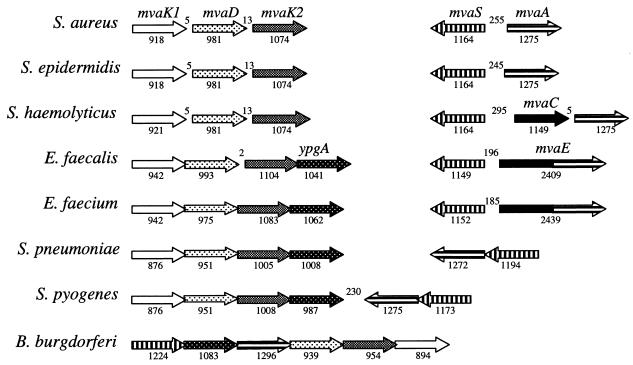
Gene organization of the mevalonate pathway.
The mevalonate pathway genes are located at two positions on the chromosome in the gram-positive bacteria examined, except in S. pyogenes, where all five genes are clustered at a single location (Fig. 4). The organization of the mvaK1DK2 genes is identical in the gram-positive cocci. A fourth gene (ypgA) is present downstream of mvaK2 in the enterococci and streptococci. Although its function is unknown, ypgA is also part of the mevalonate pathway operon in B. burgdorferi, and in Erwinia herbicola (open reading frame 6) it is clustered with genes involved in carotenoid biosynthesis (22), rather than with mevalonate pathway genes. Erwinia species are reported to possess the GAP-pyruvate pathway for IPP biosynthesis (35). A copy of ypgA is present in the staphylococci but at a different location on the chromosome. The organization of the HMG-CoA synthase (mvaS) and reductase (mvaA) genes is less well conserved, although in all cases they are either adjacent to each other or separated by an acetyl-CoA acetyltransferase (mvaC) coding region. mvaS and mvaA are divergently transcribed in the staphylococci and enterococci but transcribed in the same direction in the streptococci.
FIG. 4.
Organization of the mevalonate pathway genes in gram-positive cocci and B. burgdorferi. Numbers above the genes indicate the number of nucleotides between genes, and numbers below indicate the length of each gene. ypgA has homology to a gene clustered with carotenoid biosynthetic genes in E. herbicola (22).
Essentiality of the mevalonate pathway genes in S. pneumoniae R6.
DNA constructs containing an erythromycin resistance gene flanked by DNA adjacent to the target gene were generated for the allelic replacement of mvaS, mvaA, mvaK1, mvaK2, and mvaD and used to transform S. pneumoniae R6. It was not possible to obtain allelic exchange mutants for any of the genes in the absence of added mevalonate, suggesting that all of the genes are essential for the in vitro growth of S. pneumoniae under the standard growth conditions.
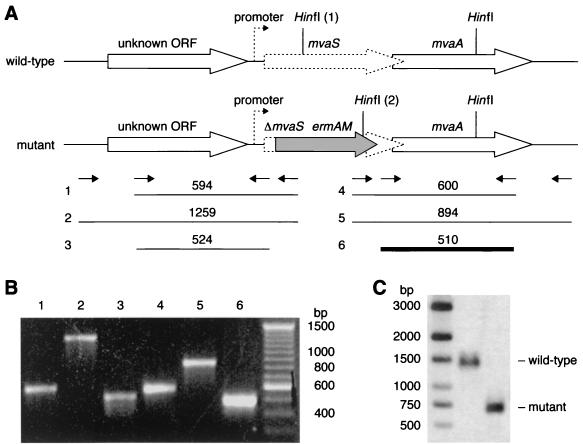
It was predicted from the pathway that mutants auxotrophic for mevalonate could be obtained. HMG-CoA, mevalonate phosphate, mevalonate diphosphate, and IPP were not expected to penetrate the cell and were not included in complementation studies. When the transformation and growth media were supplemented with 10 mM mevalonate, mvaS auxotrophic mutants were readily obtained (data not shown), and the allelic replacement was confirmed using Southern blot analysis and diagnostic PCR (Fig. 5). Surprisingly, it was not possible to obtain a mvaA auxotrophic mutant in multiple experiments under similar conditions, although double allelic replacement of both mvaA and mvaS was readily achieved and these mutants were auxotrophic for mevalonate.
FIG. 5.
Chromosomal analysis of the S. pneumoniae mvaS null mutant. (A) The chromosome of the mvaS null mutant contains an allelic replacement of mvaS (dotted block arrow, 1,194 bp) from nucleotides 166 to 1161 with the ermAM gene (gray block arrow). The translationally coupled downstream gene (mvaA) and the putative upstream promoter (dotted arrow) remain intact in the mutant. Oligonucleotide primers (small arrows) were designed to hybridize within the ermAM gene and to regions flanking mvaS and were used in PCRs against chromosomal DNA template prepared from the mutant. (B) Reactions 1, 2, 4, and 5 generated characteristic PCR products (shown), which could not be amplified with chromosomal DNA from the wild-type parent strain (not shown). Reactions 3 and 6 generated control fragments, which could be amplified in the mutant and parent strains. (C) Wild-type and mutant chromosomal DNA was restricted with HinfI, subjected to agarose gel electrophoresis, and probed with labeled PCR fragment 6 (bold line). The predicted band sizes are 696 bp in the mutant and 1396 bp in the wild-type strain, since the left HinfI site (labeled 1 in panel A) is deleted in the mutant and replaced by the ermAM gene containing another HinfI site (labeled 2).
Investigation of mevalonate auxotrophy of the mvaS null mutant.
A minimum concentration of 3 mM mevalonate was required by the mvaS null mutant for growth in Todd-Hewitt broth plus 5% (wt/vol) yeast extract (data not shown). An overnight culture of the mvaS null mutant grown in the presence of 3 mM mevalonate was diluted (104-fold) into fresh medium with and without mevalonate, and the number of viable cells over 8 h was determined by plating on medium containing mevalonate (data not shown). In the unsupplemented medium, the mvaS null mutant was unable to grow, although cells remained viable. The mutant's viable count increased by 3 log units over the same period in the presence of 3 mM mevalonate.
Virulence attenuation of the mvaS null mutant.
The mvaS mutation was transferred to the pathogenic strain S. pneumoniae 0100993 by chromosomal transformation, and the mutants were auxotrophic for mevalonate. Mice were inoculated intranasally with either S. pneumoniae 0100993 or the mvaS null mutant. Viable S. pneumoniae 0100993 harboring the deletion could not be recovered from the lungs (i.e., below the limit of detection of 1.60 log10 CFU/mouse) after 12 or 48 h when plated in the presence of mevalonate, in contrast to the wild-type S. pneumoniae 0100993, which was present at 5.67 ± 0.11 and 7.39 ± 0.75 log10 CFU/mouse at 12 and 48 h, respectively (mean ± standard deviation for five mice). The results indicate that virulence in the mutant is severely attenuated at both early and late stages of respiratory tract infection.
DISCUSSION
The low-G+C gram-positive cocci are major pathogens, and novel targets for drug intervention are required to circumvent the resistance mechanisms which compromise current antibacterial agents. In a search for such novel drug targets, five full-length genes encoding enzymes that are predicted to catalyze consecutive steps in the mevalonate pathway for IPP biosynthesis were identified in seven species of gram-positive cocci. The nucleotide and derived amino acid sequences of HMG-CoA synthase, mevalonate kinase, phosphomevalonate kinase, and mevalonate diphosphate decarboxylase have not previously been identified in bacteria other than Borrelia burgdorferi, and the amino acid sequences of streptococcal biosynthetic HMG-CoA reductases have been reported only recently (7). None of the species possessed homologs of the genes identified in the GAP-pyruvate pathway for IPP synthesis other than ygbP, suggesting that isoprenoids are synthesized in the streptococci, staphylococci, and enterococci solely via the mevalonate pathway.
Based on amino acid sequence analysis, Bochar et al. (7) suggested that there are two distinct classes of HMG-CoA reductase: class I, found in most archaea, fungi, and eukaryotes and in Streptomyces CL190 (47), and class II, found in some bacteria and the archaeon Archaeoglobus fulgidus. HMG-CoA reductases identified in staphylococci, streptococci, and enterococci are of the class II type (Fig. 3) and show a high degree of sequence homology to the Pseudomonas mevalonii enzyme (Fig. 2). Residues involved in the catalysis of HMG-CoA reductases have been identified through crystallography (27, 46) and mutagenesis (12, 53), and these residues are conserved in the enzymes of the gram-positive cocci. The product of the Staphylococcus aureus mvaA gene has been isolated and shown to be an HMG-CoA reductase (unpublished results). The enterococci are uniquely predicted to synthesize a bifunctional protein which possesses both HMG-CoA reductase and acetyl-CoA acetyltransferase activities. These enzymes catalyze the first and third steps in the production of IPP, respectively (Fig. 1). No other genes encoding polypeptides with putative acetyl-CoA acetyltransferase activity were identified in the enterococci, although the genomes are incomplete. The other enzymes of the mevalonate pathway are less well studied, although several catalytic residues have been identified and are conserved in the enzymes of the gram-positive cocci (Fig. 2).
Our analysis suggests that gram-positive cocci and B. burgdorferi probably acquired all the IPP biosynthesis pathway genes through one or two horizontal gene transfer events. A primitive eukaryotic genome probably was the single source for the mevalonate pathway genes, since the archaeal species are missing identifiable mvaK1 and mvaD genes and since the phosphomevalonate kinase of higher eukaryotes (11) shows no sequence homology to that from bacteria or Saccharomyces cerevisiae (51). Furthermore, with the exception of HMG-CoA reductase, all the phylogenies suggest that the bacterial enzymes are more closely related to those of eukaryotes than to those of the archaea. Horizontal gene transfers from eukaryotes to bacteria of genes essential for cell viability have been reported previously (9, 10). In nearly all instances, phylogenetic analyses suggest that gene transfers occurred very early in eukaryotic evolution, prior to the divergence of plants, animals, and fungi (8).
The consistent organization of mevalonate pathway genes in the gram-positive cocci is notable and may indicate a common mechanism for the regulation of the pathway; it might also provide clues to the mode of gene acquisition. mvaK1, mvaD, and mvaK2 are organized in an identical operon in all species. mvaS and mvaA (or mvaE) are adjacent on the chromosome but are some distance from the other mevalonate pathway genes, except in S. pyogenes, where the genes are clustered. Gram-positive cocci probably acquired all of the genes involved in IPP biosynthesis via one or two horizontal transfer events. A possible scenario is suggested by the highly conserved genomic organization of the IPP biosynthetic genes. The ancestral gram-positive coccal species might have first acquired mvaA and initially utilized HMG-CoA reductase for mevalonate catabolism similar to P. mevalonii. Subsequently, mvaS and mvaK1DK2 were acquired, permitting the biosynthesis of IPP via mevalonate. In B. burgdorferi, the clustering of mevalonate pathway genes is more tightly linked, suggesting that this organism participated in a separate genetic exchange with a eukaryotic cell. The tight linkage of mevalonate pathway genes in eubacteria further supports recent theories that genes in operons encoding single metabolic functions are highly favored for horizontal transfer between organisms since the recipient bacteria would acquire a novel metabolic function conferring a selective advantage (25, 26).
All five genes encoding enzymes of the mevalonate pathway were essential for the growth of S. pneumoniae in vitro under standard conditions and are envisaged to be essential in the other gram-positive cocci, since they also lack genes predicted to encode the enzymes of the GAP-pyruvate pathway. S. pneumoniae R6 mvaS null mutants grew as well as the wild type in the presence of mevalonate, although no mvaA null mutants could be obtained under similar conditions. Since both mvaS and mvaA could be replaced simultaneously in a double knockout, it is possible that deletion of mvaA alone led to a lethal intracellular accumulation of HMG-CoA. Removal of mevalonate from the medium prevented growth of the mvaS null mutant, although the mutant remained viable. The dependency of these allelic replacement mutants on mevalonate for growth supports the proposed functions of the genes. Inhibitors of HMG-CoA synthase are predicted to have a bacteriostatic rather than a bacteriocidal effect, although inhibitors of HMG-CoA reductase would probably have a bacteriocidal effect on S. pneumoniae. The mvaS null mutant was severely attenuated in the murine respiratory tract infection model, and no bacteria were recovered at either the early or late stages of infection, suggesting that the bacteria are rapidly cleared within the first 12 h. The mevalonate content of plasma in humans and rats has been estimated to be 0.02 to 0.08 μM and 0.08 to 0.50 μM, respectively (34), both of which are well below the concentration of mevalonate required to support the growth of the mvaS null mutant on rich medium in vitro.
This study shows that, unlike B. subtilis and the gram-negative bacteria except B. burgdorferi, the gram-positive cocci possess the mevalonate pathway for IPP biosynthesis (Table 1). The essentiality of the mevalonate pathway for growth and the high sequence divergence from eukaryotic homologs identifies its components as potential antibacterial targets and exemplifies the use of comparative genomics to define novel targets for drug intervention against life-threatening, multidrug-resistant gram-positive cocci.
TABLE 1.
Distribution of mevalonate and GAP-pyruvate pathways for IPP biosynthesis
| Organism | Presence of:
|
Reference(s) | |
|---|---|---|---|
| Mevaolonate pathway | GAP-pyruvate pathway | ||
| Gram-negative bacteria | |||
| Enterobacteriaceae, Haemophilus influenzae | − | + | 21, 35, 37 |
| B. burgdorferi, M. fulvus, C. ethylica | + | − | 21, 35 |
| Gram-positive bacteria | |||
| Mycobacteria, B. subtilis | − | + | 35, 52 |
| Streptomyces spp.a | + | + | 41 |
| Low-G+C gram-positive cocci | + | − | This study |
| Archaea | + | − | 5, 6 |
| Fungi | + | − | 28 |
| Higher plants | |||
| Cytoplasmic | + | − | 17 |
| Plastid | − | + | 29 |
| Algae | +b | +c | 14, 40, 43 |
| Animals | + | − | 17 |
Some species of Streptomyces possess both pathways simultaneously.
Including species such as S. obliquus, Chlorella fusca, and Chlamydomonas reinhardtii.
Including species such as E. gracilis.
ACKNOWLEDGMENTS
Preliminary sequence data for the mevalonate pathway gene project were obtained from The Institute for Genomic Research (http://www.tigr.org) and from Incyte Pharmaceutical Inc's PathoSeq™ sequence database. We thank the SmithKline Beecham sequencing and oligonucleotide biosynthesis unit for providing oligonucleotide primers and conducting the sequencing reactions. We also thank Earl May, Paul Hoffman, and Dan Gentry for their helpful advice on the manuscript.
REFERENCES
- 1.Alberts A W, Chen J, Kuron G, Hunt V, Huff J, Hoffman C, Rothrock J, Lopez M, Joshua H, Harris E, Patchett A, Monaghan R, Currie S, Stapley E, Albers-Schonberg G, Hensens O, Hirshfield J, Hoogsteen K, Liesch J, Springer J. Mevinolin: a highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc Natl Acad Sci USA. 1980;77:3957–3961. doi: 10.1073/pnas.77.7.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avery O T, MacLeod C M, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beach M J, Rodwell V W. Cloning, sequencing, and overexpression of mvaA, which encodes Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Bacteriol. 1989;171:2994–3001. doi: 10.1128/jb.171.6.2994-3001.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischoff K M, Rodwell V W. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase from Haloferax volcanii: purification, characterization, and expression in Escherichia coli. J Bacteriol. 1996;178:19–23. doi: 10.1128/jb.178.1.19-23.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bochar D A, Brown J R, Doolittle W F, Klenk H P, Lam W, Schenk M E, Stauffacher C V, Rodwell V W. 3-Hydroxy-3-methylglutaryl coenzyme A reductase of Sulfolobus solfataricus: DNA sequence, phylogeny, expression in Escherichia coli of the hmgA gene and purification and kinetic characterization of the gene product. J Bacteriol. 1997;179:3632–3638. doi: 10.1128/jb.179.11.3632-3638.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bochar D A, Stauffacher C V, Rodwell V W. Sequence comparisons reveal two classes of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol Gen Metab. 1999;66:122–127. doi: 10.1006/mgme.1998.2786. [DOI] [PubMed] [Google Scholar]
- 8.Brown J R, Doolittle W F. Archaea and the prokaryote-eukaryote transition. Microbiol Mol Biol Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown J R, Doolittle W F. Gene descent, duplication and horizontal transfer in the evolution of glutamyl- and glutaminyl-tRNA synthetases. J Mol Evol. 1999;49:485–495. doi: 10.1007/pl00006571. [DOI] [PubMed] [Google Scholar]
- 10.Brown J R, Zhang J, Hodgson J E. A bacterial antibiotic resistance gene with eukaryotic origins. Curr Biol. 1998;8:R365–R367. doi: 10.1016/s0960-9822(98)70238-6. [DOI] [PubMed] [Google Scholar]
- 11.Chambliss K L, Slaughter C A, Schreiner R, Hoffmann G F, Gibson K M. Molecular cloning of human phosphomevalonate kinase and identification of a consensus peroxisomal targeting sequence. J Biol Chem. 1996;271:17330–17334. doi: 10.1074/jbc.271.29.17330. [DOI] [PubMed] [Google Scholar]
- 12.Darnay B G, Wang Y, Rodwell V W. Identification of the catalytically important histidine of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 1992;267:15064–15070. [PubMed] [Google Scholar]
- 13.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olson G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 14.Disch A, Schwender J, Müller C, Lichtenthaler H K, Rohmer M. Distribution of the mevalonate and glyceraldehyde phosphate/pyruvate pathways for isoprenoid biosynthesis in unicellular algae and the cyanobacterium Synechocystis PCC 6714. Biochem J. 1998;333:381–388. doi: 10.1042/bj3330381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doolittle W F, Lodgson J M., Jr Archaeal genomics: do archaea have a mixed heritage? Curr Biol. 1998;6:R209–R211. doi: 10.1016/s0960-9822(98)70127-7. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.57c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 17.Goldstein J L, Brown M S. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 18.Hammond R K, White D C. Carotenoid formation by Staphylococcus aureus. J Bacteriol. 1970;103:191–198. doi: 10.1128/jb.103.1.191-198.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henikoff S, Henikoff J G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horbach S, Sahm H, Welle R. Isoprenoid biosynthesis in bacteria: two different pathways? FEMS Microbiol Lett. 1993;111:135–140. doi: 10.1111/j.1574-6968.1993.tb06375.x. [DOI] [PubMed] [Google Scholar]
- 22.Hundle B, Alberti M, Nievelstein V, Beyer P, Kleinig H, Armstrong G A, Burke D H, Hearst J E. Functional assignment of Erwinia herbicola Eho10 carotenoid genes expressed in Escherichia coli. Mol Gen Genet. 1994;245:406–416. doi: 10.1007/BF00302252. [DOI] [PubMed] [Google Scholar]
- 23.Johnson E A, Schroeder W A. Microbial carotenoids. Adv Biochem Eng Biotechnol. 1996;53:119–178. doi: 10.1007/BFb0102327. [DOI] [PubMed] [Google Scholar]
- 24.Lacks S. Integration efficiency and genetic recombination in pneumococcal transformation. Genetics. 1966;53:207–235. doi: 10.1093/genetics/53.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence J G. Selfish operons and speciation by gene transfer. Trends Microbiol. 1997;5:355–359. doi: 10.1016/S0966-842X(97)01110-4. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence J G, Roth J R. Selfish operons—horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence C M, Rodwell V W, Stauffacher C V. Crystal structure of Pseudomonas mevalonii HMG-CoA reductase at 3.0 angstrom resolution. Science. 1995;268:1758–1762. doi: 10.1126/science.7792601. [DOI] [PubMed] [Google Scholar]
- 28.Lees N D, Bard M, Kirsch D R. Biochemistry and molecular biology of sterol synthesis in Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 1999;34:33–47. [PubMed] [Google Scholar]
- 29.Lichtenthaler H K, Schwender J, Disch A, Rohmer M. Biosynthesis of isoprenoids in higher plant chloroplasts proceeds via a mevalonate-independent pathway. FEBS Lett. 1997;400:271–274. doi: 10.1016/s0014-5793(96)01404-4. [DOI] [PubMed] [Google Scholar]
- 30.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 31.Martin B, Alloing G, Mejean V, Claverys J-P. Constitutive expression of the erythromycin resistance mediated by the ermAM determinant of plasmid pAMβ1 results from deletion of 5′ leader peptide sequences. Plasmid. 1987;18:250–253. doi: 10.1016/0147-619x(87)90068-0. [DOI] [PubMed] [Google Scholar]
- 32.Meganathan R. Biosynthesis of the isoprenoid quinones menaquinone (vitamin K2) and ubiquinone (coenzyme Q) In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. I. Washington, D.C.: American Society for Microbiology; 1996. pp. 642–656. [Google Scholar]
- 33.Ohnoki S, Suzue G, Tanaka S. 5-Phosphomevalonic acid as the intermediate of the enzymatic synthesis of bacterial phytoene. J Biochem. 1962;52:423–427. doi: 10.1093/oxfordjournals.jbchem.a127638. [DOI] [PubMed] [Google Scholar]
- 34.Popják G, Boehm G, Parker T S, Edmond J, Edwards P A, Fogelman A M. Determination of mevalonate in blood plasma in man and rat. Mevalonate “tolerance” tests in man. J Lipid Res. 1979;20:716–728. [PubMed] [Google Scholar]
- 35.Putra S R, Disch A, Bravo J-M, Rohmer M. Distribution of mevalonate and glyceraldehyde-3-phosphate/pyruvate routes for isoprenoid biosynthesis in some Gram-negative bacteria and mycobacteria. FEMS Microbiol Lett. 1998;164:169–175. doi: 10.1111/j.1574-6968.1998.tb13082.x. [DOI] [PubMed] [Google Scholar]
- 36.Reusch V M., Jr Lipopolymers, isoprenoids, and the assembly of the gram-positive cell wall. Crit Rev Microbiol. 1984;11:129–155. doi: 10.3109/10408418409105475. [DOI] [PubMed] [Google Scholar]
- 37.Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner S, Herz S, Kis K, Eisenrich W, Bacher A, Zenk M H. Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc Natl Acad Sci USA. 1999;96:11758–11763. doi: 10.1073/pnas.96.21.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohmer M, Knani M, Simonin P, Sutter B, Sahm H. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentyl diphosphate. Biochem J. 1993;295:517–524. doi: 10.1042/bj2950517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Schwender J, Seeman M, Lichtenthaler H K, Rohmer M. Biosynthesis of isoprenoids (carotenoids, sterols, prenyl side-chains of chlorophylls and plastoquinone) via a novel pyruvate/glyceraldehyde 3-phosphate non-mevalonate pathway in the green alga Scenedesmus obliquus. Biochem J. 1996;316:73–80. doi: 10.1042/bj3160073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seto H, Watanabe H, Furihata K. Simultaneous operation of the mevalonate and non-mevalonate pathways in the biosynthesis of isopentenyl diphosphate in Streptomyces aeriouvifer. Tetrahedron Lett. 1996;37:7979–7982. [Google Scholar]
- 42.Spurgeon S L, Porter J W. Introduction. In: Porter J W, Spurgeon S L, editors. Biosynthesis of isoprenoid compounds. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1981. pp. 1–46. [Google Scholar]
- 43.Steele W J, Gurin S. Biosynthesis of β-carotene in Euglena gracilis. J Biol Chem. 1960;235:2778–2785. [Google Scholar]
- 44.Stephens R S, Kalman S, Lammel C J, Fan J, Marathe R, Aravind L, Mitchell W P, Olinger L, Tatusov R L, Zho Q, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 45.Swofford D L. PAUP*. Phylogenetic analysis using parsimony (* and other methods), version 4. Sunderland, Mass: Sinauer Associates; 1999. [Google Scholar]
- 46.Tabernero L, Bochar D A, Rodwell V W, Stauffacher C V. Substrate-induced closure of the flap domain in the ternary complex structures provides insights into the mechanism of catalysis by 3-hydroxy-3-methylglutaryl-CoA reductase. Proc Natl Acad Sci USA. 1999;96:7167–7171. doi: 10.1073/pnas.96.13.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi S, Kuzuyama T, Seto H. Purification, characterization and cloning of a eubacterial 3-hydroxy-3-methylglutaryl coenzyme A reductase, a key enzyme involved in biosynthesis of terpenoids. J Bacteriol. 1999;181:1256–1263. doi: 10.1128/jb.181.4.1256-1263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson J D, Higgins D G, Gibson T J. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thorne K J, Kodicek E. The structure of bactoprenol, a lipid formed by lactobacilli from mevalonic acid. Biochem J. 1966;99:123–127. doi: 10.1042/bj0990123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thorne K J I. Incorporation of radioactive mevalonate into C50 and C55 prenols by Streptococcus mutans. Biochem J. 1973;135:567–568. doi: 10.1042/bj1350567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsay Y H, Robinson G W. Cloning and characterization of ERG8, an essential gene of Saccharomyces cerevisiae that encodes phosphomevalonate kinase. Mol Cell Biol. 1991;11:620–631. doi: 10.1128/mcb.11.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner W P, Helmig D, Fall R. Isoprene biosynthesis in Bacillus subtilis via the methylerythritol phosphate pathway. J Nat Prod. 2000;63:37–70. doi: 10.1021/np990286p. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Darnay B G, Rodwell V W. Identification of the principal catalytically important acidic residue of 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Biol Chem. 1990;265:21634–21641. [PubMed] [Google Scholar]