Abstract
The family of intracellular lipid binding proteins (iLBPs) is comprised of 16 members of structurally related binding proteins that have ubiquitous tissue expression in humans. iLBPs collectively bind diverse essential endogenous lipids and xenobiotics. iLBPs solubilize and traffic lipophilic ligands through the aqueous milieu of the cell. Their expression is correlated with increased rates of ligand uptake into tissues and altered ligand metabolism. The importance of iLBPs in maintaining lipid homeostasis is well established. Fatty acid binding proteins (FABPs) make up the majority of iLBPs and are expressed in major organs relevant to xenobiotic absorption, distribution, and metabolism. FABPs bind a variety of xenobiotics including nonsteroidal anti-inflammatory drugs, psychoactive cannabinoids, benzodiazepines, antinociceptives, and peroxisome proliferators. FABP function is also associated with metabolic disease, making FABPs currently a target for drug development. Yet the potential contribution of FABP binding to distribution of xenobiotics into tissues and the mechanistic impact iLBPs may have on xenobiotic metabolism are largely undefined. This review examines the tissue-specific expression and functions of iLBPs, the ligand binding characteristics of iLBPs, their known endogenous and xenobiotic ligands, methods for measuring ligand binding, and mechanisms of ligand delivery from iLBPs to membranes and enzymes. Current knowledge of the importance of iLBPs in affecting disposition of xenobiotics is collectively described.
SIGNIFICANCE STATEMENT
The data reviewed here show that FABPs bind many drugs and suggest that binding of drugs to FABPs in various tissues will affect drug distribution into tissues. The extensive work and findings with endogenous ligands suggest that FABPs may also alter the metabolism and transport of drugs. This review illustrates the potential significance of this understudied area.
Introduction
Intracellular lipid binding proteins (iLBPs) are a family of ubiquitous proteins in animals that solubilize essential cellular lipids (Schaap et al., 2002; Storch and Corsico, 2008; Smathers and Petersen, 2011; Napoli, 2017). Together with avidins and lipocalins, iLBPs belong to the calycin superfamily of structurally related binding proteins. Despite low amino acid sequence homology (<10%), avidins, lipocalins and iLBPs share a common β-barrel structural fold that makes up their ligand binding cavity (Flower et al., 2000; Schaap et al., 2002; Smathers and Petersen, 2011). Avidins and lipocalins are found in both prokaryotic and eukaryotic organisms, but iLBPs are only present in vertebrate and invertebrate animals (Schaap et al., 2002). The ancestral iLBP gene evolved after animals diverged from plants and fungi, and individual isoforms arose through gene duplication and diversification (Schaap et al., 2002; Haunerland and Spener, 2004; Smathers and Petersen, 2011). The primary amino acid sequence identity for the 16 known human iLBPs ranges from 21% to 77% (Fig. 1A). Generally, the amino acid sequence identity for specific iLBPs across different species is greater than the sequence identity of all fatty acid binding proteins (FABPs) within the same species. For example, fatty acid binding protein 1 (FABP1) has >60% amino acid sequence identity across 18 different species (Zhang et al., 2020), but the sequence identity of all FABPs in humans is as low as 21%.
Fig. 1.
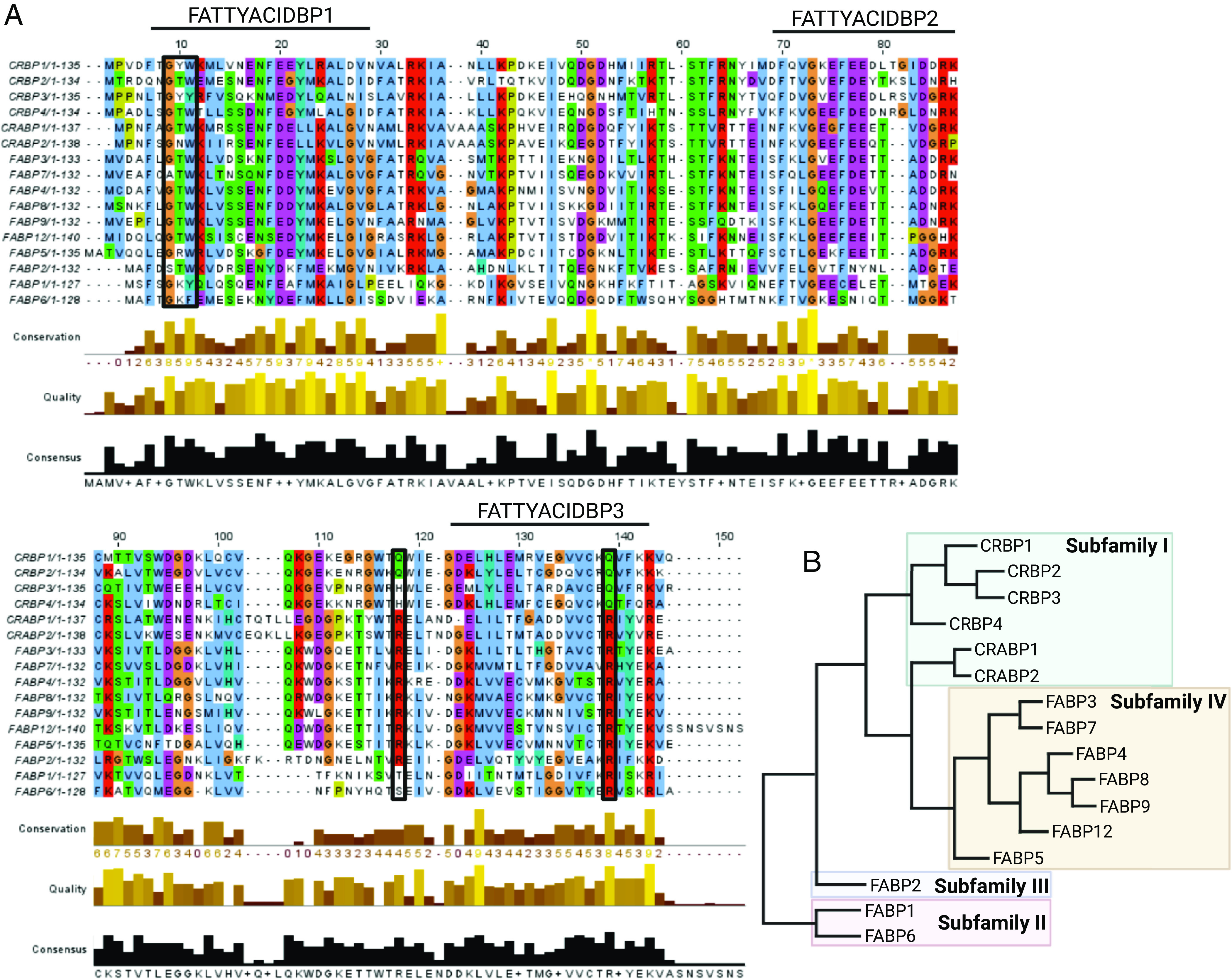
Sequence alignment (A) and phylogenetic tree (B) of human iLBPs. The primary amino acid sequences for all human iLBP family members were collected from the National Center for Biotechnology Information protein database (https://www.ncbi.nlm.nih.gov/protein/). The accession numbers for the amino acid sequences used were P09455.2 (CRBP1), P50120.3 (CRBP2), NP_113679.1 (CRBP3), Q96R05.1 (CRBP4), P29762.2 (CRABP1), P29373.2 (CRABP2), P07148.1 9 (FABP1), P12104.2 (FABP2), P05413.4 (FABP3), P15090.3 (FABP4), Q01469.3 (FABP5), P51161.2 (FABP6), O15540.3 (FABP7), P02689.3 (FABP8), Q0Z7S8.1 (FABP9), A6NFH5.2 (FAPB12). The sequences were aligned using Clustal Omega Multiple Sequence Alignment (https://www.ebi.ac.uk/Tools/msa/clustalo/) (Sievers et al., 2011) and visualized using JalView (Waterhouse et al., 2009). The black bars above the sequence alignment show the three motifs (FATTYACIDBP1-3) that make up the highly conserved fingerprint common to all iLBPs. The colored residues indicate conserved residues based on thresholds set by the Clustal X Color Scheme (https://www.jalview.org/help/html/colourSchemes/clustal.html). Red indicates positively charged residues, blue residues are hydrophobic, magenta are negatively charged, green are polar, orange are glycines, yellow are prolines, and cyan are aromatic. Boxed residues indicate locations of a highly conserved G-x-W triplet common to iLBPs and lipocalins and highly conserved residues involved in ionic interactions with hydroxy and carbonyl groups of ligands. The phylogenetic tree shown in (B) was calculated using the UPGMA clustering method in Simple Phylogeny (https://www.ebi.ac.uk/Tools/phylogeny/simple_phylogeny/) using the multiple sequence alignment data for human iLBPs. Evolutionary distances and phylogenetic relationships should not be inferred from this tree. (Figure created with BioRender.com.)
The human iLBPs are divided into four subfamilies (Fig. 1B) based on phylogenetic analysis and amino acid sequences (Schaap et al., 2002; Liu et al., 2008; Smathers and Petersen, 2011; Ragona et al., 2014). Subfamily I is comprised of the cellular retinol binding proteins (CRBPs) and cellular retinoic acid binding proteins (CRABPs). Subfamily II contains liver FABP (FABP1) and ileal FABP (FABP6, also called I-BABP). Intestinal FABP (FABP2) is the lone iLBP to make up subfamily III and heart (FABP3), adipocyte (FABP4), epidermal (FABP5), brain (FABP7), myelin (FABP8), and testis (FABP9) FABPs, and FABP12 make up subfamily IV (Schaap et al., 2002; Smathers and Petersen, 2011). The FABPs were originally named after the organs from which they were cloned but have been later found to have broader expression.
The human iLBP genes are located in several different chromosomes (Table 1) and, like most iLBP genes in animals, have four exons with three introns (Schaap et al., 2002; Babin, 2009; Smathers and Petersen, 2011; Zhang et al., 2020). The second and third exons are conserved in nearly all FABP genes (Zhang et al., 2020). Phylogenetic studies suggest that FABP genes evolved from a common ancestor likely through tandem duplication (Babin, 2009; Zhang et al., 2020). FABP4, 5, 8, 9, and 12 form a gene cluster on the same chromosome in humans and several other mammals. Some of these FABP genes also form clusters in aves, amphibians, and reptiles. This supports the hypothesis that vertebrate FABP genes may have arisen through continuous tandem duplication from a common ancestor (Zhang et al., 2020).
Table 1.
Tissue expression patterns, genomic localization, and endogenous ligands of iLBPs
| Gene | Protein Name | Molecular Weight (kDa)a | Human Gene Locusb | Tissue Expression | Known Endogenous Ligands |
|---|---|---|---|---|---|
| Subfamily I | |||||
| RBP1 | CRBP1 | 15.9 | 3q23 | Adipose, brain, heart, kidney, liver, lung, mammary gland, muscle, ovary, pituitary, spinal cord, skin, spleen, stomach, testis | Retinol, retinaldehyde |
| RBP2 | CRBP2 | 15.7 | 3q23 | Heart, muscle, small intestine, placenta | Retinol, retinaldehyde, monoacylglycerols |
| RBP5 | CRBP3 | 15.9 | 12p13.31 | Adipose, heart, muscle | Retinol, retinaldehyde |
| RBP7 | CRBP4 | 15.5 | 1p36.22 | Large intestine, heart, kidney | Retinol, retinaldehyde |
| CRABP1 | CRABP1 | 15.6 | 15q25.1 | Adipose, adrenal, brain, eye, kidney, liver, lung, lymph nodes, muscle, pancreas, skin, small intestine, spleen, stomach, testis, thymus | Retinoic acid, retinoic acid metabolites |
| CRABP2 | CRABP2 | 15.7 | 1q23.1 | Skin, testis | Retinoic acid, retinoic acid metabolites |
| Subfamily II | |||||
| FABP1 | Liver FABP | 14.2 | 2p11.2 | Kidney, liver, lung, pancreas, small intestine, stomach | LCFAs, fatty acyl CoA, fatty acyl-carnitines, monoacylglycerols, lysophospholipids, bile acids, cholesterol, heme, bilirubin, retinoic acid, endocannabinoids, prostaglandins, vitamin K |
| FABP6 | Ileal FABP | 14.4 | 5q33.3 | Adrenal, ovary, small intestine, stomach | Bile acids, LCFAs |
| Subfamily III | |||||
| FABP2 | Intestinal FABP | 15.2 | 4q26 | Small intestine, liver | LCFAs |
| Subfamily IV | |||||
| FABP3 | Heart FABP | 14.9 | 1p35.2 | Adipose, adrenal, brain, heart, kidney, liver, lung, mammary gland, muscle, ovary, placenta, spleen, stomach, testis | LCFAs, EETs, HETEs, DHETs |
| FABP4 | Adipose FABP | 14.7 | 8q21.13 | Adipose, dendritic cells, macrophages, muscle | LCFAs, retinoic acid |
| FABP5 | Epidermal FABP | 15.2 | 8q21.13 | Adipose, brain, dendritic cells, eye, heart, kidney, liver, lung, macrophages, mammary gland, muscle, placenta, skin, small intestine, spleen, stomach, testis, tongue | LCFAs, endocannabinoids, retinoic acid |
| FABP7 | Brain FABP | 14.9 | 6q22.31 | Brain, central nervous, mammary gland, muscle, system glial cells, eye | LCFAs |
| FABP8 | Myelin FABP | 14.9 | 8q21.13 | Peripheral nervous system myelin | Cholesterol, LCFAs |
| FABP9 | Testis FABP | 15.1 | 8q21.13 | Mammary gland, salivary gland, testis | LCFAs |
| FABP12 | FABP12 | 15.6 | 8q21.13 | Retina, testis | LCFAs |
aMolecular weight determined by Expasy ProtParam (Gasteiger et al., 2005) based on reference amino acid sequence.
bGene location obtained from the National Center of Biotechnology and Information gene database.
DHET, dihydroxyeicosatrienoic acids; EET, epoxyeicosatrienoic acid; HETE, hydroxyeicosatetraenoic acid.
The complete physiologic functions of iLBPs have yet to be defined, but iLBPs appear to facilitate the efficient uptake of endogenous lipids into tissues, acting as carriers to shuttle ligands through the cytosol and modulating rates of ligand metabolism (Kushlan et al., 1981; Luxon and Weisiger, 1993; Martin et al., 2003; Kaczocha et al., 2009; Yu et al., 2014; Gajda and Storch, 2015). Altered iLBP function and expression have been associated with dyslipidemia, metabolic syndrome, obesity, diabetes, atherosclerosis, and inflammation (Furuhashi and Hotamisligil, 2008; Atshaves et al., 2010; Peng et al., 2012; Wang et al., 2016; Furuhashi, 2019; Valizadeh et al., 2021). Several iLBP isoforms also bind xenobiotics (Chuang et al., 2008; Trevaskis et al., 2011; Velkov, 2013; Lee et al., 2015; Huang et al., 2018; Elmes et al., 2019). Based on their high and ubiquitous expression in tissues, iLPBs may be determinants of xenobiotic distribution and uptake into tissues. This review focuses on ligand binding to iLBPs, tissue expression of iLBPs, methods to determine ligand binding, and the biochemical roles of iLBPs as they relate to the potential of iLBPs to be determinants of drug disposition.
Intracellular Lipid Binding Protein Structures and Endogenous Ligand Binding
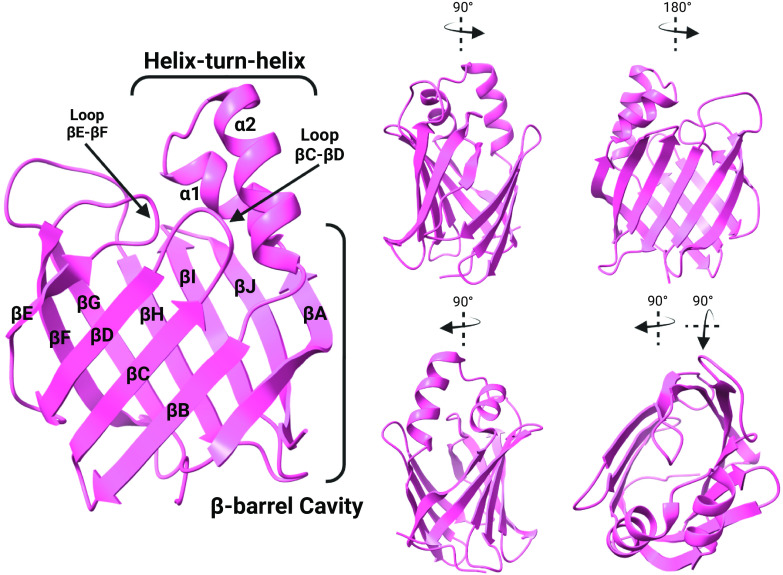
The tertiary structures of iLBPs are virtually superimposable and have two characteristic structural features, a β-barrel domain and helix-turn-helix motif (Fig. 2). Ten anti-parallel β-strands fold into two β-sheets to form the β-“clam-like” cavity of the iLBPs (Fig. 2) (Furuhashi and Hotamisligil, 2008; Storch and McDermott, 2009; Ferrolino et al., 2013; Napoli, 2016). The two alpha-helices along with nearby loops form a portal region for ligand entry and egress into the interior binding cavity (Fig. 2). The iLBPs have a characteristic fingerprint composed of three separate motifs termed FATTYACIDBP1-3 (Fig. 1A). The G-x-W triplet in the first FATTYACIDBP1 motif is highly conserved between iLBP members (Fig. 1A) and homologous with a similar motif in lipocalin family of binding proteins (Smathers and Petersen, 2011). FABP5 is unique in the iLBP family in that it is the only FABP known to form an intramolecular disulfide bond (C120-C127) (Hohoff et al., 1999). The dynamics of iLBP structures and consequences on ligand binding have been extensively studied, and several comprehensive reviews are available on this topic (Storch and McDermott, 2009; Atshaves et al., 2010; Smathers and Petersen, 2011; Ragona et al., 2014).
Fig. 2.
The crystal structure of human holo-CRABP2 (PDB 1CBS) showing the overall structural features of iLBPs. The figure shows the β-barrel cavity composed of 10 β-strands (βA-βJ), the helix-turn-helix cap consisting of the alpha helices (α1 and α2) and the portal to the ligand binding domain with the neighboring loops (loop βC-βD and βE-βF). Two β-sheets, each made up of five β-strands, fold to form the β-clam of the iLBP structure. (Structures generated from PDB using ChimeraX; figure created with BioRender.com.)
iLBP Structures
Crystal structures and NMR solution structures of iLBPs show that ligands are stabilized within the binding cavity by ionic interactions, hydrogen bonding networks with water molecules, and interactions with hydrophobic regions (Kleywegt et al., 1994; Cai et al., 2012; Nossoni et al., 2014; Silvaroli et al., 2016). Hydrophobic interactions between the ligand and amino acid sidechains that line the iLBP binding cavity are important for ligand binding (Thumser et al., 1996). The residues identified as part of the hydrophobic interaction network are shown for representative iLBPs in Fig. 3A. The importance of the hydrophobic interactions is also illustrated in the general observation that binding affinities with FABPs correlate with increasing hydrophobicity (Storch and Corsico, 2008; Smathers and Petersen, 2011). Ionic and hydrogen bonding interactions for ligand binding typically involve charged residues. The arginine residue in the FATTYACIDBP3 (Fig. 1A) is highly conserved in the iLBPs that bind acidic ligands (R122 in FABP1, R126 in FABP3 and FABP4, R129 in FABP5 and R132 in CRABP2) and is located on the βJ strand of these proteins (Fig. 3A). For CRBP1 that binds nonacidic ligands all-trans-retinol and all-trans-retinal, the Q128 appears to be the corresponding residue important for ligand binding (Silvaroli et al., 2016). For CRABP1 and CRABP2, the amino acids R132 and Y134 coordinate with the carboxylic acid of all-trans-retinoic acid (atRA) and R111 appears to coordinate with the carboxylic acid via an ordered water molecule (Fig. 3C) (Kleywegt et al., 1994). Analogous amino acids in some FABPs also coordinate with the carboxylate of fatty acids bound to FABPs (Hanhoff et al., 2002; Smathers and Petersen, 2011). However, these residues are not essential for ligand binding in all iLBPs. Mutations of the conserved arginine in the βJ strand confer different effects on ligand binding depending on the iLBP isoform and the ligand in question. A single R132A or R132Q mutation completely abolishes binding of atRA to CRABP2 (Chen et al., 1995). Similarly, an R126Q mutation on the analogous residue in FABP4 reduces the binding affinity for cis-parinaric acid by >10-fold (Sha et al., 1993). In contrast, an R122Q mutation in FABP1 only moderately decreases fatty acid binding and increases binding of bulkier ligands (Thumser et al., 1996). Charged or polar residues in the βH strand also interact with hydroxy and carbonyl head groups (Fig. 3A), except for subfamily II FABPs (FABP1 and FABP6), and likely contribute to ligand binding. The hydroxy group of all-trans-retinol interacts with Q108 in CRBP1 (Fig. 3A) and Q109 in CRBP2. Ligands appear to interact with the conserved residue R111 in CRABPs (Figs. 1A and 3A). In addition, ligands interact with R106 in FABP2, and this residue is also conserved in subfamily IV FABPs (Figs. 1A and 3A).
Fig. 3.
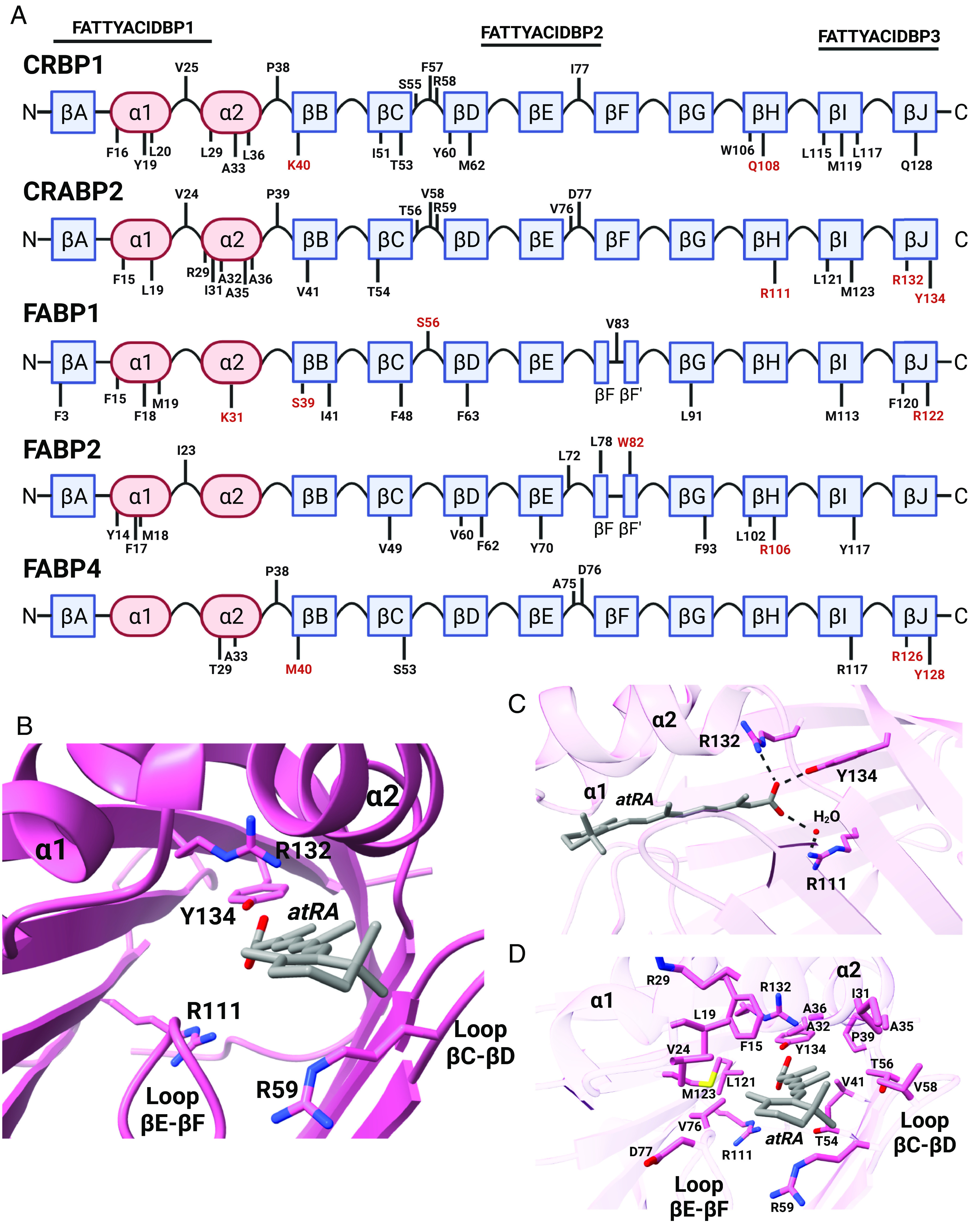
Binding characteristics of endogenous ligands with iLBPs. (A) The distribution of residues shown to interact with endogenous ligands for iLBPs are depicted along their structural features. Residues labeled in red font are involved in coordinating with the hydroxy or carbonyl groups of endogenous ligands via ionic interactions. (B) A top-down perspective into the atRA binding site of hCRABP2 (PDB 1CBS) with side chains that interact with atRA labeled. The position of R111, R132, and Y134 residues that coordinate with the carboxyl group of atRA are shown along with the position of R59 which interacts with the β-ionone ring. (C) Side view and positions of residues R111, R132, and Y134 are shown relative to the carboxylate of atRA along with hydrogen bonding interactions. (D) The amino acid sidechains that interact with atRA and form the atRA binding pocket in CRABP2 are shown. (Structures generated from PDB using ChimeraX; figure created with BioRender.com.)
The helix-turn-helix motif, in conjunction with nearby βC-βD and βE-βF loops, form the portal region of the iLBP that permits ligand entry and egress from the interior binding cavity (Fig. 2) (Vaezeslami et al., 2006; Storch and Corsico, 2008; Silvaroli et al., 2016). Different hypotheses describe the extent of the dynamics and flexibility of the portal region. Early observations from NMR solution structures of FABP2 showed disorder and flexibility in the portal region leading to the “dynamic portal hypothesis” (Hodsdon and Cistola, 1997a,b). This hypothesis suggests that the disordered portal region in the apo-protein could undergo large structural fluctuations to permit ligand entry but shifts to an ordered closed state upon ligand binding. Processes that destabilize the helical cap, such as interactions with cationic membranes, would then shift the protein toward the disordered state and, hence, facilitate ligand release. Later studies with FABP1 supported this hypothesis and showed the apo- and holo-protein structures to have an open and closed “helix cap”, respectively (He et al., 2007). However, the dynamic portal hypothesis is not sufficient to reconcile observations that some FABPs have similar structures between ligand bound and unbound forms (Vaezeslami et al., 2006; Gillilan et al., 2007; Cai et al., 2012).
Internal protein dynamics may also have a major role in influencing ligand accessibility as major fluctuations in the portal region are not observed in structural studies with retinoid binding proteins and some FABPs (Vaezeslami et al., 2006; Cai et al., 2012; Ragona et al., 2014). In CRABP2, the portal appears large enough to allow entry of all-trans-retinoic acid (atRA) in both the apo- and holo-structures with little change in the overall protein backbone (Vaezeslami et al., 2006). However, the sidechain of R59 (Fig. 3), which is located at the entry of the portal region (βC-βD loop) in the apo-protein, appears to rotate in the holo-protein to form stabilizing interactions with atRA (Fig. 3). An analogous residue (F57) in FABP4 appears to have a similar function and supports the importance of sidechain dynamics in the internal binding cavity. Structural studies with FABP4 suggest that locking the internalized ligand in the holo-protein is controlled by the F57 sidechain on the βC-βD loop that rotates into the binding cavity in the holo-conformation (Gillilan et al., 2007) despite little conformational change between apo- and holo-proteins. Indeed, ligand binding kinetics with the fluorescent ligand 8-anilinonaphthalene-1-sulfonic acid (ANS) are faster in an FABP4 portal mutant (V32G, F57G, K58G), which has an enlarged portal region, than with wildtype (WT) FABP4 (Jenkins et al., 2002).
Solution NMR studies show that the backbone dynamics in the portal region in apo-proteins vary between iLBP isoforms. FABP6 has a relatively rigid portal region while FABP1, 3, and 4 portal regions are more flexible, and the FABP2 portal is virtually disordered (Ragona et al., 2014). However, in general the changes in the backbone dynamics of FABPs upon ligand binding are consistent with disordered to ordered stabilizing interactions.
Endogenous Ligand Binding in iLBPs
The divergence of ligand specificity in iLBPs arises from nuanced differences in the β-barrel cavity and portal regions. Figure 4 shows representative structures of CRBP1, FABP1, FABP2, and FABP4 bound with their endogenous ligands illustrating general features of ligand binding with iLBPs. Historically protein fractionation, gel filtration, ion-exchange chromatography, and electrophoresis techniques were used to isolate and identify iLBPs in tissues and tissue homogenates, and bound ligands were identified from this isolated protein (Bashor et al., 1973; Ockner and Manning, 1974; Maatman et al., 1991; Veerkamp and Maatman, 1995). Subsequent characterization of ligand binding has been largely done with fluorescent probes or radiolabeled ligands in tissue homogenates or with purified recombinant iLBPs (MacDonald and Ong, 1987; Giguère et al., 1990; Nemecz et al., 1991; Sanquer and Gilchrest, 1994; Folli et al., 2001) and with x-ray crystallography and NMR (Kleywegt et al., 1994; LaLonde et al., 1994; Thompson et al., 1997; Hohoff et al., 1999).
Fig. 4.
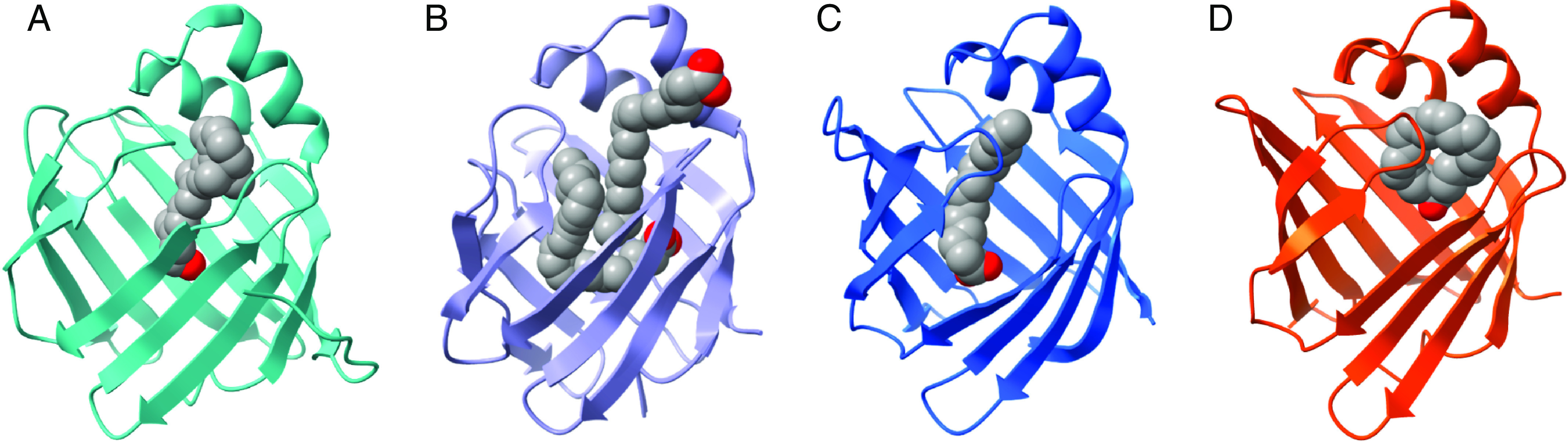
Binding orientations of endogenous ligands in the binding cavity of intracellular lipid binding proteins for (A) hCRBP1 with all-trans-retinol (PDB 5H8T), (B) hFABP1 with two oleate molecules (PDB 2LKK), (C) rFABP2 with myristate (PDB 1ICM), and (D) mFABP4 with arachidonic acid (PDB 1ADL). (Structures generated from PDB using ChimeraX; figure created with BioRender.com.)
The known endogenous ligands of iLBPs are summarized in Table 1. The retinoid binding proteins appear to be selective toward vitamin A and its metabolites while all FABPs bind long chain fatty acids (LCFAs) (Table 1). Some FABPs also bind a variety of other endogenous ligands (Table 1). It is important to note that the list of known ligands is limited to those ligands that have been explicitly tested for their binding and may not be comprehensive for all endogenous ligands. Several studies have explored synthetic derivatives of the endogenous ligands of FABPs (Wang et al., 2016; Floresta et al., 2017), but these synthetic derivatives are not discussed in this review. Additionally, the summary here includes binding data from species other than human proteins, as many ligand binding studies with iLBPs were done with rat, mouse, and bovine recombinant protein.
Endogenous Ligands of Subfamily I
Vitamin A (retinol) or its biologically important metabolites retinaldehyde and the pharmacologically active atRA bind the proteins in subfamily I with high affinity. Notably, all proteins in this subfamily are intracellular, in contrast to the lipocalin retinol binding protein 4 (RBP4), which is the circulating carrier for retinol. all-trans-retinol, all-trans-retinaldehyde, and their 13-cis and 9-cis isomers bind to CRBP1 (Fig. 4A) and CRBP2 with nanomolar affinity. Yet, neither atRA nor its 13-cis and 9-cis-isomers bind to CRBPs (Kane et al., 2011; Napoli, 2016, 2017; Menozzi et al., 2017). all-trans-retinol binds to CRBP3 (Folli et al., 2001) and all-trans-retinol along with 13-cis and 9-cis retinol bind to CRBP4 (Vogel et al., 2001; Folli et al., 2002). Although CRBPs appear to be specific for retinol and retinal ligands, monoacylglycerols were also recently shown to bind to CRBPs (Lee et al., 2020). This suggests CRBPs and CRABPs may have broader ligand specificity than previously assumed. atRA and its isomers and metabolites bind specifically to CRABP1 and CRABP2 with atRA having higher binding affinity toward CRABP1 and CRABP2 (Kd = 0.4–39 nM) (Fiorella and Napoli, 1991; Fogh et al., 1993; Norris et al., 1994; Wang et al., 1997) than 9-cis-RA (Kd = 51–69 nM) (Norris et al., 1994) or 13-cis-RA (Kd = 156–238 nM) (Fiorella et al., 1993). Generally, atRA appears to bind slightly tighter to CRABP1 than CRABP2 (Fiorella et al., 1993; Dong et al., 1999; Yabut and Isoherranen, 2022). Retinol or retinal isomers do not bind to CRABP1 or CRABP2 (Fiorella and Napoli, 1991; Fiorella et al., 1993; Napoli, 2017).
Endogenous Ligands of Subfamily II
FABP1 and FABP6 make up subfamily II. Generally, bulky ligands in addition to LCFA bind to FABP1 and FABP6 (Smathers and Petersen, 2011). The binding pockets of FABP1 and FABP6 are larger (≥639 and 460 Å3, respectively) (Lücke et al., 2000) than other FABPs that have solvent accessible binding pockets of approximately 230 to 330 Å3 (Smathers and Petersen, 2011). FABP1 and FABP6 can accommodate larger ligands found in the liver such as bile acids, cholesterol, bilirubin, and heme (Bernlohr et al., 1997; Smathers and Petersen, 2011). Other ligands of FABP1 include branched fatty acids, endocannabinoids, acyl-CoA, prostaglandins, and vitamin K (Khan and Sorof, 1990; Thumser and Wilton, 1996; Martin et al., 2003; Storch and Corsico, 2008; Atshaves et al., 2010; Smathers and Petersen, 2011). While fatty acid ligands appear to bind to all other FABPs in a 1:1 ratio, two fatty acids can bind to FABP1 simultaneously (Fig. 4B) (Bernlohr et al., 1997; Thompson et al., 1997; Cai et al., 2012). FABP1 has a high-affinity fatty acid binding site (Kd = 4–60 nM) located deep within its interior cavity and a low affinity site (0.3–12 μM) closer in proximity to the alpha-helical domain and opening of the portal region (Fig. 4B) (Atshaves et al., 2010; Smathers and Petersen, 2011; Cai et al., 2012). With larger ligands such as bile acids, this stoichiometry appears to be reduced (1:1) along with reduced binding affinities (Kd 4–50 μM) (Richieri et al., 1995). FABP6 is structurally similar to FABP1, but due to differences in interior amino acid side chains between the two proteins, preferential ligands of FABP6 include bile acids over LCFAs (Lücke et al., 2000). Due to the size of bile acids, only a single ligand is typically observed in the FABP6 binding cavity.
Endogenous Ligands of Subfamily III
FABP2 is the sole member of subfamily III. In contrast to the iLBPs in subfamily II, FABP2 has a small solvent accessible binding pocket (234 Å3) (Smathers and Petersen, 2011), and its preferential ligands include saturated LCFAs (Fig. 4C) (Lowe et al., 1987; Richieri et al., 1994; Velkov et al., 2005; Smathers and Petersen, 2011). Measured fatty acid binding affinities with FABP2 range between 0.02 and 1.5 µM based on fluorescence displacement assays (Nemecz et al., 1991; Velkov et al., 2005, 2007).
Endogenous Ligands of Subfamily IV
The seven members of subfamily IV collectively bind diverse lipids. The size of the solvent accessible binding pockets of subfamily IV FABPs appear to be larger than subfamily III (FABP2) but smaller than subfamily II (FABP1 and FABP6). FABP3, 4 and 8 have 323, 310, and 330 Å3 binding pockets, respectively (Smathers and Petersen, 2011). Saturated and unsaturated fatty acids bind to FABP3 with nanomolar affinity, and oxygenated fatty acids (epoxyeicosatrienoic acid, hydroxyeicosatetraenoic acid, dihydroxyeicosatrienoic acid) bind to FABP3 with Kd values from 0.4 to 14 µM (Widstrom et al., 2001; Smathers and Petersen, 2011). FABP4 appears to be more ligand selective, and only LCFAs bind to FABP4 with nanomolar affinity (Kd = 22–196 nM) (Fig. 4D) (Richieri et al., 1994; Gillilan et al., 2007; Storch and Corsico, 2008; Smathers and Petersen, 2011). However, other ligands such as atRA also bind to FABP4 but with a considerably lower binding affinity (Kd = 50 µM) (Matarese and Bernlohr, 1988; Veerkamp et al., 1999).
With FABP5, stearic acid and docosahexaenoic acid have nanomolar affinity to FABP5 (Kd = 0.17–0.29 and 0.16 µM, respectively) and oleic acid, lauric acid, and arachidonic acid binding affinity range from nanomolar to micromolar (Kd = 0.15–1.6, 2.5 and 0.12–1.7 µM, respectively) (Hohoff et al., 1999; Smathers and Petersen, 2011; Kaczocha et al., 2012; Pan et al., 2015; Lee et al., 2018). atRA has also been reported to bind to FABP5 in fluorescence displacement assays with ANS (Kd = 35 nM) (Schug et al., 2007). However, FABP5 did not to sequester atRA from metabolism by cytochrome P450 (CYP) enzymes, suggesting binding may not be as tight as suggested by the displacement assay (Yabut and Isoherranen, 2022). FABP7 prefers polyunsaturated fatty acids with longer chains (docosahexaenoic acid, eicosapentaenoic acid, arachidonic acid), and these fatty acids bind to FABP7 with affinities ranging from 27 to 250 nM (Smathers and Petersen, 2011).
In addition to the fatty acid ligands of FABPs, FABP3, FABP5, and FABP7 have also been shown to bind the endocannabinoids 2-archidonylglycerol and anandamide (AEA), and FABPs have been proposed to have a role in modulating endocannabinoid metabolism and signaling. 2-archidonylglycerol and AEA bind to FABP7 with higher affinity (Kd = 0.2 and 0.8 µM, respectively) than to FABP3 (Kd = 1.63 and 3.07 µM, respectively) and to FABP5 (Kd = 1.45 and 1.26 µM, respectively) (Kaczocha et al., 2012; Elmes et al., 2015). FABP8, 9, and 12 have not been extensively studied, and the binding of their endogenous ligands is not well characterized (Storch and Corsico, 2008; Smathers and Petersen, 2011).
Tissue Distribution and Expression of iLBPs
The tissue distribution of iLBPs is broad and expression patterns have been studied in several mammalian species including rat, mice, pig, and human (Paulussen et al., 1988, 1990; Gong et al., 1994; Sanquer and Gilchrest, 1994). However, species differences in tissue expression have not been comprehensively compared for all iLBPs. The following is a summary of the tissue expression of iLBPs in adult mammals determined using a combination of techniques including western and northern blot analysis, immunohistochemistry, enzyme-linked immunosorbent assay, reverse transcriptase-quantitative polymerase chain reaction, and binding assays with radiolabeled ligands. Some iLBPs are expressed in multiple tissues while others are expressed in specific tissues and cell types that may be indicative of specialized biologic functions (Storch and Corsico, 2008). The expression pattern of the FABPs is sometimes evident from the original name of the FABP, as FABPs were named after the tissues from where they were first identified. However, multiple FABPs are often expressed in the same tissues, and the expression patterns are typically broader than what is implied from the original names of the FABPs. Hence, early studies identifying FABPs in tissues often required confirming the specificity of antisera against multiple FABPs (Paulussen et al., 1990; Maatman et al., 1991; Gong et al., 1994).
Although the iLBPs are generally considered to be intracellular, FABP1, 2, 3, 4, and 5 have also been measured in plasma in humans (0.3–13 ng/mL) (Pelsers et al., 2003; Ishimura et al., 2013). Yet their concentrations are much lower than other circulating proteins such as albumin that bind fatty acids in plasma, and the importance of the circulating FABPs is unknown. FABP4 is the only isoform shown to be secreted from tissues (adipose) into circulation (Hotamisligil and Bernlohr, 2015; Shrestha et al., 2018; Villeneuve et al., 2018). For this review only CRABPs and those FABPs that xenobiotics have been shown to bind to are discussed, but the tissue expression for all iLBPs is summarized in Table 1.
CRABP1 protein is found in various tissues including liver, kidney, stomach, lymph, eye, and brain, but it is most abundant in skin and reproductive tissues (seminal vesicles, vas deferens, and testis) (Kato et al., 1985). CRABP2 protein expression appears to be limited to skin (Giguère et al., 1990).
FABP1, or liver FABP, is the major FABP in the liver and the intestine but is also found in the kidney, lung, pancreas, and stomach (Besnard et al., 2002; Pelsers et al., 2003; Gajda and Storch, 2015; Wang et al., 2015). FABP1 is most abundant in the liver and comprises 2% to 11% of all cytosolic protein in the liver (Wang et al., 2015; Schroeder et al., 2016). Expression of FABP1 in the liver is zonal, possibly indicating a unique role in specific areas of the liver (Bass et al., 1989). Peroxisome proliferators, female sex steroids, retinoids, and a diet high in fat increase the expression of FABP1 messenger RNA (mRNA) and protein in the liver (Poirier et al., 1997; Hung et al., 2003; Trevaskis et al., 2011; Velkov, 2013). Interestingly, FABP1 mRNA and protein expression are decreased after dexamethasone treatment, likely due to altered lipid metabolism and concentrations (Foucaud et al., 1998). In the gut, FABP1 mRNA is expressed throughout the length of the small intestines but is highest in the duodenum and jejunum (Agellon et al., 2002; Gajda and Storch, 2015). The expression pattern of FABP1 in the liver and intestines suggests FABP1 may also impact drug metabolism in the liver and drug absorption in the intestines. Additionally, FABP1 expression and function may have a role in metabolic disease progression as FABP1 polymorphisms in humans are associated with dyslipidemia, nonalcoholic fatty liver disease, and hepatocellular carcinoma (Peng et al., 2012; Schroeder et al., 2016; McKillop et al., 2019; Valizadeh et al., 2021). For example, the T94A mutation (allele frequency 26%–38%) in FABP1 alters FABP1 expression, ligand binding characteristics, protein structure and stability, and protein function (Schroeder et al., 2016). The T94A single nucleotide polymorphism is associated with elevated triglycerides, low-density lipoprotein cholesterol, and altered response to fenofibrate (Schroeder et al., 2016).
FABP2, also called intestinal FABP, is solely expressed in the gut, and its expression appears to be similar to FABP1 in rodent intestine but lower than FABP1 in human intestine. FABP2 mRNA is expressed throughout the length of the small intestine, and its expression is highest in the jejunum (Sacchettini et al., 1990; Gajda and Storch, 2015). Along with FABP1, FABP2 expression is highest in the villi of enterocytes, and it is not expressed in the crypt. FABP2 expression in enterocytes may be regulated by the gut peptide tyrosine tyrosine (Halldén and Aponte, 1997). FABP2 expression appears to be diffused throughout enterocytes but localized to the apical side in a fasted state (Alpers et al., 2000). Similar to FABP1, an A54T polymorphism in FABP2 appears to be associated with dyslipidemia, insulin resistance, obesity, and cardiovascular disease and may increase the risk of colorectal cancer (McKillop et al., 2019; Huang et al., 2022). FABP2 has been proposed as a potential biomarker for disruption of intestinal epithelial integrity as FABP2 is released to circulation when intestinal epithelium is compromised (Huang et al., 2022).
FABP3, or heart FABP, protein has been found in the heart, skeletal muscle, brain, kidney, liver, lung, spleen, and placenta (Paulussen et al., 1990). FABP3 is most abundant in the heart where its expression is nearly twofold greater than in skeletal muscle. Protein abundance in the kidney and brain is about half of that in the muscle and even less in the liver and placenta. FABP3 is also found to circulate at elevated levels in plasma in response to myocardial injury, presumably due to release from the heart. As such, it may be a potential biomarker for cardiovascular disease (Pelsers et al., 2005). In the kidney, FABP3 is found to be expressed in the distal and proximal convoluted tubules (Maatman et al., 1991), suggesting FABP3 could play a role in renal handling of drugs and xenobiotics.
FABP4, known as adipocyte FABP, is abundantly expressed in adipose tissue and is also the major FABP found in macrophages (Pelton et al., 1999; Furuhashi and Hotamisligil, 2008). FABP4 is the most abundant FABP in circulation (Ishimura et al., 2013) and is secreted from adipocytes via a membrane-bound pathway independent of the canonical endoplasmic reticulum-Golgi-plasma membrane secretion pathway (Villeneuve et al., 2018). Secreted FABP4 may serve as an adipokine, and lipolysis increases secretion of FABP4 from adipocytes (Furuhashi et al., 2015). Exogenous FABP4 influences hepatocyte glucose production, insulin secretion by pancreatic β cells, and cellular functions of cardiomyocytes and smooth muscle cells (Furuhashi et al., 2015). Indeed, circulating FABP4 levels are associated with the development of insulin resistance, diabetes, atherosclerosis, cardiac dysfunction, and inflammation (Furuhashi and Hotamisligil, 2008; Ishimura et al., 2013; Furuhashi et al., 2015; Saito et al., 2021). Reduced FABP4 appears to reduce the risk of metabolic and cardiovascular disease (Hotamisligil et al., 1996; Furuhashi and Hotamisligil, 2008; Furuhashi et al., 2015), and, hence, FABP4 has been explored as a potential therapeutic target (Floresta et al., 2017). Due to its small size, FABP4 found in circulation is subject to glomerular filtration, but it accumulates in the kidney via megalin-mediated reabsorption from the tubular lumen (Shrestha et al., 2018). Notably, circulating FABP4 levels also showed a sex difference with females having higher concentrations than males (Ishimura et al., 2013).
FABP5, epidermal FABP, is the major FABP found in the epidermis, but FABP5 tissue expression is broad and not restricted to the skin (Table 1). FABP5 mRNA along with FABP3 and FABP4 mRNAs are found in human brain endothelial cells (hCMEC/D3), and FABP5 protein appears to be more abundant than FABP3 and FABP4 in these cells (Lee et al., 2015).
FABP7, brain FABP, is largely expressed in the brain and central nervous system but is also found in the skeletal muscle (Shimizu et al., 1997; Veerkamp and Zimmerman, 2001; Owada et al., 2006) with diurnal variation in its expression (Gerstner et al., 2008). FABP7 mRNA in the brain increases during light periods and declines in the dark period. This leads to an accumulation of FABP7 protein in dark periods and a decrease in protein in the light period. Yet the biologic role of this diurnal variation has not been defined.
Xenobiotic Ligands of iLBPs and Methods to Characterize Ligand Binding
Known Xenobiotic Ligands of iLBPs and Their Binding Characteristics
The literature is rich with binding and structural studies of endogenous ligands of iLBPs, but binding of xenobiotics to iLBPs has not been as extensively studied. This is despite clear evidence of xenobiotics binding to iLBPs. For example, synthetic retinoid drugs [agonists of retinoic acid receptors (RAR) and retinoid X receptors] bind to retinoid binding proteins (Ferreira et al., 2020), but the clinical relevance of the binding is not known. Whether retinoid binding proteins bind other classes of therapeutic drugs has not been explored. The majority of xenobiotic binding studies with iLBPs have been done with FABPs, likely due to their broad ligand specificity and high expression in tissues relevant to drug disposition and pharmacological activity. Of the 10 FABPs, xenobiotics have been shown to bind to FABP1–5 and FABP7 in vitro (Table 2). Xenobiotic binding to FABP6, 8, 9, and 12 has not been reported to our knowledge.
TABLE 2.
Binding affinities of xenobiotic ligands with different FABPs
| Range of Reported Ki and Kd Values (µM) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Ligand | FABP1 | FABP2 | FABP3 | FABP4 | FABP5 | FABP7 | ||
| Acifluorfen | 4.2-8.9 | |||||||
| ANS | 1.1-6.0 1.1a | 12b | 3.0-31c | 0.6 | 0.03-32 | 0.07-1.3 | ||
| Aspirin | 348a | 3780b | 300-460 | |||||
| Atenolol | 717 | NBc | ||||||
| Benzafibrate | NBa | 44.4b | 26-100c | NB | 12 | NB | ||
| Benzilic acid | 110.8-200c | |||||||
| Benzyl salicylate | NB | |||||||
| Bifenox | NB | |||||||
| BMS309403 | 0.9 | |||||||
| CBD | 4.0 | 1.7 | 1.9 | 1.5 | ||||
| Ciprofibrate | 24-72c | |||||||
| Clofibrate | 6.9 | |||||||
| Clofibric acid | 17.7-110c | NB | 17 | NB | ||||
| Cortexolone | 1600-1900 | |||||||
| DAUDA | 0.4-1.4 | 0.3-0.7c | ||||||
| Dexamethasone | 22.1a | 41.3b | 1100-1200 | |||||
| Diazepam | 0.5a | 115b | 1980-2200 NBc | NB | NB | 325 | ||
| Diclofenac | 3.2a | 35b | 86.3-520c | |||||
| Dilitiazem | NBc | |||||||
| Fenbofibric acid | 1-1.6, 0.3a | 27.5b | 1-6.1c | 33 | 24 | 3.3 | ||
| Fenofibrate | 2.9 0.02a |
0.4b | 0.8 NBc |
|||||
| Fenoprofen | 14-64c | |||||||
| Flufenamic acid | 3.7-15.5 | |||||||
| (R/S) Flurbiprofen | 1.2a | 222b | 20-70 | |||||
| Gemfibrozil | 1.9a | 179b | 110.5-121.3c | NB | 3.8 | 6.1 | ||
| GW7647 | 0.3-0.6 | 1.3c | 25 | 7.6 | 0.7-8.9 | |||
| (R/S) Ibuprofen | 47.6a | 448b | 32.2-263c | 325 | 2.6 | 138 | ||
| 3-indolacetic acid | 93-200 | |||||||
| Indole-3-butyric acid | 72-170 | |||||||
| Indoprofen | 1.27a | 161b | 129-520.1 | |||||
| Jasmonic acid | 140-350 | |||||||
| Ketoprofen | 24-82.4c | |||||||
| Ketorolac | 11.6a | 119b | 9.4-2300c | |||||
| Lorazepam | 12.9a | 140b | 2100-2500 | |||||
| Meclofenamic acid | 0.4a | 0.3b | 8.9-21c | |||||
| Mefenamic acid | 63-110 | 5.8 | 1.1 | 4.3 | ||||
| Mepronil | NB | |||||||
| Midazolam | 7.9 | 12 | ||||||
| Nabumetone | NBc | |||||||
| Nadolol | 2310 | |||||||
| Nalidixic acid | NB | |||||||
| 2-naphthoxyacetic acid | 7.2-14 | |||||||
| (S)-(-)-Naproxen | 0.06a,c | 2.8b,c | 56-180 | |||||
| Nitrazepam | 1200-2300c | 28 | 36 | 20 | ||||
| Perfluorononanoic acid | 1.3-3.1c | |||||||
| Perfluorooctanoic acid | 2.4-6.5c | |||||||
| Phenytoin | 0.2a NB |
4.7b | ||||||
| Pioglitazone | 33 | NB | 11 | |||||
| Prednisolone | 2.7a | 101b | 95-113 | |||||
| Progesterone | 0.03 | 20-32 | ||||||
| Propanolol | NB | NB | ||||||
| Pyrilamine | NB | |||||||
| Rosiglitazone | 2.8 | NB | NB | 28.8 | ||||
| Sulfinpyrazone | 0.1c | 8.2c | ||||||
| THC | 0.1-2.9 | 2.0 | 3.1 | 1 | ||||
| 11-COOH-THC | 11.2 NB | |||||||
| 11-COOH-THC-glucuronide | NB | |||||||
| 11-OH-THC | 5-7.2 NB | |||||||
| Tolfenamic acid | 2.8-8.2c | 1.9 | 0.1 | 2.9 | ||||
| Tolmetin | 1300-2200 | |||||||
| Torsemide | 0.2c | 12.3c | 0.8 | |||||
| Troglitazone | 1.7 | 11 | 0.02-16 | 1 | ||||
| Valproate | 240-470 | |||||||
| Verapamil | NB | |||||||
aAffinity for first, high affinity binding site.
bAffinity for second, low affinity binding site respectively, determined in the same study as in a.
cIncludes studies where binding affinities were determined by SPR, ITC, or thermal shift with SYPRO Orange.
CBD, cannabidiol; DAUDA, 11-(dansylamino)undecanoic acid; THC, Δ9-tetrahydrocannabinol.
NB indicates that binding was tested but no binding was observed in at least one study.
References Thumser et al., 1996; Veerkamp et al., 1999; Velkov et al., 2005, 2007, 2009; Gillilan et al., 2007; Chuang et al., 2008; Rowland et al., 2009, 2015; Trevaskis et al., 2011; Kaczocha et al., 2012; Velkov, 2013; Patil et al., 2014; Yu et al., 2014; Elmes et al., 2015, 2019; Lee et al., 2015; Sheng et al., 2016; Huang et al., 2018.
Initial binding studies of lipophilic drugs to FABP1 and FABP2 were done to explore the potential of FABP1 and 2 to facilitate drug absorption into enterocytes (Velkov et al., 2005, 2007; Chuang et al., 2008). A broad range of therapeutic drugs such as nonsteroidal anti-inflammatory drugs, peroxisome proliferator-activated receptor (PPAR) agonists (fibrates and glitazones), and benzodiazapines were shown to bind to FABP1 and FABP2 (Table 2). Additionally, due to the high expression of FABP1 in the liver, the role of FABP1 binding as the rate-limiting step in hepatocyte uptake has been explored (Rowland et al., 2015). FABP3, 4, and 5 were found to be expressed in brain endothelial cell lines, and hence, the potential of drugs to bind to these FABPs at the blood-brain-barrier was evaluated (Lee et al., 2015). Similar drugs were shown to bind to FABP3, 4, and 5 as to FABP1 and FABP2. Xenobiotic cannabinoids Δ9-tetrahydrocannabinol and cannabidiol also bind to FABP1 and the brain FABPs, FABP3, 5, and 7 (Elmes et al., 2015, 2019; Huang et al., 2018). Due to its role in metabolic disease, FABP4 has become a potential therapeutic target, and a variety of inhibitor ligands have been developed and their binding to FABP4 characterized (Floresta et al., 2017).
FABP1 has been a focus of binding and structural studies with PPAR agonists (fibrates, glitazones, and synthetic agonists) to probe PPAR binding specificities as they relate to interactions with residues within the binding cavity of FABP1. Ester and carboxylic acid fibrates showed distinct differences in chemical shift perturbations (CSPs) within the FABP1 binding cavity in NMR studies (Chuang et al., 2009). Carboxylic acid fibrates showed significant CSPs with residues S39, R122, S124 in FABP1 that directly interact with the carboxylate of fatty acids while ester fibrates showed far less CSPs at these residues. Additionally, thermodynamic analysis showed that binding of carboxylic acid fibrates to the high affinity site of FABP1 was mainly driven by enthalpic interactions while ester fibrate binding had a much larger entropic component (Chuang et al., 2009). These data suggest that while ionic interactions play a role in the recognition and binding specificity of nonfatty acid ligands, they are not essential for ligand binding. Hydrophobic interactions are a large component of xenobiotic binding to FABP1.
The importance of hydrophobic interactions in ligand binding is evident in structural studies with FABP4 and (S)-ibuprofen (Fig. 5A). (S)-ibuprofen binding to FABP4 is stabilized by both ionic and edge-to-face aromatic interactions with FABP4 sidechains (Fig. 5, B and C) (González and Fisher, 2015). Similar to binding of endogenous ligands, internal protein dynamics also appear to play an important role in xenobiotic binding to FABPs. NMR solution structures of FABP1 with the synthetic PPAR agonist GW7647 bound (Fig. 5D) demonstrate that significant sidechain conformational changes occur within the binding cavity of holo-FABP1 upon ligand binding. This is despite there being little change in the overall backbone structure with ligand binding (Patil et al., 2019). Knowledge of the structures and ligand binding characteristics of individual FABPs can aid in designing FABP isoform specific ligands. For example, the synthetical FABP4 ligand BMS309403 binds to FABP4 (Fig. 5E) with a binding affinity of less than 2 nM but binds to FABP3 and FABP5 with >100-fold weaker affinity (Sulsky et al., 2007).
Fig. 5.
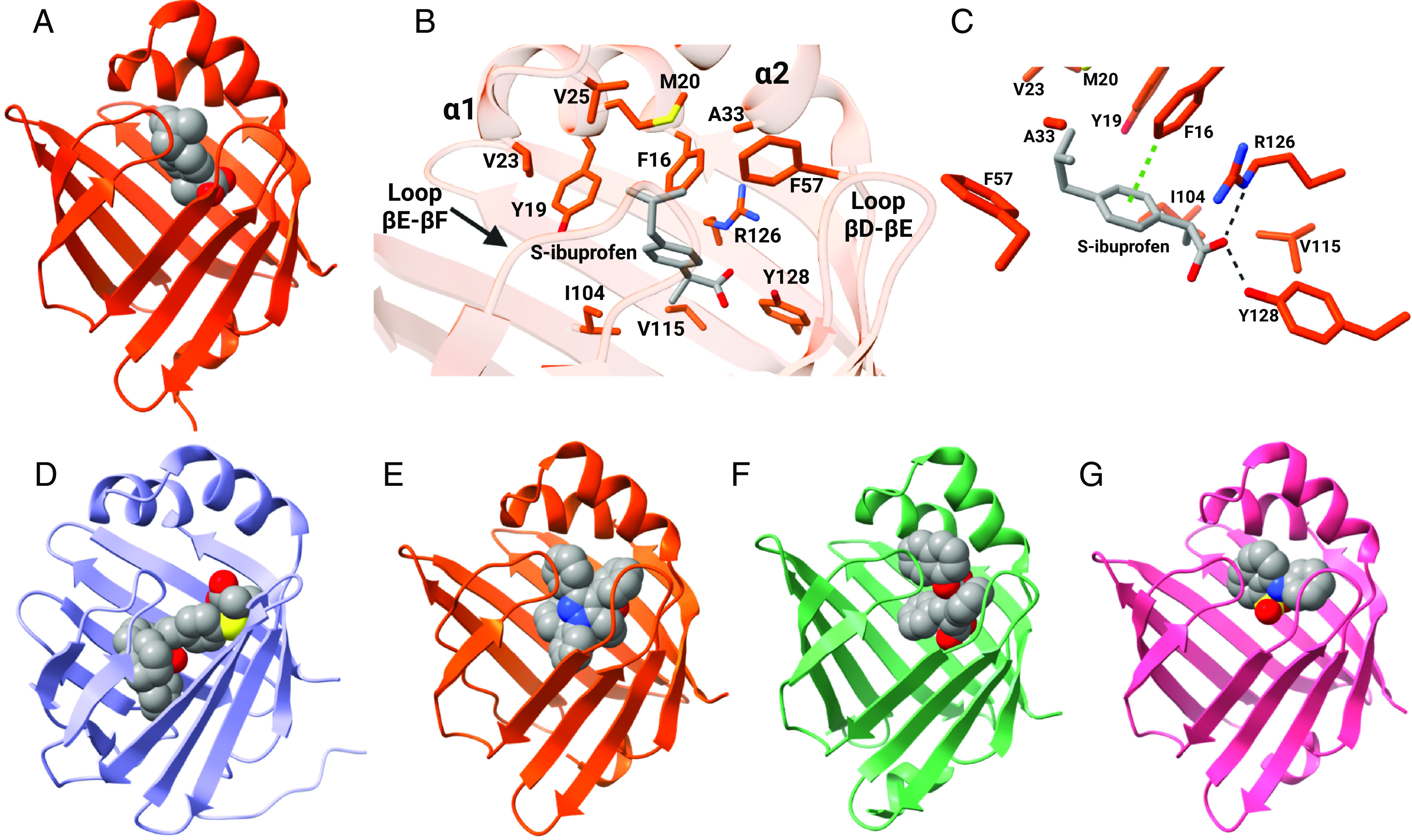
Binding characteristics of xenobiotic ligands of FABPs. (A) Crystal structure of hFABP4 complexed with (S)-ibuprofen (PDB 3P6H). (B) Amino acid side chains that line the binding pocket of (S)-ibuprofen in FABP4. (C) (S)-ibuprofen is stabilized in the binding pocket via ionic interactions between its carboxylate group and R126 and Y128 and edge-to-face aromatic interactions with residue F16 in FABP4. Structures for (D) hFABP1 in complex with PPARα agonist GW7647 (PDB 6DRG), (E) hFABP4 complexed with the inhibitor BMS309403 (PDB 2NNQ), (F) hFABP5 complexed with the antinociceptive SBFI-26 (PDB 5URA), and (G) hFABP3 complexed with ANS (PDB 3WBG). (Structures generated from PDB using ChimeraX; figure created with BioRender.com.)
Methods to Measure Ligand Binding with iLBPs
Historically, measuring free ligand concentrations using separation techniques such as Lipidex-1000 (Glatz and Veerkamp, 1983; Vork et al., 1990) was used to determine ligand binding affinities to iLBPs that were isolated from tissue homogenates or recombinantly expressed and purified. However, as the concentration of free ligand is decreased via partitioning to Lipidex, these techniques generally disturb the equilibrium between ligand and iLBP, and the binding affinities are generally underestimated (apparent Kd > true Kd) using this technique (Kane and Bernlohr, 1996; Veerkamp et al., 1999). Most of the recent work to characterize xenobiotic binding to iLBPs has been done using in vitro spectrophotometric assays. The following is a brief description of the direct and indirect spectrophotometric approaches to determine xenobiotic equilibrium binding affinities along with potential caveats associated with these methods.
Direct Binding Assays
Binding affinities (Kd) of retinoids with retinoid binding proteins are typically determined via direct fluorescence titration assays. These monitor either the increase in retinoid fluorescence upon binding to the binding protein or the quenching of intrinsic protein fluorescence (from tryptophan or tyrosine) as a result of retinoid binding (Fiorella et al., 1993; Norris et al., 1994; Wang et al., 1997; Dong et al., 1999; Vogel et al., 2001; Folli et al., 2002; Yabut and Isoherranen, 2022). These methods work well for retinoid binding proteins such as CRBP1, CRABP1, and CRABP2, which have five or more fluorescent (tryptophan and tyrosine) amino acids in their primary sequence. The intrinsic fluorescence spectra of tryptophan and tyrosine (excitation peak 280–290 nm, emission peak 330–355 nm) and the fluorescence spectra of retinoids (excitation peak 348–360 nm and emission peak 450–480 nm) (MacDonald and Ong, 1987; Fiorella and Napoli, 1991; Dong et al., 1999; Herr et al., 1999; Folli et al., 2002) are amenable for monitoring binding via fluorescence resonance energy transfer from protein to retinoid ligands (Peterson and Rask, 1971).
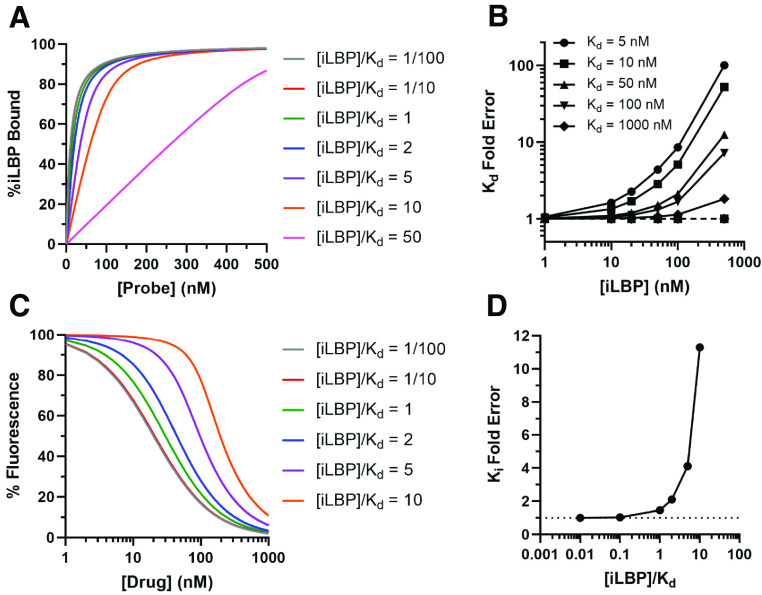
Because retinoids bind to retinoid binding proteins tightly (nanomolar affinities), performing the fluorescence titrations under steady state assumptions can be challenging. Relatively low concentrations of protein (ideally subnanomolar) are necessary to obtain accurate Kd value estimates, and hence protein fluorescence signal and instrument (fluorimeter) sensitivity can become a limitation. Therefore, retinoid binding assays are often done with protein concentrations that are much greater than the estimated Kd values. This approach may lead to inaccuracies in Kd estimates. These inaccuracies may be compounded by the use of kinetic binding models that assume steady state and that ligand binding to the binding protein does not alter free ligand concentrations in solution ([L]total ≈ [L]free). These experimental challenges likely partially explain the wide range of binding affinities reported in the literature (Norris et al., 1994; Napoli, 2016). The impact of protein concentrations and model fitting on the error in determination of the Kd values is illustrated in Fig. 6. For an iLBP-ligand interaction with a true Kd of 10 nM, using 100 nM iLBP protein (10-fold >Kd) in the experiment can result in an error as high as fivefold when a simple binding model ( ) is fit to the data (Fig. 6B). The error becomes negligible when the quadratic binding equation ( ) is fit to the data as it accounts for ligand depletion when the iLBP concentration ranges from 0.01 to 10 times the true Kd value (Fig. 6B). However, caution should be used when using the quadratic binding equation for tight binding ligands as the dependence of Kd for the model fit becomes negligible when [P] ≫ Kd, and hence, a Kd estimate may not be meaningful (Jarmoskaite et al., 2020). As such, even when the quadratic equation is used, the Kd values estimated should be assumed to be the upper limit of the true Kd value if the iLBP concentration in the assay exceeds the determined Kd value. Nevertheless, direct fluorescence titration assays have provided extensive information on the ligand binding characteristics and binding specificities of iLBPs.
Fig. 6.
Impact of experimental conditions and model fitting on determination of ligand binding affinities. (A) Simulated binding curves for a hypothetical probe with varying iLBP concentrations for direct titrations. (B) Simulation of the fold error in Kd determination for a hypothetical ligand-iLBP interaction with increasing [iLBP] concentrations used in the binding experiments in relation to different Kd values (symbols) for the ligand. The solid lines show simulated Kd values obtained using a one-site simplified hyperbolic binding equation ( ) while the dotted line shows the Kd values obtained with a one-site ligand depletion quadratic binding equation ( ) fit to the simulated data. (C) Simulated fluorescence displacement data shown with varying [iLBP] concentrations in relation to the Kd of the fluorescent probe. (D) Simulations of the fold error in Ki determination with varying [iLBP] concentrations relative to the Kd of the fluorescent probe. The Kd used in the simulations for direct binding titrations in (A) was 10 nM. The Kd of the probe and Ki of the drug used in simulations for (C) were 10 and 10 nM, respectively. (Figure created with BioRender.com.) MATLAB code used for simulations is provided in Supplementary Material.
Fluorescence Displacement Assays
Measuring direct protein fluorescence is not always feasible due to a lack of fluorescent amino acids or lack of fluorescence of the ligand. For example, FABP1 has no tryptophan residues and only one tyrosine residue, preventing the use of direct fluorescence measurements in evaluating ligand binding to FABP1. Hence, one approach for measuring direct ligand binding to FABP1 is to introduce tryptophan mutations to increase intrinsic protein fluorescence (Thumser and Wilton, 1994). However, such mutations may also affect ligand binding, and hence indirect fluorescence displacement assays are more commonly used.
Indirect measurements of ligand binding have been a common approach for estimating binding affinities for FABPs (Schug et al., 2007; Smathers and Petersen, 2011; Kaczocha et al., 2012; Elmes et al., 2015, 2019; Huang et al., 2016, 2018; Schroeder et al., 2016). Fluorescence displacement assays using fluorescent probes such as ANS or fluorophore conjugated fatty acids such as 11-(dansylamino)undecanoic acid and nitrobenzoxadiazole-stearate are commonly used due to the low intrinsic fluorescence of FABPs and the lack of measurable fluorescence from fatty acid ligands upon FABP binding. In these assays, a fluorescent probe is first bound to the FABP at a predetermined concentration, and the shift in the fluorescence of the ligand upon protein binding is measured. The drug of interest is then titrated into the sample, and the decrease in the probe fluorescence due to probe displacement by the drug is measured. Because displacement of the probe is assumed to be a purely competitive interaction, inhibitory constants (Ki) are determined either from a direct fit of a competitive binding model to the fluorescence data or are calculated from IC50 values assuming competitive inhibition (Velkov et al., 2005; Chuang et al., 2008; Lee et al., 2015; Elmes et al., 2019). Interfering fluorescence at similar wavelengths as the probe from the ligand of interest should be considered in these assays as this may confound the binding data. For example, atRA has a similar fluorescence emission peak (475 nm) as the fluorescent probe ANS (480 nm) with excitation wavelengths at 350 and 380 nm, respectively (Fiorella et al., 1993; Huang et al., 2014; Vogler, 2015). This fluorescence overlap may affect the interpretation of ANS displacement data for atRA binding to FABPs.
For most FABPs the assumption of competitive binding is likely appropriate as only one ligand appears to bind at a time to FABPs (Figs. 4 and 5). However, with FABP1, which can have multiple ligands bound to it simultaneously (Fig. 4B), a simple competitive binding model may not be appropriate, and EC50 or IC50 values determined with ligand displacement assays with FABP1 should not be directly translated to Kd or Ki values. It is possible that a ligand can bind simultaneously with a fluorescent probe to the FABP1 or that the binding of one fluorescent probe molecule affects the binding affinity of the ligand tested that may bind to a second binding site.
Similar concerns may be relevant for FABP2, although endogenous ligands appear to bind to FABP2 with 1:1 stoichiometry, possibly due to the size of the ligands. In FABP2, ANS and ketorolac have been shown to bind to different binding sites based on fluorescence and isothermal titration calorimetry data (Patil et al., 2014). This suggests that the two ligands could also bind simultaneously. However, there is currently no structural evidence that two ligands bind to FABP2 simultaneously. In the case of FABP1 and FABP2, it is unclear whether it is possible for a ligand to bind to the second binding site without affecting the binding of the fluorescent ligand in the first, high affinity binding site. The probe fluorescence intensity or the wavelength maxima of the probe fluorescence may be allosterically affected by binding of a ligand at an additional binding site, or complete displacement of the fluorescent probe may only occur when all binding sites are occupied by the xenobiotic ligand. Hence, alternative kinetic binding models may be more appropriate than a purely competitive one.
Like the direct binding assays described here, limitations with fluorescence signal, instrument sensitivity, and protein and probe concentrations should be carefully considered with displacement assays. Figure 6C shows simulated fluorescence displacement assay where the true affinity for the probe (Kd) and test ligand (Ki) are the same (10 nM). Using an iLBP concentration of 100 nM (10-fold greater than the Kd) can result in a >10-fold error in the estimated Ki value, illustrating the potential confounding effects of experimental design on the data collected (Fig. 6, C and D).
Impact of iLBPs on Ligand Distribution and Metabolism
One of the biologic functions of iLBPs is to serve as lipid carriers that bind, solubilize, and shuttle their ligands to relevant cellular compartments (Storch and Corsico, 2008; Storch and Thumser, 2010). iLBPs may simply bind their often unstable or toxic ligands to stabilize the ligand or prevent ligand interactions with nonspecific proteins in a cell. However, iLBPs have been shown to interact with phospholipid membranes, associate with cellular compartments such as mitochondria and lysosomes, and interact with different metabolic enzymes and nuclear receptors, suggesting more broad functions in a cell. Three different mechanisms for the impact of iLBPs on ligand disposition have been proposed (Fig. 7) (Smith and Storch, 1999; Storch and Corsico, 2008). In the first model, the iLBPs release their ligands to solution (diffusional model). Alternatively, iLBPs may interact with the phospholipid membranes via direct protein-membrane interactions to accept their ligands from or release their ligands directly to the membrane (collisional model). Finally, iLBPs may participate in direct protein-protein interactions and channel their ligands directly to catalytic enzymes or transporters. Since FABPs are highly expressed in tissues relevant to xenobiotic disposition, it is likely that xenobiotics bind to FABPs in these tissues, and the three models of iLBP functions may also be relevant for xenobiotic disposition. The following sections summarize various studies on the impact of iLBPs on ligand distribution and metabolism and the possible mechanisms of ligand delivery.
Fig. 7.
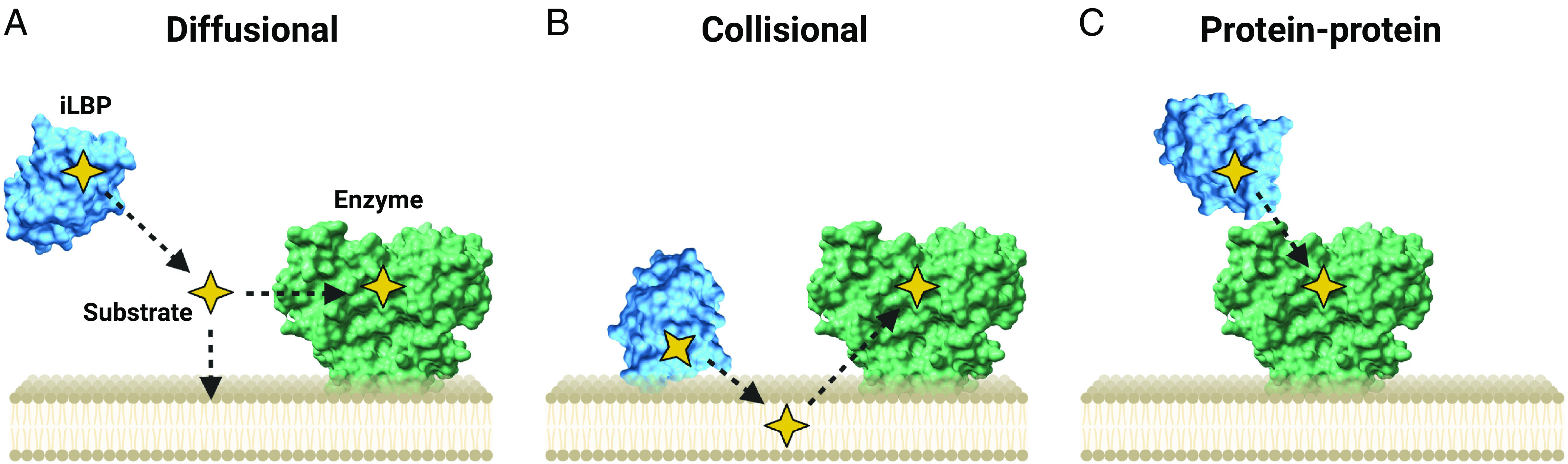
Three proposed models of ligand delivery by iLBPs. (A) The diffusional model requiring ligand release into solution, (B) the collisional model where iLBPs directly interact with phospholipids to transfer substrates to organelle membranes, and (C) direct transfer of substrates to metabolic enzyme via direct protein-protein interactions. (Figure created with BioRender.com.)
Impact of FABPs on Ligand Uptake into Tissues
The role FABPs have in regulating the uptake and tissue distribution of their endogenous ligands has been studied for many FABPs to which xenobiotic ligands also bind. It is well established that FABP1 facilitates lipid uptake into the liver, and FABP1 expression in the liver correlates with uptake of fatty acids (Kushlan et al., 1981; Hung et al., 2003; Newberry et al., 2003). Induction of FABP1 expression by PPAR agonists in HepG2 cells resulted in increased rates of oleate uptake, while knocking down FABP1 expression significantly reduced rates of uptake (Wolfrum et al., 1999). Changes in uptake appear to also alter lipid metabolic products. FABP1-knockout mice have decreased rates of [H]3oleate uptake to the liver, which corresponds to decreased fatty acid β-oxidation and incorporation of [H]3oleate into triglycerides (Newberry et al., 2003). In rat perfused livers, higher expression of FABP1 in the liver correlated with greater palmitate clearance and higher retention of palmitate and its metabolites in the liver (Hung et al., 2003). Similarly, FABP5 and FABP7 that are expressed in the brain appear to enhance endocannabinoid uptake into cells and endocannabinoid metabolism. The cellular uptake of AEA and subsequent metabolism by fatty acid amide hydrolase was greater in N18TG2 (mouse neuroblastoma) and COS-7 cells transfected with FABP5 and FABP7 when compared to mock transfected cells while FABP3 had no effect (Kaczocha et al., 2009).
FABP2 appears to play a role in the cellular uptake and distribution of xenobiotic ligands in the gut, and many orally administered drugs bind to FABP2 (Table 2). The potential role of FABP2 in modulating apical and basolateral transport of drugs in the intestine was studied in the parallel artificial membrane permeability assays where an artificial phospholipid membrane separates donor and acceptor reservoirs (Velkov et al., 2007). These studies were designed to test the effect of FABP2 on the rates of diffusion across an artificial phospholipid membrane mimicking the apical membrane of enterocytes. For apical membrane permeability, drugs were added to the donor side, and physiological concentrations of FABP2 in the enterocytes (0.33 mM) were present on the acceptor side. The rates of drug uptake from the apical membrane were increased for drugs that bound to FABP2, with tighter binding drugs showing higher rates of uptake. This suggests FABP2 may facilitate drug absorption in the small intestine. In support of these findings, FABPs appear to also increase rates of drug uptake in perfused rat intestines. In rats, FABP1 and FABP2 mRNA expression in the gut increased approximately 1.5- to twofold by feeding a high-fat diet. Compared with control-fed rats, this higher expression of FABP1 and FABP2 correlated with nearly twofold higher rates of disappearance of ibuprofen (disappearance Papp 158 versus 97 × 106 cm/s) and midazolam (disappearance Papp 239 versus 143 × 106 cm/s) from intestinal perfusate and increased accumulation of the drugs in the intestinal tissue (Trevaskis et al., 2011). This suggests that FABPs may facilitate the uptake of these drugs into the enterocytes (Trevaskis et al., 2011). Interestingly, significantly less 4-hydroxy-midazolam was quantified in mesenteric blood in animals with elevated FABP, and the extraction ratio of midazolam by the intestine was decreased from 11% to 7% in rats with higher FABP1 expression compared with control-fed rats. This suggests that midazolam likely bound to FABP1 in the enterocytes, altering midazolam metabolism in the enterocytes (Trevaskis et al., 2011). Despite these findings, the contribution of FABPs to rate and extent of drug absorption has received relatively little attention.
The potential impact of FABP binding on drug distribution is clear from the high expression of FABPs in different tissues and the capacity of FABPs for drug binding in a variety of tissues throughout the body. Binding of numerous drugs that target the central nervous system to FABP3, FABP4, and FABP5, which are expressed in the brain, was proposed to impact the distribution of drugs across the blood-brain barrier (Lee et al., 2015). This process is similar to the regulation of endogenous docosahexaenoic acid concentrations in the brain by FABP5 (Pan et al., 2015, 2016). Current models (Utsey et al., 2020) for predicting tissue distribution of drugs and tissue partition coefficients (Kp values) do not account for specific protein binding sinks in tissues, and hence extensive FABP binding in any tissue is not considered when distribution kinetics are modeled. As physiologically based pharmacokinetic models of drug distribution become more mainstream, incorporation of FABP binding into tissue distribution models and considering FABP binding when rates of distribution are considered will become increasingly important.
Ligand Delivery to Membranes
It remains unclear whether the observed changes in lipid metabolism that correlate with FABP expression are simply because FABPs provide an intracellular reservoir to increase uptake of their ligands to cells and, hence, the availability of ligands to sites of metabolism (diffusional model, Fig. 7A) or if FABPs deliver ligands via specific interactions to enzymes or to cellular membranes. The mechanism of ligand transfer from FABPs to model phospholipid membranes has been studied for FABPs 1–5 using fluorescent fatty acids as ligands (Storch and Thumser, 2000). The rates of ligand transfer from FABPs to phospholipid membranes could be measured via quenching of fluorescence upon fatty acid ligand incorporation into the phospholipid membrane. The rate of ligand transfer from FABP1 to acceptor membranes was not affected by the concentrations or composition of phospholipid. However, FABP1 transfer rates were substantially impacted by the ionic strength of the surrounding aqueous medium. These data suggest that FABP1 does not interact directly with phospholipid membranes and that ligand delivery to model membranes requires release of ligand into solution (Hsu and Storch, 1996).
In contrast, the transfer rates of FABPs 2–5 were proportional to phospholipid concentration in acceptor membranes and affected by membrane phospholipid composition (Kim and Storch, 1992; Wootan et al., 1993; Hsu and Storch, 1996; Storch and Thumser, 2000). FABP2 was shown to also accept fatty acids from donor membranes. Faster fatty acid transfer rates from donor membranes to FABP2 were observed with negatively charged membranes compared with zwitterionic membranes (Thumser and Storch, 2000). Taken together, these data suggest that FABPs 2–5 directly deliver their ligands to phospholipid membranes via ionic interactions. The structural basis for this interaction has been elucidated with mutagenesis studies with FABP2. These studies suggest that the alpha-helical domain in FABP2 is important for these interactions (Corsico et al., 1998; Falomir-Lockhart et al., 2006). The rate of fatty acid transfer from a helix-less FABP2 variant to phospholipid model membranes was unaffected by increasing phospholipid concentration compared with WT, suggesting the loss of the helices also eliminates the membrane interactions (Corsico et al., 1998). Additionally, WT FABP2 could outcompete cytochrome c interactions with anionic membranes, but this function was severely disrupted with the helix-less variant. These findings were corroborated with later mutational studies showing that charged lysine residues in the alpha-helical region are critical for efficient fatty acid transfer (Falomir-Lockhart et al., 2006). The significance of this protein-membrane interaction in vivo is unknown but may play a role in the uptake and targeting of ligands to specific cellular organelles (Hsu and Storch, 1996).
Ligand Delivery by iLBPs to Enzymes and Receptors
The role of iLBPs in delivering their ligands to metabolic enzymes or receptors via protein-protein interactions and substrate channeling has been most extensively studied with the retinoid binding proteins (Napoli, 2017). Possibly due to the reactivity and potential toxicity of retinoids, the retinoid binding proteins appear to modulate and direct retinoid metabolism and signaling via a network of protein-protein interactions. Extensive kinetic and metabolic studies have been conducted (Napoli, 2016, 2017) in rat and human intestinal and liver microsomes with holo-CRBPs. In these studies, despite the tight binding of the ligands with the CRBPs, the apparent Km values for the total ligand are often significantly decreased or unaltered when the ligand is entirely bound to the CRBP in comparison with free ligand (Ong et al., 1987; Herr et al., 1999; Napoli, 2016, 2017). This kinetic data cannot be explained by the diffusional model (free drug hypothesis) and have been interpreted through protein-protein interactions between the CRBPs and retinoid metabolizing enzymes. Consistent with the protein-protein interaction model, apo-CRBP1 appears to also inhibit retinol esterification by lecithin retinol acyltransferase enzyme suggesting a function of the apo-CRBP1 in regulating metabolism even in the absence of its ligand. These data suggest that the ratio of apo- to holo-CRBP1 or the ratio of CRBP1 to its ligand may have an important role in regulating vitamin A homeostasis in the cell. This concept is illustrated via kinetic simulations in Fig. 8. Yet these observations are limited to endogenous retinoids and their specified metabolic enzymes, and the importance to drug metabolism by major drug-metabolizing enzymes is unknown.
Fig. 8.
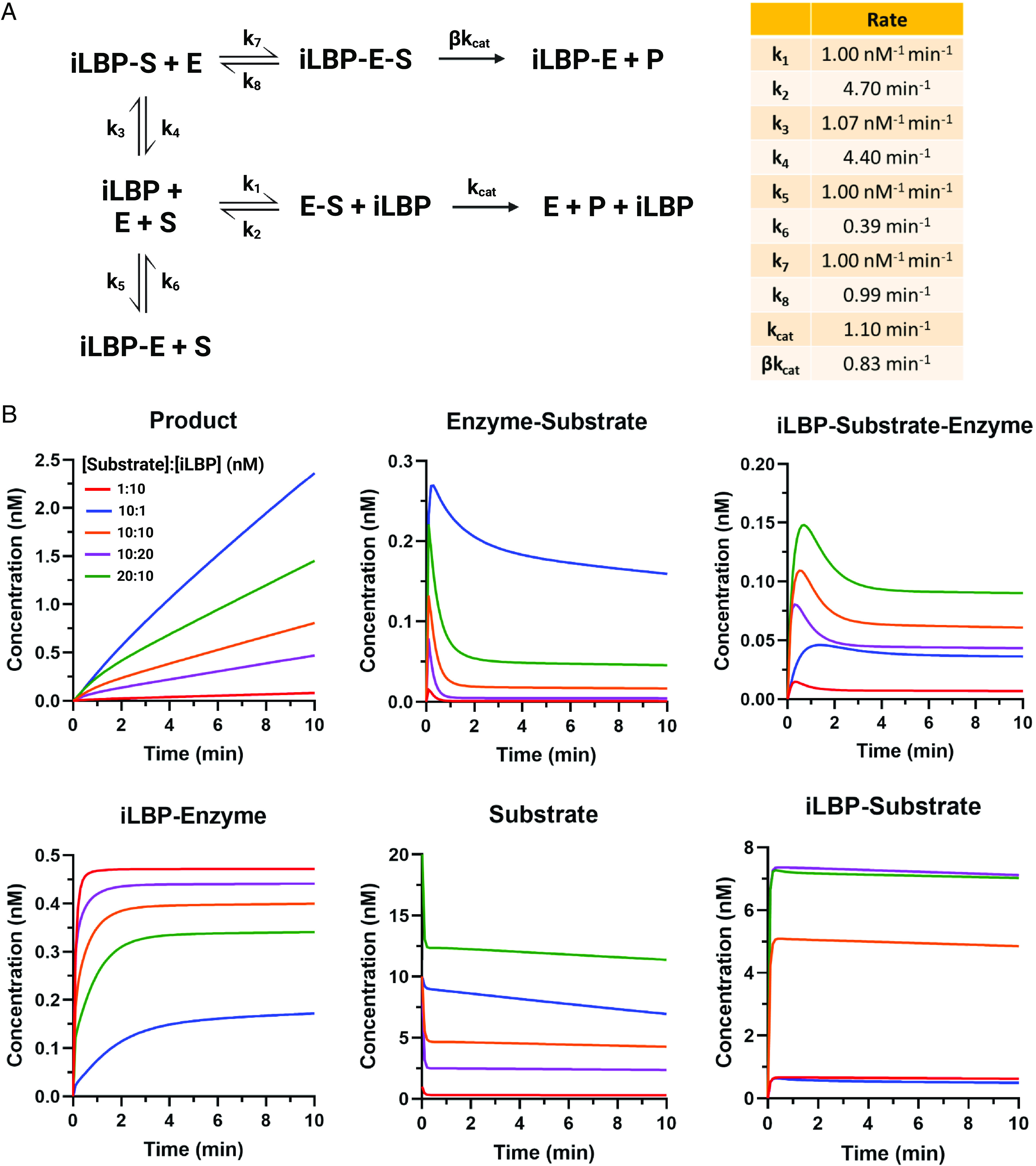
Simulation of the impact of varying ligand to iLBP ratios on metabolic enzyme activity under the circumstances that the iLBP interacts directly with the metabolic enzyme. (A) An enzyme kinetic scheme showing the overall model used for the simulations where apo-iLBP directly inhibits the enzyme and holo-iLBP can deliver substrate to the enzyme via protein-protein interactions. (B) Simulated concentrations of the metabolite (product) formation, enzyme-substrate complex concentrations, concentrations of the ternary iLBP-substrate-enzyme complex, iLBP-enzyme complex, free substrate in solution, and iLBP-substrate complex as a function of time when the ratio of the substrate concentration to binding protein concentration is varied, and all the processes are simulated according to the scheme in A. The substrate concentrations were 1 nM (red line), 10 nM, (blue, orange, and purple lines) and 20 nM (green line). The iLBP concentration was either 1 nM (blue), 10 nM (green, orange, and red lines), or 20 nM (purple line). The kinetic and catalytic rate constants used in the simulations are listed in a table in (A). The enzyme concentration in all simulations was 0.5 nM. (Figure created with BioRender.com.) MATLAB code used for simulations is provided in Supplementary Material.
Protein-protein interactions between CRABPs and nuclear RARs have also been extensively studied (Dong et al., 1999; Budhu et al., 2001; Schug et al., 2007; Majumdar et al., 2011). Expression of CRABP2 but not CRABP1 in Cos7 cells enhances RAR transactivation (Dong et al., 1999), and the transfer rate of atRA from CRABP2 to RAR appears to be dependent on RAR acceptor concentration while transfer rates from CRABP1 are unaffected by changes in RAR concentration. Holo-CRABP2 also appears to translocate to the nucleus via a SUMOylation dependent mechanism to channel atRA directly to RAR (Majumdar et al., 2011). These findings demonstrate the potential role that iLBPs may have in cellular targeting of their ligands and delivery of their ligands to target receptors, and suggest that iLBP interaction may be protein specific.
The impact of the CRABPs on atRA hydroxylation has also been studied in rodent microsomes (Napoli et al., 1991; Fiorella and Napoli, 1994), with recombinant drug metabolizing CYPs, CYP3A4, and CYP2C8 and with the atRA hydroxylases CYP26A1, CYP26B1, and CYP26C1 (Nelson et al., 2016; Zhong et al., 2018; Yabut and Isoherranen, 2022). Recently holo-CRABP2 was also shown to be a substrate of CYP27C1, a retinoid desaturase in the skin (Glass and Guengerich, 2021). As expected from the tight binding of atRA to CRABP1 and CRABP2, CYP3A4 and CYP2C8 mediated metabolism of atRA was nearly completely abolished when atRA was bound to the CRABPs, consistent with the free drug hypothesis (Nelson et al., 2016; Yabut and Isoherranen, 2022). However, with the CYP26 enzymes, efficient atRA formation was observed also when atRA was completely bound to CRABPs. The apparent Km values for holo-CRABPs were either unchanged or decreased when compared with free ligand in solution. Surprisingly the kcat values for atRA hydroxylation were also significantly decreased in the presence of CRABPs for all three CYP26 enzymes. This suggests that apo-CRABPs inhibit CYP26 enzymes via a noncompetitive mechanism similar to the inhibition observed between CRBPs and lecithin retinol acyltransferase. The observed kinetics could be explained using a substrate channeling model incorporating direct protein-protein interactions between CYP26 and apo- and holo-CRABPs (Nelson et al., 2016; Yabut and Isoherranen, 2022).
The binding protein (CRBP, CRABP)-enzyme interactions may be critical modulators of ligand metabolism and vitamin A homeostasis in cells in a ligand concentration dependent manner, and the phenomenon may be important for other iLBPs as well. This hypothesis was explored via kinetic simulations of the effect of the binding protein-ligand ratio on the metabolic rates and ligand clearance in a cell (Fig. 8). The simulations show how altered expression of the binding proteins will change ligand metabolism and concentrations through direct protein-protein interactions between the apo- and holo-binding protein and the metabolic enzyme. When substrate is in excess to the binding protein, the substrate is relatively freely metabolized (Fig. 8, blue and green lines) allowing for homeostasis to be maintained. However, under circumstances of substrate deficiency when the binding protein is in excess to substrate, nearly all of the enzyme is bound by the apo-binding protein, severely inhibiting metabolism (Fig. 8, red and purple lines).
In addition to the retinoid binding proteins, the FABPs have also been shown to directly interact with nuclear receptors and metabolic enzymes. Similar to holo-CRABP2 channeling atRA to RARs, FABP1, FABP4, and FABP5 have been shown to translocate to the nucleus upon ligand binding to enhance PPAR transactivation (Wolfrum et al., 2001; Tan et al., 2002; Schug et al., 2007; Hostetler et al., 2009; Velkov, 2013). Physical interactions between FABPs and PPARs have been demonstrated using biochemical and biophysical assays (coimmunoprecipitation, circular dichroism, fluorescence resonance energy transfer, and NMR). These studies suggest that FABP-PPAR interactions are protein specific. FABP1, FABP4, and FABP5 specifically activate and interact with PPARα, PPARγ, and PPARβ, respectively, and the extent of transactivation appears to be ligand dependent.
FABP4 and FABP5 have been shown to directly interact with hormone sensitive lipase (HSL) (Jenkins-Kruchten et al., 2003; Storch and Corsico, 2008; Storch and Thumser, 2010) to promote the liberation of free fatty acids from triglycerides in times of fatty acid scarcity. FABP4 and FABP5 showed ligand dependent interactions with HSL in isothermal titration calorimetry experiments and increased HSL catalytic activity by approximately twofold (Jenkins-Kruchten et al., 2003). Similarly, FABP1 has been shown to interact with carnitine palmitoyl transferase I (CPTI), a key mitochondrial enzyme for fatty acid β-oxidation (Hostetler et al., 2011). Significant deviation from the theoretical circular dichroism (CD) spectra of the C-terminal and active domain of CPTI was observed in the presence of FABP1. The affinity (Kd) between FABP1 and CPTI was 2.5 nM as determined by fluorescence resonance energy transfer binding assays. Notably, FABP1 enhanced CPTI activity to metabolize LCFA-CoA to LCFA-carnitine demonstrating facilitation of the rate-limiting step in fatty acid β-oxidation. Given the broad binding specificity of FABP1 and FABP2 for various xenobiotics and their high abundance in the liver and intestine, it is likely that FABPs also impact drug metabolism via similar mechanisms in vivo. This hypothesis is supported by the finding that FABP1 binds Δ9-tetrahydrocannabinol and the rate of Δ9-tetrahydrocannabinol metabolism is altered in FABP1-knockout mice (Elmes et al., 2015).
While FABP interactions with transporters have not been extensively studied, several groups have reported that FABP4 directly interacts with the fatty acid uptake transporter CD36 to mediate fatty acid metabolism (Spitsberg et al., 1995; Glatz and Luiken, 2018; Gyamfi et al., 2021). CD36 appears to act as an intracellular docking site for FABP4 to facilitate fatty acid transfer to the cytoplasm where FABP4 may then shuttle fatty acids to the peroxisomes or mitochondria for fatty acid metabolism.
Conclusions
iLBPs are ubiquitously expressed small binding proteins in tissues, which bind a variety of lipophilic compounds and facilitate the cellular uptake, diffusion, and subsequent metabolism of their endogenous ligands. Yet despite the plethora of work that exists to define biochemical functions of iLBPs, their impact on xenobiotic disposition is poorly defined, and very few studies have explored the binding characteristics of various drugs with FABPs. Many xenobiotics also bind to FABPs that are highly expressed in major organs that govern drug absorption and clearance with micromolar to submicromolar affinity in vitro. Based on the high expression of FABPs (up to 11% of all cytosolic protein), it is likely that FABPs also bind xenobiotics in vivo.
The importance of FABP binding in drug disposition is not understood; however, limited studies have shown that absorption and clearance of drugs that bind to FABPs is linked to FABP expression in animal models. These findings suggest that FABPs have the potential to be determinants of xenobiotic disposition. Variability in FABP binding/expression may explain some intra- and interindividual variability in drug disposition as FABP expression changes with diet, disease states, and administration of other therapeutics. Whether FABPs directly affect xenobiotic access to drug-metabolizing enzymes remains a knowledge gap. It is unclear if FABPs may simply provide an intracellular “sink” to increase the partitioning and availability of free drug accessible for metabolism within cells or if FABPs directly interact with metabolic enzymes to alter rates of drug metabolism. Further studies are needed to elucidate these mechanisms, which would provide insight into how FABPs may regulate xenobiotic disposition.
Data Availability
The authors declare that all the data supporting the findings of this study are available within the paper and its Supplemental Material.
Abbreviations
- AEA
anandamide
- ANS
8-anilinonaphthalene-1-sulfonic acid
- atRA
all-trans-retinoic acid
- CD
circular dichroism
- CSP
chemical shift perturbation
- CPTI
carnitine palmitoyltransferase I
- CRABP
cellular retinoic acid binding protein
- CRBP
cellular retinol binding protein
- CYP
cytochrome P450
- FABP
fatty acid binding protein
- HSL
hormone sensitive lipase
- iLBP
intracellular lipid binding protein
- LCFA
long chain fatty acid
- LRAT
lecithin retinol acyltransferase
- mRNA
messenger RNA
- PPAR
peroxisome proliferator-activated receptor
- RAR
retinoic acid receptor
- WT
wildtype
Authorship Contributions
Participated in research design: Yabut, Isoherranen.
Participated in data collection: Yabut.
Performed data analysis: Yabut, Isoherranen.
Wrote or contributed to the writing of the manuscript: Yabut, Isoherranen.
Footnotes
This work was supported in part by grants from the National Institutes of Health National Institute of General Medical Sciences [Grants T32-GM007750 and R01-GM111772] and National Institute on Drug Abuse [Grant P01-DA032507].
No author has an actual or perceived conflict of interest with the contents of this article.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Agellon LB, Toth MJ, Thomson ABR (2002) Intracellular lipid binding proteins of the small intestine, in Cellular Lipid Binding Proteins (Glatz JFC, ed) pp 79–82, Springer US, Boston, MA. [PubMed] [Google Scholar]
- Alpers DH, Bass NM, Engle MJ, DeSchryver-Kecskemeti K (2000) Intestinal fatty acid binding protein may favor differential apical fatty acid binding in the intestine. Biochim Biophys Acta 1483:352–362. [DOI] [PubMed] [Google Scholar]
- Atshaves BP, Martin GG, Hostetler HA, McIntosh AL, Kier AB, Schroeder F (2010) Liver fatty acid-binding protein and obesity. J Nutr Biochem 21:1015–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babin PJ (2009) Molecular Evolution of Vertebrate Fatty-Acid Binding Proteins, Transworld Research Network, Trivandrum, India. [Google Scholar]
- Bashor MM, Toft DO, Chytil F (1973) In vitro binding of retinol to rat-tissue components. Proc Natl Acad Sci USA 70:3483–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass NM, Barker ME, Manning JA, Jones AL, Ockner RK (1989) Acinar heterogeneity of fatty acid binding protein expression in the livers of male, female and clofibrate-treated rats. Hepatology 9:12–21. [DOI] [PubMed] [Google Scholar]
- Bernlohr DA, Simpson MA, Hertzel AV, Banaszak LJ (1997) Intracellular lipid-binding proteins and their genes. Annu Rev Nutr 17:277–303. [DOI] [PubMed] [Google Scholar]
- Besnard P, Niot I, Poirier H, Clément L, Bernard A (2002) New insights into the fatty acid-binding protein (FABP) family in the small intestine. Mol Cell Biochem 239:139–147. [PubMed] [Google Scholar]
- Budhu A, Gillilan R, Noy N (2001) Localization of the RAR interaction domain of cellular retinoic acid binding protein-II. J Mol Biol 305:939–949. [DOI] [PubMed] [Google Scholar]
- Cai J, Lücke C, Chen Z, Qiao Y, Klimtchuk E, Hamilton JA (2012) Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited. Biophys J 102:2585–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LX, Zhang ZP, Scafonas A, Cavalli RC, Gabriel JL, Soprano KJ, Soprano DR (1995) Arginine 132 of cellular retinoic acid-binding protein (type II) is important for binding of retinoic acid. J Biol Chem 270:4518–4525. [DOI] [PubMed] [Google Scholar]
- Chuang S, Velkov T, Horne J, Porter CJH, Scanlon MJ (2008) Characterization of the drug binding specificity of rat liver fatty acid binding protein. J Med Chem 51:3755–3764. [DOI] [PubMed] [Google Scholar]
- Chuang S, Velkov T, Horne J, Wielens J, Chalmers DK, Porter CJH, Scanlon MJ (2009) Probing the fibrate binding specificity of rat liver fatty acid binding protein. J Med Chem 52:5344–5355. [DOI] [PubMed] [Google Scholar]
- Corsico B, Cistola DP, Frieden C, Storch J (1998) The helical domain of intestinal fatty acid binding protein is critical for collisional transfer of fatty acids to phospholipid membranes. Proc Natl Acad Sci USA 95:12174–12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Ruuska SE, Levinthal DJ, Noy N (1999) Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J Biol Chem 274:23695–23698. [DOI] [PubMed] [Google Scholar]
- Elmes MWKaczocha MBerger WTLeung KRalph BPWang LSweeney JMMiyauchi JTTsirka SEOjima I, et al. (2015) Fatty acid-binding proteins (FABPs) are intracellular carriers for Δ9-tetrahydrocannabinol (THC) and cannabidiol (CBD). J Biol Chem 290:8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmes MWPrentis LEMcGoldrick LLGiuliano CJSweeney JMJoseph OMChe JCarbonetti GSStudholme KDeutsch DG, et al. (2019) FABP1 controls hepatic transport and biotransformation of Δ9-THC. Sci Rep 9:7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falomir-Lockhart LJ, Laborde L, Kahn PC, Storch J, Córsico B (2006) Protein-membrane interaction and fatty acid transfer from intestinal fatty acid-binding protein to membranes. Support for a multistep process. J Biol Chem 281:13979–13989. [DOI] [PubMed] [Google Scholar]
- Ferreira R, Napoli J, Enver T, Bernardino L, Ferreira L (2020) Advances and challenges in retinoid delivery systems in regenerative and therapeutic medicine. Nat Commun 11:4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrolino MC, Zhuravleva A, Budyak IL, Krishnan B, Gierasch LM (2013) Delicate balance between functionally required flexibility and aggregation risk in a β-rich protein. Biochemistry 52:8843–8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorella PD, Giguère V, Napoli JL (1993) Expression of cellular retinoic acid-binding protein (type II) in Escherichia coli. Characterization and comparison to cellular retinoic acid-binding protein (type I). J Biol Chem 268:21545–21552. [PubMed] [Google Scholar]
- Fiorella PD, Napoli JL (1991) Expression of cellular retinoic acid binding protein (CRABP) in Escherichia coli. Characterization and evidence that holo-CRABP is a substrate in retinoic acid metabolism. J Biol Chem 266:16572–16579. [PubMed] [Google Scholar]
- Fiorella PD, Napoli JL (1994) Microsomal retinoic acid metabolism. Effects of cellular retinoic acid-binding protein (type I) and C18-hydroxylation as an initial step. J Biol Chem 269:10538–10544. [PubMed] [Google Scholar]
- Floresta G, Pistarà V, Amata E, Dichiara M, Marrazzo A, Prezzavento O, Rescifina A (2017) Adipocyte fatty acid binding protein 4 (FABP4) inhibitors. A comprehensive systematic review. Eur J Med Chem 138:854–873. [DOI] [PubMed] [Google Scholar]
- Flower DR, North ACT, Sansom CE (2000) The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta 1482:9–24. [DOI] [PubMed] [Google Scholar]
- Fogh K, Voorhees JJ, Aström A (1993) Expression, purification, and binding properties of human cellular retinoic acid-binding protein type I and type II. Arch Biochem Biophys 300:751–755. [DOI] [PubMed] [Google Scholar]
- Folli C, Calderone V, Ottonello S, Bolchi A, Zanotti G, Stoppini M, Berni R (2001) Identification, retinoid binding, and x-ray analysis of a human retinol-binding protein. Proc Natl Acad Sci USA 98:3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folli C, Calderone V, Ramazzina I, Zanotti G, Berni R (2002) Ligand binding and structural analysis of a human putative cellular retinol-binding protein. J Biol Chem 277:41970–41977. [DOI] [PubMed] [Google Scholar]
- Foucaud L, Niot I, Kanda T, Besnard P (1998) Indirect dexamethasone down-regulation of the liver fatty acid-binding protein expression in rat liver. Biochim Biophys Acta 1391 :204–212. [DOI] [PubMed] [Google Scholar]
- Furuhashi M (2019) Fatty acid-binding protein 4 in cardiovascular and metabolic diseases. J Atheroscler Thromb 26:216–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi M, Hotamisligil GS (2008) Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov 7:489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi M, Saitoh S, Shimamoto K, Miura T (2015) Fatty acid-binding protein 4 (FABP4): pathophysiological insights and potent clinical biomarker of metabolic and cardiovascular diseases. Clin Med Insights Cardiol 8(Suppl 3):23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajda AM, Storch J (2015) Enterocyte fatty acid-binding proteins (FABPs): different functions of liver and intestinal FABPs in the intestine. Prostaglandins Leukot Essent Fatty Acids 93:9–16 Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server, in The Proteomics Protocols Handbook (Walker JM, ed) pp 571–607, Humana Press, Totowa, NJ. [Google Scholar]
- Gerstner JR, Bremer QZ, Vander Heyden WM, Lavaute TM, Yin JC, Landry CF (2008) Brain fatty acid binding protein (Fabp7) is diurnally regulated in astrocytes and hippocampal granule cell precursors in adult rodent brain. PLoS One 3:e1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguère V, Lyn S, Yip P, Siu CH, Amin S (1990) Molecular cloning of cDNA encoding a second cellular retinoic acid-binding protein. Proc Natl Acad Sci USA 87:6233–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillilan RE, Ayers SD, Noy N (2007) Structural basis for activation of fatty acid-binding protein 4. J Mol Biol 372:1246–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass SM, Guengerich FP (2021) Cellular retinoid-binding proteins transfer retinoids to human cytochrome P450 27C1 for desaturation. J Biol Chem 297:101142 Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatz JFC, Luiken JJFP (2018) Dynamic role of the transmembrane glycoprotein CD36 (SR-B2) in cellular fatty acid uptake and utilization. J Lipid Res 59:1084–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatz JFC, Veerkamp JH (1983) A radiochemical procedure for the assay of fatty acid binding by proteins. Anal Biochem 132:89–95. [DOI] [PubMed] [Google Scholar]
- Gong YZ, Everett ET, Schwartz DA, Norris JS, Wilson FA (1994) Molecular cloning, tissue distribution, and expression of a 14-kDa bile acid-binding protein from rat ileal cytosol. Proc Natl Acad Sci USA 91:4741–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JM, Fisher SZ (2015) Structural analysis of ibuprofen binding to human adipocyte fatty-acid binding protein (FABP4). Acta Crystallogr F Struct Biol Commun 71:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyamfi J, Yeo JH, Kwon D, Min BS, Cha YJ, Koo JS, Jeong J, Lee J, Choi J (2021) Interaction between CD36 and FABP4 modulates adipocyte-induced fatty acid import and metabolism in breast cancer. NPJ Breast Cancer 7:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halldén G, Aponte GW (1997) Evidence for a role of the gut hormone PYY in the regulation of intestinal fatty acid-binding protein transcripts in differentiated subpopulations of intestinal epithelial cell hybrids. J Biol Chem 272:12591–12600. [DOI] [PubMed] [Google Scholar]
- Hanhoff T, Lücke C, Spener F (2002) Insights into binding of fatty acids by fatty acid binding proteins. Mol Cell Biochem 239:45–54. [PubMed] [Google Scholar]
- Haunerland NH, Spener F (2004) Fatty acid-binding proteins—insights from genetic manipulations. Prog Lipid Res 43:328–349. [DOI] [PubMed] [Google Scholar]
- He Y, Yang X, Wang H, Estephan R, Francis F, Kodukula S, Storch J, Stark RE (2007) Solution-state molecular structure of apo and oleate-liganded liver fatty acid-binding protein. Biochemistry 46:12543–12556. [DOI] [PubMed] [Google Scholar]
- Herr FM, Li E, Weinberg RB, Cook VR, Storch J (1999) Differential mechanisms of retinoid transfer from cellular retinol binding proteins types I and II to phospholipid membranes. J Biol Chem 274:9556–9563 Elsevier. [DOI] [PubMed] [Google Scholar]
- Hodsdon ME, Cistola DP (1997a) Discrete backbone disorder in the nuclear magnetic resonance structure of apo intestinal fatty acid-binding protein: implications for the mechanism of ligand entry. Biochemistry 36:1450–1460. [DOI] [PubMed] [Google Scholar]
- Hodsdon ME, Cistola DP (1997b) Ligand binding alters the backbone mobility of intestinal fatty acid-binding protein as monitored by 15N NMR relaxation and 1H exchange. Biochemistry 36:2278–2290. [DOI] [PubMed] [Google Scholar]
- Hohoff C, Börchers T, Rüstow B, Spener F, van Tilbeurgh H (1999) Expression, purification, and crystal structure determination of recombinant human epidermal-type fatty acid binding protein. Biochemistry 38:12229–12239. [DOI] [PubMed] [Google Scholar]
- Hostetler HA, Lupas D, Tan Y, Dai J, Kelzer MS, Martin GG, Woldegiorgis G, Kier AB, Schroeder F (2011) Acyl-CoA binding proteins interact with the acyl-CoA binding domain of mitochondrial carnitine palmitoyl transferase I. Mol Cell Biochem 355:135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostetler HA, McIntosh AL, Atshaves BP, Storey SM, Payne HR, Kier AB, Schroeder F (2009) L-FABP directly interacts with PPARalpha in cultured primary hepatocytes. J Lipid Res 50:1663–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS, Bernlohr DA (2015) Metabolic functions of FABPs—mechanisms and therapeutic implications. Nat Rev Endocrinol 11:592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS, Johnson RS, Distel RJ, Ellis R, Papaioannou VE, Spiegelman BM (1996) Uncoupling of obesity from insulin resistance through a targeted mutation in aP2, the adipocyte fatty acid binding protein. Science 274:1377–1379. [DOI] [PubMed] [Google Scholar]
- Hsu K-T, Storch J (1996) Fatty acid transfer from liver and intestinal fatty acid-binding proteins to membranes occurs by different mechanisms. J Biol Chem 271:13317–13323. [DOI] [PubMed] [Google Scholar]
- Huang H, McIntosh AL, Martin GG, Dangott LJ, Kier AB, Schroeder F (2018) Structural and functional interaction of δ9-tetrahydrocannabinol with liver fatty acid binding protein (FABP1). Biochemistry 57:6027–6042. [DOI] [PubMed] [Google Scholar]
- Huang H, McIntosh AL, Martin GG, Landrock D, Chung S, Landrock KK, Dangott LJ, Li S, Kier AB, Schroeder F (2016) FABP1: a novel hepatic endocannabinoid and cannabinoid binding protein. Biochemistry 55:5243–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, McIntosh AL, Martin GG, Landrock KK, Landrock D, Gupta S, Atshaves BP, Kier AB, Schroeder F (2014) Structural and functional interaction of fatty acids with human liver fatty acid-binding protein (L-FABP) T94A variant. FEBS J 281:2266–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhou Y, Sun Y, Wang Q (2022) Intestinal fatty acid binding protein: a rising therapeutic target in lipid metabolism. Prog Lipid Res 87:101178. [DOI] [PubMed] [Google Scholar]
- Hung DY, Burczynski FJ, Chang P, Lewis A, Masci PP, Siebert GA, Anissimov YG, Roberts MS (2003) Fatty acid binding protein is a major determinant of hepatic pharmacokinetics of palmitate and its metabolites. Am J Physiol Gastrointest Liver Physiol 284:G423–G433. [DOI] [PubMed] [Google Scholar]
- Ishimura SFuruhashi MWatanabe YHoshina KFuseya TMita TOkazaki YKoyama MTanaka MAkasaka H, et al. (2013) Circulating levels of fatty acid-binding protein family and metabolic phenotype in the general population. PLoS One 8:e81318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, AlSadhan I, Vaidyanathan PP, Herschlag D (2020) How to measure and evaluate binding affinities. eLife 9:e57264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins AE, Hockenberry JA, Nguyen T, Bernlohr DA (2002) Testing of the portal hypothesis: analysis of a V32G, F57G, K58G mutant of the fatty acid binding protein of the murine adipocyte. Biochemistry 41:2022–2027. [DOI] [PubMed] [Google Scholar]
- Jenkins-Kruchten AE, Bennaars-Eiden A, Ross JR, Shen W-J, Kraemer FB, Bernlohr DA (2003) Fatty acid-binding protein-hormone-sensitive lipase interaction. Fatty acid dependence on binding. J Biol Chem 278:47636–47643. [DOI] [PubMed] [Google Scholar]
- Kaczocha M, Glaser ST, Deutsch DG (2009) Identification of intracellular carriers for the endocannabinoid anandamide. Proc Natl Acad Sci USA 106:6375–6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczocha M, Vivieca S, Sun J, Glaser ST, Deutsch DG (2012) Fatty acid-binding proteins transport N-acylethanolamines to nuclear receptors and are targets of endocannabinoid transport inhibitors. J Biol Chem 287:3415–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane CD, Bernlohr DA (1996) A simple assay for intracellular lipid-binding proteins using displacement of 1-anilinonaphthalene 8-sulfonic acid. Anal Biochem 233:197–204. [DOI] [PubMed] [Google Scholar]
- Kane MA, Bright FV, Napoli JL (2011) Binding affinities of CRBPI and CRBPII for 9-cis-retinoids. Biochim Biophys Acta 1810:514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Blaner WS, Mertz JR, Das K, Kato K, Goodman DS (1985) Influence of retinoid nutritional status on cellular retinol- and cellular retinoic acid-binding protein concentrations in various rat tissues. J Biol Chem 260:4832–4838. [PubMed] [Google Scholar]
- Khan SH, Sorof S (1990) Preferential binding of growth inhibitory prostaglandins by the target protein of a carcinogen. Proc Natl Acad Sci USA 87:9401–9405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Storch J (1992) Mechanism of free fatty acid transfer from rat heart fatty acid-binding protein to phospholipid membranes. Evidence for a collisional process. J Biol Chem 267:20051–20056. [PubMed] [Google Scholar]
- Kleywegt GJ, Bergfors T, Senn H, Le Motte P, Gsell B, Shudo K, Jones TA (1994) Crystal structures of cellular retinoic acid binding proteins I and II in complex with all-trans-retinoic acid and a synthetic retinoid. Structure 2:1241–1258. [DOI] [PubMed] [Google Scholar]
- Kushlan MC, Gollan JL, Ma WL, Ockner RK (1981) Sex differences in hepatic uptake of long chain fatty acids in single-pass perfused rat liver. J Lipid Res 22:431–436. [PubMed] [Google Scholar]
- LaLonde JM, Levenson MA, Roe JJ, Bernlohr DA, Banaszak LJ (1994) Adipocyte lipid-binding protein complexed with arachidonic acid. Titration calorimetry and X-ray crystallographic studies. J Biol Chem 269:25339–25347. [DOI] [PubMed] [Google Scholar]
- Lee GS, Kappler K, Porter CJH, Scanlon MJ, Nicolazzo JA (2015) Fatty acid binding proteins expressed at the human blood-brain barrier bind drugs in an isoform-specific manner. Pharm Res 32:3432–3446. [DOI] [PubMed] [Google Scholar]
- Lee GS, Pan Y, Scanlon MJ, Porter CJH, Nicolazzo JA (2018) Fatty acid-binding protein 5 mediates the uptake of fatty acids, but not drugs, into human brain endothelial cells. J Pharm Sci 107:1185–1193. [DOI] [PubMed] [Google Scholar]
- Lee S-AYang KJZBrun P-JSilvaroli JAYuen JJShmarakov IJiang HFeranil JBLi XLackey AI, et al. (2020) Retinol-binding protein 2 (RBP2) binds monoacylglycerols and modulates gut endocrine signaling and body weight. Sci Adv 6:eaay8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R-Z, Li X, Godbout R (2008) A novel fatty acid-binding protein (FABP) gene resulting from tandem gene duplication in mammals: transcription in rat retina and testis. Genomics 92:436–445. [DOI] [PubMed] [Google Scholar]
- Lowe JB, Sacchettini JC, Laposata M, McQuillan JJ, Gordon JI (1987) Expression of rat intestinal fatty acid-binding protein in Escherichia coli. Purification and comparison of ligand binding characteristics with that of Escherichia coli-derived rat liver fatty acid-binding protein. J Biol Chem 262:5931–5937. [PubMed] [Google Scholar]
- Lücke C, Zhang F, Hamilton JA, Sacchettini JC, Rüterjans H (2000) Solution structure of ileal lipid binding protein in complex with glycocholate. Eur J Biochem 267:2929–2938. [DOI] [PubMed] [Google Scholar]
- Luxon BA, Weisiger RA (1993) Sex differences in intracellular fatty acid transport: role of cytoplasmic binding proteins. Am J Physiol 265:G831–G841. [DOI] [PubMed] [Google Scholar]
- Maatman RG, Van Kuppevelt TH, Veerkamp JH (1991) Two types of fatty acid-binding protein in human kidney. Isolation, characterization and localization. Biochem J 273:759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald PN, Ong DE (1987) Binding specificities of cellular retinol-binding protein and cellular retinol-binding protein, type II. J Biol Chem 262:10550–10556. [PubMed] [Google Scholar]
- Majumdar A, Petrescu AD, Xiong Y, Noy N (2011) Nuclear translocation of cellular retinoic acid-binding protein II is regulated by retinoic acid-controlled SUMOylation. J Biol Chem 286:42749–42757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GG, Danneberg H, Kumar LS, Atshaves BP, Erol E, Bader M, Schroeder F, Binas B (2003) Decreased liver fatty acid binding capacity and altered liver lipid distribution in mice lacking the liver fatty acid-binding protein gene. J Biol Chem 278:21429–21438. [DOI] [PubMed] [Google Scholar]
- Matarese V, Bernlohr DA (1988) Purification of murine adipocyte lipid-binding protein. Characterization as a fatty acid- and retinoic acid-binding protein. J Biol Chem 263:14544–14551. [PubMed] [Google Scholar]
- McKillop IH, Girardi CA, Thompson KJ (2019) Role of fatty acid binding proteins (FABPs) in cancer development and progression. Cell Signal 62:109336. [DOI] [PubMed] [Google Scholar]
- Menozzi I, Vallese F, Polverini E, Folli C, Berni R, Zanotti G (2017) Structural and molecular determinants affecting the interaction of retinol with human CRBP1. J Struct Biol 197:330–339. [DOI] [PubMed] [Google Scholar]
- Napoli JL, Posch KP, Fiorella PD, Boerman MH (1991) Physiological occurrence, biosynthesis and metabolism of retinoic acid: evidence for roles of cellular retinol-binding protein (CRBP) and cellular retinoic acid-binding protein (CRABP) in the pathway of retinoic acid homeostasis. Biomed Pharmacother 45:131–143. [DOI] [PubMed] [Google Scholar]
- Napoli JL (2017) Cellular retinoid binding-proteins, CRBP, CRABP, FABP5: effects on retinoid metabolism, function and related diseases. Pharmacol Ther 173:19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli JL (2016) Functions of intracellular retinoid binding-proteins. Subcell Biochem 81:21–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CH, Peng C-C, Lutz JD, Yeung CK, Zelter A, Isoherranen N (2016) Direct protein-protein interactions and substrate channeling between cellular retinoic acid binding proteins and CYP26B1. FEBS Lett 590:2527–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecz G, Hubbell T, Jefferson JR, Lowe JB, Schroeder F (1991) Interaction of fatty acids with recombinant rat intestinal and liver fatty acid-binding proteins. Arch Biochem Biophys 286:300–309. [DOI] [PubMed] [Google Scholar]
- Newberry EP, Xie Y, Kennedy S, Han X, Buhman KK, Luo J, Gross RW, Davidson NO (2003) Decreased hepatic triglyceride accumulation and altered fatty acid uptake in mice with deletion of the liver fatty acid-binding protein gene. J Biol Chem 278:51664–51672. [DOI] [PubMed] [Google Scholar]
- Noiri E, Doi K, Negishi K, Tanaka T, Hamasaki Y, Fujita T, Portilla D, Sugaya T (2009) Urinary fatty acid-binding protein 1: an early predictive biomarker of kidney injury. Am J Physiol Renal Physiol 296:F669–F679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris AW, Cheng L, Giguère V, Rosenberger M, Li E (1994) Measurement of subnanomolar retinoic acid binding affinities for cellular retinoic acid binding proteins by fluorometric titration. Biochim Biophys Acta 1209:10–18. [DOI] [PubMed] [Google Scholar]
- Nossoni Z, Assar Z, Yapici I, Nosrati M, Wang W, Berbasova T, Vasileiou C, Borhan B, Geiger J (2014) Structures of holo wild-type human cellular retinol-binding protein II (hCRBPII) bound to retinol and retinal. Acta Crystallogr D Biol Crystallogr 70:3226–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ockner RK, Manning JA (1974) Fatty acid-binding protein in small intestine. Identification, isolation, and evidence for its role in cellular fatty acid transport. J Clin Invest 54:326–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong DE, Kakkad B, MacDonald PN (1987) Acyl-CoA-independent esterification of retinol bound to cellular retinol-binding protein (type II) by microsomes from rat small intestine. J Biol Chem 262:2729–2736. [PubMed] [Google Scholar]
- Owada YAbdelwahab SAKitanaka NSakagami HTakano HSugitani YSugawara MKawashima HKiso YMobarakeh JI, et al. (2006) Altered emotional behavioral responses in mice lacking brain-type fatty acid-binding protein gene. Eur J Neurosci 24:175–187. [DOI] [PubMed] [Google Scholar]
- Pan Y, Scanlon MJ, Owada Y, Yamamoto Y, Porter CJH, Nicolazzo JA (2015) Fatty acid-binding protein 5 facilitates the blood-brain barrier transport of docosahexaenoic acid. Mol Pharm 12:4375–4385. [DOI] [PubMed] [Google Scholar]
- Pan Y, Short JL, Choy KHC, Zeng AX, Marriott PJ, Owada Y, Scanlon MJ, Porter CJH, Nicolazzo JA (2016) Fatty acid-binding protein 5 at the blood-brain barrier regulates endogenous brain docosahexaenoic acid levels and cognitive function. J Neurosci 36:11755–11767 Society for Neuroscience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil R, Laguerre A, Wielens J, Headey SJ, Williams ML, Hughes MLR, Mohanty B, Porter CJH, Scanlon MJ (2014) Characterization of two distinct modes of drug binding to human intestinal fatty acid binding protein. ACS Chem Biol 9:2526–2534. [DOI] [PubMed] [Google Scholar]
- Patil RMohanty BLiu BChandrashekaran IRHeadey SJWilliams MLClements CSIlyichova ODoak BCGenissel P, et al. (2019) A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor α agonists. J Biol Chem 294:3720–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulussen RJ, van der Logt CPE, Veerkamp JH (1988) Characterization and binding properties of fatty acid-binding proteins from human, pig, and rat heart. Arch Biochem Biophys 264:533–545. [DOI] [PubMed] [Google Scholar]
- Paulussen RJ, van Moerkerk HT, Veerkamp JH (1990) Immunochemical quantitation of fatty acid-binding proteins. Tissue distribution of liver and heart FABP types in human and porcine tissues. Int J Biochem 22:393–398. [DOI] [PubMed] [Google Scholar]
- Pelsers MMAL, Hermens WT, Glatz JFC (2005) Fatty acid-binding proteins as plasma markers of tissue injury. Clin Chim Acta 352:15–35. [DOI] [PubMed] [Google Scholar]
- Pelsers MMAL, Namiot Z, Kisielewski W, Namiot A, Januszkiewicz M, Hermens WT, Glatz JFC (2003) Intestinal-type and liver-type fatty acid-binding protein in the intestine. Tissue distribution and clinical utility. Clin Biochem 36:529–535. [DOI] [PubMed] [Google Scholar]
- Pelton PD, Zhou L, Demarest KT, Burris TP (1999) PPARgamma activation induces the expression of the adipocyte fatty acid binding protein gene in human monocytes. Biochem Biophys Res Commun 261:456–458. [DOI] [PubMed] [Google Scholar]
- Peng X-E, Wu Y-L, Lu Q-Q, Hu Z-J, Lin X (2012) Two genetic variants in FABP1 and susceptibility to non-alcohol fatty liver disease in a Chinese population. Gene 500:54–58. [DOI] [PubMed] [Google Scholar]
- Peterson PA, Rask L (1971) Studies on the fluorescence of the human vitamin A-transporting plasma protein complex and its individual components. J Biol Chem 246:7544–7550. [PubMed] [Google Scholar]
- Poirier H, Braissant O, Niot I, Wahli W, Besnard P (1997) 9-cis-retinoic acid enhances fatty acid-induced expression of the liver fatty acid-binding protein gene. FEBS Lett 412:480–484. [DOI] [PubMed] [Google Scholar]
- Ragona L, Pagano K, Tomaselli S, Favretto F, Ceccon A, Zanzoni S, D’Onofrio M, Assfalg M, Molinari H (2014) The role of dynamics in modulating ligand exchange in intracellular lipid binding proteins. Biochim Biophys Acta 1844:1268–1278. [DOI] [PubMed] [Google Scholar]
- Rezar RJirak PGschwandtner MDerler RFelder TKHaslinger MKopp KSeelmaier CGranitz CHoppe UC, et al. (2020) Heart-type fatty acid-binding protein (H-FABP) and its role as a biomarker in heart failure: what do we know so far? J Clin Med 9:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richieri GV, Ogata RT, Kleinfeld AM (1994) Equilibrium constants for the binding of fatty acids with fatty acid-binding proteins from adipocyte, intestine, heart, and liver measured with the fluorescent probe ADIFAB. J Biol Chem 269:23918–23930. [PubMed] [Google Scholar]
- Richieri GV, Ogata RT, Kleinfeld AM (1995) Thermodynamics of fatty acid binding to fatty acid-binding proteins and fatty acid partition between water and membranes measured using the fluorescent probe ADIFAB. J Biol Chem 270:15076–15084. [DOI] [PubMed] [Google Scholar]
- Rowland A, Hallifax D, Nussio MR, Shapter JG, Mackenzie PI, Brian Houston J, Knights KM, Miners JO (2015) Characterization of the comparative drug binding to intra- (liver fatty acid binding protein) and extra- (human serum albumin) cellular proteins. Xenobiotica 45:847–857. [DOI] [PubMed] [Google Scholar]
- Rowland A, Knights KM, Mackenzie PI, Miners JO (2009) Characterization of the binding of drugs to human intestinal fatty acid binding protein (IFABP): potential role of IFABP as an alternative to albumin for in vitro-in vivo extrapolation of drug kinetic parameters. Drug Metab Dispos 37:1395–1403. [DOI] [PubMed] [Google Scholar]
- Sacchettini JC, Hauft SM, Van Camp SL, Cistola DP, Gordon JI (1990) Developmental and structural studies of an intracellular lipid binding protein expressed in the ileal epithelium. J Biol Chem 265:19199–19207. [PubMed] [Google Scholar]
- Saito NFuruhashi MKoyama MHigashiura YAkasaka HTanaka MMoniwa NOhnishi HSaitoh SUra N, et al. (2021) Elevated circulating FABP4 concentration predicts cardiovascular death in a general population: a 12-year prospective study. Sci Rep 11:4008 Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanquer S, Gilchrest BA (1994) Characterization of human cellular retinoic acid-binding proteins-I and -II: ligand binding affinities and distribution in skin. Arch Biochem Biophys 311:86–94. [DOI] [PubMed] [Google Scholar]
- Schaap FG, van der Vusse GJ, Glatz JFC (2002) Evolution of the family of intracellular lipid binding proteins in vertebrates. Mol Cell Biochem 239:69–77. [PubMed] [Google Scholar]
- Schroeder FMcIntosh ALMartin GGHuang HLandrock DChung SLandrock KKDangott LJLi SKaczocha M, et al. (2016) Fatty acid binding protein-1 (FABP1) and the human FABP1 T94A variant: roles in the endocannabinoid system and dyslipidemias. Lipids 51:655–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug TT, Berry DC, Shaw NS, Travis SN, Noy N (2007) Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell 129:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha RS, Kane CD, Xu Z, Banaszak LJ, Bernlohr DA (1993) Modulation of ligand binding affinity of the adipocyte lipid-binding protein by selective mutation. Analysis in vitro and in situ. J Biol Chem 268:7885–7892. [PubMed] [Google Scholar]
- Sheng N, Li J, Liu H, Zhang A, Dai J (2016) Interaction of perfluoroalkyl acids with human liver fatty acid-binding protein. Arch Toxicol 90:217–227. [DOI] [PubMed] [Google Scholar]
- Shimizu F, Watanabe TK, Shinomiya H, Nakamura Y, Fujiwara T (1997) Isolation and expression of a cDNA for human brain fatty acid-binding protein B-FABP. Biochim Biophys Acta 1354:24–28. [DOI] [PubMed] [Google Scholar]
- Shrestha SSunaga HHanaoka HYamaguchi AKuwahara SUmbarawan YNakajima KMachida TMurakami MSaito A, et al. (2018) Circulating FABP4 is eliminated by the kidney via glomerular filtration followed by megalin-mediated reabsorption. Sci Rep 8:16451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers FWilm ADineen DGibson TJKarplus KLi WLopez RMcWilliam HRemmert MSöding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvaroli JA, Arne JM, Chelstowska S, Kiser PD, Banerjee S, Golczak M (2016) Ligand binding induces conformational changes in human cellular retinol-binding protein 1 (CRBP1) revealed by atomic resolution crystal structures. J Biol Chem 291:8528–8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smathers RL, Petersen DR (2011) The human fatty acid-binding protein family: evolutionary divergences and functions. Hum Genomics 5:170–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Storch J (1999) The adipocyte fatty acid-binding protein binds to membranes by electrostatic interactions. J Biol Chem 274:35325–35330. [DOI] [PubMed] [Google Scholar]
- Spitsberg VL, Matitashvili E, Gorewit RC (1995) Association and coexpression of fatty-acid-binding protein and glycoprotein CD36 in the bovine mammary gland. Eur J Biochem 230:872–878. [DOI] [PubMed] [Google Scholar]
- Storch J, Corsico B (2008) The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu Rev Nutr 28:73–95. [DOI] [PubMed] [Google Scholar]
- Storch J, McDermott L (2009) Structural and functional analysis of fatty acid-binding proteins. J Lipid Res 50(Suppl):S126–S131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch J, Thumser AE (2010) Tissue-specific functions in the fatty acid-binding protein family. J Biol Chem 285:32679–32683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch J, Thumser AEA (2000) The fatty acid transport function of fatty acid-binding proteins. Biochim Biophys Acta 1486:28–44. [DOI] [PubMed] [Google Scholar]
- Sulsky RMagnin DRHuang YSimpkins LTaunk PPatel MZhu YStouch TRBassolino-Klimas DParker R, et al. (2007) Potent and selective biphenyl azole inhibitors of adipocyte fatty acid binding protein (aFABP). Bioorg Med Chem Lett 17:3511–3515. [DOI] [PubMed] [Google Scholar]
- Tan N-S, Shaw NS, Vinckenbosch N, Liu P, Yasmin R, Desvergne B, Wahli W, Noy N (2002) Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol Cell Biol 22:5114–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J, Winter N, Terwey D, Bratt J, Banaszak L (1997) The crystal structure of the liver fatty acid-binding protein. A complex with two bound oleates. J Biol Chem 272:7140–7150. [DOI] [PubMed] [Google Scholar]
- Thumser AE, Voysey J, Wilton DC (1996) Mutations of recombinant rat liver fatty acid-binding protein at residues 102 and 122 alter its structural integrity and affinity for physiological ligands. Biochem J 314:943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumser AE, Wilton DC (1994) Characterization of binding and structural properties of rat liver fatty-acid-binding protein using tryptophan mutants. Biochem J 300:827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumser AE, Wilton DC (1996) The binding of cholesterol and bile salts to recombinant rat liver fatty acid-binding protein. Biochem J 320:729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumser AEA, Storch J (2000) Liver and intestinal fatty acid-binding proteins obtain fatty acids from phospholipid membranes by different mechanisms. J Lipid Res 41:647–656. [PubMed] [Google Scholar]
- Trevaskis NL, Nguyen G, Scanlon MJ, Porter CJH (2011) Fatty acid binding proteins: potential chaperones of cytosolic drug transport in the enterocyte? Pharm Res 28:2176–2190. [DOI] [PubMed] [Google Scholar]
- Utsey K, Gastonguay MS, Russell S, Freling R, Riggs MM, Elmokadem A (2020) Quantification of the impact of partition coefficient prediction methods on physiologically Based pharmacokinetic model output using a standardized tissue composition. Drug Metab Dispos 48:903–916. [DOI] [PubMed] [Google Scholar]
- Vaezeslami S, Mathes E, Vasileiou C, Borhan B, Geiger JH (2006) The structure of Apo-wild-type cellular retinoic acid binding protein II at 1.4 A and its relationship to ligand binding and nuclear translocation. J Mol Biol 363:687–701. [DOI] [PubMed] [Google Scholar]
- Valizadeh MAghasizadeh MNemati MHashemi MAghaee-Bakhtiari SHZare-Feyzabadi REsmaily HGhazizdaeh HSahebi RAhangari N, et al. (2021) The association between a fatty acid binding protein 1 (FABP1) gene polymorphism and serum lipid abnormalities in the MASHAD cohort study. Prostaglandins Leukot Essent Fatty Acids 172:102324. [DOI] [PubMed] [Google Scholar]
- Veerkamp JH, Maatman RGHJ (1995) Cytoplasmic fatty acid-binding proteins: their structure and genes. Prog Lipid Res 34:17–52. [DOI] [PubMed] [Google Scholar]
- Veerkamp JH, van Moerkerk HTB, Prinsen CFM, van Kuppevelt TH (1999) Structural and functional studies on different human FABP types. Mol Cell Biochem 192:137–142. [PubMed] [Google Scholar]
- Veerkamp JH, Zimmerman AW (2001) Fatty acid-binding proteins of nervous tissue. J Mol Neurosci 16:133–142, discussion 151–157. [DOI] [PubMed] [Google Scholar]
- Velkov T (2013) Interactions between human liver fatty acid binding protein and peroxisome proliferator activated receptor selective drugs. PPAR Res 2013:938401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velkov T, Chuang S, Wielens J, Sakellaris H, Charman WN, Porter CJH, Scanlon MJ (2005) The interaction of lipophilic drugs with intestinal fatty acid-binding protein. J Biol Chem 280:17769–17776. [DOI] [PubMed] [Google Scholar]
- Velkov T, Horne J, Laguerre A, Jones E, Scanlon MJ, Porter CJH (2007) Examination of the role of intestinal fatty acid-binding protein in drug absorption using a parallel artificial membrane permeability assay. Chem Biol 14:453–465. [DOI] [PubMed] [Google Scholar]
- Velkov T, Lim MLR, Horne J, Simpson JS, Porter CJH, Scanlon MJ (2009) Characterization of lipophilic drug binding to rat intestinal fatty acid binding protein. Mol Cell Biochem 326:87–95. [DOI] [PubMed] [Google Scholar]
- Villeneuve J, Bassaganyas L, Lepreux S, Chiritoiu M, Costet P, Ripoche J, Malhotra V, Schekman R (2018) Unconventional secretion of FABP4 by endosomes and secretory lysosomes. J Cell Biol 217:649–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel S, Mendelsohn CL, Mertz JR, Piantedosi R, Waldburger C, Gottesman ME, Blaner WS (2001) Characterization of a new member of the fatty acid-binding protein family that binds all-trans-retinol. J Biol Chem 276:1353–1360. [DOI] [PubMed] [Google Scholar]
- Vogler A (2015) Fluorescence of retinoic acid in the presence of metal salts. Inorg Chem Commun 57:69–71. [Google Scholar]
- Vork MM, Glatz JFC, Surtel DAM, van der Vusse GJ (1990) Assay of the binding of fatty acids by proteins: evaluation of the Lipidex 1000 procedure. Mol Cell Biochem 98:111–117. [DOI] [PubMed] [Google Scholar]
- Wang G, Bonkovsky HL, de Lemos A, Burczynski FJ (2015) Recent insights into the biological functions of liver fatty acid binding protein 1. J Lipid Res 56:2238–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li Y, Yan H (1997) Structure-function relationships of cellular retinoic acid-binding proteins. Quantitative analysis of the ligand binding properties of the wild-type proteins and site-directed mutants. J Biol Chem 272:1541–1547. [DOI] [PubMed] [Google Scholar]
- Wang Y, Law W-K, Hu J-S, Lin H-Q, Ip T-M, Wan DC-C (2014) Discovery of FDA-approved drugs as inhibitors of fatty acid binding protein 4 using molecular docking screening. J Chem Inf Model 54:3046–3050. [DOI] [PubMed] [Google Scholar]
- Wang Y-T, Liu C-H, Zhu H-L (2016) Fatty acid binding protein (FABP) inhibitors: a patent review (2012-2015). Expert Opin Ther Pat 26:767–776. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widstrom RL, Norris AW, Spector AA (2001) Binding of cytochrome P450 monooxygenase and lipoxygenase pathway products by heart fatty acid-binding protein. Biochemistry 40:1070–1076. [DOI] [PubMed] [Google Scholar]
- Wolfrum C, Borrmann CM, Börchers T, Spener F (2001) Fatty acids and hypolipidemic drugs regulate peroxisome proliferator-activated receptors α - and γ-mediated gene expression via liver fatty acid binding protein: a signaling path to the nucleus. Proc Natl Acad Sci USA 98:2323–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum C, Buhlmann C, Rolf B, Börchers T, Spener F (1999) Variation of liver-type fatty acid binding protein content in the human hepatoma cell line HepG2 by peroxisome proliferators and antisense RNA affects the rate of fatty acid uptake. Biochim Biophys Acta 1437:194–201. [DOI] [PubMed] [Google Scholar]
- Wootan MG, Bernlohr DA, Storch J (1993) Mechanism of fluorescent fatty acid transfer from adipocyte fatty acid binding protein to membranes. Biochemistry 32:8622–8627. [DOI] [PubMed] [Google Scholar]
- Xueping E, Zhang L, Lu J, Tso P, Blaner WS, Levin MS, Li E (2002) Increased neonatal mortality in mice lacking cellular retinol-binding protein II. J Biol Chem 277:36617–36623. [DOI] [PubMed] [Google Scholar]
- Yabut KCB, Isoherranen N (2022) CRABPs alter all-trans-retinoic acid metabolism by CYP26A1 via protein-protein interactions. Nutrients 14:1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Levi L, Casadesus G, Kunos G, Noy N (2014) Fatty acid-binding protein 5 (FABP5) regulates cognitive function both by decreasing anandamide levels and by activating the nuclear receptor peroxisome proliferator-activated receptor β/δ (PPARβ/δ) in the brain. J Biol Chem 289:12748–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Ren Y, Lu R, Yang L, Nie G (2020) Tracing the evolution of fatty acid-binding proteins (FABPs) in organisms with a heterogeneous fat distribution. FEBS Open Bio 10:861–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong G, Ortiz D, Zelter A, Nath A, Isoherranen N (2018) CYP26C1 Is a hydroxylase of multiple active retinoids and interacts with cellular retinoic acid binding proteins. Mol Pharmacol 93:489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]