Abstract
Mordellistena A. Costa, 1854, the most species-rich genus of tumbling flower beetles comprises more than 800 species worldwide and more than 150 reported from Europe. Here, a new species Mordellistena(s. str.)platypoda is described from the island of Ischia in Italy. The species hypothesis is based primarily on morphological characters which are visualised using scanning electron microscopy images, high-resolution photographs, and drawings. The species hypothesis is supported by analysis of a 658 bp fragment of cytochrome c oxidase subunit I (COI). Divergences in the COI gene are evaluated using maximum likelihood and Bayesian inference analyses. The species delimitation is assessed using Assemble Species by Automatic Partitioning (ASAP) and Poisson Tree Processes (PTP) methods. Genetic distances are visualised using multidimensional scaling. Mordellistenaplatypoda Selnekovič, Goffová & Kodada, sp. nov. is recovered as a well-separated species by both molecular and morphological analyses. Our results show that M.platypoda Selnekovič, Goffová & Kodada, sp. nov. is most closely related to M.tarsata Mulsant, 1856, although the two species differ significantly in vestiture colouration, presence of lateral ctenidia on the third metatarsomere, and presence of sexual dimorphism on the protibia. The results indicate that such morphological differences, which were traditionally used to distinguish between species groups, may in fact be present between closely related species. Interestingly, examination of the numerous museum material did not reveal additional specimens of the new species, and therefore M.platypoda Selnekovič, Goffová & Kodada, sp. nov. is currently known only from the Italian island of Ischia.
Keywords: Cytochrome c oxidase subunit I (COI), DNA barcoding, integrative taxonomy, morphology, species delimitation
Introduction
More than 800 species are currently classified within Mordellistena A. Costa, 1854, making it the most species-rich genus of tumbling flower beetles. It is reported from every continent except Antarctica. However, the generic placement of many species, especially Indomalayan and Neotropical, needs to be reassessed according to the current generic classification. In Europe, the genus is represented by more than 150 species (Horák 2020), which are pollinivorous in the adult stage and are among the most frequently encountered flower-visiting beetles. Larval stages are documented in approximately 18 European species (ca. 12%) and develop in the stems of herbaceous plants, feeding on the plant tissue (e.g., Borowiec 1996; Odnosum 2010; Zemoglyadchuk and Buialska 2020). The genus was generally characterised in terms of morphology by Ermisch (1950) and Franciscolo (1957). Identification of the European species is based on the concept of the species groups defined by combinations of morphological characters (Ermisch 1956, 1977). Due to the homogeneity in the external morphology, species identification is often challenging and is possible only by examination of the genitalia and comparison with the type specimens. Essential works covering the European representatives of the genus, including identification keys and figures of diagnostic characters, were provided by Costa (1854), Mulsant (1856), Schilsky (1894, 1895, 1898), Ermisch (1956, 1963, 1969, 1977), Batten (1977), and Horák (1983, 1990, 1996). The comprehensive catalogue of Palaearctic fauna was provided by Horák (2008, 2020). The phylogeny of the genus has not yet been studied.
A recent collecting trip to the island of Ischia in Italy in June 2019 yielded more than 1,000 Mordellidae specimens representing 12 species. Within this material, we recognised a series of 52 specimens belonging to the new species described herein, Mordellistenaplatypoda Selnekovič, Goffová & Kodada, sp. nov. (Fig. 1). The aim of the present study was to describe the new species and delimit the species boundaries. To achieve this goal through an integrative approach, we tested the morphology-based species hypothesis with the results of single-gene DNA analyses. We documented the morphological characters using scanning electron microscopy, high-resolution photographs, and line drawings, and compared the new species with morphologically similar congeners. In order to analyse divergences in the mitochondrial gene that encodes cytochrome c oxidase subunit I (COI), we applied two probabilistic statistical methods (maximum likelihood and Bayesian inference) and compared the results with distance-based (Assemble Species by Automatic Partitioning) and tree-based (Poison Tree Processes) species delimitation analyses.
Figure 1.
Mordellistenaplatypoda Selnekovič, Goffová & Kodada, sp. nov. A male paratype B female paratype. Scale bar: 1.0 mm.
Materials and methods
This study is based on the examination of more than 400 specimens of the genus Mordellistena deposited at the following institutions:
HNHMMagyar Természettudományi Múzeum, Budapest, Hungary;
MNHU Museum für Naturkunde der Humboldt Universität, Berlin, Germany;
MSNGMuseo di Storia Naturale Giacomo Doria, Genova, Italy;
MZFNMuseo Zoologico dell’Università Federico II, Naples, Italy;
SDEISenckenberg Deutsches Entomologisches Institut, Müncheberg, Germany;
SNSD Senckenberg Naturhistorische Sammlungen, Dresden, Germany.
The type series of M.platypoda consist of 52 specimens collected on the island of Ischia in Italy. The specimens of the additional species included in the phylogenetic analyses were acquired from several localities in Italy, Slovakia, Spain, Cyprus, and Bulgaria (Table 1). Type specimens of M.platypoda were compared with those of the following morphologically similar species: M.austriaca Schilsky, 1898 (male lectotype and two paralectotypes, MNHU), M.balianii Franciscolo, 1942 (male holotype and two paratypes, MSNG), M.hirtipes Schilsky, 1895 (male lectotype and 35 paralectotypes, MNHU), M.micans (Germar, 1817) (two female syntypes, SDEI), M.pseudohirtipes Ermisch, 1965 (male holotype and one paratype, SNSD), M.purpurascens A. Costa, 1854 (male lectotype, MZFN), and M.tenuicornis Schilsky, 1898 (male lectotype and 17 paralectotypes, MNHU).
Table 1.
List of specimens used in DNA analyses.
| Specimen ID | GenBank | Locality | Coordinates |
|---|---|---|---|
| Mordellistenaaustriaca DSBS 60 | OM680976 | Slovakia, Virt env. | 47.760000°N, 18.340556°E |
| Mordellistenaconfinis DSBS 243 | OP586774 | Italy, Firenze, Cinipetta | 43.570000°N, 11.420556°E |
| Mordellistenaconfinis DSBS 244 | OP586776 | Italy, Firenze, Cinipetta | 43.570000°N, 11.420556°E |
| Mordellistenaconfinis DSBS 245 | OP586769 | Italy, Firenze, Cinipetta | 43.570000°N, 11.420556°E |
| Mordellistenaconfinis DSBS 250 | OP586770 | Italy, Firenze, Cinipetta | 43.570000°N, 11.420556°E |
| Mordellistenaconfinis DSBS 329 | OP586775 | Italy, Firenze, Cinipetta | 43.570000°N, 11.420556°E |
| Mordellistenahirtipes DSBS 205 | OM681007 | Cyprus, Limassol env. | 34.755278°N, 33.093333°E |
| Mordellistenahirtipes DSBS 207 | OM681008 | Cyprus, Limassol env. | 34.755278°N, 33.093333°E |
| Mordellistenahirtipes DSBS 208 | OM681009 | Cyprus, Limassol env. | 34.755278°N, 33.093333°E |
| Mordellistenalindbergi DSBS 144 | OM680979 | Cyprus, Limassol env. | 34.755278°N, 33.093333°E |
| Mordellistenalindbergi DSBS 270 | OP586772 | Cyprus, Akamas | 35.057586°N, 32.345527°E |
| Mordellistenalindbergi DSBS 271 | OP586767 | Cyprus, Akamas | 35.057586°N, 32.345527°E |
| Mordellistenalindbergi DSBS 280 | OP586771 | Cyprus, Foinikaria env. | 34.766272°N, 33.100258°E |
| Mordellistenaminima DSBS 79 | MT232550 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenaminima DSBS 172 | OM680982 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenaminima DSBS 173 | OM680983 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenaminima DSBS 215 | OP586765 | Italy, Sardinia, Castiadas | 39.206812°N, 9.562685°E |
| Mordellistenaminima DSBS 218 | OP586768 | Italy, Sardinia, Castiadas | 39.206812°N, 9.562685°E |
| Mordellistenaminima DSBS 219 | OP586766 | Italy, Sardinia, Castiadas | 39.206812°N, 9.562685°E |
| Mordellistenaplatypoda DSBS 83 | OM680977 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenaplatypoda DSBS 118 | OM680978 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenaplatypoda DSBS 194 | OM680997 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenaplatypoda DSBS 195 | OM680998 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenaplatypoda DSBS 199 | OM681002 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenaplatypoda DSBS 233 | OM681019 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenaplatypoda DSBS 235 | OM681020 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenaplatypoda DSBS 237 | OM681021 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenapseudorhenana DSBS 11 | OP586773 | Cyprus, Foinikaria env. | 34.755278°N, 33.093333°E |
| Mordellistenapseudorhenana DSBS 12 | MT232533 | Cyprus, Foinikaria env. | 34.755278°N, 33.093333°E |
| Mordellistenapseudorhenana DSBS 43 | MT232539 | Slovakia, Chotín env. | 47.806389°N, 18.198056°E |
| Mordellistenapurpurascens DSBS 82 | MT232552 | Italy, Ischia, Serrara env. | 40.721389°N, 13.883056°E |
| Mordellistenapurpurascens DSBS 111 | MT232554 | Spain, Tossa de Mar | 41.736667°N, 2.935000°W |
| Mordellistenapurpurascens DSBS 117 | MT232555 | Italy, Ischia, Serrara env. | 40.721389°N, 13.883056°E |
| Mordellistenapurpurascens DSBS 182 | OM680985 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenapurpurascens DSBS 183 | OM680986 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenapurpurascens DSBS 186 | OM680989 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenapurpurascens DSBS 187 | OM680990 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenapurpurascens DSBS 188 | OM680991 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenapurpurascens DSBS 192 | OM680995 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenapurpurascens DSBS 200 | OM681003 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenapurpurascens DSBS 201 | OM681004 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenapurpurascens DSBS 226 | OM681016 | Italy, Sardinia, Castiadas env. | 39.206812°N, 9.562685°E |
| Mordellistenapurpurascens DSBS 227 | OM681017 | Italy, Sardinia, Castiadas env. | 39.206812°N, 9.562685°E |
| Mordellistenapurpurascens DSBS 231 | OM681018 | Italy, Ischia, Serrara env. | 40.716667°N, 13.886389°E |
| Mordellistenapurpureonigrans DSBS 49 | OP575375 | Slovakia, Chotín env. | 47.806389°N, 18.198056°E |
| Mordellistenapurpureonigrans DSBS 55 | OM680974 | Slovakia, Virt env. | 47.760000°N, 18.340556°E |
| Mordellistenapurpureonigrans DSBS 56 | OM680975 | Slovakia, Virt env. | 47.760000°N, 18.340556°E |
| Mordellistenapurpureonigrans DSBS 71 | OP575374 | Slovakia, Iža env. | 47.748056°N, 18.260556°E |
| Mordellistenapurpureonigrans DSBS 73 | OP575376 | Slovakia, Iža env. | 47.748056°N, 18.260556°E |
| Mordellistenatarsata DSBS 39 | OM680971 | Slovakia, Virt env. | 47.760000°N, 18.340556°E |
| Mordellistenatarsata DSBS 41 | OM680972 | Slovakia, Virt env. | 47.760000°N, 18.340556°E |
| Mordellistenatarsata DSBS 42 | OM680973 | Slovakia, Virt env. | 47.760000°N, 18.340556°E |
| Mordellistenatarsata DSBS 209 | OM681010 | Cyprus, Skoulli env. | 34.968056°N, 32.446111°E |
| Mordellistenatarsata DSBS 210 | OM681011 | Bulgaria, Melnik env. | 41.510000°N, 23.378333°E |
| Mordellistenatarsata DSBS 211 | OM681012 | Bulgaria, Melnik env. | 41.510000°N, 23.378333°E |
| Mordellochroaabdominalis DSBS 138 | OM681022 | Slovakia, Burda | 47.847778°N, 18.789722°E |
The specimens selected for DNA isolation were killed in 96.3% ethanol and stored at -20 °C. The remaining specimens were killed with the fumes of ethyl acetate. After DNA isolation, the specimens were soaked in 5% acetic acid, dissected, and mounted on cards. The dissected genitalia were cleared in lactic acid for several days, or in 10% KOH overnight, then dehydrated in 96.3% ethanol and mounted on slides in Euparal (Paradox Co., Cracow, Poland). After examination, genitalia were mounted on the card with the respective specimen using dimethyl hydantoin formaldehyde (Entomopraxis, Barcelona, Spain). Specimens were observed under an MZ16 stereomicroscope (Leica, Wetzlar, Germany) with magnification up to 120× with diffused LED light (6400 K). The drawings were prepared using a drawing tube attached to a DM1000 compound microscope (Leica, Wetzlar, Germany) and subsequently inked with the Isograph technical pens (Rotring, Hamburg, Germany). Photographs of habitus were taken with an EOS 5D mark II camera (Canon, Tokyo, Japan) attached to an Axio Zoom V16 stereoscope (Zeiss, Oberkochen, Germany); photographs of genitalia were taken with an Axio Imager 2 (Zeiss, Oberkochen, Germany). The images were stacked in Zerene Stacker 1.4 software (https://zerenesystems.com/cms/stacker) and edited in Adobe Photoshop CC (https://www.adobe.com/products/photoshop.html) and DxO Photolab 5 (https://www.dxo.com/dxo-photolab/). For scanning electron microscopy, the body parts were disarticulated, cleaned in lactic acid for several days, dehydrated in 96.3% ethanol, coated with 25 nm thick gold layer, and examined using Quanta 250 FEG (FEI Europe B.V., Eindhoven, The Netherlands). Measurements were made with an ocular micrometre in a MZ16 stereomicroscope (Leica, Wetzlar, Germany) and are given in the text as the range followed by the arithmetic mean and standard deviation enclosed in parentheses. The measured characters are abbreviated in the text as follows:
EL elytral length from scutellar apex to elytral apices along suture;
EW maximum elytral width;
HL head length from anterior clypeal margin to occipital carina along midline;
HW maximum head width;
LPrL maximum left paramere length;
PL pronotal length along midline;
PW maximum pronotal width;
PygL maximum pygidial length;
RPrL maximum right paramere length;
TL combination of head, pronotal and elytral lengths.
The species description follows the conventional terminology as used in Franciscolo (1957), Lu et al. (1997), and Lawrence and Ślipiński (2010). Terminology for sensilla types follows Snodgrass (1935). All nomenclatorial acts follow the regulations of the International Code of Zoological Nomenclature (International Trust of Zoological Nomenclature 1999).
In total, 56 specimens were used for DNA isolation. Details on the voucher specimens, including sampling localities and GenBank accession numbers, are presented in Table 1. DNA was isolated from the entire specimens using the E.Z.N.A. Tissue DNA kit (OMEGA Biotek Inc., Norcross, GA, USA) according to the manufacturer’s protocol. The COI gene was amplified by PCR using standard primers LCO1490 and HCO2198 (Folmer et al. 1994). Individual PCR reactions were carried out in a total volume of 20 μl and included 10 μl of GoTaq Green master mix (Promega, Fitchburg, WI, USA), 0.52 μl of each primer (10 pmol/μl), 5 μl of extracted DNA, and 3.96 μl of nuclease-free water. The PCR thermocycler program was as follows: 94 °C for 120 sec, 40 cycles of 94 °C for 40 sec, 52 °C for 40 sec, 72 °C for 60 sec, and 72 °C for 10 min. PCR products were purified using EPPiC Fast (A&A Biotechnology, Gdansk, Poland) and sequenced from both sides in Macrogen Europe B.V. (Amsterdam, The Netherlands).
The consensus sequences, alignment, and final matrix were produced in Unipro UGENE 44.0 software (http://ugene.net/). Pairwise p-distances were calculated using MEGA X (Kumar et al. 2018). Maximum likelihood analysis (ML) was performed using IQ-TREE (Nguyen et al. 2015) on the web server (http://iqtree.cibiv.univie.ac.at) with the best substitution model (TIM2+F+I+G4) identified by the in-built ModelFinder according to BIC criterion. The node support values were obtained from 10,000 ultrafast bootstrap replicates (Hoang et al. 2017) and tested by the SH-aLRT branch test (Guindon et al. 2010). Bayesian inference was carried out in MrBayes 3.2.7a (Ronquist et al. 2012) on XSEDE available in CIPRES Science Gateway (https://www.phylo.org/index.php/). The number of substitution types was set to six, the nucleotide substitution model set to 4 × 4, and the among-site variation rate set to invgamma. Markov Chain Monte Carlo (MCMC) simulations included two independent runs each with four simultaneous chains, ten million generations, a sampling frequency of trees and parameters set to 1,000, and a burn-in fraction of 25%. The convergence of the MCMC analyses and an adequate sample size from the posterior distribution were confirmed using the in-built MrBayes diagnostics. Both trees were rooted subsequently in FigTree 1.4 (http://tree.bio.ed.ac.uk/software/figtree/), with Mordellochroaabdominalis (Fabricius, 1775) selected as outgroup. A distance-based species delimitation analysis, Assemble Species by Automatic Partitioning (ASAP) (Puillandre et al. 2020), was run on the ASAP web server (https://bioinfo.mnhn.fr/abi/public/asap/) using uncorrected p-distances. A tree-based species delimitation analysis, Poisson Tree Processes (PTP) (Zhang et al. 2013), was run on the bPTP web server (https://species.h-its.org) with the ML tree used as an input, in 100,000 MCMC generations with 25% burn-in fraction. The distribution of intraspecific, interspecific, and intergeneric distances was calculated from uncorrected p-distances using an Automatic Barcode Gap Discovery (ABGD) analysis (Puillandre et al. 2012) carried out on the ABGD web interface (https://bioinfo.mnhn.fr/abi/public/abgd/) in 20 steps, with Pmin = 0.001, Pmax = 0.1, and relative gap width = 1.5. Multidimensional scaling (MDS) of uncorrected p-distances was performed in IBM SPSS Statistics software (https://www.ibm.com/products/spss-statistics).
Results
COI gene analyses
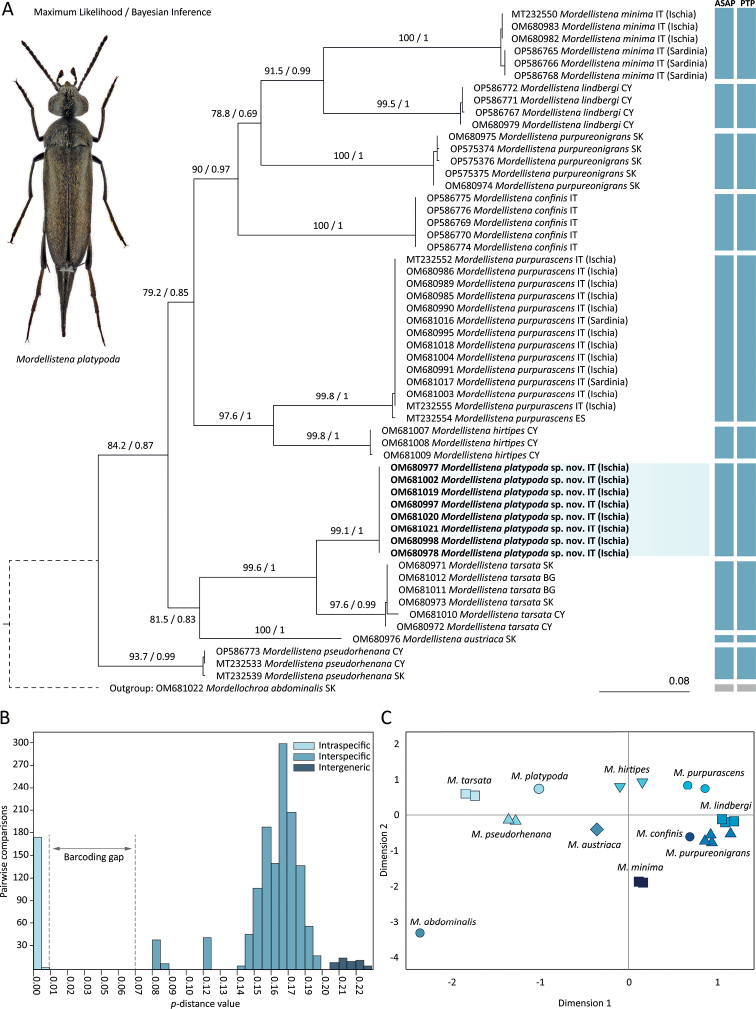
We generated and analysed a set of 56 sequences of the 658 bp COI gene fragment. The dataset comprised 237 parsimony-informative sites (36%) and 22 singleton sites (3.34%). We recovered almost identical topologies with both maximum likelihood (ML) and Bayesian inference (BI) methods, and therefore only the ML tree is presented with ML bootstrap values and BI posterior probability values (Fig. 2). ML and BI analyses revealed ten well-separated monophyletic clades (excluding outgroup) with high bootstrap (> 93) and posterior probability (> 0.99) values. Each of these clades represents a single species, initially recognised based on morphological characters. Mordellistenaplatypoda sp. nov. and M.tarsata Mulsant, 1856 were grouped in one clade with high statistical support (99.6/1.00). Another well-supported clade with high statistical support (97.6/1.00) consists of M.purpurascens and M.hirtipes. The M.micans species group which is defined by combination of morphological characters (represented here by M.austriaca, M.hirtipes, M.minima A. Costa, 1854, M.platypoda, M.pseudorhenana Ermisch, 1977, and M.purpurascens) was recovered as polyphyletic, with members of the M.tarsata species group (M.tarsata) and the M.pumila species group (M.purpureonigrans) placed within. ASAP and PTP delimitation methods recovered ten species (excluding outgroup) in congruence with morphology-based delimitation, ML, and BI analyses (Fig. 2). The lowest ASAP score was 1.5.
Figure 2.
Analyses of a 658 bp fragment of the cytochrome c oxidase subunit I gene (COI) in selected Mordellistena A. Costa, 1854 species A combined results of maximum likelihood (ML) and Bayesian inference (BI) analyses; ML bootstrap values and BI posterior probabilities are shown on ML tree. The vertical bars represent results of Assemble Species by Automatic Partitioning (ASAP) and Poisson Tree Processes (PTP) species delimitation analyses B distribution of uncorrected p-distances C multidimensional scaling of uncorrected p-distances.
The uncorrected p-distances within and between species are summarised in Appendix 1. The lowest interspecific distance within the entire data set is between M.platypoda sp. nov. (represented by a single haplotype) and M.tarsata with a range of 8.21% to 8.97%. The highest interspecific distance within the ingroup is observed between M.lindbergi and M.tarsata (19.15–19.76%). The highest intraspecific distance (1.06%) is observed in M.tarsata. The distribution of p-distances (Fig. 2) shows a distinct gap between the highest intraspecific distance (1.06%) and the lowest interspecific distance (8.21%). The multidimensional scaling of p-distances shows separate clusters for each species (Fig. 2).
Taxonomy
. Mordellistena (s. str.) platypoda
Selnekovič, Goffová & Kodada sp. nov.
993DF123-594F-5DAC-B2C6-ECEA90EF7503
https://zoobank.org/A04D9CDE-AD8B-4DC0-966D-6B72082BE9AE
Figure 7.
Habitats of Mordellistenaplatypoda Selnekovič, Goffová & Kodada, sp. nov. near Serrara village, Ischia, Italy A, B type locality, slopes with Mediterranean grassland communities (40.72138°N, 13.88305°E) C, D ruderal communities with Daucuscarota Linnaeus along road (40.71666°N, 13.88638°E).
Type locality.
Italy, Ischia, Serrara env., 40.72138°N, 13.88305°E; steep slopes with grassland communities, ca. 550 m alt. (Fig. 7).
Type material.
Holotype: italy • male; Ischia, Serrara env.; 40.72138°N, 13.88305°E; ca. 550 m alt.; 30 Jun 2019; D. Selnekovič leg.; steep slopes with grassland communities, on inflorescences of Apiaceae; GenBank: OM680978; SNSD. Paratypes: italy • 9 males, 8 females; same data as for holotype • 23 males, 11 females; Ischia, Serrara env.; 40.71666°N, 13.88638°E; 517 m alt.; 29 Jun 2019; D. Selnekovič leg.; ruderal habitat on road verge, on inflorescences of Daucuscarota; GenBank: OM680977, OM680997, OM680998, OM681002, OM681019 to OM681021; SNSD.
Differential diagnosis.
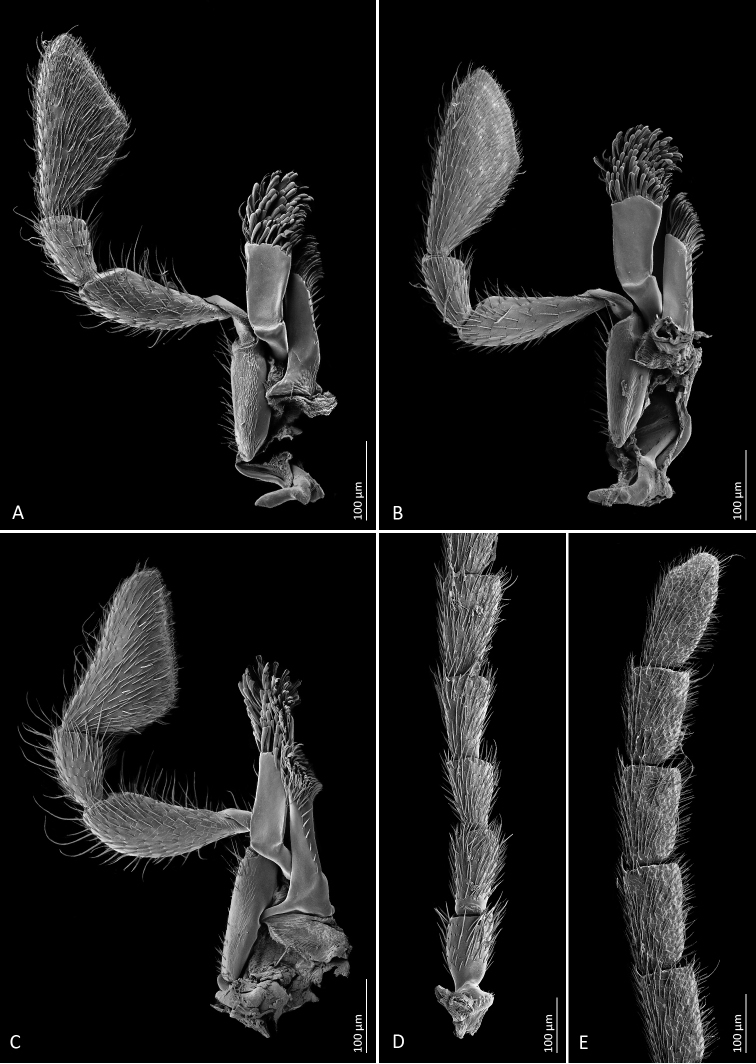
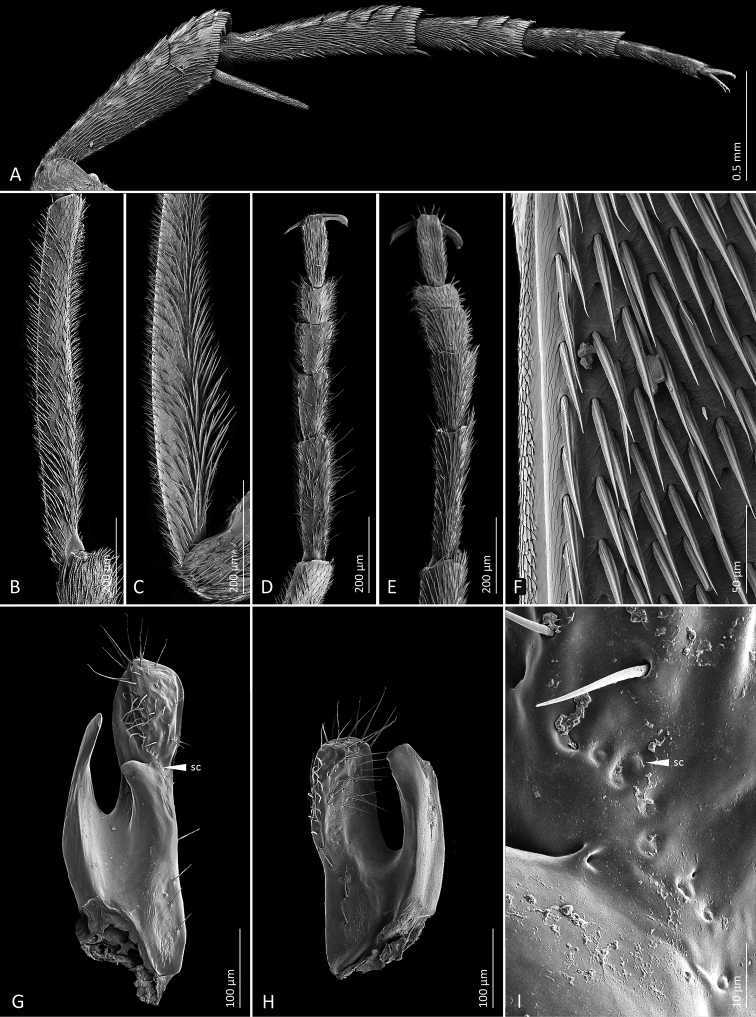
Mordellistenaplatypoda is included in the M.micans species group as defined by Batten (1977) on the basis of the following characters: the four first antennomeres are narrower and shorter than the following ones (Fig. 3D); hind tibia besides the subapical ctenidium with four lateral ctenidia that are more or less perpendicular to the dorsal edge of the tibia (Fig. 4A); only the first two metatarsomeres with lateral ctenidia (Fig. 4A); punctation of elytra not conspicuously coarse; metatibial spurs black. A rather unique character that separates the species from most of its congeners is the shape of pro- and mesotarsomeres, which are flattened and expanded. The entire male mesotarsus and the female pro- and mesotarsus are dilated apically (Figs 4D, E, 5A, B). Similarly formed tarsi are present in M.rugipennis Schilsky, 1895 and M.latitarsis Batten, 1983, both of which differ from M.platypoda in the entirely black vestiture and shape of the genitalia. Furthermore, M.platypoda is characterised by its large body dimensions (TL: ♂♂ 4.58–5.64 mm, ♀♀ 4.84–6.02 mm). Similarly large body is present within the M.micans species group only in M.purpurascens A. Costa, 1854 (TL: ♂♂ 4.27–5.76 mm, ♀♀ 4.32–5.78 mm), M.hirtipes Schilsky, 1895 (TL: ♂♂ 4.13–5.16 mm, ♀♀ 3.69–4.77 mm), and M.austriaca Schilsky, 1898 (TL: ♂♂ 3.85–5.37 mm, ♀♀ 3.67–5.33 mm). The latter species is separated from M.platypoda by almost square antennomeres 5–10, the lateral pronotal sides straight in the lateral aspect, the posterolateral pronotal angels obtuse, the elytra shorter (EL/EW ratio ≤ 2.2), and the genitalia differently shaped.
Figure 3.
Scanning electron microscope images of diagnostic characters AMordellistenaplatypoda Selnekovič, Goffová & Kodada, sp. nov., male maxilla BM.platypoda, female maxilla CM.purpurascens A. Costa, 1854, male maxilla DM.platypoda, male antenna (basal part) EM.platypoda, male antenna (apical part).
Figure 4.
Scanning electron microscope images of diagnostic characters AMordellistenaplatypoda Selnekovič, Goffová & Kodada, sp. nov., male hind leg, lateral view BM.platypoda, male protibia CM.purpurascens A. Costa, 1854, male protibia DM.platypoda, male protarsus EM.platypoda, female protarsus FM.platypoda, mesal portion of male right elytron GM.platypoda, left paramere, mesal view, white triangle points to a cluster of sensilla campaniformia shown in image I; HM.platypoda, right paramere, mesal view IM.platypoda, cluster of sensilla campaniformia on left paramere. Abbreviation: sc = sensillum campaniformium.
Figure 5.
AMordellistenaplatypoda Selnekovič, Goffová & Kodada, sp. nov., male protarsus BM.platypoda, female protarsus CM.purpurascens A. Costa, 1854, male protarsus DM.platypoda, phallobase EM.platypoda male sternite VIII FM.platypoda, male tergite VIII GM.platypoda, male sternite IX HM.platypoda, male tergites IX and X IM.platypoda, female sternite VIII JM.platypoda, female tergite VIII KM.platypoda, penis. Scale bars: 0.1 mm.
Mordellistenaplatypoda most closely resembles a sympatric species M.purpurascens but differs in paler vestiture, weakly expanded male second maxillary palpomere, with few long setae (Fig. 3A) compared to strongly expanded, with numerous long setae in M.purpurascens (Fig. 3C), weakly expanded male protibia, with few inconspicuous extended setae (Fig. 4B) compared to strongly expanded, with conspicuous long setae in M.purpurascens (Fig. 4C), protarsus expanded in both sexes and distinctly dilated apically in females (Fig. 5A, B) compared to weakly narrowed apically in M.purpurascens (Fig. 5C), elytra longer, with EL/EW ratio: ♂♂ 2.31–2.64, ♀♀ 2.22–2.40 compared to ♂♂ 1.97–2.23, ♀♀ 1.83–2.15 in M.purpurascens, and parameres distinctly shorter and differently shaped (Figs 4G, H, 5B; EL/LPrL ratio: 7.09–8.59, EL/RPrL ratio: 9.67–12.08) than in M.purpurascens (EL/LPrL ratio: 4.42–5.84, EL/RPrL ratio: 5.57–6.94; see Selnekovič and Kodada 2019: fig. 7). Mordellistenatenuicornis Schilsky, 1899 is separated from M.platypoda by the presence of two or three lateral ctenidia on the third metatarsomere, antennomeres 5–10 ca. 2.0× longer than wide, and pygidium distinctly longer and more slender.
The results of the COI gene analyses show the close relationship of M.platypoda with M.tarsata Mulsant, 1856, with p-distances of 8.21–8.97%. The two species are easily distinguished by the colouration of the dorsal vestiture, which is black with a greenish lustre in M.tarsata compared to yellowish in M.platypoda (Fig. 1), the presence of two lateral ctenidia on the third metatarsomere in M.tarsata compared to the absence of lateral ctenidia on the segment in M.platypoda, the presence of a conspicuous cluster of long setae in the proximal portion of the male protibia in M.tarsata compared to few elongated setae in in M.platypoda, and the different shape of the parameres.
Description.
Measurements (in mm; ♂♂ n = 33, ♀♀ n = 21): TL: ♂♂ 4.58–5.64 (5.25 ± 0.26), ♀♀ 4.84–6.02 (5.56 ± 0.31); HL: ♂♂ 0.81–1.01 (0.92 ± 0.05), ♀♀ 0.84–1.06 (0.97 ± 0.06); HW: ♂♂ 0.94–1.17 (1.07 ± 0.05), ♀♀ 0.95–1.19 (1.10 ± 0.06); PL: ♂♂ 0.98–1.25 (1.14 ± 0.06), ♀♀ 1.06–1.33 (1.22 ± 0.08); PW: ♂♂ 1.10–1.46 (1.32 ± 0.08), ♀♀ 1.19–1.57 (1.42 ± 0.10); EL: ♂♂ 2.75–3.50 (3.18 ± 0.18), ♀♀ 2.92–3.67 (3.37 ± 0.19); EW: ♂♂ 1.13–1.42 (1.31 ± 0.07), ♀♀ 1.25–1.54 (1.45 ± 0.08); RPrL: 0.27–0.32 (0.29–0.01); LPrL: 0.36–0.43 (0.40 ± 0.02).
Body elongated, wedge-shaped, widest in anterior half of elytra (Fig. 1). Dorsum slightly convex, venter strongly so. Entire body surface uniformly black, except for reddish brown anteclypeus. Vestiture consisting of decumbent lanceolate setae (Fig. 4F); yellowish, darkened before elytral apices and along posterior margin of ventrites 4 and 5.
Head large, transverse, moderately convex dorsally, with highest point behind middle of eyes (lateral aspect), HW/HL ratio: ♂♂ 1.09–1.23 (1.16 ± 0.03), ♀♀1.01–1.23 (1.13 ± 0.05); occipital carina convex; integument weakly microreticulate, weakly iridescent, with small round setiferous punctures. Eyes broadly oval, vertical diameter ca. 1.3× horizontal diameter; posteriorly reaching to occipital margin; finely faceted; interfacetal setae longer than facet diameter. Anterior clypeal edge weakly convex. Labrum transverse, densely setose, anterior edge weakly convex. Antenna weakly serrate (Fig. 3D, E); antennomeres 1–4 shorter and narrower than following segments; scape cylindrical, little longer than wide; pedicel cylindrical, little longer than scape; antennomere 3 little longer than wide, expanded distally; antennomere 4 little longer than 3; antennomeres 5–10 in both sexes 1.6–1.7× as long as wide; antennomere 11 oval, ca. 2.2× as long as wide; all antennomeres covered with decumbent sensilla chaetica; antennomeres 5–10 each with several long and erect sensilla trichoidea apico-laterally (Fig. 3D, E). Galea short, with spatulate sensilla and sensilla chaetica apically (Fig. 3A, B). Lacinia with numerous sensilla chaetica apically and row of sensilla chaetica along mesal margin (Fig. 3A, B). Maxillary palpomere 2 subcylindrical, weakly expanded distally, in males little wider and with somewhat longer sensilla chaetica than in females (Fig. 3A, B); terminal maxillary palpomere scalene triangular, mesal angle in middle or in anterior half, numerous decumbent sensilla chaetica and several erect sensilla chaetica over entire surface, several sensilla placoidea before distal margin.
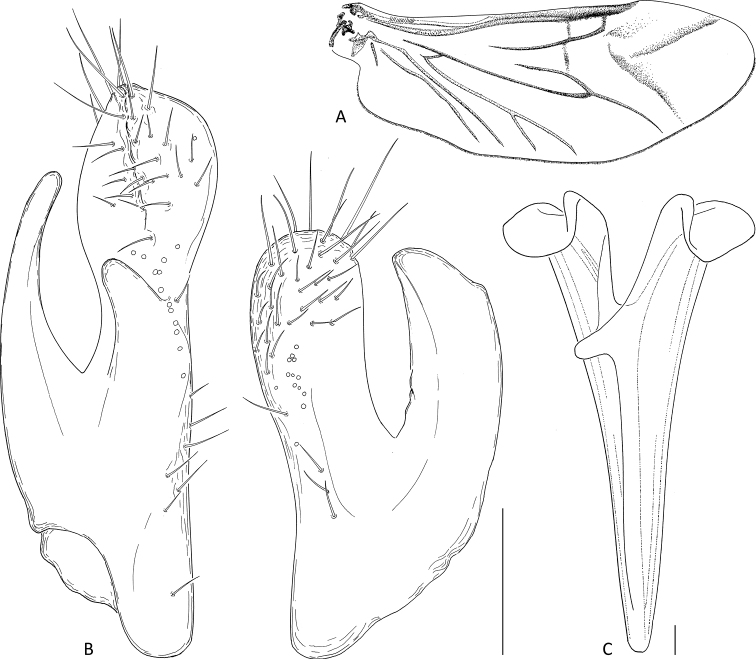
Pronotum 1.1–1.2× as wide as long, widest behind middle, moderately convex; surface microreticulate, densely covered with lanceolate setae, punctures larger than those on head; anterior edge convex in middle, anterolateral angles broadly rounded; lateral carinae sinuate in lateral aspect; posterior edge sinuate, posterolateral angles rectangular in lateral aspect. Scutellar shield triangular, densely setose. Elytra widest between first and second quarter, EL/EW ratio: ♂♂ 2.31–2.64 (2.43 ± 0.07), ♀♀ 2.22–2.40 (2.32 ± 0.05); apices separately rounded; surface microreticulate, densely covered with decumbent lanceolate setae, punctures coarser than those on pronotum. Hindwing as in Fig. 6A. Mesoventral process about as wide as mesotibia. Metaventral discrimen apparent. Metanepisternum ca. 2.3× longer than greatest width, narrowed posteriorly. Metendosternite as in Fig. 6C. Protibia ca. 1.1× longer than protarsus; in males weakly expanded proximally and with few inconspicuous extended setae (Fig. 4B). Protarsus expanded and flat in both sexes, weakly dilated distally in females (Figs 4D, E, 5A, B); protarsomere 1 little shorter than three following tarsomeres combined; penultimate protarsomere weakly expanded distally, with apical edge concave; each protarsal claw with four denticles. Mesotibia ca. 0.8× as long as mesotarsus. Mesotarsus dilated distally in both sexes; first mesotarsomere nearly as long as three subsequent tarsomeres combined. Metatibia with short subapical ctenidium and four lateral ctenidia nearly perpendicular to dorsal tibial edge, proximal ctenidium often rudimentary (Fig. 4A); outer terminal spur ca. 0.75× as long as inner one. Metatarsomere 1 with four or five lateral ctenidia; metatarsomere 2 with two lateral ctenidia; metatarsomeres 3 and 4 without lateral ctenidia (Fig. 4A).
Figure 6.
Mordellistenaplatypoda Selnekovič, Goffová & Kodada, sp. nov., paratype A male right hindwing B parameres C metendosternite. Scale bars: 0.1 mm.
Abdominal ventrite 1 longer than ventrite 2; ventrite 5 with arcuate apical edge. Pygidium long, conical, narrowly truncate at apex, EL/PygL ratio: ♂♂ 1.75–2.03 (1.87 ± 0.06), ♀♀ 2.12–2.35 (2.21 ± 0.06). Male tergite VIII deeply emarginate on posterior edge, setose apically (Fig. 6F); female tergite VIII divided by longitudinal suture basally (Fig. 5J), setose apically. Male sternite VIII strongly produced in middle of posterior edge, with long setae (Fig. 5E); female sternite VIII produced in middle of posterior edge, setose (Fig. 5H), anterior median strut short, narrowly elliptical. Male tergite IX completely divided into two parts, each with narrow basal projection (Fig. 5H). Male sternite IX narrow, strongly sclerotised at lateral edges, expanded before apex, with several sensilla trichoidea (Fig. 5G). Male tergite X divided into two parts, partly fused to tergite IX (Fig. 5H). Phallobase forming sheath around penis; tubular part short; anterior struts ca. 3.1× as long as tubular part; dorsal apodeme strongly sclerotised, lateral edges even (Fig. 5D). Parameres as in Figs 4G, H, 6B: left paramere longer than right one, EL/LPrL ratio: 7.09–8.59 (7.97 ± 0.41), dorsal process moderately dilated and obliquely truncate apically, with numerous sensilla trichoidea, ventral process shorter than dorsal one, slightly bent dorsad, narrowly rounded apically, median process short, produced ventrad, cluster of approximately nine sensilla campaniformia present above dorsal edge of median process (Figs 4I, 6B); left paramere with dorsal process subtruncate apically, with trichoid and campaniform sensilla, ventral process slightly shorter than dorsal one, bent dorsad, subtruncate at apex, EL/RPrL ratio: 9.67–12.08 (10.92 ± 0.58). Penis long, narrow, weakly expanded before apex (Fig. 5K). Ovipositor: proctiger moderately long, with sclerotised lateral baculi; paraprocts slightly shorter than gonocoxites, with sclerotised baculi; gonocoxites ventrally divided, setose, with oblique basal baculi; gonostyli attached subapically, each with two trichoid sensilla at apex.
Secondary sexual dimorphism.
Females are on average slightly larger than males. Males are more slender than females (Fig. 1). The second maxillary palpomere has longer setae in males than in females (Fig. 3A, B). Terminal maxillary palpomere is slightly narrower in females (Fig. 3A, B). The male protibia bears several elongate setae in proximal half (Fig. 4B), while the female protibia is uniformly setose. Male protarsomeres bear numerous thick setae oriented mesoventrad (Figs 4D, 5A). Protarsus and mesotarsus are more strongly dilated distally in females (Figs 4D, E, 5A, B).
DNA sequences.
Partial COI gene sequences of holotype and eight paratypes were submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/). The accession numbers are listed in Table 1.
Etymology.
The specific epithet is derived from the Greek words πλατύς (platýs), meaning wide, broad, and πόδι (pódi) meaning foot. It refers to the expanded pro- and mesotarsi, an unusual condition that separates M.platypoda from morphologically similar congeners.
Distribution.
The species is known only from the island of Ischia in Italy.
Collecting notes and habitat.
Mordellistenaplatypoda was sampled in a series of 52 specimens on 29–30 June 2019. The sampling was carried out at two nearby localities (approximately 600 m apart) near Serrara village. The type locality (40.72138°N, 13.88305°E) was characterised by the steep rocky slopes with Mediterranean grassland communities (Fig. 7A, B). The specimens were collected by sweeping the inflorescences of Apiaceae spp. At the second location (40.71666°N, 13.88638°E), the specimens were collected from the inflorescences of Daucuscarota that grew in a ruderal community along the road (Fig. 7C, D). During the same collection events, the following Mordellidae species were also collected: Mediimordabipunctata (Germar, 1823), Mordellaaculeata Linnaeus, 1758, Mordellistenaepisternalis Mulsant, 1856, M.hirtipes Schilsky, 1895, M.minima A. Costa, 1854, M.pseudorhenana Ermisch, 1977, M.purpurascens A. Costa, 1854, M.wiebesi Batten, 1977, and Variimordabasalis (A. Costa, 1854).
Discussion
Mordellistenaplatypoda is morphologically well defined. It is included in the M.micans species group defined by Batten (1977). It should be noted, however, that the results of the molecular analyses presented in this study as well as other preliminary and yet unpublished results show that, despite the similarity in morphological characters shared by its members, the M.micans species group is polyphyletic, and therefore we mention it only marginally in this study to facilitate species identification. The recognition of the new species and the species hypothesis were based primarily on the unique combination of morphological characteristics, e.g., remarkably large body, expanded and dilated pro- and mesotarsus, length of antennal segments, inconspicuous secondary sexual dimorphism in the protibia, and unique shape of the genitalia. Expanded and dilated fore and middle tarsi are present in both sexes, but the condition is more pronounced in females. It is an uncommon character state present, among the European species, only in M.rugipennis Schilsky, 1895 (Horák 1990) and M.latitarsis Batten, 1983. Both species differ from M.platypoda by the black vestiture pubescence on the dorsal surfaces. Another notable characteristic of M.platypoda is its large body that reaches up to 5.6 mm in males and 6.2 mm in females (TL). Comparably large species within the M.micans group are M.purpurascens A. Costa, 1854 and M.hirtipes Schilsky, 1895, which are sympatric with M.platypoda. Both species can be easily distinguished from M.platypoda by different shapes of the protarsus and mesotarsus, shorter antennal segments, distinct sexual dimorphism in the protibia, and different shapes of genitalia (Selnekovič and Kodada 2019).
The morphology-based species hypothesis was evaluated by analysing a 658 bp fragment of the COI gene. This standard DNA barcoding marker has frequently been used in the taxonomy of beetles not only to identify species as originally proposed (Hebert et al. 2003; DeSalle and Goldstein 2019), but also to explore species boundaries and refine morphology-based hypotheses (Pentinsaari et al. 2016; Fossen et al. 2016; Bergsten et al. 2017; Maddison and Sproul 2020). Despite the frequent use of this marker in beetle taxonomy, it has only been used once in the study of Mordellidae to identify species and interpret morphological variability (Selnekovič et al. 2021). Here, we recovered a phylogenetic tree using two probabilistic methods, maximum likelihood (ML) and Bayesian inference (BI). To assess the accuracy of the results, we compared them with those of distance-based (ASAP) and tree-based (PTP) species delimitation methods. ML and BI analyses show well-separated homogenous clades with strong statistical support for each presumed species including M.platypoda (ML bootstrap values higher than 97 and BI posterior probabilities 1.00) (Fig. 2). Mordellistenaplatypoda forms a clade with M.tarsata Mulsant, 1856 with strong statistical support (ML 97, BI 1.00). A close relationship between the two species is also supported by the interspecific divergence of 8.21%–8.87%, which is the lowest within the entire data set. Mordellistenaplatypoda and M.tarsata also have several morphological similarities, e.g., overall body shape, length of antennal segments, and shape of genitalia. However, despite the close genetic relatedness, M.platypoda and M.tarsata can be distinguished easily by the different colouration of the dorsal vestiture, which is black in M.tarsata and yellowish in M.platypoda, and by the absence of lateral ctenidia on the third metatarsomere in M.platypoda. Such characters were traditionally used to differentiate species groups among European representatives of the genus Mordellistena, for example, the M.micans group vs. M.tarsata and M.pumila group. Interestingly, our results show that such differences may, in fact, also be present among closely related species. It should be mentioned that our molecular analyses did not include species such as M.tenuicornis, M.latitarsis, and M.rugipennis, which share several morphological similarities with M.platypoda; therefore, future studies may provide new insights into the phylogenetic relationships between the aforementioned species.
The distribution of pairwise genetic distances (p-distances) shows a distinct gap between the highest intraspecific divergence (1.06%) and the lowest interspecific divergence (8.21%) (Fig. 2). Genetic distances between the analysed Mordellistena species range from 8.21% to 19.76%, similar to the previous analyses (Selnekovič et al. 2021). The multidimensional scaling of the p-distances shows separate clusters for each species (Fig. 2).
Mordellistenaplatypoda was collected in a relatively large series of 52 specimens during one collecting event on the island of Ischia in Italy. Revision of the museum specimens identified as M.micans (Germar, 1817), M.stenidea Mulsant, 1856, and M.grisea Mulsant, 1856 in the Franciscolo collection in MSNG, a series of M.micans in SDEI, and a series of M.micans in HNHM did not reveal any additional specimens of the new species. Therefore, M.platypoda can now be considered endemic to the island of Ischia. However, given the island’s proximity to the shore (ca. 30 km) and the similarity of its fauna to that of the mainland, it is possible that M.platypoda may also occur on the Italian mainland or surrounding islands.
Supplementary Material
Acknowledgements
The authors thank Győző Szél (HNHM), Bernd Jaeger (MNHU), Olaf Jäger (SNSD), Maria Tavano (MSNG), and Mandy Schröter (SDEI) for allowing us to study the material under their care. We thank Jana Poláková for her great help during the DNA isolation procedure. We thank Michal Šagát and Robert Naczi for valuable comments and suggestions during the preparation of the manuscript. The present study was supported by the Slovak Research and Development Agency under contract no. APVV-19-0076.
Appendix 1
Table A1.
Uncorrected p-distances between and within the analysed species calculated in Mega X software. Intraspecific divergences are highlighted in bold.
| M.austriaca | M.confinis | M.hirtipes | M.lindbergi | M.minima | M.platypoda | M.pseudorhenana | M.purpurascens | M.purpureonigrans | M.tarsata | M.abdominalis | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mordellistenaaustriaca | N/A | ||||||||||
| M.confinis | 0.1505 | 0.0000 | |||||||||
| M.hirtipes | 0.1581–0.1596 | 0.1581–0.1596 | 0.0000–0.0015 | ||||||||
| M.lindbergi | 0.1672–0.1687 | 0.1702–0.1733 | 0.1550–0.1581 | 0.0000–0.0030 | |||||||
| M.minima | 0.1565–0.1596 | 0.1581–0.1596 | 0.1884–0.1915 | 0.1474–0.1505 | 0.0000–0.0046 | ||||||
| M.platypoda | 0.1489 | 0.1687 | 0.1535–0.1550 | 0.1717–0.1733 | 0.1763–0.1793 | 0.0000 | |||||
| M.pseudorhenana | 0.1565 | 0.1839 | 0.1626–0.1672 | 0.1702–0.1748 | 0.1733–0.1748 | 0.1611 | 0.0000–0.0030 | ||||
| M.purpurascens | 0.1641–0.1657 | 0.1581–0.1596 | 0.1170–0.1201 | 0.1657–0.1687 | 0.1793–0.1824 | 0.1672–0.1687 | 0.1733–0.1748 | 0.0000–0.0046 | |||
| M.purpureonigrans | 0.1489–0.1535 | 0.1626–0.1672 | 0.1505–0.1550 | 0.1626–0.1702 | 0.1641–0.1687 | 0.1778–0.1824 | 0.1702–0.1733 | 0.1535–0.1581 | 0.0000–0.0046 | ||
| M.tarsata | 0.1398–0.1474 | 0.1824–0.1900 | 0.1657–0.1702 | 0.1915–0.1976 | 0.1854–0.1945 | 0.0821–0.0897 | 0.1596–0.1657 | 0.1763–0.1793 | 0.1809–0.1869 | 0.0000–0.0106 | |
| M.abdominalis | 0.1991 | 0.2112 | 0.2158–0.2173 | 0.2295–0.2325 | 0.2036 | 0.2158 | 0.2036 | 0.2264–0.2280 | 0.2112–0.2143 | 0.2112–0.2128 | N/A |
Citation
Selnekovič D, Goffová K, Šoltýs J, Kováčová E, Kodada J (2023) Mordellistena platypoda, a new species of tumbling flower beetle from the island of Ischia in Italy (Coleoptera, Mordellidae). ZooKeys 1148: 41–63. https://doi.org/10.3897/zookeys.1148.86845
References
- Batten R. (1977) Two new Mordellidae (Coleoptera) from Southern Europe, and a key to the Mordellistenamicans group. Entomologische Berichten 37: 167–176. [Google Scholar]
- Bergsten J, Weingartner E, Hájek J. (2017) Species delimitation of the Hyphydrusovatus complex in western Palaearctic with an update of species distributions (Coleoptera, Dytiscidae). ZooKeys 678: 73–96. 10.3897/zookeys.678.12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec L. (1996) Mordellidae, Miastkowate (Insecta: Coleoptera). Fauna Polski (Vol. 18). Muzeum i Instytut Zoologii, Warszawa, 190 pp. [Google Scholar]
- Costa A. (1853–1854) Coleotteri eteromeri. Famiglia de’Mordellidei–Mordellidea. Fauna del Regno di Napoli ossia enumerazione di tutti gli animali che abitano le diverse regioni di questo regno e le acque che le bagnano contenente la descrizione de’ nuovi o poco esattamente conosciuti con figure ricavate da originali viventi e dipinte al naturale di Oronzio-Gabriele Costa. Coleotteri. Parte I.a con XXIV tavole in rame. Gaetano Sautto, Naples, 1–8 (1853), 9–32 (1854). [Part I.a was published in 47 parts (1849–1854) and contains xiii + 364 pp., with each section paginated separately.]
- DeSalle R, Goldstein P. (2019) Review and interpretation of trends in DNA barcoding. Frontiers in Ecology and Evolution 7: e302. 10.3389/fevo.2019.00302 [DOI]
- Ermisch K. (1950) Die Gattungen der Mordelliden der Welt. Entomologische Blätter 45–46: 34–92.
- Ermisch K. (1956) Mordellidae. In: Horion A. (Ed.) Faunistik der Mitteleuropäischen Käfer.Band 5: Heteromera. Entomologische Arbeiten aus dem Museum G. Frey Tutzing bei München, München, 269–328.
- Ermisch K. (1963) Neue Mordelliden (Heteromera, Mordellidae) aus Deutschland und Nachträge zur Faunistik der Mitteleuropaischen Mordelliden. Entomologische Blätter 59: 1–36. [Google Scholar]
- Ermisch K. (1969) 79. Familie: Mordellidae. In: Freude H, Harde KW, Lohse GA. (Eds) Die Käfer Mitteleuropas.Band 8, Teredilia, Heteromera, Lamellicornia. Goecke & Evers, Krefeld; G. Fischer, Jena, Stuttgart, 160–196.
- Ermisch K. (1977) Die Mordellistena-Arten Ungarns und benachbarter Gebiete sowie Beschreibung einer neuen Hoshihananomia-Art aus Siebenbürgen. Folia Entomologica Hungarica 30: 151–177. [New Series] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3(5): 294–299. [PubMed] [Google Scholar]
- Fossen EI, Ekrem T, Nilsson AN, Bergsten J. (2016) Species delimitation in northern European water scavenger beetles of the genus Hydrobius (Coleoptera, Hydrophilidae). ZooKeys 564: 71–120. 10.3897/zookeys.564.6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franciscolo M. (1957) Chapter V. Coleoptera: Mordellidae. A monograph of the South African genera and species. 1. Morphology, subfamily Ctenidiinae and tribe Stenaliini. In: Hanström B, Brinck P, Rudebeck G. (Eds) South African animal life.Results of the Lund University expedition in 1950–1951 (Vol. IV). Almqvist & Wiksell, Stockholm, 207–291.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology 59(3): 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. (2003) Biological identifications through DNA barcodes. Proceedings. Biological Sciences 270(1512): 313–321. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. (2017) UFBoot2: Improving the Ultrafast Bootstrap Approximation. Molecular Biology and Evolution 35(2): 518–522. 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horák J. (1983) Revision der Mordellistena-Arten aus der pentas-Gruppe (Coleoptera, Mordellidae). Entomologische Abhandlungen 47: 1–13. [Google Scholar]
- Horák J. (1990) Typenrevision einiger wenig bekanntner Arten aus der Gattung Mordellistena Costa (Insecta, Coleoptera: Mordellidae). Entomologische Abhandlungen 53(9): 125–142. [Google Scholar]
- Horák J. (1996) Revision of some little known species of genus Mordellistena with description of two new species. Part 2. (Coleoptera: Mordellidae). Klapalekiana 32: 171–184. [Google Scholar]
- Horák J. (2008) Family Mordellidae Latreille, 1802. In: Löbl I, Smetana A. (Eds) Catalogue of Palaearctic Coleoptera (Vol.5). Tenebrionoidea. Apollo Books, Stenstrup, 87–105.
- Horák J. (2020) Family Mordellidae Latreille, 1802. In: Iwan D, Löbl I. (Eds) Catalogue of Palaearctic Coleoptera (Vol.5). Tenebrionoidea. Revised and Updated Second Edition. Brill, Leiden & Boston, 79–104. 10.1163/9789004434998 [DOI]
- International Trust of Zoological Nomenclature (1999) International Code of Zoological Nomenclature (4th edn). International Trust for Zoological Nomenclature, London. 10.5962/bhl.title.50608 [DOI]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35(6): 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JF, Ślipiński A. (2010) 11.7 Mordellidae Latreille, 1802. In: Leschen RAB, Beutel RG, Lawrence JF. (Eds) Coleoptera, Beetles (Vol.2). Morphology and Systematics (Elateroidea, Bostrichiformia, Cucujiformia partim). In: Kristensen NP, Beutel RG (Eds) Handbook of Zoology, Arthropoda: Insecta. Walter de Gruyter, Berlin & New York, 533–537. 10.1515/9783110911213.533 [DOI]
- Lu W, Jackman JA, Johnson PW. (1997) Male genitalia and phylogenetic relationships in north American Mordellidae (Coleoptera). Annals of the Entomological Society of America 90(6): 742–767. 10.1093/aesa/90.6.742 [DOI] [Google Scholar]
- Maddison DR, Sproul JS. (2020) Species delimitation, classical taxonomy and genome skimming: a review of the ground beetle genus Lionepha (Coleoptera: Carabidae). Zoological Journal of the Linnean Society 189(4): 1313–1358. 10.1093/zoolinnean/zlz167 [DOI] [Google Scholar]
- Mulsant ME. (1856) Histoire naturelle des coléoptères de France. Longipèdes. Annales de la Société Linnéenne de Lyon 3: 305–471. [Nouvelle Série] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odnosum VK. (2010) Vypusk 9, Zhuki-Gorbatki (Coleoptera, Mordellidae). Fauna Ukrainy (tom 19), Zhestokrylye. Naukova Dumka, Kiev, 263 pp. [Google Scholar]
- Pentinsaari M, Salmela H, Mutanen M, Roslin T. (2016) Molecular evolution of a widely-adopted taxonomic marker (COI) across the animal tree of life. Scientific Reports 6(1): e35275. 10.1038/srep35275 [DOI] [PMC free article] [PubMed]
- Puillandre N, Lambert A, Brouillet S, Achaz G. (2012) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology 21(8): 1864–1877. 10.1111/j.1365-294X.2011.05239.x [DOI] [PubMed] [Google Scholar]
- Puillandre N, Brouillet S, Achaz G. (2020) ASAP: Assemble species by automatic partitioning. Molecular Ecology Resources 21(2): 609–620. 10.1111/1755-0998.13281 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilsky J. (1894) Die Käfer Europa’s. XXX. Nach der Natur Beschrieben von Dr. H. C. Küster und Dr. G. Kraatz Fortgesetzt von J. Schilsky. Bauer und Raspe, Nürnberg, viii + [1–100.]
- Schilsky J. (1895) Die Käfer Europa’s. XXXI. Nach der Natur Beschrieben von Dr. H. C. Küster und Dr. G. Kraatz Fortgesetzt von J. Schilsky. Bauer und Raspe, Nürnberg, viii + [1–100.]
- Schilsky J. (1898) Die Käfer Europa’s. XXXV. Nach der Natur Beschrieben von Dr. H. C. Küster und Dr. G. Kraatz Fortgesetzt von J. Schilsky. Bauer und Raspe, Nürnberg, viii + [1–100] + 41 pp. [from A to SS].
- Selnekovič D, Kodada J. (2019) Taxonomic revision of Mordellistenahirtipes species complex with new distribution records (Insecta, Coleoptera, Mordellidae). ZooKeys 854: 89–118. 10.3897/zookeys.854.32299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selnekovič D, Goffová K, Kodada J, Improta R. (2021) Revealing the identity of Mordellistenaminima and M.pseudorhenana (Coleoptera: Mordellidae) based on re-examined type material and DNA barcodes, with new distributional records and comments on morphological variability. Canadian Entomologist 153(3): 343–367. 10.4039/tce.2021.3 [DOI] [Google Scholar]
- Snodgrass RE. (1935) Principles of Insect Morphology. McGraw-Hill Publishing Co., New York, 667 pp. [Google Scholar]
- Zemoglyadchuk A, Buialska N. (2020) Description of the larvae of three species of the genus Mordellistena (Coleoptera: Mordellidae) with notes on their ecology. Zootaxa 4743(3): 371–381. 10.11646/zootaxa.4743.3.4 [DOI] [PubMed] [Google Scholar]
- Zhang J, Kapli P, Pavlidis P, Stamatakis A. (2013) A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29(22): 2869–2876. 10.1093/bioinformatics/btt499 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.