Abstract
The genus Apiospora includes endophytes, pathogens and saprobes, with a wide host range and geographic distribution. In this paper, six Apiospora strains isolated from diseased and healthy tissues of bamboo leaves from Hainan and Shandong provinces in China were classified using a multi-locus phylogeny based on a combined dataset of ITS, LSU, tef1 and tub2, in conjunction with morphological characters, host association and ecological distribution. Two new species, Apiosporadongyingensis and A.hainanensis, and a new record of A.pseudosinensis in China, are described based on their distinct phylogenetic relationships and morphological analyses. Illustrations and descriptions of the three taxa are provided, along with comparisons with closely related taxa in the genus.
Keywords: Apiosporadongyingensis , Apiosporahainanensis , Ascomycota, bamboo, taxonomy
Introduction
Apiospora Sacc., the type genus of Apiosporaceae K.D. Hyde, J. Fröhl., Joanne E. Taylor & M.E. Barr, was introduced by Saccardo with A.montagnei Sacc. as the type species (Saccardo 1875). The sexual morphs of Apiospora are characterized by multi-locular perithecial stromata with hyaline ascospores surrounded by a thick gelatinous sheath (Dai et al. 2016, 2017; Pintos and Alvarado 2021). The asexual morphs of Apiospora are characterized by their basauxic conidiogenesis, and globose to subglobose conidia, which are usually lenticular or obovoid in the side view, and pale brown to brown in color (Kunze 1817; Hyde et al. 1998; Dai et al. 2016). Most species of Apiospora are quite similar to each other in morphology, thus it is difficult to distinguish them without molecular phylogenetic data.
Until the studies of Pintos and Alvarado (2021) and Jiang et al. (2022a), the closely related genera Apiospora, Arthrinium Kunze and Neoarthrinium Ning Jiang were considered a single taxon because of their similar morphological characteristics, especially the basauxic conidiogenesis. However, the conidia of Apiospora and Neoarthrinium are more or less rounded in the face view and lenticular in the side view, whereas the conidia of Arthrinium are variously shaped (angular, curved, fusiform, globose, polygonal, navicular). In addition, the conidiophores of several Arthrinium and Neoarthrinium species have thick blackish septa, which are rarely observed in Apiospora (Pintos and Alvarado 2021; Tian et al. 2021; Jiang et al. 2022a). Apiospora species have a worldwide distribution and can be found on various hosts, while Arthrinium species are rarely found in tropical and subtropical habitats and commonly occur on Cyperaceae Juss. and Juncaceae Juss. (Ramos et al. 2010; Dai et al. 2017; Wang et al. 2018; Hyde et al. 2020; Pintos and Alvarado 2021; Tian et al. 2021). Four Neoarthrinium species have been discovered on four hosts from three distantly related host plant families in China, Colombia and Great Britain (Jiang et al. 2022a). Most Apiospora species are associated with plants as endophytes, pathogens or saprobes (Agut and Calvo 2004; Dai et al. 2016, 2017; Tian et al. 2021). Some species are economically important plant pathogens, for example, A.arundinis causes bamboo brown culm streak, chestnut leaf spot and barley kernel blight (Martínez-Cano et al. 1992; Chen et al. 2014; Jiang et al. 2021), while A.sacchari causes damping-off of durum wheat (Mavragani et al. 2007). Some species have also been isolated from lichens, air, soil, seaweeds and animal tissues, and a few species are human pathogens which can cause cutaneous infections (Tian et al. 2021).
The aim of this study was to explore the diversity of Apiospora species in symptomatic and asymptomatic bamboo leaves collected in Hainan and Shandong provinces (China). We describe two newly discovered species, Apiosporadongyingensis and A.hainanensis, and a new record of A.pseudosinensis in China based on phylogenetic data and morphology.
Materials and methods
Isolation and morphological studies
The samples were collected at the Diaoluoshan National Nature Reserve, Hainan Province, and the Dongying Botanical Garden, Shandong Province (China). The strains of Apiospora were isolated from single spores and fungal tissue obtained from diseased and healthy bamboo leaves following the methods described by Chomnunti et al. (2014). Sampled spores were suspended in sterile distilled water, spread onto potato dextrose agar (PDA) plates, and incubated for one day at 25 °C. After germination, the spores were transferred to a new PDA plate to obtain a pure culture. Additionally, about 25 mm2 tissue fragments were taken from the margin of leaf lesions and their surface sterilized by consecutive immersions in a 75% ethanol solution for 60 s, 5% sodium hypochlorite solution for 30 s, and then rinsed in sterile distilled water for 60 s (Mu et al. 2021). The surface sterilized plant tissue was dried with sterilized paper and moved on the PDA plates. All the PDA plates were incubated at 25 °C for 3–4 days in darkness, and then hyphae were picked out of the periphery of the colonies and grown on new PDA plates (Jiang et al. 2022b).
After 7 days of incubation, the morphological characters of the colonies were recorded on PDA with a digital camera (Canon G7X). Morphological descriptions were based on cultures sporulating on water agar (WA). The size of the conidiogenous cells and conidia were shown as minimum-maximum. Color notations were done using the color charts of Rayner (1970). The micro-morphological characters of the colonies were studied using a stereomicroscope (Olympus SZX10) and a microscope (Olympus BX53), both fitted with high-definition color digital cameras. Grown cultures of Apiospora were stored in 10% sterilized glycerin and sterile water at 4 °C for further studies in the future. All specimens were deposited in the Herbarium of the Department of Plant Pathology, Shandong Agricultural University (HSAUP). Living cultures were deposited in the Shandong Agricultural University Culture Collection (SAUCC). Taxonomic information on the new taxa was submitted to MycoBank (http://www.mycobank.org).
DNA extraction and amplification
Genomic DNA was extracted from fungal mycelia grown on PDA, using a modified cetyltrimethylammonium bromide (CTAB) protocol as described in Guo et al. (2000). DNA sequences of four different loci were obtained, including the nrDNA internal transcribed spacer regions 1 and 2 with the intervening 5.8S subunit (ITS), a partial sequence of the large subunit nrDNA subunit (LSU), a partial sequence of the translation elongation factor 1-alpha gene (tef1), and a partial sequence of the beta-tubulin gene (tub2). They were all amplified with the primer pairs and polymerase chain reaction (PCR) program listed in Table 1.
Table 1.
Gene regions and respective primer pairs used in the study.
| Locus | PCR primers | PCR: thermal cycles: (Annealing temperature in bold) | Reference |
|---|---|---|---|
| ITS | ITS5/ITS4 | (94 °C: 30 s, 55 °C: 30 s, 72 °C: 45 s) × 29 cycles | White et al. 1990 |
| LSU | LR0R/LR5 | (94 °C: 30 s, 48 °C: 50 s, 72 °C: 1 min 30 s) × 35 cycles | Vilgalys and Hester 1990; Cubeta et al. 1991 |
| tef1 | EF1-728F/EF2 | (95 °C: 30 s, 51 °C: 30 s, 72 °C: 1 min) × 35 cycles | O’Donnell et al. 1998; Carbone and Kohn 1999 |
| tub2 | Bt-2a/Bt-2b | (95 °C: 30 s, 56 °C: 30 s, 72 °C: 1 min) × 35 cycles | Glass and Donaldson 1995 |
PCR was performed using an Eppendorf Master Thermocycler (Hamburg, Germany). Amplification reactions contained 12.5 μL 2× Taq Plus Master Mix II (Vazyme, Nanjing, China), 1 μL of each forward and reverse primers (10 μM) (Tsingke, Qingdao, China), 1 μL of template genomic DNA, and distilled deionized water to a total volume of 25 μL. The PCR products were visualized on 1% agarose electrophoresis gels. Bi-directional sequencing was conducted by the Tsingke Company Limited (Qingdao, China). Consensus sequences were obtained using MEGA 7.0 (Kumar et al. 2016). All sequences generated in this study were deposited in GenBank (Table 2).
Table 2.
Isolates and GenBank accession numbers used in the phylogenetic analyses.
| Species | Isolate/Strain | Host/Substrate | Origin | GenBank accession numbers | |||
|---|---|---|---|---|---|---|---|
| ITS | LSU | tef1 | tub2 | ||||
| Apiosporaacutiapica | KUMCC 20-0210 (Type) | Bambusabambos | China | MT946343 | MT946339 | MT947360 | MT947366 |
| A.agari | KUC21333 (Type) | Agarumcribrosum | Korea | MH498520 | MH498440 | MH544663 | MH498478 |
| A.aquatica | S-642 (Type) | Submerged wood | China | MK828608 | MK835806 | NA | NA |
| A.arctoscopi | KUC21331 (Type) | Egg of Arctoscopusjaponicus | Korea | MH498529 | MH498449 | MN868918 | MH498487 |
| A.arundinis | CBS 124788 | Living leaves of Fagussylvatica | Switzerland | KF144885 | KF144929 | KF145017 | KF144975 |
| A.aurea | CBS 244.83 (Type) | Air | Spain | AB220251 | KF144935 | KF145023 | KF144981 |
| A.balearica | CBS 145129 (Type) | Undetermined Poaceae | Spain | MK014869 | MK014836 | MK017946 | MK017975 |
| A.biserialis | CGMCC 3.20135 (Type) | Bamboo | China | MW481708 | MW478885 | MW522938 | MW522955 |
| A.camelliae-sinensis | LC5007 (Type) | Camelliasinensis | China | KY494704 | KY494780 | KY705103 | KY705173 |
| A.chiangraiense | MFLUCC21-0053 (Type) | Dead culms of bamboo | Thailand | MZ542520 | MZ542524 | NA | MZ546409 |
| A.chromolaenae | MFLUCC 17-1505 (Type) | Chromolaenaodorata | Thailand | MT214342 | MT214436 | NA | NA |
| A.cordylines | GUCC 10027 (Type) | Leaves of Cordylinefruticosa | China | MT040106 | NA | MT040127 | MT040148 |
| A.cyclobalanopsidis | CGMCC 3.20136 (Type) | Cyclobalanopsidisglauca | China | MW481713 | MW478892 | MW522945 | MW522962 |
| A.descalsii | CBS 145130 (Type) | Ampelodesmosmauritanicus | Spain | MK014870 | MK014837 | MK017947 | MK017976 |
| A.dichotomanthi | LC4950 (Type) | Dichotomanthustristaniaecarpa | China | KY494697 | KY494773 | KY705096 | KY705167 |
| A.dongyingensis | SAUCC 0302 (Type) | Leaf of bamboo | China | OP563375 | OP572424 | OP573264 | OP573270 |
| SAUCC 0303 | Leaf of bamboo | China | OP563374 | OP572423 | OP573263 | OP573269 | |
| A.esporlensis | CBS 145136 (Type) | Phyllostachysaurea | Spain | MK014878 | MK014845 | MK017954 | MK017983 |
| A.euphorbiae | IMI 285638b | Bambusa sp. | Bangladesh | AB220241 | AB220335 | NA | AB220288 |
| A.fermenti | KUC21289 (Type) | Seaweed | Korea | MF615226 | MF615213 | MH544667 | MF615231 |
| A.gaoyouensis | CFCC 52301 (Type) | Phragmitesaustralis | China | MH197124 | NA | MH236793 | MH236789 |
| A.garethjonesii | JHB004 (Type) | Culms of dead bamboo | China | KY356086 | KY356091 | NA | NA |
| A.gelatinosa | HKAS 111962 (Type) | Culms of dead bamboo | China | MW481706 | MW478888 | MW522941 | MW522958 |
| A.guiyangensis | HKAS 102403 (Type) | Dead culms of Poaceae | China | MW240647 | MW240577 | MW759535 | MW775604 |
| A.guizhouensis | LC5322 (Type) | Air in karst cave | China | KY494709 | KY494785 | KY705108 | KY705178 |
| A.hainanensis | SAUCC 1681 (Type) | Leaf of bamboo | China | OP563373 | OP572422 | OP573262 | OP573268 |
| SAUCC 1682 | Leaf of bamboo | China | OP563372 | OP572421 | OP573261 | OP573267 | |
| A.hispanica | IMI 326877 (Type) | Maritime sand | Spain | AB220242 | AB220336 | NA | AB220289 |
| A.hydei | CBS 114990 (Type) | Culms of Bambusatuldoides | China | KF144890 | KF144936 | KF145024 | KF144982 |
| A.hyphopodii | MFLUCC 15-0003 (Type) | Dead culms of bamboo | Thailand | KR069110 | NA | NA | NA |
| A.hysterina | ICPM 6889 (Type) | Bamboo | New Zealand | MK014874 | MK014841 | MK017951 | MK017980 |
| A.iberica | AP10118 (Type) | Arundodonax | Portugal | MK014879 | MK014846 | MK017955 | MK017984 |
| A.intestini | CBS 135835 (Type) | Gut of grasshopper | India | KR011352 | KR149063 | KR011351 | KR011350 |
| A.italica | CBS 145138 (Type) | Arundodonax | Italy | MK014880 | MK014847 | MK017956 | MK017985 |
| A.jatrophae | CBS 134262 (Type) | Jatrophapodagrica | India | JQ246355 | NA | NA | NA |
| A.jiangxiensis | LC4577 (Type) | Maesa sp. | China | KY494693 | KY494769 | KY705092 | KY705163 |
| A.kogelbergensis | CBS 113333 (Type) | Dead culms of Restionaceae | South Africa | KF144892 | KF144938 | KF145026 | KF144984 |
| A.koreana | KUC21332 (Type) | Egg of Arctoscopusjaponicus | Korea | MH498524 | MH498444 | MH544664 | MH498482 |
| A.locuta-pollinis | LC11683 (Type) | Brassicacampestris | China | MF939595 | NA | MF939616 | MF939622 |
| A.longistroma | MFLUCC 11-0481 (Type) | Culms of decaying bamboo | Thailand | KU940141 | KU863129 | NA | NA |
| A.malaysiana | CBS 102053 (Type) | Macarangahullettii stem colonised by ants | Malaysia | KF144896 | KF144942 | KF145030 | KF144988 |
| A.marianiae | AP18219 (Type) | Dead stems of Phleumpratense | Spain | ON692406 | ON692422 | ON677180 | ON677186 |
| A.marii | CBS 497.90 (Type) | Atmosphere, pharmaceutical excipients, home dust and beach sands | Spain | MH873913 | KF144947 | KF145035 | KF144993 |
| A.marina | KUC21328 (Type) | Seaweed | Korea | MH498538 | MH498458 | MH544669 | MH498496 |
| A.mediterranea | IMI 326875 (Type) | Air | Spain | AB220243 | AB220337 | NA | AB220290 |
| A.minutispora | 17E-042 (Type) | Soil | South Korea | LC517882 | NA | LC518889 | LC518888 |
| A.montagnei | AP301120 (Epitype) | Arundomicrantha | Spain | ON692408 | ON692424 | ON677182 | ON677188 |
| AP19421 | Arundomicrantha | Spain | ON692418 | ON692425 | ON677183 | ON677189 | |
| CPC 18900 | Culms of Phragmitesaustralis | Italy | KF144909 | KF144956 | KF145043 | KF145001 | |
| A.mori | MFLU 18-2514 (Type) | Dead leaves of Morusaustralis | China | MW114313 | MW114393 | NA | NA |
| A.multiloculata | MFLUCC 21-0023 (Type) | Dead culms of Bambusae | Thailand | OL873137 | OL873138 | NA | OL874718 |
| A.mytilomorpha | DAOM 214595 (Type) | Dead blades of Andropogon sp. | India | KY494685 | NA | NA | NA |
| A.neobambusae | LC7106 (Type) | Leaf of bamboo | China | KY494718 | KY494794 | KY806204 | KY705186 |
| A.neochinense | CFCC 53036 (Type) | Fargesiaqinlingensis | China | MK819291 | NA | MK818545 | MK818547 |
| A.neogarethjonesii | HKAS 102408 (Type) | Dead culms of Bambusae | China | MK070897 | MK070898 | NA | NA |
| A.neosubglobosa | JHB007 (Type) | Bamboo | China | KY356090 | KY356095 | NA | NA |
| A.obovata | LC4940 (Type) | Lithocarpus sp. | China | KY494696 | KY494772 | KY705095 | KY705166 |
| A.ovata | CBS 115042 (Type) | Arundinariahindsii | China | KF144903 | KF144950 | KF145037 | KF144995 |
| A.paraphaeosperma | MFLUCC13-0644 (Type) | Dead clumps of Bambusa sp. | Thailand | KX822128 | KX822124 | NA | NA |
| A.phyllostachydis | MFLUCC 18-1101 (Type) | Phyllostachysheteroclada | China | MK351842 | MH368077 | MK340918 | MK291949 |
| A.piptatheri | CBS 145149 (Type) | Piptatherummiliaceum | Spain | MK014893 | MK014860 | MK017969 | NA |
| A.pseudomarii | GUCC 10228 (Type) | Leaves of Aristolochiadebilis | China | MT040124 | NA | MT040145 | MT040166 |
| A.pseudoparenchymatica | LC7234 (Type) | Leaf of bamboo | China | KY494743 | KY494819 | KY705139 | KY705211 |
| A.pseudorasikravindrae | KUMCC 20-0208 (Type) | Bambusadolichoclada | China | MT946344 | NA | MT947361 | MT947367 |
| A.pseudosinensis | CPC 21546 (Type) | Leaf of bamboo | Netherlands | KF144910 | KF144957 | KF145044 | MN868936 |
| A.pseudosinensis | SAUCC 0221 | Leaf of bamboo | China | OP563377 | OP572426 | OP573266 | OP573272 |
| SAUCC 0222 | Leaf of bamboo | China | OP563376 | OP572425 | OP573265 | OP573271 | |
| A.pseudospegazzinii | CBS 102052 (Type) | Macarangahullettii stem colonized by ants | Malaysia | KF144911 | KF144958 | KF145045 | KF145002 |
| A.pterosperma | CPC 20193 (Type) | Lepidospermagladiatum | Australia | KF144913 | KF144960 | KF145046 | KF145004 |
| A.pusillisperma | KUC21321 (Type) | Seaweed | Korea | MH498533 | MH498453 | MN868930 | MH498491 |
| A.qinlingensis | CFCC 52303 (Type) | Fargesiaqinlingensis | China | MH197120 | NA | MH236795 | MH236791 |
| A.rasikravindrae | LC5449 | Soil in karst cave | China | KY494713 | KY494789 | KY705112 | KY705182 |
| A.sacchari | CBS 212.30 | Phragmitesaustralis | UK | KF144916 | KF144962 | KF145047 | KF145005 |
| A.saccharicola | CBS191.73 | Air | Netherlands | KF144920 | KF144966 | KF145051 | KF145009 |
| A.sargassi | KUC21228 (Type) | Sargassumfulvellum | Korea | KT207746 | KT207696 | MH544677 | KT207644 |
| A.sasae | CBS 146808 (Type) | Dead culms of Sasaveitchii | Netherlands | MW883402 | MW883797 | MW890104 | MW890120 |
| A.septata | CGMCC 3.20134 (Type) | Bamboo | China | MW481711 | MW478890 | MW522943 | MW522960 |
| A.serenensis | IMI 326869 (Type) | Food, pharmaceutical excipients, atmosphere and home dust | Spain | AB220250 | AB220344 | NA | AB220297 |
| A.setariae | CFCC 54041 (Type) | Decaying culms of Setariaviridis | China | MT492004 | NA | NA | NA |
| A.sichuanensis | HKAS 107008 (Type) | Dead culms of Poaceae | China | MW240648 | MW240578 | MW759536 | MW775605 |
| A.sorghi | URM 93000 (Type) | Sorghumbicolor | Brazil | MK371706 | NA | NA | MK348526 |
| A.sphaerosperma | CBS114314 | Leaf of Hordeumvulgare | Iran | KF144904 | KF144951 | KF145038 | KF144996 |
| A.stipae | CBS 146804 (Type) | Dead culm of Stipagigantea | Spain | MW883403 | MW883798 | MW890082 | MW890121 |
| A.subglobosa | MFLUCC 11-0397 (Type) | Dead bamboo culms | Thailand | KR069112 | KR069113 | NA | NA |
| A.subrosea | LC7292 (Type) | Leaf of bamboo | China | KY494752 | KY494828 | KY705148 | KY705220 |
| A.thailandica | LC5630 | Rotten wood | China | KY494714 | KF144970 | KY705113 | KY806200 |
| A.vietnamensis | IMI 99670 (Type) | Citrussinensis | Vietnam | KX986096 | KX986111 | NA | KY019466 |
| A.xenocordella | CBS 478.86 (Type) | Soil from roadway | Zimbabwe | KF144925 | KF144970 | KF145055 | KF145013 |
| A.yunnana | MFLUCC 15-0002 (Type) | Decaying bamboo culms | China | KU940147 | KU863135 | NA | NA |
| Arthriniumcaricicola | CBS 145127 | Carexericetorum | China | MK014871 | MK014838 | MK017948 | MK017977 |
Notes: Strains in this study are marked in bold. NA = not available.
Phylogenetic analyses
Newly generated ITS, LSU, tef1 and tub2 sequences from the six strains studied were aligned with all reference sequences of Apiospora and related species available in GenBank using the MAFFT v.7.11 online software (http://mafft.cbrc.jp/alignment/server/, Katoh et al. 2019) with the default settings, manually correcting the resulting alignment where necessary. Maximum likelihood (ML) and Bayesian inference (BI) phylogenetic analyses were conducted individually on each locus (ITS, LSU, tef1 and tub2) and on a combined dataset including all of them. The best-fitting evolutionary model of each partition was determined using MrModeltest v. 2.3 (Nylander 2004). ML and BI were run on the CIPRES Science Gateway portal (https://www.phylo.org/) using RaxML-HPC2 on XSEDE (8.2.12) (Miller et al. 2012; Stamatakis 2014) and MrBayes on XSEDE (3.2.7a), respectively (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003; Ronquist et al. 2012). For ML analyses the default parameters were used, while BI was carried out using a Markov chain Monte Carlo (MCMC) algorithm. BI analyses included four MCMC chains and were run for 5,000,000 generations until the average standard deviation of split frequencies was below 0.01 with trees saved every 1000 generations. The burn-in fraction was set to 0.25 and posterior probabilities (PP) were determined from the remaining trees. The resulting 50% majority-rule tree was plotted using FigTree v. 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree) and edited with Adobe Illustrator CS6.0.
Results
Phylogenetic analyses
Among the six strains of Apiospora isolated from the samples studied, two new species were discovered, and another one found for the first time in China after the combined analysis of ITS, LSU, tef1 and tub2 DNA sequences from 89 isolates of Apiospora plus Arthriniumcaricicola Kunze & J.C. Schmidt (CBS 145127) as the outgroup taxon.
A total of 2241 characters including gaps were compared in the phylogenetic analysis, viz. ITS: 1–706, LSU: 707–1513, tef1: 1514–1932, tub2: 1933–2241. Of these characters, 1436 were constant, 271 were variable and parsimony-uninformative, and 534 were parsimony-informative. For the BI and ML analyses, the substitution model GTR+I+G was selected for all partitions.
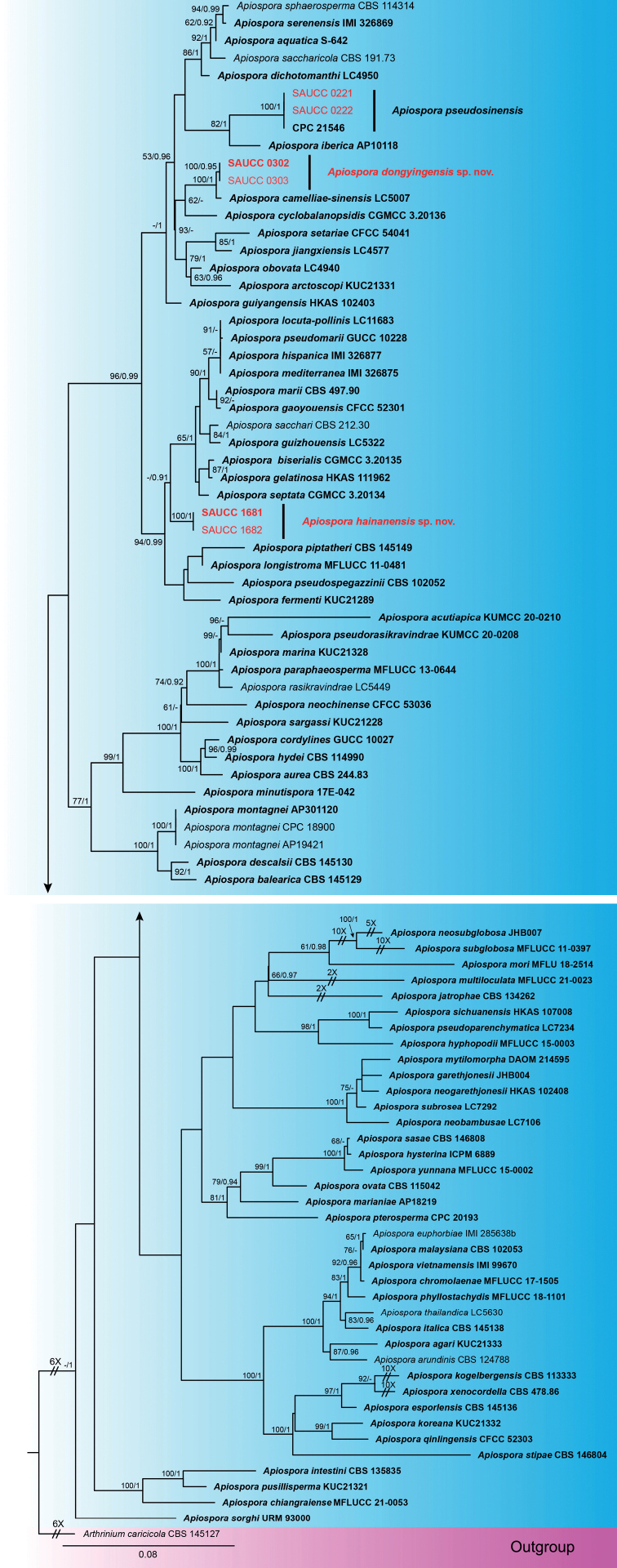
The BI analysis reached the established convergence after 3935000 generations, resulting in 39351 sampled trees, of which 29514 trees were used to calculate the posterior probabilities. The ML tree topology agreed with that obtained from the BI analysis, and therefore, only one tree (the ML) is presented (Fig. 1). The four strains (SAUCC 0302, SAUCC 0303, SAUCC 1681 and SAUCC 1682) studied in the present work represent two independent clades, interpreted as newly discovered independent species. These are described below and accommodated under the new names Apiosporadongyingensis and A.hainanensis. Another two strains (SAUCC 0221 and SAUCC 0222) clustered with A.pseudosinensis (CPC 21546) with full support (MLBS: 100% and BYPP: 1), and are therefore considered no different from this species.
Figure 1.
Phylogram of Apiospora based on combined ITS, LSU, tef1 and tub2 genes. ML bootstrap support values (MLBS ≥ 50%) and Bayesian posterior probability (BYPP ≥ 0.90) are shown as first and second position above nodes, respectively. Strains from this study are shown in red, ex-type or ex-epitype cultures are indicated in bold face. Some branches were shortened according to the indicated mulipliers.
Taxonomy
. Apiospora dongyingensis
R.Y. Liu, J.W. Xia & X.G. Zhang sp. nov.
414B86CE-7C66-5FA3-928A-9E981DC14E0F
846065
Figure 2.
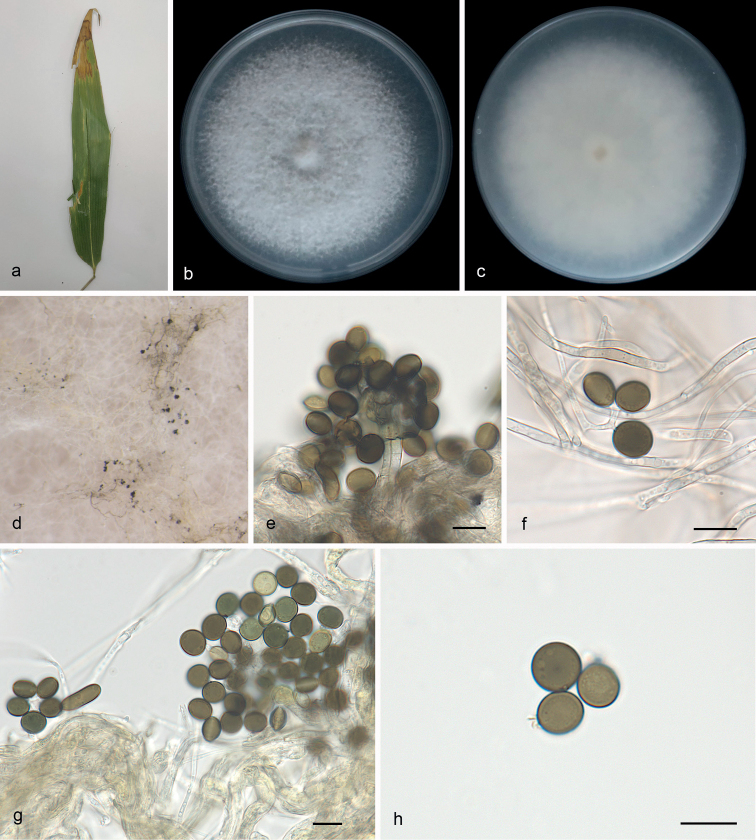
Apiosporadongyingensis (SAUCC 0302, ex-holotype culture) a leaf of host plant b, c surface (b) and reverse (c) sides of colony after incubation for 7 days on PDAd conidiomata formed in culture e, f conidiogenous cells and conidia g, h conidia. Scale bars: 10 μm (e–h).
Etymology.
Named after Dongying City (China) where the type was collected.
Type.
China, Shandong Province: Dongying Botanical Garden, on diseased leaves of bamboo, 13 July 2022, R.Y. Liu, holotype HSAUP 0302, ex-type living culture SAUCC 0302.
Description.
Asexual morph: On WA, hyphae 1.3–3.6 μm diam., hyaline, branched, septate. Conidiophores cylindrical, septate, verrucose, flexuous, sometimes reduced to conidiogenous cells. Conidiogenous cells globose to subglobose, erect, blastic, aggregated in clusters on hyphae, hyaline to pale brown, smooth, branched, 8.2–13.9 × 4.2–8.2 μm, mean ± SD: 9.6 ± 1.6 × 6.7 ± 1.1 μm (n = 40). Conidia globose, subglobose to lenticular, with a longitudinal germ slit, occasionally elongated to ellipsoidal, brown to dark brown, smooth to finely roughened, 8.0–16.5 × 5.5–9.0 μm, mean ± SD: 9.4 ± 1.9 × 7.3 ± 1.0 μm, L/W = 1.3–1.9 (n = 40). Sexual morph: Undetermined.
Culture characteristics.
Colonies on PDA flat with entire margin, aerial mycelium white to gray, floccose cottony; surface and reverse gray in the center and grayish margin. PDA attaining 78.5–86.5 mm in diameter after 7 days at 25 °C, growth rate 11.0–12.5 mm/day.
Additional specimen examined.
China, Shandong Province: Dongying Botanical Garden, on diseased leaves of bamboo, 13 July 2022, R.Y. Liu, paratype HSAUP 0303, ex-paratype living culture SAUCC 0303.
Notes.
Apiosporadongyingensis is closely related but phylogenetically distinct from A.camelliae-sinensis (M. Wang, F. Liu & L. Cai) Pintos & P. Alvarado and A.cyclobalanopsidis (Y. Feng & Jian K. Liu) X.G. Tian & Tibpromma (Fig. 1). A.dongyingensis differs from A.camelliae-sinensis by 18 nucleotides (13/518 in ITS, 2/804 in LSU, 2/374 in tef1 and 1/265 in tub2) and A.cyclobalanopsidis by 58 nucleotides (17/518 in ITS, 4/799 in LSU, 26/377 in tef1 and 11/266 in tub2). Morphologically, it differs from A.camelliae-sinensis and A.cyclobalanopsidis in its conidia (globose, subglobose to lenticular, 8.0–16.5 × 5.5–9.0 μm in A.dongyingensis vs. globose to subglobose, 9.0–13.5 × 7.0–12.0 μm in A.camelliae-sinensis and surface view globose to ellipsoid, 8–12 μm long and side view lenticular, 10–14 μm long in A.cyclobalanopsidis; Wang et al. 2018; Feng et al. 2021; Pintos and Alvarado 2021; Tian et al. 2021).
. Apiospora hainanensis
R.Y. Liu, J.W. Xia & X.G. Zhang sp. nov.
2233F929-A028-533A-BAB6-8B6EFC960F1F
846066
Figure 3.
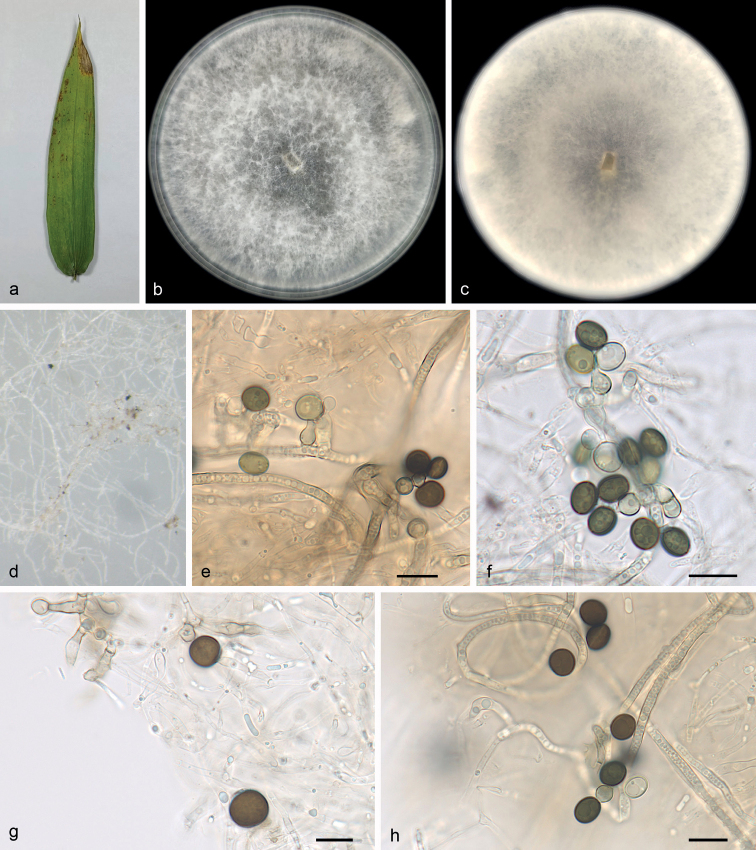
Apiosporahainanensis (SAUCC 1681, ex-holotype culture) a leaf of host plant b, c surface (b) and reverse (c) sides of colony after incubation for 7 days on PDAd conidiomata formed in culture e, f conidiogenous cells and conidia g, h conidia. Scale bars: 10 μm (e–h).
Etymology.
Named after Hainan Province (China) where the type was collected.
Type.
China, Hainan Province: Diaoluoshan National Nature Reserve, on diseased leaves of bamboo, 23 June 2021, R.Y. Liu, holotype HSAUP 1681, ex-type living culture SAUCC 1681.
Description.
Asexual morph: On WA, hyphae 1.2–3.4 μm diam., hyaline, branched, septate. Conidiophores cylindrical, septate, verrucose, flexuous, sometimes reduced to conidiogenous cells. Conidiogenous cells globose to subglobose, erect, blastic, aggregated in clusters on hyphae, hyaline to pale brown, smooth, branched, 6.4–8.8 × 5.2–7.1 μm, mean ± SD: 7.9 ± 1.1 × 6.1 ± 0.9 μm (n = 40). Conidia globose, subglobose to lenticular, with a longitudinal germ slit, occasionally elongated to ellipsoidal, brown to dark brown, smooth to finely roughened, 5.5–8.5 × 5.0–7.5 μm, mean ± SD: 6.8 ± 0.9 × 6.7 ± 0.7 μm, L/W = 1.0–1.1 (n = 40). Sexual morph: Undetermined.
Culture characteristics.
Colonies on PDA flat with entire margin, aerial mycelium white to grey, floccose cottony; reverse white to pale honey colored. PDA attaining 77.5–85.5 mm in diameter after 7 days at 25 °C, growth rate 10.5–12.5 mm/day.
Additional specimen examined.
China, Hainan Province: Diaoluoshan National Nature Reserve, on diseased leaves of bamboo, 23 June 2021, R.Y. Liu, paratype HSAUP 1682, ex-paratype living culture SAUCC 1682.
Notes.
The two strains (SAUCC 1681 and SAUCC 1682) of A.hainanensis clustered together with significant support in an isolated branch basal to A.sacchari and related species of the phaeospermum clade (Pintos and Alvarado 2022; Fig. 1). Other species in a more or less similar phylogenetic position include A.septata (Y. Feng & Jian K. Liu) X.G. Tian & Tibpromma, A.piptatheri (Pintos & P. Alvarado) Pintos & P. Alvarado, A.longistroma (D.Q. Dai & K.D. Hyde) Pintos & P. Alvarado, A.pseudospegazzinii (Crous) Pintos & Alvarado and A.fermenti (S.L. Kwon, S. Jang & J.J. Kim) S.L. Kwon & J.J. Kim. Morphologically, it differs from A.septata, A.piptatheri, A.longistroma, A.pseudospegazzinii and A.fermenti in its conidia (globose, subglobose to lenticular, 5.5–8.5 × 5.0–7.5 μm in A.hainanensis vs. surface view globose to ellipsoid, 8–13 μm long and side view lenticular, 8–14 μm long in A.septata, globose to ellipsoidal, 6–8 × 3–5 μm in A.piptatheri, asexual morph undetermined in A.longistroma, surface view globose, 7–9 μm diam. and side view lenticular, 5–6 μm diam. in A.pseudospegazzinii, surface view globose to elongate ellipsoid, 7.5–9 × 7–9 μm and side view lenticular, 6–7 μm diam. in A.fermenti; Crous and Groenewald 2013; Dai et al. 2016; Pintos et al. 2019; Feng et al. 2021; Kwon et al. 2021, 2022; Pintos and Alvarado 2021; Tian et al. 2021).
. Apiospora pseudosinensis
(Crous) Pintos & P. Alvarado, Fungal Systematics and Evolution 7: 207. (2021)
EB540E35-CA14-55C4-9F0F-D560117D8486
Figure 4.
Apiosporapseudosinensis (SAUCC 0221) a leaf of host plant b, c surface (b) and reverse (c) sides of colony after incubation for 7 days on PDAd conidiomata formed in culture e, f conidiogenous cells and conidia g, h conidia. Scale bars: 10 μm (e–h).
≡ Arthriniumpseudosinense Crous, in Crous & Groenewald, IMA Fungus 4(1): 148 (2013).
Description.
Asexual morph: On WA, hyphae 1.1–2.9 μm diam., hyaline, branched, septate. Conidiophores cylindrical, septate, verrucose, flexuous, sometimes reduced to conidiogenous cells. Conidiogenous cells globose to subglobose, erect, blastic, aggregated in clusters on hyphae, hyaline to pale brown, smooth, branched, 9.4–11.0 × 6.1–8.8 μm, mean ± SD: 10.4 ± 0.7 × 7.7 ± 1.1 μm (n = 40). Conidia globose, subglobose to lenticular, with a longitudinal germ slit, occasionally elongated to ellipsoidal, brown to dark brown, smooth to finely roughened, 7.5–11.5 × 7.0–9.5 μm, mean ± SD: 10.1 ± 1.3 × 8.3 ± 0.6 μm, L/W = 1.1–1.3 (n = 40). Sexual morph: Undetermined.
Culture characteristics.
Colonies on PDA flat with irregular margin, aerial mycelium white to pale yellow, floccose cottony; reverse pale yellow to yellow. PDA attaining 69.5–78.5 mm in diameter after 7 days at 25 °C, growth rate 9.5–11.5 mm/day.
Specimens examined.
China, Shandong Province: Dongying Botanical Garden, on diseased leaves of bamboo, 15 July 2022, R.Y. Liu, HSAUP 0221, living culture SAUCC 0221; China, Hainan Province: Diaoluoshan National Nature Reserve, on diseased leaves of bamboo, 29 June 2021, R.Y. Liu, HSAUP 0022, living culture SAUCC 0022.
Notes.
Apiosporapseudosinensis was originally described from bamboo leaves collected in the Utrecht Botanical Garden of the Netherlands (Crous and Groenewald 2013; Pintos and Alvarado 2021). In the present study, DNA sequences obtained from two strains (SAUCC 0221 and SAUCC 0222) collected also from bamboo leaves, were not significantly different from those of A.pseudosinensis (Fig. 1). Morphologically, our strains were similar to the original description (conidia 8–10 × 7–10 μm diam. in surface view, 7–8 μm diam. in side view). We therefore consider the newly found strains as A.pseudosinensis (Crous and Groenewald 2013; Pintos and Alvarado 2021).
Discussion
The family Apiosporaceae was proposed to accommodate genera with apiosporous hyaline ascospores and a basauxic, Arthrinium-like conidiogenesis (Hyde et al. 1998). Crous and Groenewald (2013) synonymized Apiospora with Arthrinium on the basis of the one fungus-one name policy (Hawksworth et al. 2011). Crous and Groenewald (2013) also resolved the genetic identity of multiple species of Arthrinium (= Apiospora), analysing ex-type collections, and confirmed that most species occur in Poaceae (R.Br.) Barnh. hosts, although some were known from many other plant host families. However, with the aid of additional genetic data from the type species of Arthrinium, Ar.caricicola, Apiospora and Arthrinium were separated again as two distinct genera (Pintos and Alvarado 2021). Arthrinium species have variously shaped conidia and inhabit Cyperaceae and Juncaceae in temperate, cold or alpine habitats. Most Apiospora species have rounded/lenticular conidia and inhabit mainly Poaceae (and many other host plant families) in a wide range of habitats, including tropical and subtropical regions (Pintos and Alvarado 2021; Samarakoon et al. 2022). An epitype for the type species of Apiospora, A.montagnei, was recently proposed by Pintos and Alvarado (2022).
There are many Apiospora species found on bamboos across the world (Table 2). Bamboos (Poaceae) are distributed in tropical and subtropical to mild temperate regions, with the heaviest concentration and largest number of species in China. Due to their abundance and economic importance, it is of great significance to study and identify the fungi growing on bamboo (Feng et al. 2021). In the present study, two new species (Apiosporadongyingensis and A.hainanensis) are introduced, and another one (A.pseudosinensis) is reported for the first time in China. All of them were collected from bamboo leaves and described based on their phylogenetic data and morphological characters. The descriptions and molecular data for species of Apiospora represent an important resource for understanding the diversity of bamboo fungi.
Supplementary Material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 31900014, U2002203 and 31750001).
Citation
Liu R, Li D, Zhang Z, Liu S, Liu X, Wang Y, Zhao H, Liu X, Zhang X, Xia J, Wang Y (2023) Morphological and phylogenetic analyses reveal two new species and a new record of Apiospora (Amphisphaeriales, Apiosporaceae) in China. MycoKeys 95: 27–45. https://doi.org/10.3897/mycokeys.95.96400
Funding Statement
This work was supported by the National Natural Science Foundation of China (no. 31900014, U2002203 and 31750001).
Contributor Information
Jiwen Xia, Email: xiajiwen1@126.com.
Yujiao Wang, Email: 18354285903@163.com.
Supplementary materials
Morphological and phylogenetic analyses reveal two new species and a new record of Apiospora (Amphisphaeriales, Apiosporaceae) in China
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Rongyu Liu, Duhua Li, Zhaoxue Zhang, Shubin Liu, Xinye Liu, Yixin Wang, Heng Zhao, Xiaoyong Liu, Xiuguo Zhang, Jiwen Xia, Yujiao Wang
Data type
phylogenetic
Explanation note
The combined ITS, LSU, tef1 and tub2 genes.
References
- Agut M, Calvo MA. (2004) In vitro conidial germination in Arthriniumaureum and Arthriniumphaeospermum. Mycopathologia 157(4): 363–367. 10.1023/B:MYCO.0000030432.08860.f3 [DOI] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91(3): 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Chen K, Wu XQ, Huang MX, Han YY. (2014) First report of brown culm streak of Phyllostachyspraecox caused by Arthriniumarundinis in Nanjing, China. Plant Disease 98(9): e1274. 10.1094/PDIS-02-14-0165-PDN [DOI] [PubMed]
- Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Peršoh D, Dhami MK, Alias AS, Xu J, Liu X, Stadler M, Hyde KD. (2014) The sooty moulds. Fungal Diversity 66(1): 1–36. 10.1007/s13225-014-0278-5 [DOI] [Google Scholar]
- Crous PW, Groenewald JZ. (2013) A phylogenetic re-evaluation of Arthrinium. IMA Fungus 4(1): 133–154. 10.5598/imafungus.2013.04.01.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeta MA, Echandi E, Abernethy T, Vilgalys R. (1991) Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology 81(11): 1395–1400. 10.1094/Phyto-81-1395 [DOI] [Google Scholar]
- Dai DQ, Jiang HB, Tang LZ, Bhat DJ. (2016) Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere 7(9): 1332–1345. 10.5943/mycosphere/7/9/7 [DOI] [Google Scholar]
- Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, Taylor JE, Hyde KD, Chukeatirote E. (2017) Bambusicolous fungi. Fungal Diversity 82(1): 1–105. 10.1007/s13225-016-0367-8 [DOI] [Google Scholar]
- Feng Y, Liu JK, Lin CG, Chen YY, Xiang MM, Liu ZY. (2021) Additions to the Genus Arthrinium (Apiosporaceae) From Bamboos in China. Frontiers in Microbiology 12: e661281. 10.3389/fmicb.2021.661281 [DOI] [PMC free article] [PubMed]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61(4): 1323–1330. 10.1128/aem.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LD, Hyde KD, Liew ECY. (2000) Identification of endophytic fungi from Livistonachinensis based on morphology and rDNA sequences. The New Phytologist 147(3): 617–630. 10.1046/j.1469-8137.2000.00716.x [DOI] [PubMed] [Google Scholar]
- Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson RA, Seifert KA, Taylor JW, Wingfield MJ, Abaci Ö, Aime C, Asan A, Bai F-Y, de Beer ZW, Begerow D, Berikten D, Boekhout T, Buchanan PK, Burgess T, Buzina W, Cai L, Cannon PF, Crane JL, Damm U, Daniel H-M, van Diepeningen AD, Druzhinina I, Dyer PS, Eberhardt U, Fell JW, Frisvad JC, Geiser DM, Geml J, Glienke C, Gräfenhan T, Groenewald JZ, Groenewald M, de Gruyter J, Guého-Kellermann E, Guo L-D, Hibbett DS, Hong S-B, de Hoog GS, Houbraken J, Huhndorf SM, Hyde KD, Ismail A, Johnston PR, Kadaifciler DG, Kirk PM, Kõljalg U, Kurtzman CP, Lagneau P-E, Lévesque CA, Liu X, Lombard L, Meyer W, Miller A, Minter DW, Najafzadeh MJ, Norvell L, Ozerskaya SM, Öziç R, Pennycook SR, Peterson SW, Pettersson OV, Quaedvlieg W, Robert VA, Ruibal C, Schnürer J, Schroers H-J, Shivas R, Slippers B, Spierenburg H, Takashima M, Taşkoin E, Thines M, Thrane U, Uztan AH, van Raak M, Varga J, Vasco A, Verkley G, Videira SIR, de Vries RP, Weir BS, Yilmaz N, Yurkov A, Zhang N. (2011) The Amsterdam declaration on fungal nomenclature. IMA Fungus 2(1): 105–112. 10.5598/imafungus.2011.02.01.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17(17): 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hyde K, Fröhlich J, Taylor J. (1998) Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia 50: 21–80. [Google Scholar]
- Hyde K, Norphanphoun C, Maharachchikumbura S, Bhat D, Jones E, Bundhun D, Chen Y, Bao D, Boonmee S, Calabon M. (2020) Refined families of Sordariomycetes. Mycosphere 11(1): 305–1059. 10.5943/mycosphere/11/1/7 [DOI] [Google Scholar]
- Jiang N, Fan XL, Tian CM. (2021) Identification and characterization of leaf-inhabiting fungi from Castanea plantations in China. Journal of Fungi 7(1): e64. 10.3390/jof7010064 [DOI] [PMC free article] [PubMed]
- Jiang N, Voglmayr H, Ma CY, Xue H, Piao CG, Li Y. (2022a) A new Arthrinium-like genus of Amphisphaeriales in China. MycoKeys 92: 27–43. 10.3897/mycokeys.92.86521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Voglmayr H, Xue H, Piao CG, Li Y. (2022b) Morphology and Phylogeny of Pestalotiopsis (Sporocadaceae, Amphisphaeriales) from Fagaceae Leaves in China. Microbiology Spectrum 10(6): 03272–22. 10.1128/spectrum.03272-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: Multiple sequence alignment,interactive sequence choice and visualization. Briefings in Bioinformatics 20(4): 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Molecular Biology and Evolution 33(7): 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G. (1817) Zehn neue Pilzgattungen. Mykol (1): 1–18.
- Kwon SL, Park MS, Jang S, Lee YM, Heo YM, Hong JH, Lee H, Jang Y, Park JH, Kim C, Kim GH, Lim YW, Kim JJ. (2021) The genus Arthrinium (Ascomycota, Sordariomycetes, Apiosporaceae) from marine habitats from Korea, with eight new species. IMA Fungus 12(1): 1–26. 10.1186/s43008-021-00065-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SL, Cho M, Lee YM, Kim C, Lee SM, Ahn BJ, Lee H, Kim JJ. (2022) Two unrecorded Apiospora species isolated from marine substrates in Korea with eight new combinations (A.piptatheri and A.rasikravindrae). Mycobiology 50(1): 46–54. 10.1080/12298093.2022.2038857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Cano C, Grey WE, Sands DC. (1992) First report of Arthriniumarundinis causing kernel blight on barley. Plant Disease 76(10): e1077. 10.1094/PD-76-1077B [DOI]
- Mavragani DC, Abdellatif L, McConkey B, Hamel C, Vujanovic V. (2007) First report of damping-off of durum wheat caused by Arthriniumsacchari in the semi-arid Saskatchewan fields. Plant Disease 91(4): e469. 10.1094/PDIS-91-4-0469A [DOI] [PubMed]
- Miller MA, Pfeiffer W, Schwartz T. (2012) The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment. Bridging from the extreme to the campus and beyond. Association for Computing Machinery 39: 1–8. 10.1145/2335755.2335836 [DOI] [Google Scholar]
- Mu TC, Zhang ZX, Liu RY, Liu SB, Li Z, Zhang XG, Xia JW. (2021) Morphological and molecular phylogenetic analyses reveal three species of Colletotrichum in Shandong province, China. MycoKeys 85: 57–71. 10.3897/mycokeys.85.75944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. (2004) MrModeltest v. 2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. (1998) Multiple evolutionary origins of the fungus causing panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95(5): 2044–2049. 10.1073/pnas.95.5.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintos Á, Alvarado P. (2021) Phylogenetic delimitation of Apiospora and Arthrinium. Fungal Systematics and Evolution 7(1): 197–221. 10.3114/fuse.2021.07.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintos Á, Alvarado P. (2022) New studies on Apiospora (Amphisphaeriales, Apiosporaceae): Epitypification of Sphaeriaapiospora, proposal of Ap.marianiae sp. nov. and description of the asexual morph of Ap.sichuanensis. MycoKeys 92: 63–78. 10.3897/mycokeys.92.87593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintos Á, Alvarado P, Planas J, Jarling R. (2019) Six new species of Arthrinium from Europe and notes about A.caricicola and other species found in Carex spp. hosts. MycoKeys 49: 15–48. 10.3897/mycokeys.49.32115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos HP, Braun GH, Pupo MT, Said S. (2010) Antimicrobial activity from endophytic fungi Arthrinium state of Apiosporamontagnei Sacc. and Papulasporaimmersa. Brazilian Archives of Biology and Technology 53(3): 629–632. 10.1590/S1516-89132010000300017 [DOI]
- Rayner RW. (1970) A Mycological Colour Chart. CMI and British Mycological Society, Kew.
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccardo P. (1875) Conspectus generum pyrenomycetum italicorum additis speciebus fungorum Venetorum novis vel criticis, systemate carpologico dispositorum. Atti della Società Veneto-Trentina di Scienze Naturali 4: 77–100. [Google Scholar]
- Samarakoon MC, Hyde KD, Maharachchikumbura SS, Stadler M, Gareth Jones EB, Promputtha I, Suwannarach N, Camporesi E, Bulgakov TS, Liu JK. (2022) Taxonomy, phylogeny, molecular dating and ancestral state reconstruction of Xylariomycetidae (Sordariomycetes). Fungal Diversity 112(1): 1–88. 10.1007/s13225-021-00495-5 [DOI] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Karunarathna SC, Mapook A, Promputtha I, Xu J, Bao D, Tibpromma S. (2021) One new species and two new host records of Apiospora from bamboo and maize in northern Thailand with thirteen new combinations. Life 11(10): e1071. 10.3390/life11101071 [DOI] [PMC free article] [PubMed]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172(8): 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tan XM, Liu F, Cai L. (2018) Eight new Arthrinium species from China. MycoKeys 1: 1–24. 10.3897/mycokeys.39.27014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphological and phylogenetic analyses reveal two new species and a new record of Apiospora (Amphisphaeriales, Apiosporaceae) in China
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Rongyu Liu, Duhua Li, Zhaoxue Zhang, Shubin Liu, Xinye Liu, Yixin Wang, Heng Zhao, Xiaoyong Liu, Xiuguo Zhang, Jiwen Xia, Yujiao Wang
Data type
phylogenetic
Explanation note
The combined ITS, LSU, tef1 and tub2 genes.