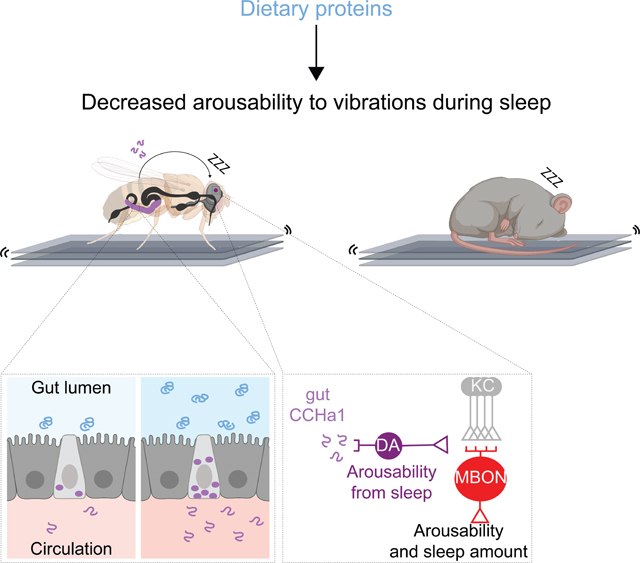
SUMMARY
Suppressing sensory arousal is critical for sleep, with deeper sleep requiring stronger sensory suppression. The mechanisms which enable sleeping animals to largely ignore their surroundings are not well understood. We show that the responsiveness of sleeping flies and mice to mechanical vibrations is better suppressed when diet is protein-rich. In flies we describe a signaling pathway through which information about ingested proteins is conveyed from the gut to the brain to help suppress arousability. Higher protein concentration in the gut leads to greater activity of enteroendocrine cells which release the peptide CCHa1. CCHa1 signals to a small group of dopamine neurons in the brain to modulate their activity; the dopaminergic activity regulates the behavioral responsiveness of animals to vibrations. The CCHa1 pathway and dietary proteins do not influence responsiveness to all sensory inputs, showing that during sleep different information streams can be gated through independent mechanisms.
Graphical Abstract
INTRODUCTION
Sleep originated early in animal evolution and has comparable characteristics across species1. Both the amount and quality of sleep are important for health2, with sleep quality depending on the timing, latency to fall asleep, frequency and duration of awakenings, and sleep depth3,4. Deep sleep, which is particularly restorative5 requires greatest sensory suppression such that most environmental signals are prevented from arousing the brain6,7. People who are awoken too easily often feel unrestored even after spending sufficient time asleep5. The mechanisms by which arousability is suppressed during sleep have not been well characterized in any model8–13.
Sleep is strongly influenced by genetics14 and multiple sleep-regulating genes have been identified using model organisms like flies15,16. Outside factors including nutrition are also important – while epidemiological studies cannot establish causation, they reveal correlation between the consumption of certain foods and changes in sleep17–19. Flies have been especially useful for studying the mechanisms through which nutrition impacts sleep but most of the work has been done in the context of starvation20, when sleep is potently suppressed. It is unclear how nutrition influences sleep in normal, non-starvation conditions.
From a screen for genes that regulate arousability from sleep we found a signaling pathway through which information about dietary proteins is conveyed from the gut to the brain, to help suppress sensory responsiveness. Ingestion of proteinaceous foods activates secretory cells in the fly gut which release the peptide CCHa1; CCHa1 signals to a group of dopamine neurons in the brain whose activity modulates responsiveness to mechanical vibrations. Higher concentration of proteins in the gut leads to higher CCHa1 levels, which enables flies to sleep through stronger vibrations, while CCHa1 depletion from the gut causes easy awakenings even when vibrations are weak. We show that mice are also less responsive to vibrations, and have better-consolidated sleep, on proteinaceous food. These results are consistent with the observations in humans which indicate that dietary proteins can suppress sensory responsiveness to promote deeper sleep21. While the CCHa1 pathway powerfully modulates responsiveness to vibrations, it does not impact responsiveness to thermal inputs, demonstrating that different sensory modalities can be gated through separate mechanisms during sleep.
RESULTS
A screen for arousal threshold mutants
We studied sensory arousability in Drosophila using a high-throughput automated setup that produces mechanical vibrations (Figures S1A,B and STAR Methods). In the setup, loudspeakers convert an electrical signal into 2-second-long vibrations every 30–40 minutes throughout the night (randomized to minimize habituation), causing the locomotor activity monitors with flies to move up and down. Flies which started moving within 3 minutes were considered awoken specifically by the stimulus (STAR Methods, spontaneous awakenings were taken into account).
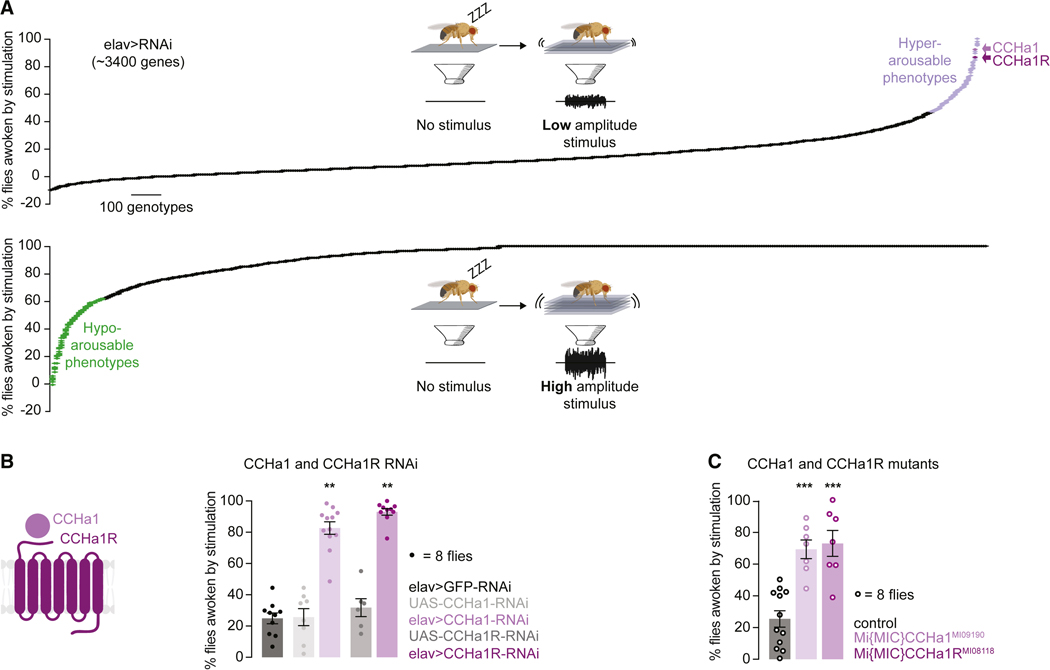
Using vibration intensities that consistently woke up many (~20%) or few (~95%) control flies, we screened for genetic modifiers of arousability (Figure 1A and STAR Methods). RNAis against ~3,400 genes (STAR Methods) were expressed using a pan-neuronal driver elav-Gal422 and 8 animals were used to generate each data point. The screen uncovered both hyper- and hypo-arousable phenotypes (Figure 1A and Table S1) which mapped to genes involved in diverse biological and molecular functions (Table S2). It is unsurprising that multiple pathways regulate arousability considering how profoundly the entire body is transformed by sleep.
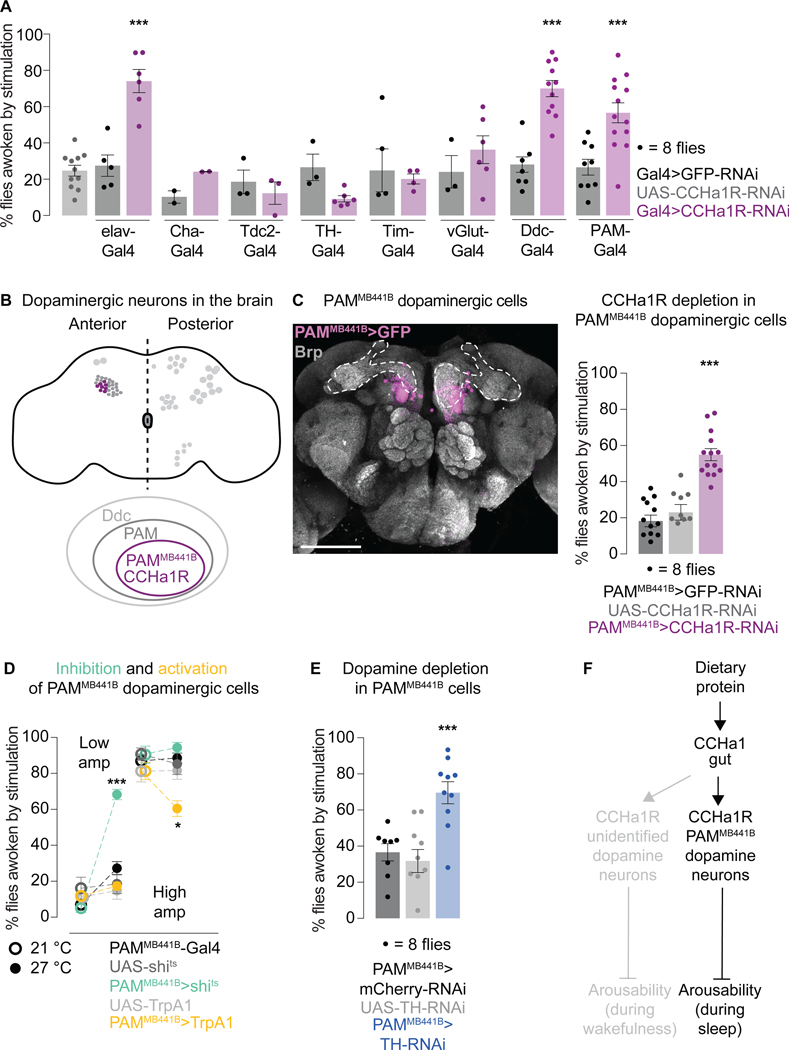
Figure 1. CCHa1 and its receptor suppress arousability from sleep.
(A) Schematized screen for genes that regulate arousability from sleep, using weak (top) and strong (bottom) mechanical vibrations. Hyper- and hypo-arousable phenotypes exceeding two standard deviations from the mean are highlighted (data in Tables S1 and S2). Data were normalized for spontaneous arousals from sleep, and negative values are seen when spontaneous arousals are more frequent than arousals after stimulation. (B) Percent of flies awoken by weak mechanical vibrations when RNAis against CCHa1 or its receptor are expressed using elav-Gal4. (C) Mutants for CCHa1 or its receptor. In B and C grays represent parental controls. Error bars, mean and S.E.M. Genotypes, sample sizes and statistical analyses, Table S3. See also Figure S1. In all figures * p<0.05, ** p<0.01, *** p<0.001.
A peptide and its receptor regulate arousability
Among strong hyper-arousability phenotypes were those produced when the neuropeptide CCHa1 or its receptor CCHa1R were depleted (Figure 1A), with ~85% of the animals waking up in response to the lower intensity stimulation (Figure 1B). CCHamides (CCHa1 and CCHa2) regulate olfactory responsiveness and circadian behavior (CCHa1) as well as feeding and growth regulation (CCHa2)23–28. The receptor for CCHa1 is homologous to the mammalian G protein-coupled receptor gastrin releasing peptide receptor (GRPR)28 which regulates itch sensation29, social interactions30,31, fear memory consolidation32–34, breathing35, food intake30,36,37 and circadian rhythms38–40. The specificity of RNAi for CCHa1 and CCHa1R (and not CCHa2 and CCHa2R) was shown with quantitative RT-PCR (Figure S1C).
Like many animals41, flies spontaneously wake up for brief periods42,43 before going back to sleep44. Some flies happened to be awake when vibrations were delivered and we analyzed their behavior (Figure S1D); animals depleted of CCHa1 or CCHa1R were more likely to increase their locomotion in response to vibrations, without being more active at baseline than the controls (Figure S1D and STAR Methods). Sleep is an extreme state of sensory suppression, but during wakefulness animals can also tune the sensory world out to varying degrees45. Our observations suggest that CCHa1 can suppress sensory arousability across sleep-wake states, similarly to the best-known regulator of arousal, dopamine46,47. We will later show that CCHa1 acts through different populations of dopaminergic cells to suppress sensory arousability when the animal is asleep vs awake. Knocking down CCHa1 or its receptor also affected several other parameters: sleep appeared more fragmented (evidenced by a slight decrease in the duration of individual sleep bouts and an increase in their frequency) (Figure S1D); latency to sleep was increased as previously described for CCHa1 depletion28; flies slept less when the CCHa1 receptor was depleted (Figure S1D). We will later explain how these different aspects of sleep can be regulated by the same peptide. The above results were confirmed with additional RNAi lines (Figures S1E and F) and mutants48 (Figures 1C and S1G). Post-developmental depletion was sufficient to cause hyper-arousability (Figure S1H), and we conclude that CCHa1 signaling likely plays an ongoing role in suppressing sensory arousal during sleep.
CCHa1 signaling originates in the gut
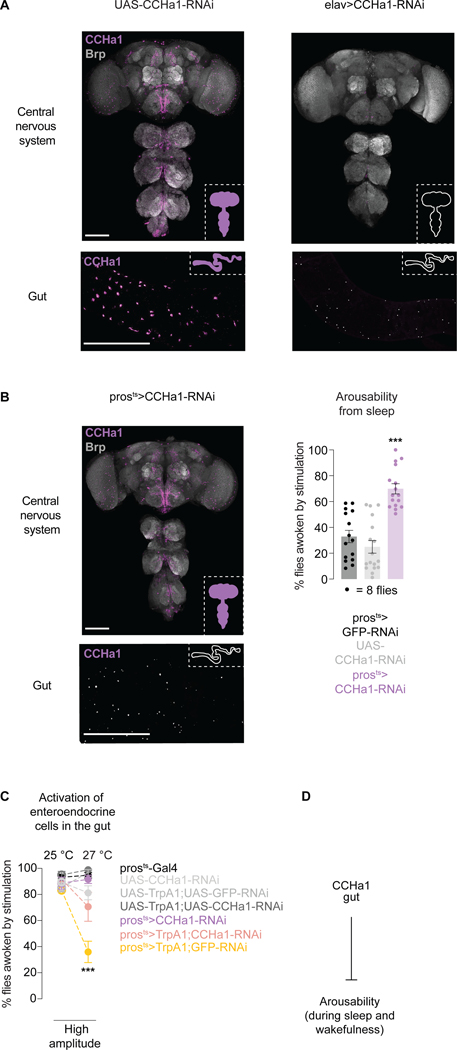
To find the cells in which CCHa1 originates, we raised antibodies against a previously validated epitope49 and expressed RNAis against the peptide using spatially-restricted Gal4s. CCHa1 has been detected in the nervous system28 and in a posterior subpopulation of enteroendocrine cells49, secretory cells sparsely distributed throughout the gut50. The observed signal was consistent with previous reports28,49,51,52 (Figures 2A and S2A) and matched the pattern of GFP expressed from the CCHa1 promoter (Figure S2B, heterozygotes left).
Figure 2. CCHa1 peptide signals from the gut to suppress arousability.
(A) Antibody staining against CCHa1 in the nervous system and the posterior midgut, in control animals (UAS-CCHa1-RNAi) and animals in which CCHa1 is depleted using elav-Gal4 (elav>CCHa1-RNAi). (B) CCHa1 staining, and arousability from sleep, when CCHa1 is conditionally depleted using pros-Gal4 (prosts>CCHa1-RNAi). In A and B, scale bars: 100 μm. (C) Arousability from sleep when enteroendocrine cells are activated (yellow), or when they are activated and simultaneously depleted of CCHa1 (pink). UAS-GFP was included to control for UAS number. (D) Schematic. Error bars, mean and S.E.M. Genotypes, sample sizes and statistical analyses, Table S3. See also Figure S2.
CCHa1 protein and mRNA were undetectable in the nervous system, and in the gut, in elav>CCHa1-RNAi flies and in CCHa1 mutants (Figures 2A, S2B homozygotes right and S2C). Neurons and enteroendocrine cells share developmental programs50, perhaps explaining why elav-Gal4, the classic pan-neuronal driver which we used in the screen, is expressed in both cell types53 (Figure S2D). We were surprised to see that depleting CCHa1 only from the gut (using a conditional version of the standard enteroendocrine driver pros-Gal4, prosts) (Figures 2B and S2E,F)54 was sufficient to cause increased arousability (Figures 2B and S2G). Unlike what we observed with elav-Gal4, there was no effect on the number of sleep episodes or latency to fall asleep (Figure S2G), indicating that CCHa1 produced in the nervous system impacts sleep consolidation and timing, while CCHa1 produced in the gut regulates arousability. We did not see any effects on feeding or food clearance when CCHa1 was depleted from the gut (Figure S2H) suggesting that the effect of CCHa1 on arousability is not due to changes in food processing. As a long-lasting state, sleep seems ideally suited for peptidergic regulation since peptides act on slower timescales than classical neurotransmitters55. It is difficult to disentangle the various functions of one peptide56, but in the case of CCHa1 gut-specific depletion affords an opportunity to study specifically how the peptide suppresses sensory arousal and promotes deeper sleep.
Secretory cells in the gut regulate arousability
The above results imply that changing the activity of enteroendocrine cells, which are electrically excitable57,58, should impact arousability. Expressing neuronal silencing tools using the enteroendocrine driver pros-Gal4 caused developmental lethality (even with conditional tools59, possibly due to leaky expression), but tools for neuronal activation could be tested. Thermogenetic activation of enteroendocrine cells using a temperature-sensitive cation channel TrpA160 led to suppressed arousability in both sleeping (Figure 2C, yellow) and awake (Figure S2I) animals. Pros-Gal4 is expressed in some neurons in addition to enteroendocrine cells (Figure S2F), so we asked to what degree the phenotype is mediated by the gut-produced CCHa1 peptide. When pros-Gal4 was used to simultaneously activate cells and deplete them of CCHa1, much of the effect on arousability was suppressed (Figure 2C and Figure S2I, pink compared to yellow). Another phenotype which is produced by activating pros-Gal4-expressing cells (slightly less sleep, Figure S2I) was however not suppressed by CCHa1 depletion (Figure S2I), suggesting that it originates either in non-CCHa1 enteroendocrine cells, or in some of the neurons labeled by pros-Gal4 (Figure S2F). The results shown so far suggest that gut-released CCHa1 suppresses arousability (Figure 2D).
Dietary proteins activate enteroendocrine cells and regulate CCHa1 signal
The origin of the CCHa1 signal raised the possibility that nutritional cues are involved in its regulation. Details are emerging in flies about the mechanisms through which nutrition influences sleep61,63, but most of the studies have been done under starvation conditions, when animals sleep less. There were no changes in sleep amount with gut-specific CCHa1 depletion (Figure S2G), indicating involvement of yet unknown mechanisms which operate under standard feeding conditions. Enteroendocrine cells found in the posterior midgut are activated by dietary proteins and amino acids58 and we asked if this is true for the CCHa1-producing subset as well, while also testing for potential effects of fats and sugars.
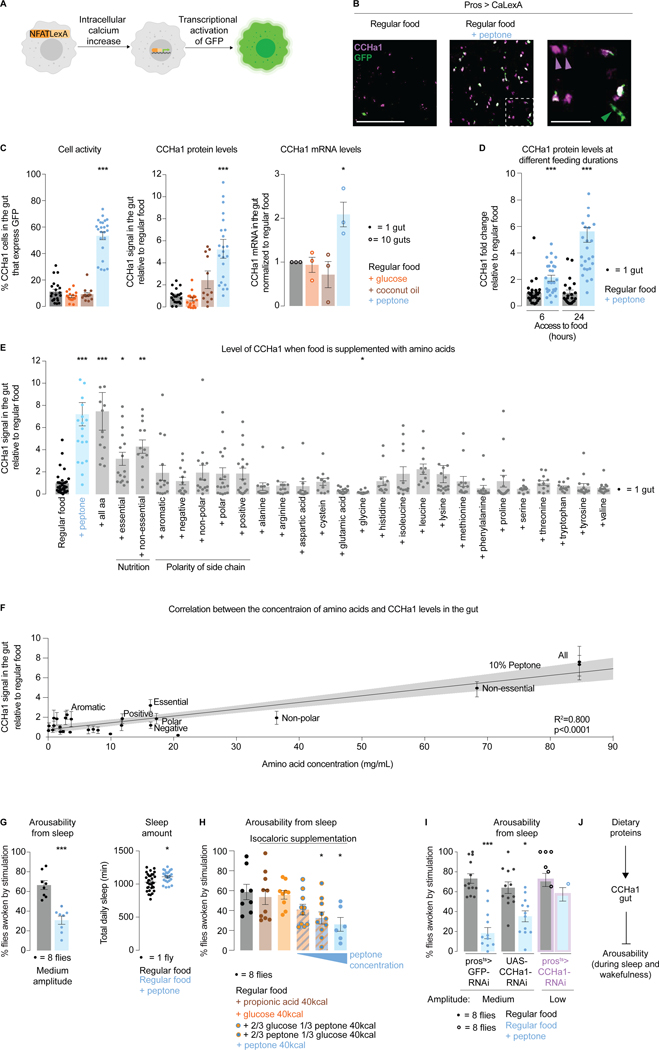
Imaging a living gut is challenging for many reasons including contractility of the organ, and potential obscurement of the epithelium by the luminal contents. CaLexA62 is a tool which relies on transcriptional changes and allows the activity of gut cells to be recorded in freely behaving animals. Increase in intracellular calcium levels, such as occurs when cell activity is increased, promotes expression of GFP which is stable for up to 24 hours64; longer or more frequent cell activation results in more accumulated GFP (Figure 3A). This tool has been shown to work in enteroendocrine cells58 in addition to in neurons65,66. CaLexA was expressed with pros-Gal454 and CCHa1-positive cells were identified through antibody staining. The percentage of cells co-expressing GFP (which indicates cell activation) and CCHa1 was quantified 24 hours after supplementing regular food with sugar, fat, or protein at several concentrations based on previous reports67–69 (STAR Methods). Three sources of sugar (glucose, galactose, fructose) and fat (coconut oil, propionic acid, hexanoic acid) were tested, while for protein supplementation we used peptone, a partially hydrolyzed dietary protein mix containing amino acids and peptides (STAR Methods).
Figure 3. CCHa1-expressing enteroendocrine cells are activated by dietary proteins and promote deeper sleep.
(A) Schematized CaLexA tool. (B) A representative image showing the effect of protein supplementation on the levels of CaLexA-dependent GFP, and CCHa1, in enteroendocrine cells. Area outlined in the second panel in magnified in the third panel. White asterisks: activated CCHa1-producing cells. Magenta arrows: non-activated cells that produce CCHa1. Green arrows: activated cells that do not produce CCHa1. Scale bars: 100 μm and 25μm (zoom-in). (C) Quantification of enteroendocrine cell activity, and their levels of CCHa1 protein and mRNA. (D) CCHa1 protein levels after 6 or 24 hours of peptone supplementation. (E) CCHa1 levels in the gut when food is supplemented with individual or combined amino acids (at their respective concentrations in peptone). (F) CCHa1 levels relative to supplemented amino acid concentration. (G) Effect of dietary proteins on arousability from sleep, and total sleep amount. (H) Effect of equicaloric supplementation with different macronutrients on arousability from sleep. (I) Arousability from sleep on regular food (gray) vs peptone-supplemented food (blue), in controls (prosts>GFP-RNAi and UAS-CCHa1-RNAi) and in flies with gut-depleted CCHa1 (prosts>CCHa1-RNAi, purple outline). (J) Schematic. For (C , D, and E), normalized to regular food (STAR Methods). Error bars, mean and S.E.M. Genotypes, sample sizes and statistical analyses, Table S3. See also Figure S3.
The activity of CCHa1-positive enteroendrocrine cells, and the levels of CCHa1 in these cells, were increased specifically with peptone supplementation (Figures 3B, 3C, and S3A). This does not seem to be due to calories since for every 100mL of regular food, peptone supplementation added 40kcal compared to 14kcal for sugar and 80kcal for fat supplementation (STAR Methods). When flies had access to a protein-supplemented diet for 24 hours, CCHa1 levels increased more than when the diet was available for 6 hours only (Figure 3D). This suggests that enteroendocrine cells can discriminate between long- and short-term dietary changes, although we cannot exclude the possibility that the dynamics of GFP production and degradation play a role. Peptone supplementation did not overtly affect food consumption or clearance from the gut (Figure S3B). We note that some of the posterior midgut cells had high CCHa1 levels without showing evidence of activation (Figure 3B, magenta arrowheads), and that some of the activated cells were CCHa1-negative (Figure 3B, green arrowhead). To test whether specific amino acids are relevant, we supplemented them at their respective peptone concentrations (alone or in combinations, STAR Methods). CCHa1 levels were increased only if the total amino acid concentration matched what is present in peptone (Figures 3E,F). This result suggests that CCHa1-producing enteroendocrine cells monitor how proteinaceous the food is overall, as well as implies that enrichment of dietary proteins should promote sensory disconnect and deeper sleep.
To test this idea, we compared responsiveness of animals fed regular and protein-rich food using a stimulus of intermediate intensity (a setting which allows alterations of arousability in either direction to be detected). Half as many flies woke up in response to vibrations if their diet was supplemented with peptone for 24 hours, compared to flies fed regular diet (Figure 3G). Peptone-supplemented animals were also slightly less responsive if awake at the time of stimulation (Figure S3C). We note that according to statistical analysis there was an effect on basal locomotor activity (Figure S3C) and sleep amount (Figure 3G) with protein supplementation, but the phenotypes were very weak and we are unsure of their biological significance.
Even though sugar and fat supplementation did not affect the levels of CCHa1 or the activity of CCHa1-producing enteroendocrine cells, we tested for their possible influence on behavioral responsiveness. Consistent with their effect on CCHa1 levels, glucose or propionic acid supplementation had no effect on how the animals responded to mechanical stimulation (Figure S3D). To additionally control for the effect of calories vs specific nutrients we made the supplementation equicaloric - in each case extra 40 kcal were provided but of different origins (either from propionic acid, glucose, peptone or from glucose-peptone combinations (2/3 calories from glucose + 1/3 from peptone, or 1/3 from glucose + 2/3 from peptone). Arousability was most potently suppressed when the majority of added calories originated from dietary proteins (Figures 3H and S3E). Based on these results we conclude that the presence of dietary proteins, and not just the caloric content, is a main factor that influences arousability.
We asked to what extent CCHa1 mediates the effects of dietary proteins on arousal by providing protein supplementation to flies lacking CCHa1 in the gut. Because CCHa1 knockdown itself causes hyper-arousability, lower amplitude stimulation had to be used for these animals in order to match their baseline (i.e. their arousability on regular food) with the parental controls; (the opposite conditions, where low intensity stimulation is used for the parental controls and medium intensity stimulation is used for the CCHa1-depleted animals were also tested (Figure S3F,G). CCHa1 depletion weakened the effect of peptone on arousal (Figures 3I and S3F), though incomplete phenotype suppression reinforces the idea that dietary proteins activate multiple signaling pathways. Taken together, our results show that ingested proteins activate CCHa1-producing cells in the gut to suppress arousability (Figure 3J).
Dietary proteins promote sensory disconnect in mammals
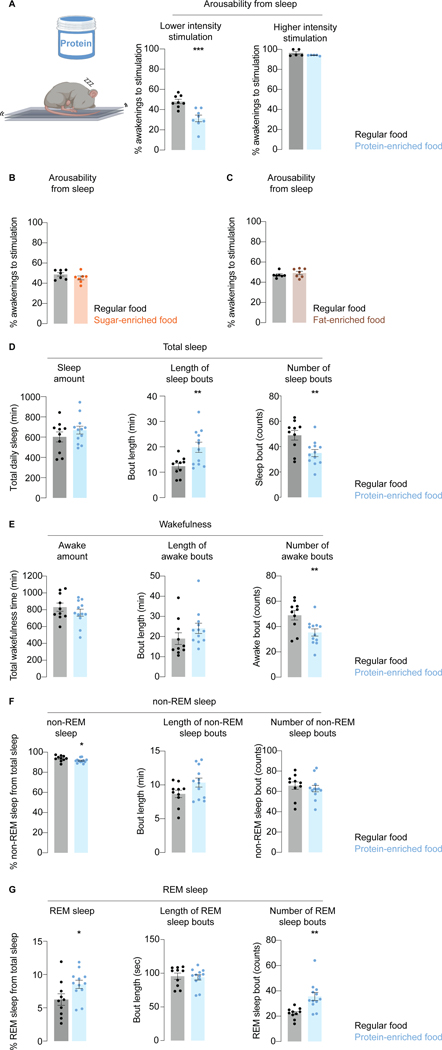
Using mice as a model we assessed the influence of dietary proteins on arousability in mammals. Individual animals were housed in sleep-recording chambers atop a platform, with shakers providing mechanical stimulation of adjustable intensity from below (STAR Methods). For two consecutive days stimulations were provided at randomized intervals every 50–70 minutes during the light phase of the circadian cycle, which is when mice sleep the most70. Responsiveness was assessed within 5 minutes of stimulus delivery using electrical activity recordings in the brain (electroencephalogram, EEG) and muscles (electromyogram, EMG). Video recordings were used for confirmation. Only events where the animal was asleep immediately prior to stimulus delivery were considered, and data were normalized for spontaneous awakenings (STAR Methods).
After determining the stimulus intensity required to wake mice up approximately 50% of the time, we asked if the responsiveness was influenced by diet. For five days prior to testing mice were fed either standard or protein-enriched diets of similar caloric values (STAR Methods) which did not affect the body weight (Figure S4A–C). In each experiment two chambers were on a single platform, one with a mouse fed regular diet and the other protein-enriched diet. Similar to flies, mice on a protein-enriched diet were harder to wake up (Figure 4A); they readily responded however when stimulation was of high intensity, ruling out general impairment (Figure 4A). No changes in arousability were seen when food was enriched with sugar (Figure 4B) or fat (Figure 4C).
Figure 4. Dietary proteins suppress arosuability from sleep in mice.
(A) Arousability from sleep in response to weak and strong vibrations in mice fed regular or protein-enriched food (equicaloric). (B,C) Arousability from sleep in response to weak vibrations in mice fed regular food, vs sugar-enriched (B) or fat-enriched (C) food (equicaloric). (D-G) Effect of dietary protein enrichment on various sleep parameters. Error bars, mean and S.E.M. Sample sizes and statistical analyses, Table S3. See also Figure S4.
Enrichment of dietary proteins had no effect on the overall sleep amount (Figure 4D), but it led to better-consolidated sleep (longer bouts of sleep that were interrupted by fewer awakenings) (Figures 4D,E and S4D). Mammalian sleep consists of rapid eye movement (REM) and non-REM phases, with non-REM sleep comprising three stages71. As characterized by decreased sensory arousability, the depth of sleep is greatest during the last stage of non-REM sleep, and REM sleep72,73. The number of REM sleep episodes was increased in animals which consumed more protein, leading to increased total time spent in the REM phase (Figures 4F,G and S4D). In addition to defining sleep depth based on behavioral responsiveness, this is also done based on the pattern of brain electrical activity as measured by EEG74. It is unclear however to what degree the EEG pattern shapes sensory responsiveness, since animals are difficult to arouse during REM sleep despite brain activity appearing awake-like75. To assess the effect of different diets on brain activity during sleep and wakefulness, we performed an EEG power analysis focusing on the lower frequency theta and delta brain waves (associated with relaxation and sleep respectively76). There were no overt effects of dietary proteins (Figure S4E,F), implying that sensory responsiveness can be regulated by mechanisms which are independent from the mechanism that regulate broad changes in brain states observed during sleep. The data were acquired with surface electrodes and we cannot discard the possibility that there are changes in some specific areas from which we did not record.
Gut-produced CCHa1 signals to dopaminergic neurons in the brain
We returned to flies to study how a gut-derived signal influences sleep. Peptides produced by the enteroendocrine cells can signal synaptically when in contact with dendritic terminals or hormonally through circulation77. Cells that produce CCHa1 reside in a poorly innervated region of the Drosophila gut50, suggesting hormonal action. To find where the peptide is received to regulate arousability, we depleted its receptor in sparse neuronal populations and tested responsiveness to vibrations. Flies were not hyper-arousable when the CCHa1 receptor was depleted from cholinergic (Cha-Gal4), octopaminergic (Tdc2-Gal4), glutamatergic (vGlut-Gal4), or circadian (Tim-Gal4) cells (Figure 5A). Though one dopaminergic driver (TH-Gal4) produced no phenotype, another one (Ddc-Gal4) led to increased responsiveness both during sleep (Figure 5A) and wakefulness (Figure S5A). Ddc-Gal4 labels the PAM cluster of dopaminergic neurons (80–90 neurons/brain hemisphere) while TH-Gal4 does not78, which suggests that these are the relevant cells. The PAM cluster is labeled by two different Gal4s expressed under the control of CCHa1R regulatory elements (Figure S5B), supporting the idea that the receptor is expressed here.
Figure 5. CCHa1 regulates arousability through dopaminergic neurons in the brain.
(A) Arousabilty from sleep when the CCHa1 receptor is depleted with various Gal4 drivers. (B) Schematized dopaminergic neurons in the fly brain, and narrowing down of the population to which CCHa1 signals. (C) PAMMB441B-Gal4 expression in the brain is visualized with membrane-targeted GFP. Presynaptic protein Brp marks the neuropil, allowing visualization of the mushroom body (dashed outline). Scale bar: 100 μm. Arousabilty from sleep when the CCHa1 receptor is depleted from PAMMB441B neurons (PAMMB441B>CCHa1-RNAi). (D) Arousability from sleep when PAMMB441B neurons are inhibited (green) or activated (yellow). (E) Arousability from sleep when tyrosine hydroxylase (TH) is depleted from PAMMB441B neurons (PAMMB441B>TH-RNAi). (F) Schematic summary. Error bars, mean and S.E.M. Genotypes, sample sizes and statistical analyses, Table S3. See also Figure S5.
Expressing RNAi against the CCHa1 receptor only in the PAM dopaminergic cluster was sufficient to make flies more arousable (Figure 5A) without affecting basal locomotion or sleep amount (Figure S5A). This implies that the various regulatory roles of the receptor are uncoupled in different neuronal populations, similarly to CCHa1 itself (Figures 2B and S2G). We note that the effect on awake responsiveness was not statistically significant with PAM-Gal4-driven CCHa1R-RNAi (Table S3), but qualitatively it appeared similar to what was seen with Ddc-Gal4 (Figure S5A). Regardless, we show below that distinct populations of dopamine neurons mediate the effects of CCHa1 on arousability during sleep and wakefulness.
Subpopulations of PAM dopaminergic neurons can be accessed thanks to a large-scale mapping and tool-generating effort79, so we further narrowed down the CCHa1 target cells. Flies were easier to wake up with vibrations if the CCHa1 receptor was depleted using PAMMB441B-Gal4 (Figure 5B,C). This driver does not label the gut (Figure S5C), and in each brain hemisphere it labels 8–10 cells (Figure 5B,C)79 which express the CCHa1 receptor according to transcriptional profiling data80. Although we cannot rule out involvement of other neurons from the PAM cluster, we focused on the PAMMB441B population because a strong phenotype maps to this small group of cells. When the CCHa1 receptor was depleted from PAMMB441B neurons, the consumption of food and its clearance from the gut were unaffected (Figure S5D), as was total sleep amount (Figure S5E), while basal locomotion was slightly decreased (Figure S5E). The responsiveness of awake animals was unaffected (Figures S5E), which was also true when the activity of PAMMB441B neurons was manipulated (Figures 5D and S5F), and when Tyrosine Hydroxylase (TH, the rate-limiting enzyme in dopamine synthesis) was depleted (Figures 5E and S5G). Based on these observations, CCHa1 is received by another subpopulation of dopaminergic neurons (which we have not identified) to suppress arousal during awake periods (Figure 5F). The results of TH manipulation argue that the effect of PAMMB441B neurons on arousal is mediated by dopamine itself. This molecule is canonically thought of as arousal-promoting46,81–83, but our data suggest that dopamine can act on specific synapses to suppress arousal, consistent with some previous observations8.
Dietary proteins activate dopaminergic neurons in the brain
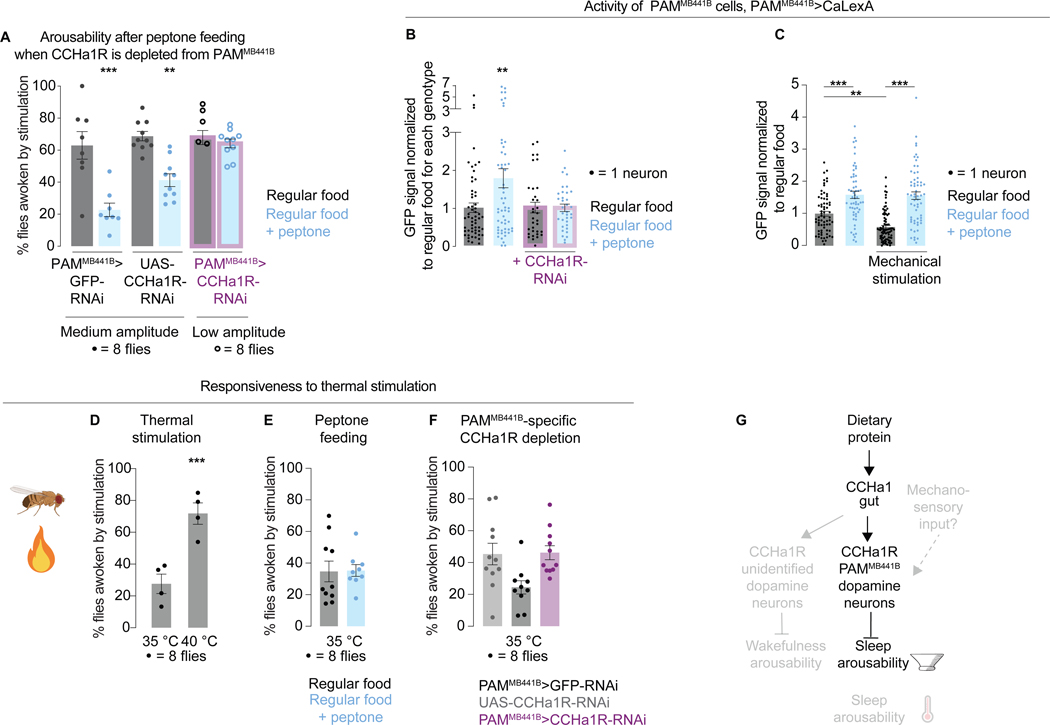
If PAMMB441B neurons are targeted by CCHa1, depleting them of the CCHa1 receptor should diminish the ability of dietary proteins to suppress arousability during sleep. This prediction was tested and validated (Figures 6A and S6A,B). Since depleting the CCHa1 receptor causes hyper-arousability, the same strategy was used as in Figure 3I, where the parental controls were exposed to medium intensity stimulation and the receptor-depleted animals were exposed to low intensity stimulation to match baseline arousabilities; the opposite conditions were also tested (Figure S6B). Peptone supplementation resulted in more PAMMB441B neurons showing evidence of activation (Figure 6B), an effect that was suppressed with PAMMB441B-specific depletion of the CCHa1 receptor (Figure 6B), supporting the idea of direct gut-to-brain signaling. Protein feeding did not cause changes in the activity of another dopaminergic population (Figure S6C), suggesting that the increase in PAMMB441B neuronal activity was not an artifact of protein feeding.
Figure 6. Dopaminergic activity is influenced by dietary proteins and CCHa1. Different sensory modalities can be gated independently.
(A) (I) Arousability from sleep on regular food (gray) vs peptone-supplemented food (blue), in controls (PAMMB441B>GFP-RNAi and UAS-CCHa1R-RNAi) and in flies with the CCHa1 receptor depleted from PAMMB441B neurons (PAMMB441B >CCHa1-RNAi, purple outline). (B) Activity of PAMMB441B neurons on regular vs protein-supplemented food. Purple outline, flies in which the CCHa1 receptor is depleted from PAMMB441B neurons. (C) Activity of PAMMB441B neurons on regular vs protein-supplemented food, with or without exposure to mechanical vibrations. (D) Arousability from sleep in response to heat. (E) Arousability by heat on regular vs peptone-supplemented food. (F) Arousability by heat in controls vs animals with the CCHa1 receptor depleted from PAMMB441B neurons. (G) Schematic. Error bars, mean and S.E.M. Genotypes, sample sizes, and statistical analyses, Table S3. See also Figure S6.
Sensory gating can be modality-specific
Our results show that a physiological signal can affect the activity of dopaminergic PAM neurons, but it is unclear from this where the sensory inputs intersect with the system. PAM neurons project to the mushroom body, a brain structure which receives sensory inputs from different sensory modalities84–90 and controls sleep amount, feeding, learning, memory, and olfaction91–96,98. Dopaminergic neurons modulate synapses between Kenyon cells and the output neurons to contextualize and drive appropriate behavioral responses91,97. Kenyon cells carry olfactory and visual information89,90,99, but there is no evidence yet that they receive mechanical inputs. The larval100,101 and adult102 PAM neurons are on the other hand known to be recipients of mechanosensory information. To estimate if and how the PAMMB441B neuronal subset responds to mechanical stimulation, we expressed CaLexA in these cells and exposed flies to a night of vibrations using the same protocol as for testing arousability (Figure S6D). The GFP signal was quantified in the PAMMB441B cell bodies 4 hours after stimulation (sufficient time for detecting GFP increase in case neurons are activated) as well as 28 hours after stimulation (sufficient time for GFP degradation in case the neuronal activity is reduced) (Figure S6D)64. CaLexA reported lower PAMMB441B neuronal activity in animals that were exposed to vibrations (Figure S6D), opposite to the effect induced by a protein-rich diet. To better understand the interaction of sensory and nutritional information, we first fed flies protein-rich food for 24 hours and then exposed them to vibrations; protein supplementation suppressed the ability of vibrations to reduce PAMMB441B neuronal activity (Figure 6C). Based on these results, it seems that information about external and internal circumstances converges on a small group of dopaminergic neurons, exerting opposite influences on the activity of these neurons. Since the level of behavioral responsiveness depends on dopaminergic activity (Figure 5D,E) this would essentially constitute a sensory gate. Given the feedback loops within the mushroom body100,103 and the fact that mushroom body cells respond to movement, we cannot discard the possibility that the changes in dopaminergic activity elicited by vibrations are also influenced by the locomotor response itself104.
During sleep all sensory inputs are attenuated62 which could occur through a central gating mechanism. In mammals sensory gating is thought to occur in the thalamus, an area through which most sensory information passes before reaching the cortex105,106. Olfactory information however gets attenuated despite bypassing the thalamus107, suggesting more specialized forms of regulation and the possibility that different sensory modalities are gated through at least partially independent mechanisms. We tested this possibility by building a setup in which heat is used instead of vibrations to wake flies up (Figures 6D and S6E and STAR Methods). Different levels of behavioral responsiveness were detected depending on the stimulus intensity (Figure 6D, 35 vs 40°C, both stimuli are perceived as hot by the fly108), but the activity of PAMMB441B neurons was unaffected by either temperature (Figure S6D). While CaLexA might have technical limitations, it could also be that thermal and mechanosensory inputs are attenuated through separate circuitry. This possibility is likely since the responsiveness of flies to heat was unaffected by either peptone supplementation or by depletion of the CCHa1 receptor from the PAMMB441B neurons (Figures 6E, F and S6F,G), in contrast to what was seen with vibrations. Although we only examined specific conditions, we conclude that thermal and mechanosensory information can be gated independently (Figure 6G), consistent with the electron microscopy data indicating that temperature information flows through the α’/β’ lobes of the mushroom body103, while the CCHa1-receiving dopamine neurons innervate the γ lobe (specifically the γ3 compartment) 79.
Mushroom Body Output Neurons might integrate information about arousability and sleep amount
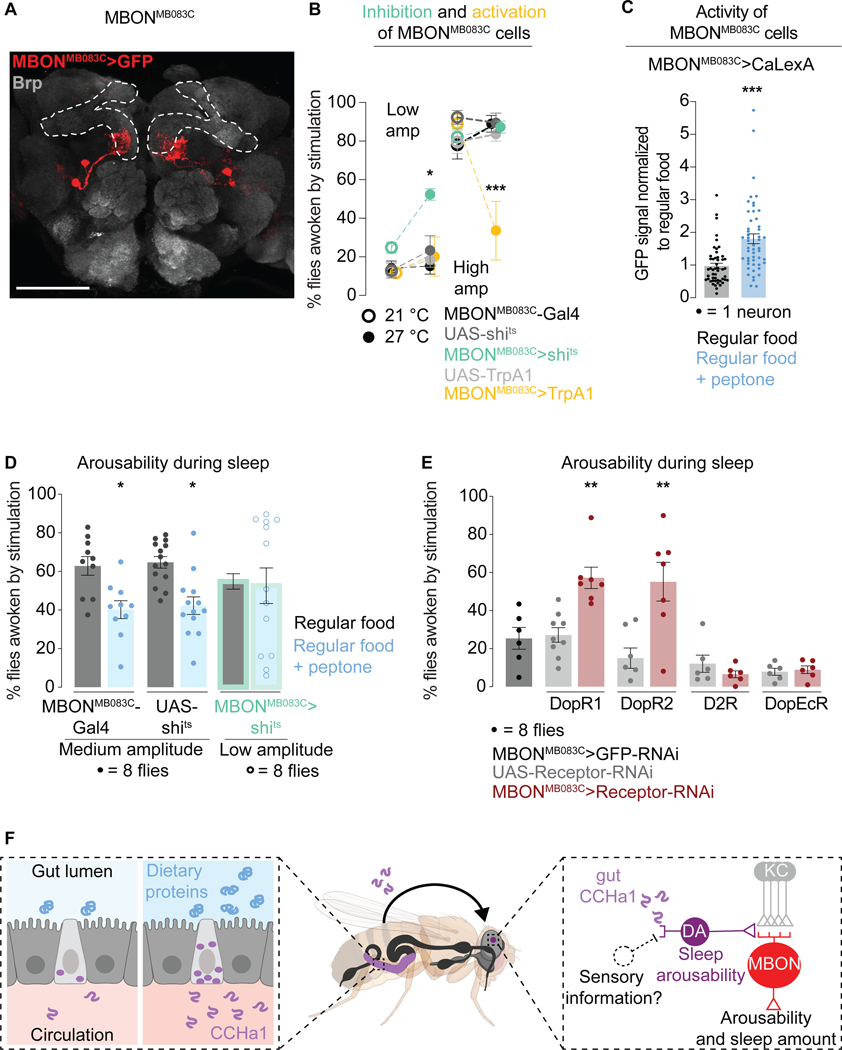
Information from the γ3 compartment is carried further by two neurons in each brain hemisphere, and these neurons are accessible with the Mushroom Body Output Neuron driver MBONMB083C-Gal479,91 (Figure 7A) which has no apparent gut expression (Figure S7A). Conditional MBONMB083C inhibition caused the flies to be easily awoken by vibrations (Figure 7B), indicating an abnormally low arousal threshold - the same phenotype observed with CCHa1 removal from the gut (Figure 2B). Conditional activation had the opposite effect (Figure 7B). These phenotypes were specific for sleep, as arousabilty during wakefulness was unaffected (Figure S7B). MBONMB083C neurons might play a role in regulating sleep amount since their activation led to sleep suppression (perhaps counterintuitively the animals slept less, but deeper) (Figure S7B). We are unsure of the biological significance of this result however, since sleep amount was unaffected by the neuronal silencing and a previous report found no effect of activation109 (though different temperature conditions were used for activation than here). Nevertheless other studies16 do implicate Kenyon cells and MBONs in regulating sleep amount, which suggests the possibility that at this locus the regulatory streams which inform sleep amount and sleep depth are integrated.
Figure 7. Mushroom body output neurons regulate arousability.
(A) The output neurons from the γ3 mushroom body compartment, visualized by MBONMB083C-Gal4-driven GFP. Brp labels synaptic terminals, allowing the mushroom body visualization (outlined). Scale bar: 100 μm. (B) Arousability from sleep when MBONMB083C are inhibited (green) or activated (yellow). (C) Activity of MBONMB083C neurons on regular vs peptone-supplemented food. (D) Arousability from sleep on regular vs protein-supplemented food. Green outline, flies in which MBONMB083C neurons are silenced. (E) Arousability from sleep when different dopamine receptors are depleted from MBONMB083C neurons. (F) Model. Error bars, mean and S.E.M. Genotypes, sample sizes, and statistical analyses, Table S3. See also Figure S7.
CaLexA revealed higher activity of MBONMB083C neurons in animals which had access to peptone-supplemented food for 24 hours compared to animals which consumed regular food (Figure 7C). When MBONMB083C neurons were silenced, the effect of peptone on arousability was suppressed (Figures 7D and S7C). To test whether MBONMB083C receive dopaminergic instructions, we depleted them of dopamine receptors and tested arousability. Depletion of either DopR1 or DopR2 but not D2R or DopEcR from MBONMB083C neurons caused increased arousability from sleep (Figures 7E and Figure S7D). This result suggests that PAMMB441B and MBONMB083C neurons communicate through dopamine to regulate arousability from sleep, and that DopR1 and DopR2 are the relevant receptors in this context. Since the Kenyon Cells also express dopamine receptors80, we cannot rule out the possibility of their involvement.
DISCUSSION
Taken together our data suggest a model where information about dietary proteins is conveyed from the gut to the brain through peptidergic signaling, to regulate arousability from sleep (Figures 7F and S7F).Sleep depth is defined based both on brain electrical activity and reduced behavioral responsiveness to stimulation75. Since sleep is thought to have originated in the earliest animals, whose nervous systems were rudimentary1, it stands to reason that there are core sleep regulatory mechanisms which involve molecular signaling and are not reflected at the level of global brain activity changes. Electrical activity is not commonly used to define and study sleep states in invertebrate models110, yet the mechanistic findings have been translating well from such models to mammals15. Through a genetic screening approach, we were led to a signaling pathway which offers several insights into the regulation of arousability and sleep depth.
Signals secreted from the gut modulate arousability
Food availability is a primary determinant of survival and animals adjust how much they sleep depending on their need for nutrients61,112. Both flies and rodents increase wakefulness at the expense of sleep during starvation111,113, and satiety can promote sleep114,115. The contribution of different nutrients to sleep regulation is unclear, but other studies together with ours indicate that dietary proteins are particularly important19. For example, postprandial sleep is promoted in flies when the meal contains proteins but not sugar, even when caloric values are comparable114. Of all the nutrients, proteins effect the highest level of satiety per calorie116 which might explain why they seem better than sugars or fats at promoting sleep and suppressing arousal.
Enteroendocrine cells are sparse but play critical roles in monitoring the contents of the gut lumen and passing the information to the rest of the body77. The extent to which these cells are neuron-like is only becoming apparent117,118. The elav-Gal4 driver used in the screen is a common neuronal driver but is also expressed in enteroendocrine cells, presumably due to the similarities between the cell types. This is how we serendipitously identified the gut as a site that regulates arousability. Enteroendocrine cells can directly synapse with neurons118 or secrete peptides77. Peptides might be ideal for regulating long-lasting states like sleep because of their slower action mode compared to classical neurotransmitters55. The receptor for CCHa1 is conserved28 and its mammalian homolog regulates itch sensation29, a form of mechanosensory processing.
The same peptide regulates multiple aspects of sleep
CCHa1 evidently regulates different aspects of sleep (latency28, amount, and depth) but our data indicate that these functions map to different cellular origins of the peptide. CCHa1 produced in the gut regulates arousability and was the focus of our study, while CCHa1 produced in the nervous system regulates sleep latency28 and amount (we note that the observed effects on arousability were much stronger than the effects on sleep amount). Identifying various components of sleep regulation is important for eventually generating therapies that can target specific sleep problems. Although the different aspects of sleep can be uncoupled there are likely to be regions in the brain where the regulatory pathways for sleep amount and depth converge, as suggested here. Arousability varies not only during sleep but also during wakefulness, and CCHa1 can act across these states. Specificity for sleep vs wakefulness is achieved by the peptide acting on different dopaminergic neuronal population. During sleep most sensory processing is attenuated, and our data suggest that this is done through at least partially independent mechanisms, as the CCHa1 signal regulates responsiveness to mechanical stimulation but not to heat. Since different types of environmental information have different meaning for animals, being able to separately tune responsiveness for each might improve the chance of survival. The modularity detected throughout the system likely helps generate a spectrum of arousal states and confers behavioral flexibility.
Dopaminergic signaling in the mushroom body regulates arousability
Much of the arousability regulation we describe takes place in the mushroom body, a brain structure with multiple compartments that are innervated by modulatory dopaminergic neurons79. Dujardin proposed in 1850 that the mushroom body is a site at which intelligence arises in insects119 after observing that bigger and more complex mushroom bodies endow animals with a greater degree of what he called free will, as opposed to reflex. Large amounts of information are received by the mushroom body, but only 34 neurons carry information to other brain areas59,91. A recent study of sensory arousal in sleeping flies also pointed to the mushroom body as a critical regulatory locus120 albeit a different compartment was implicated, presumably because a different sensory modality (olfaction) was examined; olfactory information is received by the γ5 compartment while the mechanosensory information is received by the γ3 compartment. Dopaminergic neurons exert their modulatory influence at the mushroom body output sites79,97, where the mechanisms regulating sleep amount and depth seem to converge. Dopamine is involved in seemingly all nervous system functions121, steering them according to need and circumstance by modulating synapses and receptor availability122–124, but not much is known about how the dopamine signal itself is controlled. Our study shows that the activity of dopaminergic neurons can be regulated by direct signaling from the gut.
Limitations of the study
We have not identified the subpopulation of dopaminergic neurons which regulate arousability during wakefulness, and have not determined how mechanosensory information is conveyed to the sleep system. We used CaLexA to report neuronal and enteroendocrine cell activity but we acknowledge that this tool offers an indirect measurement that depends on transcription. In our study we supplemented food with different macronutrients at different concentrations and only protein supplementation changed arousability, but we cannot rule out the possibility that other sources of macronutrients, at other concentrations, would be effective. Even though protein-enriched food decreases arousability in mice as in flies, we do not know whether this is regulated by signals from the gut and the CCHa1/CCHa1R homologues.
Closing remarks
This work provides an updated framework for thinking about sleep regulation. Sleep consumes the entire body, but most efforts to understand this state are focused on the nervous system. The observations described here, together with other emergent findings125–127, argue that a more integrative approach taking into account not only the nervous system but other organs, is needed to solve some of the biggest mysteries in biology. The centuries-old idea that digestive and nervous systems are critically linked in normal physiology and disease128 seems to merit particular attention.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dragana Rogulja (dragana_rogulja@hms.harvard.edu).
Materials Availability
The antibody generated during this study is available upon request.
Data and Code availability
Data are available from the corresponding author upon request. Codes are available on GitHub for sleep analysis (https://github.com/CrickmoreRoguljaLabs/SleepAnalysis), stimulus generation (https://github.com/IrisTitos/ATanalysis/blob/master/Goodmorning.md), and stimulus analysis (https://github.com/IrisTitos/ATanalysis/blob/master/ReadDamsPullDown, https://github.com/ArousalThreshold/Software).
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Flies
Flies were grown in 12-hour light: 12-hour dark cycles at 25 °C unless indicated otherwise. Experiments were performed in Percival Scientific Inc. (DR36VL model) incubators. Flies were raised on a cornmeal-agar medium125. Briefly, our regular food contained (measured in % weight (g) / volume (100 ml)) 1.57% (wt/vol) agar (Spectrum), 2.72% (wt/vol) yeast (MP Biomedicals), 11.96% (wt/vol) sugar (Domino) and 5.14% (wt/vol) cornmeal (Bunge). Ingredients were heated until the temperature reached 97 °C while stirring. After the temperature of the solution decreased to 75 °C, 2.5% Tegosept, 23.8 g/L Ethanol (Koptic #V1001), and water to compensate for evaporation were added.
All Drosophila stocks used in this study are listed in the key resources table. All the stocks used for behavioral experiments were outcrossed at least five times to an isogenised (iso31) genetic background. In the case of UAS-CCHa1-RNAi (BDSC #57562) and UAS-CCHa1R-RNAi (BDSC #51168), UAS-GFP-RNAi from the same library was used as a control (BDSC #41552). The two RNAis against CCHa1R target non-overlapping coding sequences: VDRC #103055 targets nucleotides 16–333, and BDSC #51168 targets nucleotides 375–397. The RNAis used for CCHa1 target partially overlapping coding sequences: VDRC #104974KK targets nucleotides 241–539, and BDSC #57562 targets nucleotides 525–547. For all the Gal4/UAS experiments, heterozygous controls were used (UAS crossed to the Gal4 genetic background, and Gal4 crossed to the UAS genetic background).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-CCHa1 | This study | N/A |
| mouse anti-Bruchpilot | DHSB (Developmental Studies Hybridoma Bank) |
Cat# nc82; RRID: AB_2314866 |
| chicken anti-GFP | Aves | Cat# GFP-1020; RRID: AB_10000240 |
| mouse anti-prospero | DSHB | Cat# pros; RRID: AB 528440 |
| mouse anti-tyrosine hydroxylase | ImmunoStar | Cat# 22941 ; RRID: AB 572268 |
| Rabbit anti-β-galactosidase | MP Biomedicals | Cat#56033 LOT#06026 |
| Alexa Fluor 488 goat anti-chicken | Life technologies | Cat# A11039; RRID: AB_142924 |
| Alexa Fluor 488 goat anti-rabbit | Life technologies | Cat# A11034; RRID: AB 2576217 |
| Alexa Fluor 647 goat anti-rabbit | Life technologies | Cat# #A21244; RRID: AB 2535812 |
| Alexa Fluor 647 donkey anti-mouse | Life technologies | Cat# A31571; RRID: AB_162542 |
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Coconut oil | Spectrum essentials | Cat# L215562P-006 |
| Propionic acid | Sigma-Aldrich | Cat# P1386; CAS: 79–09-4 |
| Hexanoic acid | Sigma-Aldrich | Cat# P21530; CAS 142–62-1 |
| Glucose | Sigma-Aldrich | Cat# 158968–1KG; CAS: 492–62-6 |
| Fructose | Sigma-Aldrich | Cat# F0127; CAS:57–48-7 |
| Galactose | Sigma-Aldrich | Cat# G0750; CAS: 59–23-4 |
| Peptone | Apex | Cat# 20–260 |
| Single amino acids | Sigma-Aldrich | Cat# LAA21–1KT |
| FD&C Blue #1 | Spectrum | Cat# FD110; CAS: 3844–45-9 |
| Triton X-100 | Amresco | Cat# M143; CAS: 9002–93-1 |
| PBS 10x | SeraCare | Cat# 5460–0030 |
| Ethanol (pure) | Koptic | Cat# V1001; CAS: 64–17-5 |
| PFA (16%) | Electron Microscopy Sciences | Cat# 15710; CAS: 30525–89-4 |
| BSA | Gemini | Cat# 700–101P; CAS: 9048–46-8 |
| Microscope glass slides | Electron Microscopy Sciences | Cat# 63421–10 |
| Microscope coverslips | Electron Microscopy Sciences | Cat# 72230–01 |
| Prolong Gold Antifade medium | Invitrogen | Cat# 1942345 |
| TRIzol | Thermo fisher | Cat# 15596026 |
| iScript cDNA Synthesis | Biorad | Cat# 1708890 |
| Sybr Green | Biorad | Cat# 170–8880 |
| Critical commercial assays | ||
| Direct-zol RNA Microprep | Zymo Research | Cat# R2061 |
| DNAse Turbo DNA-free kit | Thermo fisher | Cat# AM1907 |
| Deposited data | ||
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| wild type iso31 | Gift from Kyunghee Koh | N/A |
| elav-Gal4 | Gift from Kyunghee Koh | N/A |
| UAS-TrpA1 | Gift from Kyunghee Koh | N/A |
| UAS-Shits | Gift from Kyunghee Koh | N/A |
| elav-Gal4;UAS-Dcr2 | Gift from Nicholas Stavropoulos | N/A |
| PAM-Gal4 | BDSC (Bloomington Drosophila Stock Center) | RRID: BDSC_41347 |
| UAS-CCHa1-RNAi | BDSC | RRID: BDSC_57562 |
| UAS-CCHa1-RNAi #2 | VDRC (Vienna Drosophila Recourse Center) | RRID:SCR_013805 #104074 |
| UAS-CCHa1R-RNAi | BDSC | RRID: BDSC_51168 |
| UAS-CCHa1R-RNAi #2 | VDRC | RRID:SCR_013805 #103055 |
| UAS-GFP-RNAi | BDSC | RRID: BDSC_41552 |
| UAS-DopR1-RNAi | BDSC | RRID: BDSC_31765 |
| UAS-DopR2-RNAi | BDSC | RRID: BDSC_65997 |
| UAS-D2R-RNAi | BDSC | RRID: BDSC_36824 |
| UAS-DopEcR-RNAi | BDSC | RRID: BDSC_31981 |
| Tdc2-Gal4 | BDSC | RRID: BDSC_9313 |
| vGlut-Gal4 | BDSC | RRID: BDSC_26160 |
| PAMMB441B | BDSC | RRID: BDSC_68251 |
| MBONMB083C | BDSC | RRID: BDSC_68287 |
| CaLexA | BDSC | RRID: BDSC_66542 |
| UAS-nlsGFP | BDSC | RRID: BDSC_4776 |
| UAS-TH-RNAi | BDSC | RRID: BDSC_25796 |
| UAS-Dcr2 | BDSC | RRID: BDSC_24650 |
| CCHa1 mutant Mi{MIC}CCHa1MI09190 | BDSC | RRID: BDSC_51261 |
| CCHa1R mutant Mi{MIC}CCHa1RMI08118 | BDSC | RRID: BDSC_44750 |
| TI{2A-GAL4}CCHa1-R[2A-GAL4] | BDSC | RRID: BDSC_84601 |
| P{GMR23H07-GAL4}attP2 | BDSC | RRID: BDSC_47498 |
| UAS-CD8GFP | Gift from Michael Crickmore | N/A |
| UAS-nlsLacZ | Gift from Michael Crickmore | N/A |
| TH-Gal4 | Gift from Michael Crickmore | N/A |
| ddc-Gal4 | Gift from Michael Crickmore | N/A |
| Cha-Gal4 | Gift from Michael Crickmore | N/A |
| Tim-Gal4 | Gift from Michael Young | N/A |
| TH-D4-Gal4 | Gift from Michael Crickmore | N/A |
| pros-Gal4 | Gift from Norbert Perrimon | N/A |
| CBA/CaJ mice | Jackson Laboratories | JAX 000654 |
| Oligonucleotides | ||
| CCHa1 (F-ACTGACGTCGGACAATTTGC and R-ACACGAATGTCCGTATTCCA) | ID Technologies | Custom made |
| CCHa1R (F-GTTCCAAACACCTACATTTTATCAC and R-CGGATAATGCAGTCAGCGTA) | ID Technologies | Custom made |
| CCHa2 (F-AAACAGCAACAGCAGCAAAC and R-AGGACCACGGTGCAGATAAC) | ID Technologies | Custom made |
| CCHa2R (F-CATACCCAACACATACATTCTTTC and R-GAAAGGGCGGTCAGTGTAAA) | ID Technologies | Custom made |
| rp49 (F-ATCGGTTACGGATCGAACAA and R-GACAATCT CCTTGCGCTTCT) | ID Technologies | Custom made |
| Recombinant DNA | ||
| Software and algorithms | ||
| Sleep recording Drosophila | TriKinetics | |
| Arousal threshold stimuli delivery Drosophilas | This study | |
| Sleep analysis Drosophila | Vaccaro et al 2020 | |
| Arousal threshold analysis Drosophila | This study | |
| Sleep recording mice | Pinnacle Technology Inc |
9000-K5-S; 8200-K1-iSL |
| Sleep scoring mice | Pinnacle Technology Inc | Sirenia Sleep Pro |
| Other | ||
Mice
2-month-old male CBA/CaJ (JAX 000654) mice were housed according to standard protocols approved by the Harvard University Standing Committee on Animal Care in accordance with federal guidelines. On arrival at our animal facility, mice were housed in pathogen-free cages (max 5/cage) under 12-hour light: 12-hour dark cycles at 23 °C and 50% humidity. They were given ad libitum access to food and water.
METHOD DETAILS
Flies
Arousal threshold screen
Random RNAi lines from the Kyoto Stock Center, the Vienna Drosophila Resource Center, and the Transgenic RNAi Project (TRiP) (available at the Bloomington Drosophila Stock Center and generously shared with us by Norbert Perrimon’s laboratory) were used in the genetic screen. In addition, we screened RNAi stocks that were already being used in our laboratory related to neuropeptides, neurotransmitters, and sleep amount phenotypes. All RNAi lines were crossed to flies expressing elav-Gal4. 8 males, 3–5 days old, were tested in the system described below. In all graphs, each dot represents the average behavior of 8 flies. That is, to calculate the % arousability (either during sleep or wake), we included the data of 8 flies. Flies were exposed to low amplitude stimulation (0.2V) on the first night and high amplitude stimulation (2V) on the second night.
Analysis of screen results
To identify hits from the screen, the arousabilty values were averaged across genotypes (separately for weak and strong vibrations). Hyper-arousable hits were those with phenotypes exceeding two standard deviations from the mean (when weak vibrations were used). Hypoarousable hits were those with phenotypes exceeding two standard deviations from the mean (when strong vibrations were used). For the genes whose disruption caused hypo- or hyper-arousability phenotypes, gene ontology (GO) analysis was performed. We used the GO Term Mapper (https://go.princeton.edu/cgi-bin/GOTermMapper) to bin individual GO categories into more general terms. In order to normalize the different GO terms, we took into account their representation in the genome (i.e., how often the term appears in all the genes from the organism). For every GO term, the % of hits containing that term in our screen was normalized to the % of genes in the whole genome that contain that term. The data are represented on a log10 scale.
Locomotor activity and sleep monitoring
Locomotor activity was recorded using the Drosophila Activity Monitoring System (DAM3, TriKinetics, Waltham, MA). Flies were individually housed in 65 mm-long glass tubes (TriKinetics) filled with ~40 mm of cornmeal-agar food unless otherwise indicated. This left ~25 mm of space for flies to walk back and forth. An infrared light beam bisects this space and gets interrupted when flies walk through; this is automatically scored as a movement. Sleep was defined as five minutes of inactivity 42,43. Sleep quantification was performed with a custom MATLAB (The MathWorks, Natick, MA) software (available on GitHub at https://github.com/CrickmoreRoguljaLabs/SleepAnalysis).
The setup for probing arousal threshold with mechanical vibrations
DAMs were mounted on custom-made platforms positioned on top of large bass loudspeakers (381 mm, Delta 15 LFA, Eminence, Eminence, KY). Each speaker/platform holds up to 8 DAMs. To stimulate the flies, a custom program (Good Morning, available on GitHub at https://github.com/IrisTitos/Atanalysis/blob/master/Goodmorning.md) written in MATLAB generates a multisine signal (40 Hz – 200 Hz) using a multifunction device (USB-6211, National Instruments, Austin, TX). The signal is fed into an amplifier (SLA-1, Applied Research, and Technology, Rochester, NY) connected to the speaker. A range of frequencies was used to avoid the frequency-specificity of the response. With this setup, the mechanical vibrations of the loudspeaker translate into up and down movements of the platform on which the DAMs are mounted. Accelerometers (ADXL335, Analog Devices, Norwood, MA) measure movements of individual DAMs, at different positions on the platform to ensure that vibrations are uniform across all monitors. In our experiments, 2-second vibrations were delivered at random intervals every 30–40 minutes, starting 1.5 hours after the lights turned off (for 10 hours during the night (dark phase)). The data were processed in MATLAB together with locomotor data from the DAMs (available on GitHub at https://github.com/IrisTitos/Atanalysis/blob/master/ReadDamsPullDown, https://github.com/ArousalThreshold/Software).
Scoring arousability from sleep
Only flies that were asleep (more than 5 minutes of locomotor inactivity) when the stimulus was applied were considered when calculating arousability from sleep. If the locomotor activity was triggered within 3 minutes of vibration onset, the fly was considered awoken by the stimulus. To control for the increased chance of spontaneous awakenings in genetic manipulations that affect sleep architecture (number of bouts and/or bout length), we normalized for spontaneous awakenings: % flies awoken by stimulation = 100*(total % flies that wake up - % spontaneous awakenings) / (100 - % spontaneous awakenings). To calculate spontaneous awakenings, the activity data from the same night and the same flies were used to calculate the “total % flies that wake up”. Normalization can result in negative percentage values when spontaneous arousals occur more frequently than arousals after stimulation; this is seen more commonly with weak vibrations and is probably a result of chance. We used a modified timestamp file in which the stimulation timings are shifted by 10 minutes preceding the stimulation to calculate spontaneous awakenings. Next, we performed the same analysis previously done with the original timestamp; only flies that were asleep at the modified timestamp were considered for analysis, and flies that showed locomotor activity within the 3 minutes following the modified timestamp were considered to be awoken.
Scoring arousability during periods of wakefulness
Only flies that were awake (i.e. that showed activity during the 5 minutes prior to the stimulation) when the stimulus was applied were considered when calculating arousability during wakefulness. For those flies, we calculated the average activity during the 3 minutes prior to stimulation and the average activity during the 3 minutes after the stimulation. Flies were considered reacting to stimuli if the average activity after the stimulation was higher than prior to it.
The setup for probing arousal threshold with temperature
To probe arousability with temperature, we used a flat multi-beam activity monitor (DAM5M TriKinetics) in which the fly-containing tubes lay horizontally. A heating pad (McMaster-Carr 35765K469, Elmhurst, IL) controlled by a PID Temperature Controller (Omega CN 7800, Norwalk, CT) covers the surface of the monitor. A MATLAB custom-made software (https://github.com/ArousalThreshold/Temperature) turns on the heating pad, increasing the temperature to 40 °C (unless otherwise indicated) for 2 minutes, at random intervals (30–40 minutes) during the night. The same protocol as described for the mechanical setup was used to calculate the % of flies awoken by heat.
Feeding experiments
Male flies, 3 to 5 days old, were transferred from regular food to different food conditions approximately 10 hours after lights turned on (i.e., ZT10 in 12-hour light: 12-hour dark cycles) unless otherwise indicated. After 24 hours, they were dissected or examined for behavior. The cornmeal-agar food was melted, and different components were added to achieve these final concentrations: 10% coconut oil, 1% propionic acid (Figures 3H, S3A and S3D), 0.5% propionic acid (Figure S3E), 0.5% hexanoic acid, 200 mM glucose (Figures 3C, 3H and S3D), 566 mM glucose (Figure S3E, glucose 40 kcal), 377 mM glucose (Figure S3E, 2/3 glucose), 189 mM glucose (Figure S3E, 1/3 glucose), 200 mM fructose, 200 mM galactose, 10% peptone, 3.3% peptone (Figure S3E, 1/3 peptone), 6.7% peptone (Figure S3E, 2/3 peptone) or the same concentration as in peptone for each of the single amino acids. In order to control for the fact that liquid was added to the supplemented food, the same volume of water was added to the regular food used for the controls.
Food intake measurement
3–5 day old male flies were transferred from regular food to either regular food containing 2% (wt/vol) FD&C Blue #1 (Spectrum #FD110), or to regular food supplemented with peptone containing the same concentration of FD&C Blue #1. After 24 hours, flies were frozen at −20 °C. Frozen flies were decapitated (to avoid the eye pigment interfering with the measurement), aliquoted in groups of 5 into Eppendorf tubes containing 50 μL of 1% Triton X-100 (Amresco #M143) 1X-PBS (Phosphate Buffered Saline 10X SeraCare #5460–0030) and homogenized with a motorized pestle (Argos #9950–901). Samples were centrifuged, and the supernatant was measured at 660 nm (A660) in a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific). A standard curve was generated using serial dilutions of the blue dye, and the sample dye concentration was extrapolated from that curve.
Food clearance assay
To assess whether food digestion is altered when flies are fed a protein-rich diet or when CCHa1 or its receptor are knocked down, we performed a food excretion assay. Briefly, flies were fed food with blue dye for 24 hours, then transferred to vials with only agar for 6 hours, after which they were frozen at −20 °C. The remaining blue dye concentration present in the abdomen was quantified using the same protocol as for food intake measurements.
Immunohistochemistry
Adult males were anesthetized with carbon dioxide, washed briefly with 70% ethanol (Koptic #V1001), and dissected in 1X-PBS (Phosphate Buffered Saline 10X SeraCare #5460–0030). Guts were fixed overnight at 4 °C in PBS with 4% paraformaldehyde (PFA, Electron Microscopy Sciences #15710). Dissected brains and ventral nervous systems were fixed for 30 minutes in 4% PFA at room temperature. Tissues were washed 3 times for 20 minutes in 1X-PBS containing 0.2% Triton X-100 (Amresco #M143), and then blocked for 2 hours in 1X PBS, 0.2%Triton X-100, 2% bovine serum albumin (BSA Gemini #700–101P) at room temperature. Samples were incubated with primary antibodies overnight (except for when Bruchpilot (Brp) antibody was included, in which case the samples were incubated for ~48 hours) at 4 °C in 1X PBS, 0.2% Triton X-100, 2% BSA, at the indicated dilutions. Tissues were washed 3 times for 20 minutes in 1X-PBS, 0.2% Triton X-100 and then incubated with secondary antibodies for 2 hours in the blocking solution at room temperature at the indicated dilutions (except when Bruchpilot was included in the stainings which were incubated for ~48 hours at 4 °C). Samples were then washed 3 times for 20 minutes in 1X-PBS, 0.2% Triton X-100, and mounted between glass slides and coverslips (Electron Microscopy Sciences #72230–01 and 64321–10) in Prolong Gold Antifade medium (Invitrogen #1942345).
Rabbit anti-CCHa1 antibodies were raised against the peptide QIDADNENYSGYELT49 by Genscript, and were affinity purified by the company.
Primary antibodies used: mouse anti-Bruchpilot (1:7, DSHB nc82), rabbit anti-CCHa1 (1:50), chicken anti-GFP (1:1000, Aves #GFP-1020), mouse anti-prospero (1:50, DSHB pros) and mouse anti-tyrosine hydroxylase (TH) (1:1000 ImmunoStar #22941), rabbit anti-β-galactosidase (1:1000 MP Biomedicals #56033).
Secondary antibodies used: Alexa Fluor 488 goat anti-chicken (1:1000 Life technologies #A11039), Alexa Fluor 488 goat anti-rabbit (1:1000 Life technologies #A11034), Alexa Fluor 647 goat anti-rabbit (1:1000 Life technologies #A21244) Alexa Fluor 647 donkey anti-mouse (1:1000 Life technologies #A31571).
Fluorescence microscopy
Fluorescent images were acquired with a Leica SP8 confocal microscope. All the imaging conditions remained constant within each experiment; only the Z start and end settings were adjusted individually for each sample. The average distance between Z-stacks was 100 microns.
Image quantification
All images were scored blind.
Gut quantifications for CCHa1 levels and CaLexA experiments
Quantification of CCHa1 and GFP signals was performed with Fiji. For every image, a Z-stack summation projection of the whole gut was obtained. To quantify CCHa1 antibody staining in the gut, the CCHa1 signal was used to define the region of interest (ROI). The average pixel intensity and area of that ROI were measured. A region with a similar area adjacent to the ROI was selected to measure the average background pixel intensity. The amount of CCHa1 was quantified by subtracting the background from the CCHa1 signal and multiplying it by the CCHa1 ROI area.
For Figures 3C, 3D, 3E, 3F, and S3A, the CCHa1 signal in the gut was normalized to regular food. To do that, for each individual experiment, the mean of the CCHa1 signal in all the guts of flies exposed to regular food was calculated. That mean was then used to divide all the experimental values.
For Figures 3C and S3A, the percentage of activated CCHa1 cells in the gut was calculated based on CaLexA-dependent GFP levels. First, we quantified how many CCHa1-expressing cells were present in the posterior midgut using the “counter” function of Fiji. Then, we counted how many of those cells also had a GFP signal. To determine the % of CCHa1 cells that were activated, we used the formula: (CCHa1 and GFP-positive cells/all CCHa1-positive cells)*100.
Brain quantifications for CaLexA experiments
To quantify GFP intensity in Figures 6B, 6C, 7C, S6C and S6D we projected the summation of the Z-stacks containing the cells of interest. Each cell was manually selected, and the average pixel intensity of the GFP signal was quantified. A region next to the cells of interest but containing no GFP-positive cells was used to measure the background signal. To calculate the GFP signal, we used this formula: (GFP average pixel intensity from the cell – GFP background average pixel intensity).
Quantitative RT-PCR
For each condition, either 2 whole flies, 5 brains, or 10 guts of 3–5 days old males were dissected and immediately transferred into 100 μl of ice-cold TRIzol (Thermo fisher #15596026) one by one, flash-frozen on dry ice, and stored at −80 °C. To extract RNA, samples were homogenized with a pestle (Argos #9950–901) for 20 seconds on ice before adding 200 μl of TRIzol and 60 μl of chloroform. Samples were then centrifuged for 15 minutes at 15000 rpm, at 4 °C. The supernatant was collected and diluted in an equal amount of ethanol 99% (Koptec #V1001), mixed, and transferred to a purification column (Zymo kit Direct-zol R2061). The manufacturer’s instructions for RNA purification and subsequent DNAse treatment were followed (DNAse Turbo DNA-free kit AM1907). RNA was quantified using NanoDrop. 500ng of RNA was used for the reverse transcriptase reaction (Biorad iScript cDNA Synthesis #1708890). DNA product was diluted 1:10 and used to set up quantitative PCR reactions using Sybr Green (Biorad #170–8880). Primers were obtained from ID Technologies:
CCHa1 (F-ACTGACGTCGGACAATTTGC and R-ACACGAATGTCCGTATTCCA)51
CCHa1R (F-GTTCCAAACACCTACATTTTATCAC and R-CGGATAATGCAGTCAGCGTA)51
CCHa2 (F-AAACAGCAACAGCAGCAAAC and R- AGGACCACGGTGCAGATAAC)51 CCHa2R (F-CATACCCAACACATACATTCTTTC and R- GAAAGGGCGGTCAGTGTAAA)51 rp49 (F-ATCGGTTACGGATCGAACAA and R-GACAATCTCCTTGCGCTTCT)
Conditional experiments
Adult-specific knockdown of CCHa1 or its receptor was achieved using a temperature-sensitive allele of Gal80131, a Gal4 inhibitor (Gal80ts) combined with either elav-Gal4 or pros-Gal4. Flies were raised at 21 °C, a temperature at which Gal80ts is stable and interferes with Gal4 function. Two days after eclosion, the temperature was raised to 29 °C, the temperature at which Gal80ts is degraded, and the Gal4 driver becomes functional.
For TrpA1 and shits experiments, flies were raised at 21 °C, a temperature at which TrpA1 and shits are not active. Flies were transferred to 27 °C for behavior analysis. In Figures 2C and S2I, because the Gal4-UAS system depends on temperature (Gal4 expression is higher at higher temperatures), the experiments had to be performed above 25 °C to ensure CCHa1-RNAi expression. Since TrpA1 has been reported to be active at temperatures above 25 °C, as low as 26 °C 24, we cannot entirely discard the possibility that some mild TrpA1 activation occurs at 25 °C.
Mice
Food and diet
Upon arrival at our animal facility, mice were given standard Pico Lab Rodent Diet 20 # 5053. After surgery, 5 days prior to baseline sleep recording, their diet was changed to either ‘standard diet’ (#D11112201, Research Diets, Inc.), or ‘protein-enriched diet’ (#D1311070, Research Diets, Inc.). The recipes are calculated based on 1071.05 g of food. The standard diet is composed of: 200 g casein, 3 g L-cystine, 381 g corn starch, 110 g maltodextrin 10, 150 g dextrose, 75 g cellulose BW200, 25 g inulin, 70 g soybean oil, 10 g mineral mix S10026, 13 g dicalcium phosphate, 5.5 g calcium carbonate, 16.5 g potassium citrate, 10 g vitamin mix V10001, 2 g choline bitartrate, 0.025 g yellow dye #5 FD&C and 0.025 g blue dye #1 FD&C. The protein enriched diet is composed of: 600 g casein, 9 g L-cystine, 50 g corn starch, 35 g maltodextrin 10, 150 g dextrose, 75 g cellulose BW200, 25 g inulin, 70 g soybean oil, 10 g mineral mix S10026, 13 g dicalcium phosphate, 5.5 g calcium carbonate, 16.5 g potassium citrate, 10 g vitamin mix V10001, 2 g choline bitartrate, and 0.05 g red dye #40 FD&C. The sugar-enriched food is composed of: 200 g casein, 3 g L-cystine, 41 g maltodextrin 10, 600 g sucrose, 75 g cellulose BW200, 25 g inulin, 70 g soybean oil, 10 g mineral mix S10026, 13 g dicalcium phosphate, 5.5 g calcium carbonate, 16.5 g potassium citrate, 10 g vitamin mix V10001, 2 g choline bitartrate, 0.04 g red dye #40 FD&C and 0.01 g blue dye #1 FD&C. The fat-enriched food is composed of: 200 g casein, 3 g L-cystine, 41 g maltodextrin 10, 150 g dextrose, 75 g cellulose BW200, 25 g inulin, 25 g soybean oil, 245 g coconut oil, 10 g mineral mix S10026, 13 g dicalcium phosphate, 5.5 g calcium carbonate, 16.5 g potassium citrate, 10 g vitamin mix V10001, 2 g choline bitartrate, 0.01 g yellow dye #5 FD&C and 0.04 g red dye #40 FD&C. In all types of diet, 1071.05 g of food contained 4084 kcal. Animals remained on their respective diet up until the end of the study. Mice were weighted daily from the start of the diet change until the end of the study.
Sleep monitoring
To monitor sleep, a single male was placed in a circular cage (Pinnacle Technology 9000-K5-S) equipped with a 3-channel tethered EEG/EMG system (Pinnacle Technology 8200-K1-iSL). Head-mounted preamplifiers (Pinnacle Technology 8201-SS) were implanted surgically. For electroencephalography/electromyography (EEG/EMG), mice were anesthetized with isoflurane and mounted onto a stereotactic apparatus (Kopf) for surgery. The head was shaved, a 1.5 cm rostral-caudal incision was made starting from approximately 3.5 mm anterior to bregma, and the EEG/EMG headmount was adhered onto the surface of the dry skull with Loctite 404 Instant Adhesive (Fisher Scientific #NC0619400). Screws (Pinnacle Technology 8209, 8212) were drilled into the skull at the corresponding holes in the headmount for hippocampal and cortical EEG recordings. To insert the EMG wires, small pockets in the nuchal muscles were made. C&B Metabond Adhesive Luting Cement (Parkell) was applied to the head-mount to protect and insulate EEG/EMG leads. The skin around the headmount was sutured, and mice were allowed to recover for one week and were tested for behavior within the following week. The mice were tested for behavior within the next week. Implanted mice were tethered to the 3-channel EEG/EMG system in the sleep-recording chamber (Pinnacle Technology 9000-K5-S) and allowed to acclimate for 2 days before baseline sleep recording. Recordings were acquired for 48 hours.
Sleep scoring
Sleep recordings were scored in 10-second epochs over 48 hours with the sleep analysis software Sirenia Sleep Pro (Pinnacle Technology). Non-REM EEG epochs were scored as low frequency (0.5–4 Hz range), high amplitude (+/− 150–250 μV) delta waves with low-amplitude EMG waves. REM EEG epochs contained mixed frequencies with predominantly theta waves (5.5–8.5 Hz range), low amplitude (+/− 50–100 μV), and flat EMG waves. Wake epochs were scored as high frequency (8–50 Hz), low amplitude (+/− 50–100 μV) EEG waves with high amplitude EMG. Automated scored epochs were visually inspected to ensure that each epoch was scored according to our definitions of non-REM, REM, and Wake, and epochs that were not scored by the set rules were manually scored. Epochs that contained movement artifacts were removed.
To better compare the 24-hour sleep-related behavioral patterns for mice fed a standard and protein-enriched diet, an average hourly value for time spent in awake and sleep (non-REM, REM) stages, as well as the length and the number of wake and sleep (non-REM, REM) bouts was calculated during baseline recordings (prior to mechanical stimulation) using the Sirenia Sleep Pro software (Figure S4D). Similarly, the 24 hours average and hourly (averaged for each hour during 24 hours) delta (0.5–4 Hz) and theta power (5.5–8.5 Hz) was calculated during baseline recordings using Sirenia Sleep Pro for mice fed a standard and protein-enriched diet (Figure S4E,F). The power analysis is based on Fast Fourier transform algorithm and the EEG power represents the amount of activity in certain frequency bands of the signal129. The baseline daily and hourly graphs show the average of the 48 hour recordings. The wave power spectrum reflects the frequency content of the signal or the distribution of signal power over frequency130. For the wave power representation, we averaged the data from EEG1 and EEG2 channels present in the recording device.
Arousal threshold measurement
To probe arousability in mice, we used a mechanical setup composed of a shaker (VWR VS-2500) housed on a cart. The vibrations caused by the shaker (accelerometer data for low intensity: X=0.029, Y=0.066, Z=−0.987; high intensity: X=0.176, Y=0.32, Z=−0.878) translate into lateral movements of the cart on which sleep-recording chambers are housed. The shaker was programmed to deliver 15-second stimulations at randomized intervals centered on 50–70 minutes for two consecutive days during the light phase (that is when mice sleep the most), and for up to 11 hours. EEG/EMG recordings (please see ‘Sleep scoring’ for details) were used to score arousal and sleep. The changes in behavior were confirmed with video recordings. Only stimulations that were preceded by at least 5 minutes of sleep were considered. For each mouse, arousability was quantified as the number of events in which that animal transitioned from sleep to wakefulness within 5 minutes after mechanical stimulation. To normalize the data for differences in spontaneous awakenings, we analyzed the undisturbed behavior of individual mice 20 minutes before each stimulation. At those time points, we quantified the % of events in which the animal spontaneously transitioned from sleep to wakefulness. The % of awakenings that were due to stimulation was calculated as in flies, but in this case for individual animals: % awakenings to stimulation = 100*(total % events in which an individual animal wakes up - % spontaneous awakenings of the animal) / (100 - % spontaneous awakenings of the animal).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using GraphPad Prism software 7 (GraphPad Software Inc., San Diego, CA). Except for the screen (Figure 1A), Figures 3E, S3C, and 5A, all experiments were performed at least 3 times independently. Data were tested for normality, and all the statistical tests were two-sided. In all figures * p<0.05, ** p<0.01 and *** p<0.001. Only when the experimental condition/genotype was different from all the controls the statistical test was shown in the figures.
Graph data are presented as mean ± S.E.M. Please see Table S3 for sample sizes, tests, and p values.
Supplementary Material
Figure S1. Experimental setup and analysis of CCHa1 and CCHa1R mutants and RNAi lines, related to Figure 1. (A) Setup used for mechanical stimulation. Custom software generates 2-second-long multisine (40–200 Hz) signals that are converted into mechanical stimuli (vibrations). Vibrations are given at random intervals centered on 30–40 minutes throughout the night. Drosophila activity monitors (DAMs, TriKinetics, Waltham, MA) are mounted on custombuilt platforms on top of the loudspeakers. Accelerometers placed on top of DAMs (arrow) measure the stimulus intensities to ensure that all flies are stimulated equally regardless of their position on the platform. (B) Each fly is housed in a glass tube, with ~25 mm of space for movement. Movement is detected by the interruption of the infrared light beam and recorded every minute. (C) mRNA levels of CCHa1 and CCHa2, as measured by qRT-PCR done on whole flies, in controls and in animals which express RNAi against CCHa1 using elav-Gal4. Each dot represents 2 flies. mRNA levels of CCHa1R and CCHa2R, as measured by qRT-PCR of fly brains in controls and in animals which express RNAi against CCHa1R using elav-Gal4. Each dot represents 5 brains. (D) The first panel shows reactivity of flies which were awake at the time vibrations were delivered. Other panels show various measurements of sleep and locomotion. The last panel shows sleep amount throughout the day, in 30-minute intervals. The white portion of the bar on top indicates daytime (lights on) and the black portion indicates nighttime (lights off). (E) Measurements of arousabilty, sleep and locomotion using additional RNAis against CCHa1 and CCHa1R. (F) mRNA levels of CCHa1 and CCHa2, as measured by qRT-PCR done on whole flies, in controls and in flies which express RNAi#2 against CCHa1 using the elav-Gal4 promoter. Each dot represents 2 flies. mRNA levels of CCHa1R and CCHa2R, as measured by qRT-PCR of fly brains in controls and in flies which express RNAi#2 against CCHa1R using elav-Gal4 promoter. Each dot represents 5 brains. (G) Measurements of arousabilty, sleep, and locomotion in controls and in mutants for CCHa1 and CCHa1R. (H) Measurements of arousabilty in controls, and in flies with conditional expression of RNAi against CCHa1 and CCHa1R. Adult-specific expression was achieved by combining elav-Gal4 and a temperature-sensitive allele of the Gal4 inhibitor Gal80 (Gal80ts)131. Flies were raised at 21 °C, temperature at which Gal80ts is stable and interferes with Gal4 function. Two days after eclosion, temperature was raised to 29 °C, temperature at which Gal80ts is degraded and the Gal4 driver becomes functional. Error bars, mean and S.E.M. Genotypes, sample sizes, and statistical analyses, Table S3.
Figure S2. CCHa1 produced in the gut regulates arousability but not sleep amount, related to Figure 2. (A) Antibody staining against CCHa1 and Prospero, a marker of enteroendocrine cells, in the midgut. (B) GFP expression in Mi{MIC}CCHa1MI09190 homozygous and heterozygous animals. Mi{MIC}CCHa1MI09190 contains a GFP open reading frame downstream of the CCHa1 5’ UTR. Heterozygotes for the Mi{MIC}CCHa1MI09190 construct (left) show a GFP expression pattern that overlaps with the CCHa1 antibody; homozygotes (right) show the same GFP expression pattern as heterozygotes but no CCHa1 antibody signal since both copies of the gene are mutated. (C) mRNA levels of CCHa1 in the gut (each dot represents 10 guts) or in gutless flies (each dot represents 2 gutless flies) as measured by qRT-PCR, in controls and in animals with CCHa1 depleted using elav-Gal4. (D) Expression pattern of elav-Gal4-driven nlsLacZ in the posterior midgut, co-stained with antibodies against CCHa1. (E) mRNA levels of CCHa1 in the gut (each dot represents 10 guts) or in gutless flies (each dot represents 2 gutless flies) as measured by qRT-PCR, in controls and in animals with CCHa1 depleted using conditional prosGal4 (a combination of pros-Gal4 and TubGal80ts). (F) pros-Gal4 – driven expression pattern of nuclear GFP (nls::GFP) in the central nervous system, co-stained with antibodies against CCHa1. (G) Measurements of arousabilty, sleep and locomotion in controls and in animals with CCHa1 using conditional pros-Gal4. (H) Food intake and clearance, in controls and in flies with gut-specific knockdown of CCHa1. (I) Measurements of arousabilty, sleep, and locomotion, in controls and in animals with activated enteroendocrine cells (prosts>TrpA1), with and without depleting CCHa1. The experiment has to be performed at 25 °C to ensure CCHa1-RNAi expression (see STAR Methods for details). UAS-GFP was included to control for UAS number. Error bars, mean and S.E.M. Scale bars: 100 μm. Genotypes, ample sizes and statistical analyses, Table S3.
Figure S3. Effects of different nutrients on CCHa1 levels, related to Figure 3. (A) Activity of the enteroendocrine cells, and levels of CCHa1 in the gut, in flies fed regular food, or food supplemented with sugar (galactose, fructose) or fat (propionic acid, hexanoic acid). Normalized to regular food (STAR Methods). Each dot represents 1 gut. (B) Food intake and clearance with peptone supplementation. Each dot represents the average of three experiments, each done with 5 flies. (C) Effects or peptone supplementation on arousability of awake flies, basal locomotion, and sleep parameters. (D) Arousability from sleep and responsiveness during wakefulness on regular diet and diet supplemented with fat or sugar. (E) Effect of equicaloric diets composed of the indicated proportions of macronutrients on responsiveness of awake animals. (F) Responsiveness of awake animals, in controls and in flies with peptone-supplemented diet, with or without depletion of CCHa1 from the gut (prosts>CCHa1-RNAi). (G) Arousability in response to low amplitude vibration for the parental controls, and medium amplitude for the experimental flies, to control for the experiment shown in Figure 3I. Error bars, mean and S.E.M.. Genotypes, sample sizes and statistical analyses, Table S3.
Figure S4. The weight of mice does not differ across diet conditions; sleep stages pattern and architecture and power of delta and theta waves, related to Figure 4. (A-C) Mouse weight on regular diet or protein- (A), sugar- (B) and fat-enriched diet (C). Day 0 is when mice were transferred to standard or macronutrient-enriched food. The weight was monitored 2 days before that change in diet and for 10 days afterwards. Grey rectangles represent the days during which arousability was tested. (D) Top: time spent awake, in non-REM sleep, in REM sleep, and asleep over the course of 24 hours, in 1-hour intervals. The white portion of the bar on top indicates daytime (lights on, 12 hours) while the black portion indicates nighttime (lights off, 12 hours). Middle: the number of bouts (episodes) for wakefulness, non-REM sleep, REM sleep and total daily sleep. Bottom, duration of bouts for wakefulness, non-REM sleep, REM sleep and total daily sleep. (E) Power analysis of delta and theta brain waves during wakefulness, non-REM and REM sleep. (F) Power analysis of delta and theta waves throughout the day in 1-hour intervals. Error bars, mean and S.E.M. Genotypes, sample sizes, and statistical analyses, Table S3.
Figure S5. CCHa1 receptor in PAMMB441B neurons regulates arousability, related to Figure 5. (A) Responsiveness during awake bouts, total daily sleep and basal locomotion in animals with the CCHa1 receptor knocked down using Ddc-Gal4 or PAM-Gal4. (B) Expression of GFP driven by two different Gal4s, which are themselves expressed under the CCHa1R regulatory elements (P{GMR23H07-GAL4}attP2 and TI{2A-Gal4}CCHa1-R2A-Gal4). Top, Z-stack projections of confocal images in which the PAM clusters of dopaminergic neurons are seen. Bottom, Z-stack projections through the whole central nervous system, with outlines corresponding to the areas on top. White arrows: PAM neurons (labeled with antibodies against tyrosine hydroxylase, TH, the rate-limiting enzyme in dopamine synthesis) which also express GFP. (C) Expression pattern of PAMMB441B-Gal4 in the gut. (D) Food intake and clearance, in controls and in animals with CCHa1 depleted from the gut (PAMMB441B>CCHa1-RNAi). Each dot represents the average of three experiments, each done with 15 flies. (E) Effect of knocking down CCHa1 receptor in the PAMMB441B dopaminergic neurons on responsiveness of awake animals, and various sleep parameters. (F) Effect of activation or inhibition of PAMMB441B neurons on responsiveness of awake animals, and various sleep parameters. (G) Effect of knocking down TH in PAMMB441B neurons on responsiveness of awake animals, and various sleep parameters. Error bars, mean and S.E.M. Scale bars: 100 μm. Genotypes, sample sizes and statistical analyses, Table S3.
Figure S6. CCHa1 receptor in PAM neurons regulates arousability in response to dietary proteins, and setup to measure arousability in response to increased temperature, related to Figure 6. (A) Responsiveness of awake animals, for controls and flies in which the CCHa1 receptor was depleted from PAMMB441B neurons. (B) Arousability in response to low amplitude vibrations for the parental controls, and medium amplitude vibrations for the experimental flies, to control for the experiment shown in Figure 6A. (C) Activity of PPL1 dopaminergic neurons on regular and peptone-supplemented food, as reported by CaLexA. (D) Activity of PAMMB441B neurons following a night of exposure to high amplitude vibrations, or to heat, as reported by CaLexA. Each dot represents 1 neuron. (E) The setup for probing arosuability using thermal stimulation. (F) Responsiveness of awake animal to heat, with or without peptone supplementation (left), and with or without depletion of the CCHa1 receptor from PAMMB441B neurons (right). (G) Arousabilty in response to 40 °C stimulation, with or without peptone supplementation. Error bars, mean and S.E.M. Genotypes, sample sizes, and statistical analyses, Table S3.
Figure S7. Sleep and arousability regulation through MBONMB083C neurons, related to Figure 7. (A) Expression pattern of MBONMB083C-Gal4 driver in the gut. Scale bar: 100 μm. (B) Responsiveness of awake animals, and various sleep parameters, in animals with silenced (green) or activated (yellow) MBONMB083C neurons. (C) Peptone-induced changes in arousal during wake bouts upon inhibition of MBONMB083C neurons. (D) Responsiveness of awake animals to stimulation, for controls and flies in which various dopamine receptors are depleted fromMBONMB083C neurons.(E) Schematic: the CCHa1 pathway regulates arousability through MBONMB083C neurons. Error bars, mean and S.E.M. Genotypes, sample sizes and statistical analyses, Table S3.
Table S3. Sample sizes and statistical analyses. Related to Figures 1–7 and Supplementary Figures 1–7.
Table S1. List of genes from the screen that show hypo or hyper-arousable phenotypes when knocked down using elav-Gal4. Related to Figure 1.
Table S2. Gene ontology analysis of the genes from the screen that when knocked down using elav-Gal4 show hypo or hyper-arousable phenotypes (shown in Table S1) for biological process and molecular function. Related to Figure 1.
HIGHLIGHTS.
Enrichment of dietary proteins can make flies and mice less arousable from sleep
Dietary proteins activate cells in the fly gut to secrete the peptide CCHa1
CCHa1 signals to brain dopamine neurons to modulate sensory responsiveness
Different sensory modalities can be gated by independent mechanisms during sleep
A protein-rich diet results in the release of a peptide secreted in the gut that signals to brain dopaminergic neurons that makes flies and mice less arousable from sleep
ACKNOWLEDGEMENTS
We thank our lab and Michael Crickmore’s lab for advice and comments on the manuscript. Benjamin Sanchez developed the software used to control mechanical stimuli and analyze behavioral responsiveness. Alexa Soares assisted with the screen. Alejandra Laureano assisted with the mouse experiments. For fly stocks, we thank Michael Crickmore, Nicholas Stavropoulos, Kyunghee Koh, and Norbert Perrimon. We thank Pedro Saavedra for assistance with the quantitative RT-PCR. Schematics were created with the help of BioRender.com. I.T. was supported by the Mahoney and Brooks postdoctoral fellowships. D.R. is a New York Stem Cell - Robertson investigator. This work was supported by the New York Stem Cell Foundation, the NIH (DP2 OD022385) and the Pew Scholars Program in the Biomedical Sciences.
Footnotes
DECLARATION OF INTERESTS
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Keene AC, and Duboue ER (2018). The origins and evolution of sleep. J. Exp. Biol. 221 10.1242/jeb.159533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Someren EJ, Cirelli C, Dijk DJ, Van Cauter E, Schwartz S, and Chee MW (2015). Disrupted Sleep: From Molecules to Cognition. J Neurosci 35, 13889–13895. 10.1523/JNEUROSCI.2592-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kline C. (2013). Sleep Quality. In Encyclopedia of Behavioral Medicine (Springer New York; ), pp. 1811–1813. 10.1007/978-1-4419-1005-9_849. [DOI] [Google Scholar]
- 4.Nelson KL, Davis JE, and Corbett CF (2022). Sleep quality: An evolutionary concept analysis. Nurs. Forum 57, 144–151. 10.1111/nuf.12659. [DOI] [PubMed] [Google Scholar]
- 5.Roth T, Zammit G, Lankford A, Mayleben D, Stern T, Pitman V, Clark D, and Werth JL (2010). Nonrestorative sleep as a distinct component of insomnia. Sleep 33, 449–458. 10.1093/sleep/33.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engel-Yeger B, and Shochat T. (2012). The Relationship between Sensory Processing Patterns and Sleep Quality in Healthy Adults. Can. J. Occup. Ther 79, 134–141. 10.2182/cjot.2012.79.3.2. [DOI] [PubMed] [Google Scholar]
- 7.Hirshkowitz M, and Sharafkhaneh A. (2005). Chapter 1 The physiology of sleep. In, pp. 3–20. 10.1016/S1567-4231(09)70026-7. [DOI] [Google Scholar]
- 8.Lebestky T., Chang JS., Dankert H., Zelnik L., Kim YC., Han KA., Wolf FW., Perona P., and Anderson DJ. (2009). Two different forms of arousal in Drosophila are oppositely regulated by the dopamine D1 receptor ortholog DopR via distinct neural circuits. Neuron 64, 522–536. 10.1016/j.neuron.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson L, Petty A, Rohrscheib C, Troup M, Kirszenblat L, Eyles DW, and van Swinderen B. (2017). Transient Dysregulation of Dopamine Signaling in a Developing Drosophila Arousal Circuit Permanently Impairs Behavioral Responsiveness in Adults. Front Psychiatry 8, 22. 10.3389/fpsyt.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donlea JM, Pimentel D, Talbot CB, Kempf A, Omoto JJ, Hartenstein V, and Miesenbock G. (2018). Recurrent Circuitry for Balancing Sleep Need and Sleep. Neuron 97, 378–389 e4. 10.1016/j.neuron.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Troup M, Yap MH, Rohrscheib C, Grabowska MJ, Ertekin D, Randeniya R, Kottler B, Larkin A, Munro K, Shaw PJ, et al. (2018). Acute control of the sleep switch in Drosophila reveals a role for gap junctions in regulating behavioral responsiveness. Elife 7. 10.7554/eLife.37105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen KF, Lowe S, Lamaze A, Kratschmer P, and Jepson J. (2019). Neurocalcin regulates nighttime sleep and arousal in Drosophila. Elife 8. 10.7554/eLife.38114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayat H, Regev N, Matosevich N, Sales A, Paredes-Rodriguez E, Krom AJ, Bergman L, Li Y, Lavigne M, Kremer EJ, et al. (2020). Locus coeruleus norepinephrine activity mediates sensory-evoked awakenings from sleep. Sci. Adv 6, eaaz4232. 10.1126/sciadv.aaz4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sletten TL, Rajaratnam SMW, Wright MJ, Zhu G, Naismith S, Martin NG, and Hickie I. (2013). Genetic and environmental contributions to sleep-wake behavior in 12-year-old twins. Sleep 36, 1715–1722. 10.5665/sleep.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sehgal A, and Mignot E. (2011). Genetics of sleep and sleep disorders. Cell 146, 194–207. 10.1016/j.cell.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomita J, Ban G, and Kume K. (2017). Genes and neural circuits for sleep of the fruit fly. Neurosci Res 118, 82–91. 10.1016/j.neures.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Peuhkuri K, Sihvola N, and Korpela R. (2012). Diet promotes sleep duration and quality. Nutr. Res 32, 309–319. 10.1016/j.nutres.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Godos J., Grosso G., Castellano S., Galvano F., Caraci F., and Ferr R. (2021). Association between diet and sleep quality: A systematic review. Sleep Med. Rev 57, 101430. 10.1016/j.smrv.2021.101430. [DOI] [PubMed] [Google Scholar]
- 19.Binks H, E. Vincent G, Gupta C, Irwin C, and Khalesi S. (2020). Effects of Diet on Sleep: A Narrative Review. Nutrients 12, 936. 10.3390/nu12040936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beckwith EJ, and French AS (2019). Sleep in Drosophila and Its Context. Front Physiol 10, 1167. 10.3389/fphys.2019.01167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.St-Onge M-P, Mikic A, and Pietrolungo CE (2016). Effects of Diet on Sleep Quality. Adv. Nutr 7, 938–949. 10.3945/an.116.012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin DM, and Goodman CS (1994). Ectopic and increased expression of fasciclin II alters motoneuron growth cone guidance. Neuron 13, 507–523. 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 23.Ida T, Takahashi T, Tominaga H, Sato T, Sano H, Kume K, Ozaki M, Hiraguchi T, Shiotani H, Terajima S, et al. (2012). Isolation of the bioactive peptides CCHamide-1 and CCHamide-2 from Drosophila and their putative role in appetite regulation as ligands for G protein-coupled receptors. Front Endocrinol 3, 177. 10.3389/fendo.2012.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farhan A, Gulati J, Grobetae-Wilde E, Vogel H, Hansson BS, and Knaden M. (2013). The CCHamide 1 receptor modulates sensory perception and olfactory behavior in starved Drosophila. Sci Rep 3, 2765. 10.1038/srep02765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Torre-Muruzabal T, Søgaard KC, Ren GR, Hauser F, Engelsen SM, Pødenphanth MD, Desjardins A, and Grimmelikhuijzen CJP (2013). Expression Patterns of the Drosophila Neuropeptide CCHamide-2 and Its Receptor May Suggest Hormonal Signaling from the Gut to the Brain. PLoS One 8, e76131. 10.1371/journal.pone.0076131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sano H, Nakamura A, Texada MJ, Truman JW, Ishimoto H, Kamikouchi A, Nibu Y, Kume K, Ida T, and Kojima M. (2015). The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of Drosophila melanogaster. PLoS Genet 11, e1005209. 10.1371/journal.pgen.1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren GR, Hauser F, Rewitz KF, Kondo S, Engelbrecht AF, Didriksen AK, Schjott SR, Sembach FE, Li S, Sogaard KC, et al. (2015). CCHamide-2 Is an Orexigenic Brain-Gut Peptide in Drosophila. PLoS One 10, e0133017. 10.1371/journal.pone.0133017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujiwara Y., Hermann-Luibl C., Katsura M., Sekiguchi M., Ida T., Helfrich-Förster C., and Yoshii T. (2018). The CCHamide1 Neuropeptide Expressed in the Anterior Dorsal Neuron 1 Conveys a Circadian Signal to the Ventral Lateral Neurons in Drosophila melanogaster. Front. Physiol 9, 1276. 10.3389/fphys.2018.01276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagani M, Albisetti GW, Sivakumar N, Wildner H, Santello M, Johannssen HC, and Zeilhofer HU (2019). How Gastrin-Releasing Peptide Opens the Spinal Gate for Itch. Neuron 103, 102–117 e5. 10.1016/j.neuron.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamada K, Wada E, and Wada K. (2000). Male mice lacking the gastrin-releasing peptide receptor (GRP-R) display elevated preference for conspecific odors and increased social investigatory behaviors. Brain Res. 870, 20–26. 10.1016/S0006-8993(00)02395-7. [DOI] [PubMed] [Google Scholar]
- 31.Yamada K, Wada E, and Wada K. (2000). Bombesin-like peptides: studies on food intake and social behaviour with receptor knock-out mice. Ann. Med 32, 519–529. 10.3109/07853890008998831. [DOI] [PubMed] [Google Scholar]
- 32.Shumyatsky GP, Tsvetkov E, Malleret G, Vronskaya S, Hatton M, Hampton L, Battey JF, Dulac C, Kandel ER, and Bolshakov VY (2002). Identification of a Signaling Network in Lateral Nucleus of Amygdala Important for Inhibiting Memory Specifically Related to Learned Fear. Cell 111, 905–918. 10.1016/S0092-8674(02)01116-9. [DOI] [PubMed] [Google Scholar]
- 33.Roesler R, Lessa D, Venturella R, Vianna MRM, Luft T, Henriques JAP, Izquierdo I, and Schwartsmann G. (2004). Bombesin/gastrin-releasing peptide receptors in the basolateral amygdala regulate memory consolidation. Eur. J. Neurosci 19, 1041–1045. 10.1111/j.0953-816X.2004.03175.x. [DOI] [PubMed] [Google Scholar]
- 34.Merali Z, Bedard T, Andrews N, Davis B, McKnight AT, Gonzalez MI, Pritchard M, Kent P, and Anisman H. (2006). Bombesin Receptors as a Novel Anti-Anxiety Therapeutic Target: BB1 Receptor Actions on Anxiety through Alterations of Serotonin Activity. J. Neurosci 26, 10387–10396. 10.1523/JNEUROSCI.1219-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li P, Janczewski WA, Yackle K, Kam K, Pagliardini S, Krasnow MA, and Feldman JL (2016). The peptidergic control circuit for sighing. Nature 530, 293–297. 10.1038/nature16964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ladenheim EE, Taylor JE, Coy DH, Moore KA, and Moran TH (1996). Hindbrain GRP receptor blockade antagonizes feeding suppression by peripherally administered GRP. Am. J. Physiol 271, R180–4. 10.1152/ajpregu.1996.271.1.R180. [DOI] [PubMed] [Google Scholar]
- 37.Ladenheim EE, Behles RR, Bi S, and Moran TH (2009). Gastrin-Releasing Peptide Messenger Ribonucleic Acid Expression in the Hypothalamic Paraventricular Nucleus Is Altered by Melanocortin Receptor Stimulation and Food Deprivation. Endocrinology 150, 672–678. 10.1210/en.2008-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shinohara K., Tominaga K., Isobe Y., and Inouye ST. (1993). Photic regulation of peptides located in the ventrolateral subdivision of the suprachiasmatic nucleus of the rat: daily variations of vasoactive intestinal polypeptide, gastrin-releasing peptide, and neuropeptide Y. J. Neurosci 13, 793–800. 10.1523/JNEUROSCI.13-02-00793.1993 Free PMC article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okamura H, and Ibata Y. (1994). GRP immunoreactivity shows a day-night difference in the suprachiasmatic nuclear soma and efferent fibers: Comparison to VIP immunoreactivity. Neurosci. Lett 181, 165–168. 10.1016/0304-3940(94)90585-1. [DOI] [PubMed] [Google Scholar]
- 40.Karatsoreos IN, Romeo RD, McEwen BS, and Silver R. (2006). Diurnal regulation of the gastrin-releasing peptide receptor in the mouse circadian clock. Eur. J. Neurosci 23, 1047–1053. 10.1111/j.1460-9568.2006.04633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harbison ST (2010). Evolution of Sleep: Phylogenetic and Functional Perspectives. Patrick McNamara, Barton Robert A., and Nunn Charles L., editors. Integr. Comp. Biol 50, 685–687. 10.1093/icb/icq053. [DOI] [Google Scholar]
- 42.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, and Pack AI (2000). Rest in Drosophila is a sleep-like state. Neuron 25, 129–138. 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 43.Shaw PJ, Cirelli C, Greenspan RJ, and Tononi G. (2000). Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837. 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 44.Wiggin TD, Goodwin PR, Donelson NC, Liu C, Trinh K, Sanyal S, and Griffith LC (2020). Covert sleep-related biological processes are revealed by probabilistic analysis in Drosophila. Proc. Natl. Acad. Sci. U. S. A 117, 10024–10034. 10.1073/pnas.1917573117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirszenblat L, and van Swinderen B. (2015). The Yin and Yang of Sleep and Attention. Trends Neurosci. 38, 776–786. 10.1016/j.tins.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Swinderen B, and Andretic R. (2011). Dopamine in Drosophila: setting arousal thresholds in a miniature brain. Proc. R. Soc. B Biol. Sci 278, 906–913. 10.1098/rspb.2010.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eban-Rothschild A, Appelbaum L, and de Lecea L. (2018). Neuronal Mechanisms for Sleep/Wake Regulation and Modulatory Drive. Neuropsychopharmacology 43, 937–952. 10.1038/npp.2017.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagarkar-Jaiswal S, Lee P-T, Campbell ME, Chen K, Anguiano-Zarate S, Cantu Gutierre M., Busby T., Lin W-W., He Y., Schulze KL., et al. (2015). A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife 4. 10.7554/eLife.05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veenstra JA, and Ida T. (2014). More Drosophila enteroendocrine peptides: Orcokinin B and the CCHamides 1 and 2. Cell Tissue Res. 357, 607–621. 10.1007/s00441-014-1880-2. [DOI] [PubMed] [Google Scholar]
- 50.Miguel-Aliaga I, Jasper H, and Lemaitre B. (2018). Anatomy and Physiology of the Digestive Tract of Drosophila melanogaster. Genetics 210, 357–396. 10.1534/genetics.118.300224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S, Torre-Muruzabal T, Sogaard KC, Ren GR, Hauser F, Engelsen SM, Podenphanth MD, Desjardins A, and Grimmelikhuijzen CJ (2013). Expression patterns of the Drosophila neuropeptide CCHamide-2 and its receptor may suggest hormonal signaling from the gut to the brain. PLoS One 8, e76131. 10.1371/journal.pone.0076131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J, Reiher W, Hermann-Luibl C, Sellami A, Cognigni P, Kondo S, Helfrich-Forster C, Veenstra JA, and Wegener C. (2016). Allatostatin A Signalling in Drosophila Regulates Feeding and Sleep and Is Modulated by PDF. PLoS Genet 12, e1006346. 10.1371/journal.pgen.1006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song W, Veenstra JA, and Perrimon N. (2014). Control of lipid metabolism by tachykinin in Drosophila. Cell Rep 9, 40–47. 10.1016/j.celrep.2014.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sen A, Kuruvilla D, Pinto L, Sarin A, and Rodrigues V. (2004). Programmed cell death and context dependent activation of the EGF pathway regulate gliogenesis in the Drosophila olfactory system. Mech. Dev 121, 65–78. 10.1016/j.mod.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 55.van den Pol AN (2012). Neuropeptide transmission in brain circuits. Neuron 76, 98–115. 10.1016/j.neuron.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nässel DR, and Zandawala M. (2019). Recent advances in neuropeptide signaling in Drosophila, from genes to physiology and behavior. Prog. Neurobiol 179, 101607. 10.1016/j.pneurobio.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Rogers GJ, Tolhurst G, Ramzan A, Habib AM, Parker HE, Gribble FM, and Reimann F. (2011). Electrical activity-triggered glucagon-like peptide-1 secretion from primary murine L-cells. J Physiol 589, 1081–1093. 10.1113/jphysiol.2010.198069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park J-H, Chen J, Jang S, Ahn TJ, Kang K, Choi MS, and Kwon JY (2016). A subset of enteroendocrine cells is activated by amino acids in the Drosophila midgut. FEBS Lett. 590, 493–500. 10.1002/1873-3468.12073. [DOI] [PubMed] [Google Scholar]
- 59.Waddell S., Armstrong JD., Kitamoto T., Kaiser K., and Quinn WG. (2000). The amnesiac Gene Product Is Expressed in Two Neurons in the Drosophila Brain that Are Critical for Memory. Cell 103, 805–813. 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- 60.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, and Garrity PA (2008). An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–220. 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beckwith EJ, and French AS (2019). Sleep in Drosophila and Its Context. Front. Physiol. 10 10.3389/fphys.2019.01167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Masuyama K, Zhang Y, Rao Y, and Wang JW (2012). Mapping Neural Circuits with Activity-Dependent Nuclear Import of a Transcription Factor. J. Neurogenet 26, 89–102. 10.3109/01677063.2011.642910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown EB, Shah KD, Faville R, Kottler B, and Keene AC (2020). Drosophila insulin-like peptide 2 mediates dietary regulation of sleep intensity. PLOS Genet. 16, e1008270. 10.1371/journal.pgen.1008270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He L, Binari R, Huang J, Falo-Sanjuan J, and Perrimon N. (2019). In vivo study of gene expression with an enhanced dual-color fluorescent transcriptional timer. Elife 8. 10.7554/eLife.46181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kain P, Boyle SM, Tharadra SK, Guda T, Pham C, Dahanukar A, and Ray A. (2013). Odour receptors and neurons for DEET and new insect repellents. Nature 502, 507–512. 10.1038/nature12594. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Koh T-W, He Z, Gorur-Shandilya S, Menuz K, Larter NK, Stewart S, and Carlson JR (2014). The Drosophila IR20a Clade of Ionotropic Receptors Are Candidate Taste and Pheromone Receptors. Neuron 83, 850–865. 10.1016/j.neuron.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zandawala M, Yurgel ME, Texada MJ, Liao S, Rewitz KF, Keene AC, and Nässel DR (2018). Modulation of Drosophila post-feeding physiology and behavior by the neuropeptide leucokinin. PLOS Genet. 14, e1007767. 10.1371/journal.pgen.1007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pamboro ELS, Brown EB, and Keene AC (2020). Dietary fatty acids promote sleep through a taste-independent mechanism. Genes. Brain. Behav 19, e12629. 10.1111/gbb.12629. [DOI] [PubMed] [Google Scholar]
- 69.Heinrichsen ET, and Haddad GG (2012). Role of high-fat diet in stress response of Drosophila. PLoS One 7, e42587. 10.1371/journal.pone.0042587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soltani S, Chauvette S, Bukhtiyarova O, Lina J-M, Dubé J, Seigneur J, Carrier J, and Timofeev I. (2019). Sleep–Wake Cycle in Young and Older Mice. Front. Syst. Neurosci. 13 10.3389/fnsys.2019.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saper CB, Fuller PM, Pedersen NP, Lu J, and Scammell TE (2010). Sleep State Switching. Neuron 68, 1023–1042. 10.1016/j.neuron.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ermis U, Krakow K, and Voss U. (2010). Arousal thresholds during human tonic and phasic REM sleep. J. Sleep Res 19, 400–406. 10.1111/j.1365-2869.2010.00831.x. [DOI] [PubMed] [Google Scholar]
- 73.Neckelmann D, and Ursin R. (1993). Sleep stages and EEG power spectrum in relation to acoustical stimulus arousal threshold in the rat. Sleep 16, 467–477. [PubMed] [Google Scholar]
- 74.Dale Purves, George J Augustine, David Fitzpatrick, Lawrence C Katz, Anthony-Samuel LaMantia, James O McNamara, and S.M. W. (2001). Neuroscience 2nd editio. (Sinauer Associates; ). [Google Scholar]
- 75.Hobson JA, and Pace-Schott EF (2002). The cognitive neuroscience of sleep: neuronal systems, consciousness and learning. Nat. Rev. Neurosci 3, 679–693. 10.1038/nrn915. [DOI] [PubMed] [Google Scholar]
- 76.Abhang PA, Gawali BW, and Mehrotra SC (2016). Technological Basics of EEG Recording and Operation of Apparatus. In Introduction to EEG- and Speech-Based Emotion Recognition (Elsevier; ), pp. 19–50. 10.1016/B978-0-12-804490-2.00002-6. [DOI] [Google Scholar]
- 77.Latorre R, Sternini C, De Giorgio R, and Greenwood-Van Meerveld B. (2016). Enteroendocrine cells: a review of their role in brain-gut communication. Neurogastroenterol. Motil 28, 620–630. 10.1111/nmo.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu C, Placais PY, Yamagata N, Pfeiffer BD, Aso Y, Friedrich AB, Siwanowicz I, Rubin GM, Preat T, and Tanimoto H. (2012). A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature 488, 512–516. 10.1038/nature11304. [DOI] [PubMed] [Google Scholar]
- 79.Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H, et al. (2014). The neuronal architecture of the mushroom body provides a logic for associative learning. Elife 3, e04577. 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aso Y, Ray RP, Long X, Bushey D, Cichewicz K, Ngo TT, Sharp B, Christoforou C, Hu A, Lemire AL, et al. (2019). Nitric oxide acts as a cotransmitter in a subset of dopaminergic neurons to diversify memory dynamics. Elife 8. 10.7554/eLife.49257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Riemensperger T., Isabel G., Coulom H., Neuser K., Seugnet L., Kume K., Iche-Torres M., Cassar M., Strauss R., Preat T., et al. (2011). Behavioral consequences of dopamine deficiency in the Drosophila central nervous system. Proc Natl Acad Sci U S A 108, 834–839. 10.1073/pnas.1010930108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kume K, Kume S, Park SK, Hirsh J, and Jackson FR (2005). Dopamine is a regulator of arousal in the fruit fly. J Neurosci 25, 7377–7384. 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wisor JP (2019). Dopamine and Wakefulness: Pharmacology, Genetics, and Circuitry. Handb Exp Pharmacol 253, 321–335. 10.1007/164_2018_95. [DOI] [PubMed] [Google Scholar]
- 84.Heisenberg M, Borst A, Wagner S, and Byers D. (1985). Drosophila mushroom body mutants are deficient in olfactory learning. J Neurogenet 2, 1–30. 10.3109/01677068509100140. [DOI] [PubMed] [Google Scholar]
- 85.de Belle JS, and Heisenberg M. (1994). Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science (80-. ). 263, 692–695. 10.1126/science.8303280. [DOI] [PubMed] [Google Scholar]
- 86.Strausfeld NJ, Hansen L, Li Y, Gomez RS, and Ito K. (1998). Evolution, Discovery, and Interpretations of Arthropod Mushroom Bodies. Learn. Mem 5, 11–37. 10.1101/lm.5.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu L, Wolf R, Ernst R, and Heisenberg M. (1999). Context generalization in Drosophila visual learning requires the mushroom bodies. Nature 400, 753–756. 10.1038/23456. [DOI] [PubMed] [Google Scholar]
- 88.Hong ST, Bang S, Hyun S, Kang J, Jeong K, Paik D, Chung J, and Kim J. (2008). cAMP signalling in mushroom bodies modulates temperature preference behaviour in Drosophila. Nature 454, 771–775. 10.1038/nature07090. [DOI] [PubMed] [Google Scholar]
- 89.Vogt K, Schnaitmann C, Dylla KV, Knapek S, Aso Y, Rubin GM, and Tanimoto H. (2014). Shared mushroom body circuits underlie visual and olfactory memories in Drosophila. Elife 3, e02395. 10.7554/eLife.02395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li J, Mahoney BD, Jacob MS, and Caron SJC (2020). Visual Input into the Drosophila melanogaster Mushroom Body. Cell Rep. 32, 108138. 10.1016/j.celrep.2020.108138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aso Y, Sitaraman D, Ichinose T, Kaun KR, Vogt K, Belliart-Guerin G, Placais PY, Robie AA, Yamagata N, Schnaitmann C, et al. (2014). Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila 2014/12/24. 10.7554/eLife.04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martin JR, Ernst R, and Heisenberg M. (1998). Mushroom bodies suppress locomotor activity in Drosophila melanogaster. Learn Mem 5, 179–191. [PMC free article] [PubMed] [Google Scholar]
- 93.Dubnau J., Grady L., Kitamoto T., and Tully T. (2001). Disruption of neurotransmission in Drosophila mushroom body blocks retrieval but not acquisition of memory. Nature 411, 476–480. 10.1038/35078077. [DOI] [PubMed] [Google Scholar]
- 94.McGuire SE, Le PT, and Davis RL (2001). The role of Drosophila mushroom body signaling in olfactory memory. Science (80-. ). 293, 1330–1333. 10.1126/science.1062622. [DOI] [PubMed] [Google Scholar]
- 95.Joiner WJ, Crocker A, White BH, and Sehgal A. (2006). Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441, 757–760. 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- 96.Pitman JL, McGill JJ, Keegan KP, and Allada R. (2006). A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441, 753–756. 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- 97.Cohn R, Morantte I, and Ruta V. (2015). Coordinated and Compartmentalized Neuromodulation Shapes Sensory Processing in Drosophila. Cell 163, 1742–1755. 10.1016/j.cell.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang K, Guo JZ, Peng Y, Xi W, and Guo A. (2007). Dopamine-mushroom body circuit regulates saliency-based decision-making in Drosophila. Science (80-. ). 316, 1901–1904. 10.1126/science.1137357. [DOI] [PubMed] [Google Scholar]
- 99.Heisenberg M. (2003). Mushroom body memoir: from maps to models. Nat Rev Neurosci 4, 266–275. 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 100.Eschbach C, Fushiki A, Winding M, Schneider-Mizell CM, Shao M, Arruda R, Eichler K, Valdes-Aleman J, Ohyama T, Thum AS, et al. (2020). Recurrent architecture for adaptive regulation of learning in the insect brain. Nat. Neurosci 23, 544–555. 10.1038/s41593-020-0607-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jovanic T, Winding M, Cardona A, Truman JW, Gershow M, and Zlatic M. (2019). Neural Substrates of Drosophila Larval Anemotaxis. Curr. Biol 29, 554–566.e4. 10.1016/j.cub.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dolan M-J, Frechter S, Bates AS, Dan C, Huoviala P, Roberts RJ, Schlegel P, Dhawan S, Tabano R, Dionne H, et al. (2019). Neurogenetic dissection of the Drosophila lateral horn reveals major outputs, diverse behavioural functions, and interactions with the mushroom body. Elife 8. 10.7554/eLife.43079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li F, Lindsey JW, Marin EC, Otto N, Dreher M, Dempsey G, Stark I, Bates AS, Pleijzier MW, Schlegel P, et al. (2020). The connectome of the adult Drosophila mushroom body provides insights into function. Elife 9. 10.7554/eLife.62576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Siju KP, Štih V, Aimon S, Gjorgjieva J, Portugues R, and Grunwald Kadow IC (2020). Valence and State-Dependent Population Coding in Dopaminergic Neurons in the Fly Mushroom Body. Curr. Biol 30, 2104–2115.e4. 10.1016/j.cub.2020.04.037. [DOI] [PubMed] [Google Scholar]
- 105.Steriad M. (1997). Synchronized activities of coupled oscillators in the cerebral cortex and thalamus at different levels of vigilance [published erratum appears in Cereb Cortex 1997 Dec;7(8):779]. Cereb. Cortex 7, 583–604. 10.1093/cercor/7.6.583. [DOI] [PubMed] [Google Scholar]
- 106.Steriade M, and Llinás RR (1988). The functional states of the thalamus and the associated neuronal interplay. Physiol. Rev 68, 649–742. 10.1152/physrev.1988.68.3.649. [DOI] [PubMed] [Google Scholar]
- 107.Murakami M, Kashiwadani H, Kirino Y, and Mori K. (2005). State-Dependent Sensory Gating in Olfactory Cortex. Neuron 46, 285–296. 10.1016/j.neuron.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 108.Dillon ME, Wang G, Garrity PA, and Huey RB (2009). Thermal preference in Drosophila. J. Therm. Biol 34, 109–119. 10.1016/j.jtherbio.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sitaraman D, Aso Y, Jin X, Chen N, Felix M, Rubin GM, and Nitabach MN (2015). Propagation of Homeostatic Sleep Signals by Segregated Synaptic Microcircuits of the Drosophila Mushroom Body. Curr Biol 25, 2915–2927. 10.1016/j.cub.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shafer OT, and Keene AC (2021). The Regulation of Drosophila Sleep. Curr. Biol 31, R38–R49. 10.1016/j.cub.2020.10.082. [DOI] [PubMed] [Google Scholar]
- 111.Keene AC, Duboue ER, McDonald DM, Dus M, Suh GS, Waddell S, and Blau J. (2010). Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol 20, 1209–1215. 10.1016/j.cub.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Siegel JM (2005). Clues to the functions of mammalian sleep. Nature 437, 1264–1271. 10.1038/nature04285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hua R, Wang X, Chen X, Wang X, Huang P, Li P, Mei W, and Li H. (2018). Calretinin Neurons in the Midline Thalamus Modulate Starvation-Induced Arousal. Curr. Biol 28, 3948–3959.e4. 10.1016/j.cub.2018.11.020. [DOI] [PubMed] [Google Scholar]
- 114.Murphy KR, Deshpande SA, Yurgel ME, Quinn JP, Weissbach JL, Keene AC, Dawson-Scully K, Huber R, Tomchik SM, and Ja WW (2016). Postprandial sleep mechanics in Drosophila. Elife 5. 10.7554/eLife.19334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Danguir J, Nicoladis S, and Gerard H. (1979). Relations between feeding and sleep patterns in the rat. J. Comp. Physiol. Psychol 93, 820–830. 10.1037/h0077616. [DOI] [Google Scholar]
- 116.Westerterp-Plantenga MS, Nieuwenhuizen A, Tomé D, Soenen S, and Westerterp KR (2009). Dietary protein, weight loss, and weight maintenance. Annu. Rev. Nutr 29, 21–41. 10.1146/annurev-nutr-080508-141056. [DOI] [PubMed] [Google Scholar]
- 117.Kaelberer MM, Buchanan KL, Klein ME, Barth BB, Montoya MM, Shen X, and Bohórquez DV (2018). A gut-brain neural circuit for nutrient sensory transduction. Science 361. 10.1126/science.aat5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bohórquez DV., Samsa LA., Roholt A., Medicetty S., Chandra R., and Liddle RA. (2014). An Enteroendocrine Cell – Enteric Glia Connection Revealed by 3D Electron Microscopy. PLoS One 9, e89881. 10.1371/journal.pone.0089881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dujardin F. (1850). Memoire sur le systeme nerveux des Insectes. Ann Sci Nat Zool 14, 195–206. [Google Scholar]
- 120.French AS, Geissmann Q, Beckwith EJ, and Gilestro GF (2021). Sensory processing during sleep in Drosophila melanogaster. Nature 598, 479–482. 10.1038/s41586-021-03954-w. [DOI] [PubMed] [Google Scholar]
- 121.Klein MO, Battagello DS, Cardoso AR, Hauser DN, Bittencourt JC, and Correa RG (2019). Dopamine: Functions, Signaling, and Association with Neurological Diseases. Cell Mol Neurobiol 39, 31–59. 10.1007/s10571-018-0632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sun X, Zhao Y, and Wolf ME (2005). Dopamine Receptor Stimulation Modulates AMPA Receptor Synaptic Insertion in Prefrontal Cortex Neurons. J. Neurosci 25, 7342–7351. 10.1523/JNEUROSCI.4603-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tritsch NX, and Sabatini BL (2012). Dopaminergic Modulation of Synaptic Transmission in Cortex and Striatum. Neuron 76, 33–50. 10.1016/j.neuron.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang SX, Miner LE, Boutros CL, Rogulja D, and Crickmore MA (2018). Motivation, Perception, and Chance Converge to Make a Binary Decision. Neuron 99, 376–388 e6. 10.1016/j.neuron.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vaccaro A, Kaplan Dor Y, Nambara K, Pollina EA, Lin C, Greenberg ME, and Rogulja D. (2020). Sleep Loss Can Cause Death through Accumulation of Reactive Oxygen Species in the Gut. Cell 181, 1307–1328.e15. 10.1016/j.cell.2020.04.049. [DOI] [PubMed] [Google Scholar]
- 126.Rao M, and Gershon MD (2016). The bowel and beyond: the enteric nervous system in neurological disorders. Nat Rev Gastroenterol Hepatol 13, 517–528. 10.1038/nrgastro.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fernandes AB, Alves da Silva J, Almeida J, Cui G, Gerfen CR, Costa RM, and Oliveira-Maia AJ (2020). Postingestive Modulation of Food Seeking Depends on Vagus-Mediated Dopamine Neuron Activity. Neuron 106, 778–788.e6. 10.1016/j.neuron.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Valeur J. (2019). Gut-brain axis in history and culture. Microb. Ecol. Health Dis 29. 10.1080/16512235.2019.1602995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xiao R, Shida-Tokeshi J, Vanderbilt DL, and Smith BA (2018). Electroencephalography power and coherence changes with age and motor skill development across the first half year of life. PLoS One 13, e0190276. 10.1371/journal.pone.0190276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dressler O., Schneider G., Stockmanns G., and Kochs EF. (2004). Awareness and the EEG power spectrum: analysis of frequencies. Br. J. Anaesth 93, 806–809. 10.1093/bja/aeh270. [DOI] [PubMed] [Google Scholar]
- 131.McGuire SE, Le PT, Osborn AJ, Matsumoto K, and Davis RL (2003). Spatiotemporal Rescue of Memory Dysfunction in Drosophila. Science (80-. ). 302, 1765–1768. 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Experimental setup and analysis of CCHa1 and CCHa1R mutants and RNAi lines, related to Figure 1. (A) Setup used for mechanical stimulation. Custom software generates 2-second-long multisine (40–200 Hz) signals that are converted into mechanical stimuli (vibrations). Vibrations are given at random intervals centered on 30–40 minutes throughout the night. Drosophila activity monitors (DAMs, TriKinetics, Waltham, MA) are mounted on custombuilt platforms on top of the loudspeakers. Accelerometers placed on top of DAMs (arrow) measure the stimulus intensities to ensure that all flies are stimulated equally regardless of their position on the platform. (B) Each fly is housed in a glass tube, with ~25 mm of space for movement. Movement is detected by the interruption of the infrared light beam and recorded every minute. (C) mRNA levels of CCHa1 and CCHa2, as measured by qRT-PCR done on whole flies, in controls and in animals which express RNAi against CCHa1 using elav-Gal4. Each dot represents 2 flies. mRNA levels of CCHa1R and CCHa2R, as measured by qRT-PCR of fly brains in controls and in animals which express RNAi against CCHa1R using elav-Gal4. Each dot represents 5 brains. (D) The first panel shows reactivity of flies which were awake at the time vibrations were delivered. Other panels show various measurements of sleep and locomotion. The last panel shows sleep amount throughout the day, in 30-minute intervals. The white portion of the bar on top indicates daytime (lights on) and the black portion indicates nighttime (lights off). (E) Measurements of arousabilty, sleep and locomotion using additional RNAis against CCHa1 and CCHa1R. (F) mRNA levels of CCHa1 and CCHa2, as measured by qRT-PCR done on whole flies, in controls and in flies which express RNAi#2 against CCHa1 using the elav-Gal4 promoter. Each dot represents 2 flies. mRNA levels of CCHa1R and CCHa2R, as measured by qRT-PCR of fly brains in controls and in flies which express RNAi#2 against CCHa1R using elav-Gal4 promoter. Each dot represents 5 brains. (G) Measurements of arousabilty, sleep, and locomotion in controls and in mutants for CCHa1 and CCHa1R. (H) Measurements of arousabilty in controls, and in flies with conditional expression of RNAi against CCHa1 and CCHa1R. Adult-specific expression was achieved by combining elav-Gal4 and a temperature-sensitive allele of the Gal4 inhibitor Gal80 (Gal80ts)131. Flies were raised at 21 °C, temperature at which Gal80ts is stable and interferes with Gal4 function. Two days after eclosion, temperature was raised to 29 °C, temperature at which Gal80ts is degraded and the Gal4 driver becomes functional. Error bars, mean and S.E.M. Genotypes, sample sizes, and statistical analyses, Table S3.
Figure S2. CCHa1 produced in the gut regulates arousability but not sleep amount, related to Figure 2. (A) Antibody staining against CCHa1 and Prospero, a marker of enteroendocrine cells, in the midgut. (B) GFP expression in Mi{MIC}CCHa1MI09190 homozygous and heterozygous animals. Mi{MIC}CCHa1MI09190 contains a GFP open reading frame downstream of the CCHa1 5’ UTR. Heterozygotes for the Mi{MIC}CCHa1MI09190 construct (left) show a GFP expression pattern that overlaps with the CCHa1 antibody; homozygotes (right) show the same GFP expression pattern as heterozygotes but no CCHa1 antibody signal since both copies of the gene are mutated. (C) mRNA levels of CCHa1 in the gut (each dot represents 10 guts) or in gutless flies (each dot represents 2 gutless flies) as measured by qRT-PCR, in controls and in animals with CCHa1 depleted using elav-Gal4. (D) Expression pattern of elav-Gal4-driven nlsLacZ in the posterior midgut, co-stained with antibodies against CCHa1. (E) mRNA levels of CCHa1 in the gut (each dot represents 10 guts) or in gutless flies (each dot represents 2 gutless flies) as measured by qRT-PCR, in controls and in animals with CCHa1 depleted using conditional prosGal4 (a combination of pros-Gal4 and TubGal80ts). (F) pros-Gal4 – driven expression pattern of nuclear GFP (nls::GFP) in the central nervous system, co-stained with antibodies against CCHa1. (G) Measurements of arousabilty, sleep and locomotion in controls and in animals with CCHa1 using conditional pros-Gal4. (H) Food intake and clearance, in controls and in flies with gut-specific knockdown of CCHa1. (I) Measurements of arousabilty, sleep, and locomotion, in controls and in animals with activated enteroendocrine cells (prosts>TrpA1), with and without depleting CCHa1. The experiment has to be performed at 25 °C to ensure CCHa1-RNAi expression (see STAR Methods for details). UAS-GFP was included to control for UAS number. Error bars, mean and S.E.M. Scale bars: 100 μm. Genotypes, ample sizes and statistical analyses, Table S3.
Figure S3. Effects of different nutrients on CCHa1 levels, related to Figure 3. (A) Activity of the enteroendocrine cells, and levels of CCHa1 in the gut, in flies fed regular food, or food supplemented with sugar (galactose, fructose) or fat (propionic acid, hexanoic acid). Normalized to regular food (STAR Methods). Each dot represents 1 gut. (B) Food intake and clearance with peptone supplementation. Each dot represents the average of three experiments, each done with 5 flies. (C) Effects or peptone supplementation on arousability of awake flies, basal locomotion, and sleep parameters. (D) Arousability from sleep and responsiveness during wakefulness on regular diet and diet supplemented with fat or sugar. (E) Effect of equicaloric diets composed of the indicated proportions of macronutrients on responsiveness of awake animals. (F) Responsiveness of awake animals, in controls and in flies with peptone-supplemented diet, with or without depletion of CCHa1 from the gut (prosts>CCHa1-RNAi). (G) Arousability in response to low amplitude vibration for the parental controls, and medium amplitude for the experimental flies, to control for the experiment shown in Figure 3I. Error bars, mean and S.E.M.. Genotypes, sample sizes and statistical analyses, Table S3.
Figure S4. The weight of mice does not differ across diet conditions; sleep stages pattern and architecture and power of delta and theta waves, related to Figure 4. (A-C) Mouse weight on regular diet or protein- (A), sugar- (B) and fat-enriched diet (C). Day 0 is when mice were transferred to standard or macronutrient-enriched food. The weight was monitored 2 days before that change in diet and for 10 days afterwards. Grey rectangles represent the days during which arousability was tested. (D) Top: time spent awake, in non-REM sleep, in REM sleep, and asleep over the course of 24 hours, in 1-hour intervals. The white portion of the bar on top indicates daytime (lights on, 12 hours) while the black portion indicates nighttime (lights off, 12 hours). Middle: the number of bouts (episodes) for wakefulness, non-REM sleep, REM sleep and total daily sleep. Bottom, duration of bouts for wakefulness, non-REM sleep, REM sleep and total daily sleep. (E) Power analysis of delta and theta brain waves during wakefulness, non-REM and REM sleep. (F) Power analysis of delta and theta waves throughout the day in 1-hour intervals. Error bars, mean and S.E.M. Genotypes, sample sizes, and statistical analyses, Table S3.
Figure S5. CCHa1 receptor in PAMMB441B neurons regulates arousability, related to Figure 5. (A) Responsiveness during awake bouts, total daily sleep and basal locomotion in animals with the CCHa1 receptor knocked down using Ddc-Gal4 or PAM-Gal4. (B) Expression of GFP driven by two different Gal4s, which are themselves expressed under the CCHa1R regulatory elements (P{GMR23H07-GAL4}attP2 and TI{2A-Gal4}CCHa1-R2A-Gal4). Top, Z-stack projections of confocal images in which the PAM clusters of dopaminergic neurons are seen. Bottom, Z-stack projections through the whole central nervous system, with outlines corresponding to the areas on top. White arrows: PAM neurons (labeled with antibodies against tyrosine hydroxylase, TH, the rate-limiting enzyme in dopamine synthesis) which also express GFP. (C) Expression pattern of PAMMB441B-Gal4 in the gut. (D) Food intake and clearance, in controls and in animals with CCHa1 depleted from the gut (PAMMB441B>CCHa1-RNAi). Each dot represents the average of three experiments, each done with 15 flies. (E) Effect of knocking down CCHa1 receptor in the PAMMB441B dopaminergic neurons on responsiveness of awake animals, and various sleep parameters. (F) Effect of activation or inhibition of PAMMB441B neurons on responsiveness of awake animals, and various sleep parameters. (G) Effect of knocking down TH in PAMMB441B neurons on responsiveness of awake animals, and various sleep parameters. Error bars, mean and S.E.M. Scale bars: 100 μm. Genotypes, sample sizes and statistical analyses, Table S3.
Figure S6. CCHa1 receptor in PAM neurons regulates arousability in response to dietary proteins, and setup to measure arousability in response to increased temperature, related to Figure 6. (A) Responsiveness of awake animals, for controls and flies in which the CCHa1 receptor was depleted from PAMMB441B neurons. (B) Arousability in response to low amplitude vibrations for the parental controls, and medium amplitude vibrations for the experimental flies, to control for the experiment shown in Figure 6A. (C) Activity of PPL1 dopaminergic neurons on regular and peptone-supplemented food, as reported by CaLexA. (D) Activity of PAMMB441B neurons following a night of exposure to high amplitude vibrations, or to heat, as reported by CaLexA. Each dot represents 1 neuron. (E) The setup for probing arosuability using thermal stimulation. (F) Responsiveness of awake animal to heat, with or without peptone supplementation (left), and with or without depletion of the CCHa1 receptor from PAMMB441B neurons (right). (G) Arousabilty in response to 40 °C stimulation, with or without peptone supplementation. Error bars, mean and S.E.M. Genotypes, sample sizes, and statistical analyses, Table S3.
Figure S7. Sleep and arousability regulation through MBONMB083C neurons, related to Figure 7. (A) Expression pattern of MBONMB083C-Gal4 driver in the gut. Scale bar: 100 μm. (B) Responsiveness of awake animals, and various sleep parameters, in animals with silenced (green) or activated (yellow) MBONMB083C neurons. (C) Peptone-induced changes in arousal during wake bouts upon inhibition of MBONMB083C neurons. (D) Responsiveness of awake animals to stimulation, for controls and flies in which various dopamine receptors are depleted fromMBONMB083C neurons.(E) Schematic: the CCHa1 pathway regulates arousability through MBONMB083C neurons. Error bars, mean and S.E.M. Genotypes, sample sizes and statistical analyses, Table S3.
Table S3. Sample sizes and statistical analyses. Related to Figures 1–7 and Supplementary Figures 1–7.
Table S1. List of genes from the screen that show hypo or hyper-arousable phenotypes when knocked down using elav-Gal4. Related to Figure 1.
Table S2. Gene ontology analysis of the genes from the screen that when knocked down using elav-Gal4 show hypo or hyper-arousable phenotypes (shown in Table S1) for biological process and molecular function. Related to Figure 1.
Data Availability Statement
Data are available from the corresponding author upon request. Codes are available on GitHub for sleep analysis (https://github.com/CrickmoreRoguljaLabs/SleepAnalysis), stimulus generation (https://github.com/IrisTitos/ATanalysis/blob/master/Goodmorning.md), and stimulus analysis (https://github.com/IrisTitos/ATanalysis/blob/master/ReadDamsPullDown, https://github.com/ArousalThreshold/Software).