Summary
The transcription factor LEAFY (LFY) plays crucial roles in flower development by activating floral homeotic genes. Activation of LFY targets requires the combined action of LFY and the E3 ubiquitin ligase UFO, although the precise underlying mechanism remains unclear. Here, we show that LFY accumulates in biomolecular condensates within the cytoplasm, while recombinant LFY forms condensates with similar properties in vitro. UFO interacts with LFY within these condensates and marks it for degradation. LFY levels in the nucleus are buffered against changes in total LFY levels induced by proteasome inhibition, UFO overexpression, or mutation of lysine residues in a disordered region of LFY. Perturbation of cytoplasmic LFY condensates by 1,6-hexanediol treatment induces the relocalization of LFY to the nucleus and the subsequent activation of the LFY target AP3 in flowers. Our data suggest that nucleocytoplasmic partitioning, condensation, and ubiquitin-dependent degradation regulate LFY levels in the nucleus to control its activity.
Subject areas: Plant biochemistry, Cell biology, Plant development
Graphical abstract
Highlights
-
•
LFY accumulates in biomolecular condensates in the cytoplasm of plant cells
-
•
Recombinant LFY protein forms condensates in vitro
-
•
UFO interacts with LFY in cytoplasmic condensates and marks UFO for degradation
-
•
LFY undergoes nucleocytoplasmic partitioning, condensation, and degradation
Plant biochemistry; Cell biology; Plant development
Introduction
In Arabidopsis, LEAFY (LFY) is the master regulator of flower development. LFY orchestrates floral meristem emergence1 and regulates floral meristem identity through the activation of floral homeotic genes.2,3,4,5 Loss-of-function mutants for the LFY gene are unable to form proper flowers and often show abnormal, leaf-like structures.2,6 In contrast, ectopic LFY expression causes the conversion of lateral meristem as well as inflorescence meristem into flowers.3 LFY encodes a plant-specific transcription factor with a C-terminal DNA-binding domain7,8,9 and an N-terminal sterile alpha motif (SAM) domain, which is important for LFY oligomerization.10,11
LFY specifies the time and place of flower development, and precise control of LFY expression is crucial. LFY is weakly expressed in leaf primordia and LFY protein levels steadily increase until LFY reaches a threshold triggering flowering through direct activation of APETALA1 (AP1).4,12 To specify floral organ identity, LFY is expressed throughout the whole floral meristem,6,9 while its direct target genes, such as AP1, APETALA3 (AP3), and AGAMOUS are only expressed in distinct spatial patterns.5,13 This indicates that other factors are involved in fine-tuning LFY activity. The expression of the B-class gene AP3 is restricted to a specific region of the floral meristem immediately before the initiation of petal and stamen primordia at floral stage 3.14,15 LFY activity on the AP3 promoter is dependent on its interaction with the co-factor UNUSUAL FLOWER ORGANS (UFO).16,17,18,19,20 Ectopic expression of UFO leads to the production of more petals, caused by the ectopic expression of AP3, but this phenotype is abolished in the absence of LFY.16 In line with this, ectopic expression of LFY alone is not sufficient to trigger ectopic AP3 expression.5
UFO belongs to the large family of F box proteins,21,22 which are known to be part of the Skp1, Cullin, F box (SCF)-containing complex.23 It has been shown that UFO physically interacts via its F box with Ask1, a Skp1-homolog,21,24,25 and via its C-terminal Kelch domain with the DNA-binding domain of LFY.19 The SCF complex is an E3 ubiquitin ligase complex, known to mark substrate proteins for proteasome-dependent degradation through substrate recognition by the F box.26 Although UFO was proposed to ubiquitinate LFY (Chae et al., 2008), the precise role of UFO in regulating LFY activity and the mechanisms by which UFO potentiates LFY to drive AP3 expression remain unclear. Recently, UFO was shown to form a transcriptional complex with LFY to specify LFY binding to new cis-regulatory elements in the AP3 promoter regions.20 Interestingly, such a role of UFO appears to be independent of the role of UFO in protein degradation, since it is also observed for the UFOΔF-box variant that does not assemble a functional SCF complex. Therefore, UFO regulates nuclear LFY activity to promote AP3 gene expression.
Recently, several cellular processes have been shown to rely on the formation of biomolecular condensates that allow the compartmentalization and spatiotemporal organization of biomolecules and their activities in eukaryotic cells.27,28,29 These condensates are often composed of proteins and/or nucleic acids27 and can be formed in the nucleus or cytoplasm by liquid-liquid phase separation (LLPS), a process which separates a solution into two or more coexisting phases.28,29 To date, only few plant transcriptional regulators have been shown to undergo LLPS.30,31 Here, we uncover that LFY forms biomolecular condensates in the cytoplasm, and show that recombinant LFY forms condensates with similar properties in vitro. We show that LFY protein homeostasis is controlled by LFY sequestration in cytoplasmic condensates and UFO-mediated degradation. Our results indicate that these processes buffer the levels of LFY in the nucleus to ensure accurate LFY activity.
Results
LFY protein is localized in the nucleus and in membraneless cytoplasmic foci
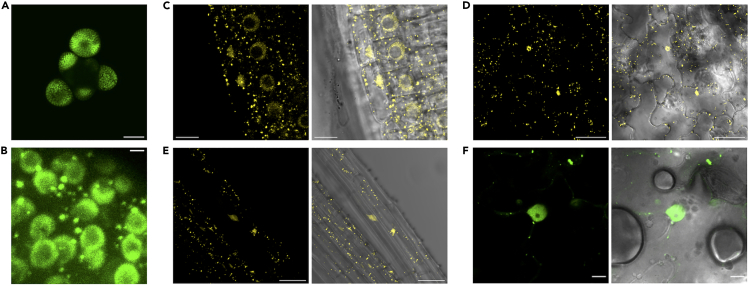
To investigate the expression pattern, subcellular localization, and regulation of the LFY protein in planta, we used a stable transgenic line of Arabidopsis thaliana expressing the functional LFY-GFP fusion protein under the control of the LFY promoter in the lfy-12 mutant background.32 Consistent with previous reports,1 imaging of the flower meristem confirmed that the LFY promoter is uniformly active in floral buds (Figure 1A). Besides being localized in the nucleus, in accordance with its transcription factor role, careful examination of LFY subcellular localization revealed the presence of cytoplasmic foci positive for LFY-GFP in flowers (Figure 1B). This indicates that when LFY is expressed at or close to endogenous levels in stable transgenic plants, a pool of LFY resides in cytoplasmic foci in flower cells. To establish if such cytoplasmic structures observed in LFY-expressing cells could also be observed when LFY is ectopically expressed, and to circumvent the difficulties of floral meristem imaging for further analyses, we imaged the 35S::LFY-YFP stable transgenic A. thaliana line33 and 35S::LFY-mCit Nicotiana benthamiana-infiltrated leaves. Analysis of the subcellular distribution of constitutively expressed LFY confirmed the ability of LFY to form cytoplasmic foci in A. thaliana root, hypocotyl, and leaf cells as well as in N. benthamiana leaf cells (Figures 1C–1F). Constitutively expressed LFY-YFP/mCit in both stable transgenic plants and in N. benthamiana leaf cells accumulated to lower levels than the functional LFY-GFP fusion driven by the LFY promoter in flowers, based on the analysis of corrected fluorescence levels (Figure S1A). This indicates that cytoplasmic foci observed for ectopically expressed LFY in roots or N. benthamiana do not result from overexpression.
Figure 1.
Localization of LFY protein in planta
(A and B) Confocal microscopy images of A. thaliana lfy-12 mutant flower carrying LFY::LFY-GFP. Scale bar = 60 μm (A) and 5 μm (B).
(C–E) Confocal microscopy images of 35S::LFY-YFP roots (C), leaves (D) and hypocotyls (E). Scale bar = 10 μm (C) and 40 μm (D, E).
(F) Transient expression of 35S::LFY-mCit in N. benthamiana leaves. Scale bar = 10 μm. The overlay between the YFP/mCit channel and transmission view is shown to the right (C–F).
The ability of LFY to trigger flower formation onto inflorescences3,6 and to reprogram root cells into flowers34,35 prompted us to investigate deeper the nature and properties of LFY cytoplasmic foci in root cells from 35S::LFY-YFP plants. Several cellular structures exist as vesicles that yield punctate cytoplasmic signals similar in size and shape to LFY-YFP foci when observed by confocal microscopy, including Golgi, trans-Golgi network/early endosomes (TGN/EE), late endosomes (LE), and autophagosomes. First, to evaluate if the cytoplasmic foci observed for LFY-YFP were of endosomal nature, we took advantage of the FM4-64 lipophilic dye that labels TGN/EE and LE. Although both LFY and FM4-64 labeled dotted structures in the cytoplasm (Figure S1B), no overlap was observed between the two signals (Pearson’s coefficient r = 0.036). This was further supported by the fact that LFY-YFP foci were not sensitive to the fungal drug Brefeldin A (BFA), which aggregates Golgi and endosomal compartments into so-called BFA bodies labeled by FM4-64 (Figure S1C). We then examined if LFY-YFP foci co-localized with a marker of autophagosome formation. To that purpose, the 35S::LFY-YFP line was crossed to the ATG8E-mCh autophagy reporter line. No significant colocalization was observed between the two signals (Pearson’s coefficient r = 0.164) (Figure S1D), even though a few LFY-YFP dots could sometimes be observed at the vicinity of ATG8E-positive structures. Altogether, these observations indicate that, in addition to its presence in the nucleus, LFY also exists in cytoplasmic membraneless foci.
LFY protein forms biomolecular condensates in vivo and in vitro
Recently, several proteins were shown to form biomolecular condensates in plant cells.29 Some of them, such as the transcriptional regulators NPR1 and ARF7/ARF19, were reported to form condensates in the cytoplasm.30,36 We therefore investigated the ability of LFY to undergo phase separation. Several features have been associated with the propensity of proteins to form phase-separated biomolecular condensates, such as their ability to establish multivalent interactions, and the presence of intrinsically disordered regions (IDR) or prion-like domains (PLD) within them. LFY is known to oligomerize, to interact with several proteins, and to be post-translationally modified, which might give it the potential to engage in multivalent interactions.10,19,33 Using the predictor of natural disordered regions (PONDR, http://www.pondr.com) algorithm, we identified two IDRs in LFY. The first IDR is located within the first 39 amino acids at the N-terminus of LFY, and the second one encompasses amino acid residues 106–240 and is located between the N-terminal SAM oligomerization and C-terminal DNA-binding domains of LFY (Figures S2A and S2B). The search for PLD in LFY using the prion-like amino acid composition algorithm (PLAAC, http://plaac.wi.mit.edu) revealed PLDs with a low score between amino acids 155 and 195, compared to the strongly predicted PLDs from ELF3 or ARF7 (Figure S2C) (Jung et al., 2020; Powers et al., 2020).
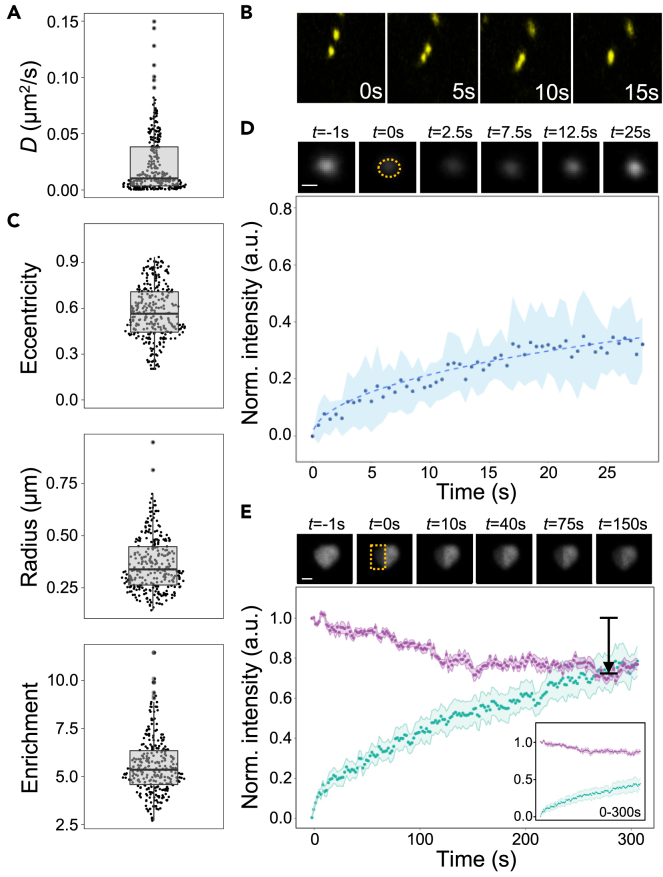
We wondered whether cytoplasmic LFY foci might correspond to phase-separated condensates, and decided to further study them by live-imaging of LFY-YFP in A. thaliana. We first acquired movies to map the dynamic properties of LFY-YFP foci. These movies showed that LFY-YFP foci were mobile, with diffusion coefficients of approximately 0.01 μm2/s (Video S1; Figure 2A). LFY-YFP foci had the ability to fuse with one another upon contact (Figure 2B), indicating that they exhibit liquid-like properties. We next quantified the size, shape, and intensity of the foci (Figure 2C). All of the three parameters showed broad but unimodal distributions, indicating that there is a single population of LFY foci. The eccentricity, which reflects the shape of the foci, shows that most foci are not spherical (eccentricity values above zero), suggesting that the shape of LFY foci is not dominated by a strong interfacial tension that would minimize their surface area and promote a spherical shape.
Figure 2.
LFY protein forms biomolecular condensates in planta
(A) Diffusion coefficients of condensates in root cells from 5-day-old 35S::LFY-YFP seedlings. n = 13.
(B) Time course analysis of condensate fusion in root cells from 5-day-old 35S::LFY-YFP seedlings.
(C) Analysis of the shape (top), size (center), and intensity enrichment relative to the nuclear intensity (bottom) of LFY-YFP condensates in root cells from 5-day-old 35S::LFY-YFP seedlings. n = 13.
(D) FRAP recovery curves after photobleaching of whole condensates in roots from 5-day-old 35S::LFY-YFP seedlings. Top: representative LFY condensate during an FRAP experiment. Scale bar = 1 μm. Bottom: FRAP recovery curve. n = 9. Error bars represent standard deviations.
(E) Half-FRAP recovery curves after photobleaching condensates in N. benthamiana leaf cells transiently expressing 35S::LFY-mCit. Top: representative LFY condensate during a half-FRAP experiment. Scale bar = 1 μm. Bottom: Half-FRAP curves corresponding to the normalized recovery of the fluorescence in the bleached half (green) and the normalized loss of fluorescence in the non-bleached half (purple). The normalized intensity decrease (dip) in the non-bleached half amounted to 27% and was significantly larger than the dip for a freely diffusing protein (n = 9; p-value = 5·10−9, one-sided t-test).37 The same curves without the correction of immobile fractions are shown in the inset. Error bars represent standard errors of the mean.
Confocal imaging time-course of 35S::LFY-YFP roots.
To better grasp the dynamic properties of LFY foci, we performed fluorescence recovery after photobleaching (FRAP) experiments in vivo. Liquid-like condensates are often characterized by a rapid fluorescent recovery on the order of seconds, while slow or no recovery is often associated with more solid-like condensates.27 After photobleaching LFY-YFP foci in the root, a ∼40% recovery plateau was reached within 30 s (Figure 3D). This indicates that LFY foci are composed of a highly dynamic LFY pool that readily exchanges between the foci and the surrounding cytoplasm, and a less dynamic LFY pool that exchanges much more slowly and appears as a ∼60% immobile fraction in the experiment. To individually assess the internal mobility of LFY within the foci and the exchange of LFY across the boundary of the foci, we performed half-FRAP in N. benthamiana leaf cells expressing 35S::LFY-mCit. For condensates assembled via multivalent interactions driving LLPS, LFY molecules are expected to preferentially move within the condensates. This is due to the fact that multiple interactions among LFY molecules have to be simultaneously broken when LFY molecules cross the interface of the condensate to enter the dilute phase, while this is not the case when LFY molecules stay within the condensate and only exchange between its two-halves. Such preferential internal mixing is reflected by a pronounced intensity decrease in the non-bleached half, whose depth scales with the strength of the barrier at the condensate interface.38,37 After photobleaching one-half of an LFY focus, we observed a rapid recovery of the intensity in the bleached half (Figure 2E) that can be caused by LFY exchange with the surrounding cytoplasm and/or internal mixing of LFY proteins between the bleached and the non-bleached half. To assess preferential internal mixing, we monitored the intensity in the non-bleached half of the foci, which showed a pronounced intensity decrease (Figure 2E). This indicates that LFY can readily move within the foci and that LFY exchange across the boundary is partially restricted. In order to quantify the interfacial barrier and the interfacial tension from the half-FRAP data, we used the recently developed MOCHA-FRAP workflow.37 The apparent interfacial barrier and the apparent interfacial tension of LFY foci amounted to 0.02 kT/molecule and 0.36 μN/m, respectively, which is similar to the values reported for other biomolecular condensates such as FUS-containing stress granules.37,39 Taken together, our results suggest that LFY foci consist of a less dynamic and a more dynamic LFY pool, with the latter one showing hallmarks of LLPS.
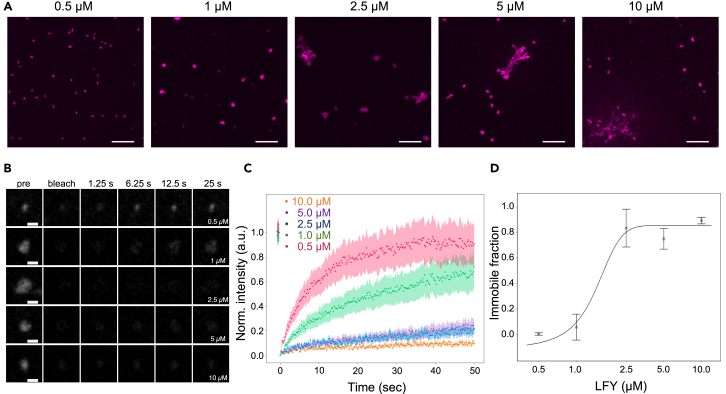
Figure 3.
LFY condensate formation in vitro is concentration-dependent
(A) In vitro condensate formation of recombinant ATTO647N-LFY-His at 0.5, 1.0, 2.5, 5.0, and 10.0 μM concentration. Scale bars = 5 μm.
(B) Snapshots of condensates of recombinant LFY-His during FRAP experiments conducted in vitro.
(C) FRAP recovery curves for LFY-His condensates formed at LFY concentrations of 0.5 (pink), 1.0 (green), 2.5 (blue), 5.0 (violet), and 10.0 μM (orange).
(D) Quantification of immobile LFY fraction in condensates formed at increasing LFY-His concentrations. Error bars represent the standard error of the mean.
The ability of LFY to form liquid-like condensates may be due to the intrinsic propensity of LFY to undergo LLPS, or through interactions between LFY and other proteins that form condensates. We therefore evaluated the in vitro behavior of recombinant LFY-His protein purified from Escherichia coli. LFY-His readily formed condensates as a function of protein concentration (Figure 3A). At low concentration, LFY formed spherical droplet-like condensates indicative of LLPS, while at higher LFY concentration, more irregularly shaped aggregate-like structures were obtained. We next performed FRAP experiments to analyze the dynamics of the different types of LFY condensates (Figures 3B and 3C). For spherical droplet-like condensates, we observed a complete recovery within less than a minute. With increasing LFY concentration, the immobile fraction increased and reached 85% for irregularly shaped condensates (Figure 3D). These results show that LFY can form condensates containing protein pools with different dynamic properties in vitro, which is reminiscent of its behavior in vivo.
UFO interacts with LFY and mediates its continuous ubiquitination and degradation
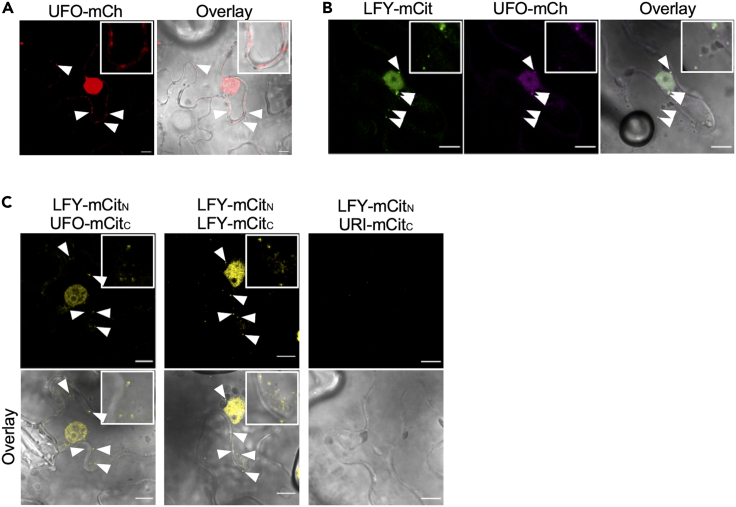
LFY is known to interact with the Kelch domain of UFO that also engages into a SCF E3 ubiquitin ligase complex in which the F box domain of UFO directly interacts with ARABIDOPSIS SKP1-LIKE proteins.21,40 We therefore set out to address the possible role of UFO in LFY regulation. Consistent with previously published results,19 we confirmed that UFO has the ability to mediate LFY ubiquitination during in vitro ubiquitination assays (Figures S3A and S3B). We then evaluated the subcellular localization of the UFO protein by transient expression of a 35S::UFO-mCh construct in N. benthamiana. UFO displayed a mostly nuclear localization, but also showed cytoplasmic foci (Figure 4A). When UFO-mCh was coexpressed with LFY-mCit, both fluorescent signals co-localized in the nucleus and also in cytoplasmic foci (Pearson’s coefficient r = 0.736) (Figure 4B). To directly examine the interaction between LFY and UFO, we took advantage of the bimolecular functional complementation (BiFC) assays. We used LFY oligomerization as positive control for BiFC and the URI transcription factor as negative control (Figure 4C). BiFC confirmed the ability of LFY and UFO to interact in the nucleus, but also highlighted their interaction in cytoplasmic foci (Figure 4C).
Figure 4.
UFO interacts with LFY in the nucleus and in cytoplasmic foci
(A) Transient expression of 35S::UFO-mCh in N. benthamiana leaves. Scale bar = 10 μm. Arrows indicate examples of cytoplasmic foci. Insets with higher magnification are shown.
(B) Transient coexpression of 35S::LFY-mCit and 35S::UFO-mCh in N. benthamiana leaves. mCit and mCh are false colored in green and magenta for better visualization of colocalization. Scale bar = 10 μm. Arrows indicate examples of cytoplasmic foci. Insets with higher magnification are shown.
(C) Bimolecular functional complementation of LFY and UFO in N. benthamiana leaves. LFY oligomerization was used as positive control, and the absence of LFY-URI interactions as negative control. Scale bar = 10 μm. In all panels, the overlay between mCitrine/mCherry fluorescence channels and the transmission view are shown. Arrows indicate examples of cytoplasmic foci. Insets with higher magnification are shown.
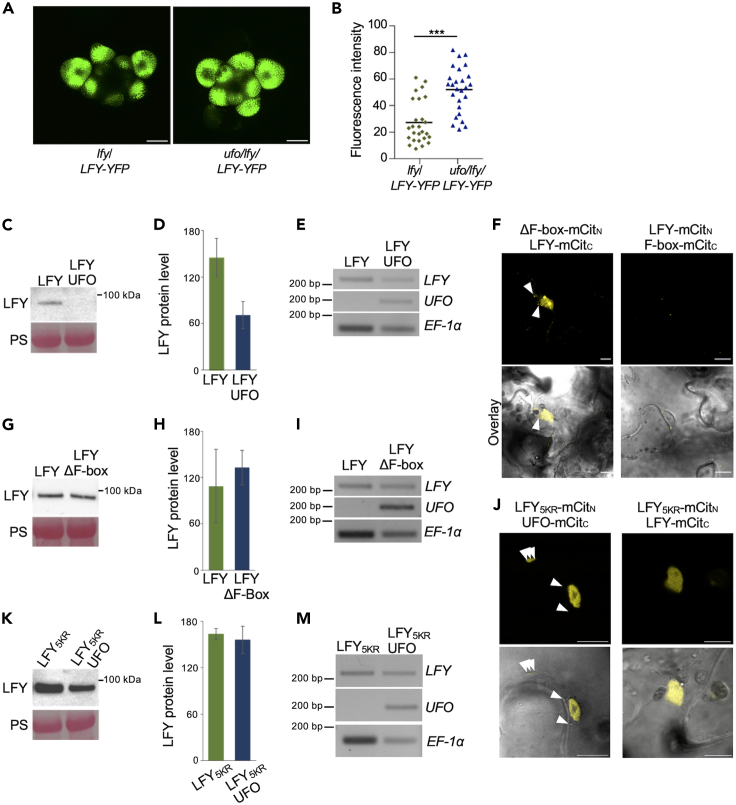
The interaction between LFY and UFO prompted us to evaluate the impact of UFO expression on LFY protein levels. Because LFY expression is largely restricted to the meristem, we could not detect LFY protein by western blot analyses in wild-type and ufo mutant backgrounds. We therefore monitored LFY protein levels by crossing lfy/LFY::LFY-YFP to ufo-2, and compared the fluorescence levels of lfy/LFY::LFY-YFP and ufo/lfy/LFY::LFY-YFP in the meristem. Confocal microscopy imaging of the meristem highlights the stronger signals obtained when LFY::LFY-YFP is expressed in the ufo mutant background, consistent with a role of UFO in LFY degradation (Figures 5A and 5B). Furthermore, we transiently expressed LFY-mCit or LFY-mCit/UFO-mCh in N. benthamiana and monitored LFY protein levels. The overexpression of UFO clearly and reproducibly reduced LFY protein accumulation as observed by western blot (Figures 5C and 5D), while LFY mRNA accumulated to similar levels between the two tested conditions (Figure 5E).
Figure 5.
UFO mediates LFY degradation and requires the UFO F box domain and lysine residues in the IDR of LFY
(A) Confocal microscopy images of lfy-12/LFY::LFY-GFP (left) and ufo-2/lfy-12/LFY::LFY-GFP (right) flowers. Scale bar = 60 μm.
(B) Quantification of fluorescence intensity from flowers as shown in (A). Each data point represents the average of fluorescence intensities from 5 LFY-GFP-positive regions of interest in independent flower buds. Independent plants were used for each data point (n = 26). Statistical significance was determined using a two-tailed Student’s t test.
(C) Total LFY protein levels in N. benthamiana leaves transiently expressing 35S::LFY-mCit or coexpressing 35S::LFY-mCit and 35S::UFO-mCh. Ponceau red staining (PS) serves as loading control.
(D) Quantification of LFY-mCit protein levels in N. benthamiana leaves as shown in (C). Results are shown as mean +/− standard error (n = 3).
(E) LFY and UFO mRNA accumulation in N. benthamiana leaves expressing 35S::LFY-mCit or coexpressing 35S::LFY-mCit and 35S::UFO-mCh.
(F) Bimolecular functional complementation of LFY and UFOΔF-box in N. benthamiana leaves. The absence of interactions between LFY and the UFO F-box domain serves as negative control. Scale bar = 10 μm. The overlays between mCitrine fluorescence channels and the transmission view are shown at the bottom. Arrows indicate examples of cytoplasmic foci.
(G) Total LFY-mCit protein levels in N. benthamiana leaves transiently expressing 35S::LFY-mCit or coexpressing 35S::LFY-mCit and 35S::ΔF-box-mCh. Ponceau red staining (PS) serves as loading control.
(H) Quantification of LFY-mCit protein levels in N. benthamiana leaves as shown in (G). Results are shown as mean +/− standard error (n = 3).
(I) LFY and UFO mRNA accumulation in N. benthamiana leaves expressing 35S::LFY-mCit or coexpressing 35S::LFY-mCit and 35S::ΔF-box-mCh.
(J) Bimolecular functional complementation of LFY5KR and UFO in N. benthamiana leaves (left). Interaction between LFY5KR and LFY is also tested (right). Scale bar = 10 μm. The overlays between mCitrine fluorescence channels and the transmission view are shown at the bottom. Arrows indicate examples of cytoplasmic foci.
(K) Total LFY5KR-mCit protein levels in N. benthamiana leaves transiently expressing 35S::LFY5KR-mCit or coexpressing 35S::LFY5KR-mCit and 35S::UFO-mCh. Ponceau red staining (PS) serves as loading control.
(L) Quantification of LFY5KR-mCit protein levels in N. benthamiana leaves as shown in (K). Results are shown as mean +/− standard error (n = 3).
(M) LFY5KR and UFO mRNA accumulation in N. benthamiana leaves expressing 35S::LFY5KR-mCit or coexpressing 35S::LFY5KR-mCit and 35S::UFO-mCh.
To gain further insight into the regulation of LFY by UFO in vivo, we expressed the UFOΔF-box version of UFO, which lacks the domain mediating its recruitment into a functional SCF E3 ligase complex. Consistently, LFY interacted with UFOΔF-box as shown by BiFC (Figure 5F), both in the nucleus and in cytoplasmic condensates. LFY protein levels were mildly increased upon UFOΔF-box expression (Figures 5G–5I), which confirms that the changes in LFY levels observed above were specific to UFO and involved UFO-mediated degradation. Next, we set out to generate an LFY mutant that is less sensitive to UFO-mediated ubiquitination and degradation. We reasoned that we might achieve this by mutating possible target lysine residues in LFY. Accordingly, we substituted 5 lysine residues in the IDR of LFY with arginine, yielding LFY5KR. Expression of LFY5KR-mCit in N. benthamiana revealed that the LFY5KR protein accumulated to higher levels than LFY (Figure S3C). LFY5KR interacted with UFO as shown by BiFC (Figure 5J), which is consistent with the LFY-UFO interaction site having been mapped to the C-terminus of LFY.19 LFY5KR levels were almost unaltered upon UFO coexpression (Figures 5K–5M), while LFY5KR ubiquitination was mildly decreased upon UFO coexpression (Figure S3D). We conclude that LFY is continuously ubiquitinated and degraded, and that the lysine residues in the LFY IDR we assessed here are involved in UFO-mediated degradation as their mutation renders LFY insensitive to UFO coexpression.
Nuclear LFY levels are buffered against cell-to-cell variations and altered LFY degradation
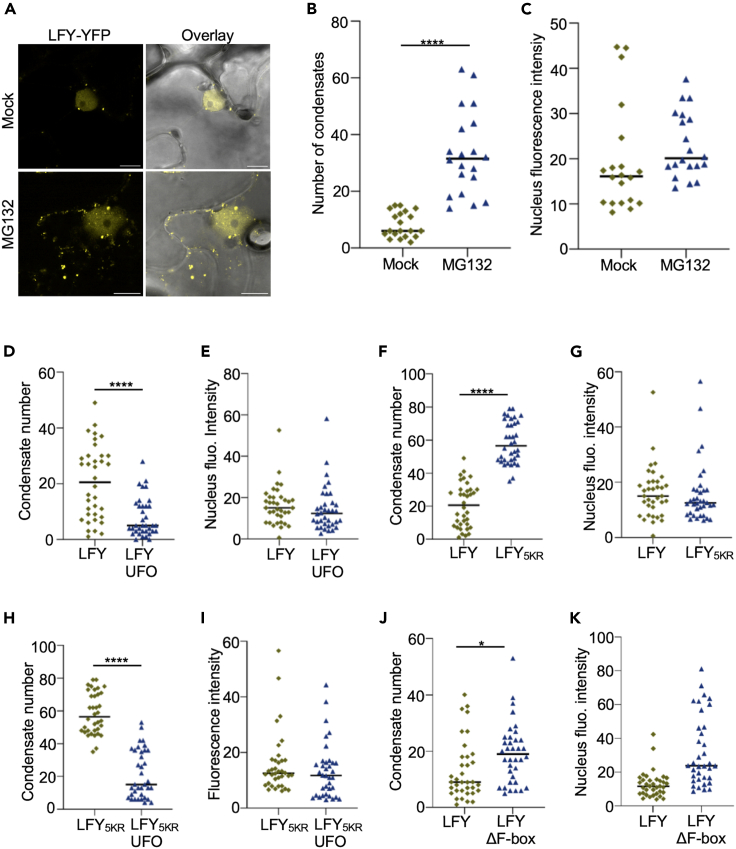
We next asked how LFY condensates in the cytoplasm and LFY levels in the nucleus react to changes in total cellular LFY levels. In particular, we wondered if the cytoplasmic LFY pool may serve as a buffer to control the effective concentration of active LFY in the nucleus. First, to investigate if LFY undergoes nucleocytoplasmic shuttling, we carried out FRAP of the whole nucleus. We observed a 15% recovery after 5 min, indicating that at least a fraction of LFY undergoes shuttling (Figure S4). However, the majority of the nuclear and cytoplasmic LFY pools did not exchange on the timescales of seconds, on which LFY turns over in cytoplasmic condensates. Next, we set out to quantify the number of cytoplasmic LFY condensates along with nuclear LFY levels. Due to the difficulties of meristem imaging, it seemed not to be feasible to robustly do this in flower primordia. To circumvent this issue, we focused our analysis on N. benthamiana leaves. To examine how nuclear and cytoplasmic LFY pools reacted to an inhibition of continuous protein degradation, we treated N. benthamiana leaves transiently expressing LFY-mCit with the MG132 proteasome inhibitor and quantified the number of cytoplasmic condensates and the fluorescence intensity in the nucleus. As opposed to mock-treated plants, we observed an increase in the number of cytoplasmic condensates following short-term MG132 treatment (Figures 6A and 6B). This was however not associated with a significant increase in nuclear LFY levels (Figure 6C), suggesting that the latter was buffered. To assess the effect of UFO-mediated degradation, we quantified the number of condensates in N. benthamiana leaves transiently expressing LFY-mCit or LFY-mCit/UFO-mCh, which exhibit different LFY levels as shown in the western blots above (Figures 5C and 5D). Notably, UFO expression reduced the number of LFY-positive cytoplasmic condensates with almost no influence on the nuclear LFY pool (Figures 6D and 6E). Next, we evaluated for both conditions the number of cytoplasmic LFY condensates across cells with different nuclear intensities (Figure S5). This analysis confirmed that both quantities were indeed independent from each other, corroborating that nuclear LFY levels were buffered. We next assessed the influence of the KR substitutions in the LFY IDR described above on LFY condensation. Confocal microscopy imaging of LFY5KR showed increased condensate numbers for LFY5KR compared to wild-type LFY (Figure 6F). Nevertheless, nuclear fluorescence signals were comparable between LFY5KR and wild-type LFY (Figure 6G). Coexpression of UFO decreased the number of LFY5KR condensates, while nuclear LFY levels remained unchanged (Figures 6H and 6I). Taken together, these results indicate that nuclear LFY levels are buffered against cell-to-cell variations, LFY accumulation upon proteasome inhibition, LFY depletion upon UFO coexpression, and mutation of lysine residues in the IDR of LFY that represent bona fide targets for ubiquitination.
Figure 6.
LFY condensate formation buffers nuclear LFY
(A) Transient expression of 35S::LFY-mCit in N. benthamiana leaves treated with mock or MG132. Scale bar = 10 μm.
(B) Number of condensates in cells of N. benthamiana leaves transiently expressing 35S::LFY-mCit. n = 20 of 2 independent experiments. Statistical significance was determined using a two-tailed Student’s t test.
(C) Quantification of fluorescence intensity in the nucleus of N. benthamiana leaves transiently expressing 35S::LFY-mCit. n = 20 of 2 independent experiments. Statistical significance was determined using a two-tailed Student’s t test.
(D) Number of condensates in cells from N. benthamiana leaves transiently expressing 35S::LFY-mCit or coexpressing 35S::LFY-mCit and 35S::UFO-mCh. n = 30 of 3 independent experiments. Statistical significance was determined using a two-tailed Student’s t test.
(E) Quantification of fluorescence intensity in the nucleus of N. benthamiana leaves transiently expressing 35S::LFY-mCit or coexpressing 35S::LFY-mCt and 35S::UFO-mCh. n = 30 of 3 independent experiments. Statistical significance was determined using a two-tailed Student’s t test.
(F) Number of condensates in cells from N. benthamiana leaves transiently expressing 35S::LFY5KR-mCit or 35S::LFY-mCit. n = 30 of 3 independent experiments. Statistical significance was determined using a two-tailed Student’s t test.
(G) Quantification of fluorescence intensity in the nucleus of N. benthamiana leaves transiently expressing 35S::LFY5KR-mCit or 35S::LFY-mCit. n = 30 of 3 independent experiments. Statistical significance was determined using a two-tailed Student’s t test.
(H) Number of condensates in cells from N. benthamiana leaves transiently expressing 35S::LFY5KR-mCit or coexpressing 35S::LFY5KR-mCit and 35S::UFO-mCh. n = 30 of 3 independent experiments. Statistical significance was determined using a two-tailed Student’s t test.
(I) Quantification of fluorescence intensity in the nucleus from N. benthamiana leaves transiently expressing 35S::LFY5KR-mCit or coexpressing 35S::LFY5KR-mCit and 35S::UFO-mCh. n = 30 of 3 independent experiments. Statistical significance was determined using a two-tailed Student’s t test.
(J) Number of condensates in cells from N. benthamiana leaves transiently expressing 35S::LFY-mCit or coexpressing 35S::LFY-mCit and 35S::ΔF-box-mCh. n = 30 of 3 independent experiments. Statistical significance was determined using a two-tailed Student’s t test.
(K) Quantification of fluorescence intensity in the nucleus of N. benthamiana leaves transiently expressing 35S::LFY-mCit or coexpressing 35S::LFY-mCit and 35S::ΔF-box-mCh. n = 30 of 3 independent experiments. Statistical significance was determined using a two-tailed Student’s t test.
Finally, we assessed the number of condensates and the nuclear LFY levels upon UFOΔF-box expression. As UFOΔF-box interacts stronger with LFY than wild-type UFO19,33 but does not mark it for degradation, it can be considered as a “substrate trap”. Coexpression of UFOΔF-box increased both the number of cytoplasmic condensates and the nuclear LFY levels (Figures 6J and 6K), suggesting that trapping LFY in stable complexes with UFOΔF-box changes the phase diagram and the resulting partitioning between cytoplasmic condensates and the nucleus.
LFY buffering via cytoplasmic condensates regulates nuclear LFY activity
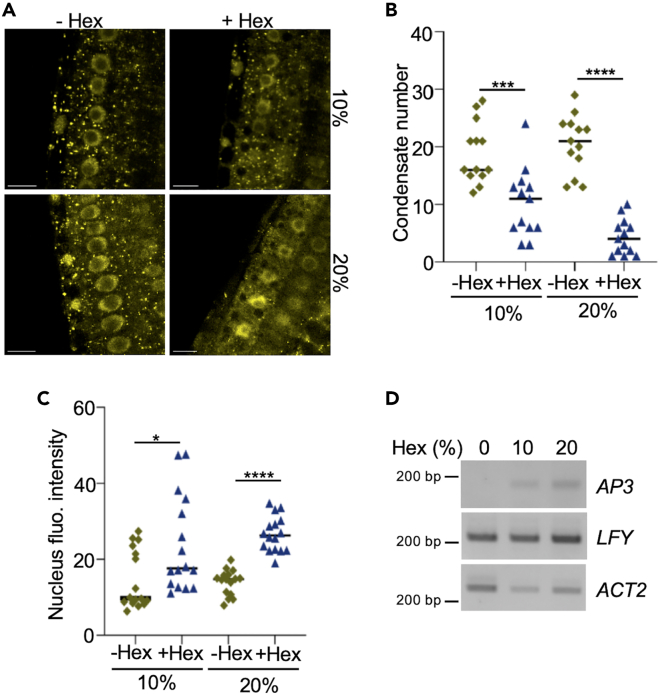
We next asked which impact the dissolution of cytoplasmic LFY condensates would have on LFY function in the nucleus. To this end, we treated plant roots with the aliphatic alcohol 1,6-hexanediol, which is known to interfere with weak hydrophobic interactions and which has often been used to dissolve condensates in vitro and in cells.41 In contrast to untreated plants, 1,6-hexanediol-treated plants showed a rapid dispersal of LFY-YFP condensates (Figures 7A and 7B), suggesting that weak hydrophobic interactions play a role in stabilizing them. Similar observations were made using transiently expressed LFY-mCit in N. benthamiana leaves (Figure S6A). Dispersal of LFY-YFP condensates upon 1,6-hexanediol treatment was associated with a strong increase in nuclear LFY levels in both LFY-YFP roots and in N. benthamiana leaves (Figures 7C, and S6B). This suggests that cytoplasmic LFY condensates sequester LFY and thereby buffer nuclear LFY levels, while the loss of these condensates triggers a misregulation of the nuclear LFY pool. To evaluate the functional relevance of LFY buffering, we monitored the expression of the AP3 LFY target gene in LFY-YFP-expressing flowers in the absence or presence of 1,6-hexanediol. Flowers treated with 1,6-hexanediol showed an upregulation of AP3 gene expression (Figure 7D), which followed the increase in nuclear LFY. These observations point to a model in which LFY-YFP cytoplasmic condensates regulate the nuclear pool of transcriptionally active LFY to allow faithful transcription of LFY target genes.
Figure 7.
LFY condensation buffers LFY activity in the nucleus
(A) Confocal microscopy images of 5-day-old 35S::LFY-YFP roots before and after treatment with 1,6-hexanediol. Scale bar = 10 μm.
(B) Number of condensates in root cells from 5-day-old 35S::LFY-YFP roots before and after treatment with 1,6-hexanediol as shown in (A). n = 14 of 3 independent experiments. Statistical significance was determined using a two-tailed Student’s t test.
(C) Quantification of fluorescence intensity in the nucleus of 5-day-old 35S::LFY-YFP roots before and after treatment with 1,6-hexanediol as shown in (A). n = 16 of 3 independent experiments. Statistical significance was determined using a two-tailed Student’s t test.
(D) Accumulation of AP3 transcripts in flowers from 35S::LFY-YFP treated with 1,6-hexanediol. ACT2 was used as loading control.
Discussion
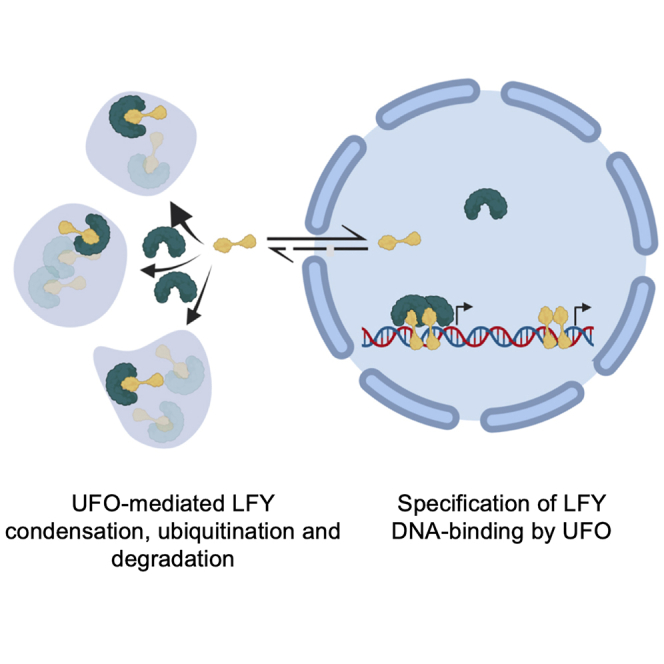
LFY has long been known to interact with the F box protein UFO to induce the localized expression of the AP3 B-class floral homeotic gene. Considering that UFO engages into an SCF E3 ubiquitin ligase complex and has the ability to ubiquitinate LFY,19,21,24,25,40,42 UFO was proposed to stimulate LFY-induced transcription through degradation of LFY. In this model, a rapid turnover of active LFY would allow the removal of “exhausted” LFY and replacement by “fresh” LFY to stimulate target gene expression,19 as proposed for other transcription factors in yeast and mammals.43 Recent evidence however suggests that the F box domain of UFO is dispensable for UFO-dependent LFY transcriptional activity.20 UFO indeed belongs to a transcriptional complex that promotes LFY recruitment to novel cis-regulatory elements within the AP3 promoter independently of UFO’s ability to mediate protein degradation. Our work now establishes an extranuclear role for UFO in LFY regulation. We demonstrate that UFO promotes LFY degradation and regulates its localization in cytoplasmic biomolecular condensates to buffer the nuclear LFY pool.
Flower meristem patterning requires the localized induction of the ABCE floral homeotic genes, which determine specific floral organ identities. LFY activates the floral homeotic genes in specific territories and thus largely controls organ specification.1,44 A precise control in time and space is therefore necessary for LFY gene expression. LFY is weakly expressed in leaves, and shows highest expression in floral meristems throughout the early primordium with earliest accumulation before cell groups have begun to separate from the inflorescence meristem.6 At later stages of floral development, LFY expression drops in the center of the flower while persisting in emerging petals, stamens, and pistil until stage 9 where its expression stops. The analysis of a large allelic series of lfy mutants highlighted the necessity for precise control of LFY activity in the transition from inflorescence to floral meristem and for proper floral patterning.6 This is further supported by the fact that strong LFY expression is sufficient to transform shoot meristems into flower meristems,3 thus acting as a developmental switch. Our observations indicate that LFY protein is found not only in the nucleus but also in cytoplasmic biomolecular condensates and that extranuclear LFY is subjected to UFO-mediated degradation. Although we could detect cytoplasmic foci in the endogenous expression territories of LFY, addressing whether LFY-mediated degradation occurs at specific stages or territories during flower development was not possible due to the complexity of high-resolution meristem imaging. Considering the importance of keeping LFY levels in check, we believe the mechanisms uncovered here likely contribute to LFY homeostasis by maintaining nuclear LFY levels that are tailored to faithfully regulate target genes.
Many transcriptional regulators are thought to form condensates, although this has been mostly described in the context of their recruitment/enrichment at specific genomic loci.45,46,47,48,49,50,51 Our findings resemble what has recently been reported for the ARF7 and ARF19 auxin signaling-related transcription factors that are kept away from the nucleus by sequestration into cytoplasmic condensates.30 However, extranuclear LFY appears not to be only sequestered but might also be subjected to degradation by the LFY-interacting UFO E3 ubiquitin ligase. Although it is clear that both processes regulate LFY homeostasis, the exact relationship between both of them remains unclear. Our data are consistent with a model in which LFY is degraded primarily outside of the nucleus, as proteasome inhibition and coexpression of the UFOΔF-box “substrate trap” variant have a much stronger effect on the extranuclear LFY pool. LFY condensates may represent the sites of LFY degradation as i) UFO co-localizes with LFY in such structures ii) UFO is an active LFY E3 ligase, iii) MG132 treatment increases the number of condensates, and iv) the degradation-inactive UFO ΔF-box variant yields increased numbers of cytoplasmic condensates. Nevertheless, as LFY can exchange between the different subcellular pools, we cannot exclude alternative models in which LFY undergoes degradation elsewhere. Pinpointing the precise localization of the proteasome in plant cells may help to distinguish between these scenarios. The mechanisms by which LFY may be protected from UFO-mediated degradation at specific subcellular locations might involve distinct sets of UFO-interacting proteins.
Our results with the LFY5KR mutant suggest that the LFY ubiquitination status can modulate LFY condensation. In particular, LFY5KR is similarly abundant as wild-type LFY but forms more condensates. Thus, the respective lysines may represent non-degradative ubiquitination sites negatively impacting condensate formation. This is reminiscent of the inhibition of UBQLN2 phase separation by ubiquitination.52 Interestingly, different ubiquitin linkage types show opposite effects on the ability of UBQLN2 to phase-separate.53 It is therefore possible that differential LFY ubiquitination by various E3 ligases controls its ability to form condensates and to be degraded. The mechanisms by which LFY5KR interferes with condensate formation and/or degradation remain unsolved for now and will be the subject of further studies.
The nuclear pool of LFY appears to be strongly buffered since interfering with the total LFY pool by UFO overexpression, MG132 treatment, or through the introduction of LFY5KR mutations had only a minor impact on nuclear LFY levels. This suggests that the extranuclear LFY pool serves to control nuclear LFY levels via LFY sequestration in condensates and LFY degradation. The levels of LFY in the nucleus only built up strongly upon treatment with 1,6-hexanediol, which dissolves cytoplasmic LFY condensates and therefore impairs buffering, leading to enhanced LFY target gene expression. The rapid hexanediol-mediated relocalization of LFY to the nucleus might also be promoted by the global effects of aliphatic alcohols on the permeability barrier of nuclear pores.54 Nevertheless, the nucleocytoplasmic shuttling of LFY we observed in untreated cells indicates that a similar (but possibly slower) LFY relocalization would be expected to occur upon condensate dissolution in cells with intact nuclear pores.
In summary, our work reveals that LFY undergoes nucleocytoplasmic partitioning to regulate LFY homeostasis via the formation of cytoplasmic biomolecular condensates and UFO-mediated degradation. This partitioning spatially separates the recently described UFO-mediated regulation of LFY transcriptional activity20 and the UFO-dependent control of cellular LFY levels, allowing cells to faithfully manage both activities without unwanted crosstalk.
Limitations of the study
Although we made sure that a functional LFY-GFP fusion forms cytoplasmic condensates when expressed in its endogenous cell territories in the lfy mutant background, the in-depth analysis of the mechanisms controlling LFY condensation and degradation were performed using constitutively expressed LFY in other cell types or in N. benthamiana. We have shown that constitutive expression of LFY in these cells does not go along with overexpression beyond the levels of the functional LFY-GFP fusion in flowers, but we cannot formally rule out that LFY degradation/condensation may be differently regulated in flowers.
All experiments performed in vivo used a transgene to express a functional fusion of LFY to a fluorescent protein. The presence of the tag may change the properties of LFY, thus impacting on LFY condensation and degradation. Besides, the contribution of UFO to LFY degradation, condensation, and activity was determined using a combination of pharmacological approaches and functional validation in N. benthamiana. Future work using stable transgenic lines would be helpful to assess the buffering hypothesis proposed in the present study.
Finally, our work did not directly address the functional relevance of LFY condensation during flower development. Identifying developmental stages or physiological conditions in which LFY degradation and partitioning in cytoplasmic foci are modulated will be one of the future challenges.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal anti-GFP horseradish peroxidase-coupled | Miltenyi Biotech | Cat# 130-091-833; RRID: AB_247003 |
| anti-ubiquitin P4D1 | Cell Signaling | Cat# 3936; RRID: AB_331292 |
| horseradish peroxidase-coupled anti-His | Sigma | Cat#A7058; RRID: AB_258326 |
| anti-FLAG | Sigma | Cat#F3165; RRID: AB_259529 |
| Bacterial and virus strains | ||
| E.coli DH5α | N/A | N/A |
| Agrobacterium tumefaciens AGL1 | N/A | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| LFY-His | Chahtane et al., 201833 | N/A |
| Ubiquitin | Ubiquigent | Cat#69-1300 |
| Human E1 ligase UBE1 | R&D Systems Europe | Cat#E-305 |
| Human E2 ligase UBE2D2 | Ubiquigent | Cat#62-0012-020 |
| Human Cul1/Rbx1/Skp1 | Ubiquigent | Cat#63-1001-025 |
| TnT SP6 High-Yield Wheat Germ Protein Expression System | Promega | Cat#L3260 |
| Anti-FLAG M2 affinity gel | Sigma | Cat#F1804 |
| 3x FLAG peptide | GenScript | Cat#RP10586 |
| GFP-Trap Magnetic beads | ChromoTek | Cat#gtma |
| ATTO 647N NHS ester | ATTO-TEC | Cat#AD 647N-31 |
| 1,6-hexanediol | Sigma | Cat#240117 |
| FM4-64 | Invitrogen | Cat#T13320 |
| RNeasy Plant Mini Kit | Qiagen | Cat# 74904 |
| M-MLV reverse transcriptase | Promega | Cat# M1701 |
| MboI | NEB | Cat# R0147S |
| PEG 20,000 | Fluka | Cat#81300 |
| Zeba MicroSpin desalting column 7K MWCO | Thermofisher | Cat#89877 |
| MG132 (Z-Leu-Leu-Leu-al) | Sigma | Cat# C2211 |
| Cycloheximide | Sigma | Cat# 01810 |
| Brefeldin A | Sigma | Cat# B5936 |
| N-ethylmaleimide | Sigma | Cat# E3876 |
| Protease inhibitor cocktail | Sigma | Cat# P9599 |
| Experimental models: Organisms/strains | ||
| A. thaliana Col-0 | Columbia | |
| ufo-1 | Wilkinson and Haughn, 199555 | N/A |
| lfy-12 | Weigel et al., 19926 | N/A |
| 35S::LFY-YFP | Chahtane et al., 201833 | N/A |
| UBI10::mCherry-ATG8E | Stephanie et al., 202056 | N/A |
| lfy-12/LFY::LFY-GFP | Wu et al., 200332 | N/A |
| Oligonucleotides | ||
| See Table S1 | Eurofins | N/A |
| Recombinant DNA | ||
| pTNT-UFO-FLAG | This paper | N/A |
| pDONR221-LFY | This paper | N/A |
| pDONR221-LFY5KR | This paper | N/A |
| pDONR-p221-UFO | This paper | N/A |
| pDONR-p221-UFOΔF-box | This paper | N/A |
| pTNT-UFO:FLAG | This paper | N/A |
| pBasta 35S::LFY:mCt-Cterm | This paper | N/A |
| pBasta 35S::LFY:mCt-Nterm | This paper | N/A |
| pBasta 35S::LFY5KR:mCt-Cterm | This paper | N/A |
| pBasta 35S::UFO:mCt-Cterm | This paper | N/A |
| pBasta 35S::URI:mCt-Cterm | This paper | N/A |
| pBasta 35S::UFO-deltaFbox:mCt-Nterm | This paper | N/A |
| pBasta 35S::UFO-Fbox:mCt-Cterm | This paper | N/A |
| pKana 35S::LFY:mCt | This paper | N/A |
| pKana 35S::LFY5KR:mCt | This paper | N/A |
| pHygro 35S::UFO:mCherry | This paper | N/A |
| pHygro 35S::UFO-deltaFbox:mCherry | This paper | N/A |
| Software and algorithms | ||
| PONDR | Open source | http://www.pondr.com |
| PLAAC | Open source | http://plaac.wi.mit.edu |
| GraphPad Prism | Open source | https://www.graphpad.com |
| R studio | Open source | https://posit.co |
| ImageJ | Open source | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Grégory Vert (Gregory.Vert@univ-tlse3.fr).
Material availability
Materials generated in this study are available upon request. For further details contact the lead contact.
Experimental model and study participant details
The Arabidopsis thaliana ecotype Col-0 was used in this study. The mutants and transgenic lines used in this study have been previously described : lfy-12,6 ufo-1,55 35S::LFY-YFP,33 UBI10::mCherry-ATG8E,56 lfy-12/LFY::LFY-GFP.32 Seeds were surface-sterilized, sown on half-strength Linsmaier & Skoog medium without sucrose containing 1% agar and stratified at 4°C for 2 days before transfer to light. Plant lines were grown under sterile conditions on vertical plates at 21°C with a 16h light/8h dark cycle. For co-localization studies, 35S::LFY-YFP was crossed with the UBI10::mCherry-ATG8E autophagy reporter line and the F1 generation was imaged. All cultivation and growth conditions relevant for Nicotiana benthamiana are included within the ‘method details’ section.
Method details
Vector construction
To generate the different transgenic lines used in this study, the coding sequences corresponding to LFY, UFO and UFOΔF-box (nucleotide 274-1349) were amplified from cDNA and cloned into pDONR221 by Gateway cloning. The LFY5KR construct was generated by site-directed mutagenesis using pDONR221-LFY as template. The UFO expression vectors were constructed using the pH7m34GW destination vector with the pDONR-P4P1r-35S, the pDONR-p221-UFO or pDONR-p221-UFOΔF-box, and the pDONR-P2rP3-mCh.
The BiFC expression vectors were constructed by multisite Gateway recombination using the pK7m34GW destination vector with the pDONR-P4P1r-35S, the pDONR-p221-LFY, pDONR-p221-LFY5KR, pDONR-p221-UFO, pDONR-p221-ΔF-box, pDONR-p221-F-box or pDONR-p221-URI for genes of interest, and pDONR-P2rP3-mCitN (amino acid 1-154) or pDONR-P2rP3-mCitC (amino acid 155-238) for the fluorescence reporter.
To produce UFO protein by in vitro transcription/translation, the coding sequence of UFO was cloned into the pTNT-FLAG vector.57 All primers used for plasmid construction are listed in Table S1.
Transient expression in N. benthamiana
For transient expression in N. benthamiana, plasmids were transformed into Agrobacterium strain AGL1 and infiltrated into wild-type leaves at OD600 of 0.5 using infiltration buffer (10mM MES, 10mM MgCl2, pH 5.6). For co-expression analysis, the Agrobacteria containing the different plasmids were used in a 1:1 ratio. Leaf discs were observed 2 days after infiltration.
Gene expression analyses and genotyping
Total RNA was extracted from N. benthamiana or A. thaliana using RNeasy Plant Mini Kit (QIAGEN). Detection of LFY, UFO, and AP3 was performed using RT-PCR on total RNA. 1μg of RNA was subjected to reverse transcription using M-MLV reverse transcriptase from Promega. PCR were performed using primers for the respective transcript detection. EF-1α for N. benthamiana or Actin2 for A. thaliana were used as a control. The ufo-1 mutant was genotyped by PCR amplification followed by restriction digestion with MboI. Primers used are listed in Table S1.
Confocal microscopy imaging
Samples were mounted in water and imaged with a 63x objective on a Leica TCS SP8 confocal laser scanning microscope. For imaging mCit/YFP, GFP and mCh, the 514-nm, 488-nm and 561-nm laser lines were used, respectively. Detection settings were kept constant in individual sets of experiments to allow for direct comparison of expression and localization of reporter proteins, unless stated otherwise. Images were taken from 5-days old A. thaliana roots, hypocotyls or cotyledons, or from N. benthamiana leaf discs 2 days post-infiltration. Flowers of A. thaliana were imaged as previously described.58
To compare fluorescence levels of LFY::LFY-GFP in flowers, 35S::LFY-YFP in roots and 35S::LFY-mCit expressed in N. benthamiana leaf cells, images were taken using the same microscope, objective, magnification, excitation (488 nm laser line, same intensity) and detection settings (500-550 nm window). Nuclear intensities were quantified using ImageJ and corrected for the molecular brightness of the respective fluorescent proteins under the respective excitation/detection conditions according to
Here, FBN is the normalized fluorescence, I is the fluorescence intensity, BM is the corrected molecular brightness, is the extinction coefficient at 488 nm (in M-1 cm-1) and is the quantum yield of the fluorophore integrated over the spectral window of detection. The following molecular brightness values were obtained for the different fluorescent proteins based on the data in FPbase (https://www.fpbase.org): BM EGFP = 24.0, BM YFP = 11.0, and BM mCit = 15.7.
For colocalization studies with FM4-64, plants were treated with 4 μM FM4-64 for 10 min and imaged 20 min after staining. For imaging FM4-64, the 488-nm laser line was used. Colocalization studies were performed using the Coloc2 plugin of ImageJ and R. Fluorescence intensity profiles were also obtained using ImageJ or R. Quantification of mobile versus immobile condensates were performed using R.
For Brefeldin A treatment, plants were pre-incubated with 100 μM cycloheximide for 30 min, exposed for an additional 30 min to 50 μM Brefeldin A and 100 μM cycloheximide, and stained with FM4-64 prior to imaging. For MG132 treatment in A. thaliana, plants were exposed to 50 μM MG132 or DMSO (mock) for 3h prior to confocal imaging. For 1,6-hexanediol treatment in A. thaliana, plants were incubated in 10% or 20% 1,6-hexanediol for 1 min prior to confocal imaging. For MG132 treatment, N. benthamiana leaves were infiltrated with 50 μM MG132 or DMSO (mock), 1 day after the initial infiltration. For 1,6-hexanediol treatment, N. benthamiana leaves were infiltrated with 20% 1,6-hexanediol and imaged immediately.
Fluorescence recovery after photobleaching (FRAP) measurements
FRAP experiments were performed on condensates from 35S::LFY-YFP root tips (full-FRAP) and N. benthamiana leaf discs transiently expressing 35S::LFY-mCit (half-FRAP) using a Zeiss LSM 710 confocal light scanning microscope (Carl Zeiss, Oberkochen, Germany) equipped with a 63x/NA 1.2 oil immersion objective. Typically, images were acquired at 128x512 pixels at a scan speed corresponding to 200 ms per image, and 300 images were acquired over 2.5 min, with an interval of 300 ms between subsequent images (or 800 ms for full-nucleus FRAP and in vitro experiments). Before photobleaching, 3 images were recorded.
Full-FRAP analysis
For full-FRAP analysis, 9 independent experiments were conducted and averaged to obtain a single curve. For each experiment, a custom R script was used to segment the image, track the condensates and retrieve the average intensity of the photobleached condensate (IB), an unbleached condensate (IREF) and the background of the image (IBG). The unbleached condensate was used as internal reference to quantify unwanted acquisition photobleaching, and the FRAP curves were calculated according to:
Then, the FRAP curves were normalized according to the number of bleached molecules:
Here, tbleach and tpre are the acquisition times of the first post-bleach and the last pre-bleach frame, respectively. Accordingly, the normalized FRAP curves are equal to unity before bleaching and zero in the first post-bleach frame. If applicable, FRAP curves were fitted with a previously described diffusion model.59
Half-FRAP analysis
For half-FRAP, 8 experiments were analyzed and averaged. For each experiment, a custom R script was used to segment the image, track the bleached condensate and retrieve the average intensity of the bleached half (IB), the non-bleached half (INB), the background of the image (IBG) and an unbleached condensate (IREF), at each frame. These intensity values were used to calculate FRAP curves for the bleached half (FRAPB(t)) and the non-bleached half (FRAPNB(t)), according to:
If unwanted photobleaching is detected in the non-bleached half in the first post-bleach frame, this contribution should be removed from the analysis. The respective molecules might have been bleached because the microscope’s focus overlapped with the non-bleached half, or because the molecules moved from the bleached to the non-bleached half during the bleaching period. The curve that is corrected for this type of unwanted photobleaching reads:
Here, tpre and tbleach are the acquisition times of the last pre-bleach and the first post-bleach frame of the experiment, respectively.
Next, FRAPB and FRAPNB were multiplied by the size of their respective ROIs (NB and NNB, respectively) to obtain curves that are proportional to the number of particles in each half:
Then, the curves were normalized with respect to the number of bleached molecules:
The resulting FRAP curves are proportional to the ROI sizes and double-normalized. Subsequently, an additive offset was applied to the signal in the non-bleached half to normalize to unity before the bleach:
The resulting curves reflect the change of the number of labeled molecules in each half. In the presence of an “immobile” fraction of molecules that do not move during the course of the experiment, the signal in both halves will not recover to the same level but there will be an offset between them that corresponds to the immobile fraction . To remove these “immobile” molecules, which do neither exchange between the two halves nor cross the boundary of the condensate, the FRAP curves were corrected according to:
The immobile fraction corresponds to the difference between the curves in the bleached and non-bleached half after both of them have reached their plateau.
Finally, the decrease of fluorescence in the non-bleached half was used to calculate the apparent energy barrier per molecule, as described previously.37 In the case of LFY condensates in vivo, this barrier amounted to 0.02 kT per molecule, which corresponds to an apparent surface tension of 0.36 μN/m according to the formula below:
Here, and are the apparent surface tension and the apparent energy barrier per molecule (in units of the thermal energy kT), respectively, and Rh is the hydrodynamic radius of LFY-YFP, which we estimated as 6.2 nm based on the LFY and YFP AlphaFold structures.
Condensate fusion and shape analysis
The fusion analysis was performed on condensates from root tips of 35S::LFY-YFP in the wild-type background on a Leica TCS SP8 confocal laser scanning microscope. Images at 512x512 or 1024x1024 pixels were acquired each 2 seconds over 2.5 minutes to generate videos of the dynamics of condensates. In total, 13 independent experiments were analyzed using a custom R script and the EBImage package.60 For each video, the nucleus and the condensates were segmented in the first frame, based on size and intensity. The eccentricity (E) of condensates was calculated as:
where Rmax and Rmin are the longest and shortest radius, respectively. Then, the position of each segmented condensate was tracked and recorded during the video. The positions of the nuclei were also recorded to correct the changes in the position of condensates due to the movement of the root, resulting in trajectories for each condensate (Figure S7A). Each trajectory was used to calculate a Mean Square Displacement (MSD) as follows:
Finally, each MSD curve was fitted to a linear equation and the diffusion coefficient was obtained from the slope of the fit curve (Figure S7B).
Bimolecular fluorescence complementation
For BiFC, the LFY-mCitN, UFO-mCitC and variants were transiently expressed in N. benthamiana and leaf discs imaged 2 days post-infiltration. LFY oligomerization was used as positive control, and the bHLH transcription factor URI as negative control.61
In vitro droplet assay
In order to prepare Atto-647N-labeled LFY for in vitro microscopy experiments, LFY at a concentration of 1 μM was labeled with ATTO 647N NHS ester (AD 647N-31, ATTO-TEC) according to the manufacturer’s instructions. Free ATTO 647N dye was removed using a Zeba MicroSpin desalting column 7K MWCO (89877, Thermofisher). LFY-ATTO 647N was spiked into unlabeled LFY at a 1:100 ratio, and phase separation was induced by mixing with 2% PEG 20,000 (813000, Fluka) at a salt concentration of 100 mM NaCl. After 15 minutes of incubation at 4°C, the samples were pipetted on a microscopy slide, which was precoated with PEG 8000. All experiments were carried out at room temperature within 20 min after pipetting.
In vitro ubiquitination assay
UFO-FLAG was expressed using TnT SP6 High-Yield Wheat Germ Protein Expression System (Promega) according to the manufacturer’s instructions. UFO-FLAG was purified using Anti-FLAG M2 affinity gel (Sigma) and eluted with 3x FLAG peptide (GenScript). Untagged ubiquitin (Ubi), the human E2 ligase UBE2D2 and the human Cul1/Rbx1/Skp1 fusion protein were all purchased from Ubiquigent (UK). The human E1 ligase UBE1 was purchased from R&D Systems Europe. The in-vitro ubiquitination assay was performed as followed: 290 nM His-LFY, 1mM Ubi, 50nM UBE1, 100nM UBE2D2, 50nM Cul1/Rbx1/Skp1 and UFO-FLAG were incubated at 30°C overnight in reaction buffer (5mM ATP, 50mM Tris-HCl pH 7.5, 200mM NaCl, 10mM MgCl2, 1 unit inorganic pyrophosphate and 1mM DTT). Reactions were stopped by adding Laemmli buffer and incubation for 5 min at 95 °C. Protein analysis was done by western blot using horseradish peroxidase-coupled anti-His (Sigma, 1/5,000), ubiquitin P4D1 (Cell Signaling, 1/2,000), anti-FLAG (Sigma, 1/2,000).
Protein extraction, immunoprecipitation and detection
Total proteins were extracted from N. benthamiana leaves 2 days after infiltration. Immunoprecipitation experiments were performed using N. benthamiana leaves 2-days after infiltration. Tissues were ground in liquid nitrogen and resuspended in ice-cold RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Igepal, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitors and 20mM N-ethylmaleimide to inhibit deubiquitinases during protein preparation. Samples were solubilized at 4°C for 30 minutes followed by centrifugation for 15 min at 14,000 x g at 4°C to eliminate cell debris. Supernatants were subjected to immunoprecipitation with the GFP-Trap Magnetic beads (ChromoTek). Immunoprecipitates were eluted off beads using Laemmli buffer. Samples were boiled for 5 minutes at 95°C and migrated on a 10% Bis-Tris NuPage Gel (Life Technologies).
Total proteins or immunoprecipitates were separated on a 10% Bis-Tris NuPage gel (Life Technologies) and transferred onto a nitrocellulose membrane. Proteins were detected using the following antibodies: Monoclonal anti-GFP horseradish peroxidase-coupled (Miltenyi Biotech 130-091-833, 1/5,000), anti-ubiquitin P4D1 (Cell Signaling, 1/2,000). Experiments were done in triplicates. Rubisco was used as loading control.
Quantification and statistical analysis
All quantification and statistical analysis details and associated citations can be found in the method details in the ‘Confocal microscopy imaging’, ‘Full-FRAP analysis’, ‘Half-FRAP analysis’ and ‘Condensate fusion and shape analysis’ sections. All experiments, if not stated differently in the figure legend were done in triplicates, n represents the number of cells. Statistical significance of the biological parameters was assessed using t-test and p-value = 5·10-9. Statistical analyses were performed with the software GraphPad Prism. The statistical parameters are reported in the figure legends.
The Predictor of Natural Disordered Regions (PONDR, http://www.pondr.com) algorithm was used for the prediction of the intrinsically disordered regions (IDRs) and the Prion-Like Amino Acid Composition algorithm (PLAAC, http://plaac.wi.mit.edu) for the prion-like domains (PLDs) in LFY protein.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: LFY (At1g78870), UFO (At1g16890), URI (AT3G19860), Actin2 (At5g59910), AP3 (AT3G54340).
Acknowledgments
We thank Y. Dagdas and François Parcy for sharing ATG8E-mCh reporter lines and LFY-related materials, respectively. We would also like to acknowledge the Imaging facilities from the Fédération de Recherche Agrobiosciences Interactions et Biodiversité of Toulouse (FRAIB) and the Center for Integrative Biology of Toulouse. This work was supported by a postdoctoral fellowship from the German Research Foundation (423452781 to U.D.) and research grants from the French National Research Agency (ANR-17-CE20-0014-02 to G.V.) and the French Laboratory of Excellence (project “TULIP” grant nos. ANR–10–LABX–41 and ANR–11–IDEX–0002–02 to G.V.).
Author contributions
Conceptualization, U.D., F.M., F.E., and G.V.; Methodology, U.D., F.M., F.E., and G.V.; Investigation, U.D., F.M., C.D., and J.N.; Writing – Original Draft, U.D., F.M., F.E., and G.V.; Writing – Review & Editing, U.D., F.M., F.E., and G.V.; Funding Acquisition, U.D. and G.V.; Resources, F.E. and G.V.; Supervision, F.E. and G.V.
Declaration of interests
The authors declare no competing interests.
Published: May 15, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106880.
Contributor Information
Fabian Erdel, Email: fabian.erdel@cnrs.fr.
Grégory Vert, Email: gregory.vert@univ-tlse3.fr.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
References
- 1.Moyroud E., Kusters E., Monniaux M., Koes R., Parcy F. LEAFY blossoms. Trends Plant Sci. 2010;15:346–352. doi: 10.1016/j.tplants.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Huala E., Sussex I.M. LEAFY interacts with floral homeotic genes to regulate Arabidopsis floral development. Plant Cell. 1992;4:901–913. doi: 10.1105/tpc.4.8.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weigel D., Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;377:495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]
- 4.Blázquez M.A., Soowal L.N., Lee I., Weigel D. LEAFY expression and flower initiation in Arabidopsis. Development. 1997;124:3835–3844. doi: 10.1242/dev.124.19.3835. [DOI] [PubMed] [Google Scholar]
- 5.Parcy F., Nilsson O., Busch M.A., Lee I., Weigel D. A genetic framework for floral patterning. Nature. 1998;395:561–566. doi: 10.1038/26903. [DOI] [PubMed] [Google Scholar]
- 6.Weigel D., Alvarez J., Smyth D.R., Yanofsky M.F., Meyerowitz E.M. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- 7.Winter C.M., Austin R.S., Blanvillain-Baufumé S., Reback M.A., Monniaux M., Wu M.F., Sang Y., Yamaguchi A., Yamaguchi N., Parker J.E., et al. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell. 2011;20:430–443. doi: 10.1016/j.devcel.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 8.Moyroud E., Minguet E.G., Ott F., Yant L., Posé D., Monniaux M., Blanchet S., Bastien O., Thévenon E., Weigel D., et al. Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell. 2011;23:1293–1306. doi: 10.1105/tpc.111.083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamès C., Ptchelkine D., Grimm C., Thevenon E., Moyroud E., Gérard F., Martiel J.L., Benlloch R., Parcy F., Müller C.W. Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins. EMBO J. 2008;27:2628–2637. doi: 10.1038/emboj.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sayou C., Nanao M.H., Jamin M., Posé D., Thévenon E., Grégoire L., Tichtinsky G., Denay G., Ott F., Peirats Llobet M., et al. A SAM oligomerization domain shapes the genomic binding landscape of the LEAFY transcription factor. Nat. Commun. 2016;7:11222. doi: 10.1038/ncomms11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siriwardana N.S., Lamb R.S. A conserved domain in the N-terminus is important for LEAFY dimerization and function in Arabidopsis thaliana. Plant J. 2012;71:736–749. doi: 10.1111/j.1365-313X.2012.05026.x. [DOI] [PubMed] [Google Scholar]
- 12.Benlloch R., Berbel A., Serrano-Mislata A., Madueño F. Floral initiation and inflorescence architecture: a comparative view. Ann. Bot. 2007;100:659–676. doi: 10.1093/aob/mcm146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner D., Sablowski R.W., Meyerowitz E.M. Transcriptional activation of APETALA1 by LEAFY. Science. 1999;285:582–584. doi: 10.1126/science.285.5427.582. [DOI] [PubMed] [Google Scholar]
- 14.Goto K., Meyerowitz E.M. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- 15.Jack T., Brockman L.L., Meyerowitz E.M. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- 16.Lee I., Wolfe D.S., Nilsson O., Weigel D. A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr. Biol. 1997;7:95–104. doi: 10.1016/s0960-9822(06)00053-4. [DOI] [PubMed] [Google Scholar]
- 17.Levin J.Z., Meyerowitz E.M. UFO: an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell. 1995;7:529–548. doi: 10.1105/tpc.7.5.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hepworth S.R., Klenz J.E., Haughn G.W. UFO in the Arabidopsis inflorescence apex is required for floral-meristem identity and bract suppression. Planta. 2006;223:769–778. doi: 10.1007/s00425-005-0138-3. [DOI] [PubMed] [Google Scholar]
- 19.Chae E., Tan Q.K.G., Hill T.A., Irish V.F. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development. 2008;135:1235–1245. doi: 10.1242/dev.015842. [DOI] [PubMed] [Google Scholar]
- 20.Rieu P., Turchi L., Thévenon E., Zarkadas E., Nanao M., Chahtane H., Tichtinsky G., Lucas J., Blanc-Mathieu R., Zubieta C., et al. The F-box cofactor UFO redirects the LEAFY floral regulator to novel <em>cis</em>-elements. bioRxiv. 2022 doi: 10.1101/2022.06.14.495942. Preprint at. [DOI] [Google Scholar]
- 21.Samach A., Klenz J.E., Kohalmi S.E., Risseeuw E., Haughn G.W., Crosby W.L. The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 1999;20:433–445. doi: 10.1046/j.1365-313x.1999.00617.x. [DOI] [PubMed] [Google Scholar]
- 22.Risseeuw E., Venglat P., Xiang D., Komendant K., Daskalchuk T., Babic V., Crosby W., Datla R. An activated form of UFO alters leaf development and produces ectopic floral and inflorescence meristems. PLoS One. 2013;8:e83807. doi: 10.1371/journal.pone.0083807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lechner E., Achard P., Vansiri A., Potuschak T., Genschik P. F-box proteins everywhere. Curr. Opin. Plant Biol. 2006;9:631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Zhao D., Yu Q., Chen M., Ma H. The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis. Development. 2001;128:2735–2746. doi: 10.1242/dev.128.14.2735. [DOI] [PubMed] [Google Scholar]
- 25.Ni W., Xie D., Hobbie L., Feng B., Zhao D., Akkara J., Ma H. Regulation of flower development in Arabidopsis by SCF complexes. Plant Physiol. 2004;134:1574–1585. doi: 10.1104/pp.103.031971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gagne J.M., Smalle J., Gingerich D.J., Walker J.M., Yoo S.D., Yanagisawa S., Vierstra R.D. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. USA. 2004;101:6803–6808. doi: 10.1073/pnas.0401698101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuevas-Velazquez C.L., Dinneny J.R. Organization out of disorder: liquid-liquid phase separation in plants. Curr. Opin. Plant Biol. 2018;45:68–74. doi: 10.1016/j.pbi.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Emenecker R.J., Holehouse A.S., Strader L.C. Emerging roles for phase separation in plants. Dev. Cell. 2020;55:69–83. doi: 10.1016/j.devcel.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powers S.K., Holehouse A.S., Korasick D.A., Schreiber K.H., Clark N.M., Jing H., Emenecker R., Han S., Tycksen E., Hwang I., et al. Nucleo-cytoplasmic partitioning of ARF proteins controls auxin responses in Arabidopsis thaliana. Mol. Cell. 2019;76:177–190.e5. doi: 10.1016/j.molcel.2019.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jing H., Korasick D.A., Emenecker R.J., Morffy N., Wilkinson E.G., Powers S.K., Strader L.C. Regulation of AUXIN RESPONSE FACTOR condensation and nucleo-cytoplasmic partitioning. Nat. Commun. 2022;13:4015. doi: 10.1038/s41467-022-31628-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X., Dinneny J.R., Crawford K.M., Rhee Y., Citovsky V., Zambryski P.C., Weigel D. Modes of intercellular transcription factor movement in the Arabidopsis apex. Development. 2003;130:3735–3745. doi: 10.1242/dev.00577. [DOI] [PubMed] [Google Scholar]
- 33.Chahtane H., Zhang B., Norberg M., LeMasson M., Thévenon E., Bakó L., Benlloch R., Holmlund M., Parcy F., Nilsson O., Vachon G. LEAFY activity is post-transcriptionally regulated by BLADE ON PETIOLE2 and CULLIN3 in Arabidopsis. New Phytol. 2018;220:579–592. doi: 10.1111/nph.15329. [DOI] [PubMed] [Google Scholar]
- 34.Gallois J.L., Nora F.R., Mizukami Y., Sablowski R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 2004;18:375–380. doi: 10.1101/gad.291204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner D., Wellmer F., Dilks K., William D., Smith M.R., Kumar P.P., Riechmann J.L., Greenland A.J., Meyerowitz E.M. Floral induction in tissue culture: a system for the analysis of LEAFY-dependent gene regulation. Plant J. 2004;39:273–282. doi: 10.1111/j.1365-313X.2004.02127.x. [DOI] [PubMed] [Google Scholar]
- 36.Zavaliev R., Mohan R., Chen T., Dong X. formation of NPR1 condensates promotes cell survival during the plant immune response. Cell. 2020;182:1093–1108.e18. doi: 10.1016/j.cell.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muzzopappa F., Hummert J., Anfossi M., Tashev S.A., Herten D.P., Erdel F. Detecting and quantifying liquid-liquid phase separation in living cells by model-free calibrated half-bleaching. Nat. Commun. 2022;13:7787. doi: 10.1038/s41467-022-35430-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erdel F., Rademacher A., Vlijm R., Tünnermann J., Frank L., Weinmann R., Schweigert E., Yserentant K., Hummert J., Bauer C., et al. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol. Cell. 2020;78:236–249.e7. doi: 10.1016/j.molcel.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H., Kelley F.M., Milovanovic D., Schuster B.S., Shi Z. Surface tension and viscosity of protein condensates quantified by micropipette aspiration. Biophys. Rep. 2021;1:100011. doi: 10.1016/j.bpr.2021.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X., Feng S., Nakayama N., Crosby W.L., Irish V., Deng X.W., Wei N. The COP9 signalosome interacts with SCF UFO and participates in Arabidopsis flower development. Plant Cell. 2003;15:1071–1082. doi: 10.1105/tpc.009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alberti S., Gladfelter A., Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao D., Yang M., Solava J., Ma H. The ASK1 gene regulates development and interacts with the UFO gene to control floral organ identity in Arabidopsis. Dev. Genet. 1999;25:209–223. doi: 10.1002/(sici)1520-6408(1999)25:3<209::Aid-dvg4>3.0.Co;2-o. [DOI] [PubMed] [Google Scholar]
- 43.Kodadek T., Sikder D., Nalley K. Keeping transcriptional activators under control. Cell. 2006;127:261–264. doi: 10.1016/j.cell.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Irish V.F. The flowering of Arabidopsis flower development. Plant J. 2010;61:1014–1028. doi: 10.1111/j.1365-313X.2009.04065.x. [DOI] [PubMed] [Google Scholar]
- 45.Boija A., Klein I.A., Sabari B.R., Dall'Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M., et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018;175:1842–1855.e16. doi: 10.1016/j.cell.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., Tjian R. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361:eaar2555. doi: 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sabari B.R., Dall'Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C., et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho W.K., Jayanth N., English B.P., Inoue T., Andrews J.O., Conway W., Grimm J.B., Spille J.H., Lavis L.D., Lionnet T., Cisse I.I. RNA Polymerase II cluster dynamics predict mRNA output in living cells. Elife. 2016;5:e13617. doi: 10.7554/eLife.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho W.K., Spille J.H., Hecht M., Lee C., Li C., Grube V., Cisse I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412–415. doi: 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu H., Yu D., Hansen A.S., Ganguly S., Liu R., Heckert A., Darzacq X., Zhou Q. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature. 2018;558:318–323. doi: 10.1038/s41586-018-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dao T.P., Kolaitis R.M., Kim H.J., O'Donovan K., Martyniak B., Colicino E., Hehnly H., Taylor J.P., Castañeda C.A. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell. 2018;69:965–978.e6. doi: 10.1016/j.molcel.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dao T.P., Yang Y., Presti M.F., Cosgrove M.S., Hopkins J.B., Ma W., Loh S.N., Castañeda C.A. Mechanistic insights into enhancement or inhibition of phase separation by different polyubiquitin chains. EMBO Rep. 2022;23:e55056. doi: 10.15252/embr.202255056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ribbeck K., Görlich D. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 2002;21:2664–2671. doi: 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilkinson M.D., Haughn G.W. UNUSUAL FLORAL ORGANS controls meristem identity and organ primordia fate in Arabidopsis. Plant Cell. 1995;7:1485–1499. doi: 10.1105/tpc.7.9.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stephani M., Picchianti L., Gajic A., Beveridge R., Skarwan E., Sanchez de Medina Hernandez V., Mohseni A., Clavel M., Zeng Y., Naumann C., et al. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. Elife. 2020;9:e58396. doi: 10.7554/eLife.58396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nito K., Wong C.C.L., Yates J.R., Chory J. Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors. Cell Rep. 2013;3:1970–1979. doi: 10.1016/j.celrep.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prunet N., Jack T.P., Meyerowitz E.M. Live confocal imaging of Arabidopsis flower buds. Dev. Biol. 2016;419:114–120. doi: 10.1016/j.ydbio.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soumpasis D.M. Theoretical analysis of fluorescence photobleaching recovery experiments. Biophys. J. 1983;41:95–97. doi: 10.1016/s0006-3495(83)84410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pau G., Fuchs F., Sklyar O., Boutros M., Huber W. EBImage--an R package for image processing with applications to cellular phenotypes. Bioinformatics. 2010;26:979–981. doi: 10.1093/bioinformatics/btq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim S.A., LaCroix I.S., Gerber S.A., Guerinot M.L. The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proc. Natl. Acad. Sci. USA. 2019;116:24933–24942. doi: 10.1073/pnas.1916892116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confocal imaging time-course of 35S::LFY-YFP roots.
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request