Abstract
Nuclear migration and positioning in Saccharomyces cerevisiae depend on long astral microtubules emanating from the spindle pole bodies (SPBs). Herein, we show by in vivo fluorescence microscopy that cells lacking Spc72, the SPB receptor of the cytoplasmic γ-tubulin complex, can only generate very short (<1 μm) and unstable astral microtubules. Consequently, nuclear migration to the bud neck and orientation of the anaphase spindle along the mother-bud axis are absent in these cells. However, SPC72 deletion is not lethal because elongated but misaligned spindles can frequently reorient in mother cells, permitting delayed but otherwise correct nuclear segregation. High-resolution time-lapse sequences revealed that this spindle reorientation was most likely accomplished by cortex interactions of the very short astral microtubules. In addition, a set of double mutants suggested that reorientation was dependent on the SPB outer plaque and the astral microtubule motor function of Kar3 but not Kip2/Kip3/Dhc1, or the cortex components Kar9/Num1. Our observations suggest that Spc72 is required for astral microtubule formation at the SPB half-bridge and for stabilization of astral microtubules at the SPB outer plaque. In addition, our data exclude involvement of Spc72 in spindle formation and elongation functions.
INTRODUCTION
In most eukaryotic organisms the position of the spindle determines the location of the cleavage furrow at cytokinesis (Hyman, 1989). In the budding yeast Saccharomyces cerevisiae this is not the case. The plane of cytokinesis is predefined by the position of an emerging bud. To ensure that both the mother and daughter cell receive a nucleus upon spindle elongation, the spindle has to be actively positioned close to the mother-daughter junction (the bud neck) and oriented along the mother-daughter axis.
These processes are accomplished by the action of dynamic forces acting on the nuclei via microtubules (reviewed by Hildebrandt and Hoyt, 2000). Microtubules can be classified as nuclear or astral (cytoplasmic) (Byers and Goetsch, 1975). Nuclear microtubules are involved in assembly of a bipolar spindle and in segregation of the chromosomes (Jacobs et al., 1988; Straight et al., 1997); astral microtubules function to position, move, and orient the spindle and thus the nucleus within the cell (Palmer et al., 1992; Sullivan and Huffaker, 1992; Carminati and Stearns, 1997; Shaw et al., 1997; Tirnauer et al., 1999). All microtubules are organized by the spindle pole body (SPB) the functional homolog of the microtubule organizing center of higher eukaryotic cells. In electron micrographs, the spindle pole body appears as a three-laminar structure embedded in the nuclear envelope and a one-sided extension of the central layer localized on top of the nuclear envelope called a half-bridge (Moens and Rapport, 1971; Byers and Goetsch, 1975).
Microtubules are nucleated on the γ-tubulin complex that consists of Spc98, Spc97, and Tub4 (Geissler et al., 1996; Knop et al., 1997; Murphy et al., 1998, Rout and Kilmartin, 1990). The γ-tubulin complex assembles in the cytoplasm and is then targeted and anchored to the inner plaque of the SPB by Spc110 and to the outer plaque and half-bridge region by Spc72 (Rout and Kilmartin, 1990; Spang et al., 1996; Knop and Schiebel, 1997, 1998; Nguyen et al., 1998). Although the interaction sites of Spc110 with the γ-tubulin complex and the SPB have been identified and its function is relatively well understood (Kilmartin and Goh, 1996; Knop and Schiebel, 1997; Nguyen et al., 1998), the function of Spc72 remains less clear. The observations that astral microtubules are localized to two different cytoplasmic SPB substructures during specific phases of the cell cycle, the outer plaque and the half-bridge, and that Spc72 is present at both substructures suggest that Spc72 may have multiple functions.
Nuclear segregation and thus cell viability have been shown to be strictly dependent on intact astral microtubules (Sullivan and Huffaker, 1992). Based on the proposed function of Spc72 as an anchor of the γ-tubulin complex and thus the site for astral microtubule formation, loss of Spc72 was expected to result in a lethal phenotype (Chen et al., 1998; Knop and Schiebel, 1998). This however was not the case in several strain backgrounds (Souès and Adams, 1998; this study). Remarkably, cells lacking Spc72 displayed defects in astral microtubule formation and in nuclear segregation, yet still were able to proliferate. This raises the question how cells with impaired or even absent astral microtubules position their spindles and segregate their nuclei?
In this study, we describe live cell imaging of green fluorescent protein (GFP)-labeled nuclei and microtubules in wild-type and spc72Δ deletion mutants. Our observations enable us to propose a model that explains the nuclear dynamics defect observed in cells lacking Spc72 and also how residual successful nuclear segregation occurs in the absence of Spc72 protein. Phenotypes of various double mutants supported the observations of the time-lapse studies and revealed an astral microtubule motor essential for cell viability in the absence of the Spc72 protein. In addition, our data exclude involvement of Spc72 in nuclear spindle functions. Finally, our results are the first dynamic analyses of cells lacking the Spc72 protein and together with data of previous studies provide a new, more coherent image of Spc72 function.
MATERIALS AND METHODS
Strains, Media, and Yeast Transformation
Yeast strains used in this study are summarized in Table 1. Yeast media were prepared as described by Guthrie and Fink (1991). The yeast transformation procedure was based on the protocol by Schiestl and Gietz (1989). After the heat shock step, cells were pelleted and resuspended in 5 ml of YPD and incubated for 2 h at 30°C. Cells were again pelleted, resuspended in 1 ml of distilled H2O, and plated on selective YPD-G418 medium (200 mg/l geneticin). The Escherichia coli strain XL1-blue (Bullock et al., 1987) was used to propagate plasmids.
Table 1.
Yeast strains used in this study
| Name | Genotype | Source |
|---|---|---|
| FY 1679 | MATa/α ura3-52/ura3-52 trp1Δ63/TRP1 leu2Δ1/LEU2 his3Δ200/HIS3 | B. Dujon |
| CEN.PK2 | MATa/α ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ1 | K.D. Entian |
| DHY6 | MATa/α HHF2∷GFP-KanMX6/HHF2 ura3-52Δ1/ura3-52Δ1 trp1Δ63/TRP1 leu2Δ1/LEU2 his3Δ200/HIS3 | This study |
| DHY19 | MATa/α HHF2∷GFP-His3MX6/HHF2 spc72Δ1∷KanMX4/spc72∷KanMX4 ura3-52/ura3-52 trp1Δ63/TRP1 leu2Δ1/LEU2 his3Δ200/HIS3 | This study |
| DHY177 | MATa/α spc72Δ1∷KanMX4/SPC72 cnm67Δ1∷klTRP1/CNM67 ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ1 | This study |
| DHY195 | MATα ura3Δ1∷GFP-TUB1-URA3(pAFS125) trp1Δ63 LEU2 HIS3 | This study |
| DHY205 | MATα ura3Δ1∷GFP-TUB1-URA3(pAFS125) spc72Δ1∷klTRP1 trp1Δ63 LEU2 HIS3 | This study |
| DHY208 | MATa/α ura3Δ1∷GFP-TUB1-URA3(pAFS125)/ura3-52Δ1∷GFP-TUB1-URA3(pAFS125) spc72Δ1∷klTRP1/spc72Δ1∷KanMX4 trp1Δ63/trp1Δ63 leu2Δ1/LEU2 his3Δ200/his3Δ200 | This study |
| DHY209 | MATa/α ura3Δ1∷GFP-TUB1-URA3(pAFS125)/ura3-52Δ1∷GFP-TUB1-URA3(pAFS125) trp1Δ63/TRP1 LEU2/LEU2 his3Δ200/HIS3 | This study |
| DHY236 | MATa/α spc72Δ1∷klTRP1/SPC72 kip2Δ1∷KanMX4/KIP2 ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ1 | This study |
| DHY237 | MATa/α spc72Δ1∷klTRP1/SPC72 kip3Δ1∷KanMX4/KIP3 ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ | This study |
| DHY238 | MATa/α spc72Δ1∷klTRP1/SPC72 kar3Δ1∷KanMX4/KAR3 ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ | This study |
| DHY239 | MATa/α spc72Δ1∷klTRP1/SPC72 num1Δ1∷KanMX4/NUM1 ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ | This study |
| DHY242 | MATa/α spc72Δ1∷klTRP1/SPC72 Δhc1Δ1∷KanMX4/ΔHCl ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his3Δ1/his3Δ1 | This study |
| DHY259 | MATα kar3Δ1∷KanMX4 ura3Δ1∷GFP-TUB1-URA3(pAFS125) trp1-289 leu2-3,112 his3Δ1 | This study |
| DHY278 | MATα spc72Δ1∷klTRP1 ura3Δ1∷GFP-TUB1-URA3(pAFS125) trp1-289 leu2-3,112∷pspc72-7-LEU2 his3Δ1 | This study |
| DHY280 | MATa spc72Δ1∷klTRP1 kar3Δ1∷KanMX4 ura3Δ1∷GFP-TUB1-URA3(pAFS125) trp1-289 leu2-3,112∷pspc72-7-LEU2 his3Δ1 | This study |
| DHY282 | MATa/α spc72Δ1∷klTRP1/SPC72 kar9Δ1∷KanMX4/KAR9 ura3-52/ura3-52 trp1-289/trp1-289 leu2-3,112/leu2-3,112 his31/his3Δ | This study |
| DHY293 | MATα cnm67Δ1∷His3MX6 ura3-52 trp1-289 leu2-3,112 his3Δ1 | This study |
| DHY317 | MATα spc72Δ1∷klTRP1 cnm67Δ1∷His3MX6 ura3Δ1∷GFP-TUB1-URA3(pAFS125) trp1-289 leu2-3,112∷pspc72-7-LEU2 his3Δ1 | This study |
DNA Manipulations and Strain Constructions
Standard DNA manipulations were performed as described by Sambrook et al. (1989). We applied a polymerase chain reaction (PCR)-based method to construct gene deletion cassettes that were used in yeast transformations (Wach et al., 1994). DNA of E. coli plasmids pFA6-KanMX4 (Wach et al., 1994), pFA6-HIS3MX6, pFA6-GFP-KanMX6, pFA6-GFP-HIS3MX6 (Wach et al., 1997), and pYM3-klTRP1 (Knop et al., 1999) served as the template for preparative PCR reactions. Correct genomic integration of the corresponding construct was verified by analytical PCR (Huxley et al., 1990; Wach et al., 1994). Yeast strains were grown on YPD-G418 (200 mg/l geneticin) to select for transformants that had integrated KanMX4, or GFP-KanMX6 cassettes. SD plates lacking histidine or tryptophane were used to select for GFP-HIS3MX6, HIS3MX6, or klTRP1 integration.
For gene deletions we followed the EUROFAN guidelines (Wach, Brachat, and Philippsen [1996], Guidelines for EUROFAN B0 program ORF deletants, plasmid tools, basic functional analyses, available at www.mips.biochem.mpg.de/proj/eurofan/index.html) for gene replacement in S. cerevisiae. We used the following oligonucleotide pairs for generation of the KanMX4, His3MX6, klTRP1 deletion cassettes with flanking homologies to the target genes: spc72Δ1: 5′-A A C A C T A A T A T C A A A A A A C T A A G C A A A C A A C A T A A G G A A A G T T A T A G C C G C T T C G T A C G C T G C A G G T C G-3′ and 5′-A G A G T G A C T G A G T G T T A C A T T A A A T A T A T T T A T A T A T A A A C G T A T G A T A T A T C A T C G A T G A A T T C G A G C T C G T T-3′ The oligonucleotide pairs used for deletion of KIP2, KIP3, KAR3, DHC1, KAR9, NUM1, and CNM67 are described by Hoepfner et al. (2000). C-Terminal fusion of the S65T variant of GFP to Hhf2 (histone H4) was performed as described by Wach et al. (1997). This label was used in one copy in diploid strains. Growth rate and morphology of these strains were indistinguishable from those of wild type. To label microtubules we integrated plasmid pAFS125 into the ura3 locus (Straight et al., 1997). Spindle and astral microtubules were clearly observable under the fluorescence microscope upon successful transformation. The suitability of this label for in vivo studies has already been demonstrated (Straight et al., 1997). SPC72 temperature-sensitive (ts) mutants were generated by integrating the linearized pspc72-7 plasmid (Knop and Schiebel, 1998) into the leu2 locus of cells deleted for SPC72. Generation of double mutants was achieved either by crossing of the single mutants followed by sporulation and dissection of the four-spored asci or by serial gene deletion with the kanMX4/His3MX6 and the klTRP1 cassette.
In Vivo Microscopy Procedures and Techniques
The video microscopy setup and in vivo time-lapse procedures with Hhf2-GFP– or GFP-Tub1–labeled strains were described by Hoepfner et al. (2000). We preferentially used diploid cells because spreading of the cells during the time-lapse experiment was better due to the bipolar budding pattern. Haploid cells overgrew each other rapidly impairing long observation of individual cells. General Hhf2-GFP acquisition settings were as follows: 1-min interval time, 0.1-s exposure time, 3% illumination transmission, one z-axis plane, and no binning. General GFP-Tub1 acquisition settings were as follows: 2-min interval time, 0.4-s exposure time, 50% illumination transmission, three z-axis planes spaced by 0.8 μm, and 2×2 binning. By using these conditions cells showed steady growth for up to 72 h. Nuclear and microtubule dynamics of individual cells could be tracked for more than eight divisions. Acquisition settings for the high-resolution GFP-tub1 studies were as follows: 15-s interval time, 1-s exposure time, 100% illumination transmission, three z-axis planes spaced by 0.8 μm, and no binning. The temperature of immersion oil on the microscope slide near the sample was ∼24°C. The z-axis stacks were merged into one plane by using the “stack arithmetic:maximum” command of MetaMorph. The stored images were then scaled and converted to 8-bit files. A red look-up table was assigned to the phase-contrast image, and a green look-up table was assigned for the fluorescence image. The phase-contrast and fluorescence 8-bit planes were then overlaid using the built-in “overlay” command with the default balance. For time-lapse analysis we then assembled the picture files to a movie in QuickTime format (Apple Computer, Cupertino, CA) with frame rate of 10 frames/s by using the Premiere 4.2 program (Adobe Systems Europe, Edinburgh, Scotland).
Acquisition of Still Images
GFP-Tub1–engineered wild-type and spc72Δ strains were grown in YPD medium to early log phase at 30°C. Three microliters of the culture was spread on a poly-l-lysine–treated slide overlaid with a coverslip, sealed with nail polish, and immediately used for microscopy. No prepared slide older than 5 min was analyzed. GFP-Tub1–engineered spc72-7 stains were grown at 23°C to early log phase and analyzed as described above. To analyze the phenotype at the nonpermissive temperature, early log phase cultures were shifted from 23 to 37°C for 3 h then 3 μl of culture was spread on a 37°C prewarmed, poly-l-lysine–treated slide and immediately used for microscopy. The microscope stage was temperature adjustable and set to 37°C. No prepared slide older than 5 min was analyzed. Acquisition settings were as follows: 1-s exposure time, 100% illumination transmission, five z-axis planes spaced by 0.8 μm, and no binning. Images were processed as described by Hoepfner et al. (2000).
RESULTS
Impaired Nuclear Positioning and Spindle Orientation in spc72Δ Cells
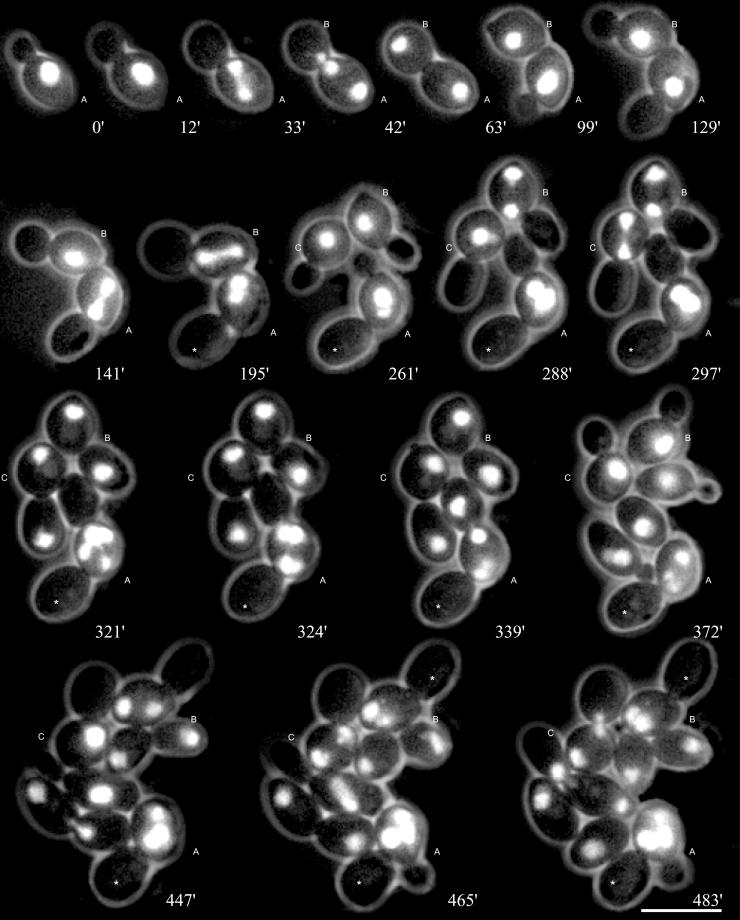
We analyzed nuclear dynamics in diploid wild-type (Movie 1A) and spc72Δ (Movie 1B) cells by in vivo time-lapse fluorescence microscopy. These cells expressed green fluorescent protein-tagged histone H4 (Hhf2-GFP), which was shown in previous studies to mark nuclei in S. cerevisiae without interfering with the nuclear cycle (Wach et al., 1997; Hoepfner et al., 2000). In contrast to wild type we observed an accumulation of bi- and multinucleate as well as anucleate cells in microcolonies of the spc72Δ mutant (Movie 1B; representative frames shown in Figure 1). To investigate the basis for this frequent failure in segregation of daughter nuclei we at first analyzed the behavior of nuclei in early steps of 220 cell cycles. In particular, we monitored nuclear positioning (movement of the nucleus to the bud neck before anaphase), nuclear orientation (alignment of the elongating nucleus along the mother-bud axis, mirroring spindle orientation), and insertion of the anaphase nucleus into the bud neck (mirroring spindle insertion) in spc72Δ cells carrying a single nucleus. Mutant cells with more nuclei will be discussed in a later section.
Figure 1.
Nuclear migration dynamics of diploid spc72Δ cells. Mitotic cell cycles were observed by in vivo time-lapse fluorescence microscopy for several generations. Representative frames of Movie 1B are presented. Compared with wild-type cells, shown in Movie 1A, the following differences can be seen. Nuclear positioning of the preanaphase nucleus to the bud-neck is absent (e.g., 0′, cell A), spindle elongation during anaphase is restricted to the mother cell and elongation of the anaphase spindle does frequently not occur along the mother-bud axis (33′, cell A). Nevertheless, delayed but successful segregation of one of the daughter nuclei into the bud is often observed (42′–63′, cell A). On the other hand, even correct spindle alignment (141′, cell A) is sometimes followed by a failure in nuclear segregation, leading to a binucleate mother cell and an anucleate bud. (195′–297′, cell A). Nuclear divisions in binucleate cells occur simultaneously (321′, cell A). After successful transit of a nucleus into the bud, cell separation can be observed by a change of the position of the bud relative to the mother cell after cytokinesis (63′–99′, cell A). Anucleate buds (marked with asterisks) are not separated from the mother cell during subsequent cell cycles. Bar, 10 μm. Movie 1A and 1B: Nuclear dynamics was observed in diploid wild-type (1A, strain DHY6) and spc72Δ cells (1B, strain DHY19). One of the four genes coding for histone H4 was engineered to express a carboxy-terminal GFP-fusion (Wach et al., 1997). Nuclear dynamics was followed for more than 9 h. Acquisition interval, 3 min; movie speed, 10 frames/s = 30 min/s. One z-axis plane fluorescence image was acquired.
Nuclear oscillations and movements typical for the G1 phase of wild-type cells (Movie 1A) were completely absent in spc72Δ mutants (Movie 1B). Occasional movements of the nucleus were only observed when the vacuole (sometimes visible in the red phase-contrast image of Movie 1B) displaced the nucleus. The nucleus normally did not move from the position at which it had been placed at the end of the previous cell cycle. A quantitative evaluation revealed that only 24% of spc72Δ cells had preanaphase nuclei positioned close to the bud neck (Table 2), not due to active movement of the nucleus toward the nascent bud, but rather due to the fact that these nuclei where positioned by chance at this site during the previous mitosis. 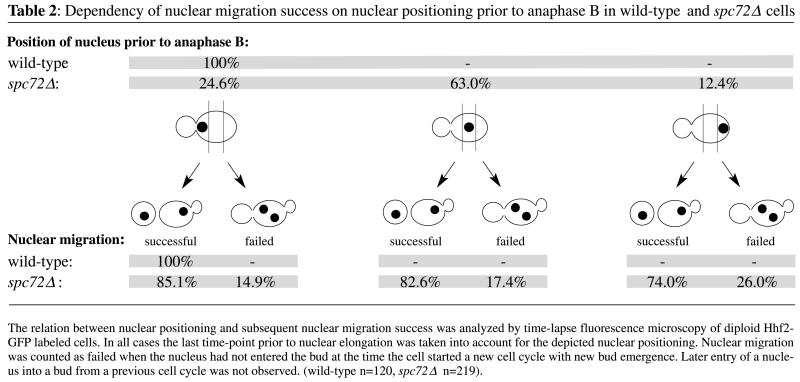
Orientation of the elongating nucleus and presumably the spindle along the mother-bud axis, which is typical for wild-type cells, frequently did not occur in the spc72Δ mutant. In 53% of all mutant cells nuclear elongation was not oriented along the mother-daughter axis (Table 3). Early insertion of the anaphase nucleus into the bud neck was never observed in spc72Δ cells, whereas this was always the case for wild-type cells (Movie 1A). Hence, elongation of the anaphase nucleus occurred in mother cells of the spc72Δ mutant. Also, rapid oscillations and occasional bending of the elongated nucleus commonly observed for anaphase in wild-type cells were completely absent in the mutant. 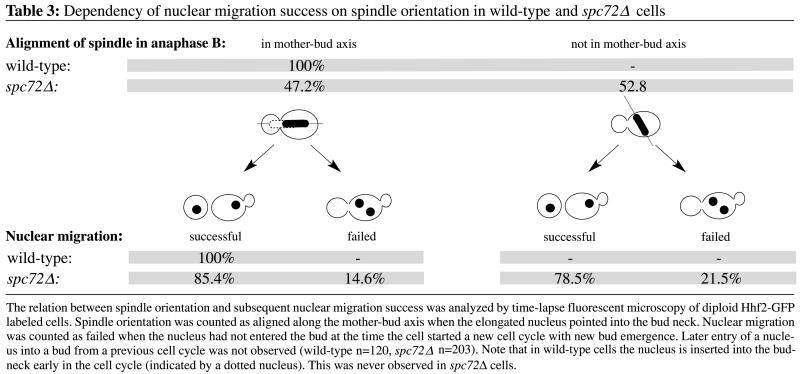
Spindle Positioning and Orientation Defects Can Be Rescued in spc72Δ Cells
In S. cerevisiae proper nuclear segregation depends on correct nuclear positioning and on the preanaphase orientation of the mitotic spindle along the mother-daughter polarity axis. Cells displaying wrongly positioned and misoriented spindles due to mutations often fail to correctly segregate daughter nuclei (Hoepfner et al., 2000; Segal et al., 2000). The observed nuclear mispositioning or spindle misorientation in spc72Δ cells did not severely impair the distribution of nuclei between mother and daughter cells in later cell cycle phases. As evident from Figure 1 and Movie 1B separation of both nuclear masses occurred entirely in the mother cells, frequently followed by migration of one daughter nucleus into the bud. Table 2 summarizes the consequences of correct or impaired nuclear positioning on nuclear segregation in >200 cells. Remarkably, 74% of cells with completely mispositioned nuclei still displayed successful nuclear segregation compared with 85% of cells with nuclei positioned at the bud neck. Table 3 summarizes a similar analysis of the consequences of misaligned spindles on nuclear segregation. Many cells (78%) with misaligned preanaphase spindle were able to segregate one nucleus into the bud compared with 85% of cells with a spindle oriented along the mother-bud axis.
This behavior of spc72Δ cells strikingly contrasts observations of cnm67Δ cells analyzed by the same method in the same strain background (Hoepfner et al., 2000). In the SPB mutant cnm67Δ impaired astral microtubule organization led to the formation of bi- and multinucleate cells within a few cell cycles. In this mutant nuclear positioning and spindle orientation defects occurred significantly less frequently than in spc72Δ cells (27% wrongly positioned nuclei and 25% misoriented spindles in cnm67Δ cells) but nuclear segregation more often failed when the preanaphase nucleus was not positioned at the neck. Remarkably, the overall ratio of single to multinucleate cells was the same in both mutants when still images with large numbers of fixed log phase cells were analyzed (spc72Δ 34%; cnm67Δ 33% bi- and multinucleate cells; n = 800). Only by using time-lapse microscopy was it possible to identify characteristic differences between the viable SPB mutants spc72Δ and cnm67Δ.
Our analyses of Hhf2-GFP–expressing cells revealed that regulated nuclear positioning was basically absent and spindle orientation was drastically impaired in spc72Δ mutants. Nevertheless, unlike in the cnm67Δ mutant, many spc72Δ cells were able to compensate for failures in early nuclear migration steps in later cell cycle stages, the mechanism of which will be investigated below.
Long Astral Microtubules Are Absent throughout Cell Cycle in spc72Δ Cells
Nuclear migration is achieved by forces acting on the nucleus via astral microtubules (Huffaker et al., 1988; Sullivan and Huffaker, 1992; Carminati and Stearns, 1997; Shaw et al., 1997). Because spc72Δ cells frequently showed successful, although delayed, nuclear migrations it was important to investigate astral microtubule organization and dynamics in spc72Δ cells. Using a GFP-Tub1 fusion (Straight et al., 1997), we constructed diploid wild-type and spc72Δ cells with fluorescently labeled microtubules. We investigated microtubule morphology over several cell cycles by time-lapse studies in a total of 76 wild-type cells (Movie 2A) and 84 spc72Δ cells (Movie 2B) and by acquisition of still images (Figure 2, A and B). Analysis of these data revealed a complete lack of long astral microtubules in the deletion mutant. This explains the observed impairment of early nuclear migration steps described above (Figure 1 and Table 2). Reduced astral microtubule arrays had already been described in previous studies by using spc72-ts and deletion mutants (Chen et al., 1998; Knop and Schiebel, 1998; Souès and Adams, 1998). Our in vivo observations supported this reported lack of long astral microtubules throughout the cell cycle. We were not able to detect residual long astral microtubules early in the cell cycle mentioned in one study (Souès and Adams, 1998).
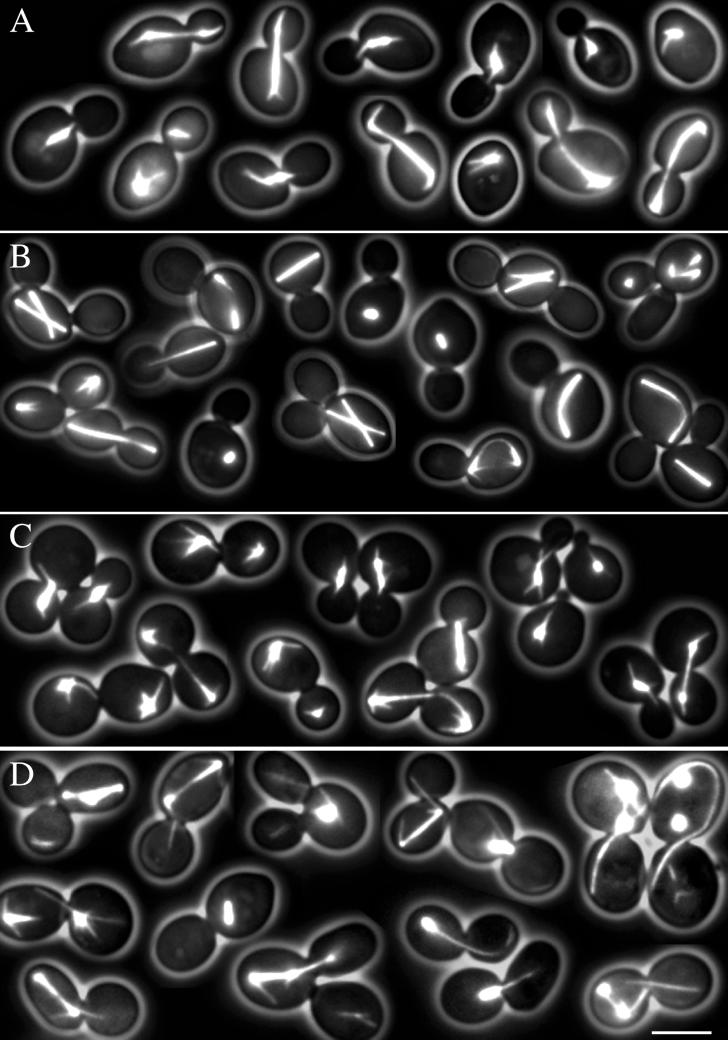
Figure 2.
Microtubule morphologies at different cell cycle stages in S. cerevisiae wild-type cells and in two spc72 mutants. Representative cells with GFP-labeled microtubules were selected from still pictures. (A) Wild type. In unbudded cells an array of astral microtubules is visible, eventually interacting with the cortex. In small budded cells the spindle poles separate and a short thick spindle is formed. Astral microtubules interact with the bud, position the nucleus, and orient the spindle. In large budded cells astral microtubules interacting with the cortex are prominent. (B) Spc72Δ mutant. Long astral microtubules are completely absent throughout the cell cycle. As in wild-type cells, spindles are clearly visible as short or long bars, but are frequently misoriented. Misoriented long spindles appear bent. Multibudded cells contain more than one spindle. Short microtubules similarly oriented as the spindle are most likely nuclear microtubules. (C) Spc72Δ spc72-7 cells grown at 23°C. Microtubule numbers, morphologies, and orientations are indistinguishable from wild type. (D) Spc72Δ spc72-7 cells grown at 37°C. Astral microtubules are clearly visible and appear significantly longer than in wild-type cells. Only very few cells show reduced or no astral microtubules. In many cells astral microtubules are detached from the SPB region. Some of the detached microtubules are extremely long. Cells with multiple and misaligned spindles are present. Cells with more than one spindle possess more than one bud. All cell images of this figure represent merged still images of five z-axis planes. Bar, 5 μm. Movie 2A. Microtubule dynamics in wild-type cells (strain DHY209) expressing GFP-Tub1 followed for 2.5 h. Acquisition interval, 2 min; movie speed, 10 frames/s = 20 min/s. Three z-axis plane fluorescence images were acquired and merged. Movie 2B. Microtubule dynamics in spc72Δ cells (strain DHY208) expressing GFP-Tub1 followed for 8 h. Acquisition interval, 2 min; movie speed, 10 frames/s = 20 min/s. Three z-axis plane fluorescence images were acquired and merged.
Active Spindle Reorientation Occurs via Very Short Astral Microtubules in spc72Δ Cells
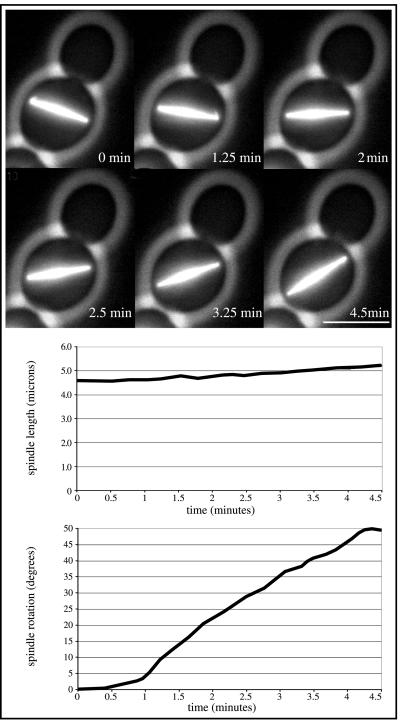
Investigations of time-lapse sequences of cells with randomly oriented spindles revealed frequent reorientation of spindles despite the absence of detectable long astral microtubules (Movie 2B). This reorientation started as soon as both spindle pole bodies of the elongating spindle were close to the cell cortex. Two models could explain such a behavior: first, passive alignment of the elongating spindle along the longest cell axis of an ellipsoid-shaped cell, or second, active reorientation mediated by forces acting on the spindle. The first possibility is very unlikely, because we observed spindle reorientation events followed by successful nuclear migration independently of the ellipsoid shape of the mother cell and of the position of the bud (Figure 1, cell B, 261–321 min). An active spindle reorientation capacity of spc72Δ cells was apparent in time-lapse studies of haploid, more spherical cells. These studies revealed directed reorientation events of >50° within 4.5 min (Figure 3 and Movie 3). During this time the spindle elongated <0.6 μm, finally ruling out a passive random spindle movement or alignment along the longest cell axis.
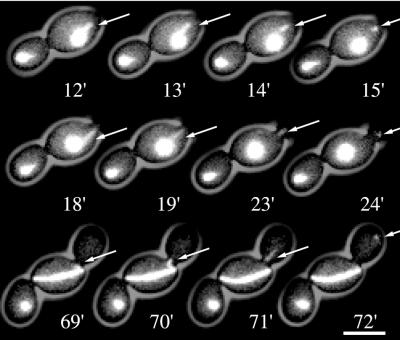
Figure 3.
Kinetics of spindle reorientation process in a spc72Δ cell (strain DHY205). Top, representative frames of Movie 3. Within 4.5 min the misoriented spindle rotates >50°C. Short astral microtubules, possibly mediating this rotation, are visible at 0 min at the left SPB and at 1.25 min at the right SPB. Bar, 5 μm. Middle, length of the rotating spindle plotted against the rotation time. Bottom, rotation angle of the spindle plotted against the rotation time. Movie 3. Time-lapse sequence of the spindle reorientation process analyzed in Figure 3. Acquisition interval, 15 s; movie speed, 10 frames/s = 3 min/s. Three z-axis plane fluorescence images were acquired and merged.
To investigate the factors involved in active spindle reorientation, we performed additional time-lapse studies with higher time resolution of 15 s and higher magnification, concentrating on the phase where directed spindle reorientation was observed (Movie 4, A and B, and representative frames shown in Figure 4). Visual inspection of these movie sequences revealed very short and unstable astral microtubules emanating from the mutant SPBs that had neither been detectable in previous time-lapse studies nor still images. These observed astral microtubules never reached lengths >1 μm (n = 28) and could often be identified in a single movie frame only. In four similar cases we could follow minor growth steps of individual astral microtubules for up to four movie frames, representing 1-min real time. In the fifth frame, the microtubules were no longer detectable, suggesting that disassembly was a very rapid process. Estimations based on the measured length and observed dynamics of these microtubules suggest slower growth, faster shrinkage, and higher catastrophy frequency in spc72Δ cells with respect to published astral microtubules dynamics found in wild-type cells (Carminati and Stearns, 1997). In addition, in 30% of the cells analyzed by high-resolution time-lapse imaging (n = 18) it was not possible to detect short astral microtubules. In these cells, the spindle did not reorient along the mother-bud axis and nuclear segregation failed. This strongly supported the view that the observed reorientation of anaphase spindles depended on the presence of residual short astral microtubules.
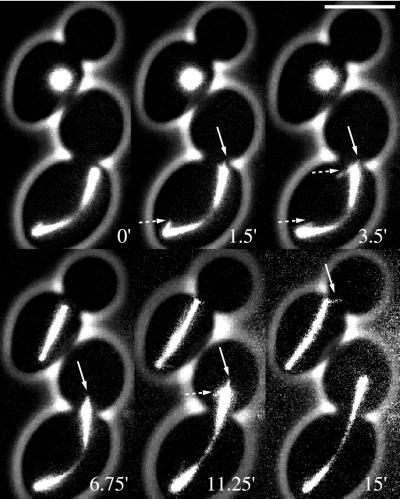
Figure 4.
Residual short and unstable astral microtubules in spc72Δ cells. Six images of the high-resolution time-lapse sequence of Movie 4A are shown. In the lower cell the already elongated spindle is misoriented and mislocalized (0′). Short astral microtubules are visible during the reorientation of the spindle. Further spindle elongation leads to penetration of the spindle through the bud neck (3.5′–15′). The upper cell enters anaphase without properly localizing the spindle to the bud neck. Although the orientation of the spindle almost points into the bud, penetration is not observed before the spindle spans the whole length of the cell. At 15′ one short astral microtubule is seen at the upper SPB pointing into the bud. Microtubules indicated with solid arrows are astral microtubules evidenced by the orientation angle relative to the spindle. Microtubules indicated with dotted arrows are either nuclear microtubules or astral microtubules with orientation angles relative to the spindle not observed in wild-type cells. Bar, 5 μm. Movie 4, A and B. Short astral microtubules and spindle reorientation in GFP-Tub1 expressing spc72Δ (strain DHY208) cells followed for 15 and 20 min, respectively. Acquisition interval, 15 s; movie speed, 10 frames/s = 2.5 min/s. Three z-axis plane fluorescence images were acquired and merged.
A very rare event that was only observed in one of 84 cells confirmed the proposed capability of spc72Δ cells to form residual microtubules in the absence of the Spc72 protein. In this cell (Movie 5, and representative frames in Figure 5) astral microtubules generated at one SPB grew much longer than observed in all other spc72Δ cells. These long astral microtubules detached after having contacted the cell cortex and were pulled into the bud. Unlike in cnm67Δ cells where detached astral microtubules (carrying Spc72p at one end) were stable for >30 min (Hoepfner et al., 2000), detached astral microtubules in the spc72Δ cell were rapidly degraded within a few minutes.
Figure 5.
Astral microtubule formation and detachment observed in one spc72Δ cell. Representative frames of Movie 5 are shown. At bud emergence one astral microtubule longer than 1 μm, interacting with the site of the nascent bud, can be seen (0′). Unlike in wild-type cells this does not result in movement of the nucleus toward the bud neck; instead, the astral microtubule detaches from the SPB region and is pulled into the small bud (13′–15′). A new astral microtubule is formed that also detaches after interaction with the bud cortex and moves into the bud (18′–24′). After spindle elongation new astral microtubules are formed that presumably try to reorient the misaligned spindle. These astral microtubules also detach (69′–72′). As apparent in the corresponding time-lapse movie no more long astral microtubule formation can be observed at later time points. The cell shows failed nuclear migration and follows all mutant characteristics observed in other spc72Δ cells. Bar, 5 μm. Movie 5. Time-lapse study of growth and detachment of astral microtubules in a spc72Δ cell (strain DHY208) followed for 3 h. Cells express an amino-terminal GFP-Tub1 fusion (Straight et al., 1997). Acquisition interval, 1min; movie speed, 10 frames/s = 10 min/s. Three z-axis plane fluorescence images were acquired and merged.
Our observations suggest that spindle poles lacking the Spc72 protein are able to nucleate only unstable astral microtubules which, with rare exception, are very short. These residual short astral microtubules are essential to reorient misoriented spindles upon cortex contact; however, they are not capable of positioning G1-phase nuclei in spc72Δ cells because longer astral microtubules that span the SPB-cortex distance would be required to perform this task.
spc72Δ Is Synthetically Lethal with Loss of Kar3 or Cnm67
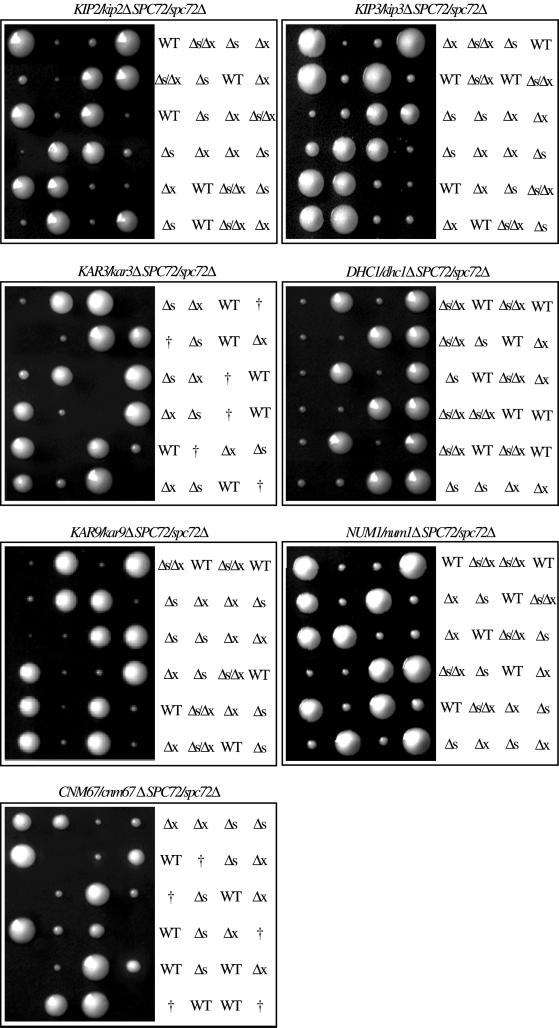
It is conceivable that directed force production via residual astral microtubules is responsible for the observed spindle reorientation in spc72Δ cells. Such mechanism would require the action of motor proteins (reviewed by Hildebrandt and Hoyt, 2000) and possibly cortical determinants (Farkasovsky and Küntzel, 1995; Miller and Rose, 1998; Yeh et al., 2000). To genetically test this hypothesis, we constructed a set of spc72Δ double mutants carrying deletions for known astral microtubule motor genes and genes of cortical determinants such as KIP2, KIP3, KAR3, DHC1, KAR9, and NUM1 (Meluh and Rose, 1990; Eshel et al., 1993; Li et al., 1993; Farkasovsky and Küntzel, 1995; Carminati and Stearns, 1997; Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Saunders et al., 1997; Miller and Rose, 1998; Yeh et al., 2000). Six different heterozygous diploids each carrying two deletions were constructed by mating of the respective haploids. Afterward, they were subjected to tetrad analysis, including verification of the deletion alleles in the growing colonies by PCR and by identification of the deletion-associated markers (Figure 6). We obtained four viable colonies from all tested heterozygous diploids except for spc72Δ/SPC72 kar3Δ/KAR3 (Figure 6). Microscopy of the nonviable spc72Δ/kar3Δ spores revealed that they germinated but then arrested as large, multibudded cells (our unpublished data). The other double mutants grew as slowly as the spc72Δ single mutants.
Figure 6.
Tetrad analysis of spc72Δ double mutants. Four astral microtubule motors (KIP2, KIP3, KAR3, and DHC1), two astral microtubule–cortex interaction mediators (KAR9 and NUM1), and one SPB component encoding genes have been deleted in a heterozygous diploid spc72Δ/SPC72 background. The cells were sporulated and the asci subjected to tetrad analysis. The genotypes of the individual spores were determined by the segregation of genetic markers and by analytical PCR. Genotypes are indicated as follows: WT, wild type; Δs, spc72Δ; Δx, other deletion mentioned in the title; †, lethal. The figure shows six representative tetrads of each strain after growth on solid medium for 4 d.
We also genetically tested the hypothesis that active spindle reorientation and thus viability depended on residual astral microtubules in spc72Δ cells. If so, further impairing the remaining astral microtubule formation at the SPB outer plaque in spc72Δ cells by deletion of CNM67 should be lethal. Loss of Cnm67 has been shown to result in the loss of the SPB outer plaque, which exclusively impaired the formation of astral microtubules at this substructure, without altering the dynamics of spindle microtubules nor the formation of astral microtubules at the half-bridge (Brachat et al., 1998; Hoepfner et al., 2000). Tetrad analysis of the heterozygous diploid spc72Δ/SPC72 cnm67Δ/CNM67 revealed that the spc72Δ/cnm67Δ double mutant was nonviable (Figure 6, bottom). Microscopy of the spc72Δ/cnm67Δ spores showed that they germinated and then arrested as multibudded cells. In summary, in a spc72Δ background, single deletion of four astral microtubule motors, two astral microtubule–cortex interaction mediators, and one other SPB outer plaque component revealed synthetic lethality of spc72Δ only in the absence of the microtubule motor Kar3 and the SPB component Cnm67, respectively. This allows the conclusion that short astral microtubules organized by the SPB outer plaque in the spc72Δ mutant are essential for reorientation of misaligned spindles.
To investigate in more detail the terminal phenotype of kar3Δ spc72Δ and cnm67Δ spc72Δ cells, we used the ts allele spc72-7 (Knop and Schiebel, 1998) to construct conditional cnm67Δ spc72Δ spc72-7 and kar3Δ spc72Δ spc72-7 mutants. Unfortunately, when shifted 37°C, we were not able to induce synthetic lethality. However, we found an interesting phenotype and astral microtubule morphology in control spc72Δ cells carrying the spc72-7 allele and the GFP-TUB1 fusion gene. At 23°C the spc72-7 allele complemented the spc72Δ phenotype; long astral microtubules emanated from the spindle poles and elongating spindles were correctly oriented (Figure 2C). After 5-h incubation at 37°C cells did not stop dividing. Long astral microtubules still emanated from the spindle poles, however, many cells contained detached long and apparently stable microtubules (Figure 2D). In addition, a substantial fraction of the spindles were misoriented, probably due to an unstable anchoring of astral microtubules at the half-bridge or outer spindle pole plaque. The detached microtubules in these cells may still be associated with the Spc72 mutant protein because microtubules detaching from the spindle pole together with the Spc72 anchor are very stable (Hoepfner et al., 2000)
Spindle Elongation Dynamics Is Unaffected by Loss of Spc72 or Kar3
As evidenced in our time-lapse sequences, final transit of a nucleus through the bud neck in spc72Δ cells, after successful reorientation via short astral microtubules, was dependent on the final elongation phase of the spindle. The first phase of spindle elongation, including separation of the nuclear masses was always restricted to the mother cell. When the spindle was as long as the diameter of the mother cell and when it continued to elongate we did observe transit of one SPB through the bud neck (n = 203). Transit of a nucleus through the bud neck after spindle breakdown in the mother cell was never observed.
Because spindle dynamics significantly contributed to nuclear migration success, it was important to verify that the observed synthetic lethality of spc72Δ cnm67Δ and spc72Δ kar3Δ double mutants was caused by astral microtubule-related functions and not by altered spindle elongation kinetics. In previous time-lapse experiments it was already shown that loss of the cytoplasmic SPB component Cnm67 did not alter spindle kinetics compared with wild-type cells (Hoepfner et al., 2000). However, genetic screens and functional analyses implicated Kar3 in spindle functions (Meluh and Rose, 1990; Page and Snyder, 1992; Cottingham et al., 1999; Manning et al., 1999) and loss of Spc72 was also suggested to interfere with spindle dynamics (Chen et al., 1998; Knop and Schiebel, 1998). Using a GFP-Tub1 label (Straight et al., 1997) and time-lapse microscopy at 1-min intervals, we measured parameters of spindle kinetics in wild-type, spc72Δ, and kar3Δ cells (Table 4). Our results of wild-type cells were very similar to published data obtained using the same GFP label (Straight et al., 1998). All five parameters measured in spc72Δ cells were similar to wild type, strongly suggesting no involvement of the Spc72 protein in nuclear microtubule functions.
Table 4.
Spindle dynamics in wild-type and mutant cells
| Strain | Unelongated spindle length | Breakdown length | Anaphase duration | Fast elongation | Slow elongation | n |
|---|---|---|---|---|---|---|
| μm | μm | min | μm/min | μm/min | ||
| Wild-type 2n | 1.98 | 13.25 | 28.2 | 0.78 | 0.23 | 30 |
| ς = 0.08 | ς = 0.42 | ς = 3.1 | ς = 0.06 | ς = 0.03 | ||
| Δspc72/Δspc72 2n | 2.01 | 12.81 | 32.1 | 0.78 | 0.24 | 25 |
| ς = 0.12 | ς = 0.51 | ς = 4.2 | ς = 0.06 | ς = 0.03 | ||
| Wild-type 1na | 2.02 | 10.50 | 21.8 | 0.73 | 0.26 | 20 |
| ς = 0.11 | ς = 0.49 | ς = 2.8 | ς = 0.07 | ς = 0.02 | ||
| Δkar3 1nb | 1.98 | 10.90 | 22.6 | 0.81 | 0.24 | 15 |
| ς = 0.14 | ς = 0.72 | ς = 2.4 | ς = 0.08 | ς = 0.09 |
In haploid wild-type cells anaphase B was observed 62 min (ς = 12) after bud emergence.
In Δkar3 cells anaphase B was observed 98 min (ς = 22 min) after bud emergence.
Similarly, kar3Δ cells showed spindle elongation and spindle breakdown kinetics comparable to wild type. However, onset of anaphase B was significantly delayed (Table 4). Time-lapse movies (our unpublished data) revealed impaired formation of short, bar-shaped spindles and many cells formed a diffuse array of nuclear microtubules as already observed in previous studies on Kar3 function (Meluh and Rose, 1990; Saunders et al., 1997; Cottingham et al., 1999). Nevertheless, all cells finally assembled a bipolar spindle and successfully executed anaphase. Thus, loss of Kar3 impairs spindle assembly, which delays onset of anaphase but does not affect the elongation kinetics per se. Because it is the spindle elongation that contributes to the viability of spc72Δ cells and because this process is not impaired by loss of the Kar3 motor it seems very likely that the observed synthetic lethality of spc72Δ kar3Δ cells is caused by loss of the astral microtubule-associated function of Kar3 and not by loss of its role in spindle assembly.
Consequences of Failed Nuclear Segregations
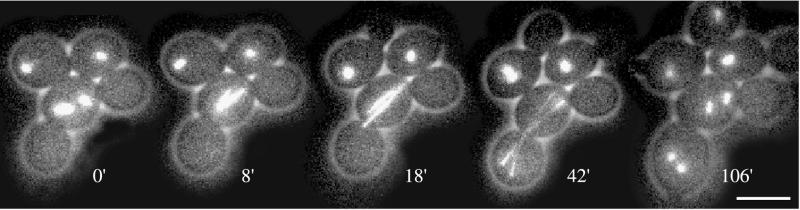
Spc72Δ cells that failed to direct one nucleus into the bud finished the cell cycle with a binucleate mother cell and an anucleate bud. These aberrant segregation events, however, did not trigger a permanent growth arrest, and the binucleate cells initiated without delay a new cell cycle as seen by new bud emergence, SPB duplication, spindle assembly, and simultaneous mitotic divisions of the two nuclei (Figure 1, cell A, 141–324 min, and Movie 2B). Further nuclear segregation failures in the pedigrees of binucleate cells led to multinucleate cells. A frequent generation of binucleate mother cells with attached anucleate bud, followed by nuclear segregation failures during new cell cycles, was already observed for cnm67Δ the outer plaque deletion mutant (Hoepfner et al., 2000). A log phase culture of spc72Δ cells showed the following distribution: 69.5% with one nucleus, 18.5% with two nuclei, 3.2% with three nuclei, 7% with four nuclei, and 8% with five or more nuclei (n = 800). The frequency of cells with even numbers of nuclei was slightly higher compared with cnm67Δ cells, although cnm67Δ cells showed the same ratio of single-to-multinucleate cells. This could be explained by the observation that multinucleate spc72Δ cells sometimes transferred more than one nucleus into a nascent bud (Figure 7 and Movie 7, A and B), leading to an initially binucleate daughter cell. Such an event was never observed in cnm67Δ cells (n = 325; Hoepfner et al., 2000) but occurred in 12% of bi- or multinucleate spc72Δ cells (n = 220). As in cnm67Δ cells, deposition of a nucleus into a still attached anucleate bud of a previous cell cycle was never detected.
Figure 7.
Simultaneous segregation of two daughter nuclei after mitosis in a binucleate spc72Δ cell. Representative frames of Movie 7A are shown. Two mispositioned and misoriented short spindles are visible in a mother cell with two buds; the emerging bud of the current cell cycle is the lower one (0′). Both spindles enter anaphase B simultaneously (8′). Reorientation of both spindles results in subsequent insertion of both spindles into the bud (18′–42′). After spindle breakdown and cytokinesis bud emergence is visible in the newborn binucleate daughter cell (106′). Bar, 5 μm. Similar translocation of two nuclei into one bud was also observed in spc720 cells with histone-GFP labeled nuclei, in Movie 7B. Movie 7A. Translocation of two daughter nuclei during one cell cycle of a binucleate spc72Δ cell. Proliferation of GFP-Tub1 expressing spc72Δ cells (strain DHY209) was followed for 5 h. Acquisition interval, 2 min; movie speed, 10 frames/s = 20 min/s. Three z-axis plane fluorescence images were acquired and merged. Movie 7B. Translocation of two daughter nuclei during one cell cycle of a binucleate spc72Δ cell. Proliferation of Hhf2-GFP expressing spc72Δ cells (strain DHY19) was followed for 6.5 h. Acquisition interval, 1 min; movie speed, 10 frames/s = 10 min/s. One z-axis plane fluorescence image was acquired.
We were also able to monitor cell separation in wild-type and spc72Δ cells, which was evidenced by a sudden change in the position of the bud relative to that of the mother cell (Figure 1, cells A and B, 63–99 min, and Movies 1 and 2, A and B). In wild-type and spc72Δ cells, separation of nucleate buds from the mother cell occurred ∼30 min after entry of the nucleus into the bud. However, anucleate buds in spc72Δ cells stayed permanently attached to their mother cells (Figure 1, cell A, 195–483 min; cell B, 372–483 min; and Movies 1B and 2B). These observations suggest a link between successful nuclear migration and cell separation in spc72Δ mutants as was already observed in cnm67Δ cells (Hoepfner et al., 2000). The formation of anucleate cells that were counted in cultures of fixed cells in other studies (Chen et al., 1998; Knop and Schiebel, 1998; Souès and Adams, 1998) could therefore not be recapitulated during in vivo time-lapse studies.
During our analyses it became apparent that the first nuclear migration event in newborn daughter cells was always successful despite the absence of long astral microtubules. Nuclear migrations in later cell cycles showed a constant failure ratio. Such an age-dependent characteristic has already been reported for cnm67Δ cells and was shown not to be related to cell size, budding pattern, or the presence of the She1 protein (Hoepfner et al., 2000). This points toward a general mother-daughter difference in nuclear migration rather than an spc72Δ- or cnm67Δ-specific characteristic.
DISCUSSION
spc72Δ Cells Organize Residual Short and Unstable Astral Microtubules
We used in vivo time-lapse analysis of cells with labeled nuclei and microtubules to compare nuclear migration and microtubule behavior of wild-type and spc72Δ cells. We examined why the absence of Spc72 is not lethal as would be predicted by its previously proposed function as anchor for the cytoplasmic γ-tubulin complex (Knop and Schiebel, 1998; Pereira et al., 1999). In this study, we could show that loss of Spc72 resulted in lack of long astral microtubules but very short and unstable astral microtubules persisted. These short astral microtubules were unable to cover long distances from the SPB to the cortex early in the cell cycle, resulting in a strong nuclear positioning and spindle orientation defect. However, upon spindle elongation and contact of the SPB with the cortex, these short astral microtubules were able to reorient the elongating spindle. As soon as the spindle had been successfully reoriented in spc72Δ cells, spindle elongation proceeded with wild-type kinetics, resulting in the penetration of one SPB through the bud neck. This frequently led to delayed but successful segregation of the chromosomal masses. Thus, our time-lapse sequences showed that successful nuclear migrations were not stochastic events as suggested in other studies but depended on forces acting via the residual astral microtubules.
Revised Model for Spc72 Function
Our findings allow new interpretations of previously published results. The residual astral microtubules in this study and the integrity of the SPB outer plaque in the absence of Spc72 suggest the presence of at least one other γ-tubulin complex binding protein at the SPB outer plaque. This hypothesis is in agreement with several other published experimental results. First, in vegetatively growing wild-type cells only a small percentage of the γ-tubulin complex directly interacts with Spc72 (Knop and Schiebel, 1998). Second, only a small percentage of Spc72 interacts with Nud1, which forms the proposed interaction site at the outer plaque (Gruneberg et al., 2000). Third, loss of Cnm67 and Nud1 but not Spc72 causes loss of the SPB outer plaque (Brachat et al., 1998; Chen et al., 1998; Souès and Adams, 1998; Adams and Kilmartin, 1999; Gruneberg et al., 2000). Finally, although loss of the SPB outer plaque is observed in mutants with N-terminal truncations of the Cnm67 protein, in 25% of the cases observed by electron microscopy, astral microtubules appeared to be still organized by the cytoplasmic face of the SPB (Schaerer et al., 2001). Together with our observation that deletion of the central region of Cnm67 is synthetic lethal in an spc72Δ background, this suggests the existence of a factor that interacts with the central region of Cnm67 and is capable of organizing astral microtubules.
Several findings support the notion that Spc72 is the only γ-tubulin complex anchor at the half-bridge. First, in the absence of Spc72 loss of the SPB outer plaque results in a lethal phenotype, indicating that the observed residual microtubules in spc72Δ cells are not organized by the half-bridge structure. Second, in the absence of the SPB outer plaque lethality is still observed in the presence of Spc72 if the first 15 amino acids of Kar1, which comprises the Spc72 interaction site at the half-bridge, are missing (Pereira et al., 1999). Thus, impairing the known Spc72 anchor site at the half-bridge apparently abolishes astral microtubule formation from this SPB substructure.
As shown recently, the formation of astral microtubules by two different SPB substructures seems to be controlled via phosphorylation of Spc72: Nud1 preferentially interacts with phosphorylated Spc72 that starts to appear at G2/S transition (Gruneberg et al., 2000). This is in agreement with our previous observation that astral microtubules and the Spc72 protein switch from the half-bridge to the outer plaque as visualized by time-lapse microscopy (Hoepfner et al., 2000). In the context of this study, the following model is in better agreement with the experimental data: in the early stages of the cell cycle Spc72 is the astral microtubule anchor at the half-bridge. On G2/S transition Spc72 is phosphorylated and astral microtubules are switched to the outer plaque where they are maintained by a second γ-tubulin anchor.
Finally, we suggest that the observed astral microtubule instability in spc72Δ cells is due to impaired organization of Stu2, an essential protein, that was shown to laterally bind microtubules and interact with Spc72 (Wang and Huffaker; 1997; Chen et al., 1998). It was speculated that the lateral binding capability of Stu2 could maintain the attachment of microtubules to the pole, even during subunit exchange at the ends (Wang and Huffaker, 1997). Involvement of Stu2 in microtubule anchoring is now supported by the observed microtubule detachment upon shift to the nonpermissive temperature in Spc72-ts cells. Astral microtubule detachment was also observed in the very rare cases where astral microtubules longer than 1 μm were formed in spc72Δ deletion mutants. These astral microtubules were very unstable and were rapidly degraded. In the previously analyzed cnm67Δ SPB mutant, detached astral microtubules appeared to be much more stable, most likely because these microtubules were still capped with the γ-tubulin complex (Hoepfner et al., 2000). Therefore, we conclude that in the absence of Spc72, Stu2-dependent microtubule anchoring to the γ-tubulin complex is impaired and microtubule stability reduced. Microtubules that are able to achieve detectable lengths detach from the γ-tubulin complex and thus the SPB and are rapidly degraded. During the reviewing process of this article, new evidence was published supporting our model. Detailed analyses of conditional Stu2 mutants revealed that Stu2 plays a prominent role in determining assembly properties of astral microtubules (Kosco et al., 2001).
Spindle Reorientation Is Dependent on Kar3 but Not Kar9/Myo2 Pathway
Our analysis of double mutants with spc72Δ revealed synthetic lethality with the microtubule motor Kar3. Based on our data, we believe that this is not due to the involvement of Kar3 in spindle assembly, but due to its function on astral microtubules. Loss of Kar3 was shown to increase the length of astral microtubules (Saunders et al., 1997). Longer astral microtubules would facilitate spindle orientation in spc72Δ cells. Because this was not the case, we believe that it is the actual motor function of Kar3 that is involved in the observed spindle reorientation in spc72Δ cells. To our surprise, no synthetic effect was observed in spc72Δ kip3Δ cells although several experiments suggest redundant functions of Kip3 and Kar3 in nuclear positioning and spindle orientation (Miller et al., 1998; Cottingham et al., 1999). Furthermore, loss of Kar9 did not additionally impair growth of spc72Δ cells despite recent findings that Kar9 is involved in a capturing process of Bim1-coated microtubules, which are then directed along actin cables into the bud by translocation of Myo2, a class V myosin (Beach et al., 2000; Miller et al., 2000; Yeh et al., 2000; Yin et al., 2000). Apparently, in the absence of Kar3, neither Kip3 nor Kar9, in conjunction with Myo2, is sufficient to perform the observed reorientation of spindles in spc72Δ cells observed in this study.
It therefore appears that Kar3 is the most efficient motor producing force on the nucleus during astral microtubule-mother cell cortex interactions. This is also reflected by the observation that cells expressing Kar3 as the only astral microtubule motor are viable (Cottingham et al., 1999). In addition, a previous study focusing on the spindle pole cnm67Δ mutant showed that deletion of Kar3 caused a much more severe nuclear migration impairment than deletion of any other astral microtubule motor or Kar9 (Hoepfner et al., 2000).
spc72Δ Cells Show Mother-Daughter Differences in Nuclear Segregation Fidelity Like cnm67Δ Cells
Long-term time-lapse investigations allowed pedigree analysis of spc72Δ cells. This revealed that the first mitosis of newborn, single-nucleate daughter cells was always successful despite absence of long astral microtubules. Later divisions of the same cells often failed. We observed the same phenomenon in cnm67Δ SPB mutants (Hoepfner et al., 2000). These observations suggest a general mother-daughter difference in nuclear migration rather than an spc72Δ-specific characteristic. Successful first division of daughter cells persisted independently of the cell size, budding pattern or the presence of the She1 protein (Hoepfner et al., 2000), suggesting a possible change in the spindle pole structure itself is affecting the mode of nuclear migration. Maturation of centrioles has been described in mammalian cells where newly assembled centrioles were shown to be unable to perform the functions of centrioles generated in the previous cell cycle (Piel et al., 2000). However, the complete lack of early spindle orientation control in spc72Δ cells as apparent in the time-lapse sequences rather suggests that spc72Δ cells have lost control over which SPB finally enters the bud. Therefore, although SPB maturation describes an exciting explanation for the mother-daughter specific differences in nuclear migration it cannot explain the behavior of the spc72Δ mutant because sometimes also the “old” SPB might be inserted into the bud.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Schiebel for the pspc72-7 plasmid and A.F. Straight for the pAFS125 plasmid. We are grateful to Robbie Loewith and Amy Gladfelter for careful reading of the manuscript. This work was supported by grants from the Swiss Federal Office for Education and Science (grant 95.0191-12) and the Swiss National Science Foundation (grant 31-55941.98).
Footnotes
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–07–0338. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–07–0338.
REFERENCES
- Adams IR, Kilmartin JV. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J Cell Biol. 1999;4:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach DL, Thibodeaux J, Maddox P, Yeh E, Bloom K. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr Biol. 2000;10:1497–1506. doi: 10.1016/s0960-9822(00)00837-x. [DOI] [PubMed] [Google Scholar]
- Bullock WO, Fernandez JM, Short JM. XLl-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques. 1987;5:376–378. [Google Scholar]
- Brachat A, Kilmartin JV, Wach A, Philippsen P. Saccharomyces cerevisiae cells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol Biol Cell. 1998;9:977–991. doi: 10.1091/mbc.9.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XP, Yin H, Huffaker TC. The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J Cell Biol. 1998;141:1169–1179. doi: 10.1083/jcb.141.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Gheber L, Miller DL, Hoyt MA. Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J Cell Biol. 1999;147:335–350. doi: 10.1083/jcb.147.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11186. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky M, Küntzel H. Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J Cell Biol. 1995;131:1003–1014. doi: 10.1083/jcb.131.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Souès S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the γ-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:14–15. [PubMed] [Google Scholar]
- Gruneberg U, Campell K, Simpson C, Grindlay J, Schiebel E. Nud1 links astral microtubule organization and the control of exit from mitosis. EMBO J. 2000;19:6475–6488. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt ER, Hoyt AM. Mitotic motors in Saccharomyces cerevisiae. Biochim Biophys Acta. 2000;1496:99–116. doi: 10.1016/s0167-4889(00)00012-4. [DOI] [PubMed] [Google Scholar]
- Hoepfner D, Brachat A, Philppsen P. Time-lapse video microscopy reveals astral microtubule detachment in the yeast spindle pole mutant. cnm67. Mol Biol Cell. 2000;11:1197–1211. doi: 10.1091/mbc.11.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker TC, Thomas JH, Botstein D. Diverse effects of beta-tubulin mutations on microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley C, Green ED, Dunham I. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 1990;6:236. doi: 10.1016/0168-9525(90)90190-h. [DOI] [PubMed] [Google Scholar]
- Hyman AA. Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J Cell Biol. 1989;109:1185–1193. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs CW, Adams AE, Szaniszlo PJ, Pringle JR. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Goh PY. Spc110p: assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO J. 1996;15:4592–4602. [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kosco KA, Pearson CG, Maddox PS, Wang PJ, Adams IR, Salmon ED, Bloom K, Huffaker TC. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol Biol Cell. 2001;12:2870–2880. doi: 10.1091/mbc.12.9.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Barrett JG, Wallace JA, Granok H, Snyder M. Differential regulation of the Kar3 kinesin regulated protein by two associated proteins, Cik1p and Vik1p. J Cell Biol. 1999;144:1219–1233. doi: 10.1083/jcb.144.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. Kar3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Miller RK, Cheng SC, Rose MD. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol Biol Cell. 2000;11:2949–2959. doi: 10.1091/mbc.11.9.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Rose MD. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Rapport E. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae. J Cell Biol. 1971;50:344–361. doi: 10.1083/jcb.50.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Urbani L, Stearns T. The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Vinh DBN, Crawford DK, Davis TN. A genetic analysis of interactions with Spc110p reveals distinct functions of Spc97p and Spc98p components of the yeast gamma-tubulin complex. Mol Biol Cell. 1998;9:2201–2216. doi: 10.1091/mbc.9.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Snyder M. CIK1: a developmentally regulated spindle pole body-associated protein important for micortubule functions in Saccharomyces cerevisiae. Genes Dev. 1992;6:1414–1429. doi: 10.1101/gad.6.8.1414. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast. Saccharomyces cerevisiae. J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Gruenenberg U, Knop M, Schiebel E. Interaction of the yeast gamma-tubulin complex-binding protein Spc72p with Kar1p is essential for microtubule function during karyogamy. EMBO J. 1999;18:4180–4195. doi: 10.1093/emboj/18.15.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Saunders W, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerer F, Morgan G, Giddings T, Winey M, Philippsen P. Cnm67p is a spacer of the Saccharomyces cerevisiae spindle pole body outer plaque. Mol Biol Cell. 2001;12:2519–2533. doi: 10.1091/mbc.12.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Segal M, Bloom K, Reed SI. Bud6 directs sequential microtubule interactions with the bud tip, and bud neck during spindle morphogenesis in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:3689–3702. doi: 10.1091/mbc.11.11.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souès S, Adams IR. SPC72: a spindle pole component required for spindle orientation in the yeast Saccharomyces cerevisiae. J Cell Sci. 1998;111:2809–2818. doi: 10.1242/jcs.111.18.2809. [DOI] [PubMed] [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. γ-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living yeast: anaphase A but not metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, O'Toole E, Berrueta L, Bierer BE, Pellman D. Yeast Bim1p promotes the G1-spcific dynamics of microtubules. J Cell Biol. 1999;145:993–1007. doi: 10.1083/jcb.145.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wang PJ, Huffaker TC. Stu2p: a microtubule-binding protein that is an essential component of the yeast spindle pole body. J Cell Biol. 1997;139:1271–1280. doi: 10.1083/jcb.139.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Pruyne D, Huffaker TC, Bretscher A. Myosin V orientates the mitotic spindle in yeast. Nature. 2000;406:1013–1015. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.