Abstract
Under conditions of sulfur deprivation, O-acetylserine (OAS) accumulates, which leads to the induction of a common set of six genes, called OAS cluster genes. These genes are induced not only under sulfur deprivation, but also under other conditions where OAS accumulates, such as shift to darkness and stress conditions leading to reactive oxygen species (ROS) or methyl-jasmonate accumulation. Using the OAS cluster genes as a query in ATTED-II, a co-expression network is derived stably spanning several hundred conditions. This allowed us not only to describe the downstream function of the OAS cluster genes but also to score for functions of the members of the co-regulated co-expression network and hence the effects of the OAS signal on the sulfate assimilation pathway and co-regulated pathways. Further, we summarized existing knowledge on the regulation of the OAS cluster and the co-expressed genes. We revealed that the known sulfate deprivation-related transcription factor EIL3/SLIM1 exhibits a prominent role, as most genes are subject to regulation by this transcription factor. The role of other transcription factors in response to OAS awaits further investigation.
Keywords: APR, ATTED-II, BGLU28, gene regulation, network, O-acetylserine, oxidative stress, ROS, SDIs, sulfur, sulfur deficiency, SLIM1
O-Acetylserine (OAS), a sulfate deprivation marker, accumulates under stress conditions. We provide information about the function and regulation of OAS cluster genes and an OAS cluster co-expression network.
Introduction
O-Acetylserine (OAS) has been shown to accumulate under sulfate-deprived growth conditions (Boxes 1, 2) and has been suggested to be a signaling molecule under these conditions due to its inverse correlation with sulfate content (Saito, 2004). External (Hirai et al., 2003) or internal application (Hubberten et al., 2012b) of OAS further supported this hypothesis. In addition to the accumulation of OAS under sulfate deprivation, further conditions could be identified that lead to the accumulation of OAS, even under sulfate-sufficient conditions (Espinoza et al., 2010; Caldana et al., 2011; Hubberten et al., 2012b). A set of six genes was identified as being induced when OAS accumulates, termed OAS cluster genes (Box 3) (Hubberten et al., 2012b). Conditions such as herbicide treatment with menadione were accompanied by the accumulation of reactive oxygen species (ROS) as a hallmark of stress. ROS accumulation leads to the induction in particular of serine acetyltranferase 2;1 (SERAT2;1) and somewhat less SERAT2;2 (Watanabe et al., 2015; De Kok et al., 2017; Watanabe and Hoefgen, 2019), while under these conditions SERAT3;2 and SERAT3;1 are not induced. Treating Arabidopsis roots with menadione resulted in the accumulation of OAS after 0.5 h and showed a peak at 6 h of treatment (Lehmann et al., 2009, 2012). OAS accumulation correlated to SERAT transcript accumulation and OASTL transcript reduction. Presumably, certain oxidative stress conditions in Arabidopsis thaliana result in OAS accumulation. Sulfur metabolism is activated in roots as a response to oxidative stress (Lehmann et al., 2009). Further, it is speculated that jasmonate biosynthesis and the sulfur pathway interact (Jost et al., 2005). The transcript profile of jasmonate-regulated genes is comparable with sulfur deprivation and OAS application experiments (Hirai et al., 2003; Saito, 2004). SERAT3 was one of the numerous genes that were induced under methyljasmonate (MeJa) treatment. OAS accumulation is thus a response facilitated through specific regulatory circuits employing different SERAT groups in order to synthesize OAS as a response to different stresses.
Box 1. O-Acetylserine and cysteine biosynthesis.
O-Acetylserine (OAS) is synthesized from serine via serine acetyltransferase (SERAT) from the nitrogen and carbon assimilation pathway, providing the backbone yielding cysteine (Hoefgen and Nikiforova, 2008). In Arabidopsis five genes encoding SERAT proteins can be found: SERAT2;1 (AT1G55920) and SERAT2;2 (AT3G13110), which are localized in plastids and mitochondria, respectively, and SERAT1;1 (AT5G56760), SERAT3;1 (AT2G17640), and SERAT3;2 (AT4G35640), which are localized and expressed in the cytosol (Watanabe et al., 2008; Krueger et al., 2009). When plants experience reduced sulfate levels, OAS accumulates concomitant with the induction of SERAT2;2 and SERAT3;2, and to a lesser extent, and mainly in roots, with that of SERAT3;1 and SERAT2;1 (Watanabe et al., 2015; De Kok et al., 2017; Watanabe and Hoefgen, 2019; Dietzen et al., 2020). The mitochondrial SERAT2;2 is the enzyme that contributes most to OAS formation while having the highest activity (Watanabe et al., 2008). It has been shown that SERAT and OAS thiol lyase (OASTL) can form a hetero-oligomeric cysteine synthase complex (CSC), which is stabilized by the presence of sulfide and can be dissociated by OAS availability (Hell and Wirtz, 2011). Thus, SERAT can affect and contribute to the control of cysteine synthesis (Hell and Wirtz, 2011; Maruyama-Nakashita, 2017). SERAT3;1 and SERAT3;2 are able to synthesize OAS independently of the CSC, presumably allowing the accumulation of OAS under conditions where the CSC is dissociated (Watanabe et al., 2015). It has been shown that in the CSC, SERAT is activated, while OASTL is inactive, functioning as a regulatory subunit for SERAT. As a result, the produced OAS dissociates the complex and is further converted to cysteine by free OASTL (Feldman-Salit et al., 2009). Cysteine feedback inhibits SERAT3;1 and SERAT3;2 activity, further supporting their specific function under sulfate deprivation as resupply of sulfate leads to synthesis of cysteine and a shutdown of the CSC-independent OAS production, thus preventing an overshoot of cysteine production (Watanabe et al., 2015).
Box 3. O-Acetylserine cluster gene abbreviations.
SDI1 SULFUR DEFICIENCY INDUCED 1
SDI2 SULFUR DEFICIENCY INDUCED 2
LSU1 RESPONSE TO LOW SULFUR 1
SHM7/MSA1 SERINE HYDROXYMETHYLTRANSFERASE 7/ MORE SULFUR ACCUMULATION1
ChaC/GGCT2;1 GAMMA-GLUTAMYL CYCLOTRANSFERASE 2;1
APR3 APS REDUCTASE 3
The downstream function of OAS cluster genes
OAS accumulation induces the expression of the core OAS cluster genes (Box 3): APR3 (AT4G21990), SDI1 (AT5G48850), SDI2 (AT1G04770), LSU1 (AT3G49580), SHM7/MSA1 (AT1G36370), and ChaC/GGCT2;1 (AT5G26220) (Hubberten et al., 2012b). To date, the functions of these genes have been partially resolved. Common to all of them is that they seem to be functionally involved not only in the sulfate deprivation response but also in other metabolic and physiological processes, probably having metabolic functions (GGCT2;1; APR3) or acting as upstream regulators (SDI1, SDI2, and MSA1), but also still other unclear functions such as being putatively involved in various processes as ethylene responses and autophagy (LSU1).
SDI1 and SDI2
Under sulfur deprivation, where OAS is strongly induced, the transcripts of OAS-responsive genes such as SDI1 and SDI2 are drastically increased (Hirai et al., 2003; Howarth et al., 2009; Hubberten et al., 2012b; Dong et al., 2017), (Fig. 1). SDIs are also induced in SERAT-overexpressing Arabidopsis plants accumulating OAS despite sulfate-sufficient growth conditions, showing that OAS alone is able to induce the OAS cluster genes, among them SDIs (Hubberten et al., 2012b) (Fig. 1). SDI proteins contain a tetratricopeptide repeat (TPR) domain, which is known to mediate protein–protein interactions (Aarabi et al., 2016). It has been shown that SDI1 and SDI2 act as major repressors of GSL biosynthesis in sulfur deprivation conditions (Aarabi et al., 2016). SDI1 is localized in the nucleus and forms a complex negatively affecting the transcription factor (TF) MYB28 (Aarabi et al., 2016) which promotes aliphatic GSL biosynthesis (Hirai et al., 2003; Gigolashvili et al., 2007b). Additionally, it was shown through transient transactivation assays that SDI1 inhibits the MYB28-mediated transactivation of the promoters of two aliphatic GSL biosynthetic genes, CYP79F1 and CYP83A1 (Aarabi et al., 2016). SDI2 lacks a nuclear localization signal, but it complements an sdi1 knockout line, indicating that it is able to move into the nucleus. Presumably its protein–protein interaction capacity might recruit a carrier protein assisting SDI2’s access to the nucleus. From RNA sequencing data available under accession number GSE157765 (Dietzen et al., 2020), it can be deduced that the basal expression of SDI1 under complete nutrient conditions is significantly lower than that of SDI2 (Fig. 1). This possibly indicates that SDI2 provides basic cellular functions while SDI1 is responsible for the strong response to stress, such as sulfur deficiency and OAS accumulation. Further research on SDI2 needs to be performed in order to determine its basal functions under non-stress conditions. Recent findings for SDI1 demonstrated that besides inhibition of GSL biosynthesis, it down-regulates, developmentally or in response to sulfate deprivation, the biosynthesis of sulfur-rich 2S seed storage proteins in Arabidopsis seeds. SDI1 forms a protein complex with MYB28 and MYC2, which binds, for example, to the At2S4 gene promoter (Aarabi et al., 2021). It can be speculated that GSL inhibition is an acquired feature in Brassicaceae, while its control of seed protein composition is a feature of all seed plants.
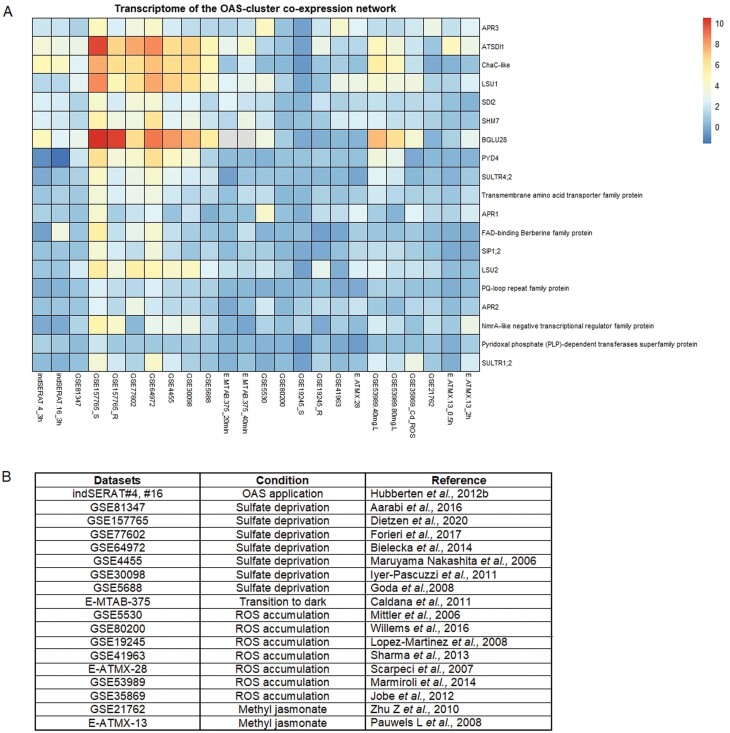
Fig. 1.
Meta-analysis depicting gene expression values of the extended OAS cluster co-expression gene network under conditions that result in OAS accumulation. (A) The data were collected from the available transcriptomic datasets (microarray and RNA-seq), using the Gene Expression Omnibus (GEO DataSets) provided by the NCBI. All the values are relative to the control—full nutrient conditions of each experiment—and log2 expression values. The heatmap was designed in R, using the Pheatmap function. No clustering was chosen. A few genes, such as those encoding hypothetical protein, SERAT3;2, and LSU3, are not included in the meta-analysis presented, since these genes were not found to have an available representative public ID in the GPL198 platform. (B) Available transcriptomic dataset IDs, which were used to design the heatmap in (A). For each dataset, not only the condition of treatment or growth is mentioned, but also the respective reference.
LSU-like family—LSU1
The LSU family in Arabidopsis consists of four members: LSU1 (AT3G49580), LSU2 (AT5G24660), LSU3 (AT3G49570), and LSU4 (AT5G24655). With the exception of LSU4, the other LSUs are induced under sulfur deprivation conditions (Hubberten et al., 2012a; Sirko et al., 2015; Maruyama-Nakashita, 2017; Li et al., 2020) (Fig. 1). LSU-like proteins are involved in plant responses to nutrient changes, such as sulfur deprivation (Dietzen et al., 2020), salt stress, or plant immune responses (Sirko et al., 2015), with their transcripts accumulating under these conditions (Garcia-Molina et al., 2017). Loss-of-function mutants of LSU proteins showed sensitivity towards nutrient deficiency, salinity, or heavy metal toxicity, indicating a widespread involvement of LSU family members in plant stress responses (Garcia-Molina et al., 2017). LSU proteins are involved in regulating cellular degradation processes and might interact with E3 ubiquitin ligases, chaperons, and the NBR1 receptor which are involved in autophagy (Sirko et al., 2015). Due to their involvement in diverse stress response pathways, further research is needed to fill the knowledge gap around LSUs.
SHM7/MSA1
SHM7 is a serine hydroxymethyltransferase 7 gene, also recently identified in a mutant screen study as ‘more sulfur accumulation 1’, MSA1 (AT1G36370). MSA1/SHM7 is among the genes that have regulatory functions under sulfur deprivation conditions and are strongly induced by OAS accumulation (Hubberten et al., 2012b) (Fig. 1). Huang et al. (2016) used β-glucuronidase (GUS) and green fluorescent protein (GFP) constructs for MSA1 to identify the tissue expression of MSA1 and its subcellular localization. MSA1 is highly expressed under sulfur deprivation in roots and leaves relative to full nutrient control conditions (Dietzen et al., 2020) (Fig. 1). MSA1 is localized in the nucleus and its localization is unaffected by the sulfur status (Huang et al., 2016). Loss-of-function mutants of MSA1 show a reduction of S-adenosylmethionine (SAM) levels, by inhibition of folate biosynthesis, which as a result reduces the DNA methylation levels, leading to a sulfur deprivation response (Huang et al., 2016). In msa1 under full nutrient conditions, genes such as SULTR1;1, SULTR1;2, APR3, and ATPS4 displayed increased expression levels, suggesting that the promoters of these genes were unmethylated, thus mimicking the state under sulfur-deprived growth conditions (Huang et al., 2016). It is further speculated that histone methylation and histone acetylation might play an important role in sulfur homeostasis (Huang et al., 2019) which would provide a gross regulation of various pathways. SHM7, unlike other members of the SHM family, does not display tetrahydrofolate biosynthetic activity. SHM7 has further been implicated to function, for example, during fruit ripening and gametogenesis, and to be localized in the nucleus (reviewed in Nogués et al., 2022). In summary, SHM7 is likely to be involved in epigenetic modifications in response to sulfate stress and OAS accumulation, probably affecting the regulation of primary sulfate metabolism, but it is also probably functional in further developmental processes.
GGCT2;1 or ChaC-like family
GGCT2;1 (AT5G26220) or ChaC-like is another member of the OAS cluster genes (Hubberten et al., 2012b), which is sharply increased upon sulfur deprivation and when OAS accumulates (Fig. 1). GGCT2;1 then displays high expression in roots compared with shoots (Joshi et al., 2019). A further study using long-term growth on reduced sulfate-containing medium revealed strong induction in roots as well in shoots (Dietzen et al., 2020). In inducible SERAT Arabidopsis plants, GGCT2;1 is significantly induced, where OAS internally accumulates (Hubberten et al., 2012b) (Fig. 1). GGCT2;1 is localized in the apoplast (Ferretti et al., 2009) where it initiates glutathione (GSH) degradation to l-glutamate, l-cysteine, and l-glycine in the γ-glutamyl cycle (Joshi et al., 2019; Ito et al., 2022). In root tips of the ggct2;1 mutant, the GSH content was increased compared with Col-0, corroborating that GGCT2;1 is involved in GSH degradation (Joshi et al., 2019). GGCT2;1 affects root architecture, in correlation with GSH degradation, as ggct2;1 under sulfur deprivation conditions demonstrates increased primary root length and less suppression of lateral root growth than Col-0 plants (Joshi et al., 2019). GGCT2;1 participates through GSH degradation in the cellular responses during abiotic stress, such as toxic metal detoxification (Paulose et al., 2013) or ROS accumulation (Dorion et al., 2021). GGCT2;1 transcripts accumulate under salinity stress (Gong et al., 2005). Recently it was shown that the cytosolic γ-glutamyl peptidases (GGP1 and GGP3) exhibit GSH-degrading activity similar to GGCT2;1. Under full nutrient conditions, the GSH concentration in ggp1-1 was significantly higher relative to Col-0 and ggct2;1, and, interestingly, this mutant accumulated OAS. This indicated that under full nutrient conditions GGP1 and probably also GGP3 degrade GSH, while surprisingly the more energy-consuming pathway via GGCT2;1 is induced under sulfate deprivation (Ito et al., 2022). It can be speculated that the resulting 5-oxoproline from the GGCT2;1 branch might be used to contribute to biotic stress responses (Fonseca et al., 2021), alleviating the reduction of GSL accompanying sulfur depletion. At the same time sulfur from GSH is recycled to primary metabolism while in parallel SDIs reduce de novo biosynthesis of GSL.
APR family—APR3
The APR family (APR1: AT4G04610, APR2: AT1G62180, and APR3: AT4G21990) are considered to be key enzymes not only for sulfate assimilation in higher plants but also for the nitrate assimilation pathway and diurnal rhythm (Kopriva et al., 1999; Lee et al., 2011). All three APR isoforms demonstrate decreased enzyme activity under darkness (Kopriva et al., 1999). APR3 is found to be exclusively localized in chloroplasts where it catalyzes the reduction of APS to sulfite by transferring two electrons (Koprivova et al., 2008). APR activity is increased by OAS (Lee et al., 2011). Further, the TF HY5 (AT5G11260) which coordinates nitrogen and sulfur assimilation, is a regulator of APR expression (Lee et al., 2011). Additionally, APR activity and mRNA levels of all three APR isoforms increased under treatment with NaCl. APR transcripts were unaffected in mutants deficient in abscisic acid (ABA) synthesis while treatment of plants with ABA did not alter the mRNA levels of APR, showing that APR is regulated by salt stress in an ABA-independent manner (Koprivova et al., 2008). In summary, APR is integrating various metabolic and stress inputs to coordinate sulfate assimilation, by potentially increasing flux through the assimilation and reduction pathway. APRs are induced under sulfur deprivation (Dietzen et al., 2020), but not significantly up-regulated by OAS application (Fig. 1) (Hirai et al., 2003; Hubberten et al., 2012b).
Additional genes potentially playing a role in the OAS response
ATTED-II is a gene co-expression database for plant species, Arabidopsis included, based on publicly available RNA-seq-derived data from the Ath-r.c5-0 platform (14 741 runs) and microarray data from Ath-m.c9-0 (12 686 chips) (Obayashi et al., 2022). Using ATTED-II, a co-expression network was built using as a query the above-mentioned ‘OAS cluster genes’ (Hubberten et al., 2012b) which comprise 22 genes (Fig. 2) being stably co-expressed under many diverse conditions. Genes are recruited to the network when co-expressed with at least two genes from the query gene list. As displayed in Table 1, the core OAS cluster genes display the highest connectivity within the network. This analysis obviously does not only identify OAS-responsive genes, which in turn might allow the discernment of co-regulation properties.
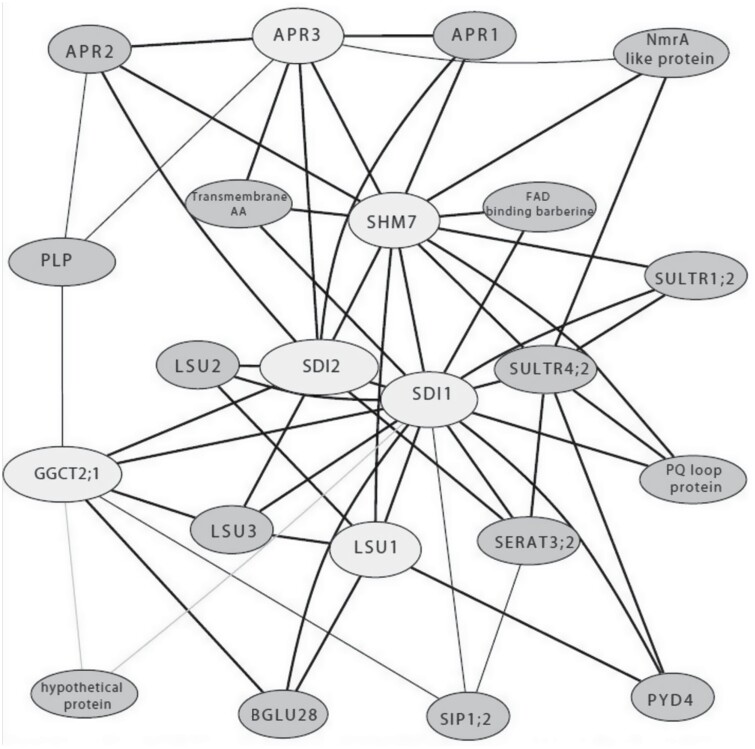
Fig. 2.
The extended OAS cluster co-expression gene network from ATTED-II. The six OAS cluster genes (light gray color) were used as query genes on the ATTED-II database. The ATTED-II contains data derived from RNA-seq (Ath-r.c5-0 platform) and microarray (Ath-m.c9-0). An additional 16 genes (dark gray color) were found to be stably co-regulated with the six core OAS cluster genes. Among them, several genes are known to be associated with sulfur metabolism and response to sulfur deprivation, such as APRs, LSUs, SULTRs, and SERAT3;2. The z-score is a factor which indicates the stability of the co-expression. The thicker line, which connects two genes, indicates a higher z-score displaying a higher degree of co-expression between the two genes. The thinner the line is, the lower the z-score is. A lower z-score indicates lower stability of co-expression of the two genes.
Table 1.
The new extended OAS cluster co-expression gene network from ATTED-II
| Gene name | AGI code | No. of genes connected | GO terms | Reference |
|---|---|---|---|---|
| FAD binding barberine | AT4G20820 | 2 | Sulfur metabolism | Depuydt and Vandepoele (2021) |
| SDI2 | AT1G04770 | 8 | Sulfur metabolism | Maruyama-Nakashita et al. (2005) |
| SULTR4;2 | AT3G12520 | 7 | Sulfur metabolism | Kataoka et al. (2004) |
| APR3 | AT4G21990 | 7 | Sulfur metabolism | Bick and Leustek. (1998) |
| LSU1 | AT3G49580 | 6 | Sulfur metabolism | Maruyama-Nakashita et al. (2005); Sirko et al. (2015) |
| LSU3 | AT3G49570 | 4 | Sulfur metabolism | Depuydt and Vandepoele (2021) |
| APR2 | AT1G62180 | 4 | Sulfur metabolism | Setya et al. (1996) |
| LSU2 | AT5G24660 | 3 | Sulfur metabolism | Sirko et al. (2015) |
| APR1 | AT4G04610 | 3 | Sulfur metabolism | Setya et al. (1996); Bick et al. (1998); Koprivova et al. (2000) |
| SULTR1;2 | AT1G78000 | 3 | Sulfur metabolism | Rouached et al. (2005, 2008) |
| ChaC-like/GGCT2;1 | AT5G26220 | 7 | Sulfur metabolism, glutathione process | Paulose et al. (2013) |
| SERAT3;2 | AT4G35640 | 4 | Sulfur metabolism, cysteine | Kawashima et al. (2005) |
| SDI1 | AT5G48850 | 16 | Sulfur metabolism, carbon metabolism, transcription | Maruyama-Nakashita et al. (2005) |
| SHM7 | AT1G36370 | 12 | Sulfur metabolism, carbon metabolism, methionine process | Huang et al. (2016) |
| NmrA-like protein | AT1G75280 | 3 | Response to oxidative stress biosynthesis | Babiychuk et al. (1995) |
| PQ loop repeat family protein | AT5G40670 | 3 | cysteine biosynthesis | Gaudet et al. (2011) |
| BGLU28 | AT2G44460 | 3 | Carbon metabolism, glucosinolate hydrolysis | Gaudet et al. (2011) |
| Pyridoxal phosphate (PLP) protein | AT1G77670 | 3 | Carbon metabolism, transaminase activity | Depuydt and Vandepoele (2021) |
| PYD4 | AT3G08860 | 3 | Response to nitrogen, transaminase activity | Zrenner et al. (2009) |
| SIP1;2 | AT5G18290 | 3 | Transporter activity | Ishikawa et al. (2005) |
| Transmembrane amino acid protein | AT3G56200 | 3 | Amino acid transporter activity | Gaudet et al. (2011) |
| hypothetical protein | AT2G32487 | 2 | Response to ABA | Depuydt and Vandepoele (2021) |
Summarizing table demonstrating information about the extended OAS cluster co-expression gene network from ATTED-II. The table provides information about the number of the genes to which each gene is connected in the network depicted in Fig. 2. SDI1 shows the highest connectivity between the network, connected with 16 genes out 22. This means that SDI1 is co-expressed with most of the genes existing in the network, throughout many transcriptomic experiments in A. thaliana. Additionally, information is provided about the GO terms of each gene. Most of the genes are involved in sulfur metabolism and sulfur response (SULTR, SERAT3;2, LSU, SDI, APR). Additionally, a few seem to be involved in carbon metabolism or GSL regulation (BGLU28), cysteine biosynthesis (PQ loop repeat family protein), oxidative stress (NmrA-like protein), or ABA response (hypothetical protein).
The OAS cluster co-expression network
A majority of the genes (14) that are present in the OAS cluster co-expression network are known to be involved in sulfur metabolism. Gene Ontology (GO) enrichment analysis (Table 1) using ATTED (Obayashi et al., 2022) and TAIR (Berardini et al., 2015) revealed biological processes except for sulfur metabolism. These include carbon and nitrogen metabolism (metabolic processes) or terms involved in stress response, such as oxidative stress or ABA (Table 1). It is known that sulfate availability affects the ABA content in plant tissues, as the ABA level is reduced in sulfate-deprived plants (Cao et al., 2014). This explains why genes involved in stresses or responsive to different ABA levels, such as the hypothetical protein (AT2G32487), are included in the OAS cluster ATTED network. The sulfate assimilation pathway and the carbon and nitrate pathways converge at the level of cysteine synthesis. In the plant cell, the pathways of carbon assimilation, through the Calvin cycle, of nitrate assimilation, and of sulfate assimilation co-influence one another (Jobe et al., 2019). The inter-relationship between those pathways is very strong since sulfate deprivation reduces nitrate uptake and carbon assimilation, and vice versa. The sensors and mechanisms of the connection of these three pathways are poorly understood (Koprivova and Kopriva, 2014). It is likely that the pathway genes could transcriptionally respond in concert if any changes occur in one of the three pathways. Hence, the GO enrichment analysis of the OAS cluster co-expression network (Table 1) displays genes involved in nitrogen response (PYD4) or carbon metabolism (PLP, BGLU28, and SDI1).
BGLU28
A similar response as the OAS cluster genes under sulfur deprivation or OAS accumulation is displayed by β-glucosidase 28 (BGLU28) (AT2G44460), which is considered to be a sulfur deprivation marker (Zhang et al., 2014). BGLU28 is involved in GSL catabolism under sulfur deprivation conditions by hydrolyzing GSL resulting in d-glucose and sulfate in order to recycle sulfate from GSL for primary metabolism (Nikiforova et al., 2004; Maruyama-Nakashita et al., 2005, 2006). Zhang et al. (2020) proved this hypothesis by using a double knockout mutant of BGLU28 and BGLU30 (AT3G60140), encoding another main β-glucosidase and likewise induced by sulfate deprivation. The double knockout bglu28/30 displayed growth retardation and reduced metabolic performance under sulfur deprivation relative to Col-0, with GSL contents being increased and other sulfur-containing compounds such as GSH, cysteine, or sulfate being reduced (Zhang et al., 2020). In support of the GSL recycling function, it could be shown that labeled 34S from GSL was allocated to primary sulfur metabolism as substrate, eventually ending up in, for example, cysteine or GSH (Sugiyama et al., 2021). These results prove not only the role of BGLU28 in GSL catabolism but also the role of GSL as a sulfur reservoir (Sugiyama et al., 2021). BGLU28 displays high connectivity within the OAS cluster co-expression network (Fig. 2) and it is strongly induced under sulfur deprivation conditions (Dietzen et al., 2020) (Fig. 1) but less induced in inducible SERAT plants or upon OAS treatment (Hirai et al., 2003; Hubberten et al., 2012b) (Fig. 1). BGLU30 is not OAS induced, though it is also a sulfate deprivation-inducible gene, and both BGLU28 and BGLU30 are controlled by SLIM1 (Dietzen et al., 2020). This again indicates that although OAS accumulates under sulfur deprivation conditions, there must be additional signals for sulfate deprivation-specific gene regulation.
SERAT family
SERAT3;2 is part of the co-expression network (Fig. 2; Table 1). Of the five SERAT genes, SERAT1;1 (At5g56760), SERAT2;1 (At1g55920), SERAT2;2 (At3g13110), SERAT3;1 (At2g17640), and SERAT3;2 (At4g35640), each has a different transcriptional response to certain conditions (Watanabe and Hoefgen, 2017). It was shown that SERAT group III genes are highly induced under sulfur deprivation, while group II is induced under oxidative stress and prolonged sulfate deprivation (Watanabe et al., 2015; Watanabe and Hoefgen, 2017; Dietzen et al., 2020); group I does not respond to tested conditions. The variability in the transcript responses of the SERAT genes under sulfur deprivation might indicate that the plant organism balances the OAS production and responds to the need for OAS transport to different cellular compartments (Watanabe and Hoefgen, 2017).
Sulfate transporters (SULTRs)
SULTR4;2 and SULTR1;2 are part of the OAS cluster co-expression network (Fig. 2; Table 1). Plants take up sulfate through their root system with the help of root high-affinity sulfate transporters SULTR1;1 (AT4G08620) and SULTR1;2 (AT1G78000) (Takahashi et al., 2000, 2011; Yoshimoto et al., 2007). SULTR1;1 and SULTR1;2 are the main transporters involved in sulfate assimilation and are increased under sulfur-deprived conditions (Fig. 1) at transcriptional and protein levels (Takahashi et al., 2000; Yoshimoto et al., 2007). SULTR2;1 (AT5G10180) and SULTR2;2 (AT1G77990) are suggested to transfer sulfate from the roots to the shoots, and SULTR2;1 also controls the sulfate transfer into the developing seeds (Takahashi et al., 2011). Group III sulfate transporters have been shown to be expressed mainly in leaves (Takahashi et al., 2000). SULTR3;1 (AT3G51895) is responsible for the sulfate uptake into the chloroplasts (Cao et al., 2014). SULTR4;1 (AT5G13550) and SULTR4;2 (AT5G13550) are tonoplast-localized transporters and coordinate the release of sulfate from the vacuoles (Takahashi et al., 2011). SULTR4;1 and SULTR4;2 are highly induced under sulfur deprivation (Takahashi et al., 2000, 2011; Cao et al., 2014) (Fig. 1). It is known that SULTRs, group I and IV, respond to OAS accumulation in inducible SERAT plants and under OAS application (Hirai et al., 2003; Hubberten et al., 2012b).
PYD4
PYD4 (AT3G08860) belongs to the aminotransferase gene family, and it functions as an alanine:glyoxylate aminotransferase/β-alanine:pyruvate aminotransferase (Parthasarathy et al., 2019). PYD4 is up-regulated in response to osmotic stress and has been shown to be putatively involved in β-alanine metabolism (Parthasarathy et al., 2019). Studies have identified that PYD4 is involved in multiple processes in plants, such as changes in light and carbon availability (Parthasarathy et al., 2019). PYD4 is localized in the mitochondria or the peroxisome (Niessen et al., 2012) and it is down-regulated in the microarray of Hubberten et al. (2012b), where OAS is induced internally; however, in sulfur deficiency microarrays and RNA-seq, PYD4 is co-regulated with the OAS cluster genes (Fig. 1). It is connected with SDI1, LSU1, and SULTR4;2 in the OAS cluster co-expression network (Fig. 2), and controlled by SLIM1 (Table 2). PYD4, like BGLU30, seems to be sulfate deprivation responsive rather than OAS responsive.
Table 2.
SLIM1 binding in the promoters and regulation of the new extended OAS cluster co-expression gene network from ATTED-II
| Gene name | AGI code | Dap-seq EIL3 |
MN 2006 EIL3 |
EIL3-Dietzen and -S |
|---|---|---|---|---|
| ATSDI1 | AT5G48850 | Yes | Yes | Yes |
| SHM7 | AT1G36370 | Yes | Yes | Yes |
| SDI2 | AT1G04770 | Yes | Yes | Yes |
| SULTR4;2 | AT3G12520 | Yes | Yes | Yes |
| ChaC-like/GGCT2;1 | AT5G26220 | Yes | Yes | Yes |
| APR3 | AT4G21990 | Yes | No | Yes |
| LSU1 | AT3G49580 | Yes | Yes | Yes |
| LSU3 | AT3G49570 | Yes | No | Yes |
| APR2 | AT1G62180 | Yes | No | Yes |
| SERAT3;2 | AT4G35640 | No | – | Yes |
| LSU2 | AT5G24660 | Yes | No | Yes |
| APR1 | AT4G04610 | Yes | No | Yes |
| SULTR1;2 | AT1G78000 | Yes | Yes | Yes |
| PYD4 | AT3G08860 | Yes | Yes | Yes |
| Transmembrane AA | AT3G56200 | Yes | No | Yes |
| BGLU28 | AT2G44460 | Yes | Yes | Yes |
| SIP1;2 | AT5G18290 | No | – | Yes |
| PQ loop repeat family | AT5G40670 | No | Yes | Yes |
| FAD binding barberine | AT4G20820 | Yes | No | Yes |
| Pyridoxal phosphate (PLP) | AT1G77670 | Yes | No | Yes |
| NmrA-like negative transcriptional regulator | AT1G75280 | No | Yes | Yes |
| hypothetical protein | AT2G32487 | Yes | No | Yes |
Information about the regulation of the extended OAS cluster co-expression gene network from ATTED-II by EIL3/SLIM1. We used three different experimental approaches to demonstrate the binding and regulation of SLIM1 at the promoters of the extended OAS cluster co-expression gene network from ATTED-II genes. First, we used the data from Dap-seq by O’Malley et al. (2016) which showed direct binding of SLIM1 at the promoters of all the genes except SERAT3;2, SIP1;2, PQ loop family protein, and NmrA like negative transcriptional regulator. Microarray analysis by Maruyama-Nakashita et al. (2006) indicates that SLIM1 regulates the majority of the genes, with few exceptions. Interestingly, the genes to which SLIM1 was found not to bind (Dap-seq), are not the same as those that seem not to be regulated by SLIM1 (MN). Moreover, RNA-seq by Dietzen et al. (2020) indicates that SLIM1 can regulate all the genes under sulfur deprivation conditions.
SIP1;2
SIP1;2 (AT5G18290) encodes an aquaporin and expressed in all Arabidopsis tissues except dry seeds. It is localized in the endoplasmic reticulum (ER) membrane and has a water channel activity (Ishikawa et al., 2005). SIP1;2 is involved in controlling the volume and morphology of the ER lumen and the concentration of ions in the ER (Ishikawa et al., 2005). SIP1;2 is slightly induced in OAS-accumulating plants (Hubberten et al., 2012b) and under sulfur deficiency (Dietzen et al., 2020) (Fig. 1).
Additional members of the co-expression network from ATTED
The OAS cluster co-expression network contains four genes that have not been studied extensively before. These are LSU3 (AT3G49570), FAD binding berberine family protein or AtBBE18 (AT4G20820), pyridoxal phosphate (PLP)-dependent transferase (AT1G77670), and the hypothetical protein (AT2G32487) (Fig. 2; Table 1). Depuydt and Vandepoele (2021) identified the functional relationship of these three genes to sulfur metabolism and/or OAS. LSU3 was found to respond to sulfur deprivation and other stresses such as salt stress, ABA, wounding, or exposure to fungi. This agrees with the previous predictions and results for the involvement of the LSU family in stresses (Sirko et al., 2015; Garcia-Molina et al., 2017). The FAD binding berberin was annotated with only one GO term, sulfur metabolic process, without any indication on its molecular or physiological function (Table 1) (Depuydt and Vandepoele, 2021). AtBBE18 has been characterized as a biomass regulator and was shown to be important for salt stress tolerance (Daniel et al., 2016). The pyridoxal phosphate (PLP)-dependent transferase protein is associated with cellular responses to stress, carboxylic acid metabolic processes, and seed development, while the hypothetical protein AT2G32487 is involved in the ABA response (Depuydt and Vandepoele, 2021). Reduced sulfate availability reduces ABA contents in plant tissues (Cao et al., 2014) while ABA accumulates under abiotic stresses such as salt or drought. High concentrations of NaCl result in sulfate content reduction (Hongqiao et al., 2021). The inclusion of LSU3, FAD binding berberine, and (PLP)-dependent transferase in the OAS cluster co-expression network indicates a relationship between the pathways of sulfate, and ABA or salt.
Functional relationships between the genes of the OAS cluster co-expression network
It is striking that the genes of the expanded OAS cluster co-expression network are involved in various metabolic processes with the major one being sulfur metabolism (Table 1). These processes are directly connected with the sulfur pathway (cysteine, methionine, GSH, and GSL) or seemingly unrelated (ABA or ROS response, and carbon, nitrogen, and amino acid metabolism). Their co-expression, however, suggests that there might be physiological and functional relationships between the genes involved in these pathways and a need for coordinated regulations. When exposed to sulfur deficiency, plants alter morphological and physiological processes, in which the above-mentioned genes are involved. The initial step of the sulfur assimilation pathway (sulfur sufficiency or deprivation) is the absorption of any sulfate molecule from the soil by the SULTR1 transporters (Li et al., 2020). After a series of enzymatic reactions (see Box 2), sulfate is incorporated into cysteine, and some of the genes shown in Fig. 2, are involved in these steps: SULTR4;2 translocates sulfur from the vacuoles where its stored in order to cover the demands of sulfate-deficient plants (Kataoka et al., 2004). In parallel, APR3 drives sulfate reduction for primary metabolism rather than sulfate processes catalyzed by SERATs (SERAT group III) (De Kok et al., 2017; Watanabe and Hoefgen, 2017). Sulfur deprivation activates recycling processes of sulfate-containing molecules such as as GSH or GSLs by GGCT2;1 or BGL28 (Paulose et al., 2013); Zhang et al., 2020; Ito et al., 2022) or reduces their biosynthesis through inhibiting the MYB28 regulatory activity by SDI genes (Aarabi et al., 2016, 2020). Further, general degradatory processes such as autophagy through LSU genes (LSU1 and LSU3) are induced (Sirko et al., 2015; Dong et al., 2022). Methylation by MSA1 very generally affects diverse processes in response to sulfate deprivation (Huang et al., 2016, 2019). Lastly, ROS and OAS accumulation often appear linked (Dorion et al., 2021) and it might be speculated that OAS serves as a signal under sulfate-deprived conditions but also under sulfate-sufficient conditions when ROS levels increase together with OAS (Hubberten et al., 2012b; Aarabi et al., 2020). As such, the co-expressed genes are presumably part of a complex regulatory network integrating diverse inputs, eventually regulating plant cell homeostasis. This is reflected by the fact that the promoters of OAS cluster genes analyzed so far contain diverse sets of cis-elements, and various TFs have been identified to affect their respective expression (Rakpenthai et al., 2022).
Box 2. Sulfate assimilation and cysteine synthesis.
Sulfate (SO42–) which has been taken up by sulfate transporters (SULTRs), is transported to the shoot via the xylem and eventually to the plastids. ATP sulfurylase (ATPS) forms 5-adenylylsulfate (APS) (Murillo and Leustek, 1995). APS provides a branchpoint and can follow different pathways. First, it can be reduced to sulfide following a two-step reaction which is catalyzed by APS reductase (APR) (Rotte and Leustek, 2000) to form sulfite and by sulfite reductase (SiR) to form sulfide (Rotte and Leustek, 2000; Saito, 2004; Hell and Wirtz, 2011; Takahashi et al., 2011; Naumann et al., 2018). Finally, sulfate will be incorporated into cysteine, the first organic form of the pathway. Sulfide and OAS are converted to cysteine by OASTL (Hell and Wirtz, 2011). Cysteine is further used as the backbone to form a huge number of sulfur-containing compounds such as glutathione (GSH), methionine, glucosinolates (GSLs), or further metabolites, vitamins, or lipids (Leustek et al., 2000; Saito, 2004; Davidian and Kopriva, 2010; Takahashi et al., 2011; Kopriva et al., 2012). Alternatively, APS can be phosphorylated to form 3ʹ-phosphoadenosine-5ʹ-phosphosulfate (PAPS) by APS kinase (APK) being involved in sulfation reactions.
OAS, sulfur metabolism, and their connection with hormones
Plant hormones are regulators of a diverse set of physiological responses in plants. Information on the interplay of sulfate metabolism, especially under conditions of sulfate deprivation, is still fragmented. Systematic studies on the dynamics and tissue specificity of hormone responses are still lacking. However, available research allows us to offer a first overview on the topic (Fig. 3). However, at this level, it is not yet possible to differentiate between the effects of sulfate deprivation and its potential direct effects, and the signals involved and those exerted by OAS. ABA is one of the key regulators of stress responses (Soma et al., 2021). Sulfur availability and especially increased cysteine levels positively affect ABA biosynthesis (Cao et al., 2014) and control stomatal closure through ABA (Batool et al., 2018). In addition to this rapid response, for example to drought conditions, stress-induced ABA also fosters adaptation processes widely affecting plant physiology (Danquah et al., 2015). Sulfate deprivation leads in roots to the induction of many regulatory genes, as described above. In particular, SNRK genes have been described to be responsive to various nutrient stresses (Iyer-Pascuzzi et al., 2011; Heyneke et al., 2015), among them SNRK2 as part of the ABA core signaling module and SNRK3.15 and SnRK3.22 as central hubs controlling ABA-responsive genes (Lumba et al., 2014). SNRK 3.15 is induced under sulfate deprivation (Iyer-Pascuzzi et al., 2011; Heyneke et al., 2015), when ABA does not accumulate (Cao et al., 2014), possibly allowing recruitment of ABA-dependent responses under sulfate deprivation, such as nutrient depletion-induced senescence (NUDIS) (Watanabe et al., 2010). OAS also accumulates upon ROS accumulation when plants are exposed to stresses (Hubberten et al., 2012b) and among those stresses also upon sulfate deprivation (Schachtman and Shin, 2007; Sachdev et al., 2021). LSU1 has been found to reduce ROS production under sulfur deprivation and promote stomatal closure (Garcia-Molina et al., 2017), though the mechanism is as yet unclear.
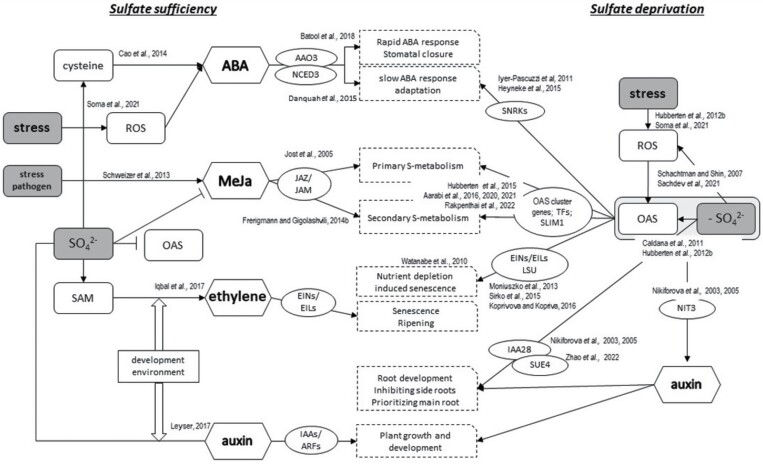
Fig. 3.
Scheme of interaction and crosstalk of sulfate metabolism and various signal hormones, such as ABA, MeJa, ethylene, and auxin. Arrows with arrowheads represent induction while arrows with blunt ends represent negative regulation. The outcome of the hormone effect is depicted in broken border boxes. References are reported in the figure with respective to each pathway.
Jasmonate application induces the expression of numerous genes involved in sulfur assimilation, methionine biosynthesis, SAM biosynthesis, and further sulfur-related processes (Jost et al., 2005) and SDI1 (Rakpenthai et al., 2022). Transcriptome analyses of plants under sulfur-deprived conditions revealed that genes of MeJa biosynthesis are induced within 24–48 h (Hirai et al., 2003; Nikiforova et al., 2003; Jost et al., 2005). Whether the jasmonate signaling pathway is activated by OAS or by signals such as ROS (Koo, 2018) remains to be validated. In the JAZ/JAM system (Fig. 3), the JASMONATE ZIM-domain (JAZ) and CORONATINE INSENSITIVE1 (COI1) protein complex inhibits jasmonate target gene expression. MeJa leads to polyubiquitination of JAZ, priming it for degradation and allowing other TFs such as MYC2 to access the promoter which, for example, leads to activation of GSL biosynthesis. MYC2 and MYB28 are TFs critical for inducing GSL synthesis (Schweizer et al., 2013; Frerigmann and Gigolashvili, 2014). SDI1, on the other hand, inhibits GSL biosythesis through interaction with MYC2 and MYB28 (Aarabi et al., 2016) which appears contradictory. SDI genes and OAS cluster genes act to prevent flux into secondary metabolites or to retrieve sulfur by degradation and induce primary sulfate metabolism (Aarabi et al., 2020), thus reducing the plant’s capacity to react against biotrophic pathogens. MeJa induces biosynthesis of secondary metabolite such as GSL which might possibly be a mechanism to redirect sulfur to secondary metabolite biosynthesis when exposed to MeJa-inducing pathogens (Schweizer et al., 2013). The exact interplay of this cross-regulatory effect needs to be further investigated. Another link between jasmonate signaling and OAS might be the basic helix–loop–helix (bHLH) TF At1g10585 that is induced by OAS and not sulfate deprivation, is a target gene of the JAZ/JAM system (Hubberten et al., 2015), and is ROS responsive (Inze et al., 2012).
Ethylene is a key regulator of leaf senescence and fruit ripening (Iqbal et al., 2017) as well as nutrient depletion-induced senescence (Watanabe et al., 2010). Ethylene is tightly linked to methionine metabolism (Moniuszko et al., 2013; Sirko et al., 2015; Koprivova and Kopriva, 2014) as SAM is the precursor of ethylene synthesis (Fig. 3). Members of the EIL (ETHYLENE-INSENSITIVE) TF family, such as EIN3, EIL1, EIL2 (Wawrzyńska et al., 2010), or EIL3/SLIM1 have been shown to regulate sulfur-responsive genes under sulfur deprivation conditions (Maruyama-Nakashita et al., 2006; Wawrzyńska and Sirko, 2016; Dietzen et al., 2020), demonstrating their involvement in ethylene-responsive and sulfur-responsive gene regulation. Under sulfur deprivation conditions, ethylene accumulates in Col-0 plants. Tobacco LSU and UP9C mutants (Sirko et al., 2015) accumulated significantly less ethylene than Col-0. Additionally, in up9c, under sulfur deprivation conditions, the transcripts of ethylene-responsive genes were significantly less expressed relative to Col-0. Hence, members of the LSU family are likely to be involved in the modulation of the ethylene signaling pathway which is crucial for the sulfur deficiency response (Sirko et al., 2015) (Fig. 3). Yet, SAM levels, and hence the precursor of ethylene, massively decreased upon prolonged sulfate deprivation due to the reduced availability of methionine (Nikiforova et al., 2005b) and despite the activity of the SAM/Met cycle (Büerstenbinder et al., 2007). Our understanding of the interplay of ethylene, OAS, and sulfate is still lacking details to provide a resolved model.
Auxin is involved in numerous aspects of plant physiology, coordinating plant growth and development by affecting transcription through the AUX/IAA–ARF system as well as polarity in organs such as roots through directed transport by the PIN system (Leyser, 2018). It is thus not surprising that auxin-related genes have been identified to be responsive to sulfate deprivation (Nikiforova et al., 2004; Dietzen et al., 2020) among them induction of nitrilase 3 (NIT3) involved in auxin biosynthesis. Further, IAA28 has been identified as a hub in network analysis under sulfate deprivation (Nikiforova et al., 2005a; Watanabe and Hoefgen, 2017) though its expression is only marginally increased (Dietzen et al., 2020). IAA28 probably inhibits lateral root development in response to sulfate deprivation (Rogg et al., 2001; Falkenberg et al., 2008). Recently SUE4 (sulfate utilization efficiency 4) was identified to be induced by sulfate starvation and to foster primary root elongation by interacting with PIN1 and targeting it for protein degradation (Zhao et al., 2023) (Fig. 3). Both genes are consistent with the known plant phenotype of lateral root repression and enhanced primary root growth upon sulfate deprivation (Hubberten et al., 2012a). This allows plants to search for sulfate-rich patches in the soil and expand the root system when exposed to sulfate.
While in general sulfate depletion or OAS accumulation might affect phytohormone accumulation, where, as stated above, sufficient data related to time and tissue distribution are missing, sulfate deprivation-derived signals or OAS directly affect hormone-related pathways (Fig. 3). Through this, existing hormone response pathways are utilized to facilitate sulfur deficiency/OAS-specific responses. In addition, it has to be noted that plant hormones interact and influence one another in a complex manner (Rubio et al., 2009).
How are OAS-responsive genes regulated?
The plant responses to sulfur deprivation conditions have been studied during the past decade (Davidian and Kopriva, 2010; Nakai and Maruyama-Nakashita, 2020; Ristova and Kopriva, 2022). Which signals are perceived and how TFs regulate these responses is still not finally resolved (Kopriva, 2006). Some progress has been achieved and suggestions provided (Maruyama-Nakashita et al., 2006; Zhang et al., 2014; Bielecka et al., 2015; Aarabi et al., 2016; Huang et al., 2016; Watanabe and Hoefgen, 2017, 2019; Forzani et al., 2018; Rakpenthai et al., 2022; Wawrzyńska et al., 2022). In addition to these analyses, available databases allow the identification of TFs possibly involved in the OAS-driven response.
In this context, we scored TFs suggested to regulate the genes of the co-expression network (Fig. 2; Table 1). The Plant Regulomics database (Ran et al., 2020) was used to identify TFs which bind to the promoters of the new network genes (Fig. 2; Table 1) in order to obtain information on whether these might be linked to the OAS signal. The dominant TF EIL3/SLIM1 binds to 18 of the 22 genes of the OAS co-expression network (Table 3).
Table 3.
Transcription factors regulating the new extended OAS cluster co-expression gene network from ATTED-II
| Transcription factor | AGI code | No. of genes regulated | Source | Regulation |
|---|---|---|---|---|
| EIL3 | AT1G73730 | 18 | DAP-seq | Ethylene and sulfate deprivation signaling |
| EIN3 | AT3G20770 | 8 | DAP-seq | Ethylene signaling |
| MYB67 | AT3G12720 | 10 | DAP-seq | Response to wounding |
| DTAF1 | AT3G45810 | 7 | ChIP-seq | NAD(P)H oxidase H2O2-forming activity, |
| ERF115 | AT5G07310 | 7 | ChIP-seq | Ethylene signaling |
| HB7 | AT2G46680 | 7 | ChIP-seq | Drought response and ABA |
| NFYB2 | AT5G47640 | 7 | ChIP-seq | Response to nutrient levels |
| E2Fa | AT2G36010 | 7 | ChIP-seq | E2F pathway |
| NRPE1 | AT2G40030 | 6 | ChIP-seq | DNA methylation, defense response to fungus |
| BBM | AT5G17430 | 6 | ChIP-seq | Lateral roof formation |
| HY5 | AT5G11260 | 6 | ChIP-seq | Anthocyanin accumulation in far-red light |
| NFYC2 | AT1G56170 | 5 | ChIP-seq | Response to nutrient levels |
| PIF4 | AT2G43010 | 6 | ChIP-seq | Shade avoidance response, response to nutrient levels |
| MYB3 | AT1G22640 | 5 | ChIP-seq | Phenylpropanoid biosynthesis gene expression |
| MYB related family | AT3G10580 | 5 | DAP-seq | – |
| RVE8 | AT3G09600 | 7 | DAP-seq | Regulation of the circadian clock by modulating the pattern of histone 3 (H3) acetylation, involved in heat shock response |
| PIF3 | AT1G09530 | 6 | ChIP-seq | Binds to anthocyanin biosynthetic genes in a light- and HY5-independent fashion, regulation of photosynthesis, light reaction |
| LHY | AT1G01060 | 6 | ChIP-seq | Circadian rhythm along with another Myb transcription factors |
| HB6 | AT2G22430 | 6 | ChIP-seq | Hormone responses in Arabidopsis such as ABA |
| CCA1 | AT2G46830 | 4 | ChIP-seq | Circadian rhythms, long-day photoperiodism, flowering |
| DTAF2 | AT5G50360 | 6 | ChIP-seq | ABA signaling |
Transcription factors (TFs) which are suggested by Plant Regulomics to bind at the promoters of the extended OAS cluster co-expression gene network from ATTED-II. The table demonstrates the number of the genes of the extended OAS cluster co-expression gene network from ATTED-II on which a particular TF is bound on their promoters. EIL3/SLIM1 is the TF which binds to the majority of the gene promoters. Another TF binding at many promoters is MYB67. Information about the binding of the TFs was also provided by Plant Regulomics. Dap-seq (DNA affinity purification sequencing) or Chip-seq (ChIP sequencing) are the molecular experiments proving the TF binding at the promoters. With the help of the TAIR tool, the conditions in which those TFs are involved or regulated by were identified and provided.
EIL3/SLIM1 is involved in the sulfate deprivation signaling pathway and is a central transcriptional regulator of plant sulfate metabolism (Maruyama-Nakashita et al., 2006; Wawrzyńska et al., 2010, 2022; Kawashima et al., 2011; Wawrzyńska and Sirko, 2014, 2016; Dietzen et al., 2020; Rakpenthai et al., 2022), by controlling many sulfate deprivation response genes. EIL3/SLIM1 binds to the UPE box, the TEBS element, and the SURE element, which are present in many sulfate- and OAS-responsive genes (Maruyama-Nakashita et al., 2005; Wawrzyńska et al., 2010; Rakpenthai et al., 2022). In addition to binding properties as displayed in the Plant Regulomics database, transcriptional regulation of OAS cluster genes and OAS network genes could be shown using transcriptome data (Maruyama-Nakashita et al., 2006; O’Malley et al., 2016; Dietzen et al., 2020) (Table 2). Interestingly, SLIM1 transcriptional levels do not alter under sulfur deprivation (Wawrzyńska et al., 2010; Rakpenthai et al., 2022) or OAS treatment (Hubberten et al., 2012b), which indicates that SLIM1’s activity is affected by post-transcriptional and/or post-translational modifications. Further, it has been indicated that EIL3/SLIM1 might conditionally also act as a repressor or as an enhancer of target gene expression, for example under sulfate deprivation or arsenic treatment (Jobe et al., 2019; Dietzen et al., 2020), which might explain the differences in the identified transcriptional control patterns between different experiments (Table 2). Despite its early detection and prominent role the functions of EIL3/SLIM1, its interactions with other regulators, and its post-transcriptional/post-translational properties are not yet entirely resolved.
Further ethylene-responsive TFs have been suggested to regulate sulfate deprivation metabolism. EIN3 is a regulator of the ethylene pathway and interacts with EIL3/SLIM1 (Wawrzyńska and Sirko, 2016). EIN3 binds to the promoters of eight genes of the co-expression network (Table 3) and has been shown to form heterodimers with EIL3/SLIM1, preventing SLIM1 binding of the UPE-box (Wawrzyńska and Sirko, 2016). EIL1 (AT2G27050) has been shown to regulate numerous sulfate deprivation genes in concert with EIL3/SLIM1 or independently (Dietzen et al., 2020). EIL1 was not identified in the Plant Regulomics dataset, leading to the conclusion that EIL1 might exert its regulatory function through protein–protein interaction with other TFs (such as EIL3/SLIM1) rather than itself binding directly to the promoter.
MYB TFs are known to be involved in numerous plant regulatory processes (Dubos et al., 2010). With respect to sulfur metabolism they have been identified to be involved in GSL biosynthesis regulation (Celenza et al., 2005; Gigolashvili et al., 2007a; Frerigmann and Gigolashvili, 2014; Frerigmann et al., 2014; Aarabi et al., 2016, 2020) and in seed storage protein regulation (Aarabi et al., 2021). In the Plant Regulomics database two new MYB TFs have been identified (Table 3) which deserve further analysis. Hitherto, MYB3 (AT1G22640) has been assigned to phenylpropanoid metabolism (Kim et al., 2022) but also to plant growth control through phytosulfokines, sulfur compounds modulating auxin responses (Badola et al., 2022). Phytosulfokines, which can be viewed as peptide hormones, are insufficiently studied in terms of their relationship to sulfate deprivation or OAS signaling (Komori et al., 2009; Kopriva et al., 2012). Under stress conditions, whether nutrient deprivation or other stresses involving ROS, phenylpropanoid biosynthesis is induced to alleviate stress symptoms. In the context of sulfate deprivation, MYB TFs have been associated with this response, among them PAP1/MYB75 (AT1G56650), which controls anthocyanin biosynthesis (Nikiforova et al., 2005a; Wulff-Zotele et al., 2010; Watanabe and Hoefgen, 2019). Interestingly, the dataset (GSE157765) of Dietzen et al. (2020) indicates jointly coordinated repression of MYB75 by EIL3/SLIM1 and EIL1, which requires an induction independent of the EIL3/SLIM1 regulatory circuit. Furthermore, in addition to MYB3, the TFs HY5 and PIF3, which regulate anthocyanin accumulation and phenyl-propanoid biosynthesis (Oyama et al., 1997; Kim et al., 2003; Shin et al., 2007; Kim et al., 2022), seem to be involved in the regulation of the OAS-responsive gene network (Table 3). For the second MYB-related TF (AT3G10580), scarce information is available to date, but as it targets five genes of the OAS cluster expression network, its future analysis is recommended. AT3G10580 has been identified to potentially interact with the above-mentioned MYB3 which controls anthocyanin and lignin biosynthesis under salt stress (Kim et al., 2022).
An additional level of regulation during sulfur deprivation is provided through epigenetic modifications associated with the OAS cluster gene MSA1 (Hubberten et al., 2012b) that is involved in DNA and other regulatory methylation reactions (Huang et al., 2016). Further, NRPE1 is a TF involved in RNA-directed DNA methylation presumably playing a role in gene control, seed development, and pathogen responses (Sasaki et al., 2019; Miao et al., 2021; Wang et al., 2021). NRPE1 binds to the promoters of six genes of the co-expression network (Table 3). Its relationship to the OAS signaling pathway or sulfate availability is unclear and needs further investigations.
Among the identified TFs are several whose link to OAS signaling and/or sulfate metabolism is still unclear. NFYB2, NFYC2, and PIF4 (Table 3) are all known to be involved in regulating genes responding to nutrient levels (Brumbarova and Ivanov, 2019). Moreover, there is a connection between sulfate and ABA since reduced sulfate availability results in reduced ABA content in the plant tissue (Cao et al., 2014). This explains the presence of three TFs, DTAF, HB6, and HB7 (Ré et al., 2014; Gaudinier et al., 2018; Ma et al., 2021) (Table 3). Moreover, TFs involved in the circadian clock and response to light were revealed. OAS has been indicated to accumulate in the middle of the night-time (Espinoza et al., 2010) or immediately after the plants are transferred from light into the dark (Caldana et al., 2011). OAS accumulation results in the regulation of numerous genes, such as the core OAS cluster genes (Hubberten et al., 2012b). TFs such as RVE8, PIF3, LHY, and CCA1 (Pérez-García et al., 2015; Wang et al., 2021; Hao et al., 2022) which regulate genes involved in light responses, might potentially regulate a significant number of genes from the OAS co-expression network (Table 3).
Conclusion
OAS has long been considered a signal within the sulfate deprivation response (Saito, 2000; Hirai et al., 2003) with increasing evidence in recent years (Hubberten et al., 2012b; Aarabi et al., 2016, 2020). OAS, though an inherent precursor of cysteine synthesis, has been shown to accumulate not only under low sulfur conditions (Maruyama-Nakashita et al., 2006; Hubberten et al., 2012a), but also under different stresses. These stresses, such as heavy metal exposure (Jalmi et al., 2018), ROS-inducing herbicide treatment (Lehmann et al., 2009, 2012), jasmonate accumulation (Jost et al., 2005), or shifts from light to darkness (Caldana et al., 2011) provoke ROS accumulation. Notably, sulfate availability is not altered under these conditions. Experimentally this has been corroborated by overproducing SERAT leading to OAS accumulation (Hubberten et al., 2012b). All these conditions lead to the induction of a core set of genes, the OAS cluster genes.
All OAS cluster core genes, except APR3, seem to be regulated, at least in response to sulfate deprivation, by the commonly accepted central regulator of plant sulfate metabolism, SLIM1/EIL3 (Tables 2, 3). SDI2 seems rather to be repressed by SLIM1 while SDI1 needs the presence of SLIM1 under sulfur-depleted conditions (Dietzen et al., 2020). The promoters of the SLIM1/EIL3-regulated genes contain the known cis-elements UPE-box and TEBs (Wawrzyńska et al., 2010, 2022), and SURE elements were recently proven to bind EIL2/SLIM1(Rakpenthai et al., 2022). In silico and in vitro analyses of the promoter regions revealed many more putative cis-elements, indicating that further TFs are potentially able to bind to the promoter regions (Table 3). This, together with the fact that APR is an OAS cluster gene but not subject to SLIM1 control, suggests that other regulatory circuits control the expression of the OAS cluster genes in response to OAS rather than SLIM1 alone. From the OAS co-expression cluster, BGLU28, BGLU30, and PYD also display features indicating independent sulfate deprivation and OAS signaling pathways, probably acting in parallel under sulfate-deprived growth conditions. Having a knockout SLIM1 line available (Wawrzyńska et al., 2022) now provides the possibility to test this experimentally. Based on the fact that stresses other than sulfate deprivation result in OAS accumulation and the common responses of the OAS cluster genes, we have to assume that OAS signaling is distinct from sulfate deprivation signaling, though both coincide under sulfate-deprived growth conditions. This decoupling of the OAS and the sulfur deprivation response is supported by the fact that in potato (Solanum tuberosum) several sulfate deprivation responses such as SULTR expression and increased sulfate uptake capacity precede OAS accumulation (Hopkins et al., 2005).
Further evidence is provided by the extended OAS cluster co-expression network (Fig. 2; Table 1) as these genes are co-expressed over a wide range of conditions but are only partially sulfate deprivation-responsive genes or induced by OAS. Hence, we have to conclude the existence of an additional regulatory pathway specific for distinct stresses and, positive as well as negative, interference of the TF by modulating their target genes in response to various stresses and signals. Such stress-specific response pathways are also indicated through the differential inducibility of the OAS-synthesizing SERAT genes. Moreover, the fact that some genes, such as APR or LSU genes, are up-regulated in sulfur deprivation conditions (Howarth et al., 2003; Dietzen et al., 2020), but not or weakly upon OAS treatment (Hubberten et al., 2012b), indicates that there are further regulatory factors affecting gene expression, other than just the signal molecule OAS, to be considered.
SDIs are not TFs but interact with them and regulate gene expression of, for example, MYB28 (Aarabi et al., 2016, 2020) or regulate the accumulation of sulfur-rich seed storage proteins in seeds (Aarabi et al., 2021). The latter might be the original function present in all plants, while GSL regulation is a Brassicaceae-specific acquisition. In addition, MSA1 and potentially NRPE1 through their methylation activity are non-TF regulators epigenetically affecting sulfur homeostasis (Huang et al., 2016), and perhaps also other processes. This leads to the conclusion that in addition to TFs, other regulators will have to be considered as regulating either the sulfate deprivation response or other, OAS-specific stress responses. In this context, while miRNA395 has been shown to affect SULTR activity and sulfate allocation under sulfate deprivation in Arabidopsis (Davidian and Kopriva, 2010), other RNA-based or proteinaceous regulators still need to be considered. To further understand the regulation of the OAS cluster genes and, hence, their downstream effects on sulfate metabolism or other metabolic pathways, the positioning and structure of cis-elements within various promoters will need more attention. There are sets of TFs targeting similar cis-elements or overlapping cis-elements for different TFs (Rakpenthai et al., 2022). What determines the priority of binding? Does binding alter existing promotor features such as palindromic structures? Could this lead to acceptance or exclusion of TFs targeting OAS cluster gene promoters?
Eventually, neither OAS signal perception nor the signal transduction pathways are reliably resolved yet.
Glossary
Abbreviations
- ABA
abscisic acid
- APK
APS kinase
- APR
APS reductase
- APS
adenosine 5ʹ-phosphosulfate
- CSC
cysteine synthase complex
- DAP-seq
DNA Affinity Purification and sequencing
- ER
endoplasmic reticulum
- GO
Gene Ontology
- GSL
glucosinolate
- GSH
glutathione
- MeJa
methyljasmonate;
- OAS
O-acetyl-serine
- OASTL
O-acetylserine-thiol-lyase
- ROS
reactive oxygen species
- SERAT
serine acetyltransferase
- TF
transcription factor
Contributor Information
Anastasia Apodiakou, Max Planck Institute of Molecular Plant Physiology, Am Mühlenberg 1, D-14476 Potsdam-Golm, Germany.
Rainer Hoefgen, Max Planck Institute of Molecular Plant Physiology, Am Mühlenberg 1, D-14476 Potsdam-Golm, Germany.
Ann Cuypers, Hasselt University, Belgium.
Funding
AA has been funded by the Deutsche Forschungsgemeinschaft (DFG) through the grant HO1916/13-1, SignalOAS, and the Max Planck Society. RH received institutional funding from the Max Planck Society. Open Access funding was enabled and organized by Project DEAL.
Conflict of interest
The authors have no conflict to declare.
Data availability
Raw data that support the findings of this study, such as ATTED results, are available from the corresponding author, upon request. Further data which were used to generate the heatmap in Fig. 1, can be found from the datasets mentioned in Fig. 1B.
References
- Aarabi F, Miyuki K, Tohge T, et al. 2016. Sulfur deficiency-induced repressor proteins optimize glucosinolate biosynthesis in plants. Science Advances 2, e1601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarabi F, Rakpenthai A, Barahimipour R, et al. 2021. Sulfur deficiency-induced genes affect seed protein accumulation and composition under sulfate deprivation. Plant Physiology 187, 2419–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarabi F, Thomas N, Fernie AR, Hoefgen R.. 2020. Coordinating sulfur pools under sulfate deprivation. Trends in Plant Science 25, 1227–1239. [DOI] [PubMed] [Google Scholar]
- Babiychuk E, Kushnir S, Belles-Boix E, Van Montagu M, Inzé D.. 1995. Arabidopsis thaliana NADPH oxidoreductase homologs confer tolerance of yeasts toward the thiol-oxidizing drug diamide. Journal of Biological Chemistry 270, 26224–26231. [DOI] [PubMed] [Google Scholar]
- Badola P, Sharma A, Gautam H, Trivedi PK.. 2022. MicroRNA858a, its encoded peptide, and phytosulfokine regulate Arabidopsis growth and development. Plant Physiology 189, 1397–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batool S, Uslu VV, Rajab H, et al. 2018. Sulfate is incorporated into cysteine to trigger ABA production and stomatal closure. The Plant Cell 30, 2973–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E.. 2015. The Arabidopsis information resource: making and mining the ‘Gold Standard’ annotated reference plant genome. Genesis 53, 474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick JA, Leustek T.. 1998. Plant sulfur metabolism-the reduction of sulfate to sulfite. Current Opinion in Plant Biology 1, 240–244. [DOI] [PubMed] [Google Scholar]
- Bielecka M, Watanabe M, Morcuende R, Scheible WR, Hawkesford MJ, Hesse H, Hoefgen R.. 2015. Transcriptome and metabolome analysis of plant sulfate starvation and resupply provides novel information on transcriptional regulation of metabolism associated with sulfur, nitrogen and phosphorus nutritional responses in Arabidopsis. Frontiers in Plant Science 5, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T, Ivanov R.. 2019. The nutrient response transcriptional regulome of Arabidopsis. IScience 19, 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerstenbinder K, Rzequski G, Wirtz M, Hell R, Sauter M.. 2007. The role of methionine recycling for ethylene synthesis in Arabidopsis. The Plant Journal 49, 238–249. [DOI] [PubMed] [Google Scholar]
- Caldana C, Degenkolbe T, Cuadros-Inostroza A, Klie S, Sulpice R, Leisse A, Steinhauser D, Fernie AR, Willmitzer L, Hannah MA.. 2011. High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. The Plant Journal 67, 869–884. [DOI] [PubMed] [Google Scholar]
- Cao MJ, Wang Z, Zhao Q, Mao JL, Speiser A, Wirtz M, Hell R, Zhu JK, Xiang CB.. 2014. Sulfate availability affects ABA levels and germination response to ABA and salt stress in Arabidopsis thaliana. The Plant Journal 77, 604–615. [DOI] [PubMed] [Google Scholar]
- Celenza JL, Quiel JA, Smolen GA, Merrikh H, Silvestro AR, Normanly J, Bende J.. 2005. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiology 137, 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel B, Wallner S, Steiner B, Oberdorfer G, Kumar P, Graaff E, Roitsch T, Sensen CW, Gruber K, Macheroux P.. 2016. Structure of a berberine bridge enzyme-like enzyme with an active site specific to the plant family Brassicaceae. PLoS One 11, e0156892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danquah A, Zelicourt A, Boudsocq M, et al. 2015. Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. The Plant Journal 82, 232–244. [DOI] [PubMed] [Google Scholar]
- Davidian JC, Kopriva S.. 2010. Regulation of sulfate uptake and assimilation—the same or not the same? Molecular Plant 3, 314–325. [DOI] [PubMed] [Google Scholar]
- De Kok LJ, Hawkesford MJ, Haneklaus S, Schnug E.. 2017. Sulfur metabolism in higher plants—fundamental, environmental and agricultural aspects. Cham: Springer. [Google Scholar]
- Depuydt T, Vandepoele K.. 2021. Multi-omics network-based functional annotation of unknown Arabidopsis genes. The Plant Journal 108, 1193–1212. [DOI] [PubMed] [Google Scholar]
- Dietzen C, Koprivova A, Whitcomb SJ, Langen G, Jobe TO, Hoefgen R, Kopriva S.. 2020. The transcription factor EIL1 participates in the regulation of sulfur-deficiency response. Plant Physiology 184, 2120–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Aref R, Forieri I, Schiel D, Leemhuis W, Meyer C, Hell R, Wirtz M.. 2022. The plant TOR kinase tunes autophagy and meristem activity for nutrient stress-induced developmental plasticity. The Plant Cell 34, 3814–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Silbermann M, Speiser A, et al. 2017. Sulfur availability regulates plant growth via glucose–TOR signaling. Nature Communications 8, 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dorion S, Ouellet JC, Rivoal J.. 2021. Glutathione metabolism in plants under stress: beyond reactive oxygen species detoxification. Metabolites 11, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L.. 2010. MYB transcription factors in Arabidopsis. Trends in Plant Science 15, 573–581. [DOI] [PubMed] [Google Scholar]
- Espinoza C, Degenkolbe T, Caldana C, Zuther E, Leisse A, Willmitzer L, Hincha DK, Hannah MA.. 2010. Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS One 5, e14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg B, Witt I, Zanor MI, Steinhauser D, Mueller-Roeber B, Hesse H, Hoefgen R.. 2008. Transcription factors relevant to auxin signalling coordinate broad-spectrum metabolic shifts including sulphur metabolism. Journal of Experimental Botany 59, 2831–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman-Salit A, Wirtz M, Hell R, Wade, RC.. 2009. A mechanistic model of the cysteine synthase complex. Journal of Molecular Biology 386, 37–59. [DOI] [PubMed] [Google Scholar]
- Ferretti M, Destro T, Tosatto S, Rocca N, Rascio N, Masi A.. 2009. Gamma-glutamyl transferase in the cell wall participates in extracellular glutathione salvage from the root apoplast. New Phytologist 181, 115–126. [DOI] [PubMed] [Google Scholar]
- Fonseca JP, Oh S, Boschiero C, Watson B, Huhman D, Mysore KS.. 2021. The Arabidopsis iron–sulfur (Fe–S) cluster gene Mfdx1 plays a role in host and nonhost disease resistance by accumulation of defense-related metabolites. International Journal of Molecular Sciences 22, 7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forzani C, Duarte GT, Meyer C.. 2018. The plant target of rapamycin kinase: a connector between sulfur and growth. Trends in Plant Science 23, 472–475. [DOI] [PubMed] [Google Scholar]
- Frerigmann H, Berger B, Gigolashvili T.. 2014. BHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiology 166, 349–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerigmann H, Gigolashvili T.. 2014. MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Molecular Plant 7, 814–828. [DOI] [PubMed] [Google Scholar]
- Garcia-Molina A, Altmann M, Alkofer A, Epple PM, Dangl JL, Braun PF.. 2017. LSU network hubs integrate abiotic and biotic stress responses via interaction with the superoxide dismutase FSD2. Journal of Experimental Botany 68, 1185–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet P, Livstone MS, Lewis SE, Thomas PD.. 2011. Phylogenetic-based propagation of functional annotations within the Gene Ontology Consortium. Briefings in Bioinformatics 5, 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudinier A, Rodriguez-Medina J, Zhang L, et al. 2018. Transcriptional regulation of nitrogen-associated metabolism and growth. Nature 563, 259–264. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Berger B, Mock HP, Müller C, Weisshaar B, Flügge UI.. 2007a. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. The Plant Journal 50, 886–901. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Yatusevich R, Berger B, Müller C, Flügge UI.. 2007b. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. The Plant Journal 51, 247–261. [DOI] [PubMed] [Google Scholar]
- Gong Q, Li P, Ma S, Rupassara SI, Bohnert HJ.. 2005. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. The Plant Journal 44, 826–839. [DOI] [PubMed] [Google Scholar]
- Hao C, Yang Y, Du J, Deng XW.. 2022. The PCY–SAG14 phytocyanin module regulated by PIFs and miR408 promotes dark-induced leaf senescence in Arabidopsis. Plant Biology 119, e2116623119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell R, Wirtz M.. 2011. Molecular biology, biochemistry and cellular physiology of cysteine metabolism in Arabidopsis thaliana. The Arabidopsis Book 9, e0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyneke E, Watanabe M, Aarabi F, Hoefgen R.. 2015. The CBL–SnRK3 network: connections to sulfur metabolism. In: De Kok L, Hawksford M, Rennenberg H, Saito K, Schnug E, eds. Molecular physiology and ecophysiology of sulfur. Proceedings of the International Plant Sulfur Workshop. Cham: Springer, 145–152 [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K.. 2003. Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. The Plant Journal 33, 651–663. [DOI] [PubMed] [Google Scholar]
- Hoefgen R, Nikiforova VJ.. 2008. Metabolomics integrated with transcriptomics: assessing systems response to sulfur-deficiency stress. Physiologia Plantarum 132, 190–198. [DOI] [PubMed] [Google Scholar]
- Hongqiao L, Suyama A, Mitani-Ueno N, Hell R, Maruyama-Nakashita A.. 2021. A low level of NaCl stimulates plant growth by improving carbon and sulfur assimilation in Arabidopsis thaliana. Plants 10, 2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins L, Parmar S, Błaszczyk A, Hesse H, Hoefgen R, Hawkesford MJ.. 2005. O-Acetylserine and the regulation of expression of genes encoding components for sulfate uptake and assimilation in potato. Plant Physiology 138, 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth JR, Domínguez-Solís JR, Gutiérrez-Alcalá G, Wray JL, Romero LC, Gotor C.. 2003. The serine acetyltransferase gene family in Arabidopsis thaliana and the regulation of its expression by cadmium. Plant Molecular Biology 51, 589–598. [DOI] [PubMed] [Google Scholar]
- Howarth JR, Parmar S, Barraclough PB, Hawkesford MJ.. 2009. A sulphur deficiency-induced gene, SDI1, involved in the utilization of stored sulphate pools under sulphur-limiting conditions has potential as a diagnostic indicator of sulphur nutritional status. Plant Biotechnology Journal 7, 200–209. [DOI] [PubMed] [Google Scholar]
- Huang XY, Chao DY, Koprivova A, et al. 2016. Nuclear localised More Sulphur Accumulation1 epigenetically regulates sulphur homeostasis in Arabidopsis thaliana. PLoS Genetics 12, e1006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Li M, Luo R, Zhao FJ, Salt DE.. 2019. Epigenetic regulation of sulfur homeostasis in plants. Journal of Experimental Botany 70, 4171–4182. [DOI] [PubMed] [Google Scholar]
- Hubberten HM, Drozd A, Tran BV, Hesse H, Hoefgen R.. 2012a. Local and systemic regulation of sulfur homeostasis in roots of Arabidopsis thaliana. The Plant Journal 72, 625–635. [DOI] [PubMed] [Google Scholar]
- Hubberten HM, Klie S, Caldana C, Degenkolbe T, Willmitzer L, Hoefgen R.. 2012b. Additional role of O-acetylserine as a sulfur status-independent regulator during plant growth. The Plant Journal 70, 666–677. [DOI] [PubMed] [Google Scholar]
- Hubberten HM, Watanabe M, Bielecka M, Heyneke E, Aarabi F, Hoefgen R.. 2015. More than a substrate: the O-acetylserine responsive transcriptome. In: De Kok LJ, Hawkesford MJ, Rennenberg H, Saito K, Schnug E, eds. Molecular physiology and ecophysiology of sulfur. Proceedings of the International Plant Sulfur Workshop. Cham: Springer, 133–143. [Google Scholar]
- Inze A, Vanderauwera S, Hoeberichts F, Vandorpe M, Gaever V, Breusegem F.. 2012. A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant, Cell & Environment 35, 308–320. [DOI] [PubMed] [Google Scholar]
- Iqbal N, Khan N, Ferrante A, Trivellini A, Francini A, Khan MIR.. 2017. Ethylene role in plant growth, development and senescence: interaction with other phytohormones. Frontiers in Plant Science 8, 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F, Suga S, Uemura T, Sato MH, Maeshima M.. 2005. Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell-specific expression in Arabidopsis thaliana. FEBS Letters 579, 58145817–58145820. [DOI] [PubMed] [Google Scholar]
- Ito T, Kitaiwa T, Nishizono K, et al. 2022. Glutathione degradation activity of γ-glutamyl peptidase 1 manifests its dual roles in primary and secondary sulfur metabolism in Arabidopsis. The Plant Journal 111, 1626–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Pascuzzi A, Jackson T, Cui H, Petricka J, Busch W, Tsukagoshi H, Benfey P.. 2011. Cell identity regulators link development and stress responses in the Arabidopsis root. Developmental Cell 21, 770–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalmi SK, Bhagat PK, Verma D, Noryang S, Tayyeba S, Singh K, Sharma D, Sinha AK.. 2018. Traversing the links between heavy metal stress and plant signaling. Frontiers in Plant Science 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe TO, Zenzen I, Karvansara PR, Kopriva S.. 2019. Integration of sulfate assimilation with carbon and nitrogen metabolism in transition from C3 to C4 photosynthesis. Journal of Experimental Botany 70, 4211–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi NC, Meyer AJ, Bangash SAK, Zheng ZL, Leustek T.. 2019. Arabidopsis γ-glutamylcyclotransferase affects glutathione content and root system architecture during sulfur starvation. New Phytologist 221, 1387–1397. [DOI] [PubMed] [Google Scholar]
- Jost R, Altschmied L, Bloem E, et al. 2005. Expression profiling of metabolic genes in response to methyl jasmonate reveals regulation of genes of primary and secondary sulfur-related pathways in Arabidopsis thaliana. Photosynthesis Research 86, 491–508. [DOI] [PubMed] [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H.. 2004. Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiology 136, 4198–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima CG, Berkowitz O, Hell R, Noji M, Saito K.. 2005. Characterization and expression analysis of a serine acetyltransferase gene family involved in a key step of the sulfur assimilation pathway in Arabidopsis. Plant Physiology 137, 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima CG, Matthewman CA, Huang S, et al. 2011. Interplay of SLIM1 and miR395 in the regulationof sulfate assimilation in Arabidopsis. The Plant Journal 66, 863–876. [DOI] [PubMed] [Google Scholar]
- Kim D, Jeon SJ, Yanders S, Park SC, Kim HS, Kim S.. 2022. MYB3 plays an important role in lignin and anthocyanin biosynthesis under salt stress condition in Arabidopsis. Plant Cell Reports 41, 1549–1560. [DOI] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS, Choi G.. 2003. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. The Plant Cell 15, 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori R, Amano Y, Ohnishi M, Matsubayashi Y.. 2009. Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 15067–15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJ. 2018. Metabolism of the plant hormone jasmonate: a sentinel for tissue damage and master regulator of stress response. Phytochemistry Reviews 17, 51–80. [Google Scholar]
- Kopriva S. 2006. Regulation of sulfate assimilation in Arabidopsis and beyond. Annals of Botany 97, 479–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Mugford SG, Baraniecka P, Lee BR, Matthewman CA, Koprivova A.. 2012. Control of sulfur partitioning between primary and secondary metabolism in Arabidopsis. Frontiers in Plant Science 3, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Muheim R, Koprivova A, Trachsel N, Catalano C, Suter M, Brunold C.. 1999. Light regulation of assimilatory sulphate reduction in Arabidopsis thaliana. The Plant Journal 20, 37–44. [DOI] [PubMed] [Google Scholar]
- Koprivova A, Kopriva S.. 2014. Molecular mechanisms of regulation of sulfate assimilation: first steps on a long road. Frontiers in Plant Science 5, 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, North KA, Kopriva S.. 2008. Complex signaling network in regulation of adenosine 5ʹ-phosphosulfate reductase by salt stress in Arabidopsis roots. Plant Physiology 146, 1408–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, Suter M, Op den Camp R, Brunold C, Kopriva S.. 2000. Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiology 122, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger S, Niehl A, Lopez Martin MC, Steinhauser D, Donath A, Hildebrandt T, Romero LC, Hoefgen R, Gotor C, Hesse H.. 2009. Analysis of cytosolic and plastidic serine acetyltransferase mutants and subcellular metabolite distributions suggests interplay of the cellular compartments for cysteine biosynthesis in Arabidopsis. Amino Acids 39, 1029–1042. [DOI] [PubMed] [Google Scholar]
- Lee BR, Koprivova A, Kopriva S.. 2011. The key enzyme of sulfate assimilation, adenosine 5ʹ-phosphosulfate reductase, is regulated by HY5 in Arabidopsis. The Plant Journal 67, 1042–1054. [DOI] [PubMed] [Google Scholar]
- Lehmann M, Laxa M, Sweetlove LJ, Fernie AR, Obata T.. 2012. Metabolic recovery of Arabidopsis thaliana roots following cessation of oxidative stress. Metabolomics 8, 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Schwarzländer M, Obata T, et al. 2009. The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Molecular Plant 2, 390–406. [DOI] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick J, Davies JP.. 2000. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Plant Physiology 51, 141–165. [DOI] [PubMed] [Google Scholar]
- Leyser O. 2018. Auxin signaling. Plant Physiology 176, 465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Gao Y, Yang A.. 2020. Sulfur homeostasis in plants. International Journal of Molecular Sciences 21, 8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumba S, Toh S, Handfield L, et al. 2014. A mesoscale abscisic acid hormone interactome reveals a dynamic signaling landscape in Arabidopsis. Developmental Cell 29, 360–372. [DOI] [PubMed] [Google Scholar]
- Ma Y, Tian H, Lin R, et al. 2021. AITRL, an evolutionarily conserved plant specific transcription repressor regulates ABA response in Arabidopsis. Scientific Reports 11, 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A. 2017. Metabolic changes sustain the plant life in low-sulfur environments. Current Opinion in Plant Biology 39, 144–151. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H.. 2006. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. The Plant Cell 18, 3235–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H.. 2005. Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. The Plant Journal 42, 305–314. [DOI] [PubMed] [Google Scholar]
- Miao W, Dai J, Wang Y, et al. 2021. Roles of Idm3 and Sdj1/2/3 in establishment and/or maintenance of DNA methylation in Arabidopsis. Plant and Cell Physiology 62, 1409–1422. [DOI] [PubMed] [Google Scholar]
- Moniuszko G, Skoneczny M, Zientara-Rytter K, Wawrzyńska A, Głów D, Cristescu SM, Harren FJ, Sirko A.. 2013. Tobacco LSU-like protein couples sulphur-deficiency response with ethylene signalling pathway. Journal of Experimental Botany 64, 5173–5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo M, Leustek T.. 1995. Adenosine-5ʹ-triphosphate-sulfurylase from Arabidopsis thaliana and Escherichia coli are functionally equivalent but structurally and kinetically divergent: nucleotide sequence of two adenosine-5ʹ-triphosphate-sulfurylase cDNAs from Arabidopsis thaliana and analysis of a recombinant enzyme. Archives of Biochemistry and Biophysics 323, 195–204. [DOI] [PubMed] [Google Scholar]
- Nakai Y, Maruyama-Nakashita A.. 2020. Biosynthesis of sulfur-containing small biomolecules in plants. International Journal of Molecular Sciences 21, 3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann M, Hubberten HM, Watanabe M, Hänsch R, Schöttler MA, Hoefgen R.. 2018. Sulfite reductase co-suppression in tobacco reveals detoxification mechanisms and downstream responses comparable to sulfate starvation. Frontiers in Plant Science 871, 1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen M, Krause K, Horst I, Staebler N, Klaus S, Gaertner S, Kebeish R, Araujo WL, Fernie AR, Peterhansel C.. 2012. Two alanine aminotranferases link mitochondrial glycolate oxidation to the major photorespiratory pathway in Arabidopsis and rice. Journal of Experimental Botany 63, 2705–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova VJ, Daub CO, Hesse H, Willmitzer L, Hoefgen R.. 2005a. Integrative gene–metabolite network with implemented causality deciphers informational fluxes of sulphur stress response. Journal of Experimental Botany 56, 1887–1896. [DOI] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, HoefgenR.. 2003.Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. The Plant Journal 33, 633–650. [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Gakière B, Kempa S, Adamik M, Willmitzer L, Hesse H, Hoefgen R.. 2004. Towards dissecting nutrient metabolism in plants: a systems biology case study on sulphur metabolism. Journal of Experimental Botany 55, 1861–1870. [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R.. 2005b. Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of arabidopsis plants. Plant Physiology 138, 304–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogués I, Sekula B, Angelaccio S, Grzechowiak M, Tramonti A, Contestabile R, Ruszkowski M.. 2022. Arabidopsis thaliana serine hydroxymethyltransferases: functions, structures, and perspectives. Plant Physiology and Biochemistry 187, 37–49. [DOI] [PubMed] [Google Scholar]
- Obayashi T, Hibara H, Kagaya Y, Aoki Y, Kinoshita K.. 2022. ATTED-II V11: a plant gene coexpression database using a sample balancing technique by subagging of principal components. Plant and Cell Physiology 63, 869–881. [DOI] [PubMed] [Google Scholar]
- O’Malley RC, Carol Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR.. 2016. Erratum: cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K.. 1997. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes and Development 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy A, Adams LE, Savka FC, Hudson AO.. 2019. The Arabidopsis thaliana gene annotated by the locus tag At3g08860 encodes alanine aminotransferase. Plant Direct 3, e00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulose B, Chhikara S, Coomey J, Jung H, Vatamaniuk O, Dhankher OP.. 2013. A γ-glutamyl cyclotransferase protects Arabidopsis plants from heavy metal toxicity by recycling glutamate to maintain glutathione homeostasis. The Plant Cell 25, 4580–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-García P, Ma Y, Yanovsky MJ, Mas P.. 2015. Time-dependent sequestration of RVE8 by LNK proteins shapes the diurnal oscillation of anthocyanin biosynthesis. Proceedings of the National Academy of Sciences, USA 112, 5249–5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakpenthai A, Apodiakou A, Whitcomb SJ, Hoefgen R.. 2022. In silico analysis of cis-elements and identification of transcription factors putatively involved in the regulation of the OAS cluster genes SDI1 and SDI2. The Plant Journal 110, 1286–1304. [DOI] [PubMed] [Google Scholar]
- Ran X, Zhao F, Wang Y, Liu J, Zhuang Y, Ye L, Qi M, Cheng J, Zhang Y.. 2020. Plant regulomics: a data-driven interface for retrieving upstream regulators from plant multi-omics data. The Plant Journal 101, 237–248. [DOI] [PubMed] [Google Scholar]
- Ré DA, Capella M, Bonaventure G, Chan RL.. 2014. Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biology 14, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristova D and Kopriva S.. 2022. Sulfur signaling and starvation response in Arabidopsis. iScience, Volume 25, Issue 5, 104242, 10.1016/j.isci.2022.104242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg L, lasswell J, Bartel B.. 2001. A gain-of-function mutation in/AA28 suppresses lateral root development. The Plant Cell 13, 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotte C, Leustek T.. 2000. Differential subcellular localization and expression of ATP sulfurylase and 5ʹ-adenylylsulfate reductase during ontogenesis of Arabidopsis leaves indicates that cytosolic and plastid forms of ATP sulfurylase may have specialized functions. Plant Physiology 124, 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H, Berthomieu P, El Kassis E, Cathala N, Catherinot V, Labesse G, Davidian JC, Fourcroy P.. 2005. Structural and functional analysis of the C-terminal STAS (sulfate transporter and anti-sigma antagonist) domain of the Arabidopsis thaliana sulfate transporter SULTR1.2. Journal of Biological Chemistry 280, 15976–15983. [DOI] [PubMed] [Google Scholar]
- Rouached H, Wirtz M, Alary R, Hell R, Arpat AB, Davidian JC, Fourcroy P, Berthomieu P.. 2008. Differential regulation of the expression of two high-affinity sulfate transporters, SULTR1.1 and SULTR1.2, in Arabidopsis. Plant Physiology 147, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Bustos R, Irigoyen ML, Lopez X, Triana M, Ares J.. 2009. Plant hormones and nutrient signaling. Plant Molecular Biology 69, 361–373. [DOI] [PubMed] [Google Scholar]
- Sachdev A, Ansari SA, Ansari MI, Fijita M, Hasanuzzaman M.. 2021. Abiotic stress and reactive oxygen species: generation, signaling, and defense mechanisms. Antioxidants 10, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K.. 2000. Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Current Opinion in Plant Biology 3, 188–195. [PubMed] [Google Scholar]
- Saito K. 2004. Sulfur assimilatory metabolism. the long and smelling road. Plant Physiology 136, 2443–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Kawakatsu T, Ecker JR, Nordborg M.. 2019. Common alleles of CMT2 and NRPE1 are major determinants of CHH methylation variation in Arabidopsis thaliana. PLoS Genetics 15, e1008492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman D, Shin R.. 2007. Nutrient sensing and signaling: NPKS. Annual Review of Plant Biology 58, 47–69. [DOI] [PubMed] [Google Scholar]
- Schweizer F, Calvo P, Zander M, Diaz M, Fonseca S, Glauser G, Lewwsey M, Ecker J, Solano R, Reymond P.. 2013. Arabidopsis basic helix–loop–helix transcription factors MYC2,MYC3, andMYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. The Plant Cell 25, 3117–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setya A, Murillo M, Leustek T.. 1996. Sulfate reduction in higher plants: molecular evidence for a novel 5ʹ-adenylylsulfate reductase. Proceedings of the National Academy of Sciences, USA 23, 13383–13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park E, Choi G.. 2007. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. The Plant Journal 49, 981–994. [DOI] [PubMed] [Google Scholar]
- Sirko A, Wawrzynska A, Rodriguez MC, Sektas P.. 2015. The family of LSU-like proteins. Frontiers in Plant Science 5, 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma F, Takahashi F, Yamaguchi-Shinozaki K, Shinozaki K.. 2021. Cellular phosphorylation signaling and gene expression in drought stress responses: ABA-dependent and ABA-independent regulatory systems. Plants 10, 756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama R, Li R, Kuwahara A, et al. 2021. Retrograde sulfur flow from glucosinolates to cysteine in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 118, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R.. 2011. Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annual Review of Plant Biology 62, 157–184. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Watanabe-Takahashi A, Smith FW, Blake-Kalff M, Hawkesford MJ, Saito K.. 2000. The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. The Plant Journal 23, 171–182. [DOI] [PubMed] [Google Scholar]
- Wang Z, Butel N, Santos-González J, Simon L, Wärdig C, Köhler C.. 2021. Transgenerational effect of mutants in the RNA-directed DNA methylation pathway on the triploid block in Arabidopsis. Genome Biology 22, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Hoefgen R.. 2017. Expression profile of the serine acetyltransferase (SERAT) and O-acetylserine (thiol)lyase (OASTL) gene families in Arabidopsis. In: De Kok LJ, Hawkesford MJ, Haneklaus S, Schnug E, eds. Sulfur metabolism in higher plants—fundamental, environmental and agricultural aspects. Cham: Springer, 31–38. [Google Scholar]
- Watanabe M, Hoefgen R.. 2019. Sulphur systems biology—making sense of omics data. Journal of Experimental Botany 70, 4155–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Hubberten H-M, Saito K, HoefgenR.. 2010. General regulatory patterns of plant mineral nutrient depletion as revealed by serat quadruple mutants disturbed in cysteine synthesis. Molecular Plant 3, 438–466. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Hubberten HM, Saito K, Hoefgen R.. 2015. Serine acetyltransferase. In: D’Mello JPF, ed. Amino acids in higher plants. Wallingford, UK: CABI Publishing, 195–218. [Google Scholar]
- Watanabe M, Mochida K, Kato T, Tabata S, Yoshimoto N, Noji M, Saitoa K.. 2008. Comparative genomics and reverse genetics analysis reveal indispensable functions of the serine acetyltransferase gene family in Arabidopsis. The Plant Cell 20, 2484–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzyńska A, Lewandowska M, Sirko A.. 2010. Nicotiana tabacum EIL2 directly regulates expression of at least one tobacco gene induced by sulphur starvation. Journal of Experimental Botany 61, 889–900. [DOI] [PubMed] [Google Scholar]
- Wawrzyńska A, Piotrowska J, Apodiakou A, Brückner F, Hoefgen R, Sirko A.. 2022. The SLIM1 transcription factor affects sugar signaling during sulfur deficiency in Arabidopsis. Journal of Experimental Botany 73, 7362–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzyńska A, Sirko A.. 2014. To control and to be controlled: understanding the Arabidopsis SLIM1 function in sulfur deficiency through comprehensive investigation of the EIL protein family. Frontiers in Plant Science 5, 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzyńska A, Sirko A.. 2016. EIN3 interferes with the sulfur deficiency signaling in Arabidopsis thaliana through direct interaction with the SLIM1 transcription factor. Plant Science 253, 50–57. [DOI] [PubMed] [Google Scholar]
- Wulff-Zotelle C, Gatzke N, Kopka J, Orellana A, Hoefgen R, Fishan J, Hesse H.. 2010. Photosynthesis and metabolism interact during acclimation of Arabidopsis thaliana to high irradiance and sulphur depletion. Plant, Cell & Environment 33, 1974–1988. [DOI] [PubMed] [Google Scholar]
- Yoshimoto N, Inoue E, Watanabe-Takahashi A, Saito K, Takahashi H.. 2007. Posttranscriptional regulation of high-affinity sulfate transporters in Arabidopsis by sulfur nutrition. Plant Physiology 145, 378387–378388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pasini R, Dan H, Joshi N, Zhao Y, Leustek T, Zheng ZL.. 2014. Aberrant gene expression in the Arabidopsis SULTR1;2 mutants suggests a possible regulatory role for this sulfate transporter in response to sulfur nutrient status. The Plant Journal 77, 185–197. [DOI] [PubMed] [Google Scholar]
- Zhang L, Kawaguchi R, Morikawa-Ichinose T, Allahham A, Kim SJ, Maruyama-Nakashita A.. 2020. Sulfur deficiency-induced glucosinolate catabolism attributed to two β-glucosidases, Bglu28 and Bglu30, is required for plant growth maintenance under sulfur deficiency. Plant and Cell Physiology 61, 803–813. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Zhao PX, Wu Y, Zhong CQ, Liao H, Li CY, Fu XD, Fang P, Xu P, Xiang CB.. 2023. SUE4, a novel PIN1-interacting membrane protein, regulates acropetal auxin transport in response to sulfur deficiency. New Phytologist 237, 78–87. [DOI] [PubMed] [Google Scholar]
- Zrenner R, Riegler H, Marquard CR, Lange PR, Geserick C, Bartosz CE, Slocum, RD.. 2009. A functional analysis of the pyrimidine catabolic pathway in Arabidopsis. New Phytologist 183, 117–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw data that support the findings of this study, such as ATTED results, are available from the corresponding author, upon request. Further data which were used to generate the heatmap in Fig. 1, can be found from the datasets mentioned in Fig. 1B.