Significance
Cell renewal is critical for tissue homeostasis and regeneration. Using a zebrafish model, we show that an increase in mitochondrial membrane potential and TCA cycle/OXPHOS are required and sufficient for the cell cycle reentry. The elevated mitochondrial activity leads to increased ATP synthesis and ROS production. Elevated mitochondrial ROS induces the mitochondrial expression of Sgk1. Sgk1 alters cell fate by modulating F1Fo-ATP synthase phosphorylation state and ATP synthesis. This signaling loop also mediates mitochondrial metabolism–dependent S phase entry of human breast cancer cells, suggesting that it is an evolutionarily conserved mechanism.
Keywords: IGF/insulin signaling, serum- and glucocorticoid-regulated kinase 1, mitochondrial membrane potential, reactive oxygen species, F1Fo-ATP synthase
Abstract
Many types of differentiated cells can reenter the cell cycle upon injury or stress. The underlying mechanisms are still poorly understood. Here, we investigated how quiescent cells are reactivated using a zebrafish model, in which a population of differentiated epithelial cells are reactivated under a physiological context. A robust and sustained increase in mitochondrial membrane potential was observed in the reactivated cells. Genetic and pharmacological perturbations show that elevated mitochondrial metabolism and ATP synthesis are critical for cell reactivation. Further analyses showed that elevated mitochondrial metabolism increases mitochondrial ROS levels, which induces Sgk1 expression in the mitochondria. Genetic deletion and inhibition of Sgk1 in zebrafish abolished epithelial cell reactivation. Similarly, ROS-dependent mitochondrial expression of SGK1 promotes S phase entry in human breast cancer cells. Mechanistically, SGK1 coordinates mitochondrial activity with ATP synthesis by phosphorylating F1Fo-ATP synthase. These findings suggest a conserved intramitochondrial signaling loop regulating epithelial cell renewal.
Tissue homeostasis and regeneration require a tight control of cell renewal. Several adult tissues, such as intestine, epidermis, and testis, are endowed with robust cell renewal capabilities (1, 2). In the mammalian small intestinal epithelium, for instance, cells are replaced every 4 to 5 d. This was initially attributed to a population of adult stem cells known as Lgr5+ crypt base columnar cells (CBCs) (3, 4). Subsequent studies revealed that other precursor cells in the crypt can revert to a Lgr5+ cell state to carry out the cell renewal function when the Lgr5+ CBS cells are ablated (3, 5). More recent studies suggested that nearly all intestinal epithelial cells, including fully differentiated cells, have the ability to reenter the active cell cycle and replace the lost cells after injuries (6–8). This phenomenon is not unique to the intestine or other tissues with rapid turnover rates, but it is also observed in tissues with very low turnover rates (9). For example, upon partial hepatectomy, mammalian hepatocytes reenter the cell cycle and proliferate to regenerate the liver (10). Another example is the adult zebrafish heart, which displays robust regeneration after injuries (11). Adult zebrafish hearts do not appear to have a stem cell pool. Rather, the newborn cardiomyocytes are derived from differentiated cardiomyocytes (12). These and other findings have led to the notion that many types of differentiated cells are endowed with the ability to reenter the active cell cycle (6). Mounting evidence suggests that unlocking cell plasticity is a major hallmark of cancer cells (13). While it is becoming evident that this cell plasticity plays a key role in tissue homeostasis, regeneration, tumorigenesis (7), the underlying mechanisms are still poorly understood. A major challenge in the field is that our current knowledge is mainly derived from injury models. Whether fully differentiated cells can dedifferentiate and reenter the cell cycle in a physiological context is still not clear (7, 9).
Recently, we have developed a zebrafish model, Tg(igfbp5a:GFP) fish (14, 15), in which Na+–K+–ATPase-rich (NaR) cells are genetically labeled by GFP (14, 15). These polarized cells are structurally and functionally similar to human intestinal and renal epithelial cells (16). While distributed in the adult gills, intestine, and kidney, NaR cells are located on the yolk sac epidermis during the larval stage (17), making them easily accessible for experimental observation and manipulation. NaR cells are one of the five types of ionocytes derived from epidermal stem cells (18). These epidermal stem cells develop into the ionocyte progenitors and keratinocyte progenitors during early embryogenesis. Ionocyte progenitors differentiate into NaR cells around 36 to 48 hours post fertilization (hpf) (15). A hallmark of differentiated NaR cells is the expression of the calcium channel Trpv6. NaR cells become quiescent after expressing Trpv6 (15). Trpv6 is a constitutively open calcium channel and mediates Ca2+ influx continuously and maintains a high level of cytoplasmic free Ca2+ ([Ca2+]c). The high [Ca2+]c activates the conserved protein phosphatase PP2A, which promotes cell quiescence by suppressing Igf1 receptor–PI3 kinase–Akt–Tor signaling. When transferred to the induction medium containing a low concentration of [Ca2+] (0.001 mM), NaR cells rapidly divide (14, 15). BrdU pulse labeling, cell cycle analysis, and in vivo real-time cell-tracing experiments have shown that the increased NaR cell division is due to the reactivation of preexisting NaR cells (14, 15). Importantly, the low calcium stress–induced cell reactivation is specific to NaR cells and there are limited changes in other ionocyte types (14). Using this model, chemical biology screens and genetic studies have elucidated a number of cell autonomous and nonautonomous factors regulating NaR cell reactivation; they all converge onto the IGF–PI3 kinase–Tor signaling pathway (18–22). These findings are in good agreement with the roles of IGF/insulin–PI3 kinase–mTOR signaling in adult stem cell and T cell reactivation in mammals and Drosophila (23–27), suggesting a general and evolutionarily conserved mechanism at work.
The IGF/insulin–PI3 kinase–mTOR signaling pathway is a nutrient-sensitive and evolutionarily ancient pathway regulating metabolism and other biological processes (28). mTOR is a major regulator of mitochondrial activity and metabolism. Although mitochondrial metabolism has traditionally been viewed as a consequence of cellular state, recent studies suggest that mitochondrial metabolism plays key roles in dictating cell fate and cell state (29–31). Two essential and interconnected functions of mitochondria are the TCA cycle and oxidative phosphorylation (OXPHOS) (29, 32). The TCA cycle is critical in ATP production and in providing metabolic intermediates for biosynthesis and epigenetic regulation (29). OXPHOS involves a directional flow of protons from the mitochondrial matrix through the ETC complexes into the intermembrane space, which forms an electrochemical gradient (33, 34). Mitochondrial membrane potential (ΔΨm) is a key indicator of the electrochemical gradient and mitochondrial activity (35). Together with the proton gradient (ΔpH), ΔΨm forms the transmembrane potential, which drives ATP synthesis (35). OXPHOS also produces reactive oxygen species (ROS). Although initially viewed as mere by-products of mitochondrial metabolism, it is now understood that ROS also function as important signaling molecules (29).
In this study, we tested the hypothesis that mitochondrial activity/metabolism plays a key role in regulating NaR cell fate and cell renewal. We provided evidence that mitochondrial activity/metabolism is crucial in promoting NaR cell reactivation and proliferation. Mechanistically, elevated mitochondrial metabolism increases ATP synthesis and elevates mitochondrial ROS production. ROS signaling induces the expression and mitochondrial localization of serum- and glucocorticoid-regulated kinase 1 (Sgk1). Sgk1 coordinates mitochondrial activity/metabolism change with ATP synthesis by phosphorylating F1Fo-ATP synthase. This signaling loop promotes both zebrafish NaR cell reactivation and human breast cancer cell S phase entry.
Results
Elevated Mitochondrial Activity Acts Downstream of IGF Signaling to Promote NaR Cell Reactivation.
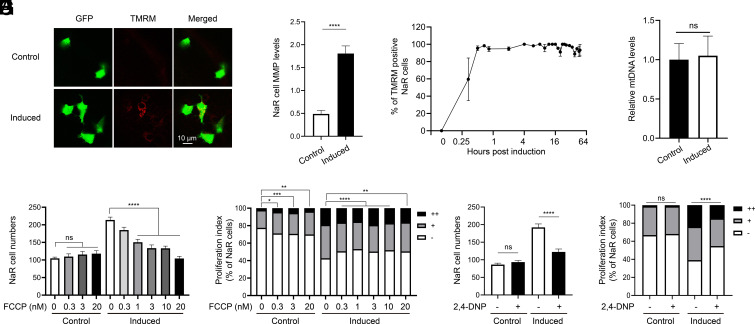
Using Tg(igfbp5a:GFP) larvae (14, 15), we measured changes in mitochondrial membrane potential (ΔΨm) by TMRM staining. A robust increase in ΔΨm was detected in reactivated NaR cells (Fig. 1 A and B). The elevation in ΔΨm was detected in nearly all NaR cells within an hour after the induction and was sustained for at least 48 h (Fig. 1C). Similar results were obtained by MitoTracker-Red staining (SI Appendix, Fig. S1 A and B). No change was detected in the mitochondrial mass (Fig. 1D). Additionally, treatment with 2,4-DNP or FCCP, two OXPHO uncouplers, impaired NaR cell reactivation and proliferation (Fig. 1 E–H). These results suggest that mitochondrial activity, but not mitochondrial mass, is elevated during NaR cell reactivation and promotes cell reactivation. This idea was tested further by inhibiting the ETC complexes which mediate the directional proton flow (33, 34). Treatment of fish larvae with ETC complexes I, III, and IV inhibitors rotenone, antimycin, and sodium azide all inhibited NaR cell reactivation (SI Appendix, Fig. S2).
Fig. 1.
Elevated mitochondrial membrane potential (MMP) promotes NaR cell reactivation. (A and B) MMP levels in the control and reactivated NaR cells. Tg(igfbp5a:GFP) larvae (3 dpf) were transferred to the control or induction medium. Two days later, MMP levels in NaR cells were measured after TMRM staining and normalized by GFP signal. Representative images are shown in A and quantified results in B. n = 221 to 447 cells from multiple fish. (C) Time course of MMP change. Tg(igfbp5a:GFP) larvae (3 dpf) were transferred to the induction medium. A subset of larvae were randomly sampled at the indicated time point and % of TMRM-positive NaR cells was measured and shown. n = 2 to 6 fish/time point. (D) Mitochondrial DNA levels. NaR cells were isolated by FACS from fish described in A and B. The levels of mitochondrial 16S rRNA gene were measured and normalized by the levels of nuclear aryl hydrocarbon receptor 2 gene. n = 4. (E−H) Tg(igfbp5a:GFP) larvae (3 dpf) were transferred to the control or induction medium containing the indicated concentration of FCCP. (E and F) or 1 μM 2,4-DNP. (G and H). Two days later, NaR cell number (E and G) and NaR cell proliferation index (F and H) were determined and shown. n = 11 to 26 fish/group. The cell proliferation index was determined by counting NaR cells that divided 0, 1, or 2 times (denoted by −, +, and ++, respectively) over the course of the experiment and presented as % of total NaR cells. All other data are shown as mean ± SEM. ns, not significant. *, **, ***, and **** indicate P < 0.05, 0.01, 0.001, and 0.0001, respectively.
We next investigated the relationship between elevated ΔΨm and IGF signaling. Previous studies have shown that NaR cell reactivation in papp-aa−/− fish, which lacks the IGF-binding protein 5 proteinase Papp-aa, is impaired due to defective IGF signaling in these cells (20). In contrast, NaR cells in trpv6−/− fish proliferate unrestrictedly due to constitutively elevated Igf1r–PI3 kinase–Akt–Tor signaling (19). While the induction medium treatment resulted in a robust increase in ΔΨm in the NaR cells of wild-type and heterozygous sibling fish, this increase was not observed in the NaR cells of pappaa−/− fish (SI Appendix, Fig. S3A). In trpv6−/− fish, NaR cells exhibited elevated ΔΨm when kept in the control medium (SI Appendix, Fig. S3B). Next, we treated Tg(igfbp5a:GFP) larvae with BMS-754807, an IGF1 receptor inhibitor (36). This drug inhibited ΔΨm increase in a dose-dependent manner (SI Appendix, Fig. S3C). Finally, rapamycin treatment abolished the NaR cell ΔΨm increase (SI Appendix, Fig. S3D). Collectively, these results suggest that elevated mitochondrial activity acts downstream of IGF and Tor signaling.
Mitochondrial Metabolism Plays an Essential Role in NaR Cell Reactivation.
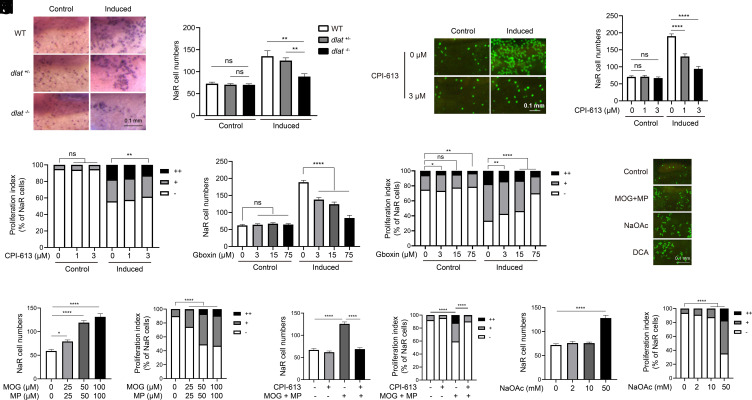
The involvement of the TCA cycle/OXPHOS was investigated using zebrafish noa−/− larvae, which lack pyruvate dehydrogenase (PDH) activity due to the deletion of the PDH E2 subunit dihydrolipoamide s-acetyltransferase (dlat) (37). While NaR cell reactivation was observed in the siblings, it was impaired in the noa−/− fish (Fig. 2 A and B). Likewise, inhibition of PDH by CPI-613 (38) (SI Appendix, Fig. S4A) reduced NaR cell reactivation in a dose-dependent manner (Fig. 2 C–E). CPI-613 treatment reduced NaR cell proliferation in trpv6−/− fish to the level of its siblings (SI Appendix, Fig. S4 B and C). Inhibition of pyruvate transport from the cytoplasm into the mitochondria by CHC (39) also impaired NaR cell reactivation (SI Appendix, Fig. S4 D and E). Gboxin, an inhibitor of OXPHOS (40), had a similar effect (Fig. 2 F and G).
Fig. 2.
Elevated mitochondrial metabolism is required and sufficient to reactivate NaR cells. (A and B) Genetic disruption of the PDH complex impairs NaR cell reactivation. The progenies of dlat+/− intercrosses were transferred to the control or induction medium at 3 dpf. Two days later, NaR cells were labeled by in situ hybridization and quantified. Fish were genotyped individually afterward. Representative images are shown in A and NaR cell number in B. n = 13 to 40 fish/group. (C–E) Effect of CPI-613. Tg(igfbp5a:GFP) larvae (3 dpf) were transferred to the control or induction medium containing the indicated doses of CPI-613. Two days later, NaR cell number (D) and proliferation index (E) were measured and shown. Representative images are shown in C. n = 25 to 31 fish/group. (F and G) Effect of Gboxin. Tg(igfbp5a:GFP) embryos were raised and treated as described in C with the indicated doses of Gboxin. NaR cell number (F) and proliferation index (G) were measured and shown. n = 24 to 33 fish/group. (H–N) Effects of dimethyl 2-oxoglutarate (MOG)+methyl pyruvate (MP), sodium acetate (NaOAc), and CPI-613 (3 μM). Tg(igfbp5a:GFP) larvae were transferred to the control medium containing the indicated chemicals at 3 dpf. Two days later, NaR cell number (I, K, and M) and proliferation index (J, L, and N) were measured and shown. n = 15 to 31 fish/group. Representative images are shown in H. The proliferation index results are shown as % of total NaR cells counted. All other data are shown as mean ± SEM. ns, not significant. *, **, ***, and **** indicate P < 0.05, 0.01, 0.001, and 0.0001, respectively.
To test whether an increase in TCA cycle/OXPHOS activity is sufficient, fish were treated with MOG and MP. MOG and MP can be cleaved to generate α-ketoglutarate and pyruvate in cells, supplying key intermediates so as to increase TCA cycle and mitochondrial activity (41). MOG+MP treatment elevated ΔΨm and increased NaR cell reactivation under the control condition (Fig. 2 H–J and SI Appendix, Fig. S1C). Cotreatment with CPI-613 abolished the MOG+MP-induced ΔΨm increase and NaR cell reactivation (Fig. 2 K and L and SI Appendix, Fig. S1C). Similarly, treatment with DCA, which indirectly activates PDH by inhibiting pyruvate dehydrogenase kinase (42), increased NaR cell reactivation (SI Appendix, Fig. S4 F and G). Finally, addition of sodium acetate, a precursor of acetyl-coA (43) into the control medium, was sufficient to induce NaR cell reactivation (Fig. 2 M and N). These results suggest that elevated TCA cycle/OXPHOS is both necessary and sufficient to promote NaR cell reactivation.
ATP Synthesis and ROS Signaling Are Involved in Cell Reactivation.
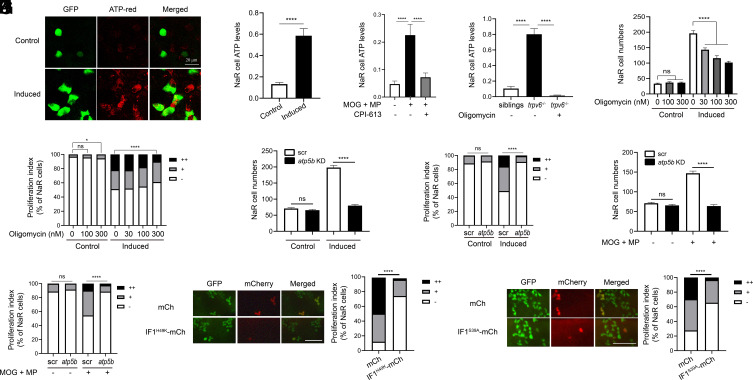
We next measured changes in NaR cell mitochondrial ATP levels. The induction medium treatment resulted in elevated mitochondrial ATP levels in nearly all GFP-positive NaR cells (Fig. 3A). Quantification results showed a fourfold increase in ATP levels in reactivated NaR cells (Fig. 3B). Increased mitochondrial ATP signal was also detected in a small number of GFP-negative cells located in the yolk sac epidermis, suggesting that this effect is predominantly, but not exclusively, in NaR cells. This finding is in line with a previous study showing low calcium stress caused a modest increase in NCC cells (14). MOG+MP treatment increased NaR cell mitochondrial ATP levels in fish kept in the control medium and this increase was abolished by CPI-613 cotreatment (Fig. 3C), but CPI-613 treatment alone had no effect (SI Appendix, Fig. S5). In continuously dividing NaR cells in trpv6−/− larvae, the mitochondrial ATP levels were eightfold greater than those in their siblings (Fig. 3D). Addition of the F1Fo-ATP synthase inhibitor oligomycin reduced the ATP levels to nearly undetectable levels (Fig. 3D). Oligomycin treatment also inhibited NaR cell reactivation in a dose-dependent manner (Fig. 3 E and F).
Fig. 3.
NaR cell reactivation requires ATP synthesis. (A and B) ATP levels. Tg(igfbp5a:GFP) larvae (3 dpf) were transferred to the control or induction medium. After 18 h, NaR cell mitochondrial ATP levels were measured. Representative images are shown in A and quantified results in B. n = 205 to 447 cells from multiple fish. (C) Effects of MOG + MP and CPI-613. Tg(igfbp5a:GFP) fish (3 dpf) were transferred to the control medium containing DMSO, 100 μM MOG + 100 μM MP, and/or 3 μM CPI-613. Five hours later, NaR cell mitochondrial ATP levels were measured and shown. n = 157 to 233 cells from multiple fish. (D) Elevated mitochondrial ATP levels in trpv6−/− NaR cells. Progeny from trpv6+/−; Tg(igfbp5a:GFP) intercrosses were raised to 3 dpf. They were transferred to the control medium with or without 200 nM oligomycin. After measuring NaR cell mitochondrial ATP levels, the genotype of each fish was determined. n = 112 to 315 cells from multiple fish. (E and F) Effect of oligomycin. Tg(igfbp5a:GFP) fish (3 dpf) were transferred to the control or induction medium containing oligomycin at the indicated concentrations. Two days later, NaR cell number (E) and proliferation index (F) were measured and shown. n = 12 to 34 fish/group. (G−J) CRISPR-Cas9-mediated atp5b deletion. Tg(igfbp5a:GFP) embryos injected with targeting gRNAs and Cas9 mRNA were raised to 3 dpf and transferred to the control or induction medium without (G and H) or with 100 μM MOG + 100 μM MP (I and J). Two days later, NaR cell number (G and I) and proliferation index (H and J) were measured and shown. n = 14 to 42 fish/group. (K and L) NaR cell–specific expression of IF1H49K (K) or IF1S39A (L). Tg(igfbp5a:GFP) embryos injected with BAC-mCherry, BAC-IF1H49K, or BAC- IF1S39A-mCherry DNA were raised to 3 dpf and transferred to the induction medium. Two days later, NaR cells expressing the transgene were identified, and cell proliferation index was measured. Representative images are shown in the left panel and quantified data in the right. Scale bar represents 0.1 mm. n = 102 to 163 in K and n = 29 to 148 cells from multiple fish in L. The proliferation index data are shown as % of total NaR cells counted. All other data are shown as mean ± SEM. ns, not significant. *, **, ***, and **** indicate P < 0.05, 0.01, 0.001, and 0.0001, respectively.
The idea that elevated ATP synthesis is required for cell reactivation was tested further using a F0 CRISPR/Cas9 gene deletion approach (44, 45). Deletion of atp5b, which encodes the β subunit of F1Fo-ATP synthase, abolished NaR cell reactivation induced by the induction medium as well as by MOG-MP treatment (Fig. 3 G−J and SI Appendix, Fig. S6). One caveat of the above manipulations is that they affect ATP synthesis in all cell types. To determine the role of endogenous ATP synthesis in NaR cells, ATPase inhibitory factor 1 (IF1) was expressed in a subset of NaR cells in Tg(igfbp5a:GFP) fish using a Tol2 transposon–mediated genetic mosaic assay (18). IF1 binds to and inhibits F1Fo-ATP synthase in a pH-dependent manner (46). IF1H49K is a pH-insensitive and constitutively active form of IF1 (47). When expressed randomly in a subset of NaR cells, IF1H49K inhibited NaR cell reactivation (Fig. 3K). NaR cell–specific expression of another constitutively active IF1 mutant, hIF1S39A (48), had similar effect (Fig. 3L).
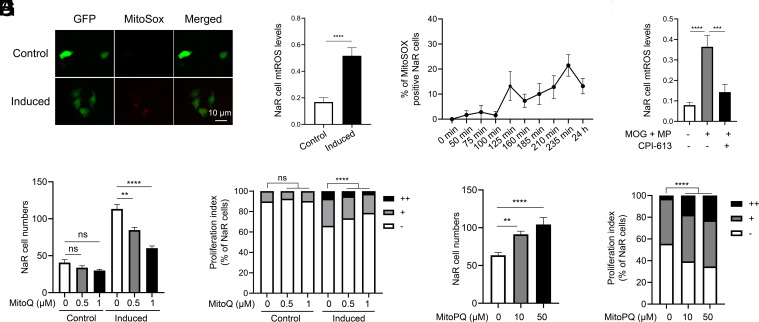
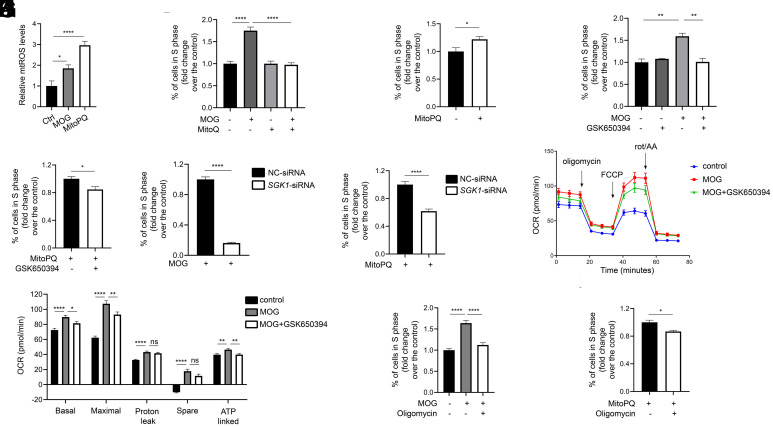
OXPHOS uses O2 as the final electron acceptor to produce ATP and this process also generates mitochondrial reactive oxygen species (ROS) (49, 50). MitoSOX Red staining detected a significant increase in mitochondrial ROS levels (mtROS) in reactivated NaR cells (Fig. 4 A and B). The signal is specific as it was abolished by the addition of an ROS scavenger, MitoQ (SI Appendix, Fig. S7A). The increase in mtROS was detected within 2 h and sustained thereafter (Fig. 4C). MOG+MP treatment increased mtROS levels in NaR cells (Fig. 4D) and CPI-613 cotreatment abolished this effect. CPI-613 alone had no effect (SI Appendix, Fig. S7B). MitoQ inhibited NaR cell reactivation in a dose-dependent manner (Fig. 4 E and F). Conversely, treatment of fish with MitoPQ, which increases mtROS (51), induced NaR cell reactivation in a dose-dependent manner (Fig. 4 G and H). These results suggest that mtROS signaling plays a crucial role in regulating NaR cell renewal.
Fig. 4.
ROS signaling promotes NaR cell reactivation. (A and B) Mitochondrial reactive oxygen species (mtROS) levels. Tg(igfbp5a:GFP) larvae were transferred to the control or induction medium at 3 dpf. One day later, NaR cell mtROS levels were measured. Representative images are shown in A and quantified results in B. n = 201 to 390 cells from multiple fish. (C) Time course changes. Tg(igfbp5a:GFP) larvae (3 dpf) were transferred to the induction medium. A subset of larvae were randomly sampled at the indicated time point and % of MitoSOX-positive NaR cells was determined and shown. n = 4 fish/group. (D) Effects of MOG+MP and CPI-613. Tg(igfbp5a:GFP) larvae (3 dpf) were transferred to the control or induction medium with or without 100 μM MOG + MP and 3 μM CPI-613. One day later, NaR cell mtROS levels were measured and shown. n= 197 to 276 cells from multiple fish. (E and F) mtROS signaling is required. Tg(igfbp5a:GFP) larvae were transferred to the control or induction medium containing the indicated dose of MitoQ at 3 dpf. Two days later, NaR cell number (E) and proliferation index (F) were measured and shown. n = 15 to 42 fish/group. (G and H) ROS signaling is sufficient. Tg(igfbp5a:GFP) larvae were raised in the control medium containing the indicated concentrations of MitoPQ from 3 to 5 dpf. NaR cell number (G) and proliferation index (H) were measured and shown. n = 15 to 42 fish/group. The proliferation index results are shown as % of total NaR cells counted. All other data are shown as mean ± SEM. ns, not significant. *, **, ***, and **** indicate P < 0.05, 0.01, 0.001, and 0.0001, respectively.
Sgk1 Coordinates ROS Signaling with ATP Synthesis by Modulating F1Fo-ATP Synthase Phosphorylation.
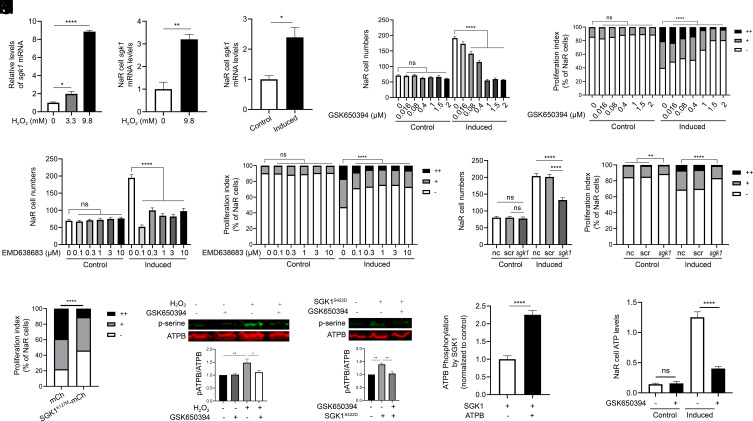
We tested the idea that Sgk1 acts as an effector molecule downstream of the ROS signaling. SGK1/Sgk1 is a protein kinase that acts downstream of the PI3 kinase (52). Previous studies demonstrated that ROS signaling increases SGK1 expression in human cell culture systems (53–55). H2O2 treatment of zebrafish larvae increased whole-body sgk1 mRNA levels in a dose-dependent manner (Fig. 5A). Next, Tg(igfbp5a:GFP) larvae were treated with H2O2, and NaR cells were isolated by FACS sorting. qRT-PCR analysis results showed a threefold increase in sgk1 mRNA levels in NaR cells (Fig. 5B). A significant increase was also detected in reactivated NaR cells. (Fig. 5C). To determine the possible role of Sgk1 in regulating NaR cell renewal, zebrafish larvae were treated with two distinct SGK1 inhibitors, GSK650394 and EMD-63868. GSK650394 inhibited NaR cell reactivation in a dose-dependent manner (Fig. 5 D and E). EMD-638683 had a similar effect (Fig. 5 F and G). CRISPR/Cas9-based deletion of sgk1 impaired NaR cell reactivation (Fig. 5 H and I and SI Appendix, Fig. S8). This action is specific because CRISPR/Cas9-mediated deletion of sgk2 had no such effect (SI Appendix, Fig. S9). Finally, SGK1K127M, a dominant-negative form of SGK1, was expressed in NaR cells specifically using the genetic mosaic assay. SGK1K127M expression impaired the reactivation in a cell-autonomous fashion (Fig. 5J).
Fig. 5.
ROS signaling–induced Sgk1 mitochondrial expression is critical. (A) Induction of sgk1 expression by H2O2. Zebrafish larvae (3 dpf) were transferred to the control medium containing the indicated doses of H2O2. After 3 h, the larvae were collected and sgk1 mRNA levels measured. n = 4. (B) Expression and ROS induction of sgk1 mRNA in NaR cells. Tg(igfbp5a:GFP) larvae were treated with H2O2 at the indicated dose for 3 h. NaR cells were isolated by FACS sorting and analyzed by qRT-PCR. n = 3. (C) Increased sgk1 mRNA levels in reactivated NaR cells. NaR cells were isolated by FACS sorting from Tg(igfbp5a:GFP) larvae treated with the control or induction medium for 18 h. sgk1 mRNA levels were measured and shown. n = 4. (D–G) Effects of SGK1 inhibitors. Tg(igfbp5a:GFP) embryos (3 dpf) were transferred to the control or induction medium containing the indicated doses of GSK-650394 (D and E) or EMD638683 (F and G). Two days later, NaR cell number (D and F) and proliferation index (E and G) were determined as described in Fig. 1 and shown. n = 15 to 41 fish/group. (H and I) CRISPR/Cas9-mediated sgk1 deletion. Tg(igfbp5a:GFP) embryos were injected with sgk1 targeting gRNAs and Cas9 mRNA at the one-cell stage. They were raised to 3 dpf and transferred to the control or induction medium. Two days later, NaR cell number (H) and proliferation index (I) were measured and shown. n = 30 to 58 fish/group. (J) Effect of SGK1K127M expression in NaR cells. Tg(igfbp5a:GFP) embryos were injected with the indicated BAC-mCherry DNA at the one-cell stage. They were raised to 3 dpf and transferred to the induction medium. Two days later, cell proliferation index in mCherry-expressing NaR cells was determined and shown. n = 45 to 211 from multiple fish. (K) HEK293 cells were treated with DMSO or GSK650394 overnight, followed by 2-h H2O2 treatment (250 μM H2O2). Cells were subjected to IP using an anti-ATP5B antibody. The IP samples were analyzed by western blot using the indicated antibodies. Representative results are shown in the top panel. Ratio of phospho- and total ATP5B is shown at the bottom. n = 4. (L) HEK293 cells were transfected with Sgk1S422D-mcherry or an empty vector, followed by GSK650394 treatment. Cells were analyzed and shown as described in K. n = 3. (M) SGK1 phosphorylates ATP5B. n = 5. Human SGK1 was incubated with or without ATP5B, and changes in ATP5B phosphorylation were measured. (N) Elevated mitochondrial ATP synthesis in reactivated NaR cells is Sgk1 dependent. Tg(igfbp5a:GFP) larvae (3 dpf) were transferred to the control or induction medium for 14 h. They were then treated with DMSO or GSK650394 for 4 h. ATP levels were analyzed and shown. n = 178 to 609 cells from multiple fish. The proliferation index results are shown as % of total NaR cells counted. All other data are shown as mean ± SEM. ns, not significant. *, **, ***, and **** indicate P < 0.05, 0.01, 0.001, and 0.0001, respectively.
Human SGK1 has a putative mitochondrial targeting sequence. This sequence is conserved in zebrafish Sgk1 but absent in zebrafish Sgk2 (SI Appendix, Figs. S9A and S10A). SGK1 was found to localize to the mitochondria in mouse mammary gland epithelial cells (54) and in human breast cancer cells under sorbitol-induced osmotic stress (SI Appendix, Fig. S10 B and C). In vitro, mouse Sgk1 was capable of increasing F1Fo-ATP synthase phosphorylation (54). We postulated that ROS signaling–induced SGK1/Sgk1 expression may increase ATP production by phosphorylating F1Fo-ATP synthase. HEK 293 cells were used to test this hypothesis because of antibody availability and the high transfection efficacy. H2O2 treatment significantly increased the levels of phosphorylated ATP5B, the F1Fo-ATP synthase β subunit (Fig. 5K). This increase was abolished by GSK650394, suggesting that this is a SGK1-dependent event (Fig. 5K). Likewise, expression of SGK1S422D, a constitutively active form of SGK1, increased the levels of phosphorylated ATP5B and this effect was abolished by GSK650394 (Fig. 5L). To determine whether SGK1 phosphorylates ATP5B directly, in vitro kinase assays were carried out. Incubation of SGK1 with ATP5B protein resulted in a significant increase in ATP5B phosphorylation (Fig. 5M). Consistent with these in vitro results, GSK650394 treatment abolished the increase in ATP levels in NaR cells under the induction condition (Fig. 5N). Collectively, these data suggest that elevated ROS signaling induces mitochondrial Sgk1 expression, which promotes cell reactivation by phosphorylating F1Fo-ATP synthase and increasing ATP synthesis.
SGK1 Mediates ROS Signaling-Induced Human Breast Cancer Cell Proliferation.
We used MDA-MB-231 breast cancer cells to investigate the role of ROS-dependent SGK1 mitochondrial expression in cell cycle regulation. These cells undergo proliferation in response to increases in mitochondrial metabolism (56). qRT-PCR analysis showed that SGK1 is the predominant form of SGKs expressed in these cells and its mRNA levels were nearly 184-fold higher than those of SGK2 (SI Appendix, Fig. S12). H2O2 treatment strongly induced SGK1 mitochondrial localization (SI Appendix, Fig. S10 B and C). MOG treatment resulted in a significant increase in mtROS levels (Fig. 6A). MOG treatment increased the percentage of cells entering the S phase and this effect was abolished by MitoQ (Fig. 6B). Conversely, treatment of cells with MitoPQ (Fig. 6A) stimulated cells to enter the S phase (Fig. 6C). Inhibition of SGK1 by GSK-650394 abolished both MOG- and MitoPQ-induced cell proliferation (Fig. 6 D and E). GSK-650394 treatment resulted in a modest but statistically significant decrease in SGK1 mRNA levels, while it did not change SGK2 mRNA levels (SI Appendix, Fig. S13). Likewise, siRNA-mediated SGK1 knockdown inhibited cell proliferation in response to MOG and MitoPQ treatments (Fig. 6 F and G and SI Appendix, Fig. S14). The SGK1 siRNA caused a fivefold, highly significant decrease in SGK1 mRNA levels, while it had no effect on SGK2 mRNA levels (SI Appendix, Fig. S14). Next, seahorse assays were carried out to measure oxygen consumption rate (ORC) changes. As shown in Fig. 6 H and I, MOG treatment increased the basal ORC, the maximal mitochondrial respiratory capacity, and ATP production. This effect was inhibited by GSK-650394 cotreatment (Fig. 6 H and I). MOG treatment also increased proton leak and the spare or reserved capacity but GSK-650394 did not have significant effect on these parameters (Fig. 6 H and I). Finally, inhibition of F1Fo-ATP synthase by oligomycin impaired MOG- and MitoPQ-stimulated cell proliferation (Fig. 6 I and J). Oligomycin alone had no such effect (SI Appendix, Fig. S15). These results suggest that SGK1 mediates the mitochondrial metabolism and ROS signaling–induced breast cancer cell S phase entry.
Fig. 6.
The mitochondrial metabolism–ROS–SGK1–ATP synthase signaling loop stimulates human breast cancer cell S phase entry. (A) MOG and MitoQ increase mtROS levels. MDA-MB-231 cells were treated with MOG (4 mM) or MitoPQ (5 μM) for 1 d. mtROS levels were measured and shown. n = 6. (B) mtROS signaling mediates MOG-induced cell proliferation. After MDA-MB-231 cells were treated with MOG (4 mM) and/or MitoQ (1 μM) for 2 d, % of cells in S phase was determined by flow cytometry and expressed as fold changes over the control group. n = 4. (C) Effect of MitoPQ. After MDA-MB-231 cells were treated with MitoPQ (5 μM) for 2 d, % of cells in S phase was determined and expressed as fold change over the control group. n = 7. (D and E) SGK1 mediates MOG- and MitoPQ-induced cell proliferation. MDA-MB-231 cells were treated with MOG (4 mM), MitoPQ (5 μM), and/or GSK650394 (20 μM) for 2 d. % of cells in S phase was determined and expressed as fold change over the control group. n = 3. (F and G) Knockdown of SGK1 abolishes MOG (F)- and MitoPQ (G)-induced cell proliferation. MDA-MB-231 cells were transfected with the control (NC-siRNA) or SGK1-siRNA. One day after transfection, MOG (4 mM) or MitoPQ (5 μM) was added. % of cells in S phase was determined 2 d later and expressed as fold change over the control group. n = 6. (H and I) Inhibition of SGK1 reduces oxygen consumption rate (OCR). MDA-MB-231 cells were treated with MOG and/or GSK650394 (20 μM) for 4 h and subjected to seahorse assays. Oligomycin, FCCP, and rotenone/AA were spiked into cells at various timepoints, and OCR was measured. Calculated parameters are shown in I. n = 8. Similar results were obtained in a separate experiment. (J and K) F1Fo-ATP synthase activity is required in MOG (J)- and ROS (K)-induced cell proliferation. After MDA-MB-231 cells were treated with MOG (4 mM), MitoPQ (5 μM), and/or oligomycin (5 μM) for 2 d, % of cells in S phase was determined and expressed as fold change over the control group. n = 6. Data are shown as mean ± SEM. ns, not significant. *, **, ***, and **** indicate P < 0.05, 0.01, 0.001, and 0.0001, respectively.
Discussion
In this study, we provided several lines of independent evidence suggesting that mitochondrial activity/metabolism plays a key role in regulating epithelial cell fate and renewal. We found that mitochondrial activity and the TCA cycle/OXPHOS are maintained at relatively low levels in quiescent NaR cells. During cell reactivation, the mitochondrial activity and the TCA cycle/OXPHOS are elevated. This leads to increased ATP synthesis and higher mtROS levels. Elevated mtROS signaling induces the expression and mitochondrial localization of Sgk1. Sgk1 links changes in mitochondrial metabolism/activity to ATP synthesis by modulating the F1Fo-ATP synthase phosphorylation state. The ROS signaling–SGK1–F1Fo-ATP synthase signaling loop not only plays a critical role in zebrafish NaR cell reactivation but also stimulates human breast cancer cell S phase entry.
Zebrafish use NaR cells to take up Ca2+ from the surrounding aquatic habitat. When facing a low [Ca2+] environment, NaR cells are reactivated and undergo robust proliferation to yield more NaR cells in order to maintain body Ca2+ homeostasis (14, 15, 17, 18). This adaptive NaR cell reactivation is regulated by the IGF–PI3 kinase–Akt–Tor signaling pathway (14, 15, 18–20). In the present study, we show that NaR cell reactivation is accompanied by a robust and sustained increase in ΔΨm. Furthermore, in vivo measurement detected a major increase in mitochondrial ATP levels in reactivated NaR cells. Given that we observed no significant changes in mitochondrial mass, these results indicate that the mitochondrial activity is elevated in reactivated cells. This increase is likely resulted from the activation of IGF–PI3 kinase–Akt–Tor signaling. This conclusion is supported by ΔΨm changes observed in pappaa−/− and trpv6−/− fish and by the fact that the ΔΨm increase was abolished by inhibition of the IGF1 receptor and Torc1/2. It is also in good agreement with previous studies showing IGF and Torc1/2 signaling as essential in NaR cell reactivation (14, 15, 18–20). Our results showed that the elevated mitochondrial activity and metabolism are critical. Dissipating the mitochondrial membrane potential by 2,4-DNP and FCCP impaired NaR cell reactivation. Treatment with inhibitors for ETC complexes I, III, and IV also impaired NaR cell reactivation. Genetic deletion and pharmacological inhibition of PDH abolished NaR cell reactivation. Conversely, increasing TCA cycle/OXPHOS activity was sufficient to promote NaR cell reactivation. These results argue strongly that mitochondrial metabolism plays a key role in NaR cell reactivation.
Most published studies on the role of mitochondrial metabolism in epithelial renewal focus on adult stem cells. Whether these genetic manipulations alter the state of differentiated epithelial cells has not been fully explored. Knockout of the transcriptional repressor YinYang 1 in mouse intestinal stem cells up-regulated ETC genes and increased OXPHOS. This resulted in stem cell reactivation, proliferation, and eventually exhaustion (57). Deletion of the glycolytic enzyme pyruvate kinase M2 isoform (Pkm2) resulted in elevated OXPHOS and ATP production in mouse intestinal stem cells, and this led to the development of colon cancer (58). Similarly, intestinal stem cells in mutant Drosophila carrying a mutation impairing ETC proliferated at a slower rate (59). These findings imply that an increase in OXPHOS is necessary to reactivate quiescent intestinal stem cells and promote their proliferation. In contrast, Rodríguez-Colman et al. reported that quiescent intestinal stem cells display high mitochondrial activity and greater OXPHOS compared with Paneth cells (60). Inhibition of mitochondrial activity impairs intestinal stem cell reactivation (60). Along the same line, genetic deletion of the mitochondrial pyruvate carrier (MPC) in mouse intestinal stem cells increased their proliferation rate, while MPC overexpression suppressed it (61). The reasons underlying these conflicting findings are not entirely clear, but these studies all used injury assays in stable mutant animals. Recent studies suggest that permanent knockouts can cause transcriptional adaptation and/or other compensatory mechanisms due to the inherent genetic robustness in multicellular organisms (62). Our model differs from these studies in that we investigated the reactivation cells under a physiological stress. Additionally, we used a combination of permanent and transient gene deletion, pharmacological inhibition/activation, and genetic mosaic assays.
A key finding made in this study is that increased mtROS levels promote cell reactivation by inducing Sgk1 expression in the mitochondria. Several lines of evidence support this conclusion. First, there is a significant increase in mtROS levels in reactivated NaR cells. The elevated mtROS are functionally important as NaR cell reactivation was abolished by treatment with an mtROS scavenger. Second, ROS treatment increased sgk1 mRNA expression in NaR cells. Third, ROS treatment resulted in a robust increase of SGK1 in the mitochondria in human breast cancer cells. Inhibition of Sgk1/SGK1 impaired NaR cell reactivation in vivo and inhibited ROS-induced human breast cancer proliferation in vitro. CRISPR/Cas9-mediated deletion of sgk1 blocked NaR cell reactivation. Likewise, siRNA-mediated knockdown of SGK1 inhibited breast cancer proliferation. Finally, dominant-negative inhibition of Sgk1 in NaR cells specifically impaired NaR cell reactivation. We postulated that Sgk1/SGK1 regulates cell reactivation by modulating F1Fo-ATP synthase phosphorylation state and ATP synthesis. In support of this idea, a robust increase in mitochondrial ATP levels was detected in reactivated NaR cells. Similarly, there was a robust increase of mitochondrial ATP levels in continuously proliferating NaR cells in trpv6−/− mutant fish. Genetic deletion and pharmacological inhibition of Sgk1 reduced mitochondrial ATP levels and blocked NaR cell reactivation. Inhibition or siRNA-mediated knockdown of SGK1 abolished ROS-stimulated S phase entry of human breast cancer cells. Finally, inhibition of F1Fo-ATP synthase by oligomycin treatment impaired NaR cell reactivation in vivo. These data strongly argue that mtROS signaling promotes cell reactivation by inducing mitochondrial Sgk1 expression. While our studies focused on NaR cell reactivation under low calcium stress, it is possible that the mtROS–Sgk1 signaling loop may operate more broadly. In addition to NaR cells, increased mitochondrial ATP signal was detected in a very small number of non-GFP cells. These cells are probably NCC cells because low calcium stress caused a modest increase in NCC proliferation (14). A recent report showed that SGK1 is required and sufficient to increase ATP production in extracellular matrix–detached (but not attached) human cancer cells, although increased GLU1-mediated glucose uptake was proposed to be the underlying mechanism (63). In this study, we provided evidence that SGK1 mediates mitochondrial metabolism and ROS signaling–induced human breast cancer cell proliferation.
Although SGK1 is structurally related to AKT, it lacks a PH domain. It was reported that SGK1 activation occurs at a distinct subcellular compartment from that of AKT (64). In this study, we found that SGK1/Sgk1 has a conserved mitochondrial targeting sequence and that SGK1 is localized in the mitochondria under elevated mitochondrial activity and ROS signaling. This is in agreement with a study reporting an increase of SGK1 expression in the mitochondria under osmotic stress (54). Several F1Fo-ATP synthase subunits are known to be posttranslationally modified by protein kinases and these posttranslational modifications are thought to be an important means of regulation of ATP synthesis (65). In this study, we found that H2O2 treatment increased ATP5B phosphorylation and elevated ATP production in a SGK1/Sgk1-dependent manner. Likewise, expression of a constitutively active form of SGK1 was sufficient to increase the levels of phosphorylated ATP5B, and this effect was abolished by an SGK1 inhibitor. Our in vitro kinase assay results provided unequivocal evidence that SGK1 directly phosphorylates ATP5B, suggesting that SGK1 affects ATP synthesis by modulating ATP5B phosphorylation state.
In summary, the results of this study indicate a previously unrecognized intramitochondrial signaling loop which links mitochondrial activity to ATP production and regulates epithelial cell renewal. A key module in this regulatory loop is Sgk1 mitochondrial expression induced by ROS signaling. Mitochondrial Sgk1 changes cell state by stimulating ATP synthesis via modulating F1Fo-ATP synthase phosphorylation, which in turn stimulates ATP synthesis. Given that approximately 90% of human cancers arise from epithelial cells and cell plasticity is a major hallmark of cancer (66), the regulation of epithelial cell renewal by Sgk1 may have broader implications. Indeed, our results suggested a critical role of SGK1 in mediating mitochondrial metabolism– and ROS signaling–regulated S phase entry of human breast cancer cells. SGK1 was first discovered in rat tumor cells as a gene regulated by serum and glucocorticoids and has been implicated in ion channels and kidney function (67–69). SGK1 was subsequently found to play a critical role in the proliferation of cancer cells harboring PI3 kinase mutations (70, 71). Knockdown of SGK1 impaired basilar smooth muscle cell progress from G0/G1 phase to S phase (72) and overexpression of SGK1 stimulated cultured colon cancer cell proliferation (73). SGK1 expression is elevated in colon cancer cells and in other cancers (74). Sgk1-/- mutant mice had reduced colon tumor formation when exposed to carcinogens (75). Future studies are needed to determine the potential involvement of the ROS signaling–SGK1–F1Fo-ATP synthase signaling loop in colon cancer and other epithelial derived cancers.
Methods
A summary of the Methods is reported below. Please refer to SI Appendix for additional details.
Mitochondrial membrane potential was measured by TMRM or Mito-Tracker Red staining. Isolation of NaR cells by FCAS and qPCR was performed as previously described (15). NaR cell number and proliferation index quantification were determined as described in previous papers (refs. 18 and 21). ATP and mtROS levels were measured with ATP-Red staining and MitoSOX staining. CRISPR/Cas9-mediated gene deletion and transposon-mediated genetic mosaic assay were performed as previously reported (18).
Plasmid transient transfection of human cells was performed using Lipofectamine 3000 and siRNA transfection using Lipofectamine RNAiMAX. IP was performed with the Dynabeads Protein G kit and analyzed by western blotting.
SGK1 kinase assays were performed using a Promega SGK1 Assay kit. The oxygen consumption rate was measured by the Seahorse XF extracellular flux assay.
Statistical analyses were performed using unpaired two-tailed t test or ANOVA when appropriate.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Dr. José M. Cuezva, Autonomous University of Madrid; Dr. Arohan R. Subramanya, University of Pittsburgh; Dr. Yonghua Sun, the Institute of Hydrobiology, Chinese Academy of Sciences; Dr. Heather Flanagan-Steet, Clemson University; and Dr. Yanzhuang Wang, University of Michigan, for providing reagents, cell lines, and fish lines. We are grateful to Dr. Chunyang Zhang for her advice on mitochondrial DNA quantification. This work was supported by NSF grant IOS-1557850 and NSF IOS-1755268 to C.D. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Y.L., C.L., and C.D. designed research; Y.L., C.L., L.R., V.S., Z.C., J.G., and C.B. performed research; Z.C. and J.L. contributed new reagents/analytic tools; Y.L., C.L., L.R., V.S., and C.D. analyzed data; and Y.L. and C.D. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Guillot C., Lecuit T., Mechanics of epithelial tissue homeostasis and morphogenesis. Science 340, 1185–1189 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Tai K., Cockburn K., Greco V., Flexibility sustains epithelial tissue homeostasis. Curr. Opin. Cell Biol. 60, 84–91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clevers H., The intestinal crypt, a prototype stem cell compartment. Cell 154, 274–284 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Metcalfe C., Kljavin N. M., Ybarra R., de Sauvage F. J., Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell 14, 149–159 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Montgomery R. K., et al. , Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl. Acad. Sci. U.S.A. 108, 179–184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Melo F. d. S., de Sauvage F. J., Cellular plasticity in intestinal homeostasis and disease. Cell Stem Cell 24, 54–64 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Post Y., Clevers H., Defining adult stem cell function at its simplest: The ability to replace lost cells through mitosis. Cell Stem Cell 25, 174–183 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Guiu J., et al. , Tracing the origin of adult intestinal stem cells. Nature 570, 107–111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clevers H., Watt F. M., Defining adult stem cells by function, not by phenotype. Annu. Rev. Biochem. 87, 1015–1027 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Gadd V. L., Aleksieva N., Forbes S. J., Epithelial plasticity during liver injury and regeneration. Cell Stem Cell 27, 557–573 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Poss K. D., Wilson L. G., Keating M. T., Heart regeneration in zebrafish. Science 298, 2188–2190 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Tzahor E., Poss K. D., Cardiac regeneration strategies: Staying young at heart. Science 356, 1035–1039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D., Hallmarks of cancer: New dimensions. Cancer Discov. 12, 31–46 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Dai W., et al. , Calcium deficiency-induced and TRP channel-regulated IGF1R-PI3K-Akt signaling regulates abnormal epithelial cell proliferation. Cell Death Differ. 21, 568–581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C., et al. , Development of a whole organism platform for phenotype-based analysis of IGF1R-PI3K-Akt-Tor action. Sci. Rep. 7, 1–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan J.-J., Hwang P.-P., Novel discoveries in acid-base regulation and osmoregulation: A review of selected hormonal actions in zebrafish and medaka. Gen. Comp. Endocrinol. 277, 20–29 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Hwang P.-P., Ion uptake and acid secretion in zebrafish (Danio rerio). J. Exp. Biol. 212, 1745–1752 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Liu C., et al. , Ca2+ concentration–dependent premature death of igfbp5a−/− fish reveals a critical role of IGF signaling in adaptive epithelial growth. Sci. Signal. 11, eaat2231 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Xin Y., et al. , Cell-autonomous regulation of epithelial cell quiescence by calcium channel Trpv6. Elife 8, e48003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C., et al. , The metalloproteinase Papp-aa controls epithelial cell quiescence-proliferation transition. Elife 9, e52322 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S., et al. , Calcium state-dependent regulation of epithelial cell quiescence by Stanniocalcin 1a. Front. Cell Dev. Biol. 9, 662915 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xin Y., Guan J., Li Y., Duan C., Regulation of cell quiescence–proliferation balance by Ca2+–CaMKK–Akt signaling J. Cell Sci. 134, jcs253807 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Chen C., et al. , TSC–mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J. Exp. Med. 205, 2397–2408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C., Liu Y., Liu Y., Zheng P., mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci. Signal. 2, ra75 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chell J. M., Brand A. H., Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell 143, 1161–1173 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziegler A. N., Levison S. W., Wood T. L., Insulin and IGF receptor signalling in neural-stem-cell homeostasis. Nat. Rev. Endocrinol. 11, 161–170 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziegler A. N., et al. , Insulin-like growth factor II: An essential adult stem cell niche constituent in brain and intestine. Stem Cell Reports 12, 816–830 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mossmann D., Park S., Hall M. N., mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 18, 744–757 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Chakrabarty R. P., Chandel N. S., Mitochondria as signaling organelles control mammalian stem cell fate. Cell Stem Cell 28, 394–408 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura-Ishizu A., Ito K., Suda T., Hematopoietic stem cell metabolism during development and aging. Dev. Cell 54, 239–255 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rath E., Urbauer E., Haller D., Mitochondrial metabolism in the intestinal stem cell niche-sensing and signaling in health and disease. Front. Cell Dev. Biol. 8, 1520 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martínez-Reyes I., et al. , TCA cycle and mitochondrial membrane potential are necessary for diverse biological functions. Mol. cell 61, 199–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell P., Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol. Rev. Camb. Philos. Soc. 41, 445–502 (1966). [DOI] [PubMed] [Google Scholar]
- 34.Morelli A. M., Ravera S., Calzia D., Panfoli I., An update of the chemiosmotic theory as suggested by possible proton currents inside the coupling membrane. Open Biol. 9, 180221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zorova L. D., et al. , Mitochondrial membrane potential. Anal. Biochem. 552, 50–59 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carboni J. M., et al. , BMS-754807, a small molecule inhibitor of insulin-like growth factor-1R/IRIGF-1R/IR. Mol. Cancer Ther. 8, 3341–3349 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Taylor M. R., Hurley J. B., Van Epps H. A., Brockerhoff S. E., A zebrafish model for pyruvate dehydrogenase deficiency: Rescue of neurological dysfunction and embryonic lethality using a ketogenic diet. Proc. Natl. Acad. Sci. U.S.A. 101, 4584–4589 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zachar Z., et al. , Non-redox-active lipoate derivates disrupt cancer cell mitochondrial metabolism and are potent anticancer agents in vivo. J. Mol. Med. 89, 1137–1148 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Del Prete E., Lutz T., Scharrer E., Inhibition of glucose oxidation by α-cyano-4-hydroxycinnamic acid stimulates feeding in rats. Physiol. Behav. 80, 489–498 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Shi Y., et al. , Gboxin is an oxidative phosphorylation inhibitor that targets glioblastoma. Nature 567, 341–346 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang R., et al. , Restraining lysosomal activity preserves hematopoietic stem cell quiescence and potency. Cell Stem Cell 26, 359–376.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stacpoole P. W., The pharmacology of dichloroacetate. Metabolism 38, 1124–1144 (1989). [DOI] [PubMed] [Google Scholar]
- 43.Tiwari S. K., Toshniwal A. G., Mandal S., Mandal L., Fatty acid β-oxidation is required for the differentiation of larval hematopoietic progenitors in Drosophila. Elife 9, e53247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu R. S., et al. , A rapid method for directed gene knockout for screening in G0 zebrafish. Dev. Cell 46, 112–125.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Kroll F., et al. , A simple and effective F0 knockout method for rapid screening of behaviour and other complex phenotypes. Elife 10, e59683 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cabezon E., Runswick M., Leslie A., Walker J., The structure of bovine IF1, the regulatory subunit of mitochondrial F-ATPase. EMBO J. 20, 6990–6996 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Formentini L., et al. , In vivo inhibition of the mitochondrial H+-ATP synthase in neurons promotes metabolic preconditioning. EMBO J. 33, 762–778 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.García-Bermúdez J., et al. , PKA phosphorylates the ATPase inhibitory factor 1 and inactivates its capacity to bind and inhibit the mitochondrial H+-ATP synthase. Cell Rep. 12, 2143–2155 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Shadel G. S., Horvath T. L., Mitochondrial ROS signaling in organismal homeostasis. Cell 163, 560–569 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rath E., Moschetta A., Haller D., Mitochondrial function—Gatekeeper of intestinal epithelial cell homeostasis. Nat. Rev. Gastroenterol. Hepatol. 15, 497–516 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Robb E. L., et al. , Selective superoxide generation within mitochondria by the targeted redox cycler MitoParaquat. Free Radic. Biol. Med. 89, 883–894 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Di Cristofano A., SGK1: The dark side of PI3K signaling. Curr. Top. Dev. Biol. 123, 49–71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi T., Deak M., Morrice N., Cohen P., Characterization of the structure and regulation of two novel isoforms of serum-and glucocorticoid-induced protein kinase. Biochem. J. 344, 189–197 (1999). [PMC free article] [PubMed] [Google Scholar]
- 54.O’Keeffe B. A., Cilia S., Maiyar A. C., Vaysberg M., Firestone G. L., The serum-and glucocorticoid-induced protein kinase-1 (Sgk-1) mitochondria connection: Identification of the IF-1 inhibitor of the F1F0-ATPase as a mitochondria-specific binding target and the stress-induced mitochondrial localization of endogenous Sgk-1. Biochimie 95, 1258–1265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang D., et al. , SGK1 attenuates oxidative stress-induced renal tubular epithelial cell injury by regulating mitochondrial function. Oxid. Med. Cell. Longev. 2019, 2013594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang C. S., et al. , Glutamine-utilizing transaminases are a metabolic vulnerability of TAZ/YAP-activated cancer cells. EMBO Rep. 19, e43577 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perekatt A. O., et al. , YY1 is indispensable for Lgr5+ intestinal stem cell renewal. Proc. Natl. Acad. Sci. U.S.A. 111, 7695–7700 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim Y., et al. , Loss of PKM2 in Lgr5+ intestinal stem cells promotes colitis-associated colorectal cancer. Sci. Rep. 9, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang F., Pirooznia M., Xu H., Mitochondria regulate intestinal stem cell proliferation and epithelial homeostasis through FOXO. Mol. Biol. Cell 31, 1538–1549 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodríguez-Colman M. J., et al. , Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 543, 424–427 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Schell J. C., et al. , Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nat. Cell Biol. 19, 1027–1036 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jakutis G., Stainier D. Y., Genotype-phenotype relationships in the context of transcriptional adaptation and genetic robustness. Annu. Rev. Genet. 55, 71–91 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Mason J. A., et al. , SGK1 signaling promotes glucose metabolism and survival in extracellular matrix detached cells. Cell Rep. 34, 108821 (2021). [DOI] [PubMed] [Google Scholar]
- 64.Gleason C. E., et al. , Phosphorylation at distinct subcellular locations underlies specificity in mTORC2-mediated activation of SGK1 and Akt. J. Cell Sci. 132, jcs224931 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nesci S., Trombetti F., Ventrella V., Pagliarani A., Post-translational modifications of the mitochondrial F1FO-ATPase. Biochim. Biophys. Acta Gen. Subj. 1861, 2902–2912 (2017). [DOI] [PubMed] [Google Scholar]
- 66.De Francesco E. M., Sotgia F., Lisanti M. P., Cancer stem cells (CSCs): Metabolic strategies for their identification and eradication. Biochem. J. 475, 1611–1634 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Webster M. K., Goya L., Ge Y., Maiyar A., Firestone G., Characterization of sgk, a novel member of the serine/threonine protein kinase gene family which is transcriptionally induced by glucocorticoids and serum. Mol. Cell. Biol. 13, 2031–2040 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Firestone G., Giampaolo J., O’Keeffe B., Stimulus-dependent regulation of serum and glucocorticoid inducible protein kinase (SGK) transcription, subcellular localization and enzymatic activity. Cell. Physiol. Biochem. 13, 1–12 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Cicenas J., et al. , SGK1 in cancer: Biomarker and drug target. Cancers 14, 2385 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Castel P., et al. , PDK1-SGK1 signaling sustains AKT-independent mTORC1 activation and confers resistance to PI3Kα inhibition. Cancer Cell 30, 229–242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Orlacchio A., et al. , SGK1 is a critical component of an AKT-independent pathway essential for PI3K-Mediated tumor development and maintenance SGK1: An integral component of the PI3K-transforming pathway. Cancer Res. 77, 6914–6926 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen B.-Y., et al. , SGK1 mediates hypotonic challenge-induced proliferation in basilar artery smooth muscle cells via promoting CREB signaling pathway. Eur. J. Pharmacol. 898, 173997 (2021). [DOI] [PubMed] [Google Scholar]
- 73.Liang X., et al. , Therapeutic inhibition of SGK1 suppresses colorectal cancer. Exp. Mol. Med. 49, e399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lang F., Perrotti N., Stournaras C., Colorectal carcinoma cells—Regulation of survival and growth by SGK1. Int. J. Biochem. Cell Biol. 42, 1571–1575 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Nasir O., et al. , Relative resistance of SGK1 knockout mice against chemical carcinogenesis. IUBMB Life 61, 768–776 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.